GISTIC identifies genomic regions that are significantly gained or lost across a set of tumors. The pipeline first filters out normal samples from the segmented copy-number data by inspecting the TCGA barcodes and then executes GISTIC version 2.0.15 (cga svn revision 32817).
There were 80 tumor samples used in this analysis: 17 significant arm-level results, 5 significant focal amplifications, and 18 significant focal deletions were found.
Figure 1. Genomic positions of amplified regions: the X-axis represents the normalized amplification signals (top) and significance by Q value (bottom). The green line represents the significance cutoff at Q value=0.25.
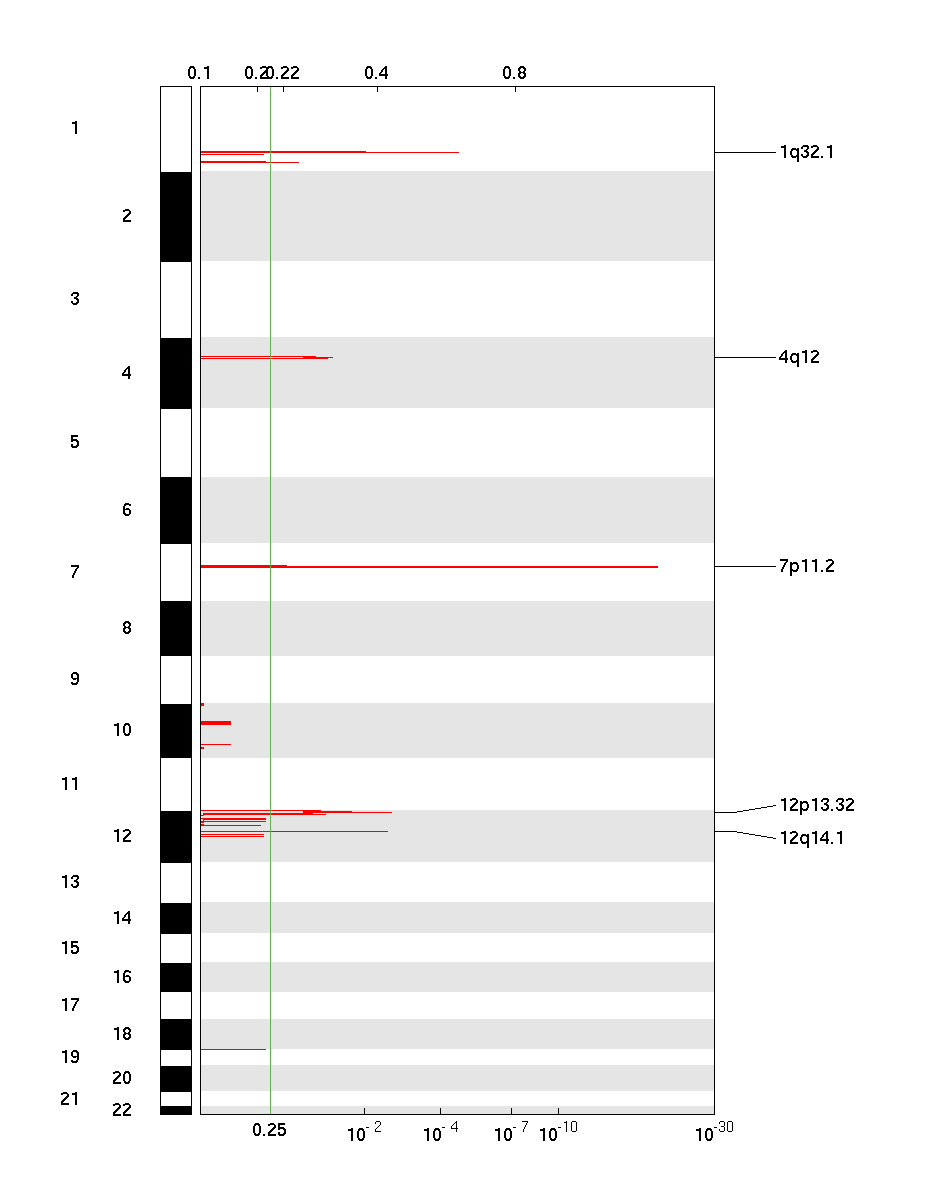
Table 1. Get Full Table Amplifications Table - 5 significant amplifications found. Click the link in the last column to view a comprehensive list of candidate genes. If no genes were identified within the peak, the nearest gene appears in brackets.
| Cytoband | Q value | Residual Q value | Wide Peak Boundaries | # Genes in Wide Peak |
|---|---|---|---|---|
| 7p11.2 | 5.3906e-21 | 5.3906e-21 | chr7:54910170-55074962 | 1 |
| 1q32.1 | 2.462e-05 | 2.462e-05 | chr1:201977214-202897544 | 16 |
| 12p13.32 | 0.0025135 | 0.0025135 | chr12:4400387-4472821 | 2 |
| 12q14.1 | 0.0031783 | 0.0031783 | chr12:56370368-56546867 | 14 |
| 4q12 | 0.037424 | 0.037424 | chr4:54050264-56186612 | 12 |
This is the comprehensive list of amplified genes in the wide peak for 7p11.2.
Table S1. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| EGFR |
This is the comprehensive list of amplified genes in the wide peak for 1q32.1.
Table S2. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| MDM4 |
| ATP2B4 |
| KISS1 |
| PIK3C2B |
| REN |
| SNRPE |
| SOX13 |
| ZC3H11A |
| LRRN2 |
| PLEKHA6 |
| LAX1 |
| ETNK2 |
| PPP1R15B |
| LOC127841 |
| GOLT1A |
| C1orf157 |
This is the comprehensive list of amplified genes in the wide peak for 12p13.32.
Table S3. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| FGF6 |
| C12orf4 |
This is the comprehensive list of amplified genes in the wide peak for 12q14.1.
Table S4. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CDK4 |
| hsa-mir-26a-2 |
| CYP27B1 |
| METTL1 |
| TSPAN31 |
| TSFM |
| CTDSP2 |
| AVIL |
| OS9 |
| FAM119B |
| MARCH9 |
| AGAP2 |
| MIR26A2 |
| LOC100130776 |
This is the comprehensive list of amplified genes in the wide peak for 4q12.
Table S5. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| KDR |
| KIT |
| PDGFRA |
| CHIC2 |
| CLOCK |
| NMU |
| TMEM165 |
| SRD5A3 |
| LNX1 |
| PDCL2 |
| GSX2 |
| RPL21P44 |
Figure 2. Genomic positions of deleted regions: the X-axis represents the normalized deletion signals (top) and significance by Q value (bottom). The green line represents the significance cutoff at Q value=0.25.
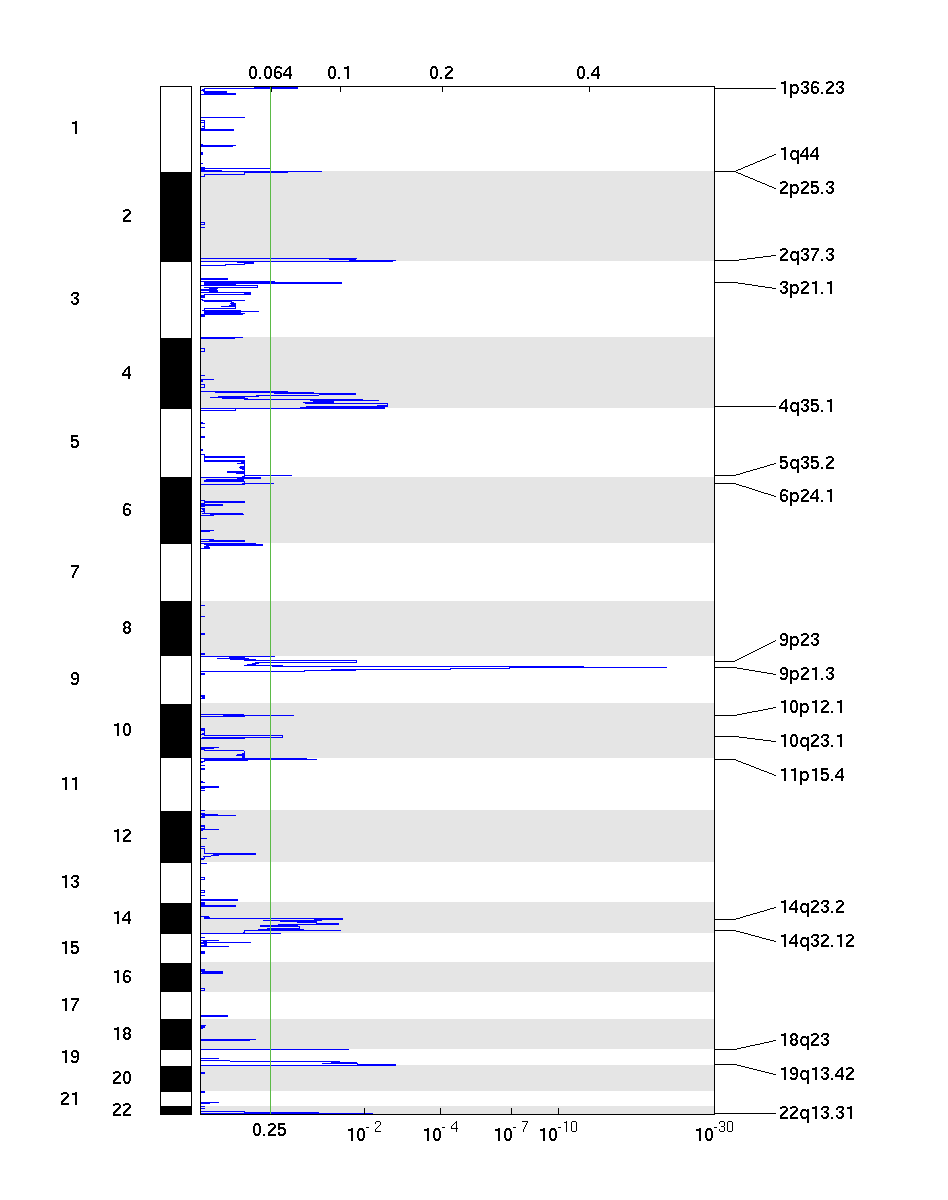
Table 2. Get Full Table Deletions Table - 18 significant deletions found. Click the link in the last column to view a comprehensive list of candidate genes. If no genes were identified within the peak, the nearest gene appears in brackets.
| Cytoband | Q value | Residual Q value | Wide Peak Boundaries | # Genes in Wide Peak |
|---|---|---|---|---|
| 9p21.3 | 2.6497e-22 | 7.9106e-21 | chr9:21981753-22438737 | 2 |
| 2q37.3 | 0.0019765 | 0.0019765 | chr2:232493600-242951149 | 107 |
| 19q13.42 | 0.0019765 | 0.0019765 | chr19:21055359-63811651 | 920 |
| 4q35.1 | 0.0032038 | 0.0032038 | chr4:167012652-191273063 | 85 |
| 22q13.31 | 0.0066855 | 0.0066855 | chr22:42722705-49691432 | 80 |
| 18q23 | 0.020502 | 0.020502 | chr18:75847612-76117153 | 4 |
| 3p21.1 | 0.02704 | 0.02704 | chr3:49545887-53099552 | 95 |
| 2p25.3 | 0.055711 | 0.055711 | chr2:1-9464621 | 29 |
| 14q32.12 | 0.028444 | 0.055711 | chr14:68406386-106368585 | 379 |
| 11p15.4 | 0.067246 | 0.073038 | chr11:1-11941429 | 264 |
| 14q23.2 | 0.025167 | 0.073038 | chr14:57939329-79751635 | 175 |
| 1p36.23 | 0.12196 | 0.12196 | chr1:1-28712888 | 437 |
| 10p12.1 | 0.13538 | 0.13403 | chr10:22746393-26919698 | 19 |
| 5q35.2 | 0.14392 | 0.14392 | chr5:104437660-180857866 | 572 |
| 1q44 | 0.16964 | 0.17801 | chr1:235015260-247249719 | 94 |
| 10q23.1 | 0.1833 | 0.17801 | chr10:63621438-90529962 | 170 |
| 6p24.1 | 0.22994 | 0.22953 | chr6:1-170899992 | 1120 |
| 9p24.1 | 0.014044 | 0.25244 | chr9:1-140273252 | 890 |
This is the comprehensive list of deleted genes in the wide peak for 9p21.3.
Table S6. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CDKN2B |
| CDKN2BAS |
This is the comprehensive list of deleted genes in the wide peak for 2q37.3.
Table S7. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-149 |
| hsa-mir-562 |
| AGXT |
| ALPI |
| ALPP |
| ALPPL2 |
| KIF1A |
| BOK |
| CHRND |
| CHRNG |
| COL6A3 |
| DTYMK |
| GBX2 |
| GPC1 |
| GPR35 |
| HDLBP |
| INPP5D |
| KCNJ13 |
| NDUFA10 |
| SEPT2 |
| NEU2 |
| NPPC |
| PDCD1 |
| PPP1R7 |
| SAG |
| SPP2 |
| DGKD |
| PER2 |
| LRRFIP1 |
| ECEL1 |
| EIF4E2 |
| HDAC4 |
| FARP2 |
| ARL4C |
| RAMP1 |
| STK25 |
| COPS8 |
| CAPN10 |
| PASK |
| ATG4B |
| SH3BP4 |
| NGEF |
| SNED1 |
| GIGYF2 |
| TRAF3IP1 |
| ANO7 |
| PRLH |
| THAP4 |
| ANKMY1 |
| SCLY |
| ASB1 |
| UGT1A10 |
| UGT1A8 |
| UGT1A7 |
| UGT1A6 |
| UGT1A5 |
| UGT1A9 |
| UGT1A4 |
| UGT1A1 |
| UGT1A3 |
| ATG16L1 |
| USP40 |
| HJURP |
| HES6 |
| CXCR7 |
| RNPEPL1 |
| GAL3ST2 |
| RAB17 |
| TRPM8 |
| MLPH |
| IQCA1 |
| C2orf54 |
| EFHD1 |
| ILKAP |
| ING5 |
| MGC16025 |
| AGAP1 |
| DIS3L2 |
| NEU4 |
| MTERFD2 |
| UBE2F |
| OTOS |
| MYEOV2 |
| OR6B3 |
| LOC151174 |
| MSL3L2 |
| TIGD1 |
| C2orf85 |
| DUSP28 |
| ESPNL |
| ECEL1P2 |
| RBM44 |
| AQP12A |
| KLHL30 |
| C2orf82 |
| OR6B2 |
| ASB18 |
| MIR149 |
| DNAJB3 |
| LOC643387 |
| PRR21 |
| AQP12B |
| SCARNA6 |
| SCARNA5 |
| D2HGDH |
| LOC728323 |
| PP14571 |
This is the comprehensive list of deleted genes in the wide peak for 19q13.42.
Table S8. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| AKT2 |
| BCL3 |
| CD79A |
| CEBPA |
| ERCC2 |
| KLK2 |
| PPP2R1A |
| CIC |
| CBLC |
| TFPT |
| ZNF331 |
| hsa-mir-1274b |
| hsa-mir-935 |
| hsa-mir-373 |
| hsa-mir-519a-2 |
| hsa-mir-643 |
| hsa-mir-125a |
| hsa-mir-150 |
| hsa-mir-220c |
| hsa-mir-769 |
| hsa-mir-642 |
| hsa-mir-330 |
| hsa-mir-641 |
| A1BG |
| ACTN4 |
| AP2A1 |
| APLP1 |
| APOC1 |
| APOC1P1 |
| APOC2 |
| APOC4 |
| APOE |
| KLK3 |
| ATP1A3 |
| ATP4A |
| AXL |
| BAX |
| BCAT2 |
| BCKDHA |
| CEACAM1 |
| BLVRB |
| C5AR1 |
| CA11 |
| CALM3 |
| CAPNS1 |
| CCNE1 |
| CD22 |
| CD33 |
| SIGLEC6 |
| CD37 |
| CEACAM5 |
| CEBPG |
| CGB |
| CEACAM3 |
| CEACAM7 |
| CEACAM8 |
| CEACAM4 |
| TBCB |
| CKM |
| AP2S1 |
| CLC |
| CLPTM1 |
| COX6B1 |
| COX7A1 |
| CRX |
| CYP2A6 |
| CYP2A7 |
| CYP2A13 |
| CYP2B6 |
| CYP2B7P1 |
| CYP2F1 |
| DBP |
| DMPK |
| DMWD |
| ECH1 |
| MEGF8 |
| EMP3 |
| ERCC1 |
| ERF |
| FBL |
| ETFB |
| ETV2 |
| FCAR |
| FCGRT |
| FKBP1P1 |
| FLT3LG |
| FOSB |
| FPR1 |
| FPR2 |
| FPR3 |
| FTL |
| FUT1 |
| FUT2 |
| GIPR |
| GPI |
| GPR4 |
| GPR32 |
| FFAR1 |
| FFAR3 |
| FFAR2 |
| GRIK5 |
| GRIN2D |
| GRLF1 |
| GSK3A |
| GYS1 |
| HAS1 |
| FOXA3 |
| HNRNPL |
| HPN |
| HRC |
| PRMT1 |
| IL11 |
| IRF3 |
| KCNA7 |
| KCNC3 |
| KCNJ14 |
| KCNN4 |
| KIR2DL1 |
| KIR2DL3 |
| KIR2DL4 |
| KIR2DS4 |
| KIR3DL1 |
| KIR3DL2 |
| KLK1 |
| LAIR1 |
| LAIR2 |
| LGALS4 |
| LGALS7 |
| LHB |
| LIG1 |
| LIM2 |
| LIPE |
| LRP3 |
| BCAM |
| MAG |
| MAP3K10 |
| MYBPC2 |
| CEACAM6 |
| NDUFA3 |
| NFKBIB |
| NKG7 |
| CNOT3 |
| NOVA2 |
| NPAS1 |
| NPHS1 |
| NTF4 |
| NUCB1 |
| PAFAH1B3 |
| PEG3 |
| PEPD |
| PLAUR |
| FXYD1 |
| FXYD3 |
| POLD1 |
| POLR2I |
| POU2F2 |
| PPP5C |
| PRKCG |
| PRRG2 |
| KLK7 |
| KLK6 |
| KLK10 |
| PSG1 |
| PSG2 |
| PSG3 |
| PSG4 |
| PSG5 |
| PSG6 |
| PSG7 |
| PSG9 |
| PSG11 |
| PSMC4 |
| PSMD8 |
| PTGIR |
| PTPRH |
| PVR |
| PVRL2 |
| RELB |
| RPL18 |
| RPL28 |
| MRPS12 |
| RPS5 |
| RPS9 |
| RPS11 |
| RPS16 |
| RPS19 |
| RRAS |
| RTN2 |
| RYR1 |
| CLEC11A |
| SCN1B |
| SEPW1 |
| SLC1A5 |
| SLC8A2 |
| SNRNP70 |
| SNRPA |
| SNRPD2 |
| SPIB |
| AURKC |
| SULT2B1 |
| SULT2A1 |
| SUPT5H |
| SYT5 |
| TGFB1 |
| TNNI3 |
| TNNT1 |
| TULP2 |
| TYROBP |
| NR1H2 |
| UQCRFS1 |
| USF2 |
| VASP |
| XRCC1 |
| ZFP36 |
| ZNF8 |
| ZNF708 |
| ZNF17 |
| ZNF28 |
| MZF1 |
| ZNF43 |
| ZNF45 |
| ZNF221 |
| ZNF91 |
| ZNF99 |
| ZNF222 |
| ZNF132 |
| ZNF134 |
| ZNF135 |
| ZNF137 |
| ZNF146 |
| ZNF154 |
| ZNF155 |
| ZNF175 |
| ZNF180 |
| ZNF208 |
| ZNF223 |
| ZNF224 |
| ZNF225 |
| ZNF226 |
| ZNF227 |
| ZFP112 |
| ZNF229 |
| ZNF230 |
| SYMPK |
| MIA |
| DPF1 |
| LTBP4 |
| TEAD2 |
| PPFIA3 |
| PLA2G4C |
| C19orf2 |
| NAPA |
| SIGLEC5 |
| FCGBP |
| PGLYRP1 |
| UBE2M |
| ARHGEF1 |
| PDCD5 |
| DYRK1B |
| NUMBL |
| CYTH2 |
| ZNF235 |
| ZNF264 |
| KCNK6 |
| NCR1 |
| NAPSA |
| ZNF254 |
| GMFG |
| KLK4 |
| ZNF432 |
| DHX34 |
| KIAA0355 |
| ZNF536 |
| MLL4 |
| UBA2 |
| SAE1 |
| TRIM28 |
| ZNF256 |
| LILRB2 |
| PAK4 |
| TMEM147 |
| TOMM40 |
| ZNF211 |
| RABAC1 |
| TRAPPC2P1 |
| SPINT2 |
| DLL3 |
| POP4 |
| ZNF234 |
| ZNF274 |
| ZNF460 |
| PPP1R13L |
| CD3EAP |
| RUVBL2 |
| LILRB1 |
| HCST |
| KDELR1 |
| LILRB5 |
| SLC27A5 |
| LILRB4 |
| KLK11 |
| LILRA1 |
| LILRB3 |
| LILRA3 |
| LILRA2 |
| UPK1A |
| HNRNPUL1 |
| SFRS16 |
| KPTN |
| SLC7A9 |
| MAP4K1 |
| KLK8 |
| PNKP |
| U2AF2 |
| ATF5 |
| ZFP30 |
| ZNF507 |
| SAPS1 |
| CARD8 |
| SIRT2 |
| SIPA1L3 |
| ZC3H4 |
| HAUS5 |
| FBXO46 |
| ETHE1 |
| RPL13A |
| SYNGR4 |
| LILRA4 |
| PRG1 |
| ZIM2 |
| NUP62 |
| HSPBP1 |
| PPP1R15A |
| PLD3 |
| EML2 |
| ZNF324 |
| KLK5 |
| ZNF345 |
| PRKD2 |
| ZNF473 |
| CLIP3 |
| LSM14A |
| KLK13 |
| CCDC9 |
| PRPF31 |
| IRF2BP1 |
| FGF21 |
| GAPDHS |
| SNORD35A |
| SNORD34 |
| SNORD33 |
| SNORD32A |
| ZNF285A |
| ZBTB32 |
| SIGLEC7 |
| LYPD3 |
| BBC3 |
| DKKL1 |
| SIGLEC9 |
| SIGLEC8 |
| GPR77 |
| CHMP2A |
| DHDH |
| ZNF544 |
| EIF3K |
| UBE2S |
| SLC6A16 |
| LGALS13 |
| CYP2S1 |
| STRN4 |
| CCDC106 |
| EPN1 |
| SERTAD3 |
| SERTAD1 |
| GLTSCR2 |
| GLTSCR1 |
| EHD2 |
| KLK14 |
| KLK12 |
| SHANK1 |
| NOSIP |
| ZNF580 |
| HSD17B14 |
| GP6 |
| VRK3 |
| ZNF571 |
| ZNF581 |
| LSR |
| PTOV1 |
| FXYD7 |
| FXYD5 |
| RAB4B |
| PAF1 |
| PPP1R12C |
| TRPM4 |
| ZNF586 |
| QPCTL |
| FAM83E |
| EPS8L1 |
| RASIP1 |
| SARS2 |
| TMEM160 |
| PIH1D1 |
| GPATCH1 |
| SAMD4B |
| ATP5SL |
| C19orf73 |
| PNMAL1 |
| TMEM143 |
| ZNF444 |
| KLK15 |
| MED29 |
| NLRP2 |
| ZNF416 |
| ZNF446 |
| ZNF701 |
| ZNF83 |
| ZNF415 |
| PSENEN |
| ZNF302 |
| LIN37 |
| C19orf61 |
| IRGC |
| SLC7A10 |
| CABP5 |
| SPHK2 |
| LGALS14 |
| EXOSC5 |
| MEIS3 |
| CEACAM19 |
| SLC17A7 |
| NAT14 |
| CD177 |
| VN1R1 |
| RCN3 |
| ZNF304 |
| TTYH1 |
| PNMAL2 |
| PRR12 |
| ZNF471 |
| ZNF492 |
| TSHZ3 |
| LRFN1 |
| GRAMD1A |
| USP29 |
| PLEKHA4 |
| ZFP14 |
| ZNF529 |
| PRX |
| SPTBN4 |
| MARK4 |
| HAMP |
| CATSPERG |
| ZNF71 |
| SCAF1 |
| PRODH2 |
| CACNG8 |
| CACNG7 |
| CACNG6 |
| ZNF350 |
| TSKS |
| ZNF667 |
| DMRTC2 |
| C19orf33 |
| ELSPBP1 |
| LIN7B |
| HIF3A |
| CHST8 |
| ZNF574 |
| PLEKHG2 |
| ZNF649 |
| ZSCAN18 |
| MGC2752 |
| TSEN34 |
| KCTD15 |
| TRAPPC6A |
| MBOAT7 |
| FKRP |
| ZSCAN5A |
| PLEKHF1 |
| LENG1 |
| LILRP2 |
| LILRA6 |
| RBM42 |
| ZNF576 |
| LRFN3 |
| ZNF329 |
| TMEM149 |
| TBC1D17 |
| ZNF419 |
| GEMIN7 |
| ISOC2 |
| MYH14 |
| ZNF665 |
| ZNF552 |
| ZNF671 |
| ZNF613 |
| ADCK4 |
| CNTD2 |
| ZNF702P |
| LOC80054 |
| ZNF606 |
| ZNF614 |
| FUZ |
| OPA3 |
| ITPKC |
| B9D2 |
| RSHL1 |
| ZNF611 |
| MED25 |
| BCL2L12 |
| C19orf12 |
| TEX101 |
| GRWD1 |
| WDR87 |
| CCDC8 |
| KIRREL2 |
| ANKRD27 |
| ZNF541 |
| SYT3 |
| PDCD2L |
| AKT1S1 |
| ZNF528 |
| BRSK1 |
| ZNF527 |
| CNFN |
| SNORD35B |
| ZNF347 |
| ZNF577 |
| ZNF607 |
| SUV420H2 |
| C19orf48 |
| NFKBID |
| ZBTB45 |
| CCDC123 |
| ZNF382 |
| ZNF587 |
| FIZ1 |
| ZNF566 |
| ALKBH6 |
| RHPN2 |
| GALP |
| SIGLEC10 |
| SIGLEC12 |
| ZNF628 |
| KIR3DX1 |
| ZNF30 |
| ZNF551 |
| CEACAM21 |
| ZNF616 |
| ZNF766 |
| CCDC97 |
| EXOC3L2 |
| ZNF468 |
| ZNF160 |
| CTU1 |
| ZNF835 |
| YIF1B |
| C19orf40 |
| TDRD12 |
| ZNF765 |
| NLRP12 |
| MYADM |
| ZNF845 |
| ZNF461 |
| ZNF585B |
| TIMM50 |
| SHKBP1 |
| DMKN |
| CCDC114 |
| DKFZp434J0226 |
| ACPT |
| CGB5 |
| DKFZp566F0947 |
| CGB7 |
| LRRC4B |
| LENG9 |
| CGB8 |
| GNG8 |
| PPP1R14A |
| EGLN2 |
| BIRC8 |
| FAM71E1 |
| RDH13 |
| PTH2 |
| ZNF257 |
| ZIM3 |
| SIGLEC11 |
| CGB1 |
| CGB2 |
| LMTK3 |
| LENG8 |
| FBXO17 |
| KIR3DL3 |
| SNX26 |
| RASGRP4 |
| ZNF526 |
| ZNF837 |
| CLDND2 |
| ZNF816A |
| ZNF543 |
| CEACAM20 |
| COX6B2 |
| OSCAR |
| ZNF813 |
| JOSD2 |
| C19orf41 |
| CPT1C |
| ALDH16A1 |
| NTN5 |
| NLRP13 |
| NLRP8 |
| NLRP5 |
| ZNF787 |
| ZNF573 |
| WDR88 |
| EID2B |
| IRGQ |
| ZNF428 |
| WTIP |
| ZNF792 |
| HSPB6 |
| RINL |
| FBXO27 |
| C19orf47 |
| ZFP28 |
| VSIG10L |
| NCRNA00085 |
| ZNF480 |
| ZNF534 |
| ZNF578 |
| C19orf18 |
| ZNF418 |
| ZNF417 |
| ZNF548 |
| FLJ40125 |
| KLC3 |
| LYPD4 |
| TMEM190 |
| HIPK4 |
| TMC4 |
| LOC147804 |
| ZNF524 |
| ZNF784 |
| CCDC155 |
| DACT3 |
| SIX5 |
| IGFL2 |
| ZNF420 |
| ZNF565 |
| NLRP4 |
| ZNF542 |
| ZNF582 |
| ZNF583 |
| FAM98C |
| CAPN12 |
| DPY19L3 |
| TTC9B |
| ZNF599 |
| FAM187B |
| C19orf55 |
| LOC148145 |
| CDC42EP5 |
| LOC148189 |
| ZNF98 |
| ZNF738 |
| ZNF714 |
| ZNF681 |
| ZNF569 |
| ZNF570 |
| ZNF836 |
| ZNF610 |
| ZNF600 |
| ZNF320 |
| ZNF497 |
| ZNF550 |
| ZNF296 |
| DEDD2 |
| ZNF579 |
| ZNF114 |
| ZNF567 |
| ZNF383 |
| ZNF781 |
| EID2 |
| ZNF780B |
| LGI4 |
| C19orf46 |
| ZNF676 |
| ZNF100 |
| ZNF540 |
| ZNF525 |
| ZNF431 |
| SPACA4 |
| ZNF675 |
| ZNF585A |
| NLRP7 |
| GGN |
| CADM4 |
| THAP8 |
| U2AF1L4 |
| C19orf76 |
| ZNF584 |
| ZSCAN4 |
| NLRP11 |
| TMEM86B |
| PRR24 |
| ZNF549 |
| NAPSB |
| IL4I1 |
| IL28A |
| IL28B |
| IL29 |
| SSC5D |
| ZNF547 |
| ZIK1 |
| ZNF776 |
| ZSCAN1 |
| ZNF780A |
| C19orf54 |
| PRR19 |
| TMEM145 |
| CXCL17 |
| ZNF575 |
| LYPD5 |
| ZNF283 |
| NKPD1 |
| TPRX1 |
| MAMSTR |
| IZUMO1 |
| C19orf63 |
| KLK9 |
| SIGLECP3 |
| C19orf75 |
| ZNF615 |
| ZNF841 |
| LOC284379 |
| SCGBL |
| WDR62 |
| ZFP82 |
| LOC284412 |
| VSTM1 |
| TMEM150B |
| FAM71E2 |
| ZNF493 |
| HKR1 |
| VN1R2 |
| VN1R4 |
| NLRP9 |
| ZNF181 |
| ZNF260 |
| ZNF546 |
| MYPOP |
| NANOS2 |
| VSTM2B |
| NCCRP1 |
| SYCN |
| LEUTX |
| ZNF404 |
| ZNF284 |
| ZNF677 |
| RFPL4A |
| ZSCAN5B |
| ZSCAN22 |
| SELV |
| ZNF530 |
| C19orf51 |
| ZNF429 |
| ZNF233 |
| LILRA5 |
| SBSN |
| ZNF829 |
| ZNF568 |
| B3GNT8 |
| IGFL1 |
| ZNF773 |
| RPSAP58 |
| RGS9BP |
| KRTDAP |
| ZNF790 |
| FLJ41856 |
| CEACAM16 |
| BLOC1S3 |
| IGFL3 |
| ZNF808 |
| ZNF761 |
| ZNF470 |
| ZNF749 |
| ZNF324B |
| NUDT19 |
| ZNF793 |
| PAPL |
| ZNF805 |
| SPRED3 |
| ZNF321 |
| LOC400696 |
| SIGLEC16 |
| FLJ26850 |
| ZNF880 |
| ZNF772 |
| IGLON5 |
| MIRLET7E |
| MIR125A |
| MIR150 |
| MIR99B |
| PSG8 |
| OLT-2 |
| MIR330 |
| MIR371 |
| MIR372 |
| MIR373 |
| IGFL4 |
| NCRNA00181 |
| DPRX |
| DUXA |
| ASPDH |
| MIR512-1 |
| MIR512-2 |
| MIR498 |
| MIR520E |
| MIR515-1 |
| MIR519E |
| MIR520F |
| MIR515-2 |
| MIR519C |
| MIR520A |
| MIR526B |
| MIR519B |
| MIR525 |
| MIR523 |
| MIR518F |
| MIR520B |
| MIR518B |
| MIR526A1 |
| MIR520C |
| MIR518C |
| MIR524 |
| MIR517A |
| MIR519D |
| MIR521-2 |
| MIR520D |
| MIR517B |
| MIR520G |
| MIR516B2 |
| MIR526A2 |
| MIR518E |
| MIR518A1 |
| MIR518D |
| MIR516B1 |
| MIR518A2 |
| MIR517C |
| MIR520H |
| MIR521-1 |
| MIR522 |
| MIR519A1 |
| MIR527 |
| MIR516A1 |
| MIR516A2 |
| MIR519A2 |
| KLKP1 |
| LOC641367 |
| TMEM91 |
| LOC643719 |
| SDHAF1 |
| SBK2 |
| PSG10 |
| LGALS7B |
| PHLDB3 |
| SEC1 |
| SNORD23 |
| SNORD88A |
| SNORD88B |
| SNORD88C |
| MIR641 |
| MIR642 |
| MIR643 |
| RPL13AP5 |
| CCDC61 |
| CEACAM18 |
| SHISA7 |
| ZNF814 |
| MIR769 |
| KIR3DP1 |
| SIGLEC14 |
| MIMT1 |
| LOC100101266 |
| MIR935 |
| SNAR-G1 |
| SNAR-F |
| SNAR-A1 |
| SNAR-A2 |
| SNAR-A12 |
| LOC100128675 |
| LOC100129935 |
| BSPH1 |
| LOC100131691 |
| LOC100134317 |
| PEG3AS |
| SNAR-A3 |
| SNAR-A5 |
| SNAR-A7 |
| SNAR-A11 |
| SNAR-A9 |
| SNAR-A4 |
| SNAR-A6 |
| SNAR-A8 |
| SNAR-A13 |
| SNAR-A10 |
| SNAR-B2 |
| SNAR-C2 |
| SNAR-C4 |
| SNAR-E |
| SNAR-C5 |
| SNAR-B1 |
| SNAR-C1 |
| SNAR-C3 |
| SNAR-D |
| SNAR-G2 |
| SRRM5 |
| C19orf69 |
| SNAR-A14 |
This is the comprehensive list of deleted genes in the wide peak for 4q35.1.
Table S9. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| DUX4 |
| hsa-mir-1305 |
| AGA |
| SLC25A4 |
| CASP3 |
| CLCN3 |
| DCTD |
| F11 |
| ACSL1 |
| FAT1 |
| FRG1 |
| GPM6A |
| HMGB2 |
| HPGD |
| ING2 |
| IRF2 |
| KLKB1 |
| MTNR1A |
| NEK1 |
| TLL1 |
| TLR3 |
| VEGFC |
| GLRA3 |
| SORBS2 |
| SAP30 |
| HAND2 |
| MFAP3L |
| MORF4 |
| ADAM29 |
| ANXA10 |
| SCRG1 |
| PALLD |
| FAM149A |
| FBXO8 |
| PDLIM3 |
| SPOCK3 |
| AADAT |
| GALNT7 |
| CLDN22 |
| C4orf27 |
| NEIL3 |
| UFSP2 |
| DDX60 |
| CDKN2AIP |
| ODZ3 |
| LRP2BP |
| TUBB4Q |
| STOX2 |
| KIAA1430 |
| SH3RF1 |
| SPCS3 |
| C4orf41 |
| MLF1IP |
| NBLA00301 |
| WWC2 |
| KIAA1712 |
| SNX25 |
| CBR4 |
| MGC45800 |
| DDX60L |
| WDR17 |
| ZFP42 |
| SPATA4 |
| ENPP6 |
| ASB5 |
| C4orf38 |
| RWDD4A |
| CCDC111 |
| TRIML2 |
| CCDC110 |
| CYP4V2 |
| LOC285501 |
| TRIML1 |
| ANKRD37 |
| HELT |
| FAM92A3 |
| C4orf47 |
| GALNTL6 |
| FRG2 |
| SLED1 |
| LOC653543 |
| LOC653544 |
| LOC653545 |
| LOC653548 |
| LOC728410 |
This is the comprehensive list of deleted genes in the wide peak for 22q13.31.
Table S10. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-let-7b |
| hsa-mir-1249 |
| ACR |
| ARSA |
| CHKB |
| CPT1B |
| TYMP |
| FBLN1 |
| PPARA |
| MAPK11 |
| MAPK12 |
| SBF1 |
| UPK3A |
| WNT7B |
| CELSR1 |
| SAPS2 |
| ZBED4 |
| SCO2 |
| PKDREJ |
| NUP50 |
| RABL2B |
| GRAMD4 |
| MLC1 |
| C22orf9 |
| MAPK8IP2 |
| PLXNB2 |
| BRD1 |
| ARHGAP8 |
| TBC1D22A |
| ATXN10 |
| FAM19A5 |
| RIBC2 |
| SMC1B |
| PARVB |
| NCAPH2 |
| GTSE1 |
| MOV10L1 |
| FAM118A |
| TTC38 |
| C22orf26 |
| MIOX |
| PRR5 |
| TRMU |
| PANX2 |
| PARVG |
| CERK |
| ALG12 |
| CRELD2 |
| ADM2 |
| TRABD |
| SELO |
| HDAC10 |
| LDOC1L |
| KIAA1644 |
| SHANK3 |
| TUBGCP6 |
| LOC90834 |
| LMF2 |
| PHF21B |
| KLHDC7B |
| LOC150381 |
| C22orf40 |
| CN5H6.4 |
| TTLL8 |
| RPL23AP82 |
| C22orf34 |
| CHKB-CPT1B |
| NCRNA00207 |
| LOC400931 |
| IL17REL |
| MIRLET7A3 |
| MIRLET7B |
| FAM116B |
| PIM3 |
| ODF3B |
| PRR5-ARHGAP8 |
| C22orf41 |
| LOC730668 |
| LOC100144603 |
| LOC100271722 |
This is the comprehensive list of deleted genes in the wide peak for 18q23.
Table S11. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| ADNP2 |
| C18orf22 |
| PARD6G |
| LOC100130522 |
This is the comprehensive list of deleted genes in the wide peak for 3p21.1.
Table S12. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BAP1 |
| PBRM1 |
| hsa-mir-135a-1 |
| hsa-let-7g |
| hsa-mir-566 |
| ACY1 |
| ALAS1 |
| APEH |
| CISH |
| DOCK3 |
| DUSP7 |
| GNAI2 |
| GNAT1 |
| GRM2 |
| HYAL1 |
| ITIH1 |
| ITIH3 |
| ITIH4 |
| MST1 |
| MST1R |
| RPL29 |
| SEMA3F |
| NEK4 |
| TNNC1 |
| UBA7 |
| IFRD2 |
| MAPKAPK3 |
| SEMA3B |
| MANF |
| HYAL3 |
| HYAL2 |
| BSN |
| RRP9 |
| CACNA2D2 |
| VPRBP |
| IP6K1 |
| PARP3 |
| RBM6 |
| RBM5 |
| TRAIP |
| TUSC4 |
| SLC38A3 |
| CYB561D2 |
| TMEM115 |
| RASSF1 |
| NISCH |
| TUSC2 |
| TWF2 |
| RAD54L2 |
| STAB1 |
| NAT6 |
| ABHD14A |
| WDR51A |
| DNAH1 |
| GNL3 |
| SPCS1 |
| RBM15B |
| GMPPB |
| C3orf18 |
| ZMYND10 |
| TEX264 |
| HEMK1 |
| SFMBT1 |
| PHF7 |
| TLR9 |
| GLT8D1 |
| SEMA3G |
| PCBP4 |
| RNF123 |
| NT5DC2 |
| CAMKV |
| WDR82 |
| MON1A |
| ABHD14B |
| GPR62 |
| IQCF1 |
| GLYCTK |
| PPM1M |
| C3orf45 |
| TMEM110 |
| AMIGO3 |
| CDH29 |
| C3orf54 |
| IQCF2 |
| IQCF5 |
| MUSTN1 |
| IQCF3 |
| MIRLET7G |
| MIR135A1 |
| IQCF6 |
| LOC440957 |
| SNORD19 |
| SNORD69 |
| SNORD19B |
| C3orf74 |
This is the comprehensive list of deleted genes in the wide peak for 2p25.3.
Table S13. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| ACP1 |
| ID2 |
| RPS7 |
| SOX11 |
| TPO |
| TSSC1 |
| PXDN |
| ASAP2 |
| RNF144A |
| MYT1L |
| SH3YL1 |
| TTC15 |
| SNTG2 |
| ADI1 |
| ALLC |
| KIDINS220 |
| COLEC11 |
| RSAD2 |
| CMPK2 |
| MBOAT2 |
| TMEM18 |
| LOC150622 |
| RNASEH1 |
| FAM150B |
| LOC339788 |
| C2orf90 |
| LOC400940 |
| FAM110C |
| LOC730811 |
This is the comprehensive list of deleted genes in the wide peak for 14q32.12.
Table S14. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| AKT1 |
| TSHR |
| TCL1A |
| TRIP11 |
| GOLGA5 |
| DICER1 |
| TCL6 |
| BCL11B |
| hsa-mir-203 |
| hsa-mir-1247 |
| hsa-mir-656 |
| hsa-mir-370 |
| hsa-mir-770 |
| hsa-mir-345 |
| hsa-mir-342 |
| hsa-mir-1260 |
| SERPINA3 |
| ACTN1 |
| ACYP1 |
| BCYRN1 |
| BDKRB1 |
| BDKRB2 |
| CALM1 |
| SERPINA6 |
| ENTPD5 |
| FOXN3 |
| CHGA |
| CKB |
| CRIP1 |
| CRIP2 |
| DIO2 |
| DIO3 |
| DLST |
| DYNC1H1 |
| EIF5 |
| EML1 |
| ERH |
| ESRRB |
| FOS |
| GALC |
| GSTZ1 |
| GTF2A1 |
| BRF1 |
| HSP90AA1 |
| IFI27 |
| ITPK1 |
| JAG2 |
| KLC1 |
| LTBP2 |
| MARK3 |
| ATXN3 |
| MAP3K9 |
| ALDH6A1 |
| NDUFB1 |
| SERPINA5 |
| PGF |
| SERPINA1 |
| SERPINA4 |
| PPP2R5C |
| LGMN |
| PSEN1 |
| PSMC1 |
| ABCD4 |
| RAGE |
| SEL1L |
| SFRS5 |
| SLC8A3 |
| SLC10A1 |
| TGFB3 |
| TNFAIP2 |
| TRAF3 |
| VRK1 |
| WARS |
| XRCC3 |
| YY1 |
| DPF3 |
| GPR68 |
| GPR65 |
| NUMB |
| ADAM21 |
| ADAM20 |
| ADAM6 |
| DLK1 |
| CCNK |
| DCAF5 |
| ALKBH1 |
| EIF2B2 |
| MTA1 |
| PNMA1 |
| RPS6KA5 |
| NRXN3 |
| SPTLC2 |
| BAG5 |
| C14orf2 |
| CDC42BPB |
| TCL1B |
| RGS6 |
| KIAA0247 |
| KIAA0125 |
| KIAA0317 |
| TECPR2 |
| MED6 |
| FBLN5 |
| BATF |
| SIVA1 |
| NPC2 |
| AHSA1 |
| CYP46A1 |
| PAPOLA |
| ACOT2 |
| TMED10 |
| PTPN21 |
| C14orf1 |
| VASH1 |
| SNW1 |
| PCNX |
| TTLL5 |
| RCOR1 |
| PACS2 |
| ANGEL1 |
| PPP1R13B |
| TTC9 |
| FLRT2 |
| SIPA1L1 |
| DCAF4 |
| KIF26A |
| C14orf109 |
| MLH3 |
| PRO1768 |
| GPR132 |
| POMT2 |
| COQ6 |
| FCF1 |
| SERPINA10 |
| GLRX5 |
| COX16 |
| EVL |
| C14orf129 |
| CINP |
| ASB2 |
| ZFYVE1 |
| CPSF2 |
| KCNK10 |
| CDCA4 |
| C14orf102 |
| ATG2B |
| UBR7 |
| EXD2 |
| C14orf115 |
| SYNJ2BP |
| SLC39A9 |
| MEG3 |
| FLVCR2 |
| C14orf118 |
| SMEK1 |
| BTBD7 |
| TDP1 |
| ZNF839 |
| SPATA7 |
| YLPM1 |
| KCNK13 |
| C14orf162 |
| C14orf132 |
| DDX24 |
| ADCK1 |
| TMEM63C |
| GALNTL1 |
| KIAA1409 |
| BEGAIN |
| PPP4R4 |
| ZNF410 |
| NGB |
| RBM25 |
| C14orf133 |
| SMOC1 |
| MOAP1 |
| DIO3OS |
| C14orf4 |
| INF2 |
| OTUB2 |
| ZFYVE21 |
| MEG8 |
| WDR25 |
| C14orf139 |
| FAM164C |
| C14orf169 |
| CLMN |
| CATSPERB |
| ZC3H14 |
| RIN3 |
| C14orf159 |
| C14orf45 |
| TMEM121 |
| AMN |
| C14orf156 |
| DNAL1 |
| RPS6KL1 |
| IFI27L2 |
| SETD3 |
| C14orf153 |
| HHIPL1 |
| C14orf142 |
| STON2 |
| KIAA1737 |
| PAPLN |
| FAM181A |
| BTBD6 |
| C14orf143 |
| C14orf43 |
| LIN52 |
| NEK9 |
| C14orf73 |
| WDR20 |
| C14orf179 |
| AHNAK2 |
| TRMT61A |
| TDRD9 |
| ANKRD9 |
| AK7 |
| IFI27L1 |
| C14orf79 |
| PLD4 |
| ADSSL1 |
| C14orf148 |
| JDP2 |
| ISCA2 |
| ACOT4 |
| TTC8 |
| TC2N |
| SLC24A4 |
| SLC25A29 |
| DEGS2 |
| C14orf72 |
| ADAM21P |
| GSC |
| SERPINA12 |
| PRIMA1 |
| LOC145474 |
| PTGR2 |
| FAM161B |
| C14orf166B |
| ISM2 |
| C14orf145 |
| TTC7B |
| C14orf49 |
| C14orf174 |
| EML5 |
| MGC23270 |
| NUDT14 |
| C14orf48 |
| SERPINA11 |
| PROX2 |
| ZDHHC22 |
| TMED8 |
| C14orf178 |
| C14orf86 |
| SNHG10 |
| C14orf177 |
| C14orf68 |
| C14orf70 |
| KIAA0284 |
| C14orf80 |
| CCDC85C |
| NCRNA00203 |
| SNORD56B |
| SERPINA9 |
| VSX2 |
| COX8C |
| ASPG |
| SERPINA13 |
| C14orf64 |
| RTL1 |
| TMEM179 |
| HEATR4 |
| FLJ44817 |
| FLJ45244 |
| C14orf180 |
| MIR127 |
| MIR134 |
| MIR136 |
| MIR154 |
| MIR203 |
| MIR299 |
| CCDC88C |
| MIR323 |
| MIR337 |
| MIR342 |
| MIR345 |
| MIR376C |
| MIR369 |
| MIR376A1 |
| MIR377 |
| MIR379 |
| MIR380 |
| MIR381 |
| MIR382 |
| MIR433 |
| MIR431 |
| MIR329-1 |
| MIR329-2 |
| MIR453 |
| MIR409 |
| MIR412 |
| MIR410 |
| MIR376B |
| MIR485 |
| MIR493 |
| MIR432 |
| MIR494 |
| MIR495 |
| MIR496 |
| MIR487A |
| ACOT1 |
| ACOT6 |
| TMEM90A |
| C14orf184 |
| MIR539 |
| MIR376A2 |
| MIR487B |
| SCARNA13 |
| SNORA28 |
| SNORA79 |
| SNORD112 |
| MIR411 |
| MIR654 |
| MIR655 |
| MIR656 |
| SNORD113-1 |
| SNORD113-2 |
| SNORD113-3 |
| SNORD113-4 |
| SNORD113-5 |
| SNORD113-6 |
| SNORD113-7 |
| SNORD113-8 |
| SNORD113-9 |
| SNORD114-1 |
| SNORD114-2 |
| SNORD114-3 |
| SNORD114-4 |
| SNORD114-5 |
| SNORD114-6 |
| SNORD114-7 |
| SNORD114-8 |
| SNORD114-9 |
| SNORD114-10 |
| SNORD114-11 |
| SNORD114-12 |
| SNORD114-13 |
| SNORD114-14 |
| SNORD114-15 |
| SNORD114-16 |
| SNORD114-17 |
| SNORD114-18 |
| SNORD114-19 |
| SNORD114-20 |
| SNORD114-21 |
| SNORD114-22 |
| SNORD114-23 |
| SNORD114-24 |
| SNORD114-25 |
| SNORD114-26 |
| SNORD114-27 |
| SNORD114-28 |
| SNORD114-29 |
| SNORD114-30 |
| SNORD114-31 |
| MIR758 |
| MIR668 |
| MIR770 |
| SNORA11B |
| MIR300 |
| MIR541 |
| MIR665 |
| MIR543 |
| MIR889 |
| ZBTB42 |
| LOC100133469 |
| LOC100289511 |
This is the comprehensive list of deleted genes in the wide peak for 11p15.4.
Table S15. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CARS |
| HRAS |
| LMO1 |
| NUP98 |
| hsa-mir-302e |
| hsa-mir-483 |
| hsa-mir-675 |
| hsa-mir-210 |
| ADM |
| AP2A2 |
| AMPD3 |
| APBB1 |
| RHOG |
| ART1 |
| ASCL2 |
| CCKBR |
| CD81 |
| CD151 |
| CDKN1C |
| TPP1 |
| CNGA4 |
| CTSD |
| DRD4 |
| DUSP8 |
| EIF4G2 |
| HBB |
| HBBP1 |
| HBD |
| HBE1 |
| HBG1 |
| HBG2 |
| HPX |
| IGF2 |
| ILK |
| INS |
| IRF7 |
| KCNQ1 |
| LSP1 |
| MUC2 |
| MUC6 |
| NAP1L4 |
| SLC22A18 |
| SLC22A18AS |
| POLR2L |
| PSMD13 |
| RNH1 |
| MRPL23 |
| RPL27A |
| RPLP2 |
| RRM1 |
| SCT |
| SMPD1 |
| TRIM21 |
| ST5 |
| STIM1 |
| TAF10 |
| TALDO1 |
| TH |
| TSPAN4 |
| TNNI2 |
| TNNT3 |
| TRPC2 |
| PHLDA2 |
| TUB |
| WEE1 |
| ZNF143 |
| ZNF195 |
| ZNF214 |
| ZNF215 |
| RASSF7 |
| PPFIBP2 |
| IFITM1 |
| OR6A2 |
| DCHS1 |
| EIF3F |
| BRSK2 |
| CTR9 |
| TRIM66 |
| TSPAN32 |
| TSSC4 |
| MRVI1 |
| TRIM22 |
| IFITM3 |
| DEAF1 |
| IPO7 |
| IFITM2 |
| TRIM3 |
| LYVE1 |
| KCNQ1OT1 |
| PKP3 |
| SWAP70 |
| DENND5A |
| RRP8 |
| SIRT3 |
| OR52A1 |
| ARFIP2 |
| OR5E1P |
| OR10A3 |
| FXC1 |
| RBMXL2 |
| PGAP2 |
| C11orf21 |
| TRPM5 |
| UBQLN3 |
| RNF141 |
| IGF2AS |
| BET1L |
| CEND1 |
| CYB5R2 |
| TRIM34 |
| MUPCDH |
| TOLLIP |
| USP47 |
| TRIM68 |
| LRDD |
| KCNQ1DN |
| MMP26 |
| C11orf17 |
| C11orf16 |
| TMEM9B |
| NRIP3 |
| ASCL3 |
| CHRNA10 |
| PNPLA2 |
| PHRF1 |
| SCUBE2 |
| ZBED5 |
| SIGIRR |
| RIC8A |
| MRPL17 |
| EPS8L2 |
| STK33 |
| CHID1 |
| OR51G1 |
| OR51B4 |
| OR51B2 |
| OR52N1 |
| RIC3 |
| SLC25A22 |
| ATHL1 |
| OR51G2 |
| OR51E2 |
| PTDSS2 |
| HCCA2 |
| SBF2 |
| FAM160A2 |
| TRIM5 |
| SYT8 |
| PRKCDBP |
| ODF3 |
| OSBPL5 |
| LRRC56 |
| MRGPRE |
| ART5 |
| TRIMP1 |
| TRIM6 |
| OR52E2 |
| OR52J3 |
| OR51L1 |
| OR51A7 |
| OR51S1 |
| OR51F2 |
| OR52R1 |
| OR52M1 |
| OR52K2 |
| OR5P2 |
| OR5P3 |
| OR2D3 |
| OR2D2 |
| OR52W1 |
| OR56A4 |
| OR56A1 |
| SYT9 |
| OR52B4 |
| C11orf40 |
| OR52I2 |
| OR51E1 |
| UBQLNL |
| LOC143666 |
| OR10A5 |
| OR2AG1 |
| DNHD1 |
| SCGB1C1 |
| C11orf42 |
| NLRP6 |
| OR56B4 |
| LOC255512 |
| OR52B2 |
| C11orf35 |
| OR51F1 |
| OR51B5 |
| FLJ46111 |
| CSNK2A1P |
| OR51V1 |
| H19 |
| EFCAB4A |
| TMEM80 |
| OR10A4 |
| OLFML1 |
| C11orf36 |
| NLRP10 |
| NLRP14 |
| ANO9 |
| LOC338651 |
| B4GALNT4 |
| OR52L1 |
| OR2AG2 |
| OR52B6 |
| OR10A2 |
| OVCH2 |
| PDDC1 |
| GALNTL4 |
| MRGPRG |
| KRTAP5-1 |
| KRTAP5-3 |
| KRTAP5-4 |
| IFITM5 |
| FAM99A |
| OR56B1 |
| GVIN1 |
| OR52K1 |
| OR52I1 |
| OR51D1 |
| OR52A4 |
| OR52A5 |
| OR51B6 |
| OR51M1 |
| OR51Q1 |
| OR51I1 |
| OR51I2 |
| OR52D1 |
| OR52H1 |
| OR52N4 |
| OR52N5 |
| OR52N2 |
| OR52E6 |
| OR52E8 |
| OR52E4 |
| OR56A3 |
| OR56A5 |
| OR10A6 |
| OR51T1 |
| OR51A4 |
| OR51A2 |
| MIR210 |
| KRTAP5-5 |
| KRTAP5-2 |
| KRTAP5-6 |
| TMEM41B |
| TRIM6-TRIM34 |
| MIR483 |
| SNORA3 |
| SNORA52 |
| LOC650368 |
| SNORA23 |
| SNORA45 |
| SNORA54 |
| SNORD97 |
| INS-IGF2 |
| MUC5B |
| MIR675 |
| FAM99B |
| LOC100133161 |
| LOC100133545 |
This is the comprehensive list of deleted genes in the wide peak for 14q23.2.
Table S16. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| RAD51L1 |
| GPHN |
| hsa-mir-1260 |
| hsa-mir-625 |
| hsa-mir-548h-1 |
| ACTN1 |
| ACYP1 |
| ARG2 |
| BCYRN1 |
| ZFP36L1 |
| ENTPD5 |
| DIO2 |
| DLST |
| EIF2S1 |
| ERH |
| ESR2 |
| ESRRB |
| FNTB |
| FOS |
| FUT8 |
| GPX2 |
| GSTZ1 |
| HIF1A |
| HSPA2 |
| LTBP2 |
| MAX |
| MAP3K9 |
| ALDH6A1 |
| MNAT1 |
| MTHFD1 |
| SIX6 |
| PGF |
| PIGH |
| PPM1A |
| PPP2R5E |
| PRKCH |
| PSEN1 |
| ABCD4 |
| RTN1 |
| SFRS5 |
| SIX1 |
| SLC8A3 |
| SLC10A1 |
| SNAPC1 |
| SPTB |
| TGFB3 |
| ZBTB25 |
| DPF3 |
| NUMB |
| ADAM21 |
| ADAM20 |
| DCAF5 |
| ALKBH1 |
| EIF2B2 |
| PNMA1 |
| NRXN3 |
| AKAP5 |
| SPTLC2 |
| RGS6 |
| KIAA0247 |
| KIAA0586 |
| KIAA0317 |
| MED6 |
| VTI1B |
| BATF |
| NPC2 |
| AHSA1 |
| ACOT2 |
| TMED10 |
| C14orf1 |
| VASH1 |
| ZBTB1 |
| SNW1 |
| PCNX |
| DAAM1 |
| TTLL5 |
| SYNE2 |
| ANGEL1 |
| ZFYVE26 |
| TTC9 |
| PLEKHG3 |
| SIPA1L1 |
| DCAF4 |
| PLEK2 |
| TIMM9 |
| MLH3 |
| KCNH5 |
| POMT2 |
| COQ6 |
| FCF1 |
| RDH11 |
| COX16 |
| DACT1 |
| ATP6V1D |
| JKAMP |
| DHRS7 |
| SIX4 |
| ZFYVE1 |
| EXD2 |
| C14orf115 |
| SYNJ2BP |
| SLC39A9 |
| FLVCR2 |
| C14orf118 |
| YLPM1 |
| C14orf162 |
| ADCK1 |
| TMEM63C |
| RHOJ |
| GALNTL1 |
| PLEKHH1 |
| TRMT5 |
| ZNF410 |
| NGB |
| RBM25 |
| C14orf133 |
| SMOC1 |
| C14orf4 |
| MPP5 |
| C14orf135 |
| GPR135 |
| FAM164C |
| C14orf169 |
| C14orf45 |
| SGPP1 |
| C14orf156 |
| DNAL1 |
| RPS6KL1 |
| SYT16 |
| KIAA1737 |
| PAPLN |
| CHURC1 |
| C14orf43 |
| LIN52 |
| NEK9 |
| C14orf179 |
| WDR89 |
| C14orf149 |
| GPHB5 |
| C14orf148 |
| JDP2 |
| ISCA2 |
| ACOT4 |
| RDH12 |
| ADAM21P |
| C14orf50 |
| SLC38A6 |
| LOC145474 |
| PTGR2 |
| FAM161B |
| C14orf166B |
| ISM2 |
| FAM71D |
| TMEM229B |
| TMEM30B |
| C14orf174 |
| PROX2 |
| ZDHHC22 |
| TMED8 |
| C14orf178 |
| C14orf39 |
| SNORD56B |
| VSX2 |
| RAB15 |
| HEATR4 |
| C14orf181 |
| FLJ44817 |
| C14orf53 |
| ACOT1 |
| ACOT6 |
| LOC645431 |
| FLJ43390 |
| TMEM90A |
| C14orf38 |
| LOC100289511 |
This is the comprehensive list of deleted genes in the wide peak for 1p36.23.
Table S17. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| PAX7 |
| RPL22 |
| SDHB |
| ARID1A |
| TNFRSF14 |
| PRDM16 |
| MDS2 |
| hsa-mir-1976 |
| hsa-mir-1256 |
| hsa-mir-1290 |
| hsa-mir-34a |
| hsa-mir-551a |
| hsa-mir-429 |
| hsa-mir-1977 |
| hsa-mir-1302-2 |
| ALPL |
| RERE |
| C1QA |
| C1QB |
| C1QC |
| CA6 |
| CAPZB |
| CASP9 |
| RUNX3 |
| TNFRSF8 |
| CDA |
| CDC2L1 |
| CDC42 |
| CD52 |
| CLCN6 |
| CLCNKA |
| CLCNKB |
| CNR2 |
| CORT |
| DDOST |
| DFFA |
| DFFB |
| DVL1 |
| E2F2 |
| ECE1 |
| MEGF6 |
| EPHA2 |
| ENO1 |
| EPHA8 |
| EPHB2 |
| EXTL1 |
| EYA3 |
| FGR |
| MTOR |
| FUCA1 |
| IFI6 |
| GABRD |
| GALE |
| GNB1 |
| SFN |
| GPR3 |
| ZBTB48 |
| HMGN2 |
| HMGCL |
| HSPG2 |
| HTR1D |
| HTR6 |
| ID3 |
| TNFRSF9 |
| STMN1 |
| MFAP2 |
| MTHFR |
| NBL1 |
| NPPA |
| NPPB |
| PAFAH2 |
| PEX10 |
| PEX14 |
| PGD |
| PIK3CD |
| PLA2G2A |
| PLA2G5 |
| PLOD1 |
| EXOSC10 |
| PPP1R8 |
| PRKCZ |
| PTAFR |
| RAP1GAP |
| RHCE |
| RHD |
| RPA2 |
| RPL11 |
| RPS6KA1 |
| RSC1A1 |
| SCNN1D |
| SKI |
| SLC2A5 |
| SLC9A1 |
| SRM |
| TCEA3 |
| TCEB3 |
| TNFRSF1B |
| TP73 |
| TNFRSF4 |
| ZBTB17 |
| SLC30A2 |
| LUZP1 |
| PRDM2 |
| SNHG3 |
| NR0B2 |
| MMP23B |
| MMP23A |
| KCNAB2 |
| FCN3 |
| AKR7A2 |
| ALDH4A1 |
| EIF4G3 |
| TNFRSF25 |
| TNFRSF18 |
| PER3 |
| MAP3K6 |
| DHRS3 |
| VAMP3 |
| C1orf38 |
| H6PD |
| ISG15 |
| PLCH2 |
| CROCC |
| KIAA0562 |
| KLHL21 |
| SLC35E2 |
| ZBTB40 |
| MFN2 |
| CELA3A |
| WASF2 |
| ANGPTL7 |
| HNRNPR |
| SRRM1 |
| CNKSR1 |
| UBE4B |
| MAD2L2 |
| PDPN |
| NUDC |
| MASP2 |
| SFRS13A |
| UTS2 |
| RER1 |
| RCAN3 |
| MSTP2 |
| MSTP9 |
| PADI2 |
| LYPLA2 |
| PARK7 |
| CTRC |
| ACOT7 |
| DNAJC8 |
| CLSTN1 |
| AKR7A3 |
| SPEN |
| KDM1A |
| WDTC1 |
| KIAA0090 |
| KIF1B |
| PLEKHM2 |
| OTUD3 |
| KIAA1026 |
| CAMTA1 |
| DNAJC16 |
| UBR4 |
| ATP13A2 |
| TARDBP |
| CELA3B |
| ICMT |
| PADI4 |
| TMEM50A |
| STX12 |
| CLIC4 |
| SYF2 |
| CHD5 |
| C1orf144 |
| LDLRAP1 |
| NOC2L |
| FBXO2 |
| FBXO6 |
| PLA2G2D |
| OR4F3 |
| HSPB7 |
| ARHGEF16 |
| AHDC1 |
| SMPDL3B |
| HSPC157 |
| SSU72 |
| UBIAD1 |
| PADI1 |
| PLA2G2E |
| WDR8 |
| SLC45A1 |
| HP1BP3 |
| CELA2B |
| ZNF593 |
| SDF4 |
| MRTO4 |
| PADI3 |
| ERRFI1 |
| WNT4 |
| FBXO42 |
| RNF186 |
| MXRA8 |
| HES2 |
| GPN2 |
| FBLIM1 |
| MED18 |
| PQLC2 |
| CASZ1 |
| CPSF3L |
| C1orf159 |
| AURKAIP1 |
| MRPL20 |
| AIM1L |
| TMEM51 |
| XKR8 |
| ARHGEF10L |
| VPS13D |
| ATAD3A |
| TMEM57 |
| PANK4 |
| CAMK2N1 |
| ASAP3 |
| PNRC2 |
| PIGV |
| NBPF1 |
| NECAP2 |
| DNAJC11 |
| RCC2 |
| AJAP1 |
| FAM54B |
| CTNNBIP1 |
| C1orf63 |
| AGTRAP |
| C1orf128 |
| MAN1C1 |
| NIPAL3 |
| SEPN1 |
| KIAA0495 |
| PLEKHG5 |
| LRRC47 |
| PTCHD2 |
| KIF17 |
| HES4 |
| GRHL3 |
| IL22RA1 |
| MIIP |
| CELA2A |
| GPATCH3 |
| PLA2G2F |
| CCDC21 |
| NMNAT1 |
| VWA1 |
| PINK1 |
| PRAMEF1 |
| PRAMEF2 |
| NADK |
| PHACTR4 |
| C1orf135 |
| EFHD2 |
| MMEL1 |
| C1orf89 |
| OR4F5 |
| MUL1 |
| NOL9 |
| LIN28 |
| AGMAT |
| NCRNA00115 |
| MORN1 |
| GRRP1 |
| DHDDS |
| GPR157 |
| SPSB1 |
| GLTPD1 |
| ZNF436 |
| TAS1R2 |
| TAS1R1 |
| OR4F16 |
| ACTL8 |
| CCNL2 |
| SH3BGRL3 |
| SESN2 |
| ESPN |
| TAS1R3 |
| ATAD3B |
| TMEM222 |
| PLEKHN1 |
| USP48 |
| NBPF3 |
| ZDHHC18 |
| SLC25A33 |
| DDI2 |
| LZIC |
| TRIM63 |
| C1orf170 |
| CROCCL1 |
| SYTL1 |
| IGSF21 |
| KIAA1751 |
| KIAA2013 |
| THAP3 |
| C1orf201 |
| UBXN11 |
| C1orf158 |
| FBXO44 |
| ATPIF1 |
| CROCCL2 |
| FHAD1 |
| LOC115110 |
| FAM46B |
| RBP7 |
| ACAP3 |
| UBE2J2 |
| C1orf172 |
| AADACL3 |
| PUSL1 |
| B3GALT6 |
| IFFO2 |
| TPRG1L |
| C1orf93 |
| MYOM3 |
| KLHDC7A |
| VWA5B1 |
| UBXN10 |
| ARHGEF19 |
| ACTRT2 |
| MIB2 |
| C1orf127 |
| SAMD11 |
| LOC148413 |
| PHF13 |
| CCDC27 |
| C1orf213 |
| PDIK1L |
| C1orf64 |
| SLC2A7 |
| CALML6 |
| IL28RA |
| FAM43B |
| PAQR7 |
| FAM76A |
| TMEM201 |
| C1orf86 |
| C1orf126 |
| ATAD3C |
| AKR7L |
| TTLL10 |
| TMCO4 |
| ZNF683 |
| NPHP4 |
| FAM41C |
| LOC284632 |
| LOC284661 |
| SLC25A34 |
| ESPNP |
| C1orf174 |
| KLHL17 |
| C1orf70 |
| TMEM52 |
| AADACL4 |
| PRAMEF5 |
| HNRNPCL1 |
| PRAMEF9 |
| PRAMEF10 |
| FAM131C |
| PADI6 |
| C1orf187 |
| SPATA21 |
| AGRN |
| APITD1 |
| CATSPER4 |
| GPR153 |
| FAM132A |
| HES5 |
| LOC388588 |
| RNF207 |
| TMEM82 |
| TRNP1 |
| CD164L2 |
| HES3 |
| PRAMEF12 |
| PRAMEF21 |
| PRAMEF8 |
| PRAMEF18 |
| PRAMEF17 |
| PLA2G2C |
| PRAMEF4 |
| PRAMEF13 |
| SH2D5 |
| C1orf130 |
| PRAMEF3 |
| LDLRAD2 |
| MIR200A |
| MIR200B |
| MIR34A |
| FLJ42875 |
| PRAMEF11 |
| PRAMEF6 |
| LOC440563 |
| UQCRHL |
| C1orf151 |
| LOC441869 |
| PRAMEF7 |
| MIR429 |
| FAM138F |
| LOC643837 |
| TMEM88B |
| C1orf200 |
| PRAMEF19 |
| PRAMEF20 |
| FAM138A |
| LOC646471 |
| LOC649330 |
| LOC653566 |
| PRAMEF22 |
| PRAMEF15 |
| WASH5P |
| PRAMEF16 |
| FAM138C |
| SCARNA1 |
| SNORA59B |
| SNORA59A |
| MIR551A |
| CDC2L2 |
| LOC728661 |
| PRAMEF14 |
| FLJ37453 |
| OR4F29 |
| LOC100128003 |
| LOC100128842 |
| LOC100129534 |
| FLJ39609 |
| LOC100132062 |
| LOC100132287 |
| LOC100133331 |
| LOC100133612 |
| LOC100288778 |
This is the comprehensive list of deleted genes in the wide peak for 10p12.1.
Table S18. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-603 |
| GAD2 |
| PIP4K2A |
| MSRB2 |
| MYO3A |
| APBB1IP |
| KIAA1217 |
| PRTFDC1 |
| GPR158 |
| ARHGAP21 |
| THNSL1 |
| ENKUR |
| ARMC3 |
| OTUD1 |
| PTF1A |
| C10orf67 |
| MIR603 |
| LOC100128811 |
| PRINS |
This is the comprehensive list of deleted genes in the wide peak for 5q35.2.
Table S19. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| APC |
| CD74 |
| EBF1 |
| ITK |
| NPM1 |
| PDGFRB |
| TLX3 |
| NSD1 |
| RANBP17 |
| hsa-mir-340 |
| hsa-mir-1229 |
| hsa-mir-1271 |
| hsa-mir-585 |
| hsa-mir-218-2 |
| hsa-mir-103-1-as |
| hsa-mir-146a |
| hsa-mir-1303 |
| hsa-mir-1294 |
| hsa-mir-378 |
| hsa-mir-145 |
| hsa-mir-584 |
| hsa-mir-874 |
| hsa-mir-886 |
| hsa-mir-1289-2 |
| hsa-mir-1244 |
| hsa-mir-548f-3 |
| ADRA1B |
| ADRB2 |
| ANXA6 |
| ATOX1 |
| ALDH7A1 |
| BNIP1 |
| CAMK4 |
| CAMK2A |
| CAMLG |
| CANX |
| CCNG1 |
| CD14 |
| CDC25C |
| CDO1 |
| CDX1 |
| AP3S1 |
| CLTB |
| CSF1R |
| CSF2 |
| CSNK1A1 |
| CSNK1G3 |
| NKX2-5 |
| CTNNA1 |
| DBN1 |
| DMXL1 |
| DIAPH1 |
| DOCK2 |
| DPYSL3 |
| DRD1 |
| SLC26A2 |
| HBEGF |
| DUSP1 |
| EFNA5 |
| EGR1 |
| ETF1 |
| F12 |
| FABP6 |
| FAT2 |
| FBN2 |
| FER |
| FGF1 |
| FGFR4 |
| FOXI1 |
| FLT4 |
| GABRA1 |
| GABRA6 |
| GABRB2 |
| GABRG2 |
| GABRP |
| GDF9 |
| GFRA3 |
| GLRA1 |
| GM2A |
| GRK6 |
| GPX3 |
| GRIA1 |
| NR3C1 |
| GRM6 |
| HARS |
| HINT1 |
| HK3 |
| HMMR |
| HNRNPAB |
| HNRNPH1 |
| HRH2 |
| HSD17B4 |
| HSPA4 |
| HSPA9 |
| NDST1 |
| HTR4 |
| IK |
| IL3 |
| IL4 |
| IL5 |
| IL9 |
| IL12B |
| IL13 |
| IRF1 |
| KCNMB1 |
| KCNN2 |
| LCP2 |
| LECT2 |
| LMNB1 |
| LOX |
| LTC4S |
| SMAD5 |
| MAN2A1 |
| MCC |
| MFAP3 |
| MGAT1 |
| MSX2 |
| NDUFA2 |
| NEUROG1 |
| NPY6R |
| PCDH1 |
| PCDHGC3 |
| PDE6A |
| PFDN1 |
| PGGT1B |
| PITX1 |
| POU4F3 |
| PPIC |
| PPP2CA |
| PPP2R2B |
| MAPK9 |
| PROP1 |
| PURA |
| RARS |
| RPS14 |
| SGCD |
| SKP1 |
| SLC6A7 |
| SLC12A2 |
| SLC34A1 |
| SLC22A4 |
| SLC22A5 |
| SLIT3 |
| SNCB |
| SNX2 |
| SPARC |
| SPINK1 |
| SPOCK1 |
| SRP19 |
| STK10 |
| TAF7 |
| TCF7 |
| ZNF354A |
| TCOF1 |
| TGFBI |
| TTC1 |
| UBE2B |
| UBE2D2 |
| VDAC1 |
| WNT8A |
| REEP5 |
| NME5 |
| PDLIM4 |
| STC2 |
| EIF4EBP3 |
| PCDHGB4 |
| CDC23 |
| ADAM19 |
| FGF18 |
| HDAC3 |
| SQSTM1 |
| P4HA2 |
| ATP6V0E1 |
| ATG12 |
| PTTG1 |
| PDLIM7 |
| C5orf13 |
| CNOT8 |
| RAB9P1 |
| HAND1 |
| MED7 |
| MYOT |
| ADAMTS2 |
| NRG2 |
| CXCL14 |
| H2AFY |
| SMAD5OS |
| RNF14 |
| SNCAIP |
| CLINT1 |
| PCDHGA8 |
| PCDHA9 |
| MATR3 |
| MAML1 |
| KIAA0141 |
| JAKMIP2 |
| PJA2 |
| DDX46 |
| GFPT2 |
| SLC23A1 |
| GNPDA1 |
| SRA1 |
| RAD50 |
| KIF20A |
| G3BP1 |
| APBB3 |
| TNIP1 |
| GNB2L1 |
| SLU7 |
| RGS14 |
| SEC24A |
| CPLX2 |
| C5orf4 |
| FAM114A2 |
| BRD8 |
| TCERG1 |
| BTNL3 |
| HNRNPA0 |
| LMAN2 |
| SPINK5 |
| SOX30 |
| KIF3A |
| MGAT4B |
| B4GALT7 |
| SYNPO |
| RNF44 |
| ABLIM3 |
| HMGXB3 |
| TBC1D9B |
| ARHGAP26 |
| FSTL4 |
| ATP10B |
| N4BP3 |
| SEPT8 |
| FAF2 |
| WWC1 |
| FBXW11 |
| ACSL6 |
| PHF15 |
| LARP1 |
| HARS2 |
| ZNF346 |
| TNFAIP8 |
| GEMIN5 |
| PCDHGA12 |
| LRRTM2 |
| CCDC69 |
| PCDHB5 |
| FBXL21 |
| KLHL3 |
| TSPAN17 |
| OR4F3 |
| HAVCR1 |
| SNORD63 |
| SNORA74A |
| CYFIP2 |
| PKD2L2 |
| UQCRQ |
| AFF4 |
| PRELID1 |
| IL17B |
| MAT2B |
| SLC27A6 |
| SNX24 |
| MRPL22 |
| PCDHB1 |
| KCNIP1 |
| ZNF354C |
| TMED7 |
| ISOC1 |
| RPL26L1 |
| SAR1B |
| C5orf45 |
| DCTN4 |
| MGC29506 |
| PAIP2 |
| CDKL3 |
| PCDH12 |
| FAM13B |
| FAM53C |
| REEP2 |
| PRR16 |
| COMMD10 |
| DDX41 |
| NOP16 |
| LARS |
| CXXC5 |
| HMP19 |
| UIMC1 |
| RAPGEF6 |
| KDM3B |
| PHAX |
| RAB24 |
| RBM27 |
| NEURL1B |
| FLJ11235 |
| FAM193B |
| PCDHB18 |
| PCDHB17 |
| TMED9 |
| ZCCHC10 |
| PCDH24 |
| WDR55 |
| ANKHD1 |
| CCDC99 |
| THG1L |
| TMCO6 |
| TRIM36 |
| GALNT10 |
| NHP2 |
| RBM22 |
| RNF130 |
| PCDHGC5 |
| PCDHGC4 |
| PCDHGB7 |
| PCDHGB6 |
| PCDHGB5 |
| PCDHGB3 |
| PCDHGB2 |
| PCDHGB1 |
| PCDHGA11 |
| PCDHGA10 |
| PCDHGA9 |
| PCDHGA7 |
| PCDHGA6 |
| PCDHGA5 |
| PCDHGA4 |
| PCDHGA3 |
| PCDHGA2 |
| PCDHGA1 |
| PCDHGB8P |
| PCDHB15 |
| PCDHB14 |
| PCDHB13 |
| PCDHB12 |
| PCDHB11 |
| PCDHB10 |
| PCDHB9 |
| PCDHB8 |
| PCDHB7 |
| PCDHB6 |
| PCDHB4 |
| PCDHB3 |
| PCDHB2 |
| PCDHAC2 |
| PCDHAC1 |
| PCDHA13 |
| PCDHA12 |
| PCDHA11 |
| PCDHA10 |
| PCDHA8 |
| PCDHA7 |
| PCDHA6 |
| PCDHA5 |
| PCDHA4 |
| PCDHA3 |
| PCDHA2 |
| PCDHA1 |
| VTRNA1-3 |
| VTRNA1-2 |
| VTRNA1-1 |
| NMUR2 |
| FEM1C |
| C5orf15 |
| CDC42SE2 |
| TRPC7 |
| KIAA1191 |
| ERGIC1 |
| CLK4 |
| ODZ2 |
| CNOT6 |
| ZNF608 |
| KCTD16 |
| SEMA6A |
| PCDHB16 |
| HMHB1 |
| C5orf54 |
| EPB41L4A |
| SIL1 |
| GMCL1L |
| ARAP3 |
| RMND5B |
| FBXL17 |
| YTHDC2 |
| GRAMD3 |
| PCYOX1L |
| CCNJL |
| SH3TC2 |
| PANK3 |
| SAP30L |
| TXNDC15 |
| BTNL8 |
| DOK3 |
| ZFP2 |
| RUFY1 |
| CPEB4 |
| PRR7 |
| NDFIP1 |
| OR4F16 |
| FBXO38 |
| YIPF5 |
| TRIM7 |
| TIGD6 |
| SPRY4 |
| MXD3 |
| SLC4A9 |
| SLC25A2 |
| TSSK1B |
| PCDHB19P |
| PCBD2 |
| PSD2 |
| THOC3 |
| C5orf32 |
| MEGF10 |
| SPINK7 |
| TRIM52 |
| HAVCR2 |
| AGXT2L2 |
| C5orf62 |
| TSLP |
| FCHSD1 |
| UNC5A |
| LYRM7 |
| TRIM41 |
| SLC25A46 |
| BOD1 |
| CDKN2AIPNL |
| COL23A1 |
| TIMD4 |
| ZNF300 |
| MYOZ3 |
| UBTD2 |
| SCGB3A1 |
| PRDM6 |
| FTMT |
| SFXN1 |
| FNIP1 |
| SLC35A4 |
| GPRIN1 |
| PWWP2A |
| C1QTNF2 |
| C5orf26 |
| MARCH3 |
| LEAP2 |
| SCGB3A2 |
| ZNF354B |
| C5orf47 |
| PPARGC1B |
| PRRC1 |
| C5orf58 |
| ZNF474 |
| OR2Y1 |
| AFAP1L1 |
| GRPEL2 |
| LSM11 |
| GPR151 |
| STARD4 |
| WDR36 |
| LOC134466 |
| NUDCD2 |
| UBLCP1 |
| ANKRD43 |
| SHROOM1 |
| C5orf24 |
| C5orf20 |
| SLC36A2 |
| SPINK5L3 |
| C5orf41 |
| CEP120 |
| LOC153328 |
| SRFBP1 |
| ZMAT2 |
| BTNL9 |
| CCDC112 |
| PPP1R2P3 |
| FAM71B |
| PRELID2 |
| SH3RF2 |
| PLAC8L1 |
| RNF145 |
| DCP2 |
| ADAMTS19 |
| HIGD2A |
| SPATA24 |
| DNAJC18 |
| FAM153B |
| LOC202181 |
| STK32A |
| LVRN |
| SLC36A1 |
| EIF4E1B |
| RASGEF1C |
| LOC257358 |
| SH3PXD2B |
| LOC285593 |
| FAM153A |
| ARL10 |
| DTWD2 |
| RELL2 |
| LOC285627 |
| LOC285629 |
| SLC36A3 |
| KIF4B |
| OR2V2 |
| ZNF454 |
| C5orf60 |
| CHSY3 |
| TMEM173 |
| FAM170A |
| LOC340074 |
| ARSI |
| PFN3 |
| ZNF879 |
| IRGM |
| FBLL1 |
| CATSPER3 |
| NIPAL4 |
| TICAM2 |
| DND1 |
| C5orf25 |
| C5orf48 |
| LOC389332 |
| LOC389333 |
| C5orf46 |
| FLJ41603 |
| FLJ44606 |
| SPINK6 |
| ANKHD1-EIF4EBP3 |
| MIR103-1 |
| MIR143 |
| MIR145 |
| MIR146A |
| MIR218-2 |
| SPINK5L2 |
| C5orf40 |
| C5orf56 |
| MIR340 |
| C5orf53 |
| TIFAB |
| CTXN3 |
| SNORD95 |
| SNORD96A |
| ECSCR |
| TMEM232 |
| GRXCR2 |
| SPINK9 |
| ZFP62 |
| FLJ33630 |
| CCNI2 |
| CBY3 |
| FAM153C |
| SNORA13 |
| SNORA74B |
| MIR548C |
| MIR548D2 |
| MIR585 |
| SNHG4 |
| LOC728264 |
| LOC728554 |
| AACSL |
| LOC729678 |
| OR4F29 |
| MIR886 |
| MIR874 |
| FAM196B |
| LOC100132062 |
| LOC100132287 |
| LOC100133331 |
| C5orf52 |
| LOC100268168 |
| LOC100286948 |
| TMED7-TICAM2 |
This is the comprehensive list of deleted genes in the wide peak for 1q44.
Table S20. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| FH |
| ADSS |
| CHML |
| CHRM3 |
| HNRNPU |
| MTR |
| RGS7 |
| RYR2 |
| ZNF124 |
| KMO |
| EXO1 |
| CEP170 |
| AKT3 |
| ZNF238 |
| SDCCAG8 |
| OPN3 |
| TRIM58 |
| AHCTF1 |
| OR1C1 |
| OR2M4 |
| OR2L2 |
| OR2L1P |
| OR2T1 |
| PPPDE1 |
| SCCPDH |
| KIF26B |
| ZNF692 |
| FMN2 |
| ZNF695 |
| ZP4 |
| TFB2M |
| GREM2 |
| SMYD3 |
| ZNF669 |
| ZNF672 |
| SH3BP5L |
| OR2G3 |
| OR2G2 |
| OR2C3 |
| EFCAB2 |
| ZNF496 |
| ZNF670 |
| NLRP3 |
| FAM36A |
| OR2M5 |
| OR2M3 |
| OR2T12 |
| OR14C36 |
| OR2T34 |
| OR2T10 |
| OR2T4 |
| OR2T11 |
| OR2B11 |
| WDR64 |
| C1orf150 |
| LOC148824 |
| LOC149134 |
| CNST |
| PLD5 |
| C1orf100 |
| OR2T6 |
| C1orf101 |
| PGBD2 |
| OR2L13 |
| OR14A16 |
| NCRNA00201 |
| VN1R5 |
| LOC339535 |
| OR6F1 |
| OR2W3 |
| OR2T8 |
| OR2T3 |
| OR2T29 |
| C1orf229 |
| OR2M1P |
| OR11L1 |
| OR2L8 |
| OR2AK2 |
| OR2L3 |
| OR2M2 |
| OR2T33 |
| OR2M7 |
| OR2G6 |
| OR2T2 |
| OR2T5 |
| OR14I1 |
| OR2T27 |
| OR2T35 |
| MAP1LC3C |
| OR2W5 |
| OR13G1 |
| LOC646627 |
| LOC731275 |
| LOC100130331 |
This is the comprehensive list of deleted genes in the wide peak for 10q23.1.
Table S21. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BMPR1A |
| PRF1 |
| PTEN |
| MYST4 |
| hsa-mir-346 |
| hsa-mir-606 |
| hsa-mir-1254 |
| hsa-mir-1296 |
| ADK |
| ANXA2P3 |
| ANXA7 |
| ANXA11 |
| CAMK2G |
| COL13A1 |
| DNA2 |
| EGR2 |
| EIF4EBP2 |
| GLUD1 |
| GRID1 |
| HK1 |
| HNRNPH3 |
| KCNMA1 |
| MAT1A |
| NODAL |
| P4HA1 |
| PCBD1 |
| PLAU |
| PPA1 |
| PPP3CB |
| SRGN |
| PSAP |
| RGR |
| RPS24 |
| SFTPD |
| SNCG |
| SUPV3L1 |
| TACR2 |
| VCL |
| VDAC2 |
| SLC25A16 |
| NDST2 |
| MBL1P1 |
| LIPF |
| SGPL1 |
| PAPSS2 |
| DDX21 |
| DLG5 |
| CHST3 |
| VPS26A |
| MINPP1 |
| SEC24C |
| SPOCK2 |
| PPIF |
| CBARA1 |
| NRG3 |
| C10orf116 |
| POLR3A |
| LDB3 |
| ECD |
| ZNF365 |
| KIAA0913 |
| WAPAL |
| DNAJC9 |
| SIRT1 |
| TSPAN15 |
| NUDT13 |
| HERC4 |
| LRIT1 |
| KIAA1279 |
| AP3M1 |
| GHITM |
| KIAA1274 |
| CTNNA3 |
| NRBF2 |
| NEUROG3 |
| ASCC1 |
| MRPS16 |
| DUSP13 |
| FAM190B |
| FAM35A |
| DDIT4 |
| DNAJB12 |
| LRRC20 |
| SLC29A3 |
| RNLS |
| H2AFY2 |
| RUFY2 |
| CCAR1 |
| DNAJC12 |
| SAR1A |
| ZMIZ1 |
| MYOZ1 |
| CDH23 |
| PBLD |
| NPFFR1 |
| C10orf54 |
| DDX50 |
| MMRN2 |
| SYNPO2L |
| C10orf57 |
| HKDC1 |
| TET1 |
| TSPAN14 |
| C10orf11 |
| C10orf58 |
| DYDC2 |
| PLA2G12B |
| MYPN |
| ZNF503 |
| AIFM2 |
| ADO |
| ATAD1 |
| LOC84989 |
| CCDC109A |
| PCDH21 |
| OPN4 |
| CHCHD1 |
| ZMYND17 |
| TTC18 |
| COMTD1 |
| AGAP11 |
| C10orf104 |
| ADAMTS14 |
| SAMD8 |
| LIPJ |
| CFLP1 |
| DYDC1 |
| EIF5AL1 |
| USP54 |
| FUT11 |
| OIT3 |
| LOC219347 |
| PLAC9 |
| ZCCHC24 |
| UNC5B |
| STOX1 |
| C10orf35 |
| TYSND1 |
| RTKN2 |
| C10orf27 |
| ATOH7 |
| REEP3 |
| JMJD1C |
| LOC283050 |
| FAM149B1 |
| DUPD1 |
| LRIT2 |
| LRRTM3 |
| SH2D4B |
| C10orf99 |
| C10orf105 |
| C10orf55 |
| LOC439994 |
| MIR346 |
| LOC642361 |
| FAM25A |
| LIPK |
| LIPN |
| LOC650623 |
| SFTPA1 |
| SNORD98 |
| FAM22A |
| FAM22D |
| LOC728190 |
| AGAP5 |
| BMS1P4 |
| SFTPA2 |
| LOC100128292 |
| C10orf41 |
| KILLIN |
This is the comprehensive list of deleted genes in the wide peak for 6p24.1.
Table S22. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| PRDM1 |
| CCND3 |
| DAXX |
| FANCE |
| HMGA1 |
| IRF4 |
| MLLT4 |
| MYB |
| PIM1 |
| POU5F1 |
| TRIM27 |
| ROS1 |
| SFRS3 |
| TNFAIP3 |
| DEK |
| TFEB |
| STL |
| HIST1H4I |
| FGFR1OP |
| GOPC |
| hsa-mir-1913 |
| hsa-mir-1202 |
| hsa-mir-548a-2 |
| hsa-mir-588 |
| hsa-mir-548b |
| hsa-mir-587 |
| hsa-mir-2113 |
| hsa-mir-30a |
| hsa-mir-133b |
| hsa-mir-586 |
| hsa-mir-1275 |
| hsa-mir-219-1 |
| hsa-mir-1236 |
| hsa-mir-877 |
| hsa-mir-548a-1 |
| ABCF1 |
| ACAT2 |
| CRISP1 |
| AGER |
| AIF1 |
| AIM1 |
| AMD1 |
| ARG1 |
| ATP6V1G2 |
| BAI3 |
| BAK1 |
| BCKDHB |
| CFB |
| BMP5 |
| BMP6 |
| DST |
| BPHL |
| BTN1A1 |
| BYSL |
| C2 |
| C4A |
| C4B |
| DDR1 |
| RUNX2 |
| CCNC |
| CDC5L |
| CDKN1A |
| CDSN |
| CGA |
| CLIC1 |
| CLPS |
| CCR6 |
| CNR1 |
| COL9A1 |
| COL10A1 |
| COL11A2 |
| COL12A1 |
| COL19A1 |
| COX7A2 |
| ATF6B |
| MAPK14 |
| CSNK2B |
| CTGF |
| CYP21A2 |
| DNAH8 |
| DOM3Z |
| DSP |
| E2F3 |
| EDN1 |
| EEF1A1 |
| SERPINB1 |
| SLC29A1 |
| EPB41L2 |
| EPHA7 |
| EYA4 |
| ESR1 |
| F13A1 |
| FABP7 |
| FKBP5 |
| FOXF2 |
| FOXC1 |
| FOXO3 |
| FRK |
| FUCA2 |
| FYN |
| GABBR1 |
| GABRR1 |
| GABRR2 |
| GCNT2 |
| GJA1 |
| GCLC |
| GLO1 |
| GLP1R |
| GMDS |
| GMPR |
| GNL1 |
| GPLD1 |
| GPR6 |
| GPR31 |
| GPX5 |
| GRIK2 |
| GRM1 |
| GRM4 |
| GSTA1 |
| GSTA2 |
| GSTA3 |
| GSTA4 |
| GTF2H4 |
| GUCA1A |
| GUCA1B |
| HIST1H1C |
| HIST1H1D |
| HIST1H1E |
| HIST1H1B |
| HIST1H1T |
| HIST1H2AE |
| HIST1H2AD |
| HIST1H2BD |
| HIST1H2BB |
| HIST1H1A |
| HCRTR2 |
| HDAC2 |
| HFE |
| HIVEP1 |
| HIVEP2 |
| HLA-A |
| HLA-B |
| HLA-C |
| HLA-DMA |
| HLA-DMB |
| HLA-DOA |
| HLA-DOB |
| HLA-DPA1 |
| HLA-DPB1 |
| HLA-DPB2 |
| HLA-DQA1 |
| HLA-DQA2 |
| HLA-DQB1 |
| HLA-DQB2 |
| HLA-DRA |
| HLA-DRB1 |
| HLA-DRB5 |
| HLA-DRB6 |
| HLA-E |
| HLA-F |
| HLA-G |
| HLA-H |
| HLA-J |
| HLA-L |
| HSF2 |
| HSPA1A |
| HSPA1B |
| HSPA1L |
| HSP90AB1 |
| HTR1B |
| HTR1E |
| ID4 |
| IFNGR1 |
| IGF2R |
| IL17A |
| IMPG1 |
| ITPR3 |
| JARID2 |
| KIFC1 |
| KIF25 |
| KPNA5 |
| LAMA2 |
| LAMA4 |
| LPA |
| LTA |
| LTB |
| MARCKS |
| MAK |
| MAN1A1 |
| MAS1 |
| MCM3 |
| MDFI |
| ME1 |
| MEA1 |
| MAP3K4 |
| MAP3K5 |
| MEP1A |
| MICA |
| MICB |
| MLN |
| MOCS1 |
| MOG |
| MSH5 |
| MUT |
| MYO6 |
| RPL10A |
| NEDD9 |
| NEU1 |
| NFKBIE |
| NFKBIL1 |
| NFYA |
| NMBR |
| NQO2 |
| NOTCH4 |
| NT5E |
| OPRM1 |
| PARK2 |
| PBX2 |
| PCMT1 |
| PDCD2 |
| ENPP1 |
| ENPP3 |
| PEX6 |
| PEX7 |
| PGC |
| PGK2 |
| PGM3 |
| PHF1 |
| SERPINB6 |
| SERPINB9 |
| PKHD1 |
| PLAGL1 |
| PLG |
| PLN |
| POLH |
| POU3F2 |
| PPARD |
| PPP1R2P1 |
| PPP1R10 |
| PPP2R5D |
| PREP |
| PRIM2 |
| PKIB |
| MAPK13 |
| PRL |
| PSMB1 |
| PSMB8 |
| PSMB9 |
| PTK7 |
| PTPRK |
| RGL2 |
| PRPH2 |
| REV3L |
| RHAG |
| RING1 |
| BRD2 |
| RNF5 |
| RPS6KA2 |
| RPS10 |
| RPS12 |
| RPS18 |
| RREB1 |
| RXRB |
| VPS52 |
| ATXN1 |
| SGK1 |
| SIM1 |
| SKIV2L |
| SLC17A1 |
| SLC22A1 |
| SLC22A3 |
| SLC22A2 |
| SMPD2 |
| SNRPC |
| SOD2 |
| SOX4 |
| SRF |
| SRPK1 |
| SSR1 |
| ELOVL4 |
| T |
| TAF11 |
| MAP3K7 |
| TAP1 |
| TAP2 |
| TAPBP |
| TBCC |
| TBP |
| TCF19 |
| TCF21 |
| TCP1 |
| TCP10 |
| TCP11 |
| TCTE3 |
| PPP1R11 |
| DYNLT1 |
| TEAD3 |
| TFAP2A |
| TFAP2B |
| THBS2 |
| NR2E1 |
| TNF |
| TNXA |
| TNXB |
| TPBG |
| TPD52L1 |
| TPMT |
| CRISP2 |
| TSPYL1 |
| TTK |
| TUBB2A |
| TULP1 |
| UTRN |
| VARS |
| VEGFA |
| EZR |
| VIP |
| ZNF76 |
| ZNF165 |
| TRIM26 |
| ZNF184 |
| ZNF187 |
| ZNF192 |
| ZNF193 |
| ZNF204 |
| PTP4A1 |
| ALDH5A1 |
| BAT2 |
| BAT3 |
| BAT4 |
| BAT1 |
| BAT5 |
| SLC39A7 |
| HSD17B8 |
| OR2H2 |
| RDBP |
| LST1 |
| PLA2G7 |
| EPM2A |
| HIST1H2AI |
| HIST1H2AK |
| HIST1H2AJ |
| HIST1H2AL |
| HIST1H2AC |
| HIST1H2AB |
| HIST1H2AM |
| HIST1H2BG |
| HIST1H2BL |
| HIST1H2BN |
| HIST1H2BM |
| HIST1H2BF |
| HIST1H2BE |
| HIST1H2BH |
| HIST1H2BI |
| HIST1H2BC |
| HIST1H2BO |
| HIST1H3A |
| HIST1H3D |
| HIST1H3C |
| HIST1H3E |
| HIST1H3I |
| HIST1H3G |
| HIST1H3J |
| HIST1H3H |
| HIST1H3B |
| HIST1H4A |
| HIST1H4D |
| HIST1H4F |
| HIST1H4K |
| HIST1H4J |
| HIST1H4C |
| HIST1H4H |
| HIST1H4B |
| HIST1H4E |
| HIST1H4L |
| HIST1H4G |
| STX7 |
| CMAH |
| DHX16 |
| SUPT3H |
| PEX3 |
| GCM1 |
| DDO |
| RNASET2 |
| KCNK5 |
| STX11 |
| B3GALT4 |
| SNX3 |
| RNGTT |
| RIPK1 |
| CD164 |
| SYNGAP1 |
| WISP3 |
| STK19 |
| IER3 |
| SYNJ2 |
| VNN2 |
| VNN1 |
| PRPF4B |
| WASF1 |
| HIST1H3F |
| HIST1H2AG |
| HIST1H2BJ |
| RNF8 |
| TAAR5 |
| MAP7 |
| TBX18 |
| LATS1 |
| GCM2 |
| WDR46 |
| ZBTB22 |
| TAAR2 |
| TAAR3 |
| CD83 |
| HMGN3 |
| PPT2 |
| CDYL |
| NCR2 |
| MED23 |
| QKI |
| LY86 |
| FHL5 |
| AKAP7 |
| ATG5 |
| MED20 |
| SLC25A27 |
| TBPL1 |
| EEF1E1 |
| BAG2 |
| POLR1C |
| MAD2L1BP |
| WTAP |
| AKAP12 |
| MDC1 |
| TRAM2 |
| KIAA0408 |
| PHACTR2 |
| FAM65B |
| ZSCAN12 |
| BCLAF1 |
| CUL7 |
| ZBTB24 |
| KIAA0319 |
| SNAP91 |
| FIG4 |
| NUP153 |
| CASP8AP2 |
| RANBP9 |
| SLC17A4 |
| UST |
| TRIM10 |
| FLOT1 |
| RCAN2 |
| SLC17A2 |
| HCG9 |
| PRSS16 |
| CRISP3 |
| TRDN |
| CITED2 |
| BTN3A3 |
| BTN2A2 |
| PECI |
| PFDN6 |
| HMGN4 |
| TRIM38 |
| CAP2 |
| SYNCRIP |
| UBD |
| AGPAT1 |
| SLC35A1 |
| SCGN |
| C6orf108 |
| C6orf10 |
| FARS2 |
| FUT9 |
| CNPY3 |
| TRAF3IP2 |
| HBS1L |
| SLC17A3 |
| RPP40 |
| FRS3 |
| PDE10A |
| SLC22A7 |
| HCP5 |
| EHMT2 |
| SMPDL3A |
| APOBEC2 |
| PNRC1 |
| ASCC3 |
| RAB32 |
| TRIM31 |
| KATNA1 |
| BTN3A2 |
| BTN3A1 |
| BTN2A1 |
| CAPN11 |
| BVES |
| NUDT3 |
| SEC63 |
| NRM |
| STK38 |
| RBM16 |
| KIAA1009 |
| ICK |
| ENPP4 |
| ANKRD6 |
| RIMS1 |
| DOPEY1 |
| ZNF292 |
| FTSJD2 |
| CDC2L6 |
| CUL9 |
| MAP3K7IP2 |
| MDN1 |
| TSPYL4 |
| ANKS1A |
| UBR2 |
| SASH1 |
| SYNE1 |
| KIAA0776 |
| SIRT5 |
| PHF3 |
| HEY2 |
| DAAM2 |
| KIAA0240 |
| DDAH2 |
| HEBP2 |
| ORC3L |
| CD2AP |
| BRD7P3 |
| MTCH1 |
| ZNF318 |
| SPDEF |
| MTO1 |
| ASF1A |
| YIPF3 |
| USP49 |
| CCDC28A |
| MTHFD1L |
| SFRS18 |
| IBTK |
| MOXD1 |
| IPCEF1 |
| ZNF451 |
| SENP6 |
| OR2B6 |
| TIAM2 |
| FBXL4 |
| C6orf123 |
| FAM50B |
| FBXO9 |
| FBXO5 |
| SLC17A5 |
| OR12D2 |
| OR11A1 |
| RGS17 |
| OR2W1 |
| OR2J2 |
| OR2H1 |
| SNORD52 |
| SNORD50A |
| SNORD48 |
| PDE7B |
| FILIP1 |
| BRPF3 |
| GNMT |
| TNFRSF21 |
| SESN1 |
| TINAG |
| DLL1 |
| OSTM1 |
| MRPS18B |
| TMEM14A |
| MRPL18 |
| NDUFAF4 |
| C6orf15 |
| MYLIP |
| ABT1 |
| DSE |
| PRICKLE4 |
| PACSIN1 |
| ZNRD1 |
| CLDN20 |
| NOX3 |
| DEF6 |
| C6orf48 |
| SLC35B3 |
| HDDC2 |
| GMNN |
| MRPL2 |
| TFB1M |
| CYB5R4 |
| TUBE1 |
| C6orf203 |
| TBC1D7 |
| NRN1 |
| CYP39A1 |
| CDC40 |
| RWDD1 |
| AIG1 |
| NOL7 |
| SNX9 |
| FAM8A1 |
| UBE2J1 |
| DCDC2 |
| ETV7 |
| TMEM14C |
| VTA1 |
| LGSN |
| TTRAP |
| CUTA |
| PPIL1 |
| BRP44L |
| HECA |
| RAB23 |
| COQ3 |
| CLIC5 |
| IL20RA |
| TREM2 |
| TREM1 |
| UNC93A |
| HCG4 |
| GFOD1 |
| HMGCLL1 |
| MTRF1L |
| CCHCR1 |
| GTPBP2 |
| BTN2A3 |
| AHI1 |
| UHRF1BP1 |
| ELOVL2 |
| CDKAL1 |
| PAK1IP1 |
| RMND1 |
| PHIP |
| SOBP |
| AKIRIN2 |
| CENPQ |
| MRPS18A |
| MRPS10 |
| LRRC1 |
| PHF10 |
| QRSL1 |
| VNN3 |
| TMEM63B |
| NCRNA00120 |
| DDX43 |
| FAM46A |
| LRRC16A |
| TBC1D22B |
| TMEM30A |
| EXOC2 |
| C6orf64 |
| C6orf70 |
| LMBRD1 |
| TRERF1 |
| ACOT13 |
| ECHDC1 |
| APOM |
| BTNL2 |
| KCNQ5 |
| TRIM39 |
| AGPAT4 |
| WRNIP1 |
| DUSP22 |
| TULP4 |
| RARS2 |
| HYMAI |
| PDSS2 |
| LYRM4 |
| C6orf162 |
| VARS2 |
| GPR126 |
| KIAA1244 |
| NHSL1 |
| LYRM2 |
| SNX14 |
| MRS2 |
| PLEKHG1 |
| ARID1B |
| LRFN2 |
| AARS2 |
| XPO5 |
| SERINC1 |
| HACE1 |
| FAM135A |
| TMEM181 |
| ZBTB2 |
| BEND3 |
| KIAA1586 |
| CPNE5 |
| LSM2 |
| C6orf47 |
| LY6G5B |
| C6orf115 |
| RRAGD |
| LY6G6D |
| ENPP5 |
| PRDM13 |
| PBOV1 |
| BACH2 |
| ELOVL5 |
| TRMT11 |
| SMAP1 |
| ZFAND3 |
| SLC22A23 |
| C6orf164 |
| MUTED |
| CCDC90A |
| GPSM3 |
| FKBPL |
| KIF13A |
| PERP |
| SMOC2 |
| POPDC3 |
| ZNF323 |
| ALDH8A1 |
| C6orf106 |
| MICAL1 |
| MRPL14 |
| DLK2 |
| LY6G6E |
| ULBP3 |
| OR2A4 |
| C6orf211 |
| OGFRL1 |
| FAM184A |
| ZDHHC14 |
| ZNF322A |
| MANEA |
| C6orf103 |
| TREML2 |
| RPP21 |
| C6orf155 |
| C6orf134 |
| FRMD1 |
| C6orf59 |
| C6orf208 |
| C6orf97 |
| MYCT1 |
| ZKSCAN3 |
| ULBP2 |
| ULBP1 |
| ZSCAN16 |
| LPAL2 |
| RNF39 |
| SLC44A4 |
| C6orf27 |
| C6orf25 |
| LY6G6C |
| LY6G5C |
| PRR3 |
| KHDC1 |
| NCRNA00171 |
| PRRT1 |
| EGFL8 |
| HCG2P7 |
| HCG4P6 |
| GPR63 |
| TXNDC5 |
| COL21A1 |
| C6orf62 |
| OR5V1 |
| OR2B2 |
| PPP1R14C |
| OR12D3 |
| SPACA1 |
| RNF146 |
| TMEM14B |
| SF3B5 |
| TAAR8 |
| SH3BGRL2 |
| RIOK1 |
| TFAP2D |
| KCNK16 |
| RSPH3 |
| TTLL2 |
| FKSG83 |
| DTNBP1 |
| ARMC2 |
| FBXO30 |
| RPF2 |
| C6orf125 |
| L3MBTL3 |
| FAM120B |
| MCHR2 |
| PGBD1 |
| C6orf168 |
| FNDC1 |
| TTBK1 |
| GJA10 |
| RTN4IP1 |
| C6orf105 |
| RSPO3 |
| LRP11 |
| LTV1 |
| SERAC1 |
| USP45 |
| REPS1 |
| HIST1H2AH |
| HIST1H2BK |
| PPIL4 |
| PAQR8 |
| C6orf114 |
| SLC22A16 |
| C6orf153 |
| KCNK17 |
| ABCC10 |
| TRIM15 |
| KLC4 |
| UBE2CBP |
| C6orf142 |
| C6orf176 |
| KIAA1919 |
| TJAP1 |
| ARHGAP18 |
| POM121L2 |
| SYTL3 |
| FOXQ1 |
| GTF3C6 |
| MRAP2 |
| RWDD2A |
| IL17F |
| FAM54A |
| SFT2D1 |
| EFHC1 |
| BTBD9 |
| KLHL32 |
| TMEM200A |
| SCAND3 |
| C6orf150 |
| FOXP4 |
| KLHDC3 |
| NUS1 |
| C6orf72 |
| SLC26A8 |
| IL22RA2 |
| MAS1L |
| C6orf192 |
| SLC16A10 |
| IP6K3 |
| TAGAP |
| TAF8 |
| ADAT2 |
| RIPPLY2 |
| IRAK1BP1 |
| CLVS2 |
| TAAR9 |
| TAAR1 |
| STXBP5 |
| NCOA7 |
| HINT3 |
| PACRG |
| B3GAT2 |
| C6orf57 |
| CD109 |
| RAET1E |
| PM20D2 |
| SFRS13B |
| C6orf141 |
| HUS1B |
| TRIM40 |
| DPCR1 |
| NRSN1 |
| LOC153910 |
| C6orf94 |
| SNRNP48 |
| CNKSR3 |
| RAET1L |
| SAMD3 |
| MGC34034 |
| SLC2A12 |
| MBOAT1 |
| HDGFL1 |
| PNLDC1 |
| RNF217 |
| NKAIN2 |
| C6orf221 |
| C6orf165 |
| C6orf195 |
| LOC154449 |
| C6orf129 |
| PRSS35 |
| LCA5 |
| OLIG3 |
| TXLNB |
| DACT2 |
| C6orf118 |
| PSORS1C1 |
| PSORS1C2 |
| KIAA1949 |
| PTCRA |
| OSTCL |
| TCTE1 |
| KHDRBS2 |
| TUBB |
| C6orf163 |
| MGC26597 |
| C6orf185 |
| AKD1 |
| NT5DC1 |
| FAM26D |
| ZUFSP |
| FAM162B |
| C6orf170 |
| BEND6 |
| GSTA5 |
| OPN5 |
| GPR115 |
| GPR116 |
| TDRD6 |
| SPATS1 |
| C6orf223 |
| RSPH9 |
| C6orf154 |
| TREML2P |
| LOC221442 |
| C6orf130 |
| KIF6 |
| TMEM217 |
| FGD2 |
| PI16 |
| C6orf89 |
| C6orf81 |
| C6orf1 |
| LEMD2 |
| ZBTB9 |
| ZBTB12 |
| C6orf136 |
| ZSCAN12L1 |
| HIST1H2AA |
| KDM1B |
| RBM24 |
| RNF182 |
| PHACTR1 |
| LOC221710 |
| SYCP2L |
| C6orf218 |
| C6orf145 |
| HS3ST5 |
| GPRC6A |
| RFX6 |
| SLC35F1 |
| FAM83B |
| GPR111 |
| TSPO2 |
| UNC5CL |
| KCTD20 |
| PXT1 |
| LHFPL5 |
| SCUBE3 |
| ZSCAN23 |
| NKAPL |
| LOC222699 |
| C6orf146 |
| VGLL2 |
| DEFB110 |
| DEFB112 |
| DEFB113 |
| DEFB114 |
| LACE1 |
| HCG27 |
| C6orf191 |
| C6orf167 |
| WDR27 |
| FAM26E |
| MCM9 |
| RNF144B |
| HIST1H2BA |
| SCML4 |
| GPX6 |
| SHPRH |
| NCR3 |
| LY6G6F |
| MDGA1 |
| GPR110 |
| ZNF311 |
| LOC285733 |
| LOC285735 |
| LOC285740 |
| C6orf182 |
| PPIL6 |
| FLJ34503 |
| DCBLD1 |
| LOC285768 |
| LOC285780 |
| CAGE1 |
| LOC285796 |
| PRR18 |
| LOC285830 |
| HCG22 |
| LOC285847 |
| PNPLA1 |
| TREML4 |
| RPL7L1 |
| RNF5P1 |
| TAAR6 |
| SLC35D3 |
| ZC3H12D |
| MYLK4 |
| DPPA5 |
| IFITM4P |
| C6orf127 |
| TREML1 |
| TREML3 |
| RSPH4A |
| ECT2L |
| EYS |
| ZNF391 |
| ZFP57 |
| TUBB2B |
| SLC35B2 |
| C6orf52 |
| NUP43 |
| HCG26 |
| C6orf58 |
| RAET1G |
| KAAG1 |
| GUSBL2 |
| GJB7 |
| NHLRC1 |
| ZKSCAN4 |
| GUSBL1 |
| SNHG5 |
| FAM136B |
| SUMO4 |
| C6orf147 |
| C6orf173 |
| C6orf174 |
| C6orf204 |
| C6orf120 |
| THEMIS |
| PSMG4 |
| SFTA2 |
| C6orf126 |
| C6orf222 |
| C6orf140 |
| GFRAL |
| LIN28B |
| SAMD5 |
| IYD |
| MUC21 |
| DKFZP686I15217 |
| FLJ22536 |
| MCCD1 |
| C6orf26 |
| C6orf227 |
| CRIP3 |
| KLHL31 |
| TCP10L2 |
| C6orf122 |
| DEFB133 |
| C6orf201 |
| GTF2H5 |
| HERV-FRD |
| MIR206 |
| MIR219-1 |
| MIR30A |
| MIR30C2 |
| HCG18 |
| C6orf226 |
| TMEM151B |
| OOEP |
| FAM26F |
| LOC441177 |
| OR2B3 |
| OR2J3 |
| OR14J1 |
| OR10C1 |
| C6orf138 |
| MCART3P |
| LOC442245 |
| RFPL4B |
| MIR133B |
| GGNBP1 |
| HCG11 |
| LOC553137 |
| SNORD101 |
| SNORD100 |
| SNORA33 |
| C6orf225 |
| TSG1 |
| CTAGE9 |
| RAET1K |
| C6orf132 |
| PPP1R3G |
| ZNF389 |
| C6orf124 |
| LYPLA2P1 |
| SNORA20 |
| SNORA29 |
| SNORA38 |
| SNORD50B |
| SNORD32B |
| SNORD84 |
| SNORD117 |
| MIR548A1 |
| MIR548B |
| MIR586 |
| LOC728402 |
| C6orf186 |
| HULC |
| LOC729176 |
| LOC729603 |
| LOC730101 |
| TDRG1 |
| TMEM170B |
| SCARNA27 |
| MIR877 |
| HGC6.3 |
| BET3L |
| KHDC1L |
| PSORS1C3 |
| C6orf217 |
| LOC100132354 |
| C6orf41 |
| TOMM6 |
| LOC100270746 |
| LOC100287718 |
| NHEG1 |
This is the comprehensive list of deleted genes in the wide peak for 9p24.1.
Table S23. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| ABL1 |
| FANCC |
| FANCG |
| GNAQ |
| JAK2 |
| MLLT3 |
| NFIB |
| NOTCH1 |
| OMD |
| PAX5 |
| RALGDS |
| SET |
| SYK |
| TAL2 |
| TSC1 |
| XPA |
| NR4A3 |
| BRD3 |
| NUP214 |
| FNBP1 |
| CD274 |
| hsa-mir-602 |
| hsa-mir-126 |
| hsa-mir-219-2 |
| hsa-mir-199b |
| hsa-mir-181b-2 |
| hsa-mir-601 |
| hsa-mir-600 |
| hsa-mir-147 |
| hsa-mir-455 |
| hsa-mir-32 |
| hsa-mir-1302-8 |
| hsa-mir-24-1 |
| hsa-let-7d |
| hsa-mir-7-1 |
| hsa-mir-204 |
| hsa-mir-1299 |
| hsa-mir-873 |
| hsa-mir-31 |
| hsa-mir-491 |
| hsa-mir-101-2 |
| hsa-mir-1302-2 |
| ABCA1 |
| ABCA2 |
| ABO |
| ACO1 |
| PLIN2 |
| AK1 |
| ALAD |
| ALDH1A1 |
| ALDH1B1 |
| ALDOB |
| AMBP |
| ANXA1 |
| ANXA2P2 |
| NUDT2 |
| APBA1 |
| AQP3 |
| AQP7 |
| ASS1 |
| AUH |
| BAAT |
| BAG1 |
| KLF9 |
| C5 |
| C8G |
| CA9 |
| CACNA1B |
| CCIN |
| CCBL1 |
| TNFSF8 |
| ENTPD2 |
| CD72 |
| CDK9 |
| CDKN2A |
| CDKN2B |
| CEL |
| CELP |
| CKS2 |
| CLTA |
| CNTFR |
| COL5A1 |
| COL15A1 |
| SLC31A1 |
| SLC31A2 |
| CRAT |
| CTSL1 |
| CTSL2 |
| CYLC2 |
| DAPK1 |
| DBC1 |
| DBH |
| SARDH |
| DNM1 |
| DMRT1 |
| ECM2 |
| TOR1A |
| LPAR1 |
| S1PR3 |
| MEGF9 |
| ELAVL2 |
| ENDOG |
| ENG |
| STOM |
| FBP1 |
| FKTN |
| FCN1 |
| FCN2 |
| FOXD4 |
| FOXE1 |
| MLANA |
| FPGS |
| FXN |
| NR5A1 |
| FUT7 |
| GALT |
| GAS1 |
| NR6A1 |
| GCNT1 |
| GGTA1 |
| B4GALT1 |
| GLDC |
| GLE1 |
| GNG10 |
| GOLGA1 |
| GOLGA2 |
| GPR21 |
| RAPGEF1 |
| GRIN1 |
| GSN |
| HNRNPK |
| HSD17B3 |
| DNAJA1 |
| HSPA5 |
| TNC |
| IARS |
| IFNA1 |
| IFNA2 |
| IFNA4 |
| IFNA5 |
| IFNA6 |
| IFNA7 |
| IFNA8 |
| IFNA10 |
| IFNA13 |
| IFNA14 |
| IFNA16 |
| IFNA17 |
| IFNA21 |
| IFNB1 |
| IFNW1 |
| IL11RA |
| INSL4 |
| LCN1 |
| LCN2 |
| LMX1B |
| MTAP |
| MUSK |
| NCBP1 |
| NDUFA8 |
| NDUFB6 |
| NFIL3 |
| NFX1 |
| NINJ1 |
| NPR2 |
| NTRK2 |
| ROR2 |
| ODF2 |
| OGN |
| ORM1 |
| ORM2 |
| PAEP |
| PAPPA |
| PDCL |
| PBX3 |
| PCSK5 |
| PGM5 |
| PHF2 |
| PPP2R4 |
| PPP3R2 |
| PPP6C |
| PRKACG |
| PRSS3 |
| PSMB7 |
| PSMD5 |
| PTCH1 |
| PTGDS |
| PTGS1 |
| PTPN3 |
| PTPRD |
| RAD23B |
| RFX3 |
| RGS3 |
| RLN1 |
| RLN2 |
| RMRP |
| RORB |
| RPL7A |
| RPL12 |
| RPS6 |
| RXRA |
| CCL19 |
| CCL21 |
| SH3GL2 |
| SHB |
| SLC1A1 |
| SMARCA2 |
| SNAPC3 |
| SNAPC4 |
| SPTAN1 |
| STXBP1 |
| SURF1 |
| SURF2 |
| SURF4 |
| MED22 |
| SURF6 |
| TEK |
| TESK1 |
| TGFBR1 |
| TLE1 |
| TLE4 |
| TLN1 |
| TLR4 |
| TMOD1 |
| TPM2 |
| TRAF1 |
| TRAF2 |
| TTF1 |
| TXN |
| TYRP1 |
| UGCG |
| VAV2 |
| VCP |
| VLDLR |
| CORO2A |
| ZFP37 |
| ZNF79 |
| ZNF189 |
| ZFAND5 |
| LHX3 |
| GFI1B |
| PIP5K1B |
| RECK |
| IKBKAP |
| CDC14B |
| TMEFF1 |
| SSNA1 |
| EDF1 |
| CTNNAL1 |
| MPDZ |
| FBP2 |
| DPM2 |
| FUBP3 |
| CLIC3 |
| PRPF4 |
| KLF4 |
| GTF3C5 |
| GTF3C4 |
| CER1 |
| LHX2 |
| PLAA |
| GRHPR |
| FAM189A2 |
| TJP2 |
| MED27 |
| PTGES |
| ATP6V1G1 |
| GABBR2 |
| GDA |
| GNA14 |
| RALGPS1 |
| ADAMTSL2 |
| RGP1 |
| TRIM14 |
| MELK |
| RUSC2 |
| KIAA0649 |
| SEC16A |
| ZBTB5 |
| KIAA0020 |
| TNFSF15 |
| ROD1 |
| GNE |
| SH2D3C |
| RCL1 |
| TOPORS |
| RABEPK |
| SIGMAR1 |
| LAMC3 |
| TUBB2C |
| UBAC1 |
| OLFM1 |
| ZER1 |
| CREB3 |
| UNC13B |
| SEMA4D |
| ANP32B |
| AGPAT2 |
| SPTLC1 |
| POMT1 |
| SMC2 |
| DMRT2 |
| RRAGA |
| ZBTB6 |
| NEK6 |
| SDCCAG3 |
| NOXA1 |
| CCL27 |
| USP20 |
| ACTL7B |
| ACTL7A |
| GADD45G |
| SPIN1 |
| SEC61B |
| SLC27A4 |
| SLC35D2 |
| CEP110 |
| WDR5 |
| C9orf9 |
| ADAMTS13 |
| C9orf7 |
| PSIP1 |
| INSL6 |
| SLC2A6 |
| PTENP1 |
| AKAP2 |
| RPL35 |
| MAN1B1 |
| DCTN3 |
| FRMPD1 |
| DOLK |
| ZNF510 |
| HABP4 |
| PTGR1 |
| TRIM32 |
| SETX |
| ERP44 |
| KDM4C |
| ZBTB43 |
| SMC5 |
| KANK1 |
| FAM120A |
| PMPCA |
| VPS13A |
| ASTN2 |
| AGTPBP1 |
| BICD2 |
| FKBP15 |
| KIAA1045 |
| KIAA0368 |
| EXOSC2 |
| FREQ |
| TDRD7 |
| SLC44A1 |
| ANGPTL2 |
| NUP188 |
| CCRK |
| DDX58 |
| RABGAP1 |
| TMEM2 |
| C9orf5 |
| C9orf4 |
| SLC24A2 |
| CIZ1 |
| DNAJB5 |
| DCAF12 |
| DFNB31 |
| COBRA1 |
| NIPSNAP3A |
| NELF |
| GPSM1 |
| LOC26102 |
| GAPVD1 |
| PHF19 |
| ZNF658 |
| FAM75A7 |
| FBXW2 |
| SPAG8 |
| OR1J4 |
| OR2K2 |
| FBXO10 |
| GBGT1 |
| LHX6 |
| OSTF1 |
| OR1L3 |
| OR1L1 |
| OR1J2 |
| SNORA65 |
| SNORD62A |
| SNORD36C |
| SNORD36B |
| SNORD36A |
| SNORD24 |
| RANBP6 |
| TRUB2 |
| DNAI1 |
| ST6GALNAC4 |
| INVS |
| NDOR1 |
| SIT1 |
| SPINK4 |
| TOR1B |
| TOR2A |
| METTL11A |
| PHPT1 |
| ANAPC2 |
| PKN3 |
| DPP7 |
| PSAT1 |
| UBQLN1 |
| SLC2A8 |
| OBP2B |
| OBP2A |
| ST6GALNAC6 |
| STOML2 |
| DEC1 |
| PCA3 |
| AK3 |
| EXOSC3 |
| FAM108B1 |
| MRPS2 |
| COQ4 |
| CERCAM |
| EGFL7 |
| C9orf53 |
| UBAP1 |
| GOLM1 |
| PRRX2 |
| C9orf114 |
| CHMP5 |
| C9orf156 |
| RAB14 |
| TMEM8B |
| C9orf78 |
| SHC3 |
| POLE3 |
| NANS |
| FBXW5 |
| MRPL50 |
| RC3H2 |
| EPB41L4B |
| C9orf11 |
| TBC1D13 |
| FAM22F |
| DIRAS2 |
| BNC2 |
| HAUS6 |
| ASPN |
| BSPRY |
| APTX |
| C9orf167 |
| CNTLN |
| TEX10 |
| LPPR1 |
| KIAA1797 |
| UBE2R2 |
| EXD3 |
| C9orf6 |
| C9orf95 |
| STX17 |
| NOL8 |
| C9orf68 |
| C9orf40 |
| TMEM38B |
| SMU1 |
| RFK |
| NIPSNAP3B |
| STRBP |
| TBC1D2 |
| HEMGN |
| KIF27 |
| CDC37L1 |
| DENND4C |
| C9orf86 |
| CDK5RAP2 |
| UBAP2 |
| C9orf46 |
| CBWD1 |
| KLHL9 |
| BARX1 |
| RNF20 |
| LRRC8A |
| INPP5E |
| NPDC1 |
| OR2S2 |
| BARHL1 |
| IFNK |
| SH3GLB2 |
| REXO4 |
| DOLPP1 |
| KIAA1161 |
| KCNT1 |
| KIAA1432 |
| KIAA1529 |
| ZBTB26 |
| GBA2 |
| DENND1A |
| GPR107 |
| SLC46A2 |
| C9orf27 |
| C9orf80 |
| ZNF462 |
| DMRT3 |
| PRDM12 |
| MAK10 |
| DMRTA1 |
| SLC28A3 |
| CARD9 |
| SUSD1 |
| POLR1E |
| IPPK |
| DDX31 |
| FAM129B |
| LRRC19 |
| MRPL41 |
| NOL6 |
| WNK2 |
| SECISBP2 |
| C9orf16 |
| MAPKAP1 |
| DCAF10 |
| ZCCHC6 |
| GALNT12 |
| EHMT1 |
| MOBKL2B |
| C9orf82 |
| CNTNAP3 |
| ERMP1 |
| SVEP1 |
| RMI1 |
| TRPM3 |
| PTGES2 |
| IFT74 |
| KIAA1539 |
| GKAP1 |
| PDCD1LG2 |
| AKNA |
| C9orf45 |
| URM1 |
| ISCA1 |
| DOCK8 |
| ARPC5L |
| HDHD3 |
| AIF1L |
| UCK1 |
| ZNF484 |
| FSD1L |
| CEP78 |
| ZCCHC7 |
| ANKRD20A1 |
| GARNL3 |
| HSDL2 |
| C9orf64 |
| C9orf89 |
| HIATL2 |
| C9orf125 |
| NTNG2 |
| HIATL1 |
| HINT2 |
| C9orf24 |
| PIGO |
| BAT2L |
| PPAPDC3 |
| C9orf70 |
| ZDHHC12 |
| FAM73B |
| C9orf100 |
| C9orf3 |
| FIBCD1 |
| KIAA1984 |
| SNHG7 |
| TMEM141 |
| C9orf37 |
| COL27A1 |
| ALG2 |
| FGD3 |
| FAM125B |
| TPD52L3 |
| WDR34 |
| C9orf140 |
| C9orf69 |
| LRSAM1 |
| IL33 |
| C9orf123 |
| C9orf30 |
| UAP1L1 |
| MCART1 |
| MRRF |
| RBM18 |
| ARRDC1 |
| WDR85 |
| ADAMTSL1 |
| LOC92973 |
| TMEM203 |
| KIF12 |
| PALM2 |
| SLC25A25 |
| WDR31 |
| ZNF618 |
| UHRF2 |
| FAM122A |
| ZMYND19 |
| GRIN3A |
| TMC1 |
| RNF183 |
| NACC2 |
| C9orf116 |
| C9orf41 |
| C9orf57 |
| C9orf85 |
| C9orf135 |
| LCN8 |
| FAM69B |
| PTRH1 |
| PIP5KL1 |
| TAF1L |
| PTPDC1 |
| ANKRD19 |
| ARID3C |
| C9orf23 |
| C9orf131 |
| OR13C5 |
| OR13C8 |
| OR13C3 |
| OR13C4 |
| OR13F1 |
| OR1L8 |
| OR1N2 |
| OR1N1 |
| ASB6 |
| TRPM6 |
| SLC34A3 |
| RNF38 |
| GLIPR2 |
| DAB2IP |
| CAMSAP1 |
| C9orf66 |
| NCRNA00032 |
| LINGO2 |
| NXNL2 |
| C9orf163 |
| MAMDC4 |
| LCN6 |
| C9orf98 |
| OR1Q1 |
| TTLL11 |
| RASEF |
| TTC39B |
| C9orf122 |
| RG9MTD3 |
| TTC16 |
| FAM120AOS |
| FAM154A |
| C9orf44 |
| FREM1 |
| KIAA2026 |
| LOC158376 |
| LOC158381 |
| ZNF483 |
| C9orf84 |
| KIAA1958 |
| TSTD2 |
| ZNF782 |
| PRUNE2 |
| C9orf96 |
| KCNV2 |
| OLFML2A |
| C9orf71 |
| QSOX2 |
| GLIS3 |
| LOC169834 |
| ZNF169 |
| C9orf21 |
| ZNF367 |
| C9orf91 |
| C9orf72 |
| C9orf93 |
| NAIF1 |
| C9orf25 |
| CCDC107 |
| ANKS6 |
| SUSD3 |
| CBWD5 |
| CDC26 |
| LOC253039 |
| PHYHD1 |
| MORN5 |
| OR1L4 |
| TXNDC8 |
| MAMDC2 |
| FRMD3 |
| C9orf43 |
| C9orf144B |
| NCRNA00094 |
| CRB2 |
| SCAI |
| C9orf117 |
| C9orf47 |
| C9orf79 |
| LOC286238 |
| LCN12 |
| C9orf142 |
| C9orf75 |
| TUSC1 |
| C9orf109 |
| FAM78A |
| C9orf150 |
| LOC286359 |
| OR13C9 |
| OR13D1 |
| LOC286367 |
| FOXD4L3 |
| IFNE |
| ZDHHC21 |
| ACER2 |
| LOC340508 |
| GPR144 |
| FLJ43950 |
| QRFP |
| OR1J1 |
| OR1B1 |
| KIF24 |
| IGFBPL1 |
| MURC |
| FOXD4L4 |
| GLT6D1 |
| ENHO |
| AQP7P1 |
| PTAR1 |
| C9orf102 |
| C9orf119 |
| C9orf50 |
| PNPLA7 |
| C9orf169 |
| ENTPD8 |
| ZNF322B |
| KGFLP1 |
| LOC389705 |
| C9orf144 |
| FAM75A6 |
| MGC21881 |
| AQP7P2 |
| FLJ43859 |
| FLJ44082 |
| FLJ46321 |
| LOC389765 |
| C9orf153 |
| LOC389791 |
| IER5L |
| C9orf171 |
| LCN15 |
| C9orf172 |
| LRRC26 |
| TMEM8C |
| C9orf128 |
| OR13J1 |
| CTSL3 |
| OR13C2 |
| OR1L6 |
| OR5C1 |
| OR1K1 |
| LCN9 |
| FAM102A |
| FLJ35024 |
| PTPLAD2 |
| TMEM215 |
| TOMM5 |
| FAM74A1 |
| FAM74A4 |
| ZNF658B |
| C9orf170 |
| CENPP |
| C9orf152 |
| SNX30 |
| WDR38 |
| LCNL1 |
| C9orf139 |
| FAM166A |
| LOC402377 |
| SOHLH1 |
| PPAPDC2 |
| ZBTB34 |
| MIRLET7A1 |
| MIRLET7D |
| MIRLET7F1 |
| MIR101-2 |
| MIR126 |
| MIR147 |
| MIR181A2 |
| MIR181B2 |
| MIR199B |
| MIR204 |
| MIR219-2 |
| MIR23B |
| MIR24-1 |
| MIR27B |
| MIR31 |
| MIR32 |
| MIR7-1 |
| C9orf106 |
| C9orf103 |
| LCN10 |
| LOC415056 |
| LOC440173 |
| LOC440839 |
| LOC440896 |
| SUGT1P |
| ANKRD20A3 |
| ANKRD20A2 |
| AQP7P3 |
| FAM75C1 |
| LOC441454 |
| LOC441455 |
| FAM22G |
| C9orf173 |
| NRARP |
| LOC442421 |
| FOXB2 |
| CBWD3 |
| C9orf129 |
| PALM2-AKAP2 |
| FAM27A |
| DNAJC25 |
| DNAJC25-GNG10 |
| LOC554202 |
| LOC572558 |
| MIR491 |
| PGM5P2 |
| MIR455 |
| FAM138F |
| FAM75A2 |
| LOC642313 |
| LOC642929 |
| FAM163B |
| FLJ40292 |
| TUBBP5 |
| CBWD6 |
| FAM138A |
| LOC645961 |
| HRCT1 |
| FAM75A1 |
| RPSAP9 |
| FOXD4L6 |
| FOXD4L5 |
| LOC653501 |
| KGFLP2 |
| FAM138C |
| SCARNA8 |
| SNORA17 |
| SNORA43 |
| SNORD62B |
| MSMP |
| SNORD90 |
| MIR548D1 |
| MIR600 |
| MIR601 |
| MIR602 |
| RNF208 |
| FAM75A3 |
| FAM75A5 |
| DNLZ |
| FAM74A3 |
| ANKRD20A4 |
| CCDC29 |
| FAM166B |
| FOXD4L2 |
| CDKN2BAS |
| SNORD121A |
| SNORD121B |
| SNORA84 |
| SNORA70C |
| MIR876 |
| MIR873 |
| LOC100128076 |
| C9orf110 |
| C9orf130 |
| LOC100129034 |
| LOC100129066 |
| LOC100130426 |
| LOC100131193 |
| FAM157B |
| FAM27C |
| FAM95B1 |
| FAM27B |
| LOC100133920 |
| RNU6ATAC |
| NCRNA00092 |
| LOC100272217 |
| WASH1 |
| LOC100289341 |
Table 3. Get Full Table Arm-level significance table - 17 significant results found.
| Arm | # Genes | Amp Frequency | Amp Z score | Amp Q value | Del Frequency | Del Z score | Del Q value |
|---|---|---|---|---|---|---|---|
| 1p | 1731 | 0.05 | 1.09 | 0.596 | 0.30 | 14.2 | 0 |
| 1q | 1572 | 0.04 | 0.305 | 0.991 | 0.05 | 0.918 | 0.411 |
| 2p | 753 | 0.03 | -1.24 | 0.991 | 0.00 | -2.17 | 0.989 |
| 2q | 1235 | 0.01 | -1.33 | 0.991 | 0.01 | -1.33 | 0.989 |
| 3p | 853 | 0.01 | -1.58 | 0.991 | 0.05 | -0.14 | 0.942 |
| 3q | 917 | 0.00 | -2.04 | 0.991 | 0.06 | 0.408 | 0.666 |
| 4p | 366 | 0.01 | -1.87 | 0.991 | 0.08 | 0.265 | 0.735 |
| 4q | 865 | 0.01 | -1.45 | 0.991 | 0.14 | 3.36 | 0.00167 |
| 5p | 207 | 0.00 | -2.38 | 0.991 | 0.10 | 0.926 | 0.411 |
| 5q | 1246 | 0.00 | -1.83 | 0.991 | 0.06 | 0.875 | 0.414 |
| 6p | 937 | 0.03 | -1.04 | 0.991 | 0.03 | -1.04 | 0.989 |
| 6q | 692 | 0.01 | -1.55 | 0.991 | 0.15 | 3.51 | 0.00109 |
| 7p | 508 | 0.29 | 8.14 | 4.33e-15 | 0.00 | -1.98 | 0.989 |
| 7q | 1071 | 0.36 | 13.2 | 0 | 0.00 | -1.6 | 0.989 |
| 8p | 495 | 0.05 | -0.453 | 0.991 | 0.04 | -0.893 | 0.989 |
| 8q | 697 | 0.15 | 3.52 | 0.0028 | 0.01 | -1.54 | 0.989 |
| 9p | 343 | 0.02 | -1.58 | 0.991 | 0.27 | 6.89 | 2.63e-11 |
| 9q | 916 | 0.03 | -1.06 | 0.991 | 0.03 | -1.06 | 0.989 |
| 10p | 312 | 0.11 | 1.23 | 0.529 | 0.22 | 5 | 1.91e-06 |
| 10q | 1050 | 0.00 | -1.78 | 0.991 | 0.23 | 7.35 | 1.24e-12 |
| 11p | 731 | 0.03 | -1.05 | 0.991 | 0.13 | 2.67 | 0.0136 |
| 11q | 1279 | 0.05 | 0.447 | 0.991 | 0.04 | -0.0996 | 0.942 |
| 12p | 484 | 0.07 | 0.0288 | 0.991 | 0.05 | -0.409 | 0.989 |
| 12q | 1162 | 0.01 | -1.31 | 0.991 | 0.08 | 1.32 | 0.251 |
| 13q | 554 | 0.01 | -1.64 | 0.991 | 0.15 | 3.26 | 0.0022 |
| 14q | 1144 | 0.00 | -1.79 | 0.991 | 0.16 | 5 | 1.91e-06 |
| 15q | 1132 | 0.03 | -0.781 | 0.991 | 0.08 | 1.3 | 0.251 |
| 16p | 719 | 0.00 | -2.18 | 0.991 | 0.04 | -0.788 | 0.989 |
| 16q | 562 | 0.01 | -1.82 | 0.991 | 0.03 | -1.37 | 0.989 |
| 17p | 575 | 0.01 | -1.84 | 0.991 | 0.00 | -2.29 | 0.989 |
| 17q | 1321 | 0.03 | -0.704 | 0.991 | 0.00 | -1.82 | 0.989 |
| 18p | 117 | 0.03 | -1.47 | 0.991 | 0.14 | 2.18 | 0.0437 |
| 18q | 340 | 0.00 | -2.26 | 0.991 | 0.14 | 2.41 | 0.0261 |
| 19p | 870 | 0.10 | 1.84 | 0.212 | 0.18 | 4.71 | 6.76e-06 |
| 19q | 1452 | 0.10 | 2.38 | 0.0675 | 0.40 | 16.7 | 0 |
| 20p | 295 | 0.11 | 1.51 | 0.367 | 0.01 | -1.86 | 0.989 |
| 20q | 627 | 0.12 | 2.4 | 0.0675 | 0.00 | -2.13 | 0.989 |
| 21q | 422 | 0.03 | -1.41 | 0.991 | 0.05 | -0.548 | 0.989 |
| 22q | 764 | 0.01 | -1.61 | 0.991 | 0.08 | 0.735 | 0.474 |
Table 4. Get Full Table First 10 out of 80 Input Tumor Samples.
| Tumor Sample Names |
|---|
| TCGA-CS-4938-01B-11D-1892-01 |
| TCGA-CS-4941-01A-01D-1466-01 |
| TCGA-CS-4942-01A-01D-1466-01 |
| TCGA-CS-4943-01A-01D-1466-01 |
| TCGA-CS-4944-01A-01D-1466-01 |
| TCGA-CS-5390-01A-02D-1466-01 |
| TCGA-CS-5393-01A-01D-1466-01 |
| TCGA-CS-5394-01A-01D-1466-01 |
| TCGA-CS-5395-01A-01D-1466-01 |
| TCGA-CS-5396-01A-02D-1466-01 |
Figure 3. Segmented copy number profiles in the input data
GISTIC identifies genomic regions that are significantly gained or lost across a set of tumors. It takes segmented copy number ratios as input, separates arm-level events from focal events, and then performs two tests: (i) identifies significantly amplified/deleted chromosome arms; and (ii) identifies regions that are significantly focally amplified or deleted. For the focal analysis, the significance levels (Q values) are calculated by comparing the observed gains/losses at each locus to those obtained by randomly permuting the events along the genome to reflect the null hypothesis that they are all 'passengers' and could have occurred anywhere. The locus-specific significance levels are then corrected for multiple hypothesis testing. The arm-level significance is calculated by comparing the frequency of gains/losses of each arm to the expected rate given its size. The method outputs genomic views of significantly amplified and deleted regions, as well as a table of genes with gain or loss scores. A more in depth discussion of the GISTIC algorithm and its utility is given in [1], [3], and [5].
Regions of the genome that are prone to germ line variations in copy number are excluded from the GISTIC analysis using a list of germ line copy number variations (CNVs). A CNV is a DNA sequence that may be found at different copy numbers in the germ line of two different individuals. Such germ line variations can confound a GISTIC analysis, which finds significant somatic copy number variations in cancer. A more in depth discussion is provided in [6]. GISTIC currently uses two CNV exclusion lists. One is based on the literature describing copy number variation, and a second one comes from an analysis of significant variations among the blood normals in the TCGA data set.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.