This report serves to describe the mutational landscape and properties of a given individual set, as well as rank genes and genesets according to mutational significance. MutSig v1.5 was used to generate the results found in this report.
Working with individual set: BLCA.
Number of patients in set: 28
The input for this pipeline is a set of individuals with the following files associated for each:
1. An annotated .maf file describing the mutations called for the respective individual, and their properties.
2. A .wig file that contains information about the coverage of the sample.
Significantly mutated genes (q ≤ 0.1): 9
Mutations seen in COSMIC: 0
Significantly mutated genes in COSMIC territory: 0
Genes with clustered mutations (&le 3 aa apart): 0
Significantly mutated genesets: 33
Significantly mutated genesets: (excluding sig. mutated genes): 2
Table 1. Get Full Table Table representing breakdown of mutations by type.
| type | count |
|---|---|
| Frame_Shift_Del | 103 |
| Frame_Shift_Ins | 48 |
| In_Frame_Del | 29 |
| In_Frame_Ins | 5 |
| Missense_Mutation | 3424 |
| Nonsense_Mutation | 305 |
| Nonstop_Mutation | 6 |
| Silent | 1300 |
| Splice_Site | 84 |
| Translation_Start_Site | 7 |
| Total | 5311 |
Table 2. Get Full Table A breakdown of mutation rates per category discovered for this individual set.
| category | n | N | rate | rate_per_mb | relative_rate |
|---|---|---|---|---|---|
| A->T | 743 | 415320976 | 1.8e-06 | 1.8 | 0.38 |
| *Np(A/T)->nonflip | 1427 | 459226432 | 3.1e-06 | 3.1 | 0.66 |
| *Np(C/G)->nonflip | 838 | 392937804 | 2.1e-06 | 2.1 | 0.45 |
| C->G | 423 | 436843260 | 9.7e-07 | 0.97 | 0.21 |
| indel+null | 566 | 852164264 | 6.6e-07 | 0.66 | 0.14 |
| double_null | 14 | 852164264 | 1.6e-08 | 0.016 | 0.0035 |
| Total | 4011 | 852164264 | 4.7e-06 | 4.7 | 1 |
The x axis represents the samples. The y axis represents the exons, one row per exon, and they are sorted by average coverage across samples. For exons with exactly the same average coverage, they are sorted next by the %GC of the exon. (The secondary sort is especially useful for the zero-coverage exons at the bottom).
Figure 1.

Figure 2.
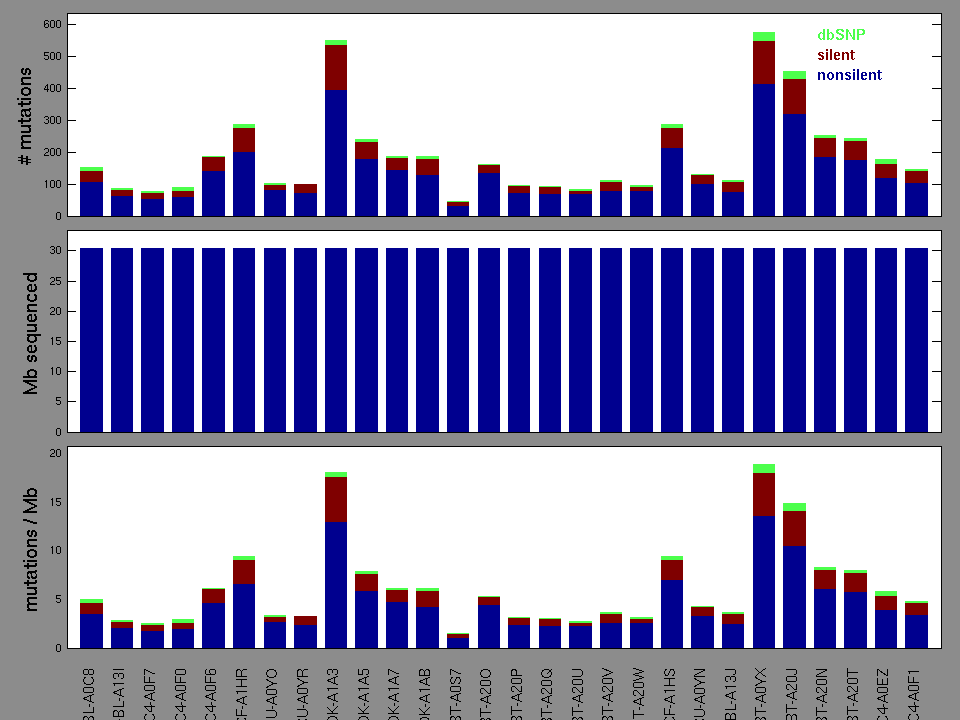
Table 3. Get Full Table A Ranked List of Significantly Mutated Genes. Number of significant genes found: 9. Number of genes displayed: 35
| rank | gene | description | N | n | npat | nsite | nsil | n1 | n2 | n3 | n4 | n5 | n6 | p | q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | TP53 | tumor protein p53 | 35896 | 12 | 9 | 12 | 0 | 4 | 4 | 1 | 0 | 3 | 0 | 3.5e-14 | 6.4e-10 |
| 2 | KDM6A | 117040 | 6 | 6 | 6 | 2 | 0 | 0 | 0 | 0 | 6 | 0 | 2.8e-07 | 0.0025 | |
| 3 | HLA-A | major histocompatibility complex, class I, A | 31640 | 3 | 3 | 3 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0.000012 | 0.053 |
| 4 | ELF3 | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) | 32144 | 3 | 3 | 3 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0.000013 | 0.053 |
| 5 | XPR1 | xenotropic and polytropic retrovirus receptor | 60228 | 4 | 4 | 4 | 1 | 0 | 0 | 1 | 1 | 2 | 0 | 0.000015 | 0.053 |
| 6 | ARID1A | AT rich interactive domain 1A (SWI-like) | 164864 | 6 | 5 | 6 | 1 | 1 | 0 | 0 | 0 | 5 | 0 | 0.000024 | 0.071 |
| 7 | OR2T35 | olfactory receptor, family 2, subfamily T, member 35 | 12964 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0.000038 | 0.097 |
| 8 | ERCC2 | excision repair cross-complementing rodent repair deficiency, complementation group 2 (xeroderma pigmentosum D) | 61460 | 4 | 4 | 4 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0.000045 | 0.1 |
| 9 | FBXW7 | F-box and WD repeat domain containing 7 | 72352 | 5 | 4 | 4 | 0 | 0 | 4 | 0 | 0 | 1 | 0 | 0.000097 | 0.19 |
| 10 | C20orf20 | chromosome 20 open reading frame 20 | 13524 | 2 | 2 | 2 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0.00016 | 0.3 |
| 11 | NAA25 | 82684 | 4 | 4 | 4 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0.00019 | 0.31 | |
| 12 | LETMD1 | LETM1 domain containing 1 | 31332 | 3 | 3 | 3 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0.00022 | 0.33 |
| 13 | CREBBP | CREB binding protein (Rubinstein-Taybi syndrome) | 201964 | 5 | 5 | 5 | 1 | 1 | 0 | 1 | 0 | 3 | 0 | 0.00024 | 0.33 |
| 14 | HCRT | hypocretin (orexin) neuropeptide precursor | 4060 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0.00027 | 0.35 |
| 15 | FAM57A | family with sequence similarity 57, member A | 17388 | 2 | 2 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0.00036 | 0.4 |
| 16 | CUL1 | cullin 1 | 67620 | 3 | 3 | 2 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0.00038 | 0.4 |
| 17 | RIMS3 | regulating synaptic membrane exocytosis 3 | 26572 | 2 | 2 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0.00039 | 0.4 |
| 18 | MTERFD2 | MTERF domain containing 2 | 31836 | 3 | 3 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0.0004 | 0.4 |
| 19 | GTF3C3 | general transcription factor IIIC, polypeptide 3, 102kDa | 76524 | 3 | 3 | 3 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0.00043 | 0.41 |
| 20 | ACN9 | ACN9 homolog (S. cerevisiae) | 10808 | 2 | 2 | 2 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0.00046 | 0.42 |
| 21 | IL34 | 20188 | 2 | 2 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0.00052 | 0.43 | |
| 22 | OTUD7A | OTU domain containing 7A | 57260 | 3 | 3 | 3 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0.00054 | 0.43 |
| 23 | CSNK1E | casein kinase 1, epsilon | 33992 | 2 | 2 | 2 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0.00058 | 0.43 |
| 24 | C7orf36 | chromosome 7 open reading frame 36 | 19404 | 2 | 2 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0.00058 | 0.43 |
| 25 | BCLAF1 | BCL2-associated transcription factor 1 | 78596 | 4 | 3 | 4 | 0 | 0 | 1 | 0 | 2 | 1 | 0 | 0.00067 | 0.46 |
| 26 | ORC3L | origin recognition complex, subunit 3-like (yeast) | 62132 | 3 | 3 | 3 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0.00068 | 0.46 |
| 27 | CAT | catalase | 45780 | 3 | 3 | 3 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0.00071 | 0.46 |
| 28 | GPS2 | G protein pathway suppressor 2 | 27468 | 2 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0.00073 | 0.46 |
| 29 | CDH22 | cadherin-like 22 | 55244 | 3 | 3 | 3 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0.00076 | 0.46 |
| 30 | NFE2L2 | nuclear factor (erythroid-derived 2)-like 2 | 50092 | 3 | 3 | 3 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0.0008 | 0.46 |
| 31 | TMCO2 | transmembrane and coiled-coil domains 2 | 15596 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0.00083 | 0.46 |
| 32 | PAK3 | p21 (CDKN1A)-activated kinase 3 | 48692 | 3 | 3 | 3 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0.00085 | 0.46 |
| 33 | LASP1 | LIM and SH3 protein 1 | 19656 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0.00087 | 0.46 |
| 34 | CD200 | CD200 molecule | 25116 | 2 | 2 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0.0009 | 0.46 |
| 35 | HOXB4 | homeobox B4 | 18256 | 2 | 2 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0.00094 | 0.46 |
Note:
N - number of sequenced bases in this gene across the individual set.
n - number of (nonsilent) mutations in this gene across the individual set.
npat - number of patients (individuals) with at least one nonsilent mutation.
nsite - number of unique sites having a non-silent mutation.
nsil - number of silent mutations in this gene across the individual set.
n1 - number of nonsilent mutations of type: A->T .
n2 - number of nonsilent mutations of type: *Np(A/T)->nonflip .
n3 - number of nonsilent mutations of type: *Np(C/G)->nonflip .
n4 - number of nonsilent mutations of type: C->G .
n5 - number of nonsilent mutations of type: indel+null .
null - mutation category that includes nonsense, frameshift, splice-site mutations
p_classic = p-value for the observed amount of nonsilent mutations being elevated in this gene
p_ns_s = p-value for the observed nonsilent/silent ratio being elevated in this gene
p = p-value (overall)
q = q-value, False Discovery Rate (Benjamini-Hochberg procedure)
In this analysis, COSMIC is used as a filter to increase power by restricting the territory of each gene. Cosmic version: v48.
Table 4. Get Full Table Significantly mutated genes (COSMIC territory only). To access the database please go to: COSMIC. Number of significant genes found: 0. Number of genes displayed: 10
| rank | gene | description | n | cos | n_cos | N_cos | cos_ev | p | q |
|---|---|---|---|---|---|---|---|---|---|
| 1 | A4GNT | alpha-1,4-N-acetylglucosaminyltransferase | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 2 | AACS | acetoacetyl-CoA synthetase | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 3 | ABCA9 | ATP-binding cassette, sub-family A (ABC1), member 9 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| 4 | ABCC10 | ATP-binding cassette, sub-family C (CFTR/MRP), member 10 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| 5 | ABCF2 | ATP-binding cassette, sub-family F (GCN20), member 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 6 | ABHD2 | abhydrolase domain containing 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 7 | ABHD4 | abhydrolase domain containing 4 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 8 | ACADS | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 9 | ACOT11 | acyl-CoA thioesterase 11 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 10 | ACRBP | acrosin binding protein | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
Note:
n - number of (nonsilent) mutations in this gene across the individual set.
cos = number of unique mutated sites in this gene in COSMIC
n_cos = overlap between n and cos.
N_cos = number of individuals times cos.
cos_ev = total evidence: number of reports in COSMIC for mutations seen in this gene.
p = p-value for seeing the observed amount of overlap in this gene)
q = q-value, False Discovery Rate (Benjamini-Hochberg procedure)
Table 5. Get Full Table Genes with Clustered Mutations
| num | gene | desc | n | mindist | npairs3 | npairs12 |
|---|---|---|---|---|---|---|
| 1762 | MXRA5 | matrix-remodelling associated 5 | 2 | 48 | 0 | 0 |
| 1 | A2M | alpha-2-macroglobulin | 2 | Inf | 0 | 0 |
| 4 | ABCA10 | ATP-binding cassette, sub-family A (ABC1), member 10 | 3 | Inf | 0 | 0 |
| 6 | ABCA13 | ATP-binding cassette, sub-family A (ABC1), member 13 | 2 | Inf | 0 | 0 |
| 10 | ABCA8 | ATP-binding cassette, sub-family A (ABC1), member 8 | 3 | Inf | 0 | 0 |
| 17 | ABCC9 | ATP-binding cassette, sub-family C (CFTR/MRP), member 9 | 2 | Inf | 0 | 0 |
| 32 | ACN9 | ACN9 homolog (S. cerevisiae) | 2 | Inf | 0 | 0 |
| 38 | ACTA2 | actin, alpha 2, smooth muscle, aorta | 2 | Inf | 0 | 0 |
| 41 | ACTN4 | actinin, alpha 4 | 3 | Inf | 0 | 0 |
| 50 | ADAMTS12 | ADAM metallopeptidase with thrombospondin type 1 motif, 12 | 4 | Inf | 0 | 0 |
Note:
n - number of mutations in this gene in the individual set.
mindist - distance (in aa) between closest pair of mutations in this gene
npairs3 - how many pairs of mutations are within 3 aa of each other.
npairs12 - how many pairs of mutations are within 12 aa of each other.
Table 6. Get Full Table A Ranked List of Significantly Mutated Genesets. (Source: MSigDB GSEA Cannonical Pathway Set).Number of significant genesets found: 33. Number of genesets displayed: 10
| rank | geneset | description | genes | N_genes | mut_tally | N | n | npat | nsite | nsil | n1 | n2 | n3 | n4 | n5 | n6 | p | q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PMLPATHWAY | Ring-shaped PML nuclear bodies regulate transcription and are required co-activators in p53- and DAXX-mediated apoptosis. | CREBBP, DAXX, HRAS, PAX3, PML, PRAM-1, RARA, RB1, SIRT1, SP100, TNF, TNFRSF1A, TNFRSF1B, TNFRSF6, TNFSF6, TP53, UBL1 | 13 | CREBBP(5), DAXX(1), PML(2), RARA(1), RB1(1), SP100(1), TNFRSF1B(1), TP53(12) | 823508 | 24 | 15 | 24 | 1 | 6 | 7 | 3 | 1 | 7 | 0 | 1.7e-08 | 1e-05 |
| 2 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. | ARF1, ARF3, CCND1, CDK2, CDK4, CDKN1A, CDKN1B, CDKN2A, CFL1, E2F1, E2F2, MDM2, NXT1, PRB1, TP53 | 15 | CCND1(1), CDKN1A(2), E2F2(1), TP53(12) | 343280 | 16 | 11 | 16 | 0 | 5 | 5 | 2 | 1 | 3 | 0 | 3.9e-08 | 0.000012 |
| 3 | P53PATHWAY | p53 induces cell cycle arrest or apoptosis under conditions of DNA damage. | APAF1, ATM, BAX, BCL2, CCND1, CCNE1, CDK2, CDK4, CDKN1A, E2F1, GADD45A, MDM2, PCNA, RB1, TIMP3, TP53 | 16 | ATM(3), BAX(1), CCND1(1), CDKN1A(2), RB1(1), TP53(12) | 778904 | 20 | 14 | 20 | 2 | 6 | 6 | 2 | 1 | 5 | 0 | 1.4e-07 | 0.000024 |
| 4 | TERTPATHWAY | hTERC, the RNA subunit of telomerase, and hTERT, the catalytic protein subunit, are required for telomerase activity and are overexpressed in many cancers. | HDAC1, MAX, MYC, SP1, SP3, TP53, WT1, ZNF42 | 7 | SP3(1), TP53(12) | 298060 | 13 | 10 | 13 | 1 | 5 | 4 | 1 | 0 | 3 | 0 | 1.6e-07 | 0.000024 |
| 5 | RBPATHWAY | The ATM protein kinase recognizes DNA damage and blocks cell cycle progression by phosphorylating chk1 and p53, which normally inhibits Rb to allow G1/S transitions. | ATM, CDC2, CDC25A, CDC25B, CDC25C, CDK2, CDK4, CHEK1, MYT1, RB1, TP53, WEE1, YWHAH | 12 | ATM(3), RB1(1), TP53(12), WEE1(1) | 748720 | 17 | 13 | 17 | 1 | 5 | 5 | 2 | 0 | 5 | 0 | 3.9e-07 | 0.000048 |
| 6 | HISTONE_METHYLTRANSFERASE | Genes with HMT activity | AOF2, KDM6A, ASH1L, ASH2L, C17orf79, CARM1, CTCFL, DOT1L, EED, EHMT1, EHMT2, EZH1, EZH2, FBXL10, FBXL11, FBXO11, HCFC1, HSF4, JMJD1A, JMJD1B, JMJD2A, JMJD2B, JMJD2C, JMJD2D, JMJD3, JMJD4, JMJD6, MEN1, MLL, MLL2, MLL3, MLL4, MLL5, NSD1, OGT, PAXIP1, PPP1CA, PPP1CB, PPP1CC, PRDM2, PRDM6, PRDM7, PRDM9, PRMT1, PRMT5, PRMT6, PRMT7, PRMT8, RBBP5, SATB1, SETD1A, SETD1B, SETD2, SETD7, SETD8, SETDB1, SETDB2, SETMAR, SMYD3, STK38, SUV39H1, SUV39H2, SUV420H1, SUV420H2, SUZ12, WHSC1, WHSC1L1 | 55 | ASH1L(3), CARM1(1), CTCFL(1), DOT1L(5), EZH1(2), HCFC1(1), KDM6A(6), MLL(6), MLL2(5), MLL3(7), MLL4(2), NSD1(1), OGT(1), PPP1CA(3), PRDM2(1), PRMT6(1), SETD1A(1), SETD2(3), SETD8(1), STK38(1), SUV420H1(2), WHSC1(2) | 5165860 | 56 | 24 | 55 | 8 | 9 | 13 | 12 | 4 | 14 | 4 | 2.2e-06 | 0.00022 |
| 7 | P53HYPOXIAPATHWAY | Hypoxia induces p53 accumulation and consequent apoptosis with p53-mediated cell cycle arrest, which is present under conditions of DNA damage. | ABCB1, AKT1, ATM, BAX, CDKN1A, CPB2, CSNK1A1, CSNK1D, FHL2, GADD45A, HIC1, HIF1A, HSPA1A, HSPCA, IGFBP3, MAPK8, MDM2, NFKBIB, NQO1, TP53 | 19 | ATM(3), BAX(1), CDKN1A(2), MAPK8(2), TP53(12) | 892864 | 20 | 13 | 20 | 1 | 6 | 6 | 2 | 2 | 4 | 0 | 6.8e-06 | 0.0006 |
| 8 | G1PATHWAY | CDK4/6-cyclin D and CDK2-cyclin E phosphorylate Rb, which allows the transcription of genes needed for the G1/S cell cycle transition. | ABL1, ATM, ATR, CCNA1, CCND1, CCNE1, CDC2, CDC25A, CDK2, CDK4, CDK6, CDKN1A, CDKN1B, CDKN2A, CDKN2B, DHFR, E2F1, GSK3B, HDAC1, MADH3, MADH4, RB1, SKP2, TFDP1, TGFB1, TGFB2, TGFB3, TP53 | 25 | ABL1(1), ATM(3), ATR(2), CCND1(1), CDK6(1), CDKN1A(2), RB1(1), TP53(12) | 1271004 | 23 | 15 | 23 | 0 | 7 | 7 | 4 | 1 | 4 | 0 | 0.000011 | 0.00088 |
| 9 | TIDPATHWAY | On ligand binding, interferon gamma receptors stimulate JAK2 kinase to phosphorylate STAT transcription factors, which promote expression of interferon responsive genes. | DNAJA3, HSPA1A, IFNG, IFNGR1, IFNGR2, IKBKB, JAK2, LIN7A, NFKB1, NFKBIA, RB1, RELA, TIP-1, TNF, TNFRSF1A, TNFRSF1B, TP53, USH1C, WT1 | 18 | IFNGR2(1), JAK2(1), RB1(1), RELA(1), TNFRSF1B(1), TP53(12), USH1C(1) | 798532 | 18 | 12 | 18 | 0 | 5 | 6 | 2 | 1 | 4 | 0 | 0.000015 | 0.001 |
| 10 | RNAPATHWAY | dsRNA-activated protein kinase phosphorylates elF2a, which generally inhibits translation, and activates NF-kB to provoke inflammation. | CHUK, DNAJC3, EIF2S1, EIF2S2, MAP3K14, NFKB1, NFKBIA, PRKR, RELA, TP53 | 9 | RELA(1), TP53(12) | 430948 | 13 | 9 | 13 | 0 | 4 | 5 | 1 | 0 | 3 | 0 | 2e-05 | 0.0011 |
Table 7. Get Full Table A Ranked List of Significantly Mutated Genesets (Excluding Significantly Mutated Genes). Number of significant genesets found: 2. Number of genesets displayed: 10
| rank | geneset | description | genes | N_genes | mut_tally | N | n | npat | nsite | nsil | n1 | n2 | n3 | n4 | n5 | n6 | p | q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HISTONE_METHYLTRANSFERASE | Genes with HMT activity | AOF2, KDM6A, ASH1L, ASH2L, C17orf79, CARM1, CTCFL, DOT1L, EED, EHMT1, EHMT2, EZH1, EZH2, FBXL10, FBXL11, FBXO11, HCFC1, HSF4, JMJD1A, JMJD1B, JMJD2A, JMJD2B, JMJD2C, JMJD2D, JMJD3, JMJD4, JMJD6, MEN1, MLL, MLL2, MLL3, MLL4, MLL5, NSD1, OGT, PAXIP1, PPP1CA, PPP1CB, PPP1CC, PRDM2, PRDM6, PRDM7, PRDM9, PRMT1, PRMT5, PRMT6, PRMT7, PRMT8, RBBP5, SATB1, SETD1A, SETD1B, SETD2, SETD7, SETD8, SETDB1, SETDB2, SETMAR, SMYD3, STK38, SUV39H1, SUV39H2, SUV420H1, SUV420H2, SUZ12, WHSC1, WHSC1L1 | 54 | ASH1L(3), CARM1(1), CTCFL(1), DOT1L(5), EZH1(2), HCFC1(1), MLL(6), MLL2(5), MLL3(7), MLL4(2), NSD1(1), OGT(1), PPP1CA(3), PRDM2(1), PRMT6(1), SETD1A(1), SETD2(3), SETD8(1), STK38(1), SUV420H1(2), WHSC1(2) | 5048820 | 50 | 23 | 49 | 6 | 9 | 13 | 12 | 4 | 8 | 4 | 0.000049 | 0.018 |
| 2 | FBW7PATHWAY | Cyclin E interacts with cell cycle checkpoint kinase cdk2 to allow transcription of genes required for S phase, including transcription of additional cyclin E. | CCNE1, CDC34, CDK2, CUL1, E2F1, FBXW7, RB1, SKP1A, TFDP1 | 8 | CUL1(3), FBXW7(5), RB1(1) | 361480 | 9 | 7 | 7 | 0 | 1 | 4 | 2 | 0 | 2 | 0 | 0.000058 | 0.018 |
| 3 | HSA00680_METHANE_METABOLISM | Genes involved in methane metabolism | ADH5, CAT, EPX, LPO, MPO, MTHFR, PRDX6, SHMT1, SHMT2, TPO | 10 | CAT(3), MPO(1), MTHFR(1), PRDX6(1), TPO(2) | 496552 | 8 | 8 | 8 | 1 | 1 | 4 | 2 | 0 | 1 | 0 | 0.00065 | 0.13 |
| 4 | PMLPATHWAY | Ring-shaped PML nuclear bodies regulate transcription and are required co-activators in p53- and DAXX-mediated apoptosis. | CREBBP, DAXX, HRAS, PAX3, PML, PRAM-1, RARA, RB1, SIRT1, SP100, TNF, TNFRSF1A, TNFRSF1B, TNFRSF6, TNFSF6, TP53, UBL1 | 12 | CREBBP(5), DAXX(1), PML(2), RARA(1), RB1(1), SP100(1), TNFRSF1B(1) | 787612 | 12 | 9 | 12 | 1 | 2 | 3 | 2 | 1 | 4 | 0 | 0.00094 | 0.15 |
| 5 | SODDPATHWAY | Some members of the tumor necrosis factor receptor family have cytoplasmic death domains that promote apoptosis when active and are repressed by silencers called SODDs. | BAG4, BIRC3, CASP8, FADD, RIPK1, TNF, TNFRSF1A, TNFRSF1B, TRADD, TRAF2 | 10 | BAG4(1), CASP8(2), FADD(1), RIPK1(1), TNFRSF1B(1), TRAF2(1) | 369068 | 7 | 6 | 7 | 1 | 0 | 5 | 0 | 2 | 0 | 0 | 0.0014 | 0.15 |
| 6 | CERAMIDEPATHWAY | Ceramide is a lipid signaling molecule that can activate proliferative or apoptotic pathways, depending on signaling context, localization, and cell type. | BAD, BAX, BCL2, CASP8, CYCS, FADD, MAP2K1, MAP2K4, MAP3K1, MAPK1, MAPK3, MAPK8, NFKB1, NSMAF, PDCD8, RAF1, RELA, RIPK1, SMPD1, TNFRSF1A, TRADD, TRAF2 | 21 | BAX(1), CASP8(2), FADD(1), MAP2K1(1), MAP3K1(1), MAPK8(2), NSMAF(1), RELA(1), RIPK1(1), SMPD1(1), TRAF2(1) | 885640 | 13 | 10 | 13 | 3 | 3 | 6 | 1 | 2 | 1 | 0 | 0.0015 | 0.15 |
| 7 | SETPATHWAY | Cytotoxic T cells release perforin, which to allow entry into target cells of granzyme B, which activates caspases, and granzyme A, which induces caspase-independent apoptosis. | ANP32A, APEX1, CREBBP, DFFA, DFFB, GZMA, GZMB, HMGB2, NME1, PRF1, SET | 11 | CREBBP(5), GZMB(1), PRF1(1) | 449904 | 7 | 6 | 7 | 3 | 1 | 1 | 2 | 0 | 3 | 0 | 0.0019 | 0.17 |
| 8 | HSA00940_PHENYLPROPANOID_BIOSYNTHESIS | Genes involved in phenylpropanoid biosynthesis | EPX, GBA, GBA3, LPO, MPO, PRDX6, TPO | 7 | GBA(1), GBA3(1), MPO(1), PRDX6(1), TPO(2) | 362236 | 6 | 6 | 6 | 2 | 2 | 2 | 0 | 1 | 1 | 0 | 0.0023 | 0.18 |
| 9 | METHANE_METABOLISM | ADH5, ATP6V0C, SHMT1, CAT, EPX, LPO, MPO, PRDX1, PRDX2, PRDX5, PRDX6, SHMT1, SHMT2, TPO | 13 | CAT(3), MPO(1), PRDX6(1), TPO(2) | 505624 | 7 | 7 | 7 | 1 | 1 | 3 | 2 | 0 | 1 | 0 | 0.0032 | 0.19 | |
| 10 | RELAPATHWAY | Acetylated NF-kB proteins are immune to IkB regulation and promote transcription until the histone deacetylase HDAC3 deacetylates the RelA subunit of NF-kB. | CHUK, CREBBP, EP300, FADD, HDAC3, IKBKB, IKBKG, NFKB1, NFKBIA, RELA, RIPK1, TNF, TNFRSF1A, TNFRSF1B, TRADD, TRAF6 | 15 | CREBBP(5), EP300(1), FADD(1), RELA(1), RIPK1(1), TNFRSF1B(1) | 962360 | 10 | 9 | 10 | 2 | 1 | 4 | 1 | 1 | 3 | 0 | 0.0033 | 0.19 |
In brief, we tabulate the number of mutations and the number of covered bases for each gene. The counts are broken down by mutation context category: four context categories that are discovered by MutSig, and one for indel and 'null' mutations, which include indels, nonsense mutations, splice-site mutations, and non-stop (read-through) mutations. For each gene, we calculate the probability of seeing the observed constellation of mutations, i.e. the product P1 x P2 x ... x Pm, or a more extreme one, given the background mutation rates calculated across the dataset.[1]
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.