This pipeline computes the correlation between cancer subtypes identified by different molecular patterns and selected clinical features.
Testing the association between subtypes identified by 2 different clustering approaches and 2 clinical features across 24 patients, no significant finding detected with P value < 0.05.
-
CNMF clustering analysis on sequencing-based miR expression data identified 2 subtypes that do not correlate to any clinical features.
-
Consensus hierarchical clustering analysis on sequencing-based miR expression data identified 2 subtypes that do not correlate to any clinical features.
Table 1. Get Full Table Overview of the association between subtypes identified by 2 different clustering approaches and 2 clinical features. Shown in the table are P values from statistical tests. Thresholded by P value < 0.05, no significant finding detected.
|
Clinical Features |
VITALSTATUS | GENDER |
| Statistical Tests | Fisher's exact test | Fisher's exact test |
| MIRseq CNMF subtypes | 0.675 | 0.0803 |
| MIRseq cHierClus subtypes | 0.675 | 0.0803 |
Table S1. Get Full Table Description of clustering approach #1: 'MIRseq CNMF subtypes'
| Cluster Labels | 1 | 2 |
|---|---|---|
| Number of samples | 9 | 15 |
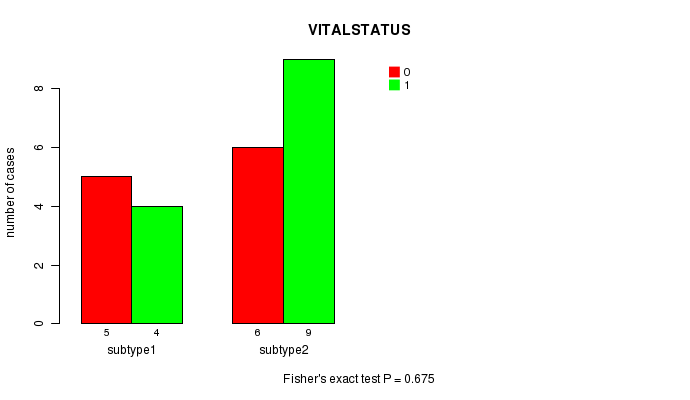
P value = 0.675 (Fisher's exact test)
Table S2. Clustering Approach #1: 'MIRseq CNMF subtypes' versus Clinical Feature #1: 'VITALSTATUS'
| nPatients | Class0 | Class1 |
|---|---|---|
| ALL | 11 | 13 |
| subtype1 | 5 | 4 |
| subtype2 | 6 | 9 |
Figure S1. Get High-res Image Clustering Approach #1: 'MIRseq CNMF subtypes' versus Clinical Feature #1: 'VITALSTATUS'
P value = 0.0803 (Fisher's exact test)
Table S3. Clustering Approach #1: 'MIRseq CNMF subtypes' versus Clinical Feature #2: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 15 | 9 |
| subtype1 | 8 | 1 |
| subtype2 | 7 | 8 |
Figure S2. Get High-res Image Clustering Approach #1: 'MIRseq CNMF subtypes' versus Clinical Feature #2: 'GENDER'
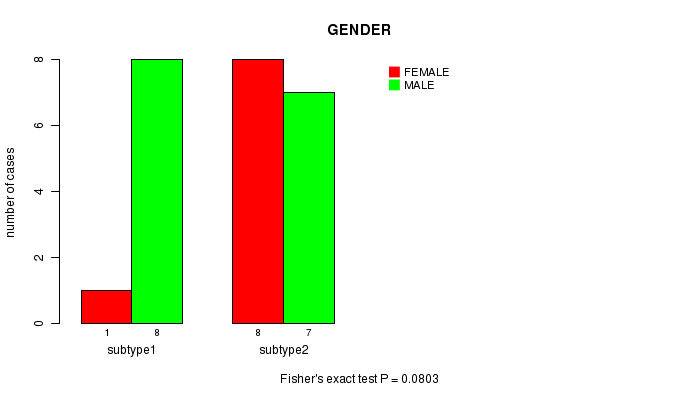
Table S4. Get Full Table Description of clustering approach #2: 'MIRseq cHierClus subtypes'
| Cluster Labels | 1 | 2 |
|---|---|---|
| Number of samples | 15 | 9 |
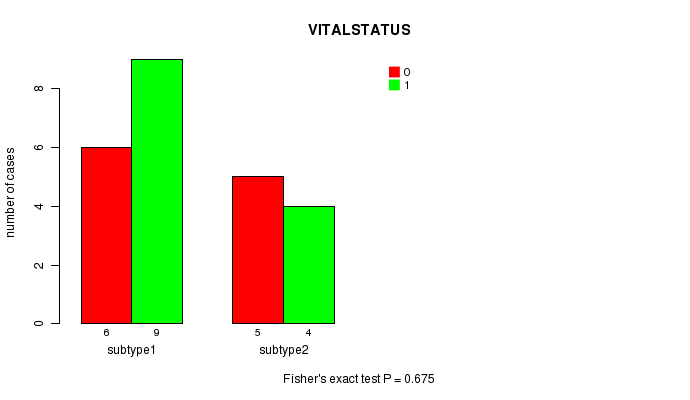
P value = 0.675 (Fisher's exact test)
Table S5. Clustering Approach #2: 'MIRseq cHierClus subtypes' versus Clinical Feature #1: 'VITALSTATUS'
| nPatients | Class0 | Class1 |
|---|---|---|
| ALL | 11 | 13 |
| subtype1 | 6 | 9 |
| subtype2 | 5 | 4 |
Figure S3. Get High-res Image Clustering Approach #2: 'MIRseq cHierClus subtypes' versus Clinical Feature #1: 'VITALSTATUS'
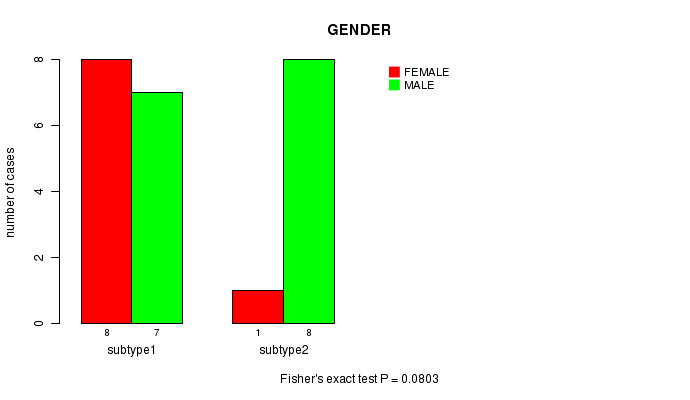
P value = 0.0803 (Fisher's exact test)
Table S6. Clustering Approach #2: 'MIRseq cHierClus subtypes' versus Clinical Feature #2: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 15 | 9 |
| subtype1 | 7 | 8 |
| subtype2 | 8 | 1 |
Figure S4. Get High-res Image Clustering Approach #2: 'MIRseq cHierClus subtypes' versus Clinical Feature #2: 'GENDER'
-
Cluster data file = LIHC.mergedcluster.txt
-
Clinical data file = LIHC.clin.merged.picked.txt
-
Number of patients = 24
-
Number of clustering approaches = 2
-
Number of selected clinical features = 2
-
Exclude small clusters that include fewer than K patients, K = 3
consensus non-negative matrix factorization clustering approach (Brunet et al. 2004)
Resampling-based clustering method (Monti et al. 2003)
For binary clinical features, two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
This is an experimental feature. Location of data archives could not be determined.