This pipeline computes the correlation between significantly recurrent gene mutations and selected clinical features.
Testing the association between mutation status of 283 genes and 5 clinical features across 248 patients, 8 significant findings detected with Q value < 0.25.
-
PTEN mutation correlated to 'HISTOLOGICAL.TYPE'.
-
TP53 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
KRAS mutation correlated to 'AGE'.
-
LRCH2 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
BRS3 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
NEFM mutation correlated to 'HISTOLOGICAL.TYPE'.
-
SIGLEC8 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
COX7B mutation correlated to 'Time to Death'.
Table 1. Get Full Table Overview of the association between mutation status of 283 genes and 5 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 8 significant findings detected.
|
Clinical Features |
Time to Death |
AGE |
HISTOLOGICAL TYPE |
RADIATIONS RADIATION REGIMENINDICATION |
NEOADJUVANT THERAPY |
||
| nMutated (%) | nWild-Type | logrank test | t-test | Chi-square test | Fisher's exact test | Fisher's exact test | |
| PTEN | 141 (57%) | 107 |
0.246 (1.00) |
0.0141 (1.00) |
2.6e-14 (3.67e-11) |
0.177 (1.00) |
0.0148 (1.00) |
| TP53 | 46 (19%) | 202 |
0.369 (1.00) |
0.000734 (1.00) |
2.48e-15 (3.51e-12) |
0.731 (1.00) |
0.00326 (1.00) |
| KRAS | 27 (11%) | 221 |
0.116 (1.00) |
3.28e-05 (0.0463) |
0.117 (1.00) |
0.672 (1.00) |
0.0116 (1.00) |
| LRCH2 | 17 (7%) | 231 |
0.655 (1.00) |
0.0283 (1.00) |
0.000157 (0.222) |
1 (1.00) |
0.415 (1.00) |
| BRS3 | 13 (5%) | 235 |
0.222 (1.00) |
0.426 (1.00) |
2.47e-05 (0.0349) |
0.142 (1.00) |
0.524 (1.00) |
| NEFM | 14 (6%) | 234 |
0.295 (1.00) |
0.171 (1.00) |
9.77e-05 (0.138) |
0.0204 (1.00) |
0.218 (1.00) |
| SIGLEC8 | 12 (5%) | 236 |
0.843 (1.00) |
0.773 (1.00) |
0.000128 (0.181) |
1 (1.00) |
0.741 (1.00) |
| COX7B | 3 (1%) | 245 |
5.16e-05 (0.0728) |
0.687 (1.00) |
0.765 (1.00) |
1 (1.00) |
0.564 (1.00) |
| CTCF | 40 (16%) | 208 |
0.563 (1.00) |
0.0108 (1.00) |
0.0205 (1.00) |
0.717 (1.00) |
1 (1.00) |
| PIK3CA | 114 (46%) | 134 |
0.0372 (1.00) |
0.329 (1.00) |
0.222 (1.00) |
0.424 (1.00) |
0.776 (1.00) |
| PIK3R1 | 77 (31%) | 171 |
0.999 (1.00) |
0.704 (1.00) |
0.000284 (0.4) |
0.248 (1.00) |
0.222 (1.00) |
| ARID1A | 77 (31%) | 171 |
0.0142 (1.00) |
0.00511 (1.00) |
0.00361 (1.00) |
0.472 (1.00) |
0.0138 (1.00) |
| CTNNB1 | 61 (25%) | 187 |
0.897 (1.00) |
0.0045 (1.00) |
0.00026 (0.366) |
0.438 (1.00) |
0.0126 (1.00) |
| FBXW7 | 26 (10%) | 222 |
0.168 (1.00) |
0.926 (1.00) |
0.176 (1.00) |
0.275 (1.00) |
0.244 (1.00) |
| FGFR2 | 25 (10%) | 223 |
0.729 (1.00) |
0.783 (1.00) |
0.289 (1.00) |
0.828 (1.00) |
0.815 (1.00) |
| PPP2R1A | 27 (11%) | 221 |
0.121 (1.00) |
0.0374 (1.00) |
0.00478 (1.00) |
0.52 (1.00) |
0.112 (1.00) |
| ARID5B | 26 (10%) | 222 |
0.333 (1.00) |
0.368 (1.00) |
0.128 (1.00) |
1 (1.00) |
0.816 (1.00) |
| SPOP | 18 (7%) | 230 |
0.753 (1.00) |
0.464 (1.00) |
0.682 (1.00) |
1 (1.00) |
0.413 (1.00) |
| CCND1 | 12 (5%) | 236 |
0.238 (1.00) |
0.142 (1.00) |
0.594 (1.00) |
0.552 (1.00) |
0.741 (1.00) |
| MUC5B | 39 (16%) | 209 |
0.0956 (1.00) |
0.892 (1.00) |
0.0164 (1.00) |
0.271 (1.00) |
0.848 (1.00) |
| ZFHX3 | 41 (17%) | 207 |
0.376 (1.00) |
0.709 (1.00) |
0.0156 (1.00) |
0.722 (1.00) |
0.254 (1.00) |
| SMTNL2 | 9 (4%) | 239 |
0.472 (1.00) |
0.53 (1.00) |
0.524 (1.00) |
1 (1.00) |
0.451 (1.00) |
| MLL4 | 28 (11%) | 220 |
0.116 (1.00) |
0.246 (1.00) |
0.135 (1.00) |
0.836 (1.00) |
0.118 (1.00) |
| BCOR | 29 (12%) | 219 |
0.115 (1.00) |
0.0278 (1.00) |
0.104 (1.00) |
0.836 (1.00) |
0.119 (1.00) |
| TNFAIP6 | 12 (5%) | 236 |
0.819 (1.00) |
0.302 (1.00) |
0.124 (1.00) |
1 (1.00) |
0.741 (1.00) |
| GIGYF2 | 21 (8%) | 227 |
0.766 (1.00) |
0.14 (1.00) |
0.0296 (1.00) |
0.00773 (1.00) |
0.452 (1.00) |
| CSMD3 | 37 (15%) | 211 |
0.784 (1.00) |
0.329 (1.00) |
0.141 (1.00) |
0.853 (1.00) |
0.237 (1.00) |
| RBMX | 13 (5%) | 235 |
0.336 (1.00) |
0.161 (1.00) |
0.00678 (1.00) |
0.142 (1.00) |
0.756 (1.00) |
| FOXA2 | 9 (4%) | 239 |
0.745 (1.00) |
0.396 (1.00) |
0.845 (1.00) |
0.722 (1.00) |
0.709 (1.00) |
| NFASC | 24 (10%) | 224 |
0.392 (1.00) |
0.579 (1.00) |
0.452 (1.00) |
1 (1.00) |
1 (1.00) |
| RASA1 | 18 (7%) | 230 |
0.224 (1.00) |
0.64 (1.00) |
0.638 (1.00) |
0.616 (1.00) |
0.786 (1.00) |
| ETV3 | 9 (4%) | 239 |
0.524 (1.00) |
0.404 (1.00) |
0.429 (1.00) |
0.497 (1.00) |
1 (1.00) |
| FAM9A | 10 (4%) | 238 |
0.44 (1.00) |
0.754 (1.00) |
0.855 (1.00) |
1 (1.00) |
0.293 (1.00) |
| NRAS | 8 (3%) | 240 |
0.393 (1.00) |
0.799 (1.00) |
0.815 (1.00) |
0.719 (1.00) |
0.452 (1.00) |
| LOC642587 | 7 (3%) | 241 |
0.679 (1.00) |
0.548 (1.00) |
0.159 (1.00) |
1 (1.00) |
0.677 (1.00) |
| NFE2L2 | 12 (5%) | 236 |
0.935 (1.00) |
0.397 (1.00) |
0.312 (1.00) |
1 (1.00) |
1 (1.00) |
| RB1 | 18 (7%) | 230 |
0.596 (1.00) |
0.144 (1.00) |
0.0185 (1.00) |
0.313 (1.00) |
0.786 (1.00) |
| ESR1 | 10 (4%) | 238 |
0.332 (1.00) |
0.441 (1.00) |
0.49 (1.00) |
0.318 (1.00) |
0.143 (1.00) |
| CHD4 | 23 (9%) | 225 |
0.784 (1.00) |
0.817 (1.00) |
0.249 (1.00) |
0.819 (1.00) |
0.629 (1.00) |
| DNER | 16 (6%) | 232 |
0.915 (1.00) |
0.0211 (1.00) |
0.00818 (1.00) |
0.182 (1.00) |
0.388 (1.00) |
| ELFN2 | 11 (4%) | 237 |
0.378 (1.00) |
0.776 (1.00) |
0.0106 (1.00) |
0.518 (1.00) |
0.298 (1.00) |
| KCNQ2 | 9 (4%) | 239 |
0.358 (1.00) |
0.254 (1.00) |
0.266 (1.00) |
0.281 (1.00) |
1 (1.00) |
| RNPC3 | 5 (2%) | 243 |
0.16 (1.00) |
0.47 (1.00) |
0.823 (1.00) |
1 (1.00) |
0.617 (1.00) |
| BCL6B | 9 (4%) | 239 |
0.487 (1.00) |
0.59 (1.00) |
0.356 (1.00) |
0.497 (1.00) |
1 (1.00) |
| CYLC1 | 15 (6%) | 233 |
0.317 (1.00) |
0.729 (1.00) |
0.117 (1.00) |
0.0955 (1.00) |
0.766 (1.00) |
| KCND2 | 17 (7%) | 231 |
0.909 (1.00) |
0.149 (1.00) |
0.0204 (1.00) |
0.432 (1.00) |
1 (1.00) |
| INPP4A | 14 (6%) | 234 |
0.24 (1.00) |
0.438 (1.00) |
0.0536 (1.00) |
0.777 (1.00) |
0.362 (1.00) |
| PPFIA2 | 19 (8%) | 229 |
0.239 (1.00) |
0.458 (1.00) |
0.025 (1.00) |
0.805 (1.00) |
0.604 (1.00) |
| SIN3A | 18 (7%) | 230 |
0.702 (1.00) |
0.0914 (1.00) |
0.000598 (0.84) |
0.44 (1.00) |
0.587 (1.00) |
| CDH10 | 19 (8%) | 229 |
0.168 (1.00) |
0.133 (1.00) |
0.0746 (1.00) |
1 (1.00) |
0.604 (1.00) |
| NCAM2 | 18 (7%) | 230 |
0.188 (1.00) |
0.253 (1.00) |
0.155 (1.00) |
0.313 (1.00) |
0.786 (1.00) |
| RPL14 | 6 (2%) | 242 |
0.571 (1.00) |
0.714 (1.00) |
0.702 (1.00) |
0.0971 (1.00) |
0.667 (1.00) |
| LCORL | 10 (4%) | 238 |
0.368 (1.00) |
0.14 (1.00) |
0.257 (1.00) |
1 (1.00) |
0.293 (1.00) |
| PASD1 | 17 (7%) | 231 |
0.188 (1.00) |
0.756 (1.00) |
0.438 (1.00) |
0.599 (1.00) |
0.573 (1.00) |
| CNPY1 | 7 (3%) | 241 |
0.449 (1.00) |
0.18 (1.00) |
0.55 (1.00) |
1 (1.00) |
0.677 (1.00) |
| ATP11C | 18 (7%) | 230 |
0.603 (1.00) |
0.237 (1.00) |
0.0688 (1.00) |
0.313 (1.00) |
0.786 (1.00) |
| MUC4 | 23 (9%) | 225 |
0.294 (1.00) |
0.305 (1.00) |
0.0878 (1.00) |
1 (1.00) |
0.629 (1.00) |
| EPHA5 | 17 (7%) | 231 |
0.218 (1.00) |
0.155 (1.00) |
0.108 (1.00) |
0.292 (1.00) |
0.166 (1.00) |
| IRX1 | 10 (4%) | 238 |
0.5 (1.00) |
0.341 (1.00) |
0.0039 (1.00) |
0.318 (1.00) |
0.732 (1.00) |
| PXDN | 21 (8%) | 227 |
0.722 (1.00) |
0.771 (1.00) |
0.0181 (1.00) |
0.811 (1.00) |
1 (1.00) |
| C5ORF38 | 6 (2%) | 242 |
0.682 (1.00) |
0.873 (1.00) |
0.0719 (1.00) |
0.415 (1.00) |
1 (1.00) |
| SGK1 | 10 (4%) | 238 |
0.647 (1.00) |
0.145 (1.00) |
0.204 (1.00) |
0.0345 (1.00) |
1 (1.00) |
| FAM47A | 15 (6%) | 233 |
0.951 (1.00) |
0.333 (1.00) |
0.125 (1.00) |
0.78 (1.00) |
0.563 (1.00) |
| ANKRD30A | 22 (9%) | 226 |
0.486 (1.00) |
0.638 (1.00) |
0.374 (1.00) |
0.639 (1.00) |
0.453 (1.00) |
| RASSF2 | 10 (4%) | 238 |
0.449 (1.00) |
0.0327 (1.00) |
0.261 (1.00) |
0.739 (1.00) |
1 (1.00) |
| NOL4 | 19 (8%) | 229 |
0.591 (1.00) |
0.218 (1.00) |
0.0452 (1.00) |
0.616 (1.00) |
0.604 (1.00) |
| SSX5 | 10 (4%) | 238 |
0.33 (1.00) |
0.108 (1.00) |
0.00338 (1.00) |
1 (1.00) |
0.732 (1.00) |
| MRPS30 | 9 (4%) | 239 |
0.655 (1.00) |
0.74 (1.00) |
0.12 (1.00) |
0.497 (1.00) |
0.709 (1.00) |
| PCDHA9 | 13 (5%) | 235 |
0.959 (1.00) |
0.195 (1.00) |
0.0027 (1.00) |
0.142 (1.00) |
0.197 (1.00) |
| SSTR1 | 9 (4%) | 239 |
0.318 (1.00) |
0.568 (1.00) |
0.429 (1.00) |
0.722 (1.00) |
1 (1.00) |
| OR10J3 | 9 (4%) | 239 |
0.618 (1.00) |
0.331 (1.00) |
0.133 (1.00) |
1 (1.00) |
0.263 (1.00) |
| DSC3 | 10 (4%) | 238 |
0.872 (1.00) |
0.542 (1.00) |
0.171 (1.00) |
1 (1.00) |
0.293 (1.00) |
| PGR | 10 (4%) | 238 |
0.298 (1.00) |
0.207 (1.00) |
0.342 (1.00) |
0.739 (1.00) |
0.732 (1.00) |
| RYR2 | 36 (15%) | 212 |
0.853 (1.00) |
0.951 (1.00) |
0.0703 (1.00) |
0.256 (1.00) |
0.227 (1.00) |
| COLEC11 | 8 (3%) | 240 |
0.708 (1.00) |
0.665 (1.00) |
0.0691 (1.00) |
0.127 (1.00) |
0.688 (1.00) |
| FATE1 | 6 (2%) | 242 |
0.607 (1.00) |
0.961 (1.00) |
0.00573 (1.00) |
0.415 (1.00) |
0.667 (1.00) |
| HELT | 7 (3%) | 241 |
0.44 (1.00) |
0.58 (1.00) |
0.192 (1.00) |
0.236 (1.00) |
0.677 (1.00) |
| MKI67 | 29 (12%) | 219 |
0.306 (1.00) |
0.607 (1.00) |
0.00204 (1.00) |
0.836 (1.00) |
0.119 (1.00) |
| OR4K2 | 10 (4%) | 238 |
0.306 (1.00) |
0.328 (1.00) |
0.0268 (1.00) |
0.739 (1.00) |
0.468 (1.00) |
| PCDHA12 | 18 (7%) | 230 |
0.83 (1.00) |
0.078 (1.00) |
0.0106 (1.00) |
1 (1.00) |
1 (1.00) |
| PDZD4 | 7 (3%) | 241 |
0.491 (1.00) |
0.731 (1.00) |
0.0368 (1.00) |
1 (1.00) |
1 (1.00) |
| SLCO4A1 | 10 (4%) | 238 |
0.504 (1.00) |
0.287 (1.00) |
0.072 (1.00) |
1 (1.00) |
0.732 (1.00) |
| SI | 22 (9%) | 226 |
0.457 (1.00) |
0.444 (1.00) |
0.521 (1.00) |
0.346 (1.00) |
0.453 (1.00) |
| KLK6 | 8 (3%) | 240 |
0.458 (1.00) |
0.5 (1.00) |
0.641 (1.00) |
1 (1.00) |
1 (1.00) |
| LSR | 7 (3%) | 241 |
0.000747 (1.00) |
0.0556 (1.00) |
0.106 (1.00) |
0.0485 (1.00) |
0.396 (1.00) |
| DCC | 21 (8%) | 227 |
0.776 (1.00) |
0.0961 (1.00) |
0.101 (1.00) |
0.229 (1.00) |
1 (1.00) |
| FAM161A | 15 (6%) | 233 |
0.179 (1.00) |
0.832 (1.00) |
0.113 (1.00) |
0.275 (1.00) |
0.369 (1.00) |
| JPH3 | 8 (3%) | 240 |
0.376 (1.00) |
0.59 (1.00) |
0.0434 (1.00) |
0.127 (1.00) |
0.22 (1.00) |
| L1TD1 | 14 (6%) | 234 |
0.307 (1.00) |
0.269 (1.00) |
0.266 (1.00) |
0.148 (1.00) |
0.764 (1.00) |
| RBBP6 | 20 (8%) | 228 |
0.702 (1.00) |
0.921 (1.00) |
0.00311 (1.00) |
1 (1.00) |
0.113 (1.00) |
| OR4A15 | 11 (4%) | 237 |
0.294 (1.00) |
0.118 (1.00) |
0.287 (1.00) |
0.753 (1.00) |
0.499 (1.00) |
| C9 | 14 (6%) | 234 |
0.259 (1.00) |
0.069 (1.00) |
0.00936 (1.00) |
1 (1.00) |
0.764 (1.00) |
| CDH4 | 13 (5%) | 235 |
0.178 (1.00) |
0.0384 (1.00) |
0.384 (1.00) |
0.769 (1.00) |
0.0223 (1.00) |
| FAM98B | 9 (4%) | 239 |
0.497 (1.00) |
0.169 (1.00) |
0.0372 (1.00) |
1 (1.00) |
1 (1.00) |
| HIST1H2BD | 5 (2%) | 243 |
0.0789 (1.00) |
0.876 (1.00) |
0.287 (1.00) |
0.342 (1.00) |
1 (1.00) |
| C1ORF212 | 4 (2%) | 244 |
0.54 (1.00) |
0.0113 (1.00) |
0.612 (1.00) |
1 (1.00) |
0.0639 (1.00) |
| LZTR1 | 12 (5%) | 236 |
0.371 (1.00) |
0.981 (1.00) |
0.0185 (1.00) |
0.349 (1.00) |
0.188 (1.00) |
| PAPD4 | 12 (5%) | 236 |
0.879 (1.00) |
0.141 (1.00) |
0.0132 (1.00) |
0.349 (1.00) |
0.021 (1.00) |
| PCGF3 | 7 (3%) | 241 |
0.272 (1.00) |
0.291 (1.00) |
0.223 (1.00) |
0.0485 (1.00) |
0.677 (1.00) |
| SESN3 | 12 (5%) | 236 |
0.294 (1.00) |
0.655 (1.00) |
0.0208 (1.00) |
0.552 (1.00) |
0.52 (1.00) |
| CNTN6 | 18 (7%) | 230 |
0.202 (1.00) |
0.607 (1.00) |
0.0147 (1.00) |
1 (1.00) |
1 (1.00) |
| MBOAT7 | 8 (3%) | 240 |
0.496 (1.00) |
0.619 (1.00) |
0.842 (1.00) |
1 (1.00) |
0.452 (1.00) |
| MUTED | 7 (3%) | 241 |
0.378 (1.00) |
0.963 (1.00) |
0.192 (1.00) |
1 (1.00) |
1 (1.00) |
| ACSM3 | 14 (6%) | 234 |
0.227 (1.00) |
0.126 (1.00) |
0.426 (1.00) |
0.777 (1.00) |
0.764 (1.00) |
| CNTNAP5 | 20 (8%) | 228 |
0.615 (1.00) |
0.893 (1.00) |
0.219 (1.00) |
1 (1.00) |
0.796 (1.00) |
| KIAA0664 | 10 (4%) | 238 |
0.582 (1.00) |
0.0249 (1.00) |
0.0422 (1.00) |
0.318 (1.00) |
0.732 (1.00) |
| PCDHA8 | 14 (6%) | 234 |
0.285 (1.00) |
0.0589 (1.00) |
0.544 (1.00) |
0.248 (1.00) |
0.764 (1.00) |
| FAM65B | 14 (6%) | 234 |
0.252 (1.00) |
0.0417 (1.00) |
0.01 (1.00) |
1 (1.00) |
1 (1.00) |
| PCDH15 | 27 (11%) | 221 |
0.371 (1.00) |
0.698 (1.00) |
0.0276 (1.00) |
1 (1.00) |
1 (1.00) |
| CDK17 | 11 (4%) | 237 |
0.907 (1.00) |
0.155 (1.00) |
0.333 (1.00) |
0.753 (1.00) |
1 (1.00) |
| SLC7A5 | 5 (2%) | 243 |
0.522 (1.00) |
0.51 (1.00) |
0.17 (1.00) |
0.342 (1.00) |
0.327 (1.00) |
| ATP6V1C2 | 10 (4%) | 238 |
0.319 (1.00) |
0.0783 (1.00) |
0.0874 (1.00) |
1 (1.00) |
0.732 (1.00) |
| VKORC1L1 | 5 (2%) | 243 |
0.562 (1.00) |
0.737 (1.00) |
0.823 (1.00) |
1 (1.00) |
1 (1.00) |
| ALS2CR11 | 15 (6%) | 233 |
0.282 (1.00) |
0.703 (1.00) |
0.0994 (1.00) |
0.275 (1.00) |
0.766 (1.00) |
| SSTR4 | 9 (4%) | 239 |
0.506 (1.00) |
0.479 (1.00) |
0.0204 (1.00) |
0.497 (1.00) |
0.451 (1.00) |
| PITX2 | 8 (3%) | 240 |
0.418 (1.00) |
0.762 (1.00) |
0.319 (1.00) |
0.451 (1.00) |
0.452 (1.00) |
| ZNF334 | 16 (6%) | 232 |
0.95 (1.00) |
0.035 (1.00) |
0.445 (1.00) |
0.79 (1.00) |
0.568 (1.00) |
| AGBL4 | 9 (4%) | 239 |
0.198 (1.00) |
0.109 (1.00) |
0.182 (1.00) |
0.171 (1.00) |
0.709 (1.00) |
| FAM9B | 7 (3%) | 241 |
0.401 (1.00) |
0.512 (1.00) |
0.56 (1.00) |
0.236 (1.00) |
0.677 (1.00) |
| GNG4 | 5 (2%) | 243 |
0.552 (1.00) |
0.879 (1.00) |
0.632 (1.00) |
0.168 (1.00) |
0.327 (1.00) |
| TCP11L2 | 12 (5%) | 236 |
0.229 (1.00) |
0.906 (1.00) |
0.312 (1.00) |
1 (1.00) |
0.52 (1.00) |
| COL5A1 | 18 (7%) | 230 |
0.88 (1.00) |
0.543 (1.00) |
0.0717 (1.00) |
0.797 (1.00) |
0.277 (1.00) |
| ZNF267 | 13 (5%) | 235 |
0.759 (1.00) |
0.576 (1.00) |
0.057 (1.00) |
1 (1.00) |
1 (1.00) |
| KCNA6 | 12 (5%) | 236 |
0.311 (1.00) |
0.0116 (1.00) |
0.12 (1.00) |
0.0253 (1.00) |
0.188 (1.00) |
| DCAF12L2 | 10 (4%) | 238 |
0.27 (1.00) |
0.35 (1.00) |
0.204 (1.00) |
1 (1.00) |
0.732 (1.00) |
| MPRIP | 11 (4%) | 237 |
0.453 (1.00) |
0.507 (1.00) |
0.199 (1.00) |
0.518 (1.00) |
0.298 (1.00) |
| NFE2L3 | 11 (4%) | 237 |
0.338 (1.00) |
0.736 (1.00) |
0.0107 (1.00) |
0.194 (1.00) |
1 (1.00) |
| FERD3L | 6 (2%) | 242 |
0.411 (1.00) |
0.659 (1.00) |
0.299 (1.00) |
0.667 (1.00) |
0.193 (1.00) |
| KCNA5 | 12 (5%) | 236 |
0.715 (1.00) |
0.143 (1.00) |
0.344 (1.00) |
0.552 (1.00) |
1 (1.00) |
| SYCP1 | 14 (6%) | 234 |
0.262 (1.00) |
0.432 (1.00) |
0.0111 (1.00) |
0.565 (1.00) |
0.764 (1.00) |
| ABCB1 | 19 (8%) | 229 |
0.24 (1.00) |
0.442 (1.00) |
0.174 (1.00) |
1 (1.00) |
1 (1.00) |
| HOXA7 | 7 (3%) | 241 |
0.487 (1.00) |
0.43 (1.00) |
0.159 (1.00) |
0.236 (1.00) |
0.677 (1.00) |
| TAB3 | 13 (5%) | 235 |
0.311 (1.00) |
0.00569 (1.00) |
0.173 (1.00) |
1 (1.00) |
0.352 (1.00) |
| UPF3B | 13 (5%) | 235 |
0.867 (1.00) |
0.843 (1.00) |
0.683 (1.00) |
1 (1.00) |
0.121 (1.00) |
| BRD3 | 9 (4%) | 239 |
0.897 (1.00) |
0.452 (1.00) |
0.524 (1.00) |
0.722 (1.00) |
0.451 (1.00) |
| HCN1 | 17 (7%) | 231 |
0.774 (1.00) |
0.198 (1.00) |
0.0149 (1.00) |
1 (1.00) |
1 (1.00) |
| EPHA10 | 12 (5%) | 236 |
0.296 (1.00) |
0.864 (1.00) |
0.0322 (1.00) |
0.552 (1.00) |
1 (1.00) |
| NAA25 | 16 (6%) | 232 |
0.339 (1.00) |
0.0781 (1.00) |
0.437 (1.00) |
1 (1.00) |
0.568 (1.00) |
| ZBTB33 | 14 (6%) | 234 |
0.337 (1.00) |
0.202 (1.00) |
0.305 (1.00) |
0.777 (1.00) |
0.764 (1.00) |
| ADAM23 | 14 (6%) | 234 |
0.266 (1.00) |
0.42 (1.00) |
0.0136 (1.00) |
0.565 (1.00) |
0.764 (1.00) |
| CHL1 | 19 (8%) | 229 |
0.0547 (1.00) |
0.128 (1.00) |
0.0565 (1.00) |
1 (1.00) |
0.604 (1.00) |
| ZNF275 | 8 (3%) | 240 |
0.624 (1.00) |
0.463 (1.00) |
0.0528 (1.00) |
1 (1.00) |
0.452 (1.00) |
| COX19 | 4 (2%) | 244 |
0.652 (1.00) |
0.269 (1.00) |
0.679 (1.00) |
1 (1.00) |
0.303 (1.00) |
| MORC4 | 14 (6%) | 234 |
0.219 (1.00) |
0.114 (1.00) |
0.202 (1.00) |
0.248 (1.00) |
0.362 (1.00) |
| INPPL1 | 14 (6%) | 234 |
0.829 (1.00) |
0.379 (1.00) |
0.09 (1.00) |
1 (1.00) |
1 (1.00) |
| THBS2 | 14 (6%) | 234 |
0.294 (1.00) |
0.275 (1.00) |
0.099 (1.00) |
0.777 (1.00) |
1 (1.00) |
| ACSL4 | 14 (6%) | 234 |
0.996 (1.00) |
0.174 (1.00) |
0.0367 (1.00) |
0.392 (1.00) |
0.362 (1.00) |
| BRCC3 | 8 (3%) | 240 |
0.716 (1.00) |
0.559 (1.00) |
0.201 (1.00) |
0.719 (1.00) |
0.452 (1.00) |
| CDKN1B | 6 (2%) | 242 |
0.423 (1.00) |
0.17 (1.00) |
0.77 (1.00) |
1 (1.00) |
0.193 (1.00) |
| UBE2E3 | 6 (2%) | 242 |
0.642 (1.00) |
0.355 (1.00) |
0.0886 (1.00) |
0.415 (1.00) |
1 (1.00) |
| FAM47B | 16 (6%) | 232 |
0.99 (1.00) |
0.0123 (1.00) |
0.007 (1.00) |
0.423 (1.00) |
0.568 (1.00) |
| FMN2 | 16 (6%) | 232 |
0.997 (1.00) |
0.589 (1.00) |
0.00824 (1.00) |
0.423 (1.00) |
0.773 (1.00) |
| MCTP1 | 12 (5%) | 236 |
0.335 (1.00) |
0.0453 (1.00) |
0.182 (1.00) |
0.349 (1.00) |
0.319 (1.00) |
| CASP8 | 11 (4%) | 237 |
0.709 (1.00) |
0.406 (1.00) |
0.0362 (1.00) |
0.753 (1.00) |
0.732 (1.00) |
| WHSC2 | 6 (2%) | 242 |
0.339 (1.00) |
0.211 (1.00) |
0.969 (1.00) |
0.415 (1.00) |
0.193 (1.00) |
| MNDA | 9 (4%) | 239 |
0.667 (1.00) |
0.204 (1.00) |
0.732 (1.00) |
0.171 (1.00) |
1 (1.00) |
| STAG2 | 18 (7%) | 230 |
0.244 (1.00) |
0.439 (1.00) |
0.419 (1.00) |
0.313 (1.00) |
1 (1.00) |
| WDR45 | 9 (4%) | 239 |
0.358 (1.00) |
0.63 (1.00) |
0.000996 (1.00) |
0.281 (1.00) |
1 (1.00) |
| ACVR1 | 9 (4%) | 239 |
0.393 (1.00) |
0.442 (1.00) |
0.429 (1.00) |
0.281 (1.00) |
0.451 (1.00) |
| CCDC104 | 10 (4%) | 238 |
0.327 (1.00) |
0.0526 (1.00) |
0.022 (1.00) |
0.501 (1.00) |
0.732 (1.00) |
| HIST1H2AM | 4 (2%) | 244 |
0.534 (1.00) |
0.847 (1.00) |
0.485 (1.00) |
0.302 (1.00) |
0.577 (1.00) |
| LEPREL2 | 9 (4%) | 239 |
0.0428 (1.00) |
0.0574 (1.00) |
0.000988 (1.00) |
1 (1.00) |
0.709 (1.00) |
| ADAMTS19 | 16 (6%) | 232 |
0.957 (1.00) |
0.283 (1.00) |
0.0463 (1.00) |
1 (1.00) |
0.773 (1.00) |
| ANKMY1 | 13 (5%) | 235 |
0.888 (1.00) |
0.294 (1.00) |
0.0565 (1.00) |
1 (1.00) |
0.756 (1.00) |
| PCDHA4 | 15 (6%) | 233 |
0.358 (1.00) |
0.99 (1.00) |
0.3 (1.00) |
1 (1.00) |
1 (1.00) |
| KIAA1967 | 12 (5%) | 236 |
0.628 (1.00) |
0.546 (1.00) |
0.0879 (1.00) |
0.552 (1.00) |
0.52 (1.00) |
| RUNX1 | 8 (3%) | 240 |
0.446 (1.00) |
0.163 (1.00) |
0.163 (1.00) |
0.719 (1.00) |
1 (1.00) |
| TRIM59 | 9 (4%) | 239 |
0.315 (1.00) |
0.949 (1.00) |
0.356 (1.00) |
1 (1.00) |
0.451 (1.00) |
| CD1C | 9 (4%) | 239 |
0.432 (1.00) |
0.907 (1.00) |
0.732 (1.00) |
0.722 (1.00) |
0.451 (1.00) |
| POU4F1 | 6 (2%) | 242 |
0.556 (1.00) |
0.497 (1.00) |
0.0886 (1.00) |
1 (1.00) |
1 (1.00) |
| TSC2 | 11 (4%) | 237 |
0.39 (1.00) |
0.546 (1.00) |
0.22 (1.00) |
1 (1.00) |
0.732 (1.00) |
| BCL2L11 | 5 (2%) | 243 |
0.812 (1.00) |
0.985 (1.00) |
0.823 (1.00) |
0.168 (1.00) |
0.327 (1.00) |
| MARCH11 | 8 (3%) | 240 |
0.39 (1.00) |
0.238 (1.00) |
0.579 (1.00) |
0.451 (1.00) |
1 (1.00) |
| IGFBP7 | 5 (2%) | 243 |
0.517 (1.00) |
0.567 (1.00) |
0.617 (1.00) |
0.663 (1.00) |
1 (1.00) |
| FBXO31 | 6 (2%) | 242 |
0.418 (1.00) |
0.955 (1.00) |
0.368 (1.00) |
0.185 (1.00) |
1 (1.00) |
| OR5AS1 | 8 (3%) | 240 |
0.455 (1.00) |
0.73 (1.00) |
0.214 (1.00) |
0.719 (1.00) |
0.111 (1.00) |
| PON1 | 11 (4%) | 237 |
0.303 (1.00) |
0.0454 (1.00) |
0.367 (1.00) |
0.753 (1.00) |
1 (1.00) |
| ITGB2 | 10 (4%) | 238 |
0.582 (1.00) |
0.428 (1.00) |
0.367 (1.00) |
0.739 (1.00) |
0.293 (1.00) |
| GALNT13 | 13 (5%) | 235 |
0.294 (1.00) |
0.407 (1.00) |
0.000521 (0.732) |
0.769 (1.00) |
1 (1.00) |
| TRPM2 | 13 (5%) | 235 |
0.897 (1.00) |
0.319 (1.00) |
0.0012 (1.00) |
0.769 (1.00) |
0.756 (1.00) |
| GFOD1 | 7 (3%) | 241 |
0.587 (1.00) |
0.569 (1.00) |
0.959 (1.00) |
1 (1.00) |
0.677 (1.00) |
| DCLK1 | 14 (6%) | 234 |
0.228 (1.00) |
0.046 (1.00) |
0.121 (1.00) |
0.565 (1.00) |
1 (1.00) |
| TMC4 | 8 (3%) | 240 |
0.38 (1.00) |
0.625 (1.00) |
0.699 (1.00) |
0.451 (1.00) |
1 (1.00) |
| ZC3H18 | 12 (5%) | 236 |
0.237 (1.00) |
0.97 (1.00) |
0.647 (1.00) |
0.552 (1.00) |
0.52 (1.00) |
| TAF1 | 25 (10%) | 223 |
0.442 (1.00) |
0.89 (1.00) |
0.0107 (1.00) |
0.514 (1.00) |
0.346 (1.00) |
| FLNA | 22 (9%) | 226 |
0.698 (1.00) |
0.847 (1.00) |
0.0423 (1.00) |
0.0572 (1.00) |
0.325 (1.00) |
| BMP7 | 9 (4%) | 239 |
0.321 (1.00) |
0.599 (1.00) |
0.266 (1.00) |
1 (1.00) |
0.119 (1.00) |
| DSG1 | 11 (4%) | 237 |
0.288 (1.00) |
0.67 (1.00) |
0.0397 (1.00) |
1 (1.00) |
0.732 (1.00) |
| HPD | 7 (3%) | 241 |
0.43 (1.00) |
0.0538 (1.00) |
0.55 (1.00) |
1 (1.00) |
1 (1.00) |
| KCNE2 | 6 (2%) | 242 |
0.641 (1.00) |
0.171 (1.00) |
0.0886 (1.00) |
0.415 (1.00) |
1 (1.00) |
| LIMCH1 | 14 (6%) | 234 |
0.993 (1.00) |
0.571 (1.00) |
0.0488 (1.00) |
1 (1.00) |
1 (1.00) |
| MAP7D2 | 16 (6%) | 232 |
0.237 (1.00) |
0.0182 (1.00) |
0.0389 (1.00) |
0.79 (1.00) |
1 (1.00) |
| SATB1 | 12 (5%) | 236 |
0.27 (1.00) |
0.422 (1.00) |
0.397 (1.00) |
1 (1.00) |
0.52 (1.00) |
| SHOX2 | 7 (3%) | 241 |
0.391 (1.00) |
0.428 (1.00) |
0.21 (1.00) |
0.694 (1.00) |
0.677 (1.00) |
| TPTE2 | 12 (5%) | 236 |
0.299 (1.00) |
0.822 (1.00) |
0.023 (1.00) |
1 (1.00) |
0.741 (1.00) |
| CIRBP | 4 (2%) | 244 |
0.641 (1.00) |
0.726 (1.00) |
0.282 (1.00) |
0.0132 (1.00) |
1 (1.00) |
| DCAF8L1 | 11 (4%) | 237 |
0.308 (1.00) |
0.0727 (1.00) |
0.000692 (0.97) |
1 (1.00) |
0.499 (1.00) |
| DLGAP2 | 11 (4%) | 237 |
0.977 (1.00) |
0.76 (1.00) |
0.185 (1.00) |
1 (1.00) |
0.732 (1.00) |
| GK | 12 (5%) | 236 |
0.269 (1.00) |
0.903 (1.00) |
0.000413 (0.581) |
0.0253 (1.00) |
1 (1.00) |
| GK2 | 15 (6%) | 233 |
0.276 (1.00) |
0.277 (1.00) |
0.16 (1.00) |
0.0955 (1.00) |
0.766 (1.00) |
| NAP1L5 | 3 (1%) | 245 |
0.881 (1.00) |
0.743 (1.00) |
0.851 (1.00) |
0.271 (1.00) |
1 (1.00) |
| SAFB | 10 (4%) | 238 |
0.3 (1.00) |
0.433 (1.00) |
0.445 (1.00) |
1 (1.00) |
0.732 (1.00) |
| FOXP2 | 13 (5%) | 235 |
0.402 (1.00) |
0.747 (1.00) |
0.0896 (1.00) |
0.551 (1.00) |
0.524 (1.00) |
| GYPB | 4 (2%) | 244 |
0.589 (1.00) |
0.817 (1.00) |
0.864 (1.00) |
0.302 (1.00) |
1 (1.00) |
| KLK7 | 4 (2%) | 244 |
0.506 (1.00) |
0.389 (1.00) |
0.95 (1.00) |
0.118 (1.00) |
1 (1.00) |
| SDK2 | 18 (7%) | 230 |
0.224 (1.00) |
0.982 (1.00) |
0.0382 (1.00) |
0.0689 (1.00) |
0.587 (1.00) |
| SSX3 | 7 (3%) | 241 |
0.391 (1.00) |
0.499 (1.00) |
0.55 (1.00) |
1 (1.00) |
1 (1.00) |
| MTPAP | 12 (5%) | 236 |
0.294 (1.00) |
0.257 (1.00) |
0.235 (1.00) |
0.229 (1.00) |
0.52 (1.00) |
| KHDRBS2 | 8 (3%) | 240 |
0.4 (1.00) |
0.176 (1.00) |
0.0577 (1.00) |
0.719 (1.00) |
0.688 (1.00) |
| CACNA1E | 25 (10%) | 223 |
0.433 (1.00) |
0.835 (1.00) |
0.0168 (1.00) |
0.514 (1.00) |
0.482 (1.00) |
| TRIM67 | 11 (4%) | 237 |
0.378 (1.00) |
0.684 (1.00) |
0.0488 (1.00) |
0.518 (1.00) |
1 (1.00) |
| GRIA3 | 16 (6%) | 232 |
0.759 (1.00) |
0.405 (1.00) |
0.0306 (1.00) |
0.423 (1.00) |
0.0775 (1.00) |
| ATP6V1B1 | 10 (4%) | 238 |
0.38 (1.00) |
0.719 (1.00) |
0.342 (1.00) |
1 (1.00) |
0.732 (1.00) |
| SMARCA1 | 14 (6%) | 234 |
0.282 (1.00) |
0.892 (1.00) |
0.352 (1.00) |
0.148 (1.00) |
0.121 (1.00) |
| SOCS2 | 6 (2%) | 242 |
0.529 (1.00) |
0.155 (1.00) |
0.445 (1.00) |
0.667 (1.00) |
0.667 (1.00) |
| BTNL8 | 7 (3%) | 241 |
0.453 (1.00) |
0.773 (1.00) |
0.613 (1.00) |
0.694 (1.00) |
1 (1.00) |
| ADARB2 | 7 (3%) | 241 |
0.609 (1.00) |
0.59 (1.00) |
0.613 (1.00) |
0.236 (1.00) |
1 (1.00) |
| CYLC2 | 9 (4%) | 239 |
0.875 (1.00) |
0.97 (1.00) |
0.5 (1.00) |
0.722 (1.00) |
0.263 (1.00) |
| ZNF648 | 9 (4%) | 239 |
0.357 (1.00) |
0.513 (1.00) |
0.317 (1.00) |
0.281 (1.00) |
1 (1.00) |
| POU4F2 | 7 (3%) | 241 |
0.462 (1.00) |
0.643 (1.00) |
0.623 (1.00) |
1 (1.00) |
0.677 (1.00) |
| CCDC75 | 5 (2%) | 243 |
0.474 (1.00) |
0.367 (1.00) |
0.412 (1.00) |
0.663 (1.00) |
1 (1.00) |
| MED12 | 21 (8%) | 227 |
0.207 (1.00) |
0.0181 (1.00) |
0.0996 (1.00) |
0.811 (1.00) |
0.803 (1.00) |
| HIST1H1E | 4 (2%) | 244 |
0.512 (1.00) |
0.354 (1.00) |
0.679 (1.00) |
0.118 (1.00) |
0.0639 (1.00) |
| PHRF1 | 10 (4%) | 238 |
0.748 (1.00) |
0.134 (1.00) |
0.0438 (1.00) |
0.318 (1.00) |
0.468 (1.00) |
| CDH19 | 15 (6%) | 233 |
0.231 (1.00) |
0.0434 (1.00) |
0.0399 (1.00) |
1 (1.00) |
1 (1.00) |
| GDF10 | 8 (3%) | 240 |
0.32 (1.00) |
0.513 (1.00) |
0.641 (1.00) |
0.451 (1.00) |
0.452 (1.00) |
| INSC | 10 (4%) | 238 |
0.392 (1.00) |
0.546 (1.00) |
0.766 (1.00) |
1 (1.00) |
0.293 (1.00) |
| NKAP | 9 (4%) | 239 |
0.653 (1.00) |
0.314 (1.00) |
0.017 (1.00) |
0.497 (1.00) |
0.709 (1.00) |
| P2RY11 | 8 (3%) | 240 |
0.47 (1.00) |
0.398 (1.00) |
0.272 (1.00) |
0.451 (1.00) |
0.688 (1.00) |
| HIST1H2AG | 5 (2%) | 243 |
0.647 (1.00) |
0.837 (1.00) |
0.788 (1.00) |
0.0483 (1.00) |
1 (1.00) |
| NDN | 6 (2%) | 242 |
0.535 (1.00) |
0.487 (1.00) |
0.975 (1.00) |
1 (1.00) |
0.667 (1.00) |
| BMP5 | 11 (4%) | 237 |
0.224 (1.00) |
0.289 (1.00) |
0.208 (1.00) |
0.753 (1.00) |
0.732 (1.00) |
| FAT1 | 30 (12%) | 218 |
0.122 (1.00) |
0.861 (1.00) |
0.0217 (1.00) |
1 (1.00) |
0.827 (1.00) |
| MTM1 | 12 (5%) | 236 |
0.307 (1.00) |
0.773 (1.00) |
0.566 (1.00) |
0.756 (1.00) |
0.188 (1.00) |
| MYPN | 19 (8%) | 229 |
0.667 (1.00) |
0.403 (1.00) |
0.0015 (1.00) |
0.218 (1.00) |
0.294 (1.00) |
| NRXN1 | 18 (7%) | 230 |
0.674 (1.00) |
0.587 (1.00) |
0.0219 (1.00) |
0.44 (1.00) |
0.413 (1.00) |
| MED15 | 10 (4%) | 238 |
0.405 (1.00) |
0.41 (1.00) |
0.127 (1.00) |
0.739 (1.00) |
0.293 (1.00) |
| ARHGAP36 | 12 (5%) | 236 |
0.285 (1.00) |
0.252 (1.00) |
0.33 (1.00) |
1 (1.00) |
1 (1.00) |
| GABRA3 | 10 (4%) | 238 |
0.334 (1.00) |
0.557 (1.00) |
0.19 (1.00) |
0.318 (1.00) |
0.732 (1.00) |
| CSDE1 | 15 (6%) | 233 |
0.296 (1.00) |
0.192 (1.00) |
0.0664 (1.00) |
0.589 (1.00) |
0.369 (1.00) |
| WDR64 | 14 (6%) | 234 |
0.277 (1.00) |
0.528 (1.00) |
0.35 (1.00) |
0.148 (1.00) |
0.764 (1.00) |
| PORCN | 10 (4%) | 238 |
0.382 (1.00) |
0.131 (1.00) |
0.364 (1.00) |
1 (1.00) |
0.732 (1.00) |
| FLJ43860 | 12 (5%) | 236 |
0.589 (1.00) |
0.159 (1.00) |
0.101 (1.00) |
0.115 (1.00) |
0.52 (1.00) |
| DIS3L2 | 12 (5%) | 236 |
0.859 (1.00) |
0.0572 (1.00) |
0.0237 (1.00) |
0.552 (1.00) |
0.021 (1.00) |
| GPR142 | 11 (4%) | 237 |
0.367 (1.00) |
0.204 (1.00) |
0.0653 (1.00) |
1 (1.00) |
1 (1.00) |
| MED24 | 12 (5%) | 236 |
0.25 (1.00) |
0.167 (1.00) |
0.0973 (1.00) |
0.115 (1.00) |
0.52 (1.00) |
| TMEM132D | 12 (5%) | 236 |
0.269 (1.00) |
0.782 (1.00) |
0.892 (1.00) |
1 (1.00) |
0.741 (1.00) |
| RPL39 | 3 (1%) | 245 |
0.538 (1.00) |
0.427 (1.00) |
0.851 (1.00) |
0.553 (1.00) |
1 (1.00) |
| ATF6 | 12 (5%) | 236 |
0.275 (1.00) |
0.472 (1.00) |
0.0154 (1.00) |
0.552 (1.00) |
1 (1.00) |
| CPB1 | 8 (3%) | 240 |
0.012 (1.00) |
0.687 (1.00) |
0.716 (1.00) |
0.719 (1.00) |
1 (1.00) |
| KRTAP19-8 | 3 (1%) | 245 |
0.541 (1.00) |
0.563 (1.00) |
0.464 (1.00) |
0.271 (1.00) |
1 (1.00) |
| ATM | 25 (10%) | 223 |
0.136 (1.00) |
0.2 (1.00) |
0.0321 (1.00) |
1 (1.00) |
0.0189 (1.00) |
| CDC73 | 12 (5%) | 236 |
0.25 (1.00) |
0.235 (1.00) |
0.0817 (1.00) |
1 (1.00) |
0.741 (1.00) |
| NPEPL1 | 6 (2%) | 242 |
0.59 (1.00) |
0.713 (1.00) |
0.669 (1.00) |
1 (1.00) |
1 (1.00) |
| SLC16A2 | 10 (4%) | 238 |
0.59 (1.00) |
0.846 (1.00) |
0.509 (1.00) |
0.501 (1.00) |
0.732 (1.00) |
| DMD | 30 (12%) | 218 |
0.51 (1.00) |
0.978 (1.00) |
0.687 (1.00) |
0.416 (1.00) |
0.668 (1.00) |
| OR5M11 | 7 (3%) | 241 |
0.4 (1.00) |
0.729 (1.00) |
0.0585 (1.00) |
1 (1.00) |
1 (1.00) |
| AGAP2 | 12 (5%) | 236 |
0.425 (1.00) |
0.493 (1.00) |
0.166 (1.00) |
0.756 (1.00) |
1 (1.00) |
| C16ORF45 | 5 (2%) | 243 |
0.451 (1.00) |
0.355 (1.00) |
0.17 (1.00) |
1 (1.00) |
0.617 (1.00) |
| KCNH2 | 10 (4%) | 238 |
0.432 (1.00) |
0.597 (1.00) |
0.109 (1.00) |
0.0959 (1.00) |
0.143 (1.00) |
| CPXM2 | 12 (5%) | 236 |
0.344 (1.00) |
0.804 (1.00) |
0.0185 (1.00) |
0.552 (1.00) |
0.52 (1.00) |
| GPA33 | 9 (4%) | 239 |
0.948 (1.00) |
0.214 (1.00) |
0.0146 (1.00) |
0.722 (1.00) |
0.709 (1.00) |
| CDH1 | 11 (4%) | 237 |
0.951 (1.00) |
0.0392 (1.00) |
0.334 (1.00) |
0.753 (1.00) |
0.499 (1.00) |
| ENPP5 | 10 (4%) | 238 |
0.386 (1.00) |
0.181 (1.00) |
0.445 (1.00) |
0.501 (1.00) |
1 (1.00) |
| LTF | 10 (4%) | 238 |
0.368 (1.00) |
0.618 (1.00) |
0.0127 (1.00) |
0.318 (1.00) |
0.732 (1.00) |
| MOGAT3 | 7 (3%) | 241 |
0.437 (1.00) |
0.562 (1.00) |
0.223 (1.00) |
0.694 (1.00) |
0.092 (1.00) |
| NRG1 | 14 (6%) | 234 |
0.281 (1.00) |
0.725 (1.00) |
0.0205 (1.00) |
0.565 (1.00) |
1 (1.00) |
| CHD3 | 22 (9%) | 226 |
0.199 (1.00) |
0.638 (1.00) |
0.00487 (1.00) |
1 (1.00) |
0.208 (1.00) |
| OR5D13 | 8 (3%) | 240 |
0.872 (1.00) |
0.845 (1.00) |
0.0732 (1.00) |
0.451 (1.00) |
0.688 (1.00) |
| PDLIM2 | 4 (2%) | 244 |
0.702 (1.00) |
0.588 (1.00) |
0.612 (1.00) |
0.608 (1.00) |
1 (1.00) |
| SLC48A1 | 3 (1%) | 245 |
0.691 (1.00) |
0.354 (1.00) |
0.353 (1.00) |
1 (1.00) |
1 (1.00) |
| STEAP4 | 10 (4%) | 238 |
0.288 (1.00) |
0.533 (1.00) |
0.0755 (1.00) |
1 (1.00) |
0.732 (1.00) |
| BMF | 3 (1%) | 245 |
0.00143 (1.00) |
0.266 (1.00) |
0.597 (1.00) |
0.553 (1.00) |
1 (1.00) |
| ZC3H13 | 20 (8%) | 228 |
0.152 (1.00) |
0.236 (1.00) |
0.00876 (1.00) |
0.329 (1.00) |
0.796 (1.00) |
| CFL2 | 6 (2%) | 242 |
0.42 (1.00) |
0.194 (1.00) |
0.0719 (1.00) |
0.415 (1.00) |
1 (1.00) |
| RRAS2 | 4 (2%) | 244 |
0.752 (1.00) |
0.0717 (1.00) |
0.864 (1.00) |
0.608 (1.00) |
0.0639 (1.00) |
| CDCA7L | 9 (4%) | 239 |
0.441 (1.00) |
0.0696 (1.00) |
0.726 (1.00) |
0.722 (1.00) |
0.119 (1.00) |
| KCMF1 | 9 (4%) | 239 |
0.472 (1.00) |
0.617 (1.00) |
0.000996 (1.00) |
0.497 (1.00) |
1 (1.00) |
| THEG | 6 (2%) | 242 |
0.464 (1.00) |
0.154 (1.00) |
0.547 (1.00) |
0.415 (1.00) |
0.193 (1.00) |
| PIK3R3 | 8 (3%) | 240 |
0.373 (1.00) |
0.523 (1.00) |
0.684 (1.00) |
0.27 (1.00) |
0.111 (1.00) |
| CAPS2 | 8 (3%) | 240 |
0.38 (1.00) |
0.0342 (1.00) |
0.00547 (1.00) |
0.719 (1.00) |
1 (1.00) |
| LRFN2 | 13 (5%) | 235 |
0.914 (1.00) |
0.831 (1.00) |
0.051 (1.00) |
0.378 (1.00) |
0.352 (1.00) |
| NSDHL | 9 (4%) | 239 |
0.435 (1.00) |
0.315 (1.00) |
0.0158 (1.00) |
0.281 (1.00) |
0.709 (1.00) |
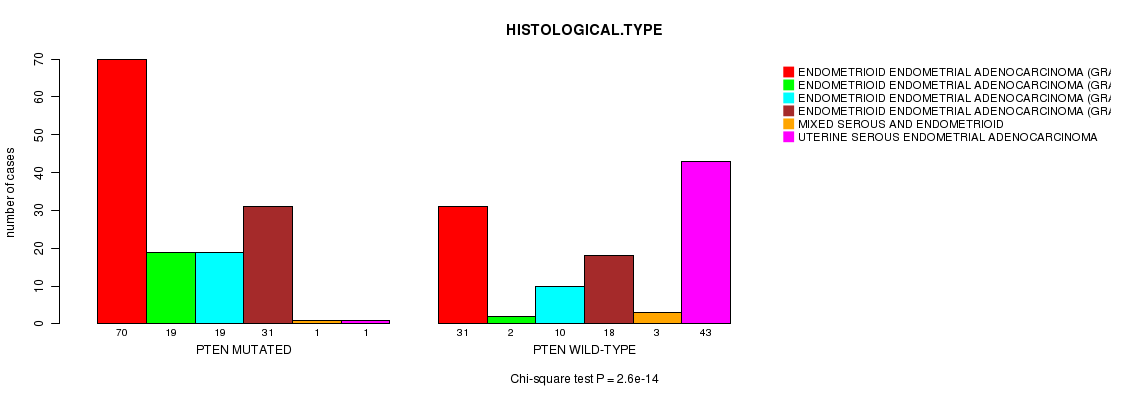
P value = 2.6e-14 (Chi-square test), Q value = 3.7e-11
Table S1. Gene #4: 'PTEN MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | UTERINE SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|
| ALL | 101 | 21 | 29 | 49 | 4 | 44 |
| PTEN MUTATED | 70 | 19 | 19 | 31 | 1 | 1 |
| PTEN WILD-TYPE | 31 | 2 | 10 | 18 | 3 | 43 |
Figure S1. Get High-res Image Gene #4: 'PTEN MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
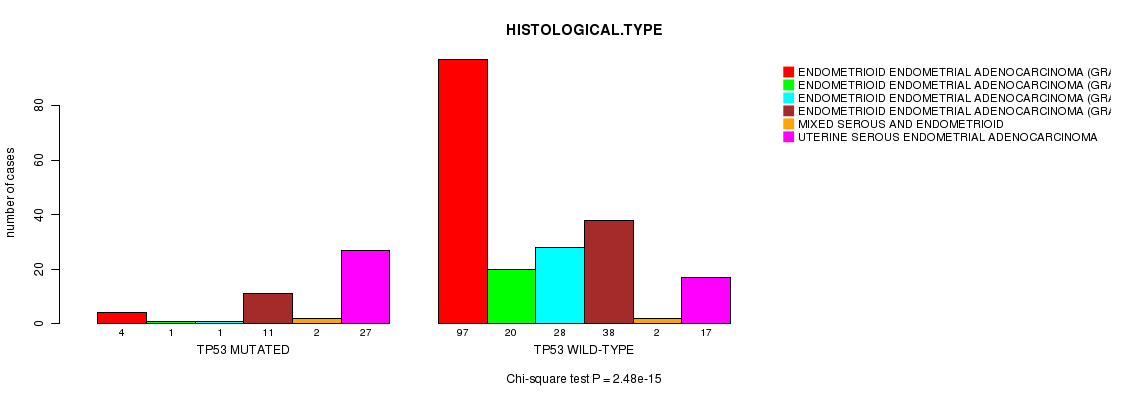
P value = 2.48e-15 (Chi-square test), Q value = 3.5e-12
Table S2. Gene #5: 'TP53 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | UTERINE SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|
| ALL | 101 | 21 | 29 | 49 | 4 | 44 |
| TP53 MUTATED | 4 | 1 | 1 | 11 | 2 | 27 |
| TP53 WILD-TYPE | 97 | 20 | 28 | 38 | 2 | 17 |
Figure S2. Get High-res Image Gene #5: 'TP53 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
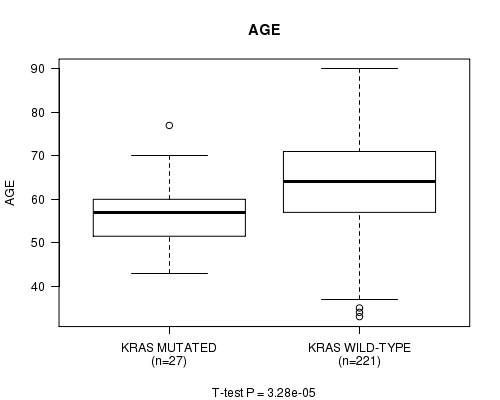
P value = 3.28e-05 (t-test), Q value = 0.046
Table S3. Gene #11: 'KRAS MUTATION STATUS' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 248 | 63.1 (11.1) |
| KRAS MUTATED | 27 | 56.3 (7.5) |
| KRAS WILD-TYPE | 221 | 63.9 (11.2) |
Figure S3. Get High-res Image Gene #11: 'KRAS MUTATION STATUS' versus Clinical Feature #2: 'AGE'
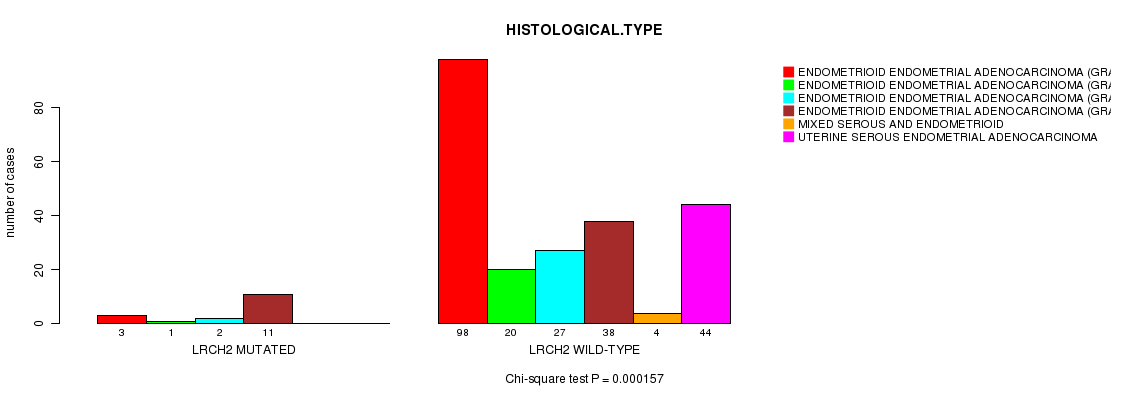
P value = 0.000157 (Chi-square test), Q value = 0.22
Table S4. Gene #34: 'LRCH2 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | UTERINE SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|
| ALL | 101 | 21 | 29 | 49 | 4 | 44 |
| LRCH2 MUTATED | 3 | 1 | 2 | 11 | 0 | 0 |
| LRCH2 WILD-TYPE | 98 | 20 | 27 | 38 | 4 | 44 |
Figure S4. Get High-res Image Gene #34: 'LRCH2 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
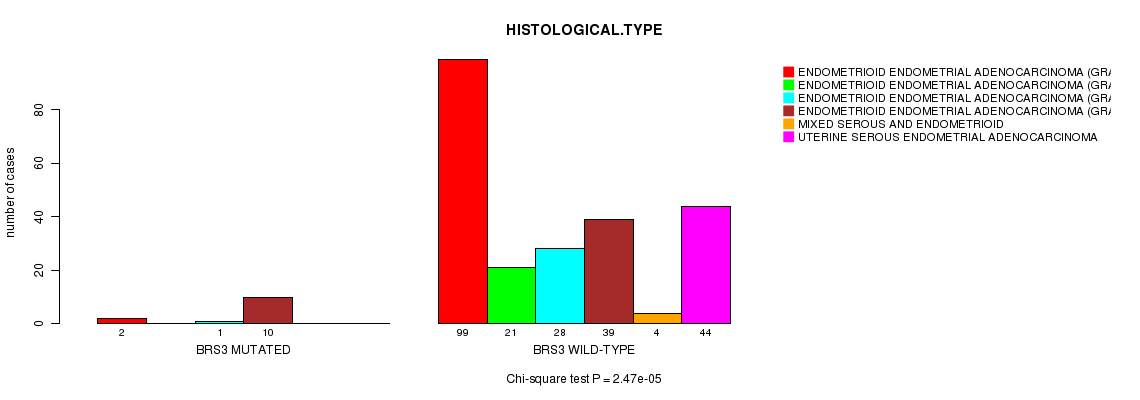
P value = 2.47e-05 (Chi-square test), Q value = 0.035
Table S5. Gene #52: 'BRS3 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | UTERINE SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|
| ALL | 101 | 21 | 29 | 49 | 4 | 44 |
| BRS3 MUTATED | 2 | 0 | 1 | 10 | 0 | 0 |
| BRS3 WILD-TYPE | 99 | 21 | 28 | 39 | 4 | 44 |
Figure S5. Get High-res Image Gene #52: 'BRS3 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
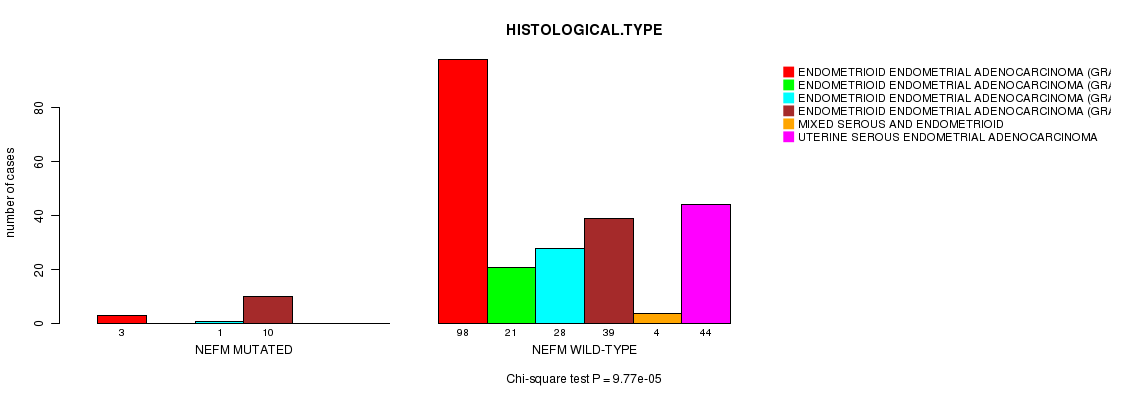
P value = 9.77e-05 (Chi-square test), Q value = 0.14
Table S6. Gene #108: 'NEFM MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | UTERINE SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|
| ALL | 101 | 21 | 29 | 49 | 4 | 44 |
| NEFM MUTATED | 3 | 0 | 1 | 10 | 0 | 0 |
| NEFM WILD-TYPE | 98 | 21 | 28 | 39 | 4 | 44 |
Figure S6. Get High-res Image Gene #108: 'NEFM MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
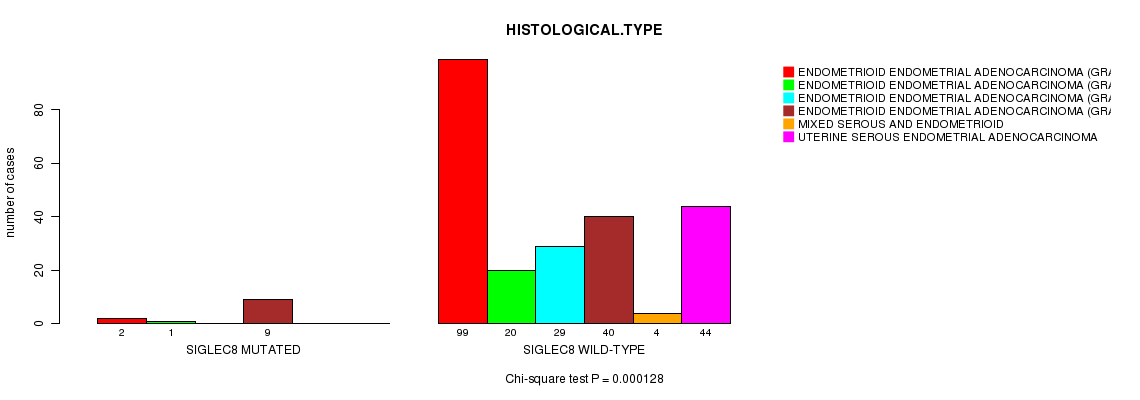
P value = 0.000128 (Chi-square test), Q value = 0.18
Table S7. Gene #136: 'SIGLEC8 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | UTERINE SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|
| ALL | 101 | 21 | 29 | 49 | 4 | 44 |
| SIGLEC8 MUTATED | 2 | 1 | 0 | 9 | 0 | 0 |
| SIGLEC8 WILD-TYPE | 99 | 20 | 29 | 40 | 4 | 44 |
Figure S7. Get High-res Image Gene #136: 'SIGLEC8 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
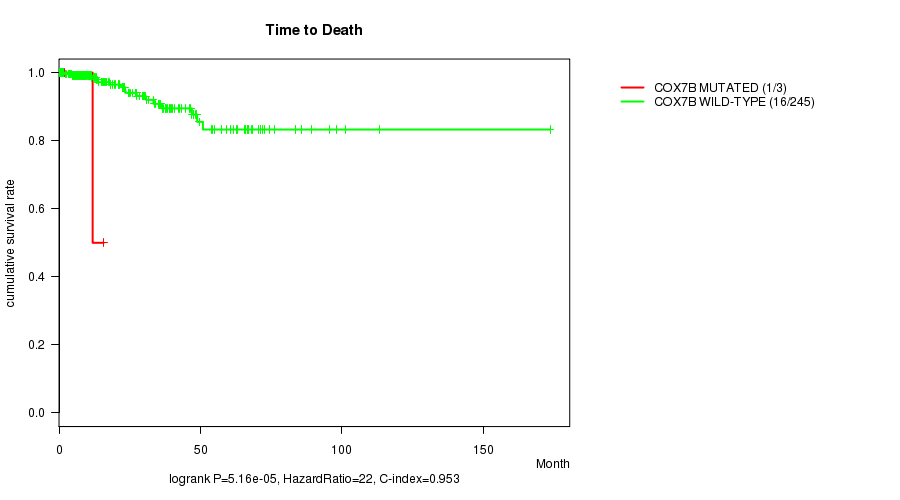
P value = 5.16e-05 (logrank test), Q value = 0.073
Table S8. Gene #187: 'COX7B MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 248 | 17 | 0.0 - 173.6 (22.2) |
| COX7B MUTATED | 3 | 1 | 1.8 - 15.8 (11.9) |
| COX7B WILD-TYPE | 245 | 16 | 0.0 - 173.6 (22.4) |
Figure S8. Get High-res Image Gene #187: 'COX7B MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
-
Mutation data file = UCEC.mutsig.cluster.txt
-
Clinical data file = UCEC.clin.merged.picked.txt
-
Number of patients = 248
-
Number of significantly mutated genes = 283
-
Number of selected clinical features = 5
-
Exclude genes that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For continuous numerical clinical features, two-tailed Student's t test with unequal variance (Lehmann and Romano 2005) was applied to compare the clinical values between tumors with and without gene mutations using 't.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. Location of data archives could not be determined.