This pipeline computes the correlation between significantly recurrent gene mutations and selected clinical features.
Testing the association between mutation status of 63 genes and 5 clinical features across 248 patients, 6 significant findings detected with Q value < 0.25.
-
OR10X1 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
ZNF69 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
TREM2 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
ZMYND10 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
CEACAM4 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
PPP2CA mutation correlated to 'HISTOLOGICAL.TYPE'.
Table 1. Get Full Table Overview of the association between mutation status of 63 genes and 5 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 6 significant findings detected.
|
Clinical Features |
Time to Death |
AGE |
HISTOLOGICAL TYPE |
RADIATIONS RADIATION REGIMENINDICATION |
NEOADJUVANT THERAPY |
||
| nMutated (%) | nWild-Type | logrank test | t-test | Chi-square test | Fisher's exact test | Fisher's exact test | |
| OR10X1 | 7 (3%) | 241 |
0.646 (1.00) |
0.036 (1.00) |
0.000147 (0.0458) |
0.428 (1.00) |
1 (1.00) |
| ZNF69 | 7 (3%) | 241 |
0.514 (1.00) |
0.0163 (1.00) |
0.000147 (0.0458) |
1 (1.00) |
0.396 (1.00) |
| TREM2 | 7 (3%) | 241 |
0.361 (1.00) |
0.197 (1.00) |
0.000147 (0.0458) |
0.236 (1.00) |
1 (1.00) |
| ZMYND10 | 7 (3%) | 241 |
0.384 (1.00) |
0.956 (1.00) |
2.1e-11 (6.62e-09) |
1 (1.00) |
1 (1.00) |
| CEACAM4 | 6 (2%) | 242 |
0.554 (1.00) |
0.0488 (1.00) |
2.07e-05 (0.0065) |
0.667 (1.00) |
0.667 (1.00) |
| PPP2CA | 6 (2%) | 242 |
0.494 (1.00) |
0.0624 (1.00) |
2.77e-05 (0.00868) |
0.667 (1.00) |
0.35 (1.00) |
| TNFAIP6 | 12 (5%) | 236 |
0.819 (1.00) |
0.302 (1.00) |
0.6 (1.00) |
1 (1.00) |
0.741 (1.00) |
| RPL14 | 7 (3%) | 241 |
0.532 (1.00) |
0.677 (1.00) |
0.991 (1.00) |
0.428 (1.00) |
0.396 (1.00) |
| IK | 8 (3%) | 240 |
0.734 (1.00) |
0.629 (1.00) |
0.809 (1.00) |
1 (1.00) |
0.22 (1.00) |
| CCDC160 | 11 (4%) | 237 |
0.271 (1.00) |
0.34 (1.00) |
0.00658 (1.00) |
1 (1.00) |
0.732 (1.00) |
| PRPF38B | 11 (4%) | 237 |
0.39 (1.00) |
0.482 (1.00) |
0.623 (1.00) |
0.753 (1.00) |
1 (1.00) |
| NKTR | 18 (7%) | 230 |
0.194 (1.00) |
0.263 (1.00) |
0.0375 (1.00) |
0.616 (1.00) |
0.786 (1.00) |
| SARNP | 5 (2%) | 243 |
0.496 (1.00) |
0.656 (1.00) |
0.461 (1.00) |
0.663 (1.00) |
1 (1.00) |
| C1ORF189 | 4 (2%) | 244 |
0.652 (1.00) |
0.2 (1.00) |
0.078 (1.00) |
1 (1.00) |
0.0639 (1.00) |
| ZNF564 | 9 (4%) | 239 |
0.32 (1.00) |
0.44 (1.00) |
0.00167 (0.515) |
0.497 (1.00) |
0.451 (1.00) |
| USP48 | 13 (5%) | 235 |
0.257 (1.00) |
0.0295 (1.00) |
0.4 (1.00) |
1 (1.00) |
0.524 (1.00) |
| LGI2 | 9 (4%) | 239 |
0.467 (1.00) |
0.882 (1.00) |
0.0573 (1.00) |
0.281 (1.00) |
1 (1.00) |
| LINGO4 | 10 (4%) | 238 |
0.407 (1.00) |
0.0353 (1.00) |
0.707 (1.00) |
0.739 (1.00) |
0.468 (1.00) |
| SERPINB9 | 8 (3%) | 240 |
0.464 (1.00) |
0.0216 (1.00) |
0.628 (1.00) |
0.451 (1.00) |
0.111 (1.00) |
| FRMD6 | 11 (4%) | 237 |
0.357 (1.00) |
0.501 (1.00) |
0.653 (1.00) |
0.518 (1.00) |
0.499 (1.00) |
| TRIM46 | 9 (4%) | 239 |
0.546 (1.00) |
0.413 (1.00) |
0.987 (1.00) |
1 (1.00) |
1 (1.00) |
| SEPT5 | 8 (3%) | 240 |
0.585 (1.00) |
0.743 (1.00) |
0.833 (1.00) |
0.719 (1.00) |
1 (1.00) |
| ABCB6 | 10 (4%) | 238 |
0.316 (1.00) |
0.913 (1.00) |
0.315 (1.00) |
1 (1.00) |
0.293 (1.00) |
| KRT32 | 10 (4%) | 238 |
0.834 (1.00) |
0.822 (1.00) |
0.00363 (1.00) |
0.739 (1.00) |
0.468 (1.00) |
| VSIG1 | 7 (3%) | 241 |
0.442 (1.00) |
0.279 (1.00) |
0.856 (1.00) |
0.694 (1.00) |
1 (1.00) |
| ZBTB33 | 15 (6%) | 233 |
0.327 (1.00) |
0.231 (1.00) |
0.827 (1.00) |
0.589 (1.00) |
0.766 (1.00) |
| CIC | 17 (7%) | 231 |
0.285 (1.00) |
0.647 (1.00) |
0.0741 (1.00) |
0.599 (1.00) |
0.166 (1.00) |
| ERBB3 | 17 (7%) | 231 |
0.311 (1.00) |
0.653 (1.00) |
0.0843 (1.00) |
0.599 (1.00) |
1 (1.00) |
| HFE | 6 (2%) | 242 |
0.547 (1.00) |
0.832 (1.00) |
0.996 (1.00) |
0.415 (1.00) |
1 (1.00) |
| MTSS1 | 11 (4%) | 237 |
0.457 (1.00) |
0.272 (1.00) |
0.404 (1.00) |
0.194 (1.00) |
0.732 (1.00) |
| OR4L1 | 10 (4%) | 238 |
0.93 (1.00) |
0.11 (1.00) |
0.456 (1.00) |
0.0959 (1.00) |
0.732 (1.00) |
| RDH8 | 8 (3%) | 240 |
0.507 (1.00) |
0.272 (1.00) |
0.518 (1.00) |
0.451 (1.00) |
0.452 (1.00) |
| BTG4 | 7 (3%) | 241 |
0.46 (1.00) |
0.855 (1.00) |
0.294 (1.00) |
0.694 (1.00) |
0.677 (1.00) |
| MLN | 6 (2%) | 242 |
0.498 (1.00) |
0.539 (1.00) |
0.897 (1.00) |
1 (1.00) |
1 (1.00) |
| RAP2C | 5 (2%) | 243 |
0.616 (1.00) |
0.797 (1.00) |
0.933 (1.00) |
0.0483 (1.00) |
0.327 (1.00) |
| PITRM1 | 13 (5%) | 235 |
0.702 (1.00) |
0.775 (1.00) |
0.015 (1.00) |
0.039 (1.00) |
0.756 (1.00) |
| OR6B1 | 6 (2%) | 242 |
0.524 (1.00) |
0.654 (1.00) |
0.897 (1.00) |
0.415 (1.00) |
1 (1.00) |
| BCL7C | 5 (2%) | 243 |
0.369 (1.00) |
0.251 (1.00) |
0.149 (1.00) |
0.663 (1.00) |
1 (1.00) |
| CPA5 | 7 (3%) | 241 |
0.399 (1.00) |
0.0488 (1.00) |
0.294 (1.00) |
1 (1.00) |
0.396 (1.00) |
| CFHR5 | 8 (3%) | 240 |
0.39 (1.00) |
0.958 (1.00) |
0.343 (1.00) |
0.719 (1.00) |
1 (1.00) |
| RGL2 | 7 (3%) | 241 |
0.636 (1.00) |
0.762 (1.00) |
0.856 (1.00) |
0.428 (1.00) |
0.677 (1.00) |
| PSG9 | 5 (2%) | 243 |
0.515 (1.00) |
0.508 (1.00) |
0.193 (1.00) |
1 (1.00) |
0.617 (1.00) |
| CD4 | 6 (2%) | 242 |
0.496 (1.00) |
0.517 (1.00) |
0.897 (1.00) |
0.667 (1.00) |
0.667 (1.00) |
| IL27 | 4 (2%) | 244 |
0.612 (1.00) |
0.919 (1.00) |
0.961 (1.00) |
0.302 (1.00) |
0.577 (1.00) |
| HOXA1 | 7 (3%) | 241 |
0.505 (1.00) |
0.457 (1.00) |
0.785 (1.00) |
0.236 (1.00) |
0.195 (1.00) |
| SLCO2A1 | 9 (4%) | 239 |
0.81 (1.00) |
0.983 (1.00) |
0.376 (1.00) |
0.722 (1.00) |
0.451 (1.00) |
| C20ORF112 | 6 (2%) | 242 |
0.165 (1.00) |
0.785 (1.00) |
0.897 (1.00) |
1 (1.00) |
1 (1.00) |
| PRKRA | 4 (2%) | 244 |
0.512 (1.00) |
0.498 (1.00) |
0.998 (1.00) |
0.608 (1.00) |
0.577 (1.00) |
| LIMA1 | 8 (3%) | 240 |
0.452 (1.00) |
0.821 (1.00) |
0.343 (1.00) |
0.719 (1.00) |
0.452 (1.00) |
| MS4A6A | 4 (2%) | 244 |
0.497 (1.00) |
0.39 (1.00) |
0.0702 (1.00) |
1 (1.00) |
1 (1.00) |
| PREB | 5 (2%) | 243 |
0.496 (1.00) |
0.252 (1.00) |
0.933 (1.00) |
0.342 (1.00) |
1 (1.00) |
| ATP6AP1 | 8 (3%) | 240 |
0.444 (1.00) |
0.74 (1.00) |
0.343 (1.00) |
0.451 (1.00) |
1 (1.00) |
| C20ORF4 | 5 (2%) | 243 |
0.482 (1.00) |
0.864 (1.00) |
0.933 (1.00) |
0.342 (1.00) |
0.617 (1.00) |
| GTSF1 | 3 (1%) | 245 |
0.648 (1.00) |
0.547 (1.00) |
0.981 (1.00) |
1 (1.00) |
0.564 (1.00) |
| SEMG2 | 11 (4%) | 237 |
0.337 (1.00) |
0.111 (1.00) |
0.354 (1.00) |
1 (1.00) |
1 (1.00) |
| CHST10 | 5 (2%) | 243 |
0.47 (1.00) |
0.985 (1.00) |
0.933 (1.00) |
0.342 (1.00) |
0.327 (1.00) |
| HINT3 | 4 (2%) | 244 |
0.67 (1.00) |
0.518 (1.00) |
0.328 (1.00) |
1 (1.00) |
0.577 (1.00) |
| OIP5 | 3 (1%) | 245 |
0.848 (1.00) |
0.642 (1.00) |
0.981 (1.00) |
0.271 (1.00) |
0.564 (1.00) |
| TMEM212 | 3 (1%) | 245 |
0.655 (1.00) |
0.396 (1.00) |
0.0134 (1.00) |
0.553 (1.00) |
0.564 (1.00) |
| NDUFB7 | 3 (1%) | 245 |
0.648 (1.00) |
0.811 (1.00) |
0.159 (1.00) |
0.0393 (1.00) |
1 (1.00) |
| NDUFAF1 | 9 (4%) | 239 |
0.351 (1.00) |
0.83 (1.00) |
0.376 (1.00) |
0.497 (1.00) |
1 (1.00) |
| CASP10 | 7 (3%) | 241 |
0.478 (1.00) |
0.416 (1.00) |
0.294 (1.00) |
1 (1.00) |
0.677 (1.00) |
| PLAC8L1 | 5 (2%) | 243 |
0.45 (1.00) |
0.545 (1.00) |
0.149 (1.00) |
0.342 (1.00) |
0.617 (1.00) |
P value = 0.000147 (Chi-square test), Q value = 0.046
Table S1. Gene #9: 'OR10X1 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| OR10X1 MUTATED | 5 | 0 | 0 | 0 | 2 | 0 | 0 |
| OR10X1 WILD-TYPE | 177 | 3 | 8 | 2 | 3 | 4 | 44 |
Figure S1. Get High-res Image Gene #9: 'OR10X1 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
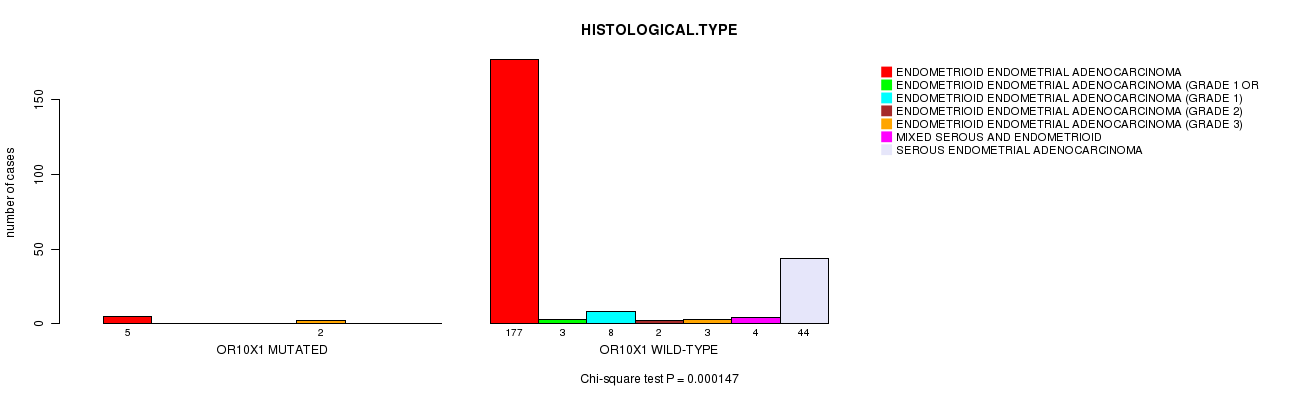
P value = 0.000147 (Chi-square test), Q value = 0.046
Table S2. Gene #22: 'ZNF69 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| ZNF69 MUTATED | 5 | 0 | 0 | 0 | 2 | 0 | 0 |
| ZNF69 WILD-TYPE | 177 | 3 | 8 | 2 | 3 | 4 | 44 |
Figure S2. Get High-res Image Gene #22: 'ZNF69 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
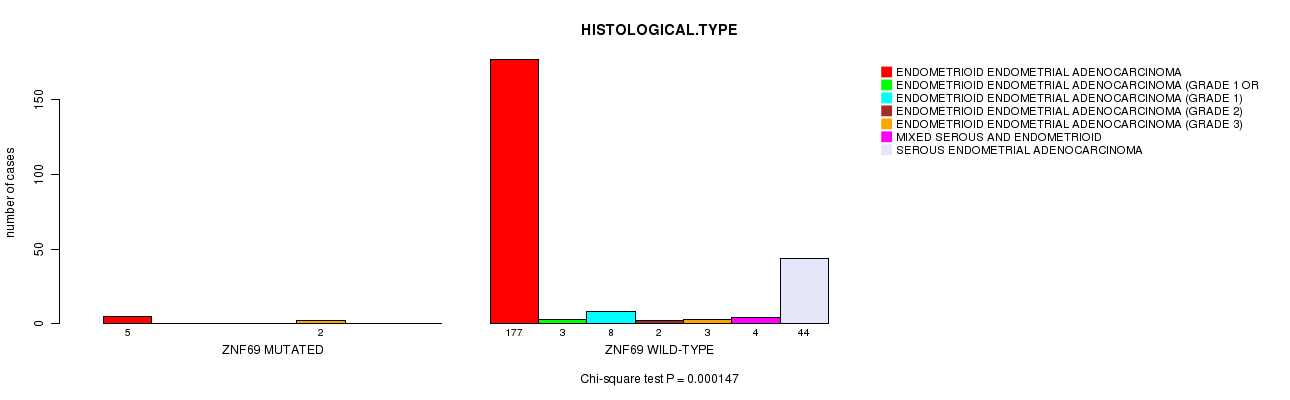
P value = 0.000147 (Chi-square test), Q value = 0.046
Table S3. Gene #29: 'TREM2 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| TREM2 MUTATED | 5 | 0 | 0 | 0 | 2 | 0 | 0 |
| TREM2 WILD-TYPE | 177 | 3 | 8 | 2 | 3 | 4 | 44 |
Figure S3. Get High-res Image Gene #29: 'TREM2 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
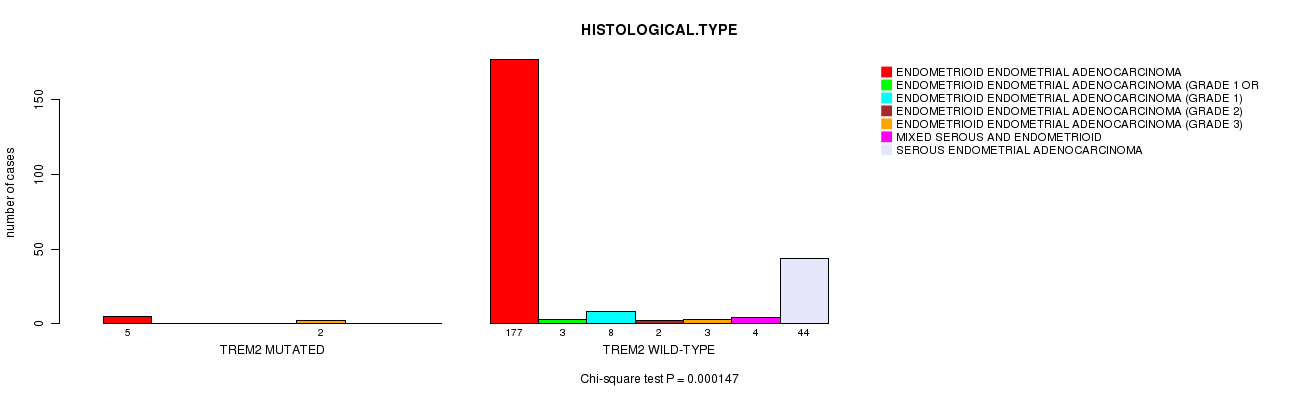
P value = 2.1e-11 (Chi-square test), Q value = 6.6e-09
Table S4. Gene #37: 'ZMYND10 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| ZMYND10 MUTATED | 4 | 0 | 0 | 0 | 3 | 0 | 0 |
| ZMYND10 WILD-TYPE | 178 | 3 | 8 | 2 | 2 | 4 | 44 |
Figure S4. Get High-res Image Gene #37: 'ZMYND10 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
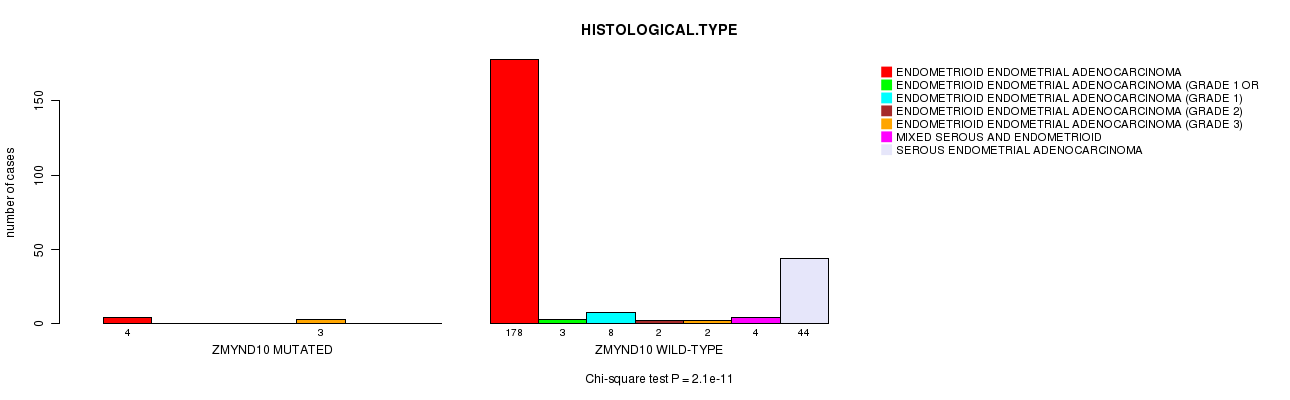
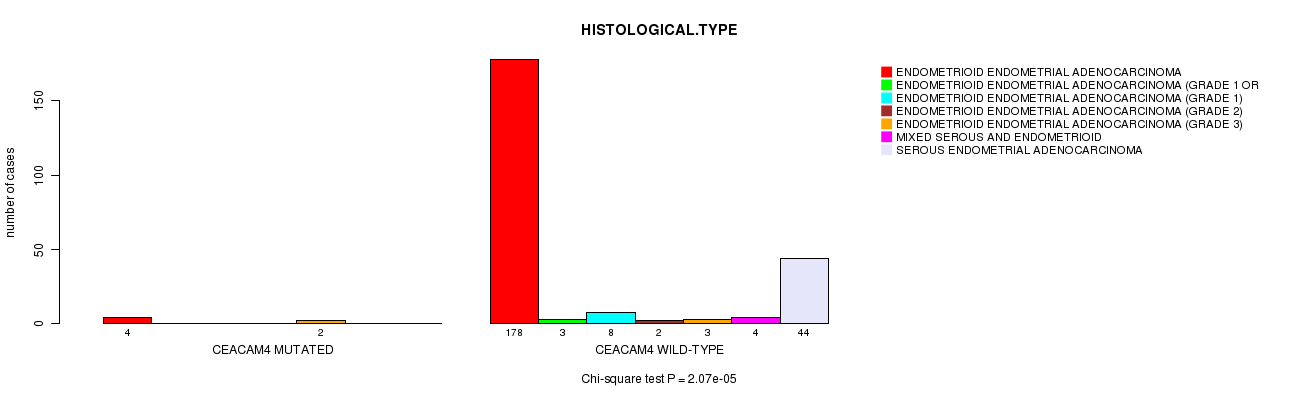
P value = 2.07e-05 (Chi-square test), Q value = 0.0065
Table S5. Gene #57: 'CEACAM4 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| CEACAM4 MUTATED | 4 | 0 | 0 | 0 | 2 | 0 | 0 |
| CEACAM4 WILD-TYPE | 178 | 3 | 8 | 2 | 3 | 4 | 44 |
Figure S5. Get High-res Image Gene #57: 'CEACAM4 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
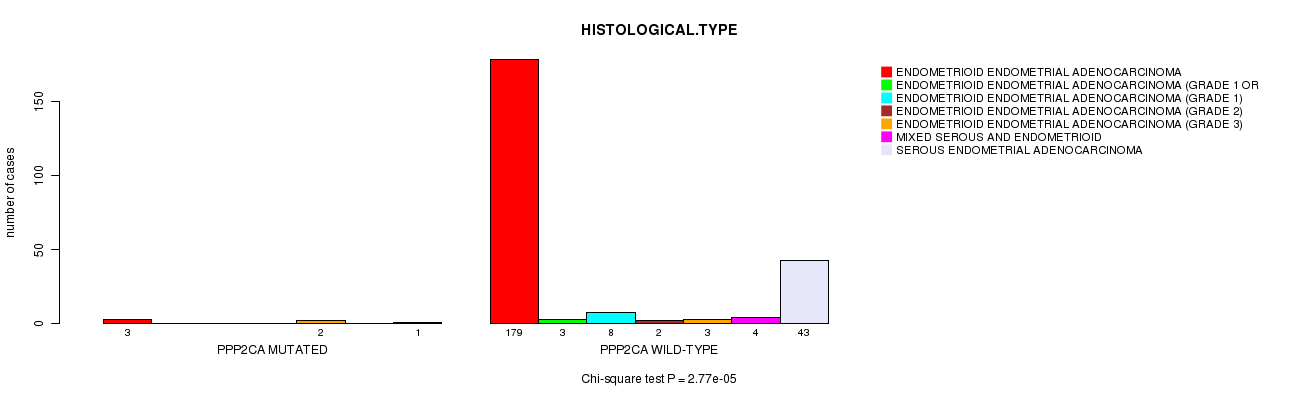
P value = 2.77e-05 (Chi-square test), Q value = 0.0087
Table S6. Gene #63: 'PPP2CA MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| PPP2CA MUTATED | 3 | 0 | 0 | 0 | 2 | 0 | 1 |
| PPP2CA WILD-TYPE | 179 | 3 | 8 | 2 | 3 | 4 | 43 |
Figure S6. Get High-res Image Gene #63: 'PPP2CA MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
-
Mutation data file = UCEC.mutsig.cluster.txt
-
Clinical data file = UCEC.clin.merged.picked.txt
-
Number of patients = 248
-
Number of significantly mutated genes = 63
-
Number of selected clinical features = 5
-
Exclude genes that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For continuous numerical clinical features, two-tailed Student's t test with unequal variance (Lehmann and Romano 2005) was applied to compare the clinical values between tumors with and without gene mutations using 't.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.