This pipeline computes the correlation between significantly recurrent gene mutations and selected clinical features.
Testing the association between mutation status of 305 genes and 5 clinical features across 248 patients, 14 significant findings detected with Q value < 0.25.
-
CTNNB1 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
PTEN mutation correlated to 'HISTOLOGICAL.TYPE'.
-
TP53 mutation correlated to 'AGE', 'HISTOLOGICAL.TYPE', and 'NEOADJUVANT.THERAPY'.
-
ANKRD31 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
SH3BGRL mutation correlated to 'HISTOLOGICAL.TYPE'.
-
S100A12 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
C4ORF33 mutation correlated to 'Time to Death'.
-
GLRX3 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
SLC16A10 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
SLC10A5 mutation correlated to 'HISTOLOGICAL.TYPE'.
-
CDKN1B mutation correlated to 'HISTOLOGICAL.TYPE'.
-
ALKBH6 mutation correlated to 'HISTOLOGICAL.TYPE'.
Table 1. Get Full Table Overview of the association between mutation status of 305 genes and 5 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 14 significant findings detected.
|
Clinical Features |
Time to Death |
AGE |
HISTOLOGICAL TYPE |
RADIATIONS RADIATION REGIMENINDICATION |
NEOADJUVANT THERAPY |
||
| nMutated (%) | nWild-Type | logrank test | t-test | Chi-square test | Fisher's exact test | Fisher's exact test | |
| TP53 | 69 (28%) | 179 |
0.325 (1.00) |
6.15e-06 (0.00936) |
3.98e-23 (6.07e-20) |
0.371 (1.00) |
6.08e-05 (0.0921) |
| CTNNB1 | 74 (30%) | 174 |
0.878 (1.00) |
0.0017 (1.00) |
1.04e-05 (0.0158) |
0.381 (1.00) |
0.0124 (1.00) |
| PTEN | 161 (65%) | 87 |
0.19 (1.00) |
0.0276 (1.00) |
5.44e-19 (8.3e-16) |
0.0503 (1.00) |
0.0016 (1.00) |
| ANKRD31 | 17 (7%) | 231 |
0.2 (1.00) |
0.964 (1.00) |
4.87e-05 (0.0738) |
0.432 (1.00) |
1 (1.00) |
| SH3BGRL | 6 (2%) | 242 |
0.501 (1.00) |
0.323 (1.00) |
2.07e-05 (0.0314) |
0.667 (1.00) |
1 (1.00) |
| S100A12 | 5 (2%) | 243 |
0.614 (1.00) |
0.00132 (1.00) |
1.18e-06 (0.00179) |
0.663 (1.00) |
1 (1.00) |
| C4ORF33 | 4 (2%) | 244 |
0.000124 (0.187) |
0.354 (1.00) |
0.998 (1.00) |
0.608 (1.00) |
0.0639 (1.00) |
| GLRX3 | 7 (3%) | 241 |
0.529 (1.00) |
0.024 (1.00) |
0.000147 (0.222) |
0.428 (1.00) |
1 (1.00) |
| SLC16A10 | 9 (4%) | 239 |
0.381 (1.00) |
0.563 (1.00) |
3.06e-05 (0.0465) |
0.171 (1.00) |
0.451 (1.00) |
| SLC10A5 | 7 (3%) | 241 |
0.825 (1.00) |
0.538 (1.00) |
1.45e-05 (0.022) |
0.694 (1.00) |
0.396 (1.00) |
| CDKN1B | 7 (3%) | 241 |
0.42 (1.00) |
0.0875 (1.00) |
0.000147 (0.222) |
1 (1.00) |
0.195 (1.00) |
| ALKBH6 | 6 (2%) | 242 |
0.468 (1.00) |
0.545 (1.00) |
2.07e-05 (0.0314) |
0.667 (1.00) |
0.667 (1.00) |
| ARID1A | 83 (33%) | 165 |
0.008 (1.00) |
0.00721 (1.00) |
0.00127 (1.00) |
0.673 (1.00) |
0.0234 (1.00) |
| CTCF | 45 (18%) | 203 |
0.405 (1.00) |
0.0198 (1.00) |
0.0021 (1.00) |
0.863 (1.00) |
1 (1.00) |
| FBXW7 | 39 (16%) | 209 |
0.772 (1.00) |
0.904 (1.00) |
0.0473 (1.00) |
0.855 (1.00) |
0.117 (1.00) |
| FGFR2 | 31 (12%) | 217 |
0.9 (1.00) |
0.998 (1.00) |
0.719 (1.00) |
1 (1.00) |
0.39 (1.00) |
| KRAS | 52 (21%) | 196 |
0.0772 (1.00) |
0.000643 (0.968) |
0.00753 (1.00) |
0.19 (1.00) |
0.0136 (1.00) |
| PIK3CA | 132 (53%) | 116 |
0.055 (1.00) |
0.195 (1.00) |
0.555 (1.00) |
0.593 (1.00) |
0.887 (1.00) |
| PIK3R1 | 83 (33%) | 165 |
0.871 (1.00) |
0.806 (1.00) |
0.000827 (1.00) |
0.395 (1.00) |
0.293 (1.00) |
| SPOP | 21 (8%) | 227 |
0.65 (1.00) |
0.527 (1.00) |
0.332 (1.00) |
0.811 (1.00) |
0.452 (1.00) |
| PPP2R1A | 27 (11%) | 221 |
0.121 (1.00) |
0.0374 (1.00) |
0.00977 (1.00) |
0.52 (1.00) |
0.112 (1.00) |
| P2RY11 | 9 (4%) | 239 |
0.395 (1.00) |
0.396 (1.00) |
0.987 (1.00) |
0.497 (1.00) |
0.709 (1.00) |
| CHD4 | 35 (14%) | 213 |
0.361 (1.00) |
0.418 (1.00) |
0.779 (1.00) |
0.565 (1.00) |
0.683 (1.00) |
| CCND1 | 15 (6%) | 233 |
0.729 (1.00) |
0.0556 (1.00) |
0.413 (1.00) |
0.4 (1.00) |
0.13 (1.00) |
| FAM9A | 14 (6%) | 234 |
0.378 (1.00) |
0.913 (1.00) |
0.89 (1.00) |
1 (1.00) |
0.121 (1.00) |
| NFE2L2 | 15 (6%) | 233 |
0.903 (1.00) |
0.191 (1.00) |
0.447 (1.00) |
0.589 (1.00) |
1 (1.00) |
| FOXA2 | 12 (5%) | 236 |
0.799 (1.00) |
0.531 (1.00) |
0.972 (1.00) |
0.229 (1.00) |
1 (1.00) |
| ARID5B | 29 (12%) | 219 |
0.642 (1.00) |
0.649 (1.00) |
0.00865 (1.00) |
1 (1.00) |
0.508 (1.00) |
| TNFAIP6 | 12 (5%) | 236 |
0.819 (1.00) |
0.302 (1.00) |
0.6 (1.00) |
1 (1.00) |
0.741 (1.00) |
| MORC4 | 20 (8%) | 228 |
0.152 (1.00) |
0.0204 (1.00) |
0.629 (1.00) |
1 (1.00) |
0.295 (1.00) |
| CYLC1 | 18 (7%) | 230 |
0.198 (1.00) |
0.765 (1.00) |
0.000601 (0.905) |
0.313 (1.00) |
0.413 (1.00) |
| RBMX | 13 (5%) | 235 |
0.336 (1.00) |
0.161 (1.00) |
0.4 (1.00) |
0.142 (1.00) |
0.756 (1.00) |
| RASA1 | 22 (9%) | 226 |
0.127 (1.00) |
0.466 (1.00) |
0.0211 (1.00) |
0.639 (1.00) |
0.803 (1.00) |
| SOX17 | 7 (3%) | 241 |
0.669 (1.00) |
0.848 (1.00) |
0.00567 (1.00) |
0.428 (1.00) |
0.677 (1.00) |
| SGK1 | 15 (6%) | 233 |
0.925 (1.00) |
0.427 (1.00) |
0.759 (1.00) |
0.158 (1.00) |
0.563 (1.00) |
| ZNF471 | 15 (6%) | 233 |
0.549 (1.00) |
0.0733 (1.00) |
0.025 (1.00) |
0.589 (1.00) |
1 (1.00) |
| SYCP1 | 16 (6%) | 232 |
0.258 (1.00) |
0.302 (1.00) |
0.0297 (1.00) |
0.423 (1.00) |
1 (1.00) |
| TCP11L2 | 14 (6%) | 234 |
0.194 (1.00) |
0.777 (1.00) |
0.0202 (1.00) |
1 (1.00) |
0.362 (1.00) |
| SIN3A | 21 (8%) | 227 |
0.574 (1.00) |
0.256 (1.00) |
0.1 (1.00) |
0.811 (1.00) |
0.609 (1.00) |
| LOC642587 | 7 (3%) | 241 |
0.679 (1.00) |
0.548 (1.00) |
0.294 (1.00) |
1 (1.00) |
0.677 (1.00) |
| CHEK2 | 13 (5%) | 235 |
0.891 (1.00) |
0.0163 (1.00) |
0.0307 (1.00) |
1 (1.00) |
0.352 (1.00) |
| MAX | 11 (4%) | 237 |
0.889 (1.00) |
0.336 (1.00) |
0.404 (1.00) |
0.753 (1.00) |
0.732 (1.00) |
| RUNDC3B | 11 (4%) | 237 |
0.704 (1.00) |
0.346 (1.00) |
0.142 (1.00) |
1 (1.00) |
0.499 (1.00) |
| TAB3 | 18 (7%) | 230 |
0.22 (1.00) |
0.107 (1.00) |
0.00686 (1.00) |
0.616 (1.00) |
1 (1.00) |
| BRS3 | 15 (6%) | 233 |
0.184 (1.00) |
0.133 (1.00) |
0.025 (1.00) |
0.4 (1.00) |
0.766 (1.00) |
| KLHL8 | 12 (5%) | 236 |
0.362 (1.00) |
0.504 (1.00) |
0.404 (1.00) |
1 (1.00) |
1 (1.00) |
| CSDE1 | 21 (8%) | 227 |
0.802 (1.00) |
0.294 (1.00) |
0.0435 (1.00) |
1 (1.00) |
0.452 (1.00) |
| DNER | 18 (7%) | 230 |
0.746 (1.00) |
0.0414 (1.00) |
0.0324 (1.00) |
0.44 (1.00) |
0.277 (1.00) |
| PAPD4 | 13 (5%) | 235 |
0.982 (1.00) |
0.181 (1.00) |
0.962 (1.00) |
0.378 (1.00) |
0.0492 (1.00) |
| NFE2L3 | 12 (5%) | 236 |
0.336 (1.00) |
0.656 (1.00) |
0.342 (1.00) |
0.115 (1.00) |
0.741 (1.00) |
| UPF3B | 16 (6%) | 232 |
0.735 (1.00) |
0.81 (1.00) |
0.395 (1.00) |
0.588 (1.00) |
0.0775 (1.00) |
| NAA15 | 14 (6%) | 234 |
0.763 (1.00) |
0.64 (1.00) |
0.00864 (1.00) |
0.248 (1.00) |
1 (1.00) |
| RPL14 | 7 (3%) | 241 |
0.532 (1.00) |
0.677 (1.00) |
0.991 (1.00) |
0.428 (1.00) |
0.396 (1.00) |
| OR4A15 | 13 (5%) | 235 |
0.213 (1.00) |
0.167 (1.00) |
0.00219 (1.00) |
1 (1.00) |
0.756 (1.00) |
| C14ORF118 | 15 (6%) | 233 |
0.268 (1.00) |
0.0138 (1.00) |
0.369 (1.00) |
0.589 (1.00) |
0.766 (1.00) |
| OR4K2 | 11 (4%) | 237 |
0.304 (1.00) |
0.241 (1.00) |
0.00658 (1.00) |
1 (1.00) |
0.499 (1.00) |
| OR5D13 | 10 (4%) | 238 |
0.895 (1.00) |
0.741 (1.00) |
0.188 (1.00) |
0.318 (1.00) |
0.468 (1.00) |
| CNPY1 | 7 (3%) | 241 |
0.449 (1.00) |
0.18 (1.00) |
0.294 (1.00) |
1 (1.00) |
0.677 (1.00) |
| COL19A1 | 21 (8%) | 227 |
0.169 (1.00) |
0.198 (1.00) |
0.0682 (1.00) |
0.345 (1.00) |
1 (1.00) |
| GYPB | 5 (2%) | 243 |
0.583 (1.00) |
0.843 (1.00) |
0.149 (1.00) |
0.168 (1.00) |
1 (1.00) |
| ASB15 | 14 (6%) | 234 |
0.298 (1.00) |
0.236 (1.00) |
0.182 (1.00) |
0.777 (1.00) |
0.764 (1.00) |
| AGBL4 | 14 (6%) | 234 |
0.615 (1.00) |
0.0925 (1.00) |
0.653 (1.00) |
0.392 (1.00) |
1 (1.00) |
| CDH10 | 22 (9%) | 226 |
0.134 (1.00) |
0.0308 (1.00) |
0.0581 (1.00) |
0.818 (1.00) |
0.453 (1.00) |
| MTM1 | 15 (6%) | 233 |
0.258 (1.00) |
0.896 (1.00) |
0.00648 (1.00) |
0.589 (1.00) |
0.0753 (1.00) |
| NRAS | 9 (4%) | 239 |
0.354 (1.00) |
0.616 (1.00) |
0.0289 (1.00) |
0.722 (1.00) |
0.451 (1.00) |
| SMTNL2 | 9 (4%) | 239 |
0.472 (1.00) |
0.53 (1.00) |
0.579 (1.00) |
1 (1.00) |
0.451 (1.00) |
| OR5AS1 | 10 (4%) | 238 |
0.675 (1.00) |
0.503 (1.00) |
0.396 (1.00) |
0.501 (1.00) |
0.732 (1.00) |
| STEAP4 | 12 (5%) | 236 |
0.286 (1.00) |
0.933 (1.00) |
0.000714 (1.00) |
0.756 (1.00) |
0.52 (1.00) |
| ZFHX3 | 44 (18%) | 204 |
0.325 (1.00) |
0.765 (1.00) |
0.147 (1.00) |
0.73 (1.00) |
0.141 (1.00) |
| GPM6A | 11 (4%) | 237 |
0.306 (1.00) |
0.502 (1.00) |
0.545 (1.00) |
1 (1.00) |
0.732 (1.00) |
| GEN1 | 16 (6%) | 232 |
0.302 (1.00) |
0.992 (1.00) |
0.019 (1.00) |
0.79 (1.00) |
1 (1.00) |
| FAM194B | 13 (5%) | 235 |
0.393 (1.00) |
0.44 (1.00) |
0.0403 (1.00) |
0.229 (1.00) |
1 (1.00) |
| CLIC2 | 10 (4%) | 238 |
0.294 (1.00) |
0.855 (1.00) |
0.00231 (1.00) |
1 (1.00) |
0.732 (1.00) |
| ZNF267 | 16 (6%) | 232 |
0.565 (1.00) |
0.207 (1.00) |
0.0275 (1.00) |
0.79 (1.00) |
1 (1.00) |
| ZMYM2 | 17 (7%) | 231 |
0.252 (1.00) |
0.0462 (1.00) |
0.0414 (1.00) |
1 (1.00) |
0.166 (1.00) |
| FMR1 | 16 (6%) | 232 |
0.304 (1.00) |
0.643 (1.00) |
0.0688 (1.00) |
0.588 (1.00) |
0.0775 (1.00) |
| OR4A5 | 10 (4%) | 238 |
0.454 (1.00) |
0.495 (1.00) |
0.00676 (1.00) |
0.501 (1.00) |
0.066 (1.00) |
| ALG8 | 10 (4%) | 238 |
0.369 (1.00) |
0.855 (1.00) |
0.635 (1.00) |
0.501 (1.00) |
1 (1.00) |
| C1ORF100 | 9 (4%) | 239 |
0.344 (1.00) |
0.246 (1.00) |
0.376 (1.00) |
0.497 (1.00) |
0.709 (1.00) |
| SNTG1 | 11 (4%) | 237 |
0.307 (1.00) |
0.506 (1.00) |
0.0644 (1.00) |
0.753 (1.00) |
1 (1.00) |
| UBE2E3 | 7 (3%) | 241 |
0.77 (1.00) |
0.993 (1.00) |
0.294 (1.00) |
0.694 (1.00) |
1 (1.00) |
| ANKRD50 | 21 (8%) | 227 |
0.484 (1.00) |
0.754 (1.00) |
0.82 (1.00) |
1 (1.00) |
0.609 (1.00) |
| CTXN3 | 6 (2%) | 242 |
0.512 (1.00) |
0.794 (1.00) |
0.548 (1.00) |
1 (1.00) |
0.193 (1.00) |
| GALNT13 | 16 (6%) | 232 |
0.258 (1.00) |
0.633 (1.00) |
0.0343 (1.00) |
1 (1.00) |
0.568 (1.00) |
| F9 | 13 (5%) | 235 |
0.861 (1.00) |
0.0227 (1.00) |
0.63 (1.00) |
0.551 (1.00) |
1 (1.00) |
| PASD1 | 17 (7%) | 231 |
0.188 (1.00) |
0.756 (1.00) |
0.325 (1.00) |
0.599 (1.00) |
0.573 (1.00) |
| GIGYF2 | 27 (11%) | 221 |
0.91 (1.00) |
0.612 (1.00) |
0.28 (1.00) |
0.0179 (1.00) |
0.362 (1.00) |
| SSX5 | 11 (4%) | 237 |
0.328 (1.00) |
0.15 (1.00) |
0.00658 (1.00) |
1 (1.00) |
0.732 (1.00) |
| RAE1 | 11 (4%) | 237 |
0.282 (1.00) |
0.603 (1.00) |
0.0127 (1.00) |
0.34 (1.00) |
0.298 (1.00) |
| UGT2B11 | 12 (5%) | 236 |
0.383 (1.00) |
0.168 (1.00) |
0.0105 (1.00) |
0.552 (1.00) |
0.741 (1.00) |
| RB1 | 20 (8%) | 228 |
0.484 (1.00) |
0.237 (1.00) |
0.000223 (0.338) |
0.465 (1.00) |
0.603 (1.00) |
| C7ORF60 | 10 (4%) | 238 |
0.31 (1.00) |
0.136 (1.00) |
0.707 (1.00) |
0.318 (1.00) |
0.732 (1.00) |
| ZNF334 | 17 (7%) | 231 |
0.95 (1.00) |
0.0232 (1.00) |
0.787 (1.00) |
1 (1.00) |
0.415 (1.00) |
| ATM | 29 (12%) | 219 |
0.0912 (1.00) |
0.139 (1.00) |
0.0783 (1.00) |
0.534 (1.00) |
0.119 (1.00) |
| CCDC104 | 10 (4%) | 238 |
0.327 (1.00) |
0.0526 (1.00) |
0.396 (1.00) |
0.501 (1.00) |
0.732 (1.00) |
| MAP3K1 | 21 (8%) | 227 |
0.0962 (1.00) |
0.813 (1.00) |
0.2 (1.00) |
1 (1.00) |
0.452 (1.00) |
| LDB2 | 10 (4%) | 238 |
0.363 (1.00) |
0.169 (1.00) |
0.00676 (1.00) |
1 (1.00) |
0.468 (1.00) |
| ING1 | 13 (5%) | 235 |
0.37 (1.00) |
0.694 (1.00) |
0.178 (1.00) |
0.142 (1.00) |
1 (1.00) |
| ZNF781 | 10 (4%) | 238 |
0.355 (1.00) |
0.227 (1.00) |
0.396 (1.00) |
0.501 (1.00) |
1 (1.00) |
| ACSL4 | 15 (6%) | 233 |
0.95 (1.00) |
0.107 (1.00) |
0.0529 (1.00) |
0.275 (1.00) |
0.766 (1.00) |
| GNPDA2 | 8 (3%) | 240 |
0.789 (1.00) |
0.175 (1.00) |
0.518 (1.00) |
0.451 (1.00) |
0.111 (1.00) |
| HIST1H2BD | 6 (2%) | 242 |
0.0836 (1.00) |
0.474 (1.00) |
0.897 (1.00) |
0.415 (1.00) |
1 (1.00) |
| CTNND1 | 19 (8%) | 229 |
0.572 (1.00) |
0.655 (1.00) |
0.0471 (1.00) |
0.46 (1.00) |
0.604 (1.00) |
| IGFBP7 | 6 (2%) | 242 |
0.406 (1.00) |
0.659 (1.00) |
0.228 (1.00) |
0.667 (1.00) |
0.667 (1.00) |
| SI | 26 (10%) | 222 |
0.387 (1.00) |
0.5 (1.00) |
0.0667 (1.00) |
0.514 (1.00) |
0.17 (1.00) |
| DCAF6 | 16 (6%) | 232 |
0.262 (1.00) |
0.448 (1.00) |
0.13 (1.00) |
1 (1.00) |
1 (1.00) |
| C12ORF50 | 11 (4%) | 237 |
0.323 (1.00) |
0.00222 (1.00) |
0.354 (1.00) |
0.104 (1.00) |
0.499 (1.00) |
| MUTED | 7 (3%) | 241 |
0.378 (1.00) |
0.963 (1.00) |
0.00841 (1.00) |
1 (1.00) |
1 (1.00) |
| TRIM59 | 9 (4%) | 239 |
0.315 (1.00) |
0.949 (1.00) |
0.0343 (1.00) |
1 (1.00) |
0.451 (1.00) |
| ANKRD30A | 22 (9%) | 226 |
0.486 (1.00) |
0.638 (1.00) |
0.1 (1.00) |
0.639 (1.00) |
0.453 (1.00) |
| METTL14 | 10 (4%) | 238 |
0.358 (1.00) |
0.833 (1.00) |
0.707 (1.00) |
1 (1.00) |
0.293 (1.00) |
| ZNF449 | 12 (5%) | 236 |
0.355 (1.00) |
0.95 (1.00) |
0.655 (1.00) |
0.552 (1.00) |
0.52 (1.00) |
| IQCF2 | 6 (2%) | 242 |
0.445 (1.00) |
0.577 (1.00) |
0.897 (1.00) |
1 (1.00) |
0.667 (1.00) |
| TIGD4 | 13 (5%) | 235 |
0.3 (1.00) |
0.0791 (1.00) |
0.015 (1.00) |
0.769 (1.00) |
0.756 (1.00) |
| BBX | 15 (6%) | 233 |
0.783 (1.00) |
0.15 (1.00) |
0.799 (1.00) |
1 (1.00) |
0.563 (1.00) |
| RBM27 | 16 (6%) | 232 |
0.215 (1.00) |
0.187 (1.00) |
0.0631 (1.00) |
1 (1.00) |
0.773 (1.00) |
| WDR45 | 11 (4%) | 237 |
0.355 (1.00) |
0.49 (1.00) |
0.157 (1.00) |
0.518 (1.00) |
0.732 (1.00) |
| ADAM23 | 14 (6%) | 234 |
0.266 (1.00) |
0.42 (1.00) |
0.0389 (1.00) |
0.565 (1.00) |
0.764 (1.00) |
| COX19 | 4 (2%) | 244 |
0.652 (1.00) |
0.269 (1.00) |
0.998 (1.00) |
1 (1.00) |
0.303 (1.00) |
| ACSM3 | 15 (6%) | 233 |
0.215 (1.00) |
0.0585 (1.00) |
0.025 (1.00) |
1 (1.00) |
1 (1.00) |
| ANKRD30B | 22 (9%) | 226 |
0.208 (1.00) |
0.159 (1.00) |
0.821 (1.00) |
0.639 (1.00) |
1 (1.00) |
| CASP8 | 17 (7%) | 231 |
0.775 (1.00) |
0.652 (1.00) |
0.000397 (0.6) |
1 (1.00) |
0.415 (1.00) |
| GK2 | 17 (7%) | 231 |
0.202 (1.00) |
0.334 (1.00) |
0.0119 (1.00) |
0.432 (1.00) |
0.415 (1.00) |
| IL24 | 7 (3%) | 241 |
0.426 (1.00) |
0.988 (1.00) |
0.00148 (1.00) |
0.428 (1.00) |
0.677 (1.00) |
| GOLGA5 | 12 (5%) | 236 |
0.32 (1.00) |
0.801 (1.00) |
0.0257 (1.00) |
1 (1.00) |
0.188 (1.00) |
| HPD | 7 (3%) | 241 |
0.43 (1.00) |
0.0538 (1.00) |
0.0585 (1.00) |
1 (1.00) |
1 (1.00) |
| NDN | 6 (2%) | 242 |
0.535 (1.00) |
0.487 (1.00) |
0.699 (1.00) |
1 (1.00) |
0.667 (1.00) |
| ZNF674 | 14 (6%) | 234 |
0.208 (1.00) |
0.0713 (1.00) |
0.00864 (1.00) |
0.565 (1.00) |
0.764 (1.00) |
| LRRC6 | 11 (4%) | 237 |
0.846 (1.00) |
0.636 (1.00) |
0.00658 (1.00) |
0.518 (1.00) |
0.732 (1.00) |
| TPTE | 15 (6%) | 233 |
0.273 (1.00) |
0.167 (1.00) |
0.00648 (1.00) |
1 (1.00) |
0.369 (1.00) |
| NUSAP1 | 11 (4%) | 237 |
0.306 (1.00) |
0.838 (1.00) |
0.00884 (1.00) |
0.518 (1.00) |
0.179 (1.00) |
| CCDC160 | 11 (4%) | 237 |
0.271 (1.00) |
0.34 (1.00) |
0.00658 (1.00) |
1 (1.00) |
0.732 (1.00) |
| CCDC73 | 12 (5%) | 236 |
0.271 (1.00) |
0.205 (1.00) |
0.00139 (1.00) |
0.229 (1.00) |
1 (1.00) |
| FOXP2 | 16 (6%) | 232 |
0.339 (1.00) |
0.222 (1.00) |
0.0639 (1.00) |
0.588 (1.00) |
0.568 (1.00) |
| IL20 | 7 (3%) | 241 |
0.404 (1.00) |
0.206 (1.00) |
0.294 (1.00) |
0.236 (1.00) |
0.396 (1.00) |
| RBBP6 | 22 (9%) | 226 |
0.661 (1.00) |
0.97 (1.00) |
0.00138 (1.00) |
0.818 (1.00) |
0.208 (1.00) |
| TAP1 | 8 (3%) | 240 |
0.588 (1.00) |
0.877 (1.00) |
0.343 (1.00) |
1 (1.00) |
0.452 (1.00) |
| ATF6 | 14 (6%) | 234 |
0.237 (1.00) |
0.216 (1.00) |
0.65 (1.00) |
0.565 (1.00) |
0.764 (1.00) |
| CDK17 | 14 (6%) | 234 |
0.856 (1.00) |
0.0797 (1.00) |
0.368 (1.00) |
0.777 (1.00) |
0.764 (1.00) |
| FAM9B | 7 (3%) | 241 |
0.401 (1.00) |
0.512 (1.00) |
0.294 (1.00) |
0.236 (1.00) |
0.677 (1.00) |
| MAPK8 | 10 (4%) | 238 |
0.462 (1.00) |
0.777 (1.00) |
0.396 (1.00) |
0.501 (1.00) |
0.293 (1.00) |
| PAPOLB | 15 (6%) | 233 |
0.253 (1.00) |
0.37 (1.00) |
0.025 (1.00) |
0.589 (1.00) |
0.369 (1.00) |
| SLITRK4 | 17 (7%) | 231 |
0.231 (1.00) |
0.721 (1.00) |
0.0843 (1.00) |
0.794 (1.00) |
0.786 (1.00) |
| TIAL1 | 11 (4%) | 237 |
0.337 (1.00) |
0.475 (1.00) |
0.404 (1.00) |
0.753 (1.00) |
0.732 (1.00) |
| ZCCHC5 | 12 (5%) | 236 |
0.978 (1.00) |
0.0933 (1.00) |
0.0105 (1.00) |
0.552 (1.00) |
0.52 (1.00) |
| ZNF300 | 15 (6%) | 233 |
0.222 (1.00) |
0.338 (1.00) |
0.135 (1.00) |
0.275 (1.00) |
0.766 (1.00) |
| FCER1A | 9 (4%) | 239 |
0.379 (1.00) |
0.77 (1.00) |
0.968 (1.00) |
0.497 (1.00) |
0.709 (1.00) |
| PATE1 | 3 (1%) | 245 |
0.776 (1.00) |
0.443 (1.00) |
0.981 (1.00) |
1 (1.00) |
1 (1.00) |
| PLSCR2 | 9 (4%) | 239 |
0.88 (1.00) |
0.163 (1.00) |
0.00167 (1.00) |
0.722 (1.00) |
0.263 (1.00) |
| ZNF611 | 12 (5%) | 236 |
0.935 (1.00) |
0.262 (1.00) |
0.404 (1.00) |
1 (1.00) |
0.741 (1.00) |
| CFL2 | 7 (3%) | 241 |
0.42 (1.00) |
0.114 (1.00) |
0.294 (1.00) |
0.694 (1.00) |
0.677 (1.00) |
| DEPDC1B | 11 (4%) | 237 |
0.951 (1.00) |
0.951 (1.00) |
0.129 (1.00) |
0.518 (1.00) |
1 (1.00) |
| DCLK1 | 17 (7%) | 231 |
0.166 (1.00) |
0.226 (1.00) |
0.0119 (1.00) |
1 (1.00) |
1 (1.00) |
| CETN3 | 5 (2%) | 243 |
0.396 (1.00) |
0.98 (1.00) |
0.0126 (1.00) |
1 (1.00) |
1 (1.00) |
| MPPED2 | 10 (4%) | 238 |
0.381 (1.00) |
0.297 (1.00) |
0.62 (1.00) |
0.318 (1.00) |
0.732 (1.00) |
| NCAM2 | 20 (8%) | 228 |
0.185 (1.00) |
0.218 (1.00) |
0.0207 (1.00) |
0.808 (1.00) |
0.603 (1.00) |
| NGFRAP1 | 5 (2%) | 243 |
0.652 (1.00) |
0.142 (1.00) |
0.149 (1.00) |
0.663 (1.00) |
1 (1.00) |
| DUSP16 | 11 (4%) | 237 |
0.338 (1.00) |
0.611 (1.00) |
0.286 (1.00) |
0.518 (1.00) |
0.298 (1.00) |
| C9ORF102 | 16 (6%) | 232 |
0.245 (1.00) |
0.35 (1.00) |
0.0343 (1.00) |
0.588 (1.00) |
0.568 (1.00) |
| PPM1N | 7 (3%) | 241 |
0.355 (1.00) |
0.79 (1.00) |
0.294 (1.00) |
1 (1.00) |
1 (1.00) |
| OR2M3 | 10 (4%) | 238 |
0.315 (1.00) |
0.822 (1.00) |
0.707 (1.00) |
0.739 (1.00) |
0.066 (1.00) |
| MNDA | 13 (5%) | 235 |
0.239 (1.00) |
0.29 (1.00) |
0.397 (1.00) |
0.229 (1.00) |
0.756 (1.00) |
| NDUFS1 | 12 (5%) | 236 |
0.361 (1.00) |
0.0218 (1.00) |
0.0105 (1.00) |
0.552 (1.00) |
0.741 (1.00) |
| OR4C12 | 10 (4%) | 238 |
0.292 (1.00) |
0.266 (1.00) |
0.248 (1.00) |
1 (1.00) |
1 (1.00) |
| ATF7IP | 17 (7%) | 231 |
0.207 (1.00) |
0.276 (1.00) |
0.401 (1.00) |
0.432 (1.00) |
0.415 (1.00) |
| FBXO8 | 11 (4%) | 237 |
0.305 (1.00) |
0.628 (1.00) |
0.00488 (1.00) |
1 (1.00) |
0.732 (1.00) |
| NAA30 | 8 (3%) | 240 |
0.565 (1.00) |
0.358 (1.00) |
0.518 (1.00) |
0.719 (1.00) |
0.688 (1.00) |
| PGR | 12 (5%) | 236 |
0.268 (1.00) |
0.228 (1.00) |
0.0105 (1.00) |
1 (1.00) |
0.52 (1.00) |
| ZNF879 | 13 (5%) | 235 |
0.34 (1.00) |
0.0926 (1.00) |
0.4 (1.00) |
1 (1.00) |
1 (1.00) |
| ZNF649 | 14 (6%) | 234 |
0.246 (1.00) |
0.288 (1.00) |
0.182 (1.00) |
0.777 (1.00) |
1 (1.00) |
| CDH12 | 17 (7%) | 231 |
0.666 (1.00) |
0.111 (1.00) |
0.0138 (1.00) |
0.432 (1.00) |
0.786 (1.00) |
| LRRIQ3 | 12 (5%) | 236 |
0.372 (1.00) |
0.00386 (1.00) |
0.00221 (1.00) |
0.229 (1.00) |
0.741 (1.00) |
| ZNF286A | 12 (5%) | 236 |
0.308 (1.00) |
0.518 (1.00) |
0.00262 (1.00) |
0.229 (1.00) |
1 (1.00) |
| C11ORF80 | 9 (4%) | 239 |
0.605 (1.00) |
0.00827 (1.00) |
0.579 (1.00) |
0.171 (1.00) |
1 (1.00) |
| LUC7L2 | 8 (3%) | 240 |
0.332 (1.00) |
0.728 (1.00) |
0.343 (1.00) |
0.27 (1.00) |
0.452 (1.00) |
| EIF2S2 | 9 (4%) | 239 |
0.409 (1.00) |
0.251 (1.00) |
0.265 (1.00) |
0.171 (1.00) |
1 (1.00) |
| HDX | 15 (6%) | 233 |
0.189 (1.00) |
0.483 (1.00) |
0.258 (1.00) |
0.78 (1.00) |
1 (1.00) |
| SERHL2 | 6 (2%) | 242 |
0.464 (1.00) |
0.666 (1.00) |
0.897 (1.00) |
0.185 (1.00) |
0.667 (1.00) |
| EIF4A2 | 8 (3%) | 240 |
0.449 (1.00) |
0.269 (1.00) |
0.205 (1.00) |
0.719 (1.00) |
0.452 (1.00) |
| HAO2 | 10 (4%) | 238 |
0.37 (1.00) |
0.89 (1.00) |
0.138 (1.00) |
0.501 (1.00) |
1 (1.00) |
| MRPS30 | 11 (4%) | 237 |
0.885 (1.00) |
0.605 (1.00) |
0.404 (1.00) |
1 (1.00) |
1 (1.00) |
| NKAIN3 | 8 (3%) | 240 |
0.312 (1.00) |
0.88 (1.00) |
0.343 (1.00) |
0.719 (1.00) |
1 (1.00) |
| CCDC102B | 10 (4%) | 238 |
0.341 (1.00) |
0.259 (1.00) |
0.00363 (1.00) |
1 (1.00) |
0.732 (1.00) |
| PIK3R3 | 8 (3%) | 240 |
0.373 (1.00) |
0.523 (1.00) |
0.000594 (0.896) |
0.27 (1.00) |
0.111 (1.00) |
| PRPF38B | 11 (4%) | 237 |
0.39 (1.00) |
0.482 (1.00) |
0.623 (1.00) |
0.753 (1.00) |
1 (1.00) |
| WAS | 10 (4%) | 238 |
0.392 (1.00) |
0.337 (1.00) |
0.00676 (1.00) |
0.501 (1.00) |
0.732 (1.00) |
| ZC3H13 | 21 (8%) | 227 |
0.148 (1.00) |
0.478 (1.00) |
0.00103 (1.00) |
0.472 (1.00) |
1 (1.00) |
| CR2 | 20 (8%) | 228 |
0.662 (1.00) |
0.342 (1.00) |
0.0961 (1.00) |
1 (1.00) |
0.603 (1.00) |
| PTGFR | 11 (4%) | 237 |
0.288 (1.00) |
0.0241 (1.00) |
0.129 (1.00) |
0.753 (1.00) |
1 (1.00) |
| OR14C36 | 11 (4%) | 237 |
0.286 (1.00) |
0.133 (1.00) |
0.00488 (1.00) |
0.753 (1.00) |
0.732 (1.00) |
| TXNDC8 | 6 (2%) | 242 |
0.439 (1.00) |
0.46 (1.00) |
0.32 (1.00) |
0.667 (1.00) |
1 (1.00) |
| TNFRSF11B | 11 (4%) | 237 |
0.329 (1.00) |
0.398 (1.00) |
0.644 (1.00) |
0.753 (1.00) |
0.499 (1.00) |
| DPP4 | 14 (6%) | 234 |
0.276 (1.00) |
0.0989 (1.00) |
0.02 (1.00) |
1 (1.00) |
1 (1.00) |
| XPA | 7 (3%) | 241 |
0.169 (1.00) |
0.9 (1.00) |
0.981 (1.00) |
0.236 (1.00) |
0.092 (1.00) |
| NAA25 | 17 (7%) | 231 |
0.336 (1.00) |
0.0537 (1.00) |
0.178 (1.00) |
0.794 (1.00) |
0.415 (1.00) |
| CCNE2 | 8 (3%) | 240 |
0.811 (1.00) |
0.215 (1.00) |
0.809 (1.00) |
0.451 (1.00) |
0.0376 (1.00) |
| CXORF65 | 7 (3%) | 241 |
0.548 (1.00) |
0.825 (1.00) |
0.991 (1.00) |
1 (1.00) |
0.396 (1.00) |
| EPS8 | 12 (5%) | 236 |
0.605 (1.00) |
0.121 (1.00) |
0.595 (1.00) |
1 (1.00) |
1 (1.00) |
| PSMC6 | 9 (4%) | 239 |
0.264 (1.00) |
0.863 (1.00) |
0.0573 (1.00) |
0.497 (1.00) |
0.119 (1.00) |
| ZNF367 | 8 (3%) | 240 |
0.449 (1.00) |
0.365 (1.00) |
0.628 (1.00) |
0.127 (1.00) |
1 (1.00) |
| ZNF766 | 12 (5%) | 236 |
0.364 (1.00) |
0.22 (1.00) |
0.404 (1.00) |
0.552 (1.00) |
1 (1.00) |
| ASPN | 9 (4%) | 239 |
0.35 (1.00) |
0.0263 (1.00) |
0.376 (1.00) |
1 (1.00) |
1 (1.00) |
| L1TD1 | 16 (6%) | 232 |
0.261 (1.00) |
0.326 (1.00) |
0.00906 (1.00) |
0.588 (1.00) |
0.568 (1.00) |
| PON1 | 11 (4%) | 237 |
0.303 (1.00) |
0.0454 (1.00) |
0.0722 (1.00) |
0.753 (1.00) |
1 (1.00) |
| SLC20A1 | 11 (4%) | 237 |
0.261 (1.00) |
0.712 (1.00) |
0.404 (1.00) |
0.753 (1.00) |
1 (1.00) |
| GC | 12 (5%) | 236 |
0.339 (1.00) |
0.647 (1.00) |
0.0105 (1.00) |
0.0631 (1.00) |
0.52 (1.00) |
| TMEM225 | 9 (4%) | 239 |
0.333 (1.00) |
0.35 (1.00) |
0.115 (1.00) |
0.497 (1.00) |
0.709 (1.00) |
| RG9MTD3 | 8 (3%) | 240 |
0.362 (1.00) |
0.6 (1.00) |
0.487 (1.00) |
0.27 (1.00) |
0.688 (1.00) |
| GNG4 | 5 (2%) | 243 |
0.552 (1.00) |
0.879 (1.00) |
0.461 (1.00) |
0.168 (1.00) |
0.327 (1.00) |
| GPHB5 | 5 (2%) | 243 |
0.716 (1.00) |
0.123 (1.00) |
0.149 (1.00) |
1 (1.00) |
0.617 (1.00) |
| KLRC4 | 5 (2%) | 243 |
0.636 (1.00) |
0.714 (1.00) |
0.933 (1.00) |
0.342 (1.00) |
0.128 (1.00) |
| OR2B2 | 9 (4%) | 239 |
0.863 (1.00) |
0.564 (1.00) |
0.00167 (1.00) |
1 (1.00) |
0.709 (1.00) |
| OR8K3 | 8 (3%) | 240 |
0.628 (1.00) |
0.151 (1.00) |
0.000594 (0.896) |
0.719 (1.00) |
1 (1.00) |
| LCE2C | 4 (2%) | 244 |
0.632 (1.00) |
0.809 (1.00) |
0.0235 (1.00) |
1 (1.00) |
0.303 (1.00) |
| ADAMTS19 | 19 (8%) | 229 |
0.779 (1.00) |
0.244 (1.00) |
0.0471 (1.00) |
1 (1.00) |
0.789 (1.00) |
| NR3C1 | 14 (6%) | 234 |
0.305 (1.00) |
0.0743 (1.00) |
0.0202 (1.00) |
0.392 (1.00) |
0.539 (1.00) |
| C3AR1 | 8 (3%) | 240 |
0.274 (1.00) |
0.554 (1.00) |
0.0085 (1.00) |
1 (1.00) |
1 (1.00) |
| GIPC2 | 9 (4%) | 239 |
0.851 (1.00) |
0.042 (1.00) |
0.376 (1.00) |
0.497 (1.00) |
0.709 (1.00) |
| NEBL | 18 (7%) | 230 |
0.646 (1.00) |
0.451 (1.00) |
0.389 (1.00) |
0.313 (1.00) |
1 (1.00) |
| ARHGEF6 | 16 (6%) | 232 |
0.4 (1.00) |
0.748 (1.00) |
0.277 (1.00) |
0.79 (1.00) |
1 (1.00) |
| ZNF645 | 10 (4%) | 238 |
0.353 (1.00) |
0.879 (1.00) |
0.0527 (1.00) |
1 (1.00) |
0.293 (1.00) |
| BCHE | 14 (6%) | 234 |
0.237 (1.00) |
0.662 (1.00) |
0.02 (1.00) |
0.392 (1.00) |
0.362 (1.00) |
| OR5H14 | 8 (3%) | 240 |
0.445 (1.00) |
0.116 (1.00) |
0.343 (1.00) |
0.27 (1.00) |
1 (1.00) |
| ACVR1 | 10 (4%) | 238 |
0.405 (1.00) |
0.739 (1.00) |
0.707 (1.00) |
0.318 (1.00) |
0.293 (1.00) |
| CCDC110 | 12 (5%) | 236 |
0.84 (1.00) |
0.409 (1.00) |
0.00877 (1.00) |
0.229 (1.00) |
1 (1.00) |
| OAZ3 | 8 (3%) | 240 |
0.393 (1.00) |
0.219 (1.00) |
0.518 (1.00) |
0.451 (1.00) |
0.688 (1.00) |
| BMP5 | 13 (5%) | 235 |
0.221 (1.00) |
0.167 (1.00) |
0.00507 (1.00) |
0.551 (1.00) |
0.524 (1.00) |
| PLA2G2E | 5 (2%) | 243 |
0.495 (1.00) |
0.307 (1.00) |
0.0343 (1.00) |
0.663 (1.00) |
1 (1.00) |
| CNBD1 | 11 (4%) | 237 |
0.269 (1.00) |
0.0411 (1.00) |
0.644 (1.00) |
0.518 (1.00) |
1 (1.00) |
| ENPP5 | 10 (4%) | 238 |
0.386 (1.00) |
0.181 (1.00) |
0.396 (1.00) |
0.501 (1.00) |
1 (1.00) |
| OAS2 | 15 (6%) | 233 |
0.263 (1.00) |
0.762 (1.00) |
0.666 (1.00) |
0.4 (1.00) |
0.563 (1.00) |
| PPIG | 16 (6%) | 232 |
0.291 (1.00) |
0.167 (1.00) |
0.401 (1.00) |
0.182 (1.00) |
0.568 (1.00) |
| SRD5A2 | 6 (2%) | 242 |
0.562 (1.00) |
0.596 (1.00) |
0.32 (1.00) |
0.667 (1.00) |
0.193 (1.00) |
| ESR1 | 11 (4%) | 237 |
0.329 (1.00) |
0.317 (1.00) |
0.653 (1.00) |
0.518 (1.00) |
0.179 (1.00) |
| OR9G4 | 8 (3%) | 240 |
0.382 (1.00) |
0.859 (1.00) |
0.343 (1.00) |
0.0537 (1.00) |
1 (1.00) |
| FAM133A | 6 (2%) | 242 |
0.562 (1.00) |
0.00857 (1.00) |
0.00195 (1.00) |
0.0971 (1.00) |
1 (1.00) |
| HSD17B11 | 8 (3%) | 240 |
0.19 (1.00) |
0.91 (1.00) |
0.982 (1.00) |
1 (1.00) |
1 (1.00) |
| OR4K1 | 8 (3%) | 240 |
0.401 (1.00) |
0.821 (1.00) |
0.982 (1.00) |
0.451 (1.00) |
1 (1.00) |
| SLC9A11 | 16 (6%) | 232 |
0.867 (1.00) |
0.391 (1.00) |
0.0976 (1.00) |
0.79 (1.00) |
0.773 (1.00) |
| OR2M7 | 8 (3%) | 240 |
0.387 (1.00) |
0.664 (1.00) |
0.518 (1.00) |
1 (1.00) |
0.688 (1.00) |
| C11ORF67 | 5 (2%) | 243 |
0.797 (1.00) |
0.38 (1.00) |
0.933 (1.00) |
0.663 (1.00) |
0.327 (1.00) |
| C20ORF152 | 11 (4%) | 237 |
0.598 (1.00) |
0.923 (1.00) |
0.644 (1.00) |
1 (1.00) |
0.499 (1.00) |
| PPFIA2 | 25 (10%) | 223 |
0.141 (1.00) |
0.52 (1.00) |
0.068 (1.00) |
1 (1.00) |
0.482 (1.00) |
| PHB | 9 (4%) | 239 |
0.392 (1.00) |
0.54 (1.00) |
0.579 (1.00) |
1 (1.00) |
0.709 (1.00) |
| BCL6B | 11 (4%) | 237 |
0.722 (1.00) |
0.788 (1.00) |
0.157 (1.00) |
1 (1.00) |
0.732 (1.00) |
| FIGNL1 | 11 (4%) | 237 |
0.379 (1.00) |
0.0642 (1.00) |
0.00658 (1.00) |
0.34 (1.00) |
1 (1.00) |
| KLK6 | 8 (3%) | 240 |
0.458 (1.00) |
0.5 (1.00) |
0.487 (1.00) |
1 (1.00) |
1 (1.00) |
| SLC48A1 | 5 (2%) | 243 |
0.601 (1.00) |
0.645 (1.00) |
0.461 (1.00) |
0.663 (1.00) |
1 (1.00) |
| CNTN6 | 21 (8%) | 227 |
0.177 (1.00) |
0.624 (1.00) |
0.0435 (1.00) |
0.638 (1.00) |
0.803 (1.00) |
| OR4C13 | 8 (3%) | 240 |
0.492 (1.00) |
0.178 (1.00) |
0.343 (1.00) |
0.719 (1.00) |
1 (1.00) |
| POTEF | 17 (7%) | 231 |
0.759 (1.00) |
0.693 (1.00) |
0.0255 (1.00) |
1 (1.00) |
1 (1.00) |
| GNPTAB | 20 (8%) | 228 |
0.219 (1.00) |
0.308 (1.00) |
0.125 (1.00) |
0.465 (1.00) |
0.603 (1.00) |
| CDH7 | 19 (8%) | 229 |
0.179 (1.00) |
0.86 (1.00) |
0.0394 (1.00) |
0.805 (1.00) |
0.604 (1.00) |
| DAPP1 | 8 (3%) | 240 |
0.568 (1.00) |
0.462 (1.00) |
0.343 (1.00) |
0.719 (1.00) |
0.688 (1.00) |
| DCBLD2 | 13 (5%) | 235 |
0.856 (1.00) |
0.112 (1.00) |
0.946 (1.00) |
0.0667 (1.00) |
0.756 (1.00) |
| LEF1 | 9 (4%) | 239 |
0.617 (1.00) |
0.0331 (1.00) |
0.759 (1.00) |
0.281 (1.00) |
1 (1.00) |
| LPL | 12 (5%) | 236 |
0.359 (1.00) |
0.126 (1.00) |
0.595 (1.00) |
1 (1.00) |
0.319 (1.00) |
| PPP1R3A | 22 (9%) | 226 |
0.151 (1.00) |
0.198 (1.00) |
0.179 (1.00) |
0.818 (1.00) |
0.803 (1.00) |
| BRDT | 14 (6%) | 234 |
0.275 (1.00) |
0.135 (1.00) |
0.0043 (1.00) |
0.392 (1.00) |
0.764 (1.00) |
| MS4A1 | 6 (2%) | 242 |
0.587 (1.00) |
0.59 (1.00) |
0.897 (1.00) |
0.019 (1.00) |
1 (1.00) |
| ZBBX | 12 (5%) | 236 |
0.319 (1.00) |
0.0217 (1.00) |
0.0105 (1.00) |
0.756 (1.00) |
0.52 (1.00) |
| CACNB4 | 11 (4%) | 237 |
0.416 (1.00) |
0.368 (1.00) |
0.404 (1.00) |
0.34 (1.00) |
0.499 (1.00) |
| FAM172A | 10 (4%) | 238 |
0.392 (1.00) |
0.395 (1.00) |
0.00676 (1.00) |
0.739 (1.00) |
0.732 (1.00) |
| OR5H6 | 7 (3%) | 241 |
0.356 (1.00) |
0.204 (1.00) |
0.294 (1.00) |
0.694 (1.00) |
0.677 (1.00) |
| RUNX1T1 | 15 (6%) | 233 |
0.279 (1.00) |
0.286 (1.00) |
0.025 (1.00) |
0.78 (1.00) |
0.369 (1.00) |
| POLDIP2 | 7 (3%) | 241 |
0.442 (1.00) |
0.249 (1.00) |
0.601 (1.00) |
0.428 (1.00) |
0.396 (1.00) |
| ABHD4 | 10 (4%) | 238 |
0.576 (1.00) |
0.437 (1.00) |
0.396 (1.00) |
0.739 (1.00) |
0.293 (1.00) |
| ATP6V1C2 | 12 (5%) | 236 |
0.314 (1.00) |
0.0315 (1.00) |
0.00877 (1.00) |
1 (1.00) |
0.52 (1.00) |
| C13ORF33 | 7 (3%) | 241 |
0.458 (1.00) |
0.751 (1.00) |
0.856 (1.00) |
1 (1.00) |
1 (1.00) |
| CASC4 | 8 (3%) | 240 |
0.561 (1.00) |
0.251 (1.00) |
0.518 (1.00) |
0.719 (1.00) |
1 (1.00) |
| EXOSC9 | 10 (4%) | 238 |
0.368 (1.00) |
0.893 (1.00) |
0.396 (1.00) |
1 (1.00) |
0.732 (1.00) |
| FATE1 | 6 (2%) | 242 |
0.607 (1.00) |
0.961 (1.00) |
0.32 (1.00) |
0.415 (1.00) |
0.667 (1.00) |
| FRA10AC1 | 7 (3%) | 241 |
0.334 (1.00) |
0.866 (1.00) |
0.294 (1.00) |
0.694 (1.00) |
1 (1.00) |
| HIST1H2AG | 7 (3%) | 241 |
0.542 (1.00) |
0.416 (1.00) |
0.991 (1.00) |
0.0485 (1.00) |
1 (1.00) |
| ITM2B | 6 (2%) | 242 |
0.52 (1.00) |
0.688 (1.00) |
0.897 (1.00) |
1 (1.00) |
1 (1.00) |
| LINGO2 | 12 (5%) | 236 |
0.288 (1.00) |
0.0178 (1.00) |
0.0209 (1.00) |
0.229 (1.00) |
0.52 (1.00) |
| SARNP | 5 (2%) | 243 |
0.496 (1.00) |
0.656 (1.00) |
0.461 (1.00) |
0.663 (1.00) |
1 (1.00) |
| SLK | 18 (7%) | 230 |
0.19 (1.00) |
0.0952 (1.00) |
0.0538 (1.00) |
0.616 (1.00) |
1 (1.00) |
| SPINK7 | 5 (2%) | 243 |
0.514 (1.00) |
0.153 (1.00) |
0.933 (1.00) |
0.663 (1.00) |
1 (1.00) |
| TBC1D15 | 12 (5%) | 236 |
0.262 (1.00) |
0.234 (1.00) |
0.166 (1.00) |
1 (1.00) |
0.52 (1.00) |
| CSGALNACT1 | 13 (5%) | 235 |
0.258 (1.00) |
0.997 (1.00) |
0.102 (1.00) |
0.551 (1.00) |
1 (1.00) |
| TECRL | 9 (4%) | 239 |
0.363 (1.00) |
0.556 (1.00) |
0.987 (1.00) |
0.281 (1.00) |
0.709 (1.00) |
| ZBTB6 | 8 (3%) | 240 |
0.412 (1.00) |
0.398 (1.00) |
0.0873 (1.00) |
0.451 (1.00) |
0.688 (1.00) |
| SLMO1 | 6 (2%) | 242 |
0.429 (1.00) |
0.328 (1.00) |
0.228 (1.00) |
1 (1.00) |
1 (1.00) |
| PGBD2 | 13 (5%) | 235 |
0.261 (1.00) |
0.755 (1.00) |
0.4 (1.00) |
0.378 (1.00) |
1 (1.00) |
| GCET2 | 6 (2%) | 242 |
0.542 (1.00) |
0.698 (1.00) |
0.897 (1.00) |
0.415 (1.00) |
0.667 (1.00) |
| SLC16A7 | 12 (5%) | 236 |
0.287 (1.00) |
0.595 (1.00) |
0.0914 (1.00) |
0.756 (1.00) |
1 (1.00) |
| HINT3 | 4 (2%) | 244 |
0.67 (1.00) |
0.518 (1.00) |
0.328 (1.00) |
1 (1.00) |
0.577 (1.00) |
| BRCC3 | 8 (3%) | 240 |
0.716 (1.00) |
0.559 (1.00) |
0.000594 (0.896) |
0.719 (1.00) |
0.452 (1.00) |
| DYRK1A | 12 (5%) | 236 |
0.939 (1.00) |
0.386 (1.00) |
0.595 (1.00) |
0.552 (1.00) |
1 (1.00) |
| DUXA | 7 (3%) | 241 |
0.426 (1.00) |
0.41 (1.00) |
0.294 (1.00) |
0.694 (1.00) |
0.677 (1.00) |
| FRMD3 | 12 (5%) | 236 |
0.273 (1.00) |
0.479 (1.00) |
0.00139 (1.00) |
1 (1.00) |
1 (1.00) |
| G6PC2 | 10 (4%) | 238 |
0.306 (1.00) |
0.859 (1.00) |
0.0913 (1.00) |
1 (1.00) |
0.293 (1.00) |
| MIPOL1 | 11 (4%) | 237 |
0.308 (1.00) |
0.291 (1.00) |
0.404 (1.00) |
0.753 (1.00) |
0.499 (1.00) |
| MMP10 | 10 (4%) | 238 |
0.479 (1.00) |
0.573 (1.00) |
0.00231 (1.00) |
1 (1.00) |
0.066 (1.00) |
| ORAI1 | 7 (3%) | 241 |
0.439 (1.00) |
0.578 (1.00) |
0.432 (1.00) |
0.236 (1.00) |
0.396 (1.00) |
| RAB3GAP1 | 16 (6%) | 232 |
0.98 (1.00) |
0.596 (1.00) |
0.35 (1.00) |
1 (1.00) |
0.773 (1.00) |
| PSIP1 | 12 (5%) | 236 |
0.145 (1.00) |
0.0166 (1.00) |
0.016 (1.00) |
0.229 (1.00) |
0.741 (1.00) |
| RRAS2 | 4 (2%) | 244 |
0.752 (1.00) |
0.0717 (1.00) |
0.961 (1.00) |
0.608 (1.00) |
0.0639 (1.00) |
| CEP290 | 26 (10%) | 222 |
0.465 (1.00) |
0.5 (1.00) |
0.108 (1.00) |
0.514 (1.00) |
0.17 (1.00) |
| SEPT10 | 8 (3%) | 240 |
0.383 (1.00) |
0.545 (1.00) |
0.343 (1.00) |
0.27 (1.00) |
0.111 (1.00) |
| ZFP90 | 12 (5%) | 236 |
0.28 (1.00) |
0.265 (1.00) |
0.0105 (1.00) |
1 (1.00) |
1 (1.00) |
| NOL4 | 23 (9%) | 225 |
0.421 (1.00) |
0.218 (1.00) |
0.247 (1.00) |
1 (1.00) |
0.629 (1.00) |
| OR51G1 | 9 (4%) | 239 |
0.463 (1.00) |
0.215 (1.00) |
0.00296 (1.00) |
0.497 (1.00) |
1 (1.00) |
| VBP1 | 7 (3%) | 241 |
0.558 (1.00) |
0.0833 (1.00) |
0.856 (1.00) |
0.694 (1.00) |
1 (1.00) |
P value = 1.04e-05 (Chi-square test), Q value = 0.016
Table S1. Gene #3: 'CTNNB1 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| CTNNB1 MUTATED | 69 | 1 | 2 | 2 | 0 | 0 | 0 |
| CTNNB1 WILD-TYPE | 113 | 2 | 6 | 0 | 5 | 4 | 44 |
Figure S1. Get High-res Image Gene #3: 'CTNNB1 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'

P value = 5.44e-19 (Chi-square test), Q value = 8.3e-16
Table S2. Gene #9: 'PTEN MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| PTEN MUTATED | 146 | 2 | 6 | 1 | 4 | 1 | 1 |
| PTEN WILD-TYPE | 36 | 1 | 2 | 1 | 1 | 3 | 43 |
Figure S2. Get High-res Image Gene #9: 'PTEN MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'

P value = 6.15e-06 (t-test), Q value = 0.0094
Table S3. Gene #10: 'TP53 MUTATION STATUS' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 248 | 63.1 (11.1) |
| TP53 MUTATED | 69 | 67.9 (9.4) |
| TP53 WILD-TYPE | 179 | 61.3 (11.2) |
Figure S3. Get High-res Image Gene #10: 'TP53 MUTATION STATUS' versus Clinical Feature #2: 'AGE'

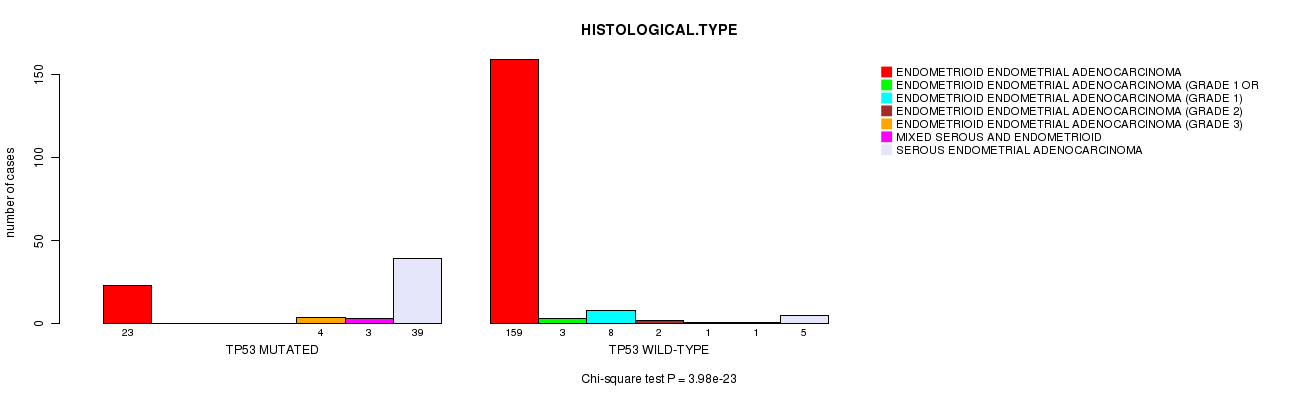
P value = 3.98e-23 (Chi-square test), Q value = 6.1e-20
Table S4. Gene #10: 'TP53 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| TP53 MUTATED | 23 | 0 | 0 | 0 | 4 | 3 | 39 |
| TP53 WILD-TYPE | 159 | 3 | 8 | 2 | 1 | 1 | 5 |
Figure S4. Get High-res Image Gene #10: 'TP53 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
P value = 6.08e-05 (Fisher's exact test), Q value = 0.092
Table S5. Gene #10: 'TP53 MUTATION STATUS' versus Clinical Feature #5: 'NEOADJUVANT.THERAPY'
| nPatients | NO | YES |
|---|---|---|
| ALL | 68 | 180 |
| TP53 MUTATED | 32 | 37 |
| TP53 WILD-TYPE | 36 | 143 |
Figure S5. Get High-res Image Gene #10: 'TP53 MUTATION STATUS' versus Clinical Feature #5: 'NEOADJUVANT.THERAPY'

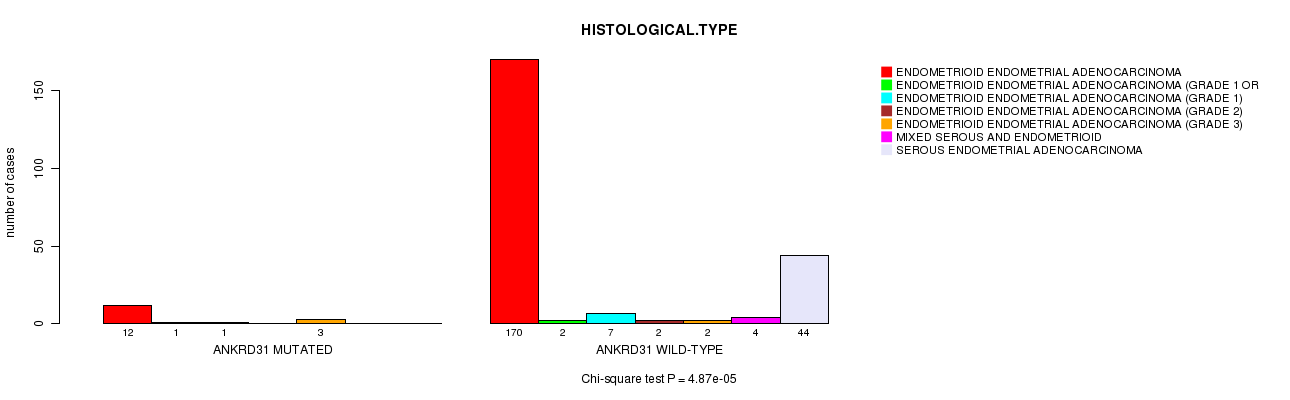
P value = 4.87e-05 (Chi-square test), Q value = 0.074
Table S6. Gene #28: 'ANKRD31 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| ANKRD31 MUTATED | 12 | 1 | 1 | 0 | 3 | 0 | 0 |
| ANKRD31 WILD-TYPE | 170 | 2 | 7 | 2 | 2 | 4 | 44 |
Figure S6. Get High-res Image Gene #28: 'ANKRD31 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
P value = 2.07e-05 (Chi-square test), Q value = 0.031
Table S7. Gene #188: 'SH3BGRL MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| SH3BGRL MUTATED | 4 | 0 | 0 | 0 | 2 | 0 | 0 |
| SH3BGRL WILD-TYPE | 178 | 3 | 8 | 2 | 3 | 4 | 44 |
Figure S7. Get High-res Image Gene #188: 'SH3BGRL MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'

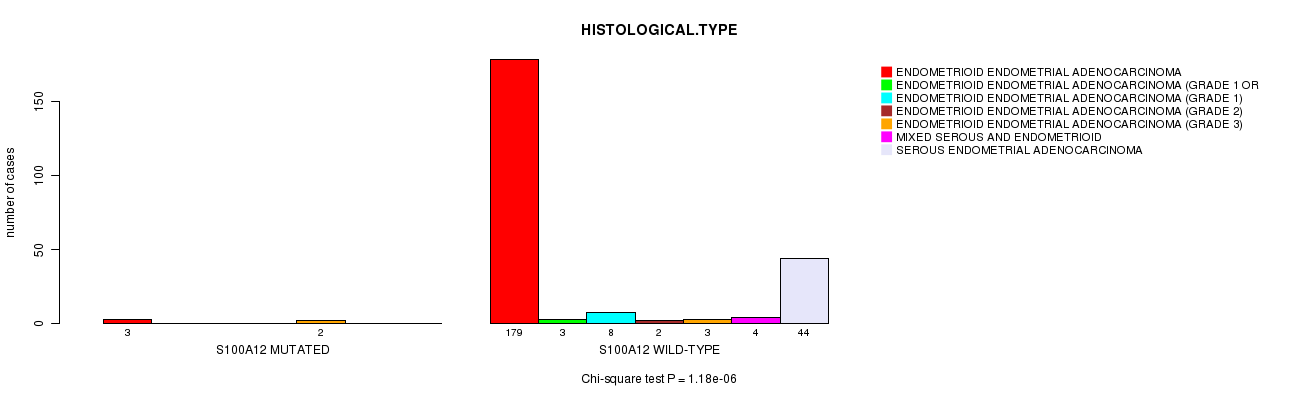
P value = 1.18e-06 (Chi-square test), Q value = 0.0018
Table S8. Gene #207: 'S100A12 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| S100A12 MUTATED | 3 | 0 | 0 | 0 | 2 | 0 | 0 |
| S100A12 WILD-TYPE | 179 | 3 | 8 | 2 | 3 | 4 | 44 |
Figure S8. Get High-res Image Gene #207: 'S100A12 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
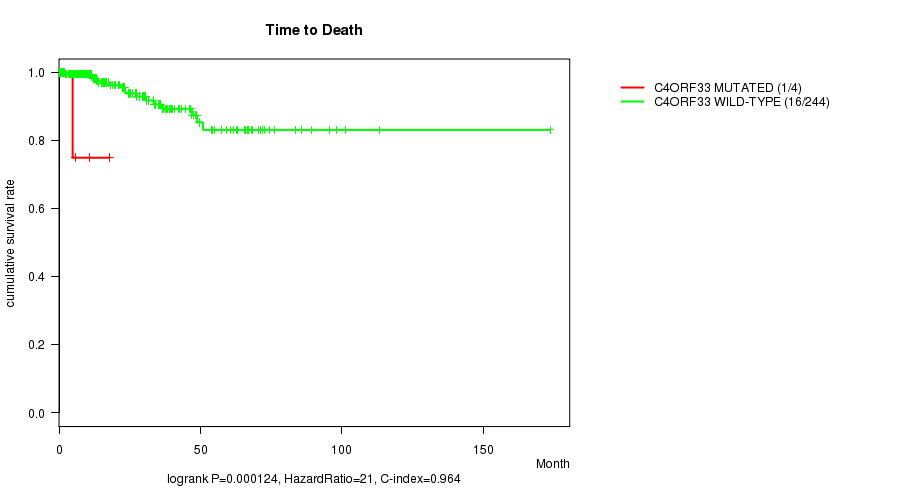
P value = 0.000124 (logrank test), Q value = 0.19
Table S9. Gene #223: 'C4ORF33 MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 248 | 17 | 0.0 - 173.6 (22.2) |
| C4ORF33 MUTATED | 4 | 1 | 4.8 - 17.8 (8.2) |
| C4ORF33 WILD-TYPE | 244 | 16 | 0.0 - 173.6 (22.5) |
Figure S9. Get High-res Image Gene #223: 'C4ORF33 MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
P value = 0.000147 (Chi-square test), Q value = 0.22
Table S10. Gene #226: 'GLRX3 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| GLRX3 MUTATED | 5 | 0 | 0 | 0 | 2 | 0 | 0 |
| GLRX3 WILD-TYPE | 177 | 3 | 8 | 2 | 3 | 4 | 44 |
Figure S10. Get High-res Image Gene #226: 'GLRX3 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'

P value = 3.06e-05 (Chi-square test), Q value = 0.046
Table S11. Gene #248: 'SLC16A10 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| SLC16A10 MUTATED | 5 | 1 | 1 | 0 | 2 | 0 | 0 |
| SLC16A10 WILD-TYPE | 177 | 2 | 7 | 2 | 3 | 4 | 44 |
Figure S11. Get High-res Image Gene #248: 'SLC16A10 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'

P value = 1.45e-05 (Chi-square test), Q value = 0.022
Table S12. Gene #258: 'SLC10A5 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| SLC10A5 MUTATED | 3 | 1 | 2 | 0 | 1 | 0 | 0 |
| SLC10A5 WILD-TYPE | 179 | 2 | 6 | 2 | 4 | 4 | 44 |
Figure S12. Get High-res Image Gene #258: 'SLC10A5 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
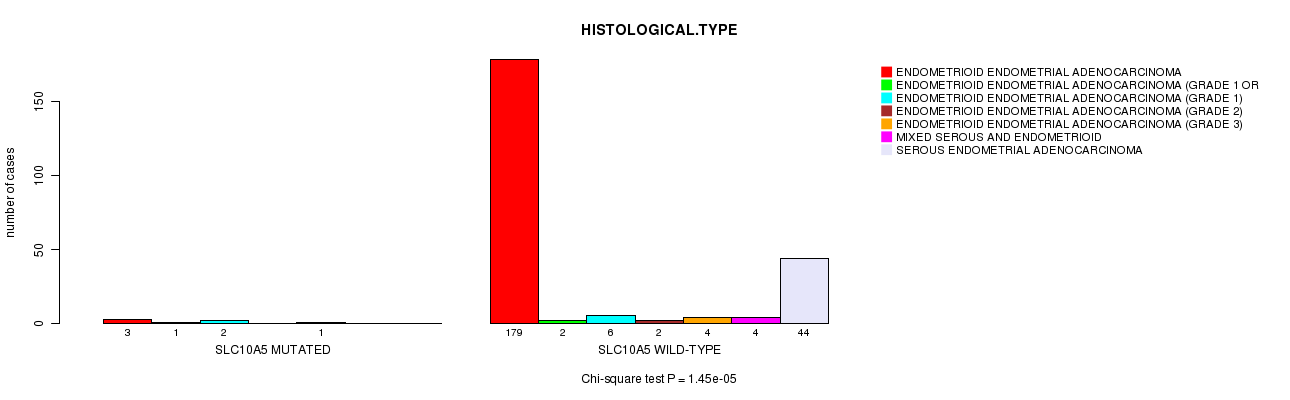
P value = 0.000147 (Chi-square test), Q value = 0.22
Table S13. Gene #261: 'CDKN1B MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| CDKN1B MUTATED | 5 | 0 | 0 | 0 | 2 | 0 | 0 |
| CDKN1B WILD-TYPE | 177 | 3 | 8 | 2 | 3 | 4 | 44 |
Figure S13. Get High-res Image Gene #261: 'CDKN1B MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'

P value = 2.07e-05 (Chi-square test), Q value = 0.031
Table S14. Gene #301: 'ALKBH6 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1 OR 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 1) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 2) | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA (GRADE 3) | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|---|---|---|---|
| ALL | 182 | 3 | 8 | 2 | 5 | 4 | 44 |
| ALKBH6 MUTATED | 4 | 0 | 0 | 0 | 2 | 0 | 0 |
| ALKBH6 WILD-TYPE | 178 | 3 | 8 | 2 | 3 | 4 | 44 |
Figure S14. Get High-res Image Gene #301: 'ALKBH6 MUTATION STATUS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
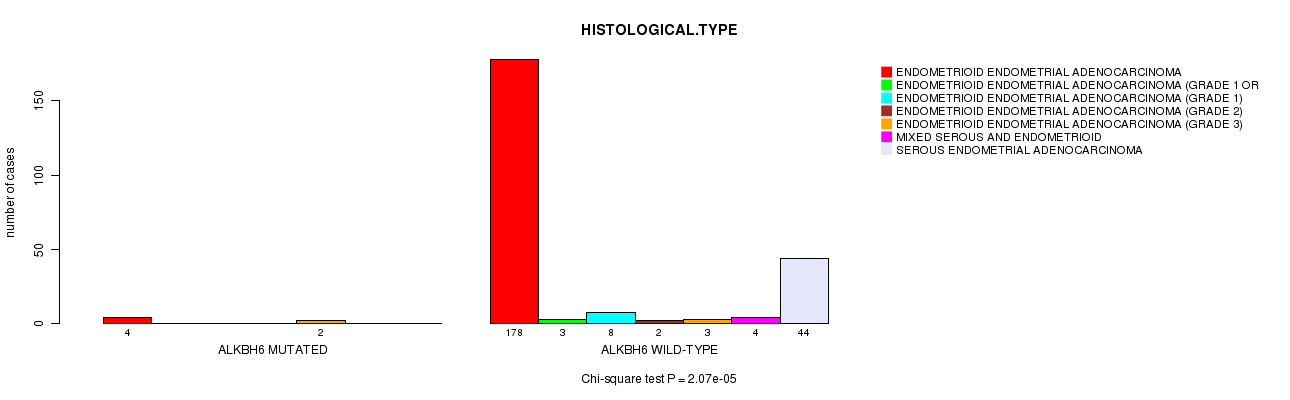
-
Mutation data file = UCEC.mutsig.cluster.txt
-
Clinical data file = UCEC.clin.merged.picked.txt
-
Number of patients = 248
-
Number of significantly mutated genes = 305
-
Number of selected clinical features = 5
-
Exclude genes that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For continuous numerical clinical features, two-tailed Student's t test with unequal variance (Lehmann and Romano 2005) was applied to compare the clinical values between tumors with and without gene mutations using 't.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.