This pipeline computes the correlation between significantly recurrent gene mutations and selected clinical features.
Testing the association between mutation status of 50 genes and 9 clinical features across 293 patients, 2 significant findings detected with Q value < 0.25.
-
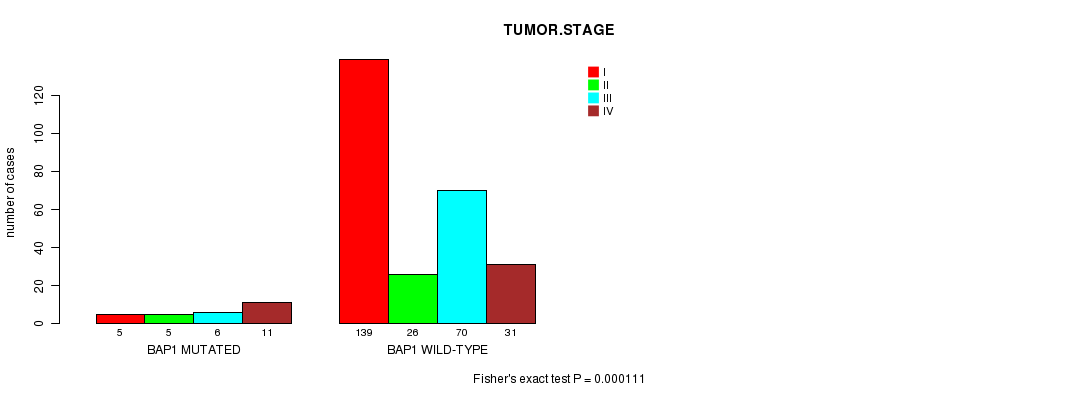
BAP1 mutation correlated to 'TUMOR.STAGE'.
-
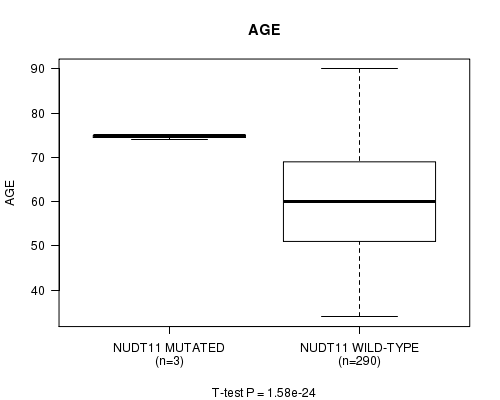
NUDT11 mutation correlated to 'AGE'.
Table 1. Get Full Table Overview of the association between mutation status of 50 genes and 9 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 2 significant findings detected.
|
Clinical Features |
Time to Death |
AGE | GENDER |
KARNOFSKY PERFORMANCE SCORE |
PATHOLOGY T |
PATHOLOGY N |
PATHOLOGICSPREAD(M) |
TUMOR STAGE |
NEOADJUVANT THERAPY |
||
| nMutated (%) | nWild-Type | logrank test | t-test | Fisher's exact test | t-test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| BAP1 | 27 (9%) | 266 |
0.0131 (1.00) |
0.828 (1.00) |
0.00961 (1.00) |
0.00295 (1.00) |
0.627 (1.00) |
0.000895 (0.355) |
0.000111 (0.044) |
0.253 (1.00) |
|
| NUDT11 | 3 (1%) | 290 |
0.6 (1.00) |
1.58e-24 (6.29e-22) |
0.554 (1.00) |
0.718 (1.00) |
1 (1.00) |
0.479 (1.00) |
1 (1.00) |
||
| SETD2 | 32 (11%) | 261 |
0.949 (1.00) |
0.11 (1.00) |
0.244 (1.00) |
0.229 (1.00) |
0.605 (1.00) |
0.0519 (1.00) |
0.184 (1.00) |
0.294 (1.00) |
|
| SV2C | 3 (1%) | 290 |
0.731 (1.00) |
0.0205 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
|
| TOR1A | 3 (1%) | 290 |
0.0532 (1.00) |
0.704 (1.00) |
1 (1.00) |
1 (1.00) |
0.0753 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
|
| VHL | 146 (50%) | 147 |
0.82 (1.00) |
0.0114 (1.00) |
0.0868 (1.00) |
0.359 (1.00) |
0.438 (1.00) |
0.76 (1.00) |
0.731 (1.00) |
0.509 (1.00) |
1 (1.00) |
| PBRM1 | 98 (33%) | 195 |
0.444 (1.00) |
0.339 (1.00) |
0.0698 (1.00) |
0.35 (1.00) |
0.731 (1.00) |
1 (1.00) |
0.201 (1.00) |
0.218 (1.00) |
0.26 (1.00) |
| KDM5C | 18 (6%) | 275 |
0.0803 (1.00) |
0.206 (1.00) |
0.00486 (1.00) |
0.622 (1.00) |
0.516 (1.00) |
1 (1.00) |
0.856 (1.00) |
1 (1.00) |
|
| PTEN | 15 (5%) | 278 |
0.87 (1.00) |
0.104 (1.00) |
1 (1.00) |
0.489 (1.00) |
0.0657 (1.00) |
1 (1.00) |
0.592 (1.00) |
0.146 (1.00) |
|
| STAG3L2 | 6 (2%) | 287 |
0.252 (1.00) |
0.115 (1.00) |
1 (1.00) |
0.415 (1.00) |
1 (1.00) |
0.579 (1.00) |
0.334 (1.00) |
1 (1.00) |
|
| PIK3CA | 14 (5%) | 279 |
0.797 (1.00) |
0.387 (1.00) |
0.57 (1.00) |
0.152 (1.00) |
0.555 (1.00) |
1 (1.00) |
0.251 (1.00) |
1 (1.00) |
|
| MTOR | 24 (8%) | 269 |
0.123 (1.00) |
0.203 (1.00) |
0.119 (1.00) |
0.298 (1.00) |
0.0602 (1.00) |
1 (1.00) |
0.346 (1.00) |
1 (1.00) |
|
| EBPL | 6 (2%) | 287 |
0.48 (1.00) |
0.527 (1.00) |
0.00161 (0.637) |
0.117 (1.00) |
1 (1.00) |
1 (1.00) |
0.0646 (1.00) |
1 (1.00) |
|
| FAM174B | 3 (1%) | 290 |
0.476 (1.00) |
0.668 (1.00) |
1 (1.00) |
0.0917 (1.00) |
1 (1.00) |
0.0724 (1.00) |
1 (1.00) |
||
| VCX2 | 3 (1%) | 290 |
0.293 (1.00) |
0.058 (1.00) |
1 (1.00) |
0.143 (1.00) |
1 (1.00) |
0.35 (1.00) |
0.117 (1.00) |
1 (1.00) |
|
| RRAD | 4 (1%) | 289 |
0.0836 (1.00) |
0.806 (1.00) |
0.123 (1.00) |
0.0613 (1.00) |
0.211 (1.00) |
0.437 (1.00) |
0.0702 (1.00) |
1 (1.00) |
|
| ANKRD36 | 10 (3%) | 283 |
0.284 (1.00) |
0.339 (1.00) |
1 (1.00) |
0.391 (1.00) |
1 (1.00) |
1 (1.00) |
0.637 (1.00) |
1 (1.00) |
|
| KANK3 | 6 (2%) | 287 |
0.733 (1.00) |
0.513 (1.00) |
1 (1.00) |
0.51 (1.00) |
1 (1.00) |
1 (1.00) |
0.696 (1.00) |
1 (1.00) |
|
| KRTAP1-1 | 3 (1%) | 290 |
0.296 (1.00) |
0.97 (1.00) |
0.0414 (1.00) |
0.0917 (1.00) |
0.35 (1.00) |
0.146 (1.00) |
1 (1.00) |
||
| UQCRFS1 | 3 (1%) | 290 |
0.321 (1.00) |
0.518 (1.00) |
0.278 (1.00) |
0.0452 (1.00) |
1 (1.00) |
1 (1.00) |
0.0554 (1.00) |
1 (1.00) |
|
| ZCCHC3 | 3 (1%) | 290 |
0.392 (1.00) |
0.829 (1.00) |
0.554 (1.00) |
0.374 (1.00) |
1 (1.00) |
1 (1.00) |
0.811 (1.00) |
1 (1.00) |
|
| MADCAM1 | 4 (1%) | 289 |
0.357 (1.00) |
0.751 (1.00) |
0.123 (1.00) |
0.0204 (1.00) |
1 (1.00) |
0.0871 (1.00) |
0.00475 (1.00) |
1 (1.00) |
|
| CR1 | 11 (4%) | 282 |
0.582 (1.00) |
0.936 (1.00) |
0.523 (1.00) |
0.831 (1.00) |
1 (1.00) |
0.645 (1.00) |
0.692 (1.00) |
1 (1.00) |
|
| DACH2 | 7 (2%) | 286 |
0.972 (1.00) |
0.7 (1.00) |
0.242 (1.00) |
1 (1.00) |
0.328 (1.00) |
0.236 (1.00) |
0.631 (1.00) |
0.0702 (1.00) |
|
| NBPF10 | 20 (7%) | 273 |
0.237 (1.00) |
0.321 (1.00) |
0.151 (1.00) |
0.202 (1.00) |
0.139 (1.00) |
0.0353 (1.00) |
0.134 (1.00) |
1 (1.00) |
|
| OR5H1 | 4 (1%) | 289 |
0.919 (1.00) |
0.524 (1.00) |
0.612 (1.00) |
0.606 (1.00) |
1 (1.00) |
1 (1.00) |
0.67 (1.00) |
1 (1.00) |
|
| TSPAN19 | 4 (1%) | 289 |
0.319 (1.00) |
0.467 (1.00) |
1 (1.00) |
0.458 (1.00) |
1 (1.00) |
0.437 (1.00) |
0.223 (1.00) |
1 (1.00) |
|
| WDR52 | 9 (3%) | 284 |
0.967 (1.00) |
0.395 (1.00) |
0.724 (1.00) |
0.647 (1.00) |
1 (1.00) |
0.342 (1.00) |
0.842 (1.00) |
1 (1.00) |
|
| KRT1 | 5 (2%) | 288 |
0.00506 (1.00) |
0.0322 (1.00) |
0.346 (1.00) |
0.172 (1.00) |
1 (1.00) |
1 (1.00) |
0.072 (1.00) |
1 (1.00) |
|
| MSN | 4 (1%) | 289 |
0.63 (1.00) |
0.578 (1.00) |
1 (1.00) |
0.19 (1.00) |
1 (1.00) |
1 (1.00) |
0.464 (1.00) |
1 (1.00) |
|
| PCDHGA8 | 4 (1%) | 289 |
0.251 (1.00) |
0.207 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.768 (1.00) |
1 (1.00) |
|
| ABCB1 | 8 (3%) | 285 |
0.116 (1.00) |
0.582 (1.00) |
0.718 (1.00) |
0.882 (1.00) |
1 (1.00) |
0.603 (1.00) |
0.784 (1.00) |
1 (1.00) |
|
| BAGE2 | 4 (1%) | 289 |
0.961 (1.00) |
0.667 (1.00) |
0.612 (1.00) |
0.796 (1.00) |
1 (1.00) |
1 (1.00) |
0.589 (1.00) |
1 (1.00) |
|
| POTEC | 10 (3%) | 283 |
0.386 (1.00) |
0.0352 (1.00) |
1 (1.00) |
0.81 (1.00) |
1 (1.00) |
1 (1.00) |
0.753 (1.00) |
1 (1.00) |
|
| CNTNAP4 | 9 (3%) | 284 |
0.485 (1.00) |
0.5 (1.00) |
0.724 (1.00) |
0.51 (1.00) |
1 (1.00) |
1 (1.00) |
0.513 (1.00) |
1 (1.00) |
|
| NPNT | 6 (2%) | 287 |
0.0806 (1.00) |
0.222 (1.00) |
0.668 (1.00) |
0.342 (1.00) |
1 (1.00) |
1 (1.00) |
0.219 (1.00) |
1 (1.00) |
|
| CLIC6 | 3 (1%) | 290 |
0.364 (1.00) |
0.379 (1.00) |
0.278 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
|
| SPAM1 | 5 (2%) | 288 |
0.27 (1.00) |
0.192 (1.00) |
0.661 (1.00) |
0.096 (1.00) |
1 (1.00) |
1 (1.00) |
0.288 (1.00) |
1 (1.00) |
|
| TP53 | 9 (3%) | 284 |
0.0454 (1.00) |
0.091 (1.00) |
0.285 (1.00) |
0.032 (1.00) |
0.328 (1.00) |
1 (1.00) |
0.0461 (1.00) |
1 (1.00) |
|
| NFE2L2 | 5 (2%) | 288 |
0.163 (1.00) |
0.162 (1.00) |
1 (1.00) |
0.827 (1.00) |
1 (1.00) |
0.513 (1.00) |
0.619 (1.00) |
1 (1.00) |
|
| SLC2A14 | 3 (1%) | 290 |
0.713 (1.00) |
0.897 (1.00) |
1 (1.00) |
0.519 (1.00) |
1 (1.00) |
1 (1.00) |
0.38 (1.00) |
1 (1.00) |
|
| TPTE2 | 7 (2%) | 286 |
0.3 (1.00) |
0.309 (1.00) |
1 (1.00) |
0.48 (1.00) |
1 (1.00) |
0.599 (1.00) |
0.596 (1.00) |
1 (1.00) |
|
| ADCY8 | 5 (2%) | 288 |
0.706 (1.00) |
0.582 (1.00) |
0.167 (1.00) |
0.262 (1.00) |
0.0753 (1.00) |
0.513 (1.00) |
0.0946 (1.00) |
1 (1.00) |
|
| CCNB2 | 5 (2%) | 288 |
0.784 (1.00) |
0.459 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.911 (1.00) |
1 (1.00) |
|
| LRP11 | 3 (1%) | 290 |
0.282 (1.00) |
0.0434 (1.00) |
1 (1.00) |
0.374 (1.00) |
1 (1.00) |
0.811 (1.00) |
1 (1.00) |
||
| PTPN18 | 4 (1%) | 289 |
0.416 (1.00) |
0.349 (1.00) |
1 (1.00) |
0.258 (1.00) |
1 (1.00) |
1 (1.00) |
0.407 (1.00) |
1 (1.00) |
|
| LPAR4 | 4 (1%) | 289 |
0.534 (1.00) |
0.457 (1.00) |
0.612 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.768 (1.00) |
1 (1.00) |
|
| POLDIP2 | 4 (1%) | 289 |
0.458 (1.00) |
0.705 (1.00) |
1 (1.00) |
0.458 (1.00) |
1 (1.00) |
0.437 (1.00) |
0.357 (1.00) |
1 (1.00) |
|
| ZNF800 | 6 (2%) | 287 |
0.166 (1.00) |
0.599 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.579 (1.00) |
0.806 (1.00) |
1 (1.00) |
|
| SP8 | 3 (1%) | 290 |
0.252 (1.00) |
0.0518 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
P value = 0.000111 (Fisher's exact test), Q value = 0.044
Table S1. Gene #6: 'BAP1 MUTATION STATUS' versus Clinical Feature #8: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 144 | 31 | 76 | 42 |
| BAP1 MUTATED | 5 | 5 | 6 | 11 |
| BAP1 WILD-TYPE | 139 | 26 | 70 | 31 |
Figure S1. Get High-res Image Gene #6: 'BAP1 MUTATION STATUS' versus Clinical Feature #8: 'TUMOR.STAGE'
P value = 1.58e-24 (t-test), Q value = 6.3e-22
Table S2. Gene #14: 'NUDT11 MUTATION STATUS' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 293 | 60.3 (11.9) |
| NUDT11 MUTATED | 3 | 74.7 (0.6) |
| NUDT11 WILD-TYPE | 290 | 60.2 (11.9) |
Figure S2. Get High-res Image Gene #14: 'NUDT11 MUTATION STATUS' versus Clinical Feature #2: 'AGE'
-
Mutation data file = KIRC.mutsig.cluster.txt
-
Clinical data file = KIRC.clin.merged.picked.txt
-
Number of patients = 293
-
Number of significantly mutated genes = 50
-
Number of selected clinical features = 9
-
Exclude genes that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For continuous numerical clinical features, two-tailed Student's t test with unequal variance (Lehmann and Romano 2005) was applied to compare the clinical values between tumors with and without gene mutations using 't.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.