This pipeline computes the correlation between cancer subtypes identified by different molecular patterns and selected clinical features.
Testing the association between subtypes identified by 8 different clustering approaches and 3 clinical features across 138 patients, no significant finding detected with P value < 0.05.
-
4 subtypes identified in current cancer cohort by 'CN CNMF'. These subtypes do not correlate to any clinical features.
-
3 subtypes identified in current cancer cohort by 'METHLYATION CNMF'. These subtypes do not correlate to any clinical features.
-
CNMF clustering analysis on RPPA data identified 3 subtypes that do not correlate to any clinical features.
-
Consensus hierarchical clustering analysis on RPPA data identified 3 subtypes that do not correlate to any clinical features.
-
CNMF clustering analysis on sequencing-based mRNA expression data identified 3 subtypes that do not correlate to any clinical features.
-
Consensus hierarchical clustering analysis on sequencing-based mRNA expression data identified 3 subtypes that do not correlate to any clinical features.
-
CNMF clustering analysis on sequencing-based miR expression data identified 3 subtypes that do not correlate to any clinical features.
-
Consensus hierarchical clustering analysis on sequencing-based miR expression data identified 3 subtypes that do not correlate to any clinical features.
Table 1. Get Full Table Overview of the association between subtypes identified by 8 different clustering approaches and 3 clinical features. Shown in the table are P values from statistical tests. Thresholded by P value < 0.05, no significant finding detected.
|
Clinical Features |
Time to Death |
AGE | GENDER |
| Statistical Tests | logrank test | ANOVA | Fisher's exact test |
| CN CNMF | 0.349 | 0.652 | 0.279 |
| METHLYATION CNMF | 0.247 | 0.291 | 0.283 |
| RPPA CNMF subtypes | 0.143 | 0.365 | 0.93 |
| RPPA cHierClus subtypes | 0.165 | 0.585 | 0.313 |
| RNAseq CNMF subtypes | 0.322 | 0.541 | 0.701 |
| RNAseq cHierClus subtypes | 0.507 | 0.316 | 0.192 |
| MIRseq CNMF subtypes | 0.167 | 0.526 | 0.945 |
| MIRseq cHierClus subtypes | 0.261 | 0.843 | 0.925 |
Table S1. Get Full Table Description of clustering approach #1: 'CN CNMF'
| Cluster Labels | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Number of samples | 48 | 22 | 42 | 26 |
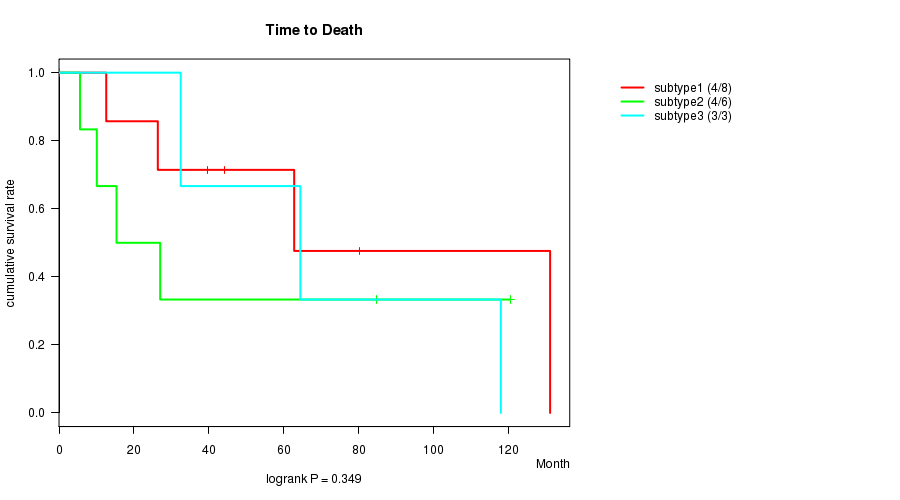
P value = 0.349 (logrank test)
Table S2. Clustering Approach #1: 'CN CNMF' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 18 | 11 | 0.2 - 131.1 (41.8) |
| subtype1 | 8 | 4 | 0.2 - 131.1 (41.8) |
| subtype2 | 6 | 4 | 5.6 - 120.5 (21.2) |
| subtype3 | 3 | 3 | 32.5 - 117.9 (64.4) |
| subtype4 | 1 | 0 | 80.0 - 80.0 (80.0) |
Figure S1. Get High-res Image Clustering Approach #1: 'CN CNMF' versus Clinical Feature #1: 'Time to Death'
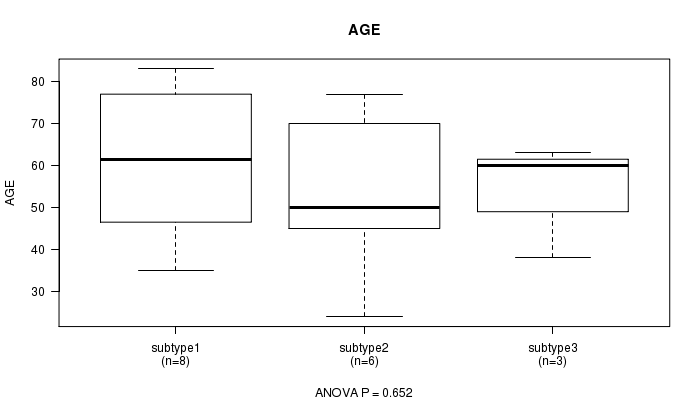
P value = 0.652 (ANOVA)
Table S3. Clustering Approach #1: 'CN CNMF' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 19 | 56.8 (16.0) |
| subtype1 | 8 | 61.0 (17.5) |
| subtype2 | 6 | 52.7 (19.0) |
| subtype3 | 3 | 53.7 (13.7) |
| subtype4 | 2 | 57.0 (1.4) |
Figure S2. Get High-res Image Clustering Approach #1: 'CN CNMF' versus Clinical Feature #2: 'AGE'
P value = 0.279 (Fisher's exact test)
Table S4. Clustering Approach #1: 'CN CNMF' versus Clinical Feature #3: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 46 | 92 |
| subtype1 | 20 | 28 |
| subtype2 | 4 | 18 |
| subtype3 | 13 | 29 |
| subtype4 | 9 | 17 |
Figure S3. Get High-res Image Clustering Approach #1: 'CN CNMF' versus Clinical Feature #3: 'GENDER'
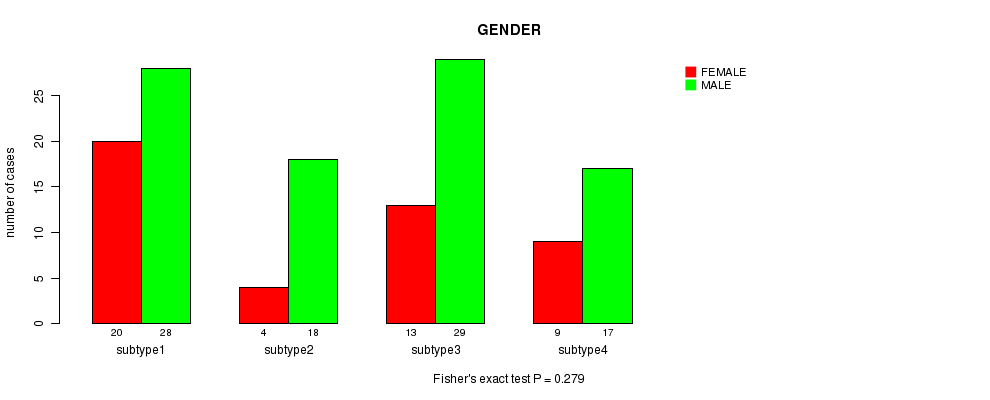
Table S5. Get Full Table Description of clustering approach #2: 'METHLYATION CNMF'
| Cluster Labels | 1 | 2 | 3 |
|---|---|---|---|
| Number of samples | 34 | 52 | 52 |
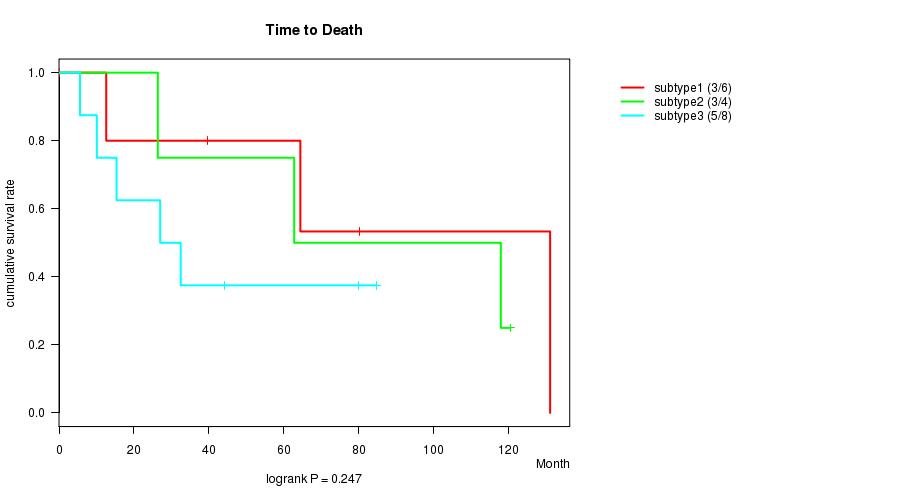
P value = 0.247 (logrank test)
Table S6. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 18 | 11 | 0.2 - 131.1 (41.8) |
| subtype1 | 6 | 3 | 0.2 - 131.1 (52.0) |
| subtype2 | 4 | 3 | 26.4 - 120.5 (90.3) |
| subtype3 | 8 | 5 | 5.6 - 84.7 (29.7) |
Figure S4. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #1: 'Time to Death'
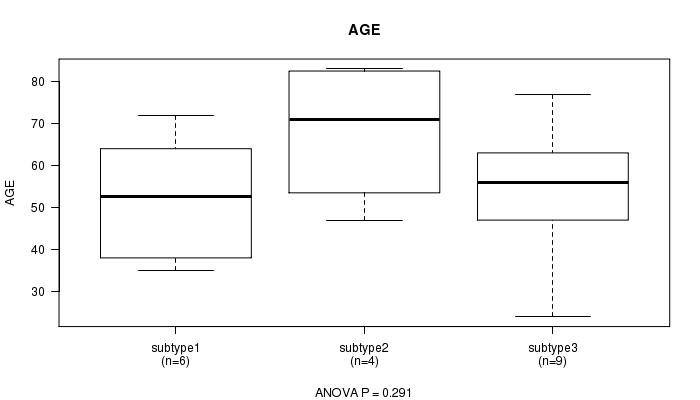
P value = 0.291 (ANOVA)
Table S7. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 19 | 56.8 (16.0) |
| subtype1 | 6 | 52.3 (14.9) |
| subtype2 | 4 | 68.0 (17.6) |
| subtype3 | 9 | 54.8 (15.5) |
Figure S5. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #2: 'AGE'
P value = 0.283 (Fisher's exact test)
Table S8. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #3: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 46 | 92 |
| subtype1 | 13 | 21 |
| subtype2 | 20 | 32 |
| subtype3 | 13 | 39 |
Figure S6. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #3: 'GENDER'
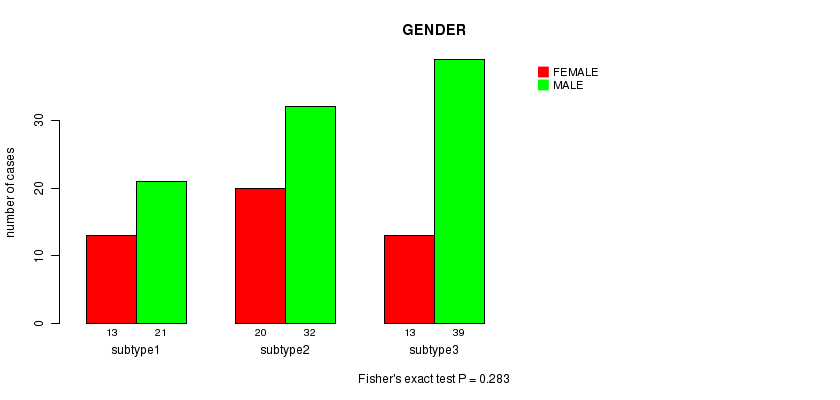
Table S9. Get Full Table Description of clustering approach #3: 'RPPA CNMF subtypes'
| Cluster Labels | 1 | 2 | 3 |
|---|---|---|---|
| Number of samples | 40 | 34 | 25 |
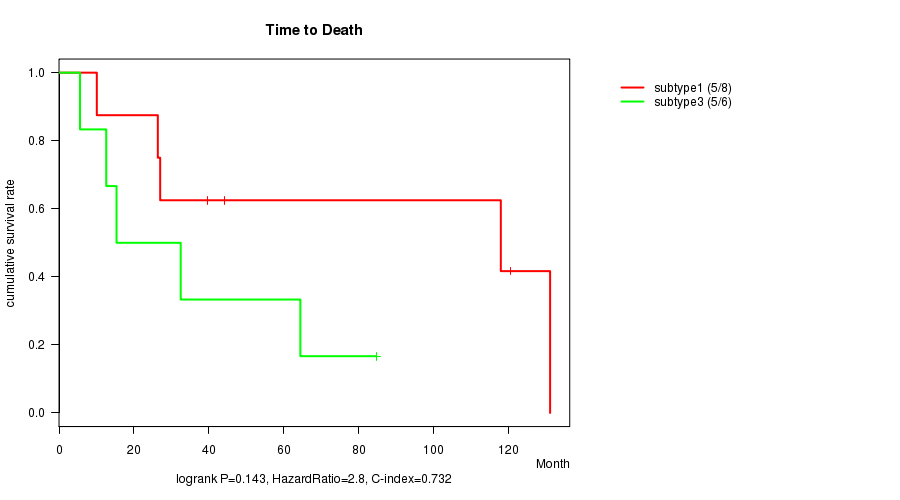
P value = 0.143 (logrank test)
Table S10. Clustering Approach #3: 'RPPA CNMF subtypes' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 16 | 11 | 5.6 - 131.1 (41.8) |
| subtype1 | 8 | 5 | 10.1 - 131.1 (41.8) |
| subtype2 | 2 | 1 | 62.8 - 80.0 (71.4) |
| subtype3 | 6 | 5 | 5.6 - 84.7 (23.9) |
Figure S7. Get High-res Image Clustering Approach #3: 'RPPA CNMF subtypes' versus Clinical Feature #1: 'Time to Death'
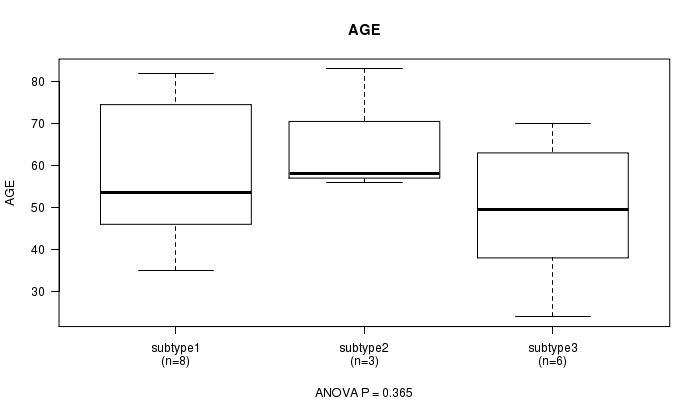
P value = 0.365 (ANOVA)
Table S11. Clustering Approach #3: 'RPPA CNMF subtypes' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 17 | 56.2 (16.9) |
| subtype1 | 8 | 58.1 (17.2) |
| subtype2 | 3 | 65.7 (15.0) |
| subtype3 | 6 | 49.0 (16.8) |
Figure S8. Get High-res Image Clustering Approach #3: 'RPPA CNMF subtypes' versus Clinical Feature #2: 'AGE'
P value = 0.93 (Fisher's exact test)
Table S12. Clustering Approach #3: 'RPPA CNMF subtypes' versus Clinical Feature #3: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 35 | 64 |
| subtype1 | 15 | 25 |
| subtype2 | 11 | 23 |
| subtype3 | 9 | 16 |
Figure S9. Get High-res Image Clustering Approach #3: 'RPPA CNMF subtypes' versus Clinical Feature #3: 'GENDER'
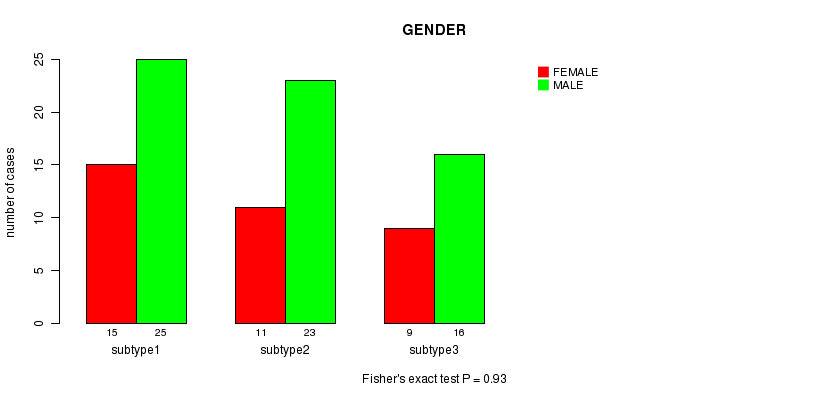
Table S13. Get Full Table Description of clustering approach #4: 'RPPA cHierClus subtypes'
| Cluster Labels | 1 | 2 | 3 |
|---|---|---|---|
| Number of samples | 36 | 40 | 23 |
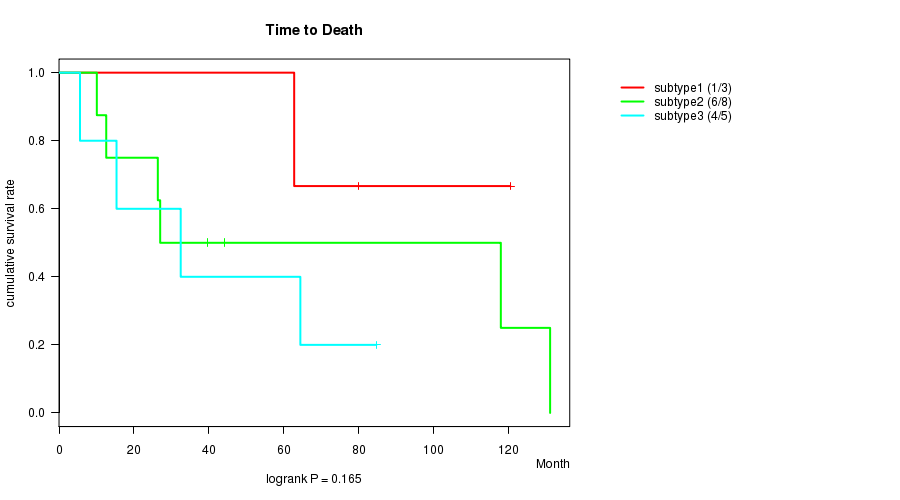
P value = 0.165 (logrank test)
Table S14. Clustering Approach #4: 'RPPA cHierClus subtypes' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 16 | 11 | 5.6 - 131.1 (41.8) |
| subtype1 | 3 | 1 | 62.8 - 120.5 (80.0) |
| subtype2 | 8 | 6 | 10.1 - 131.1 (33.3) |
| subtype3 | 5 | 4 | 5.6 - 84.7 (32.5) |
Figure S10. Get High-res Image Clustering Approach #4: 'RPPA cHierClus subtypes' versus Clinical Feature #1: 'Time to Death'
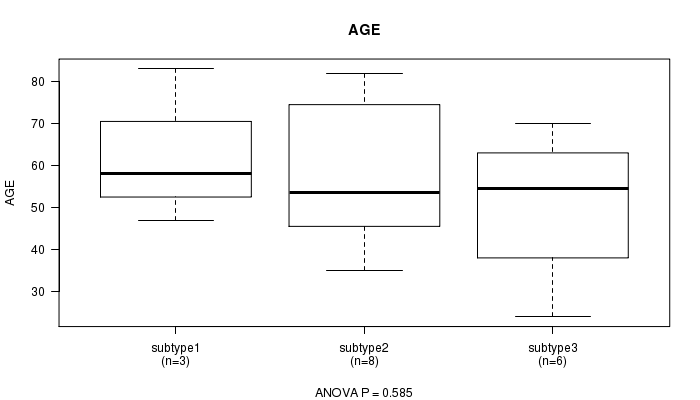
P value = 0.585 (ANOVA)
Table S15. Clustering Approach #4: 'RPPA cHierClus subtypes' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 17 | 56.2 (16.9) |
| subtype1 | 3 | 62.7 (18.4) |
| subtype2 | 8 | 58.0 (17.3) |
| subtype3 | 6 | 50.7 (16.9) |
Figure S11. Get High-res Image Clustering Approach #4: 'RPPA cHierClus subtypes' versus Clinical Feature #2: 'AGE'
P value = 0.313 (Fisher's exact test)
Table S16. Clustering Approach #4: 'RPPA cHierClus subtypes' versus Clinical Feature #3: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 35 | 64 |
| subtype1 | 14 | 22 |
| subtype2 | 16 | 24 |
| subtype3 | 5 | 18 |
Figure S12. Get High-res Image Clustering Approach #4: 'RPPA cHierClus subtypes' versus Clinical Feature #3: 'GENDER'
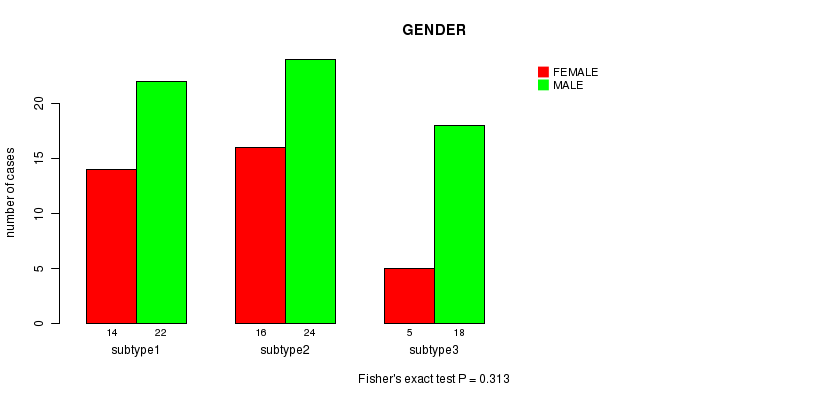
Table S17. Get Full Table Description of clustering approach #5: 'RNAseq CNMF subtypes'
| Cluster Labels | 1 | 2 | 3 |
|---|---|---|---|
| Number of samples | 46 | 41 | 47 |
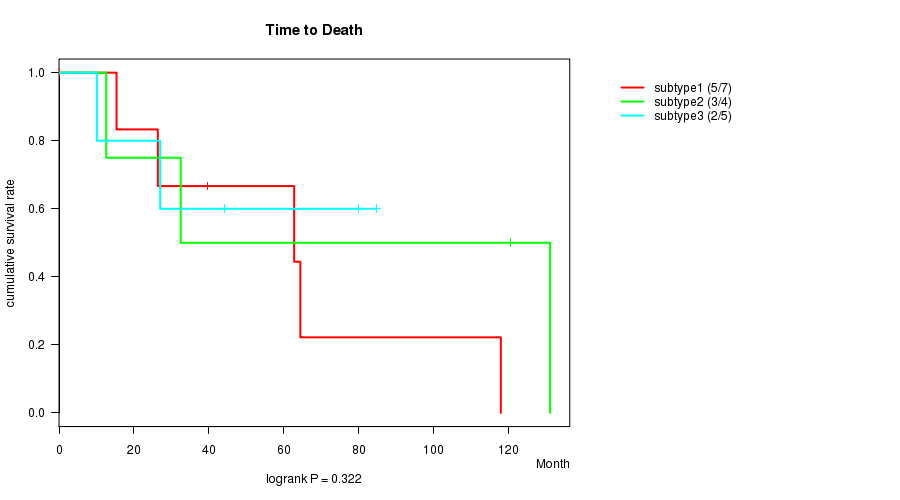
P value = 0.322 (logrank test)
Table S18. Clustering Approach #5: 'RNAseq CNMF subtypes' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 16 | 10 | 0.2 - 131.1 (41.8) |
| subtype1 | 7 | 5 | 0.2 - 117.9 (39.6) |
| subtype2 | 4 | 3 | 12.6 - 131.1 (76.5) |
| subtype3 | 5 | 2 | 10.1 - 84.7 (44.0) |
Figure S13. Get High-res Image Clustering Approach #5: 'RNAseq CNMF subtypes' versus Clinical Feature #1: 'Time to Death'
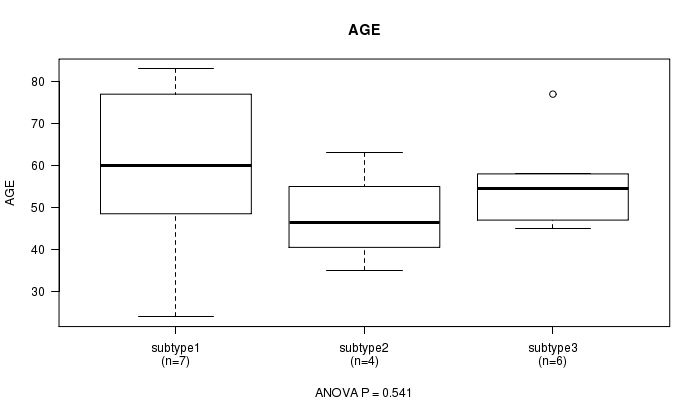
P value = 0.541 (ANOVA)
Table S19. Clustering Approach #5: 'RNAseq CNMF subtypes' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 17 | 55.6 (16.5) |
| subtype1 | 7 | 59.7 (22.1) |
| subtype2 | 4 | 47.8 (11.5) |
| subtype3 | 6 | 56.0 (11.5) |
Figure S14. Get High-res Image Clustering Approach #5: 'RNAseq CNMF subtypes' versus Clinical Feature #2: 'AGE'
P value = 0.701 (Fisher's exact test)
Table S20. Clustering Approach #5: 'RNAseq CNMF subtypes' versus Clinical Feature #3: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 45 | 89 |
| subtype1 | 14 | 32 |
| subtype2 | 16 | 25 |
| subtype3 | 15 | 32 |
Figure S15. Get High-res Image Clustering Approach #5: 'RNAseq CNMF subtypes' versus Clinical Feature #3: 'GENDER'
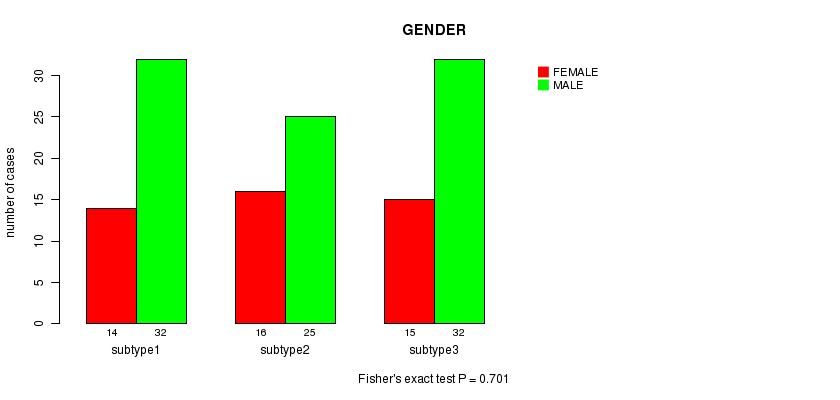
Table S21. Get Full Table Description of clustering approach #6: 'RNAseq cHierClus subtypes'
| Cluster Labels | 1 | 2 | 3 |
|---|---|---|---|
| Number of samples | 33 | 68 | 33 |
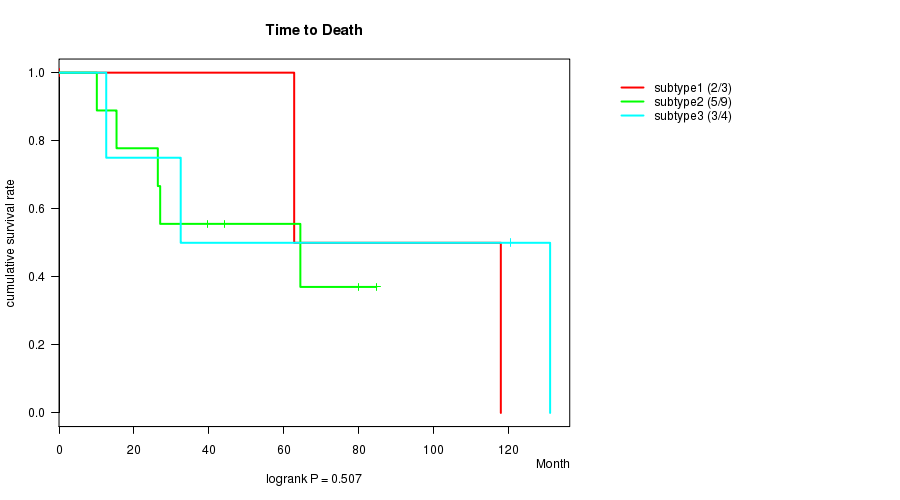
P value = 0.507 (logrank test)
Table S22. Clustering Approach #6: 'RNAseq cHierClus subtypes' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 16 | 10 | 0.2 - 131.1 (41.8) |
| subtype1 | 3 | 2 | 0.2 - 117.9 (62.8) |
| subtype2 | 9 | 5 | 10.1 - 84.7 (39.6) |
| subtype3 | 4 | 3 | 12.6 - 131.1 (76.5) |
Figure S16. Get High-res Image Clustering Approach #6: 'RNAseq cHierClus subtypes' versus Clinical Feature #1: 'Time to Death'
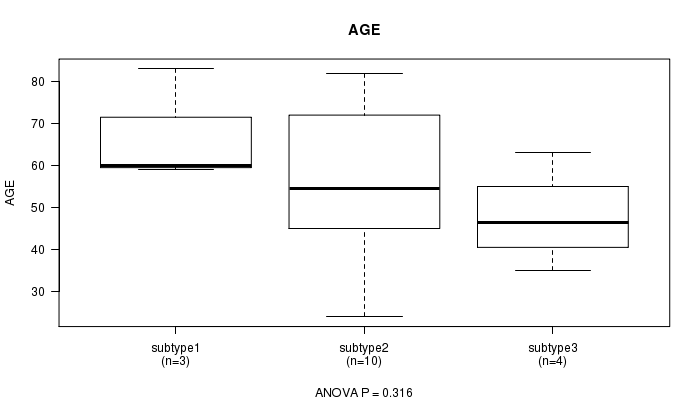
P value = 0.316 (ANOVA)
Table S23. Clustering Approach #6: 'RNAseq cHierClus subtypes' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 17 | 55.6 (16.5) |
| subtype1 | 3 | 67.3 (13.6) |
| subtype2 | 10 | 55.2 (18.0) |
| subtype3 | 4 | 47.8 (11.5) |
Figure S17. Get High-res Image Clustering Approach #6: 'RNAseq cHierClus subtypes' versus Clinical Feature #2: 'AGE'
P value = 0.192 (Fisher's exact test)
Table S24. Clustering Approach #6: 'RNAseq cHierClus subtypes' versus Clinical Feature #3: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 45 | 89 |
| subtype1 | 8 | 25 |
| subtype2 | 22 | 46 |
| subtype3 | 15 | 18 |
Figure S18. Get High-res Image Clustering Approach #6: 'RNAseq cHierClus subtypes' versus Clinical Feature #3: 'GENDER'
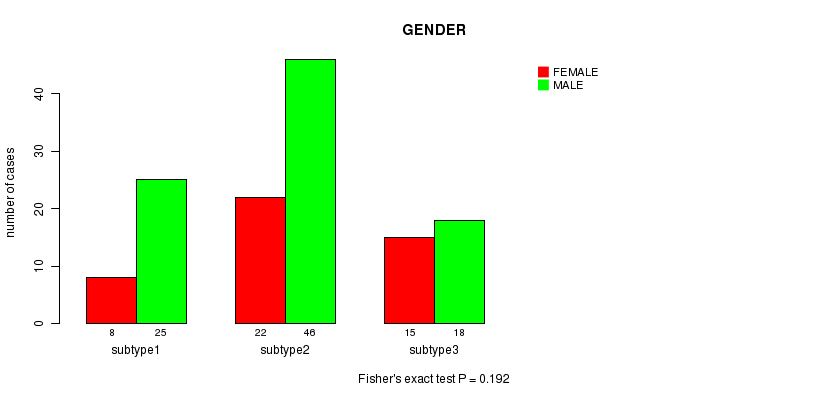
Table S25. Get Full Table Description of clustering approach #7: 'MIRseq CNMF subtypes'
| Cluster Labels | 1 | 2 | 3 |
|---|---|---|---|
| Number of samples | 57 | 45 | 31 |
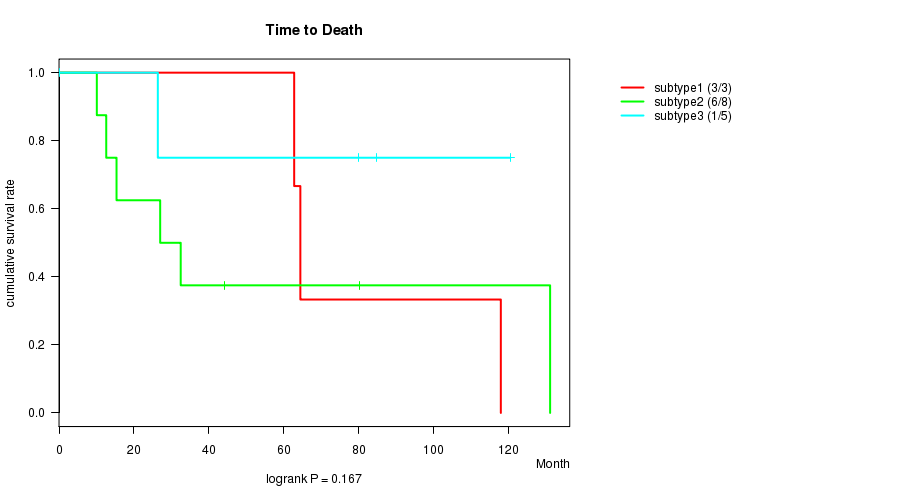
P value = 0.167 (logrank test)
Table S26. Clustering Approach #7: 'MIRseq CNMF subtypes' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 16 | 10 | 0.2 - 131.1 (53.4) |
| subtype1 | 3 | 3 | 62.8 - 117.9 (64.4) |
| subtype2 | 8 | 6 | 10.1 - 131.1 (29.7) |
| subtype3 | 5 | 1 | 0.2 - 120.5 (80.0) |
Figure S19. Get High-res Image Clustering Approach #7: 'MIRseq CNMF subtypes' versus Clinical Feature #1: 'Time to Death'
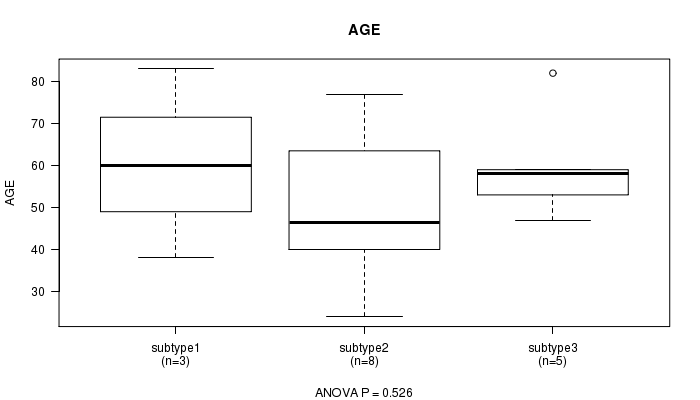
P value = 0.526 (ANOVA)
Table S27. Clustering Approach #7: 'MIRseq CNMF subtypes' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 16 | 55.1 (16.6) |
| subtype1 | 3 | 60.3 (22.5) |
| subtype2 | 8 | 50.1 (17.1) |
| subtype3 | 5 | 59.8 (13.3) |
Figure S20. Get High-res Image Clustering Approach #7: 'MIRseq CNMF subtypes' versus Clinical Feature #2: 'AGE'
P value = 0.945 (Fisher's exact test)
Table S28. Clustering Approach #7: 'MIRseq CNMF subtypes' versus Clinical Feature #3: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 44 | 89 |
| subtype1 | 19 | 38 |
| subtype2 | 14 | 31 |
| subtype3 | 11 | 20 |
Figure S21. Get High-res Image Clustering Approach #7: 'MIRseq CNMF subtypes' versus Clinical Feature #3: 'GENDER'
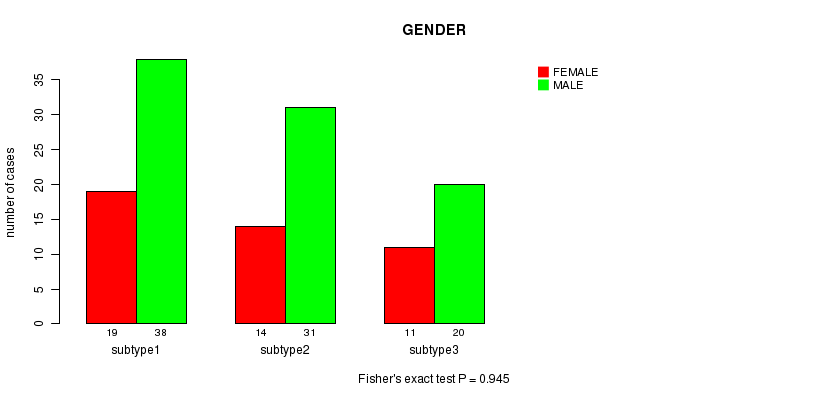
Table S29. Get Full Table Description of clustering approach #8: 'MIRseq cHierClus subtypes'
| Cluster Labels | 1 | 2 | 3 |
|---|---|---|---|
| Number of samples | 16 | 87 | 30 |
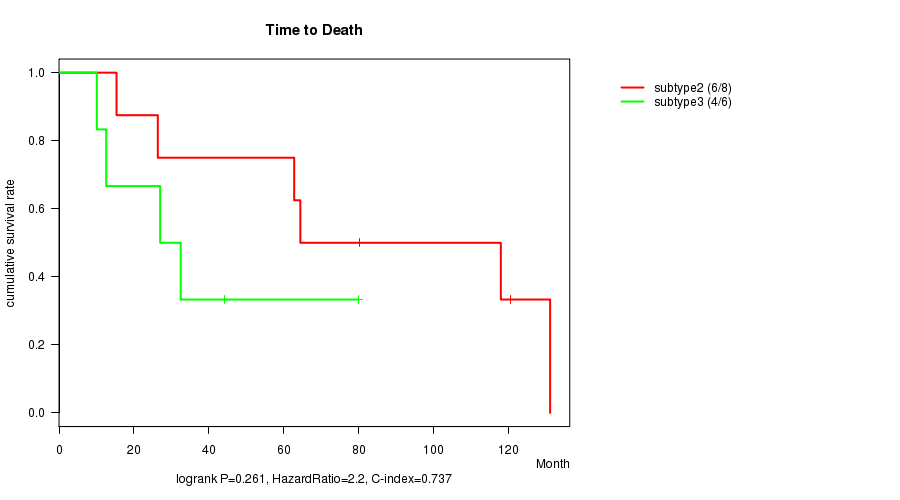
P value = 0.261 (logrank test)
Table S30. Clustering Approach #8: 'MIRseq cHierClus subtypes' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 16 | 10 | 0.2 - 131.1 (53.4) |
| subtype1 | 2 | 0 | 0.2 - 84.7 (42.5) |
| subtype2 | 8 | 6 | 15.3 - 131.1 (72.4) |
| subtype3 | 6 | 4 | 10.1 - 80.0 (29.7) |
Figure S22. Get High-res Image Clustering Approach #8: 'MIRseq cHierClus subtypes' versus Clinical Feature #1: 'Time to Death'
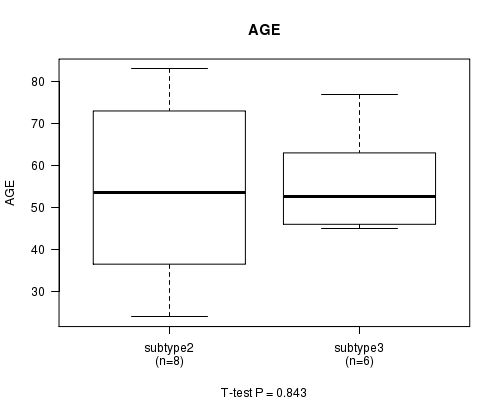
P value = 0.843 (ANOVA)
Table S31. Clustering Approach #8: 'MIRseq cHierClus subtypes' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 16 | 55.1 (16.6) |
| subtype1 | 2 | 56.0 (4.2) |
| subtype2 | 8 | 54.1 (21.8) |
| subtype3 | 6 | 56.0 (12.6) |
Figure S23. Get High-res Image Clustering Approach #8: 'MIRseq cHierClus subtypes' versus Clinical Feature #2: 'AGE'
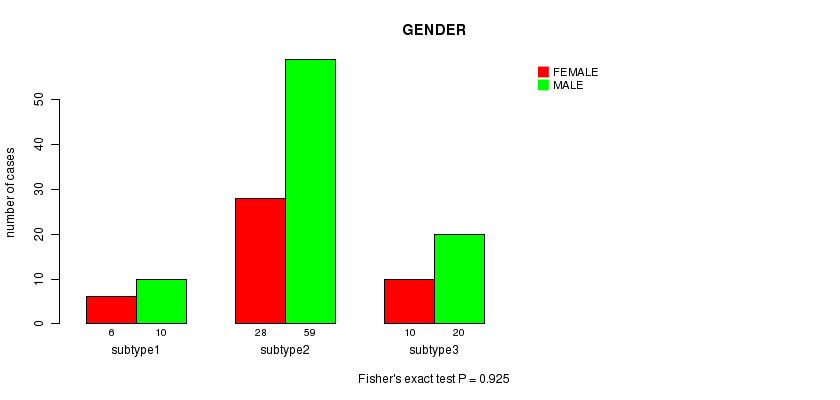
P value = 0.925 (Fisher's exact test)
Table S32. Clustering Approach #8: 'MIRseq cHierClus subtypes' versus Clinical Feature #3: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 44 | 89 |
| subtype1 | 6 | 10 |
| subtype2 | 28 | 59 |
| subtype3 | 10 | 20 |
Figure S24. Get High-res Image Clustering Approach #8: 'MIRseq cHierClus subtypes' versus Clinical Feature #3: 'GENDER'
-
Cluster data file = SKCM.mergedcluster.txt
-
Clinical data file = SKCM.clin.merged.picked.txt
-
Number of patients = 138
-
Number of clustering approaches = 8
-
Number of selected clinical features = 3
-
Exclude small clusters that include fewer than K patients, K = 3
consensus non-negative matrix factorization clustering approach (Brunet et al. 2004)
Resampling-based clustering method (Monti et al. 2003)
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For continuous numerical clinical features, one-way analysis of variance (Howell 2002) was applied to compare the clinical values between tumor subtypes using 'anova' function in R
For binary clinical features, two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.