This report serves to describe the mutational landscape and properties of a given individual set, as well as rank genes and genesets according to mutational significance. MutSig v2.0 was used to generate the results found in this report.
-
Working with individual set: PRAD-TP
-
Number of patients in set: 83
The input for this pipeline is a set of individuals with the following files associated for each:
-
An annotated .maf file describing the mutations called for the respective individual, and their properties.
-
A .wig file that contains information about the coverage of the sample.
-
MAF used for this analysis:PRAD-TP.final_analysis_set.maf
-
Significantly mutated genes (q ≤ 0.1): 24
-
Mutations seen in COSMIC: 26
-
Significantly mutated genes in COSMIC territory: 6
-
Genes with clustered mutations (≤ 3 aa apart): 10
-
Significantly mutated genesets: 2
-
Significantly mutated genesets: (excluding sig. mutated genes):0
-
Read 83 MAFs of type "Broad"
-
Total number of mutations in input MAFs: 12281
-
After removing 28 mutations outside chr1-24: 12253
-
After removing 2166 blacklisted mutations: 10087
-
After removing 5009 noncoding mutations: 5078
-
After collapsing adjacent/redundant mutations: 5056
-
Number of mutations before filtering: 5056
-
After removing 133 mutations outside gene set: 4923
-
After removing 4 mutations outside category set: 4919
Table 1. Get Full Table Table representing breakdown of mutations by type.
| type | count |
|---|---|
| Frame_Shift_Del | 145 |
| Frame_Shift_Ins | 65 |
| In_Frame_Del | 42 |
| In_Frame_Ins | 7 |
| Missense_Mutation | 3032 |
| Nonsense_Mutation | 171 |
| Nonstop_Mutation | 2 |
| Silent | 1359 |
| Splice_Site | 88 |
| Translation_Start_Site | 8 |
| Total | 4919 |
Table 2. Get Full Table A breakdown of mutation rates per category discovered for this individual set.
| category | n | N | rate | rate_per_mb | relative_rate | exp_ns_s_ratio |
|---|---|---|---|---|---|---|
| *CpG->T | 815 | 138713067 | 5.9e-06 | 5.9 | 4.1 | 2.1 |
| *Np(A/C/T)->transit | 826 | 1965699488 | 4.2e-07 | 0.42 | 0.29 | 2 |
| *ApG->G | 94 | 381313535 | 2.5e-07 | 0.25 | 0.17 | 2.1 |
| transver | 1302 | 2485726090 | 5.2e-07 | 0.52 | 0.37 | 5 |
| indel+null | 520 | 2485726090 | 2.1e-07 | 0.21 | 0.15 | NaN |
| double_null | 3 | 2485726090 | 1.2e-09 | 0.0012 | 0.00084 | NaN |
| Total | 3560 | 2485726090 | 1.4e-06 | 1.4 | 1 | 3.5 |
The x axis represents the samples. The y axis represents the exons, one row per exon, and they are sorted by average coverage across samples. For exons with exactly the same average coverage, they are sorted next by the %GC of the exon. (The secondary sort is especially useful for the zero-coverage exons at the bottom).
Figure 1.
Figure 2. Patients counts and rates file used to generate this plot: PRAD-TP.patients.counts_and_rates.txt
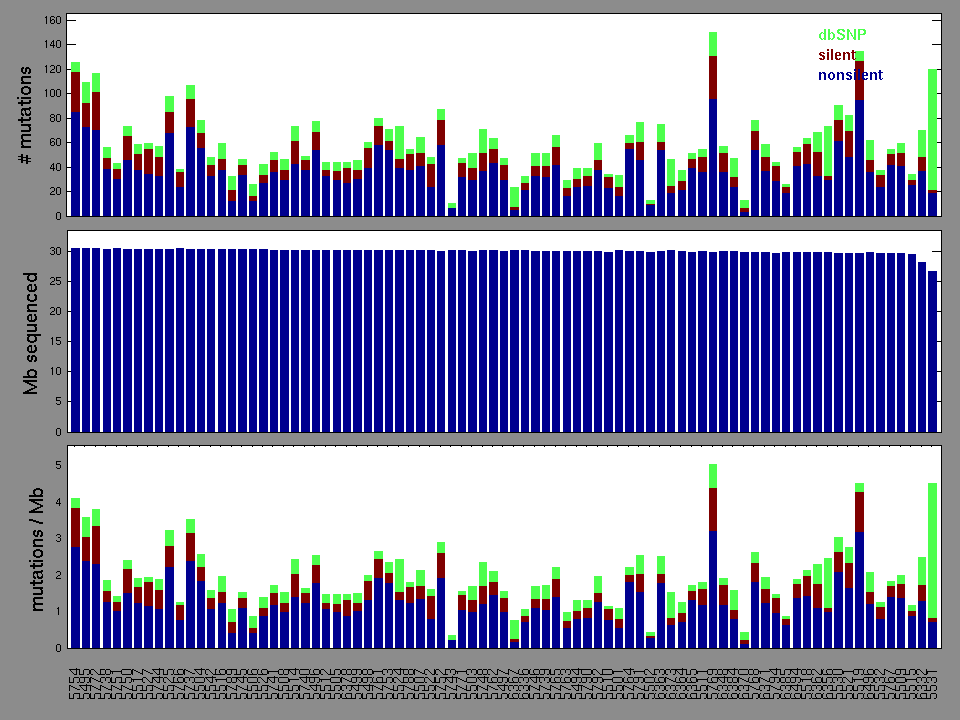
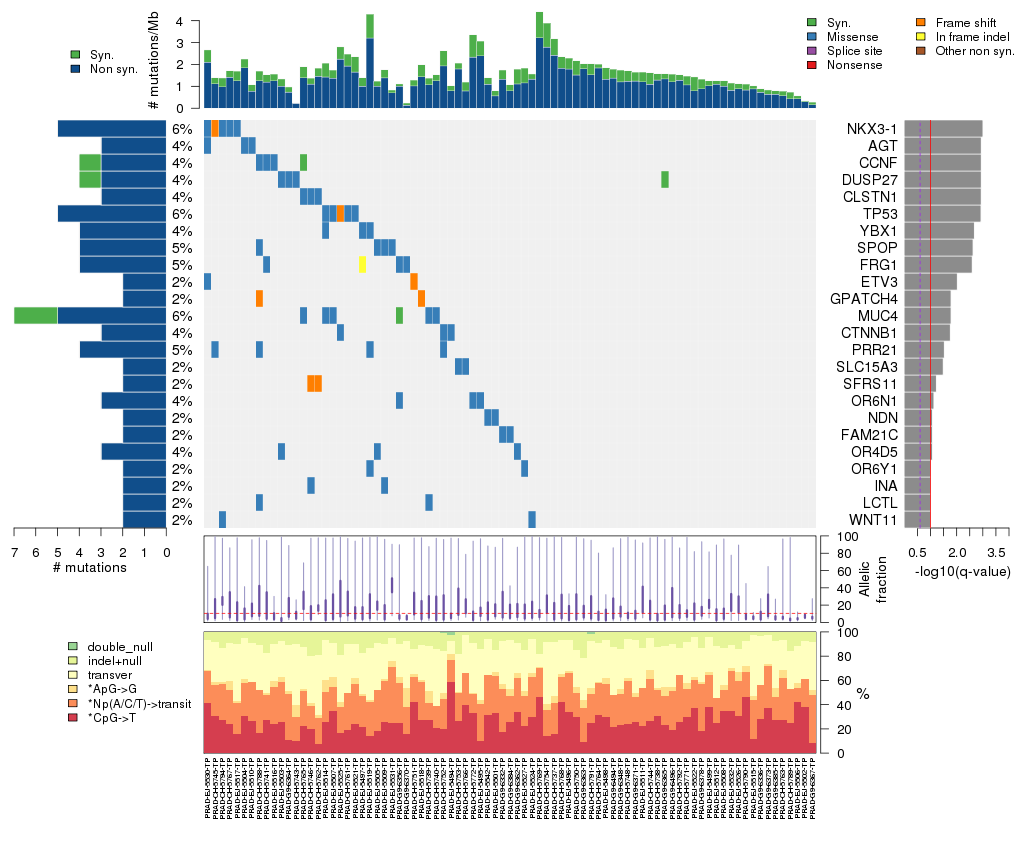
Figure 3. Get High-res Image The matrix in the center of the figure represents individual mutations in patient samples, color-coded by type of mutation, for the significantly mutated genes. The rate of synonymous and non-synonymous mutations is displayed at the top of the matrix. The barplot on the left of the matrix shows the number of mutations in each gene. The percentages represent the fraction of tumors with at least one mutation in the specified gene. The barplot to the right of the matrix displays the q-values for the most significantly mutated genes. The purple boxplots below the matrix (only displayed if required columns are present in the provided MAF) represent the distributions of allelic fractions observed in each sample. The plot at the bottom represents the base substitution distribution of individual samples, using the same categories that were used to calculate significance.
Column Descriptions:
-
N = number of sequenced bases in this gene across the individual set
-
n = number of (nonsilent) mutations in this gene across the individual set
-
npat = number of patients (individuals) with at least one nonsilent mutation
-
nsite = number of unique sites having a non-silent mutation
-
nsil = number of silent mutations in this gene across the individual set
-
n1 = number of nonsilent mutations of type: *CpG->T
-
n2 = number of nonsilent mutations of type: *Np(A/C/T)->transit
-
n3 = number of nonsilent mutations of type: *ApG->G
-
n4 = number of nonsilent mutations of type: transver
-
n5 = number of nonsilent mutations of type: indel+null
-
n6 = number of nonsilent mutations of type: double_null
-
p_classic = p-value for the observed amount of nonsilent mutations being elevated in this gene
-
p_ns_s = p-value for the observed nonsilent/silent ratio being elevated in this gene
-
p_ks = p-value for clustering of mutations (Kolmogorov-Smirnoff test)
-
p_cons = p-value for enrichment of mutations at evolutionarily most-conserved sites in gene
-
p_joint = p-value for clustering + conservation
-
p = p-value (overall)
-
q = q-value, False Discovery Rate (Benjamini-Hochberg procedure)
Table 3. Get Full Table A Ranked List of Significantly Mutated Genes. Number of significant genes found: 24. Number of genes displayed: 35. Click on a gene name to display its stick figure depicting the distribution of mutations and mutation types across the chosen gene (this feature may not be available for all significant genes).
| rank | gene | description | N | n | npat | nsite | nsil | n1 | n2 | n3 | n4 | n5 | n6 | p_classic | p_ns_s | p_ks | p_cons | p_joint | p | q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NKX3-1 | NK3 homeobox 1 | 43527 | 5 | 5 | 5 | 0 | 0 | 2 | 0 | 2 | 1 | 0 | 2.1e-09 | 0.26 | 0.044 | 0.0026 | 0.0059 | 3.3e-10 | 6e-06 |
| 2 | AGT | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | 122342 | 3 | 3 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0.0005 | 0.53 | 6e-07 | 1 | 1.8e-06 | 2e-08 | 0.00016 |
| 3 | CCNF | cyclin F | 199411 | 3 | 3 | 1 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0.0024 | 0.79 | 6e-07 | 0.023 | 6e-07 | 3.1e-08 | 0.00016 |
| 4 | DUSP27 | dual specificity phosphatase 27 (putative) | 264959 | 3 | 3 | 1 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0.0014 | 0.82 | 2e-07 | 0.92 | 1.2e-06 | 3.6e-08 | 0.00016 |
| 5 | CLSTN1 | calsyntenin 1 | 242387 | 3 | 3 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0.0056 | 0.6 | 4e-07 | 0.052 | 4e-07 | 4.6e-08 | 0.00017 |
| 6 | TP53 | tumor protein p53 | 105521 | 5 | 5 | 5 | 0 | 3 | 0 | 0 | 1 | 1 | 0 | 5.5e-07 | 0.34 | 0.069 | 0.0012 | 0.0056 | 6.3e-08 | 0.00019 |
| 7 | YBX1 | Y box binding protein 1 | 68945 | 4 | 3 | 2 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0.00012 | 0.32 | 0.0082 | 0.00043 | 0.0002 | 4.4e-07 | 0.0011 |
| 8 | SPOP | speckle-type POZ protein | 96345 | 4 | 4 | 3 | 0 | 0 | 0 | 1 | 3 | 0 | 0 | 3.8e-06 | 0.39 | 0.0072 | 0.27 | 0.0091 | 6.2e-07 | 0.0014 |
| 9 | FRG1 | FSHD region gene 1 | 67208 | 4 | 4 | 2 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 2.9e-07 | 0.65 | 0.082 | 1 | 0.15 | 7.9e-07 | 0.0016 |
| 10 | ETV3 | ets variant gene 3 | 36841 | 2 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0.00045 | 0.65 | 0.00065 | 0.42 | 0.00065 | 4.7e-06 | 0.0085 |
| 11 | GPATCH4 | G patch domain containing 4 | 97579 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0.0013 | 1 | 0.00084 | 0.28 | 0.0005 | 9.8e-06 | 0.016 |
| 12 | MUC4 | mucin 4, cell surface associated | 272535 | 5 | 5 | 4 | 2 | 1 | 2 | 0 | 2 | 0 | 0 | 0.0034 | 0.69 | 0.00036 | 0.1 | 0.0002 | 1e-05 | 0.016 |
| 13 | CTNNB1 | catenin (cadherin-associated protein), beta 1, 88kDa | 199245 | 3 | 3 | 3 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0.00098 | 0.46 | 0.00068 | 0.24 | 0.00081 | 0.000012 | 0.017 |
| 14 | PRR21 | proline rich 21 | 58941 | 4 | 4 | 4 | 0 | 1 | 1 | 0 | 2 | 0 | 0 | 2.2e-06 | 0.26 | 0.24 | 0.94 | 0.69 | 0.000022 | 0.029 |
| 15 | SLC15A3 | solute carrier family 15, member 3 | 101449 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0.0057 | 0.66 | 0.00075 | 0.066 | 0.00033 | 0.000026 | 0.032 |
| 16 | SFRS11 | splicing factor, arginine/serine-rich 11 | 124634 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0.0059 | 1 | 0.00066 | 0.8 | 0.00066 | 0.000052 | 0.059 |
| 17 | OR6N1 | olfactory receptor, family 6, subfamily N, member 1 | 78146 | 3 | 3 | 2 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0.000012 | 0.46 | 0.36 | 0.5 | 0.44 | 7e-05 | 0.075 |
| 18 | NDN | necdin homolog (mouse) | 74771 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0.0012 | 0.67 | 0.0013 | 0.5 | 0.0056 | 0.000086 | 0.083 |
| 19 | FAM21C | family with sequence similarity 21, member C | 199157 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0.022 | 0.73 | 0.00032 | 0.47 | 0.0003 | 0.000088 | 0.083 |
| 20 | OR4D5 | olfactory receptor, family 4, subfamily D, member 5 | 79763 | 3 | 3 | 3 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0.000012 | 0.46 | 0.78 | 0.29 | 0.62 | 0.000092 | 0.083 |
| 21 | OR6Y1 | olfactory receptor, family 6, subfamily Y, member 1 | 81216 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0.0044 | 0.67 | 0.0013 | 0.28 | 0.0021 | 0.00012 | 0.099 |
| 22 | INA | internexin neuronal intermediate filament protein, alpha | 84606 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0.0042 | 0.67 | 0.00085 | 0.84 | 0.0023 | 0.00012 | 0.099 |
| 23 | LCTL | lactase-like | 145584 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0.014 | 0.71 | 0.00074 | 0.16 | 0.00076 | 0.00013 | 0.099 |
| 24 | WNT11 | wingless-type MMTV integration site family, member 11 | 59955 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0.0016 | 0.69 | 0.0012 | 0.86 | 0.0066 | 0.00013 | 0.099 |
| 25 | PIM1 | pim-1 oncogene | 80153 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0.0089 | 0.69 | 0.0013 | 0.16 | 0.0013 | 0.00014 | 0.1 |
| 26 | LILRB1 | leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 | 165618 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0.006 | 0.65 | 0.00065 | 0.6 | 0.0021 | 0.00015 | 0.11 |
| 27 | BCMO1 | beta-carotene 15,15-monooxygenase 1 139964 2 2 1 0 0 2 0 0 0 0 0.00384 0.444 0.00119 0.150 0.00350 0.000164 0.106 28 GJB3 gap junction protein, beta 3, 31kDa 67808 2 2 1 0 0 0 0 2 0 0 0.00586 0.672 0.00153 0.140 0.00229 0.000164 0.106 29 MLL3 myeloid/lymphoid or mixed-lineage leukemia 3 1228308 8 7 7 0 0 1 0 2 4 1 0.000123 0.228 0.0568 0.726 0.116 0.000173 0.108 30 OR2AE1 olfactory receptor, family 2, subfamily AE, member 1 81008 2 2 2 0 1 0 0 0 1 0 0.000977 0.616 0.168 0.00693 0.0204 0.000236 0.141 31 SORBS1 sorbin and SH3 domain containing 1 336929 2 2 2 0 1 0 0 1 0 0 0.0731 0.563 0.000279 0.606 0.000279 0.000241 0.141 32 ATM ataxia telangiectasia mutated 780236 5 5 5 0 0 4 0 1 0 0 0.00180 0.177 0.136 0.0125 0.0132 0.000276 0.157 33 NEB nebulin 1722208 2 2 1 0 0 0 0 2 0 0 0.595 0.757 4.96e-05 0.322 4.96e-05 0.000337 0.185 34 C1orf116 chromosome 1 open reading frame 116 150780 2 2 2 0 0 1 0 1 0 0 0.0152 0.523 0.324 0.00211 0.00221 0.000380 0.198 35 TAF1L TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 210kDa-like 455255 3 3 3 0 1 2 0 0 0 0 0.0126 0.380 0.0309 0.00786 0.00275 0.000390 0.198 36 CNTNAP5 contactin associated protein-like 5 309961 5 5 5 0 2 0 0 2 1 0 4.99e-05 0.196 0.431 0.820 0.700 0.000394 0.198 37 MED12 mediator complex subunit 12 479645 2 2 1 0 0 2 0 0 0 0 0.0691 0.414 0.000335 0.593 0.000561 0.000433 0.212 38 OR5L2 olfactory receptor, family 5, subfamily L, member 2 78017 3 3 3 0 0 0 0 3 0 0 0.000101 0.549 0.693 0.169 0.411 0.000462 0.220 39 BCL6 B-cell CLL/lymphoma 6 (zinc finger protein 51) 178187 3 3 3 1 1 2 0 0 0 0 0.000512 0.604 0.0432 0.932 0.0843 0.000477 0.222 40 KAT2A K(lysine) acetyltransferase 2A 179164 2 1 2 0 0 1 0 1 0 0 0.0945 0.516 0.000472 0.465 0.000472 0.000491 0.223 41 SPAG16 sperm associated antigen 16 158375 2 2 2 0 1 0 1 0 0 0 0.00134 0.401 0.0878 0.127 0.0365 0.000534 0.236 42 CCDC105 coiled-coil domain containing 105 82844 2 2 2 1 1 0 0 1 0 0 0.00481 0.801 0.501 0.0144 0.0105 0.000552 0.238 43 TRIM41 tripartite motif-containing 41 157230 2 2 1 0 0 0 0 2 0 0 0.0233 0.691 0.000620 0.619 0.00235 0.000591 0.245 44 SLITRK4 SLIT and NTRK-like family, member 4 208728 3 3 2 0 0 0 0 0 3 0 0.000336 0.721 0.0459 1.000 0.164 0.000596 0.245 45 SETD5 SET domain containing 5 345156 4 1 4 1 0 3 0 1 0 0 0.266 0.540 8.16e-05 0.425 0.000252 0.000712 0.281 46 LHX3 LIM homeobox 3 74497 2 2 2 0 0 1 0 1 0 0 0.00264 0.515 0.477 0.0277 0.0255 0.000712 0.281 47 GUCY1A2 guanylate cyclase 1, soluble, alpha 2 169886 2 2 1 0 2 0 0 0 0 0 0.00322 0.523 0.0139 0.0363 0.0220 0.000747 0.288 48 PTEN phosphatase and tensin homolog (mutated in multiple advanced cancers 1) 102374 3 3 3 0 0 1 0 1 1 0 0.000158 0.404 0.774 0.114 0.467 0.000776 0.293 49 PCDHB14 protocadherin beta 14 199174 3 3 3 0 1 0 0 1 1 0 0.000464 0.273 0.139 0.538 0.191 0.000914 0.338 50 ADAMTS19 ADAM metallopeptidase with thrombospondin type 1 motif, 19 268474 2 2 2 0 1 0 0 1 0 0 0.00795 0.634 0.141 0.0249 0.0122 0.000994 0.361 51 AIM2 absent in melanoma 2 87306 3 3 3 0 0 1 0 1 1 0 0.000112 0.569 0.477 0.785 1.000 0.00113 0.388 52 ZMYM3 zinc finger, MYM-type 3 288038 4 4 4 0 0 0 0 1 3 0 0.000250 0.575 0.852 0.258 0.452 0.00114 0.388 53 LPHN3 latrophilin 3 311602 4 4 4 1 2 1 0 1 0 0 0.000335 0.613 0.136 0.501 0.339 0.00114 0.388 54 OR5B12 olfactory receptor, family 5, subfamily B, member 12 78709 2 2 2 0 1 0 0 1 0 0 0.000633 0.828 0.774 0.0932 0.185 0.00118 0.388 55 OR52A1 olfactory receptor, family 52, subfamily A, member 1 78103 2 2 2 0 2 0 0 0 0 0 0.000166 0.840 0.439 0.598 0.724 0.00121 0.388 56 GALNT4 UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) 144459 2 2 2 0 0 1 0 1 0 0 0.00304 0.563 0.505 0.0249 0.0397 0.00121 0.388 57 CROCC ciliary rootlet coiled-coil, rootletin 295882 4 4 2 1 3 0 0 1 0 0 0.000628 0.593 0.0910 0.664 0.196 0.00123 0.388 58 FAM75C1 family with sequence similarity 75, member C1 280752 2 2 2 1 0 2 0 0 0 0 0.0233 0.732 0.000569 0.980 0.00532 0.00124 0.388 59 GGTLC2 gamma-glutamyltransferase light chain 2 47824 2 2 2 1 0 0 1 1 0 0 0.000133 0.813 0.437 0.182 0.982 0.00130 0.398 60 FOXA1 forkhead box A1 92254 2 2 2 0 0 0 0 0 1 1 0.000918 0.821 0.662 0.153 0.149 0.00136 0.408 61 CYP2A6 cytochrome P450, family 2, subfamily A, polypeptide 6 126233 3 3 3 1 0 2 0 1 0 0 0.000548 0.650 0.186 0.440 0.257 0.00139 0.408 62 CRIPAK cysteine-rich PAK1 inhibitor 110493 3 3 3 1 0 1 0 2 0 0 0.000256 0.705 0.351 0.997 0.552 0.00140 0.408 63 LIMK2 LIM domain kinase 2 197664 3 3 3 0 0 0 0 3 0 0 0.00263 0.584 0.0940 0.109 0.0566 0.00146 0.420 64 XRCC6 X-ray repair complementing defective repair in Chinese hamster cells 6 (Ku autoantigen, 70kDa) 155415 2 2 2 0 1 1 0 0 0 0 0.00196 0.468 0.195 0.115 0.0868 0.00165 0.468 65 ACBD7 acyl-Coenzyme A binding domain containing 7 23420 1 1 1 0 0 0 0 1 0 0 0.00173 0.855 NaN NaN NaN 0.00173 0.483 66 GRID2 glutamate receptor, ionotropic, delta 2 256214 4 4 4 1 3 1 0 0 0 0 0.000200 0.623 0.710 0.851 1.000 0.00191 0.524 67 KIRREL kin of IRRE like (Drosophila) 161899 2 2 2 1 1 0 0 1 0 0 0.0178 0.839 0.548 0.00943 0.0118 0.00199 0.538 68 NTM neurotrimin 96778 2 2 2 0 0 0 0 1 1 0 0.00240 0.672 0.470 0.0634 0.0895 0.00203 0.542 69 LCE2D late cornified envelope 2D 27971 2 2 2 0 1 0 0 1 0 0 0.000419 0.750 0.545 0.555 0.529 0.00209 0.548 70 FAM83B family with sequence similarity 83, member B 253181 3 3 3 1 0 1 0 2 0 0 0.00342 0.760 0.773 0.0160 0.0662 0.00213 0.551 71 ZNF98 zinc finger protein 98 (F7175) 122162 3 3 3 0 0 0 0 2 1 0 0.000244 0.668 0.809 0.903 1.000 0.00228 0.573 72 BBS9 Bardet-Biedl syndrome 9 227840 3 3 2 0 0 1 0 0 2 0 0.000377 0.678 0.375 0.586 0.648 0.00228 0.573 73 HEXB hexosaminidase B (beta polypeptide) 129584 2 2 2 0 1 0 0 0 1 0 0.00128 0.693 0.181 0.156 0.198 0.00235 0.583 74 B3GNT4 UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 94944 2 2 2 0 0 1 0 0 0 1 0.000504 0.629 0.396 0.488 0.513 0.00239 0.584 75 OR2M3 olfactory receptor, family 2, subfamily M, member 3 78186 2 2 2 0 2 0 0 0 0 0 0.000359 0.694 0.325 0.836 0.727 0.00242 0.584 76 MMP12 matrix metallopeptidase 12 (macrophage elastase) 112366 2 2 2 0 0 0 0 2 0 0 0.00849 0.733 0.676 0.0153 0.0316 0.00248 0.588 77 NKX2-4 NK2 homeobox 4 23952 1 1 1 0 0 0 0 0 1 0 0.00253 1.000 NaN NaN NaN 0.00253 0.588 78 PBX4 pre-B-cell leukemia homeobox 4 79940 2 2 2 0 2 0 0 0 0 0 0.000575 0.419 0.848 0.0995 0.486 0.00257 0.588 79 KLC2 kinesin light chain 2 150347 3 3 2 1 1 0 0 2 0 0 0.000931 0.832 0.389 0.156 0.300 0.00257 0.588 80 RNF17 ring finger protein 17 415152 3 3 2 0 0 1 0 0 2 0 0.00318 0.509 0.0437 0.292 0.0890 0.00260 0.588 81 FGFR1OP2 FGFR1 oncogene partner 2 65824 2 2 2 0 0 1 0 1 0 0 0.00313 0.594 0.281 0.0850 0.0929 0.00266 0.596 82 CDKN1B cyclin-dependent kinase inhibitor 1B (p27, Kip1) 50142 2 2 2 0 0 0 0 0 2 0 0.000640 1.000 0.542 0.494 0.503 0.00291 0.626 83 LRP1B low density lipoprotein-related protein 1B (deleted in tumors) 1171940 5 5 5 1 0 0 0 4 1 0 0.0106 0.740 0.0158 0.673 0.0303 0.00291 0.626 84 TPTE2 transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 136261 3 3 3 0 1 1 0 1 0 0 0.000325 0.420 0.892 0.621 1.000 0.00293 0.626 85 BANF2 barrier to autointegration factor 2 23572 2 2 2 0 1 0 0 1 0 0 0.000325 0.434 0.919 0.844 1.000 0.00294 0.626 86 TBX18 T-box 18 132272 3 3 3 0 2 1 0 0 0 0 0.000360 0.292 0.971 0.507 0.951 0.00307 0.645 87 PLXNB2 plexin B2 418795 3 3 3 0 1 0 1 1 0 0 0.0313 0.443 0.605 0.00209 0.0111 0.00311 0.645 88 C16orf13 chromosome 16 open reading frame 13 26264 1 1 1 0 0 0 0 0 1 0 0.00313 1.000 NaN NaN NaN 0.00313 0.645 89 PLCH2 phospholipase C, eta 2 227836 3 3 3 0 0 1 0 1 1 0 0.00147 0.425 0.0622 0.778 0.245 0.00322 0.656 90 NBPF1 neuroblastoma breakpoint family, member 1 280054 3 3 3 0 1 1 0 1 0 0 0.00106 0.372 0.702 0.391 0.347 0.00328 0.661 91 RCBTB2 regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 141339 3 3 2 0 0 0 0 3 0 0 0.000627 0.582 0.375 0.900 0.597 0.00333 0.663 92 MOCOS molybdenum cofactor sulfurase 214117 3 3 3 0 0 0 0 2 1 0 0.000546 0.571 0.917 0.535 0.741 0.00356 0.693 93 LECT2 leukocyte cell-derived chemotaxin 2 39157 1 1 1 0 0 1 0 0 0 0 0.00357 0.691 NaN NaN NaN 0.00357 0.693 94 TMEM220 transmembrane protein 220 35765 1 1 1 0 1 0 0 0 0 0 0.00368 0.750 NaN NaN NaN 0.00368 0.693 95 PCDHGA2 protocadherin gamma subfamily A, 2 237411 3 3 3 1 2 0 0 1 0 0 0.00521 0.563 0.378 0.0815 0.0809 0.00370 0.693 96 C12orf63 chromosome 12 open reading frame 63 305322 3 3 3 0 1 0 0 0 2 0 0.000889 0.475 0.153 0.937 0.475 0.00370 0.693 97 PFDN5 prefoldin subunit 5 40244 1 1 1 0 0 0 1 0 0 0 0.00371 0.748 NaN NaN NaN 0.00371 0.693 98 ITGBL1 integrin, beta-like 1 (with EGF-like repeat domains) 109525 2 2 2 0 2 0 0 0 0 0 0.000659 0.447 0.415 0.686 0.692 0.00396 0.723 99 SHFM1 split hand/foot malformation (ectrodactyly) type 1 18675 1 1 1 0 0 0 0 0 1 0 0.00400 0.912 NaN NaN NaN 0.00400 0.723 100 ZNF485 zinc finger protein 485 108980 2 2 2 0 0 0 0 1 1 0 0.00480 0.747 0.0402 0.149 0.0967 0.00403 0.723 101 OR7A17 olfactory receptor, family 7, subfamily A, member 17 77177 1 1 1 0 1 0 0 0 0 0 0.00406 0.800 NaN NaN NaN 0.00406 0.723 102 KRT25 keratin 25 114954 3 3 3 0 1 0 0 1 1 0 0.000470 0.457 0.891 0.781 1.000 0.00407 0.723 103 NEUROD6 neurogenic differentiation 6 84494 2 2 2 0 1 0 1 0 0 0 0.000477 0.408 0.788 0.599 1.000 0.00412 0.726 104 SGSH N-sulfoglucosamine sulfohydrolase (sulfamidase) 107279 2 2 2 0 0 1 0 1 0 0 0.00144 0.517 0.0486 0.166 0.338 0.00419 0.728 105 LCN2 lipocalin 2 51538 2 2 2 0 0 2 0 0 0 0 0.00135 0.414 0.741 0.294 0.363 0.00421 0.728 106 AP4B1 adaptor-related protein complex 4, beta 1 subunit 187520 2 2 1 0 2 0 0 0 0 0 0.00978 0.640 0.0152 0.974 0.0505 0.00426 0.728 107 EPB41L4A erythrocyte membrane protein band 4.1 like 4A 175718 2 2 2 0 0 1 0 0 1 0 0.0128 0.682 0.482 0.0203 0.0400 0.00439 0.744 108 NBPF3 neuroblastoma breakpoint family, member 3 154947 3 3 3 0 0 1 0 2 0 0 0.00101 0.488 0.344 0.959 0.526 0.00453 0.752 109 ANO4 anoctamin 4 237399 3 3 3 0 2 0 0 1 0 0 0.00275 0.480 0.191 0.0684 0.194 0.00455 0.752 110 S100A7L2 S100 calcium binding protein A7-like 2 27473 1 1 1 0 1 0 0 0 0 0 0.00456 0.600 NaN NaN NaN 0.00456 0.752 111 NCOA4 nuclear receptor coactivator 4 157195 2 2 2 0 1 1 0 0 0 0 0.00406 0.541 0.0100 0.789 0.136 0.00468 0.765 112 RTBDN retbindin 56473 1 1 1 0 0 0 0 0 1 0 0.00475 0.740 NaN NaN NaN 0.00475 0.769 113 PIK3CA phosphoinositide-3-kinase, catalytic, alpha polypeptide 272368 2 2 2 0 0 1 1 0 0 0 0.00782 0.392 0.209 0.103 0.0735 0.00486 0.780 114 TMEM14B transmembrane protein 14B 30295 1 1 1 0 1 0 0 0 0 0 0.00499 0.556 NaN NaN NaN 0.00499 0.794 115 GNPNAT1 glucosamine-phosphate N-acetyltransferase 1 47717 1 1 1 0 0 0 0 0 1 0 0.00508 1.000 NaN NaN NaN 0.00508 0.800 116 C7orf44 chromosome 7 open reading frame 44 38171 1 1 1 0 1 0 0 0 0 0 0.00518 0.555 NaN NaN NaN 0.00518 0.810 117 CLGN calmegin 156720 3 2 3 0 1 1 0 1 0 0 0.00386 0.401 0.118 0.350 0.164 0.00528 0.819 118 CLK3 CDC-like kinase 3 125062 2 2 2 0 0 1 0 1 0 0 0.00425 0.524 0.0868 0.523 0.154 0.00545 0.837 119 CARD9 caspase recruitment domain family, member 9 112466 2 2 2 0 0 0 2 0 0 0 0.00120 0.908 0.181 0.790 0.559 0.00557 0.846 120 MRPL50 mitochondrial ribosomal protein L50 40255 1 1 1 0 0 0 0 0 1 0 0.00560 1.000 NaN NaN NaN 0.00560 0.846 121 SOD3 superoxide dismutase 3, extracellular 24149 1 1 1 0 0 1 0 0 0 0 0.00570 0.636 NaN NaN NaN 0.00570 0.847 122 PCDHGB7 protocadherin gamma subfamily B, 7 230249 2 2 2 0 0 2 0 0 0 0 0.00864 0.427 0.336 0.0752 0.0800 0.00573 0.847 123 RP1 retinitis pigmentosa 1 (autosomal dominant) 537356 4 4 4 0 0 2 0 1 1 0 0.00229 0.341 0.673 0.0873 0.303 0.00574 0.847 124 OR5AS1 olfactory receptor, family 5, subfamily AS, member 1 81080 2 2 2 0 1 1 0 0 0 0 0.00154 0.436 0.352 0.286 0.459 0.00583 0.853 125 ESCO1 establishment of cohesion 1 homolog 1 (S. cerevisiae) 211818 3 3 3 0 0 0 0 1 2 0 0.000714 0.740 0.820 0.903 1.000 0.00588 0.854 126 AP3B2 adaptor-related protein complex 3, beta 2 subunit 245769 3 3 3 0 0 0 0 2 1 0 0.00138 0.560 0.391 0.668 0.531 0.00601 0.862 127 CPEB4 cytoplasmic polyadenylation element binding protein 4 185085 3 3 3 0 0 0 0 1 2 0 0.00181 0.833 0.144 1.000 0.405 0.00603 0.862 128 TUBA3C tubulin, alpha 3c 113959 2 2 2 1 0 1 0 0 1 0 0.00623 0.700 NaN NaN NaN 0.00623 0.882 129 GPR37L1 G protein-coupled receptor 37 like 1 119958 2 2 2 0 1 1 0 0 0 0 0.00280 0.377 0.502 0.0962 0.280 0.00638 0.894 130 CCDC140 coiled-coil domain containing 140 40036 1 1 1 0 0 0 0 1 0 0 0.00646 0.822 NaN NaN NaN 0.00646 0.894 131 BARX2 BARX homeobox 2 70698 1 1 1 0 0 0 1 0 0 0 0.00648 0.777 NaN NaN NaN 0.00648 0.894 132 TMOD2 tropomodulin 2 (neuronal) 90480 1 1 1 0 0 0 1 0 0 0 0.00651 0.617 NaN NaN NaN 0.00651 0.894 133 MUC21 mucin 21, cell surface associated 142134 2 2 2 1 0 0 1 1 0 0 0.00275 0.737 0.0206 1.000 0.298 0.00664 0.904 134 PSG3 pregnancy specific beta-1-glycoprotein 3 108813 2 2 2 0 2 0 0 0 0 0 0.00139 0.506 0.593 0.370 0.598 0.00671 0.904 135 CTDSP2 CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 68706 1 1 1 0 0 0 1 0 0 0 0.00687 0.773 NaN NaN NaN 0.00687 0.904 136 FHIT fragile histidine triad gene 38456 1 1 1 0 0 1 0 0 0 0 0.00694 0.679 NaN NaN NaN 0.00694 0.904 137 COMMD4 COMM domain containing 4 48730 1 1 1 0 0 0 1 0 0 0 0.00694 0.813 NaN NaN NaN 0.00694 0.904 138 CLVS1 clavesin 1 89971 2 2 2 0 0 1 0 1 0 0 0.000869 0.544 0.763 0.227 1.000 0.00700 0.904 139 MAPRE1 microtubule-associated protein, RP/EB family, member 1 68973 1 1 1 1 0 0 0 0 1 0 0.00701 0.954 NaN NaN NaN 0.00701 0.904 140 C20orf111 chromosome 20 open reading frame 111 73923 2 2 2 0 0 1 0 0 1 0 0.00123 0.696 0.387 0.767 0.713 0.00706 0.904 141 ZNF181 zinc finger protein 181 142663 3 2 3 0 0 1 0 1 1 0 0.00491 0.511 0.264 0.154 0.179 0.00708 0.904 142 TUSC5 tumor suppressor candidate 5 45312 1 1 1 0 0 0 0 1 0 0 0.00711 0.797 NaN NaN NaN 0.00711 0.904 143 ZSCAN23 zinc finger and SCAN domain containing 23 25955 1 1 1 0 0 1 0 0 0 0 0.00722 0.587 NaN NaN NaN 0.00722 0.904 144 MORF4L1 mortality factor 4 like 1 90631 1 1 1 0 0 0 1 0 0 0 0.00736 0.645 NaN NaN NaN 0.00736 0.904 145 C20orf194 chromosome 20 open reading frame 194 292036 2 2 2 0 1 0 0 1 0 0 0.0188 0.615 0.270 0.0695 0.0499 0.00747 0.904 146 LCE1C late cornified envelope 1C 29963 1 1 1 0 0 1 0 0 0 0 0.00757 0.629 NaN NaN NaN 0.00757 0.904 147 FGA fibrinogen alpha chain 216715 3 3 3 0 0 2 0 1 0 0 0.00269 0.397 0.192 0.690 0.356 0.00761 0.904 148 SPIC Spi-C transcription factor (Spi-1/PU.1 related) 63070 1 1 1 1 0 0 1 0 0 0 0.00762 0.908 NaN NaN NaN 0.00762 0.904 149 LELP1 late cornified envelope-like proline-rich 1 24951 1 1 1 0 1 0 0 0 0 0 0.00768 0.600 NaN NaN NaN 0.00768 0.904 150 OR10P1 olfactory receptor, family 10, subfamily P, member 1 78434 1 1 1 1 1 0 0 0 0 0 0.00771 0.933 NaN NaN NaN 0.00771 0.904 151 KRT6C keratin 6C 129323 2 2 2 0 1 0 0 1 0 0 0.0100 0.587 0.128 0.163 0.0972 0.00774 0.904 152 PRM3 protamine 3 11147 1 1 1 0 1 0 0 0 0 0 0.00775 0.739 NaN NaN NaN 0.00775 0.904 153 C7orf63 chromosome 7 open reading frame 63 228478 2 2 2 0 0 2 0 0 0 0 0.0287 0.475 0.232 0.0343 0.0341 0.00776 0.904 154 ATF7IP activating transcription factor 7 interacting protein 320964 3 3 3 0 0 1 0 1 1 0 0.00201 0.470 0.508 0.0818 0.488 0.00776 0.904 155 OR2G6 olfactory receptor, family 2, subfamily G, member 6 79230 2 2 2 0 1 0 0 1 0 0 0.000979 0.515 0.390 0.938 1.000 0.00776 0.904 156 MAS1L MAS1 oncogene-like 94537 1 1 1 0 0 0 1 0 0 0 0.00782 0.774 NaN NaN NaN 0.00782 0.904 157 KRTAP3-3 keratin associated protein 3-3 24983 1 1 1 1 1 0 0 0 0 0 0.00782 0.938 NaN NaN NaN 0.00782 0.904 158 OR4X1 olfactory receptor, family 4, subfamily X, member 1 76313 1 1 1 0 0 0 1 0 0 0 0.00794 0.762 NaN NaN NaN 0.00794 0.912 159 TMEM141 transmembrane protein 141 16915 1 1 1 0 0 1 0 0 0 0 0.00800 0.677 NaN NaN NaN 0.00800 0.912 160 ZNF99 zinc finger protein 99 259638 2 2 2 0 0 0 0 1 1 0 0.0130 0.877 0.130 0.107 0.0785 0.00805 0.913 161 LRIT2 leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 138107 2 2 2 0 1 0 0 1 0 0 0.00489 0.611 0.0486 0.535 0.212 0.00816 0.917 162 RAE1 RAE1 RNA export 1 homolog (S. pombe) 95345 2 2 2 0 0 1 0 1 0 0 0.00269 0.596 0.0302 0.822 0.390 0.00823 0.917 163 DERL3 Der1-like domain family, member 3 45303 1 1 1 0 0 0 0 1 0 0 0.00830 0.799 NaN NaN NaN 0.00830 0.917 164 TMEM167B transmembrane protein 167B 18923 1 1 1 0 0 0 0 1 0 0 0.00833 0.817 NaN NaN NaN 0.00833 0.917 165 GHITM growth hormone inducible transmembrane protein 88729 1 1 1 0 0 0 0 0 1 0 0.00834 0.697 NaN NaN NaN 0.00834 0.917 166 UTP3 UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) 119744 2 2 2 0 1 0 0 0 1 0 0.00370 0.738 0.891 0.215 0.289 0.00840 0.918 167 PRB1 proline-rich protein BstNI subfamily 1 82441 2 2 2 0 0 1 0 1 0 0 0.00109 0.560 0.615 0.931 1.000 0.00856 0.929 168 CGREF1 cell growth regulator with EF-hand domain 1 84091 1 1 1 0 0 0 1 0 0 0 0.00861 0.606 NaN NaN NaN 0.00861 0.929 169 MLLT10 myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 261908 3 3 2 0 0 0 0 2 1 0 0.00298 0.685 0.276 0.620 0.375 0.00871 0.934 170 METTL3 methyltransferase like 3 148280 2 2 2 0 0 0 0 2 0 0 0.0151 0.691 0.280 0.0569 0.0755 0.00889 0.939 171 HIST1H4K histone cluster 1, H4k 26228 1 1 1 0 0 0 0 1 0 0 0.00890 0.779 NaN NaN NaN 0.00890 0.939 172 ENAM enamelin 287223 2 2 2 0 0 0 0 1 1 0 0.0429 0.845 0.810 0.0236 0.0267 0.00891 0.939 173 TFAP2D transcription factor AP-2 delta (activating enhancer binding protein 2 delta) 115264 2 2 2 0 1 1 0 0 0 0 0.00833 0.416 0.0576 0.734 0.139 0.00898 0.941 174 LDOC1L leucine zipper, down-regulated in cancer 1-like 60078 1 1 1 0 0 0 0 0 1 0 0.00915 0.711 NaN NaN NaN 0.00915 0.951 175 KCNK6 potassium channel, subfamily K, member 6 63464 1 1 1 0 0 0 0 0 1 0 0.00921 0.616 NaN NaN NaN 0.00921 0.951 176 CCT8L2 chaperonin containing TCP1, subunit 8 (theta)-like 2 139274 2 2 2 0 0 1 0 1 0 0 0.00961 0.524 0.704 0.0747 0.125 0.00929 0.951 177 TNC tenascin C (hexabrachion) 557033 2 2 2 0 0 0 0 1 1 0 0.0813 0.679 0.990 0.0139 0.0149 0.00934 0.951 178 C16orf54 chromosome 16 open reading frame 54 21394 1 1 1 0 1 0 0 0 0 0 0.00939 0.733 NaN NaN NaN 0.00939 0.951 179 SALL1 sal-like 1 (Drosophila) 327740 3 3 3 1 1 1 1 0 0 0 0.00187 0.510 0.414 0.947 0.651 0.00939 0.951 180 KRTAP5-5 keratin associated protein 5-5 58232 1 1 1 0 1 0 0 0 0 0 0.00955 0.544 NaN NaN NaN 0.00955 0.959 181 SFRS2 splicing factor, arginine/serine-rich 2 52860 2 2 2 0 0 0 0 0 2 0 0.00125 0.764 0.375 0.708 1.000 0.00961 0.959 182 CCDC121 coiled-coil domain containing 121 79597 2 2 2 0 0 1 0 1 0 0 0.00206 0.530 0.481 0.394 0.608 0.00963 0.959 183 CD1B CD1b molecule 85064 1 1 1 0 0 0 0 0 1 0 0.00968 0.764 NaN NaN NaN 0.00968 0.959 184 SFI1 Sfi1 homolog, spindle assembly associated (yeast) 305151 2 2 2 0 0 0 1 0 1 0 0.0119 0.792 0.632 0.0789 0.108 0.00982 0.968 185 TM4SF18 transmembrane 4 L six family member 18 51787 1 1 1 0 0 0 0 0 1 0 0.0100 0.830 NaN NaN NaN 0.0100 0.969 186 AFG3L2 AFG3 ATPase family gene 3-like 2 (yeast) 194024 2 2 2 0 0 0 0 2 0 0 0.0280 0.694 0.732 0.0153 0.0471 0.0101 0.969 187 TRAF3IP3 TRAF3 interacting protein 3 141557 2 2 2 0 0 0 0 2 0 0 0.00643 0.730 0.338 0.175 0.205 0.0101 0.969 188 MYL6 myosin, light chain 6, alkali, smooth muscle and non-muscle 42579 2 1 2 0 0 0 0 2 0 0 0.0337 0.724 0.0104 0.590 0.0394 0.0101 0.969 189 KCND3 potassium voltage-gated channel, Shal-related subfamily, member 3 160578 2 2 2 0 0 1 0 1 0 0 0.0194 0.514 0.731 0.0326 0.0684 0.0101 0.969 190 OR4C16 olfactory receptor, family 4, subfamily C, member 16 77521 1 1 1 0 1 0 0 0 0 0 0.0102 0.667 NaN NaN NaN 0.0102 0.969 191 NLRP1 NLR family, pyrin domain containing 1 355867 3 3 3 0 0 1 0 1 1 0 0.00236 0.524 0.950 0.308 0.586 0.0105 0.976 192 HSD17B3 hydroxysteroid (17-beta) dehydrogenase 3 80943 1 1 1 0 1 0 0 0 0 0 0.0105 0.583 NaN NaN NaN 0.0105 0.976 193 SDF2L1 stromal cell-derived factor 2-like 1 26574 1 1 1 0 0 1 0 0 0 0 0.0105 0.669 NaN NaN NaN 0.0105 0.976 194 BEND5 BEN domain containing 5 72239 1 1 1 0 0 0 0 0 1 0 0.0105 1.000 NaN NaN NaN 0.0105 0.976 195 ATP6V1F ATPase, H+ transporting, lysosomal 14kDa, V1 subunit F 30486 1 1 1 0 0 1 0 0 0 0 0.0106 0.638 NaN NaN NaN 0.0106 0.976 196 AQP7 aquaporin 7 84601 1 1 1 0 1 0 0 0 0 0 0.0106 0.609 NaN NaN NaN 0.0106 0.976 197 C21orf70 chromosome 21 open reading frame 70 38253 1 1 1 0 0 0 0 1 0 0 0.0106 0.824 NaN NaN NaN 0.0106 0.976 198 CSTB cystatin B (stefin B) 19837 1 1 1 0 0 1 0 0 0 0 0.0107 0.654 NaN NaN NaN 0.0107 0.976 199 GABRE gamma-aminobutyric acid (GABA) A receptor, epsilon 118453 2 2 2 0 0 2 0 0 0 0 0.00537 0.434 0.0981 0.910 0.266 0.0108 0.976 200 RSPO2 R-spondin 2 homolog (Xenopus laevis) 62290 1 1 1 0 0 0 0 0 1 0 0.0108 1.000 NaN NaN NaN 0.0108 0.976 201 ZNF799 zinc finger protein 799 155407 3 2 3 1 0 1 0 2 0 0 0.0109 0.866 NaN NaN NaN 0.0109 0.976 202 OR2M2 olfactory receptor, family 2, subfamily M, member 2 86818 1 1 1 0 1 0 0 0 0 0 0.0109 0.773 NaN NaN NaN 0.0109 0.976 203 SECISBP2L SECIS binding protein 2-like 265338 3 3 2 0 0 0 0 3 0 0 0.00273 0.600 0.364 0.639 0.531 0.0109 0.976 204 EMR2 egf-like module containing, mucin-like, hormone receptor-like 2 209876 2 2 2 0 0 1 0 1 0 0 0.0328 0.535 0.369 0.0420 0.0447 0.0110 0.976 205 GZMM granzyme M (lymphocyte met-ase 1) 47037 1 1 1 0 0 0 0 1 0 0 0.0111 0.782 NaN NaN NaN 0.0111 0.976 206 CHP calcineurin-like EF hand protein 1 47133 1 1 1 0 1 0 0 0 0 0 0.0111 0.872 NaN NaN NaN 0.0111 0.976 207 GPM6A glycoprotein M6A 71957 1 1 1 0 1 0 0 0 0 0 0.0112 0.656 NaN NaN NaN 0.0112 0.978 208 CABP4 calcium binding protein 4 55451 1 1 1 0 0 1 0 0 0 0 0.0112 0.633 NaN NaN NaN 0.0112 0.978 209 C10orf47 chromosome 10 open reading frame 47 48287 1 1 1 0 1 0 0 0 0 0 0.0113 0.601 NaN NaN NaN 0.0113 0.978 210 SPRR2F small proline-rich protein 2F 18509 1 1 1 0 0 1 0 0 0 0 0.0114 0.659 NaN NaN NaN 0.0114 0.978 211 MPEG1 macrophage expressed 1 178755 3 3 3 0 0 1 0 2 0 0 0.00166 0.444 0.780 0.631 0.920 0.0114 0.978 212 CCDC144A coiled-coil domain containing 144A 267958 3 3 3 0 0 1 0 1 1 0 0.00153 0.589 0.921 0.361 1.000 0.0115 0.978 213 GPR135 G protein-coupled receptor 135 61846 1 1 1 0 0 0 0 1 0 0 0.0115 0.786 NaN NaN NaN 0.0115 0.978 214 SNRNP27 small nuclear ribonucleoprotein 27kDa (U4/U6.U5) 40820 1 1 1 0 0 0 0 0 1 0 0.0116 0.653 NaN NaN NaN 0.0116 0.978 215 MAGEB3 melanoma antigen family B, 3 83560 1 1 1 1 1 0 0 0 0 0 0.0116 0.950 NaN NaN NaN 0.0116 0.978 216 LCE1A late cornified envelope 1A 27888 1 1 1 0 1 0 0 0 0 0 0.0118 0.667 NaN NaN NaN 0.0118 0.984 217 HPSE heparanase 123259 1 1 1 0 0 0 1 0 0 0 0.0118 0.631 NaN NaN NaN 0.0118 0.984 218 OR5W2 olfactory receptor, family 5, subfamily W, member 2 77525 1 1 1 0 1 0 0 0 0 0 0.0122 0.833 NaN NaN NaN 0.0122 1.000 219 GZMA granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) 67133 1 1 1 0 1 0 0 0 0 0 0.0122 0.708 NaN NaN NaN 0.0122 1.000 220 C19orf53 chromosome 19 open reading frame 53 25569 1 1 1 0 0 0 0 0 1 0 0.0125 1.000 NaN NaN NaN 0.0125 1.000 221 ZNF747 zinc finger protein 747 21312 1 1 1 0 0 0 0 1 0 0 0.0125 0.791 NaN NaN NaN 0.0125 1.000 222 DHFR dihydrofolate reductase 48122 1 1 1 0 0 0 0 0 1 0 0.0126 1.000 NaN NaN NaN 0.0126 1.000 223 NEUROD2 neurogenic differentiation 2 64200 1 1 1 0 0 0 0 0 1 0 0.0128 1.000 NaN NaN NaN 0.0128 1.000 224 NHLH2 nescient helix loop helix 2 21885 1 1 1 0 0 0 0 1 0 0 0.0128 0.809 NaN NaN NaN 0.0128 1.000 225 ZNF257 zinc finger protein 257 140925 2 2 2 1 1 0 0 1 0 0 0.00326 0.945 0.481 0.268 0.542 0.0130 1.000 226 SKIV2L2 superkiller viralicidic activity 2-like 2 (S. cerevisiae) 267765 3 3 3 0 0 0 0 2 1 0 0.00375 0.381 0.321 0.869 0.476 0.0131 1.000 227 GIMAP7 GTPase, IMAP family member 7 75104 1 1 1 0 1 0 0 0 0 0 0.0131 0.734 NaN NaN NaN 0.0131 1.000 228 KYNU kynureninase (L-kynurenine hydrolase) 122108 2 2 2 0 0 0 0 1 1 0 0.00412 0.633 0.697 0.316 0.436 0.0132 1.000 229 FAM153A family with sequence similarity 153, member A 57748 1 1 1 0 1 0 0 0 0 0 0.0133 0.696 NaN NaN NaN 0.0133 1.000 230 OR5R1 olfactory receptor, family 5, subfamily R, member 1 81060 2 2 2 0 0 1 0 1 0 0 0.00414 0.542 0.0287 0.761 0.447 0.0135 1.000 231 SPATA1 spermatogenesis associated 1 78526 1 1 1 0 0 1 0 0 0 0 0.0137 0.664 NaN NaN NaN 0.0137 1.000 232 HSPBP1 HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 69694 1 1 1 0 0 0 1 0 0 0 0.0138 0.815 NaN NaN NaN 0.0138 1.000 233 HRNR hornerin 535873 3 3 3 0 1 1 0 1 0 0 0.0284 0.384 0.637 0.0218 0.0671 0.0138 1.000 234 RIPK2 receptor-interacting serine-threonine kinase 2 135030 1 1 1 0 0 0 1 0 0 0 0.0140 0.595 NaN NaN NaN 0.0140 1.000 235 FCAMR Fc receptor, IgA, IgM, high affinity 79437 1 1 1 0 0 0 1 0 0 0 0.0141 0.755 NaN NaN NaN 0.0141 1.000 236 CD68 CD68 molecule 89297 1 1 1 0 0 0 0 0 1 0 0.0141 1.000 NaN NaN NaN 0.0141 1.000 237 ACTR8 ARP8 actin-related protein 8 homolog (yeast) 149190 2 2 2 0 0 0 0 0 2 0 0.00195 0.842 0.930 0.804 1.000 0.0141 1.000 238 CDH18 cadherin 18, type 2 200548 2 2 2 0 1 1 0 0 0 0 0.0166 0.485 0.0613 0.508 0.119 0.0142 1.000 239 MMP24 matrix metallopeptidase 24 (membrane-inserted) 109926 1 1 1 0 0 0 1 0 0 0 0.0143 0.784 NaN NaN NaN 0.0143 1.000 240 GNB3 guanine nucleotide binding protein (G protein), beta polypeptide 3 82870 1 1 1 0 0 1 0 0 0 0 0.0143 0.653 NaN NaN NaN 0.0143 1.000 241 JUND jun D proto-oncogene 38710 1 1 1 0 0 1 0 0 0 0 0.0144 0.583 NaN NaN NaN 0.0144 1.000 242 BTNL8 butyrophilin-like 8 133009 1 1 1 1 1 0 0 0 0 0 0.0144 0.928 NaN NaN NaN 0.0144 1.000 243 C1orf109 chromosome 1 open reading frame 109 52094 1 1 1 0 0 0 0 1 0 0 0.0145 0.832 NaN NaN NaN 0.0145 1.000 244 C14orf129 chromosome 14 open reading frame 129 35511 1 1 1 0 1 0 0 0 0 0 0.0146 0.625 NaN NaN NaN 0.0146 1.000 245 HBG2 hemoglobin, gamma G 32573 1 1 1 0 0 1 0 0 0 0 0.0146 0.632 NaN NaN NaN 0.0146 1.000 246 AKIRIN1 akirin 1 31076 1 1 1 0 0 0 0 1 0 0 0.0147 0.856 NaN NaN NaN 0.0147 1.000 247 TIMD4 T-cell immunoglobulin and mucin domain containing 4 97186 1 1 1 0 0 0 0 0 1 0 0.0147 1.000 NaN NaN NaN 0.0147 1.000 248 KIAA1324L KIAA1324-like 192712 2 2 2 0 0 1 1 0 0 0 0.00218 0.391 0.569 0.588 0.942 0.0148 1.000 249 OR9Q2 olfactory receptor, family 9, subfamily Q, member 2 78767 2 2 2 0 1 0 0 1 0 0 0.00569 0.544 0.879 0.190 0.361 0.0148 1.000 250 FAM78B family with sequence similarity 78, member B 60159 1 1 1 0 1 0 0 0 0 0 0.0148 0.700 NaN NaN NaN 0.0148 1.000 251 SYCE1 synaptonemal complex central element protein 1 73863 1 1 1 0 1 0 0 0 0 0 0.0148 0.793 NaN NaN NaN 0.0148 1.000 252 OR8J1 olfactory receptor, family 8, subfamily J, member 1 79086 1 1 1 0 0 1 0 0 0 0 0.0150 0.657 NaN NaN NaN 0.0150 1.000 253 OR10A4 olfactory receptor, family 10, subfamily A, member 4 79016 1 1 1 0 1 0 0 0 0 0 0.0150 0.722 NaN NaN NaN 0.0150 1.000 254 ZDHHC20 zinc finger, DHHC-type containing 20 62138 1 1 1 0 0 0 0 0 1 0 0.0151 1.000 NaN NaN NaN 0.0151 1.000 255 EXD3 exonuclease 3-5 domain containing 3 142573 2 2 2 0 0 1 0 1 0 0 0.00949 0.491 0.0698 0.412 0.225 0.0153 1.000 256 NAP1L5 nucleosome assembly protein 1-like 5 45862 1 1 1 0 0 0 0 0 1 0 0.0154 1.000 NaN NaN NaN 0.0154 1.000 257 ATG5 ATG5 autophagy related 5 homolog (S. cerevisiae) 70881 1 1 1 0 0 0 0 0 1 0 0.0154 0.779 NaN NaN NaN 0.0154 1.000 258 C22orf43 chromosome 22 open reading frame 43 60351 1 1 1 0 0 0 0 0 1 0 0.0155 1.000 NaN NaN NaN 0.0155 1.000 259 NSFL1C NSFL1 (p97) cofactor (p47) 86264 1 1 1 0 0 0 0 0 1 0 0.0155 0.684 NaN NaN NaN 0.0155 1.000 260 LACTB lactamase, beta 109267 1 1 1 0 0 1 0 0 0 0 0.0156 0.688 NaN NaN NaN 0.0156 1.000 261 C22orf29 chromosome 22 open reading frame 29 89332 1 1 1 0 0 0 0 1 0 0 0.0156 0.803 NaN NaN NaN 0.0156 1.000 262 DGKQ diacylglycerol kinase, theta 110kDa 153726 2 2 2 0 0 0 1 1 0 0 0.00331 0.677 0.208 0.443 0.665 0.0157 1.000 263 TPRN taperin 95516 2 2 2 0 2 0 0 0 0 0 0.00597 0.406 0.630 0.363 0.371 0.0157 1.000 264 SULT1E1 sulfotransferase family 1E, estrogen-preferring, member 1 75649 1 1 1 0 0 0 0 0 1 0 0.0159 0.871 NaN NaN NaN 0.0159 1.000 265 SDF2 stromal cell-derived factor 2 53784 1 1 1 0 0 1 0 0 0 0 0.0159 0.660 NaN NaN NaN 0.0159 1.000 266 KRTAP5-9 keratin associated protein 5-9 42662 1 1 1 1 0 1 0 0 0 0 0.0161 0.844 NaN NaN NaN 0.0161 1.000 267 ZCCHC7 zinc finger, CCHC domain containing 7 138036 1 1 1 0 0 0 1 0 0 0 0.0161 0.638 NaN NaN NaN 0.0161 1.000 268 ATG10 ATG10 autophagy related 10 homolog (S. cerevisiae) 57021 1 1 1 0 0 0 0 0 1 0 0.0162 0.858 NaN NaN NaN 0.0162 1.000 269 ARGFX arginine-fifty homeobox 69969 1 1 1 0 1 0 0 0 0 0 0.0162 0.812 NaN NaN NaN 0.0162 1.000 270 LRRC2 leucine rich repeat containing 2 95092 1 1 1 0 0 1 0 0 0 0 0.0162 0.648 NaN NaN NaN 0.0162 1.000 271 NOP58 NOP58 ribonucleoprotein homolog (yeast) 136737 1 1 1 1 0 0 0 0 1 0 0.0162 0.955 NaN NaN NaN 0.0162 1.000 272 OR5B17 olfactory receptor, family 5, subfamily B, member 17 78591 1 1 1 0 0 0 0 1 0 0 0.0162 0.823 NaN NaN NaN 0.0162 1.000 273 COL11A2 collagen, type XI, alpha 2 421404 2 2 2 0 1 0 0 1 0 0 0.0652 0.565 0.205 0.0374 0.0353 0.0163 1.000 274 IFNAR2 interferon (alpha, beta and omega) receptor 2 140637 1 1 1 0 0 0 0 0 1 0 0.0164 0.670 NaN NaN NaN 0.0164 1.000 275 UGT2A1 UDP glucuronosyltransferase 2 family, polypeptide A1 133244 1 1 1 0 0 1 0 0 0 0 0.0165 0.687 NaN NaN NaN 0.0165 1.000 276 MYEOV myeloma overexpressed gene (in a subset of t(11;14) positive multiple myelomas) 74611 1 1 1 1 0 0 0 0 1 0 0.0165 1.000 NaN NaN NaN 0.0165 1.000 277 TMEM165 transmembrane protein 165 65322 1 1 1 0 0 0 0 0 1 0 0.0165 1.000 NaN NaN NaN 0.0165 1.000 278 IRF2 interferon regulatory factor 2 89773 1 1 1 0 0 0 0 0 1 0 0.0166 1.000 NaN NaN NaN 0.0166 1.000 279 C19orf75 50962 1 1 1 0 0 1 0 0 0 0 0.0166 0.669 NaN NaN NaN 0.0166 1.000 280 PSRC1 proline/serine-rich coiled-coil 1 87150 2 2 2 0 2 0 0 0 0 0 0.00488 0.727 0.634 0.135 0.488 0.0168 1.000 281 NR1I2 nuclear receptor subfamily 1, group I, member 2 120177 1 1 1 0 0 0 1 0 0 0 0.0168 0.757 NaN NaN NaN 0.0168 1.000 282 CEACAM6 carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) 87565 2 2 2 0 0 0 0 2 0 0 0.00239 0.650 0.568 0.456 1.000 0.0168 1.000 283 SSR3 signal sequence receptor, gamma (translocon-associated protein gamma) 47896 1 1 1 0 1 0 0 0 0 0 0.0169 0.654 NaN NaN NaN 0.0169 1.000 284 CXCR6 chemokine (C-X-C motif) receptor 6 85739 1 1 1 0 1 0 0 0 0 0 0.0169 0.687 NaN NaN NaN 0.0169 1.000 285 GRM5 glutamate receptor, metabotropic 5 262785 2 2 2 0 2 0 0 0 0 0 0.0169 0.418 NaN NaN NaN 0.0169 1.000 286 OR10G8 olfactory receptor, family 10, subfamily G, member 8 78020 2 2 2 0 1 1 0 0 0 0 0.00261 0.382 0.449 0.812 0.930 0.0170 1.000 287 TCN1 transcobalamin I (vitamin B12 binding protein, R binder family) 111054 1 1 1 0 0 0 1 0 0 0 0.0171 0.637 NaN NaN NaN 0.0171 1.000 288 RENBP renin binding protein 93840 1 1 1 0 0 0 0 0 1 0 0.0171 0.835 NaN NaN NaN 0.0171 1.000 289 TBXA2R thromboxane A2 receptor 65696 2 2 2 1 1 0 0 0 1 0 0.00397 0.656 0.305 0.372 0.618 0.0172 1.000 290 CYR61 cysteine-rich, angiogenic inducer, 61 94287 1 1 1 0 0 1 0 0 0 0 0.0172 0.641 NaN NaN NaN 0.0172 1.000 291 JAM2 junctional adhesion molecule 2 73473 1 1 1 0 1 0 0 0 0 0 0.0172 0.840 NaN NaN NaN 0.0172 1.000 292 C7orf28B chromosome 7 open reading frame 28B 96358 1 1 1 0 0 0 0 0 1 0 0.0172 0.880 NaN NaN NaN 0.0172 1.000 293 ETV3L ets variant gene 3-like 85761 2 2 2 0 1 0 0 1 0 0 0.00362 0.564 0.725 0.311 0.680 0.0172 1.000 294 SPSB3 splA/ryanodine receptor domain and SOCS box containing 3 88448 1 1 1 0 0 0 1 0 0 0 0.0173 0.864 NaN NaN NaN 0.0173 1.000 295 HIST1H2BG histone cluster 1, H2bg 31955 1 1 1 0 0 0 0 0 1 0 0.0173 1.000 NaN NaN NaN 0.0173 1.000 296 TRIM6 tripartite motif-containing 6 131389 1 1 1 0 0 0 1 0 0 0 0.0174 0.701 NaN NaN NaN 0.0174 1.000 297 SCHIP1 schwannomin interacting protein 1 78958 1 1 1 0 0 0 0 0 1 0 0.0174 1.000 NaN NaN NaN 0.0174 1.000 298 CLEC1A C-type lectin domain family 1, member A 71410 2 2 2 0 0 0 0 2 0 0 0.00253 0.726 0.213 0.442 0.983 0.0174 1.000 299 FTCD formiminotransferase cyclodeaminase 88512 1 1 1 0 0 0 0 0 1 0 0.0175 0.813 NaN NaN NaN 0.0175 1.000 300 CCNG1 cyclin G1 75364 1 1 1 0 0 0 0 0 1 0 0.0175 0.852 NaN NaN NaN 0.0175 1.000 301 TDO2 tryptophan 2,3-dioxygenase 103823 1 1 1 0 1 0 0 0 0 0 0.0177 0.821 NaN NaN NaN 0.0177 1.000 302 AQP9 aquaporin 9 75589 1 1 1 0 1 0 0 0 0 0 0.0177 0.643 NaN NaN NaN 0.0177 1.000 303 SH3GLB2 SH3-domain GRB2-like endophilin B2 76122 1 1 1 0 0 1 0 0 0 0 0.0178 0.626 NaN NaN NaN 0.0178 1.000 304 ATG12 ATG12 autophagy related 12 homolog (S. cerevisiae) 36327 1 1 1 0 1 0 0 0 0 0 0.0178 0.591 NaN NaN NaN 0.0178 1.000 305 DMXL2 Dmx-like 2 762674 4 4 4 0 0 0 0 1 3 0 0.00422 0.212 0.885 0.121 0.608 0.0179 1.000 306 RASL11B RAS-like, family 11, member B 63221 1 1 1 0 0 0 0 0 1 0 0.0179 1.000 NaN NaN NaN 0.0179 1.000 307 IGDCC4 immunoglobulin superfamily, DCC subclass, member 4 259218 2 2 2 0 1 0 0 0 1 0 0.0153 0.401 0.144 0.618 0.169 0.0179 1.000 308 TIMM22 translocase of inner mitochondrial membrane 22 homolog (yeast) 49251 1 1 1 0 0 0 0 0 1 0 0.0182 0.812 NaN NaN NaN 0.0182 1.000 309 SLC30A4 solute carrier family 30 (zinc transporter), member 4 109252 1 1 1 0 0 0 0 0 1 0 0.0182 1.000 NaN NaN NaN 0.0182 1.000 310 TFB2M transcription factor B2, mitochondrial 101427 2 2 2 0 0 1 0 1 0 0 0.00347 0.568 0.252 0.905 0.758 0.0182 1.000 311 MAL mal, T-cell differentiation protein 33073 1 1 1 0 1 0 0 0 0 0 0.0183 0.500 NaN NaN NaN 0.0183 1.000 312 BAI3 brain-specific angiogenesis inhibitor 3 388078 2 2 2 1 0 0 0 1 1 0 0.0440 0.894 0.320 0.0276 0.0599 0.0183 1.000 313 CARD16 caspase recruitment domain family, member 16 52290 1 1 1 0 0 0 0 1 0 0 0.0184 0.849 NaN NaN NaN 0.0184 1.000 314 RNASEH2C ribonuclease H2, subunit C 33152 1 1 1 0 0 0 0 0 1 0 0.0185 1.000 NaN NaN NaN 0.0185 1.000 315 CSPG5 chondroitin sulfate proteoglycan 5 (neuroglycan C) 125906 1 1 1 0 0 0 1 0 0 0 0.0185 0.684 NaN NaN NaN 0.0185 1.000 316 CFHR5 complement factor H-related 5 143611 1 1 1 1 1 0 0 0 0 0 0.0187 0.904 NaN NaN NaN 0.0187 1.000 317 ACER3 alkaline ceramidase 3 61407 1 1 1 0 0 0 0 1 0 0 0.0187 0.848 NaN NaN NaN 0.0187 1.000 318 PCDHA6 protocadherin alpha 6 239455 3 3 2 0 1 0 0 2 0 0 0.00464 0.382 0.414 0.740 0.588 0.0188 1.000 319 FBXO4 F-box protein 4 84959 2 2 2 0 0 0 0 1 1 0 0.00309 0.858 0.585 0.878 0.884 0.0189 1.000 320 KRTDAP keratinocyte differentiation-associated protein 26842 1 1 1 0 1 0 0 0 0 0 0.0189 0.591 NaN NaN NaN 0.0189 1.000 321 CYTL1 cytokine-like 1 29528 1 1 1 0 1 0 0 0 0 0 0.0189 0.708 NaN NaN NaN 0.0189 1.000 322 ARHGDIB Rho GDP dissociation inhibitor (GDI) beta 51958 1 1 1 0 1 0 0 0 0 0 0.0191 0.615 NaN NaN NaN 0.0191 1.000 323 PELO pelota homolog (Drosophila) 96678 1 1 1 0 0 0 0 1 0 0 0.0191 0.833 NaN NaN NaN 0.0191 1.000 324 ARID5A AT rich interactive domain 5A (MRF1-like) 134331 2 2 2 0 0 2 0 0 0 0 0.00726 0.402 0.109 0.869 0.383 0.0191 1.000 325 TBX3 T-box 3 (ulnar mammary syndrome) 115107 2 2 2 0 0 0 0 1 1 0 0.00307 0.829 0.557 0.419 0.910 0.0192 1.000 326 HIST1H2BK histone cluster 1, H2bk 31955 1 1 1 0 1 0 0 0 0 0 0.0192 0.600 NaN NaN NaN 0.0192 1.000 327 SCN7A sodium channel, voltage-gated, type VII, alpha 341589 3 3 3 1 0 1 0 2 0 0 0.00371 0.838 0.788 0.532 0.757 0.0193 1.000 328 RFK riboflavin kinase 37475 1 1 1 0 0 0 0 1 0 0 0.0193 0.856 NaN NaN NaN 0.0193 1.000 329 GLRA2 glycine receptor, alpha 2 121636 1 1 1 0 0 0 0 0 1 0 0.0194 0.694 NaN NaN NaN 0.0194 1.000 330 FAM171A1 family with sequence similarity 171, member A1 215966 2 2 2 0 0 1 0 0 1 0 0.00881 0.648 0.630 0.281 0.321 0.0194 1.000 331 PLAU plasminogen activator, urokinase 113128 1 1 1 0 0 0 0 0 1 0 0.0195 1.000 NaN NaN NaN 0.0195 1.000 332 RDH8 retinol dehydrogenase 8 (all-trans) 79432 1 1 1 0 0 0 0 1 0 0 0.0196 0.795 NaN NaN NaN 0.0196 1.000 333 TUBD1 tubulin, delta 1 115682 1 1 1 0 1 0 0 0 0 0 0.0196 0.769 NaN NaN NaN 0.0196 1.000 334 C8orf46 chromosome 8 open reading frame 46 43764 1 1 1 0 0 0 0 1 0 0 0.0197 0.809 NaN NaN NaN 0.0197 1.000 335 CALML6 calmodulin-like 6 36663 1 1 1 0 0 1 0 0 0 0 0.0197 0.657 NaN NaN NaN 0.0197 1.000 336 PROKR1 prokineticin receptor 1 98604 2 2 2 1 1 0 0 1 0 0 0.0105 0.857 0.103 0.253 0.276 0.0198 1.000 337 ANG angiogenin, ribonuclease, RNase A family, 5 37184 1 1 1 0 0 1 0 0 0 0 0.0198 0.626 NaN NaN NaN 0.0198 1.000 338 PALMD palmdelphin 139837 1 1 1 0 1 0 0 0 0 0 0.0198 0.607 NaN NaN NaN 0.0198 1.000 339 DEFA1B defensin, alpha 1B 18348 1 1 1 0 0 0 0 1 0 0 0.0198 0.825 NaN NaN NaN 0.0198 1.000 340 ZNF24 zinc finger protein 24 92872 1 1 1 0 0 1 0 0 0 0 0.0198 0.663 NaN NaN NaN 0.0198 1.000 341 SLFNL1 schlafen-like 1 92399 1 1 1 0 0 1 0 0 0 0 0.0199 0.591 NaN NaN NaN 0.0199 1.000 342 ZNF283 zinc finger protein 283 158673 1 1 1 1 0 0 1 0 0 0 0.0199 0.967 NaN NaN NaN 0.0199 1.000 343 TRYX3 protease, serine, 58 61915 1 1 1 1 0 1 0 0 0 0 0.0199 0.948 NaN NaN NaN 0.0199 1.000 344 HS6ST1 heparan sulfate 6-O-sulfotransferase 1 75612 1 1 1 0 0 0 0 0 1 0 0.0199 0.659 NaN NaN NaN 0.0199 1.000 345 TBC1D13 TBC1 domain family, member 13 101537 1 1 1 0 0 0 0 0 1 0 0.0202 1.000 NaN NaN NaN 0.0202 1.000 346 PRKCSH protein kinase C substrate 80K-H 121768 1 1 1 1 0 0 1 0 0 0 0.0202 0.924 NaN NaN NaN 0.0202 1.000 347 SKA1 spindle and kinetochore associated complex subunit 1 65676 1 1 1 0 0 1 0 0 0 0 0.0203 0.676 NaN NaN NaN 0.0203 1.000 348 RTL1 retrotransposon-like 1 99791 1 1 1 0 0 1 0 0 0 0 0.0204 0.660 NaN NaN NaN 0.0204 1.000 349 LPPR5 81898 1 1 1 0 0 0 0 0 1 0 0.0204 0.606 NaN NaN NaN 0.0204 1.000 350 CILP cartilage intermediate layer protein, nucleotide pyrophosphohydrolase 297572 2 2 2 1 1 0 0 0 1 0 0.0117 0.911 0.629 0.0634 0.258 0.0205 1.000 351 DBR1 debranching enzyme homolog 1 (S. cerevisiae) 137534 1 1 1 0 0 0 0 0 1 0 0.0206 1.000 NaN NaN NaN 0.0206 1.000 352 CDCP2 CUB domain containing protein 2 113315 1 1 1 0 1 0 0 0 0 0 0.0208 0.600 NaN NaN NaN 0.0208 1.000 353 PVRIG poliovirus receptor related immunoglobulin domain containing 63218 1 1 1 0 0 1 0 0 0 0 0.0208 0.671 NaN NaN NaN 0.0208 1.000 354 OR7A5 olfactory receptor, family 7, subfamily A, member 5 80012 2 2 2 0 0 0 0 1 1 0 0.00306 0.665 0.569 0.867 1.000 0.0208 1.000 355 C14orf178 chromosome 14 open reading frame 178 21667 1 1 1 0 0 1 0 0 0 0 0.0208 0.702 NaN NaN NaN 0.0208 1.000 356 NUSAP1 nucleolar and spindle associated protein 1 94490 1 1 1 0 0 1 0 0 0 0 0.0209 0.683 NaN NaN NaN 0.0209 1.000 357 UGT2B10 UDP glucuronosyltransferase 2 family, polypeptide B10 264942 2 2 2 1 0 0 0 1 1 0 0.0196 0.856 0.114 0.187 0.158 0.0209 1.000 358 PON2 paraoxonase 2 84482 1 1 1 0 0 0 1 0 0 0 0.0210 0.558 NaN NaN NaN 0.0210 1.000 359 RPE65 retinal pigment epithelium-specific protein 65kDa 137463 2 2 2 0 0 0 0 1 1 0 0.00644 0.507 0.115 0.928 0.483 0.0211 1.000 360 INADL InaD-like (Drosophila) 462165 4 4 4 0 0 1 0 2 1 0 0.00312 0.314 0.820 0.826 1.000 0.0211 1.000 361 C1QL3 complement component 1, q subcomponent-like 3 44587 1 1 1 0 0 0 0 1 0 0 0.0212 0.827 NaN NaN NaN 0.0212 1.000 362 OR4C13 olfactory receptor, family 4, subfamily C, member 13 77506 1 1 1 0 0 1 0 0 0 0 0.0212 0.668 NaN NaN NaN 0.0212 1.000 363 ST6GALNAC2 ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 82669 1 1 1 0 0 1 0 0 0 0 0.0212 0.643 NaN NaN NaN 0.0212 1.000 364 SPC24 SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) 18508 1 1 1 0 0 0 0 1 0 0 0.0213 0.849 NaN NaN NaN 0.0213 1.000 365 XRCC3 X-ray repair complementing defective repair in Chinese hamster cells 3 49472 1 1 1 0 1 0 0 0 0 0 0.0213 0.705 NaN NaN NaN 0.0213 1.000 366 EFCAB1 EF-hand calcium binding domain 1 54723 1 1 1 0 0 1 0 0 0 0 0.0214 0.670 NaN NaN NaN 0.0214 1.000 367 MAPKAPK3 mitogen-activated protein kinase-activated protein kinase 3 94531 1 1 1 0 0 0 0 0 1 0 0.0215 1.000 NaN NaN NaN 0.0215 1.000 368 LYST lysosomal trafficking regulator 958359 2 2 2 0 1 1 0 0 0 0 0.229 0.491 0.0883 0.00848 0.0139 0.0215 1.000 369 POTED POTE ankyrin domain family, member D 40820 1 1 1 0 0 1 0 0 0 0 0.0216 0.668 NaN NaN NaN 0.0216 1.000 370 HRASLS2 HRAS-like suppressor 2 41904 1 1 1 0 1 0 0 0 0 0 0.0216 0.647 NaN NaN NaN 0.0216 1.000 371 FAM163A family with sequence similarity 163, member A 34445 1 1 1 0 0 1 0 0 0 0 0.0217 0.651 NaN NaN NaN 0.0217 1.000 372 ETV5 ets variant gene 5 (ets-related molecule) 131154 1 1 1 0 1 0 0 0 0 0 0.0217 0.726 NaN NaN NaN 0.0217 1.000 373 ZNF177 zinc finger protein 177 95379 1 1 1 0 0 0 0 1 0 0 0.0218 0.861 NaN NaN NaN 0.0218 1.000 374 ATP8B4 ATPase, class I, type 8B, member 4 305620 2 2 2 0 0 1 0 0 1 0 0.00904 0.675 0.0810 0.582 0.359 0.0218 1.000 375 ZNRF2 zinc and ring finger 2 23678 1 1 1 0 0 0 0 1 0 0 0.0220 0.856 NaN NaN NaN 0.0220 1.000 376 ZCCHC10 zinc finger, CCHC domain containing 10 43838 1 1 1 0 0 0 0 0 1 0 0.0220 1.000 NaN NaN NaN 0.0220 1.000 377 ADAMTSL1 ADAMTS-like 1 410611 2 2 2 0 0 2 0 0 0 0 0.0406 0.428 0.320 0.0354 0.0810 0.0221 1.000 378 DSN1 DSN1, MIND kinetochore complex component, homolog (S. cerevisiae) 92083 1 1 1 0 0 1 0 0 0 0 0.0221 0.683 NaN NaN NaN 0.0221 1.000 379 ZBTB34 zinc finger and BTB domain containing 34 124985 1 1 1 0 0 0 0 0 1 0 0.0223 0.644 NaN NaN NaN 0.0223 1.000 380 KIAA1704 KIAA1704 87483 1 1 1 0 0 0 0 0 1 0 0.0223 0.857 NaN NaN NaN 0.0223 1.000 381 OR13D1 olfactory receptor, family 13, subfamily D, member 1 86605 1 1 1 0 0 1 0 0 0 0 0.0223 0.649 NaN NaN NaN 0.0223 1.000 382 ADRA2A adrenergic, alpha-2A-, receptor 67894 1 1 1 0 0 0 0 1 0 0 0.0224 0.799 NaN NaN NaN 0.0224 1.000 383 ASH1L ash1 (absent, small, or homeotic)-like (Drosophila) 747177 4 4 4 0 0 0 0 2 2 0 0.00665 0.597 0.233 0.949 0.502 0.0224 1.000 384 C1QTNF7 C1q and tumor necrosis factor related protein 7 73538 1 1 1 0 1 0 0 0 0 0 0.0224 0.588 NaN NaN NaN 0.0224 1.000 385 IFNA13 interferon, alpha 13 46094 1 1 1 0 0 1 0 0 0 0 0.0225 0.632 NaN NaN NaN 0.0225 1.000 386 PATE2 prostate and testis expressed 2 29690 1 1 1 0 0 1 0 0 0 0 0.0225 0.661 NaN NaN NaN 0.0225 1.000 387 HTR5A 5-hydroxytryptamine (serotonin) receptor 5A 89806 1 1 1 0 0 1 0 0 0 0 0.0225 0.630 NaN NaN NaN 0.0225 1.000 388 ARFGAP3 ADP-ribosylation factor GTPase activating protein 3 127260 1 1 1 0 0 0 0 0 1 0 0.0226 1.000 NaN NaN NaN 0.0226 1.000 389 ZCCHC5 zinc finger, CCHC domain containing 5 113384 1 1 1 0 1 0 0 0 0 0 0.0226 0.822 NaN NaN NaN 0.0226 1.000 390 OR51D1 olfactory receptor, family 51, subfamily D, member 1 81257 1 1 1 0 0 0 0 0 1 0 0.0227 1.000 NaN NaN NaN 0.0227 1.000 391 C22orf40 31739 1 1 1 0 0 1 0 0 0 0 0.0228 0.640 NaN NaN NaN 0.0228 1.000 392 DMRT2 doublesex and mab-3 related transcription factor 2 102703 2 2 2 0 1 1 0 0 0 0 0.00366 0.437 0.379 0.759 0.936 0.0228 1.000 393 LCE1F late cornified envelope 1F 29880 1 1 1 0 0 0 0 1 0 0 0.0229 0.831 NaN NaN NaN 0.0229 1.000 394 PRIM2 primase, DNA, polypeptide 2 (58kDa) 120943 1 1 1 1 0 0 0 1 0 0 0.0229 0.955 NaN NaN NaN 0.0229 1.000 395 OSGIN1 oxidative stress induced growth inhibitor 1 122885 1 1 1 1 0 0 1 0 0 0 0.0229 0.945 NaN NaN NaN 0.0229 1.000 396 C8orf45 chromosome 8 open reading frame 45 150757 1 1 1 0 1 0 0 0 0 0 0.0229 0.773 NaN NaN NaN 0.0229 1.000 397 SALL3 sal-like 3 (Drosophila) 211699 3 3 3 1 1 0 0 2 0 0 0.00625 0.678 0.393 0.422 0.556 0.0232 1.000 398 KLK2 kallikrein-related peptidase 2 69301 1 1 1 0 0 0 0 0 1 0 0.0232 1.000 NaN NaN NaN 0.0232 1.000 399 ZNF395 zinc finger protein 395 123796 1 1 1 0 0 0 0 0 1 0 0.0232 1.000 NaN NaN NaN 0.0232 1.000 400 DEFB135 defensin, beta 135 20074 1 1 1 0 0 0 0 1 0 0 0.0232 0.832 NaN NaN NaN 0.0232 1.000 401 TCEB2 transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B) 38874 1 1 1 0 0 0 0 0 1 0 0.0232 0.639 NaN NaN NaN 0.0232 1.000 402 DACT1 dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis) 179417 2 2 2 0 1 0 1 0 0 0 0.00532 0.444 0.341 0.923 0.656 0.0232 1.000 403 C10orf99 chromosome 10 open reading frame 99 21414 1 1 1 0 0 0 0 1 0 0 0.0232 0.801 NaN NaN NaN 0.0232 1.000 404 NTNG1 netrin G1 141446 2 2 2 0 1 1 0 0 0 0 0.0104 0.413 0.584 0.241 0.338 0.0233 1.000 405 OR13C2 olfactory receptor, family 13, subfamily C, member 2 79652 1 1 1 0 0 1 0 0 0 0 0.0234 0.665 NaN NaN NaN 0.0234 1.000 406 FRG2B FSHD region gene 2 family, member B 50823 1 1 1 0 0 1 0 0 0 0 0.0235 0.657 NaN NaN NaN 0.0235 1.000 407 KLK3 kallikrein-related peptidase 3 74729 2 2 2 0 0 0 0 2 0 0 0.00454 0.655 0.465 0.671 0.780 0.0235 1.000 408 PICK1 protein interacting with PRKCA 1 98779 1 1 1 0 0 1 0 0 0 0 0.0235 0.635 NaN NaN NaN 0.0235 1.000 409 TAAR9 trace amine associated receptor 9 86216 1 1 1 0 1 0 0 0 0 0 0.0235 0.531 NaN NaN NaN 0.0235 1.000 410 KRTAP4-8 keratin associated protein 4-8 35563 1 1 1 0 0 1 0 0 0 0 0.0236 0.595 NaN NaN NaN 0.0236 1.000 411 PROZ protein Z, vitamin K-dependent plasma glycoprotein 91691 2 2 2 0 0 1 0 1 0 0 0.00756 0.545 0.211 0.451 0.470 0.0236 1.000 412 HAVCR1 hepatitis A virus cellular receptor 1 92978 1 1 1 0 0 1 0 0 0 0 0.0236 0.689 NaN NaN NaN 0.0236 1.000 413 NXN nucleoredoxin 85529 1 1 1 0 0 0 0 0 1 0 0.0236 0.823 NaN NaN NaN 0.0236 1.000 414 NUDT6 nudix (nucleoside diphosphate linked moiety X)-type motif 6 79980 2 1 2 0 1 0 1 0 0 0 0.00985 0.379 0.510 0.204 0.362 0.0237 1.000 415 ERCC2 excision repair cross-complementing rodent repair deficiency, complementation group 2 (xeroderma pigmentosum D) 178779 2 2 2 0 1 1 0 0 0 0 0.0158 0.429 0.0502 0.598 0.226 0.0237 1.000 416 TCEAL6 transcription elongation factor A (SII)-like 6 45948 1 1 1 0 1 0 0 0 0 0 0.0237 0.606 NaN NaN NaN 0.0237 1.000 417 CYP2B6 cytochrome P450, family 2, subfamily B, polypeptide 6 125356 1 1 1 0 1 0 0 0 0 0 0.0239 0.750 NaN NaN NaN 0.0239 1.000 418 ABHD10 abhydrolase domain containing 10 78086 1 1 1 0 0 0 0 0 1 0 0.0240 1.000 NaN NaN NaN 0.0240 1.000 419 PSG11 pregnancy specific beta-1-glycoprotein 11 85276 1 1 1 0 0 0 0 0 1 0 0.0240 0.821 NaN NaN NaN 0.0240 1.000 420 LILRA4 leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 4 127121 2 2 2 0 2 0 0 0 0 0 0.00607 0.394 0.952 0.301 0.599 0.0240 1.000 421 AGK acylglycerol kinase 104171 1 1 1 0 0 0 0 0 1 0 0.0241 0.836 NaN NaN NaN 0.0241 1.000 422 TAL2 T-cell acute lymphocytic leukemia 2 27473 1 1 1 0 0 0 0 1 0 0 0.0241 0.832 NaN NaN NaN 0.0241 1.000 423 WNT2 wingless-type MMTV integration site family member 2 91401 1 1 1 0 0 0 0 1 0 0 0.0243 0.839 NaN NaN NaN 0.0243 1.000 424 HOOK3 hook homolog 3 (Drosophila) 179953 1 1 1 0 0 0 1 0 0 0 0.0243 0.607 NaN NaN NaN 0.0243 1.000 425 C4orf36 chromosome 4 open reading frame 36 30365 1 1 1 0 0 0 0 1 0 0 0.0245 0.834 NaN NaN NaN 0.0245 1.000 426 ANKMY2 ankyrin repeat and MYND domain containing 2 113198 1 1 1 0 0 0 0 1 0 0 0.0246 0.859 NaN NaN NaN 0.0246 1.000 427 WBSCR27 Williams Beuren syndrome chromosome region 27 59227 1 1 1 0 0 1 0 0 0 0 0.0246 0.633 NaN NaN NaN 0.0246 1.000 428 SEPT9 septin 9 155818 2 2 2 0 1 1 0 0 0 0 0.0279 0.437 0.608 0.110 0.135 0.0248 1.000 429 PLAC1L placenta-specific 1-like 40896 1 1 1 0 0 0 0 1 0 0 0.0248 0.839 NaN NaN NaN 0.0248 1.000 430 SEC61A2 Sec61 alpha 2 subunit (S. cerevisiae) 121815 1 1 1 0 0 0 0 0 1 0 0.0248 0.655 NaN NaN NaN 0.0248 1.000 431 SRP68 signal recognition particle 68kDa 158054 1 1 1 0 0 0 0 0 1 0 0.0248 0.848 NaN NaN NaN 0.0248 1.000 432 FAM170A family with sequence similarity 170, member A 83498 1 1 1 0 0 1 0 0 0 0 0.0249 0.675 NaN NaN NaN 0.0249 1.000 433 PRSS48 protease, serine, 48 83398 2 2 2 0 0 2 0 0 0 0 0.00379 0.473 0.953 0.948 1.000 0.0249 1.000 434 HAPLN4 hyaluronan and proteoglycan link protein 4 66114 1 1 1 0 0 1 0 0 0 0 0.0250 0.643 NaN NaN NaN 0.0250 1.000 435 TPPP2 tubulin polymerization-promoting protein family member 2 43419 1 1 1 0 0 1 0 0 0 0 0.0251 0.678 NaN NaN NaN 0.0251 1.000 436 PANX3 pannexin 3 99171 1 1 1 0 1 0 0 0 0 0 0.0251 0.776 NaN NaN NaN 0.0251 1.000 437 SURF4 surfeit 4 63850 1 1 1 0 0 0 0 0 1 0 0.0252 1.000 NaN NaN NaN 0.0252 1.000 438 C1orf192 chromosome 1 open reading frame 192 45977 1 1 1 0 0 1 0 0 0 0 0.0252 0.687 NaN NaN NaN 0.0252 1.000 439 CD300LB CD300 molecule-like family member b 60832 1 1 1 0 0 0 0 0 1 0 0.0253 1.000 NaN NaN NaN 0.0253 1.000 440 PLEKHA2 pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 91959 1 1 1 0 0 0 0 0 1 0 0.0254 1.000 NaN NaN NaN 0.0254 1.000 441 IQGAP1 IQ motif containing GTPase activating protein 1 415725 2 2 2 0 0 0 0 2 0 0 0.0985 0.727 0.532 0.0366 0.0394 0.0254 1.000 442 SLC10A2 solute carrier family 10 (sodium/bile acid cotransporter family), member 2 88890 2 2 2 0 1 0 0 1 0 0 0.00633 0.433 0.831 0.446 0.614 0.0254 1.000 443 SPATA19 spermatogenesis associated 19 43824 1 1 1 0 0 0 0 0 1 0 0.0256 0.833 NaN NaN NaN 0.0256 1.000 444 SPHKAP SPHK1 interactor, AKAP domain containing 425854 3 3 3 0 2 1 0 0 0 0 0.00648 0.250 0.966 0.379 0.603 0.0256 1.000 445 AATK apoptosis-associated tyrosine kinase 129220 1 1 1 0 0 0 1 0 0 0 0.0257 0.799 NaN NaN NaN 0.0257 1.000 446 STAT3 signal transducer and activator of transcription 3 (acute-phase response factor) 198658 2 2 2 1 1 0 0 0 1 0 0.00392 0.802 0.834 0.574 1.000 0.0257 1.000 447 HN1 hematological and neurological expressed 1 41676 1 1 1 0 0 1 0 0 0 0 0.0258 0.759 NaN NaN NaN 0.0258 1.000 448 CDKN2AIPNL CDKN2A interacting protein N-terminal like 23506 1 1 1 0 1 0 0 0 0 0 0.0258 0.690 NaN NaN NaN 0.0258 1.000 449 ASPHD2 aspartate beta-hydroxylase domain containing 2 93123 2 2 2 0 2 0 0 0 0 0 0.00396 0.437 0.810 0.287 1.000 0.0258 1.000 450 IGF2BP2 insulin-like growth factor 2 mRNA binding protein 2 148402 1 1 1 0 0 0 1 0 0 0 0.0259 0.666 NaN NaN NaN 0.0259 1.000 451 MSTN myostatin 94596 1 1 1 0 0 0 0 0 1 0 0.0261 1.000 NaN NaN NaN 0.0261 1.000 452 MRPS30 mitochondrial ribosomal protein S30 108566 1 1 1 0 0 0 0 1 0 0 0.0262 0.825 NaN NaN NaN 0.0262 1.000 453 HIST1H2BN histone cluster 1, H2bn 31955 1 1 1 0 0 0 0 1 0 0 0.0263 0.815 NaN NaN NaN 0.0263 1.000 454 MUC17 mucin 17, cell surface associated 1123286 5 5 5 1 1 3 0 1 0 0 0.00667 0.433 0.299 0.694 0.605 0.0263 1.000 455 TBL1XR1 transducin (beta)-like 1 X-linked receptor 1 118320 2 2 2 0 0 0 0 0 2 0 0.00425 0.839 0.545 0.642 0.950 0.0263 1.000 456 CRIPT cysteine-rich PDZ-binding protein 25319 1 1 1 0 0 0 0 0 1 0 0.0264 1.000 NaN NaN NaN 0.0264 1.000 457 SCAMP1 secretory carrier membrane protein 1 76534 1 1 1 0 0 0 0 0 1 0 0.0265 1.000 NaN NaN NaN 0.0265 1.000 458 FAM71B family with sequence similarity 71, member B 151558 1 1 1 0 0 0 1 0 0 0 0.0265 0.584 NaN NaN NaN 0.0265 1.000 459 B3GAT3 beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) 84866 2 2 2 0 0 1 0 1 0 0 0.00409 0.480 0.937 0.620 1.000 0.0266 1.000 460 C11orf70 chromosome 11 open reading frame 70 59262 1 1 1 0 0 0 0 0 1 0 0.0266 0.857 NaN NaN NaN 0.0266 1.000 461 SIRPB1 signal-regulatory protein beta 1 134106 1 1 1 1 0 0 1 0 0 0 0.0267 0.883 NaN NaN NaN 0.0267 1.000 462 KRT4 keratin 4 151101 1 1 1 0 1 0 0 0 0 0 0.0268 0.667 NaN NaN NaN 0.0268 1.000 463 C11orf83 chromosome 11 open reading frame 83 22781 1 1 1 0 1 0 0 0 0 0 0.0269 0.574 NaN NaN NaN 0.0269 1.000 464 REEP4 receptor accessory protein 4 60750 1 1 1 0 0 0 0 0 1 0 0.0269 1.000 NaN NaN NaN 0.0269 1.000 465 SNTB1 syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) 125835 1 1 1 1 0 1 0 0 0 0 0.0270 0.937 NaN NaN NaN 0.0270 1.000 466 MYL1 myosin, light chain 1, alkali; skeletal, fast 51969 1 1 1 0 0 1 0 0 0 0 0.0271 0.695 NaN NaN NaN 0.0271 1.000 467 H1F0 H1 histone family, member 0 48873 1 1 1 0 0 1 0 0 0 0 0.0271 0.646 NaN NaN NaN 0.0271 1.000 468 KIF3B kinesin family member 3B 188559 2 2 2 0 1 0 0 0 1 0 0.00586 0.609 0.442 0.274 0.716 0.0272 1.000 469 CXorf58 chromosome X open reading frame 58 84687 1 1 1 0 0 1 0 0 0 0 0.0273 0.684 NaN NaN NaN 0.0273 1.000 470 FAM127A family with sequence similarity 127, member A 28669 1 1 1 0 0 0 0 1 0 0 0.0273 0.816 NaN NaN NaN 0.0273 1.000 471 SLC2A14 solute carrier family 2 (facilitated glucose transporter), member 14 125428 1 1 1 0 0 1 0 0 0 0 0.0274 0.666 NaN NaN NaN 0.0274 1.000 472 MRPL30 mitochondrial ribosomal protein L30 41824 1 1 1 0 0 1 0 0 0 0 0.0274 0.696 NaN NaN NaN 0.0274 1.000 473 OC90 otoconin 90 105785 1 1 1 0 1 0 0 0 0 0 0.0274 0.631 NaN NaN NaN 0.0274 1.000 474 RHBDD1 rhomboid domain containing 1 80517 2 1 2 0 0 0 0 1 1 0 0.0326 0.566 0.0478 0.238 0.130 0.0275 1.000 475 ZIC1 Zic family member 1 (odd-paired homolog, Drosophila) 111694 2 2 2 0 1 0 0 1 0 0 0.00832 0.460 0.529 0.0659 0.511 0.0275 1.000 476 DRD2 dopamine receptor D2 112826 1 1 1 0 0 1 0 0 0 0 0.0275 0.638 NaN NaN NaN 0.0275 1.000 477 KBTBD7 kelch repeat and BTB (POZ) domain containing 7 170733 1 1 1 0 0 0 1 0 0 0 0.0276 0.678 NaN NaN NaN 0.0276 1.000 478 MRPL20 mitochondrial ribosomal protein L20 36356 1 1 1 0 0 0 0 1 0 0 0.0276 0.831 NaN NaN NaN 0.0276 1.000 479 GSK3A glycogen synthase kinase 3 alpha 88503 1 1 1 0 0 0 0 1 0 0 0.0279 0.817 NaN NaN NaN 0.0279 1.000 480 GNAO1 guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O 115408 1 1 1 0 0 0 1 0 0 0 0.0279 0.752 NaN NaN NaN 0.0279 1.000 481 PHYH phytanoyl-CoA 2-hydroxylase 80752 1 1 1 0 1 0 0 0 0 0 0.0280 0.629 NaN NaN NaN 0.0280 1.000 482 B3GALNT1 beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) 82944 1 1 1 0 1 0 0 0 0 0 0.0281 0.682 NaN NaN NaN 0.0281 1.000 483 MAP9 microtubule-associated protein 9 157567 1 1 1 0 0 0 1 0 0 0 0.0281 0.508 NaN NaN NaN 0.0281 1.000 484 CDH5 cadherin 5, type 2, VE-cadherin (vascular epithelium) 189904 2 2 2 0 1 1 0 0 0 0 0.0238 0.408 0.448 0.142 0.183 0.0281 1.000 485 FAM86A family with sequence similarity 86, member A 77320 2 1 2 0 1 1 0 0 0 0 0.0450 0.421 0.0119 0.299 0.0973 0.0281 1.000 486 CRTAC1 cartilage acidic protein 1 147802 2 2 2 0 0 0 0 1 1 0 0.0135 0.580 0.293 0.391 0.324 0.0282 1.000 487 THTPA thiamine triphosphatase 57908 1 1 1 0 1 0 0 0 0 0 0.0282 0.853 NaN NaN NaN 0.0282 1.000 488 CDH19 cadherin 19, type 2 195829 1 1 1 0 1 0 0 0 0 0 0.0283 0.707 NaN NaN NaN 0.0283 1.000 489 FAM81B family with sequence similarity 81, member B 115791 1 1 1 0 1 0 0 0 0 0 0.0283 0.700 NaN NaN NaN 0.0283 1.000 490 OTOP1 otopetrin 1 135710 2 1 2 0 1 0 0 0 1 0 0.117 0.636 0.0102 0.118 0.0377 0.0283 1.000 491 ISCU iron-sulfur cluster scaffold homolog (E. coli) 37101 1 1 1 0 0 0 0 1 0 0 0.0285 0.841 NaN NaN NaN 0.0285 1.000 492 CSMD3 CUB and Sushi multiple domains 3 950868 4 4 4 1 1 1 0 0 2 0 0.0128 0.721 0.310 0.172 0.348 0.0286 1.000 493 CASP1 caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) 103827 2 2 2 0 1 0 0 1 0 0 0.00639 0.622 0.198 0.650 0.699 0.0286 1.000 494 PEMT phosphatidylethanolamine N-methyltransferase 55134 1 1 1 1 0 1 0 0 0 0 0.0286 0.844 NaN NaN NaN 0.0286 1.000 495 DKK2 dickkopf homolog 2 (Xenopus laevis) 66068 2 2 2 0 2 0 0 0 0 0 0.00448 0.504 0.522 0.124 1.000 0.0287 1.000 496 GABRA4 gamma-aminobutyric acid (GABA) A receptor, alpha 4 141044 1 1 1 0 0 0 0 1 0 0 0.0287 0.824 NaN NaN NaN 0.0287 1.000 497 SFRP2 secreted frizzled-related protein 2 74137 1 1 1 0 0 1 0 0 0 0 0.0288 0.623 NaN NaN NaN 0.0288 1.000 498 GABRR2 gamma-aminobutyric acid (GABA) receptor, rho 2 123616 2 2 1 0 2 0 0 0 0 0 0.00451 0.421 0.0177 0.0539 1.000 0.0289 1.000 499 OR9G4 olfactory receptor, family 9, subfamily G, member 4 81921 1 1 1 0 0 1 0 0 0 0 0.0289 0.657 NaN NaN NaN 0.0289 1.000 500 ZNF492 zinc finger protein 492 123622 2 2 2 0 0 1 0 1 0 0 0.00913 0.597 0.0489 0.496 0.497 0.0290 1.000 501 SH3BP5L SH3-binding domain protein 5-like 92623 1 1 1 0 0 0 0 1 0 0 0.0290 0.817 NaN NaN NaN 0.0290 1.000 502 PLA2G2A phospholipase A2, group IIA (platelets, synovial fluid) 37244 1 1 1 0 0 0 0 1 0 0 0.0290 0.861 NaN NaN NaN 0.0290 1.000 503 KLC1 kinesin light chain 1 146990 1 1 1 0 0 0 1 0 0 0 0.0293 0.689 NaN NaN NaN 0.0293 1.000 504 DAAM2 dishevelled associated activator of morphogenesis 2 220276 2 2 2 1 0 0 1 1 0 0 0.00848 0.863 0.199 0.887 0.542 0.0293 1.000 505 UQCRC1 ubiquinol-cytochrome c reductase core protein I 118734 1 1 1 0 0 1 0 0 0 0 0.0294 0.644 NaN NaN NaN 0.0294 1.000 506 LPAR1 lysophosphatidic acid receptor 1 91881 1 1 1 0 0 1 0 0 0 0 0.0295 0.664 NaN NaN NaN 0.0295 1.000 507 SPRR3 small proline-rich protein 3 42658 1 1 1 0 0 0 0 1 0 0 0.0297 0.816 NaN NaN NaN 0.0297 1.000 508 ITGB1BP1 integrin beta 1 binding protein 1 52041 1 1 1 0 0 1 0 0 0 0 0.0297 0.669 NaN NaN NaN 0.0297 1.000 509 CENPP centromere protein P 65212 1 1 1 0 0 0 0 1 0 0 0.0298 0.849 NaN NaN NaN 0.0298 1.000 510 PLCXD2 phosphatidylinositol-specific phospholipase C, X domain containing 2 77262 1 1 1 0 1 0 0 0 0 0 0.0299 0.738 NaN NaN NaN 0.0299 1.000 511 KLK1 kallikrein 1 67143 1 1 1 0 0 0 0 0 1 0 0.0299 1.000 NaN NaN NaN 0.0299 1.000 512 PTH2R parathyroid hormone 2 receptor 141284 1 1 1 1 0 0 0 0 1 0 0.0301 0.974 NaN NaN NaN 0.0301 1.000 513 KRTAP19-5 keratin associated protein 19-5 18426 1 1 1 0 1 0 0 0 0 0 0.0302 0.625 NaN NaN NaN 0.0302 1.000 514 SFRS1 splicing factor, arginine/serine-rich 1 (splicing factor 2, alternate splicing factor) 67713 1 1 1 0 0 1 0 0 0 0 0.0304 0.663 NaN NaN NaN 0.0304 1.000 515 STK35 serine/threonine kinase 35 82304 1 1 1 0 1 0 0 0 0 0 0.0304 0.676 NaN NaN NaN 0.0304 1.000 516 C5orf48 chromosome 5 open reading frame 48 34491 1 1 1 0 0 0 0 1 0 0 0.0304 0.853 NaN NaN NaN 0.0304 1.000 517 C19orf2 chromosome 19 open reading frame 2 129179 1 1 1 0 1 0 0 0 0 0 0.0305 0.736 NaN NaN NaN 0.0305 1.000 518 RNF214 ring finger protein 214 179554 1 1 1 0 0 0 0 0 1 0 0.0305 0.834 NaN NaN NaN 0.0305 1.000 519 PRSS50 protease, serine, 50 88486 2 2 2 0 2 0 0 0 0 0 0.00482 0.420 0.829 0.718 1.000 0.0305 1.000 520 ZNF782 zinc finger protein 782 175441 1 1 1 0 1 0 0 0 0 0 0.0305 0.706 NaN NaN NaN 0.0305 1.000 521 TSPAN16 tetraspanin 16 58878 1 1 1 0 0 1 0 0 0 0 0.0306 0.670 NaN NaN NaN 0.0306 1.000 522 C1QL2 complement component 1, q subcomponent-like 2 39946 1 1 1 0 0 0 0 1 0 0 0.0307 0.835 NaN NaN NaN 0.0307 1.000 523 TNIP3 TNFAIP3 interacting protein 3 82963 1 1 1 1 0 0 0 0 1 0 0.0308 0.961 NaN NaN NaN 0.0308 1.000 524 HIST1H3G histone cluster 1, H3g 34395 1 1 1 0 1 0 0 0 0 0 0.0308 0.730 NaN NaN NaN 0.0308 1.000 525 CCDC144NL coiled-coil domain containing 144 family, N-terminal like 43759 1 1 1 0 0 0 0 0 1 0 0.0308 1.000 NaN NaN NaN 0.0308 1.000 526 CST9L cystatin 9-like 37754 1 1 1 0 0 0 0 1 0 0 0.0309 0.863 NaN NaN NaN 0.0309 1.000 527 CYP2A7 cytochrome P450, family 2, subfamily A, polypeptide 7 126615 2 2 2 0 0 1 0 1 0 0 0.0108 0.528 0.539 0.313 0.454 0.0309 1.000 528 PTGER2 prostaglandin E receptor 2 (subtype EP2), 53kDa 90023 1 1 1 0 0 1 0 0 0 0 0.0310 0.620 NaN NaN NaN 0.0310 1.000 529 DNAJC5B DnaJ (Hsp40) homolog, subfamily C, member 5 beta 50883 1 1 1 0 0 0 0 1 0 0 0.0310 0.855 NaN NaN NaN 0.0310 1.000 530 PLIN3 perilipin 3 108562 1 1 1 0 0 0 0 0 1 0 0.0310 1.000 NaN NaN NaN 0.0310 1.000 531 ANKH ankylosis, progressive homolog (mouse) 126051 1 1 1 0 0 1 0 0 0 0 0.0310 0.666 NaN NaN NaN 0.0310 1.000 532 DNAJC4 DnaJ (Hsp40) homolog, subfamily C, member 4 52956 1 1 1 0 0 0 0 1 0 0 0.0310 0.831 NaN NaN NaN 0.0310 1.000 533 FASTKD2 FAST kinase domains 2 180043 2 2 2 0 0 0 0 1 1 0 0.0101 0.854 0.256 0.375 0.486 0.0311 1.000 534 LCE5A late cornified envelope 5A 29963 1 1 1 0 0 0 0 1 0 0 0.0311 0.831 NaN NaN NaN 0.0311 1.000 535 BTBD10 BTB (POZ) domain containing 10 121180 1 1 1 0 0 0 0 0 1 0 0.0311 0.839 NaN NaN NaN 0.0311 1.000 536 ARID4B AT rich interactive domain 4B (RBP1-like) 333710 3 3 3 0 0 1 0 1 1 0 0.00493 0.604 0.654 0.699 1.000 0.0311 1.000 537 HIST2H2BF histone cluster 2, H2bf 63234 1 1 1 0 0 1 0 0 0 0 0.0311 0.630 NaN NaN NaN 0.0311 1.000 538 NBPF10 neuroblastoma breakpoint family, member 10 288668 2 2 2 1 0 1 0 1 0 0 0.0428 0.816 0.921 0.0692 0.115 0.0312 1.000 539 TP53TG5 TP53 target 5 74000 1 1 1 0 1 0 0 0 0 0 0.0312 0.757 NaN NaN NaN 0.0312 1.000 540 FCRL4 Fc receptor-like 4 132198 2 2 2 0 1 0 0 0 1 0 0.00495 0.580 0.775 0.481 1.000 0.0312 1.000 541 SIPA1L3 signal-induced proliferation-associated 1 like 3 415292 3 3 3 0 1 0 1 1 0 0 0.0127 0.414 0.215 0.825 0.392 0.0313 1.000 542 C17orf77 chromosome 17 open reading frame 77 61088 1 1 1 0 0 0 0 1 0 0 0.0313 0.823 NaN NaN NaN 0.0313 1.000 543 TTLL11 tubulin tyrosine ligase-like family, member 11 83913 2 2 2 0 0 0 0 2 0 0 0.00627 0.683 0.332 0.472 0.793 0.0313 1.000 544 PFN2 profilin 2 33283 1 1 1 0 0 0 0 1 0 0 0.0314 0.830 NaN NaN NaN 0.0314 1.000 545 PDYN prodynorphin 64144 1 1 1 0 1 0 0 0 0 0 0.0314 0.775 NaN NaN NaN 0.0314 1.000 546 ZNRF4 zinc and ring finger 4 107385 1 1 1 0 0 0 0 0 1 0 0.0314 0.619 NaN NaN NaN 0.0314 1.000 547 AGAP6 ArfGAP with GTPase domain, ankyrin repeat and PH domain 6 172400 2 2 2 1 0 0 0 2 0 0 0.0179 0.879 0.129 0.914 0.279 0.0315 1.000 548 VCX3B variable charge, X-linked 3B 38415 1 1 1 0 0 1 0 0 0 0 0.0316 0.592 NaN NaN NaN 0.0316 1.000 549 U2AF1 U2 small nuclear RNA auxiliary factor 1 68096 2 2 1 0 2 0 0 0 0 0 0.00520 0.571 0.0129 0.228 0.970 0.0317 1.000 550 GABRG1 gamma-aminobutyric acid (GABA) A receptor, gamma 1 118007 2 2 2 0 1 0 0 1 0 0 0.00537 0.635 0.285 0.808 0.941 0.0318 1.000 551 ZNF167 zinc finger protein 167 183161 1 1 1 0 0 0 1 0 0 0 0.0318 0.679 NaN NaN NaN 0.0318 1.000 552 PRG3 proteoglycan 3 57397 1 1 1 0 0 1 0 0 0 0 0.0319 0.658 NaN NaN NaN 0.0319 1.000 553 TXN2 thioredoxin 2 42579 1 1 1 0 0 1 0 0 0 0 0.0320 0.647 NaN NaN NaN 0.0320 1.000 554 TPRX1 tetra-peptide repeat homeobox 1 55755 1 1 1 0 1 0 0 0 0 0 0.0320 0.634 NaN NaN NaN 0.0320 1.000 555 SAMSN1 SAM domain, SH3 domain and nuclear localization signals 1 95561 1 1 1 0 0 0 0 1 0 0 0.0320 0.865 NaN NaN NaN 0.0320 1.000 556 FARSB phenylalanyl-tRNA synthetase, beta subunit 151411 1 1 1 0 1 0 0 0 0 0 0.0321 0.733 NaN NaN NaN 0.0321 1.000 557 TAF1D TATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa 71072 1 1 1 0 0 1 0 0 0 0 0.0321 0.682 NaN NaN NaN 0.0321 1.000 558 PPP2R2B protein phosphatase 2 (formerly 2A), regulatory subunit B, beta isoform 123701 2 2 2 0 1 1 0 0 0 0 0.00512 0.476 0.525 0.684 1.000 0.0321 1.000 559 LINGO2 leucine rich repeat and Ig domain containing 2 151475 2 2 2 0 1 0 0 0 1 0 0.0128 0.779 0.443 0.343 0.399 0.0321 1.000 560 CYB5R2 cytochrome b5 reductase 2 71055 1 1 1 0 1 0 0 0 0 0 0.0322 0.633 NaN NaN NaN 0.0322 1.000 561 CBFA2T3 core-binding factor, runt domain, alpha subunit 2; translocated to, 3 100410 1 1 1 0 0 0 0 0 1 0 0.0323 0.806 NaN NaN NaN 0.0323 1.000 562 VEGFC vascular endothelial growth factor C 102844 1 1 1 0 0 0 0 1 0 0 0.0324 0.855 NaN NaN NaN 0.0324 1.000 563 NR6A1 nuclear receptor subfamily 6, group A, member 1 110840 1 1 1 0 1 0 0 0 0 0 0.0326 0.736 NaN NaN NaN 0.0326 1.000 564 EFHC1 EF-hand domain (C-terminal) containing 1 163088 1 1 1 1 0 0 0 0 1 0 0.0326 1.000 NaN NaN NaN 0.0326 1.000 565 TRAPPC6A trafficking protein particle complex 6A 36279 1 1 1 0 1 0 0 0 0 0 0.0327 0.596 NaN NaN NaN 0.0327 1.000 566 TSPAN33 tetraspanin 33 69213 1 1 1 0 0 1 0 0 0 0 0.0327 0.644 NaN NaN NaN 0.0327 1.000 567 FOXP1 forkhead box P1 188161 1 1 1 0 0 0 0 0 1 0 0.0328 1.000 NaN NaN NaN 0.0328 1.000 568 C4orf43 46626 1 1 1 0 0 0 0 1 0 0 0.0328 0.881 NaN NaN NaN 0.0328 1.000 569 SMPDL3A sphingomyelin phosphodiesterase, acid-like 3A 106062 1 1 1 0 0 1 0 0 0 0 0.0329 0.680 NaN NaN NaN 0.0329 1.000 570 FABP9 fatty acid binding protein 9, testis 34343 1 1 1 0 0 0 0 0 1 0 0.0329 0.635 NaN NaN NaN 0.0329 1.000 571 MYCT1 myc target 1 59416 1 1 1 0 1 0 0 0 0 0 0.0329 0.833 NaN NaN NaN 0.0329 1.000 572 BEND6 BEN domain containing 6 71212 1 1 1 0 0 1 0 0 0 0 0.0331 0.679 NaN NaN NaN 0.0331 1.000 573 C20orf12 chromosome 20 open reading frame 12 173608 1 1 1 0 0 0 0 0 1 0 0.0331 0.839 NaN NaN NaN 0.0331 1.000 574 WDYHV1 WDYHV motif containing 1 45181 1 1 1 0 0 1 0 0 0 0 0.0332 0.698 NaN NaN NaN 0.0332 1.000 575 EPN2 epsin 2 151846 1 1 1 0 1 0 0 0 0 0 0.0333 0.665 NaN NaN NaN 0.0333 1.000 576 C9orf125 chromosome 9 open reading frame 125 100928 1 1 1 0 0 0 0 1 0 0 0.0333 0.817 NaN NaN NaN 0.0333 1.000 577 RSPH4A radial spoke head 4 homolog A (Chlamydomonas) 179991 1 1 1 0 1 0 0 0 0 0 0.0333 0.705 NaN NaN NaN 0.0333 1.000 578 ROBO4 roundabout homolog 4, magic roundabout (Drosophila) 252042 2 2 2 0 2 0 0 0 0 0 0.00810 0.536 0.907 0.161 0.663 0.0334 1.000 579 OR13C5 olfactory receptor, family 13, subfamily C, member 5 79563 1 1 1 0 0 1 0 0 0 0 0.0335 0.653 NaN NaN NaN 0.0335 1.000 580 EOMES eomesodermin homolog (Xenopus laevis) 120356 1 1 1 0 0 0 0 0 1 0 0.0335 1.000 NaN NaN NaN 0.0335 1.000 581 DPYSL2 dihydropyrimidinase-like 2 147079 1 1 1 0 0 1 0 0 0 0 0.0335 0.672 NaN NaN NaN 0.0335 1.000 582 HDAC6 histone deacetylase 6 247426 2 2 2 0 0 1 0 1 0 0 0.0304 0.536 0.297 0.265 0.177 0.0335 1.000 583 ARL13A ADP-ribosylation factor-like 13A 64313 1 1 1 0 0 0 0 1 0 0 0.0336 0.845 NaN NaN NaN 0.0336 1.000 584 NFU1 NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) 65641 1 1 1 0 0 0 0 1 0 0 0.0336 0.840 NaN NaN NaN 0.0336 1.000 585 PLA2G4A phospholipase A2, group IVA (cytosolic, calcium-dependent) 192134 1 1 1 1 1 0 0 0 0 0 0.0337 0.908 NaN NaN NaN 0.0337 1.000 586 CENPI centromere protein I 194618 1 1 1 0 1 0 0 0 0 0 0.0337 0.763 NaN NaN NaN 0.0337 1.000 587 WDR38 WD repeat domain 38 75668 1 1 1 0 0 0 0 1 0 0 0.0338 0.813 NaN NaN NaN 0.0338 1.000 588 PLA2G5 phospholipase A2, group V 35937 1 1 1 0 0 0 0 1 0 0 0.0339 0.843 NaN NaN NaN 0.0339 1.000 589 ARRDC3 arrestin domain containing 3 105950 1 1 1 0 1 0 0 0 0 0 0.0339 0.708 NaN NaN NaN 0.0339 1.000 590 HDAC8 histone deacetylase 8 101181 1 1 1 0 0 0 0 1 0 0 0.0339 0.836 NaN NaN NaN 0.0339 1.000 591 AKAP3 A kinase (PRKA) anchor protein 3 213626 1 1 1 0 0 0 1 0 0 0 0.0343 0.603 NaN NaN NaN 0.0343 1.000 592 PLP1 proteolipid protein 1 (Pelizaeus-Merzbacher disease, spastic paraplegia 2, uncomplicated) 71467 1 1 1 0 1 0 0 0 0 0 0.0343 0.591 NaN NaN NaN 0.0343 1.000 593 JUB jub, ajuba homolog (Xenopus laevis) 98585 1 1 1 0 0 1 0 0 0 0 0.0344 0.645 NaN NaN NaN 0.0344 1.000 594 C3orf59 chromosome 3 open reading frame 59 123172 1 1 1 0 0 0 0 0 1 0 0.0344 0.841 NaN NaN NaN 0.0344 1.000 595 SKAP1 src kinase associated phosphoprotein 1 88811 1 1 1 0 1 0 0 0 0 0 0.0345 0.778 NaN NaN NaN 0.0345 1.000 596 BET1 blocked early in transport 1 homolog (S. cerevisiae) 30873 1 1 1 0 0 0 0 1 0 0 0.0345 0.852 NaN NaN NaN 0.0345 1.000 597 PPP5C protein phosphatase 5, catalytic subunit 117528 1 1 1 0 0 0 0 0 1 0 0.0345 1.000 NaN NaN NaN 0.0345 1.000 598 ZNF622 zinc finger protein 622 121014 1 1 1 0 0 0 0 0 1 0 0.0346 0.854 NaN NaN NaN 0.0346 1.000 599 C19orf41 chromosome 19 open reading frame 41 51284 1 1 1 0 0 1 0 0 0 0 0.0346 0.613 NaN NaN NaN 0.0346 1.000 600 TEKT1 tektin 1 106655 1 1 1 0 0 0 0 1 0 0 0.0347 0.850 NaN NaN NaN 0.0347 1.000 601 LRIG2 leucine-rich repeats and immunoglobulin-like domains 2 270446 2 2 2 0 0 1 0 0 1 0 0.0149 0.562 0.689 0.217 0.378 0.0348 1.000 602 GFI1B growth factor independent 1B transcription repressor 84148 1 1 1 0 0 1 0 0 0 0 0.0348 0.623 NaN NaN NaN 0.0348 1.000 603 KANK1 KN motif and ankyrin repeat domains 1 339799 2 2 2 0 0 0 1 1 0 0 0.0169 0.556 0.0561 0.951 0.334 0.0349 1.000 604 IRS2 insulin receptor substrate 2 102979 2 2 2 0 0 0 0 2 0 0 0.00775 0.641 0.683 0.307 0.728 0.0349 1.000 605 SLC2A7 solute carrier family 2 (facilitated glucose transporter), member 7 112202 1 1 1 0 1 0 0 0 0 0 0.0350 0.586 NaN NaN NaN 0.0350 1.000 606 PRRG1 proline rich Gla (G-carboxyglutamic acid) 1 54865 1 1 1 0 0 1 0 0 0 0 0.0352 0.679 NaN NaN NaN 0.0352 1.000 607 INMT indolethylamine N-methyltransferase 66568 1 1 1 0 1 0 0 0 0 0 0.0352 0.645 NaN NaN NaN 0.0352 1.000 608 KRTAP10-6 keratin associated protein 10-6 91039 2 2 2 0 0 1 0 1 0 0 0.00574 0.518 0.554 0.997 1.000 0.0354 1.000 609 LHFP lipoma HMGIC fusion partner 51041 1 1 1 0 0 1 0 0 0 0 0.0354 0.665 NaN NaN NaN 0.0354 1.000 610 IMP4 IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) 58132 1 1 1 0 0 0 0 0 1 0 0.0354 1.000 NaN NaN NaN 0.0354 1.000 611 SMR3A submaxillary gland androgen regulated protein 3A 34279 1 1 1 0 0 1 0 0 0 0 0.0357 0.716 NaN NaN NaN 0.0357 1.000 612 BTBD9 BTB (POZ) domain containing 9 156571 1 1 1 0 0 0 0 0 1 0 0.0357 0.788 NaN NaN NaN 0.0357 1.000 613 MCL1 myeloid cell leukemia sequence 1 (BCL2-related) 76777 1 1 1 0 0 1 0 0 0 0 0.0359 0.709 NaN NaN NaN 0.0359 1.000 614 GALNT6 UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) 158392 2 2 2 0 0 0 0 1 1 0 0.0202 0.835 0.674 0.171 0.290 0.0359 1.000 615 SCN11A sodium channel, voltage-gated, type XI, alpha subunit 454191 3 3 3 0 1 0 0 2 0 0 0.0194 0.526 0.166 0.763 0.303 0.0360 1.000 616 NTRK1 neurotrophic tyrosine kinase, receptor, type 1 174955 2 2 2 0 0 2 0 0 0 0 0.00755 0.392 0.947 0.346 0.780 0.0361 1.000 617 AHNAK2 AHNAK nucleoprotein 2 1403235 7 7 7 2 1 2 0 4 0 0 0.0162 0.561 0.230 0.567 0.365 0.0362 1.000 618 EXOC2 exocyst complex component 2 239018 2 2 2 0 2 0 0 0 0 0 0.00596 0.436 0.0662 0.250 1.000 0.0365 1.000 619 ATL2 atlastin GTPase 2 146507 1 1 1 0 1 0 0 0 0 0 0.0366 0.799 NaN NaN NaN 0.0366 1.000 620 RAC1 ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) 51667 1 1 1 0 0 1 0 0 0 0 0.0366 0.670 NaN NaN NaN 0.0366 1.000 621 C19orf47 chromosome 19 open reading frame 47 87788 1 1 1 0 1 0 0 0 0 0 0.0366 0.705 NaN NaN NaN 0.0366 1.000 622 CCNYL1 cyclin Y-like 1 81687 1 1 1 0 0 0 0 0 1 0 0.0367 1.000 NaN NaN NaN 0.0367 1.000 623 TCTN2 tectonic family member 2 179586 1 1 1 0 0 0 0 0 1 0 0.0368 1.000 NaN NaN NaN 0.0368 1.000 624 HIST3H3 histone cluster 3, H3 34445 1 1 1 0 1 0 0 0 0 0 0.0369 0.731 NaN NaN NaN 0.0369 1.000 625 P2RX5 purinergic receptor P2X, ligand-gated ion channel, 5 106054 1 1 1 0 0 0 0 0 1 0 0.0370 1.000 NaN NaN NaN 0.0370 1.000 626 CMA1 chymase 1, mast cell 63350 1 1 1 0 0 1 0 0 0 0 0.0371 0.658 NaN NaN NaN 0.0371 1.000 627 LRRC49 leucine rich repeat containing 49 176193 2 2 2 0 0 2 0 0 0 0 0.0137 0.433 0.0794 0.979 0.445 0.0371 1.000 628 BANP BTG3 associated nuclear protein 116048 1 1 1 0 0 0 0 0 1 0 0.0371 0.835 NaN NaN NaN 0.0371 1.000 629 MIP major intrinsic protein of lens fiber 67023 1 1 1 0 1 0 0 0 0 0 0.0372 0.659 NaN NaN NaN 0.0372 1.000 630 MFSD4 major facilitator superfamily domain containing 4 115252 1 1 1 0 0 0 0 1 0 0 0.0372 0.805 NaN NaN NaN 0.0372 1.000 631 DBC1 deleted in bladder cancer 1 191472 2 2 2 0 2 0 0 0 0 0 0.0174 0.529 0.241 0.629 0.350 0.0372 1.000 632 CSH2 chorionic somatomammotropin hormone 2 48855 1 1 1 0 0 0 0 1 0 0 0.0372 0.822 NaN NaN NaN 0.0372 1.000 633 DHX32 DEAH (Asp-Glu-Ala-His) box polypeptide 32 188783 1 1 1 0 0 0 0 0 1 0 0.0373 0.852 NaN NaN NaN 0.0373 1.000 634 TCF19 transcription factor 19 (SC1) 84300 1 1 1 0 0 0 0 1 0 0 0.0373 0.792 NaN NaN NaN 0.0373 1.000 635 ZDHHC11 zinc finger, DHHC-type containing 11 95144 1 1 1 0 0 1 0 0 0 0 0.0374 0.664 NaN NaN NaN 0.0374 1.000 636 RG9MTD2 RNA (guanine-9-) methyltransferase domain containing 2 86755 1 1 1 0 1 0 0 0 0 0 0.0374 0.869 NaN NaN NaN 0.0374 1.000 637 INHBA inhibin, beta A 106987 2 2 2 0 1 0 0 1 0 0 0.00685 0.534 0.314 0.408 0.900 0.0375 1.000 638 LAT2 linker for activation of T cells family, member 2 61840 1 1 1 0 0 0 0 1 0 0 0.0376 0.832 NaN NaN NaN 0.0376 1.000 639 HCRTR2 hypocretin (orexin) receptor 2 112917 1 1 1 0 0 1 0 0 0 0 0.0376 0.675 NaN NaN NaN 0.0376 1.000 640 CDK19 cyclin-dependent kinase 19 129534 1 1 1 0 0 1 0 0 0 0 0.0376 0.674 NaN NaN NaN 0.0376 1.000 641 ZNF735 zinc finger protein 735 96450 1 1 1 0 0 1 0 0 0 0 0.0377 0.678 NaN NaN NaN 0.0377 1.000 642 ZCCHC12 zinc finger, CCHC domain containing 12 100668 1 1 1 0 0 1 0 0 0 0 0.0378 0.656 NaN NaN NaN 0.0378 1.000 643 G2E3 G2/M-phase specific E3 ubiquitin protein ligase 180287 1 1 1 0 1 0 0 0 0 0 0.0379 0.842 NaN NaN NaN 0.0379 1.000 644 GRXCR1 glutaredoxin, cysteine rich 1 73717 1 1 1 0 1 0 0 0 0 0 0.0379 0.795 NaN NaN NaN 0.0379 1.000 645 C6orf182 chromosome 6 open reading frame 182 117254 1 1 1 0 0 0 0 0 1 0 0.0379 1.000 NaN NaN NaN 0.0379 1.000 646 USP16 ubiquitin specific peptidase 16 210411 1 1 1 0 0 0 1 0 0 0 0.0379 0.559 NaN NaN NaN 0.0379 1.000 647 DUSP7 dual specificity phosphatase 7 74555 1 1 1 0 0 1 0 0 0 0 0.0379 0.611 NaN NaN NaN 0.0379 1.000 648 SEPSECS Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase 118721 1 1 1 0 0 1 0 0 0 0 0.0379 0.687 NaN NaN NaN 0.0379 1.000 649 NGLY1 N-glycanase 1 160812 2 2 2 0 0 1 0 1 0 0 0.0154 0.586 0.267 0.384 0.405 0.0380 1.000 650 TMED9 transmembrane emp24 protein transport domain containing 9 57138 1 1 1 0 0 0 0 0 1 0 0.0380 0.733 NaN NaN NaN 0.0380 1.000 651 SERPINB9 serpin peptidase inhibitor, clade B (ovalbumin), member 9 95865 1 1 1 0 1 0 0 0 0 0 0.0380 0.561 NaN NaN NaN 0.0380 1.000 652 FAM5C family with sequence similarity 5, member C 193212 1 1 1 0 0 0 1 0 0 0 0.0381 0.711 NaN NaN NaN 0.0381 1.000 653 COL21A1 collagen, type XXI, alpha 1 215094 2 2 2 0 0 0 1 1 0 0 0.00629 0.363 0.830 0.826 1.000 0.0382 1.000 654 NFYC nuclear transcription factor Y, gamma 86521 1 1 1 0 1 0 0 0 0 0 0.0383 0.685 NaN NaN NaN 0.0383 1.000 655 RASEF RAS and EF-hand domain containing 165468 1 1 1 0 0 1 0 0 0 0 0.0383 0.670 NaN NaN NaN 0.0383 1.000 656 CDR1 cerebellar degeneration-related protein 1, 34kDa 65819 1 1 1 0 0 0 0 1 0 0 0.0384 0.876 NaN NaN NaN 0.0384 1.000 657 SLC22A4 solute carrier family 22 (organic cation/ergothioneine transporter), member 4 135280 1 1 1 0 0 0 0 0 1 0 0.0387 1.000 NaN NaN NaN 0.0387 1.000 658 PRRG4 proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) 58017 1 1 1 0 0 1 0 0 0 0 0.0388 0.681 NaN NaN NaN 0.0388 1.000 659 SIP1 survival of motor neuron protein interacting protein 1 73168 1 1 1 0 0 0 0 0 1 0 0.0390 1.000 NaN NaN NaN 0.0390 1.000 660 MGAT4C mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) 119926 1 1 1 0 1 0 0 0 0 0 0.0391 0.900 NaN NaN NaN 0.0391 1.000 661 GCK glucokinase (hexokinase 4) 125523 1 1 1 0 0 0 0 0 1 0 0.0391 0.641 NaN NaN NaN 0.0391 1.000 662 C12orf52 chromosome 12 open reading frame 52 66935 1 1 1 0 0 0 0 0 1 0 0.0392 1.000 NaN NaN NaN 0.0392 1.000 663 DUOX2 dual oxidase 2 350231 2 1 2 0 0 1 0 1 0 0 0.370 0.513 0.00984 0.0193 0.0177 0.0395 1.000 664 MID1IP1 MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) 44714 1 1 1 0 1 0 0 0 0 0 0.0395 0.584 NaN NaN NaN 0.0395 1.000 665 ANXA13 annexin A13 93071 1 1 1 0 0 0 0 0 1 0 0.0396 0.650 NaN NaN NaN 0.0396 1.000 666 DHRS13 dehydrogenase/reductase (SDR family) member 13 78837 1 1 1 0 0 1 0 0 0 0 0.0396 0.619 NaN NaN NaN 0.0396 1.000 667 OR10H3 olfactory receptor, family 10, subfamily H, member 3 79099 1 1 1 0 0 1 0 0 0 0 0.0397 0.651 NaN NaN NaN 0.0397 1.000 668 SMARCB1 SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 96471 2 2 2 0 0 0 0 2 0 0 0.00661 0.714 0.332 0.914 1.000 0.0398 1.000 669 DGAT1 diacylglycerol O-acyltransferase homolog 1 (mouse) 102990 1 1 1 0 0 1 0 0 0 0 0.0398 0.637 NaN NaN NaN 0.0398 1.000 670 KRT78 keratin 78 131767 1 1 1 0 1 0 0 0 0 0 0.0399 0.704 NaN NaN NaN 0.0399 1.000 671 APBB1 amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) 171618 1 1 1 0 1 0 0 0 0 0 0.0399 0.713 NaN NaN NaN 0.0399 1.000 672 XK X-linked Kx blood group (McLeod syndrome) 96661 1 1 1 0 0 1 0 0 0 0 0.0399 0.638 NaN NaN NaN 0.0399 1.000 673 ZNF695 zinc finger protein 695 129108 1 1 1 0 0 1 0 0 0 0 0.0400 0.666 NaN NaN NaN 0.0400 1.000 674 OR2A5 olfactory receptor, family 2, subfamily A, member 5 77937 1 1 1 0 0 1 0 0 0 0 0.0400 0.625 NaN NaN NaN 0.0400 1.000 675 NCAM2 neural cell adhesion molecule 2 213566 2 2 2 0 0 1 0 0 1 0 0.00673 0.699 0.873 0.919 0.989 0.0400 1.000 676 PHGDH phosphoglycerate dehydrogenase 134824 1 1 1 0 0 0 0 0 1 0 0.0401 1.000 NaN NaN NaN 0.0401 1.000 677 RYBP RING1 and YY1 binding protein 55508 1 1 1 0 0 1 0 0 0 0 0.0401 0.677 NaN NaN NaN 0.0401 1.000 678 HEPHL1 hephaestin-like 1 288784 2 2 2 0 0 1 0 1 0 0 0.0401 0.576 NaN NaN NaN 0.0401 1.000 679 MNDA myeloid cell nuclear differentiation antigen 103405 1 1 1 0 1 0 0 0 0 0 0.0401 0.625 NaN NaN NaN 0.0401 1.000 680 ITPK1 inositol 1,3,4-triphosphate 5/6 kinase 91781 1 1 1 0 0 1 0 0 0 0 0.0402 0.636 NaN NaN NaN 0.0402 1.000 681 SCML4 sex comb on midleg-like 4 (Drosophila) 67949 1 1 1 0 1 0 0 0 0 0 0.0402 0.624 NaN NaN NaN 0.0402 1.000 682 CD8A CD8a molecule 46770 1 1 1 0 1 0 0 0 0 0 0.0403 0.610 NaN NaN NaN 0.0403 1.000 683 ACVR2A activin A receptor, type IIA 131563 1 1 1 0 0 0 0 0 1 0 0.0403 1.000 NaN NaN NaN 0.0403 1.000 684 RAB5C RAB5C, member RAS oncogene family 54013 1 1 1 0 1 0 0 0 0 0 0.0403 0.683 NaN NaN NaN 0.0403 1.000 685 PRODH2 proline dehydrogenase (oxidase) 2 121384 1 1 1 0 1 0 0 0 0 0 0.0404 0.705 NaN NaN NaN 0.0404 1.000 686 ZNF562 zinc finger protein 562 100109 1 1 1 0 0 0 0 1 0 0 0.0404 0.861 NaN NaN NaN 0.0404 1.000 687 NOX3 NADPH oxidase 3 145762 1 1 1 0 0 0 0 0 1 0 0.0406 1.000 NaN NaN NaN 0.0406 1.000 688 EPX eosinophil peroxidase 182122 1 1 1 0 0 0 1 0 0 0 0.0407 0.789 NaN NaN NaN 0.0407 1.000 689 PCK2 phosphoenolpyruvate carboxykinase 2 (mitochondrial) 167792 1 1 1 0 0 1 0 0 0 0 0.0409 0.652 NaN NaN NaN 0.0409 1.000 690 OR5D18 olfactory receptor, family 5, subfamily D, member 18 78434 1 1 1 0 0 1 0 0 0 0 0.0409 0.631 NaN NaN NaN 0.0409 1.000 691 KRTAP5-4 keratin associated protein 5-4 52456 1 1 1 1 0 0 0 1 0 0 0.0410 0.968 NaN NaN NaN 0.0410 1.000 692 BIN2 bridging integrator 2 145212 1 1 1 0 1 0 0 0 0 0 0.0410 0.680 NaN NaN NaN 0.0410 1.000 693 DNMT1 DNA (cytosine-5-)-methyltransferase 1 408768 3 3 3 0 1 1 0 1 0 0 0.0274 0.366 0.139 0.721 0.250 0.0410 1.000 694 YARS2 tyrosyl-tRNA synthetase 2, mitochondrial 120589 1 1 1 0 0 0 0 1 0 0 0.0411 0.818 NaN NaN NaN 0.0411 1.000 695 PFKM phosphofructokinase, muscle 220100 1 1 1 0 0 0 1 0 0 0 0.0411 0.657 NaN NaN NaN 0.0411 1.000 696 FAM126B family with sequence similarity 126, member B 135468 1 1 1 0 0 0 0 0 1 0 0.0414 0.673 NaN NaN NaN 0.0414 1.000 697 MAP7D3 MAP7 domain containing 3 217258 1 1 1 0 0 1 0 0 0 0 0.0414 0.696 NaN NaN NaN 0.0414 1.000 698 TNFAIP2 tumor necrosis factor, alpha-induced protein 2 102300 1 1 1 0 0 0 0 1 0 0 0.0414 0.829 NaN NaN NaN 0.0414 1.000 699 MRPS2 mitochondrial ribosomal protein S2 70080 1 1 1 0 0 1 0 0 0 0 0.0414 0.631 NaN NaN NaN 0.0414 1.000 700 ARFGAP2 ADP-ribosylation factor GTPase activating protein 2 128480 1 1 1 0 0 1 0 0 0 0 0.0415 0.666 NaN NaN NaN 0.0415 1.000 701 VRK3 vaccinia related kinase 3 122047 1 1 1 0 0 0 0 0 1 0 0.0416 1.000 NaN NaN NaN 0.0416 1.000 702 TLK1 tousled-like kinase 1 196362 1 1 1 1 0 0 0 0 1 0 0.0416 1.000 NaN NaN NaN 0.0416 1.000 703 DLX5 distal-less homeobox 5 73206 1 1 1 0 0 0 0 1 0 0 0.0417 0.823 NaN NaN NaN 0.0417 1.000 704 RXFP1 relaxin/insulin-like family peptide receptor 1 194326 1 1 1 0 1 0 0 0 0 0 0.0419 0.680 NaN NaN NaN 0.0419 1.000 705 GGT5 gamma-glutamyltransferase 5 120408 1 1 1 1 1 0 0 0 0 0 0.0420 0.923 NaN NaN NaN 0.0420 1.000 706 SPAST spastin 139184 1 1 1 0 0 0 0 0 1 0 0.0420 0.828 NaN NaN NaN 0.0420 1.000 707 GDPD5 glycerophosphodiester phosphodiesterase domain containing 5 142629 1 1 1 0 0 0 0 0 1 0 0.0422 1.000 NaN NaN NaN 0.0422 1.000 708 LMTK3 lemur tyrosine kinase 3 145715 1 1 1 0 0 0 0 0 1 0 0.0422 1.000 NaN NaN NaN 0.0422 1.000 709 RABAC1 Rab acceptor 1 (prenylated) 43085 1 1 1 0 0 0 0 1 0 0 0.0422 0.810 NaN NaN NaN 0.0422 1.000 710 OSBPL10 oxysterol binding protein-like 10 174069 1 1 1 0 0 0 0 0 1 0 0.0423 0.664 NaN NaN NaN 0.0423 1.000 711 RWDD4A RWD domain containing 4A 47546 1 1 1 0 0 0 0 1 0 0 0.0423 0.859 NaN NaN NaN 0.0423 1.000 712 EIF2B3 eukaryotic translation initiation factor 2B, subunit 3 gamma, 58kDa 119445 1 1 1 0 0 0 0 1 0 0 0.0424 0.839 NaN NaN NaN 0.0424 1.000 713 SNTG2 syntrophin, gamma 2 99571 2 2 2 0 0 1 0 1 0 0 0.00856 0.566 0.185 0.193 0.834 0.0424 1.000 714 C1orf110 chromosome 1 open reading frame 110 76171 1 1 1 0 0 1 0 0 0 0 0.0424 0.660 NaN NaN NaN 0.0424 1.000 715 GYPC glycophorin C (Gerbich blood group) 30709 1 1 1 0 0 0 0 1 0 0 0.0424 0.811 NaN NaN NaN 0.0424 1.000 716 HERC5 hect domain and RLD 5 240237 1 1 1 0 0 0 0 1 0 0 0.0424 0.864 NaN NaN NaN 0.0424 1.000 717 MYLIP myosin regulatory light chain interacting protein 106286 2 2 2 0 0 1 0 1 0 0 0.00715 0.541 0.877 0.950 1.000 0.0425 1.000 718 KLF10 Kruppel-like factor 10 121218 1 1 1 0 0 0 0 0 1 0 0.0426 0.820 NaN NaN NaN 0.0426 1.000 719 KAAG1 kidney associated antigen 1 21497 1 1 1 0 1 0 0 0 0 0 0.0430 0.810 NaN NaN NaN 0.0430 1.000 720 CSNK1A1L casein kinase 1, alpha 1-like 84494 1 1 1 0 0 0 0 1 0 0 0.0430 0.858 NaN NaN NaN 0.0430 1.000 721 C2orf78 chromosome 2 open reading frame 78 221448 1 1 1 0 0 0 0 1 0 0 0.0430 0.820 NaN NaN NaN 0.0430 1.000 722 LRCH1 leucine-rich repeats and calponin homology (CH) domain containing 1 166157 1 1 1 0 0 0 0 0 1 0 0.0433 0.845 NaN NaN NaN 0.0433 1.000 723 GLTSCR1 glioma tumor suppressor candidate region gene 1 97049 2 2 2 0 0 1 0 1 0 0 0.00734 0.509 0.912 0.706 1.000 0.0434 1.000 724 CCDC71 coiled-coil domain containing 71 116554 1 1 1 0 1 0 0 0 0 0 0.0435 0.910 NaN NaN NaN 0.0435 1.000 725 ANAPC7 anaphase promoting complex subunit 7 144004 1 1 1 0 0 0 0 0 1 0 0.0435 0.846 NaN NaN NaN 0.0435 1.000 726 FAM57A family with sequence similarity 57, member A 49551 1 1 1 0 0 0 0 1 0 0 0.0438 0.815 NaN NaN NaN 0.0438 1.000 727 MKRN1 makorin, ring finger protein, 1 107503 1 1 1 0 0 0 0 0 1 0 0.0438 1.000 NaN NaN NaN 0.0438 1.000 728 CYP2D6 cytochrome P450, family 2, subfamily D, polypeptide 6 101528 1 1 1 0 0 1 0 0 0 0 0.0438 0.610 NaN NaN NaN 0.0438 1.000 729 TMPRSS11D transmembrane protease, serine 11D 107534 1 1 1 0 0 0 0 0 1 0 0.0439 1.000 NaN NaN NaN 0.0439 1.000 730 TMEM198 transmembrane protein 198 85504 1 1 1 0 0 1 0 0 0 0 0.0439 0.602 NaN NaN NaN 0.0439 1.000 731 BRE brain and reproductive organ-expressed (TNFRSF1A modulator) 116759 1 1 1 0 0 0 0 0 1 0 0.0440 0.763 NaN NaN NaN 0.0440 1.000 732 FAM169A family with sequence similarity 169, member A 170941 1 1 1 0 0 0 0 0 1 0 0.0440 0.693 NaN NaN NaN 0.0440 1.000 733 ZNF678 zinc finger protein 678 134858 1 1 1 0 0 0 0 0 1 0 0.0440 0.667 NaN NaN NaN 0.0440 1.000 734 COL8A2 collagen, type VIII, alpha 2 117575 1 1 1 0 0 0 0 0 1 0 0.0441 0.759 NaN NaN NaN 0.0441 1.000 735 CACYBP calcyclin binding protein 58610 1 1 1 0 0 1 0 0 0 0 0.0441 0.685 NaN NaN NaN 0.0441 1.000 736 OR2C1 olfactory receptor, family 2, subfamily C, member 1 78091 1 1 1 0 1 0 0 0 0 0 0.0441 0.696 NaN NaN NaN 0.0441 1.000 737 ANXA2 annexin A2 89868 1 1 1 0 0 0 0 1 0 0 0.0442 0.854 NaN NaN NaN 0.0442 1.000 738 MSMP microseminoprotein, prostate associated 35829 1 1 1 0 0 0 0 1 0 0 0.0443 0.825 NaN NaN NaN 0.0443 1.000 739 CFH complement factor H 314157 2 2 2 0 0 0 0 2 0 0 0.0531 0.725 0.506 0.116 0.141 0.0443 1.000 740 PCDHB7 protocadherin beta 7 198033 2 2 2 1 0 0 0 1 1 0 0.00760 0.731 0.342 0.614 0.991 0.0443 1.000 741 PSMD6 proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 95615 1 1 1 0 0 0 0 1 0 0 0.0444 0.853 NaN NaN NaN 0.0444 1.000 742 ANKRD56 ankyrin repeat domain 56 153148 2 2 2 0 1 0 0 0 1 0 0.00982 0.691 0.246 0.857 0.769 0.0445 1.000 743 RORB RAR-related orphan receptor B 117807 1 1 1 0 1 0 0 0 0 0 0.0446 0.640 NaN NaN NaN 0.0446 1.000 744 SMC2 structural maintenance of chromosomes 2 305701 2 2 2 0 0 1 0 1 0 0 0.0318 0.593 0.0513 0.254 0.238 0.0446 1.000 745 TPPP tubulin polymerization promoting protein 53971 1 1 1 0 0 1 0 0 0 0 0.0449 0.648 NaN NaN NaN 0.0449 1.000 746 TPR translocated promoter region (to activated MET oncogene) 604176 3 3 3 0 0 1 0 1 1 0 0.0158 0.583 0.241 0.994 0.484 0.0449 1.000 747 OPN5 opsin 5 89591 1 1 1 0 0 1 0 0 0 0 0.0449 0.675 NaN NaN NaN 0.0449 1.000 748 METTL2B methyltransferase like 2B 97000 1 1 1 0 0 1 0 0 0 0 0.0450 0.662 NaN NaN NaN 0.0450 1.000 749 PLCXD3 phosphatidylinositol-specific phospholipase C, X domain containing 3 81104 1 1 1 0 0 0 0 0 1 0 0.0450 0.643 NaN NaN NaN 0.0450 1.000 750 PI16 peptidase inhibitor 16 110280 1 1 1 1 1 0 0 0 0 0 0.0451 0.863 NaN NaN NaN 0.0451 1.000 751 FBXL4 F-box and leucine-rich repeat protein 4 157065 1 1 1 0 0 0 0 1 0 0 0.0452 0.840 NaN NaN NaN 0.0452 1.000 752 VGLL4 vestigial like 4 (Drosophila) 69156 1 1 1 0 1 0 0 0 0 0 0.0453 0.633 NaN NaN NaN 0.0453 1.000 753 ABCB6 ATP-binding cassette, sub-family B (MDR/TAP), member 6 208004 1 1 1 0 0 0 1 0 0 0 0.0453 0.749 NaN NaN NaN 0.0453 1.000 754 MRGPRX4 MAS-related GPR, member X4 80759 1 1 1 0 0 1 0 0 0 0 0.0454 0.597 NaN NaN NaN 0.0454 1.000 755 ALDH1A2 aldehyde dehydrogenase 1 family, member A2 133408 1 1 1 0 0 0 0 1 0 0 0.0455 0.831 NaN NaN NaN 0.0455 1.000 756 ZFAND5 zinc finger, AN1-type domain 5 54906 1 1 1 0 0 0 0 1 0 0 0.0455 0.845 NaN NaN NaN 0.0455 1.000 757 LASS6 LAG1 homolog, ceramide synthase 6 99179 1 1 1 0 0 0 0 0 1 0 0.0457 1.000 NaN NaN NaN 0.0457 1.000 758 FAM166A family with sequence similarity 166, member A 81442 1 1 1 0 0 1 0 0 0 0 0.0459 0.642 NaN NaN NaN 0.0459 1.000 759 ZNF536 zinc finger protein 536 312681 3 3 3 1 0 1 1 1 0 0 0.00785 0.678 0.676 0.778 1.000 0.0459 1.000 760 C10orf129 chromosome 10 open reading frame 129 57519 1 1 1 0 0 0 0 0 1 0 0.0461 0.833 NaN NaN NaN 0.0461 1.000 761 PRICKLE2 prickle homolog 2 (Drosophila) 212729 1 1 1 0 0 0 0 0 1 0 0.0462 0.723 NaN NaN NaN 0.0462 1.000 762 DHTKD1 dehydrogenase E1 and transketolase domain containing 1 230625 1 1 1 0 0 1 0 0 0 0 0.0462 0.658 NaN NaN NaN 0.0462 1.000 763 CCNC cyclin C 69851 1 1 1 0 0 0 0 1 0 0 0.0465 0.873 NaN NaN NaN 0.0465 1.000 764 TRIM37 tripartite motif-containing 37 247554 1 1 1 0 0 0 0 0 1 0 0.0466 1.000 NaN NaN NaN 0.0466 1.000 765 VSIG8 V-set and immunoglobulin domain containing 8 74402 1 1 1 0 1 0 0 0 0 0 0.0466 0.671 NaN NaN NaN 0.0466 1.000 766 TFAM transcription factor A, mitochondrial 55330 1 1 1 0 0 1 0 0 0 0 0.0467 0.691 NaN NaN NaN 0.0467 1.000 767 TNNI3K TNNI3 interacting kinase 245563 1 1 1 0 0 1 0 0 0 0 0.0467 0.680 NaN NaN NaN 0.0467 1.000 768 CLEC18B C-type lectin domain family 18, member B 103573 1 1 1 0 1 0 0 0 0 0 0.0468 0.654 NaN NaN NaN 0.0468 1.000 769 ENTPD8 ectonucleoside triphosphate diphosphohydrolase 8 122684 1 1 1 0 0 0 0 0 1 0 0.0470 0.808 NaN NaN NaN 0.0470 1.000 770 C14orf149 chromosome 14 open reading frame 149 81222 1 1 1 0 0 0 0 1 0 0 0.0470 0.809 NaN NaN NaN 0.0470 1.000 771 ACTBL2 actin, beta-like 2 94205 1 1 1 0 0 0 0 1 0 0 0.0470 0.821 NaN NaN NaN 0.0470 1.000 772 LGR5 leucine-rich repeat-containing G protein-coupled receptor 5 231903 1 1 1 0 0 1 0 0 0 0 0.0472 0.645 NaN NaN NaN 0.0472 1.000 773 FAM154A family with sequence similarity 154, member A 119562 1 1 1 0 1 0 0 0 0 0 0.0472 0.735 NaN NaN NaN 0.0472 1.000 774 SLC13A5 solute carrier family 13 (sodium-dependent citrate transporter), member 5 145179 1 1 1 0 0 1 0 0 0 0 0.0473 0.655 NaN NaN NaN 0.0473 1.000 775 OR3A1 olfactory receptor, family 3, subfamily A, member 1 79016 1 1 1 0 1 0 0 0 0 0 0.0474 0.763 NaN NaN NaN 0.0474 1.000 776 OR1S1 olfactory receptor, family 1, subfamily S, member 1 81423 1 1 1 0 0 1 0 0 0 0 0.0474 0.643 NaN NaN NaN 0.0474 1.000 777 FZD9 frizzled homolog 9 (Drosophila) 112852 1 1 1 0 1 0 0 0 0 0 0.0475 0.600 NaN NaN NaN 0.0475 1.000 778 NFIL3 nuclear factor, interleukin 3 regulated 115619 1 1 1 0 0 0 0 0 1 0 0.0475 0.840 NaN NaN NaN 0.0475 1.000 779 KIFC1 kinesin family member C1 171440 1 1 1 0 0 0 0 0 1 0 0.0476 0.623 NaN NaN NaN 0.0476 1.000 780 SILV silver homolog (mouse) 168490 1 1 1 0 0 0 0 0 1 0 0.0478 1.000 NaN NaN NaN 0.0478 1.000 781 EXOSC4 exosome component 4 54531 1 1 1 0 0 0 0 1 0 0 0.0479 0.787 NaN NaN NaN 0.0479 1.000 782 GSTA2 glutathione S-transferase A2 57504 1 1 1 0 0 0 0 1 0 0 0.0479 0.858 NaN NaN NaN 0.0479 1.000 783 SLC28A1 solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 165597 2 2 2 0 1 0 0 1 0 0 0.0162 0.507 0.372 0.413 0.510 0.0479 1.000 784 C14orf101 chromosome 14 open reading frame 101 167805 1 1 1 0 1 0 0 0 0 0 0.0480 0.667 NaN NaN NaN 0.0480 1.000 785 FOXA2 forkhead box A2 89504 2 2 2 0 1 0 0 1 0 0 0.00828 0.471 0.858 0.833 1.000 0.0480 1.000 786 FAM192A family with sequence similarity 192, member A 65422 1 1 1 0 0 1 0 0 0 0 0.0481 0.654 NaN NaN NaN 0.0481 1.000 787 NOVA1 neuro-oncological ventral antigen 1 120561 1 1 1 1 0 0 0 1 0 0 0.0481 0.940 NaN NaN NaN 0.0481 1.000 788 PIGA phosphatidylinositol glycan anchor biosynthesis, class A (paroxysmal nocturnal hemoglobinuria) 122395 1 1 1 0 0 1 0 0 0 0 0.0481 0.682 NaN NaN NaN 0.0481 1.000 789 UBB ubiquitin B 57574 1 1 1 1 0 0 0 1 0 0 0.0481 0.933 NaN NaN NaN 0.0481 1.000 790 FIGN fidgetin 189898 2 2 2 0 0 1 0 1 0 0 0.00831 0.556 0.888 0.340 1.000 0.0481 1.000 791 HOXB13 homeobox B13 71629 1 1 1 0 0 1 0 0 0 0 0.0482 0.661 NaN NaN NaN 0.0482 1.000 792 BMP6 bone morphogenetic protein 6 103356 1 1 1 0 0 1 0 0 0 0 0.0483 0.651 NaN NaN NaN 0.0483 1.000 793 BRD9 bromodomain containing 9 118057 1 1 1 0 0 1 0 0 0 0 0.0483 0.691 NaN NaN NaN 0.0483 1.000 794 TMCC2 transmembrane and coiled-coil domain family 2 174955 1 1 1 0 0 0 0 0 1 0 0.0485 1.000 NaN NaN NaN 0.0485 1.000 795 PLXNA2 plexin A2 481166 3 3 3 0 2 1 0 0 0 0 0.0250 0.284 0.192 0.416 0.337 0.0487 1.000 796 CRYAA crystallin, alpha A 44298 1 1 1 0 1 0 0 0 0 0 0.0488 0.663 NaN NaN NaN 0.0488 1.000 797 OR10G2 olfactory receptor, family 10, subfamily G, member 2 77249 1 1 1 0 0 0 0 1 0 0 0.0489 0.799 NaN NaN NaN 0.0489 1.000 798 LRCH4 leucine-rich repeats and calponin homology (CH) domain containing 4 160096 1 1 1 0 0 1 0 0 0 0 0.0489 0.609 NaN NaN NaN 0.0489 1.000 799 ZFPM2 zinc finger protein, multitype 2 284117 2 2 2 0 0 2 0 0 0 0 0.00848 0.457 0.472 0.934 1.000 0.0489 1.000 800 SNAP25 synaptosomal-associated protein, 25kDa 63586 1 1 1 0 1 0 0 0 0 0 0.0490 0.742 NaN NaN NaN 0.0490 1.000 801 KREMEN1 kringle containing transmembrane protein 1 113129 1 1 1 0 0 1 0 0 0 0 0.0490 0.671 NaN NaN NaN 0.0490 1.000 802 NF2 neurofibromin 2 (merlin) 139050 1 1 1 0 0 1 0 0 0 0 0.0491 0.651 NaN NaN NaN 0.0491 1.000 803 C10orf71 chromosome 10 open reading frame 71 172963 1 1 1 0 0 0 0 1 0 0 0.0491 0.833 NaN NaN NaN 0.0491 1.000 804 TGOLN2 trans-golgi network protein 2 101113 1 1 1 0 0 1 0 0 0 0 0.0492 0.694 NaN NaN NaN 0.0492 1.000 805 CRNN cornulin 124168 1 1 1 0 1 0 0 0 0 0 0.0492 0.604 NaN NaN NaN 0.0492 1.000 806 UNC5B unc-5 homolog B (C. elegans) 231101 2 2 2 0 0 1 0 1 0 0 0.0186 0.513 0.0686 0.914 0.460 0.0493 1.000 807 CA8 carbonic anhydrase VIII 73026 1 1 1 0 1 0 0 0 0 0 0.0494 0.694 NaN NaN NaN 0.0494 1.000 808 CLEC2D C-type lectin domain family 2, member D 56304 1 1 1 0 0 0 0 1 0 0 0.0494 0.873 NaN NaN NaN 0.0494 1.000 809 FAM108B1 family with sequence similarity 108, member B1 75430 1 1 1 0 0 1 0 0 0 0 0.0495 0.683 NaN NaN NaN 0.0495 1.000 810 BMP4 bone morphogenetic protein 4 102505 1 1 1 0 0 1 0 0 0 0 0.0496 0.628 NaN NaN NaN 0.0496 1.000 811 MSH6 mutS homolog 6 (E. coli) 333869 2 2 2 0 0 0 1 1 0 0 0.0112 0.500 0.381 0.970 0.768 0.0496 1.000 812 GARS glycyl-tRNA synthetase 175577 1 1 1 0 0 0 0 0 1 0 0.0496 1.000 NaN NaN NaN 0.0496 1.000 813 C17orf70 chromosome 17 open reading frame 70 172627 1 1 1 0 0 0 0 0 1 0 0.0497 1.000 NaN NaN NaN 0.0497 1.000 814 MCM6 minichromosome maintenance complex component 6 209648 1 1 1 0 1 0 0 0 0 0 0.0497 0.762 NaN NaN NaN 0.0497 1.000 815 CYC1 cytochrome c-1 72116 1 1 1 1 0 0 0 0 1 0 0.0498 1.000 NaN NaN NaN 0.0498 1.000 816 LTBR lymphotoxin beta receptor (TNFR superfamily, member 3) 103355 1 1 1 0 0 1 0 0 0 0 0.0501 0.661 NaN NaN NaN 0.0501 1.000 817 PCDHA13 protocadherin alpha 13 240515 3 3 3 0 2 0 0 1 0 0 0.00873 0.265 0.644 0.680 1.000 0.0501 1.000 818 PIKFYVE phosphoinositide kinase, FYVE finger containing 536975 3 3 3 0 0 1 0 1 1 0 0.0146 0.449 0.396 0.844 0.600 0.0503 1.000 819 STK19 serine/threonine kinase 19 90374 1 1 1 1 0 0 0 0 1 0 0.0504 1.000 NaN NaN NaN 0.0504 1.000 820 CCDC7 coiled-coil domain containing 7 119894 1 1 1 0 0 1 0 0 0 0 0.0505 0.684 NaN NaN NaN 0.0505 1.000 821 SLC6A15 solute carrier family 6, member 15 194894 2 2 2 0 0 0 0 2 0 0 0.0193 0.704 0.395 0.342 0.457 0.0506 1.000 822 ARHGAP32 Rho GTPase activating protein 32 516762 3 3 3 0 1 1 0 0 1 0 0.0309 0.397 0.792 0.0614 0.286 0.0506 1.000 823 CHERP calcium homeostasis endoplasmic reticulum protein 173068 1 1 1 0 0 0 0 0 1 0 0.0506 1.000 NaN NaN NaN 0.0506 1.000 824 RSPH6A radial spoke head 6 homolog A (Chlamydomonas) 176369 1 1 1 0 0 0 0 0 1 0 0.0507 1.000 NaN NaN NaN 0.0507 1.000 825 RABGAP1 RAB GTPase activating protein 1 273667 1 1 1 0 0 0 0 0 1 0 0.0507 0.608 NaN NaN NaN 0.0507 1.000 826 REG3G regenerating islet-derived 3 gamma 45484 1 1 1 0 0 0 0 1 0 0 0.0509 0.842 NaN NaN NaN 0.0509 1.000 827 SLC36A2 solute carrier family 36 (proton/amino acid symporter), member 2 123595 1 1 1 0 0 1 0 0 0 0 0.0510 0.632 NaN NaN NaN 0.0510 1.000 828 HLA-DMB major histocompatibility complex, class II, DM beta 67125 1 1 1 0 0 0 0 1 0 0 0.0511 0.815 NaN NaN NaN 0.0511 1.000 829 ZFP36L1 zinc finger protein 36, C3H type-like 1 85033 1 1 1 0 0 1 0 0 0 0 0.0513 0.613 NaN NaN NaN 0.0513 1.000 830 ZC3H11A zinc finger CCCH-type containing 11A 206858 1 1 1 1 0 0 0 0 1 0 0.0513 1.000 NaN NaN NaN 0.0513 1.000 831 NOTCH2NL Notch homolog 2 (Drosophila) N-terminal like 60118 1 1 1 0 0 1 0 0 0 0 0.0513 0.666 NaN NaN NaN 0.0513 1.000 832 STK17A serine/threonine kinase 17a 92060 1 1 1 0 0 0 0 0 1 0 0.0514 0.745 NaN NaN NaN 0.0514 1.000 833 ABHD2 abhydrolase domain containing 2 109059 1 1 1 0 0 1 0 0 0 0 0.0515 0.650 NaN NaN NaN 0.0515 1.000 834 LGTN ligatin 150338 1 1 1 0 1 0 0 0 0 0 0.0515 0.667 NaN NaN NaN 0.0515 1.000 835 FCGR3B Fc fragment of IgG, low affinity IIIb, receptor (CD16b) 58999 1 1 1 0 0 0 0 1 0 0 0.0516 0.836 NaN NaN NaN 0.0516 1.000 836 TPTE transmembrane phosphatase with tensin homology 144342 2 2 2 0 0 0 0 2 0 0 0.0169 0.717 0.473 0.661 0.537 0.0516 1.000 837 IFNA10 interferon, alpha 10 47642 1 1 1 0 0 0 0 1 0 0 0.0516 0.845 NaN NaN NaN 0.0516 1.000 838 OR5D16 olfactory receptor, family 5, subfamily D, member 16 81942 1 1 1 1 0 0 0 1 0 0 0.0516 0.933 NaN NaN NaN 0.0516 1.000 839 POF1B premature ovarian failure, 1B 149298 1 1 1 0 0 0 0 0 1 0 0.0517 0.868 NaN NaN NaN 0.0517 1.000 840 CTSC cathepsin C 116886 1 1 1 0 0 0 0 1 0 0 0.0518 0.847 NaN NaN NaN 0.0518 1.000 841 FZD2 frizzled homolog 2 (Drosophila) 132861 4 1 4 4 0 1 0 2 1 0 0.0552 0.944 0.0933 0.939 0.165 0.0519 1.000 842 INHBC inhibin, beta C 88556 1 1 1 0 0 1 0 0 0 0 0.0519 0.647 NaN NaN NaN 0.0519 1.000 843 GCC2 GRIP and coiled-coil domain containing 2 425096 2 2 2 0 0 1 0 0 1 0 0.0709 0.671 0.793 0.0437 0.129 0.0519 1.000 844 KPRP keratinocyte proline-rich protein 144752 1 1 1 1 1 0 0 0 0 0 0.0521 0.975 NaN NaN NaN 0.0521 1.000 845 FCGBP Fc fragment of IgG binding protein 929727 5 5 4 1 0 2 0 3 0 0 0.0135 0.463 0.363 0.358 0.679 0.0521 1.000 846 SPRYD4 SPRY domain containing 4 52456 1 1 1 0 0 1 0 0 0 0 0.0521 0.631 NaN NaN NaN 0.0521 1.000 847 FGGY FGGY carbohydrate kinase domain containing 148237 1 1 1 0 0 0 0 0 1 0 0.0522 1.000 NaN NaN NaN 0.0522 1.000 848 RAG1 recombination activating gene 1 260288 1 1 1 0 0 1 0 0 0 0 0.0525 0.655 NaN NaN NaN 0.0525 1.000 849 FAM134B family with sequence similarity 134, member B 106300 1 1 1 0 0 1 0 0 0 0 0.0525 0.674 NaN NaN NaN 0.0525 1.000 850 HIST1H2BH histone cluster 1, H2bh 31955 1 1 1 0 0 0 0 1 0 0 0.0526 0.815 NaN NaN NaN 0.0526 1.000 851 CHAT choline acetyltransferase 167629 2 2 2 0 1 0 0 1 0 0 0.0271 0.547 0.224 0.276 0.342 0.0526 1.000 852 OR1M1 olfactory receptor, family 1, subfamily M, member 1 78435 1 1 1 0 0 1 0 0 0 0 0.0526 0.636 NaN NaN NaN 0.0526 1.000 853 SLC25A2 solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 2 75479 1 1 1 0 0 1 0 0 0 0 0.0527 0.678 NaN NaN NaN 0.0527 1.000 854 EEF1E1 eukaryotic translation elongation factor 1 epsilon 1 44839 1 1 1 0 0 1 0 0 0 0 0.0528 0.648 NaN NaN NaN 0.0528 1.000 855 SGIP1 SH3-domain GRB2-like (endophilin) interacting protein 1 214609 1 1 1 0 0 0 0 1 0 0 0.0528 0.815 NaN NaN NaN 0.0528 1.000 856 PRAMEF2 PRAME family member 2 119242 2 2 2 1 0 0 0 2 0 0 0.00933 0.875 0.857 0.758 1.000 0.0529 1.000 857 HOMER1 homer homolog 1 (Drosophila) 91304 1 1 1 0 0 0 0 1 0 0 0.0529 0.856 NaN NaN NaN 0.0529 1.000 858 MED1 mediator complex subunit 1 399450 2 2 2 0 1 0 0 0 1 0 0.0146 0.694 0.131 0.863 0.640 0.0529 1.000 859 CPSF2 cleavage and polyadenylation specific factor 2, 100kDa 199313 1 1 1 0 0 0 0 0 1 0 0.0530 0.686 NaN NaN NaN 0.0530 1.000 860 ACCN2 amiloride-sensitive cation channel 2, neuronal 144442 1 1 1 1 0 1 0 0 0 0 0.0530 0.940 NaN NaN NaN 0.0530 1.000 861 CD48 CD48 molecule 62063 1 1 1 0 0 0 0 1 0 0 0.0531 0.841 NaN NaN NaN 0.0531 1.000 862 UNC45B unc-45 homolog B (C. elegans) 229483 1 1 1 0 0 0 1 0 0 0 0.0532 0.784 NaN NaN NaN 0.0532 1.000 863 KCTD16 potassium channel tetramerisation domain containing 16 107485 1 1 1 0 1 0 0 0 0 0 0.0533 0.750 NaN NaN NaN 0.0533 1.000 864 CCDC3 coiled-coil domain containing 3 44217 1 1 1 0 1 0 0 0 0 0 0.0534 0.669 NaN NaN NaN 0.0534 1.000 865 HERPUD1 homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 94768 1 1 1 0 0 1 0 0 0 0 0.0535 0.689 NaN NaN NaN 0.0535 1.000 866 CLIC2 chloride intracellular channel 2 63512 1 1 1 0 0 0 0 1 0 0 0.0537 0.860 NaN NaN NaN 0.0537 1.000 867 AMAC1L2 acyl-malonyl condensing enzyme 1-like 2 84743 1 1 1 0 1 0 0 0 0 0 0.0537 0.661 NaN NaN NaN 0.0537 1.000 868 MAGEA6 melanoma antigen family A, 6 66711 1 1 1 1 1 0 0 0 0 0 0.0538 0.831 NaN NaN NaN 0.0538 1.000 869 CTNNAL1 catenin (cadherin-associated protein), alpha-like 1 174866 1 1 1 0 0 1 0 0 0 0 0.0539 0.683 NaN NaN NaN 0.0539 1.000 870 RTP3 receptor (chemosensory) transporter protein 3 58681 1 1 1 0 0 1 0 0 0 0 0.0539 0.656 NaN NaN NaN 0.0539 1.000 871 SLC39A4 solute carrier family 39 (zinc transporter), member 4 123864 1 1 1 0 0 1 0 0 0 0 0.0541 0.606 NaN NaN NaN 0.0541 1.000 872 VEZF1 vascular endothelial zinc finger 1 128899 1 1 1 0 0 0 0 0 1 0 0.0542 1.000 NaN NaN NaN 0.0542 1.000 873 ANKRD13C ankyrin repeat domain 13C 136040 1 1 1 0 0 1 0 0 0 0 0.0542 0.678 NaN NaN NaN 0.0542 1.000 874 SLC45A1 solute carrier family 45, member 1 185966 1 1 1 0 0 0 0 0 1 0 0.0543 0.802 NaN NaN NaN 0.0543 1.000 875 MRPL15 mitochondrial ribosomal protein L15 66280 1 1 1 0 0 0 0 1 0 0 0.0544 0.831 NaN NaN NaN 0.0544 1.000 876 MBD6 methyl-CpG binding domain protein 6 253237 1 1 1 0 0 0 0 0 1 0 0.0545 1.000 NaN NaN NaN 0.0545 1.000 877 QKI quaking homolog, KH domain RNA binding (mouse) 96315 1 1 1 0 0 1 0 0 0 0 0.0546 0.700 NaN NaN NaN 0.0546 1.000 878 XIRP2 xin actin-binding repeat containing 2 1014377 2 2 2 1 1 1 0 0 0 0 0.145 0.848 0.795 0.0350 0.0667 0.0546 1.000 879 ZNF844 zinc finger protein 844 116918 1 1 1 0 0 1 0 0 0 0 0.0547 0.682 NaN NaN NaN 0.0547 1.000 880 ETFA electron-transfer-flavoprotein, alpha polypeptide (glutaric aciduria II) 83253 1 1 1 0 0 1 0 0 0 0 0.0547 0.696 NaN NaN NaN 0.0547 1.000 881 FBXO5 F-box protein 5 104562 1 1 1 0 0 1 0 0 0 0 0.0548 0.657 NaN NaN NaN 0.0548 1.000 882 MYOCD myocardin 238721 1 1 1 0 0 0 0 0 1 0 0.0548 1.000 NaN NaN NaN 0.0548 1.000 883 PARP16 poly (ADP-ribose) polymerase family, member 16 77393 1 1 1 0 1 0 0 0 0 0 0.0549 0.653 NaN NaN NaN 0.0549 1.000 884 SLC38A1 solute carrier family 38, member 1 125965 1 1 1 0 1 0 0 0 0 0 0.0550 0.620 NaN NaN NaN 0.0550 1.000 885 NTN5 netrin 5 74927 1 1 1 0 1 0 0 0 0 0 0.0550 0.753 NaN NaN NaN 0.0550 1.000 886 ANLN anillin, actin binding protein 287207 2 2 2 0 0 0 0 2 0 0 0.0478 0.701 0.0575 0.408 0.205 0.0551 1.000 887 C1orf163 chromosome 1 open reading frame 163 58764 1 1 1 0 0 1 0 0 0 0 0.0551 0.630 NaN NaN NaN 0.0551 1.000 888 GPR32 G protein-coupled receptor 32 89142 1 1 1 0 1 0 0 0 0 0 0.0552 0.698 NaN NaN NaN 0.0552 1.000 889 AK7 adenylate kinase 7 185534 1 1 1 0 0 1 0 0 0 0 0.0552 0.681 NaN NaN NaN 0.0552 1.000 890 TMEM184C transmembrane protein 184C 112608 1 1 1 0 0 0 0 1 0 0 0.0554 0.841 NaN NaN NaN 0.0554 1.000 891 USP38 ubiquitin specific peptidase 38 262677 1 1 1 0 0 0 1 0 0 0 0.0554 0.617 NaN NaN NaN 0.0554 1.000 892 TMEM110 transmembrane protein 110 62441 1 1 1 0 0 0 0 1 0 0 0.0554 0.838 NaN NaN NaN 0.0554 1.000 893 NAA20 N(alpha)-acetyltransferase 20, NatB catalytic subunit 47415 1 1 1 0 0 1 0 0 0 0 0.0555 0.712 NaN NaN NaN 0.0555 1.000 894 CLDN12 claudin 12 61337 1 1 1 0 0 1 0 0 0 0 0.0555 0.644 NaN NaN NaN 0.0555 1.000 895 TLX1 T-cell leukemia homeobox 1 56391 1 1 1 0 0 0 0 1 0 0 0.0556 0.819 NaN NaN NaN 0.0556 1.000 896 UBTF upstream binding transcription factor, RNA polymerase I 189579 1 1 1 1 0 1 0 0 0 0 0.0558 0.873 NaN NaN NaN 0.0558 1.000 897 SLC5A2 solute carrier family 5 (sodium/glucose cotransporter), member 2 170230 2 2 2 0 0 0 0 0 2 0 0.00998 0.515 0.0838 0.937 1.000 0.0560 1.000 898 ART4 ADP-ribosyltransferase 4 (Dombrock blood group) 79130 1 1 1 0 0 0 0 0 1 0 0.0560 0.842 NaN NaN NaN 0.0560 1.000 899 ZNF441 zinc finger protein 441 167626 1 1 1 0 0 1 0 0 0 0 0.0561 0.681 NaN NaN NaN 0.0561 1.000 900 ST8SIA3 ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 96138 1 1 1 0 0 0 0 1 0 0 0.0562 0.846 NaN NaN NaN 0.0562 1.000 901 SBK2 SH3-binding domain kinase family, member 2 51969 1 1 1 0 1 0 0 0 0 0 0.0564 0.616 NaN NaN NaN 0.0564 1.000 902 C4orf33 chromosome 4 open reading frame 33 51250 1 1 1 0 0 1 0 0 0 0 0.0567 0.705 NaN NaN NaN 0.0567 1.000 903 TCERG1 transcription elongation regulator 1 280058 1 1 1 0 0 0 0 0 1 0 0.0567 0.831 NaN NaN NaN 0.0567 1.000 904 SRPRB signal recognition particle receptor, B subunit 70052 1 1 1 0 0 0 0 1 0 0 0.0567 0.827 NaN NaN NaN 0.0567 1.000 905 GIPC3 GIPC PDZ domain containing family, member 3 58202 1 1 1 0 0 0 0 1 0 0 0.0568 0.828 NaN NaN NaN 0.0568 1.000 906 TLL1 tolloid-like 1 259335 2 2 2 1 1 0 0 0 1 0 0.0102 0.908 0.547 0.419 1.000 0.0570 1.000 907 PPARG peroxisome proliferator-activated receptor gamma 128229 1 1 1 0 0 1 0 0 0 0 0.0570 0.659 NaN NaN NaN 0.0570 1.000 908 FKBP4 FK506 binding protein 4, 59kDa 112352 2 2 2 0 0 0 0 2 0 0 0.0102 0.720 0.177 0.888 1.000 0.0571 1.000 909 KIFC2 kinesin family member C2 154127 1 1 1 0 0 0 0 1 0 0 0.0572 0.787 NaN NaN NaN 0.0572 1.000 910 LOXL4 lysyl oxidase-like 4 188777 1 1 1 0 0 0 0 0 1 0 0.0574 0.826 NaN NaN NaN 0.0574 1.000 911 GBP1 guanylate binding protein 1, interferon-inducible, 67kDa 150886 1 1 1 0 0 1 0 0 0 0 0.0574 0.647 NaN NaN NaN 0.0574 1.000 912 PCMTD1 protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 90702 1 1 1 0 0 1 0 0 0 0 0.0575 0.684 NaN NaN NaN 0.0575 1.000 913 EFEMP2 EGF-containing fibulin-like extracellular matrix protein 2 99872 1 1 1 0 0 0 0 1 0 0 0.0575 0.841 NaN NaN NaN 0.0575 1.000 914 COL9A2 collagen, type IX, alpha 2 126744 1 1 1 0 0 0 0 0 1 0 0.0575 1.000 NaN NaN NaN 0.0575 1.000 915 GPRIN2 G protein regulated inducer of neurite outgrowth 2 109021 1 1 1 0 1 0 0 0 0 0 0.0575 0.699 NaN NaN NaN 0.0575 1.000 916 ERMAP erythroblast membrane-associated protein (Scianna blood group) 117631 1 1 1 0 1 0 0 0 0 0 0.0576 0.743 NaN NaN NaN 0.0576 1.000 917 OR5H14 olfactory receptor, family 5, subfamily H, member 14 77655 1 1 1 0 0 0 0 0 1 0 0.0576 1.000 NaN NaN NaN 0.0576 1.000 918 GALNT13 UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13) 142209 1 1 1 1 0 0 0 0 1 0 0.0576 1.000 NaN NaN NaN 0.0576 1.000 919 SPG20 spastic paraplegia 20 (Troyer syndrome) 168738 1 1 1 0 0 1 0 0 0 0 0.0577 0.703 NaN NaN NaN 0.0577 1.000 920 CCNA1 cyclin A1 118928 1 1 1 0 0 1 0 0 0 0 0.0578 0.680 NaN NaN NaN 0.0578 1.000 921 ZNF621 zinc finger protein 621 96815 1 1 1 0 0 1 0 0 0 0 0.0578 0.670 NaN NaN NaN 0.0578 1.000 922 LIMCH1 LIM and calponin homology domains 1 268511 2 2 2 1 1 0 0 0 1 0 0.0335 0.895 0.120 0.861 0.311 0.0579 1.000 923 PITPNC1 phosphatidylinositol transfer protein, cytoplasmic 1 86897 1 1 1 0 1 0 0 0 0 0 0.0579 0.717 NaN NaN NaN 0.0579 1.000 924 OR4A16 olfactory receptor, family 4, subfamily A, member 16 82009 1 1 1 0 0 0 0 1 0 0 0.0580 0.828 NaN NaN NaN 0.0580 1.000 925 VKORC1L1 vitamin K epoxide reductase complex, subunit 1-like 1 36026 1 1 1 0 1 0 0 0 0 0 0.0580 0.659 NaN NaN NaN 0.0580 1.000 926 PRKAG3 protein kinase, AMP-activated, gamma 3 non-catalytic subunit 121188 1 1 1 0 1 0 0 0 0 0 0.0581 0.705 NaN NaN NaN 0.0581 1.000 927 POC1A POC1 centriolar protein homolog A (Chlamydomonas) 103322 1 1 1 0 0 0 0 1 0 0 0.0581 0.823 NaN NaN NaN 0.0581 1.000 928 CCDC15 coiled-coil domain containing 15 211866 1 1 1 0 0 0 0 0 1 0 0.0581 0.662 NaN NaN NaN 0.0581 1.000 929 OR5K1 olfactory receptor, family 5, subfamily K, member 1 77252 1 1 1 0 0 0 0 1 0 0 0.0581 0.837 NaN NaN NaN 0.0581 1.000 930 TPCN1 two pore segment channel 1 219653 1 1 1 1 0 0 0 0 1 0 0.0583 1.000 NaN NaN NaN 0.0583 1.000 931 KIAA1467 KIAA1467 155948 1 1 1 0 1 0 0 0 0 0 0.0584 0.682 NaN NaN NaN 0.0584 1.000 932 HLA-F major histocompatibility complex, class I, F 104793 1 1 1 0 0 1 0 0 0 0 0.0585 0.653 NaN NaN NaN 0.0585 1.000 933 ZPBP2 zona pellucida binding protein 2 87019 1 1 1 0 0 1 0 0 0 0 0.0586 0.691 NaN NaN NaN 0.0586 1.000 934 C5orf45 chromosome 5 open reading frame 45 81898 1 1 1 0 0 1 0 0 0 0 0.0586 0.661 NaN NaN NaN 0.0586 1.000 935 TTC39C tetratricopeptide repeat domain 39C 135405 1 1 1 0 0 1 0 0 0 0 0.0587 0.664 NaN NaN NaN 0.0587 1.000 936 AGFG1 ArfGAP with FG repeats 1 141405 1 1 1 0 0 0 0 1 0 0 0.0587 0.820 NaN NaN NaN 0.0587 1.000 937 FRMD7 FERM domain containing 7 181864 1 1 1 1 0 1 0 0 0 0 0.0590 0.886 NaN NaN NaN 0.0590 1.000 938 CA2 carbonic anhydrase II 64148 1 1 1 0 0 0 0 1 0 0 0.0590 0.842 NaN NaN NaN 0.0590 1.000 939 SLITRK5 SLIT and NTRK-like family, member 5 234660 2 2 2 0 1 0 1 0 0 0 0.0107 0.466 0.615 0.772 1.000 0.0591 1.000 940 RWDD2A RWD domain containing 2A 73592 1 1 1 0 0 0 0 1 0 0 0.0591 0.860 NaN NaN NaN 0.0591 1.000 941 TAF15 TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa 151300 1 1 1 0 0 0 0 0 1 0 0.0591 0.700 NaN NaN NaN 0.0591 1.000 942 C9orf131 chromosome 9 open reading frame 131 269580 1 1 1 0 0 0 0 0 1 0 0.0591 0.667 NaN NaN NaN 0.0591 1.000 943 PACRGL PARK2 co-regulated-like 57493 1 1 1 0 0 0 0 1 0 0 0.0592 0.830 NaN NaN NaN 0.0592 1.000 944 SV2A synaptic vesicle glycoprotein 2A 188412 1 1 1 2 0 0 0 0 1 0 0.0592 0.970 NaN NaN NaN 0.0592 1.000 945 OR52E6 olfactory receptor, family 52, subfamily E, member 6 78351 1 1 1 0 0 0 0 1 0 0 0.0593 0.830 NaN NaN NaN 0.0593 1.000 946 MAMLD1 mastermind-like domain containing 1 196871 1 1 1 0 1 0 0 0 0 0 0.0593 0.612 NaN NaN NaN 0.0593 1.000 947 RASAL1 RAS protein activator like 1 (GAP1 like) 178133 2 2 2 0 0 0 0 2 0 0 0.0266 0.667 0.870 0.273 0.403 0.0594 1.000 948 ZNF800 zinc finger protein 800 167181 1 1 1 0 1 0 0 0 0 0 0.0595 0.783 NaN NaN NaN 0.0595 1.000 949 KRTAP4-11 keratin associated protein 4-11 42353 2 1 2 1 0 1 0 1 0 0 0.0210 0.768 0.179 0.841 0.512 0.0595 1.000 950 FAM83H family with sequence similarity 83, member H 195410 1 1 1 0 0 0 1 0 0 0 0.0595 0.811 NaN NaN NaN 0.0595 1.000 951 C3orf38 chromosome 3 open reading frame 38 83110 1 1 1 0 0 1 0 0 0 0 0.0595 0.672 NaN NaN NaN 0.0595 1.000 952 CAPZA3 capping protein (actin filament) muscle Z-line, alpha 3 75023 1 1 1 0 0 0 0 1 0 0 0.0596 0.862 NaN NaN NaN 0.0596 1.000 953 GPR141 G protein-coupled receptor 141 76509 1 1 1 0 0 0 0 1 0 0 0.0596 0.839 NaN NaN NaN 0.0596 1.000 954 BMP2K BMP2 inducible kinase 282100 2 2 2 1 0 1 0 0 1 0 0.0140 0.952 0.174 0.799 0.771 0.0596 1.000 955 GALNTL5 UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 113156 1 1 1 0 0 1 0 0 0 0 0.0598 0.670 NaN NaN NaN 0.0598 1.000 956 PRPS1L1 phosphoribosyl pyrophosphate synthetase 1-like 1 79763 1 1 1 0 1 0 0 0 0 0 0.0600 0.737 NaN NaN NaN 0.0600 1.000 957 ZFP3 zinc finger protein 3 homolog (mouse) 125414 1 1 1 0 1 0 0 0 0 0 0.0601 0.850 NaN NaN NaN 0.0601 1.000 958 ANKRD30B ankyrin repeat domain 30B 194526 1 1 1 1 0 0 0 1 0 0 0.0602 0.981 NaN NaN NaN 0.0602 1.000 959 RTN4R reticulon 4 receptor 104122 1 1 1 0 0 0 0 1 0 0 0.0603 0.781 NaN NaN NaN 0.0603 1.000 960 OR4A15 olfactory receptor, family 4, subfamily A, member 15 86154 1 1 1 0 0 0 0 1 0 0 0.0603 0.830 NaN NaN NaN 0.0603 1.000 961 COIL coilin 142720 1 1 1 0 0 1 0 0 0 0 0.0605 0.679 NaN NaN NaN 0.0605 1.000 962 AMDHD2 amidohydrolase domain containing 2 143221 1 1 1 0 0 1 0 0 0 0 0.0606 0.667 NaN NaN NaN 0.0606 1.000 963 SYT16 synaptotagmin XVI 160589 1 1 1 0 1 0 0 0 0 0 0.0607 0.717 NaN NaN NaN 0.0607 1.000 964 ZNF600 zinc finger protein 600 180359 1 1 1 0 0 0 0 0 1 0 0.0607 0.872 NaN NaN NaN 0.0607 1.000 965 TSSK2 testis-specific serine kinase 2 89723 1 1 1 0 0 0 0 1 0 0 0.0607 0.834 NaN NaN NaN 0.0607 1.000 966 GOT1L1 glutamic-oxaloacetic transaminase 1-like 1 90419 1 1 1 0 0 1 0 0 0 0 0.0608 0.658 NaN NaN NaN 0.0608 1.000 967 AMBN ameloblastin (enamel matrix protein) 115695 1 1 1 1 0 1 0 0 0 0 0.0609 0.949 NaN NaN NaN 0.0609 1.000 968 ACTG1 actin, gamma 1 95284 1 1 1 0 0 1 0 0 0 0 0.0609 0.652 NaN NaN NaN 0.0609 1.000 969 DZIP3 DAZ interacting protein 3, zinc finger 305634 1 1 1 1 0 0 1 0 0 0 0.0611 0.827 NaN NaN NaN 0.0611 1.000 970 EDN2 endothelin 2 40300 1 1 1 0 0 0 0 1 0 0 0.0611 0.802 NaN NaN NaN 0.0611 1.000 971 RNF26 ring finger protein 26 108398 1 1 1 0 0 0 0 1 0 0 0.0611 0.790 NaN NaN NaN 0.0611 1.000 972 KRTAP10-3 keratin associated protein 10-3 55604 1 1 1 0 1 0 0 0 0 0 0.0611 0.593 NaN NaN NaN 0.0611 1.000 973 C14orf166B chromosome 14 open reading frame 166B 117745 2 2 2 0 0 0 0 2 0 0 0.0115 0.713 0.331 0.895 0.965 0.0611 1.000 974 CSF2RA colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) 117489 1 1 1 0 0 0 0 1 0 0 0.0613 0.857 NaN NaN NaN 0.0613 1.000 975 FUK fucokinase 219396 1 1 1 1 0 0 0 0 1 0 0.0614 0.924 NaN NaN NaN 0.0614 1.000 976 LARP4 La ribonucleoprotein domain family, member 4 186067 1 1 1 0 0 0 0 0 1 0 0.0614 1.000 NaN NaN NaN 0.0614 1.000 977 LRRC43 leucine rich repeat containing 43 151964 1 1 1 0 0 1 0 0 0 0 0.0615 0.630 NaN NaN NaN 0.0615 1.000 978 AKNAD1 AKNA domain containing 1 213186 1 1 1 0 1 0 0 0 0 0 0.0615 0.600 NaN NaN NaN 0.0615 1.000 979 ZNF182 zinc finger protein 182 155283 1 1 1 0 0 0 0 1 0 0 0.0615 0.866 NaN NaN NaN 0.0615 1.000 980 CCDC8 coiled-coil domain containing 8 134534 1 1 1 0 0 1 0 0 0 0 0.0616 0.659 NaN NaN NaN 0.0616 1.000 981 FUT10 fucosyltransferase 10 (alpha (1,3) fucosyltransferase) 120745 1 1 1 0 0 1 0 0 0 0 0.0616 0.654 NaN NaN NaN 0.0616 1.000 982 CCDC135 coiled-coil domain containing 135 222900 1 1 1 0 0 0 1 0 0 0 0.0620 0.873 NaN NaN NaN 0.0620 1.000 983 CCDC34 coiled-coil domain containing 34 92161 1 1 1 0 0 1 0 0 0 0 0.0620 0.675 NaN NaN NaN 0.0620 1.000 984 TALDO1 transaldolase 1 80405 1 1 1 0 0 0 0 1 0 0 0.0621 0.831 NaN NaN NaN 0.0621 1.000 985 MTHFR 5,10-methylenetetrahydrofolate reductase (NADPH) 167067 1 1 1 0 0 1 0 0 0 0 0.0622 0.637 NaN NaN NaN 0.0622 1.000 986 KLF11 Kruppel-like factor 11 125197 1 1 1 0 1 0 0 0 0 0 0.0622 0.697 NaN NaN NaN 0.0622 1.000 987 FAM53C family with sequence similarity 53, member C 99185 1 1 1 0 1 0 0 0 0 0 0.0622 0.850 NaN NaN NaN 0.0622 1.000 988 SLC22A7 solute carrier family 22 (organic anion transporter), member 7 138834 1 1 1 0 1 0 0 0 0 0 0.0623 0.698 NaN NaN NaN 0.0623 1.000 989 KRT10 keratin 10 (epidermolytic hyperkeratosis; keratosis palmaris et plantaris) 126897 1 1 1 0 1 0 0 0 0 0 0.0625 0.694 NaN NaN NaN 0.0625 1.000 990 NXNL1 nucleoredoxin-like 1 36940 1 1 1 0 1 0 0 0 0 0 0.0627 0.661 NaN NaN NaN 0.0627 1.000 991 MRPL1 mitochondrial ribosomal protein L1 84072 1 1 1 0 0 0 0 1 0 0 0.0627 0.856 NaN NaN NaN 0.0627 1.000 992 DSCC1 defective in sister chromatid cohesion 1 homolog (S. cerevisiae) 87668 1 1 1 0 0 0 0 0 1 0 0.0627 1.000 NaN NaN NaN 0.0627 1.000 993 KRT73 keratin 73 137672 1 1 1 0 1 0 0 0 0 0 0.0627 0.683 NaN NaN NaN 0.0627 1.000 994 PRSS55 protease, serine, 55 89128 1 1 1 0 0 1 0 0 0 0 0.0627 0.671 NaN NaN NaN 0.0627 1.000 995 FAM123C family with sequence similarity 123C 200915 1 1 1 0 0 1 0 0 0 0 0.0629 0.639 NaN NaN NaN 0.0629 1.000 996 ZMYND12 zinc finger, MYND-type containing 12 93736 1 1 1 0 0 0 0 1 0 0 0.0629 0.861 NaN NaN NaN 0.0629 1.000 997 RC3H2 ring finger and CCCH-type zinc finger domains 2 309738 1 1 1 0 0 0 1 0 0 0 0.0629 0.571 NaN NaN NaN 0.0629 1.000 998 MX1 myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) 169401 1 1 1 0 1 0 0 0 0 0 0.0629 0.645 NaN NaN NaN 0.0629 1.000 999 PUS3 pseudouridylate synthase 3 121014 1 1 1 0 0 0 0 1 0 0 0.0629 0.851 NaN NaN NaN 0.0629 1.000 1000 HPS1 Hermansky-Pudlak syndrome 1 160997 1 1 1 0 1 0 0 0 0 0 0.0629 0.669 NaN NaN NaN 0.0629 1.000 1001 TSPAN2 tetraspanin 2 49351 1 1 1 0 0 0 0 1 0 0 0.0630 0.821 NaN NaN NaN 0.0630 1.000 1002 GALNT11 UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) 155285 1 1 1 0 0 1 0 0 0 0 0.0631 0.669 NaN NaN NaN 0.0631 1.000 1003 MSC musculin (activated B-cell factor-1) 43066 1 1 1 0 1 0 0 0 0 0 0.0631 0.642 NaN NaN NaN 0.0631 1.000 1004 SELENBP1 selenium binding protein 1 116051 1 1 1 0 1 0 0 0 0 0 0.0632 0.688 NaN NaN NaN 0.0632 1.000 1005 ROR2 receptor tyrosine kinase-like orphan receptor 2 226351 2 2 2 1 1 1 0 0 0 0 0.0277 0.704 0.486 0.325 0.419 0.0633 1.000 1006 HFM1 HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) 364168 2 2 2 1 1 0 0 0 1 0 0.0116 0.922 0.761 0.270 1.000 0.0635 1.000 1007 SLC25A20 solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 74033 1 1 1 0 0 0 0 1 0 0 0.0635 0.820 NaN NaN NaN 0.0635 1.000 1008 ELMO3 engulfment and cell motility 3 195347 1 1 1 0 0 0 0 0 1 0 0.0635 0.596 NaN NaN NaN 0.0635 1.000 1009 NCKAP1L NCK-associated protein 1-like 291032 1 1 1 0 1 0 0 0 0 0 0.0635 0.833 NaN NaN NaN 0.0635 1.000 1010 ITK IL2-inducible T-cell kinase 160168 1 1 1 0 1 0 0 0 0 0 0.0636 0.787 NaN NaN NaN 0.0636 1.000 1011 TNKS tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 338407 3 3 3 0 0 1 0 2 0 0 0.0117 0.462 0.795 0.655 1.000 0.0636 1.000 1012 VSIG4 V-set and immunoglobulin domain containing 4 96096 1 1 1 0 0 1 0 0 0 0 0.0636 0.655 NaN NaN NaN 0.0636 1.000 1013 SPDYE6 speedy homolog E6 (Xenopus laevis) 42805 1 1 1 0 1 0 0 0 0 0 0.0638 0.868 NaN NaN NaN 0.0638 1.000 1014 PRRX1 paired related homeobox 1 64691 1 1 1 0 1 0 0 0 0 0 0.0638 0.640 NaN NaN NaN 0.0638 1.000 1015 ZNF528 zinc finger protein 528 157908 1 1 1 0 0 0 0 1 0 0 0.0641 0.870 NaN NaN NaN 0.0641 1.000 1016 ARHGAP11A Rho GTPase activating protein 11A 260324 1 1 1 0 0 0 0 0 1 0 0.0642 1.000 NaN NaN NaN 0.0642 1.000 1017 C9orf98 chromosome 9 open reading frame 98 102806 1 1 1 0 0 0 0 1 0 0 0.0644 0.841 NaN NaN NaN 0.0644 1.000 1018 DDX28 DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 123462 1 1 1 0 0 0 0 1 0 0 0.0644 0.789 NaN NaN NaN 0.0644 1.000 1019 HGF hepatocyte growth factor (hepapoietin A; scatter factor) 182098 2 2 2 0 0 1 0 1 0 0 0.0144 0.587 0.379 0.142 0.823 0.0644 1.000 1020 SLAIN2 SLAIN motif family, member 2 113840 1 1 1 0 0 0 0 0 1 0 0.0645 0.693 NaN NaN NaN 0.0645 1.000 1021 OR8D2 olfactory receptor, family 8, subfamily D, member 2 77937 1 1 1 0 0 0 0 1 0 0 0.0647 0.817 NaN NaN NaN 0.0647 1.000 1022 CLMN calmin (calponin-like, transmembrane) 253791 2 2 2 0 2 0 0 0 0 0 0.0130 0.368 0.363 0.761 0.920 0.0648 1.000 1023 DCST2 DC-STAMP domain containing 2 195771 1 1 1 0 0 0 0 0 1 0 0.0650 1.000 NaN NaN NaN 0.0650 1.000 1024 FKBP5 FK506 binding protein 5 122155 1 1 1 0 0 1 0 0 0 0 0.0650 0.704 NaN NaN NaN 0.0650 1.000 1025 MSGN1 mesogenin 1 48555 1 1 1 0 0 0 0 1 0 0 0.0652 0.809 NaN NaN NaN 0.0652 1.000 1026 SLC37A4 solute carrier family 37 (glucose-6-phosphate transporter), member 4 98585 1 1 1 0 0 0 0 1 0 0 0.0652 0.809 NaN NaN NaN 0.0652 1.000 1027 ZPBP zona pellucida binding protein 83352 1 1 1 0 0 0 0 1 0 0 0.0652 0.847 NaN NaN NaN 0.0652 1.000 1028 GRIA4 glutamate receptor, ionotrophic, AMPA 4 251527 1 1 1 0 1 0 0 0 0 0 0.0653 0.650 NaN NaN NaN 0.0653 1.000 1029 CCDC39 coiled-coil domain containing 39 183048 1 1 1 0 0 0 0 0 1 0 0.0655 1.000 NaN NaN NaN 0.0655 1.000 1030 SLC5A1 solute carrier family 5 (sodium/glucose cotransporter), member 1 170534 1 1 1 0 1 0 0 0 0 0 0.0656 0.634 NaN NaN NaN 0.0656 1.000 1031 MTHFD1 methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase 241655 1 1 1 0 0 0 0 1 0 0 0.0657 0.822 NaN NaN NaN 0.0657 1.000 1032 WWC2 WW and C2 domain containing 2 257292 1 1 1 0 0 0 1 0 0 0 0.0657 0.647 NaN NaN NaN 0.0657 1.000 1033 CCDC74A coiled-coil domain containing 74A 89555 1 1 1 0 0 0 0 1 0 0 0.0658 0.824 NaN NaN NaN 0.0658 1.000 1034 SPATS2 spermatogenesis associated, serine-rich 2 139081 1 1 1 0 0 0 0 1 0 0 0.0658 0.837 NaN NaN NaN 0.0658 1.000 1035 VPS33B vacuolar protein sorting 33 homolog B (yeast) 156126 1 1 1 0 0 0 0 0 1 0 0.0659 0.848 NaN NaN NaN 0.0659 1.000 1036 ATP5F1 ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1 66240 1 1 1 0 0 1 0 0 0 0 0.0661 0.668 NaN NaN NaN 0.0661 1.000 1037 NAP1L1 nucleosome assembly protein 1-like 1 102089 1 1 1 0 0 1 0 0 0 0 0.0661 0.709 NaN NaN NaN 0.0661 1.000 1038 RPL19 ribosomal protein L19 50356 1 1 1 0 1 0 0 0 0 0 0.0661 0.926 NaN NaN NaN 0.0661 1.000 1039 ITFG1 integrin alpha FG-GAP repeat containing 1 152389 1 1 1 0 0 0 0 0 1 0 0.0661 1.000 NaN NaN NaN 0.0661 1.000 1040 S100PBP S100P binding protein 103472 1 1 1 0 0 1 0 0 0 0 0.0662 0.677 NaN NaN NaN 0.0662 1.000 1041 MED17 mediator complex subunit 17 153942 1 1 1 0 0 0 0 0 1 0 0.0662 1.000 NaN NaN NaN 0.0662 1.000 1042 MKRN3 makorin, ring finger protein, 3 126606 1 1 1 0 0 0 0 1 0 0 0.0662 0.827 NaN NaN NaN 0.0662 1.000 1043 TXNDC11 thioredoxin domain containing 11 221676 1 1 1 1 0 0 0 0 1 0 0.0663 1.000 NaN NaN NaN 0.0663 1.000 1044 AASDH aminoadipate-semialdehyde dehydrogenase 277598 1 1 1 1 1 0 0 0 0 0 0.0663 0.878 NaN NaN NaN 0.0663 1.000 1045 WNT9A wingless-type MMTV integration site family, member 9A 74204 1 1 1 0 0 1 0 0 0 0 0.0664 0.607 NaN NaN NaN 0.0664 1.000 1046 LHCGR luteinizing hormone/choriogonadotropin receptor 162981 1 1 1 0 0 0 0 0 1 0 0.0666 0.834 NaN NaN NaN 0.0666 1.000 1047 ZNF187 zinc finger protein 187 96316 1 1 1 0 0 1 0 0 0 0 0.0666 0.646 NaN NaN NaN 0.0666 1.000 1048 FAM69B family with sequence similarity 69, member B 85875 1 1 1 0 1 0 0 0 0 0 0.0666 0.654 NaN NaN NaN 0.0666 1.000 1049 GPR174 G protein-coupled receptor 174 82146 1 1 1 0 0 1 0 0 0 0 0.0668 0.651 NaN NaN NaN 0.0668 1.000 1050 MOXD1 monooxygenase, DBH-like 1 137857 2 2 2 0 0 1 0 1 0 0 0.0124 0.574 0.762 0.658 1.000 0.0668 1.000 1051 SLC14A2 solute carrier family 14 (urea transporter), member 2 235407 1 1 1 0 1 0 0 0 0 0 0.0669 0.562 NaN NaN NaN 0.0669 1.000 1052 LASS3 LAG1 homolog, ceramide synthase 3 98392 1 1 1 0 0 0 0 1 0 0 0.0670 0.869 NaN NaN NaN 0.0670 1.000 1053 KIAA0586 KIAA0586 322764 1 1 1 0 0 0 0 0 1 0 0.0670 0.500 NaN NaN NaN 0.0670 1.000 1054 GRIP1 glutamate receptor interacting protein 1 276011 1 1 1 0 1 0 0 0 0 0 0.0671 0.697 NaN NaN NaN 0.0671 1.000 1055 ALKBH3 alkB, alkylation repair homolog 3 (E. coli) 74297 1 1 1 0 0 0 0 1 0 0 0.0671 0.839 NaN NaN NaN 0.0671 1.000 1056 JAK1 Janus kinase 1 (a protein tyrosine kinase) 295284 3 2 3 0 0 1 0 1 1 0 0.0301 0.486 0.253 0.820 0.415 0.0671 1.000 1057 MTX1 metaxin 1 58927 1 1 1 0 0 1 0 0 0 0 0.0673 0.644 NaN NaN NaN 0.0673 1.000 1058 IFT80 intraflagellar transport 80 homolog (Chlamydomonas) 199510 1 1 1 0 0 1 0 0 0 0 0.0673 0.695 NaN NaN NaN 0.0673 1.000 1059 RNLS renalase, FAD-dependent amine oxidase 93440 1 1 1 0 0 1 0 0 0 0 0.0676 0.682 NaN NaN NaN 0.0676 1.000 1060 CRB2 crumbs homolog 2 (Drosophila) 232873 1 1 1 0 0 0 0 1 0 0 0.0677 0.801 NaN NaN NaN 0.0677 1.000 1061 CTGF connective tissue growth factor 64156 1 1 1 0 0 0 0 1 0 0 0.0677 0.846 NaN NaN NaN 0.0677 1.000 1062 CUL2 cullin 2 191344 1 1 1 0 0 1 0 0 0 0 0.0678 0.684 NaN NaN NaN 0.0678 1.000 1063 STON1-GTF2A1L STON1-GTF2A1L 297702 2 2 2 0 0 1 0 1 0 0 0.0253 0.573 0.139 0.956 0.499 0.0678 1.000 1064 STK32C serine/threonine kinase 32C 94476 1 1 1 0 0 1 0 0 0 0 0.0680 0.633 NaN NaN NaN 0.0680 1.000 1065 VSTM2A V-set and transmembrane domain containing 2A 52014 1 1 1 0 0 0 0 1 0 0 0.0680 0.837 NaN NaN NaN 0.0680 1.000 1066 SNX31 sorting nexin 31 109560 1 1 1 0 0 0 0 0 1 0 0.0681 0.652 NaN NaN NaN 0.0681 1.000 1067 GRM4 glutamate receptor, metabotropic 4 230254 1 1 1 1 1 0 0 0 0 0 0.0681 0.887 NaN NaN NaN 0.0681 1.000 1068 GSTM2 glutathione S-transferase M2 (muscle) 58191 1 1 1 1 0 0 0 1 0 0 0.0682 0.982 NaN NaN NaN 0.0682 1.000 1069 NOMO3 NODAL modulator 3 128284 1 1 1 0 0 1 0 0 0 0 0.0682 0.680 NaN NaN NaN 0.0682 1.000 1070 PIGQ phosphatidylinositol glycan anchor biosynthesis, class Q 174037 1 1 1 0 0 1 0 0 0 0 0.0682 0.627 NaN NaN NaN 0.0682 1.000 1071 HEXIM1 hexamethylene bis-acetamide inducible 1 87986 1 1 1 0 0 0 0 0 1 0 0.0682 1.000 NaN NaN NaN 0.0682 1.000 1072 ADAM23 ADAM metallopeptidase domain 23 197510 1 1 1 0 1 0 0 0 0 0 0.0684 0.704 NaN NaN NaN 0.0684 1.000 1073 TNFSF12 tumor necrosis factor (ligand) superfamily, member 12 46855 1 1 1 0 0 0 0 1 0 0 0.0684 0.795 NaN NaN NaN 0.0684 1.000 1074 CLDN9 claudin 9 54614 1 1 1 0 0 0 0 1 0 0 0.0685 0.768 NaN NaN NaN 0.0685 1.000 1075 KCNG1 potassium voltage-gated channel, subfamily G, member 1 110489 2 2 2 0 1 0 0 1 0 0 0.0128 0.515 0.0811 0.684 1.000 0.0686 1.000 1076 SHOX short stature homeobox 50024 1 1 1 0 1 0 0 0 0 0 0.0686 0.625 NaN NaN NaN 0.0686 1.000 1077 KLHL4 kelch-like 4 (Drosophila) 186814 1 1 1 0 1 0 0 0 0 0 0.0686 0.741 NaN NaN NaN 0.0686 1.000 1078 MLH1 mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) 194787 1 1 1 0 0 1 0 0 0 0 0.0687 0.668 NaN NaN NaN 0.0687 1.000 1079 HYAL3 hyaluronoglucosaminidase 3 105073 1 1 1 0 0 0 0 1 0 0 0.0689 0.805 NaN NaN NaN 0.0689 1.000 1080 CCDC102A coiled-coil domain containing 102A 94916 1 1 1 0 0 0 0 1 0 0 0.0690 0.836 NaN NaN NaN 0.0690 1.000 1081 HLA-B major histocompatibility complex, class I, B 86328 1 1 1 0 0 0 0 1 0 0 0.0690 0.812 NaN NaN NaN 0.0690 1.000 1082 OR10R2 olfactory receptor, family 10, subfamily R, member 2 83903 1 1 1 0 0 0 0 1 0 0 0.0691 0.819 NaN NaN NaN 0.0691 1.000 1083 DGKI diacylglycerol kinase, iota 244415 1 1 1 0 0 0 0 0 1 0 0.0692 0.839 NaN NaN NaN 0.0692 1.000 1084 CXCR1 chemokine (C-X-C motif) receptor 1 87731 1 1 1 0 1 0 0 0 0 0 0.0694 0.778 NaN NaN NaN 0.0694 1.000 1085 ALDOC aldolase C, fructose-bisphosphate 93541 1 1 1 0 0 0 0 1 0 0 0.0694 0.812 NaN NaN NaN 0.0694 1.000 1086 LRGUK leucine-rich repeats and guanylate kinase domain containing 212198 1 1 1 0 0 1 0 0 0 0 0.0695 0.665 NaN NaN NaN 0.0695 1.000 1087 DNAJB2 DnaJ (Hsp40) homolog, subfamily B, member 2 83665 1 1 1 0 1 0 0 0 0 0 0.0697 0.811 NaN NaN NaN 0.0697 1.000 1088 SRFBP1 serum response factor binding protein 1 109256 1 1 1 0 0 0 0 0 1 0 0.0698 1.000 NaN NaN NaN 0.0698 1.000 1089 STS steroid sulfatase (microsomal), isozyme S 145889 1 1 1 0 1 0 0 0 0 0 0.0699 0.633 NaN NaN NaN 0.0699 1.000 1090 CORIN corin, serine peptidase 266737 1 1 1 0 1 0 0 0 0 0 0.0700 0.655 NaN NaN NaN 0.0700 1.000 1091 OVGP1 oviductal glycoprotein 1, 120kDa (mucin 9, oviductin) 172528 1 1 1 0 0 1 0 0 0 0 0.0700 0.680 NaN NaN NaN 0.0700 1.000 1092 SUPT5H suppressor of Ty 5 homolog (S. cerevisiae) 267714 2 2 2 1 0 0 1 1 0 0 0.0133 0.910 0.768 0.283 0.988 0.0700 1.000 1093 SMARCE1 SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 105720 1 1 1 0 0 1 0 0 0 0 0.0703 0.695 NaN NaN NaN 0.0703 1.000 1094 OR2T3 olfactory receptor, family 2, subfamily T, member 3 78812 1 1 1 0 0 0 0 1 0 0 0.0704 0.818 NaN NaN NaN 0.0704 1.000 1095 CNDP1 carnosine dipeptidase 1 (metallopeptidase M20 family) 130377 1 1 1 0 0 1 0 0 0 0 0.0704 0.675 NaN NaN NaN 0.0704 1.000 1096 CCDC13 coiled-coil domain containing 13 176018 1 1 1 0 1 0 0 0 0 0 0.0704 0.728 NaN NaN NaN 0.0704 1.000 1097 CAPN13 calpain 13 157969 1 1 1 0 0 1 0 0 0 0 0.0704 0.662 NaN NaN NaN 0.0704 1.000 1098 OR52R1 olfactory receptor, family 52, subfamily R, member 1 78878 1 1 1 0 0 0 0 1 0 0 0.0706 0.811 NaN NaN NaN 0.0706 1.000 1099 TTC4 tetratricopeptide repeat domain 4 93902 1 1 1 0 0 0 0 0 1 0 0.0706 1.000 NaN NaN NaN 0.0706 1.000 1100 EMID1 EMI domain containing 1 87449 1 1 1 0 0 0 0 1 0 0 0.0708 0.781 NaN NaN NaN 0.0708 1.000 1101 DENND3 DENN/MADD domain containing 3 300905 2 2 2 0 0 0 0 1 1 0 0.0729 0.704 0.683 0.0977 0.183 0.0708 1.000 1102 FCGR3A Fc fragment of IgG, low affinity IIIa, receptor (CD16a) 73983 1 1 1 0 0 0 0 0 1 0 0.0709 0.653 NaN NaN NaN 0.0709 1.000 1103 INTS12 integrator complex subunit 12 117100 1 1 1 0 0 1 0 0 0 0 0.0710 0.695 NaN NaN NaN 0.0710 1.000 1104 CNGA4 cyclic nucleotide gated channel alpha 4 145152 1 1 1 0 1 0 0 0 0 0 0.0711 0.739 NaN NaN NaN 0.0711 1.000 1105 RAB11A RAB11A, member RAS oncogene family 55317 1 1 1 0 0 0 0 1 0 0 0.0711 0.835 NaN NaN NaN 0.0711 1.000 1106 PCDHGB3 protocadherin gamma subfamily B, 3 234371 2 2 2 0 1 0 0 0 1 0 0.0135 0.605 0.126 0.589 0.991 0.0712 1.000 1107 VASN vasorin 115450 1 1 1 0 0 0 0 1 0 0 0.0712 0.774 NaN NaN NaN 0.0712 1.000 1108 GIGYF2 GRB10 interacting GYF protein 2 313749 1 1 1 0 0 0 1 0 0 0 0.0712 0.588 NaN NaN NaN 0.0712 1.000 1109 FOXL1 forkhead box L1 59389 1 1 1 0 1 0 0 0 0 0 0.0713 0.611 NaN NaN NaN 0.0713 1.000 1110 TNFRSF11B tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) 99697 1 1 1 0 0 0 0 1 0 0 0.0713 0.868 NaN NaN NaN 0.0713 1.000 1111 GH2 growth hormone 2 83898 1 1 1 0 1 0 0 0 0 0 0.0713 0.734 NaN NaN NaN 0.0713 1.000 1112 TSHR thyroid stimulating hormone receptor 205160 1 1 1 0 1 0 0 0 0 0 0.0714 0.630 NaN NaN NaN 0.0714 1.000 1113 FSCN2 fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) 42821 1 1 1 0 0 0 0 1 0 0 0.0714 0.811 NaN NaN NaN 0.0714 1.000 1114 KCNB2 potassium voltage-gated channel, Shab-related subfamily, member 2 227752 2 2 2 0 1 0 0 1 0 0 0.0387 0.520 0.280 0.113 0.348 0.0714 1.000 1115 C19orf57 chromosome 19 open reading frame 57 158444 1 1 1 0 0 1 0 0 0 0 0.0716 0.670 NaN NaN NaN 0.0716 1.000 1116 OR5I1 olfactory receptor, family 5, subfamily I, member 1 78199 1 1 1 0 0 0 0 1 0 0 0.0717 0.834 NaN NaN NaN 0.0717 1.000 1117 IL6ST interleukin 6 signal transducer (gp130, oncostatin M receptor) 233374 1 1 1 0 0 0 0 0 1 0 0.0718 1.000 NaN NaN NaN 0.0718 1.000 1118 BTBD11 BTB (POZ) domain containing 11 217545 1 1 1 0 0 0 0 0 1 0 0.0720 1.000 NaN NaN NaN 0.0720 1.000 1119 LRRC47 leucine rich repeat containing 47 98450 1 1 1 0 0 0 0 1 0 0 0.0723 0.814 NaN NaN NaN 0.0723 1.000 1120 FILIP1 filamin A interacting protein 1 303943 2 2 2 0 0 1 0 1 0 0 0.0574 0.567 0.683 0.0658 0.238 0.0723 1.000 1121 ULK4 unc-51-like kinase 4 (C. elegans) 328534 2 2 2 0 0 1 0 1 0 0 0.0505 0.551 0.455 0.192 0.271 0.0724 1.000 1122 ZNF134 zinc finger protein 134 106941 1 1 1 0 0 1 0 0 0 0 0.0724 0.666 NaN NaN NaN 0.0724 1.000 1123 TPD52L1 tumor protein D52-like 1 52823 1 1 1 0 1 0 0 0 0 0 0.0724 0.660 NaN NaN NaN 0.0724 1.000 1124 GPR27 G protein-coupled receptor 27 39825 1 1 1 0 0 0 0 1 0 0 0.0724 0.784 NaN NaN NaN 0.0724 1.000 1125 PON1 paraoxonase 1 91546 1 1 1 0 0 0 0 1 0 0 0.0725 0.843 NaN NaN NaN 0.0725 1.000 1126 PBX2 pre-B-cell leukemia homeobox 2 101171 1 1 1 1 0 0 0 1 0 0 0.0725 0.967 NaN NaN NaN 0.0725 1.000 1127 VWC2L von Willebrand factor C domain containing protein 2-like 56338 1 1 1 0 0 0 0 1 0 0 0.0725 0.867 NaN NaN NaN 0.0725 1.000 1128 ATG4C ATG4 autophagy related 4 homolog C (S. cerevisiae) 117187 1 1 1 0 0 1 0 0 0 0 0.0725 0.706 NaN NaN NaN 0.0725 1.000 1129 UBQLN2 ubiquilin 2 126753 1 1 1 0 1 0 0 0 0 0 0.0726 0.589 NaN NaN NaN 0.0726 1.000 1130 GRHL3 grainyhead-like 3 (Drosophila) 155356 1 1 1 0 0 0 0 0 1 0 0.0727 0.843 NaN NaN NaN 0.0727 1.000 1131 STARD3 StAR-related lipid transfer (START) domain containing 3 114469 1 1 1 0 0 1 0 0 0 0 0.0727 0.671 NaN NaN NaN 0.0727 1.000 1132 ABCB5 ATP-binding cassette, sub-family B (MDR/TAP), member 5 312538 1 1 1 0 1 0 0 0 0 0 0.0728 0.691 NaN NaN NaN 0.0728 1.000 1133 C16orf46 chromosome 16 open reading frame 46 101592 1 1 1 0 0 1 0 0 0 0 0.0728 0.666 NaN NaN NaN 0.0728 1.000 1134 SGOL2 shugoshin-like 2 (S. pombe) 317574 1 1 1 0 0 0 1 0 0 0 0.0729 0.579 NaN NaN NaN 0.0729 1.000 1135 PLEKHA3 pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 77479 1 1 1 0 0 0 0 0 1 0 0.0730 1.000 NaN NaN NaN 0.0730 1.000 1136 UBR3 ubiquitin protein ligase E3 component n-recognin 3 200388 1 1 1 1 0 1 0 0 0 0 0.0731 0.895 NaN NaN NaN 0.0731 1.000 1137 NEUROG3 neurogenin 3 44798 1 1 1 0 0 0 0 1 0 0 0.0731 0.781 NaN NaN NaN 0.0731 1.000 1138 SSSCA1 Sjogren syndrome/scleroderma autoantigen 1 51095 1 1 1 0 1 0 0 0 0 0 0.0731 0.686 NaN NaN NaN 0.0731 1.000 1139 MFAP4 microfibrillar-associated protein 4 57493 1 1 1 0 0 1 0 0 0 0 0.0731 0.638 NaN NaN NaN 0.0731 1.000 1140 SRRD SRR1 domain containing 69305 1 1 1 0 0 0 0 1 0 0 0.0731 0.858 NaN NaN NaN 0.0731 1.000 1141 ZFAT zinc finger and AT hook domain containing 286933 1 1 1 0 0 0 0 0 1 0 0.0732 0.844 NaN NaN NaN 0.0732 1.000 1142 ATP6V1C2 ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C2 110227 1 1 1 0 0 0 0 1 0 0 0.0732 0.839 NaN NaN NaN 0.0732 1.000 1143 TSPAN7 tetraspanin 7 57477 1 1 1 0 0 0 0 1 0 0 0.0733 0.821 NaN NaN NaN 0.0733 1.000 1144 PSG6 pregnancy specific beta-1-glycoprotein 6 107817 1 1 1 1 0 1 0 0 0 0 0.0733 0.884 NaN NaN NaN 0.0733 1.000 1145 AGPAT2 1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) 63530 1 1 1 0 0 0 0 1 0 0 0.0735 0.805 NaN NaN NaN 0.0735 1.000 1146 DCDC1 doublecortin domain containing 1 90644 1 1 1 0 0 0 0 1 0 0 0.0736 0.845 NaN NaN NaN 0.0736 1.000 1147 TRAM2 translocation associated membrane protein 2 95543 1 1 1 0 0 0 0 0 1 0 0.0737 0.841 NaN NaN NaN 0.0737 1.000 1148 FAM13B family with sequence similarity 13, member B 239744 1 1 1 0 0 0 0 0 1 0 0.0738 0.593 NaN NaN NaN 0.0738 1.000 1149 ZNF781 zinc finger protein 781 81971 1 1 1 0 0 0 0 1 0 0 0.0740 0.858 NaN NaN NaN 0.0740 1.000 1150 RPS6KB2 ribosomal protein S6 kinase, 70kDa, polypeptide 2 121457 1 1 1 0 1 0 0 0 0 0 0.0740 0.682 NaN NaN NaN 0.0740 1.000 1151 TTC29 tetratricopeptide repeat domain 29 109115 1 1 1 0 0 1 0 0 0 0 0.0740 0.670 NaN NaN NaN 0.0740 1.000 1152 KIAA0494 KIAA0494 127025 1 1 1 0 0 1 0 0 0 0 0.0741 0.664 NaN NaN NaN 0.0741 1.000 1153 ANKRD29 ankyrin repeat domain 29 75711 1 1 1 0 0 0 0 1 0 0 0.0743 0.813 NaN NaN NaN 0.0743 1.000 1154 ACAT1 acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) 104213 1 1 1 0 0 0 0 1 0 0 0.0745 0.823 NaN NaN NaN 0.0745 1.000 1155 HIST1H1A histone cluster 1, H1a 54115 1 1 1 0 1 0 0 0 0 0 0.0745 0.534 NaN NaN NaN 0.0745 1.000 1156 TTYH3 tweety homolog 3 (Drosophila) 77841 1 1 1 0 0 0 0 1 0 0 0.0746 0.818 NaN NaN NaN 0.0746 1.000 1157 TMPRSS2 transmembrane protease, serine 2 128805 1 1 1 0 1 0 0 0 0 0 0.0746 0.547 NaN NaN NaN 0.0746 1.000 1158 A2BP1 RNA binding protein, fox-1 homolog (C. elegans) 1 118539 2 2 2 1 1 0 0 0 1 0 0.0149 0.772 0.664 0.408 0.955 0.0746 1.000 1159 DDX19B DEAD (Asp-Glu-Ala-As) box polypeptide 19B 119290 1 1 1 0 0 1 0 0 0 0 0.0747 0.663 NaN NaN NaN 0.0747 1.000 1160 TPX2 TPX2, microtubule-associated, homolog (Xenopus laevis) 191447 1 1 1 0 0 0 0 0 1 0 0.0747 0.851 NaN NaN NaN 0.0747 1.000 1161 TEX13B testis expressed 13B 78578 1 1 1 0 0 1 0 0 0 0 0.0747 0.662 NaN NaN NaN 0.0747 1.000 1162 OR4N2 olfactory receptor, family 4, subfamily N, member 2 76941 1 1 1 0 0 0 0 1 0 0 0.0747 0.816 NaN NaN NaN 0.0747 1.000 1163 ZNF333 zinc finger protein 333 169242 1 1 1 1 0 0 0 0 1 0 0.0748 1.000 NaN NaN NaN 0.0748 1.000 1164 FBLIM1 filamin binding LIM protein 1 101111 1 1 1 0 0 0 0 1 0 0 0.0748 0.817 NaN NaN NaN 0.0748 1.000 1165 LYPD5 LY6/PLAUR domain containing 5 51907 1 1 1 0 0 0 0 1 0 0 0.0749 0.809 NaN NaN NaN 0.0749 1.000 1166 CEP68 centrosomal protein 68kDa 190402 1 1 1 0 1 0 0 0 0 0 0.0749 0.708 NaN NaN NaN 0.0749 1.000 1167 GJA5 gap junction protein, alpha 5, 40kDa 89723 1 1 1 0 1 0 0 0 0 0 0.0749 0.743 NaN NaN NaN 0.0749 1.000 1168 PBXIP1 pre-B-cell leukemia homeobox interacting protein 1 184843 1 1 1 0 1 0 0 0 0 0 0.0750 0.811 NaN NaN NaN 0.0750 1.000 1169 RTN4RL1 reticulon 4 receptor-like 1 93082 1 1 1 1 1 0 0 0 0 0 0.0751 0.853 NaN NaN NaN 0.0751 1.000 1170 CRISP2 cysteine-rich secretory protein 2 63054 1 1 1 0 0 0 0 1 0 0 0.0751 0.864 NaN NaN NaN 0.0751 1.000 1171 FST follistatin 80784 1 1 1 0 0 0 0 1 0 0 0.0752 0.854 NaN NaN NaN 0.0752 1.000 1172 C9orf100 chromosome 9 open reading frame 100 71328 1 1 1 0 0 0 0 1 0 0 0.0753 0.821 NaN NaN NaN 0.0753 1.000 1173 RAB3B RAB3B, member RAS oncogene family 56090 1 1 1 0 1 0 0 0 0 0 0.0753 0.711 NaN NaN NaN 0.0753 1.000 1174 SPDYC speedy homolog C (Drosophila) 75415 1 1 1 0 0 1 0 0 0 0 0.0754 0.638 NaN NaN NaN 0.0754 1.000 1175 FER1L5 fer-1-like 5 (C. elegans) 181498 1 1 1 0 0 1 0 0 0 0 0.0755 0.652 NaN NaN NaN 0.0755 1.000 1176 ANGPT1 angiopoietin 1 127145 1 1 1 0 0 0 0 0 1 0 0.0755 1.000 NaN NaN NaN 0.0755 1.000 1177 DVL2 dishevelled, dsh homolog 2 (Drosophila) 185974 1 1 1 0 0 0 0 0 1 0 0.0755 1.000 NaN NaN NaN 0.0755 1.000 1178 SNRPN small nuclear ribonucleoprotein polypeptide N 62227 1 1 1 0 0 0 0 0 1 0 0.0756 1.000 NaN NaN NaN 0.0756 1.000 1179 SOX17 SRY (sex determining region Y)-box 17 45174 1 1 1 0 1 0 0 0 0 0 0.0756 0.615 NaN NaN NaN 0.0756 1.000 1180 GRIN3B glutamate receptor, ionotropic, N-methyl-D-aspartate 3B 129156 1 1 1 0 0 0 0 1 0 0 0.0761 0.798 NaN NaN NaN 0.0761 1.000 1181 MLEC malectin 65136 1 1 1 0 0 0 0 1 0 0 0.0761 0.840 NaN NaN NaN 0.0761 1.000 1182 EPS15 epidermal growth factor receptor pathway substrate 15 237132 1 1 1 0 0 0 0 0 1 0 0.0761 0.688 NaN NaN NaN 0.0761 1.000 1183 SMARCA1 SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 265391 1 1 1 0 0 0 0 0 1 0 0.0761 0.856 NaN NaN NaN 0.0761 1.000 1184 MMP3 matrix metallopeptidase 3 (stromelysin 1, progelatinase) 122211 1 1 1 0 0 1 0 0 0 0 0.0763 0.681 NaN NaN NaN 0.0763 1.000 1185 SIX1 SIX homeobox 1 71581 1 1 1 0 1 0 0 0 0 0 0.0763 0.625 NaN NaN NaN 0.0763 1.000 1186 FBN1 fibrillin 1 736185 3 3 3 0 0 2 0 1 0 0 0.0284 0.392 0.487 0.355 0.514 0.0763 1.000 1187 GRK7 G protein-coupled receptor kinase 7 139074 1 1 1 0 1 0 0 0 0 0 0.0764 0.664 NaN NaN NaN 0.0764 1.000 1188 SFRS4 splicing factor, arginine/serine-rich 4 125167 1 1 1 0 0 0 0 0 1 0 0.0765 1.000 NaN NaN NaN 0.0765 1.000 1189 KRT222 keratin 222 75562 1 1 1 0 0 0 0 1 0 0 0.0765 0.855 NaN NaN NaN 0.0765 1.000 1190 CEBPZ CCAAT/enhancer binding protein zeta 267371 1 1 1 0 1 0 0 0 0 0 0.0766 0.700 NaN NaN NaN 0.0766 1.000 1191 OR5T1 olfactory receptor, family 5, subfamily T, member 1 81755 1 1 1 1 0 0 0 1 0 0 0.0766 0.943 NaN NaN NaN 0.0766 1.000 1192 TMPRSS7 transmembrane protease, serine 7 153548 1 1 1 0 1 0 0 0 0 0 0.0766 0.712 NaN NaN NaN 0.0766 1.000 1193 HIPK1 homeodomain interacting protein kinase 1 314963 1 1 1 1 0 0 1 0 0 0 0.0767 0.871 NaN NaN NaN 0.0767 1.000 1194 DNM1L dynamin 1-like 189502 1 1 1 0 0 1 0 0 0 0 0.0767 0.678 NaN NaN NaN 0.0767 1.000 1195 CHL1 cell adhesion molecule with homology to L1CAM (close homolog of L1) 313214 1 1 1 0 1 0 0 0 0 0 0.0767 0.650 NaN NaN NaN 0.0767 1.000 1196 AMY2B amylase, alpha 2B (pancreatic) 130808 1 1 1 0 0 0 0 1 0 0 0.0768 0.848 NaN NaN NaN 0.0768 1.000 1197 B3GALT5 UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 77771 1 1 1 0 0 0 0 1 0 0 0.0769 0.848 NaN NaN NaN 0.0769 1.000 1198 OR11L1 olfactory receptor, family 11, subfamily L, member 1 80759 1 1 1 0 0 0 0 1 0 0 0.0769 0.815 NaN NaN NaN 0.0769 1.000 1199 ISX intestine-specific homeobox 59783 1 1 1 0 0 0 0 1 0 0 0.0770 0.814 NaN NaN NaN 0.0770 1.000 1200 C17orf28 chromosome 17 open reading frame 28 167234 1 1 1 0 0 0 0 0 1 0 0.0770 0.824 NaN NaN NaN 0.0770 1.000 1201 GZMH granzyme H (cathepsin G-like 2, protein h-CCPX) 63163 1 1 1 0 0 0 0 1 0 0 0.0773 0.827 NaN NaN NaN 0.0773 1.000 1202 ADA adenosine deaminase 84359 1 1 1 0 0 0 0 1 0 0 0.0773 0.835 NaN NaN NaN 0.0773 1.000 1203 LILRB5 leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 5 146467 1 1 1 1 1 0 0 0 0 0 0.0774 0.928 NaN NaN NaN 0.0774 1.000 1204 HOXB3 homeobox B3 105359 1 1 1 0 0 0 0 1 0 0 0.0774 0.804 NaN NaN NaN 0.0774 1.000 1205 SLC5A8 solute carrier family 5 (iodide transporter), member 8 156129 1 1 1 0 1 0 0 0 0 0 0.0775 0.575 NaN NaN NaN 0.0775 1.000 1206 IDH1 isocitrate dehydrogenase 1 (NADP+), soluble 105613 1 1 1 0 1 0 0 0 0 0 0.0776 0.695 NaN NaN NaN 0.0776 1.000 1207 GSDMD gasdermin D 99148 1 1 1 1 0 0 0 1 0 0 0.0776 0.930 NaN NaN NaN 0.0776 1.000 1208 FAM5B family with sequence similarity 5, member B 197315 1 1 1 0 1 0 0 0 0 0 0.0776 0.790 NaN NaN NaN 0.0776 1.000 1209 ARHGAP11B Rho GTPase activating protein 11B 68637 1 1 1 0 0 0 0 1 0 0 0.0778 0.841 NaN NaN NaN 0.0778 1.000 1210 TWF1 twinfilin, actin-binding protein, homolog 1 (Drosophila) 87847 1 1 1 0 0 0 0 1 0 0 0.0778 0.855 NaN NaN NaN 0.0778 1.000 1211 MS4A12 membrane-spanning 4-domains, subfamily A, member 12 68724 1 1 1 0 0 0 0 1 0 0 0.0779 0.817 NaN NaN NaN 0.0779 1.000 1212 ADRA1D adrenergic, alpha-1D-, receptor 76251 1 1 1 0 1 0 0 0 0 0 0.0779 0.604 NaN NaN NaN 0.0779 1.000 1213 TMEM56 transmembrane protein 56 67716 1 1 1 0 0 0 0 1 0 0 0.0779 0.843 NaN NaN NaN 0.0779 1.000 1214 MSLNL mesothelin-like 188083 2 2 2 0 1 1 0 0 0 0 0.0316 0.405 0.862 0.369 0.474 0.0779 1.000 1215 C14orf106 chromosome 14 open reading frame 106 272750 1 1 1 0 0 0 0 0 1 0 0.0780 1.000 NaN NaN NaN 0.0780 1.000 1216 PPP1R3B protein phosphatase 1, regulatory (inhibitor) subunit 3B 71543 1 1 1 0 0 1 0 0 0 0 0.0780 0.657 NaN NaN NaN 0.0780 1.000 1217 MFN2 mitofusin 2 193918 1 1 1 0 0 0 0 0 1 0 0.0781 0.837 NaN NaN NaN 0.0781 1.000 1218 PPP1R3A protein phosphatase 1, regulatory (inhibitor) subunit 3A 280856 1 1 1 0 0 0 0 0 1 0 0.0781 1.000 NaN NaN NaN 0.0781 1.000 1219 OR6X1 olfactory receptor, family 6, subfamily X, member 1 78103 1 1 1 0 0 0 0 1 0 0 0.0781 0.810 NaN NaN NaN 0.0781 1.000 1220 PCDH18 protocadherin 18 284186 1 1 1 0 1 0 0 0 0 0 0.0782 0.679 NaN NaN NaN 0.0782 1.000 1221 TUBB8 tubulin, beta 8 class VIII 100606 1 1 1 1 0 1 0 0 0 0 0.0782 0.871 NaN NaN NaN 0.0782 1.000 1222 TM9SF2 transmembrane 9 superfamily member 2 170882 1 1 1 0 0 1 0 0 0 0 0.0784 0.682 NaN NaN NaN 0.0784 1.000 1223 ABCB1 ATP-binding cassette, sub-family B (MDR/TAP), member 1 327746 2 2 2 0 1 1 0 0 0 0 0.0151 0.532 0.794 0.958 1.000 0.0784 1.000 1224 PSPC1 paraspeckle component 1 133335 1 1 1 0 1 0 0 0 0 0 0.0785 0.717 NaN NaN NaN 0.0785 1.000 1225 SGSM2 small G protein signaling modulator 2 250900 1 1 1 0 0 0 0 1 0 0 0.0786 0.834 NaN NaN NaN 0.0786 1.000 1226 ZNF676 zinc finger protein 676 147582 1 1 1 1 0 1 0 0 0 0 0.0788 0.890 NaN NaN NaN 0.0788 1.000 1227 EFHB EF-hand domain family, member B 206850 1 1 1 0 0 0 0 1 0 0 0.0788 0.842 NaN NaN NaN 0.0788 1.000 1228 LGSN lengsin, lens protein with glutamine synthetase domain 128291 1 1 1 0 0 1 0 0 0 0 0.0790 0.698 NaN NaN NaN 0.0790 1.000 1229 RXRB retinoid X receptor, beta 104946 1 1 1 0 1 0 0 0 0 0 0.0791 0.781 NaN NaN NaN 0.0791 1.000 1230 ABCG1 ATP-binding cassette, sub-family G (WHITE), member 1 175780 1 1 1 0 0 0 0 1 0 0 0.0794 0.824 NaN NaN NaN 0.0794 1.000 1231 OR4C3 olfactory receptor, family 4, subfamily C, member 3 82336 1 1 1 0 0 0 0 1 0 0 0.0795 0.820 NaN NaN NaN 0.0795 1.000 1232 GSG1L GSG1-like 53484 1 1 1 1 0 0 0 1 0 0 0.0795 0.974 NaN NaN NaN 0.0795 1.000 1233 ASB5 ankyrin repeat and SOCS box-containing 5 84382 1 1 1 1 0 0 0 1 0 0 0.0796 0.944 NaN NaN NaN 0.0796 1.000 1234 WDR77 WD repeat domain 77 77227 1 1 1 0 0 0 0 1 0 0 0.0796 0.814 NaN NaN NaN 0.0796 1.000 1235 ADAM29 ADAM metallopeptidase domain 29 204690 2 2 2 1 0 1 0 0 1 0 0.0154 0.827 0.452 0.822 1.000 0.0796 1.000 1236 CHD1 chromodomain helicase DNA binding protein 1 432240 2 2 2 1 1 0 0 1 0 0 0.0380 0.890 0.672 0.148 0.405 0.0797 1.000 1237 KRT15 keratin 15 116439 1 1 1 0 1 0 0 0 0 0 0.0797 0.698 NaN NaN NaN 0.0797 1.000 1238 ZNF212 zinc finger protein 212 122651 1 1 1 0 0 1 0 0 0 0 0.0797 0.607 NaN NaN NaN 0.0797 1.000 1239 HRASLS5 HRAS-like suppressor family, member 5 66012 1 1 1 0 0 0 0 1 0 0 0.0797 0.844 NaN NaN NaN 0.0797 1.000 1240 ABP1 amiloride binding protein 1 (amine oxidase (copper-containing)) 188026 1 1 1 0 1 0 0 0 0 0 0.0798 0.665 NaN NaN NaN 0.0798 1.000 1241 CNTN6 contactin 6 263175 2 2 2 0 0 1 0 0 1 0 0.0155 0.692 0.970 0.900 0.996 0.0799 1.000 1242 POTEE POTE ankyrin domain family, member E 196207 2 2 2 0 0 1 0 1 0 0 0.0162 0.562 0.365 0.240 0.951 0.0799 1.000 1243 PDK1 pyruvate dehydrogenase kinase, isozyme 1 99121 1 1 1 0 0 0 0 1 0 0 0.0799 0.843 NaN NaN NaN 0.0799 1.000 1244 ADAM10 ADAM metallopeptidase domain 10 186816 1 1 1 0 0 1 0 0 0 0 0.0799 0.689 NaN NaN NaN 0.0799 1.000 1245 ARHGAP36 Rho GTPase activating protein 36 139525 1 1 1 0 1 0 0 0 0 0 0.0799 0.787 NaN NaN NaN 0.0799 1.000 1246 GPRIN3 GPRIN family member 3 193805 2 2 2 0 0 2 0 0 0 0 0.0218 0.468 0.0409 0.950 0.709 0.0800 1.000 1247 OR6K6 olfactory receptor, family 6, subfamily K, member 6 85988 1 1 1 0 0 0 0 1 0 0 0.0800 0.828 NaN NaN NaN 0.0800 1.000 1248 DFNA5 deafness, autosomal dominant 5 126681 1 1 1 0 1 0 0 0 0 0 0.0800 0.575 NaN NaN NaN 0.0800 1.000 1249 QRFPR pyroglutamylated RFamide peptide receptor 106324 1 1 1 0 0 0 0 1 0 0 0.0801 0.833 NaN NaN NaN 0.0801 1.000 1250 HLA-C major histocompatibility complex, class I, C 93266 1 1 1 1 0 0 0 1 0 0 0.0802 0.935 NaN NaN NaN 0.0802 1.000 1251 PTCD3 Pentatricopeptide repeat domain 3 179561 1 1 1 0 0 1 0 0 0 0 0.0805 0.679 NaN NaN NaN 0.0805 1.000 1252 KLHL21 kelch-like 21 (Drosophila) 68459 1 1 1 0 1 0 0 0 0 0 0.0806 0.607 NaN NaN NaN 0.0806 1.000 1253 MRGPRX3 MAS-related GPR, member X3 80759 1 1 1 0 1 0 0 0 0 0 0.0807 0.649 NaN NaN NaN 0.0807 1.000 1254 C2orf3 chromosome 2 open reading frame 3 181068 1 1 1 0 0 1 0 0 0 0 0.0807 0.678 NaN NaN NaN 0.0807 1.000 1255 PBX1 pre-B-cell leukemia homeobox 1 93987 1 1 1 0 0 0 0 1 0 0 0.0807 0.833 NaN NaN NaN 0.0807 1.000 1256 ENPP2 ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) 236192 1 1 1 0 0 1 0 0 0 0 0.0808 0.681 NaN NaN NaN 0.0808 1.000 1257 PPARGC1B peroxisome proliferator-activated receptor gamma, coactivator 1 beta 241840 1 1 1 0 0 0 0 1 0 0 0.0809 0.824 NaN NaN NaN 0.0809 1.000 1258 KCNA4 potassium voltage-gated channel, shaker-related subfamily, member 4 163175 1 1 1 0 1 0 0 0 0 0 0.0809 0.768 NaN NaN NaN 0.0809 1.000 1259 WDR52 WD repeat domain 52 251145 1 1 1 0 0 0 0 0 1 0 0.0809 0.856 NaN NaN NaN 0.0809 1.000 1260 SLCO6A1 solute carrier organic anion transporter family, member 6A1 183365 1 1 1 1 1 0 0 0 0 0 0.0809 0.907 NaN NaN NaN 0.0809 1.000 1261 PHTF2 putative homeodomain transcription factor 2 179005 1 1 1 0 0 1 0 0 0 0 0.0810 0.686 NaN NaN NaN 0.0810 1.000 1262 PACS1 phosphofurin acidic cluster sorting protein 1 217496 1 1 1 0 0 0 0 0 1 0 0.0810 1.000 NaN NaN NaN 0.0810 1.000 1263 CR1L complement component (3b/4b) receptor 1-like 144977 2 2 2 0 0 0 0 2 0 0 0.0157 0.694 0.737 0.951 1.000 0.0810 1.000 1264 BRMS1L breast cancer metastasis-suppressor 1-like 81162 1 1 1 0 0 0 0 1 0 0 0.0810 0.875 NaN NaN NaN 0.0810 1.000 1265 NUBPL nucleotide binding protein-like 82510 1 1 1 0 0 1 0 0 0 0 0.0811 0.700 NaN NaN NaN 0.0811 1.000 1266 ATP5A1 ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle 141748 1 1 1 0 0 0 0 0 1 0 0.0812 1.000 NaN NaN NaN 0.0812 1.000 1267 EML4 echinoderm microtubule associated protein like 4 250955 2 2 2 0 0 1 0 1 0 0 0.0158 0.584 0.582 0.948 1.000 0.0812 1.000 1268 KIF11 kinesin family member 11 267119 1 1 1 0 0 1 0 0 0 0 0.0813 0.675 NaN NaN NaN 0.0813 1.000 1269 LRRC3B leucine rich repeat containing 3B 64964 1 1 1 0 0 0 0 1 0 0 0.0814 0.844 NaN NaN NaN 0.0814 1.000 1270 STAT4 signal transducer and activator of transcription 4 193617 1 1 1 0 0 0 0 0 1 0 0.0814 0.707 NaN NaN NaN 0.0814 1.000 1271 KCNQ1 potassium voltage-gated channel, KQT-like subfamily, member 1 126897 1 1 1 0 0 0 0 1 0 0 0.0815 0.811 NaN NaN NaN 0.0815 1.000 1272 FGD6 FYVE, RhoGEF and PH domain containing 6 363134 1 1 1 0 0 0 1 0 0 0 0.0816 0.598 NaN NaN NaN 0.0816 1.000 1273 AVPR1B arginine vasopressin receptor 1B 105939 1 1 1 0 0 0 0 1 0 0 0.0817 0.791 NaN NaN NaN 0.0817 1.000 1274 BRCA2 breast cancer 2, early onset 857905 3 3 3 0 0 0 0 1 2 0 0.0643 0.859 0.203 0.467 0.248 0.0820 1.000 1275 DND1 dead end homolog 1 (zebrafish) 50940 1 1 1 1 1 0 0 0 0 0 0.0821 0.875 NaN NaN NaN 0.0821 1.000 1276 DHRS7 dehydrogenase/reductase (SDR family) member 7 83048 1 1 1 0 0 0 0 1 0 0 0.0821 0.830 NaN NaN NaN 0.0821 1.000 1277 SLC16A4 solute carrier family 16, member 4 (monocarboxylic acid transporter 5) 124135 1 1 1 0 0 0 0 1 0 0 0.0822 0.830 NaN NaN NaN 0.0822 1.000 1278 PGLYRP4 peptidoglycan recognition protein 4 95659 1 1 1 0 1 0 0 0 0 0 0.0823 0.675 NaN NaN NaN 0.0823 1.000 1279 TTC30A tetratricopeptide repeat domain 30A 158126 1 1 1 0 1 0 0 0 0 0 0.0824 0.693 NaN NaN NaN 0.0824 1.000 1280 FAM189B family with sequence similarity 189, member B 128041 1 1 1 0 0 1 0 0 0 0 0.0824 0.638 NaN NaN NaN 0.0824 1.000 1281 SLC6A9 solute carrier family 6 (neurotransmitter transporter, glycine), member 9 183467 1 1 1 0 0 0 0 0 1 0 0.0824 1.000 NaN NaN NaN 0.0824 1.000 1282 PNLIPRP2 pancreatic lipase-related protein 2 93425 2 1 2 1 0 1 0 0 1 0 0.0806 0.831 0.00934 0.810 0.199 0.0824 1.000 1283 LYAR Ly1 antibody reactive homolog (mouse) 97198 1 1 1 0 0 0 0 1 0 0 0.0825 0.880 NaN NaN NaN 0.0825 1.000 1284 FAM98C family with sequence similarity 98, member C 65935 1 1 1 0 0 0 0 1 0 0 0.0825 0.794 NaN NaN NaN 0.0825 1.000 1285 NIPA2 non imprinted in Prader-Willi/Angelman syndrome 2 91253 1 1 1 0 0 0 0 1 0 0 0.0828 0.820 NaN NaN NaN 0.0828 1.000 1286 FAM124A family with sequence similarity 124A 120105 1 1 1 0 0 0 0 1 0 0 0.0828 0.807 NaN NaN NaN 0.0828 1.000 1287 DNASE2B deoxyribonuclease II beta 89915 1 1 1 0 0 0 0 0 1 0 0.0829 1.000 NaN NaN NaN 0.0829 1.000 1288 PLK3 polo-like kinase 3 (Drosophila) 141056 1 1 1 0 0 0 0 1 0 0 0.0830 0.817 NaN NaN NaN 0.0830 1.000 1289 FCN1 ficolin (collagen/fibrinogen domain containing) 1 84040 1 1 1 0 0 1 0 0 0 0 0.0830 0.667 NaN NaN NaN 0.0830 1.000 1290 GPR78 G protein-coupled receptor 78 80435 1 1 1 0 1 0 0 0 0 0 0.0830 0.633 NaN NaN NaN 0.0830 1.000 1291 MEF2C myocyte enhancer factor 2C 116051 1 1 1 0 0 1 0 0 0 0 0.0830 0.678 NaN NaN NaN 0.0830 1.000 1292 GLIS2 GLIS family zinc finger 2 94893 1 1 1 0 1 0 0 0 0 0 0.0831 0.678 NaN NaN NaN 0.0831 1.000 1293 KLHL13 kelch-like 13 (Drosophila) 165482 2 2 2 0 0 0 0 2 0 0 0.0162 0.703 0.424 0.177 1.000 0.0831 1.000 1294 CD1E CD1e molecule 98814 1 1 1 0 0 0 0 1 0 0 0.0831 0.845 NaN NaN NaN 0.0831 1.000 1295 SSB Sjogren syndrome antigen B (autoantigen La) 105136 1 1 1 0 0 0 0 1 0 0 0.0832 0.878 NaN NaN NaN 0.0832 1.000 1296 SFRS7 splicing factor, arginine/serine-rich 7, 35kDa 62157 1 1 1 0 0 0 0 1 0 0 0.0832 0.793 NaN NaN NaN 0.0832 1.000 1297 LOXL3 lysyl oxidase-like 3 190539 1 1 1 0 0 1 0 0 0 0 0.0832 0.649 NaN NaN NaN 0.0832 1.000 1298 FASLG Fas ligand (TNF superfamily, member 6) 70909 1 1 1 0 0 0 0 1 0 0 0.0832 0.824 NaN NaN NaN 0.0832 1.000 1299 COL6A3 collagen, type VI, alpha 3 808862 2 2 2 1 2 0 0 0 0 0 0.346 0.769 0.222 0.0360 0.0470 0.0832 1.000 1300 JAKMIP3 janus kinase and microtubule interacting protein 3 160309 1 1 1 0 0 1 0 0 0 0 0.0833 0.601 NaN NaN NaN 0.0833 1.000 1301 DPP6 dipeptidyl-peptidase 6 182487 2 2 2 1 1 1 0 0 0 0 0.0241 0.673 0.0940 0.433 0.676 0.0833 1.000 1302 C1orf173 chromosome 1 open reading frame 173 376623 2 2 2 1 0 0 0 2 0 0 0.0799 0.932 0.425 0.294 0.204 0.0834 1.000 1303 CUL5 cullin 5 200407 1 1 1 0 0 0 0 0 1 0 0.0835 1.000 NaN NaN NaN 0.0835 1.000 1304 CADM2 cell adhesion molecule 2 112283 1 1 1 0 0 0 0 1 0 0 0.0835 0.827 NaN NaN NaN 0.0835 1.000 1305 CA14 carbonic anhydrase XIV 87751 1 1 1 0 0 0 0 1 0 0 0.0838 0.830 NaN NaN NaN 0.0838 1.000 1306 CKB creatine kinase, brain 73757 1 1 1 0 0 0 0 1 0 0 0.0838 0.825 NaN NaN NaN 0.0838 1.000 1307 PABPC4 poly(A) binding protein, cytoplasmic 4 (inducible form) 169411 1 1 1 0 0 1 0 0 0 0 0.0839 0.681 NaN NaN NaN 0.0839 1.000 1308 IFIT1 interferon-induced protein with tetratricopeptide repeats 1 119882 1 1 1 1 0 0 0 1 0 0 0.0839 0.954 NaN NaN NaN 0.0839 1.000 1309 SDAD1 SDA1 domain containing 1 178479 2 2 2 0 0 0 0 1 1 0 0.0164 0.869 0.648 0.718 1.000 0.0840 1.000 1310 OR9K2 olfactory receptor, family 9, subfamily K, member 2 83903 1 1 1 0 0 0 0 1 0 0 0.0840 0.823 NaN NaN NaN 0.0840 1.000 1311 HNF1A HNF1 homeobox A 157862 1 1 1 0 0 1 0 0 0 0 0.0841 0.631 NaN NaN NaN 0.0841 1.000 1312 CRK v-crk sarcoma virus CT10 oncogene homolog (avian) 75019 1 1 1 0 0 0 0 1 0 0 0.0841 0.829 NaN NaN NaN 0.0841 1.000 1313 MTSS1 metastasis suppressor 1 188683 1 1 1 0 1 0 0 0 0 0 0.0841 0.712 NaN NaN NaN 0.0841 1.000 1314 TRIP13 thyroid hormone receptor interactor 13 106767 1 1 1 0 1 0 0 0 0 0 0.0843 0.614 NaN NaN NaN 0.0843 1.000 1315 APOF apolipoprotein F 81188 1 1 1 0 0 1 0 0 0 0 0.0844 0.652 NaN NaN NaN 0.0844 1.000 1316 C1orf201 chromosome 1 open reading frame 201 74026 1 1 1 0 0 0 0 1 0 0 0.0845 0.832 NaN NaN NaN 0.0845 1.000 1317 CD209 CD209 molecule 103169 1 1 1 0 0 0 0 1 0 0 0.0845 0.840 NaN NaN NaN 0.0845 1.000 1318 OR2T33 olfactory receptor, family 2, subfamily T, member 33 80166 1 1 1 0 1 0 0 0 0 0 0.0846 0.667 NaN NaN NaN 0.0846 1.000 1319 MRPS9 mitochondrial ribosomal protein S9 102303 1 1 1 0 0 0 0 0 1 0 0.0846 0.849 NaN NaN NaN 0.0846 1.000 1320 POU2F3 POU class 2 homeobox 3 111418 1 1 1 0 0 1 0 0 0 0 0.0848 0.649 NaN NaN NaN 0.0848 1.000 1321 DHX40 DEAH (Asp-Glu-Ala-His) box polypeptide 40 176586 1 1 1 0 1 0 0 0 0 0 0.0848 0.744 NaN NaN NaN 0.0848 1.000 1322 MAP3K10 mitogen-activated protein kinase kinase kinase 10 152567 1 1 1 0 0 1 0 0 0 0 0.0849 0.616 NaN NaN NaN 0.0849 1.000 1323 ZHX1 zinc fingers and homeoboxes 1 217958 1 1 1 0 0 1 0 0 0 0 0.0849 0.695 NaN NaN NaN 0.0849 1.000 1324 FMNL3 formin-like 3 259548 1 1 1 0 0 0 0 0 1 0 0.0849 1.000 NaN NaN NaN 0.0849 1.000 1325 CARS2 cysteinyl-tRNA synthetase 2, mitochondrial (putative) 115177 1 1 1 1 1 0 0 0 0 0 0.0850 0.837 NaN NaN NaN 0.0850 1.000 1326 URGCP upregulator of cell proliferation 229941 1 1 1 0 0 0 0 1 0 0 0.0852 0.827 NaN NaN NaN 0.0852 1.000 1327 OR4D9 olfactory receptor, family 4, subfamily D, member 9 78595 1 1 1 1 0 0 0 1 0 0 0.0853 0.941 NaN NaN NaN 0.0853 1.000 1328 SEZ6L seizure related 6 homolog (mouse)-like 250522 1 1 1 1 0 0 0 0 1 0 0.0853 0.928 NaN NaN NaN 0.0853 1.000 1329 TMEM144 transmembrane protein 144 89775 1 1 1 0 0 0 0 1 0 0 0.0854 0.832 NaN NaN NaN 0.0854 1.000 1330 BBS10 Bardet-Biedl syndrome 10 179314 1 1 1 0 0 1 0 0 0 0 0.0855 0.674 NaN NaN NaN 0.0855 1.000 1331 SEC61A1 Sec61 alpha 1 subunit (S. cerevisiae) 122360 1 1 1 0 0 1 0 0 0 0 0.0856 0.670 NaN NaN NaN 0.0856 1.000 1332 GLOD4 glyoxalase domain containing 4 76810 1 1 1 0 0 1 0 0 0 0 0.0856 0.677 NaN NaN NaN 0.0856 1.000 1333 ITFG3 integrin alpha FG-GAP repeat containing 3 139759 1 1 1 0 0 1 0 0 0 0 0.0857 0.644 NaN NaN NaN 0.0857 1.000 1334 KRTAP10-4 keratin associated protein 10-4 100274 1 1 1 0 0 1 0 0 0 0 0.0859 0.631 NaN NaN NaN 0.0859 1.000 1335 EBF2 early B-cell factor 2 143577 1 1 1 1 1 0 0 0 0 0 0.0860 0.932 NaN NaN NaN 0.0860 1.000 1336 PUM1 pumilio homolog 1 (Drosophila) 302600 1 1 1 0 0 0 0 0 1 0 0.0862 1.000 NaN NaN NaN 0.0862 1.000 1337 FAM75A6 family with sequence similarity 75, member A6 247166 2 2 2 1 1 0 1 0 0 0 0.0196 0.722 0.741 0.493 0.868 0.0862 1.000 1338 CYP1A1 cytochrome P450, family 1, subfamily A, polypeptide 1 129658 1 1 1 0 0 1 0 0 0 0 0.0863 0.623 NaN NaN NaN 0.0863 1.000 1339 PKD2L1 polycystic kidney disease 2-like 1 205910 1 1 1 0 0 1 0 0 0 0 0.0865 0.637 NaN NaN NaN 0.0865 1.000 1340 NEK1 NIMA (never in mitosis gene a)-related kinase 1 279692 1 1 1 0 1 0 0 0 0 0 0.0865 0.762 NaN NaN NaN 0.0865 1.000 1341 PSMA1 proteasome (prosome, macropain) subunit, alpha type, 1 75034 1 1 1 0 0 0 0 1 0 0 0.0866 0.846 NaN NaN NaN 0.0866 1.000 1342 SPRY3 sprouty homolog 3 (Drosophila) 72293 1 1 1 0 0 0 0 1 0 0 0.0867 0.822 NaN NaN NaN 0.0867 1.000 1343 FAM90A1 family with sequence similarity 90, member A1 116307 1 1 1 0 0 1 0 0 0 0 0.0867 0.658 NaN NaN NaN 0.0867 1.000 1344 L1TD1 LINE-1 type transposase domain containing 1 115069 1 1 1 0 0 0 0 1 0 0 0.0868 0.869 NaN NaN NaN 0.0868 1.000 1345 MYNN myoneurin 154190 1 1 1 0 0 1 0 0 0 0 0.0868 0.678 NaN NaN NaN 0.0868 1.000 1346 CDKL3 cyclin-dependent kinase-like 3 123599 1 1 1 0 0 1 0 0 0 0 0.0868 0.687 NaN NaN NaN 0.0868 1.000 1347 EHHADH enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase 182538 1 1 1 0 0 1 0 0 0 0 0.0869 0.675 NaN NaN NaN 0.0869 1.000 1348 TBX5 T-box 5 136461 2 2 2 0 0 1 0 1 0 0 0.0172 0.563 0.435 0.744 1.000 0.0869 1.000 1349 GALNT8 UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 8 (GalNAc-T8) 162230 1 1 1 1 0 0 0 1 0 0 0.0871 0.955 NaN NaN NaN 0.0871 1.000 1350 CHAF1A chromatin assembly factor 1, subunit A (p150) 229806 2 2 2 0 1 1 0 0 0 0 0.0214 0.414 0.418 0.732 0.803 0.0871 1.000 1351 MOCS1 molybdenum cofactor synthesis 1 99071 1 1 1 0 0 0 0 1 0 0 0.0872 0.817 NaN NaN NaN 0.0872 1.000 1352 TNS1 tensin 1 431569 2 2 2 1 0 1 0 1 0 0 0.0683 0.804 0.914 0.0882 0.253 0.0873 1.000 1353 KCNK9 potassium channel, subfamily K, member 9 91310 1 1 1 0 1 0 0 0 0 0 0.0873 0.616 NaN NaN NaN 0.0873 1.000 1354 PAX5 paired box 5 93181 1 1 1 0 0 0 0 1 0 0 0.0873 0.810 NaN NaN NaN 0.0873 1.000 1355 TPH1 tryptophan hydroxylase 1 (tryptophan 5-monooxygenase) 113974 1 1 1 0 0 0 0 1 0 0 0.0874 0.860 NaN NaN NaN 0.0874 1.000 1356 OR4M2 olfactory receptor, family 4, subfamily M, member 2 78506 1 1 1 0 0 0 0 1 0 0 0.0876 0.827 NaN NaN NaN 0.0876 1.000 1357 LRRC17 leucine rich repeat containing 17 112344 1 1 1 0 0 0 0 1 0 0 0.0877 0.872 NaN NaN NaN 0.0877 1.000 1358 SLC39A9 solute carrier family 39 (zinc transporter), member 9 78992 1 1 1 0 0 0 0 1 0 0 0.0879 0.805 NaN NaN NaN 0.0879 1.000 1359 ZBTB17 zinc finger and BTB domain containing 17 173509 1 1 1 0 0 0 0 1 0 0 0.0880 0.832 NaN NaN NaN 0.0880 1.000 1360 ERCC6 excision repair cross-complementing rodent repair deficiency, complementation group 6 378388 2 2 2 0 0 0 0 0 2 0 0.0176 0.727 0.981 0.941 0.992 0.0881 1.000 1361 GPR183 G protein-coupled receptor 183 90470 1 1 1 0 0 0 0 1 0 0 0.0881 0.834 NaN NaN NaN 0.0881 1.000 1362 SUSD1 sushi domain containing 1 182791 1 1 1 0 1 0 0 0 0 0 0.0883 0.667 NaN NaN NaN 0.0883 1.000 1363 ZNF276 zinc finger protein 276 138248 1 1 1 0 0 1 0 0 0 0 0.0884 0.646 NaN NaN NaN 0.0884 1.000 1364 FZD6 frizzled homolog 6 (Drosophila) 177902 1 1 1 0 0 1 0 0 0 0 0.0884 0.681 NaN NaN NaN 0.0884 1.000 1365 LYSMD3 LysM, putative peptidoglycan-binding, domain containing 3 77094 1 1 1 0 0 0 0 1 0 0 0.0885 0.847 NaN NaN NaN 0.0885 1.000 1366 TRIM55 tripartite motif-containing 55 147364 1 1 1 0 0 1 0 0 0 0 0.0886 0.691 NaN NaN NaN 0.0886 1.000 1367 ARHGAP22 Rho GTPase activating protein 22 118402 1 1 1 0 0 0 0 1 0 0 0.0886 0.827 NaN NaN NaN 0.0886 1.000 1368 FERMT2 fermitin family homolog 2 (Drosophila) 175096 1 1 1 0 0 0 0 1 0 0 0.0887 0.857 NaN NaN NaN 0.0887 1.000 1369 SNCAIP synuclein, alpha interacting protein (synphilin) 232278 2 2 2 0 1 0 0 1 0 0 0.0177 0.548 0.831 0.575 1.000 0.0890 1.000 1370 GRK1 G protein-coupled receptor kinase 1 72825 1 1 1 0 0 0 0 1 0 0 0.0890 0.845 NaN NaN NaN 0.0890 1.000 1371 ACAP1 ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 171803 1 1 1 0 0 1 0 0 0 0 0.0890 0.635 NaN NaN NaN 0.0890 1.000 1372 GGA3 golgi associated, gamma adaptin ear containing, ARF binding protein 3 160375 1 1 1 0 1 0 0 0 0 0 0.0891 0.622 NaN NaN NaN 0.0891 1.000 1373 ZNF500 zinc finger protein 500 102868 1 1 1 0 0 0 0 1 0 0 0.0893 0.820 NaN NaN NaN 0.0893 1.000 1374 AGPAT3 1-acylglycerol-3-phosphate O-acyltransferase 3 96358 1 1 1 0 0 0 0 1 0 0 0.0893 0.825 NaN NaN NaN 0.0893 1.000 1375 SLC25A3 solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 102689 1 1 1 0 0 0 0 1 0 0 0.0894 0.823 NaN NaN NaN 0.0894 1.000 1376 PRAME preferentially expressed antigen in melanoma 128297 1 1 1 0 0 1 0 0 0 0 0.0894 0.609 NaN NaN NaN 0.0894 1.000 1377 OR13F1 olfactory receptor, family 13, subfamily F, member 1 79846 1 1 1 0 0 0 0 1 0 0 0.0894 0.819 NaN NaN NaN 0.0894 1.000 1378 CCDC57 coiled-coil domain containing 57 189029 1 1 1 0 0 1 0 0 0 0 0.0895 0.619 NaN NaN NaN 0.0895 1.000 1379 RIOK1 RIO kinase 1 (yeast) 132720 1 1 1 0 0 1 0 0 0 0 0.0895 0.701 NaN NaN NaN 0.0895 1.000 1380 TXLNG taxilin gamma 125102 1 1 1 0 0 0 0 1 0 0 0.0896 0.871 NaN NaN NaN 0.0896 1.000 1381 UGP2 UDP-glucose pyrophosphorylase 2 129952 1 1 1 0 0 0 0 1 0 0 0.0896 0.843 NaN NaN NaN 0.0896 1.000 1382 POTEF POTE ankyrin domain family, member F 213709 1 1 1 1 0 1 0 0 0 0 0.0897 0.885 NaN NaN NaN 0.0897 1.000 1383 TMEM132A transmembrane protein 132A 239812 1 1 1 0 0 0 0 1 0 0 0.0897 0.777 NaN NaN NaN 0.0897 1.000 1384 TTLL2 tubulin tyrosine ligase-like family, member 2 144673 1 1 1 0 0 1 0 0 0 0 0.0898 0.653 NaN NaN NaN 0.0898 1.000 1385 AKR1A1 aldo-keto reductase family 1, member A1 (aldehyde reductase) 83673 1 1 1 0 0 0 0 1 0 0 0.0898 0.826 NaN NaN NaN 0.0898 1.000 1386 ACY3 aspartoacylase (aminocyclase) 3 68855 1 1 1 0 1 0 0 0 0 0 0.0899 0.631 NaN NaN NaN 0.0899 1.000 1387 HMGCS2 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (mitochondrial) 129584 1 1 1 0 1 0 0 0 0 0 0.0899 0.797 NaN NaN NaN 0.0899 1.000 1388 RPGRIP1 retinitis pigmentosa GTPase regulator interacting protein 1 298534 1 1 1 0 0 0 0 0 1 0 0.0899 0.848 NaN NaN NaN 0.0899 1.000 1389 VIPAR VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog 129250 1 1 1 0 1 0 0 0 0 0 0.0899 0.800 NaN NaN NaN 0.0899 1.000 1390 EVX1 even-skipped homeobox 1 71148 1 1 1 0 1 0 0 0 0 0 0.0900 0.589 NaN NaN NaN 0.0900 1.000 1391 GLTSCR2 glioma tumor suppressor candidate region gene 2 77829 1 1 1 0 0 0 0 1 0 0 0.0901 0.813 NaN NaN NaN 0.0901 1.000 1392 C18orf34 chromosome 18 open reading frame 34 219420 1 1 1 1 0 1 0 0 0 0 0.0901 0.930 NaN NaN NaN 0.0901 1.000 1393 SORCS2 sortilin-related VPS10 domain containing receptor 2 195639 1 1 1 0 1 0 0 0 0 0 0.0902 0.618 NaN NaN NaN 0.0902 1.000 1394 BTNL3 butyrophilin-like 3 109672 1 1 1 0 1 0 0 0 0 0 0.0903 0.629 NaN NaN NaN 0.0903 1.000 1395 TM7SF2 transmembrane 7 superfamily member 2 82876 1 1 1 0 0 1 0 0 0 0 0.0904 0.633 NaN NaN NaN 0.0904 1.000 1396 NT5C1A 5-nucleotidase, cytosolic IA | 93870 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0.09 | 0.82 | NaN | NaN | NaN | 0.09 | 1 |
| 1397 | DNMT3A | DNA (cytosine-5-)-methyltransferase 3 alpha | 228174 | 1 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0.09 | 0.96 | NaN | NaN | NaN | 0.09 | 1 |
| 1398 | A1BG | alpha-1-B glycoprotein | 94049 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0.091 | 0.86 | NaN | NaN | NaN | 0.091 | 1 |
| 1399 | GPR133 | G protein-coupled receptor 133 | 216796 | 2 | 2 | 2 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0.018 | 0.62 | 0.84 | 0.72 | 1 | 0.091 | 1 |
| 1400 | C2orf67 | chromosome 2 open reading frame 67 | 250615 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0.091 | 0.98 | NaN | NaN | NaN | 0.091 | 1 |
| 1401 | LILRB3 | leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 | 120480 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0.091 | 0.94 | NaN | NaN | NaN | 0.091 | 1 |
| 1402 | RBM15 | RNA binding motif protein 15 | 243697 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0.091 | 0.8 | NaN | NaN | NaN | 0.091 | 1 |
| 1403 | NEDD4L | neural precursor cell expressed, developmentally down-regulated 4-like | 217352 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0.091 | 1 | NaN | NaN | NaN | 0.091 | 1 |
| 1404 | MSH3 | mutS homolog 3 (E. coli) | 283752 | 2 | 2 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0.026 | 0.58 | 0.15 | 0.58 | 0.7 | 0.091 | 1 |
Figure S1. This figure depicts the distribution of mutations and mutation types across the NKX3-1 significant gene.
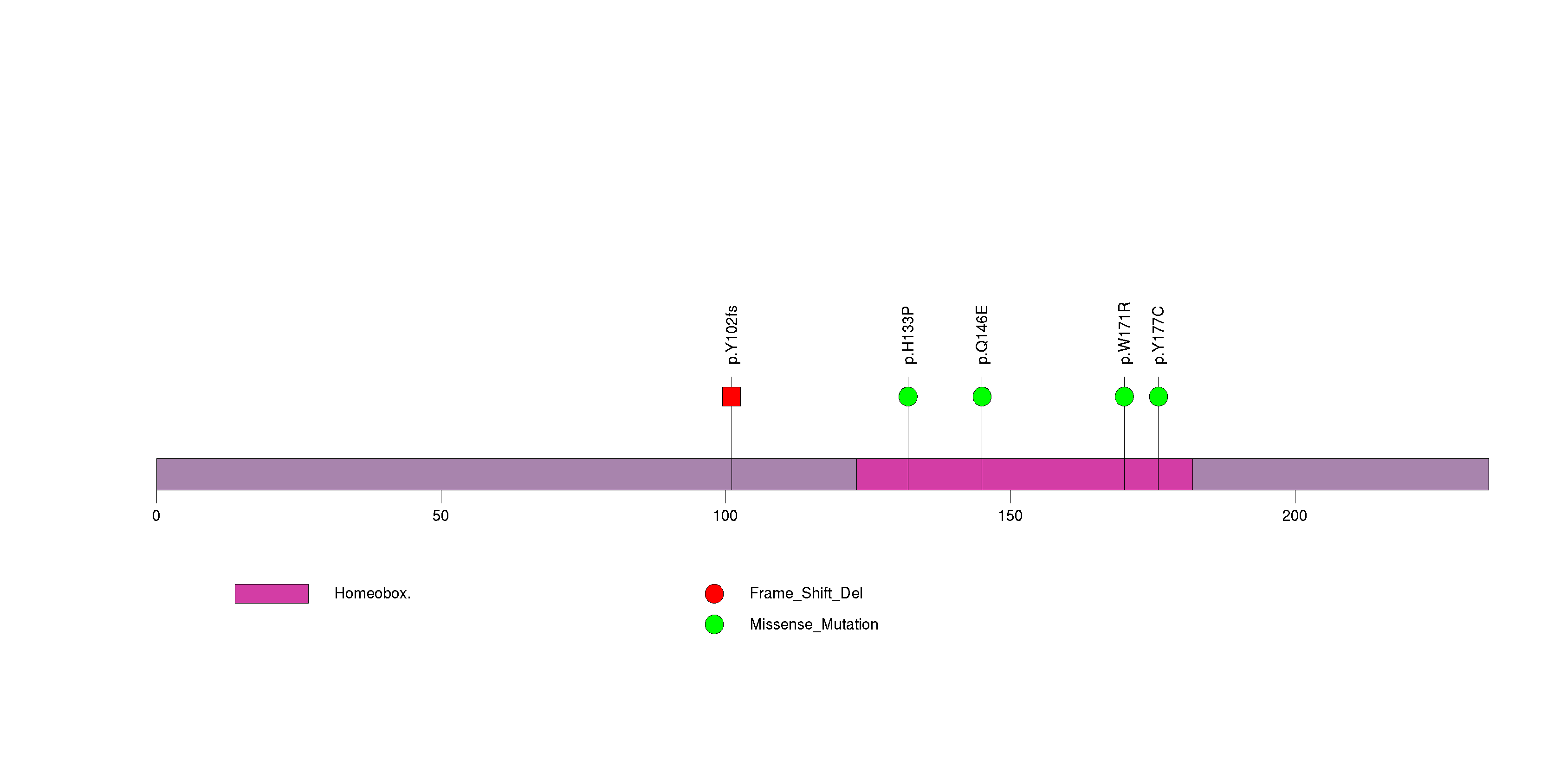
Figure S2. This figure depicts the distribution of mutations and mutation types across the AGT significant gene.

Figure S3. This figure depicts the distribution of mutations and mutation types across the CCNF significant gene.
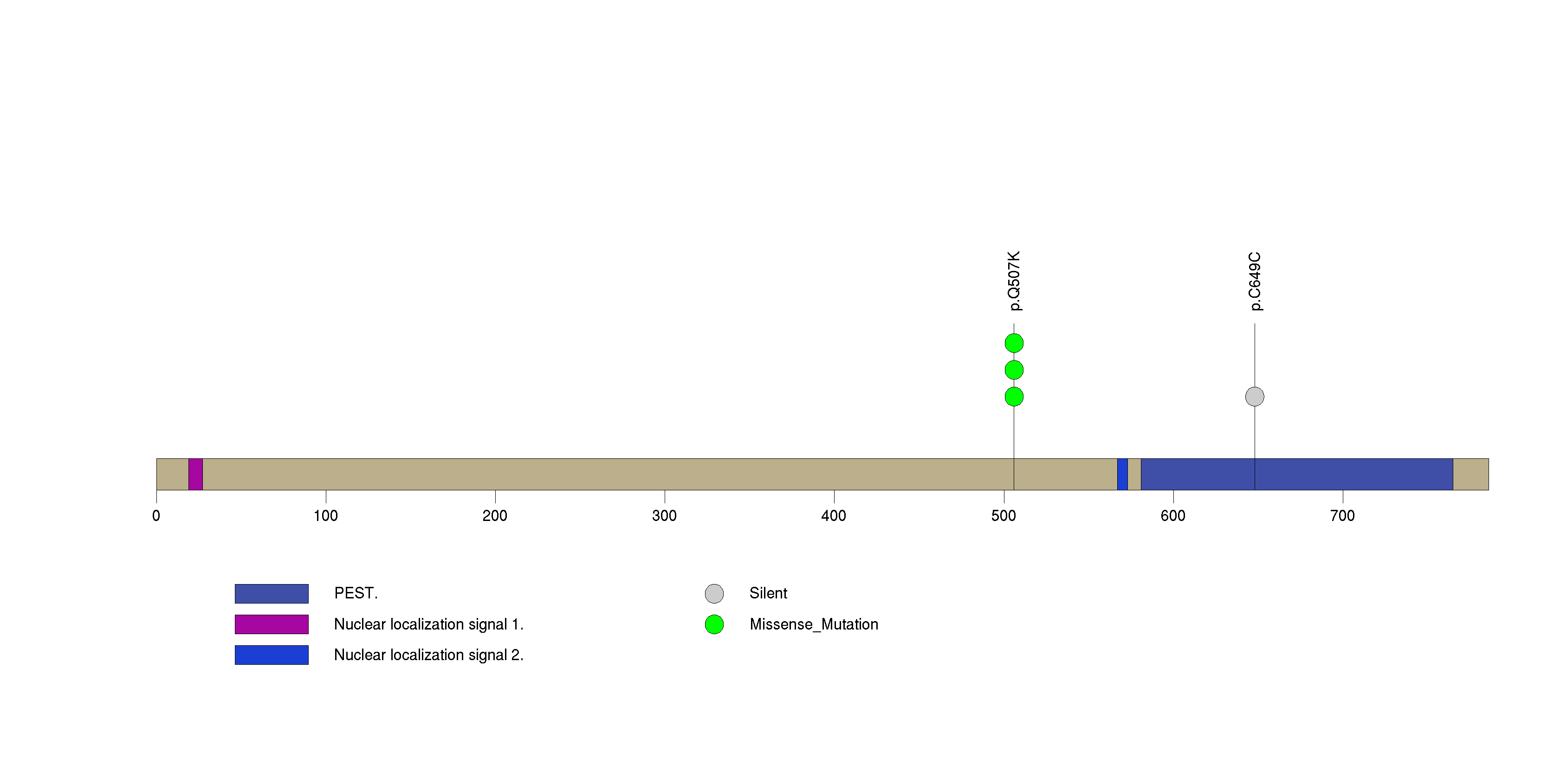
Figure S4. This figure depicts the distribution of mutations and mutation types across the DUSP27 significant gene.

Figure S5. This figure depicts the distribution of mutations and mutation types across the CLSTN1 significant gene.
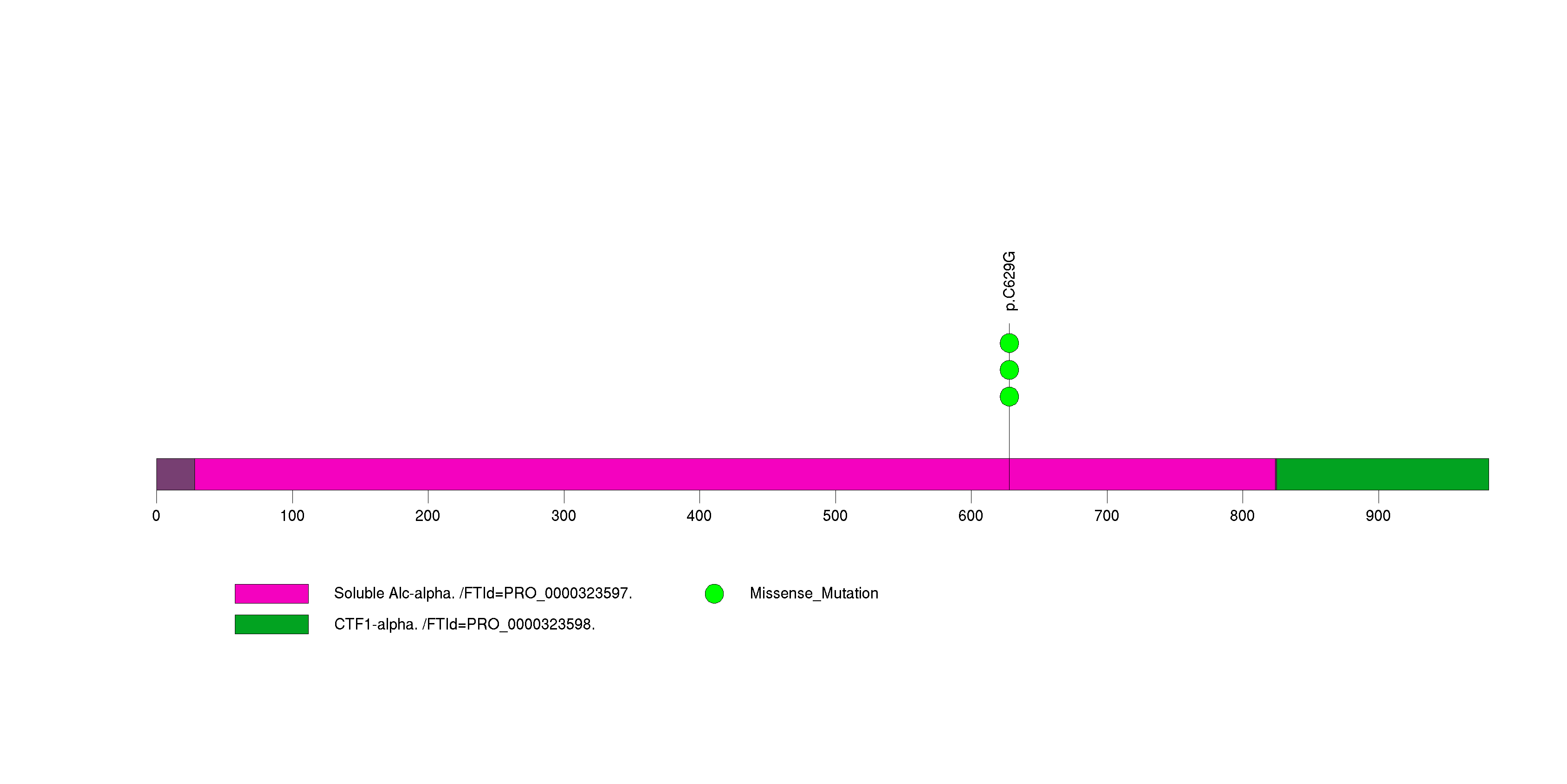
Figure S6. This figure depicts the distribution of mutations and mutation types across the TP53 significant gene.

Figure S7. This figure depicts the distribution of mutations and mutation types across the YBX1 significant gene.
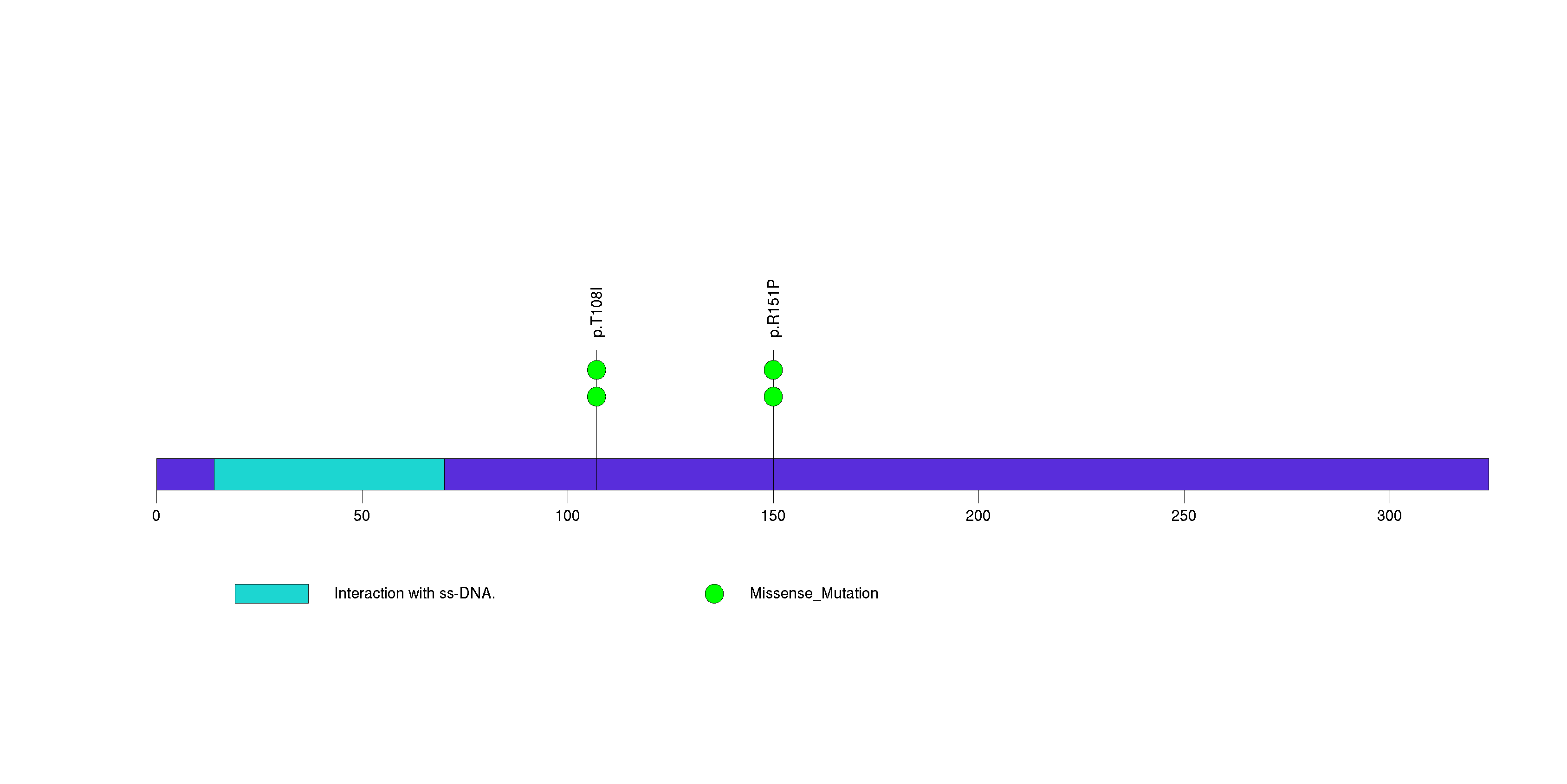
Figure S8. This figure depicts the distribution of mutations and mutation types across the SPOP significant gene.

Figure S9. This figure depicts the distribution of mutations and mutation types across the FRG1 significant gene.

Figure S10. This figure depicts the distribution of mutations and mutation types across the ETV3 significant gene.

Figure S11. This figure depicts the distribution of mutations and mutation types across the GPATCH4 significant gene.

Figure S12. This figure depicts the distribution of mutations and mutation types across the MUC4 significant gene.
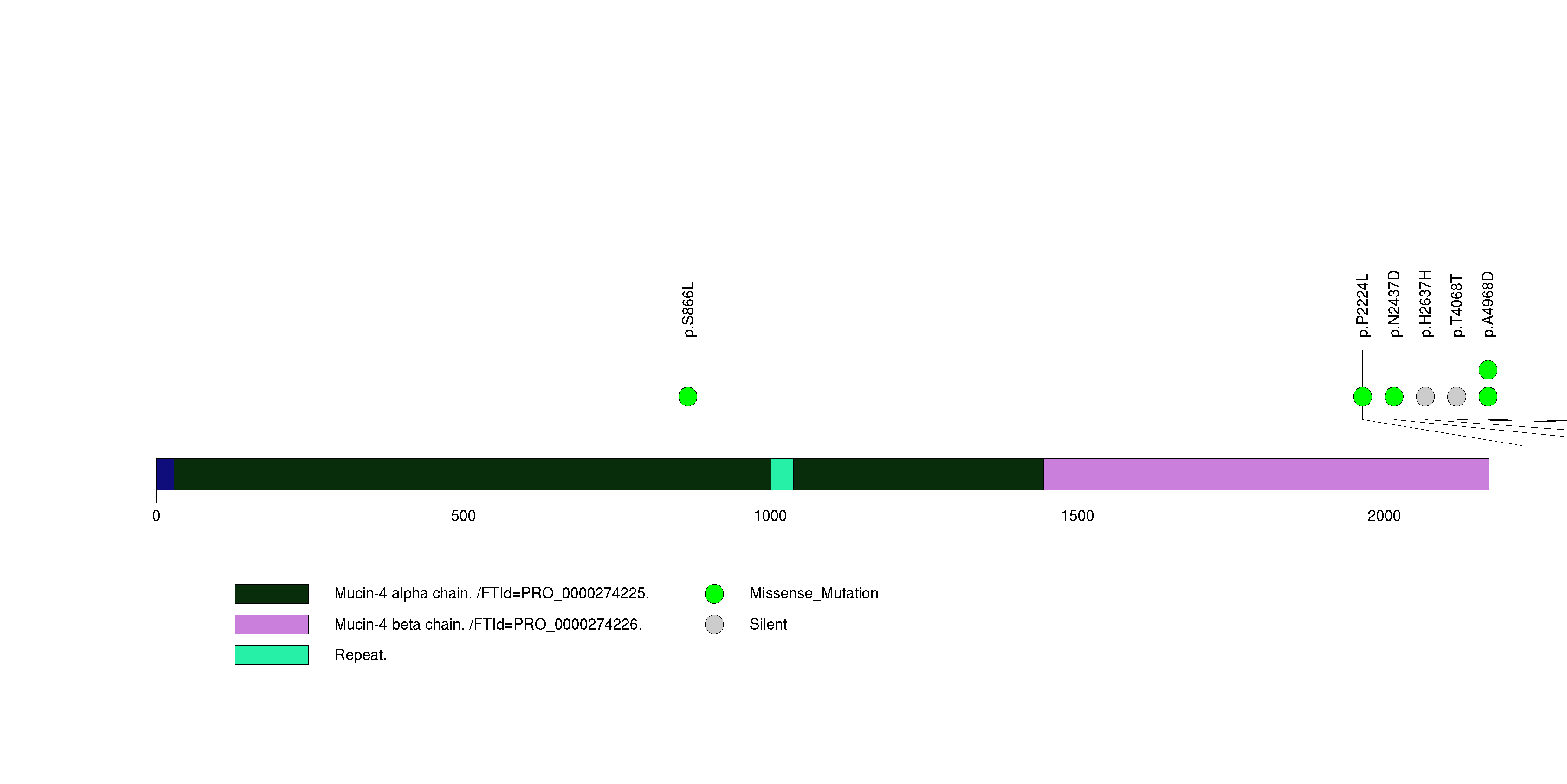
Figure S13. This figure depicts the distribution of mutations and mutation types across the CTNNB1 significant gene.
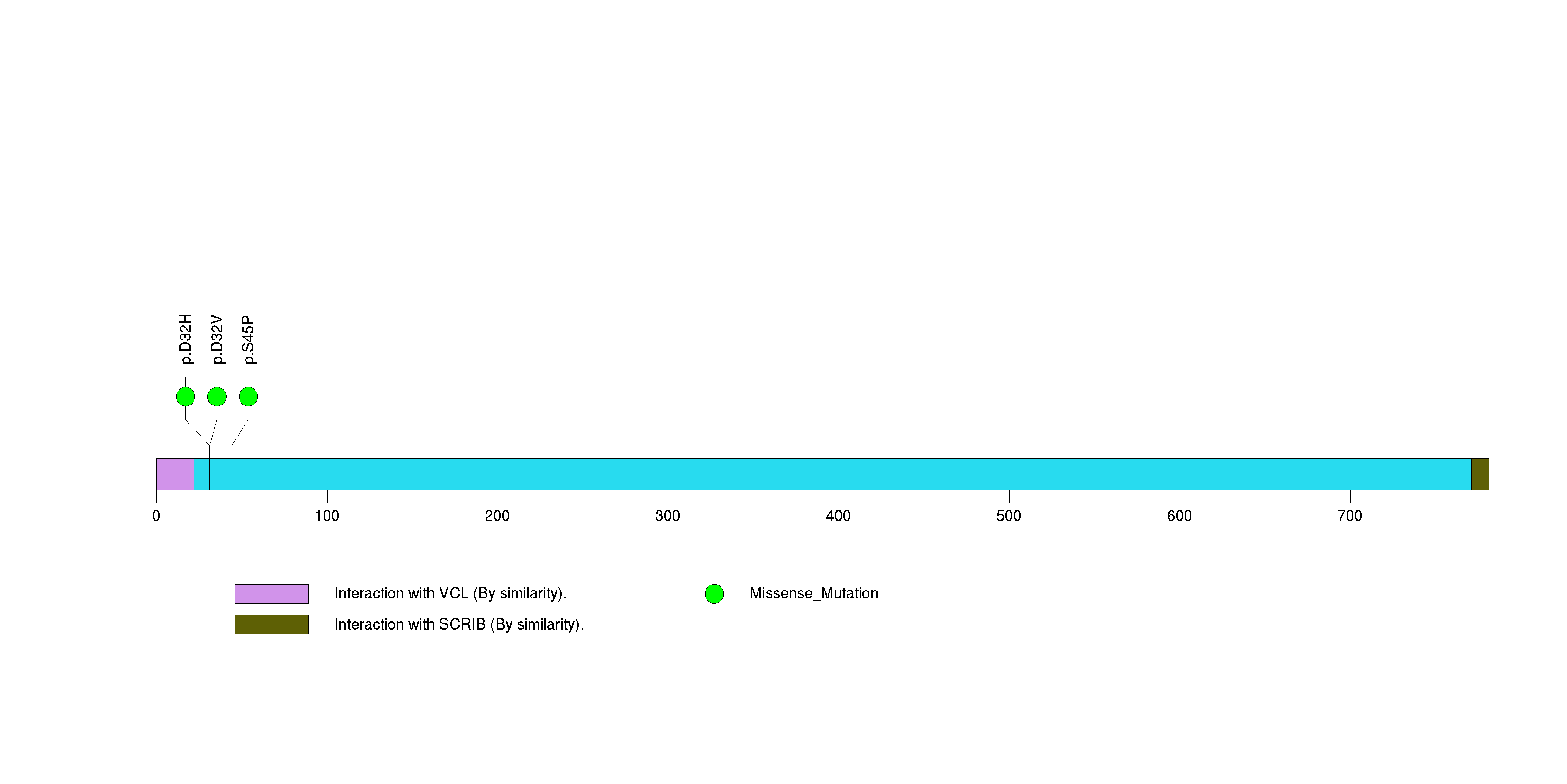
Figure S14. This figure depicts the distribution of mutations and mutation types across the PRR21 significant gene.

Figure S15. This figure depicts the distribution of mutations and mutation types across the SLC15A3 significant gene.

Figure S16. This figure depicts the distribution of mutations and mutation types across the SFRS11 significant gene.
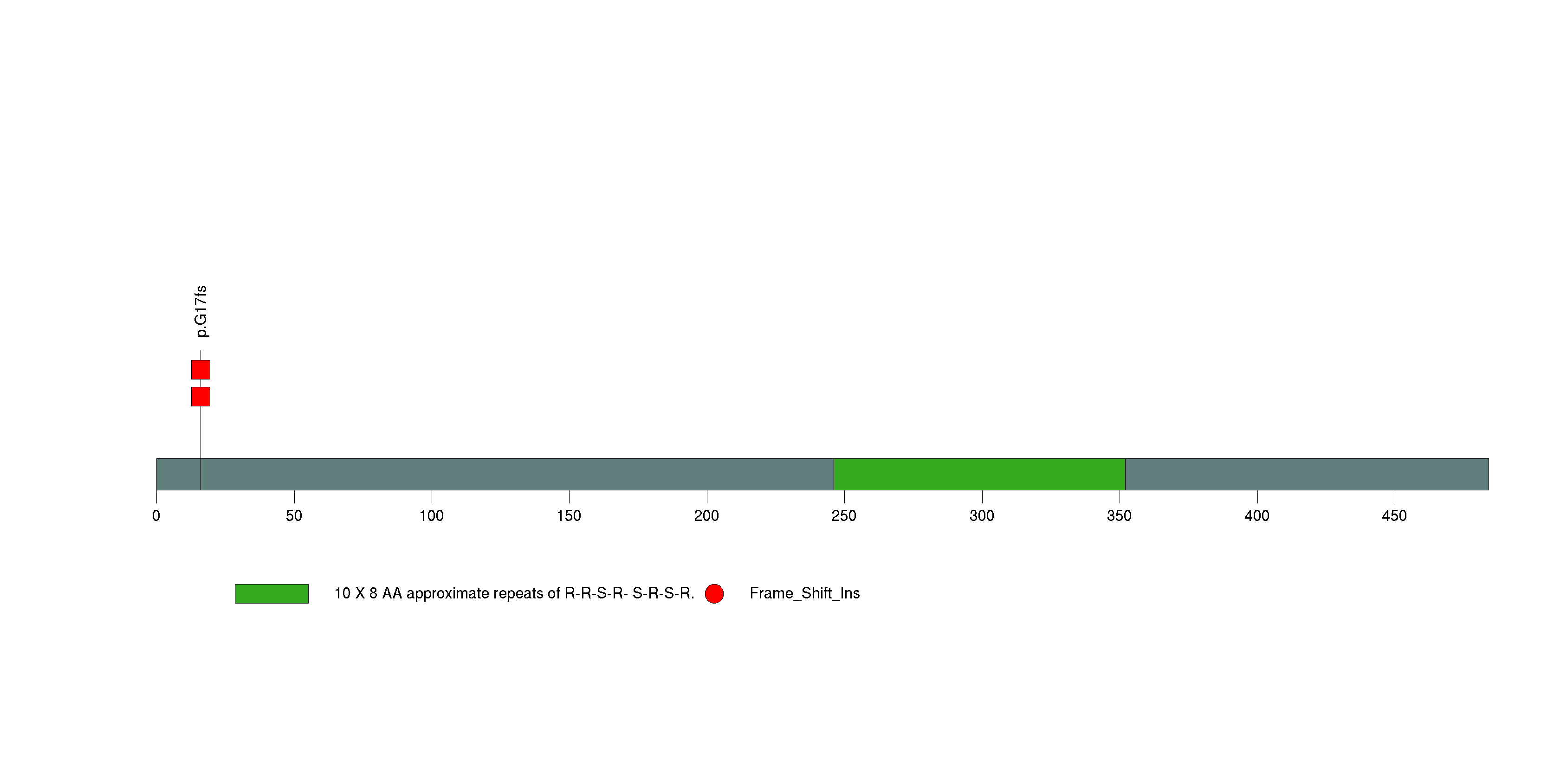
Figure S17. This figure depicts the distribution of mutations and mutation types across the OR6N1 significant gene.
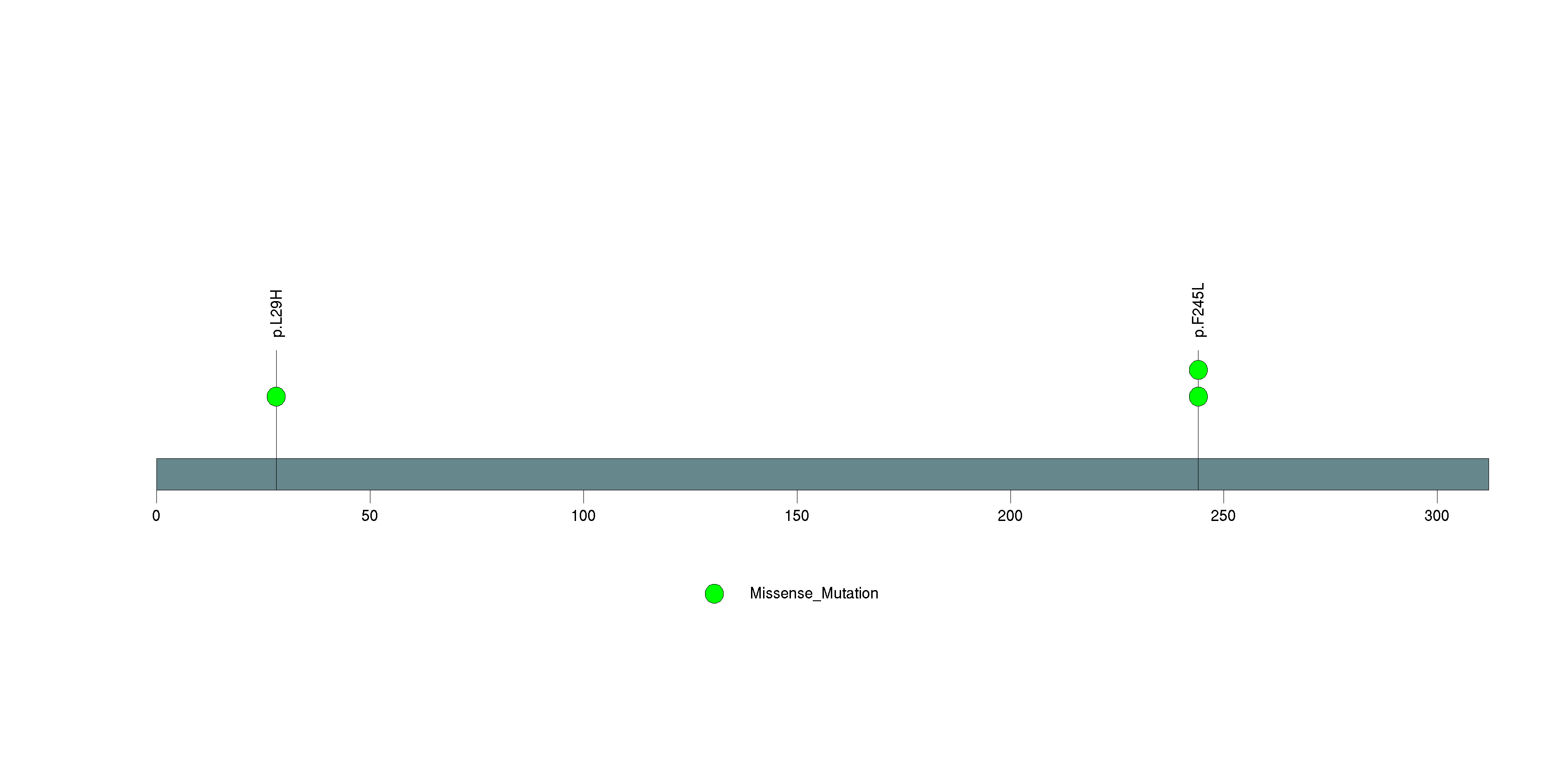
Figure S18. This figure depicts the distribution of mutations and mutation types across the NDN significant gene.
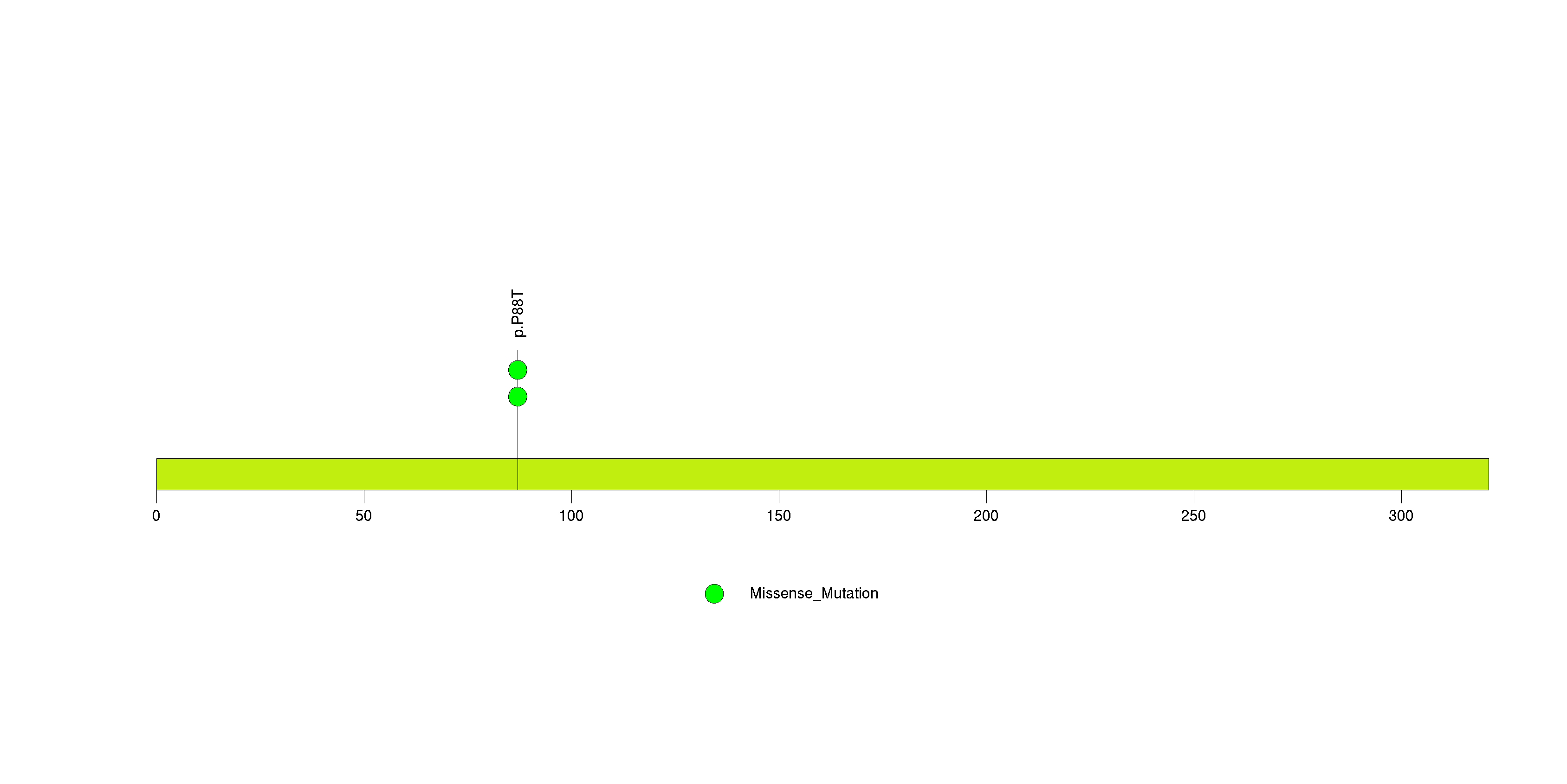
Figure S19. This figure depicts the distribution of mutations and mutation types across the OR4D5 significant gene.
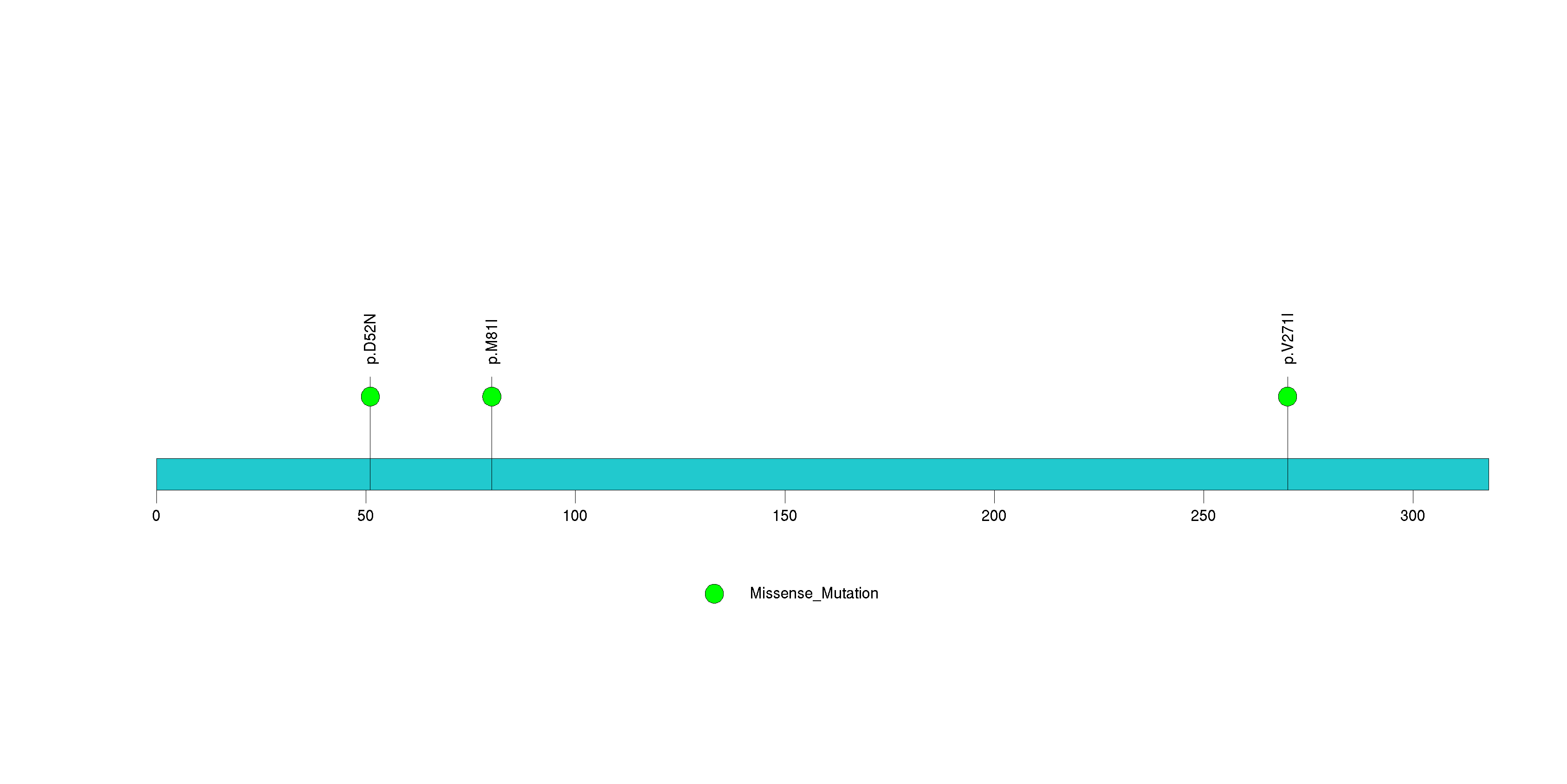
Figure S20. This figure depicts the distribution of mutations and mutation types across the OR6Y1 significant gene.
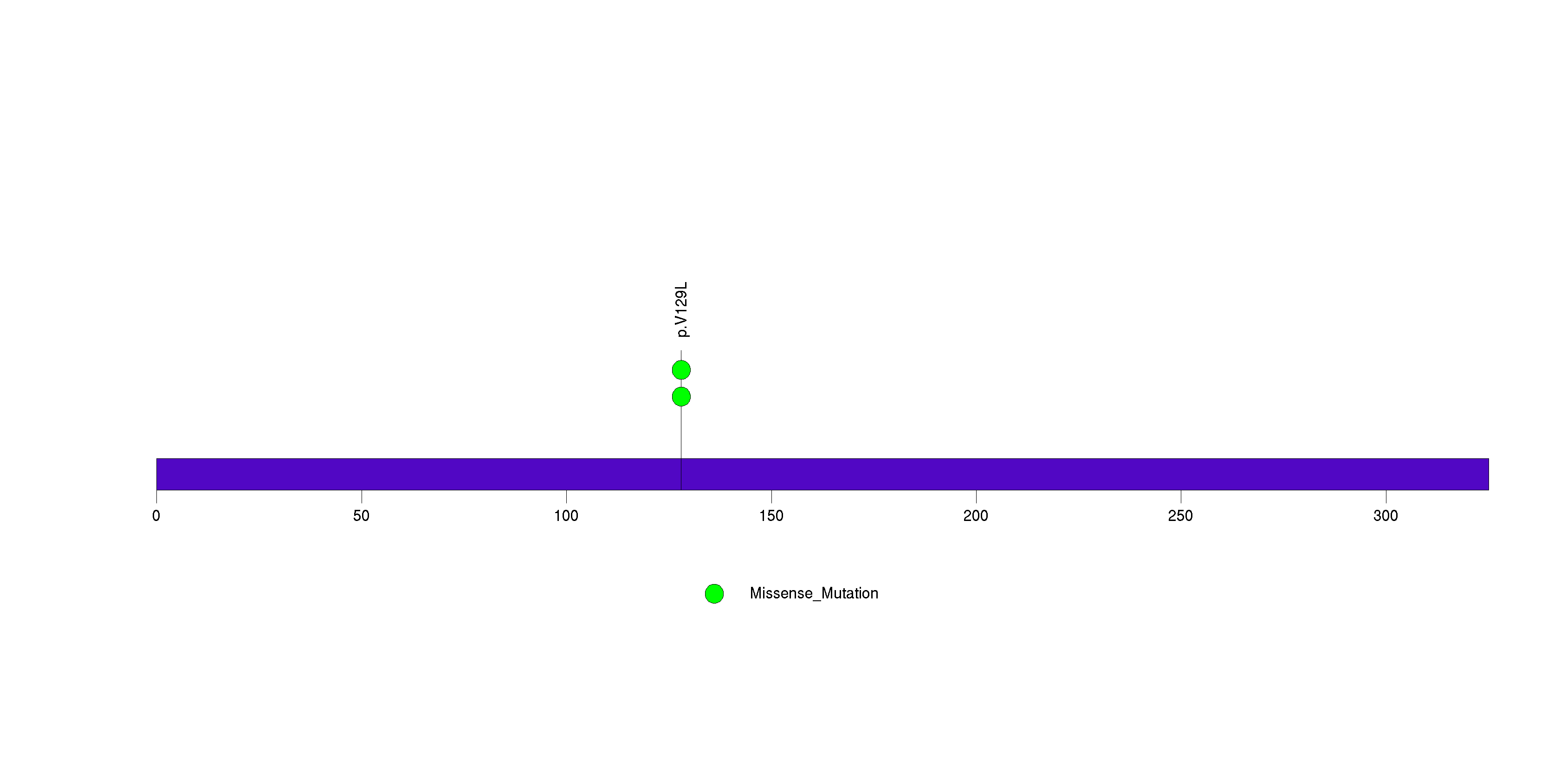
Figure S21. This figure depicts the distribution of mutations and mutation types across the INA significant gene.
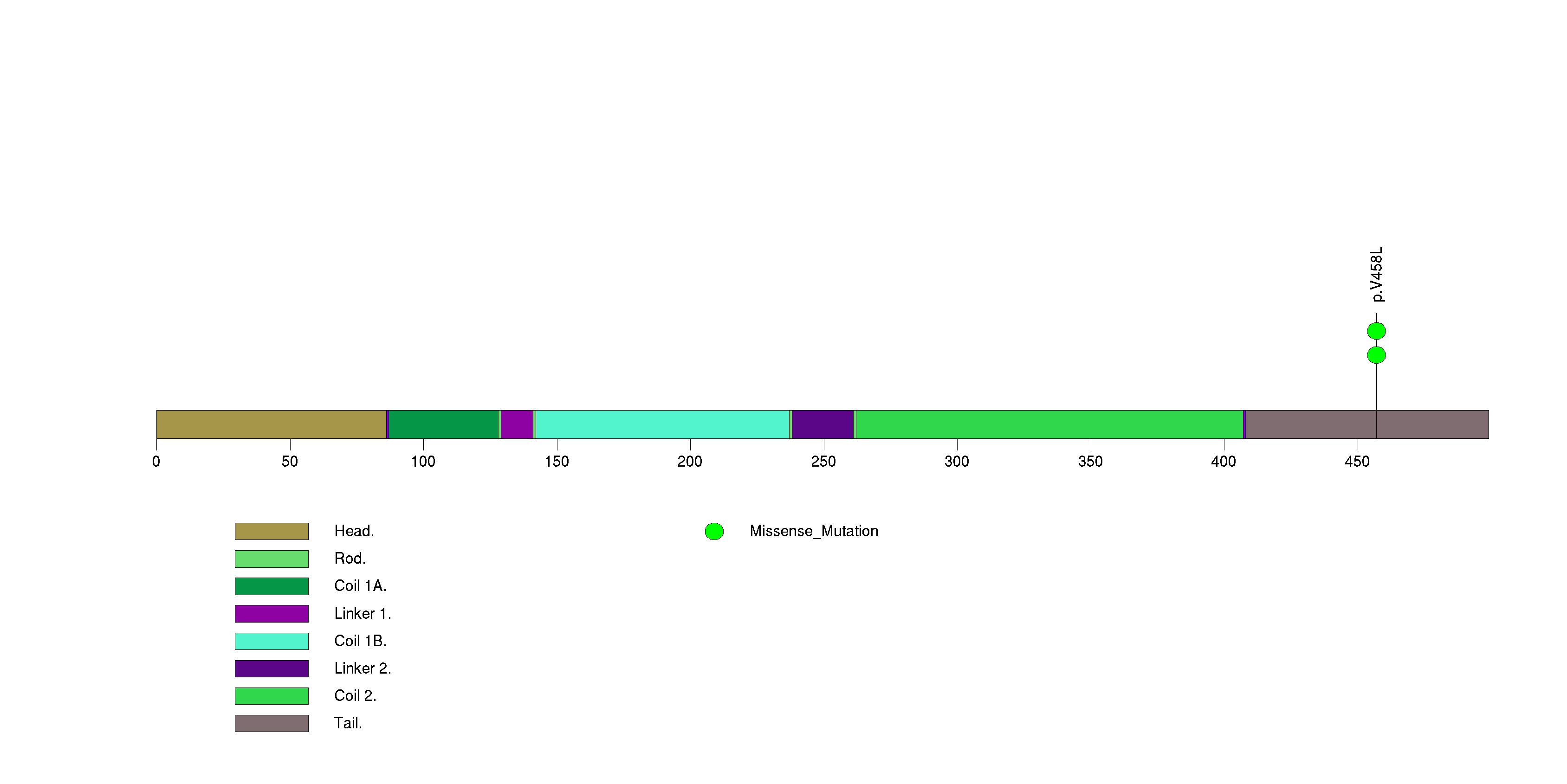
Figure S22. This figure depicts the distribution of mutations and mutation types across the LCTL significant gene.
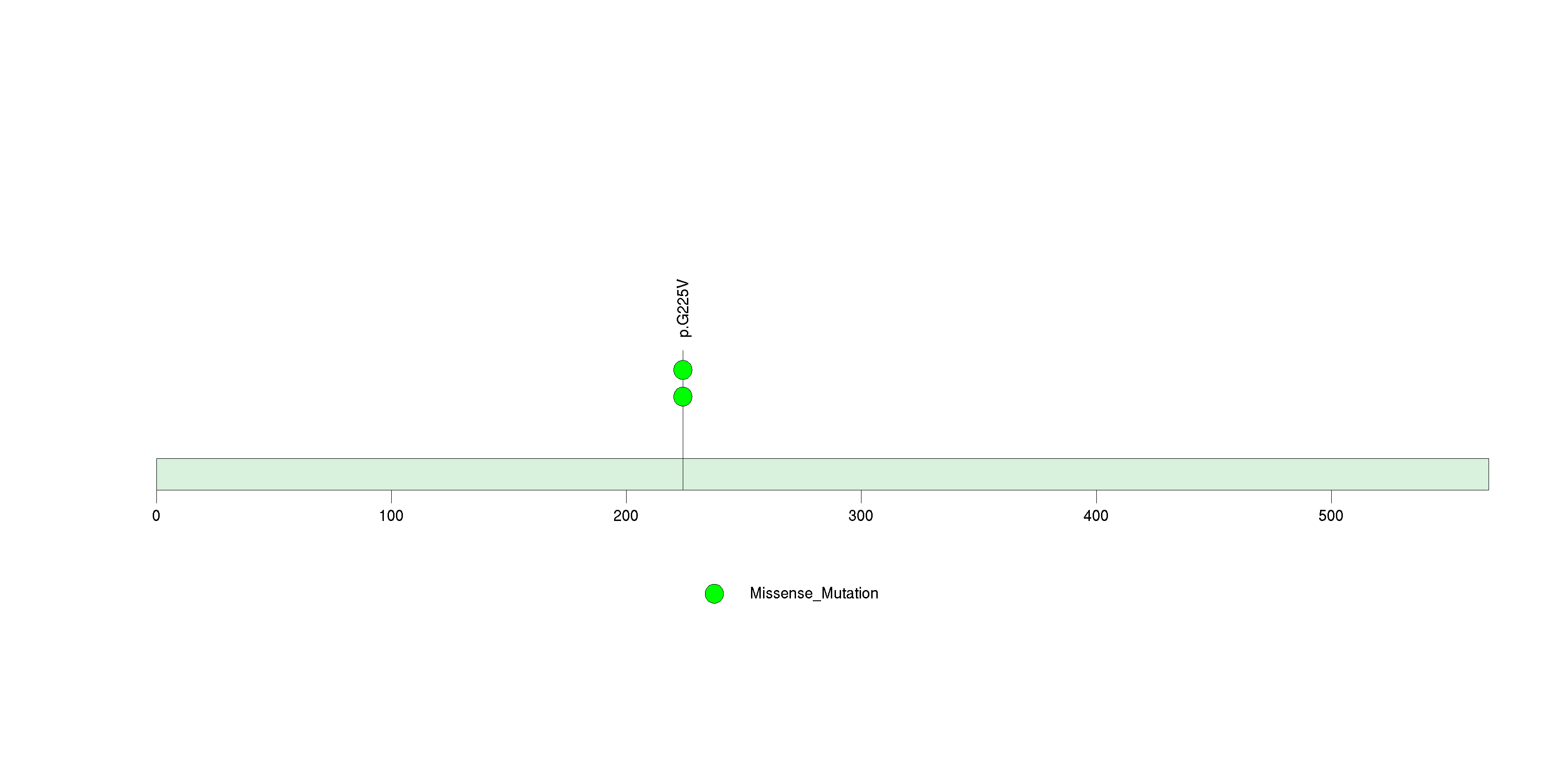
In this analysis, COSMIC is used as a filter to increase power by restricting the territory of each gene. Cosmic version: v48.
Table 4. Get Full Table Significantly mutated genes (COSMIC territory only). To access the database please go to: COSMIC. Number of significant genes found: 6. Number of genes displayed: 10
| rank | gene | description | n | cos | n_cos | N_cos | cos_ev | p | q |
|---|---|---|---|---|---|---|---|---|---|
| 1 | TP53 | tumor protein p53 | 5 | 308 | 5 | 25564 | 1608 | 5.3e-10 | 2.4e-06 |
| 2 | CTNNB1 | catenin (cadherin-associated protein), beta 1, 88kDa | 3 | 101 | 3 | 8383 | 1229 | 2.9e-07 | 0.00065 |
| 3 | PTEN | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 3 | 728 | 3 | 60424 | 15 | 0.0001 | 0.09 |
| 4 | ACSM2B | acyl-CoA synthetase medium-chain family member 2B | 1 | 1 | 1 | 83 | 1 | 0.00012 | 0.09 |
| 5 | BRE | brain and reproductive organ-expressed (TNFRSF1A modulator) | 1 | 1 | 1 | 83 | 1 | 0.00012 | 0.09 |
| 6 | KCNH1 | potassium voltage-gated channel, subfamily H (eag-related), member 1 | 1 | 1 | 1 | 83 | 1 | 0.00012 | 0.09 |
| 7 | PIK3CA | phosphoinositide-3-kinase, catalytic, alpha polypeptide | 2 | 184 | 2 | 15272 | 355 | 0.00024 | 0.12 |
| 8 | CHAT | choline acetyltransferase | 2 | 2 | 1 | 166 | 1 | 0.00024 | 0.12 |
| 9 | CYP4F2 | cytochrome P450, family 4, subfamily F, polypeptide 2 | 1 | 2 | 1 | 166 | 2 | 0.00024 | 0.12 |
| 10 | ACVR2A | activin A receptor, type IIA | 1 | 3 | 1 | 249 | 1 | 0.00036 | 0.15 |
Note:
n - number of (nonsilent) mutations in this gene across the individual set.
cos = number of unique mutated sites in this gene in COSMIC
n_cos = overlap between n and cos.
N_cos = number of individuals times cos.
cos_ev = total evidence: number of reports in COSMIC for mutations seen in this gene.
p = p-value for seeing the observed amount of overlap in this gene)
q = q-value, False Discovery Rate (Benjamini-Hochberg procedure)
Table 5. Get Full Table Genes with Clustered Mutations
| num | gene | desc | n | mindist | nmuts0 | nmuts3 | nmuts12 | npairs0 | npairs3 | npairs12 |
|---|---|---|---|---|---|---|---|---|---|---|
| 88 | AGT | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | 3 | 0 | 3 | 3 | 3 | 3 | 3 | 3 |
| 447 | CCNF | cyclin F | 3 | 0 | 3 | 3 | 3 | 3 | 3 | 3 |
| 540 | CLSTN1 | calsyntenin 1 | 3 | 0 | 3 | 3 | 3 | 3 | 3 | 3 |
| 595 | CROCC | ciliary rootlet coiled-coil, rootletin | 4 | 0 | 3 | 3 | 3 | 3 | 3 | 3 |
| 743 | DUSP27 | dual specificity phosphatase 27 (putative) | 3 | 0 | 3 | 3 | 3 | 3 | 3 | 3 |
| 932 | FRG1 | FSHD region gene 1 | 4 | 0 | 3 | 3 | 3 | 3 | 3 | 3 |
| 2724 | YBX1 | Y box binding protein 1 | 4 | 0 | 2 | 2 | 2 | 2 | 2 | 2 |
| 2350 | SPOP | speckle-type POZ protein | 4 | 0 | 1 | 3 | 3 | 1 | 3 | 3 |
| 1516 | MLL3 | myeloid/lymphoid or mixed-lineage leukemia 3 | 8 | 0 | 1 | 1 | 3 | 1 | 1 | 3 |
| 151 | AP4B1 | adaptor-related protein complex 4, beta 1 subunit | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
Note:
n - number of mutations in this gene in the individual set.
mindist - distance (in aa) between closest pair of mutations in this gene
npairs3 - how many pairs of mutations are within 3 aa of each other.
npairs12 - how many pairs of mutations are within 12 aa of each other.
Table 6. Get Full Table A Ranked List of Significantly Mutated Genesets. (Source: MSigDB GSEA Cannonical Pathway Set).Number of significant genesets found: 2. Number of genesets displayed: 10
| rank | geneset | description | genes | N_genes | mut_tally | N | n | npat | nsite | nsil | n1 | n2 | n3 | n4 | n5 | n6 | p_ns_s | p | q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. | ARF1, ARF3, CCND1, CDK2, CDK4, CDKN1A, CDKN1B, CDKN2A, CFL1, E2F1, E2F2, MDM2, NXT1, PRB1, TP53 | 15 | CDKN1B(2), PRB1(2), TP53(5) | 1062488 | 9 | 9 | 9 | 0 | 3 | 1 | 0 | 2 | 3 | 0 | 0.15 | 2e-05 | 0.012 |
| 2 | P53HYPOXIAPATHWAY | Hypoxia induces p53 accumulation and consequent apoptosis with p53-mediated cell cycle arrest, which is present under conditions of DNA damage. | ABCB1, AKT1, ATM, BAX, CDKN1A, CPB2, CSNK1A1, CSNK1D, FHL2, GADD45A, HIC1, HIF1A, HSPA1A, HSPCA, IGFBP3, MAPK8, MDM2, NFKBIB, NQO1, TP53 | 19 | ABCB1(2), AKT1(1), ATM(5), TP53(5) | 2602579 | 13 | 12 | 13 | 0 | 4 | 6 | 0 | 2 | 1 | 0 | 0.015 | 0.00015 | 0.047 |
| 3 | P53PATHWAY | p53 induces cell cycle arrest or apoptosis under conditions of DNA damage. | APAF1, ATM, BAX, BCL2, CCND1, CCNE1, CDK2, CDK4, CDKN1A, E2F1, GADD45A, MDM2, PCNA, RB1, TIMP3, TP53 | 16 | APAF1(1), ATM(5), TP53(5) | 2266716 | 11 | 10 | 11 | 0 | 3 | 5 | 0 | 2 | 1 | 0 | 0.034 | 0.00079 | 0.16 |
| 4 | TERTPATHWAY | hTERC, the RNA subunit of telomerase, and hTERT, the catalytic protein subunit, are required for telomerase activity and are overexpressed in many cancers. | HDAC1, MAX, MYC, SP1, SP3, TP53, WT1, ZNF42 | 7 | SP1(1), TP53(5) | 879118 | 6 | 6 | 6 | 0 | 3 | 1 | 0 | 1 | 1 | 0 | 0.16 | 0.0015 | 0.23 |
| 5 | RBPATHWAY | The ATM protein kinase recognizes DNA damage and blocks cell cycle progression by phosphorylating chk1 and p53, which normally inhibits Rb to allow G1/S transitions. | ATM, CDC2, CDC25A, CDC25B, CDC25C, CDK2, CDK4, CHEK1, MYT1, RB1, TP53, WEE1, YWHAH | 12 | ATM(5), TP53(5) | 2195302 | 10 | 9 | 10 | 0 | 3 | 4 | 0 | 2 | 1 | 0 | 0.058 | 0.0019 | 0.23 |
| 6 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. | CCNA1, CCNA2, CCND1, CCNE1, CCNE2, CDK2, CDK4, CDKN1B, CDKN2A, E2F1, E2F2, E2F4, PRB1 | 13 | CCNA1(1), CCNE2(1), CDKN1B(2), PRB1(2) | 1130432 | 6 | 6 | 6 | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0.3 | 0.0026 | 0.24 |
| 7 | G1PATHWAY | CDK4/6-cyclin D and CDK2-cyclin E phosphorylate Rb, which allows the transcription of genes needed for the G1/S cell cycle transition. | ABL1, ATM, ATR, CCNA1, CCND1, CCNE1, CDC2, CDC25A, CDK2, CDK4, CDK6, CDKN1A, CDKN1B, CDKN2A, CDKN2B, DHFR, E2F1, GSK3B, HDAC1, MADH3, MADH4, RB1, SKP2, TFDP1, TGFB1, TGFB2, TGFB3, TP53 | 25 | ATM(5), CCNA1(1), CDKN1B(2), DHFR(1), TP53(5) | 3787641 | 14 | 12 | 14 | 0 | 3 | 5 | 0 | 2 | 4 | 0 | 0.032 | 0.003 | 0.24 |
| 8 | CHEMICALPATHWAY | DNA damage promotes Bid cleavage, which stimulates mitochondrial cytochrome c release and consequent caspase activation, resulting in apoptosis. | ADPRT, AKT1, APAF1, ATM, BAD, BAX, BCL2, BCL2L1, BID, CASP3, CASP6, CASP7, CASP9, CYCS, EIF2S1, PRKCA, PRKCB1, PTK2, PXN, STAT1, TLN1, TP53 | 20 | AKT1(1), APAF1(1), ATM(5), TLN1(1), TP53(5) | 3433242 | 13 | 12 | 13 | 0 | 3 | 6 | 0 | 3 | 1 | 0 | 0.019 | 0.0031 | 0.24 |
| 9 | SMALL_LIGAND_GPCRS | C9orf47, CNR1, CNR2, DNMT1, EDG1, EDG2, EDG5, EDG6, MTNR1A, MTNR1B, PTAFR, PTGDR, PTGER1, PTGER2, PTGER4, PTGFR, PTGIR, TBXA2R | 13 | DNMT1(3), MTNR1A(1), PTGER2(1), TBXA2R(2) | 1410721 | 7 | 7 | 7 | 1 | 3 | 2 | 0 | 1 | 1 | 0 | 0.19 | 0.0075 | 0.44 | |
| 10 | PTENPATHWAY | PTEN suppresses AKT-induced cell proliferation and antagonizes the action of PI3K. | AKT1, BCAR1, CDKN1B, FOXO3A, GRB2, ILK, ITGB1, MAPK1, MAPK3, PDK2, PDPK1, PIK3CA, PIK3R1, PTEN, PTK2, SHC1, SOS1, TNFSF6 | 16 | AKT1(1), CDKN1B(2), PIK3CA(2), PTEN(3) | 2419782 | 8 | 8 | 8 | 1 | 0 | 3 | 1 | 1 | 3 | 0 | 0.36 | 0.0075 | 0.44 |
Table 7. Get Full Table A Ranked List of Significantly Mutated Genesets (Excluding Significantly Mutated Genes). Number of significant genesets found: 0. Number of genesets displayed: 10
| rank | geneset | description | genes | N_genes | mut_tally | N | n | npat | nsite | nsil | n1 | n2 | n3 | n4 | n5 | n6 | p_ns_s | p | q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. | CCNA1, CCNA2, CCND1, CCNE1, CCNE2, CDK2, CDK4, CDKN1B, CDKN2A, E2F1, E2F2, E2F4, PRB1 | 13 | CCNA1(1), CCNE2(1), CDKN1B(2), PRB1(2) | 1130432 | 6 | 6 | 6 | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0.3 | 0.0026 | 1 |
| 2 | SMALL_LIGAND_GPCRS | C9orf47, CNR1, CNR2, DNMT1, EDG1, EDG2, EDG5, EDG6, MTNR1A, MTNR1B, PTAFR, PTGDR, PTGER1, PTGER2, PTGER4, PTGFR, PTGIR, TBXA2R | 13 | DNMT1(3), MTNR1A(1), PTGER2(1), TBXA2R(2) | 1410721 | 7 | 7 | 7 | 1 | 3 | 2 | 0 | 1 | 1 | 0 | 0.19 | 0.0075 | 1 | |
| 3 | PTENPATHWAY | PTEN suppresses AKT-induced cell proliferation and antagonizes the action of PI3K. | AKT1, BCAR1, CDKN1B, FOXO3A, GRB2, ILK, ITGB1, MAPK1, MAPK3, PDK2, PDPK1, PIK3CA, PIK3R1, PTEN, PTK2, SHC1, SOS1, TNFSF6 | 16 | AKT1(1), CDKN1B(2), PIK3CA(2), PTEN(3) | 2419782 | 8 | 8 | 8 | 1 | 0 | 3 | 1 | 1 | 3 | 0 | 0.36 | 0.0075 | 1 |
| 4 | HSA00830_RETINOL_METABOLISM | Genes involved in retinol metabolism | ALDH1A1, ALDH1A2, BCMO1, RDH5 | 4 | ALDH1A2(1), BCMO1(2) | 483292 | 3 | 3 | 2 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0.37 | 0.0079 | 1 |
| 5 | P53HYPOXIAPATHWAY | Hypoxia induces p53 accumulation and consequent apoptosis with p53-mediated cell cycle arrest, which is present under conditions of DNA damage. | ABCB1, AKT1, ATM, BAX, CDKN1A, CPB2, CSNK1A1, CSNK1D, FHL2, GADD45A, HIC1, HIF1A, HSPA1A, HSPCA, IGFBP3, MAPK8, MDM2, NFKBIB, NQO1, TP53 | 18 | ABCB1(2), AKT1(1), ATM(5) | 2497058 | 8 | 8 | 8 | 0 | 1 | 6 | 0 | 1 | 0 | 0 | 0.053 | 0.012 | 1 |
| 6 | HSA00564_GLYCEROPHOSPHOLIPID_METABOLISM | Genes involved in glycerophospholipid metabolism | ACHE, AGPAT1, AGPAT2, AGPAT3, AGPAT4, AGPAT6, ARD1A, CDIPT, CDS1, CDS2, CHAT, CHKA, CHKB, CHPT1, CRLS1, DGKA, DGKB, DGKD, DGKE, DGKG, DGKH, DGKI, DGKQ, DGKZ, ESCO1, ESCO2, ETNK1, ETNK2, GNPAT, GPAM, GPD1, GPD1L, GPD2, LCAT, LYCAT, LYPLA1, LYPLA2, LYPLA3, MYST3, MYST4, NAT5, NAT6, PCYT1A, PCYT1B, PEMT, PHOSPHO1, PISD, PLA2G10, PLA2G12A, PLA2G12B, PLA2G1B, PLA2G2A, PLA2G2D, PLA2G2E, PLA2G2F, PLA2G3, PLA2G4A, PLA2G5, PLA2G6, PLD1, PLD2, PNPLA3, PPAP2A, PPAP2B, PPAP2C, PTDSS1, PTDSS2, SH3GLB1 | 64 | AGPAT2(1), AGPAT3(1), CHAT(2), DGKD(1), DGKH(1), DGKI(1), DGKQ(2), ESCO1(3), GNPAT(1), MYST3(1), PEMT(1), PLA2G2A(1), PLA2G4A(1), PLA2G5(1), PLD1(1), PTDSS1(1) | 8241571 | 20 | 19 | 20 | 2 | 2 | 4 | 1 | 9 | 4 | 0 | 0.11 | 0.014 | 1 |
| 7 | LONGEVITYPATHWAY | Caloric restriction in animals often increases lifespan, which may occur via decreased IGF receptor expression and consequent expression of stress-resistance proteins. | AKT1, CAT, FOXO3A, GH1, GHR, HRAS, IGF1, IGF1R, PIK3CA, PIK3R1, SHC1, SOD1, SOD2, SOD3 | 13 | AKT1(1), GHR(1), IGF1R(1), PIK3CA(2), SOD3(1) | 1657357 | 6 | 6 | 6 | 0 | 0 | 4 | 1 | 1 | 0 | 0 | 0.12 | 0.016 | 1 |
| 8 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. | ARF1, ARF3, CCND1, CDK2, CDK4, CDKN1A, CDKN1B, CDKN2A, CFL1, E2F1, E2F2, MDM2, NXT1, PRB1, TP53 | 14 | CDKN1B(2), PRB1(2) | 956967 | 4 | 4 | 4 | 0 | 0 | 1 | 0 | 1 | 2 | 0 | 0.54 | 0.019 | 1 |
| 9 | TRKAPATHWAY | Nerve growth factor (NGF) promotes neuronal survival and proliferation by binding its receptor TrkA, which activates PI3K/AKT, Ras, and the MAP kinase pathway. | AKT1, DPM2, GRB2, HRAS, KLK2, NGFB, NTRK1, PIK3CA, PIK3R1, PLCG1, PRKCA, PRKCB1, SHC1, SOS1 | 12 | AKT1(1), KLK2(1), NTRK1(2), PIK3CA(2) | 1940556 | 6 | 6 | 6 | 0 | 0 | 4 | 1 | 0 | 1 | 0 | 0.13 | 0.021 | 1 |
| 10 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | The phosphoinositide-3 kinase pathway produces the lipid second messenger PIP3 and regulates cell growth, survival, and movement. | A1BG, AKT1, AKT2, AKT3, BAD, BTK, CDKN2A, CSL4, DAF, DAPP1, FOXO1A, GRB2, GSK3A, GSK3B, IARS, IGFBP1, INPP5D, P14, PDK1, PIK3CA, PPP1R13B, PSCD3, PTEN, RPS6KA1, RPS6KA2, RPS6KA3, RPS6KB1, SFN, SHC1, SOS1, SOS2, TEC, YWHAB, YWHAE, YWHAG, YWHAH, YWHAQ, YWHAZ | 33 | A1BG(1), AKT1(1), BTK(1), GSK3A(1), PDK1(1), PIK3CA(2), PPP1R13B(1), PTEN(3) | 4518736 | 11 | 11 | 11 | 1 | 1 | 3 | 1 | 4 | 2 | 0 | 0.16 | 0.022 | 1 |
In brief, we tabulate the number of mutations and the number of covered bases for each gene. The counts are broken down by mutation context category: four context categories that are discovered by MutSig, and one for indel and 'null' mutations, which include indels, nonsense mutations, splice-site mutations, and non-stop (read-through) mutations. For each gene, we calculate the probability of seeing the observed constellation of mutations, i.e. the product P1 x P2 x ... x Pm, or a more extreme one, given the background mutation rates calculated across the dataset. [1]
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.