This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 94 genes and 8 molecular subtypes across 306 patients, 11 significant findings detected with P value < 0.05 and Q value < 0.25.
-
NOTCH1 mutation correlated to 'MRNASEQ_CHIERARCHICAL' and 'MIRSEQ_CNMF'.
-
HRAS mutation correlated to 'CN_CNMF'.
-
NFE2L2 mutation correlated to 'METHLYATION_CNMF'.
-
TP53 mutation correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
CASP8 mutation correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
NSD1 mutation correlated to 'METHLYATION_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between mutation status of 94 genes and 8 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 11 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Chi-square test | Chi-square test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| CASP8 | 27 (9%) | 279 |
0.00102 (0.733) |
7.22e-06 (0.00524) |
1 (1.00) |
0.778 (1.00) |
3e-07 (0.000219) |
5.28e-07 (0.000385) |
0.0199 (1.00) |
0.404 (1.00) |
| NOTCH1 | 57 (19%) | 249 |
0.183 (1.00) |
0.000746 (0.537) |
0.0786 (1.00) |
0.362 (1.00) |
0.000428 (0.309) |
2.54e-06 (0.00185) |
4.53e-06 (0.0033) |
0.0837 (1.00) |
| TP53 | 214 (70%) | 92 |
7.17e-11 (5.25e-08) |
3.14e-18 (2.3e-15) |
1 (1.00) |
0.608 (1.00) |
0.0113 (1.00) |
0.00143 (1.00) |
0.132 (1.00) |
0.00148 (1.00) |
| NSD1 | 33 (11%) | 273 |
0.361 (1.00) |
3.04e-23 (2.23e-20) |
0.168 (1.00) |
0.224 (1.00) |
0.000411 (0.297) |
6.41e-06 (0.00466) |
0.0212 (1.00) |
0.0341 (1.00) |
| HRAS | 10 (3%) | 296 |
6.93e-05 (0.0503) |
0.0612 (1.00) |
0.424 (1.00) |
0.69 (1.00) |
0.00339 (1.00) |
0.0011 (0.787) |
0.142 (1.00) |
1 (1.00) |
| NFE2L2 | 17 (6%) | 289 |
0.688 (1.00) |
0.000297 (0.215) |
0.821 (1.00) |
0.828 (1.00) |
1 (1.00) |
0.464 (1.00) |
0.492 (1.00) |
0.146 (1.00) |
| CDKN2A | 65 (21%) | 241 |
0.00337 (1.00) |
0.00306 (1.00) |
0.395 (1.00) |
0.459 (1.00) |
0.201 (1.00) |
0.206 (1.00) |
0.484 (1.00) |
0.124 (1.00) |
| PIK3CA | 64 (21%) | 242 |
0.0486 (1.00) |
0.0554 (1.00) |
0.372 (1.00) |
0.856 (1.00) |
0.119 (1.00) |
0.398 (1.00) |
0.0993 (1.00) |
0.606 (1.00) |
| FAT1 | 72 (24%) | 234 |
0.325 (1.00) |
0.227 (1.00) |
0.301 (1.00) |
0.487 (1.00) |
0.152 (1.00) |
0.288 (1.00) |
0.294 (1.00) |
0.172 (1.00) |
| JUB | 18 (6%) | 288 |
0.161 (1.00) |
0.0799 (1.00) |
0.407 (1.00) |
0.0806 (1.00) |
0.305 (1.00) |
0.221 (1.00) |
0.325 (1.00) |
1 (1.00) |
| MLL2 | 56 (18%) | 250 |
0.00286 (1.00) |
0.0471 (1.00) |
0.174 (1.00) |
0.311 (1.00) |
0.0532 (1.00) |
0.00795 (1.00) |
0.122 (1.00) |
0.00373 (1.00) |
| FBXW7 | 15 (5%) | 291 |
0.777 (1.00) |
0.134 (1.00) |
0.0527 (1.00) |
0.478 (1.00) |
0.881 (1.00) |
0.636 (1.00) |
1 (1.00) |
1 (1.00) |
| BAGE2 | 11 (4%) | 295 |
0.522 (1.00) |
0.0999 (1.00) |
0.573 (1.00) |
0.21 (1.00) |
0.253 (1.00) |
0.291 (1.00) |
0.339 (1.00) |
0.139 (1.00) |
| POTEC | 15 (5%) | 291 |
0.167 (1.00) |
0.0364 (1.00) |
0.639 (1.00) |
0.907 (1.00) |
0.17 (1.00) |
0.254 (1.00) |
0.6 (1.00) |
0.169 (1.00) |
| ZNF750 | 13 (4%) | 293 |
0.897 (1.00) |
0.159 (1.00) |
0.325 (1.00) |
0.248 (1.00) |
0.119 (1.00) |
0.128 (1.00) |
0.88 (1.00) |
1 (1.00) |
| EP300 | 25 (8%) | 281 |
0.433 (1.00) |
0.151 (1.00) |
0.939 (1.00) |
0.302 (1.00) |
0.00955 (1.00) |
0.00467 (1.00) |
0.249 (1.00) |
0.856 (1.00) |
| POM121L12 | 14 (5%) | 292 |
0.195 (1.00) |
0.304 (1.00) |
0.105 (1.00) |
0.109 (1.00) |
0.658 (1.00) |
0.657 (1.00) |
0.481 (1.00) |
0.207 (1.00) |
| RHOA | 4 (1%) | 302 |
0.114 (1.00) |
0.54 (1.00) |
0.231 (1.00) |
0.424 (1.00) |
0.245 (1.00) |
0.141 (1.00) |
0.237 (1.00) |
0.395 (1.00) |
| B2M | 7 (2%) | 299 |
0.547 (1.00) |
0.0618 (1.00) |
0.515 (1.00) |
0.517 (1.00) |
0.571 (1.00) |
0.767 (1.00) |
||
| RAC1 | 9 (3%) | 297 |
0.83 (1.00) |
0.585 (1.00) |
1 (1.00) |
0.379 (1.00) |
0.195 (1.00) |
0.192 (1.00) |
0.324 (1.00) |
0.497 (1.00) |
| ZNF804B | 21 (7%) | 285 |
0.482 (1.00) |
0.487 (1.00) |
0.939 (1.00) |
0.915 (1.00) |
0.812 (1.00) |
0.746 (1.00) |
0.959 (1.00) |
0.759 (1.00) |
| KCNT2 | 17 (6%) | 289 |
0.148 (1.00) |
0.235 (1.00) |
0.727 (1.00) |
0.478 (1.00) |
0.05 (1.00) |
0.121 (1.00) |
0.0579 (1.00) |
0.0117 (1.00) |
| RASA1 | 14 (5%) | 292 |
0.306 (1.00) |
0.24 (1.00) |
0.485 (1.00) |
0.786 (1.00) |
0.357 (1.00) |
0.271 (1.00) |
0.272 (1.00) |
0.0425 (1.00) |
| PRIM2 | 9 (3%) | 297 |
0.214 (1.00) |
0.305 (1.00) |
1 (1.00) |
0.5 (1.00) |
0.4 (1.00) |
0.395 (1.00) |
0.53 (1.00) |
0.497 (1.00) |
| CDH10 | 23 (8%) | 283 |
0.171 (1.00) |
0.531 (1.00) |
0.593 (1.00) |
0.558 (1.00) |
0.0743 (1.00) |
0.265 (1.00) |
0.242 (1.00) |
0.483 (1.00) |
| CNTNAP5 | 15 (5%) | 291 |
0.0655 (1.00) |
0.115 (1.00) |
0.37 (1.00) |
1 (1.00) |
0.0101 (1.00) |
0.0354 (1.00) |
0.0164 (1.00) |
0.169 (1.00) |
| POTEG | 10 (3%) | 296 |
0.0959 (1.00) |
0.0869 (1.00) |
0.0285 (1.00) |
0.741 (1.00) |
0.16 (1.00) |
0.143 (1.00) |
0.79 (1.00) |
0.436 (1.00) |
| ZNF99 | 22 (7%) | 284 |
0.687 (1.00) |
0.126 (1.00) |
0.233 (1.00) |
0.436 (1.00) |
0.0343 (1.00) |
0.0116 (1.00) |
0.0131 (1.00) |
0.00332 (1.00) |
| OR2T12 | 11 (4%) | 295 |
0.106 (1.00) |
0.12 (1.00) |
0.884 (1.00) |
0.69 (1.00) |
0.552 (1.00) |
0.502 (1.00) |
0.64 (1.00) |
0.68 (1.00) |
| REG1A | 8 (3%) | 298 |
0.375 (1.00) |
0.102 (1.00) |
1 (1.00) |
0.339 (1.00) |
0.058 (1.00) |
0.00131 (0.94) |
0.0765 (1.00) |
0.0515 (1.00) |
| PABPC5 | 9 (3%) | 297 |
0.698 (1.00) |
0.407 (1.00) |
0.843 (1.00) |
0.602 (1.00) |
0.089 (1.00) |
0.237 (1.00) |
0.324 (1.00) |
0.81 (1.00) |
| IL32 | 4 (1%) | 302 |
0.0781 (1.00) |
0.00177 (1.00) |
0.561 (1.00) |
1 (1.00) |
0.474 (1.00) |
1 (1.00) |
||
| PRAMEF11 | 10 (3%) | 296 |
0.421 (1.00) |
0.438 (1.00) |
1 (1.00) |
0.874 (1.00) |
0.669 (1.00) |
0.606 (1.00) |
0.853 (1.00) |
1 (1.00) |
| EPHA2 | 14 (5%) | 292 |
0.296 (1.00) |
0.00256 (1.00) |
0.639 (1.00) |
1 (1.00) |
0.499 (1.00) |
0.57 (1.00) |
0.348 (1.00) |
0.186 (1.00) |
| FCRL4 | 13 (4%) | 293 |
0.0784 (1.00) |
0.0179 (1.00) |
0.525 (1.00) |
1 (1.00) |
0.405 (1.00) |
0.106 (1.00) |
0.281 (1.00) |
0.724 (1.00) |
| LCP1 | 12 (4%) | 294 |
0.133 (1.00) |
0.000851 (0.612) |
0.555 (1.00) |
0.0359 (1.00) |
0.873 (1.00) |
0.758 (1.00) |
1 (1.00) |
0.244 (1.00) |
| LRFN5 | 13 (4%) | 293 |
0.0175 (1.00) |
0.07 (1.00) |
0.521 (1.00) |
0.855 (1.00) |
0.279 (1.00) |
0.0765 (1.00) |
0.0332 (1.00) |
0.387 (1.00) |
| MAPK1 | 4 (1%) | 302 |
0.787 (1.00) |
0.924 (1.00) |
0.842 (1.00) |
1 (1.00) |
0.691 (1.00) |
0.686 (1.00) |
0.825 (1.00) |
1 (1.00) |
| PEG3 | 23 (8%) | 283 |
0.101 (1.00) |
0.022 (1.00) |
0.371 (1.00) |
0.0892 (1.00) |
0.0217 (1.00) |
0.053 (1.00) |
0.124 (1.00) |
0.296 (1.00) |
| PRSS1 | 8 (3%) | 298 |
0.788 (1.00) |
0.478 (1.00) |
0.842 (1.00) |
1 (1.00) |
1 (1.00) |
0.89 (1.00) |
1 (1.00) |
1 (1.00) |
| DOK6 | 8 (3%) | 298 |
0.235 (1.00) |
0.582 (1.00) |
0.447 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.605 (1.00) |
0.786 (1.00) |
| GABRB3 | 11 (4%) | 295 |
0.559 (1.00) |
0.0147 (1.00) |
0.469 (1.00) |
1 (1.00) |
0.253 (1.00) |
0.192 (1.00) |
0.339 (1.00) |
0.68 (1.00) |
| STEAP4 | 10 (3%) | 296 |
0.187 (1.00) |
0.636 (1.00) |
0.653 (1.00) |
0.633 (1.00) |
0.0813 (1.00) |
0.0997 (1.00) |
0.0687 (1.00) |
1 (1.00) |
| ASXL3 | 21 (7%) | 285 |
0.712 (1.00) |
0.632 (1.00) |
0.409 (1.00) |
0.331 (1.00) |
0.96 (1.00) |
0.714 (1.00) |
1 (1.00) |
0.359 (1.00) |
| NTM | 7 (2%) | 299 |
0.948 (1.00) |
0.24 (1.00) |
0.128 (1.00) |
0.0226 (1.00) |
0.182 (1.00) |
0.0877 (1.00) |
0.141 (1.00) |
0.405 (1.00) |
| FAM155A | 11 (4%) | 295 |
0.253 (1.00) |
0.0585 (1.00) |
0.332 (1.00) |
0.077 (1.00) |
0.233 (1.00) |
0.175 (1.00) |
0.146 (1.00) |
0.277 (1.00) |
| HLA-A | 9 (3%) | 297 |
0.586 (1.00) |
0.335 (1.00) |
0.44 (1.00) |
1 (1.00) |
0.072 (1.00) |
0.0311 (1.00) |
0.296 (1.00) |
0.386 (1.00) |
| KRTAP1-5 | 3 (1%) | 303 |
0.838 (1.00) |
0.807 (1.00) |
0.78 (1.00) |
0.63 (1.00) |
0.0525 (1.00) |
1 (1.00) |
||
| CTCF | 11 (4%) | 295 |
0.386 (1.00) |
0.401 (1.00) |
0.121 (1.00) |
0.617 (1.00) |
0.636 (1.00) |
0.632 (1.00) |
0.929 (1.00) |
1 (1.00) |
| BRWD3 | 16 (5%) | 290 |
0.891 (1.00) |
0.532 (1.00) |
0.205 (1.00) |
1 (1.00) |
0.256 (1.00) |
0.136 (1.00) |
0.316 (1.00) |
0.0456 (1.00) |
| CNPY3 | 3 (1%) | 303 |
0.336 (1.00) |
0.48 (1.00) |
0.5 (1.00) |
0.499 (1.00) |
0.38 (1.00) |
0.61 (1.00) |
||
| FAM101A | 4 (1%) | 302 |
0.477 (1.00) |
0.374 (1.00) |
0.691 (1.00) |
1 (1.00) |
0.825 (1.00) |
0.692 (1.00) |
||
| HLA-B | 9 (3%) | 297 |
0.253 (1.00) |
0.0957 (1.00) |
0.0877 (1.00) |
0.69 (1.00) |
0.00341 (1.00) |
0.00425 (1.00) |
0.0708 (1.00) |
0.16 (1.00) |
| PSG8 | 9 (3%) | 297 |
0.165 (1.00) |
0.618 (1.00) |
0.703 (1.00) |
0.571 (1.00) |
0.483 (1.00) |
0.484 (1.00) |
0.918 (1.00) |
0.386 (1.00) |
| OR56A1 | 8 (3%) | 298 |
0.37 (1.00) |
0.0158 (1.00) |
0.751 (1.00) |
0.812 (1.00) |
0.357 (1.00) |
0.352 (1.00) |
0.201 (1.00) |
0.287 (1.00) |
| OR2M2 | 8 (3%) | 298 |
0.00675 (1.00) |
0.126 (1.00) |
0.181 (1.00) |
0.0118 (1.00) |
0.498 (1.00) |
0.0946 (1.00) |
||
| OR4M2 | 8 (3%) | 298 |
0.799 (1.00) |
0.88 (1.00) |
0.34 (1.00) |
0.379 (1.00) |
0.643 (1.00) |
0.639 (1.00) |
0.403 (1.00) |
0.603 (1.00) |
| CPXCR1 | 7 (2%) | 299 |
0.622 (1.00) |
0.536 (1.00) |
0.206 (1.00) |
0.192 (1.00) |
0.345 (1.00) |
0.498 (1.00) |
0.723 (1.00) |
0.131 (1.00) |
| EYA1 | 10 (3%) | 296 |
0.105 (1.00) |
0.636 (1.00) |
0.191 (1.00) |
0.831 (1.00) |
0.0274 (1.00) |
0.0263 (1.00) |
0.919 (1.00) |
0.187 (1.00) |
| LINGO2 | 10 (3%) | 296 |
0.268 (1.00) |
0.82 (1.00) |
0.00359 (1.00) |
1 (1.00) |
0.015 (1.00) |
0.0144 (1.00) |
0.0894 (1.00) |
0.117 (1.00) |
| SEMA5A | 19 (6%) | 287 |
0.000736 (0.531) |
0.00318 (1.00) |
1 (1.00) |
1 (1.00) |
0.00661 (1.00) |
0.0234 (1.00) |
0.379 (1.00) |
0.0635 (1.00) |
| TGFBR2 | 10 (3%) | 296 |
0.353 (1.00) |
0.201 (1.00) |
0.903 (1.00) |
1 (1.00) |
0.313 (1.00) |
0.785 (1.00) |
0.666 (1.00) |
1 (1.00) |
| PLSCR1 | 5 (2%) | 301 |
0.0057 (1.00) |
0.23 (1.00) |
0.802 (1.00) |
1 (1.00) |
0.524 (1.00) |
0.526 (1.00) |
0.737 (1.00) |
0.494 (1.00) |
| LIN28B | 6 (2%) | 300 |
0.409 (1.00) |
0.938 (1.00) |
0.802 (1.00) |
1 (1.00) |
0.737 (1.00) |
0.739 (1.00) |
0.514 (1.00) |
0.515 (1.00) |
| AGTR1 | 8 (3%) | 298 |
0.616 (1.00) |
0.401 (1.00) |
0.169 (1.00) |
0.106 (1.00) |
0.181 (1.00) |
0.2 (1.00) |
0.0765 (1.00) |
0.603 (1.00) |
| C6 | 12 (4%) | 294 |
0.734 (1.00) |
0.591 (1.00) |
0.157 (1.00) |
1 (1.00) |
0.0762 (1.00) |
0.222 (1.00) |
0.534 (1.00) |
0.582 (1.00) |
| MS4A14 | 9 (3%) | 297 |
0.397 (1.00) |
0.418 (1.00) |
0.0584 (1.00) |
1 (1.00) |
0.4 (1.00) |
0.484 (1.00) |
0.268 (1.00) |
0.497 (1.00) |
| HIST1H4E | 5 (2%) | 301 |
0.732 (1.00) |
0.731 (1.00) |
0.156 (1.00) |
0.741 (1.00) |
0.866 (1.00) |
0.386 (1.00) |
0.193 (1.00) |
0.224 (1.00) |
| LBP | 7 (2%) | 299 |
0.76 (1.00) |
0.837 (1.00) |
0.775 (1.00) |
0.771 (1.00) |
1 (1.00) |
0.565 (1.00) |
||
| RGS17 | 5 (2%) | 301 |
0.603 (1.00) |
0.81 (1.00) |
0.157 (1.00) |
0.155 (1.00) |
0.193 (1.00) |
0.224 (1.00) |
||
| LILRB1 | 12 (4%) | 294 |
0.302 (1.00) |
0.166 (1.00) |
0.653 (1.00) |
0.812 (1.00) |
0.861 (1.00) |
0.929 (1.00) |
0.754 (1.00) |
0.423 (1.00) |
| MAGEL2 | 12 (4%) | 294 |
0.209 (1.00) |
0.00566 (1.00) |
0.469 (1.00) |
0.352 (1.00) |
0.538 (1.00) |
0.341 (1.00) |
0.659 (1.00) |
0.582 (1.00) |
| OR8D4 | 6 (2%) | 300 |
0.992 (1.00) |
0.119 (1.00) |
0.51 (1.00) |
0.42 (1.00) |
0.387 (1.00) |
1 (1.00) |
0.444 (1.00) |
0.748 (1.00) |
| HIST1H1B | 7 (2%) | 299 |
0.429 (1.00) |
0.356 (1.00) |
0.447 (1.00) |
0.812 (1.00) |
0.802 (1.00) |
0.717 (1.00) |
0.723 (1.00) |
0.131 (1.00) |
| C3ORF59 | 8 (3%) | 298 |
0.142 (1.00) |
0.476 (1.00) |
0.169 (1.00) |
0.00687 (1.00) |
1 (1.00) |
1 (1.00) |
0.82 (1.00) |
1 (1.00) |
| C8ORF34 | 7 (2%) | 299 |
0.847 (1.00) |
0.0892 (1.00) |
0.177 (1.00) |
0.42 (1.00) |
0.802 (1.00) |
1 (1.00) |
0.429 (1.00) |
0.565 (1.00) |
| OR8J1 | 9 (3%) | 297 |
0.224 (1.00) |
0.319 (1.00) |
0.128 (1.00) |
0.0226 (1.00) |
0.245 (1.00) |
0.265 (1.00) |
0.268 (1.00) |
0.207 (1.00) |
| RAB32 | 3 (1%) | 303 |
0.453 (1.00) |
0.825 (1.00) |
0.613 (1.00) |
0.106 (1.00) |
0.114 (1.00) |
0.116 (1.00) |
0.492 (1.00) |
0.342 (1.00) |
| TMEM195 | 8 (3%) | 298 |
0.713 (1.00) |
0.448 (1.00) |
0.531 (1.00) |
0.571 (1.00) |
0.821 (1.00) |
0.905 (1.00) |
1 (1.00) |
1 (1.00) |
| PCDH11X | 21 (7%) | 285 |
0.764 (1.00) |
0.71 (1.00) |
0.401 (1.00) |
0.0572 (1.00) |
0.346 (1.00) |
0.377 (1.00) |
0.882 (1.00) |
0.759 (1.00) |
| SLITRK4 | 12 (4%) | 294 |
0.468 (1.00) |
0.178 (1.00) |
0.587 (1.00) |
0.596 (1.00) |
0.092 (1.00) |
0.0896 (1.00) |
0.273 (1.00) |
0.2 (1.00) |
| FRG2B | 6 (2%) | 300 |
0.76 (1.00) |
0.692 (1.00) |
1 (1.00) |
0.664 (1.00) |
0.291 (1.00) |
0.289 (1.00) |
0.0943 (1.00) |
0.748 (1.00) |
| OR2L13 | 7 (2%) | 299 |
0.452 (1.00) |
0.101 (1.00) |
0.843 (1.00) |
1 (1.00) |
0.345 (1.00) |
0.498 (1.00) |
0.723 (1.00) |
0.131 (1.00) |
| CUL3 | 10 (3%) | 296 |
0.423 (1.00) |
0.124 (1.00) |
0.903 (1.00) |
0.518 (1.00) |
0.403 (1.00) |
0.852 (1.00) |
0.853 (1.00) |
0.52 (1.00) |
| FOSL2 | 7 (2%) | 299 |
0.178 (1.00) |
0.611 (1.00) |
0.843 (1.00) |
1 (1.00) |
0.345 (1.00) |
0.717 (1.00) |
0.0869 (1.00) |
0.565 (1.00) |
| GPR158 | 12 (4%) | 294 |
0.635 (1.00) |
0.131 (1.00) |
0.469 (1.00) |
0.855 (1.00) |
0.538 (1.00) |
0.531 (1.00) |
0.754 (1.00) |
0.582 (1.00) |
| PTPRT | 19 (6%) | 287 |
0.202 (1.00) |
0.0925 (1.00) |
1 (1.00) |
0.749 (1.00) |
0.0996 (1.00) |
0.693 (1.00) |
0.501 (1.00) |
0.199 (1.00) |
| POTEH | 7 (2%) | 299 |
0.211 (1.00) |
0.55 (1.00) |
0.102 (1.00) |
0.602 (1.00) |
0.123 (1.00) |
0.103 (1.00) |
0.647 (1.00) |
0.767 (1.00) |
| OR5D13 | 7 (2%) | 299 |
0.591 (1.00) |
0.16 (1.00) |
0.685 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.767 (1.00) |
| PRB1 | 7 (2%) | 299 |
0.554 (1.00) |
0.177 (1.00) |
0.613 (1.00) |
0.664 (1.00) |
0.89 (1.00) |
0.639 (1.00) |
0.141 (1.00) |
0.186 (1.00) |
| NEUROD6 | 7 (2%) | 299 |
0.668 (1.00) |
0.862 (1.00) |
0.502 (1.00) |
0.563 (1.00) |
1 (1.00) |
1 (1.00) |
||
| DPP10 | 16 (5%) | 290 |
0.161 (1.00) |
0.12 (1.00) |
0.783 (1.00) |
0.21 (1.00) |
0.17 (1.00) |
0.0574 (1.00) |
0.0154 (1.00) |
0.0456 (1.00) |
| DPPA4 | 7 (2%) | 299 |
0.0108 (1.00) |
0.0035 (1.00) |
0.575 (1.00) |
0.119 (1.00) |
0.123 (1.00) |
0.0461 (1.00) |
0.723 (1.00) |
0.767 (1.00) |
| EPDR1 | 6 (2%) | 300 |
0.584 (1.00) |
0.799 (1.00) |
0.277 (1.00) |
0.602 (1.00) |
0.327 (1.00) |
0.675 (1.00) |
0.774 (1.00) |
0.748 (1.00) |
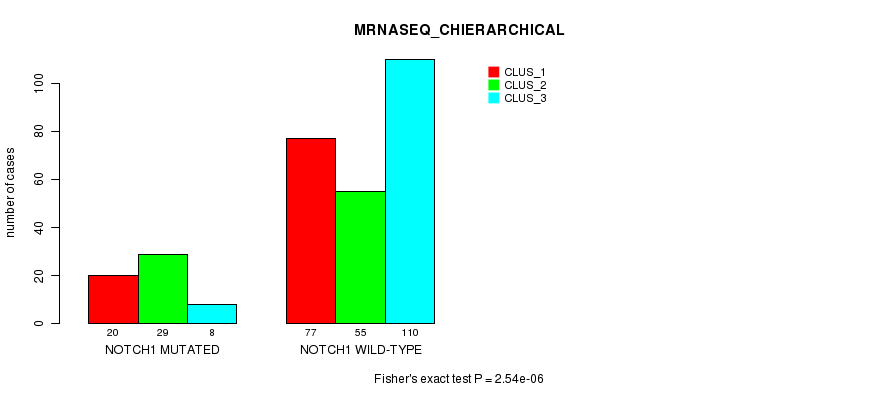
P value = 2.54e-06 (Fisher's exact test), Q value = 0.0019
Table S1. Gene #1: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 97 | 84 | 118 |
| NOTCH1 MUTATED | 20 | 29 | 8 |
| NOTCH1 WILD-TYPE | 77 | 55 | 110 |
Figure S1. Get High-res Image Gene #1: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
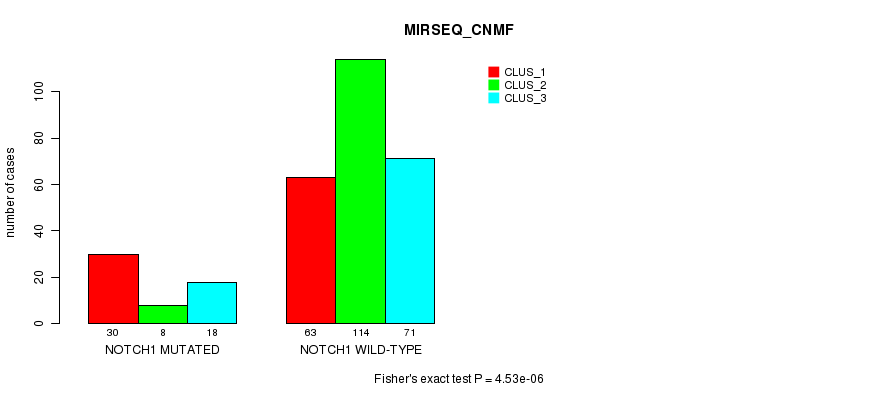
P value = 4.53e-06 (Fisher's exact test), Q value = 0.0033
Table S2. Gene #1: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 93 | 122 | 89 |
| NOTCH1 MUTATED | 30 | 8 | 18 |
| NOTCH1 WILD-TYPE | 63 | 114 | 71 |
Figure S2. Get High-res Image Gene #1: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
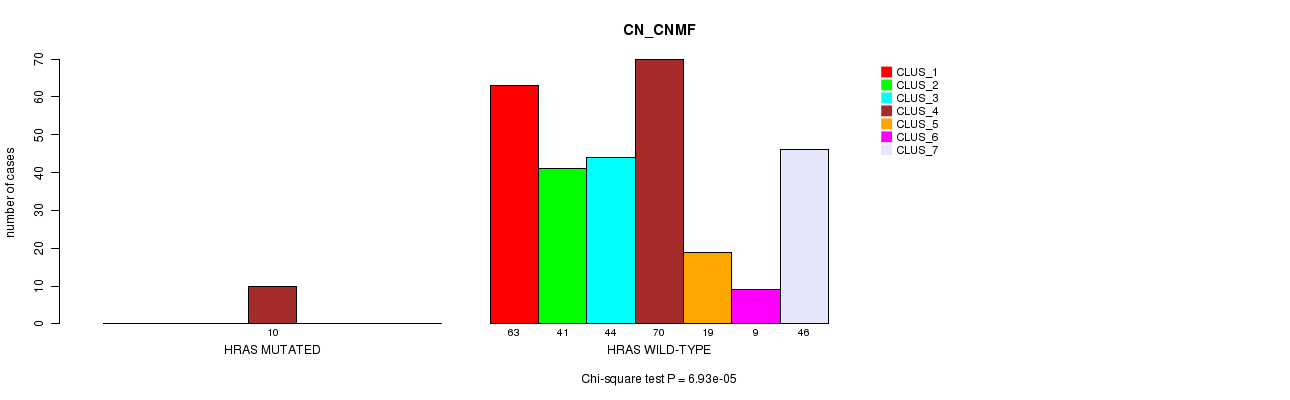
P value = 6.93e-05 (Chi-square test), Q value = 0.05
Table S3. Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 63 | 41 | 44 | 80 | 19 | 9 | 46 |
| HRAS MUTATED | 0 | 0 | 0 | 10 | 0 | 0 | 0 |
| HRAS WILD-TYPE | 63 | 41 | 44 | 70 | 19 | 9 | 46 |
Figure S3. Get High-res Image Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #1: 'CN_CNMF'
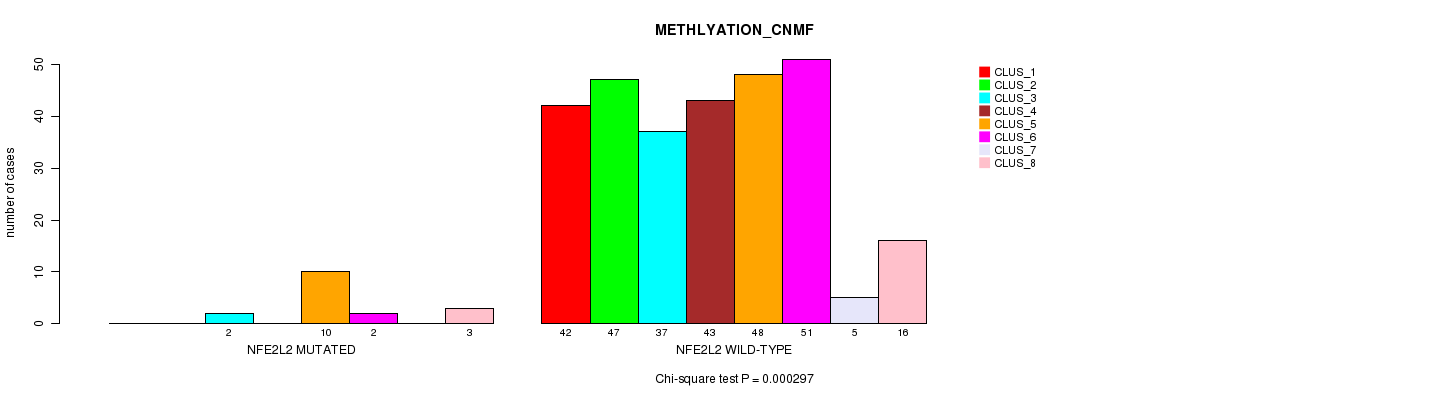
P value = 0.000297 (Chi-square test), Q value = 0.21
Table S4. Gene #4: 'NFE2L2 MUTATION STATUS' versus Clinical Feature #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 | CLUS_8 |
|---|---|---|---|---|---|---|---|---|
| ALL | 42 | 47 | 39 | 43 | 58 | 53 | 5 | 19 |
| NFE2L2 MUTATED | 0 | 0 | 2 | 0 | 10 | 2 | 0 | 3 |
| NFE2L2 WILD-TYPE | 42 | 47 | 37 | 43 | 48 | 51 | 5 | 16 |
Figure S4. Get High-res Image Gene #4: 'NFE2L2 MUTATION STATUS' versus Clinical Feature #2: 'METHLYATION_CNMF'
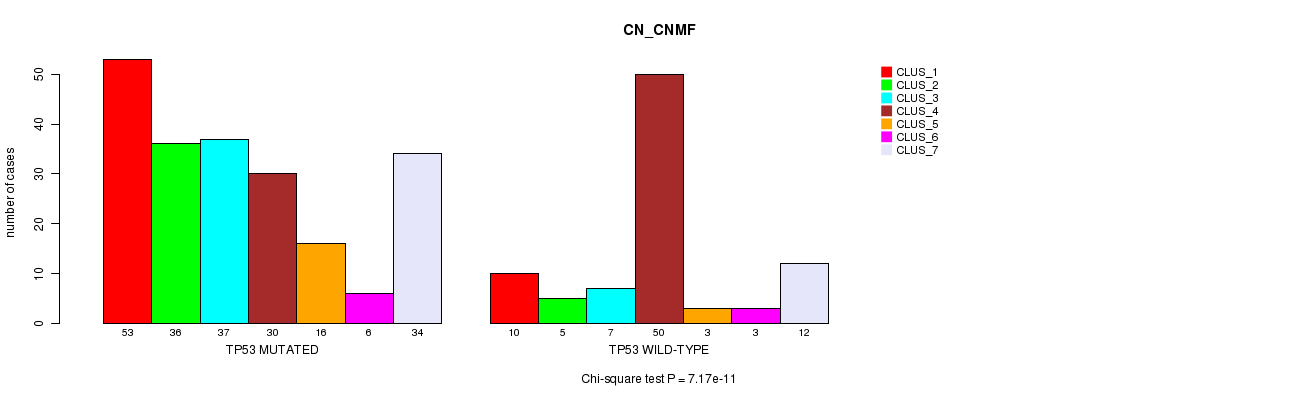
P value = 7.17e-11 (Chi-square test), Q value = 5.2e-08
Table S5. Gene #6: 'TP53 MUTATION STATUS' versus Clinical Feature #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 63 | 41 | 44 | 80 | 19 | 9 | 46 |
| TP53 MUTATED | 53 | 36 | 37 | 30 | 16 | 6 | 34 |
| TP53 WILD-TYPE | 10 | 5 | 7 | 50 | 3 | 3 | 12 |
Figure S5. Get High-res Image Gene #6: 'TP53 MUTATION STATUS' versus Clinical Feature #1: 'CN_CNMF'
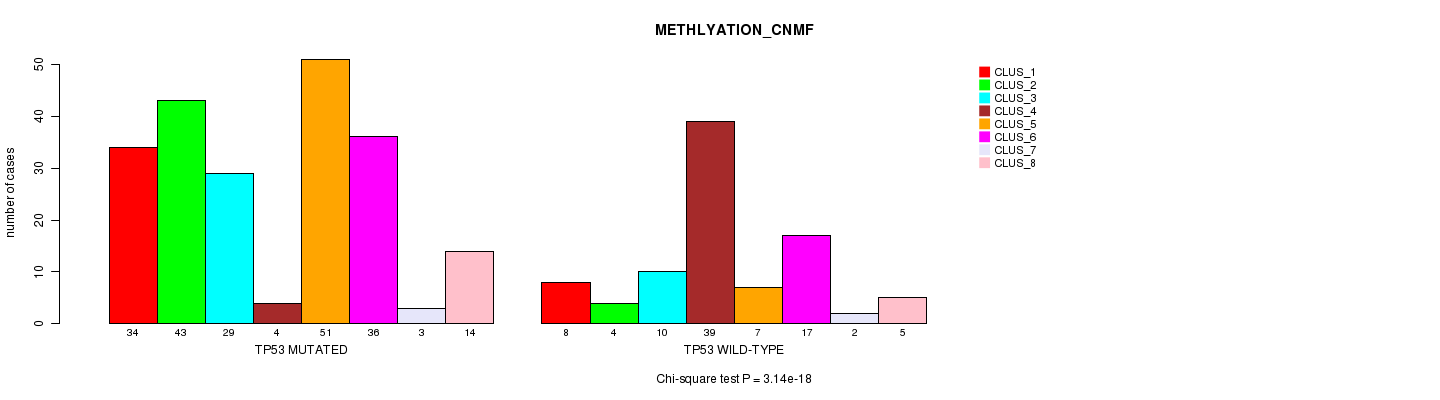
P value = 3.14e-18 (Chi-square test), Q value = 2.3e-15
Table S6. Gene #6: 'TP53 MUTATION STATUS' versus Clinical Feature #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 | CLUS_8 |
|---|---|---|---|---|---|---|---|---|
| ALL | 42 | 47 | 39 | 43 | 58 | 53 | 5 | 19 |
| TP53 MUTATED | 34 | 43 | 29 | 4 | 51 | 36 | 3 | 14 |
| TP53 WILD-TYPE | 8 | 4 | 10 | 39 | 7 | 17 | 2 | 5 |
Figure S6. Get High-res Image Gene #6: 'TP53 MUTATION STATUS' versus Clinical Feature #2: 'METHLYATION_CNMF'
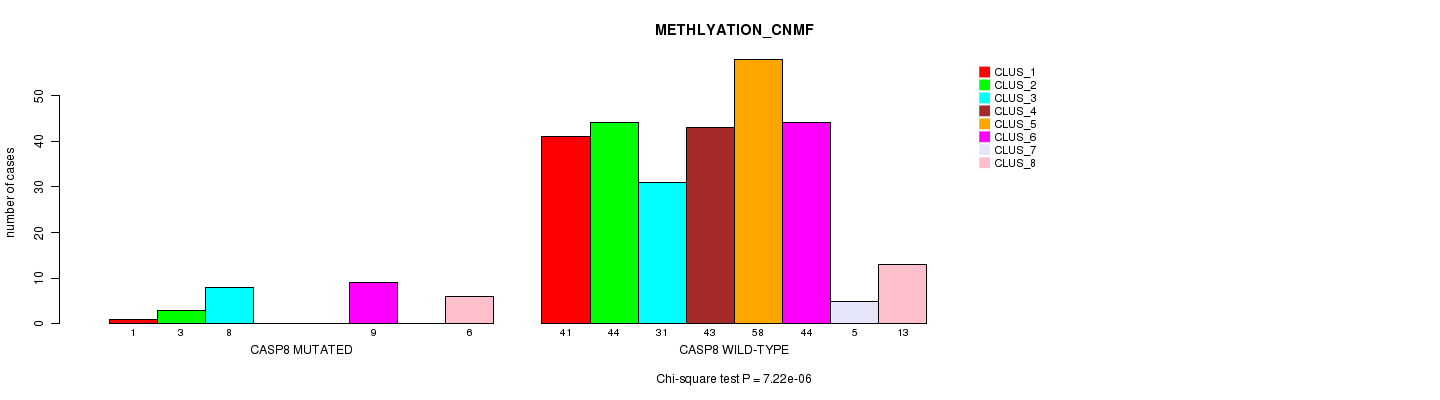
P value = 7.22e-06 (Chi-square test), Q value = 0.0052
Table S7. Gene #7: 'CASP8 MUTATION STATUS' versus Clinical Feature #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 | CLUS_8 |
|---|---|---|---|---|---|---|---|---|
| ALL | 42 | 47 | 39 | 43 | 58 | 53 | 5 | 19 |
| CASP8 MUTATED | 1 | 3 | 8 | 0 | 0 | 9 | 0 | 6 |
| CASP8 WILD-TYPE | 41 | 44 | 31 | 43 | 58 | 44 | 5 | 13 |
Figure S7. Get High-res Image Gene #7: 'CASP8 MUTATION STATUS' versus Clinical Feature #2: 'METHLYATION_CNMF'
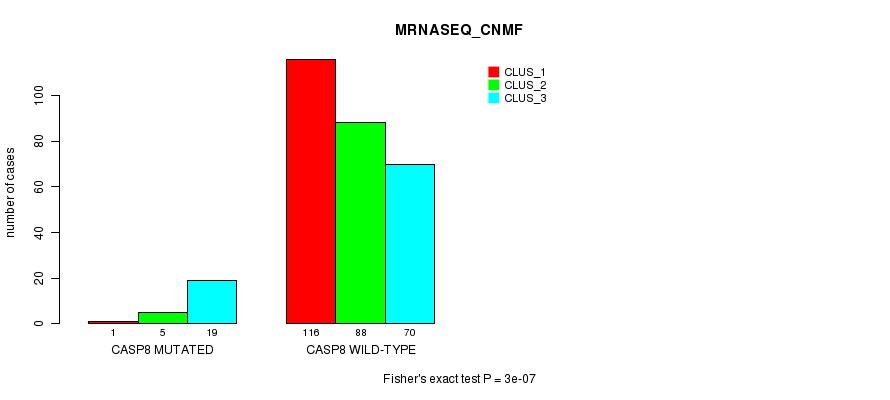
P value = 3e-07 (Fisher's exact test), Q value = 0.00022
Table S8. Gene #7: 'CASP8 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 117 | 93 | 89 |
| CASP8 MUTATED | 1 | 5 | 19 |
| CASP8 WILD-TYPE | 116 | 88 | 70 |
Figure S8. Get High-res Image Gene #7: 'CASP8 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
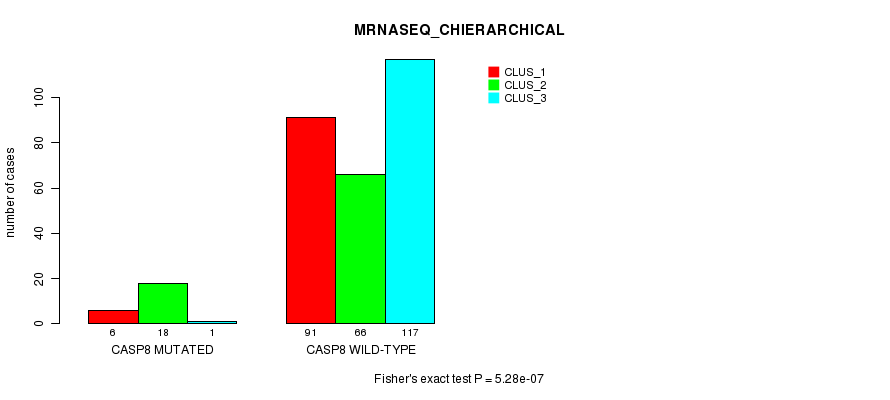
P value = 5.28e-07 (Fisher's exact test), Q value = 0.00039
Table S9. Gene #7: 'CASP8 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 97 | 84 | 118 |
| CASP8 MUTATED | 6 | 18 | 1 |
| CASP8 WILD-TYPE | 91 | 66 | 117 |
Figure S9. Get High-res Image Gene #7: 'CASP8 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
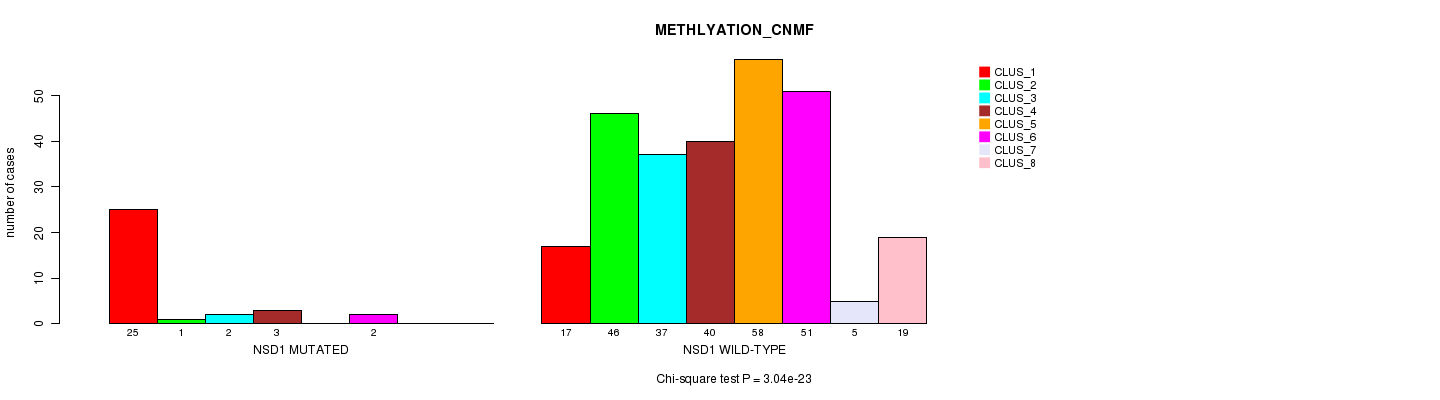
P value = 3.04e-23 (Chi-square test), Q value = 2.2e-20
Table S10. Gene #12: 'NSD1 MUTATION STATUS' versus Clinical Feature #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 | CLUS_8 |
|---|---|---|---|---|---|---|---|---|
| ALL | 42 | 47 | 39 | 43 | 58 | 53 | 5 | 19 |
| NSD1 MUTATED | 25 | 1 | 2 | 3 | 0 | 2 | 0 | 0 |
| NSD1 WILD-TYPE | 17 | 46 | 37 | 40 | 58 | 51 | 5 | 19 |
Figure S10. Get High-res Image Gene #12: 'NSD1 MUTATION STATUS' versus Clinical Feature #2: 'METHLYATION_CNMF'
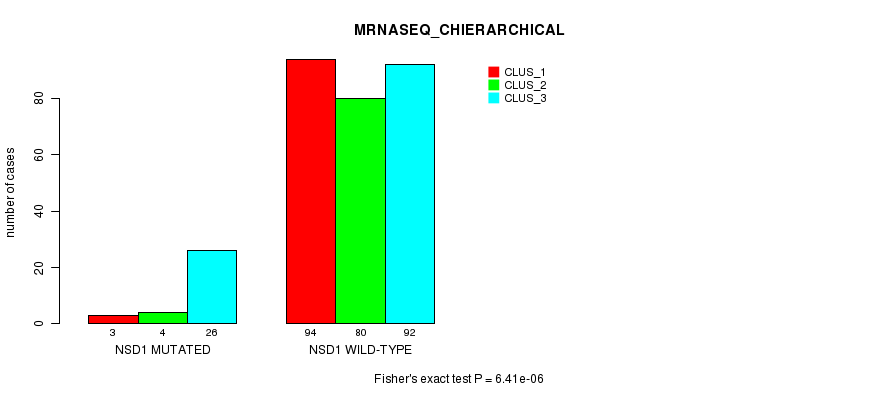
P value = 6.41e-06 (Fisher's exact test), Q value = 0.0047
Table S11. Gene #12: 'NSD1 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 97 | 84 | 118 |
| NSD1 MUTATED | 3 | 4 | 26 |
| NSD1 WILD-TYPE | 94 | 80 | 92 |
Figure S11. Get High-res Image Gene #12: 'NSD1 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
-
Mutation data file = HNSC-TP.mutsig.cluster.txt
-
Molecular subtypes file = HNSC-TP.transferedmergedcluster.txt
-
Number of patients = 306
-
Number of significantly mutated genes = 94
-
Number of Molecular subtypes = 8
-
Exclude genes that fewer than K tumors have mutations, K = 3
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.