This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 33 genes and 8 molecular subtypes across 170 patients, 36 significant findings detected with P value < 0.05 and Q value < 0.25.
-
IDH1 mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CNMF'.
-
TP53 mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CNMF'.
-
PIK3CA mutation correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
ATRX mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CNMF'.
-
CIC mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CNMF'.
-
FUBP1 mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CNMF'.
-
NOTCH1 mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
PTEN mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CNMF'.
-
NF1 mutation correlated to 'METHLYATION_CNMF' and 'MRNASEQ_CNMF'.
-
EGFR mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CNMF'.
Table 1. Get Full Table Overview of the association between mutation status of 33 genes and 8 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 36 significant findings detected.
|
Clinical Features |
MRNA CNMF |
MRNA CHIERARCHICAL |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Chi-square test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| TP53 | 88 (52%) | 82 |
0.136 (1.00) |
0.188 (1.00) |
8.29e-19 (1.72e-16) |
1.8e-32 (3.78e-30) |
5.91e-12 (1.19e-09) |
1.65e-10 (3.26e-08) |
2.46e-05 (0.00456) |
0.377 (1.00) |
| ATRX | 73 (43%) | 97 |
0.0421 (1.00) |
0.0271 (1.00) |
1.44e-10 (2.86e-08) |
1.97e-21 (4.1e-19) |
1.76e-12 (3.58e-10) |
5.04e-07 (9.73e-05) |
1.32e-05 (0.00248) |
0.465 (1.00) |
| CIC | 35 (21%) | 135 |
0.0764 (1.00) |
0.118 (1.00) |
6.08e-15 (1.24e-12) |
2.65e-18 (5.47e-16) |
4.68e-10 (9.23e-08) |
5.81e-12 (1.17e-09) |
2.72e-07 (5.31e-05) |
0.15 (1.00) |
| FUBP1 | 19 (11%) | 151 |
0.0862 (1.00) |
0.0919 (1.00) |
3.3e-08 (6.47e-06) |
3.75e-06 (0.000717) |
3.36e-05 (0.00618) |
1.6e-05 (0.00297) |
0.000268 (0.0487) |
0.922 (1.00) |
| IDH1 | 131 (77%) | 39 |
0.179 (1.00) |
0.272 (1.00) |
4.15e-11 (8.31e-09) |
2.28e-26 (4.77e-24) |
1.16e-16 (2.38e-14) |
0.064 (1.00) |
0.148 (1.00) |
0.0741 (1.00) |
| NOTCH1 | 16 (9%) | 154 |
0.502 (1.00) |
0.463 (1.00) |
0.000671 (0.119) |
0.000265 (0.0485) |
0.0188 (1.00) |
0.00063 (0.112) |
0.00675 (1.00) |
0.043 (1.00) |
| PTEN | 8 (5%) | 162 |
5.51e-06 (0.00105) |
7.94e-06 (0.00149) |
3.34e-07 (6.47e-05) |
0.0227 (1.00) |
0.00448 (0.78) |
0.235 (1.00) |
||
| EGFR | 8 (5%) | 162 |
5.51e-06 (0.00105) |
8.8e-07 (0.000169) |
0.00121 (0.213) |
0.382 (1.00) |
0.0688 (1.00) |
1 (1.00) |
||
| PIK3CA | 15 (9%) | 155 |
0.000481 (0.0866) |
0.000482 (0.0866) |
0.0362 (1.00) |
0.215 (1.00) |
0.573 (1.00) |
0.305 (1.00) |
||
| NF1 | 11 (6%) | 159 |
0.0489 (1.00) |
0.000388 (0.0703) |
0.00123 (0.216) |
0.306 (1.00) |
0.29 (1.00) |
0.763 (1.00) |
||
| IDH2 | 6 (4%) | 164 |
0.24 (1.00) |
0.281 (1.00) |
0.133 (1.00) |
0.00734 (1.00) |
0.0436 (1.00) |
0.787 (1.00) |
||
| PIK3R1 | 12 (7%) | 158 |
0.0331 (1.00) |
0.378 (1.00) |
0.78 (1.00) |
0.915 (1.00) |
0.867 (1.00) |
0.79 (1.00) |
||
| ZNF844 | 5 (3%) | 165 |
0.451 (1.00) |
0.838 (1.00) |
0.376 (1.00) |
0.711 (1.00) |
0.414 (1.00) |
0.382 (1.00) |
||
| IL32 | 4 (2%) | 166 |
0.128 (1.00) |
0.106 (1.00) |
0.233 (1.00) |
1 (1.00) |
0.826 (1.00) |
0.239 (1.00) |
||
| TIMD4 | 6 (4%) | 164 |
0.447 (1.00) |
0.68 (1.00) |
0.699 (1.00) |
0.737 (1.00) |
0.558 (1.00) |
1 (1.00) |
||
| CREBZF | 4 (2%) | 166 |
0.177 (1.00) |
0.0547 (1.00) |
0.347 (1.00) |
0.0594 (1.00) |
0.313 (1.00) |
0.0527 (1.00) |
||
| ZNF57 | 6 (4%) | 164 |
0.134 (1.00) |
0.201 (1.00) |
0.0959 (1.00) |
0.0725 (1.00) |
0.0297 (1.00) |
0.272 (1.00) |
||
| ARID1A | 11 (6%) | 159 |
0.646 (1.00) |
0.504 (1.00) |
0.304 (1.00) |
0.25 (1.00) |
0.307 (1.00) |
0.265 (1.00) |
||
| TCF12 | 6 (4%) | 164 |
1 (1.00) |
0.247 (1.00) |
0.871 (1.00) |
0.322 (1.00) |
0.34 (1.00) |
0.366 (1.00) |
||
| NOX4 | 5 (3%) | 165 |
0.141 (1.00) |
1 (1.00) |
0.589 (1.00) |
0.248 (1.00) |
0.623 (1.00) |
0.219 (1.00) |
||
| ZBTB20 | 7 (4%) | 163 |
0.386 (1.00) |
0.232 (1.00) |
0.626 (1.00) |
1 (1.00) |
0.951 (1.00) |
0.416 (1.00) |
||
| MUC7 | 5 (3%) | 165 |
0.174 (1.00) |
0.231 (1.00) |
0.323 (1.00) |
0.577 (1.00) |
0.923 (1.00) |
0.382 (1.00) |
||
| ZNF845 | 6 (4%) | 164 |
0.285 (1.00) |
0.821 (1.00) |
0.699 (1.00) |
0.552 (1.00) |
0.836 (1.00) |
0.0872 (1.00) |
||
| ANKRD30A | 7 (4%) | 163 |
0.615 (1.00) |
0.436 (1.00) |
0.473 (1.00) |
0.869 (1.00) |
0.19 (1.00) |
0.151 (1.00) |
||
| SPDYE5 | 4 (2%) | 166 |
0.0598 (1.00) |
0.0445 (1.00) |
0.0616 (1.00) |
0.295 (1.00) |
0.0693 (1.00) |
0.329 (1.00) |
||
| SCAF1 | 4 (2%) | 166 |
1 (1.00) |
1 (1.00) |
0.456 (1.00) |
0.394 (1.00) |
0.118 (1.00) |
1 (1.00) |
||
| PRDM9 | 5 (3%) | 165 |
0.845 (1.00) |
0.362 (1.00) |
0.64 (1.00) |
0.711 (1.00) |
0.196 (1.00) |
0.562 (1.00) |
||
| PRAMEF11 | 5 (3%) | 165 |
0.845 (1.00) |
0.838 (1.00) |
0.293 (1.00) |
0.464 (1.00) |
0.414 (1.00) |
0.0231 (1.00) |
||
| ZCCHC12 | 3 (2%) | 167 |
0.0628 (1.00) |
0.0703 (1.00) |
0.556 (1.00) |
0.152 (1.00) |
0.505 (1.00) |
0.147 (1.00) |
||
| ZNF91 | 5 (3%) | 165 |
0.369 (1.00) |
0.062 (1.00) |
0.369 (1.00) |
0.388 (1.00) |
1 (1.00) |
1 (1.00) |
||
| C3ORF35 | 3 (2%) | 167 |
1 (1.00) |
0.845 (1.00) |
0.566 (1.00) |
0.581 (1.00) |
0.774 (1.00) |
0.147 (1.00) |
||
| RPTN | 5 (3%) | 165 |
0.248 (1.00) |
0.0988 (1.00) |
0.549 (1.00) |
0.823 (1.00) |
0.809 (1.00) |
0.562 (1.00) |
||
| DDX5 | 5 (3%) | 165 |
1 (1.00) |
1 (1.00) |
0.437 (1.00) |
0.108 (1.00) |
0.414 (1.00) |
0.219 (1.00) |
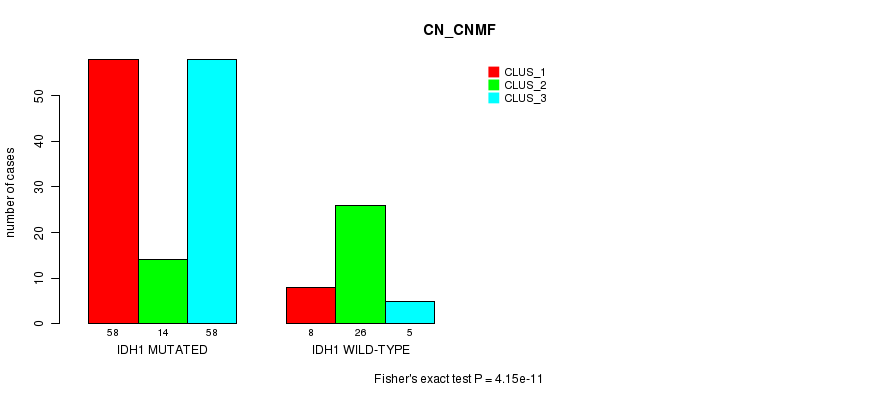
P value = 4.15e-11 (Fisher's exact test), Q value = 8.3e-09
Table S1. Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 40 | 63 |
| IDH1 MUTATED | 58 | 14 | 58 |
| IDH1 WILD-TYPE | 8 | 26 | 5 |
Figure S1. Get High-res Image Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
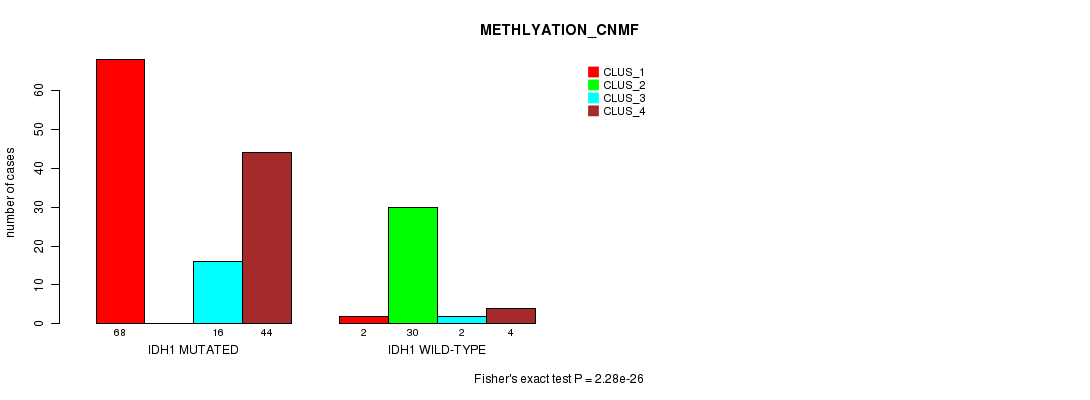
P value = 2.28e-26 (Fisher's exact test), Q value = 4.8e-24
Table S2. Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 30 | 18 | 48 |
| IDH1 MUTATED | 68 | 0 | 16 | 44 |
| IDH1 WILD-TYPE | 2 | 30 | 2 | 4 |
Figure S2. Get High-res Image Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
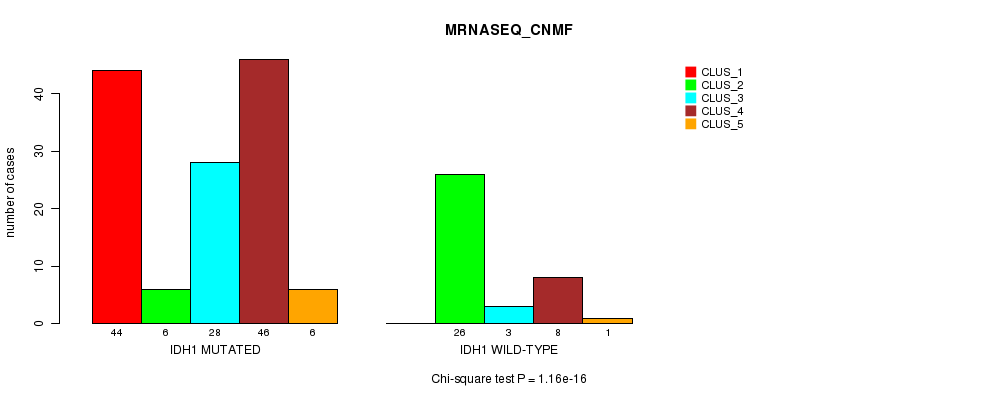
P value = 1.16e-16 (Chi-square test), Q value = 2.4e-14
Table S3. Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 44 | 32 | 31 | 54 | 7 |
| IDH1 MUTATED | 44 | 6 | 28 | 46 | 6 |
| IDH1 WILD-TYPE | 0 | 26 | 3 | 8 | 1 |
Figure S3. Get High-res Image Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
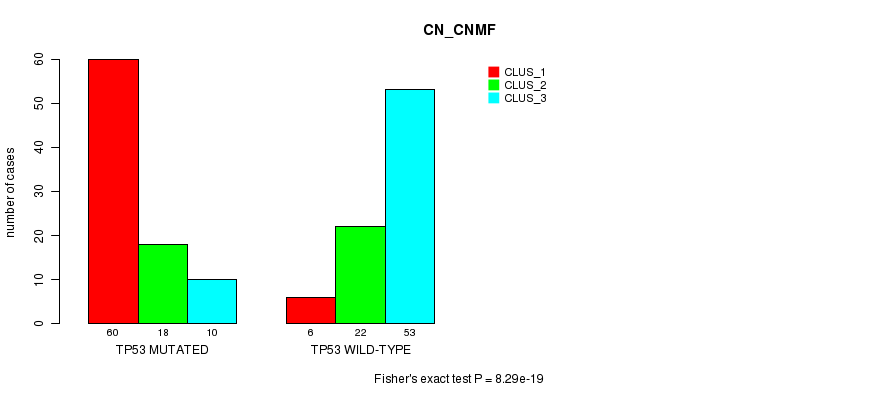
P value = 8.29e-19 (Fisher's exact test), Q value = 1.7e-16
Table S4. Gene #3: 'TP53 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 40 | 63 |
| TP53 MUTATED | 60 | 18 | 10 |
| TP53 WILD-TYPE | 6 | 22 | 53 |
Figure S4. Get High-res Image Gene #3: 'TP53 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
P value = 1.8e-32 (Fisher's exact test), Q value = 3.8e-30
Table S5. Gene #3: 'TP53 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 30 | 18 | 48 |
| TP53 MUTATED | 69 | 6 | 11 | 2 |
| TP53 WILD-TYPE | 1 | 24 | 7 | 46 |
Figure S5. Get High-res Image Gene #3: 'TP53 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
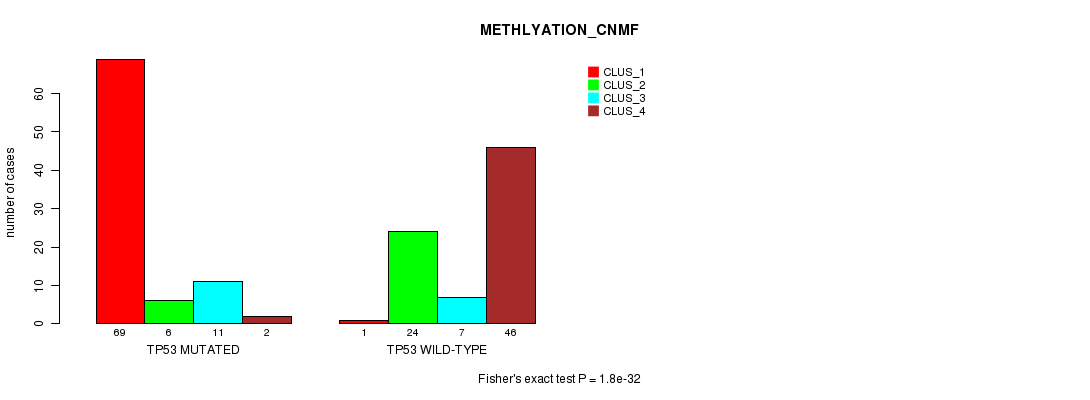
P value = 5.91e-12 (Chi-square test), Q value = 1.2e-09
Table S6. Gene #3: 'TP53 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 44 | 32 | 31 | 54 | 7 |
| TP53 MUTATED | 42 | 12 | 3 | 27 | 3 |
| TP53 WILD-TYPE | 2 | 20 | 28 | 27 | 4 |
Figure S6. Get High-res Image Gene #3: 'TP53 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
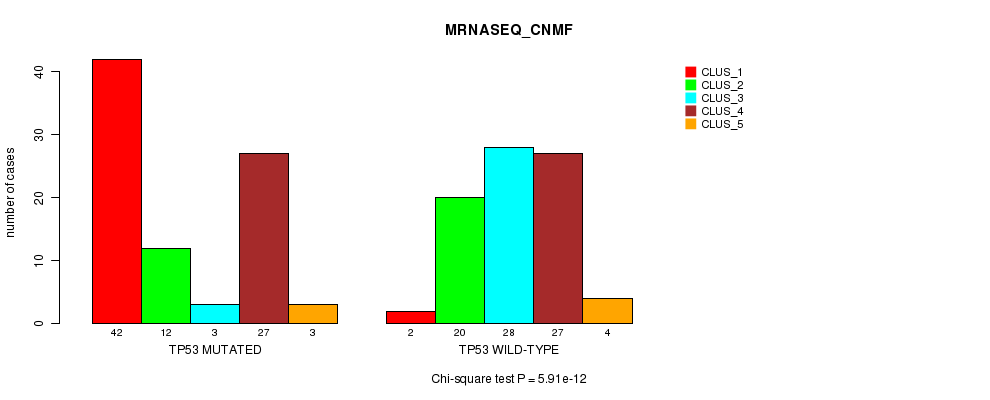
P value = 1.65e-10 (Fisher's exact test), Q value = 3.3e-08
Table S7. Gene #3: 'TP53 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 88 | 51 |
| TP53 MUTATED | 1 | 62 | 24 |
| TP53 WILD-TYPE | 28 | 26 | 27 |
Figure S7. Get High-res Image Gene #3: 'TP53 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'

P value = 2.46e-05 (Fisher's exact test), Q value = 0.0046
Table S8. Gene #3: 'TP53 MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 38 | 27 | 65 |
| TP53 MUTATED | 26 | 28 | 5 | 29 |
| TP53 WILD-TYPE | 14 | 10 | 22 | 36 |
Figure S8. Get High-res Image Gene #3: 'TP53 MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
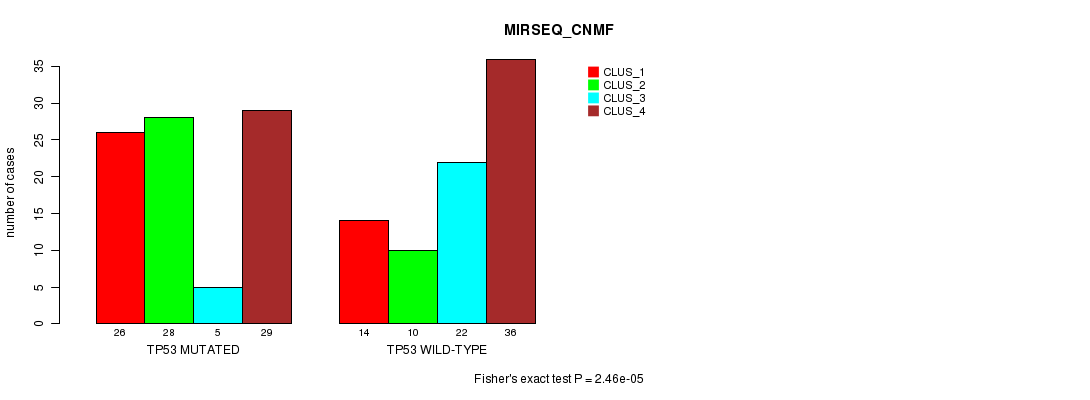
P value = 0.000481 (Fisher's exact test), Q value = 0.087
Table S9. Gene #4: 'PIK3CA MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 40 | 63 |
| PIK3CA MUTATED | 0 | 4 | 11 |
| PIK3CA WILD-TYPE | 66 | 36 | 52 |
Figure S9. Get High-res Image Gene #4: 'PIK3CA MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
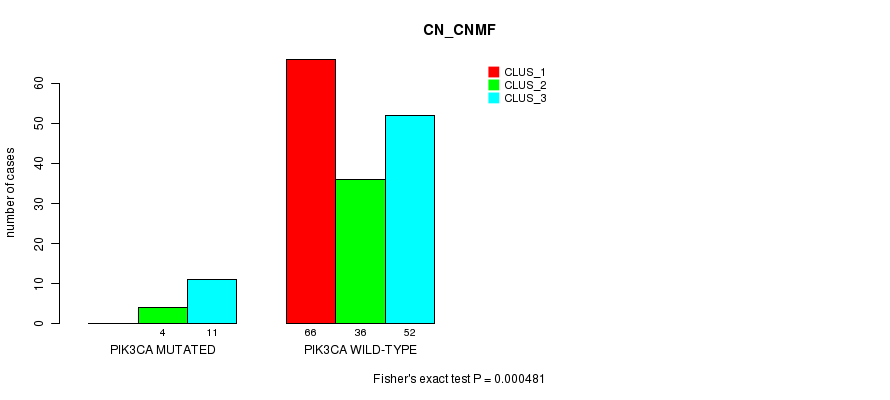
P value = 0.000482 (Fisher's exact test), Q value = 0.087
Table S10. Gene #4: 'PIK3CA MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 30 | 18 | 48 |
| PIK3CA MUTATED | 0 | 4 | 2 | 9 |
| PIK3CA WILD-TYPE | 70 | 26 | 16 | 39 |
Figure S10. Get High-res Image Gene #4: 'PIK3CA MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
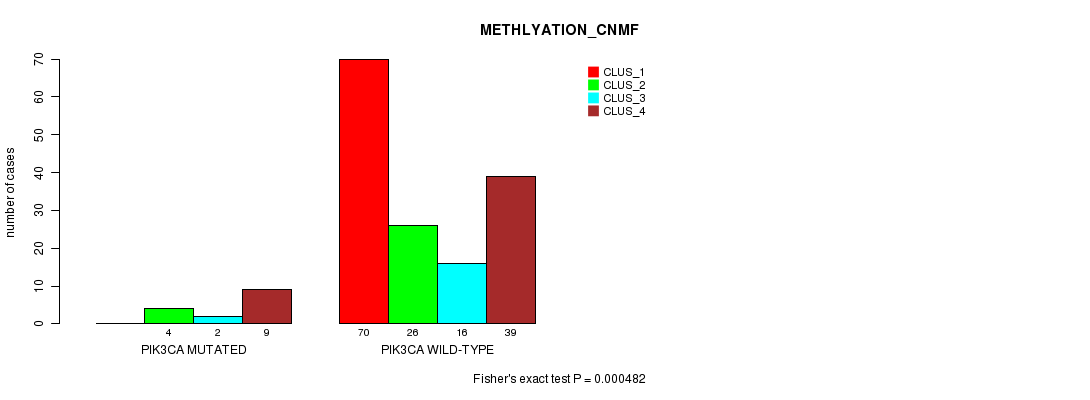
P value = 1.44e-10 (Fisher's exact test), Q value = 2.9e-08
Table S11. Gene #5: 'ATRX MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 40 | 63 |
| ATRX MUTATED | 48 | 15 | 10 |
| ATRX WILD-TYPE | 18 | 25 | 53 |
Figure S11. Get High-res Image Gene #5: 'ATRX MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
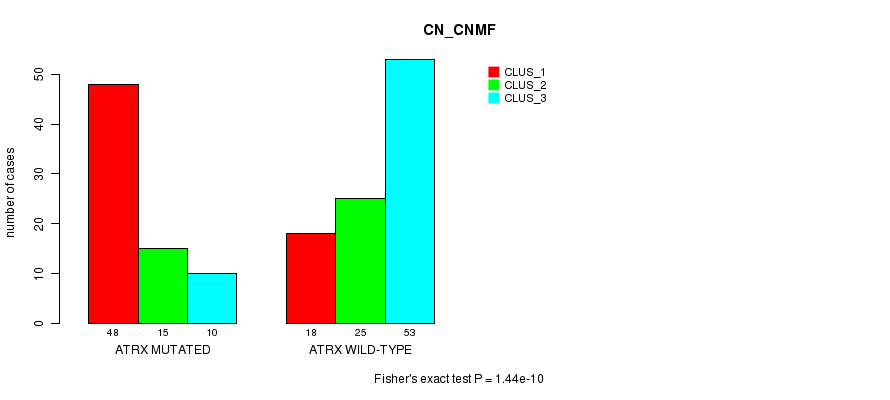
P value = 1.97e-21 (Fisher's exact test), Q value = 4.1e-19
Table S12. Gene #5: 'ATRX MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 30 | 18 | 48 |
| ATRX MUTATED | 58 | 4 | 9 | 2 |
| ATRX WILD-TYPE | 12 | 26 | 9 | 46 |
Figure S12. Get High-res Image Gene #5: 'ATRX MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
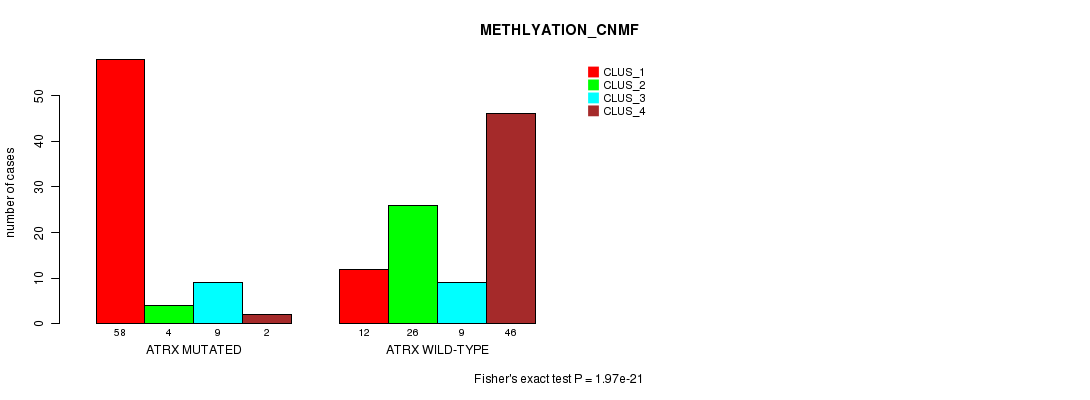
P value = 1.76e-12 (Chi-square test), Q value = 3.6e-10
Table S13. Gene #5: 'ATRX MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 44 | 32 | 31 | 54 | 7 |
| ATRX MUTATED | 39 | 8 | 2 | 22 | 1 |
| ATRX WILD-TYPE | 5 | 24 | 29 | 32 | 6 |
Figure S13. Get High-res Image Gene #5: 'ATRX MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
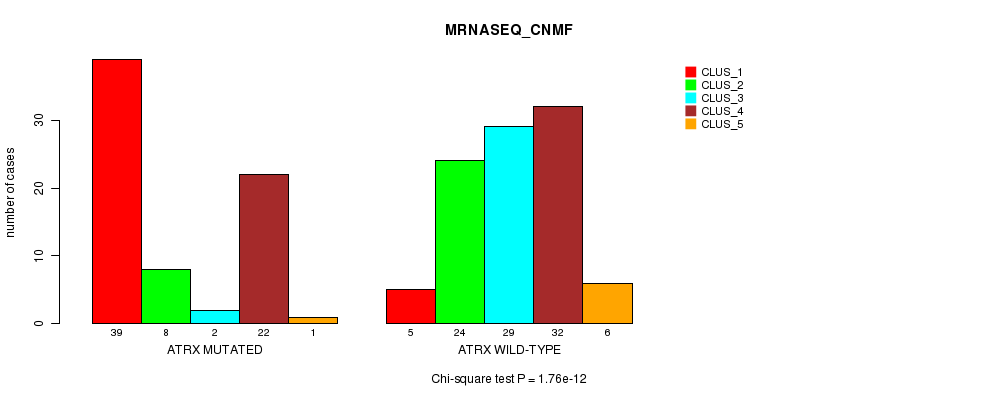
P value = 5.04e-07 (Fisher's exact test), Q value = 9.7e-05
Table S14. Gene #5: 'ATRX MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 88 | 51 |
| ATRX MUTATED | 1 | 49 | 22 |
| ATRX WILD-TYPE | 28 | 39 | 29 |
Figure S14. Get High-res Image Gene #5: 'ATRX MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
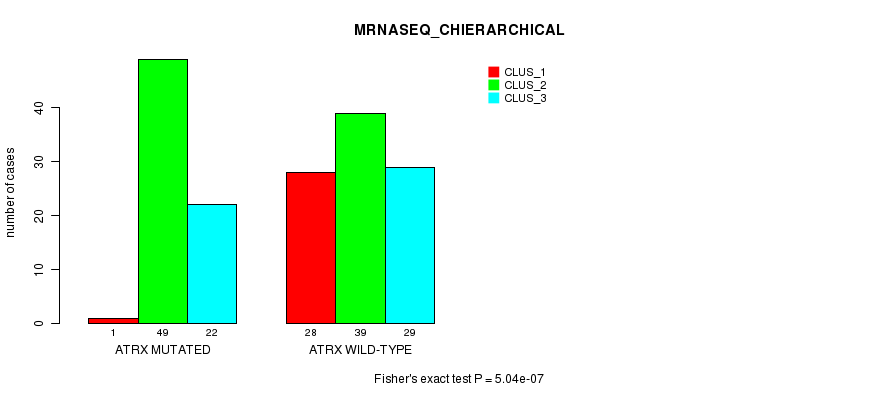
P value = 1.32e-05 (Fisher's exact test), Q value = 0.0025
Table S15. Gene #5: 'ATRX MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 38 | 27 | 65 |
| ATRX MUTATED | 18 | 27 | 3 | 25 |
| ATRX WILD-TYPE | 22 | 11 | 24 | 40 |
Figure S15. Get High-res Image Gene #5: 'ATRX MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
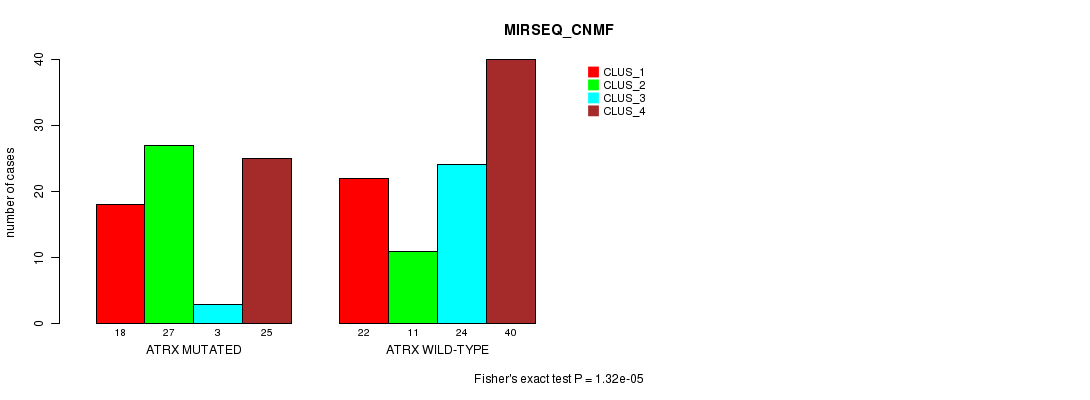
P value = 6.08e-15 (Fisher's exact test), Q value = 1.2e-12
Table S16. Gene #6: 'CIC MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 40 | 63 |
| CIC MUTATED | 1 | 1 | 33 |
| CIC WILD-TYPE | 65 | 39 | 30 |
Figure S16. Get High-res Image Gene #6: 'CIC MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
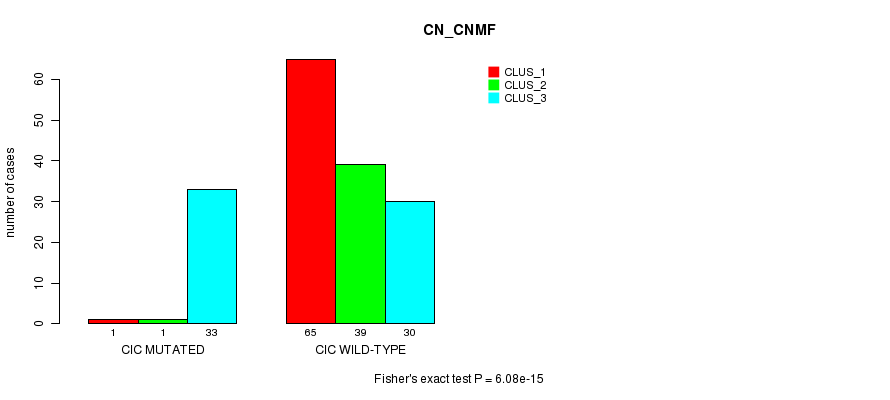
P value = 2.65e-18 (Fisher's exact test), Q value = 5.5e-16
Table S17. Gene #6: 'CIC MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 30 | 18 | 48 |
| CIC MUTATED | 0 | 1 | 2 | 31 |
| CIC WILD-TYPE | 70 | 29 | 16 | 17 |
Figure S17. Get High-res Image Gene #6: 'CIC MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
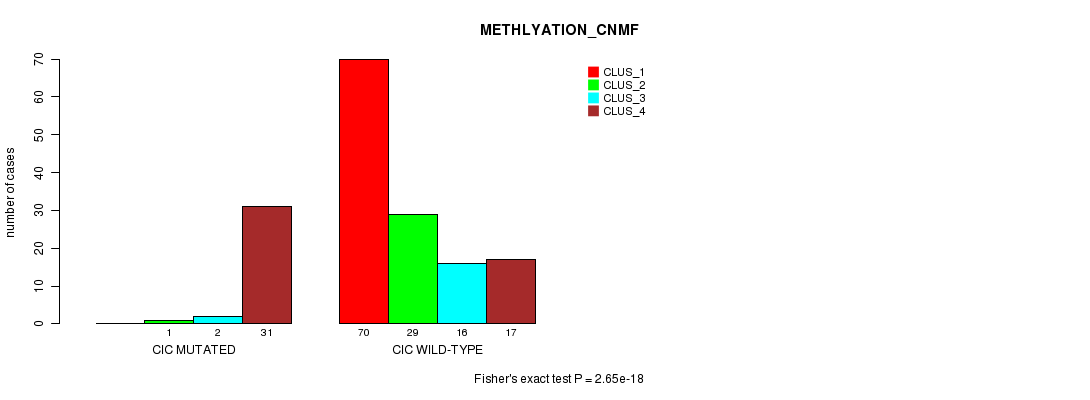
P value = 4.68e-10 (Chi-square test), Q value = 9.2e-08
Table S18. Gene #6: 'CIC MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 44 | 32 | 31 | 54 | 7 |
| CIC MUTATED | 0 | 1 | 19 | 14 | 1 |
| CIC WILD-TYPE | 44 | 31 | 12 | 40 | 6 |
Figure S18. Get High-res Image Gene #6: 'CIC MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
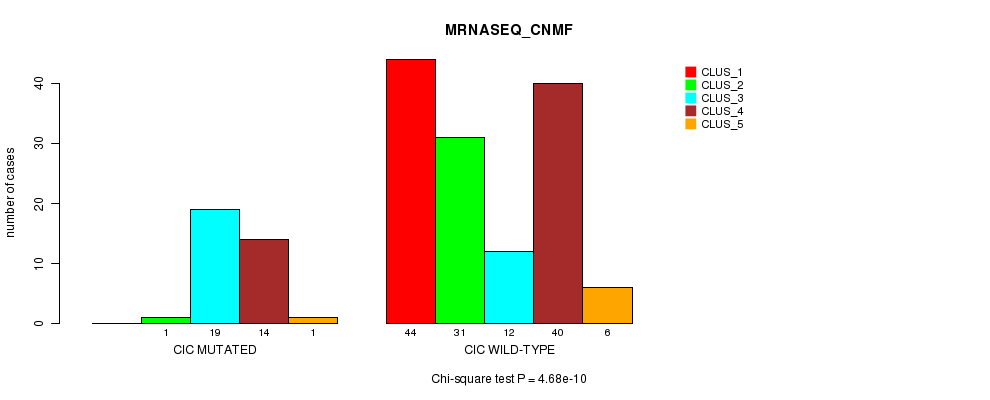
P value = 5.81e-12 (Fisher's exact test), Q value = 1.2e-09
Table S19. Gene #6: 'CIC MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 88 | 51 |
| CIC MUTATED | 19 | 3 | 13 |
| CIC WILD-TYPE | 10 | 85 | 38 |
Figure S19. Get High-res Image Gene #6: 'CIC MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'

P value = 2.72e-07 (Fisher's exact test), Q value = 5.3e-05
Table S20. Gene #6: 'CIC MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 38 | 27 | 65 |
| CIC MUTATED | 4 | 1 | 16 | 14 |
| CIC WILD-TYPE | 36 | 37 | 11 | 51 |
Figure S20. Get High-res Image Gene #6: 'CIC MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
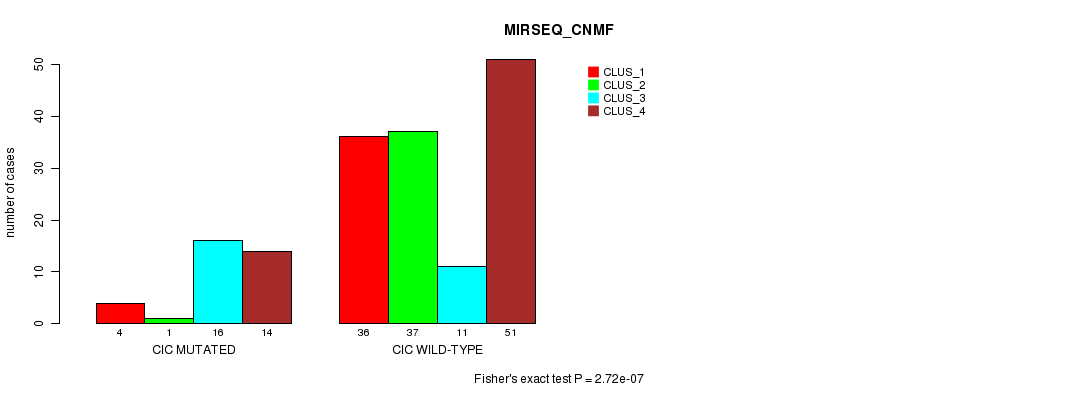
P value = 3.3e-08 (Fisher's exact test), Q value = 6.5e-06
Table S21. Gene #7: 'FUBP1 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 40 | 63 |
| FUBP1 MUTATED | 0 | 1 | 18 |
| FUBP1 WILD-TYPE | 66 | 39 | 45 |
Figure S21. Get High-res Image Gene #7: 'FUBP1 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
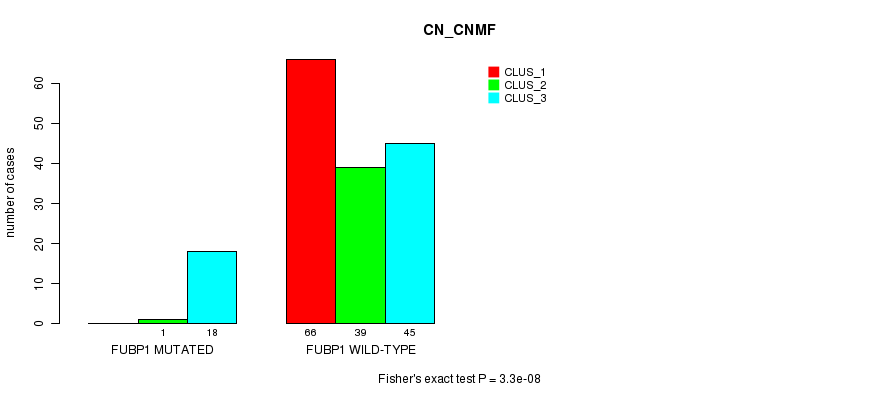
P value = 3.75e-06 (Fisher's exact test), Q value = 0.00072
Table S22. Gene #7: 'FUBP1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 30 | 18 | 48 |
| FUBP1 MUTATED | 0 | 1 | 2 | 13 |
| FUBP1 WILD-TYPE | 70 | 29 | 16 | 35 |
Figure S22. Get High-res Image Gene #7: 'FUBP1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
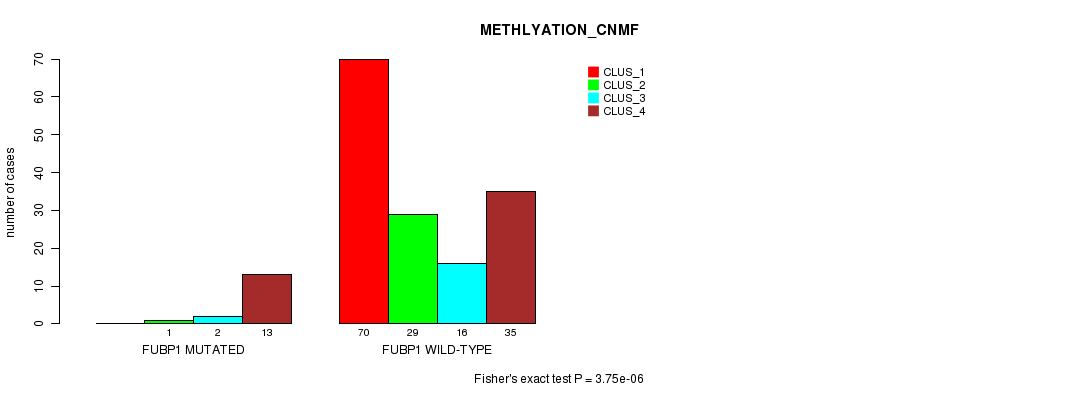
P value = 3.36e-05 (Chi-square test), Q value = 0.0062
Table S23. Gene #7: 'FUBP1 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 44 | 32 | 31 | 54 | 7 |
| FUBP1 MUTATED | 0 | 1 | 11 | 6 | 1 |
| FUBP1 WILD-TYPE | 44 | 31 | 20 | 48 | 6 |
Figure S23. Get High-res Image Gene #7: 'FUBP1 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
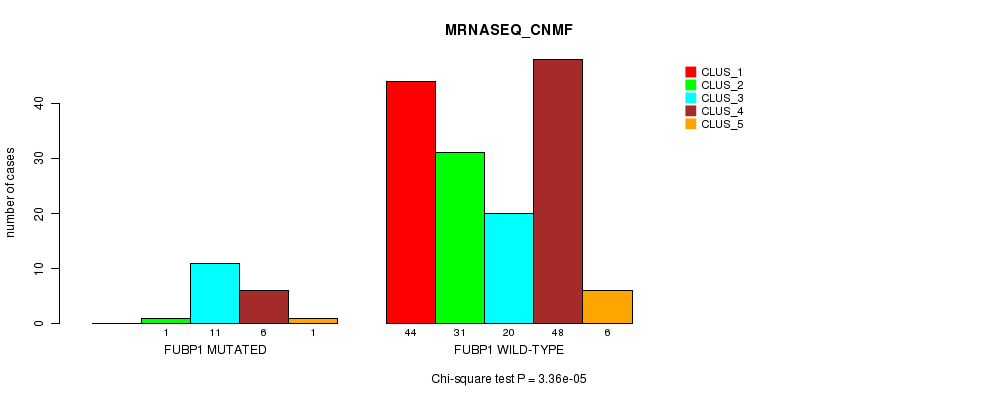
P value = 1.6e-05 (Fisher's exact test), Q value = 0.003
Table S24. Gene #7: 'FUBP1 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 88 | 51 |
| FUBP1 MUTATED | 11 | 3 | 5 |
| FUBP1 WILD-TYPE | 18 | 85 | 46 |
Figure S24. Get High-res Image Gene #7: 'FUBP1 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
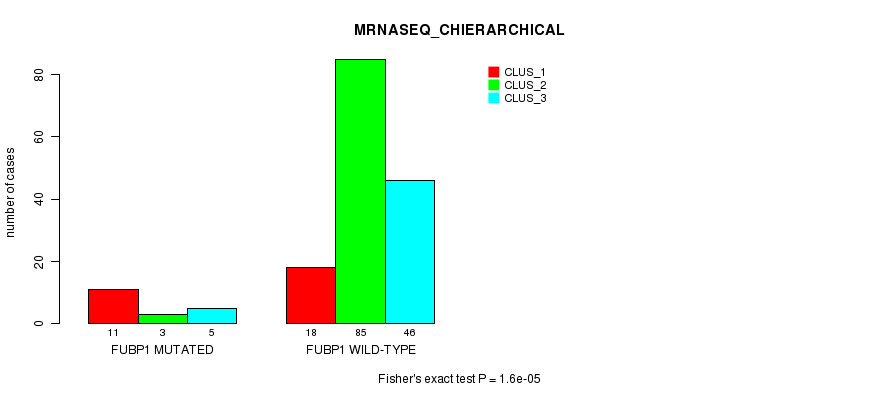
P value = 0.000268 (Fisher's exact test), Q value = 0.049
Table S25. Gene #7: 'FUBP1 MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 38 | 27 | 65 |
| FUBP1 MUTATED | 2 | 1 | 10 | 6 |
| FUBP1 WILD-TYPE | 38 | 37 | 17 | 59 |
Figure S25. Get High-res Image Gene #7: 'FUBP1 MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
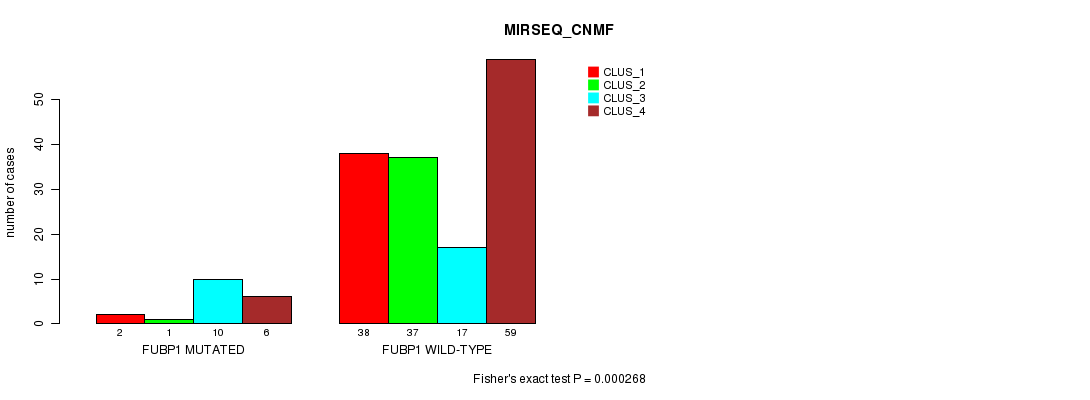
P value = 0.000671 (Fisher's exact test), Q value = 0.12
Table S26. Gene #9: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 40 | 63 |
| NOTCH1 MUTATED | 1 | 2 | 13 |
| NOTCH1 WILD-TYPE | 65 | 38 | 50 |
Figure S26. Get High-res Image Gene #9: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
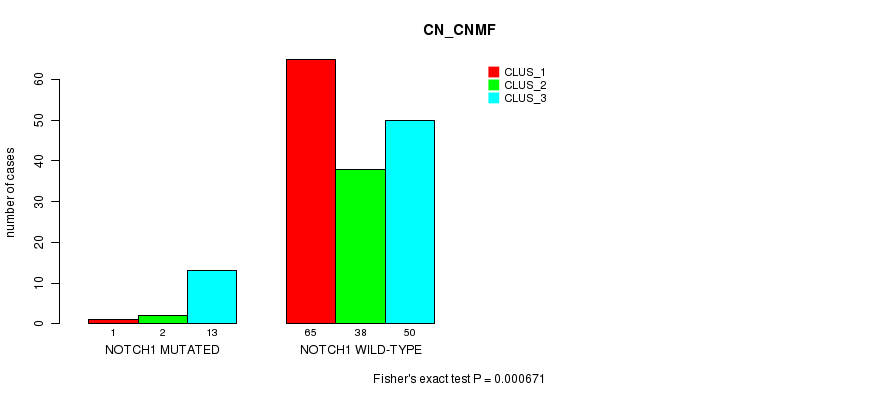
P value = 0.000265 (Fisher's exact test), Q value = 0.048
Table S27. Gene #9: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 30 | 18 | 48 |
| NOTCH1 MUTATED | 2 | 1 | 0 | 12 |
| NOTCH1 WILD-TYPE | 68 | 29 | 18 | 36 |
Figure S27. Get High-res Image Gene #9: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
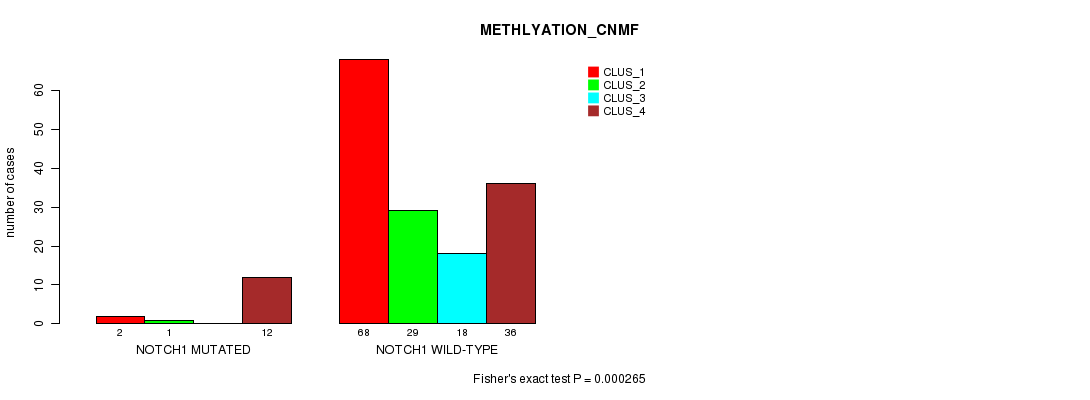
P value = 0.00063 (Fisher's exact test), Q value = 0.11
Table S28. Gene #9: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 88 | 51 |
| NOTCH1 MUTATED | 7 | 2 | 7 |
| NOTCH1 WILD-TYPE | 22 | 86 | 44 |
Figure S28. Get High-res Image Gene #9: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
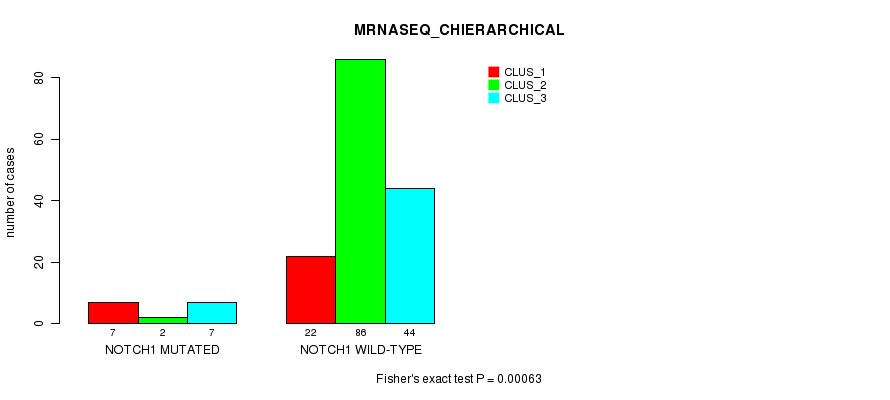
P value = 5.51e-06 (Fisher's exact test), Q value = 0.001
Table S29. Gene #12: 'PTEN MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 40 | 63 |
| PTEN MUTATED | 0 | 8 | 0 |
| PTEN WILD-TYPE | 66 | 32 | 63 |
Figure S29. Get High-res Image Gene #12: 'PTEN MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
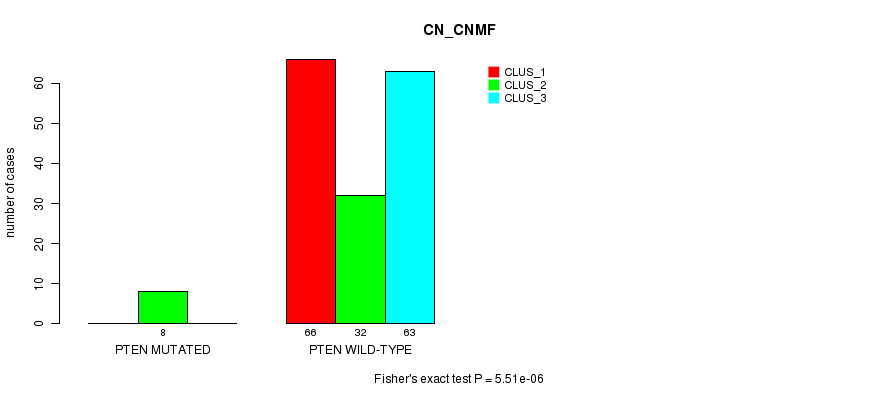
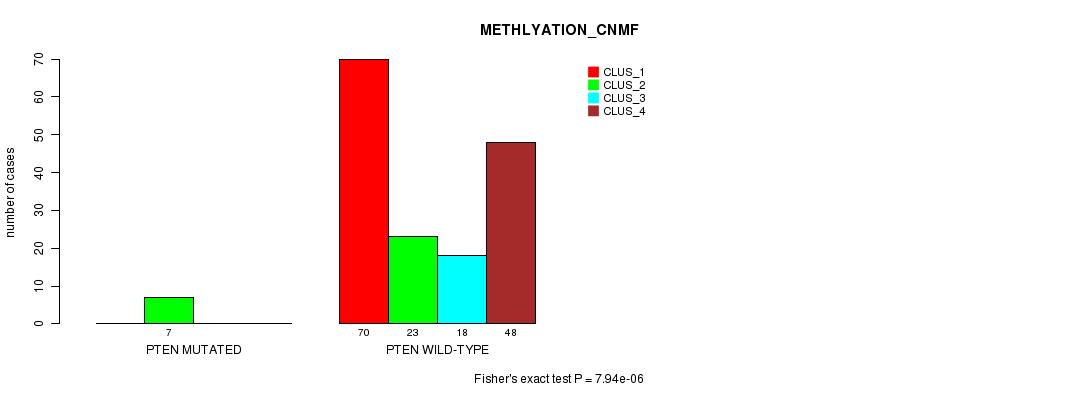
P value = 7.94e-06 (Fisher's exact test), Q value = 0.0015
Table S30. Gene #12: 'PTEN MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 30 | 18 | 48 |
| PTEN MUTATED | 0 | 7 | 0 | 0 |
| PTEN WILD-TYPE | 70 | 23 | 18 | 48 |
Figure S30. Get High-res Image Gene #12: 'PTEN MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
P value = 3.34e-07 (Chi-square test), Q value = 6.5e-05
Table S31. Gene #12: 'PTEN MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 44 | 32 | 31 | 54 | 7 |
| PTEN MUTATED | 0 | 8 | 0 | 0 | 0 |
| PTEN WILD-TYPE | 44 | 24 | 31 | 54 | 7 |
Figure S31. Get High-res Image Gene #12: 'PTEN MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
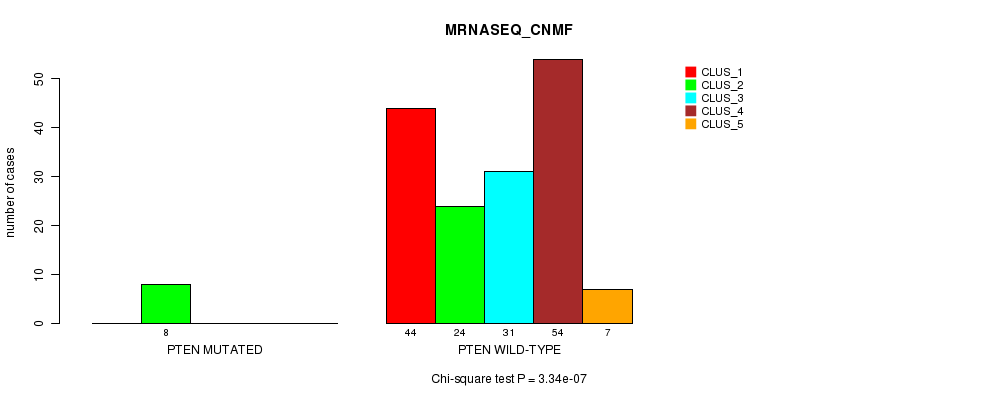
P value = 0.000388 (Fisher's exact test), Q value = 0.07
Table S32. Gene #17: 'NF1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 30 | 18 | 48 |
| NF1 MUTATED | 1 | 6 | 0 | 0 |
| NF1 WILD-TYPE | 69 | 24 | 18 | 48 |
Figure S32. Get High-res Image Gene #17: 'NF1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
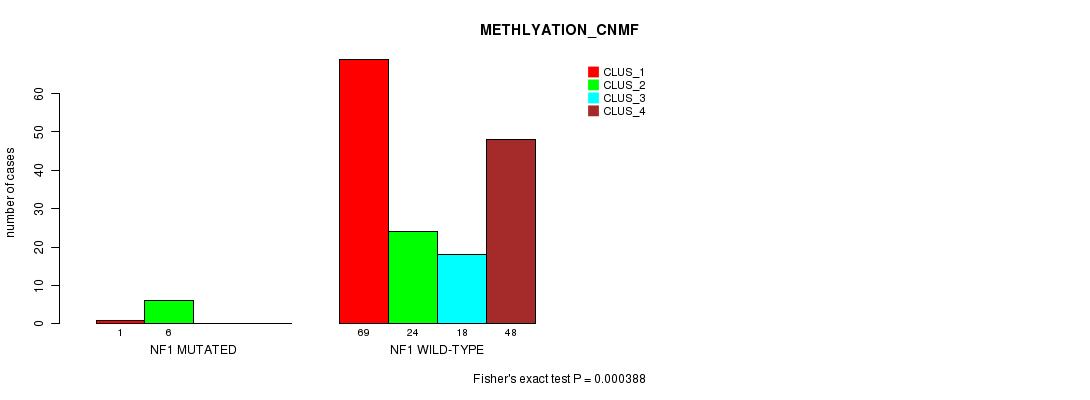
P value = 0.00123 (Chi-square test), Q value = 0.22
Table S33. Gene #17: 'NF1 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 44 | 32 | 31 | 54 | 7 |
| NF1 MUTATED | 0 | 7 | 2 | 1 | 1 |
| NF1 WILD-TYPE | 44 | 25 | 29 | 53 | 6 |
Figure S33. Get High-res Image Gene #17: 'NF1 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
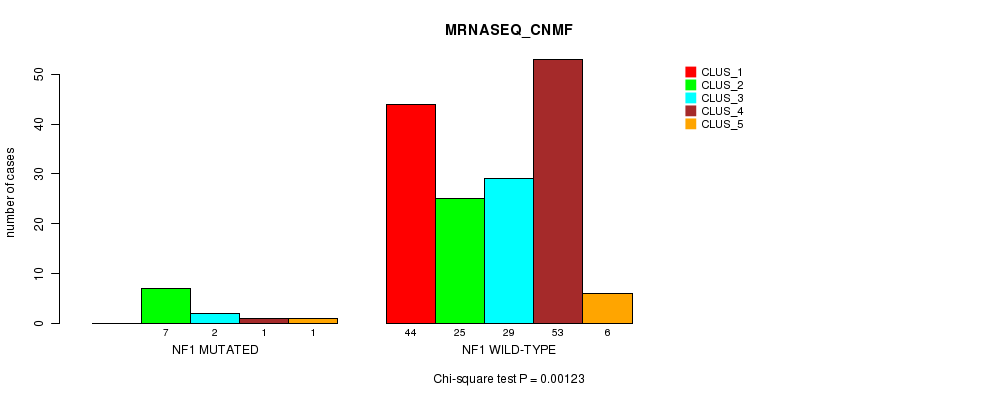
P value = 5.51e-06 (Fisher's exact test), Q value = 0.001
Table S34. Gene #20: 'EGFR MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 40 | 63 |
| EGFR MUTATED | 0 | 8 | 0 |
| EGFR WILD-TYPE | 66 | 32 | 63 |
Figure S34. Get High-res Image Gene #20: 'EGFR MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
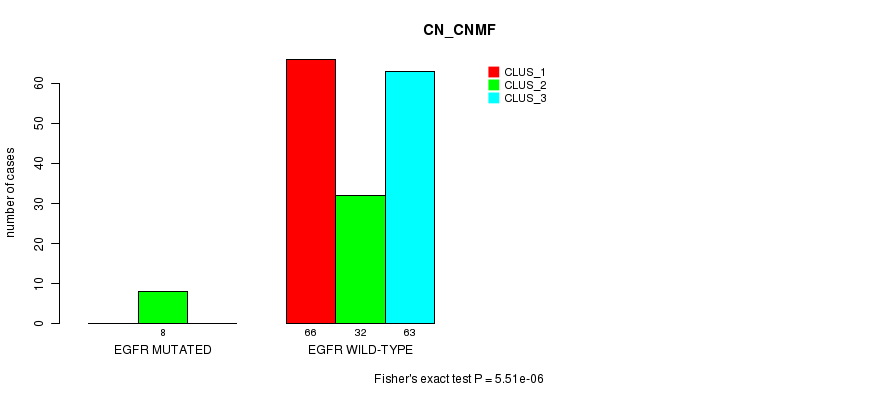
P value = 8.8e-07 (Fisher's exact test), Q value = 0.00017
Table S35. Gene #20: 'EGFR MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 30 | 18 | 48 |
| EGFR MUTATED | 0 | 8 | 0 | 0 |
| EGFR WILD-TYPE | 70 | 22 | 18 | 48 |
Figure S35. Get High-res Image Gene #20: 'EGFR MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
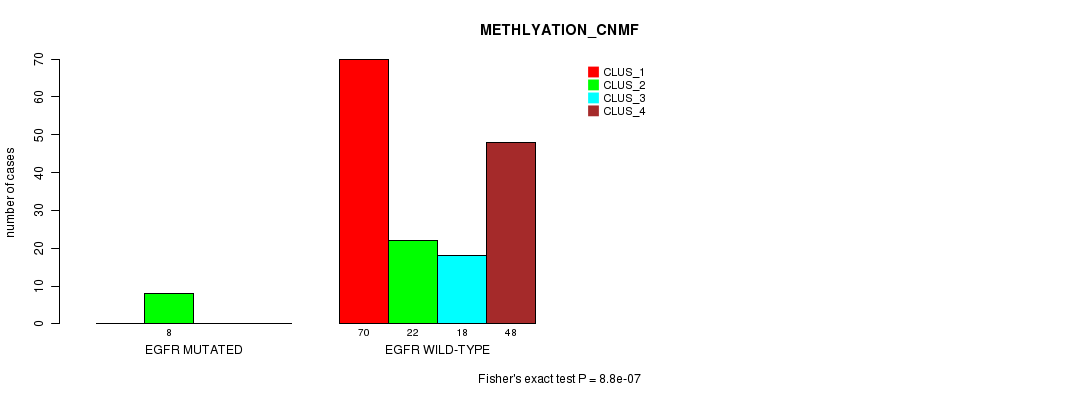
P value = 0.00121 (Chi-square test), Q value = 0.21
Table S36. Gene #20: 'EGFR MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 44 | 32 | 31 | 54 | 7 |
| EGFR MUTATED | 0 | 6 | 0 | 2 | 0 |
| EGFR WILD-TYPE | 44 | 26 | 31 | 52 | 7 |
Figure S36. Get High-res Image Gene #20: 'EGFR MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
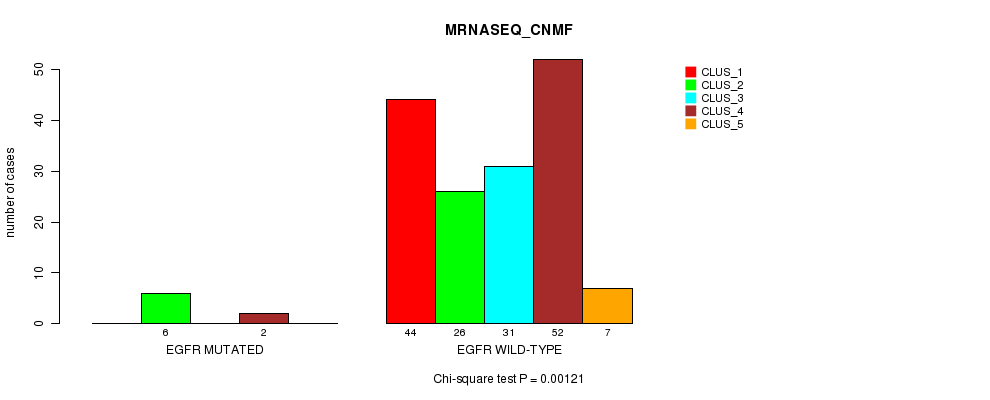
-
Mutation data file = LGG-TP.mutsig.cluster.txt
-
Molecular subtypes file = LGG-TP.transferedmergedcluster.txt
-
Number of patients = 170
-
Number of significantly mutated genes = 33
-
Number of Molecular subtypes = 8
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.