(primary solid tumor cohort)
This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 120 genes and 7 molecular subtypes across 155 patients, 12 significant findings detected with P value < 0.05 and Q value < 0.25.
-
BRAF mutation correlated to 'MRNA_CNMF', 'MRNA_CHIERARCHICAL', and 'CN_CNMF'.
-
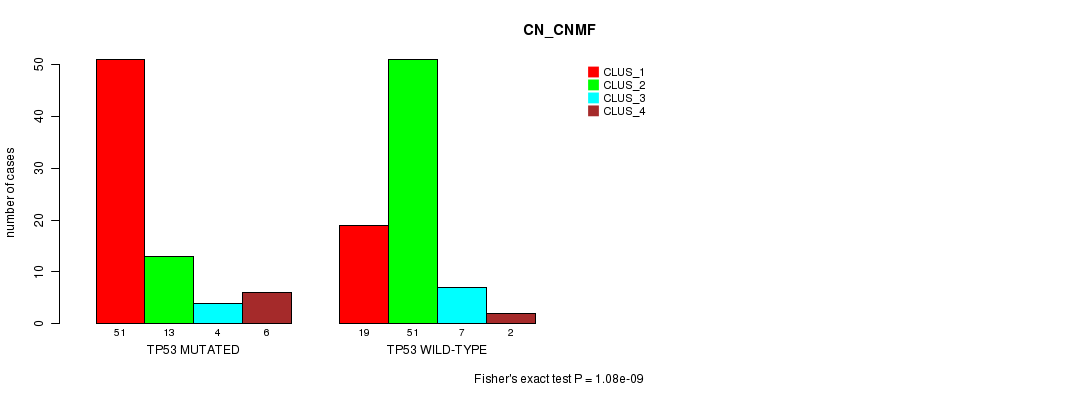
TP53 mutation correlated to 'CN_CNMF'.
-
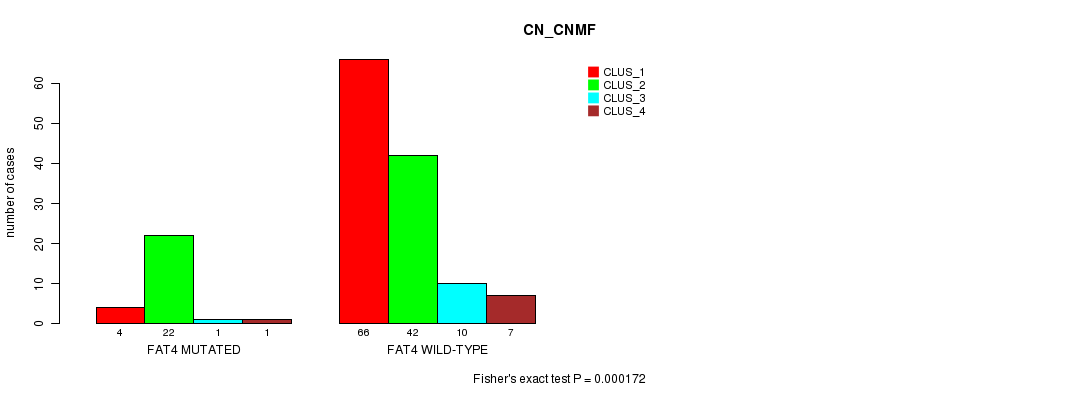
FAT4 mutation correlated to 'CN_CNMF'.
-
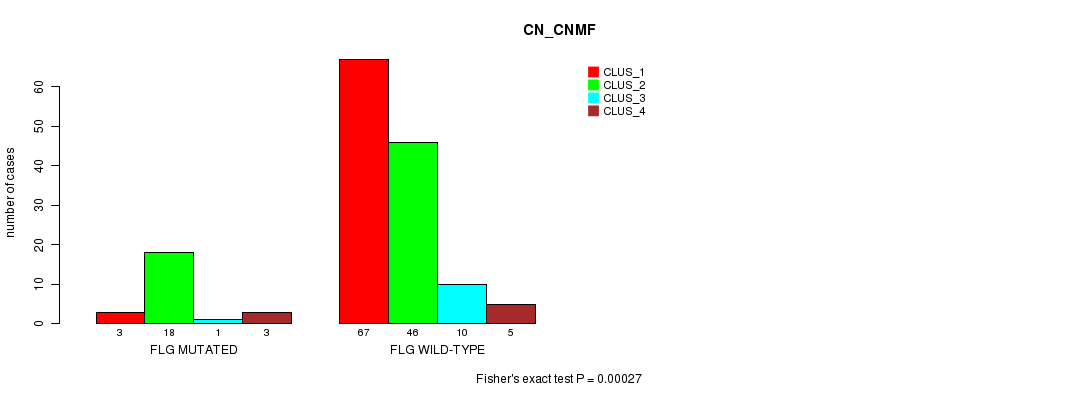
FLG mutation correlated to 'MRNA_CNMF' and 'CN_CNMF'.
-
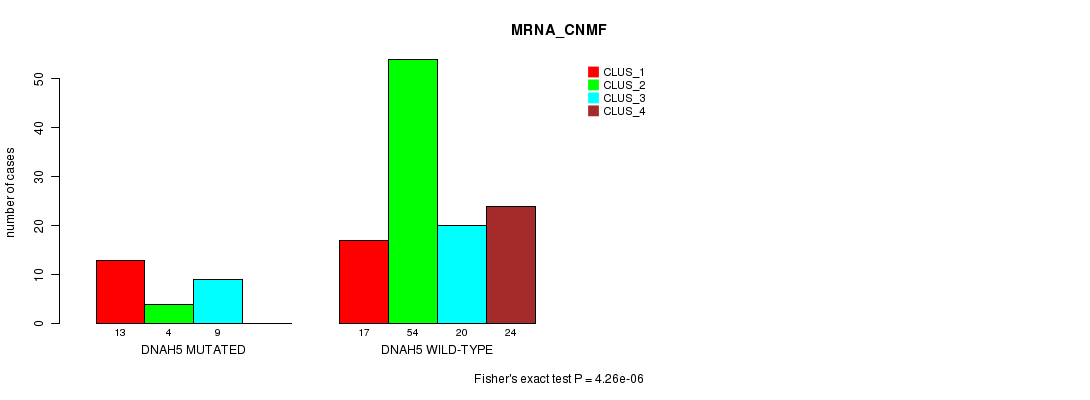
DNAH5 mutation correlated to 'MRNA_CNMF'.
-
CASP8 mutation correlated to 'MRNA_CHIERARCHICAL'.
-
ZC3H13 mutation correlated to 'MRNA_CHIERARCHICAL'.
-
SYNE1 mutation correlated to 'MRNA_CNMF' and 'CN_CNMF'.
Table 1. Get Full Table Overview of the association between mutation status of 120 genes and 7 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 12 significant findings detected.
|
Clinical Features |
MRNA CNMF |
MRNA CHIERARCHICAL |
CN CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| BRAF | 20 (13%) | 135 |
5.58e-06 (0.00427) |
8.43e-08 (6.47e-05) |
8.5e-05 (0.0648) |
0.844 (1.00) |
0.536 (1.00) |
0.145 (1.00) |
1 (1.00) |
| FLG | 26 (17%) | 129 |
0.000232 (0.176) |
0.00927 (1.00) |
0.00027 (0.205) |
0.0772 (1.00) |
0.528 (1.00) |
1 (1.00) |
0.61 (1.00) |
| SYNE1 | 37 (24%) | 118 |
0.000146 (0.111) |
0.000472 (0.356) |
0.000289 (0.218) |
0.49 (1.00) |
0.676 (1.00) |
1 (1.00) |
0.181 (1.00) |
| TP53 | 75 (48%) | 80 |
0.00371 (1.00) |
0.00883 (1.00) |
1.08e-09 (8.29e-07) |
0.896 (1.00) |
0.862 (1.00) |
0.366 (1.00) |
0.807 (1.00) |
| FAT4 | 29 (19%) | 126 |
0.0176 (1.00) |
0.0068 (1.00) |
0.000172 (0.131) |
0.271 (1.00) |
0.743 (1.00) |
0.241 (1.00) |
0.0218 (1.00) |
| DNAH5 | 28 (18%) | 127 |
4.26e-06 (0.00326) |
0.00131 (0.974) |
0.0134 (1.00) |
0.31 (1.00) |
0.72 (1.00) |
0.585 (1.00) |
0.642 (1.00) |
| CASP8 | 10 (6%) | 145 |
0.000518 (0.39) |
1.21e-05 (0.00921) |
0.00174 (1.00) |
0.726 (1.00) |
1 (1.00) |
1 (1.00) |
0.255 (1.00) |
| ZC3H13 | 17 (11%) | 138 |
0.00144 (1.00) |
0.000266 (0.202) |
0.00682 (1.00) |
0.692 (1.00) |
0.443 (1.00) |
1 (1.00) |
0.41 (1.00) |
| APC | 103 (66%) | 52 |
0.0438 (1.00) |
0.0286 (1.00) |
0.101 (1.00) |
0.164 (1.00) |
0.219 (1.00) |
1 (1.00) |
1 (1.00) |
| FBXW7 | 29 (19%) | 126 |
0.176 (1.00) |
0.109 (1.00) |
0.000825 (0.618) |
0.0419 (1.00) |
0.0719 (1.00) |
0.583 (1.00) |
0.657 (1.00) |
| KRAS | 58 (37%) | 97 |
0.00324 (1.00) |
0.000947 (0.708) |
0.15 (1.00) |
0.613 (1.00) |
0.465 (1.00) |
1 (1.00) |
0.613 (1.00) |
| NRAS | 15 (10%) | 140 |
0.131 (1.00) |
0.408 (1.00) |
0.531 (1.00) |
0.473 (1.00) |
0.536 (1.00) |
1 (1.00) |
1 (1.00) |
| PIK3CA | 26 (17%) | 129 |
0.252 (1.00) |
0.385 (1.00) |
0.0178 (1.00) |
0.0171 (1.00) |
0.0751 (1.00) |
1 (1.00) |
0.592 (1.00) |
| SMAD4 | 18 (12%) | 137 |
0.212 (1.00) |
0.581 (1.00) |
0.01 (1.00) |
0.793 (1.00) |
0.126 (1.00) |
1 (1.00) |
1 (1.00) |
| FAM123B | 19 (12%) | 136 |
0.168 (1.00) |
0.0336 (1.00) |
0.019 (1.00) |
0.363 (1.00) |
0.453 (1.00) |
0.498 (1.00) |
0.498 (1.00) |
| TTN | 69 (45%) | 86 |
0.00599 (1.00) |
0.00803 (1.00) |
0.0011 (0.818) |
0.29 (1.00) |
0.921 (1.00) |
0.655 (1.00) |
0.793 (1.00) |
| WBSCR17 | 17 (11%) | 138 |
0.48 (1.00) |
0.376 (1.00) |
0.388 (1.00) |
0.422 (1.00) |
0.357 (1.00) |
0.109 (1.00) |
0.109 (1.00) |
| ACVR1B | 13 (8%) | 142 |
0.27 (1.00) |
0.46 (1.00) |
0.00744 (1.00) |
0.309 (1.00) |
0.0729 (1.00) |
1 (1.00) |
0.386 (1.00) |
| TNFRSF10C | 6 (4%) | 149 |
0.426 (1.00) |
1 (1.00) |
0.0811 (1.00) |
0.133 (1.00) |
0.127 (1.00) |
1 (1.00) |
1 (1.00) |
| SMAD2 | 10 (6%) | 145 |
0.028 (1.00) |
0.233 (1.00) |
0.0333 (1.00) |
0.809 (1.00) |
0.565 (1.00) |
0.283 (1.00) |
1 (1.00) |
| MAP2K4 | 9 (6%) | 146 |
0.485 (1.00) |
0.133 (1.00) |
0.0512 (1.00) |
0.341 (1.00) |
0.602 (1.00) |
1 (1.00) |
0.255 (1.00) |
| PCBP1 | 4 (3%) | 151 |
0.385 (1.00) |
0.386 (1.00) |
0.486 (1.00) |
1 (1.00) |
1 (1.00) |
||
| LRP1B | 30 (19%) | 125 |
0.00437 (1.00) |
0.00102 (0.765) |
0.00112 (0.836) |
0.549 (1.00) |
0.867 (1.00) |
1 (1.00) |
0.00789 (1.00) |
| NRXN1 | 17 (11%) | 138 |
0.21 (1.00) |
0.208 (1.00) |
0.0134 (1.00) |
0.773 (1.00) |
1 (1.00) |
1 (1.00) |
0.455 (1.00) |
| GRIA1 | 14 (9%) | 141 |
0.228 (1.00) |
0.103 (1.00) |
0.329 (1.00) |
1 (1.00) |
0.000411 (0.31) |
||
| SDK1 | 25 (16%) | 130 |
0.0158 (1.00) |
0.00187 (1.00) |
0.31 (1.00) |
0.904 (1.00) |
0.638 (1.00) |
1 (1.00) |
0.575 (1.00) |
| SOX9 | 9 (6%) | 146 |
0.255 (1.00) |
0.436 (1.00) |
0.585 (1.00) |
0.857 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| DMD | 27 (17%) | 128 |
0.0222 (1.00) |
0.00627 (1.00) |
0.00855 (1.00) |
0.559 (1.00) |
0.222 (1.00) |
1 (1.00) |
0.0998 (1.00) |
| CNTN6 | 15 (10%) | 140 |
0.126 (1.00) |
0.162 (1.00) |
0.329 (1.00) |
0.726 (1.00) |
0.674 (1.00) |
1 (1.00) |
0.361 (1.00) |
| GABRA5 | 8 (5%) | 147 |
0.663 (1.00) |
1 (1.00) |
1 (1.00) |
0.197 (1.00) |
0.197 (1.00) |
||
| OR2M4 | 8 (5%) | 147 |
0.00569 (1.00) |
0.259 (1.00) |
0.0771 (1.00) |
0.726 (1.00) |
0.511 (1.00) |
1 (1.00) |
0.255 (1.00) |
| OR6N1 | 8 (5%) | 147 |
0.00206 (1.00) |
0.00822 (1.00) |
0.00971 (1.00) |
0.226 (1.00) |
1 (1.00) |
||
| PCDH9 | 16 (10%) | 139 |
0.28 (1.00) |
0.711 (1.00) |
0.354 (1.00) |
0.875 (1.00) |
0.757 (1.00) |
1 (1.00) |
0.41 (1.00) |
| TXNDC3 | 10 (6%) | 145 |
0.0949 (1.00) |
0.288 (1.00) |
0.0548 (1.00) |
0.453 (1.00) |
0.049 (1.00) |
1 (1.00) |
0.283 (1.00) |
| MGC26647 | 7 (5%) | 148 |
0.274 (1.00) |
0.796 (1.00) |
0.887 (1.00) |
0.367 (1.00) |
0.511 (1.00) |
1 (1.00) |
0.166 (1.00) |
| FAM22F | 8 (5%) | 147 |
0.918 (1.00) |
0.272 (1.00) |
0.906 (1.00) |
0.875 (1.00) |
0.602 (1.00) |
1 (1.00) |
1 (1.00) |
| ACVR2A | 8 (5%) | 147 |
0.0188 (1.00) |
0.337 (1.00) |
0.098 (1.00) |
0.166 (1.00) |
0.406 (1.00) |
1 (1.00) |
0.255 (1.00) |
| TCF7L2 | 11 (7%) | 144 |
0.116 (1.00) |
0.815 (1.00) |
0.0875 (1.00) |
0.624 (1.00) |
1 (1.00) |
1 (1.00) |
0.283 (1.00) |
| AFF2 | 15 (10%) | 140 |
0.0104 (1.00) |
0.0293 (1.00) |
0.648 (1.00) |
0.161 (1.00) |
0.111 (1.00) |
0.41 (1.00) |
0.41 (1.00) |
| CNBD1 | 6 (4%) | 149 |
0.472 (1.00) |
0.773 (1.00) |
0.312 (1.00) |
1 (1.00) |
0.197 (1.00) |
||
| OTOL1 | 7 (5%) | 148 |
0.385 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.226 (1.00) |
||
| KCNA4 | 10 (6%) | 145 |
0.0778 (1.00) |
0.0895 (1.00) |
0.123 (1.00) |
0.166 (1.00) |
0.139 (1.00) |
1 (1.00) |
1 (1.00) |
| ISL1 | 8 (5%) | 147 |
0.0987 (1.00) |
0.389 (1.00) |
0.756 (1.00) |
0.445 (1.00) |
1 (1.00) |
1 (1.00) |
0.255 (1.00) |
| MGC42105 | 10 (6%) | 145 |
0.00859 (1.00) |
0.103 (1.00) |
0.162 (1.00) |
0.231 (1.00) |
0.111 (1.00) |
1 (1.00) |
1 (1.00) |
| ACOT4 | 3 (2%) | 152 |
0.676 (1.00) |
0.396 (1.00) |
0.736 (1.00) |
1 (1.00) |
1 (1.00) |
||
| CDH11 | 13 (8%) | 142 |
0.132 (1.00) |
0.168 (1.00) |
0.258 (1.00) |
1 (1.00) |
1 (1.00) |
0.336 (1.00) |
1 (1.00) |
| ATM | 21 (14%) | 134 |
0.116 (1.00) |
0.435 (1.00) |
0.00208 (1.00) |
0.469 (1.00) |
1 (1.00) |
1 (1.00) |
0.0702 (1.00) |
| TPTE | 11 (7%) | 144 |
0.0132 (1.00) |
0.288 (1.00) |
0.129 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.31 (1.00) |
| BTNL8 | 3 (2%) | 152 |
0.343 (1.00) |
0.396 (1.00) |
0.317 (1.00) |
0.166 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| KLHL4 | 12 (8%) | 143 |
0.00859 (1.00) |
0.202 (1.00) |
0.00618 (1.00) |
0.0614 (1.00) |
1 (1.00) |
0.283 (1.00) |
0.283 (1.00) |
| ROBO1 | 14 (9%) | 141 |
0.119 (1.00) |
0.0543 (1.00) |
0.0866 (1.00) |
0.0787 (1.00) |
0.796 (1.00) |
1 (1.00) |
0.386 (1.00) |
| GGT1 | 3 (2%) | 152 |
0.317 (1.00) |
1 (1.00) |
1 (1.00) |
||||
| PCDHB2 | 13 (8%) | 142 |
0.0404 (1.00) |
0.0108 (1.00) |
0.0912 (1.00) |
0.773 (1.00) |
1 (1.00) |
1 (1.00) |
0.336 (1.00) |
| POSTN | 11 (7%) | 144 |
0.27 (1.00) |
0.501 (1.00) |
0.0759 (1.00) |
0.269 (1.00) |
0.406 (1.00) |
1 (1.00) |
1 (1.00) |
| OR51V1 | 9 (6%) | 146 |
0.00622 (1.00) |
0.00439 (1.00) |
0.00436 (1.00) |
0.692 (1.00) |
1 (1.00) |
1 (1.00) |
0.283 (1.00) |
| GRIK3 | 14 (9%) | 141 |
0.33 (1.00) |
0.0396 (1.00) |
0.476 (1.00) |
0.549 (1.00) |
0.536 (1.00) |
1 (1.00) |
1 (1.00) |
| LRRN3 | 9 (6%) | 146 |
0.0188 (1.00) |
0.0772 (1.00) |
0.288 (1.00) |
0.166 (1.00) |
0.139 (1.00) |
0.226 (1.00) |
1 (1.00) |
| PRDM9 | 11 (7%) | 144 |
0.00152 (1.00) |
0.501 (1.00) |
0.129 (1.00) |
0.127 (1.00) |
0.743 (1.00) |
1 (1.00) |
0.336 (1.00) |
| ADAM29 | 9 (6%) | 146 |
0.809 (1.00) |
0.354 (1.00) |
0.268 (1.00) |
1 (1.00) |
1 (1.00) |
||
| KIAA1486 | 9 (6%) | 146 |
0.817 (1.00) |
0.631 (1.00) |
0.469 (1.00) |
0.624 (1.00) |
1 (1.00) |
1 (1.00) |
0.255 (1.00) |
| PSG8 | 8 (5%) | 147 |
0.406 (1.00) |
0.511 (1.00) |
0.197 (1.00) |
0.226 (1.00) |
0.102 (1.00) |
||
| NCKAP5 | 13 (8%) | 142 |
0.166 (1.00) |
0.0505 (1.00) |
0.143 (1.00) |
0.0172 (1.00) |
1 (1.00) |
1 (1.00) |
0.361 (1.00) |
| C8B | 9 (6%) | 146 |
0.194 (1.00) |
0.174 (1.00) |
0.853 (1.00) |
0.809 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| CCDC160 | 5 (3%) | 150 |
0.521 (1.00) |
0.831 (1.00) |
0.506 (1.00) |
1 (1.00) |
0.166 (1.00) |
||
| DCAF4L2 | 9 (6%) | 146 |
0.0567 (1.00) |
0.00439 (1.00) |
0.232 (1.00) |
1 (1.00) |
1 (1.00) |
||
| DKK2 | 5 (3%) | 150 |
0.00642 (1.00) |
0.0245 (1.00) |
0.506 (1.00) |
0.445 (1.00) |
0.602 (1.00) |
1 (1.00) |
1 (1.00) |
| EPHA3 | 13 (8%) | 142 |
0.00394 (1.00) |
0.0543 (1.00) |
0.258 (1.00) |
0.143 (1.00) |
0.678 (1.00) |
1 (1.00) |
0.361 (1.00) |
| ERBB4 | 14 (9%) | 141 |
0.112 (1.00) |
0.165 (1.00) |
0.0538 (1.00) |
0.913 (1.00) |
0.743 (1.00) |
1 (1.00) |
0.361 (1.00) |
| GUCY1A3 | 10 (6%) | 145 |
0.171 (1.00) |
0.233 (1.00) |
0.227 (1.00) |
1 (1.00) |
0.283 (1.00) |
||
| IFT80 | 7 (5%) | 148 |
0.155 (1.00) |
0.027 (1.00) |
0.135 (1.00) |
0.197 (1.00) |
1 (1.00) |
||
| INHBA | 7 (5%) | 148 |
0.113 (1.00) |
0.25 (1.00) |
0.312 (1.00) |
0.166 (1.00) |
0.166 (1.00) |
||
| KLK2 | 3 (2%) | 152 |
1 (1.00) |
0.198 (1.00) |
0.736 (1.00) |
1 (1.00) |
|||
| LPPR4 | 11 (7%) | 144 |
0.132 (1.00) |
0.0111 (1.00) |
0.283 (1.00) |
0.0908 (1.00) |
0.757 (1.00) |
1 (1.00) |
1 (1.00) |
| OR7C1 | 6 (4%) | 149 |
0.0296 (1.00) |
0.0379 (1.00) |
0.744 (1.00) |
1 (1.00) |
0.197 (1.00) |
||
| SCN7A | 12 (8%) | 143 |
0.00115 (0.858) |
0.0116 (1.00) |
0.161 (1.00) |
0.308 (1.00) |
0.743 (1.00) |
1 (1.00) |
0.336 (1.00) |
| TGFBR2 | 7 (5%) | 148 |
0.0293 (1.00) |
0.23 (1.00) |
0.0609 (1.00) |
0.197 (1.00) |
0.197 (1.00) |
||
| TRPS1 | 13 (8%) | 142 |
0.0891 (1.00) |
0.233 (1.00) |
0.000547 (0.411) |
1 (1.00) |
0.336 (1.00) |
||
| OR8J1 | 4 (3%) | 151 |
0.0844 (1.00) |
0.319 (1.00) |
0.619 (1.00) |
1 (1.00) |
0.103 (1.00) |
||
| RIMS2 | 12 (8%) | 143 |
0.00804 (1.00) |
0.282 (1.00) |
0.0429 (1.00) |
0.624 (1.00) |
1 (1.00) |
1 (1.00) |
0.336 (1.00) |
| SFMBT2 | 12 (8%) | 143 |
0.45 (1.00) |
0.259 (1.00) |
0.21 (1.00) |
0.857 (1.00) |
1 (1.00) |
1 (1.00) |
0.361 (1.00) |
| IL18R1 | 8 (5%) | 147 |
0.286 (1.00) |
0.259 (1.00) |
0.294 (1.00) |
0.624 (1.00) |
1 (1.00) |
1 (1.00) |
0.255 (1.00) |
| PCDH17 | 14 (9%) | 141 |
0.347 (1.00) |
0.927 (1.00) |
0.673 (1.00) |
0.624 (1.00) |
0.406 (1.00) |
1 (1.00) |
0.361 (1.00) |
| FLRT2 | 11 (7%) | 144 |
0.68 (1.00) |
0.789 (1.00) |
0.367 (1.00) |
0.171 (1.00) |
1 (1.00) |
1 (1.00) |
0.283 (1.00) |
| PTEN | 4 (3%) | 151 |
0.017 (1.00) |
0.688 (1.00) |
0.132 (1.00) |
1 (1.00) |
0.135 (1.00) |
||
| ZFHX4 | 17 (11%) | 138 |
0.516 (1.00) |
0.376 (1.00) |
0.455 (1.00) |
0.875 (1.00) |
1 (1.00) |
1 (1.00) |
0.0452 (1.00) |
| NRXN3 | 15 (10%) | 140 |
0.00109 (0.816) |
0.0385 (1.00) |
0.0506 (1.00) |
1 (1.00) |
0.263 (1.00) |
1 (1.00) |
0.386 (1.00) |
| LIFR | 13 (8%) | 142 |
0.0325 (1.00) |
0.578 (1.00) |
0.355 (1.00) |
1 (1.00) |
0.565 (1.00) |
1 (1.00) |
0.0253 (1.00) |
| ZNF429 | 7 (5%) | 148 |
0.0908 (1.00) |
0.511 (1.00) |
0.887 (1.00) |
1 (1.00) |
0.197 (1.00) |
||
| OR2M2 | 6 (4%) | 149 |
0.406 (1.00) |
1 (1.00) |
0.574 (1.00) |
1 (1.00) |
0.166 (1.00) |
||
| TMEM132D | 15 (10%) | 140 |
0.000353 (0.267) |
0.000781 (0.587) |
0.0272 (1.00) |
0.692 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| SAT1 | 4 (3%) | 151 |
0.00392 (1.00) |
0.071 (1.00) |
0.619 (1.00) |
0.453 (1.00) |
0.0756 (1.00) |
1 (1.00) |
1 (1.00) |
| CDH18 | 10 (6%) | 145 |
0.0179 (1.00) |
0.165 (1.00) |
0.0471 (1.00) |
0.285 (1.00) |
0.678 (1.00) |
1 (1.00) |
0.283 (1.00) |
| P2RY10 | 6 (4%) | 149 |
0.328 (1.00) |
0.442 (1.00) |
0.744 (1.00) |
1 (1.00) |
1 (1.00) |
||
| SYT16 | 6 (4%) | 149 |
0.0293 (1.00) |
0.101 (1.00) |
0.0596 (1.00) |
0.453 (1.00) |
0.406 (1.00) |
0.197 (1.00) |
1 (1.00) |
| DKK4 | 4 (3%) | 151 |
0.071 (1.00) |
0.071 (1.00) |
0.188 (1.00) |
1 (1.00) |
1 (1.00) |
||
| NALCN | 14 (9%) | 141 |
0.012 (1.00) |
0.092 (1.00) |
0.0129 (1.00) |
0.755 (1.00) |
0.82 (1.00) |
1 (1.00) |
0.361 (1.00) |
| SULT1C4 | 3 (2%) | 152 |
0.0124 (1.00) |
0.027 (1.00) |
0.0684 (1.00) |
1 (1.00) |
|||
| UMOD | 7 (5%) | 148 |
0.117 (1.00) |
0.136 (1.00) |
0.166 (1.00) |
0.453 (1.00) |
0.565 (1.00) |
1 (1.00) |
0.135 (1.00) |
| CPXCR1 | 6 (4%) | 149 |
0.071 (1.00) |
0.151 (1.00) |
0.574 (1.00) |
0.197 (1.00) |
0.0141 (1.00) |
||
| PDZRN4 | 12 (8%) | 143 |
0.00509 (1.00) |
0.00295 (1.00) |
0.00593 (1.00) |
0.624 (1.00) |
1 (1.00) |
0.31 (1.00) |
0.31 (1.00) |
| USP17L2 | 7 (5%) | 148 |
0.329 (1.00) |
0.337 (1.00) |
0.744 (1.00) |
1 (1.00) |
0.602 (1.00) |
0.197 (1.00) |
0.197 (1.00) |
| ESR1 | 11 (7%) | 144 |
0.135 (1.00) |
0.0736 (1.00) |
0.105 (1.00) |
0.453 (1.00) |
0.565 (1.00) |
1 (1.00) |
0.283 (1.00) |
| FAM5C | 10 (6%) | 145 |
0.118 (1.00) |
0.389 (1.00) |
0.0471 (1.00) |
0.269 (1.00) |
0.565 (1.00) |
0.283 (1.00) |
0.283 (1.00) |
| MIER3 | 10 (6%) | 145 |
0.000875 (0.656) |
0.00803 (1.00) |
0.00303 (1.00) |
0.773 (1.00) |
1 (1.00) |
1 (1.00) |
0.31 (1.00) |
| NLRP5 | 10 (6%) | 145 |
0.0259 (1.00) |
0.732 (1.00) |
0.162 (1.00) |
1 (1.00) |
0.255 (1.00) |
||
| OR6T1 | 6 (4%) | 149 |
0.934 (1.00) |
0.59 (1.00) |
0.744 (1.00) |
0.166 (1.00) |
1 (1.00) |
||
| PCDH10 | 14 (9%) | 141 |
0.136 (1.00) |
0.162 (1.00) |
0.237 (1.00) |
0.467 (1.00) |
1 (1.00) |
1 (1.00) |
0.361 (1.00) |
| PCDHB5 | 12 (8%) | 143 |
0.087 (1.00) |
0.282 (1.00) |
0.161 (1.00) |
0.549 (1.00) |
0.168 (1.00) |
1 (1.00) |
0.31 (1.00) |
| TRPC6 | 8 (5%) | 147 |
0.105 (1.00) |
0.796 (1.00) |
0.35 (1.00) |
1 (1.00) |
0.197 (1.00) |
||
| RHOA | 4 (3%) | 151 |
0.619 (1.00) |
0.135 (1.00) |
1 (1.00) |
||||
| CYTL1 | 4 (3%) | 151 |
0.287 (1.00) |
0.776 (1.00) |
0.619 (1.00) |
1 (1.00) |
0.135 (1.00) |
||
| CHRM2 | 8 (5%) | 147 |
0.472 (1.00) |
0.25 (1.00) |
0.138 (1.00) |
0.857 (1.00) |
1 (1.00) |
0.226 (1.00) |
0.226 (1.00) |
| GALNT14 | 9 (6%) | 146 |
0.177 (1.00) |
0.541 (1.00) |
0.0807 (1.00) |
0.0332 (1.00) |
0.0642 (1.00) |
1 (1.00) |
0.226 (1.00) |
| LRRIQ3 | 7 (5%) | 148 |
0.635 (1.00) |
0.433 (1.00) |
0.47 (1.00) |
0.166 (1.00) |
0.565 (1.00) |
1 (1.00) |
0.226 (1.00) |
| NAALAD2 | 11 (7%) | 144 |
0.0262 (1.00) |
0.0381 (1.00) |
0.0127 (1.00) |
0.624 (1.00) |
0.406 (1.00) |
1 (1.00) |
0.283 (1.00) |
| PCDH11X | 11 (7%) | 144 |
0.102 (1.00) |
0.501 (1.00) |
0.129 (1.00) |
1 (1.00) |
0.31 (1.00) |
||
| TFAP2D | 5 (3%) | 150 |
0.582 (1.00) |
0.864 (1.00) |
0.506 (1.00) |
0.726 (1.00) |
0.511 (1.00) |
1 (1.00) |
1 (1.00) |
| TNR | 15 (10%) | 140 |
0.0139 (1.00) |
0.0253 (1.00) |
0.00125 (0.931) |
0.341 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| CNTNAP4 | 10 (6%) | 145 |
0.0102 (1.00) |
0.688 (1.00) |
0.0226 (1.00) |
0.346 (1.00) |
1 (1.00) |
0.283 (1.00) |
0.0325 (1.00) |
| DOCK2 | 16 (10%) | 139 |
0.00245 (1.00) |
0.00388 (1.00) |
0.00754 (1.00) |
0.334 (1.00) |
0.536 (1.00) |
1 (1.00) |
0.433 (1.00) |
P value = 5.58e-06 (Fisher's exact test), Q value = 0.0043
Table S1. Gene #2: 'BRAF MUTATION STATUS' versus Clinical Feature #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 30 | 58 | 29 | 24 |
| BRAF MUTATED | 12 | 1 | 4 | 1 |
| BRAF WILD-TYPE | 18 | 57 | 25 | 23 |
Figure S1. Get High-res Image Gene #2: 'BRAF MUTATION STATUS' versus Clinical Feature #1: 'MRNA_CNMF'

P value = 8.43e-08 (Fisher's exact test), Q value = 6.5e-05
Table S2. Gene #2: 'BRAF MUTATION STATUS' versus Clinical Feature #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 43 | 45 |
| BRAF MUTATED | 1 | 16 | 1 |
| BRAF WILD-TYPE | 52 | 27 | 44 |
Figure S2. Get High-res Image Gene #2: 'BRAF MUTATION STATUS' versus Clinical Feature #2: 'MRNA_CHIERARCHICAL'

P value = 8.5e-05 (Fisher's exact test), Q value = 0.065
Table S3. Gene #2: 'BRAF MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 64 | 11 | 8 |
| BRAF MUTATED | 2 | 18 | 0 | 0 |
| BRAF WILD-TYPE | 68 | 46 | 11 | 8 |
Figure S3. Get High-res Image Gene #2: 'BRAF MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'

P value = 1.08e-09 (Fisher's exact test), Q value = 8.3e-07
Table S4. Gene #7: 'TP53 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 64 | 11 | 8 |
| TP53 MUTATED | 51 | 13 | 4 | 6 |
| TP53 WILD-TYPE | 19 | 51 | 7 | 2 |
Figure S4. Get High-res Image Gene #7: 'TP53 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
P value = 0.000172 (Fisher's exact test), Q value = 0.13
Table S5. Gene #17: 'FAT4 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 64 | 11 | 8 |
| FAT4 MUTATED | 4 | 22 | 1 | 1 |
| FAT4 WILD-TYPE | 66 | 42 | 10 | 7 |
Figure S5. Get High-res Image Gene #17: 'FAT4 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
P value = 0.000232 (Fisher's exact test), Q value = 0.18
Table S6. Gene #48: 'FLG MUTATION STATUS' versus Clinical Feature #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 30 | 58 | 29 | 24 |
| FLG MUTATED | 10 | 2 | 8 | 2 |
| FLG WILD-TYPE | 20 | 56 | 21 | 22 |
Figure S6. Get High-res Image Gene #48: 'FLG MUTATION STATUS' versus Clinical Feature #1: 'MRNA_CNMF'

P value = 0.00027 (Fisher's exact test), Q value = 0.2
Table S7. Gene #48: 'FLG MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 64 | 11 | 8 |
| FLG MUTATED | 3 | 18 | 1 | 3 |
| FLG WILD-TYPE | 67 | 46 | 10 | 5 |
Figure S7. Get High-res Image Gene #48: 'FLG MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
P value = 4.26e-06 (Fisher's exact test), Q value = 0.0033
Table S8. Gene #60: 'DNAH5 MUTATION STATUS' versus Clinical Feature #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 30 | 58 | 29 | 24 |
| DNAH5 MUTATED | 13 | 4 | 9 | 0 |
| DNAH5 WILD-TYPE | 17 | 54 | 20 | 24 |
Figure S8. Get High-res Image Gene #60: 'DNAH5 MUTATION STATUS' versus Clinical Feature #1: 'MRNA_CNMF'
P value = 1.21e-05 (Fisher's exact test), Q value = 0.0092
Table S9. Gene #61: 'CASP8 MUTATION STATUS' versus Clinical Feature #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 43 | 45 |
| CASP8 MUTATED | 0 | 9 | 0 |
| CASP8 WILD-TYPE | 53 | 34 | 45 |
Figure S9. Get High-res Image Gene #61: 'CASP8 MUTATION STATUS' versus Clinical Feature #2: 'MRNA_CHIERARCHICAL'
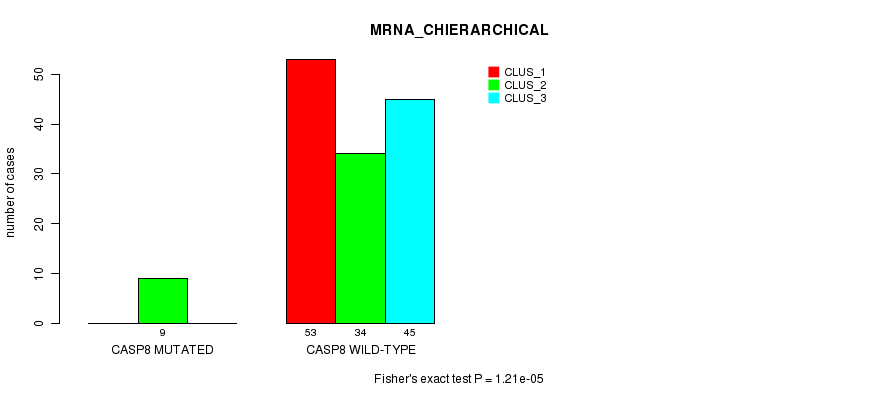
P value = 0.000266 (Fisher's exact test), Q value = 0.2
Table S10. Gene #82: 'ZC3H13 MUTATION STATUS' versus Clinical Feature #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 43 | 45 |
| ZC3H13 MUTATED | 3 | 12 | 1 |
| ZC3H13 WILD-TYPE | 50 | 31 | 44 |
Figure S10. Get High-res Image Gene #82: 'ZC3H13 MUTATION STATUS' versus Clinical Feature #2: 'MRNA_CHIERARCHICAL'

P value = 0.000146 (Fisher's exact test), Q value = 0.11
Table S11. Gene #110: 'SYNE1 MUTATION STATUS' versus Clinical Feature #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 30 | 58 | 29 | 24 |
| SYNE1 MUTATED | 13 | 7 | 14 | 3 |
| SYNE1 WILD-TYPE | 17 | 51 | 15 | 21 |
Figure S11. Get High-res Image Gene #110: 'SYNE1 MUTATION STATUS' versus Clinical Feature #1: 'MRNA_CNMF'

P value = 0.000289 (Fisher's exact test), Q value = 0.22
Table S12. Gene #110: 'SYNE1 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 70 | 64 | 11 | 8 |
| SYNE1 MUTATED | 9 | 26 | 0 | 1 |
| SYNE1 WILD-TYPE | 61 | 38 | 11 | 7 |
Figure S12. Get High-res Image Gene #110: 'SYNE1 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'

-
Mutation data file = COAD-TP.mutsig.cluster.txt
-
Molecular subtypes file = COAD-TP.transferedmergedcluster.txt
-
Number of patients = 155
-
Number of significantly mutated genes = 120
-
Number of Molecular subtypes = 7
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.