(primary solid tumor cohort)
This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and selected clinical features.
Testing the association between copy number variation 79 arm-level results and 5 clinical features across 447 patients, 53 significant findings detected with Q value < 0.25.
-
3q gain cnv correlated to 'HISTOLOGICAL.TYPE'.
-
5p gain cnv correlated to 'Time to Death' and 'HISTOLOGICAL.TYPE'.
-
13q gain cnv correlated to 'HISTOLOGICAL.TYPE'.
-
17p gain cnv correlated to 'Time to Death'.
-
18p gain cnv correlated to 'AGE' and 'HISTOLOGICAL.TYPE'.
-
19p gain cnv correlated to 'AGE' and 'HISTOLOGICAL.TYPE'.
-
19q gain cnv correlated to 'AGE' and 'HISTOLOGICAL.TYPE'.
-
20p gain cnv correlated to 'HISTOLOGICAL.TYPE'.
-
20q gain cnv correlated to 'HISTOLOGICAL.TYPE'.
-
21q gain cnv correlated to 'HISTOLOGICAL.TYPE'.
-
3p loss cnv correlated to 'HISTOLOGICAL.TYPE'.
-
4p loss cnv correlated to 'HISTOLOGICAL.TYPE'.
-
4q loss cnv correlated to 'HISTOLOGICAL.TYPE'.
-
5p loss cnv correlated to 'HISTOLOGICAL.TYPE'.
-
5q loss cnv correlated to 'HISTOLOGICAL.TYPE'.
-
7p loss cnv correlated to 'HISTOLOGICAL.TYPE'.
-
7q loss cnv correlated to 'HISTOLOGICAL.TYPE'.
-
8p loss cnv correlated to 'AGE' and 'HISTOLOGICAL.TYPE'.
-
8q loss cnv correlated to 'HISTOLOGICAL.TYPE'.
-
9p loss cnv correlated to 'AGE' and 'HISTOLOGICAL.TYPE'.
-
9q loss cnv correlated to 'AGE' and 'HISTOLOGICAL.TYPE'.
-
11p loss cnv correlated to 'AGE' and 'HISTOLOGICAL.TYPE'.
-
11q loss cnv correlated to 'Time to Death', 'AGE', and 'HISTOLOGICAL.TYPE'.
-
12p loss cnv correlated to 'HISTOLOGICAL.TYPE'.
-
13q loss cnv correlated to 'HISTOLOGICAL.TYPE'.
-
14q loss cnv correlated to 'AGE' and 'HISTOLOGICAL.TYPE'.
-
15q loss cnv correlated to 'AGE' and 'HISTOLOGICAL.TYPE'.
-
16p loss cnv correlated to 'AGE' and 'HISTOLOGICAL.TYPE'.
-
16q loss cnv correlated to 'AGE' and 'HISTOLOGICAL.TYPE'.
-
17p loss cnv correlated to 'AGE' and 'HISTOLOGICAL.TYPE'.
-
17q loss cnv correlated to 'AGE' and 'HISTOLOGICAL.TYPE'.
-
18q loss cnv correlated to 'HISTOLOGICAL.TYPE'.
-
19p loss cnv correlated to 'HISTOLOGICAL.TYPE'.
-
19q loss cnv correlated to 'HISTOLOGICAL.TYPE'.
-
21q loss cnv correlated to 'HISTOLOGICAL.TYPE'.
-
22q loss cnv correlated to 'AGE' and 'HISTOLOGICAL.TYPE'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 79 arm-level results and 5 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 53 significant findings detected.
|
Clinical Features |
Time to Death |
AGE |
HISTOLOGICAL TYPE |
RADIATIONS RADIATION REGIMENINDICATION |
COMPLETENESS OF RESECTION |
||
| nCNV (%) | nWild-Type | logrank test | t-test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 11q loss | 39 (9%) | 408 |
0.000215 (0.075) |
0.00012 (0.0424) |
8.75e-07 (0.000327) |
0.205 (1.00) |
0.0705 (1.00) |
| 5p gain | 29 (6%) | 418 |
2.85e-05 (0.0103) |
0.102 (1.00) |
7.3e-07 (0.000274) |
0.675 (1.00) |
0.0603 (1.00) |
| 18p gain | 32 (7%) | 415 |
0.0966 (1.00) |
0.000394 (0.136) |
2.78e-08 (1.06e-05) |
0.689 (1.00) |
0.024 (1.00) |
| 19p gain | 25 (6%) | 422 |
0.0523 (1.00) |
4.84e-05 (0.0174) |
0.000217 (0.0757) |
0.175 (1.00) |
0.0904 (1.00) |
| 19q gain | 22 (5%) | 425 |
0.00798 (1.00) |
1.14e-05 (0.00413) |
4.97e-07 (0.000187) |
0.814 (1.00) |
0.169 (1.00) |
| 8p loss | 41 (9%) | 406 |
0.513 (1.00) |
8.76e-05 (0.0312) |
8.24e-17 (3.24e-14) |
0.591 (1.00) |
0.49 (1.00) |
| 9p loss | 65 (15%) | 382 |
0.85 (1.00) |
2.7e-06 (0.000996) |
1.1e-14 (4.32e-12) |
1 (1.00) |
0.291 (1.00) |
| 9q loss | 82 (18%) | 365 |
0.186 (1.00) |
4.23e-10 (1.63e-07) |
4.82e-13 (1.88e-10) |
0.423 (1.00) |
0.669 (1.00) |
| 11p loss | 47 (11%) | 400 |
0.000924 (0.314) |
0.000108 (0.0383) |
2.01e-07 (7.57e-05) |
0.399 (1.00) |
0.529 (1.00) |
| 14q loss | 37 (8%) | 410 |
0.284 (1.00) |
0.000234 (0.0813) |
4.84e-09 (1.85e-06) |
1 (1.00) |
0.0805 (1.00) |
| 15q loss | 65 (15%) | 382 |
0.757 (1.00) |
3.28e-05 (0.0118) |
2.38e-11 (9.22e-09) |
0.241 (1.00) |
0.833 (1.00) |
| 16p loss | 56 (13%) | 391 |
0.0279 (1.00) |
6.56e-05 (0.0235) |
7.49e-09 (2.86e-06) |
0.643 (1.00) |
0.0317 (1.00) |
| 16q loss | 93 (21%) | 354 |
0.0373 (1.00) |
2.36e-06 (0.000874) |
2.22e-14 (8.67e-12) |
0.254 (1.00) |
0.0513 (1.00) |
| 17p loss | 86 (19%) | 361 |
0.163 (1.00) |
1.53e-06 (0.000571) |
1.9e-20 (7.5e-18) |
0.431 (1.00) |
0.167 (1.00) |
| 17q loss | 57 (13%) | 390 |
0.977 (1.00) |
0.000598 (0.205) |
2.12e-06 (0.000786) |
0.162 (1.00) |
0.288 (1.00) |
| 22q loss | 73 (16%) | 374 |
0.00769 (1.00) |
0.000208 (0.0729) |
2.79e-09 (1.07e-06) |
0.33 (1.00) |
0.847 (1.00) |
| 3q gain | 43 (10%) | 404 |
0.0467 (1.00) |
0.163 (1.00) |
0.000664 (0.227) |
0.602 (1.00) |
0.0542 (1.00) |
| 13q gain | 19 (4%) | 428 |
0.447 (1.00) |
0.000813 (0.277) |
9.07e-06 (0.00333) |
0.456 (1.00) |
0.353 (1.00) |
| 17p gain | 6 (1%) | 441 |
0.00055 (0.189) |
0.773 (1.00) |
0.458 (1.00) |
0.369 (1.00) |
0.0754 (1.00) |
| 20p gain | 52 (12%) | 395 |
0.0121 (1.00) |
0.00541 (1.00) |
1.84e-06 (0.000683) |
0.0154 (1.00) |
0.0689 (1.00) |
| 20q gain | 61 (14%) | 386 |
0.0683 (1.00) |
0.0043 (1.00) |
1.14e-10 (4.43e-08) |
0.0154 (1.00) |
0.0251 (1.00) |
| 21q gain | 20 (4%) | 427 |
0.838 (1.00) |
0.00859 (1.00) |
0.000426 (0.147) |
1 (1.00) |
0.934 (1.00) |
| 3p loss | 26 (6%) | 421 |
0.118 (1.00) |
0.0183 (1.00) |
8.24e-05 (0.0294) |
0.828 (1.00) |
0.82 (1.00) |
| 4p loss | 49 (11%) | 398 |
0.054 (1.00) |
0.00183 (0.609) |
1.18e-11 (4.6e-09) |
0.32 (1.00) |
0.0467 (1.00) |
| 4q loss | 46 (10%) | 401 |
0.0356 (1.00) |
0.00949 (1.00) |
1.39e-08 (5.3e-06) |
0.0609 (1.00) |
0.602 (1.00) |
| 5p loss | 21 (5%) | 426 |
0.424 (1.00) |
0.89 (1.00) |
1.77e-05 (0.00642) |
0.807 (1.00) |
0.152 (1.00) |
| 5q loss | 35 (8%) | 412 |
0.94 (1.00) |
0.0199 (1.00) |
1.49e-09 (5.72e-07) |
0.338 (1.00) |
0.187 (1.00) |
| 7p loss | 25 (6%) | 422 |
0.36 (1.00) |
0.0105 (1.00) |
0.000117 (0.0416) |
1 (1.00) |
0.268 (1.00) |
| 7q loss | 23 (5%) | 424 |
0.839 (1.00) |
0.000941 (0.319) |
3.73e-08 (1.41e-05) |
1 (1.00) |
0.286 (1.00) |
| 8q loss | 7 (2%) | 440 |
0.694 (1.00) |
0.609 (1.00) |
7.65e-06 (0.00281) |
0.68 (1.00) |
0.229 (1.00) |
| 12p loss | 19 (4%) | 428 |
0.178 (1.00) |
0.00777 (1.00) |
9.07e-06 (0.00333) |
0.608 (1.00) |
0.929 (1.00) |
| 13q loss | 52 (12%) | 395 |
0.114 (1.00) |
0.00144 (0.482) |
0.000175 (0.0614) |
0.519 (1.00) |
0.553 (1.00) |
| 18q loss | 41 (9%) | 406 |
0.514 (1.00) |
0.0492 (1.00) |
1.04e-05 (0.0038) |
0.287 (1.00) |
0.323 (1.00) |
| 19p loss | 29 (6%) | 418 |
0.34 (1.00) |
0.861 (1.00) |
1.97e-07 (7.44e-05) |
1 (1.00) |
0.112 (1.00) |
| 19q loss | 22 (5%) | 425 |
0.563 (1.00) |
0.673 (1.00) |
0.000203 (0.0714) |
0.814 (1.00) |
0.103 (1.00) |
| 21q loss | 26 (6%) | 421 |
0.248 (1.00) |
0.449 (1.00) |
1.22e-05 (0.00442) |
0.376 (1.00) |
0.281 (1.00) |
| 1p gain | 17 (4%) | 430 |
0.0625 (1.00) |
0.0554 (1.00) |
0.0529 (1.00) |
0.417 (1.00) |
0.849 (1.00) |
| 1q gain | 129 (29%) | 318 |
0.575 (1.00) |
0.0491 (1.00) |
0.00106 (0.356) |
0.569 (1.00) |
0.0897 (1.00) |
| 2p gain | 47 (11%) | 400 |
0.0961 (1.00) |
0.0406 (1.00) |
0.0822 (1.00) |
0.614 (1.00) |
0.64 (1.00) |
| 2q gain | 37 (8%) | 410 |
0.114 (1.00) |
0.208 (1.00) |
0.14 (1.00) |
0.574 (1.00) |
0.26 (1.00) |
| 3p gain | 24 (5%) | 423 |
0.487 (1.00) |
0.413 (1.00) |
0.557 (1.00) |
0.0666 (1.00) |
0.377 (1.00) |
| 4p gain | 5 (1%) | 442 |
0.594 (1.00) |
0.0246 (1.00) |
0.00913 (1.00) |
0.637 (1.00) |
1 (1.00) |
| 5q gain | 8 (2%) | 439 |
0.322 (1.00) |
0.929 (1.00) |
0.0109 (1.00) |
1 (1.00) |
0.0374 (1.00) |
| 6p gain | 33 (7%) | 414 |
0.682 (1.00) |
0.404 (1.00) |
0.00566 (1.00) |
1 (1.00) |
0.742 (1.00) |
| 6q gain | 28 (6%) | 419 |
0.971 (1.00) |
0.411 (1.00) |
0.0158 (1.00) |
0.523 (1.00) |
0.612 (1.00) |
| 7p gain | 40 (9%) | 407 |
0.322 (1.00) |
0.714 (1.00) |
0.103 (1.00) |
0.102 (1.00) |
0.757 (1.00) |
| 7q gain | 38 (9%) | 409 |
0.0185 (1.00) |
0.606 (1.00) |
0.883 (1.00) |
0.138 (1.00) |
0.48 (1.00) |
| 8p gain | 79 (18%) | 368 |
0.00123 (0.414) |
0.383 (1.00) |
0.723 (1.00) |
0.588 (1.00) |
0.2 (1.00) |
| 8q gain | 103 (23%) | 344 |
0.00652 (1.00) |
0.871 (1.00) |
0.0111 (1.00) |
0.714 (1.00) |
0.112 (1.00) |
| 9p gain | 11 (2%) | 436 |
0.793 (1.00) |
0.784 (1.00) |
0.21 (1.00) |
0.0388 (1.00) |
0.11 (1.00) |
| 9q gain | 3 (1%) | 444 |
0.797 (1.00) |
0.783 (1.00) |
0.0473 (1.00) |
0.558 (1.00) |
|
| 10p gain | 82 (18%) | 365 |
0.701 (1.00) |
0.513 (1.00) |
0.427 (1.00) |
0.894 (1.00) |
0.989 (1.00) |
| 10q gain | 77 (17%) | 370 |
0.848 (1.00) |
0.681 (1.00) |
0.41 (1.00) |
1 (1.00) |
0.413 (1.00) |
| 11p gain | 5 (1%) | 442 |
0.446 (1.00) |
0.648 (1.00) |
0.0782 (1.00) |
0.637 (1.00) |
0.43 (1.00) |
| 11q gain | 6 (1%) | 441 |
0.556 (1.00) |
0.705 (1.00) |
0.458 (1.00) |
1 (1.00) |
0.528 (1.00) |
| 12p gain | 33 (7%) | 414 |
0.586 (1.00) |
0.68 (1.00) |
0.00667 (1.00) |
0.845 (1.00) |
0.513 (1.00) |
| 12q gain | 26 (6%) | 421 |
0.719 (1.00) |
0.485 (1.00) |
0.335 (1.00) |
1 (1.00) |
0.344 (1.00) |
| 14q gain | 16 (4%) | 431 |
0.198 (1.00) |
0.634 (1.00) |
0.0428 (1.00) |
0.0476 (1.00) |
0.916 (1.00) |
| 15q gain | 5 (1%) | 442 |
0.0101 (1.00) |
0.508 (1.00) |
1 (1.00) |
0.328 (1.00) |
0.0365 (1.00) |
| 16p gain | 11 (2%) | 436 |
0.317 (1.00) |
0.126 (1.00) |
0.0168 (1.00) |
0.519 (1.00) |
0.553 (1.00) |
| 16q gain | 4 (1%) | 443 |
0.67 (1.00) |
0.482 (1.00) |
0.626 (1.00) |
0.323 (1.00) |
0.0288 (1.00) |
| 17q gain | 10 (2%) | 437 |
0.0784 (1.00) |
0.163 (1.00) |
0.605 (1.00) |
0.493 (1.00) |
0.0477 (1.00) |
| 18q gain | 16 (4%) | 431 |
0.00527 (1.00) |
0.0679 (1.00) |
0.0033 (1.00) |
0.413 (1.00) |
0.353 (1.00) |
| 22q gain | 6 (1%) | 441 |
0.521 (1.00) |
0.0184 (1.00) |
0.0305 (1.00) |
1 (1.00) |
0.299 (1.00) |
| Xq gain | 7 (2%) | 440 |
0.0893 (1.00) |
0.343 (1.00) |
0.0641 (1.00) |
1 (1.00) |
0.434 (1.00) |
| 1p loss | 11 (2%) | 436 |
0.266 (1.00) |
0.26 (1.00) |
0.013 (1.00) |
0.316 (1.00) |
0.443 (1.00) |
| 1q loss | 4 (1%) | 443 |
0.0129 (1.00) |
0.182 (1.00) |
0.0224 (1.00) |
0.323 (1.00) |
0.101 (1.00) |
| 2p loss | 6 (1%) | 441 |
0.53 (1.00) |
0.071 (1.00) |
0.686 (1.00) |
0.186 (1.00) |
0.0522 (1.00) |
| 2q loss | 7 (2%) | 440 |
0.348 (1.00) |
0.0304 (1.00) |
0.52 (1.00) |
0.109 (1.00) |
0.0943 (1.00) |
| 3q loss | 12 (3%) | 435 |
0.228 (1.00) |
0.147 (1.00) |
0.0254 (1.00) |
0.523 (1.00) |
0.519 (1.00) |
| 6p loss | 7 (2%) | 440 |
0.208 (1.00) |
0.169 (1.00) |
0.0641 (1.00) |
0.43 (1.00) |
1 (1.00) |
| 6q loss | 9 (2%) | 438 |
0.671 (1.00) |
0.182 (1.00) |
0.0197 (1.00) |
0.461 (1.00) |
1 (1.00) |
| 10p loss | 17 (4%) | 430 |
0.0173 (1.00) |
0.0658 (1.00) |
0.000964 (0.326) |
1 (1.00) |
0.279 (1.00) |
| 10q loss | 16 (4%) | 431 |
0.364 (1.00) |
0.0313 (1.00) |
0.0606 (1.00) |
0.787 (1.00) |
0.897 (1.00) |
| 12q loss | 12 (3%) | 435 |
0.454 (1.00) |
0.0255 (1.00) |
0.00159 (0.531) |
1 (1.00) |
1 (1.00) |
| 18p loss | 28 (6%) | 419 |
0.227 (1.00) |
0.138 (1.00) |
0.00159 (0.531) |
0.673 (1.00) |
0.127 (1.00) |
| 20p loss | 13 (3%) | 434 |
0.647 (1.00) |
0.0805 (1.00) |
0.0143 (1.00) |
0.22 (1.00) |
0.519 (1.00) |
| 20q loss | 6 (1%) | 441 |
0.479 (1.00) |
0.238 (1.00) |
0.0138 (1.00) |
0.369 (1.00) |
1 (1.00) |
| Xq loss | 13 (3%) | 434 |
0.628 (1.00) |
0.164 (1.00) |
0.0143 (1.00) |
0.12 (1.00) |
0.127 (1.00) |
P value = 0.000664 (Fisher's exact test), Q value = 0.23
Table S1. Gene #6: '3q gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 3Q GAIN MUTATED | 24 | 2 | 17 |
| 3Q GAIN WILD-TYPE | 326 | 16 | 62 |
Figure S1. Get High-res Image Gene #6: '3q gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
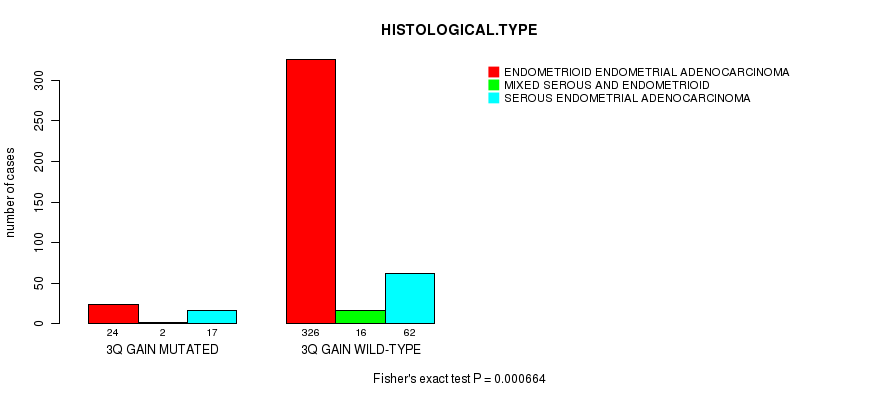
P value = 2.85e-05 (logrank test), Q value = 0.01
Table S2. Gene #8: '5p gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 442 | 41 | 0.0 - 187.1 (15.9) |
| 5P GAIN MUTATED | 28 | 8 | 0.1 - 59.0 (11.1) |
| 5P GAIN WILD-TYPE | 414 | 33 | 0.0 - 187.1 (16.2) |
Figure S2. Get High-res Image Gene #8: '5p gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
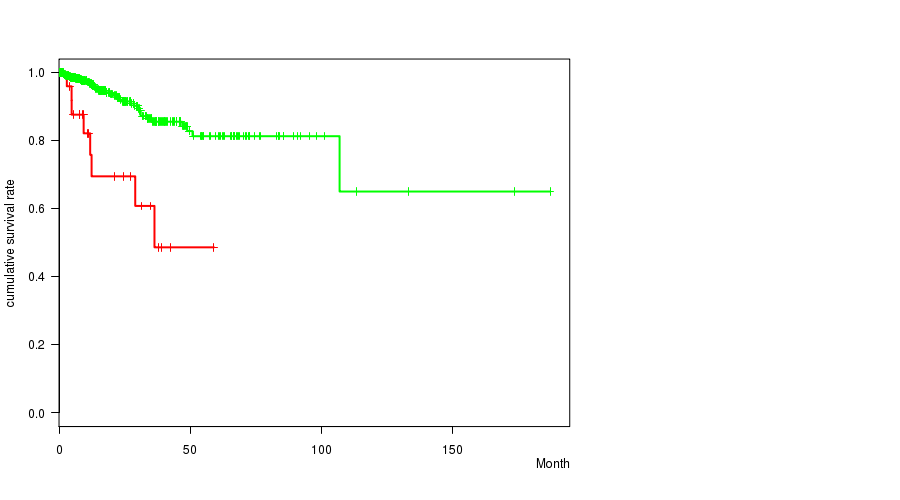
P value = 7.3e-07 (Fisher's exact test), Q value = 0.00027
Table S3. Gene #8: '5p gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 5P GAIN MUTATED | 11 | 1 | 17 |
| 5P GAIN WILD-TYPE | 339 | 17 | 62 |
Figure S3. Get High-res Image Gene #8: '5p gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
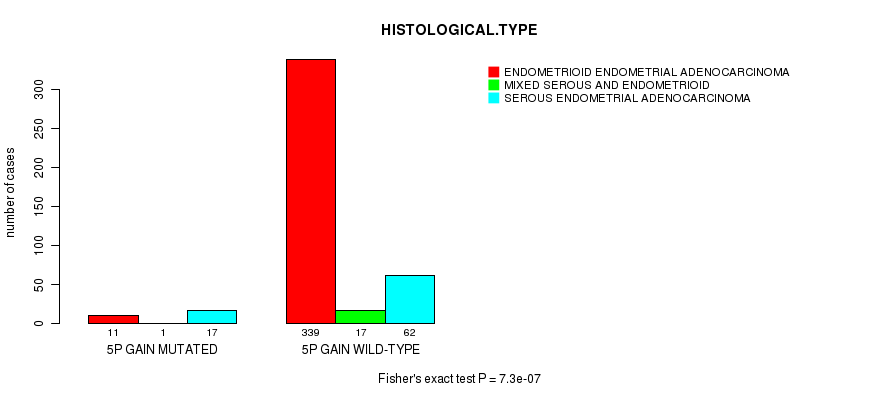
P value = 9.07e-06 (Fisher's exact test), Q value = 0.0033
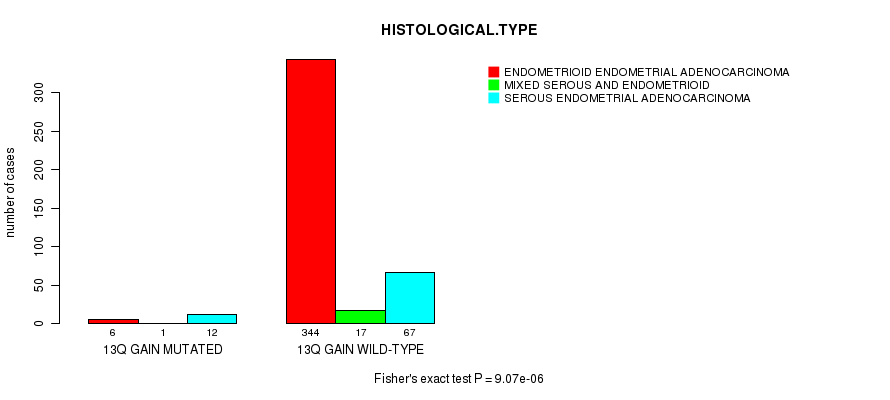
Table S4. Gene #24: '13q gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 13Q GAIN MUTATED | 6 | 1 | 12 |
| 13Q GAIN WILD-TYPE | 344 | 17 | 67 |
Figure S4. Get High-res Image Gene #24: '13q gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
P value = 0.00055 (logrank test), Q value = 0.19
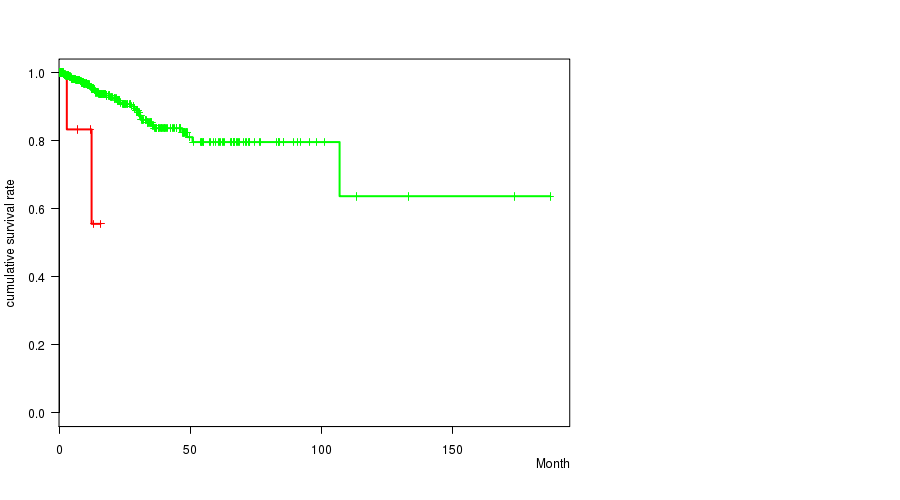
Table S5. Gene #29: '17p gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 442 | 41 | 0.0 - 187.1 (15.9) |
| 17P GAIN MUTATED | 6 | 2 | 3.0 - 15.8 (12.1) |
| 17P GAIN WILD-TYPE | 436 | 39 | 0.0 - 187.1 (16.2) |
Figure S5. Get High-res Image Gene #29: '17p gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
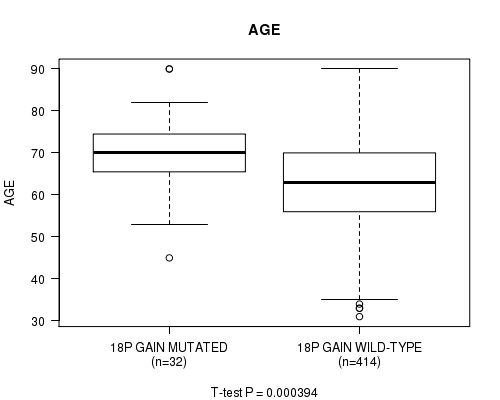
P value = 0.000394 (t-test), Q value = 0.14
Table S6. Gene #31: '18p gain mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 18P GAIN MUTATED | 32 | 69.8 (9.2) |
| 18P GAIN WILD-TYPE | 414 | 63.1 (11.3) |
Figure S6. Get High-res Image Gene #31: '18p gain mutation analysis' versus Clinical Feature #2: 'AGE'
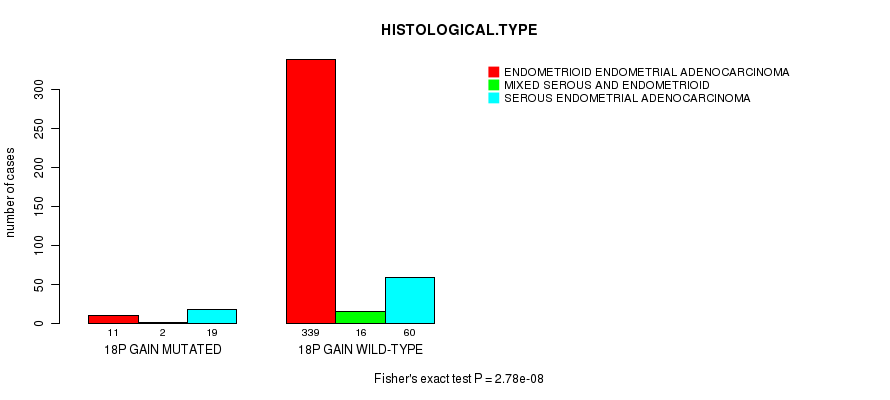
P value = 2.78e-08 (Fisher's exact test), Q value = 1.1e-05
Table S7. Gene #31: '18p gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 18P GAIN MUTATED | 11 | 2 | 19 |
| 18P GAIN WILD-TYPE | 339 | 16 | 60 |
Figure S7. Get High-res Image Gene #31: '18p gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
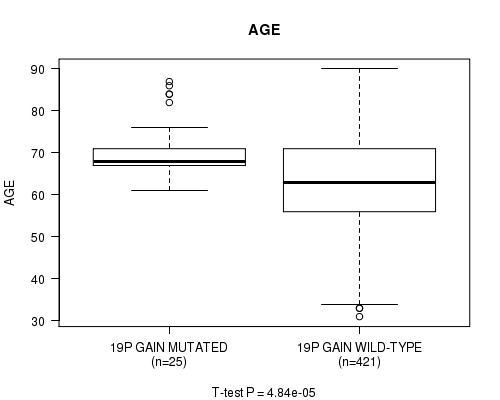
P value = 4.84e-05 (t-test), Q value = 0.017
Table S8. Gene #33: '19p gain mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 19P GAIN MUTATED | 25 | 70.9 (7.7) |
| 19P GAIN WILD-TYPE | 421 | 63.1 (11.3) |
Figure S8. Get High-res Image Gene #33: '19p gain mutation analysis' versus Clinical Feature #2: 'AGE'
P value = 0.000217 (Fisher's exact test), Q value = 0.076
Table S9. Gene #33: '19p gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 19P GAIN MUTATED | 12 | 0 | 13 |
| 19P GAIN WILD-TYPE | 338 | 18 | 66 |
Figure S9. Get High-res Image Gene #33: '19p gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
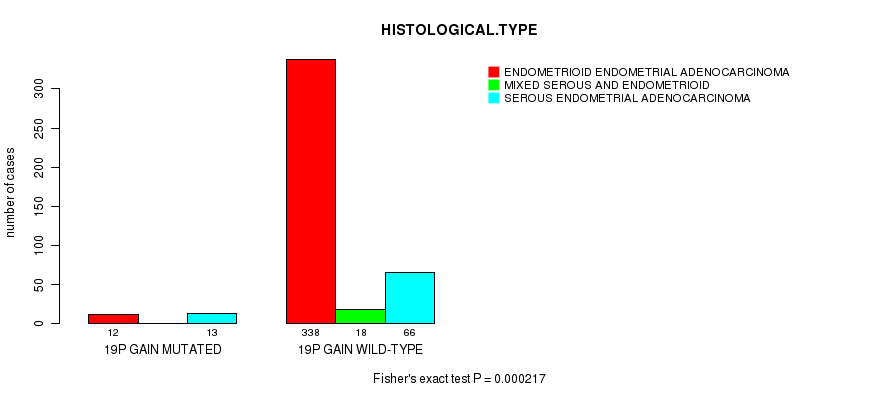
P value = 1.14e-05 (t-test), Q value = 0.0041
Table S10. Gene #34: '19q gain mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 19Q GAIN MUTATED | 22 | 72.4 (7.6) |
| 19Q GAIN WILD-TYPE | 424 | 63.1 (11.2) |
Figure S10. Get High-res Image Gene #34: '19q gain mutation analysis' versus Clinical Feature #2: 'AGE'
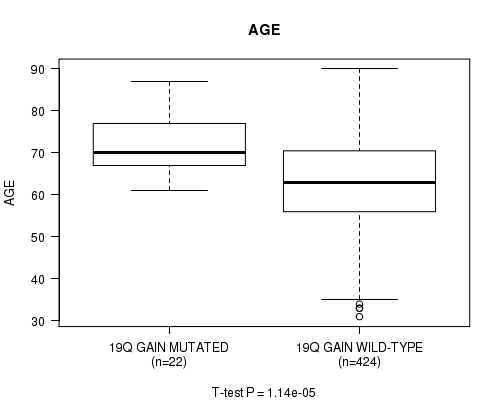
P value = 4.97e-07 (Fisher's exact test), Q value = 0.00019
Table S11. Gene #34: '19q gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 19Q GAIN MUTATED | 7 | 0 | 15 |
| 19Q GAIN WILD-TYPE | 343 | 18 | 64 |
Figure S11. Get High-res Image Gene #34: '19q gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
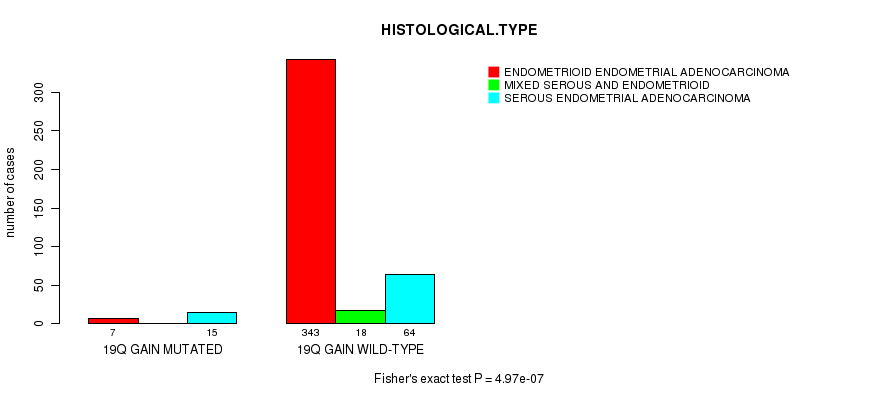
P value = 1.84e-06 (Fisher's exact test), Q value = 0.00068
Table S12. Gene #35: '20p gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 20P GAIN MUTATED | 26 | 3 | 23 |
| 20P GAIN WILD-TYPE | 324 | 15 | 56 |
Figure S12. Get High-res Image Gene #35: '20p gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
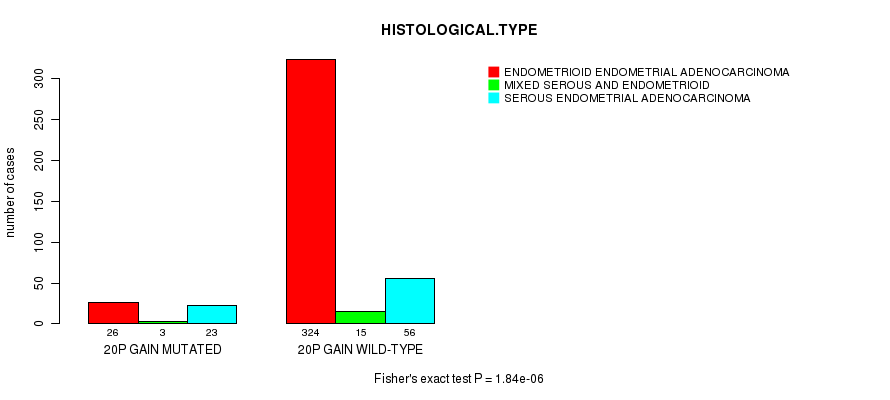
P value = 1.14e-10 (Fisher's exact test), Q value = 4.4e-08
Table S13. Gene #36: '20q gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 20Q GAIN MUTATED | 27 | 3 | 31 |
| 20Q GAIN WILD-TYPE | 323 | 15 | 48 |
Figure S13. Get High-res Image Gene #36: '20q gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
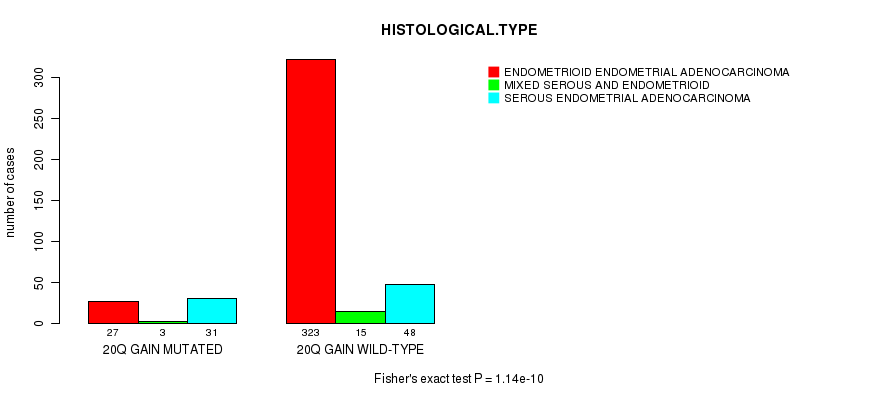
P value = 0.000426 (Fisher's exact test), Q value = 0.15
Table S14. Gene #37: '21q gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 21Q GAIN MUTATED | 9 | 0 | 11 |
| 21Q GAIN WILD-TYPE | 341 | 18 | 68 |
Figure S14. Get High-res Image Gene #37: '21q gain mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
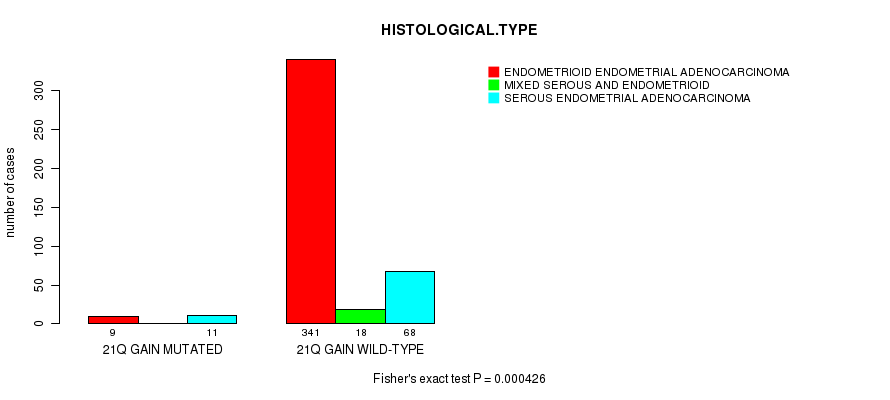
P value = 8.24e-05 (Fisher's exact test), Q value = 0.029
Table S15. Gene #44: '3p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 3P LOSS MUTATED | 11 | 3 | 12 |
| 3P LOSS WILD-TYPE | 339 | 15 | 67 |
Figure S15. Get High-res Image Gene #44: '3p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
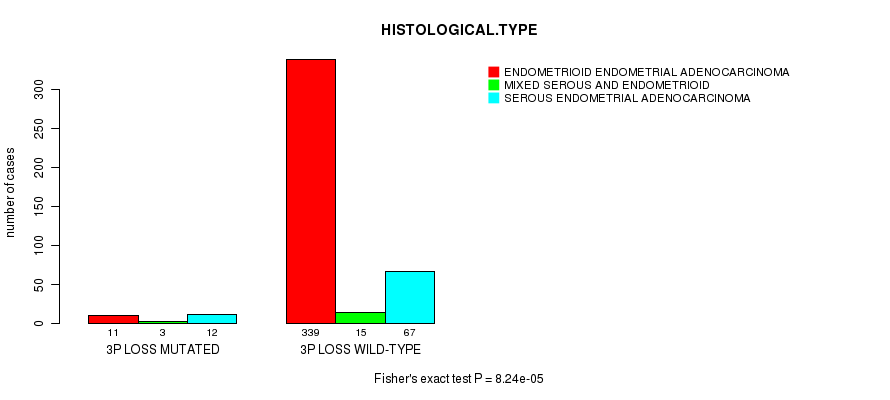
P value = 1.18e-11 (Fisher's exact test), Q value = 4.6e-09
Table S16. Gene #46: '4p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 4P LOSS MUTATED | 18 | 3 | 28 |
| 4P LOSS WILD-TYPE | 332 | 15 | 51 |
Figure S16. Get High-res Image Gene #46: '4p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
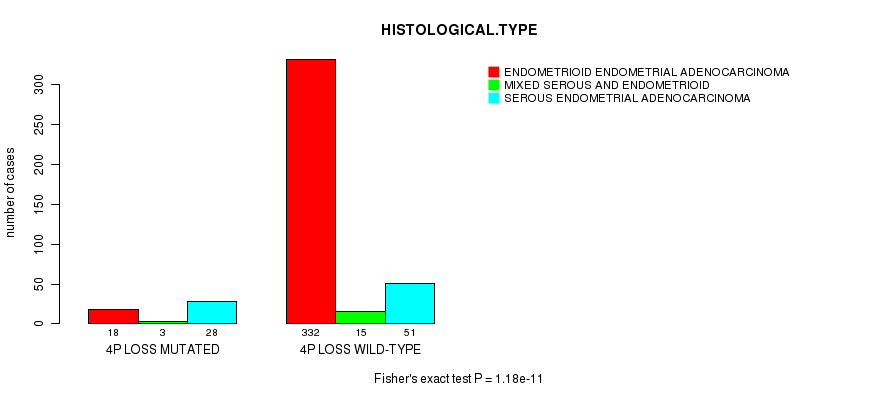
P value = 1.39e-08 (Fisher's exact test), Q value = 5.3e-06
Table S17. Gene #47: '4q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 4Q LOSS MUTATED | 19 | 4 | 23 |
| 4Q LOSS WILD-TYPE | 331 | 14 | 56 |
Figure S17. Get High-res Image Gene #47: '4q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
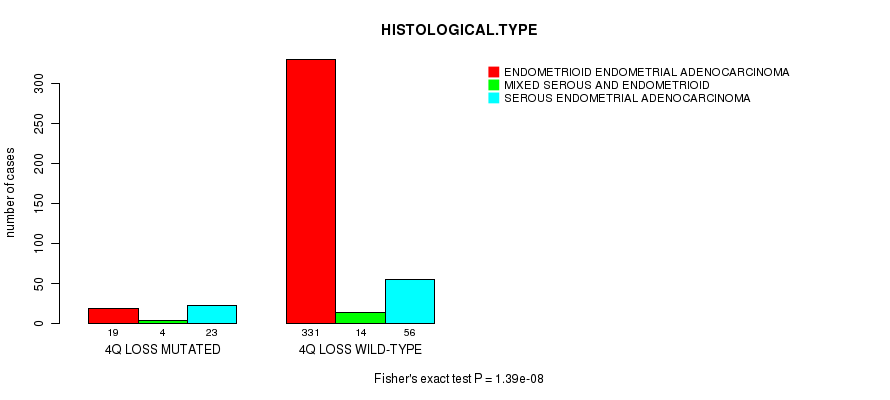
P value = 1.77e-05 (Fisher's exact test), Q value = 0.0064
Table S18. Gene #48: '5p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 5P LOSS MUTATED | 8 | 0 | 13 |
| 5P LOSS WILD-TYPE | 342 | 18 | 66 |
Figure S18. Get High-res Image Gene #48: '5p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
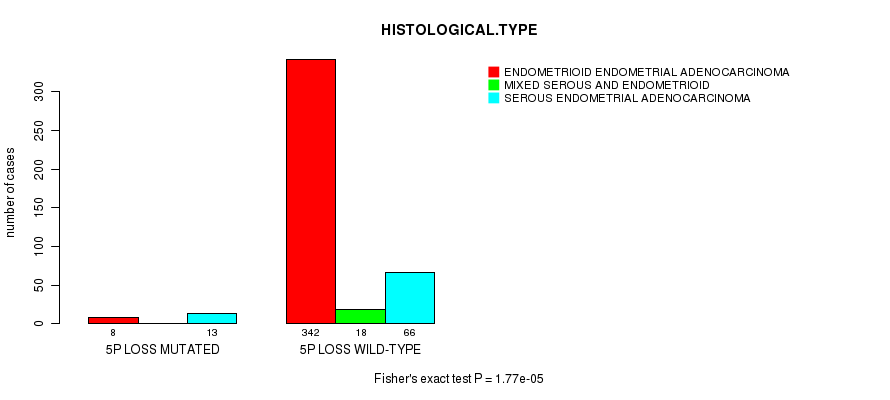
P value = 1.49e-09 (Fisher's exact test), Q value = 5.7e-07
Table S19. Gene #49: '5q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 5Q LOSS MUTATED | 12 | 1 | 22 |
| 5Q LOSS WILD-TYPE | 338 | 17 | 57 |
Figure S19. Get High-res Image Gene #49: '5q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
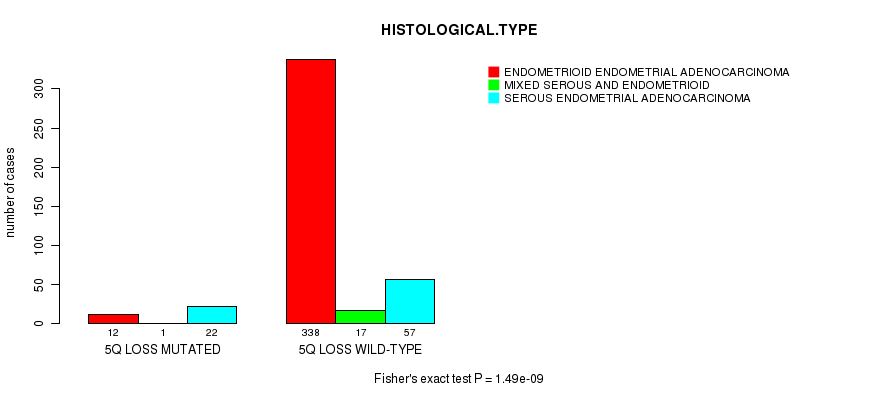
P value = 0.000117 (Fisher's exact test), Q value = 0.042
Table S20. Gene #52: '7p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 7P LOSS MUTATED | 11 | 1 | 13 |
| 7P LOSS WILD-TYPE | 339 | 17 | 66 |
Figure S20. Get High-res Image Gene #52: '7p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
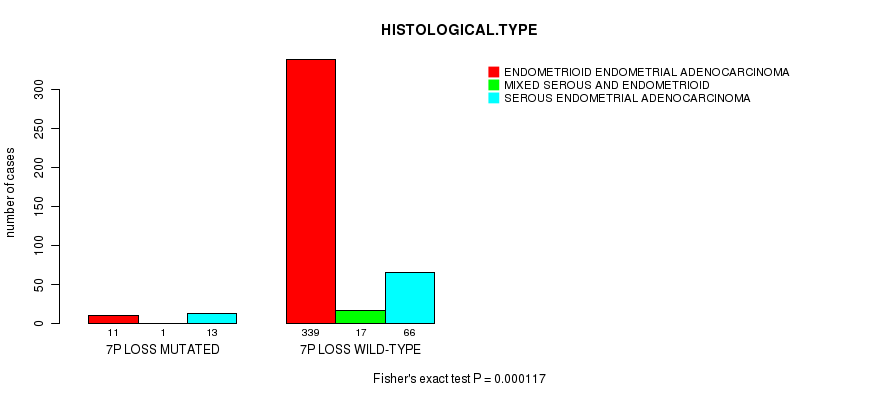
P value = 3.73e-08 (Fisher's exact test), Q value = 1.4e-05
Table S21. Gene #53: '7q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 7Q LOSS MUTATED | 6 | 1 | 16 |
| 7Q LOSS WILD-TYPE | 344 | 17 | 63 |
Figure S21. Get High-res Image Gene #53: '7q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'

P value = 8.76e-05 (t-test), Q value = 0.031
Table S22. Gene #54: '8p loss mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 8P LOSS MUTATED | 41 | 68.3 (7.0) |
| 8P LOSS WILD-TYPE | 405 | 63.1 (11.5) |
Figure S22. Get High-res Image Gene #54: '8p loss mutation analysis' versus Clinical Feature #2: 'AGE'
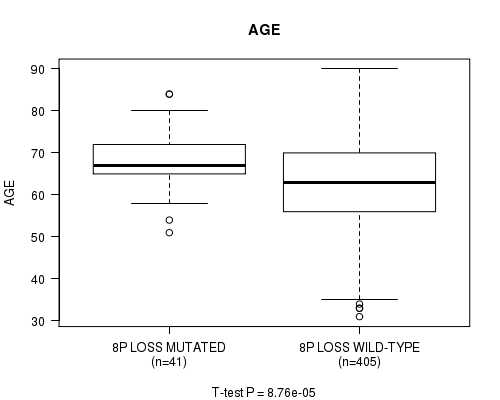
P value = 8.24e-17 (Fisher's exact test), Q value = 3.2e-14
Table S23. Gene #54: '8p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 8P LOSS MUTATED | 9 | 2 | 30 |
| 8P LOSS WILD-TYPE | 341 | 16 | 49 |
Figure S23. Get High-res Image Gene #54: '8p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
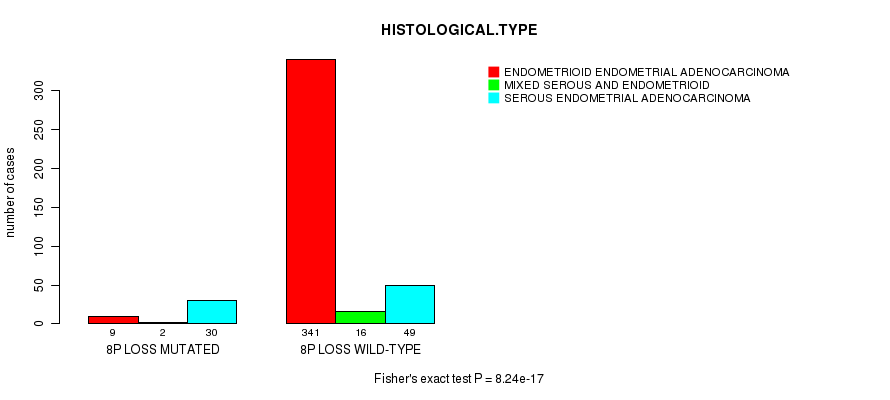
P value = 7.65e-06 (Fisher's exact test), Q value = 0.0028
Table S24. Gene #55: '8q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 8Q LOSS MUTATED | 0 | 0 | 7 |
| 8Q LOSS WILD-TYPE | 350 | 18 | 72 |
Figure S24. Get High-res Image Gene #55: '8q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
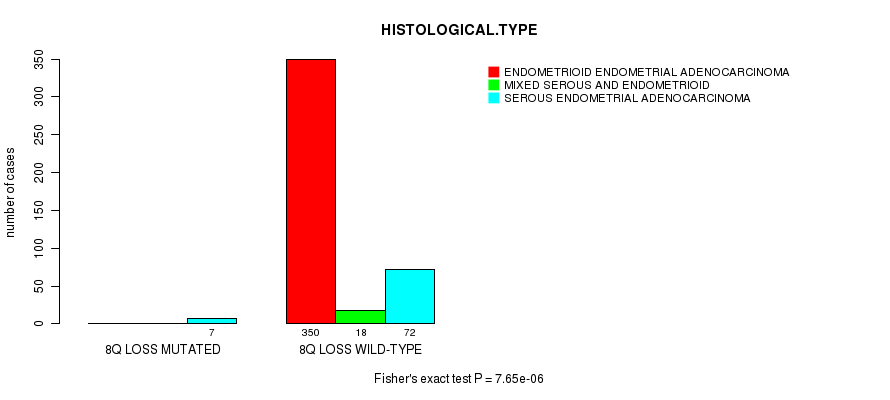
P value = 2.7e-06 (t-test), Q value = 0.001
Table S25. Gene #56: '9p loss mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 9P LOSS MUTATED | 65 | 68.9 (8.9) |
| 9P LOSS WILD-TYPE | 381 | 62.7 (11.4) |
Figure S25. Get High-res Image Gene #56: '9p loss mutation analysis' versus Clinical Feature #2: 'AGE'
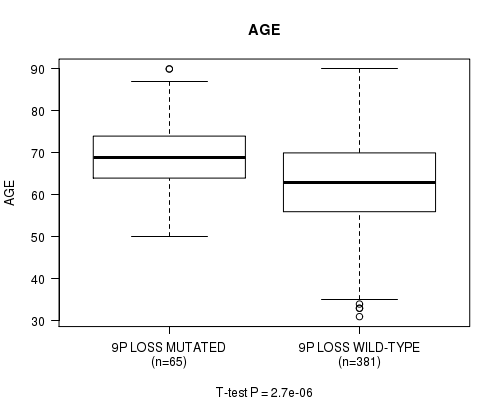
P value = 1.1e-14 (Fisher's exact test), Q value = 4.3e-12
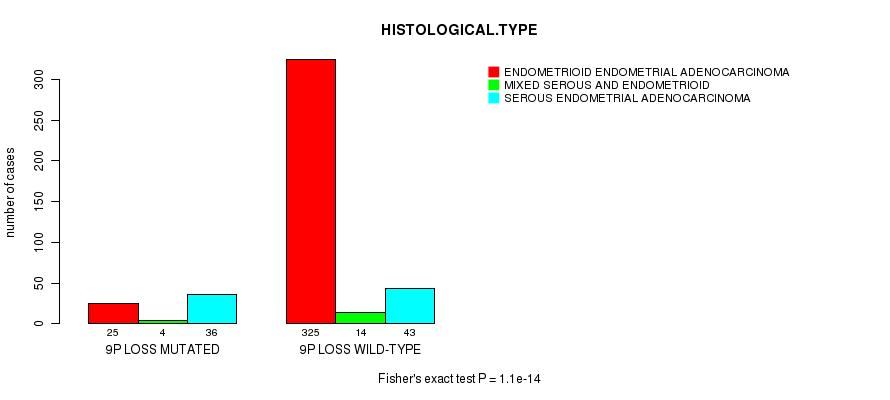
Table S26. Gene #56: '9p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 9P LOSS MUTATED | 25 | 4 | 36 |
| 9P LOSS WILD-TYPE | 325 | 14 | 43 |
Figure S26. Get High-res Image Gene #56: '9p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
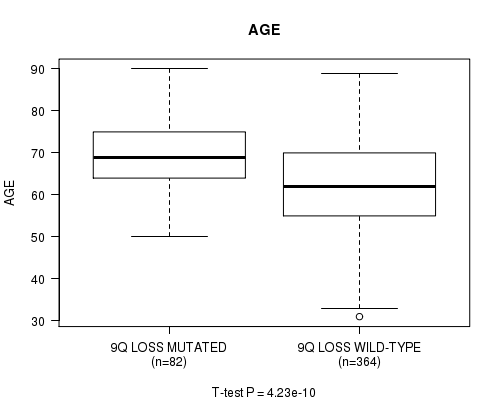
P value = 4.23e-10 (t-test), Q value = 1.6e-07
Table S27. Gene #57: '9q loss mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 9Q LOSS MUTATED | 82 | 69.7 (8.7) |
| 9Q LOSS WILD-TYPE | 364 | 62.2 (11.3) |
Figure S27. Get High-res Image Gene #57: '9q loss mutation analysis' versus Clinical Feature #2: 'AGE'
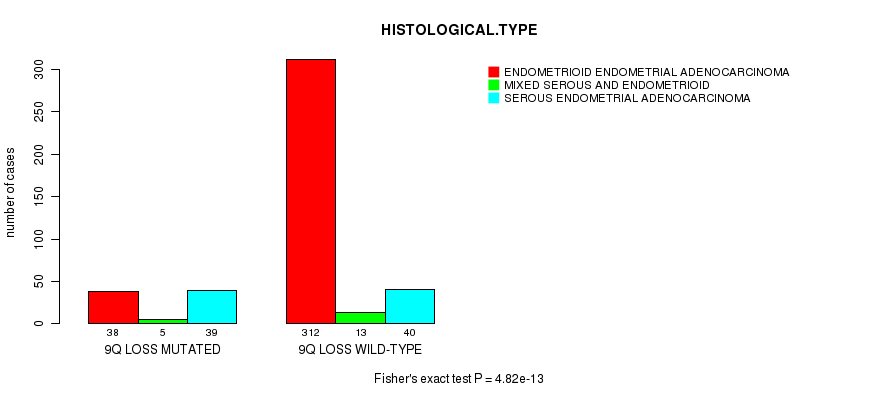
P value = 4.82e-13 (Fisher's exact test), Q value = 1.9e-10
Table S28. Gene #57: '9q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 9Q LOSS MUTATED | 38 | 5 | 39 |
| 9Q LOSS WILD-TYPE | 312 | 13 | 40 |
Figure S28. Get High-res Image Gene #57: '9q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
P value = 0.000108 (t-test), Q value = 0.038
Table S29. Gene #60: '11p loss mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 11P LOSS MUTATED | 47 | 69.2 (9.7) |
| 11P LOSS WILD-TYPE | 399 | 62.9 (11.3) |
Figure S29. Get High-res Image Gene #60: '11p loss mutation analysis' versus Clinical Feature #2: 'AGE'
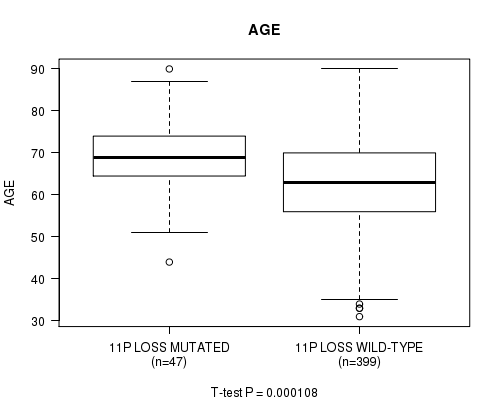
P value = 2.01e-07 (Fisher's exact test), Q value = 7.6e-05
Table S30. Gene #60: '11p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 11P LOSS MUTATED | 21 | 4 | 22 |
| 11P LOSS WILD-TYPE | 329 | 14 | 57 |
Figure S30. Get High-res Image Gene #60: '11p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
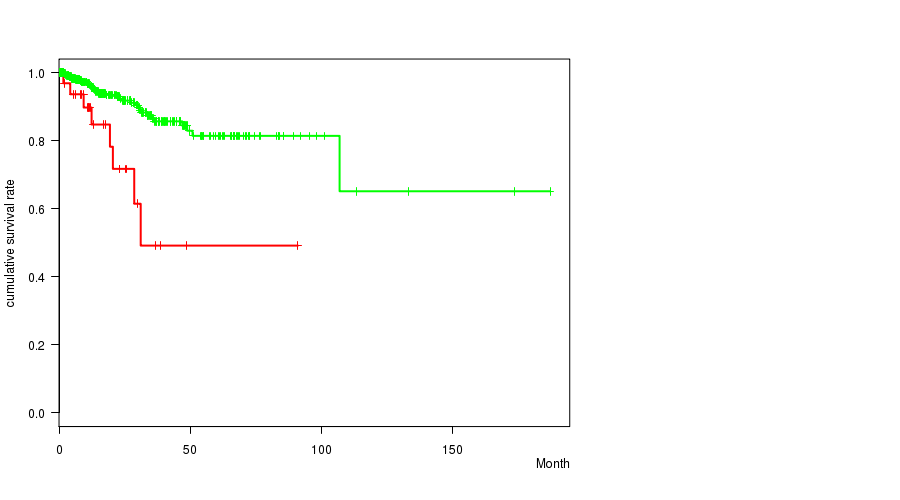
P value = 0.000215 (logrank test), Q value = 0.075
Table S31. Gene #61: '11q loss mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 442 | 41 | 0.0 - 187.1 (15.9) |
| 11Q LOSS MUTATED | 37 | 8 | 0.0 - 91.0 (11.8) |
| 11Q LOSS WILD-TYPE | 405 | 33 | 0.0 - 187.1 (16.3) |
Figure S31. Get High-res Image Gene #61: '11q loss mutation analysis' versus Clinical Feature #1: 'Time to Death'
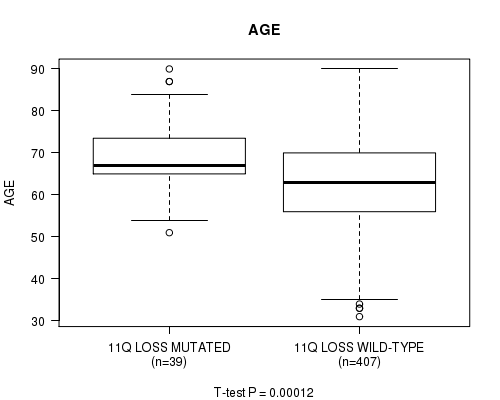
P value = 0.00012 (t-test), Q value = 0.042
Table S32. Gene #61: '11q loss mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 11Q LOSS MUTATED | 39 | 69.7 (9.3) |
| 11Q LOSS WILD-TYPE | 407 | 63.0 (11.3) |
Figure S32. Get High-res Image Gene #61: '11q loss mutation analysis' versus Clinical Feature #2: 'AGE'
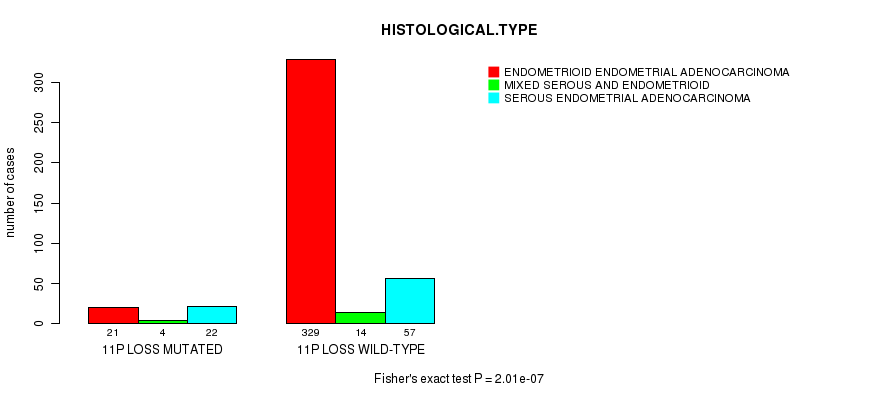
P value = 8.75e-07 (Fisher's exact test), Q value = 0.00033
Table S33. Gene #61: '11q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 11Q LOSS MUTATED | 17 | 5 | 17 |
| 11Q LOSS WILD-TYPE | 333 | 13 | 62 |
Figure S33. Get High-res Image Gene #61: '11q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
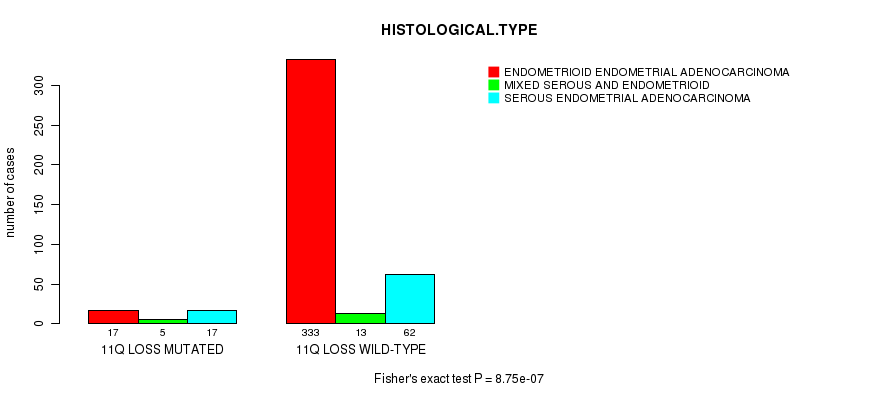
P value = 9.07e-06 (Fisher's exact test), Q value = 0.0033
Table S34. Gene #62: '12p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 12P LOSS MUTATED | 6 | 1 | 12 |
| 12P LOSS WILD-TYPE | 344 | 17 | 67 |
Figure S34. Get High-res Image Gene #62: '12p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
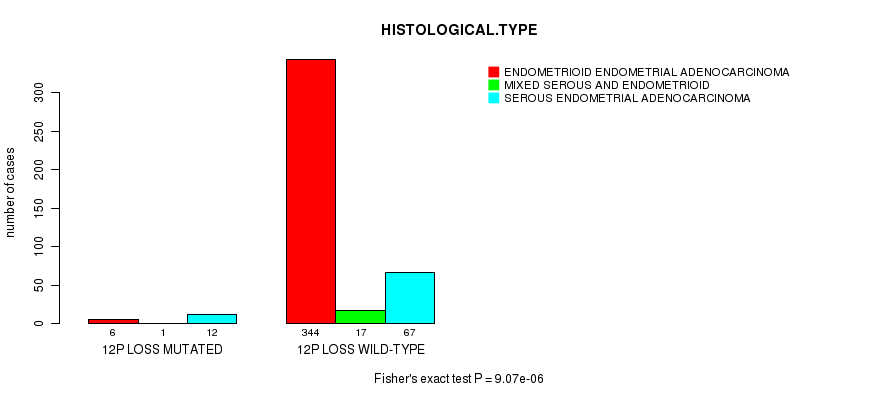
P value = 0.000175 (Fisher's exact test), Q value = 0.061
Table S35. Gene #64: '13q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 13Q LOSS MUTATED | 29 | 3 | 20 |
| 13Q LOSS WILD-TYPE | 321 | 15 | 59 |
Figure S35. Get High-res Image Gene #64: '13q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
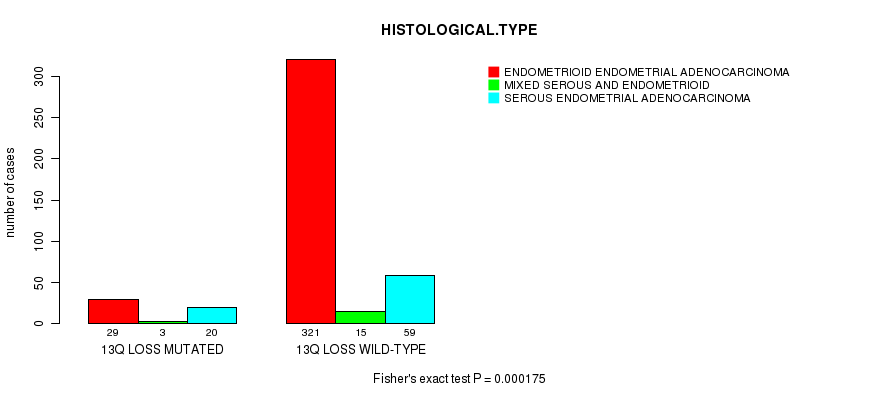
P value = 0.000234 (t-test), Q value = 0.081
Table S36. Gene #65: '14q loss mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 14Q LOSS MUTATED | 37 | 69.2 (8.7) |
| 14Q LOSS WILD-TYPE | 409 | 63.1 (11.3) |
Figure S36. Get High-res Image Gene #65: '14q loss mutation analysis' versus Clinical Feature #2: 'AGE'
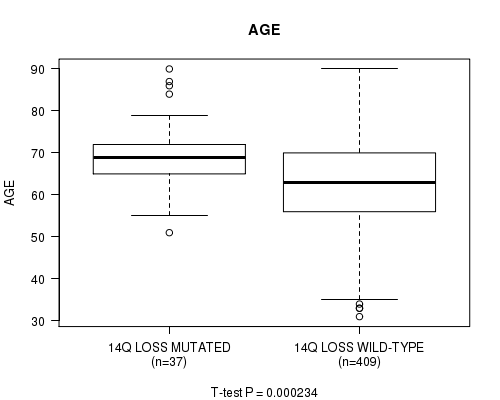
P value = 4.84e-09 (Fisher's exact test), Q value = 1.9e-06
Table S37. Gene #65: '14q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 14Q LOSS MUTATED | 13 | 3 | 21 |
| 14Q LOSS WILD-TYPE | 337 | 15 | 58 |
Figure S37. Get High-res Image Gene #65: '14q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
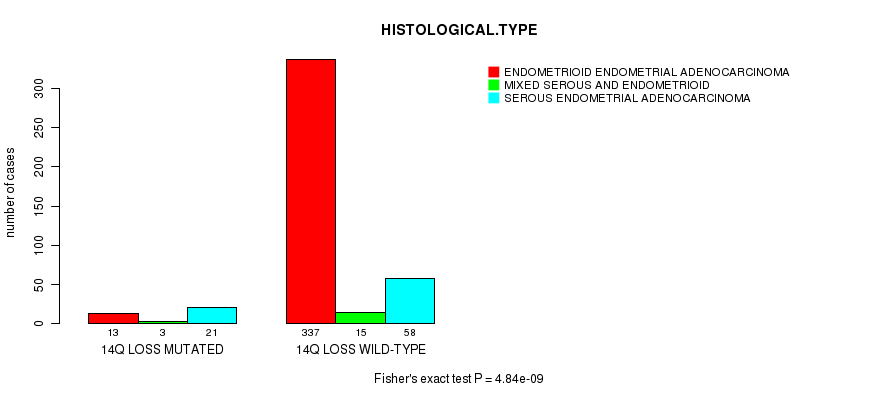
P value = 3.28e-05 (t-test), Q value = 0.012
Table S38. Gene #66: '15q loss mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 15Q LOSS MUTATED | 65 | 69.0 (10.8) |
| 15Q LOSS WILD-TYPE | 381 | 62.6 (11.1) |
Figure S38. Get High-res Image Gene #66: '15q loss mutation analysis' versus Clinical Feature #2: 'AGE'
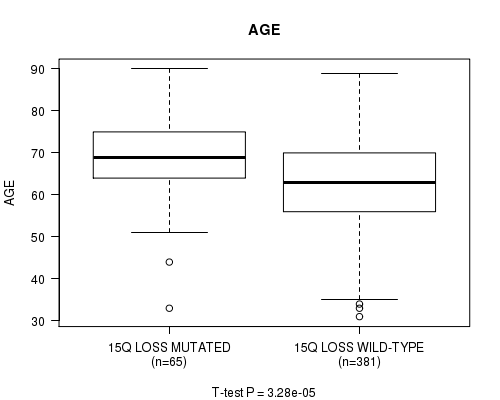
P value = 2.38e-11 (Fisher's exact test), Q value = 9.2e-09
Table S39. Gene #66: '15q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 15Q LOSS MUTATED | 28 | 6 | 31 |
| 15Q LOSS WILD-TYPE | 322 | 12 | 48 |
Figure S39. Get High-res Image Gene #66: '15q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
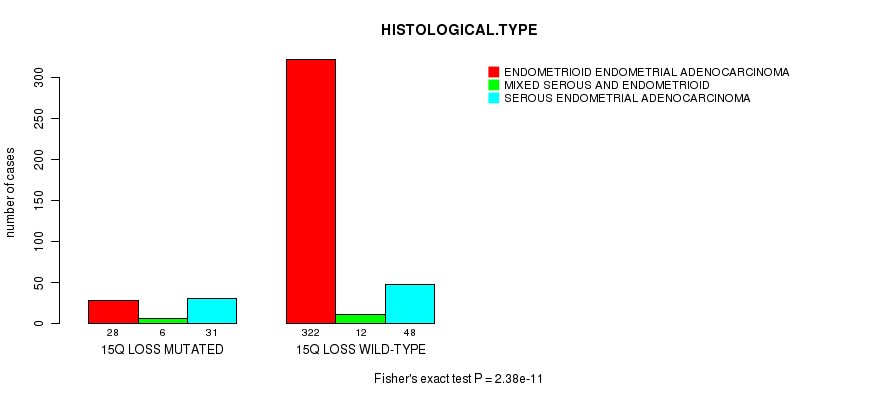
P value = 6.56e-05 (t-test), Q value = 0.023
Table S40. Gene #67: '16p loss mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 16P LOSS MUTATED | 56 | 68.9 (9.8) |
| 16P LOSS WILD-TYPE | 390 | 62.8 (11.3) |
Figure S40. Get High-res Image Gene #67: '16p loss mutation analysis' versus Clinical Feature #2: 'AGE'
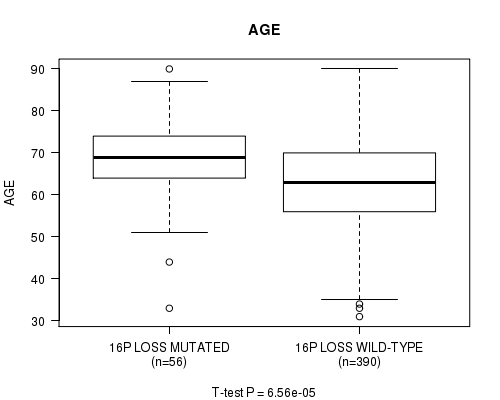
P value = 7.49e-09 (Fisher's exact test), Q value = 2.9e-06
Table S41. Gene #67: '16p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 16P LOSS MUTATED | 25 | 5 | 26 |
| 16P LOSS WILD-TYPE | 325 | 13 | 53 |
Figure S41. Get High-res Image Gene #67: '16p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
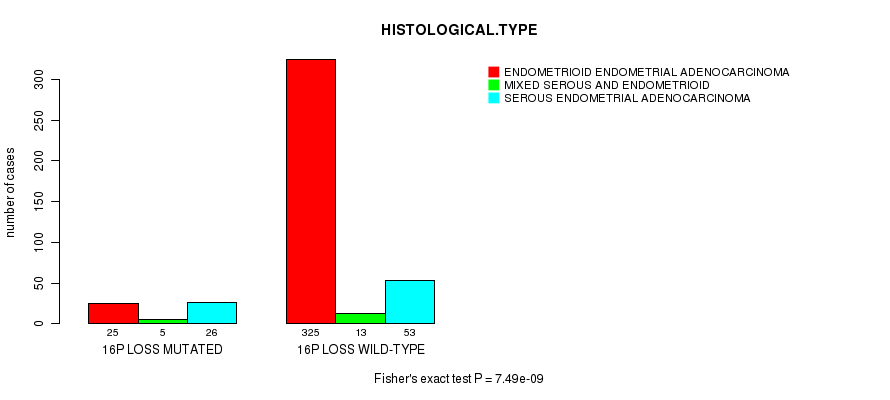
P value = 2.36e-06 (t-test), Q value = 0.00087
Table S42. Gene #68: '16q loss mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 16Q LOSS MUTATED | 93 | 68.0 (9.4) |
| 16Q LOSS WILD-TYPE | 353 | 62.4 (11.4) |
Figure S42. Get High-res Image Gene #68: '16q loss mutation analysis' versus Clinical Feature #2: 'AGE'
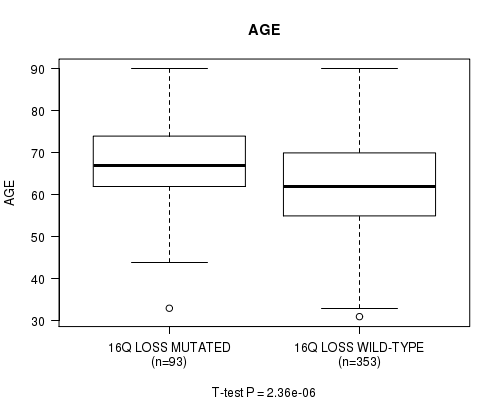
P value = 2.22e-14 (Fisher's exact test), Q value = 8.7e-12
Table S43. Gene #68: '16q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 16Q LOSS MUTATED | 44 | 6 | 43 |
| 16Q LOSS WILD-TYPE | 306 | 12 | 36 |
Figure S43. Get High-res Image Gene #68: '16q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
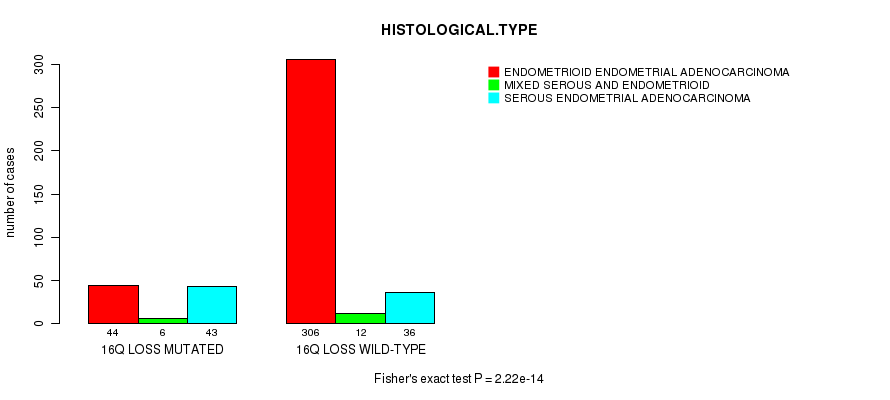
P value = 1.53e-06 (t-test), Q value = 0.00057
Table S44. Gene #69: '17p loss mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 17P LOSS MUTATED | 86 | 68.4 (9.5) |
| 17P LOSS WILD-TYPE | 360 | 62.4 (11.3) |
Figure S44. Get High-res Image Gene #69: '17p loss mutation analysis' versus Clinical Feature #2: 'AGE'
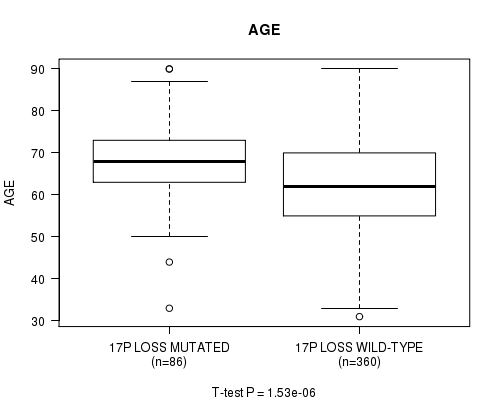
P value = 1.9e-20 (Fisher's exact test), Q value = 7.5e-18
Table S45. Gene #69: '17p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 17P LOSS MUTATED | 33 | 7 | 46 |
| 17P LOSS WILD-TYPE | 317 | 11 | 33 |
Figure S45. Get High-res Image Gene #69: '17p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
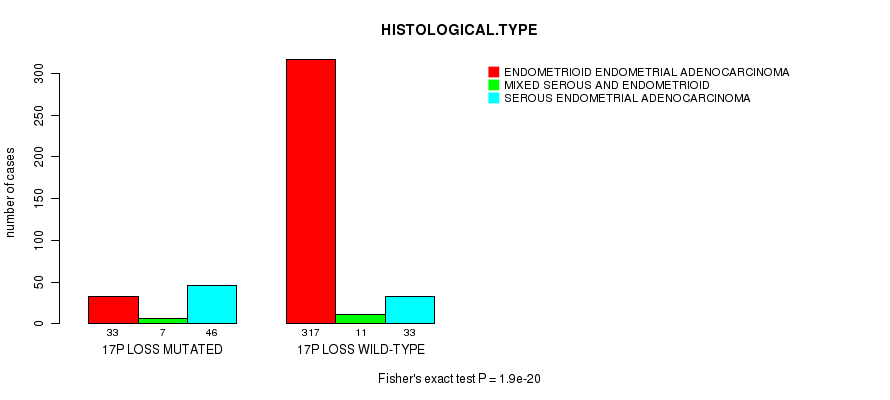
P value = 0.000598 (t-test), Q value = 0.21
Table S46. Gene #70: '17q loss mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 17Q LOSS MUTATED | 57 | 68.4 (10.8) |
| 17Q LOSS WILD-TYPE | 389 | 62.9 (11.2) |
Figure S46. Get High-res Image Gene #70: '17q loss mutation analysis' versus Clinical Feature #2: 'AGE'
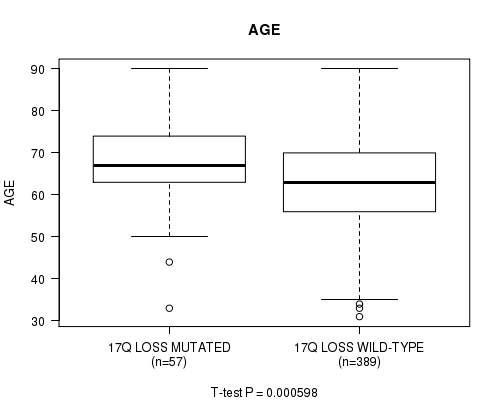
P value = 2.12e-06 (Fisher's exact test), Q value = 0.00079
Table S47. Gene #70: '17q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 17Q LOSS MUTATED | 29 | 5 | 23 |
| 17Q LOSS WILD-TYPE | 321 | 13 | 56 |
Figure S47. Get High-res Image Gene #70: '17q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
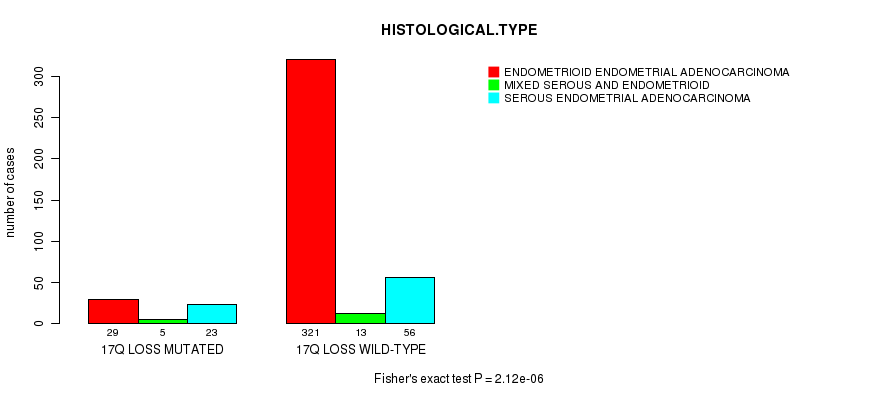
P value = 1.04e-05 (Fisher's exact test), Q value = 0.0038
Table S48. Gene #72: '18q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 18Q LOSS MUTATED | 20 | 6 | 15 |
| 18Q LOSS WILD-TYPE | 330 | 12 | 64 |
Figure S48. Get High-res Image Gene #72: '18q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
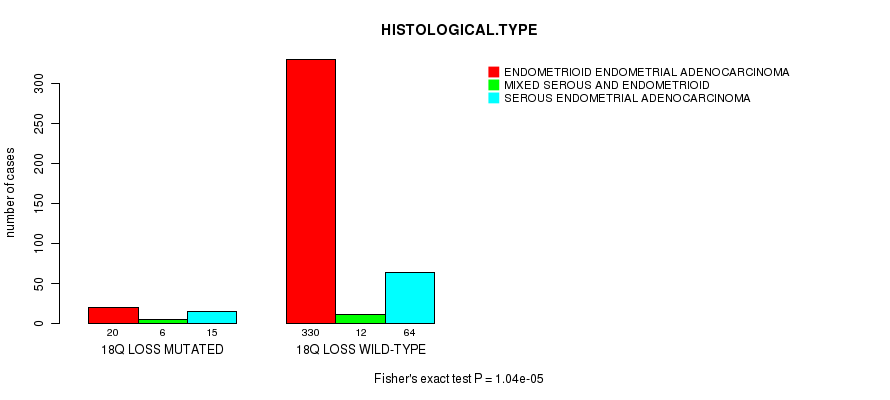
P value = 1.97e-07 (Fisher's exact test), Q value = 7.4e-05
Table S49. Gene #73: '19p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 19P LOSS MUTATED | 10 | 2 | 17 |
| 19P LOSS WILD-TYPE | 340 | 16 | 62 |
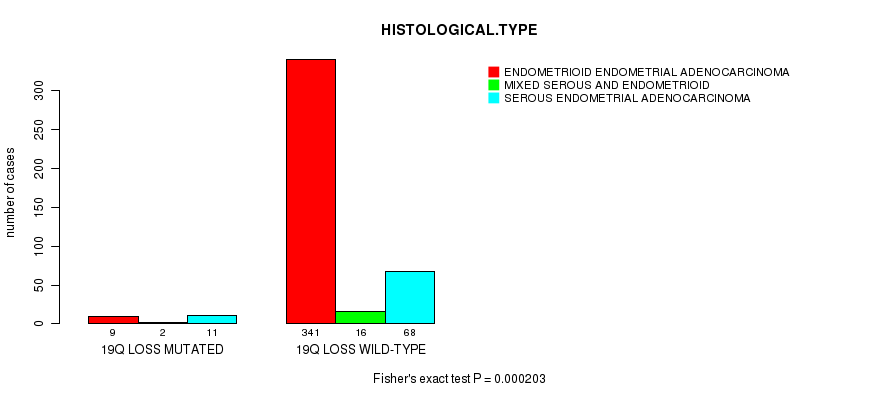
Figure S49. Get High-res Image Gene #73: '19p loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
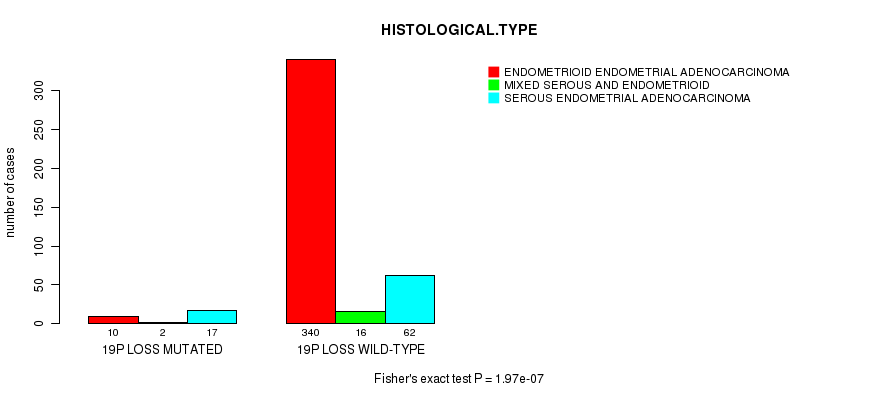
P value = 0.000203 (Fisher's exact test), Q value = 0.071
Table S50. Gene #74: '19q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 19Q LOSS MUTATED | 9 | 2 | 11 |
| 19Q LOSS WILD-TYPE | 341 | 16 | 68 |
Figure S50. Get High-res Image Gene #74: '19q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
P value = 1.22e-05 (Fisher's exact test), Q value = 0.0044
Table S51. Gene #77: '21q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 21Q LOSS MUTATED | 10 | 3 | 13 |
| 21Q LOSS WILD-TYPE | 340 | 15 | 66 |
Figure S51. Get High-res Image Gene #77: '21q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
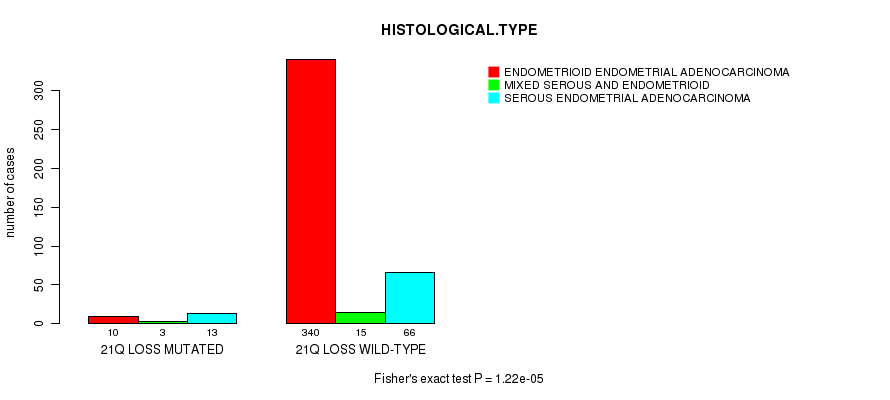
P value = 0.000208 (t-test), Q value = 0.073
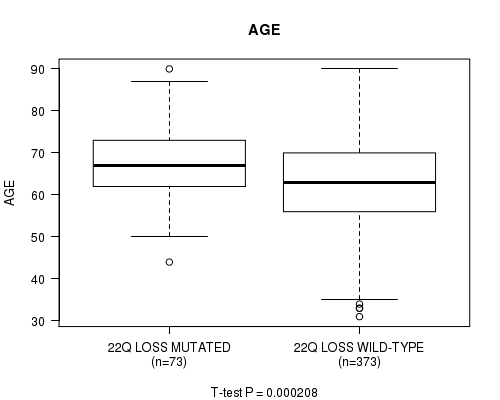
Table S52. Gene #78: '22q loss mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 446 | 63.6 (11.3) |
| 22Q LOSS MUTATED | 73 | 67.5 (9.1) |
| 22Q LOSS WILD-TYPE | 373 | 62.8 (11.5) |
Figure S52. Get High-res Image Gene #78: '22q loss mutation analysis' versus Clinical Feature #2: 'AGE'
P value = 2.79e-09 (Fisher's exact test), Q value = 1.1e-06
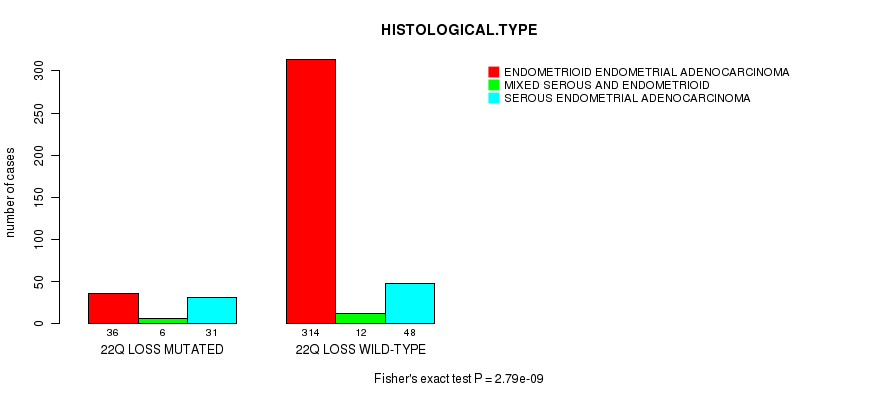
Table S53. Gene #78: '22q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | MIXED SEROUS AND ENDOMETRIOID | SEROUS ENDOMETRIAL ADENOCARCINOMA |
|---|---|---|---|
| ALL | 350 | 18 | 79 |
| 22Q LOSS MUTATED | 36 | 6 | 31 |
| 22Q LOSS WILD-TYPE | 314 | 12 | 48 |
Figure S53. Get High-res Image Gene #78: '22q loss mutation analysis' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
-
Mutation data file = broad_values_by_arm.mutsig.cluster.txt
-
Clinical data file = UCEC-TP.clin.merged.picked.txt
-
Number of patients = 447
-
Number of significantly arm-level cnvs = 79
-
Number of selected clinical features = 5
-
Exclude genes that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For continuous numerical clinical features, two-tailed Student's t test with unequal variance (Lehmann and Romano 2005) was applied to compare the clinical values between tumors with and without gene mutations using 't.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.