This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and subtypes.
Testing the association between copy number variation 57 arm-level results and 6 molecular subtypes across 66 patients, 51 significant findings detected with Q value < 0.25.
-
4p gain cnv correlated to 'MRNASEQ_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
4q gain cnv correlated to 'MRNASEQ_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
11p gain cnv correlated to 'MRNASEQ_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
11q gain cnv correlated to 'MRNASEQ_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
14q gain cnv correlated to 'MRNASEQ_CNMF'.
-
16p gain cnv correlated to 'MRNASEQ_CNMF'.
-
16q gain cnv correlated to 'MRNASEQ_CNMF'.
-
Xq gain cnv correlated to 'MRNASEQ_CNMF'.
-
1p loss cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
1q loss cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
2p loss cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
2q loss cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
6p loss cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
6q loss cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
8p loss cnv correlated to 'CN_CNMF'.
-
8q loss cnv correlated to 'CN_CNMF'.
-
10p loss cnv correlated to 'MRNASEQ_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
10q loss cnv correlated to 'MRNASEQ_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
13q loss cnv correlated to 'MRNASEQ_CHIERARCHICAL'.
-
16q loss cnv correlated to 'MRNASEQ_CHIERARCHICAL'.
-
17p loss cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
17q loss cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
Xq loss cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 57 arm-level results and 6 molecular subtypes. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 51 significant findings detected.
|
Molecular subtypes |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Chi-square test | Fisher's exact test | Fisher's exact test | |
| 2p loss | 0 (0%) | 23 |
0.0828 (1.00) |
4.57e-06 (0.00148) |
8.31e-09 (2.82e-06) |
2.26e-07 (7.58e-05) |
0.0679 (1.00) |
0.000592 (0.174) |
| 2q loss | 0 (0%) | 23 |
0.0828 (1.00) |
4.57e-06 (0.00148) |
8.31e-09 (2.82e-06) |
2.26e-07 (7.58e-05) |
0.0679 (1.00) |
0.000592 (0.174) |
| 17p loss | 0 (0%) | 21 |
0.158 (1.00) |
1.31e-06 (0.00043) |
2.61e-09 (8.91e-07) |
3e-07 (1e-04) |
0.0256 (1.00) |
0.000255 (0.0761) |
| 17q loss | 0 (0%) | 21 |
0.158 (1.00) |
1.31e-06 (0.00043) |
2.61e-09 (8.91e-07) |
3e-07 (1e-04) |
0.0256 (1.00) |
0.000255 (0.0761) |
| 1p loss | 0 (0%) | 18 |
0.342 (1.00) |
5.89e-06 (0.00189) |
8.26e-06 (0.00264) |
1.79e-05 (0.0057) |
0.0572 (1.00) |
0.00103 (0.296) |
| 1q loss | 0 (0%) | 19 |
0.32 (1.00) |
1.06e-05 (0.00337) |
3.11e-05 (0.00972) |
6.59e-05 (0.0205) |
0.127 (1.00) |
0.00157 (0.443) |
| 6p loss | 0 (0%) | 19 |
0.32 (1.00) |
0.000132 (0.0401) |
1.26e-06 (0.000417) |
2.67e-05 (0.00844) |
0.127 (1.00) |
0.00157 (0.443) |
| 6q loss | 0 (0%) | 19 |
0.32 (1.00) |
0.000132 (0.0401) |
1.26e-06 (0.000417) |
2.67e-05 (0.00844) |
0.127 (1.00) |
0.00157 (0.443) |
| Xq loss | 0 (0%) | 33 |
0.713 (1.00) |
2.45e-05 (0.00777) |
7.96e-05 (0.0247) |
0.000217 (0.0648) |
0.019 (1.00) |
0.00208 (0.575) |
| 4p gain | 0 (0%) | 48 |
0.383 (1.00) |
0.813 (1.00) |
4.56e-05 (0.0142) |
0.000161 (0.0486) |
0.239 (1.00) |
0.1 (1.00) |
| 4q gain | 0 (0%) | 49 |
0.362 (1.00) |
0.701 (1.00) |
0.000173 (0.0519) |
0.000784 (0.228) |
0.144 (1.00) |
0.0977 (1.00) |
| 11p gain | 0 (0%) | 53 |
0.517 (1.00) |
0.271 (1.00) |
0.000287 (0.0851) |
0.000156 (0.0472) |
0.5 (1.00) |
0.186 (1.00) |
| 11q gain | 0 (0%) | 52 |
0.606 (1.00) |
0.273 (1.00) |
8.33e-05 (0.0257) |
2.78e-05 (0.00874) |
0.698 (1.00) |
0.186 (1.00) |
| 10p loss | 0 (0%) | 22 |
0.261 (1.00) |
0.00198 (0.553) |
2.51e-06 (0.00082) |
3.03e-06 (0.000983) |
0.391 (1.00) |
0.0506 (1.00) |
| 10q loss | 0 (0%) | 22 |
0.261 (1.00) |
0.00198 (0.553) |
2.51e-06 (0.00082) |
3.03e-06 (0.000983) |
0.391 (1.00) |
0.0506 (1.00) |
| 14q gain | 0 (0%) | 50 |
0.34 (1.00) |
0.554 (1.00) |
0.000706 (0.206) |
0.00332 (0.903) |
0.261 (1.00) |
0.436 (1.00) |
| 16p gain | 0 (0%) | 51 |
0.556 (1.00) |
0.8 (1.00) |
9.21e-05 (0.0282) |
0.000936 (0.271) |
0.137 (1.00) |
0.106 (1.00) |
| 16q gain | 0 (0%) | 51 |
0.556 (1.00) |
0.8 (1.00) |
9.21e-05 (0.0282) |
0.000936 (0.271) |
0.137 (1.00) |
0.106 (1.00) |
| Xq gain | 0 (0%) | 57 |
1 (1.00) |
0.503 (1.00) |
8.84e-05 (0.0271) |
0.00417 (1.00) |
0.294 (1.00) |
0.341 (1.00) |
| 8p loss | 0 (0%) | 57 |
8.11e-08 (2.73e-05) |
0.0576 (1.00) |
0.0531 (1.00) |
0.0036 (0.974) |
0.148 (1.00) |
0.341 (1.00) |
| 8q loss | 0 (0%) | 58 |
5.86e-07 (0.000195) |
0.0381 (1.00) |
0.134 (1.00) |
0.00434 (1.00) |
0.291 (1.00) |
0.586 (1.00) |
| 13q loss | 0 (0%) | 29 |
0.116 (1.00) |
0.00923 (1.00) |
0.00123 (0.351) |
0.000506 (0.149) |
0.743 (1.00) |
0.036 (1.00) |
| 16q loss | 0 (0%) | 62 |
0.685 (1.00) |
0.653 (1.00) |
0.00219 (0.601) |
8.58e-05 (0.0264) |
0.678 (1.00) |
0.452 (1.00) |
| 3p gain | 0 (0%) | 61 |
0.721 (1.00) |
0.698 (1.00) |
1 (1.00) |
0.942 (1.00) |
0.15 (1.00) |
0.134 (1.00) |
| 3q gain | 0 (0%) | 61 |
0.721 (1.00) |
0.698 (1.00) |
1 (1.00) |
0.942 (1.00) |
0.15 (1.00) |
0.134 (1.00) |
| 5p gain | 0 (0%) | 61 |
0.549 (1.00) |
0.698 (1.00) |
1 (1.00) |
0.839 (1.00) |
0.857 (1.00) |
0.134 (1.00) |
| 5q gain | 0 (0%) | 61 |
0.549 (1.00) |
0.698 (1.00) |
1 (1.00) |
0.839 (1.00) |
0.857 (1.00) |
0.134 (1.00) |
| 7p gain | 0 (0%) | 48 |
0.383 (1.00) |
0.384 (1.00) |
0.00906 (1.00) |
0.0191 (1.00) |
0.197 (1.00) |
1 (1.00) |
| 7q gain | 0 (0%) | 48 |
0.383 (1.00) |
0.384 (1.00) |
0.00906 (1.00) |
0.0191 (1.00) |
0.197 (1.00) |
1 (1.00) |
| 8p gain | 0 (0%) | 53 |
0.274 (1.00) |
0.0865 (1.00) |
0.0276 (1.00) |
0.0848 (1.00) |
0.177 (1.00) |
1 (1.00) |
| 8q gain | 0 (0%) | 52 |
0.157 (1.00) |
0.105 (1.00) |
0.00994 (1.00) |
0.0317 (1.00) |
0.387 (1.00) |
1 (1.00) |
| 9p gain | 0 (0%) | 58 |
1 (1.00) |
0.609 (1.00) |
0.911 (1.00) |
0.841 (1.00) |
0.804 (1.00) |
0.298 (1.00) |
| 9q gain | 0 (0%) | 58 |
1 (1.00) |
0.609 (1.00) |
0.911 (1.00) |
0.841 (1.00) |
0.804 (1.00) |
0.298 (1.00) |
| 12p gain | 0 (0%) | 52 |
0.688 (1.00) |
0.437 (1.00) |
0.0129 (1.00) |
0.0542 (1.00) |
0.698 (1.00) |
1 (1.00) |
| 12q gain | 0 (0%) | 53 |
0.764 (1.00) |
0.709 (1.00) |
0.0135 (1.00) |
0.0191 (1.00) |
0.86 (1.00) |
0.675 (1.00) |
| 15q gain | 0 (0%) | 52 |
0.224 (1.00) |
0.249 (1.00) |
0.00113 (0.324) |
0.00646 (1.00) |
0.287 (1.00) |
0.671 (1.00) |
| 18p gain | 0 (0%) | 52 |
0.475 (1.00) |
0.294 (1.00) |
0.0062 (1.00) |
0.0191 (1.00) |
0.806 (1.00) |
0.386 (1.00) |
| 18q gain | 0 (0%) | 53 |
0.671 (1.00) |
0.581 (1.00) |
0.00409 (1.00) |
0.0183 (1.00) |
0.926 (1.00) |
1 (1.00) |
| 19p gain | 0 (0%) | 55 |
0.867 (1.00) |
0.83 (1.00) |
0.00206 (0.571) |
0.00219 (0.601) |
0.29 (1.00) |
0.337 (1.00) |
| 19q gain | 0 (0%) | 57 |
0.713 (1.00) |
0.72 (1.00) |
0.00269 (0.734) |
0.00372 (1.00) |
0.397 (1.00) |
0.341 (1.00) |
| 20p gain | 0 (0%) | 52 |
0.224 (1.00) |
0.672 (1.00) |
0.00468 (1.00) |
0.00377 (1.00) |
0.124 (1.00) |
0.671 (1.00) |
| 20q gain | 0 (0%) | 51 |
0.282 (1.00) |
0.397 (1.00) |
0.00713 (1.00) |
0.00743 (1.00) |
0.17 (1.00) |
1 (1.00) |
| 21q gain | 0 (0%) | 63 |
0.646 (1.00) |
0.327 (1.00) |
0.333 (1.00) |
0.194 (1.00) |
0.361 (1.00) |
|
| 22q gain | 0 (0%) | 51 |
0.882 (1.00) |
0.116 (1.00) |
0.0748 (1.00) |
0.196 (1.00) |
0.466 (1.00) |
1 (1.00) |
| 3p loss | 0 (0%) | 59 |
0.0826 (1.00) |
1 (1.00) |
0.252 (1.00) |
0.00554 (1.00) |
0.481 (1.00) |
0.581 (1.00) |
| 3q loss | 0 (0%) | 60 |
0.0326 (1.00) |
0.532 (1.00) |
0.135 (1.00) |
0.00126 (0.358) |
0.354 (1.00) |
0.585 (1.00) |
| 5p loss | 0 (0%) | 59 |
0.302 (1.00) |
0.175 (1.00) |
0.0329 (1.00) |
0.00478 (1.00) |
0.886 (1.00) |
0.581 (1.00) |
| 5q loss | 0 (0%) | 59 |
0.302 (1.00) |
0.175 (1.00) |
0.0329 (1.00) |
0.00478 (1.00) |
0.886 (1.00) |
0.581 (1.00) |
| 9p loss | 0 (0%) | 58 |
1 (1.00) |
1 (1.00) |
0.171 (1.00) |
0.579 (1.00) |
1 (1.00) |
0.298 (1.00) |
| 9q loss | 0 (0%) | 58 |
1 (1.00) |
1 (1.00) |
0.171 (1.00) |
0.579 (1.00) |
1 (1.00) |
0.298 (1.00) |
| 11p loss | 0 (0%) | 61 |
1 (1.00) |
0.593 (1.00) |
0.129 (1.00) |
0.408 (1.00) |
0.12 (1.00) |
1 (1.00) |
| 11q loss | 0 (0%) | 61 |
1 (1.00) |
0.593 (1.00) |
0.129 (1.00) |
0.408 (1.00) |
0.12 (1.00) |
1 (1.00) |
| 16p loss | 0 (0%) | 63 |
1 (1.00) |
0.417 (1.00) |
0.0126 (1.00) |
0.00148 (0.418) |
1 (1.00) |
1 (1.00) |
| 18p loss | 0 (0%) | 61 |
0.257 (1.00) |
0.118 (1.00) |
0.0134 (1.00) |
0.0012 (0.343) |
0.12 (1.00) |
1 (1.00) |
| 18q loss | 0 (0%) | 59 |
0.222 (1.00) |
0.175 (1.00) |
0.0481 (1.00) |
0.0178 (1.00) |
0.203 (1.00) |
1 (1.00) |
| 21q loss | 0 (0%) | 38 |
1 (1.00) |
0.0183 (1.00) |
0.23 (1.00) |
0.0379 (1.00) |
0.575 (1.00) |
0.722 (1.00) |
| 22q loss | 0 (0%) | 61 |
0.721 (1.00) |
0.118 (1.00) |
0.745 (1.00) |
0.38 (1.00) |
1 (1.00) |
1 (1.00) |
P value = 4.56e-05 (Fisher's exact test), Q value = 0.014
Table S1. Gene #3: '4p gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 4P GAIN CNV | 3 | 14 | 1 | 0 |
| 4P GAIN WILD-TYPE | 16 | 8 | 14 | 10 |
Figure S1. Get High-res Image Gene #3: '4p gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
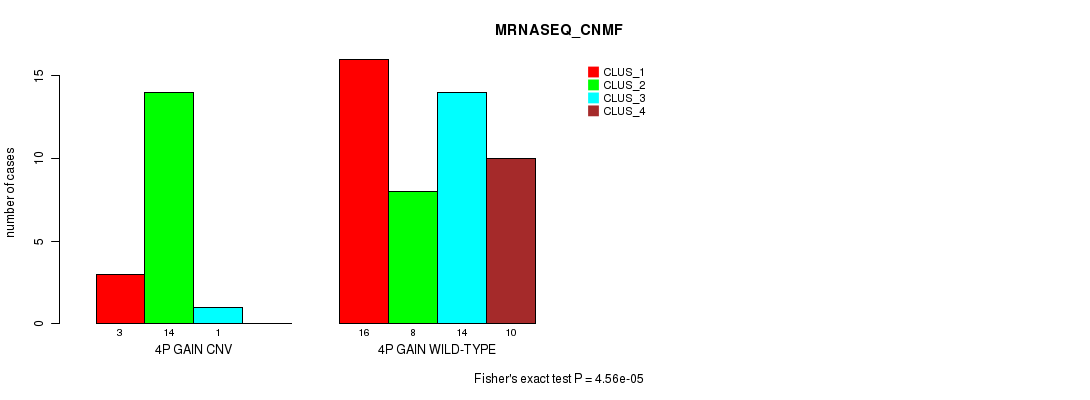
P value = 0.000161 (Chi-square test), Q value = 0.049
Table S2. Gene #3: '4p gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 4P GAIN CNV | 1 | 1 | 12 | 0 | 4 |
| 4P GAIN WILD-TYPE | 9 | 9 | 5 | 6 | 19 |
Figure S2. Get High-res Image Gene #3: '4p gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
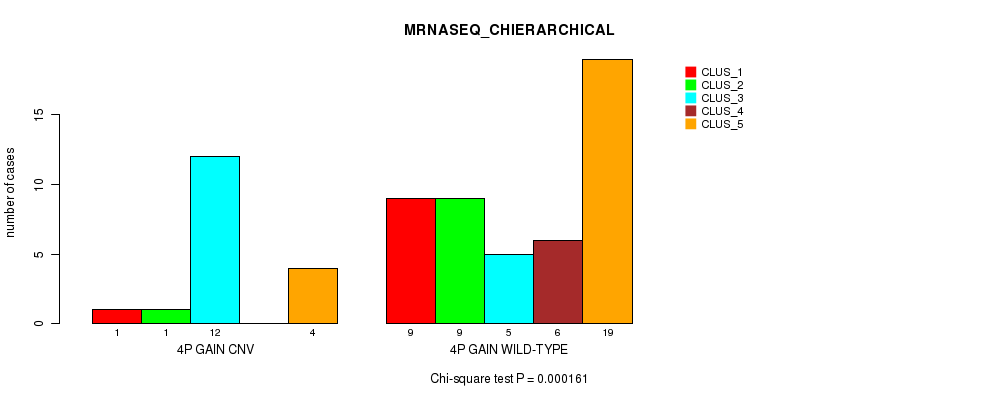
P value = 0.000173 (Fisher's exact test), Q value = 0.052
Table S3. Gene #4: '4q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 4Q GAIN CNV | 3 | 13 | 1 | 0 |
| 4Q GAIN WILD-TYPE | 16 | 9 | 14 | 10 |
Figure S3. Get High-res Image Gene #4: '4q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
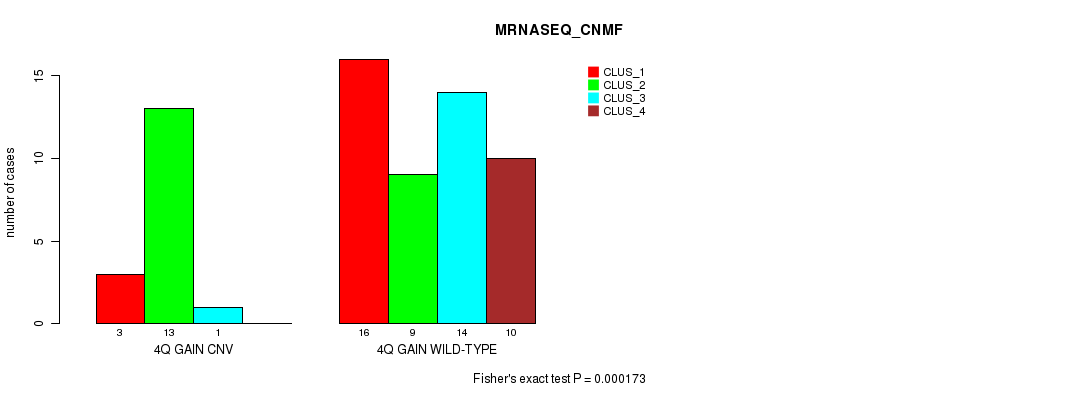
P value = 0.000784 (Chi-square test), Q value = 0.23
Table S4. Gene #4: '4q gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 4Q GAIN CNV | 1 | 1 | 11 | 0 | 4 |
| 4Q GAIN WILD-TYPE | 9 | 9 | 6 | 6 | 19 |
Figure S4. Get High-res Image Gene #4: '4q gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
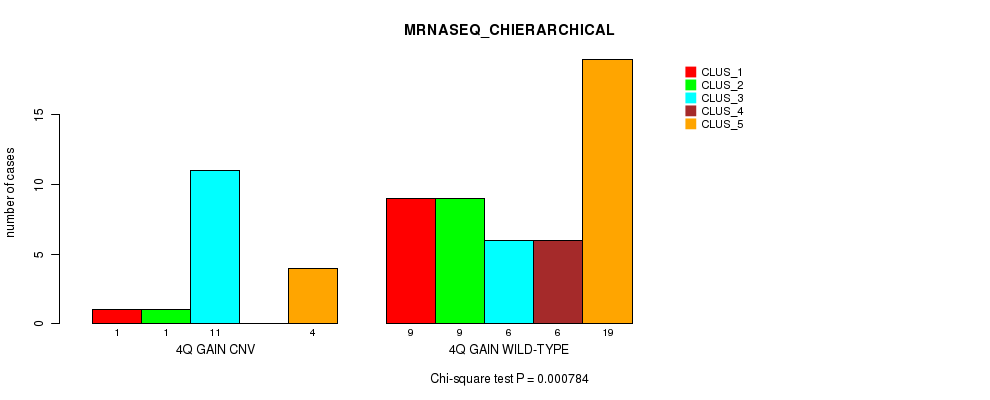
P value = 0.000287 (Fisher's exact test), Q value = 0.085
Table S5. Gene #13: '11p gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 11P GAIN CNV | 1 | 11 | 1 | 0 |
| 11P GAIN WILD-TYPE | 18 | 11 | 14 | 10 |
Figure S5. Get High-res Image Gene #13: '11p gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
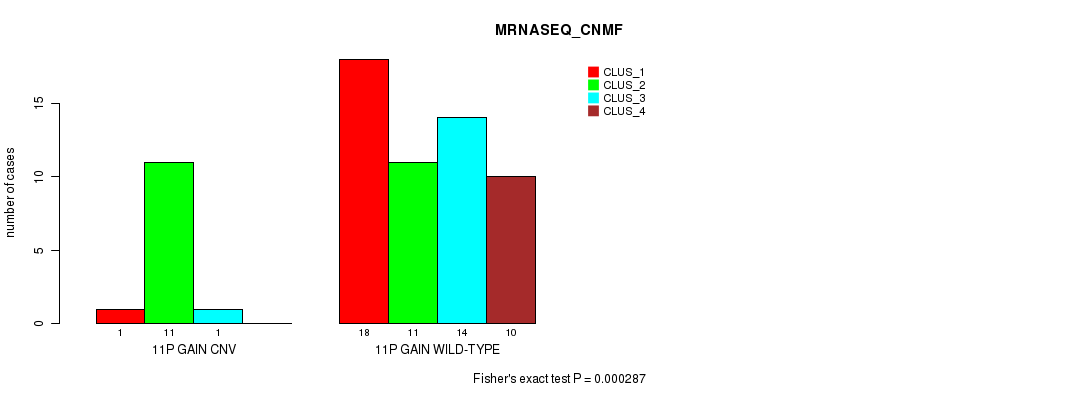
P value = 0.000156 (Chi-square test), Q value = 0.047
Table S6. Gene #13: '11p gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 11P GAIN CNV | 1 | 1 | 10 | 0 | 1 |
| 11P GAIN WILD-TYPE | 9 | 9 | 7 | 6 | 22 |
Figure S6. Get High-res Image Gene #13: '11p gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
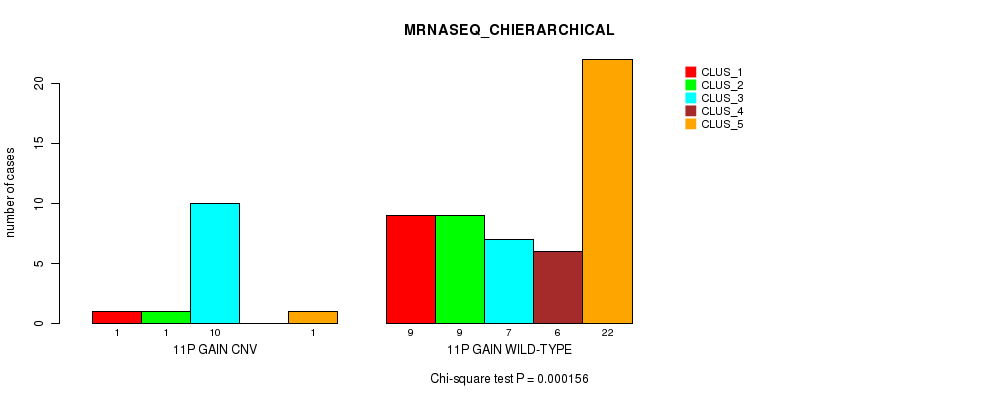
P value = 8.33e-05 (Fisher's exact test), Q value = 0.026
Table S7. Gene #14: '11q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 11Q GAIN CNV | 1 | 12 | 1 | 0 |
| 11Q GAIN WILD-TYPE | 18 | 10 | 14 | 10 |
Figure S7. Get High-res Image Gene #14: '11q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
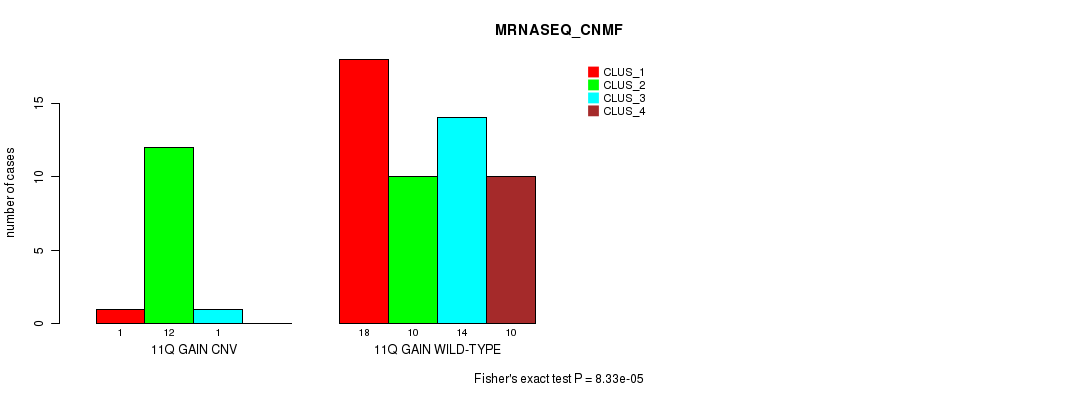
P value = 2.78e-05 (Chi-square test), Q value = 0.0087
Table S8. Gene #14: '11q gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 11Q GAIN CNV | 1 | 1 | 11 | 0 | 1 |
| 11Q GAIN WILD-TYPE | 9 | 9 | 6 | 6 | 22 |
Figure S8. Get High-res Image Gene #14: '11q gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
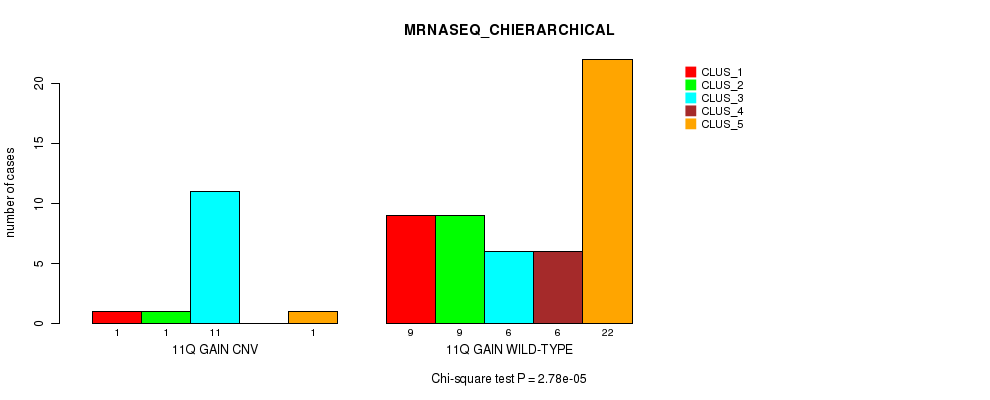
P value = 0.000706 (Fisher's exact test), Q value = 0.21
Table S9. Gene #17: '14q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 14Q GAIN CNV | 3 | 12 | 1 | 0 |
| 14Q GAIN WILD-TYPE | 16 | 10 | 14 | 10 |
Figure S9. Get High-res Image Gene #17: '14q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
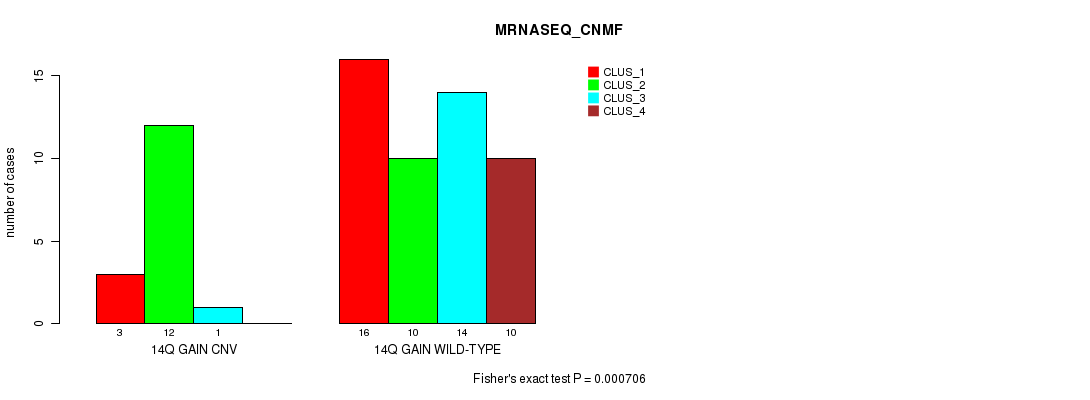
P value = 9.21e-05 (Fisher's exact test), Q value = 0.028
Table S10. Gene #19: '16p gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 16P GAIN CNV | 3 | 12 | 0 | 0 |
| 16P GAIN WILD-TYPE | 16 | 10 | 15 | 10 |
Figure S10. Get High-res Image Gene #19: '16p gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
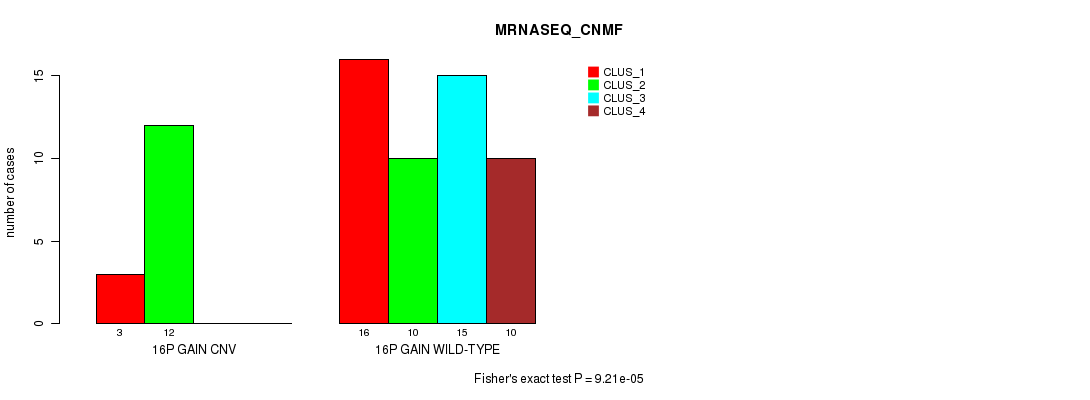
P value = 9.21e-05 (Fisher's exact test), Q value = 0.028
Table S11. Gene #20: '16q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 16Q GAIN CNV | 3 | 12 | 0 | 0 |
| 16Q GAIN WILD-TYPE | 16 | 10 | 15 | 10 |
Figure S11. Get High-res Image Gene #20: '16q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
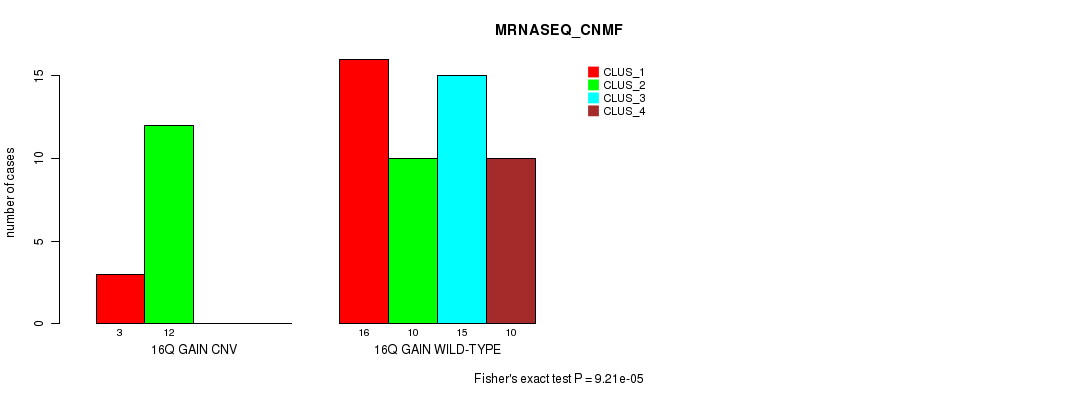
P value = 8.84e-05 (Fisher's exact test), Q value = 0.027
Table S12. Gene #29: 'Xq gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| XQ GAIN CNV | 0 | 9 | 0 | 0 |
| XQ GAIN WILD-TYPE | 19 | 13 | 15 | 10 |
Figure S12. Get High-res Image Gene #29: 'Xq gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
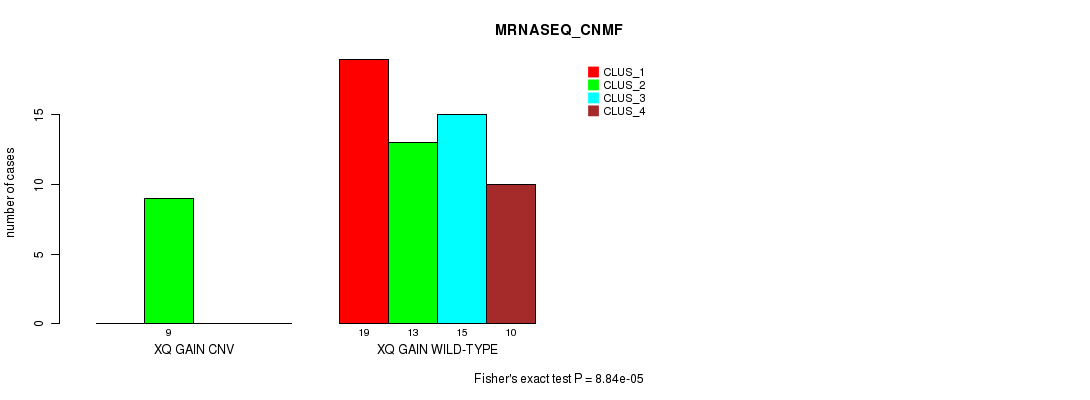
P value = 5.89e-06 (Fisher's exact test), Q value = 0.0019
Table S13. Gene #30: '1p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 34 | 14 |
| 1P LOSS CNV | 16 | 29 | 3 |
| 1P LOSS WILD-TYPE | 1 | 5 | 11 |
Figure S13. Get High-res Image Gene #30: '1p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
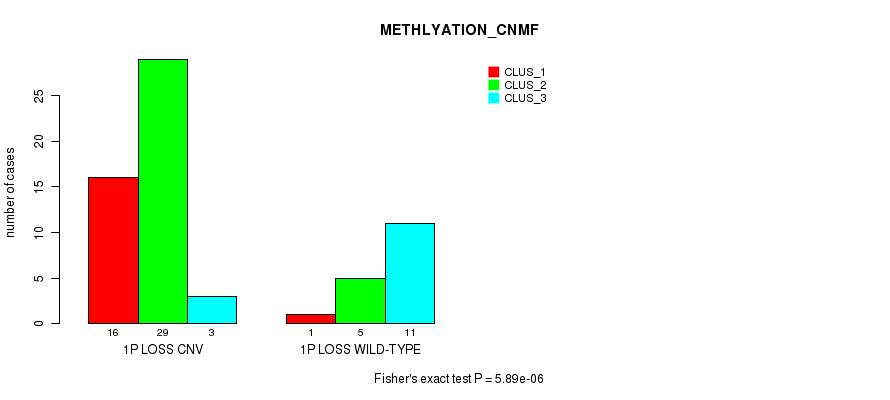
P value = 8.26e-06 (Fisher's exact test), Q value = 0.0026
Table S14. Gene #30: '1p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 1P LOSS CNV | 19 | 12 | 14 | 3 |
| 1P LOSS WILD-TYPE | 0 | 10 | 1 | 7 |
Figure S14. Get High-res Image Gene #30: '1p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
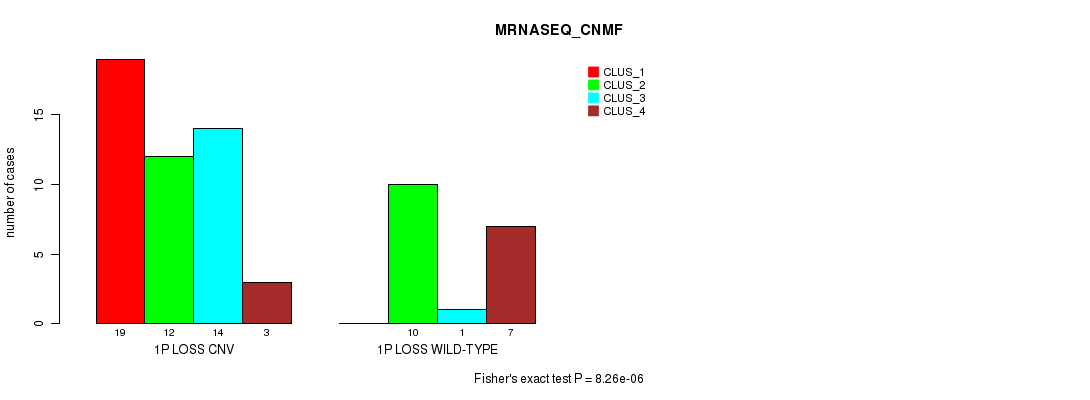
P value = 1.79e-05 (Chi-square test), Q value = 0.0057
Table S15. Gene #30: '1p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 1P LOSS CNV | 9 | 2 | 9 | 6 | 22 |
| 1P LOSS WILD-TYPE | 1 | 8 | 8 | 0 | 1 |
Figure S15. Get High-res Image Gene #30: '1p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
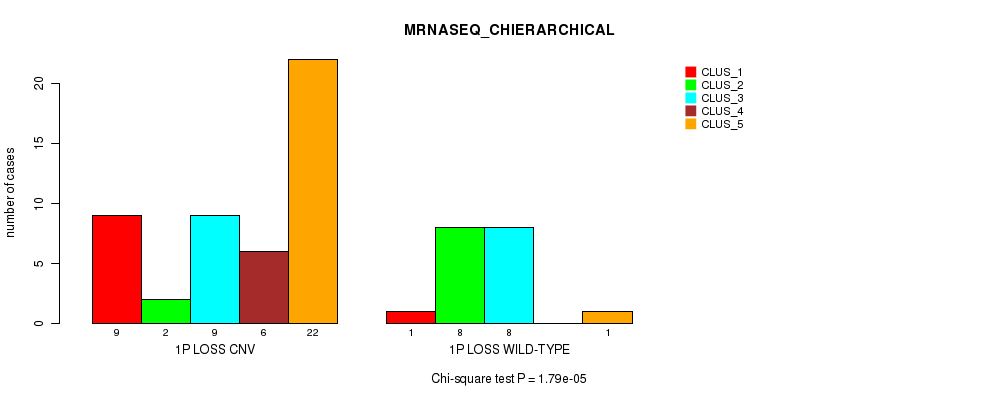
P value = 1.06e-05 (Fisher's exact test), Q value = 0.0034
Table S16. Gene #31: '1q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 34 | 14 |
| 1Q LOSS CNV | 16 | 28 | 3 |
| 1Q LOSS WILD-TYPE | 1 | 6 | 11 |
Figure S16. Get High-res Image Gene #31: '1q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
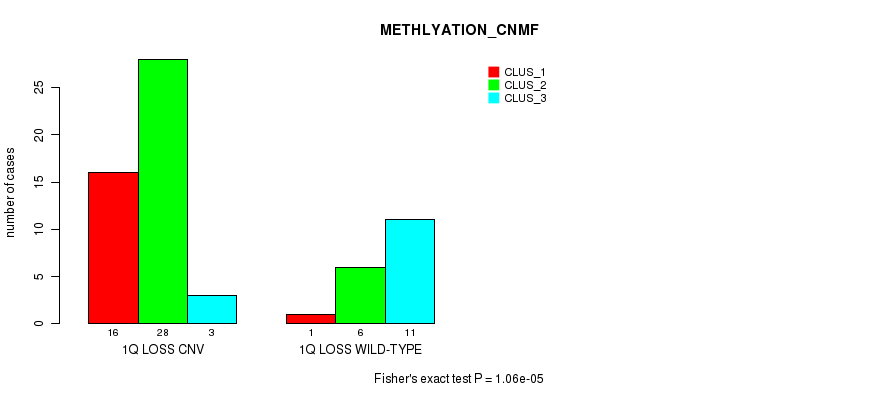
P value = 3.11e-05 (Fisher's exact test), Q value = 0.0097
Table S17. Gene #31: '1q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 1Q LOSS CNV | 19 | 12 | 13 | 3 |
| 1Q LOSS WILD-TYPE | 0 | 10 | 2 | 7 |
Figure S17. Get High-res Image Gene #31: '1q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
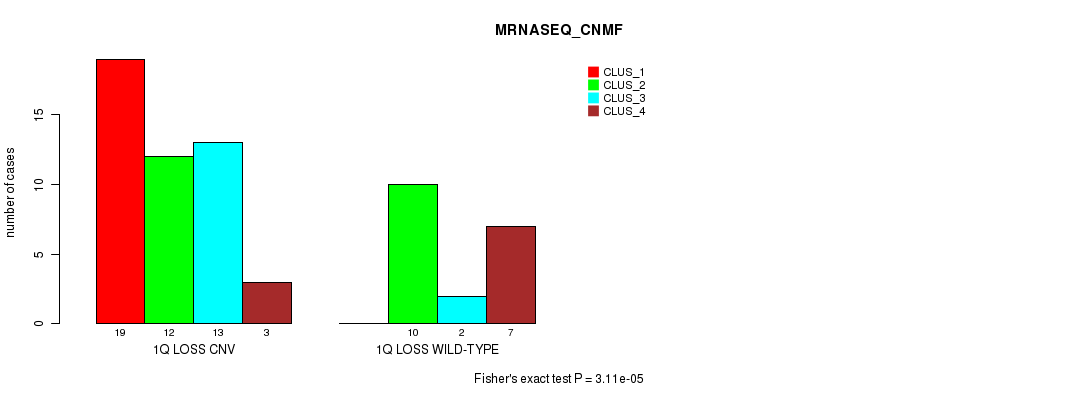
P value = 6.59e-05 (Chi-square test), Q value = 0.021
Table S18. Gene #31: '1q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 1Q LOSS CNV | 9 | 2 | 9 | 5 | 22 |
| 1Q LOSS WILD-TYPE | 1 | 8 | 8 | 1 | 1 |
Figure S18. Get High-res Image Gene #31: '1q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
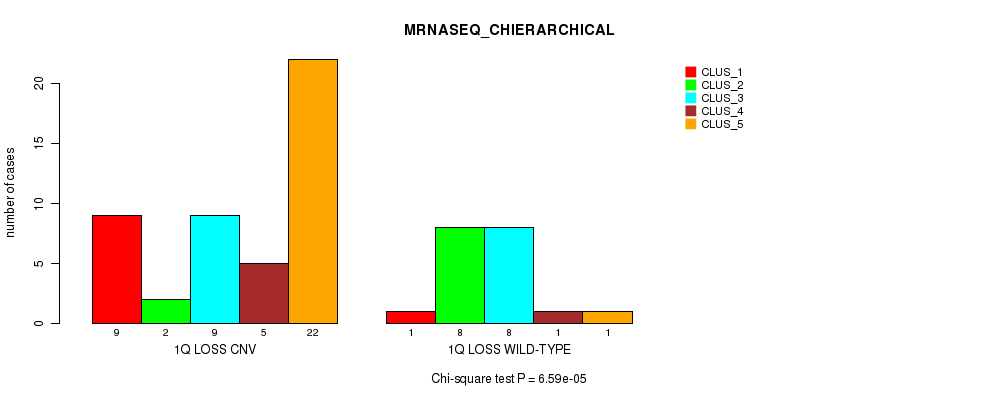
P value = 4.57e-06 (Fisher's exact test), Q value = 0.0015
Table S19. Gene #32: '2p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 34 | 14 |
| 2P LOSS CNV | 16 | 25 | 2 |
| 2P LOSS WILD-TYPE | 1 | 9 | 12 |
Figure S19. Get High-res Image Gene #32: '2p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
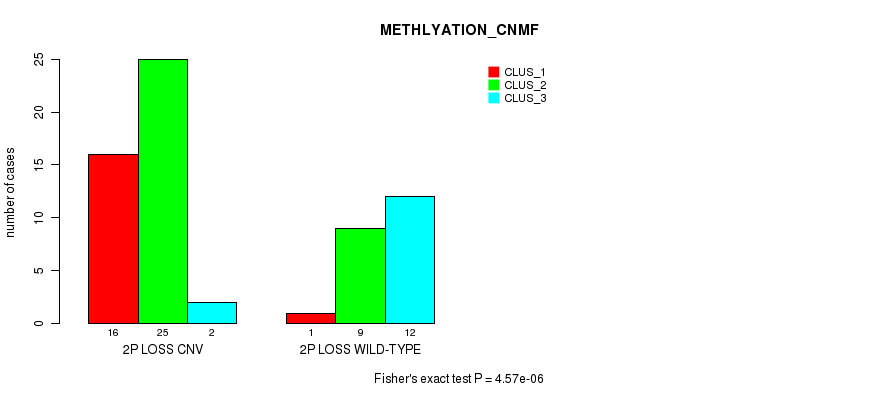
P value = 8.31e-09 (Fisher's exact test), Q value = 2.8e-06
Table S20. Gene #32: '2p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 2P LOSS CNV | 19 | 11 | 13 | 0 |
| 2P LOSS WILD-TYPE | 0 | 11 | 2 | 10 |
Figure S20. Get High-res Image Gene #32: '2p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
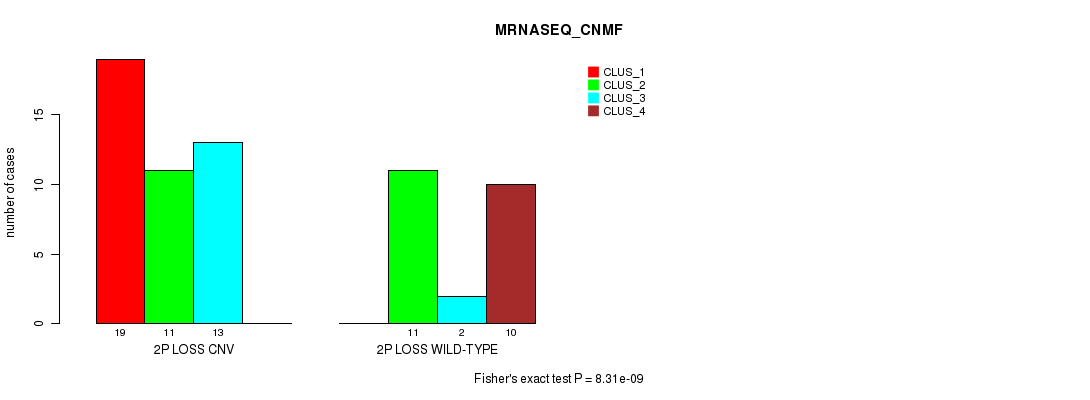
P value = 2.26e-07 (Chi-square test), Q value = 7.6e-05
Table S21. Gene #32: '2p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 2P LOSS CNV | 10 | 0 | 8 | 3 | 22 |
| 2P LOSS WILD-TYPE | 0 | 10 | 9 | 3 | 1 |
Figure S21. Get High-res Image Gene #32: '2p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
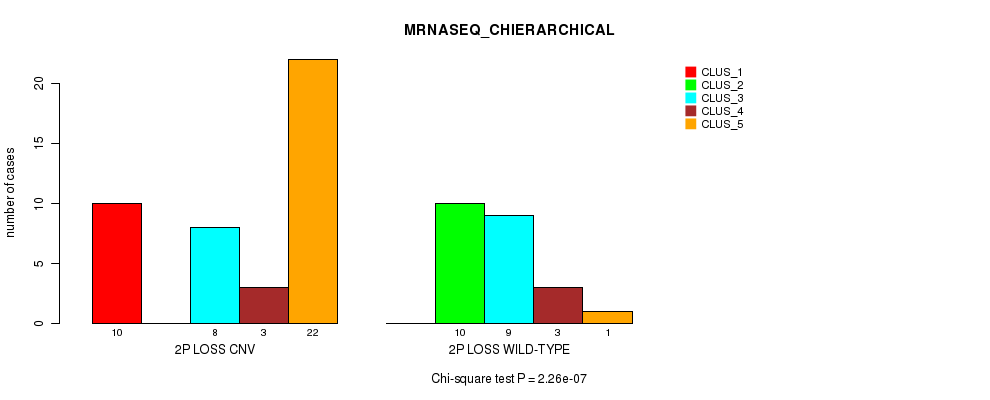
P value = 0.000592 (Fisher's exact test), Q value = 0.17
Table S22. Gene #32: '2p loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 9 | 57 |
| 2P LOSS CNV | 1 | 42 |
| 2P LOSS WILD-TYPE | 8 | 15 |
Figure S22. Get High-res Image Gene #32: '2p loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
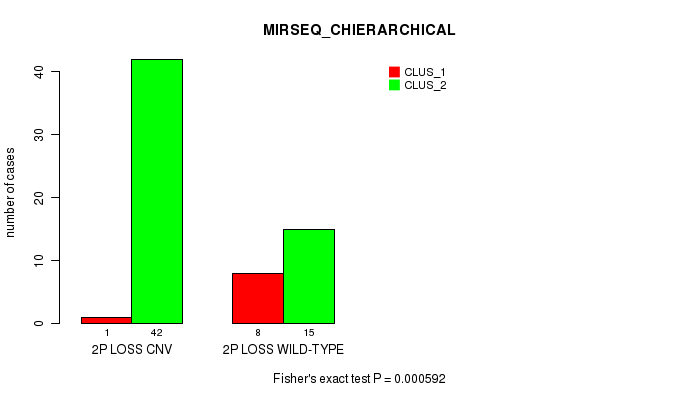
P value = 4.57e-06 (Fisher's exact test), Q value = 0.0015
Table S23. Gene #33: '2q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 34 | 14 |
| 2Q LOSS CNV | 16 | 25 | 2 |
| 2Q LOSS WILD-TYPE | 1 | 9 | 12 |
Figure S23. Get High-res Image Gene #33: '2q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
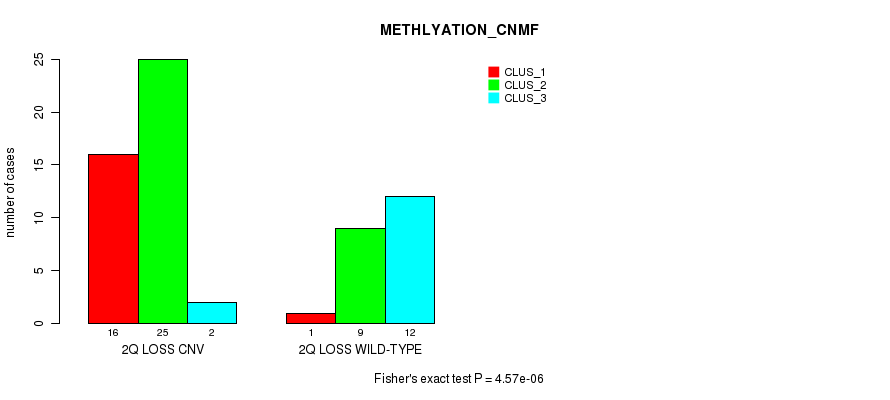
P value = 8.31e-09 (Fisher's exact test), Q value = 2.8e-06
Table S24. Gene #33: '2q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 2Q LOSS CNV | 19 | 11 | 13 | 0 |
| 2Q LOSS WILD-TYPE | 0 | 11 | 2 | 10 |
Figure S24. Get High-res Image Gene #33: '2q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
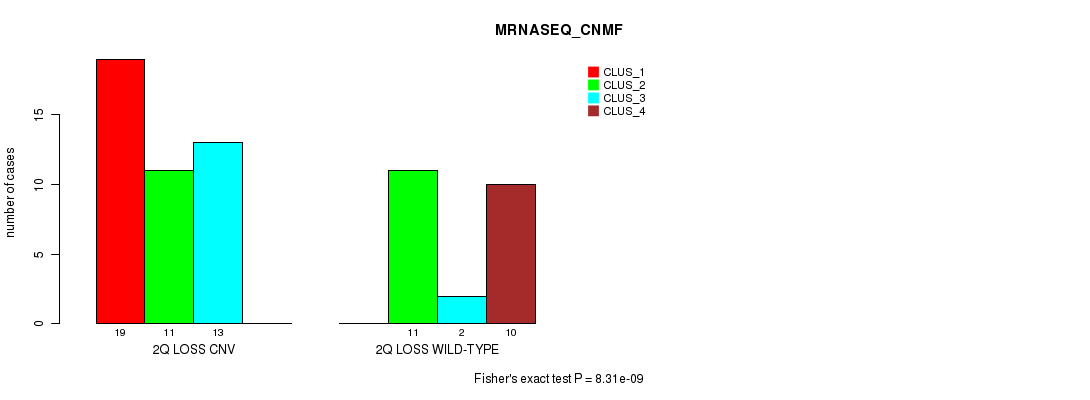
P value = 2.26e-07 (Chi-square test), Q value = 7.6e-05
Table S25. Gene #33: '2q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 2Q LOSS CNV | 10 | 0 | 8 | 3 | 22 |
| 2Q LOSS WILD-TYPE | 0 | 10 | 9 | 3 | 1 |
Figure S25. Get High-res Image Gene #33: '2q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
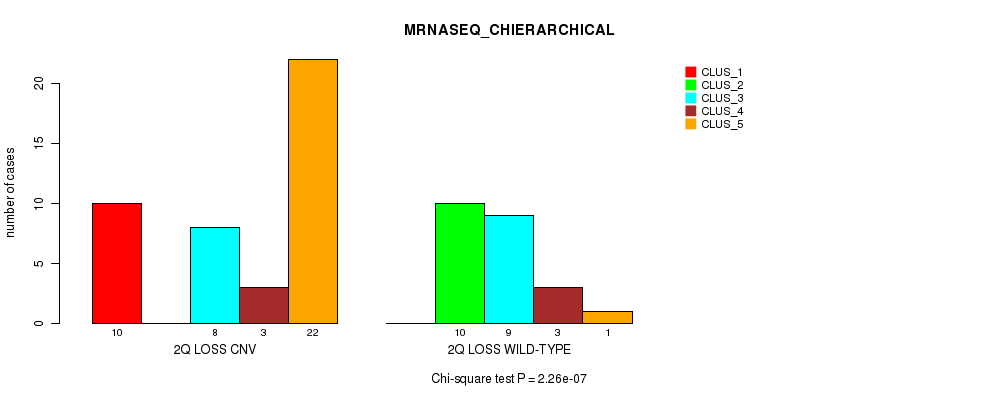
P value = 0.000592 (Fisher's exact test), Q value = 0.17
Table S26. Gene #33: '2q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 9 | 57 |
| 2Q LOSS CNV | 1 | 42 |
| 2Q LOSS WILD-TYPE | 8 | 15 |
Figure S26. Get High-res Image Gene #33: '2q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
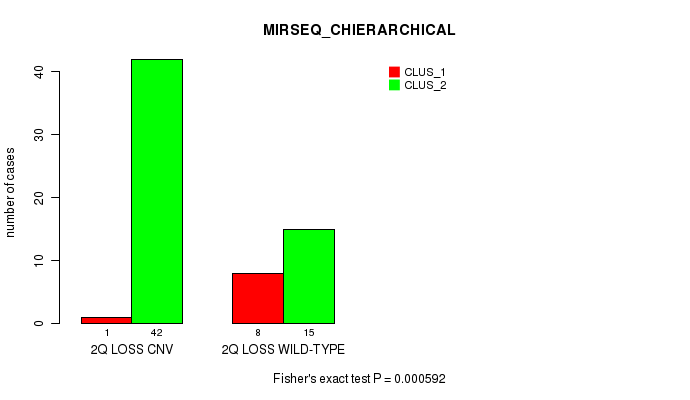
P value = 0.000132 (Fisher's exact test), Q value = 0.04
Table S27. Gene #38: '6p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 34 | 14 |
| 6P LOSS CNV | 16 | 27 | 4 |
| 6P LOSS WILD-TYPE | 1 | 7 | 10 |
Figure S27. Get High-res Image Gene #38: '6p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
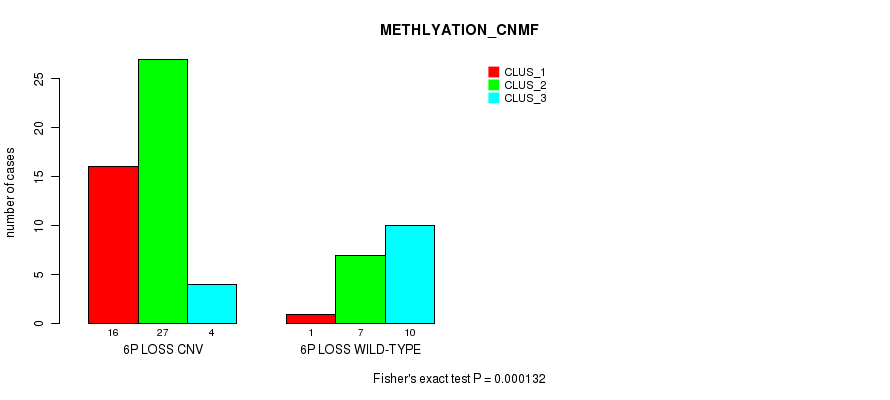
P value = 1.26e-06 (Fisher's exact test), Q value = 0.00042
Table S28. Gene #38: '6p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 6P LOSS CNV | 19 | 12 | 14 | 2 |
| 6P LOSS WILD-TYPE | 0 | 10 | 1 | 8 |
Figure S28. Get High-res Image Gene #38: '6p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
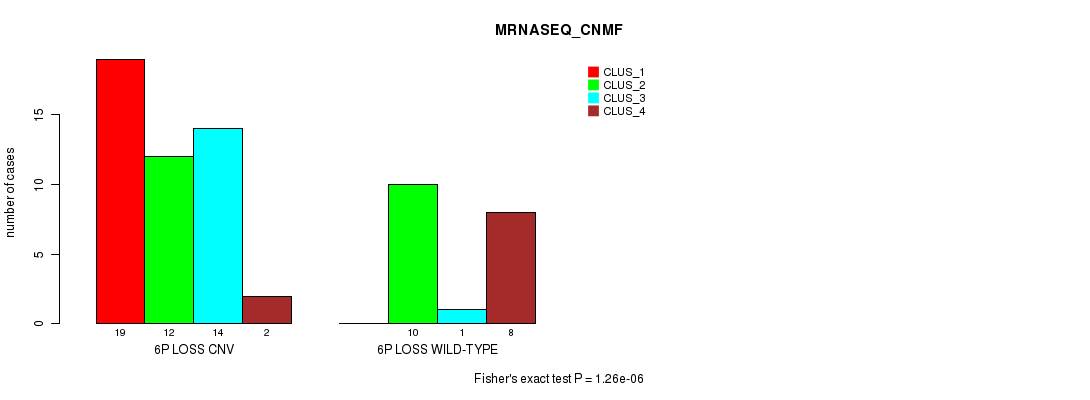
P value = 2.67e-05 (Chi-square test), Q value = 0.0084
Table S29. Gene #38: '6p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 6P LOSS CNV | 10 | 2 | 9 | 4 | 22 |
| 6P LOSS WILD-TYPE | 0 | 8 | 8 | 2 | 1 |
Figure S29. Get High-res Image Gene #38: '6p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
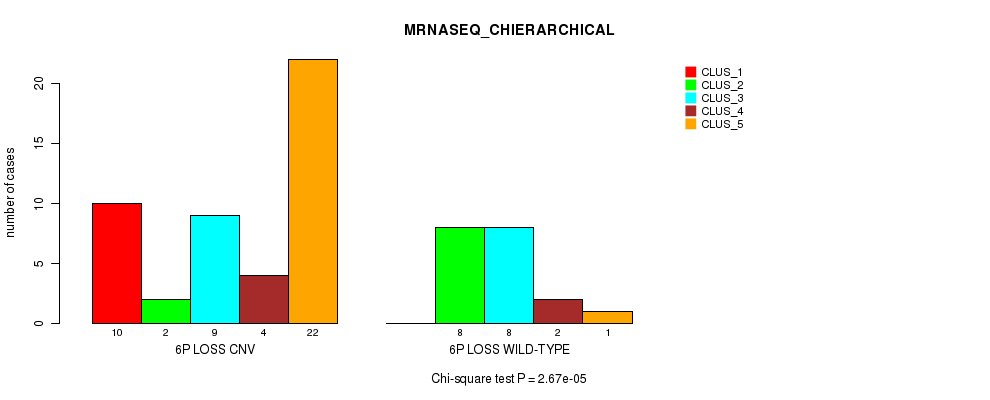
P value = 0.000132 (Fisher's exact test), Q value = 0.04
Table S30. Gene #39: '6q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 34 | 14 |
| 6Q LOSS CNV | 16 | 27 | 4 |
| 6Q LOSS WILD-TYPE | 1 | 7 | 10 |
Figure S30. Get High-res Image Gene #39: '6q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
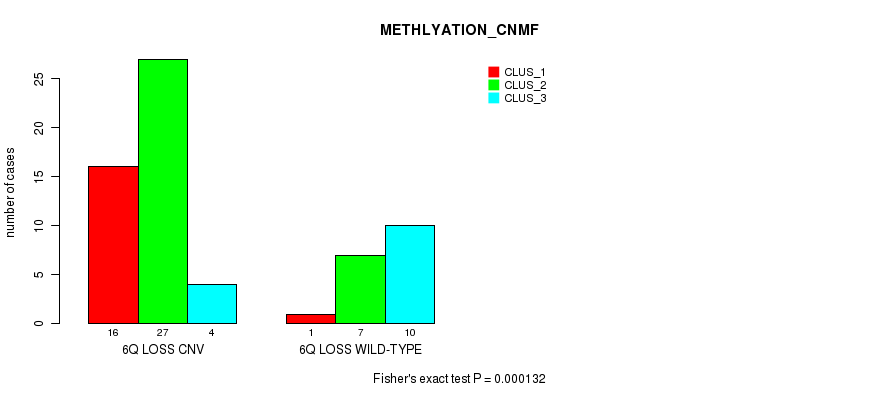
P value = 1.26e-06 (Fisher's exact test), Q value = 0.00042
Table S31. Gene #39: '6q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 6Q LOSS CNV | 19 | 12 | 14 | 2 |
| 6Q LOSS WILD-TYPE | 0 | 10 | 1 | 8 |
Figure S31. Get High-res Image Gene #39: '6q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
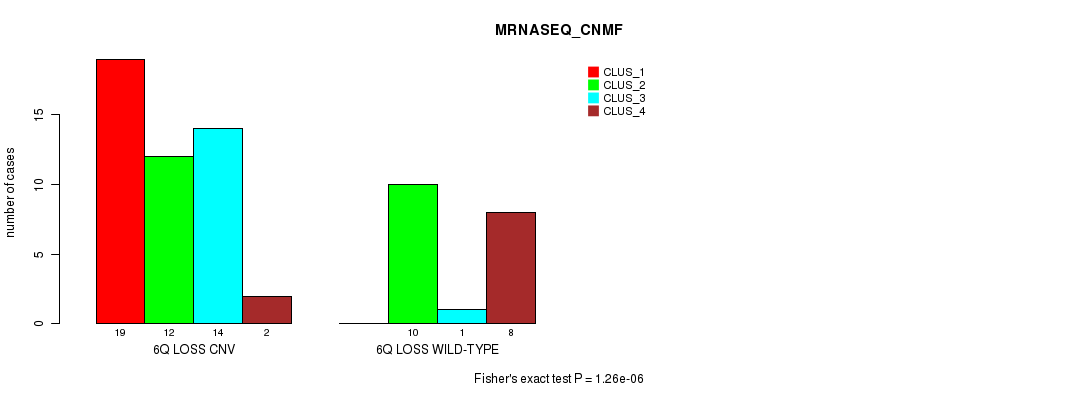
P value = 2.67e-05 (Chi-square test), Q value = 0.0084
Table S32. Gene #39: '6q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 6Q LOSS CNV | 10 | 2 | 9 | 4 | 22 |
| 6Q LOSS WILD-TYPE | 0 | 8 | 8 | 2 | 1 |
Figure S32. Get High-res Image Gene #39: '6q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
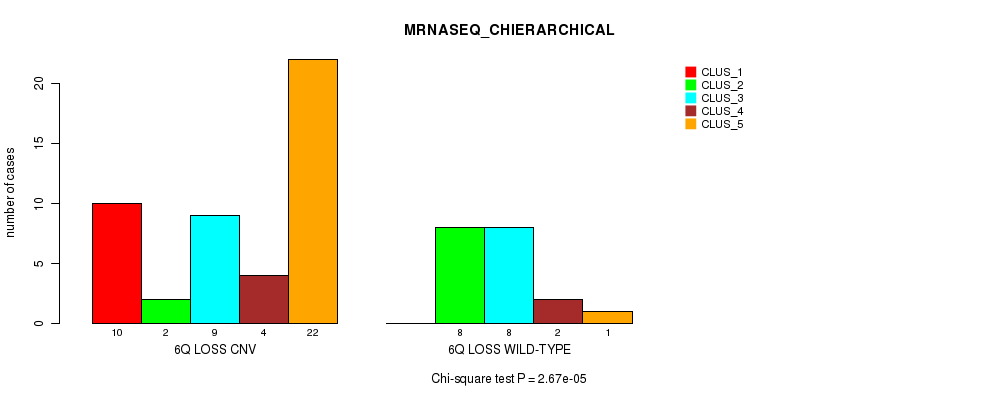
P value = 8.11e-08 (Fisher's exact test), Q value = 2.7e-05
Table S33. Gene #40: '8p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 5 | 47 | 14 |
| 8P LOSS CNV | 0 | 0 | 9 |
| 8P LOSS WILD-TYPE | 5 | 47 | 5 |
Figure S33. Get High-res Image Gene #40: '8p loss' versus Molecular Subtype #1: 'CN_CNMF'
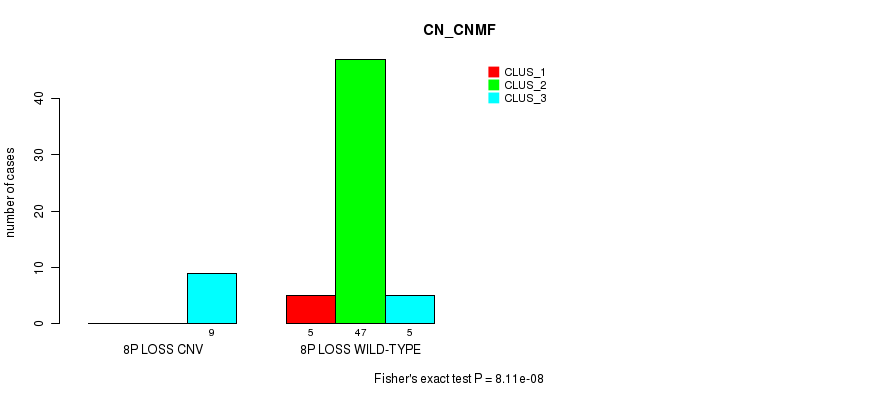
P value = 5.86e-07 (Fisher's exact test), Q value = 0.00019
Table S34. Gene #41: '8q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 5 | 47 | 14 |
| 8Q LOSS CNV | 0 | 0 | 8 |
| 8Q LOSS WILD-TYPE | 5 | 47 | 6 |
Figure S34. Get High-res Image Gene #41: '8q loss' versus Molecular Subtype #1: 'CN_CNMF'
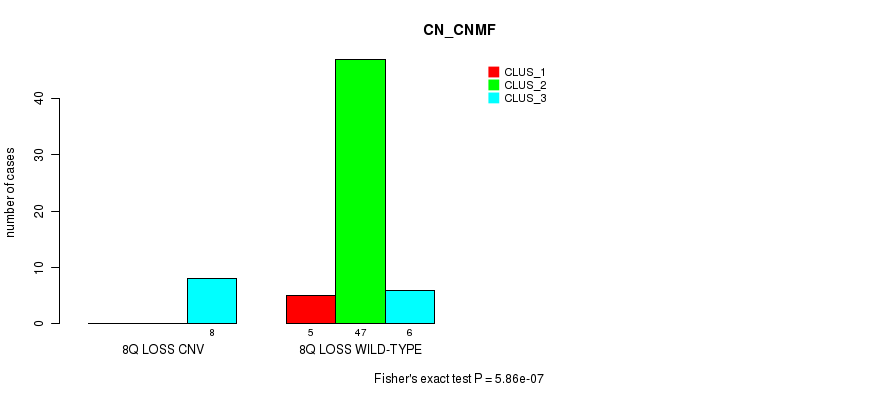
P value = 2.51e-06 (Fisher's exact test), Q value = 0.00082
Table S35. Gene #44: '10p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 10P LOSS CNV | 19 | 9 | 13 | 3 |
| 10P LOSS WILD-TYPE | 0 | 13 | 2 | 7 |
Figure S35. Get High-res Image Gene #44: '10p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
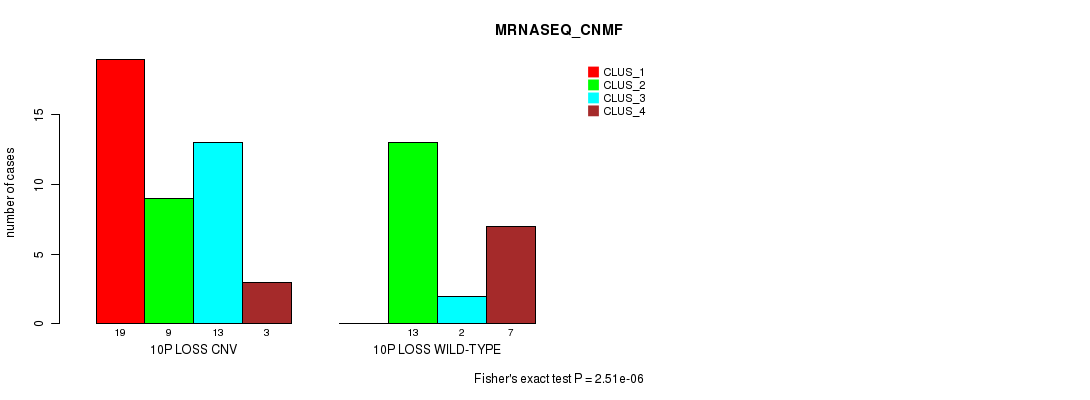
P value = 3.03e-06 (Chi-square test), Q value = 0.00098
Table S36. Gene #44: '10p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 10P LOSS CNV | 10 | 2 | 6 | 4 | 22 |
| 10P LOSS WILD-TYPE | 0 | 8 | 11 | 2 | 1 |
Figure S36. Get High-res Image Gene #44: '10p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
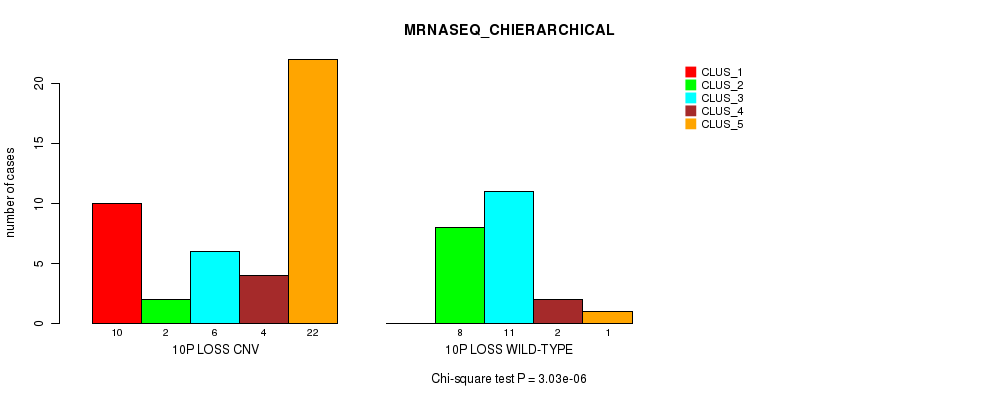
P value = 2.51e-06 (Fisher's exact test), Q value = 0.00082
Table S37. Gene #45: '10q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 10Q LOSS CNV | 19 | 9 | 13 | 3 |
| 10Q LOSS WILD-TYPE | 0 | 13 | 2 | 7 |
Figure S37. Get High-res Image Gene #45: '10q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
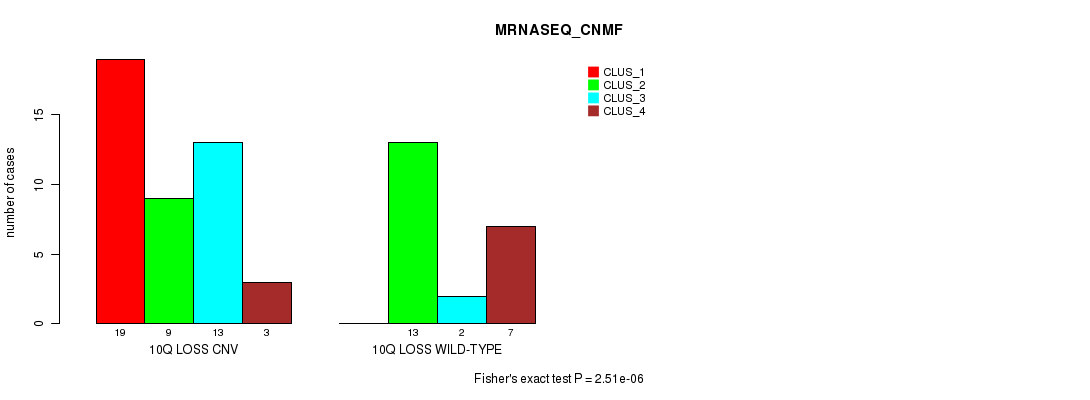
P value = 3.03e-06 (Chi-square test), Q value = 0.00098
Table S38. Gene #45: '10q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 10Q LOSS CNV | 10 | 2 | 6 | 4 | 22 |
| 10Q LOSS WILD-TYPE | 0 | 8 | 11 | 2 | 1 |
Figure S38. Get High-res Image Gene #45: '10q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
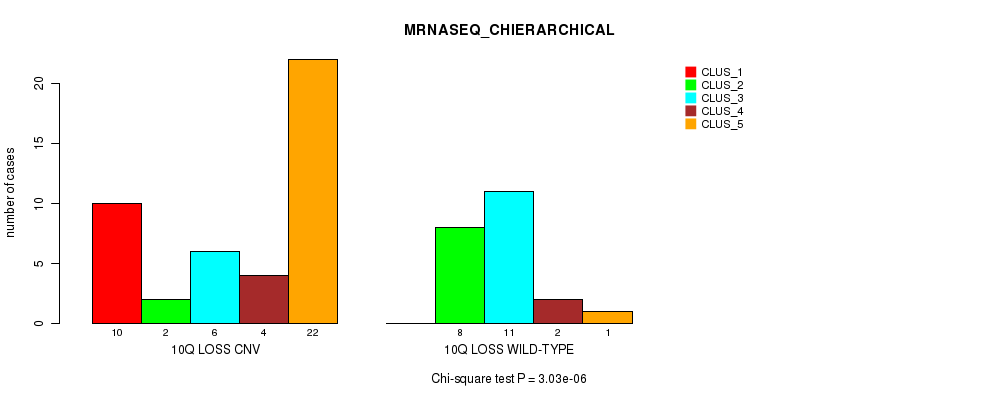
P value = 0.000506 (Chi-square test), Q value = 0.15
Table S39. Gene #48: '13q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 13Q LOSS CNV | 10 | 1 | 8 | 2 | 16 |
| 13Q LOSS WILD-TYPE | 0 | 9 | 9 | 4 | 7 |
Figure S39. Get High-res Image Gene #48: '13q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
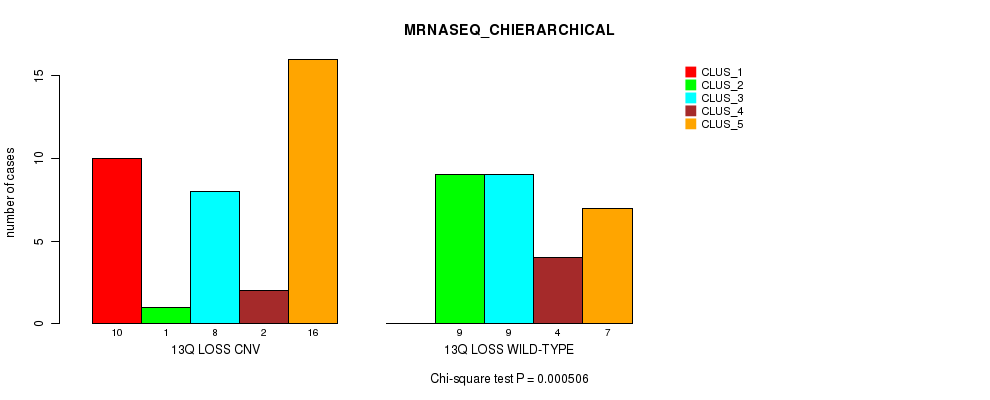
P value = 8.58e-05 (Chi-square test), Q value = 0.026
Table S40. Gene #50: '16q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 16Q LOSS CNV | 4 | 0 | 0 | 0 | 0 |
| 16Q LOSS WILD-TYPE | 6 | 10 | 17 | 6 | 23 |
Figure S40. Get High-res Image Gene #50: '16q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
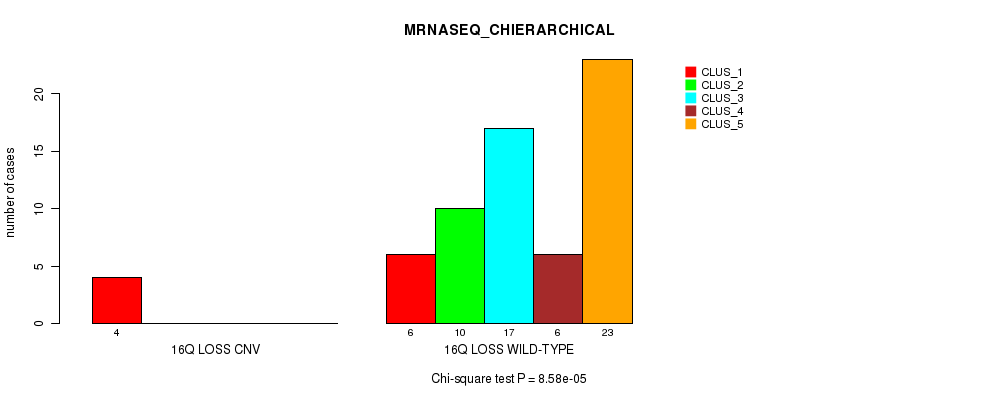
P value = 1.31e-06 (Fisher's exact test), Q value = 0.00043
Table S41. Gene #51: '17p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 34 | 14 |
| 17P LOSS CNV | 16 | 27 | 2 |
| 17P LOSS WILD-TYPE | 1 | 7 | 12 |
Figure S41. Get High-res Image Gene #51: '17p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
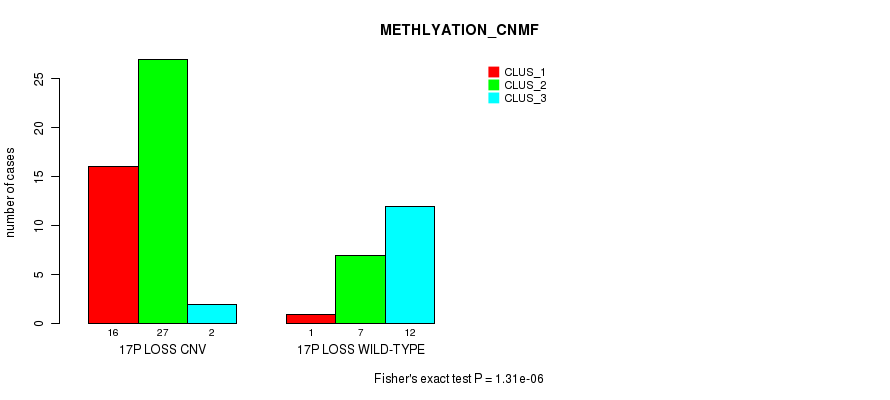
P value = 2.61e-09 (Fisher's exact test), Q value = 8.9e-07
Table S42. Gene #51: '17p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 17P LOSS CNV | 19 | 12 | 14 | 0 |
| 17P LOSS WILD-TYPE | 0 | 10 | 1 | 10 |
Figure S42. Get High-res Image Gene #51: '17p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
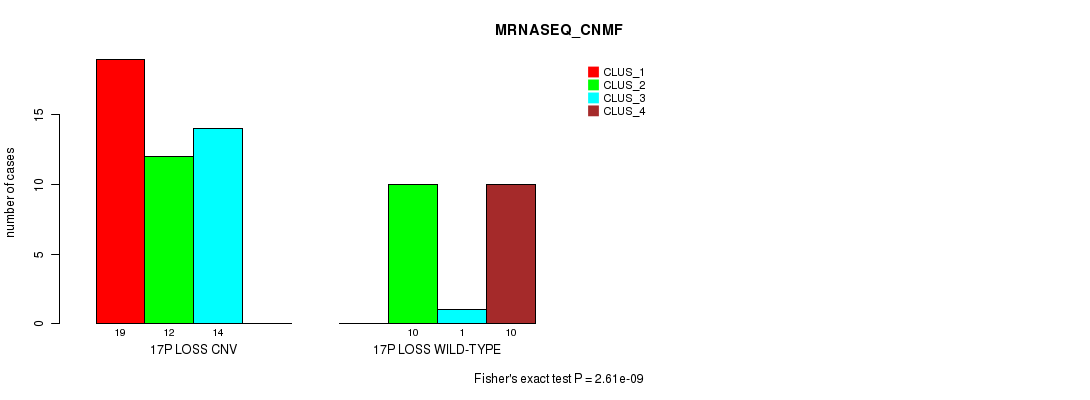
P value = 3e-07 (Chi-square test), Q value = 1e-04
Table S43. Gene #51: '17p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 17P LOSS CNV | 10 | 0 | 9 | 4 | 22 |
| 17P LOSS WILD-TYPE | 0 | 10 | 8 | 2 | 1 |
Figure S43. Get High-res Image Gene #51: '17p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
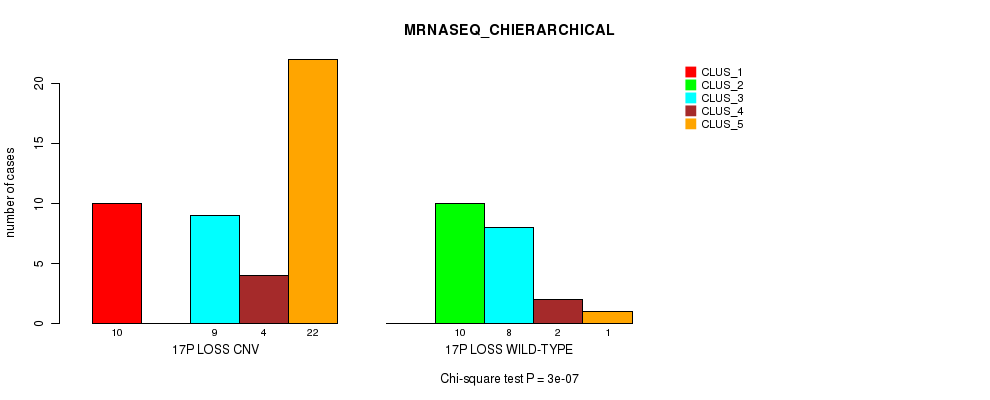
P value = 0.000255 (Fisher's exact test), Q value = 0.076
Table S44. Gene #51: '17p loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 9 | 57 |
| 17P LOSS CNV | 1 | 44 |
| 17P LOSS WILD-TYPE | 8 | 13 |
Figure S44. Get High-res Image Gene #51: '17p loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
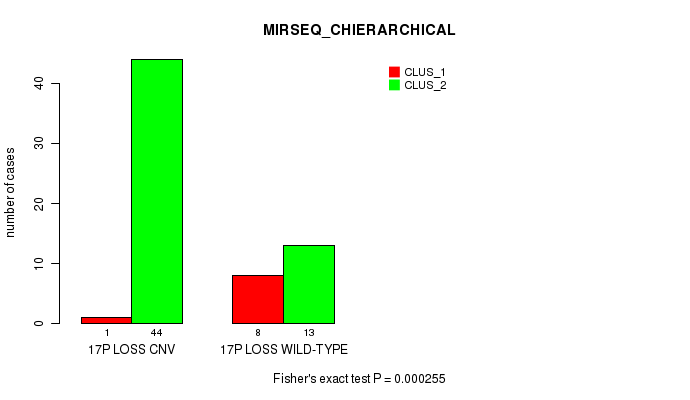
P value = 1.31e-06 (Fisher's exact test), Q value = 0.00043
Table S45. Gene #52: '17q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 34 | 14 |
| 17Q LOSS CNV | 16 | 27 | 2 |
| 17Q LOSS WILD-TYPE | 1 | 7 | 12 |
Figure S45. Get High-res Image Gene #52: '17q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
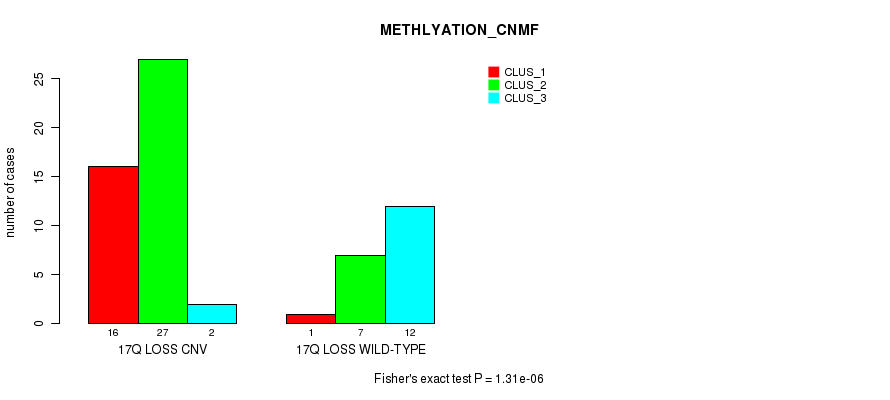
P value = 2.61e-09 (Fisher's exact test), Q value = 8.9e-07
Table S46. Gene #52: '17q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 17Q LOSS CNV | 19 | 12 | 14 | 0 |
| 17Q LOSS WILD-TYPE | 0 | 10 | 1 | 10 |
Figure S46. Get High-res Image Gene #52: '17q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
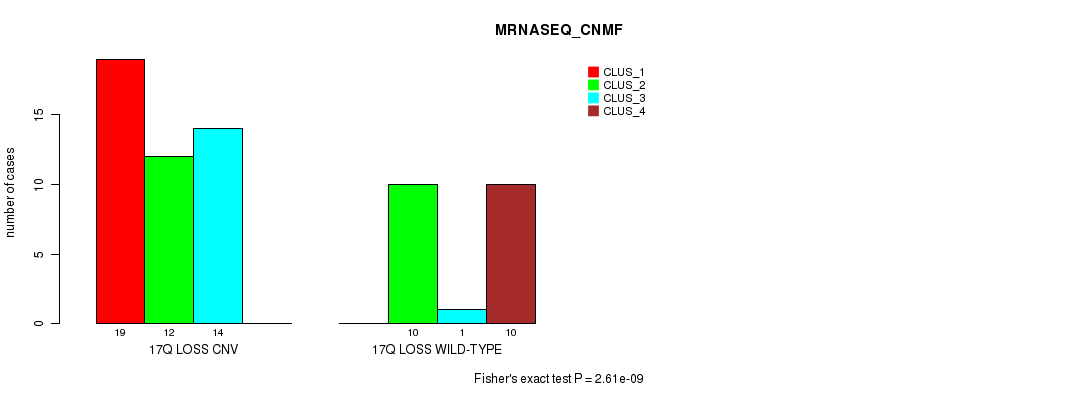
P value = 3e-07 (Chi-square test), Q value = 1e-04
Table S47. Gene #52: '17q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 17Q LOSS CNV | 10 | 0 | 9 | 4 | 22 |
| 17Q LOSS WILD-TYPE | 0 | 10 | 8 | 2 | 1 |
Figure S47. Get High-res Image Gene #52: '17q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
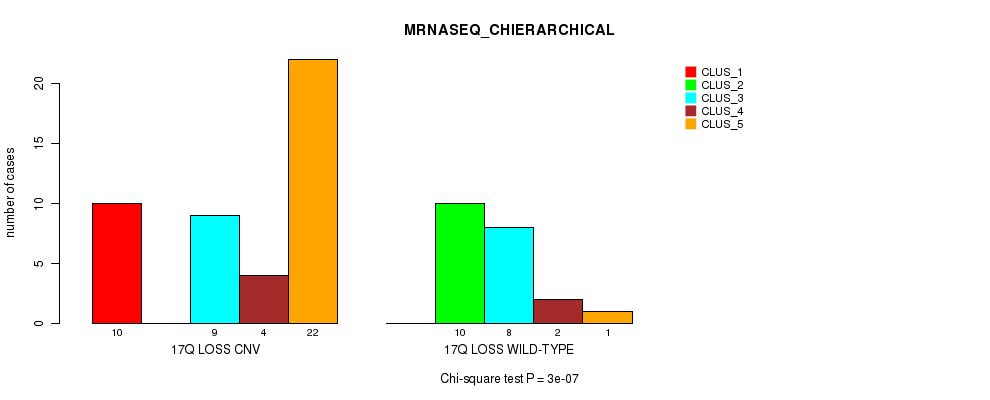
P value = 0.000255 (Fisher's exact test), Q value = 0.076
Table S48. Gene #52: '17q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 9 | 57 |
| 17Q LOSS CNV | 1 | 44 |
| 17Q LOSS WILD-TYPE | 8 | 13 |
Figure S48. Get High-res Image Gene #52: '17q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
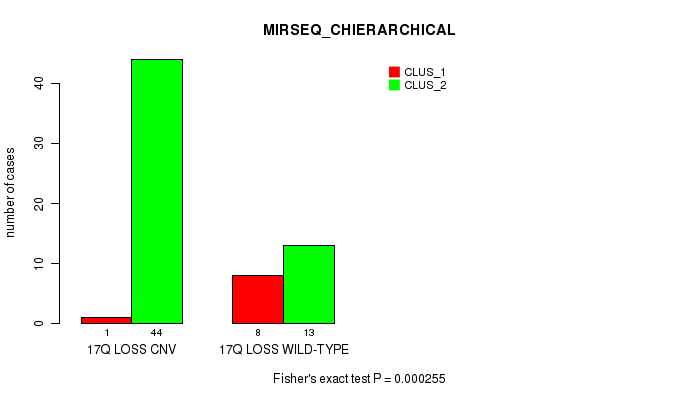
P value = 2.45e-05 (Fisher's exact test), Q value = 0.0078
Table S49. Gene #57: 'Xq loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 34 | 14 |
| XQ LOSS CNV | 12 | 21 | 0 |
| XQ LOSS WILD-TYPE | 5 | 13 | 14 |
Figure S49. Get High-res Image Gene #57: 'Xq loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
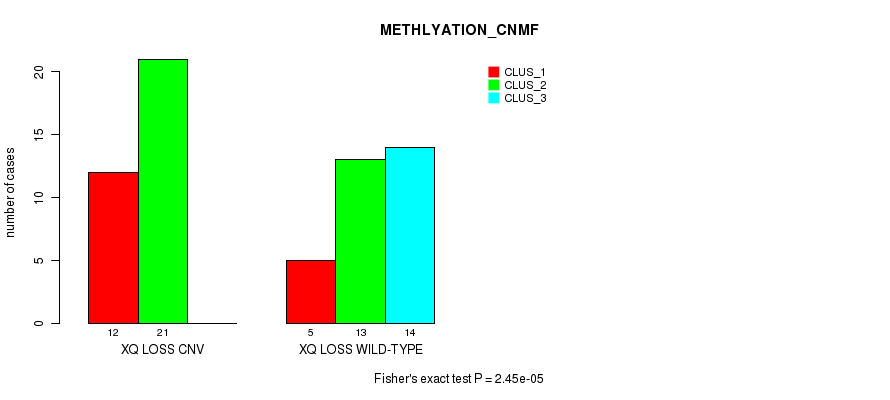
P value = 7.96e-05 (Fisher's exact test), Q value = 0.025
Table S50. Gene #57: 'Xq loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| XQ LOSS CNV | 15 | 8 | 10 | 0 |
| XQ LOSS WILD-TYPE | 4 | 14 | 5 | 10 |
Figure S50. Get High-res Image Gene #57: 'Xq loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
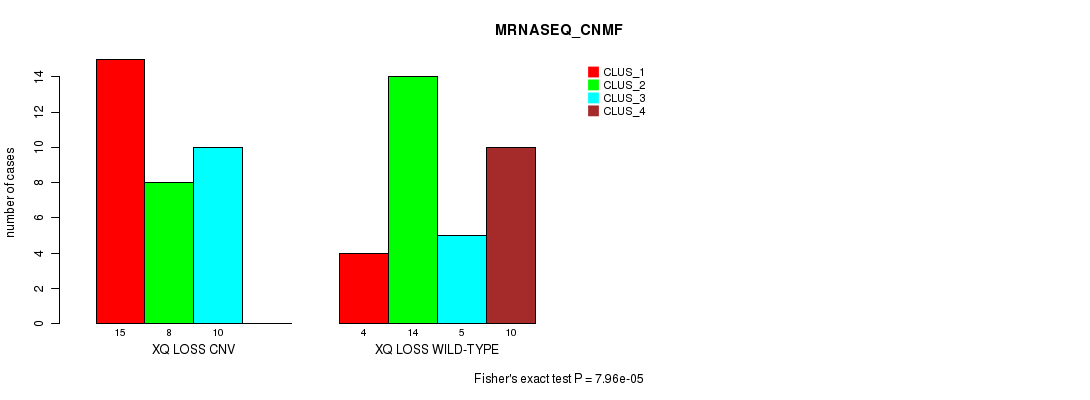
P value = 0.000217 (Chi-square test), Q value = 0.065
Table S51. Gene #57: 'Xq loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| XQ LOSS CNV | 7 | 0 | 5 | 3 | 18 |
| XQ LOSS WILD-TYPE | 3 | 10 | 12 | 3 | 5 |
Figure S51. Get High-res Image Gene #57: 'Xq loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
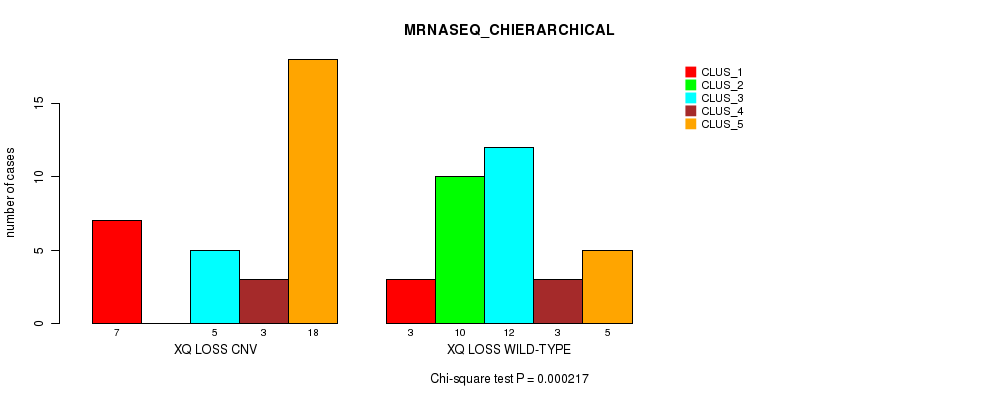
-
Mutation data file = broad_values_by_arm.mutsig.cluster.txt
-
Molecular subtypes file = KICH-TP.transferedmergedcluster.txt
-
Number of patients = 66
-
Number of significantly arm-level cnvs = 57
-
Number of molecular subtypes = 6
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.