This pipeline computes the correlation between significant copy number variation (cnv focal) genes and selected clinical features.
Testing the association between copy number variation 62 arm-level results and 11 clinical features across 162 patients, 2 significant findings detected with Q value < 0.25.
-
Del Peak 6(3q26.31) cnv correlated to 'Time to Death'.
-
Del Peak 30(18q21.2) cnv correlated to 'NUMBER.OF.LYMPH.NODES'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 62 arm-level results and 11 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 2 significant findings detected.
|
Clinical Features |
Time to Death |
AGE | GENDER |
HISTOLOGICAL TYPE |
PATHOLOGY T |
PATHOLOGY N |
PATHOLOGICSPREAD(M) |
TUMOR STAGE |
RADIATIONS RADIATION REGIMENINDICATION |
COMPLETENESS OF RESECTION |
NUMBER OF LYMPH NODES |
||
| nCNV (%) | nWild-Type | logrank test | t-test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | t-test | |
| Del Peak 6(3q26 31) | 0 (0%) | 145 |
8.16e-05 (0.0556) |
0.705 (1.00) |
0.0036 (1.00) |
0.0303 (1.00) |
0.216 (1.00) |
0.573 (1.00) |
0.2 (1.00) |
0.00346 (1.00) |
0.121 (1.00) |
0.0387 (1.00) |
0.251 (1.00) |
| Del Peak 30(18q21 2) | 0 (0%) | 19 |
0.653 (1.00) |
0.443 (1.00) |
1 (1.00) |
1 (1.00) |
0.00589 (1.00) |
0.0568 (1.00) |
0.208 (1.00) |
0.0231 (1.00) |
1 (1.00) |
0.673 (1.00) |
0.000153 (0.104) |
| Amp Peak 1(1q21 3) | 0 (0%) | 114 |
0.538 (1.00) |
0.951 (1.00) |
0.863 (1.00) |
1 (1.00) |
0.121 (1.00) |
0.441 (1.00) |
0.106 (1.00) |
0.835 (1.00) |
0.0639 (1.00) |
0.691 (1.00) |
0.227 (1.00) |
| Amp Peak 2(1q32 2) | 0 (0%) | 114 |
0.0492 (1.00) |
0.645 (1.00) |
1 (1.00) |
1 (1.00) |
0.0297 (1.00) |
0.264 (1.00) |
0.357 (1.00) |
0.604 (1.00) |
0.362 (1.00) |
0.64 (1.00) |
0.0174 (1.00) |
| Amp Peak 3(5q22 3) | 0 (0%) | 143 |
0.529 (1.00) |
0.383 (1.00) |
0.81 (1.00) |
0.37 (1.00) |
0.881 (1.00) |
0.772 (1.00) |
0.743 (1.00) |
0.784 (1.00) |
1 (1.00) |
0.806 (1.00) |
0.372 (1.00) |
| Amp Peak 4(6p21 1) | 0 (0%) | 110 |
0.178 (1.00) |
0.528 (1.00) |
0.4 (1.00) |
0.0626 (1.00) |
0.428 (1.00) |
0.00855 (1.00) |
0.00762 (1.00) |
0.0299 (1.00) |
0.0844 (1.00) |
0.0957 (1.00) |
0.0836 (1.00) |
| Amp Peak 5(8p11 23) | 0 (0%) | 110 |
0.0852 (1.00) |
0.264 (1.00) |
0.0635 (1.00) |
0.222 (1.00) |
0.216 (1.00) |
0.457 (1.00) |
0.305 (1.00) |
0.223 (1.00) |
1 (1.00) |
0.0229 (1.00) |
0.289 (1.00) |
| Amp Peak 6(8p11 21) | 0 (0%) | 93 |
0.478 (1.00) |
0.541 (1.00) |
0.637 (1.00) |
0.154 (1.00) |
0.408 (1.00) |
0.814 (1.00) |
0.191 (1.00) |
0.074 (1.00) |
0.241 (1.00) |
0.00288 (1.00) |
0.574 (1.00) |
| Amp Peak 7(8q24 21) | 0 (0%) | 55 |
0.693 (1.00) |
0.228 (1.00) |
0.868 (1.00) |
0.0326 (1.00) |
0.49 (1.00) |
0.671 (1.00) |
0.533 (1.00) |
0.284 (1.00) |
0.665 (1.00) |
0.372 (1.00) |
0.763 (1.00) |
| Amp Peak 8(10q22 2) | 0 (0%) | 151 |
0.306 (1.00) |
0.686 (1.00) |
0.551 (1.00) |
0.592 (1.00) |
0.935 (1.00) |
0.914 (1.00) |
0.743 (1.00) |
0.969 (1.00) |
0.349 (1.00) |
1 (1.00) |
0.939 (1.00) |
| Amp Peak 9(11p15 5) | 0 (0%) | 119 |
0.607 (1.00) |
0.969 (1.00) |
1 (1.00) |
0.188 (1.00) |
0.907 (1.00) |
0.195 (1.00) |
0.803 (1.00) |
0.258 (1.00) |
0.656 (1.00) |
0.938 (1.00) |
0.685 (1.00) |
| Amp Peak 10(11q13 3) | 0 (0%) | 136 |
0.456 (1.00) |
0.0577 (1.00) |
1 (1.00) |
0.697 (1.00) |
0.494 (1.00) |
0.361 (1.00) |
0.288 (1.00) |
0.104 (1.00) |
0.591 (1.00) |
0.66 (1.00) |
0.0272 (1.00) |
| Amp Peak 11(12p13 33) | 0 (0%) | 121 |
0.984 (1.00) |
0.246 (1.00) |
0.589 (1.00) |
0.308 (1.00) |
0.418 (1.00) |
0.546 (1.00) |
0.105 (1.00) |
0.633 (1.00) |
0.171 (1.00) |
0.274 (1.00) |
0.339 (1.00) |
| Amp Peak 12(12p12 1) | 0 (0%) | 122 |
0.702 (1.00) |
0.334 (1.00) |
0.716 (1.00) |
0.191 (1.00) |
0.373 (1.00) |
0.516 (1.00) |
0.0872 (1.00) |
0.446 (1.00) |
0.161 (1.00) |
0.219 (1.00) |
0.102 (1.00) |
| Amp Peak 13(13q12 13) | 0 (0%) | 41 |
0.0417 (1.00) |
0.113 (1.00) |
0.718 (1.00) |
0.00387 (1.00) |
0.068 (1.00) |
0.0187 (1.00) |
0.254 (1.00) |
0.258 (1.00) |
0.171 (1.00) |
1 (1.00) |
0.524 (1.00) |
| Amp Peak 14(13q12 2) | 0 (0%) | 42 |
0.052 (1.00) |
0.0514 (1.00) |
0.369 (1.00) |
0.00467 (1.00) |
0.0663 (1.00) |
0.027 (1.00) |
0.306 (1.00) |
0.307 (1.00) |
0.181 (1.00) |
1 (1.00) |
0.524 (1.00) |
| Amp Peak 15(13q22 1) | 0 (0%) | 45 |
0.202 (1.00) |
0.337 (1.00) |
0.482 (1.00) |
0.00132 (0.894) |
0.202 (1.00) |
0.139 (1.00) |
0.289 (1.00) |
0.433 (1.00) |
0.67 (1.00) |
0.801 (1.00) |
0.861 (1.00) |
| Amp Peak 16(15q26 1) | 0 (0%) | 147 |
0.854 (1.00) |
0.944 (1.00) |
0.106 (1.00) |
0.362 (1.00) |
0.221 (1.00) |
0.423 (1.00) |
0.446 (1.00) |
0.572 (1.00) |
0.0967 (1.00) |
0.375 (1.00) |
0.487 (1.00) |
| Amp Peak 17(16p11 2) | 0 (0%) | 117 |
0.183 (1.00) |
0.323 (1.00) |
0.862 (1.00) |
0.0205 (1.00) |
0.607 (1.00) |
0.146 (1.00) |
0.0837 (1.00) |
0.371 (1.00) |
0.67 (1.00) |
0.0597 (1.00) |
0.907 (1.00) |
| Amp Peak 18(17q12) | 0 (0%) | 119 |
0.653 (1.00) |
0.676 (1.00) |
0.215 (1.00) |
0.517 (1.00) |
0.725 (1.00) |
0.364 (1.00) |
0.335 (1.00) |
0.429 (1.00) |
0.343 (1.00) |
0.173 (1.00) |
0.31 (1.00) |
| Amp Peak 19(17q24 1) | 0 (0%) | 125 |
0.659 (1.00) |
0.423 (1.00) |
1 (1.00) |
0.0401 (1.00) |
0.258 (1.00) |
0.266 (1.00) |
0.192 (1.00) |
0.204 (1.00) |
0.338 (1.00) |
0.221 (1.00) |
0.206 (1.00) |
| Amp Peak 20(19p13 2) | 0 (0%) | 130 |
0.0245 (1.00) |
0.451 (1.00) |
0.693 (1.00) |
0.0717 (1.00) |
0.228 (1.00) |
0.545 (1.00) |
0.578 (1.00) |
0.709 (1.00) |
0.599 (1.00) |
0.511 (1.00) |
0.554 (1.00) |
| Amp Peak 21(20q11 21) | 0 (0%) | 19 |
0.209 (1.00) |
0.099 (1.00) |
1 (1.00) |
0.000738 (0.499) |
0.55 (1.00) |
0.0956 (1.00) |
0.547 (1.00) |
0.0969 (1.00) |
0.533 (1.00) |
0.581 (1.00) |
0.486 (1.00) |
| Amp Peak 22(20q11 23) | 0 (0%) | 18 |
0.211 (1.00) |
0.179 (1.00) |
1 (1.00) |
0.000498 (0.338) |
0.603 (1.00) |
0.0293 (1.00) |
0.433 (1.00) |
0.0857 (1.00) |
0.513 (1.00) |
0.581 (1.00) |
0.486 (1.00) |
| Amp Peak 23(20q13 31) | 0 (0%) | 17 |
0.217 (1.00) |
0.239 (1.00) |
0.8 (1.00) |
0.000498 (0.338) |
0.614 (1.00) |
0.0489 (1.00) |
0.493 (1.00) |
0.139 (1.00) |
0.492 (1.00) |
0.55 (1.00) |
0.446 (1.00) |
| Amp Peak 24(Xp22 2) | 0 (0%) | 129 |
0.945 (1.00) |
0.427 (1.00) |
0.0771 (1.00) |
0.303 (1.00) |
0.699 (1.00) |
0.654 (1.00) |
0.465 (1.00) |
0.598 (1.00) |
0.348 (1.00) |
0.273 (1.00) |
0.43 (1.00) |
| Amp Peak 25(Xq27 3) | 0 (0%) | 124 |
0.459 (1.00) |
0.225 (1.00) |
0.356 (1.00) |
0.3 (1.00) |
0.35 (1.00) |
0.743 (1.00) |
0.667 (1.00) |
0.645 (1.00) |
0.337 (1.00) |
0.685 (1.00) |
0.806 (1.00) |
| Del Peak 1(1p36 11) | 0 (0%) | 91 |
0.939 (1.00) |
0.617 (1.00) |
0.751 (1.00) |
0.56 (1.00) |
0.102 (1.00) |
0.215 (1.00) |
0.393 (1.00) |
0.643 (1.00) |
1 (1.00) |
0.327 (1.00) |
0.0468 (1.00) |
| Del Peak 2(1p33) | 0 (0%) | 113 |
0.522 (1.00) |
0.652 (1.00) |
1 (1.00) |
0.11 (1.00) |
0.528 (1.00) |
0.229 (1.00) |
0.76 (1.00) |
0.637 (1.00) |
0.368 (1.00) |
0.547 (1.00) |
0.649 (1.00) |
| Del Peak 3(1p13 2) | 0 (0%) | 111 |
0.944 (1.00) |
0.646 (1.00) |
0.735 (1.00) |
0.347 (1.00) |
0.147 (1.00) |
0.779 (1.00) |
0.499 (1.00) |
0.182 (1.00) |
0.38 (1.00) |
0.688 (1.00) |
0.245 (1.00) |
| Del Peak 4(2p12) | 0 (0%) | 145 |
0.498 (1.00) |
0.175 (1.00) |
0.8 (1.00) |
1 (1.00) |
0.364 (1.00) |
0.653 (1.00) |
0.285 (1.00) |
0.252 (1.00) |
0.492 (1.00) |
1 (1.00) |
0.471 (1.00) |
| Del Peak 5(3p14 2) | 0 (0%) | 133 |
0.052 (1.00) |
0.811 (1.00) |
0.013 (1.00) |
0.456 (1.00) |
0.196 (1.00) |
0.943 (1.00) |
0.285 (1.00) |
0.0402 (1.00) |
0.292 (1.00) |
0.0853 (1.00) |
0.367 (1.00) |
| Del Peak 7(4p13) | 0 (0%) | 104 |
0.215 (1.00) |
0.877 (1.00) |
0.0996 (1.00) |
0.38 (1.00) |
0.748 (1.00) |
0.327 (1.00) |
0.961 (1.00) |
0.673 (1.00) |
0.00177 (1.00) |
0.399 (1.00) |
0.675 (1.00) |
| Del Peak 8(4q22 1) | 0 (0%) | 90 |
0.107 (1.00) |
0.378 (1.00) |
0.153 (1.00) |
0.393 (1.00) |
0.287 (1.00) |
0.714 (1.00) |
0.857 (1.00) |
0.622 (1.00) |
0.0892 (1.00) |
0.215 (1.00) |
0.078 (1.00) |
| Del Peak 9(4q35 1) | 0 (0%) | 90 |
0.294 (1.00) |
0.645 (1.00) |
0.428 (1.00) |
0.15 (1.00) |
0.445 (1.00) |
0.56 (1.00) |
0.957 (1.00) |
0.791 (1.00) |
0.0892 (1.00) |
0.118 (1.00) |
0.306 (1.00) |
| Del Peak 10(5q11 2) | 0 (0%) | 113 |
0.708 (1.00) |
0.836 (1.00) |
0.233 (1.00) |
0.347 (1.00) |
0.927 (1.00) |
0.871 (1.00) |
0.336 (1.00) |
0.253 (1.00) |
0.01 (1.00) |
0.888 (1.00) |
0.708 (1.00) |
| Del Peak 11(5q22 2) | 0 (0%) | 107 |
0.58 (1.00) |
0.56 (1.00) |
0.405 (1.00) |
0.222 (1.00) |
0.495 (1.00) |
0.423 (1.00) |
0.903 (1.00) |
0.565 (1.00) |
0.0176 (1.00) |
1 (1.00) |
0.374 (1.00) |
| Del Peak 12(6p25 3) | 0 (0%) | 138 |
0.409 (1.00) |
0.391 (1.00) |
0.825 (1.00) |
0.386 (1.00) |
0.213 (1.00) |
0.38 (1.00) |
0.67 (1.00) |
0.08 (1.00) |
0.217 (1.00) |
0.66 (1.00) |
0.979 (1.00) |
| Del Peak 13(6q26) | 0 (0%) | 132 |
0.4 (1.00) |
0.852 (1.00) |
0.158 (1.00) |
0.0138 (1.00) |
0.392 (1.00) |
0.0813 (1.00) |
0.677 (1.00) |
0.533 (1.00) |
0.308 (1.00) |
0.588 (1.00) |
0.217 (1.00) |
| Del Peak 14(7q31 1) | 0 (0%) | 158 |
0.174 (1.00) |
1 (1.00) |
0.296 (1.00) |
1 (1.00) |
0.528 (1.00) |
1 (1.00) |
0.919 (1.00) |
1 (1.00) |
1 (1.00) |
0.548 (1.00) |
|
| Del Peak 15(8p23 2) | 0 (0%) | 62 |
0.71 (1.00) |
0.494 (1.00) |
0.872 (1.00) |
0.564 (1.00) |
0.201 (1.00) |
0.537 (1.00) |
0.799 (1.00) |
0.79 (1.00) |
0.0831 (1.00) |
0.386 (1.00) |
0.112 (1.00) |
| Del Peak 16(8p11 21) | 0 (0%) | 138 |
0.847 (1.00) |
0.184 (1.00) |
0.506 (1.00) |
1 (1.00) |
0.755 (1.00) |
0.812 (1.00) |
0.0829 (1.00) |
0.328 (1.00) |
0.217 (1.00) |
0.742 (1.00) |
0.123 (1.00) |
| Del Peak 17(10q21 1) | 0 (0%) | 125 |
0.498 (1.00) |
0.261 (1.00) |
0.574 (1.00) |
0.179 (1.00) |
0.828 (1.00) |
0.568 (1.00) |
0.107 (1.00) |
0.197 (1.00) |
0.0249 (1.00) |
0.109 (1.00) |
0.806 (1.00) |
| Del Peak 18(10q25 2) | 0 (0%) | 115 |
0.422 (1.00) |
0.369 (1.00) |
0.864 (1.00) |
0.0535 (1.00) |
0.148 (1.00) |
0.904 (1.00) |
0.563 (1.00) |
0.298 (1.00) |
0.00819 (1.00) |
0.204 (1.00) |
0.914 (1.00) |
| Del Peak 19(11q22 3) | 0 (0%) | 125 |
0.298 (1.00) |
0.453 (1.00) |
0.71 (1.00) |
0.0322 (1.00) |
0.635 (1.00) |
0.941 (1.00) |
0.0617 (1.00) |
0.272 (1.00) |
0.133 (1.00) |
0.164 (1.00) |
0.225 (1.00) |
| Del Peak 20(12p13 1) | 0 (0%) | 129 |
0.0863 (1.00) |
0.894 (1.00) |
0.845 (1.00) |
0.465 (1.00) |
0.891 (1.00) |
0.136 (1.00) |
0.28 (1.00) |
0.754 (1.00) |
1 (1.00) |
0.568 (1.00) |
0.488 (1.00) |
| Del Peak 21(12q21 33) | 0 (0%) | 134 |
0.626 (1.00) |
0.292 (1.00) |
1 (1.00) |
0.469 (1.00) |
0.697 (1.00) |
0.851 (1.00) |
0.677 (1.00) |
0.254 (1.00) |
0.277 (1.00) |
0.314 (1.00) |
0.707 (1.00) |
| Del Peak 22(14q11 2) | 0 (0%) | 95 |
0.23 (1.00) |
0.427 (1.00) |
0.427 (1.00) |
0.0746 (1.00) |
0.617 (1.00) |
0.291 (1.00) |
0.724 (1.00) |
0.334 (1.00) |
0.232 (1.00) |
0.101 (1.00) |
0.501 (1.00) |
| Del Peak 23(14q31 1) | 0 (0%) | 93 |
0.607 (1.00) |
0.84 (1.00) |
0.27 (1.00) |
0.00787 (1.00) |
0.16 (1.00) |
0.0566 (1.00) |
0.606 (1.00) |
0.265 (1.00) |
1 (1.00) |
0.436 (1.00) |
0.518 (1.00) |
| Del Peak 24(15q22 33) | 0 (0%) | 87 |
0.558 (1.00) |
0.86 (1.00) |
0.344 (1.00) |
1 (1.00) |
0.0115 (1.00) |
0.262 (1.00) |
0.741 (1.00) |
0.0732 (1.00) |
0.0966 (1.00) |
0.575 (1.00) |
0.0338 (1.00) |
| Del Peak 25(16p13 3) | 0 (0%) | 149 |
0.00338 (1.00) |
0.433 (1.00) |
1 (1.00) |
0.263 (1.00) |
0.788 (1.00) |
0.181 (1.00) |
0.797 (1.00) |
0.135 (1.00) |
1 (1.00) |
1 (1.00) |
0.309 (1.00) |
| Del Peak 26(16q23 1) | 0 (0%) | 139 |
0.925 (1.00) |
0.585 (1.00) |
0.826 (1.00) |
1 (1.00) |
0.606 (1.00) |
0.0144 (1.00) |
0.841 (1.00) |
0.327 (1.00) |
1 (1.00) |
1 (1.00) |
0.072 (1.00) |
| Del Peak 27(17p12) | 0 (0%) | 52 |
0.4 (1.00) |
0.0943 (1.00) |
0.866 (1.00) |
0.122 (1.00) |
0.0153 (1.00) |
0.893 (1.00) |
0.88 (1.00) |
0.211 (1.00) |
1 (1.00) |
0.854 (1.00) |
0.73 (1.00) |
| Del Peak 28(17p11 2) | 0 (0%) | 60 |
0.775 (1.00) |
0.201 (1.00) |
0.514 (1.00) |
0.0783 (1.00) |
0.0832 (1.00) |
0.693 (1.00) |
0.357 (1.00) |
0.598 (1.00) |
0.671 (1.00) |
0.862 (1.00) |
0.689 (1.00) |
| Del Peak 29(17q25 3) | 0 (0%) | 138 |
0.103 (1.00) |
0.696 (1.00) |
0.119 (1.00) |
1 (1.00) |
0.366 (1.00) |
0.828 (1.00) |
0.13 (1.00) |
0.369 (1.00) |
0.217 (1.00) |
0.102 (1.00) |
0.627 (1.00) |
| Del Peak 31(18q22 1) | 0 (0%) | 24 |
0.653 (1.00) |
0.227 (1.00) |
0.825 (1.00) |
0.397 (1.00) |
0.0492 (1.00) |
0.337 (1.00) |
0.567 (1.00) |
0.126 (1.00) |
0.593 (1.00) |
0.585 (1.00) |
0.00206 (1.00) |
| Del Peak 32(19p13 3) | 0 (0%) | 124 |
0.816 (1.00) |
0.795 (1.00) |
1 (1.00) |
0.734 (1.00) |
0.297 (1.00) |
0.562 (1.00) |
0.251 (1.00) |
0.736 (1.00) |
1 (1.00) |
0.778 (1.00) |
0.693 (1.00) |
| Del Peak 33(20p12 1) | 0 (0%) | 102 |
0.131 (1.00) |
0.23 (1.00) |
0.257 (1.00) |
1 (1.00) |
0.85 (1.00) |
0.88 (1.00) |
0.473 (1.00) |
0.839 (1.00) |
0.0855 (1.00) |
0.466 (1.00) |
0.557 (1.00) |
| Del Peak 34(20p12 1) | 0 (0%) | 102 |
0.131 (1.00) |
0.23 (1.00) |
0.257 (1.00) |
1 (1.00) |
0.85 (1.00) |
0.88 (1.00) |
0.473 (1.00) |
0.839 (1.00) |
0.0855 (1.00) |
0.466 (1.00) |
0.557 (1.00) |
| Del Peak 35(21q11 2) | 0 (0%) | 92 |
0.996 (1.00) |
0.454 (1.00) |
0.633 (1.00) |
0.24 (1.00) |
0.139 (1.00) |
0.322 (1.00) |
0.764 (1.00) |
0.594 (1.00) |
1 (1.00) |
0.501 (1.00) |
0.295 (1.00) |
| Del Peak 36(22q13 31) | 0 (0%) | 96 |
0.0572 (1.00) |
0.255 (1.00) |
0.339 (1.00) |
0.563 (1.00) |
0.28 (1.00) |
0.00822 (1.00) |
0.207 (1.00) |
0.331 (1.00) |
0.688 (1.00) |
0.246 (1.00) |
0.425 (1.00) |
| Del Peak 37(Xp22 33) | 0 (0%) | 129 |
0.626 (1.00) |
0.681 (1.00) |
0.845 (1.00) |
0.716 (1.00) |
0.699 (1.00) |
0.856 (1.00) |
0.596 (1.00) |
0.894 (1.00) |
1 (1.00) |
0.682 (1.00) |
0.787 (1.00) |
P value = 8.16e-05 (logrank test), Q value = 0.056
Table S1. Gene #31: 'Del Peak 6(3q26.31)' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 125 | 11 | 0.2 - 121.1 (6.3) |
| DEL PEAK 6(3Q26.31) CNV | 15 | 3 | 0.2 - 38.9 (2.8) |
| DEL PEAK 6(3Q26.31) WILD-TYPE | 110 | 8 | 0.4 - 121.1 (7.0) |
Figure S1. Get High-res Image Gene #31: 'Del Peak 6(3q26.31)' versus Clinical Feature #1: 'Time to Death'
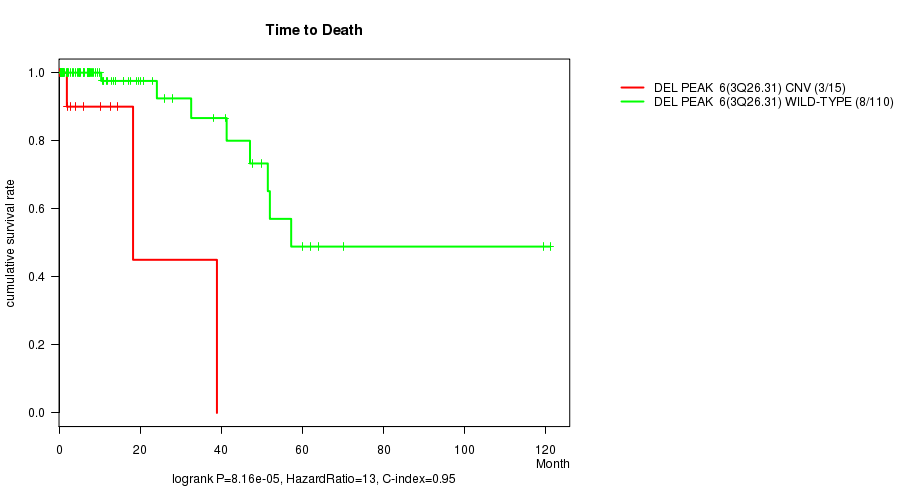
P value = 0.000153 (t-test), Q value = 0.1
Table S2. Gene #55: 'Del Peak 30(18q21.2)' versus Clinical Feature #11: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 149 | 2.7 (5.4) |
| DEL PEAK 30(18Q21.2) CNV | 130 | 2.9 (5.7) |
| DEL PEAK 30(18Q21.2) WILD-TYPE | 19 | 0.6 (1.4) |
Figure S2. Get High-res Image Gene #55: 'Del Peak 30(18q21.2)' versus Clinical Feature #11: 'NUMBER.OF.LYMPH.NODES'
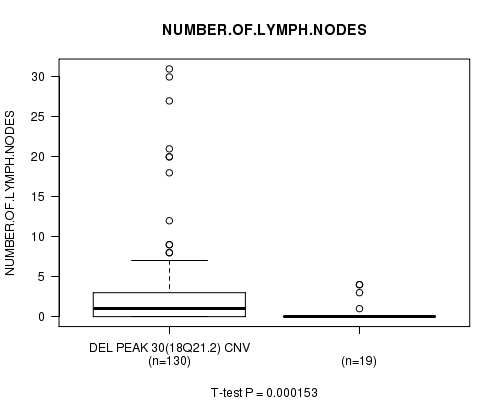
-
Mutation data file = all_lesions.conf_99.cnv.cluster.txt
-
Clinical data file = READ-TP.clin.merged.picked.txt
-
Number of patients = 162
-
Number of significantly arm-level cnvs = 62
-
Number of selected clinical features = 11
-
Exclude genes that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For continuous numerical clinical features, two-tailed Student's t test with unequal variance (Lehmann and Romano 2005) was applied to compare the clinical values between tumors with and without gene mutations using 't.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.