This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and subtypes.
Testing the association between copy number variation 78 arm-level results and 12 molecular subtypes across 575 patients, 103 significant findings detected with Q value < 0.25.
-
2p gain cnv correlated to 'MRNASEQ_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
2q gain cnv correlated to 'MRNA_CHIERARCHICAL', 'METHLYATION_CNMF', and 'MRNASEQ_CNMF'.
-
3p gain cnv correlated to 'CN_CNMF'.
-
3q gain cnv correlated to 'CN_CNMF'.
-
6p gain cnv correlated to 'CN_CNMF'.
-
6q gain cnv correlated to 'CN_CNMF'.
-
7p gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
7q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
8p gain cnv correlated to 'CN_CNMF'.
-
8q gain cnv correlated to 'CN_CNMF'.
-
11p gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
11q gain cnv correlated to 'CN_CNMF'.
-
13q gain cnv correlated to 'MRNA_CNMF', 'MRNA_CHIERARCHICAL', 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
16p gain cnv correlated to 'CN_CNMF'.
-
16q gain cnv correlated to 'CN_CNMF'.
-
17q gain cnv correlated to 'CN_CNMF' and 'MRNASEQ_CNMF'.
-
19p gain cnv correlated to 'CN_CNMF'.
-
20p gain cnv correlated to 'MRNA_CNMF', 'MRNA_CHIERARCHICAL', 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
20q gain cnv correlated to 'MRNA_CNMF', 'MRNA_CHIERARCHICAL', 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'RPPA_CHIERARCHICAL', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
1p loss cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
4p loss cnv correlated to 'CN_CNMF'.
-
4q loss cnv correlated to 'CN_CNMF'.
-
5q loss cnv correlated to 'CN_CNMF'.
-
8p loss cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
9p loss cnv correlated to 'CN_CNMF'.
-
9q loss cnv correlated to 'CN_CNMF'.
-
10q loss cnv correlated to 'CN_CNMF'.
-
12p loss cnv correlated to 'CN_CNMF'.
-
12q loss cnv correlated to 'CN_CNMF'.
-
14q loss cnv correlated to 'MRNA_CNMF', 'MRNA_CHIERARCHICAL', 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
15q loss cnv correlated to 'CN_CNMF', 'MRNASEQ_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
17p loss cnv correlated to 'MRNA_CNMF', 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MIRSEQ_MATURE_CNMF'.
-
18p loss cnv correlated to 'MRNA_CNMF', 'MRNA_CHIERARCHICAL', 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
18q loss cnv correlated to 'MRNA_CNMF', 'MRNA_CHIERARCHICAL', 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
19p loss cnv correlated to 'CN_CNMF'.
-
19q loss cnv correlated to 'CN_CNMF'.
-
20p loss cnv correlated to 'CN_CNMF'.
-
21q loss cnv correlated to 'CN_CNMF'.
-
22q loss cnv correlated to 'CN_CNMF'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 78 arm-level results and 12 molecular subtypes. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 103 significant findings detected.
|
Molecular subtypes |
MRNA CNMF |
MRNA CHIERARCHICAL |
CN CNMF |
METHLYATION CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Chi-square test | |
| 20q gain | 0 (0%) | 172 |
4.38e-17 (3.89e-14) |
4.38e-16 (3.88e-13) |
2.22e-46 (2.01e-43) |
1.68e-12 (1.48e-09) |
0.000311 (0.25) |
0.000133 (0.109) |
9.03e-18 (8.04e-15) |
1.13e-13 (9.97e-11) |
0.463 (1.00) |
0.101 (1.00) |
2.65e-09 (2.3e-06) |
1.39e-21 (1.25e-18) |
| 13q gain | 0 (0%) | 261 |
4.38e-07 (0.000373) |
0.000109 (0.089) |
1.62e-37 (1.46e-34) |
2.86e-08 (2.47e-05) |
0.0129 (1.00) |
0.0683 (1.00) |
8.54e-09 (7.4e-06) |
6.4e-07 (0.000542) |
0.01 (1.00) |
0.45 (1.00) |
2.85e-07 (0.000244) |
1.21e-10 (1.05e-07) |
| 18p loss | 0 (0%) | 251 |
1.24e-13 (1.09e-10) |
3.4e-12 (2.99e-09) |
4.26e-47 (3.85e-44) |
1.35e-18 (1.2e-15) |
0.012 (1.00) |
0.015 (1.00) |
5.39e-16 (4.77e-13) |
2.73e-11 (2.39e-08) |
0.065 (1.00) |
0.787 (1.00) |
1.08e-10 (9.36e-08) |
7.83e-12 (6.85e-09) |
| 18q loss | 0 (0%) | 223 |
1.36e-12 (1.19e-09) |
4.31e-12 (3.77e-09) |
6.8e-51 (6.15e-48) |
1.99e-19 (1.78e-16) |
0.0241 (1.00) |
0.0948 (1.00) |
3.96e-16 (3.51e-13) |
3.68e-09 (3.19e-06) |
0.00938 (1.00) |
0.378 (1.00) |
1.07e-09 (9.26e-07) |
5.16e-12 (4.51e-09) |
| 20p gain | 0 (0%) | 287 |
0.000145 (0.118) |
1.65e-07 (0.000141) |
4.25e-22 (3.81e-19) |
0.000106 (0.0866) |
0.00725 (1.00) |
0.0222 (1.00) |
3.2e-07 (0.000273) |
3.2e-07 (0.000273) |
0.625 (1.00) |
0.472 (1.00) |
0.000742 (0.587) |
6.81e-08 (5.87e-05) |
| 14q loss | 0 (0%) | 432 |
6.52e-05 (0.0537) |
2.84e-07 (0.000243) |
1.12e-31 (1.01e-28) |
1.63e-07 (0.00014) |
0.000364 (0.291) |
0.012 (1.00) |
1.73e-07 (0.000148) |
0.000705 (0.559) |
0.114 (1.00) |
0.709 (1.00) |
1.51e-06 (0.00127) |
0.000145 (0.118) |
| 7p gain | 0 (0%) | 285 |
0.00152 (1.00) |
0.00193 (1.00) |
1.89e-21 (1.7e-18) |
8.55e-08 (7.35e-05) |
0.22 (1.00) |
0.0313 (1.00) |
3.72e-08 (3.21e-05) |
9.54e-05 (0.0783) |
0.0703 (1.00) |
0.0304 (1.00) |
0.00865 (1.00) |
4.41e-08 (3.8e-05) |
| 7q gain | 0 (0%) | 320 |
0.0019 (1.00) |
0.00889 (1.00) |
6.95e-16 (6.15e-13) |
5.98e-06 (0.005) |
0.777 (1.00) |
0.18 (1.00) |
8.79e-07 (0.000744) |
1.24e-05 (0.0103) |
0.245 (1.00) |
0.155 (1.00) |
0.0465 (1.00) |
4.73e-05 (0.0391) |
| 17p loss | 0 (0%) | 323 |
0.000277 (0.223) |
0.00176 (1.00) |
5.49e-22 (4.92e-19) |
1.43e-06 (0.00121) |
0.317 (1.00) |
0.486 (1.00) |
3.25e-06 (0.00272) |
0.000795 (0.628) |
0.027 (1.00) |
0.497 (1.00) |
6.54e-05 (0.0538) |
0.000313 (0.251) |
| 2q gain | 0 (0%) | 516 |
0.0178 (1.00) |
8.52e-05 (0.07) |
0.00101 (0.792) |
1.71e-05 (0.0142) |
0.209 (1.00) |
0.806 (1.00) |
0.000234 (0.189) |
0.000824 (0.65) |
0.251 (1.00) |
0.0504 (1.00) |
0.919 (1.00) |
0.0116 (1.00) |
| 11p gain | 0 (0%) | 536 |
0.0615 (1.00) |
0.275 (1.00) |
1.71e-05 (0.0142) |
0.000204 (0.165) |
0.826 (1.00) |
0.764 (1.00) |
0.0491 (1.00) |
0.000153 (0.125) |
0.246 (1.00) |
0.882 (1.00) |
0.142 (1.00) |
0.00713 (1.00) |
| 15q loss | 0 (0%) | 419 |
0.005 (1.00) |
0.00311 (1.00) |
2.09e-24 (1.88e-21) |
0.0131 (1.00) |
0.303 (1.00) |
0.832 (1.00) |
1.19e-05 (0.00992) |
0.000572 (0.455) |
0.368 (1.00) |
0.246 (1.00) |
0.00873 (1.00) |
0.000158 (0.129) |
| 2p gain | 0 (0%) | 515 |
0.0608 (1.00) |
0.000322 (0.258) |
0.00816 (1.00) |
0.000508 (0.405) |
0.0735 (1.00) |
0.357 (1.00) |
0.000224 (0.181) |
0.000137 (0.112) |
0.216 (1.00) |
0.0209 (1.00) |
0.445 (1.00) |
0.00282 (1.00) |
| 17q gain | 0 (0%) | 508 |
0.0504 (1.00) |
0.035 (1.00) |
0.000269 (0.216) |
0.00271 (1.00) |
0.649 (1.00) |
0.0748 (1.00) |
1.47e-05 (0.0122) |
0.00195 (1.00) |
0.0785 (1.00) |
0.354 (1.00) |
0.474 (1.00) |
0.257 (1.00) |
| 1p loss | 0 (0%) | 507 |
0.245 (1.00) |
0.00443 (1.00) |
7.55e-06 (0.0063) |
0.000189 (0.153) |
0.984 (1.00) |
0.821 (1.00) |
0.0764 (1.00) |
0.00823 (1.00) |
0.193 (1.00) |
0.62 (1.00) |
0.973 (1.00) |
0.0367 (1.00) |
| 8p loss | 0 (0%) | 430 |
0.00406 (1.00) |
0.00186 (1.00) |
1.14e-18 (1.02e-15) |
0.000109 (0.089) |
0.28 (1.00) |
0.0393 (1.00) |
0.11 (1.00) |
0.152 (1.00) |
0.515 (1.00) |
0.73 (1.00) |
0.00232 (1.00) |
0.00767 (1.00) |
| 3p gain | 0 (0%) | 539 |
0.133 (1.00) |
0.593 (1.00) |
0.000181 (0.147) |
0.148 (1.00) |
0.374 (1.00) |
0.77 (1.00) |
0.26 (1.00) |
0.885 (1.00) |
0.208 (1.00) |
0.366 (1.00) |
0.571 (1.00) |
0.0609 (1.00) |
| 3q gain | 0 (0%) | 522 |
0.235 (1.00) |
0.0891 (1.00) |
2.49e-06 (0.00209) |
0.0159 (1.00) |
0.215 (1.00) |
0.655 (1.00) |
0.075 (1.00) |
0.588 (1.00) |
0.111 (1.00) |
0.544 (1.00) |
0.312 (1.00) |
0.133 (1.00) |
| 6p gain | 0 (0%) | 498 |
0.185 (1.00) |
0.00295 (1.00) |
1.55e-06 (0.0013) |
0.278 (1.00) |
0.704 (1.00) |
0.12 (1.00) |
0.0227 (1.00) |
0.227 (1.00) |
0.848 (1.00) |
0.392 (1.00) |
0.952 (1.00) |
0.216 (1.00) |
| 6q gain | 0 (0%) | 504 |
0.309 (1.00) |
0.0401 (1.00) |
7.47e-07 (0.000633) |
0.112 (1.00) |
0.317 (1.00) |
0.0418 (1.00) |
0.0607 (1.00) |
0.575 (1.00) |
0.749 (1.00) |
0.251 (1.00) |
0.372 (1.00) |
0.0608 (1.00) |
| 8p gain | 0 (0%) | 467 |
0.963 (1.00) |
0.717 (1.00) |
1e-08 (8.68e-06) |
0.454 (1.00) |
0.28 (1.00) |
0.215 (1.00) |
0.181 (1.00) |
0.44 (1.00) |
0.49 (1.00) |
0.891 (1.00) |
0.187 (1.00) |
0.898 (1.00) |
| 8q gain | 0 (0%) | 340 |
0.145 (1.00) |
0.0172 (1.00) |
1.06e-33 (9.52e-31) |
0.0135 (1.00) |
0.15 (1.00) |
0.33 (1.00) |
0.0011 (0.862) |
0.0265 (1.00) |
0.459 (1.00) |
0.633 (1.00) |
0.0345 (1.00) |
0.147 (1.00) |
| 11q gain | 0 (0%) | 536 |
0.0956 (1.00) |
0.0299 (1.00) |
1.33e-06 (0.00112) |
0.000725 (0.575) |
0.497 (1.00) |
0.651 (1.00) |
0.0356 (1.00) |
0.00526 (1.00) |
0.559 (1.00) |
0.406 (1.00) |
0.00378 (1.00) |
0.0582 (1.00) |
| 16p gain | 0 (0%) | 479 |
0.0882 (1.00) |
0.165 (1.00) |
1.26e-16 (1.12e-13) |
0.00119 (0.932) |
0.941 (1.00) |
0.0295 (1.00) |
0.0228 (1.00) |
0.136 (1.00) |
0.577 (1.00) |
0.347 (1.00) |
0.631 (1.00) |
0.00678 (1.00) |
| 16q gain | 0 (0%) | 479 |
0.0988 (1.00) |
0.174 (1.00) |
3.02e-18 (2.69e-15) |
0.000413 (0.33) |
0.991 (1.00) |
0.0182 (1.00) |
0.0198 (1.00) |
0.0602 (1.00) |
0.465 (1.00) |
0.277 (1.00) |
0.214 (1.00) |
0.00166 (1.00) |
| 19p gain | 0 (0%) | 523 |
0.194 (1.00) |
0.146 (1.00) |
5.84e-05 (0.0482) |
0.0188 (1.00) |
0.598 (1.00) |
0.0285 (1.00) |
0.423 (1.00) |
0.474 (1.00) |
0.718 (1.00) |
0.745 (1.00) |
0.965 (1.00) |
0.0747 (1.00) |
| 4p loss | 0 (0%) | 453 |
0.227 (1.00) |
0.208 (1.00) |
5.5e-11 (4.79e-08) |
0.00276 (1.00) |
0.693 (1.00) |
0.889 (1.00) |
0.0062 (1.00) |
0.0572 (1.00) |
0.045 (1.00) |
0.769 (1.00) |
0.0802 (1.00) |
0.0273 (1.00) |
| 4q loss | 0 (0%) | 462 |
0.213 (1.00) |
0.134 (1.00) |
3.18e-12 (2.79e-09) |
0.00888 (1.00) |
0.606 (1.00) |
0.81 (1.00) |
0.00213 (1.00) |
0.0535 (1.00) |
0.0122 (1.00) |
0.564 (1.00) |
0.0346 (1.00) |
0.048 (1.00) |
| 5q loss | 0 (0%) | 515 |
0.0144 (1.00) |
0.0106 (1.00) |
5.53e-07 (0.00047) |
0.17 (1.00) |
0.186 (1.00) |
0.812 (1.00) |
0.0211 (1.00) |
0.0985 (1.00) |
0.963 (1.00) |
0.579 (1.00) |
0.518 (1.00) |
0.0399 (1.00) |
| 9p loss | 0 (0%) | 536 |
0.911 (1.00) |
0.286 (1.00) |
2.9e-05 (0.024) |
0.00499 (1.00) |
0.247 (1.00) |
0.147 (1.00) |
0.102 (1.00) |
0.0221 (1.00) |
0.127 (1.00) |
1 (1.00) |
0.284 (1.00) |
0.0515 (1.00) |
| 9q loss | 0 (0%) | 538 |
0.241 (1.00) |
0.0097 (1.00) |
1.95e-06 (0.00164) |
0.000468 (0.373) |
0.692 (1.00) |
0.209 (1.00) |
0.0214 (1.00) |
0.0167 (1.00) |
0.0491 (1.00) |
0.82 (1.00) |
0.122 (1.00) |
0.0228 (1.00) |
| 10q loss | 0 (0%) | 529 |
0.335 (1.00) |
0.419 (1.00) |
1.44e-06 (0.00121) |
0.148 (1.00) |
0.0204 (1.00) |
0.772 (1.00) |
0.102 (1.00) |
0.231 (1.00) |
0.714 (1.00) |
0.811 (1.00) |
0.398 (1.00) |
0.916 (1.00) |
| 12p loss | 0 (0%) | 542 |
0.0938 (1.00) |
0.228 (1.00) |
5.42e-07 (0.000461) |
0.271 (1.00) |
0.694 (1.00) |
0.373 (1.00) |
0.201 (1.00) |
0.46 (1.00) |
0.541 (1.00) |
0.222 (1.00) |
0.0764 (1.00) |
0.633 (1.00) |
| 12q loss | 0 (0%) | 547 |
0.352 (1.00) |
0.0834 (1.00) |
1.11e-06 (0.000936) |
0.521 (1.00) |
0.885 (1.00) |
0.619 (1.00) |
0.143 (1.00) |
0.189 (1.00) |
0.616 (1.00) |
0.681 (1.00) |
0.132 (1.00) |
0.736 (1.00) |
| 19p loss | 0 (0%) | 555 |
0.74 (1.00) |
0.431 (1.00) |
1.46e-05 (0.0122) |
0.0476 (1.00) |
0.579 (1.00) |
0.705 (1.00) |
0.00155 (1.00) |
0.00289 (1.00) |
0.235 (1.00) |
0.371 (1.00) |
0.0514 (1.00) |
0.558 (1.00) |
| 19q loss | 0 (0%) | 557 |
0.74 (1.00) |
0.431 (1.00) |
0.000235 (0.19) |
0.128 (1.00) |
0.625 (1.00) |
0.912 (1.00) |
0.00155 (1.00) |
0.00289 (1.00) |
0.424 (1.00) |
0.0976 (1.00) |
0.0542 (1.00) |
0.408 (1.00) |
| 20p loss | 0 (0%) | 528 |
0.0412 (1.00) |
0.184 (1.00) |
4.56e-07 (0.000388) |
0.117 (1.00) |
0.77 (1.00) |
0.365 (1.00) |
0.577 (1.00) |
0.391 (1.00) |
0.554 (1.00) |
0.486 (1.00) |
0.805 (1.00) |
0.686 (1.00) |
| 21q loss | 0 (0%) | 461 |
0.305 (1.00) |
0.487 (1.00) |
3.64e-05 (0.0301) |
0.0782 (1.00) |
0.328 (1.00) |
0.31 (1.00) |
0.235 (1.00) |
0.392 (1.00) |
0.118 (1.00) |
0.342 (1.00) |
0.383 (1.00) |
0.166 (1.00) |
| 22q loss | 0 (0%) | 453 |
0.0148 (1.00) |
0.00606 (1.00) |
7.8e-14 (6.89e-11) |
0.278 (1.00) |
0.419 (1.00) |
0.264 (1.00) |
0.0676 (1.00) |
0.0646 (1.00) |
0.0662 (1.00) |
0.1 (1.00) |
0.00226 (1.00) |
0.0954 (1.00) |
| 1p gain | 0 (0%) | 560 |
0.408 (1.00) |
0.375 (1.00) |
0.932 (1.00) |
0.567 (1.00) |
0.497 (1.00) |
0.128 (1.00) |
0.178 (1.00) |
0.33 (1.00) |
0.111 (1.00) |
0.911 (1.00) |
0.485 (1.00) |
0.0368 (1.00) |
| 1q gain | 0 (0%) | 495 |
0.497 (1.00) |
0.96 (1.00) |
0.0158 (1.00) |
0.825 (1.00) |
0.255 (1.00) |
0.15 (1.00) |
0.128 (1.00) |
0.0758 (1.00) |
0.0729 (1.00) |
0.978 (1.00) |
0.159 (1.00) |
0.0036 (1.00) |
| 4p gain | 0 (0%) | 569 |
0.0622 (1.00) |
0.226 (1.00) |
0.184 (1.00) |
0.339 (1.00) |
0.137 (1.00) |
0.0525 (1.00) |
0.385 (1.00) |
0.226 (1.00) |
||||
| 4q gain | 0 (0%) | 568 |
0.707 (1.00) |
0.719 (1.00) |
0.25 (1.00) |
0.787 (1.00) |
0.732 (1.00) |
0.645 (1.00) |
0.383 (1.00) |
0.809 (1.00) |
||||
| 5p gain | 0 (0%) | 515 |
0.138 (1.00) |
0.661 (1.00) |
0.0015 (1.00) |
0.174 (1.00) |
0.795 (1.00) |
0.863 (1.00) |
0.401 (1.00) |
0.4 (1.00) |
0.784 (1.00) |
0.354 (1.00) |
0.853 (1.00) |
0.863 (1.00) |
| 5q gain | 0 (0%) | 542 |
0.527 (1.00) |
0.826 (1.00) |
0.00356 (1.00) |
0.0956 (1.00) |
0.742 (1.00) |
0.557 (1.00) |
0.73 (1.00) |
0.446 (1.00) |
0.818 (1.00) |
0.483 (1.00) |
0.488 (1.00) |
0.849 (1.00) |
| 9p gain | 0 (0%) | 496 |
0.503 (1.00) |
1 (1.00) |
0.178 (1.00) |
0.791 (1.00) |
0.91 (1.00) |
0.204 (1.00) |
0.539 (1.00) |
0.855 (1.00) |
0.78 (1.00) |
0.781 (1.00) |
0.813 (1.00) |
0.379 (1.00) |
| 9q gain | 0 (0%) | 511 |
0.385 (1.00) |
0.854 (1.00) |
0.214 (1.00) |
0.582 (1.00) |
0.612 (1.00) |
0.0336 (1.00) |
0.989 (1.00) |
0.691 (1.00) |
0.408 (1.00) |
0.739 (1.00) |
1 (1.00) |
0.525 (1.00) |
| 10p gain | 0 (0%) | 547 |
0.196 (1.00) |
0.628 (1.00) |
0.00981 (1.00) |
0.0135 (1.00) |
0.673 (1.00) |
0.857 (1.00) |
0.617 (1.00) |
0.0446 (1.00) |
0.616 (1.00) |
0.496 (1.00) |
0.00697 (1.00) |
0.46 (1.00) |
| 10q gain | 0 (0%) | 562 |
0.887 (1.00) |
0.277 (1.00) |
0.193 (1.00) |
0.0574 (1.00) |
0.597 (1.00) |
0.288 (1.00) |
0.448 (1.00) |
0.0762 (1.00) |
0.694 (1.00) |
0.902 (1.00) |
0.174 (1.00) |
0.693 (1.00) |
| 12p gain | 0 (0%) | 482 |
0.383 (1.00) |
0.631 (1.00) |
0.256 (1.00) |
0.0205 (1.00) |
0.629 (1.00) |
0.205 (1.00) |
0.0232 (1.00) |
0.0134 (1.00) |
0.22 (1.00) |
0.112 (1.00) |
0.00132 (1.00) |
0.0114 (1.00) |
| 12q gain | 0 (0%) | 497 |
0.328 (1.00) |
0.763 (1.00) |
0.0986 (1.00) |
0.00237 (1.00) |
0.91 (1.00) |
0.339 (1.00) |
0.0353 (1.00) |
0.0154 (1.00) |
0.222 (1.00) |
0.166 (1.00) |
0.00136 (1.00) |
0.00673 (1.00) |
| 14q gain | 0 (0%) | 556 |
0.519 (1.00) |
0.251 (1.00) |
0.0301 (1.00) |
0.709 (1.00) |
0.724 (1.00) |
0.397 (1.00) |
0.0605 (1.00) |
0.048 (1.00) |
1 (1.00) |
0.371 (1.00) |
0.11 (1.00) |
0.076 (1.00) |
| 15q gain | 0 (0%) | 568 |
0.515 (1.00) |
0.882 (1.00) |
0.133 (1.00) |
0.131 (1.00) |
1 (1.00) |
0.244 (1.00) |
0.87 (1.00) |
0.374 (1.00) |
0.105 (1.00) |
0.866 (1.00) |
||
| 17p gain | 0 (0%) | 562 |
0.78 (1.00) |
0.00703 (1.00) |
1 (1.00) |
0.922 (1.00) |
0.552 (1.00) |
0.721 (1.00) |
0.0157 (1.00) |
0.009 (1.00) |
0.694 (1.00) |
0.902 (1.00) |
0.361 (1.00) |
0.645 (1.00) |
| 18p gain | 0 (0%) | 554 |
0.207 (1.00) |
0.0592 (1.00) |
0.00061 (0.484) |
0.0103 (1.00) |
0.514 (1.00) |
0.897 (1.00) |
0.133 (1.00) |
0.391 (1.00) |
0.568 (1.00) |
0.397 (1.00) |
0.55 (1.00) |
0.346 (1.00) |
| 18q gain | 0 (0%) | 563 |
0.172 (1.00) |
0.00739 (1.00) |
0.648 (1.00) |
0.468 (1.00) |
0.122 (1.00) |
0.858 (1.00) |
0.105 (1.00) |
0.134 (1.00) |
0.611 (1.00) |
0.349 (1.00) |
||
| 19q gain | 0 (0%) | 514 |
0.0157 (1.00) |
0.0412 (1.00) |
0.0122 (1.00) |
0.0777 (1.00) |
0.233 (1.00) |
0.0383 (1.00) |
0.507 (1.00) |
0.512 (1.00) |
0.832 (1.00) |
0.495 (1.00) |
0.42 (1.00) |
0.163 (1.00) |
| 21q gain | 0 (0%) | 556 |
0.456 (1.00) |
0.715 (1.00) |
0.00268 (1.00) |
0.709 (1.00) |
0.48 (1.00) |
0.0895 (1.00) |
0.619 (1.00) |
0.834 (1.00) |
0.686 (1.00) |
0.143 (1.00) |
0.926 (1.00) |
0.617 (1.00) |
| 22q gain | 0 (0%) | 568 |
0.691 (1.00) |
0.00703 (1.00) |
0.234 (1.00) |
0.573 (1.00) |
0.685 (1.00) |
0.47 (1.00) |
0.689 (1.00) |
0.499 (1.00) |
0.459 (1.00) |
0.596 (1.00) |
||
| Xq gain | 0 (0%) | 558 |
0.178 (1.00) |
0.446 (1.00) |
0.0167 (1.00) |
0.34 (1.00) |
0.244 (1.00) |
0.313 (1.00) |
0.846 (1.00) |
1 (1.00) |
0.482 (1.00) |
0.705 (1.00) |
0.521 (1.00) |
0.881 (1.00) |
| 1q loss | 0 (0%) | 550 |
0.559 (1.00) |
0.0258 (1.00) |
0.0823 (1.00) |
0.155 (1.00) |
0.825 (1.00) |
0.949 (1.00) |
0.158 (1.00) |
0.324 (1.00) |
0.425 (1.00) |
0.725 (1.00) |
1 (1.00) |
0.202 (1.00) |
| 2p loss | 0 (0%) | 565 |
1 (1.00) |
0.739 (1.00) |
0.0735 (1.00) |
0.0168 (1.00) |
0.658 (1.00) |
0.0151 (1.00) |
0.0133 (1.00) |
|||||
| 2q loss | 0 (0%) | 567 |
0.74 (1.00) |
0.614 (1.00) |
0.548 (1.00) |
0.157 (1.00) |
0.299 (1.00) |
0.0584 (1.00) |
0.0455 (1.00) |
|||||
| 3p loss | 0 (0%) | 540 |
1 (1.00) |
0.292 (1.00) |
0.831 (1.00) |
0.834 (1.00) |
0.703 (1.00) |
0.536 (1.00) |
0.441 (1.00) |
0.157 (1.00) |
0.105 (1.00) |
0.729 (1.00) |
0.588 (1.00) |
0.302 (1.00) |
| 3q loss | 0 (0%) | 558 |
0.45 (1.00) |
0.0138 (1.00) |
0.757 (1.00) |
0.0198 (1.00) |
0.0903 (1.00) |
0.398 (1.00) |
0.385 (1.00) |
0.184 (1.00) |
0.74 (1.00) |
0.772 (1.00) |
0.903 (1.00) |
0.411 (1.00) |
| 5p loss | 0 (0%) | 541 |
0.269 (1.00) |
0.45 (1.00) |
0.000544 (0.433) |
0.0928 (1.00) |
0.556 (1.00) |
0.449 (1.00) |
0.234 (1.00) |
0.0352 (1.00) |
0.969 (1.00) |
0.829 (1.00) |
0.727 (1.00) |
0.0884 (1.00) |
| 6p loss | 0 (0%) | 557 |
0.132 (1.00) |
0.186 (1.00) |
0.026 (1.00) |
0.278 (1.00) |
0.611 (1.00) |
0.641 (1.00) |
0.89 (1.00) |
0.601 (1.00) |
0.602 (1.00) |
0.923 (1.00) |
1 (1.00) |
0.646 (1.00) |
| 6q loss | 0 (0%) | 542 |
0.784 (1.00) |
0.621 (1.00) |
0.0983 (1.00) |
0.939 (1.00) |
0.132 (1.00) |
0.145 (1.00) |
0.737 (1.00) |
0.677 (1.00) |
0.262 (1.00) |
0.298 (1.00) |
0.679 (1.00) |
0.799 (1.00) |
| 7q loss | 0 (0%) | 572 |
0.212 (1.00) |
0.834 (1.00) |
1 (1.00) |
0.764 (1.00) |
1 (1.00) |
|||||||
| 8q loss | 0 (0%) | 563 |
0.669 (1.00) |
0.667 (1.00) |
0.0828 (1.00) |
0.58 (1.00) |
0.0237 (1.00) |
0.533 (1.00) |
1 (1.00) |
0.828 (1.00) |
0.159 (1.00) |
0.271 (1.00) |
1 (1.00) |
0.692 (1.00) |
| 10p loss | 0 (0%) | 535 |
0.72 (1.00) |
0.49 (1.00) |
0.000862 (0.679) |
0.434 (1.00) |
0.053 (1.00) |
0.682 (1.00) |
0.409 (1.00) |
0.532 (1.00) |
0.132 (1.00) |
0.591 (1.00) |
1 (1.00) |
0.973 (1.00) |
| 11p loss | 0 (0%) | 535 |
0.73 (1.00) |
0.336 (1.00) |
0.00977 (1.00) |
0.705 (1.00) |
0.322 (1.00) |
0.201 (1.00) |
0.0716 (1.00) |
0.0838 (1.00) |
0.113 (1.00) |
0.359 (1.00) |
0.0803 (1.00) |
0.0629 (1.00) |
| 11q loss | 0 (0%) | 524 |
0.827 (1.00) |
0.464 (1.00) |
0.00166 (1.00) |
0.322 (1.00) |
0.731 (1.00) |
0.292 (1.00) |
0.0997 (1.00) |
0.509 (1.00) |
0.129 (1.00) |
0.835 (1.00) |
0.0742 (1.00) |
0.00583 (1.00) |
| 13q loss | 0 (0%) | 563 |
0.297 (1.00) |
0.282 (1.00) |
0.0495 (1.00) |
0.181 (1.00) |
0.0716 (1.00) |
0.108 (1.00) |
0.236 (1.00) |
0.668 (1.00) |
0.726 (1.00) |
0.403 (1.00) |
0.438 (1.00) |
0.445 (1.00) |
| 16p loss | 0 (0%) | 560 |
0.266 (1.00) |
0.184 (1.00) |
0.0995 (1.00) |
0.181 (1.00) |
0.561 (1.00) |
0.646 (1.00) |
0.589 (1.00) |
0.601 (1.00) |
1 (1.00) |
0.911 (1.00) |
0.602 (1.00) |
0.605 (1.00) |
| 16q loss | 0 (0%) | 558 |
0.0284 (1.00) |
0.243 (1.00) |
0.00221 (1.00) |
0.493 (1.00) |
0.459 (1.00) |
0.748 (1.00) |
0.0911 (1.00) |
0.121 (1.00) |
0.134 (1.00) |
0.257 (1.00) |
0.386 (1.00) |
0.34 (1.00) |
| 17q loss | 0 (0%) | 530 |
0.844 (1.00) |
0.487 (1.00) |
0.0023 (1.00) |
0.926 (1.00) |
0.576 (1.00) |
0.587 (1.00) |
0.101 (1.00) |
0.105 (1.00) |
0.116 (1.00) |
0.635 (1.00) |
0.198 (1.00) |
0.304 (1.00) |
| Xq loss | 0 (0%) | 564 |
0.765 (1.00) |
1 (1.00) |
0.339 (1.00) |
0.507 (1.00) |
0.108 (1.00) |
0.495 (1.00) |
0.0157 (1.00) |
0.0674 (1.00) |
0.515 (1.00) |
0.652 (1.00) |
0.247 (1.00) |
0.176 (1.00) |
P value = 0.000224 (Fisher's exact test), Q value = 0.18
Table S1. Gene #3: '2p gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 38 | 81 | 56 | 66 |
| 2P GAIN CNV | 1 | 20 | 4 | 3 |
| 2P GAIN WILD-TYPE | 37 | 61 | 52 | 63 |
Figure S1. Get High-res Image Gene #3: '2p gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
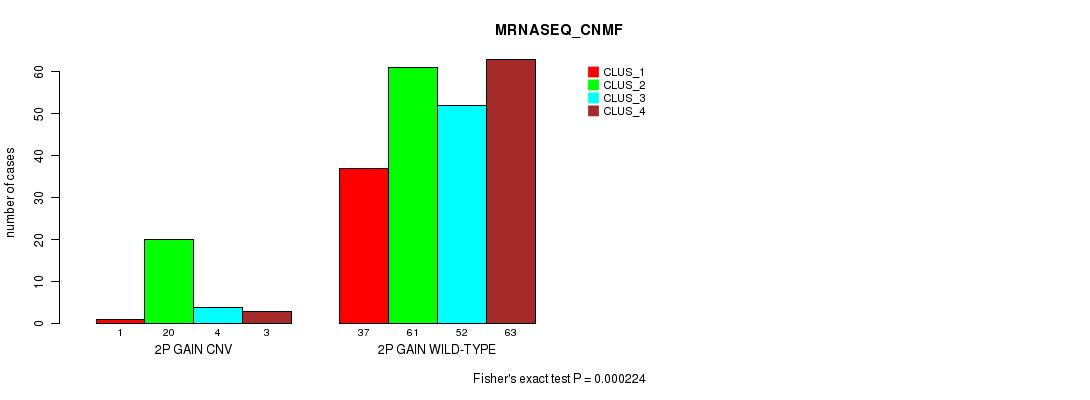
P value = 0.000137 (Fisher's exact test), Q value = 0.11
Table S2. Gene #3: '2p gain' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 95 | 67 | 79 |
| 2P GAIN CNV | 9 | 1 | 18 |
| 2P GAIN WILD-TYPE | 86 | 66 | 61 |
Figure S2. Get High-res Image Gene #3: '2p gain' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
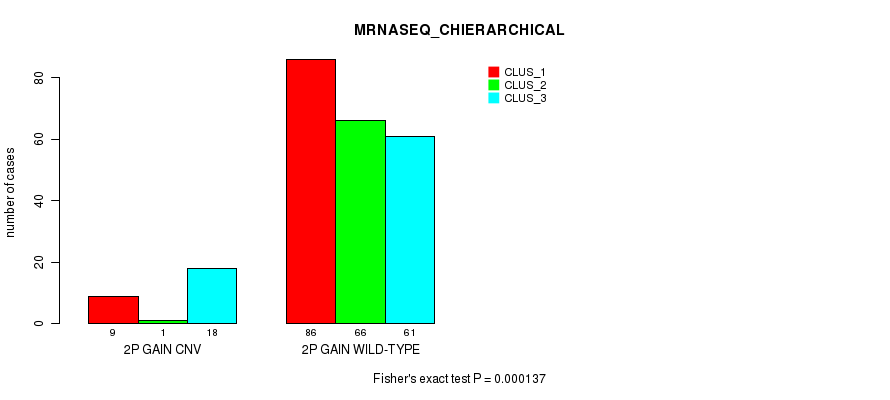
P value = 8.52e-05 (Fisher's exact test), Q value = 0.07
Table S3. Gene #4: '2q gain' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 50 | 63 | 60 |
| 2Q GAIN CNV | 7 | 1 | 0 | 10 |
| 2Q GAIN WILD-TYPE | 35 | 49 | 63 | 50 |
Figure S3. Get High-res Image Gene #4: '2q gain' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
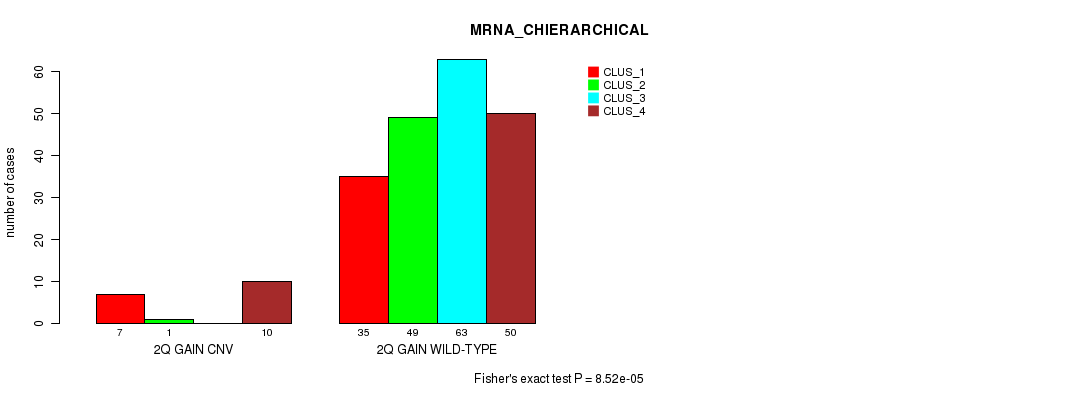
P value = 1.71e-05 (Fisher's exact test), Q value = 0.014
Table S4. Gene #4: '2q gain' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 142 | 114 | 91 |
| 2Q GAIN CNV | 27 | 9 | 1 |
| 2Q GAIN WILD-TYPE | 115 | 105 | 90 |
Figure S4. Get High-res Image Gene #4: '2q gain' versus Molecular Subtype #4: 'METHLYATION_CNMF'
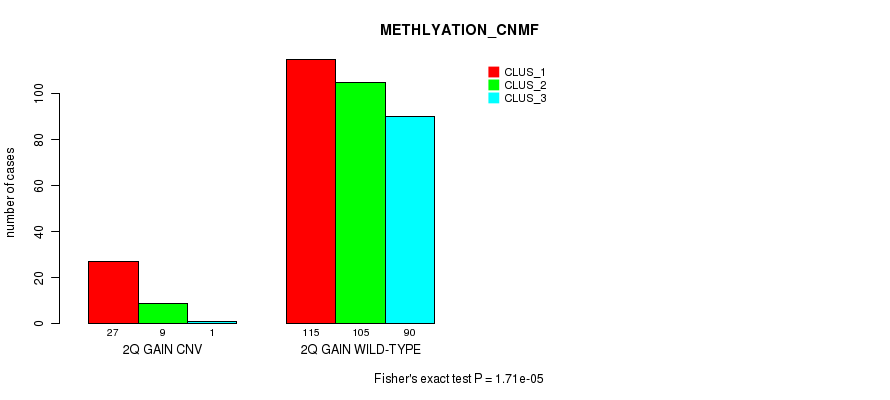
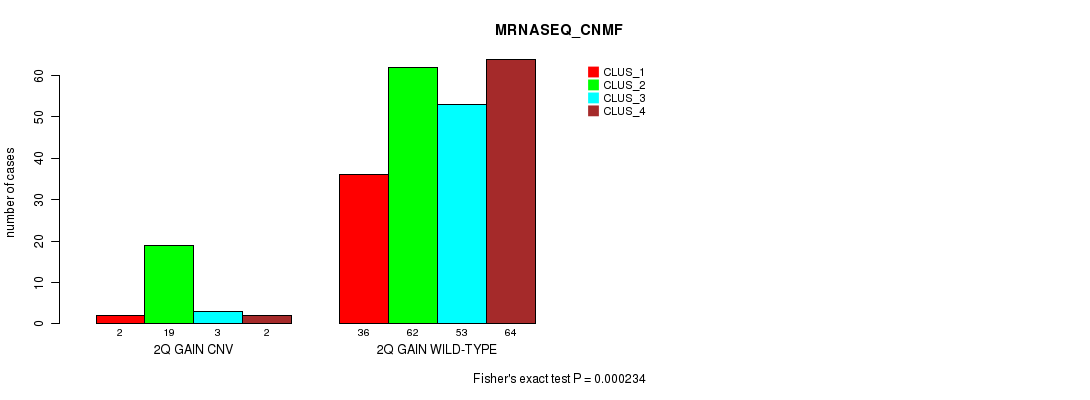
P value = 0.000234 (Fisher's exact test), Q value = 0.19
Table S5. Gene #4: '2q gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 38 | 81 | 56 | 66 |
| 2Q GAIN CNV | 2 | 19 | 3 | 2 |
| 2Q GAIN WILD-TYPE | 36 | 62 | 53 | 64 |
Figure S5. Get High-res Image Gene #4: '2q gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
P value = 0.000181 (Fisher's exact test), Q value = 0.15
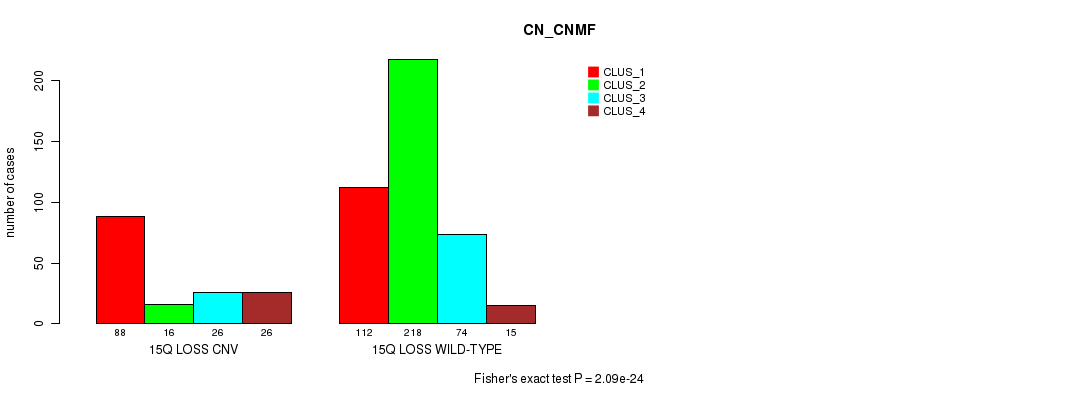
Table S6. Gene #5: '3p gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 3P GAIN CNV | 25 | 7 | 4 | 0 |
| 3P GAIN WILD-TYPE | 175 | 227 | 96 | 41 |
Figure S6. Get High-res Image Gene #5: '3p gain' versus Molecular Subtype #3: 'CN_CNMF'
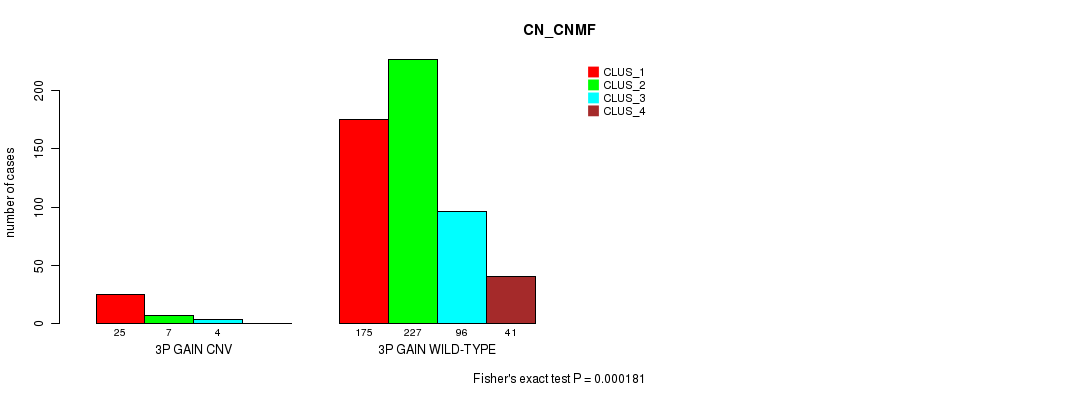
P value = 2.49e-06 (Fisher's exact test), Q value = 0.0021
Table S7. Gene #6: '3q gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 3Q GAIN CNV | 35 | 7 | 9 | 2 |
| 3Q GAIN WILD-TYPE | 165 | 227 | 91 | 39 |
Figure S7. Get High-res Image Gene #6: '3q gain' versus Molecular Subtype #3: 'CN_CNMF'
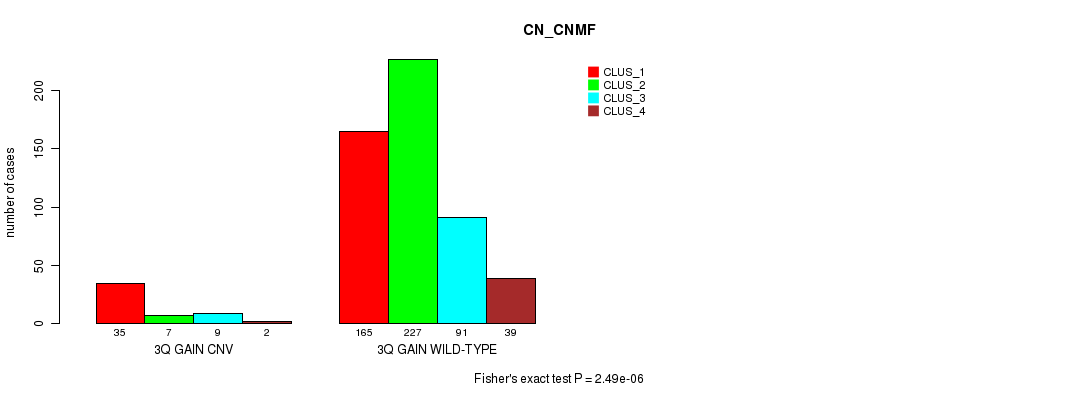
P value = 1.55e-06 (Fisher's exact test), Q value = 0.0013
Table S8. Gene #11: '6p gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 6P GAIN CNV | 46 | 15 | 8 | 8 |
| 6P GAIN WILD-TYPE | 154 | 219 | 92 | 33 |
Figure S8. Get High-res Image Gene #11: '6p gain' versus Molecular Subtype #3: 'CN_CNMF'
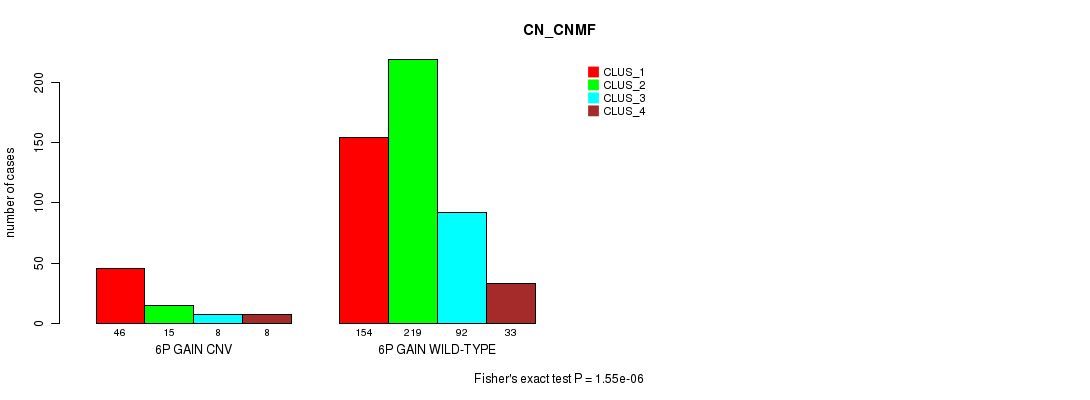
P value = 7.47e-07 (Fisher's exact test), Q value = 0.00063
Table S9. Gene #12: '6q gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 6Q GAIN CNV | 43 | 12 | 8 | 8 |
| 6Q GAIN WILD-TYPE | 157 | 222 | 92 | 33 |
Figure S9. Get High-res Image Gene #12: '6q gain' versus Molecular Subtype #3: 'CN_CNMF'
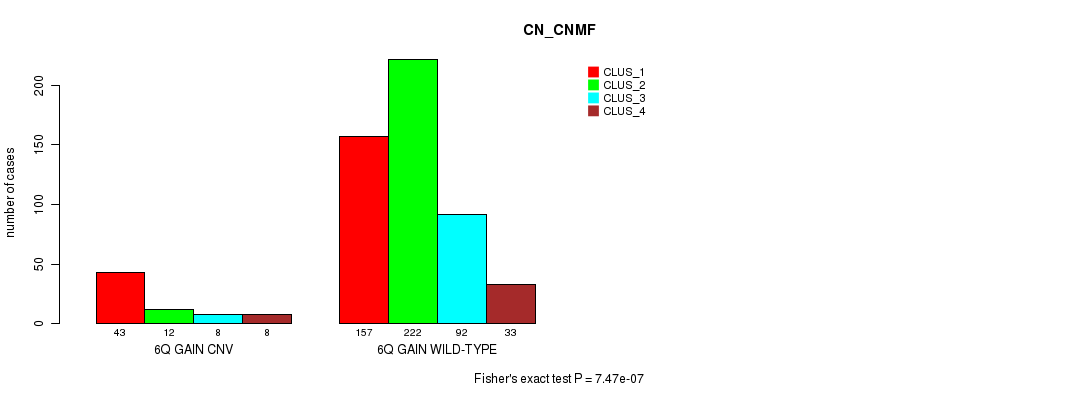
P value = 1.89e-21 (Fisher's exact test), Q value = 1.7e-18
Table S10. Gene #13: '7p gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 7P GAIN CNV | 144 | 62 | 59 | 25 |
| 7P GAIN WILD-TYPE | 56 | 172 | 41 | 16 |
Figure S10. Get High-res Image Gene #13: '7p gain' versus Molecular Subtype #3: 'CN_CNMF'
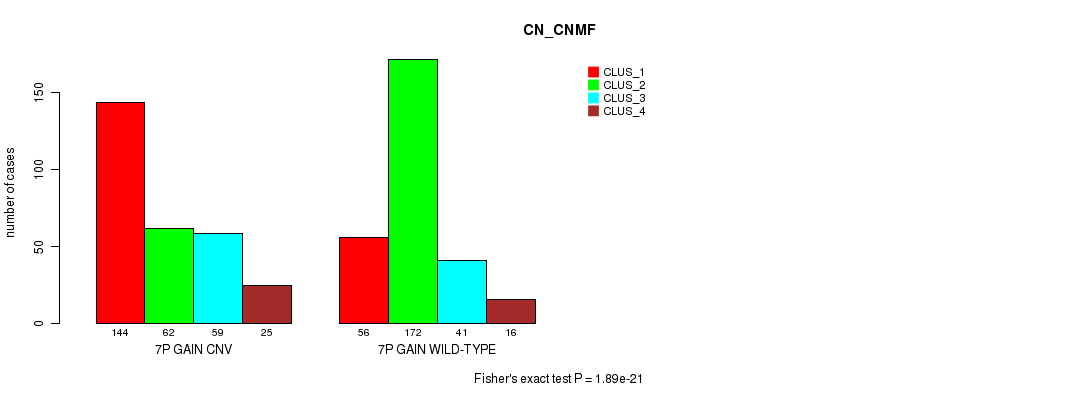
P value = 8.55e-08 (Fisher's exact test), Q value = 7.4e-05
Table S11. Gene #13: '7p gain' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 142 | 114 | 91 |
| 7P GAIN CNV | 103 | 52 | 35 |
| 7P GAIN WILD-TYPE | 39 | 62 | 56 |
Figure S11. Get High-res Image Gene #13: '7p gain' versus Molecular Subtype #4: 'METHLYATION_CNMF'
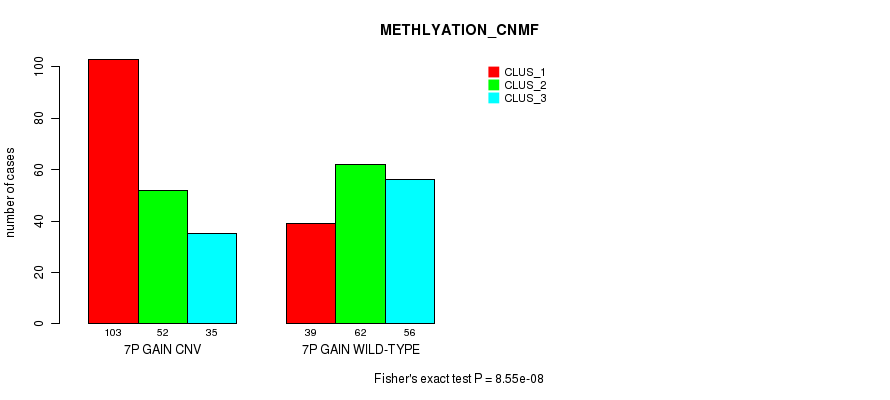
P value = 3.72e-08 (Fisher's exact test), Q value = 3.2e-05
Table S12. Gene #13: '7p gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 38 | 81 | 56 | 66 |
| 7P GAIN CNV | 22 | 61 | 31 | 17 |
| 7P GAIN WILD-TYPE | 16 | 20 | 25 | 49 |
Figure S12. Get High-res Image Gene #13: '7p gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
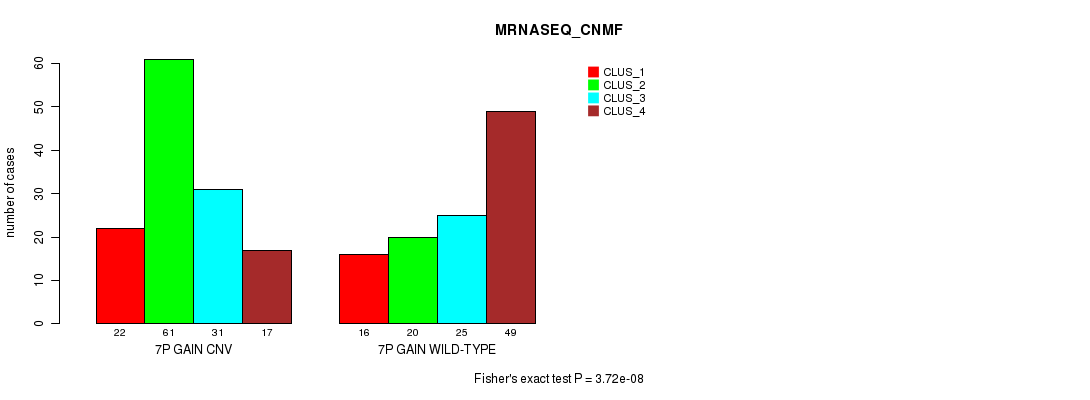
P value = 9.54e-05 (Fisher's exact test), Q value = 0.078
Table S13. Gene #13: '7p gain' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 95 | 67 | 79 |
| 7P GAIN CNV | 53 | 23 | 55 |
| 7P GAIN WILD-TYPE | 42 | 44 | 24 |
Figure S13. Get High-res Image Gene #13: '7p gain' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
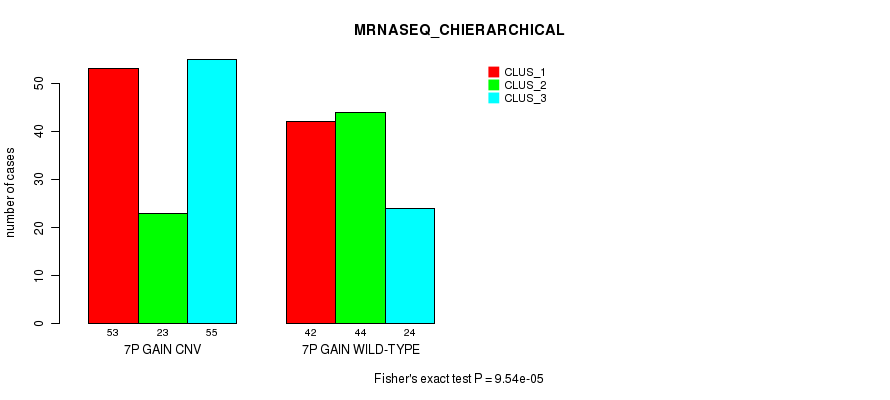
P value = 4.41e-08 (Chi-square test), Q value = 3.8e-05
Table S14. Gene #13: '7p gain' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 30 | 23 | 62 | 65 | 46 | 69 |
| 7P GAIN CNV | 13 | 13 | 41 | 42 | 33 | 15 |
| 7P GAIN WILD-TYPE | 17 | 10 | 21 | 23 | 13 | 54 |
Figure S14. Get High-res Image Gene #13: '7p gain' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
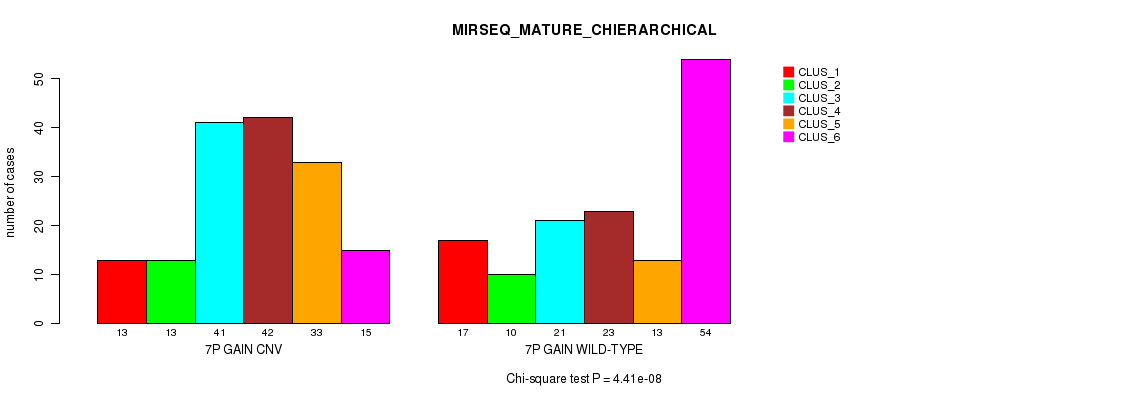
P value = 6.95e-16 (Fisher's exact test), Q value = 6.1e-13
Table S15. Gene #14: '7q gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 7Q GAIN CNV | 126 | 56 | 52 | 21 |
| 7Q GAIN WILD-TYPE | 74 | 178 | 48 | 20 |
Figure S15. Get High-res Image Gene #14: '7q gain' versus Molecular Subtype #3: 'CN_CNMF'
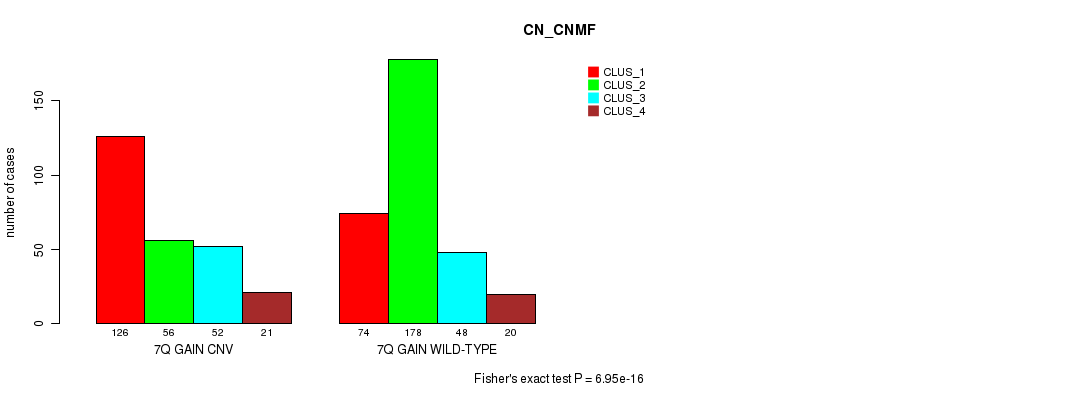
P value = 5.98e-06 (Fisher's exact test), Q value = 0.005
Table S16. Gene #14: '7q gain' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 142 | 114 | 91 |
| 7Q GAIN CNV | 88 | 47 | 28 |
| 7Q GAIN WILD-TYPE | 54 | 67 | 63 |
Figure S16. Get High-res Image Gene #14: '7q gain' versus Molecular Subtype #4: 'METHLYATION_CNMF'
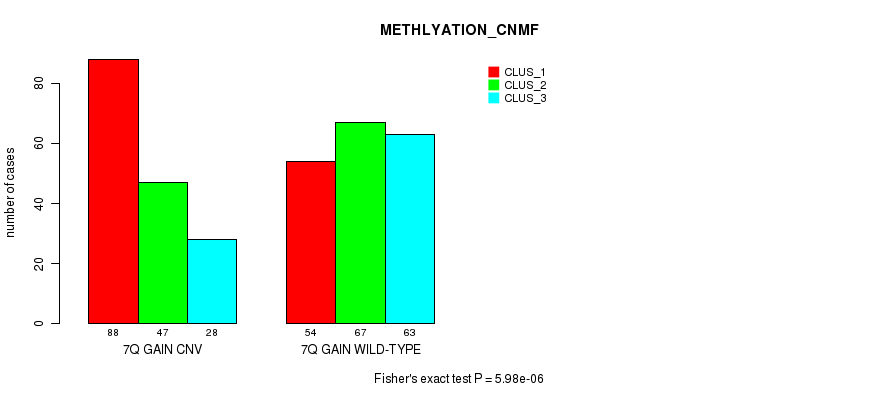
P value = 8.79e-07 (Fisher's exact test), Q value = 0.00074
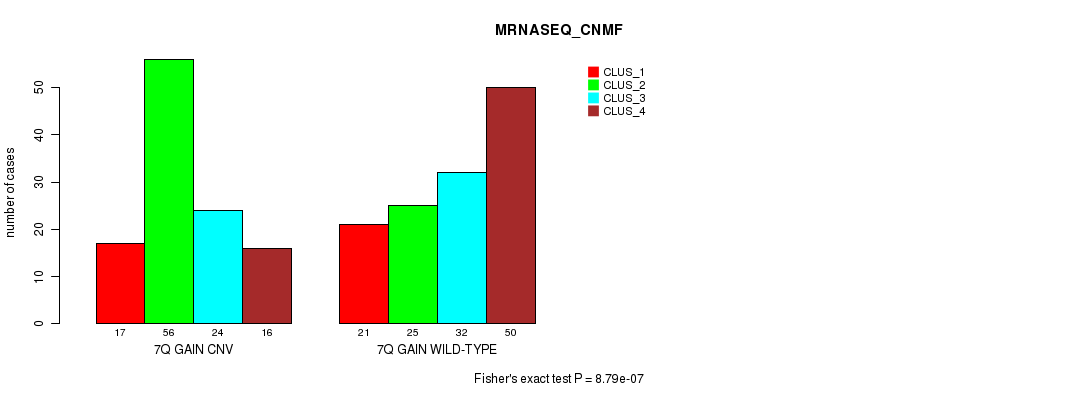
Table S17. Gene #14: '7q gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 38 | 81 | 56 | 66 |
| 7Q GAIN CNV | 17 | 56 | 24 | 16 |
| 7Q GAIN WILD-TYPE | 21 | 25 | 32 | 50 |
Figure S17. Get High-res Image Gene #14: '7q gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
P value = 1.24e-05 (Fisher's exact test), Q value = 0.01
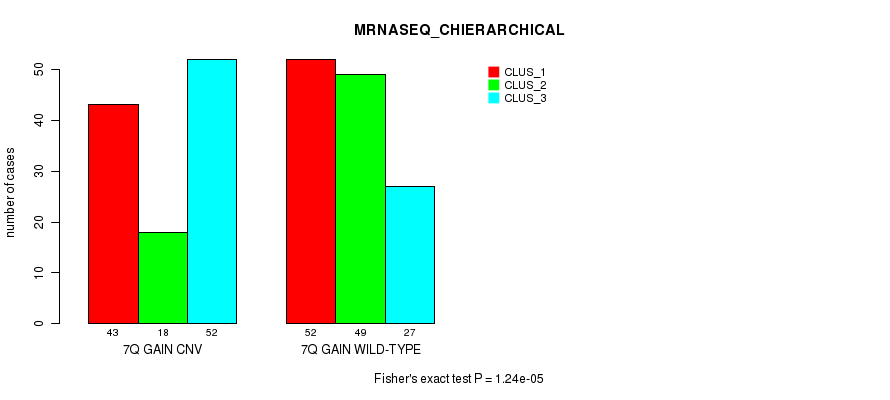
Table S18. Gene #14: '7q gain' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 95 | 67 | 79 |
| 7Q GAIN CNV | 43 | 18 | 52 |
| 7Q GAIN WILD-TYPE | 52 | 49 | 27 |
Figure S18. Get High-res Image Gene #14: '7q gain' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
P value = 4.73e-05 (Chi-square test), Q value = 0.039
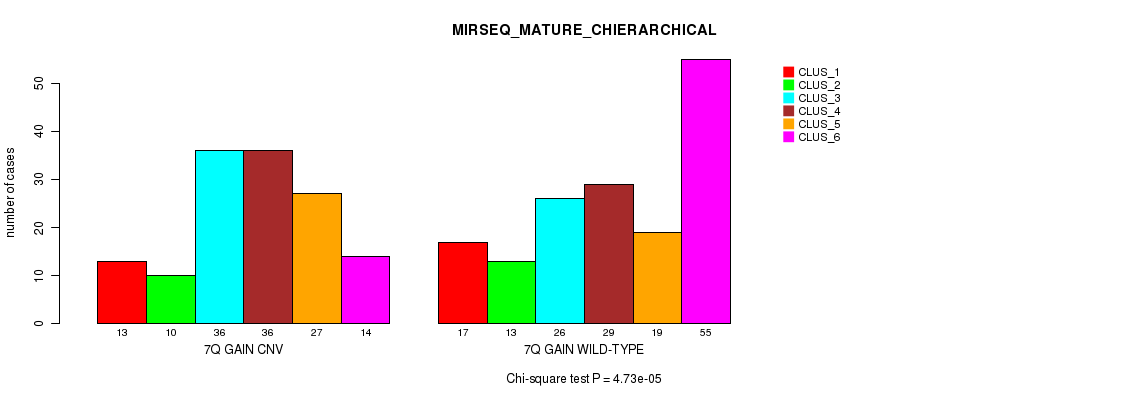
Table S19. Gene #14: '7q gain' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 30 | 23 | 62 | 65 | 46 | 69 |
| 7Q GAIN CNV | 13 | 10 | 36 | 36 | 27 | 14 |
| 7Q GAIN WILD-TYPE | 17 | 13 | 26 | 29 | 19 | 55 |
Figure S19. Get High-res Image Gene #14: '7q gain' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
P value = 1e-08 (Fisher's exact test), Q value = 8.7e-06
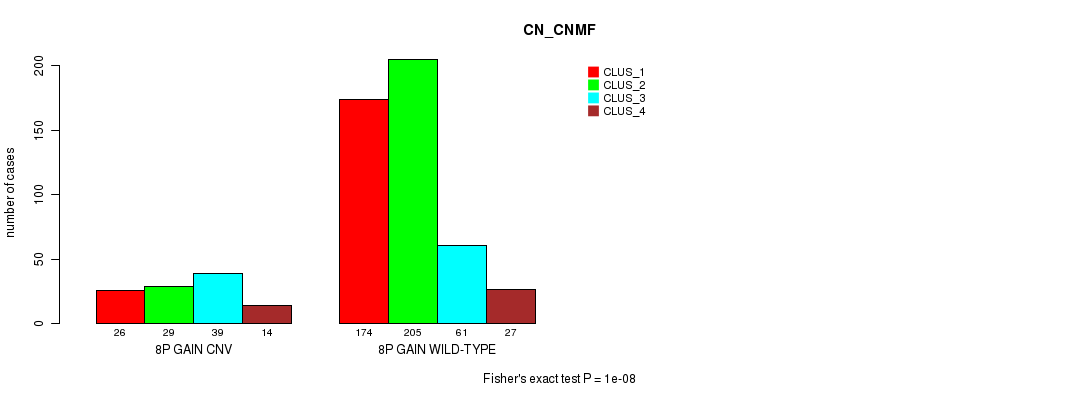
Table S20. Gene #15: '8p gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 8P GAIN CNV | 26 | 29 | 39 | 14 |
| 8P GAIN WILD-TYPE | 174 | 205 | 61 | 27 |
Figure S20. Get High-res Image Gene #15: '8p gain' versus Molecular Subtype #3: 'CN_CNMF'
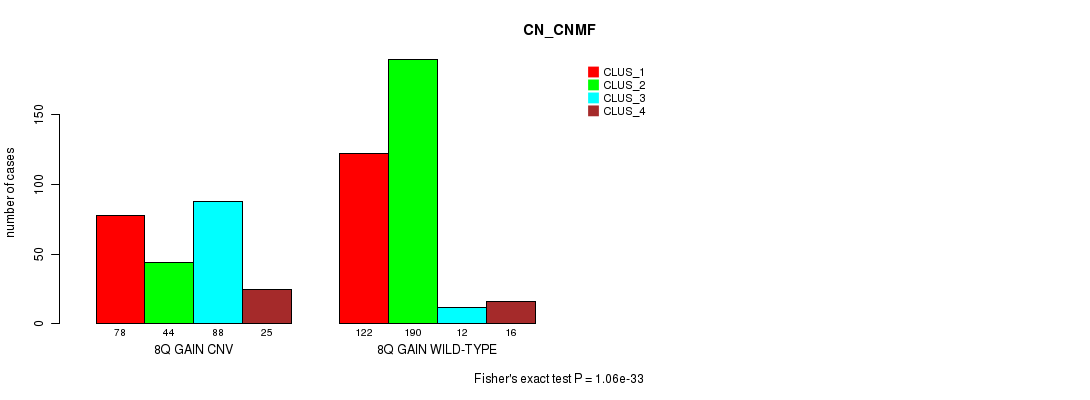
P value = 1.06e-33 (Fisher's exact test), Q value = 9.5e-31
Table S21. Gene #16: '8q gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 8Q GAIN CNV | 78 | 44 | 88 | 25 |
| 8Q GAIN WILD-TYPE | 122 | 190 | 12 | 16 |
Figure S21. Get High-res Image Gene #16: '8q gain' versus Molecular Subtype #3: 'CN_CNMF'
P value = 1.71e-05 (Fisher's exact test), Q value = 0.014
Table S22. Gene #21: '11p gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 11P GAIN CNV | 26 | 4 | 5 | 4 |
| 11P GAIN WILD-TYPE | 174 | 230 | 95 | 37 |
Figure S22. Get High-res Image Gene #21: '11p gain' versus Molecular Subtype #3: 'CN_CNMF'
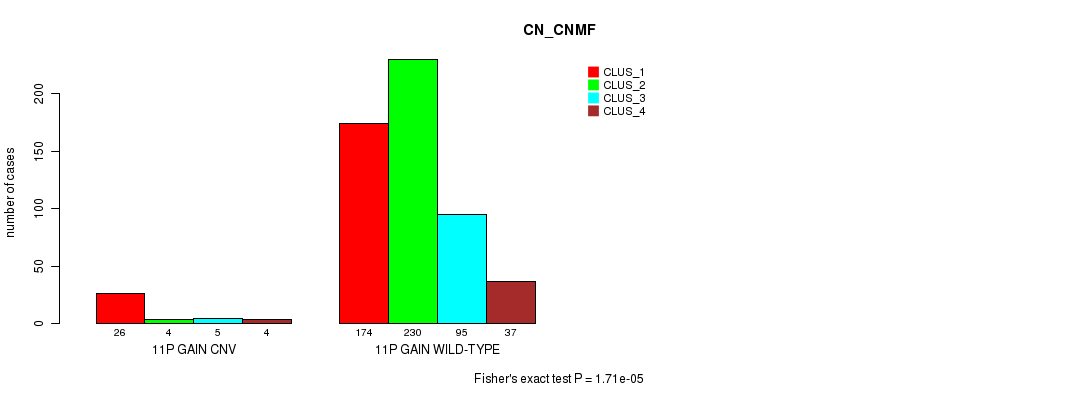
P value = 0.000204 (Fisher's exact test), Q value = 0.17
Table S23. Gene #21: '11p gain' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 142 | 114 | 91 |
| 11P GAIN CNV | 20 | 2 | 3 |
| 11P GAIN WILD-TYPE | 122 | 112 | 88 |
Figure S23. Get High-res Image Gene #21: '11p gain' versus Molecular Subtype #4: 'METHLYATION_CNMF'
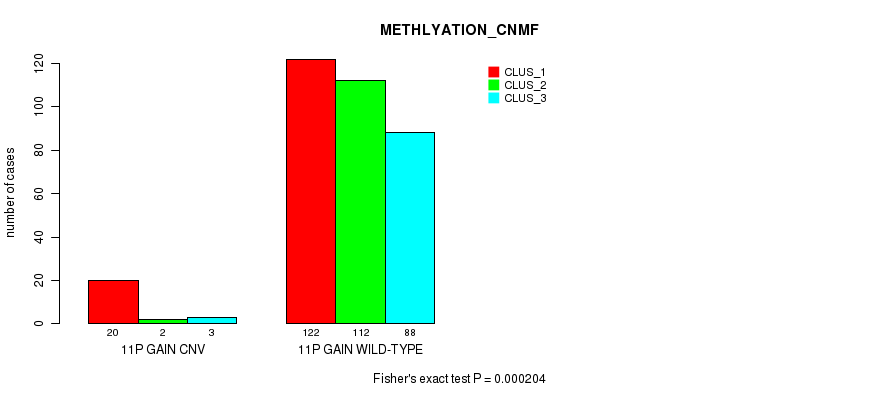
P value = 0.000153 (Fisher's exact test), Q value = 0.12
Table S24. Gene #21: '11p gain' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 95 | 67 | 79 |
| 11P GAIN CNV | 3 | 0 | 12 |
| 11P GAIN WILD-TYPE | 92 | 67 | 67 |
Figure S24. Get High-res Image Gene #21: '11p gain' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
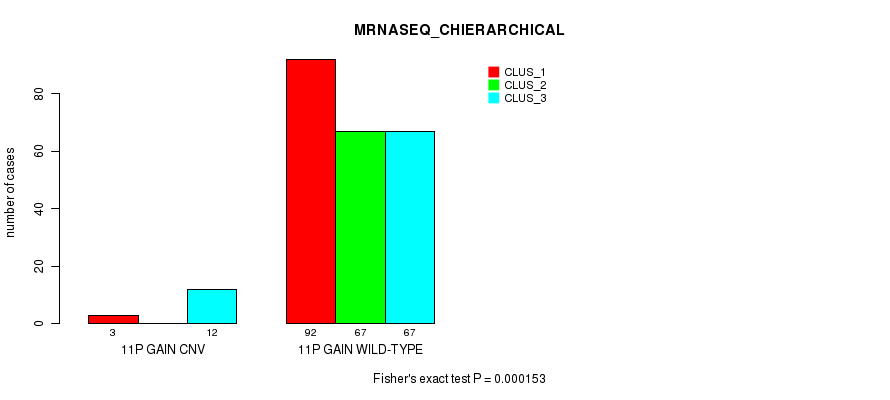
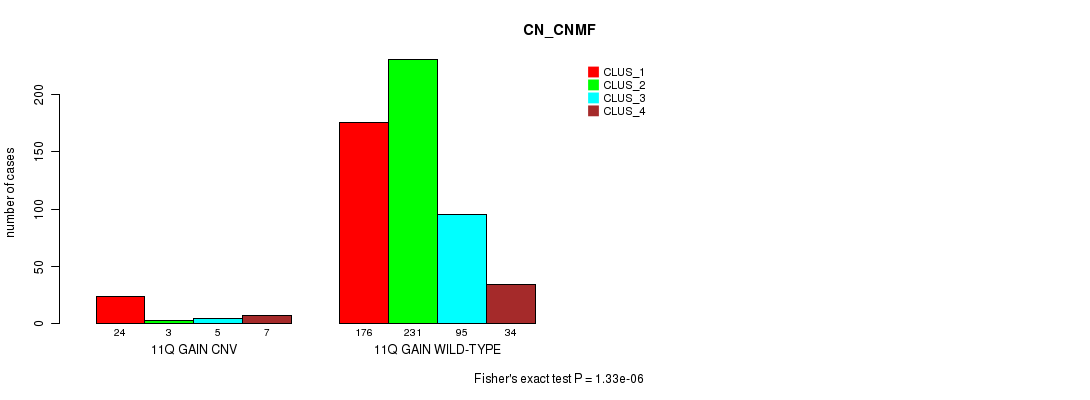
P value = 1.33e-06 (Fisher's exact test), Q value = 0.0011
Table S25. Gene #22: '11q gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 11Q GAIN CNV | 24 | 3 | 5 | 7 |
| 11Q GAIN WILD-TYPE | 176 | 231 | 95 | 34 |
Figure S25. Get High-res Image Gene #22: '11q gain' versus Molecular Subtype #3: 'CN_CNMF'
P value = 4.38e-07 (Fisher's exact test), Q value = 0.00037
Table S26. Gene #25: '13q gain' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 62 | 69 | 41 |
| 13Q GAIN CNV | 12 | 45 | 48 | 15 |
| 13Q GAIN WILD-TYPE | 31 | 17 | 21 | 26 |
Figure S26. Get High-res Image Gene #25: '13q gain' versus Molecular Subtype #1: 'MRNA_CNMF'
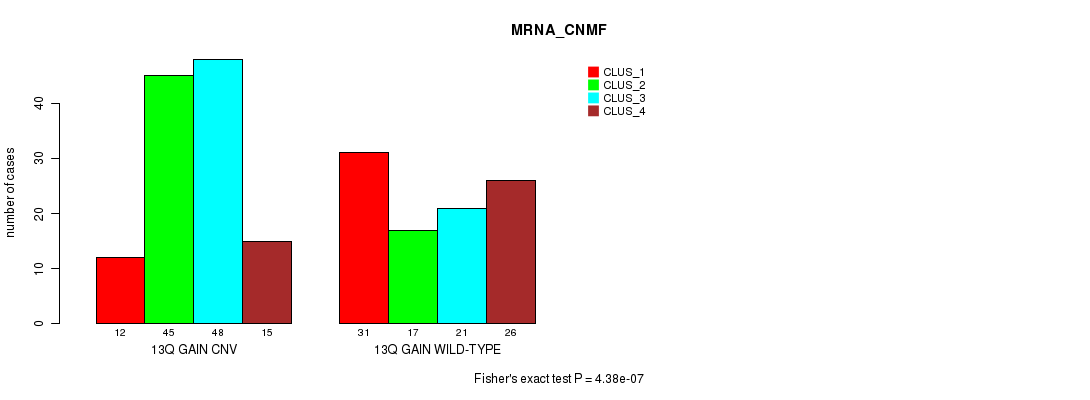
P value = 0.000109 (Fisher's exact test), Q value = 0.089
Table S27. Gene #25: '13q gain' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 50 | 63 | 60 |
| 13Q GAIN CNV | 27 | 33 | 20 | 40 |
| 13Q GAIN WILD-TYPE | 15 | 17 | 43 | 20 |
Figure S27. Get High-res Image Gene #25: '13q gain' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
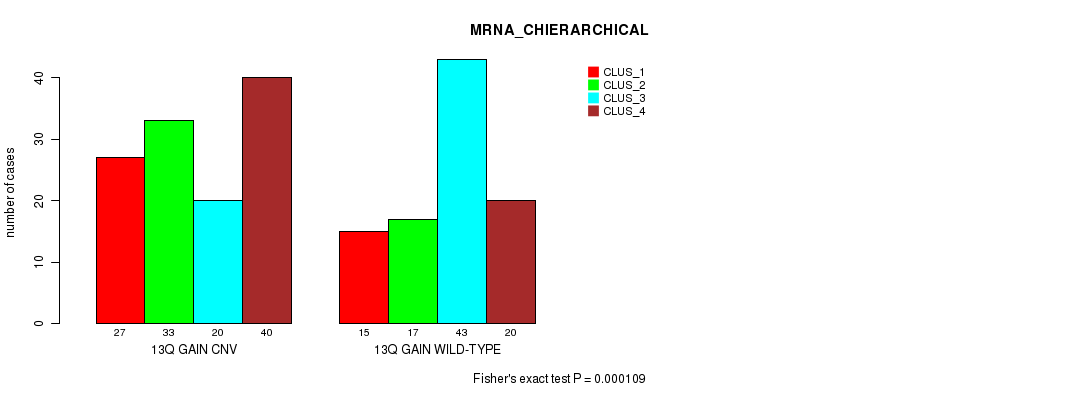
P value = 1.62e-37 (Fisher's exact test), Q value = 1.5e-34
Table S28. Gene #25: '13q gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 13Q GAIN CNV | 175 | 64 | 56 | 19 |
| 13Q GAIN WILD-TYPE | 25 | 170 | 44 | 22 |
Figure S28. Get High-res Image Gene #25: '13q gain' versus Molecular Subtype #3: 'CN_CNMF'
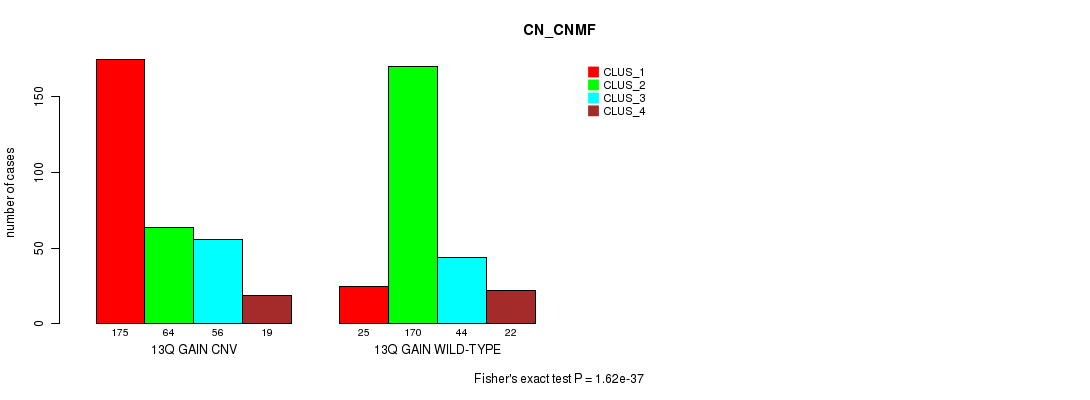
P value = 2.86e-08 (Fisher's exact test), Q value = 2.5e-05
Table S29. Gene #25: '13q gain' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 142 | 114 | 91 |
| 13Q GAIN CNV | 105 | 47 | 40 |
| 13Q GAIN WILD-TYPE | 37 | 67 | 51 |
Figure S29. Get High-res Image Gene #25: '13q gain' versus Molecular Subtype #4: 'METHLYATION_CNMF'
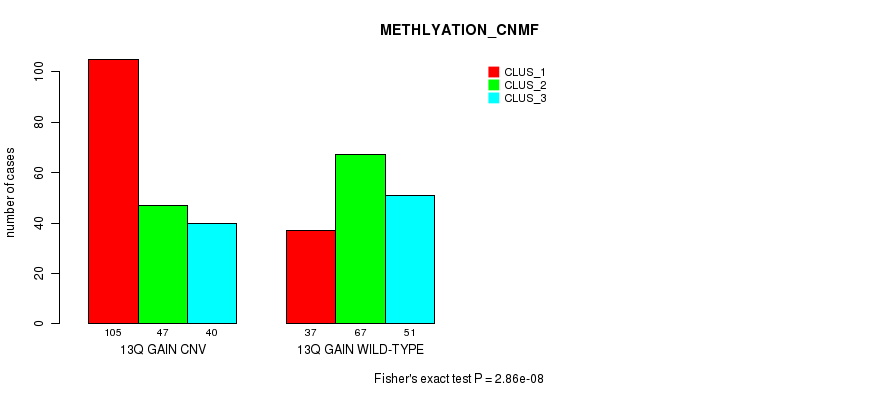
P value = 8.54e-09 (Fisher's exact test), Q value = 7.4e-06
Table S30. Gene #25: '13q gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 38 | 81 | 56 | 66 |
| 13Q GAIN CNV | 25 | 57 | 28 | 14 |
| 13Q GAIN WILD-TYPE | 13 | 24 | 28 | 52 |
Figure S30. Get High-res Image Gene #25: '13q gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
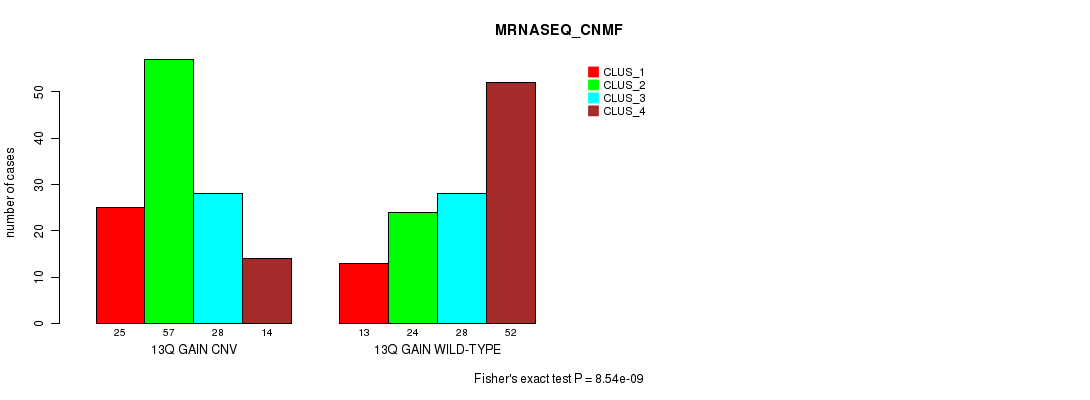
P value = 6.4e-07 (Fisher's exact test), Q value = 0.00054
Table S31. Gene #25: '13q gain' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 95 | 67 | 79 |
| 13Q GAIN CNV | 53 | 17 | 54 |
| 13Q GAIN WILD-TYPE | 42 | 50 | 25 |
Figure S31. Get High-res Image Gene #25: '13q gain' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
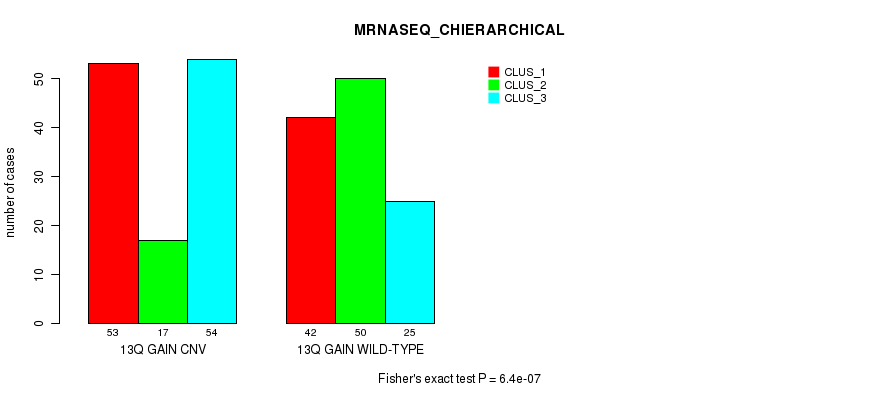
P value = 2.85e-07 (Fisher's exact test), Q value = 0.00024
Table S32. Gene #25: '13q gain' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 105 | 105 | 85 |
| 13Q GAIN CNV | 39 | 78 | 50 |
| 13Q GAIN WILD-TYPE | 66 | 27 | 35 |
Figure S32. Get High-res Image Gene #25: '13q gain' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
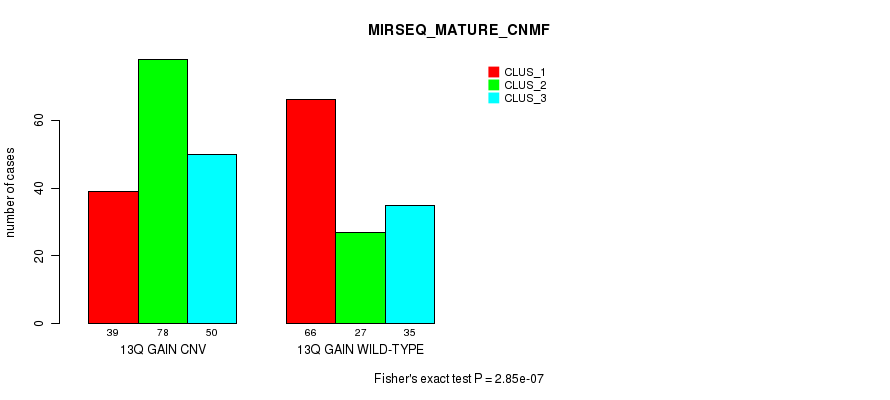
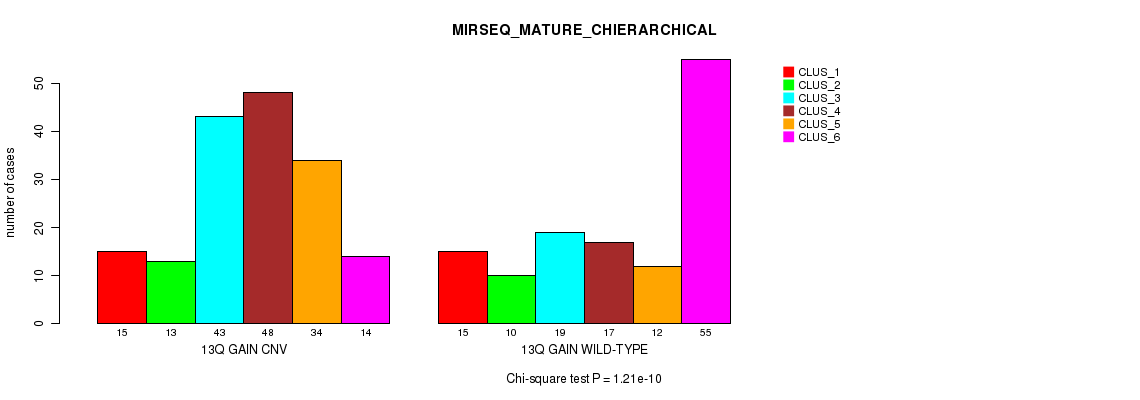
P value = 1.21e-10 (Chi-square test), Q value = 1.1e-07
Table S33. Gene #25: '13q gain' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 30 | 23 | 62 | 65 | 46 | 69 |
| 13Q GAIN CNV | 15 | 13 | 43 | 48 | 34 | 14 |
| 13Q GAIN WILD-TYPE | 15 | 10 | 19 | 17 | 12 | 55 |
Figure S33. Get High-res Image Gene #25: '13q gain' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
P value = 1.26e-16 (Fisher's exact test), Q value = 1.1e-13
Table S34. Gene #28: '16p gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 16P GAIN CNV | 66 | 7 | 16 | 7 |
| 16P GAIN WILD-TYPE | 134 | 227 | 84 | 34 |
Figure S34. Get High-res Image Gene #28: '16p gain' versus Molecular Subtype #3: 'CN_CNMF'
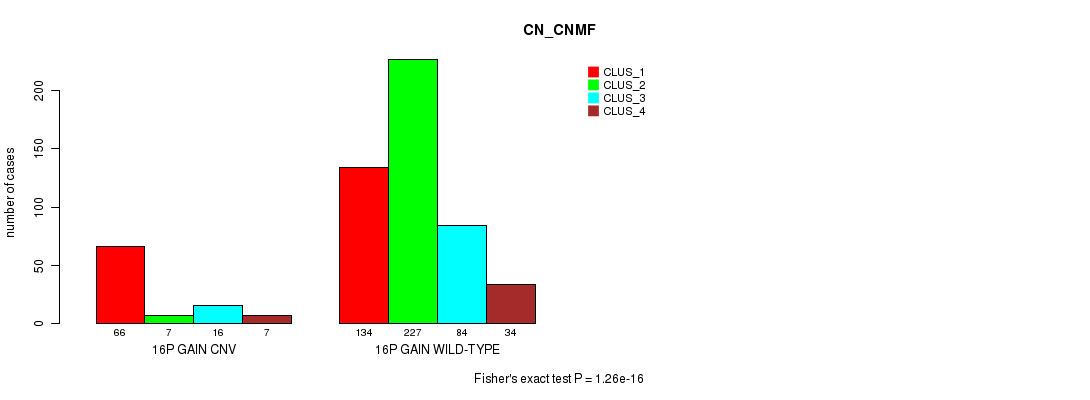
P value = 3.02e-18 (Fisher's exact test), Q value = 2.7e-15
Table S35. Gene #29: '16q gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 16Q GAIN CNV | 68 | 6 | 16 | 6 |
| 16Q GAIN WILD-TYPE | 132 | 228 | 84 | 35 |
Figure S35. Get High-res Image Gene #29: '16q gain' versus Molecular Subtype #3: 'CN_CNMF'
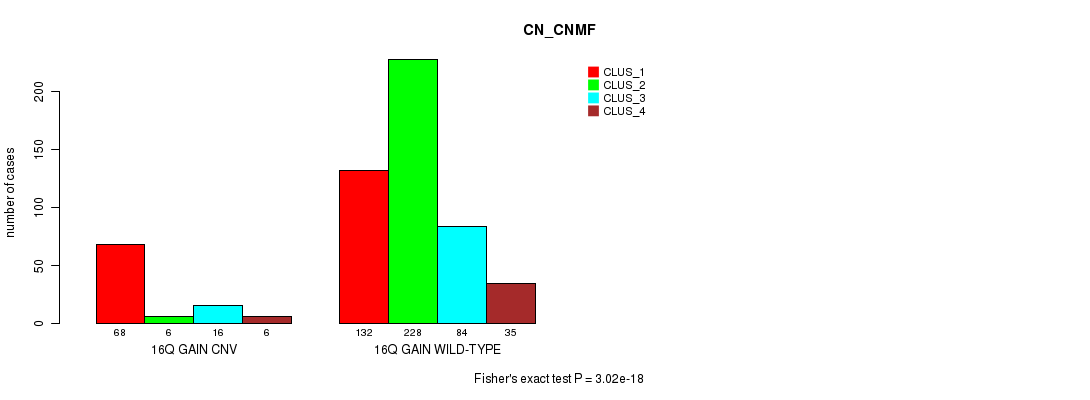
P value = 0.000269 (Fisher's exact test), Q value = 0.22
Table S36. Gene #31: '17q gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 17Q GAIN CNV | 36 | 13 | 15 | 3 |
| 17Q GAIN WILD-TYPE | 164 | 221 | 85 | 38 |
Figure S36. Get High-res Image Gene #31: '17q gain' versus Molecular Subtype #3: 'CN_CNMF'
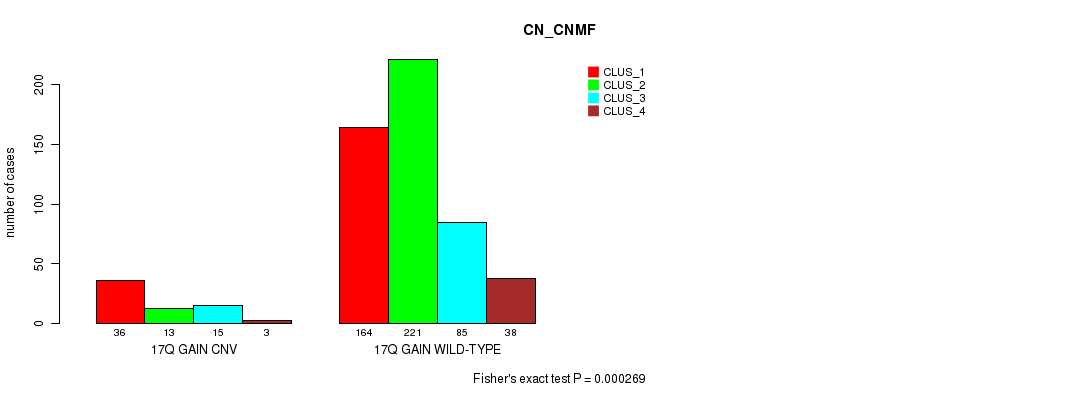
P value = 1.47e-05 (Fisher's exact test), Q value = 0.012
Table S37. Gene #31: '17q gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 38 | 81 | 56 | 66 |
| 17Q GAIN CNV | 5 | 22 | 2 | 2 |
| 17Q GAIN WILD-TYPE | 33 | 59 | 54 | 64 |
Figure S37. Get High-res Image Gene #31: '17q gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
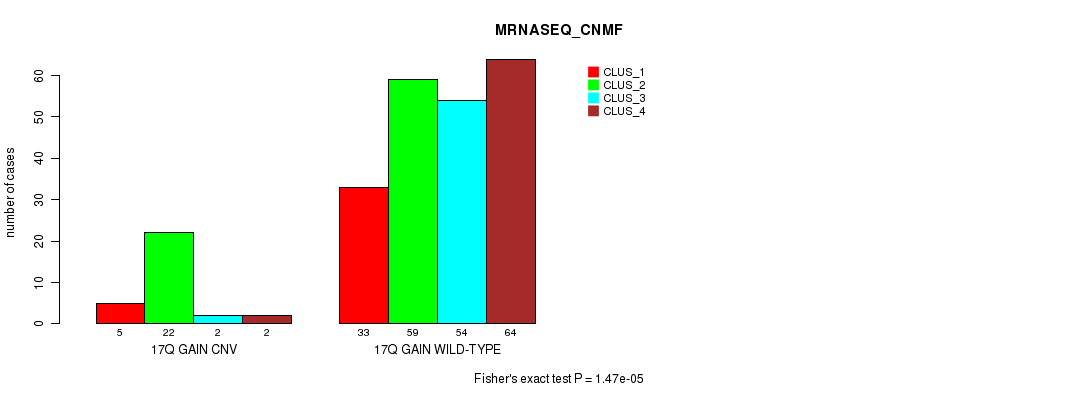
P value = 5.84e-05 (Fisher's exact test), Q value = 0.048
Table S38. Gene #34: '19p gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 19P GAIN CNV | 29 | 7 | 10 | 6 |
| 19P GAIN WILD-TYPE | 171 | 227 | 90 | 35 |
Figure S38. Get High-res Image Gene #34: '19p gain' versus Molecular Subtype #3: 'CN_CNMF'
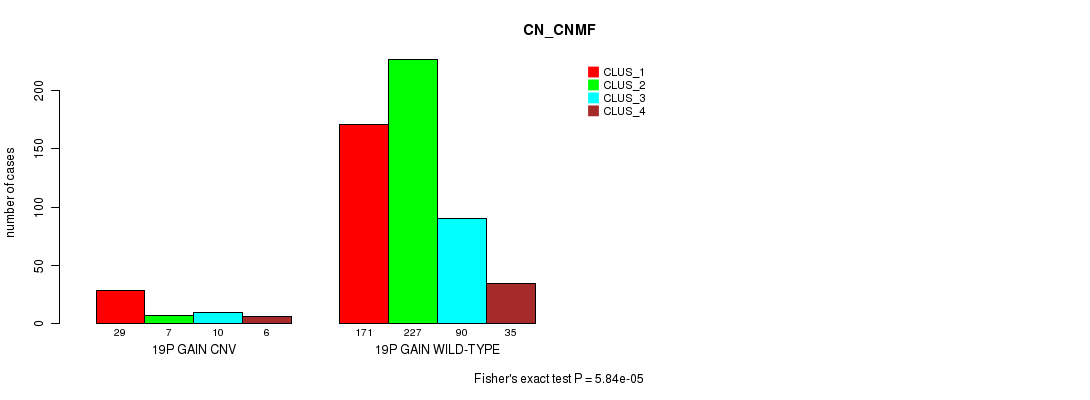
P value = 0.000145 (Fisher's exact test), Q value = 0.12
Table S39. Gene #36: '20p gain' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 62 | 69 | 41 |
| 20P GAIN CNV | 13 | 38 | 47 | 16 |
| 20P GAIN WILD-TYPE | 30 | 24 | 22 | 25 |
Figure S39. Get High-res Image Gene #36: '20p gain' versus Molecular Subtype #1: 'MRNA_CNMF'
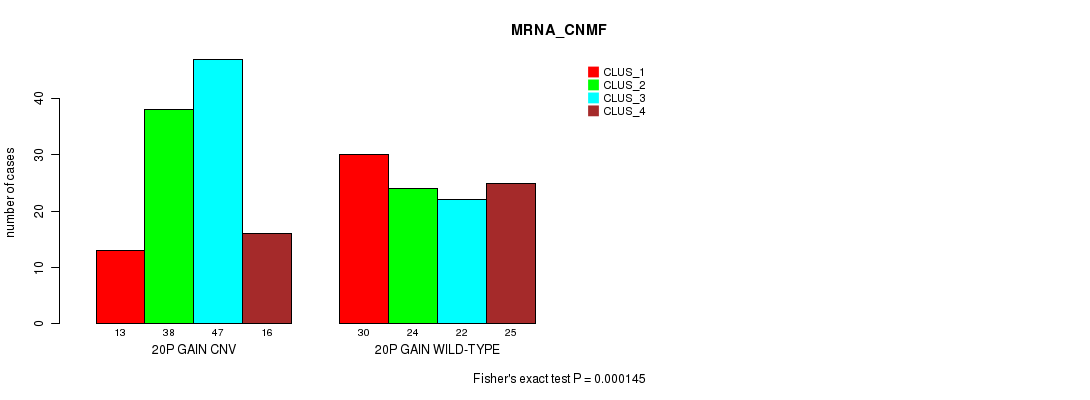
P value = 1.65e-07 (Fisher's exact test), Q value = 0.00014
Table S40. Gene #36: '20p gain' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 50 | 63 | 60 |
| 20P GAIN CNV | 32 | 30 | 15 | 37 |
| 20P GAIN WILD-TYPE | 10 | 20 | 48 | 23 |
Figure S40. Get High-res Image Gene #36: '20p gain' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
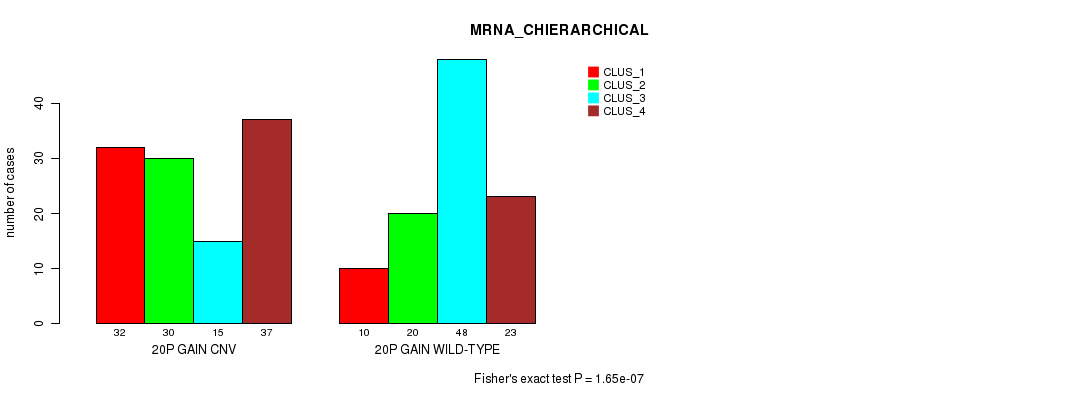
P value = 4.25e-22 (Fisher's exact test), Q value = 3.8e-19
Table S41. Gene #36: '20p gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 20P GAIN CNV | 130 | 67 | 52 | 39 |
| 20P GAIN WILD-TYPE | 70 | 167 | 48 | 2 |
Figure S41. Get High-res Image Gene #36: '20p gain' versus Molecular Subtype #3: 'CN_CNMF'
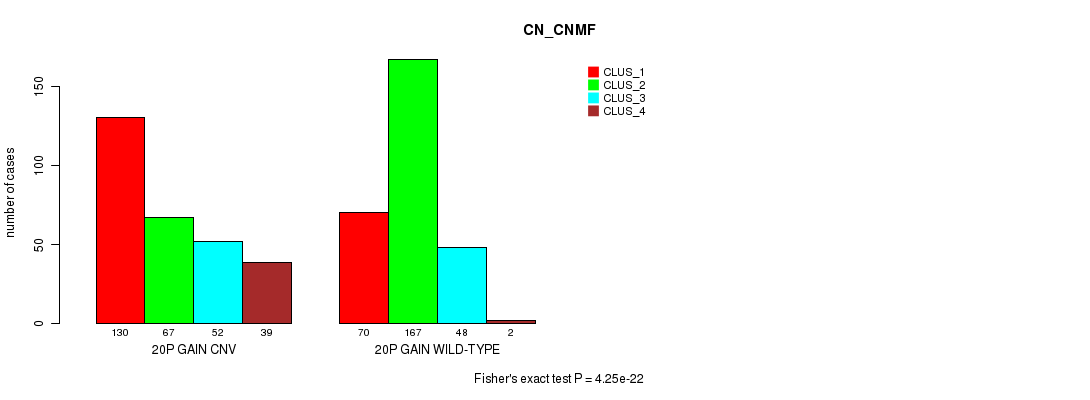
P value = 0.000106 (Fisher's exact test), Q value = 0.087
Table S42. Gene #36: '20p gain' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 142 | 114 | 91 |
| 20P GAIN CNV | 88 | 42 | 38 |
| 20P GAIN WILD-TYPE | 54 | 72 | 53 |
Figure S42. Get High-res Image Gene #36: '20p gain' versus Molecular Subtype #4: 'METHLYATION_CNMF'
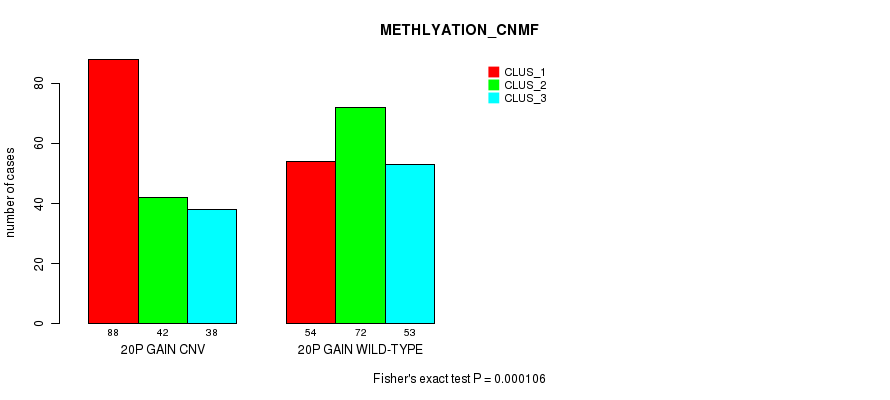
P value = 3.2e-07 (Fisher's exact test), Q value = 0.00027
Table S43. Gene #36: '20p gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 38 | 81 | 56 | 66 |
| 20P GAIN CNV | 19 | 48 | 26 | 10 |
| 20P GAIN WILD-TYPE | 19 | 33 | 30 | 56 |
Figure S43. Get High-res Image Gene #36: '20p gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
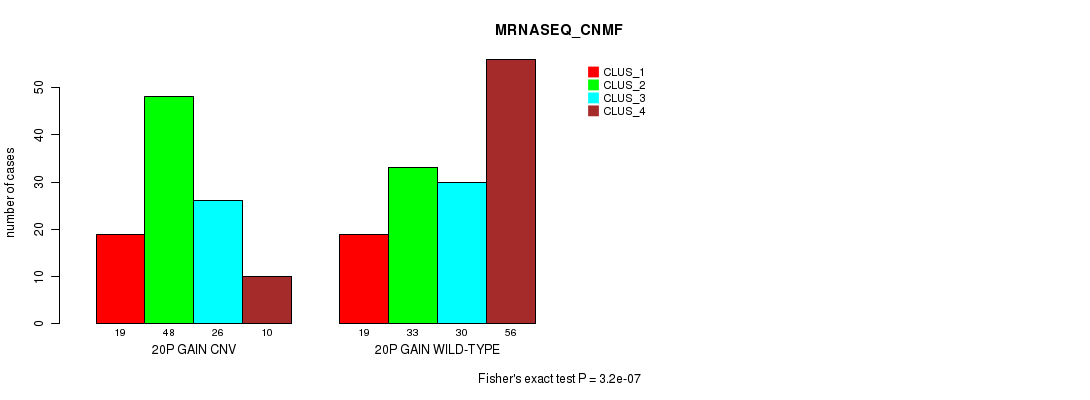
P value = 3.2e-07 (Fisher's exact test), Q value = 0.00027
Table S44. Gene #36: '20p gain' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 95 | 67 | 79 |
| 20P GAIN CNV | 46 | 11 | 46 |
| 20P GAIN WILD-TYPE | 49 | 56 | 33 |
Figure S44. Get High-res Image Gene #36: '20p gain' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
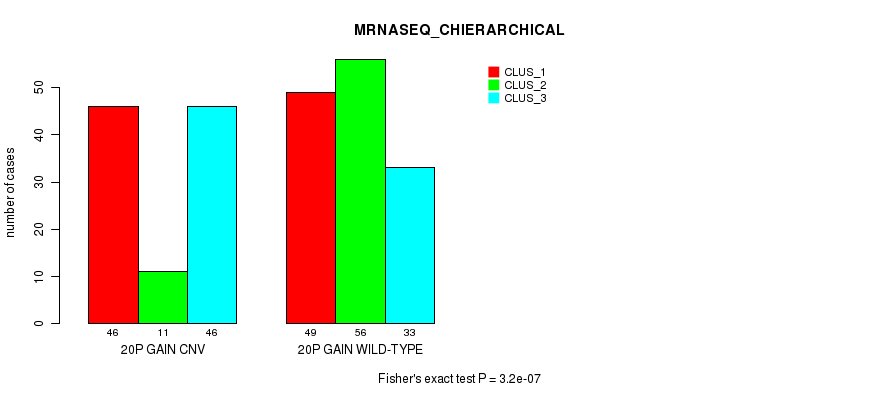
P value = 6.81e-08 (Chi-square test), Q value = 5.9e-05
Table S45. Gene #36: '20p gain' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 30 | 23 | 62 | 65 | 46 | 69 |
| 20P GAIN CNV | 16 | 13 | 42 | 40 | 20 | 12 |
| 20P GAIN WILD-TYPE | 14 | 10 | 20 | 25 | 26 | 57 |
Figure S45. Get High-res Image Gene #36: '20p gain' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
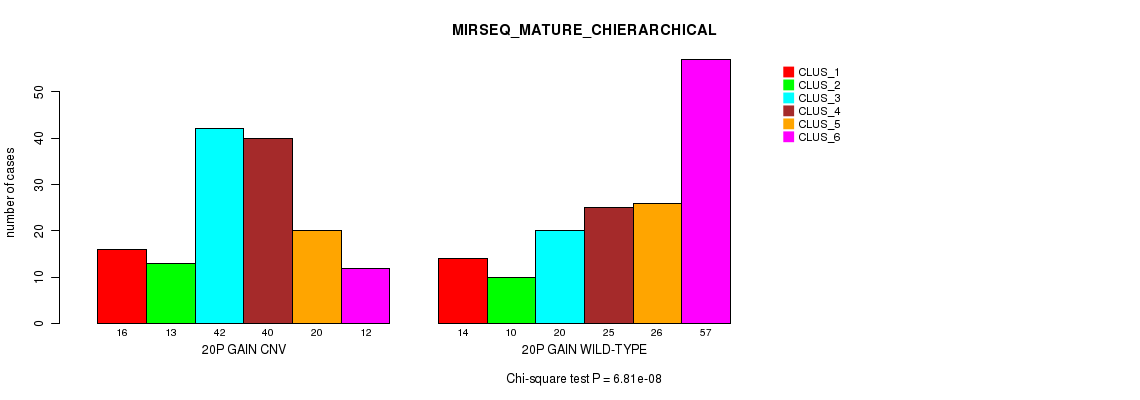
P value = 4.38e-17 (Fisher's exact test), Q value = 3.9e-14
Table S46. Gene #37: '20q gain' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 62 | 69 | 41 |
| 20Q GAIN CNV | 14 | 60 | 62 | 20 |
| 20Q GAIN WILD-TYPE | 29 | 2 | 7 | 21 |
Figure S46. Get High-res Image Gene #37: '20q gain' versus Molecular Subtype #1: 'MRNA_CNMF'
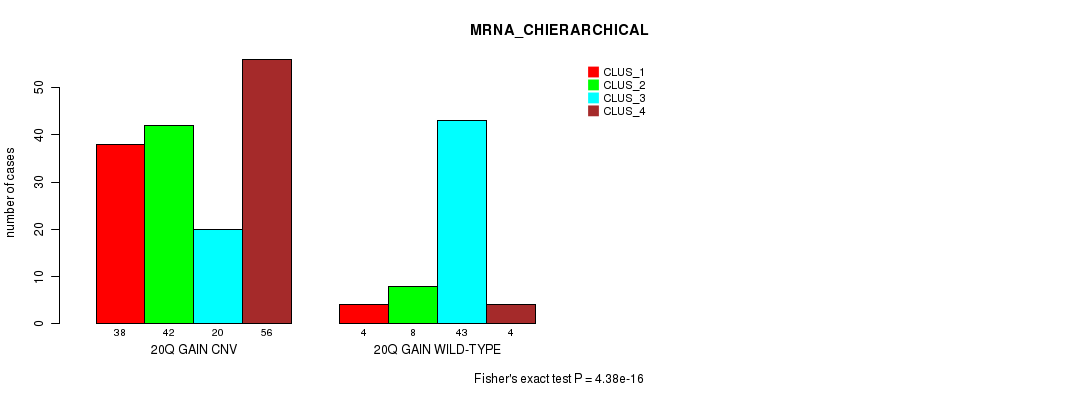
P value = 4.38e-16 (Fisher's exact test), Q value = 3.9e-13
Table S47. Gene #37: '20q gain' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 50 | 63 | 60 |
| 20Q GAIN CNV | 38 | 42 | 20 | 56 |
| 20Q GAIN WILD-TYPE | 4 | 8 | 43 | 4 |
Figure S47. Get High-res Image Gene #37: '20q gain' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
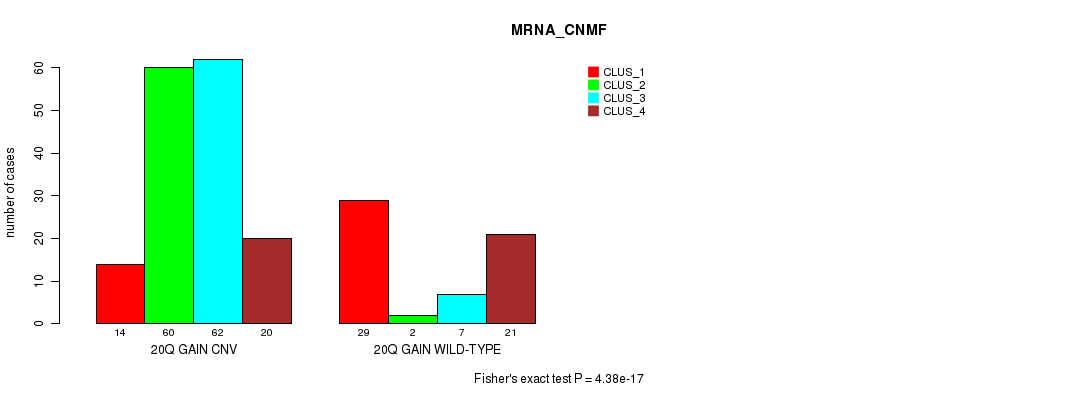
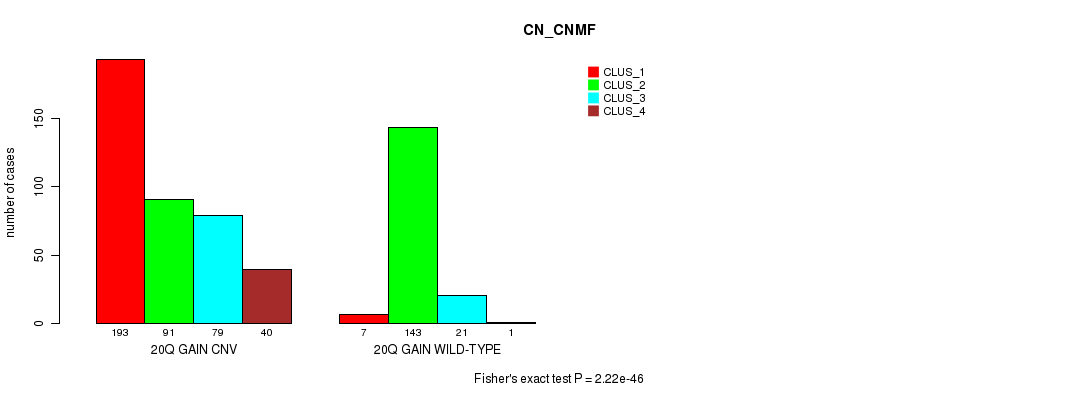
P value = 2.22e-46 (Fisher's exact test), Q value = 2e-43
Table S48. Gene #37: '20q gain' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 20Q GAIN CNV | 193 | 91 | 79 | 40 |
| 20Q GAIN WILD-TYPE | 7 | 143 | 21 | 1 |
Figure S48. Get High-res Image Gene #37: '20q gain' versus Molecular Subtype #3: 'CN_CNMF'
P value = 1.68e-12 (Fisher's exact test), Q value = 1.5e-09
Table S49. Gene #37: '20q gain' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 142 | 114 | 91 |
| 20Q GAIN CNV | 127 | 58 | 53 |
| 20Q GAIN WILD-TYPE | 15 | 56 | 38 |
Figure S49. Get High-res Image Gene #37: '20q gain' versus Molecular Subtype #4: 'METHLYATION_CNMF'
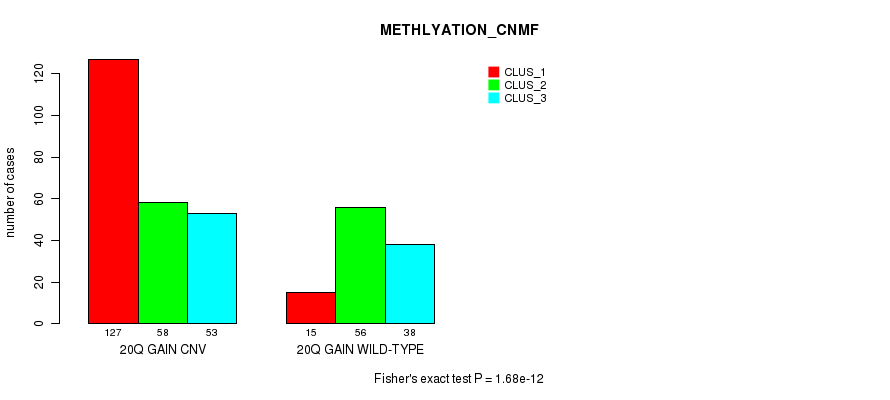
P value = 0.000311 (Fisher's exact test), Q value = 0.25
Table S50. Gene #37: '20q gain' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 115 | 184 | 33 | 119 |
| 20Q GAIN CNV | 85 | 141 | 23 | 64 |
| 20Q GAIN WILD-TYPE | 30 | 43 | 10 | 55 |
Figure S50. Get High-res Image Gene #37: '20q gain' versus Molecular Subtype #5: 'RPPA_CNMF'
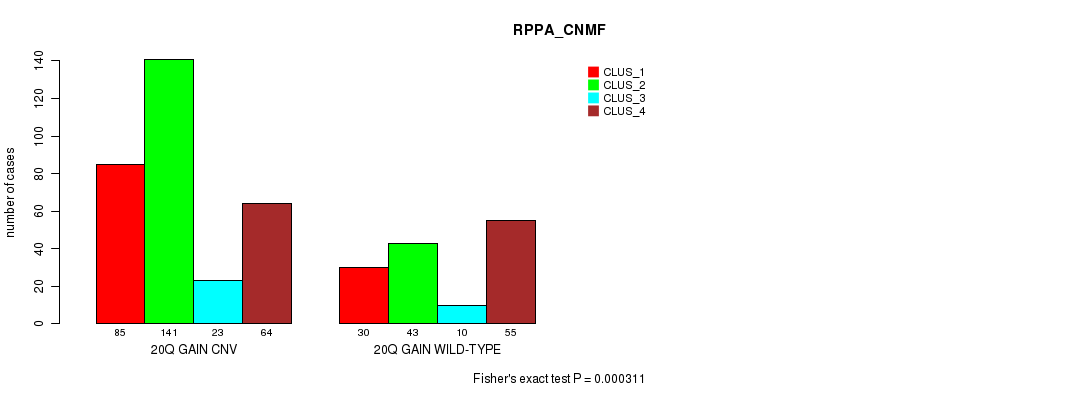
P value = 0.000133 (Fisher's exact test), Q value = 0.11
Table S51. Gene #37: '20q gain' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 76 | 248 | 92 | 35 |
| 20Q GAIN CNV | 62 | 182 | 50 | 19 |
| 20Q GAIN WILD-TYPE | 14 | 66 | 42 | 16 |
Figure S51. Get High-res Image Gene #37: '20q gain' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
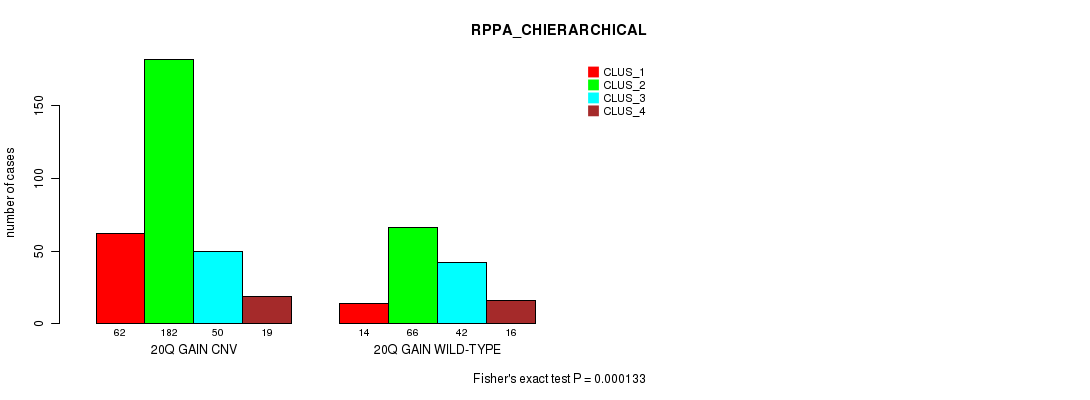
P value = 9.03e-18 (Fisher's exact test), Q value = 8e-15
Table S52. Gene #37: '20q gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 38 | 81 | 56 | 66 |
| 20Q GAIN CNV | 27 | 74 | 41 | 15 |
| 20Q GAIN WILD-TYPE | 11 | 7 | 15 | 51 |
Figure S52. Get High-res Image Gene #37: '20q gain' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
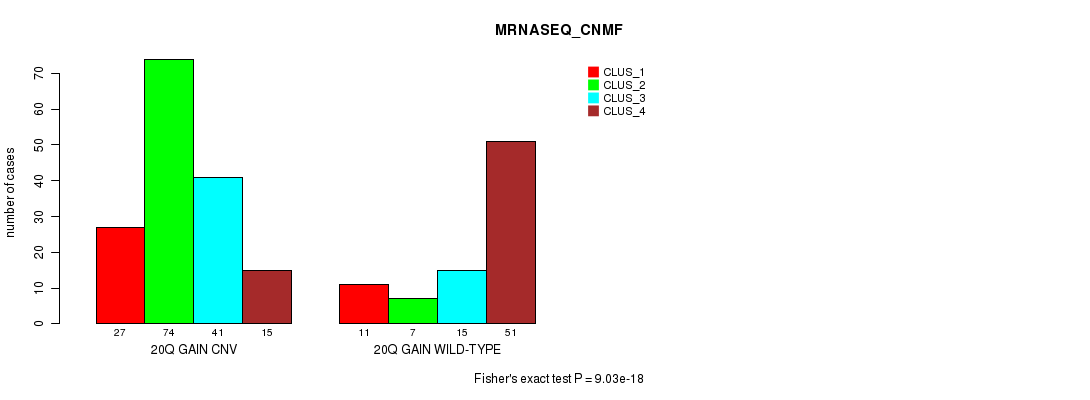
P value = 1.13e-13 (Fisher's exact test), Q value = 1e-10
Table S53. Gene #37: '20q gain' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 95 | 67 | 79 |
| 20Q GAIN CNV | 69 | 19 | 69 |
| 20Q GAIN WILD-TYPE | 26 | 48 | 10 |
Figure S53. Get High-res Image Gene #37: '20q gain' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
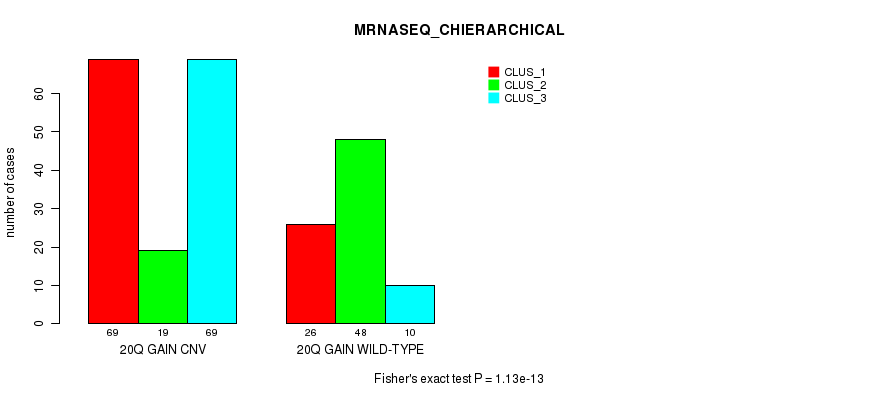
P value = 2.65e-09 (Fisher's exact test), Q value = 2.3e-06
Table S54. Gene #37: '20q gain' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 105 | 105 | 85 |
| 20Q GAIN CNV | 49 | 90 | 64 |
| 20Q GAIN WILD-TYPE | 56 | 15 | 21 |
Figure S54. Get High-res Image Gene #37: '20q gain' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
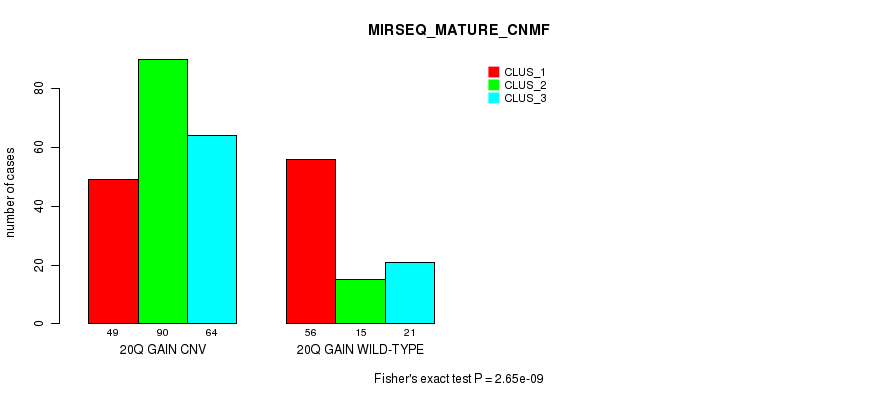
P value = 1.39e-21 (Chi-square test), Q value = 1.2e-18
Table S55. Gene #37: '20q gain' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 30 | 23 | 62 | 65 | 46 | 69 |
| 20Q GAIN CNV | 27 | 18 | 53 | 57 | 35 | 13 |
| 20Q GAIN WILD-TYPE | 3 | 5 | 9 | 8 | 11 | 56 |
Figure S55. Get High-res Image Gene #37: '20q gain' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
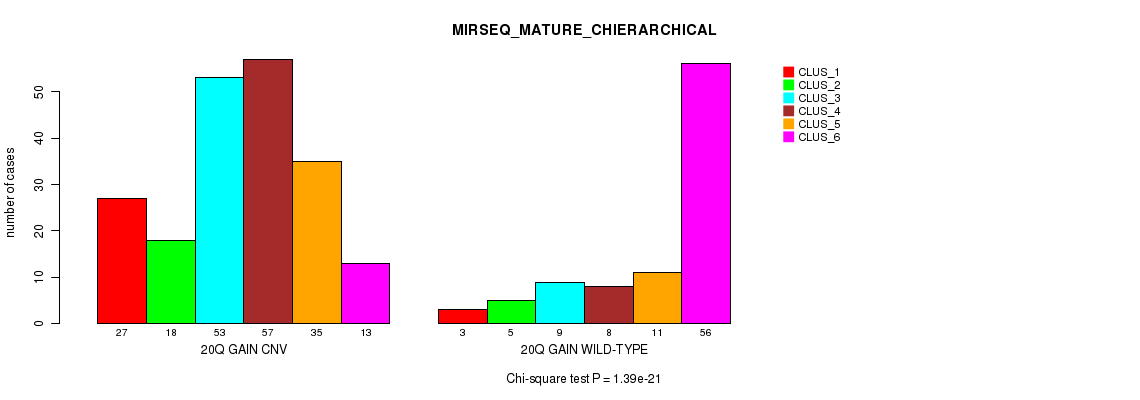
P value = 7.55e-06 (Fisher's exact test), Q value = 0.0063
Table S56. Gene #41: '1p loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 1P LOSS CNV | 39 | 14 | 6 | 9 |
| 1P LOSS WILD-TYPE | 161 | 220 | 94 | 32 |
Figure S56. Get High-res Image Gene #41: '1p loss' versus Molecular Subtype #3: 'CN_CNMF'
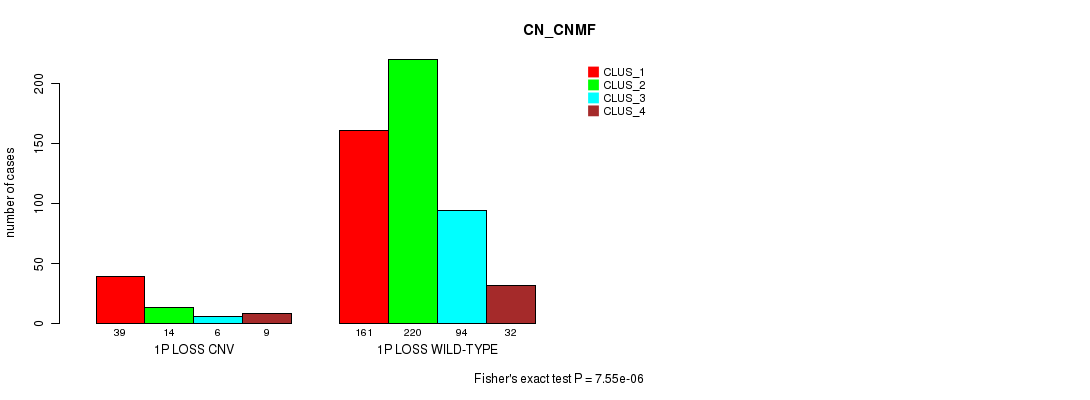
P value = 0.000189 (Fisher's exact test), Q value = 0.15
Table S57. Gene #41: '1p loss' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 142 | 114 | 91 |
| 1P LOSS CNV | 28 | 5 | 6 |
| 1P LOSS WILD-TYPE | 114 | 109 | 85 |
Figure S57. Get High-res Image Gene #41: '1p loss' versus Molecular Subtype #4: 'METHLYATION_CNMF'
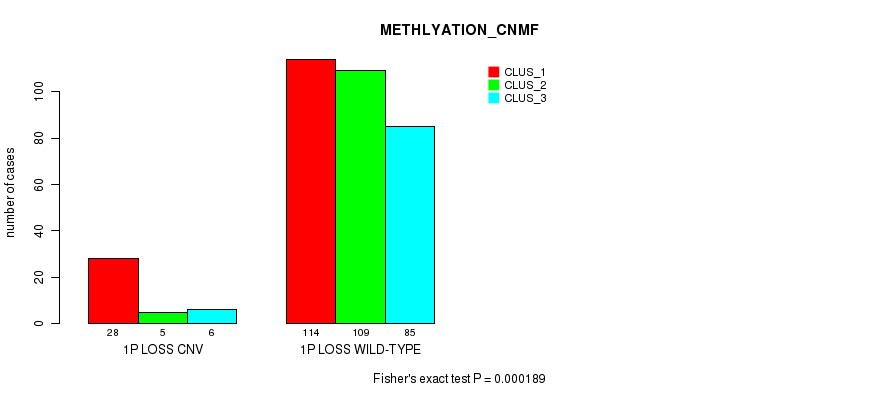
P value = 5.5e-11 (Fisher's exact test), Q value = 4.8e-08
Table S58. Gene #47: '4p loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 4P LOSS CNV | 70 | 21 | 17 | 14 |
| 4P LOSS WILD-TYPE | 130 | 213 | 83 | 27 |
Figure S58. Get High-res Image Gene #47: '4p loss' versus Molecular Subtype #3: 'CN_CNMF'
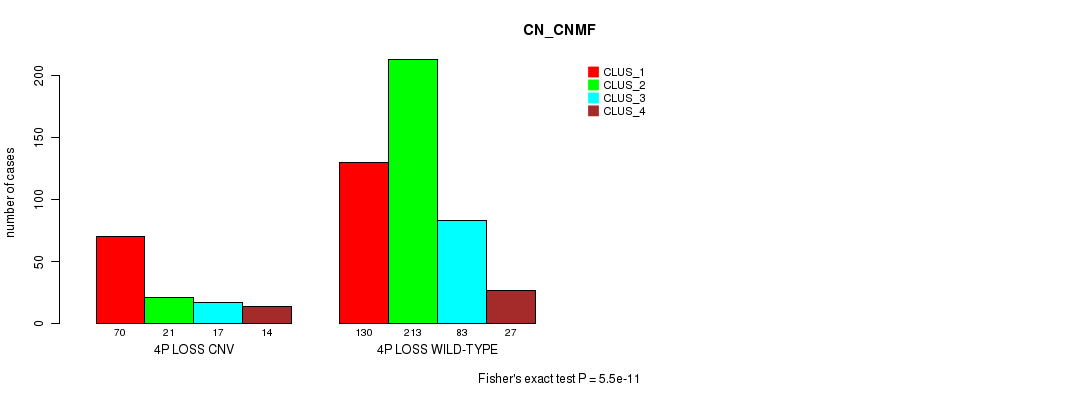
P value = 3.18e-12 (Fisher's exact test), Q value = 2.8e-09
Table S59. Gene #48: '4q loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 4Q LOSS CNV | 68 | 16 | 17 | 12 |
| 4Q LOSS WILD-TYPE | 132 | 218 | 83 | 29 |
Figure S59. Get High-res Image Gene #48: '4q loss' versus Molecular Subtype #3: 'CN_CNMF'
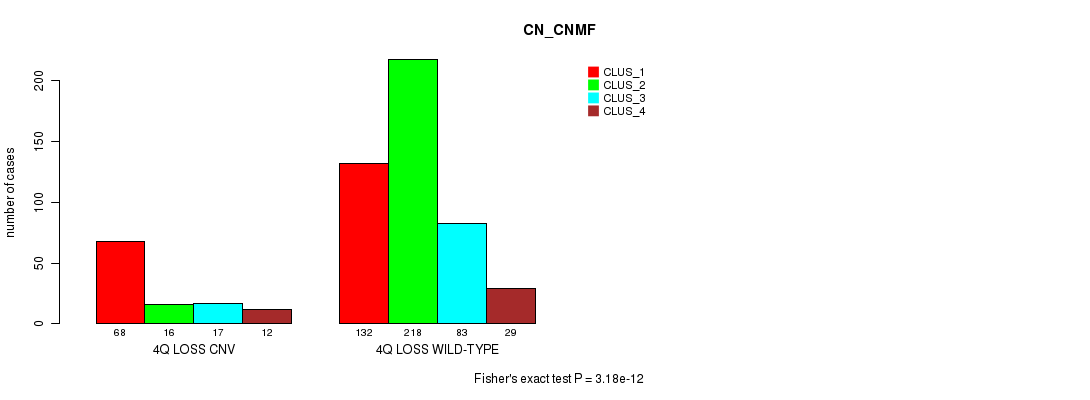
P value = 5.53e-07 (Fisher's exact test), Q value = 0.00047
Table S60. Gene #50: '5q loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 5Q LOSS CNV | 31 | 6 | 17 | 6 |
| 5Q LOSS WILD-TYPE | 169 | 228 | 83 | 35 |
Figure S60. Get High-res Image Gene #50: '5q loss' versus Molecular Subtype #3: 'CN_CNMF'
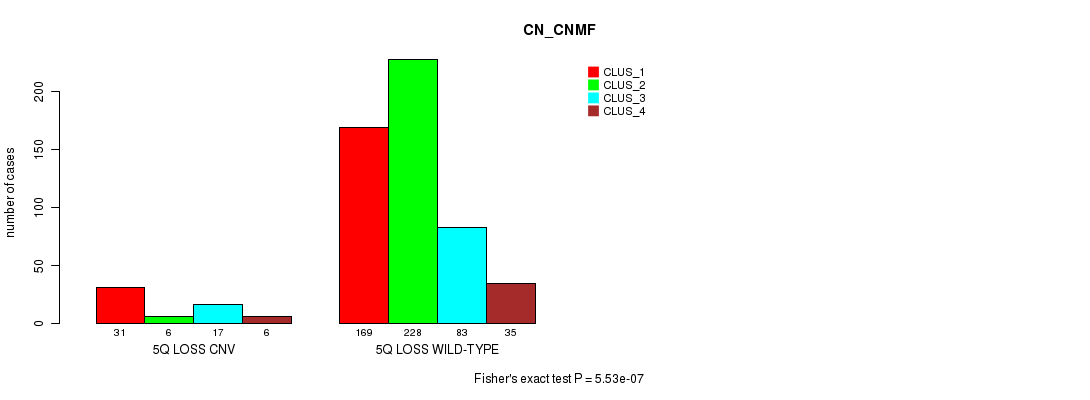
P value = 1.14e-18 (Fisher's exact test), Q value = 1e-15
Table S61. Gene #54: '8p loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 8P LOSS CNV | 88 | 17 | 31 | 9 |
| 8P LOSS WILD-TYPE | 112 | 217 | 69 | 32 |
Figure S61. Get High-res Image Gene #54: '8p loss' versus Molecular Subtype #3: 'CN_CNMF'
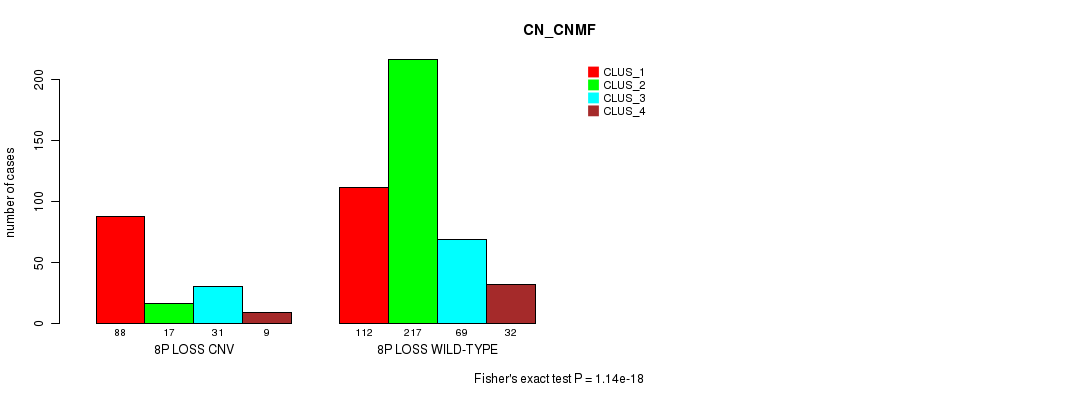
P value = 0.000109 (Fisher's exact test), Q value = 0.089
Table S62. Gene #54: '8p loss' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 142 | 114 | 91 |
| 8P LOSS CNV | 50 | 17 | 14 |
| 8P LOSS WILD-TYPE | 92 | 97 | 77 |
Figure S62. Get High-res Image Gene #54: '8p loss' versus Molecular Subtype #4: 'METHLYATION_CNMF'
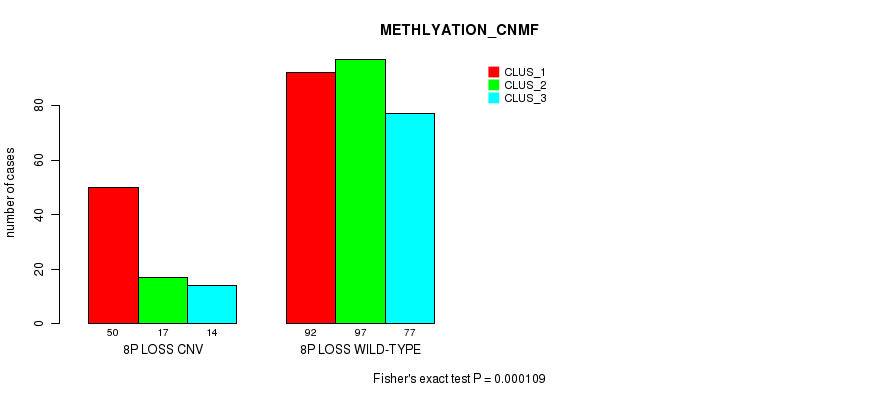
P value = 2.9e-05 (Fisher's exact test), Q value = 0.024
Table S63. Gene #56: '9p loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 9P LOSS CNV | 26 | 4 | 7 | 2 |
| 9P LOSS WILD-TYPE | 174 | 230 | 93 | 39 |
Figure S63. Get High-res Image Gene #56: '9p loss' versus Molecular Subtype #3: 'CN_CNMF'
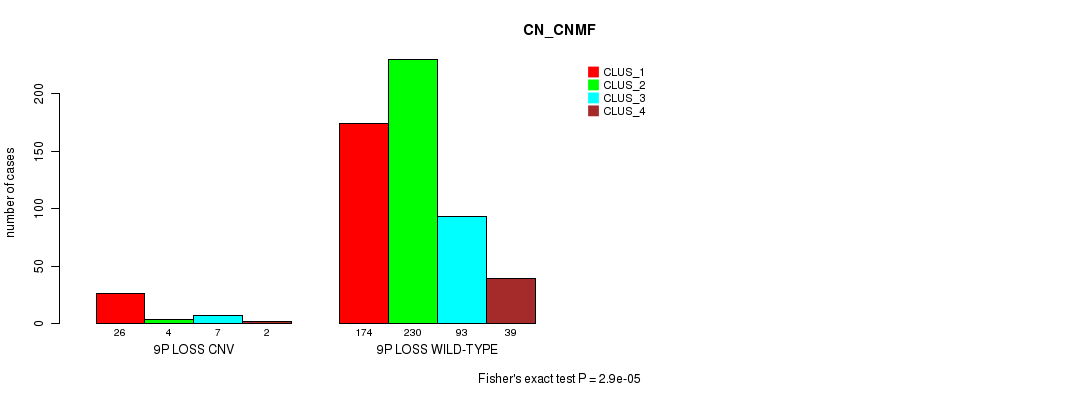
P value = 1.95e-06 (Fisher's exact test), Q value = 0.0016
Table S64. Gene #57: '9q loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 9Q LOSS CNV | 28 | 4 | 3 | 2 |
| 9Q LOSS WILD-TYPE | 172 | 230 | 97 | 39 |
Figure S64. Get High-res Image Gene #57: '9q loss' versus Molecular Subtype #3: 'CN_CNMF'
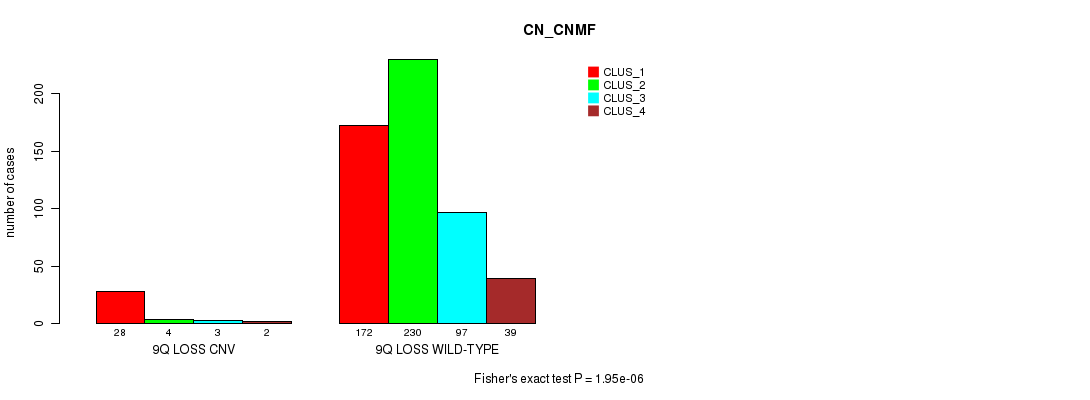
P value = 1.44e-06 (Fisher's exact test), Q value = 0.0012
Table S65. Gene #59: '10q loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 10Q LOSS CNV | 16 | 6 | 13 | 11 |
| 10Q LOSS WILD-TYPE | 184 | 228 | 87 | 30 |
Figure S65. Get High-res Image Gene #59: '10q loss' versus Molecular Subtype #3: 'CN_CNMF'
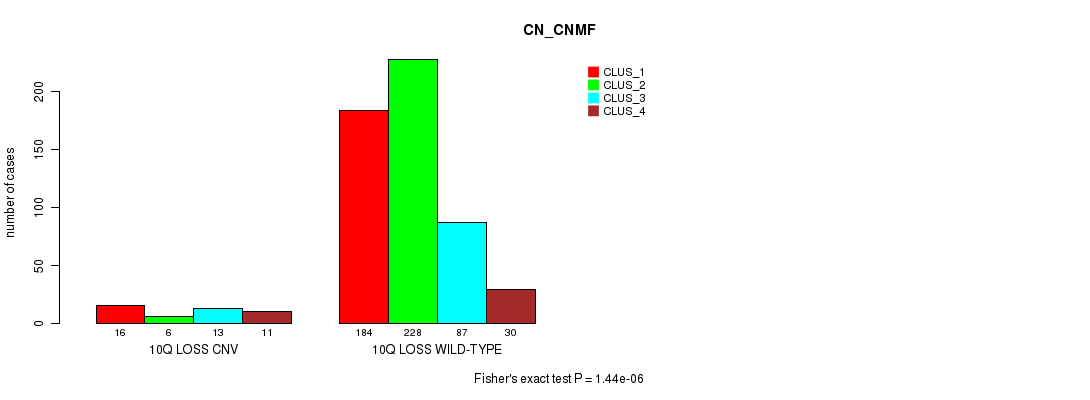
P value = 5.42e-07 (Fisher's exact test), Q value = 0.00046
Table S66. Gene #62: '12p loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 12P LOSS CNV | 21 | 2 | 3 | 7 |
| 12P LOSS WILD-TYPE | 179 | 232 | 97 | 34 |
Figure S66. Get High-res Image Gene #62: '12p loss' versus Molecular Subtype #3: 'CN_CNMF'
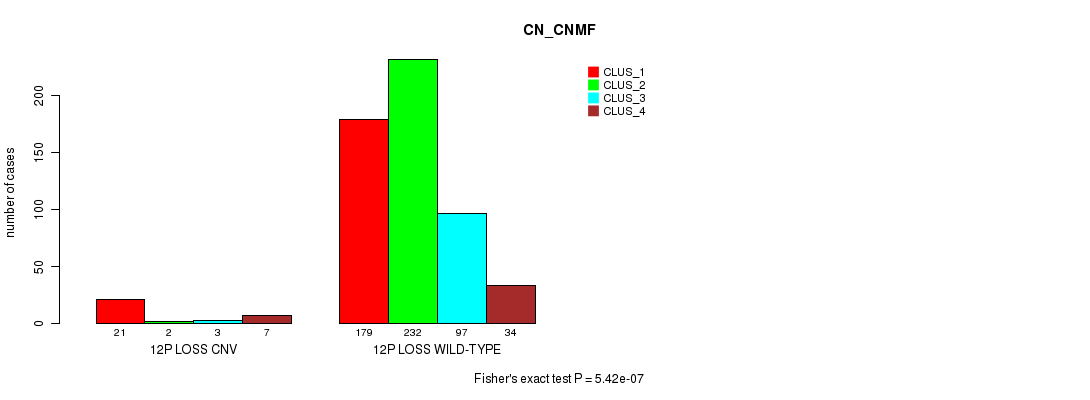
P value = 1.11e-06 (Fisher's exact test), Q value = 0.00094
Table S67. Gene #63: '12q loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 12Q LOSS CNV | 19 | 2 | 1 | 6 |
| 12Q LOSS WILD-TYPE | 181 | 232 | 99 | 35 |
Figure S67. Get High-res Image Gene #63: '12q loss' versus Molecular Subtype #3: 'CN_CNMF'
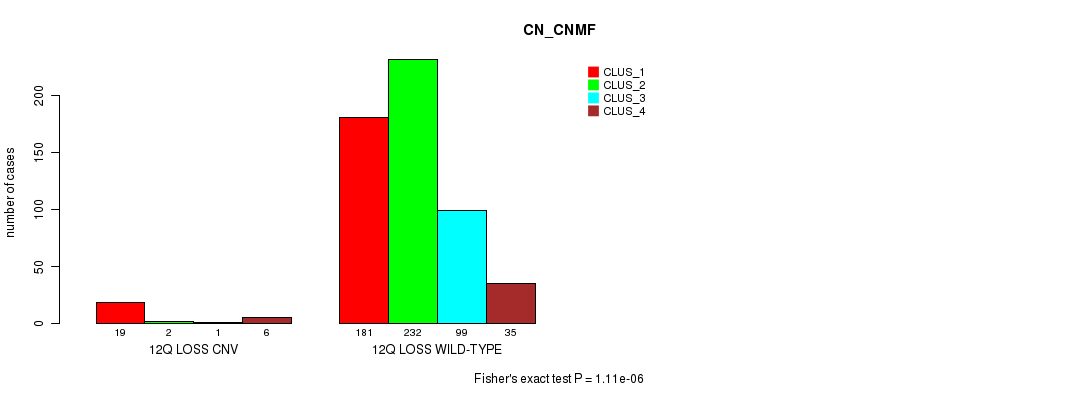
P value = 6.52e-05 (Fisher's exact test), Q value = 0.054
Table S68. Gene #65: '14q loss' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 62 | 69 | 41 |
| 14Q LOSS CNV | 6 | 27 | 26 | 4 |
| 14Q LOSS WILD-TYPE | 37 | 35 | 43 | 37 |
Figure S68. Get High-res Image Gene #65: '14q loss' versus Molecular Subtype #1: 'MRNA_CNMF'
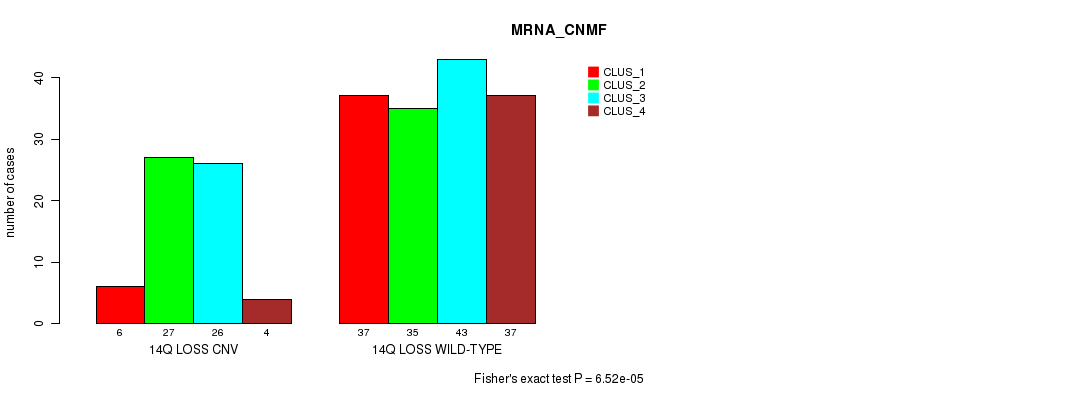
P value = 2.84e-07 (Fisher's exact test), Q value = 0.00024
Table S69. Gene #65: '14q loss' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 50 | 63 | 60 |
| 14Q LOSS CNV | 20 | 17 | 3 | 23 |
| 14Q LOSS WILD-TYPE | 22 | 33 | 60 | 37 |
Figure S69. Get High-res Image Gene #65: '14q loss' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
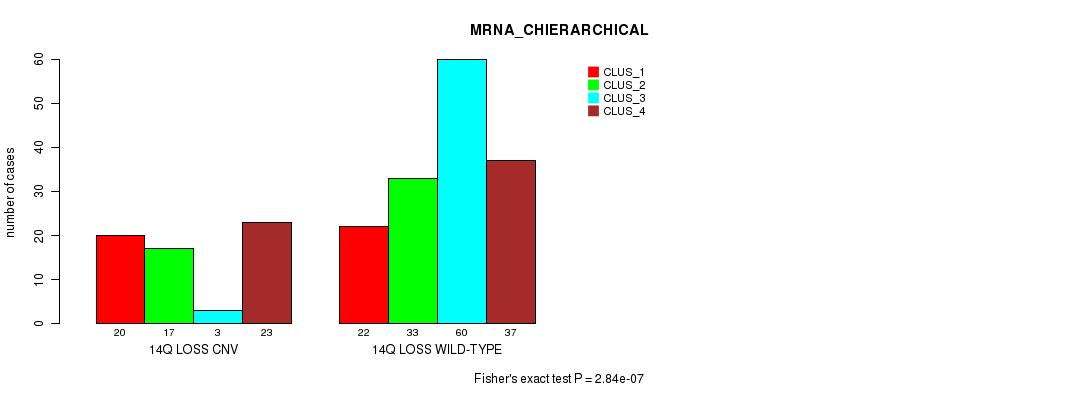
P value = 1.12e-31 (Fisher's exact test), Q value = 1e-28
Table S70. Gene #65: '14q loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 14Q LOSS CNV | 109 | 20 | 6 | 8 |
| 14Q LOSS WILD-TYPE | 91 | 214 | 94 | 33 |
Figure S70. Get High-res Image Gene #65: '14q loss' versus Molecular Subtype #3: 'CN_CNMF'
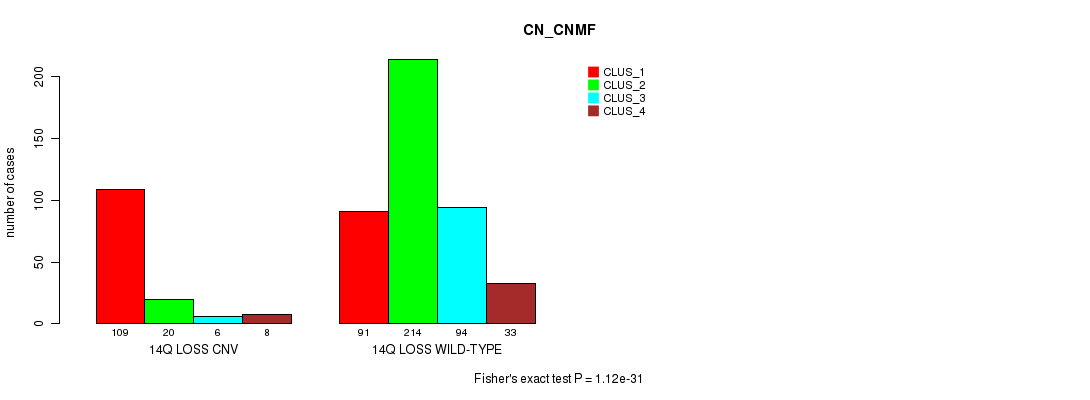
P value = 1.63e-07 (Fisher's exact test), Q value = 0.00014
Table S71. Gene #65: '14q loss' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 142 | 114 | 91 |
| 14Q LOSS CNV | 53 | 13 | 11 |
| 14Q LOSS WILD-TYPE | 89 | 101 | 80 |
Figure S71. Get High-res Image Gene #65: '14q loss' versus Molecular Subtype #4: 'METHLYATION_CNMF'
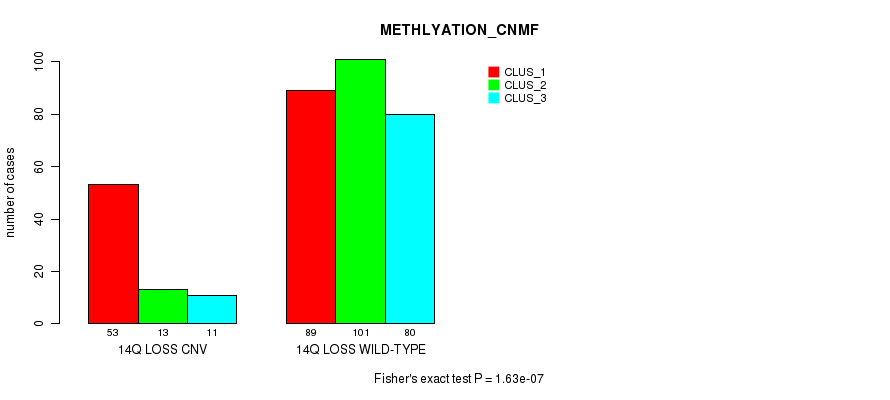
P value = 1.73e-07 (Fisher's exact test), Q value = 0.00015
Table S72. Gene #65: '14q loss' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 38 | 81 | 56 | 66 |
| 14Q LOSS CNV | 9 | 33 | 10 | 2 |
| 14Q LOSS WILD-TYPE | 29 | 48 | 46 | 64 |
Figure S72. Get High-res Image Gene #65: '14q loss' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
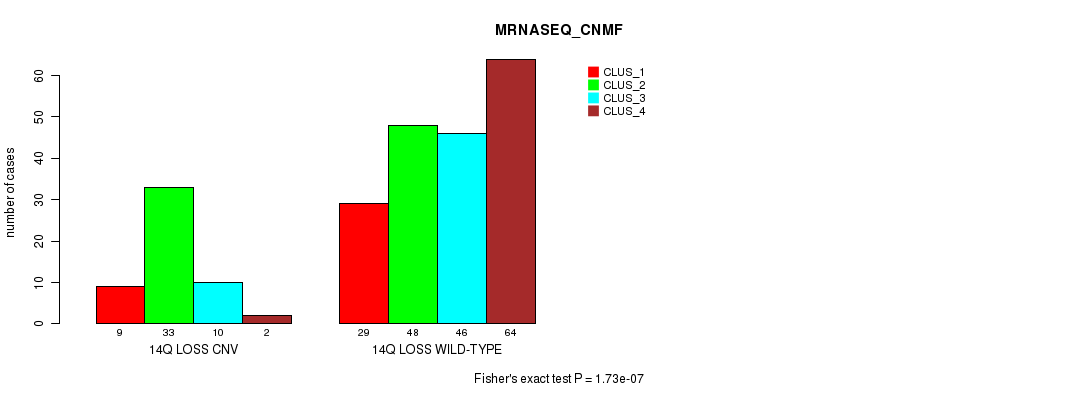
P value = 1.51e-06 (Fisher's exact test), Q value = 0.0013
Table S73. Gene #65: '14q loss' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 105 | 105 | 85 |
| 14Q LOSS CNV | 10 | 41 | 16 |
| 14Q LOSS WILD-TYPE | 95 | 64 | 69 |
Figure S73. Get High-res Image Gene #65: '14q loss' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
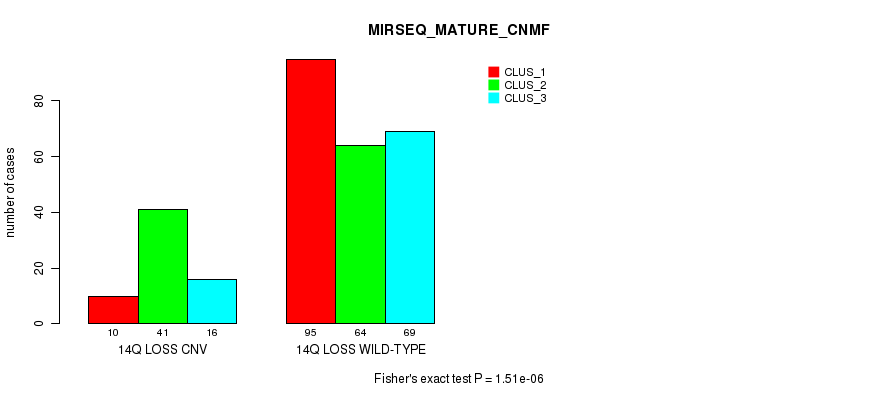
P value = 0.000145 (Chi-square test), Q value = 0.12
Table S74. Gene #65: '14q loss' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 30 | 23 | 62 | 65 | 46 | 69 |
| 14Q LOSS CNV | 11 | 5 | 18 | 19 | 13 | 1 |
| 14Q LOSS WILD-TYPE | 19 | 18 | 44 | 46 | 33 | 68 |
Figure S74. Get High-res Image Gene #65: '14q loss' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
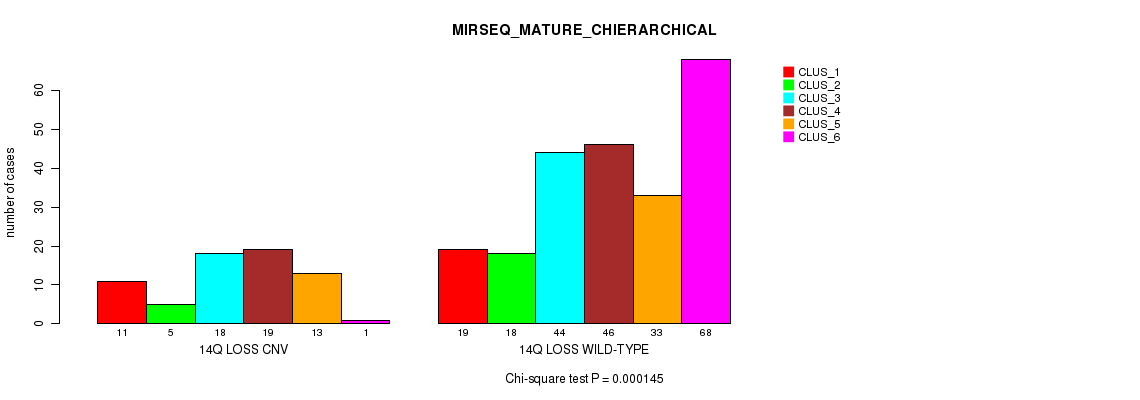
P value = 2.09e-24 (Fisher's exact test), Q value = 1.9e-21
Table S75. Gene #66: '15q loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 15Q LOSS CNV | 88 | 16 | 26 | 26 |
| 15Q LOSS WILD-TYPE | 112 | 218 | 74 | 15 |
Figure S75. Get High-res Image Gene #66: '15q loss' versus Molecular Subtype #3: 'CN_CNMF'
P value = 1.19e-05 (Fisher's exact test), Q value = 0.0099
Table S76. Gene #66: '15q loss' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 38 | 81 | 56 | 66 |
| 15Q LOSS CNV | 11 | 30 | 13 | 3 |
| 15Q LOSS WILD-TYPE | 27 | 51 | 43 | 63 |
Figure S76. Get High-res Image Gene #66: '15q loss' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
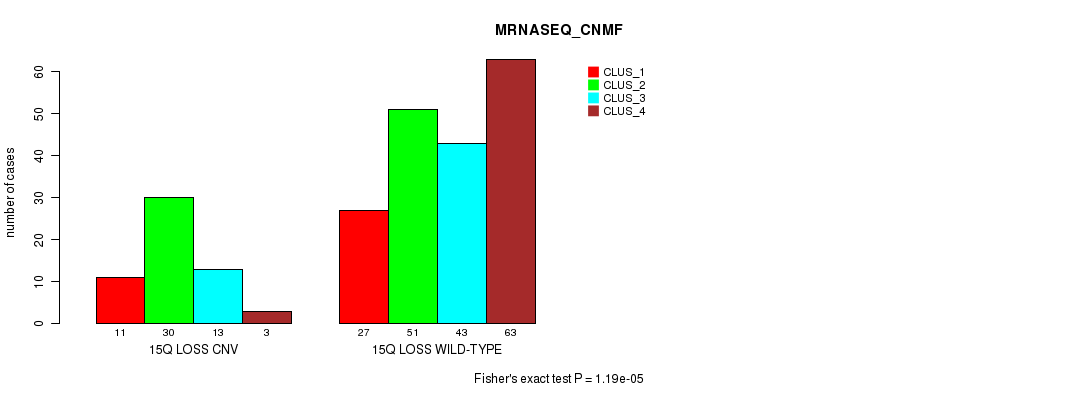
P value = 0.000158 (Chi-square test), Q value = 0.13
Table S77. Gene #66: '15q loss' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 30 | 23 | 62 | 65 | 46 | 69 |
| 15Q LOSS CNV | 8 | 1 | 20 | 22 | 14 | 4 |
| 15Q LOSS WILD-TYPE | 22 | 22 | 42 | 43 | 32 | 65 |
Figure S77. Get High-res Image Gene #66: '15q loss' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
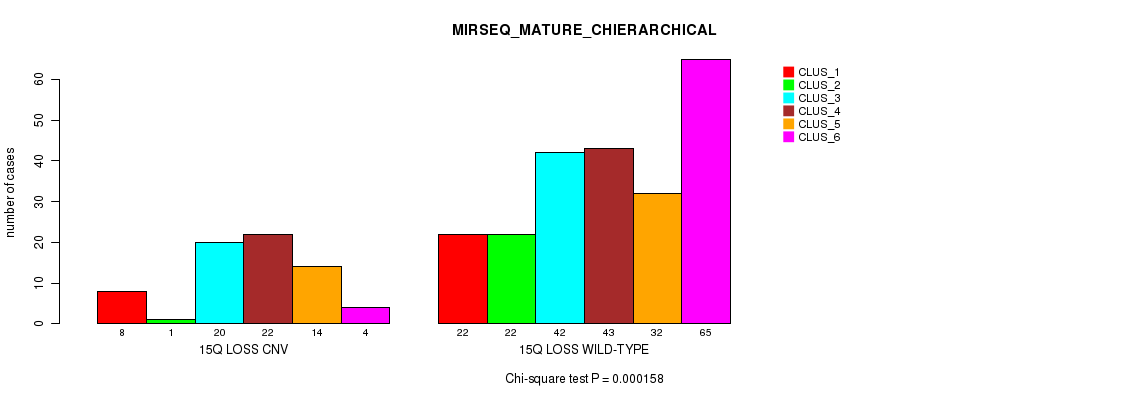
P value = 0.000277 (Fisher's exact test), Q value = 0.22
Table S78. Gene #69: '17p loss' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 62 | 69 | 41 |
| 17P LOSS CNV | 12 | 36 | 41 | 12 |
| 17P LOSS WILD-TYPE | 31 | 26 | 28 | 29 |
Figure S78. Get High-res Image Gene #69: '17p loss' versus Molecular Subtype #1: 'MRNA_CNMF'
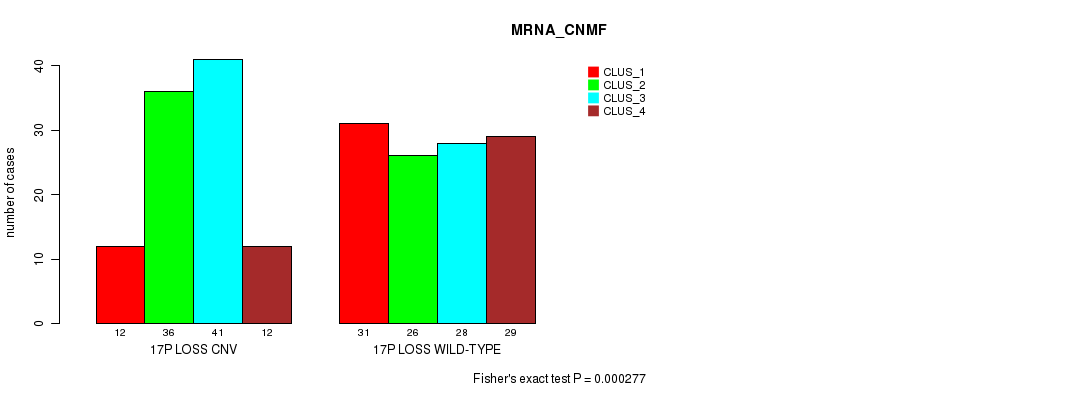
P value = 5.49e-22 (Fisher's exact test), Q value = 4.9e-19
Table S79. Gene #69: '17p loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 17P LOSS CNV | 128 | 46 | 57 | 21 |
| 17P LOSS WILD-TYPE | 72 | 188 | 43 | 20 |
Figure S79. Get High-res Image Gene #69: '17p loss' versus Molecular Subtype #3: 'CN_CNMF'
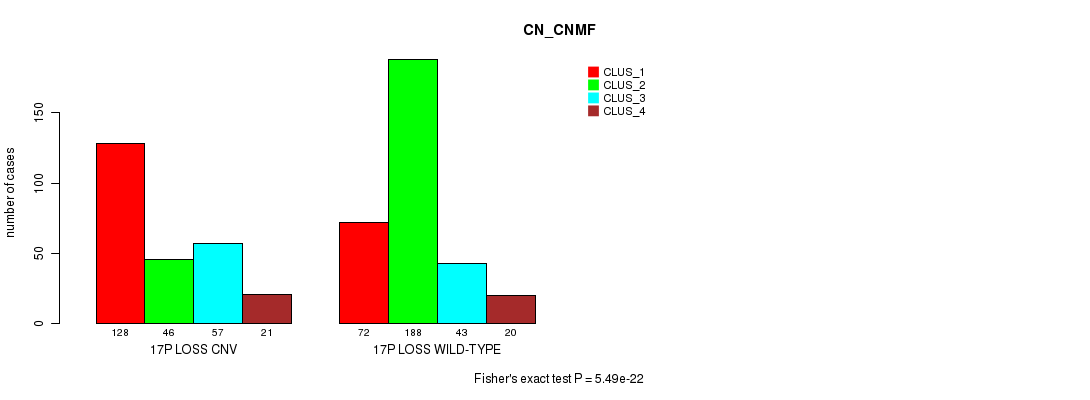
P value = 1.43e-06 (Fisher's exact test), Q value = 0.0012
Table S80. Gene #69: '17p loss' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 142 | 114 | 91 |
| 17P LOSS CNV | 84 | 37 | 27 |
| 17P LOSS WILD-TYPE | 58 | 77 | 64 |
Figure S80. Get High-res Image Gene #69: '17p loss' versus Molecular Subtype #4: 'METHLYATION_CNMF'
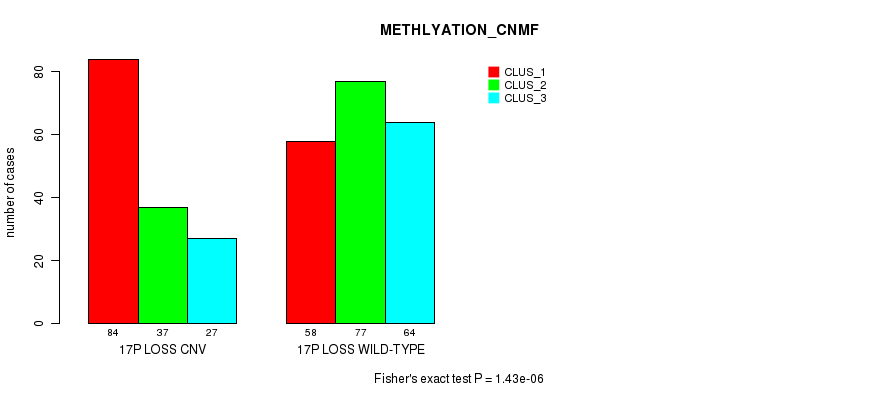
P value = 3.25e-06 (Fisher's exact test), Q value = 0.0027
Table S81. Gene #69: '17p loss' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 38 | 81 | 56 | 66 |
| 17P LOSS CNV | 21 | 45 | 21 | 11 |
| 17P LOSS WILD-TYPE | 17 | 36 | 35 | 55 |
Figure S81. Get High-res Image Gene #69: '17p loss' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
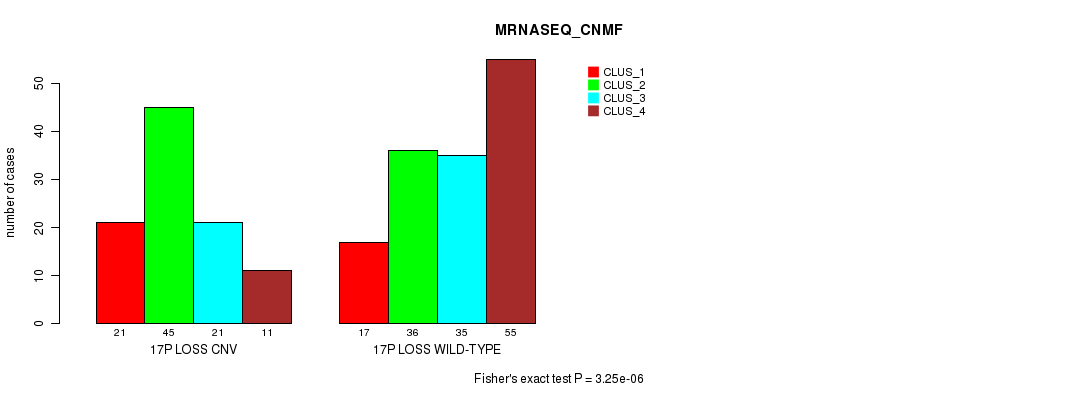
P value = 6.54e-05 (Fisher's exact test), Q value = 0.054
Table S82. Gene #69: '17p loss' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 105 | 105 | 85 |
| 17P LOSS CNV | 29 | 60 | 33 |
| 17P LOSS WILD-TYPE | 76 | 45 | 52 |
Figure S82. Get High-res Image Gene #69: '17p loss' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
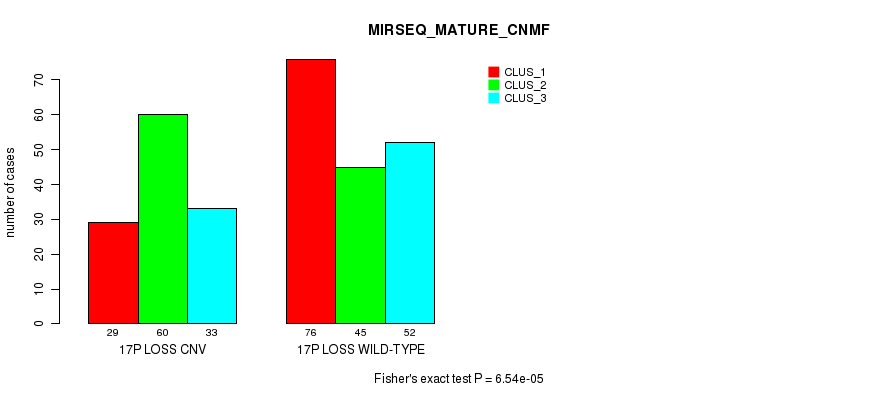
P value = 1.24e-13 (Fisher's exact test), Q value = 1.1e-10
Table S83. Gene #71: '18p loss' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 62 | 69 | 41 |
| 18P LOSS CNV | 10 | 52 | 52 | 12 |
| 18P LOSS WILD-TYPE | 33 | 10 | 17 | 29 |
Figure S83. Get High-res Image Gene #71: '18p loss' versus Molecular Subtype #1: 'MRNA_CNMF'
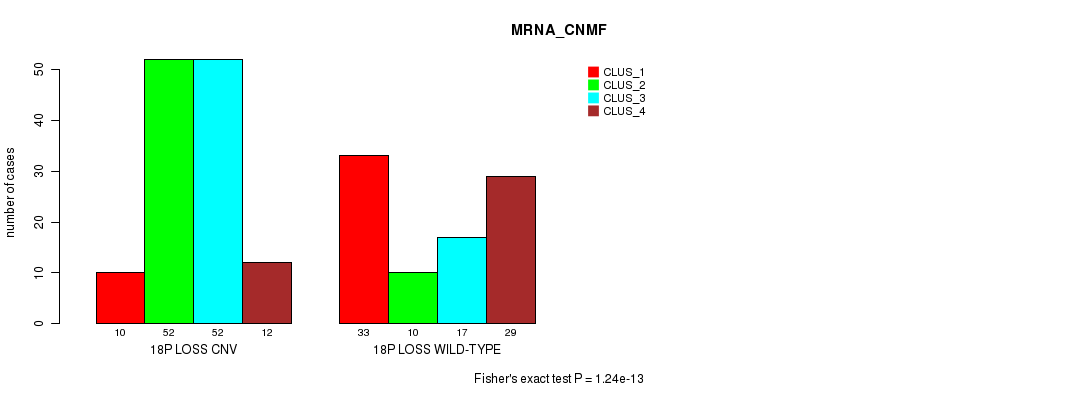
P value = 3.4e-12 (Fisher's exact test), Q value = 3e-09
Table S84. Gene #71: '18p loss' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 50 | 63 | 60 |
| 18P LOSS CNV | 35 | 30 | 14 | 47 |
| 18P LOSS WILD-TYPE | 7 | 20 | 49 | 13 |
Figure S84. Get High-res Image Gene #71: '18p loss' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
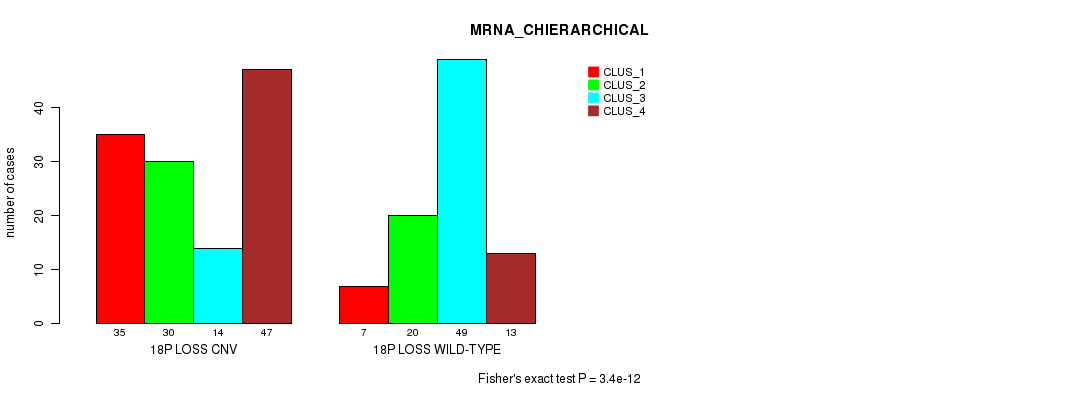
P value = 4.26e-47 (Fisher's exact test), Q value = 3.9e-44
Table S85. Gene #71: '18p loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 18P LOSS CNV | 176 | 53 | 62 | 33 |
| 18P LOSS WILD-TYPE | 24 | 181 | 38 | 8 |
Figure S85. Get High-res Image Gene #71: '18p loss' versus Molecular Subtype #3: 'CN_CNMF'
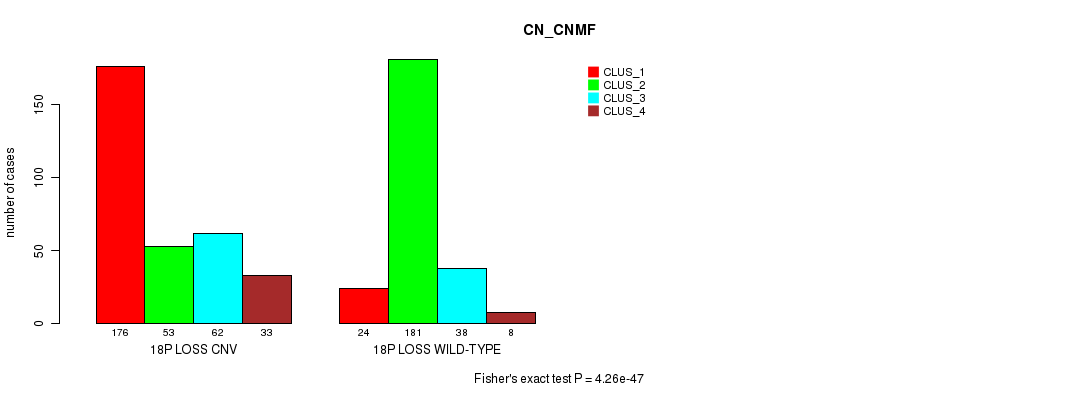
P value = 1.35e-18 (Fisher's exact test), Q value = 1.2e-15
Table S86. Gene #71: '18p loss' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 142 | 114 | 91 |
| 18P LOSS CNV | 119 | 40 | 34 |
| 18P LOSS WILD-TYPE | 23 | 74 | 57 |
Figure S86. Get High-res Image Gene #71: '18p loss' versus Molecular Subtype #4: 'METHLYATION_CNMF'
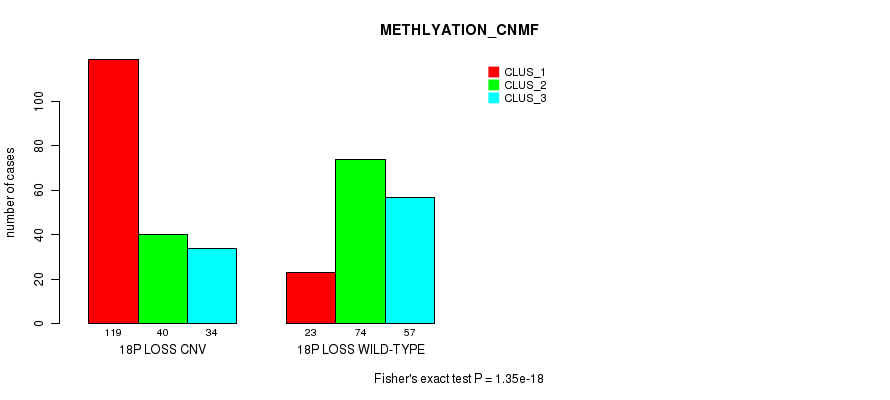
P value = 5.39e-16 (Fisher's exact test), Q value = 4.8e-13
Table S87. Gene #71: '18p loss' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 38 | 81 | 56 | 66 |
| 18P LOSS CNV | 20 | 66 | 33 | 9 |
| 18P LOSS WILD-TYPE | 18 | 15 | 23 | 57 |
Figure S87. Get High-res Image Gene #71: '18p loss' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
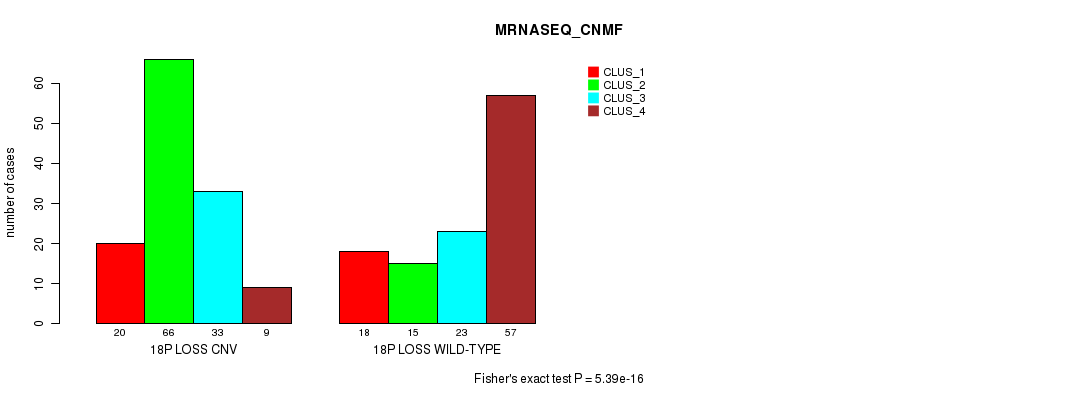
P value = 2.73e-11 (Fisher's exact test), Q value = 2.4e-08
Table S88. Gene #71: '18p loss' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 95 | 67 | 79 |
| 18P LOSS CNV | 56 | 13 | 59 |
| 18P LOSS WILD-TYPE | 39 | 54 | 20 |
Figure S88. Get High-res Image Gene #71: '18p loss' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
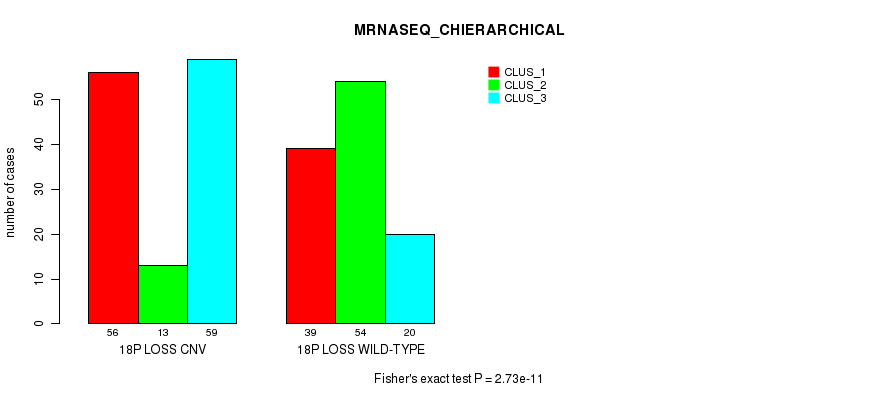
P value = 1.08e-10 (Fisher's exact test), Q value = 9.4e-08
Table S89. Gene #71: '18p loss' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 105 | 105 | 85 |
| 18P LOSS CNV | 33 | 81 | 47 |
| 18P LOSS WILD-TYPE | 72 | 24 | 38 |
Figure S89. Get High-res Image Gene #71: '18p loss' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
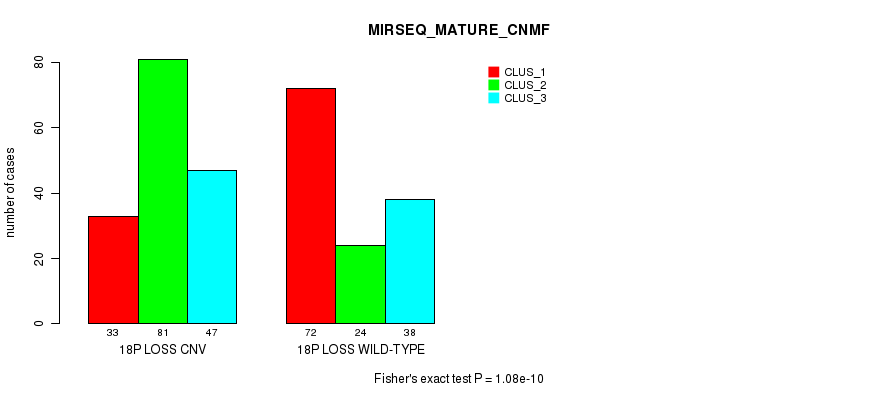
P value = 7.83e-12 (Chi-square test), Q value = 6.8e-09
Table S90. Gene #71: '18p loss' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 30 | 23 | 62 | 65 | 46 | 69 |
| 18P LOSS CNV | 22 | 12 | 42 | 44 | 31 | 10 |
| 18P LOSS WILD-TYPE | 8 | 11 | 20 | 21 | 15 | 59 |
Figure S90. Get High-res Image Gene #71: '18p loss' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
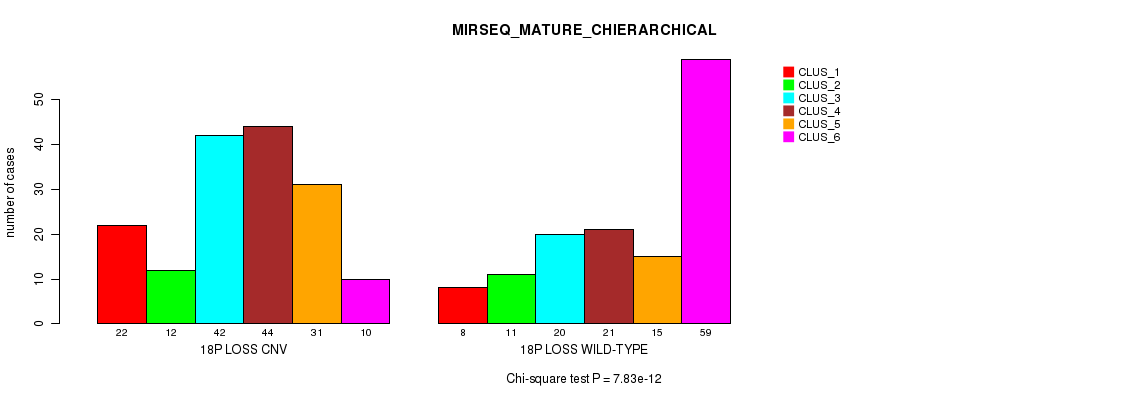
P value = 1.36e-12 (Fisher's exact test), Q value = 1.2e-09
Table S91. Gene #72: '18q loss' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 62 | 69 | 41 |
| 18Q LOSS CNV | 13 | 54 | 54 | 14 |
| 18Q LOSS WILD-TYPE | 30 | 8 | 15 | 27 |
Figure S91. Get High-res Image Gene #72: '18q loss' versus Molecular Subtype #1: 'MRNA_CNMF'
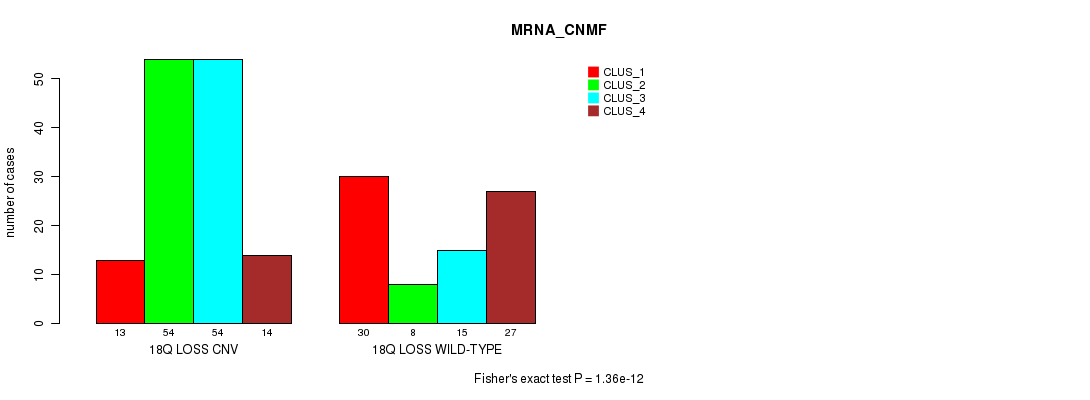
P value = 4.31e-12 (Fisher's exact test), Q value = 3.8e-09
Table S92. Gene #72: '18q loss' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 50 | 63 | 60 |
| 18Q LOSS CNV | 36 | 32 | 17 | 50 |
| 18Q LOSS WILD-TYPE | 6 | 18 | 46 | 10 |
Figure S92. Get High-res Image Gene #72: '18q loss' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
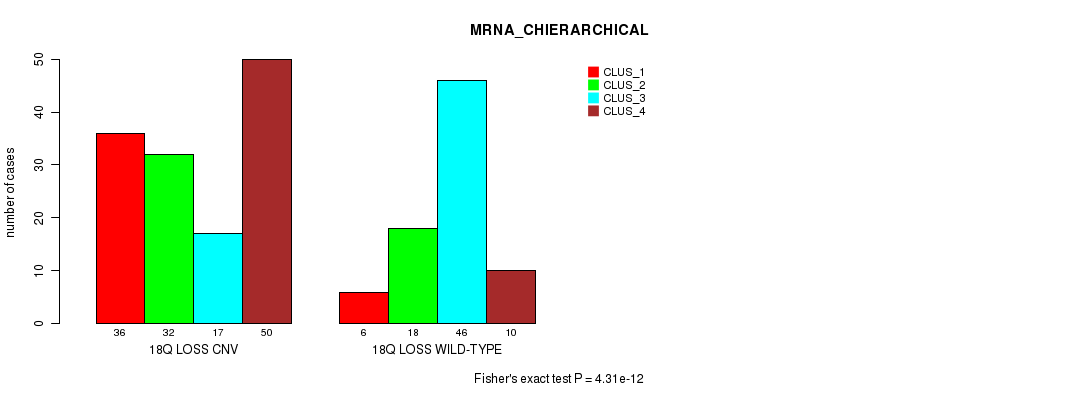
P value = 6.8e-51 (Fisher's exact test), Q value = 6.2e-48
Table S93. Gene #72: '18q loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 18Q LOSS CNV | 180 | 59 | 77 | 36 |
| 18Q LOSS WILD-TYPE | 20 | 175 | 23 | 5 |
Figure S93. Get High-res Image Gene #72: '18q loss' versus Molecular Subtype #3: 'CN_CNMF'
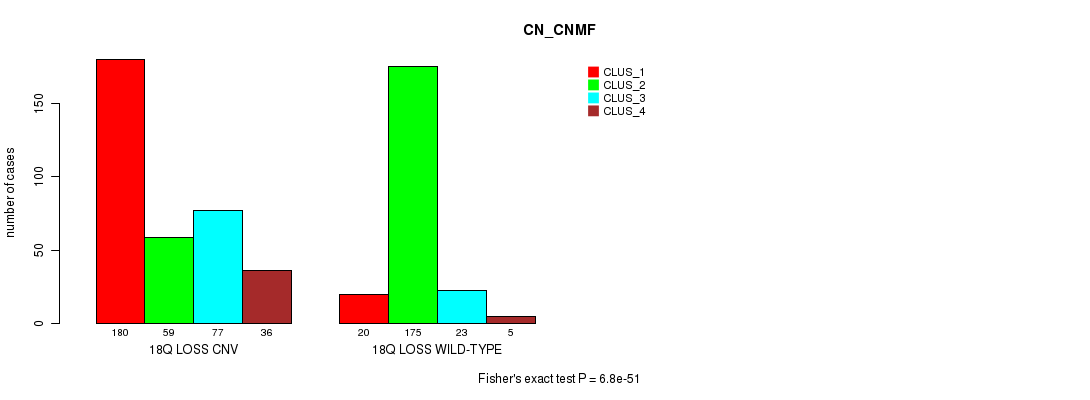
P value = 1.99e-19 (Fisher's exact test), Q value = 1.8e-16
Table S94. Gene #72: '18q loss' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 142 | 114 | 91 |
| 18Q LOSS CNV | 126 | 47 | 38 |
| 18Q LOSS WILD-TYPE | 16 | 67 | 53 |
Figure S94. Get High-res Image Gene #72: '18q loss' versus Molecular Subtype #4: 'METHLYATION_CNMF'
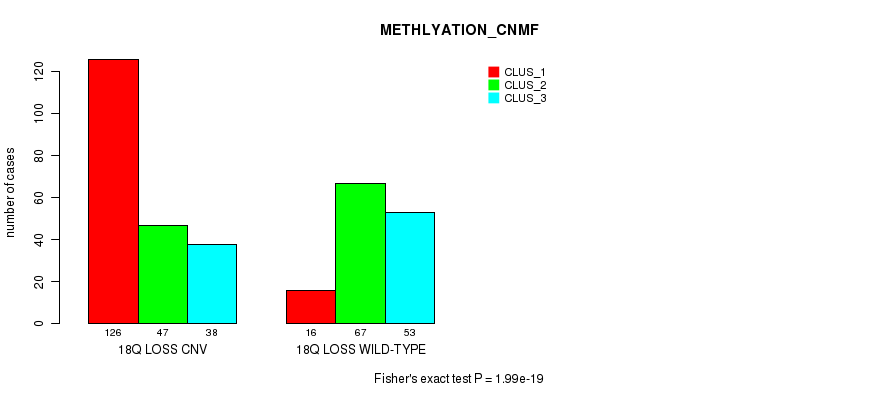
P value = 3.96e-16 (Fisher's exact test), Q value = 3.5e-13
Table S95. Gene #72: '18q loss' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 38 | 81 | 56 | 66 |
| 18Q LOSS CNV | 26 | 68 | 34 | 11 |
| 18Q LOSS WILD-TYPE | 12 | 13 | 22 | 55 |
Figure S95. Get High-res Image Gene #72: '18q loss' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
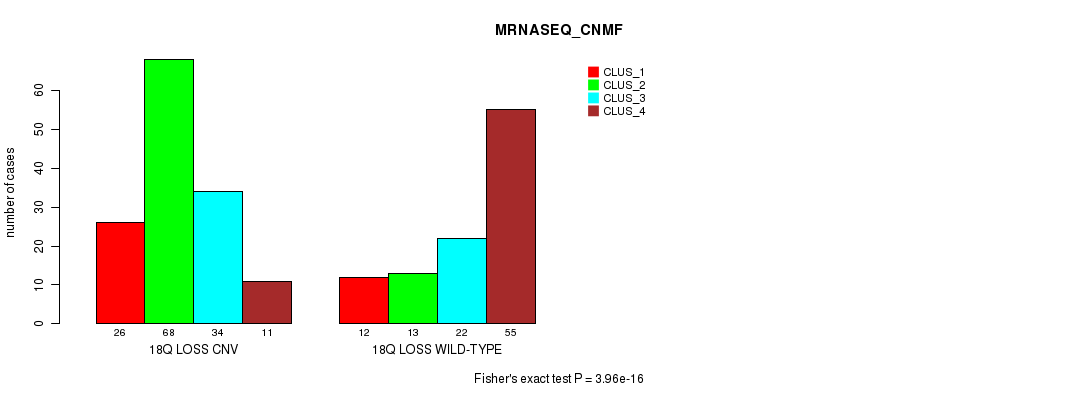
P value = 3.68e-09 (Fisher's exact test), Q value = 3.2e-06
Table S96. Gene #72: '18q loss' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 95 | 67 | 79 |
| 18Q LOSS CNV | 58 | 19 | 62 |
| 18Q LOSS WILD-TYPE | 37 | 48 | 17 |
Figure S96. Get High-res Image Gene #72: '18q loss' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
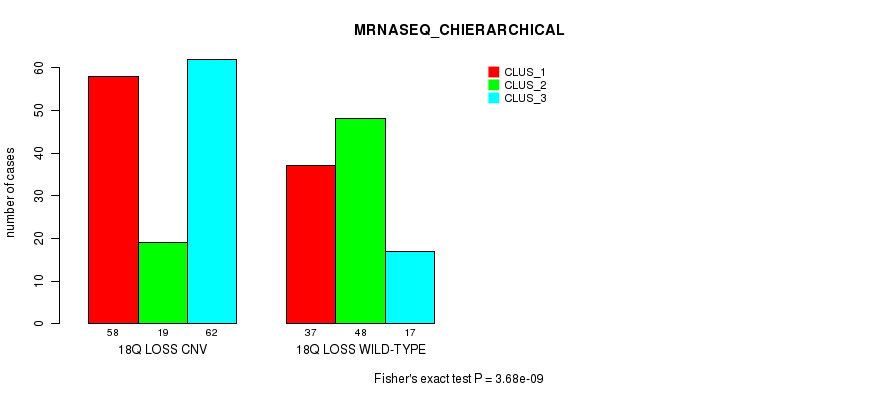
P value = 1.07e-09 (Fisher's exact test), Q value = 9.3e-07
Table S97. Gene #72: '18q loss' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 105 | 105 | 85 |
| 18Q LOSS CNV | 40 | 85 | 50 |
| 18Q LOSS WILD-TYPE | 65 | 20 | 35 |
Figure S97. Get High-res Image Gene #72: '18q loss' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
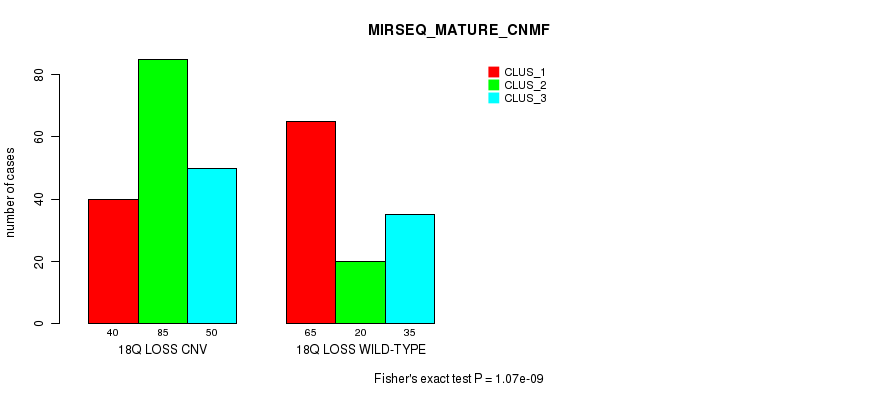
P value = 5.16e-12 (Chi-square test), Q value = 4.5e-09
Table S98. Gene #72: '18q loss' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 30 | 23 | 62 | 65 | 46 | 69 |
| 18Q LOSS CNV | 20 | 16 | 46 | 46 | 34 | 13 |
| 18Q LOSS WILD-TYPE | 10 | 7 | 16 | 19 | 12 | 56 |
Figure S98. Get High-res Image Gene #72: '18q loss' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
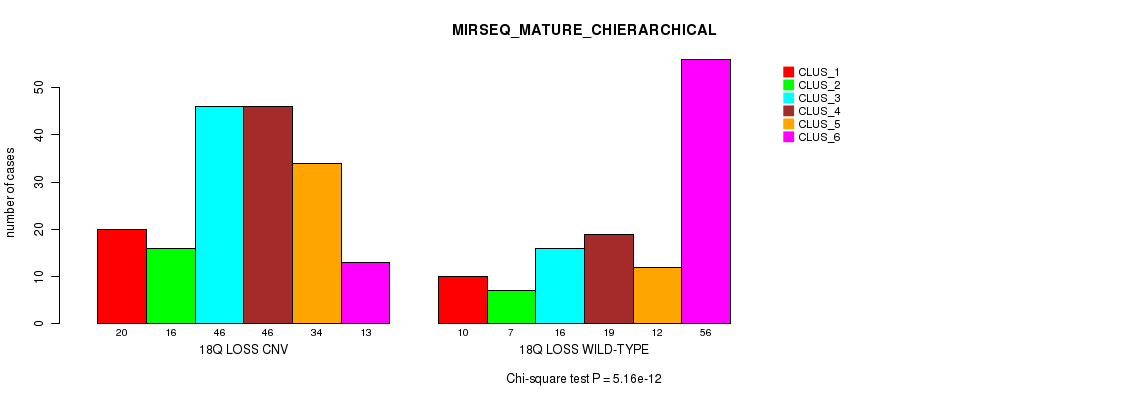
P value = 1.46e-05 (Fisher's exact test), Q value = 0.012
Table S99. Gene #73: '19p loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 19P LOSS CNV | 7 | 1 | 5 | 7 |
| 19P LOSS WILD-TYPE | 193 | 233 | 95 | 34 |
Figure S99. Get High-res Image Gene #73: '19p loss' versus Molecular Subtype #3: 'CN_CNMF'
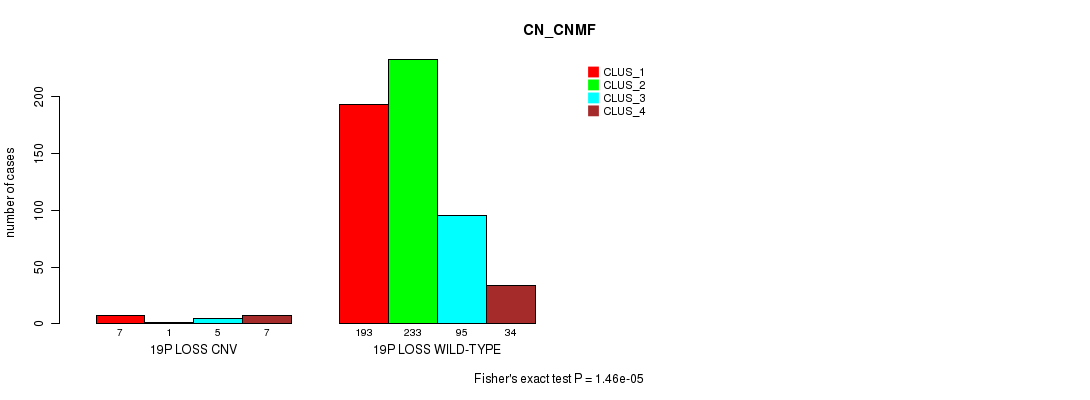
P value = 0.000235 (Fisher's exact test), Q value = 0.19
Table S100. Gene #74: '19q loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 19Q LOSS CNV | 6 | 1 | 6 | 5 |
| 19Q LOSS WILD-TYPE | 194 | 233 | 94 | 36 |
Figure S100. Get High-res Image Gene #74: '19q loss' versus Molecular Subtype #3: 'CN_CNMF'
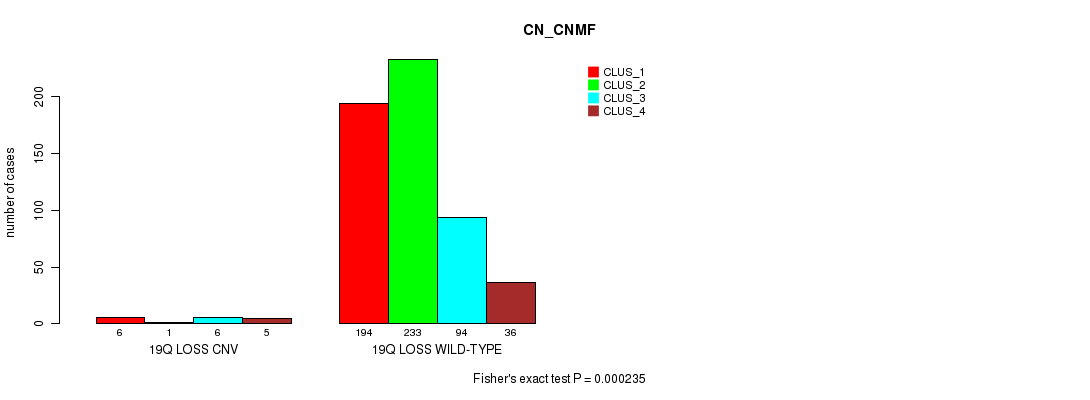
P value = 4.56e-07 (Fisher's exact test), Q value = 0.00039
Table S101. Gene #75: '20p loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 20P LOSS CNV | 27 | 5 | 15 | 0 |
| 20P LOSS WILD-TYPE | 173 | 229 | 85 | 41 |
Figure S101. Get High-res Image Gene #75: '20p loss' versus Molecular Subtype #3: 'CN_CNMF'
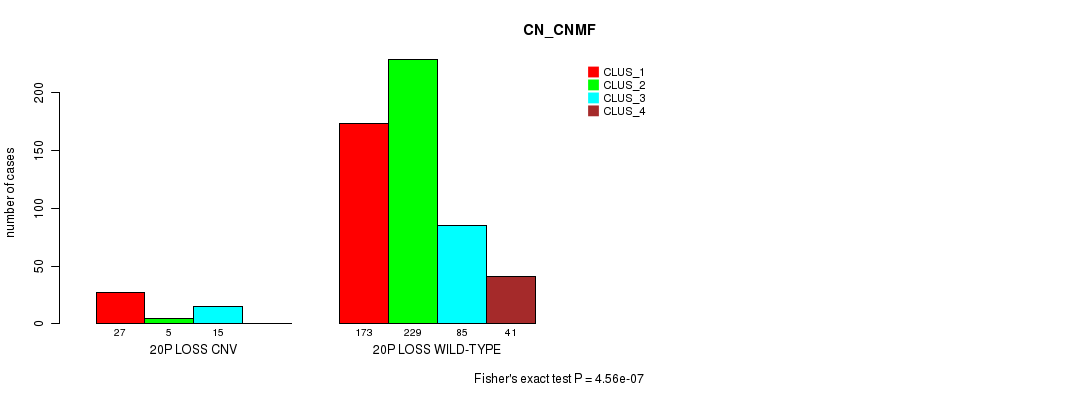
P value = 3.64e-05 (Fisher's exact test), Q value = 0.03
Table S102. Gene #76: '21q loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 21Q LOSS CNV | 58 | 28 | 16 | 12 |
| 21Q LOSS WILD-TYPE | 142 | 206 | 84 | 29 |
Figure S102. Get High-res Image Gene #76: '21q loss' versus Molecular Subtype #3: 'CN_CNMF'
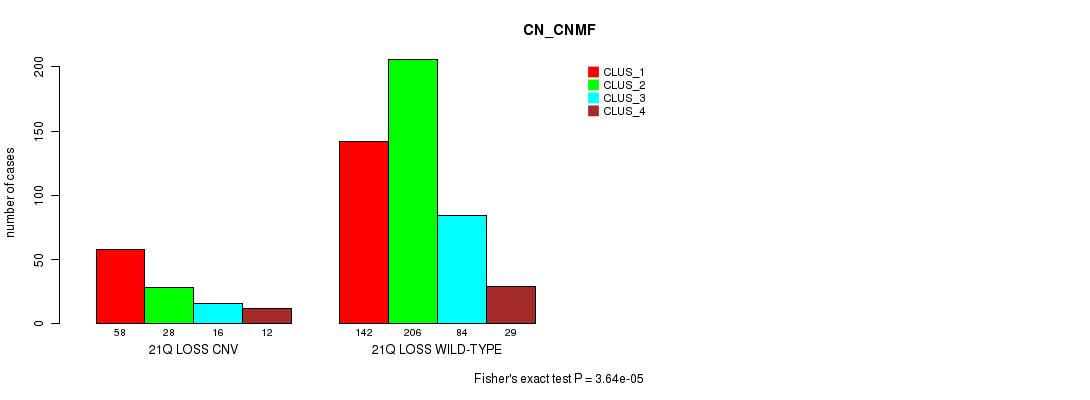
P value = 7.8e-14 (Fisher's exact test), Q value = 6.9e-11
Table S103. Gene #77: '22q loss' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 200 | 234 | 100 | 41 |
| 22Q LOSS CNV | 76 | 20 | 13 | 13 |
| 22Q LOSS WILD-TYPE | 124 | 214 | 87 | 28 |
Figure S103. Get High-res Image Gene #77: '22q loss' versus Molecular Subtype #3: 'CN_CNMF'
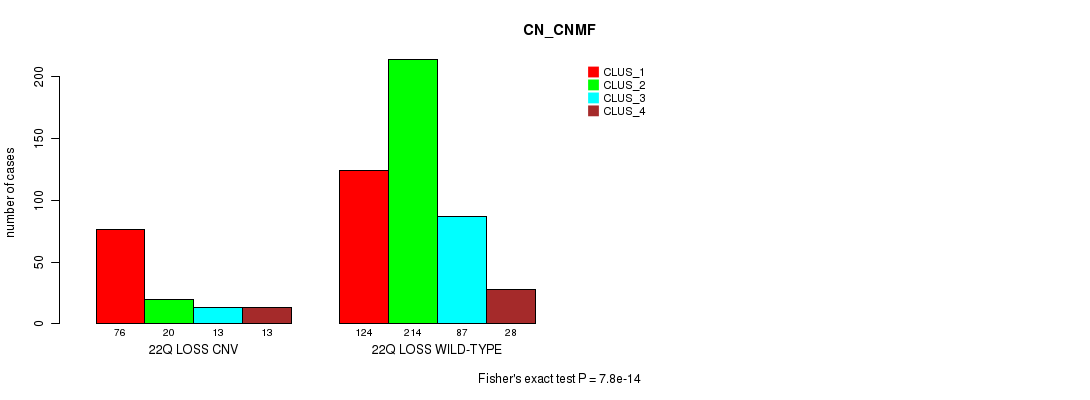
-
Mutation data file = broad_values_by_arm.mutsig.cluster.txt
-
Molecular subtypes file = COADREAD-TP.transferedmergedcluster.txt
-
Number of patients = 575
-
Number of significantly arm-level cnvs = 78
-
Number of molecular subtypes = 12
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.