This pipeline computes the correlation between significant copy number variation (cnv focal) genes and selected clinical features.
Testing the association between copy number variation 32 arm-level results and 8 clinical features across 493 patients, 19 significant findings detected with Q value < 0.25.
-
Amp Peak 3(3q26.32) cnv correlated to 'DISTANT.METASTASIS' and 'NEOPLASM.DISEASESTAGE'.
-
Amp Peak 7(8q24.22) cnv correlated to 'NEOPLASM.DISEASESTAGE'.
-
Del Peak 9(3q11.2) cnv correlated to 'GENDER'.
-
Del Peak 10(3q21.3) cnv correlated to 'GENDER'.
-
Del Peak 11(4q34.3) cnv correlated to 'Time to Death' and 'NEOPLASM.DISEASESTAGE'.
-
Del Peak 15(9p23) cnv correlated to 'Time to Death', 'DISTANT.METASTASIS', and 'NEOPLASM.DISEASESTAGE'.
-
Del Peak 16(9p21.3) cnv correlated to 'Time to Death', 'GENDER', 'DISTANT.METASTASIS', and 'NEOPLASM.DISEASESTAGE'.
-
Del Peak 18(13q13.3) cnv correlated to 'Time to Death' and 'NEOPLASM.DISEASESTAGE'.
-
Del Peak 19(14q31.1) cnv correlated to 'DISTANT.METASTASIS', 'LYMPH.NODE.METASTASIS', and 'NEOPLASM.DISEASESTAGE'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 32 arm-level results and 8 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 19 significant findings detected.
|
Clinical Features |
Time to Death |
AGE | GENDER |
KARNOFSKY PERFORMANCE SCORE |
DISTANT METASTASIS |
LYMPH NODE METASTASIS |
TUMOR STAGECODE |
NEOPLASM DISEASESTAGE |
||
| nCNV (%) | nWild-Type | logrank test | t-test | Fisher's exact test | t-test | Fisher's exact test | Fisher's exact test | t-test | Fisher's exact test | |
| Del Peak 16(9p21 3) | 0 (0%) | 341 |
2.03e-06 (0.000445) |
0.0261 (1.00) |
0.00069 (0.144) |
0.656 (1.00) |
6.93e-05 (0.0148) |
0.105 (1.00) |
1.02e-08 (2.27e-06) |
|
| Del Peak 15(9p23) | 0 (0%) | 349 |
0.000138 (0.0292) |
0.0136 (1.00) |
0.0018 (0.362) |
0.656 (1.00) |
0.000876 (0.18) |
0.122 (1.00) |
1.64e-06 (0.000361) |
|
| Del Peak 19(14q31 1) | 0 (0%) | 275 |
0.00179 (0.361) |
0.055 (1.00) |
0.296 (1.00) |
0.0872 (1.00) |
0.00101 (0.208) |
0.000371 (0.0779) |
1.48e-05 (0.0032) |
|
| Amp Peak 3(3q26 32) | 0 (0%) | 413 |
0.156 (1.00) |
0.903 (1.00) |
0.442 (1.00) |
0.152 (1.00) |
7.09e-05 (0.0151) |
0.00823 (1.00) |
2.97e-05 (0.00639) |
|
| Del Peak 11(4q34 3) | 0 (0%) | 417 |
0.000557 (0.117) |
0.568 (1.00) |
0.433 (1.00) |
0.0261 (1.00) |
0.00862 (1.00) |
0.354 (1.00) |
2.41e-06 (0.000526) |
|
| Del Peak 18(13q13 3) | 0 (0%) | 417 |
0.000848 (0.175) |
0.884 (1.00) |
0.794 (1.00) |
0.748 (1.00) |
0.0377 (1.00) |
0.601 (1.00) |
0.000129 (0.0274) |
|
| Amp Peak 7(8q24 22) | 0 (0%) | 420 |
0.0527 (1.00) |
0.0214 (1.00) |
0.11 (1.00) |
0.297 (1.00) |
0.053 (1.00) |
0.923 (1.00) |
0.00122 (0.249) |
|
| Del Peak 9(3q11 2) | 0 (0%) | 343 |
0.635 (1.00) |
0.0111 (1.00) |
3.95e-08 (8.73e-06) |
0.749 (1.00) |
0.591 (1.00) |
0.562 (1.00) |
0.613 (1.00) |
|
| Del Peak 10(3q21 3) | 0 (0%) | 390 |
0.846 (1.00) |
0.00205 (0.41) |
7.86e-06 (0.00171) |
0.0875 (1.00) |
0.879 (1.00) |
0.185 (1.00) |
0.424 (1.00) |
|
| Amp Peak 1(1q24 1) | 0 (0%) | 433 |
0.2 (1.00) |
0.164 (1.00) |
0.666 (1.00) |
0.499 (1.00) |
0.339 (1.00) |
0.111 (1.00) |
0.339 (1.00) |
|
| Amp Peak 2(1q32 1) | 0 (0%) | 434 |
0.437 (1.00) |
0.0725 (1.00) |
0.771 (1.00) |
0.499 (1.00) |
0.703 (1.00) |
0.653 (1.00) |
0.234 (1.00) |
|
| Amp Peak 4(4q32 1) | 0 (0%) | 479 |
0.0584 (1.00) |
0.416 (1.00) |
0.258 (1.00) |
0.411 (1.00) |
0.462 (1.00) |
0.0499 (1.00) |
0.431 (1.00) |
|
| Amp Peak 5(5q35 1) | 0 (0%) | 182 |
0.0454 (1.00) |
0.0923 (1.00) |
0.141 (1.00) |
0.944 (1.00) |
0.366 (1.00) |
0.499 (1.00) |
0.557 (1.00) |
|
| Amp Peak 6(7q36 3) | 0 (0%) | 330 |
0.724 (1.00) |
0.806 (1.00) |
0.0352 (1.00) |
0.57 (1.00) |
0.00523 (1.00) |
0.428 (1.00) |
0.0198 (1.00) |
|
| Amp Peak 8(10p14) | 0 (0%) | 477 |
0.234 (1.00) |
0.423 (1.00) |
0.594 (1.00) |
0.887 (1.00) |
0.288 (1.00) |
0.0682 (1.00) |
0.129 (1.00) |
|
| Amp Peak 9(17q24 3) | 0 (0%) | 456 |
0.0512 (1.00) |
0.0662 (1.00) |
0.858 (1.00) |
0.471 (1.00) |
0.244 (1.00) |
0.535 (1.00) |
0.739 (1.00) |
|
| Amp Peak 10(Xp22 2) | 0 (0%) | 458 |
0.468 (1.00) |
0.676 (1.00) |
0.854 (1.00) |
0.726 (1.00) |
0.0897 (1.00) |
0.177 (1.00) |
0.0176 (1.00) |
|
| Amp Peak 11(Xp11 4) | 0 (0%) | 457 |
0.759 (1.00) |
0.712 (1.00) |
0.857 (1.00) |
0.726 (1.00) |
0.0509 (1.00) |
0.276 (1.00) |
0.00905 (1.00) |
|
| Amp Peak 12(Xq11 2) | 0 (0%) | 461 |
0.803 (1.00) |
0.145 (1.00) |
0.706 (1.00) |
0.726 (1.00) |
0.31 (1.00) |
0.216 (1.00) |
0.103 (1.00) |
|
| Amp Peak 13(Xq28) | 0 (0%) | 456 |
0.631 (1.00) |
0.805 (1.00) |
0.474 (1.00) |
0.726 (1.00) |
0.34 (1.00) |
0.053 (1.00) |
0.352 (1.00) |
|
| Del Peak 1(1p36 23) | 0 (0%) | 395 |
0.0495 (1.00) |
0.123 (1.00) |
0.123 (1.00) |
0.284 (1.00) |
0.0844 (1.00) |
0.829 (1.00) |
0.202 (1.00) |
|
| Del Peak 2(1p31 1) | 0 (0%) | 421 |
0.219 (1.00) |
0.0726 (1.00) |
0.016 (1.00) |
0.284 (1.00) |
0.294 (1.00) |
0.765 (1.00) |
0.428 (1.00) |
|
| Del Peak 3(1q43) | 0 (0%) | 455 |
0.704 (1.00) |
0.335 (1.00) |
1 (1.00) |
1 (1.00) |
0.589 (1.00) |
0.694 (1.00) |
||
| Del Peak 4(2q37 3) | 0 (0%) | 445 |
0.673 (1.00) |
0.63 (1.00) |
0.153 (1.00) |
0.402 (1.00) |
0.952 (1.00) |
0.622 (1.00) |
||
| Del Peak 5(3p25 3) | 0 (0%) | 61 |
0.783 (1.00) |
0.788 (1.00) |
0.569 (1.00) |
0.0817 (1.00) |
0.707 (1.00) |
0.553 (1.00) |
0.0266 (1.00) |
|
| Del Peak 6(3p21 32) | 0 (0%) | 58 |
0.498 (1.00) |
0.704 (1.00) |
0.772 (1.00) |
0.0817 (1.00) |
0.333 (1.00) |
0.631 (1.00) |
0.00141 (0.286) |
|
| Del Peak 7(3p12 3) | 0 (0%) | 182 |
0.69 (1.00) |
0.762 (1.00) |
0.00441 (0.874) |
0.944 (1.00) |
0.608 (1.00) |
0.609 (1.00) |
0.405 (1.00) |
|
| Del Peak 8(3p12 2) | 0 (0%) | 210 |
0.614 (1.00) |
0.761 (1.00) |
0.00221 (0.441) |
0.741 (1.00) |
0.53 (1.00) |
0.779 (1.00) |
0.609 (1.00) |
|
| Del Peak 12(6q26) | 0 (0%) | 351 |
0.565 (1.00) |
0.287 (1.00) |
0.012 (1.00) |
0.256 (1.00) |
0.41 (1.00) |
0.0069 (1.00) |
0.281 (1.00) |
|
| Del Peak 13(6q26) | 0 (0%) | 351 |
0.589 (1.00) |
0.226 (1.00) |
0.00648 (1.00) |
0.256 (1.00) |
0.41 (1.00) |
0.0069 (1.00) |
0.281 (1.00) |
|
| Del Peak 14(8p23 2) | 0 (0%) | 347 |
0.0385 (1.00) |
0.139 (1.00) |
0.256 (1.00) |
0.893 (1.00) |
0.055 (1.00) |
0.48 (1.00) |
0.119 (1.00) |
|
| Del Peak 17(10q23 31) | 0 (0%) | 403 |
0.217 (1.00) |
0.209 (1.00) |
0.0272 (1.00) |
0.764 (1.00) |
0.333 (1.00) |
0.549 (1.00) |
0.0187 (1.00) |
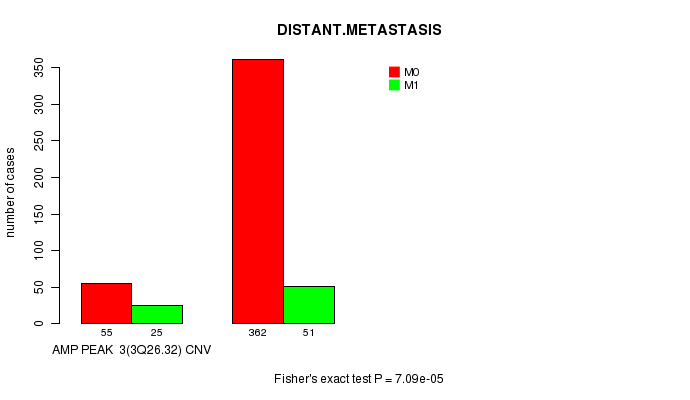
P value = 7.09e-05 (Fisher's exact test), Q value = 0.015
Table S1. Gene #3: 'Amp Peak 3(3q26.32)' versus Clinical Feature #5: 'DISTANT.METASTASIS'
| nPatients | M0 | M1 |
|---|---|---|
| ALL | 417 | 76 |
| AMP PEAK 3(3Q26.32) CNV | 55 | 25 |
| AMP PEAK 3(3Q26.32) WILD-TYPE | 362 | 51 |
Figure S1. Get High-res Image Gene #3: 'Amp Peak 3(3q26.32)' versus Clinical Feature #5: 'DISTANT.METASTASIS'
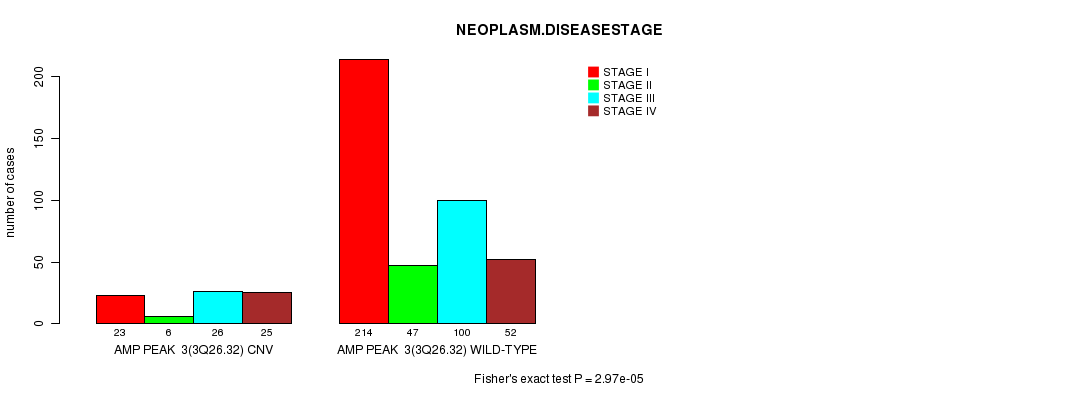
P value = 2.97e-05 (Fisher's exact test), Q value = 0.0064
Table S2. Gene #3: 'Amp Peak 3(3q26.32)' versus Clinical Feature #8: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IV |
|---|---|---|---|---|
| ALL | 237 | 53 | 126 | 77 |
| AMP PEAK 3(3Q26.32) CNV | 23 | 6 | 26 | 25 |
| AMP PEAK 3(3Q26.32) WILD-TYPE | 214 | 47 | 100 | 52 |
Figure S2. Get High-res Image Gene #3: 'Amp Peak 3(3q26.32)' versus Clinical Feature #8: 'NEOPLASM.DISEASESTAGE'
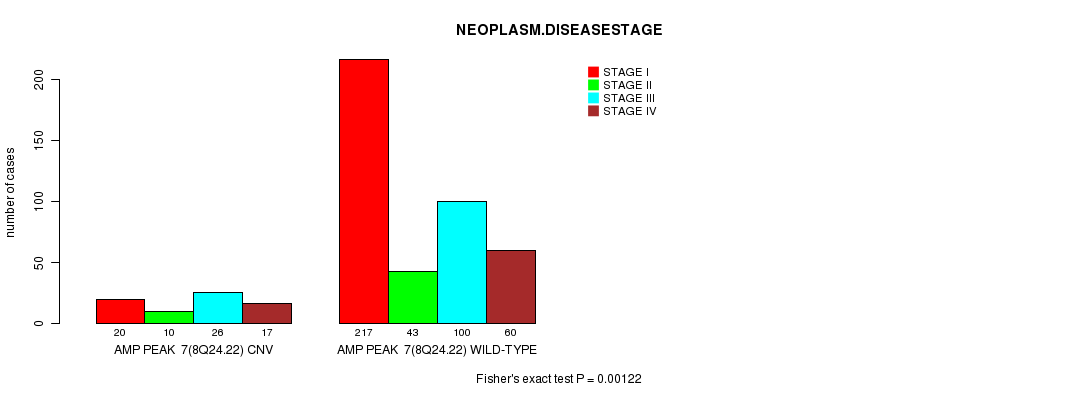
P value = 0.00122 (Fisher's exact test), Q value = 0.25
Table S3. Gene #7: 'Amp Peak 7(8q24.22)' versus Clinical Feature #8: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IV |
|---|---|---|---|---|
| ALL | 237 | 53 | 126 | 77 |
| AMP PEAK 7(8Q24.22) CNV | 20 | 10 | 26 | 17 |
| AMP PEAK 7(8Q24.22) WILD-TYPE | 217 | 43 | 100 | 60 |
Figure S3. Get High-res Image Gene #7: 'Amp Peak 7(8q24.22)' versus Clinical Feature #8: 'NEOPLASM.DISEASESTAGE'
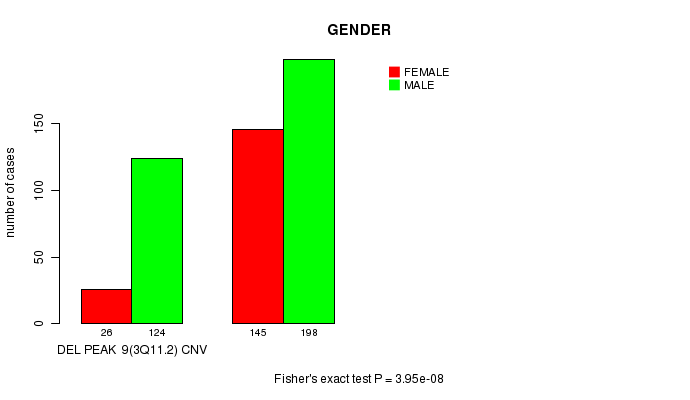
P value = 3.95e-08 (Fisher's exact test), Q value = 8.7e-06
Table S4. Gene #22: 'Del Peak 9(3q11.2)' versus Clinical Feature #3: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 171 | 322 |
| DEL PEAK 9(3Q11.2) CNV | 26 | 124 |
| DEL PEAK 9(3Q11.2) WILD-TYPE | 145 | 198 |
Figure S4. Get High-res Image Gene #22: 'Del Peak 9(3q11.2)' versus Clinical Feature #3: 'GENDER'
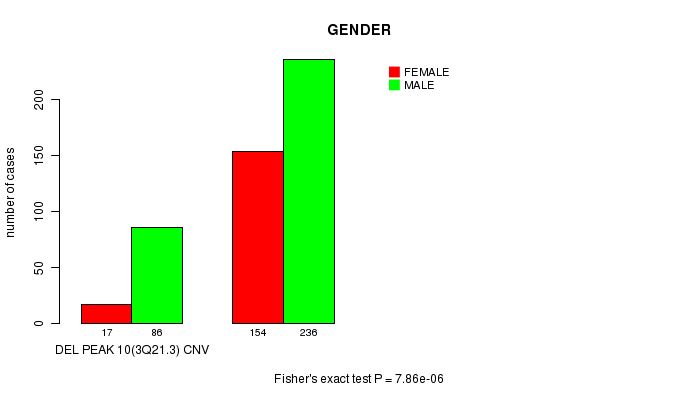
P value = 7.86e-06 (Fisher's exact test), Q value = 0.0017
Table S5. Gene #23: 'Del Peak 10(3q21.3)' versus Clinical Feature #3: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 171 | 322 |
| DEL PEAK 10(3Q21.3) CNV | 17 | 86 |
| DEL PEAK 10(3Q21.3) WILD-TYPE | 154 | 236 |
Figure S5. Get High-res Image Gene #23: 'Del Peak 10(3q21.3)' versus Clinical Feature #3: 'GENDER'
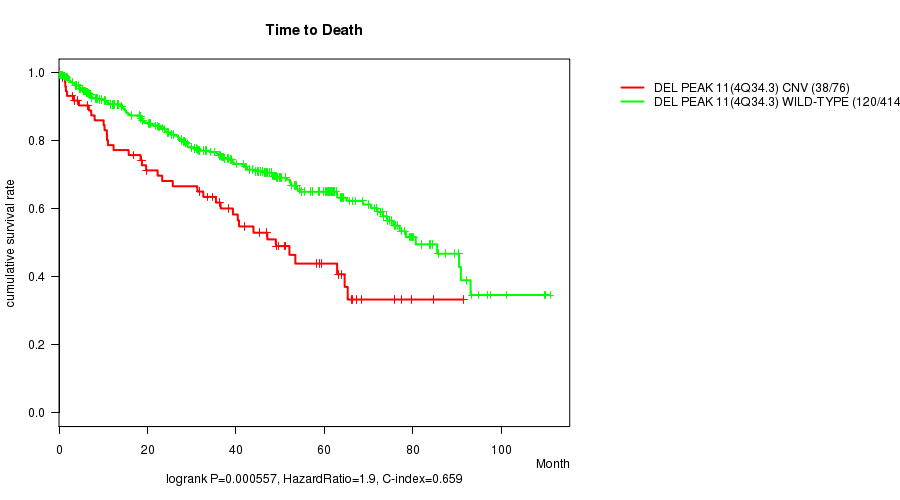
P value = 0.000557 (logrank test), Q value = 0.12
Table S6. Gene #24: 'Del Peak 11(4q34.3)' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 490 | 158 | 0.1 - 111.0 (35.2) |
| DEL PEAK 11(4Q34.3) CNV | 76 | 38 | 0.2 - 91.4 (35.1) |
| DEL PEAK 11(4Q34.3) WILD-TYPE | 414 | 120 | 0.1 - 111.0 (35.2) |
Figure S6. Get High-res Image Gene #24: 'Del Peak 11(4q34.3)' versus Clinical Feature #1: 'Time to Death'
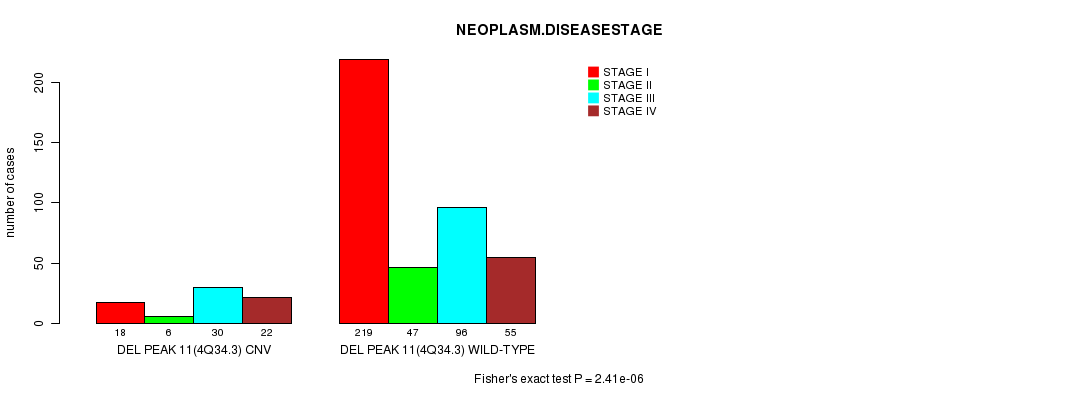
P value = 2.41e-06 (Fisher's exact test), Q value = 0.00053
Table S7. Gene #24: 'Del Peak 11(4q34.3)' versus Clinical Feature #8: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IV |
|---|---|---|---|---|
| ALL | 237 | 53 | 126 | 77 |
| DEL PEAK 11(4Q34.3) CNV | 18 | 6 | 30 | 22 |
| DEL PEAK 11(4Q34.3) WILD-TYPE | 219 | 47 | 96 | 55 |
Figure S7. Get High-res Image Gene #24: 'Del Peak 11(4q34.3)' versus Clinical Feature #8: 'NEOPLASM.DISEASESTAGE'
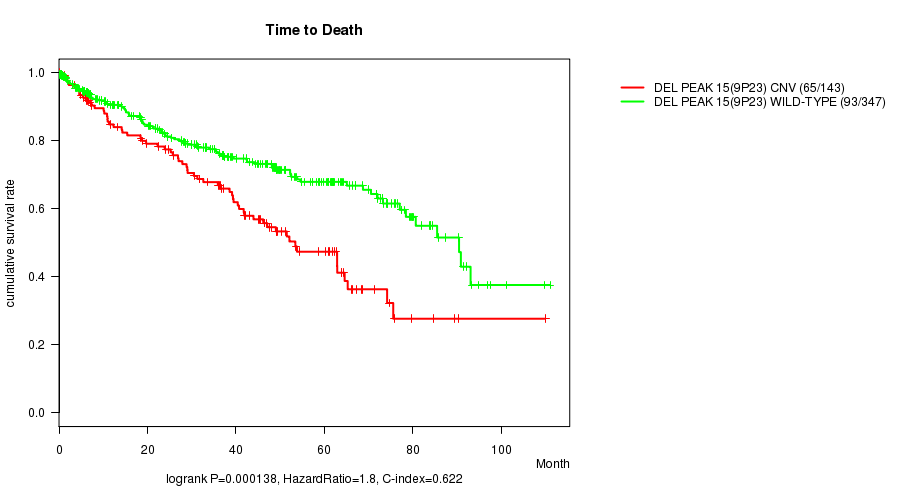
P value = 0.000138 (logrank test), Q value = 0.029
Table S8. Gene #28: 'Del Peak 15(9p23)' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 490 | 158 | 0.1 - 111.0 (35.2) |
| DEL PEAK 15(9P23) CNV | 143 | 65 | 0.2 - 109.9 (36.1) |
| DEL PEAK 15(9P23) WILD-TYPE | 347 | 93 | 0.1 - 111.0 (34.6) |
Figure S8. Get High-res Image Gene #28: 'Del Peak 15(9p23)' versus Clinical Feature #1: 'Time to Death'
P value = 0.000876 (Fisher's exact test), Q value = 0.18
Table S9. Gene #28: 'Del Peak 15(9p23)' versus Clinical Feature #5: 'DISTANT.METASTASIS'
| nPatients | M0 | M1 |
|---|---|---|
| ALL | 417 | 76 |
| DEL PEAK 15(9P23) CNV | 109 | 35 |
| DEL PEAK 15(9P23) WILD-TYPE | 308 | 41 |
Figure S9. Get High-res Image Gene #28: 'Del Peak 15(9p23)' versus Clinical Feature #5: 'DISTANT.METASTASIS'
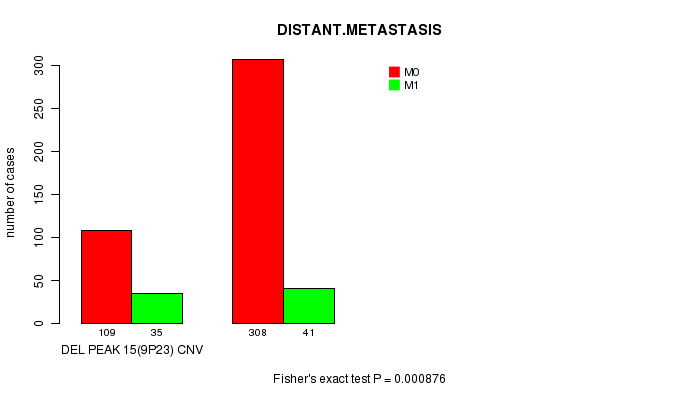
P value = 1.64e-06 (Fisher's exact test), Q value = 0.00036
Table S10. Gene #28: 'Del Peak 15(9p23)' versus Clinical Feature #8: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IV |
|---|---|---|---|---|
| ALL | 237 | 53 | 126 | 77 |
| DEL PEAK 15(9P23) CNV | 47 | 11 | 50 | 36 |
| DEL PEAK 15(9P23) WILD-TYPE | 190 | 42 | 76 | 41 |
Figure S10. Get High-res Image Gene #28: 'Del Peak 15(9p23)' versus Clinical Feature #8: 'NEOPLASM.DISEASESTAGE'
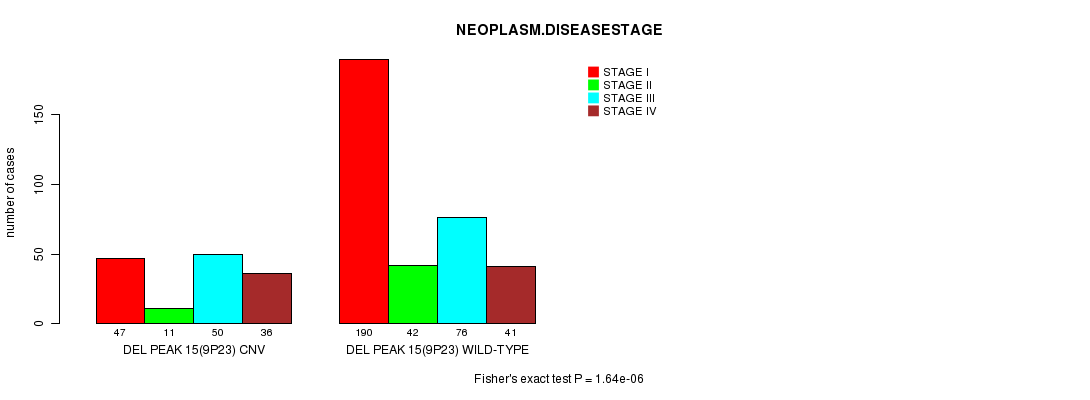
P value = 2.03e-06 (logrank test), Q value = 0.00044
Table S11. Gene #29: 'Del Peak 16(9p21.3)' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 490 | 158 | 0.1 - 111.0 (35.2) |
| DEL PEAK 16(9P21.3) CNV | 152 | 72 | 0.2 - 109.9 (31.5) |
| DEL PEAK 16(9P21.3) WILD-TYPE | 338 | 86 | 0.1 - 111.0 (36.2) |
Figure S11. Get High-res Image Gene #29: 'Del Peak 16(9p21.3)' versus Clinical Feature #1: 'Time to Death'
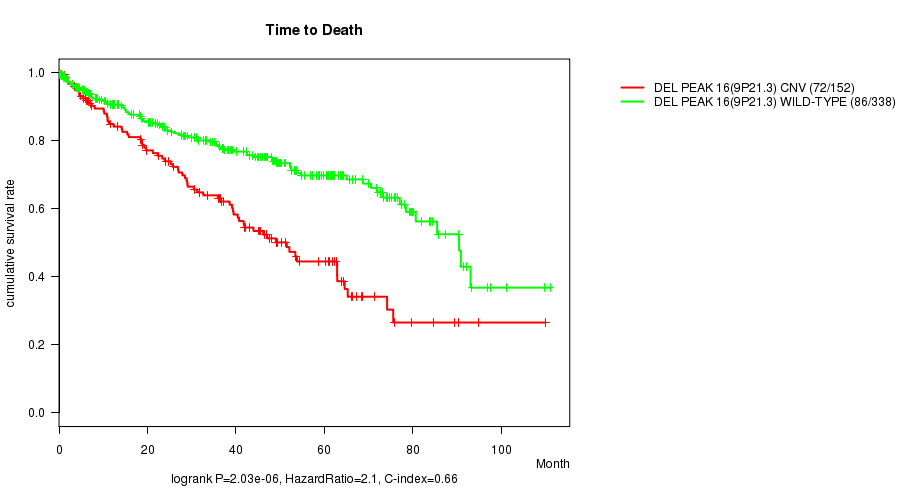
P value = 0.00069 (Fisher's exact test), Q value = 0.14
Table S12. Gene #29: 'Del Peak 16(9p21.3)' versus Clinical Feature #3: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 171 | 322 |
| DEL PEAK 16(9P21.3) CNV | 36 | 116 |
| DEL PEAK 16(9P21.3) WILD-TYPE | 135 | 206 |
Figure S12. Get High-res Image Gene #29: 'Del Peak 16(9p21.3)' versus Clinical Feature #3: 'GENDER'
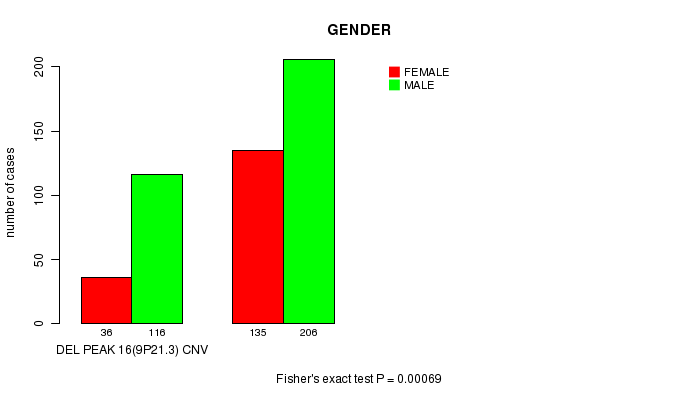
P value = 6.93e-05 (Fisher's exact test), Q value = 0.015
Table S13. Gene #29: 'Del Peak 16(9p21.3)' versus Clinical Feature #5: 'DISTANT.METASTASIS'
| nPatients | M0 | M1 |
|---|---|---|
| ALL | 417 | 76 |
| DEL PEAK 16(9P21.3) CNV | 113 | 39 |
| DEL PEAK 16(9P21.3) WILD-TYPE | 304 | 37 |
Figure S13. Get High-res Image Gene #29: 'Del Peak 16(9p21.3)' versus Clinical Feature #5: 'DISTANT.METASTASIS'
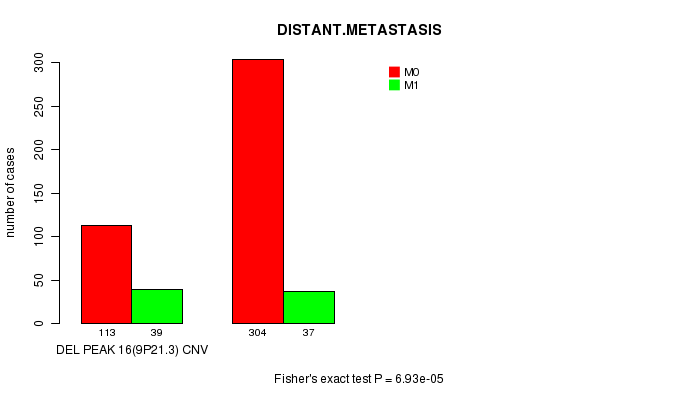
P value = 1.02e-08 (Fisher's exact test), Q value = 2.3e-06
Table S14. Gene #29: 'Del Peak 16(9p21.3)' versus Clinical Feature #8: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IV |
|---|---|---|---|---|
| ALL | 237 | 53 | 126 | 77 |
| DEL PEAK 16(9P21.3) CNV | 48 | 10 | 54 | 40 |
| DEL PEAK 16(9P21.3) WILD-TYPE | 189 | 43 | 72 | 37 |
Figure S14. Get High-res Image Gene #29: 'Del Peak 16(9p21.3)' versus Clinical Feature #8: 'NEOPLASM.DISEASESTAGE'
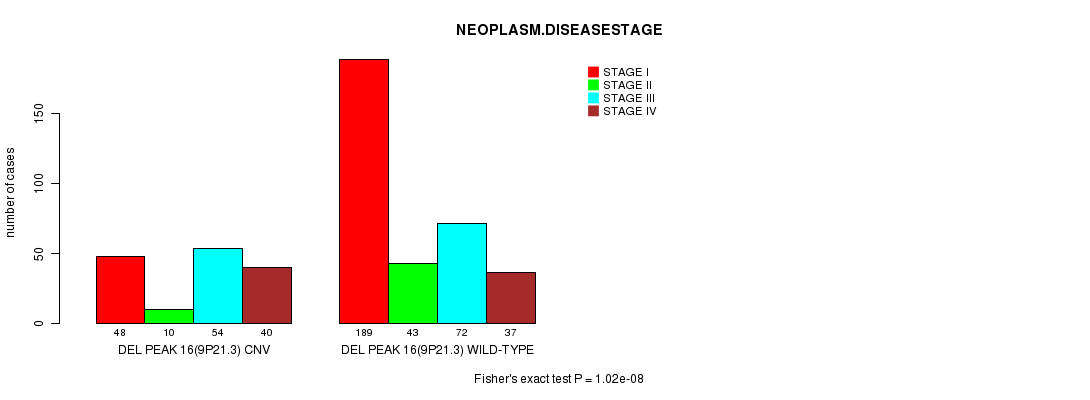
P value = 0.000848 (logrank test), Q value = 0.18
Table S15. Gene #31: 'Del Peak 18(13q13.3)' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 490 | 158 | 0.1 - 111.0 (35.2) |
| DEL PEAK 18(13Q13.3) CNV | 75 | 34 | 0.1 - 89.4 (28.5) |
| DEL PEAK 18(13Q13.3) WILD-TYPE | 415 | 124 | 0.1 - 111.0 (36.3) |
Figure S15. Get High-res Image Gene #31: 'Del Peak 18(13q13.3)' versus Clinical Feature #1: 'Time to Death'
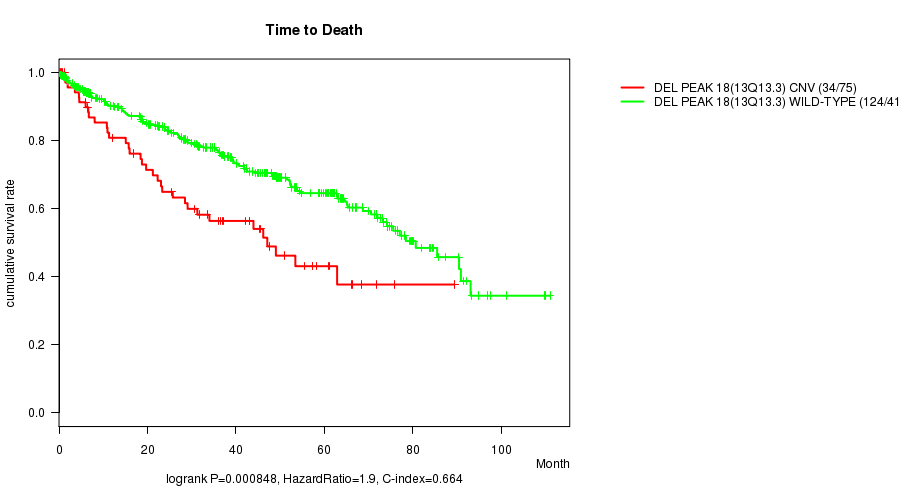
P value = 0.000129 (Fisher's exact test), Q value = 0.027
Table S16. Gene #31: 'Del Peak 18(13q13.3)' versus Clinical Feature #8: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IV |
|---|---|---|---|---|
| ALL | 237 | 53 | 126 | 77 |
| DEL PEAK 18(13Q13.3) CNV | 26 | 2 | 28 | 20 |
| DEL PEAK 18(13Q13.3) WILD-TYPE | 211 | 51 | 98 | 57 |
Figure S16. Get High-res Image Gene #31: 'Del Peak 18(13q13.3)' versus Clinical Feature #8: 'NEOPLASM.DISEASESTAGE'
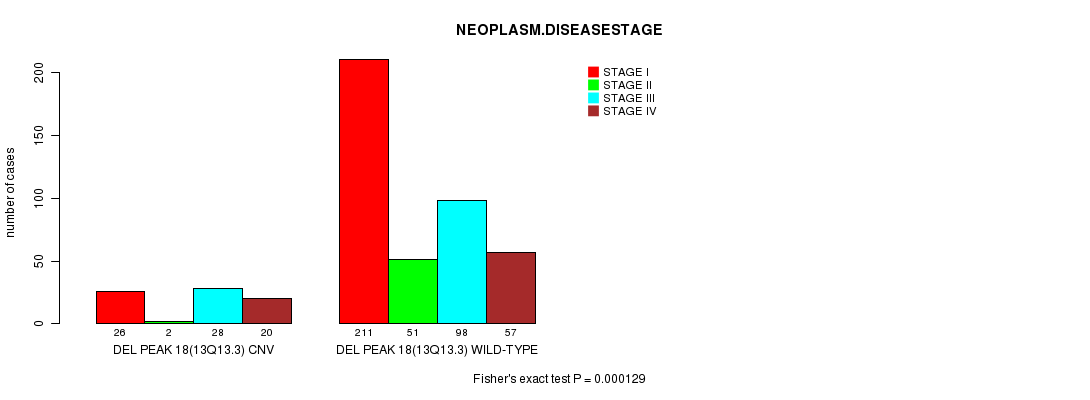
P value = 0.00101 (Fisher's exact test), Q value = 0.21
Table S17. Gene #32: 'Del Peak 19(14q31.1)' versus Clinical Feature #5: 'DISTANT.METASTASIS'
| nPatients | M0 | M1 |
|---|---|---|
| ALL | 417 | 76 |
| DEL PEAK 19(14Q31.1) CNV | 171 | 47 |
| DEL PEAK 19(14Q31.1) WILD-TYPE | 246 | 29 |
Figure S17. Get High-res Image Gene #32: 'Del Peak 19(14q31.1)' versus Clinical Feature #5: 'DISTANT.METASTASIS'
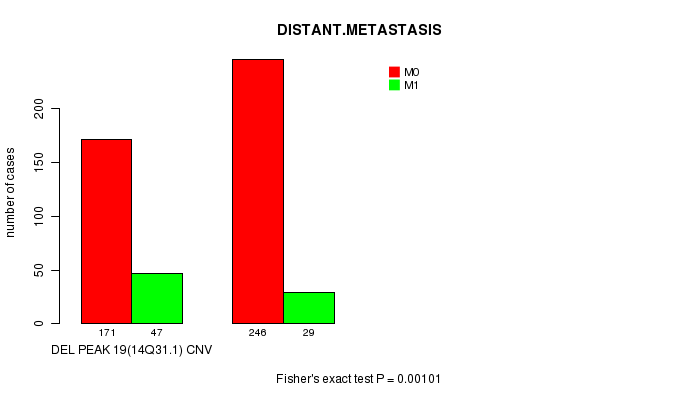
P value = 0.000371 (Fisher's exact test), Q value = 0.078
Table S18. Gene #32: 'Del Peak 19(14q31.1)' versus Clinical Feature #6: 'LYMPH.NODE.METASTASIS'
| nPatients | N0 | N1 | NX |
|---|---|---|---|
| ALL | 228 | 18 | 247 |
| DEL PEAK 19(14Q31.1) CNV | 96 | 16 | 106 |
| DEL PEAK 19(14Q31.1) WILD-TYPE | 132 | 2 | 141 |
Figure S18. Get High-res Image Gene #32: 'Del Peak 19(14q31.1)' versus Clinical Feature #6: 'LYMPH.NODE.METASTASIS'
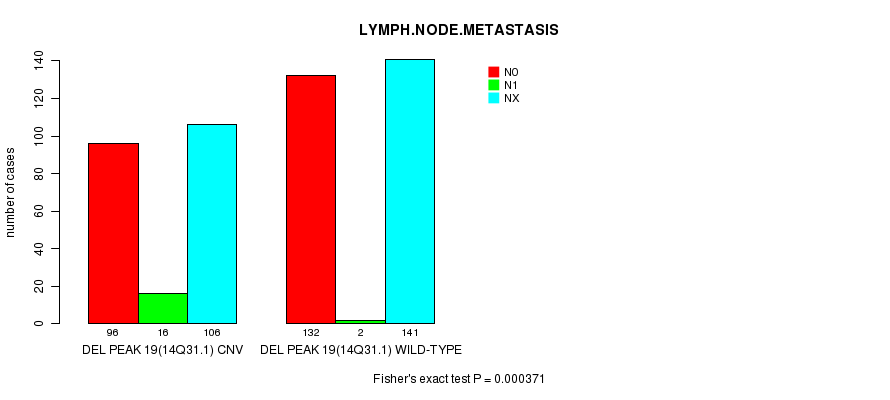
P value = 1.48e-05 (Fisher's exact test), Q value = 0.0032
Table S19. Gene #32: 'Del Peak 19(14q31.1)' versus Clinical Feature #8: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IV |
|---|---|---|---|---|
| ALL | 237 | 53 | 126 | 77 |
| DEL PEAK 19(14Q31.1) CNV | 78 | 27 | 67 | 46 |
| DEL PEAK 19(14Q31.1) WILD-TYPE | 159 | 26 | 59 | 31 |
Figure S19. Get High-res Image Gene #32: 'Del Peak 19(14q31.1)' versus Clinical Feature #8: 'NEOPLASM.DISEASESTAGE'
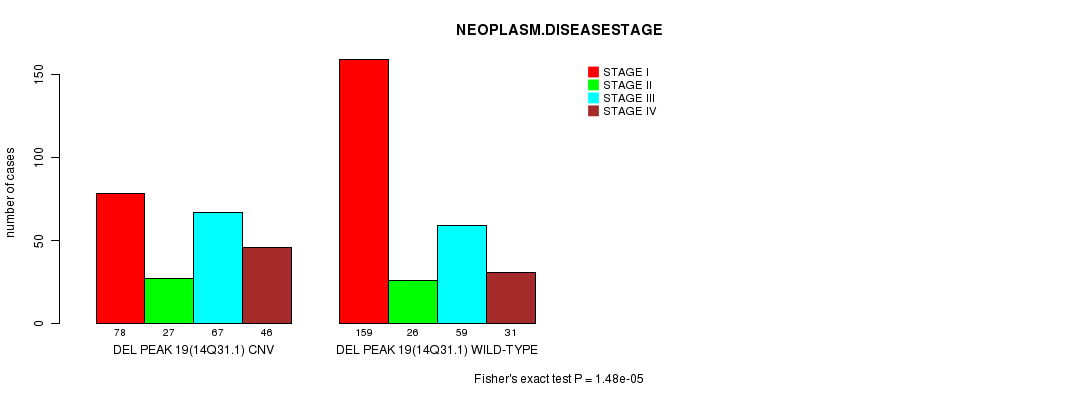
-
Mutation data file = all_lesions.conf_99.cnv.cluster.txt
-
Clinical data file = KIRC-TP.clin.merged.picked.txt
-
Number of patients = 493
-
Number of significantly arm-level cnvs = 32
-
Number of selected clinical features = 8
-
Exclude genes that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For continuous numerical clinical features, two-tailed Student's t test with unequal variance (Lehmann and Romano 2005) was applied to compare the clinical values between tumors with and without gene mutations using 't.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.