This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and selected clinical features.
Testing the association between copy number variation 80 arm-level events and 8 clinical features across 117 patients, one significant finding detected with Q value < 0.25.
-
11Q GAIN MUTATION ANALYSIS cnv correlated to 'NEOPLASM.DISEASESTAGE'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 80 arm-level events and 8 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, one significant finding detected.
|
Clinical Features |
Time to Death |
AGE |
NEOPLASM DISEASESTAGE |
PATHOLOGY T STAGE |
PATHOLOGY N STAGE |
PATHOLOGY M STAGE |
GENDER |
COMPLETENESS OF RESECTION |
||
| nCNV (%) | nWild-Type | logrank test | t-test | Chi-square test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 11Q GAIN MUTATION ANALYSIS | 4 (3%) | 113 |
0.109 (1.00) |
0.79 (1.00) |
0.000293 (0.188) |
0.57 (1.00) |
1 (1.00) |
0.0971 (1.00) |
0.296 (1.00) |
0.0275 (1.00) |
| 1P GAIN MUTATION ANALYSIS | 17 (15%) | 100 |
0.587 (1.00) |
0.658 (1.00) |
0.802 (1.00) |
0.147 (1.00) |
1 (1.00) |
0.652 (1.00) |
0.591 (1.00) |
0.271 (1.00) |
| 1Q GAIN MUTATION ANALYSIS | 64 (55%) | 53 |
0.167 (1.00) |
0.827 (1.00) |
0.0574 (1.00) |
0.0763 (1.00) |
0.106 (1.00) |
0.63 (1.00) |
0.13 (1.00) |
1 (1.00) |
| 2P GAIN MUTATION ANALYSIS | 18 (15%) | 99 |
0.773 (1.00) |
0.421 (1.00) |
0.0853 (1.00) |
0.379 (1.00) |
0.0519 (1.00) |
0.368 (1.00) |
0.114 (1.00) |
0.127 (1.00) |
| 2Q GAIN MUTATION ANALYSIS | 15 (13%) | 102 |
0.853 (1.00) |
0.477 (1.00) |
0.152 (1.00) |
0.817 (1.00) |
0.345 (1.00) |
0.31 (1.00) |
0.0846 (1.00) |
0.271 (1.00) |
| 3P GAIN MUTATION ANALYSIS | 8 (7%) | 109 |
0.0367 (1.00) |
0.771 (1.00) |
0.843 (1.00) |
0.362 (1.00) |
0.15 (1.00) |
1 (1.00) |
0.708 (1.00) |
0.587 (1.00) |
| 3Q GAIN MUTATION ANALYSIS | 9 (8%) | 108 |
0.0122 (1.00) |
0.689 (1.00) |
0.843 (1.00) |
0.578 (1.00) |
0.15 (1.00) |
0.494 (1.00) |
1 (1.00) |
0.64 (1.00) |
| 4P GAIN MUTATION ANALYSIS | 8 (7%) | 109 |
0.389 (1.00) |
0.327 (1.00) |
0.998 (1.00) |
0.935 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.427 (1.00) |
| 4Q GAIN MUTATION ANALYSIS | 3 (3%) | 114 |
0.871 (1.00) |
0.501 (1.00) |
0.99 (1.00) |
0.668 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.0928 (1.00) |
| 5P GAIN MUTATION ANALYSIS | 47 (40%) | 70 |
0.904 (1.00) |
0.184 (1.00) |
0.552 (1.00) |
0.347 (1.00) |
0.286 (1.00) |
0.409 (1.00) |
0.847 (1.00) |
1 (1.00) |
| 5Q GAIN MUTATION ANALYSIS | 36 (31%) | 81 |
0.989 (1.00) |
0.342 (1.00) |
0.711 (1.00) |
0.798 (1.00) |
0.553 (1.00) |
0.909 (1.00) |
0.311 (1.00) |
0.719 (1.00) |
| 6P GAIN MUTATION ANALYSIS | 29 (25%) | 88 |
0.203 (1.00) |
0.545 (1.00) |
0.413 (1.00) |
1 (1.00) |
1 (1.00) |
0.0908 (1.00) |
1 (1.00) |
0.458 (1.00) |
| 6Q GAIN MUTATION ANALYSIS | 17 (15%) | 100 |
0.229 (1.00) |
0.245 (1.00) |
0.0785 (1.00) |
0.814 (1.00) |
1 (1.00) |
0.0172 (1.00) |
0.591 (1.00) |
0.0981 (1.00) |
| 7P GAIN MUTATION ANALYSIS | 37 (32%) | 80 |
0.979 (1.00) |
0.0481 (1.00) |
0.563 (1.00) |
0.381 (1.00) |
0.195 (1.00) |
0.828 (1.00) |
0.685 (1.00) |
0.613 (1.00) |
| 7Q GAIN MUTATION ANALYSIS | 39 (33%) | 78 |
0.855 (1.00) |
0.163 (1.00) |
0.418 (1.00) |
0.365 (1.00) |
0.211 (1.00) |
0.535 (1.00) |
0.842 (1.00) |
0.898 (1.00) |
| 8P GAIN MUTATION ANALYSIS | 22 (19%) | 95 |
0.821 (1.00) |
0.149 (1.00) |
0.468 (1.00) |
0.647 (1.00) |
1 (1.00) |
0.307 (1.00) |
0.0499 (1.00) |
0.0912 (1.00) |
| 8Q GAIN MUTATION ANALYSIS | 56 (48%) | 61 |
0.897 (1.00) |
0.773 (1.00) |
0.288 (1.00) |
0.514 (1.00) |
1 (1.00) |
0.686 (1.00) |
0.00797 (1.00) |
0.83 (1.00) |
| 9P GAIN MUTATION ANALYSIS | 4 (3%) | 113 |
0.819 (1.00) |
0.99 (1.00) |
0.369 (1.00) |
0.745 (1.00) |
1 (1.00) |
1 (1.00) |
0.631 (1.00) |
0.321 (1.00) |
| 9Q GAIN MUTATION ANALYSIS | 4 (3%) | 113 |
0.16 (1.00) |
0.855 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.149 (1.00) |
0.321 (1.00) |
| 10P GAIN MUTATION ANALYSIS | 17 (15%) | 100 |
0.408 (1.00) |
0.197 (1.00) |
0.337 (1.00) |
0.564 (1.00) |
1 (1.00) |
0.291 (1.00) |
1 (1.00) |
1 (1.00) |
| 10Q GAIN MUTATION ANALYSIS | 10 (9%) | 107 |
0.786 (1.00) |
0.677 (1.00) |
0.196 (1.00) |
0.648 (1.00) |
1 (1.00) |
0.196 (1.00) |
0.499 (1.00) |
0.686 (1.00) |
| 11P GAIN MUTATION ANALYSIS | 5 (4%) | 112 |
0.14 (1.00) |
0.519 (1.00) |
0.00393 (1.00) |
0.633 (1.00) |
1 (1.00) |
0.0561 (1.00) |
0.155 (1.00) |
0.0396 (1.00) |
| 12P GAIN MUTATION ANALYSIS | 8 (7%) | 109 |
0.76 (1.00) |
0.241 (1.00) |
0.809 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.528 (1.00) |
| 12Q GAIN MUTATION ANALYSIS | 12 (10%) | 105 |
0.461 (1.00) |
0.152 (1.00) |
0.109 (1.00) |
0.882 (1.00) |
0.315 (1.00) |
0.564 (1.00) |
0.13 (1.00) |
0.699 (1.00) |
| 13Q GAIN MUTATION ANALYSIS | 9 (8%) | 108 |
0.161 (1.00) |
0.255 (1.00) |
0.762 (1.00) |
0.942 (1.00) |
0.15 (1.00) |
0.729 (1.00) |
0.727 (1.00) |
0.587 (1.00) |
| 14Q GAIN MUTATION ANALYSIS | 8 (7%) | 109 |
0.999 (1.00) |
0.994 (1.00) |
0.086 (1.00) |
1 (1.00) |
1 (1.00) |
0.0972 (1.00) |
0.708 (1.00) |
0.0677 (1.00) |
| 15Q GAIN MUTATION ANALYSIS | 10 (9%) | 107 |
0.862 (1.00) |
0.4 (1.00) |
0.963 (1.00) |
0.273 (1.00) |
1 (1.00) |
0.541 (1.00) |
1 (1.00) |
1 (1.00) |
| 16P GAIN MUTATION ANALYSIS | 11 (9%) | 106 |
0.906 (1.00) |
0.52 (1.00) |
0.218 (1.00) |
0.621 (1.00) |
1 (1.00) |
0.756 (1.00) |
1 (1.00) |
1 (1.00) |
| 16Q GAIN MUTATION ANALYSIS | 4 (3%) | 113 |
0.795 (1.00) |
0.371 (1.00) |
0.891 (1.00) |
0.438 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| 17P GAIN MUTATION ANALYSIS | 10 (9%) | 107 |
0.321 (1.00) |
0.82 (1.00) |
0.0654 (1.00) |
0.906 (1.00) |
1 (1.00) |
0.541 (1.00) |
0.741 (1.00) |
0.407 (1.00) |
| 17Q GAIN MUTATION ANALYSIS | 36 (31%) | 81 |
0.183 (1.00) |
0.454 (1.00) |
0.052 (1.00) |
0.195 (1.00) |
1 (1.00) |
0.137 (1.00) |
0.68 (1.00) |
0.231 (1.00) |
| 18P GAIN MUTATION ANALYSIS | 9 (8%) | 108 |
0.345 (1.00) |
0.349 (1.00) |
0.0672 (1.00) |
0.556 (1.00) |
1 (1.00) |
0.168 (1.00) |
1 (1.00) |
0.0501 (1.00) |
| 18Q GAIN MUTATION ANALYSIS | 9 (8%) | 108 |
0.345 (1.00) |
0.349 (1.00) |
0.0672 (1.00) |
0.556 (1.00) |
1 (1.00) |
0.168 (1.00) |
1 (1.00) |
0.0501 (1.00) |
| 19P GAIN MUTATION ANALYSIS | 27 (23%) | 90 |
0.482 (1.00) |
0.739 (1.00) |
0.00294 (1.00) |
0.0108 (1.00) |
0.532 (1.00) |
0.0595 (1.00) |
0.258 (1.00) |
0.261 (1.00) |
| 19Q GAIN MUTATION ANALYSIS | 29 (25%) | 88 |
0.491 (1.00) |
0.929 (1.00) |
0.00634 (1.00) |
0.029 (1.00) |
0.532 (1.00) |
0.0689 (1.00) |
0.19 (1.00) |
0.231 (1.00) |
| 20P GAIN MUTATION ANALYSIS | 34 (29%) | 83 |
0.226 (1.00) |
0.557 (1.00) |
0.641 (1.00) |
0.966 (1.00) |
1 (1.00) |
0.122 (1.00) |
0.403 (1.00) |
0.441 (1.00) |
| 20Q GAIN MUTATION ANALYSIS | 36 (31%) | 81 |
0.285 (1.00) |
0.565 (1.00) |
0.722 (1.00) |
0.955 (1.00) |
1 (1.00) |
0.103 (1.00) |
0.544 (1.00) |
0.302 (1.00) |
| 21Q GAIN MUTATION ANALYSIS | 10 (9%) | 107 |
0.563 (1.00) |
0.756 (1.00) |
0.425 (1.00) |
0.629 (1.00) |
1 (1.00) |
1 (1.00) |
0.174 (1.00) |
0.216 (1.00) |
| 22Q GAIN MUTATION ANALYSIS | 16 (14%) | 101 |
0.915 (1.00) |
0.528 (1.00) |
0.78 (1.00) |
0.681 (1.00) |
0.374 (1.00) |
0.812 (1.00) |
1 (1.00) |
0.201 (1.00) |
| XQ GAIN MUTATION ANALYSIS | 24 (21%) | 93 |
0.195 (1.00) |
0.0205 (1.00) |
0.659 (1.00) |
0.323 (1.00) |
1 (1.00) |
0.862 (1.00) |
0.479 (1.00) |
0.917 (1.00) |
| 1P LOSS MUTATION ANALYSIS | 28 (24%) | 89 |
0.749 (1.00) |
0.796 (1.00) |
0.15 (1.00) |
0.944 (1.00) |
0.149 (1.00) |
0.0979 (1.00) |
0.512 (1.00) |
0.0361 (1.00) |
| 1Q LOSS MUTATION ANALYSIS | 12 (10%) | 105 |
0.332 (1.00) |
0.16 (1.00) |
0.214 (1.00) |
0.705 (1.00) |
0.0272 (1.00) |
0.776 (1.00) |
0.762 (1.00) |
0.289 (1.00) |
| 2P LOSS MUTATION ANALYSIS | 8 (7%) | 109 |
0.289 (1.00) |
0.902 (1.00) |
0.837 (1.00) |
0.118 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.427 (1.00) |
| 2Q LOSS MUTATION ANALYSIS | 10 (9%) | 107 |
0.888 (1.00) |
0.829 (1.00) |
0.924 (1.00) |
0.425 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.266 (1.00) |
| 3P LOSS MUTATION ANALYSIS | 14 (12%) | 103 |
0.0909 (1.00) |
0.931 (1.00) |
0.109 (1.00) |
0.138 (1.00) |
0.374 (1.00) |
0.228 (1.00) |
0.381 (1.00) |
0.662 (1.00) |
| 3Q LOSS MUTATION ANALYSIS | 6 (5%) | 111 |
0.0392 (1.00) |
0.0969 (1.00) |
0.652 (1.00) |
0.627 (1.00) |
0.185 (1.00) |
1 (1.00) |
0.671 (1.00) |
1 (1.00) |
| 4P LOSS MUTATION ANALYSIS | 28 (24%) | 89 |
1 (1.00) |
0.765 (1.00) |
0.162 (1.00) |
0.919 (1.00) |
0.121 (1.00) |
0.0979 (1.00) |
0.512 (1.00) |
0.367 (1.00) |
| 4Q LOSS MUTATION ANALYSIS | 37 (32%) | 80 |
0.85 (1.00) |
0.856 (1.00) |
0.459 (1.00) |
0.779 (1.00) |
0.228 (1.00) |
0.177 (1.00) |
0.838 (1.00) |
0.31 (1.00) |
| 5P LOSS MUTATION ANALYSIS | 9 (8%) | 108 |
0.442 (1.00) |
0.911 (1.00) |
0.387 (1.00) |
0.763 (1.00) |
0.219 (1.00) |
1 (1.00) |
0.48 (1.00) |
0.491 (1.00) |
| 5Q LOSS MUTATION ANALYSIS | 13 (11%) | 104 |
0.174 (1.00) |
0.714 (1.00) |
0.621 (1.00) |
0.329 (1.00) |
0.252 (1.00) |
1 (1.00) |
1 (1.00) |
0.636 (1.00) |
| 6P LOSS MUTATION ANALYSIS | 13 (11%) | 104 |
0.697 (1.00) |
0.983 (1.00) |
0.977 (1.00) |
1 (1.00) |
0.374 (1.00) |
0.0911 (1.00) |
1 (1.00) |
1 (1.00) |
| 6Q LOSS MUTATION ANALYSIS | 35 (30%) | 82 |
0.668 (1.00) |
0.0536 (1.00) |
0.5 (1.00) |
0.881 (1.00) |
0.195 (1.00) |
0.337 (1.00) |
0.298 (1.00) |
0.941 (1.00) |
| 7P LOSS MUTATION ANALYSIS | 8 (7%) | 109 |
0.427 (1.00) |
0.615 (1.00) |
0.85 (1.00) |
0.381 (1.00) |
1 (1.00) |
1 (1.00) |
0.0513 (1.00) |
0.587 (1.00) |
| 7Q LOSS MUTATION ANALYSIS | 11 (9%) | 106 |
0.367 (1.00) |
0.845 (1.00) |
0.876 (1.00) |
0.345 (1.00) |
1 (1.00) |
0.55 (1.00) |
0.0192 (1.00) |
0.378 (1.00) |
| 8P LOSS MUTATION ANALYSIS | 56 (48%) | 61 |
0.0881 (1.00) |
0.00114 (0.726) |
0.319 (1.00) |
0.258 (1.00) |
0.0555 (1.00) |
0.375 (1.00) |
0.567 (1.00) |
0.321 (1.00) |
| 8Q LOSS MUTATION ANALYSIS | 14 (12%) | 103 |
0.74 (1.00) |
0.408 (1.00) |
0.0244 (1.00) |
0.00297 (1.00) |
0.252 (1.00) |
0.105 (1.00) |
0.143 (1.00) |
1 (1.00) |
| 9P LOSS MUTATION ANALYSIS | 36 (31%) | 81 |
0.265 (1.00) |
0.87 (1.00) |
0.0724 (1.00) |
0.235 (1.00) |
0.245 (1.00) |
0.0886 (1.00) |
0.311 (1.00) |
0.0625 (1.00) |
| 9Q LOSS MUTATION ANALYSIS | 37 (32%) | 80 |
0.06 (1.00) |
0.898 (1.00) |
0.0517 (1.00) |
0.603 (1.00) |
0.0277 (1.00) |
0.0325 (1.00) |
0.838 (1.00) |
0.0542 (1.00) |
| 10P LOSS MUTATION ANALYSIS | 13 (11%) | 104 |
0.84 (1.00) |
0.0441 (1.00) |
0.0847 (1.00) |
0.355 (1.00) |
0.315 (1.00) |
0.787 (1.00) |
0.365 (1.00) |
1 (1.00) |
| 10Q LOSS MUTATION ANALYSIS | 26 (22%) | 91 |
0.0887 (1.00) |
0.721 (1.00) |
0.355 (1.00) |
0.325 (1.00) |
0.483 (1.00) |
1 (1.00) |
0.495 (1.00) |
0.549 (1.00) |
| 11P LOSS MUTATION ANALYSIS | 20 (17%) | 97 |
0.902 (1.00) |
0.00271 (1.00) |
0.321 (1.00) |
0.469 (1.00) |
0.0519 (1.00) |
0.84 (1.00) |
0.805 (1.00) |
0.668 (1.00) |
| 11Q LOSS MUTATION ANALYSIS | 22 (19%) | 95 |
0.978 (1.00) |
0.0154 (1.00) |
0.286 (1.00) |
0.326 (1.00) |
0.0722 (1.00) |
0.715 (1.00) |
1 (1.00) |
0.277 (1.00) |
| 12P LOSS MUTATION ANALYSIS | 21 (18%) | 96 |
0.147 (1.00) |
0.672 (1.00) |
0.345 (1.00) |
0.862 (1.00) |
0.0833 (1.00) |
0.247 (1.00) |
1 (1.00) |
0.151 (1.00) |
| 12Q LOSS MUTATION ANALYSIS | 12 (10%) | 105 |
0.615 (1.00) |
0.302 (1.00) |
0.136 (1.00) |
1 (1.00) |
1 (1.00) |
0.3 (1.00) |
1 (1.00) |
0.105 (1.00) |
| 13Q LOSS MUTATION ANALYSIS | 40 (34%) | 77 |
0.786 (1.00) |
0.0861 (1.00) |
0.3 (1.00) |
0.0423 (1.00) |
1 (1.00) |
0.417 (1.00) |
0.841 (1.00) |
0.233 (1.00) |
| 14Q LOSS MUTATION ANALYSIS | 38 (32%) | 79 |
0.848 (1.00) |
0.768 (1.00) |
0.116 (1.00) |
0.0415 (1.00) |
1 (1.00) |
0.466 (1.00) |
0.685 (1.00) |
0.174 (1.00) |
| 15Q LOSS MUTATION ANALYSIS | 22 (19%) | 95 |
0.399 (1.00) |
0.508 (1.00) |
0.513 (1.00) |
0.357 (1.00) |
0.0952 (1.00) |
0.208 (1.00) |
0.333 (1.00) |
0.912 (1.00) |
| 16P LOSS MUTATION ANALYSIS | 33 (28%) | 84 |
0.083 (1.00) |
0.368 (1.00) |
0.149 (1.00) |
0.41 (1.00) |
0.245 (1.00) |
0.203 (1.00) |
0.834 (1.00) |
0.0722 (1.00) |
| 16Q LOSS MUTATION ANALYSIS | 43 (37%) | 74 |
0.208 (1.00) |
0.31 (1.00) |
0.439 (1.00) |
0.526 (1.00) |
0.573 (1.00) |
0.275 (1.00) |
0.554 (1.00) |
0.0737 (1.00) |
| 17P LOSS MUTATION ANALYSIS | 58 (50%) | 59 |
0.346 (1.00) |
0.158 (1.00) |
0.134 (1.00) |
0.515 (1.00) |
0.251 (1.00) |
0.271 (1.00) |
0.849 (1.00) |
0.0781 (1.00) |
| 17Q LOSS MUTATION ANALYSIS | 9 (8%) | 108 |
0.573 (1.00) |
0.0433 (1.00) |
0.0353 (1.00) |
0.502 (1.00) |
0.15 (1.00) |
1 (1.00) |
0.727 (1.00) |
1 (1.00) |
| 18P LOSS MUTATION ANALYSIS | 18 (15%) | 99 |
0.242 (1.00) |
0.365 (1.00) |
0.966 (1.00) |
0.915 (1.00) |
0.374 (1.00) |
0.828 (1.00) |
0.599 (1.00) |
1 (1.00) |
| 18Q LOSS MUTATION ANALYSIS | 19 (16%) | 98 |
0.481 (1.00) |
0.161 (1.00) |
0.487 (1.00) |
0.978 (1.00) |
0.374 (1.00) |
0.32 (1.00) |
0.195 (1.00) |
1 (1.00) |
| 19P LOSS MUTATION ANALYSIS | 14 (12%) | 103 |
0.0753 (1.00) |
0.705 (1.00) |
0.956 (1.00) |
0.732 (1.00) |
0.315 (1.00) |
0.792 (1.00) |
0.564 (1.00) |
0.0712 (1.00) |
| 19Q LOSS MUTATION ANALYSIS | 10 (9%) | 107 |
0.293 (1.00) |
0.616 (1.00) |
0.948 (1.00) |
0.906 (1.00) |
0.252 (1.00) |
0.739 (1.00) |
0.741 (1.00) |
0.178 (1.00) |
| 20P LOSS MUTATION ANALYSIS | 7 (6%) | 110 |
0.765 (1.00) |
0.237 (1.00) |
0.512 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| 20Q LOSS MUTATION ANALYSIS | 4 (3%) | 113 |
0.246 (1.00) |
0.911 (1.00) |
0.0631 (1.00) |
0.656 (1.00) |
1 (1.00) |
0.604 (1.00) |
0.631 (1.00) |
1 (1.00) |
| 21Q LOSS MUTATION ANALYSIS | 29 (25%) | 88 |
0.0522 (1.00) |
0.503 (1.00) |
0.0816 (1.00) |
0.313 (1.00) |
0.135 (1.00) |
0.103 (1.00) |
0.0141 (1.00) |
0.31 (1.00) |
| 22Q LOSS MUTATION ANALYSIS | 20 (17%) | 97 |
0.196 (1.00) |
0.741 (1.00) |
0.92 (1.00) |
0.225 (1.00) |
1 (1.00) |
0.696 (1.00) |
0.217 (1.00) |
0.0175 (1.00) |
| XQ LOSS MUTATION ANALYSIS | 16 (14%) | 101 |
0.437 (1.00) |
0.716 (1.00) |
0.71 (1.00) |
0.122 (1.00) |
0.345 (1.00) |
0.64 (1.00) |
0.782 (1.00) |
0.269 (1.00) |
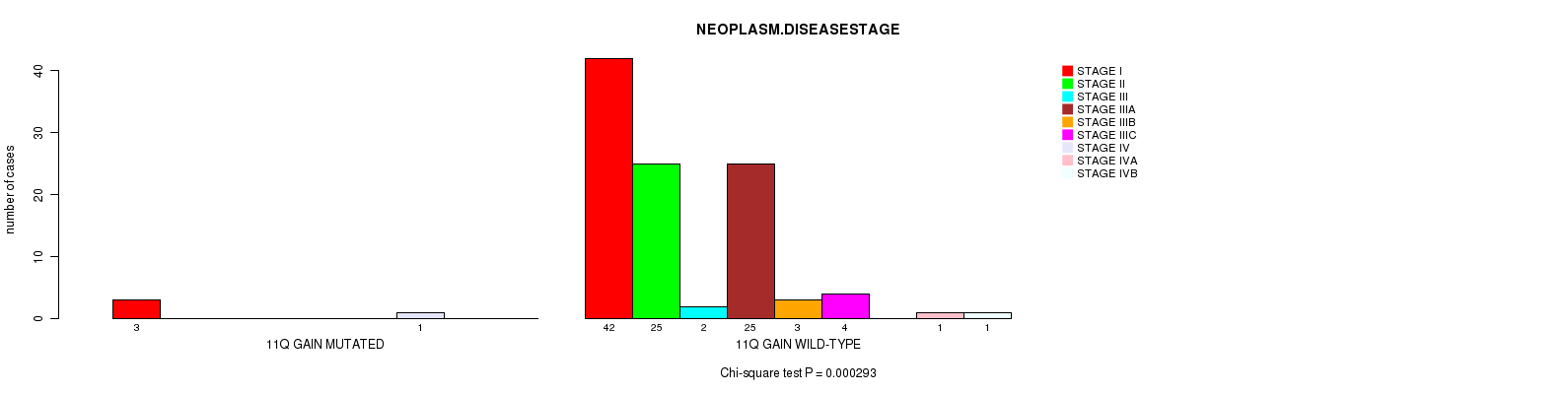
P value = 0.000293 (Chi-square test), Q value = 0.19
Table S1. Gene #22: '11Q GAIN MUTATION STATUS' versus Clinical Feature #3: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IIIA | STAGE IIIB | STAGE IIIC | STAGE IV | STAGE IVA | STAGE IVB |
|---|---|---|---|---|---|---|---|---|---|
| ALL | 45 | 25 | 2 | 25 | 3 | 4 | 1 | 1 | 1 |
| 11Q GAIN MUTATED | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 11Q GAIN WILD-TYPE | 42 | 25 | 2 | 25 | 3 | 4 | 0 | 1 | 1 |
Figure S1. Get High-res Image Gene #22: '11Q GAIN MUTATION STATUS' versus Clinical Feature #3: 'NEOPLASM.DISEASESTAGE'
-
Copy number data file = transformed.cor.cli.txt
-
Clinical data file = LIHC-TP.clin.merged.picked.txt
-
Number of patients = 117
-
Number of significantly arm-level cnvs = 80
-
Number of selected clinical features = 8
-
Exclude regions that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For continuous numerical clinical features, two-tailed Student's t test with unequal variance (Lehmann and Romano 2005) was applied to compare the clinical values between tumors with and without gene mutations using 't.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.