This pipeline computes the correlation between significant copy number variation (cnv) genes and molecular subtypes.
Testing the association between copy number variation of 44 peak regions and 8 molecular subtypes across 65 patients, 61 significant findings detected with Q value < 0.25.
-
Amp Peak 3(7q22.1) cnvs correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
Amp Peak 4(8q24.21) cnvs correlated to 'CN_CNMF' and 'MRNASEQ_CNMF'.
-
Amp Peak 7(12p13.33) cnvs correlated to 'CN_CNMF'.
-
Amp Peak 8(12p11.21) cnvs correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
Amp Peak 12(18q11.2) cnvs correlated to 'CN_CNMF', 'MRNASEQ_CNMF', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
Amp Peak 14(20q13.2) cnvs correlated to 'CN_CNMF'.
-
Del Peak 2(1p36.11) cnvs correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
Del Peak 3(1p21.2) cnvs correlated to 'CN_CNMF'.
-
Del Peak 4(5q14.2) cnvs correlated to 'CN_CNMF'.
-
Del Peak 5(5q15) cnvs correlated to 'CN_CNMF'.
-
Del Peak 6(6p25.2) cnvs correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CNMF'.
-
Del Peak 7(6p22.3) cnvs correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CNMF'.
-
Del Peak 8(6q22.33) cnvs correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
Del Peak 10(8p21.3) cnvs correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_MATURE_CNMF'.
-
Del Peak 12(9p21.3) cnvs correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
Del Peak 13(10q23.2) cnvs correlated to 'CN_CNMF'.
-
Del Peak 15(12p12.3) cnvs correlated to 'CN_CNMF'.
-
Del Peak 16(12q12) cnvs correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
Del Peak 17(12q24.31) cnvs correlated to 'METHLYATION_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CNMF'.
-
Del Peak 18(15q15.1) cnvs correlated to 'CN_CNMF'.
-
Del Peak 19(17p11.2) cnvs correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CNMF'.
-
Del Peak 20(17q22) cnvs correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
Del Peak 28(Xq21.1) cnvs correlated to 'CN_CNMF'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 44 regions and 8 molecular subtypes. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 61 significant findings detected.
|
Molecular subtypes |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Chi-square test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| Del Peak 12(9p21 3) | 0 (0%) | 24 |
1.43e-06 (0.000496) |
9.11e-08 (3.2e-05) |
2.16e-05 (0.00726) |
4.09e-06 (0.00141) |
2.95e-05 (0.00987) |
4.02e-05 (0.0133) |
2.78e-07 (9.69e-05) |
4.88e-06 (0.00167) |
| Del Peak 8(6q22 33) | 0 (0%) | 34 |
0.000728 (0.216) |
3.79e-05 (0.0126) |
0.00218 (0.607) |
4.39e-06 (0.00151) |
0.000693 (0.207) |
0.000148 (0.0477) |
0.000163 (0.052) |
0.000466 (0.143) |
| Del Peak 10(8p21 3) | 0 (0%) | 46 |
2.52e-05 (0.00847) |
0.000211 (0.0666) |
8.93e-05 (0.0292) |
0.000538 (0.165) |
0.000789 (0.233) |
0.00201 (0.561) |
0.000163 (0.052) |
0.00115 (0.328) |
| Del Peak 19(17p11 2) | 0 (0%) | 39 |
5.43e-05 (0.0179) |
1.13e-08 (3.99e-06) |
0.000809 (0.238) |
0.000824 (0.241) |
0.00233 (0.643) |
0.000332 (0.104) |
0.000246 (0.0774) |
0.00165 (0.467) |
| Amp Peak 12(18q11 2) | 0 (0%) | 46 |
0.000293 (0.0919) |
0.00369 (0.983) |
0.000369 (0.115) |
0.00807 (1.00) |
0.00589 (1.00) |
0.00109 (0.313) |
0.000478 (0.147) |
0.000556 (0.17) |
| Del Peak 6(6p25 2) | 0 (0%) | 39 |
4.12e-05 (0.0136) |
1.76e-05 (0.00596) |
0.000193 (0.0611) |
0.00724 (1.00) |
0.0226 (1.00) |
0.00597 (1.00) |
0.00737 (1.00) |
0.018 (1.00) |
| Del Peak 7(6p22 3) | 0 (0%) | 40 |
0.000854 (0.249) |
6.32e-05 (0.0207) |
0.000119 (0.0386) |
0.00228 (0.63) |
0.00633 (1.00) |
0.00252 (0.694) |
0.00497 (1.00) |
0.0134 (1.00) |
| Del Peak 17(12q24 31) | 0 (0%) | 49 |
0.000948 (0.275) |
9.01e-05 (0.0294) |
0.0138 (1.00) |
0.000169 (0.0538) |
0.00107 (0.31) |
0.00541 (1.00) |
0.000454 (0.14) |
0.00402 (1.00) |
| Amp Peak 3(7q22 1) | 0 (0%) | 47 |
0.000766 (0.227) |
0.000101 (0.0328) |
0.27 (1.00) |
0.191 (1.00) |
0.125 (1.00) |
0.218 (1.00) |
0.109 (1.00) |
0.162 (1.00) |
| Amp Peak 4(8q24 21) | 0 (0%) | 40 |
7.91e-06 (0.0027) |
0.00287 (0.778) |
0.000579 (0.176) |
0.00333 (0.895) |
0.0284 (1.00) |
0.0744 (1.00) |
0.00983 (1.00) |
0.0358 (1.00) |
| Amp Peak 8(12p11 21) | 0 (0%) | 51 |
1.52e-05 (0.00514) |
0.000696 (0.208) |
0.00328 (0.885) |
0.0824 (1.00) |
0.00428 (1.00) |
0.017 (1.00) |
0.00584 (1.00) |
0.0135 (1.00) |
| Del Peak 2(1p36 11) | 0 (0%) | 47 |
0.00068 (0.205) |
0.000619 (0.187) |
0.00953 (1.00) |
0.26 (1.00) |
0.0801 (1.00) |
0.0769 (1.00) |
0.00911 (1.00) |
0.0518 (1.00) |
| Del Peak 16(12q12) | 0 (0%) | 55 |
0.00014 (0.0453) |
3.91e-05 (0.013) |
0.0244 (1.00) |
0.0185 (1.00) |
0.108 (1.00) |
0.0614 (1.00) |
0.0138 (1.00) |
0.0413 (1.00) |
| Del Peak 20(17q22) | 0 (0%) | 47 |
9.68e-06 (0.00329) |
3.54e-05 (0.0118) |
0.0403 (1.00) |
0.00272 (0.74) |
0.0142 (1.00) |
0.0299 (1.00) |
0.00822 (1.00) |
0.0183 (1.00) |
| Amp Peak 7(12p13 33) | 0 (0%) | 55 |
2.52e-07 (8.8e-05) |
0.00565 (1.00) |
0.0587 (1.00) |
0.0562 (1.00) |
0.0436 (1.00) |
0.113 (1.00) |
0.0158 (1.00) |
0.0791 (1.00) |
| Amp Peak 14(20q13 2) | 0 (0%) | 48 |
2.88e-06 (0.000992) |
0.121 (1.00) |
0.053 (1.00) |
0.191 (1.00) |
0.146 (1.00) |
0.218 (1.00) |
0.0368 (1.00) |
0.162 (1.00) |
| Del Peak 3(1p21 2) | 0 (0%) | 54 |
0.000152 (0.0489) |
0.0382 (1.00) |
0.153 (1.00) |
0.252 (1.00) |
0.0668 (1.00) |
0.316 (1.00) |
0.0137 (1.00) |
0.209 (1.00) |
| Del Peak 4(5q14 2) | 0 (0%) | 53 |
0.000601 (0.182) |
0.329 (1.00) |
0.234 (1.00) |
0.245 (1.00) |
0.184 (1.00) |
0.5 (1.00) |
0.0517 (1.00) |
0.467 (1.00) |
| Del Peak 5(5q15) | 0 (0%) | 51 |
0.000688 (0.206) |
0.0877 (1.00) |
0.546 (1.00) |
0.113 (1.00) |
0.124 (1.00) |
0.331 (1.00) |
0.029 (1.00) |
0.256 (1.00) |
| Del Peak 13(10q23 2) | 0 (0%) | 51 |
1.2e-07 (4.2e-05) |
0.0371 (1.00) |
0.0892 (1.00) |
0.0209 (1.00) |
0.0421 (1.00) |
0.0149 (1.00) |
0.00118 (0.336) |
0.0107 (1.00) |
| Del Peak 15(12p12 3) | 0 (0%) | 54 |
0.000408 (0.127) |
0.00266 (0.729) |
0.125 (1.00) |
0.134 (1.00) |
0.0436 (1.00) |
0.00989 (1.00) |
0.0181 (1.00) |
0.00739 (1.00) |
| Del Peak 18(15q15 1) | 0 (0%) | 51 |
1.29e-06 (0.000446) |
0.00359 (0.962) |
0.0414 (1.00) |
0.00942 (1.00) |
0.0194 (1.00) |
0.0346 (1.00) |
0.000994 (0.287) |
0.0159 (1.00) |
| Del Peak 28(Xq21 1) | 0 (0%) | 58 |
0.00041 (0.127) |
0.0299 (1.00) |
0.365 (1.00) |
0.782 (1.00) |
0.252 (1.00) |
0.554 (1.00) |
0.0704 (1.00) |
0.396 (1.00) |
| Amp Peak 1(1p34 2) | 0 (0%) | 60 |
0.173 (1.00) |
0.729 (1.00) |
1 (1.00) |
1 (1.00) |
0.656 (1.00) |
1 (1.00) |
0.821 (1.00) |
1 (1.00) |
| Amp Peak 2(1p12) | 0 (0%) | 54 |
0.0445 (1.00) |
0.251 (1.00) |
0.952 (1.00) |
0.746 (1.00) |
0.889 (1.00) |
0.885 (1.00) |
0.474 (1.00) |
0.791 (1.00) |
| Amp Peak 5(9p13 3) | 0 (0%) | 58 |
0.077 (1.00) |
0.404 (1.00) |
0.133 (1.00) |
0.119 (1.00) |
0.305 (1.00) |
0.226 (1.00) |
0.161 (1.00) |
0.166 (1.00) |
| Amp Peak 6(11p11 2) | 0 (0%) | 59 |
0.0888 (1.00) |
0.059 (1.00) |
0.314 (1.00) |
0.195 (1.00) |
0.571 (1.00) |
0.195 (1.00) |
0.312 (1.00) |
0.206 (1.00) |
| Amp Peak 9(12q15) | 0 (0%) | 57 |
0.00549 (1.00) |
0.0311 (1.00) |
0.237 (1.00) |
0.0355 (1.00) |
0.0225 (1.00) |
0.0131 (1.00) |
0.00463 (1.00) |
0.00874 (1.00) |
| Amp Peak 10(17p11 2) | 0 (0%) | 62 |
0.635 (1.00) |
0.338 (1.00) |
0.885 (1.00) |
0.683 (1.00) |
0.577 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| Amp Peak 11(17q21 33) | 0 (0%) | 58 |
0.0162 (1.00) |
0.239 (1.00) |
0.609 (1.00) |
0.72 (1.00) |
1 (1.00) |
0.756 (1.00) |
0.67 (1.00) |
0.768 (1.00) |
| Amp Peak 13(19q13 13) | 0 (0%) | 51 |
0.0229 (1.00) |
0.0745 (1.00) |
0.309 (1.00) |
0.0443 (1.00) |
0.286 (1.00) |
0.22 (1.00) |
0.286 (1.00) |
0.434 (1.00) |
| Amp Peak 15(Xp22 11) | 0 (0%) | 61 |
0.248 (1.00) |
0.667 (1.00) |
0.885 (1.00) |
0.312 (1.00) |
1 (1.00) |
0.818 (1.00) |
0.821 (1.00) |
0.816 (1.00) |
| Amp Peak 16(Xq27 1) | 0 (0%) | 59 |
0.0165 (1.00) |
0.873 (1.00) |
0.391 (1.00) |
0.615 (1.00) |
0.463 (1.00) |
1 (1.00) |
0.624 (1.00) |
0.744 (1.00) |
| Del Peak 1(1p36 22) | 0 (0%) | 51 |
0.0207 (1.00) |
0.00361 (0.964) |
0.0753 (1.00) |
0.134 (1.00) |
0.0339 (1.00) |
0.186 (1.00) |
0.0686 (1.00) |
0.107 (1.00) |
| Del Peak 9(7q36 1) | 0 (0%) | 58 |
0.00189 (0.533) |
0.0439 (1.00) |
0.133 (1.00) |
0.119 (1.00) |
0.305 (1.00) |
0.74 (1.00) |
0.461 (1.00) |
0.643 (1.00) |
| Del Peak 11(8q12 1) | 0 (0%) | 60 |
0.0349 (1.00) |
0.077 (1.00) |
0.314 (1.00) |
0.257 (1.00) |
0.571 (1.00) |
0.195 (1.00) |
0.312 (1.00) |
0.206 (1.00) |
| Del Peak 14(11p15 4) | 0 (0%) | 53 |
0.0304 (1.00) |
0.467 (1.00) |
0.761 (1.00) |
0.592 (1.00) |
0.525 (1.00) |
1 (1.00) |
0.732 (1.00) |
1 (1.00) |
| Del Peak 21(18p11 32) | 0 (0%) | 44 |
0.124 (1.00) |
1 (1.00) |
0.542 (1.00) |
1 (1.00) |
0.218 (1.00) |
0.935 (1.00) |
0.664 (1.00) |
1 (1.00) |
| Del Peak 22(18q12 1) | 0 (0%) | 28 |
0.02 (1.00) |
0.0844 (1.00) |
0.113 (1.00) |
0.338 (1.00) |
0.0835 (1.00) |
0.0889 (1.00) |
0.0632 (1.00) |
0.141 (1.00) |
| Del Peak 23(18q21 2) | 0 (0%) | 20 |
0.00267 (0.729) |
0.000946 (0.275) |
0.00907 (1.00) |
0.0226 (1.00) |
0.00861 (1.00) |
0.00197 (0.551) |
0.00115 (0.328) |
0.00436 (1.00) |
| Del Peak 24(19q13 33) | 0 (0%) | 58 |
0.00189 (0.533) |
0.0851 (1.00) |
0.819 (1.00) |
1 (1.00) |
0.848 (1.00) |
0.74 (1.00) |
0.461 (1.00) |
0.643 (1.00) |
| Del Peak 25(21q22 11) | 0 (0%) | 43 |
0.0192 (1.00) |
0.158 (1.00) |
0.154 (1.00) |
0.00376 (0.996) |
0.0379 (1.00) |
0.265 (1.00) |
0.0685 (1.00) |
0.181 (1.00) |
| Del Peak 26(21q22 11) | 0 (0%) | 43 |
0.0192 (1.00) |
0.158 (1.00) |
0.154 (1.00) |
0.00376 (0.996) |
0.0379 (1.00) |
0.265 (1.00) |
0.0685 (1.00) |
0.181 (1.00) |
| Del Peak 27(22q13 31) | 0 (0%) | 49 |
0.006 (1.00) |
0.15 (1.00) |
0.358 (1.00) |
0.26 (1.00) |
0.38 (1.00) |
0.424 (1.00) |
0.286 (1.00) |
0.326 (1.00) |
P value = 0.000766 (Chi-square test), Q value = 0.23
Table S1. Gene #3: 'Amp Peak 3(7q22.1)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| AMP PEAK 3(7Q22.1) CNV | 4 | 4 | 7 | 2 | 1 |
| AMP PEAK 3(7Q22.1) WILD-TYPE | 7 | 31 | 2 | 7 | 0 |
Figure S1. Get High-res Image Gene #3: 'Amp Peak 3(7q22.1)' versus Molecular Subtype #1: 'CN_CNMF'
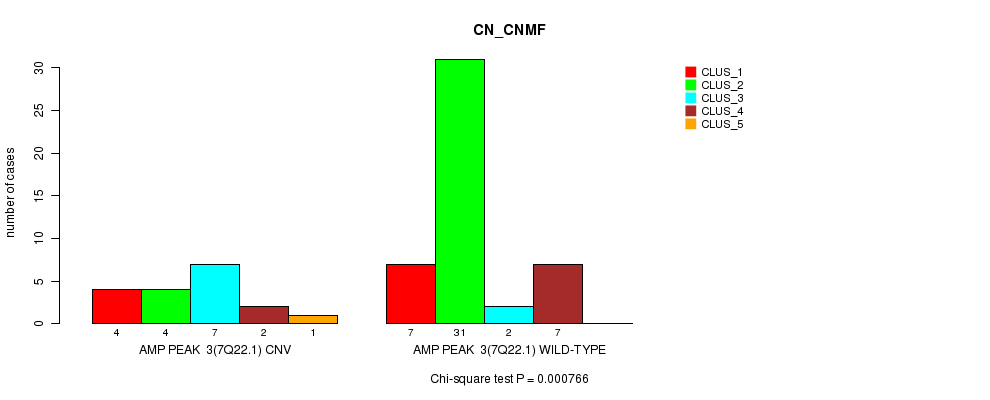
P value = 0.000101 (Fisher's exact test), Q value = 0.033
Table S2. Gene #3: 'Amp Peak 3(7q22.1)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 25 | 15 |
| AMP PEAK 3(7Q22.1) CNV | 10 | 0 | 7 |
| AMP PEAK 3(7Q22.1) WILD-TYPE | 14 | 25 | 8 |
Figure S2. Get High-res Image Gene #3: 'Amp Peak 3(7q22.1)' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 7.91e-06 (Chi-square test), Q value = 0.0027
Table S3. Gene #4: 'Amp Peak 4(8q24.21)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| AMP PEAK 4(8Q24.21) CNV | 11 | 6 | 6 | 2 | 0 |
| AMP PEAK 4(8Q24.21) WILD-TYPE | 0 | 29 | 3 | 7 | 1 |
Figure S3. Get High-res Image Gene #4: 'Amp Peak 4(8q24.21)' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.000579 (Fisher's exact test), Q value = 0.18
Table S4. Gene #4: 'Amp Peak 4(8q24.21)' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 21 | 12 | 12 | 10 |
| AMP PEAK 4(8Q24.21) CNV | 14 | 5 | 2 | 0 |
| AMP PEAK 4(8Q24.21) WILD-TYPE | 7 | 7 | 10 | 10 |
Figure S4. Get High-res Image Gene #4: 'Amp Peak 4(8q24.21)' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 2.52e-07 (Chi-square test), Q value = 8.8e-05
Table S5. Gene #7: 'Amp Peak 7(12p13.33)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| AMP PEAK 7(12P13.33) CNV | 8 | 0 | 2 | 0 | 0 |
| AMP PEAK 7(12P13.33) WILD-TYPE | 3 | 35 | 7 | 9 | 1 |
Figure S5. Get High-res Image Gene #7: 'Amp Peak 7(12p13.33)' versus Molecular Subtype #1: 'CN_CNMF'
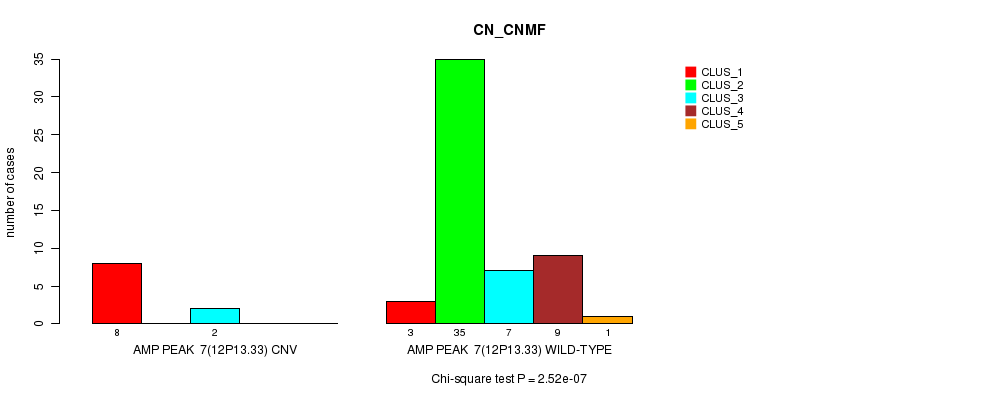
P value = 1.52e-05 (Chi-square test), Q value = 0.0051
Table S6. Gene #8: 'Amp Peak 8(12p11.21)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| AMP PEAK 8(12P11.21) CNV | 7 | 0 | 2 | 4 | 1 |
| AMP PEAK 8(12P11.21) WILD-TYPE | 4 | 35 | 7 | 5 | 0 |
Figure S6. Get High-res Image Gene #8: 'Amp Peak 8(12p11.21)' versus Molecular Subtype #1: 'CN_CNMF'
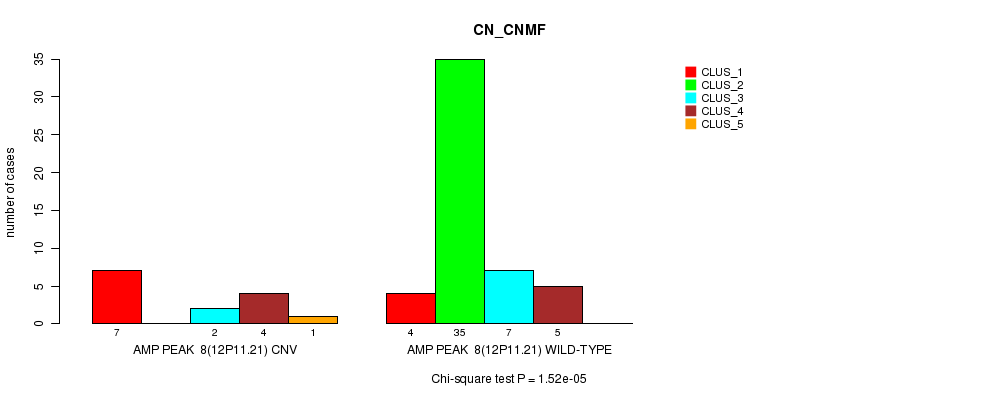
P value = 0.000696 (Fisher's exact test), Q value = 0.21
Table S7. Gene #8: 'Amp Peak 8(12p11.21)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 25 | 15 |
| AMP PEAK 8(12P11.21) CNV | 8 | 0 | 6 |
| AMP PEAK 8(12P11.21) WILD-TYPE | 16 | 25 | 9 |
Figure S7. Get High-res Image Gene #8: 'Amp Peak 8(12p11.21)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
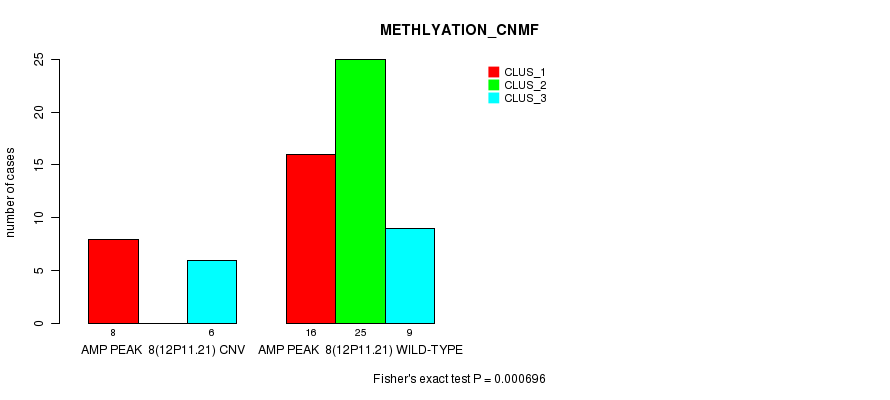
P value = 0.000293 (Chi-square test), Q value = 0.092
Table S8. Gene #12: 'Amp Peak 12(18q11.2)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| AMP PEAK 12(18Q11.2) CNV | 4 | 3 | 5 | 7 | 0 |
| AMP PEAK 12(18Q11.2) WILD-TYPE | 7 | 32 | 4 | 2 | 1 |
Figure S8. Get High-res Image Gene #12: 'Amp Peak 12(18q11.2)' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.000369 (Fisher's exact test), Q value = 0.11
Table S9. Gene #12: 'Amp Peak 12(18q11.2)' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 21 | 12 | 12 | 10 |
| AMP PEAK 12(18Q11.2) CNV | 13 | 3 | 0 | 1 |
| AMP PEAK 12(18Q11.2) WILD-TYPE | 8 | 9 | 12 | 9 |
Figure S9. Get High-res Image Gene #12: 'Amp Peak 12(18q11.2)' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 0.000478 (Fisher's exact test), Q value = 0.15
Table S10. Gene #12: 'Amp Peak 12(18q11.2)' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 25 | 10 | 23 |
| AMP PEAK 12(18Q11.2) CNV | 14 | 1 | 2 |
| AMP PEAK 12(18Q11.2) WILD-TYPE | 11 | 9 | 21 |
Figure S10. Get High-res Image Gene #12: 'Amp Peak 12(18q11.2)' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'

P value = 0.000556 (Fisher's exact test), Q value = 0.17
Table S11. Gene #12: 'Amp Peak 12(18q11.2)' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 12 | 26 | 20 |
| AMP PEAK 12(18Q11.2) CNV | 0 | 14 | 3 |
| AMP PEAK 12(18Q11.2) WILD-TYPE | 12 | 12 | 17 |
Figure S11. Get High-res Image Gene #12: 'Amp Peak 12(18q11.2)' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
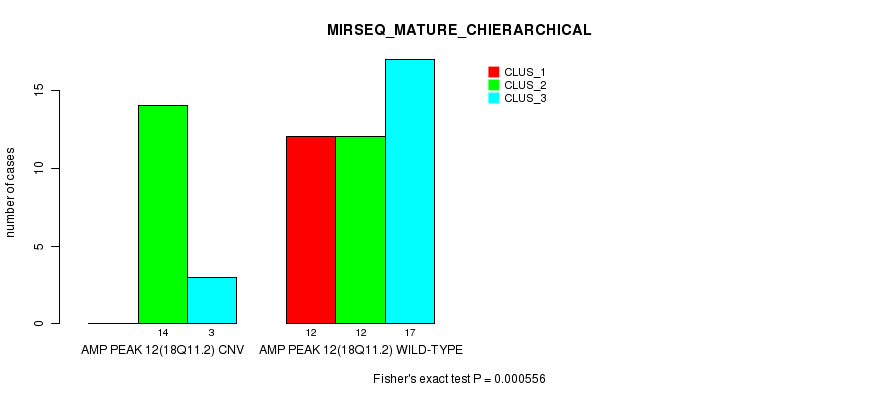
P value = 2.88e-06 (Chi-square test), Q value = 0.00099
Table S12. Gene #14: 'Amp Peak 14(20q13.2)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| AMP PEAK 14(20Q13.2) CNV | 10 | 3 | 3 | 1 | 0 |
| AMP PEAK 14(20Q13.2) WILD-TYPE | 1 | 32 | 6 | 8 | 1 |
Figure S12. Get High-res Image Gene #14: 'Amp Peak 14(20q13.2)' versus Molecular Subtype #1: 'CN_CNMF'
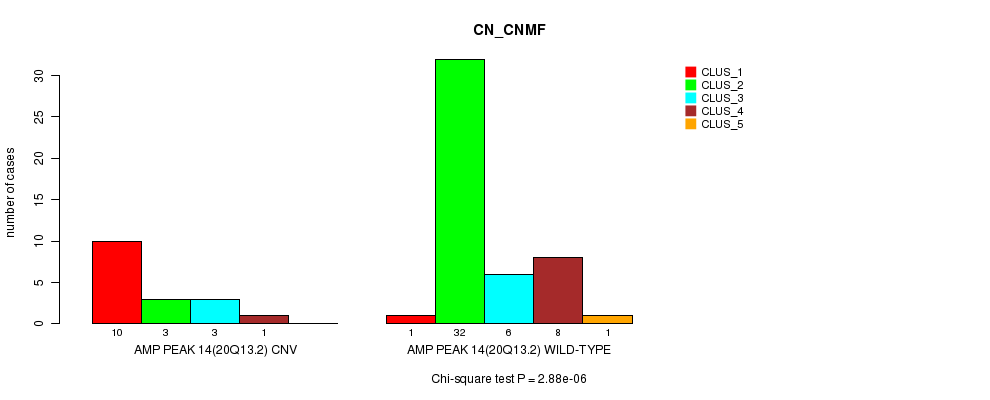
P value = 0.00068 (Chi-square test), Q value = 0.2
Table S13. Gene #18: 'Del Peak 2(1p36.11)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 2(1P36.11) CNV | 8 | 3 | 3 | 4 | 0 |
| DEL PEAK 2(1P36.11) WILD-TYPE | 3 | 32 | 6 | 5 | 1 |
Figure S13. Get High-res Image Gene #18: 'Del Peak 2(1p36.11)' versus Molecular Subtype #1: 'CN_CNMF'
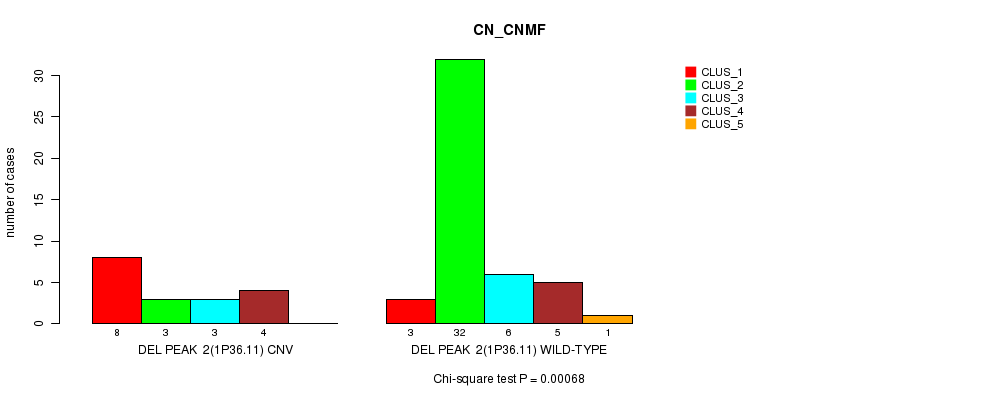
P value = 0.000619 (Fisher's exact test), Q value = 0.19
Table S14. Gene #18: 'Del Peak 2(1p36.11)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 25 | 15 |
| DEL PEAK 2(1P36.11) CNV | 9 | 1 | 8 |
| DEL PEAK 2(1P36.11) WILD-TYPE | 15 | 24 | 7 |
Figure S14. Get High-res Image Gene #18: 'Del Peak 2(1p36.11)' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.000152 (Chi-square test), Q value = 0.049
Table S15. Gene #19: 'Del Peak 3(1p21.2)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 3(1P21.2) CNV | 7 | 1 | 2 | 1 | 0 |
| DEL PEAK 3(1P21.2) WILD-TYPE | 4 | 34 | 7 | 8 | 1 |
Figure S15. Get High-res Image Gene #19: 'Del Peak 3(1p21.2)' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.000601 (Chi-square test), Q value = 0.18
Table S16. Gene #20: 'Del Peak 4(5q14.2)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 4(5Q14.2) CNV | 6 | 2 | 4 | 0 | 0 |
| DEL PEAK 4(5Q14.2) WILD-TYPE | 5 | 33 | 5 | 9 | 1 |
Figure S16. Get High-res Image Gene #20: 'Del Peak 4(5q14.2)' versus Molecular Subtype #1: 'CN_CNMF'
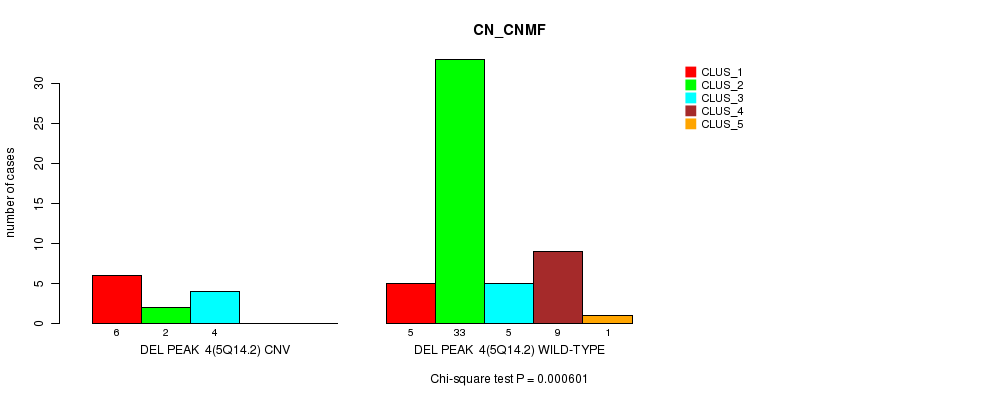
P value = 0.000688 (Chi-square test), Q value = 0.21
Table S17. Gene #21: 'Del Peak 5(5q15)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 5(5Q15) CNV | 6 | 2 | 5 | 1 | 0 |
| DEL PEAK 5(5Q15) WILD-TYPE | 5 | 33 | 4 | 8 | 1 |
Figure S17. Get High-res Image Gene #21: 'Del Peak 5(5q15)' versus Molecular Subtype #1: 'CN_CNMF'

P value = 4.12e-05 (Chi-square test), Q value = 0.014
Table S18. Gene #22: 'Del Peak 6(6p25.2)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 6(6P25.2) CNV | 9 | 6 | 8 | 3 | 0 |
| DEL PEAK 6(6P25.2) WILD-TYPE | 2 | 29 | 1 | 6 | 1 |
Figure S18. Get High-res Image Gene #22: 'Del Peak 6(6p25.2)' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1.76e-05 (Fisher's exact test), Q value = 0.006
Table S19. Gene #22: 'Del Peak 6(6p25.2)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 25 | 15 |
| DEL PEAK 6(6P25.2) CNV | 17 | 2 | 6 |
| DEL PEAK 6(6P25.2) WILD-TYPE | 7 | 23 | 9 |
Figure S19. Get High-res Image Gene #22: 'Del Peak 6(6p25.2)' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.000193 (Fisher's exact test), Q value = 0.061
Table S20. Gene #22: 'Del Peak 6(6p25.2)' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 21 | 12 | 12 | 10 |
| DEL PEAK 6(6P25.2) CNV | 15 | 4 | 2 | 0 |
| DEL PEAK 6(6P25.2) WILD-TYPE | 6 | 8 | 10 | 10 |
Figure S20. Get High-res Image Gene #22: 'Del Peak 6(6p25.2)' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 0.000854 (Chi-square test), Q value = 0.25
Table S21. Gene #23: 'Del Peak 7(6p22.3)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 7(6P22.3) CNV | 8 | 6 | 7 | 4 | 0 |
| DEL PEAK 7(6P22.3) WILD-TYPE | 3 | 29 | 2 | 5 | 1 |
Figure S21. Get High-res Image Gene #23: 'Del Peak 7(6p22.3)' versus Molecular Subtype #1: 'CN_CNMF'

P value = 6.32e-05 (Fisher's exact test), Q value = 0.021
Table S22. Gene #23: 'Del Peak 7(6p22.3)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 25 | 15 |
| DEL PEAK 7(6P22.3) CNV | 16 | 2 | 6 |
| DEL PEAK 7(6P22.3) WILD-TYPE | 8 | 23 | 9 |
Figure S22. Get High-res Image Gene #23: 'Del Peak 7(6p22.3)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
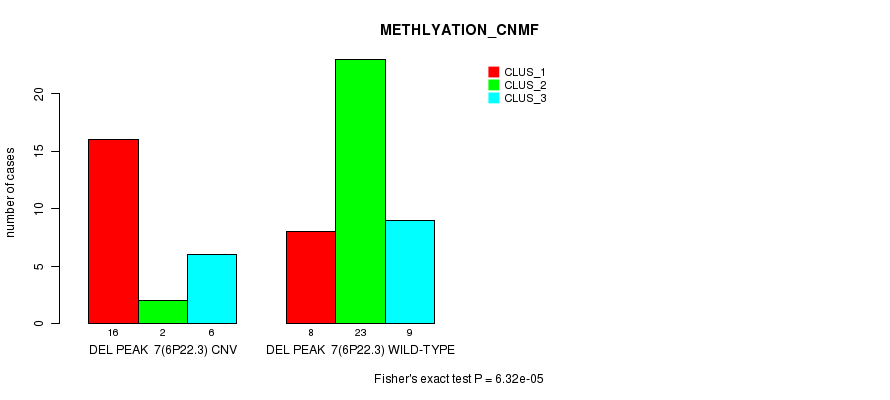
P value = 0.000119 (Fisher's exact test), Q value = 0.039
Table S23. Gene #23: 'Del Peak 7(6p22.3)' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 21 | 12 | 12 | 10 |
| DEL PEAK 7(6P22.3) CNV | 15 | 3 | 2 | 0 |
| DEL PEAK 7(6P22.3) WILD-TYPE | 6 | 9 | 10 | 10 |
Figure S23. Get High-res Image Gene #23: 'Del Peak 7(6p22.3)' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 0.000728 (Chi-square test), Q value = 0.22
Table S24. Gene #24: 'Del Peak 8(6q22.33)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 8(6Q22.33) CNV | 9 | 9 | 8 | 5 | 0 |
| DEL PEAK 8(6Q22.33) WILD-TYPE | 2 | 26 | 1 | 4 | 1 |
Figure S24. Get High-res Image Gene #24: 'Del Peak 8(6q22.33)' versus Molecular Subtype #1: 'CN_CNMF'
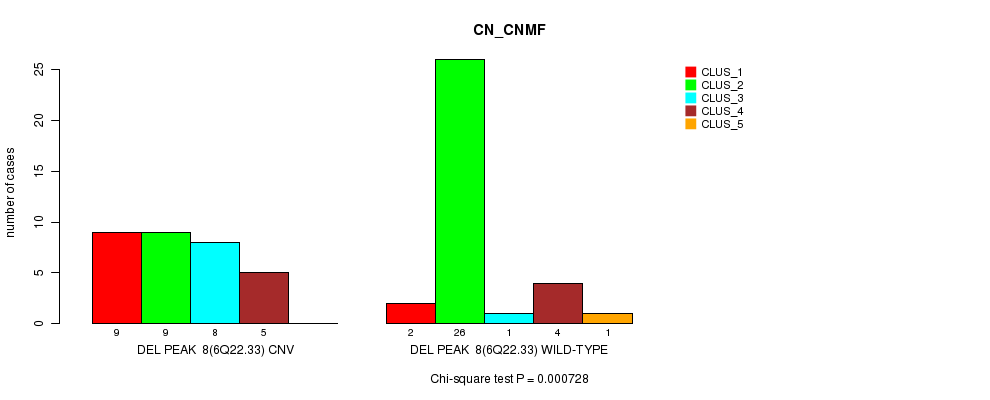
P value = 3.79e-05 (Fisher's exact test), Q value = 0.013
Table S25. Gene #24: 'Del Peak 8(6q22.33)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 25 | 15 |
| DEL PEAK 8(6Q22.33) CNV | 19 | 4 | 7 |
| DEL PEAK 8(6Q22.33) WILD-TYPE | 5 | 21 | 8 |
Figure S25. Get High-res Image Gene #24: 'Del Peak 8(6q22.33)' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 4.39e-06 (Fisher's exact test), Q value = 0.0015
Table S26. Gene #24: 'Del Peak 8(6q22.33)' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 5 | 22 | 28 |
| DEL PEAK 8(6Q22.33) CNV | 0 | 18 | 6 |
| DEL PEAK 8(6Q22.33) WILD-TYPE | 5 | 4 | 22 |
Figure S26. Get High-res Image Gene #24: 'Del Peak 8(6q22.33)' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 0.000693 (Fisher's exact test), Q value = 0.21
Table S27. Gene #24: 'Del Peak 8(6q22.33)' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 11 | 18 |
| DEL PEAK 8(6Q22.33) CNV | 20 | 3 | 3 |
| DEL PEAK 8(6Q22.33) WILD-TYPE | 9 | 8 | 15 |
Figure S27. Get High-res Image Gene #24: 'Del Peak 8(6q22.33)' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
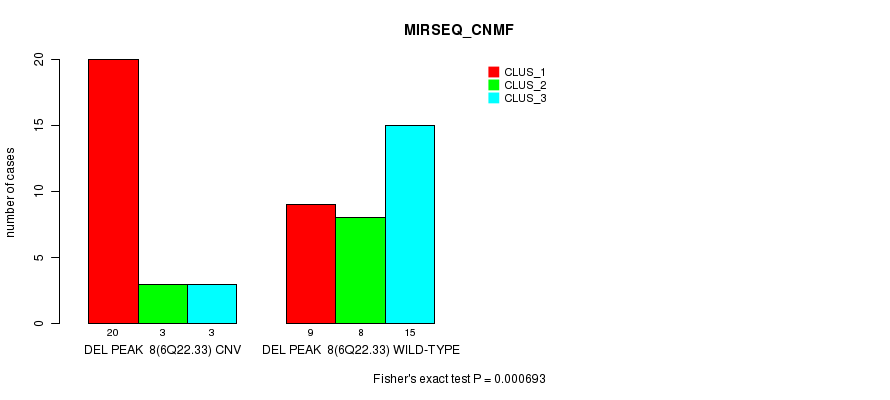
P value = 0.000148 (Fisher's exact test), Q value = 0.048
Table S28. Gene #24: 'Del Peak 8(6q22.33)' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 11 | 20 | 27 |
| DEL PEAK 8(6Q22.33) CNV | 2 | 4 | 20 |
| DEL PEAK 8(6Q22.33) WILD-TYPE | 9 | 16 | 7 |
Figure S28. Get High-res Image Gene #24: 'Del Peak 8(6q22.33)' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 0.000163 (Fisher's exact test), Q value = 0.052
Table S29. Gene #24: 'Del Peak 8(6q22.33)' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 25 | 10 | 23 |
| DEL PEAK 8(6Q22.33) CNV | 19 | 2 | 5 |
| DEL PEAK 8(6Q22.33) WILD-TYPE | 6 | 8 | 18 |
Figure S29. Get High-res Image Gene #24: 'Del Peak 8(6q22.33)' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'

P value = 0.000466 (Fisher's exact test), Q value = 0.14
Table S30. Gene #24: 'Del Peak 8(6q22.33)' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 12 | 26 | 20 |
| DEL PEAK 8(6Q22.33) CNV | 3 | 19 | 4 |
| DEL PEAK 8(6Q22.33) WILD-TYPE | 9 | 7 | 16 |
Figure S30. Get High-res Image Gene #24: 'Del Peak 8(6q22.33)' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 2.52e-05 (Chi-square test), Q value = 0.0085
Table S31. Gene #26: 'Del Peak 10(8p21.3)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 10(8P21.3) CNV | 9 | 2 | 4 | 4 | 0 |
| DEL PEAK 10(8P21.3) WILD-TYPE | 2 | 33 | 5 | 5 | 1 |
Figure S31. Get High-res Image Gene #26: 'Del Peak 10(8p21.3)' versus Molecular Subtype #1: 'CN_CNMF'
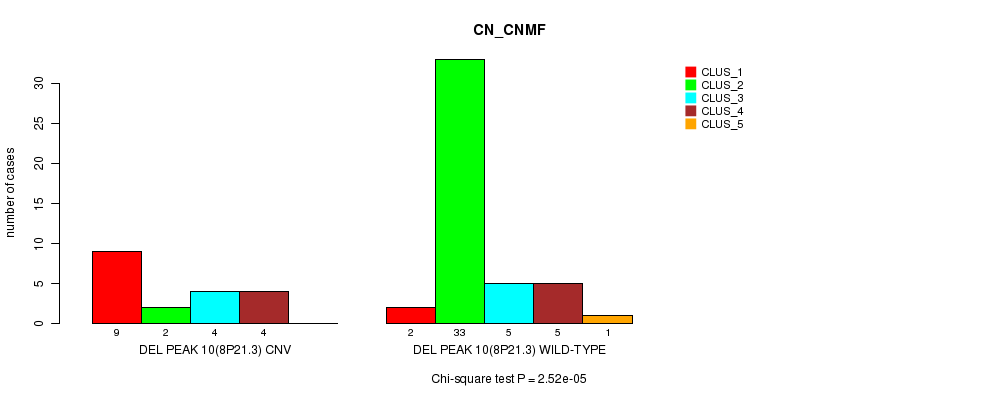
P value = 0.000211 (Fisher's exact test), Q value = 0.067
Table S32. Gene #26: 'Del Peak 10(8p21.3)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 25 | 15 |
| DEL PEAK 10(8P21.3) CNV | 13 | 1 | 4 |
| DEL PEAK 10(8P21.3) WILD-TYPE | 11 | 24 | 11 |
Figure S32. Get High-res Image Gene #26: 'Del Peak 10(8p21.3)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
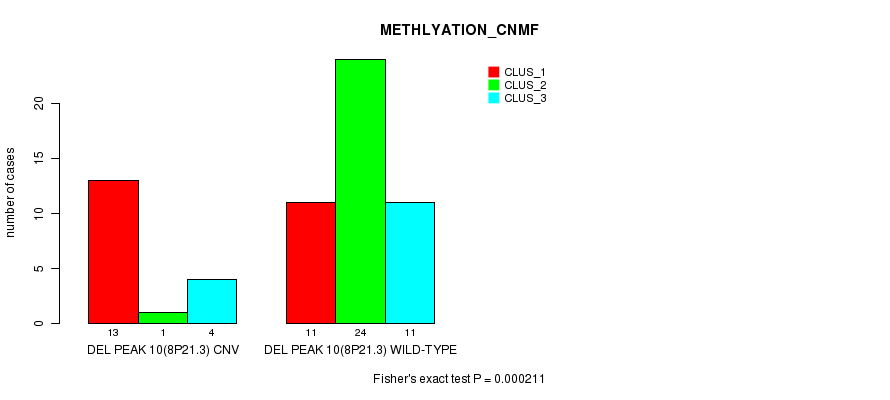
P value = 8.93e-05 (Fisher's exact test), Q value = 0.029
Table S33. Gene #26: 'Del Peak 10(8p21.3)' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 21 | 12 | 12 | 10 |
| DEL PEAK 10(8P21.3) CNV | 13 | 1 | 1 | 0 |
| DEL PEAK 10(8P21.3) WILD-TYPE | 8 | 11 | 11 | 10 |
Figure S33. Get High-res Image Gene #26: 'Del Peak 10(8p21.3)' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 0.000538 (Fisher's exact test), Q value = 0.16
Table S34. Gene #26: 'Del Peak 10(8p21.3)' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 5 | 22 | 28 |
| DEL PEAK 10(8P21.3) CNV | 1 | 12 | 2 |
| DEL PEAK 10(8P21.3) WILD-TYPE | 4 | 10 | 26 |
Figure S34. Get High-res Image Gene #26: 'Del Peak 10(8p21.3)' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
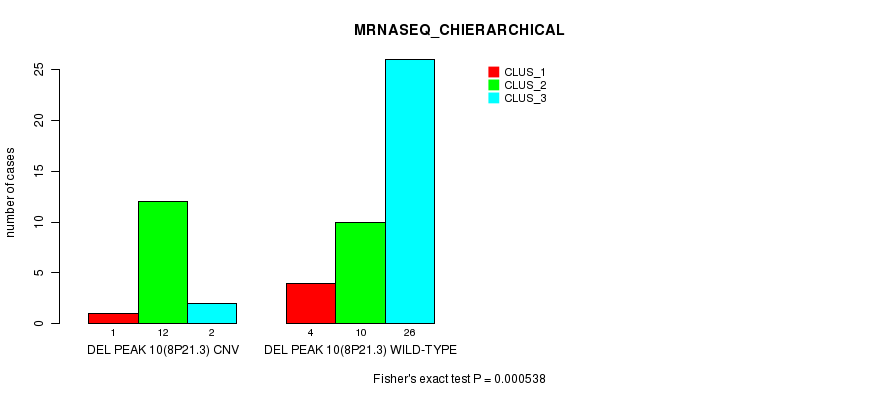
P value = 0.000789 (Fisher's exact test), Q value = 0.23
Table S35. Gene #26: 'Del Peak 10(8p21.3)' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 11 | 18 |
| DEL PEAK 10(8P21.3) CNV | 15 | 1 | 1 |
| DEL PEAK 10(8P21.3) WILD-TYPE | 14 | 10 | 17 |
Figure S35. Get High-res Image Gene #26: 'Del Peak 10(8p21.3)' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
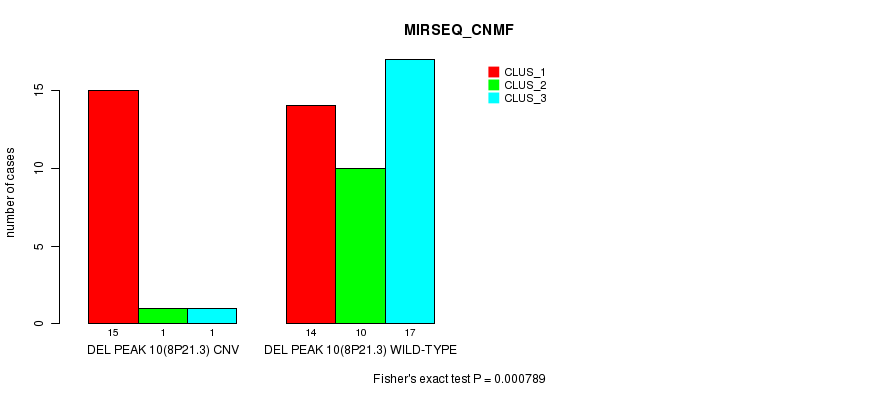
P value = 0.000163 (Fisher's exact test), Q value = 0.052
Table S36. Gene #26: 'Del Peak 10(8p21.3)' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 25 | 10 | 23 |
| DEL PEAK 10(8P21.3) CNV | 14 | 2 | 1 |
| DEL PEAK 10(8P21.3) WILD-TYPE | 11 | 8 | 22 |
Figure S36. Get High-res Image Gene #26: 'Del Peak 10(8p21.3)' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'

P value = 1.43e-06 (Chi-square test), Q value = 5e-04
Table S37. Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 12(9P21.3) CNV | 11 | 11 | 9 | 9 | 1 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 0 | 24 | 0 | 0 | 0 |
Figure S37. Get High-res Image Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #1: 'CN_CNMF'
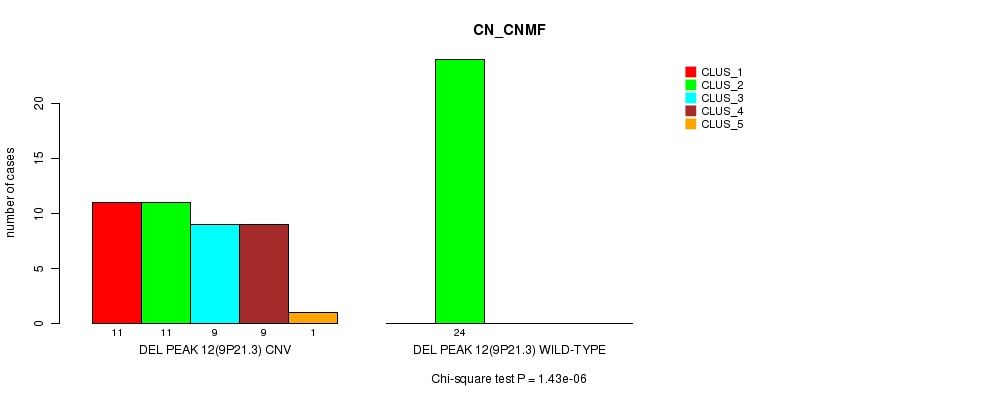
P value = 9.11e-08 (Fisher's exact test), Q value = 3.2e-05
Table S38. Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 25 | 15 |
| DEL PEAK 12(9P21.3) CNV | 24 | 7 | 9 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 0 | 18 | 6 |
Figure S38. Get High-res Image Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
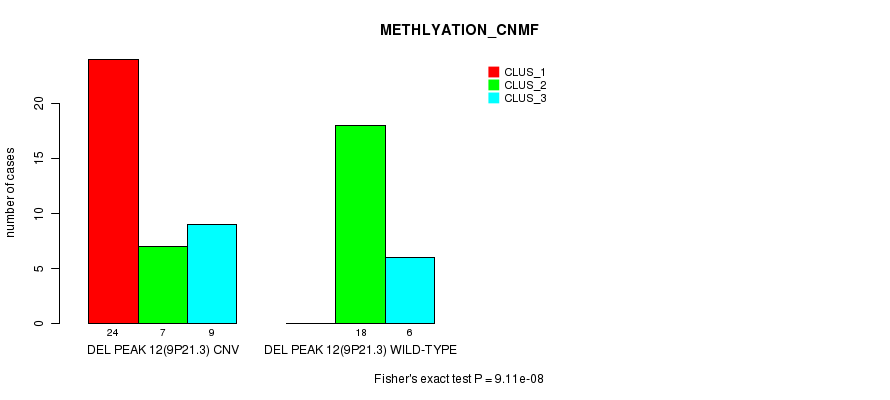
P value = 2.16e-05 (Fisher's exact test), Q value = 0.0073
Table S39. Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 21 | 12 | 12 | 10 |
| DEL PEAK 12(9P21.3) CNV | 20 | 6 | 3 | 3 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 1 | 6 | 9 | 7 |
Figure S39. Get High-res Image Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
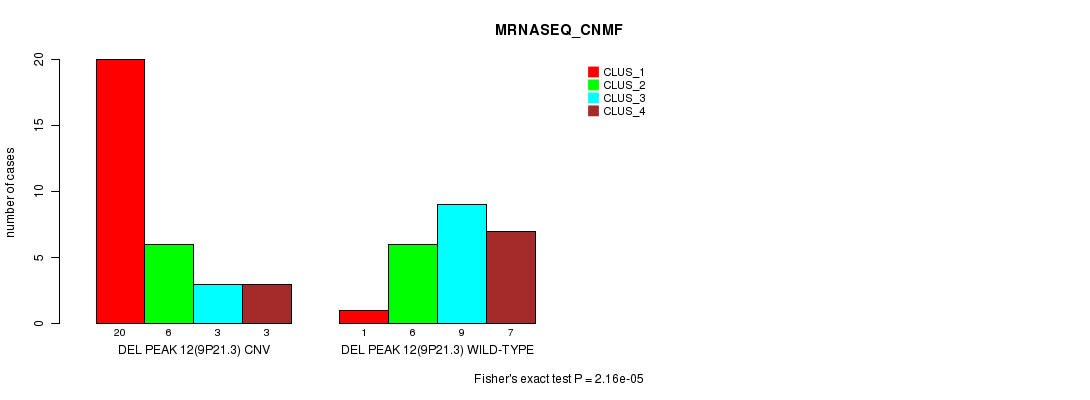
P value = 4.09e-06 (Fisher's exact test), Q value = 0.0014
Table S40. Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 5 | 22 | 28 |
| DEL PEAK 12(9P21.3) CNV | 1 | 21 | 10 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 4 | 1 | 18 |
Figure S40. Get High-res Image Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 2.95e-05 (Fisher's exact test), Q value = 0.0099
Table S41. Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 11 | 18 |
| DEL PEAK 12(9P21.3) CNV | 25 | 5 | 4 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 4 | 6 | 14 |
Figure S41. Get High-res Image Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 4.02e-05 (Fisher's exact test), Q value = 0.013
Table S42. Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 11 | 20 | 27 |
| DEL PEAK 12(9P21.3) CNV | 3 | 7 | 24 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 8 | 13 | 3 |
Figure S42. Get High-res Image Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
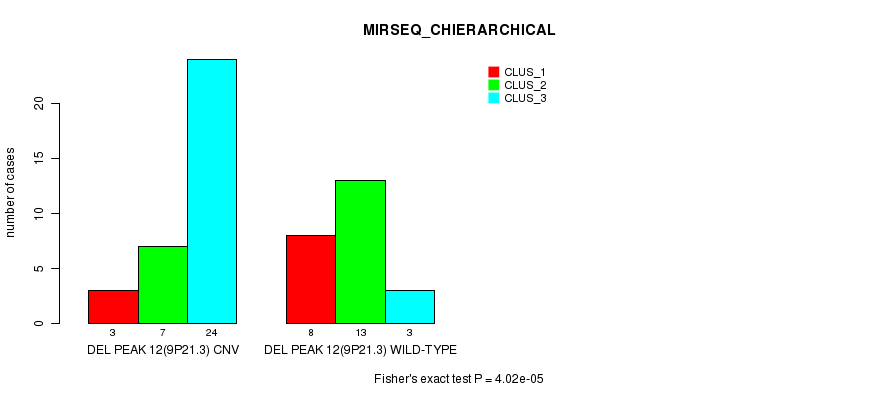
P value = 2.78e-07 (Fisher's exact test), Q value = 9.7e-05
Table S43. Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 25 | 10 | 23 |
| DEL PEAK 12(9P21.3) CNV | 24 | 4 | 6 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 1 | 6 | 17 |
Figure S43. Get High-res Image Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'

P value = 4.88e-06 (Fisher's exact test), Q value = 0.0017
Table S44. Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 12 | 26 | 20 |
| DEL PEAK 12(9P21.3) CNV | 3 | 24 | 7 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 9 | 2 | 13 |
Figure S44. Get High-res Image Gene #28: 'Del Peak 12(9p21.3)' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
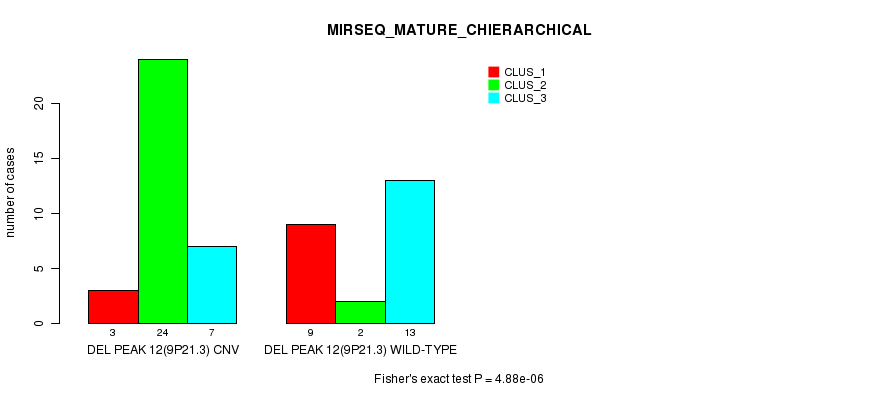
P value = 1.2e-07 (Chi-square test), Q value = 4.2e-05
Table S45. Gene #29: 'Del Peak 13(10q23.2)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 13(10Q23.2) CNV | 5 | 1 | 8 | 0 | 0 |
| DEL PEAK 13(10Q23.2) WILD-TYPE | 6 | 34 | 1 | 9 | 1 |
Figure S45. Get High-res Image Gene #29: 'Del Peak 13(10q23.2)' versus Molecular Subtype #1: 'CN_CNMF'
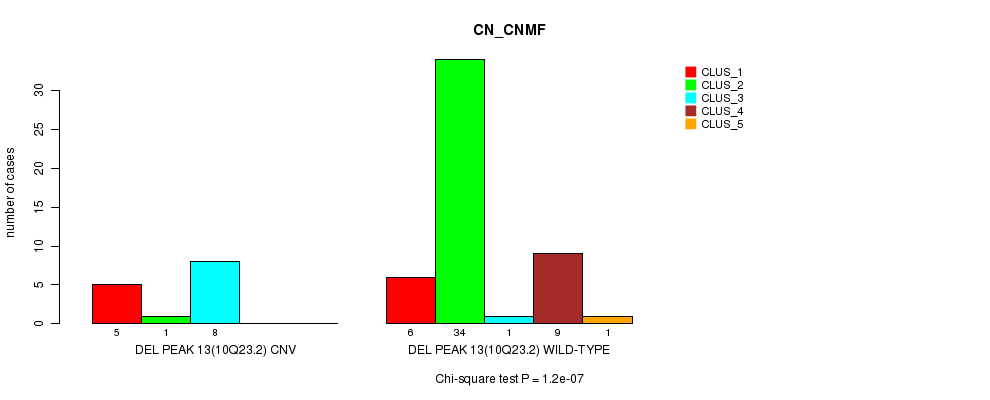
P value = 0.000408 (Chi-square test), Q value = 0.13
Table S46. Gene #31: 'Del Peak 15(12p12.3)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 15(12P12.3) CNV | 3 | 1 | 1 | 5 | 1 |
| DEL PEAK 15(12P12.3) WILD-TYPE | 8 | 34 | 8 | 4 | 0 |
Figure S46. Get High-res Image Gene #31: 'Del Peak 15(12p12.3)' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00014 (Chi-square test), Q value = 0.045
Table S47. Gene #32: 'Del Peak 16(12q12)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 16(12Q12) CNV | 2 | 1 | 6 | 1 | 0 |
| DEL PEAK 16(12Q12) WILD-TYPE | 9 | 34 | 3 | 8 | 1 |
Figure S47. Get High-res Image Gene #32: 'Del Peak 16(12q12)' versus Molecular Subtype #1: 'CN_CNMF'
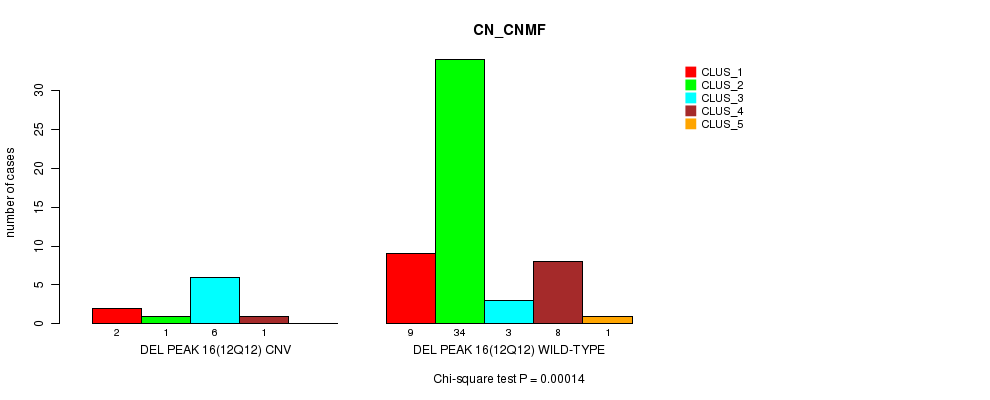
P value = 3.91e-05 (Fisher's exact test), Q value = 0.013
Table S48. Gene #32: 'Del Peak 16(12q12)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 25 | 15 |
| DEL PEAK 16(12Q12) CNV | 10 | 0 | 0 |
| DEL PEAK 16(12Q12) WILD-TYPE | 14 | 25 | 15 |
Figure S48. Get High-res Image Gene #32: 'Del Peak 16(12q12)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
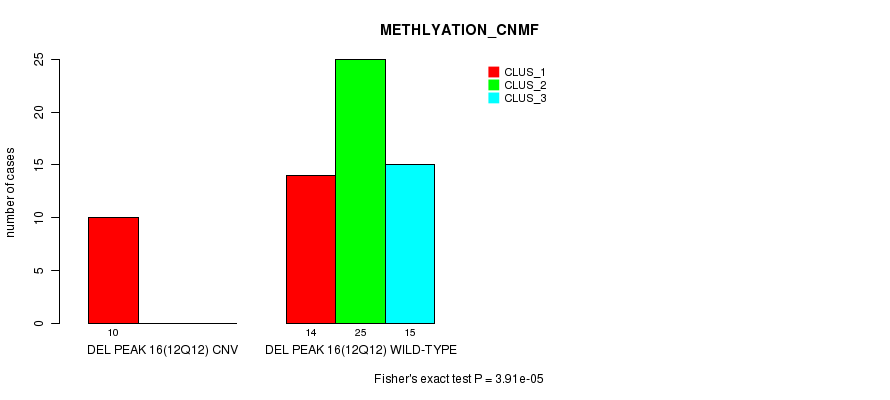
P value = 9.01e-05 (Fisher's exact test), Q value = 0.029
Table S49. Gene #33: 'Del Peak 17(12q24.31)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 25 | 15 |
| DEL PEAK 17(12Q24.31) CNV | 13 | 1 | 2 |
| DEL PEAK 17(12Q24.31) WILD-TYPE | 11 | 24 | 13 |
Figure S49. Get High-res Image Gene #33: 'Del Peak 17(12q24.31)' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.000169 (Fisher's exact test), Q value = 0.054
Table S50. Gene #33: 'Del Peak 17(12q24.31)' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 5 | 22 | 28 |
| DEL PEAK 17(12Q24.31) CNV | 0 | 11 | 1 |
| DEL PEAK 17(12Q24.31) WILD-TYPE | 5 | 11 | 27 |
Figure S50. Get High-res Image Gene #33: 'Del Peak 17(12q24.31)' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
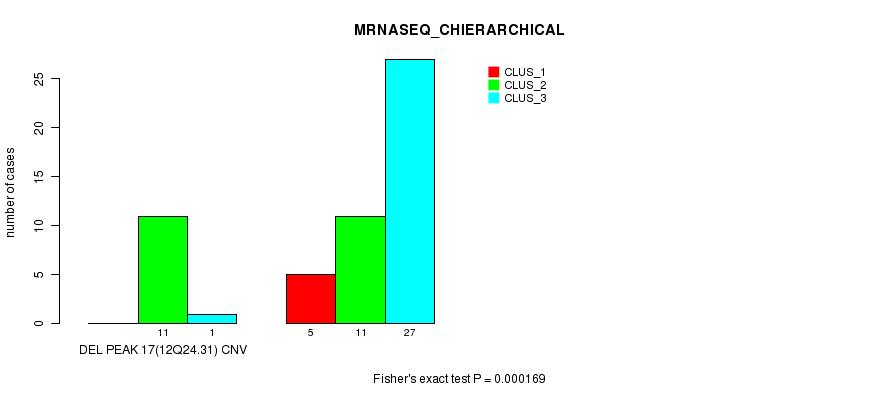
P value = 0.000454 (Fisher's exact test), Q value = 0.14
Table S51. Gene #33: 'Del Peak 17(12q24.31)' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 25 | 10 | 23 |
| DEL PEAK 17(12Q24.31) CNV | 11 | 2 | 0 |
| DEL PEAK 17(12Q24.31) WILD-TYPE | 14 | 8 | 23 |
Figure S51. Get High-res Image Gene #33: 'Del Peak 17(12q24.31)' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
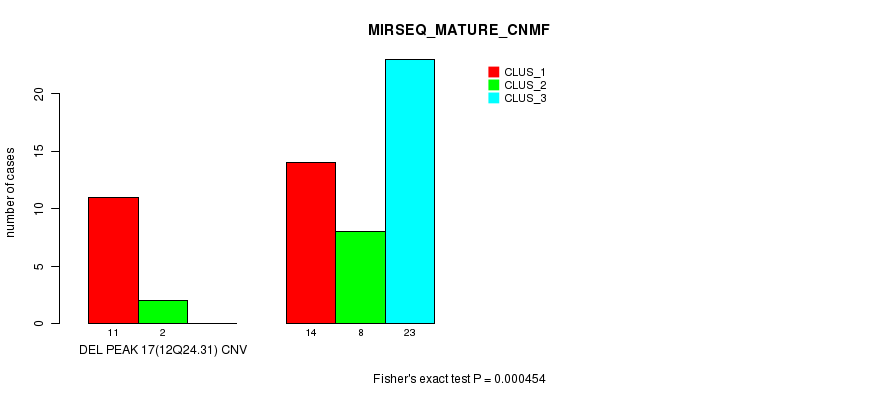
P value = 1.29e-06 (Chi-square test), Q value = 0.00045
Table S52. Gene #34: 'Del Peak 18(15q15.1)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 18(15Q15.1) CNV | 7 | 0 | 6 | 1 | 0 |
| DEL PEAK 18(15Q15.1) WILD-TYPE | 4 | 35 | 3 | 8 | 1 |
Figure S52. Get High-res Image Gene #34: 'Del Peak 18(15q15.1)' versus Molecular Subtype #1: 'CN_CNMF'
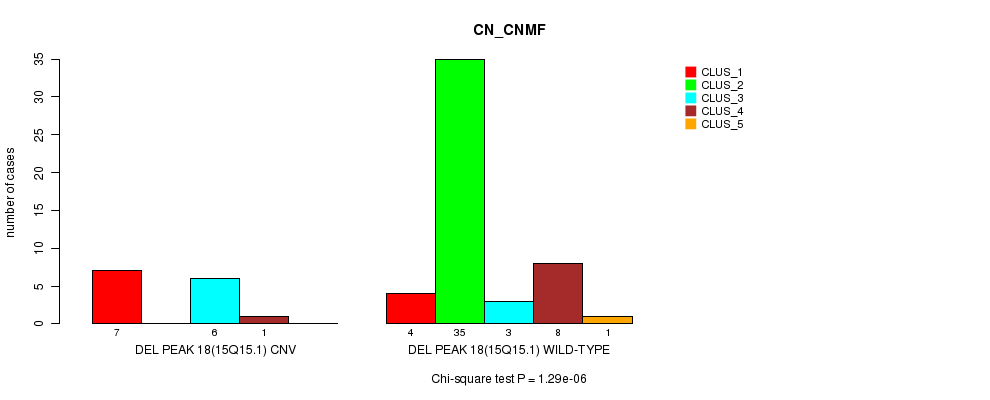
P value = 5.43e-05 (Chi-square test), Q value = 0.018
Table S53. Gene #35: 'Del Peak 19(17p11.2)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 19(17P11.2) CNV | 10 | 5 | 5 | 5 | 1 |
| DEL PEAK 19(17P11.2) WILD-TYPE | 1 | 30 | 4 | 4 | 0 |
Figure S53. Get High-res Image Gene #35: 'Del Peak 19(17p11.2)' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1.13e-08 (Fisher's exact test), Q value = 4e-06
Table S54. Gene #35: 'Del Peak 19(17p11.2)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 25 | 15 |
| DEL PEAK 19(17P11.2) CNV | 18 | 0 | 8 |
| DEL PEAK 19(17P11.2) WILD-TYPE | 6 | 25 | 7 |
Figure S54. Get High-res Image Gene #35: 'Del Peak 19(17p11.2)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
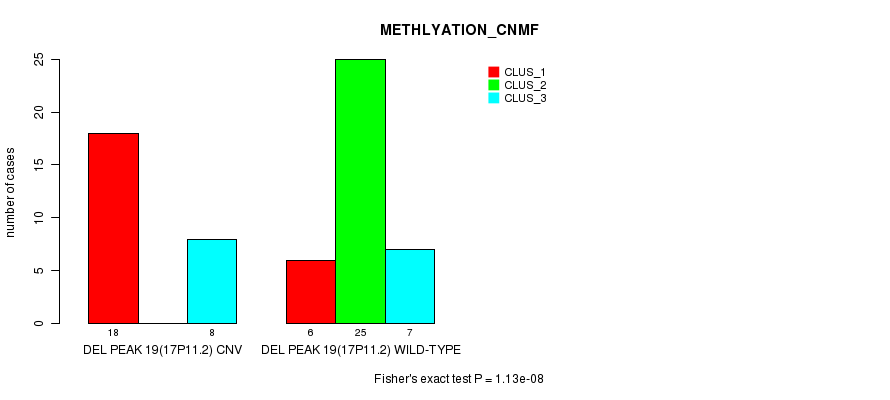
P value = 0.000809 (Fisher's exact test), Q value = 0.24
Table S55. Gene #35: 'Del Peak 19(17p11.2)' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 21 | 12 | 12 | 10 |
| DEL PEAK 19(17P11.2) CNV | 15 | 3 | 2 | 1 |
| DEL PEAK 19(17P11.2) WILD-TYPE | 6 | 9 | 10 | 9 |
Figure S55. Get High-res Image Gene #35: 'Del Peak 19(17p11.2)' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 0.000824 (Fisher's exact test), Q value = 0.24
Table S56. Gene #35: 'Del Peak 19(17p11.2)' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 5 | 22 | 28 |
| DEL PEAK 19(17P11.2) CNV | 1 | 15 | 5 |
| DEL PEAK 19(17P11.2) WILD-TYPE | 4 | 7 | 23 |
Figure S56. Get High-res Image Gene #35: 'Del Peak 19(17p11.2)' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 0.000332 (Fisher's exact test), Q value = 0.1
Table S57. Gene #35: 'Del Peak 19(17p11.2)' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 11 | 20 | 27 |
| DEL PEAK 19(17P11.2) CNV | 1 | 4 | 18 |
| DEL PEAK 19(17P11.2) WILD-TYPE | 10 | 16 | 9 |
Figure S57. Get High-res Image Gene #35: 'Del Peak 19(17p11.2)' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 0.000246 (Fisher's exact test), Q value = 0.077
Table S58. Gene #35: 'Del Peak 19(17p11.2)' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 25 | 10 | 23 |
| DEL PEAK 19(17P11.2) CNV | 17 | 3 | 3 |
| DEL PEAK 19(17P11.2) WILD-TYPE | 8 | 7 | 20 |
Figure S58. Get High-res Image Gene #35: 'Del Peak 19(17p11.2)' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'

P value = 9.68e-06 (Chi-square test), Q value = 0.0033
Table S59. Gene #36: 'Del Peak 20(17q22)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 20(17Q22) CNV | 9 | 2 | 2 | 4 | 1 |
| DEL PEAK 20(17Q22) WILD-TYPE | 2 | 33 | 7 | 5 | 0 |
Figure S59. Get High-res Image Gene #36: 'Del Peak 20(17q22)' versus Molecular Subtype #1: 'CN_CNMF'
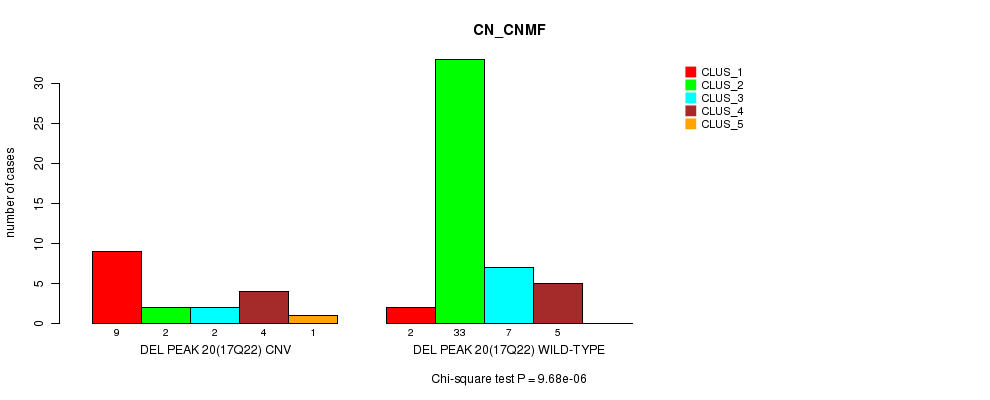
P value = 3.54e-05 (Fisher's exact test), Q value = 0.012
Table S60. Gene #36: 'Del Peak 20(17q22)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 25 | 15 |
| DEL PEAK 20(17Q22) CNV | 12 | 0 | 6 |
| DEL PEAK 20(17Q22) WILD-TYPE | 12 | 25 | 9 |
Figure S60. Get High-res Image Gene #36: 'Del Peak 20(17q22)' versus Molecular Subtype #2: 'METHLYATION_CNMF'
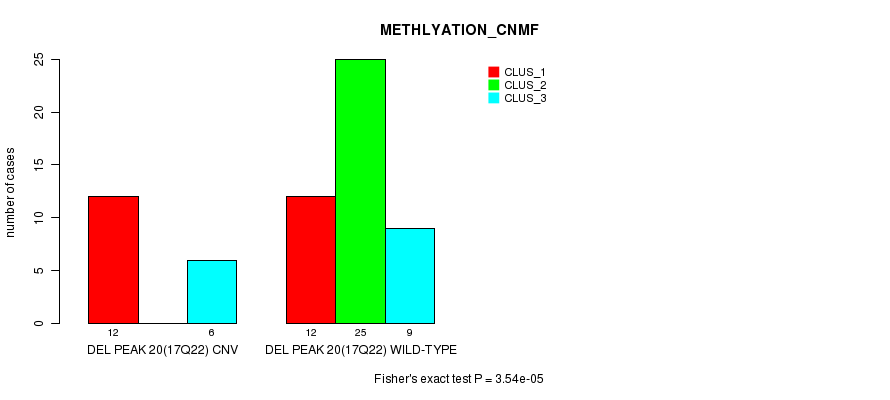
P value = 0.00041 (Chi-square test), Q value = 0.13
Table S61. Gene #44: 'Del Peak 28(Xq21.1)' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 11 | 35 | 9 | 9 | 1 |
| DEL PEAK 28(XQ21.1) CNV | 5 | 0 | 2 | 0 | 0 |
| DEL PEAK 28(XQ21.1) WILD-TYPE | 6 | 35 | 7 | 9 | 1 |
Figure S61. Get High-res Image Gene #44: 'Del Peak 28(Xq21.1)' versus Molecular Subtype #1: 'CN_CNMF'

-
Copy number data file = All Lesions File (all_lesions.conf_##.txt, where ## is the confidence level). The all lesions file is from GISTIC pipeline and summarizes the results from the GISTIC run. It contains data about the significant regions of amplification and deletion as well as which samples are amplified or deleted in each of these regions. The identified regions are listed down the first column, and the samples are listed across the first row, starting in column 10.
-
Molecular subtype file = PAAD-TP.transferedmergedcluster.txt
-
Number of patients = 65
-
Number of copy number variation regions = 44
-
Number of molecular subtypes = 8
-
Exclude regions that fewer than K tumors have alterations, K = 3
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.