This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and subtypes.
Testing the association between copy number variation 78 arm-level results and 10 molecular subtypes across 305 patients, 112 significant findings detected with Q value < 0.25.
-
1q gain cnv correlated to 'CN_CNMF' and 'MRNASEQ_CNMF'.
-
2p gain cnv correlated to 'CN_CNMF' and 'MIRSEQ_MATURE_CNMF'.
-
2q gain cnv correlated to 'CN_CNMF'.
-
5p gain cnv correlated to 'CN_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
6p gain cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
6q gain cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
7p gain cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
7q gain cnv correlated to 'CN_CNMF'.
-
8p gain cnv correlated to 'MIRSEQ_MATURE_CNMF'.
-
8q gain cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MIRSEQ_CNMF', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
10p gain cnv correlated to 'CN_CNMF'.
-
10q gain cnv correlated to 'CN_CNMF'.
-
13q gain cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
18p gain cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
20p gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
20q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
1p loss cnv correlated to 'CN_CNMF'.
-
3p loss cnv correlated to 'CN_CNMF'.
-
3q loss cnv correlated to 'CN_CNMF'.
-
4p loss cnv correlated to 'CN_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
4q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
5p loss cnv correlated to 'CN_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
5q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
7q loss cnv correlated to 'CN_CNMF'.
-
8p loss cnv correlated to 'CN_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_MATURE_CNMF'.
-
9p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CNMF'.
-
9q loss cnv correlated to 'CN_CNMF'.
-
10q loss cnv correlated to 'CN_CNMF'.
-
11p loss cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
11q loss cnv correlated to 'CN_CNMF'.
-
12p loss cnv correlated to 'CN_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
14q loss cnv correlated to 'CN_CNMF'.
-
15q loss cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
16p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_MATURE_CNMF'.
-
16q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
17p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CNMF'.
-
17q loss cnv correlated to 'CN_CNMF'.
-
18q loss cnv correlated to 'CN_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
19p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
19q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
20p loss cnv correlated to 'CN_CNMF'.
-
21q loss cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
22q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', and 'MRNASEQ_CNMF'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 78 arm-level results and 10 molecular subtypes. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 112 significant findings detected.
|
Molecular subtypes |
CN CNMF |
METHLYATION CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Chi-square test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 20p gain | 0 (0%) | 177 |
7.59e-18 (5.85e-15) |
5.66e-09 (4.25e-06) |
0.0126 (1.00) |
0.115 (1.00) |
7.19e-07 (0.000531) |
2.41e-06 (0.00176) |
2.16e-05 (0.0153) |
5.86e-07 (0.000434) |
0.00112 (0.711) |
3.99e-06 (0.00289) |
| 20q gain | 0 (0%) | 148 |
3.08e-26 (2.38e-23) |
7.34e-14 (5.61e-11) |
0.00347 (1.00) |
0.252 (1.00) |
1.83e-10 (1.38e-07) |
7.23e-10 (5.45e-07) |
3.03e-07 (0.000225) |
5.79e-12 (4.4e-09) |
0.00282 (1.00) |
1.51e-10 (1.14e-07) |
| 5q loss | 0 (0%) | 270 |
2.21e-13 (1.68e-10) |
2.47e-06 (0.0018) |
0.671 (1.00) |
0.818 (1.00) |
4.12e-06 (0.00299) |
5.94e-06 (0.00429) |
0.00607 (1.00) |
0.000273 (0.183) |
0.0117 (1.00) |
8.44e-05 (0.0575) |
| 16p loss | 0 (0%) | 276 |
5.37e-13 (4.08e-10) |
1.89e-05 (0.0135) |
0.336 (1.00) |
0.181 (1.00) |
2.39e-06 (0.00175) |
5.43e-06 (0.00392) |
1.31e-05 (0.00937) |
0.00949 (1.00) |
5.43e-05 (0.0376) |
0.0148 (1.00) |
| 19p loss | 0 (0%) | 272 |
2.51e-13 (1.91e-10) |
4.4e-06 (0.00318) |
0.0262 (1.00) |
0.305 (1.00) |
3.15e-09 (2.37e-06) |
1.24e-05 (0.00891) |
0.0202 (1.00) |
1.62e-06 (0.00119) |
0.0116 (1.00) |
7.33e-05 (0.0502) |
| 8q gain | 0 (0%) | 183 |
0.231 (1.00) |
2.44e-07 (0.000181) |
0.0175 (1.00) |
0.0506 (1.00) |
0.000343 (0.227) |
0.0733 (1.00) |
0.000171 (0.115) |
0.00361 (1.00) |
4.46e-05 (0.031) |
1.89e-05 (0.0134) |
| 4q loss | 0 (0%) | 254 |
2.28e-14 (1.74e-11) |
4.26e-05 (0.0297) |
0.0365 (1.00) |
0.197 (1.00) |
0.000242 (0.162) |
0.000226 (0.152) |
0.00142 (0.895) |
6.4e-05 (0.044) |
0.00496 (1.00) |
0.0029 (1.00) |
| 4p loss | 0 (0%) | 250 |
2.31e-11 (1.75e-08) |
0.000389 (0.257) |
0.249 (1.00) |
0.199 (1.00) |
8.51e-05 (0.0579) |
6.56e-05 (0.045) |
0.00462 (1.00) |
9.16e-05 (0.0622) |
0.00597 (1.00) |
0.000922 (0.591) |
| 16q loss | 0 (0%) | 273 |
4.29e-10 (3.24e-07) |
3.51e-06 (0.00255) |
0.119 (1.00) |
0.00647 (1.00) |
5.92e-05 (0.0409) |
6.15e-05 (0.0424) |
0.000978 (0.624) |
0.0863 (1.00) |
0.00352 (1.00) |
0.177 (1.00) |
| 19q loss | 0 (0%) | 282 |
1.33e-08 (9.97e-06) |
1.26e-05 (0.00899) |
0.0555 (1.00) |
0.436 (1.00) |
4.22e-07 (0.000313) |
0.00467 (1.00) |
0.0649 (1.00) |
1.25e-05 (0.00892) |
0.0923 (1.00) |
0.000574 (0.373) |
| 22q loss | 0 (0%) | 265 |
6.63e-16 (5.1e-13) |
1.9e-06 (0.00139) |
2.34e-05 (0.0165) |
0.16 (1.00) |
5.87e-05 (0.0406) |
0.00625 (1.00) |
0.00163 (1.00) |
0.00105 (0.667) |
0.011 (1.00) |
0.00532 (1.00) |
| 5p gain | 0 (0%) | 262 |
3.19e-06 (0.00232) |
0.00218 (1.00) |
0.338 (1.00) |
0.155 (1.00) |
6.25e-05 (0.043) |
0.000367 (0.242) |
0.00134 (0.845) |
0.0124 (1.00) |
0.00213 (1.00) |
0.00255 (1.00) |
| 8p loss | 0 (0%) | 275 |
2.68e-06 (0.00195) |
0.0321 (1.00) |
0.758 (1.00) |
0.0644 (1.00) |
0.0122 (1.00) |
0.0209 (1.00) |
0.000323 (0.215) |
0.0215 (1.00) |
9.73e-05 (0.0658) |
0.329 (1.00) |
| 9p loss | 0 (0%) | 254 |
1.64e-13 (1.25e-10) |
2.64e-05 (0.0186) |
0.015 (1.00) |
0.529 (1.00) |
0.000171 (0.115) |
0.0015 (0.938) |
0.0501 (1.00) |
0.0205 (1.00) |
0.515 (1.00) |
0.0373 (1.00) |
| 12p loss | 0 (0%) | 284 |
2.3e-05 (0.0162) |
0.000447 (0.293) |
0.0324 (1.00) |
0.242 (1.00) |
0.000163 (0.109) |
0.000127 (0.0858) |
0.0233 (1.00) |
0.0161 (1.00) |
0.141 (1.00) |
0.00205 (1.00) |
| 17p loss | 0 (0%) | 241 |
6.43e-15 (4.93e-12) |
8.72e-07 (0.000643) |
0.671 (1.00) |
0.897 (1.00) |
0.000141 (0.0954) |
0.00103 (0.656) |
0.00872 (1.00) |
0.000645 (0.417) |
0.00174 (1.00) |
0.00299 (1.00) |
| 1q gain | 0 (0%) | 263 |
1.75e-06 (0.00128) |
0.000395 (0.26) |
0.00222 (1.00) |
0.00297 (1.00) |
8.31e-05 (0.0567) |
0.000934 (0.598) |
0.00112 (0.709) |
0.026 (1.00) |
0.0152 (1.00) |
0.00865 (1.00) |
| 2p gain | 0 (0%) | 281 |
2.12e-05 (0.0151) |
0.00148 (0.932) |
0.012 (1.00) |
0.874 (1.00) |
0.167 (1.00) |
0.116 (1.00) |
0.000635 (0.411) |
0.0631 (1.00) |
0.000278 (0.185) |
0.0127 (1.00) |
| 6p gain | 0 (0%) | 279 |
1.07e-06 (0.000788) |
6.02e-05 (0.0416) |
0.928 (1.00) |
0.743 (1.00) |
0.00286 (1.00) |
0.0235 (1.00) |
0.000417 (0.274) |
0.019 (1.00) |
0.000748 (0.482) |
0.0035 (1.00) |
| 6q gain | 0 (0%) | 283 |
1.08e-05 (0.00775) |
0.000353 (0.233) |
0.88 (1.00) |
0.866 (1.00) |
0.00588 (1.00) |
0.00253 (1.00) |
0.00244 (1.00) |
0.101 (1.00) |
0.0052 (1.00) |
0.0148 (1.00) |
| 7p gain | 0 (0%) | 210 |
3.91e-15 (3.01e-12) |
3.85e-05 (0.0269) |
0.382 (1.00) |
0.36 (1.00) |
0.00898 (1.00) |
0.0704 (1.00) |
0.188 (1.00) |
0.0653 (1.00) |
0.0066 (1.00) |
0.00339 (1.00) |
| 13q gain | 0 (0%) | 240 |
1.3e-07 (9.69e-05) |
0.000325 (0.216) |
0.266 (1.00) |
0.377 (1.00) |
0.0108 (1.00) |
0.386 (1.00) |
0.00385 (1.00) |
0.0331 (1.00) |
0.0018 (1.00) |
0.0281 (1.00) |
| 18p gain | 0 (0%) | 280 |
0.000284 (0.189) |
3.87e-05 (0.027) |
0.0855 (1.00) |
0.241 (1.00) |
0.135 (1.00) |
0.257 (1.00) |
0.548 (1.00) |
0.112 (1.00) |
0.668 (1.00) |
0.089 (1.00) |
| 5p loss | 0 (0%) | 288 |
1.74e-06 (0.00128) |
0.00392 (1.00) |
0.534 (1.00) |
0.414 (1.00) |
0.00375 (1.00) |
0.000329 (0.218) |
0.164 (1.00) |
0.0421 (1.00) |
0.13 (1.00) |
0.0153 (1.00) |
| 11p loss | 0 (0%) | 286 |
3.13e-05 (0.0219) |
5.39e-05 (0.0374) |
0.00671 (1.00) |
0.443 (1.00) |
0.208 (1.00) |
0.0226 (1.00) |
0.161 (1.00) |
0.149 (1.00) |
0.00714 (1.00) |
0.318 (1.00) |
| 15q loss | 0 (0%) | 280 |
3.08e-08 (2.3e-05) |
2.34e-05 (0.0165) |
0.0102 (1.00) |
0.0317 (1.00) |
0.00641 (1.00) |
0.00117 (0.741) |
0.00183 (1.00) |
0.000712 (0.46) |
0.0496 (1.00) |
0.0036 (1.00) |
| 18q loss | 0 (0%) | 253 |
2.54e-05 (0.0178) |
0.0206 (1.00) |
0.0365 (1.00) |
0.0939 (1.00) |
0.00255 (1.00) |
9.55e-05 (0.0648) |
0.0244 (1.00) |
0.0257 (1.00) |
0.0306 (1.00) |
0.0422 (1.00) |
| 21q loss | 0 (0%) | 244 |
1.96e-08 (1.47e-05) |
6.83e-06 (0.00492) |
0.311 (1.00) |
0.74 (1.00) |
0.109 (1.00) |
0.000966 (0.618) |
0.0319 (1.00) |
0.681 (1.00) |
0.126 (1.00) |
0.404 (1.00) |
| 2q gain | 0 (0%) | 282 |
3.73e-05 (0.0261) |
0.0108 (1.00) |
0.0379 (1.00) |
0.771 (1.00) |
0.0375 (1.00) |
0.0172 (1.00) |
0.00648 (1.00) |
0.127 (1.00) |
0.00672 (1.00) |
0.0264 (1.00) |
| 7q gain | 0 (0%) | 228 |
9.67e-15 (7.41e-12) |
0.000562 (0.366) |
0.398 (1.00) |
0.771 (1.00) |
0.16 (1.00) |
0.337 (1.00) |
0.255 (1.00) |
0.429 (1.00) |
0.261 (1.00) |
0.0539 (1.00) |
| 8p gain | 0 (0%) | 214 |
0.0394 (1.00) |
0.00044 (0.289) |
0.0527 (1.00) |
0.0487 (1.00) |
0.0501 (1.00) |
0.0386 (1.00) |
0.00762 (1.00) |
0.0496 (1.00) |
2e-05 (0.0142) |
0.00194 (1.00) |
| 10p gain | 0 (0%) | 260 |
5.75e-11 (4.36e-08) |
0.00204 (1.00) |
0.822 (1.00) |
0.765 (1.00) |
0.0529 (1.00) |
0.0134 (1.00) |
0.585 (1.00) |
0.0173 (1.00) |
0.532 (1.00) |
0.00467 (1.00) |
| 10q gain | 0 (0%) | 279 |
6.15e-07 (0.000455) |
0.0481 (1.00) |
0.718 (1.00) |
0.501 (1.00) |
0.471 (1.00) |
0.64 (1.00) |
0.661 (1.00) |
0.782 (1.00) |
0.574 (1.00) |
0.183 (1.00) |
| 1p loss | 0 (0%) | 291 |
8.32e-06 (0.00598) |
0.0945 (1.00) |
0.151 (1.00) |
0.5 (1.00) |
0.408 (1.00) |
0.158 (1.00) |
0.835 (1.00) |
0.652 (1.00) |
0.225 (1.00) |
0.794 (1.00) |
| 3p loss | 0 (0%) | 281 |
1.31e-06 (0.000966) |
0.00152 (0.95) |
0.182 (1.00) |
0.508 (1.00) |
0.000568 (0.369) |
0.0234 (1.00) |
0.00155 (0.969) |
0.112 (1.00) |
0.0346 (1.00) |
0.0211 (1.00) |
| 3q loss | 0 (0%) | 291 |
7.61e-05 (0.0521) |
0.0197 (1.00) |
0.613 (1.00) |
0.927 (1.00) |
0.000459 (0.3) |
0.0965 (1.00) |
0.000747 (0.482) |
0.284 (1.00) |
0.00978 (1.00) |
0.265 (1.00) |
| 7q loss | 0 (0%) | 292 |
2.12e-05 (0.0151) |
0.0414 (1.00) |
0.307 (1.00) |
0.479 (1.00) |
0.0156 (1.00) |
0.265 (1.00) |
0.0177 (1.00) |
0.103 (1.00) |
0.0178 (1.00) |
0.21 (1.00) |
| 9q loss | 0 (0%) | 280 |
8.9e-08 (6.64e-05) |
0.00415 (1.00) |
0.265 (1.00) |
0.0634 (1.00) |
0.0695 (1.00) |
0.042 (1.00) |
0.247 (1.00) |
0.621 (1.00) |
0.712 (1.00) |
0.122 (1.00) |
| 10q loss | 0 (0%) | 289 |
1.63e-05 (0.0116) |
0.00307 (1.00) |
0.547 (1.00) |
0.553 (1.00) |
0.012 (1.00) |
0.0536 (1.00) |
0.00374 (1.00) |
0.0639 (1.00) |
0.26 (1.00) |
0.0129 (1.00) |
| 11q loss | 0 (0%) | 289 |
3.36e-05 (0.0235) |
0.00892 (1.00) |
0.399 (1.00) |
0.811 (1.00) |
0.422 (1.00) |
0.225 (1.00) |
0.358 (1.00) |
0.579 (1.00) |
0.0127 (1.00) |
0.527 (1.00) |
| 14q loss | 0 (0%) | 277 |
7.85e-08 (5.86e-05) |
0.000905 (0.581) |
0.526 (1.00) |
0.672 (1.00) |
0.00125 (0.786) |
0.00364 (1.00) |
0.0342 (1.00) |
0.0483 (1.00) |
0.567 (1.00) |
0.0209 (1.00) |
| 17q loss | 0 (0%) | 286 |
8.04e-05 (0.0549) |
0.0245 (1.00) |
0.772 (1.00) |
0.53 (1.00) |
0.0175 (1.00) |
0.0122 (1.00) |
0.0455 (1.00) |
0.00451 (1.00) |
0.111 (1.00) |
0.156 (1.00) |
| 20p loss | 0 (0%) | 296 |
0.000145 (0.0976) |
0.017 (1.00) |
0.69 (1.00) |
0.074 (1.00) |
0.000993 (0.632) |
0.173 (1.00) |
0.438 (1.00) |
0.512 (1.00) |
0.17 (1.00) |
0.415 (1.00) |
| 1p gain | 0 (0%) | 291 |
0.0016 (0.995) |
0.507 (1.00) |
0.0327 (1.00) |
0.204 (1.00) |
0.13 (1.00) |
0.0928 (1.00) |
0.277 (1.00) |
0.0324 (1.00) |
0.149 (1.00) |
0.275 (1.00) |
| 3p gain | 0 (0%) | 292 |
1 (1.00) |
0.202 (1.00) |
0.142 (1.00) |
0.0144 (1.00) |
0.336 (1.00) |
0.362 (1.00) |
0.166 (1.00) |
0.823 (1.00) |
0.109 (1.00) |
0.512 (1.00) |
| 3q gain | 0 (0%) | 274 |
0.00815 (1.00) |
0.271 (1.00) |
1 (1.00) |
0.46 (1.00) |
0.0224 (1.00) |
0.0231 (1.00) |
0.4 (1.00) |
0.138 (1.00) |
0.0834 (1.00) |
0.117 (1.00) |
| 4p gain | 0 (0%) | 300 |
0.0117 (1.00) |
0.286 (1.00) |
0.844 (1.00) |
1 (1.00) |
0.111 (1.00) |
0.444 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.789 (1.00) |
| 5q gain | 0 (0%) | 295 |
0.0436 (1.00) |
0.95 (1.00) |
1 (1.00) |
0.13 (1.00) |
0.125 (1.00) |
0.545 (1.00) |
0.282 (1.00) |
0.921 (1.00) |
0.687 (1.00) |
0.127 (1.00) |
| 9p gain | 0 (0%) | 283 |
0.122 (1.00) |
0.139 (1.00) |
0.309 (1.00) |
0.346 (1.00) |
0.439 (1.00) |
0.309 (1.00) |
0.729 (1.00) |
0.0345 (1.00) |
0.668 (1.00) |
0.0566 (1.00) |
| 9q gain | 0 (0%) | 276 |
0.0384 (1.00) |
0.113 (1.00) |
0.117 (1.00) |
0.24 (1.00) |
0.583 (1.00) |
0.252 (1.00) |
0.97 (1.00) |
0.447 (1.00) |
0.823 (1.00) |
0.152 (1.00) |
| 11p gain | 0 (0%) | 293 |
0.784 (1.00) |
0.115 (1.00) |
0.84 (1.00) |
0.911 (1.00) |
0.537 (1.00) |
0.183 (1.00) |
0.0863 (1.00) |
0.05 (1.00) |
0.521 (1.00) |
0.0258 (1.00) |
| 11q gain | 0 (0%) | 285 |
0.186 (1.00) |
0.0338 (1.00) |
0.949 (1.00) |
0.523 (1.00) |
0.382 (1.00) |
0.0634 (1.00) |
0.0571 (1.00) |
0.0303 (1.00) |
0.0849 (1.00) |
0.00818 (1.00) |
| 12p gain | 0 (0%) | 270 |
0.00281 (1.00) |
0.178 (1.00) |
0.124 (1.00) |
0.327 (1.00) |
0.0616 (1.00) |
0.152 (1.00) |
0.00352 (1.00) |
0.453 (1.00) |
0.00192 (1.00) |
0.022 (1.00) |
| 12q gain | 0 (0%) | 277 |
0.11 (1.00) |
0.168 (1.00) |
0.336 (1.00) |
0.243 (1.00) |
0.819 (1.00) |
0.635 (1.00) |
0.334 (1.00) |
0.26 (1.00) |
0.0164 (1.00) |
0.0453 (1.00) |
| 14q gain | 0 (0%) | 300 |
0.0536 (1.00) |
0.283 (1.00) |
0.0359 (1.00) |
0.532 (1.00) |
0.28 (1.00) |
0.612 (1.00) |
0.43 (1.00) |
0.34 (1.00) |
||
| 15q gain | 0 (0%) | 295 |
0.00736 (1.00) |
0.543 (1.00) |
0.0364 (1.00) |
0.824 (1.00) |
0.0723 (1.00) |
0.00699 (1.00) |
0.724 (1.00) |
0.153 (1.00) |
0.336 (1.00) |
0.0613 (1.00) |
| 16p gain | 0 (0%) | 292 |
0.0453 (1.00) |
0.116 (1.00) |
0.782 (1.00) |
0.916 (1.00) |
0.0407 (1.00) |
0.814 (1.00) |
0.726 (1.00) |
0.0964 (1.00) |
0.113 (1.00) |
0.674 (1.00) |
| 16q gain | 0 (0%) | 293 |
0.0187 (1.00) |
0.826 (1.00) |
0.569 (1.00) |
0.278 (1.00) |
0.17 (1.00) |
0.581 (1.00) |
0.757 (1.00) |
0.121 (1.00) |
0.572 (1.00) |
1 (1.00) |
| 17p gain | 0 (0%) | 298 |
0.011 (1.00) |
0.457 (1.00) |
0.626 (1.00) |
0.135 (1.00) |
0.539 (1.00) |
1 (1.00) |
0.636 (1.00) |
0.89 (1.00) |
0.0726 (1.00) |
0.56 (1.00) |
| 17q gain | 0 (0%) | 290 |
0.00908 (1.00) |
0.664 (1.00) |
0.598 (1.00) |
0.333 (1.00) |
0.172 (1.00) |
0.674 (1.00) |
1 (1.00) |
0.439 (1.00) |
0.135 (1.00) |
0.253 (1.00) |
| 18q gain | 0 (0%) | 291 |
0.0496 (1.00) |
0.0615 (1.00) |
0.221 (1.00) |
0.0943 (1.00) |
0.316 (1.00) |
0.189 (1.00) |
0.112 (1.00) |
0.116 (1.00) |
0.0397 (1.00) |
0.478 (1.00) |
| 19p gain | 0 (0%) | 292 |
0.636 (1.00) |
0.0219 (1.00) |
0.685 (1.00) |
0.217 (1.00) |
0.857 (1.00) |
0.399 (1.00) |
0.493 (1.00) |
0.935 (1.00) |
0.495 (1.00) |
0.552 (1.00) |
| 19q gain | 0 (0%) | 273 |
0.0875 (1.00) |
0.382 (1.00) |
0.487 (1.00) |
0.268 (1.00) |
0.245 (1.00) |
0.0624 (1.00) |
0.139 (1.00) |
0.218 (1.00) |
0.249 (1.00) |
0.431 (1.00) |
| 21q gain | 0 (0%) | 302 |
0.105 (1.00) |
0.86 (1.00) |
0.529 (1.00) |
0.411 (1.00) |
0.112 (1.00) |
0.343 (1.00) |
0.311 (1.00) |
0.195 (1.00) |
||
| 22q gain | 0 (0%) | 299 |
0.0208 (1.00) |
0.181 (1.00) |
0.866 (1.00) |
0.135 (1.00) |
0.741 (1.00) |
0.35 (1.00) |
0.253 (1.00) |
0.244 (1.00) |
0.185 (1.00) |
0.308 (1.00) |
| Xq gain | 0 (0%) | 300 |
0.361 (1.00) |
0.223 (1.00) |
0.552 (1.00) |
1 (1.00) |
0.135 (1.00) |
0.265 (1.00) |
0.134 (1.00) |
0.00969 (1.00) |
0.816 (1.00) |
0.0193 (1.00) |
| 1q loss | 0 (0%) | 302 |
0.74 (1.00) |
0.86 (1.00) |
0.724 (1.00) |
0.196 (1.00) |
0.516 (1.00) |
0.26 (1.00) |
0.194 (1.00) |
0.436 (1.00) |
||
| 2p loss | 0 (0%) | 299 |
0.00486 (1.00) |
0.4 (1.00) |
0.067 (1.00) |
0.422 (1.00) |
0.108 (1.00) |
0.0273 (1.00) |
0.385 (1.00) |
0.0471 (1.00) |
0.508 (1.00) |
0.712 (1.00) |
| 2q loss | 0 (0%) | 300 |
0.0117 (1.00) |
0.68 (1.00) |
0.745 (1.00) |
0.728 (1.00) |
0.279 (1.00) |
0.0737 (1.00) |
0.631 (1.00) |
0.0791 (1.00) |
0.816 (1.00) |
0.34 (1.00) |
| 6p loss | 0 (0%) | 291 |
0.00837 (1.00) |
0.29 (1.00) |
0.269 (1.00) |
0.126 (1.00) |
0.165 (1.00) |
0.454 (1.00) |
0.835 (1.00) |
0.345 (1.00) |
0.494 (1.00) |
0.527 (1.00) |
| 6q loss | 0 (0%) | 287 |
0.00283 (1.00) |
0.107 (1.00) |
0.108 (1.00) |
0.0653 (1.00) |
0.0299 (1.00) |
0.203 (1.00) |
0.511 (1.00) |
0.711 (1.00) |
1 (1.00) |
0.885 (1.00) |
| 7p loss | 0 (0%) | 301 |
0.0273 (1.00) |
0.217 (1.00) |
0.106 (1.00) |
0.109 (1.00) |
0.444 (1.00) |
0.557 (1.00) |
0.287 (1.00) |
|||
| 8q loss | 0 (0%) | 297 |
0.00513 (1.00) |
0.268 (1.00) |
1 (1.00) |
0.497 (1.00) |
0.61 (1.00) |
0.89 (1.00) |
0.144 (1.00) |
0.0583 (1.00) |
0.185 (1.00) |
0.75 (1.00) |
| 10p loss | 0 (0%) | 289 |
0.000506 (0.33) |
0.00808 (1.00) |
0.123 (1.00) |
0.254 (1.00) |
0.0317 (1.00) |
0.0237 (1.00) |
0.02 (1.00) |
0.0169 (1.00) |
0.183 (1.00) |
0.011 (1.00) |
| 12q loss | 0 (0%) | 293 |
0.0015 (0.938) |
0.00679 (1.00) |
0.0734 (1.00) |
0.839 (1.00) |
0.0432 (1.00) |
0.225 (1.00) |
1 (1.00) |
0.154 (1.00) |
0.91 (1.00) |
0.0147 (1.00) |
| 13q loss | 0 (0%) | 294 |
0.000478 (0.312) |
0.106 (1.00) |
0.619 (1.00) |
0.638 (1.00) |
0.519 (1.00) |
0.225 (1.00) |
0.213 (1.00) |
0.794 (1.00) |
0.46 (1.00) |
0.897 (1.00) |
| 18p loss | 0 (0%) | 265 |
0.000837 (0.538) |
0.013 (1.00) |
0.244 (1.00) |
0.85 (1.00) |
0.026 (1.00) |
0.00499 (1.00) |
0.0528 (1.00) |
0.113 (1.00) |
0.00277 (1.00) |
0.144 (1.00) |
| Xq loss | 0 (0%) | 290 |
0.00391 (1.00) |
0.0701 (1.00) |
0.616 (1.00) |
0.389 (1.00) |
0.274 (1.00) |
0.116 (1.00) |
0.00731 (1.00) |
0.269 (1.00) |
0.00672 (1.00) |
0.331 (1.00) |
P value = 1.75e-06 (Fisher's exact test), Q value = 0.0013
Table S1. Gene #2: '1q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 1Q GAIN CNV | 6 | 10 | 26 |
| 1Q GAIN WILD-TYPE | 144 | 36 | 83 |
Figure S1. Get High-res Image Gene #2: '1q gain' versus Molecular Subtype #1: 'CN_CNMF'
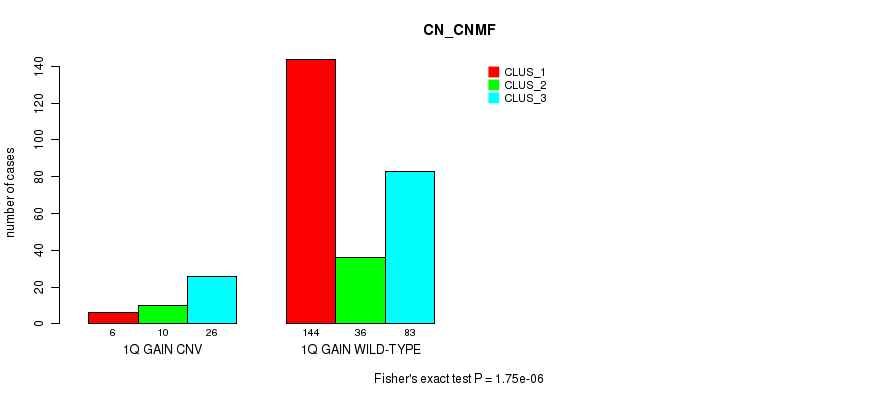
P value = 8.31e-05 (Chi-square test), Q value = 0.057
Table S2. Gene #2: '1q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 1Q GAIN CNV | 7 | 1 | 4 | 6 | 19 |
| 1Q GAIN WILD-TYPE | 54 | 62 | 39 | 36 | 43 |
Figure S2. Get High-res Image Gene #2: '1q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
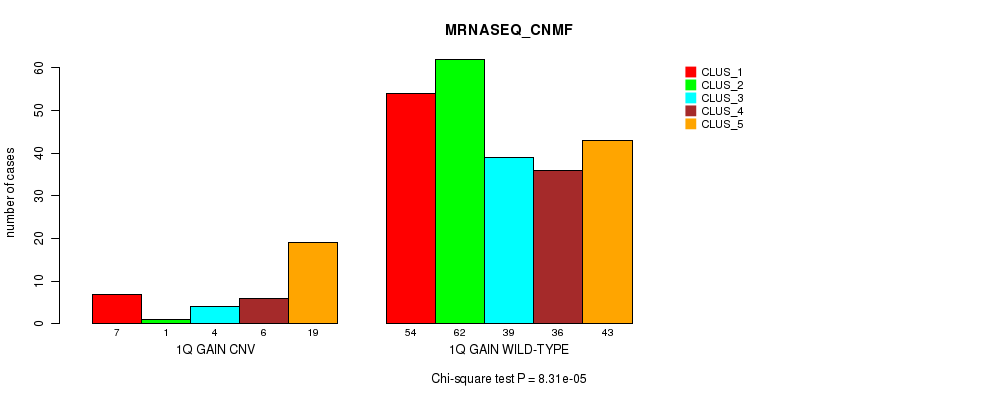
P value = 2.12e-05 (Fisher's exact test), Q value = 0.015
Table S3. Gene #3: '2p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 2P GAIN CNV | 2 | 8 | 14 |
| 2P GAIN WILD-TYPE | 148 | 38 | 95 |
Figure S3. Get High-res Image Gene #3: '2p gain' versus Molecular Subtype #1: 'CN_CNMF'
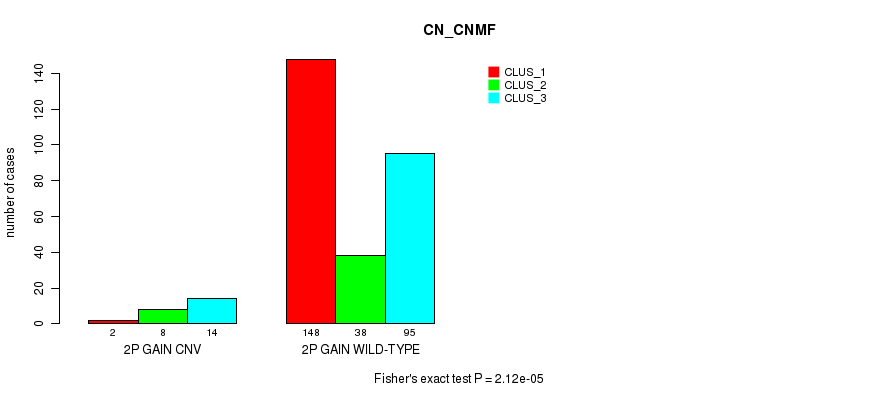
P value = 0.000278 (Fisher's exact test), Q value = 0.19
Table S4. Gene #3: '2p gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 54 | 78 | 127 |
| 2P GAIN CNV | 4 | 0 | 18 |
| 2P GAIN WILD-TYPE | 50 | 78 | 109 |
Figure S4. Get High-res Image Gene #3: '2p gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
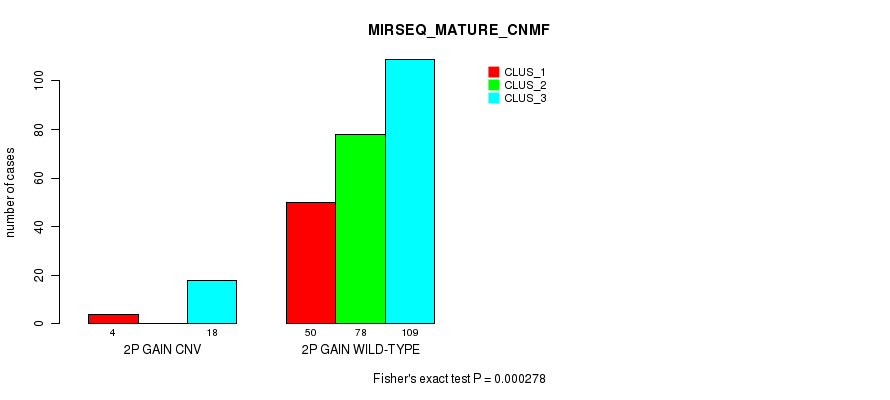
P value = 3.73e-05 (Fisher's exact test), Q value = 0.026
Table S5. Gene #4: '2q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 2Q GAIN CNV | 2 | 8 | 13 |
| 2Q GAIN WILD-TYPE | 148 | 38 | 96 |
Figure S5. Get High-res Image Gene #4: '2q gain' versus Molecular Subtype #1: 'CN_CNMF'
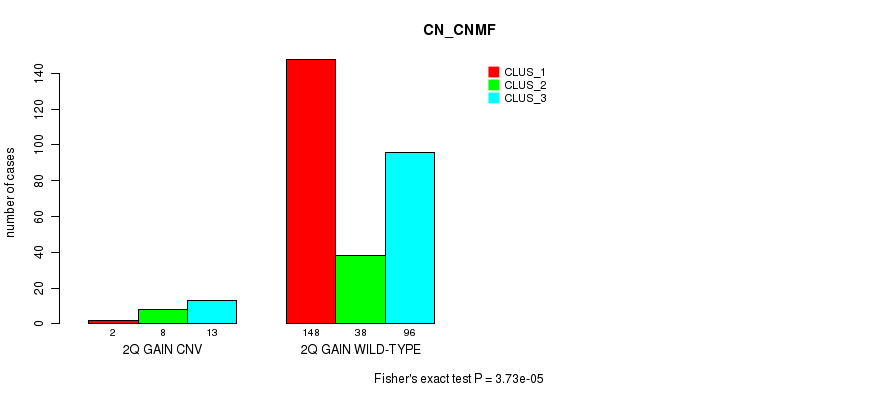
P value = 3.19e-06 (Fisher's exact test), Q value = 0.0023
Table S6. Gene #8: '5p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 5P GAIN CNV | 7 | 8 | 28 |
| 5P GAIN WILD-TYPE | 143 | 38 | 81 |
Figure S6. Get High-res Image Gene #8: '5p gain' versus Molecular Subtype #1: 'CN_CNMF'
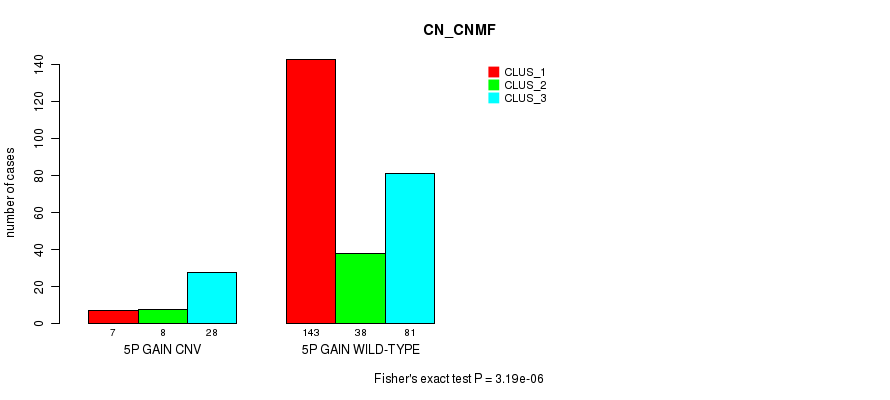
P value = 6.25e-05 (Chi-square test), Q value = 0.043
Table S7. Gene #8: '5p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 5P GAIN CNV | 7 | 2 | 4 | 5 | 20 |
| 5P GAIN WILD-TYPE | 54 | 61 | 39 | 37 | 42 |
Figure S7. Get High-res Image Gene #8: '5p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
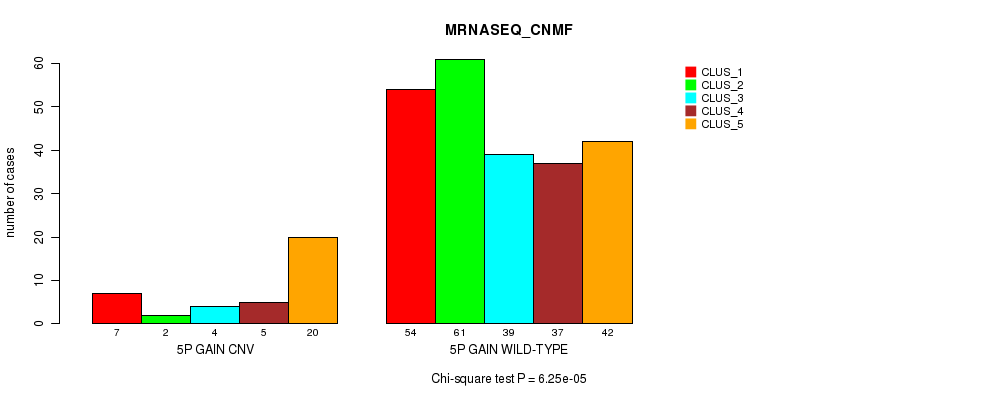
P value = 0.000367 (Fisher's exact test), Q value = 0.24
Table S8. Gene #8: '5p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 141 | 64 |
| 5P GAIN CNV | 2 | 19 | 17 |
| 5P GAIN WILD-TYPE | 64 | 122 | 47 |
Figure S8. Get High-res Image Gene #8: '5p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
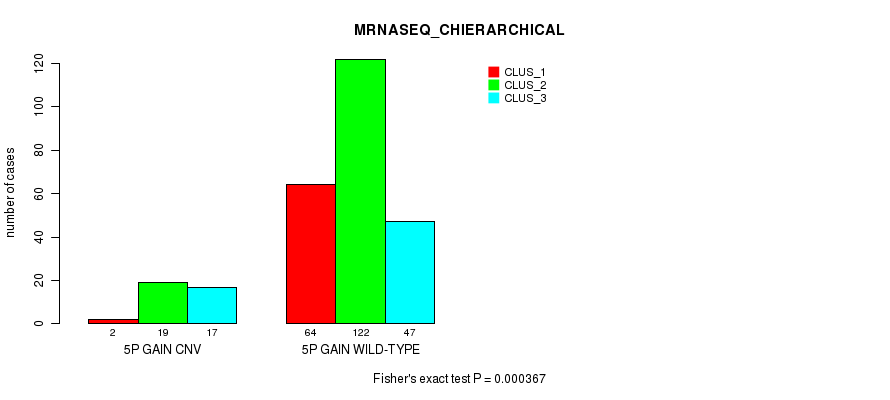
P value = 1.07e-06 (Fisher's exact test), Q value = 0.00079
Table S9. Gene #10: '6p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 6P GAIN CNV | 2 | 3 | 21 |
| 6P GAIN WILD-TYPE | 148 | 43 | 88 |
Figure S9. Get High-res Image Gene #10: '6p gain' versus Molecular Subtype #1: 'CN_CNMF'
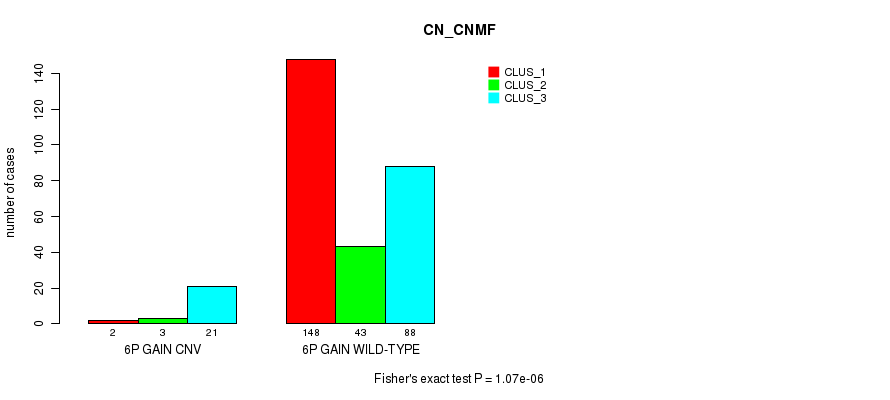
P value = 6.02e-05 (Fisher's exact test), Q value = 0.042
Table S10. Gene #10: '6p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 6P GAIN CNV | 0 | 7 | 0 | 14 |
| 6P GAIN WILD-TYPE | 37 | 73 | 65 | 62 |
Figure S10. Get High-res Image Gene #10: '6p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
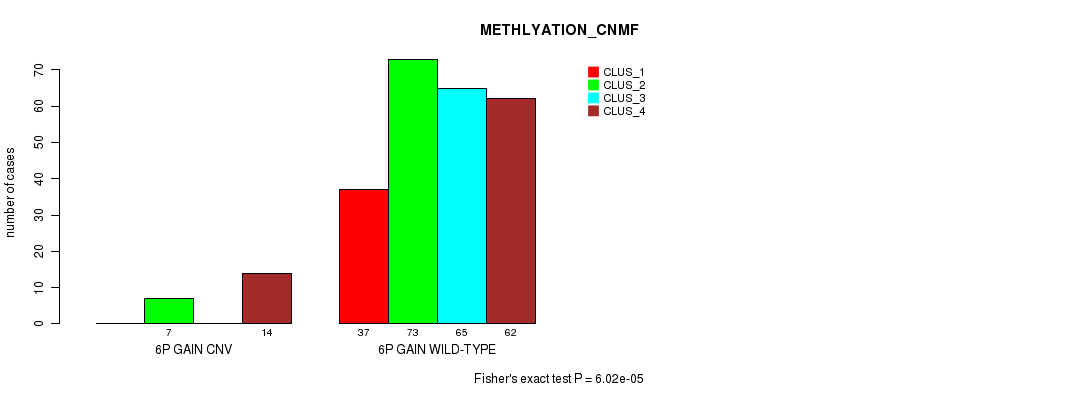
P value = 1.08e-05 (Fisher's exact test), Q value = 0.0077
Table S11. Gene #11: '6q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 6Q GAIN CNV | 2 | 2 | 18 |
| 6Q GAIN WILD-TYPE | 148 | 44 | 91 |
Figure S11. Get High-res Image Gene #11: '6q gain' versus Molecular Subtype #1: 'CN_CNMF'
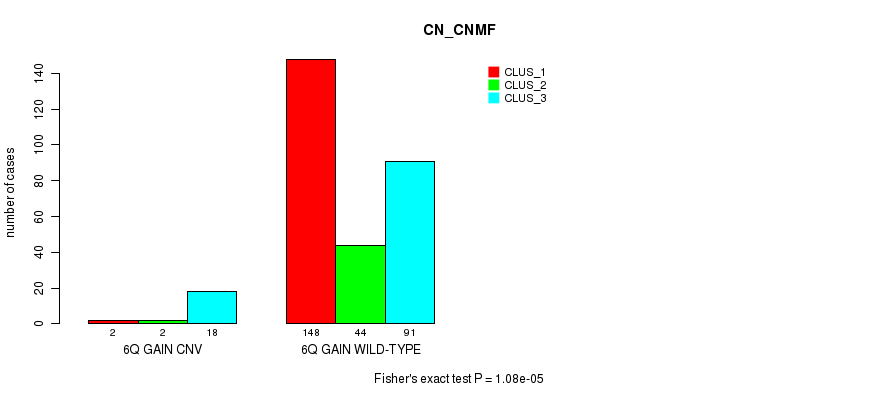
P value = 0.000353 (Fisher's exact test), Q value = 0.23
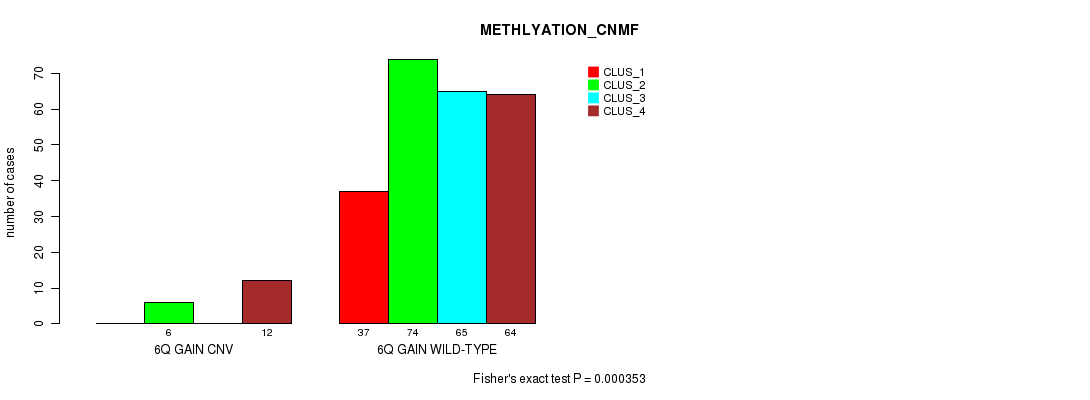
Table S12. Gene #11: '6q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 6Q GAIN CNV | 0 | 6 | 0 | 12 |
| 6Q GAIN WILD-TYPE | 37 | 74 | 65 | 64 |
Figure S12. Get High-res Image Gene #11: '6q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
P value = 3.91e-15 (Fisher's exact test), Q value = 3e-12
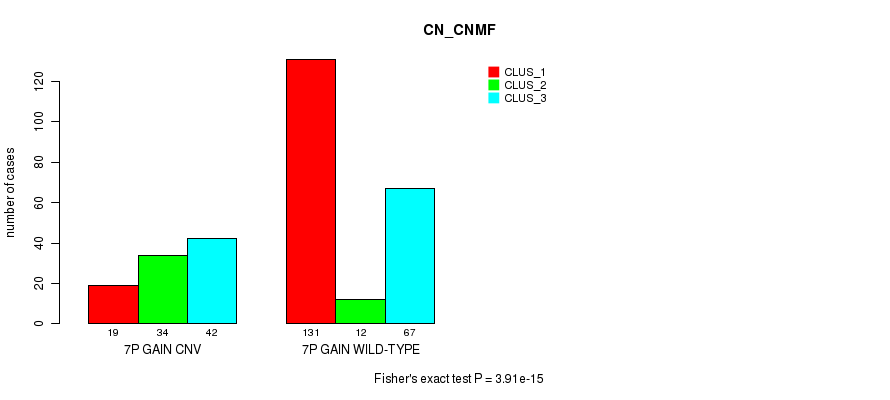
Table S13. Gene #12: '7p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 7P GAIN CNV | 19 | 34 | 42 |
| 7P GAIN WILD-TYPE | 131 | 12 | 67 |
Figure S13. Get High-res Image Gene #12: '7p gain' versus Molecular Subtype #1: 'CN_CNMF'
P value = 3.85e-05 (Fisher's exact test), Q value = 0.027
Table S14. Gene #12: '7p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 7P GAIN CNV | 11 | 30 | 6 | 32 |
| 7P GAIN WILD-TYPE | 26 | 50 | 59 | 44 |
Figure S14. Get High-res Image Gene #12: '7p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
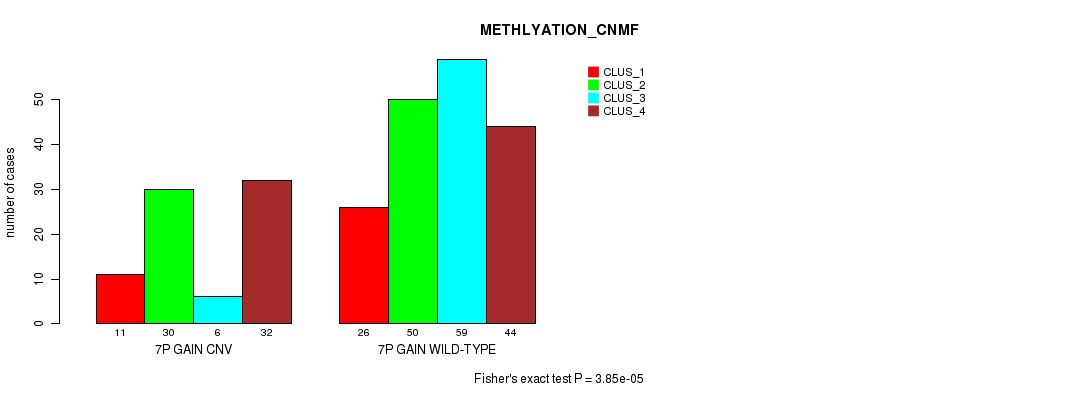
P value = 9.67e-15 (Fisher's exact test), Q value = 7.4e-12
Table S15. Gene #13: '7q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 7Q GAIN CNV | 18 | 34 | 25 |
| 7Q GAIN WILD-TYPE | 132 | 12 | 84 |
Figure S15. Get High-res Image Gene #13: '7q gain' versus Molecular Subtype #1: 'CN_CNMF'
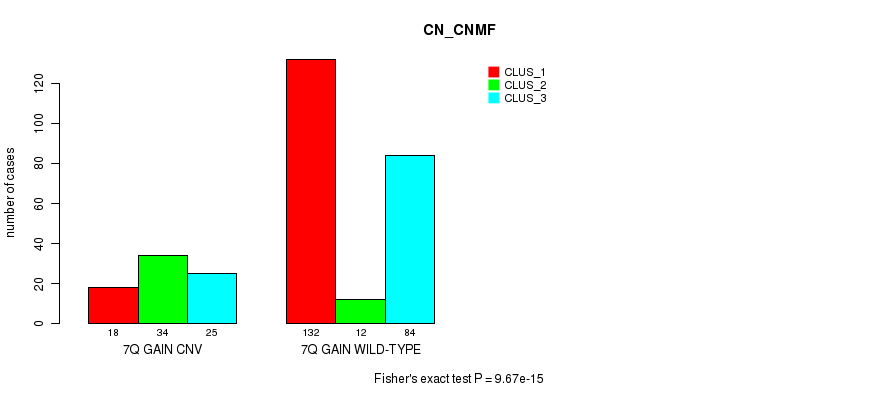
P value = 2e-05 (Fisher's exact test), Q value = 0.014
Table S16. Gene #14: '8p gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 54 | 78 | 127 |
| 8P GAIN CNV | 20 | 7 | 44 |
| 8P GAIN WILD-TYPE | 34 | 71 | 83 |
Figure S16. Get High-res Image Gene #14: '8p gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
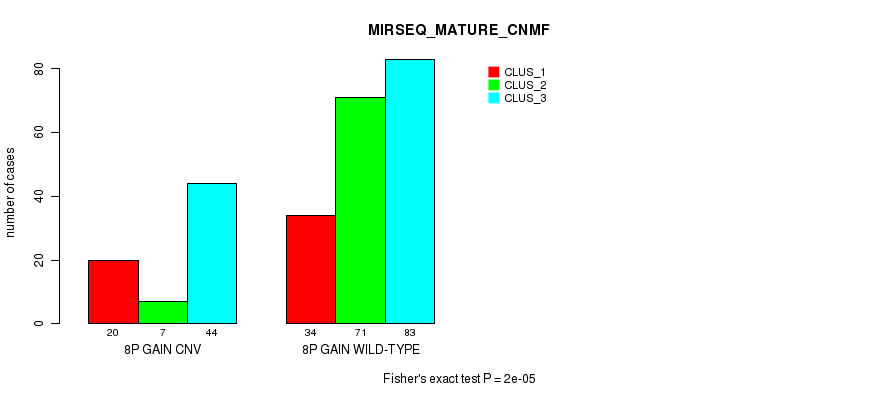
P value = 2.44e-07 (Fisher's exact test), Q value = 0.00018
Table S17. Gene #15: '8q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 8Q GAIN CNV | 13 | 39 | 7 | 39 |
| 8Q GAIN WILD-TYPE | 24 | 41 | 58 | 37 |
Figure S17. Get High-res Image Gene #15: '8q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
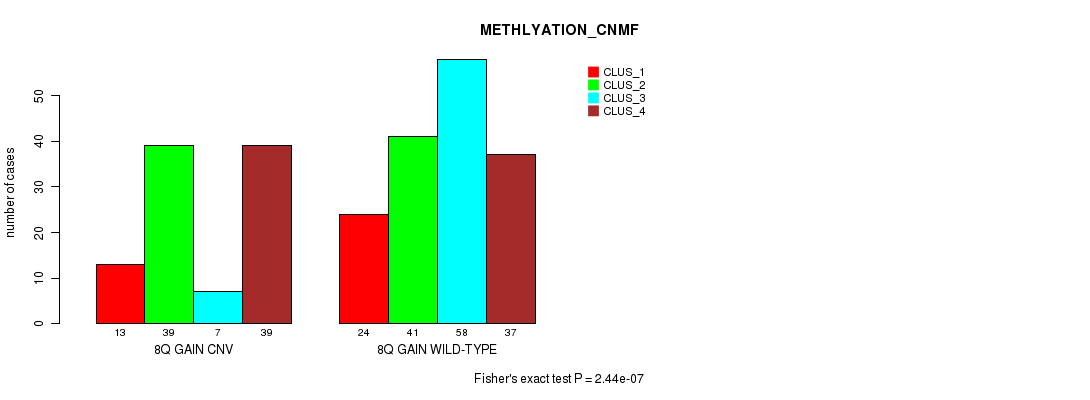
P value = 0.000343 (Chi-square test), Q value = 0.23
Table S18. Gene #15: '8q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 8Q GAIN CNV | 24 | 14 | 11 | 21 | 35 |
| 8Q GAIN WILD-TYPE | 37 | 49 | 32 | 21 | 27 |
Figure S18. Get High-res Image Gene #15: '8q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
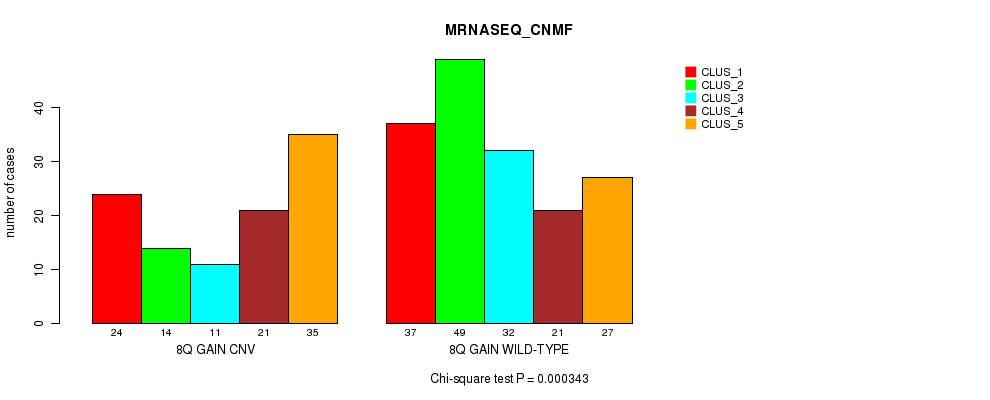
P value = 0.000171 (Fisher's exact test), Q value = 0.12
Table S19. Gene #15: '8q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 91 | 114 |
| 8Q GAIN CNV | 43 | 21 | 58 |
| 8Q GAIN WILD-TYPE | 57 | 70 | 56 |
Figure S19. Get High-res Image Gene #15: '8q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
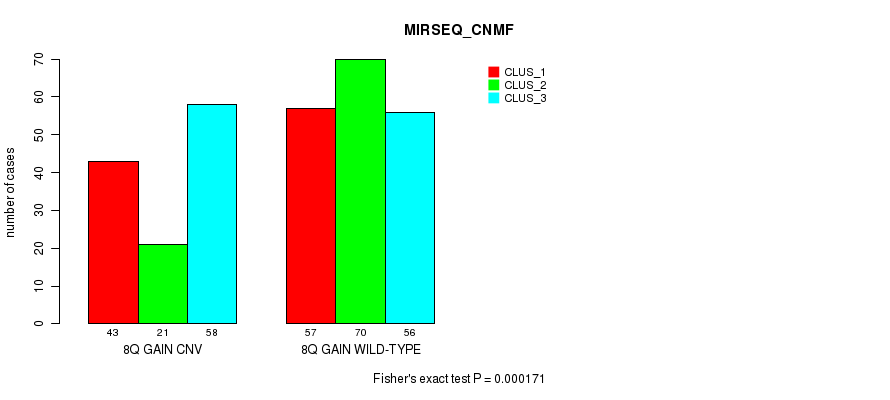
P value = 4.46e-05 (Fisher's exact test), Q value = 0.031
Table S20. Gene #15: '8q gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 54 | 78 | 127 |
| 8Q GAIN CNV | 24 | 14 | 60 |
| 8Q GAIN WILD-TYPE | 30 | 64 | 67 |
Figure S20. Get High-res Image Gene #15: '8q gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
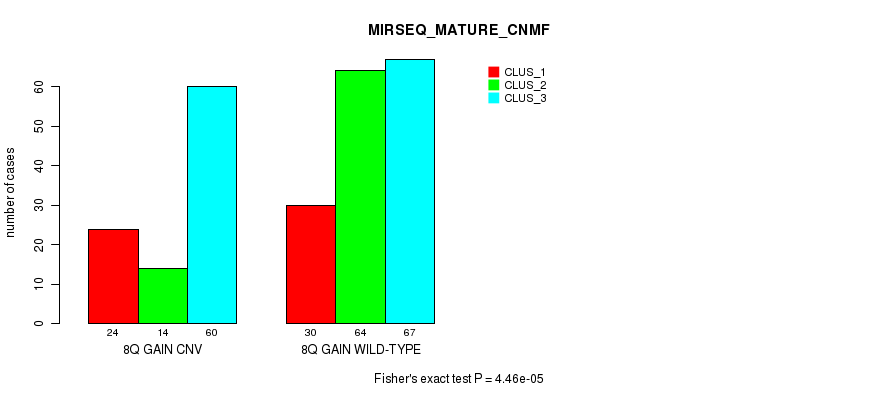
P value = 1.89e-05 (Fisher's exact test), Q value = 0.013
Table S21. Gene #15: '8q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 57 | 72 | 130 |
| 8Q GAIN CNV | 30 | 12 | 56 |
| 8Q GAIN WILD-TYPE | 27 | 60 | 74 |
Figure S21. Get High-res Image Gene #15: '8q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 5.75e-11 (Fisher's exact test), Q value = 4.4e-08
Table S22. Gene #18: '10p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 10P GAIN CNV | 3 | 9 | 33 |
| 10P GAIN WILD-TYPE | 147 | 37 | 76 |
Figure S22. Get High-res Image Gene #18: '10p gain' versus Molecular Subtype #1: 'CN_CNMF'
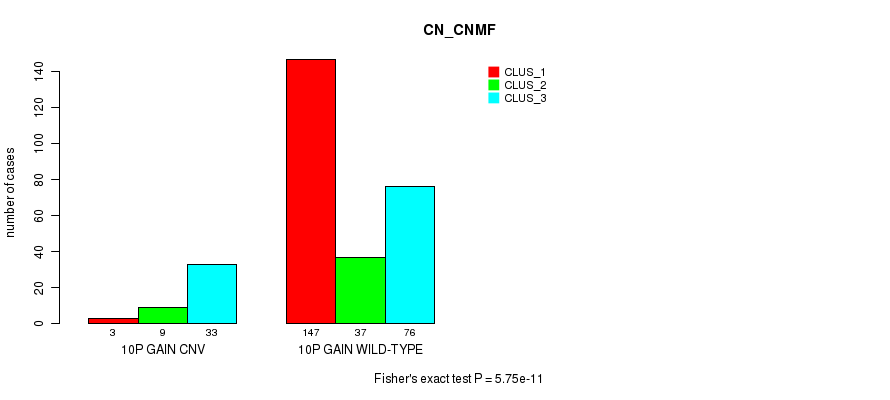
P value = 6.15e-07 (Fisher's exact test), Q value = 0.00045
Table S23. Gene #19: '10q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 10Q GAIN CNV | 3 | 1 | 22 |
| 10Q GAIN WILD-TYPE | 147 | 45 | 87 |
Figure S23. Get High-res Image Gene #19: '10q gain' versus Molecular Subtype #1: 'CN_CNMF'
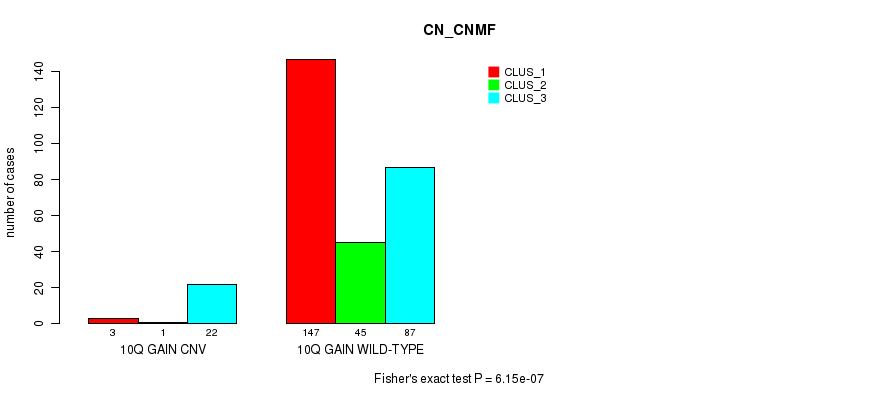
P value = 1.3e-07 (Fisher's exact test), Q value = 9.7e-05
Table S24. Gene #24: '13q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 13Q GAIN CNV | 14 | 21 | 30 |
| 13Q GAIN WILD-TYPE | 136 | 25 | 79 |
Figure S24. Get High-res Image Gene #24: '13q gain' versus Molecular Subtype #1: 'CN_CNMF'
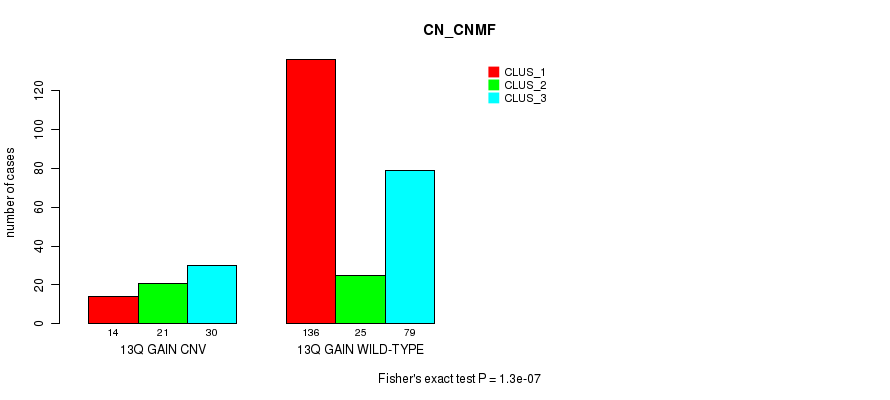
P value = 0.000325 (Fisher's exact test), Q value = 0.22
Table S25. Gene #24: '13q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 13Q GAIN CNV | 7 | 15 | 5 | 28 |
| 13Q GAIN WILD-TYPE | 30 | 65 | 60 | 48 |
Figure S25. Get High-res Image Gene #24: '13q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
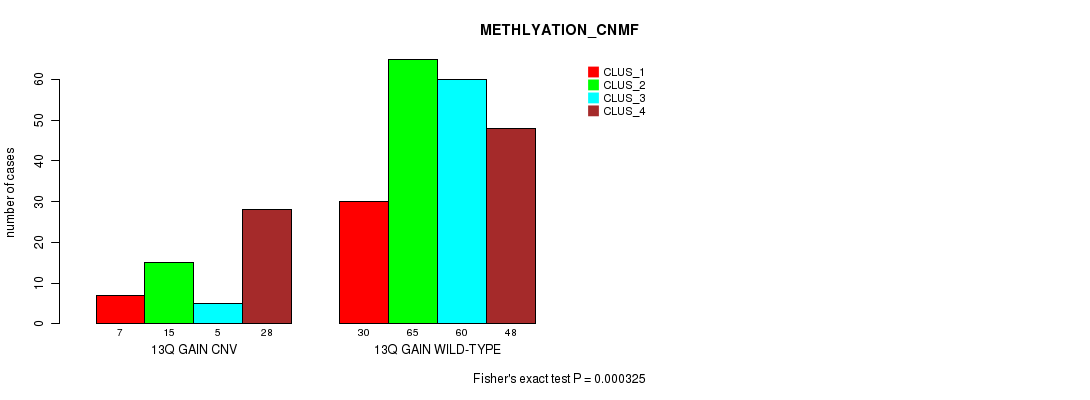
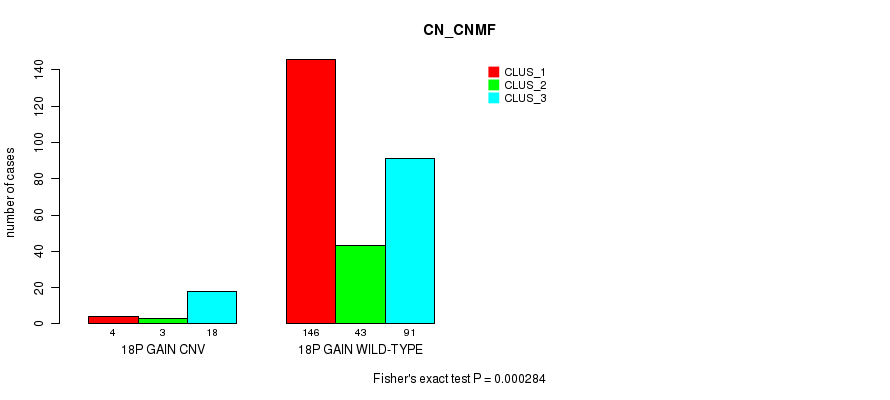
P value = 0.000284 (Fisher's exact test), Q value = 0.19
Table S26. Gene #31: '18p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 18P GAIN CNV | 4 | 3 | 18 |
| 18P GAIN WILD-TYPE | 146 | 43 | 91 |
Figure S26. Get High-res Image Gene #31: '18p gain' versus Molecular Subtype #1: 'CN_CNMF'
P value = 3.87e-05 (Fisher's exact test), Q value = 0.027
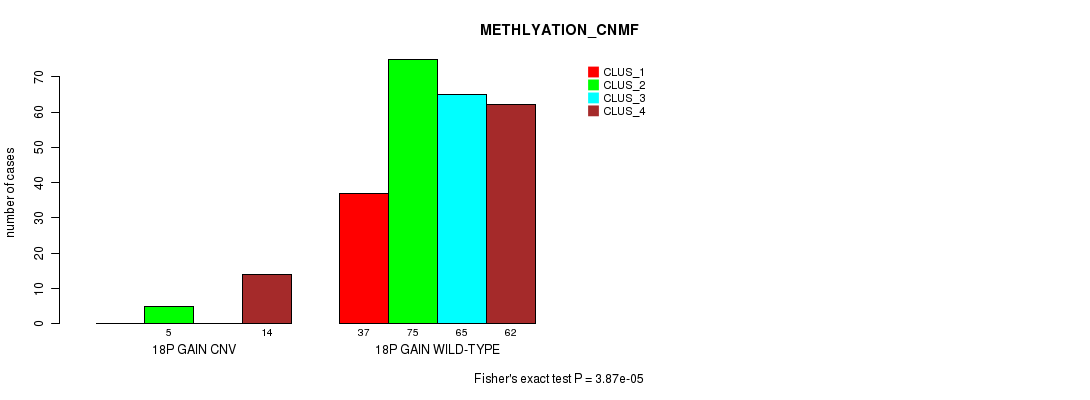
Table S27. Gene #31: '18p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 18P GAIN CNV | 0 | 5 | 0 | 14 |
| 18P GAIN WILD-TYPE | 37 | 75 | 65 | 62 |
Figure S27. Get High-res Image Gene #31: '18p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
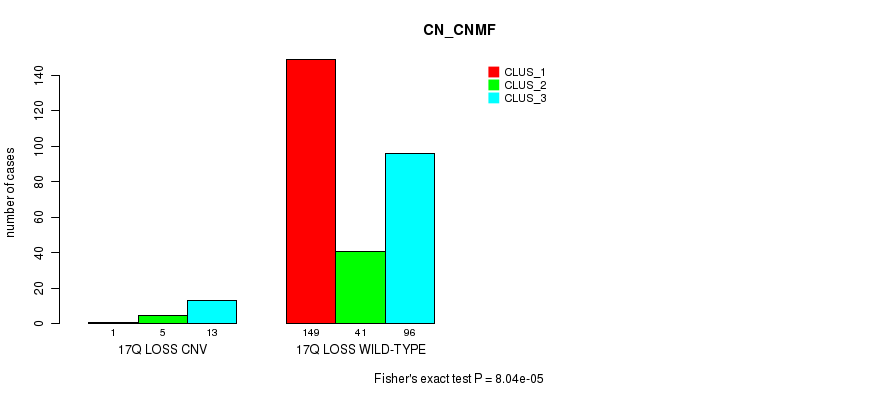
P value = 7.59e-18 (Fisher's exact test), Q value = 5.8e-15
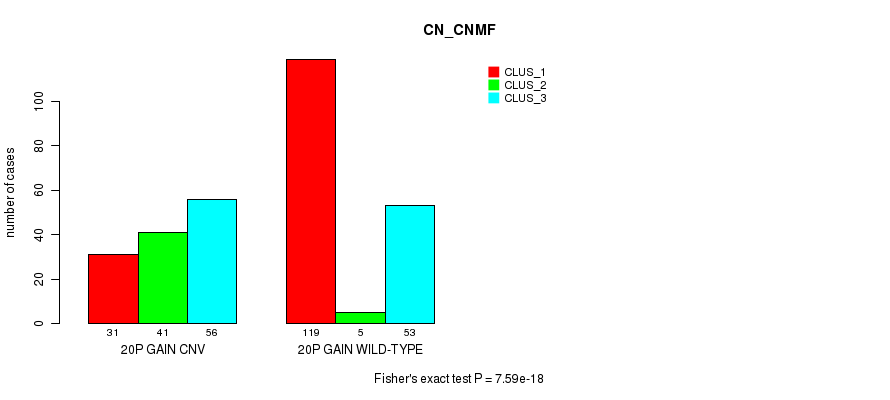
Table S28. Gene #35: '20p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 20P GAIN CNV | 31 | 41 | 56 |
| 20P GAIN WILD-TYPE | 119 | 5 | 53 |
Figure S28. Get High-res Image Gene #35: '20p gain' versus Molecular Subtype #1: 'CN_CNMF'
P value = 5.66e-09 (Fisher's exact test), Q value = 4.3e-06
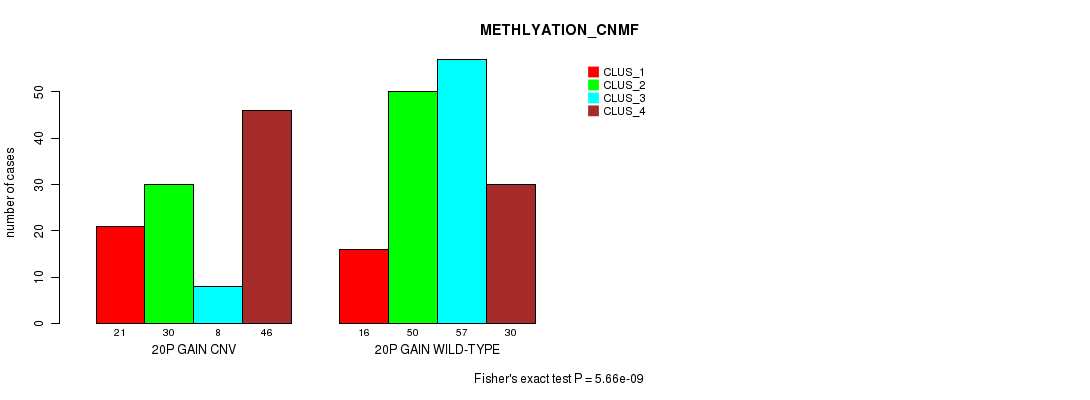
Table S29. Gene #35: '20p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 20P GAIN CNV | 21 | 30 | 8 | 46 |
| 20P GAIN WILD-TYPE | 16 | 50 | 57 | 30 |
Figure S29. Get High-res Image Gene #35: '20p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
P value = 7.19e-07 (Chi-square test), Q value = 0.00053
Table S30. Gene #35: '20p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 20P GAIN CNV | 19 | 13 | 14 | 23 | 41 |
| 20P GAIN WILD-TYPE | 42 | 50 | 29 | 19 | 21 |
Figure S30. Get High-res Image Gene #35: '20p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
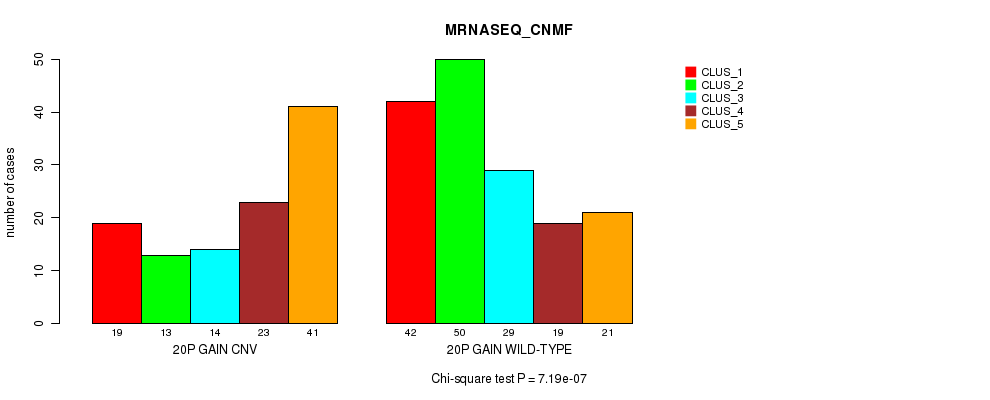
P value = 2.41e-06 (Fisher's exact test), Q value = 0.0018
Table S31. Gene #35: '20p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 141 | 64 |
| 20P GAIN CNV | 12 | 59 | 39 |
| 20P GAIN WILD-TYPE | 54 | 82 | 25 |
Figure S31. Get High-res Image Gene #35: '20p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
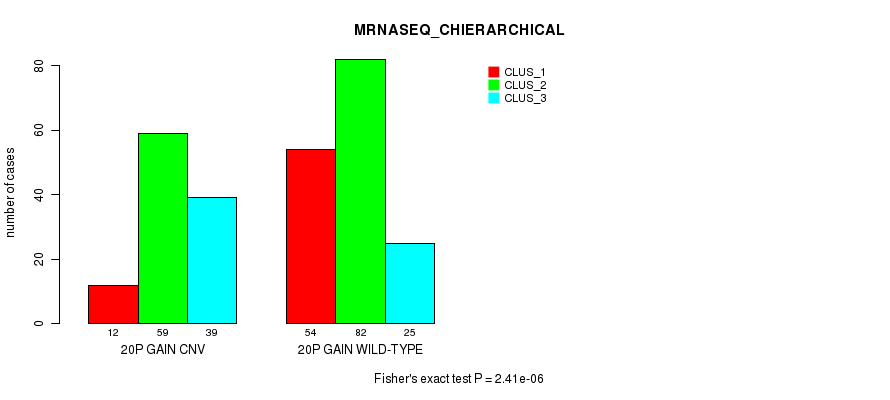
P value = 2.16e-05 (Fisher's exact test), Q value = 0.015
Table S32. Gene #35: '20p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 91 | 114 |
| 20P GAIN CNV | 40 | 23 | 65 |
| 20P GAIN WILD-TYPE | 60 | 68 | 49 |
Figure S32. Get High-res Image Gene #35: '20p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
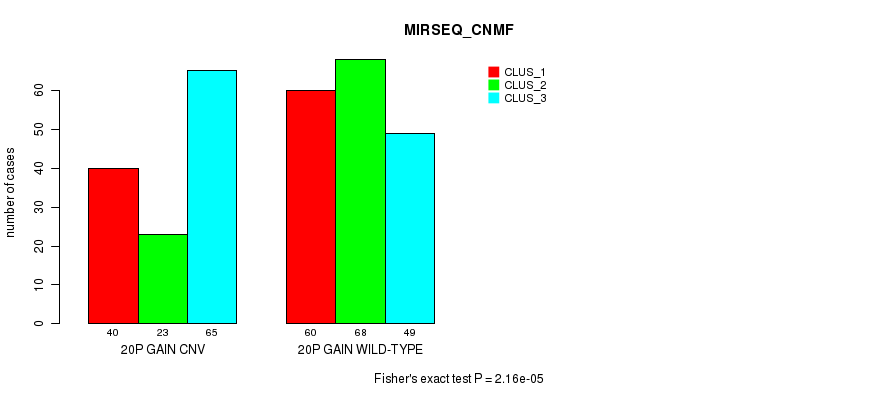
P value = 5.86e-07 (Fisher's exact test), Q value = 0.00043
Table S33. Gene #35: '20p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 133 | 77 | 95 |
| 20P GAIN CNV | 52 | 51 | 25 |
| 20P GAIN WILD-TYPE | 81 | 26 | 70 |
Figure S33. Get High-res Image Gene #35: '20p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
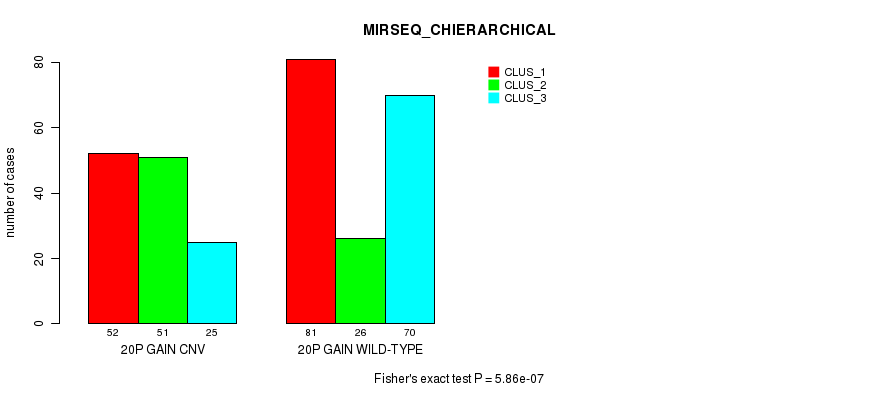
P value = 3.99e-06 (Fisher's exact test), Q value = 0.0029
Table S34. Gene #35: '20p gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 57 | 72 | 130 |
| 20P GAIN CNV | 38 | 17 | 50 |
| 20P GAIN WILD-TYPE | 19 | 55 | 80 |
Figure S34. Get High-res Image Gene #35: '20p gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
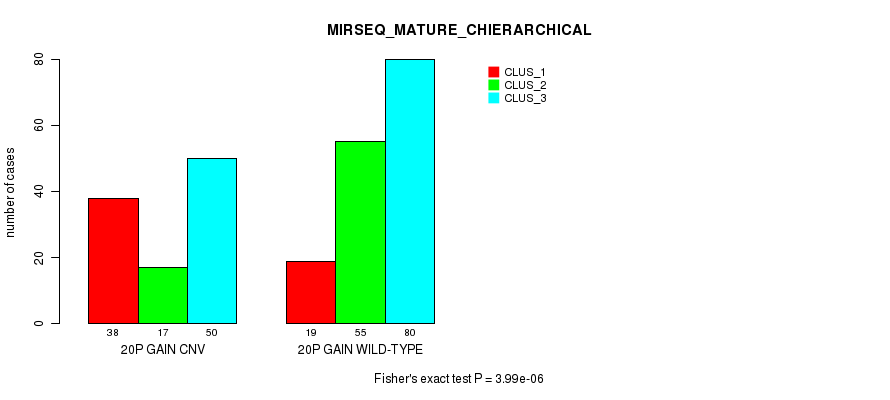
P value = 3.08e-26 (Fisher's exact test), Q value = 2.4e-23
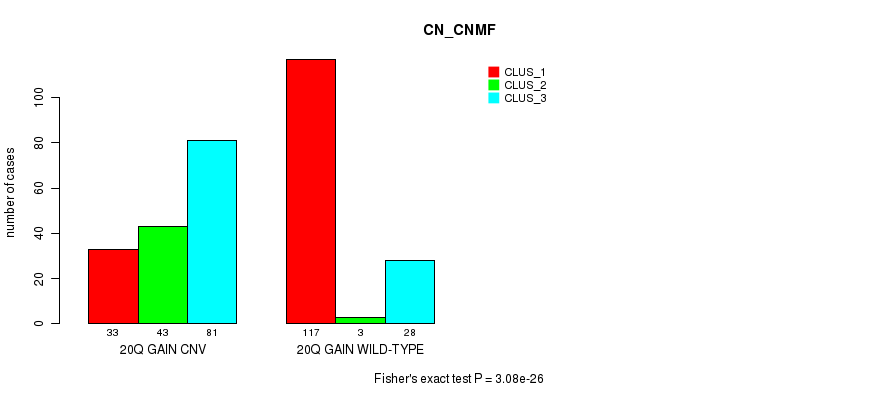
Table S35. Gene #36: '20q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 20Q GAIN CNV | 33 | 43 | 81 |
| 20Q GAIN WILD-TYPE | 117 | 3 | 28 |
Figure S35. Get High-res Image Gene #36: '20q gain' versus Molecular Subtype #1: 'CN_CNMF'
P value = 7.34e-14 (Fisher's exact test), Q value = 5.6e-11
Table S36. Gene #36: '20q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 20Q GAIN CNV | 21 | 37 | 12 | 63 |
| 20Q GAIN WILD-TYPE | 16 | 43 | 53 | 13 |
Figure S36. Get High-res Image Gene #36: '20q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
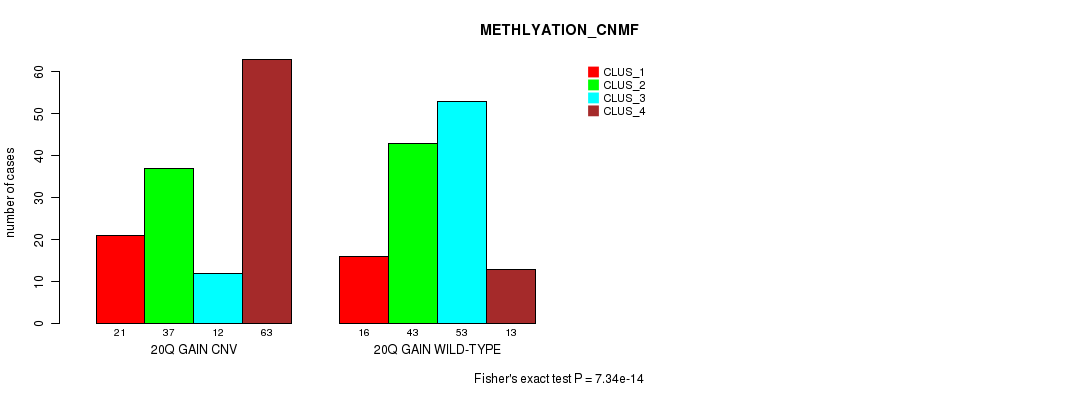
P value = 1.83e-10 (Chi-square test), Q value = 1.4e-07
Table S37. Gene #36: '20q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 20Q GAIN CNV | 22 | 17 | 19 | 25 | 53 |
| 20Q GAIN WILD-TYPE | 39 | 46 | 24 | 17 | 9 |
Figure S37. Get High-res Image Gene #36: '20q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
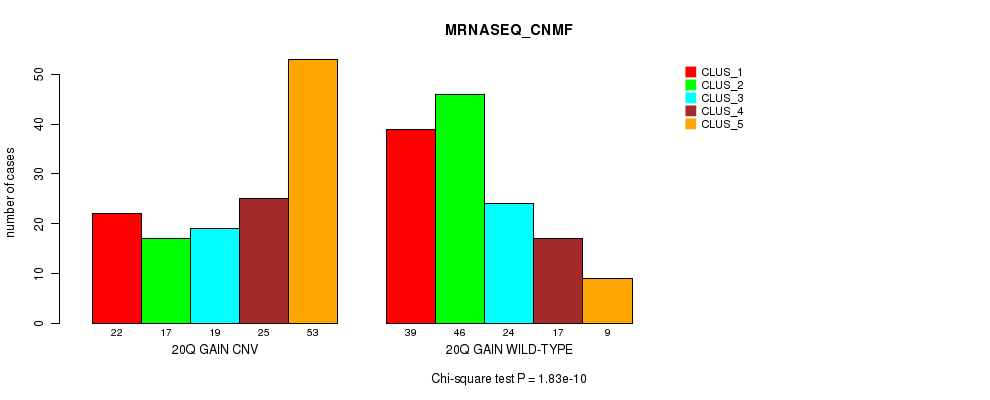
P value = 7.23e-10 (Fisher's exact test), Q value = 5.4e-07
Table S38. Gene #36: '20q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 141 | 64 |
| 20Q GAIN CNV | 16 | 69 | 51 |
| 20Q GAIN WILD-TYPE | 50 | 72 | 13 |
Figure S38. Get High-res Image Gene #36: '20q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
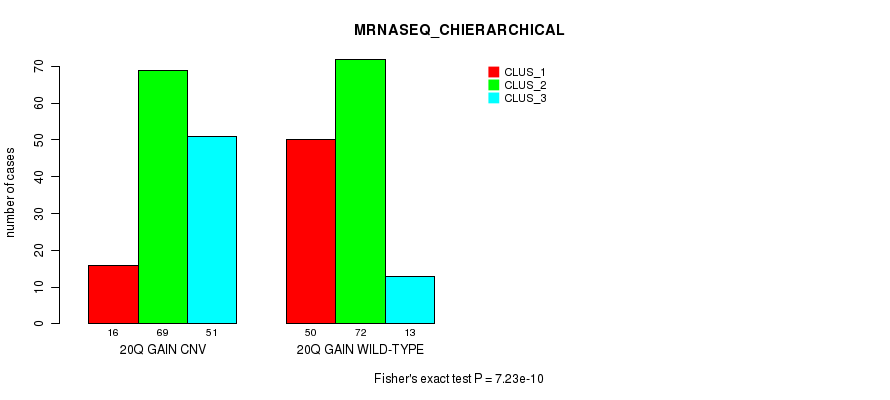
P value = 3.03e-07 (Fisher's exact test), Q value = 0.00022
Table S39. Gene #36: '20q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 91 | 114 |
| 20Q GAIN CNV | 44 | 32 | 81 |
| 20Q GAIN WILD-TYPE | 56 | 59 | 33 |
Figure S39. Get High-res Image Gene #36: '20q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
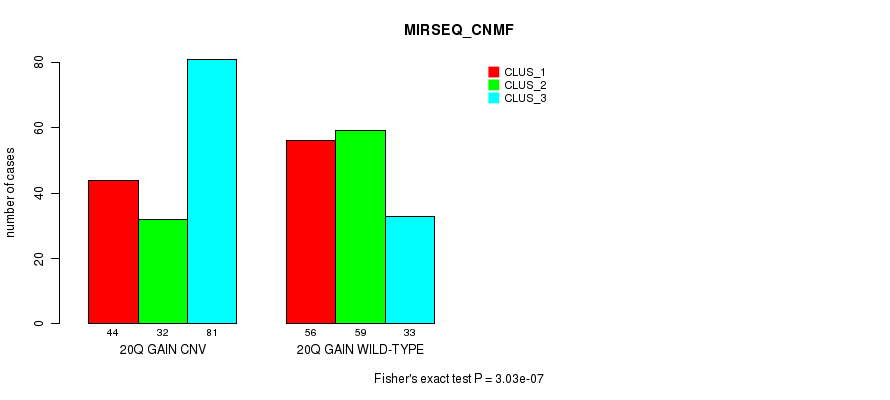
P value = 5.79e-12 (Fisher's exact test), Q value = 4.4e-09
Table S40. Gene #36: '20q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 133 | 77 | 95 |
| 20Q GAIN CNV | 61 | 65 | 31 |
| 20Q GAIN WILD-TYPE | 72 | 12 | 64 |
Figure S40. Get High-res Image Gene #36: '20q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
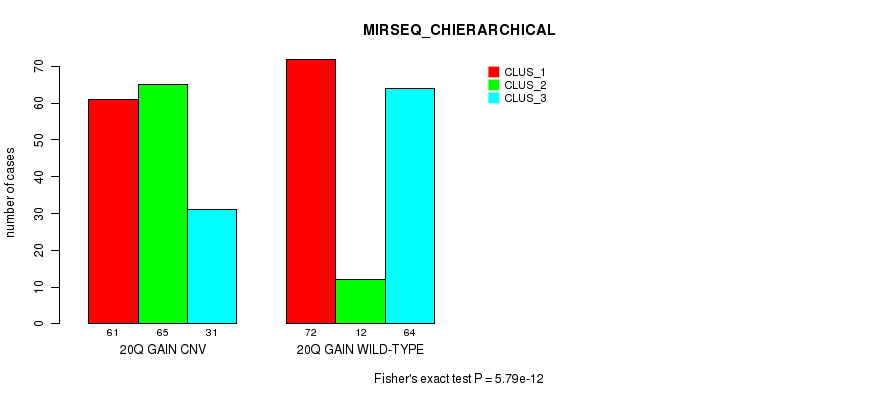
P value = 1.51e-10 (Fisher's exact test), Q value = 1.1e-07
Table S41. Gene #36: '20q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 57 | 72 | 130 |
| 20Q GAIN CNV | 48 | 19 | 66 |
| 20Q GAIN WILD-TYPE | 9 | 53 | 64 |
Figure S41. Get High-res Image Gene #36: '20q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
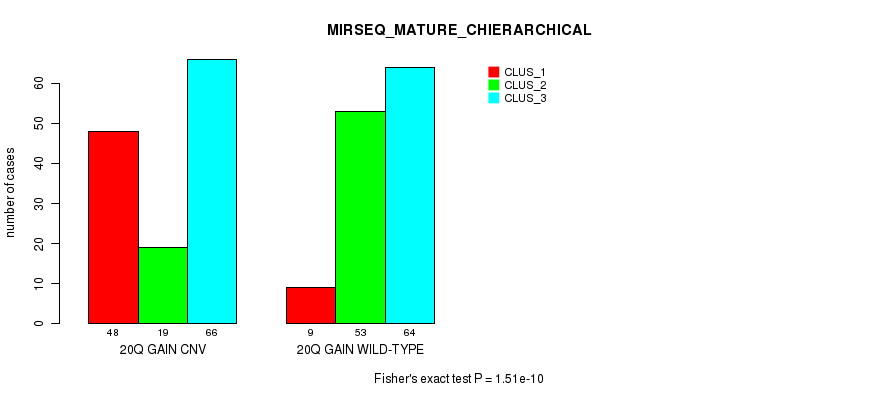
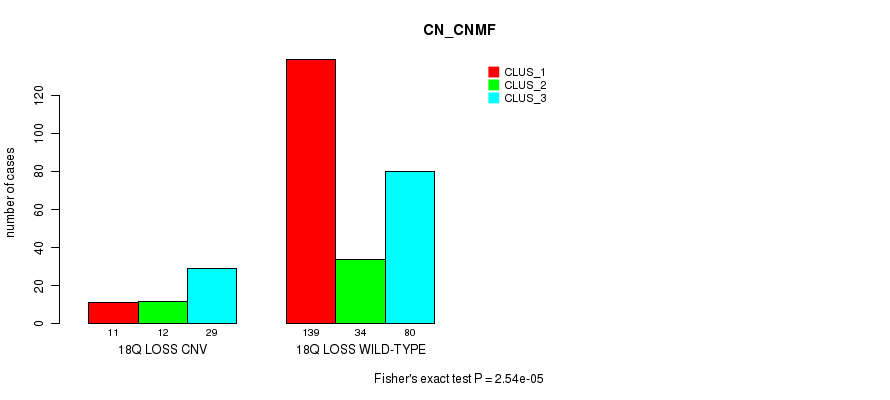
P value = 8.32e-06 (Fisher's exact test), Q value = 0.006
Table S42. Gene #40: '1p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 1P LOSS CNV | 0 | 1 | 13 |
| 1P LOSS WILD-TYPE | 150 | 45 | 96 |
Figure S42. Get High-res Image Gene #40: '1p loss' versus Molecular Subtype #1: 'CN_CNMF'
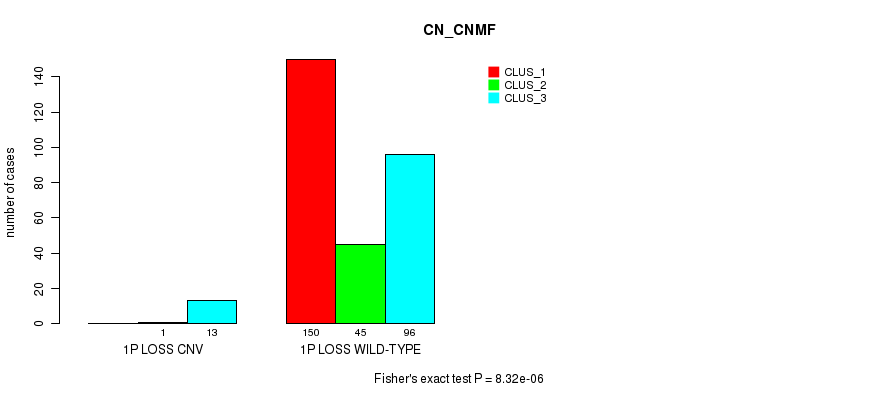
P value = 1.31e-06 (Fisher's exact test), Q value = 0.00097
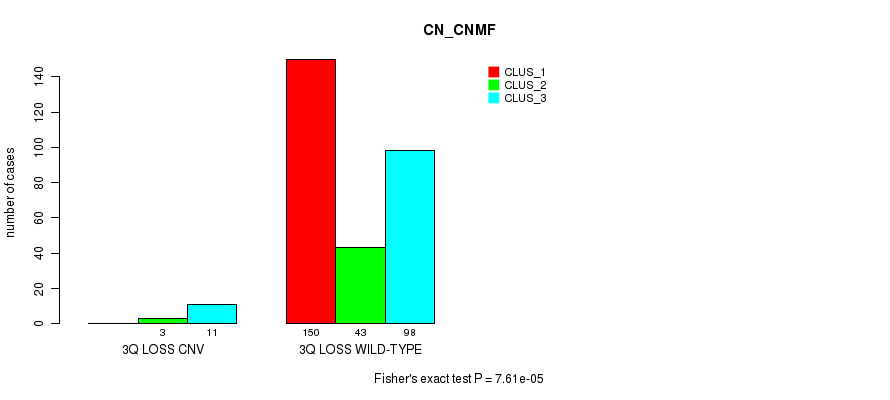
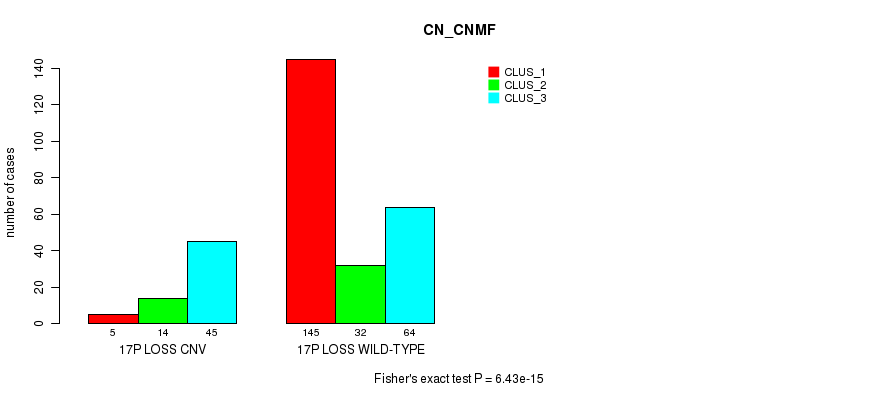
Table S43. Gene #44: '3p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 3P LOSS CNV | 1 | 5 | 18 |
| 3P LOSS WILD-TYPE | 149 | 41 | 91 |
Figure S43. Get High-res Image Gene #44: '3p loss' versus Molecular Subtype #1: 'CN_CNMF'
P value = 7.61e-05 (Fisher's exact test), Q value = 0.052
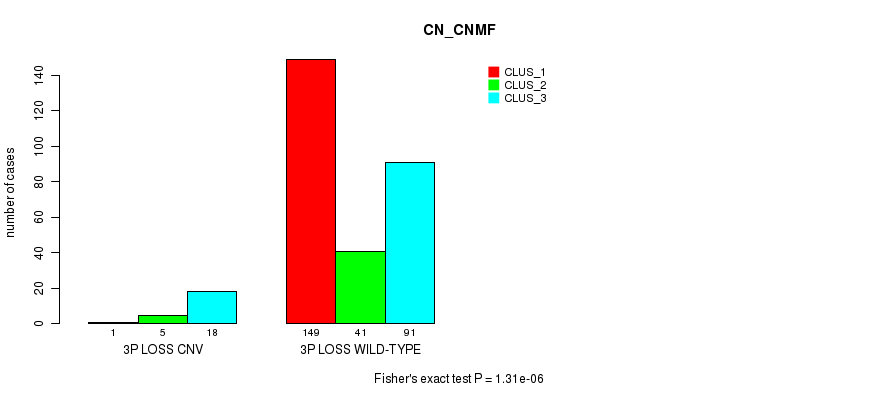
Table S44. Gene #45: '3q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 3Q LOSS CNV | 0 | 3 | 11 |
| 3Q LOSS WILD-TYPE | 150 | 43 | 98 |
Figure S44. Get High-res Image Gene #45: '3q loss' versus Molecular Subtype #1: 'CN_CNMF'
P value = 2.31e-11 (Fisher's exact test), Q value = 1.7e-08
Table S45. Gene #46: '4p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 4P LOSS CNV | 6 | 9 | 40 |
| 4P LOSS WILD-TYPE | 144 | 37 | 69 |
Figure S45. Get High-res Image Gene #46: '4p loss' versus Molecular Subtype #1: 'CN_CNMF'
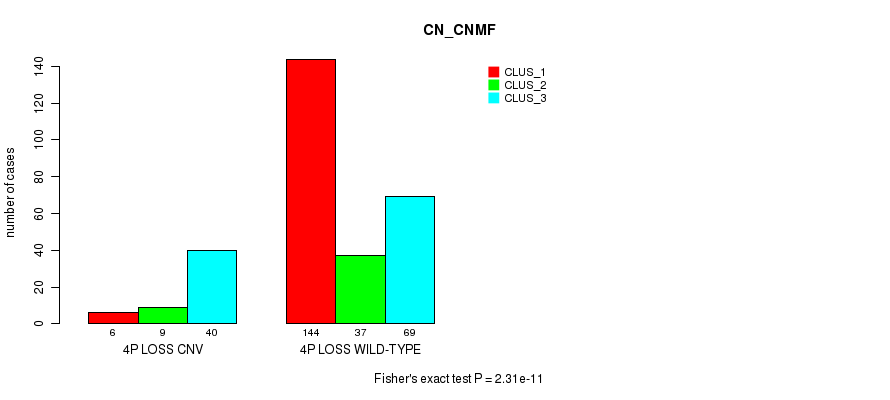
P value = 8.51e-05 (Chi-square test), Q value = 0.058
Table S46. Gene #46: '4p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 4P LOSS CNV | 7 | 5 | 5 | 6 | 23 |
| 4P LOSS WILD-TYPE | 54 | 58 | 38 | 36 | 39 |
Figure S46. Get High-res Image Gene #46: '4p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
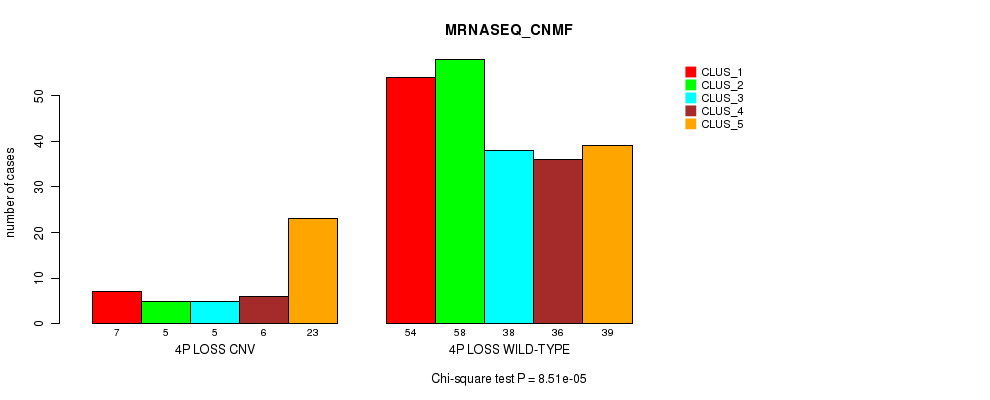
P value = 6.56e-05 (Fisher's exact test), Q value = 0.045
Table S47. Gene #46: '4p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 141 | 64 |
| 4P LOSS CNV | 6 | 17 | 23 |
| 4P LOSS WILD-TYPE | 60 | 124 | 41 |
Figure S47. Get High-res Image Gene #46: '4p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
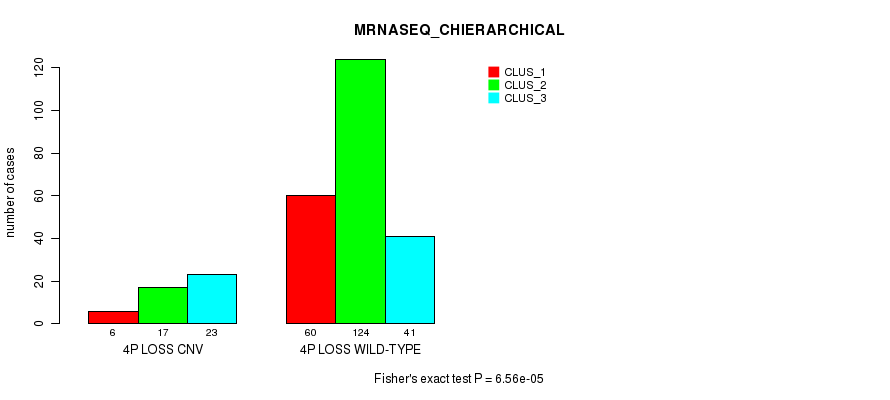
P value = 9.16e-05 (Fisher's exact test), Q value = 0.062
Table S48. Gene #46: '4p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 133 | 77 | 95 |
| 4P LOSS CNV | 15 | 27 | 13 |
| 4P LOSS WILD-TYPE | 118 | 50 | 82 |
Figure S48. Get High-res Image Gene #46: '4p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
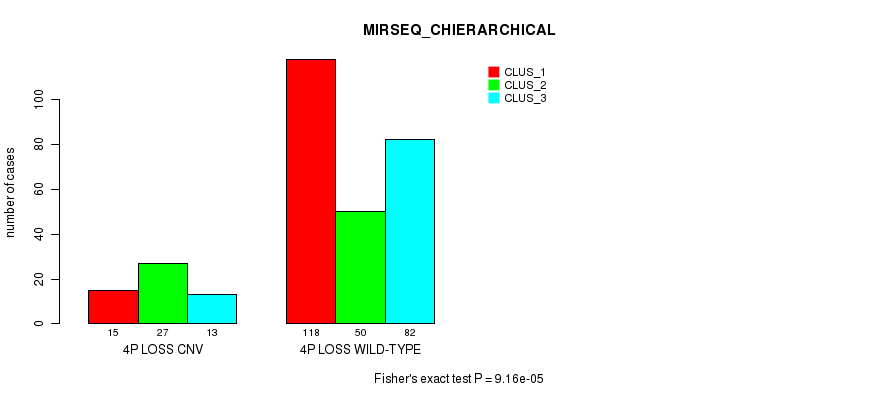
P value = 2.28e-14 (Fisher's exact test), Q value = 1.7e-11
Table S49. Gene #47: '4q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 4Q LOSS CNV | 3 | 7 | 41 |
| 4Q LOSS WILD-TYPE | 147 | 39 | 68 |
Figure S49. Get High-res Image Gene #47: '4q loss' versus Molecular Subtype #1: 'CN_CNMF'
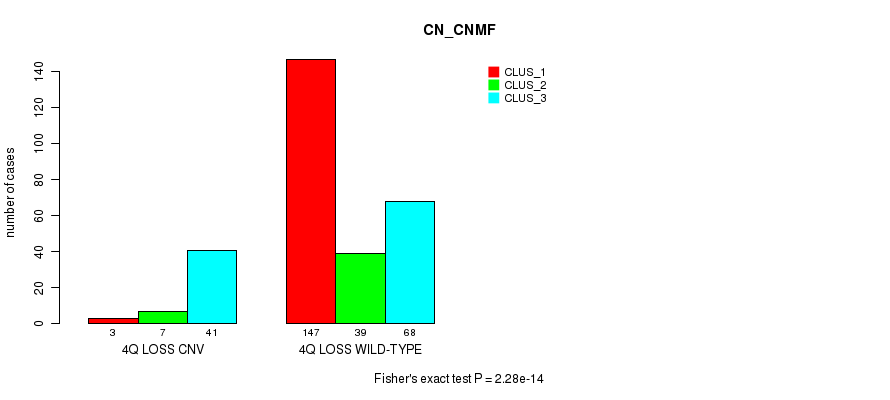
P value = 4.26e-05 (Fisher's exact test), Q value = 0.03
Table S50. Gene #47: '4q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 4Q LOSS CNV | 1 | 18 | 4 | 23 |
| 4Q LOSS WILD-TYPE | 36 | 62 | 61 | 53 |
Figure S50. Get High-res Image Gene #47: '4q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
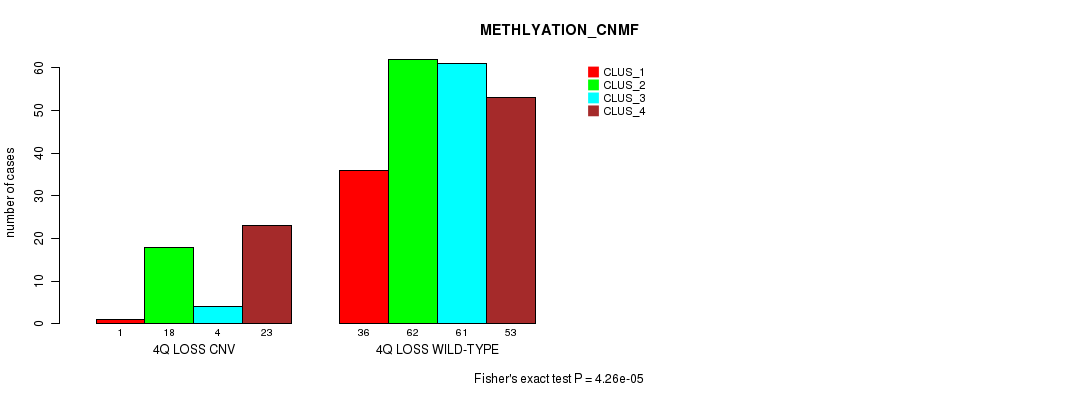
P value = 0.000242 (Chi-square test), Q value = 0.16
Table S51. Gene #47: '4q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 4Q LOSS CNV | 6 | 4 | 5 | 7 | 21 |
| 4Q LOSS WILD-TYPE | 55 | 59 | 38 | 35 | 41 |
Figure S51. Get High-res Image Gene #47: '4q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
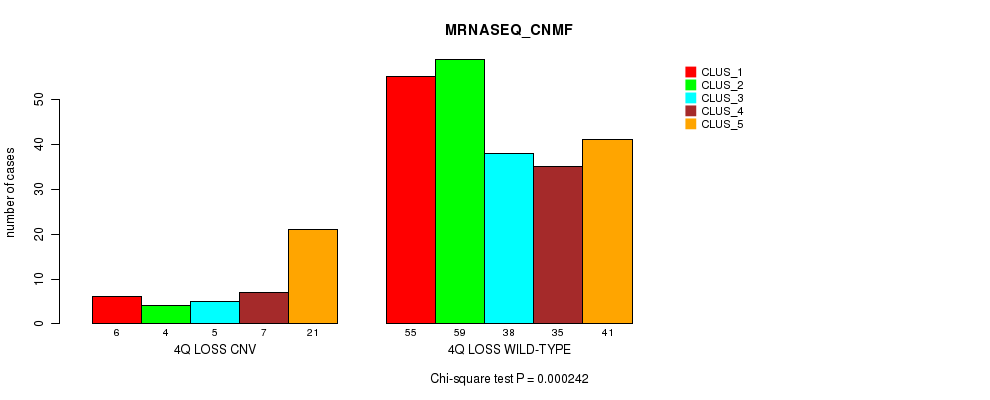
P value = 0.000226 (Fisher's exact test), Q value = 0.15
Table S52. Gene #47: '4q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 141 | 64 |
| 4Q LOSS CNV | 5 | 17 | 21 |
| 4Q LOSS WILD-TYPE | 61 | 124 | 43 |
Figure S52. Get High-res Image Gene #47: '4q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
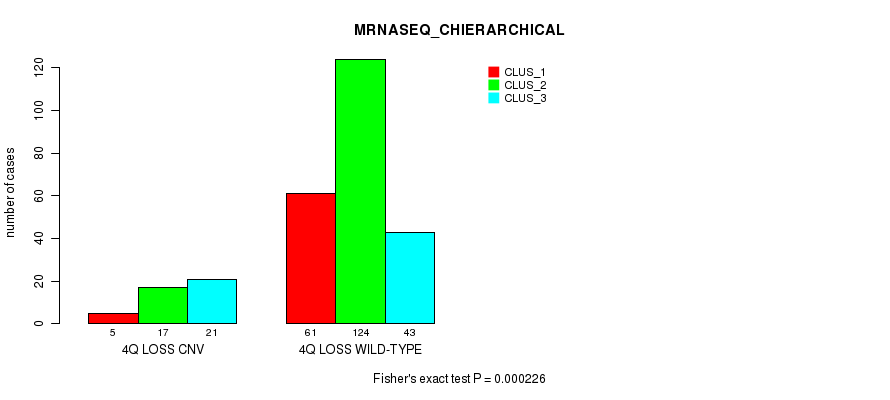
P value = 6.4e-05 (Fisher's exact test), Q value = 0.044
Table S53. Gene #47: '4q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 133 | 77 | 95 |
| 4Q LOSS CNV | 15 | 26 | 10 |
| 4Q LOSS WILD-TYPE | 118 | 51 | 85 |
Figure S53. Get High-res Image Gene #47: '4q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
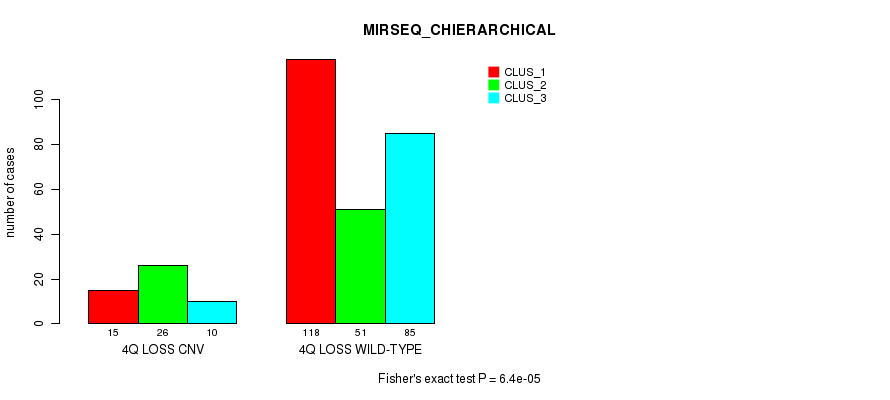
P value = 1.74e-06 (Fisher's exact test), Q value = 0.0013
Table S54. Gene #48: '5p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 5P LOSS CNV | 0 | 2 | 15 |
| 5P LOSS WILD-TYPE | 150 | 44 | 94 |
Figure S54. Get High-res Image Gene #48: '5p loss' versus Molecular Subtype #1: 'CN_CNMF'
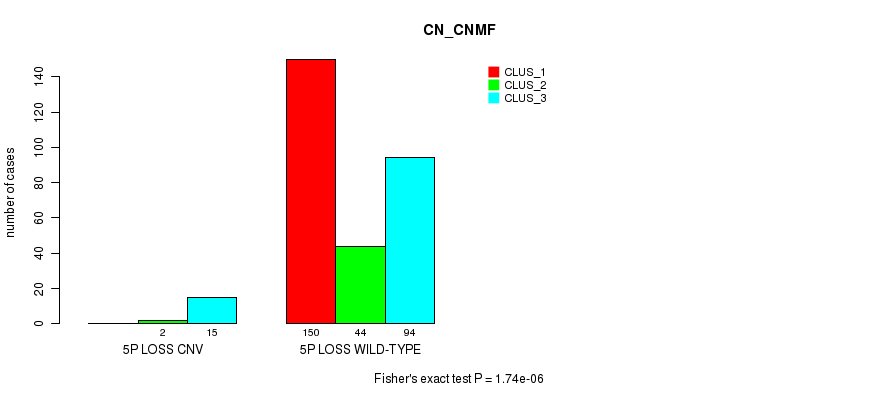
P value = 0.000329 (Fisher's exact test), Q value = 0.22
Table S55. Gene #48: '5p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 141 | 64 |
| 5P LOSS CNV | 1 | 3 | 10 |
| 5P LOSS WILD-TYPE | 65 | 138 | 54 |
Figure S55. Get High-res Image Gene #48: '5p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
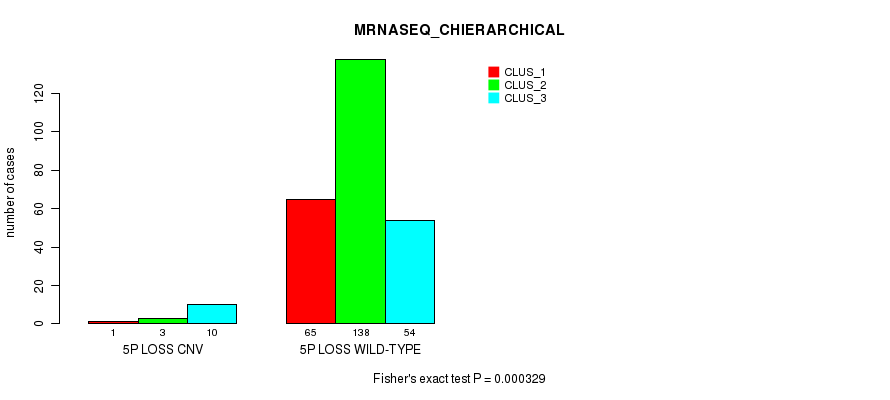
P value = 2.21e-13 (Fisher's exact test), Q value = 1.7e-10
Table S56. Gene #49: '5q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 5Q LOSS CNV | 0 | 4 | 31 |
| 5Q LOSS WILD-TYPE | 150 | 42 | 78 |
Figure S56. Get High-res Image Gene #49: '5q loss' versus Molecular Subtype #1: 'CN_CNMF'
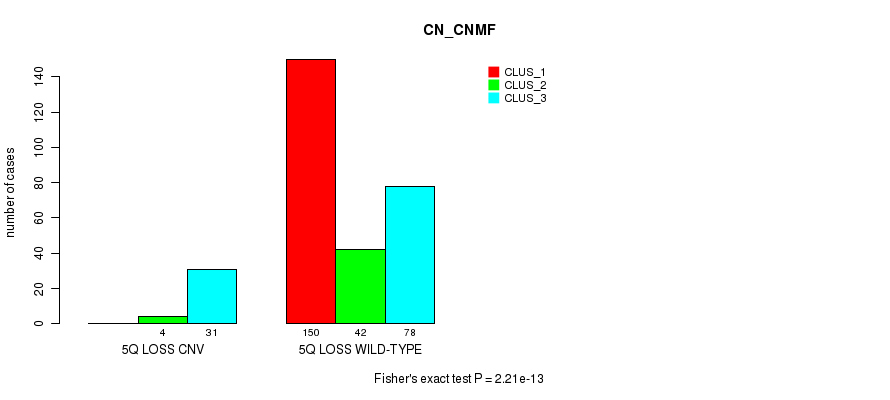
P value = 2.47e-06 (Fisher's exact test), Q value = 0.0018
Table S57. Gene #49: '5q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 5Q LOSS CNV | 0 | 11 | 1 | 20 |
| 5Q LOSS WILD-TYPE | 37 | 69 | 64 | 56 |
Figure S57. Get High-res Image Gene #49: '5q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
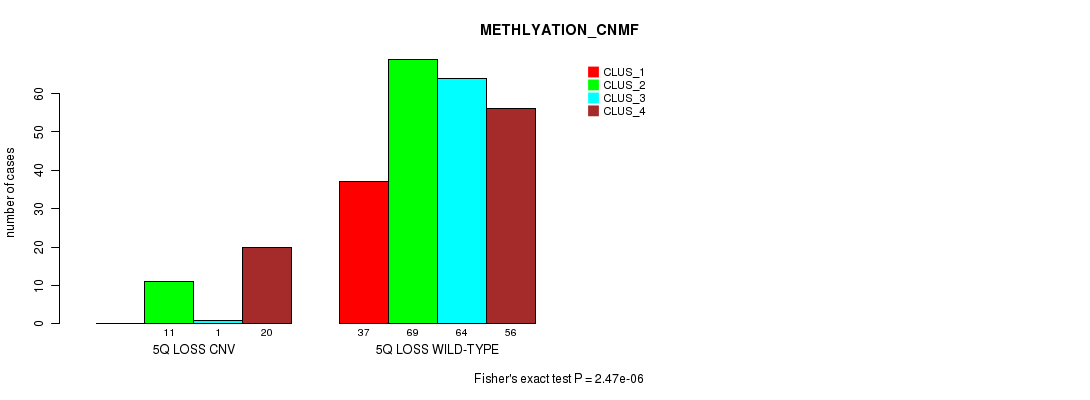
P value = 4.12e-06 (Chi-square test), Q value = 0.003
Table S58. Gene #49: '5q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 5Q LOSS CNV | 4 | 2 | 2 | 4 | 19 |
| 5Q LOSS WILD-TYPE | 57 | 61 | 41 | 38 | 43 |
Figure S58. Get High-res Image Gene #49: '5q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
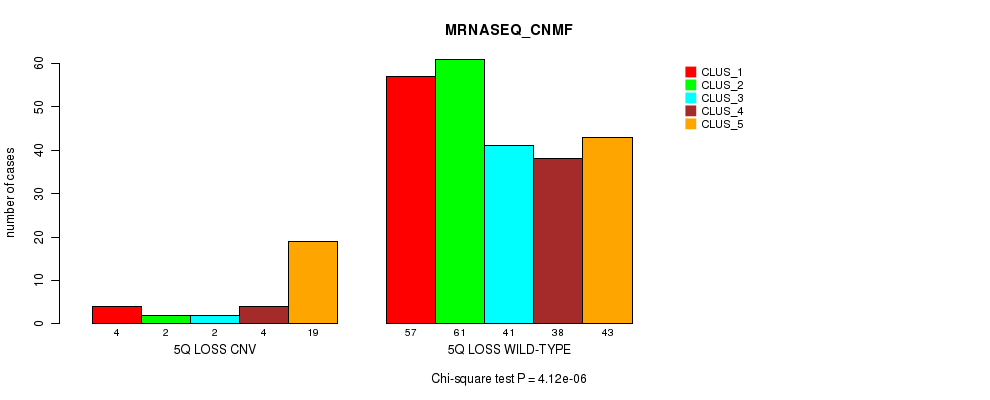
P value = 5.94e-06 (Fisher's exact test), Q value = 0.0043
Table S59. Gene #49: '5q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 141 | 64 |
| 5Q LOSS CNV | 3 | 9 | 19 |
| 5Q LOSS WILD-TYPE | 63 | 132 | 45 |
Figure S59. Get High-res Image Gene #49: '5q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
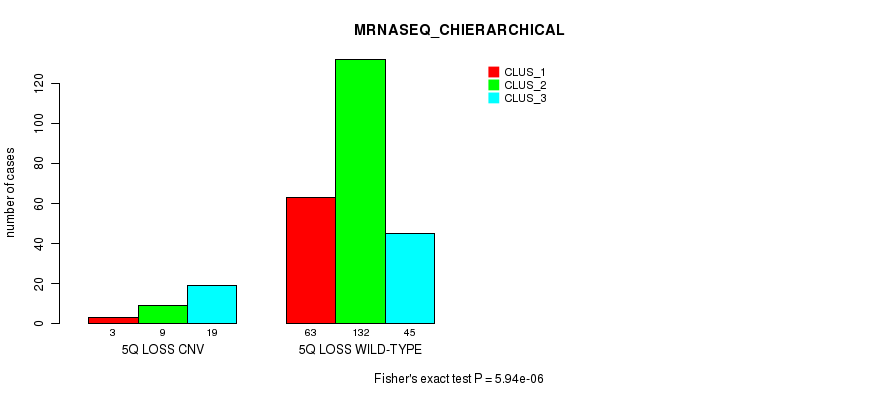
P value = 0.000273 (Fisher's exact test), Q value = 0.18
Table S60. Gene #49: '5q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 133 | 77 | 95 |
| 5Q LOSS CNV | 11 | 19 | 5 |
| 5Q LOSS WILD-TYPE | 122 | 58 | 90 |
Figure S60. Get High-res Image Gene #49: '5q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
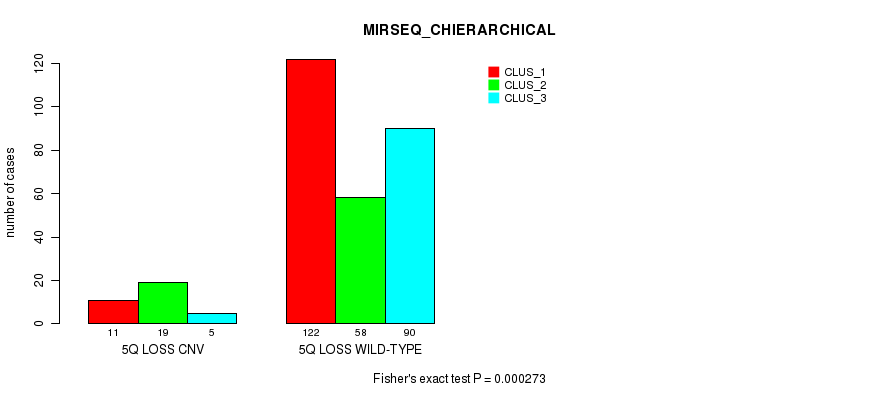
P value = 8.44e-05 (Fisher's exact test), Q value = 0.057
Table S61. Gene #49: '5q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 57 | 72 | 130 |
| 5Q LOSS CNV | 17 | 3 | 13 |
| 5Q LOSS WILD-TYPE | 40 | 69 | 117 |
Figure S61. Get High-res Image Gene #49: '5q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
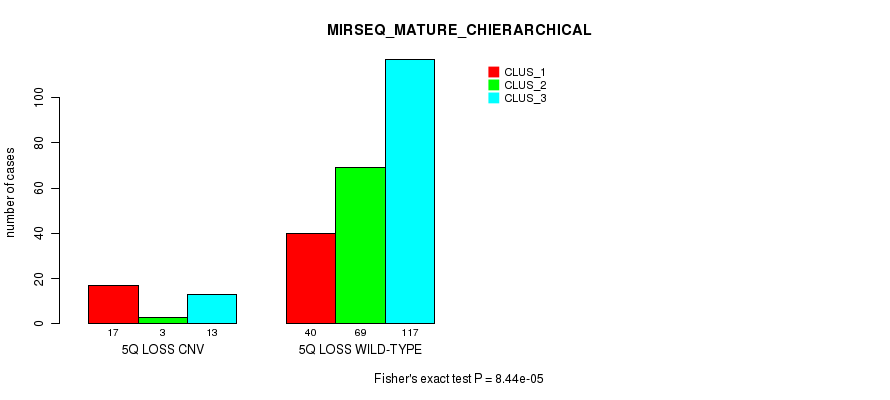
P value = 2.12e-05 (Fisher's exact test), Q value = 0.015
Table S62. Gene #53: '7q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 7Q LOSS CNV | 0 | 1 | 12 |
| 7Q LOSS WILD-TYPE | 150 | 45 | 97 |
Figure S62. Get High-res Image Gene #53: '7q loss' versus Molecular Subtype #1: 'CN_CNMF'
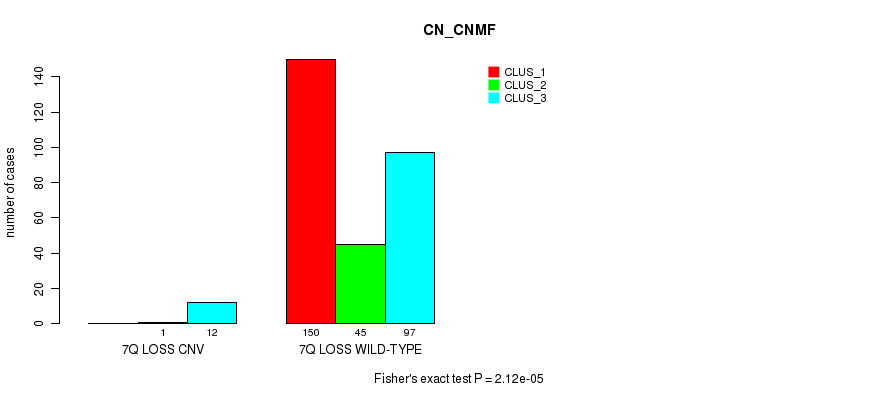
P value = 2.68e-06 (Fisher's exact test), Q value = 0.002
Table S63. Gene #54: '8p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 8P LOSS CNV | 3 | 5 | 22 |
| 8P LOSS WILD-TYPE | 147 | 41 | 87 |
Figure S63. Get High-res Image Gene #54: '8p loss' versus Molecular Subtype #1: 'CN_CNMF'
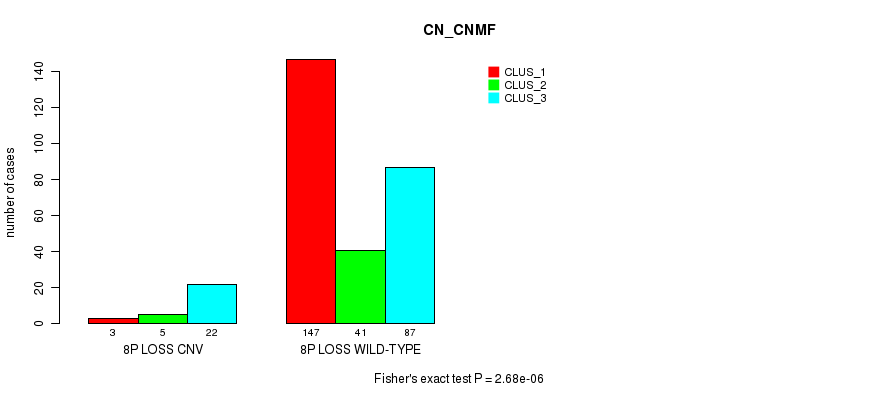
P value = 0.000323 (Fisher's exact test), Q value = 0.21
Table S64. Gene #54: '8p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 91 | 114 |
| 8P LOSS CNV | 10 | 1 | 19 |
| 8P LOSS WILD-TYPE | 90 | 90 | 95 |
Figure S64. Get High-res Image Gene #54: '8p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'

P value = 9.73e-05 (Fisher's exact test), Q value = 0.066
Table S65. Gene #54: '8p loss' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 54 | 78 | 127 |
| 8P LOSS CNV | 2 | 1 | 22 |
| 8P LOSS WILD-TYPE | 52 | 77 | 105 |
Figure S65. Get High-res Image Gene #54: '8p loss' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
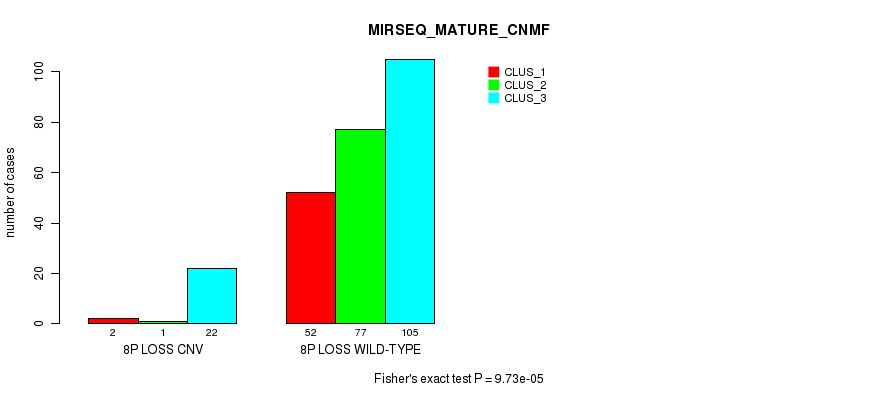
P value = 1.64e-13 (Fisher's exact test), Q value = 1.3e-10
Table S66. Gene #56: '9p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 9P LOSS CNV | 4 | 6 | 41 |
| 9P LOSS WILD-TYPE | 146 | 40 | 68 |
Figure S66. Get High-res Image Gene #56: '9p loss' versus Molecular Subtype #1: 'CN_CNMF'
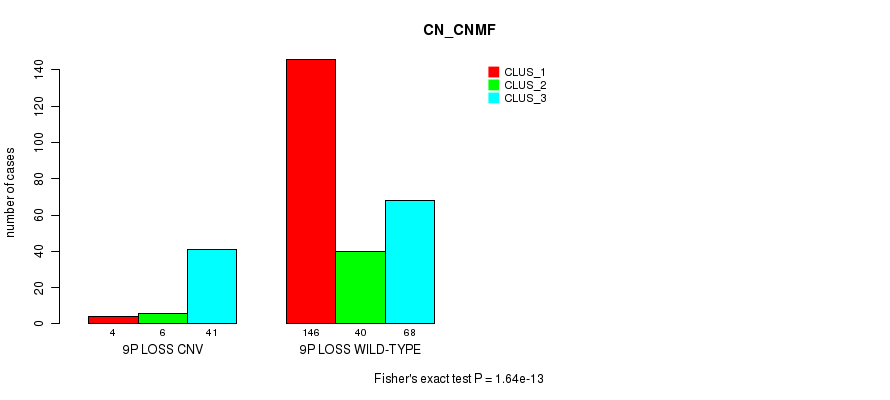
P value = 2.64e-05 (Fisher's exact test), Q value = 0.019
Table S67. Gene #56: '9p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 9P LOSS CNV | 0 | 13 | 6 | 24 |
| 9P LOSS WILD-TYPE | 37 | 67 | 59 | 52 |
Figure S67. Get High-res Image Gene #56: '9p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
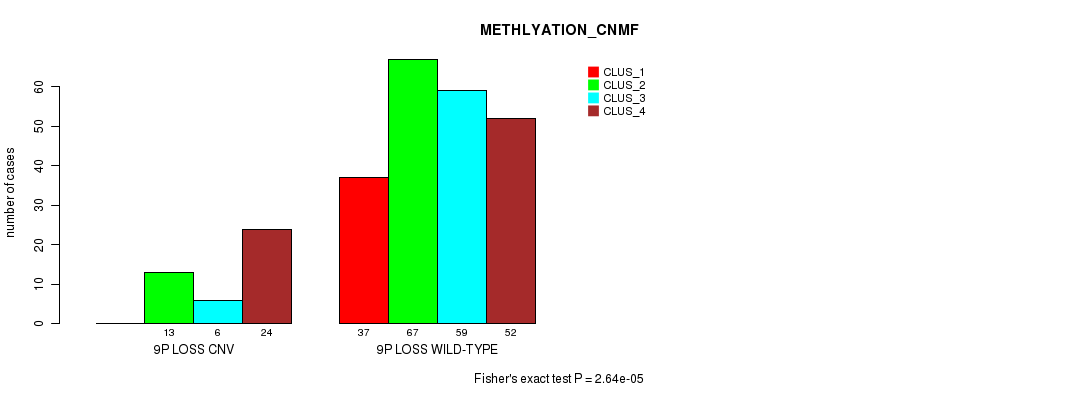
P value = 0.000171 (Chi-square test), Q value = 0.12
Table S68. Gene #56: '9p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 9P LOSS CNV | 5 | 5 | 9 | 6 | 22 |
| 9P LOSS WILD-TYPE | 56 | 58 | 34 | 36 | 40 |
Figure S68. Get High-res Image Gene #56: '9p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
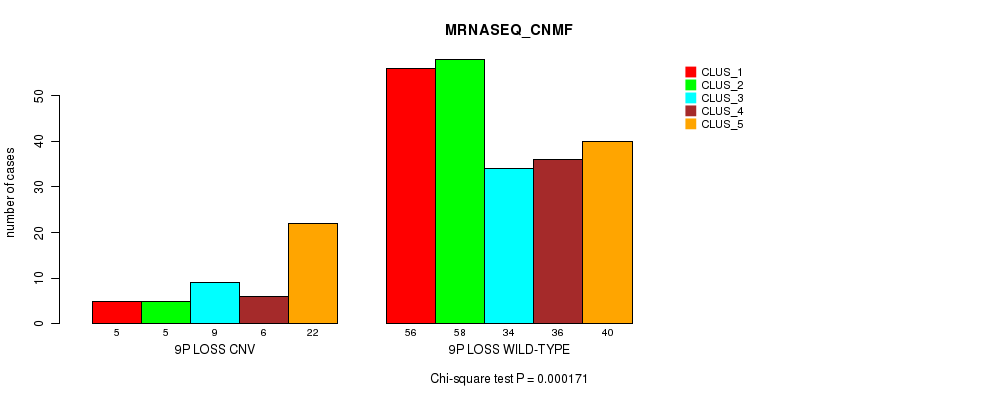
P value = 8.9e-08 (Fisher's exact test), Q value = 6.6e-05
Table S69. Gene #57: '9q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 9Q LOSS CNV | 2 | 1 | 22 |
| 9Q LOSS WILD-TYPE | 148 | 45 | 87 |
Figure S69. Get High-res Image Gene #57: '9q loss' versus Molecular Subtype #1: 'CN_CNMF'
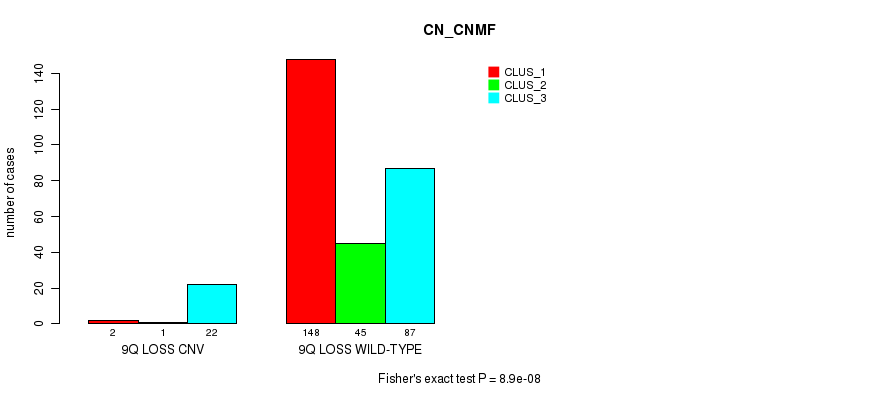
P value = 1.63e-05 (Fisher's exact test), Q value = 0.012
Table S70. Gene #59: '10q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 10Q LOSS CNV | 0 | 6 | 10 |
| 10Q LOSS WILD-TYPE | 150 | 40 | 99 |
Figure S70. Get High-res Image Gene #59: '10q loss' versus Molecular Subtype #1: 'CN_CNMF'
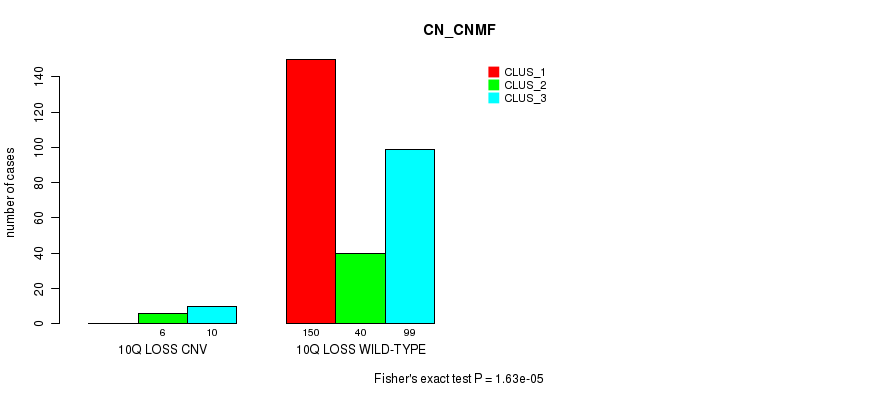
P value = 3.13e-05 (Fisher's exact test), Q value = 0.022
Table S71. Gene #60: '11p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 11P LOSS CNV | 2 | 1 | 16 |
| 11P LOSS WILD-TYPE | 148 | 45 | 93 |
Figure S71. Get High-res Image Gene #60: '11p loss' versus Molecular Subtype #1: 'CN_CNMF'
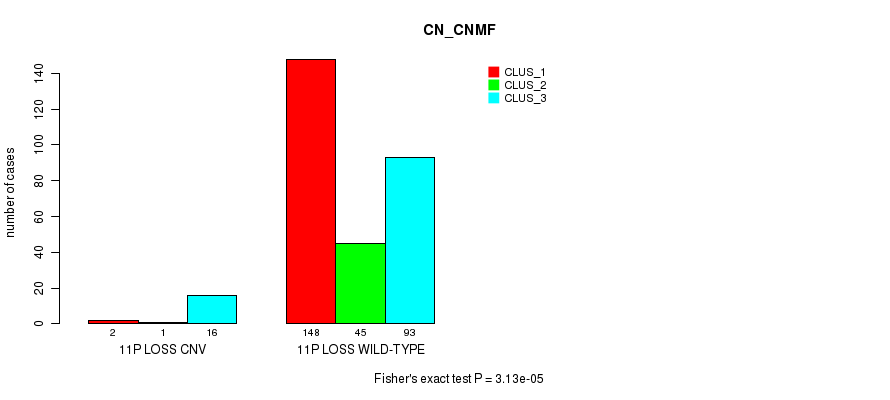
P value = 5.39e-05 (Fisher's exact test), Q value = 0.037
Table S72. Gene #60: '11p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 11P LOSS CNV | 0 | 2 | 0 | 12 |
| 11P LOSS WILD-TYPE | 37 | 78 | 65 | 64 |
Figure S72. Get High-res Image Gene #60: '11p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
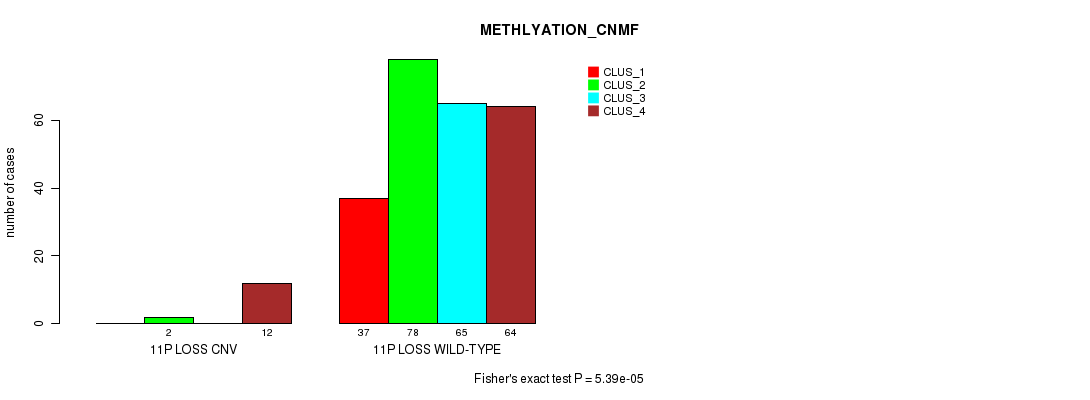
P value = 3.36e-05 (Fisher's exact test), Q value = 0.024
Table S73. Gene #61: '11q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 11Q LOSS CNV | 1 | 1 | 14 |
| 11Q LOSS WILD-TYPE | 149 | 45 | 95 |
Figure S73. Get High-res Image Gene #61: '11q loss' versus Molecular Subtype #1: 'CN_CNMF'
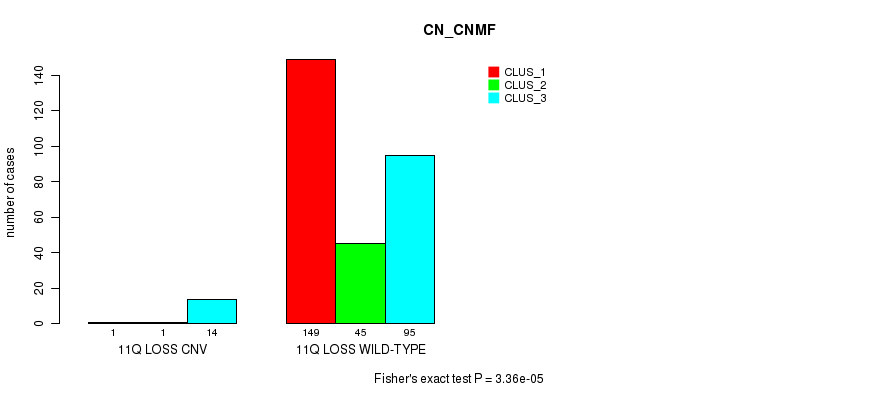
P value = 2.3e-05 (Fisher's exact test), Q value = 0.016
Table S74. Gene #62: '12p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 12P LOSS CNV | 2 | 2 | 17 |
| 12P LOSS WILD-TYPE | 148 | 44 | 92 |
Figure S74. Get High-res Image Gene #62: '12p loss' versus Molecular Subtype #1: 'CN_CNMF'
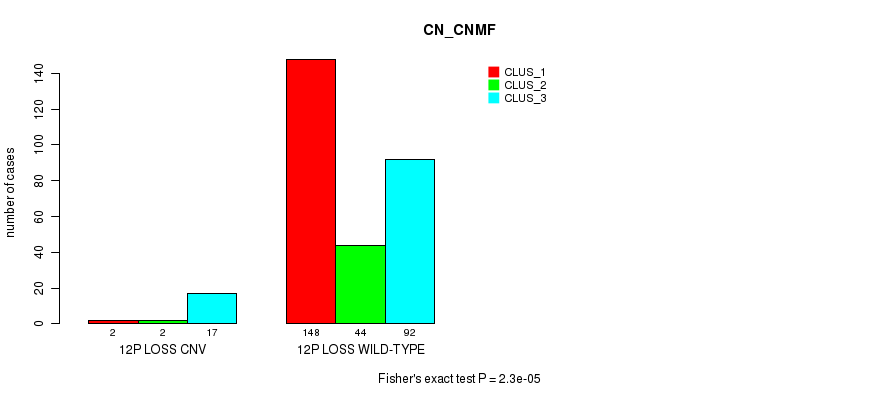
P value = 0.000163 (Chi-square test), Q value = 0.11
Table S75. Gene #62: '12p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 12P LOSS CNV | 1 | 1 | 3 | 1 | 12 |
| 12P LOSS WILD-TYPE | 60 | 62 | 40 | 41 | 50 |
Figure S75. Get High-res Image Gene #62: '12p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
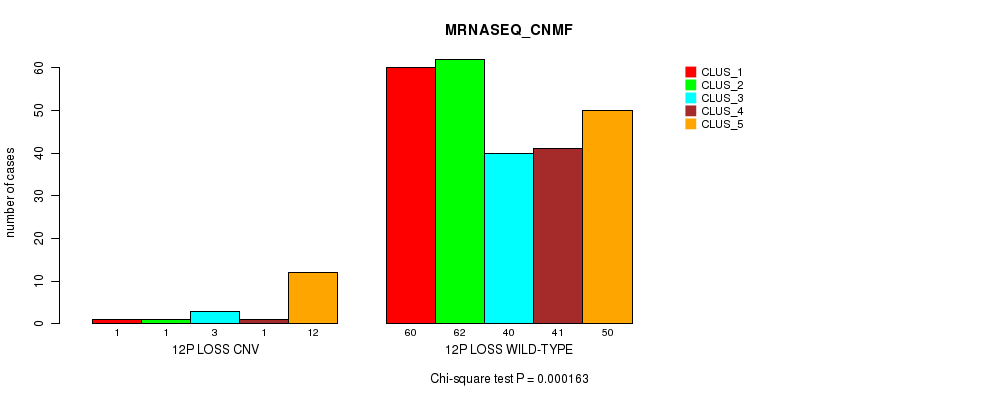
P value = 0.000127 (Fisher's exact test), Q value = 0.086
Table S76. Gene #62: '12p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 141 | 64 |
| 12P LOSS CNV | 1 | 5 | 12 |
| 12P LOSS WILD-TYPE | 65 | 136 | 52 |
Figure S76. Get High-res Image Gene #62: '12p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
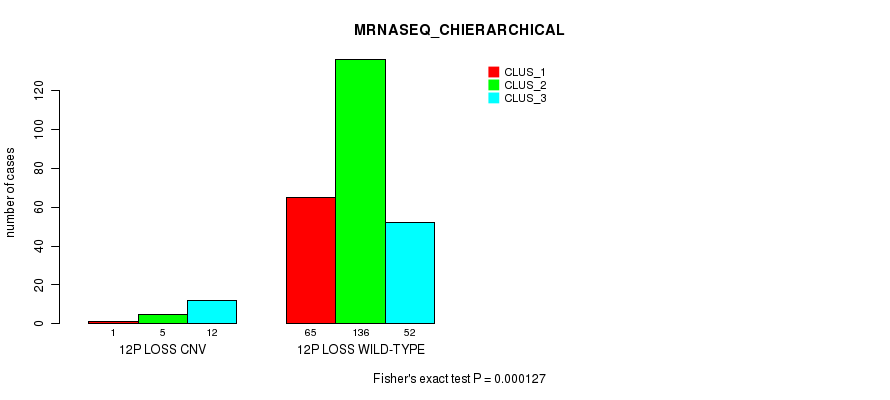
P value = 7.85e-08 (Fisher's exact test), Q value = 5.9e-05
Table S77. Gene #65: '14q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 14Q LOSS CNV | 3 | 1 | 24 |
| 14Q LOSS WILD-TYPE | 147 | 45 | 85 |
Figure S77. Get High-res Image Gene #65: '14q loss' versus Molecular Subtype #1: 'CN_CNMF'
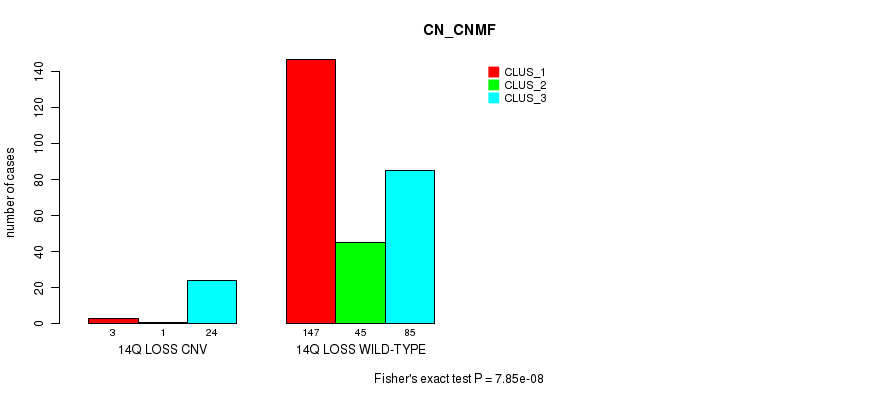
P value = 3.08e-08 (Fisher's exact test), Q value = 2.3e-05
Table S78. Gene #66: '15q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 15Q LOSS CNV | 0 | 7 | 18 |
| 15Q LOSS WILD-TYPE | 150 | 39 | 91 |
Figure S78. Get High-res Image Gene #66: '15q loss' versus Molecular Subtype #1: 'CN_CNMF'
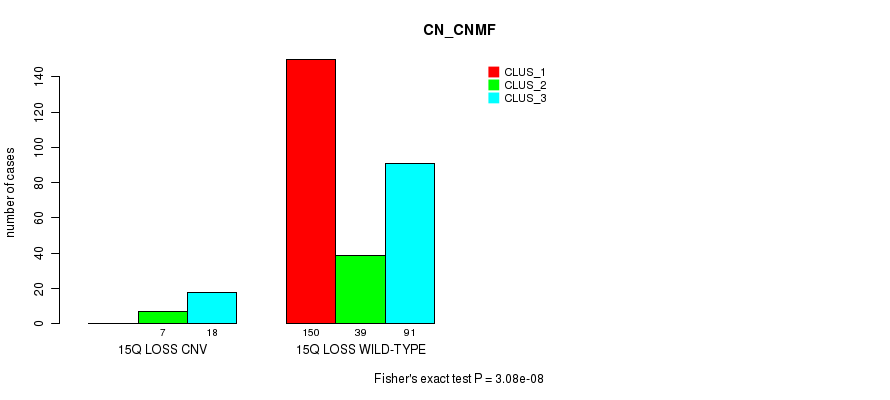
P value = 2.34e-05 (Fisher's exact test), Q value = 0.017
Table S79. Gene #66: '15q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 15Q LOSS CNV | 0 | 7 | 0 | 15 |
| 15Q LOSS WILD-TYPE | 37 | 73 | 65 | 61 |
Figure S79. Get High-res Image Gene #66: '15q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
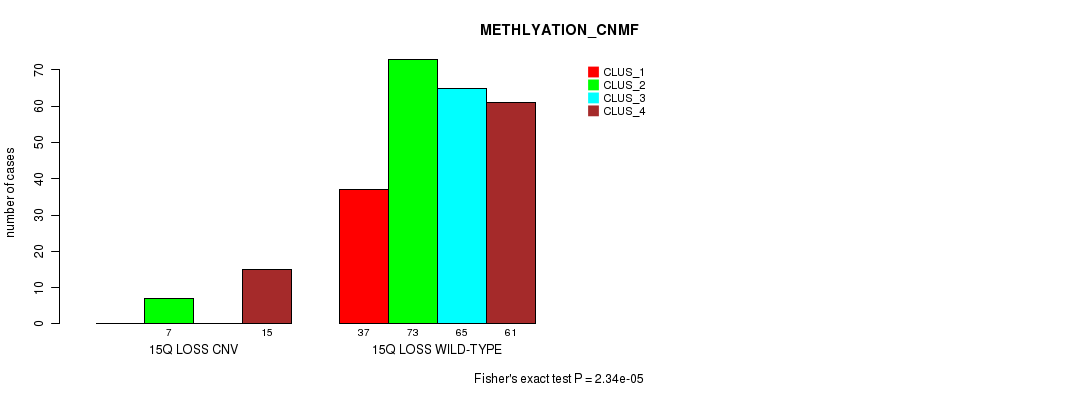
P value = 5.37e-13 (Fisher's exact test), Q value = 4.1e-10
Table S80. Gene #67: '16p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 16P LOSS CNV | 0 | 1 | 28 |
| 16P LOSS WILD-TYPE | 150 | 45 | 81 |
Figure S80. Get High-res Image Gene #67: '16p loss' versus Molecular Subtype #1: 'CN_CNMF'
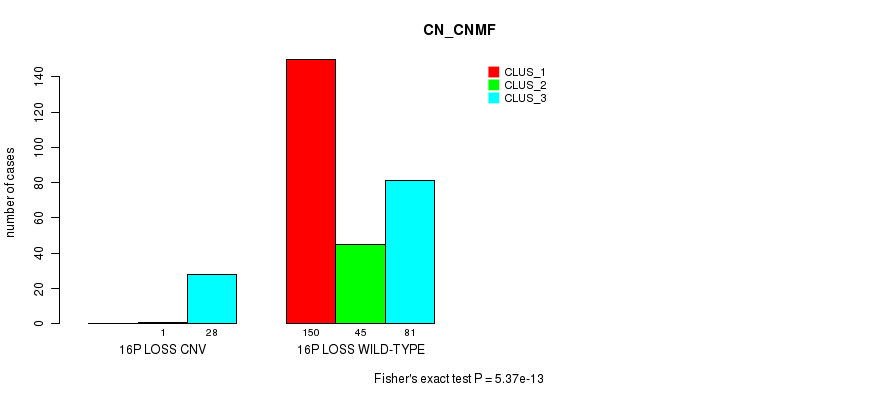
P value = 1.89e-05 (Fisher's exact test), Q value = 0.013
Table S81. Gene #67: '16p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 16P LOSS CNV | 0 | 10 | 0 | 15 |
| 16P LOSS WILD-TYPE | 37 | 70 | 65 | 61 |
Figure S81. Get High-res Image Gene #67: '16p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
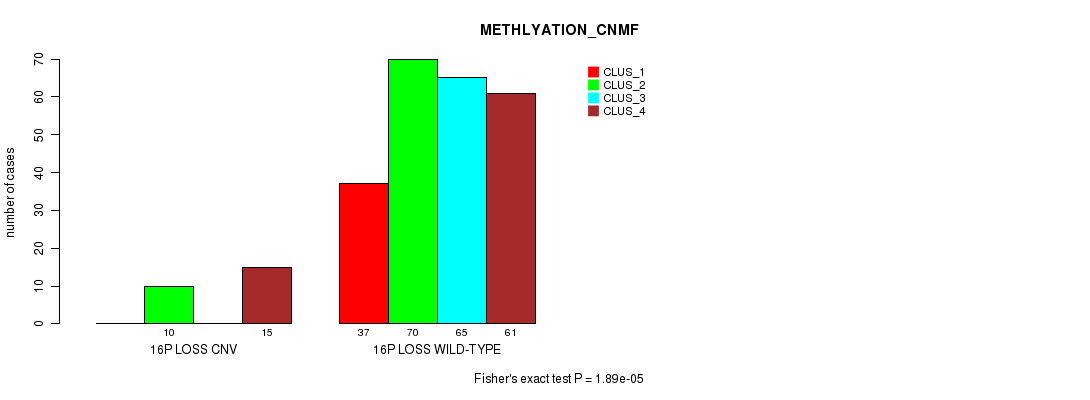
P value = 2.39e-06 (Chi-square test), Q value = 0.0017
Table S82. Gene #67: '16p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 16P LOSS CNV | 3 | 1 | 1 | 4 | 17 |
| 16P LOSS WILD-TYPE | 58 | 62 | 42 | 38 | 45 |
Figure S82. Get High-res Image Gene #67: '16p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
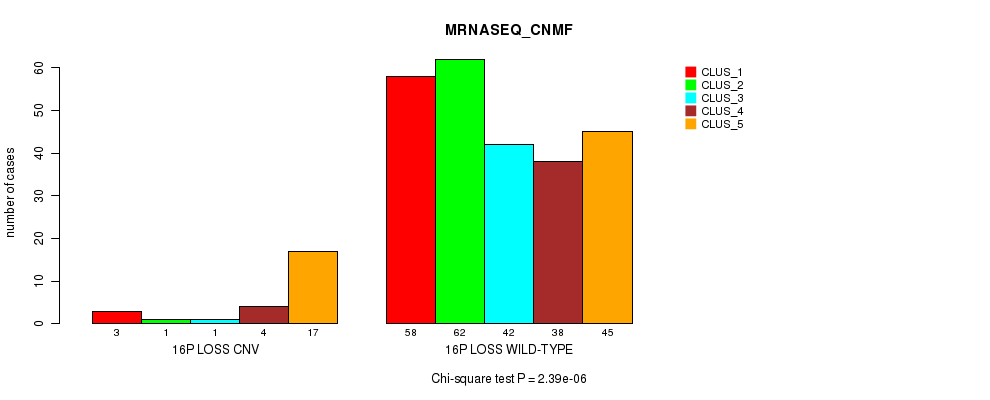
P value = 5.43e-06 (Fisher's exact test), Q value = 0.0039
Table S83. Gene #67: '16p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 141 | 64 |
| 16P LOSS CNV | 2 | 7 | 17 |
| 16P LOSS WILD-TYPE | 64 | 134 | 47 |
Figure S83. Get High-res Image Gene #67: '16p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
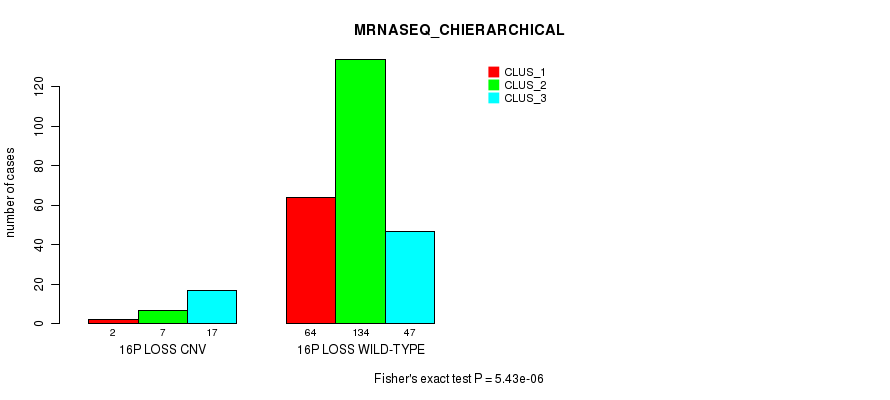
P value = 1.31e-05 (Fisher's exact test), Q value = 0.0094
Table S84. Gene #67: '16p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 91 | 114 |
| 16P LOSS CNV | 6 | 1 | 22 |
| 16P LOSS WILD-TYPE | 94 | 90 | 92 |
Figure S84. Get High-res Image Gene #67: '16p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
P value = 5.43e-05 (Fisher's exact test), Q value = 0.038
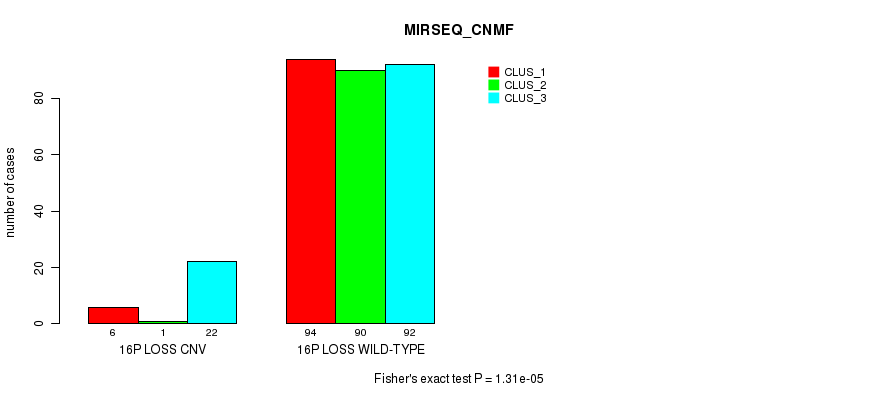
Table S85. Gene #67: '16p loss' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 54 | 78 | 127 |
| 16P LOSS CNV | 2 | 1 | 23 |
| 16P LOSS WILD-TYPE | 52 | 77 | 104 |
Figure S85. Get High-res Image Gene #67: '16p loss' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
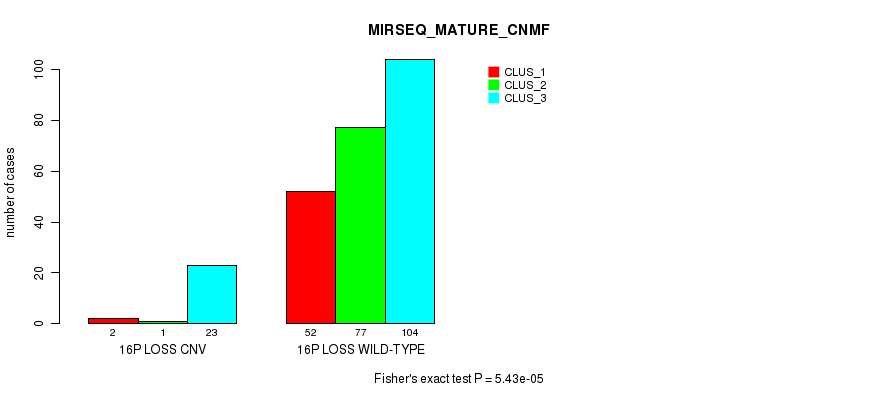
P value = 4.29e-10 (Fisher's exact test), Q value = 3.2e-07
Table S86. Gene #68: '16q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 16Q LOSS CNV | 2 | 2 | 28 |
| 16Q LOSS WILD-TYPE | 148 | 44 | 81 |
Figure S86. Get High-res Image Gene #68: '16q loss' versus Molecular Subtype #1: 'CN_CNMF'
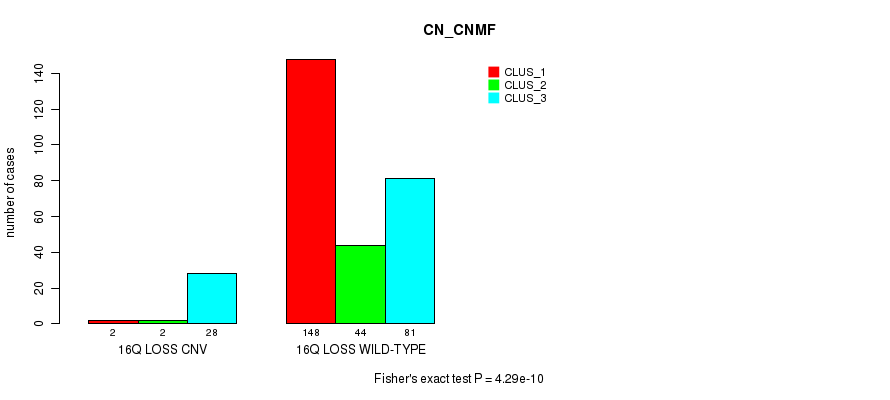
P value = 3.51e-06 (Fisher's exact test), Q value = 0.0026
Table S87. Gene #68: '16q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 16Q LOSS CNV | 0 | 9 | 0 | 17 |
| 16Q LOSS WILD-TYPE | 37 | 71 | 65 | 59 |
Figure S87. Get High-res Image Gene #68: '16q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
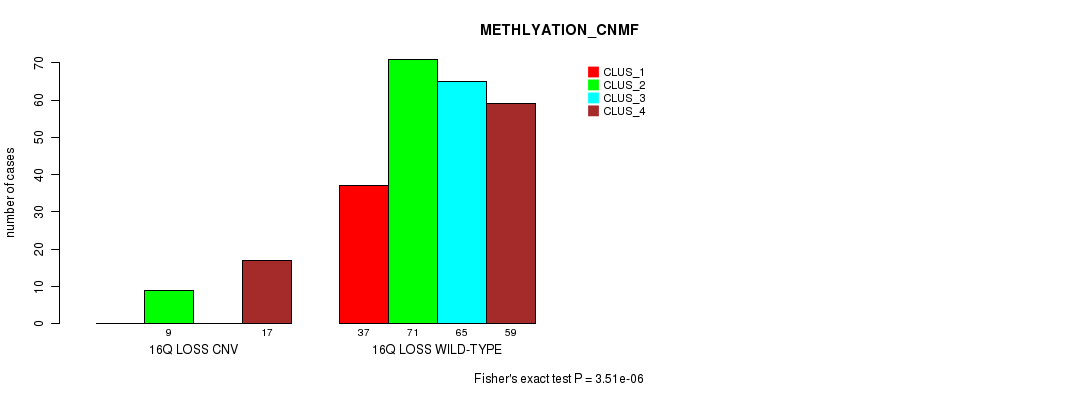
P value = 5.92e-05 (Chi-square test), Q value = 0.041
Table S88. Gene #68: '16q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 16Q LOSS CNV | 3 | 2 | 4 | 3 | 17 |
| 16Q LOSS WILD-TYPE | 58 | 61 | 39 | 39 | 45 |
Figure S88. Get High-res Image Gene #68: '16q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
P value = 6.15e-05 (Fisher's exact test), Q value = 0.042
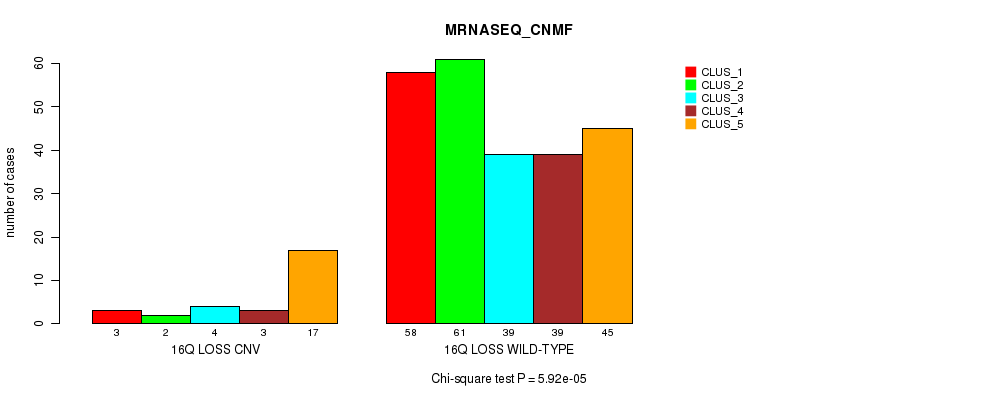
Table S89. Gene #68: '16q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 141 | 64 |
| 16Q LOSS CNV | 3 | 9 | 17 |
| 16Q LOSS WILD-TYPE | 63 | 132 | 47 |
Figure S89. Get High-res Image Gene #68: '16q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
P value = 6.43e-15 (Fisher's exact test), Q value = 4.9e-12
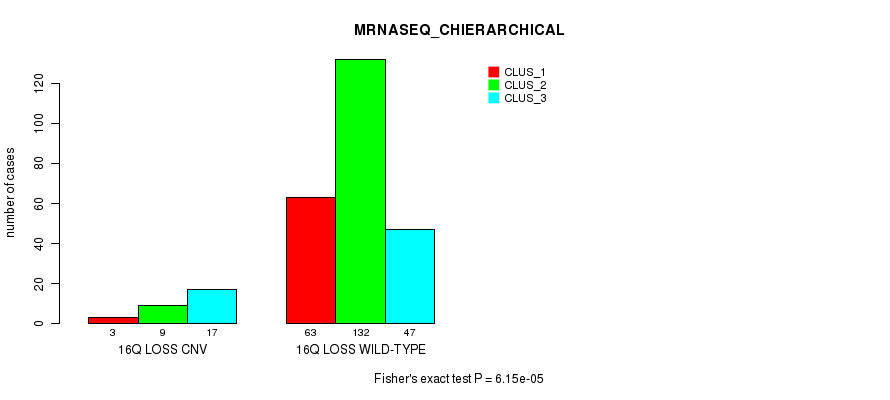
Table S90. Gene #69: '17p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 17P LOSS CNV | 5 | 14 | 45 |
| 17P LOSS WILD-TYPE | 145 | 32 | 64 |
Figure S90. Get High-res Image Gene #69: '17p loss' versus Molecular Subtype #1: 'CN_CNMF'
P value = 8.72e-07 (Fisher's exact test), Q value = 0.00064
Table S91. Gene #69: '17p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 17P LOSS CNV | 2 | 18 | 4 | 30 |
| 17P LOSS WILD-TYPE | 35 | 62 | 61 | 46 |
Figure S91. Get High-res Image Gene #69: '17p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
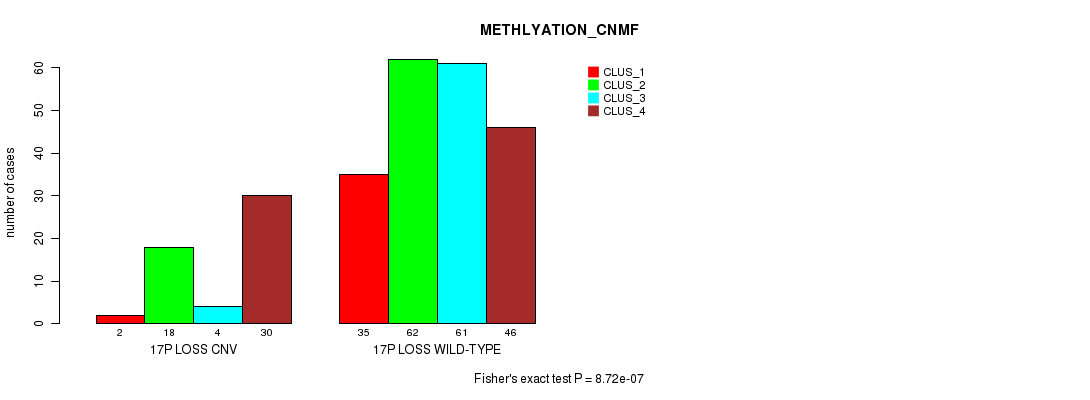
P value = 0.000141 (Chi-square test), Q value = 0.095
Table S92. Gene #69: '17p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 17P LOSS CNV | 6 | 5 | 12 | 8 | 23 |
| 17P LOSS WILD-TYPE | 55 | 58 | 31 | 34 | 39 |
Figure S92. Get High-res Image Gene #69: '17p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
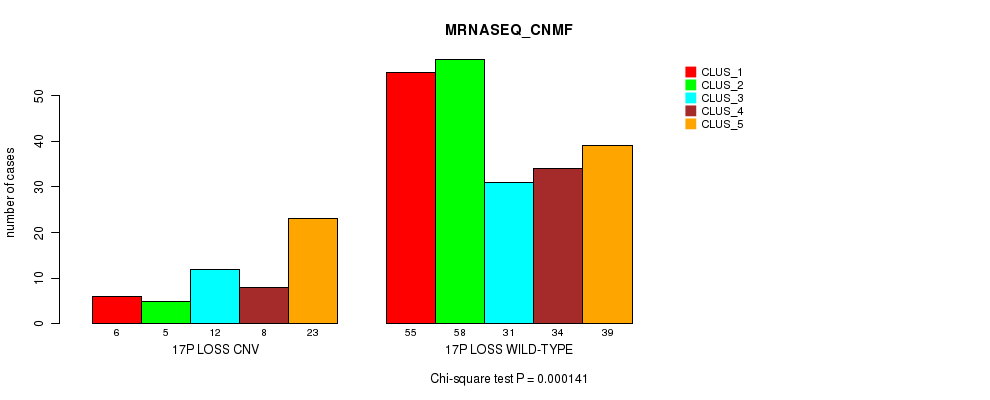
P value = 8.04e-05 (Fisher's exact test), Q value = 0.055
Table S93. Gene #70: '17q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 17Q LOSS CNV | 1 | 5 | 13 |
| 17Q LOSS WILD-TYPE | 149 | 41 | 96 |
Figure S93. Get High-res Image Gene #70: '17q loss' versus Molecular Subtype #1: 'CN_CNMF'
P value = 2.54e-05 (Fisher's exact test), Q value = 0.018
Table S94. Gene #72: '18q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 18Q LOSS CNV | 11 | 12 | 29 |
| 18Q LOSS WILD-TYPE | 139 | 34 | 80 |
Figure S94. Get High-res Image Gene #72: '18q loss' versus Molecular Subtype #1: 'CN_CNMF'
P value = 9.55e-05 (Fisher's exact test), Q value = 0.065
Table S95. Gene #72: '18q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 141 | 64 |
| 18Q LOSS CNV | 2 | 23 | 19 |
| 18Q LOSS WILD-TYPE | 64 | 118 | 45 |
Figure S95. Get High-res Image Gene #72: '18q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
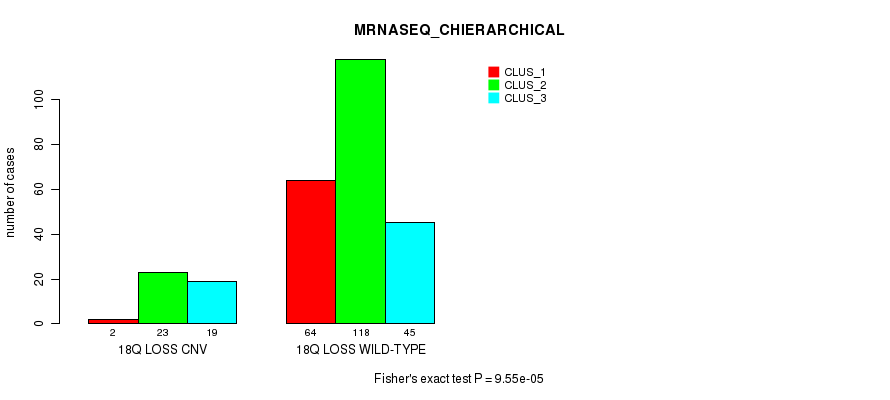
P value = 2.51e-13 (Fisher's exact test), Q value = 1.9e-10
Table S96. Gene #73: '19p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 19P LOSS CNV | 0 | 3 | 30 |
| 19P LOSS WILD-TYPE | 150 | 43 | 79 |
Figure S96. Get High-res Image Gene #73: '19p loss' versus Molecular Subtype #1: 'CN_CNMF'
P value = 4.4e-06 (Fisher's exact test), Q value = 0.0032
Table S97. Gene #73: '19p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 19P LOSS CNV | 0 | 7 | 1 | 19 |
| 19P LOSS WILD-TYPE | 37 | 73 | 64 | 57 |
Figure S97. Get High-res Image Gene #73: '19p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
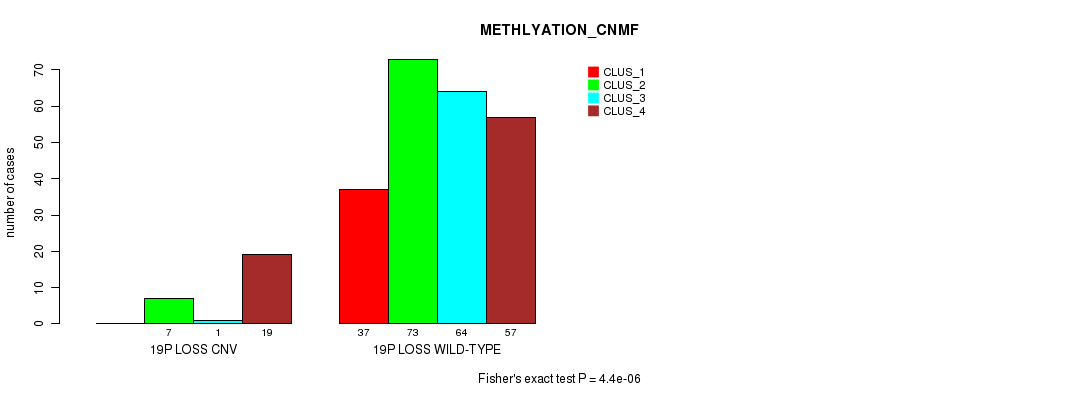
P value = 3.15e-09 (Chi-square test), Q value = 2.4e-06
Table S98. Gene #73: '19p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 19P LOSS CNV | 2 | 1 | 1 | 3 | 20 |
| 19P LOSS WILD-TYPE | 59 | 62 | 42 | 39 | 42 |
Figure S98. Get High-res Image Gene #73: '19p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
P value = 1.24e-05 (Fisher's exact test), Q value = 0.0089
Table S99. Gene #73: '19p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 66 | 141 | 64 |
| 19P LOSS CNV | 2 | 8 | 17 |
| 19P LOSS WILD-TYPE | 64 | 133 | 47 |
Figure S99. Get High-res Image Gene #73: '19p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
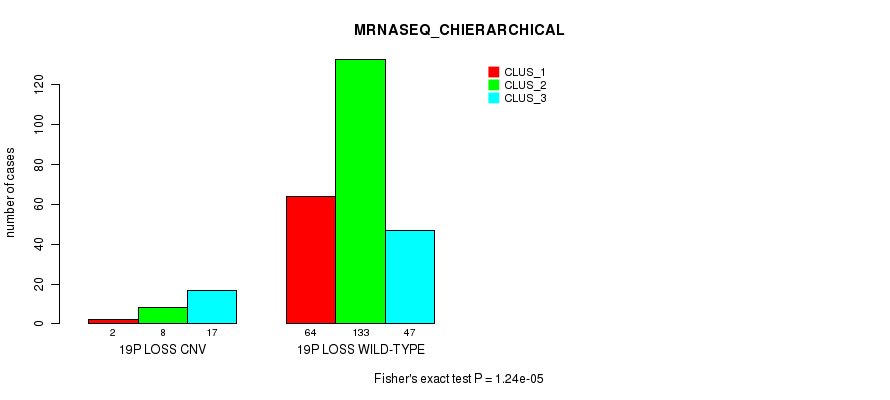
P value = 1.62e-06 (Fisher's exact test), Q value = 0.0012
Table S100. Gene #73: '19p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 133 | 77 | 95 |
| 19P LOSS CNV | 9 | 21 | 3 |
| 19P LOSS WILD-TYPE | 124 | 56 | 92 |
Figure S100. Get High-res Image Gene #73: '19p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
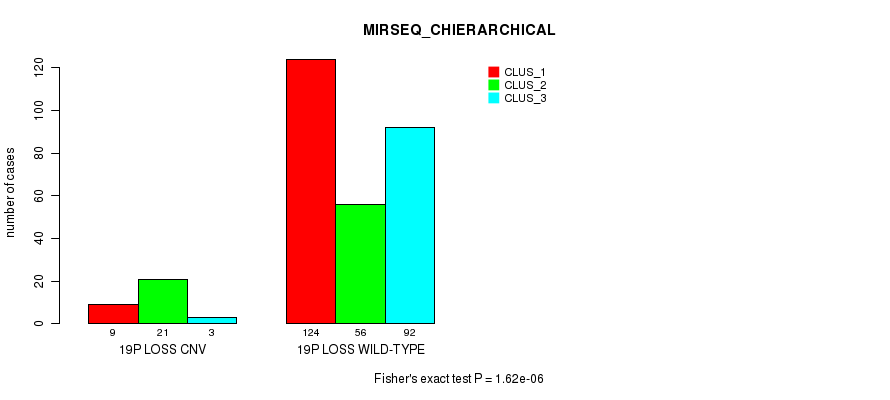
P value = 7.33e-05 (Fisher's exact test), Q value = 0.05
Table S101. Gene #73: '19p loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 57 | 72 | 130 |
| 19P LOSS CNV | 15 | 2 | 10 |
| 19P LOSS WILD-TYPE | 42 | 70 | 120 |
Figure S101. Get High-res Image Gene #73: '19p loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
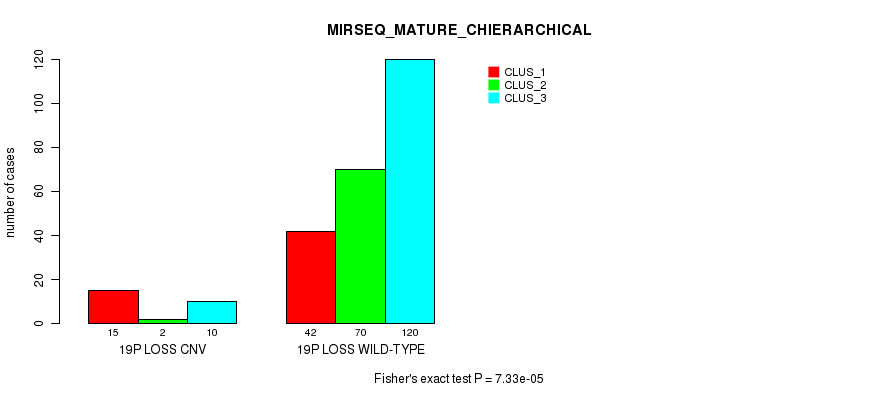
P value = 1.33e-08 (Fisher's exact test), Q value = 1e-05
Table S102. Gene #74: '19q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 19Q LOSS CNV | 0 | 3 | 20 |
| 19Q LOSS WILD-TYPE | 150 | 43 | 89 |
Figure S102. Get High-res Image Gene #74: '19q loss' versus Molecular Subtype #1: 'CN_CNMF'
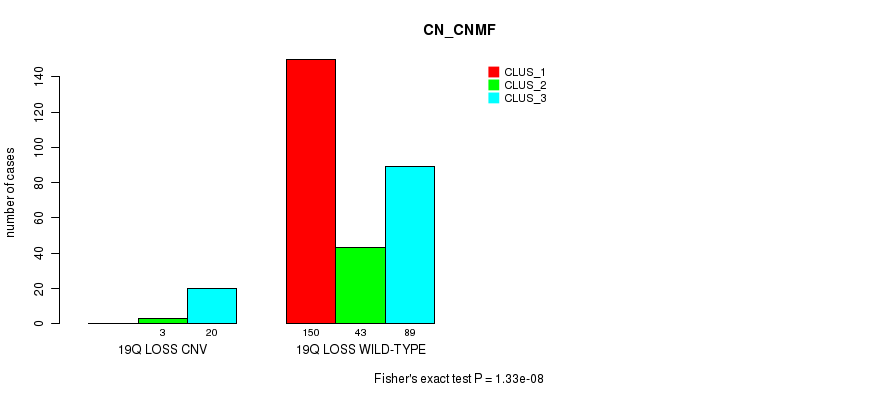
P value = 1.26e-05 (Fisher's exact test), Q value = 0.009
Table S103. Gene #74: '19q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 19Q LOSS CNV | 0 | 2 | 1 | 15 |
| 19Q LOSS WILD-TYPE | 37 | 78 | 64 | 61 |
Figure S103. Get High-res Image Gene #74: '19q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
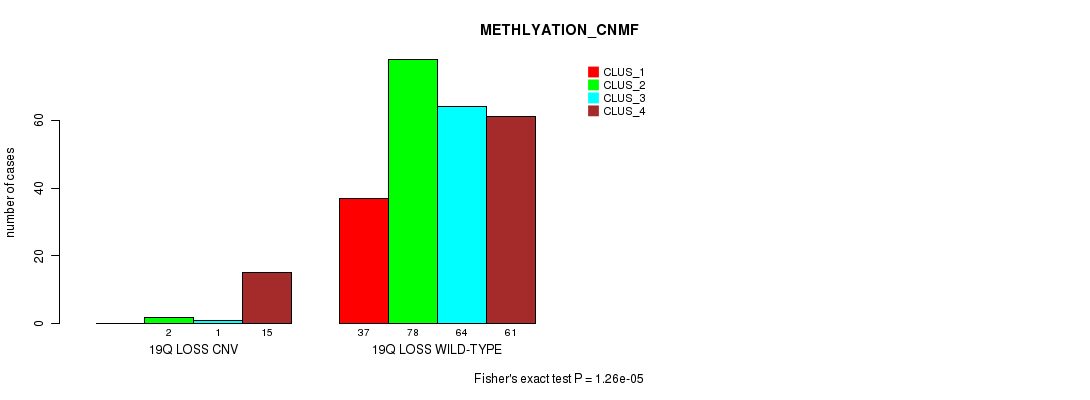
P value = 4.22e-07 (Chi-square test), Q value = 0.00031
Table S104. Gene #74: '19q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 19Q LOSS CNV | 0 | 1 | 1 | 3 | 15 |
| 19Q LOSS WILD-TYPE | 61 | 62 | 42 | 39 | 47 |
Figure S104. Get High-res Image Gene #74: '19q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
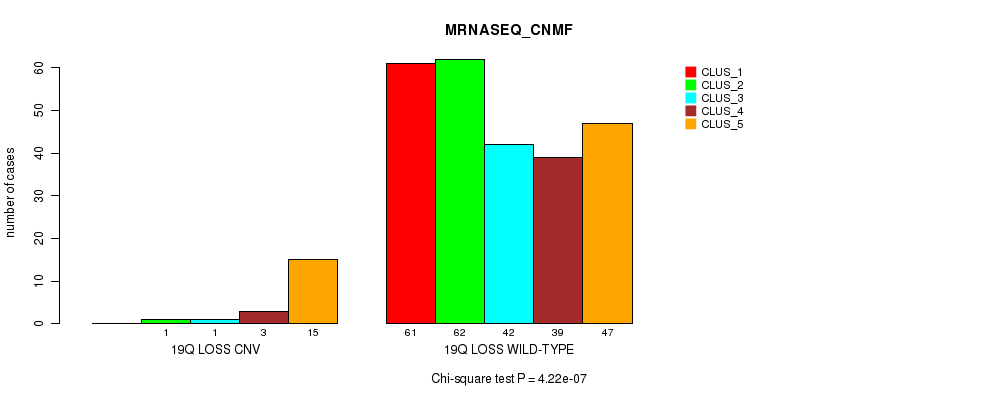
P value = 1.25e-05 (Fisher's exact test), Q value = 0.0089
Table S105. Gene #74: '19q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 133 | 77 | 95 |
| 19Q LOSS CNV | 4 | 16 | 3 |
| 19Q LOSS WILD-TYPE | 129 | 61 | 92 |
Figure S105. Get High-res Image Gene #74: '19q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
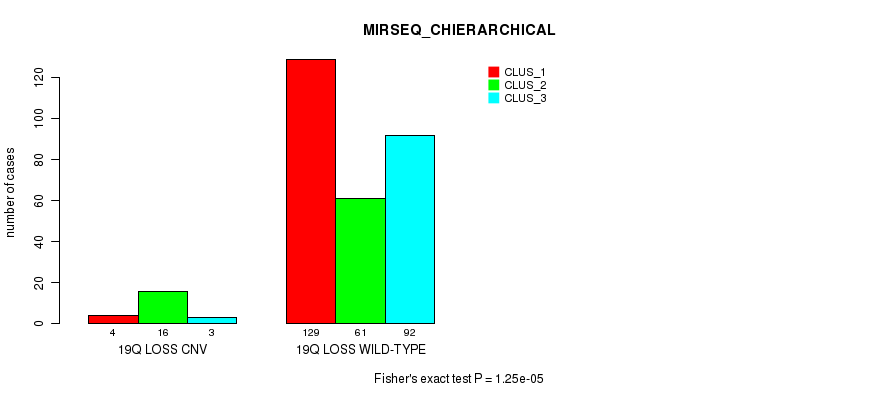
P value = 0.000145 (Fisher's exact test), Q value = 0.098
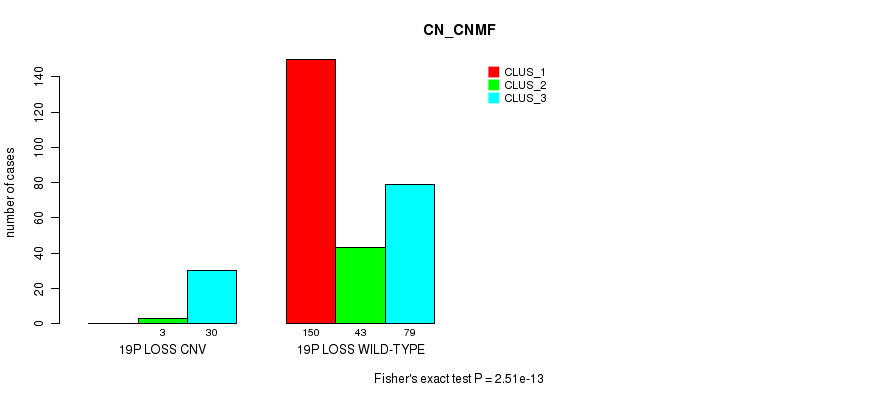
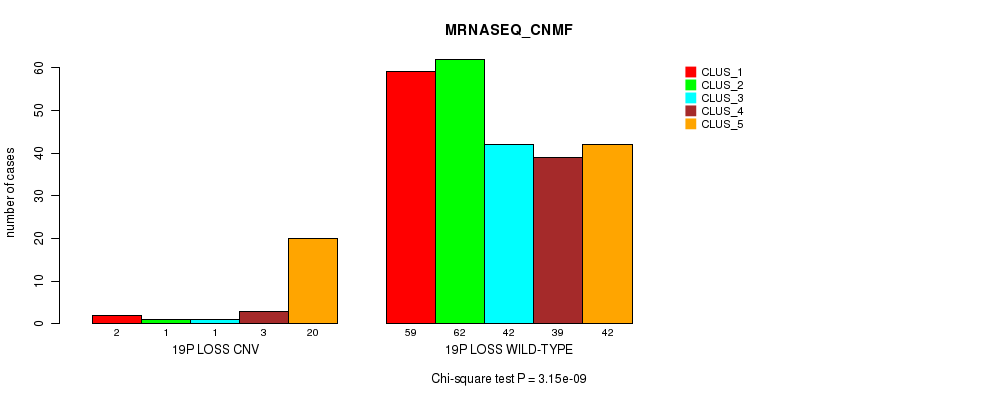
Table S106. Gene #75: '20p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 20P LOSS CNV | 0 | 0 | 9 |
| 20P LOSS WILD-TYPE | 150 | 46 | 100 |
Figure S106. Get High-res Image Gene #75: '20p loss' versus Molecular Subtype #1: 'CN_CNMF'
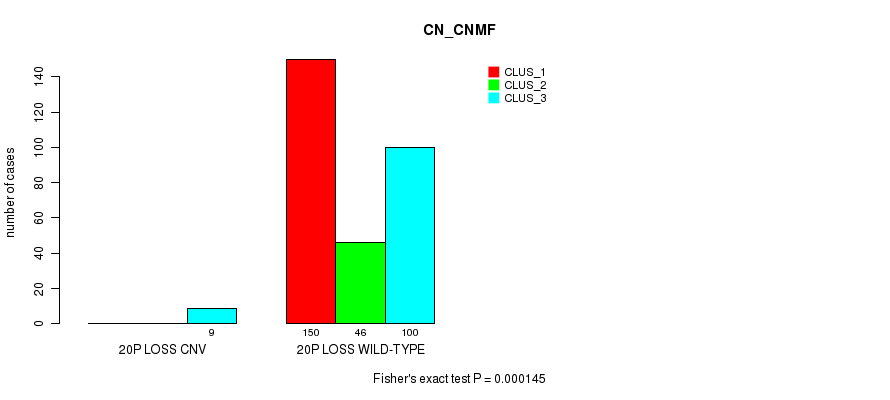
P value = 1.96e-08 (Fisher's exact test), Q value = 1.5e-05
Table S107. Gene #76: '21q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 21Q LOSS CNV | 11 | 10 | 40 |
| 21Q LOSS WILD-TYPE | 139 | 36 | 69 |
Figure S107. Get High-res Image Gene #76: '21q loss' versus Molecular Subtype #1: 'CN_CNMF'
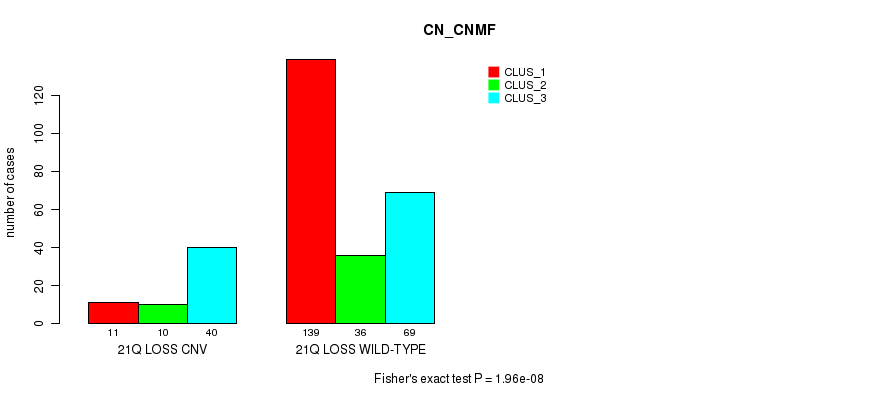
P value = 6.83e-06 (Fisher's exact test), Q value = 0.0049
Table S108. Gene #76: '21q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 21Q LOSS CNV | 1 | 28 | 5 | 18 |
| 21Q LOSS WILD-TYPE | 36 | 52 | 60 | 58 |
Figure S108. Get High-res Image Gene #76: '21q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
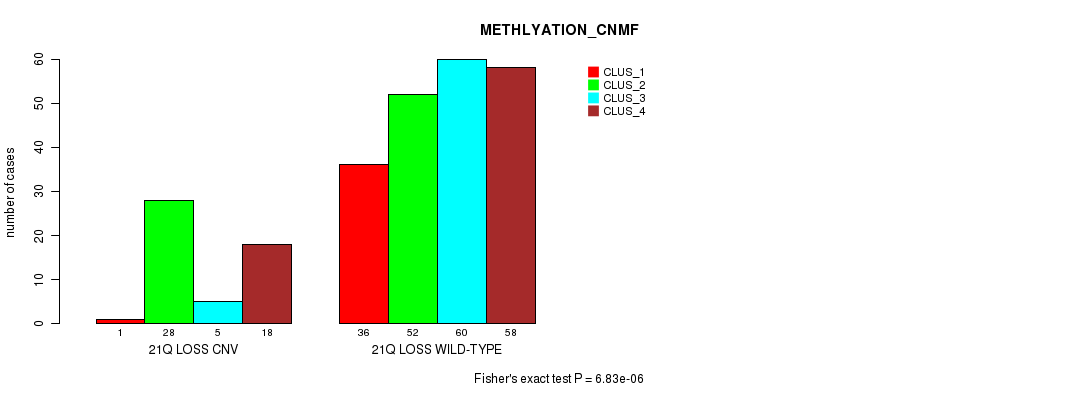
P value = 6.63e-16 (Fisher's exact test), Q value = 5.1e-13
Table S109. Gene #77: '22q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 150 | 46 | 109 |
| 22Q LOSS CNV | 0 | 4 | 36 |
| 22Q LOSS WILD-TYPE | 150 | 42 | 73 |
Figure S109. Get High-res Image Gene #77: '22q loss' versus Molecular Subtype #1: 'CN_CNMF'
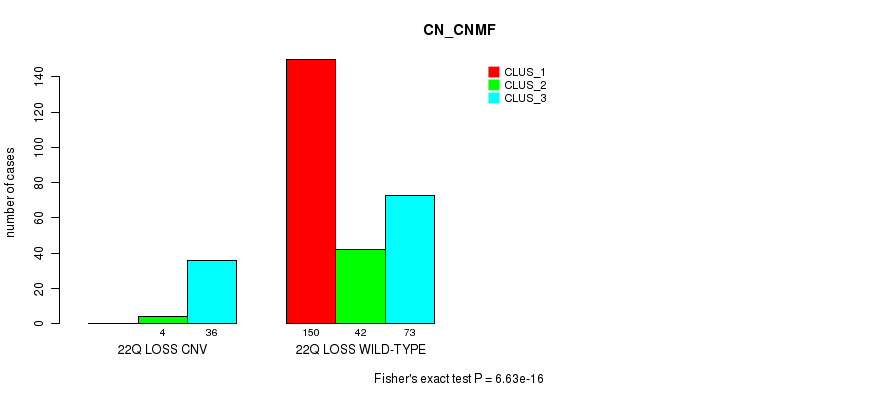
P value = 1.9e-06 (Fisher's exact test), Q value = 0.0014
Table S110. Gene #77: '22q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 80 | 65 | 76 |
| 22Q LOSS CNV | 0 | 9 | 2 | 22 |
| 22Q LOSS WILD-TYPE | 37 | 71 | 63 | 54 |
Figure S110. Get High-res Image Gene #77: '22q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
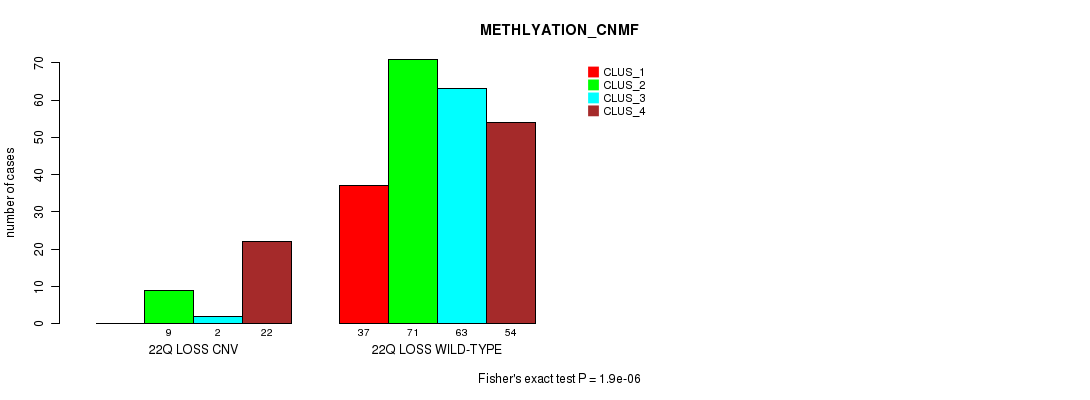
P value = 2.34e-05 (Fisher's exact test), Q value = 0.017
Table S111. Gene #77: '22q loss' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 81 | 90 | 91 |
| 22Q LOSS CNV | 4 | 7 | 25 |
| 22Q LOSS WILD-TYPE | 77 | 83 | 66 |
Figure S111. Get High-res Image Gene #77: '22q loss' versus Molecular Subtype #3: 'RPPA_CNMF'
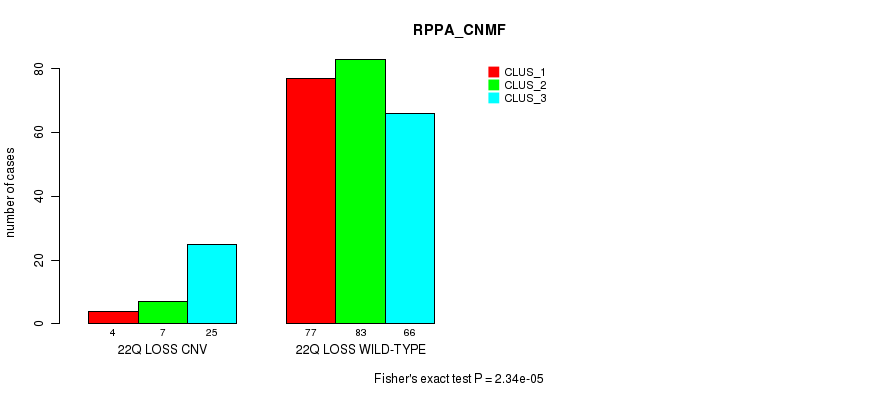
P value = 5.87e-05 (Chi-square test), Q value = 0.041
Table S112. Gene #77: '22q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 61 | 63 | 43 | 42 | 62 |
| 22Q LOSS CNV | 4 | 2 | 5 | 3 | 18 |
| 22Q LOSS WILD-TYPE | 57 | 61 | 38 | 39 | 44 |
Figure S112. Get High-res Image Gene #77: '22q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
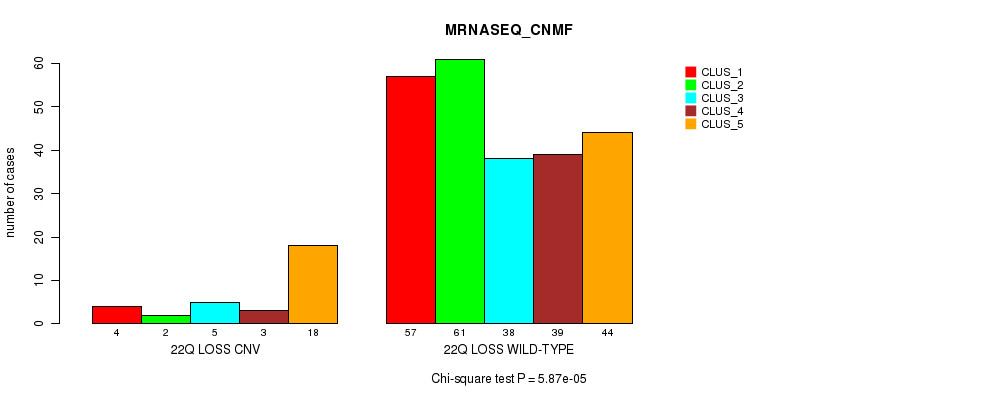
-
Mutation data file = broad_values_by_arm.mutsig.cluster.txt
-
Molecular subtypes file = STAD-TP.transferedmergedcluster.txt
-
Number of patients = 305
-
Number of significantly arm-level cnvs = 78
-
Number of molecular subtypes = 10
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.