This pipeline computes the correlation between significant copy number variation (cnv focal) genes and molecular subtypes.
Testing the association between copy number variation 40 focal events and 8 molecular subtypes across 66 patients, 43 significant findings detected with P value < 0.05 and Q value < 0.25.
-
1p cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
1q cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
2p cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
2q cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
4p cnv correlated to 'MRNASEQ_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
4q cnv correlated to 'MRNASEQ_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
6p cnv correlated to 'METHLYATION_CNMF' and 'MRNASEQ_CNMF'.
-
6q cnv correlated to 'METHLYATION_CNMF' and 'MRNASEQ_CNMF'.
-
8p cnv correlated to 'CN_CNMF'.
-
10p cnv correlated to 'MRNASEQ_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
10q cnv correlated to 'MRNASEQ_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
11q cnv correlated to 'MRNASEQ_CHIERARCHICAL'.
-
13q cnv correlated to 'METHLYATION_CNMF'.
-
16p cnv correlated to 'MRNASEQ_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
16q cnv correlated to 'MRNASEQ_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
17p cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
17q cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 40 focal events and 8 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 43 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Chi-square test | Fisher's exact test | Fisher's exact test | Chi-square test | Fisher's exact test | |
| 17p | 50 (76%) | 16 |
0.299 (1.00) |
4.75e-07 (0.000152) |
5.74e-07 (0.000181) |
5.55e-07 (0.000177) |
0.0506 (1.00) |
0.000396 (0.114) |
0.05 (1.00) |
0.000108 (0.0323) |
| 17q | 50 (76%) | 16 |
0.299 (1.00) |
4.75e-07 (0.000152) |
5.74e-07 (0.000181) |
5.55e-07 (0.000177) |
0.0506 (1.00) |
0.000396 (0.114) |
0.05 (1.00) |
0.000108 (0.0323) |
| 2p | 48 (73%) | 18 |
0.165 (1.00) |
3.89e-05 (0.0119) |
2.21e-06 (0.000694) |
2.68e-06 (0.000836) |
0.119 (1.00) |
0.00103 (0.284) |
0.0242 (1.00) |
0.00018 (0.0531) |
| 2q | 48 (73%) | 18 |
0.165 (1.00) |
3.89e-05 (0.0119) |
2.21e-06 (0.000694) |
2.68e-06 (0.000836) |
0.119 (1.00) |
0.00103 (0.284) |
0.0242 (1.00) |
0.00018 (0.0531) |
| 1p | 54 (82%) | 12 |
0.657 (1.00) |
5.29e-06 (0.00164) |
5.95e-05 (0.018) |
1.16e-05 (0.00358) |
0.315 (1.00) |
0.00742 (1.00) |
0.276 (1.00) |
0.00427 (1.00) |
| 1q | 53 (80%) | 13 |
0.671 (1.00) |
2.22e-05 (0.00684) |
0.000362 (0.105) |
4.71e-05 (0.0143) |
0.422 (1.00) |
0.0113 (1.00) |
0.201 (1.00) |
0.00486 (1.00) |
| 4p | 25 (38%) | 41 |
0.0622 (1.00) |
0.779 (1.00) |
7.61e-05 (0.0229) |
0.000406 (0.116) |
0.941 (1.00) |
0.464 (1.00) |
0.817 (1.00) |
0.0608 (1.00) |
| 4q | 25 (38%) | 41 |
0.0622 (1.00) |
0.779 (1.00) |
7.61e-05 (0.0229) |
0.000406 (0.116) |
0.941 (1.00) |
0.464 (1.00) |
0.817 (1.00) |
0.0608 (1.00) |
| 6p | 52 (79%) | 14 |
0.475 (1.00) |
0.000506 (0.144) |
0.000255 (0.0744) |
0.0021 (0.566) |
0.544 (1.00) |
0.0166 (1.00) |
0.319 (1.00) |
0.00559 (1.00) |
| 6q | 52 (79%) | 14 |
0.475 (1.00) |
0.000506 (0.144) |
0.000255 (0.0744) |
0.0021 (0.566) |
0.544 (1.00) |
0.0166 (1.00) |
0.319 (1.00) |
0.00559 (1.00) |
| 10p | 52 (79%) | 14 |
0.157 (1.00) |
0.00496 (1.00) |
0.000255 (0.0744) |
4.55e-05 (0.0138) |
0.24 (1.00) |
0.0166 (1.00) |
0.0525 (1.00) |
0.00559 (1.00) |
| 10q | 51 (77%) | 15 |
0.33 (1.00) |
0.00128 (0.349) |
0.00018 (0.0531) |
3.39e-05 (0.0104) |
0.355 (1.00) |
0.0233 (1.00) |
0.0872 (1.00) |
0.00491 (1.00) |
| 16p | 25 (38%) | 41 |
0.166 (1.00) |
0.909 (1.00) |
0.000589 (0.166) |
0.000597 (0.167) |
0.367 (1.00) |
0.137 (1.00) |
0.969 (1.00) |
0.111 (1.00) |
| 16q | 26 (39%) | 40 |
0.147 (1.00) |
0.818 (1.00) |
0.000842 (0.234) |
0.00078 (0.217) |
0.611 (1.00) |
0.465 (1.00) |
0.964 (1.00) |
0.214 (1.00) |
| 8p | 26 (39%) | 40 |
9.22e-05 (0.0276) |
0.896 (1.00) |
0.0434 (1.00) |
0.19 (1.00) |
0.918 (1.00) |
0.465 (1.00) |
0.399 (1.00) |
0.351 (1.00) |
| 11q | 22 (33%) | 44 |
0.677 (1.00) |
0.648 (1.00) |
0.00286 (0.767) |
0.000592 (0.166) |
0.202 (1.00) |
0.253 (1.00) |
0.969 (1.00) |
0.0611 (1.00) |
| 13q | 45 (68%) | 21 |
0.344 (1.00) |
0.000206 (0.0603) |
0.00121 (0.332) |
0.0186 (1.00) |
0.548 (1.00) |
0.0242 (1.00) |
0.998 (1.00) |
0.163 (1.00) |
| 3p | 17 (26%) | 49 |
0.362 (1.00) |
0.559 (1.00) |
0.507 (1.00) |
0.428 (1.00) |
0.896 (1.00) |
1 (1.00) |
0.398 (1.00) |
0.421 (1.00) |
| 3q | 16 (24%) | 50 |
0.34 (1.00) |
0.217 (1.00) |
0.346 (1.00) |
0.278 (1.00) |
1 (1.00) |
1 (1.00) |
0.376 (1.00) |
0.362 (1.00) |
| 5p | 18 (27%) | 48 |
0.342 (1.00) |
0.118 (1.00) |
0.0354 (1.00) |
0.0193 (1.00) |
0.549 (1.00) |
0.696 (1.00) |
0.455 (1.00) |
0.136 (1.00) |
| 5q | 18 (27%) | 48 |
0.342 (1.00) |
0.118 (1.00) |
0.0354 (1.00) |
0.0193 (1.00) |
0.549 (1.00) |
0.696 (1.00) |
0.455 (1.00) |
0.136 (1.00) |
| 7p | 25 (38%) | 41 |
0.166 (1.00) |
0.909 (1.00) |
0.00579 (1.00) |
0.0294 (1.00) |
0.652 (1.00) |
1 (1.00) |
0.78 (1.00) |
0.314 (1.00) |
| 7q | 25 (38%) | 41 |
0.166 (1.00) |
0.909 (1.00) |
0.00579 (1.00) |
0.0294 (1.00) |
0.652 (1.00) |
1 (1.00) |
0.78 (1.00) |
0.314 (1.00) |
| 8q | 26 (39%) | 40 |
0.00091 (0.252) |
0.896 (1.00) |
0.0194 (1.00) |
0.0845 (1.00) |
0.918 (1.00) |
0.465 (1.00) |
0.791 (1.00) |
0.102 (1.00) |
| 9p | 20 (30%) | 46 |
0.445 (1.00) |
1 (1.00) |
0.291 (1.00) |
0.739 (1.00) |
0.722 (1.00) |
0.437 (1.00) |
0.228 (1.00) |
0.736 (1.00) |
| 9q | 20 (30%) | 46 |
0.445 (1.00) |
1 (1.00) |
0.291 (1.00) |
0.739 (1.00) |
0.722 (1.00) |
0.437 (1.00) |
0.228 (1.00) |
0.736 (1.00) |
| 11p | 22 (33%) | 44 |
0.677 (1.00) |
0.648 (1.00) |
0.0184 (1.00) |
0.00623 (1.00) |
0.503 (1.00) |
0.253 (1.00) |
0.845 (1.00) |
0.206 (1.00) |
| 12p | 21 (32%) | 45 |
0.344 (1.00) |
0.565 (1.00) |
0.131 (1.00) |
0.219 (1.00) |
0.929 (1.00) |
0.45 (1.00) |
0.678 (1.00) |
0.0822 (1.00) |
| 12q | 21 (32%) | 45 |
0.409 (1.00) |
0.394 (1.00) |
0.167 (1.00) |
0.0723 (1.00) |
0.829 (1.00) |
0.45 (1.00) |
0.91 (1.00) |
0.182 (1.00) |
| 14q | 23 (35%) | 43 |
0.151 (1.00) |
0.949 (1.00) |
0.00985 (1.00) |
0.0101 (1.00) |
0.906 (1.00) |
1 (1.00) |
0.989 (1.00) |
0.217 (1.00) |
| 15q | 23 (35%) | 43 |
0.151 (1.00) |
0.949 (1.00) |
0.00937 (1.00) |
0.0282 (1.00) |
0.906 (1.00) |
0.478 (1.00) |
0.946 (1.00) |
0.339 (1.00) |
| 18p | 25 (38%) | 41 |
0.799 (1.00) |
0.575 (1.00) |
0.00476 (1.00) |
0.00369 (0.974) |
0.576 (1.00) |
0.721 (1.00) |
0.377 (1.00) |
0.098 (1.00) |
| 18q | 26 (39%) | 40 |
0.677 (1.00) |
0.469 (1.00) |
0.0165 (1.00) |
0.00963 (1.00) |
0.401 (1.00) |
0.73 (1.00) |
0.237 (1.00) |
0.156 (1.00) |
| 19p | 20 (30%) | 46 |
0.445 (1.00) |
0.504 (1.00) |
0.00939 (1.00) |
0.00346 (0.919) |
0.697 (1.00) |
0.712 (1.00) |
0.851 (1.00) |
0.0993 (1.00) |
| 19q | 20 (30%) | 46 |
0.445 (1.00) |
0.504 (1.00) |
0.00939 (1.00) |
0.00346 (0.919) |
0.697 (1.00) |
0.712 (1.00) |
0.851 (1.00) |
0.0993 (1.00) |
| 20p | 24 (36%) | 42 |
0.68 (1.00) |
0.673 (1.00) |
0.0999 (1.00) |
0.0354 (1.00) |
0.921 (1.00) |
0.469 (1.00) |
0.639 (1.00) |
0.851 (1.00) |
| 20q | 24 (36%) | 42 |
0.23 (1.00) |
0.312 (1.00) |
0.179 (1.00) |
0.057 (1.00) |
0.545 (1.00) |
1 (1.00) |
0.388 (1.00) |
0.687 (1.00) |
| 21q | 39 (59%) | 27 |
0.694 (1.00) |
0.0787 (1.00) |
0.461 (1.00) |
0.215 (1.00) |
1 (1.00) |
1 (1.00) |
0.96 (1.00) |
0.373 (1.00) |
| 22q | 27 (41%) | 39 |
0.776 (1.00) |
0.677 (1.00) |
0.00172 (0.466) |
0.00159 (0.432) |
0.81 (1.00) |
0.29 (1.00) |
0.912 (1.00) |
0.296 (1.00) |
| xq | 45 (68%) | 21 |
1 (1.00) |
0.188 (1.00) |
0.0471 (1.00) |
0.0188 (1.00) |
0.0322 (1.00) |
0.00337 (0.899) |
0.00606 (1.00) |
0.0446 (1.00) |
P value = 5.29e-06 (Fisher's exact test), Q value = 0.0016
Table S1. Gene #1: '1p' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 35 | 13 |
| 1P MUTATED | 17 | 33 | 4 |
| 1P WILD-TYPE | 1 | 2 | 9 |
Figure S1. Get High-res Image Gene #1: '1p' versus Molecular Subtype #2: 'METHLYATION_CNMF'
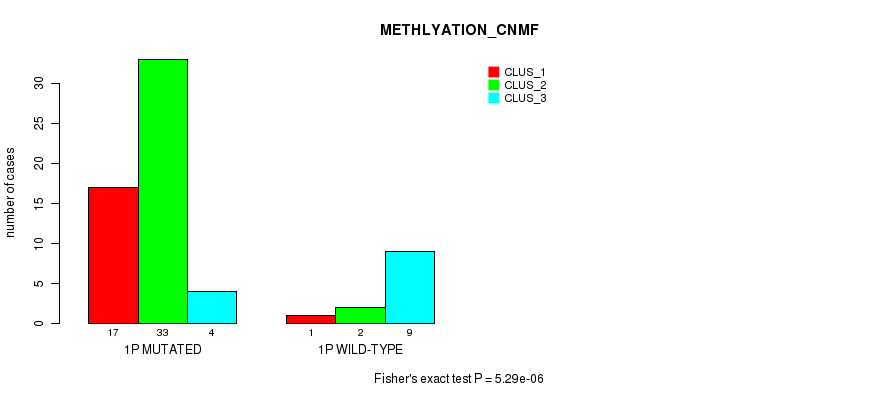
P value = 5.95e-05 (Fisher's exact test), Q value = 0.018
Table S2. Gene #1: '1p' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 1P MUTATED | 19 | 16 | 15 | 4 |
| 1P WILD-TYPE | 0 | 6 | 0 | 6 |
Figure S2. Get High-res Image Gene #1: '1p' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
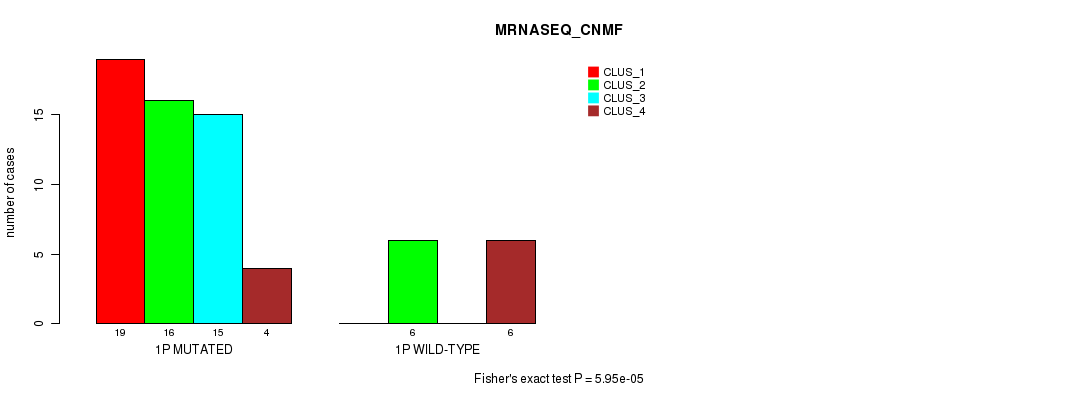
P value = 1.16e-05 (Chi-square test), Q value = 0.0036
Table S3. Gene #1: '1p' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 1P MUTATED | 10 | 3 | 12 | 6 | 23 |
| 1P WILD-TYPE | 0 | 7 | 5 | 0 | 0 |
Figure S3. Get High-res Image Gene #1: '1p' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
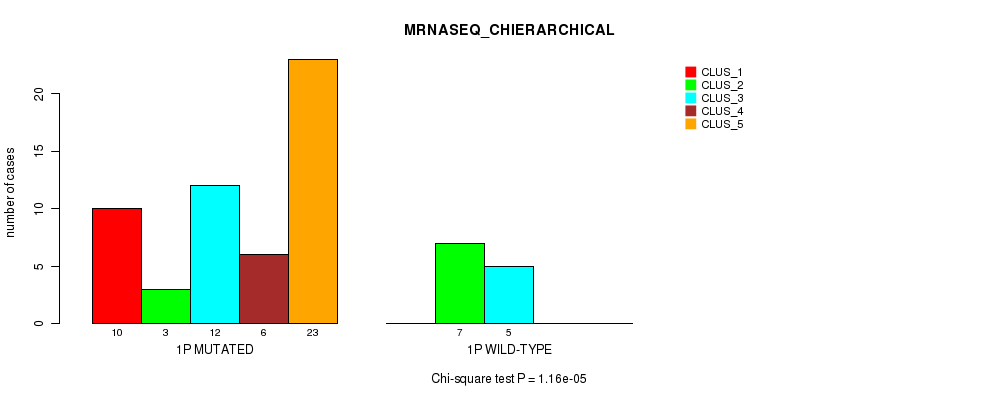
P value = 2.22e-05 (Fisher's exact test), Q value = 0.0068
Table S4. Gene #2: '1q' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 35 | 13 |
| 1Q MUTATED | 17 | 32 | 4 |
| 1Q WILD-TYPE | 1 | 3 | 9 |
Figure S4. Get High-res Image Gene #2: '1q' versus Molecular Subtype #2: 'METHLYATION_CNMF'
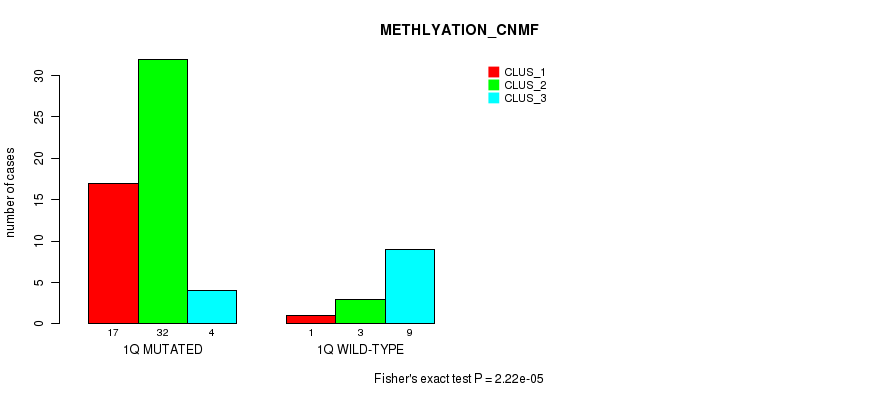
P value = 0.000362 (Fisher's exact test), Q value = 0.1
Table S5. Gene #2: '1q' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 1Q MUTATED | 19 | 16 | 14 | 4 |
| 1Q WILD-TYPE | 0 | 6 | 1 | 6 |
Figure S5. Get High-res Image Gene #2: '1q' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
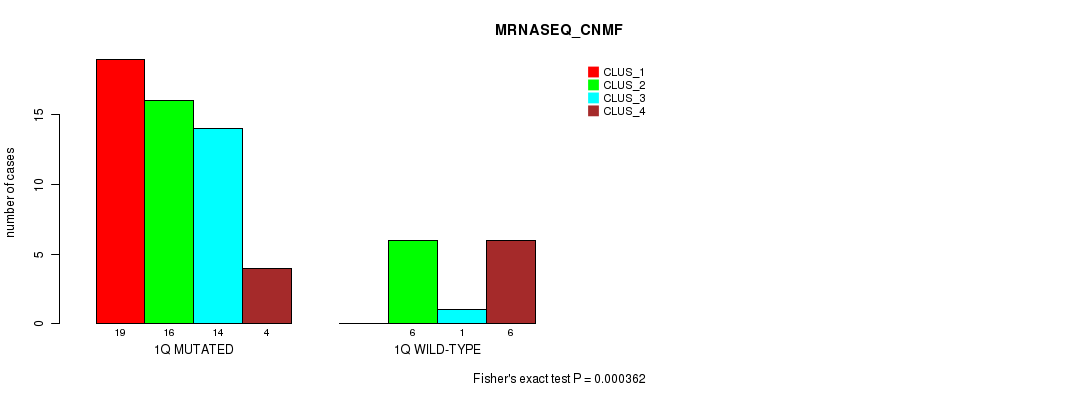
P value = 4.71e-05 (Chi-square test), Q value = 0.014
Table S6. Gene #2: '1q' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 1Q MUTATED | 10 | 3 | 12 | 5 | 23 |
| 1Q WILD-TYPE | 0 | 7 | 5 | 1 | 0 |
Figure S6. Get High-res Image Gene #2: '1q' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
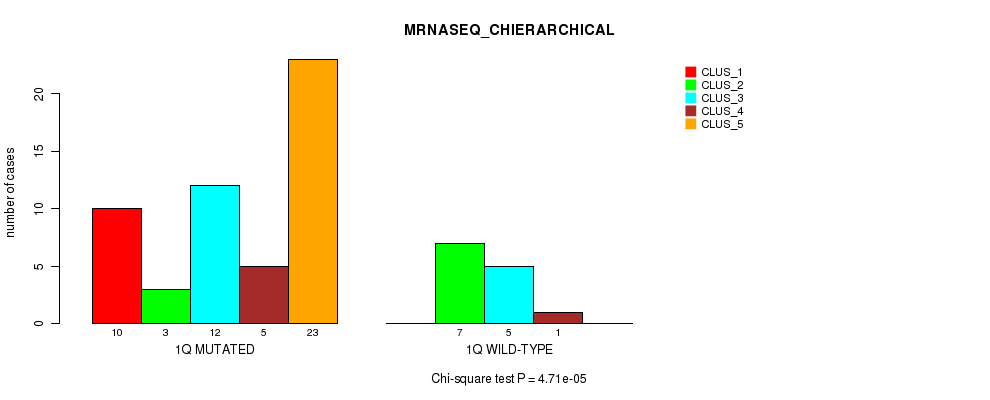
P value = 3.89e-05 (Fisher's exact test), Q value = 0.012
Table S7. Gene #3: '2p' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 35 | 13 |
| 2P MUTATED | 17 | 28 | 3 |
| 2P WILD-TYPE | 1 | 7 | 10 |
Figure S7. Get High-res Image Gene #3: '2p' versus Molecular Subtype #2: 'METHLYATION_CNMF'
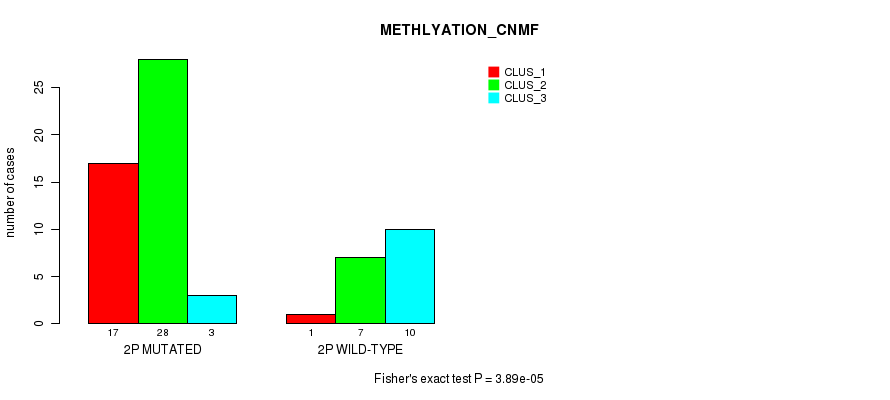
P value = 2.21e-06 (Fisher's exact test), Q value = 0.00069
Table S8. Gene #3: '2p' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 2P MUTATED | 19 | 15 | 13 | 1 |
| 2P WILD-TYPE | 0 | 7 | 2 | 9 |
Figure S8. Get High-res Image Gene #3: '2p' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
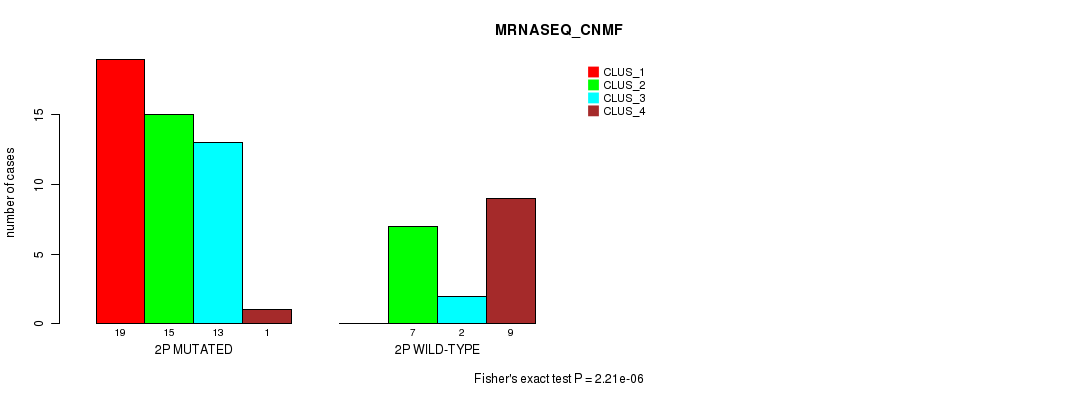
P value = 2.68e-06 (Chi-square test), Q value = 0.00084
Table S9. Gene #3: '2p' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 2P MUTATED | 10 | 1 | 12 | 3 | 22 |
| 2P WILD-TYPE | 0 | 9 | 5 | 3 | 1 |
Figure S9. Get High-res Image Gene #3: '2p' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
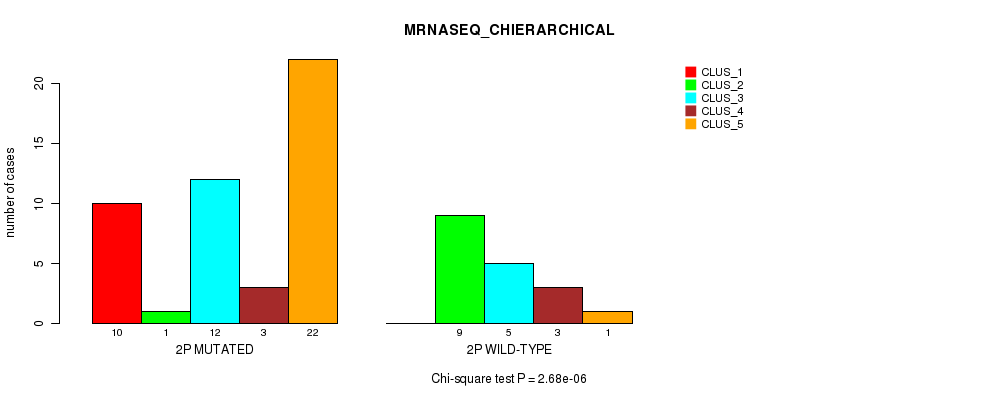
P value = 0.00018 (Fisher's exact test), Q value = 0.053
Table S10. Gene #3: '2p' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 12 | 8 | 25 | 21 |
| 2P MUTATED | 4 | 5 | 18 | 21 |
| 2P WILD-TYPE | 8 | 3 | 7 | 0 |
Figure S10. Get High-res Image Gene #3: '2p' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
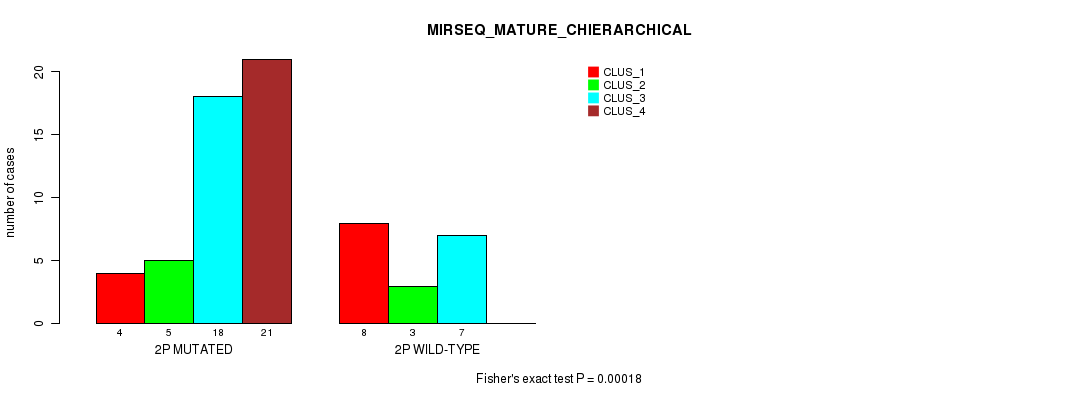
P value = 3.89e-05 (Fisher's exact test), Q value = 0.012
Table S11. Gene #4: '2q' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 35 | 13 |
| 2Q MUTATED | 17 | 28 | 3 |
| 2Q WILD-TYPE | 1 | 7 | 10 |
Figure S11. Get High-res Image Gene #4: '2q' versus Molecular Subtype #2: 'METHLYATION_CNMF'
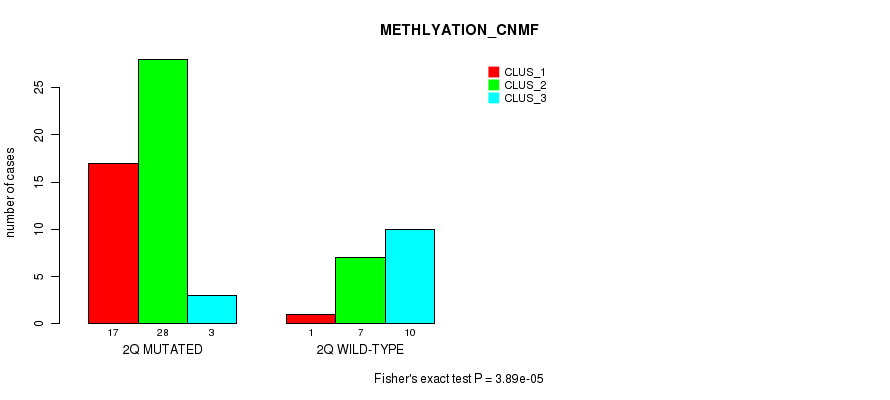
P value = 2.21e-06 (Fisher's exact test), Q value = 0.00069
Table S12. Gene #4: '2q' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 2Q MUTATED | 19 | 15 | 13 | 1 |
| 2Q WILD-TYPE | 0 | 7 | 2 | 9 |
Figure S12. Get High-res Image Gene #4: '2q' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
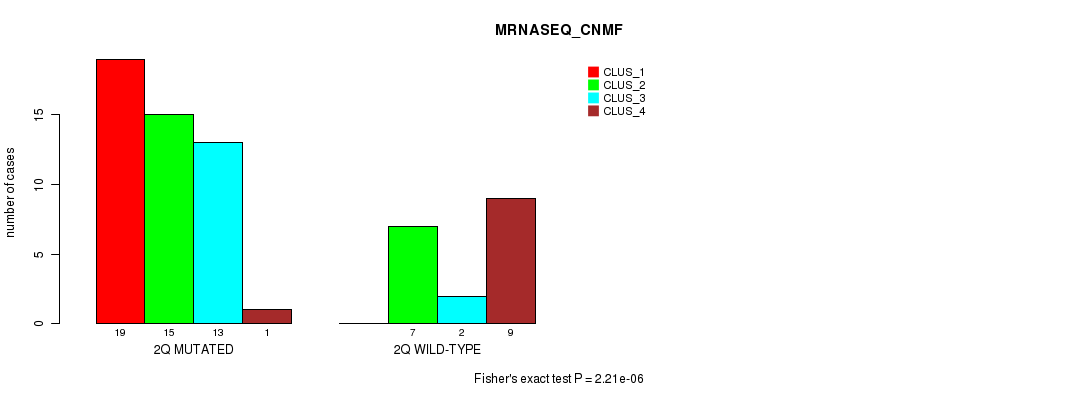
P value = 2.68e-06 (Chi-square test), Q value = 0.00084
Table S13. Gene #4: '2q' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 2Q MUTATED | 10 | 1 | 12 | 3 | 22 |
| 2Q WILD-TYPE | 0 | 9 | 5 | 3 | 1 |
Figure S13. Get High-res Image Gene #4: '2q' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
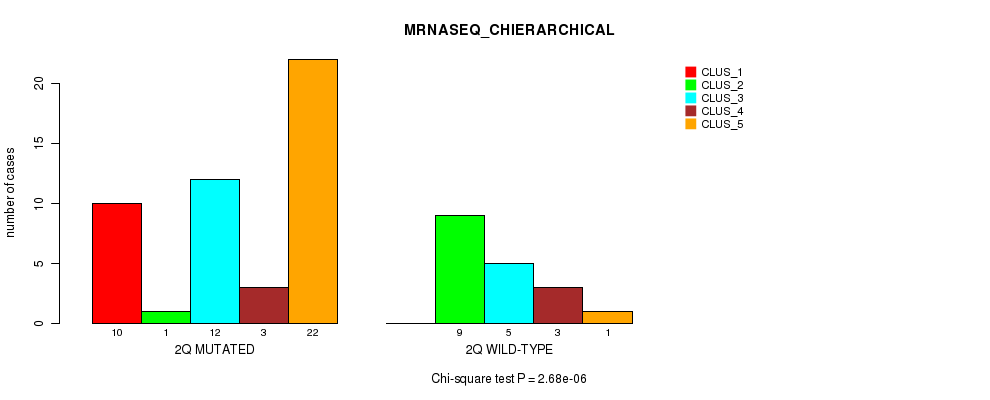
P value = 0.00018 (Fisher's exact test), Q value = 0.053
Table S14. Gene #4: '2q' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 12 | 8 | 25 | 21 |
| 2Q MUTATED | 4 | 5 | 18 | 21 |
| 2Q WILD-TYPE | 8 | 3 | 7 | 0 |
Figure S14. Get High-res Image Gene #4: '2q' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
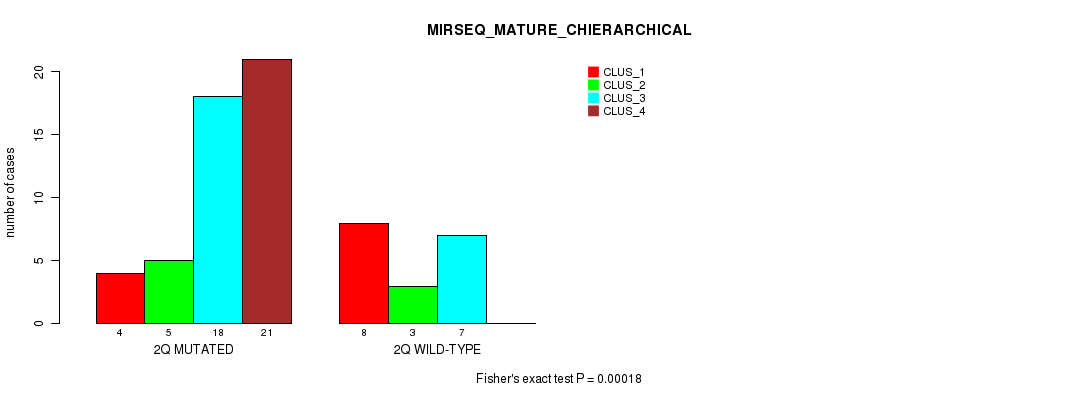
P value = 7.61e-05 (Fisher's exact test), Q value = 0.023
Table S15. Gene #7: '4p' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 4P MUTATED | 4 | 17 | 2 | 2 |
| 4P WILD-TYPE | 15 | 5 | 13 | 8 |
Figure S15. Get High-res Image Gene #7: '4p' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
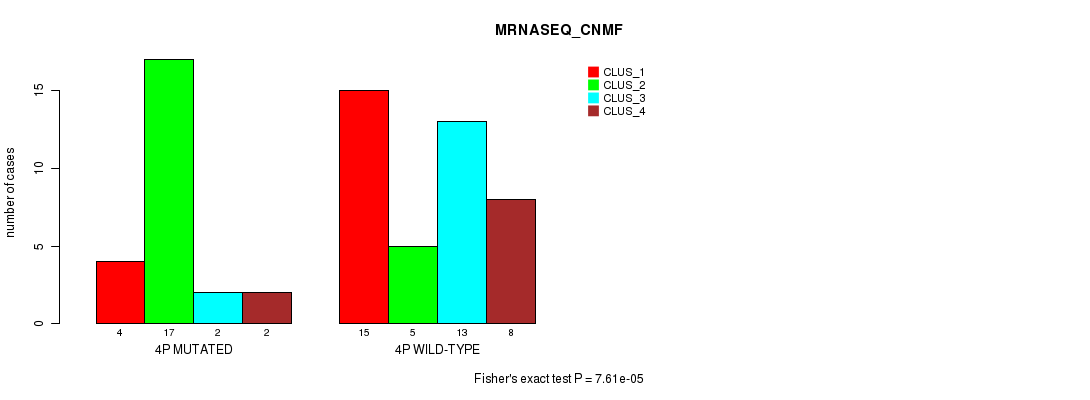
P value = 0.000406 (Chi-square test), Q value = 0.12
Table S16. Gene #7: '4p' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 4P MUTATED | 1 | 3 | 14 | 2 | 5 |
| 4P WILD-TYPE | 9 | 7 | 3 | 4 | 18 |
Figure S16. Get High-res Image Gene #7: '4p' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
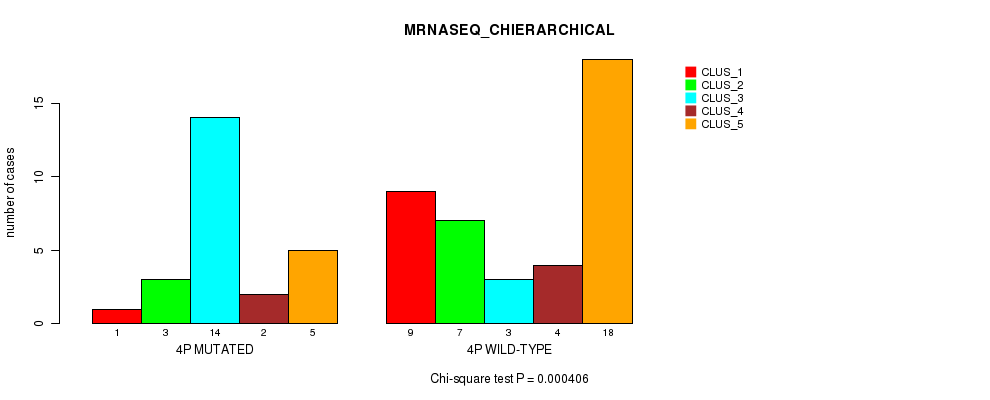
P value = 7.61e-05 (Fisher's exact test), Q value = 0.023
Table S17. Gene #8: '4q' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 4Q MUTATED | 4 | 17 | 2 | 2 |
| 4Q WILD-TYPE | 15 | 5 | 13 | 8 |
Figure S17. Get High-res Image Gene #8: '4q' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
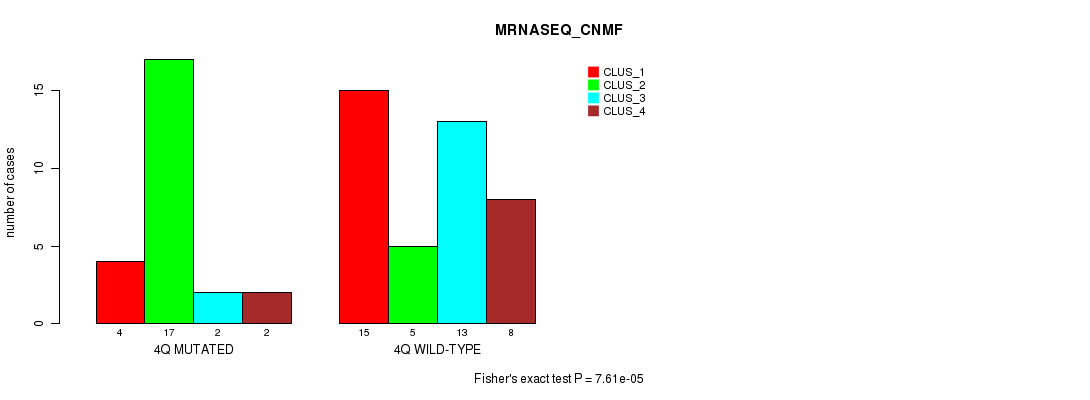
P value = 0.000406 (Chi-square test), Q value = 0.12
Table S18. Gene #8: '4q' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 4Q MUTATED | 1 | 3 | 14 | 2 | 5 |
| 4Q WILD-TYPE | 9 | 7 | 3 | 4 | 18 |
Figure S18. Get High-res Image Gene #8: '4q' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
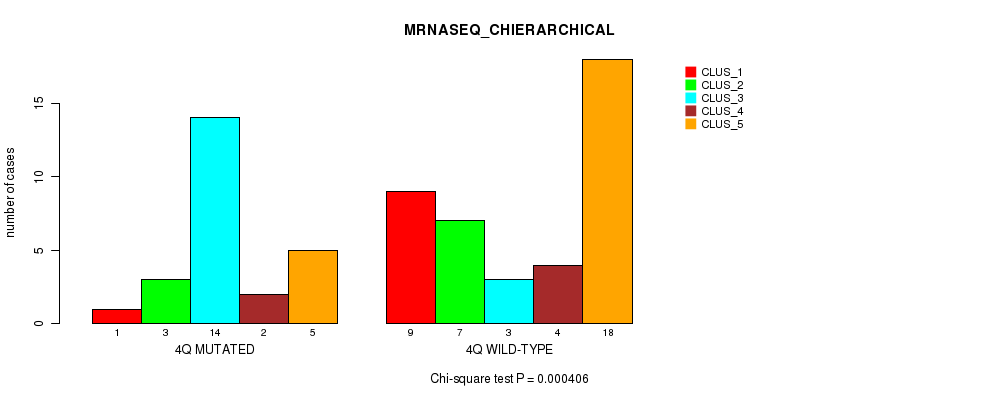
P value = 0.000506 (Fisher's exact test), Q value = 0.14
Table S19. Gene #11: '6p' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 35 | 13 |
| 6P MUTATED | 17 | 30 | 5 |
| 6P WILD-TYPE | 1 | 5 | 8 |
Figure S19. Get High-res Image Gene #11: '6p' versus Molecular Subtype #2: 'METHLYATION_CNMF'
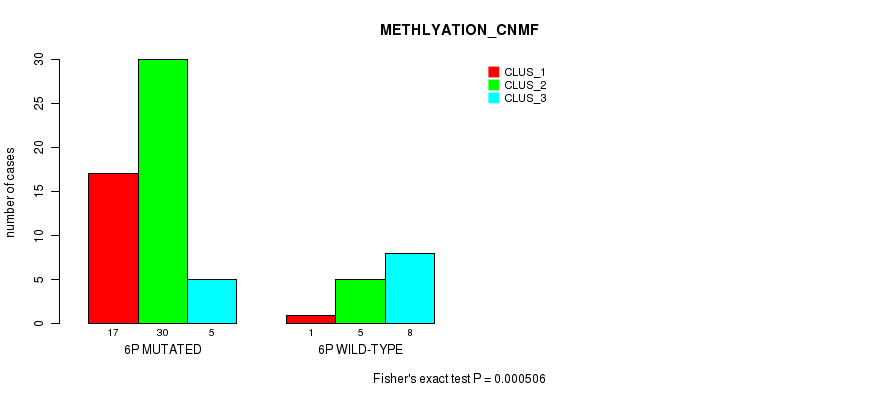
P value = 0.000255 (Fisher's exact test), Q value = 0.074
Table S20. Gene #11: '6p' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 6P MUTATED | 19 | 15 | 14 | 4 |
| 6P WILD-TYPE | 0 | 7 | 1 | 6 |
Figure S20. Get High-res Image Gene #11: '6p' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
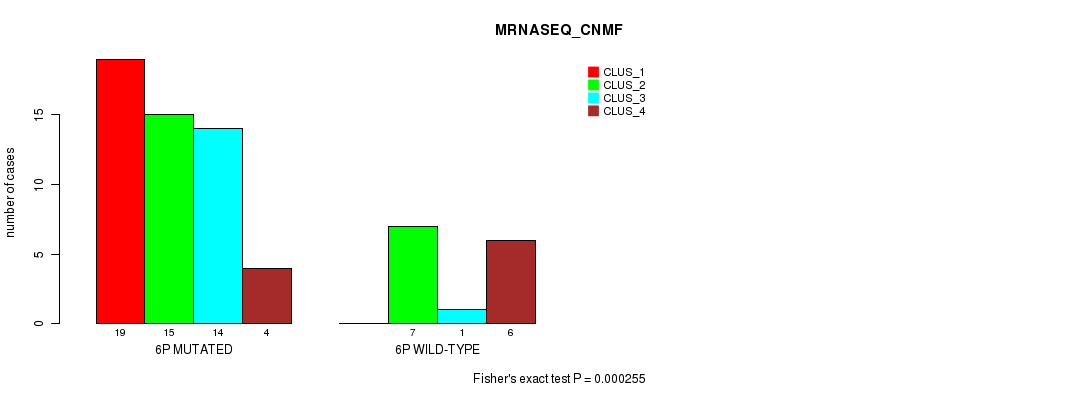
P value = 0.000506 (Fisher's exact test), Q value = 0.14
Table S21. Gene #12: '6q' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 35 | 13 |
| 6Q MUTATED | 17 | 30 | 5 |
| 6Q WILD-TYPE | 1 | 5 | 8 |
Figure S21. Get High-res Image Gene #12: '6q' versus Molecular Subtype #2: 'METHLYATION_CNMF'
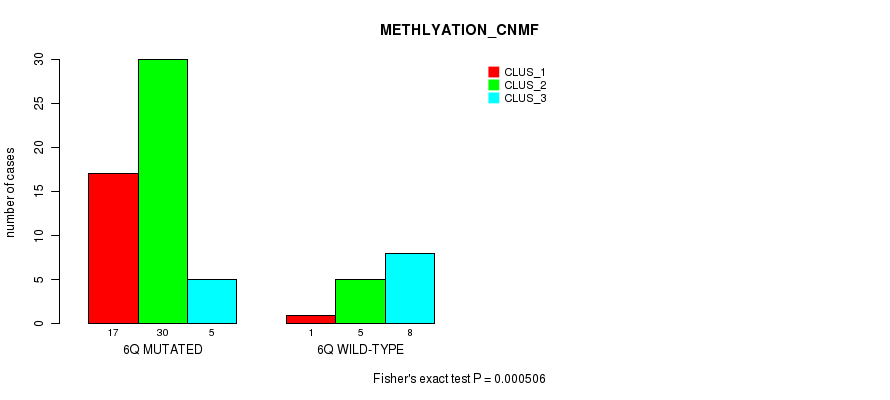
P value = 0.000255 (Fisher's exact test), Q value = 0.074
Table S22. Gene #12: '6q' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 6Q MUTATED | 19 | 15 | 14 | 4 |
| 6Q WILD-TYPE | 0 | 7 | 1 | 6 |
Figure S22. Get High-res Image Gene #12: '6q' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
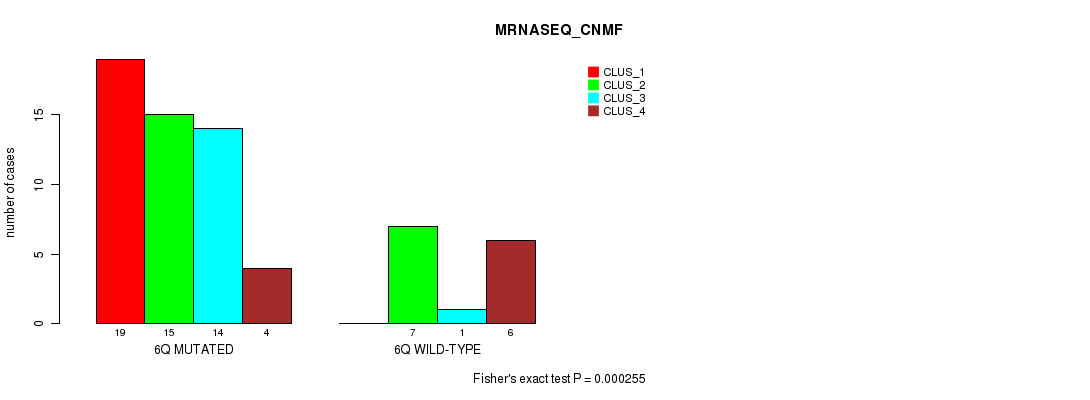
P value = 9.22e-05 (Fisher's exact test), Q value = 0.028
Table S23. Gene #15: '8p' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 5 | 47 | 14 |
| 8P MUTATED | 0 | 14 | 12 |
| 8P WILD-TYPE | 5 | 33 | 2 |
Figure S23. Get High-res Image Gene #15: '8p' versus Molecular Subtype #1: 'CN_CNMF'
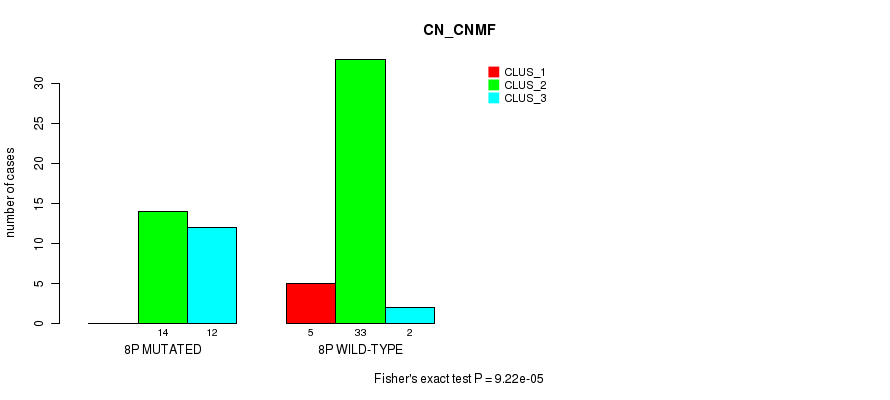
P value = 0.000255 (Fisher's exact test), Q value = 0.074
Table S24. Gene #19: '10p' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 10P MUTATED | 19 | 15 | 14 | 4 |
| 10P WILD-TYPE | 0 | 7 | 1 | 6 |
Figure S24. Get High-res Image Gene #19: '10p' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
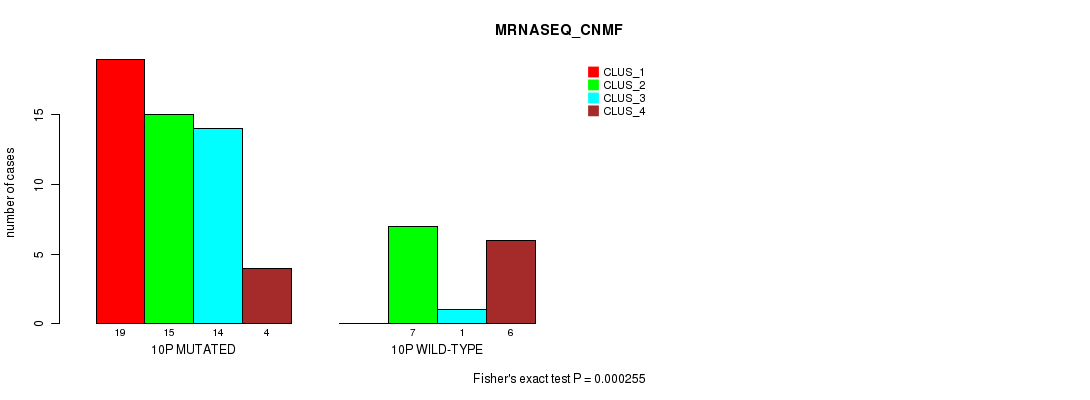
P value = 4.55e-05 (Chi-square test), Q value = 0.014
Table S25. Gene #19: '10p' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 10P MUTATED | 10 | 3 | 11 | 5 | 23 |
| 10P WILD-TYPE | 0 | 7 | 6 | 1 | 0 |
Figure S25. Get High-res Image Gene #19: '10p' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
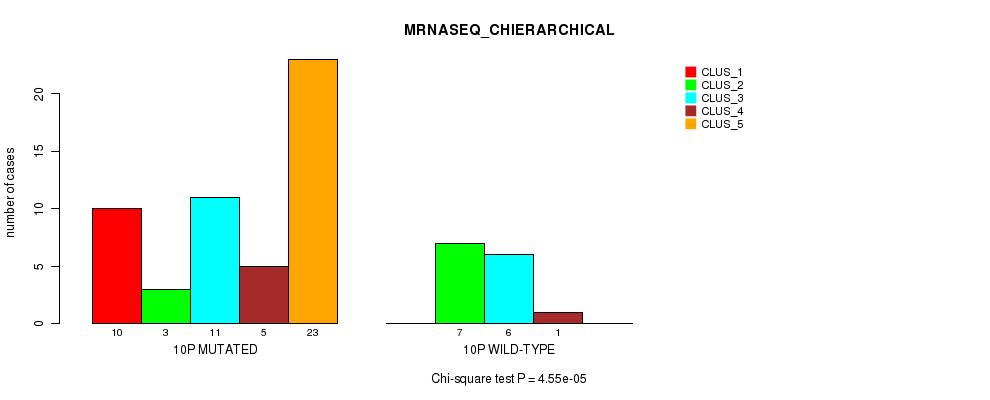
P value = 0.00018 (Fisher's exact test), Q value = 0.053
Table S26. Gene #20: '10q' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 10Q MUTATED | 19 | 14 | 14 | 4 |
| 10Q WILD-TYPE | 0 | 8 | 1 | 6 |
Figure S26. Get High-res Image Gene #20: '10q' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
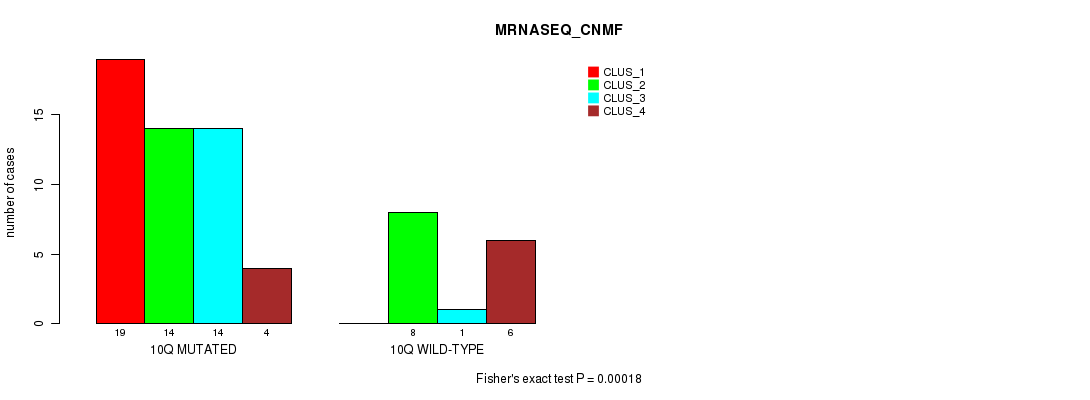
P value = 3.39e-05 (Chi-square test), Q value = 0.01
Table S27. Gene #20: '10q' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 10Q MUTATED | 10 | 3 | 10 | 5 | 23 |
| 10Q WILD-TYPE | 0 | 7 | 7 | 1 | 0 |
Figure S27. Get High-res Image Gene #20: '10q' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
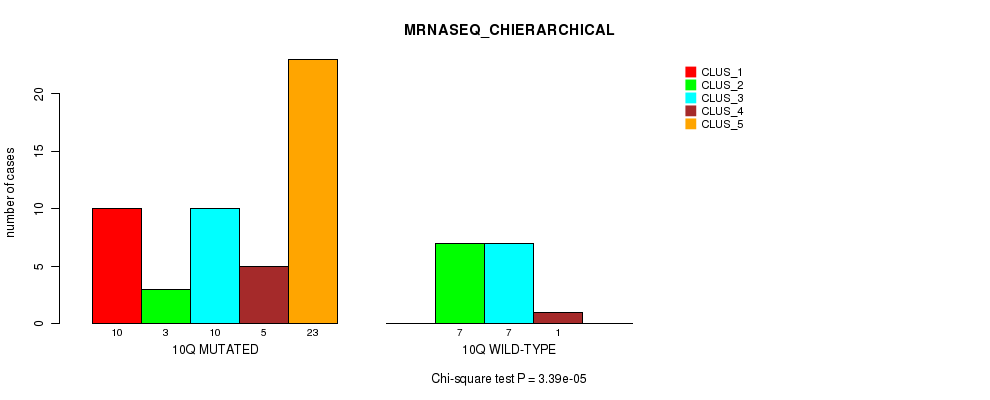
P value = 0.000592 (Chi-square test), Q value = 0.17
Table S28. Gene #22: '11q' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 11Q MUTATED | 1 | 2 | 13 | 1 | 5 |
| 11Q WILD-TYPE | 9 | 8 | 4 | 5 | 18 |
Figure S28. Get High-res Image Gene #22: '11q' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
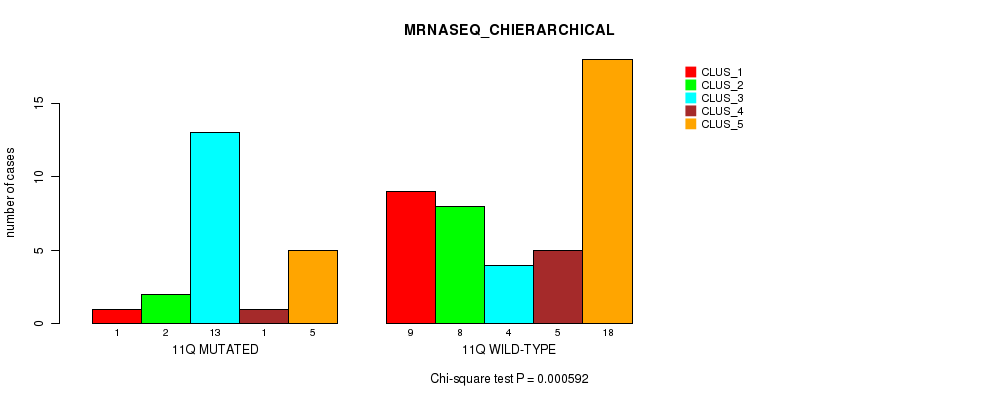
P value = 0.000206 (Fisher's exact test), Q value = 0.06
Table S29. Gene #25: '13q' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 35 | 13 |
| 13Q MUTATED | 12 | 30 | 3 |
| 13Q WILD-TYPE | 6 | 5 | 10 |
Figure S29. Get High-res Image Gene #25: '13q' versus Molecular Subtype #2: 'METHLYATION_CNMF'
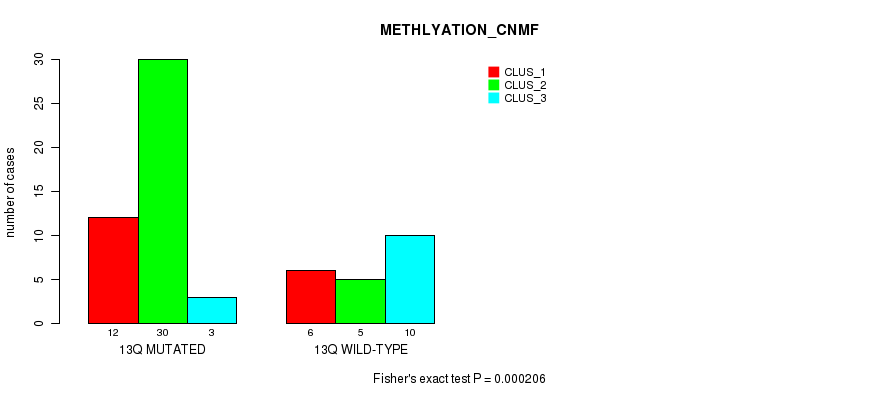
P value = 0.000589 (Fisher's exact test), Q value = 0.17
Table S30. Gene #28: '16p' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 16P MUTATED | 4 | 16 | 4 | 1 |
| 16P WILD-TYPE | 15 | 6 | 11 | 9 |
Figure S30. Get High-res Image Gene #28: '16p' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
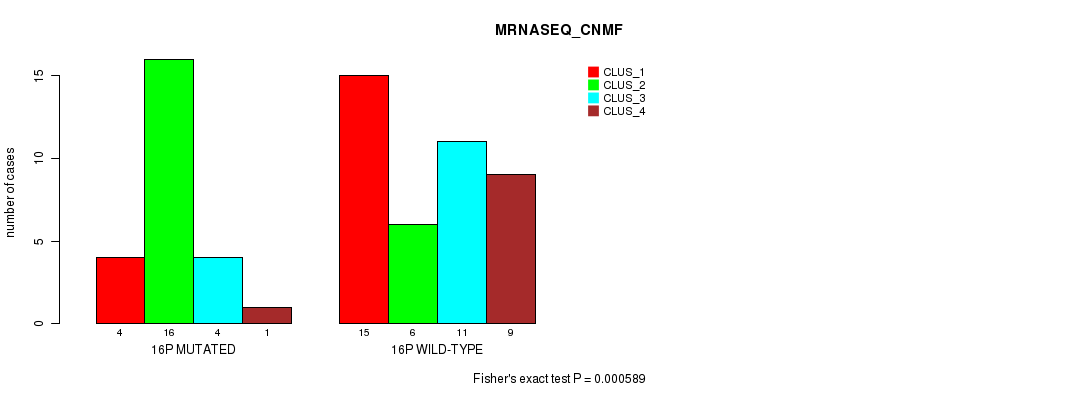
P value = 0.000597 (Chi-square test), Q value = 0.17
Table S31. Gene #28: '16p' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 16P MUTATED | 3 | 2 | 14 | 1 | 5 |
| 16P WILD-TYPE | 7 | 8 | 3 | 5 | 18 |
Figure S31. Get High-res Image Gene #28: '16p' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
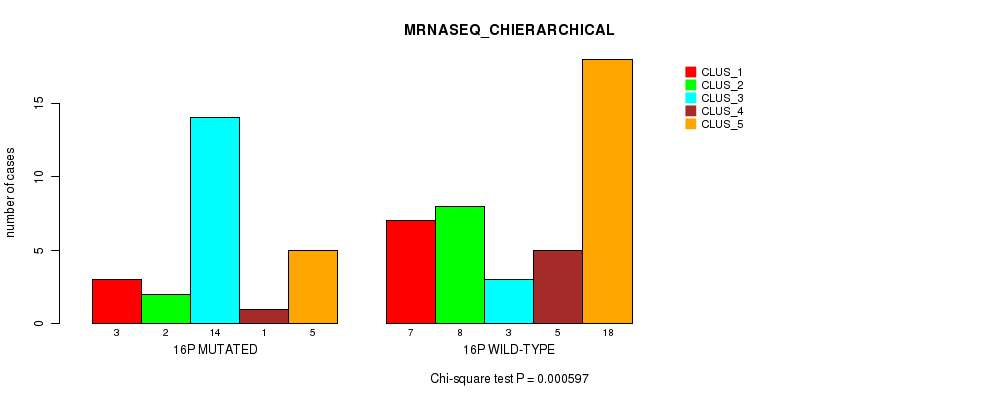
P value = 0.000842 (Fisher's exact test), Q value = 0.23
Table S32. Gene #29: '16q' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 16Q MUTATED | 4 | 16 | 5 | 1 |
| 16Q WILD-TYPE | 15 | 6 | 10 | 9 |
Figure S32. Get High-res Image Gene #29: '16q' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
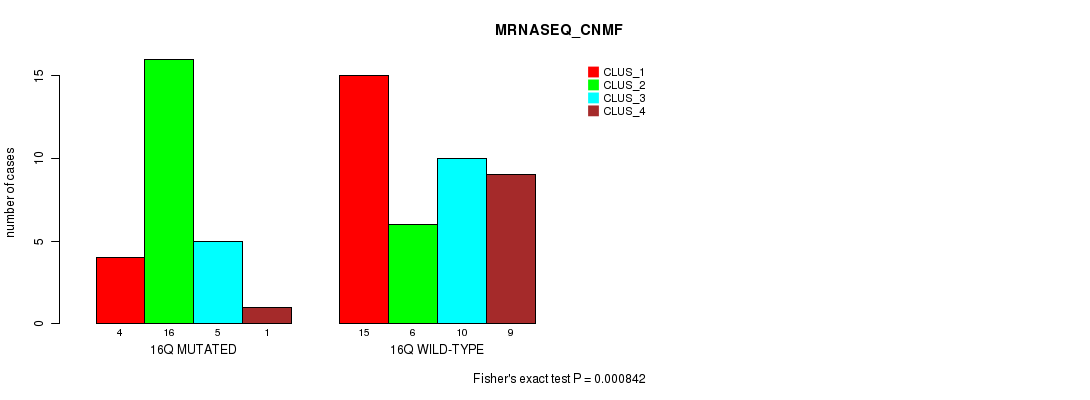
P value = 0.00078 (Chi-square test), Q value = 0.22
Table S33. Gene #29: '16q' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 16Q MUTATED | 4 | 2 | 14 | 1 | 5 |
| 16Q WILD-TYPE | 6 | 8 | 3 | 5 | 18 |
Figure S33. Get High-res Image Gene #29: '16q' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
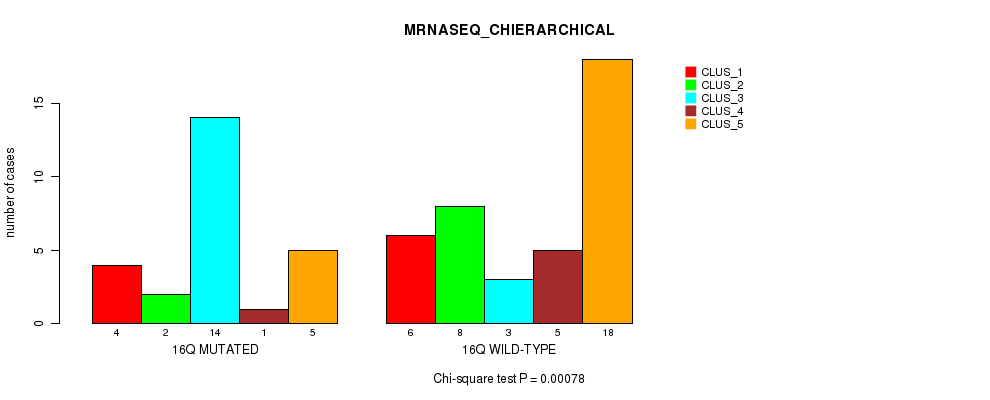
P value = 4.75e-07 (Fisher's exact test), Q value = 0.00015
Table S34. Gene #30: '17p' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 35 | 13 |
| 17P MUTATED | 17 | 31 | 2 |
| 17P WILD-TYPE | 1 | 4 | 11 |
Figure S34. Get High-res Image Gene #30: '17p' versus Molecular Subtype #2: 'METHLYATION_CNMF'
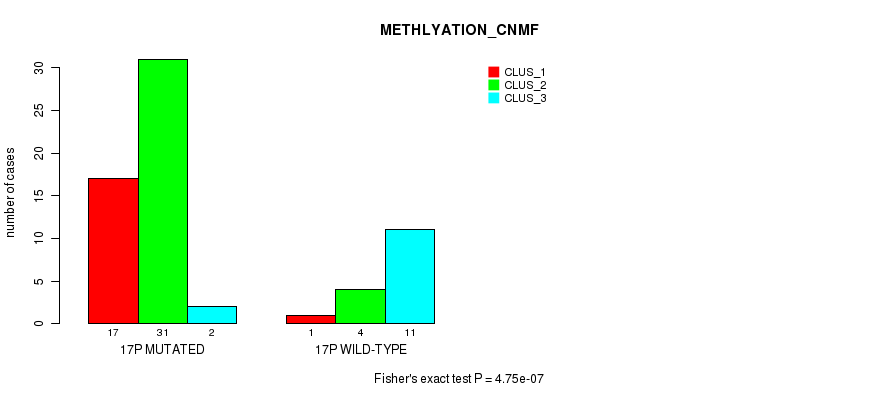
P value = 5.74e-07 (Fisher's exact test), Q value = 0.00018
Table S35. Gene #30: '17p' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 17P MUTATED | 19 | 16 | 14 | 1 |
| 17P WILD-TYPE | 0 | 6 | 1 | 9 |
Figure S35. Get High-res Image Gene #30: '17p' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
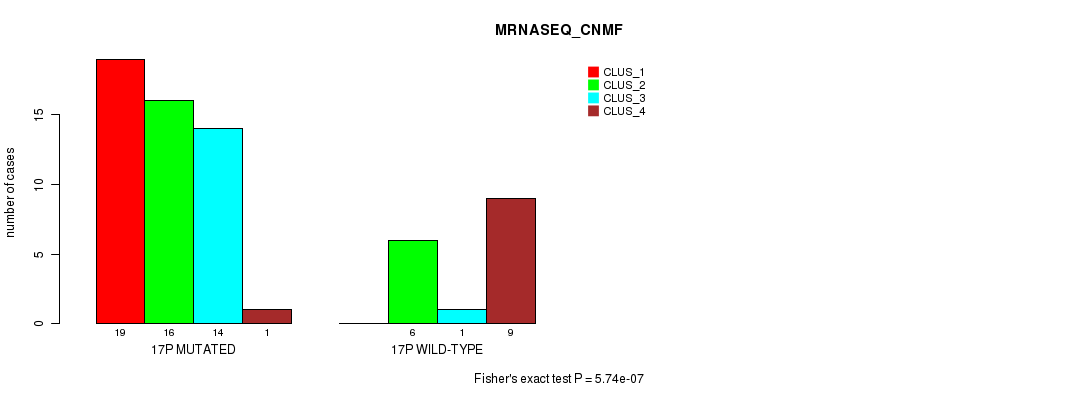
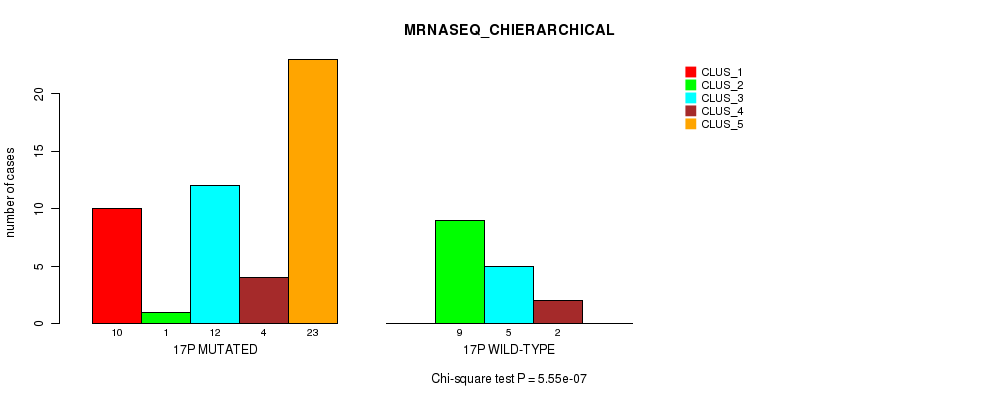
P value = 5.55e-07 (Chi-square test), Q value = 0.00018
Table S36. Gene #30: '17p' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 17P MUTATED | 10 | 1 | 12 | 4 | 23 |
| 17P WILD-TYPE | 0 | 9 | 5 | 2 | 0 |
Figure S36. Get High-res Image Gene #30: '17p' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
P value = 0.000396 (Fisher's exact test), Q value = 0.11
Table S37. Gene #30: '17p' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 9 | 57 |
| 17P MUTATED | 2 | 48 |
| 17P WILD-TYPE | 7 | 9 |
Figure S37. Get High-res Image Gene #30: '17p' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
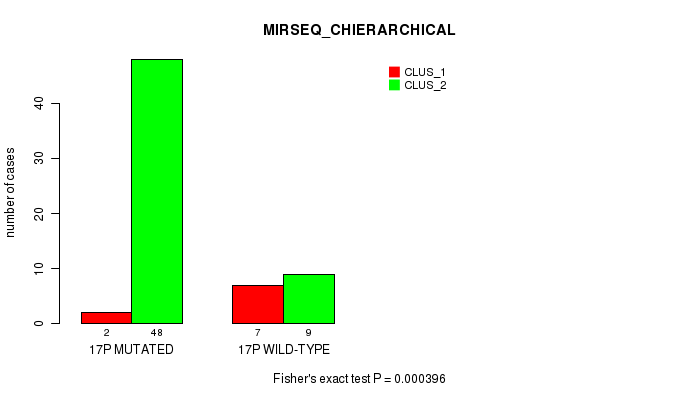
P value = 0.000108 (Fisher's exact test), Q value = 0.032
Table S38. Gene #30: '17p' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 12 | 8 | 25 | 21 |
| 17P MUTATED | 4 | 5 | 20 | 21 |
| 17P WILD-TYPE | 8 | 3 | 5 | 0 |
Figure S38. Get High-res Image Gene #30: '17p' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
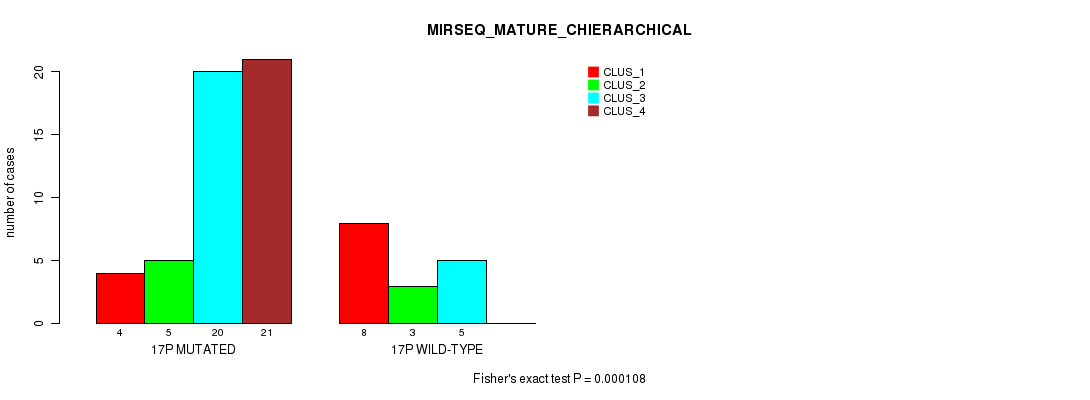
P value = 4.75e-07 (Fisher's exact test), Q value = 0.00015
Table S39. Gene #31: '17q' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 35 | 13 |
| 17Q MUTATED | 17 | 31 | 2 |
| 17Q WILD-TYPE | 1 | 4 | 11 |
Figure S39. Get High-res Image Gene #31: '17q' versus Molecular Subtype #2: 'METHLYATION_CNMF'
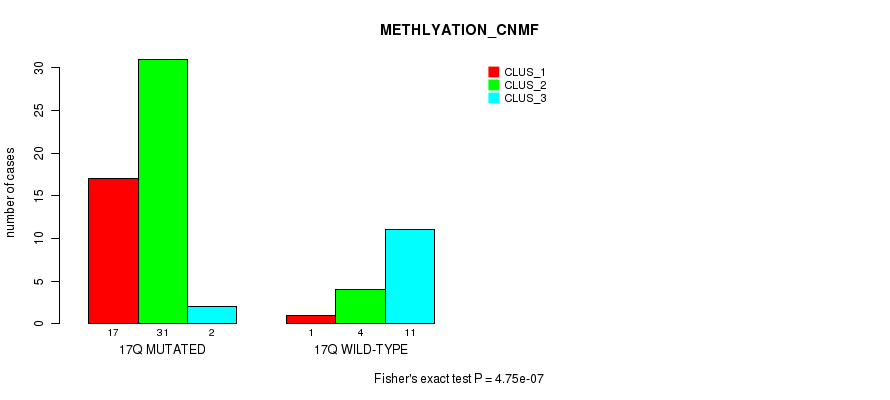
P value = 5.74e-07 (Fisher's exact test), Q value = 0.00018
Table S40. Gene #31: '17q' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| 17Q MUTATED | 19 | 16 | 14 | 1 |
| 17Q WILD-TYPE | 0 | 6 | 1 | 9 |
Figure S40. Get High-res Image Gene #31: '17q' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
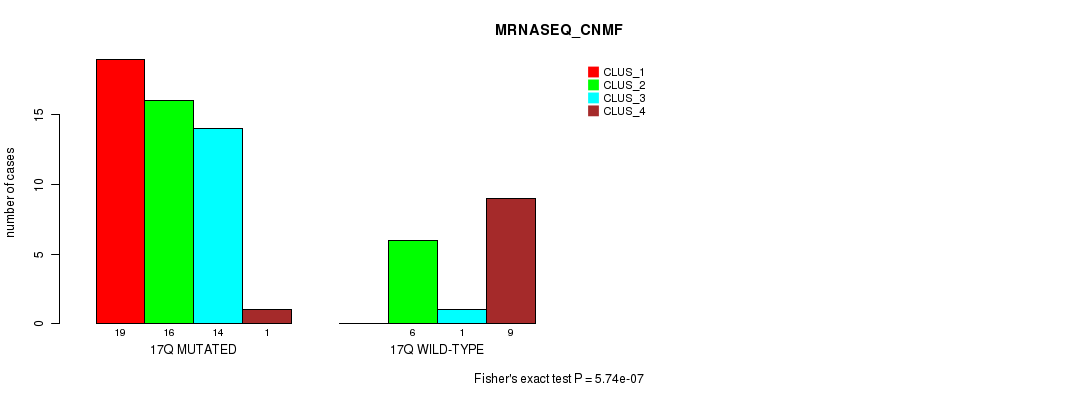
P value = 5.55e-07 (Chi-square test), Q value = 0.00018
Table S41. Gene #31: '17q' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 10 | 10 | 17 | 6 | 23 |
| 17Q MUTATED | 10 | 1 | 12 | 4 | 23 |
| 17Q WILD-TYPE | 0 | 9 | 5 | 2 | 0 |
Figure S41. Get High-res Image Gene #31: '17q' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
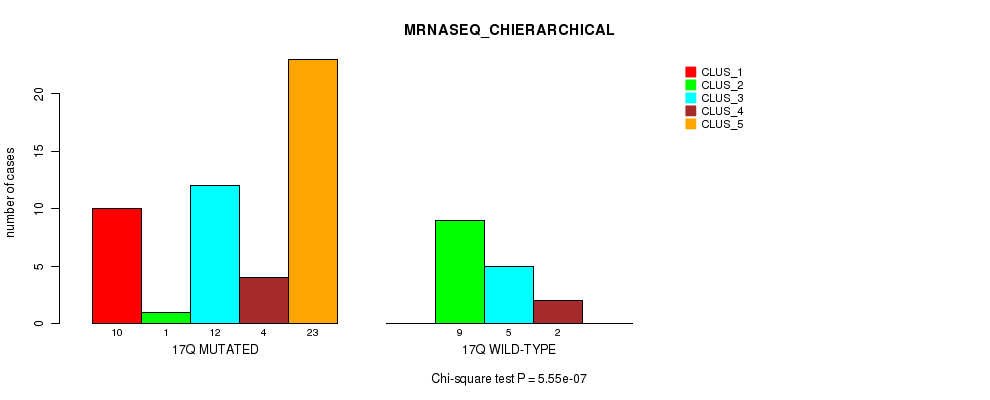
P value = 0.000396 (Fisher's exact test), Q value = 0.11
Table S42. Gene #31: '17q' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 9 | 57 |
| 17Q MUTATED | 2 | 48 |
| 17Q WILD-TYPE | 7 | 9 |
Figure S42. Get High-res Image Gene #31: '17q' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
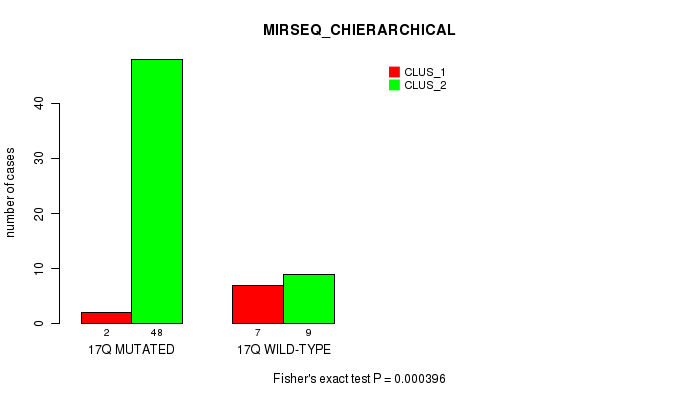
P value = 0.000108 (Fisher's exact test), Q value = 0.032
Table S43. Gene #31: '17q' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 12 | 8 | 25 | 21 |
| 17Q MUTATED | 4 | 5 | 20 | 21 |
| 17Q WILD-TYPE | 8 | 3 | 5 | 0 |
Figure S43. Get High-res Image Gene #31: '17q' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
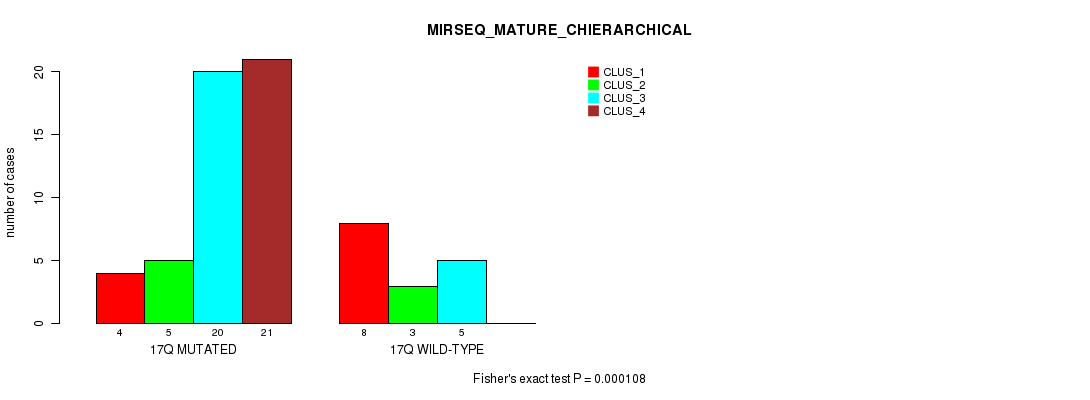
-
Copy number data file = transformed.cor.cli.txt
-
Molecular subtype file = KICH-TP.transferedmergedcluster.txt
-
Number of patients = 66
-
Number of significantly focal cnvs = 40
-
Number of molecular subtypes = 8
-
Exclude genes that fewer than K tumors have alterations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.