This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 10 genes and 10 molecular subtypes across 261 patients, 7 significant findings detected with P value < 0.05 and Q value < 0.25.
-
SPOP mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
TP53 mutation correlated to 'CN_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
FOXA1 mutation correlated to 'METHLYATION_CNMF'.
Table 1. Get Full Table Overview of the association between mutation status of 10 genes and 10 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 7 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Chi-square test | Fisher's exact test | Chi-square test | Fisher's exact test | |
| SPOP | 26 (10%) | 235 |
0.000169 (0.0155) |
1.55e-08 (1.49e-06) |
0.324 (1.00) |
0.743 (1.00) |
0.000141 (0.0131) |
5.74e-07 (5.4e-05) |
0.418 (1.00) |
0.915 (1.00) |
0.133 (1.00) |
0.0934 (1.00) |
| TP53 | 23 (9%) | 238 |
3.89e-07 (3.69e-05) |
0.271 (1.00) |
0.00775 (0.658) |
0.0187 (1.00) |
0.0181 (1.00) |
0.00039 (0.0351) |
0.825 (1.00) |
0.42 (1.00) |
0.355 (1.00) |
0.529 (1.00) |
| FOXA1 | 12 (5%) | 249 |
0.0228 (1.00) |
0.000178 (0.0162) |
1 (1.00) |
0.714 (1.00) |
0.0459 (1.00) |
0.0434 (1.00) |
0.345 (1.00) |
0.0702 (1.00) |
0.492 (1.00) |
0.187 (1.00) |
| KIDINS220 | 3 (1%) | 258 |
0.54 (1.00) |
0.389 (1.00) |
1 (1.00) |
1 (1.00) |
0.794 (1.00) |
1 (1.00) |
0.968 (1.00) |
1 (1.00) |
||
| CTNNB1 | 9 (3%) | 252 |
0.0573 (1.00) |
0.0426 (1.00) |
0.04 (1.00) |
0.0379 (1.00) |
0.0758 (1.00) |
0.306 (1.00) |
0.442 (1.00) |
0.292 (1.00) |
0.772 (1.00) |
0.539 (1.00) |
| PTEN | 13 (5%) | 248 |
0.0351 (1.00) |
0.287 (1.00) |
0.197 (1.00) |
0.598 (1.00) |
0.151 (1.00) |
0.0618 (1.00) |
0.252 (1.00) |
0.124 (1.00) |
0.29 (1.00) |
0.519 (1.00) |
| LMOD2 | 3 (1%) | 258 |
1 (1.00) |
0.638 (1.00) |
1 (1.00) |
1 (1.00) |
0.786 (1.00) |
0.778 (1.00) |
0.0811 (1.00) |
0.596 (1.00) |
||
| IDH1 | 4 (2%) | 257 |
0.114 (1.00) |
0.00643 (0.566) |
0.0609 (1.00) |
0.024 (1.00) |
0.0357 (1.00) |
0.0102 (0.856) |
0.00646 (0.566) |
0.00604 (0.537) |
0.00649 (0.566) |
0.134 (1.00) |
| NKX3-1 | 5 (2%) | 256 |
0.358 (1.00) |
0.632 (1.00) |
0.0286 (1.00) |
0.0697 (1.00) |
1 (1.00) |
0.871 (1.00) |
0.0204 (1.00) |
0.429 (1.00) |
0.0244 (1.00) |
0.0113 (0.935) |
| CDKN1B | 4 (2%) | 257 |
0.114 (1.00) |
0.104 (1.00) |
0.243 (1.00) |
0.387 (1.00) |
0.329 (1.00) |
0.273 (1.00) |
0.733 (1.00) |
0.0358 (1.00) |
0.898 (1.00) |
0.102 (1.00) |
P value = 0.54 (Fisher's exact test), Q value = 1
Table S1. Gene #1: 'KIDINS220 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 35 | 144 | 79 |
| KIDINS220 MUTATED | 0 | 1 | 2 |
| KIDINS220 WILD-TYPE | 35 | 143 | 77 |
P value = 0.389 (Fisher's exact test), Q value = 1
Table S2. Gene #1: 'KIDINS220 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 75 | 89 | 97 |
| KIDINS220 MUTATED | 1 | 2 | 0 |
| KIDINS220 WILD-TYPE | 74 | 87 | 97 |
P value = 1 (Fisher's exact test), Q value = 1
Table S3. Gene #1: 'KIDINS220 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 89 | 81 | 88 |
| KIDINS220 MUTATED | 1 | 1 | 1 |
| KIDINS220 WILD-TYPE | 88 | 80 | 87 |
P value = 1 (Fisher's exact test), Q value = 1
Table S4. Gene #1: 'KIDINS220 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 83 | 84 | 91 |
| KIDINS220 MUTATED | 1 | 1 | 1 |
| KIDINS220 WILD-TYPE | 82 | 83 | 90 |
P value = 0.794 (Chi-square test), Q value = 1
Table S5. Gene #1: 'KIDINS220 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 63 | 39 | 80 | 16 | 61 |
| KIDINS220 MUTATED | 1 | 1 | 1 | 0 | 0 |
| KIDINS220 WILD-TYPE | 62 | 38 | 79 | 16 | 61 |
P value = 1 (Fisher's exact test), Q value = 1
Table S6. Gene #1: 'KIDINS220 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 54 | 89 | 73 |
| KIDINS220 MUTATED | 0 | 1 | 1 | 1 |
| KIDINS220 WILD-TYPE | 43 | 53 | 88 | 72 |
P value = 0.968 (Chi-square test), Q value = 1
Table S7. Gene #1: 'KIDINS220 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 81 | 16 | 85 | 59 | 18 |
| KIDINS220 MUTATED | 1 | 0 | 1 | 1 | 0 |
| KIDINS220 WILD-TYPE | 80 | 16 | 84 | 58 | 18 |
P value = 1 (Fisher's exact test), Q value = 1
Table S8. Gene #1: 'KIDINS220 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 67 | 32 | 78 | 82 |
| KIDINS220 MUTATED | 1 | 0 | 1 | 1 |
| KIDINS220 WILD-TYPE | 66 | 32 | 77 | 81 |
P value = 0.0573 (Fisher's exact test), Q value = 1
Table S9. Gene #2: 'CTNNB1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 35 | 144 | 79 |
| CTNNB1 MUTATED | 3 | 2 | 4 |
| CTNNB1 WILD-TYPE | 32 | 142 | 75 |
P value = 0.0426 (Fisher's exact test), Q value = 1
Table S10. Gene #2: 'CTNNB1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 75 | 89 | 97 |
| CTNNB1 MUTATED | 3 | 0 | 6 |
| CTNNB1 WILD-TYPE | 72 | 89 | 91 |
Figure S1. Get High-res Image Gene #2: 'CTNNB1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
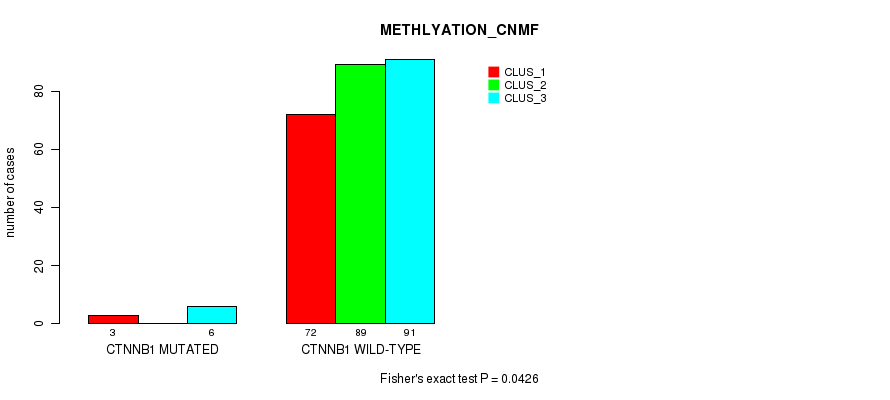
P value = 0.04 (Fisher's exact test), Q value = 1
Table S11. Gene #2: 'CTNNB1 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 42 | 59 | 63 |
| CTNNB1 MUTATED | 0 | 0 | 4 |
| CTNNB1 WILD-TYPE | 42 | 59 | 59 |
Figure S2. Get High-res Image Gene #2: 'CTNNB1 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
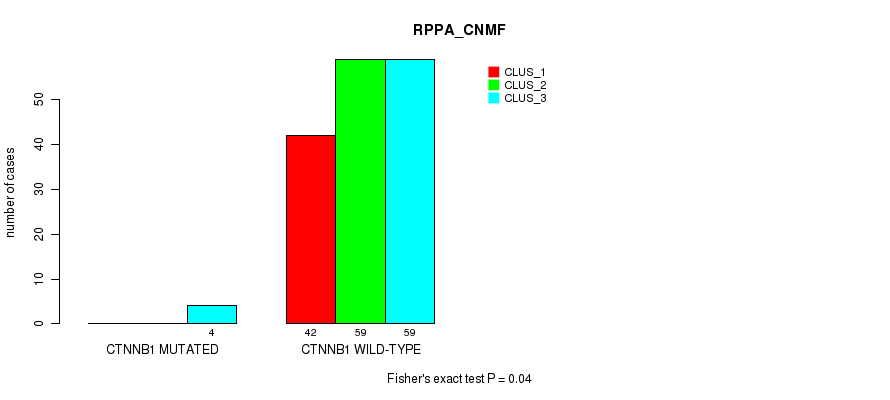
P value = 0.0379 (Fisher's exact test), Q value = 1
Table S12. Gene #2: 'CTNNB1 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 52 | 64 | 48 |
| CTNNB1 MUTATED | 0 | 4 | 0 |
| CTNNB1 WILD-TYPE | 52 | 60 | 48 |
Figure S3. Get High-res Image Gene #2: 'CTNNB1 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
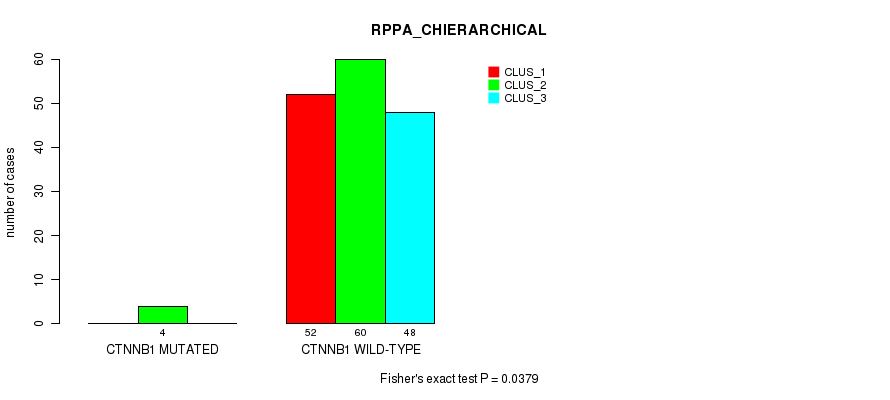
P value = 0.0758 (Fisher's exact test), Q value = 1
Table S13. Gene #2: 'CTNNB1 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 89 | 81 | 88 |
| CTNNB1 MUTATED | 4 | 0 | 5 |
| CTNNB1 WILD-TYPE | 85 | 81 | 83 |
P value = 0.306 (Fisher's exact test), Q value = 1
Table S14. Gene #2: 'CTNNB1 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 83 | 84 | 91 |
| CTNNB1 MUTATED | 3 | 1 | 5 |
| CTNNB1 WILD-TYPE | 80 | 83 | 86 |
P value = 0.442 (Chi-square test), Q value = 1
Table S15. Gene #2: 'CTNNB1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 63 | 39 | 80 | 16 | 61 |
| CTNNB1 MUTATED | 2 | 0 | 3 | 0 | 4 |
| CTNNB1 WILD-TYPE | 61 | 39 | 77 | 16 | 57 |
P value = 0.292 (Fisher's exact test), Q value = 1
Table S16. Gene #2: 'CTNNB1 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 54 | 89 | 73 |
| CTNNB1 MUTATED | 0 | 4 | 3 | 2 |
| CTNNB1 WILD-TYPE | 43 | 50 | 86 | 71 |
P value = 0.772 (Chi-square test), Q value = 1
Table S17. Gene #2: 'CTNNB1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 81 | 16 | 85 | 59 | 18 |
| CTNNB1 MUTATED | 3 | 1 | 2 | 3 | 0 |
| CTNNB1 WILD-TYPE | 78 | 15 | 83 | 56 | 18 |
P value = 0.539 (Fisher's exact test), Q value = 1
Table S18. Gene #2: 'CTNNB1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 67 | 32 | 78 | 82 |
| CTNNB1 MUTATED | 2 | 0 | 2 | 5 |
| CTNNB1 WILD-TYPE | 65 | 32 | 76 | 77 |
P value = 0.000169 (Fisher's exact test), Q value = 0.016
Table S19. Gene #3: 'SPOP MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 35 | 144 | 79 |
| SPOP MUTATED | 10 | 6 | 9 |
| SPOP WILD-TYPE | 25 | 138 | 70 |
Figure S4. Get High-res Image Gene #3: 'SPOP MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
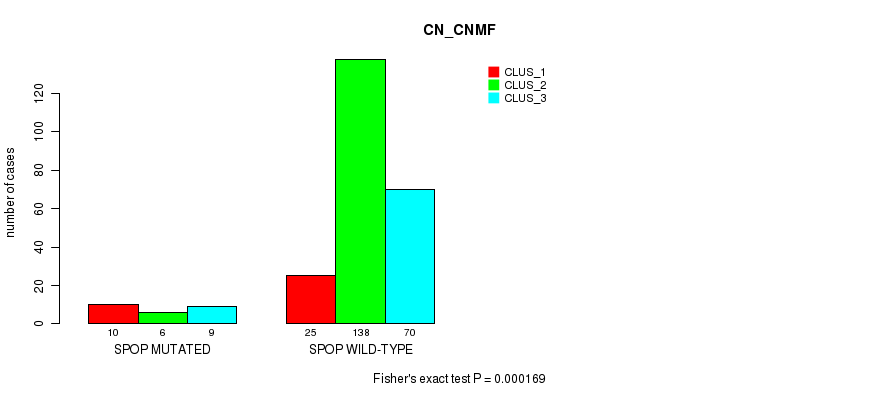
P value = 1.55e-08 (Fisher's exact test), Q value = 1.5e-06
Table S20. Gene #3: 'SPOP MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 75 | 89 | 97 |
| SPOP MUTATED | 21 | 4 | 1 |
| SPOP WILD-TYPE | 54 | 85 | 96 |
Figure S5. Get High-res Image Gene #3: 'SPOP MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
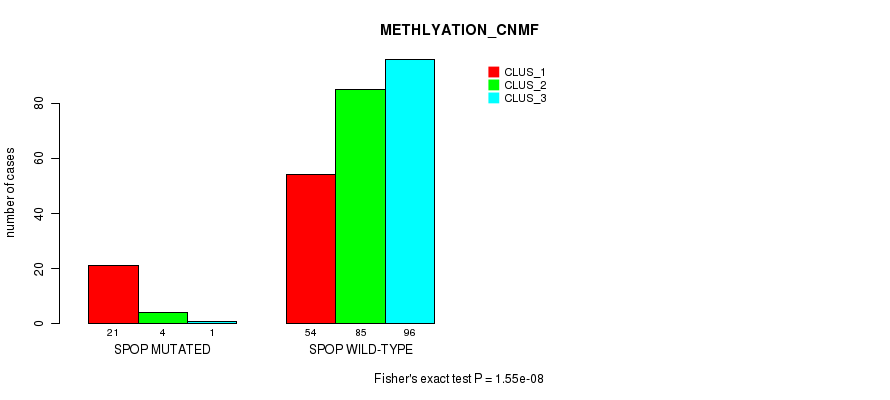
P value = 0.324 (Fisher's exact test), Q value = 1
Table S21. Gene #3: 'SPOP MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 42 | 59 | 63 |
| SPOP MUTATED | 4 | 3 | 8 |
| SPOP WILD-TYPE | 38 | 56 | 55 |
P value = 0.743 (Fisher's exact test), Q value = 1
Table S22. Gene #3: 'SPOP MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 52 | 64 | 48 |
| SPOP MUTATED | 5 | 7 | 3 |
| SPOP WILD-TYPE | 47 | 57 | 45 |
P value = 0.000141 (Fisher's exact test), Q value = 0.013
Table S23. Gene #3: 'SPOP MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 89 | 81 | 88 |
| SPOP MUTATED | 17 | 7 | 1 |
| SPOP WILD-TYPE | 72 | 74 | 87 |
Figure S6. Get High-res Image Gene #3: 'SPOP MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
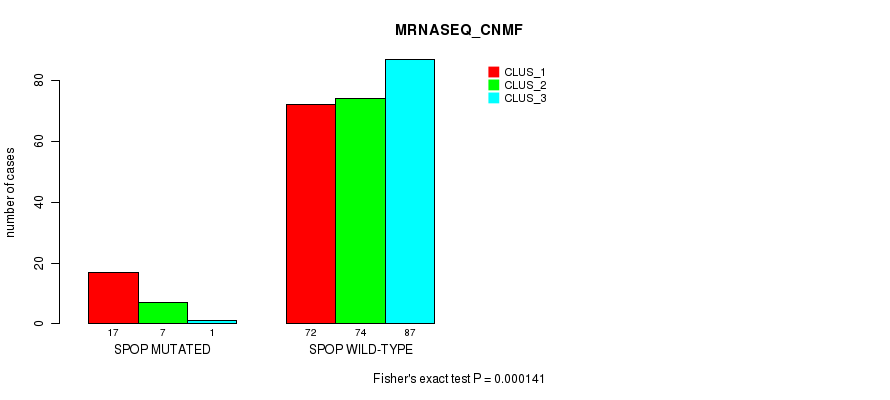
P value = 5.74e-07 (Fisher's exact test), Q value = 5.4e-05
Table S24. Gene #3: 'SPOP MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 83 | 84 | 91 |
| SPOP MUTATED | 20 | 4 | 1 |
| SPOP WILD-TYPE | 63 | 80 | 90 |
Figure S7. Get High-res Image Gene #3: 'SPOP MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
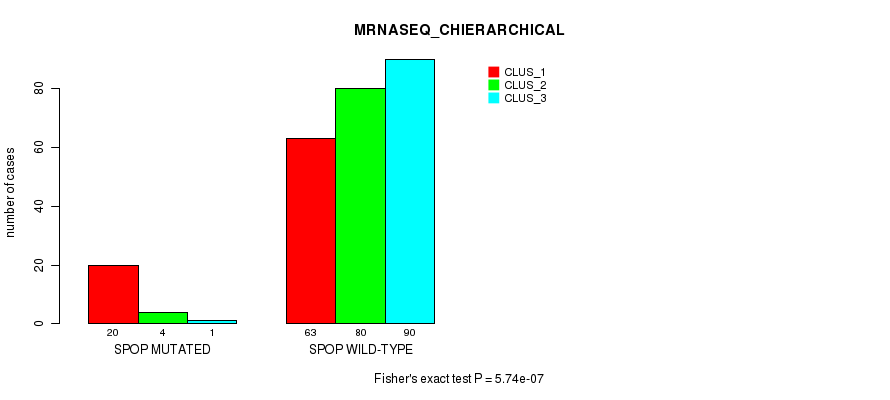
P value = 0.418 (Chi-square test), Q value = 1
Table S25. Gene #3: 'SPOP MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 63 | 39 | 80 | 16 | 61 |
| SPOP MUTATED | 4 | 4 | 10 | 0 | 8 |
| SPOP WILD-TYPE | 59 | 35 | 70 | 16 | 53 |
P value = 0.915 (Fisher's exact test), Q value = 1
Table S26. Gene #3: 'SPOP MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 54 | 89 | 73 |
| SPOP MUTATED | 3 | 6 | 9 | 8 |
| SPOP WILD-TYPE | 40 | 48 | 80 | 65 |
P value = 0.133 (Chi-square test), Q value = 1
Table S27. Gene #3: 'SPOP MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 81 | 16 | 85 | 59 | 18 |
| SPOP MUTATED | 4 | 0 | 10 | 9 | 3 |
| SPOP WILD-TYPE | 77 | 16 | 75 | 50 | 15 |
P value = 0.0934 (Fisher's exact test), Q value = 1
Table S28. Gene #3: 'SPOP MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 67 | 32 | 78 | 82 |
| SPOP MUTATED | 3 | 1 | 11 | 11 |
| SPOP WILD-TYPE | 64 | 31 | 67 | 71 |
P value = 3.89e-07 (Fisher's exact test), Q value = 3.7e-05
Table S29. Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 35 | 144 | 79 |
| TP53 MUTATED | 0 | 4 | 19 |
| TP53 WILD-TYPE | 35 | 140 | 60 |
Figure S8. Get High-res Image Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
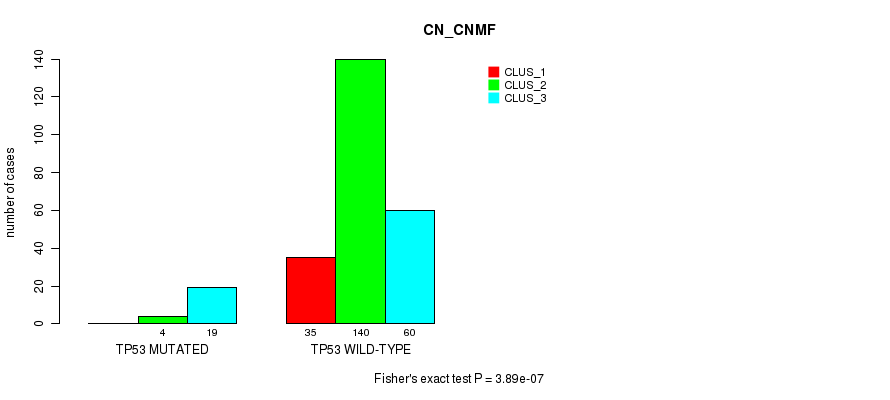
P value = 0.271 (Fisher's exact test), Q value = 1
Table S30. Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 75 | 89 | 97 |
| TP53 MUTATED | 6 | 5 | 12 |
| TP53 WILD-TYPE | 69 | 84 | 85 |
P value = 0.00775 (Fisher's exact test), Q value = 0.66
Table S31. Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 42 | 59 | 63 |
| TP53 MUTATED | 0 | 3 | 10 |
| TP53 WILD-TYPE | 42 | 56 | 53 |
Figure S9. Get High-res Image Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
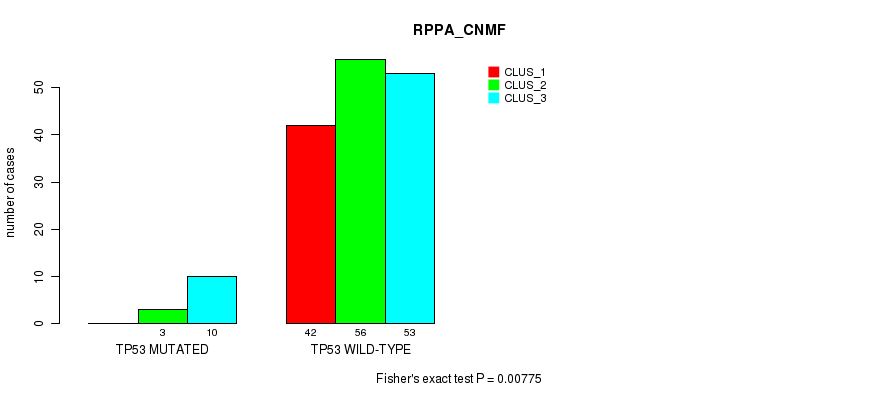
P value = 0.0187 (Fisher's exact test), Q value = 1
Table S32. Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 52 | 64 | 48 |
| TP53 MUTATED | 1 | 10 | 2 |
| TP53 WILD-TYPE | 51 | 54 | 46 |
Figure S10. Get High-res Image Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
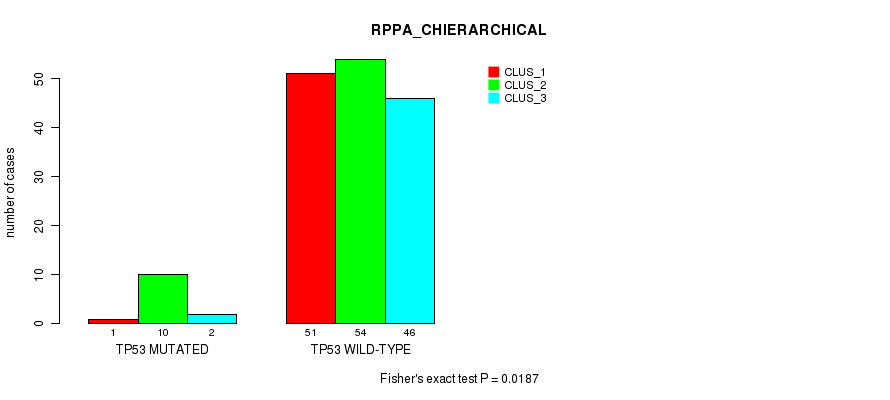
P value = 0.0181 (Fisher's exact test), Q value = 1
Table S33. Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 89 | 81 | 88 |
| TP53 MUTATED | 6 | 3 | 14 |
| TP53 WILD-TYPE | 83 | 78 | 74 |
Figure S11. Get High-res Image Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
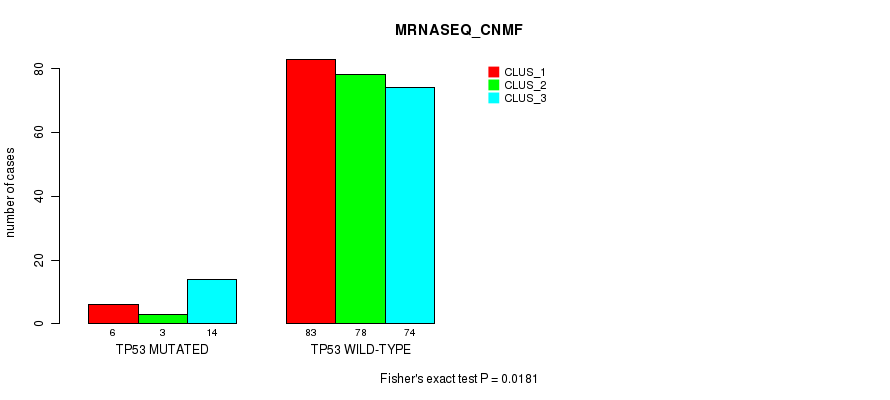
P value = 0.00039 (Fisher's exact test), Q value = 0.035
Table S34. Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 83 | 84 | 91 |
| TP53 MUTATED | 4 | 2 | 17 |
| TP53 WILD-TYPE | 79 | 82 | 74 |
Figure S12. Get High-res Image Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
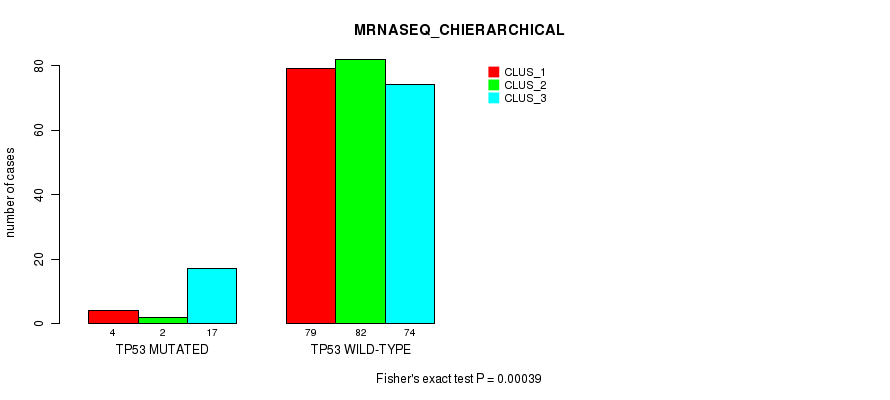
P value = 0.825 (Chi-square test), Q value = 1
Table S35. Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 63 | 39 | 80 | 16 | 61 |
| TP53 MUTATED | 5 | 2 | 9 | 1 | 6 |
| TP53 WILD-TYPE | 58 | 37 | 71 | 15 | 55 |
P value = 0.42 (Fisher's exact test), Q value = 1
Table S36. Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 54 | 89 | 73 |
| TP53 MUTATED | 2 | 7 | 6 | 8 |
| TP53 WILD-TYPE | 41 | 47 | 83 | 65 |
P value = 0.355 (Chi-square test), Q value = 1
Table S37. Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 81 | 16 | 85 | 59 | 18 |
| TP53 MUTATED | 5 | 0 | 11 | 6 | 1 |
| TP53 WILD-TYPE | 76 | 16 | 74 | 53 | 17 |
P value = 0.529 (Fisher's exact test), Q value = 1
Table S38. Gene #4: 'TP53 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 67 | 32 | 78 | 82 |
| TP53 MUTATED | 4 | 2 | 10 | 7 |
| TP53 WILD-TYPE | 63 | 30 | 68 | 75 |
P value = 0.0351 (Fisher's exact test), Q value = 1
Table S39. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 35 | 144 | 79 |
| PTEN MUTATED | 3 | 3 | 7 |
| PTEN WILD-TYPE | 32 | 141 | 72 |
Figure S13. Get High-res Image Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
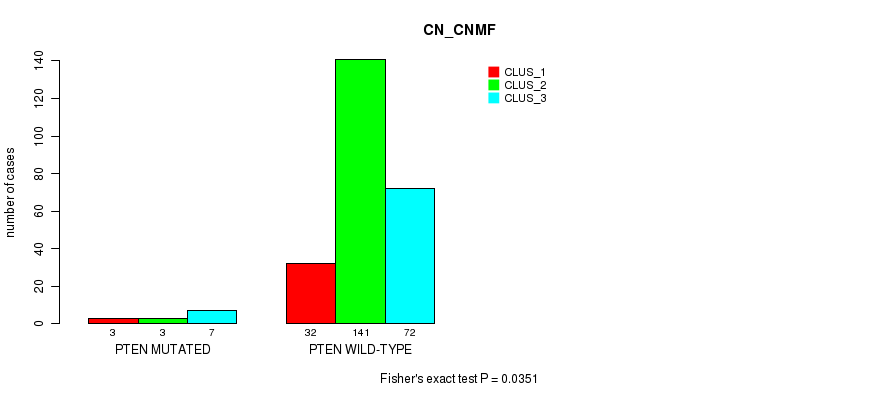
P value = 0.287 (Fisher's exact test), Q value = 1
Table S40. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 75 | 89 | 97 |
| PTEN MUTATED | 4 | 2 | 7 |
| PTEN WILD-TYPE | 71 | 87 | 90 |
P value = 0.197 (Fisher's exact test), Q value = 1
Table S41. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 42 | 59 | 63 |
| PTEN MUTATED | 4 | 1 | 3 |
| PTEN WILD-TYPE | 38 | 58 | 60 |
P value = 0.598 (Fisher's exact test), Q value = 1
Table S42. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 52 | 64 | 48 |
| PTEN MUTATED | 4 | 2 | 2 |
| PTEN WILD-TYPE | 48 | 62 | 46 |
P value = 0.151 (Fisher's exact test), Q value = 1
Table S43. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 89 | 81 | 88 |
| PTEN MUTATED | 6 | 1 | 6 |
| PTEN WILD-TYPE | 83 | 80 | 82 |
P value = 0.0618 (Fisher's exact test), Q value = 1
Table S44. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 83 | 84 | 91 |
| PTEN MUTATED | 4 | 1 | 8 |
| PTEN WILD-TYPE | 79 | 83 | 83 |
P value = 0.252 (Chi-square test), Q value = 1
Table S45. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 63 | 39 | 80 | 16 | 61 |
| PTEN MUTATED | 3 | 0 | 3 | 1 | 6 |
| PTEN WILD-TYPE | 60 | 39 | 77 | 15 | 55 |
P value = 0.124 (Fisher's exact test), Q value = 1
Table S46. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 54 | 89 | 73 |
| PTEN MUTATED | 1 | 6 | 2 | 4 |
| PTEN WILD-TYPE | 42 | 48 | 87 | 69 |
P value = 0.29 (Chi-square test), Q value = 1
Table S47. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 81 | 16 | 85 | 59 | 18 |
| PTEN MUTATED | 3 | 1 | 3 | 6 | 0 |
| PTEN WILD-TYPE | 78 | 15 | 82 | 53 | 18 |
P value = 0.519 (Fisher's exact test), Q value = 1
Table S48. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 67 | 32 | 78 | 82 |
| PTEN MUTATED | 3 | 0 | 4 | 6 |
| PTEN WILD-TYPE | 64 | 32 | 74 | 76 |
P value = 0.0228 (Fisher's exact test), Q value = 1
Table S49. Gene #6: 'FOXA1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 35 | 144 | 79 |
| FOXA1 MUTATED | 5 | 4 | 3 |
| FOXA1 WILD-TYPE | 30 | 140 | 76 |
Figure S14. Get High-res Image Gene #6: 'FOXA1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
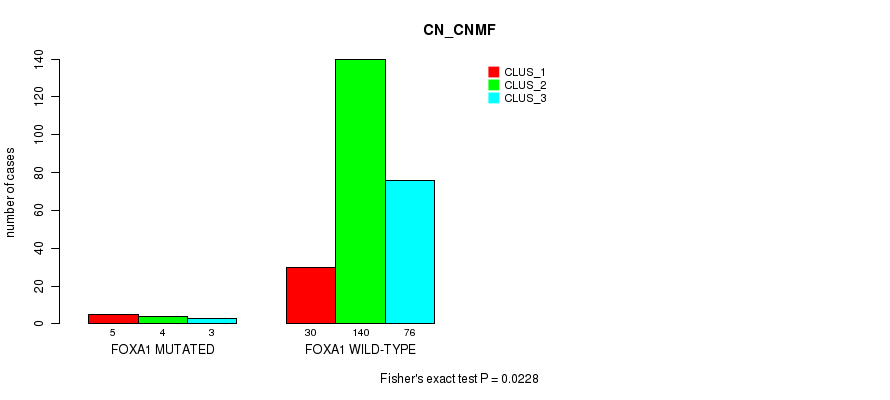
P value = 0.000178 (Fisher's exact test), Q value = 0.016
Table S50. Gene #6: 'FOXA1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 75 | 89 | 97 |
| FOXA1 MUTATED | 10 | 1 | 1 |
| FOXA1 WILD-TYPE | 65 | 88 | 96 |
Figure S15. Get High-res Image Gene #6: 'FOXA1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
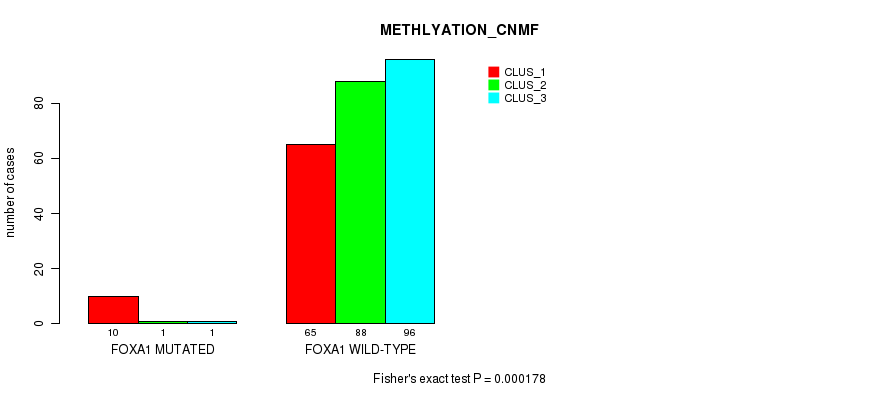
P value = 1 (Fisher's exact test), Q value = 1
Table S51. Gene #6: 'FOXA1 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 42 | 59 | 63 |
| FOXA1 MUTATED | 2 | 2 | 3 |
| FOXA1 WILD-TYPE | 40 | 57 | 60 |
P value = 0.714 (Fisher's exact test), Q value = 1
Table S52. Gene #6: 'FOXA1 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 52 | 64 | 48 |
| FOXA1 MUTATED | 3 | 3 | 1 |
| FOXA1 WILD-TYPE | 49 | 61 | 47 |
P value = 0.0459 (Fisher's exact test), Q value = 1
Table S53. Gene #6: 'FOXA1 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 89 | 81 | 88 |
| FOXA1 MUTATED | 8 | 3 | 1 |
| FOXA1 WILD-TYPE | 81 | 78 | 87 |
Figure S16. Get High-res Image Gene #6: 'FOXA1 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
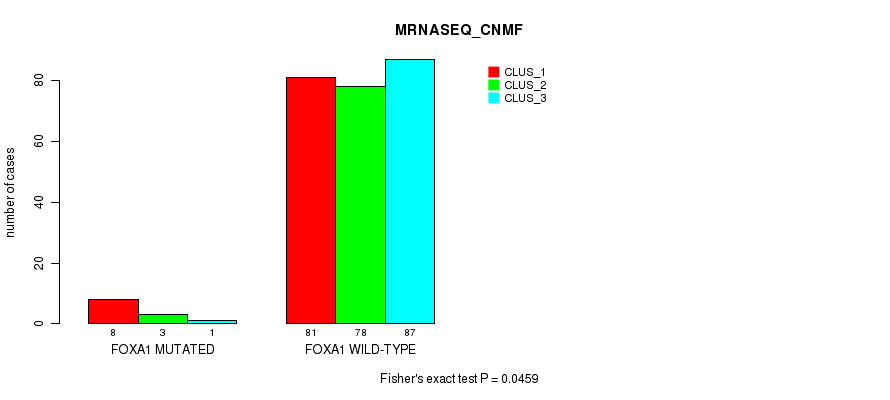
P value = 0.0434 (Fisher's exact test), Q value = 1
Table S54. Gene #6: 'FOXA1 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 83 | 84 | 91 |
| FOXA1 MUTATED | 8 | 2 | 2 |
| FOXA1 WILD-TYPE | 75 | 82 | 89 |
Figure S17. Get High-res Image Gene #6: 'FOXA1 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
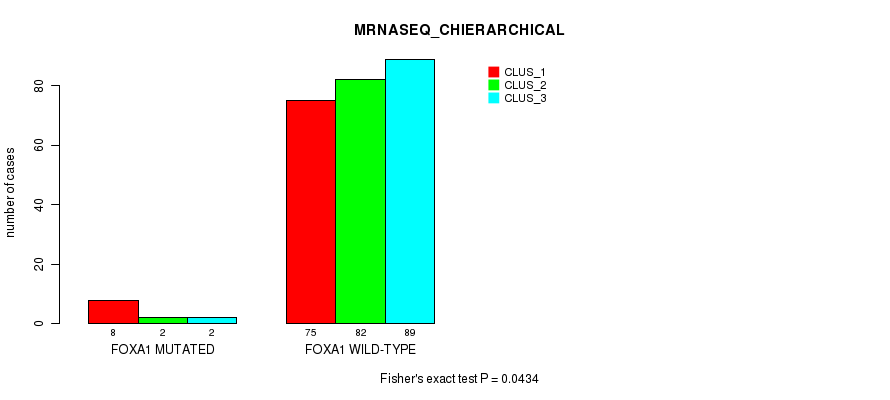
P value = 0.345 (Chi-square test), Q value = 1
Table S55. Gene #6: 'FOXA1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 63 | 39 | 80 | 16 | 61 |
| FOXA1 MUTATED | 1 | 1 | 6 | 0 | 4 |
| FOXA1 WILD-TYPE | 62 | 38 | 74 | 16 | 57 |
P value = 0.0702 (Fisher's exact test), Q value = 1
Table S56. Gene #6: 'FOXA1 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 54 | 89 | 73 |
| FOXA1 MUTATED | 1 | 5 | 1 | 5 |
| FOXA1 WILD-TYPE | 42 | 49 | 88 | 68 |
P value = 0.492 (Chi-square test), Q value = 1
Table S57. Gene #6: 'FOXA1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 81 | 16 | 85 | 59 | 18 |
| FOXA1 MUTATED | 1 | 1 | 6 | 3 | 1 |
| FOXA1 WILD-TYPE | 80 | 15 | 79 | 56 | 17 |
P value = 0.187 (Fisher's exact test), Q value = 1
Table S58. Gene #6: 'FOXA1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 67 | 32 | 78 | 82 |
| FOXA1 MUTATED | 1 | 0 | 5 | 6 |
| FOXA1 WILD-TYPE | 66 | 32 | 73 | 76 |
P value = 1 (Fisher's exact test), Q value = 1
Table S59. Gene #7: 'LMOD2 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 35 | 144 | 79 |
| LMOD2 MUTATED | 0 | 2 | 1 |
| LMOD2 WILD-TYPE | 35 | 142 | 78 |
P value = 0.638 (Fisher's exact test), Q value = 1
Table S60. Gene #7: 'LMOD2 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 75 | 89 | 97 |
| LMOD2 MUTATED | 0 | 2 | 1 |
| LMOD2 WILD-TYPE | 75 | 87 | 96 |
P value = 1 (Fisher's exact test), Q value = 1
Table S61. Gene #7: 'LMOD2 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 89 | 81 | 88 |
| LMOD2 MUTATED | 1 | 1 | 1 |
| LMOD2 WILD-TYPE | 88 | 80 | 87 |
P value = 1 (Fisher's exact test), Q value = 1
Table S62. Gene #7: 'LMOD2 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 83 | 84 | 91 |
| LMOD2 MUTATED | 1 | 1 | 1 |
| LMOD2 WILD-TYPE | 82 | 83 | 90 |
P value = 0.786 (Chi-square test), Q value = 1
Table S63. Gene #7: 'LMOD2 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 63 | 39 | 80 | 16 | 61 |
| LMOD2 MUTATED | 0 | 1 | 1 | 0 | 1 |
| LMOD2 WILD-TYPE | 63 | 38 | 79 | 16 | 60 |
P value = 0.778 (Fisher's exact test), Q value = 1
Table S64. Gene #7: 'LMOD2 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 54 | 89 | 73 |
| LMOD2 MUTATED | 1 | 0 | 1 | 1 |
| LMOD2 WILD-TYPE | 42 | 54 | 88 | 72 |
P value = 0.0811 (Chi-square test), Q value = 1
Table S65. Gene #7: 'LMOD2 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 81 | 16 | 85 | 59 | 18 |
| LMOD2 MUTATED | 0 | 1 | 1 | 0 | 1 |
| LMOD2 WILD-TYPE | 81 | 15 | 84 | 59 | 17 |
P value = 0.596 (Fisher's exact test), Q value = 1
Table S66. Gene #7: 'LMOD2 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 67 | 32 | 78 | 82 |
| LMOD2 MUTATED | 0 | 1 | 1 | 1 |
| LMOD2 WILD-TYPE | 67 | 31 | 77 | 81 |
P value = 0.114 (Fisher's exact test), Q value = 1
Table S67. Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 35 | 144 | 79 |
| IDH1 MUTATED | 2 | 1 | 1 |
| IDH1 WILD-TYPE | 33 | 143 | 78 |
P value = 0.00643 (Fisher's exact test), Q value = 0.57
Table S68. Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 75 | 89 | 97 |
| IDH1 MUTATED | 4 | 0 | 0 |
| IDH1 WILD-TYPE | 71 | 89 | 97 |
Figure S18. Get High-res Image Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
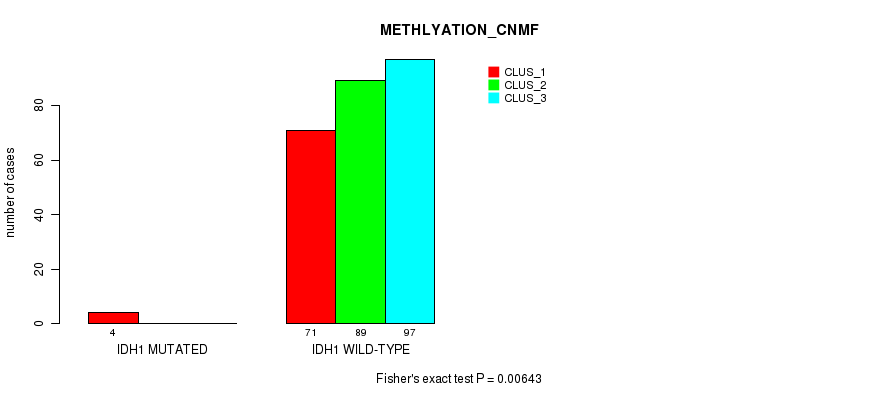
P value = 0.0609 (Fisher's exact test), Q value = 1
Table S69. Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 42 | 59 | 63 |
| IDH1 MUTATED | 0 | 3 | 0 |
| IDH1 WILD-TYPE | 42 | 56 | 63 |
P value = 0.024 (Fisher's exact test), Q value = 1
Table S70. Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 52 | 64 | 48 |
| IDH1 MUTATED | 0 | 0 | 3 |
| IDH1 WILD-TYPE | 52 | 64 | 45 |
Figure S19. Get High-res Image Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
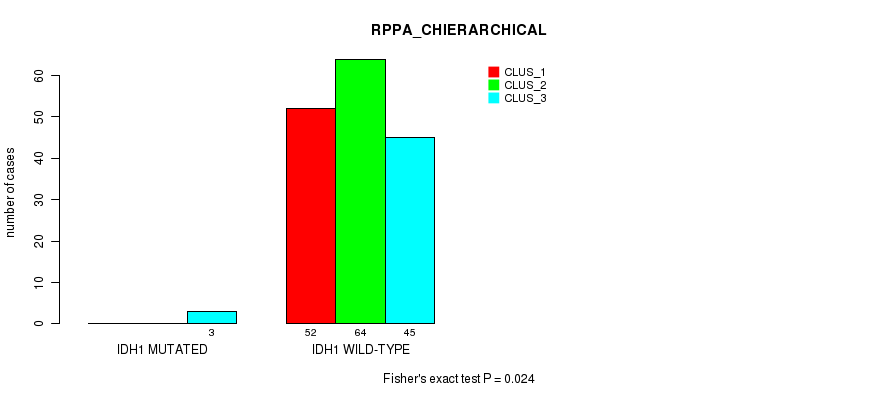
P value = 0.0357 (Fisher's exact test), Q value = 1
Table S71. Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 89 | 81 | 88 |
| IDH1 MUTATED | 4 | 0 | 0 |
| IDH1 WILD-TYPE | 85 | 81 | 88 |
Figure S20. Get High-res Image Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
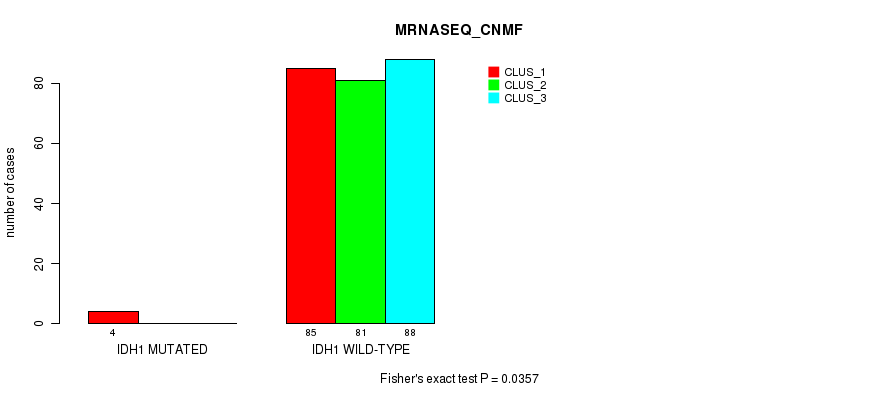
P value = 0.0102 (Fisher's exact test), Q value = 0.86
Table S72. Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 83 | 84 | 91 |
| IDH1 MUTATED | 4 | 0 | 0 |
| IDH1 WILD-TYPE | 79 | 84 | 91 |
Figure S21. Get High-res Image Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
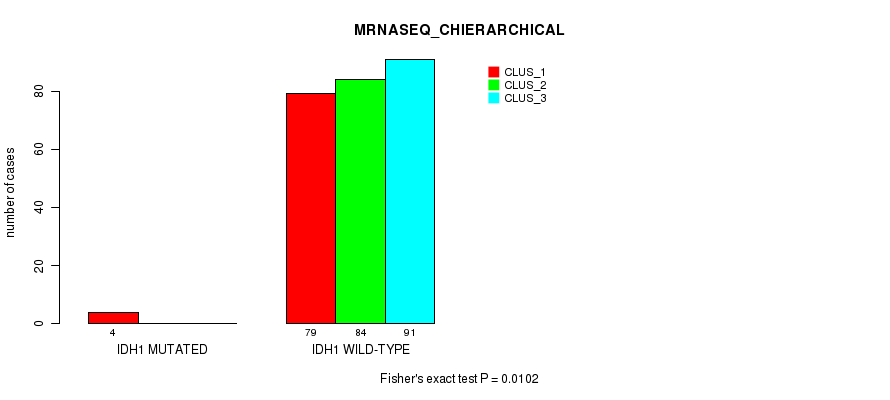
P value = 0.00646 (Chi-square test), Q value = 0.57
Table S73. Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 63 | 39 | 80 | 16 | 61 |
| IDH1 MUTATED | 0 | 0 | 1 | 2 | 1 |
| IDH1 WILD-TYPE | 63 | 39 | 79 | 14 | 60 |
Figure S22. Get High-res Image Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
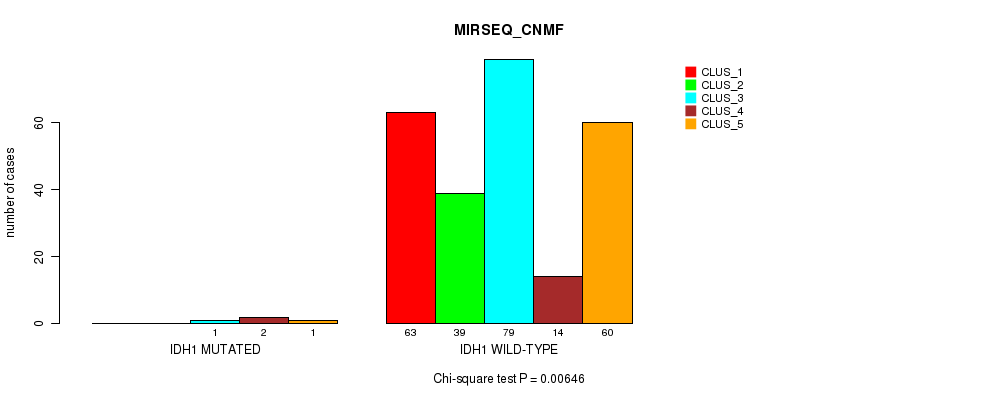
P value = 0.00604 (Fisher's exact test), Q value = 0.54
Table S74. Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 54 | 89 | 73 |
| IDH1 MUTATED | 3 | 1 | 0 | 0 |
| IDH1 WILD-TYPE | 40 | 53 | 89 | 73 |
Figure S23. Get High-res Image Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
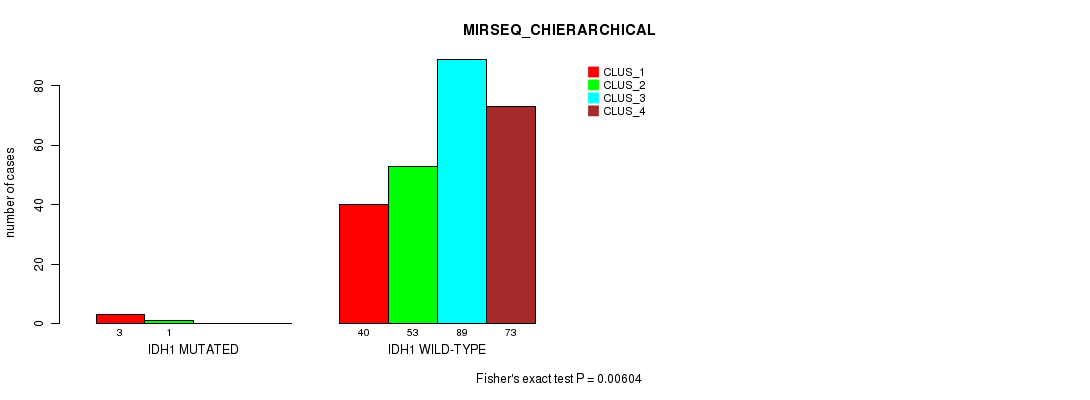
P value = 0.00649 (Chi-square test), Q value = 0.57
Table S75. Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 81 | 16 | 85 | 59 | 18 |
| IDH1 MUTATED | 0 | 2 | 1 | 1 | 0 |
| IDH1 WILD-TYPE | 81 | 14 | 84 | 58 | 18 |
Figure S24. Get High-res Image Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
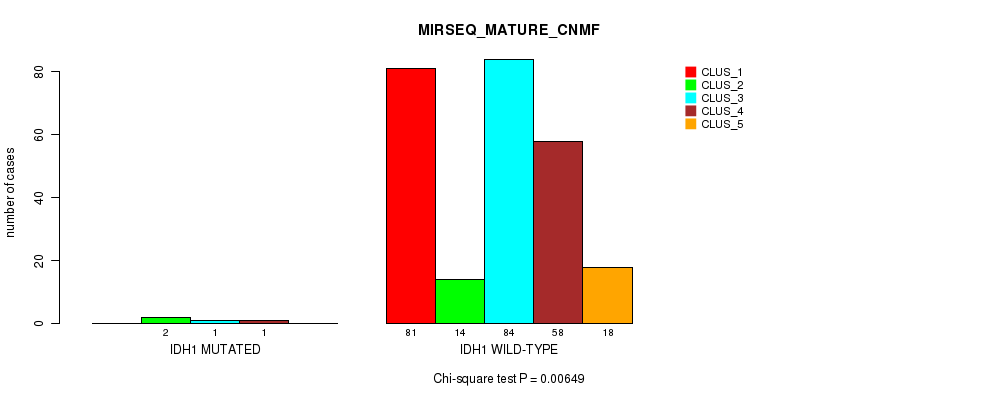
P value = 0.134 (Fisher's exact test), Q value = 1
Table S76. Gene #8: 'IDH1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 67 | 32 | 78 | 82 |
| IDH1 MUTATED | 0 | 2 | 1 | 1 |
| IDH1 WILD-TYPE | 67 | 30 | 77 | 81 |
P value = 0.358 (Fisher's exact test), Q value = 1
Table S77. Gene #9: 'NKX3-1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 35 | 144 | 79 |
| NKX3-1 MUTATED | 1 | 3 | 0 |
| NKX3-1 WILD-TYPE | 34 | 141 | 79 |
P value = 0.632 (Fisher's exact test), Q value = 1
Table S78. Gene #9: 'NKX3-1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 75 | 89 | 97 |
| NKX3-1 MUTATED | 1 | 1 | 3 |
| NKX3-1 WILD-TYPE | 74 | 88 | 94 |
P value = 0.0286 (Fisher's exact test), Q value = 1
Table S79. Gene #9: 'NKX3-1 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 42 | 59 | 63 |
| NKX3-1 MUTATED | 4 | 0 | 1 |
| NKX3-1 WILD-TYPE | 38 | 59 | 62 |
Figure S25. Get High-res Image Gene #9: 'NKX3-1 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
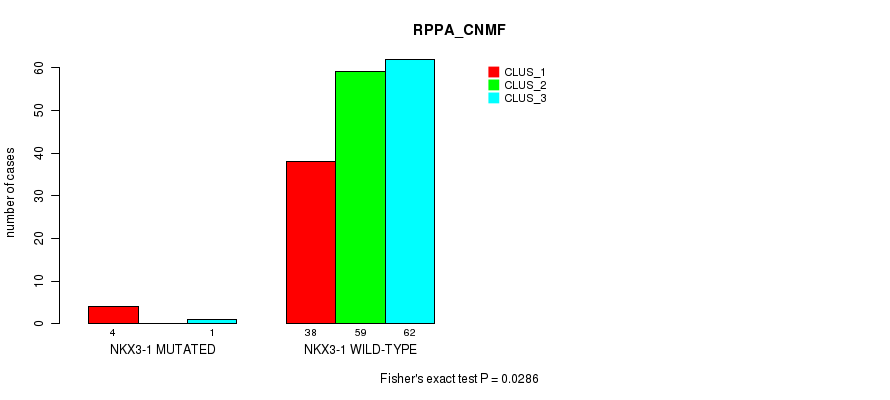
P value = 0.0697 (Fisher's exact test), Q value = 1
Table S80. Gene #9: 'NKX3-1 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 52 | 64 | 48 |
| NKX3-1 MUTATED | 4 | 1 | 0 |
| NKX3-1 WILD-TYPE | 48 | 63 | 48 |
P value = 1 (Fisher's exact test), Q value = 1
Table S81. Gene #9: 'NKX3-1 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 89 | 81 | 88 |
| NKX3-1 MUTATED | 2 | 1 | 2 |
| NKX3-1 WILD-TYPE | 87 | 80 | 86 |
P value = 0.871 (Fisher's exact test), Q value = 1
Table S82. Gene #9: 'NKX3-1 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 83 | 84 | 91 |
| NKX3-1 MUTATED | 2 | 1 | 2 |
| NKX3-1 WILD-TYPE | 81 | 83 | 89 |
P value = 0.0204 (Chi-square test), Q value = 1
Table S83. Gene #9: 'NKX3-1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 63 | 39 | 80 | 16 | 61 |
| NKX3-1 MUTATED | 4 | 0 | 0 | 1 | 0 |
| NKX3-1 WILD-TYPE | 59 | 39 | 80 | 15 | 61 |
Figure S26. Get High-res Image Gene #9: 'NKX3-1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
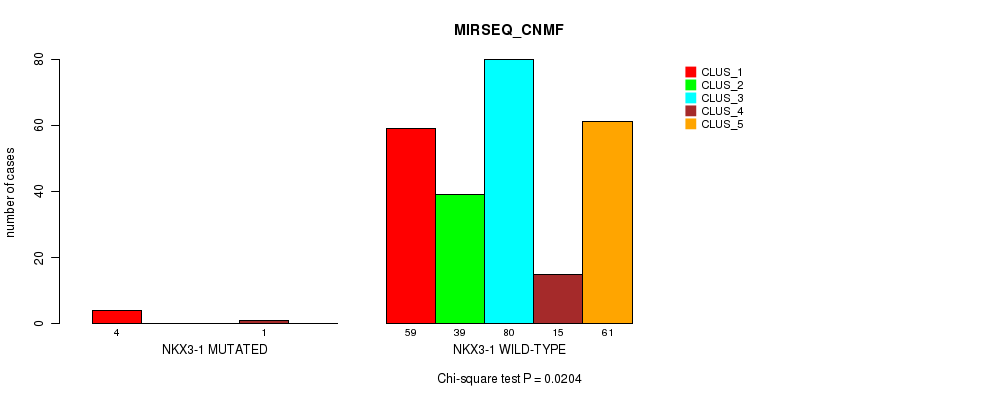
P value = 0.429 (Fisher's exact test), Q value = 1
Table S84. Gene #9: 'NKX3-1 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 54 | 89 | 73 |
| NKX3-1 MUTATED | 2 | 0 | 2 | 1 |
| NKX3-1 WILD-TYPE | 41 | 54 | 87 | 72 |
P value = 0.0244 (Chi-square test), Q value = 1
Table S85. Gene #9: 'NKX3-1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 81 | 16 | 85 | 59 | 18 |
| NKX3-1 MUTATED | 5 | 0 | 0 | 0 | 0 |
| NKX3-1 WILD-TYPE | 76 | 16 | 85 | 59 | 18 |
Figure S27. Get High-res Image Gene #9: 'NKX3-1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
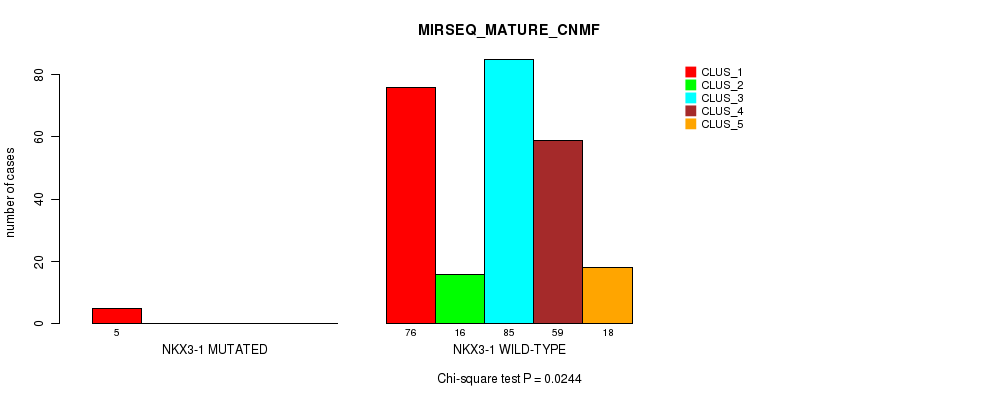
P value = 0.0113 (Fisher's exact test), Q value = 0.93
Table S86. Gene #9: 'NKX3-1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 67 | 32 | 78 | 82 |
| NKX3-1 MUTATED | 3 | 2 | 0 | 0 |
| NKX3-1 WILD-TYPE | 64 | 30 | 78 | 82 |
Figure S28. Get High-res Image Gene #9: 'NKX3-1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
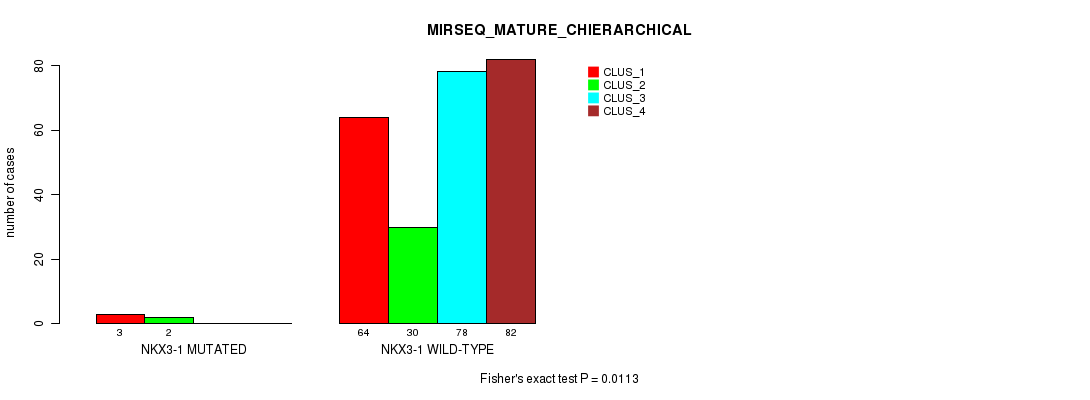
P value = 0.114 (Fisher's exact test), Q value = 1
Table S87. Gene #10: 'CDKN1B MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 35 | 144 | 79 |
| CDKN1B MUTATED | 2 | 1 | 1 |
| CDKN1B WILD-TYPE | 33 | 143 | 78 |
P value = 0.104 (Fisher's exact test), Q value = 1
Table S88. Gene #10: 'CDKN1B MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 75 | 89 | 97 |
| CDKN1B MUTATED | 3 | 0 | 1 |
| CDKN1B WILD-TYPE | 72 | 89 | 96 |
P value = 0.243 (Fisher's exact test), Q value = 1
Table S89. Gene #10: 'CDKN1B MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 42 | 59 | 63 |
| CDKN1B MUTATED | 1 | 0 | 3 |
| CDKN1B WILD-TYPE | 41 | 59 | 60 |
P value = 0.387 (Fisher's exact test), Q value = 1
Table S90. Gene #10: 'CDKN1B MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 52 | 64 | 48 |
| CDKN1B MUTATED | 1 | 3 | 0 |
| CDKN1B WILD-TYPE | 51 | 61 | 48 |
P value = 0.329 (Fisher's exact test), Q value = 1
Table S91. Gene #10: 'CDKN1B MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 89 | 81 | 88 |
| CDKN1B MUTATED | 3 | 0 | 1 |
| CDKN1B WILD-TYPE | 86 | 81 | 87 |
P value = 0.273 (Fisher's exact test), Q value = 1
Table S92. Gene #10: 'CDKN1B MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 83 | 84 | 91 |
| CDKN1B MUTATED | 1 | 0 | 3 |
| CDKN1B WILD-TYPE | 82 | 84 | 88 |
P value = 0.733 (Chi-square test), Q value = 1
Table S93. Gene #10: 'CDKN1B MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 63 | 39 | 80 | 16 | 61 |
| CDKN1B MUTATED | 2 | 0 | 1 | 0 | 1 |
| CDKN1B WILD-TYPE | 61 | 39 | 79 | 16 | 60 |
P value = 0.0358 (Fisher's exact test), Q value = 1
Table S94. Gene #10: 'CDKN1B MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 43 | 54 | 89 | 73 |
| CDKN1B MUTATED | 2 | 2 | 0 | 0 |
| CDKN1B WILD-TYPE | 41 | 52 | 89 | 73 |
Figure S29. Get High-res Image Gene #10: 'CDKN1B MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
P value = 0.898 (Chi-square test), Q value = 1
Table S95. Gene #10: 'CDKN1B MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 81 | 16 | 85 | 59 | 18 |
| CDKN1B MUTATED | 2 | 0 | 1 | 1 | 0 |
| CDKN1B WILD-TYPE | 79 | 16 | 84 | 58 | 18 |
P value = 0.102 (Fisher's exact test), Q value = 1
Table S96. Gene #10: 'CDKN1B MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 67 | 32 | 78 | 82 |
| CDKN1B MUTATED | 1 | 2 | 0 | 1 |
| CDKN1B WILD-TYPE | 66 | 30 | 78 | 81 |
-
Mutation data file = transformed.cor.cli.txt
-
Molecular subtypes file = PRAD-TP.transferedmergedcluster.txt
-
Number of patients = 261
-
Number of significantly mutated genes = 10
-
Number of Molecular subtypes = 10
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.