This pipeline computes the correlation between cancer subtypes identified by different molecular patterns and selected clinical features.
Testing the association between subtypes identified by 8 different clustering approaches and 45 clinical features across 250 patients, 16 significant findings detected with P value < 0.05 and Q value < 0.25.
-
3 subtypes identified in current cancer cohort by 'Copy Number Ratio CNMF subtypes'. These subtypes do not correlate to any clinical features.
-
6 subtypes identified in current cancer cohort by 'METHLYATION CNMF'. These subtypes correlate to 'HISTOLOGICAL.TYPE'.
-
CNMF clustering analysis on sequencing-based mRNA expression data identified 3 subtypes that correlate to 'HISTOLOGICAL.TYPE'.
-
Consensus hierarchical clustering analysis on sequencing-based mRNA expression data identified 3 subtypes that correlate to 'HISTOLOGICAL.TYPE'.
-
3 subtypes identified in current cancer cohort by 'MIRSEQ CNMF'. These subtypes correlate to 'HISTOLOGICAL.TYPE'.
-
5 subtypes identified in current cancer cohort by 'MIRSEQ CHIERARCHICAL'. These subtypes correlate to 'AGE', 'HISTOLOGICAL.TYPE', 'AGE_AT_DIAGNOSIS', and 'STAGE_EVENT.CLINICAL_STAGE'.
-
3 subtypes identified in current cancer cohort by 'MIRseq Mature CNMF subtypes'. These subtypes correlate to 'HISTOLOGICAL.TYPE', 'RADIATIONS.RADIATION.REGIMENINDICATION', 'RADIATION_THERAPY_TYPE', and 'INITIAL_PATHOLOGIC_DX_YEAR'.
-
4 subtypes identified in current cancer cohort by 'MIRseq Mature cHierClus subtypes'. These subtypes correlate to 'HISTOLOGICAL.TYPE', 'RADIATIONS.RADIATION.REGIMENINDICATION', 'RADIATION_THERAPY_TYPE', and 'INITIAL_PATHOLOGIC_DX_YEAR'.
Table 1. Get Full Table Overview of the association between subtypes identified by 8 different clustering approaches and 45 clinical features. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 16 significant findings detected.
|
Clinical Features |
Statistical Tests |
Copy Number Ratio CNMF subtypes |
METHLYATION CNMF |
RNAseq CNMF subtypes |
RNAseq cHierClus subtypes |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRseq Mature CNMF subtypes |
MIRseq Mature cHierClus subtypes |
| Time to Death | logrank test |
0.364 (1.00) |
0.0878 (1.00) |
0.152 (1.00) |
0.00602 (1.00) |
0.436 (1.00) |
0.519 (1.00) |
0.765 (1.00) |
0.981 (1.00) |
| AGE | Kruskal-Wallis (anova) |
0.032 (1.00) |
0.00769 (1.00) |
0.0018 (0.616) |
0.0161 (1.00) |
0.171 (1.00) |
8.5e-06 (0.00304) |
0.415 (1.00) |
0.546 (1.00) |
| PATHOLOGY T STAGE | Fisher's exact test |
0.166 (1.00) |
0.995 (1.00) |
0.777 (1.00) |
0.946 (1.00) |
0.932 (1.00) |
0.00149 (0.511) |
0.892 (1.00) |
0.825 (1.00) |
| PATHOLOGY N STAGE | Fisher's exact test |
0.246 (1.00) |
0.415 (1.00) |
0.0774 (1.00) |
0.387 (1.00) |
0.832 (1.00) |
0.354 (1.00) |
0.339 (1.00) |
0.102 (1.00) |
| PATHOLOGY M STAGE | Fisher's exact test |
0.0537 (1.00) |
0.571 (1.00) |
0.197 (1.00) |
0.0499 (1.00) |
0.267 (1.00) |
0.00764 (1.00) |
0.124 (1.00) |
0.22 (1.00) |
| HISTOLOGICAL TYPE | Fisher's exact test |
0.16 (1.00) |
1e-05 (0.00357) |
1e-05 (0.00357) |
1e-05 (0.00357) |
1e-05 (0.00357) |
1e-05 (0.00357) |
8e-05 (0.0277) |
1e-05 (0.00357) |
| RADIATIONS RADIATION REGIMENINDICATION | Fisher's exact test |
0.878 (1.00) |
0.171 (1.00) |
0.438 (1.00) |
0.255 (1.00) |
0.251 (1.00) |
0.0434 (1.00) |
1e-05 (0.00357) |
1e-05 (0.00357) |
| NUMBERPACKYEARSSMOKED | Kruskal-Wallis (anova) |
0.536 (1.00) |
0.635 (1.00) |
0.461 (1.00) |
0.628 (1.00) |
0.147 (1.00) |
0.324 (1.00) |
0.408 (1.00) |
0.238 (1.00) |
| NUMBER OF LYMPH NODES | Kruskal-Wallis (anova) |
0.248 (1.00) |
0.601 (1.00) |
0.0389 (1.00) |
0.0689 (1.00) |
0.397 (1.00) |
0.228 (1.00) |
0.446 (1.00) |
0.118 (1.00) |
| RACE | Fisher's exact test |
0.0975 (1.00) |
0.32 (1.00) |
0.153 (1.00) |
0.469 (1.00) |
0.339 (1.00) |
0.59 (1.00) |
0.099 (1.00) |
0.499 (1.00) |
| ETHNICITY | Fisher's exact test |
0.839 (1.00) |
0.31 (1.00) |
0.496 (1.00) |
0.518 (1.00) |
0.26 (1.00) |
0.178 (1.00) |
0.0475 (1.00) |
0.294 (1.00) |
| WEIGHT KG AT DIAGNOSIS | Kruskal-Wallis (anova) |
0.462 (1.00) |
0.799 (1.00) |
0.332 (1.00) |
0.481 (1.00) |
0.476 (1.00) |
0.765 (1.00) |
0.889 (1.00) |
0.679 (1.00) |
| TUMOR STATUS | Fisher's exact test |
0.609 (1.00) |
0.84 (1.00) |
0.74 (1.00) |
0.606 (1.00) |
0.362 (1.00) |
0.88 (1.00) |
0.566 (1.00) |
0.399 (1.00) |
| TUMOR SAMPLE PROCUREMENT COUNTRY | Fisher's exact test |
0.381 (1.00) |
0.497 (1.00) |
0.766 (1.00) |
0.453 (1.00) |
0.0295 (1.00) |
0.239 (1.00) |
0.0764 (1.00) |
0.0207 (1.00) |
| NEOPLASMHISTOLOGICGRADE | Fisher's exact test |
0.123 (1.00) |
0.839 (1.00) |
0.00964 (1.00) |
0.0353 (1.00) |
0.0292 (1.00) |
0.0646 (1.00) |
0.0342 (1.00) |
0.0459 (1.00) |
| TOBACCO SMOKING YEAR STOPPED | Kruskal-Wallis (anova) |
0.866 (1.00) |
0.801 (1.00) |
0.975 (1.00) |
0.478 (1.00) |
0.12 (1.00) |
0.0569 (1.00) |
0.317 (1.00) |
0.0617 (1.00) |
| TOBACCO SMOKING PACK YEARS SMOKED | Kruskal-Wallis (anova) |
0.536 (1.00) |
0.635 (1.00) |
0.461 (1.00) |
0.628 (1.00) |
0.147 (1.00) |
0.324 (1.00) |
0.408 (1.00) |
0.238 (1.00) |
| TOBACCO SMOKING HISTORY | Fisher's exact test |
0.738 (1.00) |
0.584 (1.00) |
0.0998 (1.00) |
0.355 (1.00) |
0.238 (1.00) |
0.49 (1.00) |
0.124 (1.00) |
0.459 (1.00) |
| PATIENT AGEBEGANSMOKINGINYEARS | Kruskal-Wallis (anova) |
0.204 (1.00) |
0.93 (1.00) |
0.489 (1.00) |
0.368 (1.00) |
0.796 (1.00) |
0.705 (1.00) |
0.936 (1.00) |
0.998 (1.00) |
| RADIATION TOTAL DOSE | Kruskal-Wallis (anova) |
0.82 (1.00) |
0.974 (1.00) |
0.311 (1.00) |
0.897 (1.00) |
0.68 (1.00) |
0.415 (1.00) |
0.0236 (1.00) |
0.297 (1.00) |
| RADIATION THERAPY TYPE | Fisher's exact test |
0.929 (1.00) |
0.193 (1.00) |
0.556 (1.00) |
0.672 (1.00) |
0.12 (1.00) |
0.243 (1.00) |
2e-05 (0.00696) |
1e-05 (0.00357) |
| RADIATION THERAPY STATUS | Fisher's exact test |
0.262 (1.00) |
0.886 (1.00) |
0.454 (1.00) |
0.658 (1.00) |
0.396 (1.00) |
0.86 (1.00) |
1 (1.00) |
0.458 (1.00) |
| RADIATION THERAPY SITE | Fisher's exact test |
0.0858 (1.00) |
0.0606 (1.00) |
0.0669 (1.00) |
0.117 (1.00) |
0.0861 (1.00) |
0.151 (1.00) |
0.797 (1.00) |
0.504 (1.00) |
| RADIATION ADJUVANT UNITS | Fisher's exact test |
1 (1.00) |
0.928 (1.00) |
0.74 (1.00) |
1 (1.00) |
0.413 (1.00) |
0.881 (1.00) |
1 (1.00) |
0.761 (1.00) |
| PREGNANCIES COUNT TOTAL | Kruskal-Wallis (anova) |
0.479 (1.00) |
0.291 (1.00) |
0.0112 (1.00) |
0.0973 (1.00) |
0.461 (1.00) |
0.106 (1.00) |
0.436 (1.00) |
0.115 (1.00) |
| PREGNANCIES COUNT STILLBIRTH | Kruskal-Wallis (anova) |
0.246 (1.00) |
0.00484 (1.00) |
0.153 (1.00) |
0.578 (1.00) |
0.848 (1.00) |
0.477 (1.00) |
0.218 (1.00) |
0.216 (1.00) |
| PATIENT PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT | Kruskal-Wallis (anova) |
0.735 (1.00) |
0.423 (1.00) |
0.933 (1.00) |
0.0932 (1.00) |
0.00877 (1.00) |
0.307 (1.00) |
0.279 (1.00) |
0.357 (1.00) |
| PREGNANCIES COUNT LIVE BIRTH | Kruskal-Wallis (anova) |
0.619 (1.00) |
0.114 (1.00) |
0.0455 (1.00) |
0.0515 (1.00) |
0.326 (1.00) |
0.026 (1.00) |
0.341 (1.00) |
0.036 (1.00) |
| PATIENT PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT | Kruskal-Wallis (anova) |
0.896 (1.00) |
0.151 (1.00) |
0.849 (1.00) |
0.801 (1.00) |
0.396 (1.00) |
0.915 (1.00) |
0.555 (1.00) |
0.64 (1.00) |
| PREGNANCIES COUNT ECTOPIC | Kruskal-Wallis (anova) |
0.162 (1.00) |
0.769 (1.00) |
0.975 (1.00) |
0.984 (1.00) |
0.686 (1.00) |
0.794 (1.00) |
0.849 (1.00) |
0.658 (1.00) |
| POS LYMPH NODE LOCATION | Fisher's exact test |
0.862 (1.00) |
0.888 (1.00) |
0.988 (1.00) |
0.993 (1.00) |
0.809 (1.00) |
0.899 (1.00) |
0.646 (1.00) |
0.7 (1.00) |
| MENOPAUSE STATUS | Fisher's exact test |
0.0142 (1.00) |
0.0477 (1.00) |
0.0193 (1.00) |
0.026 (1.00) |
0.407 (1.00) |
0.00307 (1.00) |
0.414 (1.00) |
0.396 (1.00) |
| LYMPHOVASCULAR INVOLVEMENT | Fisher's exact test |
0.164 (1.00) |
0.592 (1.00) |
0.482 (1.00) |
0.596 (1.00) |
0.666 (1.00) |
0.445 (1.00) |
0.715 (1.00) |
0.0762 (1.00) |
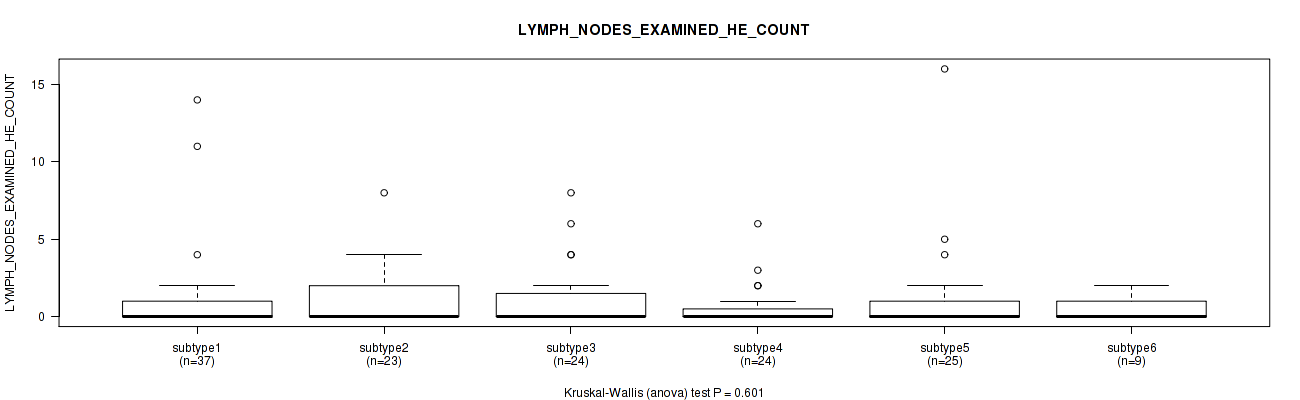
| LYMPH NODES EXAMINED HE COUNT | Kruskal-Wallis (anova) |
0.248 (1.00) |
0.601 (1.00) |
0.0389 (1.00) |
0.0689 (1.00) |
0.397 (1.00) |
0.228 (1.00) |
0.446 (1.00) |
0.118 (1.00) |
| LYMPH NODES EXAMINED | Kruskal-Wallis (anova) |
0.193 (1.00) |
0.588 (1.00) |
0.987 (1.00) |
0.595 (1.00) |
0.644 (1.00) |
0.533 (1.00) |
0.18 (1.00) |
0.641 (1.00) |
| KERATINIZATION SQUAMOUS CELL | Fisher's exact test |
0.838 (1.00) |
0.00981 (1.00) |
0.0619 (1.00) |
0.0717 (1.00) |
0.0298 (1.00) |
0.0905 (1.00) |
0.0458 (1.00) |
0.0192 (1.00) |
| INITIAL PATHOLOGIC DX YEAR | Kruskal-Wallis (anova) |
0.656 (1.00) |
0.386 (1.00) |
0.227 (1.00) |
0.154 (1.00) |
0.0327 (1.00) |
0.127 (1.00) |
2.69e-05 (0.00933) |
1.22e-06 (0.000438) |
| HISTORY HORMONAL CONTRACEPTIVES USE | Fisher's exact test |
0.617 (1.00) |
0.759 (1.00) |
0.0854 (1.00) |
0.03 (1.00) |
0.679 (1.00) |
0.416 (1.00) |
0.9 (1.00) |
0.395 (1.00) |
| HEIGHT CM AT DIAGNOSIS | Kruskal-Wallis (anova) |
0.0625 (1.00) |
0.275 (1.00) |
0.657 (1.00) |
0.418 (1.00) |
0.407 (1.00) |
0.685 (1.00) |
0.533 (1.00) |
0.753 (1.00) |
| CORPUS INVOLVEMENT | Fisher's exact test |
0.352 (1.00) |
0.694 (1.00) |
0.0605 (1.00) |
0.274 (1.00) |
0.803 (1.00) |
0.318 (1.00) |
0.755 (1.00) |
0.486 (1.00) |
| CHEMO CONCURRENT TYPE | Fisher's exact test |
0.538 (1.00) |
0.21 (1.00) |
1 (1.00) |
0.256 (1.00) |
1 (1.00) |
0.442 (1.00) |
0.206 (1.00) |
0.14 (1.00) |
| CERVIX SUV RESULTS | Kruskal-Wallis (anova) |
0.0166 (1.00) |
0.0821 (1.00) |
0.0298 (1.00) |
0.0961 (1.00) |
0.508 (1.00) |
0.207 (1.00) |
0.348 (1.00) |
0.302 (1.00) |
| AJCC TUMOR PATHOLOGIC PT | Fisher's exact test |
0.15 (1.00) |
0.444 (1.00) |
0.222 (1.00) |
0.114 (1.00) |
0.392 (1.00) |
0.00142 (0.488) |
0.235 (1.00) |
0.829 (1.00) |
| AGE AT DIAGNOSIS | Kruskal-Wallis (anova) |
0.0189 (1.00) |
0.0102 (1.00) |
0.00227 (0.773) |
0.0125 (1.00) |
0.21 (1.00) |
6.47e-06 (0.00232) |
0.37 (1.00) |
0.611 (1.00) |
| STAGE EVENT CLINICAL STAGE | Fisher's exact test |
0.0865 (1.00) |
0.105 (1.00) |
0.00183 (0.624) |
0.273 (1.00) |
0.0469 (1.00) |
0.00057 (0.197) |
0.00658 (1.00) |
0.193 (1.00) |
Table S1. Description of clustering approach #1: 'Copy Number Ratio CNMF subtypes'
| Cluster Labels | 1 | 2 | 3 |
|---|---|---|---|
| Number of samples | 105 | 57 | 76 |
P value = 0.364 (logrank test), Q value = 1
Table S2. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 233 | 50 | 0.0 - 195.8 (15.9) |
| subtype1 | 105 | 19 | 0.0 - 195.8 (14.5) |
| subtype2 | 54 | 16 | 0.1 - 147.3 (17.1) |
| subtype3 | 74 | 15 | 0.0 - 173.3 (17.8) |
Figure S1. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #1: 'Time to Death'

P value = 0.032 (Kruskal-Wallis (anova)), Q value = 1
Table S3. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 234 | 47.5 (13.5) |
| subtype1 | 105 | 46.5 (14.4) |
| subtype2 | 56 | 45.6 (13.5) |
| subtype3 | 73 | 50.2 (11.9) |
Figure S2. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #2: 'AGE'

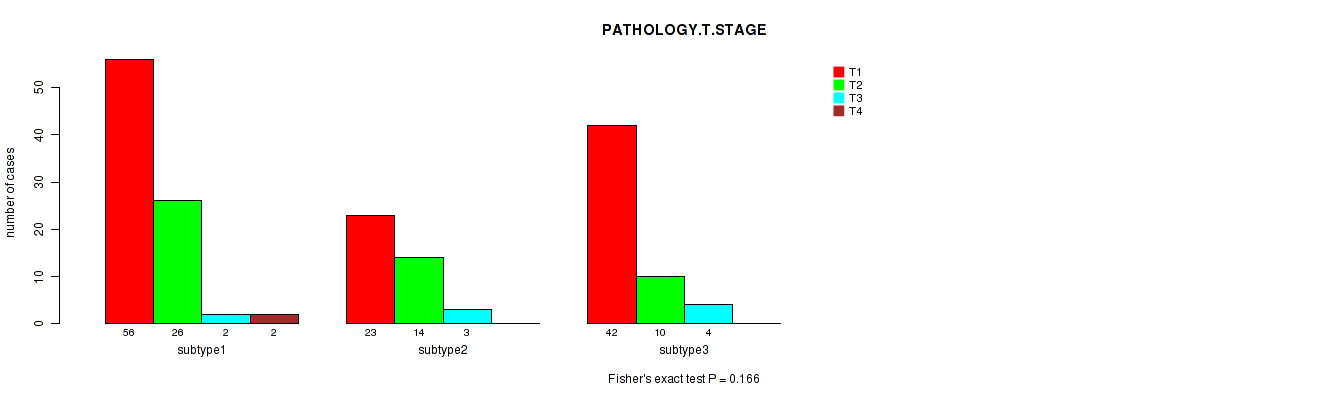
P value = 0.166 (Fisher's exact test), Q value = 1
Table S4. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'
| nPatients | T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 121 | 50 | 9 | 2 |
| subtype1 | 56 | 26 | 2 | 2 |
| subtype2 | 23 | 14 | 3 | 0 |
| subtype3 | 42 | 10 | 4 | 0 |
Figure S3. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'
P value = 0.246 (Fisher's exact test), Q value = 1
Table S5. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'
| nPatients | 0 | 1 |
|---|---|---|
| ALL | 111 | 47 |
| subtype1 | 58 | 18 |
| subtype2 | 21 | 13 |
| subtype3 | 32 | 16 |
Figure S4. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'

P value = 0.0537 (Fisher's exact test), Q value = 1
Table S6. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 91 | 8 | 93 |
| subtype1 | 36 | 4 | 50 |
| subtype2 | 18 | 3 | 22 |
| subtype3 | 37 | 1 | 21 |
Figure S5. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'

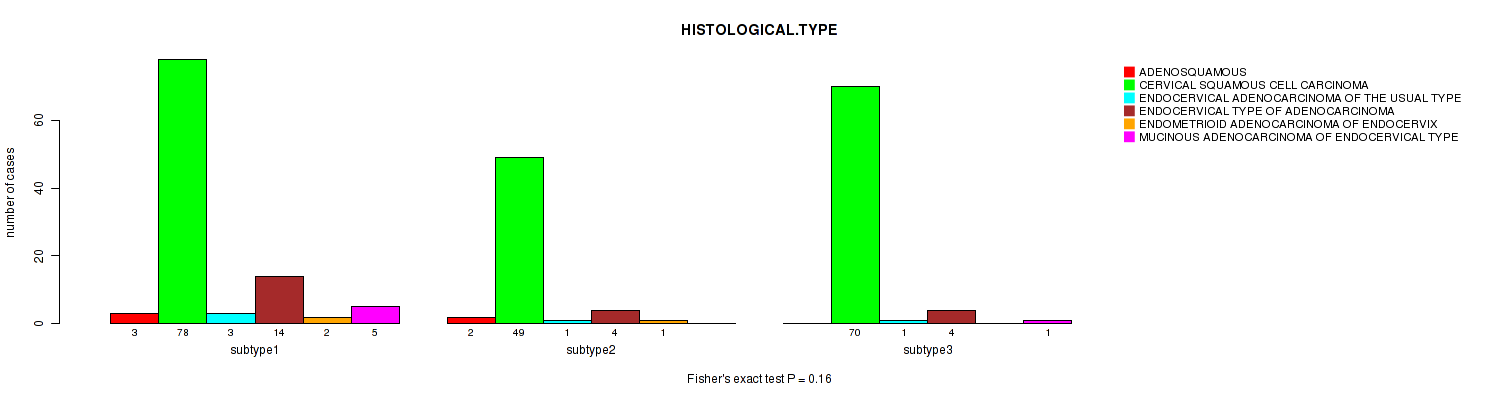
P value = 0.16 (Fisher's exact test), Q value = 1
Table S7. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | ADENOSQUAMOUS | CERVICAL SQUAMOUS CELL CARCINOMA | ENDOCERVICAL ADENOCARCINOMA OF THE USUAL TYPE | ENDOCERVICAL TYPE OF ADENOCARCINOMA | ENDOMETRIOID ADENOCARCINOMA OF ENDOCERVIX | MUCINOUS ADENOCARCINOMA OF ENDOCERVICAL TYPE |
|---|---|---|---|---|---|---|
| ALL | 5 | 197 | 5 | 22 | 3 | 6 |
| subtype1 | 3 | 78 | 3 | 14 | 2 | 5 |
| subtype2 | 2 | 49 | 1 | 4 | 1 | 0 |
| subtype3 | 0 | 70 | 1 | 4 | 0 | 1 |
Figure S6. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
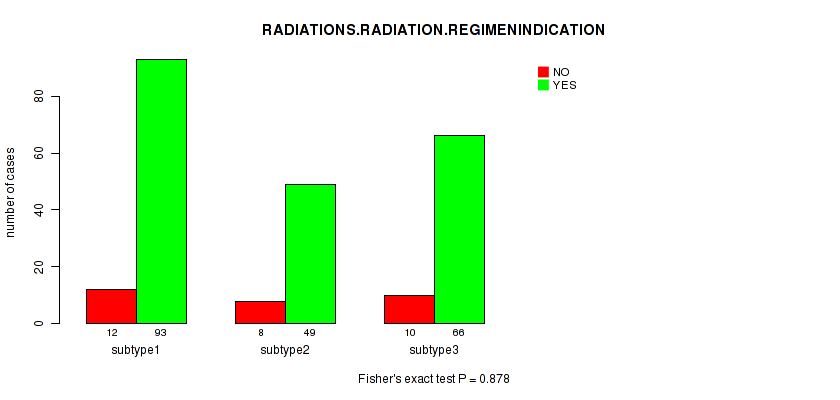
P value = 0.878 (Fisher's exact test), Q value = 1
Table S8. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 30 | 208 |
| subtype1 | 12 | 93 |
| subtype2 | 8 | 49 |
| subtype3 | 10 | 66 |
Figure S7. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'
P value = 0.536 (Kruskal-Wallis (anova)), Q value = 1
Table S9. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 73 | 19.0 (14.0) |
| subtype1 | 34 | 16.8 (11.5) |
| subtype2 | 16 | 21.0 (12.9) |
| subtype3 | 23 | 20.8 (17.7) |
Figure S8. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'

P value = 0.248 (Kruskal-Wallis (anova)), Q value = 1
Table S10. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 135 | 0.9 (2.1) |
| subtype1 | 67 | 1.0 (2.6) |
| subtype2 | 26 | 1.0 (1.8) |
| subtype3 | 42 | 0.8 (1.3) |
Figure S9. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'

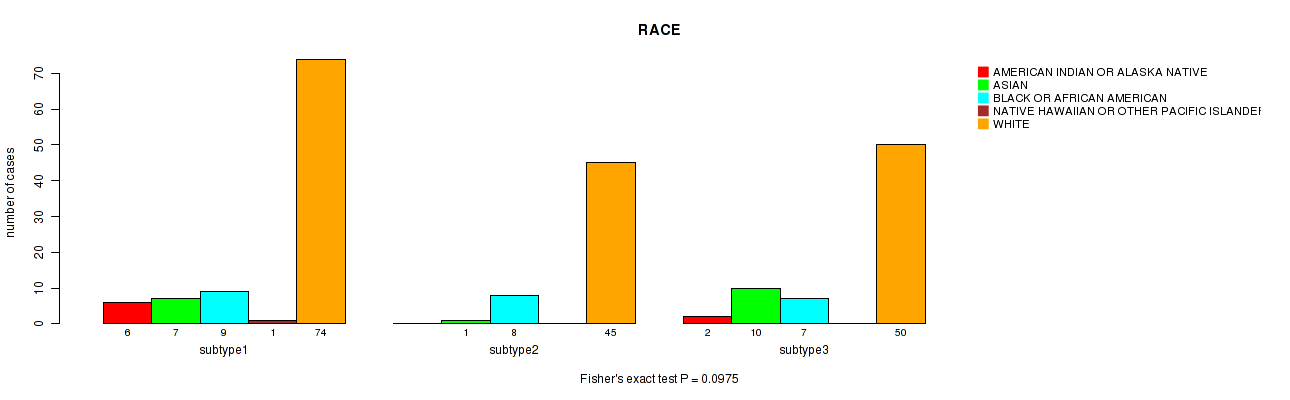
P value = 0.0975 (Fisher's exact test), Q value = 1
Table S11. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER | WHITE |
|---|---|---|---|---|---|
| ALL | 8 | 18 | 24 | 1 | 169 |
| subtype1 | 6 | 7 | 9 | 1 | 74 |
| subtype2 | 0 | 1 | 8 | 0 | 45 |
| subtype3 | 2 | 10 | 7 | 0 | 50 |
Figure S10. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #10: 'RACE'
P value = 0.839 (Fisher's exact test), Q value = 1
Table S12. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #11: 'ETHNICITY'
| nPatients | HISPANIC OR LATINO | NOT HISPANIC OR LATINO |
|---|---|---|
| ALL | 16 | 149 |
| subtype1 | 8 | 67 |
| subtype2 | 4 | 34 |
| subtype3 | 4 | 48 |
Figure S11. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #11: 'ETHNICITY'

P value = 0.462 (Kruskal-Wallis (anova)), Q value = 1
Table S13. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 216 | 75.1 (22.4) |
| subtype1 | 90 | 75.8 (19.0) |
| subtype2 | 54 | 71.8 (16.9) |
| subtype3 | 72 | 76.7 (29.1) |
Figure S12. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'

P value = 0.609 (Fisher's exact test), Q value = 1
Table S14. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #13: 'TUMOR_STATUS'
| nPatients | TUMOR FREE | WITH TUMOR |
|---|---|---|
| ALL | 71 | 25 |
| subtype1 | 32 | 9 |
| subtype2 | 16 | 8 |
| subtype3 | 23 | 8 |
Figure S13. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #13: 'TUMOR_STATUS'

P value = 0.381 (Fisher's exact test), Q value = 1
Table S15. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'
| nPatients | BRAZIL | CANADA | NIGERIA | RUSSIA | UKRAINE | UNITED STATES | VIETNAM |
|---|---|---|---|---|---|---|---|
| ALL | 23 | 5 | 1 | 10 | 7 | 178 | 14 |
| subtype1 | 7 | 3 | 0 | 3 | 4 | 83 | 5 |
| subtype2 | 8 | 1 | 1 | 2 | 1 | 43 | 1 |
| subtype3 | 8 | 1 | 0 | 5 | 2 | 52 | 8 |
Figure S14. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'
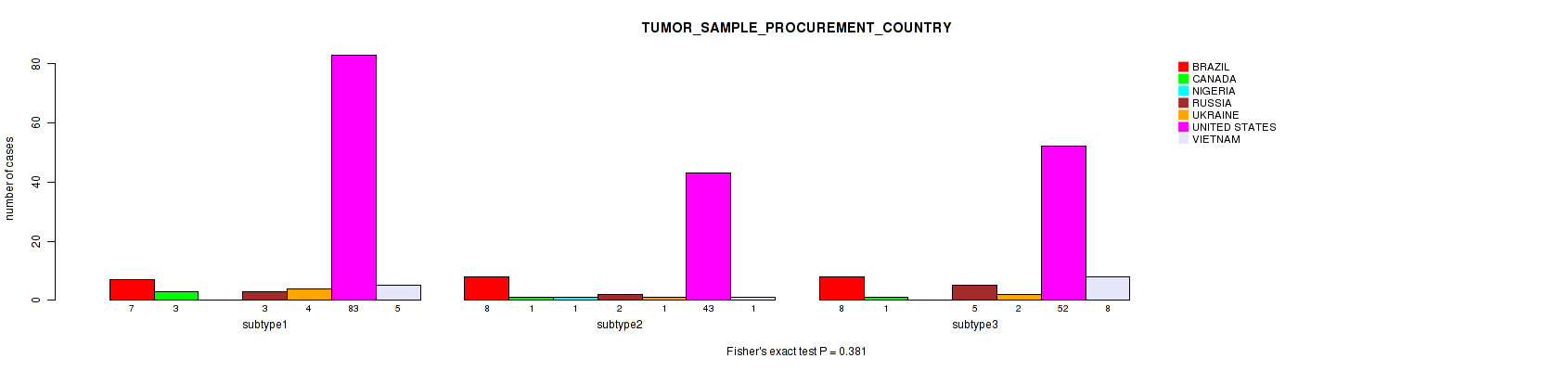
P value = 0.123 (Fisher's exact test), Q value = 1
Table S16. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'
| nPatients | G1 | G2 | G3 | G4 | GX |
|---|---|---|---|---|---|
| ALL | 17 | 106 | 97 | 1 | 14 |
| subtype1 | 9 | 49 | 40 | 0 | 6 |
| subtype2 | 2 | 17 | 31 | 1 | 4 |
| subtype3 | 6 | 40 | 26 | 0 | 4 |
Figure S15. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'

P value = 0.866 (Kruskal-Wallis (anova)), Q value = 1
Table S17. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 34 | 2000.4 (11.2) |
| subtype1 | 14 | 2000.6 (11.7) |
| subtype2 | 10 | 2001.2 (11.7) |
| subtype3 | 10 | 1999.4 (11.2) |
Figure S16. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'

P value = 0.536 (Kruskal-Wallis (anova)), Q value = 1
Table S18. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 73 | 19.0 (14.0) |
| subtype1 | 34 | 16.8 (11.5) |
| subtype2 | 16 | 21.0 (12.9) |
| subtype3 | 23 | 20.8 (17.7) |
Figure S17. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'
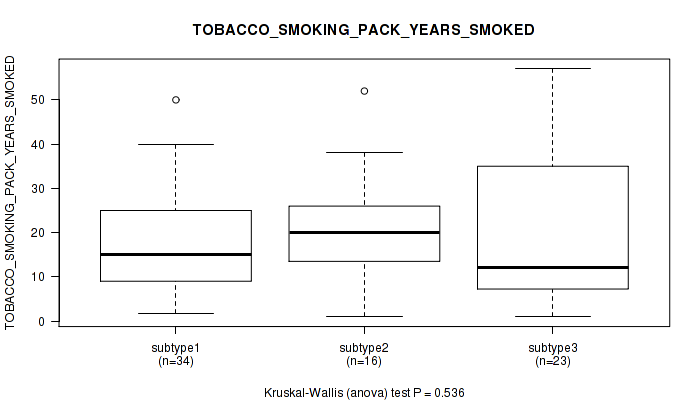
P value = 0.738 (Fisher's exact test), Q value = 1
Table S19. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'
| nPatients | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT REFORMED SMOKER, DURATION NOT SPECIFIED | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|
| ALL | 31 | 8 | 2 | 52 | 112 |
| subtype1 | 13 | 3 | 0 | 24 | 56 |
| subtype2 | 9 | 1 | 1 | 11 | 21 |
| subtype3 | 9 | 4 | 1 | 17 | 35 |
Figure S18. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'

P value = 0.204 (Kruskal-Wallis (anova)), Q value = 1
Table S20. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 67 | 21.5 (7.8) |
| subtype1 | 30 | 20.1 (6.2) |
| subtype2 | 16 | 20.7 (9.0) |
| subtype3 | 21 | 24.2 (8.7) |
Figure S19. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'

P value = 0.82 (Kruskal-Wallis (anova)), Q value = 1
Table S21. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 103 | 3861.7 (1648.6) |
| subtype1 | 41 | 3987.6 (1645.6) |
| subtype2 | 28 | 3543.8 (1858.6) |
| subtype3 | 34 | 3971.7 (1474.4) |
Figure S20. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'
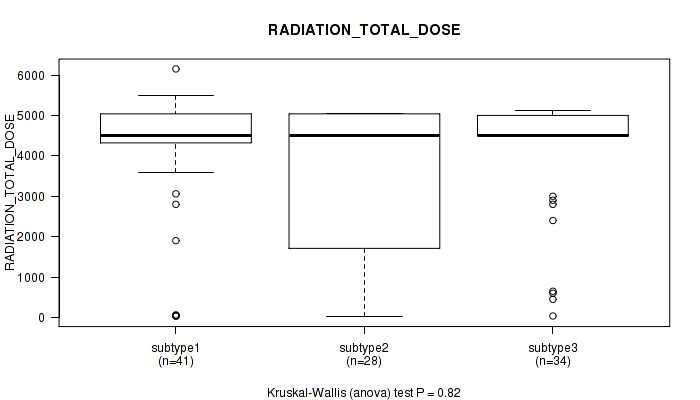
P value = 0.929 (Fisher's exact test), Q value = 1
Table S22. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'
| nPatients | COMBINATION | EXTERNAL | EXTERNAL BEAM | INTERNAL |
|---|---|---|---|---|
| ALL | 19 | 75 | 12 | 12 |
| subtype1 | 7 | 33 | 5 | 4 |
| subtype2 | 4 | 19 | 4 | 4 |
| subtype3 | 8 | 23 | 3 | 4 |
Figure S21. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'
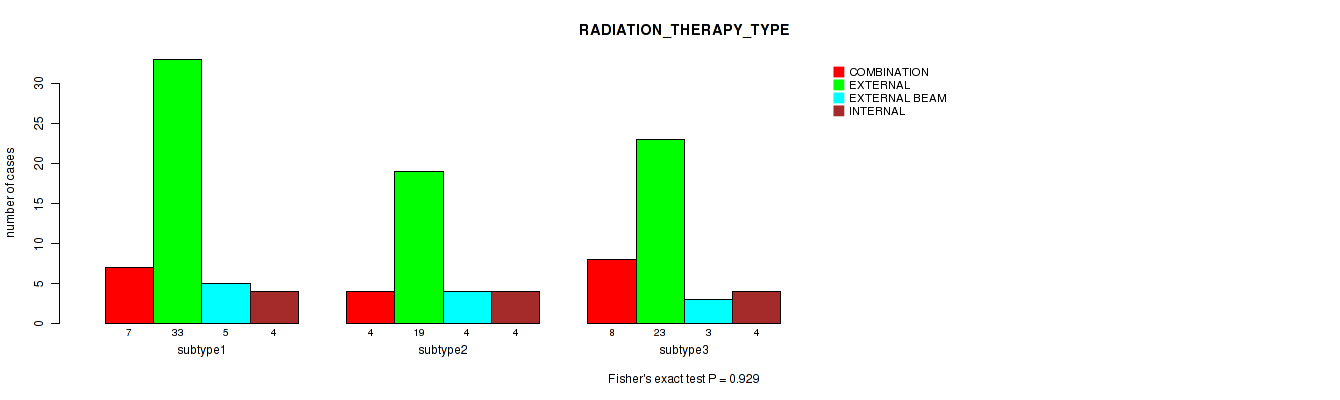
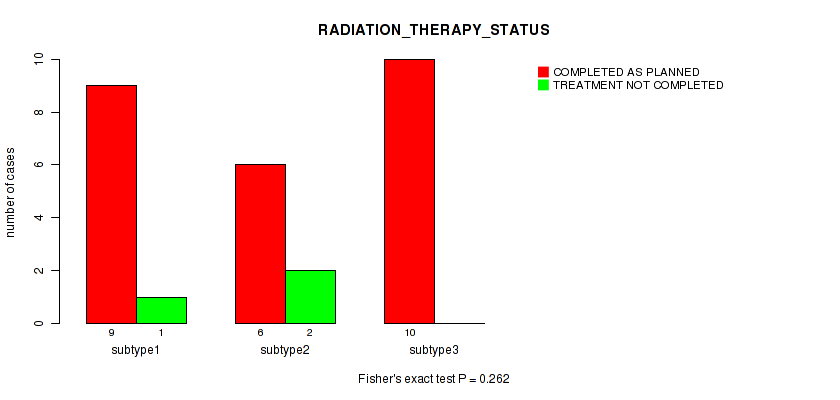
P value = 0.262 (Fisher's exact test), Q value = 1
Table S23. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'
| nPatients | COMPLETED AS PLANNED | TREATMENT NOT COMPLETED |
|---|---|---|
| ALL | 25 | 3 |
| subtype1 | 9 | 1 |
| subtype2 | 6 | 2 |
| subtype3 | 10 | 0 |
Figure S22. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'
P value = 0.0858 (Fisher's exact test), Q value = 1
Table S24. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'
| nPatients | DISTANT RECURRENCE | LOCAL RECURRENCE | PRIMARY TUMOR FIELD | REGIONAL SITE |
|---|---|---|---|---|
| ALL | 2 | 2 | 28 | 16 |
| subtype1 | 2 | 0 | 12 | 3 |
| subtype2 | 0 | 2 | 9 | 5 |
| subtype3 | 0 | 0 | 7 | 8 |
Figure S23. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'

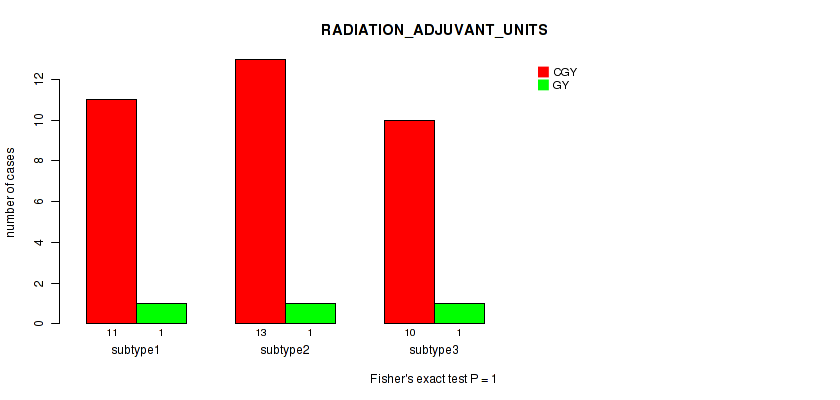
P value = 1 (Fisher's exact test), Q value = 1
Table S25. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'
| nPatients | CGY | GY |
|---|---|---|
| ALL | 34 | 3 |
| subtype1 | 11 | 1 |
| subtype2 | 13 | 1 |
| subtype3 | 10 | 1 |
Figure S24. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'
P value = 0.479 (Kruskal-Wallis (anova)), Q value = 1
Table S26. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 207 | 3.5 (2.4) |
| subtype1 | 92 | 3.4 (2.1) |
| subtype2 | 49 | 3.4 (2.6) |
| subtype3 | 66 | 3.8 (2.7) |
Figure S25. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'

P value = 0.246 (Kruskal-Wallis (anova)), Q value = 1
Table S27. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 103 | 0.1 (0.4) |
| subtype1 | 52 | 0.1 (0.3) |
| subtype2 | 18 | 0.0 (0.0) |
| subtype3 | 33 | 0.1 (0.5) |
Figure S26. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'

P value = 0.735 (Kruskal-Wallis (anova)), Q value = 1
Table S28. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 128 | 0.5 (0.8) |
| subtype1 | 63 | 0.4 (0.8) |
| subtype2 | 24 | 0.5 (0.7) |
| subtype3 | 41 | 0.5 (1.0) |
Figure S27. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'

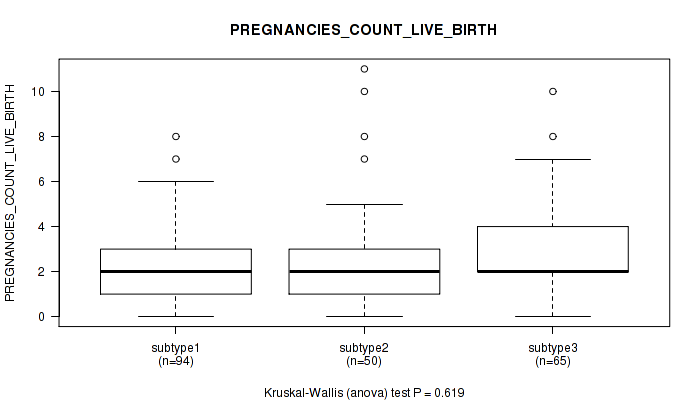
P value = 0.619 (Kruskal-Wallis (anova)), Q value = 1
Table S29. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 209 | 2.6 (1.9) |
| subtype1 | 94 | 2.5 (1.6) |
| subtype2 | 50 | 2.8 (2.3) |
| subtype3 | 65 | 2.7 (1.8) |
Figure S28. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'
P value = 0.896 (Kruskal-Wallis (anova)), Q value = 1
Table S30. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 113 | 0.8 (1.7) |
| subtype1 | 57 | 0.7 (1.1) |
| subtype2 | 22 | 0.7 (1.0) |
| subtype3 | 34 | 1.2 (2.6) |
Figure S29. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'

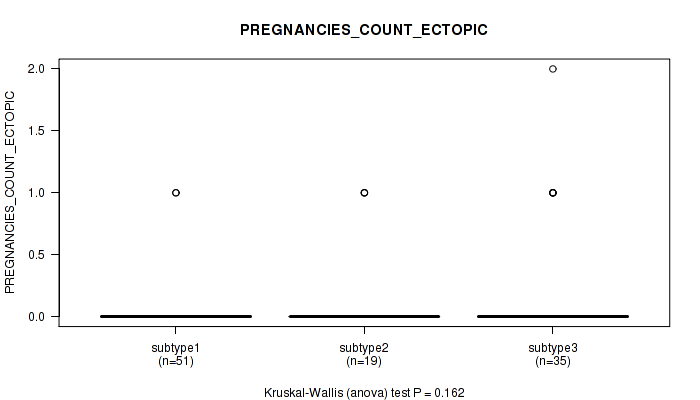
P value = 0.162 (Kruskal-Wallis (anova)), Q value = 1
Table S31. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 105 | 0.1 (0.3) |
| subtype1 | 51 | 0.0 (0.2) |
| subtype2 | 19 | 0.2 (0.4) |
| subtype3 | 35 | 0.2 (0.5) |
Figure S30. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'
P value = 0.862 (Fisher's exact test), Q value = 1
Table S32. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'
| nPatients | MACROSCOPIC PARAMETRIAL INVOLVEMENT | MICROSCOPIC PARAMETRIAL INVOLVEMENT | OTHER LOCATION, SPECIFY | POSITIVE BLADDER MARGIN | POSITIVE VAGINAL MARGIN |
|---|---|---|---|---|---|
| ALL | 2 | 6 | 29 | 1 | 7 |
| subtype1 | 1 | 1 | 12 | 1 | 3 |
| subtype2 | 0 | 3 | 7 | 0 | 1 |
| subtype3 | 1 | 2 | 10 | 0 | 3 |
Figure S31. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'
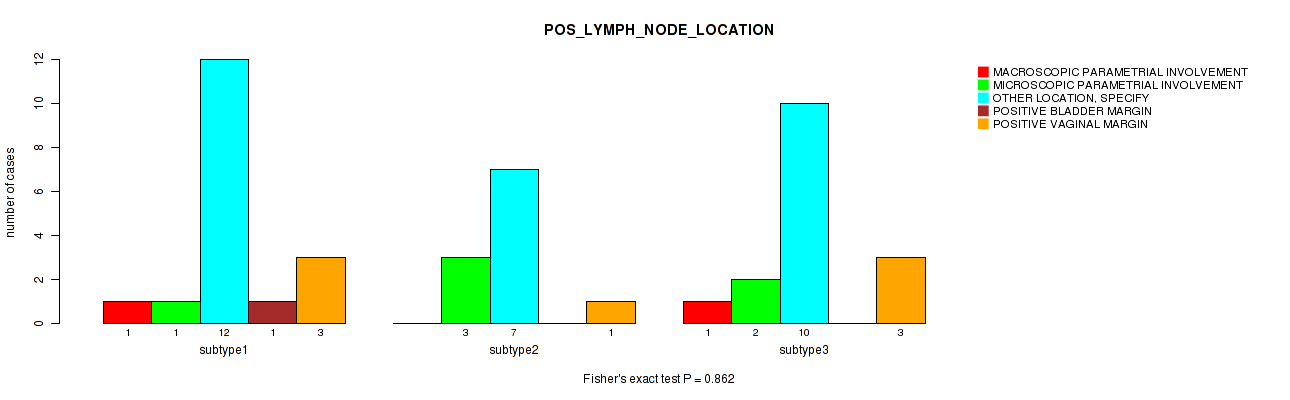
P value = 0.0142 (Fisher's exact test), Q value = 1
Table S33. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #32: 'MENOPAUSE_STATUS'
| nPatients | INDETERMINATE (NEITHER PRE OR POSTMENOPAUSAL) | PERI (6-12 MONTHS SINCE LAST MENSTRUAL PERIOD) | POST (PRIOR BILATERAL OVARIECTOMY OR >12 MO SINCE LMP WITH NO PRIOR HYSTERECTOMY) | PRE (<6 MONTHS SINCE LMP AND NO PRIOR BILATERAL OVARIECTOMY AND NOT ON ESTROGEN REPLACEMENT) |
|---|---|---|---|---|
| ALL | 2 | 16 | 68 | 105 |
| subtype1 | 0 | 4 | 29 | 53 |
| subtype2 | 0 | 2 | 12 | 27 |
| subtype3 | 2 | 10 | 27 | 25 |
Figure S32. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #32: 'MENOPAUSE_STATUS'

P value = 0.164 (Fisher's exact test), Q value = 1
Table S34. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 68 | 70 |
| subtype1 | 37 | 30 |
| subtype2 | 9 | 18 |
| subtype3 | 22 | 22 |
Figure S33. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'

P value = 0.248 (Kruskal-Wallis (anova)), Q value = 1
Table S35. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 135 | 0.9 (2.1) |
| subtype1 | 67 | 1.0 (2.6) |
| subtype2 | 26 | 1.0 (1.8) |
| subtype3 | 42 | 0.8 (1.3) |
Figure S34. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'

P value = 0.193 (Kruskal-Wallis (anova)), Q value = 1
Table S36. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 156 | 21.7 (12.6) |
| subtype1 | 75 | 20.9 (12.4) |
| subtype2 | 33 | 25.1 (12.7) |
| subtype3 | 48 | 20.7 (12.9) |
Figure S35. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'

P value = 0.838 (Fisher's exact test), Q value = 1
Table S37. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'
| nPatients | KERATINIZING SQUAMOUS CELL CARCINOMA | NON-KERATINIZING SQUAMOUS CELL CARCINOMA |
|---|---|---|
| ALL | 43 | 94 |
| subtype1 | 19 | 42 |
| subtype2 | 8 | 21 |
| subtype3 | 16 | 31 |
Figure S36. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'

P value = 0.656 (Kruskal-Wallis (anova)), Q value = 1
Table S38. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 236 | 2007.9 (4.9) |
| subtype1 | 103 | 2008.1 (5.0) |
| subtype2 | 57 | 2007.8 (4.6) |
| subtype3 | 76 | 2007.7 (5.0) |
Figure S37. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'

P value = 0.617 (Fisher's exact test), Q value = 1
Table S39. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'
| nPatients | CURRENT USER | FORMER USER | NEVER USED |
|---|---|---|---|
| ALL | 10 | 49 | 57 |
| subtype1 | 5 | 23 | 23 |
| subtype2 | 3 | 12 | 11 |
| subtype3 | 2 | 14 | 23 |
Figure S38. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'
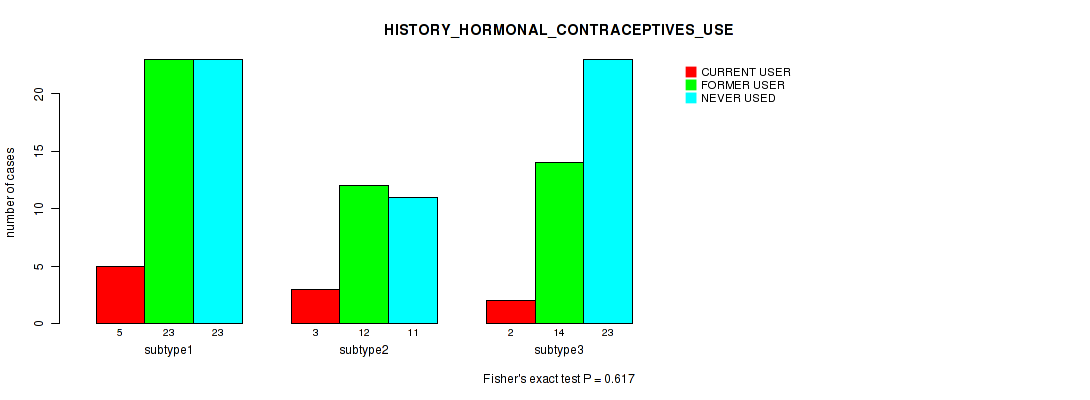
P value = 0.0625 (Kruskal-Wallis (anova)), Q value = 1
Table S40. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 207 | 161.8 (6.5) |
| subtype1 | 89 | 162.3 (7.4) |
| subtype2 | 49 | 163.0 (6.3) |
| subtype3 | 69 | 160.4 (5.3) |
Figure S39. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'

P value = 0.352 (Fisher's exact test), Q value = 1
Table S41. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 89 | 16 |
| subtype1 | 44 | 6 |
| subtype2 | 14 | 5 |
| subtype3 | 31 | 5 |
Figure S40. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'

P value = 0.538 (Fisher's exact test), Q value = 1
Table S42. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'
| nPatients | CARBOPLATIN | CISPLATIN | OTHER |
|---|---|---|---|
| ALL | 1 | 15 | 1 |
| subtype1 | 1 | 4 | 1 |
| subtype2 | 0 | 7 | 0 |
| subtype3 | 0 | 4 | 0 |
Figure S41. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'

P value = 0.0166 (Kruskal-Wallis (anova)), Q value = 1
Table S43. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 13 | 12.4 (5.5) |
| subtype1 | 7 | 9.1 (2.9) |
| subtype2 | 3 | 20.1 (5.2) |
| subtype3 | 3 | 12.5 (1.7) |
Figure S42. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'

P value = 0.15 (Fisher's exact test), Q value = 1
Table S44. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'
| nPatients | T1A | T1A1 | T1B | T1B1 | T1B2 | T2 | T2A | T2A1 | T2A2 | T2B | T3 | T3B | T4 | TIS | TX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 1 | 1 | 30 | 65 | 24 | 4 | 8 | 7 | 9 | 22 | 1 | 8 | 2 | 1 | 14 |
| subtype1 | 0 | 1 | 13 | 29 | 13 | 2 | 5 | 3 | 7 | 9 | 1 | 1 | 2 | 0 | 6 |
| subtype2 | 0 | 0 | 7 | 13 | 3 | 0 | 0 | 4 | 2 | 8 | 0 | 3 | 0 | 1 | 2 |
| subtype3 | 1 | 0 | 10 | 23 | 8 | 2 | 3 | 0 | 0 | 5 | 0 | 4 | 0 | 0 | 6 |
Figure S43. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'

P value = 0.0189 (Kruskal-Wallis (anova)), Q value = 1
Table S45. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 238 | 47.6 (13.5) |
| subtype1 | 105 | 46.5 (14.4) |
| subtype2 | 57 | 45.7 (13.3) |
| subtype3 | 76 | 50.5 (11.7) |
Figure S44. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'

P value = 0.0865 (Fisher's exact test), Q value = 1
Table S46. Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'
| nPatients | STAGE I | STAGE IA | STAGE IA1 | STAGE IA2 | STAGE IB | STAGE IB1 | STAGE IB2 | STAGE II | STAGE IIA | STAGE IIA1 | STAGE IIA2 | STAGE IIB | STAGE III | STAGE IIIB | STAGE IVA | STAGE IVB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 4 | 2 | 1 | 1 | 31 | 69 | 32 | 3 | 6 | 5 | 7 | 27 | 1 | 34 | 3 | 7 |
| subtype1 | 1 | 0 | 1 | 0 | 12 | 35 | 15 | 0 | 4 | 2 | 5 | 9 | 1 | 14 | 1 | 5 |
| subtype2 | 1 | 0 | 0 | 1 | 7 | 13 | 7 | 0 | 2 | 3 | 2 | 12 | 0 | 8 | 0 | 0 |
| subtype3 | 2 | 2 | 0 | 0 | 12 | 21 | 10 | 3 | 0 | 0 | 0 | 6 | 0 | 12 | 2 | 2 |
Figure S45. Get High-res Image Clustering Approach #1: 'Copy Number Ratio CNMF subtypes' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'

Table S47. Description of clustering approach #2: 'METHLYATION CNMF'
| Cluster Labels | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Number of samples | 56 | 41 | 44 | 51 | 37 | 14 |
P value = 0.0878 (logrank test), Q value = 1
Table S48. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 238 | 53 | 0.0 - 195.8 (15.9) |
| subtype1 | 55 | 12 | 0.1 - 137.2 (15.0) |
| subtype2 | 41 | 6 | 0.1 - 195.8 (15.6) |
| subtype3 | 44 | 10 | 0.0 - 182.9 (19.7) |
| subtype4 | 49 | 19 | 0.0 - 154.3 (17.2) |
| subtype5 | 35 | 5 | 0.0 - 134.3 (20.9) |
| subtype6 | 14 | 1 | 0.1 - 53.2 (5.5) |
Figure S46. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #1: 'Time to Death'

P value = 0.00769 (Kruskal-Wallis (anova)), Q value = 1
Table S49. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 239 | 47.4 (13.6) |
| subtype1 | 54 | 45.3 (11.6) |
| subtype2 | 40 | 50.5 (14.3) |
| subtype3 | 44 | 46.7 (13.9) |
| subtype4 | 50 | 42.6 (12.9) |
| subtype5 | 37 | 53.0 (13.1) |
| subtype6 | 14 | 50.9 (14.8) |
Figure S47. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #2: 'AGE'

P value = 0.995 (Fisher's exact test), Q value = 1
Table S50. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'
| nPatients | T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 125 | 48 | 8 | 4 |
| subtype1 | 33 | 13 | 1 | 1 |
| subtype2 | 20 | 7 | 2 | 1 |
| subtype3 | 24 | 10 | 1 | 1 |
| subtype4 | 21 | 10 | 2 | 1 |
| subtype5 | 20 | 5 | 2 | 0 |
| subtype6 | 7 | 3 | 0 | 0 |
Figure S48. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'
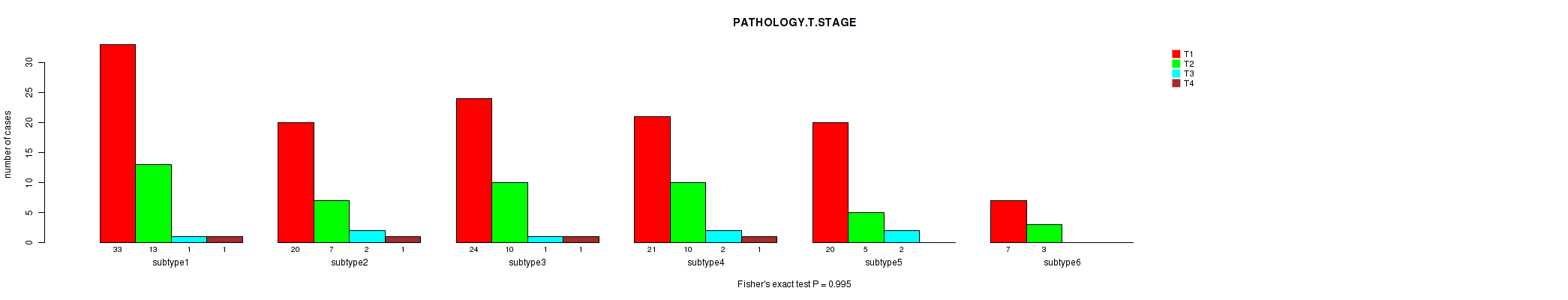
P value = 0.415 (Fisher's exact test), Q value = 1
Table S51. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'
| nPatients | 0 | 1 |
|---|---|---|
| ALL | 115 | 50 |
| subtype1 | 30 | 13 |
| subtype2 | 15 | 12 |
| subtype3 | 22 | 11 |
| subtype4 | 23 | 5 |
| subtype5 | 18 | 6 |
| subtype6 | 7 | 3 |
Figure S49. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'
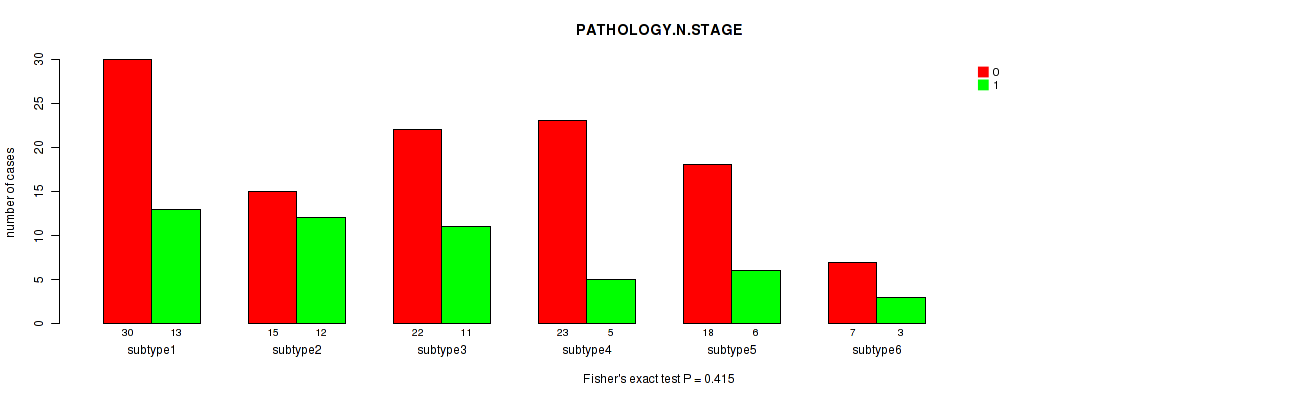
P value = 0.571 (Fisher's exact test), Q value = 1
Table S52. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 99 | 8 | 89 |
| subtype1 | 23 | 5 | 21 |
| subtype2 | 17 | 0 | 16 |
| subtype3 | 21 | 1 | 16 |
| subtype4 | 19 | 2 | 16 |
| subtype5 | 16 | 0 | 13 |
| subtype6 | 3 | 0 | 7 |
Figure S50. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'

P value = 1e-05 (Fisher's exact test), Q value = 0.0036
Table S53. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | ADENOSQUAMOUS | CERVICAL SQUAMOUS CELL CARCINOMA | ENDOCERVICAL ADENOCARCINOMA OF THE USUAL TYPE | ENDOCERVICAL TYPE OF ADENOCARCINOMA | ENDOMETRIOID ADENOCARCINOMA OF ENDOCERVIX | MUCINOUS ADENOCARCINOMA OF ENDOCERVICAL TYPE |
|---|---|---|---|---|---|---|
| ALL | 4 | 204 | 5 | 22 | 2 | 6 |
| subtype1 | 4 | 20 | 4 | 20 | 2 | 6 |
| subtype2 | 0 | 39 | 0 | 2 | 0 | 0 |
| subtype3 | 0 | 44 | 0 | 0 | 0 | 0 |
| subtype4 | 0 | 51 | 0 | 0 | 0 | 0 |
| subtype5 | 0 | 37 | 0 | 0 | 0 | 0 |
| subtype6 | 0 | 13 | 1 | 0 | 0 | 0 |
Figure S51. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 0.171 (Fisher's exact test), Q value = 1
Table S54. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 35 | 208 |
| subtype1 | 4 | 52 |
| subtype2 | 3 | 38 |
| subtype3 | 8 | 36 |
| subtype4 | 11 | 40 |
| subtype5 | 7 | 30 |
| subtype6 | 2 | 12 |
Figure S52. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'

P value = 0.635 (Kruskal-Wallis (anova)), Q value = 1
Table S55. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 75 | 18.4 (13.4) |
| subtype1 | 15 | 15.1 (11.4) |
| subtype2 | 14 | 20.2 (16.7) |
| subtype3 | 14 | 15.1 (8.0) |
| subtype4 | 15 | 17.8 (14.0) |
| subtype5 | 13 | 22.5 (12.8) |
| subtype6 | 4 | 24.5 (22.3) |
Figure S53. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'
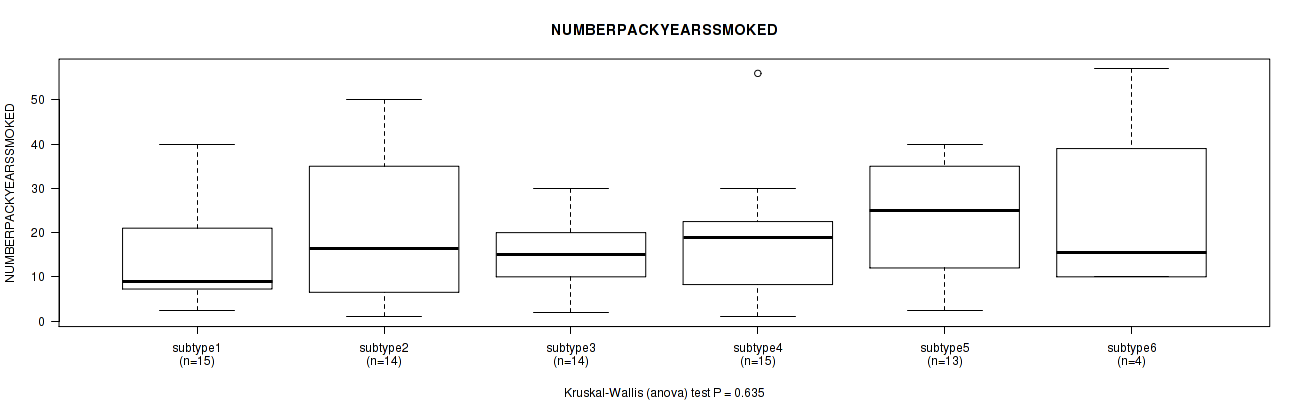
P value = 0.601 (Kruskal-Wallis (anova)), Q value = 1
Table S56. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 142 | 1.0 (2.4) |
| subtype1 | 37 | 1.1 (2.9) |
| subtype2 | 23 | 1.3 (2.0) |
| subtype3 | 24 | 1.2 (2.2) |
| subtype4 | 24 | 0.6 (1.4) |
| subtype5 | 25 | 1.2 (3.3) |
| subtype6 | 9 | 0.6 (0.9) |
Figure S54. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'

P value = 0.32 (Fisher's exact test), Q value = 1
Table S57. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER | WHITE |
|---|---|---|---|---|---|
| ALL | 8 | 19 | 26 | 1 | 172 |
| subtype1 | 1 | 4 | 3 | 0 | 43 |
| subtype2 | 1 | 4 | 4 | 0 | 29 |
| subtype3 | 1 | 6 | 6 | 0 | 28 |
| subtype4 | 4 | 3 | 8 | 1 | 34 |
| subtype5 | 1 | 0 | 2 | 0 | 29 |
| subtype6 | 0 | 2 | 3 | 0 | 9 |
Figure S55. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #10: 'RACE'

P value = 0.31 (Fisher's exact test), Q value = 1
Table S58. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #11: 'ETHNICITY'
| nPatients | HISPANIC OR LATINO | NOT HISPANIC OR LATINO |
|---|---|---|
| ALL | 17 | 155 |
| subtype1 | 6 | 35 |
| subtype2 | 4 | 25 |
| subtype3 | 1 | 33 |
| subtype4 | 5 | 29 |
| subtype5 | 1 | 22 |
| subtype6 | 0 | 11 |
Figure S56. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #11: 'ETHNICITY'

P value = 0.799 (Kruskal-Wallis (anova)), Q value = 1
Table S59. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 221 | 75.0 (22.6) |
| subtype1 | 50 | 75.6 (15.1) |
| subtype2 | 41 | 75.4 (23.1) |
| subtype3 | 38 | 71.0 (21.0) |
| subtype4 | 44 | 74.9 (20.5) |
| subtype5 | 35 | 76.8 (29.4) |
| subtype6 | 13 | 77.9 (35.4) |
Figure S57. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'

P value = 0.84 (Fisher's exact test), Q value = 1
Table S60. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #13: 'TUMOR_STATUS'
| nPatients | TUMOR FREE | WITH TUMOR |
|---|---|---|
| ALL | 75 | 28 |
| subtype1 | 18 | 7 |
| subtype2 | 10 | 5 |
| subtype3 | 18 | 4 |
| subtype4 | 15 | 8 |
| subtype5 | 9 | 3 |
| subtype6 | 5 | 1 |
Figure S58. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #13: 'TUMOR_STATUS'

P value = 0.497 (Fisher's exact test), Q value = 1
Table S61. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'
| nPatients | BRAZIL | CANADA | NIGERIA | RUSSIA | UKRAINE | UNITED STATES | VIETNAM |
|---|---|---|---|---|---|---|---|
| ALL | 20 | 5 | 1 | 11 | 7 | 185 | 14 |
| subtype1 | 4 | 3 | 1 | 2 | 1 | 43 | 2 |
| subtype2 | 4 | 0 | 0 | 2 | 2 | 30 | 3 |
| subtype3 | 3 | 1 | 0 | 3 | 0 | 33 | 4 |
| subtype4 | 7 | 0 | 0 | 0 | 3 | 38 | 3 |
| subtype5 | 2 | 1 | 0 | 4 | 1 | 29 | 0 |
| subtype6 | 0 | 0 | 0 | 0 | 0 | 12 | 2 |
Figure S59. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'

P value = 0.839 (Fisher's exact test), Q value = 1
Table S62. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'
| nPatients | G1 | G2 | G3 | G4 | GX |
|---|---|---|---|---|---|
| ALL | 15 | 112 | 98 | 1 | 14 |
| subtype1 | 6 | 24 | 23 | 0 | 3 |
| subtype2 | 1 | 21 | 18 | 0 | 1 |
| subtype3 | 2 | 22 | 17 | 0 | 3 |
| subtype4 | 1 | 22 | 22 | 1 | 3 |
| subtype5 | 4 | 18 | 11 | 0 | 3 |
| subtype6 | 1 | 5 | 7 | 0 | 1 |
Figure S60. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'

P value = 0.801 (Kruskal-Wallis (anova)), Q value = 1
Table S63. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 34 | 2000.4 (11.2) |
| subtype1 | 6 | 1999.0 (14.1) |
| subtype2 | 8 | 1998.8 (14.6) |
| subtype3 | 6 | 1999.8 (5.8) |
| subtype4 | 4 | 2002.0 (5.6) |
| subtype5 | 6 | 1999.2 (14.0) |
| subtype6 | 4 | 2007.0 (8.3) |
Figure S61. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'
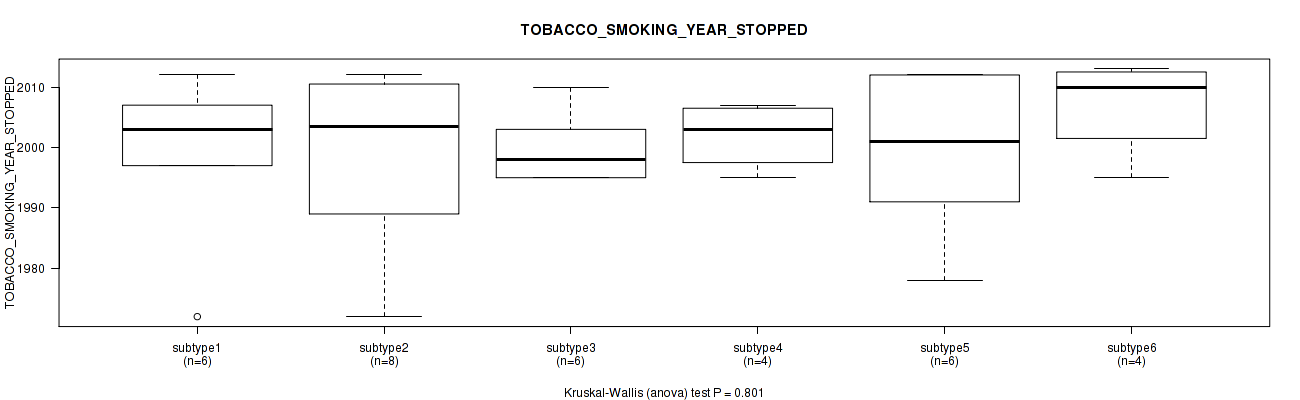
P value = 0.635 (Kruskal-Wallis (anova)), Q value = 1
Table S64. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 75 | 18.4 (13.4) |
| subtype1 | 15 | 15.1 (11.4) |
| subtype2 | 14 | 20.2 (16.7) |
| subtype3 | 14 | 15.1 (8.0) |
| subtype4 | 15 | 17.8 (14.0) |
| subtype5 | 13 | 22.5 (12.8) |
| subtype6 | 4 | 24.5 (22.3) |
Figure S62. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'

P value = 0.584 (Fisher's exact test), Q value = 1
Table S65. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'
| nPatients | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT REFORMED SMOKER, DURATION NOT SPECIFIED | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|
| ALL | 33 | 8 | 3 | 53 | 108 |
| subtype1 | 6 | 1 | 0 | 10 | 31 |
| subtype2 | 7 | 2 | 1 | 8 | 18 |
| subtype3 | 6 | 2 | 0 | 9 | 22 |
| subtype4 | 5 | 0 | 1 | 14 | 21 |
| subtype5 | 5 | 3 | 1 | 9 | 11 |
| subtype6 | 4 | 0 | 0 | 3 | 5 |
Figure S63. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'
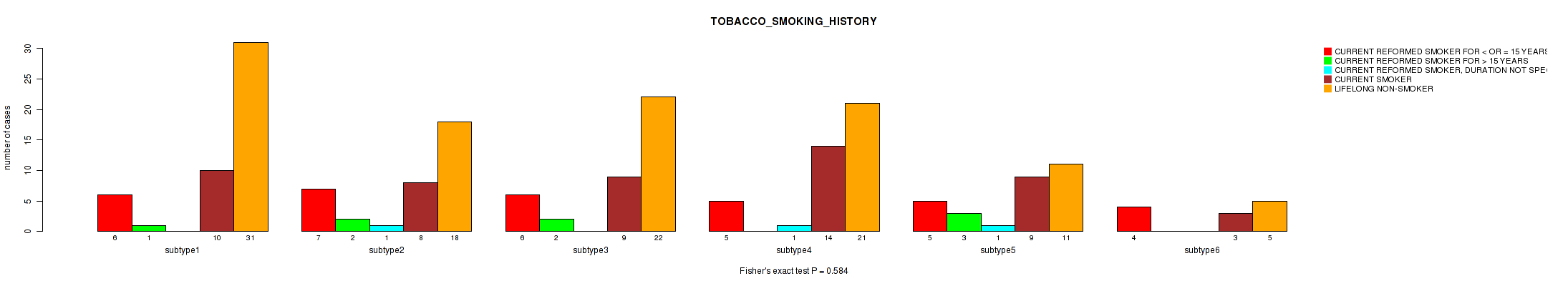
P value = 0.93 (Kruskal-Wallis (anova)), Q value = 1
Table S66. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 67 | 21.5 (7.9) |
| subtype1 | 14 | 19.3 (4.4) |
| subtype2 | 15 | 21.9 (10.1) |
| subtype3 | 11 | 20.1 (5.8) |
| subtype4 | 13 | 23.1 (9.5) |
| subtype5 | 10 | 23.5 (8.8) |
| subtype6 | 4 | 21.0 (6.2) |
Figure S64. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'

P value = 0.974 (Kruskal-Wallis (anova)), Q value = 1
Table S67. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 104 | 3933.6 (1618.2) |
| subtype1 | 22 | 3930.1 (1721.5) |
| subtype2 | 15 | 3355.2 (2028.8) |
| subtype3 | 18 | 4321.4 (1175.0) |
| subtype4 | 28 | 3842.7 (1787.4) |
| subtype5 | 16 | 4331.2 (851.0) |
| subtype6 | 5 | 3525.0 (2117.0) |
Figure S65. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'

P value = 0.193 (Fisher's exact test), Q value = 1
Table S68. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'
| nPatients | COMBINATION | EXTERNAL | EXTERNAL BEAM | IMPLANTS | INTERNAL |
|---|---|---|---|---|---|
| ALL | 20 | 72 | 15 | 1 | 11 |
| subtype1 | 3 | 20 | 1 | 0 | 1 |
| subtype2 | 3 | 11 | 0 | 1 | 2 |
| subtype3 | 5 | 13 | 3 | 0 | 0 |
| subtype4 | 6 | 16 | 5 | 0 | 4 |
| subtype5 | 2 | 8 | 5 | 0 | 4 |
| subtype6 | 1 | 4 | 1 | 0 | 0 |
Figure S66. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'
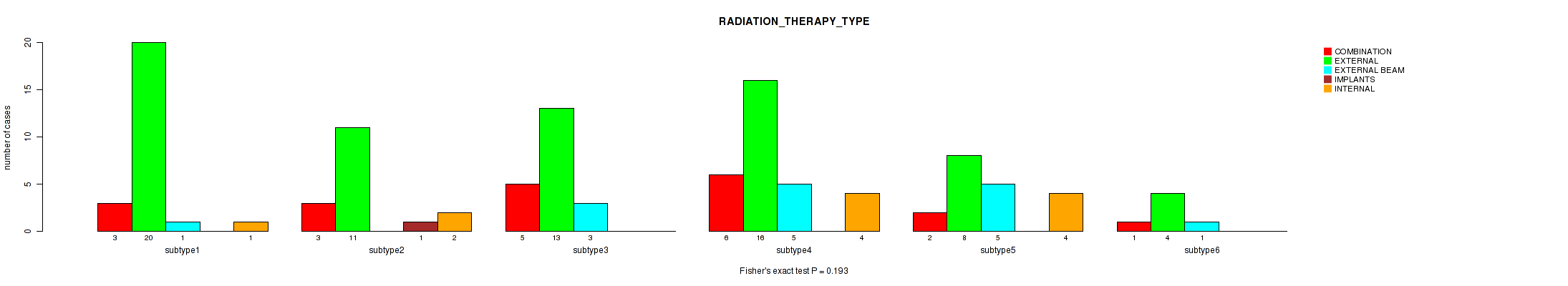
P value = 0.886 (Fisher's exact test), Q value = 1
Table S69. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'
| nPatients | COMPLETED AS PLANNED | TREATMENT NOT COMPLETED |
|---|---|---|
| ALL | 29 | 3 |
| subtype1 | 4 | 0 |
| subtype2 | 2 | 0 |
| subtype3 | 7 | 1 |
| subtype4 | 8 | 2 |
| subtype5 | 7 | 0 |
| subtype6 | 1 | 0 |
Figure S67. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'

P value = 0.0606 (Fisher's exact test), Q value = 1
Table S70. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'
| nPatients | DISTANT RECURRENCE | LOCAL RECURRENCE | PRIMARY TUMOR FIELD | REGIONAL SITE |
|---|---|---|---|---|
| ALL | 2 | 2 | 28 | 13 |
| subtype1 | 1 | 0 | 3 | 1 |
| subtype2 | 0 | 0 | 1 | 5 |
| subtype3 | 0 | 1 | 7 | 0 |
| subtype4 | 1 | 1 | 9 | 4 |
| subtype5 | 0 | 0 | 7 | 3 |
| subtype6 | 0 | 0 | 1 | 0 |
Figure S68. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'

P value = 0.928 (Fisher's exact test), Q value = 1
Table S71. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'
| nPatients | CGY | GY |
|---|---|---|
| ALL | 31 | 3 |
| subtype1 | 5 | 0 |
| subtype2 | 4 | 0 |
| subtype3 | 5 | 1 |
| subtype4 | 10 | 2 |
| subtype5 | 6 | 0 |
| subtype6 | 1 | 0 |
Figure S69. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'

P value = 0.291 (Kruskal-Wallis (anova)), Q value = 1
Table S72. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 212 | 3.6 (2.5) |
| subtype1 | 51 | 2.9 (2.1) |
| subtype2 | 35 | 3.8 (2.5) |
| subtype3 | 39 | 3.8 (2.5) |
| subtype4 | 43 | 3.8 (2.9) |
| subtype5 | 33 | 3.9 (2.5) |
| subtype6 | 11 | 2.8 (2.1) |
Figure S70. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'

P value = 0.00484 (Kruskal-Wallis (anova)), Q value = 1
Table S73. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 107 | 0.1 (0.4) |
| subtype1 | 24 | 0.0 (0.2) |
| subtype2 | 21 | 0.0 (0.0) |
| subtype3 | 21 | 0.3 (0.7) |
| subtype4 | 21 | 0.0 (0.0) |
| subtype5 | 16 | 0.0 (0.0) |
| subtype6 | 4 | 0.0 (0.0) |
Figure S71. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'

P value = 0.423 (Kruskal-Wallis (anova)), Q value = 1
Table S74. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 133 | 0.5 (0.8) |
| subtype1 | 26 | 0.5 (0.6) |
| subtype2 | 24 | 0.4 (1.2) |
| subtype3 | 29 | 0.5 (0.7) |
| subtype4 | 28 | 0.6 (1.0) |
| subtype5 | 21 | 0.3 (0.5) |
| subtype6 | 5 | 0.4 (0.5) |
Figure S72. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'

P value = 0.114 (Kruskal-Wallis (anova)), Q value = 1
Table S75. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 214 | 2.6 (1.8) |
| subtype1 | 50 | 2.1 (1.8) |
| subtype2 | 36 | 3.3 (2.2) |
| subtype3 | 41 | 2.5 (1.6) |
| subtype4 | 42 | 2.8 (1.9) |
| subtype5 | 34 | 2.4 (1.3) |
| subtype6 | 11 | 2.5 (1.6) |
Figure S73. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'

P value = 0.151 (Kruskal-Wallis (anova)), Q value = 1
Table S76. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 117 | 0.9 (1.8) |
| subtype1 | 26 | 0.7 (1.0) |
| subtype2 | 22 | 0.3 (0.8) |
| subtype3 | 23 | 1.1 (2.3) |
| subtype4 | 23 | 0.8 (1.6) |
| subtype5 | 18 | 1.6 (3.1) |
| subtype6 | 5 | 0.2 (0.4) |
Figure S74. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'

P value = 0.769 (Kruskal-Wallis (anova)), Q value = 1
Table S77. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 109 | 0.1 (0.3) |
| subtype1 | 23 | 0.1 (0.3) |
| subtype2 | 22 | 0.0 (0.2) |
| subtype3 | 21 | 0.0 (0.2) |
| subtype4 | 21 | 0.1 (0.4) |
| subtype5 | 17 | 0.2 (0.5) |
| subtype6 | 5 | 0.2 (0.4) |
Figure S75. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'

P value = 0.888 (Fisher's exact test), Q value = 1
Table S78. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'
| nPatients | MACROSCOPIC PARAMETRIAL INVOLVEMENT | MICROSCOPIC PARAMETRIAL INVOLVEMENT | OTHER LOCATION, SPECIFY | POSITIVE BLADDER MARGIN | POSITIVE VAGINAL MARGIN |
|---|---|---|---|---|---|
| ALL | 2 | 7 | 33 | 1 | 9 |
| subtype1 | 1 | 1 | 7 | 0 | 2 |
| subtype2 | 0 | 1 | 3 | 0 | 2 |
| subtype3 | 0 | 0 | 8 | 0 | 2 |
| subtype4 | 0 | 2 | 7 | 1 | 1 |
| subtype5 | 1 | 3 | 5 | 0 | 2 |
| subtype6 | 0 | 0 | 3 | 0 | 0 |
Figure S76. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'

P value = 0.0477 (Fisher's exact test), Q value = 1
Table S79. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #32: 'MENOPAUSE_STATUS'
| nPatients | INDETERMINATE (NEITHER PRE OR POSTMENOPAUSAL) | PERI (6-12 MONTHS SINCE LAST MENSTRUAL PERIOD) | POST (PRIOR BILATERAL OVARIECTOMY OR >12 MO SINCE LMP WITH NO PRIOR HYSTERECTOMY) | PRE (<6 MONTHS SINCE LMP AND NO PRIOR BILATERAL OVARIECTOMY AND NOT ON ESTROGEN REPLACEMENT) |
|---|---|---|---|---|
| ALL | 2 | 15 | 68 | 105 |
| subtype1 | 0 | 4 | 14 | 29 |
| subtype2 | 1 | 3 | 15 | 14 |
| subtype3 | 0 | 3 | 14 | 20 |
| subtype4 | 0 | 2 | 4 | 26 |
| subtype5 | 1 | 2 | 15 | 11 |
| subtype6 | 0 | 1 | 6 | 5 |
Figure S77. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #32: 'MENOPAUSE_STATUS'

P value = 0.592 (Fisher's exact test), Q value = 1
Table S80. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 68 | 74 |
| subtype1 | 20 | 20 |
| subtype2 | 8 | 14 |
| subtype3 | 13 | 13 |
| subtype4 | 12 | 9 |
| subtype5 | 13 | 12 |
| subtype6 | 2 | 6 |
Figure S78. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'
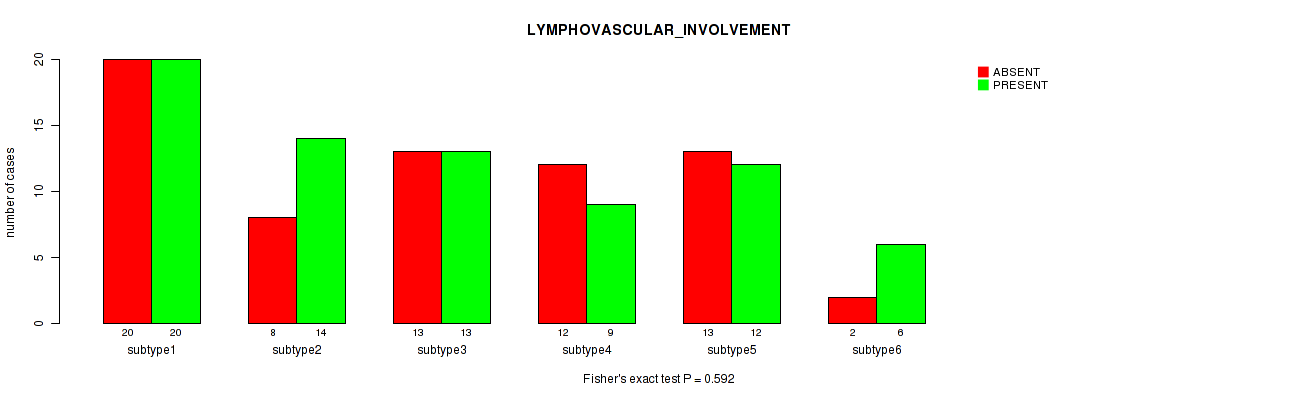
P value = 0.601 (Kruskal-Wallis (anova)), Q value = 1
Table S81. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 142 | 1.0 (2.4) |
| subtype1 | 37 | 1.1 (2.9) |
| subtype2 | 23 | 1.3 (2.0) |
| subtype3 | 24 | 1.2 (2.2) |
| subtype4 | 24 | 0.6 (1.4) |
| subtype5 | 25 | 1.2 (3.3) |
| subtype6 | 9 | 0.6 (0.9) |
Figure S79. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'
P value = 0.588 (Kruskal-Wallis (anova)), Q value = 1
Table S82. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 163 | 21.7 (12.3) |
| subtype1 | 42 | 20.8 (9.0) |
| subtype2 | 25 | 24.4 (15.8) |
| subtype3 | 29 | 23.8 (13.8) |
| subtype4 | 29 | 20.5 (11.7) |
| subtype5 | 27 | 21.0 (11.0) |
| subtype6 | 11 | 17.6 (15.5) |
Figure S80. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'

P value = 0.00981 (Fisher's exact test), Q value = 1
Table S83. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'
| nPatients | KERATINIZING SQUAMOUS CELL CARCINOMA | NON-KERATINIZING SQUAMOUS CELL CARCINOMA |
|---|---|---|
| ALL | 49 | 96 |
| subtype1 | 1 | 15 |
| subtype2 | 6 | 21 |
| subtype3 | 10 | 22 |
| subtype4 | 15 | 18 |
| subtype5 | 15 | 13 |
| subtype6 | 2 | 7 |
Figure S81. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'
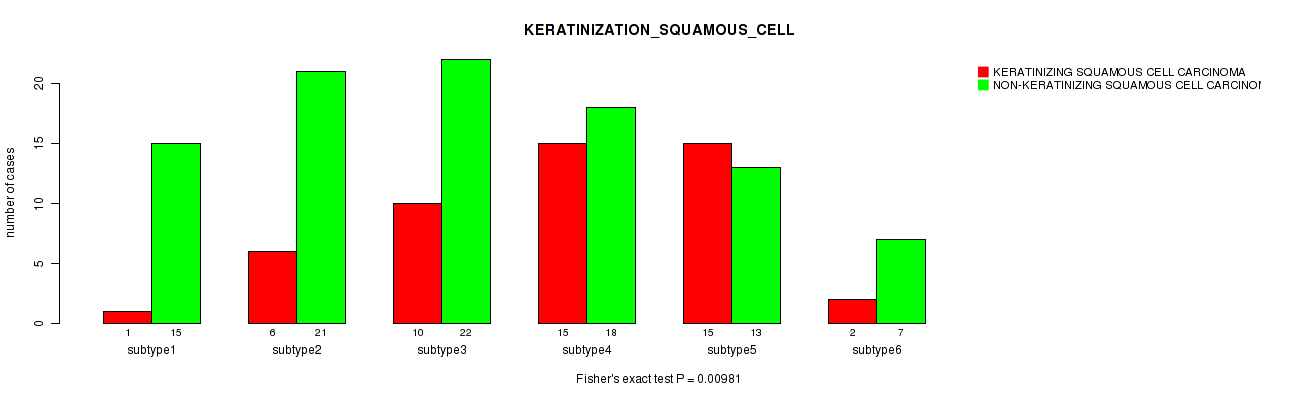
P value = 0.386 (Kruskal-Wallis (anova)), Q value = 1
Table S84. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 241 | 2007.7 (5.0) |
| subtype1 | 55 | 2008.6 (4.1) |
| subtype2 | 41 | 2007.3 (5.8) |
| subtype3 | 44 | 2006.8 (5.4) |
| subtype4 | 51 | 2007.0 (5.3) |
| subtype5 | 36 | 2007.9 (5.0) |
| subtype6 | 14 | 2009.6 (3.5) |
Figure S82. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'

P value = 0.759 (Fisher's exact test), Q value = 1
Table S85. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'
| nPatients | CURRENT USER | FORMER USER | NEVER USED |
|---|---|---|---|
| ALL | 10 | 47 | 58 |
| subtype1 | 5 | 15 | 11 |
| subtype2 | 1 | 8 | 11 |
| subtype3 | 1 | 7 | 12 |
| subtype4 | 2 | 10 | 10 |
| subtype5 | 1 | 4 | 10 |
| subtype6 | 0 | 3 | 4 |
Figure S83. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'
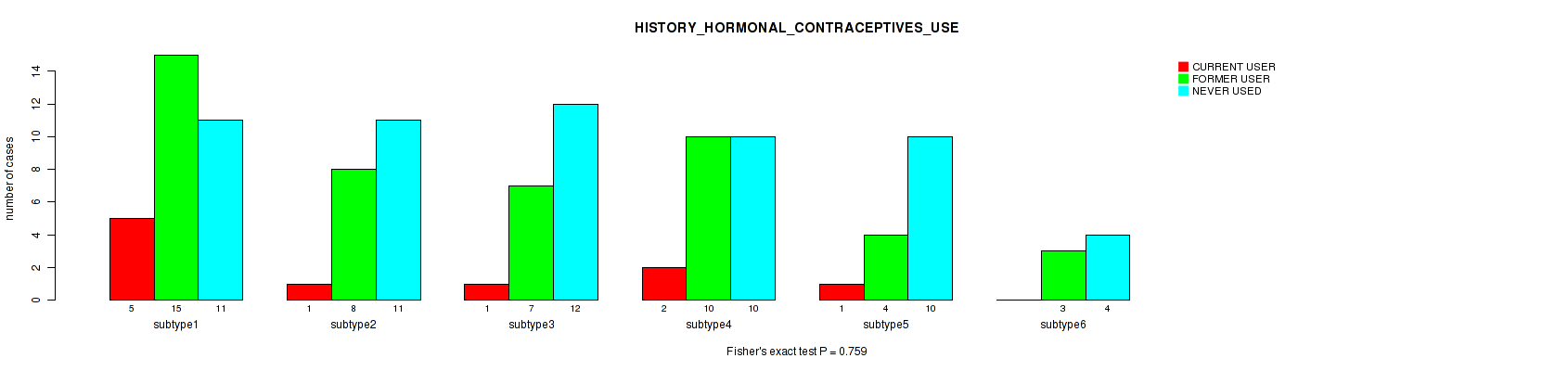
P value = 0.275 (Kruskal-Wallis (anova)), Q value = 1
Table S86. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 213 | 161.7 (7.0) |
| subtype1 | 49 | 162.9 (6.7) |
| subtype2 | 39 | 162.1 (6.8) |
| subtype3 | 38 | 159.3 (7.8) |
| subtype4 | 42 | 161.6 (6.8) |
| subtype5 | 32 | 161.6 (6.9) |
| subtype6 | 13 | 163.6 (6.1) |
Figure S84. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'

P value = 0.694 (Fisher's exact test), Q value = 1
Table S87. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 95 | 17 |
| subtype1 | 29 | 5 |
| subtype2 | 15 | 2 |
| subtype3 | 15 | 4 |
| subtype4 | 15 | 1 |
| subtype5 | 16 | 5 |
| subtype6 | 5 | 0 |
Figure S85. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'

P value = 0.21 (Fisher's exact test), Q value = 1
Table S88. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'
| nPatients | CARBOPLATIN | CISPLATIN | OTHER |
|---|---|---|---|
| ALL | 3 | 18 | 1 |
| subtype1 | 0 | 2 | 0 |
| subtype2 | 1 | 1 | 0 |
| subtype3 | 1 | 5 | 1 |
| subtype4 | 0 | 8 | 0 |
| subtype5 | 1 | 2 | 0 |
| subtype6 | 0 | 0 | 0 |
Figure S86. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'

P value = 0.0821 (Kruskal-Wallis (anova)), Q value = 1
Table S89. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 13 | 12.4 (5.5) |
| subtype1 | 1 | 7.1 (NA) |
| subtype2 | 1 | 11.1 (NA) |
| subtype3 | 3 | 8.4 (1.8) |
| subtype4 | 4 | 18.0 (6.0) |
| subtype5 | 3 | 10.8 (4.4) |
| subtype6 | 1 | 13.8 (NA) |
Figure S87. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'

P value = 0.444 (Fisher's exact test), Q value = 1
Table S90. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'
| nPatients | T1A | T1A1 | T1B | T1B1 | T1B2 | T2 | T2A | T2A1 | T2A2 | T2B | T3 | T3B | T4 | TIS | TX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 1 | 1 | 32 | 66 | 25 | 4 | 8 | 7 | 10 | 19 | 1 | 7 | 4 | 1 | 15 |
| subtype1 | 0 | 0 | 3 | 23 | 7 | 0 | 2 | 3 | 3 | 5 | 0 | 1 | 1 | 0 | 1 |
| subtype2 | 1 | 0 | 4 | 10 | 5 | 2 | 1 | 0 | 2 | 2 | 1 | 1 | 1 | 0 | 5 |
| subtype3 | 0 | 0 | 8 | 14 | 2 | 2 | 1 | 2 | 1 | 4 | 0 | 1 | 1 | 0 | 2 |
| subtype4 | 0 | 0 | 11 | 7 | 3 | 0 | 1 | 1 | 2 | 6 | 0 | 2 | 1 | 1 | 4 |
| subtype5 | 0 | 1 | 6 | 8 | 5 | 0 | 2 | 1 | 1 | 1 | 0 | 2 | 0 | 0 | 2 |
| subtype6 | 0 | 0 | 0 | 4 | 3 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
Figure S88. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'

P value = 0.0102 (Kruskal-Wallis (anova)), Q value = 1
Table S91. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 243 | 47.5 (13.5) |
| subtype1 | 56 | 45.5 (11.4) |
| subtype2 | 41 | 50.7 (14.2) |
| subtype3 | 44 | 46.7 (13.9) |
| subtype4 | 51 | 43.0 (13.1) |
| subtype5 | 37 | 53.0 (13.1) |
| subtype6 | 14 | 50.9 (14.8) |
Figure S89. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'

P value = 0.105 (Fisher's exact test), Q value = 1
Table S92. Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'
| nPatients | STAGE I | STAGE IA | STAGE IA1 | STAGE IA2 | STAGE IB | STAGE IB1 | STAGE IB2 | STAGE II | STAGE IIA | STAGE IIA1 | STAGE IIA2 | STAGE IIB | STAGE III | STAGE IIIB | STAGE IVA | STAGE IVB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 4 | 2 | 1 | 1 | 35 | 68 | 34 | 4 | 7 | 5 | 7 | 23 | 1 | 33 | 5 | 7 |
| subtype1 | 0 | 0 | 0 | 0 | 3 | 25 | 10 | 0 | 1 | 1 | 2 | 5 | 0 | 5 | 0 | 4 |
| subtype2 | 1 | 1 | 0 | 0 | 6 | 8 | 5 | 2 | 0 | 0 | 1 | 2 | 1 | 7 | 2 | 1 |
| subtype3 | 0 | 1 | 0 | 0 | 7 | 14 | 3 | 1 | 1 | 2 | 0 | 3 | 0 | 10 | 1 | 1 |
| subtype4 | 1 | 0 | 0 | 0 | 13 | 9 | 5 | 0 | 4 | 1 | 1 | 8 | 0 | 6 | 1 | 1 |
| subtype5 | 2 | 0 | 1 | 1 | 5 | 9 | 7 | 1 | 1 | 1 | 1 | 4 | 0 | 3 | 1 | 0 |
| subtype6 | 0 | 0 | 0 | 0 | 1 | 3 | 4 | 0 | 0 | 0 | 2 | 1 | 0 | 2 | 0 | 0 |
Figure S90. Get High-res Image Clustering Approach #2: 'METHLYATION CNMF' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'

Table S93. Description of clustering approach #3: 'RNAseq CNMF subtypes'
| Cluster Labels | 1 | 2 | 3 |
|---|---|---|---|
| Number of samples | 77 | 100 | 62 |
P value = 0.152 (logrank test), Q value = 1
Table S94. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 234 | 51 | 0.0 - 195.8 (15.9) |
| subtype1 | 75 | 14 | 0.1 - 147.4 (15.0) |
| subtype2 | 98 | 18 | 0.0 - 195.8 (17.3) |
| subtype3 | 61 | 19 | 0.0 - 154.3 (15.6) |
Figure S91. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #1: 'Time to Death'
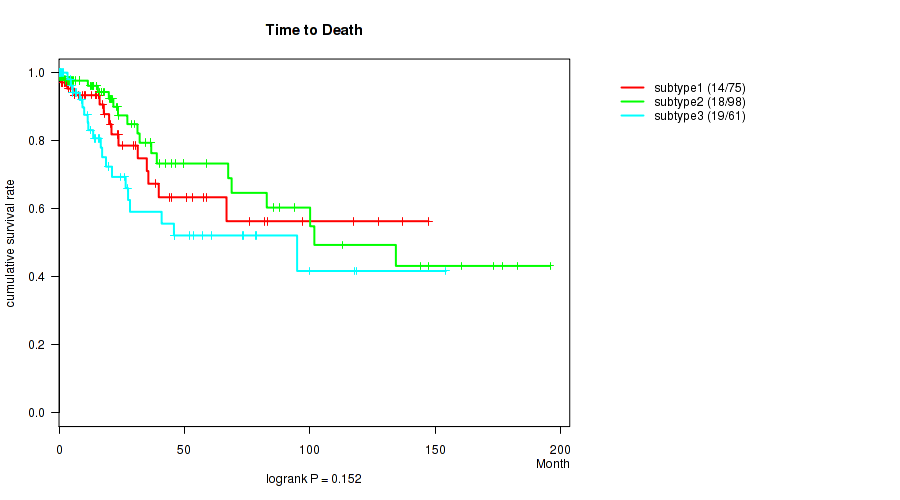
P value = 0.0018 (Kruskal-Wallis (anova)), Q value = 0.62
Table S95. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 235 | 47.2 (13.4) |
| subtype1 | 75 | 46.3 (12.0) |
| subtype2 | 99 | 50.7 (14.0) |
| subtype3 | 61 | 42.5 (12.6) |
Figure S92. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #2: 'AGE'

P value = 0.777 (Fisher's exact test), Q value = 1
Table S96. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'
| nPatients | T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 123 | 47 | 8 | 3 |
| subtype1 | 46 | 16 | 1 | 1 |
| subtype2 | 48 | 21 | 5 | 2 |
| subtype3 | 29 | 10 | 2 | 0 |
Figure S93. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'
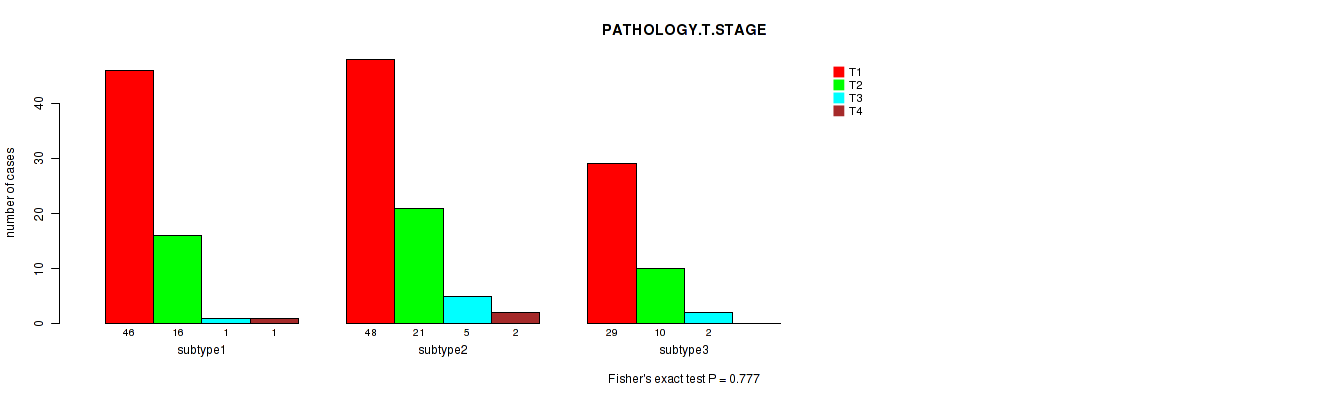
P value = 0.0774 (Fisher's exact test), Q value = 1
Table S97. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'
| nPatients | 0 | 1 |
|---|---|---|
| ALL | 112 | 49 |
| subtype1 | 45 | 14 |
| subtype2 | 40 | 27 |
| subtype3 | 27 | 8 |
Figure S94. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'

P value = 0.197 (Fisher's exact test), Q value = 1
Table S98. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 96 | 8 | 88 |
| subtype1 | 35 | 5 | 25 |
| subtype2 | 37 | 1 | 43 |
| subtype3 | 24 | 2 | 20 |
Figure S95. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'

P value = 1e-05 (Fisher's exact test), Q value = 0.0036
Table S99. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | ADENOSQUAMOUS | CERVICAL SQUAMOUS CELL CARCINOMA | ENDOCERVICAL ADENOCARCINOMA OF THE USUAL TYPE | ENDOCERVICAL TYPE OF ADENOCARCINOMA | ENDOMETRIOID ADENOCARCINOMA OF ENDOCERVIX | MUCINOUS ADENOCARCINOMA OF ENDOCERVICAL TYPE |
|---|---|---|---|---|---|---|
| ALL | 4 | 200 | 5 | 22 | 2 | 6 |
| subtype1 | 4 | 38 | 5 | 22 | 2 | 6 |
| subtype2 | 0 | 100 | 0 | 0 | 0 | 0 |
| subtype3 | 0 | 62 | 0 | 0 | 0 | 0 |
Figure S96. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 0.438 (Fisher's exact test), Q value = 1
Table S100. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 33 | 206 |
| subtype1 | 8 | 69 |
| subtype2 | 14 | 86 |
| subtype3 | 11 | 51 |
Figure S97. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'

P value = 0.461 (Kruskal-Wallis (anova)), Q value = 1
Table S101. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 75 | 18.4 (13.4) |
| subtype1 | 21 | 15.9 (12.0) |
| subtype2 | 40 | 20.6 (15.1) |
| subtype3 | 14 | 15.7 (9.0) |
Figure S98. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'

P value = 0.0389 (Kruskal-Wallis (anova)), Q value = 1
Table S102. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 139 | 1.0 (2.4) |
| subtype1 | 52 | 0.8 (2.5) |
| subtype2 | 60 | 1.2 (1.9) |
| subtype3 | 27 | 0.9 (3.1) |
Figure S99. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'

P value = 0.153 (Fisher's exact test), Q value = 1
Table S103. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER | WHITE |
|---|---|---|---|---|---|
| ALL | 7 | 18 | 26 | 1 | 170 |
| subtype1 | 1 | 7 | 4 | 0 | 59 |
| subtype2 | 2 | 7 | 11 | 0 | 70 |
| subtype3 | 4 | 4 | 11 | 1 | 41 |
Figure S100. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #10: 'RACE'
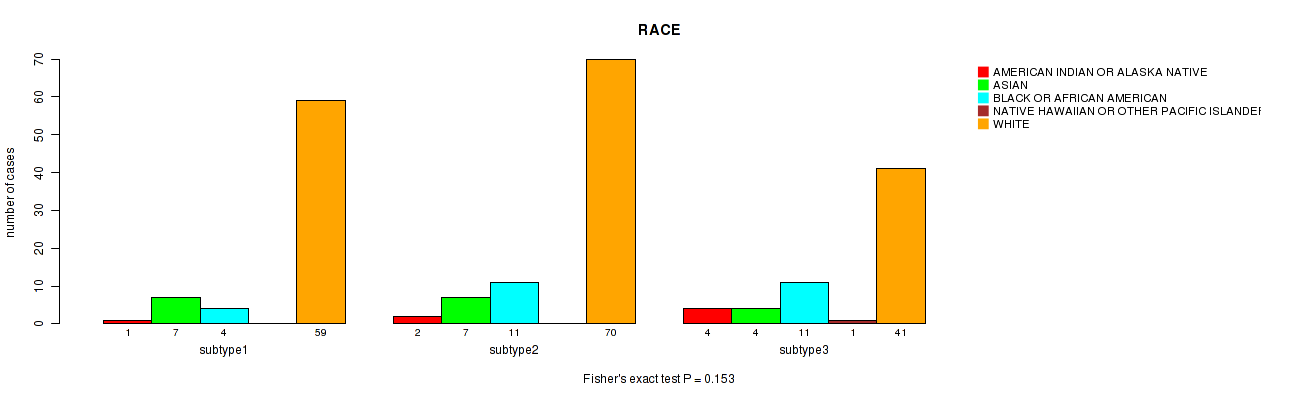
P value = 0.496 (Fisher's exact test), Q value = 1
Table S104. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #11: 'ETHNICITY'
| nPatients | HISPANIC OR LATINO | NOT HISPANIC OR LATINO |
|---|---|---|
| ALL | 17 | 151 |
| subtype1 | 6 | 50 |
| subtype2 | 5 | 63 |
| subtype3 | 6 | 38 |
Figure S101. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #11: 'ETHNICITY'
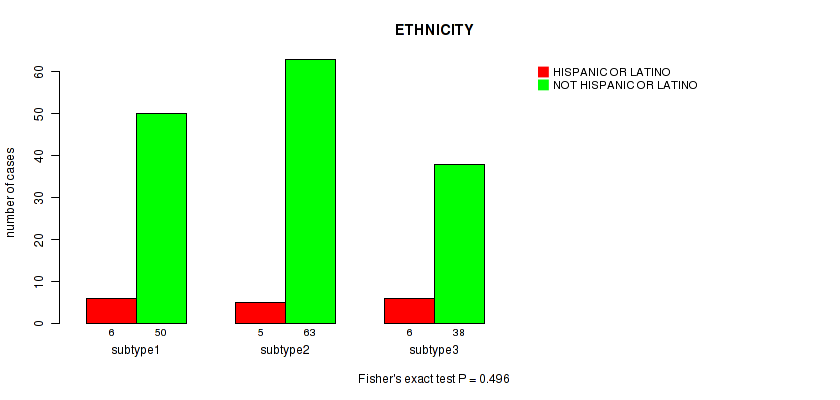
P value = 0.332 (Kruskal-Wallis (anova)), Q value = 1
Table S105. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 217 | 74.9 (22.2) |
| subtype1 | 71 | 77.3 (21.1) |
| subtype2 | 92 | 74.2 (25.2) |
| subtype3 | 54 | 72.9 (17.9) |
Figure S102. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'

P value = 0.74 (Fisher's exact test), Q value = 1
Table S106. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #13: 'TUMOR_STATUS'
| nPatients | TUMOR FREE | WITH TUMOR |
|---|---|---|
| ALL | 74 | 26 |
| subtype1 | 26 | 8 |
| subtype2 | 29 | 9 |
| subtype3 | 19 | 9 |
Figure S103. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #13: 'TUMOR_STATUS'

P value = 0.766 (Fisher's exact test), Q value = 1
Table S107. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'
| nPatients | BRAZIL | CANADA | NIGERIA | RUSSIA | UKRAINE | UNITED STATES | VIETNAM |
|---|---|---|---|---|---|---|---|
| ALL | 20 | 5 | 1 | 11 | 7 | 181 | 14 |
| subtype1 | 4 | 3 | 1 | 3 | 2 | 59 | 5 |
| subtype2 | 10 | 2 | 0 | 7 | 3 | 72 | 6 |
| subtype3 | 6 | 0 | 0 | 1 | 2 | 50 | 3 |
Figure S104. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'

P value = 0.00964 (Fisher's exact test), Q value = 1
Table S108. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'
| nPatients | G1 | G2 | G3 | G4 | GX |
|---|---|---|---|---|---|
| ALL | 15 | 110 | 96 | 1 | 14 |
| subtype1 | 6 | 30 | 35 | 0 | 5 |
| subtype2 | 7 | 60 | 29 | 0 | 4 |
| subtype3 | 2 | 20 | 32 | 1 | 5 |
Figure S105. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'

P value = 0.975 (Kruskal-Wallis (anova)), Q value = 1
Table S109. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 34 | 2000.4 (11.2) |
| subtype1 | 11 | 1999.7 (12.2) |
| subtype2 | 19 | 2000.5 (11.9) |
| subtype3 | 4 | 2002.0 (5.6) |
Figure S106. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'

P value = 0.461 (Kruskal-Wallis (anova)), Q value = 1
Table S110. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 75 | 18.4 (13.4) |
| subtype1 | 21 | 15.9 (12.0) |
| subtype2 | 40 | 20.6 (15.1) |
| subtype3 | 14 | 15.7 (9.0) |
Figure S107. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'

P value = 0.0998 (Fisher's exact test), Q value = 1
Table S111. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'
| nPatients | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT REFORMED SMOKER, DURATION NOT SPECIFIED | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|
| ALL | 33 | 8 | 3 | 52 | 106 |
| subtype1 | 12 | 1 | 0 | 14 | 40 |
| subtype2 | 16 | 7 | 2 | 25 | 37 |
| subtype3 | 5 | 0 | 1 | 13 | 29 |
Figure S108. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'

P value = 0.489 (Kruskal-Wallis (anova)), Q value = 1
Table S112. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 67 | 21.5 (7.9) |
| subtype1 | 19 | 19.3 (3.9) |
| subtype2 | 37 | 23.0 (8.9) |
| subtype3 | 11 | 20.3 (8.9) |
Figure S109. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'

P value = 0.311 (Kruskal-Wallis (anova)), Q value = 1
Table S113. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 102 | 3950.7 (1603.4) |
| subtype1 | 30 | 4026.2 (1702.4) |
| subtype2 | 39 | 3933.7 (1509.6) |
| subtype3 | 33 | 3902.2 (1665.4) |
Figure S110. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'

P value = 0.556 (Fisher's exact test), Q value = 1
Table S114. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'
| nPatients | COMBINATION | EXTERNAL | EXTERNAL BEAM | INTERNAL |
|---|---|---|---|---|
| ALL | 19 | 72 | 15 | 11 |
| subtype1 | 6 | 24 | 2 | 1 |
| subtype2 | 8 | 27 | 7 | 6 |
| subtype3 | 5 | 21 | 6 | 4 |
Figure S111. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'

P value = 0.454 (Fisher's exact test), Q value = 1
Table S115. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'
| nPatients | COMPLETED AS PLANNED | TREATMENT NOT COMPLETED |
|---|---|---|
| ALL | 28 | 3 |
| subtype1 | 8 | 0 |
| subtype2 | 12 | 1 |
| subtype3 | 8 | 2 |
Figure S112. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'

P value = 0.0669 (Fisher's exact test), Q value = 1
Table S116. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'
| nPatients | DISTANT RECURRENCE | LOCAL RECURRENCE | PRIMARY TUMOR FIELD | REGIONAL SITE |
|---|---|---|---|---|
| ALL | 2 | 2 | 28 | 13 |
| subtype1 | 1 | 0 | 4 | 1 |
| subtype2 | 0 | 1 | 10 | 10 |
| subtype3 | 1 | 1 | 14 | 2 |
Figure S113. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'
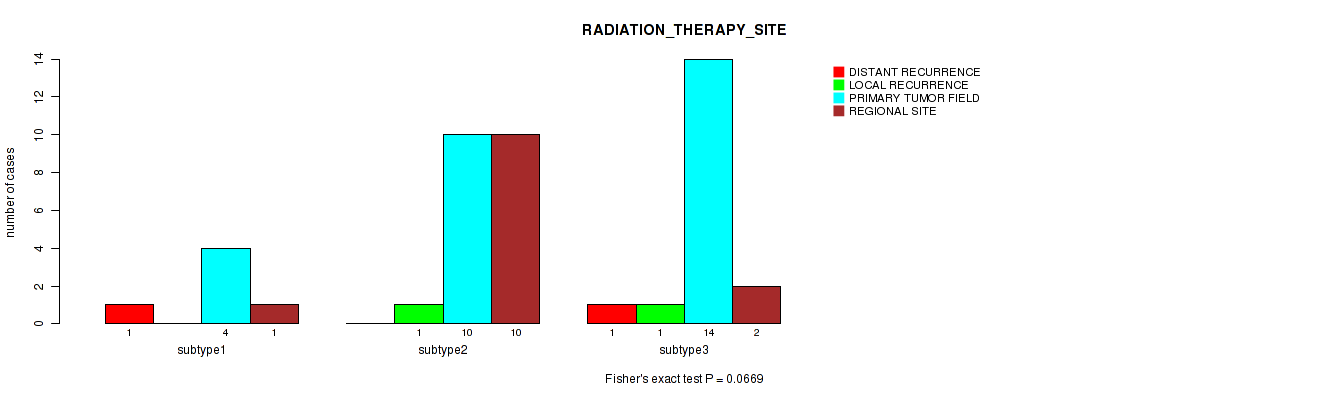
P value = 0.74 (Fisher's exact test), Q value = 1
Table S117. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'
| nPatients | CGY | GY |
|---|---|---|
| ALL | 31 | 3 |
| subtype1 | 5 | 0 |
| subtype2 | 15 | 1 |
| subtype3 | 11 | 2 |
Figure S114. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'

P value = 0.0112 (Kruskal-Wallis (anova)), Q value = 1
Table S118. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 208 | 3.5 (2.5) |
| subtype1 | 67 | 2.9 (1.9) |
| subtype2 | 89 | 4.0 (2.7) |
| subtype3 | 52 | 3.7 (2.6) |
Figure S115. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'

P value = 0.153 (Kruskal-Wallis (anova)), Q value = 1
Table S119. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 104 | 0.1 (0.4) |
| subtype1 | 33 | 0.0 (0.2) |
| subtype2 | 48 | 0.1 (0.5) |
| subtype3 | 23 | 0.0 (0.0) |
Figure S116. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'

P value = 0.933 (Kruskal-Wallis (anova)), Q value = 1
Table S120. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 130 | 0.5 (0.8) |
| subtype1 | 35 | 0.4 (0.6) |
| subtype2 | 62 | 0.5 (0.9) |
| subtype3 | 33 | 0.5 (0.9) |
Figure S117. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'

P value = 0.0455 (Kruskal-Wallis (anova)), Q value = 1
Table S121. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 210 | 2.6 (1.8) |
| subtype1 | 66 | 2.2 (1.7) |
| subtype2 | 91 | 2.8 (1.8) |
| subtype3 | 53 | 2.8 (2.0) |
Figure S118. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'

P value = 0.849 (Kruskal-Wallis (anova)), Q value = 1
Table S122. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 114 | 0.9 (1.9) |
| subtype1 | 36 | 0.6 (1.0) |
| subtype2 | 53 | 1.2 (2.5) |
| subtype3 | 25 | 0.6 (1.1) |
Figure S119. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'
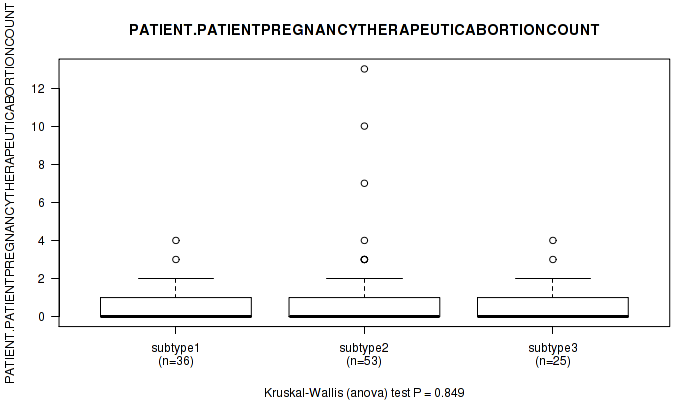
P value = 0.975 (Kruskal-Wallis (anova)), Q value = 1
Table S123. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 106 | 0.1 (0.3) |
| subtype1 | 33 | 0.1 (0.3) |
| subtype2 | 50 | 0.1 (0.4) |
| subtype3 | 23 | 0.1 (0.3) |
Figure S120. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'

P value = 0.988 (Fisher's exact test), Q value = 1
Table S124. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'
| nPatients | MACROSCOPIC PARAMETRIAL INVOLVEMENT | MICROSCOPIC PARAMETRIAL INVOLVEMENT | OTHER LOCATION, SPECIFY | POSITIVE BLADDER MARGIN | POSITIVE VAGINAL MARGIN |
|---|---|---|---|---|---|
| ALL | 2 | 6 | 32 | 1 | 8 |
| subtype1 | 1 | 2 | 10 | 0 | 3 |
| subtype2 | 1 | 2 | 13 | 1 | 4 |
| subtype3 | 0 | 2 | 9 | 0 | 1 |
Figure S121. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'
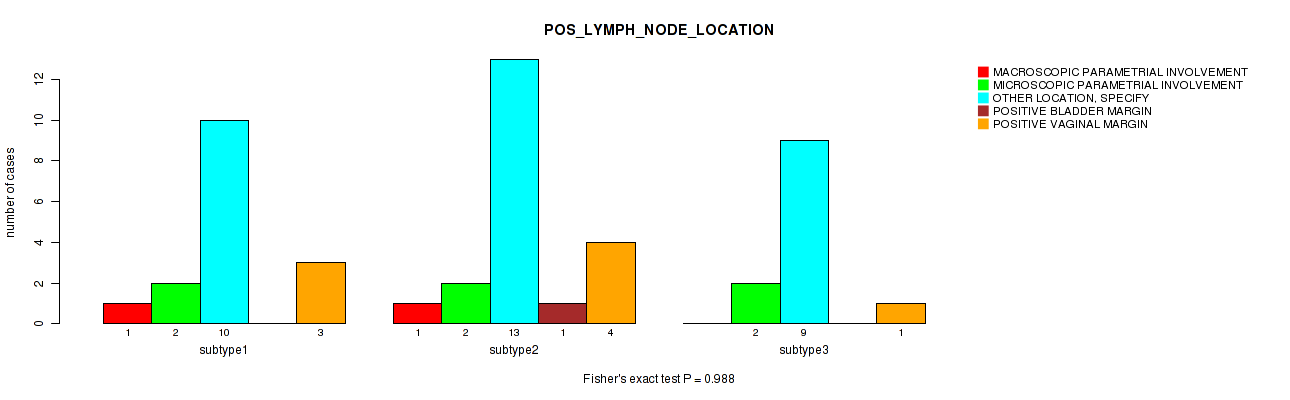
P value = 0.0193 (Fisher's exact test), Q value = 1
Table S125. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #32: 'MENOPAUSE_STATUS'
| nPatients | INDETERMINATE (NEITHER PRE OR POSTMENOPAUSAL) | PERI (6-12 MONTHS SINCE LAST MENSTRUAL PERIOD) | POST (PRIOR BILATERAL OVARIECTOMY OR >12 MO SINCE LMP WITH NO PRIOR HYSTERECTOMY) | PRE (<6 MONTHS SINCE LMP AND NO PRIOR BILATERAL OVARIECTOMY AND NOT ON ESTROGEN REPLACEMENT) |
|---|---|---|---|---|
| ALL | 2 | 15 | 65 | 105 |
| subtype1 | 0 | 5 | 22 | 36 |
| subtype2 | 2 | 8 | 36 | 37 |
| subtype3 | 0 | 2 | 7 | 32 |
Figure S122. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #32: 'MENOPAUSE_STATUS'

P value = 0.482 (Fisher's exact test), Q value = 1
Table S126. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 67 | 71 |
| subtype1 | 25 | 29 |
| subtype2 | 27 | 32 |
| subtype3 | 15 | 10 |
Figure S123. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'
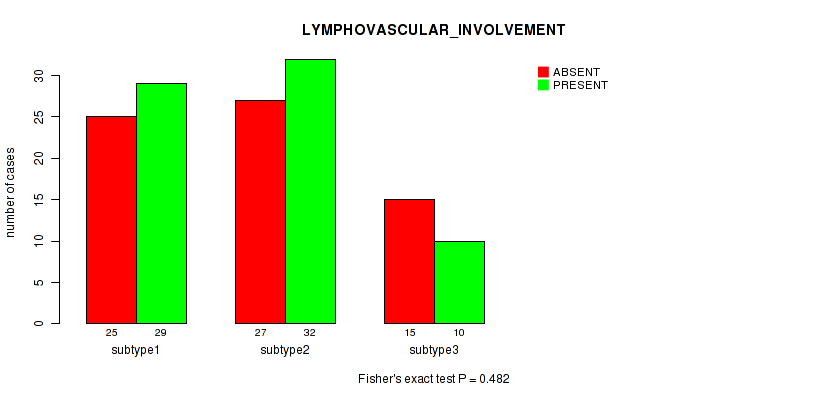
P value = 0.0389 (Kruskal-Wallis (anova)), Q value = 1
Table S127. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 139 | 1.0 (2.4) |
| subtype1 | 52 | 0.8 (2.5) |
| subtype2 | 60 | 1.2 (1.9) |
| subtype3 | 27 | 0.9 (3.1) |
Figure S124. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'

P value = 0.987 (Kruskal-Wallis (anova)), Q value = 1
Table S128. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 159 | 21.8 (12.4) |
| subtype1 | 58 | 21.3 (11.0) |
| subtype2 | 67 | 22.7 (14.5) |
| subtype3 | 34 | 20.7 (10.0) |
Figure S125. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'

P value = 0.0619 (Fisher's exact test), Q value = 1
Table S129. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'
| nPatients | KERATINIZING SQUAMOUS CELL CARCINOMA | NON-KERATINIZING SQUAMOUS CELL CARCINOMA |
|---|---|---|
| ALL | 48 | 94 |
| subtype1 | 4 | 23 |
| subtype2 | 28 | 44 |
| subtype3 | 16 | 27 |
Figure S126. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'

P value = 0.227 (Kruskal-Wallis (anova)), Q value = 1
Table S130. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 237 | 2007.7 (5.0) |
| subtype1 | 76 | 2008.6 (4.1) |
| subtype2 | 99 | 2007.5 (5.6) |
| subtype3 | 62 | 2007.1 (5.1) |
Figure S127. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'
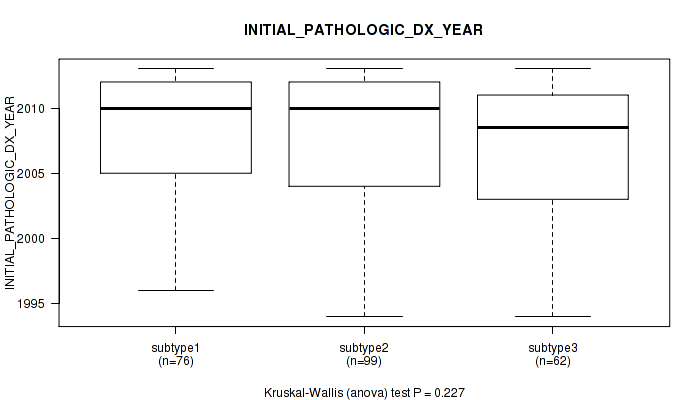
P value = 0.0854 (Fisher's exact test), Q value = 1
Table S131. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'
| nPatients | CURRENT USER | FORMER USER | NEVER USED |
|---|---|---|---|
| ALL | 10 | 47 | 56 |
| subtype1 | 6 | 20 | 15 |
| subtype2 | 1 | 16 | 28 |
| subtype3 | 3 | 11 | 13 |
Figure S128. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'

P value = 0.657 (Kruskal-Wallis (anova)), Q value = 1
Table S132. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 209 | 161.8 (6.7) |
| subtype1 | 67 | 162.6 (6.6) |
| subtype2 | 90 | 161.4 (6.8) |
| subtype3 | 52 | 161.4 (6.5) |
Figure S129. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'

P value = 0.0605 (Fisher's exact test), Q value = 1
Table S133. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 93 | 16 |
| subtype1 | 39 | 7 |
| subtype2 | 33 | 9 |
| subtype3 | 21 | 0 |
Figure S130. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'

P value = 1 (Fisher's exact test), Q value = 1
Table S134. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'
| nPatients | CARBOPLATIN | CISPLATIN | OTHER |
|---|---|---|---|
| ALL | 2 | 17 | 1 |
| subtype1 | 0 | 4 | 0 |
| subtype2 | 1 | 6 | 0 |
| subtype3 | 1 | 7 | 1 |
Figure S131. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'

P value = 0.0298 (Kruskal-Wallis (anova)), Q value = 1
Table S135. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 13 | 12.4 (5.5) |
| subtype1 | 2 | 10.8 (5.2) |
| subtype2 | 7 | 9.7 (2.9) |
| subtype3 | 4 | 18.0 (6.0) |
Figure S132. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'
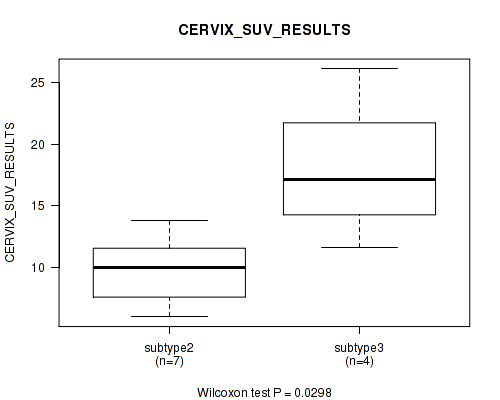
P value = 0.222 (Fisher's exact test), Q value = 1
Table S136. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'
| nPatients | T1A | T1A1 | T1B | T1B1 | T1B2 | T2 | T2A | T2A1 | T2A2 | T2B | T3 | T3B | T4 | TIS | TX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 1 | 1 | 32 | 64 | 25 | 4 | 8 | 7 | 10 | 18 | 1 | 7 | 3 | 1 | 15 |
| subtype1 | 0 | 0 | 4 | 32 | 10 | 2 | 3 | 3 | 3 | 5 | 0 | 1 | 1 | 0 | 2 |
| subtype2 | 1 | 1 | 17 | 20 | 9 | 1 | 4 | 2 | 5 | 9 | 1 | 4 | 2 | 0 | 7 |
| subtype3 | 0 | 0 | 11 | 12 | 6 | 1 | 1 | 2 | 2 | 4 | 0 | 2 | 0 | 1 | 6 |
Figure S133. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'

P value = 0.00227 (Kruskal-Wallis (anova)), Q value = 0.77
Table S137. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 239 | 47.3 (13.3) |
| subtype1 | 77 | 46.4 (11.9) |
| subtype2 | 100 | 50.8 (13.9) |
| subtype3 | 62 | 42.9 (12.7) |
Figure S134. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'

P value = 0.00183 (Fisher's exact test), Q value = 0.62
Table S138. Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'
| nPatients | STAGE I | STAGE IA | STAGE IA1 | STAGE IA2 | STAGE IB | STAGE IB1 | STAGE IB2 | STAGE II | STAGE IIA | STAGE IIA1 | STAGE IIA2 | STAGE IIB | STAGE III | STAGE IIIB | STAGE IVA | STAGE IVB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 4 | 2 | 1 | 1 | 34 | 67 | 34 | 4 | 6 | 5 | 7 | 23 | 1 | 33 | 4 | 7 |
| subtype1 | 1 | 0 | 0 | 0 | 4 | 37 | 14 | 1 | 1 | 1 | 2 | 6 | 0 | 6 | 0 | 4 |
| subtype2 | 2 | 2 | 1 | 1 | 15 | 19 | 10 | 3 | 2 | 2 | 4 | 9 | 1 | 19 | 4 | 2 |
| subtype3 | 1 | 0 | 0 | 0 | 15 | 11 | 10 | 0 | 3 | 2 | 1 | 8 | 0 | 8 | 0 | 1 |
Figure S135. Get High-res Image Clustering Approach #3: 'RNAseq CNMF subtypes' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'

Table S139. Description of clustering approach #4: 'RNAseq cHierClus subtypes'
| Cluster Labels | 1 | 2 | 3 |
|---|---|---|---|
| Number of samples | 55 | 150 | 34 |
P value = 0.00602 (logrank test), Q value = 1
Table S140. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 234 | 51 | 0.0 - 195.8 (15.9) |
| subtype1 | 54 | 11 | 0.1 - 137.2 (14.9) |
| subtype2 | 147 | 28 | 0.0 - 195.8 (17.9) |
| subtype3 | 33 | 12 | 0.0 - 99.9 (11.4) |
Figure S136. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #1: 'Time to Death'

P value = 0.0161 (Kruskal-Wallis (anova)), Q value = 1
Table S141. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 235 | 47.2 (13.4) |
| subtype1 | 53 | 46.4 (11.3) |
| subtype2 | 148 | 48.9 (13.8) |
| subtype3 | 34 | 41.1 (12.8) |
Figure S137. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #2: 'AGE'

P value = 0.946 (Fisher's exact test), Q value = 1
Table S142. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'
| nPatients | T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 123 | 47 | 8 | 3 |
| subtype1 | 32 | 13 | 1 | 1 |
| subtype2 | 77 | 27 | 6 | 2 |
| subtype3 | 14 | 7 | 1 | 0 |
Figure S138. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'

P value = 0.387 (Fisher's exact test), Q value = 1
Table S143. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'
| nPatients | 0 | 1 |
|---|---|---|
| ALL | 112 | 49 |
| subtype1 | 31 | 11 |
| subtype2 | 65 | 34 |
| subtype3 | 16 | 4 |
Figure S139. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'
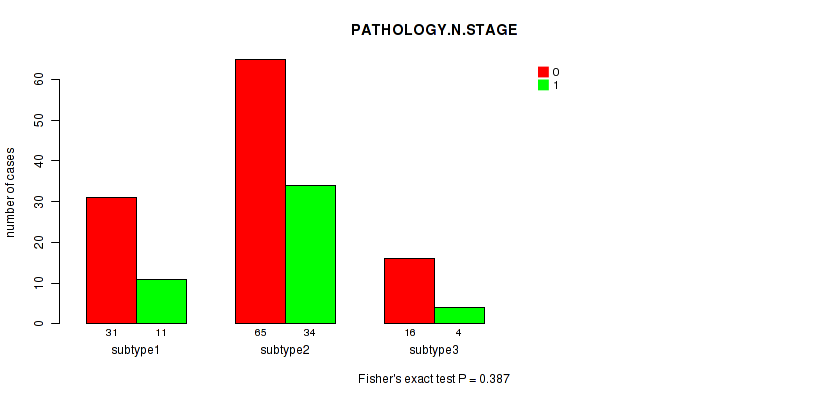
P value = 0.0499 (Fisher's exact test), Q value = 1
Table S144. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 96 | 8 | 88 |
| subtype1 | 23 | 5 | 21 |
| subtype2 | 61 | 1 | 56 |
| subtype3 | 12 | 2 | 11 |
Figure S140. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'
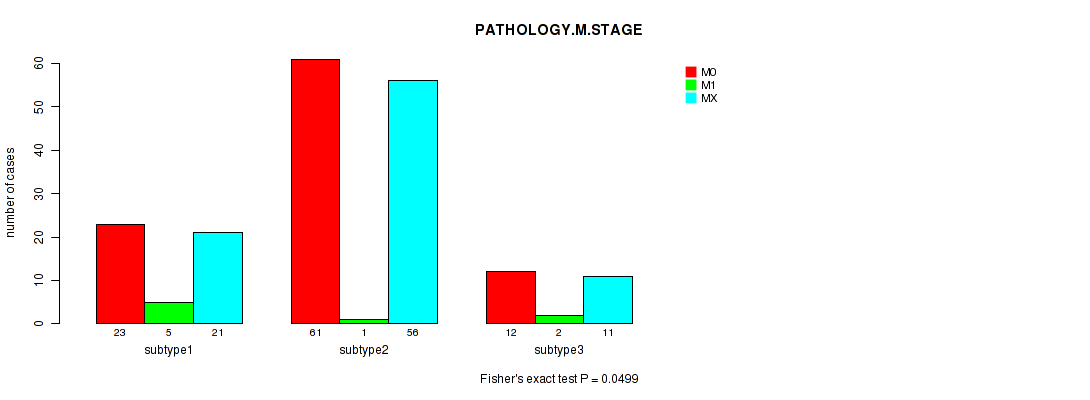
P value = 1e-05 (Fisher's exact test), Q value = 0.0036
Table S145. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | ADENOSQUAMOUS | CERVICAL SQUAMOUS CELL CARCINOMA | ENDOCERVICAL ADENOCARCINOMA OF THE USUAL TYPE | ENDOCERVICAL TYPE OF ADENOCARCINOMA | ENDOMETRIOID ADENOCARCINOMA OF ENDOCERVIX | MUCINOUS ADENOCARCINOMA OF ENDOCERVICAL TYPE |
|---|---|---|---|---|---|---|
| ALL | 4 | 200 | 5 | 22 | 2 | 6 |
| subtype1 | 3 | 19 | 4 | 22 | 2 | 5 |
| subtype2 | 1 | 147 | 1 | 0 | 0 | 1 |
| subtype3 | 0 | 34 | 0 | 0 | 0 | 0 |
Figure S141. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
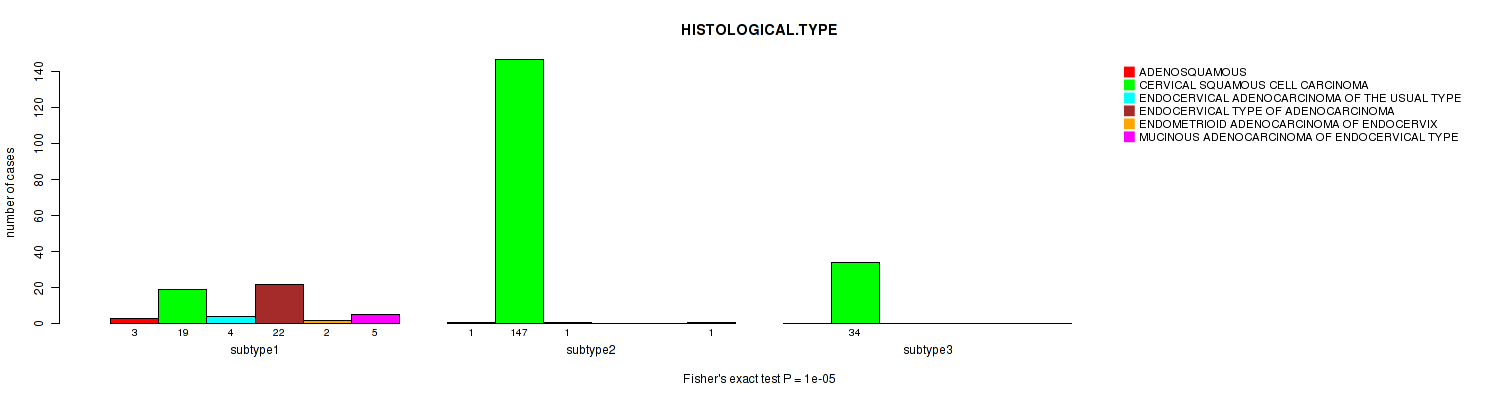
P value = 0.255 (Fisher's exact test), Q value = 1
Table S146. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 33 | 206 |
| subtype1 | 4 | 51 |
| subtype2 | 24 | 126 |
| subtype3 | 5 | 29 |
Figure S142. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'

P value = 0.628 (Kruskal-Wallis (anova)), Q value = 1
Table S147. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 75 | 18.4 (13.4) |
| subtype1 | 14 | 15.8 (11.6) |
| subtype2 | 51 | 19.7 (14.3) |
| subtype3 | 10 | 15.3 (10.0) |
Figure S143. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'
P value = 0.0689 (Kruskal-Wallis (anova)), Q value = 1
Table S148. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 139 | 1.0 (2.4) |
| subtype1 | 37 | 0.7 (2.4) |
| subtype2 | 86 | 1.3 (2.5) |
| subtype3 | 16 | 0.2 (0.6) |
Figure S144. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'

P value = 0.469 (Fisher's exact test), Q value = 1
Table S149. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER | WHITE |
|---|---|---|---|---|---|
| ALL | 7 | 18 | 26 | 1 | 170 |
| subtype1 | 1 | 5 | 3 | 0 | 41 |
| subtype2 | 4 | 12 | 16 | 1 | 105 |
| subtype3 | 2 | 1 | 7 | 0 | 24 |
Figure S145. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #10: 'RACE'
P value = 0.518 (Fisher's exact test), Q value = 1
Table S150. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #11: 'ETHNICITY'
| nPatients | HISPANIC OR LATINO | NOT HISPANIC OR LATINO |
|---|---|---|
| ALL | 17 | 151 |
| subtype1 | 6 | 34 |
| subtype2 | 9 | 97 |
| subtype3 | 2 | 20 |
Figure S146. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #11: 'ETHNICITY'

P value = 0.481 (Kruskal-Wallis (anova)), Q value = 1
Table S151. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 217 | 74.9 (22.2) |
| subtype1 | 51 | 75.7 (15.4) |
| subtype2 | 138 | 75.2 (25.2) |
| subtype3 | 28 | 72.1 (16.9) |
Figure S147. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'

P value = 0.606 (Fisher's exact test), Q value = 1
Table S152. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #13: 'TUMOR_STATUS'
| nPatients | TUMOR FREE | WITH TUMOR |
|---|---|---|
| ALL | 74 | 26 |
| subtype1 | 19 | 6 |
| subtype2 | 49 | 16 |
| subtype3 | 6 | 4 |
Figure S148. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #13: 'TUMOR_STATUS'

P value = 0.453 (Fisher's exact test), Q value = 1
Table S153. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'
| nPatients | BRAZIL | CANADA | NIGERIA | RUSSIA | UKRAINE | UNITED STATES | VIETNAM |
|---|---|---|---|---|---|---|---|
| ALL | 20 | 5 | 1 | 11 | 7 | 181 | 14 |
| subtype1 | 4 | 3 | 1 | 2 | 2 | 40 | 3 |
| subtype2 | 12 | 2 | 0 | 9 | 3 | 114 | 10 |
| subtype3 | 4 | 0 | 0 | 0 | 2 | 27 | 1 |
Figure S149. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'
P value = 0.0353 (Fisher's exact test), Q value = 1
Table S154. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'
| nPatients | G1 | G2 | G3 | G4 | GX |
|---|---|---|---|---|---|
| ALL | 15 | 110 | 96 | 1 | 14 |
| subtype1 | 6 | 24 | 21 | 0 | 4 |
| subtype2 | 8 | 77 | 54 | 0 | 8 |
| subtype3 | 1 | 9 | 21 | 1 | 2 |
Figure S150. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'

P value = 0.478 (Kruskal-Wallis (anova)), Q value = 1
Table S155. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 34 | 2000.4 (11.2) |
| subtype1 | 6 | 1996.2 (12.6) |
| subtype2 | 25 | 2001.4 (11.5) |
| subtype3 | 3 | 2000.3 (5.5) |
Figure S151. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'

P value = 0.628 (Kruskal-Wallis (anova)), Q value = 1
Table S156. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 75 | 18.4 (13.4) |
| subtype1 | 14 | 15.8 (11.6) |
| subtype2 | 51 | 19.7 (14.3) |
| subtype3 | 10 | 15.3 (10.0) |
Figure S152. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'
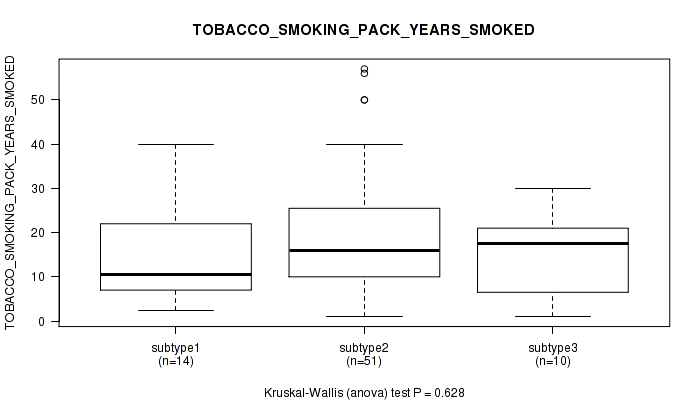
P value = 0.355 (Fisher's exact test), Q value = 1
Table S157. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'
| nPatients | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT REFORMED SMOKER, DURATION NOT SPECIFIED | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|
| ALL | 33 | 8 | 3 | 52 | 106 |
| subtype1 | 6 | 1 | 0 | 9 | 32 |
| subtype2 | 24 | 7 | 2 | 34 | 60 |
| subtype3 | 3 | 0 | 1 | 9 | 14 |
Figure S153. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'

P value = 0.368 (Kruskal-Wallis (anova)), Q value = 1
Table S158. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 67 | 21.5 (7.9) |
| subtype1 | 13 | 19.6 (4.5) |
| subtype2 | 44 | 22.4 (8.3) |
| subtype3 | 10 | 19.7 (9.2) |
Figure S154. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'

P value = 0.897 (Kruskal-Wallis (anova)), Q value = 1
Table S159. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 102 | 3950.7 (1603.4) |
| subtype1 | 21 | 3877.2 (1745.6) |
| subtype2 | 63 | 4032.5 (1518.2) |
| subtype3 | 18 | 3750.5 (1791.5) |
Figure S155. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'

P value = 0.672 (Fisher's exact test), Q value = 1
Table S160. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'
| nPatients | COMBINATION | EXTERNAL | EXTERNAL BEAM | INTERNAL |
|---|---|---|---|---|
| ALL | 19 | 72 | 15 | 11 |
| subtype1 | 3 | 17 | 1 | 1 |
| subtype2 | 13 | 41 | 12 | 7 |
| subtype3 | 3 | 14 | 2 | 3 |
Figure S156. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'

P value = 0.658 (Fisher's exact test), Q value = 1
Table S161. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'
| nPatients | COMPLETED AS PLANNED | TREATMENT NOT COMPLETED |
|---|---|---|
| ALL | 28 | 3 |
| subtype1 | 4 | 0 |
| subtype2 | 20 | 2 |
| subtype3 | 4 | 1 |
Figure S157. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'
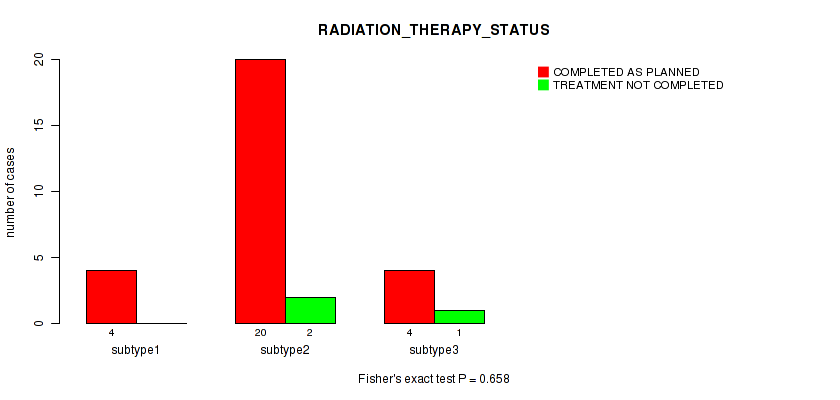
P value = 0.117 (Fisher's exact test), Q value = 1
Table S162. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'
| nPatients | DISTANT RECURRENCE | LOCAL RECURRENCE | PRIMARY TUMOR FIELD | REGIONAL SITE |
|---|---|---|---|---|
| ALL | 2 | 2 | 28 | 13 |
| subtype1 | 1 | 0 | 3 | 1 |
| subtype2 | 0 | 1 | 16 | 11 |
| subtype3 | 1 | 1 | 9 | 1 |
Figure S158. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'
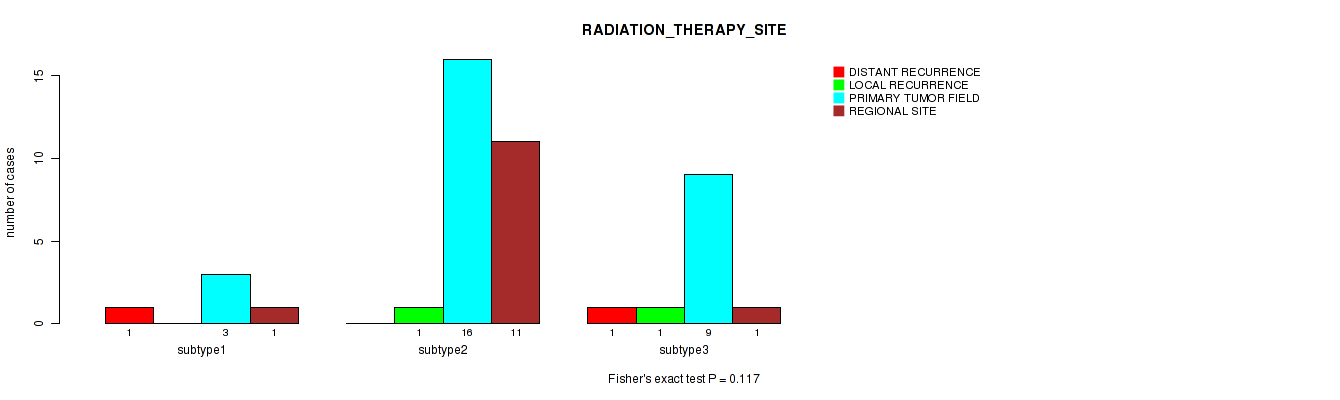
P value = 1 (Fisher's exact test), Q value = 1
Table S163. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'
| nPatients | CGY | GY |
|---|---|---|
| ALL | 31 | 3 |
| subtype1 | 5 | 0 |
| subtype2 | 18 | 2 |
| subtype3 | 8 | 1 |
Figure S159. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'

P value = 0.0973 (Kruskal-Wallis (anova)), Q value = 1
Table S164. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 208 | 3.5 (2.5) |
| subtype1 | 48 | 2.9 (2.1) |
| subtype2 | 131 | 3.7 (2.6) |
| subtype3 | 29 | 3.8 (2.6) |
Figure S160. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'

P value = 0.578 (Kruskal-Wallis (anova)), Q value = 1
Table S165. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 104 | 0.1 (0.4) |
| subtype1 | 23 | 0.0 (0.2) |
| subtype2 | 69 | 0.1 (0.4) |
| subtype3 | 12 | 0.0 (0.0) |
Figure S161. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'
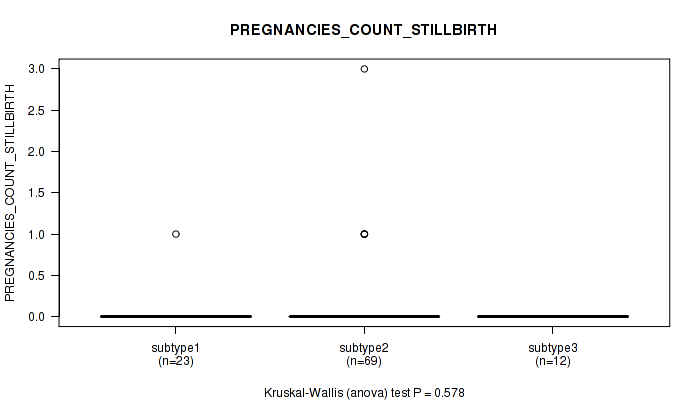
P value = 0.0932 (Kruskal-Wallis (anova)), Q value = 1
Table S166. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 130 | 0.5 (0.8) |
| subtype1 | 25 | 0.5 (0.6) |
| subtype2 | 86 | 0.4 (0.8) |
| subtype3 | 19 | 0.7 (1.0) |
Figure S162. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'
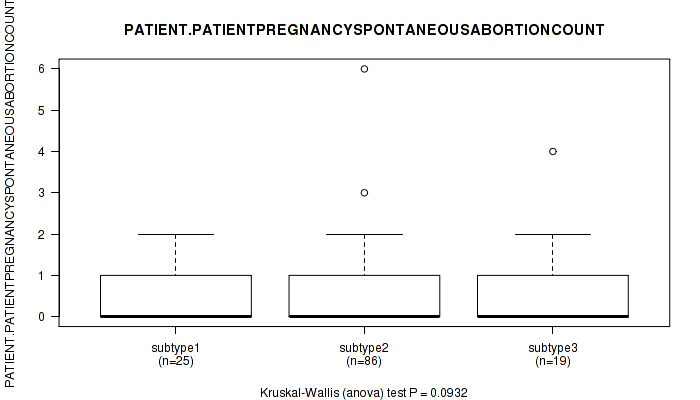
P value = 0.0515 (Kruskal-Wallis (anova)), Q value = 1
Table S167. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 210 | 2.6 (1.8) |
| subtype1 | 48 | 2.1 (1.8) |
| subtype2 | 133 | 2.7 (1.8) |
| subtype3 | 29 | 2.9 (2.1) |
Figure S163. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'

P value = 0.801 (Kruskal-Wallis (anova)), Q value = 1
Table S168. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 114 | 0.9 (1.9) |
| subtype1 | 25 | 0.7 (1.0) |
| subtype2 | 75 | 1.0 (2.2) |
| subtype3 | 14 | 0.7 (1.0) |
Figure S164. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'

P value = 0.984 (Kruskal-Wallis (anova)), Q value = 1
Table S169. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 106 | 0.1 (0.3) |
| subtype1 | 22 | 0.1 (0.3) |
| subtype2 | 72 | 0.1 (0.4) |
| subtype3 | 12 | 0.1 (0.3) |
Figure S165. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'

P value = 0.993 (Fisher's exact test), Q value = 1
Table S170. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'
| nPatients | MACROSCOPIC PARAMETRIAL INVOLVEMENT | MICROSCOPIC PARAMETRIAL INVOLVEMENT | OTHER LOCATION, SPECIFY | POSITIVE BLADDER MARGIN | POSITIVE VAGINAL MARGIN |
|---|---|---|---|---|---|
| ALL | 2 | 6 | 32 | 1 | 8 |
| subtype1 | 1 | 1 | 7 | 0 | 2 |
| subtype2 | 1 | 4 | 20 | 1 | 5 |
| subtype3 | 0 | 1 | 5 | 0 | 1 |
Figure S166. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'

P value = 0.026 (Fisher's exact test), Q value = 1
Table S171. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #32: 'MENOPAUSE_STATUS'
| nPatients | INDETERMINATE (NEITHER PRE OR POSTMENOPAUSAL) | PERI (6-12 MONTHS SINCE LAST MENSTRUAL PERIOD) | POST (PRIOR BILATERAL OVARIECTOMY OR >12 MO SINCE LMP WITH NO PRIOR HYSTERECTOMY) | PRE (<6 MONTHS SINCE LMP AND NO PRIOR BILATERAL OVARIECTOMY AND NOT ON ESTROGEN REPLACEMENT) |
|---|---|---|---|---|
| ALL | 2 | 15 | 65 | 105 |
| subtype1 | 0 | 4 | 16 | 27 |
| subtype2 | 2 | 10 | 48 | 60 |
| subtype3 | 0 | 1 | 1 | 18 |
Figure S167. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #32: 'MENOPAUSE_STATUS'

P value = 0.596 (Fisher's exact test), Q value = 1
Table S172. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 67 | 71 |
| subtype1 | 21 | 19 |
| subtype2 | 37 | 45 |
| subtype3 | 9 | 7 |
Figure S168. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'
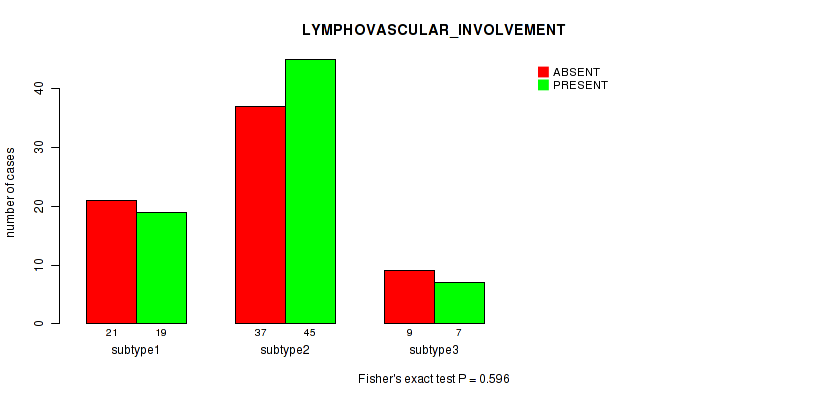
P value = 0.0689 (Kruskal-Wallis (anova)), Q value = 1
Table S173. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 139 | 1.0 (2.4) |
| subtype1 | 37 | 0.7 (2.4) |
| subtype2 | 86 | 1.3 (2.5) |
| subtype3 | 16 | 0.2 (0.6) |
Figure S169. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'

P value = 0.595 (Kruskal-Wallis (anova)), Q value = 1
Table S174. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 159 | 21.8 (12.4) |
| subtype1 | 41 | 20.3 (9.5) |
| subtype2 | 99 | 22.0 (13.5) |
| subtype3 | 19 | 23.8 (11.9) |
Figure S170. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'

P value = 0.0717 (Fisher's exact test), Q value = 1
Table S175. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'
| nPatients | KERATINIZING SQUAMOUS CELL CARCINOMA | NON-KERATINIZING SQUAMOUS CELL CARCINOMA |
|---|---|---|
| ALL | 48 | 94 |
| subtype1 | 1 | 13 |
| subtype2 | 40 | 67 |
| subtype3 | 7 | 14 |
Figure S171. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'

P value = 0.154 (Kruskal-Wallis (anova)), Q value = 1
Table S176. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 237 | 2007.7 (5.0) |
| subtype1 | 54 | 2008.9 (3.8) |
| subtype2 | 149 | 2007.7 (5.1) |
| subtype3 | 34 | 2006.3 (6.0) |
Figure S172. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'

P value = 0.03 (Fisher's exact test), Q value = 1
Table S177. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'
| nPatients | CURRENT USER | FORMER USER | NEVER USED |
|---|---|---|---|
| ALL | 10 | 47 | 56 |
| subtype1 | 5 | 15 | 12 |
| subtype2 | 2 | 29 | 36 |
| subtype3 | 3 | 3 | 8 |
Figure S173. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'

P value = 0.418 (Kruskal-Wallis (anova)), Q value = 1
Table S178. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 209 | 161.8 (6.7) |
| subtype1 | 50 | 162.3 (6.6) |
| subtype2 | 133 | 161.3 (6.7) |
| subtype3 | 26 | 163.0 (6.5) |
Figure S174. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'
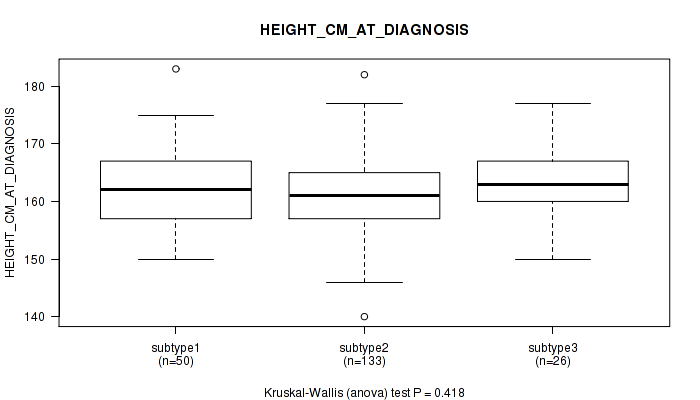
P value = 0.274 (Fisher's exact test), Q value = 1
Table S179. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 93 | 16 |
| subtype1 | 31 | 4 |
| subtype2 | 52 | 12 |
| subtype3 | 10 | 0 |
Figure S175. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'
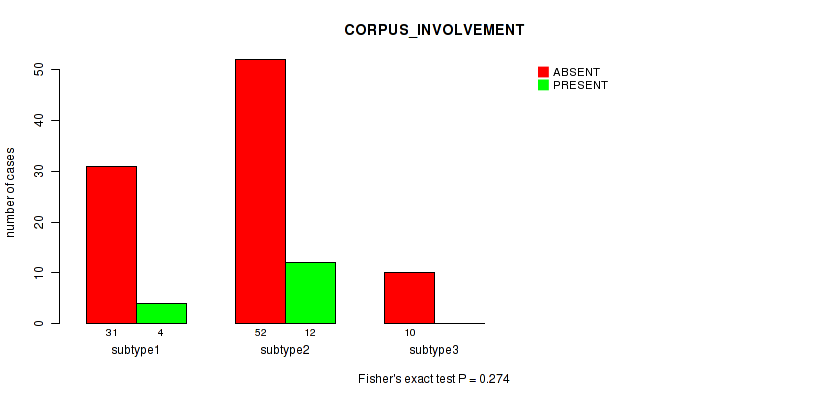
P value = 0.256 (Fisher's exact test), Q value = 1
Table S180. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'
| nPatients | CARBOPLATIN | CISPLATIN | OTHER |
|---|---|---|---|
| ALL | 2 | 17 | 1 |
| subtype1 | 0 | 2 | 0 |
| subtype2 | 2 | 12 | 0 |
| subtype3 | 0 | 3 | 1 |
Figure S176. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'

P value = 0.0961 (Kruskal-Wallis (anova)), Q value = 1
Table S181. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 13 | 12.4 (5.5) |
| subtype1 | 1 | 7.1 (NA) |
| subtype2 | 9 | 11.0 (3.7) |
| subtype3 | 3 | 18.4 (7.3) |
Figure S177. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'

P value = 0.114 (Fisher's exact test), Q value = 1
Table S182. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'
| nPatients | T1A | T1A1 | T1B | T1B1 | T1B2 | T2 | T2A | T2A1 | T2A2 | T2B | T3 | T3B | T4 | TIS | TX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 1 | 1 | 32 | 64 | 25 | 4 | 8 | 7 | 10 | 18 | 1 | 7 | 3 | 1 | 15 |
| subtype1 | 0 | 0 | 3 | 23 | 6 | 1 | 2 | 2 | 3 | 5 | 0 | 1 | 1 | 0 | 2 |
| subtype2 | 1 | 1 | 20 | 39 | 16 | 3 | 5 | 2 | 6 | 11 | 1 | 5 | 2 | 0 | 10 |
| subtype3 | 0 | 0 | 9 | 2 | 3 | 0 | 1 | 3 | 1 | 2 | 0 | 1 | 0 | 1 | 3 |
Figure S178. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'

P value = 0.0125 (Kruskal-Wallis (anova)), Q value = 1
Table S183. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 239 | 47.3 (13.3) |
| subtype1 | 55 | 46.6 (11.2) |
| subtype2 | 150 | 49.0 (13.8) |
| subtype3 | 34 | 41.1 (12.8) |
Figure S179. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'

P value = 0.273 (Fisher's exact test), Q value = 1
Table S184. Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'
| nPatients | STAGE I | STAGE IA | STAGE IA1 | STAGE IA2 | STAGE IB | STAGE IB1 | STAGE IB2 | STAGE II | STAGE IIA | STAGE IIA1 | STAGE IIA2 | STAGE IIB | STAGE III | STAGE IIIB | STAGE IVA | STAGE IVB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 4 | 2 | 1 | 1 | 34 | 67 | 34 | 4 | 6 | 5 | 7 | 23 | 1 | 33 | 4 | 7 |
| subtype1 | 1 | 0 | 0 | 0 | 3 | 24 | 8 | 1 | 1 | 1 | 2 | 5 | 0 | 5 | 0 | 4 |
| subtype2 | 3 | 2 | 1 | 1 | 23 | 37 | 18 | 3 | 3 | 2 | 4 | 15 | 1 | 25 | 4 | 3 |
| subtype3 | 0 | 0 | 0 | 0 | 8 | 6 | 8 | 0 | 2 | 2 | 1 | 3 | 0 | 3 | 0 | 0 |
Figure S180. Get High-res Image Clustering Approach #4: 'RNAseq cHierClus subtypes' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'

Table S185. Description of clustering approach #5: 'MIRSEQ CNMF'
| Cluster Labels | 1 | 2 | 3 |
|---|---|---|---|
| Number of samples | 79 | 68 | 103 |
P value = 0.436 (logrank test), Q value = 1
Table S186. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 245 | 54 | 0.0 - 195.8 (16.1) |
| subtype1 | 79 | 16 | 0.1 - 160.4 (15.6) |
| subtype2 | 65 | 11 | 0.0 - 182.9 (15.0) |
| subtype3 | 101 | 27 | 0.0 - 195.8 (17.6) |
Figure S181. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #1: 'Time to Death'

P value = 0.171 (Kruskal-Wallis (anova)), Q value = 1
Table S187. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 246 | 47.6 (13.5) |
| subtype1 | 77 | 46.3 (11.6) |
| subtype2 | 68 | 50.0 (13.2) |
| subtype3 | 101 | 46.9 (14.9) |
Figure S182. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #2: 'AGE'
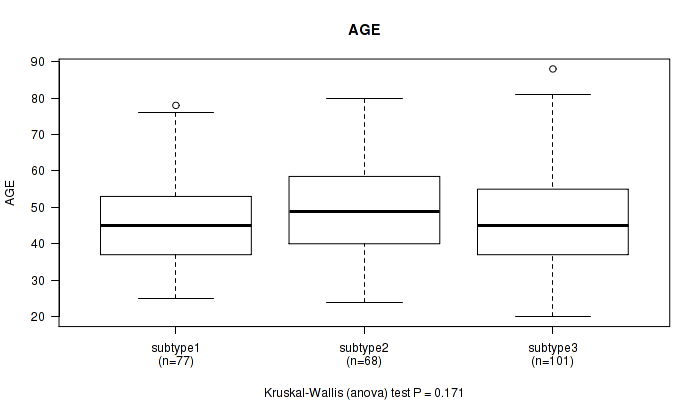
P value = 0.932 (Fisher's exact test), Q value = 1
Table S188. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'
| nPatients | T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 127 | 52 | 9 | 4 |
| subtype1 | 45 | 20 | 4 | 1 |
| subtype2 | 38 | 14 | 1 | 1 |
| subtype3 | 44 | 18 | 4 | 2 |
Figure S183. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'

P value = 0.832 (Fisher's exact test), Q value = 1
Table S189. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'
| nPatients | 0 | 1 |
|---|---|---|
| ALL | 117 | 51 |
| subtype1 | 42 | 17 |
| subtype2 | 35 | 18 |
| subtype3 | 40 | 16 |
Figure S184. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'

P value = 0.267 (Fisher's exact test), Q value = 1
Table S190. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 99 | 8 | 96 |
| subtype1 | 34 | 6 | 33 |
| subtype2 | 30 | 1 | 25 |
| subtype3 | 35 | 1 | 38 |
Figure S185. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'

P value = 1e-05 (Fisher's exact test), Q value = 0.0036
Table S191. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | ADENOSQUAMOUS | CERVICAL SQUAMOUS CELL CARCINOMA | ENDOCERVICAL ADENOCARCINOMA OF THE USUAL TYPE | ENDOCERVICAL TYPE OF ADENOCARCINOMA | ENDOMETRIOID ADENOCARCINOMA OF ENDOCERVIX | MUCINOUS ADENOCARCINOMA OF ENDOCERVICAL TYPE |
|---|---|---|---|---|---|---|
| ALL | 5 | 209 | 5 | 22 | 3 | 6 |
| subtype1 | 4 | 47 | 4 | 17 | 3 | 4 |
| subtype2 | 1 | 60 | 1 | 5 | 0 | 1 |
| subtype3 | 0 | 102 | 0 | 0 | 0 | 1 |
Figure S186. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 0.251 (Fisher's exact test), Q value = 1
Table S192. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 35 | 215 |
| subtype1 | 8 | 71 |
| subtype2 | 8 | 60 |
| subtype3 | 19 | 84 |
Figure S187. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'

P value = 0.147 (Kruskal-Wallis (anova)), Q value = 1
Table S193. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 76 | 18.8 (13.8) |
| subtype1 | 18 | 14.8 (10.4) |
| subtype2 | 24 | 23.2 (14.8) |
| subtype3 | 34 | 17.8 (14.2) |
Figure S188. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'

P value = 0.397 (Kruskal-Wallis (anova)), Q value = 1
Table S194. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 144 | 1.1 (2.5) |
| subtype1 | 51 | 0.8 (2.3) |
| subtype2 | 48 | 1.5 (2.6) |
| subtype3 | 45 | 0.9 (2.5) |
Figure S189. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'

P value = 0.339 (Fisher's exact test), Q value = 1
Table S195. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER | WHITE |
|---|---|---|---|---|---|
| ALL | 8 | 19 | 26 | 1 | 178 |
| subtype1 | 1 | 8 | 7 | 0 | 54 |
| subtype2 | 1 | 7 | 6 | 0 | 48 |
| subtype3 | 6 | 4 | 13 | 1 | 76 |
Figure S190. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #10: 'RACE'

P value = 0.26 (Fisher's exact test), Q value = 1
Table S196. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #11: 'ETHNICITY'
| nPatients | HISPANIC OR LATINO | NOT HISPANIC OR LATINO |
|---|---|---|
| ALL | 17 | 157 |
| subtype1 | 7 | 49 |
| subtype2 | 2 | 47 |
| subtype3 | 8 | 61 |
Figure S191. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #11: 'ETHNICITY'
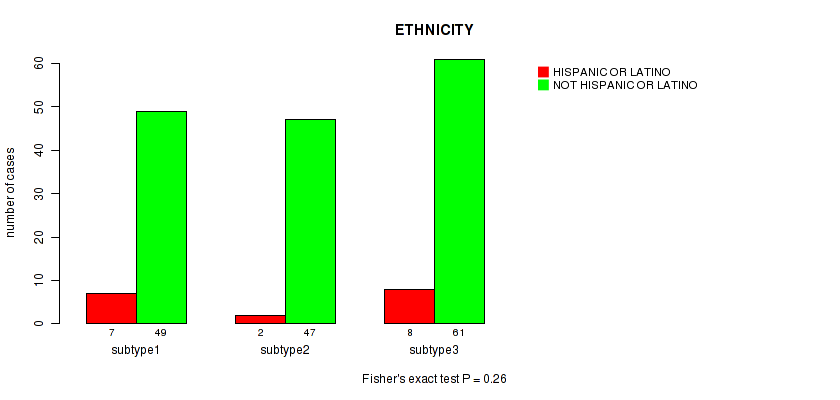
P value = 0.476 (Kruskal-Wallis (anova)), Q value = 1
Table S197. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 228 | 74.8 (22.4) |
| subtype1 | 75 | 76.5 (20.2) |
| subtype2 | 59 | 75.4 (29.1) |
| subtype3 | 94 | 73.0 (19.1) |
Figure S192. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'

P value = 0.362 (Fisher's exact test), Q value = 1
Table S198. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #13: 'TUMOR_STATUS'
| nPatients | TUMOR FREE | WITH TUMOR |
|---|---|---|
| ALL | 75 | 28 |
| subtype1 | 26 | 9 |
| subtype2 | 22 | 5 |
| subtype3 | 27 | 14 |
Figure S193. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #13: 'TUMOR_STATUS'

P value = 0.0295 (Fisher's exact test), Q value = 1
Table S199. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'
| nPatients | BRAZIL | CANADA | NIGERIA | RUSSIA | UKRAINE | UNITED STATES | VIETNAM |
|---|---|---|---|---|---|---|---|
| ALL | 24 | 5 | 1 | 11 | 7 | 188 | 14 |
| subtype1 | 11 | 3 | 1 | 2 | 3 | 54 | 5 |
| subtype2 | 1 | 2 | 0 | 5 | 2 | 52 | 6 |
| subtype3 | 12 | 0 | 0 | 4 | 2 | 82 | 3 |
Figure S194. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'

P value = 0.0292 (Fisher's exact test), Q value = 1
Table S200. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'
| nPatients | G1 | G2 | G3 | G4 | GX |
|---|---|---|---|---|---|
| ALL | 17 | 113 | 102 | 1 | 14 |
| subtype1 | 7 | 30 | 35 | 0 | 7 |
| subtype2 | 8 | 34 | 23 | 1 | 1 |
| subtype3 | 2 | 49 | 44 | 0 | 6 |
Figure S195. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'
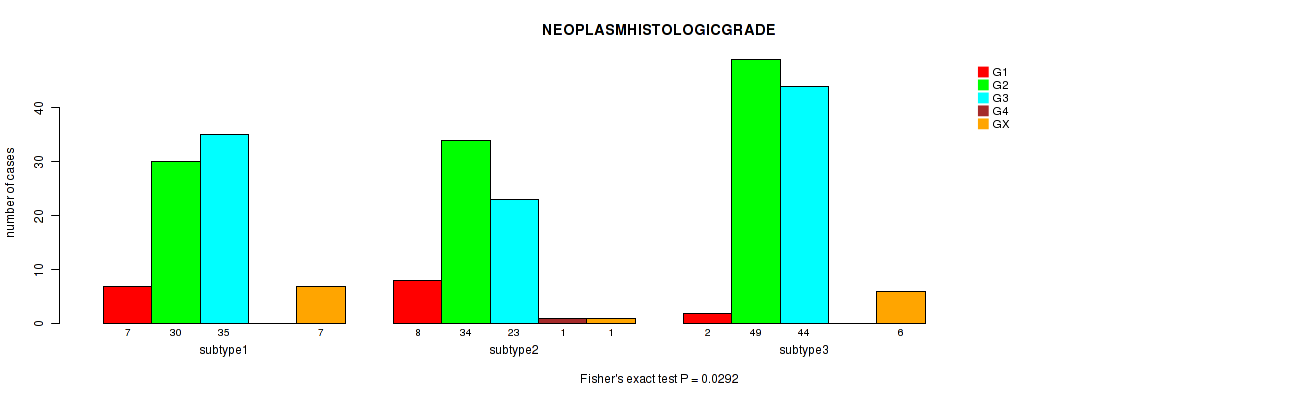
P value = 0.12 (Kruskal-Wallis (anova)), Q value = 1
Table S201. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 34 | 2000.4 (11.2) |
| subtype1 | 8 | 1999.9 (12.7) |
| subtype2 | 9 | 1995.1 (11.5) |
| subtype3 | 17 | 2003.5 (9.9) |
Figure S196. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'

P value = 0.147 (Kruskal-Wallis (anova)), Q value = 1
Table S202. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 76 | 18.8 (13.8) |
| subtype1 | 18 | 14.8 (10.4) |
| subtype2 | 24 | 23.2 (14.8) |
| subtype3 | 34 | 17.8 (14.2) |
Figure S197. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'

P value = 0.238 (Fisher's exact test), Q value = 1
Table S203. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'
| nPatients | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT REFORMED SMOKER, DURATION NOT SPECIFIED | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|
| ALL | 33 | 8 | 3 | 54 | 114 |
| subtype1 | 9 | 1 | 0 | 13 | 46 |
| subtype2 | 8 | 3 | 2 | 18 | 29 |
| subtype3 | 16 | 4 | 1 | 23 | 39 |
Figure S198. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'

P value = 0.796 (Kruskal-Wallis (anova)), Q value = 1
Table S204. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 68 | 21.4 (7.8) |
| subtype1 | 16 | 20.4 (7.1) |
| subtype2 | 21 | 21.5 (6.4) |
| subtype3 | 31 | 21.9 (9.1) |
Figure S199. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'

P value = 0.68 (Kruskal-Wallis (anova)), Q value = 1
Table S205. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 110 | 3890.9 (1636.4) |
| subtype1 | 36 | 3934.4 (1684.4) |
| subtype2 | 24 | 3955.9 (1661.7) |
| subtype3 | 50 | 3828.3 (1620.7) |
Figure S200. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'

P value = 0.12 (Fisher's exact test), Q value = 1
Table S206. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'
| nPatients | COMBINATION | EXTERNAL | EXTERNAL BEAM | IMPLANTS | INTERNAL |
|---|---|---|---|---|---|
| ALL | 20 | 77 | 15 | 1 | 12 |
| subtype1 | 6 | 30 | 2 | 0 | 2 |
| subtype2 | 2 | 18 | 6 | 1 | 3 |
| subtype3 | 12 | 29 | 7 | 0 | 7 |
Figure S201. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'

P value = 0.396 (Fisher's exact test), Q value = 1
Table S207. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'
| nPatients | COMPLETED AS PLANNED | TREATMENT NOT COMPLETED |
|---|---|---|
| ALL | 29 | 3 |
| subtype1 | 8 | 0 |
| subtype2 | 6 | 0 |
| subtype3 | 15 | 3 |
Figure S202. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'

P value = 0.0861 (Fisher's exact test), Q value = 1
Table S208. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'
| nPatients | DISTANT RECURRENCE | LOCAL RECURRENCE | PRIMARY TUMOR FIELD | REGIONAL SITE |
|---|---|---|---|---|
| ALL | 2 | 2 | 30 | 16 |
| subtype1 | 1 | 0 | 3 | 7 |
| subtype2 | 0 | 1 | 8 | 3 |
| subtype3 | 1 | 1 | 19 | 6 |
Figure S203. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'

P value = 0.413 (Fisher's exact test), Q value = 1
Table S209. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'
| nPatients | CGY | GY |
|---|---|---|
| ALL | 36 | 3 |
| subtype1 | 11 | 0 |
| subtype2 | 8 | 0 |
| subtype3 | 17 | 3 |
Figure S204. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'
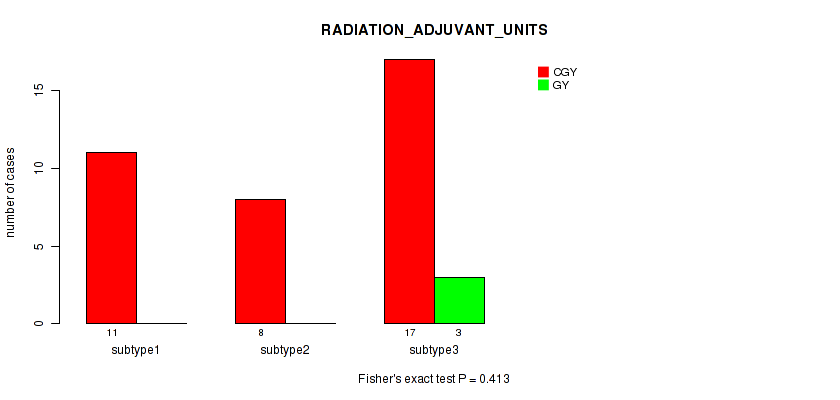
P value = 0.461 (Kruskal-Wallis (anova)), Q value = 1
Table S210. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 219 | 3.6 (2.5) |
| subtype1 | 69 | 3.4 (2.7) |
| subtype2 | 62 | 3.6 (2.4) |
| subtype3 | 88 | 3.6 (2.5) |
Figure S205. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'

P value = 0.848 (Kruskal-Wallis (anova)), Q value = 1
Table S211. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 109 | 0.1 (0.4) |
| subtype1 | 29 | 0.0 (0.2) |
| subtype2 | 32 | 0.1 (0.6) |
| subtype3 | 48 | 0.1 (0.2) |
Figure S206. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'

P value = 0.00877 (Kruskal-Wallis (anova)), Q value = 1
Table S212. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 135 | 0.5 (0.8) |
| subtype1 | 36 | 0.8 (1.1) |
| subtype2 | 40 | 0.3 (0.5) |
| subtype3 | 59 | 0.4 (0.8) |
Figure S207. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'
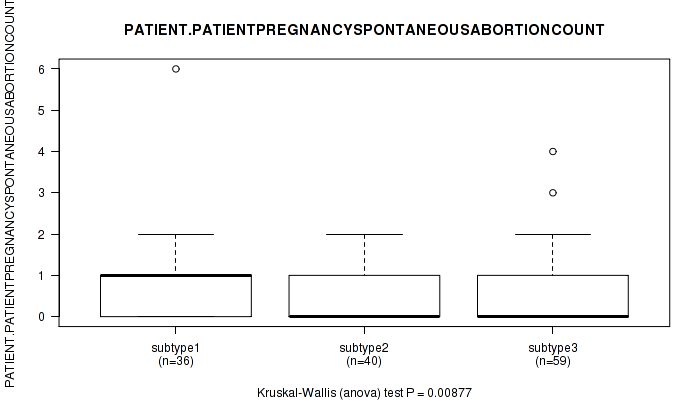
P value = 0.326 (Kruskal-Wallis (anova)), Q value = 1
Table S213. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 220 | 2.6 (1.9) |
| subtype1 | 67 | 2.6 (2.3) |
| subtype2 | 62 | 2.7 (1.7) |
| subtype3 | 91 | 2.7 (1.7) |
Figure S208. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'

P value = 0.396 (Kruskal-Wallis (anova)), Q value = 1
Table S214. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 119 | 0.9 (1.8) |
| subtype1 | 32 | 0.8 (1.5) |
| subtype2 | 36 | 1.1 (2.3) |
| subtype3 | 51 | 0.7 (1.6) |
Figure S209. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'

P value = 0.686 (Kruskal-Wallis (anova)), Q value = 1
Table S215. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 111 | 0.1 (0.3) |
| subtype1 | 29 | 0.1 (0.3) |
| subtype2 | 32 | 0.2 (0.4) |
| subtype3 | 50 | 0.1 (0.3) |
Figure S210. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'

P value = 0.809 (Fisher's exact test), Q value = 1
Table S216. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'
| nPatients | MACROSCOPIC PARAMETRIAL INVOLVEMENT | MICROSCOPIC PARAMETRIAL INVOLVEMENT | OTHER LOCATION, SPECIFY | POSITIVE BLADDER MARGIN | POSITIVE VAGINAL MARGIN |
|---|---|---|---|---|---|
| ALL | 2 | 7 | 33 | 1 | 9 |
| subtype1 | 1 | 2 | 8 | 0 | 2 |
| subtype2 | 1 | 2 | 11 | 1 | 5 |
| subtype3 | 0 | 3 | 14 | 0 | 2 |
Figure S211. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'

P value = 0.407 (Fisher's exact test), Q value = 1
Table S217. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #32: 'MENOPAUSE_STATUS'
| nPatients | INDETERMINATE (NEITHER PRE OR POSTMENOPAUSAL) | PERI (6-12 MONTHS SINCE LAST MENSTRUAL PERIOD) | POST (PRIOR BILATERAL OVARIECTOMY OR >12 MO SINCE LMP WITH NO PRIOR HYSTERECTOMY) | PRE (<6 MONTHS SINCE LMP AND NO PRIOR BILATERAL OVARIECTOMY AND NOT ON ESTROGEN REPLACEMENT) |
|---|---|---|---|---|
| ALL | 2 | 16 | 70 | 109 |
| subtype1 | 1 | 8 | 19 | 37 |
| subtype2 | 1 | 3 | 23 | 26 |
| subtype3 | 0 | 5 | 28 | 46 |
Figure S212. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #32: 'MENOPAUSE_STATUS'
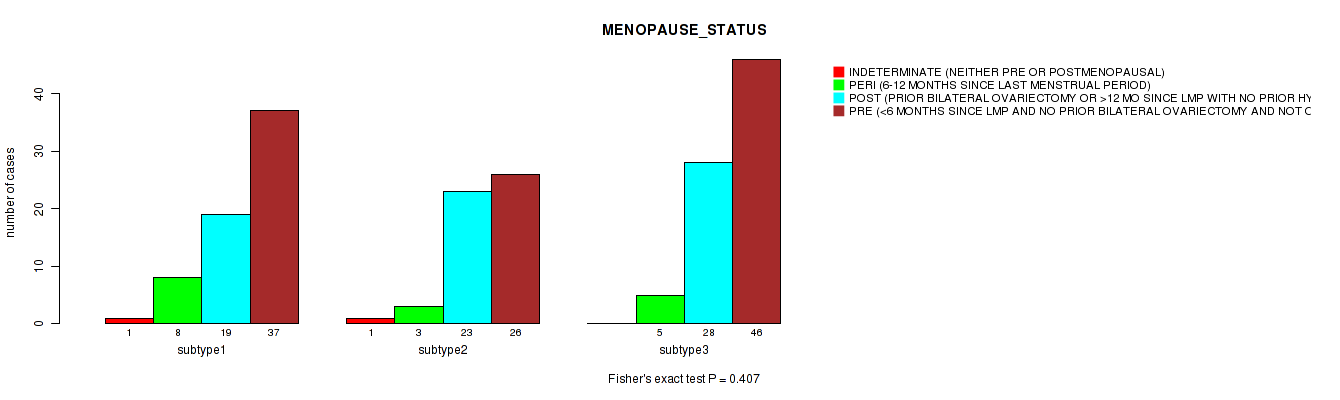
P value = 0.666 (Fisher's exact test), Q value = 1
Table S218. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 69 | 76 |
| subtype1 | 24 | 29 |
| subtype2 | 21 | 26 |
| subtype3 | 24 | 21 |
Figure S213. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'

P value = 0.397 (Kruskal-Wallis (anova)), Q value = 1
Table S219. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 144 | 1.1 (2.5) |
| subtype1 | 51 | 0.8 (2.3) |
| subtype2 | 48 | 1.5 (2.6) |
| subtype3 | 45 | 0.9 (2.5) |
Figure S214. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'

P value = 0.644 (Kruskal-Wallis (anova)), Q value = 1
Table S220. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 166 | 21.7 (12.4) |
| subtype1 | 56 | 20.3 (10.8) |
| subtype2 | 55 | 24.0 (14.4) |
| subtype3 | 55 | 21.0 (11.6) |
Figure S215. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'

P value = 0.0298 (Fisher's exact test), Q value = 1
Table S221. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'
| nPatients | KERATINIZING SQUAMOUS CELL CARCINOMA | NON-KERATINIZING SQUAMOUS CELL CARCINOMA |
|---|---|---|
| ALL | 49 | 97 |
| subtype1 | 4 | 25 |
| subtype2 | 18 | 25 |
| subtype3 | 27 | 47 |
Figure S216. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'

P value = 0.0327 (Kruskal-Wallis (anova)), Q value = 1
Table S222. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 248 | 2007.7 (5.0) |
| subtype1 | 78 | 2008.8 (4.7) |
| subtype2 | 67 | 2007.5 (4.9) |
| subtype3 | 103 | 2007.1 (5.2) |
Figure S217. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'

P value = 0.679 (Fisher's exact test), Q value = 1
Table S223. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'
| nPatients | CURRENT USER | FORMER USER | NEVER USED |
|---|---|---|---|
| ALL | 10 | 49 | 62 |
| subtype1 | 5 | 18 | 20 |
| subtype2 | 2 | 15 | 15 |
| subtype3 | 3 | 16 | 27 |
Figure S218. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'

P value = 0.407 (Kruskal-Wallis (anova)), Q value = 1
Table S224. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 218 | 161.6 (7.1) |
| subtype1 | 70 | 162.1 (6.9) |
| subtype2 | 55 | 162.5 (7.5) |
| subtype3 | 93 | 160.7 (6.9) |
Figure S219. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'

P value = 0.803 (Fisher's exact test), Q value = 1
Table S225. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 96 | 17 |
| subtype1 | 34 | 7 |
| subtype2 | 31 | 6 |
| subtype3 | 31 | 4 |
Figure S220. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'

P value = 1 (Fisher's exact test), Q value = 1
Table S226. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'
| nPatients | CARBOPLATIN | CISPLATIN | OTHER |
|---|---|---|---|
| ALL | 3 | 18 | 1 |
| subtype1 | 0 | 3 | 0 |
| subtype2 | 1 | 3 | 0 |
| subtype3 | 2 | 12 | 1 |
Figure S221. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'
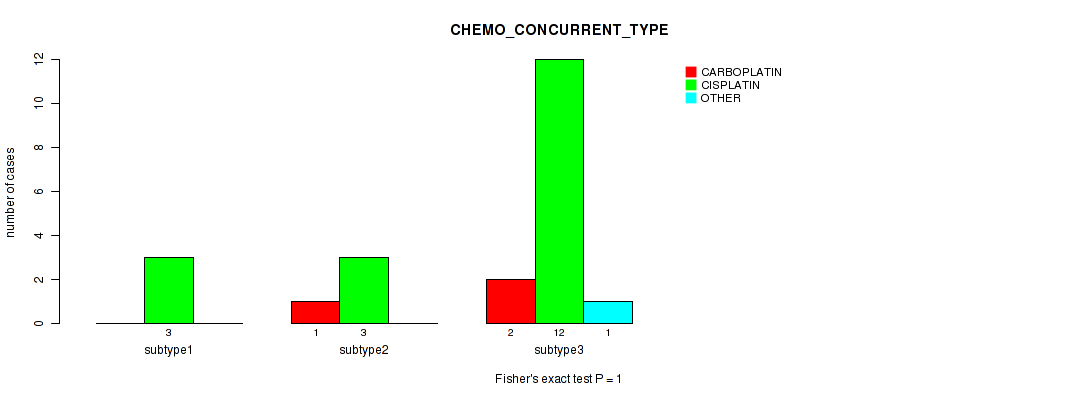
P value = 0.508 (Kruskal-Wallis (anova)), Q value = 1
Table S227. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 13 | 12.4 (5.5) |
| subtype1 | 2 | 10.8 (5.2) |
| subtype2 | 4 | 10.2 (3.3) |
| subtype3 | 7 | 14.1 (6.6) |
Figure S222. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'
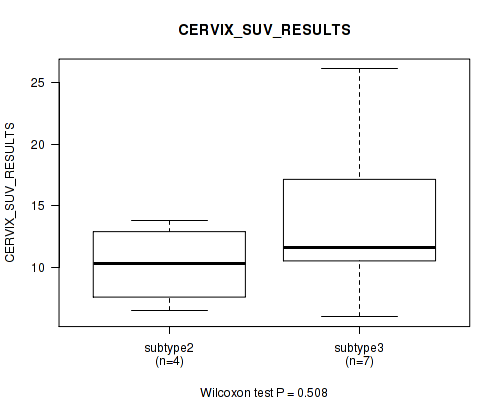
P value = 0.392 (Fisher's exact test), Q value = 1
Table S228. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'
| nPatients | T1A | T1A1 | T1B | T1B1 | T1B2 | T2 | T2A | T2A1 | T2A2 | T2B | T3 | T3B | T4 | TIS | TX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 1 | 1 | 32 | 68 | 25 | 4 | 8 | 7 | 10 | 23 | 1 | 8 | 4 | 1 | 15 |
| subtype1 | 0 | 0 | 8 | 29 | 8 | 2 | 3 | 3 | 6 | 6 | 0 | 4 | 1 | 0 | 3 |
| subtype2 | 0 | 1 | 6 | 23 | 8 | 1 | 3 | 2 | 2 | 6 | 0 | 1 | 1 | 0 | 3 |
| subtype3 | 1 | 0 | 18 | 16 | 9 | 1 | 2 | 2 | 2 | 11 | 1 | 3 | 2 | 1 | 9 |
Figure S223. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'

P value = 0.21 (Kruskal-Wallis (anova)), Q value = 1
Table S229. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 250 | 47.7 (13.5) |
| subtype1 | 79 | 46.4 (11.5) |
| subtype2 | 68 | 50.0 (13.2) |
| subtype3 | 103 | 47.1 (14.9) |
Figure S224. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'

P value = 0.0469 (Fisher's exact test), Q value = 1
Table S230. Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'
| nPatients | STAGE I | STAGE IA | STAGE IA1 | STAGE IA2 | STAGE IB | STAGE IB1 | STAGE IB2 | STAGE II | STAGE IIA | STAGE IIA1 | STAGE IIA2 | STAGE IIB | STAGE III | STAGE IIIB | STAGE IVA | STAGE IVB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 4 | 2 | 1 | 1 | 35 | 70 | 34 | 4 | 7 | 5 | 7 | 27 | 1 | 34 | 5 | 7 |
| subtype1 | 2 | 0 | 0 | 0 | 7 | 28 | 13 | 1 | 1 | 1 | 5 | 5 | 0 | 11 | 1 | 4 |
| subtype2 | 0 | 1 | 1 | 1 | 7 | 24 | 9 | 1 | 3 | 2 | 2 | 5 | 0 | 7 | 2 | 1 |
| subtype3 | 2 | 1 | 0 | 0 | 21 | 18 | 12 | 2 | 3 | 2 | 0 | 17 | 1 | 16 | 2 | 2 |
Figure S225. Get High-res Image Clustering Approach #5: 'MIRSEQ CNMF' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'
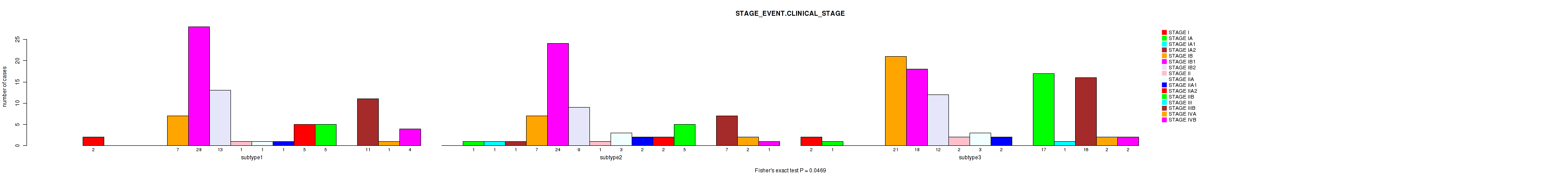
Table S231. Description of clustering approach #6: 'MIRSEQ CHIERARCHICAL'
| Cluster Labels | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Number of samples | 48 | 79 | 64 | 20 | 39 |
P value = 0.519 (logrank test), Q value = 1
Table S232. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 245 | 54 | 0.0 - 195.8 (16.1) |
| subtype1 | 47 | 9 | 0.1 - 137.2 (14.9) |
| subtype2 | 79 | 16 | 0.0 - 160.4 (16.1) |
| subtype3 | 63 | 13 | 0.0 - 182.9 (21.7) |
| subtype4 | 18 | 5 | 0.1 - 195.8 (13.7) |
| subtype5 | 38 | 11 | 0.1 - 144.2 (14.3) |
Figure S226. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #1: 'Time to Death'
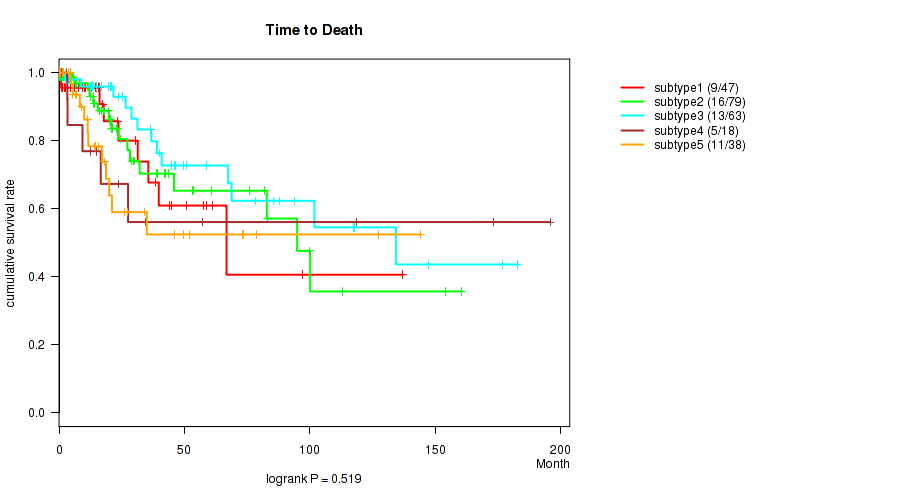
P value = 8.5e-06 (Kruskal-Wallis (anova)), Q value = 0.003
Table S233. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 246 | 47.6 (13.5) |
| subtype1 | 46 | 44.1 (11.8) |
| subtype2 | 78 | 49.9 (13.7) |
| subtype3 | 64 | 51.2 (14.2) |
| subtype4 | 19 | 51.7 (9.1) |
| subtype5 | 39 | 39.1 (11.5) |
Figure S227. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #2: 'AGE'

P value = 0.00149 (Fisher's exact test), Q value = 0.51
Table S234. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'
| nPatients | T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 127 | 52 | 9 | 4 |
| subtype1 | 31 | 8 | 0 | 0 |
| subtype2 | 28 | 23 | 4 | 2 |
| subtype3 | 44 | 8 | 0 | 1 |
| subtype4 | 8 | 5 | 2 | 1 |
| subtype5 | 16 | 8 | 3 | 0 |
Figure S228. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'

P value = 0.354 (Fisher's exact test), Q value = 1
Table S235. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'
| nPatients | 0 | 1 |
|---|---|---|
| ALL | 117 | 51 |
| subtype1 | 28 | 9 |
| subtype2 | 27 | 18 |
| subtype3 | 34 | 17 |
| subtype4 | 10 | 2 |
| subtype5 | 18 | 5 |
Figure S229. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'

P value = 0.00764 (Fisher's exact test), Q value = 1
Table S236. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 99 | 8 | 96 |
| subtype1 | 17 | 2 | 22 |
| subtype2 | 21 | 1 | 39 |
| subtype3 | 33 | 1 | 21 |
| subtype4 | 12 | 1 | 4 |
| subtype5 | 16 | 3 | 10 |
Figure S230. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'
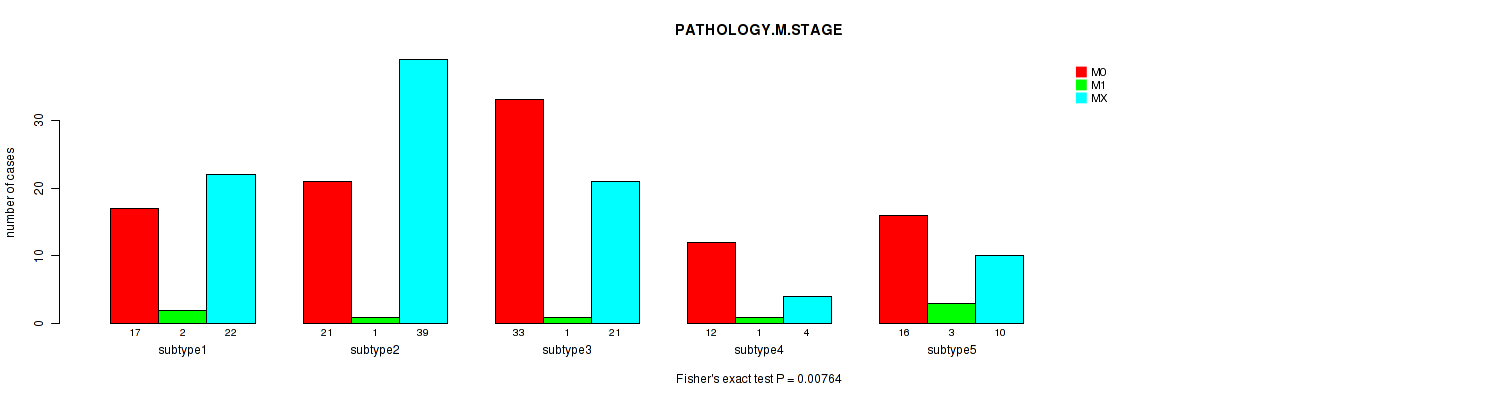
P value = 1e-05 (Fisher's exact test), Q value = 0.0036
Table S237. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | ADENOSQUAMOUS | CERVICAL SQUAMOUS CELL CARCINOMA | ENDOCERVICAL ADENOCARCINOMA OF THE USUAL TYPE | ENDOCERVICAL TYPE OF ADENOCARCINOMA | ENDOMETRIOID ADENOCARCINOMA OF ENDOCERVIX | MUCINOUS ADENOCARCINOMA OF ENDOCERVICAL TYPE |
|---|---|---|---|---|---|---|
| ALL | 5 | 209 | 5 | 22 | 3 | 6 |
| subtype1 | 4 | 11 | 4 | 20 | 3 | 6 |
| subtype2 | 1 | 76 | 1 | 1 | 0 | 0 |
| subtype3 | 0 | 64 | 0 | 0 | 0 | 0 |
| subtype4 | 0 | 20 | 0 | 0 | 0 | 0 |
| subtype5 | 0 | 38 | 0 | 1 | 0 | 0 |
Figure S231. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
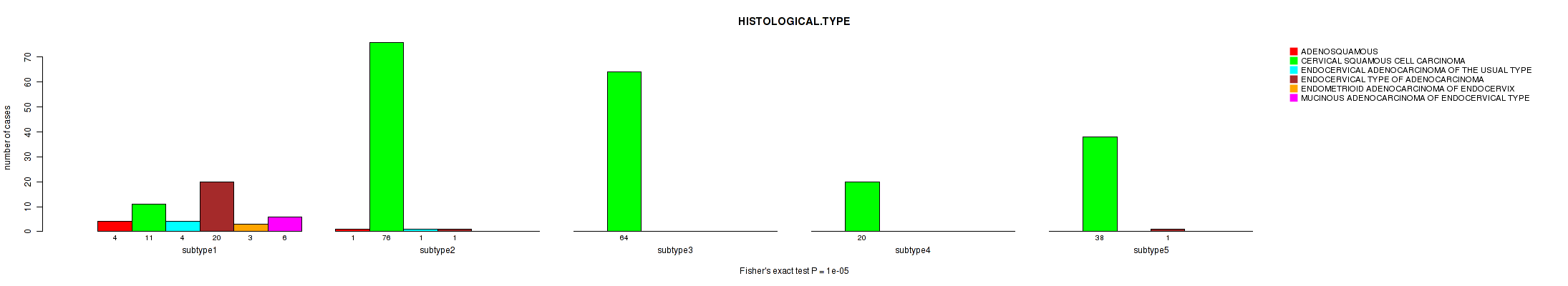
P value = 0.0434 (Fisher's exact test), Q value = 1
Table S238. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 35 | 215 |
| subtype1 | 3 | 45 |
| subtype2 | 7 | 72 |
| subtype3 | 12 | 52 |
| subtype4 | 3 | 17 |
| subtype5 | 10 | 29 |
Figure S232. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'

P value = 0.324 (Kruskal-Wallis (anova)), Q value = 1
Table S239. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 76 | 18.8 (13.8) |
| subtype1 | 15 | 15.1 (11.4) |
| subtype2 | 29 | 21.6 (17.6) |
| subtype3 | 19 | 20.6 (10.9) |
| subtype4 | 2 | 16.0 (5.7) |
| subtype5 | 11 | 13.8 (9.5) |
Figure S233. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'

P value = 0.228 (Kruskal-Wallis (anova)), Q value = 1
Table S240. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 144 | 1.1 (2.5) |
| subtype1 | 35 | 0.7 (2.0) |
| subtype2 | 37 | 1.8 (3.2) |
| subtype3 | 45 | 1.0 (1.8) |
| subtype4 | 8 | 0.4 (0.7) |
| subtype5 | 19 | 0.9 (3.2) |
Figure S234. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'

P value = 0.59 (Fisher's exact test), Q value = 1
Table S241. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER | WHITE |
|---|---|---|---|---|---|
| ALL | 8 | 19 | 26 | 1 | 178 |
| subtype1 | 0 | 4 | 2 | 0 | 37 |
| subtype2 | 3 | 5 | 11 | 0 | 53 |
| subtype3 | 1 | 6 | 6 | 0 | 46 |
| subtype4 | 1 | 2 | 3 | 0 | 14 |
| subtype5 | 3 | 2 | 4 | 1 | 28 |
Figure S235. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #10: 'RACE'

P value = 0.178 (Fisher's exact test), Q value = 1
Table S242. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #11: 'ETHNICITY'
| nPatients | HISPANIC OR LATINO | NOT HISPANIC OR LATINO |
|---|---|---|
| ALL | 17 | 157 |
| subtype1 | 4 | 30 |
| subtype2 | 7 | 46 |
| subtype3 | 1 | 48 |
| subtype4 | 2 | 14 |
| subtype5 | 3 | 19 |
Figure S236. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #11: 'ETHNICITY'

P value = 0.765 (Kruskal-Wallis (anova)), Q value = 1
Table S243. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 228 | 74.8 (22.4) |
| subtype1 | 43 | 75.1 (14.8) |
| subtype2 | 75 | 74.5 (21.5) |
| subtype3 | 58 | 76.4 (29.8) |
| subtype4 | 18 | 68.3 (17.5) |
| subtype5 | 34 | 75.5 (20.3) |
Figure S237. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'
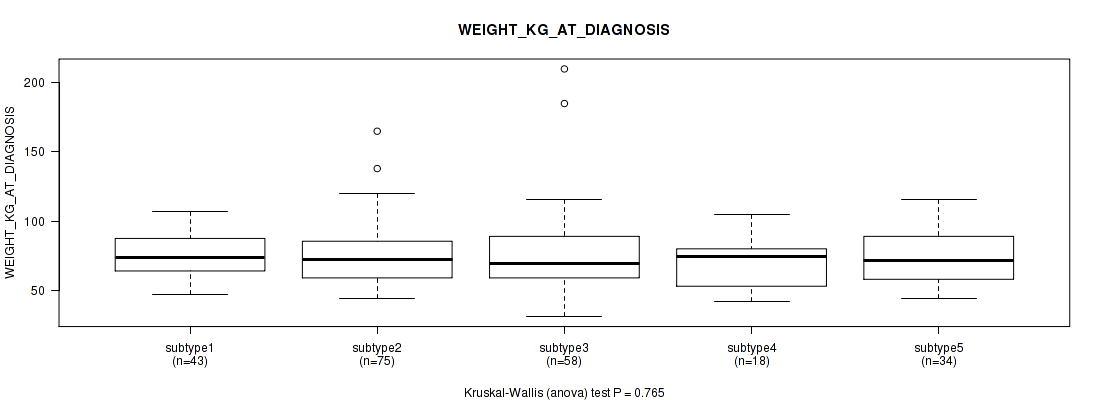
P value = 0.88 (Fisher's exact test), Q value = 1
Table S244. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #13: 'TUMOR_STATUS'
| nPatients | TUMOR FREE | WITH TUMOR |
|---|---|---|
| ALL | 75 | 28 |
| subtype1 | 17 | 5 |
| subtype2 | 15 | 8 |
| subtype3 | 26 | 8 |
| subtype4 | 5 | 2 |
| subtype5 | 12 | 5 |
Figure S238. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #13: 'TUMOR_STATUS'

P value = 0.239 (Fisher's exact test), Q value = 1
Table S245. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'
| nPatients | BRAZIL | CANADA | NIGERIA | RUSSIA | UKRAINE | UNITED STATES | VIETNAM |
|---|---|---|---|---|---|---|---|
| ALL | 24 | 5 | 1 | 11 | 7 | 188 | 14 |
| subtype1 | 3 | 3 | 0 | 1 | 2 | 37 | 2 |
| subtype2 | 12 | 0 | 0 | 4 | 2 | 57 | 4 |
| subtype3 | 2 | 2 | 0 | 5 | 1 | 50 | 4 |
| subtype4 | 2 | 0 | 1 | 1 | 0 | 14 | 2 |
| subtype5 | 5 | 0 | 0 | 0 | 2 | 30 | 2 |
Figure S239. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'

P value = 0.0646 (Fisher's exact test), Q value = 1
Table S246. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'
| nPatients | G1 | G2 | G3 | G4 | GX |
|---|---|---|---|---|---|
| ALL | 17 | 113 | 102 | 1 | 14 |
| subtype1 | 6 | 21 | 17 | 0 | 4 |
| subtype2 | 3 | 40 | 30 | 0 | 4 |
| subtype3 | 6 | 31 | 26 | 0 | 1 |
| subtype4 | 0 | 12 | 7 | 0 | 1 |
| subtype5 | 2 | 9 | 22 | 1 | 4 |
Figure S240. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'

P value = 0.0569 (Kruskal-Wallis (anova)), Q value = 1
Table S247. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 34 | 2000.4 (11.2) |
| subtype1 | 6 | 1999.0 (14.1) |
| subtype2 | 12 | 2005.6 (10.3) |
| subtype3 | 7 | 1993.4 (11.4) |
| subtype4 | 2 | 1992.5 (6.4) |
| subtype5 | 7 | 2002.0 (7.7) |
Figure S241. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'

P value = 0.324 (Kruskal-Wallis (anova)), Q value = 1
Table S248. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 76 | 18.8 (13.8) |
| subtype1 | 15 | 15.1 (11.4) |
| subtype2 | 29 | 21.6 (17.6) |
| subtype3 | 19 | 20.6 (10.9) |
| subtype4 | 2 | 16.0 (5.7) |
| subtype5 | 11 | 13.8 (9.5) |
Figure S242. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'

P value = 0.49 (Fisher's exact test), Q value = 1
Table S249. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'
| nPatients | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT REFORMED SMOKER, DURATION NOT SPECIFIED | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|
| ALL | 33 | 8 | 3 | 54 | 114 |
| subtype1 | 6 | 1 | 0 | 10 | 26 |
| subtype2 | 12 | 2 | 1 | 18 | 31 |
| subtype3 | 4 | 5 | 1 | 16 | 31 |
| subtype4 | 3 | 0 | 0 | 2 | 11 |
| subtype5 | 8 | 0 | 1 | 8 | 15 |
Figure S243. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'
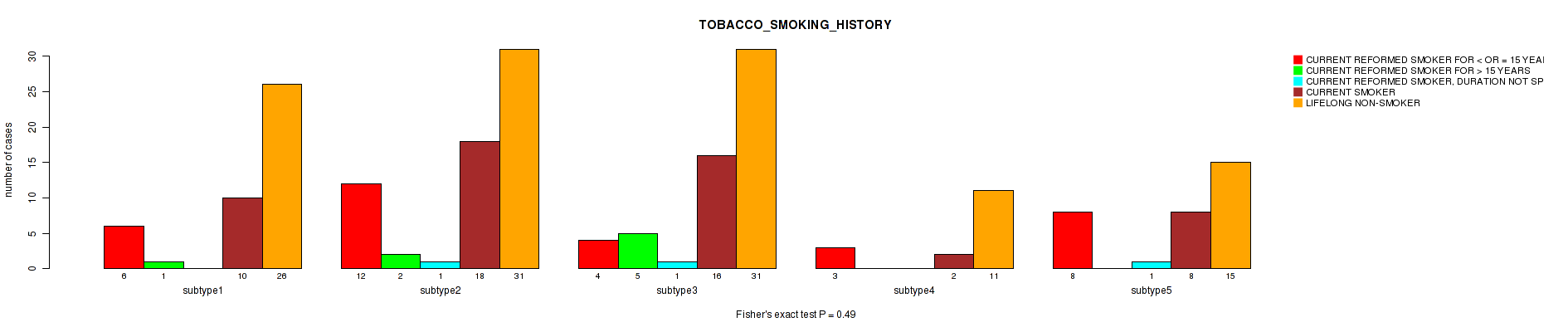
P value = 0.705 (Kruskal-Wallis (anova)), Q value = 1
Table S250. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 68 | 21.4 (7.8) |
| subtype1 | 14 | 19.3 (4.4) |
| subtype2 | 25 | 23.1 (9.5) |
| subtype3 | 14 | 22.1 (7.2) |
| subtype4 | 2 | 20.0 (5.7) |
| subtype5 | 13 | 20.0 (8.2) |
Figure S244. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'
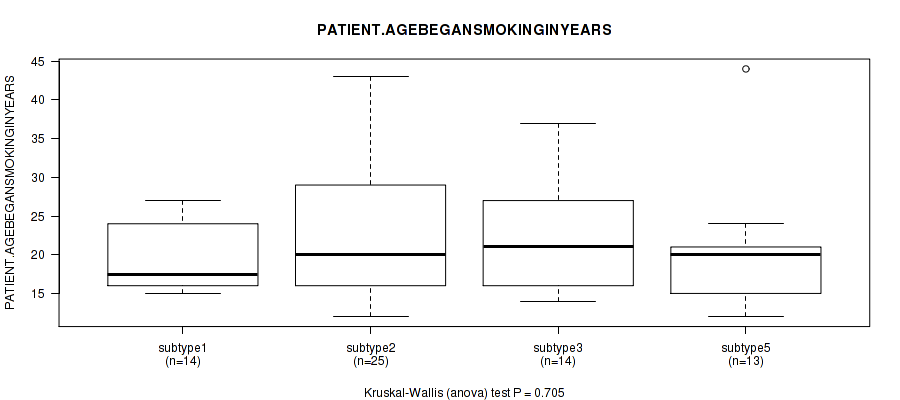
P value = 0.415 (Kruskal-Wallis (anova)), Q value = 1
Table S251. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 110 | 3890.9 (1636.4) |
| subtype1 | 18 | 4214.9 (1570.1) |
| subtype2 | 37 | 3608.0 (1701.3) |
| subtype3 | 23 | 4340.9 (1163.1) |
| subtype4 | 9 | 3488.2 (1690.4) |
| subtype5 | 23 | 3799.9 (1930.5) |
Figure S245. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'

P value = 0.243 (Fisher's exact test), Q value = 1
Table S252. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'
| nPatients | COMBINATION | EXTERNAL | EXTERNAL BEAM | IMPLANTS | INTERNAL |
|---|---|---|---|---|---|
| ALL | 20 | 77 | 15 | 1 | 12 |
| subtype1 | 3 | 16 | 0 | 0 | 1 |
| subtype2 | 4 | 27 | 3 | 0 | 7 |
| subtype3 | 7 | 15 | 5 | 1 | 1 |
| subtype4 | 1 | 5 | 2 | 0 | 1 |
| subtype5 | 5 | 14 | 5 | 0 | 2 |
Figure S246. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'
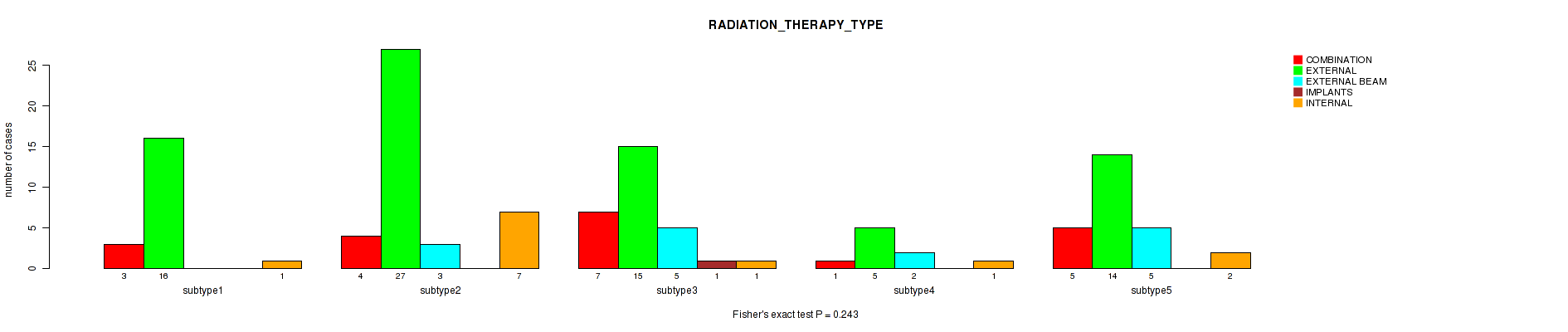
P value = 0.86 (Fisher's exact test), Q value = 1
Table S253. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'
| nPatients | COMPLETED AS PLANNED | TREATMENT NOT COMPLETED |
|---|---|---|
| ALL | 29 | 3 |
| subtype1 | 3 | 0 |
| subtype2 | 7 | 0 |
| subtype3 | 9 | 1 |
| subtype4 | 2 | 0 |
| subtype5 | 8 | 2 |
Figure S247. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'
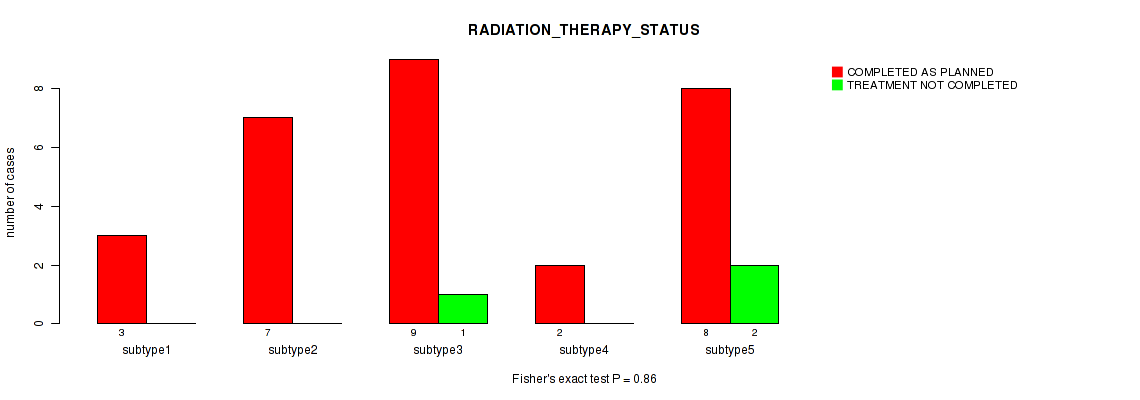
P value = 0.151 (Fisher's exact test), Q value = 1
Table S254. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'
| nPatients | DISTANT RECURRENCE | LOCAL RECURRENCE | PRIMARY TUMOR FIELD | REGIONAL SITE |
|---|---|---|---|---|
| ALL | 2 | 2 | 30 | 16 |
| subtype1 | 1 | 0 | 3 | 1 |
| subtype2 | 0 | 0 | 13 | 11 |
| subtype3 | 0 | 1 | 6 | 1 |
| subtype4 | 0 | 0 | 2 | 2 |
| subtype5 | 1 | 1 | 6 | 1 |
Figure S248. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'

P value = 0.881 (Fisher's exact test), Q value = 1
Table S255. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'
| nPatients | CGY | GY |
|---|---|---|
| ALL | 36 | 3 |
| subtype1 | 5 | 0 |
| subtype2 | 16 | 1 |
| subtype3 | 5 | 1 |
| subtype4 | 3 | 0 |
| subtype5 | 7 | 1 |
Figure S249. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'

P value = 0.106 (Kruskal-Wallis (anova)), Q value = 1
Table S256. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 219 | 3.6 (2.5) |
| subtype1 | 42 | 2.8 (1.9) |
| subtype2 | 69 | 4.1 (3.1) |
| subtype3 | 57 | 3.7 (2.0) |
| subtype4 | 15 | 3.7 (2.5) |
| subtype5 | 36 | 3.2 (2.4) |
Figure S250. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'

P value = 0.477 (Kruskal-Wallis (anova)), Q value = 1
Table S257. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 109 | 0.1 (0.4) |
| subtype1 | 19 | 0.1 (0.2) |
| subtype2 | 26 | 0.0 (0.2) |
| subtype3 | 38 | 0.2 (0.5) |
| subtype4 | 9 | 0.0 (0.0) |
| subtype5 | 17 | 0.0 (0.0) |
Figure S251. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'

P value = 0.307 (Kruskal-Wallis (anova)), Q value = 1
Table S258. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 135 | 0.5 (0.8) |
| subtype1 | 20 | 0.6 (0.6) |
| subtype2 | 35 | 0.5 (1.1) |
| subtype3 | 45 | 0.4 (0.7) |
| subtype4 | 12 | 0.3 (0.7) |
| subtype5 | 23 | 0.5 (0.7) |
Figure S252. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'
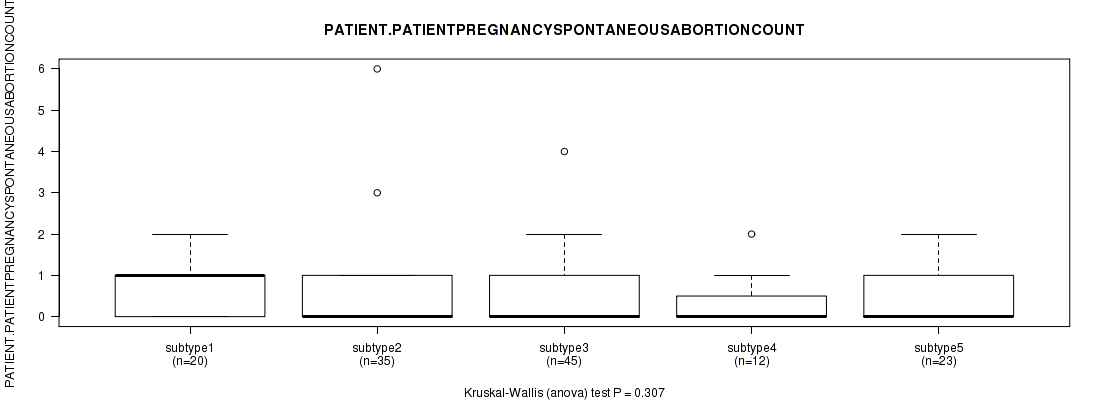
P value = 0.026 (Kruskal-Wallis (anova)), Q value = 1
Table S259. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 220 | 2.6 (1.9) |
| subtype1 | 42 | 2.0 (1.6) |
| subtype2 | 66 | 3.0 (2.2) |
| subtype3 | 60 | 2.7 (1.6) |
| subtype4 | 16 | 3.1 (2.1) |
| subtype5 | 36 | 2.4 (1.8) |
Figure S253. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'

P value = 0.915 (Kruskal-Wallis (anova)), Q value = 1
Table S260. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 119 | 0.9 (1.8) |
| subtype1 | 21 | 0.6 (0.7) |
| subtype2 | 30 | 1.6 (3.1) |
| subtype3 | 40 | 0.7 (1.1) |
| subtype4 | 9 | 0.7 (1.4) |
| subtype5 | 19 | 0.6 (0.9) |
Figure S254. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'
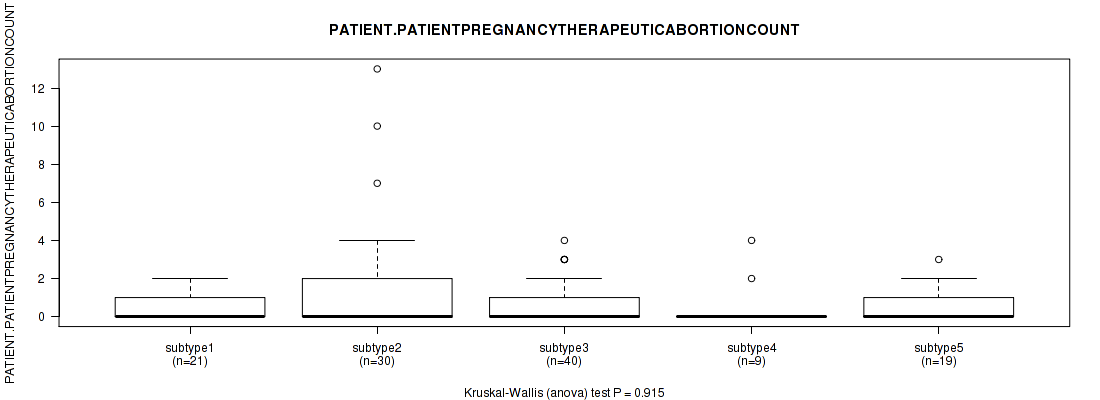
P value = 0.794 (Kruskal-Wallis (anova)), Q value = 1
Table S261. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 111 | 0.1 (0.3) |
| subtype1 | 18 | 0.1 (0.3) |
| subtype2 | 29 | 0.1 (0.4) |
| subtype3 | 38 | 0.1 (0.4) |
| subtype4 | 9 | 0.1 (0.3) |
| subtype5 | 17 | 0.1 (0.2) |
Figure S255. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'

P value = 0.899 (Fisher's exact test), Q value = 1
Table S262. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'
| nPatients | MACROSCOPIC PARAMETRIAL INVOLVEMENT | MICROSCOPIC PARAMETRIAL INVOLVEMENT | OTHER LOCATION, SPECIFY | POSITIVE BLADDER MARGIN | POSITIVE VAGINAL MARGIN |
|---|---|---|---|---|---|
| ALL | 2 | 7 | 33 | 1 | 9 |
| subtype1 | 1 | 0 | 5 | 0 | 2 |
| subtype2 | 0 | 2 | 7 | 1 | 3 |
| subtype3 | 1 | 3 | 12 | 0 | 3 |
| subtype4 | 0 | 0 | 2 | 0 | 0 |
| subtype5 | 0 | 2 | 7 | 0 | 1 |
Figure S256. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'

P value = 0.00307 (Fisher's exact test), Q value = 1
Table S263. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #32: 'MENOPAUSE_STATUS'
| nPatients | INDETERMINATE (NEITHER PRE OR POSTMENOPAUSAL) | PERI (6-12 MONTHS SINCE LAST MENSTRUAL PERIOD) | POST (PRIOR BILATERAL OVARIECTOMY OR >12 MO SINCE LMP WITH NO PRIOR HYSTERECTOMY) | PRE (<6 MONTHS SINCE LMP AND NO PRIOR BILATERAL OVARIECTOMY AND NOT ON ESTROGEN REPLACEMENT) |
|---|---|---|---|---|
| ALL | 2 | 16 | 70 | 109 |
| subtype1 | 0 | 2 | 13 | 25 |
| subtype2 | 1 | 7 | 23 | 31 |
| subtype3 | 1 | 4 | 28 | 21 |
| subtype4 | 0 | 2 | 4 | 7 |
| subtype5 | 0 | 1 | 2 | 25 |
Figure S257. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #32: 'MENOPAUSE_STATUS'

P value = 0.445 (Fisher's exact test), Q value = 1
Table S264. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 69 | 76 |
| subtype1 | 20 | 15 |
| subtype2 | 17 | 22 |
| subtype3 | 18 | 28 |
| subtype4 | 5 | 3 |
| subtype5 | 9 | 8 |
Figure S258. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'
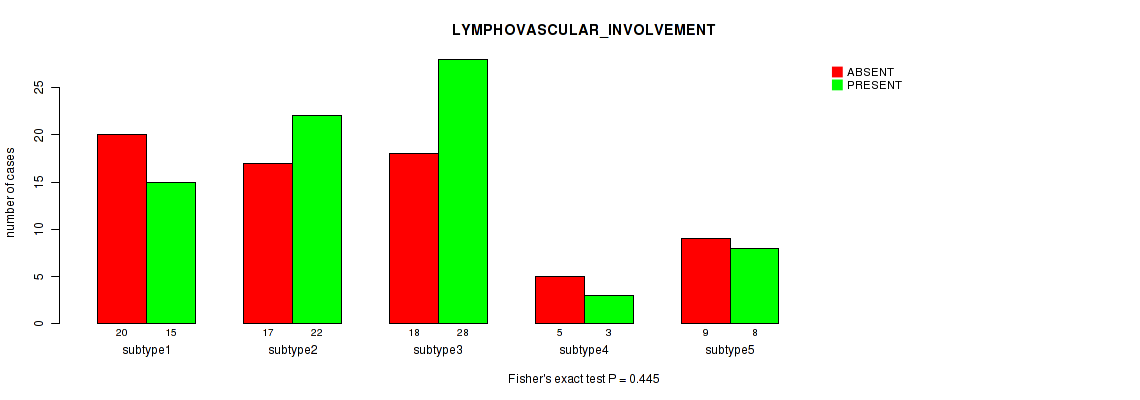
P value = 0.228 (Kruskal-Wallis (anova)), Q value = 1
Table S265. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 144 | 1.1 (2.5) |
| subtype1 | 35 | 0.7 (2.0) |
| subtype2 | 37 | 1.8 (3.2) |
| subtype3 | 45 | 1.0 (1.8) |
| subtype4 | 8 | 0.4 (0.7) |
| subtype5 | 19 | 0.9 (3.2) |
Figure S259. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'

P value = 0.533 (Kruskal-Wallis (anova)), Q value = 1
Table S266. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 166 | 21.7 (12.4) |
| subtype1 | 38 | 21.4 (10.6) |
| subtype2 | 45 | 20.7 (12.3) |
| subtype3 | 51 | 23.8 (14.5) |
| subtype4 | 10 | 15.9 (8.3) |
| subtype5 | 22 | 22.4 (11.7) |
Figure S260. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'

P value = 0.0905 (Fisher's exact test), Q value = 1
Table S267. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'
| nPatients | KERATINIZING SQUAMOUS CELL CARCINOMA | NON-KERATINIZING SQUAMOUS CELL CARCINOMA |
|---|---|---|
| ALL | 49 | 97 |
| subtype1 | 0 | 10 |
| subtype2 | 18 | 34 |
| subtype3 | 19 | 27 |
| subtype4 | 3 | 11 |
| subtype5 | 9 | 15 |
Figure S261. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'

P value = 0.127 (Kruskal-Wallis (anova)), Q value = 1
Table S268. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 248 | 2007.7 (5.0) |
| subtype1 | 47 | 2008.8 (4.2) |
| subtype2 | 79 | 2008.3 (4.6) |
| subtype3 | 63 | 2006.7 (5.2) |
| subtype4 | 20 | 2007.2 (6.5) |
| subtype5 | 39 | 2007.1 (5.4) |
Figure S262. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'
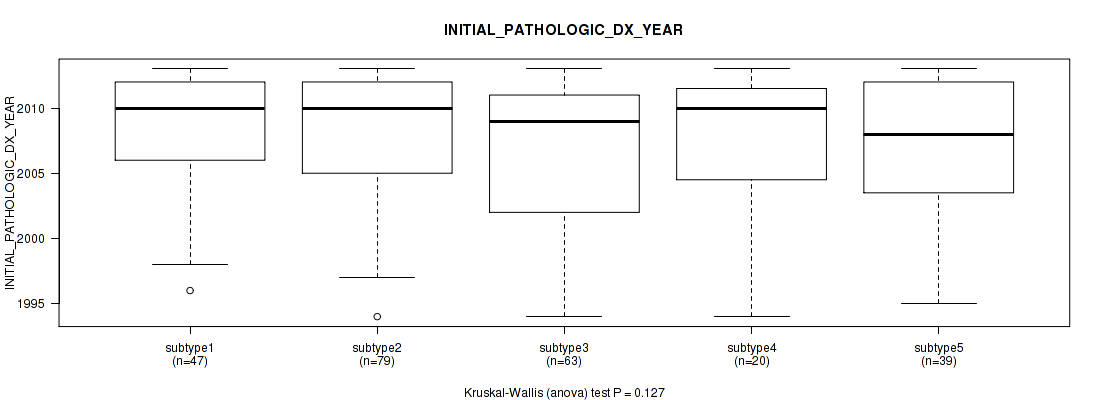
P value = 0.416 (Fisher's exact test), Q value = 1
Table S269. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'
| nPatients | CURRENT USER | FORMER USER | NEVER USED |
|---|---|---|---|
| ALL | 10 | 49 | 62 |
| subtype1 | 4 | 13 | 10 |
| subtype2 | 2 | 15 | 22 |
| subtype3 | 1 | 13 | 17 |
| subtype4 | 0 | 4 | 4 |
| subtype5 | 3 | 4 | 9 |
Figure S263. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'

P value = 0.685 (Kruskal-Wallis (anova)), Q value = 1
Table S270. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 218 | 161.6 (7.1) |
| subtype1 | 41 | 162.5 (6.8) |
| subtype2 | 72 | 161.0 (7.0) |
| subtype3 | 56 | 161.2 (7.5) |
| subtype4 | 17 | 160.7 (8.0) |
| subtype5 | 32 | 162.8 (6.2) |
Figure S264. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'

P value = 0.318 (Fisher's exact test), Q value = 1
Table S271. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 96 | 17 |
| subtype1 | 27 | 3 |
| subtype2 | 21 | 5 |
| subtype3 | 31 | 7 |
| subtype4 | 5 | 2 |
| subtype5 | 12 | 0 |
Figure S265. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'

P value = 0.442 (Fisher's exact test), Q value = 1
Table S272. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'
| nPatients | CARBOPLATIN | CISPLATIN | OTHER |
|---|---|---|---|
| ALL | 3 | 18 | 1 |
| subtype1 | 0 | 1 | 0 |
| subtype2 | 1 | 2 | 0 |
| subtype3 | 2 | 7 | 0 |
| subtype4 | 0 | 1 | 0 |
| subtype5 | 0 | 7 | 1 |
Figure S266. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'

P value = 0.207 (Kruskal-Wallis (anova)), Q value = 1
Table S273. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 13 | 12.4 (5.5) |
| subtype1 | 1 | 7.1 (NA) |
| subtype2 | 3 | 10.5 (4.3) |
| subtype3 | 6 | 11.3 (3.7) |
| subtype5 | 3 | 18.4 (7.3) |
Figure S267. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'

P value = 0.00142 (Fisher's exact test), Q value = 0.49
Table S274. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'
| nPatients | T1A | T1A1 | T1B | T1B1 | T1B2 | T2 | T2A | T2A1 | T2A2 | T2B | T3 | T3B | T4 | TIS | TX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 1 | 1 | 32 | 68 | 25 | 4 | 8 | 7 | 10 | 23 | 1 | 8 | 4 | 1 | 15 |
| subtype1 | 0 | 0 | 3 | 23 | 5 | 1 | 2 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 2 |
| subtype2 | 1 | 0 | 6 | 13 | 8 | 2 | 2 | 0 | 6 | 13 | 0 | 4 | 2 | 0 | 7 |
| subtype3 | 0 | 1 | 12 | 24 | 7 | 0 | 3 | 1 | 0 | 4 | 0 | 0 | 1 | 0 | 3 |
| subtype4 | 0 | 0 | 3 | 3 | 2 | 0 | 0 | 1 | 2 | 2 | 1 | 1 | 1 | 0 | 1 |
| subtype5 | 0 | 0 | 8 | 5 | 3 | 1 | 1 | 2 | 1 | 3 | 0 | 3 | 0 | 1 | 2 |
Figure S268. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'

P value = 6.47e-06 (Kruskal-Wallis (anova)), Q value = 0.0023
Table S275. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 250 | 47.7 (13.5) |
| subtype1 | 48 | 44.4 (11.6) |
| subtype2 | 79 | 50.0 (13.7) |
| subtype3 | 64 | 51.2 (14.2) |
| subtype4 | 20 | 52.3 (9.2) |
| subtype5 | 39 | 39.1 (11.5) |
Figure S269. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'

P value = 0.00057 (Fisher's exact test), Q value = 0.2
Table S276. Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'
| nPatients | STAGE I | STAGE IA | STAGE IA1 | STAGE IA2 | STAGE IB | STAGE IB1 | STAGE IB2 | STAGE II | STAGE IIA | STAGE IIA1 | STAGE IIA2 | STAGE IIB | STAGE III | STAGE IIIB | STAGE IVA | STAGE IVB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 4 | 2 | 1 | 1 | 35 | 70 | 34 | 4 | 7 | 5 | 7 | 27 | 1 | 34 | 5 | 7 |
| subtype1 | 1 | 0 | 0 | 0 | 2 | 25 | 8 | 1 | 1 | 1 | 0 | 2 | 0 | 5 | 0 | 2 |
| subtype2 | 2 | 1 | 0 | 0 | 7 | 15 | 10 | 2 | 1 | 0 | 3 | 13 | 0 | 15 | 3 | 2 |
| subtype3 | 0 | 1 | 1 | 1 | 13 | 21 | 8 | 1 | 2 | 1 | 1 | 2 | 0 | 8 | 2 | 1 |
| subtype4 | 1 | 0 | 0 | 0 | 6 | 1 | 1 | 0 | 0 | 1 | 2 | 2 | 1 | 3 | 0 | 2 |
| subtype5 | 0 | 0 | 0 | 0 | 7 | 8 | 7 | 0 | 3 | 2 | 1 | 8 | 0 | 3 | 0 | 0 |
Figure S270. Get High-res Image Clustering Approach #6: 'MIRSEQ CHIERARCHICAL' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'

Table S277. Description of clustering approach #7: 'MIRseq Mature CNMF subtypes'
| Cluster Labels | 1 | 2 | 3 |
|---|---|---|---|
| Number of samples | 99 | 68 | 72 |
P value = 0.765 (logrank test), Q value = 1
Table S278. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 235 | 52 | 0.0 - 182.9 (16.6) |
| subtype1 | 98 | 16 | 0.0 - 182.9 (15.6) |
| subtype2 | 65 | 13 | 0.1 - 147.4 (14.1) |
| subtype3 | 72 | 23 | 0.1 - 177.0 (28.6) |
Figure S271. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #1: 'Time to Death'

P value = 0.415 (Kruskal-Wallis (anova)), Q value = 1
Table S279. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 236 | 47.7 (13.6) |
| subtype1 | 97 | 47.6 (12.8) |
| subtype2 | 67 | 49.3 (14.0) |
| subtype3 | 72 | 46.3 (14.3) |
Figure S272. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #2: 'AGE'

P value = 0.892 (Fisher's exact test), Q value = 1
Table S280. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'
| nPatients | T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 122 | 49 | 7 | 4 |
| subtype1 | 46 | 23 | 4 | 2 |
| subtype2 | 34 | 13 | 1 | 1 |
| subtype3 | 42 | 13 | 2 | 1 |
Figure S273. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'
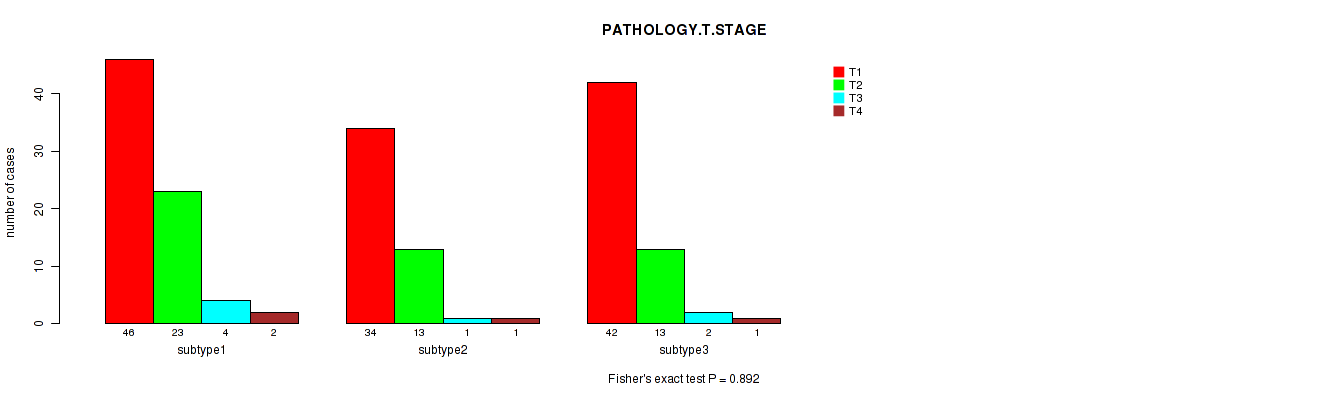
P value = 0.339 (Fisher's exact test), Q value = 1
Table S281. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'
| nPatients | 0 | 1 |
|---|---|---|
| ALL | 112 | 48 |
| subtype1 | 43 | 15 |
| subtype2 | 36 | 13 |
| subtype3 | 33 | 20 |
Figure S274. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'

P value = 0.124 (Fisher's exact test), Q value = 1
Table S282. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 96 | 7 | 90 |
| subtype1 | 35 | 6 | 41 |
| subtype2 | 27 | 1 | 24 |
| subtype3 | 34 | 0 | 25 |
Figure S275. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'

P value = 8e-05 (Fisher's exact test), Q value = 0.028
Table S283. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | ADENOSQUAMOUS | CERVICAL SQUAMOUS CELL CARCINOMA | ENDOCERVICAL ADENOCARCINOMA OF THE USUAL TYPE | ENDOCERVICAL TYPE OF ADENOCARCINOMA | ENDOMETRIOID ADENOCARCINOMA OF ENDOCERVIX | MUCINOUS ADENOCARCINOMA OF ENDOCERVICAL TYPE |
|---|---|---|---|---|---|---|
| ALL | 5 | 199 | 5 | 21 | 3 | 6 |
| subtype1 | 4 | 70 | 3 | 14 | 3 | 5 |
| subtype2 | 1 | 57 | 2 | 7 | 0 | 1 |
| subtype3 | 0 | 72 | 0 | 0 | 0 | 0 |
Figure S276. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
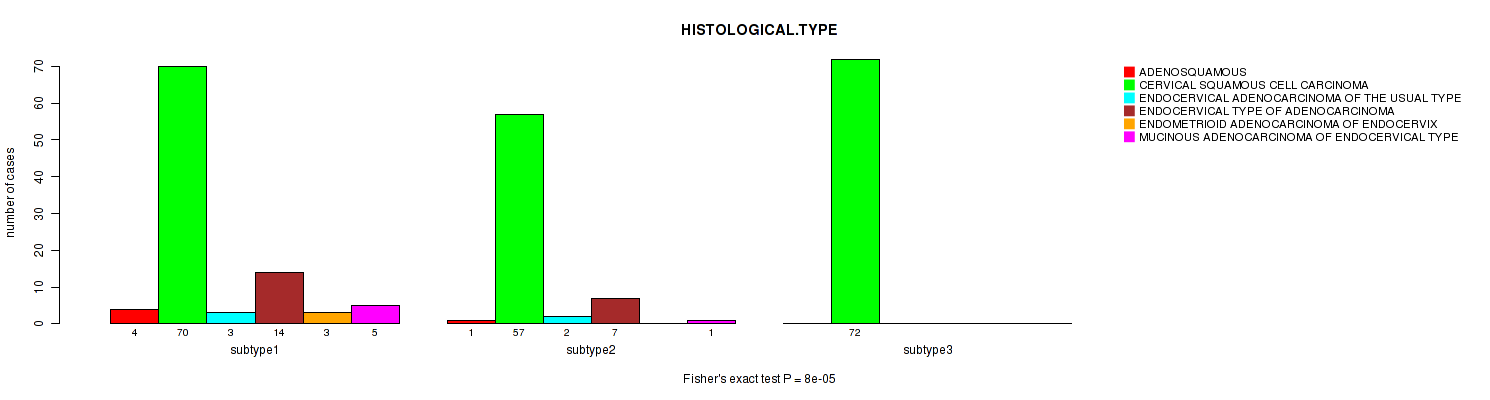
P value = 1e-05 (Fisher's exact test), Q value = 0.0036
Table S284. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 33 | 206 |
| subtype1 | 3 | 96 |
| subtype2 | 6 | 62 |
| subtype3 | 24 | 48 |
Figure S277. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'

P value = 0.408 (Kruskal-Wallis (anova)), Q value = 1
Table S285. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 71 | 18.7 (13.6) |
| subtype1 | 22 | 16.4 (15.2) |
| subtype2 | 23 | 19.6 (12.2) |
| subtype3 | 26 | 19.7 (13.8) |
Figure S278. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'

P value = 0.446 (Kruskal-Wallis (anova)), Q value = 1
Table S286. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 140 | 1.0 (2.4) |
| subtype1 | 46 | 0.7 (2.2) |
| subtype2 | 43 | 1.2 (2.3) |
| subtype3 | 51 | 1.2 (2.7) |
Figure S279. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'

P value = 0.099 (Fisher's exact test), Q value = 1
Table S287. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER | WHITE |
|---|---|---|---|---|---|
| ALL | 7 | 18 | 23 | 1 | 172 |
| subtype1 | 5 | 9 | 13 | 0 | 64 |
| subtype2 | 1 | 6 | 2 | 0 | 54 |
| subtype3 | 1 | 3 | 8 | 1 | 54 |
Figure S280. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #10: 'RACE'

P value = 0.0475 (Fisher's exact test), Q value = 1
Table S288. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #11: 'ETHNICITY'
| nPatients | HISPANIC OR LATINO | NOT HISPANIC OR LATINO |
|---|---|---|
| ALL | 17 | 148 |
| subtype1 | 12 | 58 |
| subtype2 | 2 | 46 |
| subtype3 | 3 | 44 |
Figure S281. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #11: 'ETHNICITY'

P value = 0.889 (Kruskal-Wallis (anova)), Q value = 1
Table S289. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 217 | 74.6 (22.3) |
| subtype1 | 91 | 73.8 (19.8) |
| subtype2 | 62 | 76.0 (28.6) |
| subtype3 | 64 | 74.4 (18.8) |
Figure S282. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'

P value = 0.566 (Fisher's exact test), Q value = 1
Table S290. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #13: 'TUMOR_STATUS'
| nPatients | TUMOR FREE | WITH TUMOR |
|---|---|---|
| ALL | 72 | 27 |
| subtype1 | 25 | 9 |
| subtype2 | 17 | 4 |
| subtype3 | 30 | 14 |
Figure S283. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #13: 'TUMOR_STATUS'
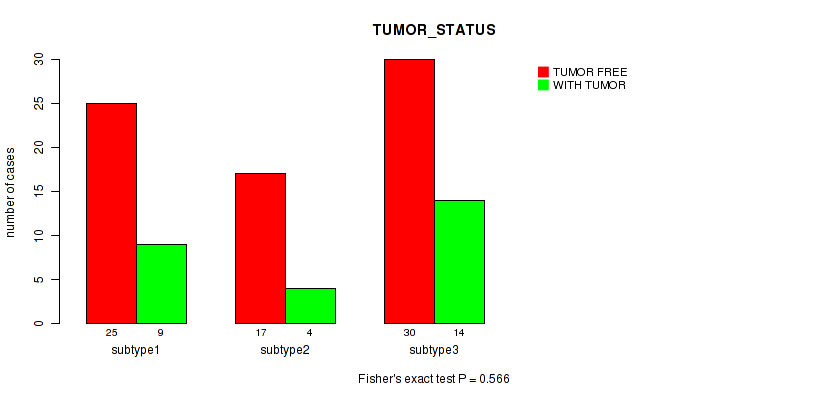
P value = 0.0764 (Fisher's exact test), Q value = 1
Table S291. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'
| nPatients | BRAZIL | CANADA | NIGERIA | RUSSIA | UKRAINE | UNITED STATES | VIETNAM |
|---|---|---|---|---|---|---|---|
| ALL | 24 | 5 | 1 | 11 | 6 | 179 | 13 |
| subtype1 | 15 | 3 | 1 | 2 | 4 | 68 | 6 |
| subtype2 | 3 | 1 | 0 | 5 | 2 | 51 | 6 |
| subtype3 | 6 | 1 | 0 | 4 | 0 | 60 | 1 |
Figure S284. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'

P value = 0.0342 (Fisher's exact test), Q value = 1
Table S292. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'
| nPatients | G1 | G2 | G3 | G4 | GX |
|---|---|---|---|---|---|
| ALL | 15 | 106 | 100 | 1 | 14 |
| subtype1 | 6 | 36 | 49 | 0 | 8 |
| subtype2 | 8 | 35 | 22 | 0 | 2 |
| subtype3 | 1 | 35 | 29 | 1 | 4 |
Figure S285. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'

P value = 0.317 (Kruskal-Wallis (anova)), Q value = 1
Table S293. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 32 | 2000.8 (11.4) |
| subtype1 | 11 | 2005.9 (5.9) |
| subtype2 | 9 | 1996.2 (16.2) |
| subtype3 | 12 | 1999.6 (10.1) |
Figure S286. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'

P value = 0.408 (Kruskal-Wallis (anova)), Q value = 1
Table S294. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 71 | 18.7 (13.6) |
| subtype1 | 22 | 16.4 (15.2) |
| subtype2 | 23 | 19.6 (12.2) |
| subtype3 | 26 | 19.7 (13.8) |
Figure S287. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'
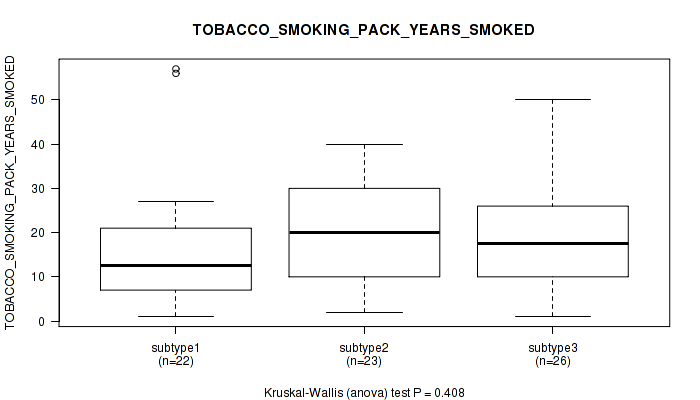
P value = 0.124 (Fisher's exact test), Q value = 1
Table S295. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'
| nPatients | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT REFORMED SMOKER, DURATION NOT SPECIFIED | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|
| ALL | 32 | 7 | 3 | 51 | 112 |
| subtype1 | 12 | 1 | 1 | 14 | 53 |
| subtype2 | 7 | 3 | 2 | 18 | 29 |
| subtype3 | 13 | 3 | 0 | 19 | 30 |
Figure S288. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'

P value = 0.936 (Kruskal-Wallis (anova)), Q value = 1
Table S296. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 63 | 21.1 (7.7) |
| subtype1 | 22 | 21.6 (7.7) |
| subtype2 | 19 | 20.7 (7.2) |
| subtype3 | 22 | 21.0 (8.3) |
Figure S289. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'

P value = 0.0236 (Kruskal-Wallis (anova)), Q value = 1
Table S297. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 107 | 3911.4 (1611.9) |
| subtype1 | 44 | 3495.4 (1847.3) |
| subtype2 | 25 | 3885.5 (1601.3) |
| subtype3 | 38 | 4410.1 (1162.8) |
Figure S290. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'

P value = 2e-05 (Fisher's exact test), Q value = 0.007
Table S298. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'
| nPatients | COMBINATION | EXTERNAL | EXTERNAL BEAM | IMPLANTS | INTERNAL |
|---|---|---|---|---|---|
| ALL | 19 | 74 | 15 | 1 | 12 |
| subtype1 | 2 | 39 | 1 | 0 | 6 |
| subtype2 | 2 | 18 | 5 | 0 | 4 |
| subtype3 | 15 | 17 | 9 | 1 | 2 |
Figure S291. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'

P value = 1 (Fisher's exact test), Q value = 1
Table S299. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'
| nPatients | COMPLETED AS PLANNED | TREATMENT NOT COMPLETED |
|---|---|---|
| ALL | 27 | 3 |
| subtype1 | 3 | 0 |
| subtype2 | 5 | 0 |
| subtype3 | 19 | 3 |
Figure S292. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'
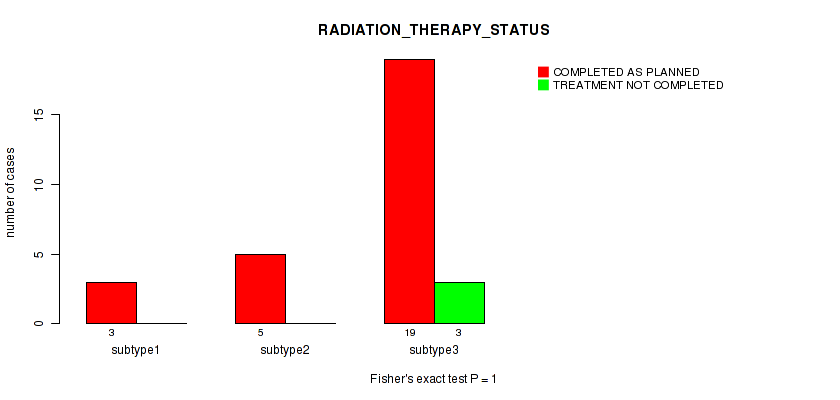
P value = 0.797 (Fisher's exact test), Q value = 1
Table S300. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'
| nPatients | DISTANT RECURRENCE | LOCAL RECURRENCE | PRIMARY TUMOR FIELD | REGIONAL SITE |
|---|---|---|---|---|
| ALL | 2 | 2 | 29 | 16 |
| subtype1 | 1 | 0 | 12 | 8 |
| subtype2 | 1 | 1 | 8 | 5 |
| subtype3 | 0 | 1 | 9 | 3 |
Figure S293. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'

P value = 1 (Fisher's exact test), Q value = 1
Table S301. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'
| nPatients | CGY | GY |
|---|---|---|
| ALL | 35 | 3 |
| subtype1 | 17 | 1 |
| subtype2 | 9 | 1 |
| subtype3 | 9 | 1 |
Figure S294. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'

P value = 0.436 (Kruskal-Wallis (anova)), Q value = 1
Table S302. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 210 | 3.6 (2.5) |
| subtype1 | 83 | 3.4 (2.5) |
| subtype2 | 62 | 3.7 (2.3) |
| subtype3 | 65 | 3.6 (2.6) |
Figure S295. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'

P value = 0.218 (Kruskal-Wallis (anova)), Q value = 1
Table S303. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 104 | 0.1 (0.3) |
| subtype1 | 31 | 0.0 (0.2) |
| subtype2 | 27 | 0.0 (0.0) |
| subtype3 | 46 | 0.1 (0.5) |
Figure S296. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'

P value = 0.279 (Kruskal-Wallis (anova)), Q value = 1
Table S304. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 131 | 0.5 (0.8) |
| subtype1 | 44 | 0.7 (1.1) |
| subtype2 | 37 | 0.4 (0.5) |
| subtype3 | 50 | 0.4 (0.7) |
Figure S297. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'

P value = 0.341 (Kruskal-Wallis (anova)), Q value = 1
Table S305. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 211 | 2.7 (1.9) |
| subtype1 | 83 | 2.7 (2.1) |
| subtype2 | 62 | 2.8 (1.7) |
| subtype3 | 66 | 2.4 (1.8) |
Figure S298. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'

P value = 0.555 (Kruskal-Wallis (anova)), Q value = 1
Table S306. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 113 | 0.8 (1.8) |
| subtype1 | 34 | 0.5 (0.9) |
| subtype2 | 32 | 1.2 (2.5) |
| subtype3 | 47 | 0.8 (1.7) |
Figure S299. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'

P value = 0.849 (Kruskal-Wallis (anova)), Q value = 1
Table S307. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 107 | 0.1 (0.3) |
| subtype1 | 34 | 0.1 (0.3) |
| subtype2 | 27 | 0.1 (0.4) |
| subtype3 | 46 | 0.1 (0.3) |
Figure S300. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'

P value = 0.646 (Fisher's exact test), Q value = 1
Table S308. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'
| nPatients | MACROSCOPIC PARAMETRIAL INVOLVEMENT | MICROSCOPIC PARAMETRIAL INVOLVEMENT | OTHER LOCATION, SPECIFY | POSITIVE BLADDER MARGIN | POSITIVE VAGINAL MARGIN |
|---|---|---|---|---|---|
| ALL | 1 | 6 | 32 | 1 | 9 |
| subtype1 | 0 | 1 | 7 | 0 | 1 |
| subtype2 | 1 | 1 | 8 | 1 | 4 |
| subtype3 | 0 | 4 | 17 | 0 | 4 |
Figure S301. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'

P value = 0.414 (Fisher's exact test), Q value = 1
Table S309. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #32: 'MENOPAUSE_STATUS'
| nPatients | INDETERMINATE (NEITHER PRE OR POSTMENOPAUSAL) | PERI (6-12 MONTHS SINCE LAST MENSTRUAL PERIOD) | POST (PRIOR BILATERAL OVARIECTOMY OR >12 MO SINCE LMP WITH NO PRIOR HYSTERECTOMY) | PRE (<6 MONTHS SINCE LMP AND NO PRIOR BILATERAL OVARIECTOMY AND NOT ON ESTROGEN REPLACEMENT) |
|---|---|---|---|---|
| ALL | 2 | 16 | 65 | 104 |
| subtype1 | 0 | 9 | 23 | 44 |
| subtype2 | 0 | 4 | 21 | 27 |
| subtype3 | 2 | 3 | 21 | 33 |
Figure S302. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #32: 'MENOPAUSE_STATUS'

P value = 0.715 (Fisher's exact test), Q value = 1
Table S310. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 66 | 70 |
| subtype1 | 23 | 22 |
| subtype2 | 22 | 21 |
| subtype3 | 21 | 27 |
Figure S303. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'

P value = 0.446 (Kruskal-Wallis (anova)), Q value = 1
Table S311. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 140 | 1.0 (2.4) |
| subtype1 | 46 | 0.7 (2.2) |
| subtype2 | 43 | 1.2 (2.3) |
| subtype3 | 51 | 1.2 (2.7) |
Figure S304. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'

P value = 0.18 (Kruskal-Wallis (anova)), Q value = 1
Table S312. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 158 | 22.2 (12.5) |
| subtype1 | 55 | 20.8 (12.0) |
| subtype2 | 49 | 25.0 (13.0) |
| subtype3 | 54 | 21.0 (12.4) |
Figure S305. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'

P value = 0.0458 (Fisher's exact test), Q value = 1
Table S313. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'
| nPatients | KERATINIZING SQUAMOUS CELL CARCINOMA | NON-KERATINIZING SQUAMOUS CELL CARCINOMA |
|---|---|---|
| ALL | 46 | 92 |
| subtype1 | 8 | 35 |
| subtype2 | 17 | 25 |
| subtype3 | 21 | 32 |
Figure S306. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'

P value = 2.69e-05 (Kruskal-Wallis (anova)), Q value = 0.0093
Table S314. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 237 | 2007.7 (5.0) |
| subtype1 | 98 | 2009.0 (4.5) |
| subtype2 | 67 | 2007.9 (4.8) |
| subtype3 | 72 | 2005.7 (5.3) |
Figure S307. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'
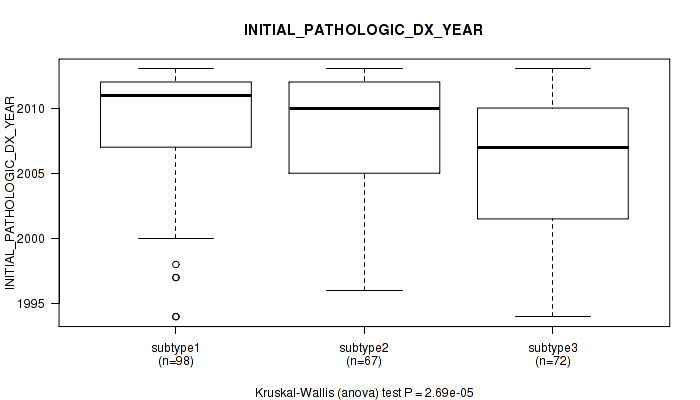
P value = 0.9 (Fisher's exact test), Q value = 1
Table S315. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'
| nPatients | CURRENT USER | FORMER USER | NEVER USED |
|---|---|---|---|
| ALL | 10 | 49 | 58 |
| subtype1 | 5 | 19 | 26 |
| subtype2 | 3 | 15 | 14 |
| subtype3 | 2 | 15 | 18 |
Figure S308. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'

P value = 0.533 (Kruskal-Wallis (anova)), Q value = 1
Table S316. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 208 | 161.4 (7.0) |
| subtype1 | 88 | 161.1 (6.3) |
| subtype2 | 58 | 162.0 (7.0) |
| subtype3 | 62 | 161.3 (8.0) |
Figure S309. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'

P value = 0.755 (Fisher's exact test), Q value = 1
Table S317. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 92 | 17 |
| subtype1 | 29 | 4 |
| subtype2 | 28 | 5 |
| subtype3 | 35 | 8 |
Figure S310. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'

P value = 0.206 (Fisher's exact test), Q value = 1
Table S318. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'
| nPatients | CARBOPLATIN | CISPLATIN | OTHER |
|---|---|---|---|
| ALL | 3 | 17 | 1 |
| subtype1 | 0 | 2 | 0 |
| subtype2 | 0 | 2 | 1 |
| subtype3 | 3 | 13 | 0 |
Figure S311. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'

P value = 0.348 (Kruskal-Wallis (anova)), Q value = 1
Table S319. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 12 | 12.0 (5.6) |
| subtype1 | 3 | 14.8 (10.0) |
| subtype2 | 5 | 9.4 (3.4) |
| subtype3 | 4 | 13.2 (3.1) |
Figure S312. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'

P value = 0.235 (Fisher's exact test), Q value = 1
Table S320. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'
| nPatients | T1A | T1B | T1B1 | T1B2 | T2 | T2A | T2A1 | T2A2 | T2B | T3B | T4 | TIS | TX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 1 | 31 | 66 | 24 | 4 | 8 | 7 | 9 | 21 | 7 | 4 | 1 | 15 |
| subtype1 | 1 | 9 | 26 | 10 | 2 | 4 | 2 | 5 | 10 | 4 | 2 | 1 | 8 |
| subtype2 | 0 | 4 | 21 | 9 | 0 | 4 | 3 | 2 | 4 | 1 | 1 | 0 | 5 |
| subtype3 | 0 | 18 | 19 | 5 | 2 | 0 | 2 | 2 | 7 | 2 | 1 | 0 | 2 |
Figure S313. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'

P value = 0.37 (Kruskal-Wallis (anova)), Q value = 1
Table S321. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 239 | 47.7 (13.5) |
| subtype1 | 99 | 47.6 (12.7) |
| subtype2 | 68 | 49.5 (13.9) |
| subtype3 | 72 | 46.3 (14.3) |
Figure S314. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'

P value = 0.00658 (Fisher's exact test), Q value = 1
Table S322. Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'
| nPatients | STAGE I | STAGE IA | STAGE IA2 | STAGE IB | STAGE IB1 | STAGE IB2 | STAGE II | STAGE IIA | STAGE IIA1 | STAGE IIA2 | STAGE IIB | STAGE IIIB | STAGE IVA | STAGE IVB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 4 | 2 | 1 | 34 | 68 | 32 | 4 | 6 | 5 | 7 | 26 | 33 | 5 | 6 |
| subtype1 | 3 | 1 | 0 | 8 | 32 | 14 | 1 | 2 | 0 | 4 | 13 | 13 | 1 | 4 |
| subtype2 | 0 | 1 | 1 | 4 | 22 | 11 | 2 | 3 | 3 | 2 | 6 | 6 | 3 | 1 |
| subtype3 | 1 | 0 | 0 | 22 | 14 | 7 | 1 | 1 | 2 | 1 | 7 | 14 | 1 | 1 |
Figure S315. Get High-res Image Clustering Approach #7: 'MIRseq Mature CNMF subtypes' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'

Table S323. Description of clustering approach #8: 'MIRseq Mature cHierClus subtypes'
| Cluster Labels | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Number of samples | 50 | 42 | 31 | 116 |
P value = 0.981 (logrank test), Q value = 1
Table S324. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 235 | 52 | 0.0 - 182.9 (16.6) |
| subtype1 | 49 | 9 | 0.1 - 137.2 (15.0) |
| subtype2 | 40 | 7 | 0.1 - 147.4 (14.5) |
| subtype3 | 31 | 12 | 1.2 - 177.0 (35.6) |
| subtype4 | 115 | 24 | 0.0 - 182.9 (16.1) |
Figure S316. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #1: 'Time to Death'

P value = 0.546 (Kruskal-Wallis (anova)), Q value = 1
Table S325. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 236 | 47.7 (13.6) |
| subtype1 | 48 | 45.8 (11.8) |
| subtype2 | 42 | 49.9 (14.3) |
| subtype3 | 31 | 48.9 (14.4) |
| subtype4 | 115 | 47.3 (13.8) |
Figure S317. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #2: 'AGE'

P value = 0.825 (Fisher's exact test), Q value = 1
Table S326. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'
| nPatients | T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 122 | 49 | 7 | 4 |
| subtype1 | 28 | 13 | 0 | 1 |
| subtype2 | 22 | 7 | 1 | 1 |
| subtype3 | 20 | 5 | 1 | 0 |
| subtype4 | 52 | 24 | 5 | 2 |
Figure S318. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #3: 'PATHOLOGY.T.STAGE'

P value = 0.102 (Fisher's exact test), Q value = 1
Table S327. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'
| nPatients | 0 | 1 |
|---|---|---|
| ALL | 112 | 48 |
| subtype1 | 30 | 8 |
| subtype2 | 21 | 8 |
| subtype3 | 13 | 13 |
| subtype4 | 48 | 19 |
Figure S319. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #4: 'PATHOLOGY.N.STAGE'

P value = 0.22 (Fisher's exact test), Q value = 1
Table S328. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 96 | 7 | 90 |
| subtype1 | 20 | 4 | 20 |
| subtype2 | 18 | 1 | 13 |
| subtype3 | 17 | 0 | 9 |
| subtype4 | 41 | 2 | 48 |
Figure S320. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #5: 'PATHOLOGY.M.STAGE'
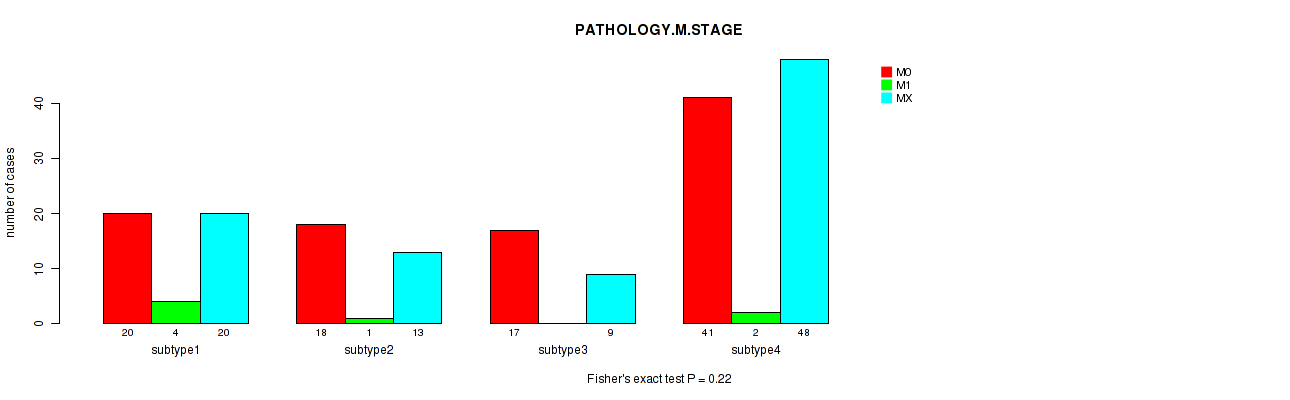
P value = 1e-05 (Fisher's exact test), Q value = 0.0036
Table S329. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | ADENOSQUAMOUS | CERVICAL SQUAMOUS CELL CARCINOMA | ENDOCERVICAL ADENOCARCINOMA OF THE USUAL TYPE | ENDOCERVICAL TYPE OF ADENOCARCINOMA | ENDOMETRIOID ADENOCARCINOMA OF ENDOCERVIX | MUCINOUS ADENOCARCINOMA OF ENDOCERVICAL TYPE |
|---|---|---|---|---|---|---|
| ALL | 5 | 199 | 5 | 21 | 3 | 6 |
| subtype1 | 3 | 14 | 4 | 20 | 3 | 6 |
| subtype2 | 1 | 41 | 0 | 0 | 0 | 0 |
| subtype3 | 0 | 30 | 0 | 1 | 0 | 0 |
| subtype4 | 1 | 114 | 1 | 0 | 0 | 0 |
Figure S321. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1e-05 (Fisher's exact test), Q value = 0.0036
Table S330. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 33 | 206 |
| subtype1 | 1 | 49 |
| subtype2 | 3 | 39 |
| subtype3 | 19 | 12 |
| subtype4 | 10 | 106 |
Figure S322. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #7: 'RADIATIONS.RADIATION.REGIMENINDICATION'
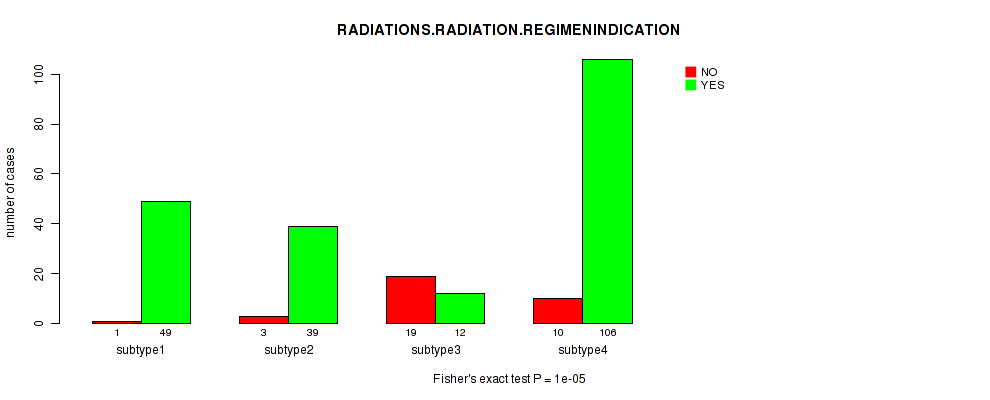
P value = 0.238 (Kruskal-Wallis (anova)), Q value = 1
Table S331. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 71 | 18.7 (13.6) |
| subtype1 | 13 | 14.2 (10.6) |
| subtype2 | 12 | 23.2 (11.6) |
| subtype3 | 10 | 19.5 (9.9) |
| subtype4 | 36 | 18.5 (15.9) |
Figure S323. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #8: 'NUMBERPACKYEARSSMOKED'

P value = 0.118 (Kruskal-Wallis (anova)), Q value = 1
Table S332. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 140 | 1.0 (2.4) |
| subtype1 | 34 | 0.6 (2.0) |
| subtype2 | 24 | 1.1 (1.8) |
| subtype3 | 25 | 2.4 (4.5) |
| subtype4 | 57 | 0.6 (1.1) |
Figure S324. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #9: 'NUMBER.OF.LYMPH.NODES'

P value = 0.499 (Fisher's exact test), Q value = 1
Table S333. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDER | WHITE |
|---|---|---|---|---|---|
| ALL | 7 | 18 | 23 | 1 | 172 |
| subtype1 | 1 | 5 | 3 | 0 | 36 |
| subtype2 | 1 | 5 | 1 | 0 | 31 |
| subtype3 | 0 | 2 | 3 | 0 | 26 |
| subtype4 | 5 | 6 | 16 | 1 | 79 |
Figure S325. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #10: 'RACE'

P value = 0.294 (Fisher's exact test), Q value = 1
Table S334. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #11: 'ETHNICITY'
| nPatients | HISPANIC OR LATINO | NOT HISPANIC OR LATINO |
|---|---|---|
| ALL | 17 | 148 |
| subtype1 | 6 | 31 |
| subtype2 | 1 | 29 |
| subtype3 | 1 | 22 |
| subtype4 | 9 | 66 |
Figure S326. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #11: 'ETHNICITY'

P value = 0.679 (Kruskal-Wallis (anova)), Q value = 1
Table S335. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 217 | 74.6 (22.3) |
| subtype1 | 45 | 75.4 (14.9) |
| subtype2 | 39 | 78.0 (34.3) |
| subtype3 | 28 | 71.8 (17.7) |
| subtype4 | 105 | 73.8 (20.6) |
Figure S327. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #12: 'WEIGHT_KG_AT_DIAGNOSIS'

P value = 0.399 (Fisher's exact test), Q value = 1
Table S336. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #13: 'TUMOR_STATUS'
| nPatients | TUMOR FREE | WITH TUMOR |
|---|---|---|
| ALL | 72 | 27 |
| subtype1 | 17 | 4 |
| subtype2 | 11 | 2 |
| subtype3 | 16 | 10 |
| subtype4 | 28 | 11 |
Figure S328. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #13: 'TUMOR_STATUS'
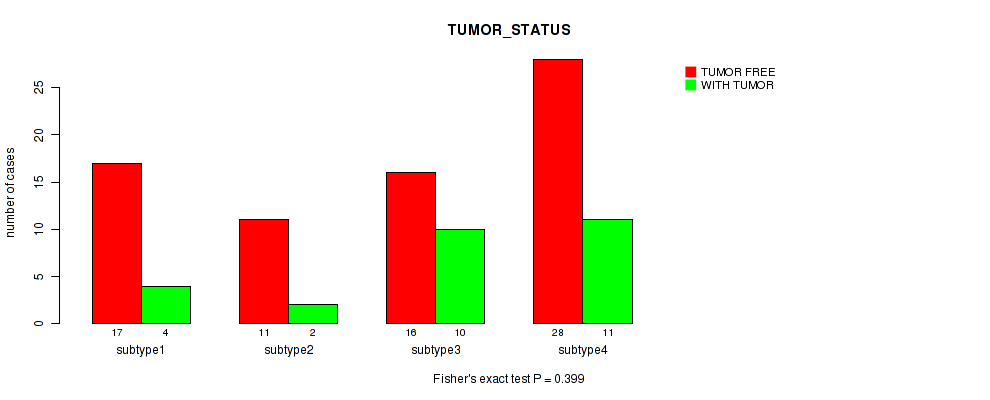
P value = 0.0207 (Fisher's exact test), Q value = 1
Table S337. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'
| nPatients | BRAZIL | CANADA | NIGERIA | RUSSIA | UKRAINE | UNITED STATES | VIETNAM |
|---|---|---|---|---|---|---|---|
| ALL | 24 | 5 | 1 | 11 | 6 | 179 | 13 |
| subtype1 | 4 | 3 | 1 | 2 | 1 | 36 | 3 |
| subtype2 | 3 | 1 | 0 | 5 | 2 | 26 | 5 |
| subtype3 | 0 | 0 | 0 | 1 | 0 | 30 | 0 |
| subtype4 | 17 | 1 | 0 | 3 | 3 | 87 | 5 |
Figure S329. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #14: 'TUMOR_SAMPLE_PROCUREMENT_COUNTRY'

P value = 0.0459 (Fisher's exact test), Q value = 1
Table S338. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'
| nPatients | G1 | G2 | G3 | G4 | GX |
|---|---|---|---|---|---|
| ALL | 15 | 106 | 100 | 1 | 14 |
| subtype1 | 6 | 22 | 18 | 0 | 4 |
| subtype2 | 5 | 23 | 11 | 0 | 2 |
| subtype3 | 0 | 17 | 14 | 0 | 0 |
| subtype4 | 4 | 44 | 57 | 1 | 8 |
Figure S330. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #15: 'NEOPLASMHISTOLOGICGRADE'

P value = 0.0617 (Kruskal-Wallis (anova)), Q value = 1
Table S339. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 32 | 2000.8 (11.4) |
| subtype1 | 5 | 1996.4 (14.1) |
| subtype2 | 5 | 1995.4 (15.9) |
| subtype3 | 5 | 1995.6 (5.4) |
| subtype4 | 17 | 2005.2 (9.4) |
Figure S331. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #16: 'TOBACCO_SMOKING_YEAR_STOPPED'
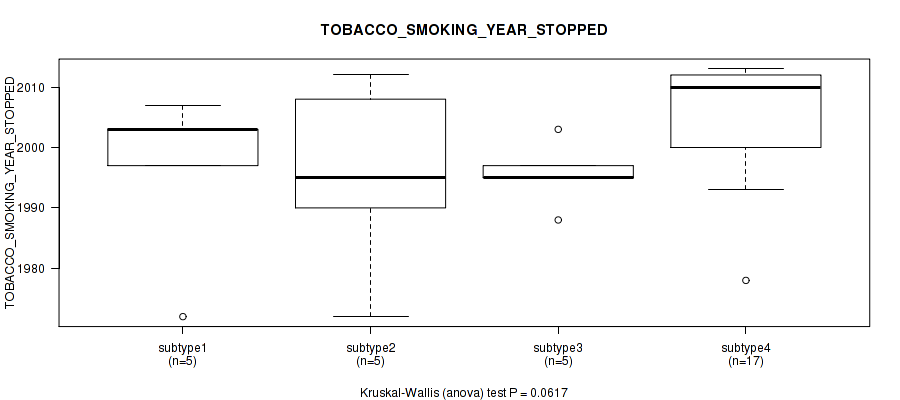
P value = 0.238 (Kruskal-Wallis (anova)), Q value = 1
Table S340. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 71 | 18.7 (13.6) |
| subtype1 | 13 | 14.2 (10.6) |
| subtype2 | 12 | 23.2 (11.6) |
| subtype3 | 10 | 19.5 (9.9) |
| subtype4 | 36 | 18.5 (15.9) |
Figure S332. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #17: 'TOBACCO_SMOKING_PACK_YEARS_SMOKED'

P value = 0.459 (Fisher's exact test), Q value = 1
Table S341. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'
| nPatients | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT REFORMED SMOKER, DURATION NOT SPECIFIED | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|
| ALL | 32 | 7 | 3 | 51 | 112 |
| subtype1 | 5 | 1 | 0 | 9 | 29 |
| subtype2 | 3 | 2 | 1 | 10 | 19 |
| subtype3 | 8 | 2 | 0 | 5 | 14 |
| subtype4 | 16 | 2 | 2 | 27 | 50 |
Figure S333. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #18: 'TOBACCO_SMOKING_HISTORY'

P value = 0.998 (Kruskal-Wallis (anova)), Q value = 1
Table S342. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 63 | 21.1 (7.7) |
| subtype1 | 12 | 19.7 (4.6) |
| subtype2 | 9 | 20.1 (5.3) |
| subtype3 | 8 | 19.9 (5.2) |
| subtype4 | 34 | 22.2 (9.4) |
Figure S334. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #19: 'PATIENT.AGEBEGANSMOKINGINYEARS'

P value = 0.297 (Kruskal-Wallis (anova)), Q value = 1
Table S343. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 107 | 3911.4 (1611.9) |
| subtype1 | 18 | 3763.4 (1871.6) |
| subtype2 | 13 | 3685.2 (1875.1) |
| subtype3 | 19 | 4549.5 (914.6) |
| subtype4 | 57 | 3797.0 (1631.5) |
Figure S335. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #20: 'RADIATION_TOTAL_DOSE'
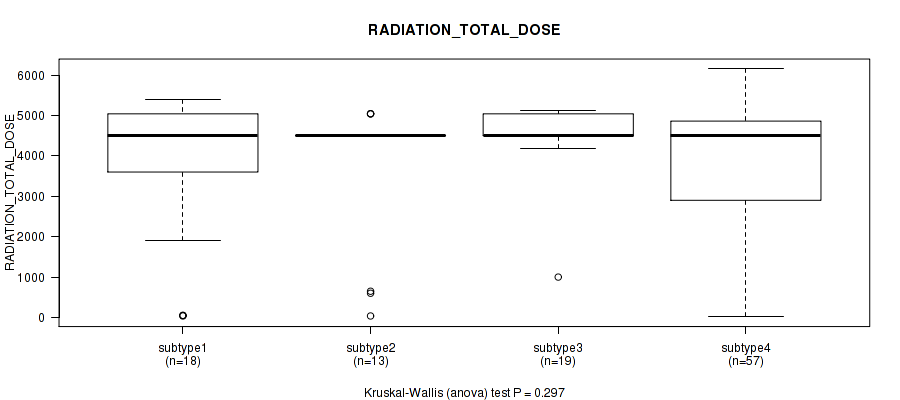
P value = 1e-05 (Fisher's exact test), Q value = 0.0036
Table S344. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'
| nPatients | COMBINATION | EXTERNAL | EXTERNAL BEAM | IMPLANTS | INTERNAL |
|---|---|---|---|---|---|
| ALL | 19 | 74 | 15 | 1 | 12 |
| subtype1 | 1 | 17 | 0 | 0 | 1 |
| subtype2 | 0 | 11 | 3 | 0 | 3 |
| subtype3 | 13 | 1 | 7 | 1 | 0 |
| subtype4 | 5 | 45 | 5 | 0 | 8 |
Figure S336. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #21: 'RADIATION_THERAPY_TYPE'

P value = 0.458 (Fisher's exact test), Q value = 1
Table S345. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'
| nPatients | COMPLETED AS PLANNED | TREATMENT NOT COMPLETED |
|---|---|---|
| ALL | 27 | 3 |
| subtype1 | 1 | 0 |
| subtype2 | 2 | 0 |
| subtype3 | 17 | 1 |
| subtype4 | 7 | 2 |
Figure S337. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #22: 'RADIATION_THERAPY_STATUS'

P value = 0.504 (Fisher's exact test), Q value = 1
Table S346. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'
| nPatients | DISTANT RECURRENCE | LOCAL RECURRENCE | PRIMARY TUMOR FIELD | REGIONAL SITE |
|---|---|---|---|---|
| ALL | 2 | 2 | 29 | 16 |
| subtype1 | 1 | 0 | 3 | 1 |
| subtype2 | 0 | 1 | 5 | 4 |
| subtype3 | 0 | 0 | 2 | 0 |
| subtype4 | 1 | 1 | 19 | 11 |
Figure S338. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #23: 'RADIATION_THERAPY_SITE'

P value = 0.761 (Fisher's exact test), Q value = 1
Table S347. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'
| nPatients | CGY | GY |
|---|---|---|
| ALL | 35 | 3 |
| subtype1 | 5 | 0 |
| subtype2 | 6 | 1 |
| subtype3 | 2 | 0 |
| subtype4 | 22 | 2 |
Figure S339. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #24: 'RADIATION_ADJUVANT_UNITS'
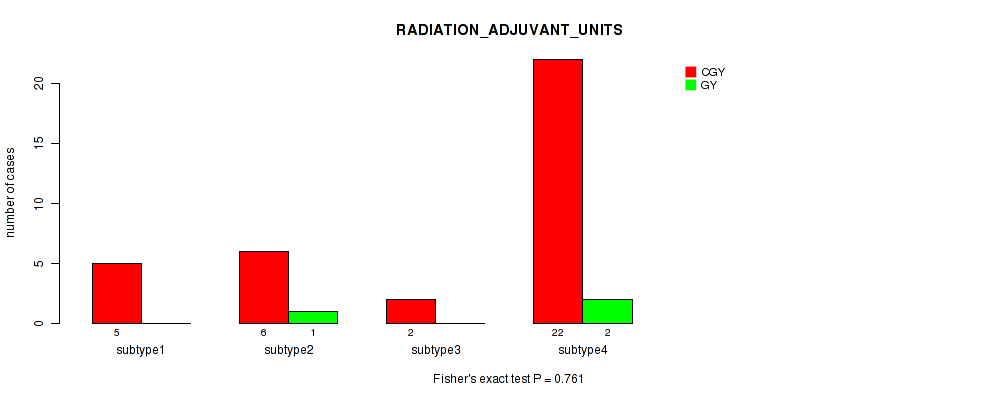
P value = 0.115 (Kruskal-Wallis (anova)), Q value = 1
Table S348. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 210 | 3.6 (2.5) |
| subtype1 | 44 | 2.9 (2.1) |
| subtype2 | 37 | 3.5 (2.5) |
| subtype3 | 30 | 3.3 (1.9) |
| subtype4 | 99 | 4.0 (2.7) |
Figure S340. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #25: 'PREGNANCIES_COUNT_TOTAL'

P value = 0.216 (Kruskal-Wallis (anova)), Q value = 1
Table S349. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 104 | 0.1 (0.3) |
| subtype1 | 20 | 0.0 (0.0) |
| subtype2 | 17 | 0.0 (0.0) |
| subtype3 | 26 | 0.2 (0.6) |
| subtype4 | 41 | 0.0 (0.2) |
Figure S341. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #26: 'PREGNANCIES_COUNT_STILLBIRTH'

P value = 0.357 (Kruskal-Wallis (anova)), Q value = 1
Table S350. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 131 | 0.5 (0.8) |
| subtype1 | 23 | 0.5 (0.5) |
| subtype2 | 23 | 0.3 (0.4) |
| subtype3 | 27 | 0.4 (0.7) |
| subtype4 | 58 | 0.6 (1.1) |
Figure S342. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #27: 'PATIENT.PATIENTPREGNANCYSPONTANEOUSABORTIONCOUNT'

P value = 0.036 (Kruskal-Wallis (anova)), Q value = 1
Table S351. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 211 | 2.7 (1.9) |
| subtype1 | 43 | 2.2 (1.9) |
| subtype2 | 37 | 2.6 (1.3) |
| subtype3 | 31 | 2.2 (1.6) |
| subtype4 | 100 | 3.0 (2.2) |
Figure S343. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #28: 'PREGNANCIES_COUNT_LIVE_BIRTH'

P value = 0.64 (Kruskal-Wallis (anova)), Q value = 1
Table S352. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 113 | 0.8 (1.8) |
| subtype1 | 22 | 0.7 (1.0) |
| subtype2 | 21 | 1.5 (2.9) |
| subtype3 | 25 | 0.6 (1.0) |
| subtype4 | 45 | 0.7 (1.7) |
Figure S344. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #29: 'PATIENT.PATIENTPREGNANCYTHERAPEUTICABORTIONCOUNT'

P value = 0.658 (Kruskal-Wallis (anova)), Q value = 1
Table S353. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 107 | 0.1 (0.3) |
| subtype1 | 20 | 0.1 (0.2) |
| subtype2 | 17 | 0.1 (0.5) |
| subtype3 | 26 | 0.1 (0.3) |
| subtype4 | 44 | 0.1 (0.3) |
Figure S345. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #30: 'PREGNANCIES_COUNT_ECTOPIC'

P value = 0.7 (Fisher's exact test), Q value = 1
Table S354. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'
| nPatients | MACROSCOPIC PARAMETRIAL INVOLVEMENT | MICROSCOPIC PARAMETRIAL INVOLVEMENT | OTHER LOCATION, SPECIFY | POSITIVE BLADDER MARGIN | POSITIVE VAGINAL MARGIN |
|---|---|---|---|---|---|
| ALL | 1 | 6 | 32 | 1 | 9 |
| subtype1 | 0 | 1 | 5 | 0 | 2 |
| subtype2 | 1 | 1 | 4 | 1 | 1 |
| subtype3 | 0 | 2 | 13 | 0 | 2 |
| subtype4 | 0 | 2 | 10 | 0 | 4 |
Figure S346. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #31: 'POS_LYMPH_NODE_LOCATION'
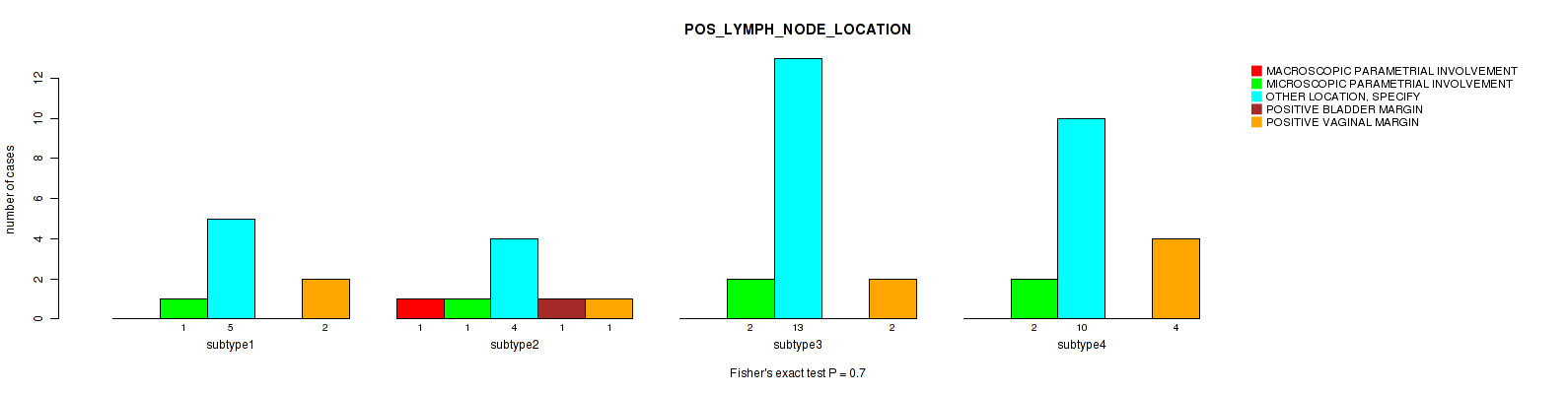
P value = 0.396 (Fisher's exact test), Q value = 1
Table S355. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #32: 'MENOPAUSE_STATUS'
| nPatients | INDETERMINATE (NEITHER PRE OR POSTMENOPAUSAL) | PERI (6-12 MONTHS SINCE LAST MENSTRUAL PERIOD) | POST (PRIOR BILATERAL OVARIECTOMY OR >12 MO SINCE LMP WITH NO PRIOR HYSTERECTOMY) | PRE (<6 MONTHS SINCE LMP AND NO PRIOR BILATERAL OVARIECTOMY AND NOT ON ESTROGEN REPLACEMENT) |
|---|---|---|---|---|
| ALL | 2 | 16 | 65 | 104 |
| subtype1 | 0 | 3 | 14 | 25 |
| subtype2 | 0 | 3 | 13 | 16 |
| subtype3 | 2 | 1 | 12 | 14 |
| subtype4 | 0 | 9 | 26 | 49 |
Figure S347. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #32: 'MENOPAUSE_STATUS'
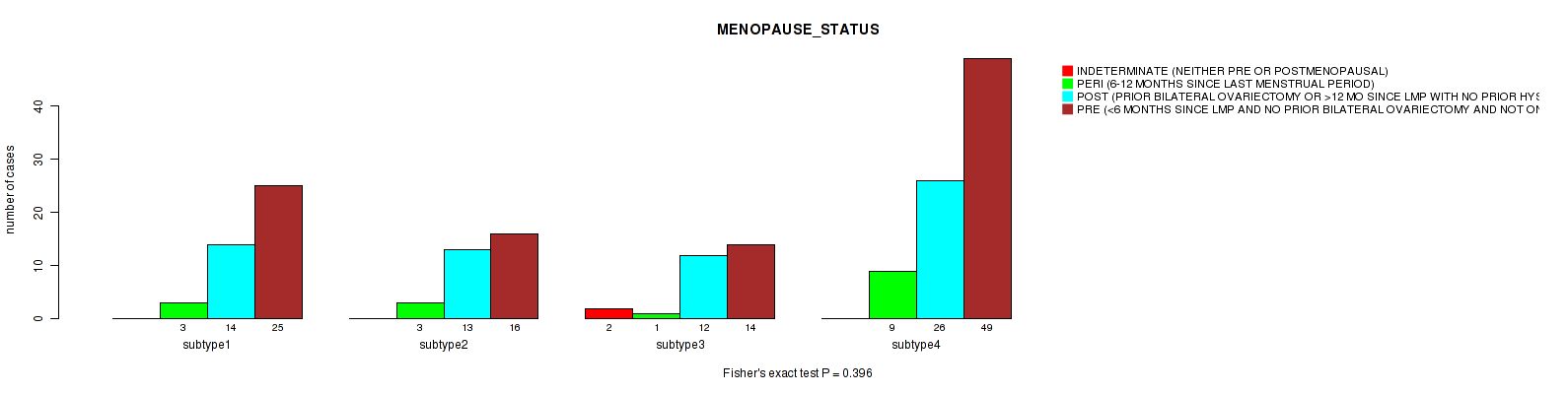
P value = 0.0762 (Fisher's exact test), Q value = 1
Table S356. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 66 | 70 |
| subtype1 | 20 | 15 |
| subtype2 | 13 | 13 |
| subtype3 | 6 | 18 |
| subtype4 | 27 | 24 |
Figure S348. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #33: 'LYMPHOVASCULAR_INVOLVEMENT'
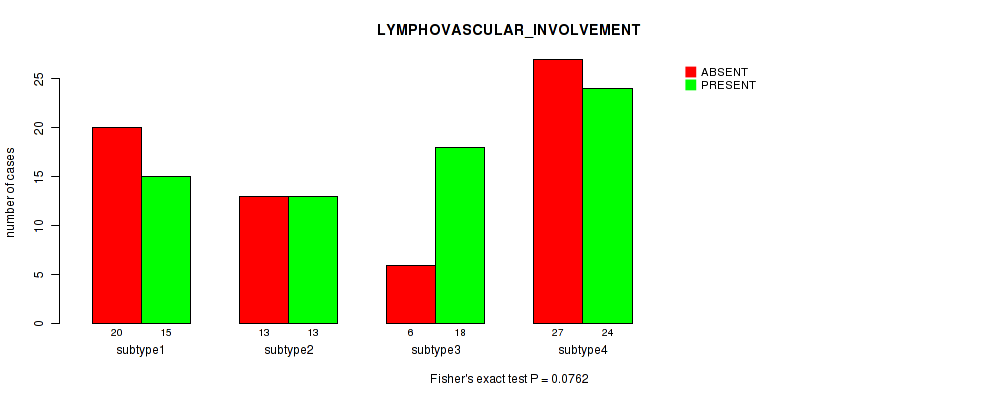
P value = 0.118 (Kruskal-Wallis (anova)), Q value = 1
Table S357. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 140 | 1.0 (2.4) |
| subtype1 | 34 | 0.6 (2.0) |
| subtype2 | 24 | 1.1 (1.8) |
| subtype3 | 25 | 2.4 (4.5) |
| subtype4 | 57 | 0.6 (1.1) |
Figure S349. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #34: 'LYMPH_NODES_EXAMINED_HE_COUNT'

P value = 0.641 (Kruskal-Wallis (anova)), Q value = 1
Table S358. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 158 | 22.2 (12.5) |
| subtype1 | 38 | 20.8 (10.5) |
| subtype2 | 29 | 21.2 (12.5) |
| subtype3 | 25 | 26.1 (15.2) |
| subtype4 | 66 | 21.9 (12.4) |
Figure S350. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #35: 'LYMPH_NODES_EXAMINED'
P value = 0.0192 (Fisher's exact test), Q value = 1
Table S359. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'
| nPatients | KERATINIZING SQUAMOUS CELL CARCINOMA | NON-KERATINIZING SQUAMOUS CELL CARCINOMA |
|---|---|---|
| ALL | 46 | 92 |
| subtype1 | 0 | 11 |
| subtype2 | 13 | 15 |
| subtype3 | 9 | 12 |
| subtype4 | 24 | 54 |
Figure S351. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #36: 'KERATINIZATION_SQUAMOUS_CELL'

P value = 1.22e-06 (Kruskal-Wallis (anova)), Q value = 0.00044
Table S360. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 237 | 2007.7 (5.0) |
| subtype1 | 49 | 2009.0 (3.8) |
| subtype2 | 41 | 2008.9 (4.1) |
| subtype3 | 31 | 2003.0 (5.1) |
| subtype4 | 116 | 2007.9 (5.1) |
Figure S352. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #37: 'INITIAL_PATHOLOGIC_DX_YEAR'

P value = 0.395 (Fisher's exact test), Q value = 1
Table S361. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'
| nPatients | CURRENT USER | FORMER USER | NEVER USED |
|---|---|---|---|
| ALL | 10 | 49 | 58 |
| subtype1 | 5 | 13 | 12 |
| subtype2 | 0 | 9 | 9 |
| subtype3 | 0 | 5 | 10 |
| subtype4 | 5 | 22 | 27 |
Figure S353. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #38: 'HISTORY_HORMONAL_CONTRACEPTIVES_USE'

P value = 0.753 (Kruskal-Wallis (anova)), Q value = 1
Table S362. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 208 | 161.4 (7.0) |
| subtype1 | 44 | 162.2 (6.9) |
| subtype2 | 37 | 162.5 (7.8) |
| subtype3 | 25 | 160.8 (8.0) |
| subtype4 | 102 | 160.9 (6.6) |
Figure S354. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #39: 'HEIGHT_CM_AT_DIAGNOSIS'

P value = 0.486 (Fisher's exact test), Q value = 1
Table S363. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'
| nPatients | ABSENT | PRESENT |
|---|---|---|
| ALL | 92 | 17 |
| subtype1 | 27 | 4 |
| subtype2 | 16 | 3 |
| subtype3 | 17 | 6 |
| subtype4 | 32 | 4 |
Figure S355. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #40: 'CORPUS_INVOLVEMENT'
P value = 0.14 (Fisher's exact test), Q value = 1
Table S364. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'
| nPatients | CARBOPLATIN | CISPLATIN | OTHER |
|---|---|---|---|
| ALL | 3 | 17 | 1 |
| subtype1 | 0 | 0 | 0 |
| subtype2 | 0 | 1 | 0 |
| subtype3 | 3 | 9 | 0 |
| subtype4 | 0 | 7 | 1 |
Figure S356. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #41: 'CHEMO_CONCURRENT_TYPE'

P value = 0.302 (Kruskal-Wallis (anova)), Q value = 1
Table S365. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 12 | 12.0 (5.6) |
| subtype1 | 1 | 7.1 (NA) |
| subtype2 | 4 | 9.6 (3.9) |
| subtype3 | 3 | 11.0 (3.0) |
| subtype4 | 4 | 16.4 (7.0) |
Figure S357. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #42: 'CERVIX_SUV_RESULTS'

P value = 0.829 (Fisher's exact test), Q value = 1
Table S366. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'
| nPatients | T1A | T1B | T1B1 | T1B2 | T2 | T2A | T2A1 | T2A2 | T2B | T3B | T4 | TIS | TX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 1 | 31 | 66 | 24 | 4 | 8 | 7 | 9 | 21 | 7 | 4 | 1 | 15 |
| subtype1 | 0 | 3 | 20 | 5 | 1 | 2 | 2 | 3 | 5 | 0 | 1 | 0 | 2 |
| subtype2 | 0 | 6 | 12 | 4 | 0 | 3 | 1 | 1 | 2 | 1 | 1 | 0 | 2 |
| subtype3 | 0 | 8 | 9 | 3 | 1 | 0 | 1 | 0 | 3 | 1 | 0 | 0 | 0 |
| subtype4 | 1 | 14 | 25 | 12 | 2 | 3 | 3 | 5 | 11 | 5 | 2 | 1 | 11 |
Figure S358. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #43: 'AJCC_TUMOR_PATHOLOGIC_PT'

P value = 0.611 (Kruskal-Wallis (anova)), Q value = 1
Table S367. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 239 | 47.7 (13.5) |
| subtype1 | 50 | 46.0 (11.6) |
| subtype2 | 42 | 49.9 (14.3) |
| subtype3 | 31 | 48.9 (14.4) |
| subtype4 | 116 | 47.4 (13.8) |
Figure S359. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #44: 'AGE_AT_DIAGNOSIS'

P value = 0.193 (Fisher's exact test), Q value = 1
Table S368. Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'
| nPatients | STAGE I | STAGE IA | STAGE IA2 | STAGE IB | STAGE IB1 | STAGE IB2 | STAGE II | STAGE IIA | STAGE IIA1 | STAGE IIA2 | STAGE IIB | STAGE IIIB | STAGE IVA | STAGE IVB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 4 | 2 | 1 | 34 | 68 | 32 | 4 | 6 | 5 | 7 | 26 | 33 | 5 | 6 |
| subtype1 | 1 | 0 | 0 | 3 | 22 | 8 | 1 | 1 | 1 | 2 | 4 | 4 | 0 | 3 |
| subtype2 | 0 | 1 | 1 | 4 | 14 | 4 | 1 | 2 | 1 | 1 | 5 | 4 | 2 | 1 |
| subtype3 | 0 | 0 | 0 | 12 | 5 | 4 | 0 | 1 | 1 | 0 | 2 | 6 | 0 | 0 |
| subtype4 | 3 | 1 | 0 | 15 | 27 | 16 | 2 | 2 | 2 | 4 | 15 | 19 | 3 | 2 |
Figure S360. Get High-res Image Clustering Approach #8: 'MIRseq Mature cHierClus subtypes' versus Clinical Feature #45: 'STAGE_EVENT.CLINICAL_STAGE'

-
Cluster data file = CESC-TP.mergedcluster.txt
-
Clinical data file = CESC-TP.merged_data.txt
-
Number of patients = 250
-
Number of clustering approaches = 8
-
Number of selected clinical features = 45
-
Exclude small clusters that include fewer than K patients, K = 3
consensus non-negative matrix factorization clustering approach (Brunet et al. 2004)
Resampling-based clustering method (Monti et al. 2003)
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For binary clinical features, two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.