This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 4 genes and 8 molecular subtypes across 66 patients, no significant finding detected with P value < 0.05 and Q value < 0.25.
-
No gene mutations related to molecuar subtypes.
Table 1. Get Full Table Overview of the association between mutation status of 4 genes and 8 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, no significant finding detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| TP53 | 22 (33%) | 44 |
0.615 (1.00) |
0.0891 (1.00) |
0.0657 (1.00) |
0.016 (0.495) |
0.643 (1.00) |
0.662 (1.00) |
1 (1.00) |
0.688 (1.00) |
| PTEN | 6 (9%) | 60 |
0.0996 (1.00) |
0.322 (1.00) |
0.352 (1.00) |
0.475 (1.00) |
0.353 (1.00) |
0.0674 (1.00) |
0.372 (1.00) |
0.374 (1.00) |
| PABPC1 | 7 (11%) | 59 |
0.489 (1.00) |
0.236 (1.00) |
0.204 (1.00) |
0.74 (1.00) |
0.61 (1.00) |
0.308 (1.00) |
0.286 (1.00) |
0.143 (1.00) |
| URGCP | 3 (5%) | 63 |
0.648 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.195 (1.00) |
0.542 (1.00) |
0.152 (1.00) |
P value = 0.615 (Fisher's exact test), Q value = 1
Table S1. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 5 | 47 | 14 |
| TP53 MUTATED | 2 | 14 | 6 |
| TP53 WILD-TYPE | 3 | 33 | 8 |
P value = 0.0891 (Fisher's exact test), Q value = 1
Table S2. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 35 | 13 |
| TP53 MUTATED | 3 | 16 | 3 |
| TP53 WILD-TYPE | 15 | 19 | 10 |
P value = 0.0657 (Fisher's exact test), Q value = 1
Table S3. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| TP53 MUTATED | 6 | 6 | 9 | 1 |
| TP53 WILD-TYPE | 13 | 16 | 6 | 9 |
P value = 0.016 (Fisher's exact test), Q value = 0.49
Table S4. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 19 | 16 | 7 | 6 | 10 | 4 | 4 |
| TP53 MUTATED | 5 | 6 | 1 | 0 | 7 | 3 | 0 |
| TP53 WILD-TYPE | 14 | 10 | 6 | 6 | 3 | 1 | 4 |
Figure S1. Get High-res Image Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
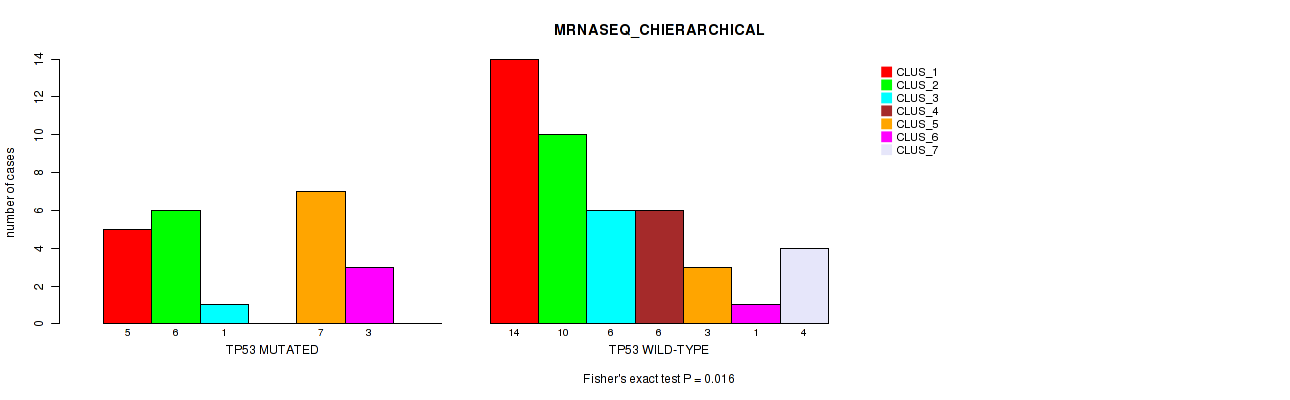
P value = 0.643 (Fisher's exact test), Q value = 1
Table S5. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 20 | 24 | 22 |
| TP53 MUTATED | 5 | 9 | 8 |
| TP53 WILD-TYPE | 15 | 15 | 14 |
P value = 0.662 (Fisher's exact test), Q value = 1
Table S6. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 18 | 11 | 12 | 6 | 5 | 5 | 9 |
| TP53 MUTATED | 7 | 5 | 3 | 1 | 1 | 3 | 2 |
| TP53 WILD-TYPE | 11 | 6 | 9 | 5 | 4 | 2 | 7 |
P value = 1 (Fisher's exact test), Q value = 1
Table S7. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 19 | 29 |
| TP53 MUTATED | 7 | 11 |
| TP53 WILD-TYPE | 12 | 18 |
P value = 0.688 (Fisher's exact test), Q value = 1
Table S8. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 | CLUS_8 |
|---|---|---|---|---|---|---|---|---|
| ALL | 10 | 6 | 5 | 8 | 4 | 6 | 5 | 4 |
| TP53 MUTATED | 4 | 3 | 2 | 2 | 1 | 3 | 3 | 0 |
| TP53 WILD-TYPE | 6 | 3 | 3 | 6 | 3 | 3 | 2 | 4 |
P value = 0.0996 (Fisher's exact test), Q value = 1
Table S9. Gene #2: 'PTEN MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 5 | 47 | 14 |
| PTEN MUTATED | 2 | 3 | 1 |
| PTEN WILD-TYPE | 3 | 44 | 13 |
P value = 0.322 (Fisher's exact test), Q value = 1
Table S10. Gene #2: 'PTEN MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 35 | 13 |
| PTEN MUTATED | 3 | 3 | 0 |
| PTEN WILD-TYPE | 15 | 32 | 13 |
P value = 0.352 (Fisher's exact test), Q value = 1
Table S11. Gene #2: 'PTEN MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| PTEN MUTATED | 2 | 1 | 3 | 0 |
| PTEN WILD-TYPE | 17 | 21 | 12 | 10 |
P value = 0.475 (Fisher's exact test), Q value = 1
Table S12. Gene #2: 'PTEN MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 19 | 16 | 7 | 6 | 10 | 4 | 4 |
| PTEN MUTATED | 2 | 1 | 0 | 0 | 3 | 0 | 0 |
| PTEN WILD-TYPE | 17 | 15 | 7 | 6 | 7 | 4 | 4 |
P value = 0.353 (Fisher's exact test), Q value = 1
Table S13. Gene #2: 'PTEN MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 20 | 24 | 22 |
| PTEN MUTATED | 1 | 4 | 1 |
| PTEN WILD-TYPE | 19 | 20 | 21 |
P value = 0.0674 (Fisher's exact test), Q value = 1
Table S14. Gene #2: 'PTEN MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 18 | 11 | 12 | 6 | 5 | 5 | 9 |
| PTEN MUTATED | 5 | 0 | 0 | 0 | 0 | 1 | 0 |
| PTEN WILD-TYPE | 13 | 11 | 12 | 6 | 5 | 4 | 9 |
P value = 0.372 (Fisher's exact test), Q value = 1
Table S15. Gene #2: 'PTEN MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 19 | 29 |
| PTEN MUTATED | 3 | 2 |
| PTEN WILD-TYPE | 16 | 27 |
P value = 0.374 (Fisher's exact test), Q value = 1
Table S16. Gene #2: 'PTEN MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 | CLUS_8 |
|---|---|---|---|---|---|---|---|---|
| ALL | 10 | 6 | 5 | 8 | 4 | 6 | 5 | 4 |
| PTEN MUTATED | 3 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| PTEN WILD-TYPE | 7 | 6 | 4 | 8 | 4 | 6 | 4 | 4 |
P value = 0.489 (Fisher's exact test), Q value = 1
Table S17. Gene #3: 'PABPC1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 5 | 47 | 14 |
| PABPC1 MUTATED | 1 | 4 | 2 |
| PABPC1 WILD-TYPE | 4 | 43 | 12 |
P value = 0.236 (Fisher's exact test), Q value = 1
Table S18. Gene #3: 'PABPC1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 35 | 13 |
| PABPC1 MUTATED | 2 | 2 | 3 |
| PABPC1 WILD-TYPE | 16 | 33 | 10 |
P value = 0.204 (Fisher's exact test), Q value = 1
Table S19. Gene #3: 'PABPC1 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| PABPC1 MUTATED | 2 | 1 | 1 | 3 |
| PABPC1 WILD-TYPE | 17 | 21 | 14 | 7 |
P value = 0.74 (Fisher's exact test), Q value = 1
Table S20. Gene #3: 'PABPC1 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 19 | 16 | 7 | 6 | 10 | 4 | 4 |
| PABPC1 MUTATED | 2 | 1 | 2 | 1 | 1 | 0 | 0 |
| PABPC1 WILD-TYPE | 17 | 15 | 5 | 5 | 9 | 4 | 4 |
P value = 0.61 (Fisher's exact test), Q value = 1
Table S21. Gene #3: 'PABPC1 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 20 | 24 | 22 |
| PABPC1 MUTATED | 3 | 3 | 1 |
| PABPC1 WILD-TYPE | 17 | 21 | 21 |
P value = 0.308 (Fisher's exact test), Q value = 1
Table S22. Gene #3: 'PABPC1 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 18 | 11 | 12 | 6 | 5 | 5 | 9 |
| PABPC1 MUTATED | 2 | 1 | 0 | 1 | 0 | 0 | 3 |
| PABPC1 WILD-TYPE | 16 | 10 | 12 | 5 | 5 | 5 | 6 |
P value = 0.286 (Fisher's exact test), Q value = 1
Table S23. Gene #3: 'PABPC1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 19 | 29 |
| PABPC1 MUTATED | 3 | 1 |
| PABPC1 WILD-TYPE | 16 | 28 |
P value = 0.143 (Fisher's exact test), Q value = 1
Table S24. Gene #3: 'PABPC1 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 | CLUS_8 |
|---|---|---|---|---|---|---|---|---|
| ALL | 10 | 6 | 5 | 8 | 4 | 6 | 5 | 4 |
| PABPC1 MUTATED | 0 | 0 | 1 | 0 | 0 | 2 | 1 | 0 |
| PABPC1 WILD-TYPE | 10 | 6 | 4 | 8 | 4 | 4 | 4 | 4 |
P value = 0.648 (Fisher's exact test), Q value = 1
Table S25. Gene #4: 'URGCP MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 5 | 47 | 14 |
| URGCP MUTATED | 0 | 2 | 1 |
| URGCP WILD-TYPE | 5 | 45 | 13 |
P value = 1 (Fisher's exact test), Q value = 1
Table S26. Gene #4: 'URGCP MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 35 | 13 |
| URGCP MUTATED | 1 | 2 | 0 |
| URGCP WILD-TYPE | 17 | 33 | 13 |
P value = 1 (Fisher's exact test), Q value = 1
Table S27. Gene #4: 'URGCP MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 19 | 22 | 15 | 10 |
| URGCP MUTATED | 1 | 1 | 1 | 0 |
| URGCP WILD-TYPE | 18 | 21 | 14 | 10 |
P value = 1 (Fisher's exact test), Q value = 1
Table S28. Gene #4: 'URGCP MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 19 | 16 | 7 | 6 | 10 | 4 | 4 |
| URGCP MUTATED | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| URGCP WILD-TYPE | 18 | 15 | 7 | 6 | 9 | 4 | 4 |
P value = 0.195 (Fisher's exact test), Q value = 1
Table S29. Gene #4: 'URGCP MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 20 | 24 | 22 |
| URGCP MUTATED | 2 | 0 | 1 |
| URGCP WILD-TYPE | 18 | 24 | 21 |
P value = 0.542 (Fisher's exact test), Q value = 1
Table S30. Gene #4: 'URGCP MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 18 | 11 | 12 | 6 | 5 | 5 | 9 |
| URGCP MUTATED | 1 | 0 | 0 | 1 | 0 | 0 | 1 |
| URGCP WILD-TYPE | 17 | 11 | 12 | 5 | 5 | 5 | 8 |
P value = 0.152 (Fisher's exact test), Q value = 1
Table S31. Gene #4: 'URGCP MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 19 | 29 |
| URGCP MUTATED | 2 | 0 |
| URGCP WILD-TYPE | 17 | 29 |
-
Mutation data file = transformed.cor.cli.txt
-
Molecular subtypes file = KICH-TP.transferedmergedcluster.txt
-
Number of patients = 66
-
Number of significantly mutated genes = 4
-
Number of Molecular subtypes = 8
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.