This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and molecular subtypes.
Testing the association between copy number variation 49 arm-level events and 8 molecular subtypes across 80 patients, 34 significant findings detected with P value < 0.05 and Q value < 0.25.
-
6p gain cnv correlated to 'CN_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
6q gain cnv correlated to 'MRNASEQ_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
8q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
3p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
3q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
6q loss cnv correlated to 'CN_CNMF'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 49 arm-level events and 8 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 34 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 8q gain | 53 (66%) | 27 |
1e-05 (0.00392) |
1e-05 (0.00392) |
4e-05 (0.0148) |
1e-05 (0.00392) |
5e-05 (0.0183) |
1e-05 (0.00392) |
1e-05 (0.00392) |
5e-05 (0.0183) |
| 3p loss | 43 (54%) | 37 |
1e-05 (0.00392) |
1e-05 (0.00392) |
1e-05 (0.00392) |
1e-05 (0.00392) |
1e-05 (0.00392) |
1e-05 (0.00392) |
1e-05 (0.00392) |
1e-05 (0.00392) |
| 3q loss | 43 (54%) | 37 |
1e-05 (0.00392) |
1e-05 (0.00392) |
1e-05 (0.00392) |
1e-05 (0.00392) |
1e-05 (0.00392) |
1e-05 (0.00392) |
1e-05 (0.00392) |
1e-05 (0.00392) |
| 6p gain | 39 (49%) | 41 |
0.00026 (0.0941) |
0.0039 (1.00) |
0.00026 (0.0941) |
0.00027 (0.0972) |
1e-05 (0.00392) |
3e-05 (0.0111) |
0.0031 (1.00) |
5e-05 (0.0183) |
| 6q gain | 16 (20%) | 64 |
0.00579 (1.00) |
0.00251 (0.884) |
0.00037 (0.133) |
0.00088 (0.315) |
4e-05 (0.0148) |
0.00015 (0.0544) |
0.00623 (1.00) |
0.00088 (0.315) |
| 6q loss | 17 (21%) | 63 |
7e-05 (0.0255) |
0.00327 (1.00) |
0.0508 (1.00) |
0.00502 (1.00) |
0.11 (1.00) |
0.0294 (1.00) |
0.00433 (1.00) |
0.00252 (0.885) |
| 1q gain | 8 (10%) | 72 |
0.715 (1.00) |
0.714 (1.00) |
0.701 (1.00) |
0.803 (1.00) |
0.206 (1.00) |
0.156 (1.00) |
0.314 (1.00) |
0.826 (1.00) |
| 2p gain | 10 (12%) | 70 |
0.315 (1.00) |
1 (1.00) |
0.0377 (1.00) |
0.116 (1.00) |
0.117 (1.00) |
0.199 (1.00) |
0.658 (1.00) |
0.00819 (1.00) |
| 2q gain | 8 (10%) | 72 |
0.393 (1.00) |
0.715 (1.00) |
0.0549 (1.00) |
0.111 (1.00) |
0.312 (1.00) |
0.0163 (1.00) |
0.422 (1.00) |
0.00247 (0.872) |
| 4p gain | 7 (9%) | 73 |
0.0273 (1.00) |
0.282 (1.00) |
0.151 (1.00) |
0.12 (1.00) |
0.775 (1.00) |
0.548 (1.00) |
0.365 (1.00) |
0.296 (1.00) |
| 4q gain | 4 (5%) | 76 |
0.159 (1.00) |
0.271 (1.00) |
0.398 (1.00) |
0.344 (1.00) |
0.329 (1.00) |
0.808 (1.00) |
0.512 (1.00) |
0.302 (1.00) |
| 5p gain | 3 (4%) | 77 |
0.161 (1.00) |
0.442 (1.00) |
0.784 (1.00) |
0.108 (1.00) |
1 (1.00) |
0.0756 (1.00) |
1 (1.00) |
0.842 (1.00) |
| 5q gain | 3 (4%) | 77 |
0.163 (1.00) |
0.439 (1.00) |
0.782 (1.00) |
0.108 (1.00) |
1 (1.00) |
0.0762 (1.00) |
1 (1.00) |
0.838 (1.00) |
| 7p gain | 9 (11%) | 71 |
0.163 (1.00) |
0.127 (1.00) |
1 (1.00) |
0.742 (1.00) |
0.658 (1.00) |
0.221 (1.00) |
0.895 (1.00) |
0.433 (1.00) |
| 7q gain | 8 (10%) | 72 |
0.0556 (1.00) |
0.0303 (1.00) |
0.89 (1.00) |
0.448 (1.00) |
0.273 (1.00) |
0.359 (1.00) |
0.689 (1.00) |
0.268 (1.00) |
| 8p gain | 33 (41%) | 47 |
0.00111 (0.395) |
0.00111 (0.395) |
0.00253 (0.885) |
0.00435 (1.00) |
0.0407 (1.00) |
0.0202 (1.00) |
0.0322 (1.00) |
0.0257 (1.00) |
| 9p gain | 6 (8%) | 74 |
0.233 (1.00) |
0.176 (1.00) |
0.402 (1.00) |
0.638 (1.00) |
0.741 (1.00) |
0.907 (1.00) |
0.087 (1.00) |
0.937 (1.00) |
| 9q gain | 5 (6%) | 75 |
0.51 (1.00) |
0.0378 (1.00) |
0.838 (1.00) |
0.839 (1.00) |
0.342 (1.00) |
0.974 (1.00) |
0.00797 (1.00) |
0.719 (1.00) |
| 11p gain | 9 (11%) | 71 |
0.166 (1.00) |
0.539 (1.00) |
1 (1.00) |
0.74 (1.00) |
0.272 (1.00) |
0.245 (1.00) |
0.546 (1.00) |
0.281 (1.00) |
| 11q gain | 10 (12%) | 70 |
0.317 (1.00) |
0.829 (1.00) |
0.909 (1.00) |
0.83 (1.00) |
0.146 (1.00) |
0.172 (1.00) |
0.544 (1.00) |
0.285 (1.00) |
| 12p gain | 3 (4%) | 77 |
0.162 (1.00) |
0.442 (1.00) |
0.0906 (1.00) |
0.0102 (1.00) |
1 (1.00) |
0.00996 (1.00) |
1 (1.00) |
0.125 (1.00) |
| 12q gain | 3 (4%) | 77 |
0.161 (1.00) |
0.44 (1.00) |
0.0885 (1.00) |
0.00952 (1.00) |
1 (1.00) |
0.00994 (1.00) |
1 (1.00) |
0.125 (1.00) |
| 13q gain | 6 (8%) | 74 |
0.105 (1.00) |
1 (1.00) |
0.737 (1.00) |
0.753 (1.00) |
0.645 (1.00) |
0.105 (1.00) |
1 (1.00) |
0.0605 (1.00) |
| 14q gain | 3 (4%) | 77 |
0.0378 (1.00) |
0.441 (1.00) |
0.589 (1.00) |
0.329 (1.00) |
1 (1.00) |
0.185 (1.00) |
1 (1.00) |
0.729 (1.00) |
| 16p gain | 4 (5%) | 76 |
1 (1.00) |
0.439 (1.00) |
1 (1.00) |
0.438 (1.00) |
0.413 (1.00) |
0.0264 (1.00) |
0.38 (1.00) |
1 (1.00) |
| 17p gain | 8 (10%) | 72 |
0.0809 (1.00) |
0.803 (1.00) |
0.389 (1.00) |
0.22 (1.00) |
0.392 (1.00) |
0.00993 (1.00) |
0.543 (1.00) |
0.283 (1.00) |
| 17q gain | 9 (11%) | 71 |
0.218 (1.00) |
1 (1.00) |
0.422 (1.00) |
0.311 (1.00) |
0.271 (1.00) |
0.0179 (1.00) |
0.412 (1.00) |
0.353 (1.00) |
| 20p gain | 8 (10%) | 72 |
0.00937 (1.00) |
0.569 (1.00) |
0.39 (1.00) |
0.219 (1.00) |
0.31 (1.00) |
0.18 (1.00) |
0.317 (1.00) |
0.695 (1.00) |
| 20q gain | 9 (11%) | 71 |
0.00203 (0.719) |
0.539 (1.00) |
0.144 (1.00) |
0.0566 (1.00) |
0.247 (1.00) |
0.247 (1.00) |
0.414 (1.00) |
0.911 (1.00) |
| 21q gain | 14 (18%) | 66 |
0.00309 (1.00) |
0.196 (1.00) |
0.104 (1.00) |
0.0129 (1.00) |
0.926 (1.00) |
0.165 (1.00) |
0.527 (1.00) |
0.162 (1.00) |
| 22q gain | 6 (8%) | 74 |
0.106 (1.00) |
0.644 (1.00) |
0.153 (1.00) |
0.221 (1.00) |
0.645 (1.00) |
0.00656 (1.00) |
0.729 (1.00) |
0.059 (1.00) |
| xq gain | 10 (12%) | 70 |
0.39 (1.00) |
0.0934 (1.00) |
0.108 (1.00) |
0.139 (1.00) |
0.157 (1.00) |
0.0556 (1.00) |
0.367 (1.00) |
0.366 (1.00) |
| 1p loss | 19 (24%) | 61 |
0.0044 (1.00) |
0.0523 (1.00) |
0.781 (1.00) |
0.663 (1.00) |
0.318 (1.00) |
0.00897 (1.00) |
0.461 (1.00) |
0.052 (1.00) |
| 1q loss | 3 (4%) | 77 |
0.0383 (1.00) |
0.169 (1.00) |
0.217 (1.00) |
0.322 (1.00) |
0.42 (1.00) |
0.492 (1.00) |
0.0685 (1.00) |
0.0732 (1.00) |
| 4q loss | 3 (4%) | 77 |
1 (1.00) |
0.791 (1.00) |
0.592 (1.00) |
0.602 (1.00) |
1 (1.00) |
0.612 (1.00) |
0.379 (1.00) |
0.622 (1.00) |
| 5q loss | 3 (4%) | 77 |
0.45 (1.00) |
0.109 (1.00) |
0.0883 (1.00) |
0.17 (1.00) |
0.595 (1.00) |
0.185 (1.00) |
0.204 (1.00) |
0.841 (1.00) |
| 8p loss | 9 (11%) | 71 |
0.0303 (1.00) |
0.344 (1.00) |
0.0371 (1.00) |
0.023 (1.00) |
0.199 (1.00) |
0.163 (1.00) |
0.139 (1.00) |
0.0313 (1.00) |
| 8q loss | 3 (4%) | 77 |
0.786 (1.00) |
0.602 (1.00) |
1 (1.00) |
0.791 (1.00) |
0.795 (1.00) |
1 (1.00) |
1 (1.00) |
0.839 (1.00) |
| 9p loss | 8 (10%) | 72 |
0.109 (1.00) |
0.0415 (1.00) |
0.344 (1.00) |
0.151 (1.00) |
0.184 (1.00) |
0.254 (1.00) |
0.151 (1.00) |
0.399 (1.00) |
| 9q loss | 7 (9%) | 73 |
0.167 (1.00) |
0.0749 (1.00) |
0.593 (1.00) |
0.239 (1.00) |
0.0911 (1.00) |
0.401 (1.00) |
0.0867 (1.00) |
0.155 (1.00) |
| 11p loss | 3 (4%) | 77 |
0.0373 (1.00) |
0.236 (1.00) |
0.0432 (1.00) |
0.108 (1.00) |
0.223 (1.00) |
0.249 (1.00) |
0.283 (1.00) |
0.188 (1.00) |
| 12p loss | 3 (4%) | 77 |
0.788 (1.00) |
0.604 (1.00) |
1 (1.00) |
0.789 (1.00) |
0.792 (1.00) |
1 (1.00) |
0.769 (1.00) |
1 (1.00) |
| 13q loss | 3 (4%) | 77 |
1 (1.00) |
0.17 (1.00) |
0.593 (1.00) |
0.603 (1.00) |
0.794 (1.00) |
0.61 (1.00) |
0.559 (1.00) |
0.626 (1.00) |
| 15q loss | 4 (5%) | 76 |
0.155 (1.00) |
0.0492 (1.00) |
0.83 (1.00) |
0.826 (1.00) |
1 (1.00) |
0.662 (1.00) |
0.814 (1.00) |
0.565 (1.00) |
| 16p loss | 3 (4%) | 77 |
1 (1.00) |
0.437 (1.00) |
0.591 (1.00) |
0.606 (1.00) |
0.794 (1.00) |
0.771 (1.00) |
0.561 (1.00) |
0.625 (1.00) |
| 16q loss | 16 (20%) | 64 |
0.0212 (1.00) |
0.132 (1.00) |
0.0961 (1.00) |
0.013 (1.00) |
0.933 (1.00) |
0.00625 (1.00) |
0.358 (1.00) |
0.242 (1.00) |
| 19p loss | 3 (4%) | 77 |
0.452 (1.00) |
0.604 (1.00) |
0.782 (1.00) |
1 (1.00) |
0.598 (1.00) |
0.00287 (0.999) |
0.561 (1.00) |
0.281 (1.00) |
| 19q loss | 3 (4%) | 77 |
0.454 (1.00) |
0.603 (1.00) |
0.782 (1.00) |
1 (1.00) |
0.597 (1.00) |
0.00257 (0.897) |
0.56 (1.00) |
0.279 (1.00) |
| xq loss | 12 (15%) | 68 |
0.339 (1.00) |
0.0384 (1.00) |
0.321 (1.00) |
0.266 (1.00) |
0.392 (1.00) |
0.0489 (1.00) |
0.102 (1.00) |
0.0725 (1.00) |
P value = 0.00026 (Fisher's exact test), Q value = 0.094
Table S1. Gene #8: '6p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 36 | 22 |
| 6P GAIN MUTATED | 9 | 26 | 4 |
| 6P GAIN WILD-TYPE | 13 | 10 | 18 |
Figure S1. Get High-res Image Gene #8: '6p gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00026 (Fisher's exact test), Q value = 0.094
Table S2. Gene #8: '6p gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 15 | 35 |
| 6P GAIN MUTATED | 8 | 5 | 26 |
| 6P GAIN WILD-TYPE | 22 | 10 | 9 |
Figure S2. Get High-res Image Gene #8: '6p gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 0.00027 (Fisher's exact test), Q value = 0.097
Table S3. Gene #8: '6p gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 33 | 18 |
| 6P GAIN MUTATED | 9 | 25 | 5 |
| 6P GAIN WILD-TYPE | 20 | 8 | 13 |
Figure S3. Get High-res Image Gene #8: '6p gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S4. Gene #8: '6p gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 16 | 29 |
| 6P GAIN MUTATED | 10 | 4 | 24 |
| 6P GAIN WILD-TYPE | 24 | 12 | 5 |
Figure S4. Get High-res Image Gene #8: '6p gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 3e-05 (Fisher's exact test), Q value = 0.011
Table S5. Gene #8: '6p gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 8 | 12 | 18 | 24 | 10 | 7 |
| 6P GAIN MUTATED | 2 | 5 | 16 | 5 | 8 | 2 |
| 6P GAIN WILD-TYPE | 6 | 7 | 2 | 19 | 2 | 5 |
Figure S5. Get High-res Image Gene #8: '6p gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 5e-05 (Fisher's exact test), Q value = 0.018
Table S6. Gene #8: '6p gain' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 11 | 32 | 10 |
| 6P GAIN MUTATED | 2 | 4 | 23 | 6 |
| 6P GAIN WILD-TYPE | 18 | 7 | 9 | 4 |
Figure S6. Get High-res Image Gene #8: '6p gain' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 0.00037 (Fisher's exact test), Q value = 0.13
Table S7. Gene #9: '6q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 15 | 35 |
| 6Q GAIN MUTATED | 1 | 1 | 14 |
| 6Q GAIN WILD-TYPE | 29 | 14 | 21 |
Figure S7. Get High-res Image Gene #9: '6q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 4e-05 (Fisher's exact test), Q value = 0.015
Table S8. Gene #9: '6q gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 16 | 29 |
| 6Q GAIN MUTATED | 1 | 1 | 13 |
| 6Q GAIN WILD-TYPE | 33 | 15 | 16 |
Figure S8. Get High-res Image Gene #9: '6q gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 0.00015 (Fisher's exact test), Q value = 0.054
Table S9. Gene #9: '6q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 8 | 12 | 18 | 24 | 10 | 7 |
| 6Q GAIN MUTATED | 0 | 2 | 10 | 0 | 2 | 1 |
| 6Q GAIN WILD-TYPE | 8 | 10 | 8 | 24 | 8 | 6 |
Figure S9. Get High-res Image Gene #9: '6q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S10. Gene #13: '8q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 36 | 22 |
| 8Q GAIN MUTATED | 21 | 12 | 20 |
| 8Q GAIN WILD-TYPE | 1 | 24 | 2 |
Figure S10. Get High-res Image Gene #13: '8q gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S11. Gene #13: '8q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 18 | 29 |
| 8Q GAIN MUTATED | 31 | 12 | 10 |
| 8Q GAIN WILD-TYPE | 2 | 6 | 19 |
Figure S11. Get High-res Image Gene #13: '8q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 4e-05 (Fisher's exact test), Q value = 0.015
Table S12. Gene #13: '8q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 15 | 35 |
| 8Q GAIN MUTATED | 27 | 12 | 14 |
| 8Q GAIN WILD-TYPE | 3 | 3 | 21 |
Figure S12. Get High-res Image Gene #13: '8q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
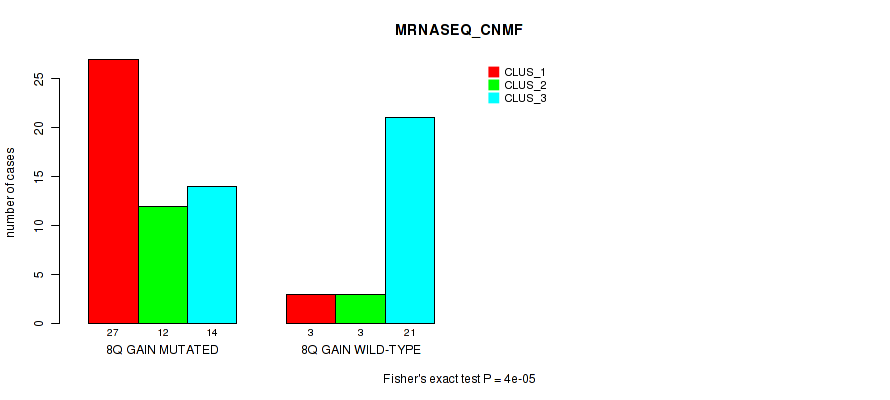
P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S13. Gene #13: '8q gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 33 | 18 |
| 8Q GAIN MUTATED | 27 | 12 | 14 |
| 8Q GAIN WILD-TYPE | 2 | 21 | 4 |
Figure S13. Get High-res Image Gene #13: '8q gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 5e-05 (Fisher's exact test), Q value = 0.018
Table S14. Gene #13: '8q gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 16 | 29 |
| 8Q GAIN MUTATED | 31 | 8 | 13 |
| 8Q GAIN WILD-TYPE | 3 | 8 | 16 |
Figure S14. Get High-res Image Gene #13: '8q gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S15. Gene #13: '8q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 8 | 12 | 18 | 24 | 10 | 7 |
| 8Q GAIN MUTATED | 8 | 2 | 10 | 22 | 4 | 6 |
| 8Q GAIN WILD-TYPE | 0 | 10 | 8 | 2 | 6 | 1 |
Figure S15. Get High-res Image Gene #13: '8q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S16. Gene #13: '8q gain' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 29 | 12 |
| 8Q GAIN MUTATED | 30 | 10 | 10 |
| 8Q GAIN WILD-TYPE | 2 | 19 | 2 |
Figure S16. Get High-res Image Gene #13: '8q gain' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'

P value = 5e-05 (Fisher's exact test), Q value = 0.018
Table S17. Gene #13: '8q gain' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 11 | 32 | 10 |
| 8Q GAIN MUTATED | 18 | 9 | 13 | 10 |
| 8Q GAIN WILD-TYPE | 2 | 2 | 19 | 0 |
Figure S17. Get High-res Image Gene #13: '8q gain' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S18. Gene #32: '3p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 36 | 22 |
| 3P LOSS MUTATED | 22 | 2 | 19 |
| 3P LOSS WILD-TYPE | 0 | 34 | 3 |
Figure S18. Get High-res Image Gene #32: '3p loss' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S19. Gene #32: '3p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 18 | 29 |
| 3P LOSS MUTATED | 33 | 9 | 1 |
| 3P LOSS WILD-TYPE | 0 | 9 | 28 |
Figure S19. Get High-res Image Gene #32: '3p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S20. Gene #32: '3p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 15 | 35 |
| 3P LOSS MUTATED | 30 | 10 | 3 |
| 3P LOSS WILD-TYPE | 0 | 5 | 32 |
Figure S20. Get High-res Image Gene #32: '3p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S21. Gene #32: '3p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 33 | 18 |
| 3P LOSS MUTATED | 29 | 2 | 12 |
| 3P LOSS WILD-TYPE | 0 | 31 | 6 |
Figure S21. Get High-res Image Gene #32: '3p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
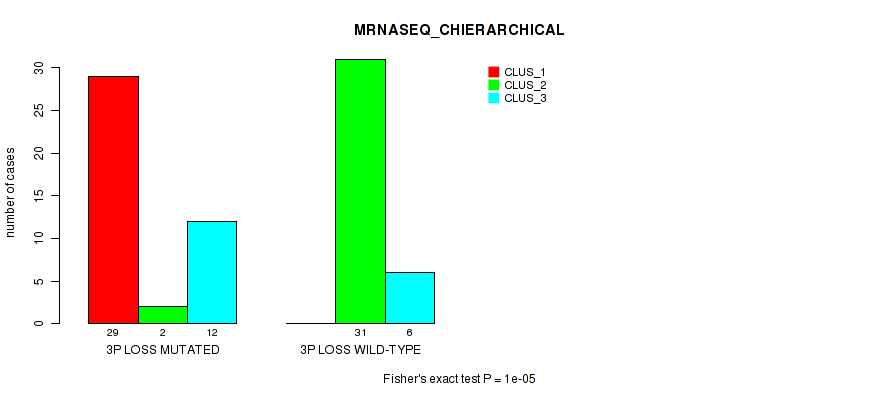
P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S22. Gene #32: '3p loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 16 | 29 |
| 3P LOSS MUTATED | 30 | 11 | 2 |
| 3P LOSS WILD-TYPE | 4 | 5 | 27 |
Figure S22. Get High-res Image Gene #32: '3p loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
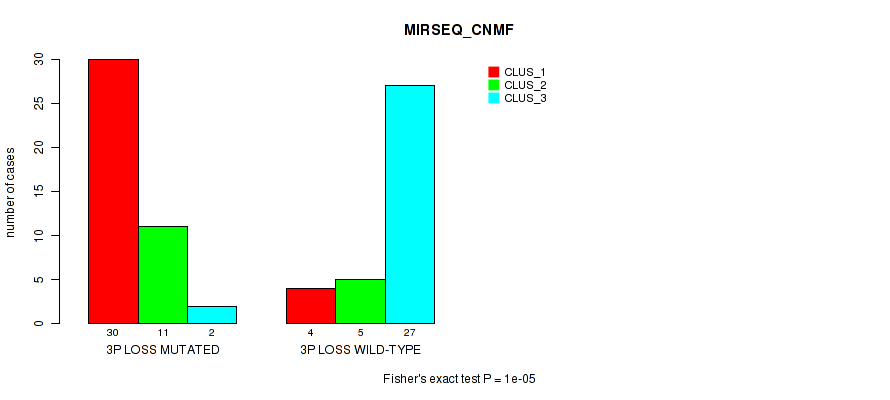
P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S23. Gene #32: '3p loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 8 | 12 | 18 | 24 | 10 | 7 |
| 3P LOSS MUTATED | 8 | 1 | 1 | 24 | 2 | 7 |
| 3P LOSS WILD-TYPE | 0 | 11 | 17 | 0 | 8 | 0 |
Figure S23. Get High-res Image Gene #32: '3p loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
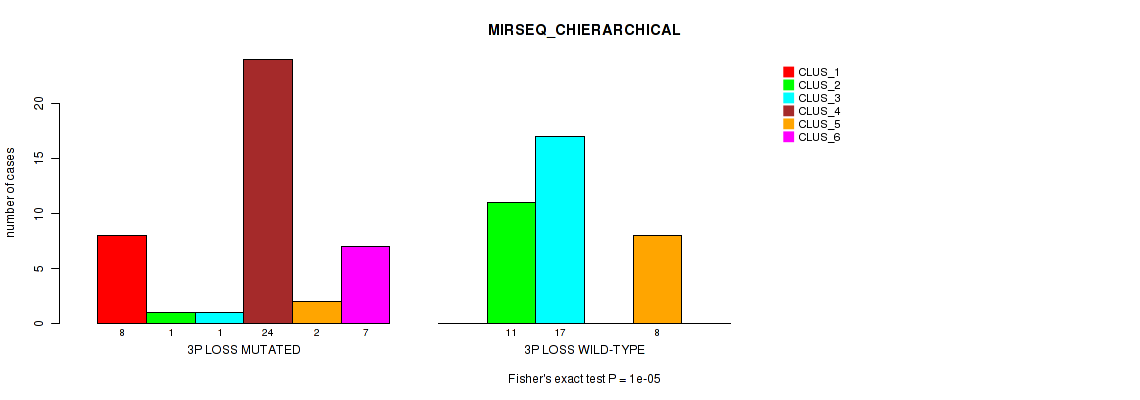
P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S24. Gene #32: '3p loss' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 29 | 12 |
| 3P LOSS MUTATED | 30 | 4 | 8 |
| 3P LOSS WILD-TYPE | 2 | 25 | 4 |
Figure S24. Get High-res Image Gene #32: '3p loss' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
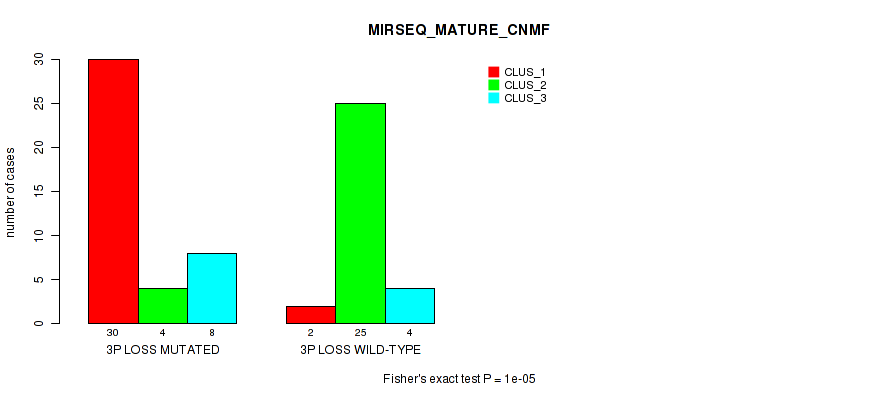
P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S25. Gene #32: '3p loss' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 11 | 32 | 10 |
| 3P LOSS MUTATED | 20 | 7 | 5 | 10 |
| 3P LOSS WILD-TYPE | 0 | 4 | 27 | 0 |
Figure S25. Get High-res Image Gene #32: '3p loss' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S26. Gene #33: '3q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 36 | 22 |
| 3Q LOSS MUTATED | 22 | 2 | 19 |
| 3Q LOSS WILD-TYPE | 0 | 34 | 3 |
Figure S26. Get High-res Image Gene #33: '3q loss' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S27. Gene #33: '3q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 18 | 29 |
| 3Q LOSS MUTATED | 33 | 9 | 1 |
| 3Q LOSS WILD-TYPE | 0 | 9 | 28 |
Figure S27. Get High-res Image Gene #33: '3q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S28. Gene #33: '3q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 15 | 35 |
| 3Q LOSS MUTATED | 30 | 10 | 3 |
| 3Q LOSS WILD-TYPE | 0 | 5 | 32 |
Figure S28. Get High-res Image Gene #33: '3q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
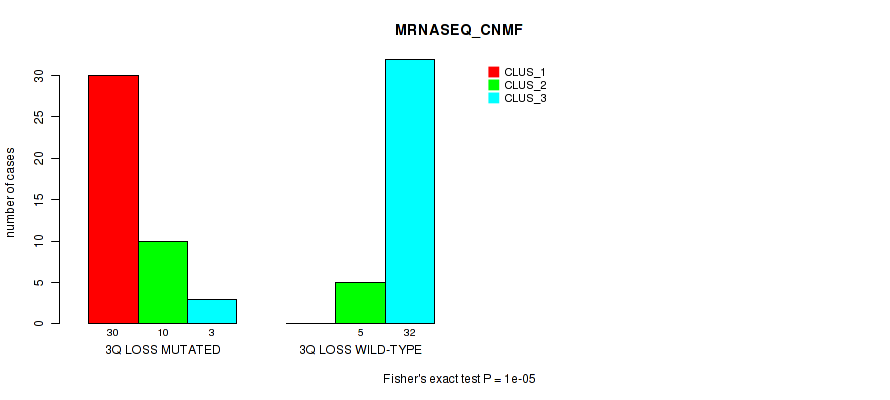
P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S29. Gene #33: '3q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 33 | 18 |
| 3Q LOSS MUTATED | 29 | 2 | 12 |
| 3Q LOSS WILD-TYPE | 0 | 31 | 6 |
Figure S29. Get High-res Image Gene #33: '3q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S30. Gene #33: '3q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 16 | 29 |
| 3Q LOSS MUTATED | 30 | 11 | 2 |
| 3Q LOSS WILD-TYPE | 4 | 5 | 27 |
Figure S30. Get High-res Image Gene #33: '3q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S31. Gene #33: '3q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 8 | 12 | 18 | 24 | 10 | 7 |
| 3Q LOSS MUTATED | 8 | 1 | 1 | 24 | 2 | 7 |
| 3Q LOSS WILD-TYPE | 0 | 11 | 17 | 0 | 8 | 0 |
Figure S31. Get High-res Image Gene #33: '3q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S32. Gene #33: '3q loss' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 29 | 12 |
| 3Q LOSS MUTATED | 30 | 4 | 8 |
| 3Q LOSS WILD-TYPE | 2 | 25 | 4 |
Figure S32. Get High-res Image Gene #33: '3q loss' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
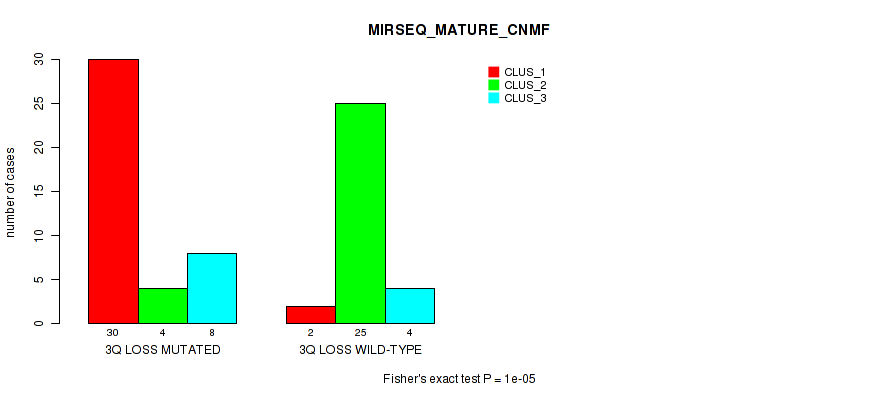
P value = 1e-05 (Fisher's exact test), Q value = 0.0039
Table S33. Gene #33: '3q loss' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 11 | 32 | 10 |
| 3Q LOSS MUTATED | 20 | 7 | 5 | 10 |
| 3Q LOSS WILD-TYPE | 0 | 4 | 27 | 0 |
Figure S33. Get High-res Image Gene #33: '3q loss' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 7e-05 (Fisher's exact test), Q value = 0.025
Table S34. Gene #36: '6q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 36 | 22 |
| 6Q LOSS MUTATED | 12 | 4 | 1 |
| 6Q LOSS WILD-TYPE | 10 | 32 | 21 |
Figure S34. Get High-res Image Gene #36: '6q loss' versus Molecular Subtype #1: 'CN_CNMF'
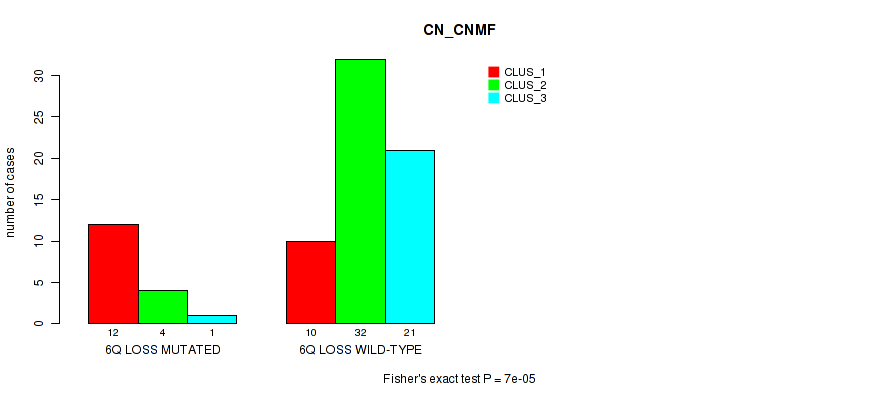
-
Copy number data file = transformed.cor.cli.txt
-
Molecular subtypes file = UVM-TP.transferedmergedcluster.txt
-
Number of patients = 80
-
Number of significantly arm-level cnvs = 49
-
Number of molecular subtypes = 8
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.