This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and selected clinical features.
Testing the association between copy number variation 52 arm-level events and 6 clinical features across 47 patients, one significant finding detected with Q value < 0.25.
-
9p gain cnv correlated to 'HISTOLOGICAL_TYPE'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 52 arm-level events and 6 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, one significant finding detected.
|
Clinical Features |
Time to Death |
YEARS TO BIRTH |
GENDER |
HISTOLOGICAL TYPE |
RACE | ETHNICITY | ||
| nCNV (%) | nWild-Type | logrank test | Wilcoxon-test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 9p gain | 7 (15%) | 40 |
0.975 (1.00) |
0.16 (0.712) |
0.684 (1.00) |
0.00053 (0.165) |
0.494 (0.964) |
0.35 (0.84) |
| 1q gain | 5 (11%) | 42 |
0.265 (0.785) |
0.277 (0.785) |
1 (1.00) |
1 (1.00) |
0.232 (0.785) |
1 (1.00) |
| 2p gain | 6 (13%) | 41 |
0.984 (1.00) |
0.1 (0.631) |
1 (1.00) |
0.111 (0.631) |
0.71 (1.00) |
0.164 (0.712) |
| 2q gain | 6 (13%) | 41 |
0.984 (1.00) |
0.1 (0.631) |
1 (1.00) |
0.111 (0.631) |
0.709 (1.00) |
0.164 (0.712) |
| 3p gain | 10 (21%) | 37 |
0.0935 (0.631) |
0.979 (1.00) |
1 (1.00) |
0.786 (1.00) |
0.591 (1.00) |
0.251 (0.785) |
| 3q gain | 13 (28%) | 34 |
0.0687 (0.596) |
0.568 (1.00) |
0.746 (1.00) |
1 (1.00) |
0.811 (1.00) |
0.269 (0.785) |
| 5p gain | 7 (15%) | 40 |
0.636 (1.00) |
0.317 (0.84) |
1 (1.00) |
0.0132 (0.415) |
0.735 (1.00) |
0.0595 (0.573) |
| 5q gain | 6 (13%) | 41 |
0.995 (1.00) |
0.156 (0.712) |
0.678 (1.00) |
0.0071 (0.313) |
1 (1.00) |
0.0301 (0.5) |
| 6p gain | 6 (13%) | 41 |
0.275 (0.785) |
0.823 (1.00) |
0.204 (0.785) |
0.641 (1.00) |
0.709 (1.00) |
0.637 (1.00) |
| 6q gain | 4 (9%) | 43 |
0.439 (0.961) |
0.954 (1.00) |
0.117 (0.631) |
1 (1.00) |
0.347 (0.84) |
1 (1.00) |
| 7p gain | 15 (32%) | 32 |
0.131 (0.692) |
0.723 (1.00) |
0.758 (1.00) |
0.217 (0.785) |
1 (1.00) |
0.481 (0.962) |
| 7q gain | 13 (28%) | 34 |
0.282 (0.792) |
0.695 (1.00) |
0.746 (1.00) |
0.318 (0.84) |
0.809 (1.00) |
0.713 (1.00) |
| 8p gain | 7 (15%) | 40 |
0.757 (1.00) |
0.0391 (0.555) |
0.436 (0.961) |
0.15 (0.712) |
0.335 (0.84) |
0.00802 (0.313) |
| 8q gain | 8 (17%) | 39 |
0.672 (1.00) |
0.0282 (0.5) |
0.715 (1.00) |
0.0209 (0.435) |
0.272 (0.785) |
0.00184 (0.287) |
| 9q gain | 7 (15%) | 40 |
0.919 (1.00) |
0.139 (0.712) |
0.217 (0.785) |
0.0133 (0.415) |
0.494 (0.964) |
0.35 (0.84) |
| 10p gain | 4 (9%) | 43 |
0.439 (0.961) |
0.954 (1.00) |
0.117 (0.631) |
1 (1.00) |
0.67 (1.00) |
1 (1.00) |
| 10q gain | 4 (9%) | 43 |
0.439 (0.961) |
0.457 (0.961) |
0.617 (1.00) |
1 (1.00) |
0.668 (1.00) |
0.266 (0.785) |
| 11p gain | 9 (19%) | 38 |
0.691 (1.00) |
0.695 (1.00) |
0.0305 (0.5) |
0.0663 (0.591) |
0.555 (1.00) |
0.0352 (0.523) |
| 11q gain | 13 (28%) | 34 |
0.71 (1.00) |
0.475 (0.962) |
0.102 (0.631) |
0.232 (0.785) |
0.655 (1.00) |
0.269 (0.785) |
| 12p gain | 7 (15%) | 40 |
0.23 (0.785) |
0.354 (0.843) |
1 (1.00) |
0.0575 (0.573) |
1 (1.00) |
0.35 (0.84) |
| 12q gain | 9 (19%) | 38 |
0.0624 (0.573) |
0.57 (1.00) |
0.486 (0.964) |
0.0203 (0.435) |
0.774 (1.00) |
0.205 (0.785) |
| 13q gain | 5 (11%) | 42 |
0.606 (1.00) |
0.342 (0.84) |
1 (1.00) |
1 (1.00) |
0.431 (0.961) |
0.59 (1.00) |
| 16p gain | 7 (15%) | 40 |
0.248 (0.785) |
0.754 (1.00) |
1 (1.00) |
0.275 (0.785) |
1 (1.00) |
1 (1.00) |
| 16q gain | 7 (15%) | 40 |
0.0943 (0.631) |
0.964 (1.00) |
0.684 (1.00) |
0.277 (0.785) |
1 (1.00) |
1 (1.00) |
| 17q gain | 3 (6%) | 44 |
0.215 (0.785) |
0.24 (0.785) |
0.242 (0.785) |
0.39 (0.914) |
0.114 (0.631) |
0.56 (1.00) |
| 18p gain | 13 (28%) | 34 |
0.0587 (0.573) |
0.295 (0.815) |
0.746 (1.00) |
0.804 (1.00) |
0.112 (0.631) |
0.713 (1.00) |
| 18q gain | 14 (30%) | 33 |
0.218 (0.785) |
0.522 (1.00) |
1 (1.00) |
0.804 (1.00) |
0.0591 (0.573) |
0.731 (1.00) |
| 19p gain | 3 (6%) | 44 |
0.459 (0.961) |
0.0895 (0.631) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.156 (0.712) |
| 19q gain | 3 (6%) | 44 |
0.459 (0.961) |
0.0895 (0.631) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.156 (0.712) |
| 20p gain | 5 (11%) | 42 |
0.791 (1.00) |
0.641 (1.00) |
0.0561 (0.573) |
0.573 (1.00) |
0.433 (0.961) |
0.59 (1.00) |
| 20q gain | 4 (9%) | 43 |
0.6 (1.00) |
0.26 (0.785) |
0.117 (0.631) |
0.488 (0.964) |
0.669 (1.00) |
0.266 (0.785) |
| 21q gain | 10 (21%) | 37 |
0.842 (1.00) |
0.317 (0.84) |
0.475 (0.962) |
0.285 (0.795) |
0.777 (1.00) |
0.7 (1.00) |
| xp gain | 6 (13%) | 41 |
0.985 (1.00) |
0.655 (1.00) |
1 (1.00) |
0.0189 (0.435) |
1 (1.00) |
0.0301 (0.5) |
| xq gain | 6 (13%) | 41 |
0.924 (1.00) |
0.632 (1.00) |
1 (1.00) |
0.0193 (0.435) |
0.708 (1.00) |
0.164 (0.712) |
| 1p loss | 3 (6%) | 44 |
0.459 (0.961) |
0.384 (0.907) |
0.579 (1.00) |
0.198 (0.785) |
1 (1.00) |
1 (1.00) |
| 3p loss | 5 (11%) | 42 |
0.444 (0.961) |
0.262 (0.785) |
1 (1.00) |
0.154 (0.712) |
0.23 (0.785) |
0.0971 (0.631) |
| 3q loss | 4 (9%) | 43 |
0.444 (0.961) |
0.35 (0.84) |
1 (1.00) |
0.0985 (0.631) |
0.217 (0.785) |
0.0459 (0.573) |
| 4p loss | 3 (6%) | 44 |
0.349 (0.84) |
0.242 (0.785) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
|
| 4q loss | 4 (9%) | 43 |
0.534 (1.00) |
0.469 (0.962) |
0.117 (0.631) |
1 (1.00) |
0.67 (1.00) |
1 (1.00) |
| 6q loss | 7 (15%) | 40 |
0.769 (1.00) |
0.811 (1.00) |
1 (1.00) |
0.0884 (0.631) |
0.336 (0.84) |
0.0595 (0.573) |
| 8p loss | 8 (17%) | 39 |
0.955 (1.00) |
0.799 (1.00) |
0.269 (0.785) |
0.342 (0.84) |
0.747 (1.00) |
0.403 (0.931) |
| 8q loss | 4 (9%) | 43 |
0.459 (0.961) |
0.153 (0.712) |
0.617 (1.00) |
0.266 (0.785) |
1 (1.00) |
0.266 (0.785) |
| 13q loss | 3 (6%) | 44 |
0.571 (1.00) |
0.879 (1.00) |
1 (1.00) |
0.0533 (0.573) |
0.313 (0.84) |
0.156 (0.712) |
| 15q loss | 7 (15%) | 40 |
0.257 (0.785) |
0.765 (1.00) |
0.112 (0.631) |
1 (1.00) |
1 (1.00) |
0.659 (1.00) |
| 16q loss | 4 (9%) | 43 |
0.478 (0.962) |
0.909 (1.00) |
0.117 (0.631) |
1 (1.00) |
0.346 (0.84) |
0.56 (1.00) |
| 17p loss | 9 (19%) | 38 |
0.201 (0.785) |
0.871 (1.00) |
0.16 (0.712) |
0.0204 (0.435) |
0.554 (1.00) |
0.0048 (0.299) |
| 17q loss | 4 (9%) | 43 |
0.473 (0.962) |
0.504 (0.977) |
1 (1.00) |
0.0039 (0.299) |
0.216 (0.785) |
0.00278 (0.289) |
| 18p loss | 5 (11%) | 42 |
0.648 (1.00) |
0.863 (1.00) |
0.644 (1.00) |
0.332 (0.84) |
1 (1.00) |
0.59 (1.00) |
| 18q loss | 4 (9%) | 43 |
0.399 (0.929) |
0.0327 (0.51) |
0.617 (1.00) |
0.266 (0.785) |
0.216 (0.785) |
0.0459 (0.573) |
| 22q loss | 3 (6%) | 44 |
0.527 (1.00) |
0.794 (1.00) |
0.242 (0.785) |
1 (1.00) |
0.115 (0.631) |
0.56 (1.00) |
| xp loss | 5 (11%) | 42 |
0.0624 (0.573) |
0.468 (0.962) |
0.0561 (0.573) |
1 (1.00) |
0.434 (0.961) |
1 (1.00) |
| xq loss | 4 (9%) | 43 |
0.00594 (0.309) |
1 (1.00) |
0.117 (0.631) |
1 (1.00) |
0.345 (0.84) |
0.56 (1.00) |
P value = 0.00053 (Fisher's exact test), Q value = 0.17
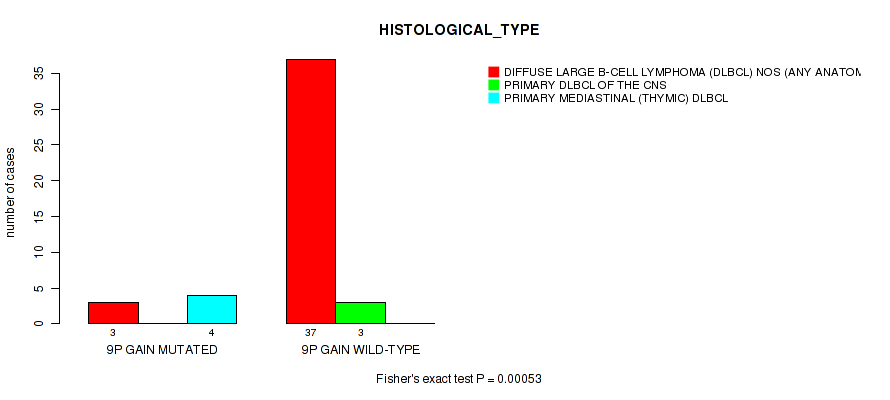
Table S1. Gene #14: '9p gain' versus Clinical Feature #4: 'HISTOLOGICAL_TYPE'
| nPatients | DIFFUSE LARGE B-CELL LYMPHOMA (DLBCL) NOS (ANY ANATOMIC SITE NODAL OR EXTRANODAL) | PRIMARY DLBCL OF THE CNS | PRIMARY MEDIASTINAL (THYMIC) DLBCL |
|---|---|---|---|
| ALL | 40 | 3 | 4 |
| 9P GAIN MUTATED | 3 | 0 | 4 |
| 9P GAIN WILD-TYPE | 37 | 3 | 0 |
Figure S1. Get High-res Image Gene #14: '9p gain' versus Clinical Feature #4: 'HISTOLOGICAL_TYPE'
-
Copy number data file = broad_values_by_arm.txt from GISTIC pipeline
-
Processed Copy number data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_Correlate_Genomic_Events_Preprocess/DLBC-TP/15082594/transformed.cor.cli.txt
-
Clinical data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/Append_Data/DLBC-TP/15078461/DLBC-TP.merged_data.txt
-
Number of patients = 47
-
Number of significantly arm-level cnvs = 52
-
Number of selected clinical features = 6
-
Exclude regions that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.