This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and molecular subtypes.
Testing the association between copy number variation 32 arm-level events and 6 molecular subtypes across 191 patients, 58 significant findings detected with P value < 0.05 and Q value < 0.25.
-
1p gain cnv correlated to 'MIRSEQ_CNMF'.
-
4p gain cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
4q gain cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
8p gain cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
8q gain cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
11p gain cnv correlated to 'MIRSEQ_CNMF' and 'MIRSEQ_CHIERARCHICAL'.
-
11q gain cnv correlated to 'MIRSEQ_CNMF' and 'MIRSEQ_CHIERARCHICAL'.
-
13q gain cnv correlated to 'METHLYATION_CNMF' and 'MIRSEQ_CNMF'.
-
21q gain cnv correlated to 'MIRSEQ_CNMF'.
-
22q gain cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
xp gain cnv correlated to 'MIRSEQ_CHIERARCHICAL'.
-
xq gain cnv correlated to 'MIRSEQ_CHIERARCHICAL'.
-
3p loss cnv correlated to 'CN_CNMF'.
-
3q loss cnv correlated to 'CN_CNMF'.
-
5q loss cnv correlated to 'METHLYATION_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
7p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
7q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
16q loss cnv correlated to 'CN_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
17p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
17q loss cnv correlated to 'MIRSEQ_CHIERARCHICAL'.
-
18p loss cnv correlated to 'CN_CNMF'.
-
xp loss cnv correlated to 'CN_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
xq loss cnv correlated to 'CN_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 32 arm-level events and 6 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 58 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 7p loss | 17 (9%) | 174 |
1e-05 (0.000384) |
0.00042 (0.0134) |
0.00111 (0.0194) |
1e-05 (0.000384) |
0.00658 (0.0486) |
0.00293 (0.0316) |
| 7q loss | 20 (10%) | 171 |
1e-05 (0.000384) |
1e-05 (0.000384) |
0.00978 (0.0626) |
1e-05 (0.000384) |
0.00296 (0.0316) |
0.0008 (0.0171) |
| 8p gain | 22 (12%) | 169 |
0.959 (1.00) |
0.00223 (0.0268) |
0.0273 (0.109) |
0.00076 (0.0171) |
0.0683 (0.212) |
0.0165 (0.0835) |
| 8q gain | 23 (12%) | 168 |
0.886 (0.989) |
0.00558 (0.045) |
0.0141 (0.08) |
0.00182 (0.0233) |
0.0618 (0.198) |
0.0142 (0.08) |
| 22q gain | 9 (5%) | 182 |
0.239 (0.455) |
0.0121 (0.0749) |
0.148 (0.345) |
0.0163 (0.0835) |
0.00067 (0.0171) |
0.0136 (0.08) |
| 17p loss | 13 (7%) | 178 |
0.0171 (0.0843) |
0.00356 (0.0344) |
0.171 (0.377) |
0.00157 (0.0233) |
0.195 (0.407) |
0.00358 (0.0344) |
| 5q loss | 6 (3%) | 185 |
0.302 (0.523) |
0.0223 (0.101) |
0.228 (0.452) |
0.632 (0.795) |
0.0325 (0.122) |
0.032 (0.122) |
| 16q loss | 4 (2%) | 187 |
0.0357 (0.128) |
0.161 (0.361) |
0.789 (0.925) |
1 (1.00) |
0.0237 (0.101) |
0.023 (0.101) |
| xp loss | 5 (3%) | 186 |
0.00926 (0.0626) |
0.308 (0.523) |
0.23 (0.452) |
0.633 (0.795) |
0.00563 (0.045) |
0.00504 (0.044) |
| xq loss | 5 (3%) | 186 |
0.00945 (0.0626) |
0.307 (0.523) |
0.228 (0.452) |
0.631 (0.795) |
0.00598 (0.0459) |
0.00495 (0.044) |
| 4p gain | 4 (2%) | 187 |
0.0359 (0.128) |
0.0259 (0.108) |
0.592 (0.758) |
0.835 (0.96) |
||
| 4q gain | 4 (2%) | 187 |
0.0354 (0.128) |
0.0265 (0.108) |
0.593 (0.758) |
0.833 (0.96) |
||
| 11p gain | 5 (3%) | 186 |
0.443 (0.645) |
0.306 (0.523) |
0.231 (0.452) |
0.235 (0.452) |
0.0151 (0.0807) |
0.0321 (0.122) |
| 11q gain | 7 (4%) | 184 |
0.233 (0.452) |
0.0728 (0.221) |
0.0767 (0.223) |
0.0747 (0.221) |
0.00177 (0.0233) |
0.015 (0.0807) |
| 13q gain | 6 (3%) | 185 |
0.423 (0.629) |
0.0238 (0.101) |
0.149 (0.345) |
0.0832 (0.235) |
0.00182 (0.0233) |
0.0736 (0.221) |
| 1p gain | 3 (2%) | 188 |
0.56 (0.742) |
0.336 (0.555) |
0.261 (0.486) |
0.588 (0.758) |
0.00105 (0.0194) |
0.0919 (0.245) |
| 21q gain | 8 (4%) | 183 |
0.126 (0.31) |
0.215 (0.445) |
0.149 (0.345) |
0.525 (0.725) |
0.0498 (0.165) |
0.409 (0.62) |
| xp gain | 3 (2%) | 188 |
0.794 (0.925) |
0.479 (0.672) |
0.443 (0.645) |
0.046 (0.159) |
||
| xq gain | 3 (2%) | 188 |
0.795 (0.925) |
0.478 (0.672) |
0.442 (0.645) |
0.047 (0.159) |
||
| 3p loss | 3 (2%) | 188 |
0.0227 (0.101) |
0.338 (0.555) |
0.787 (0.925) |
1 (1.00) |
0.101 (0.255) |
0.0921 (0.245) |
| 3q loss | 3 (2%) | 188 |
0.0217 (0.101) |
0.336 (0.555) |
0.785 (0.925) |
1 (1.00) |
0.101 (0.255) |
0.0932 (0.245) |
| 17q loss | 7 (4%) | 184 |
0.0858 (0.239) |
0.149 (0.345) |
0.0782 (0.224) |
0.236 (0.452) |
0.119 (0.297) |
0.0471 (0.159) |
| 18p loss | 5 (3%) | 186 |
0.00909 (0.0626) |
0.0647 (0.204) |
1 (1.00) |
0.865 (0.972) |
0.269 (0.486) |
0.185 (0.392) |
| 10q gain | 3 (2%) | 188 |
0.447 (0.645) |
0.337 (0.555) |
0.262 (0.486) |
0.587 (0.758) |
0.159 (0.361) |
0.093 (0.245) |
| 17q gain | 3 (2%) | 188 |
0.795 (0.925) |
0.783 (0.925) |
1 (1.00) |
0.589 (0.758) |
0.664 (0.827) |
0.303 (0.523) |
| 19p gain | 5 (3%) | 186 |
0.399 (0.62) |
0.265 (0.486) |
1 (1.00) |
0.865 (0.972) |
0.409 (0.62) |
1 (1.00) |
| 19q gain | 5 (3%) | 186 |
0.397 (0.62) |
0.267 (0.486) |
1 (1.00) |
0.866 (0.972) |
0.41 (0.62) |
1 (1.00) |
| 12p loss | 4 (2%) | 187 |
0.538 (0.728) |
0.502 (0.698) |
0.381 (0.614) |
0.389 (0.62) |
0.398 (0.62) |
0.184 (0.392) |
| 15q loss | 4 (2%) | 187 |
0.539 (0.728) |
0.45 (0.645) |
0.0968 (0.251) |
0.369 (0.601) |
0.143 (0.345) |
0.185 (0.392) |
| 18q loss | 4 (2%) | 187 |
0.538 (0.728) |
0.162 (0.361) |
0.475 (0.672) |
0.864 (0.972) |
0.0617 (0.198) |
0.186 (0.392) |
| 19p loss | 3 (2%) | 188 |
0.56 (0.742) |
0.783 (0.925) |
0.787 (0.925) |
1 (1.00) |
0.283 (0.503) |
0.413 (0.62) |
| 19q loss | 3 (2%) | 188 |
0.559 (0.742) |
0.783 (0.925) |
0.787 (0.925) |
1 (1.00) |
0.283 (0.503) |
0.413 (0.62) |
P value = 0.00105 (Fisher's exact test), Q value = 0.019
Table S1. Gene #1: '1p gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 57 | 31 | 39 | 33 | 19 |
| 1P GAIN MUTATED | 0 | 0 | 0 | 0 | 3 |
| 1P GAIN WILD-TYPE | 57 | 31 | 39 | 33 | 16 |
Figure S1. Get High-res Image Gene #1: '1p gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
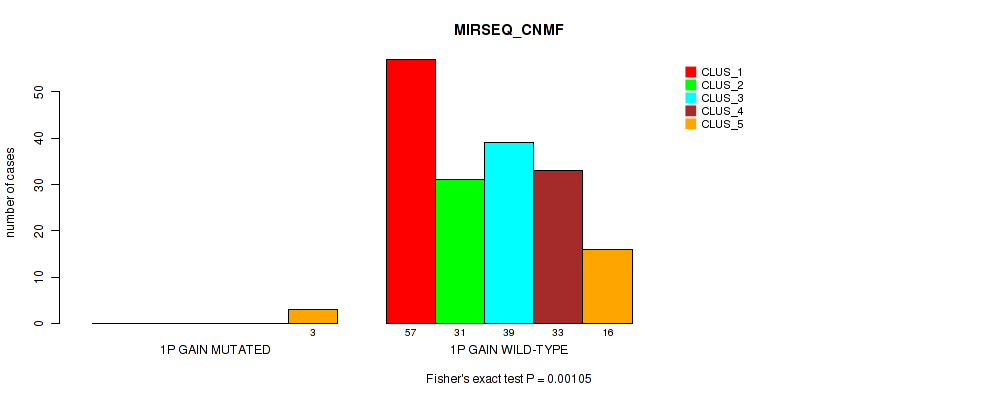
P value = 0.0359 (Fisher's exact test), Q value = 0.13
Table S2. Gene #2: '4p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 44 | 55 | 43 |
| 4P GAIN MUTATED | 0 | 3 | 0 | 1 |
| 4P GAIN WILD-TYPE | 49 | 41 | 55 | 42 |
Figure S2. Get High-res Image Gene #2: '4p gain' versus Molecular Subtype #1: 'CN_CNMF'
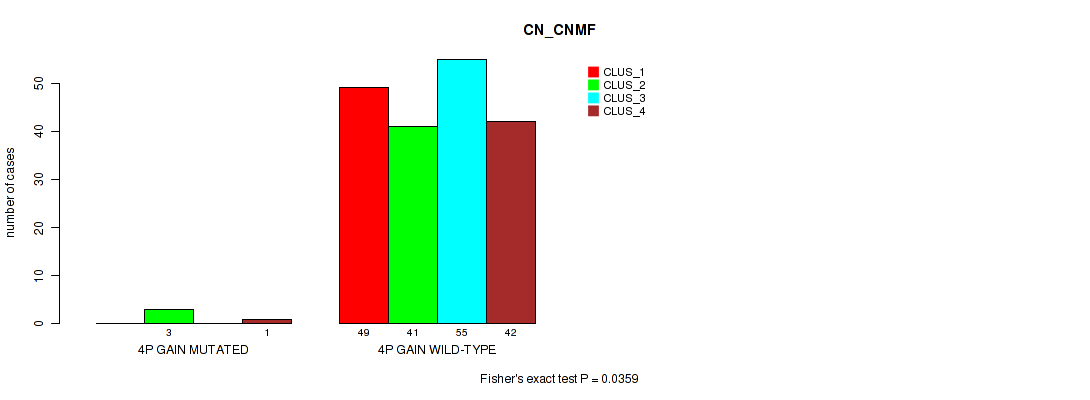
P value = 0.0259 (Fisher's exact test), Q value = 0.11
Table S3. Gene #2: '4p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 23 | 25 | 60 | 35 | 42 |
| 4P GAIN MUTATED | 2 | 0 | 0 | 2 | 0 |
| 4P GAIN WILD-TYPE | 21 | 25 | 60 | 33 | 42 |
Figure S3. Get High-res Image Gene #2: '4p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
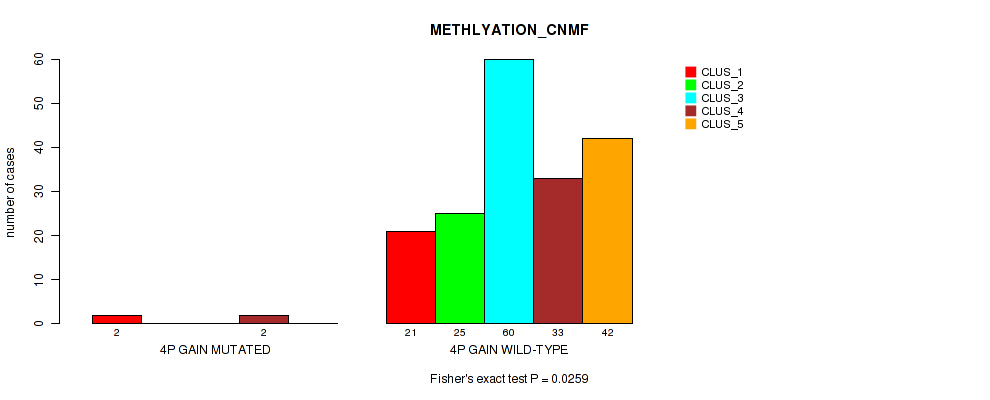
P value = 0.0354 (Fisher's exact test), Q value = 0.13
Table S4. Gene #3: '4q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 44 | 55 | 43 |
| 4Q GAIN MUTATED | 0 | 3 | 0 | 1 |
| 4Q GAIN WILD-TYPE | 49 | 41 | 55 | 42 |
Figure S4. Get High-res Image Gene #3: '4q gain' versus Molecular Subtype #1: 'CN_CNMF'
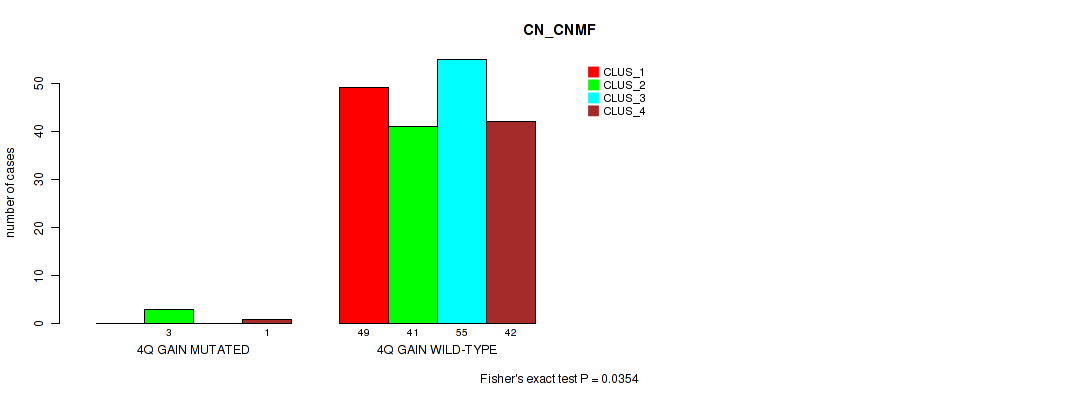
P value = 0.0265 (Fisher's exact test), Q value = 0.11
Table S5. Gene #3: '4q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 23 | 25 | 60 | 35 | 42 |
| 4Q GAIN MUTATED | 2 | 0 | 0 | 2 | 0 |
| 4Q GAIN WILD-TYPE | 21 | 25 | 60 | 33 | 42 |
Figure S5. Get High-res Image Gene #3: '4q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
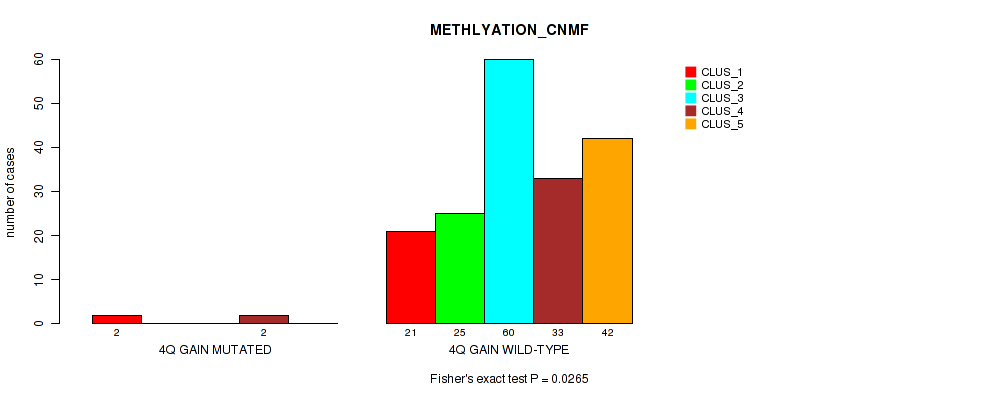
P value = 0.00223 (Fisher's exact test), Q value = 0.027
Table S6. Gene #4: '8p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 23 | 25 | 60 | 35 | 42 |
| 8P GAIN MUTATED | 2 | 2 | 15 | 1 | 1 |
| 8P GAIN WILD-TYPE | 21 | 23 | 45 | 34 | 41 |
Figure S6. Get High-res Image Gene #4: '8p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
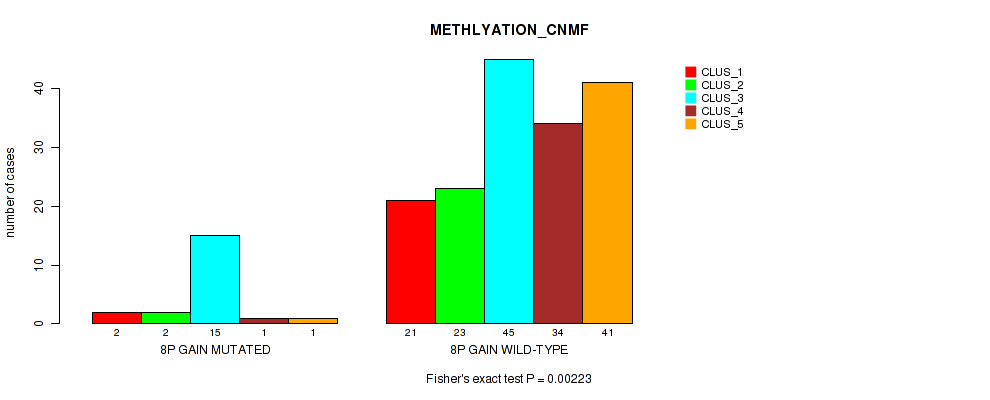
P value = 0.0273 (Fisher's exact test), Q value = 0.11
Table S7. Gene #4: '8p gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 71 | 50 | 45 |
| 8P GAIN MUTATED | 13 | 2 | 3 |
| 8P GAIN WILD-TYPE | 58 | 48 | 42 |
Figure S7. Get High-res Image Gene #4: '8p gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
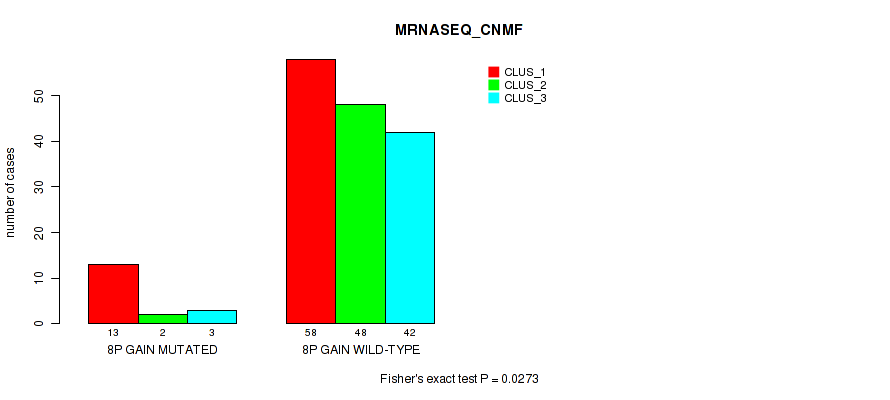
P value = 0.00076 (Fisher's exact test), Q value = 0.017
Table S8. Gene #4: '8p gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| 8P GAIN MUTATED | 1 | 0 | 0 | 15 | 1 | 1 |
| 8P GAIN WILD-TYPE | 15 | 13 | 14 | 40 | 25 | 41 |
Figure S8. Get High-res Image Gene #4: '8p gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
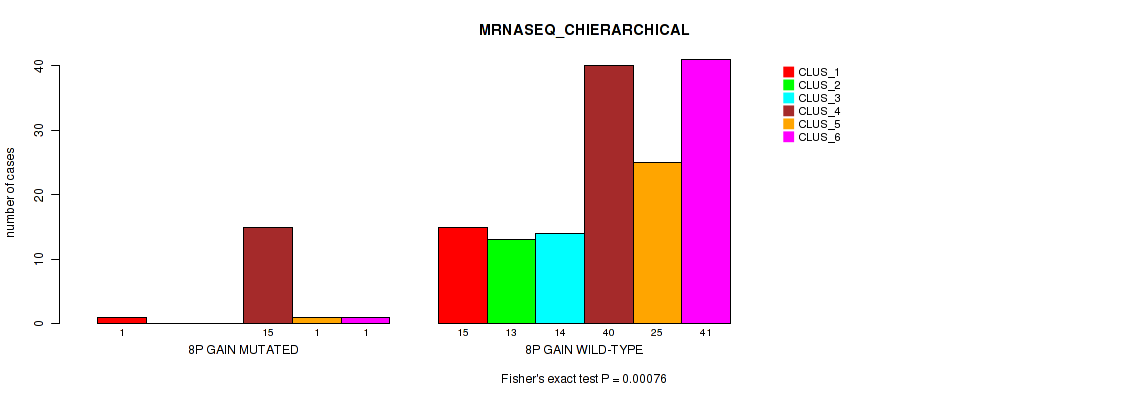
P value = 0.0165 (Fisher's exact test), Q value = 0.083
Table S9. Gene #4: '8p gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 8P GAIN MUTATED | 3 | 8 | 10 |
| 8P GAIN WILD-TYPE | 64 | 26 | 68 |
Figure S9. Get High-res Image Gene #4: '8p gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
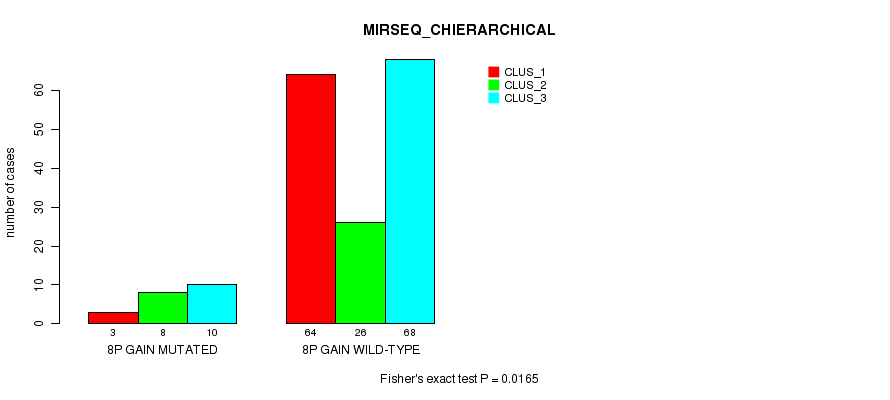
P value = 0.00558 (Fisher's exact test), Q value = 0.045
Table S10. Gene #5: '8q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 23 | 25 | 60 | 35 | 42 |
| 8Q GAIN MUTATED | 2 | 2 | 15 | 2 | 1 |
| 8Q GAIN WILD-TYPE | 21 | 23 | 45 | 33 | 41 |
Figure S10. Get High-res Image Gene #5: '8q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
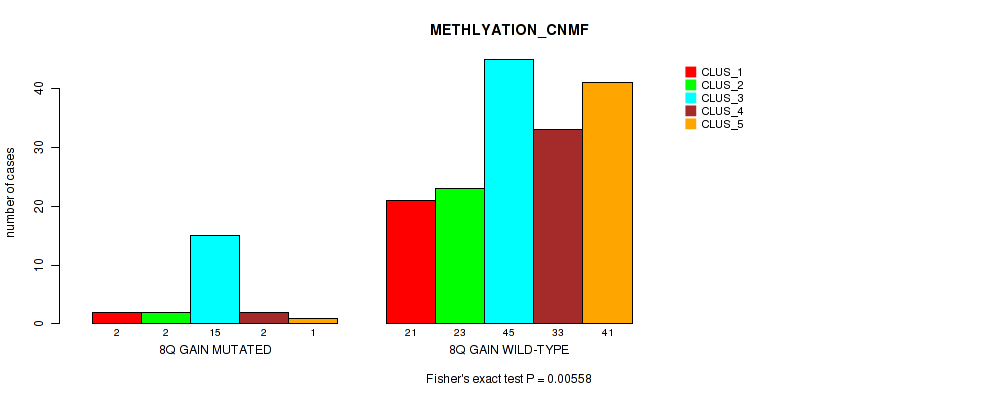
P value = 0.0141 (Fisher's exact test), Q value = 0.08
Table S11. Gene #5: '8q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 71 | 50 | 45 |
| 8Q GAIN MUTATED | 14 | 2 | 3 |
| 8Q GAIN WILD-TYPE | 57 | 48 | 42 |
Figure S11. Get High-res Image Gene #5: '8q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
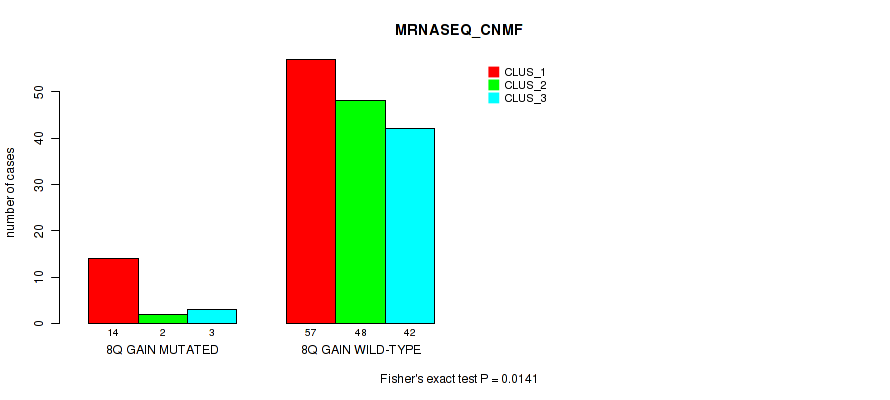
P value = 0.00182 (Fisher's exact test), Q value = 0.023
Table S12. Gene #5: '8q gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| 8Q GAIN MUTATED | 1 | 0 | 1 | 15 | 1 | 1 |
| 8Q GAIN WILD-TYPE | 15 | 13 | 13 | 40 | 25 | 41 |
Figure S12. Get High-res Image Gene #5: '8q gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
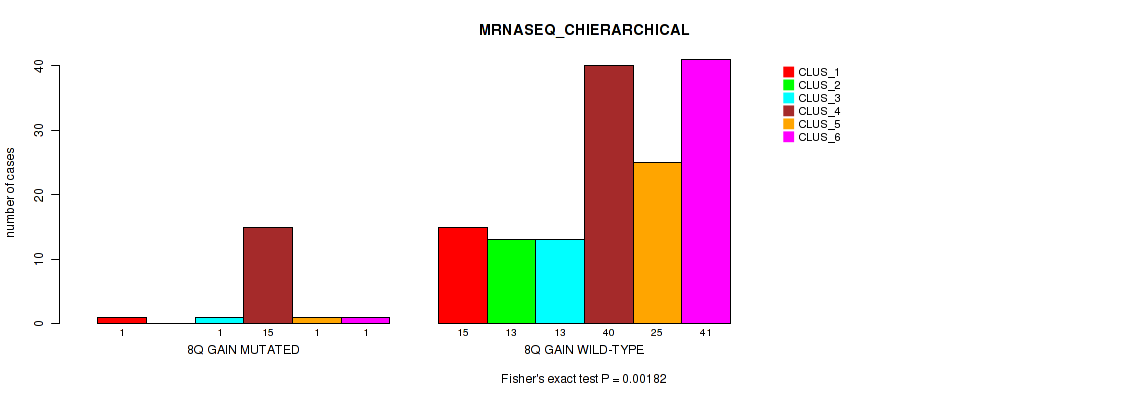
P value = 0.0142 (Fisher's exact test), Q value = 0.08
Table S13. Gene #5: '8q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 8Q GAIN MUTATED | 3 | 8 | 11 |
| 8Q GAIN WILD-TYPE | 64 | 26 | 67 |
Figure S13. Get High-res Image Gene #5: '8q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
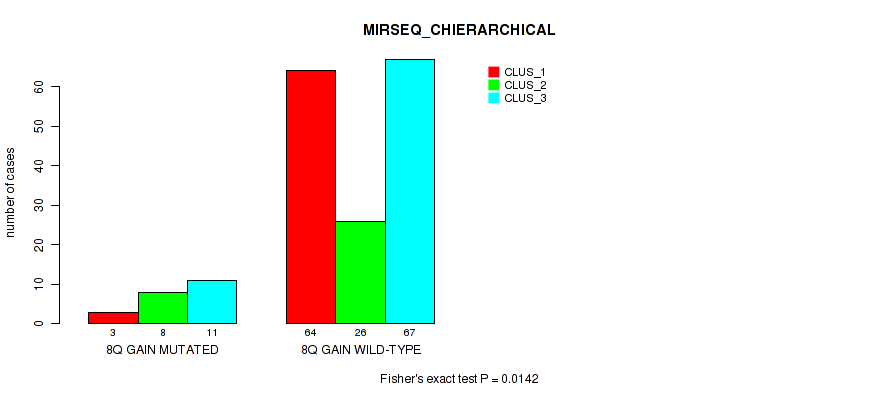
P value = 0.0151 (Fisher's exact test), Q value = 0.081
Table S14. Gene #7: '11p gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 57 | 31 | 39 | 33 | 19 |
| 11P GAIN MUTATED | 1 | 1 | 0 | 0 | 3 |
| 11P GAIN WILD-TYPE | 56 | 30 | 39 | 33 | 16 |
Figure S14. Get High-res Image Gene #7: '11p gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
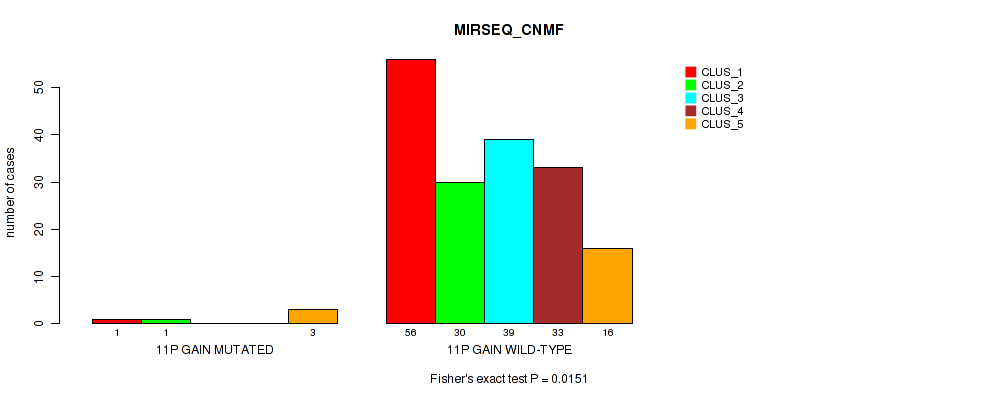
P value = 0.0321 (Fisher's exact test), Q value = 0.12
Table S15. Gene #7: '11p gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 11P GAIN MUTATED | 0 | 3 | 2 |
| 11P GAIN WILD-TYPE | 67 | 31 | 76 |
Figure S15. Get High-res Image Gene #7: '11p gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
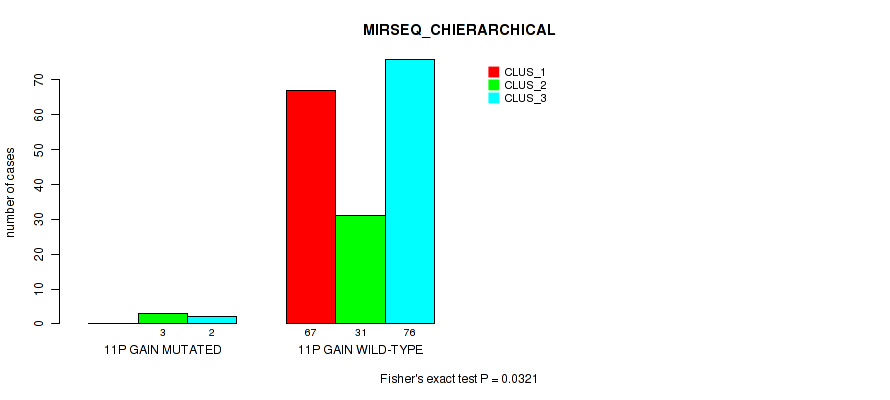
P value = 0.00177 (Fisher's exact test), Q value = 0.023
Table S16. Gene #8: '11q gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 57 | 31 | 39 | 33 | 19 |
| 11Q GAIN MUTATED | 1 | 2 | 0 | 0 | 4 |
| 11Q GAIN WILD-TYPE | 56 | 29 | 39 | 33 | 15 |
Figure S16. Get High-res Image Gene #8: '11q gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
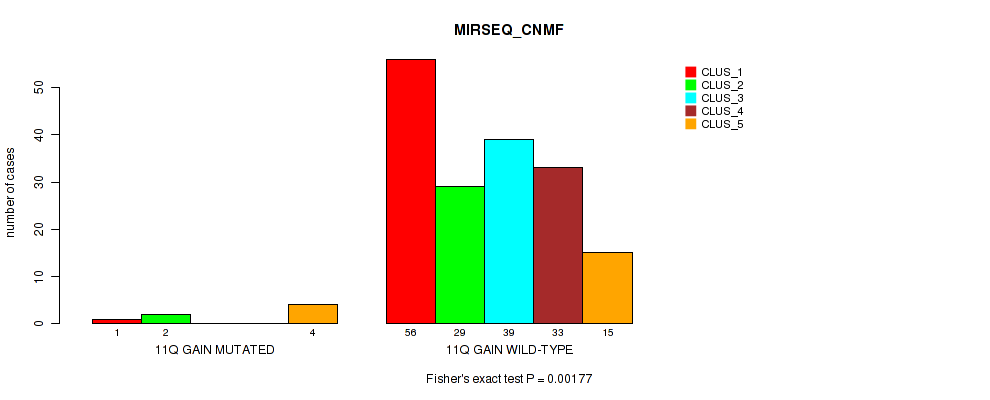
P value = 0.015 (Fisher's exact test), Q value = 0.081
Table S17. Gene #8: '11q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 11Q GAIN MUTATED | 0 | 4 | 3 |
| 11Q GAIN WILD-TYPE | 67 | 30 | 75 |
Figure S17. Get High-res Image Gene #8: '11q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
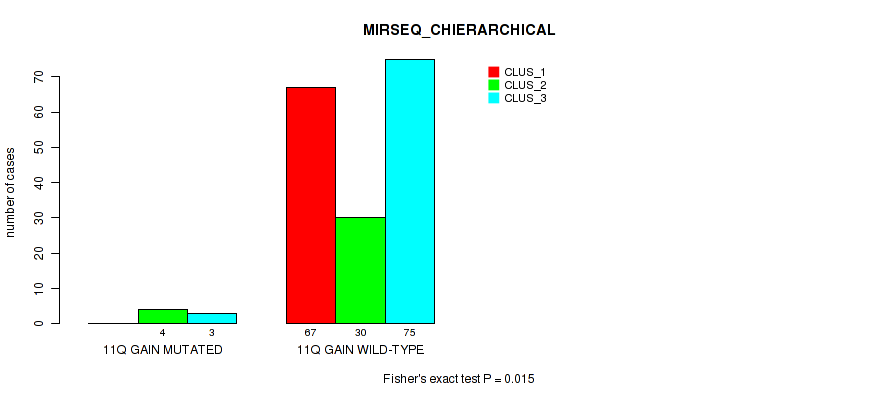
P value = 0.0238 (Fisher's exact test), Q value = 0.1
Table S18. Gene #9: '13q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 23 | 25 | 60 | 35 | 42 |
| 13Q GAIN MUTATED | 0 | 0 | 6 | 0 | 0 |
| 13Q GAIN WILD-TYPE | 23 | 25 | 54 | 35 | 42 |
Figure S18. Get High-res Image Gene #9: '13q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
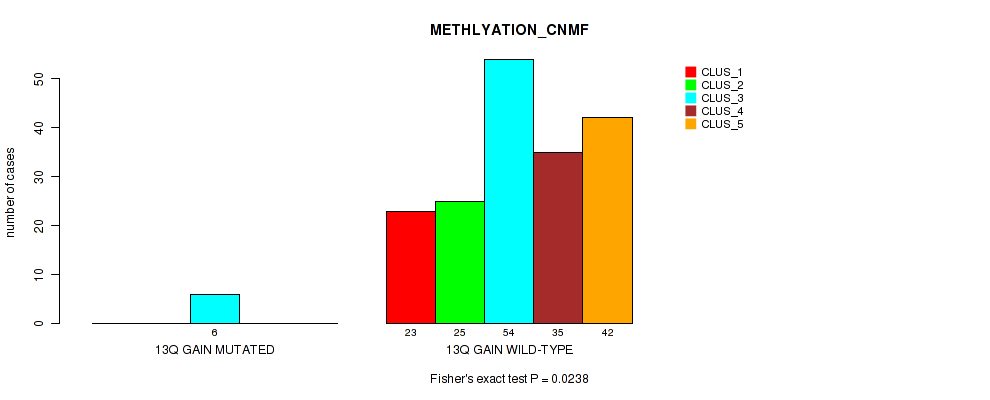
P value = 0.00182 (Fisher's exact test), Q value = 0.023
Table S19. Gene #9: '13q gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 57 | 31 | 39 | 33 | 19 |
| 13Q GAIN MUTATED | 1 | 1 | 0 | 0 | 4 |
| 13Q GAIN WILD-TYPE | 56 | 30 | 39 | 33 | 15 |
Figure S19. Get High-res Image Gene #9: '13q gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
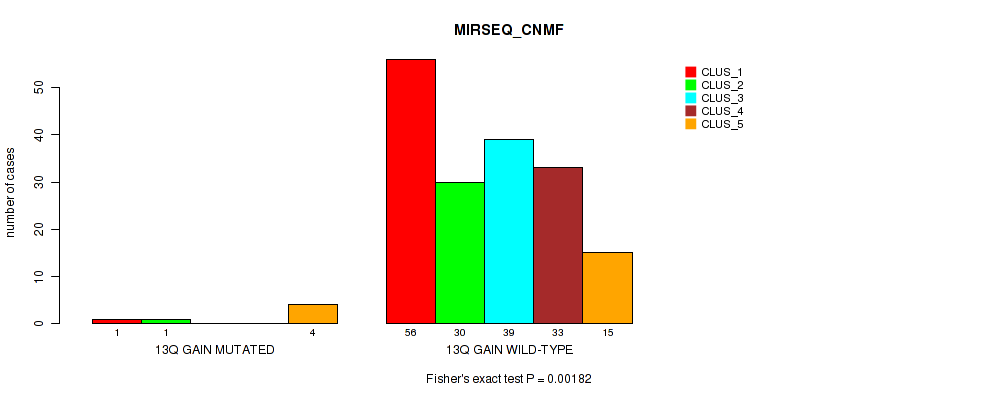
P value = 0.0498 (Fisher's exact test), Q value = 0.16
Table S20. Gene #13: '21q gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 57 | 31 | 39 | 33 | 19 |
| 21Q GAIN MUTATED | 1 | 2 | 0 | 4 | 0 |
| 21Q GAIN WILD-TYPE | 56 | 29 | 39 | 29 | 19 |
Figure S20. Get High-res Image Gene #13: '21q gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
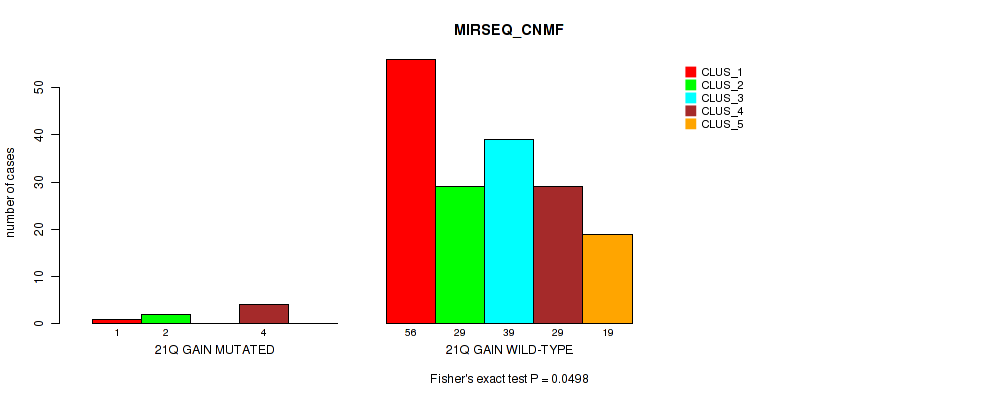
P value = 0.0121 (Fisher's exact test), Q value = 0.075
Table S21. Gene #14: '22q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 23 | 25 | 60 | 35 | 42 |
| 22Q GAIN MUTATED | 0 | 0 | 8 | 1 | 0 |
| 22Q GAIN WILD-TYPE | 23 | 25 | 52 | 34 | 42 |
Figure S21. Get High-res Image Gene #14: '22q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
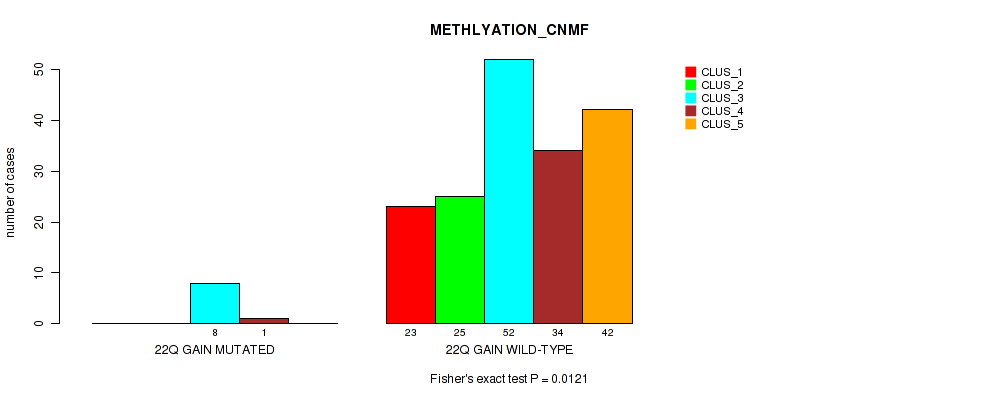
P value = 0.0163 (Fisher's exact test), Q value = 0.083
Table S22. Gene #14: '22q gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| 22Q GAIN MUTATED | 0 | 1 | 0 | 8 | 0 | 0 |
| 22Q GAIN WILD-TYPE | 16 | 12 | 14 | 47 | 26 | 42 |
Figure S22. Get High-res Image Gene #14: '22q gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
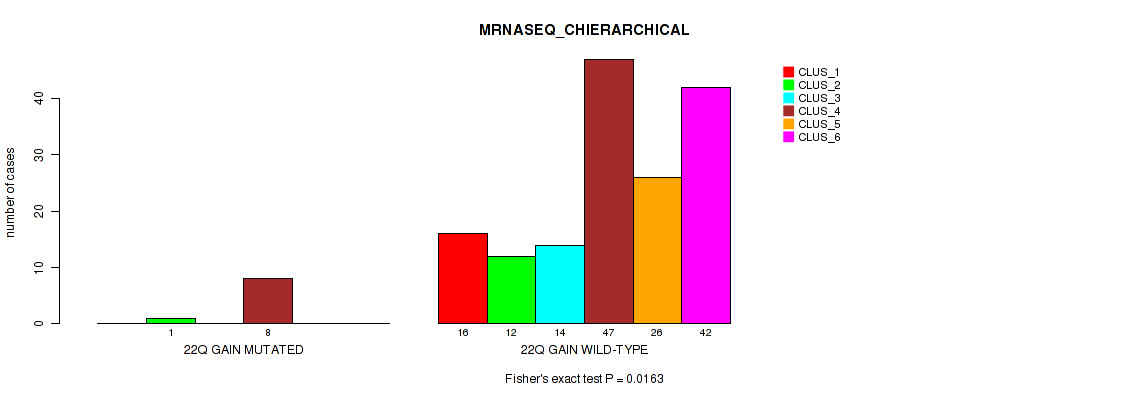
P value = 0.00067 (Fisher's exact test), Q value = 0.017
Table S23. Gene #14: '22q gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 57 | 31 | 39 | 33 | 19 |
| 22Q GAIN MUTATED | 0 | 2 | 1 | 1 | 5 |
| 22Q GAIN WILD-TYPE | 57 | 29 | 38 | 32 | 14 |
Figure S23. Get High-res Image Gene #14: '22q gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
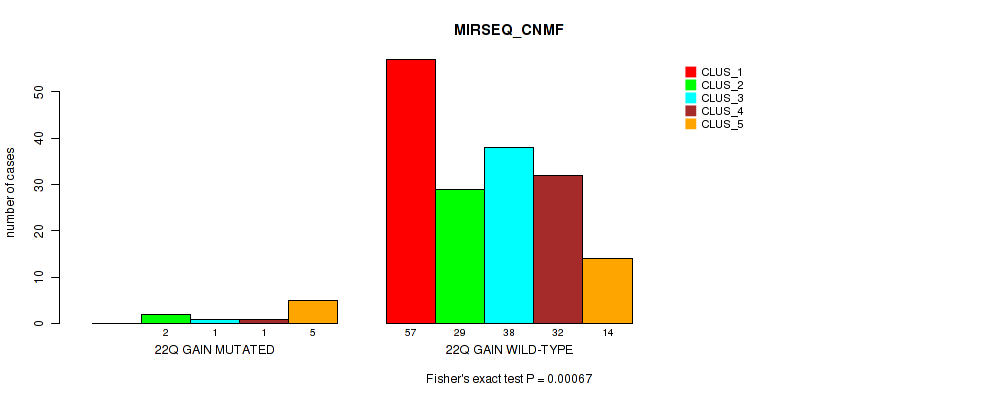
P value = 0.0136 (Fisher's exact test), Q value = 0.08
Table S24. Gene #14: '22q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 22Q GAIN MUTATED | 0 | 4 | 5 |
| 22Q GAIN WILD-TYPE | 67 | 30 | 73 |
Figure S24. Get High-res Image Gene #14: '22q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
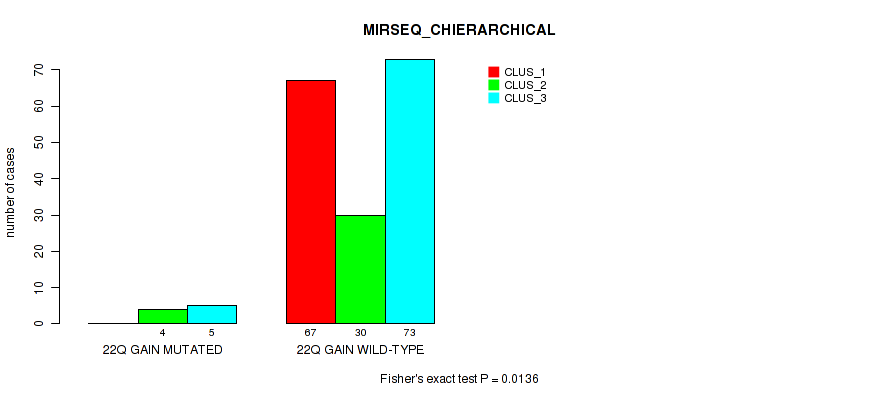
P value = 0.046 (Fisher's exact test), Q value = 0.16
Table S25. Gene #15: 'xp gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| XP GAIN MUTATED | 1 | 2 | 0 |
| XP GAIN WILD-TYPE | 66 | 32 | 78 |
Figure S25. Get High-res Image Gene #15: 'xp gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
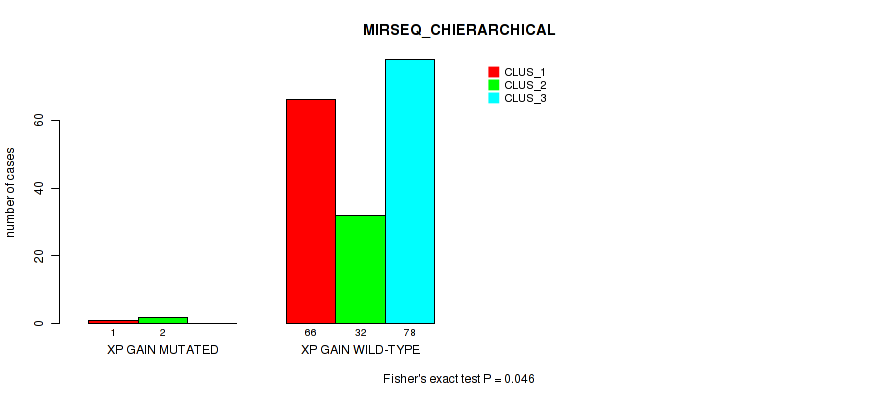
P value = 0.047 (Fisher's exact test), Q value = 0.16
Table S26. Gene #16: 'xq gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| XQ GAIN MUTATED | 1 | 2 | 0 |
| XQ GAIN WILD-TYPE | 66 | 32 | 78 |
Figure S26. Get High-res Image Gene #16: 'xq gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
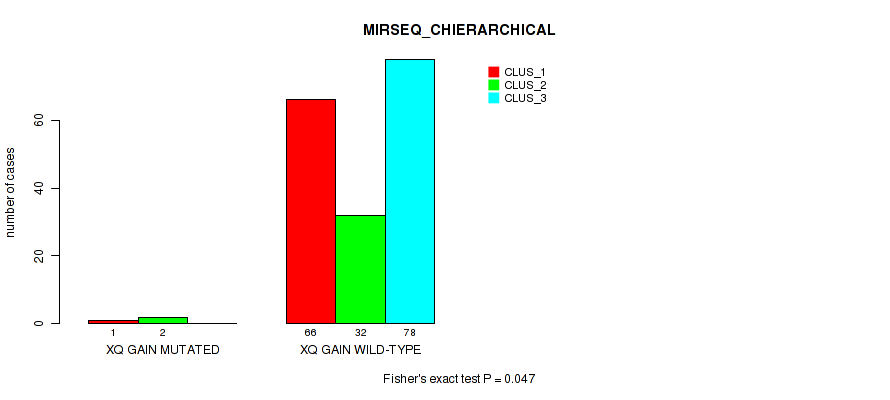
P value = 0.0227 (Fisher's exact test), Q value = 0.1
Table S27. Gene #17: '3p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 44 | 55 | 43 |
| 3P LOSS MUTATED | 0 | 3 | 0 | 0 |
| 3P LOSS WILD-TYPE | 49 | 41 | 55 | 43 |
Figure S27. Get High-res Image Gene #17: '3p loss' versus Molecular Subtype #1: 'CN_CNMF'
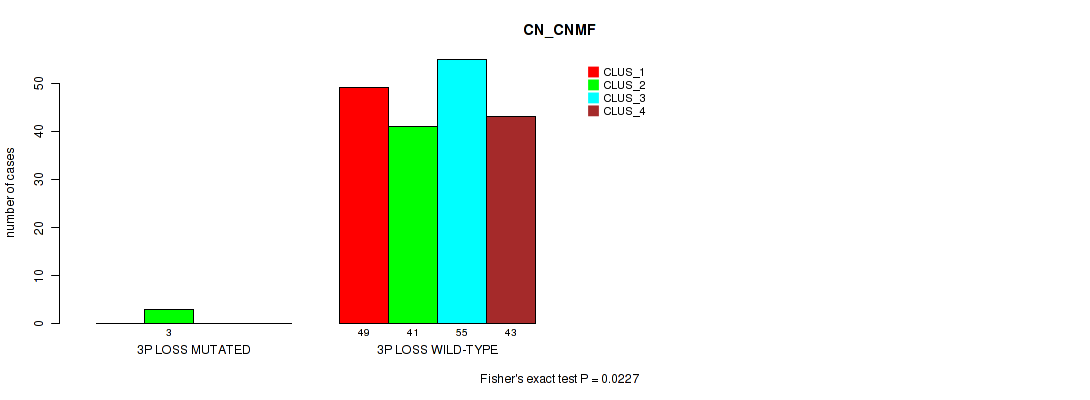
P value = 0.0217 (Fisher's exact test), Q value = 0.1
Table S28. Gene #18: '3q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 44 | 55 | 43 |
| 3Q LOSS MUTATED | 0 | 3 | 0 | 0 |
| 3Q LOSS WILD-TYPE | 49 | 41 | 55 | 43 |
Figure S28. Get High-res Image Gene #18: '3q loss' versus Molecular Subtype #1: 'CN_CNMF'
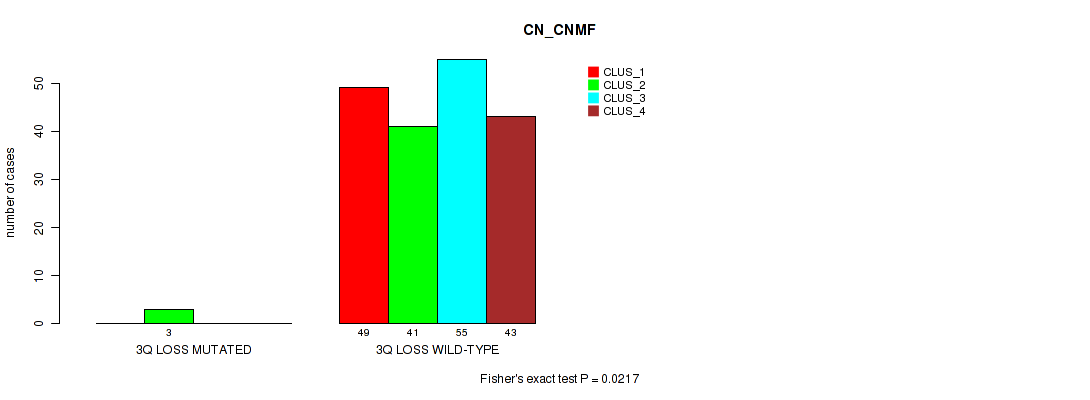
P value = 0.0223 (Fisher's exact test), Q value = 0.1
Table S29. Gene #19: '5q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 23 | 25 | 60 | 35 | 42 |
| 5Q LOSS MUTATED | 0 | 0 | 6 | 0 | 0 |
| 5Q LOSS WILD-TYPE | 23 | 25 | 54 | 35 | 42 |
Figure S29. Get High-res Image Gene #19: '5q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
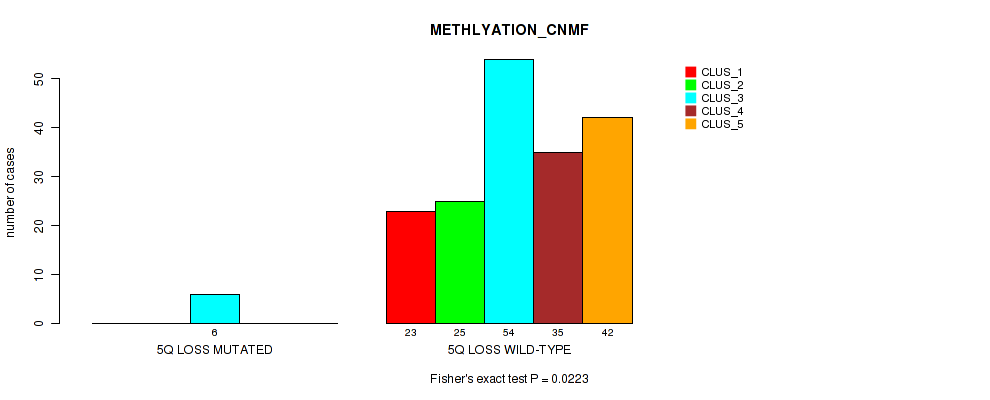
P value = 0.0325 (Fisher's exact test), Q value = 0.12
Table S30. Gene #19: '5q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 57 | 31 | 39 | 33 | 19 |
| 5Q LOSS MUTATED | 0 | 3 | 0 | 1 | 1 |
| 5Q LOSS WILD-TYPE | 57 | 28 | 39 | 32 | 18 |
Figure S30. Get High-res Image Gene #19: '5q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
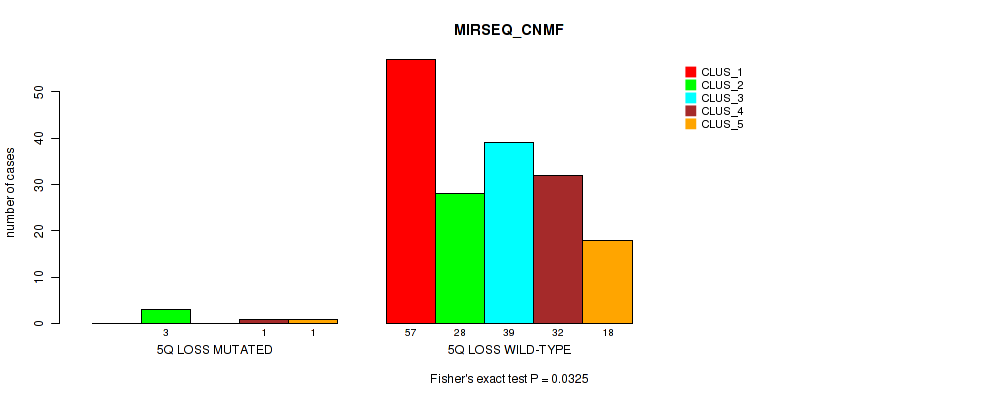
P value = 0.032 (Fisher's exact test), Q value = 0.12
Table S31. Gene #19: '5q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 5Q LOSS MUTATED | 0 | 3 | 2 |
| 5Q LOSS WILD-TYPE | 67 | 31 | 76 |
Figure S31. Get High-res Image Gene #19: '5q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
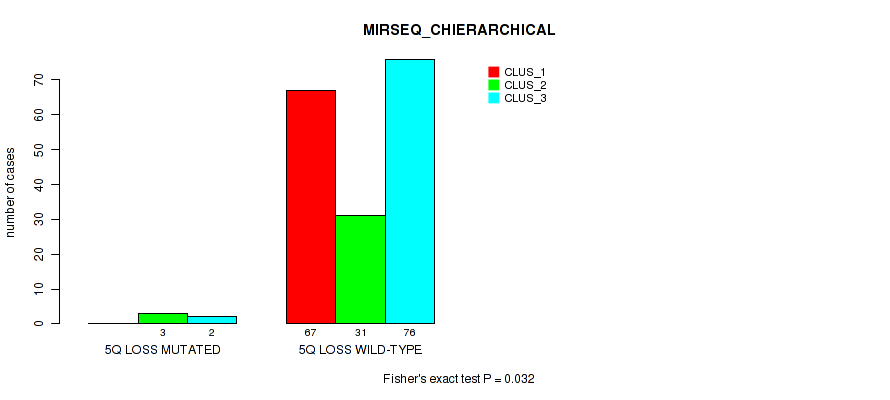
P value = 1e-05 (Fisher's exact test), Q value = 0.00038
Table S32. Gene #20: '7p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 44 | 55 | 43 |
| 7P LOSS MUTATED | 0 | 5 | 0 | 12 |
| 7P LOSS WILD-TYPE | 49 | 39 | 55 | 31 |
Figure S32. Get High-res Image Gene #20: '7p loss' versus Molecular Subtype #1: 'CN_CNMF'
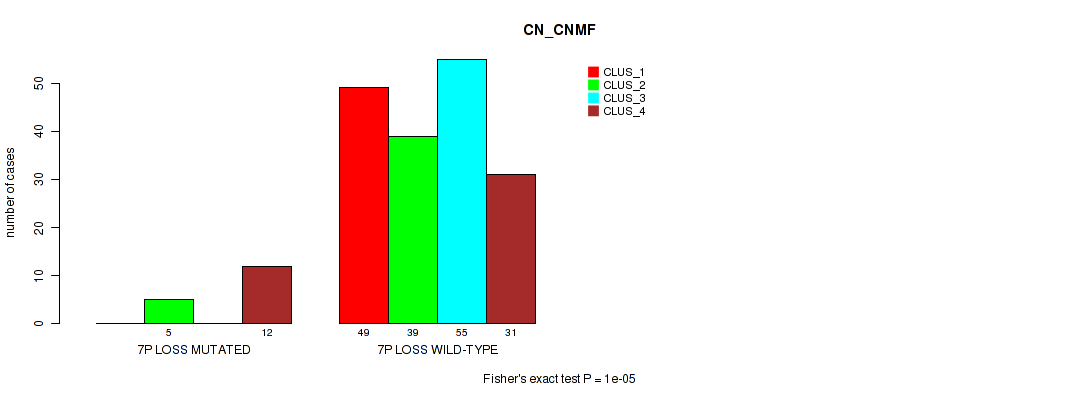
P value = 0.00042 (Fisher's exact test), Q value = 0.013
Table S33. Gene #20: '7p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 23 | 25 | 60 | 35 | 42 |
| 7P LOSS MUTATED | 1 | 1 | 14 | 0 | 1 |
| 7P LOSS WILD-TYPE | 22 | 24 | 46 | 35 | 41 |
Figure S33. Get High-res Image Gene #20: '7p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
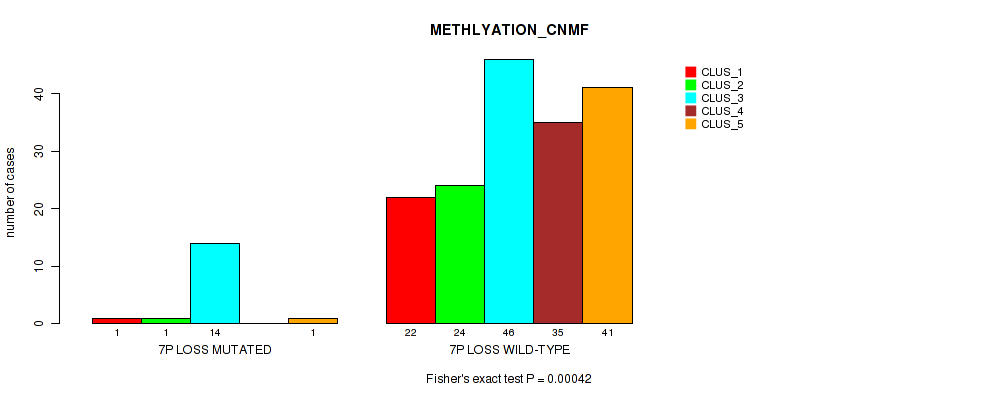
P value = 0.00111 (Fisher's exact test), Q value = 0.019
Table S34. Gene #20: '7p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 71 | 50 | 45 |
| 7P LOSS MUTATED | 13 | 0 | 3 |
| 7P LOSS WILD-TYPE | 58 | 50 | 42 |
Figure S34. Get High-res Image Gene #20: '7p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
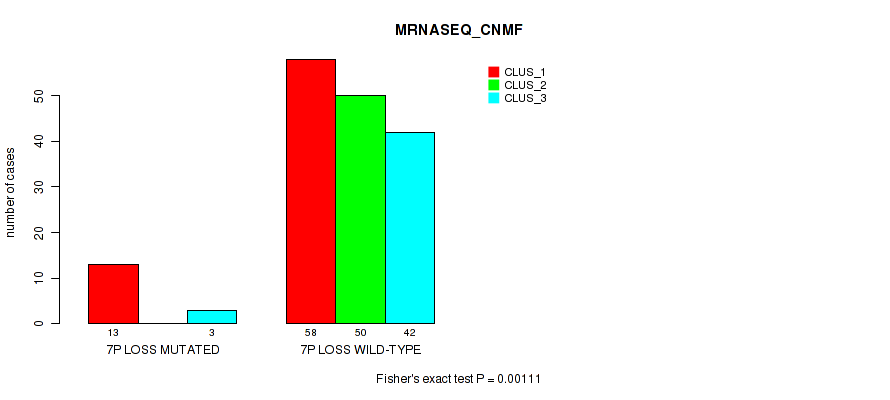
P value = 1e-05 (Fisher's exact test), Q value = 0.00038
Table S35. Gene #20: '7p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| 7P LOSS MUTATED | 0 | 0 | 0 | 16 | 0 | 0 |
| 7P LOSS WILD-TYPE | 16 | 13 | 14 | 39 | 26 | 42 |
Figure S35. Get High-res Image Gene #20: '7p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
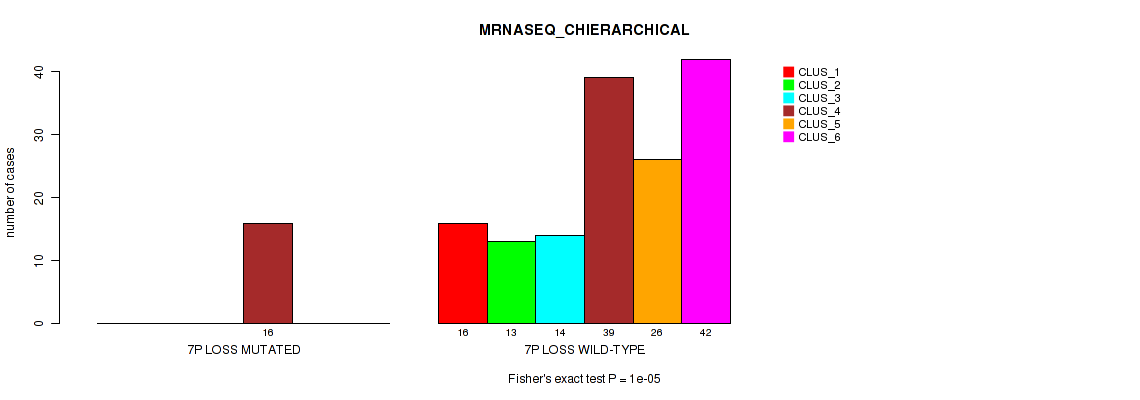
P value = 0.00658 (Fisher's exact test), Q value = 0.049
Table S36. Gene #20: '7p loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 57 | 31 | 39 | 33 | 19 |
| 7P LOSS MUTATED | 1 | 8 | 4 | 2 | 2 |
| 7P LOSS WILD-TYPE | 56 | 23 | 35 | 31 | 17 |
Figure S36. Get High-res Image Gene #20: '7p loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
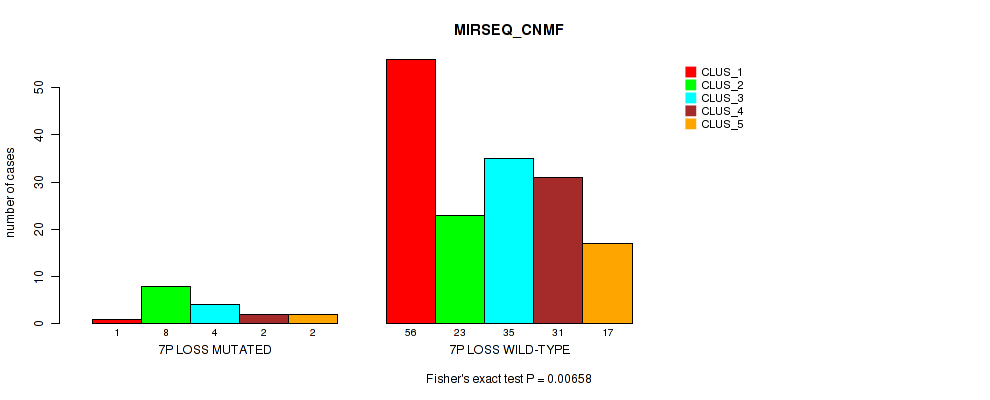
P value = 0.00293 (Fisher's exact test), Q value = 0.032
Table S37. Gene #20: '7p loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 7P LOSS MUTATED | 1 | 7 | 9 |
| 7P LOSS WILD-TYPE | 66 | 27 | 69 |
Figure S37. Get High-res Image Gene #20: '7p loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
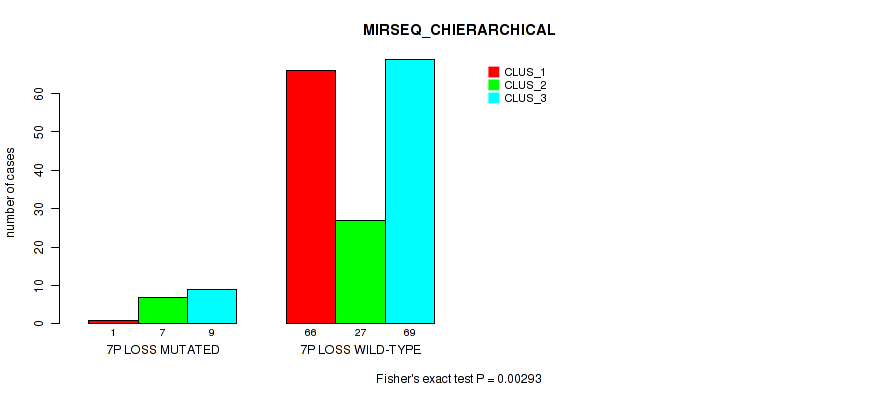
P value = 1e-05 (Fisher's exact test), Q value = 0.00038
Table S38. Gene #21: '7q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 44 | 55 | 43 |
| 7Q LOSS MUTATED | 1 | 6 | 0 | 13 |
| 7Q LOSS WILD-TYPE | 48 | 38 | 55 | 30 |
Figure S38. Get High-res Image Gene #21: '7q loss' versus Molecular Subtype #1: 'CN_CNMF'
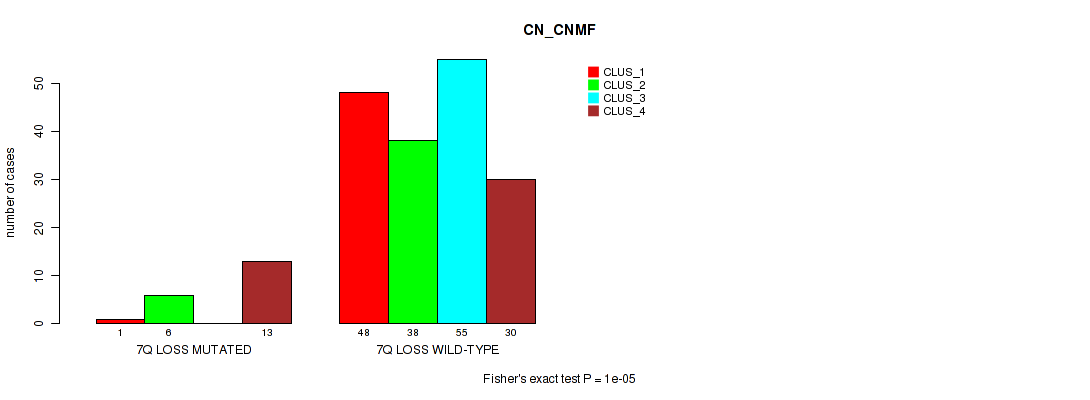
P value = 1e-05 (Fisher's exact test), Q value = 0.00038
Table S39. Gene #21: '7q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 23 | 25 | 60 | 35 | 42 |
| 7Q LOSS MUTATED | 1 | 1 | 17 | 0 | 1 |
| 7Q LOSS WILD-TYPE | 22 | 24 | 43 | 35 | 41 |
Figure S39. Get High-res Image Gene #21: '7q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
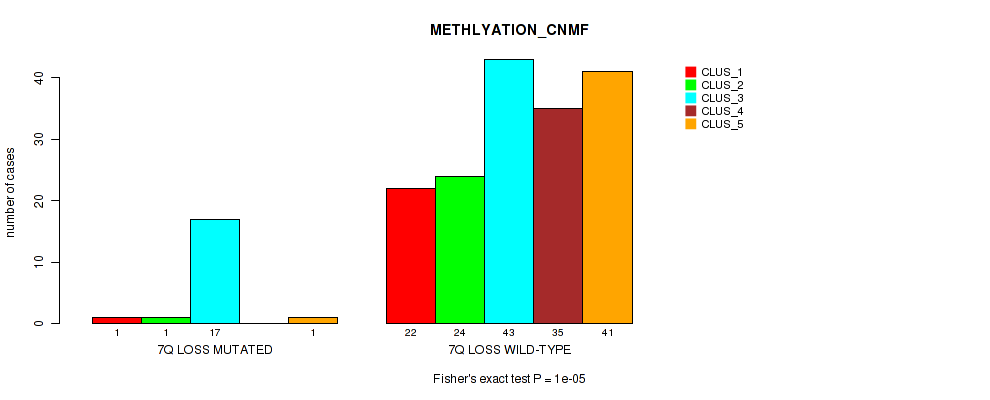
P value = 0.00978 (Fisher's exact test), Q value = 0.063
Table S40. Gene #21: '7q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 71 | 50 | 45 |
| 7Q LOSS MUTATED | 13 | 1 | 3 |
| 7Q LOSS WILD-TYPE | 58 | 49 | 42 |
Figure S40. Get High-res Image Gene #21: '7q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
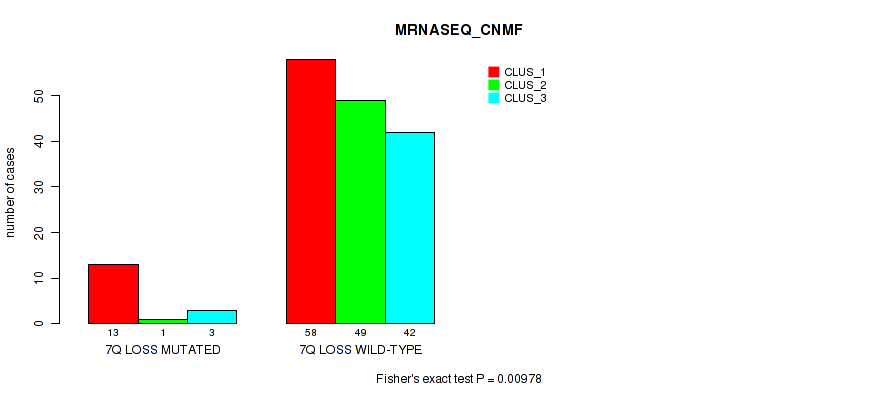
P value = 1e-05 (Fisher's exact test), Q value = 0.00038
Table S41. Gene #21: '7q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| 7Q LOSS MUTATED | 0 | 0 | 0 | 17 | 0 | 0 |
| 7Q LOSS WILD-TYPE | 16 | 13 | 14 | 38 | 26 | 42 |
Figure S41. Get High-res Image Gene #21: '7q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
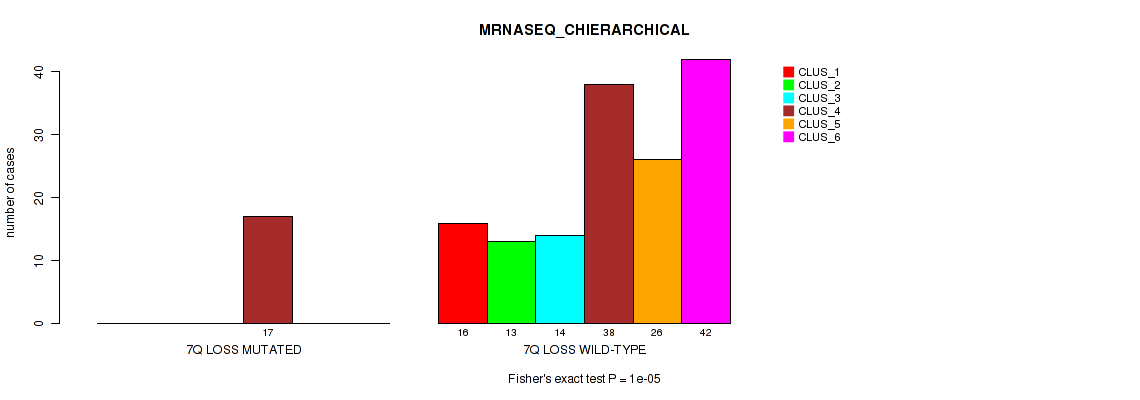
P value = 0.00296 (Fisher's exact test), Q value = 0.032
Table S42. Gene #21: '7q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 57 | 31 | 39 | 33 | 19 |
| 7Q LOSS MUTATED | 1 | 9 | 4 | 3 | 2 |
| 7Q LOSS WILD-TYPE | 56 | 22 | 35 | 30 | 17 |
Figure S42. Get High-res Image Gene #21: '7q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
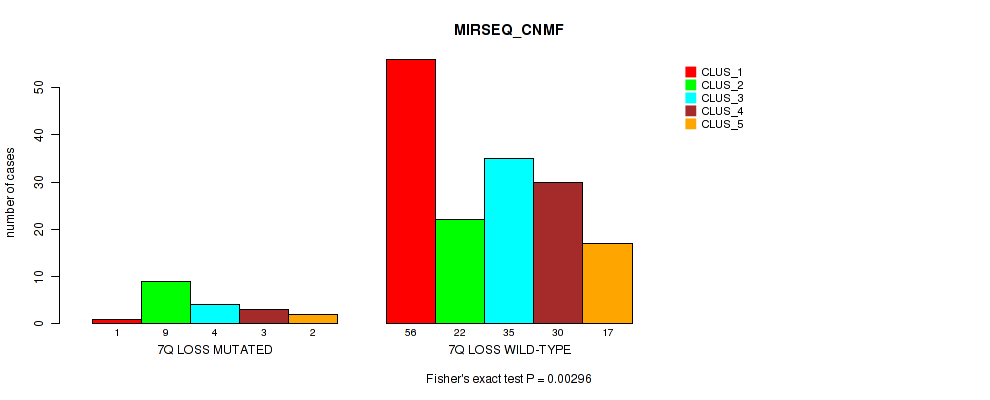
P value = 8e-04 (Fisher's exact test), Q value = 0.017
Table S43. Gene #21: '7q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 7Q LOSS MUTATED | 1 | 8 | 10 |
| 7Q LOSS WILD-TYPE | 66 | 26 | 68 |
Figure S43. Get High-res Image Gene #21: '7q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
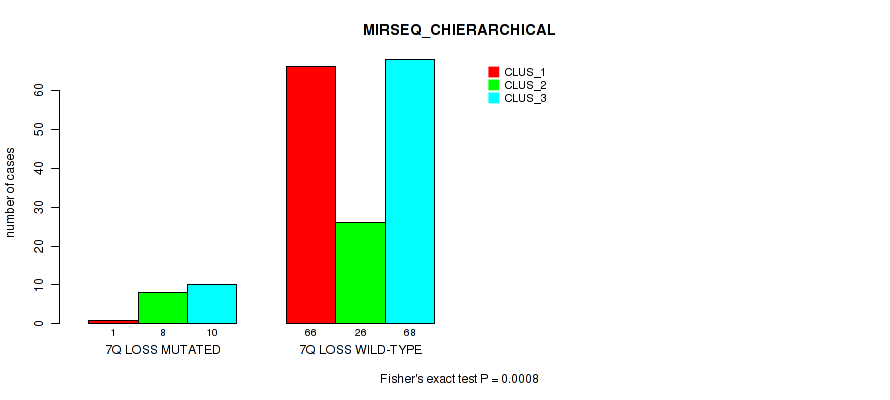
P value = 0.0357 (Fisher's exact test), Q value = 0.13
Table S44. Gene #24: '16q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 44 | 55 | 43 |
| 16Q LOSS MUTATED | 0 | 3 | 0 | 1 |
| 16Q LOSS WILD-TYPE | 49 | 41 | 55 | 42 |
Figure S44. Get High-res Image Gene #24: '16q loss' versus Molecular Subtype #1: 'CN_CNMF'
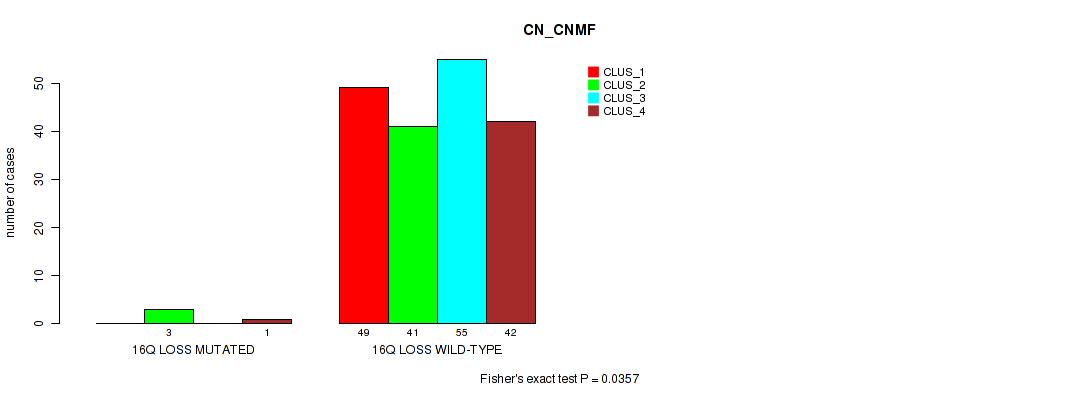
P value = 0.0237 (Fisher's exact test), Q value = 0.1
Table S45. Gene #24: '16q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 57 | 31 | 39 | 33 | 19 |
| 16Q LOSS MUTATED | 0 | 3 | 0 | 1 | 0 |
| 16Q LOSS WILD-TYPE | 57 | 28 | 39 | 32 | 19 |
Figure S45. Get High-res Image Gene #24: '16q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
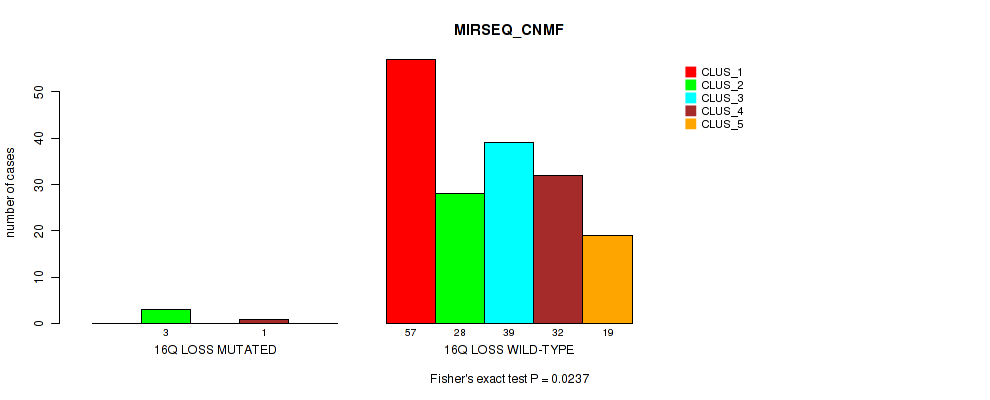
P value = 0.023 (Fisher's exact test), Q value = 0.1
Table S46. Gene #24: '16q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 16Q LOSS MUTATED | 0 | 3 | 1 |
| 16Q LOSS WILD-TYPE | 67 | 31 | 77 |
Figure S46. Get High-res Image Gene #24: '16q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
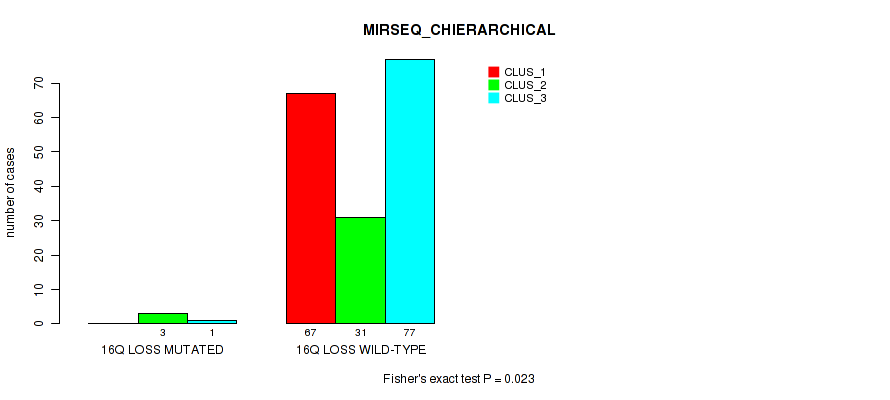
P value = 0.0171 (Fisher's exact test), Q value = 0.084
Table S47. Gene #25: '17p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 44 | 55 | 43 |
| 17P LOSS MUTATED | 0 | 7 | 3 | 3 |
| 17P LOSS WILD-TYPE | 49 | 37 | 52 | 40 |
Figure S47. Get High-res Image Gene #25: '17p loss' versus Molecular Subtype #1: 'CN_CNMF'
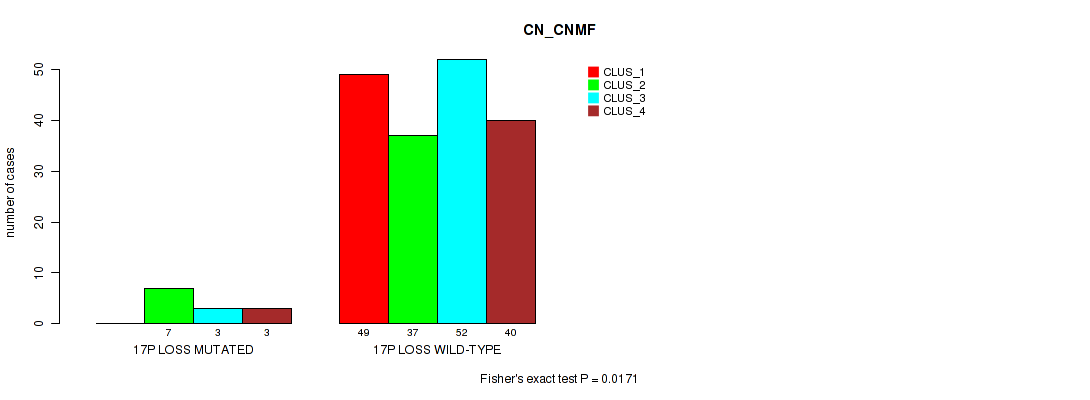
P value = 0.00356 (Fisher's exact test), Q value = 0.034
Table S48. Gene #25: '17p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 23 | 25 | 60 | 35 | 42 |
| 17P LOSS MUTATED | 0 | 0 | 10 | 3 | 0 |
| 17P LOSS WILD-TYPE | 23 | 25 | 50 | 32 | 42 |
Figure S48. Get High-res Image Gene #25: '17p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
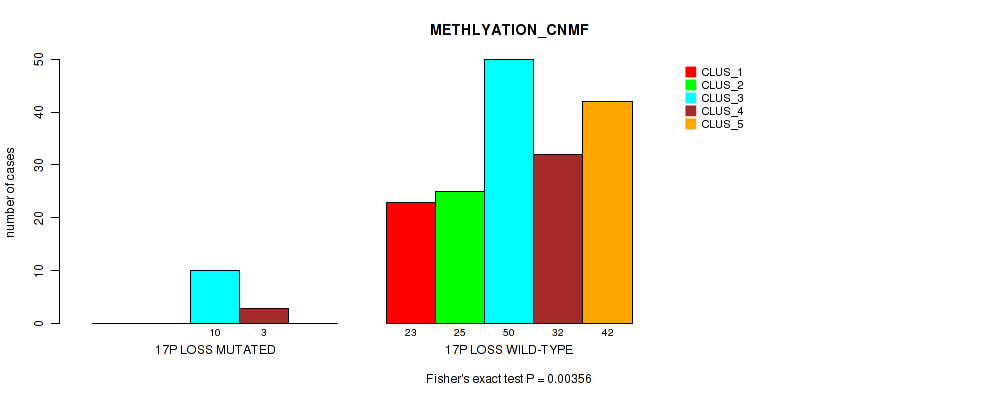
P value = 0.00157 (Fisher's exact test), Q value = 0.023
Table S49. Gene #25: '17p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| 17P LOSS MUTATED | 0 | 0 | 0 | 12 | 0 | 1 |
| 17P LOSS WILD-TYPE | 16 | 13 | 14 | 43 | 26 | 41 |
Figure S49. Get High-res Image Gene #25: '17p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
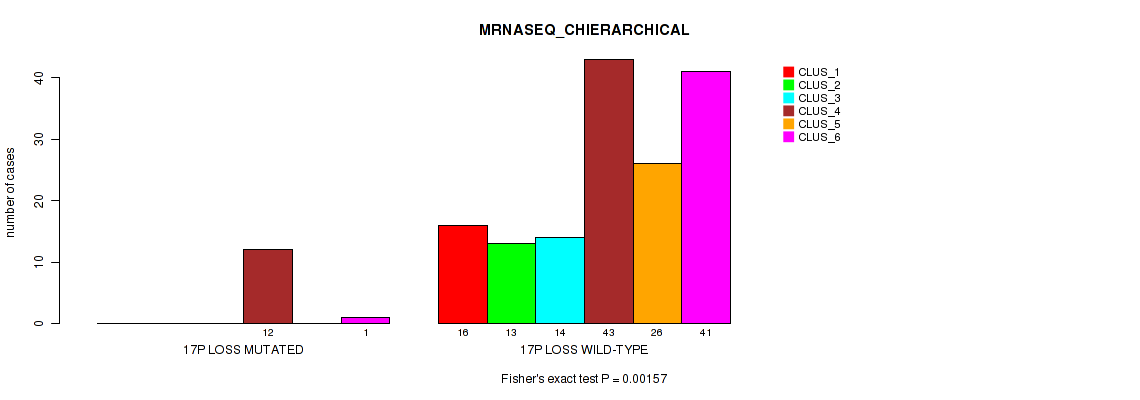
P value = 0.00358 (Fisher's exact test), Q value = 0.034
Table S50. Gene #25: '17p loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 17P LOSS MUTATED | 0 | 4 | 9 |
| 17P LOSS WILD-TYPE | 67 | 30 | 69 |
Figure S50. Get High-res Image Gene #25: '17p loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
P value = 0.0471 (Fisher's exact test), Q value = 0.16
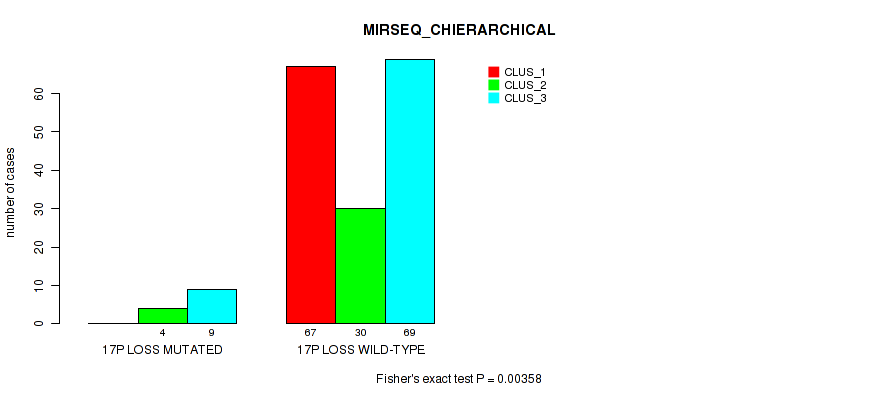
Table S51. Gene #26: '17q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 17Q LOSS MUTATED | 0 | 3 | 4 |
| 17Q LOSS WILD-TYPE | 67 | 31 | 74 |
Figure S51. Get High-res Image Gene #26: '17q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
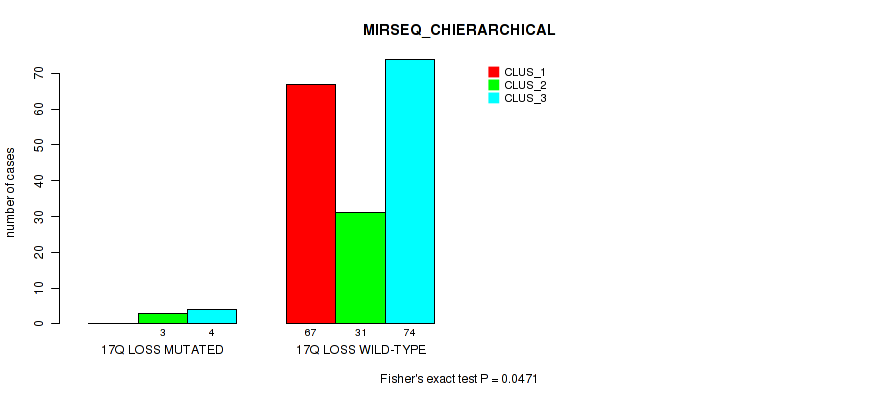
P value = 0.00909 (Fisher's exact test), Q value = 0.063
Table S52. Gene #27: '18p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 44 | 55 | 43 |
| 18P LOSS MUTATED | 0 | 4 | 0 | 1 |
| 18P LOSS WILD-TYPE | 49 | 40 | 55 | 42 |
Figure S52. Get High-res Image Gene #27: '18p loss' versus Molecular Subtype #1: 'CN_CNMF'
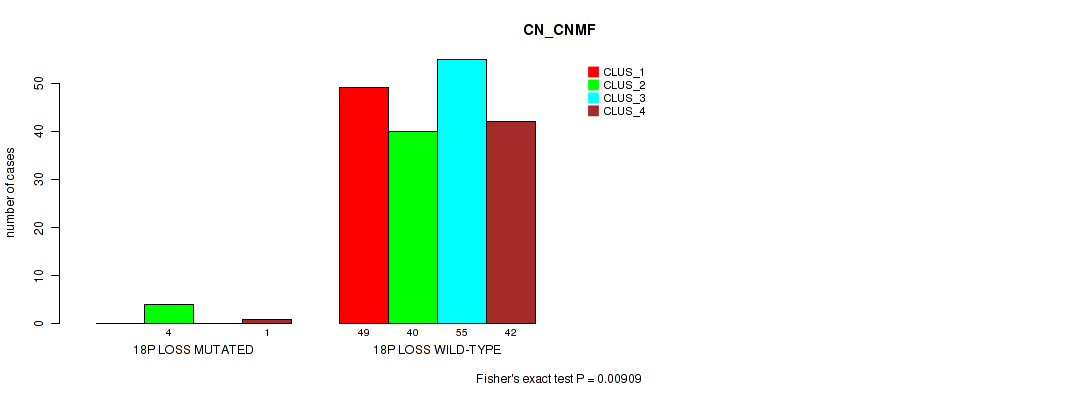
P value = 0.00926 (Fisher's exact test), Q value = 0.063
Table S53. Gene #31: 'xp loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 44 | 55 | 43 |
| XP LOSS MUTATED | 0 | 4 | 0 | 1 |
| XP LOSS WILD-TYPE | 49 | 40 | 55 | 42 |
Figure S53. Get High-res Image Gene #31: 'xp loss' versus Molecular Subtype #1: 'CN_CNMF'
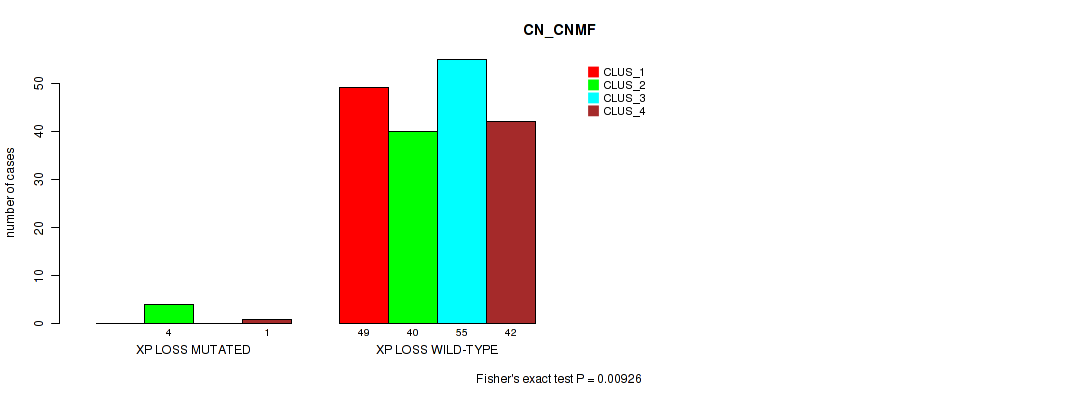
P value = 0.00563 (Fisher's exact test), Q value = 0.045
Table S54. Gene #31: 'xp loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 57 | 31 | 39 | 33 | 19 |
| XP LOSS MUTATED | 0 | 4 | 0 | 1 | 0 |
| XP LOSS WILD-TYPE | 57 | 27 | 39 | 32 | 19 |
Figure S54. Get High-res Image Gene #31: 'xp loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
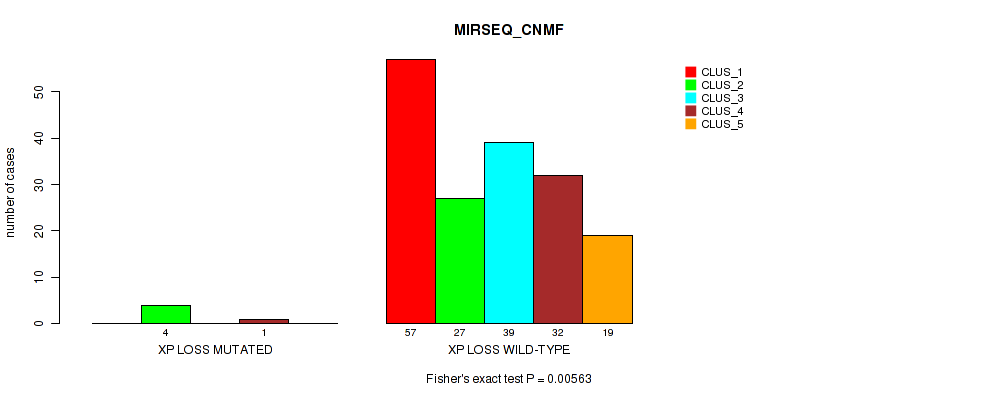
P value = 0.00504 (Fisher's exact test), Q value = 0.044
Table S55. Gene #31: 'xp loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| XP LOSS MUTATED | 0 | 4 | 1 |
| XP LOSS WILD-TYPE | 67 | 30 | 77 |
Figure S55. Get High-res Image Gene #31: 'xp loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
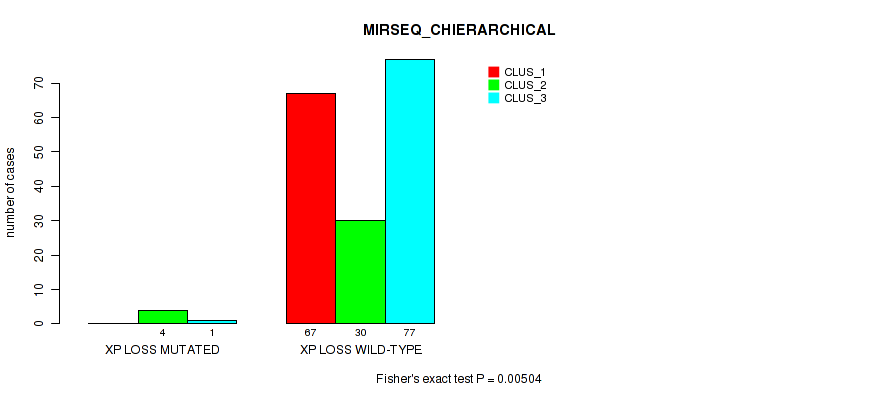
P value = 0.00945 (Fisher's exact test), Q value = 0.063
Table S56. Gene #32: 'xq loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 44 | 55 | 43 |
| XQ LOSS MUTATED | 0 | 4 | 0 | 1 |
| XQ LOSS WILD-TYPE | 49 | 40 | 55 | 42 |
Figure S56. Get High-res Image Gene #32: 'xq loss' versus Molecular Subtype #1: 'CN_CNMF'
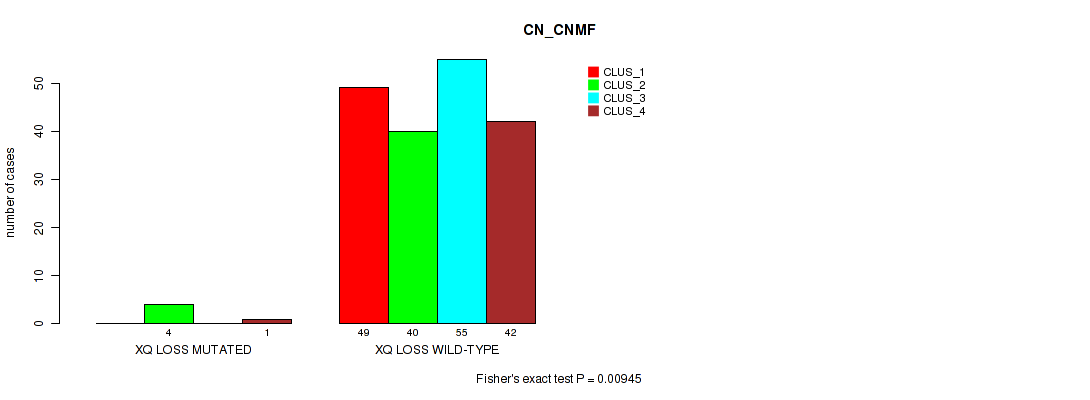
P value = 0.00598 (Fisher's exact test), Q value = 0.046
Table S57. Gene #32: 'xq loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 57 | 31 | 39 | 33 | 19 |
| XQ LOSS MUTATED | 0 | 4 | 0 | 1 | 0 |
| XQ LOSS WILD-TYPE | 57 | 27 | 39 | 32 | 19 |
Figure S57. Get High-res Image Gene #32: 'xq loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
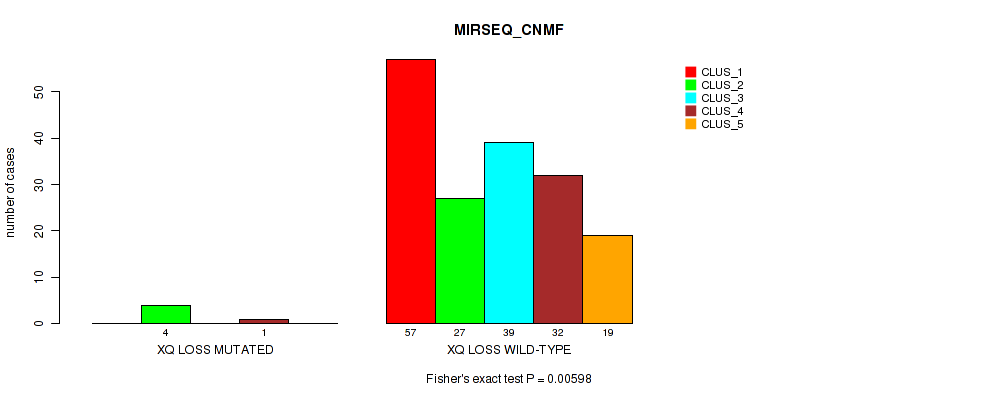
P value = 0.00495 (Fisher's exact test), Q value = 0.044
Table S58. Gene #32: 'xq loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| XQ LOSS MUTATED | 0 | 4 | 1 |
| XQ LOSS WILD-TYPE | 67 | 30 | 77 |
Figure S58. Get High-res Image Gene #32: 'xq loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
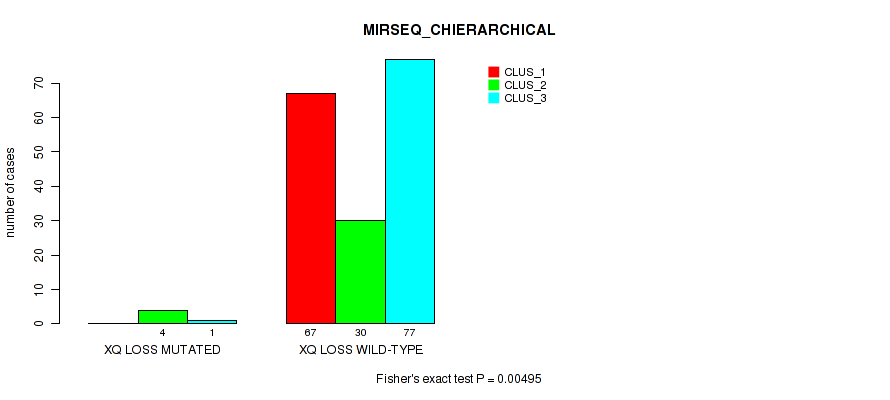
-
Copy number data file = broad_values_by_arm.txt from GISTIC pipeline
-
Processed Copy number data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_Correlate_Genomic_Events_Preprocess/LAML-TB/22529564/transformed.cor.cli.txt
-
Molecular subtypes file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_mergedClustering/LAML-TB/22541002/LAML-TB.transferedmergedcluster.txt
-
Number of patients = 191
-
Number of significantly arm-level cnvs = 32
-
Number of molecular subtypes = 6
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.