This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and selected clinical features.
Testing the association between copy number variation 79 arm-level events and 5 clinical features across 56 patients, one significant finding detected with Q value < 0.25.
-
21q loss cnv correlated to 'RADIATION_THERAPY'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 79 arm-level events and 5 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, one significant finding detected.
|
Clinical Features |
Time to Death |
YEARS TO BIRTH |
RADIATION THERAPY |
HISTOLOGICAL TYPE |
RACE | ||
| nCNV (%) | nWild-Type | logrank test | Wilcoxon-test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 21q loss | 17 (30%) | 39 |
0.195 (1.00) |
0.175 (1.00) |
0.000482 (0.19) |
0.708 (1.00) |
1 (1.00) |
| 1p gain | 24 (43%) | 32 |
0.757 (1.00) |
0.285 (1.00) |
0.579 (1.00) |
0.45 (1.00) |
0.452 (1.00) |
| 1q gain | 31 (55%) | 25 |
0.0585 (1.00) |
0.723 (1.00) |
0.786 (1.00) |
0.699 (1.00) |
0.167 (1.00) |
| 2p gain | 23 (41%) | 33 |
0.119 (1.00) |
0.526 (1.00) |
0.0558 (1.00) |
0.69 (1.00) |
1 (1.00) |
| 2q gain | 21 (38%) | 35 |
0.77 (1.00) |
0.635 (1.00) |
0.0481 (1.00) |
0.884 (1.00) |
0.873 (1.00) |
| 3p gain | 10 (18%) | 46 |
0.522 (1.00) |
0.578 (1.00) |
0.482 (1.00) |
0.454 (1.00) |
0.129 (1.00) |
| 3q gain | 22 (39%) | 34 |
0.415 (1.00) |
0.271 (1.00) |
1 (1.00) |
0.609 (1.00) |
0.755 (1.00) |
| 4p gain | 10 (18%) | 46 |
0.482 (1.00) |
0.822 (1.00) |
0.715 (1.00) |
0.322 (1.00) |
0.0687 (1.00) |
| 4q gain | 3 (5%) | 53 |
0.952 (1.00) |
0.985 (1.00) |
0.0864 (1.00) |
0.45 (1.00) |
1 (1.00) |
| 5p gain | 23 (41%) | 33 |
0.625 (1.00) |
0.683 (1.00) |
1 (1.00) |
1 (1.00) |
0.878 (1.00) |
| 5q gain | 8 (14%) | 48 |
0.782 (1.00) |
0.833 (1.00) |
0.444 (1.00) |
0.103 (1.00) |
0.24 (1.00) |
| 6p gain | 31 (55%) | 25 |
0.971 (1.00) |
0.108 (1.00) |
1 (1.00) |
0.655 (1.00) |
0.449 (1.00) |
| 6q gain | 27 (48%) | 29 |
0.707 (1.00) |
0.178 (1.00) |
1 (1.00) |
0.277 (1.00) |
0.602 (1.00) |
| 7p gain | 21 (38%) | 35 |
0.709 (1.00) |
0.192 (1.00) |
0.78 (1.00) |
0.178 (1.00) |
0.237 (1.00) |
| 7q gain | 18 (32%) | 38 |
0.788 (1.00) |
0.409 (1.00) |
0.766 (1.00) |
0.0567 (1.00) |
1 (1.00) |
| 8p gain | 18 (32%) | 38 |
0.273 (1.00) |
0.673 (1.00) |
0.235 (1.00) |
0.0745 (1.00) |
0.417 (1.00) |
| 8q gain | 30 (54%) | 26 |
0.154 (1.00) |
0.336 (1.00) |
0.00582 (1.00) |
0.0905 (1.00) |
0.449 (1.00) |
| 9p gain | 6 (11%) | 50 |
0.874 (1.00) |
0.353 (1.00) |
1 (1.00) |
0.0217 (1.00) |
0.703 (1.00) |
| 10p gain | 20 (36%) | 36 |
0.386 (1.00) |
0.837 (1.00) |
0.401 (1.00) |
0.822 (1.00) |
0.439 (1.00) |
| 10q gain | 17 (30%) | 39 |
0.287 (1.00) |
0.865 (1.00) |
0.766 (1.00) |
0.813 (1.00) |
0.353 (1.00) |
| 11p gain | 5 (9%) | 51 |
0.967 (1.00) |
0.752 (1.00) |
1 (1.00) |
0.84 (1.00) |
1 (1.00) |
| 11q gain | 7 (12%) | 49 |
0.942 (1.00) |
0.457 (1.00) |
0.688 (1.00) |
0.582 (1.00) |
1 (1.00) |
| 12p gain | 22 (39%) | 34 |
0.116 (1.00) |
0.45 (1.00) |
0.582 (1.00) |
0.344 (1.00) |
0.282 (1.00) |
| 12q gain | 11 (20%) | 45 |
0.93 (1.00) |
0.584 (1.00) |
1 (1.00) |
0.688 (1.00) |
0.394 (1.00) |
| 13q gain | 15 (27%) | 41 |
0.248 (1.00) |
0.505 (1.00) |
0.535 (1.00) |
0.469 (1.00) |
0.731 (1.00) |
| 14q gain | 7 (12%) | 49 |
0.0241 (1.00) |
0.766 (1.00) |
1 (1.00) |
0.771 (1.00) |
0.56 (1.00) |
| 15q gain | 4 (7%) | 52 |
0.574 (1.00) |
0.166 (1.00) |
1 (1.00) |
0.672 (1.00) |
0.204 (1.00) |
| 16p gain | 10 (18%) | 46 |
0.584 (1.00) |
0.416 (1.00) |
1 (1.00) |
0.406 (1.00) |
0.397 (1.00) |
| 16q gain | 6 (11%) | 50 |
0.39 (1.00) |
0.853 (1.00) |
0.678 (1.00) |
0.465 (1.00) |
0.493 (1.00) |
| 17p gain | 9 (16%) | 47 |
0.24 (1.00) |
0.396 (1.00) |
0.444 (1.00) |
0.268 (1.00) |
0.284 (1.00) |
| 17q gain | 18 (32%) | 38 |
0.903 (1.00) |
0.765 (1.00) |
1 (1.00) |
0.819 (1.00) |
0.753 (1.00) |
| 18p gain | 18 (32%) | 38 |
0.334 (1.00) |
0.854 (1.00) |
0.557 (1.00) |
0.154 (1.00) |
0.655 (1.00) |
| 18q gain | 13 (23%) | 43 |
0.408 (1.00) |
0.93 (1.00) |
0.751 (1.00) |
0.395 (1.00) |
0.404 (1.00) |
| 19p gain | 24 (43%) | 32 |
0.612 (1.00) |
1 (1.00) |
0.161 (1.00) |
0.942 (1.00) |
0.877 (1.00) |
| 19q gain | 29 (52%) | 27 |
0.611 (1.00) |
0.538 (1.00) |
0.0136 (1.00) |
0.221 (1.00) |
0.523 (1.00) |
| 20p gain | 36 (64%) | 20 |
0.491 (1.00) |
0.202 (1.00) |
0.16 (1.00) |
0.938 (1.00) |
0.335 (1.00) |
| 20q gain | 43 (77%) | 13 |
0.959 (1.00) |
0.676 (1.00) |
1 (1.00) |
0.287 (1.00) |
0.252 (1.00) |
| 21q gain | 18 (32%) | 38 |
0.645 (1.00) |
0.752 (1.00) |
0.384 (1.00) |
0.934 (1.00) |
0.257 (1.00) |
| 22q gain | 8 (14%) | 48 |
0.0711 (1.00) |
0.038 (1.00) |
1 (1.00) |
0.627 (1.00) |
0.278 (1.00) |
| xp gain | 18 (32%) | 38 |
0.572 (1.00) |
0.0382 (1.00) |
0.372 (1.00) |
0.874 (1.00) |
0.751 (1.00) |
| xq gain | 15 (27%) | 41 |
0.407 (1.00) |
0.63 (1.00) |
1 (1.00) |
0.929 (1.00) |
0.0878 (1.00) |
| 1p loss | 10 (18%) | 46 |
0.268 (1.00) |
0.275 (1.00) |
0.715 (1.00) |
0.144 (1.00) |
0.651 (1.00) |
| 1q loss | 10 (18%) | 46 |
0.142 (1.00) |
0.199 (1.00) |
0.269 (1.00) |
0.113 (1.00) |
0.645 (1.00) |
| 3p loss | 22 (39%) | 34 |
0.324 (1.00) |
0.731 (1.00) |
0.16 (1.00) |
0.829 (1.00) |
0.46 (1.00) |
| 3q loss | 15 (27%) | 41 |
0.563 (1.00) |
0.331 (1.00) |
0.338 (1.00) |
0.636 (1.00) |
0.435 (1.00) |
| 4p loss | 30 (54%) | 26 |
0.307 (1.00) |
0.974 (1.00) |
0.786 (1.00) |
0.0474 (1.00) |
0.881 (1.00) |
| 4q loss | 34 (61%) | 22 |
0.419 (1.00) |
0.626 (1.00) |
0.573 (1.00) |
0.162 (1.00) |
0.156 (1.00) |
| 5p loss | 9 (16%) | 47 |
0.0784 (1.00) |
0.118 (1.00) |
0.269 (1.00) |
0.894 (1.00) |
0.608 (1.00) |
| 5q loss | 18 (32%) | 38 |
0.495 (1.00) |
0.533 (1.00) |
0.235 (1.00) |
0.933 (1.00) |
0.751 (1.00) |
| 6p loss | 5 (9%) | 51 |
0.1 (1.00) |
0.508 (1.00) |
0.649 (1.00) |
0.84 (1.00) |
0.404 (1.00) |
| 6q loss | 7 (12%) | 49 |
0.218 (1.00) |
0.738 (1.00) |
1 (1.00) |
1 (1.00) |
0.181 (1.00) |
| 7p loss | 13 (23%) | 43 |
0.864 (1.00) |
0.734 (1.00) |
0.75 (1.00) |
0.719 (1.00) |
1 (1.00) |
| 7q loss | 12 (21%) | 44 |
0.132 (1.00) |
0.905 (1.00) |
0.187 (1.00) |
0.709 (1.00) |
0.469 (1.00) |
| 8p loss | 24 (43%) | 32 |
0.783 (1.00) |
0.967 (1.00) |
0.786 (1.00) |
0.482 (1.00) |
0.357 (1.00) |
| 8q loss | 9 (16%) | 47 |
0.951 (1.00) |
0.577 (1.00) |
0.0623 (1.00) |
0.268 (1.00) |
0.283 (1.00) |
| 9p loss | 35 (62%) | 21 |
0.734 (1.00) |
0.0673 (1.00) |
0.566 (1.00) |
0.0337 (1.00) |
0.677 (1.00) |
| 9q loss | 40 (71%) | 16 |
0.34 (1.00) |
0.1 (1.00) |
0.547 (1.00) |
0.287 (1.00) |
0.466 (1.00) |
| 10p loss | 24 (43%) | 32 |
0.6 (1.00) |
0.246 (1.00) |
0.278 (1.00) |
0.736 (1.00) |
0.148 (1.00) |
| 10q loss | 20 (36%) | 36 |
0.182 (1.00) |
0.804 (1.00) |
0.772 (1.00) |
0.207 (1.00) |
0.235 (1.00) |
| 11p loss | 28 (50%) | 28 |
0.0193 (1.00) |
0.21 (1.00) |
0.17 (1.00) |
0.123 (1.00) |
1 (1.00) |
| 11q loss | 26 (46%) | 30 |
0.337 (1.00) |
0.537 (1.00) |
0.783 (1.00) |
0.207 (1.00) |
0.882 (1.00) |
| 12p loss | 13 (23%) | 43 |
0.435 (1.00) |
0.907 (1.00) |
0.534 (1.00) |
1 (1.00) |
0.303 (1.00) |
| 12q loss | 14 (25%) | 42 |
0.33 (1.00) |
0.191 (1.00) |
1 (1.00) |
0.673 (1.00) |
0.731 (1.00) |
| 13q loss | 26 (46%) | 30 |
0.767 (1.00) |
0.831 (1.00) |
0.783 (1.00) |
0.888 (1.00) |
1 (1.00) |
| 14q loss | 27 (48%) | 29 |
0.252 (1.00) |
0.533 (1.00) |
0.098 (1.00) |
0.0866 (1.00) |
0.0858 (1.00) |
| 15q loss | 33 (59%) | 23 |
0.353 (1.00) |
0.647 (1.00) |
0.154 (1.00) |
0.573 (1.00) |
0.222 (1.00) |
| 16p loss | 32 (57%) | 24 |
0.517 (1.00) |
0.868 (1.00) |
0.416 (1.00) |
0.544 (1.00) |
0.516 (1.00) |
| 16q loss | 37 (66%) | 19 |
0.134 (1.00) |
0.965 (1.00) |
0.146 (1.00) |
0.302 (1.00) |
0.326 (1.00) |
| 17p loss | 34 (61%) | 22 |
0.18 (1.00) |
0.574 (1.00) |
0.573 (1.00) |
0.371 (1.00) |
0.243 (1.00) |
| 17q loss | 17 (30%) | 39 |
0.0663 (1.00) |
0.562 (1.00) |
1 (1.00) |
0.464 (1.00) |
0.741 (1.00) |
| 18p loss | 18 (32%) | 38 |
0.821 (1.00) |
0.699 (1.00) |
0.145 (1.00) |
0.0971 (1.00) |
0.259 (1.00) |
| 18q loss | 20 (36%) | 36 |
0.309 (1.00) |
0.515 (1.00) |
0.78 (1.00) |
0.171 (1.00) |
0.0372 (1.00) |
| 19p loss | 15 (27%) | 41 |
0.97 (1.00) |
0.897 (1.00) |
0.76 (1.00) |
0.862 (1.00) |
1 (1.00) |
| 19q loss | 12 (21%) | 44 |
0.99 (1.00) |
0.442 (1.00) |
0.482 (1.00) |
0.353 (1.00) |
0.706 (1.00) |
| 20p loss | 8 (14%) | 48 |
0.638 (1.00) |
0.797 (1.00) |
0.436 (1.00) |
1 (1.00) |
0.444 (1.00) |
| 20q loss | 5 (9%) | 51 |
0.53 (1.00) |
0.943 (1.00) |
0.617 (1.00) |
0.706 (1.00) |
0.682 (1.00) |
| 22q loss | 33 (59%) | 23 |
0.31 (1.00) |
0.683 (1.00) |
0.278 (1.00) |
0.614 (1.00) |
0.102 (1.00) |
| xp loss | 15 (27%) | 41 |
0.795 (1.00) |
0.604 (1.00) |
1 (1.00) |
0.737 (1.00) |
1 (1.00) |
| xq loss | 18 (32%) | 38 |
0.453 (1.00) |
0.611 (1.00) |
0.384 (1.00) |
0.874 (1.00) |
0.497 (1.00) |
P value = 0.000482 (Fisher's exact test), Q value = 0.19
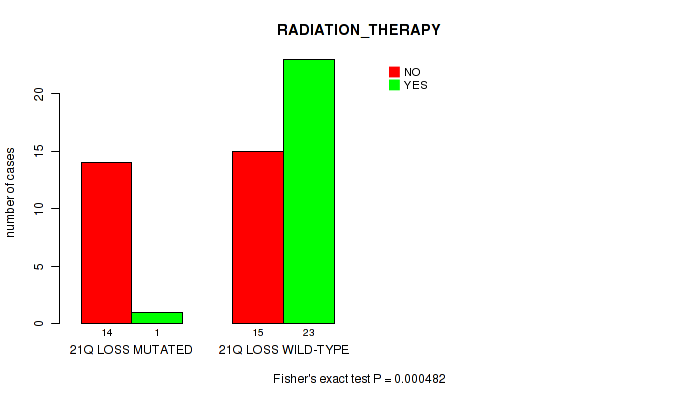
Table S1. Gene #76: '21q loss' versus Clinical Feature #3: 'RADIATION_THERAPY'
| nPatients | NO | YES |
|---|---|---|
| ALL | 29 | 24 |
| 21Q LOSS MUTATED | 14 | 1 |
| 21Q LOSS WILD-TYPE | 15 | 23 |
Figure S1. Get High-res Image Gene #76: '21q loss' versus Clinical Feature #3: 'RADIATION_THERAPY'
-
Copy number data file = broad_values_by_arm.txt from GISTIC pipeline
-
Processed Copy number data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_Correlate_Genomic_Events_Preprocess/UCS-TP/22533738/transformed.cor.cli.txt
-
Clinical data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/Append_Data/UCS-TP/22507158/UCS-TP.merged_data.txt
-
Number of patients = 56
-
Number of significantly arm-level cnvs = 79
-
Number of selected clinical features = 5
-
Exclude regions that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.