This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 7 genes and 8 molecular subtypes across 80 patients, 27 significant findings detected with P value < 0.05 and Q value < 0.25.
-
GNAQ mutation correlated to 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
EIF1AX mutation correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
SF3B1 mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
BAP1 mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
CYSLTR2 mutation correlated to 'CN_CNMF'.
-
SFRS2 mutation correlated to 'MIRSEQ_MATURE_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between mutation status of 7 genes and 8 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 27 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| SF3B1 | 18 (22%) | 62 |
0.0364 (0.0841) |
0.00011 (0.000684) |
9e-05 (0.00063) |
0.00183 (0.00932) |
0.0325 (0.0829) |
0.0184 (0.0596) |
0.00793 (0.0313) |
0.0166 (0.0582) |
| BAP1 | 22 (28%) | 58 |
3e-05 (0.00028) |
1e-05 (0.000112) |
1e-05 (0.000112) |
1e-05 (0.000112) |
1e-05 (0.000112) |
1e-05 (0.000112) |
0.00277 (0.0129) |
7e-05 (0.00056) |
| EIF1AX | 10 (12%) | 70 |
0.221 (0.326) |
0.00022 (0.00123) |
0.0192 (0.0596) |
0.00839 (0.0313) |
0.0253 (0.0746) |
0.00498 (0.0215) |
0.215 (0.325) |
0.036 (0.0841) |
| GNAQ | 40 (50%) | 40 |
0.373 (0.435) |
0.505 (0.555) |
0.134 (0.215) |
0.0375 (0.0841) |
0.0448 (0.0929) |
0.0899 (0.168) |
0.099 (0.179) |
0.0324 (0.0829) |
| CYSLTR2 | 3 (4%) | 77 |
0.0275 (0.0771) |
1 (1.00) |
0.0599 (0.116) |
0.107 (0.184) |
0.24 (0.344) |
0.274 (0.357) |
0.6 (0.646) |
0.367 (0.435) |
| SFRS2 | 3 (4%) | 77 |
0.258 (0.357) |
0.182 (0.283) |
0.0595 (0.116) |
0.109 (0.184) |
0.402 (0.45) |
0.381 (0.435) |
0.818 (0.833) |
0.0416 (0.0896) |
| GNA11 | 36 (45%) | 44 |
0.269 (0.357) |
0.726 (0.753) |
0.35 (0.426) |
0.134 (0.215) |
0.669 (0.707) |
0.268 (0.357) |
0.324 (0.404) |
0.307 (0.391) |
P value = 0.373 (Fisher's exact test), Q value = 0.43
Table S1. Gene #1: 'GNAQ MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 22 | 7 | 10 | 31 | 6 | 4 |
| GNAQ MUTATED | 10 | 2 | 4 | 20 | 3 | 1 |
| GNAQ WILD-TYPE | 12 | 5 | 6 | 11 | 3 | 3 |
P value = 0.505 (Fisher's exact test), Q value = 0.55
Table S2. Gene #1: 'GNAQ MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 28 | 14 | 14 | 24 |
| GNAQ MUTATED | 11 | 8 | 7 | 14 |
| GNAQ WILD-TYPE | 17 | 6 | 7 | 10 |
P value = 0.134 (Fisher's exact test), Q value = 0.22
Table S3. Gene #1: 'GNAQ MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 18 | 15 | 32 | 15 |
| GNAQ MUTATED | 8 | 5 | 21 | 6 |
| GNAQ WILD-TYPE | 10 | 10 | 11 | 9 |
P value = 0.0375 (Fisher's exact test), Q value = 0.084
Table S4. Gene #1: 'GNAQ MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 33 | 18 |
| GNAQ MUTATED | 10 | 22 | 8 |
| GNAQ WILD-TYPE | 19 | 11 | 10 |
Figure S1. Get High-res Image Gene #1: 'GNAQ MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
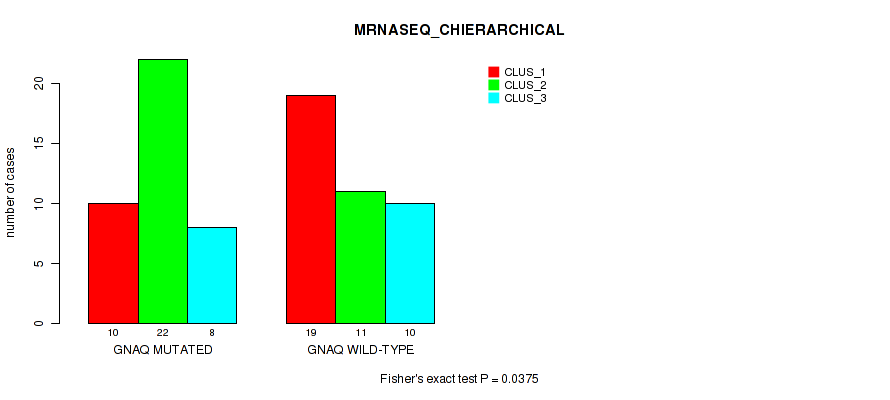
P value = 0.0448 (Fisher's exact test), Q value = 0.093
Table S5. Gene #1: 'GNAQ MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 8 | 8 | 11 | 25 | 23 | 5 |
| GNAQ MUTATED | 2 | 1 | 7 | 17 | 10 | 3 |
| GNAQ WILD-TYPE | 6 | 7 | 4 | 8 | 13 | 2 |
Figure S2. Get High-res Image Gene #1: 'GNAQ MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
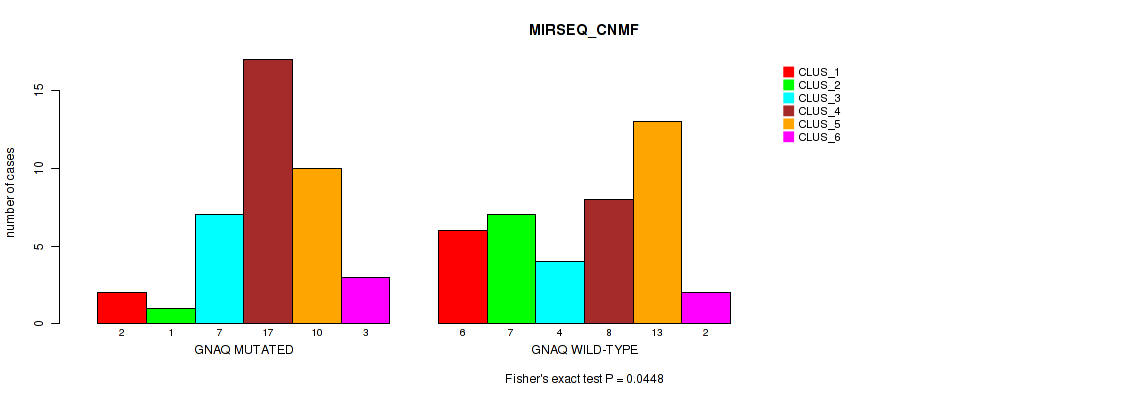
P value = 0.0899 (Fisher's exact test), Q value = 0.17
Table S6. Gene #1: 'GNAQ MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 17 | 13 | 28 | 22 |
| GNAQ MUTATED | 5 | 8 | 18 | 9 |
| GNAQ WILD-TYPE | 12 | 5 | 10 | 13 |
P value = 0.099 (Fisher's exact test), Q value = 0.18
Table S7. Gene #1: 'GNAQ MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 20 | 7 | 17 | 9 | 8 | 7 | 6 |
| GNAQ MUTATED | 9 | 0 | 11 | 5 | 4 | 5 | 3 |
| GNAQ WILD-TYPE | 11 | 7 | 6 | 4 | 4 | 2 | 3 |
P value = 0.0324 (Fisher's exact test), Q value = 0.083
Table S8. Gene #1: 'GNAQ MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 18 | 28 | 16 | 12 |
| GNAQ MUTATED | 7 | 20 | 5 | 5 |
| GNAQ WILD-TYPE | 11 | 8 | 11 | 7 |
Figure S3. Get High-res Image Gene #1: 'GNAQ MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
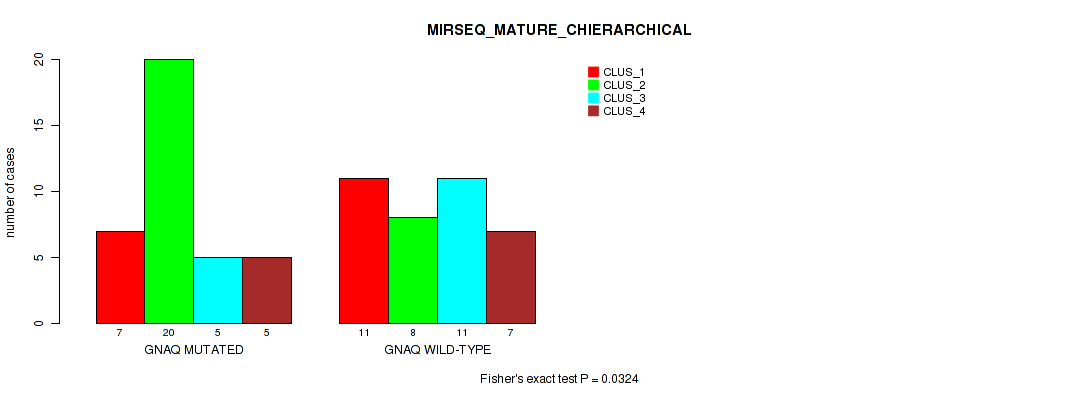
P value = 0.269 (Fisher's exact test), Q value = 0.36
Table S9. Gene #2: 'GNA11 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 22 | 7 | 10 | 31 | 6 | 4 |
| GNA11 MUTATED | 12 | 5 | 3 | 11 | 2 | 3 |
| GNA11 WILD-TYPE | 10 | 2 | 7 | 20 | 4 | 1 |
P value = 0.726 (Fisher's exact test), Q value = 0.75
Table S10. Gene #2: 'GNA11 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 28 | 14 | 14 | 24 |
| GNA11 MUTATED | 15 | 6 | 6 | 9 |
| GNA11 WILD-TYPE | 13 | 8 | 8 | 15 |
P value = 0.35 (Fisher's exact test), Q value = 0.43
Table S11. Gene #2: 'GNA11 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 18 | 15 | 32 | 15 |
| GNA11 MUTATED | 11 | 7 | 11 | 7 |
| GNA11 WILD-TYPE | 7 | 8 | 21 | 8 |
P value = 0.134 (Fisher's exact test), Q value = 0.22
Table S12. Gene #2: 'GNA11 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 33 | 18 |
| GNA11 MUTATED | 17 | 11 | 8 |
| GNA11 WILD-TYPE | 12 | 22 | 10 |
P value = 0.669 (Fisher's exact test), Q value = 0.71
Table S13. Gene #2: 'GNA11 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 8 | 8 | 11 | 25 | 23 | 5 |
| GNA11 MUTATED | 5 | 5 | 4 | 9 | 11 | 2 |
| GNA11 WILD-TYPE | 3 | 3 | 7 | 16 | 12 | 3 |
P value = 0.268 (Fisher's exact test), Q value = 0.36
Table S14. Gene #2: 'GNA11 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 17 | 13 | 28 | 22 |
| GNA11 MUTATED | 10 | 4 | 10 | 12 |
| GNA11 WILD-TYPE | 7 | 9 | 18 | 10 |
P value = 0.324 (Fisher's exact test), Q value = 0.4
Table S15. Gene #2: 'GNA11 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 20 | 7 | 17 | 9 | 8 | 7 | 6 |
| GNA11 MUTATED | 10 | 6 | 6 | 3 | 4 | 2 | 2 |
| GNA11 WILD-TYPE | 10 | 1 | 11 | 6 | 4 | 5 | 4 |
P value = 0.307 (Fisher's exact test), Q value = 0.39
Table S16. Gene #2: 'GNA11 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 18 | 28 | 16 | 12 |
| GNA11 MUTATED | 10 | 9 | 9 | 5 |
| GNA11 WILD-TYPE | 8 | 19 | 7 | 7 |
P value = 0.221 (Fisher's exact test), Q value = 0.33
Table S17. Gene #3: 'EIF1AX MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 22 | 7 | 10 | 31 | 6 | 4 |
| EIF1AX MUTATED | 2 | 0 | 0 | 8 | 0 | 0 |
| EIF1AX WILD-TYPE | 20 | 7 | 10 | 23 | 6 | 4 |
P value = 0.00022 (Fisher's exact test), Q value = 0.0012
Table S18. Gene #3: 'EIF1AX MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 28 | 14 | 14 | 24 |
| EIF1AX MUTATED | 0 | 6 | 0 | 4 |
| EIF1AX WILD-TYPE | 28 | 8 | 14 | 20 |
Figure S4. Get High-res Image Gene #3: 'EIF1AX MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
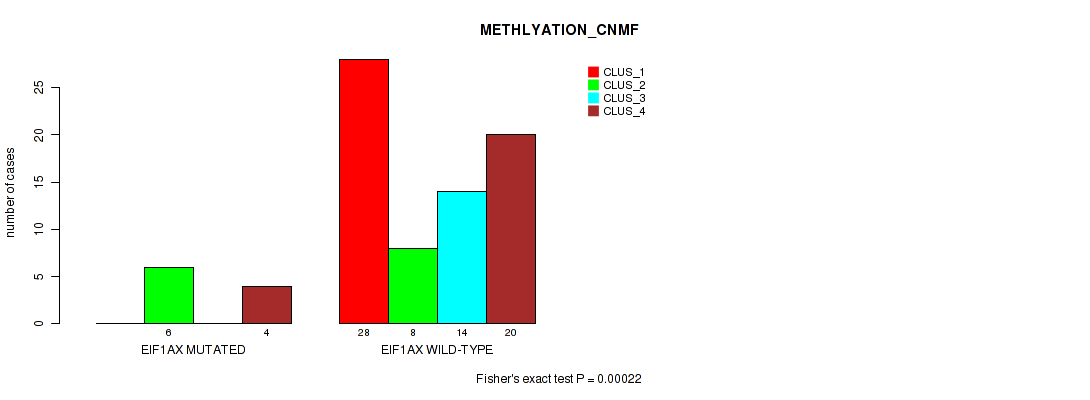
P value = 0.0192 (Fisher's exact test), Q value = 0.06
Table S19. Gene #3: 'EIF1AX MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 18 | 15 | 32 | 15 |
| EIF1AX MUTATED | 0 | 2 | 8 | 0 |
| EIF1AX WILD-TYPE | 18 | 13 | 24 | 15 |
Figure S5. Get High-res Image Gene #3: 'EIF1AX MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
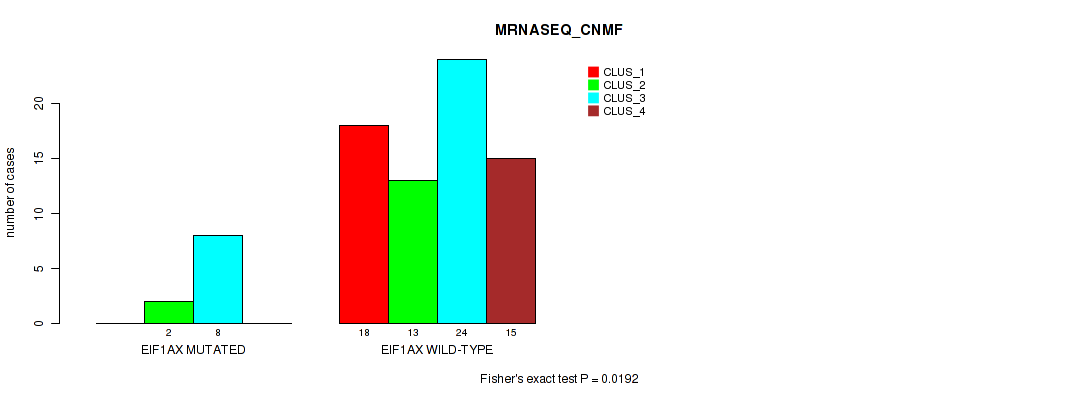
P value = 0.00839 (Fisher's exact test), Q value = 0.031
Table S20. Gene #3: 'EIF1AX MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 33 | 18 |
| EIF1AX MUTATED | 0 | 8 | 2 |
| EIF1AX WILD-TYPE | 29 | 25 | 16 |
Figure S6. Get High-res Image Gene #3: 'EIF1AX MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
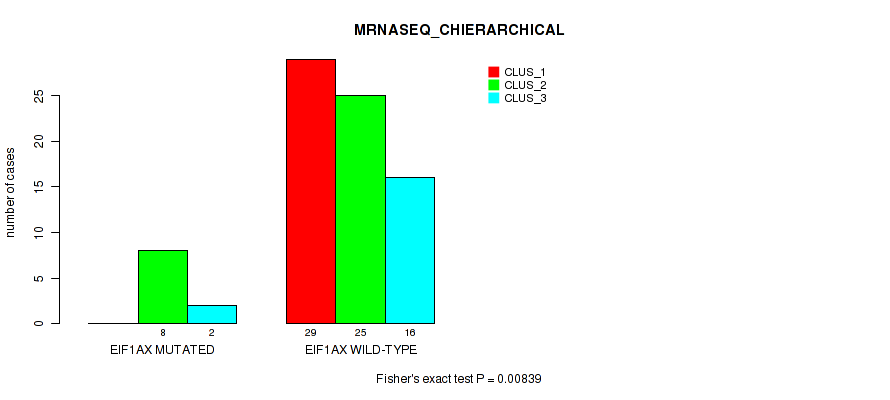
P value = 0.0253 (Fisher's exact test), Q value = 0.075
Table S21. Gene #3: 'EIF1AX MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 8 | 8 | 11 | 25 | 23 | 5 |
| EIF1AX MUTATED | 0 | 0 | 3 | 6 | 0 | 1 |
| EIF1AX WILD-TYPE | 8 | 8 | 8 | 19 | 23 | 4 |
Figure S7. Get High-res Image Gene #3: 'EIF1AX MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
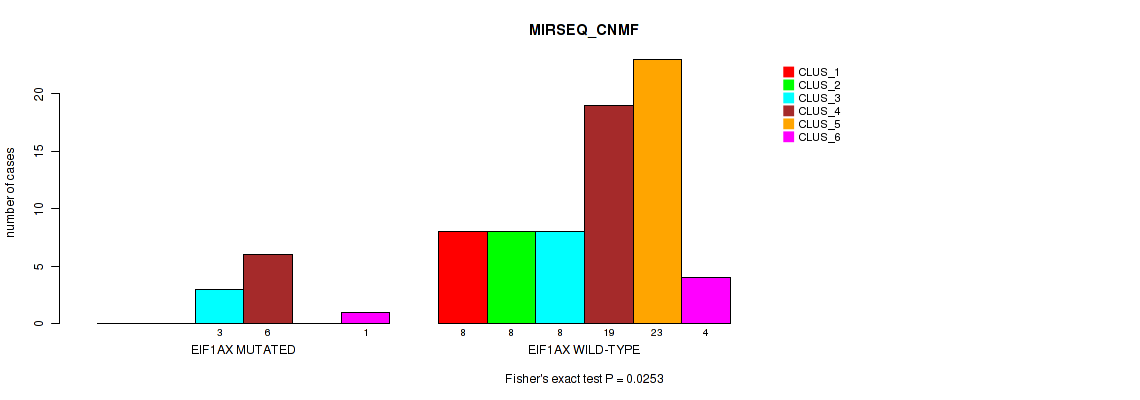
P value = 0.00498 (Fisher's exact test), Q value = 0.021
Table S22. Gene #3: 'EIF1AX MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 17 | 13 | 28 | 22 |
| EIF1AX MUTATED | 0 | 3 | 7 | 0 |
| EIF1AX WILD-TYPE | 17 | 10 | 21 | 22 |
Figure S8. Get High-res Image Gene #3: 'EIF1AX MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 0.215 (Fisher's exact test), Q value = 0.33
Table S23. Gene #3: 'EIF1AX MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 20 | 7 | 17 | 9 | 8 | 7 | 6 |
| EIF1AX MUTATED | 0 | 1 | 4 | 1 | 1 | 0 | 1 |
| EIF1AX WILD-TYPE | 20 | 6 | 13 | 8 | 7 | 7 | 5 |
P value = 0.036 (Fisher's exact test), Q value = 0.084
Table S24. Gene #3: 'EIF1AX MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 18 | 28 | 16 | 12 |
| EIF1AX MUTATED | 0 | 6 | 0 | 2 |
| EIF1AX WILD-TYPE | 18 | 22 | 16 | 10 |
Figure S9. Get High-res Image Gene #3: 'EIF1AX MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 0.0364 (Fisher's exact test), Q value = 0.084
Table S25. Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 22 | 7 | 10 | 31 | 6 | 4 |
| SF3B1 MUTATED | 3 | 0 | 1 | 13 | 0 | 1 |
| SF3B1 WILD-TYPE | 19 | 7 | 9 | 18 | 6 | 3 |
Figure S10. Get High-res Image Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
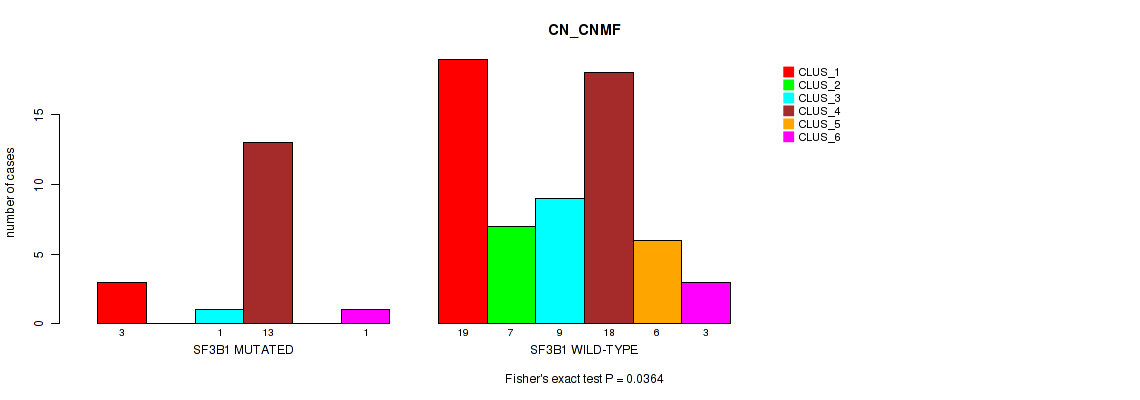
P value = 0.00011 (Fisher's exact test), Q value = 0.00068
Table S26. Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 28 | 14 | 14 | 24 |
| SF3B1 MUTATED | 0 | 2 | 5 | 11 |
| SF3B1 WILD-TYPE | 28 | 12 | 9 | 13 |
Figure S11. Get High-res Image Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
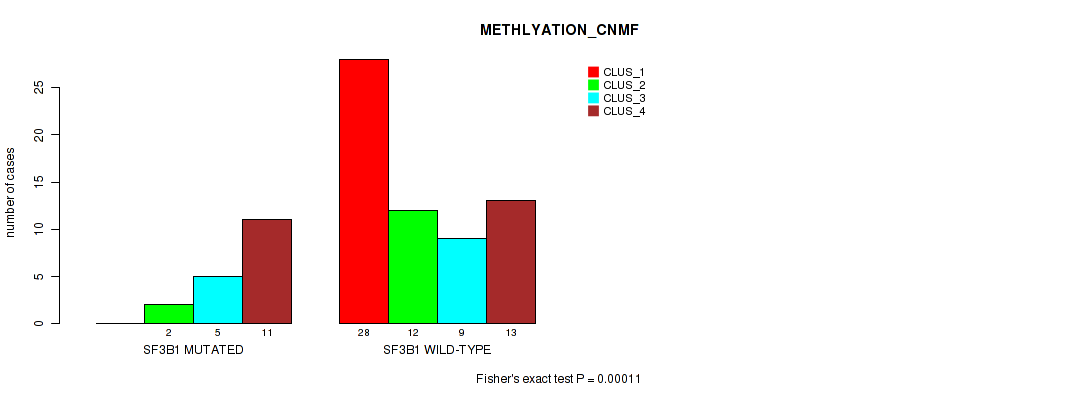
P value = 9e-05 (Fisher's exact test), Q value = 0.00063
Table S27. Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 18 | 15 | 32 | 15 |
| SF3B1 MUTATED | 3 | 0 | 15 | 0 |
| SF3B1 WILD-TYPE | 15 | 15 | 17 | 15 |
Figure S12. Get High-res Image Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 0.00183 (Fisher's exact test), Q value = 0.0093
Table S28. Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 33 | 18 |
| SF3B1 MUTATED | 2 | 14 | 2 |
| SF3B1 WILD-TYPE | 27 | 19 | 16 |
Figure S13. Get High-res Image Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
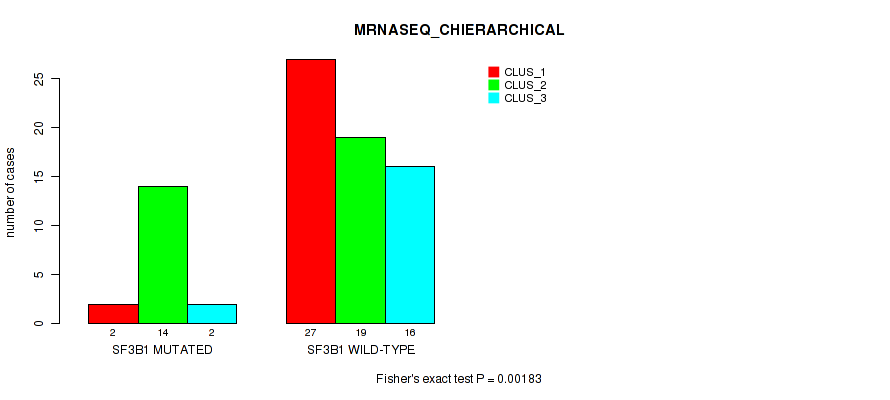
P value = 0.0325 (Fisher's exact test), Q value = 0.083
Table S29. Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 8 | 8 | 11 | 25 | 23 | 5 |
| SF3B1 MUTATED | 1 | 1 | 3 | 10 | 1 | 2 |
| SF3B1 WILD-TYPE | 7 | 7 | 8 | 15 | 22 | 3 |
Figure S14. Get High-res Image Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
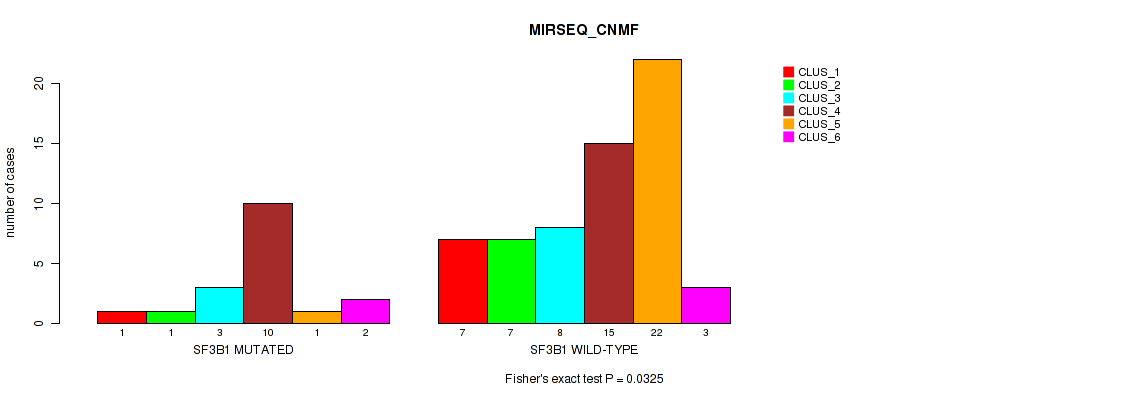
P value = 0.0184 (Fisher's exact test), Q value = 0.06
Table S30. Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 17 | 13 | 28 | 22 |
| SF3B1 MUTATED | 1 | 5 | 10 | 2 |
| SF3B1 WILD-TYPE | 16 | 8 | 18 | 20 |
Figure S15. Get High-res Image Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
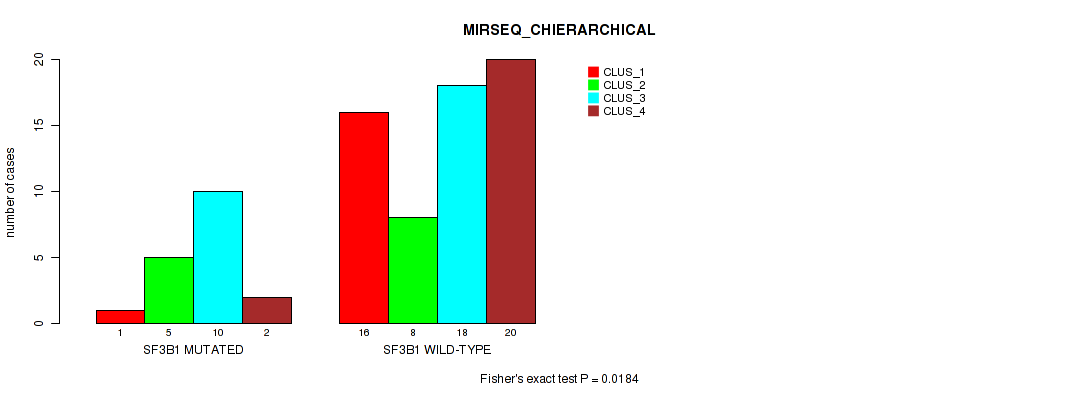
P value = 0.00793 (Fisher's exact test), Q value = 0.031
Table S31. Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 20 | 7 | 17 | 9 | 8 | 7 | 6 |
| SF3B1 MUTATED | 0 | 0 | 7 | 3 | 3 | 2 | 2 |
| SF3B1 WILD-TYPE | 20 | 7 | 10 | 6 | 5 | 5 | 4 |
Figure S16. Get High-res Image Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
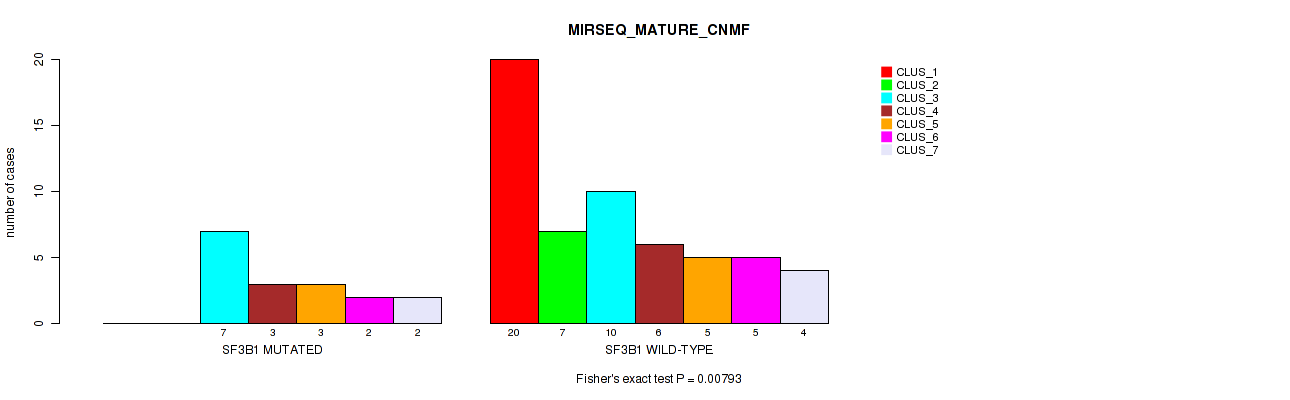
P value = 0.0166 (Fisher's exact test), Q value = 0.058
Table S32. Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 18 | 28 | 16 | 12 |
| SF3B1 MUTATED | 1 | 12 | 2 | 2 |
| SF3B1 WILD-TYPE | 17 | 16 | 14 | 10 |
Figure S17. Get High-res Image Gene #4: 'SF3B1 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 3e-05 (Fisher's exact test), Q value = 0.00028
Table S33. Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 22 | 7 | 10 | 31 | 6 | 4 |
| BAP1 MUTATED | 8 | 5 | 5 | 0 | 3 | 1 |
| BAP1 WILD-TYPE | 14 | 2 | 5 | 31 | 3 | 3 |
Figure S18. Get High-res Image Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S34. Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 28 | 14 | 14 | 24 |
| BAP1 MUTATED | 14 | 1 | 7 | 0 |
| BAP1 WILD-TYPE | 14 | 13 | 7 | 24 |
Figure S19. Get High-res Image Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
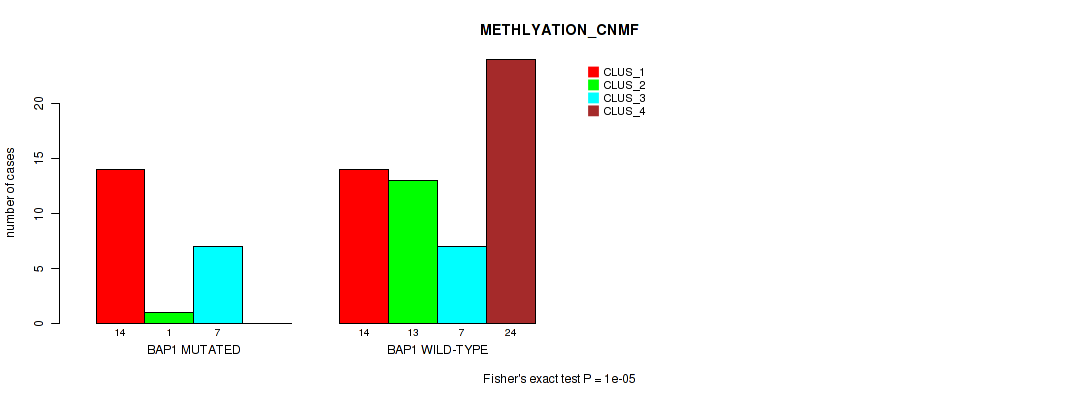
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S35. Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 18 | 15 | 32 | 15 |
| BAP1 MUTATED | 11 | 6 | 0 | 5 |
| BAP1 WILD-TYPE | 7 | 9 | 32 | 10 |
Figure S20. Get High-res Image Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S36. Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 33 | 18 |
| BAP1 MUTATED | 14 | 0 | 8 |
| BAP1 WILD-TYPE | 15 | 33 | 10 |
Figure S21. Get High-res Image Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
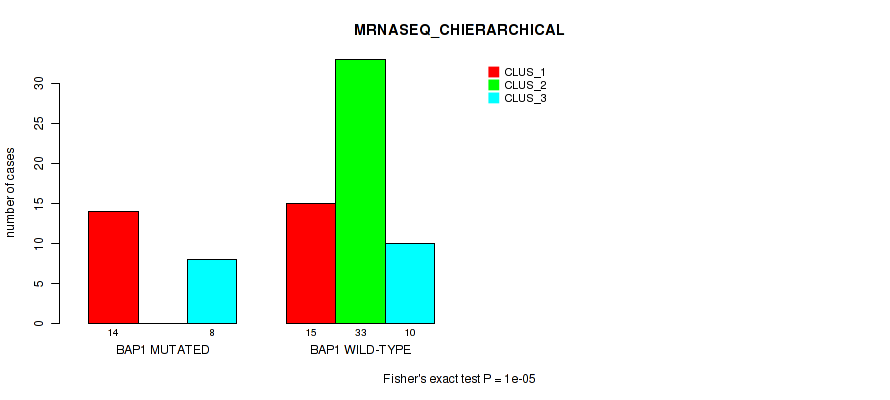
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S37. Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 8 | 8 | 11 | 25 | 23 | 5 |
| BAP1 MUTATED | 6 | 2 | 0 | 1 | 12 | 1 |
| BAP1 WILD-TYPE | 2 | 6 | 11 | 24 | 11 | 4 |
Figure S22. Get High-res Image Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
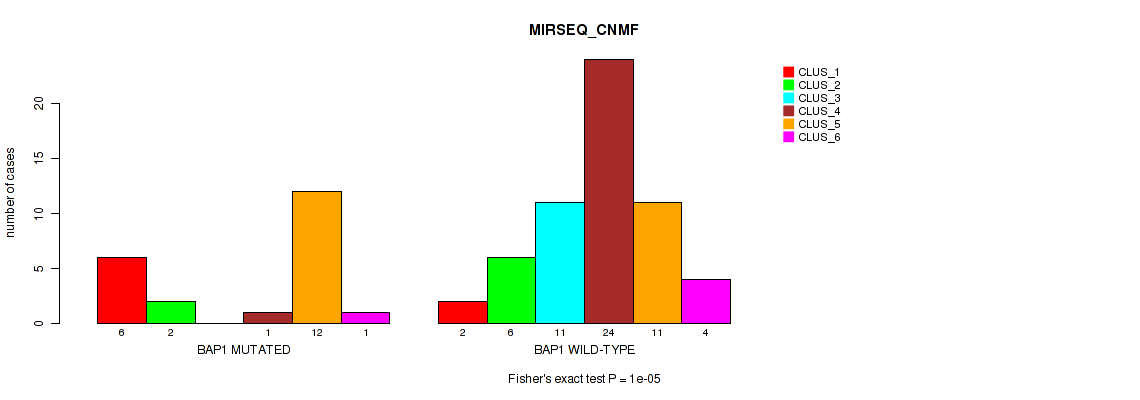
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S38. Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 17 | 13 | 28 | 22 |
| BAP1 MUTATED | 9 | 0 | 1 | 12 |
| BAP1 WILD-TYPE | 8 | 13 | 27 | 10 |
Figure S23. Get High-res Image Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
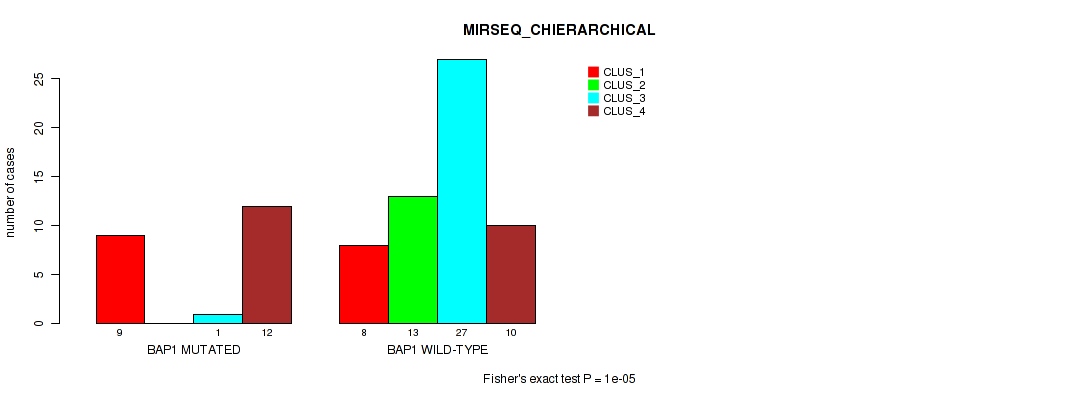
P value = 0.00277 (Fisher's exact test), Q value = 0.013
Table S39. Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 20 | 7 | 17 | 9 | 8 | 7 | 6 |
| BAP1 MUTATED | 10 | 3 | 0 | 4 | 3 | 1 | 0 |
| BAP1 WILD-TYPE | 10 | 4 | 17 | 5 | 5 | 6 | 6 |
Figure S24. Get High-res Image Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
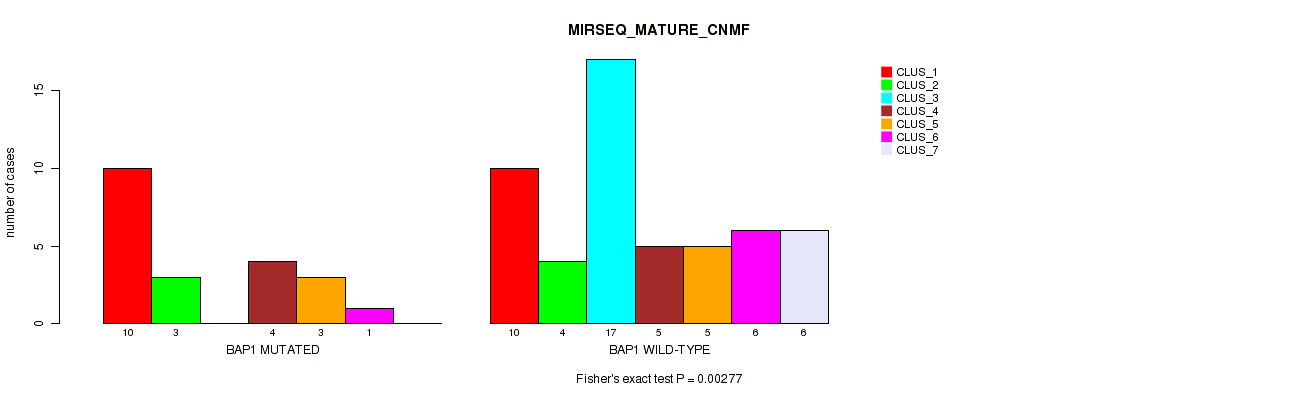
P value = 7e-05 (Fisher's exact test), Q value = 0.00056
Table S40. Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 18 | 28 | 16 | 12 |
| BAP1 MUTATED | 11 | 1 | 6 | 3 |
| BAP1 WILD-TYPE | 7 | 27 | 10 | 9 |
Figure S25. Get High-res Image Gene #5: 'BAP1 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 0.0275 (Fisher's exact test), Q value = 0.077
Table S41. Gene #6: 'CYSLTR2 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 22 | 7 | 10 | 31 | 6 | 4 |
| CYSLTR2 MUTATED | 0 | 0 | 2 | 0 | 1 | 0 |
| CYSLTR2 WILD-TYPE | 22 | 7 | 8 | 31 | 5 | 4 |
Figure S26. Get High-res Image Gene #6: 'CYSLTR2 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1 (Fisher's exact test), Q value = 1
Table S42. Gene #6: 'CYSLTR2 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 28 | 14 | 14 | 24 |
| CYSLTR2 MUTATED | 1 | 0 | 1 | 1 |
| CYSLTR2 WILD-TYPE | 27 | 14 | 13 | 23 |
P value = 0.0599 (Fisher's exact test), Q value = 0.12
Table S43. Gene #6: 'CYSLTR2 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 18 | 15 | 32 | 15 |
| CYSLTR2 MUTATED | 0 | 2 | 0 | 1 |
| CYSLTR2 WILD-TYPE | 18 | 13 | 32 | 14 |
P value = 0.107 (Fisher's exact test), Q value = 0.18
Table S44. Gene #6: 'CYSLTR2 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 33 | 18 |
| CYSLTR2 MUTATED | 1 | 0 | 2 |
| CYSLTR2 WILD-TYPE | 28 | 33 | 16 |
P value = 0.24 (Fisher's exact test), Q value = 0.34
Table S45. Gene #6: 'CYSLTR2 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 8 | 8 | 11 | 25 | 23 | 5 |
| CYSLTR2 MUTATED | 1 | 1 | 0 | 0 | 1 | 0 |
| CYSLTR2 WILD-TYPE | 7 | 7 | 11 | 25 | 22 | 5 |
P value = 0.274 (Fisher's exact test), Q value = 0.36
Table S46. Gene #6: 'CYSLTR2 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 17 | 13 | 28 | 22 |
| CYSLTR2 MUTATED | 2 | 0 | 1 | 0 |
| CYSLTR2 WILD-TYPE | 15 | 13 | 27 | 22 |
P value = 0.6 (Fisher's exact test), Q value = 0.65
Table S47. Gene #6: 'CYSLTR2 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 20 | 7 | 17 | 9 | 8 | 7 | 6 |
| CYSLTR2 MUTATED | 1 | 0 | 0 | 1 | 0 | 1 | 0 |
| CYSLTR2 WILD-TYPE | 19 | 7 | 17 | 8 | 8 | 6 | 6 |
P value = 0.367 (Fisher's exact test), Q value = 0.43
Table S48. Gene #6: 'CYSLTR2 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 18 | 28 | 16 | 12 |
| CYSLTR2 MUTATED | 1 | 0 | 1 | 1 |
| CYSLTR2 WILD-TYPE | 17 | 28 | 15 | 11 |
P value = 0.258 (Fisher's exact test), Q value = 0.36
Table S49. Gene #7: 'SFRS2 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 22 | 7 | 10 | 31 | 6 | 4 |
| SFRS2 MUTATED | 3 | 0 | 0 | 0 | 0 | 0 |
| SFRS2 WILD-TYPE | 19 | 7 | 10 | 31 | 6 | 4 |
P value = 0.182 (Fisher's exact test), Q value = 0.28
Table S50. Gene #7: 'SFRS2 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 28 | 14 | 14 | 24 |
| SFRS2 MUTATED | 1 | 2 | 0 | 0 |
| SFRS2 WILD-TYPE | 27 | 12 | 14 | 24 |
P value = 0.0595 (Fisher's exact test), Q value = 0.12
Table S51. Gene #7: 'SFRS2 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 18 | 15 | 32 | 15 |
| SFRS2 MUTATED | 0 | 2 | 0 | 1 |
| SFRS2 WILD-TYPE | 18 | 13 | 32 | 14 |
P value = 0.109 (Fisher's exact test), Q value = 0.18
Table S52. Gene #7: 'SFRS2 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 29 | 33 | 18 |
| SFRS2 MUTATED | 1 | 0 | 2 |
| SFRS2 WILD-TYPE | 28 | 33 | 16 |
P value = 0.402 (Fisher's exact test), Q value = 0.45
Table S53. Gene #7: 'SFRS2 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 8 | 8 | 11 | 25 | 23 | 5 |
| SFRS2 MUTATED | 1 | 0 | 1 | 0 | 1 | 0 |
| SFRS2 WILD-TYPE | 7 | 8 | 10 | 25 | 22 | 5 |
P value = 0.381 (Fisher's exact test), Q value = 0.44
Table S54. Gene #7: 'SFRS2 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 17 | 13 | 28 | 22 |
| SFRS2 MUTATED | 1 | 1 | 0 | 1 |
| SFRS2 WILD-TYPE | 16 | 12 | 28 | 21 |
P value = 0.818 (Fisher's exact test), Q value = 0.83
Table S55. Gene #7: 'SFRS2 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 20 | 7 | 17 | 9 | 8 | 7 | 6 |
| SFRS2 MUTATED | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| SFRS2 WILD-TYPE | 19 | 6 | 16 | 9 | 8 | 7 | 6 |
P value = 0.0416 (Fisher's exact test), Q value = 0.09
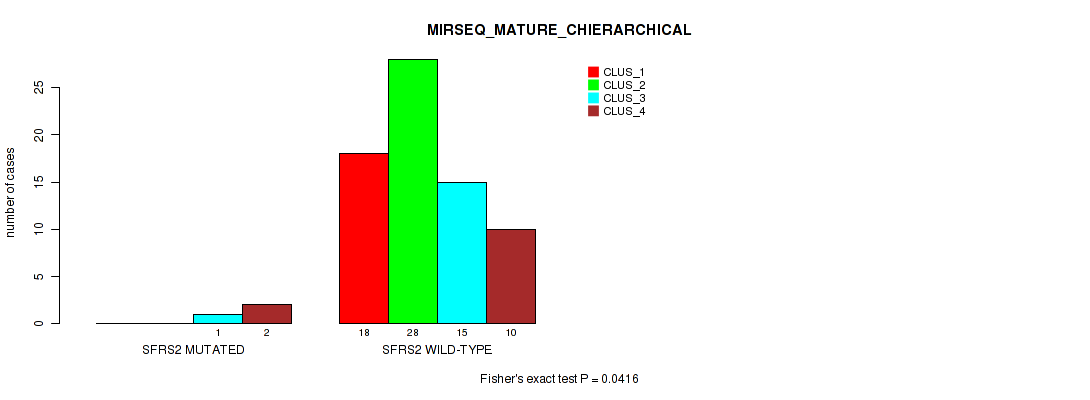
Table S56. Gene #7: 'SFRS2 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 18 | 28 | 16 | 12 |
| SFRS2 MUTATED | 0 | 0 | 1 | 2 |
| SFRS2 WILD-TYPE | 18 | 28 | 15 | 10 |
Figure S27. Get High-res Image Gene #7: 'SFRS2 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
-
Mutation data file = sample_sig_gene_table.txt from Mutsig_2CV pipeline
-
Processed Mutation data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_Correlate_Genomic_Events_Preprocess/UVM-TP/22572046/transformed.cor.cli.txt
-
Molecular subtypes file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_mergedClustering/UVM-TP/22542449/UVM-TP.transferedmergedcluster.txt
-
Number of patients = 80
-
Number of significantly mutated genes = 7
-
Number of Molecular subtypes = 8
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.