This pipeline infers putative direct gene targets of miRs based on miR and gene expression profiles across multiple samples.
This pipeline use a relevance network approach to infer putative miR:mRNA regulatory connections. All miR:mRNA pairs that have correlations < -0.3 and have predicted interactions in three sequence prediction databases (Miranda, Pictar, Targetscan) define the final network.
Figure 1. Get High-res Image All miR hubs with their strong anti-correlated genes and predicted interactions in three sequence prediction databases.
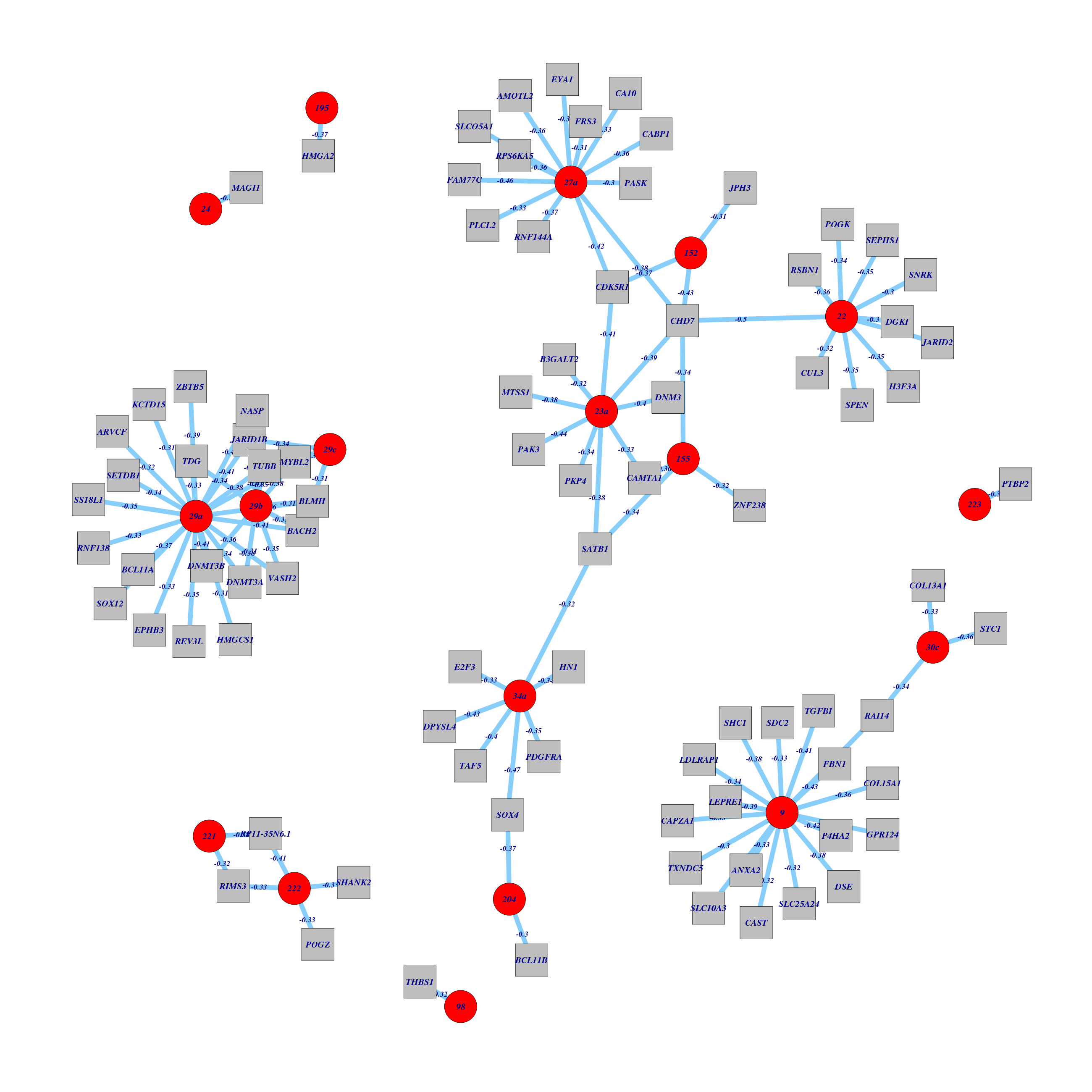
Table 1. Get Full Table List of miR:gene pairs with corr < -0.30 and predicted interactions in three sequence prediction databases.
| miR | gene | Corr | prediction databases | miranda | pictar | targetscan | total |
|---|---|---|---|---|---|---|---|
| hsa-mir-22 | CHD7 | -0.497 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-mir-34a | SOX4 | -0.471 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-mir-27a | FAM77C | -0.465 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-mir-23a | PAK3 | -0.441 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-mir-9 | FBN1 | -0.430 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-mir-152 | CHD7 | -0.429 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-mir-34a | DPYSL4 | -0.429 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-mir-27a | CDK5R1 | -0.424 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-mir-9 | P4HA2 | -0.418 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-mir-9 | TGFBI | -0.414 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
Table 2. Get Full Table All miR hubs with their associated genes in the putative direct target network.
| Mir | Number.of.Genes | Genes |
|---|---|---|
| hsa-mir-29a | 21 | ARVCF, BACH2, BCL11A, BLMH, DNMT3A, DNMT3B, EPHB3, HMGCS1, JARID1B, KCTD15, MYBL2, NASP, REV3L, RNF138, SOX12, SS18L1, TDG, TUBB, VASH2, ZBTB5,SETDB1 |
| hsa-mir-9 | 17 | ANXA2, CAPZA1, CAST, COL15A1, DSE, FBN1, GPR124, LDLRAP1, LEPRE1, P4HA2, RAI14, SHC1, SLC10A3, SLC25A24, TGFBI, TXNDC5,SDC2 |
| hsa-mir-27a | 13 | AMOTL2, CA10, CABP1, CDK5R1, CHD7, EYA1, FAM77C, FRS3, PASK, PLCL2, RNF144A, SLCO5A1,RPS6KA5 |
| hsa-mir-22 | 10 | CHD7, CUL3, DGKI, H3F3A, JARID2, POGK, RSBN1, SNRK, SPEN,SEPHS1 |
| hsa-mir-29b | 10 | BACH2, BLMH, DNMT3A, DNMT3B, JARID1B, MYBL2, TDG, TUBB, VASH2,NASP |
| hsa-mir-23a | 9 | B3GALT2, CDK5R1, CHD7, DNM3, MTSS1, PAK3, PKP4, SATB1,CAMTA1 |
| hsa-mir-34a | 7 | E2F3, HN1, PDGFRA, SATB1, SOX4, TAF5,DPYSL4 |
| hsa-mir-29c | 5 | BLMH, JARID1B, MYBL2, TUBB,BACH2 |
| hsa-mir-155 | 4 | CHD7, SATB1, ZNF238,CAMTA1 |
| hsa-mir-222 | 4 | POGZ, RIMS3, SHANK2,RP11-35N6.1 |
Table 3. Get Full Table All gene hubs with their associated miRs in the putative direct target network.
| Gene | Number.of.Mirs | Mirs |
|---|---|---|
| CHD7 | 5 | hsa-mir-155, hsa-mir-22, hsa-mir-23a, hsa-mir-27a,hsa-mir-152 |
| BLMH | 3 | hsa-mir-29b, hsa-mir-29c,hsa-mir-29a |
| TUBB | 3 | hsa-mir-29b, hsa-mir-29c,hsa-mir-29a |
| CDK5R1 | 3 | hsa-mir-23a, hsa-mir-27a,hsa-mir-152 |
| MYBL2 | 3 | hsa-mir-29b, hsa-mir-29c,hsa-mir-29a |
| JARID1B | 3 | hsa-mir-29b, hsa-mir-29c,hsa-mir-29a |
| SATB1 | 3 | hsa-mir-23a, hsa-mir-34a,hsa-mir-155 |
| BACH2 | 3 | hsa-mir-29b, hsa-mir-29c,hsa-mir-29a |
| TDG | 2 | hsa-mir-29b,hsa-mir-29a |
| NASP | 2 | hsa-mir-29b,hsa-mir-29a |
This section should list the files that were used as input.
-
Level 3 miR expression file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_MergeDataFilesPipeline/GBMLGG-TP/22217161/GDAC_MergeDataFiles_12184404/GBMLGG-TP.mirna__h_mirna_8x15k__unc_edu__Level_3__unc_DWD_Batch_adjusted__data.data.txt
-
Level 3 gene expression file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_MergeDataFilesPipeline/GBMLGG-TP/22217225/GDAC_MergeDataFiles_12184409/GBMLGG-TP.transcriptome__ht_hg_u133a__broad_mit_edu__Level_3__gene_rma__data.data.txt
-
miR:gene predicted interactions file = /xchip/cga/reference/miR_predictions/human_interactions.predicted.v2.txt
-
Miranda = microrna.org Aug 2010 release, Microcosm version 5
-
Pictar = version 1
-
Targetscan = release 5.2
Pairwise Pearson correlations coefficients between all miR:gene pairs are first computed. All genes that have correlation values less than the user-defined threshold (-0.3) with a particular miR and have been predicted as targets of that miR in three sequence based prediction databases: Miranda[1][2] Pictar[3][4], TargetScan [5][6][7] are identified as putative direct targets of that miR. We infer a direct target miR:gene network which comprises all such putative direct associations.
-
threshold = -0.3
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.