This pipeline infers putative direct gene targets of miRs based on miRseq and mRNAseq expression profiles across multiple samples. This pipeline has the following steps:
-
The CLR approach[8] is applied to infer putative miR:gene regulatory connections.
-
Filtering miR:gene pairs based on Pearson correlation (<= -0.3 ).
-
Filtering miR:gene pairs based on predicted interactions in three sequence prediction databases (Miranda, Pictar, Targetscan)
The CLR algorithm was applied on 761 miRs and 18694 mRNAs across 370 samples. After 2 filtering steps, the number of 154 miR:genes pairs were detected.
Figure 1. Get High-res Image All miR hubs with their strong anti-correlated genes and predicted interactions in three sequence prediction databases.
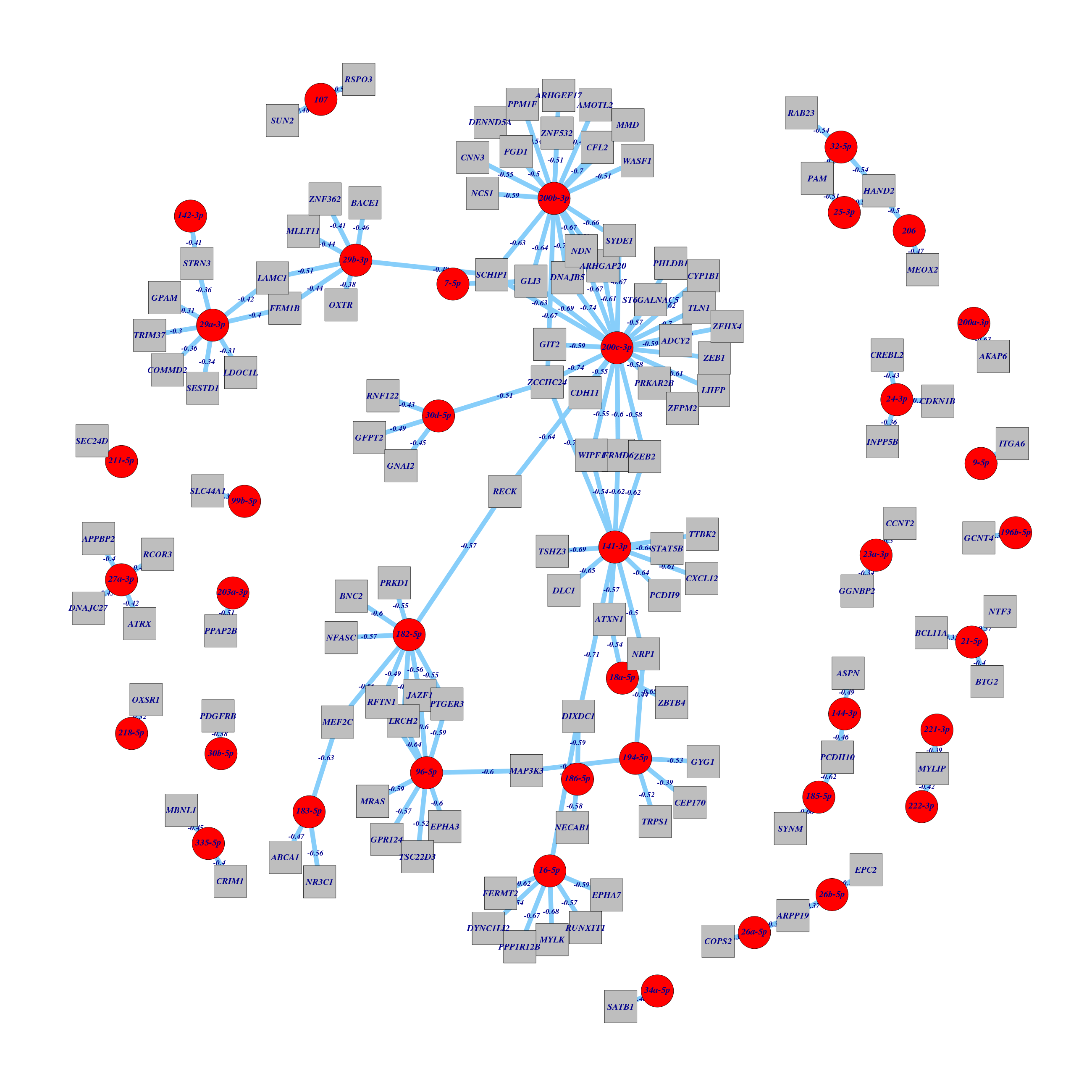
Table 1. Get Full Table List of miR:gene pairs with corr < -0.30 and predicted interactions in three sequence prediction databases.
| Mir | Gene | Corr | p | q | Prediction.DBs | Miranda | Pictar | Targetscan | Total |
|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-107 | RSPO3 | -0.54 | 1.6e-06 | 2e-06 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-miR-107 | SUN2 | -0.48 | 1.2e-06 | 1.5e-06 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-miR-141-3p | ZCCHC24 | -0.75 | 2.8e-12 | 1.5e-11 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-miR-141-3p | DIXDC1 | -0.71 | 2.8e-06 | 3.1e-06 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-miR-141-3p | TSHZ3 | -0.69 | 2.2e-11 | 1e-10 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-miR-141-3p | DLC1 | -0.65 | 2e-09 | 5.7e-09 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-miR-141-3p | PCDH9 | -0.64 | 2.6e-06 | 2.9e-06 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-miR-141-3p | STAT5B | -0.64 | 1.4e-08 | 3.2e-08 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-miR-141-3p | FRMD6 | -0.62 | 1.2e-08 | 2.8e-08 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
| hsa-miR-141-3p | ZEB2 | -0.62 | 1.5e-10 | 5.7e-10 | miranda,pictar,targetscan | 1 | 1 | 1 | 3 |
Table 2. Get Full Table All miR hubs with their associated genes in the putative direct target network.
| Mir | Number.of.Genes | Genes |
|---|---|---|
| hsa-miR-200c-3p | 23 | ARHGAP20, CDH11, CYP1B1, DNAJB5, FRMD6, GIT2, GLI3, LHFP, NDN, PHLDB1, PRKAR2B, RECK, SCHIP1, ST6GALNAC5, SYDE1, TLN1, WIPF1, ZCCHC24, ZEB1, ZEB2, ZFHX4, ZFPM2,ADCY2 |
| hsa-miR-200b-3p | 18 | ARHGAP20, ARHGEF17, CFL2, CNN3, DENND5A, DNAJB5, FGD1, GLI3, MMD, NCS1, NDN, PPM1F, SCHIP1, SYDE1, WASF1, ZCCHC24, ZNF532,AMOTL2 |
| hsa-miR-141-3p | 13 | CXCL12, DIXDC1, DLC1, FRMD6, NRP1, PCDH9, STAT5B, TSHZ3, TTBK2, WIPF1, ZCCHC24, ZEB2,ATXN1 |
| hsa-miR-96-5p | 9 | GPR124, JAZF1, LRCH2, MAP3K3, MRAS, PTGER3, RFTN1, TSC22D3,EPHA3 |
| hsa-miR-182-5p | 9 | JAZF1, LRCH2, MEF2C, NFASC, PRKD1, PTGER3, RECK, RFTN1,BNC2 |
| hsa-miR-29a-3p | 8 | FEM1B, GPAM, LAMC1, LDOC1L, SESTD1, STRN3, TRIM37,COMMD2 |
| hsa-miR-29b-3p | 7 | FEM1B, LAMC1, MLLT11, OXTR, SCHIP1, ZNF362,BACE1 |
| hsa-miR-16-5p | 7 | DYNC1LI2, EPHA7, FERMT2, MYLK, PPP1R12B, RUNX1T1,DIXDC1 |
| hsa-miR-194-5p | 5 | GYG1, MAP3K3, NRP1, TRPS1,CEP170 |
| hsa-miR-30d-5p | 4 | GNAI2, RNF122, ZCCHC24,GFPT2 |
Table 3. Get Full Table All gene hubs with their associated miRs in the putative direct target network.
| Gene | Number.of.Mirs | Mirs |
|---|---|---|
| ZCCHC24 | 4 | hsa-miR-141-3p, hsa-miR-200b-3p, hsa-miR-200c-3p,hsa-miR-30d-5p |
| HAND2 | 3 | hsa-miR-206, hsa-miR-32-5p,hsa-miR-25-3p |
| DIXDC1 | 3 | hsa-miR-141-3p, hsa-miR-186-5p,hsa-miR-16-5p |
| GLI3 | 3 | hsa-miR-200b-3p, hsa-miR-200c-3p,hsa-miR-7-5p |
| SCHIP1 | 3 | hsa-miR-200b-3p, hsa-miR-200c-3p,hsa-miR-29b-3p |
| PTGER3 | 2 | hsa-miR-182-5p,hsa-miR-96-5p |
| STRN3 | 2 | hsa-miR-142-3p,hsa-miR-29a-3p |
| ARHGAP20 | 2 | hsa-miR-200c-3p,hsa-miR-200b-3p |
| SYDE1 | 2 | hsa-miR-200c-3p,hsa-miR-200b-3p |
| PAM | 2 | hsa-miR-32-5p,hsa-miR-25-3p |
This section should list the files that were used as input.
-
miRseq (at precursor level) of RPM value (reads per million reads aligned to miRBase precursor) with log2 transformed = /xchip/cga/gdac-prod/tcga-gdac/jobResults/miRseq_mature_preprocess/STAD-TP/22312697/STAD-TP.miRseq_mature_RPM_log2.txt
-
mRNAseq of RSEM/RPKM value with log2 transformed = /xchip/cga/gdac-prod/tcga-gdac/jobResults/mRNAseq_preprocessor/STAD-TP/22315237/STAD-TP.uncv2.mRNAseq_RSEM_normalized_log2.txt
-
miR:gene predicted interactions file = /xchip/cga_home/hailei/FH/CLR/human_interactions.predicted.v2.mirbase21.txt
-
Miranda = microrna.org Aug 2010 release, Microcosm version 5
-
Pictar = version 1
-
Targetscan = release 5.2
The CLR (Context Likelihood of Relatedness) algorithm builds upon the relevance network strategies, by applying a background correction step. After computing the mutual information between regulators and their potential target genes, CLR calculates the statistical likelihood of each mutual information value within its network context. The algorithm compares the mutual information between a miR/gene pair to the background distribution of mutual information scores for all possible miR/gene pairs that include either the miR or its target. After this background correction, the most probable interactions are those whose mutual information scores stand significantly above the background distribution of mutual information scores[8] .
Pairwise Pearson correlations coefficients between all miR:gene pairs are first computed. All genes that have correlation values less than the user-defined threshold (-0.3) with a particular miR and have been predicted as targets of that miR in three sequence based prediction databases: Miranda[1][2] Pictar[3][4], TargetScan [5][6][7] are identified as putative direct targets of that miR. We infer a direct target miR:gene network which comprises all such putative direct associations.
-
threshold = -0.3
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.