This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 11 genes and 10 molecular subtypes across 57 patients, 8 significant findings detected with P value < 0.05 and Q value < 0.25.
-
KRAS mutation correlated to 'METHLYATION_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
PTEN mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CHIERARCHICAL', 'MRNASEQ_CNMF', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between mutation status of 11 genes and 10 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 8 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| PTEN | 11 (19%) | 46 |
0.00877 (0.204) |
0.00121 (0.104) |
0.0515 (0.388) |
0.0182 (0.25) |
0.0111 (0.204) |
0.0447 (0.378) |
0.0653 (0.392) |
0.06 (0.388) |
0.0093 (0.204) |
0.0134 (0.211) |
| KRAS | 7 (12%) | 50 |
0.14 (0.532) |
0.00189 (0.104) |
0.611 (0.884) |
0.201 (0.583) |
0.0301 (0.331) |
0.00587 (0.204) |
0.875 (1.00) |
0.0928 (0.464) |
0.169 (0.532) |
0.0592 (0.388) |
| TP53 | 51 (89%) | 6 |
0.631 (0.89) |
0.645 (0.892) |
0.418 (0.825) |
0.138 (0.532) |
1 (1.00) |
0.513 (0.841) |
1 (1.00) |
0.912 (1.00) |
0.801 (0.977) |
1 (1.00) |
| FBXW7 | 22 (39%) | 35 |
0.0886 (0.464) |
0.775 (0.958) |
0.0703 (0.392) |
0.166 (0.532) |
0.54 (0.848) |
0.104 (0.475) |
0.508 (0.841) |
0.932 (1.00) |
0.914 (1.00) |
0.774 (0.958) |
| PPP2R1A | 16 (28%) | 41 |
0.649 (0.892) |
0.808 (0.977) |
0.583 (0.862) |
0.501 (0.841) |
0.775 (0.958) |
0.672 (0.911) |
0.364 (0.792) |
0.316 (0.772) |
0.588 (0.862) |
0.235 (0.662) |
| RB1 | 6 (11%) | 51 |
1 (1.00) |
0.368 (0.792) |
0.471 (0.841) |
0.313 (0.772) |
0.0712 (0.392) |
0.0977 (0.467) |
0.871 (1.00) |
0.863 (1.00) |
0.522 (0.841) |
1 (1.00) |
| ZBTB7B | 6 (11%) | 51 |
0.527 (0.841) |
1 (1.00) |
0.144 (0.532) |
0.375 (0.792) |
1 (1.00) |
0.915 (1.00) |
0.157 (0.532) |
0.148 (0.532) |
0.427 (0.825) |
0.16 (0.532) |
| PIK3R1 | 6 (11%) | 51 |
0.871 (1.00) |
1 (1.00) |
0.506 (0.841) |
0.773 (0.958) |
0.571 (0.862) |
0.706 (0.935) |
1 (1.00) |
0.501 (0.841) |
0.423 (0.825) |
0.305 (0.772) |
| ARHGAP35 | 6 (11%) | 51 |
1 (1.00) |
0.458 (0.841) |
0.551 (0.853) |
0.344 (0.792) |
0.572 (0.862) |
0.398 (0.811) |
0.502 (0.841) |
0.192 (0.57) |
0.445 (0.841) |
0.174 (0.532) |
| PIK3CA | 20 (35%) | 37 |
0.389 (0.808) |
0.156 (0.532) |
0.715 (0.937) |
0.294 (0.771) |
0.275 (0.738) |
0.369 (0.792) |
0.0233 (0.284) |
0.339 (0.792) |
0.173 (0.532) |
0.269 (0.738) |
| MAMLD1 | 4 (7%) | 53 |
1 (1.00) |
0.137 (0.532) |
0.0396 (0.363) |
0.036 (0.36) |
0.626 (0.89) |
0.769 (0.958) |
0.0593 (0.388) |
0.679 (0.911) |
0.493 (0.841) |
0.356 (0.792) |
P value = 0.631 (Fisher's exact test), Q value = 0.89
Table S1. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 21 | 22 | 13 |
| TP53 MUTATED | 20 | 19 | 11 |
| TP53 WILD-TYPE | 1 | 3 | 2 |
P value = 0.645 (Fisher's exact test), Q value = 0.89
Table S2. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 13 | 13 | 11 |
| TP53 MUTATED | 19 | 11 | 12 | 9 |
| TP53 WILD-TYPE | 1 | 2 | 1 | 2 |
P value = 0.418 (Fisher's exact test), Q value = 0.82
Table S3. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 12 | 6 | 9 |
| TP53 MUTATED | 12 | 8 | 10 | 6 | 7 |
| TP53 WILD-TYPE | 0 | 1 | 2 | 0 | 2 |
P value = 0.138 (Fisher's exact test), Q value = 0.53
Table S4. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 9 | 8 | 7 | 7 | 7 | 5 | 5 |
| TP53 MUTATED | 9 | 8 | 7 | 6 | 6 | 4 | 3 |
| TP53 WILD-TYPE | 0 | 0 | 0 | 1 | 1 | 1 | 2 |
P value = 1 (Fisher's exact test), Q value = 1
Table S5. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 16 | 19 | 12 | 10 |
| TP53 MUTATED | 14 | 17 | 11 | 9 |
| TP53 WILD-TYPE | 2 | 2 | 1 | 1 |
P value = 0.513 (Fisher's exact test), Q value = 0.84
Table S6. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 11 | 4 | 8 | 6 | 8 | 6 | 14 |
| TP53 MUTATED | 11 | 3 | 8 | 5 | 7 | 5 | 12 |
| TP53 WILD-TYPE | 0 | 1 | 0 | 1 | 1 | 1 | 2 |
P value = 1 (Fisher's exact test), Q value = 1
Table S7. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 24 | 17 |
| TP53 MUTATED | 14 | 22 | 15 |
| TP53 WILD-TYPE | 1 | 2 | 2 |
P value = 0.912 (Fisher's exact test), Q value = 1
Table S8. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 24 | 8 | 18 | 6 |
| TP53 MUTATED | 21 | 8 | 16 | 6 |
| TP53 WILD-TYPE | 3 | 0 | 2 | 0 |
P value = 0.801 (Fisher's exact test), Q value = 0.98
Table S9. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 10 | 14 | 14 | 18 |
| TP53 MUTATED | 10 | 13 | 12 | 16 |
| TP53 WILD-TYPE | 0 | 1 | 2 | 2 |
P value = 1 (Fisher's exact test), Q value = 1
Table S10. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 25 | 7 | 17 | 7 |
| TP53 MUTATED | 22 | 7 | 15 | 7 |
| TP53 WILD-TYPE | 3 | 0 | 2 | 0 |
P value = 0.0886 (Fisher's exact test), Q value = 0.46
Table S11. Gene #2: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 21 | 22 | 13 |
| FBXW7 MUTATED | 11 | 9 | 2 |
| FBXW7 WILD-TYPE | 10 | 13 | 11 |
P value = 0.775 (Fisher's exact test), Q value = 0.96
Table S12. Gene #2: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 13 | 13 | 11 |
| FBXW7 MUTATED | 9 | 4 | 4 | 5 |
| FBXW7 WILD-TYPE | 11 | 9 | 9 | 6 |
P value = 0.0703 (Fisher's exact test), Q value = 0.39
Table S13. Gene #2: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 12 | 6 | 9 |
| FBXW7 MUTATED | 5 | 6 | 7 | 1 | 1 |
| FBXW7 WILD-TYPE | 7 | 3 | 5 | 5 | 8 |
P value = 0.166 (Fisher's exact test), Q value = 0.53
Table S14. Gene #2: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 9 | 8 | 7 | 7 | 7 | 5 | 5 |
| FBXW7 MUTATED | 4 | 4 | 1 | 3 | 5 | 3 | 0 |
| FBXW7 WILD-TYPE | 5 | 4 | 6 | 4 | 2 | 2 | 5 |
P value = 0.54 (Fisher's exact test), Q value = 0.85
Table S15. Gene #2: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 16 | 19 | 12 | 10 |
| FBXW7 MUTATED | 7 | 7 | 6 | 2 |
| FBXW7 WILD-TYPE | 9 | 12 | 6 | 8 |
P value = 0.104 (Fisher's exact test), Q value = 0.48
Table S16. Gene #2: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 11 | 4 | 8 | 6 | 8 | 6 | 14 |
| FBXW7 MUTATED | 4 | 4 | 5 | 2 | 2 | 2 | 3 |
| FBXW7 WILD-TYPE | 7 | 0 | 3 | 4 | 6 | 4 | 11 |
P value = 0.508 (Fisher's exact test), Q value = 0.84
Table S17. Gene #2: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 24 | 17 |
| FBXW7 MUTATED | 4 | 10 | 8 |
| FBXW7 WILD-TYPE | 11 | 14 | 9 |
P value = 0.932 (Fisher's exact test), Q value = 1
Table S18. Gene #2: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 24 | 8 | 18 | 6 |
| FBXW7 MUTATED | 9 | 4 | 7 | 2 |
| FBXW7 WILD-TYPE | 15 | 4 | 11 | 4 |
P value = 0.914 (Fisher's exact test), Q value = 1
Table S19. Gene #2: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 10 | 14 | 14 | 18 |
| FBXW7 MUTATED | 5 | 5 | 5 | 7 |
| FBXW7 WILD-TYPE | 5 | 9 | 9 | 11 |
P value = 0.774 (Fisher's exact test), Q value = 0.96
Table S20. Gene #2: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 25 | 7 | 17 | 7 |
| FBXW7 MUTATED | 9 | 4 | 6 | 3 |
| FBXW7 WILD-TYPE | 16 | 3 | 11 | 4 |
P value = 0.649 (Fisher's exact test), Q value = 0.89
Table S21. Gene #3: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 21 | 22 | 13 |
| PPP2R1A MUTATED | 5 | 8 | 3 |
| PPP2R1A WILD-TYPE | 16 | 14 | 10 |
P value = 0.808 (Fisher's exact test), Q value = 0.98
Table S22. Gene #3: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 13 | 13 | 11 |
| PPP2R1A MUTATED | 7 | 4 | 3 | 2 |
| PPP2R1A WILD-TYPE | 13 | 9 | 10 | 9 |
P value = 0.583 (Fisher's exact test), Q value = 0.86
Table S23. Gene #3: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 12 | 6 | 9 |
| PPP2R1A MUTATED | 5 | 2 | 4 | 1 | 1 |
| PPP2R1A WILD-TYPE | 7 | 7 | 8 | 5 | 8 |
P value = 0.501 (Fisher's exact test), Q value = 0.84
Table S24. Gene #3: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 9 | 8 | 7 | 7 | 7 | 5 | 5 |
| PPP2R1A MUTATED | 3 | 1 | 3 | 3 | 1 | 2 | 0 |
| PPP2R1A WILD-TYPE | 6 | 7 | 4 | 4 | 6 | 3 | 5 |
P value = 0.775 (Fisher's exact test), Q value = 0.96
Table S25. Gene #3: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 16 | 19 | 12 | 10 |
| PPP2R1A MUTATED | 6 | 4 | 3 | 3 |
| PPP2R1A WILD-TYPE | 10 | 15 | 9 | 7 |
P value = 0.672 (Fisher's exact test), Q value = 0.91
Table S26. Gene #3: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 11 | 4 | 8 | 6 | 8 | 6 | 14 |
| PPP2R1A MUTATED | 4 | 1 | 4 | 1 | 2 | 2 | 2 |
| PPP2R1A WILD-TYPE | 7 | 3 | 4 | 5 | 6 | 4 | 12 |
P value = 0.364 (Fisher's exact test), Q value = 0.79
Table S27. Gene #3: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 24 | 17 |
| PPP2R1A MUTATED | 6 | 7 | 3 |
| PPP2R1A WILD-TYPE | 9 | 17 | 14 |
P value = 0.316 (Fisher's exact test), Q value = 0.77
Table S28. Gene #3: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 24 | 8 | 18 | 6 |
| PPP2R1A MUTATED | 8 | 4 | 3 | 1 |
| PPP2R1A WILD-TYPE | 16 | 4 | 15 | 5 |
P value = 0.588 (Fisher's exact test), Q value = 0.86
Table S29. Gene #3: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 10 | 14 | 14 | 18 |
| PPP2R1A MUTATED | 3 | 6 | 3 | 4 |
| PPP2R1A WILD-TYPE | 7 | 8 | 11 | 14 |
P value = 0.235 (Fisher's exact test), Q value = 0.66
Table S30. Gene #3: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 25 | 7 | 17 | 7 |
| PPP2R1A MUTATED | 8 | 4 | 3 | 1 |
| PPP2R1A WILD-TYPE | 17 | 3 | 14 | 6 |
P value = 0.14 (Fisher's exact test), Q value = 0.53
Table S31. Gene #4: 'KRAS MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 21 | 22 | 13 |
| KRAS MUTATED | 2 | 5 | 0 |
| KRAS WILD-TYPE | 19 | 17 | 13 |
P value = 0.00189 (Fisher's exact test), Q value = 0.1
Table S32. Gene #4: 'KRAS MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 13 | 13 | 11 |
| KRAS MUTATED | 0 | 1 | 1 | 5 |
| KRAS WILD-TYPE | 20 | 12 | 12 | 6 |
Figure S1. Get High-res Image Gene #4: 'KRAS MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
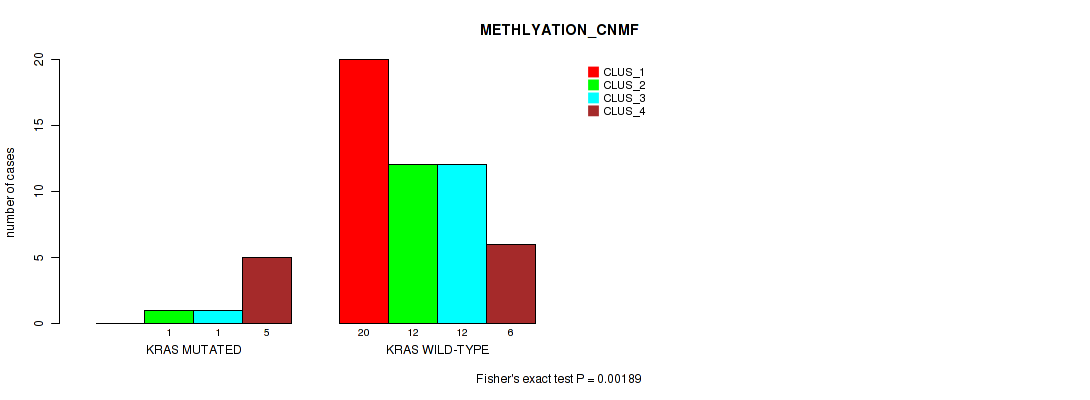
P value = 0.611 (Fisher's exact test), Q value = 0.88
Table S33. Gene #4: 'KRAS MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 12 | 6 | 9 |
| KRAS MUTATED | 1 | 2 | 2 | 0 | 0 |
| KRAS WILD-TYPE | 11 | 7 | 10 | 6 | 9 |
P value = 0.201 (Fisher's exact test), Q value = 0.58
Table S34. Gene #4: 'KRAS MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 9 | 8 | 7 | 7 | 7 | 5 | 5 |
| KRAS MUTATED | 0 | 0 | 2 | 1 | 2 | 0 | 0 |
| KRAS WILD-TYPE | 9 | 8 | 5 | 6 | 5 | 5 | 5 |
P value = 0.0301 (Fisher's exact test), Q value = 0.33
Table S35. Gene #4: 'KRAS MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 16 | 19 | 12 | 10 |
| KRAS MUTATED | 0 | 3 | 4 | 0 |
| KRAS WILD-TYPE | 16 | 16 | 8 | 10 |
Figure S2. Get High-res Image Gene #4: 'KRAS MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
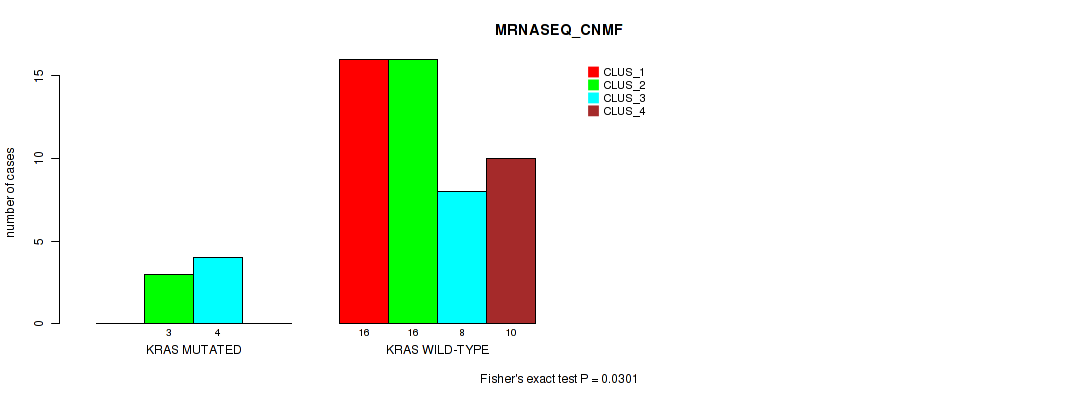
P value = 0.00587 (Fisher's exact test), Q value = 0.2
Table S36. Gene #4: 'KRAS MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 11 | 4 | 8 | 6 | 8 | 6 | 14 |
| KRAS MUTATED | 0 | 0 | 1 | 0 | 0 | 4 | 2 |
| KRAS WILD-TYPE | 11 | 4 | 7 | 6 | 8 | 2 | 12 |
Figure S3. Get High-res Image Gene #4: 'KRAS MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
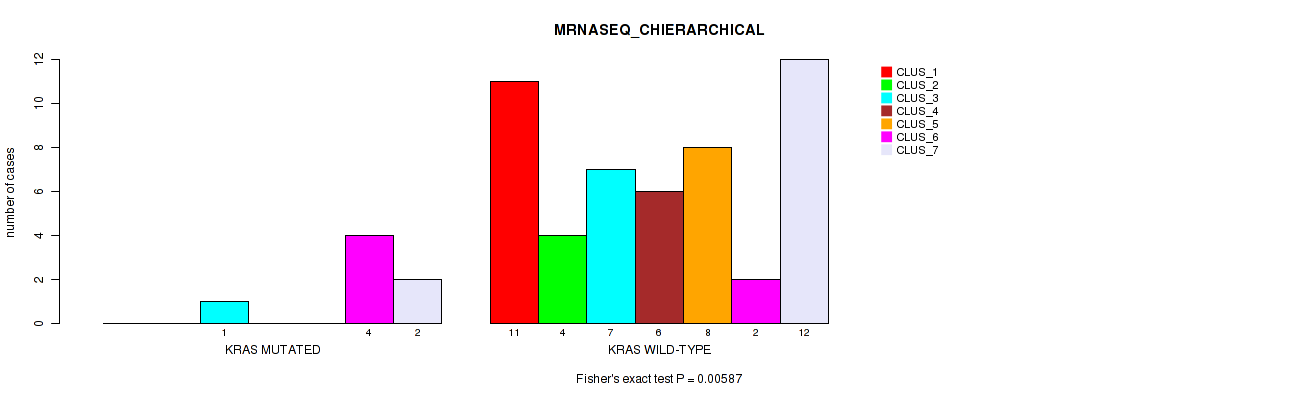
P value = 0.875 (Fisher's exact test), Q value = 1
Table S37. Gene #4: 'KRAS MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 24 | 17 |
| KRAS MUTATED | 1 | 4 | 2 |
| KRAS WILD-TYPE | 14 | 20 | 15 |
P value = 0.0928 (Fisher's exact test), Q value = 0.46
Table S38. Gene #4: 'KRAS MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 24 | 8 | 18 | 6 |
| KRAS MUTATED | 1 | 0 | 5 | 1 |
| KRAS WILD-TYPE | 23 | 8 | 13 | 5 |
P value = 0.169 (Fisher's exact test), Q value = 0.53
Table S39. Gene #4: 'KRAS MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 10 | 14 | 14 | 18 |
| KRAS MUTATED | 0 | 2 | 4 | 1 |
| KRAS WILD-TYPE | 10 | 12 | 10 | 17 |
P value = 0.0592 (Fisher's exact test), Q value = 0.39
Table S40. Gene #4: 'KRAS MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 25 | 7 | 17 | 7 |
| KRAS MUTATED | 1 | 0 | 5 | 1 |
| KRAS WILD-TYPE | 24 | 7 | 12 | 6 |
P value = 0.00877 (Fisher's exact test), Q value = 0.2
Table S41. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 21 | 22 | 13 |
| PTEN MUTATED | 1 | 9 | 1 |
| PTEN WILD-TYPE | 20 | 13 | 12 |
Figure S4. Get High-res Image Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
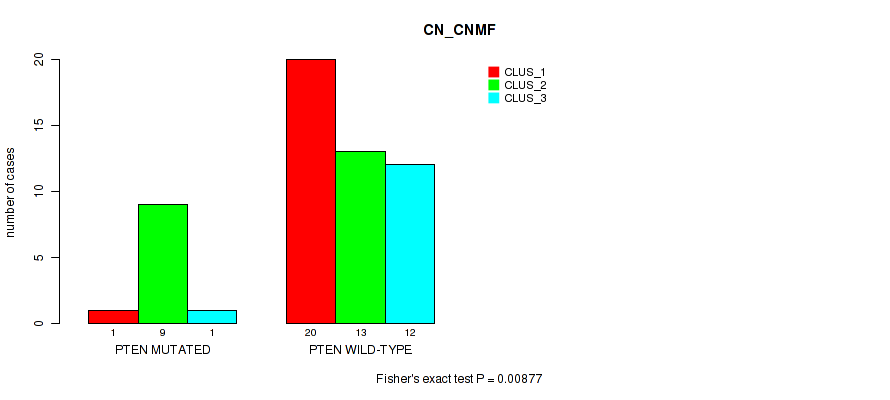
P value = 0.00121 (Fisher's exact test), Q value = 0.1
Table S42. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 13 | 13 | 11 |
| PTEN MUTATED | 1 | 7 | 3 | 0 |
| PTEN WILD-TYPE | 19 | 6 | 10 | 11 |
Figure S5. Get High-res Image Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
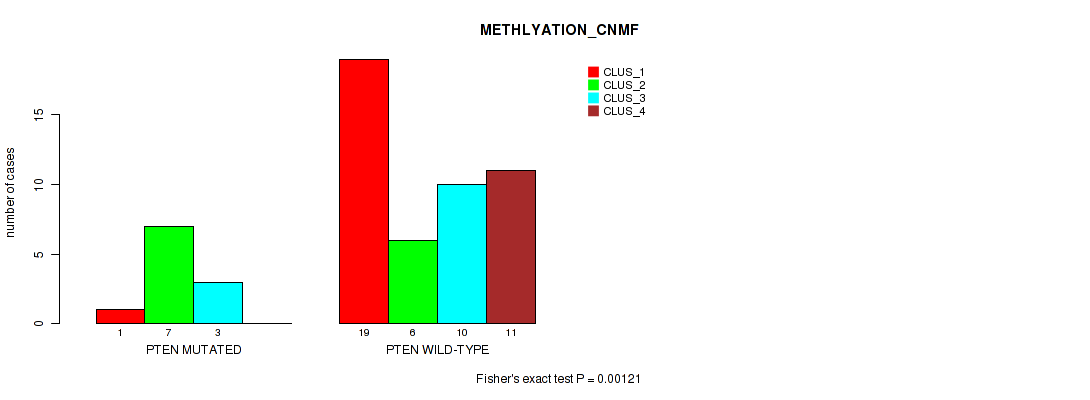
P value = 0.0515 (Fisher's exact test), Q value = 0.39
Table S43. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 12 | 6 | 9 |
| PTEN MUTATED | 0 | 0 | 3 | 2 | 3 |
| PTEN WILD-TYPE | 12 | 9 | 9 | 4 | 6 |
P value = 0.0182 (Fisher's exact test), Q value = 0.25
Table S44. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 9 | 8 | 7 | 7 | 7 | 5 | 5 |
| PTEN MUTATED | 0 | 2 | 0 | 1 | 0 | 3 | 2 |
| PTEN WILD-TYPE | 9 | 6 | 7 | 6 | 7 | 2 | 3 |
Figure S6. Get High-res Image Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
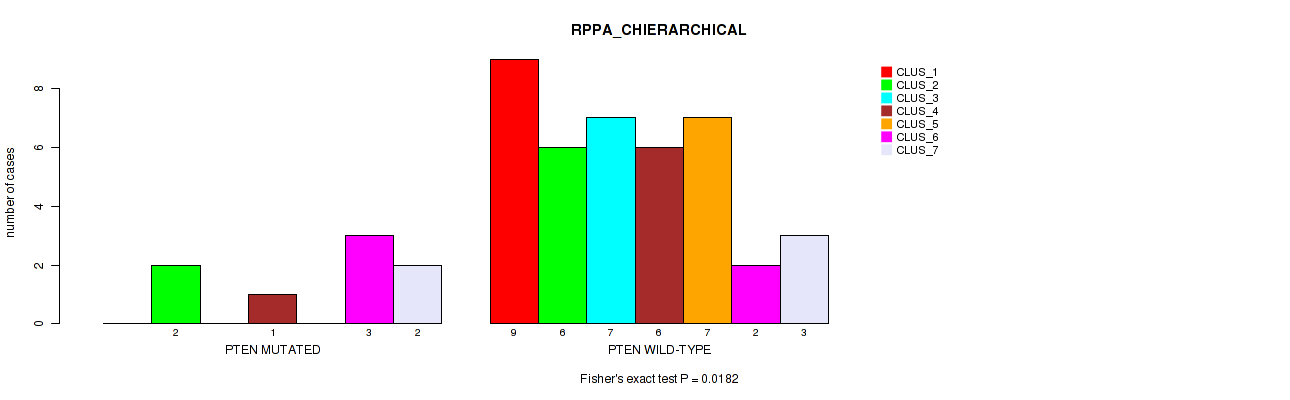
P value = 0.0111 (Fisher's exact test), Q value = 0.2
Table S45. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 16 | 19 | 12 | 10 |
| PTEN MUTATED | 1 | 8 | 0 | 2 |
| PTEN WILD-TYPE | 15 | 11 | 12 | 8 |
Figure S7. Get High-res Image Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
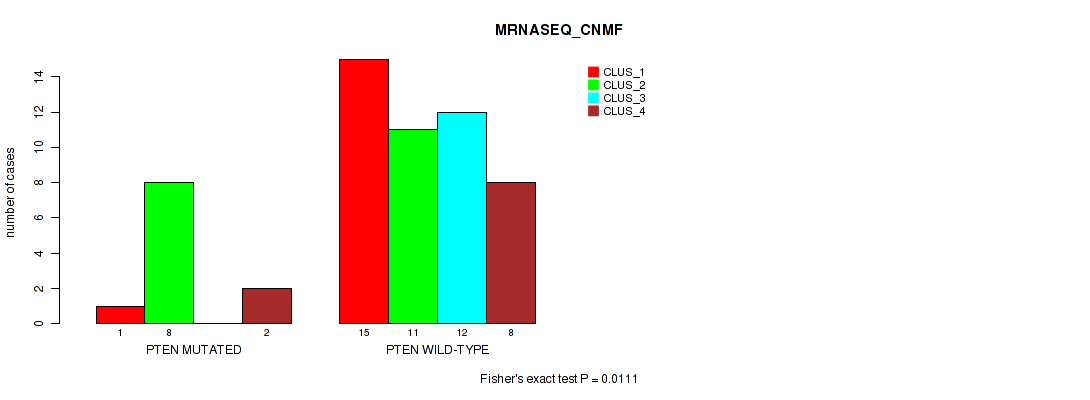
P value = 0.0447 (Fisher's exact test), Q value = 0.38
Table S46. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 11 | 4 | 8 | 6 | 8 | 6 | 14 |
| PTEN MUTATED | 0 | 0 | 3 | 0 | 3 | 0 | 5 |
| PTEN WILD-TYPE | 11 | 4 | 5 | 6 | 5 | 6 | 9 |
Figure S8. Get High-res Image Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
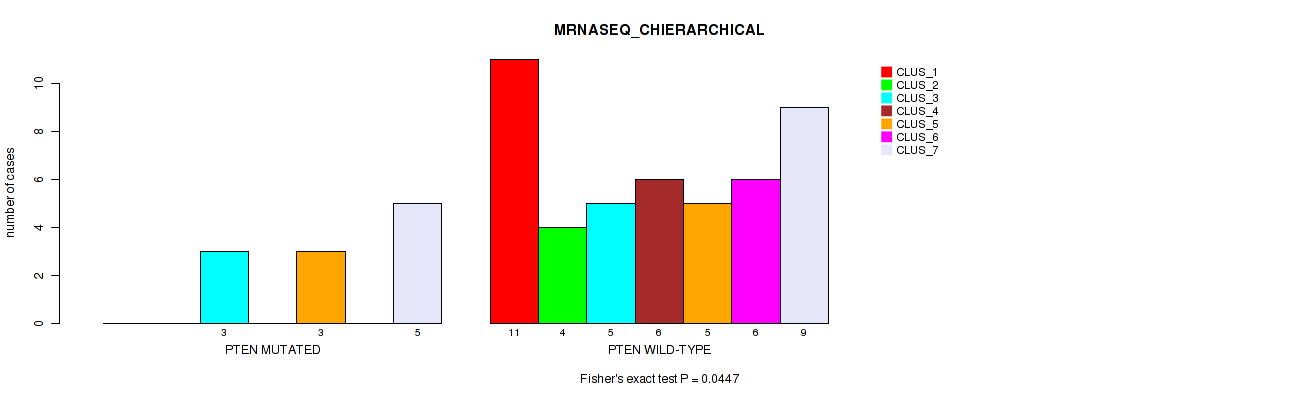
P value = 0.0653 (Fisher's exact test), Q value = 0.39
Table S47. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 24 | 17 |
| PTEN MUTATED | 4 | 1 | 5 |
| PTEN WILD-TYPE | 11 | 23 | 12 |
P value = 0.06 (Fisher's exact test), Q value = 0.39
Table S48. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 24 | 8 | 18 | 6 |
| PTEN MUTATED | 8 | 0 | 1 | 1 |
| PTEN WILD-TYPE | 16 | 8 | 17 | 5 |
P value = 0.0093 (Fisher's exact test), Q value = 0.2
Table S49. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 10 | 14 | 14 | 18 |
| PTEN MUTATED | 0 | 3 | 0 | 7 |
| PTEN WILD-TYPE | 10 | 11 | 14 | 11 |
Figure S9. Get High-res Image Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
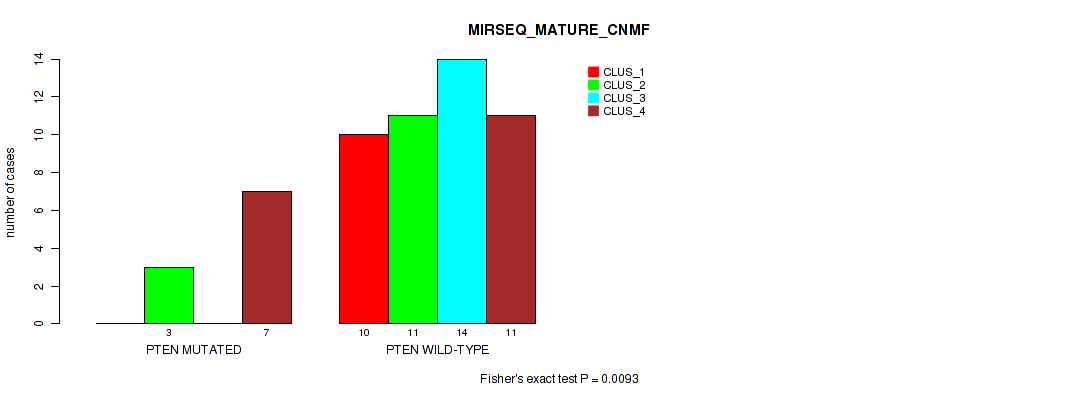
P value = 0.0134 (Fisher's exact test), Q value = 0.21
Table S50. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 25 | 7 | 17 | 7 |
| PTEN MUTATED | 8 | 0 | 0 | 2 |
| PTEN WILD-TYPE | 17 | 7 | 17 | 5 |
Figure S10. Get High-res Image Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
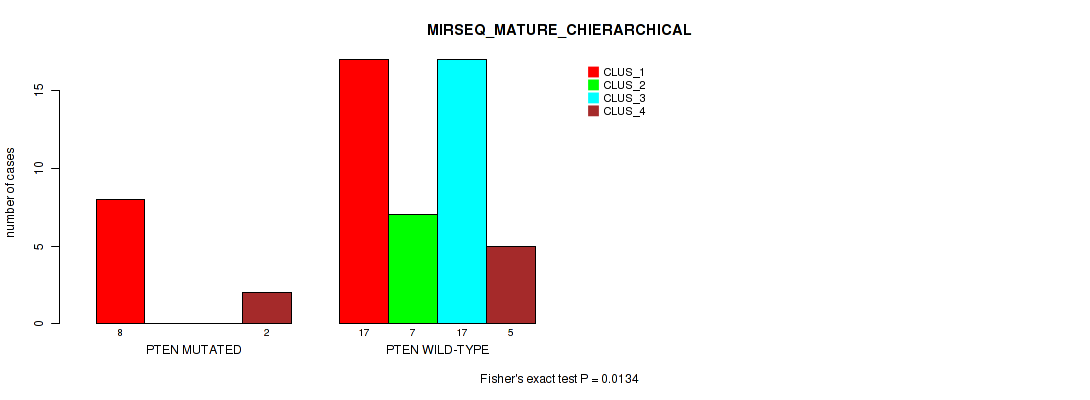
P value = 1 (Fisher's exact test), Q value = 1
Table S51. Gene #6: 'RB1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 21 | 22 | 13 |
| RB1 MUTATED | 2 | 3 | 1 |
| RB1 WILD-TYPE | 19 | 19 | 12 |
P value = 0.368 (Fisher's exact test), Q value = 0.79
Table S52. Gene #6: 'RB1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 13 | 13 | 11 |
| RB1 MUTATED | 2 | 3 | 1 | 0 |
| RB1 WILD-TYPE | 18 | 10 | 12 | 11 |
P value = 0.471 (Fisher's exact test), Q value = 0.84
Table S53. Gene #6: 'RB1 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 12 | 6 | 9 |
| RB1 MUTATED | 0 | 2 | 1 | 1 | 1 |
| RB1 WILD-TYPE | 12 | 7 | 11 | 5 | 8 |
P value = 0.313 (Fisher's exact test), Q value = 0.77
Table S54. Gene #6: 'RB1 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 9 | 8 | 7 | 7 | 7 | 5 | 5 |
| RB1 MUTATED | 0 | 1 | 0 | 2 | 0 | 1 | 1 |
| RB1 WILD-TYPE | 9 | 7 | 7 | 5 | 7 | 4 | 4 |
P value = 0.0712 (Fisher's exact test), Q value = 0.39
Table S55. Gene #6: 'RB1 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 16 | 19 | 12 | 10 |
| RB1 MUTATED | 0 | 4 | 0 | 2 |
| RB1 WILD-TYPE | 16 | 15 | 12 | 8 |
P value = 0.0977 (Fisher's exact test), Q value = 0.47
Table S56. Gene #6: 'RB1 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 11 | 4 | 8 | 6 | 8 | 6 | 14 |
| RB1 MUTATED | 0 | 0 | 2 | 0 | 3 | 0 | 1 |
| RB1 WILD-TYPE | 11 | 4 | 6 | 6 | 5 | 6 | 13 |
P value = 0.871 (Fisher's exact test), Q value = 1
Table S57. Gene #6: 'RB1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 24 | 17 |
| RB1 MUTATED | 2 | 2 | 2 |
| RB1 WILD-TYPE | 13 | 22 | 15 |
P value = 0.863 (Fisher's exact test), Q value = 1
Table S58. Gene #6: 'RB1 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 24 | 8 | 18 | 6 |
| RB1 MUTATED | 3 | 0 | 2 | 1 |
| RB1 WILD-TYPE | 21 | 8 | 16 | 5 |
P value = 0.522 (Fisher's exact test), Q value = 0.84
Table S59. Gene #6: 'RB1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 10 | 14 | 14 | 18 |
| RB1 MUTATED | 0 | 3 | 1 | 2 |
| RB1 WILD-TYPE | 10 | 11 | 13 | 16 |
P value = 1 (Fisher's exact test), Q value = 1
Table S60. Gene #6: 'RB1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 25 | 7 | 17 | 7 |
| RB1 MUTATED | 3 | 0 | 2 | 1 |
| RB1 WILD-TYPE | 22 | 7 | 15 | 6 |
P value = 0.527 (Fisher's exact test), Q value = 0.84
Table S61. Gene #7: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 21 | 22 | 13 |
| ZBTB7B MUTATED | 2 | 1 | 2 |
| ZBTB7B WILD-TYPE | 19 | 21 | 11 |
P value = 1 (Fisher's exact test), Q value = 1
Table S62. Gene #7: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 13 | 13 | 11 |
| ZBTB7B MUTATED | 2 | 1 | 2 | 1 |
| ZBTB7B WILD-TYPE | 18 | 12 | 11 | 10 |
P value = 0.144 (Fisher's exact test), Q value = 0.53
Table S63. Gene #7: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 12 | 6 | 9 |
| ZBTB7B MUTATED | 2 | 0 | 0 | 2 | 1 |
| ZBTB7B WILD-TYPE | 10 | 9 | 12 | 4 | 8 |
P value = 0.375 (Fisher's exact test), Q value = 0.79
Table S64. Gene #7: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 9 | 8 | 7 | 7 | 7 | 5 | 5 |
| ZBTB7B MUTATED | 2 | 2 | 0 | 0 | 0 | 0 | 1 |
| ZBTB7B WILD-TYPE | 7 | 6 | 7 | 7 | 7 | 5 | 4 |
P value = 1 (Fisher's exact test), Q value = 1
Table S65. Gene #7: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 16 | 19 | 12 | 10 |
| ZBTB7B MUTATED | 2 | 2 | 1 | 1 |
| ZBTB7B WILD-TYPE | 14 | 17 | 11 | 9 |
P value = 0.915 (Fisher's exact test), Q value = 1
Table S66. Gene #7: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 11 | 4 | 8 | 6 | 8 | 6 | 14 |
| ZBTB7B MUTATED | 2 | 0 | 0 | 0 | 1 | 1 | 2 |
| ZBTB7B WILD-TYPE | 9 | 4 | 8 | 6 | 7 | 5 | 12 |
P value = 0.157 (Fisher's exact test), Q value = 0.53
Table S67. Gene #7: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 24 | 17 |
| ZBTB7B MUTATED | 1 | 1 | 4 |
| ZBTB7B WILD-TYPE | 14 | 23 | 13 |
P value = 0.148 (Fisher's exact test), Q value = 0.53
Table S68. Gene #7: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 24 | 8 | 18 | 6 |
| ZBTB7B MUTATED | 5 | 1 | 0 | 0 |
| ZBTB7B WILD-TYPE | 19 | 7 | 18 | 6 |
P value = 0.427 (Fisher's exact test), Q value = 0.82
Table S69. Gene #7: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 10 | 14 | 14 | 18 |
| ZBTB7B MUTATED | 2 | 2 | 0 | 2 |
| ZBTB7B WILD-TYPE | 8 | 12 | 14 | 16 |
P value = 0.16 (Fisher's exact test), Q value = 0.53
Table S70. Gene #7: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 25 | 7 | 17 | 7 |
| ZBTB7B MUTATED | 5 | 1 | 0 | 0 |
| ZBTB7B WILD-TYPE | 20 | 6 | 17 | 7 |
P value = 0.871 (Fisher's exact test), Q value = 1
Table S71. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 21 | 22 | 13 |
| PIK3R1 MUTATED | 3 | 2 | 1 |
| PIK3R1 WILD-TYPE | 18 | 20 | 12 |
P value = 1 (Fisher's exact test), Q value = 1
Table S72. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 13 | 13 | 11 |
| PIK3R1 MUTATED | 3 | 1 | 1 | 1 |
| PIK3R1 WILD-TYPE | 17 | 12 | 12 | 10 |
P value = 0.506 (Fisher's exact test), Q value = 0.84
Table S73. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 12 | 6 | 9 |
| PIK3R1 MUTATED | 2 | 2 | 0 | 1 | 1 |
| PIK3R1 WILD-TYPE | 10 | 7 | 12 | 5 | 8 |
P value = 0.773 (Fisher's exact test), Q value = 0.96
Table S74. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 9 | 8 | 7 | 7 | 7 | 5 | 5 |
| PIK3R1 MUTATED | 2 | 2 | 0 | 1 | 1 | 0 | 0 |
| PIK3R1 WILD-TYPE | 7 | 6 | 7 | 6 | 6 | 5 | 5 |
P value = 0.571 (Fisher's exact test), Q value = 0.86
Table S75. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 16 | 19 | 12 | 10 |
| PIK3R1 MUTATED | 3 | 2 | 1 | 0 |
| PIK3R1 WILD-TYPE | 13 | 17 | 11 | 10 |
P value = 0.706 (Fisher's exact test), Q value = 0.94
Table S76. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 11 | 4 | 8 | 6 | 8 | 6 | 14 |
| PIK3R1 MUTATED | 2 | 1 | 1 | 0 | 0 | 1 | 1 |
| PIK3R1 WILD-TYPE | 9 | 3 | 7 | 6 | 8 | 5 | 13 |
P value = 1 (Fisher's exact test), Q value = 1
Table S77. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 24 | 17 |
| PIK3R1 MUTATED | 1 | 3 | 2 |
| PIK3R1 WILD-TYPE | 14 | 21 | 15 |
P value = 0.501 (Fisher's exact test), Q value = 0.84
Table S78. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 24 | 8 | 18 | 6 |
| PIK3R1 MUTATED | 2 | 2 | 2 | 0 |
| PIK3R1 WILD-TYPE | 22 | 6 | 16 | 6 |
P value = 0.423 (Fisher's exact test), Q value = 0.82
Table S79. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 10 | 14 | 14 | 18 |
| PIK3R1 MUTATED | 2 | 0 | 2 | 2 |
| PIK3R1 WILD-TYPE | 8 | 14 | 12 | 16 |
P value = 0.305 (Fisher's exact test), Q value = 0.77
Table S80. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 25 | 7 | 17 | 7 |
| PIK3R1 MUTATED | 2 | 2 | 1 | 1 |
| PIK3R1 WILD-TYPE | 23 | 5 | 16 | 6 |
P value = 1 (Fisher's exact test), Q value = 1
Table S81. Gene #9: 'ARHGAP35 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 21 | 22 | 13 |
| ARHGAP35 MUTATED | 2 | 3 | 1 |
| ARHGAP35 WILD-TYPE | 19 | 19 | 12 |
P value = 0.458 (Fisher's exact test), Q value = 0.84
Table S82. Gene #9: 'ARHGAP35 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 13 | 13 | 11 |
| ARHGAP35 MUTATED | 2 | 2 | 0 | 2 |
| ARHGAP35 WILD-TYPE | 18 | 11 | 13 | 9 |
P value = 0.551 (Fisher's exact test), Q value = 0.85
Table S83. Gene #9: 'ARHGAP35 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 12 | 6 | 9 |
| ARHGAP35 MUTATED | 2 | 0 | 3 | 0 | 1 |
| ARHGAP35 WILD-TYPE | 10 | 9 | 9 | 6 | 8 |
P value = 0.344 (Fisher's exact test), Q value = 0.79
Table S84. Gene #9: 'ARHGAP35 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 9 | 8 | 7 | 7 | 7 | 5 | 5 |
| ARHGAP35 MUTATED | 2 | 0 | 0 | 0 | 2 | 1 | 1 |
| ARHGAP35 WILD-TYPE | 7 | 8 | 7 | 7 | 5 | 4 | 4 |
P value = 0.572 (Fisher's exact test), Q value = 0.86
Table S85. Gene #9: 'ARHGAP35 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 16 | 19 | 12 | 10 |
| ARHGAP35 MUTATED | 3 | 2 | 1 | 0 |
| ARHGAP35 WILD-TYPE | 13 | 17 | 11 | 10 |
P value = 0.398 (Fisher's exact test), Q value = 0.81
Table S86. Gene #9: 'ARHGAP35 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 11 | 4 | 8 | 6 | 8 | 6 | 14 |
| ARHGAP35 MUTATED | 2 | 0 | 2 | 0 | 1 | 1 | 0 |
| ARHGAP35 WILD-TYPE | 9 | 4 | 6 | 6 | 7 | 5 | 14 |
P value = 0.502 (Fisher's exact test), Q value = 0.84
Table S87. Gene #9: 'ARHGAP35 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 24 | 17 |
| ARHGAP35 MUTATED | 0 | 3 | 2 |
| ARHGAP35 WILD-TYPE | 15 | 21 | 15 |
P value = 0.192 (Fisher's exact test), Q value = 0.57
Table S88. Gene #9: 'ARHGAP35 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 24 | 8 | 18 | 6 |
| ARHGAP35 MUTATED | 1 | 2 | 1 | 1 |
| ARHGAP35 WILD-TYPE | 23 | 6 | 17 | 5 |
P value = 0.445 (Fisher's exact test), Q value = 0.84
Table S89. Gene #9: 'ARHGAP35 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 10 | 14 | 14 | 18 |
| ARHGAP35 MUTATED | 2 | 0 | 1 | 2 |
| ARHGAP35 WILD-TYPE | 8 | 14 | 13 | 16 |
P value = 0.174 (Fisher's exact test), Q value = 0.53
Table S90. Gene #9: 'ARHGAP35 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 25 | 7 | 17 | 7 |
| ARHGAP35 MUTATED | 1 | 2 | 1 | 1 |
| ARHGAP35 WILD-TYPE | 24 | 5 | 16 | 6 |
P value = 0.389 (Fisher's exact test), Q value = 0.81
Table S91. Gene #10: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 21 | 22 | 13 |
| PIK3CA MUTATED | 6 | 10 | 3 |
| PIK3CA WILD-TYPE | 15 | 12 | 10 |
P value = 0.156 (Fisher's exact test), Q value = 0.53
Table S92. Gene #10: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 13 | 13 | 11 |
| PIK3CA MUTATED | 6 | 8 | 4 | 2 |
| PIK3CA WILD-TYPE | 14 | 5 | 9 | 9 |
P value = 0.715 (Fisher's exact test), Q value = 0.94
Table S93. Gene #10: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 12 | 6 | 9 |
| PIK3CA MUTATED | 3 | 3 | 6 | 3 | 3 |
| PIK3CA WILD-TYPE | 9 | 6 | 6 | 3 | 6 |
P value = 0.294 (Fisher's exact test), Q value = 0.77
Table S94. Gene #10: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 9 | 8 | 7 | 7 | 7 | 5 | 5 |
| PIK3CA MUTATED | 2 | 3 | 2 | 1 | 3 | 4 | 3 |
| PIK3CA WILD-TYPE | 7 | 5 | 5 | 6 | 4 | 1 | 2 |
P value = 0.275 (Fisher's exact test), Q value = 0.74
Table S95. Gene #10: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 16 | 19 | 12 | 10 |
| PIK3CA MUTATED | 5 | 10 | 3 | 2 |
| PIK3CA WILD-TYPE | 11 | 9 | 9 | 8 |
P value = 0.369 (Fisher's exact test), Q value = 0.79
Table S96. Gene #10: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 11 | 4 | 8 | 6 | 8 | 6 | 14 |
| PIK3CA MUTATED | 3 | 3 | 4 | 2 | 1 | 1 | 6 |
| PIK3CA WILD-TYPE | 8 | 1 | 4 | 4 | 7 | 5 | 8 |
P value = 0.0233 (Fisher's exact test), Q value = 0.28
Table S97. Gene #10: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 24 | 17 |
| PIK3CA MUTATED | 6 | 4 | 10 |
| PIK3CA WILD-TYPE | 9 | 20 | 7 |
Figure S11. Get High-res Image Gene #10: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
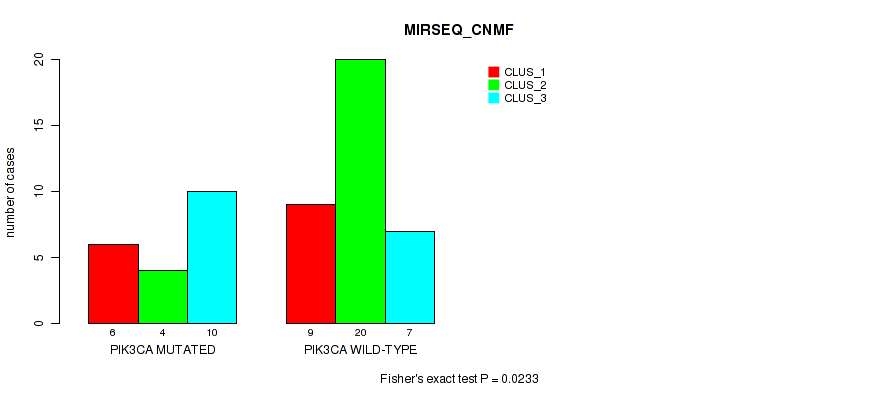
P value = 0.339 (Fisher's exact test), Q value = 0.79
Table S98. Gene #10: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 24 | 8 | 18 | 6 |
| PIK3CA MUTATED | 11 | 2 | 4 | 3 |
| PIK3CA WILD-TYPE | 13 | 6 | 14 | 3 |
P value = 0.173 (Fisher's exact test), Q value = 0.53
Table S99. Gene #10: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 10 | 14 | 14 | 18 |
| PIK3CA MUTATED | 3 | 6 | 2 | 9 |
| PIK3CA WILD-TYPE | 7 | 8 | 12 | 9 |
P value = 0.269 (Fisher's exact test), Q value = 0.74
Table S100. Gene #10: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 25 | 7 | 17 | 7 |
| PIK3CA MUTATED | 12 | 1 | 4 | 3 |
| PIK3CA WILD-TYPE | 13 | 6 | 13 | 4 |
P value = 1 (Fisher's exact test), Q value = 1
Table S101. Gene #11: 'MAMLD1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 21 | 22 | 13 |
| MAMLD1 MUTATED | 1 | 2 | 1 |
| MAMLD1 WILD-TYPE | 20 | 20 | 12 |
P value = 0.137 (Fisher's exact test), Q value = 0.53
Table S102. Gene #11: 'MAMLD1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 13 | 13 | 11 |
| MAMLD1 MUTATED | 0 | 2 | 2 | 0 |
| MAMLD1 WILD-TYPE | 20 | 11 | 11 | 11 |
P value = 0.0396 (Fisher's exact test), Q value = 0.36
Table S103. Gene #11: 'MAMLD1 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 12 | 6 | 9 |
| MAMLD1 MUTATED | 0 | 0 | 2 | 2 | 0 |
| MAMLD1 WILD-TYPE | 12 | 9 | 10 | 4 | 9 |
Figure S12. Get High-res Image Gene #11: 'MAMLD1 MUTATION STATUS' versus Molecular Subtype #3: 'RPPA_CNMF'
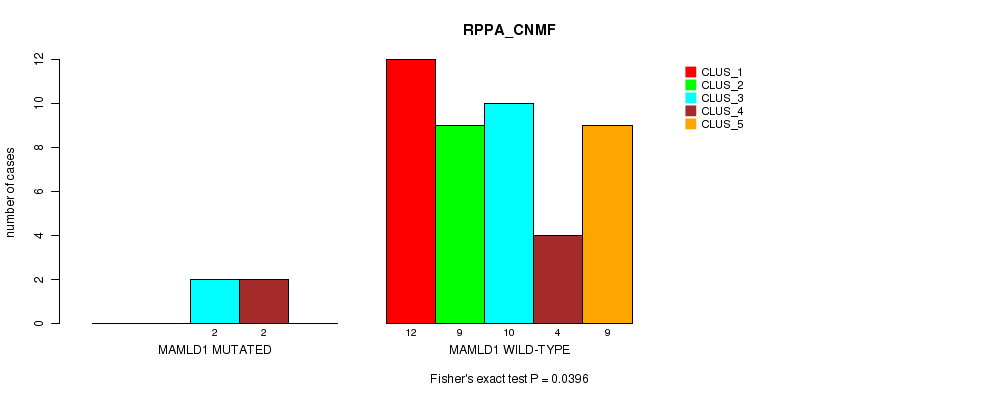
P value = 0.036 (Fisher's exact test), Q value = 0.36
Table S104. Gene #11: 'MAMLD1 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 9 | 8 | 7 | 7 | 7 | 5 | 5 |
| MAMLD1 MUTATED | 0 | 2 | 0 | 0 | 0 | 2 | 0 |
| MAMLD1 WILD-TYPE | 9 | 6 | 7 | 7 | 7 | 3 | 5 |
Figure S13. Get High-res Image Gene #11: 'MAMLD1 MUTATION STATUS' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
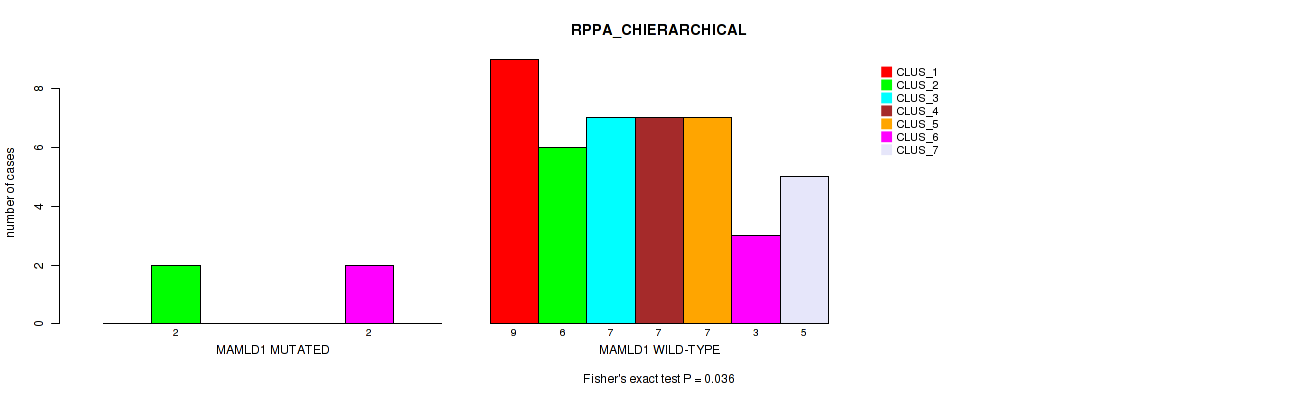
P value = 0.626 (Fisher's exact test), Q value = 0.89
Table S105. Gene #11: 'MAMLD1 MUTATION STATUS' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 16 | 19 | 12 | 10 |
| MAMLD1 MUTATED | 0 | 2 | 1 | 1 |
| MAMLD1 WILD-TYPE | 16 | 17 | 11 | 9 |
P value = 0.769 (Fisher's exact test), Q value = 0.96
Table S106. Gene #11: 'MAMLD1 MUTATION STATUS' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 11 | 4 | 8 | 6 | 8 | 6 | 14 |
| MAMLD1 MUTATED | 0 | 0 | 1 | 1 | 1 | 0 | 1 |
| MAMLD1 WILD-TYPE | 11 | 4 | 7 | 5 | 7 | 6 | 13 |
P value = 0.0593 (Fisher's exact test), Q value = 0.39
Table S107. Gene #11: 'MAMLD1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 24 | 17 |
| MAMLD1 MUTATED | 1 | 0 | 3 |
| MAMLD1 WILD-TYPE | 14 | 24 | 14 |
P value = 0.679 (Fisher's exact test), Q value = 0.91
Table S108. Gene #11: 'MAMLD1 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 24 | 8 | 18 | 6 |
| MAMLD1 MUTATED | 2 | 0 | 1 | 1 |
| MAMLD1 WILD-TYPE | 22 | 8 | 17 | 5 |
P value = 0.493 (Fisher's exact test), Q value = 0.84
Table S109. Gene #11: 'MAMLD1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 10 | 14 | 14 | 18 |
| MAMLD1 MUTATED | 0 | 2 | 0 | 2 |
| MAMLD1 WILD-TYPE | 10 | 12 | 14 | 16 |
P value = 0.356 (Fisher's exact test), Q value = 0.79
Table S110. Gene #11: 'MAMLD1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 25 | 7 | 17 | 7 |
| MAMLD1 MUTATED | 3 | 0 | 0 | 1 |
| MAMLD1 WILD-TYPE | 22 | 7 | 17 | 6 |
-
Mutation data file = sample_sig_gene_table.txt from Mutsig_2CV pipeline
-
Processed Mutation data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_Correlate_Genomic_Events_Preprocess/UCS-TP/22569431/transformed.cor.cli.txt
-
Molecular subtypes file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_mergedClustering/UCS-TP/22541488/UCS-TP.transferedmergedcluster.txt
-
Number of patients = 57
-
Number of significantly mutated genes = 11
-
Number of Molecular subtypes = 10
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.