(NRAS_Hotspot_Mutants cohort)
This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and subtypes.
Testing the association between copy number variation 67 arm-level results and 8 molecular subtypes across 62 patients, 8 significant findings detected with Q value < 0.25.
-
6p gain cnv correlated to 'METHLYATION_CNMF'.
-
8p gain cnv correlated to 'MRNASEQ_CNMF'.
-
8q gain cnv correlated to 'MRNASEQ_CNMF'.
-
13q gain cnv correlated to 'METHLYATION_CNMF' and 'MRNASEQ_CNMF'.
-
16p gain cnv correlated to 'MRNASEQ_CNMF'.
-
10p loss cnv correlated to 'CN_CNMF'.
-
10q loss cnv correlated to 'CN_CNMF'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 67 arm-level results and 8 molecular subtypes. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 8 significant findings detected.
|
Molecular subtypes |
CN CNMF |
METHLYATION CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Chi-square test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 13q gain | 18 (29%) | 44 |
0.0282 (1.00) |
0.000223 (0.115) |
0.716 (1.00) |
0.398 (1.00) |
0.000484 (0.246) |
0.018 (1.00) |
0.0797 (1.00) |
0.102 (1.00) |
| 6p gain | 28 (45%) | 34 |
0.00131 (0.664) |
0.000279 (0.143) |
1 (1.00) |
0.395 (1.00) |
0.0887 (1.00) |
0.201 (1.00) |
0.0566 (1.00) |
0.0478 (1.00) |
| 8p gain | 13 (21%) | 49 |
0.313 (1.00) |
0.0253 (1.00) |
1 (1.00) |
0.267 (1.00) |
9.88e-05 (0.0508) |
0.0606 (1.00) |
0.252 (1.00) |
1 (1.00) |
| 8q gain | 21 (34%) | 41 |
0.0343 (1.00) |
0.00327 (1.00) |
0.301 (1.00) |
0.244 (1.00) |
5.82e-05 (0.03) |
0.0286 (1.00) |
0.626 (1.00) |
0.92 (1.00) |
| 16p gain | 7 (11%) | 55 |
0.033 (1.00) |
0.0231 (1.00) |
1 (1.00) |
0.000386 (0.197) |
0.014 (1.00) |
0.677 (1.00) |
0.851 (1.00) |
|
| 10p loss | 25 (40%) | 37 |
6.88e-06 (0.00355) |
0.00949 (1.00) |
1 (1.00) |
0.045 (1.00) |
0.0133 (1.00) |
0.129 (1.00) |
0.334 (1.00) |
0.581 (1.00) |
| 10q loss | 27 (44%) | 35 |
0.000302 (0.154) |
0.0533 (1.00) |
1 (1.00) |
0.045 (1.00) |
0.0168 (1.00) |
0.225 (1.00) |
0.669 (1.00) |
0.425 (1.00) |
| 1p gain | 8 (13%) | 54 |
0.429 (1.00) |
0.804 (1.00) |
0.661 (1.00) |
0.479 (1.00) |
0.17 (1.00) |
0.712 (1.00) |
0.867 (1.00) |
0.728 (1.00) |
| 1q gain | 29 (47%) | 33 |
0.00828 (1.00) |
0.352 (1.00) |
1 (1.00) |
0.403 (1.00) |
0.11 (1.00) |
0.204 (1.00) |
0.968 (1.00) |
0.928 (1.00) |
| 2p gain | 10 (16%) | 52 |
0.361 (1.00) |
0.57 (1.00) |
1 (1.00) |
0.0797 (1.00) |
0.112 (1.00) |
0.307 (1.00) |
0.964 (1.00) |
0.867 (1.00) |
| 2q gain | 10 (16%) | 52 |
0.361 (1.00) |
0.57 (1.00) |
1 (1.00) |
0.0797 (1.00) |
0.112 (1.00) |
0.307 (1.00) |
0.964 (1.00) |
0.867 (1.00) |
| 3p gain | 6 (10%) | 56 |
0.0176 (1.00) |
0.492 (1.00) |
0.106 (1.00) |
0.721 (1.00) |
0.081 (1.00) |
0.0991 (1.00) |
0.531 (1.00) |
0.408 (1.00) |
| 3q gain | 7 (11%) | 55 |
0.114 (1.00) |
0.537 (1.00) |
0.106 (1.00) |
0.721 (1.00) |
0.134 (1.00) |
0.453 (1.00) |
0.301 (1.00) |
0.111 (1.00) |
| 4p gain | 5 (8%) | 57 |
0.073 (1.00) |
0.093 (1.00) |
1 (1.00) |
0.532 (1.00) |
1 (1.00) |
0.625 (1.00) |
0.189 (1.00) |
0.47 (1.00) |
| 4q gain | 5 (8%) | 57 |
0.073 (1.00) |
0.093 (1.00) |
1 (1.00) |
0.532 (1.00) |
1 (1.00) |
0.625 (1.00) |
0.189 (1.00) |
0.47 (1.00) |
| 5p gain | 3 (5%) | 59 |
0.215 (1.00) |
0.502 (1.00) |
0.185 (1.00) |
1 (1.00) |
1 (1.00) |
0.157 (1.00) |
||
| 6q gain | 3 (5%) | 59 |
0.111 (1.00) |
0.282 (1.00) |
0.487 (1.00) |
0.625 (1.00) |
0.787 (1.00) |
0.764 (1.00) |
1 (1.00) |
|
| 7p gain | 11 (18%) | 51 |
0.00601 (1.00) |
0.0152 (1.00) |
0.342 (1.00) |
0.281 (1.00) |
0.121 (1.00) |
1 (1.00) |
0.0653 (1.00) |
0.224 (1.00) |
| 7q gain | 9 (15%) | 53 |
0.00131 (0.665) |
0.255 (1.00) |
0.605 (1.00) |
0.267 (1.00) |
0.0294 (1.00) |
0.537 (1.00) |
0.549 (1.00) |
0.859 (1.00) |
| 9q gain | 4 (6%) | 58 |
0.23 (1.00) |
0.445 (1.00) |
1 (1.00) |
0.0995 (1.00) |
0.245 (1.00) |
0.161 (1.00) |
0.909 (1.00) |
0.407 (1.00) |
| 11p gain | 5 (8%) | 57 |
0.805 (1.00) |
0.727 (1.00) |
1 (1.00) |
0.729 (1.00) |
1 (1.00) |
0.422 (1.00) |
1 (1.00) |
|
| 11q gain | 5 (8%) | 57 |
0.805 (1.00) |
0.727 (1.00) |
1 (1.00) |
0.729 (1.00) |
1 (1.00) |
0.422 (1.00) |
1 (1.00) |
|
| 12p gain | 4 (6%) | 58 |
0.103 (1.00) |
1 (1.00) |
0.487 (1.00) |
0.245 (1.00) |
0.668 (1.00) |
0.137 (1.00) |
0.0925 (1.00) |
|
| 14q gain | 5 (8%) | 57 |
0.171 (1.00) |
0.852 (1.00) |
1 (1.00) |
0.515 (1.00) |
0.835 (1.00) |
0.125 (1.00) |
1 (1.00) |
|
| 15q gain | 7 (11%) | 55 |
0.712 (1.00) |
1 (1.00) |
1 (1.00) |
0.533 (1.00) |
0.141 (1.00) |
0.849 (1.00) |
0.712 (1.00) |
|
| 16q gain | 6 (10%) | 56 |
0.0938 (1.00) |
0.0669 (1.00) |
1 (1.00) |
0.00142 (0.716) |
0.0437 (1.00) |
0.792 (1.00) |
0.668 (1.00) |
|
| 17p gain | 3 (5%) | 59 |
0.215 (1.00) |
1 (1.00) |
1 (1.00) |
0.0587 (1.00) |
0.117 (1.00) |
0.084 (1.00) |
0.263 (1.00) |
|
| 17q gain | 9 (15%) | 53 |
0.379 (1.00) |
0.283 (1.00) |
0.407 (1.00) |
0.556 (1.00) |
0.204 (1.00) |
0.083 (1.00) |
0.549 (1.00) |
1 (1.00) |
| 18p gain | 4 (6%) | 58 |
0.321 (1.00) |
0.251 (1.00) |
1 (1.00) |
0.83 (1.00) |
1 (1.00) |
0.289 (1.00) |
1 (1.00) |
|
| 18q gain | 4 (6%) | 58 |
0.321 (1.00) |
0.251 (1.00) |
1 (1.00) |
0.83 (1.00) |
1 (1.00) |
0.289 (1.00) |
1 (1.00) |
|
| 19p gain | 6 (10%) | 56 |
0.0938 (1.00) |
0.145 (1.00) |
0.605 (1.00) |
0.749 (1.00) |
0.081 (1.00) |
0.422 (1.00) |
0.426 (1.00) |
0.322 (1.00) |
| 19q gain | 7 (11%) | 55 |
0.579 (1.00) |
0.269 (1.00) |
0.605 (1.00) |
0.22 (1.00) |
0.0358 (1.00) |
0.2 (1.00) |
0.573 (1.00) |
0.198 (1.00) |
| 20p gain | 13 (21%) | 49 |
0.0917 (1.00) |
0.0878 (1.00) |
1 (1.00) |
0.0546 (1.00) |
0.00261 (1.00) |
0.00697 (1.00) |
0.899 (1.00) |
0.893 (1.00) |
| 20q gain | 18 (29%) | 44 |
0.108 (1.00) |
0.119 (1.00) |
1 (1.00) |
0.354 (1.00) |
0.104 (1.00) |
0.281 (1.00) |
0.676 (1.00) |
0.91 (1.00) |
| 21q gain | 9 (15%) | 53 |
0.304 (1.00) |
0.601 (1.00) |
1 (1.00) |
0.0995 (1.00) |
0.0294 (1.00) |
0.0108 (1.00) |
0.741 (1.00) |
0.649 (1.00) |
| 22q gain | 11 (18%) | 51 |
0.0066 (1.00) |
0.536 (1.00) |
0.661 (1.00) |
0.134 (1.00) |
0.121 (1.00) |
0.584 (1.00) |
0.417 (1.00) |
0.547 (1.00) |
| 2q loss | 3 (5%) | 59 |
0.406 (1.00) |
0.282 (1.00) |
0.487 (1.00) |
0.625 (1.00) |
0.117 (1.00) |
0.226 (1.00) |
1 (1.00) |
|
| 3p loss | 4 (6%) | 58 |
0.354 (1.00) |
0.829 (1.00) |
1 (1.00) |
0.473 (1.00) |
1 (1.00) |
1 (1.00) |
0.26 (1.00) |
0.407 (1.00) |
| 3q loss | 4 (6%) | 58 |
0.904 (1.00) |
0.376 (1.00) |
1 (1.00) |
0.473 (1.00) |
1 (1.00) |
1 (1.00) |
0.676 (1.00) |
0.407 (1.00) |
| 4p loss | 9 (15%) | 53 |
0.0497 (1.00) |
0.736 (1.00) |
0.0915 (1.00) |
0.262 (1.00) |
0.00958 (1.00) |
0.0293 (1.00) |
0.0204 (1.00) |
0.0552 (1.00) |
| 4q loss | 10 (16%) | 52 |
0.0115 (1.00) |
1 (1.00) |
0.0436 (1.00) |
0.121 (1.00) |
0.00311 (1.00) |
0.0163 (1.00) |
0.00475 (1.00) |
0.0202 (1.00) |
| 5p loss | 14 (23%) | 48 |
0.374 (1.00) |
0.0733 (1.00) |
1 (1.00) |
0.297 (1.00) |
0.478 (1.00) |
0.455 (1.00) |
0.247 (1.00) |
0.652 (1.00) |
| 5q loss | 15 (24%) | 47 |
0.306 (1.00) |
0.00756 (1.00) |
1 (1.00) |
0.316 (1.00) |
0.0615 (1.00) |
0.0483 (1.00) |
0.975 (1.00) |
0.658 (1.00) |
| 6p loss | 7 (11%) | 55 |
0.444 (1.00) |
0.0769 (1.00) |
1 (1.00) |
0.0519 (1.00) |
0.271 (1.00) |
0.329 (1.00) |
0.851 (1.00) |
|
| 6q loss | 28 (45%) | 34 |
0.00306 (1.00) |
0.731 (1.00) |
0.111 (1.00) |
0.589 (1.00) |
0.111 (1.00) |
0.319 (1.00) |
0.236 (1.00) |
0.796 (1.00) |
| 7p loss | 3 (5%) | 59 |
0.78 (1.00) |
0.282 (1.00) |
1 (1.00) |
0.625 (1.00) |
0.117 (1.00) |
0.764 (1.00) |
1 (1.00) |
|
| 8p loss | 11 (18%) | 51 |
0.6 (1.00) |
0.379 (1.00) |
0.0197 (1.00) |
0.971 (1.00) |
0.714 (1.00) |
0.584 (1.00) |
0.00157 (0.791) |
0.224 (1.00) |
| 8q loss | 3 (5%) | 59 |
0.882 (1.00) |
0.104 (1.00) |
0.231 (1.00) |
0.658 (1.00) |
0.371 (1.00) |
0.436 (1.00) |
0.084 (1.00) |
1 (1.00) |
| 9p loss | 38 (61%) | 24 |
0.257 (1.00) |
0.118 (1.00) |
0.205 (1.00) |
0.353 (1.00) |
0.312 (1.00) |
0.752 (1.00) |
0.714 (1.00) |
0.532 (1.00) |
| 9q loss | 30 (48%) | 32 |
0.41 (1.00) |
0.367 (1.00) |
0.748 (1.00) |
0.814 (1.00) |
0.656 (1.00) |
0.579 (1.00) |
0.513 (1.00) |
0.863 (1.00) |
| 11p loss | 19 (31%) | 43 |
0.632 (1.00) |
0.018 (1.00) |
0.301 (1.00) |
0.273 (1.00) |
0.372 (1.00) |
0.686 (1.00) |
0.979 (1.00) |
1 (1.00) |
| 11q loss | 22 (35%) | 40 |
0.921 (1.00) |
0.447 (1.00) |
0.741 (1.00) |
0.292 (1.00) |
0.754 (1.00) |
0.791 (1.00) |
0.949 (1.00) |
0.658 (1.00) |
| 12p loss | 8 (13%) | 54 |
0.811 (1.00) |
0.253 (1.00) |
1 (1.00) |
0.236 (1.00) |
0.89 (1.00) |
0.89 (1.00) |
0.743 (1.00) |
1 (1.00) |
| 12q loss | 11 (18%) | 51 |
0.484 (1.00) |
0.148 (1.00) |
0.661 (1.00) |
0.18 (1.00) |
1 (1.00) |
1 (1.00) |
0.909 (1.00) |
1 (1.00) |
| 13q loss | 6 (10%) | 56 |
0.938 (1.00) |
0.224 (1.00) |
1 (1.00) |
0.602 (1.00) |
0.65 (1.00) |
1 (1.00) |
0.274 (1.00) |
0.829 (1.00) |
| 14q loss | 13 (21%) | 49 |
0.00157 (0.793) |
0.202 (1.00) |
1 (1.00) |
0.653 (1.00) |
0.676 (1.00) |
0.67 (1.00) |
0.722 (1.00) |
0.103 (1.00) |
| 15q loss | 6 (10%) | 56 |
0.0471 (1.00) |
0.0669 (1.00) |
1 (1.00) |
0.871 (1.00) |
1 (1.00) |
0.621 (1.00) |
0.322 (1.00) |
|
| 16p loss | 3 (5%) | 59 |
0.215 (1.00) |
0.502 (1.00) |
0.267 (1.00) |
1 (1.00) |
0.583 (1.00) |
0.696 (1.00) |
||
| 16q loss | 12 (19%) | 50 |
0.655 (1.00) |
0.567 (1.00) |
1 (1.00) |
0.26 (1.00) |
0.225 (1.00) |
1 (1.00) |
0.423 (1.00) |
0.123 (1.00) |
| 17p loss | 20 (32%) | 42 |
0.269 (1.00) |
0.341 (1.00) |
0.301 (1.00) |
0.326 (1.00) |
0.661 (1.00) |
0.732 (1.00) |
0.705 (1.00) |
0.604 (1.00) |
| 17q loss | 4 (6%) | 58 |
0.0754 (1.00) |
1 (1.00) |
0.487 (1.00) |
0.245 (1.00) |
0.161 (1.00) |
0.26 (1.00) |
0.407 (1.00) |
|
| 18p loss | 12 (19%) | 50 |
0.125 (1.00) |
0.781 (1.00) |
1 (1.00) |
0.601 (1.00) |
0.518 (1.00) |
0.172 (1.00) |
0.609 (1.00) |
0.488 (1.00) |
| 18q loss | 11 (18%) | 51 |
0.14 (1.00) |
0.843 (1.00) |
1 (1.00) |
0.54 (1.00) |
0.714 (1.00) |
0.323 (1.00) |
0.8 (1.00) |
0.547 (1.00) |
| 19p loss | 8 (13%) | 54 |
0.0635 (1.00) |
1 (1.00) |
1 (1.00) |
0.866 (1.00) |
0.89 (1.00) |
1 (1.00) |
0.17 (1.00) |
0.728 (1.00) |
| 19q loss | 6 (10%) | 56 |
0.237 (1.00) |
0.868 (1.00) |
1 (1.00) |
0.435 (1.00) |
0.0331 (1.00) |
0.422 (1.00) |
0.792 (1.00) |
1 (1.00) |
| 21q loss | 6 (10%) | 56 |
1 (1.00) |
0.66 (1.00) |
0.605 (1.00) |
0.175 (1.00) |
0.157 (1.00) |
0.633 (1.00) |
0.589 (1.00) |
0.186 (1.00) |
| 22q loss | 8 (13%) | 54 |
0.168 (1.00) |
0.451 (1.00) |
0.661 (1.00) |
0.717 (1.00) |
0.286 (1.00) |
0.0909 (1.00) |
0.804 (1.00) |
0.859 (1.00) |
P value = 0.000279 (Fisher's exact test), Q value = 0.14
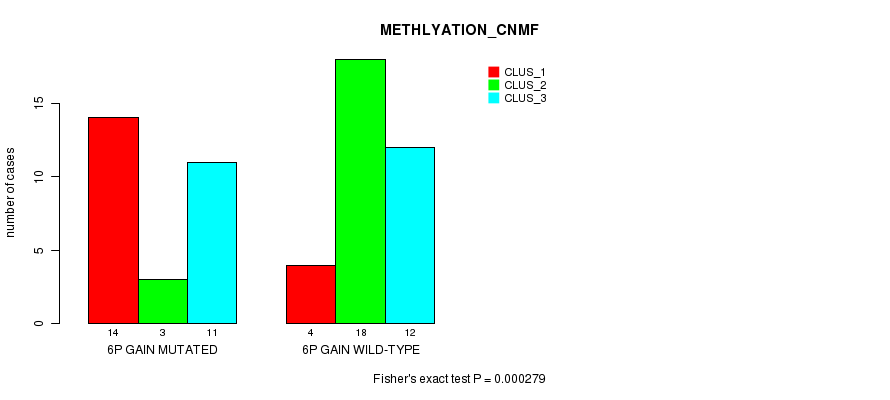
Table S1. Gene #10: '6p gain mutation analysis' versus Clinical Feature #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 21 | 23 |
| 6P GAIN MUTATED | 14 | 3 | 11 |
| 6P GAIN WILD-TYPE | 4 | 18 | 12 |
Figure S1. Get High-res Image Gene #10: '6p gain mutation analysis' versus Clinical Feature #2: 'METHLYATION_CNMF'
P value = 9.88e-05 (Fisher's exact test), Q value = 0.051
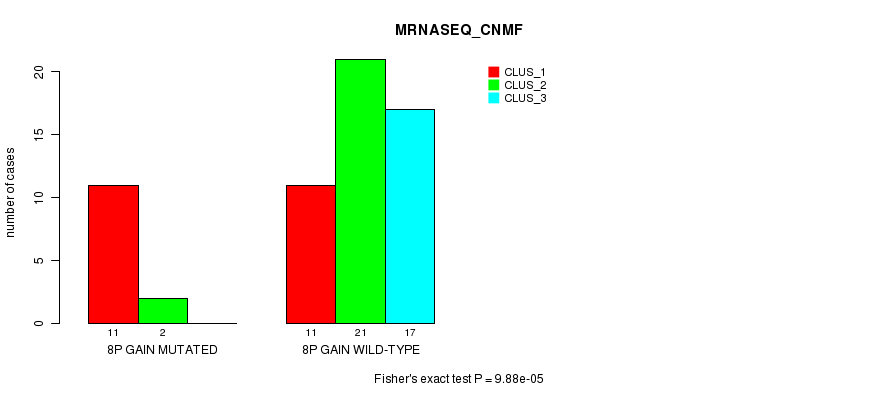
Table S2. Gene #14: '8p gain mutation analysis' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 23 | 17 |
| 8P GAIN MUTATED | 11 | 2 | 0 |
| 8P GAIN WILD-TYPE | 11 | 21 | 17 |
Figure S2. Get High-res Image Gene #14: '8p gain mutation analysis' versus Clinical Feature #5: 'MRNASEQ_CNMF'
P value = 5.82e-05 (Fisher's exact test), Q value = 0.03
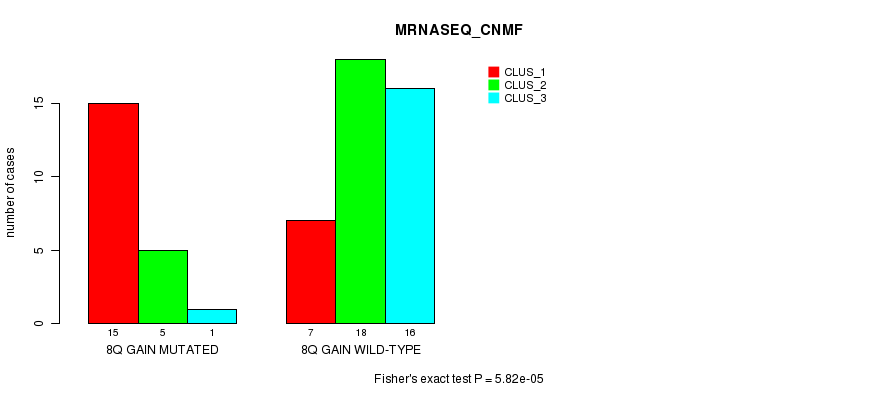
Table S3. Gene #15: '8q gain mutation analysis' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 23 | 17 |
| 8Q GAIN MUTATED | 15 | 5 | 1 |
| 8Q GAIN WILD-TYPE | 7 | 18 | 16 |
Figure S3. Get High-res Image Gene #15: '8q gain mutation analysis' versus Clinical Feature #5: 'MRNASEQ_CNMF'
P value = 0.000223 (Fisher's exact test), Q value = 0.11
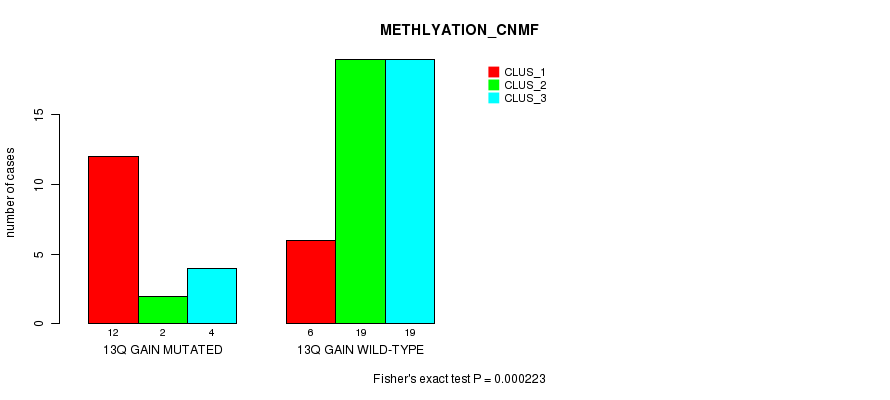
Table S4. Gene #20: '13q gain mutation analysis' versus Clinical Feature #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 18 | 21 | 23 |
| 13Q GAIN MUTATED | 12 | 2 | 4 |
| 13Q GAIN WILD-TYPE | 6 | 19 | 19 |
Figure S4. Get High-res Image Gene #20: '13q gain mutation analysis' versus Clinical Feature #2: 'METHLYATION_CNMF'
P value = 0.000484 (Fisher's exact test), Q value = 0.25
Table S5. Gene #20: '13q gain mutation analysis' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 23 | 17 |
| 13Q GAIN MUTATED | 13 | 2 | 3 |
| 13Q GAIN WILD-TYPE | 9 | 21 | 14 |
Figure S5. Get High-res Image Gene #20: '13q gain mutation analysis' versus Clinical Feature #5: 'MRNASEQ_CNMF'
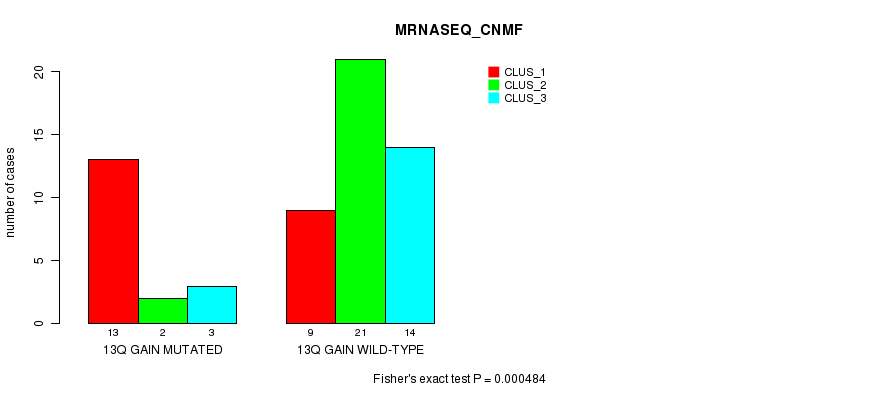
P value = 0.000386 (Fisher's exact test), Q value = 0.2
Table S6. Gene #23: '16p gain mutation analysis' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 23 | 17 |
| 16P GAIN MUTATED | 7 | 0 | 0 |
| 16P GAIN WILD-TYPE | 15 | 23 | 17 |
Figure S6. Get High-res Image Gene #23: '16p gain mutation analysis' versus Clinical Feature #5: 'MRNASEQ_CNMF'
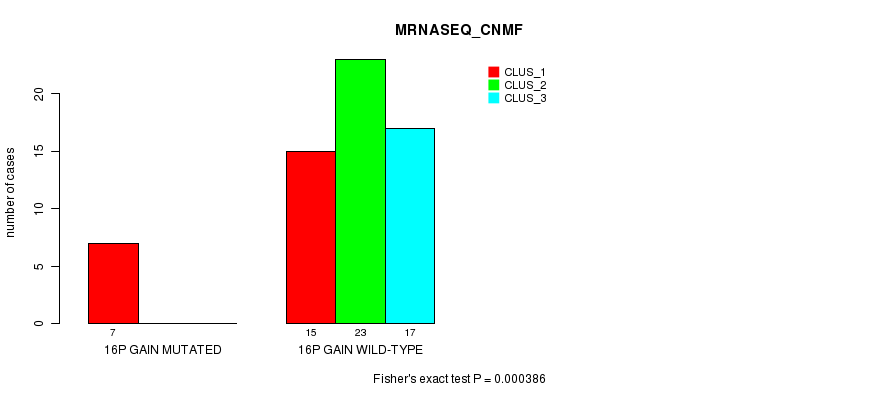
P value = 6.88e-06 (Fisher's exact test), Q value = 0.0035
Table S7. Gene #49: '10p loss mutation analysis' versus Clinical Feature #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 16 | 12 | 20 | 14 |
| 10P LOSS MUTATED | 9 | 9 | 0 | 7 |
| 10P LOSS WILD-TYPE | 7 | 3 | 20 | 7 |
Figure S7. Get High-res Image Gene #49: '10p loss mutation analysis' versus Clinical Feature #1: 'CN_CNMF'
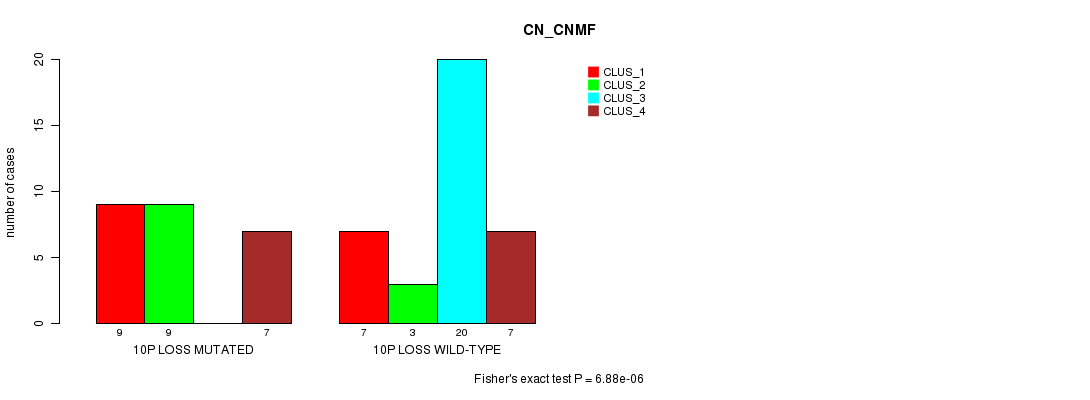
P value = 0.000302 (Fisher's exact test), Q value = 0.15
Table S8. Gene #50: '10q loss mutation analysis' versus Clinical Feature #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 16 | 12 | 20 | 14 |
| 10Q LOSS MUTATED | 8 | 10 | 2 | 7 |
| 10Q LOSS WILD-TYPE | 8 | 2 | 18 | 7 |
Figure S8. Get High-res Image Gene #50: '10q loss mutation analysis' versus Clinical Feature #1: 'CN_CNMF'
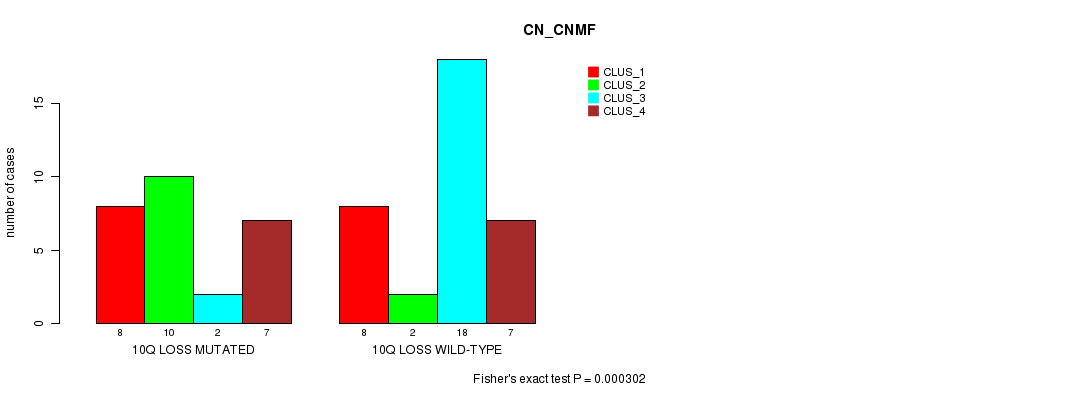
-
Mutation data file = broad_values_by_arm.mutsig.cluster.txt
-
Molecular subtypes file = SKCM-NRAS_Hotspot_Mutants.transferedmergedcluster.txt
-
Number of patients = 62
-
Number of significantly arm-level cnvs = 67
-
Number of molecular subtypes = 8
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.