Mutational signature discovery is a process of deconvoluting cancer somatic mutation counts, stratified by mutation context or biologically meaningful subgroup, into a set of characteristic patterns (signatures) and inferring the activity of each of the discovered signatures across samples.
This pipeline detected mutational signatures across samples based on Bayesian non-negative matrix factorization (BayesNMF) method [1],[2].
Our analysis idenfied 1 solution(s) of mutational signatures across 113 samples by BayesNMF method.
Figure 1. Get High-res Image Trinucleotide sequence motif: 3 distinct mutational signatures were identified.
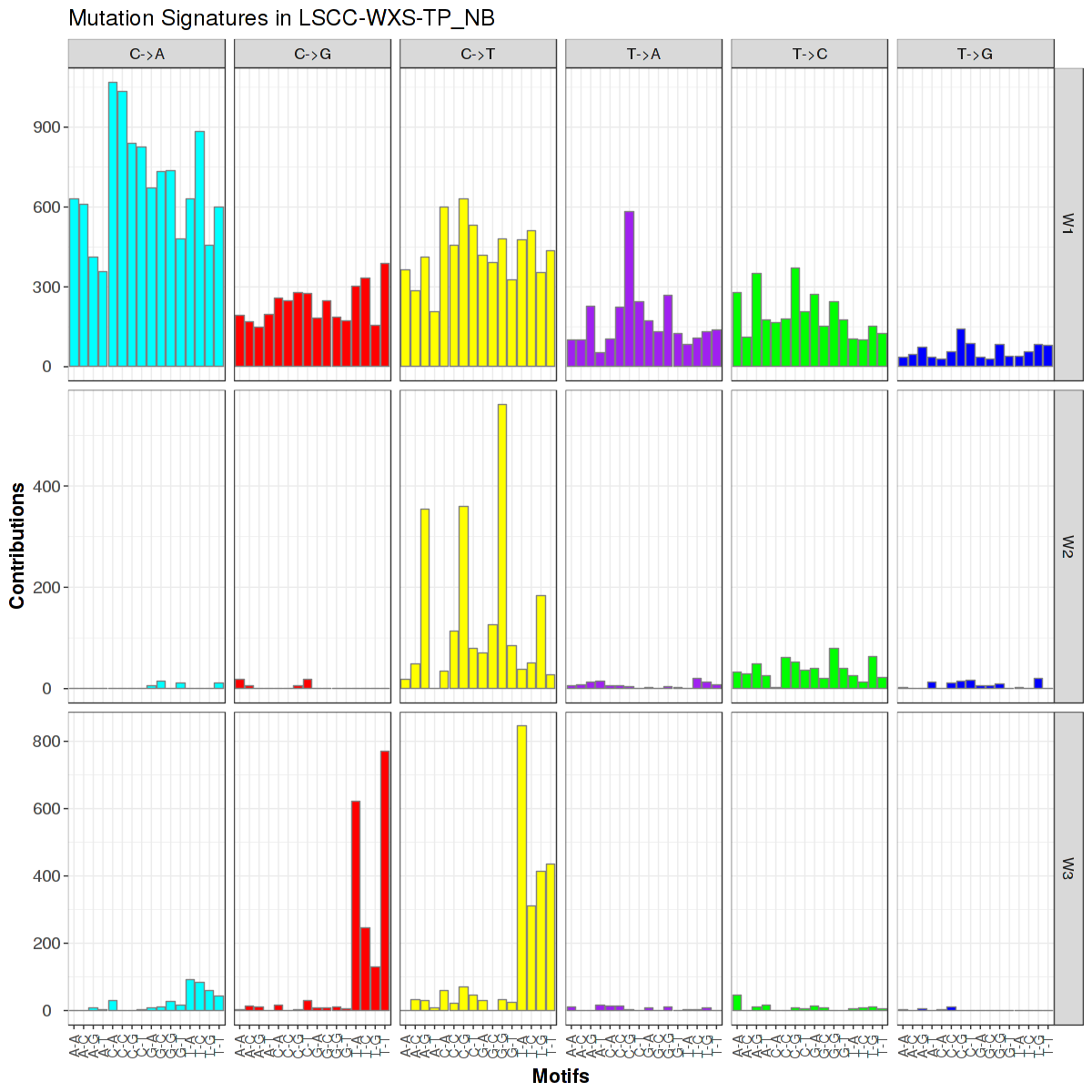
Figure 2. Get High-res Image The activity of discovered 3 mutational processes across samples. In the legend, the 'C' followed by a number represents the COSMIC signature, and the following piece of information is our annotation on that COSMIC signature. For more information on each COSMIC signature, see full table in the Methods & Data section.
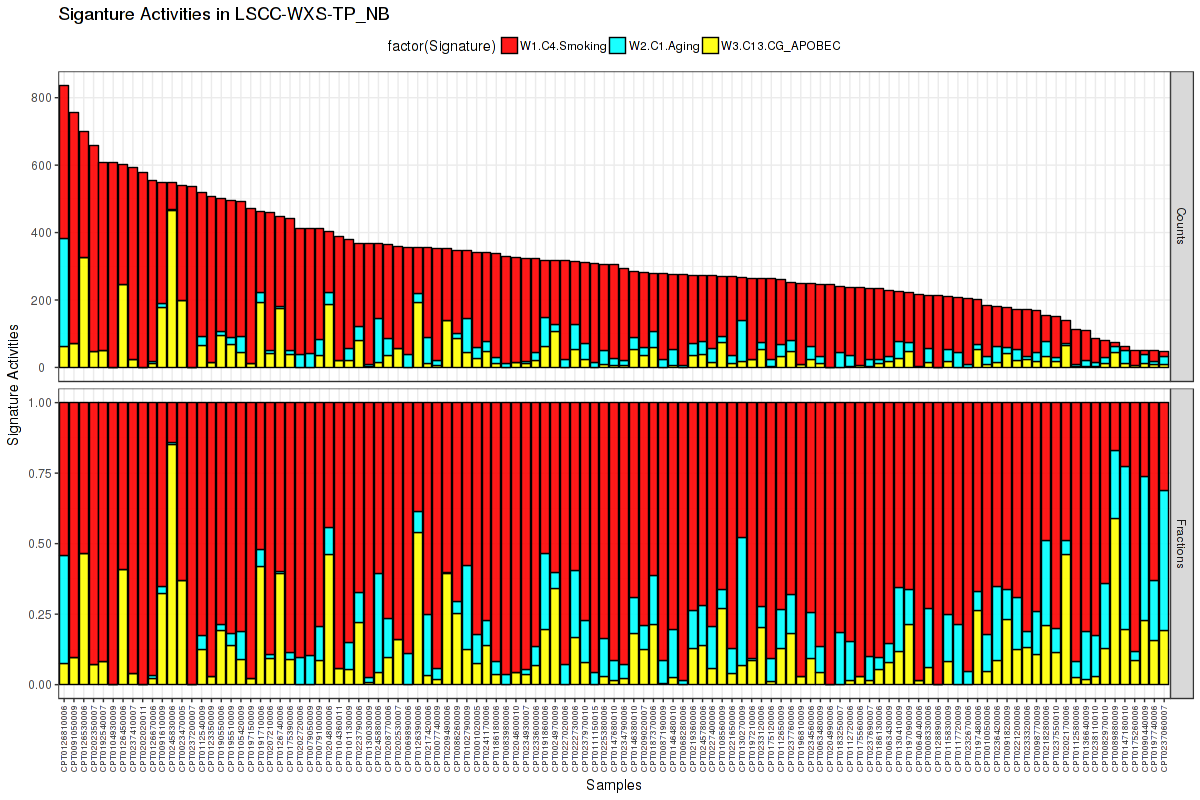
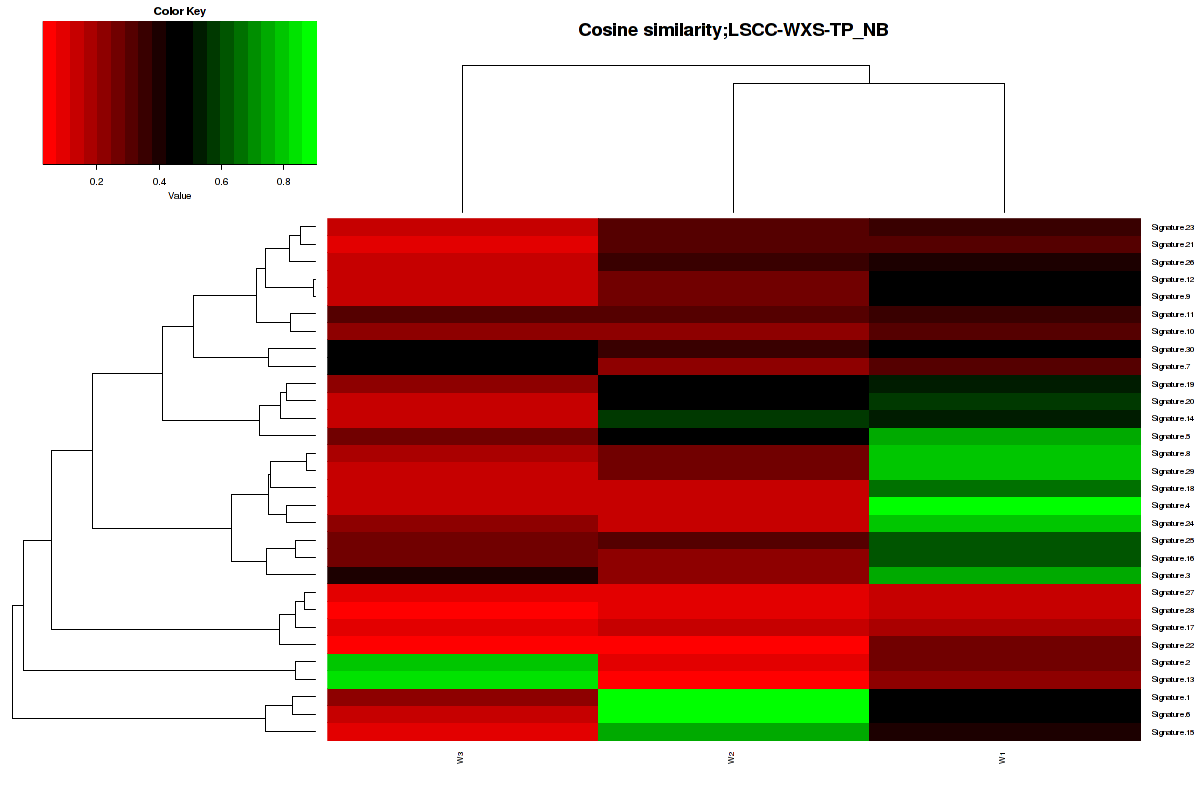
Figure 3. Get High-res Image The cosine similarity between the 3 mutational signatures and Sanger COSMIC signatures [3].
The input maf is the original maf file.
-
Input file for mutation signature pipeline = LSCC-WXS-TP_NB.final_analysis_set.maf
Output files from mutation signature pipeline :
-
*.WH.RData : a R object storing W and H matrix generated by BayesNMF. WH[[1]] is the W matrix containing the normalized profiles of discovered mutation signatures along 96 tri-nucleotide mutation contexts, which is used to generate the mutation signature bar plot. WH[[2]] is the activity matrix of mutations signatures across samples (# of signatures by # of samples) and WH[[3]] is the normalized activity matrix. The activity means the number of mutations assigned to each signature.
-
*.summary.*.RData : This R object (data frame) is a summary table listing the raw and normalized activity, and SNVs across samples.
-
*_solution_*.maf : MAF file annotated with the specified solution.
Characterizing underlying mutational processes with correct inferences for signature activities across samples provide a key understanding on cancer initiation and progression. However, the number of mutation processes K* is highly variable across patients even in a single tumor type and its accurate estimation is a non-trivial task due to a different duration and intensity of exposure to a specific mutational process. Non-negative matrix factorization (NMF) has been widely used in deciphering mutations signatures in cancer somatic mutations stratified by 96 base substitutions in tri-nucleotide sequence contexts. In contrast to conventional NMF requiring K* as an input parameter, BayesNMF exploits the automatic relevance determination technique to infer the optimal K* from data itself at a balance between the data fidelity (likelihood) and the model complexity (regularization)[1],[2].
The formula of V1[i]%*%V2[j]/sqrt(sum(V1[i]^2))/sqrt(sum(V2[j]^2)) was used to calculate to similarity between two vectors. In our cases, the value will range from 0 to 1. 1 meaning exactly the same and 0 indicating orthogonality [4].
Table 1. Get Full Table COSMIC signature table taken from https://cancer.sanger.ac.uk/cosmic/signatures on 2018-05-09 with Keyword annotations added.
| COSMIC_Signatures | Keyword | Proposed_Aetiology | Comments |
|---|---|---|---|
| 1 | Aging | Signature 1 is the result of an endogenous mutational process initiated by spontaneous deamination of 5-methylcytosine. | The number of Signature 1 mutations correlates with age of cancer diagnosis. |
| 2 | CT_APOBEC | Signature 2 has been attributed to activity of the AID/APOBEC family of cytidine deaminases. On the basis of similarities in the sequence context of cytosine mutations caused by APOBEC enzymes in experimental systems, a role for APOBEC1, APOBEC3A and/or APOBEC3B in human cancer appears more likely than for other members of the family. | Signature 2 is usually found in the same samples as Signature 13. It has been proposed that activation of AID/APOBEC cytidine deaminases is due to viral infection, retrotransposon jumping or to tissue inflammation. Currently, there is limited evidence to support these hypotheses. A germline deletion polymorphism involving APOBEC3A and APOBEC3B is associated with the presence of large numbers of Signature 2 and 13 mutations and with predisposition to breast cancer. Mutations of similar patterns to Signatures 2 and 13 are commonly found in the phenomenon of local hypermutation present in some cancers, known as kataegis, potentially implicating AID/APOBEC enzymes in this process as well. |
| 3 | BRCA_Hrdefect | Signature 3 is associated with failure of DNA double-strand break-repair by homologous recombination. | Signature 3 is strongly associated with germline and somatic BRCA1 and BRCA2 mutations in breast, pancreatic, and ovarian cancers. In pancreatic cancer, responders to platinum therapy usually exhibit Signature 3 mutations. |
| 4 | Smoking | Signature 4 is associated with smoking and its profile is similar to the mutational pattern observed in experimental systems exposed to tobacco carcinogens (e.g., benzo[a]pyrene). Signature 4 is likely due to tobacco mutagens. | Signature 29 is found in cancers associated with tobacco chewing and appears different from Signature 4. |
| 5 | Unknown_ERCC2 | The aetiology of Signature 5 is unknown. | N/A |
| 6 | MSI | Signature 6 is associated with defective DNA mismatch repair and is found in microsatellite unstable tumours. | Signature 6 is one of four mutational signatures associated with defective DNA mismatch repair and is often found in the same samples as Signatures 15, 20, and 26. |
| 7 | UV | Based on its prevalence in ultraviolet exposed areas and the similarity of the mutational pattern to that observed in experimental systems exposed to ultraviolet light Signature 7 is likely due to ultraviolet light exposure. | N/A |
| 8 | Unknown | The aetiology of Signature 8 remains unknown. | N/A |
| 9 | Non-canonical AID | Signature 9 is characterized by a pattern of mutations that has been attributed to polymerase η, which is implicated with the activity of AID during somatic hypermutation. | Chronic lymphocytic leukaemias that possess immunoglobulin gene hypermutation (IGHV-mutated) have elevated numbers of mutations attributed to Signature 9 compared to those that do not have immunoglobulin gene hypermutation. |
| 10 | POLE | It has been proposed that the mutational process underlying this signature is altered activity of the error-prone polymerase POLE. The presence of large numbers of Signature 10 mutations is associated with recurrent POLE somatic mutations, viz., Pro286Arg and Val411Leu. | Signature 10 is associated with some of most mutated cancer samples. Samples exhibiting this mutational signature have been termed ultra-hypermutators. |
| 11 | Alkylating | Signature 11 exhibits a mutational pattern resembling that of alkylating agents. Patient histories have revealed an association between treatments with the alkylating agent temozolomide and Signature 11 mutations. | N/A |
| 12 | Unknown | The aetiology of Signature 12 remains unknown. | Signature 12 usually contributes a small percentage (<20%) of the mutations observed in a liver cancer sample. |
| 13 | CG_APOBEC | Signature 13 has been attributed to activity of the AID/APOBEC family of cytidine deaminases converting cytosine to uracil. On the basis of similarities in the sequence context of cytosine mutations caused by APOBEC enzymes in experimental systems, a role for APOBEC1, APOBEC3A and/or APOBEC3B in human cancer appears more likely than for other members of the family. Signature 13 causes predominantly C>G mutations. This may be due to generation of abasic sites after removal of uracil by base excision repair and replication over these abasic sites by REV1. | Signature 2 is usually found in the same samples as Signature 13. It has been proposed that activation of AID/APOBEC cytidine deaminases is due to viral infection, retrotransposon jumping or to tissue inflammation. Currently, there is limited evidence to support these hypotheses. A germline deletion polymorphism involving APOBEC3A and APOBEC3B is associated with the presence of large numbers of Signature 2 and 13 mutations and with predisposition to breast cancer. Mutations of similar patterns to Signatures 2 and 13 are commonly found in the phenomenon of local hypermutation present in some cancers, known as kataegis, potentially implicating AID/APOBEC enzymes in this process as well. |
| 14 | MSI_POLE | The aetiology of Signature 14 remains unknown. | Signature 14 generates very high numbers of somatic mutations (>200 mutations per MB) in all samples in which it has been observed. |
| 15 | MSI | Signature 15 is associated with defective DNA mismatch repair. | Signature 15 is one of four mutational signatures associated with defective DNA mismatch repair and is often found in the same samples as Signatures 6, 20, and 26. |
| 16 | Unknown | The aetiology of Signature 16 remains unknown. | N/A |
| 17 | Unknown | The aetiology of Signature 17 remains unknown. | N/A |
| 18 | Unknown_Oxidative_MUTYH | The aetiology of Signature 18 remains unknown. | N/A |
| 19 | Unknown | The aetiology of Signature 19 remains unknown. | N/A |
| 20 | MSI_POLD | Signature 20 is believed to be associated with defective DNA mismatch repair. | Signature 20 is one of four mutational signatures associated with defective DNA mismatch repair and is often found in the same samples as Signatures 6, 15, and 26. |
| 21 | MSI | The aetiology of Signature 21 remains unknown. | Signature 21 is found only in four samples all generated by the same sequencing centre. The mutational pattern of Signature 21 is somewhat similar to the one of Signature 26. Additionally, Signature 21 is found only in samples that also have Signatures 15 and 20. As such, Signature 21 is probably also related to microsatellite unstable tumours. |
| 22 | Aristolochic Acid | Signature 22 has been found in cancer samples with known exposures to aristolochic acid. Additionally, the pattern of mutations exhibited by the signature is consistent with the one previous observed in experimental systems exposed to aristolochic acid. | Signature 22 has a very high mutational burden in urothelial carcinoma; however, its mutational burden is much lower in liver cancers. |
| 23 | Unknown | The aetiology of Signature 23 remains unknown. | N/A |
| 24 | Aflatoxin | Signature 24 has been found in cancer samples with known exposures to aflatoxin. Additionally, the pattern of mutations exhibited by the signature is consistent with that previous observed in experimental systems exposed to aflatoxin. | N/A |
| 25 | Unknown_Lymphoma | The aetiology of Signature 25 remains unknown. | This signature has only been identified in Hodgkin’s cell lines. Data is not available from primary Hodgkin lymphomas. |
| 26 | MSI | Signature 26 is believed to be associated with defective DNA mismatch repair. | Signature 26 is one of four mutational signatures associated with defective DNA mismatch repair and is often found in the same samples as Signatures 6, 15 and 20. |
| 27 | Unknown_Artifact | The aetiology of Signature 27 remains unknown. | N/A |
| 28 | Unknwon_Stomach | The aetiology of Signature 28 remains unknown. | N/A |
| 29 | Chewing_tobacco | Signature 29 has been found in cancer samples from individuals with a tobacco chewing habit. | The Signature 29 pattern of C>A mutations due to tobacco chewing appears different from the pattern of mutations due to tobacco smoking reflected by Signature 4. |
| 30 | Unknwon_NTHL1 | The aetiology of Signature 30 remains unknown. | N/A |