GISTIC identifies genomic regions that are significantly gained or lost across a set of tumors. The pipeline first filters out normal samples from the segmented copy-number data by inspecting the TCGA barcodes and then executes GISTIC version 2.0.15 (cga svn revision 32817).
There were 36 tumor samples used in this analysis: 14 significant arm-level results, 13 significant focal amplifications, and 17 significant focal deletions were found.
Figure 1. Genomic positions of amplified regions: the X-axis represents the normalized amplification signals (top) and significance by Q value (bottom). The green line represents the significance cutoff at Q value=0.25.
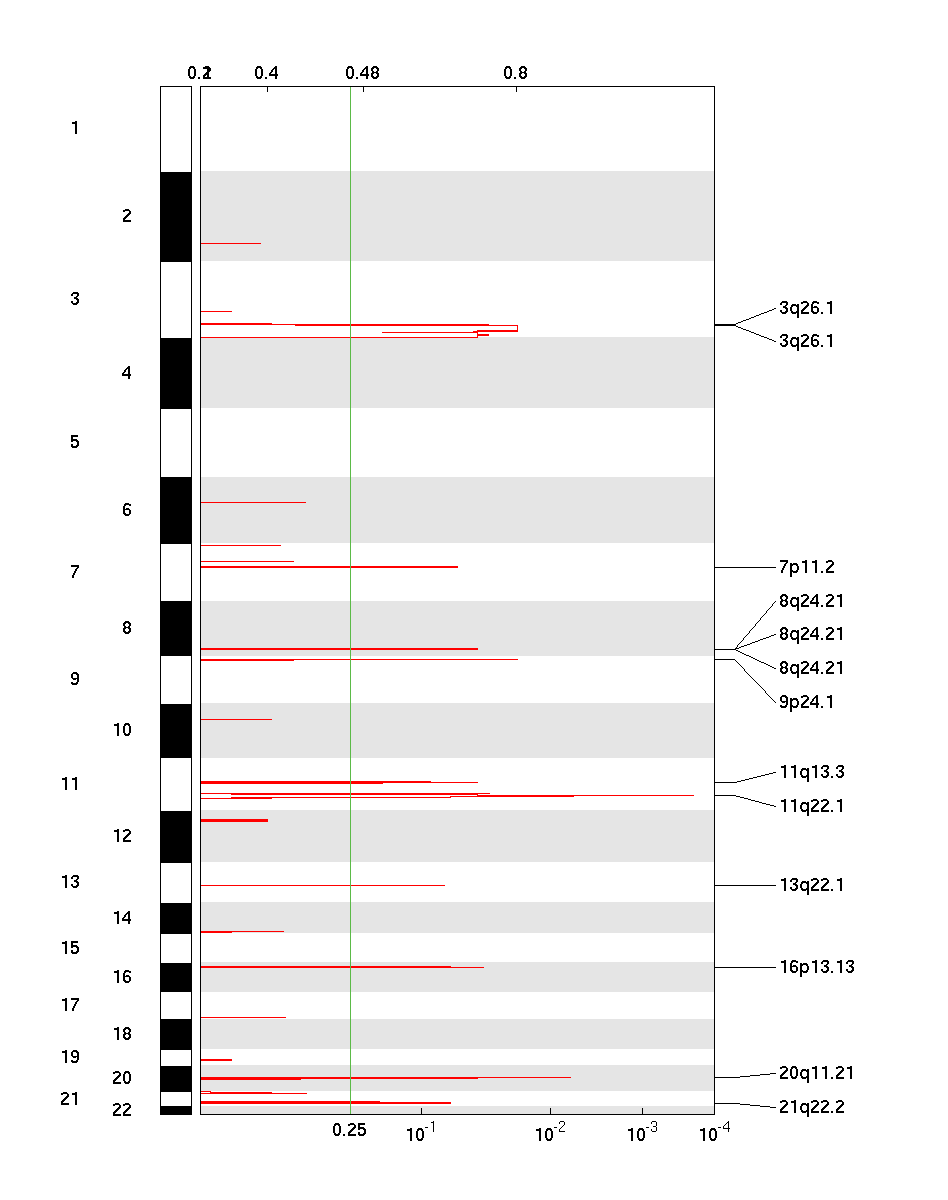
Table 1. Get Full Table Amplifications Table - 13 significant amplifications found. Click the link in the last column to view a comprehensive list of candidate genes. If no genes were identified within the peak, the nearest gene appears in brackets.
| Cytoband | Q value | Residual Q value | Wide Peak Boundaries | # Genes in Wide Peak |
|---|---|---|---|---|
| 11q22.1 | 0.00020212 | 0.0027819 | chr11:101256134-101818609 | 7 |
| 20q11.21 | 0.00647 | 0.00647 | chr20:26020583-29820251 | 20 |
| 9p24.1 | 0.019988 | 0.019988 | chr9:5649828-5709211 | 1 |
| 16p13.13 | 0.037252 | 0.037252 | chr16:11785794-11835481 | 1 |
| 3q26.1 | 0.019988 | 0.041082 | chr3:166992045-199501827 | 210 |
| 11q13.3 | 0.041082 | 0.041082 | chr11:69516254-70078801 | 6 |
| 7p11.2 | 0.057281 | 0.057281 | chr7:55018826-55426994 | 2 |
| 21q22.2 | 0.064467 | 0.064467 | chr21:38710567-39610234 | 5 |
| 13q22.1 | 0.071279 | 0.071279 | chr13:73093889-73119658 | 0 [KLF12] |
| 8q24.21 | 0.045931 | 0.074639 | chr8:129098497-129100915 | 1 |
| 3q26.1 | 0.034255 | 0.10186 | chr3:163643792-163719647 | 0 [OTOL1] |
| 8q24.21 | 0.081818 | 0.17435 | chr8:129524135-129592226 | 0 [hsa-mir-1208] |
| 8q24.21 | 0.041082 | 0.99915 | chr8:1-146274826 | 745 |
This is the comprehensive list of amplified genes in the wide peak for 11q22.1.
Table S1. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BIRC3 |
| BIRC2 |
| YAP1 |
| KIAA1377 |
| C11orf70 |
| TMEM123 |
| ANGPTL5 |
This is the comprehensive list of amplified genes in the wide peak for 20q11.21.
Table S2. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-663 |
| BCL2L1 |
| ID1 |
| TPX2 |
| REM1 |
| HM13 |
| COX4I2 |
| DEFB118 |
| NCRNA00028 |
| C20orf191 |
| DEFB115 |
| DEFB116 |
| DEFB119 |
| DEFB121 |
| DEFB122 |
| DEFB123 |
| DEFB124 |
| FRG1B |
| MIR663 |
| PSIMCT-1 |
This is the comprehensive list of amplified genes in the wide peak for 9p24.1.
Table S3. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| KIAA1432 |
This is the comprehensive list of amplified genes in the wide peak for 16p13.13.
Table S4. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BCAR4 |
This is the comprehensive list of amplified genes in the wide peak for 3q26.1.
Table S5. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BCL6 |
| EIF4A2 |
| ETV5 |
| LPP |
| PIK3CA |
| SOX2 |
| TFRC |
| hsa-mir-922 |
| hsa-mir-570 |
| hsa-mir-944 |
| hsa-mir-28 |
| hsa-mir-1248 |
| hsa-mir-1224 |
| hsa-mir-569 |
| hsa-mir-551b |
| ACTL6A |
| AHSG |
| APOD |
| BCHE |
| BDH1 |
| AP2M1 |
| CLCN2 |
| CPN2 |
| CRYGS |
| DGKG |
| DLG1 |
| DVL3 |
| ECT2 |
| EHHADH |
| EIF4G1 |
| EPHB3 |
| MECOM |
| FGF12 |
| GHSR |
| GP5 |
| HRG |
| HES1 |
| IL1RAP |
| KNG1 |
| MFI2 |
| MUC4 |
| NDUFB5 |
| OPA1 |
| CLDN11 |
| PAK2 |
| PCYT1A |
| SERPINI1 |
| SERPINI2 |
| PLD1 |
| POLR2H |
| PPP1R2 |
| PRKCI |
| MASP1 |
| PSMD2 |
| RFC4 |
| SNORA63 |
| RPL35A |
| TRA2B |
| ST6GAL1 |
| SKIL |
| SLC2A2 |
| SST |
| TERC |
| THPO |
| SEC62 |
| FXR1 |
| TP63 |
| CHRD |
| TNFSF10 |
| EIF2B5 |
| USP13 |
| CLDN1 |
| MAP3K13 |
| ADIPOQ |
| KIAA0226 |
| ECE2 |
| ABCC5 |
| TNK2 |
| ALG3 |
| KCNMB2 |
| IGF2BP2 |
| CLDN16 |
| PDCD10 |
| NLGN1 |
| NCBP2 |
| TNIK |
| MCF2L2 |
| ATP11B |
| VPS8 |
| ACAP2 |
| UBXN7 |
| GPR160 |
| FETUB |
| LAMP3 |
| KCNMB3 |
| GOLIM4 |
| ZNF639 |
| PEX5L |
| DNAJB11 |
| DCUN1D1 |
| KLHL24 |
| PIGX |
| TBCCD1 |
| LEPREL1 |
| ABCF3 |
| LSG1 |
| PARL |
| MFN1 |
| YEATS2 |
| MYNN |
| EIF5A2 |
| MCCC1 |
| HRASLS |
| MRPL47 |
| NCEH1 |
| SLC7A14 |
| SENP2 |
| GNB4 |
| RTP4 |
| MAGEF1 |
| ZMAT3 |
| FNDC3B |
| ATP13A3 |
| TBL1XR1 |
| ZBBX |
| LRRC31 |
| MAP6D1 |
| PHC3 |
| PIGZ |
| SPATA16 |
| B3GNT5 |
| IQCG |
| ATP13A4 |
| FYTTD1 |
| ARPM1 |
| MGC2889 |
| LRCH3 |
| C3orf34 |
| LMLN |
| KLHL6 |
| VWA5B2 |
| TMEM41A |
| TMEM44 |
| C3orf50 |
| CAMK2N2 |
| TM4SF19 |
| RPL39L |
| DNAJC19 |
| FAM131A |
| ZDHHC19 |
| LRRC15 |
| FAM43A |
| TMEM207 |
| RTP1 |
| TTC14 |
| WDR49 |
| LRRC34 |
| C3orf59 |
| C3orf21 |
| CCDC50 |
| PYDC2 |
| LOC152217 |
| RNF168 |
| HTR3C |
| LIPH |
| HTR3D |
| RPL22L1 |
| OSTalpha |
| FBXO45 |
| MUC20 |
| SENP5 |
| LOC220729 |
| NAALADL2 |
| TCTEX1D2 |
| C3orf43 |
| SDHALP1 |
| UTS2D |
| HTR3E |
| C3orf70 |
| TPRG1 |
| CCDC39 |
| LRRIQ4 |
| SAMD7 |
| RTP2 |
| OSTN |
| ATP13A5 |
| SOX2OT |
| WDR53 |
| LOC348840 |
| LRRC33 |
| TMEM212 |
| FLJ42393 |
| SNORD2 |
| SNORA4 |
| C3orf65 |
| LOC647309 |
| SNORA81 |
| SNORD66 |
| MIR551B |
| MIR569 |
| MIR570 |
| SDHALP2 |
| FAM157A |
| MIR922 |
| MIR944 |
| LOC100128023 |
| LOC100128164 |
| LOC100131551 |
| SNAR-I |
| MIR1224 |
This is the comprehensive list of amplified genes in the wide peak for 11q13.3.
Table S6. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-548k |
| CTTN |
| PPFIA1 |
| FADD |
| SHANK2 |
| ANO1 |
This is the comprehensive list of amplified genes in the wide peak for 7p11.2.
Table S7. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| EGFR |
| LANCL2 |
This is the comprehensive list of amplified genes in the wide peak for 21q22.2.
Table S8. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| ERG |
| ETS2 |
| PSMG1 |
| BRWD1 |
| NCRNA00114 |
This is the comprehensive list of amplified genes in the wide peak for 8q24.21.
Table S9. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| PVT1 |
This is the comprehensive list of amplified genes in the wide peak for 8q24.21.
Table S10. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| COX6C |
| EXT1 |
| FGFR1 |
| MYC |
| PCM1 |
| PLAG1 |
| TCEA1 |
| WRN |
| RECQL4 |
| NCOA2 |
| WHSC1L1 |
| CHCHD7 |
| HOOK3 |
| hsa-mir-1234 |
| hsa-mir-661 |
| hsa-mir-1302-7 |
| hsa-mir-151 |
| hsa-mir-30d |
| hsa-mir-1208 |
| hsa-mir-1206 |
| hsa-mir-1204 |
| hsa-mir-548d-1 |
| hsa-mir-2053 |
| hsa-mir-548a-3 |
| hsa-mir-1273 |
| hsa-mir-875 |
| hsa-mir-2052 |
| hsa-mir-124-2 |
| hsa-mir-486 |
| hsa-mir-548h-4 |
| hsa-mir-320a |
| hsa-mir-383 |
| hsa-mir-598 |
| hsa-mir-1322 |
| hsa-mir-124-1 |
| hsa-mir-597 |
| hsa-mir-548i-3 |
| hsa-mir-596 |
| NAT1 |
| NAT2 |
| ADCY8 |
| ADRA1A |
| ADRB3 |
| ANGPT1 |
| ANGPT2 |
| ANK1 |
| ANXA13 |
| ASAH1 |
| ASPH |
| ATP6V1B2 |
| ATP6V1C1 |
| BAI1 |
| BLK |
| BMP1 |
| POLR3D |
| BNIP3L |
| OSGIN2 |
| CA1 |
| CA2 |
| CA3 |
| CA8 |
| CALB1 |
| RUNX1T1 |
| CDH17 |
| CEBPD |
| CHRNA2 |
| CHRNB3 |
| CLU |
| CRH |
| CTSB |
| CYC1 |
| CYP7A1 |
| CYP11B1 |
| CYP11B2 |
| ADAM3A |
| DECR1 |
| DEFA1 |
| DEFA3 |
| DEFA4 |
| DEFA5 |
| DEFA6 |
| DEFB1 |
| DEFB4 |
| DPYS |
| DPYSL2 |
| DUSP4 |
| E2F5 |
| EEF1D |
| EGR3 |
| EIF4EBP1 |
| EPB49 |
| EPHX2 |
| CLN8 |
| EXTL3 |
| EYA1 |
| FABP4 |
| FABP5 |
| PTK2B |
| FDFT1 |
| FGL1 |
| FNTA |
| ADAM2 |
| GATA4 |
| GEM |
| GFRA2 |
| GLI4 |
| GML |
| GNRH1 |
| NPBWR1 |
| GPR20 |
| GPT |
| GRINA |
| GSR |
| GTF2E2 |
| HAS2 |
| NRG1 |
| HNF4G |
| HSF1 |
| IKBKB |
| IL7 |
| IMPA1 |
| IDO1 |
| EIF3E |
| KCNQ3 |
| KCNS2 |
| LOXL2 |
| LPL |
| LY6E |
| LY6H |
| LYN |
| MATN2 |
| MCM4 |
| MMP16 |
| MOS |
| MSR1 |
| MSRA |
| MYBL1 |
| NBN |
| NDUFB9 |
| NEFM |
| NEFL |
| NFKBIL2 |
| NKX3-1 |
| NOV |
| ODF1 |
| TNFRSF11B |
| OPRK1 |
| PDE7A |
| PDGFRL |
| ENPP2 |
| PENK |
| PLAT |
| PLEC1 |
| PNOC |
| PMP2 |
| POLB |
| POLR2K |
| POU5F1B |
| PPP2CB |
| PPP2R2A |
| PPP3CC |
| PKIA |
| PRKDC |
| PTK2 |
| PVT1 |
| PXMP3 |
| RAB2A |
| RAD21 |
| RP1 |
| RPL7 |
| RPL8 |
| RPL30 |
| RPS20 |
| SDC2 |
| SDCBP |
| SFRP1 |
| SFTPC |
| ST3GAL1 |
| SLA |
| SLC7A2 |
| SLC18A1 |
| SLC20A2 |
| SNAI2 |
| SNTB1 |
| SPAG1 |
| SQLE |
| STAR |
| STC1 |
| STK3 |
| TACC1 |
| TAF2 |
| TCEB1 |
| TERF1 |
| TG |
| KLF10 |
| TPD52 |
| TRHR |
| TRPS1 |
| TSTA3 |
| TTPA |
| UBE2V2 |
| COL14A1 |
| UQCRB |
| VDAC3 |
| YWHAZ |
| ZNF7 |
| ZNF16 |
| FZD3 |
| TUSC3 |
| UBXN8 |
| MYST3 |
| PSCA |
| FZD6 |
| NSMAF |
| LY6D |
| RGS20 |
| JRK |
| TNKS |
| EIF3H |
| DGAT1 |
| GPAA1 |
| ADAM18 |
| ADAM9 |
| ADAM7 |
| RIPK2 |
| TNFRSF10D |
| TNFRSF10C |
| TNFRSF10B |
| TNFRSF10A |
| FGF17 |
| GGH |
| WISP1 |
| CPNE3 |
| FOXH1 |
| CHRNA6 |
| TRPA1 |
| DOK2 |
| ASH2L |
| MTMR7 |
| CCNE2 |
| EBAG9 |
| MYOM2 |
| DLGAP2 |
| MSC |
| MFHAS1 |
| KCNB2 |
| CYP7B1 |
| BAG4 |
| ENTPD4 |
| ARHGEF10 |
| MTFR1 |
| LRRC14 |
| TTC35 |
| RIMS2 |
| ST18 |
| TOX |
| MTSS1 |
| PTDSS1 |
| PHYHIP |
| RB1CC1 |
| ZNF623 |
| KIAA0196 |
| KBTBD11 |
| HHLA1 |
| SORBS3 |
| TRIB1 |
| HRSP12 |
| NPM2 |
| DLC1 |
| NDRG1 |
| PGCP |
| SPAG11B |
| LYPLA1 |
| ARFGEF1 |
| COLEC10 |
| KHDRBS3 |
| DCTN6 |
| PNMA2 |
| ADAM28 |
| POP1 |
| AP3M2 |
| COPS5 |
| RBPMS |
| WWP1 |
| STMN2 |
| PTP4A3 |
| ERLIN2 |
| LZTS1 |
| PROSC |
| RNF139 |
| ZHX1 |
| PUF60 |
| ZHX2 |
| DENND3 |
| XPO7 |
| TRIM35 |
| ZC3H3 |
| EFR3A |
| RRS1 |
| SULF1 |
| RHOBTB2 |
| ARC |
| BOP1 |
| DDHD2 |
| KIF13B |
| PSD3 |
| ZFPM2 |
| HEY1 |
| TRAM1 |
| LEPROTL1 |
| SCRIB |
| KIAA0146 |
| SLC39A14 |
| LRRC6 |
| LY96 |
| SGK3 |
| RAD54B |
| DCAF13 |
| RNF19A |
| GPR124 |
| KIAA1429 |
| C8orf71 |
| RGS22 |
| FBXL6 |
| PTTG3P |
| FBXO25 |
| FGF20 |
| SNORA72 |
| SNORD54 |
| OPLAH |
| PABPC1 |
| KCNV1 |
| STAU2 |
| MTBP |
| DKK4 |
| EIF2C2 |
| LSM1 |
| ADAMDEC1 |
| BHLHE22 |
| MRPS28 |
| COMMD5 |
| MRPL13 |
| ATAD2 |
| DDEF1IT1 |
| MRPL15 |
| CNOT7 |
| CPSF1 |
| PURG |
| LRP12 |
| RRM2B |
| CYHR1 |
| ASAP1 |
| MTERFD1 |
| PI15 |
| FAM135B |
| FAM164A |
| PHF20L1 |
| LACTB2 |
| FAM82B |
| ZNF706 |
| GOLGA7 |
| VPS28 |
| ZDHHC2 |
| C8orf30A |
| KCNK9 |
| SLC25A37 |
| C8orf55 |
| UBR5 |
| SCARA3 |
| FAM49B |
| AZIN1 |
| ATP6V1H |
| OTUD6B |
| TMEM66 |
| CHRAC1 |
| SNTG1 |
| GDAP1 |
| EXOSC4 |
| PDP1 |
| CNGB3 |
| LY6K |
| KCTD9 |
| ESRP1 |
| IMPAD1 |
| TMEM70 |
| PINX1 |
| TRMT12 |
| OXR1 |
| WDYHV1 |
| PIWIL2 |
| ELP3 |
| THAP1 |
| ARMC1 |
| INTS10 |
| CCDC25 |
| UBE2W |
| BRF2 |
| AGPAT5 |
| LAPTM4B |
| C8orf39 |
| TMEM55A |
| SLC39A4 |
| CHD7 |
| GOLSYN |
| INTS8 |
| INTS9 |
| CSGALNACT1 |
| HR |
| PAG1 |
| PBK |
| ZNF395 |
| DEFB103A |
| BIN3 |
| TEX15 |
| GSDMC |
| C8orf44 |
| JPH1 |
| C8orf4 |
| ENY2 |
| CPA6 |
| SLURP1 |
| SLC45A4 |
| MTUS1 |
| C8orf79 |
| ZFAT |
| KIAA1967 |
| ZNF250 |
| PLEKHA2 |
| SH2D4A |
| PRDM14 |
| SNX16 |
| NECAB1 |
| PDLIM2 |
| SOX17 |
| CSMD1 |
| EBF2 |
| FAM160B2 |
| DEPDC6 |
| PYCRL |
| C8orf33 |
| ZBTB10 |
| LYNX1 |
| MTMR9 |
| DUSP26 |
| C8orf51 |
| DSCC1 |
| DERL1 |
| GPR172A |
| HMBOX1 |
| EFCAB1 |
| MCPH1 |
| PPP1R3B |
| PLEKHF2 |
| ZMAT4 |
| ZFAND1 |
| ZFHX4 |
| GSDMD |
| NIPAL2 |
| RNF122 |
| CSPP1 |
| BAALC |
| NUDT18 |
| ZNF696 |
| GRHL2 |
| DOCK5 |
| FLJ14107 |
| VCPIP1 |
| ZNF703 |
| C8orf41 |
| RAB11FIP1 |
| PREX2 |
| REEP4 |
| KIAA1688 |
| ZNF34 |
| SLC25A32 |
| TM7SF4 |
| STMN4 |
| RNF170 |
| SLCO5A1 |
| SHARPIN |
| EPPK1 |
| SCRT1 |
| SOX7 |
| FAM167A |
| AMAC1L2 |
| C8orf12 |
| CRISPLD1 |
| TRAPPC9 |
| TM2D2 |
| TATDN1 |
| NACAP1 |
| NCALD |
| SGK196 |
| MAF1 |
| UTP23 |
| GINS4 |
| PPAPDC1B |
| MAK16 |
| TRIM55 |
| FUT10 |
| PARP10 |
| C8orf76 |
| TIGD5 |
| NUDCD1 |
| FAM83A |
| PPP1R16A |
| FAM86B1 |
| LRRCC1 |
| TSPYL5 |
| DNAJC5B |
| PSKH2 |
| FAM110B |
| MED30 |
| ERI1 |
| ZNF251 |
| KIFC2 |
| TMEM67 |
| LONRF1 |
| CHMP7 |
| MTDH |
| CHMP4C |
| PKHD1L1 |
| NAPRT1 |
| WDR67 |
| HPYR1 |
| RP1L1 |
| TP53INP1 |
| TGS1 |
| MFSD3 |
| MAL2 |
| XKR4 |
| CSMD3 |
| RHPN1 |
| FBXO32 |
| C8orf40 |
| SLC26A7 |
| PCMTD1 |
| CTHRC1 |
| OSR2 |
| C8orf34 |
| TOP1MT |
| CLDN23 |
| ZNF572 |
| GOT1L1 |
| FAM92A1 |
| VPS37A |
| C8orf38 |
| TMEM68 |
| ABRA |
| LYPD2 |
| NKX2-6 |
| TMEM71 |
| SGCZ |
| ADHFE1 |
| UBXN2B |
| PXDNL |
| AGPAT6 |
| UNC5D |
| LETM2 |
| DCAF4L2 |
| RALYL |
| HGSNAT |
| DEFB104A |
| PRAGMIN |
| PEBP4 |
| CDCA2 |
| TMEM65 |
| LOC157381 |
| RDH10 |
| C8orf56 |
| ANKRD46 |
| ESCO2 |
| FBXO16 |
| LOC157627 |
| FAM84B |
| C8orf37 |
| VPS13B |
| C8orf42 |
| ERICH1 |
| SLC7A13 |
| TDH |
| TMEM74 |
| FAM91A1 |
| C8orf48 |
| C8orf45 |
| CLVS1 |
| NKX6-3 |
| KCNU1 |
| C8orf84 |
| CNBD1 |
| SLC30A8 |
| COL22A1 |
| SNX31 |
| TMEM64 |
| ZNF596 |
| IDO2 |
| SDR16C5 |
| ADCK5 |
| TSNARE1 |
| R3HCC1 |
| T-SP1 |
| C8orf74 |
| HTRA4 |
| ADAM32 |
| C8orf47 |
| LGI3 |
| MAPK15 |
| DEFB105A |
| DEFB106A |
| DEFB107A |
| DEFB109P1 |
| DEFB130 |
| ATP6V0D2 |
| NEIL2 |
| YTHDF3 |
| C8orf46 |
| REXO1L1 |
| ADAM5P |
| FLJ10661 |
| XKR6 |
| NSMCE2 |
| ZNF707 |
| BREA2 |
| FAM83H |
| LOC286094 |
| EFHA2 |
| ZNF252 |
| TMED10P |
| C8orf77 |
| C8orf31 |
| ZFP41 |
| SCARA5 |
| LOC286135 |
| RNF5P1 |
| C8orf83 |
| DPY19L4 |
| FBXO43 |
| NKAIN3 |
| LRRC67 |
| GPIHBP1 |
| LOC340357 |
| KLHL38 |
| NRBP2 |
| ZNF517 |
| KIAA1875 |
| C8ORFK29 |
| RSPO2 |
| POTEA |
| SLC10A5 |
| LOC349196 |
| SPATC1 |
| USP17L2 |
| CA13 |
| XKR5 |
| C8orf80 |
| C8orf86 |
| FAM150A |
| XKR9 |
| RBM12B |
| FLJ43860 |
| MAFA |
| LOC392196 |
| GDF6 |
| LOC401463 |
| C8orf59 |
| SAMD12 |
| MIR124-1 |
| MIR124-2 |
| MIR30B |
| MIR30D |
| MIR320A |
| DEFB103B |
| MGC70857 |
| OR4F21 |
| FAM90A13 |
| FAM90A5 |
| FAM90A7 |
| FAM90A8 |
| FAM90A18 |
| FAM90A9 |
| FAM90A10 |
| REXO1L2P |
| C8orf85 |
| LRRC24 |
| SDR16C6 |
| DEFA10P |
| C8orf22 |
| MIR383 |
| BEYLA |
| DEFB107B |
| DEFB104B |
| DEFB106B |
| DEFB105B |
| C8orf58 |
| ZFATAS |
| HAS2AS |
| DEFB135 |
| DEFB136 |
| DEFB134 |
| ZNF704 |
| C8orf75 |
| MBOAT4 |
| NCRNA00051 |
| MIR486 |
| DEFB109P1B |
| SNHG6 |
| SNORD87 |
| C8orf73 |
| SCXB |
| LOC643763 |
| RPL23AP53 |
| FAM90A14 |
| FABP9 |
| FABP12 |
| FAM86B2 |
| SPAG11A |
| FER1L6 |
| MIR596 |
| MIR597 |
| MIR598 |
| MIR599 |
| MIR661 |
| LOC727677 |
| HEATR7A |
| LOC728024 |
| DEFA1B |
| FAM90A20 |
| FAM90A19 |
| ZNF705D |
| OC90 |
| LOC731779 |
| MIR875 |
| MIR937 |
| MIR939 |
| SCXA |
| LOC100130274 |
| LRRC69 |
| LOC100131726 |
| ZNF705G |
| FAM66E |
| FAM66D |
| FAM66A |
| SBF1P1 |
| LOC100133669 |
| LOC100192378 |
Figure 2. Genomic positions of deleted regions: the X-axis represents the normalized deletion signals (top) and significance by Q value (bottom). The green line represents the significance cutoff at Q value=0.25.
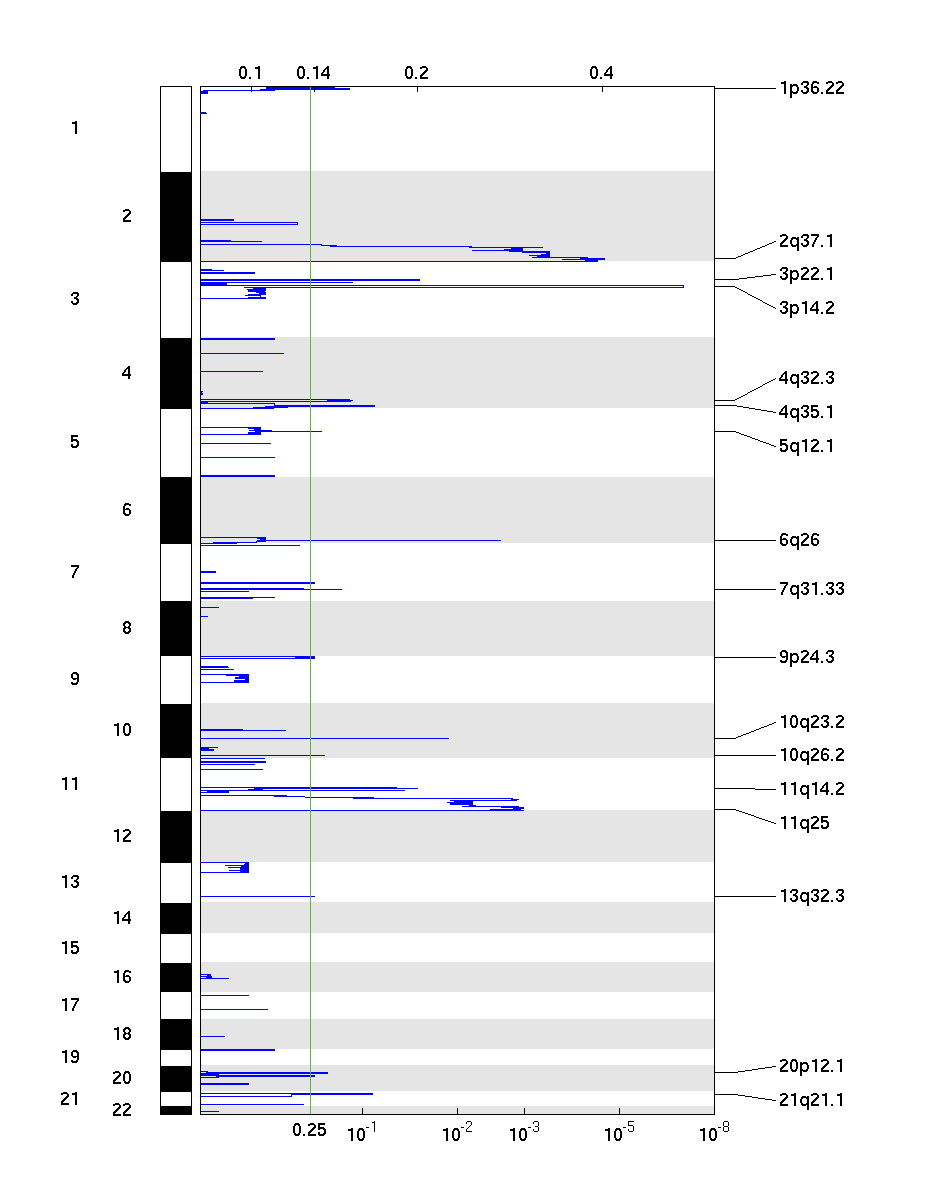
Table 2. Get Full Table Deletions Table - 17 significant deletions found. Click the link in the last column to view a comprehensive list of candidate genes. If no genes were identified within the peak, the nearest gene appears in brackets.
| Cytoband | Q value | Residual Q value | Wide Peak Boundaries | # Genes in Wide Peak |
|---|---|---|---|---|
| 3p14.2 | 1.2157e-07 | 1.2157e-07 | chr3:59009803-61522370 | 1 |
| 2q37.1 | 2.2714e-05 | 2.2714e-05 | chr2:213855358-242951149 | 247 |
| 6q26 | 0.0024205 | 0.0024205 | chr6:161613089-163073197 | 1 |
| 11q24.3 | 0.0010424 | 0.0024205 | chr11:17358096-134452384 | 1067 |
| 10q23.2 | 0.012697 | 0.012697 | chr10:89302988-89611040 | 3 |
| 3p22.1 | 0.079247 | 0.079247 | chr3:40553687-42109081 | 2 |
| 21q21.1 | 0.081912 | 0.079247 | chr21:18174997-25682320 | 4 |
| 11q14.1 | 0.02917 | 0.1053 | chr11:78825689-90243998 | 38 |
| 1p36.23 | 0.12708 | 0.12708 | chr1:1-222417645 | 2009 |
| 4q35.1 | 0.077278 | 0.15258 | chr4:174672203-191273063 | 67 |
| 7q31.33 | 0.14687 | 0.15258 | chr7:109385398-154370149 | 312 |
| 4q32.3 | 0.12708 | 0.16217 | chr4:167241131-174329125 | 13 |
| 20p12.1 | 0.18818 | 0.17837 | chr20:6695556-29311252 | 100 |
| 10q26.2 | 0.19982 | 0.19982 | chr10:97355027-135374737 | 308 |
| 5q12.1 | 0.20931 | 0.21755 | chr5:1-180857866 | 961 |
| 9p24.3 | 0.23217 | 0.23217 | chr9:1-140273252 | 890 |
| 13q32.3 | 0.23222 | 0.23217 | chr13:1-114142980 | 363 |
This is the comprehensive list of deleted genes in the wide peak for 3p14.2.
Table S11. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| FHIT |
This is the comprehensive list of deleted genes in the wide peak for 2q37.1.
Table S12. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| ATIC |
| ACSL3 |
| PAX3 |
| FEV |
| hsa-mir-149 |
| hsa-mir-562 |
| hsa-mir-1471 |
| hsa-mir-1244 |
| hsa-mir-153-1 |
| hsa-mir-375 |
| hsa-mir-26b |
| AAMP |
| AGXT |
| ALPI |
| ALPP |
| ALPPL2 |
| KIF1A |
| BARD1 |
| BCS1L |
| BOK |
| CHRND |
| CHRNG |
| COL4A3 |
| COL4A4 |
| COL6A3 |
| CRYBA2 |
| CYP27A1 |
| DES |
| DTYMK |
| EPHA4 |
| FN1 |
| GBX2 |
| GPC1 |
| GPR35 |
| HDLBP |
| AGFG1 |
| DNAJB2 |
| HTR2B |
| SP110 |
| IGFBP2 |
| IGFBP5 |
| IHH |
| IL8RA |
| IL8RB |
| IL8RBP |
| INHA |
| INPP5D |
| IRS1 |
| KCNJ13 |
| NCL |
| NDUFA10 |
| SEPT2 |
| NEU2 |
| NPPC |
| PDCD1 |
| PDE6D |
| SERPINE2 |
| PPP1R7 |
| PSMD1 |
| PTMA |
| PTPRN |
| SNORD20 |
| RPL37A |
| SAG |
| CCL20 |
| SLC4A3 |
| SLC11A1 |
| SP100 |
| SPP2 |
| TNP1 |
| TNS1 |
| TUBA4A |
| VIL1 |
| WNT6 |
| XRCC5 |
| ZNF142 |
| SCG2 |
| CUL3 |
| DGKD |
| STK16 |
| PER2 |
| CDK5R2 |
| RQCD1 |
| LRRFIP1 |
| GPR55 |
| TRIP12 |
| ECEL1 |
| EIF4E2 |
| TTLL4 |
| HDAC4 |
| FARP2 |
| FARSB |
| ABCB6 |
| ARPC2 |
| ARL4C |
| RAMP1 |
| SPEG |
| NMUR1 |
| STK25 |
| COPS8 |
| CAPN10 |
| SP140 |
| PASK |
| ATG4B |
| OBSL1 |
| DNPEP |
| SH3BP4 |
| KCNE4 |
| NGEF |
| SNORD82 |
| PNKD |
| SNED1 |
| GIGYF2 |
| TRAF3IP1 |
| ABCA12 |
| C2orf24 |
| STK36 |
| GMPPA |
| SMARCAL1 |
| ANO7 |
| PRLH |
| THAP4 |
| ANKMY1 |
| SCLY |
| ASB1 |
| CAB39 |
| PRKAG3 |
| UGT1A10 |
| UGT1A8 |
| UGT1A7 |
| UGT1A6 |
| UGT1A5 |
| UGT1A9 |
| UGT1A4 |
| UGT1A1 |
| UGT1A3 |
| PID1 |
| ATG16L1 |
| ANKZF1 |
| USP40 |
| HJURP |
| HES6 |
| ACCN4 |
| DOCK10 |
| MREG |
| PECR |
| C2orf83 |
| MFF |
| CXCR7 |
| RNPEPL1 |
| MARCH4 |
| WDFY1 |
| KIAA1486 |
| USP37 |
| CTDSP1 |
| GAL3ST2 |
| TMBIM1 |
| RAB17 |
| RNF25 |
| COPS7B |
| MRPL44 |
| TRPM8 |
| ATG9A |
| MLPH |
| FAM134A |
| GLB1L |
| SPAG16 |
| CHPF |
| IQCA1 |
| NHEJ1 |
| FAM124B |
| TM4SF20 |
| C2orf54 |
| TUBA4B |
| ARMC9 |
| EFHD1 |
| SPHKAP |
| WNT10A |
| SLC19A3 |
| ILKAP |
| ITM2C |
| RHBDD1 |
| ING5 |
| PLCD4 |
| MGC16025 |
| TMEM169 |
| DNER |
| B3GNT7 |
| SP140L |
| STK11IP |
| MOGAT1 |
| AGAP1 |
| DIS3L2 |
| NEU4 |
| AP1S3 |
| SGPP2 |
| SPATA3 |
| TMEM198 |
| ZFAND2B |
| FBXO36 |
| MTERFD2 |
| UBE2F |
| OTOS |
| MYEOV2 |
| OR6B3 |
| LOC151174 |
| CCDC140 |
| SLC23A3 |
| LOC151300 |
| GPBAR1 |
| SLC16A14 |
| C2orf52 |
| MSL3L2 |
| WDR69 |
| C2orf57 |
| TIGD1 |
| CCDC108 |
| C2orf72 |
| C2orf85 |
| RUFY4 |
| DUSP28 |
| ESPNL |
| ECEL1P2 |
| C2orf62 |
| RBM44 |
| AQP12A |
| KLHL30 |
| RESP18 |
| C2orf82 |
| OR6B2 |
| ASB18 |
| VWC2L |
| MIR149 |
| MIR153-1 |
| MIR26B |
| DNAJB3 |
| MIR375 |
| LOC643387 |
| PRR21 |
| AQP12B |
| SNORA75 |
| SCARNA6 |
| SCARNA5 |
| D2HGDH |
| LOC728323 |
| DIRC3 |
| PP14571 |
This is the comprehensive list of deleted genes in the wide peak for 6q26.
Table S13. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| PARK2 |
This is the comprehensive list of deleted genes in the wide peak for 11q24.3.
Table S14. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BIRC3 |
| ATM |
| CCND1 |
| CBL |
| DDB2 |
| DDX6 |
| DDX10 |
| EXT2 |
| FANCF |
| FLI1 |
| LMO2 |
| MEN1 |
| MLL |
| NUMA1 |
| PAFAH1B2 |
| POU2AF1 |
| SDHD |
| WT1 |
| PICALM |
| PCSK7 |
| ARHGEF12 |
| MAML2 |
| CREB3L1 |
| hsa-mir-100 |
| hsa-mir-125b-1 |
| hsa-mir-34c |
| hsa-mir-548l |
| hsa-mir-1304 |
| hsa-mir-1261 |
| hsa-mir-708 |
| hsa-mir-326 |
| hsa-mir-139 |
| hsa-mir-548k |
| hsa-mir-612 |
| hsa-mir-194-2 |
| hsa-mir-1237 |
| hsa-mir-1908 |
| hsa-mir-130a |
| hsa-mir-129-2 |
| hsa-mir-610 |
| ACAT1 |
| ACP2 |
| ACRV1 |
| ACTN3 |
| ADRBK1 |
| APLNR |
| ALDH3B1 |
| ALDH3B2 |
| BIRC2 |
| APLP2 |
| APOA1 |
| APOA4 |
| APOC3 |
| ARCN1 |
| ARHGAP1 |
| PHOX2A |
| ARL2 |
| ARRB1 |
| FXYD2 |
| BAD |
| BBS1 |
| BDNF |
| CXCR5 |
| SERPING1 |
| CAPN5 |
| C11orf2 |
| MRPL49 |
| ZNHIT2 |
| MPPED2 |
| C11orf9 |
| C11orf10 |
| DAGLA |
| CAPN1 |
| CASP1 |
| CASP4 |
| CASP5 |
| CAT |
| SERPINH1 |
| CD3D |
| CD3E |
| CD3G |
| CD5 |
| CD6 |
| MS4A1 |
| MS4A3 |
| CD44 |
| CD59 |
| CFL1 |
| CTSC |
| CHEK1 |
| CHKA |
| CHRM1 |
| CHRM4 |
| CLNS1A |
| CNTF |
| COX8A |
| CPT1A |
| CRY2 |
| CRYAB |
| CST6 |
| CSTF3 |
| CTNND1 |
| CTSW |
| DDB1 |
| DHCR7 |
| DLAT |
| DLG2 |
| DPAGT1 |
| DRD2 |
| EEF1G |
| ELF5 |
| MARK2 |
| CTTN |
| ESRRA |
| ETS1 |
| F2 |
| FAU |
| MS4A2 |
| FDX1 |
| FEN1 |
| FGF3 |
| FGF4 |
| FKBP2 |
| FOLH1 |
| FOLR1 |
| FOLR2 |
| FOLR3 |
| FSHB |
| FTH1 |
| FUT4 |
| SLC37A4 |
| LRRC32 |
| GAS2 |
| GIF |
| GNG3 |
| GRIA4 |
| GRIK4 |
| GRM5 |
| GSTP1 |
| GTF2H1 |
| GUCY1A2 |
| H2AFX |
| HMBS |
| SLC29A2 |
| HSPA8 |
| HSPB2 |
| DNAJC4 |
| HTR3A |
| IGHMBP2 |
| IL10RA |
| IL18 |
| INCENP |
| INPPL1 |
| STT3A |
| CD82 |
| KCNA4 |
| KCNC1 |
| KCNJ1 |
| KCNJ5 |
| KCNJ11 |
| KRTAP5-9 |
| LDHA |
| LDHC |
| FADS1 |
| FADS3 |
| VWA5A |
| LRP4 |
| LRP5 |
| LTBP3 |
| CAPRIN1 |
| MAP6 |
| MCAM |
| MDK |
| SCGB2A1 |
| SCGB2A2 |
| MAP3K11 |
| MMP1 |
| MMP3 |
| MMP7 |
| MMP8 |
| MMP10 |
| MMP12 |
| MMP13 |
| MRE11A |
| MTNR1B |
| MYBPC3 |
| MYO7A |
| MYOD1 |
| NCAM1 |
| NDUFC2 |
| NDUFS3 |
| NDUFV1 |
| NDUFS8 |
| NELL1 |
| NFRKB |
| NNMT |
| NPAT |
| NRGN |
| OMP |
| OPCML |
| OSBP |
| OVOL1 |
| P2RX3 |
| P2RY2 |
| P2RY6 |
| PAK1 |
| PAX6 |
| PC |
| PDE2A |
| PGA5 |
| PGR |
| PLCB3 |
| POLR2G |
| PPP1CA |
| PPP2R1B |
| PPP2R5B |
| PRCP |
| PRG2 |
| PRKRIR |
| PSMC3 |
| PTPRCAP |
| PTPRJ |
| PTS |
| PVRL1 |
| PYGM |
| RAB3IL1 |
| RAB6A |
| MAP4K2 |
| RAD9A |
| RAG1 |
| RAG2 |
| RAPSN |
| RARRES3 |
| RBM4 |
| RCN1 |
| RDX |
| RELA |
| DPF2 |
| SNORD15A |
| ROM1 |
| RPS3 |
| RPS6KB2 |
| RPS25 |
| SAA1 |
| SAA2 |
| SAA3P |
| SAA4 |
| SC5DL |
| SCN2B |
| SCN4B |
| ST3GAL4 |
| SIPA1 |
| SLC1A2 |
| SLC3A2 |
| SLN |
| SORL1 |
| SPI1 |
| SPTBN2 |
| SRPR |
| SSRP1 |
| ST14 |
| STX3 |
| STX5 |
| ABCC8 |
| TAGLN |
| TCN1 |
| TECTA |
| THRSP |
| THY1 |
| TM7SF2 |
| TPH1 |
| TRAF6 |
| TRPC6 |
| TSG101 |
| TYR |
| UCP2 |
| UCP3 |
| SCGB1A1 |
| UPK2 |
| UVRAG |
| VEGFB |
| BEST1 |
| WNT11 |
| SF1 |
| ZFPL1 |
| ZBTB16 |
| ZNF202 |
| CSRP3 |
| PDHX |
| FOSL1 |
| CUL5 |
| FZD4 |
| BBOX1 |
| PPFIA1 |
| SLC43A1 |
| DGKZ |
| CHST1 |
| BARX2 |
| API5 |
| MADD |
| JRKL |
| CTSF |
| EED |
| FADD |
| BANF1 |
| ZNF259 |
| MTMR2 |
| RPS6KA4 |
| AIP |
| SYT7 |
| SART1 |
| USP2 |
| SLC6A5 |
| FIBP |
| HTR3B |
| ZW10 |
| MTA2 |
| UBE2L6 |
| SNORD29 |
| SNORD31 |
| SNORD30 |
| SNORD28 |
| SNORD27 |
| SNORD26 |
| SNORD25 |
| SNORD22 |
| MMP20 |
| UBE4A |
| SLC22A6 |
| SLC22A8 |
| NRXN2 |
| LPXN |
| PEX16 |
| FADS2 |
| MED17 |
| MAPK8IP1 |
| TP53I11 |
| EI24 |
| PITPNM1 |
| RIN1 |
| MTL5 |
| FEZ1 |
| CEP57 |
| RICS |
| KIAA0652 |
| SPCS2 |
| CKAP5 |
| ARHGEF17 |
| GAB2 |
| C2CD2L |
| FCHSD2 |
| FGF19 |
| CCS |
| RCE1 |
| NAALAD2 |
| NAALADL1 |
| KCNE3 |
| NR1H3 |
| IL18BP |
| DPP3 |
| USH1C |
| KCNK7 |
| HIPK3 |
| RBM7 |
| PRMT3 |
| MPZL2 |
| RASGRP2 |
| GLYAT |
| CDK2AP2 |
| TCIRG1 |
| RTN3 |
| PRG3 |
| YAP1 |
| RBM14 |
| CDC42EP2 |
| EIF3M |
| NXF1 |
| KAT5 |
| HYOU1 |
| SSSCA1 |
| HTATIP2 |
| DRAP1 |
| TAF6L |
| ATP5L |
| SCGB1D2 |
| SCGB1D1 |
| CUGBP1 |
| POLD3 |
| OR5I1 |
| STARD10 |
| NEU3 |
| ME3 |
| GPR83 |
| YIF1A |
| SFRS2B |
| EHD1 |
| STIP1 |
| CLP1 |
| SF3B2 |
| CCDC85B |
| B3GNT1 |
| ADAMTS8 |
| PRSS23 |
| PLA2G16 |
| TREH |
| HPS5 |
| GPR44 |
| SLCO2B1 |
| CEP164 |
| ZP1 |
| SHANK2 |
| KDM2A |
| IGSF9B |
| ENDOD1 |
| EXPH5 |
| ATG2A |
| PHLDB1 |
| GANAB |
| FAM168A |
| DTX4 |
| SIK2 |
| NUP160 |
| NCAPD3 |
| FNBP4 |
| SIK3 |
| CLCF1 |
| VSIG2 |
| BACE1 |
| FAM89B |
| SNHG1 |
| POLA2 |
| TRIM29 |
| RAB38 |
| CADM1 |
| FLRT1 |
| MTCH2 |
| PANX1 |
| FJX1 |
| C11orf41 |
| PRDX5 |
| POU2F3 |
| ABTB2 |
| BRMS1 |
| C11orf20 |
| CHRDL2 |
| PAMR1 |
| C11orf51 |
| ZDHHC5 |
| ATL3 |
| TSKU |
| HINFP |
| REXO2 |
| C2CD3 |
| DAK |
| ODZ4 |
| DKFZP434K028 |
| B3GAT3 |
| FBXO3 |
| SERGEF |
| EHF |
| OR5L2 |
| PPP1R14B |
| OR8G2 |
| OR8B8 |
| OR8G1 |
| TIMM10 |
| TIMM8B |
| MYEOV |
| BSCL2 |
| OR8B2 |
| ELP4 |
| CHORDC1 |
| ACAD8 |
| B3GAT1 |
| RAB30 |
| PRPF19 |
| DCPS |
| C11orf54 |
| C11orf67 |
| MACROD1 |
| SLC43A3 |
| ZBTB44 |
| THYN1 |
| COMMD9 |
| DDX25 |
| PACSIN3 |
| SAC3D1 |
| SNX15 |
| RHOD |
| EFEMP2 |
| NOX4 |
| KCNK4 |
| NTM |
| CDON |
| UBXN1 |
| APIP |
| TMX2 |
| GAL |
| SIDT2 |
| SUV420H1 |
| HSD17B12 |
| TMEM216 |
| CHCHD8 |
| SLC15A3 |
| PHF21A |
| MS4A4A |
| WIT1 |
| TRAPPC4 |
| PPME1 |
| CABP2 |
| C11orf73 |
| CWC15 |
| TRMT112 |
| TMEM138 |
| PCF11 |
| MRPL48 |
| RSF1 |
| SPA17 |
| FXYD6 |
| C11orf24 |
| CNTN5 |
| SIAE |
| C11orf71 |
| ZDHHC13 |
| ROBO4 |
| WDR74 |
| SLC35F2 |
| RAB39 |
| TRIM44 |
| BTG4 |
| FAM55D |
| SYTL2 |
| ANKRD49 |
| MS4A12 |
| MRPL16 |
| SDHAF2 |
| SSH3 |
| TTC12 |
| TMEM132A |
| HRASLS2 |
| C11orf59 |
| VPS37C |
| ANO1 |
| NADSYN1 |
| FAM86C |
| C11orf57 |
| NAT10 |
| CCDC87 |
| SAPS3 |
| UEVLD |
| RNF121 |
| LIN7C |
| ACER3 |
| SLC35C1 |
| TCP11L1 |
| LGR4 |
| ELMOD1 |
| CDC42BPG |
| FOXRED1 |
| OTUB1 |
| AMBRA1 |
| PACS1 |
| KDM4D |
| KBTBD4 |
| TTC17 |
| SCN3B |
| VPS11 |
| TMEM126B |
| SLC22A11 |
| TEX12 |
| CRTAM |
| TMPRSS4 |
| GPR137 |
| C11orf60 |
| C11orf75 |
| C11orf30 |
| PRDM10 |
| PRDM11 |
| CABP4 |
| SLC17A6 |
| TRIM49 |
| CD248 |
| CORO1B |
| SCYL1 |
| DSCAML1 |
| GRAMD1B |
| USP35 |
| KIAA1377 |
| ARHGAP20 |
| ATPGD1 |
| SYT13 |
| USP28 |
| LRRC4C |
| POLD4 |
| KRTAP5-8 |
| PLEKHB1 |
| MS4A7 |
| CREBZF |
| CARD18 |
| CCDC90B |
| CCDC81 |
| AASDHPPT |
| ALX4 |
| PKNOX2 |
| FAM111A |
| TP53AIP1 |
| ANO3 |
| MMP27 |
| ABCG4 |
| ROBO3 |
| MS4A6A |
| MS4A5 |
| C11orf1 |
| KLC2 |
| TUT1 |
| MRPL11 |
| TMEM135 |
| KCTD14 |
| C11orf95 |
| LRFN4 |
| AHNAK |
| ALG8 |
| PRRG4 |
| TMEM223 |
| TMEM109 |
| CCDC86 |
| C11orf48 |
| C11orf49 |
| TRIM48 |
| TAF1D |
| RNF26 |
| FAM118B |
| DYNC2H1 |
| NLRX1 |
| C11orf61 |
| C11orf80 |
| NARS2 |
| E2F8 |
| CCDC82 |
| ALG9 |
| ZNF408 |
| ASAM |
| NAT11 |
| QSER1 |
| AGBL2 |
| ZBTB3 |
| PDZD3 |
| C11orf63 |
| CPSF7 |
| PRR5L |
| CCDC15 |
| ASRGL1 |
| MOGAT2 |
| TMEM134 |
| MUS81 |
| PAAF1 |
| PDGFD |
| INTS5 |
| ZFP91 |
| TMPRSS5 |
| OR8J3 |
| OR4P4 |
| OR4C15 |
| OR4A5 |
| OR4A16 |
| OR4A15 |
| OR10W1 |
| GDPD5 |
| CLPB |
| UNC93B1 |
| RAB1B |
| KIF18A |
| PUS3 |
| MFRP |
| C11orf68 |
| MS4A8B |
| JAM3 |
| FERMT3 |
| TRPT1 |
| RBM4B |
| FRMD8 |
| BCO2 |
| TMEM133 |
| TMPRSS13 |
| RNASEH2C |
| TMEM126A |
| DCUN1D5 |
| EIF1AD |
| NUDT22 |
| ARFGAP2 |
| KIAA1826 |
| SYVN1 |
| KIRREL3 |
| DGAT2 |
| ACCS |
| MS4A14 |
| SPRYD5 |
| BUD13 |
| TMEM25 |
| PTPN5 |
| RPUSD4 |
| TBRG1 |
| RELT |
| UBASH3B |
| C11orf70 |
| LGALS12 |
| TNKS1BP1 |
| DIXDC1 |
| KIAA1731 |
| ZC3H12C |
| GAL3ST3 |
| NAV2 |
| ATG16L2 |
| GLB1L2 |
| TSPAN18 |
| ESAM |
| DKFZp761E198 |
| CCDC34 |
| SLC39A13 |
| DEPDC7 |
| SYT12 |
| ACY3 |
| ALKBH8 |
| FDXACB1 |
| C11orf52 |
| INTS4 |
| GLYATL1 |
| VPS26B |
| GLB1L3 |
| CDCA5 |
| RPLP0P2 |
| SAAL1 |
| SLC22A9 |
| SNORD15B |
| TIRAP |
| CARD16 |
| C1QTNF4 |
| C1QTNF5 |
| TMEM123 |
| PTPMT1 |
| BATF2 |
| SLC22A12 |
| PANX3 |
| MRGPRD |
| APOA5 |
| MRGPRF |
| ARAP1 |
| CATSPER1 |
| MRGPRX2 |
| MRGPRX3 |
| MRGPRX4 |
| HRASLS5 |
| C11orf74 |
| OR4C46 |
| OR4X2 |
| OR4B1 |
| GYLTL1B |
| SLC36A4 |
| FAT3 |
| TRIM64 |
| TMEM45B |
| DBX1 |
| CASP12 |
| C11orf93 |
| PIH1D2 |
| FAM55A |
| FAM55B |
| AMICA1 |
| DNAJC24 |
| C11orf46 |
| OR8I2 |
| LDLRAD3 |
| XRRA1 |
| MUC15 |
| C11orf94 |
| FAM76B |
| SESN3 |
| PIWIL4 |
| FLJ32810 |
| KBTBD3 |
| CWF19L2 |
| KDELC2 |
| LAYN |
| TTC36 |
| C11orf84 |
| SPTY2D1 |
| TMEM86A |
| AMOTL1 |
| SLC5A12 |
| CCDC67 |
| PATE1 |
| C11orf65 |
| LDHAL6A |
| GPHA2 |
| ADAMTS15 |
| B3GNT6 |
| METT5D1 |
| MPZL3 |
| IMMP1L |
| ANO5 |
| OR8U1 |
| OR4C16 |
| OR4C11 |
| OR4S2 |
| OR4C6 |
| OR5D14 |
| OR5L1 |
| OR5D18 |
| OR7E5P |
| OR5AS1 |
| OR8K5 |
| OR5T2 |
| OR8H1 |
| OR8K3 |
| OR8J1 |
| OR5R1 |
| OR5M3 |
| OR5M8 |
| OR5M11 |
| OR5AR1 |
| LRRC55 |
| SMTNL1 |
| YPEL4 |
| MED19 |
| FOLH1B |
| C11orf45 |
| HYLS1 |
| TMEM218 |
| SLC37A2 |
| OR8B12 |
| OR8G5 |
| OR10G8 |
| OR10G9 |
| OR10S1 |
| OR6T1 |
| OR4D5 |
| TBCEL |
| TMEM136 |
| MRPL21 |
| TPCN2 |
| SPATA19 |
| OR6Q1 |
| OR9I1 |
| OR9Q1 |
| OR9Q2 |
| OR1S2 |
| OR1S1 |
| OR10Q1 |
| OR5B17 |
| OR5B21 |
| GLYATL2 |
| MPEG1 |
| OR5A2 |
| OR5A1 |
| OR4D6 |
| OR4D11 |
| PATL1 |
| PLAC1L |
| MS4A15 |
| VWCE |
| CYBASC3 |
| C11orf66 |
| GDPD4 |
| C11orf82 |
| CCDC83 |
| ORAOV1 |
| LRTOMT |
| HEPACAM |
| OAF |
| TIGD3 |
| FAM181B |
| CCDC89 |
| LRRN4CL |
| HNRNPUL2 |
| ALKBH3 |
| LOC221122 |
| MS4A6E |
| DEFB108B |
| PELI3 |
| ANGPTL5 |
| EHBP1L1 |
| SNX32 |
| TSGA10IP |
| RNF169 |
| CNIH2 |
| ZDHHC24 |
| C11orf86 |
| NUDT8 |
| ANKK1 |
| OR4C3 |
| OR4S1 |
| EML3 |
| TMEM151A |
| RNF214 |
| SVIP |
| MRGPRX1 |
| NPAS4 |
| C11orf31 |
| AQP11 |
| OR10AG1 |
| OR5J2 |
| OR4C13 |
| OR4C12 |
| C11orf85 |
| SLC25A45 |
| NEAT1 |
| BCL9L |
| FOXR1 |
| CCDC153 |
| OR8D1 |
| OR8D2 |
| OR8B4 |
| LOC283174 |
| OR9G4 |
| C11orf64 |
| P4HA3 |
| PGM2L1 |
| KLHL35 |
| KCTD21 |
| CCDC88B |
| TTC9C |
| SLC22A24 |
| RCOR2 |
| HARBI1 |
| LOC283267 |
| IGSF22 |
| LUZP2 |
| CCDC84 |
| TMEM225 |
| OR8D4 |
| OR5F1 |
| OR5AP2 |
| ANKRD13D |
| ANKRD42 |
| DCDC1 |
| C11orf53 |
| LOC341056 |
| MS4A10 |
| OR2AT4 |
| HEPHL1 |
| TBX10 |
| C11orf34 |
| RTN4RL2 |
| DKFZp779M0652 |
| FAM111B |
| TMEM179B |
| TBC1D10C |
| DNAJB13 |
| MALAT1 |
| ZFP91-CNTF |
| KRTAP5-10 |
| SLC22A25 |
| FIBIN |
| LOC387763 |
| SLC22A10 |
| SPDYC |
| LIPT2 |
| C11orf90 |
| ACCSL |
| OR4X1 |
| OR5D13 |
| OR5D16 |
| OR5W2 |
| OR8H2 |
| OR8H3 |
| OR5T3 |
| OR5T1 |
| OR8K1 |
| OR5M9 |
| OR5M10 |
| OR5M1 |
| OR9G1 |
| OR5AK2 |
| OR5B2 |
| OR5B12 |
| OR5AN1 |
| OR4D10 |
| OR4D9 |
| OR10V1 |
| LRRC10B |
| GPR152 |
| GUCY2E |
| TRIM77 |
| FOLR4 |
| KDM4DL |
| BSX |
| OR6X1 |
| OR6M1 |
| OR10G4 |
| OR10G7 |
| OR8B3 |
| OR8A1 |
| FAM180B |
| PCNXL3 |
| C11orf87 |
| C11orf92 |
| C11orf88 |
| LOC399959 |
| PATE2 |
| PATE4 |
| SNX19 |
| OR4A47 |
| OR4C45 |
| SCGB1D4 |
| MIRLET7A2 |
| MIR100 |
| MIR125B1 |
| MIR129-2 |
| MIR130A |
| MIR139 |
| MIR192 |
| MIR194-2 |
| MIR34B |
| MIR34C |
| DDI1 |
| BLID |
| LOC440040 |
| SLC22A20 |
| KRTAP5-7 |
| KRTAP5-11 |
| CARD17 |
| NCRNA00167 |
| LOC441601 |
| OR5B3 |
| MIR326 |
| CCDC73 |
| LOC494141 |
| BDNFOS |
| MS4A13 |
| OR8U8 |
| OR9G9 |
| SCARNA9 |
| HEPN1 |
| TRIM64B |
| TRIM53 |
| UBTFL1 |
| BTBD18 |
| PGA3 |
| PGA4 |
| LOC643923 |
| CLDN25 |
| LOC645332 |
| LOC646813 |
| RPL23AP64 |
| SNORA8 |
| SNORA1 |
| SNORA18 |
| SNORA40 |
| SNORA25 |
| SNORA32 |
| SNORD5 |
| SNORD6 |
| SNORD67 |
| SNORA57 |
| MIR610 |
| MIR611 |
| MIR612 |
| LOC729384 |
| LOC729799 |
| METTL12 |
| C11orf83 |
| LOC100126784 |
| LOC100128239 |
| LOC100130987 |
| C11orf91 |
| LOC100133315 |
| PATE3 |
This is the comprehensive list of deleted genes in the wide peak for 10q23.2.
Table S15. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| PAPSS2 |
| ATAD1 |
| CFLP1 |
This is the comprehensive list of deleted genes in the wide peak for 3p22.1.
Table S16. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CTNNB1 |
| ULK4 |
This is the comprehensive list of deleted genes in the wide peak for 21q21.1.
Table S17. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| NCAM2 |
| PRSS7 |
| CHODL |
| C21orf131 |
This is the comprehensive list of deleted genes in the wide peak for 11q14.1.
Table S18. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| PICALM |
| hsa-mir-1261 |
| CTSC |
| DLG2 |
| GRM5 |
| PRCP |
| TYR |
| FZD4 |
| EED |
| NAALAD2 |
| ME3 |
| PRSS23 |
| RAB38 |
| CHORDC1 |
| RAB30 |
| NOX4 |
| C11orf73 |
| PCF11 |
| SYTL2 |
| TMEM126B |
| TRIM49 |
| CREBZF |
| CCDC90B |
| CCDC81 |
| TMEM135 |
| TMEM126A |
| TRIM64 |
| FOLH1B |
| C11orf82 |
| CCDC83 |
| FAM181B |
| CCDC89 |
| ANKRD42 |
| TRIM77 |
| TRIM64B |
| TRIM53 |
| UBTFL1 |
| LOC729384 |
This is the comprehensive list of deleted genes in the wide peak for 1p36.23.
Table S19. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| ABL2 |
| ARNT |
| BCL9 |
| CDKN2C |
| ELK4 |
| EPS15 |
| FCGR2B |
| JAK1 |
| JUN |
| LCK |
| MDM4 |
| MPL |
| MUC1 |
| MUTYH |
| MYCL1 |
| NOTCH2 |
| NRAS |
| NTRK1 |
| PAX7 |
| PBX1 |
| PRCC |
| RPL22 |
| SDHB |
| SDHC |
| SFPQ |
| TAL1 |
| TPM3 |
| TPR |
| ARID1A |
| TNFRSF14 |
| BCL10 |
| PDE4DIP |
| THRAP3 |
| TRIM33 |
| PRDM16 |
| RBM15 |
| SLC45A3 |
| MDS2 |
| hsa-mir-664 |
| hsa-mir-194-1 |
| hsa-mir-205 |
| hsa-mir-29b-2 |
| hsa-mir-135b |
| hsa-mir-1231 |
| hsa-mir-181a-1 |
| hsa-mir-1278 |
| hsa-mir-488 |
| hsa-mir-199a-2 |
| hsa-mir-1295 |
| hsa-mir-557 |
| hsa-mir-1255b-2 |
| hsa-mir-921 |
| hsa-mir-556 |
| hsa-mir-765 |
| hsa-mir-9-1 |
| hsa-mir-555 |
| hsa-mir-92b |
| hsa-mir-190b |
| hsa-mir-554 |
| hsa-mir-942 |
| hsa-mir-320b-1 |
| hsa-mir-197 |
| hsa-mir-553 |
| hsa-mir-137 |
| hsa-mir-760 |
| hsa-mir-186 |
| hsa-mir-1262 |
| hsa-mir-101-1 |
| hsa-mir-30c-1 |
| hsa-mir-552 |
| hsa-mir-1976 |
| hsa-mir-1256 |
| hsa-mir-1290 |
| hsa-mir-34a |
| hsa-mir-551a |
| hsa-mir-429 |
| hsa-mir-1977 |
| hsa-mir-1302-2 |
| ABCA4 |
| ACADM |
| ADAR |
| ADORA1 |
| ADORA3 |
| AGL |
| AK2 |
| AK3L1 |
| ALDH9A1 |
| ALPL |
| ALX3 |
| AMPD1 |
| AMPD2 |
| AMY1A |
| AMY1B |
| AMY1C |
| AMY2A |
| AMY2B |
| APCS |
| APOA2 |
| FASLG |
| RHOC |
| ASTN1 |
| SERPINC1 |
| ATF3 |
| RERE |
| ATP1A1 |
| ATP1A2 |
| ATP1A4 |
| ATP1B1 |
| ATP2B4 |
| ATP5F1 |
| ATP6V0B |
| AVPR1B |
| BAI2 |
| BGLAP |
| BMP8B |
| BRDT |
| C1QA |
| C1QB |
| C1QC |
| C4BPA |
| C4BPB |
| C8A |
| C8B |
| CA6 |
| CACNA1E |
| CACNA1S |
| CAPN2 |
| CAPZA1 |
| CAPZB |
| CASP9 |
| CASQ1 |
| CASQ2 |
| RUNX3 |
| CD1A |
| CD1B |
| CD1C |
| CD1D |
| CD1E |
| CD2 |
| CD247 |
| CD5L |
| TNFRSF8 |
| CD34 |
| CD48 |
| CD53 |
| CD58 |
| CDA |
| CDC2L1 |
| CDC20 |
| CDC42 |
| CD52 |
| CENPF |
| RCC1 |
| CHI3L1 |
| CHI3L2 |
| CHIT1 |
| CHRNB2 |
| CKS1B |
| CLCA1 |
| CLCN6 |
| CLCNKA |
| CLCNKB |
| CLK2 |
| PLK3 |
| CNN3 |
| CNR2 |
| COL8A2 |
| COL9A2 |
| COL11A1 |
| COL16A1 |
| COPA |
| CORT |
| CPT2 |
| CR1 |
| CR1L |
| CR2 |
| CRABP2 |
| CRP |
| CRYZ |
| CSF1 |
| CSF3R |
| CSRP1 |
| CTBS |
| CTH |
| CTPS |
| CTSE |
| CTSK |
| CTSS |
| CYP2J2 |
| CYP4A11 |
| CYP4B1 |
| DAB1 |
| CD55 |
| DBT |
| GADD45A |
| DDOST |
| DHX9 |
| DFFA |
| DFFB |
| DHCR24 |
| DIO1 |
| DPH2 |
| DPT |
| DPYD |
| DR1 |
| DVL1 |
| E2F2 |
| ECE1 |
| ECM1 |
| S1PR1 |
| EDN2 |
| PHC2 |
| LGTN |
| EFNA1 |
| EFNA3 |
| EFNA4 |
| CELSR2 |
| MEGF6 |
| EPHA2 |
| ELAVL4 |
| ELF3 |
| ENO1 |
| ENSA |
| EPB41 |
| EPHA8 |
| EPHB2 |
| EPRS |
| ESRRG |
| ETV3 |
| EXTL1 |
| EXTL2 |
| EYA3 |
| F3 |
| F5 |
| F13B |
| FAAH |
| FABP3 |
| FCER1A |
| FCER1G |
| FCGR1A |
| FCGR1B |
| FCGR2A |
| FCGR3A |
| FCGR3B |
| FDPS |
| FGR |
| FHL3 |
| FOXE3 |
| FOXD2 |
| FLG |
| FMO1 |
| FMO2 |
| FMO3 |
| FMO4 |
| FMO5 |
| FMOD |
| MTOR |
| NR5A2 |
| FUCA1 |
| DARC |
| IFI6 |
| GABRD |
| GALE |
| GBA |
| GBAP |
| GBP1 |
| GBP2 |
| GBP3 |
| GFI1 |
| GJA4 |
| GJA5 |
| GJA8 |
| GJB3 |
| GJB5 |
| GCLM |
| GLUL |
| GNAI3 |
| GNAT2 |
| GNB1 |
| GNG5 |
| SFN |
| GPR3 |
| GPR25 |
| GPX7 |
| GRIK3 |
| GSTM1 |
| GSTM2 |
| GSTM3 |
| GSTM4 |
| GSTM5 |
| GTF2B |
| GUCA2A |
| GUCA2B |
| HCRTR1 |
| HDAC1 |
| HDGF |
| CFH |
| CFHR1 |
| CFHR2 |
| ZBTB48 |
| MR1 |
| HLX |
| HMGN2 |
| HMGCL |
| HMGCS2 |
| HPCA |
| HSD3B1 |
| HSD3B2 |
| HSD11B1 |
| HSPA6 |
| HSPA7 |
| IGSF3 |
| HSPG2 |
| HTR1D |
| HTR6 |
| ID3 |
| IFI16 |
| CYR61 |
| IL6R |
| IL10 |
| IL12RB2 |
| TNFRSF9 |
| ILF2 |
| INPP5B |
| INSRR |
| IPP |
| IRF6 |
| IVL |
| KCNA2 |
| KCNA3 |
| KCNA10 |
| KCNC4 |
| KCND3 |
| KCNH1 |
| KCNJ9 |
| KCNJ10 |
| KCNK2 |
| KCNN3 |
| KISS1 |
| LAD1 |
| LAMB3 |
| LAMC1 |
| LAMC2 |
| STMN1 |
| LEPR |
| LMNA |
| LMX1A |
| LOR |
| LY9 |
| TACSTD2 |
| MAGOH |
| MARK1 |
| MATN1 |
| MCL1 |
| CD46 |
| SMCP |
| MEF2D |
| MFAP2 |
| MGST3 |
| MNDA |
| MOV10 |
| MPZ |
| MSH4 |
| MTF1 |
| MTHFR |
| MTX1 |
| MYBPH |
| MYOC |
| MYOG |
| PPP1R12B |
| NASP |
| NBL1 |
| NCF2 |
| NDUFS2 |
| NDUFS5 |
| NEK2 |
| NFIA |
| NFYC |
| NGF |
| NHLH1 |
| NHLH2 |
| NIT1 |
| NPPA |
| NPPB |
| NPR1 |
| NRD1 |
| YBX1 |
| ROR1 |
| DDR2 |
| OPRD1 |
| ORC1L |
| OVGP1 |
| PAFAH2 |
| PRDX1 |
| PCTK3 |
| PDC |
| PDE4B |
| PDZK1 |
| PEX10 |
| PEX14 |
| PFDN2 |
| PFKFB2 |
| PGD |
| PGM1 |
| PIGC |
| PIGR |
| PIK3C2B |
| PIK3CD |
| PI4KB |
| PIN1L |
| PKLR |
| PKP1 |
| PLA2G2A |
| PLA2G4A |
| PLA2G5 |
| PLOD1 |
| PLXNA2 |
| EXOSC10 |
| PRRX1 |
| POU2F1 |
| POU3F1 |
| PPOX |
| PPP1R8 |
| PPP2R5A |
| PPT1 |
| PRELP |
| PRKAA2 |
| PRKAB2 |
| PRKACB |
| PKN2 |
| PRKCZ |
| PROX1 |
| PSMA5 |
| PSMB2 |
| PSMB4 |
| PSMD4 |
| PTAFR |
| PTGER3 |
| PTGFR |
| PTGFRN |
| PTGS2 |
| QSOX1 |
| PTPN7 |
| PTPN14 |
| PTPRC |
| PTPRF |
| PEX19 |
| ABCD3 |
| RAB3B |
| RAB13 |
| RABGGTB |
| RABIF |
| RAP1A |
| RAP1GAP |
| RBBP4 |
| RBBP5 |
| REN |
| RFX5 |
| RGS1 |
| RGS2 |
| RGS4 |
| RGS13 |
| RGS16 |
| RHCE |
| RHD |
| RIT1 |
| RLF |
| RNASEL |
| RNF2 |
| RNPEP |
| SNORD21 |
| RORC |
| RPA2 |
| RPE65 |
| RPL5 |
| RPL11 |
| RPS6KA1 |
| RPS8 |
| RPS27 |
| RSC1A1 |
| RXRG |
| S100A1 |
| SORT1 |
| S100A2 |
| S100A3 |
| S100A4 |
| S100A5 |
| S100A6 |
| S100A7 |
| S100A8 |
| S100A9 |
| S100A10 |
| S100A11 |
| S100A12 |
| S100A13 |
| SARS |
| SCNN1D |
| SCP2 |
| XCL1 |
| SELE |
| SELL |
| SELP |
| SFRS4 |
| SHC1 |
| ST3GAL3 |
| STIL |
| SKI |
| SLAMF1 |
| SLC1A7 |
| SLC2A1 |
| SLC2A5 |
| SLC6A9 |
| SLC9A1 |
| SLC16A1 |
| SNRPE |
| SOAT1 |
| UAP1 |
| SPRR1A |
| SPRR1B |
| SPRR2A |
| SPRR2B |
| SPRR2C |
| SPRR2D |
| SPRR2E |
| SPRR2F |
| SPRR2G |
| SPRR3 |
| SPTA1 |
| SRM |
| TROVE2 |
| SSR2 |
| AURKAPS1 |
| STXBP3 |
| XCL2 |
| SYCP1 |
| TAF12 |
| TAF13 |
| CNTN2 |
| TBX15 |
| TCEA3 |
| TCEB3 |
| VPS72 |
| TGFB2 |
| TGFBR3 |
| THBS3 |
| TCHH |
| TIE1 |
| TLR5 |
| TNFRSF1B |
| TNNI1 |
| TNNT2 |
| TNR |
| TOP1P1 |
| TP53BP2 |
| TP73 |
| TRAF5 |
| CCT3 |
| TSHB |
| TTC4 |
| TUFT1 |
| TNFSF4 |
| TNFRSF4 |
| UCK2 |
| UQCRH |
| UROD |
| USF1 |
| USP1 |
| USH2A |
| VCAM1 |
| WNT2B |
| ZSCAN20 |
| ZBTB17 |
| SLC30A1 |
| SLC30A2 |
| LUZP1 |
| PRDM2 |
| DNALI1 |
| LRP8 |
| LAPTM5 |
| BSND |
| CSDE1 |
| EVI5 |
| DAP3 |
| NPHS2 |
| BTG2 |
| PTP4A2 |
| CDC7 |
| HIST2H2AA3 |
| HIST2H2AC |
| HIST2H2BE |
| HIST2H4A |
| PIP5K1A |
| TAGLN2 |
| BCAR3 |
| ANXA9 |
| SNHG3 |
| NR0B2 |
| RAD54L |
| DYRK3 |
| TTF2 |
| RGS5 |
| PPFIA4 |
| PIK3R3 |
| MMP23B |
| MMP23A |
| KCNAB2 |
| ITGA10 |
| LMO4 |
| FCN3 |
| BLZF1 |
| CDC14A |
| YARS |
| MKNK1 |
| AKR7A2 |
| PPAP2B |
| RTCD1 |
| PTCH2 |
| ALDH4A1 |
| EIF3I |
| EIF4G3 |
| VAMP4 |
| PEA15 |
| B4GALT3 |
| B4GALT2 |
| B3GALT2 |
| TNFRSF25 |
| ADAM15 |
| PABPC4 |
| TNFRSF18 |
| FPGT |
| PEX11B |
| CREG1 |
| CD84 |
| PER3 |
| FUBP1 |
| EIF2B3 |
| RAB7L1 |
| SELENBP1 |
| TNFSF18 |
| TAF1A |
| MPZL1 |
| SH2D2A |
| ARTN |
| MAP3K6 |
| ANGPTL1 |
| DIRAS3 |
| TBX19 |
| FCGR2C |
| SLC16A4 |
| PRPF3 |
| KCNQ4 |
| ARHGEF2 |
| DEDD |
| ZMYM4 |
| ZMYM6 |
| XPR1 |
| FAIM3 |
| DHRS3 |
| MAPKAPK2 |
| GPR37L1 |
| GPR52 |
| SFRS11 |
| VAMP3 |
| ZFYVE9 |
| CD101 |
| SEP15 |
| ZRANB2 |
| SNRNP40 |
| ARHGAP29 |
| AIM2 |
| RASAL2 |
| C1orf38 |
| ADAMTS4 |
| TMEM59 |
| SEC22B |
| CHD1L |
| H6PD |
| SOX13 |
| PRDX6 |
| CLCA3P |
| CLCA2 |
| ISG15 |
| IKBKE |
| PLCH2 |
| HS2ST1 |
| IPO13 |
| SDC3 |
| SLC25A44 |
| KIAA0040 |
| KDM4A |
| CROCC |
| PUM1 |
| NOS1AP |
| KIAA0562 |
| RIMS3 |
| KIAA0494 |
| ARHGEF11 |
| DNAJC6 |
| CEP350 |
| LRIG2 |
| SETDB1 |
| ZC3H11A |
| SMG7 |
| LPPR4 |
| UBAP2L |
| SV2A |
| KLHL21 |
| SLC35E2 |
| DENND4B |
| RABGAP1L |
| TMCC2 |
| FAM20B |
| ZBTB40 |
| LPGAT1 |
| MFN2 |
| KIF14 |
| RBM8A |
| NR1I3 |
| INSL5 |
| PIGK |
| SCAMP3 |
| PTPRU |
| ARPC5 |
| TSPAN2 |
| TSPAN1 |
| CELA3A |
| PDZK1IP1 |
| WASF2 |
| INADL |
| PRG4 |
| ANGPTL7 |
| GPA33 |
| STX6 |
| HNRNPR |
| SRRM1 |
| CNKSR1 |
| SF3B4 |
| ZMPSTE24 |
| UBE4B |
| BCAS2 |
| AKR1A1 |
| WARS2 |
| BPNT1 |
| CEPT1 |
| PIAS3 |
| TESK2 |
| TIMM17A |
| LRRN2 |
| PPIE |
| VAV3 |
| HAX1 |
| MAD2L2 |
| PPIH |
| C1orf61 |
| CAP1 |
| LRRC41 |
| SEMA6C |
| HBXIP |
| SLC19A2 |
| IFI44 |
| POLR3C |
| IVNS1ABP |
| TXNIP |
| PDPN |
| PMVK |
| KHDRBS1 |
| GMEB1 |
| FAM189B |
| AP4B1 |
| NUDC |
| PHTF1 |
| MASP2 |
| NES |
| KDM5B |
| AHCYL1 |
| SFRS13A |
| CFHR4 |
| CFHR3 |
| WDR3 |
| OCLM |
| JTB |
| MTMR11 |
| MAN1A2 |
| UTS2 |
| SF3A3 |
| MLLT11 |
| IFI44L |
| EBNA1BP2 |
| SLC27A3 |
| KIF2C |
| IL24 |
| TDRKH |
| RER1 |
| DNAJB4 |
| ADAM30 |
| RCAN3 |
| FAF1 |
| CD160 |
| GLMN |
| HHLA3 |
| TNRC4 |
| MSTP2 |
| DDX20 |
| DUSP10 |
| MSTP9 |
| PADI2 |
| PMF1 |
| DUSP12 |
| VPS45 |
| LYPLA2 |
| PARK7 |
| CTRC |
| ACOT7 |
| CLCA4 |
| MTF2 |
| DNAJC8 |
| NTNG1 |
| PLEKHA6 |
| CLSTN1 |
| FOXJ3 |
| KIAA0907 |
| WDR47 |
| KIFAP3 |
| ATF6 |
| SCMH1 |
| AKR7A3 |
| TTC39A |
| SPEN |
| KDM1A |
| USP33 |
| WDTC1 |
| KIF21B |
| NMNAT2 |
| KIAA0090 |
| KIF1B |
| NFASC |
| POGZ |
| GLT25D2 |
| MAST2 |
| NCDN |
| CLCC1 |
| SLC35D1 |
| RGL1 |
| PLEKHM2 |
| SYT11 |
| BAT2D1 |
| FBXO28 |
| RPRD2 |
| OTUD3 |
| KIAA1026 |
| CAMTA1 |
| LPHN2 |
| CAMSAP1L1 |
| KIAA1107 |
| ZCCHC11 |
| KIAA0467 |
| DNAJC16 |
| UBR4 |
| USP24 |
| SRGAP2 |
| SMG5 |
| NCSTN |
| ATP13A2 |
| CRB1 |
| ITGB3BP |
| GPR161 |
| TARDBP |
| CELA3B |
| SLC35A3 |
| ICMT |
| MACF1 |
| LRRC8B |
| ZNF281 |
| SNAPIN |
| LPAR3 |
| PADI4 |
| DDAH1 |
| TMEM50A |
| PHLDA3 |
| RUSC1 |
| CA14 |
| KPNA6 |
| SSBP3 |
| STX12 |
| DSTYK |
| RAB3GAP2 |
| CCDC19 |
| LMOD1 |
| NBPF14 |
| BRP44 |
| INTS7 |
| OLFML2B |
| C1orf43 |
| CLIC4 |
| NSL1 |
| SYF2 |
| RWDD3 |
| PARS2 |
| MMACHC |
| ZZZ3 |
| ACOT11 |
| CHD5 |
| DNM3 |
| TOR1AIP1 |
| C1orf77 |
| C1orf144 |
| LDLRAP1 |
| SERBP1 |
| NOC2L |
| PTPN22 |
| PHGDH |
| FBXO2 |
| LCE2B |
| OPTC |
| FBXO6 |
| PLA2G2D |
| AK5 |
| MYCBP |
| OR10J1 |
| HEYL |
| EIF2C1 |
| OR4F3 |
| RPS6KC1 |
| SNORD81 |
| SNORD79 |
| SNORD80 |
| SNORA66 |
| SNORD47 |
| SNORD45B |
| SNORD45A |
| SNORD44 |
| SNORD55 |
| RNU11 |
| RNF11 |
| USP21 |
| FOXD3 |
| C1orf107 |
| TRAPPC3 |
| CACYBP |
| HSPB7 |
| CHIA |
| SLC39A1 |
| ARHGEF16 |
| AHDC1 |
| RNF115 |
| KLHL20 |
| TEKT2 |
| SMPDL3B |
| ANGPTL3 |
| ROBLD3 |
| FLVCR1 |
| PRO0611 |
| UBE2T |
| HSPC157 |
| SSU72 |
| TMOD4 |
| GNL2 |
| GPSM2 |
| UBIAD1 |
| NME7 |
| ALG6 |
| NENF |
| PADI1 |
| IL19 |
| LASS2 |
| SLC25A24 |
| PLA2G2E |
| DNTTIP2 |
| WDR8 |
| CRNN |
| G0S2 |
| IL20 |
| SLC45A1 |
| DCAF8 |
| HP1BP3 |
| F11R |
| TMED5 |
| RRP15 |
| GLRX2 |
| BOLA1 |
| CELA2B |
| ZNF593 |
| ZBTB7B |
| ZNF691 |
| TXNDC12 |
| TNNI3K |
| C1orf66 |
| ADIPOR1 |
| SH3GLB1 |
| MECR |
| APH1A |
| UTP11L |
| KCTD3 |
| SDF4 |
| MRTO4 |
| PLEKHO1 |
| HAO2 |
| ACP6 |
| TMEM69 |
| MRPL37 |
| IER5 |
| HOOK1 |
| SNX7 |
| UCHL5 |
| C1orf9 |
| HPCAL4 |
| YTHDF2 |
| GPR89B |
| HSD17B7 |
| UFC1 |
| DTL |
| ZCCHC17 |
| METTL13 |
| DPH5 |
| HSPB11 |
| OAZ3 |
| PADI3 |
| CYB5R1 |
| CMPK1 |
| CD244 |
| GPR88 |
| ERRFI1 |
| DPM3 |
| WNT4 |
| YIPF1 |
| FBXO42 |
| MRPS21 |
| CCDC76 |
| TMCO1 |
| ADAMTSL4 |
| CRCT1 |
| RNF186 |
| SPATA6 |
| MXRA8 |
| L1TD1 |
| HES2 |
| RSBN1 |
| ZNHIT6 |
| GPN2 |
| LEPROT |
| FBLIM1 |
| MED18 |
| TRIT1 |
| GIPC2 |
| C1orf26 |
| GDAP2 |
| FAM46C |
| GON4L |
| GPATCH4 |
| PALMD |
| FNBP1L |
| ST7L |
| PQLC2 |
| CASZ1 |
| LAX1 |
| RHBDL2 |
| DUSP23 |
| ADPRHL2 |
| TRNAU1AP |
| C1orf27 |
| C1orf109 |
| C1orf56 |
| CPSF3L |
| C1orf123 |
| C1orf159 |
| MOSC2 |
| AURKAIP1 |
| TTC22 |
| MRPL20 |
| AIM1L |
| SUSD4 |
| TMEM51 |
| RALGPS2 |
| GPATCH2 |
| BSDC1 |
| XKR8 |
| TMEM39B |
| PRPF38B |
| CDCA8 |
| LRRC8D |
| MSTO1 |
| DARS2 |
| ARHGEF10L |
| PRMT6 |
| RNF220 |
| VPS13D |
| FAM176B |
| GOLPH3L |
| ATAD3A |
| TMEM57 |
| KLHDC8A |
| TRIM62 |
| ETNK2 |
| RAVER2 |
| PANK4 |
| KIRREL |
| TMEM206 |
| YY1AP1 |
| ECHDC2 |
| FGGY |
| MCOLN3 |
| SLC22A15 |
| YOD1 |
| CAMK2N1 |
| BATF3 |
| SLC30A10 |
| UBE2Q1 |
| RNPC3 |
| ITLN1 |
| ASAP3 |
| POMGNT1 |
| PNRC2 |
| LRRC40 |
| DEPDC1 |
| PIGV |
| NBPF1 |
| IARS2 |
| MAP7D1 |
| IPO9 |
| TMEM48 |
| NECAP2 |
| IQCC |
| C1orf112 |
| HHAT |
| DNAJC11 |
| RCOR3 |
| C1orf106 |
| C1orf103 |
| FAM63A |
| ADCY10 |
| DCAF6 |
| ASH1L |
| LENEP |
| CTTNBP2NL |
| RCC2 |
| C1orf183 |
| DMAP1 |
| AJAP1 |
| GNG12 |
| RAG1AP1 |
| C1orf91 |
| FAM54B |
| SERTAD4 |
| CCBL2 |
| SLAMF8 |
| CDC42SE1 |
| UBQLN4 |
| TMEM167B |
| OLFML3 |
| SMYD2 |
| LHX9 |
| OTUD7B |
| CTNNBIP1 |
| C1orf63 |
| AGTRAP |
| C1orf128 |
| RAB25 |
| PGLYRP4 |
| RHBG |
| MAN1C1 |
| SCYL3 |
| CAMK1G |
| NIPAL3 |
| SEPN1 |
| ATP8B2 |
| KIAA0495 |
| VANGL2 |
| PBXIP1 |
| S100A14 |
| PLEKHG5 |
| GATAD2B |
| AMIGO1 |
| LRRC47 |
| ODF2L |
| CGN |
| KIAA1324 |
| PTCHD2 |
| IGSF9 |
| LRRC7 |
| KIF17 |
| ZNF687 |
| ZSWIM5 |
| POGK |
| KIAA1522 |
| HCN3 |
| CACHD1 |
| MIER1 |
| KIAA1614 |
| FAM5B |
| HES4 |
| C1orf114 |
| GRHL3 |
| SLAMF7 |
| CADM3 |
| PTBP2 |
| PRUNE |
| DNASE2B |
| DLGAP3 |
| IL22RA1 |
| HIVEP3 |
| KLHL12 |
| LGR6 |
| GPBP1L1 |
| HAPLN2 |
| MIIP |
| GAS5 |
| PAPPA2 |
| CELA2A |
| BCAN |
| GPATCH3 |
| TNN |
| MRPS14 |
| DMRTB1 |
| DMRTA2 |
| CLSPN |
| OXCT2 |
| RRAGC |
| ELTD1 |
| TINAGL1 |
| SPATA1 |
| LEPRE1 |
| SEMA4A |
| TOR3A |
| RFWD2 |
| RGS18 |
| PLA2G2F |
| HIAT1 |
| NUCKS1 |
| SMAP2 |
| ATPAF1 |
| MOSC1 |
| S100PBP |
| MEAF6 |
| DEM1 |
| CCDC21 |
| NMNAT1 |
| ELOVL1 |
| AIDA |
| VWA1 |
| DCLRE1B |
| MRPS15 |
| MRPL9 |
| PINK1 |
| MARCKSL1 |
| PRAMEF1 |
| PRAMEF2 |
| INTS3 |
| NADK |
| ZNF643 |
| C1orf163 |
| PHACTR4 |
| C1orf135 |
| SCNM1 |
| ERI3 |
| C1orf50 |
| WDR77 |
| C1orf116 |
| MGC4473 |
| CCDC28B |
| EFHD2 |
| MMEL1 |
| C1orf89 |
| FCRL2 |
| DLEU2L |
| OR4F5 |
| NKAIN1 |
| EPS8L3 |
| CDC73 |
| MRPL24 |
| MUL1 |
| TNFAIP8L2 |
| C1orf54 |
| TMEM53 |
| AKIRIN1 |
| HECTD3 |
| BEND5 |
| VTCN1 |
| YRDC |
| ZYG11B |
| NOL9 |
| PPCS |
| LIN28 |
| C1orf113 |
| TTLL7 |
| SNIP1 |
| C1orf115 |
| HHIPL2 |
| VASH2 |
| AGMAT |
| WDR78 |
| ZMYM1 |
| NCRNA00115 |
| RPAP2 |
| MORN1 |
| GRRP1 |
| KIAA0319L |
| DHDDS |
| PAQR6 |
| DENND2D |
| GPR177 |
| GPR157 |
| TRIM46 |
| C1orf129 |
| RPF1 |
| SIKE1 |
| ZC3H12A |
| SPSB1 |
| TARS2 |
| TRIM45 |
| EDEM3 |
| FLAD1 |
| TRAF3IP3 |
| GLTPD1 |
| ZNF436 |
| TAS1R2 |
| TAS1R1 |
| NPL |
| GJA9 |
| OR4F16 |
| OR6N2 |
| OR6K2 |
| SYNC |
| CFHR5 |
| C1orf21 |
| ACTL8 |
| ANKRD13C |
| PVRL4 |
| SNX27 |
| ANP32E |
| C1orf14 |
| C1orf25 |
| TSSK3 |
| CCNL2 |
| NUAK2 |
| VANGL1 |
| ST6GALNAC5 |
| ISG20L2 |
| HYI |
| FCRL5 |
| FCRL4 |
| SH3BGRL3 |
| DDX59 |
| NUF2 |
| RASSF5 |
| SESN2 |
| ESPN |
| TAS1R3 |
| ATAD3B |
| HMCN1 |
| GPR61 |
| STK40 |
| TM2D1 |
| FCAMR |
| REG4 |
| TMEM222 |
| C1orf49 |
| PLEKHN1 |
| HORMAD1 |
| TOMM40L |
| SYDE2 |
| ZNF644 |
| USP48 |
| ZMYND12 |
| NBPF3 |
| LRRC8C |
| ZDHHC18 |
| SGIP1 |
| POLR3GL |
| SLC25A33 |
| TMEM79 |
| DDI2 |
| ACBD6 |
| LZIC |
| PROK1 |
| EFCAB7 |
| ZBTB37 |
| NT5C1A |
| LCE3D |
| TRIM63 |
| PSRC1 |
| FAM167B |
| C1orf97 |
| MGC12982 |
| C1orf170 |
| CROCCL1 |
| FCRLA |
| ANKRD36BL1 |
| HPDL |
| C1orf203 |
| AGBL4 |
| MFSD2A |
| PPP1R15B |
| ATG4C |
| MAEL |
| PRPF38A |
| SYTL1 |
| IGSF21 |
| LSM10 |
| C1orf94 |
| DISP1 |
| SNHG12 |
| FAM40A |
| RGS8 |
| DOCK7 |
| KIAA1751 |
| NAV1 |
| SEC16B |
| AQP10 |
| LHX4 |
| SLAMF9 |
| KIAA2013 |
| THAP3 |
| C1orf201 |
| PYGO2 |
| ANGEL2 |
| SPOCD1 |
| ZNF697 |
| IGFN1 |
| NUP210L |
| BTF3L4 |
| UBXN11 |
| NEXN |
| CENPL |
| DUSP27 |
| RCSD1 |
| MEX3A |
| C1orf156 |
| GORAB |
| C1orf105 |
| TMEM183A |
| PIGM |
| IGSF8 |
| C1orf158 |
| LEMD1 |
| FBXO44 |
| ATPIF1 |
| SNORD46 |
| SNORD38A |
| SNORD38B |
| C1orf85 |
| MED8 |
| KTI12 |
| C1orf212 |
| ADC |
| TMEM54 |
| C1orf59 |
| TOE1 |
| ERMAP |
| PGLYRP3 |
| CSMD2 |
| MYSM1 |
| GNRHR2 |
| CROCCL2 |
| FHAD1 |
| SLAMF6 |
| OSBPL9 |
| SLC26A9 |
| LOC115110 |
| OMA1 |
| RAB42 |
| FCRL1 |
| FCRL3 |
| LRRC42 |
| GBP4 |
| GBP5 |
| FAM46B |
| FMO9P |
| RBP7 |
| TSEN15 |
| FAM129A |
| ACAP3 |
| TADA1 |
| THEM4 |
| SH2D1B |
| SSX2IP |
| UBE2J2 |
| OLFM3 |
| GABPB2 |
| TCHHL1 |
| RPTN |
| C1orf231 |
| TDRD10 |
| SHE |
| C1orf172 |
| AADACL3 |
| PUSL1 |
| B3GALT6 |
| WDR63 |
| KLHDC9 |
| C1orf125 |
| C1orf161 |
| IFFO2 |
| HIST2H3C |
| SLC44A3 |
| ATXN7L2 |
| C1orf194 |
| LYPLAL1 |
| ATP6V1G3 |
| ASB17 |
| TYW3 |
| C1orf173 |
| LRRIQ3 |
| TPRG1L |
| C1orf93 |
| MYOM3 |
| DMBX1 |
| OR10J5 |
| TMCO2 |
| ZNF684 |
| C1orf83 |
| PODN |
| LRRC39 |
| GJB4 |
| HMGB4 |
| RNF19B |
| DCST2 |
| ZNF648 |
| TEDDM1 |
| C1orf122 |
| OSCP1 |
| C1orf216 |
| KLHDC7A |
| VWA5B1 |
| UBXN10 |
| C1orf87 |
| ARL8A |
| SYT2 |
| LOC127841 |
| GOLT1A |
| UHMK1 |
| FCRLB |
| LIX1L |
| SPATA17 |
| KLF17 |
| TMEM125 |
| C1orf182 |
| IQGAP3 |
| APOA1BP |
| ARHGEF19 |
| DRAM2 |
| C1orf88 |
| C1orf162 |
| OR10T2 |
| OR6P1 |
| OR10X1 |
| OR10Z1 |
| OR6K6 |
| OR6N1 |
| TATDN3 |
| S100A16 |
| NEK7 |
| ACTRT2 |
| MIB2 |
| ITLN2 |
| SYT6 |
| C1orf74 |
| CREB3L4 |
| C1orf127 |
| C1orf58 |
| SAMD11 |
| LOC148413 |
| SAMD13 |
| C1orf52 |
| PHF13 |
| C1orf51 |
| TMEM56 |
| NBPF4 |
| UBE2U |
| LOC148696 |
| LOC148709 |
| PTPRV |
| HFE2 |
| ANKRD35 |
| FAM163A |
| MFSD4 |
| PM20D1 |
| SLC30A7 |
| CCDC27 |
| C1orf213 |
| KNCN |
| MOBKL2C |
| GLIS1 |
| LELP1 |
| RC3H1 |
| MGC27382 |
| DCDC2B |
| ZNF362 |
| DCST1 |
| MANEAL |
| IL23R |
| METTL11B |
| FAM78B |
| SHISA4 |
| PDIK1L |
| BNIPL |
| CLDN19 |
| WDR65 |
| C1orf210 |
| C1orf84 |
| CCDC24 |
| BTBD19 |
| CCDC17 |
| C1orf92 |
| C1orf64 |
| LOC149620 |
| PYHIN1 |
| C1orf227 |
| FAM71A |
| SLC2A7 |
| DENND2C |
| GBP6 |
| LPPR5 |
| FNDC7 |
| DENND1B |
| TDRD5 |
| TOR1AIP2 |
| CALML6 |
| IL28RA |
| CYP4Z2P |
| CITED4 |
| FLJ40434 |
| C1orf177 |
| SPRR4 |
| KANK4 |
| SASS6 |
| FAM43B |
| PPIAL4A |
| HFM1 |
| PAQR7 |
| TTC24 |
| C1orf65 |
| UBL4B |
| PDIA3P |
| EIF2C3 |
| EIF2C4 |
| LCE4A |
| ALG14 |
| FAM76A |
| C1orf168 |
| TMEM201 |
| TMEM61 |
| CYP4Z1 |
| C1orf86 |
| CDCP2 |
| SLC5A9 |
| CC2D1B |
| LOC200030 |
| NUDT17 |
| FLJ23867 |
| TXLNA |
| TCTEX1D1 |
| SPAG17 |
| SLFNL1 |
| KRTCAP2 |
| CRTC2 |
| C1orf126 |
| HIPK1 |
| SLC44A5 |
| ATAD3C |
| AKR7L |
| AFARP1 |
| TMEM9 |
| FNDC5 |
| EPHX4 |
| TTLL10 |
| AKNAD1 |
| SLC41A1 |
| LCE5A |
| TMCO4 |
| MCOLN2 |
| COL24A1 |
| PCSK9 |
| SYT14 |
| ST6GALNAC3 |
| ZNF683 |
| ARHGAP30 |
| C1orf192 |
| NEGR1 |
| ASPM |
| CYP4X1 |
| MAGI3 |
| TIPRL |
| NPHP4 |
| BEST4 |
| FAM19A3 |
| C1orf230 |
| THEM5 |
| SLC9A11 |
| CYP4A22 |
| C1orf185 |
| LOC284551 |
| NBPF15 |
| C1orf157 |
| LOC284578 |
| FAM41C |
| FAM102B |
| SYPL2 |
| CYB561D1 |
| ANKRD34A |
| C1orf104 |
| LOC284632 |
| RSPO1 |
| EPHA10 |
| LOC284661 |
| C1orf204 |
| C1orf111 |
| LOC284688 |
| ZNF326 |
| BTBD8 |
| RIMKLA |
| SLC25A34 |
| ESPNP |
| HIST2H2AB |
| PPM1J |
| HIST2H3A |
| HIST2H2BA |
| FAM151A |
| S100A7A |
| LINGO4 |
| RXFP4 |
| ANKRD45 |
| C1orf174 |
| KLHL17 |
| C1orf70 |
| TMEM52 |
| FAM5C |
| MTMR9L |
| ZBTB8OS |
| TFAP2E |
| C1orf110 |
| FAM58B |
| LOC339524 |
| C1orf228 |
| ZNF642 |
| RD3 |
| AADACL4 |
| PRAMEF5 |
| HNRNPCL1 |
| PRAMEF9 |
| PRAMEF10 |
| CCDC18 |
| MYBPHL |
| OR10R2 |
| FCRL6 |
| KCNT2 |
| BARHL2 |
| HSP90B3P |
| NBPF7 |
| TCTEX1D4 |
| SERINC2 |
| FAM159A |
| FAM131C |
| LCE1A |
| LCE1B |
| LCE1C |
| LCE1D |
| LCE1E |
| LCE1F |
| LCE2A |
| LCE2C |
| LCE2D |
| LCE3A |
| LCE3B |
| LCE3C |
| LCE3E |
| PADI6 |
| RGSL1 |
| BMP8A |
| ZBTB41 |
| CYCSP52 |
| C1orf187 |
| SPATA21 |
| CCDC23 |
| C1orf223 |
| C1orf175 |
| FAM73A |
| PEAR1 |
| SFT2D2 |
| MIA3 |
| AGRN |
| RPS10P7 |
| APITD1 |
| CATSPER4 |
| NSUN4 |
| GPR153 |
| ILDR2 |
| FAM132A |
| HES5 |
| LOC388588 |
| RNF207 |
| TMEM82 |
| TRNP1 |
| CD164L2 |
| LDLRAD1 |
| GBP7 |
| C1orf146 |
| FAM69A |
| SLC6A17 |
| NOTCH2NL |
| FLJ39739 |
| LOC388692 |
| LYSMD1 |
| HRNR |
| FLG2 |
| C1orf189 |
| FMO6P |
| C1orf53 |
| TMEM81 |
| CAPN8 |
| HES3 |
| PRAMEF12 |
| PRAMEF21 |
| PRAMEF8 |
| PRAMEF18 |
| PRAMEF17 |
| PLA2G2C |
| SKINTL |
| UOX |
| FRRS1 |
| VHLL |
| OR10K2 |
| OR10K1 |
| OR6Y1 |
| OR6K3 |
| VSIG8 |
| TMEM200B |
| PRAMEF4 |
| PRAMEF13 |
| SH2D5 |
| C1orf130 |
| LOC400752 |
| C1orf141 |
| LOC400759 |
| C1orf226 |
| LOC400794 |
| C1orf220 |
| LOC400804 |
| NBPF20 |
| FAM177B |
| PRAMEF3 |
| LDLRAD2 |
| APOBEC4 |
| MIR101-1 |
| MIR137 |
| MIR181B1 |
| MIR186 |
| MIR194-1 |
| MIR197 |
| MIR199A2 |
| MIR200A |
| MIR200B |
| MIR205 |
| MIR181A1 |
| MIR214 |
| MIR215 |
| MIR29B2 |
| MIR29C |
| MIR30C1 |
| MIR30E |
| MIR34A |
| MIR9-1 |
| RGS21 |
| LHX8 |
| C1orf180 |
| FLJ42875 |
| PRAMEF11 |
| PRAMEF6 |
| LOC440563 |
| UQCRHL |
| C1orf151 |
| FAM183A |
| ZYG11A |
| BCL2L15 |
| HIST2H2BF |
| ETV3L |
| LRRC52 |
| C1orf186 |
| LOC441869 |
| PRAMEF7 |
| LOC441897 |
| OR10J3 |
| MIR135B |
| KPRP |
| LCE6A |
| SUMO1P3 |
| LOC492303 |
| RBMXL1 |
| C1orf190 |
| PEF1 |
| MIR429 |
| HIST2H4B |
| C1orf133 |
| MIR488 |
| CYB5RL |
| SNORD74 |
| RPL31P11 |
| FAM138F |
| LOC642502 |
| LOC642587 |
| LQK1 |
| CYMP |
| KIAA0754 |
| LOC643837 |
| TMEM88B |
| PPIAL4G |
| C1orf200 |
| PPIAL4D |
| LOC645166 |
| PRAMEF19 |
| PRAMEF20 |
| FAM138A |
| LOC645676 |
| S100A7L2 |
| LOC646471 |
| LOC647121 |
| LOC648740 |
| LOC649330 |
| ZBTB8A |
| NBPF6 |
| PPIAL4B |
| GPR89A |
| LOC653566 |
| PPIAL4C |
| HIST2H3D |
| PRAMEF22 |
| PRAMEF15 |
| WASH5P |
| TMEM183B |
| FAM72B |
| PRAMEF16 |
| PCP4L1 |
| FAM138C |
| SCARNA3 |
| SCARNA2 |
| SCARNA4 |
| SCARNA1 |
| SNORA36B |
| SNORA42 |
| SNORA44 |
| SNORA55 |
| SNORA61 |
| SNORA77 |
| SNORA59B |
| SNORA59A |
| SNORA16A |
| SNORD45C |
| SNORA16B |
| SNORD75 |
| SNORD76 |
| SNORD77 |
| SNORD78 |
| SNORD85 |
| SNORD99 |
| SNORD103A |
| MIR551A |
| MIR553 |
| MIR554 |
| MIR555 |
| MIR556 |
| MIR557 |
| MIR92B |
| HIST2H2AA4 |
| ZBTB8B |
| LOC728448 |
| CCDC30 |
| CDC2L2 |
| LOC728661 |
| LOC728855 |
| LOC728875 |
| NBPF11 |
| GPR89C |
| NBPF16 |
| PDZK1P1 |
| PPIAL4F |
| RPS15AP10 |
| LOC728989 |
| LOC729467 |
| PRAMEF14 |
| FAM72A |
| FLJ37453 |
| OR4F29 |
| PPIAL4E |
| SNHG3-RCC1 |
| C1orf152 |
| MIR765 |
| MIR942 |
| MIR190B |
| MIR760 |
| MIR921 |
| LOC100128003 |
| LOC100128071 |
| LOC100128842 |
| C1orf68 |
| MSTO2P |
| LOC100129534 |
| FLJ39609 |
| LOC100130557 |
| TSTD1 |
| LOC100132062 |
| LOC100132111 |
| LOC100132287 |
| NBPF10 |
| FCGR1C |
| LOC100133331 |
| LOC100133612 |
| C2CD4D |
| LOC100286793 |
| LOC100288778 |
| LOC100302401 |
This is the comprehensive list of deleted genes in the wide peak for 4q35.1.
Table S20. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| DUX4 |
| hsa-mir-1305 |
| AGA |
| SLC25A4 |
| CASP3 |
| DCTD |
| F11 |
| ACSL1 |
| FAT1 |
| FRG1 |
| GPM6A |
| HPGD |
| ING2 |
| IRF2 |
| KLKB1 |
| MTNR1A |
| TLR3 |
| VEGFC |
| GLRA3 |
| SORBS2 |
| HAND2 |
| MORF4 |
| ADAM29 |
| FAM149A |
| FBXO8 |
| PDLIM3 |
| CLDN22 |
| NEIL3 |
| UFSP2 |
| CDKN2AIP |
| ODZ3 |
| LRP2BP |
| TUBB4Q |
| STOX2 |
| KIAA1430 |
| SPCS3 |
| C4orf41 |
| MLF1IP |
| NBLA00301 |
| WWC2 |
| KIAA1712 |
| SNX25 |
| MGC45800 |
| WDR17 |
| ZFP42 |
| SPATA4 |
| ENPP6 |
| ASB5 |
| C4orf38 |
| RWDD4A |
| CCDC111 |
| TRIML2 |
| CCDC110 |
| CYP4V2 |
| LOC285501 |
| TRIML1 |
| ANKRD37 |
| HELT |
| FAM92A3 |
| C4orf47 |
| FRG2 |
| SLED1 |
| LOC653543 |
| LOC653544 |
| LOC653545 |
| LOC653548 |
| LOC728410 |
This is the comprehensive list of deleted genes in the wide peak for 7q31.33.
Table S21. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BRAF |
| EZH2 |
| MET |
| SMO |
| KIAA1549 |
| MLL3 |
| CREB3L2 |
| hsa-mir-671 |
| hsa-mir-1975 |
| hsa-mir-548f-4 |
| hsa-mir-490 |
| hsa-mir-29b-1 |
| hsa-mir-335 |
| hsa-mir-183 |
| hsa-mir-129-1 |
| hsa-mir-593 |
| hsa-mir-592 |
| ABP1 |
| AKR1B1 |
| ARF5 |
| OPN1SW |
| BPGM |
| CALD1 |
| CALU |
| CAPZA2 |
| CASP2 |
| CAV1 |
| CAV2 |
| CDK5 |
| CFTR |
| CHRM2 |
| CLCN1 |
| CPA1 |
| CPA2 |
| DPP6 |
| EPHA1 |
| EPHB6 |
| FLNC |
| GBX1 |
| GPR37 |
| GRM8 |
| IFRD1 |
| IMPDH1 |
| IRF5 |
| KCND2 |
| KCNH2 |
| KEL |
| LEP |
| MEST |
| MKLN1 |
| NDUFA5 |
| NDUFB2 |
| NOS3 |
| CNOT4 |
| NRF1 |
| PAX4 |
| PIP |
| PODXL |
| PPP1R3A |
| PRSS1 |
| PRSS2 |
| TAS2R38 |
| PTN |
| PTPRZ1 |
| RARRES2 |
| RHEB |
| SLC4A2 |
| SLC13A1 |
| SMARCD3 |
| SPAM1 |
| AKR1D1 |
| SSBP1 |
| TBXAS1 |
| UBE2H |
| WNT2 |
| XRCC2 |
| ZYX |
| ST7 |
| ARHGEF5 |
| ZNF212 |
| ZNF282 |
| CUL1 |
| TRIM24 |
| MGAM |
| WASL |
| DGKI |
| ATP6V1F |
| ACCN3 |
| PDIA4 |
| FAM131B |
| DOCK4 |
| FAM115A |
| ABCF2 |
| AASS |
| FAM3C |
| FASTK |
| ZNF277 |
| ABCB8 |
| TFEC |
| KLHDC10 |
| SSPO |
| NUP205 |
| AHCYL2 |
| TNPO3 |
| HYAL4 |
| TSPAN12 |
| CLEC5A |
| MKRN1 |
| POT1 |
| CNTNAP2 |
| HYALP1 |
| TES |
| GIMAP2 |
| OR2F1 |
| SLC13A4 |
| COPG2 |
| TPK1 |
| SND1 |
| C7orf54 |
| DENND2A |
| ZNF777 |
| TMEM176B |
| HIPK2 |
| WDR91 |
| REPIN1 |
| C7orf68 |
| MDFIC |
| FSCN3 |
| ATP6V0A4 |
| TAS2R3 |
| TAS2R4 |
| TAS2R16 |
| CPA4 |
| WNT16 |
| PRKAG2 |
| ZC3HC1 |
| LUC7L2 |
| MRPS33 |
| NUB1 |
| LSM8 |
| GPR85 |
| TAS2R5 |
| CHPF2 |
| ING3 |
| LRRN3 |
| CHCHD3 |
| RBM28 |
| TMEM140 |
| GIMAP4 |
| GIMAP5 |
| TMEM176A |
| TRPV6 |
| AGK |
| METTL2B |
| TRPV5 |
| ANKRD7 |
| ZC3HAV1 |
| AKR1B10 |
| ACTR3B |
| KIAA1147 |
| FAM40B |
| ZNF398 |
| EXOC4 |
| GALNT11 |
| LRRC4 |
| TMEM168 |
| CCDC136 |
| PARP12 |
| LRRC61 |
| C7orf49 |
| GCC1 |
| ZNF767 |
| C7orf58 |
| TTC26 |
| JHDM1D |
| TMUB1 |
| IMMP2L |
| CTTNBP2 |
| SLC37A3 |
| KRBA1 |
| FAM71F1 |
| SLC35B4 |
| TMEM209 |
| OR6W1P |
| ADCK2 |
| PLXNA4 |
| ZC3HAV1L |
| LOC93432 |
| ST7OT1 |
| ST7OT2 |
| ST7OT3 |
| CADPS2 |
| CPA5 |
| FOXP2 |
| TSGA14 |
| C7orf29 |
| TSGA13 |
| AGAP3 |
| OR9A4 |
| OR9A2 |
| C7orf34 |
| TMEM139 |
| NOBOX |
| OR2A14 |
| OR6B1 |
| OR2F2 |
| ZNF786 |
| PRSS37 |
| KLF14 |
| C7orf45 |
| SVOPL |
| MTPN |
| LRGUK |
| ASB10 |
| TRYX3 |
| MOXD2 |
| ASZ1 |
| ASB15 |
| C7orf60 |
| TRY6 |
| LOC154761 |
| CLEC2L |
| C7orf55 |
| IQUB |
| TMEM213 |
| GIMAP8 |
| CRYGN |
| ZNF425 |
| ZNF783 |
| ZNF746 |
| ATP6V0E2 |
| GALNTL5 |
| RNF133 |
| GIMAP7 |
| ZNF467 |
| ZNF800 |
| GIMAP1 |
| C7orf33 |
| FABP5L3 |
| UBN2 |
| TAS2R39 |
| TAS2R40 |
| TAS2R41 |
| FLJ40852 |
| FAM115C |
| ZNF775 |
| ATG9B |
| C7orf53 |
| LOC286016 |
| MESTIT1 |
| ST7OT4 |
| TAS2R60 |
| CTAGE6 |
| TSPAN33 |
| AGBL3 |
| OR6V1 |
| OR2A12 |
| OR2A1 |
| FAM71F2 |
| STRA8 |
| KLRG2 |
| WDR86 |
| GSTK1 |
| KCP |
| FLJ43663 |
| RNF148 |
| FEZF1 |
| FAM180A |
| OR2A25 |
| OR2A5 |
| LOC401397 |
| PRRT4 |
| RAB19 |
| OR2A7 |
| OR2A20P |
| LOC401431 |
| OR2A42 |
| FLJ45340 |
| MIR129-1 |
| MIR182 |
| MIR183 |
| MIR29A |
| MIR29B1 |
| MIR96 |
| LOC407835 |
| AKR1B15 |
| LOC441294 |
| OR2A9P |
| OR2A2 |
| EIF3IP1 |
| LMOD2 |
| MIR335 |
| ARHGEF5L |
| GIMAP6 |
| WEE2 |
| MIR490 |
| ZNF862 |
| PL-5283 |
| ACTR3C |
| MIR592 |
| MIR593 |
| LOC728743 |
| TMEM229A |
| LUZP6 |
| MIR671 |
| LOC100124692 |
| LOC100128542 |
| CTAGE4 |
| LOC100128822 |
| LOC100132707 |
| LOC100134229 |
| LOC100134713 |
This is the comprehensive list of deleted genes in the wide peak for 4q32.3.
Table S22. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CLCN3 |
| NEK1 |
| MFAP3L |
| ANXA10 |
| PALLD |
| SPOCK3 |
| AADAT |
| C4orf27 |
| DDX60 |
| SH3RF1 |
| CBR4 |
| DDX60L |
| GALNTL6 |
This is the comprehensive list of deleted genes in the wide peak for 20p12.1.
Table S23. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-663 |
| JAG1 |
| BFSP1 |
| BMP2 |
| ENTPD6 |
| CST1 |
| CST2 |
| CST3 |
| CST4 |
| CST5 |
| FOXA2 |
| INSM1 |
| NKX2-2 |
| PAX1 |
| PCSK2 |
| PLCB4 |
| PYGB |
| RRBP1 |
| SNAP25 |
| SNRPB2 |
| SSTR4 |
| THBD |
| ZNF133 |
| MKKS |
| CST7 |
| GINS1 |
| CST8 |
| SEC23B |
| POLR3F |
| RBBP9 |
| DSTN |
| XRN2 |
| BTBD3 |
| CD93 |
| NINL |
| PLCB1 |
| FLRT3 |
| C20orf103 |
| C20orf26 |
| ABHD12 |
| ZNF337 |
| SNX5 |
| HSPC072 |
| NXT1 |
| VSX1 |
| NAT5 |
| CRNKL1 |
| ESF1 |
| HAO1 |
| RIN2 |
| C20orf12 |
| SPTLC3 |
| KIF16B |
| TASP1 |
| PLK1S1 |
| TMX4 |
| OTOR |
| C20orf3 |
| PAK7 |
| RALGAPA2 |
| CSRP2BP |
| SLC24A3 |
| OVOL2 |
| NAPB |
| ANKRD5 |
| GZF1 |
| C20orf7 |
| TMEM90B |
| SEL1L2 |
| ACSS1 |
| GGTLC1 |
| C20orf72 |
| DTD1 |
| C20orf94 |
| CSTL1 |
| CST9L |
| CST9 |
| MACROD2 |
| C20orf56 |
| BANF2 |
| NANP |
| C20orf79 |
| ISM1 |
| CST11 |
| C20orf191 |
| CSTT |
| DEFB115 |
| LOC284788 |
| LOC284798 |
| FAM182A |
| FRG1B |
| LOC388789 |
| NKX2-4 |
| SNORD17 |
| MIR663 |
| FAM182B |
| LOC100130264 |
| LOC100134868 |
| LOC100270804 |
| PET117 |
This is the comprehensive list of deleted genes in the wide peak for 10q26.2.
Table S24. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| FGFR2 |
| TLX1 |
| NFKB2 |
| DUX4 |
| SUFU |
| hsa-mir-202 |
| hsa-mir-2110 |
| hsa-mir-548e |
| hsa-mir-609 |
| hsa-mir-936 |
| hsa-mir-1307 |
| hsa-mir-146b |
| hsa-mir-608 |
| hsa-mir-1287 |
| hsa-mir-607 |
| ACADSB |
| ADAM8 |
| ADD3 |
| ADRA2A |
| ADRB1 |
| ARL3 |
| BNIP3 |
| CASP7 |
| ENTPD1 |
| CHUK |
| ABCC2 |
| COL17A1 |
| COX15 |
| CPN1 |
| CTBP2 |
| CYP2E1 |
| CYP17A1 |
| DMBT1 |
| DNTT |
| DOCK1 |
| DUSP5 |
| ECHS1 |
| EMX2 |
| FGF8 |
| GFRA1 |
| GOT1 |
| PRLHR |
| GPR26 |
| GRK5 |
| HABP2 |
| HMX2 |
| HPS1 |
| INPP5A |
| ABLIM1 |
| MGMT |
| MKI67 |
| MXI1 |
| NDUFB8 |
| NRAP |
| OAT |
| PAX2 |
| PGAM1 |
| PITX3 |
| PNLIP |
| PNLIPRP1 |
| PNLIPRP2 |
| HTRA1 |
| PSD |
| PTPRE |
| ALDH18A1 |
| RGS10 |
| SCD |
| SFRP5 |
| FBXW4 |
| SLC18A2 |
| SLIT1 |
| TAF5 |
| TCF7L2 |
| TECTB |
| TIAL1 |
| TLL2 |
| UROS |
| WNT8B |
| XPNPEP1 |
| SHOC2 |
| ADAM12 |
| UTF1 |
| EIF3A |
| GBF1 |
| LDB1 |
| BTRC |
| PKD2L1 |
| INA |
| SMC3 |
| NEURL |
| BUB3 |
| NOLC1 |
| GSTO1 |
| BAG3 |
| SH3PXD2A |
| FAM53B |
| SLK |
| ZNF518A |
| DCLRE1A |
| FRAT1 |
| ACTR1A |
| SMNDC1 |
| NPM3 |
| GLRX3 |
| DPYSL4 |
| TACC2 |
| ERLIN1 |
| LBX1 |
| MGEA5 |
| TUBGCP2 |
| PRDX3 |
| VAX1 |
| ATE1 |
| SEC23IP |
| RAB11FIP2 |
| INPP5F |
| NT5C2 |
| PDCD11 |
| SORCS3 |
| PPRC1 |
| FAM175B |
| RRP12 |
| DNMBP |
| FRAT2 |
| RP11-529I10.4 |
| SEC31B |
| ATRNL1 |
| C10orf137 |
| TCTN3 |
| C10orf12 |
| ANKRD2 |
| CNNM1 |
| PDCD4 |
| VENTX |
| C10orf28 |
| POLL |
| BLNK |
| KCNIP2 |
| CUZD1 |
| CALY |
| EXOSC1 |
| CALHM2 |
| CUTC |
| CHST15 |
| ACSL5 |
| CCNJ |
| ZRANB1 |
| NSMCE4A |
| CNNM2 |
| C10orf26 |
| C10orf118 |
| CRTAC1 |
| CWF19L1 |
| PI4K2A |
| HIF1AN |
| BRWD2 |
| FAM178A |
| DHX32 |
| PPP2R2D |
| FAM45B |
| TDRD1 |
| BCCIP |
| C10orf2 |
| TM9SF3 |
| ENTPD7 |
| AS3MT |
| GPAM |
| KIAA1598 |
| FAM160B1 |
| SEMA4G |
| PLEKHA1 |
| AVPI1 |
| HPSE2 |
| C10orf84 |
| LHPP |
| MMS19 |
| IKZF5 |
| ZDHHC6 |
| CUEDC2 |
| FBXL15 |
| C10orf76 |
| HPS6 |
| TMEM180 |
| C10orf119 |
| C10orf95 |
| C10orf81 |
| PDZD7 |
| OBFC1 |
| C10orf88 |
| UBTD1 |
| C10orf79 |
| LRRC27 |
| TRIM8 |
| KAZALD1 |
| SFXN3 |
| SLC25A28 |
| ELOVL3 |
| MARVELD1 |
| PCGF6 |
| LOXL4 |
| ZDHHC16 |
| GPR123 |
| LZTS2 |
| LCOR |
| NKX6-2 |
| MRPL43 |
| AFAP1L2 |
| PYROXD2 |
| USMG5 |
| ARHGAP19 |
| KNDC1 |
| ITPRIP |
| C10orf75 |
| MTG1 |
| NCRNA00081 |
| FANK1 |
| OPALIN |
| SYCE1 |
| DHDPSL |
| SORCS1 |
| PRAP1 |
| ZNF511 |
| C10orf90 |
| BTBD16 |
| FAM24A |
| PSTK |
| PIK3AP1 |
| MORN4 |
| ZFYVE27 |
| MMP21 |
| SFXN2 |
| PDZD8 |
| C10orf32 |
| GSTO2 |
| C10orf78 |
| CALHM3 |
| CLRN3 |
| PNLIPRP3 |
| SFXN4 |
| CPXM2 |
| TRUB1 |
| VTI1A |
| LOC143188 |
| C10orf82 |
| C10orf46 |
| NKX2-3 |
| CCDC147 |
| C10orf91 |
| PWWP2B |
| EMX2OS |
| PPAPDC1A |
| PAOX |
| FAM24B |
| EBF3 |
| CALHM1 |
| CASC2 |
| C10orf93 |
| TCERG1L |
| HSPA12A |
| C10orf125 |
| JAKMIP3 |
| STK32C |
| BLOC1S2 |
| RBM20 |
| LOC282997 |
| KCNK18 |
| VWA2 |
| NANOS1 |
| HMX3 |
| NHLRC2 |
| C10orf96 |
| FLJ46361 |
| CC2D2B |
| ARMS2 |
| C10orf122 |
| GUCY2G |
| NKX1-2 |
| FLJ41350 |
| C10orf120 |
| LOC399815 |
| METTL10 |
| FOXI2 |
| GOLGA7B |
| FAM45A |
| C10orf62 |
| FRG2B |
| SPRN |
| MIR146B |
| MIR202 |
| NPS |
| LOC619207 |
| SNORA19 |
| FAM196A |
| RPL13AP6 |
| LOC653543 |
| LOC653544 |
| LOC653545 |
| LOC653548 |
| SNORA12 |
| MIR608 |
| MIR609 |
| LOC728410 |
| LOC729020 |
| TLX1NB |
| C10orf131 |
| LOC100169752 |
| NCRNA00093 |
| LOC100270710 |
This is the comprehensive list of deleted genes in the wide peak for 5q12.1.
Table S25. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| APC |
| CD74 |
| EBF1 |
| IL6ST |
| ITK |
| LIFR |
| NPM1 |
| PDGFRB |
| PIK3R1 |
| TLX3 |
| NSD1 |
| RANBP17 |
| hsa-mir-340 |
| hsa-mir-1229 |
| hsa-mir-1271 |
| hsa-mir-585 |
| hsa-mir-218-2 |
| hsa-mir-103-1-as |
| hsa-mir-146a |
| hsa-mir-1303 |
| hsa-mir-1294 |
| hsa-mir-378 |
| hsa-mir-145 |
| hsa-mir-584 |
| hsa-mir-874 |
| hsa-mir-886 |
| hsa-mir-1289-2 |
| hsa-mir-1244 |
| hsa-mir-548f-3 |
| hsa-mir-548p |
| hsa-mir-583 |
| hsa-mir-1974 |
| hsa-mir-9-2 |
| hsa-mir-582 |
| hsa-mir-449b |
| hsa-mir-581 |
| hsa-mir-1274a |
| hsa-mir-580 |
| hsa-mir-579 |
| hsa-mir-887 |
| ADCY2 |
| ADRA1B |
| ADRB2 |
| ANXA6 |
| TRIM23 |
| ARSB |
| ATOX1 |
| ALDH7A1 |
| BHMT |
| BNIP1 |
| BTF3 |
| C6 |
| C7 |
| C9 |
| CAMK4 |
| CAMK2A |
| CAMLG |
| CANX |
| CAST |
| CCNB1 |
| CCNG1 |
| CCNH |
| CD14 |
| CDC25C |
| CDH6 |
| CDH9 |
| CDH10 |
| CDH12 |
| CDH18 |
| CDK7 |
| CDO1 |
| CDX1 |
| CETN3 |
| CHD1 |
| CKMT2 |
| ERCC8 |
| AP3S1 |
| CLTB |
| COX7C |
| CRHBP |
| HAPLN1 |
| CSF1R |
| CSF2 |
| CSNK1A1 |
| CSNK1G3 |
| VCAN |
| NKX2-5 |
| CTNNA1 |
| CTNND2 |
| DAB2 |
| DAP |
| DBN1 |
| DMXL1 |
| DHFR |
| DIAPH1 |
| DNAH5 |
| DOCK2 |
| DPYSL3 |
| DRD1 |
| SLC26A2 |
| HBEGF |
| DUSP1 |
| EFNA5 |
| EGR1 |
| ETF1 |
| F2R |
| F2RL1 |
| F2RL2 |
| F12 |
| FABP6 |
| FAT2 |
| FBN2 |
| FER |
| FGF1 |
| FGF10 |
| FGFR4 |
| FOXD1 |
| FOXI1 |
| FLT4 |
| FYB |
| GABRA1 |
| GABRA6 |
| GABRB2 |
| GABRG2 |
| GABRP |
| GDF9 |
| GDNF |
| GFRA3 |
| GHR |
| GLRA1 |
| GLRX |
| GM2A |
| GRK6 |
| GPX3 |
| GRIA1 |
| NR3C1 |
| GRM6 |
| GTF2H2 |
| GZMA |
| GZMK |
| HARS |
| HEXB |
| HINT1 |
| HK3 |
| HMGCR |
| HMGCS1 |
| HMMR |
| HNRNPAB |
| HNRNPH1 |
| HRH2 |
| HSD17B4 |
| HSPA4 |
| HSPA9 |
| NDST1 |
| HTR1A |
| HTR4 |
| IK |
| IL3 |
| IL4 |
| IL5 |
| IL7R |
| IL9 |
| IL12B |
| IL13 |
| IRF1 |
| ISL1 |
| ITGA1 |
| ITGA2 |
| KCNMB1 |
| KCNN2 |
| KIF2A |
| TNPO1 |
| LCP2 |
| LECT2 |
| LMNB1 |
| LNPEP |
| LOX |
| LTC4S |
| CD180 |
| SMAD5 |
| MAN2A1 |
| MAP1B |
| MCC |
| MEF2C |
| MAP3K1 |
| MFAP3 |
| MGAT1 |
| MOCS2 |
| MSH3 |
| MSX2 |
| MTRR |
| MYO10 |
| NAIP |
| NDUFA2 |
| NDUFS4 |
| NDUFS6 |
| NEUROG1 |
| NPR3 |
| NPY6R |
| OCLN |
| OXCT1 |
| PAM |
| PCDH1 |
| PCDHGC3 |
| PCSK1 |
| PDE4D |
| PDE6A |
| PFDN1 |
| PGGT1B |
| PITX1 |
| PMCHL1 |
| PMCHL2 |
| POU4F3 |
| PPIC |
| PPP2CA |
| PPP2R2B |
| PRKAA1 |
| MAPK9 |
| PRLR |
| PROP1 |
| PTGER4 |
| RAD1 |
| PURA |
| RAD17 |
| RARS |
| RASA1 |
| RASGRF2 |
| RPL37 |
| RPS14 |
| RPS23 |
| SDHA |
| SEPP1 |
| SGCD |
| SKP1 |
| SKP2 |
| SLC1A3 |
| SLC6A3 |
| SLC6A7 |
| SLC9A3 |
| SLC12A2 |
| SLC34A1 |
| SLC22A4 |
| SLC22A5 |
| SLIT3 |
| SMN1 |
| SMN2 |
| SNCB |
| SNX2 |
| SPARC |
| SPINK1 |
| SPOCK1 |
| SRD5A1 |
| SRP19 |
| STK10 |
| TAF7 |
| TAF9 |
| TARS |
| TBCA |
| TCF7 |
| ZNF354A |
| TCOF1 |
| TERT |
| NR2F1 |
| TGFBI |
| THBS4 |
| TRIO |
| TTC1 |
| UBE2B |
| UBE2D2 |
| VDAC1 |
| WNT8A |
| XRCC4 |
| ZNF131 |
| ST8SIA4 |
| REEP5 |
| SERF1A |
| NME5 |
| ENC1 |
| AP3B1 |
| PDLIM4 |
| PPAP2A |
| STC2 |
| PDE8B |
| EIF4EBP3 |
| PCDHGB4 |
| CDC23 |
| ADAM19 |
| FGF18 |
| HDAC3 |
| SQSTM1 |
| P4HA2 |
| HSPB3 |
| ATP6V0E1 |
| SEMA5A |
| ATG12 |
| OSMR |
| PTTG1 |
| PDLIM7 |
| C5orf13 |
| TRIP13 |
| CNOT8 |
| RAB9P1 |
| HAND1 |
| MED7 |
| HOMER1 |
| MYOT |
| ADAMTS2 |
| SCAMP1 |
| NRG2 |
| CXCL14 |
| H2AFY |
| SMAD5OS |
| RNF14 |
| CARTPT |
| SNCAIP |
| NUP155 |
| TTC37 |
| CLINT1 |
| PCDHGA8 |
| PCDHA9 |
| ZFYVE16 |
| MATR3 |
| MAML1 |
| KIAA0141 |
| JAKMIP2 |
| PJA2 |
| DDX46 |
| GFPT2 |
| SLC23A1 |
| GNPDA1 |
| SRA1 |
| PDCD6 |
| EDIL3 |
| COL4A3BP |
| RAD50 |
| KIF20A |
| G3BP1 |
| LHFPL2 |
| SDCCAG10 |
| MARCH6 |
| APBB3 |
| CCNO |
| TNIP1 |
| GNB2L1 |
| BASP1 |
| NSA2 |
| FST |
| SLU7 |
| PAIP1 |
| POLR3G |
| RGS14 |
| SLC12A7 |
| PLK2 |
| IQGAP2 |
| SEC24A |
| CPLX2 |
| C5orf4 |
| FAM114A2 |
| MRPS30 |
| BRD8 |
| TCERG1 |
| BTNL3 |
| SUB1 |
| HNRNPA0 |
| LMAN2 |
| SPINK5 |
| POLS |
| SOX30 |
| TPPP |
| ESM1 |
| KIF3A |
| ADAMTS6 |
| MGAT4B |
| B4GALT7 |
| EXOC3 |
| SYNPO |
| RHOBTB3 |
| RNF44 |
| ABLIM3 |
| ELL2 |
| CCT5 |
| SV2C |
| HMGXB3 |
| PDZD2 |
| TBC1D9B |
| ARHGAP26 |
| FSTL4 |
| MRPS27 |
| ATP10B |
| N4BP3 |
| SEPT8 |
| FBXL7 |
| FAF2 |
| HISPPD1 |
| WWC1 |
| FBXW11 |
| ACSL6 |
| PHF15 |
| LARP1 |
| KIAA0947 |
| PPWD1 |
| HARS2 |
| OTP |
| SKIV2L2 |
| NNT |
| TTC33 |
| ZNF346 |
| AMACR |
| SSBP2 |
| BHMT2 |
| TNFAIP8 |
| NIPBL |
| LOC25845 |
| PART1 |
| GEMIN5 |
| PCDHGA12 |
| LRRTM2 |
| FAM169A |
| RAI14 |
| CCDC69 |
| PCDHB5 |
| FBXL21 |
| KLHL3 |
| TSPAN17 |
| FBXO4 |
| OR4F3 |
| HAVCR1 |
| SNORD63 |
| SNORA74A |
| CYFIP2 |
| PKD2L2 |
| UQCRQ |
| AFF4 |
| PRELID1 |
| IL17B |
| DIMT1L |
| MAT2B |
| SLC27A6 |
| SNX24 |
| MRPL22 |
| RNASEN |
| PCDHB1 |
| DMGDH |
| KCNIP1 |
| ZNF354C |
| IRX4 |
| TAS2R1 |
| TMED7 |
| ISOC1 |
| RPL26L1 |
| SAR1B |
| C5orf45 |
| SLC45A2 |
| DCTN4 |
| IPO11 |
| MGC29506 |
| PAIP2 |
| CDKL3 |
| RXFP3 |
| PCDH12 |
| GCNT4 |
| FAM13B |
| FAM53C |
| REEP2 |
| PRR16 |
| COMMD10 |
| POLK |
| DDX41 |
| NOP16 |
| LARS |
| CXXC5 |
| HMP19 |
| ZFR |
| UIMC1 |
| RAPGEF6 |
| ERAP1 |
| KDM3B |
| PHAX |
| RAB24 |
| PELO |
| RBM27 |
| FAM134B |
| FAM105A |
| NEURL1B |
| DHX29 |
| FLJ11235 |
| DDX4 |
| FAM193B |
| MTMR12 |
| SGTB |
| ARL15 |
| PCDHB18 |
| PCDHB17 |
| TMED9 |
| ZCCHC10 |
| PCDH24 |
| GIN1 |
| WDR55 |
| ANKHD1 |
| NSUN2 |
| CCDC99 |
| THG1L |
| WDR70 |
| AGGF1 |
| WDR41 |
| BRIX1 |
| C5orf22 |
| TMCO6 |
| TRIM36 |
| GALNT10 |
| NHP2 |
| RBM22 |
| CEP72 |
| RIOK2 |
| DEPDC1B |
| BDP1 |
| RNF130 |
| ERBB2IP |
| PCDHGC5 |
| PCDHGC4 |
| PCDHGB7 |
| PCDHGB6 |
| PCDHGB5 |
| PCDHGB3 |
| PCDHGB2 |
| PCDHGB1 |
| PCDHGA11 |
| PCDHGA10 |
| PCDHGA9 |
| PCDHGA7 |
| PCDHGA6 |
| PCDHGA5 |
| PCDHGA4 |
| PCDHGA3 |
| PCDHGA2 |
| PCDHGA1 |
| PCDHGB8P |
| PCDHB15 |
| PCDHB14 |
| PCDHB13 |
| PCDHB12 |
| PCDHB11 |
| PCDHB10 |
| PCDHB9 |
| PCDHB8 |
| PCDHB7 |
| PCDHB6 |
| PCDHB4 |
| PCDHB3 |
| PCDHB2 |
| PCDHAC2 |
| PCDHAC1 |
| PCDHA13 |
| PCDHA12 |
| PCDHA11 |
| PCDHA10 |
| PCDHA8 |
| PCDHA7 |
| PCDHA6 |
| PCDHA5 |
| PCDHA4 |
| PCDHA3 |
| PCDHA2 |
| PCDHA1 |
| ANKH |
| CCL28 |
| VTRNA1-3 |
| VTRNA1-2 |
| VTRNA1-1 |
| NMUR2 |
| FEM1C |
| C5orf15 |
| PRDM9 |
| CDC42SE2 |
| TRPC7 |
| KIAA1191 |
| ERGIC1 |
| CLK4 |
| ODZ2 |
| CNOT6 |
| NLN |
| AHRR |
| ZNF608 |
| KCTD16 |
| SEMA6A |
| ARRDC3 |
| ZSWIM6 |
| PCDHB16 |
| ANKRA2 |
| HMHB1 |
| C5orf54 |
| GOLPH3 |
| MCCC2 |
| EPB41L4A |
| CENPK |
| ERAP2 |
| RGNEF |
| SIL1 |
| GMCL1L |
| ARAP3 |
| C5orf28 |
| RMND5B |
| FBXL17 |
| YTHDC2 |
| AGXT2 |
| SLC30A5 |
| CENPH |
| MRPL36 |
| GPBP1 |
| C5orf42 |
| BRD9 |
| GRAMD3 |
| PCYOX1L |
| FASTKD3 |
| IRX1 |
| C5orf23 |
| CCNJL |
| SH3TC2 |
| PANK3 |
| PARP8 |
| SAP30L |
| ANKRD55 |
| TXNDC15 |
| MCTP1 |
| PTCD2 |
| ZDHHC11 |
| LPCAT1 |
| BTNL8 |
| SPEF2 |
| DOK3 |
| ELOVL7 |
| C5orf44 |
| ZFP2 |
| RUFY1 |
| CPEB4 |
| PRR7 |
| NDFIP1 |
| CLPTM1L |
| OR4F16 |
| FBXO38 |
| YIPF5 |
| TRIM7 |
| TIGD6 |
| ADAMTS12 |
| SPRY4 |
| MXD3 |
| NUDT12 |
| SLC4A9 |
| ATG10 |
| ROPN1L |
| SLC25A2 |
| SPATA9 |
| TSSK1B |
| FAM172A |
| PCDHB19P |
| GPR98 |
| PCBD2 |
| UTP15 |
| ZCCHC9 |
| MED10 |
| PSD2 |
| ANKRD32 |
| THOC3 |
| ZBED3 |
| GFM2 |
| C5orf32 |
| MEGF10 |
| SPINK7 |
| SPZ1 |
| CARD6 |
| TRIM52 |
| HAVCR2 |
| AGXT2L2 |
| C5orf62 |
| NKD2 |
| TSLP |
| FCHSD1 |
| UNC5A |
| FAM105B |
| C5orf30 |
| ZNF622 |
| LYRM7 |
| TRIM41 |
| SLC25A46 |
| BOD1 |
| CDKN2AIPNL |
| COL23A1 |
| TIMD4 |
| NDUFAF2 |
| ZNF300 |
| MYOZ3 |
| UBTD2 |
| LMBRD2 |
| MRPS36 |
| ATP6AP1L |
| SCGB3A1 |
| PRDM6 |
| FTMT |
| SFXN1 |
| FNIP1 |
| SNX18 |
| SLC35A4 |
| GPRIN1 |
| PWWP2A |
| C1QTNF2 |
| C1QTNF3 |
| C5orf26 |
| MARCH3 |
| FCHO2 |
| RAB3C |
| LYSMD3 |
| C5orf55 |
| LEAP2 |
| SCGB3A2 |
| ZNF354B |
| C5orf35 |
| IL31RA |
| EMB |
| SLCO6A1 |
| C5orf47 |
| PPARGC1B |
| HEATR7B2 |
| EGFLAM |
| PRRC1 |
| C5orf33 |
| UGT3A1 |
| CAPSL |
| JMY |
| C5orf58 |
| ZNF474 |
| CCDC127 |
| OR2Y1 |
| UBE2QL1 |
| C5orf49 |
| FAM173B |
| CMBL |
| POU5F2 |
| DNAJC21 |
| AFAP1L1 |
| GRPEL2 |
| TMEM171 |
| TMEM174 |
| LSM11 |
| C5orf37 |
| GPR151 |
| STARD4 |
| WDR36 |
| LOC134466 |
| NUDCD2 |
| UBLCP1 |
| ACOT12 |
| ANKRD43 |
| SHROOM1 |
| C5orf24 |
| SFRS12 |
| C5orf20 |
| SLC38A9 |
| SLC36A2 |
| SPINK5L3 |
| C5orf41 |
| CEP120 |
| LOC153328 |
| TMEM167A |
| MBLAC2 |
| TMEM161B |
| SRFBP1 |
| PLEKHG4B |
| ZMAT2 |
| MARVELD2 |
| C5orf38 |
| IRX2 |
| BTNL9 |
| ARSK |
| FAM81B |
| TTC23L |
| LOC153684 |
| CCDC112 |
| PPP1R2P3 |
| FAM71B |
| PRELID2 |
| SH3RF2 |
| PLAC8L1 |
| RNF145 |
| MIER3 |
| CDC20B |
| UGT3A2 |
| PAPD4 |
| DCP2 |
| MGC42105 |
| LIX1 |
| ZNF366 |
| FAM151B |
| S100Z |
| ADAMTS16 |
| ADAMTS19 |
| HIGD2A |
| SPATA24 |
| DNAJC18 |
| FAM153B |
| RANBP3L |
| LOC202181 |
| CCDC125 |
| C5orf27 |
| GAPT |
| CMYA5 |
| STK32A |
| LVRN |
| SLC36A1 |
| RICTOR |
| EIF4E1B |
| LOC255167 |
| RASGEF1C |
| ANKRD31 |
| SERINC5 |
| LOC257358 |
| SH3PXD2B |
| LOC285593 |
| FAM153A |
| ARL10 |
| C5orf36 |
| GPR150 |
| DTWD2 |
| RELL2 |
| NBPF22P |
| LOC285627 |
| LOC285629 |
| C5orf51 |
| SLC36A3 |
| KIF4B |
| OR2V2 |
| FLJ37543 |
| RNF180 |
| SFRS12IP1 |
| ZNF454 |
| C5orf60 |
| LOC285692 |
| LOC285696 |
| RGMB |
| RFESD |
| CHSY3 |
| SLC6A19 |
| TMEM173 |
| FAM170A |
| LOC340074 |
| ARSI |
| LOC340094 |
| ANKRD34B |
| PFN3 |
| ZNF879 |
| PLCXD3 |
| IRGM |
| FBLL1 |
| ACTBL2 |
| FAM174A |
| MTX3 |
| CATSPER3 |
| SLC6A18 |
| NIPAL4 |
| HCN1 |
| SLCO4C1 |
| TICAM2 |
| DND1 |
| C5orf34 |
| MAST4 |
| C5orf25 |
| RNF138P1 |
| LRRC14B |
| C5orf39 |
| ISCA1L |
| C5orf48 |
| LOC389332 |
| LOC389333 |
| C5orf46 |
| FLJ41603 |
| FLJ33360 |
| RGS7BP |
| FLJ44606 |
| SPINK6 |
| ANKHD1-EIF4EBP3 |
| MIR103-1 |
| MIR143 |
| MIR145 |
| MIR146A |
| MIR218-2 |
| SPINK5L2 |
| C5orf40 |
| MARCH11 |
| LOC441089 |
| FLJ42709 |
| C5orf56 |
| MIR340 |
| C5orf53 |
| GPX8 |
| TIFAB |
| MIR449A |
| CTXN3 |
| SNORD72 |
| SNORD95 |
| SNORD96A |
| ECSCR |
| TMEM232 |
| C5orf43 |
| GRXCR2 |
| SPINK9 |
| ZFP62 |
| FLJ33630 |
| LOC644936 |
| CCNI2 |
| LOC645323 |
| CBY3 |
| LOC647859 |
| ANKRD33B |
| GUSBP3 |
| GTF2H2B |
| FAM153C |
| LOC653391 |
| SNORA13 |
| SCARNA18 |
| SNORA47 |
| SNORA74B |
| MIR449B |
| MIR548C |
| MIR548D2 |
| MIR580 |
| MIR581 |
| MIR583 |
| MIR585 |
| SNHG4 |
| LOC728264 |
| GTF2H2C |
| LOC728411 |
| SERF1B |
| LOC728554 |
| SDHAP3 |
| LOC728613 |
| LOC728723 |
| AACSL |
| LOC729678 |
| OR4F29 |
| GTF2H2D |
| SNORD123 |
| MIR886 |
| MIR874 |
| LOC100129716 |
| CCDC152 |
| LRRC70 |
| FAM196B |
| LOC100132062 |
| LOC100132287 |
| FAM159B |
| LOC100133050 |
| LOC100133331 |
| LOC100170939 |
| C5orf52 |
| LOC100268168 |
| LOC100272216 |
| LOC100286948 |
| TMED7-TICAM2 |
| NCRUPAR |
| LOC100303749 |
| LOC100310782 |
This is the comprehensive list of deleted genes in the wide peak for 9p24.3.
Table S26. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| ABL1 |
| FANCC |
| FANCG |
| GNAQ |
| JAK2 |
| MLLT3 |
| NFIB |
| NOTCH1 |
| OMD |
| PAX5 |
| RALGDS |
| SET |
| SYK |
| TAL2 |
| TSC1 |
| XPA |
| NR4A3 |
| BRD3 |
| NUP214 |
| FNBP1 |
| CD274 |
| hsa-mir-602 |
| hsa-mir-126 |
| hsa-mir-219-2 |
| hsa-mir-199b |
| hsa-mir-181b-2 |
| hsa-mir-601 |
| hsa-mir-600 |
| hsa-mir-147 |
| hsa-mir-455 |
| hsa-mir-32 |
| hsa-mir-1302-8 |
| hsa-mir-24-1 |
| hsa-let-7d |
| hsa-mir-7-1 |
| hsa-mir-204 |
| hsa-mir-1299 |
| hsa-mir-873 |
| hsa-mir-31 |
| hsa-mir-491 |
| hsa-mir-101-2 |
| hsa-mir-1302-2 |
| ABCA1 |
| ABCA2 |
| ABO |
| ACO1 |
| PLIN2 |
| AK1 |
| ALAD |
| ALDH1A1 |
| ALDH1B1 |
| ALDOB |
| AMBP |
| ANXA1 |
| ANXA2P2 |
| NUDT2 |
| APBA1 |
| AQP3 |
| AQP7 |
| ASS1 |
| AUH |
| BAAT |
| BAG1 |
| KLF9 |
| C5 |
| C8G |
| CA9 |
| CACNA1B |
| CCIN |
| CCBL1 |
| TNFSF8 |
| ENTPD2 |
| CD72 |
| CDK9 |
| CDKN2A |
| CDKN2B |
| CEL |
| CELP |
| CKS2 |
| CLTA |
| CNTFR |
| COL5A1 |
| COL15A1 |
| SLC31A1 |
| SLC31A2 |
| CRAT |
| CTSL1 |
| CTSL2 |
| CYLC2 |
| DAPK1 |
| DBC1 |
| DBH |
| SARDH |
| DNM1 |
| DMRT1 |
| ECM2 |
| TOR1A |
| LPAR1 |
| S1PR3 |
| MEGF9 |
| ELAVL2 |
| ENDOG |
| ENG |
| STOM |
| FBP1 |
| FKTN |
| FCN1 |
| FCN2 |
| FOXD4 |
| FOXE1 |
| MLANA |
| FPGS |
| FXN |
| NR5A1 |
| FUT7 |
| GALT |
| GAS1 |
| NR6A1 |
| GCNT1 |
| GGTA1 |
| B4GALT1 |
| GLDC |
| GLE1 |
| GNG10 |
| GOLGA1 |
| GOLGA2 |
| GPR21 |
| RAPGEF1 |
| GRIN1 |
| GSN |
| HNRNPK |
| HSD17B3 |
| DNAJA1 |
| HSPA5 |
| TNC |
| IARS |
| IFNA1 |
| IFNA2 |
| IFNA4 |
| IFNA5 |
| IFNA6 |
| IFNA7 |
| IFNA8 |
| IFNA10 |
| IFNA13 |
| IFNA14 |
| IFNA16 |
| IFNA17 |
| IFNA21 |
| IFNB1 |
| IFNW1 |
| IL11RA |
| INSL4 |
| LCN1 |
| LCN2 |
| LMX1B |
| MTAP |
| MUSK |
| NCBP1 |
| NDUFA8 |
| NDUFB6 |
| NFIL3 |
| NFX1 |
| NINJ1 |
| NPR2 |
| NTRK2 |
| ROR2 |
| ODF2 |
| OGN |
| ORM1 |
| ORM2 |
| PAEP |
| PAPPA |
| PDCL |
| PBX3 |
| PCSK5 |
| PGM5 |
| PHF2 |
| PPP2R4 |
| PPP3R2 |
| PPP6C |
| PRKACG |
| PRSS3 |
| PSMB7 |
| PSMD5 |
| PTCH1 |
| PTGDS |
| PTGS1 |
| PTPN3 |
| PTPRD |
| RAD23B |
| RFX3 |
| RGS3 |
| RLN1 |
| RLN2 |
| RMRP |
| RORB |
| RPL7A |
| RPL12 |
| RPS6 |
| RXRA |
| CCL19 |
| CCL21 |
| SH3GL2 |
| SHB |
| SLC1A1 |
| SMARCA2 |
| SNAPC3 |
| SNAPC4 |
| SPTAN1 |
| STXBP1 |
| SURF1 |
| SURF2 |
| SURF4 |
| MED22 |
| SURF6 |
| TEK |
| TESK1 |
| TGFBR1 |
| TLE1 |
| TLE4 |
| TLN1 |
| TLR4 |
| TMOD1 |
| TPM2 |
| TRAF1 |
| TRAF2 |
| TTF1 |
| TXN |
| TYRP1 |
| UGCG |
| VAV2 |
| VCP |
| VLDLR |
| CORO2A |
| ZFP37 |
| ZNF79 |
| ZNF189 |
| ZFAND5 |
| LHX3 |
| GFI1B |
| PIP5K1B |
| RECK |
| IKBKAP |
| CDC14B |
| TMEFF1 |
| SSNA1 |
| EDF1 |
| CTNNAL1 |
| MPDZ |
| FBP2 |
| DPM2 |
| FUBP3 |
| CLIC3 |
| PRPF4 |
| KLF4 |
| GTF3C5 |
| GTF3C4 |
| CER1 |
| LHX2 |
| PLAA |
| GRHPR |
| FAM189A2 |
| TJP2 |
| MED27 |
| PTGES |
| ATP6V1G1 |
| GABBR2 |
| GDA |
| GNA14 |
| RALGPS1 |
| ADAMTSL2 |
| RGP1 |
| TRIM14 |
| MELK |
| RUSC2 |
| KIAA0649 |
| SEC16A |
| ZBTB5 |
| KIAA0020 |
| TNFSF15 |
| ROD1 |
| GNE |
| SH2D3C |
| RCL1 |
| TOPORS |
| RABEPK |
| SIGMAR1 |
| LAMC3 |
| TUBB2C |
| UBAC1 |
| OLFM1 |
| ZER1 |
| CREB3 |
| UNC13B |
| SEMA4D |
| ANP32B |
| AGPAT2 |
| SPTLC1 |
| POMT1 |
| SMC2 |
| DMRT2 |
| RRAGA |
| ZBTB6 |
| NEK6 |
| SDCCAG3 |
| NOXA1 |
| CCL27 |
| USP20 |
| ACTL7B |
| ACTL7A |
| GADD45G |
| SPIN1 |
| SEC61B |
| SLC27A4 |
| SLC35D2 |
| CEP110 |
| WDR5 |
| C9orf9 |
| ADAMTS13 |
| C9orf7 |
| PSIP1 |
| INSL6 |
| SLC2A6 |
| PTENP1 |
| AKAP2 |
| RPL35 |
| MAN1B1 |
| DCTN3 |
| FRMPD1 |
| DOLK |
| ZNF510 |
| HABP4 |
| PTGR1 |
| TRIM32 |
| SETX |
| ERP44 |
| KDM4C |
| ZBTB43 |
| SMC5 |
| KANK1 |
| FAM120A |
| PMPCA |
| VPS13A |
| ASTN2 |
| AGTPBP1 |
| BICD2 |
| FKBP15 |
| KIAA1045 |
| KIAA0368 |
| EXOSC2 |
| FREQ |
| TDRD7 |
| SLC44A1 |
| ANGPTL2 |
| NUP188 |
| CCRK |
| DDX58 |
| RABGAP1 |
| TMEM2 |
| C9orf5 |
| C9orf4 |
| SLC24A2 |
| CIZ1 |
| DNAJB5 |
| DCAF12 |
| DFNB31 |
| COBRA1 |
| NIPSNAP3A |
| NELF |
| GPSM1 |
| LOC26102 |
| GAPVD1 |
| PHF19 |
| ZNF658 |
| FAM75A7 |
| FBXW2 |
| SPAG8 |
| OR1J4 |
| OR2K2 |
| FBXO10 |
| GBGT1 |
| LHX6 |
| OSTF1 |
| OR1L3 |
| OR1L1 |
| OR1J2 |
| SNORA65 |
| SNORD62A |
| SNORD36C |
| SNORD36B |
| SNORD36A |
| SNORD24 |
| RANBP6 |
| TRUB2 |
| DNAI1 |
| ST6GALNAC4 |
| INVS |
| NDOR1 |
| SIT1 |
| SPINK4 |
| TOR1B |
| TOR2A |
| METTL11A |
| PHPT1 |
| ANAPC2 |
| PKN3 |
| DPP7 |
| PSAT1 |
| UBQLN1 |
| SLC2A8 |
| OBP2B |
| OBP2A |
| ST6GALNAC6 |
| STOML2 |
| DEC1 |
| PCA3 |
| AK3 |
| EXOSC3 |
| FAM108B1 |
| MRPS2 |
| COQ4 |
| CERCAM |
| EGFL7 |
| C9orf53 |
| UBAP1 |
| GOLM1 |
| PRRX2 |
| C9orf114 |
| CHMP5 |
| C9orf156 |
| RAB14 |
| TMEM8B |
| C9orf78 |
| SHC3 |
| POLE3 |
| NANS |
| FBXW5 |
| MRPL50 |
| RC3H2 |
| EPB41L4B |
| C9orf11 |
| TBC1D13 |
| FAM22F |
| DIRAS2 |
| BNC2 |
| HAUS6 |
| ASPN |
| BSPRY |
| APTX |
| C9orf167 |
| CNTLN |
| TEX10 |
| LPPR1 |
| KIAA1797 |
| UBE2R2 |
| EXD3 |
| C9orf6 |
| C9orf95 |
| STX17 |
| NOL8 |
| C9orf68 |
| C9orf40 |
| TMEM38B |
| SMU1 |
| RFK |
| NIPSNAP3B |
| STRBP |
| TBC1D2 |
| HEMGN |
| KIF27 |
| CDC37L1 |
| DENND4C |
| C9orf86 |
| CDK5RAP2 |
| UBAP2 |
| C9orf46 |
| CBWD1 |
| KLHL9 |
| BARX1 |
| RNF20 |
| LRRC8A |
| INPP5E |
| NPDC1 |
| OR2S2 |
| BARHL1 |
| IFNK |
| SH3GLB2 |
| REXO4 |
| DOLPP1 |
| KIAA1161 |
| KCNT1 |
| KIAA1432 |
| KIAA1529 |
| ZBTB26 |
| GBA2 |
| DENND1A |
| GPR107 |
| SLC46A2 |
| C9orf27 |
| C9orf80 |
| ZNF462 |
| DMRT3 |
| PRDM12 |
| MAK10 |
| DMRTA1 |
| SLC28A3 |
| CARD9 |
| SUSD1 |
| POLR1E |
| IPPK |
| DDX31 |
| FAM129B |
| LRRC19 |
| MRPL41 |
| NOL6 |
| WNK2 |
| SECISBP2 |
| C9orf16 |
| MAPKAP1 |
| DCAF10 |
| ZCCHC6 |
| GALNT12 |
| EHMT1 |
| MOBKL2B |
| C9orf82 |
| CNTNAP3 |
| ERMP1 |
| SVEP1 |
| RMI1 |
| TRPM3 |
| PTGES2 |
| IFT74 |
| KIAA1539 |
| GKAP1 |
| PDCD1LG2 |
| AKNA |
| C9orf45 |
| URM1 |
| ISCA1 |
| DOCK8 |
| ARPC5L |
| HDHD3 |
| AIF1L |
| UCK1 |
| ZNF484 |
| FSD1L |
| CEP78 |
| ZCCHC7 |
| ANKRD20A1 |
| GARNL3 |
| HSDL2 |
| C9orf64 |
| C9orf89 |
| HIATL2 |
| C9orf125 |
| NTNG2 |
| HIATL1 |
| HINT2 |
| C9orf24 |
| PIGO |
| BAT2L |
| PPAPDC3 |
| C9orf70 |
| ZDHHC12 |
| FAM73B |
| C9orf100 |
| C9orf3 |
| FIBCD1 |
| KIAA1984 |
| SNHG7 |
| TMEM141 |
| C9orf37 |
| COL27A1 |
| ALG2 |
| FGD3 |
| FAM125B |
| TPD52L3 |
| WDR34 |
| C9orf140 |
| C9orf69 |
| LRSAM1 |
| IL33 |
| C9orf123 |
| C9orf30 |
| UAP1L1 |
| MCART1 |
| MRRF |
| RBM18 |
| ARRDC1 |
| WDR85 |
| ADAMTSL1 |
| LOC92973 |
| TMEM203 |
| KIF12 |
| PALM2 |
| SLC25A25 |
| WDR31 |
| ZNF618 |
| UHRF2 |
| FAM122A |
| ZMYND19 |
| GRIN3A |
| TMC1 |
| RNF183 |
| NACC2 |
| C9orf116 |
| C9orf41 |
| C9orf57 |
| C9orf85 |
| C9orf135 |
| LCN8 |
| FAM69B |
| PTRH1 |
| PIP5KL1 |
| TAF1L |
| PTPDC1 |
| ANKRD19 |
| ARID3C |
| C9orf23 |
| C9orf131 |
| OR13C5 |
| OR13C8 |
| OR13C3 |
| OR13C4 |
| OR13F1 |
| OR1L8 |
| OR1N2 |
| OR1N1 |
| ASB6 |
| TRPM6 |
| SLC34A3 |
| RNF38 |
| GLIPR2 |
| DAB2IP |
| CAMSAP1 |
| C9orf66 |
| NCRNA00032 |
| LINGO2 |
| NXNL2 |
| C9orf163 |
| MAMDC4 |
| LCN6 |
| C9orf98 |
| OR1Q1 |
| TTLL11 |
| RASEF |
| TTC39B |
| C9orf122 |
| RG9MTD3 |
| TTC16 |
| FAM120AOS |
| FAM154A |
| C9orf44 |
| FREM1 |
| KIAA2026 |
| LOC158376 |
| LOC158381 |
| ZNF483 |
| C9orf84 |
| KIAA1958 |
| TSTD2 |
| ZNF782 |
| PRUNE2 |
| C9orf96 |
| KCNV2 |
| OLFML2A |
| C9orf71 |
| QSOX2 |
| GLIS3 |
| LOC169834 |
| ZNF169 |
| C9orf21 |
| ZNF367 |
| C9orf91 |
| C9orf72 |
| C9orf93 |
| NAIF1 |
| C9orf25 |
| CCDC107 |
| ANKS6 |
| SUSD3 |
| CBWD5 |
| CDC26 |
| LOC253039 |
| PHYHD1 |
| MORN5 |
| OR1L4 |
| TXNDC8 |
| MAMDC2 |
| FRMD3 |
| C9orf43 |
| C9orf144B |
| NCRNA00094 |
| CRB2 |
| SCAI |
| C9orf117 |
| C9orf47 |
| C9orf79 |
| LOC286238 |
| LCN12 |
| C9orf142 |
| C9orf75 |
| TUSC1 |
| C9orf109 |
| FAM78A |
| C9orf150 |
| LOC286359 |
| OR13C9 |
| OR13D1 |
| LOC286367 |
| FOXD4L3 |
| IFNE |
| ZDHHC21 |
| ACER2 |
| LOC340508 |
| GPR144 |
| FLJ43950 |
| QRFP |
| OR1J1 |
| OR1B1 |
| KIF24 |
| IGFBPL1 |
| MURC |
| FOXD4L4 |
| GLT6D1 |
| ENHO |
| AQP7P1 |
| PTAR1 |
| C9orf102 |
| C9orf119 |
| C9orf50 |
| PNPLA7 |
| C9orf169 |
| ENTPD8 |
| ZNF322B |
| KGFLP1 |
| LOC389705 |
| C9orf144 |
| FAM75A6 |
| MGC21881 |
| AQP7P2 |
| FLJ43859 |
| FLJ44082 |
| FLJ46321 |
| LOC389765 |
| C9orf153 |
| LOC389791 |
| IER5L |
| C9orf171 |
| LCN15 |
| C9orf172 |
| LRRC26 |
| TMEM8C |
| C9orf128 |
| OR13J1 |
| CTSL3 |
| OR13C2 |
| OR1L6 |
| OR5C1 |
| OR1K1 |
| LCN9 |
| FAM102A |
| FLJ35024 |
| PTPLAD2 |
| TMEM215 |
| TOMM5 |
| FAM74A1 |
| FAM74A4 |
| ZNF658B |
| C9orf170 |
| CENPP |
| C9orf152 |
| SNX30 |
| WDR38 |
| LCNL1 |
| C9orf139 |
| FAM166A |
| LOC402377 |
| SOHLH1 |
| PPAPDC2 |
| ZBTB34 |
| MIRLET7A1 |
| MIRLET7D |
| MIRLET7F1 |
| MIR101-2 |
| MIR126 |
| MIR147 |
| MIR181A2 |
| MIR181B2 |
| MIR199B |
| MIR204 |
| MIR219-2 |
| MIR23B |
| MIR24-1 |
| MIR27B |
| MIR31 |
| MIR32 |
| MIR7-1 |
| C9orf106 |
| C9orf103 |
| LCN10 |
| LOC415056 |
| LOC440173 |
| LOC440839 |
| LOC440896 |
| SUGT1P |
| ANKRD20A3 |
| ANKRD20A2 |
| AQP7P3 |
| FAM75C1 |
| LOC441454 |
| LOC441455 |
| FAM22G |
| C9orf173 |
| NRARP |
| LOC442421 |
| FOXB2 |
| CBWD3 |
| C9orf129 |
| PALM2-AKAP2 |
| FAM27A |
| DNAJC25 |
| DNAJC25-GNG10 |
| LOC554202 |
| LOC572558 |
| MIR491 |
| PGM5P2 |
| MIR455 |
| FAM138F |
| FAM75A2 |
| LOC642313 |
| LOC642929 |
| FAM163B |
| FLJ40292 |
| TUBBP5 |
| CBWD6 |
| FAM138A |
| LOC645961 |
| HRCT1 |
| FAM75A1 |
| RPSAP9 |
| FOXD4L6 |
| FOXD4L5 |
| LOC653501 |
| KGFLP2 |
| FAM138C |
| SCARNA8 |
| SNORA17 |
| SNORA43 |
| SNORD62B |
| MSMP |
| SNORD90 |
| MIR548D1 |
| MIR600 |
| MIR601 |
| MIR602 |
| RNF208 |
| FAM75A3 |
| FAM75A5 |
| DNLZ |
| FAM74A3 |
| ANKRD20A4 |
| CCDC29 |
| FAM166B |
| FOXD4L2 |
| CDKN2BAS |
| SNORD121A |
| SNORD121B |
| SNORA84 |
| SNORA70C |
| MIR876 |
| MIR873 |
| LOC100128076 |
| C9orf110 |
| C9orf130 |
| LOC100129034 |
| LOC100129066 |
| LOC100130426 |
| LOC100131193 |
| FAM157B |
| FAM27C |
| FAM95B1 |
| FAM27B |
| LOC100133920 |
| RNU6ATAC |
| NCRNA00092 |
| LOC100272217 |
| WASH1 |
| LOC100289341 |
This is the comprehensive list of deleted genes in the wide peak for 13q32.3.
Table S27. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BRCA2 |
| CDX2 |
| ERCC5 |
| FLT3 |
| LCP1 |
| RB1 |
| LHFP |
| hsa-mir-1267 |
| hsa-mir-623 |
| hsa-mir-92a-1 |
| hsa-mir-622 |
| hsa-mir-1297 |
| hsa-mir-15a |
| hsa-mir-621 |
| hsa-mir-320d-1 |
| PARP4 |
| ALOX5AP |
| ATP12A |
| ATP4B |
| ATP7B |
| KLF5 |
| BTF3L1 |
| CDK8 |
| RCBTB2 |
| CLN5 |
| COL4A1 |
| COL4A2 |
| CPB2 |
| DACH1 |
| DCT |
| GPR183 |
| EDNRB |
| EFNB2 |
| ELF1 |
| ESD |
| F7 |
| F10 |
| FGF9 |
| FGF14 |
| GPC5 |
| FOXO1 |
| FLT1 |
| GAS6 |
| GJA3 |
| GJB2 |
| GPR12 |
| GPR18 |
| MLNR |
| GTF2F2 |
| GTF3A |
| GUCY1B2 |
| HMGB1 |
| HTR2A |
| ING1 |
| PDX1 |
| KPNA3 |
| IPO5 |
| LAMP1 |
| LIG4 |
| LMO7 |
| MAB21L1 |
| SMAD9 |
| MIPEP |
| NEK3 |
| PABPC3 |
| PCCA |
| PCDH8 |
| PCDH9 |
| UBL3 |
| POU4F1 |
| DNAJC3 |
| RAP2A |
| RFC3 |
| RFXAP |
| GRK1 |
| RNF6 |
| RPL21 |
| ATXN8OS |
| SGCG |
| SLC7A1 |
| SLC10A2 |
| SLC15A1 |
| SOX1 |
| TFDP1 |
| TPP2 |
| TPT1 |
| TRPC4 |
| TUBA3C |
| UCHL3 |
| ZIC2 |
| ZMYM2 |
| IFT88 |
| STK24 |
| CUL4A |
| TNFSF11 |
| IRS2 |
| SCEL |
| SUCLA2 |
| DLEU2 |
| TSC22D1 |
| PROZ |
| ARHGEF7 |
| CDC16 |
| CCNA1 |
| CLDN10 |
| MTMR6 |
| DCLK1 |
| ZMYM5 |
| ITGBL1 |
| KL |
| TM9SF2 |
| ITM2B |
| MTRF1 |
| UTP14C |
| NUPL1 |
| TBC1D4 |
| GPC6 |
| FRY |
| MBNL2 |
| FARP1 |
| LPAR6 |
| SLC25A15 |
| TRIM13 |
| USPL1 |
| MRPS31 |
| SPRY2 |
| ABCC4 |
| SAP18 |
| DLEU1 |
| TUBGCP3 |
| N4BP2L2 |
| PIBF1 |
| OLFM4 |
| POSTN |
| TNFSF13B |
| GJB6 |
| HSPH1 |
| WASF3 |
| SUGT1 |
| LECT1 |
| SOX21 |
| WBP4 |
| AKAP11 |
| KLF12 |
| EXOSC8 |
| RASA3 |
| FNDC3A |
| DZIP1 |
| DIS3 |
| MYO16 |
| PDS5B |
| MYCBP2 |
| KIAA0564 |
| ZC3H13 |
| SPG20 |
| LRCH1 |
| ATP11A |
| MCF2L |
| MTUS2 |
| DOCK9 |
| TGDS |
| SLITRK5 |
| FBXL3 |
| SACS |
| INTS6 |
| LATS2 |
| CKAP2 |
| OR7E37P |
| NUFIP1 |
| SNORD102 |
| NBEA |
| OXGR1 |
| PCDH17 |
| C13orf15 |
| MED4 |
| DNAJC15 |
| ALG5 |
| VPS36 |
| POLR1D |
| CRYL1 |
| PHF11 |
| POMP |
| UFM1 |
| ATP8A2 |
| IL17D |
| NDFIP2 |
| MPHOSPH8 |
| BIVM |
| SOHLH2 |
| TMCO3 |
| ENOX1 |
| ARGLU1 |
| DCUN1D2 |
| RCBTB1 |
| PSPC1 |
| NUDT15 |
| KIAA1704 |
| TNFRSF19 |
| FAM48A |
| ANKRD10 |
| RAB20 |
| CARKD |
| UGGT2 |
| PCID2 |
| CENPJ |
| THSD1 |
| RNF17 |
| CYSLTR2 |
| C13orf1 |
| COG6 |
| KLHL1 |
| RBM26 |
| XPO4 |
| PCDH20 |
| UPF3A |
| MRP63 |
| KDELC1 |
| CARS2 |
| RNF219 |
| NARG1L |
| RNASEH2B |
| DHRS12 |
| GRTP1 |
| C13orf34 |
| C13orf18 |
| C13orf23 |
| TDRD3 |
| CDADC1 |
| CAB39L |
| DIAPH3 |
| CCDC70 |
| COG3 |
| SETDB2 |
| KATNAL1 |
| KBTBD7 |
| SLITRK6 |
| EBPL |
| TMTC4 |
| C13orf33 |
| ABHD13 |
| ZIC5 |
| A2LD1 |
| KBTBD6 |
| STARD13 |
| N4BP2L1 |
| C13orf27 |
| TPTE2 |
| EPSTI1 |
| ADPRHL1 |
| SLITRK1 |
| KCTD12 |
| ARL11 |
| WDFY2 |
| CG030 |
| C13orf16 |
| LOC121838 |
| LOC121952 |
| CSNK1A1L |
| RXFP2 |
| C13orf26 |
| SLAIN1 |
| PRR20A |
| C13orf28 |
| RNF113B |
| LOC144776 |
| C13orf30 |
| C13orf31 |
| HNRNPA1L2 |
| FAM10A4 |
| B3GALTL |
| DGKH |
| CCDC122 |
| GPR180 |
| STOML3 |
| COMMD6 |
| CLYBL |
| C13orf39 |
| EEF1DP3 |
| FAM123A |
| USP12 |
| MTIF3 |
| GSX1 |
| FAM194B |
| SPERT |
| DLEU7 |
| FAM124A |
| LOC220115 |
| LOC220429 |
| N6AMT2 |
| SKA3 |
| EFHA1 |
| SPATA13 |
| LNX2 |
| SLC25A30 |
| ZDHHC20 |
| PAN3 |
| NALCN |
| DKFZp686A1627 |
| HS6ST3 |
| DAOA |
| C13orf29 |
| ZNF828 |
| OR7E156P |
| SUGT1L1 |
| SIAH3 |
| KCNRG |
| FLJ37307 |
| SLC46A3 |
| LOC284232 |
| UBAC2 |
| C1QTNF9 |
| FREM2 |
| NEK5 |
| FAM70B |
| LOC348021 |
| LOC374491 |
| THSD1P |
| KCTD4 |
| RASL11A |
| C1QTNF9B |
| SHISA2 |
| NHLRC3 |
| SERP2 |
| C13orf36 |
| C13orf35 |
| MIR15A |
| MIR16-1 |
| MIR17 |
| MIR18A |
| MIR19A |
| MIR19B1 |
| MIR20A |
| MIR92A1 |
| MIR17HG |
| ATP5EP2 |
| ALG11 |
| C13orf37 |
| PCOTH |
| SNORA27 |
| FLJ44054 |
| LOC643677 |
| LOC646405 |
| PRHOXNB |
| ZAR1L |
| LOC646982 |
| SERPINE3 |
| LOC647288 |
| SNORA31 |
| MIR548A2 |
| MIR621 |
| MIR623 |
| FAM155A |
| C13orf38 |
| PRR20B |
| PRR20C |
| PRR20D |
| PRR20E |
| LOC100101938 |
| RPL21P28 |
| FKSG29 |
| LOC100188949 |
| LOC100190939 |
| LOC100288730 |
Table 3. Get Full Table Arm-level significance table - 14 significant results found.
| Arm | # Genes | Amp Frequency | Amp Z score | Amp Q value | Del Frequency | Del Z score | Del Q value |
|---|---|---|---|---|---|---|---|
| 1p | 1731 | 0.34 | 3.13 | 0.0106 | 0.04 | -1.51 | 0.991 |
| 1q | 1572 | 0.56 | 6.73 | 1.67e-10 | 0.00 | -1.7 | 0.991 |
| 2p | 753 | 0.14 | -0.2 | 0.987 | 0.00 | -2.35 | 0.991 |
| 2q | 1235 | 0.09 | -1.03 | 0.987 | 0.06 | -1.46 | 0.991 |
| 3p | 853 | 0.30 | 1.86 | 0.175 | 0.53 | 5.85 | 9.73e-08 |
| 3q | 917 | 0.61 | 7.18 | 1.39e-11 | 0.29 | 1.65 | 0.243 |
| 4p | 366 | 0.00 | -1.88 | 0.987 | 0.44 | 4.95 | 4.94e-06 |
| 4q | 865 | 0.00 | -2.23 | 0.987 | 0.22 | 1.19 | 0.35 |
| 5p | 207 | 0.30 | 2.31 | 0.0823 | 0.22 | 1.06 | 0.379 |
| 5q | 1246 | 0.09 | -0.867 | 0.987 | 0.38 | 3.75 | 0.000876 |
| 6p | 937 | 0.28 | 2.05 | 0.13 | 0.15 | -0.0443 | 0.968 |
| 6q | 692 | 0.24 | 1.36 | 0.373 | 0.24 | 1.36 | 0.336 |
| 7p | 508 | 0.06 | -1.34 | 0.987 | 0.15 | -0.0538 | 0.968 |
| 7q | 1071 | 0.09 | -0.911 | 0.987 | 0.12 | -0.485 | 0.991 |
| 8p | 495 | 0.16 | 0.135 | 0.987 | 0.34 | 3.06 | 0.00716 |
| 8q | 697 | 0.26 | 1.67 | 0.231 | 0.18 | 0.412 | 0.737 |
| 9p | 343 | 0.06 | -1.39 | 0.987 | 0.12 | -0.529 | 0.991 |
| 9q | 916 | 0.03 | -1.89 | 0.987 | 0.11 | -0.609 | 0.991 |
| 10p | 312 | 0.11 | -0.635 | 0.987 | 0.24 | 1.49 | 0.297 |
| 10q | 1050 | 0.07 | -1.24 | 0.987 | 0.21 | 0.886 | 0.431 |
| 11p | 731 | 0.04 | -1.55 | 0.987 | 0.31 | 2.7 | 0.0192 |
| 11q | 1279 | 0.05 | -1.27 | 0.987 | 0.46 | 5.03 | 4.74e-06 |
| 12p | 484 | 0.14 | -0.186 | 0.987 | 0.22 | 1.08 | 0.379 |
| 12q | 1162 | 0.03 | -1.9 | 0.987 | 0.11 | -0.616 | 0.991 |
| 13q | 554 | 0.00 | -2.23 | 0.987 | 0.22 | 1.2 | 0.35 |
| 14q | 1144 | 0.09 | -1.03 | 0.987 | 0.06 | -1.46 | 0.991 |
| 15q | 1132 | 0.09 | -1.03 | 0.987 | 0.06 | -1.46 | 0.991 |
| 16p | 719 | 0.06 | -1.5 | 0.987 | 0.06 | -1.5 | 0.991 |
| 16q | 562 | 0.06 | -1.49 | 0.987 | 0.06 | -1.49 | 0.991 |
| 17p | 575 | 0.04 | -1.49 | 0.987 | 0.34 | 3.18 | 0.00568 |
| 17q | 1321 | 0.06 | -1.56 | 0.987 | 0.03 | -1.99 | 0.991 |
| 18p | 117 | 0.10 | -0.697 | 0.987 | 0.21 | 1.01 | 0.382 |
| 18q | 340 | 0.00 | -2.22 | 0.987 | 0.22 | 1.21 | 0.35 |
| 19p | 870 | 0.13 | -0.343 | 0.987 | 0.16 | 0.0818 | 0.959 |
| 19q | 1452 | 0.21 | 0.871 | 0.748 | 0.07 | -1.25 | 0.991 |
| 20p | 295 | 0.19 | 0.748 | 0.806 | 0.00 | -2.26 | 0.991 |
| 20q | 627 | 0.33 | 3.07 | 0.0106 | 0.00 | -2.06 | 0.991 |
| 21q | 422 | 0.12 | -0.466 | 0.987 | 0.09 | -0.894 | 0.991 |
| 22q | 764 | 0.03 | -1.89 | 0.987 | 0.11 | -0.605 | 0.991 |
Table 4. Get Full Table First 10 out of 36 Input Tumor Samples.
| Tumor Sample Names |
|---|
| TCGA-BI-A0VR-01A-11D-A10T-01 |
| TCGA-BI-A0VS-01A-11D-A10T-01 |
| TCGA-BI-A20A-01A-11D-A14V-01 |
| TCGA-C5-A0TN-01A-21D-A14V-01 |
| TCGA-C5-A1BE-01B-11D-A13V-01 |
| TCGA-C5-A1BF-01B-11D-A13V-01 |
| TCGA-C5-A1BI-01B-11D-A13V-01 |
| TCGA-C5-A1BJ-01A-11D-A13V-01 |
| TCGA-C5-A1BK-01B-11D-A13V-01 |
| TCGA-C5-A1BL-01A-11D-A13V-01 |
Figure 3. Segmented copy number profiles in the input data
GISTIC identifies genomic regions that are significantly gained or lost across a set of tumors. It takes segmented copy number ratios as input, separates arm-level events from focal events, and then performs two tests: (i) identifies significantly amplified/deleted chromosome arms; and (ii) identifies regions that are significantly focally amplified or deleted. For the focal analysis, the significance levels (Q values) are calculated by comparing the observed gains/losses at each locus to those obtained by randomly permuting the events along the genome to reflect the null hypothesis that they are all 'passengers' and could have occurred anywhere. The locus-specific significance levels are then corrected for multiple hypothesis testing. The arm-level significance is calculated by comparing the frequency of gains/losses of each arm to the expected rate given its size. The method outputs genomic views of significantly amplified and deleted regions, as well as a table of genes with gain or loss scores. A more in depth discussion of the GISTIC algorithm and its utility is given in [1], [3], and [5].
Regions of the genome that are prone to germ line variations in copy number are excluded from the GISTIC analysis using a list of germ line copy number variations (CNVs). A CNV is a DNA sequence that may be found at different copy numbers in the germ line of two different individuals. Such germ line variations can confound a GISTIC analysis, which finds significant somatic copy number variations in cancer. A more in depth discussion is provided in [6]. GISTIC currently uses two CNV exclusion lists. One is based on the literature describing copy number variation, and a second one comes from an analysis of significant variations among the blood normals in the TCGA data set.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.