This report serves to describe the mutational landscape and properties of a given individual set, as well as rank genes and genesets according to mutational significance. MutSig v1.5 was used to generate the results found in this report.
Working with individual set: LUSC.
Number of patients in set: 178
The input for this pipeline is a set of individuals with the following files associated for each:
1. An annotated .maf file describing the mutations called for the respective individual, and their properties.
2. A .wig file that contains information about the coverage of the sample.
Significantly mutated genes (q ≤ 0.1): 405
Mutations seen in COSMIC: 1
Significantly mutated genes in COSMIC territory: 0
Genes with clustered mutations (&le 3 aa apart): 1
Significantly mutated genesets: 100
Significantly mutated genesets: (excluding sig. mutated genes): 5
Table 1. Get Full Table Table representing breakdown of mutations by type.
| type | count |
|---|---|
| De_novo_Start_InFrame | 3 |
| De_novo_Start_OutOfFrame | 13 |
| Frame_Shift_Del | 524 |
| Frame_Shift_Ins | 132 |
| In_Frame_Del | 85 |
| In_Frame_Ins | 19 |
| Indel | 9 |
| Missense_Mutation | 34224 |
| Nonsense_Mutation | 2598 |
| Nonstop_Mutation | 42 |
| Silent | 13568 |
| Splice_Site_DNP | 36 |
| Splice_Site_Del | 14 |
| Splice_Site_Ins | 10 |
| Splice_Site_SNP | 837 |
| Start_Codon_Del | 3 |
| Total | 52117 |
Table 2. Get Full Table A breakdown of mutation rates per category discovered for this individual set.
| category | n | N | rate | rate_per_mb | relative_rate |
|---|---|---|---|---|---|
| A->T | 9734 | 2640254776 | 3.7e-06 | 3.7 | 0.52 |
| C->(A/T) | 14492 | 2777075010 | 5.2e-06 | 5.2 | 0.73 |
| A->(C/G) | 7317 | 2640254776 | 2.8e-06 | 2.8 | 0.39 |
| C->G | 2664 | 2777075010 | 9.6e-07 | 0.96 | 0.13 |
| indel+null | 4313 | 5417329964 | 8e-07 | 0.8 | 0.11 |
| double_null | 29 | 5417329964 | 5.4e-09 | 0.0054 | 0.00075 |
| Total | 38549 | 5417329964 | 7.1e-06 | 7.1 | 1 |
The x axis represents the samples. The y axis represents the exons, one row per exon, and they are sorted by average coverage across samples. For exons with exactly the same average coverage, they are sorted next by the %GC of the exon. (The secondary sort is especially useful for the zero-coverage exons at the bottom).
Figure 1.

Figure 2.
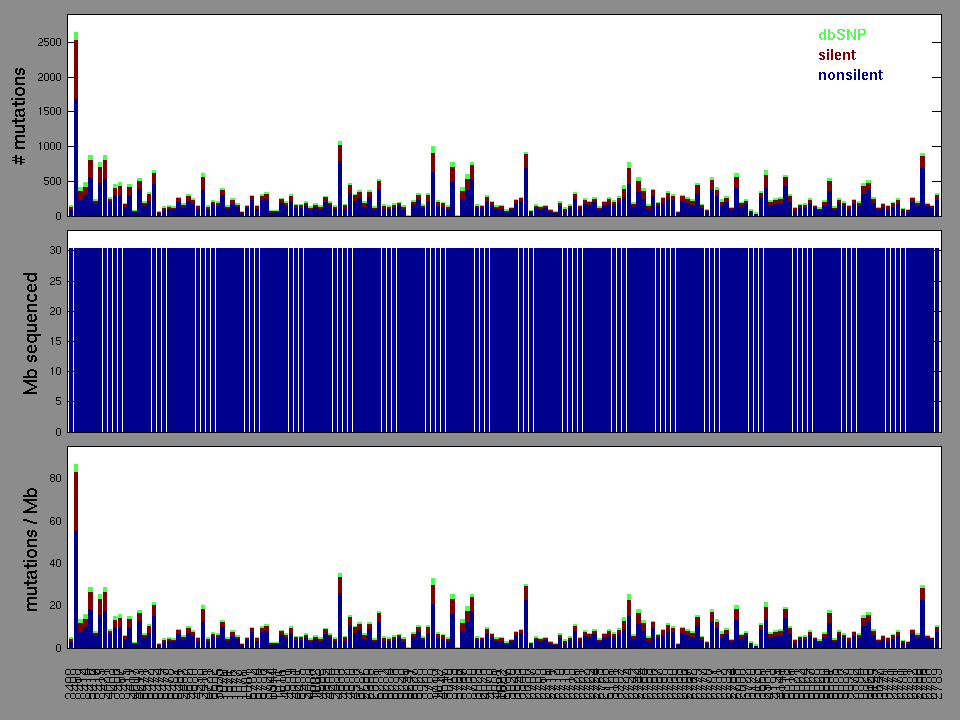
Table 3. Get Full Table A Ranked List of Significantly Mutated Genes. Number of significant genes found: 405. Number of genes displayed: 35
| rank | gene | description | N | n | npat | nsite | nsil | n1 | n2 | n3 | n4 | n5 | n6 | p | q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | TP53 | tumor protein p53 | 228196 | 119 | 117 | 82 | 7 | 23 | 20 | 20 | 13 | 43 | 0 | <1.00e-15 | <1.80e-11 |
| 2 | RYR2 | ryanodine receptor 2 (cardiac) | 2630306 | 88 | 56 | 86 | 22 | 28 | 29 | 18 | 4 | 9 | 0 | 4.22e-15 | 2.80e-11 |
| 3 | CSMD3 | CUB and Sushi multiple domains 3 | 2041660 | 77 | 58 | 77 | 16 | 21 | 22 | 11 | 5 | 16 | 2 | 4.66e-15 | 2.80e-11 |
| 4 | LRP1B | low density lipoprotein-related protein 1B (deleted in tumors) | 2521192 | 67 | 50 | 65 | 12 | 19 | 19 | 18 | 4 | 7 | 0 | 3.79e-14 | 9.71e-11 |
| 5 | CDKN2A | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 83838 | 23 | 23 | 20 | 1 | 1 | 5 | 5 | 0 | 12 | 0 | 3.81e-14 | 9.71e-11 |
| 6 | NFE2L2 | nuclear factor (erythroid-derived 2)-like 2 | 318442 | 21 | 20 | 12 | 0 | 6 | 0 | 13 | 1 | 1 | 0 | 4.60e-14 | 9.71e-11 |
| 7 | CDH10 | cadherin 10, type 2 (T2-cadherin) | 429158 | 27 | 25 | 27 | 7 | 8 | 9 | 3 | 2 | 5 | 0 | 4.70e-14 | 9.71e-11 |
| 8 | HCN1 | hyperpolarization activated cyclic nucleotide-gated potassium channel 1 | 441618 | 27 | 25 | 25 | 11 | 5 | 13 | 2 | 3 | 4 | 0 | 4.75e-14 | 9.71e-11 |
| 9 | ZFHX4 | zinc finger homeobox 4 | 1745824 | 65 | 48 | 65 | 18 | 22 | 24 | 9 | 5 | 5 | 0 | 5.32e-14 | 9.71e-11 |
| 10 | FAM5C | family with sequence similarity 5, member C | 414562 | 24 | 23 | 24 | 3 | 6 | 13 | 4 | 1 | 0 | 0 | 5.38e-14 | 9.71e-11 |
| 11 | FAM135B | family with sequence similarity 135, member B | 764866 | 35 | 29 | 34 | 9 | 11 | 16 | 6 | 0 | 2 | 0 | 8.89e-14 | 1.46e-10 |
| 12 | USH2A | Usher syndrome 2A (autosomal recessive, mild) | 2831446 | 63 | 51 | 63 | 12 | 16 | 25 | 12 | 5 | 5 | 0 | 2.26e-12 | 3.40e-09 |
| 13 | KEAP1 | kelch-like ECH-associated protein 1 | 337310 | 18 | 18 | 16 | 0 | 3 | 10 | 2 | 1 | 2 | 0 | 3.03e-12 | 4.20e-09 |
| 14 | ADAMTS12 | ADAM metallopeptidase with thrombospondin type 1 motif, 12 | 868818 | 30 | 28 | 29 | 6 | 9 | 12 | 5 | 1 | 3 | 0 | 1.05e-11 | 1.35e-08 |
| 15 | TTN | titin | 19694276 | 263 | 124 | 260 | 91 | 69 | 86 | 69 | 17 | 16 | 6 | 1.90e-11 | 2.29e-08 |
| 16 | SI | sucrase-isomaltase (alpha-glucosidase) | 1009616 | 33 | 29 | 33 | 2 | 9 | 13 | 7 | 2 | 2 | 0 | 3.26e-11 | 3.68e-08 |
| 17 | C1orf173 | chromosome 1 open reading frame 173 | 819156 | 27 | 24 | 27 | 8 | 8 | 8 | 6 | 2 | 3 | 0 | 2.97e-10 | 2.98e-07 |
| 18 | OR10G9 | olfactory receptor, family 10, subfamily G, member 9 | 167320 | 13 | 13 | 12 | 3 | 1 | 10 | 1 | 0 | 1 | 0 | 2.98e-10 | 2.98e-07 |
| 19 | CDH12 | cadherin 12, type 2 (N-cadherin 2) | 432362 | 19 | 18 | 19 | 6 | 4 | 8 | 4 | 0 | 3 | 0 | 3.79e-10 | 3.60e-07 |
| 20 | CNTNAP5 | contactin associated protein-like 5 | 680316 | 25 | 22 | 25 | 6 | 5 | 9 | 6 | 2 | 3 | 0 | 4.14e-10 | 3.73e-07 |
| 21 | ZNF804B | zinc finger protein 804B | 723748 | 22 | 21 | 21 | 8 | 7 | 1 | 6 | 4 | 4 | 0 | 5.59e-10 | 4.80e-07 |
| 22 | NAV3 | neuron navigator 3 | 1290144 | 32 | 30 | 32 | 10 | 6 | 16 | 6 | 2 | 2 | 0 | 5.94e-10 | 4.87e-07 |
| 23 | REG1A | regenerating islet-derived 1 alpha (pancreatic stone protein, pancreatic thread protein) | 92738 | 11 | 10 | 11 | 3 | 0 | 6 | 3 | 1 | 1 | 0 | 8.17e-10 | 6.41e-07 |
| 24 | LRRC4C | leucine rich repeat containing 4C | 343006 | 18 | 17 | 18 | 1 | 11 | 3 | 3 | 0 | 1 | 0 | 9.70e-10 | 7.29e-07 |
| 25 | FMN2 | formin 2 | 828412 | 24 | 23 | 22 | 8 | 5 | 11 | 4 | 1 | 3 | 0 | 1.48e-09 | 1.01e-06 |
| 26 | ZIC1 | Zic family member 1 (odd-paired homolog, Drosophila) | 241368 | 16 | 13 | 16 | 4 | 5 | 4 | 6 | 0 | 1 | 0 | 1.49e-09 | 1.01e-06 |
| 27 | PCDH11X | protocadherin 11 X-linked | 731758 | 26 | 23 | 25 | 3 | 10 | 9 | 4 | 2 | 1 | 0 | 1.51e-09 | 1.01e-06 |
| 28 | OR4M2 | olfactory receptor, family 4, subfamily M, member 2 | 168388 | 11 | 11 | 11 | 1 | 3 | 3 | 2 | 2 | 1 | 0 | 3.53e-09 | 2.28e-06 |
| 29 | SPTA1 | spectrin, alpha, erythrocytic 1 (elliptocytosis 2) | 1329304 | 36 | 29 | 36 | 12 | 6 | 16 | 6 | 4 | 4 | 0 | 3.90e-09 | 2.43e-06 |
| 30 | PTEN | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 222144 | 11 | 11 | 11 | 0 | 2 | 1 | 1 | 1 | 6 | 0 | 4.86e-09 | 2.92e-06 |
| 31 | OR5D18 | olfactory receptor, family 5, subfamily D, member 18 | 168388 | 11 | 11 | 11 | 4 | 4 | 4 | 2 | 0 | 1 | 0 | 6.55e-09 | 3.81e-06 |
| 32 | COL11A1 | collagen, type XI, alpha 1 | 1040588 | 31 | 25 | 31 | 7 | 8 | 10 | 7 | 0 | 6 | 0 | 9.01e-09 | 5.08e-06 |
| 33 | ELTD1 | EGF, latrophilin and seven transmembrane domain containing 1 | 375046 | 15 | 15 | 15 | 1 | 4 | 3 | 1 | 2 | 5 | 0 | 1.14e-08 | 6.24e-06 |
| 34 | REG3A | regenerating islet-derived 3 alpha | 97544 | 9 | 9 | 9 | 2 | 2 | 3 | 3 | 0 | 1 | 0 | 1.18e-08 | 6.26e-06 |
| 35 | PIK3CA | phosphoinositide-3-kinase, catalytic, alpha polypeptide | 585086 | 21 | 20 | 10 | 1 | 13 | 3 | 5 | 0 | 0 | 0 | 1.38e-08 | 7.13e-06 |
Note:
N - number of sequenced bases in this gene across the individual set.
n - number of (nonsilent) mutations in this gene across the individual set.
npat - number of patients (individuals) with at least one nonsilent mutation.
nsite - number of unique sites having a non-silent mutation.
nsil - number of silent mutations in this gene across the individual set.
n1 - number of nonsilent mutations of type: A->T .
n2 - number of nonsilent mutations of type: C->(A/T) .
n3 - number of nonsilent mutations of type: A->(C/G) .
n4 - number of nonsilent mutations of type: C->G .
n5 - number of nonsilent mutations of type: indel+null .
null - mutation category that includes nonsense, frameshift, splice-site mutations
p_classic = p-value for the observed amount of nonsilent mutations being elevated in this gene
p_ns_s = p-value for the observed nonsilent/silent ratio being elevated in this gene
p = p-value (overall)
q = q-value, False Discovery Rate (Benjamini-Hochberg procedure)
In this analysis, COSMIC is used as a filter to increase power by restricting the territory of each gene. Cosmic version: v48.
Table 4. Get Full Table Significantly mutated genes (COSMIC territory only). To access the database please go to: COSMIC. Number of significant genes found: 0. Number of genes displayed: 10
| rank | gene | description | n | cos | n_cos | N_cos | cos_ev | p | q |
|---|---|---|---|---|---|---|---|---|---|
| 1 | SYNE1 | spectrin repeat containing, nuclear envelope 1 | 45 | 22 | 1 | 3916 | 3 | 0.027 | 1 |
| 2 | A4GNT | alpha-1,4-N-acetylglucosaminyltransferase | 5 | 0 | 0 | 0 | 0 | 1 | 1 |
| 3 | AACS | acetoacetyl-CoA synthetase | 2 | 0 | 0 | 0 | 0 | 1 | 1 |
| 4 | ABCA9 | ATP-binding cassette, sub-family A (ABC1), member 9 | 8 | 0 | 0 | 0 | 0 | 1 | 1 |
| 5 | ABCC10 | ATP-binding cassette, sub-family C (CFTR/MRP), member 10 | 4 | 0 | 0 | 0 | 0 | 1 | 1 |
| 6 | ABCF2 | ATP-binding cassette, sub-family F (GCN20), member 2 | 6 | 0 | 0 | 0 | 0 | 1 | 1 |
| 7 | ABHD2 | abhydrolase domain containing 2 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| 8 | ABHD4 | abhydrolase domain containing 4 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| 9 | ACADS | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 3 | 0 | 0 | 0 | 0 | 1 | 1 |
| 10 | ACOT11 | acyl-CoA thioesterase 11 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
Note:
n - number of (nonsilent) mutations in this gene across the individual set.
cos = number of unique mutated sites in this gene in COSMIC
n_cos = overlap between n and cos.
N_cos = number of individuals times cos.
cos_ev = total evidence: number of reports in COSMIC for mutations seen in this gene.
p = p-value for seeing the observed amount of overlap in this gene)
q = q-value, False Discovery Rate (Benjamini-Hochberg procedure)
Table 5. Get Full Table Genes with Clustered Mutations
| num | gene | desc | n | mindist | npairs3 | npairs12 |
|---|---|---|---|---|---|---|
| 1374 | C20orf26 | chromosome 20 open reading frame 26 | 18 | 0 | 1 | 1 |
| 2643 | CUBN | cubilin (intrinsic factor-cobalamin receptor) | 22 | 4 | 0 | 1 |
| 27 | ABCA13 | ATP-binding cassette, sub-family A (ABC1), member 13 | 27 | 8 | 0 | 1 |
| 2693 | CYP11B2 | cytochrome P450, family 11, subfamily B, polypeptide 2 | 6 | 10 | 0 | 1 |
| 6712 | MUC2 | mucin 2, oligomeric mucus/gel-forming | 9 | 11 | 0 | 1 |
| 4450 | GPR98 | G protein-coupled receptor 98 | 28 | 12 | 0 | 0 |
| 11716 | WNK1 | WNK lysine deficient protein kinase 1 | 15 | 23 | 0 | 0 |
| 10440 | SYNE1 | spectrin repeat containing, nuclear envelope 1 | 45 | 34 | 0 | 0 |
| 11089 | TRANK1 | 7 | 47 | 0 | 0 | |
| 6717 | MUC6 | mucin 6, oligomeric mucus/gel-forming | 10 | 59 | 0 | 0 |
Note:
n - number of mutations in this gene in the individual set.
mindist - distance (in aa) between closest pair of mutations in this gene
npairs3 - how many pairs of mutations are within 3 aa of each other.
npairs12 - how many pairs of mutations are within 12 aa of each other.
Table 6. Get Full Table A Ranked List of Significantly Mutated Genesets. (Source: MSigDB GSEA Cannonical Pathway Set).Number of significant genesets found: 100. Number of genesets displayed: 10
| rank | geneset | description | genes | N_genes | mut_tally | N | n | npat | nsite | nsil | n1 | n2 | n3 | n4 | n5 | n6 | p | q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HSA04210_APOPTOSIS | Genes involved in apoptosis | AIFM1, AKT1, AKT2, AKT3, APAF1, ATM, BAD, BAX, BCL2, BCL2L1, BID, BIRC2, BIRC3, BIRC4, CAPN1, CAPN2, CASP10, CASP3, CASP6, CASP7, CASP8, CASP9, CFLAR, CHP, CHUK, CSF2RB, CYCS, DFFA, DFFB, ENDOG, FADD, FAS, FASLG, IKBKB, IKBKG, IL1A, IL1B, IL1R1, IL1RAP, IL3, IL3RA, IRAK1, IRAK2, IRAK3, IRAK4, MAP3K14, MYD88, NFKB1, NFKB2, NFKBIA, NGFB, NTRK1, PIK3CA, PIK3CB, PIK3CD, PIK3CG, PIK3R1, PIK3R2, PIK3R3, PIK3R5, PPP3CA, PPP3CB, PPP3CC, PPP3R1, PPP3R2, PRKACA, PRKACB, PRKACG, PRKAR1A, PRKAR1B, PRKAR2A, PRKAR2B, RELA, RIPK1, TNF, TNFRSF10A, TNFRSF10B, TNFRSF10C, TNFRSF10D, TNFRSF1A, TNFSF10, TP53, TRADD, TRAF2 | 80 | AIFM1(2), AKT1(1), AKT3(2), APAF1(1), ATM(7), BAX(1), BCL2L1(2), BID(1), BIRC3(3), CAPN1(2), CAPN2(2), CASP10(4), CASP6(1), CASP7(2), CASP8(1), CASP9(1), CFLAR(1), CHP(2), CHUK(2), CSF2RB(5), DFFA(1), DFFB(2), FADD(1), FAS(1), FASLG(1), IKBKB(3), IL1B(1), IL1RAP(2), IRAK1(4), IRAK2(1), IRAK3(3), MAP3K14(1), NFKB1(2), NFKB2(2), NTRK1(3), PIK3CA(21), PIK3CB(1), PIK3CD(3), PIK3CG(10), PIK3R1(1), PIK3R2(4), PIK3R3(2), PIK3R5(2), PPP3CA(2), PPP3CB(5), PPP3CC(2), PPP3R1(1), PPP3R2(3), PRKACA(1), PRKACB(2), PRKACG(1), PRKAR1A(3), PRKAR1B(1), PRKAR2B(3), RELA(3), TNFRSF10A(1), TNFRSF10B(1), TNFRSF10D(1), TNFRSF1A(2), TP53(119), TRADD(1) | 22576808 | 267 | 147 | 219 | 57 | 74 | 68 | 35 | 30 | 60 | 0 | <1.00e-15 | <1.93e-13 |
| 2 | RNAPATHWAY | dsRNA-activated protein kinase phosphorylates elF2a, which generally inhibits translation, and activates NF-kB to provoke inflammation. | CHUK, DNAJC3, EIF2S1, EIF2S2, MAP3K14, NFKB1, NFKBIA, PRKR, RELA, TP53 | 9 | CHUK(2), MAP3K14(1), NFKB1(2), RELA(3), TP53(119) | 2739598 | 127 | 118 | 90 | 9 | 24 | 22 | 21 | 14 | 46 | 0 | 2.55e-15 | 1.93e-13 |
| 3 | ST_JNK_MAPK_PATHWAY | JNKs are MAP kinases regulated by several levels of kinases (MAPKK, MAPKKK) and phosphorylate transcription factors and regulatory proteins. | AKT1, ATF2, CDC42, DLD, DUSP10, DUSP4, DUSP8, GAB1, GADD45A, GCK, IL1R1, JUN, MAP2K4, MAP2K5, MAP2K7, MAP3K1, MAP3K10, MAP3K11, MAP3K12, MAP3K13, MAP3K2, MAP3K3, MAP3K4, MAP3K5, MAP3K7, MAP3K7IP1, MAP3K7IP2, MAP3K9, MAPK10, MAPK7, MAPK8, MAPK9, MYEF2, NFATC3, NR2C2, PAPPA, SHC1, TP53, TRAF6, ZAK | 38 | AKT1(1), ATF2(2), CDC42(1), DLD(2), DUSP10(3), DUSP4(1), GAB1(2), GADD45A(1), GCK(1), JUN(2), MAP2K5(2), MAP3K1(3), MAP3K11(2), MAP3K12(2), MAP3K13(5), MAP3K4(4), MAP3K5(3), MAP3K7(3), MAP3K9(4), MAPK10(1), MAPK7(1), MAPK8(2), MAPK9(1), MYEF2(3), NFATC3(3), NR2C2(1), PAPPA(8), SHC1(1), TP53(119), TRAF6(1), ZAK(1) | 13764918 | 186 | 133 | 149 | 32 | 41 | 46 | 33 | 19 | 47 | 0 | 2.78e-15 | 1.93e-13 |
| 4 | PLK3PATHWAY | Active Plk3 phosphorylates CDC25c, blocking the G2/M transition, and phosphorylates p53 to induce apoptosis. | ATM, ATR, CDC25C, CHEK1, CHEK2, CNK, TP53, YWHAH | 7 | ATM(7), ATR(11), CDC25C(1), CHEK1(4), CHEK2(3), TP53(119) | 4293716 | 145 | 124 | 108 | 8 | 29 | 26 | 25 | 18 | 47 | 0 | 2.89e-15 | 1.93e-13 |
| 5 | APOPTOSIS | APAF1, BAD, BAK1, BCL2L7P1, BAX, BCL2, BCL2L1, BCL2L11, BID, BIRC2, BIRC3, BIRC4, BIRC5, BNIP3L, CASP1, CASP10, CASP1, COPl, CASP2, CASP3, CASP4, CASP6, CASP7, CASP8, CASP9, CHUK, CYCS, DFFA, DFFB, FADD, FAS, FASLG, GZMB, HELLS, HRK, IKBKB, IKBKG, IRF1, IRF2, IRF3, IRF4, IRF5, IRF6, IRF7, JUN, LTA, MAP2K4, MAP3K1, MAPK10, MDM2, MYC, NFKB1, NFKBIA, NFKBIB, NFKBIE, PRF1, RELA, RIPK1, TNF, TNFRSF10B, TNFRSF1A, TNFRSF1B, TNFRSF21, TNFRSF25, TNFRSF25, PLEKHG5, TNFSF10, TP53, TP73, TRADD, TRAF1, TRAF2, TRAF3 | 66 | APAF1(1), BAK1(1), BAX(1), BCL2L1(2), BID(1), BIRC3(3), BNIP3L(1), CASP1(2), CASP10(4), CASP2(1), CASP4(3), CASP6(1), CASP7(2), CASP8(1), CASP9(1), CHUK(2), DFFA(1), DFFB(2), FADD(1), FAS(1), FASLG(1), GZMB(1), IKBKB(3), IRF1(1), IRF3(1), IRF4(3), IRF6(5), JUN(2), LTA(3), MAP3K1(3), MAPK10(1), MDM2(1), MYC(1), NFKB1(2), NFKBIB(1), PLEKHG5(1), PRF1(2), RELA(3), TNFRSF10B(1), TNFRSF1A(2), TNFRSF1B(1), TNFRSF21(3), TP53(119), TRADD(1), TRAF1(4), TRAF3(1) | 15688386 | 199 | 135 | 162 | 41 | 39 | 46 | 33 | 23 | 58 | 0 | 3.89e-15 | 1.93e-13 | |
| 6 | G1_TO_S_CELL_CYCLE_REACTOME | ATM, CCNA1, CCNB1, CCND1, CCND2, CCND3, CCNE1, CCNE2, CCNG2, CCNH, CDC25A, CDC45L, CDK2, CDK4, CDK7, CDKN1A, CDKN1B, CDKN1C, CDKN2A, CDKN2B, CDKN2C, CDKN2D, CREB3, CREB3L1, CREB3L3, CREB3L4, CREBL1, CREBL1, TNXB, E2F1, E2F2, E2F3, E2F4, E2F5, E2F6, FLJ14001, GADD45A, GBA2, MCM2, MCM3, MCM4, MCM5, MCM6, MCM7, MDM2, MNAT1, MYC, MYT1, NACA, NACA, FKSG17, ORC1L, ORC2L, ORC3L, ORC4L, ORC5L, ORC6L, PCNA, POLA2, POLE, POLE2, PRIM1, PRIM2A, RB1, RBL1, RPA1, RPA2, RPA3, TFDP1, TFDP2, TP53, WEE1 | 65 | ATM(7), CCNA1(1), CCNB1(1), CCND1(1), CCND2(3), CCND3(1), CCNE2(2), CDK2(1), CDK7(1), CDKN2A(23), CDKN2C(1), CDKN2D(1), CREB3(2), CREB3L3(1), CREB3L4(2), E2F1(3), GADD45A(1), GBA2(2), MCM2(1), MCM3(1), MCM4(4), MCM5(6), MCM6(2), MCM7(3), MDM2(1), MYC(1), ORC1L(1), ORC2L(1), ORC3L(1), ORC5L(1), POLA2(3), POLE2(2), PRIM1(2), RB1(7), RBL1(6), TFDP2(1), TNXB(16), TP53(119), WEE1(2) | 20927638 | 235 | 137 | 195 | 36 | 45 | 63 | 39 | 20 | 68 | 0 | 4.00e-15 | 1.93e-13 | |
| 7 | ARFPATHWAY | Cyclin-dependent kinase inhibitor 2A is a tumor suppressor that induces G1 arrest and can activate the p53 pathway, leading to G2/M arrest. | ABL1, CDKN2A, E2F1, MDM2, MYC, PIK3CA, PIK3R1, POLR1A, POLR1B, POLR1C, POLR1D, RAC1, RB1, TBX2, TP53, TWIST1 | 16 | ABL1(2), CDKN2A(23), E2F1(3), MDM2(1), MYC(1), PIK3CA(21), PIK3R1(1), POLR1A(4), POLR1B(4), POLR1C(1), POLR1D(1), RAC1(2), RB1(7), TBX2(3), TP53(119), TWIST1(1) | 5437544 | 194 | 133 | 143 | 19 | 44 | 41 | 30 | 17 | 62 | 0 | 4.00e-15 | 1.93e-13 |
| 8 | ATMPATHWAY | The tumor-suppressing protein kinase ATM responds to radiation-induced DNA damage by blocking cell-cycle progression and activating DNA repair. | ABL1, ATM, BRCA1, CDKN1A, CHEK1, CHEK2, GADD45A, JUN, MAPK8, MDM2, MRE11A, NBS1, NFKB1, NFKBIA, RAD50, RAD51, RBBP8, RELA, TP53, TP73 | 19 | ABL1(2), ATM(7), BRCA1(6), CHEK1(4), CHEK2(3), GADD45A(1), JUN(2), MAPK8(2), MDM2(1), MRE11A(2), NFKB1(2), RAD50(3), RAD51(1), RBBP8(1), RELA(3), TP53(119) | 8101670 | 159 | 122 | 122 | 16 | 30 | 35 | 24 | 21 | 49 | 0 | 4.00e-15 | 1.93e-13 |
| 9 | TERTPATHWAY | hTERC, the RNA subunit of telomerase, and hTERT, the catalytic protein subunit, are required for telomerase activity and are overexpressed in many cancers. | HDAC1, MAX, MYC, SP1, SP3, TP53, WT1, ZNF42 | 7 | HDAC1(2), MYC(1), SP1(1), SP3(1), TP53(119), WT1(4) | 1894810 | 128 | 122 | 91 | 11 | 25 | 21 | 24 | 15 | 43 | 0 | 4.33e-15 | 1.93e-13 |
| 10 | RBPATHWAY | The ATM protein kinase recognizes DNA damage and blocks cell cycle progression by phosphorylating chk1 and p53, which normally inhibits Rb to allow G1/S transitions. | ATM, CDC2, CDC25A, CDC25B, CDC25C, CDK2, CDK4, CHEK1, MYT1, RB1, TP53, WEE1, YWHAH | 12 | ATM(7), CDC25B(3), CDC25C(1), CDK2(1), CHEK1(4), RB1(7), TP53(119), WEE1(2) | 4759720 | 144 | 124 | 107 | 10 | 27 | 30 | 22 | 14 | 51 | 0 | 4.77e-15 | 1.93e-13 |
Table 7. Get Full Table A Ranked List of Significantly Mutated Genesets (Excluding Significantly Mutated Genes). Number of significant genesets found: 5. Number of genesets displayed: 10
| rank | geneset | description | genes | N_genes | mut_tally | N | n | npat | nsite | nsil | n1 | n2 | n3 | n4 | n5 | n6 | p | q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HSA04080_NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION | Genes involved in neuroactive ligand-receptor interaction | ADCYAP1R1, ADORA1, ADORA2A, ADORA2B, ADORA3, ADRA1A, ADRA1B, ADRA2A, ADRA2B, ADRA2C, ADRB1, ADRB2, ADRB3, AGTR1, AGTR2, AGTRL1, AVPR1A, AVPR1B, AVPR2, BDKRB1, BDKRB2, BRS3, C3AR1, C5AR1, CALCR, CALCRL, CCKAR, CCKBR, CGA, CHRM1, CHRM2, CHRM3, CHRM4, CHRM5, CNR1, CNR2, CRHR1, CRHR2, CTSG, CYSLTR1, CYSLTR2, DRD1, DRD2, DRD3, DRD4, DRD5, EDG1, EDG2, EDG3, EDG4, EDG5, EDG6, EDG7, EDG8, EDNRA, EDNRB, F2, F2R, F2RL1, F2RL2, F2RL3, FPR1, FPRL1, FPRL2, FSHB, FSHR, GABBR1, GABBR2, GABRA1, GABRA2, GABRA3, GABRA4, GABRA5, GABRA6, GABRB1, GABRB2, GABRB3, GABRD, GABRE, GABRG1, GABRG2, GABRG3, GABRP, GABRQ, GABRR1, GABRR2, GALR1, GALR2, GALR3, GCGR, GH1, GH2, GHR, GHRHR, GHSR, GIPR, GLP1R, GLP2R, GLRA1, GLRA2, GLRA3, GLRB, GNRHR, GPR156, GPR23, GPR35, GPR50, GPR63, GPR83, GRIA1, GRIA2, GRIA3, GRIA4, GRID1, GRID2, GRIK1, GRIK2, GRIK3, GRIK4, GRIK5, GRIN1, GRIN2A, GRIN2B, GRIN2C, GRIN2D, GRIN3A, GRIN3B, GRM1, GRM2, GRM3, GRM4, GRM5, GRM6, GRM7, GRM8, GRPR, GZMA, HCRTR1, HCRTR2, HRH1, HRH2, HRH3, HRH4, HTR1A, HTR1B, HTR1D, HTR1E, HTR1F, HTR2A, HTR2B, HTR2C, HTR4, HTR5A, HTR6, HTR7, KISS1R, LEP, LEPR, LHB, LHCGR, LTB4R, LTB4R2, MAS1, MC1R, MC2R, MC3R, MC4R, MC5R, MCHR1, MCHR2, MLNR, MTNR1A, MTNR1B, NMBR, NMUR1, NMUR2, NPBWR1, NPBWR2, NPFFR1, NPFFR2, NPY1R, NPY2R, NPY5R, NR3C1, NTSR1, NTSR2, OPRD1, OPRK1, OPRL1, OPRM1, OXTR, P2RX1, P2RX2, P2RX3, P2RX4, P2RX5, P2RX7, P2RXL1, P2RY1, P2RY10, P2RY11, P2RY13, P2RY14, P2RY2, P2RY4, P2RY5, P2RY6, P2RY8, PARD3, PPYR1, PRL, PRLHR, PRLR, PRSS1, PRSS2, PRSS3, PTAFR, PTGDR, PTGER1, PTGER2, PTGER3, PTGER4, PTGFR, PTGIR, PTH2R, PTHR1, RXFP1, RXFP2, SCTR, SSTR1, SSTR2, SSTR3, SSTR4, SSTR5, TAAR1, TAAR2, TAAR5, TAAR6, TAAR8, TAAR9, TACR1, TACR2, TACR3, TBXA2R, THRA, THRB, TRHR, TRPV1, TSHB, TSHR, TSPO, UTS2R, VIPR1, VIPR2 | 222 | ADORA1(4), ADORA2A(6), ADORA2B(1), ADRA1A(2), ADRA1B(4), ADRA2B(4), ADRA2C(2), ADRB2(3), AGTR1(3), AGTR2(4), AVPR1A(3), AVPR1B(2), AVPR2(1), BDKRB1(3), BDKRB2(1), C3AR1(1), C5AR1(1), CALCR(6), CALCRL(4), CCKAR(2), CCKBR(1), CHRM2(6), CHRM3(5), CHRM4(1), CHRM5(2), CNR1(3), CNR2(3), CRHR1(4), CRHR2(3), CTSG(4), CYSLTR1(3), CYSLTR2(1), DRD1(1), DRD2(2), DRD3(5), DRD4(1), DRD5(3), EDNRA(2), EDNRB(5), F2R(2), F2RL1(3), F2RL2(1), F2RL3(2), FPR1(1), FSHB(1), GABBR1(3), GABBR2(3), GABRA1(6), GABRA3(5), GABRA4(3), GABRA5(4), GABRB1(3), GABRB2(6), GABRD(2), GABRE(4), GABRG2(5), GABRP(2), GABRQ(6), GABRR1(2), GABRR2(2), GALR1(3), GH1(1), GH2(2), GHR(2), GHRHR(3), GHSR(3), GLP1R(1), GLP2R(4), GLRA1(2), GLRA2(5), GLRA3(3), GLRB(3), GPR156(2), GPR35(1), GPR50(4), GPR63(2), GPR83(2), GRIA1(6), GRIA2(3), GRIA3(9), GRIA4(7), GRID2(12), GRIK1(4), GRIK2(8), GRIK3(7), GRIK4(4), GRIK5(1), GRIN1(1), GRIN2A(13), GRIN2C(2), GRIN2D(3), GRIN3A(8), GRIN3B(4), GRM2(3), GRM4(1), GRM6(4), GRM7(8), GRM8(9), GRPR(2), GZMA(4), HCRTR2(3), HRH1(3), HRH2(2), HRH3(1), HRH4(3), HTR1B(3), HTR1D(1), HTR1E(5), HTR1F(2), HTR2A(1), HTR2B(2), HTR2C(3), HTR4(3), HTR5A(3), HTR6(1), HTR7(3), LEPR(10), LHB(1), LHCGR(8), LTB4R2(1), MAS1(3), MC2R(2), MC3R(3), MC4R(1), MC5R(3), MCHR1(2), MCHR2(3), MTNR1A(1), NMBR(2), NMUR2(2), NPBWR1(1), NPY1R(3), NPY2R(2), NPY5R(2), NTSR1(3), NTSR2(3), OPRD1(2), OPRK1(1), OPRM1(2), OXTR(1), P2RX2(1), P2RX3(5), P2RX4(2), P2RX7(1), P2RY1(1), P2RY10(2), P2RY13(1), P2RY14(3), P2RY2(2), P2RY4(1), P2RY6(2), PARD3(5), PPYR1(3), PRL(1), PRLHR(1), PRLR(5), PRSS1(3), PRSS3(1), PTAFR(3), PTGDR(5), PTGER2(1), PTGER3(5), PTGER4(2), PTGFR(2), PTGIR(1), PTH2R(5), RXFP1(7), RXFP2(1), SSTR1(1), SSTR2(2), SSTR3(1), SSTR4(2), SSTR5(1), TAAR1(4), TAAR2(2), TAAR5(5), TAAR6(4), TAAR8(2), TAAR9(2), TACR1(2), TACR3(3), TBXA2R(2), THRA(2), THRB(1), TRHR(1), TRPV1(2), TSHR(1) | 55037422 | 557 | 157 | 554 | 207 | 157 | 205 | 106 | 28 | 61 | 0 | 2.7e-07 | 0.00017 |
| 2 | GPCRDB_CLASS_A_RHODOPSIN_LIKE | ADORA1, ADORA2A, ADORA2B, ADORA3, ADRA1A, ADRA1B, ADRA1D, ADRA2A, ADRA2C, ADRB1, ADRB2, ADRB3, AGTR1, AGTR2, AGTRL1, AVPR1A, AVPR1B, AVPR2, BDKRB1, BDKRB2, BLR1, BRS3, C3AR1, C5R1, CCBP2, CCKAR, CCKBR, CCR1, CCR10, CCR2, CCR3, CCR4, CCR5, CCR6, CCR7, CCR8, CCR9, CCRL1, CCRL2, CHML, CHRM1, CHRM2, CHRM3, CHRM4, CHRM5, CMKLR1, CMKOR1, CNR1, CNR2, CX3CR1, CXCR3, CXCR4, DRD1, DRD2, DRD3, DRD4, DRD5, EDNRA, EDNRB, ELA3A, F2R, F2RL1, F2RL2, F2RL3, FPR1, FPRL1, FPRL2, FSHR, GALR1, GALR2, GALR3, GALT, GHSR, GNB2L1, GPR10, GPR147, GPR17, GPR173, GPR174, GPR23, GPR24, GPR27, GPR3, GPR30, GPR35, GPR37, GPR37L1, GPR4, GPR44, GPR50, GPR6, GPR63, GPR74, GPR77, GPR83, GPR85, GPR87, GPR92, GRPR, HCRTR1, HCRTR2, HRH1, HRH2, HRH3, HTR1A, HTR1B, HTR1D, HTR1E, HTR1F, HTR2A, HTR2B, HTR2C, HTR4, HTR5A, HTR6, HTR7, HTR7, LOC93164, IL8RA, IL8RB, LHCGR, LTB4R, MAS1, MC1R, MC3R, MC4R, MC5R, MLNR, MTNR1A, MTNR1B, NMBR, NMUR1, NMUR2, NPY1R, NPY2R, NPY5R, NPY6R, NTSR1, NTSR2, OPN1SW, OPN3, OPRD1, OPRK1, OPRL1, OPRM1, OR10A5, OR11A1, OR12D3, OR1C1, OR1F1, OR1Q1, OR2H1, OR5V1, OR5V1, OR12D3, OR7A5, OR7C1, OR8B8, OXTR, P2RY1, P2RY10, P2RY11, P2RY12, P2RY13, P2RY14, P2RY2, P2RY5, P2RY6, PPYR1, PTAFR, PTGDR, PTGER1, PTGER2, PTGER4, PTGFR, PTGIR, Rgr, RGR, RHO, RRH, SSTR1, SSTR2, SSTR3, SSTR4, SUCNR1, TBXA2R, TRHR | 159 | ADORA1(4), ADORA2A(6), ADORA2B(1), ADRA1A(2), ADRA1B(4), ADRA1D(2), ADRA2C(2), ADRB2(3), AGTR1(3), AGTR2(4), AVPR1A(3), AVPR1B(2), AVPR2(1), BDKRB1(3), BDKRB2(1), C3AR1(1), CCBP2(4), CCKAR(2), CCKBR(1), CCR1(2), CCR10(1), CCR3(1), CCR4(3), CCR6(2), CCR8(3), CCRL1(3), CCRL2(1), CHML(1), CHRM2(6), CHRM3(5), CHRM4(1), CHRM5(2), CMKLR1(1), CNR1(3), CNR2(3), CX3CR1(3), CXCR3(1), DRD1(1), DRD2(2), DRD3(5), DRD4(1), DRD5(3), EDNRA(2), EDNRB(5), F2R(2), F2RL1(3), F2RL2(1), F2RL3(2), FPR1(1), GALR1(3), GALT(1), GHSR(3), GPR17(5), GPR174(4), GPR27(1), GPR35(1), GPR37(2), GPR37L1(2), GPR50(4), GPR6(2), GPR63(2), GPR77(4), GPR83(2), GPR85(2), GPR87(6), GRPR(2), HCRTR2(3), HRH1(3), HRH2(2), HRH3(1), HTR1B(3), HTR1D(1), HTR1E(5), HTR1F(2), HTR2A(1), HTR2B(2), HTR2C(3), HTR4(3), HTR5A(3), HTR6(1), HTR7(3), LHCGR(8), MAS1(3), MC3R(3), MC4R(1), MC5R(3), MTNR1A(1), NMBR(2), NMUR2(2), NPY1R(3), NPY2R(2), NPY5R(2), NTSR1(3), NTSR2(3), OPN1SW(1), OPN3(1), OPRD1(2), OPRK1(1), OPRM1(2), OR10A5(2), OR11A1(4), OR12D3(4), OR1F1(1), OR1Q1(3), OR2H1(1), OR5V1(1), OR7A5(1), OR8B8(5), OXTR(1), P2RY1(1), P2RY10(2), P2RY13(1), P2RY14(3), P2RY2(2), P2RY6(2), PPYR1(3), PTAFR(3), PTGDR(5), PTGER2(1), PTGER4(2), PTGFR(2), PTGIR(1), RGR(2), SSTR1(1), SSTR2(2), SSTR3(1), SSTR4(2), SUCNR1(2), TBXA2R(2), TRHR(1) | 31942456 | 308 | 137 | 307 | 112 | 86 | 113 | 58 | 14 | 37 | 0 | 0.000019 | 0.0059 | |
| 3 | C21_STEROID_HORMONE_METABOLISM | AKR1C4, AKR1D1, CYP11A1, CYP11B1, CYP11B2, CYP17A1, CYP21A2, HSD11B1, HSD11B2, HSD3B1, HSD3B2 | 10 | AKR1C4(1), AKR1D1(4), CYP11A1(2), CYP11B2(6), CYP17A1(1), CYP21A2(1), HSD11B1(3), HSD11B2(1), HSD3B1(4), HSD3B2(5) | 2150774 | 28 | 26 | 27 | 3 | 5 | 12 | 4 | 5 | 2 | 0 | 0.0006 | 0.091 | |
| 4 | HSA00140_C21_STEROID_HORMONE_METABOLISM | Genes involved in C21-steroid hormone metabolism | AKR1C4, AKR1D1, CYP11A1, CYP11B1, CYP11B2, CYP17A1, CYP21A2, HSD11B1, HSD11B2, HSD3B1, HSD3B2 | 10 | AKR1C4(1), AKR1D1(4), CYP11A1(2), CYP11B2(6), CYP17A1(1), CYP21A2(1), HSD11B1(3), HSD11B2(1), HSD3B1(4), HSD3B2(5) | 2150774 | 28 | 26 | 27 | 3 | 5 | 12 | 4 | 5 | 2 | 0 | 0.0006 | 0.091 |
| 5 | BLOOD_CLOTTING_CASCADE | F10, F11, F12, F13B, F2, F5, F7, F8, F8A1, F9, FGA, FGB, FGG, LPA, PLG, PLAT, PLAU, PLG, SERPINB2, SERPINE1, SERPINF2, VWF | 20 | F10(3), F11(4), F12(1), F13B(7), F5(15), F8(10), F9(4), FGA(5), FGB(3), FGG(1), LPA(11), PLAT(1), PLAU(2), PLG(2), SERPINB2(3), SERPINE1(2), SERPINF2(3), VWF(16) | 9480458 | 93 | 69 | 92 | 24 | 19 | 37 | 23 | 6 | 8 | 0 | 0.00074 | 0.091 | |
| 6 | FLUMAZENILPATHWAY | Flumazenil is a benzodiazepine receptor antagonist that may induce protective preconditioning in ischemic cardiomyocytes. | GABRA1, GABRA2, GABRA3, GABRA4, GABRA5, GABRA6, GPX1, PRKCE, SOD1 | 7 | GABRA1(6), GABRA3(5), GABRA4(3), GABRA5(4), PRKCE(5) | 1635108 | 23 | 23 | 23 | 5 | 9 | 10 | 3 | 0 | 1 | 0 | 0.0012 | 0.12 |
| 7 | HSA01430_CELL_COMMUNICATION | Genes involved in cell communication | ACTB, ACTG1, CHAD, COL11A1, COL11A2, COL17A1, COL1A1, COL1A2, COL2A1, COL3A1, COL4A1, COL4A2, COL4A4, COL4A6, COL5A1, COL5A2, COL5A3, COL6A1, COL6A2, COL6A3, COL6A6, COMP, DES, DSC1, DSC2, DSC3, DSG1, DSG2, DSG3, DSG4, FN1, GJA1, GJA10, GJA3, GJA4, GJA5, GJA8, GJA9, GJB1, GJB2, GJB3, GJB4, GJB5, GJB6, GJB7, GJC1, GJC2, GJC3, GJD2, GJD3, GJD4, IBSP, INA, ITGA6, ITGB4, KRT1, KRT10, KRT12, KRT13, KRT14, KRT15, KRT16, KRT17, KRT18, KRT19, KRT2, KRT20, KRT23, KRT24, KRT25, KRT27, KRT28, KRT3, KRT31, KRT32, KRT33A, KRT33B, KRT34, KRT35, KRT36, KRT37, KRT38, KRT39, KRT4, KRT40, KRT5, KRT6A, KRT6B, KRT6C, KRT7, KRT71, KRT72, KRT73, KRT74, KRT75, KRT76, KRT77, KRT78, KRT79, KRT8, KRT81, KRT82, KRT83, KRT84, KRT85, KRT86, KRT9, LAMA1, LAMA2, LAMA3, LAMA4, LAMA5, LAMB1, LAMB2, LAMB3, LAMB4, LAMC1, LAMC2, LAMC3, LMNA, LMNB1, LMNB2, LOC728760, NES, PRPH, RELN, SPP1, THBS1, THBS2, THBS3, THBS4, TNC, TNN, TNR, TNXB, VIM, VTN, VWF | 130 | ACTB(1), COL11A2(10), COL17A1(4), COL1A1(8), COL1A2(15), COL2A1(8), COL3A1(10), COL4A1(11), COL4A2(9), COL4A4(9), COL4A6(9), COL5A1(5), COL5A2(12), COL5A3(11), COL6A3(12), COMP(2), DES(2), DSC1(4), DSC2(1), DSC3(7), DSG1(8), DSG2(2), DSG3(2), DSG4(5), FN1(17), GJA1(4), GJA10(5), GJA8(2), GJB2(1), GJB4(1), GJB5(1), GJC1(2), GJC2(1), GJC3(1), GJD2(2), GJD4(2), IBSP(4), INA(2), ITGA6(6), ITGB4(9), KRT1(1), KRT10(1), KRT12(3), KRT13(2), KRT14(1), KRT16(3), KRT17(2), KRT18(2), KRT2(4), KRT20(3), KRT23(2), KRT24(4), KRT27(3), KRT28(2), KRT3(2), KRT31(1), KRT32(2), KRT33A(1), KRT35(1), KRT36(3), KRT37(2), KRT4(1), KRT5(1), KRT6A(2), KRT6B(4), KRT6C(2), KRT71(2), KRT72(2), KRT73(7), KRT74(1), KRT75(2), KRT76(2), KRT77(3), KRT78(1), KRT79(4), KRT81(3), KRT82(1), KRT83(1), KRT84(1), KRT85(1), KRT86(1), KRT9(1), LAMA1(14), LAMA2(17), LAMA3(7), LAMA4(15), LAMA5(7), LAMB1(6), LAMB2(4), LAMB3(1), LAMB4(3), LAMC1(8), LAMC2(4), LAMC3(1), NES(11), PRPH(2), RELN(26), THBS1(1), THBS3(4), TNC(16), TNXB(16), VIM(5), VWF(16) | 63492600 | 499 | 148 | 498 | 162 | 140 | 188 | 92 | 32 | 47 | 0 | 0.0028 | 0.24 |
| 8 | HSA04614_RENIN_ANGIOTENSIN_SYSTEM | Genes involved in renin-angiotensin system | ACE, ACE2, AGT, AGTR1, AGTR2, ANPEP, CMA1, CPA3, CTSA, CTSG, ENPEP, LNPEP, MAS1, MME, NLN, REN, THOP1 | 17 | ACE(4), ACE2(3), AGT(1), AGTR1(3), AGTR2(4), ANPEP(5), CMA1(4), CPA3(4), CTSA(2), CTSG(4), ENPEP(7), LNPEP(2), MAS1(3), MME(6), NLN(2), THOP1(1) | 5741924 | 55 | 46 | 55 | 22 | 11 | 23 | 8 | 4 | 9 | 0 | 0.0037 | 0.28 |
| 9 | GLUCOCORTICOID_MINERALOCORTICOID_METABOLISM | CPN2, CYP11A1, CYP11B2, CYP17A1, HSD11B1, HSD11B2, HSD3B1, HSD3B2 | 8 | CPN2(1), CYP11A1(2), CYP11B2(6), CYP17A1(1), HSD11B1(3), HSD11B2(1), HSD3B1(4), HSD3B2(5) | 1860812 | 23 | 21 | 22 | 5 | 3 | 11 | 4 | 4 | 1 | 0 | 0.006 | 0.39 | |
| 10 | HSA04810_REGULATION_OF_ACTIN_CYTOSKELETON | Genes involved in regulation of actin cytoskeleton | ABI2, ACTN1, ACTN2, ACTN3, ACTN4, APC, APC2, ARAF, ARHGEF1, ARHGEF12, ARHGEF4, ARHGEF6, ARHGEF7, ARPC1A, ARPC1B, ARPC2, ARPC3, ARPC4, ARPC5, ARPC5L, BAIAP2, BCAR1, BDKRB1, BDKRB2, BRAF, C3orf10, CD14, CDC42, CFL1, CFL2, CHRM1, CHRM2, CHRM3, CHRM4, CHRM5, CRK, CRKL, CSK, CYFIP1, CYFIP2, DIAPH1, DIAPH2, DIAPH3, DOCK1, EGF, EGFR, EZR, F2, F2R, FGD1, FGD3, FGF1, FGF10, FGF11, FGF12, FGF13, FGF14, FGF16, FGF17, FGF18, FGF19, FGF2, FGF20, FGF21, FGF22, FGF23, FGF3, FGF4, FGF5, FGF6, FGF7, FGF8, FGF9, FGFR1, FGFR2, FGFR3, FGFR4, FN1, GIT1, GNA12, GNA13, GNG12, GRLF1, GSN, HRAS, INS, IQGAP1, IQGAP2, IQGAP3, ITGA1, ITGA10, ITGA11, ITGA2, ITGA2B, ITGA3, ITGA4, ITGA5, ITGA6, ITGA7, ITGA8, ITGA9, ITGAD, ITGAE, ITGAL, ITGAM, ITGAV, ITGAX, ITGB1, ITGB2, ITGB3, ITGB4, ITGB5, ITGB6, ITGB7, ITGB8, KRAS, LIMK1, LIMK2, LOC200025, LOC645126, LOC653888, MAP2K1, MAP2K2, MAPK1, MAPK3, MLCK, MOS, MRAS, MRCL3, MRLC2, MSN, MYH10, MYH14, MYH9, MYL2, MYL5, MYL7, MYL8P, MYL9, MYLC2PL, MYLK, MYLK2, MYLPF, NCKAP1, NCKAP1L, NRAS, PAK1, PAK2, PAK3, PAK4, PAK6, PAK7, PDGFA, PDGFB, PDGFRA, PDGFRB, PFN1, PFN2, PFN3, PFN4, PIK3CA, PIK3CB, PIK3CD, PIK3CG, PIK3R1, PIK3R2, PIK3R3, PIK3R5, PIP4K2A, PIP4K2B, PIP4K2C, PIP5K1A, PIP5K1B, PIP5K1C, PIP5K3, PPP1CA, PPP1CB, PPP1CC, PPP1R12A, PPP1R12B, PTK2, PXN, RAC1, RAC2, RAC3, RAF1, RDX, RHOA, ROCK1, ROCK2, RRAS, RRAS2, SCIN, SLC9A1, SOS1, SOS2, SSH1, SSH2, SSH3, TIAM1, TIAM2, TMSB4X, TMSB4Y, TMSL3, VAV1, VAV2, VAV3, VCL, WAS, WASF1, WASF2, WASL | 202 | ACTN1(2), ACTN2(6), ACTN3(2), ACTN4(5), APC(9), APC2(2), ARAF(3), ARHGEF1(2), ARHGEF12(3), ARHGEF4(3), ARHGEF6(2), ARHGEF7(3), ARPC1A(2), ARPC1B(1), BCAR1(5), BDKRB1(3), BDKRB2(1), BRAF(7), CD14(1), CDC42(1), CFL1(3), CFL2(1), CHRM2(6), CHRM3(5), CHRM4(1), CHRM5(2), CRKL(1), CYFIP1(6), CYFIP2(7), DIAPH1(1), DIAPH3(5), DOCK1(3), EGF(6), EGFR(9), EZR(1), F2R(2), FGD1(3), FGD3(3), FGF10(2), FGF12(2), FGF14(1), FGF2(2), FGF23(1), FGF3(1), FGF5(3), FGF6(2), FGF8(1), FGFR1(3), FGFR2(6), FGFR3(2), FGFR4(1), FN1(17), GIT1(1), GNA12(1), GNA13(1), GNG12(1), GRLF1(11), GSN(2), HRAS(2), IQGAP1(6), IQGAP2(6), IQGAP3(11), ITGA1(4), ITGA10(6), ITGA11(3), ITGA2(5), ITGA2B(1), ITGA3(2), ITGA4(5), ITGA5(1), ITGA6(6), ITGA7(3), ITGA8(9), ITGA9(1), ITGAD(6), ITGAE(3), ITGAL(1), ITGAM(3), ITGAV(3), ITGAX(9), ITGB1(4), ITGB2(3), ITGB3(3), ITGB4(9), ITGB5(2), ITGB6(2), ITGB7(1), ITGB8(3), KRAS(1), LIMK1(1), LIMK2(1), MAP2K1(2), MAP2K2(2), MAPK1(1), MAPK3(2), MOS(4), MRAS(3), MSN(3), MYH10(3), MYH14(4), MYH9(14), MYL2(1), MYLK(7), MYLK2(2), MYLPF(1), NCKAP1(5), NCKAP1L(9), PAK1(3), PAK2(1), PAK3(3), PAK4(1), PAK6(2), PAK7(4), PDGFB(1), PDGFRA(8), PDGFRB(5), PFN3(1), PIK3CB(1), PIK3CD(3), PIK3CG(10), PIK3R1(1), PIK3R2(4), PIK3R3(2), PIK3R5(2), PIP4K2A(2), PIP4K2B(3), PIP4K2C(3), PIP5K1A(4), PIP5K1B(1), PIP5K1C(2), PPP1CA(1), PPP1CB(1), PPP1CC(1), PPP1R12B(3), PTK2(3), PXN(3), RAC1(2), RAF1(1), RDX(1), ROCK1(5), ROCK2(3), RRAS2(2), SCIN(1), SLC9A1(3), SOS1(6), SOS2(4), SSH1(1), SSH2(7), TIAM1(9), TIAM2(4), VAV1(7), VAV2(4), VAV3(1), VCL(2), WAS(5), WASL(2) | 75493894 | 527 | 153 | 523 | 165 | 122 | 198 | 93 | 47 | 67 | 0 | 0.0064 | 0.39 |
In brief, we tabulate the number of mutations and the number of covered bases for each gene. The counts are broken down by mutation context category: four context categories that are discovered by MutSig, and one for indel and 'null' mutations, which include indels, nonsense mutations, splice-site mutations, and non-stop (read-through) mutations. For each gene, we calculate the probability of seeing the observed constellation of mutations, i.e. the product P1 x P2 x ... x Pm, or a more extreme one, given the background mutation rates calculated across the dataset.[1]
This is an experimental feature. Location of data archives could not be determined.