GISTIC identifies genomic regions that are significantly gained or lost across a set of tumors. The pipeline first filters out normal samples from the segmented copy-number data by inspecting the TCGA barcodes and then executes GISTIC version 2.0.15 (cga svn revision 34144).
There were 211 tumor samples used in this analysis: 25 significant arm-level results, 28 significant focal amplifications, and 46 significant focal deletions were found.
Figure 1. Genomic positions of amplified regions: the X-axis represents the normalized amplification signals (top) and significance by Q value (bottom). The green line represents the significance cutoff at Q value=0.25.
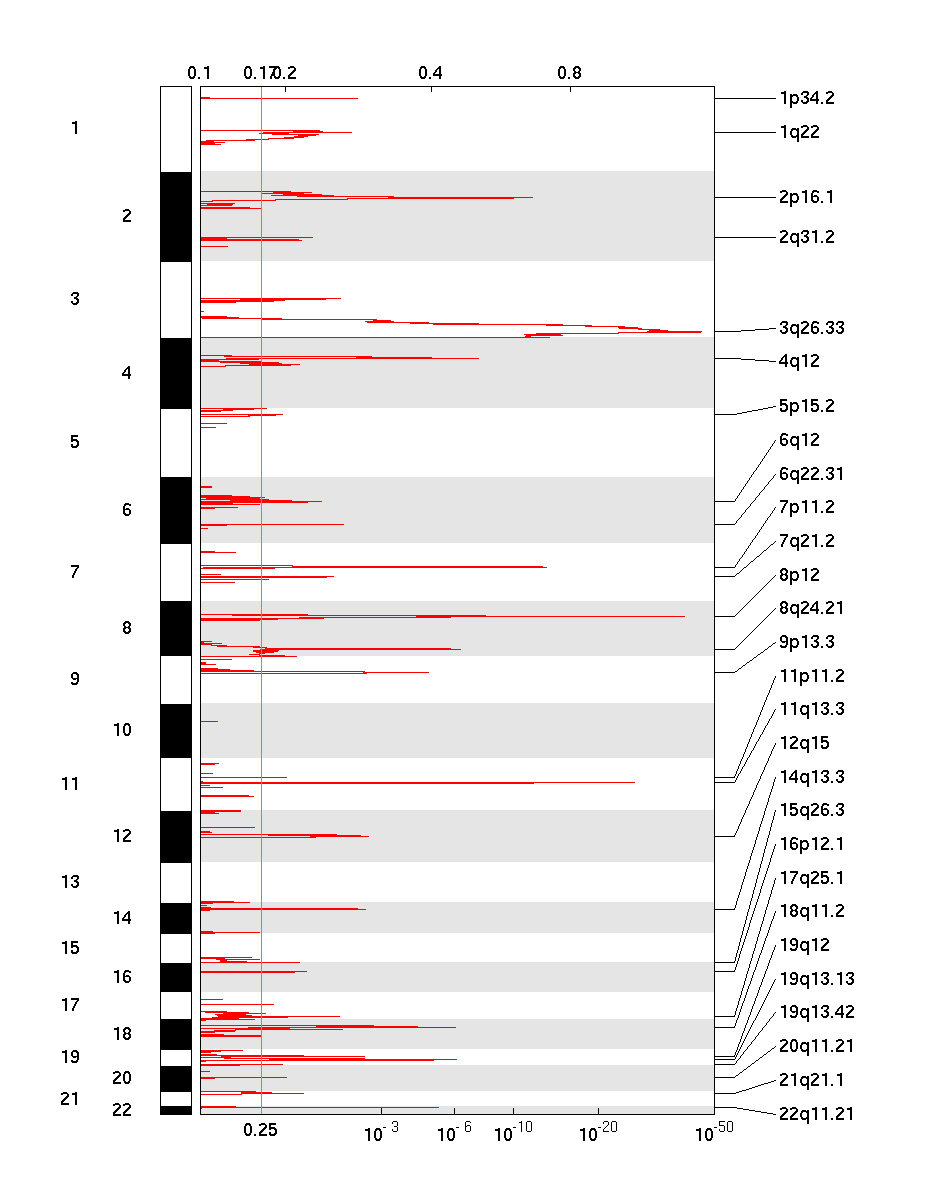
Table 1. Get Full Table Amplifications Table - 28 significant amplifications found. Click the link in the last column to view a comprehensive list of candidate genes. If no genes were identified within the peak, the nearest gene appears in brackets.
| Cytoband | Q value | Residual Q value | Wide Peak Boundaries | # Genes in Wide Peak |
|---|---|---|---|---|
| 3q26.33 | 3.7589e-46 | 3.7589e-46 | chr3:184069946-184285567 | 3 |
| 8p12 | 1.944e-40 | 1.944e-40 | chr8:38287525-38370893 | 2 |
| 11q13.3 | 1.7587e-27 | 1.9545e-27 | chr11:69408710-69588487 | 0 [ANO1] |
| 7p11.2 | 6.9009e-14 | 6.9009e-14 | chr7:54749279-55481516 | 3 |
| 2p16.1 | 2.0079e-12 | 2.0079e-12 | chr2:60382630-61008146 | 3 |
| 4q12 | 3.66e-08 | 3.66e-08 | chr4:55960066-56202497 | 4 |
| 8q24.21 | 4.547e-07 | 4.547e-07 | chr8:128071511-128693092 | 2 |
| 18q11.2 | 8.2941e-07 | 8.2941e-07 | chr18:22007290-22278528 | 2 |
| 22q11.21 | 6.588e-06 | 6.588e-06 | chr22:19200757-19740490 | 17 |
| 9p13.3 | 1.7518e-05 | 1.7518e-05 | chr9:34997337-35036417 | 1 |
| 19q13.13 | 8.0263e-07 | 0.00010246 | chr19:43029012-43993845 | 22 |
| 12q15 | 0.0023772 | 0.0023772 | chr12:65424056-70146921 | 26 |
| 14q13.3 | 0.0028692 | 0.0028692 | chr14:35852736-37332424 | 8 |
| 1p34.2 | 0.004513 | 0.004513 | chr1:39525916-40423375 | 18 |
| 1q22 | 0.0063428 | 0.0063428 | chr1:121045320-171649835 | 519 |
| 6q22.31 | 0.0098908 | 0.0098908 | chr6:124765242-125815717 | 5 |
| 17q25.1 | 0.012558 | 0.012558 | chr17:68709195-69657523 | 7 |
| 7q21.2 | 0.016778 | 0.016778 | chr7:91312491-93095567 | 21 |
| 6q12 | 0.028804 | 0.028804 | chr6:63725884-66170398 | 4 |
| 2q31.2 | 0.043752 | 0.043752 | chr2:177780568-177863084 | 3 |
| 16p12.1 | 0.055567 | 0.055567 | chr16:24972464-26907093 | 4 |
| 19q12 | 0.0054439 | 0.062079 | chr19:33389816-35460315 | 8 |
| 21q21.1 | 0.062079 | 0.062079 | chr21:1-19807858 | 28 |
| 15q26.3 | 0.072938 | 0.072938 | chr15:86853989-100338915 | 91 |
| 20q11.21 | 0.11496 | 0.11496 | chr20:26020583-30093702 | 27 |
| 5p15.2 | 0.12985 | 0.12985 | chr5:1-18514934 | 71 |
| 11p11.2 | 0.11725 | 0.13869 | chr11:45871288-46949201 | 19 |
| 19q13.42 | 0.13255 | 0.21221 | chr19:60244636-61435811 | 47 |
This is the comprehensive list of amplified genes in the wide peak for 3q26.33.
Table S1. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| ATP11B |
| DCUN1D1 |
| MCCC1 |
This is the comprehensive list of amplified genes in the wide peak for 8p12.
Table S2. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| WHSC1L1 |
| LETM2 |
This is the comprehensive list of amplified genes in the wide peak for 7p11.2.
Table S3. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| EGFR |
| SEC61G |
| LANCL2 |
This is the comprehensive list of amplified genes in the wide peak for 2p16.1.
Table S4. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| REL |
| BCL11A |
| PAPOLG |
This is the comprehensive list of amplified genes in the wide peak for 4q12.
Table S5. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CLOCK |
| NMU |
| TMEM165 |
| PDCL2 |
This is the comprehensive list of amplified genes in the wide peak for 8q24.21.
Table S6. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| POU5F1B |
| LOC727677 |
This is the comprehensive list of amplified genes in the wide peak for 18q11.2.
Table S7. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| TAF4B |
| PSMA8 |
This is the comprehensive list of amplified genes in the wide peak for 22q11.21.
Table S8. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-649 |
| CRKL |
| SERPIND1 |
| PI4KA |
| SLC7A4 |
| LZTR1 |
| P2RX6 |
| SNAP29 |
| MED15 |
| THAP7 |
| TMEM191A |
| MGC16703 |
| AIFM3 |
| POM121L4P |
| LOC400891 |
| FLJ39582 |
| P2RX6P |
This is the comprehensive list of amplified genes in the wide peak for 9p13.3.
Table S9. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| C9orf131 |
This is the comprehensive list of amplified genes in the wide peak for 19q13.13.
Table S10. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| ACTN4 |
| LGALS4 |
| LGALS7 |
| PSMD8 |
| RYR1 |
| DPF1 |
| KCNK6 |
| SPINT2 |
| MAP4K1 |
| SIPA1L3 |
| EIF3K |
| CATSPERG |
| C19orf33 |
| WDR87 |
| YIF1B |
| PPP1R14A |
| RASGRP4 |
| FAM98C |
| CAPN12 |
| GGN |
| SPRED3 |
| LGALS7B |
This is the comprehensive list of amplified genes in the wide peak for 12q15.
Table S11. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| MDM2 |
| hsa-mir-1279 |
| CPM |
| IFNG |
| LYZ |
| CNOT2 |
| PTPRB |
| PTPRR |
| RAP1B |
| TSPAN8 |
| YEATS4 |
| DYRK2 |
| LGR5 |
| CCT2 |
| FRS2 |
| CPSF6 |
| KCNMB4 |
| IL22 |
| SLC35E3 |
| IL26 |
| CAND1 |
| MDM1 |
| NUP107 |
| RAB3IP |
| BEST3 |
| LRRC10 |
This is the comprehensive list of amplified genes in the wide peak for 14q13.3.
Table S12. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| NKX2-1 |
| FOXA1 |
| PAX9 |
| NKX2-8 |
| MBIP |
| SLC25A21 |
| MIPOL1 |
| SFTA3 |
This is the comprehensive list of amplified genes in the wide peak for 1p34.2.
Table S13. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| MYCL1 |
| BMP8B |
| PPT1 |
| RLF |
| PABPC4 |
| PPIE |
| CAP1 |
| MACF1 |
| HEYL |
| HPCAL4 |
| TRIT1 |
| OXCT2 |
| NT5C1A |
| MFSD2A |
| BMP8A |
| KIAA0754 |
| SNORA55 |
| LOC728448 |
This is the comprehensive list of amplified genes in the wide peak for 1q22.
Table S14. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| ARNT |
| BCL9 |
| FCGR2B |
| MUC1 |
| NTRK1 |
| PBX1 |
| PRCC |
| SDHC |
| TPM3 |
| PDE4DIP |
| hsa-mir-199a-2 |
| hsa-mir-1295 |
| hsa-mir-557 |
| hsa-mir-1255b-2 |
| hsa-mir-921 |
| hsa-mir-556 |
| hsa-mir-765 |
| hsa-mir-9-1 |
| hsa-mir-555 |
| hsa-mir-92b |
| hsa-mir-190b |
| hsa-mir-554 |
| ADAR |
| ALDH9A1 |
| APCS |
| APOA2 |
| FASLG |
| ATP1A2 |
| ATP1A4 |
| ATP1B1 |
| BGLAP |
| CASQ1 |
| CD1A |
| CD1B |
| CD1C |
| CD1D |
| CD1E |
| CD247 |
| CD5L |
| CD48 |
| CHRNB2 |
| CKS1B |
| CLK2 |
| COPA |
| CRABP2 |
| CRP |
| CTSK |
| CTSS |
| DPT |
| ECM1 |
| EFNA1 |
| EFNA3 |
| EFNA4 |
| ENSA |
| ETV3 |
| F5 |
| FCER1A |
| FCER1G |
| FCGR1A |
| FCGR2A |
| FCGR3A |
| FCGR3B |
| FDPS |
| FLG |
| FMO1 |
| FMO2 |
| FMO3 |
| FMO4 |
| FMO5 |
| DARC |
| GBA |
| GBAP |
| GJA5 |
| GJA8 |
| HDGF |
| HSPA6 |
| HSPA7 |
| IFI16 |
| IL6R |
| ILF2 |
| INSRR |
| IVL |
| KCNJ9 |
| KCNJ10 |
| KCNN3 |
| LMNA |
| LMX1A |
| LOR |
| LY9 |
| MCL1 |
| SMCP |
| MEF2D |
| MGST3 |
| MNDA |
| MPZ |
| MTX1 |
| MYOC |
| NDUFS2 |
| NHLH1 |
| NIT1 |
| NPR1 |
| DDR2 |
| PDZK1 |
| PFDN2 |
| PIGC |
| PI4KB |
| PKLR |
| PRRX1 |
| POU2F1 |
| PPOX |
| PRKAB2 |
| PSMB4 |
| PSMD4 |
| PEX19 |
| RAB13 |
| RFX5 |
| RGS4 |
| RIT1 |
| RORC |
| RPS27 |
| RXRG |
| S100A1 |
| S100A2 |
| S100A3 |
| S100A4 |
| S100A5 |
| S100A6 |
| S100A7 |
| S100A8 |
| S100A9 |
| S100A10 |
| S100A11 |
| S100A12 |
| S100A13 |
| XCL1 |
| SELE |
| SELL |
| SELP |
| SHC1 |
| SLAMF1 |
| UAP1 |
| SPRR1A |
| SPRR1B |
| SPRR2A |
| SPRR2B |
| SPRR2C |
| SPRR2D |
| SPRR2E |
| SPRR2F |
| SPRR2G |
| SPRR3 |
| SPTA1 |
| SSR2 |
| XCL2 |
| VPS72 |
| THBS3 |
| TCHH |
| TOP1P1 |
| CCT3 |
| TUFT1 |
| TNFSF4 |
| UCK2 |
| USF1 |
| DAP3 |
| HIST2H2AA3 |
| HIST2H2AC |
| HIST2H2BE |
| HIST2H4A |
| PIP5K1A |
| TAGLN2 |
| ANXA9 |
| RGS5 |
| ITGA10 |
| BLZF1 |
| VAMP4 |
| PEA15 |
| B4GALT3 |
| ADAM15 |
| PEX11B |
| CREG1 |
| CD84 |
| SELENBP1 |
| TNFSF18 |
| MPZL1 |
| SH2D2A |
| TBX19 |
| FCGR2C |
| PRPF3 |
| ARHGEF2 |
| DEDD |
| AIM2 |
| ADAMTS4 |
| SEC22B |
| CHD1L |
| SLC25A44 |
| NOS1AP |
| ARHGEF11 |
| SETDB1 |
| UBAP2L |
| SV2A |
| DENND4B |
| RBM8A |
| NR1I3 |
| SCAMP3 |
| GPA33 |
| SF3B4 |
| PIAS3 |
| HAX1 |
| C1orf61 |
| SEMA6C |
| SLC19A2 |
| POLR3C |
| TXNIP |
| PMVK |
| FAM189B |
| NES |
| JTB |
| MTMR11 |
| MLLT11 |
| SLC27A3 |
| TDRKH |
| CD160 |
| TNRC4 |
| PMF1 |
| DUSP12 |
| VPS45 |
| KIAA0907 |
| KIFAP3 |
| ATF6 |
| POGZ |
| SYT11 |
| BAT2D1 |
| RPRD2 |
| SMG5 |
| NCSTN |
| GPR161 |
| SNAPIN |
| RUSC1 |
| CA14 |
| CCDC19 |
| NBPF14 |
| BRP44 |
| OLFML2B |
| C1orf43 |
| DNM3 |
| C1orf77 |
| LCE2B |
| OR10J1 |
| USP21 |
| SLC39A1 |
| RNF115 |
| ROBLD3 |
| TMOD4 |
| NME7 |
| LASS2 |
| CRNN |
| DCAF8 |
| F11R |
| BOLA1 |
| ZBTB7B |
| C1orf66 |
| APH1A |
| PLEKHO1 |
| ACP6 |
| C1orf9 |
| GPR89B |
| HSD17B7 |
| UFC1 |
| METTL13 |
| OAZ3 |
| CD244 |
| DPM3 |
| MRPS21 |
| TMCO1 |
| ADAMTSL4 |
| CRCT1 |
| GON4L |
| GPATCH4 |
| DUSP23 |
| C1orf56 |
| MSTO1 |
| GOLPH3L |
| KIRREL |
| YY1AP1 |
| UBE2Q1 |
| ITLN1 |
| C1orf112 |
| FAM63A |
| ADCY10 |
| DCAF6 |
| ASH1L |
| LENEP |
| RAG1AP1 |
| SLAMF8 |
| CDC42SE1 |
| UBQLN4 |
| OTUD7B |
| RAB25 |
| PGLYRP4 |
| RHBG |
| SCYL3 |
| ATP8B2 |
| VANGL2 |
| PBXIP1 |
| S100A14 |
| GATAD2B |
| CGN |
| IGSF9 |
| ZNF687 |
| POGK |
| HCN3 |
| C1orf114 |
| SLAMF7 |
| CADM3 |
| PRUNE |
| HAPLN2 |
| BCAN |
| SEMA4A |
| MRPL9 |
| INTS3 |
| SCNM1 |
| MGC4473 |
| FCRL2 |
| MRPL24 |
| TNFAIP8L2 |
| C1orf54 |
| PAQR6 |
| TRIM46 |
| C1orf129 |
| TARS2 |
| FLAD1 |
| OR6N2 |
| OR6K2 |
| PVRL4 |
| SNX27 |
| ANP32E |
| ISG20L2 |
| FCRL5 |
| FCRL4 |
| NUF2 |
| HORMAD1 |
| TOMM40L |
| POLR3GL |
| TMEM79 |
| LCE3D |
| FCRLA |
| ANKRD36BL1 |
| MAEL |
| AQP10 |
| SLAMF9 |
| PYGO2 |
| NUP210L |
| DUSP27 |
| RCSD1 |
| MEX3A |
| C1orf156 |
| GORAB |
| C1orf105 |
| PIGM |
| IGSF8 |
| C1orf85 |
| PGLYRP3 |
| GNRHR2 |
| SLAMF6 |
| FCRL1 |
| FCRL3 |
| FMO9P |
| TADA1 |
| THEM4 |
| SH2D1B |
| GABPB2 |
| TCHHL1 |
| RPTN |
| TDRD10 |
| SHE |
| KLHDC9 |
| HIST2H3C |
| OR10J5 |
| DCST2 |
| UHMK1 |
| FCRLB |
| LIX1L |
| C1orf182 |
| IQGAP3 |
| APOA1BP |
| OR10T2 |
| OR6P1 |
| OR10X1 |
| OR10Z1 |
| OR6K6 |
| OR6N1 |
| S100A16 |
| ITLN2 |
| CREB3L4 |
| C1orf51 |
| HFE2 |
| ANKRD35 |
| LELP1 |
| DCST1 |
| METTL11B |
| FAM78B |
| BNIPL |
| C1orf92 |
| PYHIN1 |
| SPRR4 |
| PPIAL4A |
| TTC24 |
| PDIA3P |
| LCE4A |
| LOC200030 |
| NUDT17 |
| KRTCAP2 |
| CRTC2 |
| LCE5A |
| ARHGAP30 |
| C1orf192 |
| TIPRL |
| C1orf230 |
| THEM5 |
| NBPF15 |
| ANKRD34A |
| C1orf104 |
| C1orf204 |
| C1orf111 |
| LOC284688 |
| HIST2H2AB |
| HIST2H3A |
| S100A7A |
| LINGO4 |
| RXFP4 |
| C1orf110 |
| OR10R2 |
| FCRL6 |
| LCE1A |
| LCE1B |
| LCE1C |
| LCE1D |
| LCE1E |
| LCE1F |
| LCE2A |
| LCE2C |
| LCE2D |
| LCE3A |
| LCE3B |
| LCE3C |
| LCE3E |
| CYCSP52 |
| PEAR1 |
| SFT2D2 |
| ILDR2 |
| NOTCH2NL |
| FLJ39739 |
| LOC388692 |
| LYSMD1 |
| HRNR |
| FLG2 |
| C1orf189 |
| FMO6P |
| VHLL |
| OR10K2 |
| OR10K1 |
| OR6Y1 |
| OR6K3 |
| VSIG8 |
| C1orf226 |
| LOC400794 |
| NBPF20 |
| MIR199A2 |
| MIR214 |
| MIR9-1 |
| HIST2H2BF |
| ETV3L |
| LRRC52 |
| OR10J3 |
| KPRP |
| LCE6A |
| SUMO1P3 |
| HIST2H4B |
| RPL31P11 |
| LOC642502 |
| PPIAL4G |
| PPIAL4D |
| LOC645166 |
| LOC645676 |
| S100A7L2 |
| PPIAL4B |
| GPR89A |
| PPIAL4C |
| HIST2H3D |
| PCP4L1 |
| SCARNA4 |
| SNORA42 |
| MIR554 |
| MIR555 |
| MIR556 |
| MIR557 |
| MIR92B |
| HIST2H2AA4 |
| LOC728855 |
| LOC728875 |
| NBPF11 |
| GPR89C |
| NBPF16 |
| PDZK1P1 |
| PPIAL4F |
| LOC728989 |
| PPIAL4E |
| C1orf152 |
| MIR765 |
| MIR190B |
| MIR921 |
| C1orf68 |
| MSTO2P |
| TSTD1 |
| LOC100132111 |
| NBPF10 |
| FCGR1C |
| C2CD4D |
| LOC100286793 |
This is the comprehensive list of amplified genes in the wide peak for 6q22.31.
Table S15. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| STL |
| TPD52L1 |
| HDDC2 |
| RNF217 |
| NKAIN2 |
This is the comprehensive list of amplified genes in the wide peak for 17q25.1.
Table S16. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| COG1 |
| CDC42EP4 |
| SDK2 |
| C17orf80 |
| FAM104A |
| C17orf54 |
| CPSF4L |
This is the comprehensive list of amplified genes in the wide peak for 7q21.2.
Table S17. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CDK6 |
| AKAP9 |
| hsa-mir-489 |
| hsa-mir-1285-1 |
| CALCR |
| KRIT1 |
| CYP51A1 |
| PEX1 |
| MTERF |
| ANKIB1 |
| SAMD9 |
| CCDC132 |
| GATAD1 |
| C7orf64 |
| MGC16142 |
| SAMD9L |
| HEPACAM2 |
| FAM133B |
| LOC401387 |
| MIR489 |
| MIR653 |
This is the comprehensive list of amplified genes in the wide peak for 6q12.
Table S18. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| PTP4A1 |
| PHF3 |
| LGSN |
| EYS |
This is the comprehensive list of amplified genes in the wide peak for 2q31.2.
Table S19. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| NFE2L2 |
| HNRNPA3 |
| LOC100130691 |
This is the comprehensive list of amplified genes in the wide peak for 16p12.1.
Table S20. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| AQP8 |
| HS3ST4 |
| LCMT1 |
| ZKSCAN2 |
This is the comprehensive list of amplified genes in the wide peak for 19q12.
Table S21. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CCNE1 |
| UQCRFS1 |
| C19orf2 |
| POP4 |
| PLEKHF1 |
| C19orf12 |
| LOC148145 |
| VSTM2B |
This is the comprehensive list of amplified genes in the wide peak for 21q21.1.
Table S22. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-125b-2 |
| BAGE |
| CXADR |
| PRSS7 |
| HSPA13 |
| TPTE |
| NRIP1 |
| BTG3 |
| USP25 |
| RBM11 |
| NCRNA00157 |
| C21orf15 |
| C21orf91 |
| SAMSN1 |
| BAGE5 |
| BAGE4 |
| BAGE3 |
| BAGE2 |
| CHODL |
| C21orf99 |
| LIPI |
| ABCC13 |
| POTED |
| C21orf34 |
| C21orf81 |
| MIRLET7C |
| MIR125B2 |
| MIR99A |
This is the comprehensive list of amplified genes in the wide peak for 15q26.3.
Table S23. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BLM |
| IDH2 |
| CRTC3 |
| hsa-mir-1302-2 |
| hsa-mir-1469 |
| hsa-mir-9-3 |
| hsa-mir-7-2 |
| ACAN |
| ALDH1A3 |
| ANPEP |
| CHD2 |
| FES |
| IGF1R |
| ISG20 |
| MAN2A2 |
| MEF2A |
| MFGE8 |
| FURIN |
| PCSK6 |
| PLIN1 |
| POLG |
| RLBP1 |
| SNRPA1 |
| NR2F2 |
| ST8SIA2 |
| PEX11A |
| IQGAP1 |
| PRC1 |
| SV2B |
| AP3S2 |
| SEMA4B |
| CIB1 |
| ABHD2 |
| CHSY1 |
| SYNM |
| GABARAPL3 |
| VPS33B |
| OR4F4 |
| SLCO3A1 |
| NGRN |
| RHCG |
| DET1 |
| LINS1 |
| FANCI |
| MCTP2 |
| SELS |
| MESP1 |
| UNC45A |
| RGMA |
| WDR93 |
| AEN |
| TTC23 |
| LRRK1 |
| TM2D3 |
| C15orf42 |
| RCCD1 |
| ARRDC4 |
| LOC91948 |
| TARSL2 |
| LRRC28 |
| ASB7 |
| LYSMD4 |
| LOC145814 |
| LOC145820 |
| C15orf32 |
| HAPLN3 |
| MESP2 |
| SPATA8 |
| ADAMTS17 |
| C15orf51 |
| LASS3 |
| LOC254559 |
| LOC283761 |
| FAM169B |
| ZNF774 |
| C15orf38 |
| KIF7 |
| ZNF710 |
| HDDC3 |
| WASH3P |
| FLJ42289 |
| C15orf58 |
| OR4F6 |
| OR4F15 |
| FAM174B |
| MIR7-2 |
| MIR9-3 |
| TTLL13 |
| FAM138E |
| LOC100144604 |
| LOC100288778 |
This is the comprehensive list of amplified genes in the wide peak for 20q11.21.
Table S24. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-663 |
| BCL2L1 |
| FOXS1 |
| ID1 |
| TPX2 |
| REM1 |
| HM13 |
| PDRG1 |
| COX4I2 |
| MYLK2 |
| DEFB118 |
| DUSP15 |
| C20orf160 |
| NCRNA00028 |
| C20orf191 |
| TTLL9 |
| DEFB115 |
| DEFB116 |
| DEFB119 |
| DEFB121 |
| DEFB122 |
| DEFB123 |
| DEFB124 |
| FRG1B |
| XKR7 |
| MIR663 |
| PSIMCT-1 |
This is the comprehensive list of amplified genes in the wide peak for 5p15.2.
Table S25. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-887 |
| ADCY2 |
| CTNND2 |
| DAP |
| DNAH5 |
| MTRR |
| MYO10 |
| NDUFS6 |
| SDHA |
| SLC6A3 |
| SLC9A3 |
| SRD5A1 |
| TERT |
| TRIO |
| SEMA5A |
| TRIP13 |
| PDCD6 |
| MARCH6 |
| BASP1 |
| SLC12A7 |
| POLS |
| TPPP |
| EXOC3 |
| CCT5 |
| FBXL7 |
| KIAA0947 |
| LOC25845 |
| IRX4 |
| TAS2R1 |
| FAM134B |
| FAM105A |
| NSUN2 |
| CEP72 |
| ANKH |
| AHRR |
| MRPL36 |
| BRD9 |
| FASTKD3 |
| IRX1 |
| ZDHHC11 |
| LPCAT1 |
| CLPTM1L |
| ROPN1L |
| MED10 |
| NKD2 |
| FAM105B |
| ZNF622 |
| C5orf55 |
| CCDC127 |
| UBE2QL1 |
| C5orf49 |
| FAM173B |
| CMBL |
| PLEKHG4B |
| C5orf38 |
| IRX2 |
| ADAMTS16 |
| LOC255167 |
| LOC285692 |
| LOC285696 |
| SLC6A19 |
| LOC340094 |
| SLC6A18 |
| LRRC14B |
| FLJ33360 |
| MARCH11 |
| ANKRD33B |
| SDHAP3 |
| LOC728613 |
| SNORD123 |
| LOC100310782 |
This is the comprehensive list of amplified genes in the wide peak for 11p11.2.
Table S26. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CREB3L1 |
| ARHGAP1 |
| CHRM4 |
| F2 |
| LRP4 |
| MDK |
| DGKZ |
| PEX16 |
| MAPK8IP1 |
| KIAA0652 |
| CKAP5 |
| PHF21A |
| AMBRA1 |
| C11orf49 |
| ZNF408 |
| GYLTL1B |
| C11orf94 |
| HARBI1 |
| SNORD67 |
This is the comprehensive list of amplified genes in the wide peak for 19q13.42.
Table S27. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| IL11 |
| PTPRH |
| RPL28 |
| SYT5 |
| TNNI3 |
| TNNT1 |
| U2AF2 |
| SAPS1 |
| HSPBP1 |
| UBE2S |
| CCDC106 |
| EPN1 |
| ZNF580 |
| ZNF581 |
| PPP1R12C |
| EPS8L1 |
| ZNF444 |
| NAT14 |
| ZSCAN5A |
| ISOC2 |
| BRSK1 |
| SUV420H2 |
| FIZ1 |
| GALP |
| ZNF628 |
| RDH13 |
| COX6B2 |
| NLRP13 |
| NLRP8 |
| NLRP5 |
| ZNF787 |
| TMEM190 |
| ZNF524 |
| ZNF784 |
| NLRP4 |
| ZNF579 |
| NLRP11 |
| TMEM86B |
| SSC5D |
| TMEM150B |
| FAM71E2 |
| NLRP9 |
| RFPL4A |
| ZSCAN5B |
| C19orf51 |
| SBK2 |
| SHISA7 |
Figure 2. Genomic positions of deleted regions: the X-axis represents the normalized deletion signals (top) and significance by Q value (bottom). The green line represents the significance cutoff at Q value=0.25.
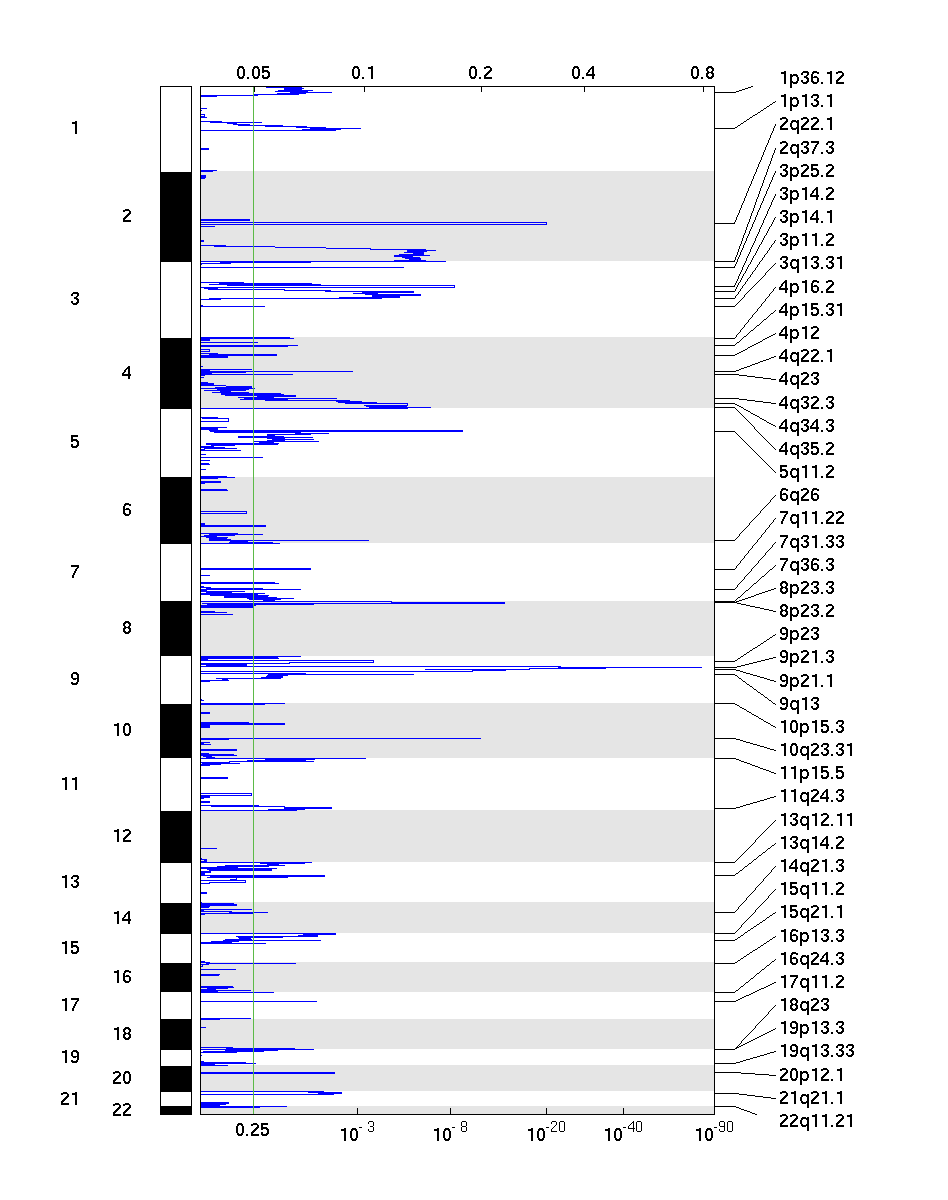
Table 2. Get Full Table Deletions Table - 46 significant deletions found. Click the link in the last column to view a comprehensive list of candidate genes. If no genes were identified within the peak, the nearest gene appears in brackets.
| Cytoband | Q value | Residual Q value | Wide Peak Boundaries | # Genes in Wide Peak |
|---|---|---|---|---|
| 9p21.3 | 3.7363e-81 | 2.8121e-72 | chr9:21855498-21986995 | 2 |
| 2q22.1 | 1.3951e-20 | 8.2635e-19 | chr2:139248662-143354308 | 1 |
| 8p23.2 | 3.5585e-14 | 9.2216e-13 | chr8:2080589-6255647 | 1 |
| 10q23.31 | 1.7926e-11 | 1.7926e-11 | chr10:89607138-90024018 | 2 |
| 5q11.2 | 9.2963e-10 | 9.2963e-10 | chr5:58296055-59823742 | 3 |
| 3p14.2 | 4.2538e-09 | 1.1952e-07 | chr3:59009803-61522370 | 1 |
| 2q37.3 | 2.5851e-08 | 1.7041e-05 | chr2:208390444-242951149 | 268 |
| 6q26 | 0.00040723 | 0.00040723 | chr6:161613089-163073197 | 1 |
| 11p15.5 | 0.00053733 | 0.00052942 | chr11:1-5098845 | 139 |
| 3p25.2 | 1.0527e-05 | 0.0010295 | chr3:11568528-11808472 | 1 |
| 9p21.2 | 2.7332e-14 | 0.0017708 | chr9:27562512-32375942 | 4 |
| 9p13.1 | 3.1206e-06 | 0.0023592 | chr9:38609152-78983232 | 85 |
| 4q35.2 | 2.9442e-07 | 0.0027227 | chr4:183326192-191273063 | 49 |
| 21q21.1 | 0.0033784 | 0.0034209 | chr21:1-21293802 | 29 |
| 1p13.1 | 0.00082933 | 0.004129 | chr1:103339163-144126225 | 168 |
| 20p12.1 | 0.0054788 | 0.0054788 | chr20:14250876-15984135 | 2 |
| 11q24.3 | 0.0067742 | 0.0067742 | chr11:126375629-134452384 | 32 |
| 3p11.2 | 0.00017846 | 0.0090642 | chr3:77779180-95076656 | 11 |
| 3p14.1 | 3.9639e-06 | 0.012183 | chr3:68676953-87070815 | 28 |
| 17q11.2 | 0.016666 | 0.016438 | chr17:26307851-26673293 | 6 |
| 4q22.1 | 0.0015066 | 0.018621 | chr4:91092757-93459528 | 2 |
| 18q23 | 0.01992 | 0.01992 | chr18:71130474-76117153 | 18 |
| 15q11.2 | 0.005207 | 0.02927 | chr15:1-47070162 | 311 |
| 1p36.12 | 0.0065654 | 0.032616 | chr1:1-33741020 | 511 |
| 7q11.22 | 0.023229 | 0.035345 | chr7:66423810-70239151 | 1 |
| 4q34.3 | 6.3706e-06 | 0.043818 | chr4:171246607-191273063 | 72 |
| 16p13.3 | 0.048546 | 0.048356 | chr16:1-7328344 | 209 |
| 4p15.31 | 0.045061 | 0.05196 | chr4:20336563-21999843 | 2 |
| 7q31.33 | 0.038137 | 0.056388 | chr7:108310780-147750120 | 244 |
| 13q12.11 | 0.021354 | 0.064195 | chr13:1-28497722 | 70 |
| 19p13.3 | 0.064992 | 0.06566 | chr19:1-13721363 | 419 |
| 22q11.21 | 0.076337 | 0.072813 | chr22:1-18213813 | 47 |
| 10p15.3 | 0.079927 | 0.080027 | chr10:1-4683335 | 15 |
| 4p16.2 | 0.053991 | 0.093143 | chr4:1-5067215 | 70 |
| 4q32.3 | 0.0049654 | 0.093143 | chr4:143166997-191273063 | 178 |
| 4p12 | 0.10907 | 0.12228 | chr4:41847185-52409225 | 27 |
| 16q24.3 | 0.12228 | 0.12228 | chr16:70168187-88827254 | 153 |
| 14q21.3 | 0.15584 | 0.15584 | chr14:1-61292206 | 309 |
| 8p23.3 | 4.0183e-06 | 0.16538 | chr8:1-2780982 | 13 |
| 9p24.1 | 0.00025666 | 0.17605 | chr9:4850981-14995054 | 27 |
| 13q14.2 | 0.010666 | 0.17605 | chr13:1-72205527 | 234 |
| 15q21.1 | 0.013256 | 0.1901 | chr15:1-52708651 | 350 |
| 4q23 | 0.056622 | 0.22405 | chr4:95893013-164308535 | 245 |
| 7q36.3 | 0.077406 | 0.22405 | chr7:130833759-158821424 | 217 |
| 19q13.33 | 0.23816 | 0.23419 | chr19:42572590-63811651 | 775 |
| 3q13.31 | 0.17245 | 0.24658 | chr3:1-199501827 | 1158 |
This is the comprehensive list of deleted genes in the wide peak for 9p21.3.
Table S28. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CDKN2A |
| C9orf53 |
This is the comprehensive list of deleted genes in the wide peak for 2q22.1.
Table S29. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| LRP1B |
This is the comprehensive list of deleted genes in the wide peak for 8p23.2.
Table S30. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CSMD1 |
This is the comprehensive list of deleted genes in the wide peak for 10q23.31.
Table S31. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| PTEN |
| KILLIN |
This is the comprehensive list of deleted genes in the wide peak for 5q11.2.
Table S32. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-582 |
| PDE4D |
| PART1 |
This is the comprehensive list of deleted genes in the wide peak for 3p14.2.
Table S33. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| FHIT |
This is the comprehensive list of deleted genes in the wide peak for 2q37.3.
Table S34. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| ATIC |
| ACSL3 |
| IDH1 |
| PAX3 |
| FEV |
| hsa-mir-149 |
| hsa-mir-562 |
| hsa-mir-1471 |
| hsa-mir-1244 |
| hsa-mir-153-1 |
| hsa-mir-375 |
| hsa-mir-26b |
| hsa-mir-548f-2 |
| AAMP |
| ACADL |
| AGXT |
| ALPI |
| ALPP |
| ALPPL2 |
| KIF1A |
| BARD1 |
| BCS1L |
| BOK |
| CHRND |
| CHRNG |
| COL4A3 |
| COL4A4 |
| COL6A3 |
| CPS1 |
| CRYBA2 |
| CRYGA |
| CRYGB |
| CRYGC |
| CRYGD |
| CYP27A1 |
| DES |
| DTYMK |
| EPHA4 |
| ERBB4 |
| FN1 |
| GBX2 |
| GPC1 |
| GPR35 |
| HDLBP |
| AGFG1 |
| DNAJB2 |
| HTR2B |
| SP110 |
| IGFBP2 |
| IGFBP5 |
| IHH |
| IL8RA |
| IL8RB |
| IL8RBP |
| INHA |
| INPP5D |
| IRS1 |
| KCNJ13 |
| MAP2 |
| MYL1 |
| NCL |
| NDUFA10 |
| SEPT2 |
| NEU2 |
| NPPC |
| PDCD1 |
| PDE6D |
| SERPINE2 |
| PPP1R7 |
| PSMD1 |
| PTH2R |
| PTMA |
| PTPRN |
| SNORD20 |
| RPE |
| RPL37A |
| SAG |
| CCL20 |
| SLC4A3 |
| SLC11A1 |
| SP100 |
| SPP2 |
| TNP1 |
| TNS1 |
| TUBA4A |
| VIL1 |
| WNT6 |
| XRCC5 |
| ZNF142 |
| SCG2 |
| CUL3 |
| DGKD |
| STK16 |
| PER2 |
| CDK5R2 |
| RQCD1 |
| LRRFIP1 |
| GPR55 |
| TRIP12 |
| ECEL1 |
| EIF4E2 |
| TTLL4 |
| HDAC4 |
| FARP2 |
| FARSB |
| ABCB6 |
| ARPC2 |
| ARL4C |
| RAMP1 |
| SPEG |
| LANCL1 |
| NMUR1 |
| STK25 |
| COPS8 |
| CAPN10 |
| SP140 |
| IKZF2 |
| PASK |
| ATG4B |
| OBSL1 |
| DNPEP |
| SH3BP4 |
| KCNE4 |
| NGEF |
| SNORD82 |
| PNKD |
| SNED1 |
| GIGYF2 |
| TRAF3IP1 |
| ABCA12 |
| C2orf24 |
| STK36 |
| LOC29034 |
| GMPPA |
| SMARCAL1 |
| ANO7 |
| PRLH |
| THAP4 |
| ANKMY1 |
| SCLY |
| ASB1 |
| CAB39 |
| PRKAG3 |
| UGT1A10 |
| UGT1A8 |
| UGT1A7 |
| UGT1A6 |
| UGT1A5 |
| UGT1A9 |
| UGT1A4 |
| UGT1A1 |
| UGT1A3 |
| PID1 |
| ATG16L1 |
| ANKZF1 |
| USP40 |
| HJURP |
| HES6 |
| ACCN4 |
| DOCK10 |
| MREG |
| PECR |
| C2orf83 |
| MFF |
| CXCR7 |
| RNPEPL1 |
| MARCH4 |
| WDFY1 |
| KIAA1486 |
| USP37 |
| CTDSP1 |
| GAL3ST2 |
| TMBIM1 |
| RAB17 |
| RNF25 |
| COPS7B |
| MRPL44 |
| TRPM8 |
| ATG9A |
| MLPH |
| FAM134A |
| GLB1L |
| SPAG16 |
| CHPF |
| IQCA1 |
| NHEJ1 |
| FAM124B |
| TM4SF20 |
| C2orf54 |
| TUBA4B |
| ARMC9 |
| EFHD1 |
| SPHKAP |
| WNT10A |
| SLC19A3 |
| ILKAP |
| ITM2C |
| RHBDD1 |
| ING5 |
| PLCD4 |
| MGC16025 |
| TMEM169 |
| DNER |
| B3GNT7 |
| SP140L |
| STK11IP |
| MOGAT1 |
| AGAP1 |
| DIS3L2 |
| NEU4 |
| AP1S3 |
| SGPP2 |
| SPATA3 |
| TMEM198 |
| ZFAND2B |
| FBXO36 |
| MTERFD2 |
| UBE2F |
| OTOS |
| MYEOV2 |
| OR6B3 |
| C2orf67 |
| LOC151174 |
| CCDC140 |
| SLC23A3 |
| LOC151300 |
| GPBAR1 |
| SLC16A14 |
| C2orf52 |
| MSL3L2 |
| WDR69 |
| C2orf57 |
| PIKFYVE |
| TIGD1 |
| CCDC108 |
| C2orf72 |
| C2orf85 |
| UNC80 |
| RUFY4 |
| DUSP28 |
| ESPNL |
| ECEL1P2 |
| C2orf62 |
| RBM44 |
| AQP12A |
| KLHL30 |
| PLEKHM3 |
| C2orf80 |
| RESP18 |
| C2orf82 |
| OR6B2 |
| ASB18 |
| VWC2L |
| MIR149 |
| MIR153-1 |
| MIR26B |
| DNAJB3 |
| MIR375 |
| LOC643387 |
| PRR21 |
| AQP12B |
| SNORA75 |
| SCARNA6 |
| SCARNA5 |
| D2HGDH |
| LOC728323 |
| DIRC3 |
| PP14571 |
This is the comprehensive list of deleted genes in the wide peak for 6q26.
Table S35. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| PARK2 |
This is the comprehensive list of deleted genes in the wide peak for 11p15.5.
Table S36. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CARS |
| HRAS |
| NUP98 |
| hsa-mir-483 |
| hsa-mir-675 |
| hsa-mir-210 |
| AP2A2 |
| RHOG |
| ART1 |
| ASCL2 |
| CD81 |
| CD151 |
| CDKN1C |
| CTSD |
| DRD4 |
| DUSP8 |
| IGF2 |
| INS |
| IRF7 |
| KCNQ1 |
| LSP1 |
| MUC2 |
| MUC6 |
| NAP1L4 |
| SLC22A18 |
| SLC22A18AS |
| POLR2L |
| PSMD13 |
| RNH1 |
| MRPL23 |
| RPLP2 |
| RRM1 |
| SCT |
| TRIM21 |
| STIM1 |
| TALDO1 |
| TH |
| TSPAN4 |
| TNNI2 |
| TNNT3 |
| TRPC2 |
| PHLDA2 |
| ZNF195 |
| RASSF7 |
| IFITM1 |
| BRSK2 |
| TSPAN32 |
| TSSC4 |
| IFITM3 |
| DEAF1 |
| IFITM2 |
| KCNQ1OT1 |
| PKP3 |
| SIRT3 |
| PGAP2 |
| C11orf21 |
| TRPM5 |
| IGF2AS |
| BET1L |
| CEND1 |
| MUPCDH |
| TOLLIP |
| TRIM68 |
| LRDD |
| KCNQ1DN |
| MMP26 |
| CHRNA10 |
| PNPLA2 |
| PHRF1 |
| SIGIRR |
| RIC8A |
| EPS8L2 |
| CHID1 |
| OR51G1 |
| SLC25A22 |
| ATHL1 |
| OR51G2 |
| OR51E2 |
| PTDSS2 |
| HCCA2 |
| SYT8 |
| ODF3 |
| OSBPL5 |
| LRRC56 |
| MRGPRE |
| ART5 |
| OR52E2 |
| OR52J3 |
| OR51L1 |
| OR51A7 |
| OR51S1 |
| OR51F2 |
| OR52R1 |
| OR52M1 |
| OR52K2 |
| OR52B4 |
| C11orf40 |
| OR52I2 |
| OR51E1 |
| LOC143666 |
| SCGB1C1 |
| NLRP6 |
| LOC255512 |
| C11orf35 |
| OR51F1 |
| H19 |
| EFCAB4A |
| TMEM80 |
| C11orf36 |
| ANO9 |
| LOC338651 |
| B4GALNT4 |
| PDDC1 |
| MRGPRG |
| KRTAP5-1 |
| KRTAP5-3 |
| KRTAP5-4 |
| IFITM5 |
| FAM99A |
| OR52K1 |
| OR52I1 |
| OR51D1 |
| OR51T1 |
| OR51A4 |
| OR51A2 |
| MIR210 |
| KRTAP5-5 |
| KRTAP5-2 |
| KRTAP5-6 |
| MIR483 |
| SNORA52 |
| LOC650368 |
| SNORA54 |
| INS-IGF2 |
| MUC5B |
| MIR675 |
| FAM99B |
| LOC100133161 |
| LOC100133545 |
This is the comprehensive list of deleted genes in the wide peak for 3p25.2.
Table S37. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| VGLL4 |
This is the comprehensive list of deleted genes in the wide peak for 9p21.2.
Table S38. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-873 |
| LINGO2 |
| MIR876 |
| MIR873 |
This is the comprehensive list of deleted genes in the wide peak for 9p13.1.
Table S39. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-204 |
| hsa-mir-1299 |
| ALDH1A1 |
| ANXA1 |
| APBA1 |
| KLF9 |
| FXN |
| GCNT1 |
| PCSK5 |
| PGM5 |
| PRKACG |
| RORB |
| ZFAND5 |
| PIP5K1B |
| FAM189A2 |
| TJP2 |
| GDA |
| SMC5 |
| TMEM2 |
| ZNF658 |
| FAM75A7 |
| OSTF1 |
| PCA3 |
| FAM108B1 |
| C9orf95 |
| C9orf40 |
| RFK |
| CNTNAP3 |
| TRPM3 |
| ANKRD20A1 |
| FAM122A |
| TMC1 |
| C9orf41 |
| C9orf57 |
| C9orf85 |
| C9orf135 |
| TRPM6 |
| C9orf122 |
| PRUNE2 |
| C9orf71 |
| CBWD5 |
| MAMDC2 |
| FOXD4L3 |
| FOXD4L4 |
| AQP7P1 |
| PTAR1 |
| KGFLP1 |
| FAM75A6 |
| MGC21881 |
| AQP7P2 |
| FAM74A1 |
| FAM74A4 |
| ZNF658B |
| MIR204 |
| LOC440839 |
| LOC440896 |
| ANKRD20A3 |
| ANKRD20A2 |
| AQP7P3 |
| LOC442421 |
| FOXB2 |
| CBWD3 |
| FAM27A |
| LOC572558 |
| PGM5P2 |
| FAM75A2 |
| LOC642929 |
| CBWD6 |
| FAM75A1 |
| RPSAP9 |
| FOXD4L6 |
| FOXD4L5 |
| LOC653501 |
| KGFLP2 |
| FAM75A3 |
| FAM75A5 |
| FAM74A3 |
| ANKRD20A4 |
| CCDC29 |
| FOXD4L2 |
| LOC100130426 |
| FAM27C |
| FAM95B1 |
| FAM27B |
| LOC100133920 |
This is the comprehensive list of deleted genes in the wide peak for 4q35.2.
Table S40. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| DUX4 |
| hsa-mir-1305 |
| SLC25A4 |
| CASP3 |
| DCTD |
| F11 |
| ACSL1 |
| FAT1 |
| FRG1 |
| ING2 |
| IRF2 |
| KLKB1 |
| MTNR1A |
| TLR3 |
| SORBS2 |
| FAM149A |
| PDLIM3 |
| CLDN22 |
| UFSP2 |
| CDKN2AIP |
| ODZ3 |
| LRP2BP |
| TUBB4Q |
| STOX2 |
| KIAA1430 |
| C4orf41 |
| MLF1IP |
| WWC2 |
| SNX25 |
| ZFP42 |
| ENPP6 |
| C4orf38 |
| RWDD4A |
| CCDC111 |
| TRIML2 |
| CCDC110 |
| CYP4V2 |
| TRIML1 |
| ANKRD37 |
| HELT |
| FAM92A3 |
| C4orf47 |
| FRG2 |
| SLED1 |
| LOC653543 |
| LOC653544 |
| LOC653545 |
| LOC653548 |
| LOC728410 |
This is the comprehensive list of deleted genes in the wide peak for 21q21.1.
Table S41. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-125b-2 |
| BAGE |
| CXADR |
| PRSS7 |
| HSPA13 |
| TPTE |
| NRIP1 |
| BTG3 |
| USP25 |
| RBM11 |
| NCRNA00157 |
| C21orf15 |
| C21orf91 |
| SAMSN1 |
| BAGE5 |
| BAGE4 |
| BAGE3 |
| BAGE2 |
| CHODL |
| C21orf99 |
| LIPI |
| ABCC13 |
| POTED |
| C21orf131 |
| C21orf34 |
| C21orf81 |
| MIRLET7C |
| MIR125B2 |
| MIR99A |
This is the comprehensive list of deleted genes in the wide peak for 1p13.1.
Table S42. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| NOTCH2 |
| NRAS |
| PDE4DIP |
| TRIM33 |
| RBM15 |
| hsa-mir-942 |
| hsa-mir-320b-1 |
| hsa-mir-197 |
| ADORA3 |
| ALX3 |
| AMPD1 |
| AMPD2 |
| AMY1A |
| AMY1B |
| AMY1C |
| AMY2A |
| AMY2B |
| RHOC |
| ATP1A1 |
| ATP5F1 |
| CAPZA1 |
| CASQ2 |
| CD2 |
| CD53 |
| CD58 |
| CHI3L2 |
| CSF1 |
| CELSR2 |
| FCGR1B |
| GNAI3 |
| GNAT2 |
| GSTM1 |
| GSTM2 |
| GSTM3 |
| GSTM4 |
| GSTM5 |
| HMGCS2 |
| HSD3B1 |
| HSD3B2 |
| IGSF3 |
| KCNA2 |
| KCNA3 |
| KCNA10 |
| KCNC4 |
| KCND3 |
| MOV10 |
| NGF |
| NHLH2 |
| OVGP1 |
| PSMA5 |
| PTGFRN |
| RAP1A |
| SORT1 |
| SARS |
| SLC16A1 |
| STXBP3 |
| SYCP1 |
| TAF13 |
| TBX15 |
| TSHB |
| WNT2B |
| CSDE1 |
| TTF2 |
| SLC16A4 |
| CD101 |
| SEC22B |
| LRIG2 |
| TSPAN2 |
| BCAS2 |
| WARS2 |
| CEPT1 |
| VAV3 |
| HBXIP |
| AP4B1 |
| PHTF1 |
| AHCYL1 |
| WDR3 |
| MAN1A2 |
| ADAM30 |
| DDX20 |
| NTNG1 |
| WDR47 |
| CLCC1 |
| PTPN22 |
| PHGDH |
| CHIA |
| GPSM2 |
| SLC25A24 |
| HAO2 |
| RSBN1 |
| GDAP2 |
| FAM46C |
| ST7L |
| PRPF38B |
| PRMT6 |
| SLC22A15 |
| RNPC3 |
| C1orf103 |
| CTTNBP2NL |
| C1orf183 |
| TMEM167B |
| OLFML3 |
| AMIGO1 |
| KIAA1324 |
| DCLRE1B |
| WDR77 |
| EPS8L3 |
| VTCN1 |
| DENND2D |
| SIKE1 |
| TRIM45 |
| VANGL1 |
| GPR61 |
| REG4 |
| PROK1 |
| PSRC1 |
| C1orf203 |
| FAM40A |
| ZNF697 |
| C1orf59 |
| C1orf161 |
| ATXN7L2 |
| C1orf194 |
| DRAM2 |
| C1orf88 |
| C1orf162 |
| SYT6 |
| NBPF4 |
| LOC149620 |
| DENND2C |
| FNDC7 |
| PPIAL4A |
| UBL4B |
| SPAG17 |
| HIPK1 |
| AFARP1 |
| AKNAD1 |
| MAGI3 |
| FAM19A3 |
| FAM102B |
| SYPL2 |
| CYB561D1 |
| PPM1J |
| HIST2H2BA |
| MYBPHL |
| NBPF7 |
| SLC6A17 |
| NOTCH2NL |
| FLJ39739 |
| NBPF20 |
| MIR197 |
| BCL2L15 |
| LOC441897 |
| CYMP |
| PPIAL4G |
| LOC647121 |
| LOC648740 |
| NBPF6 |
| PPIAL4B |
| PPIAL4C |
| FAM72B |
| SCARNA2 |
| LOC728855 |
| LOC728875 |
| C1orf152 |
| MIR942 |
| NBPF10 |
| LOC100286793 |
This is the comprehensive list of deleted genes in the wide peak for 20p12.1.
Table S43. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| FLRT3 |
| MACROD2 |
This is the comprehensive list of deleted genes in the wide peak for 11q24.3.
Table S44. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| FLI1 |
| APLP2 |
| ETS1 |
| KCNJ1 |
| KCNJ5 |
| NFRKB |
| OPCML |
| ST14 |
| BARX2 |
| RICS |
| ADAMTS8 |
| IGSF9B |
| NCAPD3 |
| ACAD8 |
| B3GAT1 |
| ZBTB44 |
| THYN1 |
| NTM |
| PRDM10 |
| TP53AIP1 |
| JAM3 |
| GLB1L2 |
| VPS26B |
| GLB1L3 |
| TMEM45B |
| ADAMTS15 |
| C11orf45 |
| SPATA19 |
| LOC283174 |
| SNX19 |
| NCRNA00167 |
| LOC100128239 |
This is the comprehensive list of deleted genes in the wide peak for 3p11.2.
Table S45. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| EPHA3 |
| GBE1 |
| HTR1F |
| POU1F1 |
| ROBO1 |
| CGGBP1 |
| CHMP2B |
| ZNF654 |
| CADM2 |
| C3orf38 |
| VGLL3 |
This is the comprehensive list of deleted genes in the wide peak for 3p14.1.
Table S46. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| MITF |
| FOXP1 |
| hsa-mir-1324 |
| hsa-mir-1284 |
| GBE1 |
| GPR27 |
| CNTN3 |
| ROBO1 |
| ROBO2 |
| TMF1 |
| UBA3 |
| ARL6IP5 |
| PDZRN3 |
| FRMD4B |
| RYBP |
| FLJ10213 |
| SHQ1 |
| LMOD3 |
| PROK2 |
| FAM19A4 |
| PPP4R2 |
| CADM2 |
| C3orf64 |
| EIF4E3 |
| FAM86D |
| GLT8D4 |
| ZNF717 |
| FRG2C |
This is the comprehensive list of deleted genes in the wide peak for 17q11.2.
Table S47. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| NF1 |
| EVI2A |
| EVI2B |
| OMG |
| RNF135 |
| DPRXP4 |
This is the comprehensive list of deleted genes in the wide peak for 4q22.1.
Table S48. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| TMSL3 |
| FAM190A |
This is the comprehensive list of deleted genes in the wide peak for 18q23.
Table S49. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| GALR1 |
| MBP |
| NFATC1 |
| ZNF236 |
| CTDP1 |
| ZNF516 |
| TXNL4A |
| ADNP2 |
| KCNG2 |
| SALL3 |
| C18orf22 |
| PQLC1 |
| PARD6G |
| C18orf62 |
| LOC284276 |
| ATP9B |
| HSBP1L1 |
| LOC100130522 |
This is the comprehensive list of deleted genes in the wide peak for 15q11.2.
Table S50. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BUB1B |
| C15orf55 |
| C15orf21 |
| hsa-mir-147b |
| hsa-mir-1282 |
| hsa-mir-627 |
| hsa-mir-626 |
| hsa-mir-1233 |
| hsa-mir-1233 |
| hsa-mir-211 |
| hsa-mir-1268 |
| ACTC1 |
| APBA2 |
| B2M |
| BCL8 |
| CAPN3 |
| CHRM5 |
| CHRNA7 |
| CKMT1B |
| DUT |
| EPB42 |
| FBN1 |
| GABRA5 |
| GABRB3 |
| GABRG3 |
| GANC |
| GATM |
| GCHFR |
| PDIA3 |
| IPW |
| ITPKA |
| IVD |
| LTK |
| MAP1A |
| MEIS2 |
| MFAP1 |
| TRPM1 |
| NDN |
| OCA2 |
| PLCB2 |
| RAD51 |
| RYR3 |
| SCG5 |
| SLC12A1 |
| SNRPN |
| SORD |
| SPINT1 |
| SRP14 |
| THBS1 |
| TJP1 |
| TP53BP1 |
| TYRO3 |
| UBE3A |
| MKRN3 |
| SLC30A4 |
| PAR5 |
| EIF3J |
| JMJD7-PLA2G4B |
| SNAP23 |
| HERC2 |
| SNURF |
| SLC28A2 |
| TGM5 |
| HISPPD2A |
| AQR |
| ARHGAP11A |
| LCMT2 |
| SLC12A6 |
| RASGRP1 |
| SERF2 |
| GPR176 |
| CHP |
| OIP5 |
| BAHD1 |
| MTMR15 |
| CEP152 |
| MAPKBP1 |
| GOLGA8A |
| RTF1 |
| CYFIP1 |
| MGA |
| VPS39 |
| FAM189A1 |
| CCNDBP1 |
| EID1 |
| C15orf2 |
| C15orf63 |
| TMEM87A |
| RPAP1 |
| DKFZP434L187 |
| PLDN |
| GREM1 |
| RPUSD2 |
| TUBGCP4 |
| EHD4 |
| DUOX2 |
| MYEF2 |
| NDUFAF1 |
| NUSAP1 |
| TMEM85 |
| SPTBN5 |
| CTDSPL2 |
| KLF13 |
| DUOX1 |
| MAGEL2 |
| DLL4 |
| INO80 |
| PPP1R14D |
| MTMR10 |
| ZNF770 |
| HAUS2 |
| FAM82A2 |
| DNAJC17 |
| NOP10 |
| NDNL2 |
| C15orf24 |
| PAK6 |
| CASC5 |
| AVEN |
| ATP10A |
| GJD2 |
| VPS18 |
| SQRDL |
| ZFP106 |
| SPATA5L1 |
| CHAC1 |
| C15orf29 |
| WDR76 |
| TMEM62 |
| SEMA6D |
| SPG11 |
| ELL3 |
| NIPA2 |
| C15orf48 |
| C15orf41 |
| ZFYVE19 |
| FRMD5 |
| DISP2 |
| CHRFAM7A |
| FAM7A3 |
| FAM7A1 |
| ARHGAP11B |
| ATPBD4 |
| C15orf57 |
| C15orf23 |
| BMF |
| SHF |
| DUOXA1 |
| SNORD107 |
| CHST14 |
| CASC4 |
| TUBGCP5 |
| TGM7 |
| CATSPER2 |
| NIPA1 |
| PLA2G4E |
| TRIM69 |
| PAR1 |
| C15orf43 |
| LOC145845 |
| TMCO5A |
| ZSCAN29 |
| TTBK2 |
| CDAN1 |
| STRC |
| CSNK1A1P |
| OTUD7A |
| SPRED1 |
| PGBD4 |
| ADAL |
| EXD1 |
| FSIP1 |
| RHOV |
| UBR1 |
| PATL2 |
| LPCAT4 |
| PLA2G4F |
| LRRC57 |
| SLC24A5 |
| OR4N4 |
| FAM98B |
| PLA2G4D |
| GOLGA6L1 |
| GOLGA8G |
| GOLGA9P |
| SNORD108 |
| SNORD109A |
| SNORD109B |
| SNORD115-1 |
| WHAMML1 |
| POTEB |
| FMN1 |
| SNORD64 |
| PAR4 |
| PAR-SN |
| MRPL42P5 |
| C15orf52 |
| GOLGA8E |
| OR4M2 |
| OR4N3P |
| SHC4 |
| CTXN2 |
| HERC2P2 |
| C15orf53 |
| C15orf54 |
| DUOXA2 |
| MIR211 |
| NF1P1 |
| WHAMML2 |
| GOLGA8B |
| EIF2AK4 |
| CATSPER2P1 |
| CKMT1A |
| SERINC4 |
| C15orf62 |
| C15orf56 |
| PHGR1 |
| LOC646214 |
| CXADRP2 |
| MIR548A3 |
| MIR626 |
| MIR627 |
| LOC723972 |
| SNORD116-19 |
| GOLGA6L6 |
| LOC727924 |
| LOC728758 |
| LOC729082 |
| GOLGA8C |
| PWRN1 |
| PWRN2 |
| SNORD116-1 |
| SNORD116-2 |
| SNORD116-3 |
| SNORD116-4 |
| SNORD116-5 |
| SNORD116-6 |
| SNORD116-7 |
| SNORD116-8 |
| SNORD116-9 |
| SNORD116-10 |
| SNORD116-11 |
| SNORD116-12 |
| SNORD116-13 |
| SNORD116-14 |
| SNORD116-15 |
| SNORD116-16 |
| SNORD116-17 |
| SNORD116-18 |
| SNORD116-20 |
| SNORD116-21 |
| SNORD116-22 |
| SNORD116-23 |
| SNORD116-24 |
| SNORD116-25 |
| SNORD115-2 |
| SNORD116-26 |
| SNORD116-27 |
| SNORD115-3 |
| SNORD115-4 |
| SNORD115-5 |
| SNORD115-6 |
| SNORD115-7 |
| SNORD115-8 |
| SNORD115-9 |
| SNORD115-10 |
| SNORD115-11 |
| SNORD115-12 |
| SNORD115-13 |
| SNORD115-14 |
| SNORD115-15 |
| SNORD115-16 |
| SNORD115-17 |
| SNORD115-18 |
| SNORD115-19 |
| SNORD115-20 |
| SNORD115-21 |
| SNORD115-22 |
| SNORD115-23 |
| SNORD115-25 |
| SNORD115-26 |
| SNORD115-29 |
| SNORD115-30 |
| SNORD115-31 |
| SNORD115-32 |
| SNORD115-33 |
| SNORD115-34 |
| SNORD115-35 |
| SNORD115-36 |
| SNORD115-37 |
| SNORD115-38 |
| SNORD115-39 |
| SNORD115-40 |
| SNORD115-41 |
| SNORD115-42 |
| SNORD115-43 |
| SNORD115-44 |
| SNORD116-28 |
| SNORD116-29 |
| SNORD115-48 |
| HBII-52-24 |
| HBII-52-27 |
| HBII-52-28 |
| HBII-52-45 |
| HBII-52-46 |
| MIR147B |
| GOLGA8F |
| GOLGA8D |
| JMJD7 |
| PLA2G4B |
| FAM7A2 |
This is the comprehensive list of deleted genes in the wide peak for 1p36.12.
Table S51. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| LCK |
| PAX7 |
| RPL22 |
| SDHB |
| ARID1A |
| TNFRSF14 |
| PRDM16 |
| MDS2 |
| hsa-mir-1976 |
| hsa-mir-1256 |
| hsa-mir-1290 |
| hsa-mir-34a |
| hsa-mir-551a |
| hsa-mir-429 |
| hsa-mir-1977 |
| hsa-mir-1302-2 |
| AK2 |
| ALPL |
| RERE |
| BAI2 |
| C1QA |
| C1QB |
| C1QC |
| CA6 |
| CAPZB |
| CASP9 |
| RUNX3 |
| TNFRSF8 |
| CDA |
| CDC2L1 |
| CDC42 |
| CD52 |
| RCC1 |
| CLCN6 |
| CLCNKA |
| CLCNKB |
| CNR2 |
| COL16A1 |
| CORT |
| DDOST |
| DFFA |
| DFFB |
| DVL1 |
| E2F2 |
| ECE1 |
| PHC2 |
| MEGF6 |
| EPHA2 |
| ENO1 |
| EPB41 |
| EPHA8 |
| EPHB2 |
| EXTL1 |
| EYA3 |
| FABP3 |
| FGR |
| MTOR |
| FUCA1 |
| IFI6 |
| GABRD |
| GALE |
| GNB1 |
| SFN |
| GPR3 |
| HCRTR1 |
| HDAC1 |
| ZBTB48 |
| HMGN2 |
| HMGCL |
| HPCA |
| HSPG2 |
| HTR1D |
| HTR6 |
| ID3 |
| TNFRSF9 |
| STMN1 |
| MATN1 |
| MFAP2 |
| MTHFR |
| NBL1 |
| NPPA |
| NPPB |
| OPRD1 |
| PAFAH2 |
| PEX10 |
| PEX14 |
| PGD |
| PIK3CD |
| PLA2G2A |
| PLA2G5 |
| PLOD1 |
| EXOSC10 |
| PPP1R8 |
| PRKCZ |
| PTAFR |
| RAP1GAP |
| RBBP4 |
| RHCE |
| RHD |
| RPA2 |
| RPL11 |
| RPS6KA1 |
| RSC1A1 |
| SCNN1D |
| SFRS4 |
| SKI |
| SLC2A5 |
| SLC9A1 |
| SRM |
| TAF12 |
| TCEA3 |
| TCEB3 |
| TNFRSF1B |
| TP73 |
| TNFRSF4 |
| ZSCAN20 |
| ZBTB17 |
| SLC30A2 |
| LUZP1 |
| PRDM2 |
| LAPTM5 |
| PTP4A2 |
| SNHG3 |
| NR0B2 |
| MMP23B |
| MMP23A |
| KCNAB2 |
| FCN3 |
| YARS |
| AKR7A2 |
| ALDH4A1 |
| EIF3I |
| EIF4G3 |
| TNFRSF25 |
| TNFRSF18 |
| PER3 |
| MAP3K6 |
| DHRS3 |
| VAMP3 |
| SNRNP40 |
| C1orf38 |
| H6PD |
| ISG15 |
| PLCH2 |
| SDC3 |
| CROCC |
| PUM1 |
| KIAA0562 |
| KLHL21 |
| SLC35E2 |
| ZBTB40 |
| MFN2 |
| PTPRU |
| CELA3A |
| WASF2 |
| ANGPTL7 |
| HNRNPR |
| SRRM1 |
| CNKSR1 |
| UBE4B |
| MAD2L2 |
| PDPN |
| KHDRBS1 |
| GMEB1 |
| NUDC |
| MASP2 |
| SFRS13A |
| UTS2 |
| RER1 |
| RCAN3 |
| MSTP2 |
| MSTP9 |
| PADI2 |
| LYPLA2 |
| PARK7 |
| CTRC |
| ACOT7 |
| DNAJC8 |
| CLSTN1 |
| AKR7A3 |
| SPEN |
| KDM1A |
| WDTC1 |
| KIAA0090 |
| KIF1B |
| PLEKHM2 |
| OTUD3 |
| KIAA1026 |
| CAMTA1 |
| DNAJC16 |
| UBR4 |
| ATP13A2 |
| TARDBP |
| CELA3B |
| ICMT |
| PADI4 |
| TMEM50A |
| KPNA6 |
| STX12 |
| CLIC4 |
| SYF2 |
| CHD5 |
| C1orf144 |
| LDLRAP1 |
| NOC2L |
| FBXO2 |
| FBXO6 |
| PLA2G2D |
| OR4F3 |
| RNU11 |
| HSPB7 |
| ARHGEF16 |
| AHDC1 |
| SMPDL3B |
| PRO0611 |
| HSPC157 |
| SSU72 |
| UBIAD1 |
| PADI1 |
| PLA2G2E |
| WDR8 |
| SLC45A1 |
| HP1BP3 |
| CELA2B |
| ZNF593 |
| MECR |
| SDF4 |
| MRTO4 |
| YTHDF2 |
| ZCCHC17 |
| PADI3 |
| ERRFI1 |
| WNT4 |
| FBXO42 |
| RNF186 |
| MXRA8 |
| HES2 |
| GPN2 |
| FBLIM1 |
| MED18 |
| PQLC2 |
| CASZ1 |
| TRNAU1AP |
| CPSF3L |
| C1orf159 |
| AURKAIP1 |
| MRPL20 |
| AIM1L |
| TMEM51 |
| BSDC1 |
| XKR8 |
| TMEM39B |
| ARHGEF10L |
| VPS13D |
| ATAD3A |
| TMEM57 |
| TRIM62 |
| PANK4 |
| CAMK2N1 |
| ASAP3 |
| PNRC2 |
| PIGV |
| NBPF1 |
| NECAP2 |
| IQCC |
| DNAJC11 |
| RCC2 |
| AJAP1 |
| C1orf91 |
| FAM54B |
| CTNNBIP1 |
| C1orf63 |
| AGTRAP |
| C1orf128 |
| MAN1C1 |
| NIPAL3 |
| SEPN1 |
| KIAA0495 |
| PLEKHG5 |
| LRRC47 |
| PTCHD2 |
| KIF17 |
| KIAA1522 |
| HES4 |
| GRHL3 |
| IL22RA1 |
| MIIP |
| CELA2A |
| GPATCH3 |
| TINAGL1 |
| PLA2G2F |
| S100PBP |
| CCDC21 |
| NMNAT1 |
| VWA1 |
| PINK1 |
| MARCKSL1 |
| PRAMEF1 |
| PRAMEF2 |
| NADK |
| PHACTR4 |
| C1orf135 |
| CCDC28B |
| EFHD2 |
| MMEL1 |
| C1orf89 |
| OR4F5 |
| NKAIN1 |
| MUL1 |
| NOL9 |
| LIN28 |
| AGMAT |
| NCRNA00115 |
| MORN1 |
| GRRP1 |
| DHDDS |
| GPR157 |
| SPSB1 |
| GLTPD1 |
| ZNF436 |
| TAS1R2 |
| TAS1R1 |
| OR4F16 |
| SYNC |
| ACTL8 |
| TSSK3 |
| CCNL2 |
| SH3BGRL3 |
| SESN2 |
| ESPN |
| TAS1R3 |
| ATAD3B |
| TMEM222 |
| PLEKHN1 |
| USP48 |
| NBPF3 |
| ZDHHC18 |
| SLC25A33 |
| DDI2 |
| LZIC |
| TRIM63 |
| FAM167B |
| C1orf170 |
| CROCCL1 |
| SYTL1 |
| IGSF21 |
| SNHG12 |
| KIAA1751 |
| KIAA2013 |
| THAP3 |
| C1orf201 |
| SPOCD1 |
| UBXN11 |
| C1orf158 |
| FBXO44 |
| ATPIF1 |
| ADC |
| TMEM54 |
| CROCCL2 |
| FHAD1 |
| LOC115110 |
| RAB42 |
| FAM46B |
| RBP7 |
| ACAP3 |
| UBE2J2 |
| C1orf172 |
| AADACL3 |
| PUSL1 |
| B3GALT6 |
| IFFO2 |
| TPRG1L |
| C1orf93 |
| MYOM3 |
| RNF19B |
| KLHDC7A |
| VWA5B1 |
| UBXN10 |
| ARHGEF19 |
| ACTRT2 |
| MIB2 |
| C1orf127 |
| SAMD11 |
| LOC148413 |
| PHF13 |
| CCDC27 |
| C1orf213 |
| DCDC2B |
| ZNF362 |
| PDIK1L |
| C1orf64 |
| SLC2A7 |
| CALML6 |
| IL28RA |
| FAM43B |
| PAQR7 |
| FAM76A |
| TMEM201 |
| C1orf86 |
| TXLNA |
| C1orf126 |
| ATAD3C |
| AKR7L |
| FNDC5 |
| TTLL10 |
| TMCO4 |
| ZNF683 |
| NPHP4 |
| LOC284551 |
| FAM41C |
| LOC284632 |
| LOC284661 |
| SLC25A34 |
| ESPNP |
| C1orf174 |
| KLHL17 |
| C1orf70 |
| TMEM52 |
| MTMR9L |
| ZBTB8OS |
| AADACL4 |
| PRAMEF5 |
| HNRNPCL1 |
| PRAMEF9 |
| PRAMEF10 |
| SERINC2 |
| FAM131C |
| PADI6 |
| C1orf187 |
| SPATA21 |
| AGRN |
| APITD1 |
| CATSPER4 |
| GPR153 |
| FAM132A |
| HES5 |
| LOC388588 |
| RNF207 |
| TMEM82 |
| TRNP1 |
| CD164L2 |
| HES3 |
| PRAMEF12 |
| PRAMEF21 |
| PRAMEF8 |
| PRAMEF18 |
| PRAMEF17 |
| PLA2G2C |
| TMEM200B |
| PRAMEF4 |
| PRAMEF13 |
| SH2D5 |
| C1orf130 |
| PRAMEF3 |
| LDLRAD2 |
| MIR200A |
| MIR200B |
| MIR34A |
| FLJ42875 |
| PRAMEF11 |
| PRAMEF6 |
| LOC440563 |
| UQCRHL |
| C1orf151 |
| LOC441869 |
| PRAMEF7 |
| PEF1 |
| MIR429 |
| FAM138F |
| LOC643837 |
| TMEM88B |
| C1orf200 |
| PRAMEF19 |
| PRAMEF20 |
| FAM138A |
| LOC646471 |
| LOC649330 |
| ZBTB8A |
| LOC653566 |
| PRAMEF22 |
| PRAMEF15 |
| WASH5P |
| PRAMEF16 |
| FAM138C |
| SCARNA1 |
| SNORA44 |
| SNORA61 |
| SNORA59B |
| SNORA59A |
| SNORA16A |
| SNORD85 |
| SNORD99 |
| SNORD103A |
| MIR551A |
| ZBTB8B |
| CDC2L2 |
| LOC728661 |
| PRAMEF14 |
| FLJ37453 |
| OR4F29 |
| SNHG3-RCC1 |
| LOC100128003 |
| LOC100128071 |
| LOC100128842 |
| LOC100129534 |
| FLJ39609 |
| LOC100132062 |
| LOC100132287 |
| LOC100133331 |
| LOC100133612 |
| LOC100288778 |
This is the comprehensive list of deleted genes in the wide peak for 7q11.22.
Table S52. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| AUTS2 |
This is the comprehensive list of deleted genes in the wide peak for 4q34.3.
Table S53. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| DUX4 |
| hsa-mir-1305 |
| AGA |
| SLC25A4 |
| CASP3 |
| DCTD |
| F11 |
| ACSL1 |
| FAT1 |
| FRG1 |
| GPM6A |
| HMGB2 |
| HPGD |
| ING2 |
| IRF2 |
| KLKB1 |
| MTNR1A |
| TLR3 |
| VEGFC |
| GLRA3 |
| SORBS2 |
| SAP30 |
| HAND2 |
| MORF4 |
| ADAM29 |
| SCRG1 |
| FAM149A |
| FBXO8 |
| PDLIM3 |
| GALNT7 |
| CLDN22 |
| NEIL3 |
| UFSP2 |
| CDKN2AIP |
| ODZ3 |
| LRP2BP |
| TUBB4Q |
| STOX2 |
| KIAA1430 |
| SPCS3 |
| C4orf41 |
| MLF1IP |
| NBLA00301 |
| WWC2 |
| KIAA1712 |
| SNX25 |
| MGC45800 |
| WDR17 |
| ZFP42 |
| SPATA4 |
| ENPP6 |
| ASB5 |
| C4orf38 |
| RWDD4A |
| CCDC111 |
| TRIML2 |
| CCDC110 |
| CYP4V2 |
| LOC285501 |
| TRIML1 |
| ANKRD37 |
| HELT |
| FAM92A3 |
| C4orf47 |
| GALNTL6 |
| FRG2 |
| SLED1 |
| LOC653543 |
| LOC653544 |
| LOC653545 |
| LOC653548 |
| LOC728410 |
This is the comprehensive list of deleted genes in the wide peak for 16p13.3.
Table S54. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CREBBP |
| TSC2 |
| hsa-mir-940 |
| hsa-mir-1225 |
| hsa-mir-662 |
| ABCA3 |
| ADCY9 |
| ARHGDIG |
| ATP6V0C |
| CCNF |
| CLCN7 |
| DCI |
| DNASE1 |
| DNASE1L2 |
| E4F1 |
| GFER |
| HAGH |
| HBA1 |
| HBA2 |
| HBM |
| HBQ1 |
| HBZ |
| HMOX2 |
| IGFALS |
| MEFV |
| MPG |
| NDUFB10 |
| NME3 |
| NME4 |
| NTHL1 |
| NTN3 |
| OR1F1 |
| OR2C1 |
| PDPK1 |
| PKD1 |
| PPL |
| RPL3L |
| RPS2 |
| SRL |
| SOLH |
| SSTR5 |
| TCEB2 |
| TFAP4 |
| TPSAB1 |
| UBE2I |
| ZNF75A |
| ZNF174 |
| ZNF200 |
| ZNF205 |
| ZNF213 |
| C16orf35 |
| AXIN1 |
| RGS11 |
| CACNA1H |
| BAIAP3 |
| RHBDL1 |
| CLDN6 |
| CLDN9 |
| PKMYT1 |
| PIGQ |
| DNAJA3 |
| SYNGR3 |
| IL32 |
| SLC9A3R2 |
| SEC14L5 |
| RAB11FIP3 |
| IFT140 |
| TELO2 |
| NUBP2 |
| ZNF263 |
| TRAP1 |
| MSLN |
| STUB1 |
| MRPL28 |
| TBL3 |
| RNPS1 |
| PRSS21 |
| CLUAP1 |
| MAPK8IP3 |
| MGRN1 |
| TPSD1 |
| SRRM2 |
| TPSG1 |
| RAB26 |
| ZNF500 |
| DECR2 |
| OR1F2P |
| SNORA64 |
| SNORD60 |
| UBN1 |
| C16orf5 |
| SOX8 |
| AMDHD2 |
| Magmas |
| NAGPA |
| TNFRSF12A |
| POLR3K |
| SEPX1 |
| GNG13 |
| KCTD5 |
| ZNF434 |
| HCFC1R1 |
| LUC7L |
| ALG1 |
| NMRAL1 |
| TBC1D24 |
| CASKIN1 |
| CRAMP1L |
| RAB40C |
| TMEM8A |
| CHTF18 |
| PRSS22 |
| MLST8 |
| RHBDF1 |
| MMP25 |
| NARFL |
| C16orf10 |
| TPSB2 |
| HS3ST6 |
| PDIA2 |
| UNKL |
| LMF1 |
| FAM173A |
| MRPS34 |
| METRN |
| THOC6 |
| KREMEN2 |
| CORO7 |
| SNRNP25 |
| ROGDI |
| TMEM204 |
| NAT15 |
| C16orf59 |
| MGC3771 |
| FAHD1 |
| PRSS27 |
| ITFG3 |
| WDR24 |
| TRAF7 |
| FLYWCH1 |
| HAGHL |
| NUDT16L1 |
| C16orf13 |
| FAM195A |
| BTBD12 |
| GNPTG |
| GLYR1 |
| GLIS2 |
| ZSCAN10 |
| RHOT2 |
| ZNF598 |
| HN1L |
| SPSB3 |
| TIGD7 |
| RPUSD1 |
| FLYWCH2 |
| VASN |
| C16orf42 |
| WFIKKN1 |
| NOXO1 |
| CCDC78 |
| ZG16B |
| TMPRSS8 |
| PAQR4 |
| ANKS3 |
| FAM100A |
| SEPT12 |
| RNF151 |
| C16orf11 |
| FBXL16 |
| LOC146336 |
| ZNF597 |
| CCDC64B |
| C16orf89 |
| C16orf71 |
| FAM86A |
| WDR90 |
| EME2 |
| NLRC3 |
| C16orf73 |
| PRSS33 |
| NPW |
| C16orf79 |
| PGP |
| NHLRC4 |
| C16orf91 |
| JMJD8 |
| LOC342346 |
| TESSP1 |
| PRR25 |
| C1QTNF8 |
| C16orf38 |
| MSLNL |
| LOC440335 |
| SNORA10 |
| FLJ42627 |
| CCDC154 |
| C16orf90 |
| ABCA17P |
| LOC652276 |
| SNORA78 |
| MIR662 |
| SNHG9 |
| CEMP1 |
| MIR940 |
| LOC100128788 |
| LOC100134368 |
| MIR1225 |
| LOC100288778 |
This is the comprehensive list of deleted genes in the wide peak for 4p15.31.
Table S55. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| KCNIP4 |
| NCRNA00099 |
This is the comprehensive list of deleted genes in the wide peak for 7q31.33.
Table S56. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BRAF |
| MET |
| SMO |
| KIAA1549 |
| CREB3L2 |
| hsa-mir-548f-4 |
| hsa-mir-490 |
| hsa-mir-29b-1 |
| hsa-mir-335 |
| hsa-mir-183 |
| hsa-mir-129-1 |
| hsa-mir-593 |
| hsa-mir-592 |
| AKR1B1 |
| ARF5 |
| OPN1SW |
| BPGM |
| CALD1 |
| CALU |
| CAPZA2 |
| CASP2 |
| CAV1 |
| CAV2 |
| CFTR |
| CHRM2 |
| CLCN1 |
| CPA1 |
| CPA2 |
| EPHA1 |
| EPHB6 |
| FLNC |
| GPR37 |
| GRM8 |
| IFRD1 |
| IMPDH1 |
| IRF5 |
| KCND2 |
| KEL |
| LEP |
| MEST |
| MKLN1 |
| NDUFA5 |
| NDUFB2 |
| CNOT4 |
| NRF1 |
| PAX4 |
| PIP |
| PODXL |
| PPP1R3A |
| PRSS1 |
| PRSS2 |
| TAS2R38 |
| PTN |
| PTPRZ1 |
| SLC13A1 |
| SPAM1 |
| AKR1D1 |
| SSBP1 |
| TBXAS1 |
| UBE2H |
| WNT2 |
| ZYX |
| ST7 |
| ARHGEF5 |
| TRIM24 |
| MGAM |
| WASL |
| DGKI |
| ATP6V1F |
| FAM131B |
| DOCK4 |
| FAM115A |
| AASS |
| FAM3C |
| ZNF277 |
| TFEC |
| KLHDC10 |
| NUP205 |
| AHCYL2 |
| TNPO3 |
| HYAL4 |
| TSPAN12 |
| CLEC5A |
| MKRN1 |
| POT1 |
| CNTNAP2 |
| HYALP1 |
| TES |
| OR2F1 |
| SLC13A4 |
| COPG2 |
| TPK1 |
| SND1 |
| C7orf54 |
| DENND2A |
| HIPK2 |
| WDR91 |
| C7orf68 |
| MDFIC |
| FSCN3 |
| ATP6V0A4 |
| TAS2R3 |
| TAS2R4 |
| TAS2R16 |
| CPA4 |
| WNT16 |
| ZC3HC1 |
| LUC7L2 |
| MRPS33 |
| LSM8 |
| GPR85 |
| TAS2R5 |
| ING3 |
| LRRN3 |
| CHCHD3 |
| RBM28 |
| TMEM140 |
| TRPV6 |
| AGK |
| METTL2B |
| TRPV5 |
| ANKRD7 |
| ZC3HAV1 |
| AKR1B10 |
| KIAA1147 |
| FAM40B |
| EXOC4 |
| LRRC4 |
| TMEM168 |
| CCDC136 |
| PARP12 |
| C7orf49 |
| GCC1 |
| C7orf58 |
| TTC26 |
| JHDM1D |
| IMMP2L |
| CTTNBP2 |
| SLC37A3 |
| FAM71F1 |
| SLC35B4 |
| TMEM209 |
| OR6W1P |
| ADCK2 |
| PLXNA4 |
| ZC3HAV1L |
| LOC93432 |
| ST7OT1 |
| ST7OT2 |
| ST7OT3 |
| CADPS2 |
| CPA5 |
| FOXP2 |
| TSGA14 |
| TSGA13 |
| OR9A4 |
| OR9A2 |
| C7orf34 |
| TMEM139 |
| NOBOX |
| OR2A14 |
| OR6B1 |
| OR2F2 |
| PRSS37 |
| KLF14 |
| C7orf45 |
| SVOPL |
| MTPN |
| LRGUK |
| TRYX3 |
| MOXD2 |
| ASZ1 |
| ASB15 |
| C7orf60 |
| TRY6 |
| LOC154761 |
| CLEC2L |
| C7orf55 |
| IQUB |
| C7orf66 |
| TMEM213 |
| RNF133 |
| ZNF800 |
| UBN2 |
| TAS2R39 |
| TAS2R40 |
| TAS2R41 |
| FLJ40852 |
| FAM115C |
| C7orf53 |
| LOC286016 |
| MESTIT1 |
| ST7OT4 |
| TAS2R60 |
| CTAGE6 |
| TSPAN33 |
| AGBL3 |
| OR6V1 |
| OR2A12 |
| OR2A1 |
| FAM71F2 |
| STRA8 |
| KLRG2 |
| GSTK1 |
| KCP |
| FLJ43663 |
| RNF148 |
| FEZF1 |
| FAM180A |
| OR2A25 |
| OR2A5 |
| LOC401397 |
| PRRT4 |
| RAB19 |
| OR2A7 |
| OR2A20P |
| OR2A42 |
| FLJ45340 |
| MIR129-1 |
| MIR182 |
| MIR183 |
| MIR29A |
| MIR29B1 |
| MIR96 |
| LOC407835 |
| AKR1B15 |
| LOC441294 |
| OR2A9P |
| OR2A2 |
| EIF3IP1 |
| LMOD2 |
| MIR335 |
| ARHGEF5L |
| WEE2 |
| MIR490 |
| PL-5283 |
| MIR592 |
| MIR593 |
| TMEM229A |
| LUZP6 |
| LOC100124692 |
| CTAGE4 |
| LOC100134229 |
| LOC100134713 |
This is the comprehensive list of deleted genes in the wide peak for 13q12.11.
Table S57. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CDX2 |
| FLT3 |
| PARP4 |
| ATP12A |
| CDK8 |
| FGF9 |
| FLT1 |
| GJA3 |
| GJB2 |
| GPR12 |
| GTF3A |
| PDX1 |
| MIPEP |
| PABPC3 |
| RNF6 |
| RPL21 |
| SGCG |
| TUBA3C |
| ZMYM2 |
| IFT88 |
| MTMR6 |
| ZMYM5 |
| NUPL1 |
| SAP18 |
| GJB6 |
| WASF3 |
| SACS |
| LATS2 |
| SNORD102 |
| POLR1D |
| CRYL1 |
| POMP |
| ATP8A2 |
| IL17D |
| MPHOSPH8 |
| PSPC1 |
| TNFRSF19 |
| CENPJ |
| RNF17 |
| XPO4 |
| MRP63 |
| TPTE2 |
| FAM123A |
| USP12 |
| MTIF3 |
| GSX1 |
| N6AMT2 |
| SKA3 |
| EFHA1 |
| SPATA13 |
| LNX2 |
| ZDHHC20 |
| PAN3 |
| DKFZp686A1627 |
| SLC46A3 |
| LOC284232 |
| C1QTNF9 |
| LOC348021 |
| LOC374491 |
| RASL11A |
| C1QTNF9B |
| SHISA2 |
| ATP5EP2 |
| PCOTH |
| SNORA27 |
| LOC646405 |
| PRHOXNB |
| LOC100101938 |
| RPL21P28 |
| LOC100288730 |
This is the comprehensive list of deleted genes in the wide peak for 19p13.3.
Table S58. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| GNA11 |
| LYL1 |
| MLLT1 |
| SH3GL1 |
| SMARCA4 |
| STK11 |
| TCF3 |
| FSTL3 |
| hsa-mir-199a-1 |
| hsa-mir-638 |
| hsa-mir-1238 |
| hsa-mir-1181 |
| hsa-mir-220b |
| hsa-mir-7-3 |
| hsa-mir-637 |
| hsa-mir-1227 |
| hsa-mir-1909 |
| hsa-mir-1302-2 |
| ACP5 |
| AES |
| AMH |
| ASNA1 |
| ATP5D |
| AZU1 |
| HCN2 |
| BSG |
| C3 |
| CACNA1A |
| CALR |
| CAPS |
| CD70 |
| CDC34 |
| CDKN2D |
| CIRBP |
| CNN1 |
| CNN2 |
| CSNK1G2 |
| DAPK3 |
| CFD |
| DHPS |
| DNASE2 |
| DNM2 |
| DNMT1 |
| ARID3A |
| EEF2 |
| EFNA2 |
| ELANE |
| ELAVL1 |
| ELAVL3 |
| EMR1 |
| EPOR |
| FARSA |
| FCER2 |
| FUT3 |
| FUT5 |
| FUT6 |
| GAMT |
| GCDH |
| GNA15 |
| GNG7 |
| MKNK2 |
| GPX4 |
| GTF2F1 |
| GZMM |
| ICAM1 |
| ICAM3 |
| ICAM4 |
| ILF3 |
| INSR |
| JUNB |
| LDLR |
| MAN2B1 |
| MATK |
| MYO1F |
| GADD45B |
| HNRNPM |
| NDUFA7 |
| NFIC |
| NFIX |
| NRTN |
| OAZ1 |
| P2RY11 |
| PALM |
| PDE4A |
| PIN1 |
| POLR2E |
| POLRMT |
| PRKCSH |
| MAP2K2 |
| MAP2K7 |
| PSPN |
| PRTN3 |
| PTBP1 |
| PTPRS |
| RAD23A |
| RFX2 |
| RPS15 |
| RPS28 |
| SAFB |
| CCL25 |
| SGTA |
| SNAPC2 |
| STXBP2 |
| TBXA2R |
| PRDX2 |
| THOP1 |
| ICAM5 |
| TLE2 |
| TYK2 |
| VAV1 |
| ZNF20 |
| ZNF69 |
| ZNF121 |
| ZNF136 |
| ZNF177 |
| MADCAM1 |
| SF3A2 |
| CLPP |
| RANBP3 |
| KHSRP |
| PPAP2C |
| EIF3G |
| STX10 |
| S1PR4 |
| TNFSF14 |
| TNFSF9 |
| AP3D1 |
| RAB11B |
| S1PR2 |
| TRIP10 |
| LONP1 |
| RAB3D |
| APBA3 |
| IER2 |
| SAFB2 |
| KEAP1 |
| MED16 |
| CHAF1A |
| SH2D3A |
| AP1M2 |
| EBI3 |
| ZNF443 |
| PLIN3 |
| APC2 |
| CLEC4M |
| ABCA7 |
| HMG20B |
| TUBB4 |
| TIMM44 |
| CARM1 |
| SEMA6B |
| RNASEH2A |
| KLF1 |
| ZNF266 |
| PNPLA6 |
| UQCR |
| TMED1 |
| CDC37 |
| SBNO2 |
| MAST1 |
| KDM4B |
| ZFR2 |
| ARHGEF18 |
| PIP5K1C |
| HMHA1 |
| SHC2 |
| RPL36 |
| KANK2 |
| TIMM13 |
| TSPAN16 |
| DAZAP1 |
| OR7E24 |
| SNORD41 |
| SNORD37 |
| FGF22 |
| TJP3 |
| ITGB1BP3 |
| UHRF1 |
| HOOK2 |
| SLC39A3 |
| TNPO2 |
| CD209 |
| COL5A3 |
| RDH8 |
| MRPL4 |
| ANGPTL4 |
| MARCH2 |
| CD320 |
| ECSIT |
| THEG |
| ZBTB7A |
| FZR1 |
| C19orf56 |
| SIRT6 |
| PIAS4 |
| LSM7 |
| ZNF44 |
| MBD3 |
| S1PR5 |
| MIER2 |
| PCSK4 |
| ZNF562 |
| BEST2 |
| FBXL12 |
| C19orf24 |
| PLEKHJ1 |
| C19orf66 |
| FEM1A |
| ZNF823 |
| STAP2 |
| TRMT1 |
| BTBD2 |
| RNF126 |
| CCDC94 |
| LOC55908 |
| C19orf10 |
| PPAN |
| RETN |
| NCLN |
| GPR108 |
| SPPL2B |
| DUS3L |
| XAB2 |
| SHD |
| RGL3 |
| SLC44A2 |
| MCOLN1 |
| WDR18 |
| REXO1 |
| ZNF490 |
| DOCK6 |
| KIAA1543 |
| ZNF317 |
| ZNF77 |
| C19orf29 |
| UBL5 |
| BRUNOL5 |
| LPPR2 |
| KRI1 |
| YIPF2 |
| C19orf43 |
| SLC25A23 |
| ZNF426 |
| FSD1 |
| ZNF557 |
| LASS4 |
| TLE6 |
| LPPR3 |
| DENND1C |
| ZNF442 |
| ZNF556 |
| LRRC8E |
| UBXN6 |
| OR4F17 |
| ACSBG2 |
| ADAMTS10 |
| QTRT1 |
| FAM108A1 |
| DOHH |
| RTBDN |
| ANGPTL6 |
| KLF16 |
| PRAM1 |
| FBXW9 |
| ALKBH7 |
| MORG1 |
| ZNF414 |
| ELOF1 |
| DOT1L |
| FBN3 |
| ZNF559 |
| KISS1R |
| CREB3L3 |
| HDGF2 |
| LMNB2 |
| RAX2 |
| MUM1 |
| MPND |
| ATG4D |
| ATCAY |
| MBD3L1 |
| MIDN |
| GADD45GIP1 |
| ZNF799 |
| C19orf52 |
| ZNF625 |
| ZNF700 |
| ZNF439 |
| DPP9 |
| C19orf22 |
| C19orf6 |
| C19orf20 |
| CRB3 |
| REEP6 |
| PEX11G |
| ZNF561 |
| OLFM2 |
| MUC16 |
| FDX1L |
| NACC1 |
| C19orf36 |
| SCAMP4 |
| ADAT3 |
| ZNF554 |
| EVI5L |
| CCDC151 |
| ZNF653 |
| GRIN3B |
| MRPL54 |
| LRG1 |
| RAVER1 |
| OR7D4 |
| OR7G1 |
| OR1M1 |
| ACER1 |
| C19orf70 |
| MBD3L2 |
| TRAPPC5 |
| PCP2 |
| ZNF441 |
| ZNF491 |
| ZNF440 |
| C19orf39 |
| CCDC159 |
| TMIGD2 |
| TNFAIP8L1 |
| ZNF57 |
| JSRP1 |
| MOBKL2A |
| C19orf28 |
| GIPC3 |
| NDUFA11 |
| C19orf21 |
| PLK5P |
| C2CD4C |
| ZNF358 |
| LOC147727 |
| ZNF560 |
| ZNF563 |
| SPC24 |
| TICAM1 |
| C19orf23 |
| ZNRF4 |
| ZNF558 |
| C19orf25 |
| ATP8B3 |
| DIRAS1 |
| ZNF555 |
| ZNF846 |
| OR7D2 |
| ZNF791 |
| ZNF564 |
| ZNF709 |
| ZNF433 |
| PRR22 |
| ANKRD24 |
| C19orf59 |
| ZNF627 |
| DAND5 |
| C19orf26 |
| C19orf34 |
| C19orf38 |
| SYCE2 |
| KANK3 |
| PLAC2 |
| TMEM146 |
| ACTL9 |
| OR2Z1 |
| ZNF763 |
| ZNF844 |
| C19orf77 |
| C19orf30 |
| SLC25A41 |
| MBD3L5 |
| ODF3L2 |
| EMR4P |
| ADAMTSL5 |
| CLEC4G |
| TMPRSS9 |
| NDUFS7 |
| C19orf35 |
| HSD11B1L |
| C19orf45 |
| ZNF699 |
| TMEM205 |
| C3P1 |
| ZNF788 |
| ONECUT3 |
| OR7G2 |
| OR7G3 |
| MEX3D |
| FLJ45445 |
| PRSSL1 |
| VMAC |
| ZNF833 |
| CTXN1 |
| MIR199A1 |
| MIR7-3 |
| PLIN5 |
| CLEC4GP1 |
| FLJ25758 |
| FAM138F |
| LINGO3 |
| ARRDC5 |
| FAM138A |
| MBD3L4 |
| MBD3L3 |
| FAM138C |
| SNORD105 |
| PPAN-P2RY11 |
| MIR637 |
| MIR638 |
| PLIN4 |
| ZNF878 |
| SNORD105B |
| ZGLP1 |
| MIR220B |
| C19orf71 |
| LOC100128573 |
This is the comprehensive list of deleted genes in the wide peak for 22q11.21.
Table S59. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CLTCL1 |
| hsa-mir-648 |
| ATP6V1E1 |
| BID |
| GGT3P |
| GP1BB |
| GSC2 |
| SEPT5 |
| PRODH |
| SLC25A1 |
| TBX1 |
| CLDN5 |
| HIRA |
| UFD1L |
| DGCR6 |
| DGCR14 |
| CDC45L |
| DGCR2 |
| USP18 |
| TSSK2 |
| IL17RA |
| POTEH |
| BCL2L13 |
| DGCR11 |
| DGCR9 |
| DGCR5 |
| DGCR10 |
| HSFYP1 |
| CECR6 |
| CECR5 |
| CECR2 |
| TUBA8 |
| CECR1 |
| PEX26 |
| MICAL3 |
| MRPL40 |
| OR11H1 |
| SLC25A18 |
| GAB4 |
| C22orf39 |
| CCT8L2 |
| XKR3 |
| LOC150185 |
| psiTPTE22 |
| CECR7 |
| CECR4 |
| FLJ41941 |
This is the comprehensive list of deleted genes in the wide peak for 10p15.3.
Table S60. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| ADARB2 |
| KLF6 |
| IDI1 |
| PFKP |
| PITRM1 |
| ZMYND11 |
| WDR37 |
| DIP2C |
| LARP4B |
| GTPBP4 |
| C10orf110 |
| IDI2 |
| TUBB8 |
| NCRNA00200 |
| C10orf108 |
This is the comprehensive list of deleted genes in the wide peak for 4p16.2.
Table S61. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| FGFR3 |
| WHSC1 |
| hsa-mir-943 |
| hsa-mir-571 |
| ADD1 |
| ADRA2C |
| ATP5I |
| CTBP1 |
| DGKQ |
| GAK |
| GRK4 |
| HTT |
| HGFAC |
| IDUA |
| LETM1 |
| LRPAP1 |
| MSX1 |
| MYL5 |
| PDE6B |
| RGS12 |
| RNF4 |
| SH3BP2 |
| WHSC2 |
| ZNF141 |
| SLBP |
| NOP14 |
| FAM193A |
| MFSD10 |
| MAEA |
| PCGF3 |
| SPON2 |
| TACC3 |
| MXD4 |
| CPLX1 |
| SLC26A1 |
| D4S234E |
| STX18 |
| FGFRL1 |
| PIGG |
| LYAR |
| KIAA1530 |
| ZFYVE28 |
| TNIP2 |
| HAUS3 |
| ABCA11P |
| MFSD7 |
| TMEM175 |
| TMEM128 |
| C4orf42 |
| TMEM129 |
| OTOP1 |
| ZNF595 |
| FAM53A |
| ZNF509 |
| ZNF721 |
| ZNF718 |
| CRIPAK |
| DOK7 |
| RNF212 |
| C4orf10 |
| NAT8L |
| C4orf44 |
| LOC348926 |
| POLN |
| C4orf48 |
| ZNF876P |
| ZNF732 |
| SCARNA22 |
| MIR943 |
| LOC100130872 |
This is the comprehensive list of deleted genes in the wide peak for 4q32.3.
Table S62. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| DUX4 |
| FBXW7 |
| hsa-mir-1305 |
| hsa-mir-1979 |
| hsa-mir-548g |
| AGA |
| SLC25A4 |
| ANXA2P1 |
| CASP3 |
| LRBA |
| CLCN3 |
| CPE |
| CTSO |
| DCTD |
| EDNRA |
| ETFDH |
| F11 |
| ACSL1 |
| FAT1 |
| FGA |
| FGB |
| FGG |
| FRG1 |
| GAB1 |
| GK3P |
| GLRB |
| GPM6A |
| GRIA2 |
| GUCY1A3 |
| GUCY1B3 |
| GYPA |
| GYPB |
| GYPE |
| HMGB2 |
| HPGD |
| ING2 |
| IRF2 |
| KLKB1 |
| SMAD1 |
| NR3C2 |
| MTNR1A |
| NEK1 |
| NPY1R |
| NPY2R |
| NPY5R |
| PET112L |
| PLRG1 |
| POU4F2 |
| PPID |
| ABCE1 |
| RPS3A |
| SC4MOL |
| SFRP2 |
| TDO2 |
| TLL1 |
| TLR2 |
| TLR3 |
| VEGFC |
| GLRA3 |
| SMARCA5 |
| SORBS2 |
| SAP30 |
| INPP4B |
| SNORD73A |
| LRAT |
| HAND2 |
| RAPGEF2 |
| MFAP3L |
| ANAPC10 |
| MAB21L2 |
| MORF4 |
| ADAM29 |
| LSM6 |
| ANXA10 |
| KLHL2 |
| SCRG1 |
| PALLD |
| KIAA0922 |
| TRIM2 |
| ANP32C |
| FAM149A |
| FBXO8 |
| ARFIP1 |
| PDLIM3 |
| SPOCK3 |
| AADAT |
| FAM198B |
| ACCN5 |
| GALNT7 |
| CLDN22 |
| OTUD4 |
| DCHS2 |
| C4orf27 |
| MARCH1 |
| NEIL3 |
| TMEM144 |
| C4orf43 |
| UFSP2 |
| DDX60 |
| CDKN2AIP |
| ODZ3 |
| TMEM184C |
| LRP2BP |
| PDGFC |
| TUBB4Q |
| FSTL5 |
| STOX2 |
| KIAA1430 |
| FNIP2 |
| SH3RF1 |
| RXFP1 |
| SPCS3 |
| C4orf41 |
| HHIP |
| ARHGAP10 |
| MLF1IP |
| NBLA00301 |
| MAP9 |
| WWC2 |
| KIAA1712 |
| SNX25 |
| TTC29 |
| MND1 |
| SLC10A7 |
| TKTL2 |
| USP38 |
| CBR4 |
| FHDC1 |
| MGC45800 |
| PRMT10 |
| DDX60L |
| NAF1 |
| WDR17 |
| ZFP42 |
| SPATA4 |
| ENPP6 |
| ASB5 |
| ZNF827 |
| SH3D19 |
| C4orf38 |
| C4orf39 |
| C4orf45 |
| DCLK2 |
| TRIM60 |
| MMAA |
| RBM46 |
| C4orf46 |
| TIGD4 |
| TMEM154 |
| TMEM192 |
| RWDD4A |
| CCDC111 |
| TRIML2 |
| CCDC110 |
| CYP4V2 |
| LOC285501 |
| RNF175 |
| TRIML1 |
| LOC340017 |
| PRSS48 |
| ANKRD37 |
| TRIM61 |
| TRIM75 |
| HELT |
| FAM92A3 |
| LOC441046 |
| C4orf47 |
| GALNTL6 |
| FRG2 |
| SLED1 |
| C4orf51 |
| LOC653543 |
| LOC653544 |
| LOC653545 |
| LOC653548 |
| MIR578 |
| LOC728410 |
| FAM160A1 |
This is the comprehensive list of deleted genes in the wide peak for 4p12.
Table S63. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CNGA1 |
| GABRA2 |
| GABRA4 |
| GABRB1 |
| GABRG1 |
| TEC |
| TXK |
| ATP8A1 |
| CORIN |
| OCIAD1 |
| COMMD8 |
| ATP10D |
| SLAIN2 |
| GUF1 |
| CWH43 |
| OCIAD2 |
| GNPDA2 |
| NFXL1 |
| NIPAL1 |
| SHISA3 |
| COX7B2 |
| SLC10A4 |
| YIPF7 |
| FRYL |
| ZAR1 |
| KCTD8 |
| GRXCR1 |
This is the comprehensive list of deleted genes in the wide peak for 16q24.3.
Table S64. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CBFA2T3 |
| FANCA |
| MAF |
| hsa-mir-1910 |
| AP1G1 |
| AFG3L1 |
| APRT |
| ZFHX3 |
| C16orf3 |
| CA5A |
| CDH13 |
| CDH15 |
| COX4I1 |
| CTRB1 |
| CYBA |
| DHODH |
| DPEP1 |
| FOXF1 |
| FOXL1 |
| FOXC2 |
| GALNS |
| GAS8 |
| GCSH |
| GLG1 |
| HP |
| HPR |
| HSBP1 |
| HSD17B2 |
| IRF8 |
| KARS |
| MC1R |
| CHST6 |
| MVD |
| CHMP1A |
| PLCG2 |
| PSMD7 |
| RPL13 |
| SPG7 |
| GAN |
| SLC7A5 |
| CDK10 |
| MBTPS1 |
| TAF1C |
| USP10 |
| BCAR1 |
| C16orf7 |
| KIAA0513 |
| FAM38A |
| DHX38 |
| KIAA0174 |
| ATP2C2 |
| CLEC3A |
| MPHOSPH6 |
| COX4NB |
| TUBB3 |
| CFDP1 |
| PRDM7 |
| GABARAPL2 |
| MON1B |
| TCF25 |
| PHLPP2 |
| ZCCHC14 |
| KIAA0182 |
| ATMIN |
| COTL1 |
| MLYCD |
| ADAT1 |
| CHST5 |
| CPNE7 |
| IL17C |
| ANKRD11 |
| OSGIN1 |
| GINS2 |
| TRAPPC2L |
| WWOX |
| BCMO1 |
| TERF2IP |
| NECAB2 |
| KLHDC4 |
| DEF8 |
| TXNL4B |
| BANP |
| RFWD3 |
| ZNF821 |
| ZDHHC7 |
| CENPN |
| C16orf61 |
| JPH3 |
| VAT1L |
| KIAA1609 |
| WFDC1 |
| MTHFSD |
| DBNDD1 |
| FA2H |
| TMEM231 |
| WDR59 |
| KLHL36 |
| FBXO31 |
| CMIP |
| CDT1 |
| MAP1LC3B |
| PMFBP1 |
| DYNLRB2 |
| HSDL1 |
| CRISPLD2 |
| SPIRE2 |
| ZNF469 |
| ZNRF1 |
| CNTNAP4 |
| MARVELD3 |
| CENPBD1 |
| ZNF276 |
| KCNG4 |
| SDR42E1 |
| PKD1L2 |
| RNF166 |
| C16orf46 |
| LRRC50 |
| SPATA2L |
| C16orf55 |
| ZC3H18 |
| CDYL2 |
| TMEM170A |
| SLC38A8 |
| FLJ30679 |
| ZFPM1 |
| ADAD2 |
| ZFP1 |
| ADAMTS18 |
| MGC23284 |
| LDHD |
| MLKL |
| ZNF778 |
| ACSF3 |
| C16orf81 |
| HTA |
| LOC283922 |
| NUDT7 |
| SNAI3 |
| FAM92B |
| ATXN1L |
| PKD1L3 |
| CTU2 |
| LOC390748 |
| C16orf74 |
| CTRB2 |
| CLEC18B |
| SNORD68 |
| SNORD71 |
| LOC732275 |
| LOC100129637 |
| LOC100130015 |
| SYCE1L |
This is the comprehensive list of deleted genes in the wide peak for 14q21.3.
Table S65. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| KTN1 |
| NKX2-1 |
| NIN |
| CCNB1IP1 |
| hsa-mir-624 |
| hsa-mir-208b |
| hsa-mir-208a |
| hsa-mir-1201 |
| ANG |
| APEX1 |
| ARF6 |
| ARHGAP5 |
| BCL2L2 |
| BMP4 |
| CDKN3 |
| CEBPE |
| CFL2 |
| CMA1 |
| LTB4R |
| CTSG |
| DAD1 |
| COCH |
| FKBP3 |
| FOXG1 |
| GCH1 |
| GMFB |
| GZMH |
| GZMB |
| HIF1A |
| FOXA1 |
| HNRNPC |
| LGALS3 |
| MGAT2 |
| CTAGE5 |
| MMP14 |
| MNAT1 |
| MYH6 |
| MYH7 |
| NEDD8 |
| NFATC4 |
| NFKBIA |
| NOVA1 |
| NP |
| NRL |
| SIX6 |
| OTX2 |
| OXA1L |
| PAX9 |
| PCK2 |
| PNN |
| POLE2 |
| PPM1A |
| PRKCH |
| PRKD1 |
| PSMA3 |
| PSMA6 |
| PSMB5 |
| PSMC6 |
| PSME1 |
| PSME2 |
| PTGDR |
| PTGER2 |
| PYGL |
| RABGGTA |
| ARID4A |
| RNASE1 |
| RNASE2 |
| RNASE3 |
| RNASE4 |
| RNASE6 |
| RPL36AL |
| RPS29 |
| RTN1 |
| SALL2 |
| SIX1 |
| SOS2 |
| SRP54 |
| SSTR1 |
| STYX |
| TEP1 |
| TGM1 |
| PABPN1 |
| SIP1 |
| CDKL1 |
| AP1G2 |
| SLC7A7 |
| SDCCAG1 |
| CPNE6 |
| AKAP6 |
| KIAA0391 |
| KIAA0586 |
| DLGAP5 |
| TOX4 |
| REC8 |
| PARP2 |
| CNIH |
| DHRS2 |
| EFS |
| IRF9 |
| PRMT5 |
| SEC23A |
| TM9SF1 |
| EXOC5 |
| CGRRF1 |
| FAM12A |
| DHRS4 |
| FERMT2 |
| RIPK3 |
| AP4S1 |
| WDHD1 |
| BAZ1A |
| MAP4K5 |
| SUPT16H |
| NID2 |
| KIAA0831 |
| ACIN1 |
| DAAM1 |
| SAMD4A |
| FAM179B |
| SCFD1 |
| KHNYN |
| SLC7A8 |
| KLHDC2 |
| HECTD1 |
| HEATR5A |
| NGDN |
| LRP10 |
| NKX2-8 |
| TINF2 |
| TIMM9 |
| OR10G3 |
| OR10G2 |
| OR4E2 |
| ATP5S |
| CIDEB |
| CHMP4A |
| STXBP6 |
| STRN3 |
| SLC39A2 |
| ERO1L |
| FAM158A |
| ATL1 |
| ZNF219 |
| GMPR2 |
| SLC22A17 |
| DACT1 |
| JKAMP |
| MBIP |
| DHRS7 |
| C14orf166 |
| SIX4 |
| GNG2 |
| KLHL28 |
| C14orf101 |
| HAUS4 |
| PPP2R3C |
| PRPF39 |
| C14orf119 |
| FBXO34 |
| RNF31 |
| RBM23 |
| C14orf104 |
| C14orf105 |
| C14orf106 |
| C14orf167 |
| G2E3 |
| OSGEP |
| FLJ10357 |
| MUDENG |
| EAPP |
| ACTR10 |
| METTL3 |
| LTB4R2 |
| SDR39U1 |
| RPGRIP1 |
| PELI2 |
| NDRG2 |
| NYNRIN |
| TXNDC16 |
| TRMT5 |
| HOMEZ |
| CHD8 |
| FANCM |
| SNX6 |
| SAV1 |
| C14orf93 |
| ABHD4 |
| NPAS3 |
| FAM12B |
| CDH24 |
| C14orf135 |
| GPR135 |
| METT11D1 |
| IL25 |
| GNPNAT1 |
| THTPA |
| OR4K5 |
| OR4K1 |
| C14orf138 |
| IPO4 |
| L2HGDH |
| NUBPL |
| DCAF11 |
| DDHD1 |
| OR4K15 |
| TMX1 |
| FSCB |
| BRMS1L |
| JPH4 |
| RNASE7 |
| INSM2 |
| C14orf128 |
| RAB2B |
| JUB |
| RPPH1 |
| SLC25A21 |
| LRRC16B |
| PPP1R3E |
| TMEM55B |
| TTC5 |
| MAPK1IP1L |
| EGLN3 |
| C14orf126 |
| C14orf149 |
| TRIM9 |
| DHRS1 |
| CMTM5 |
| MIA2 |
| C14orf28 |
| TRAPPC6B |
| RNASE11 |
| TPPP2 |
| RNASE8 |
| MRPL52 |
| PSMB11 |
| OR4K14 |
| OR4L1 |
| OR11H6 |
| PPIL5 |
| KLHDC1 |
| FRMD6 |
| SOCS4 |
| NAT12 |
| KLHL33 |
| RPL10L |
| MIPOL1 |
| SLC38A6 |
| C14orf37 |
| ABHD12B |
| MDP1 |
| LRFN5 |
| CLEC14A |
| FITM1 |
| REM2 |
| TMEM30B |
| MDGA2 |
| C14orf21 |
| C14orf147 |
| ADCY4 |
| C14orf183 |
| RALGAPA1 |
| SFTA3 |
| FBXO33 |
| C14orf19 |
| C14orf182 |
| GPR137C |
| TSSK4 |
| FAM177A1 |
| DHRS4L2 |
| C14orf39 |
| SNORD8 |
| RNASE10 |
| OR6S1 |
| SLC35F4 |
| FLJ31306 |
| TBPL2 |
| C14orf23 |
| TOMM20L |
| OR4N2 |
| OR4K2 |
| OR4K13 |
| OR4K17 |
| OR4N5 |
| OR11G2 |
| OR11H4 |
| RNASE9 |
| OR5AU1 |
| POTEG |
| MIR208A |
| C14orf165 |
| OR11H12 |
| RNASE13 |
| OR4Q3 |
| OR4M1 |
| RNASE12 |
| P704P |
| C14orf176 |
| CBLN3 |
| RPL13AP3 |
| C14orf34 |
| SNORD9 |
| MIR624 |
| DHRS4L1 |
| C14orf38 |
| SNORD127 |
| SNORD126 |
| MIR208B |
| C14orf33 |
| OTX2OS1 |
This is the comprehensive list of deleted genes in the wide peak for 8p23.3.
Table S66. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-596 |
| CLN8 |
| MYOM2 |
| DLGAP2 |
| ARHGEF10 |
| KBTBD11 |
| FBXO25 |
| C8orf42 |
| ERICH1 |
| ZNF596 |
| OR4F21 |
| RPL23AP53 |
| MIR596 |
This is the comprehensive list of deleted genes in the wide peak for 9p24.1.
Table S67. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| JAK2 |
| NFIB |
| CD274 |
| MLANA |
| GLDC |
| INSL4 |
| PTPRD |
| RLN1 |
| RLN2 |
| TYRP1 |
| MPDZ |
| CER1 |
| INSL6 |
| KDM4C |
| RANBP6 |
| C9orf46 |
| KIAA1432 |
| ERMP1 |
| PDCD1LG2 |
| TPD52L3 |
| IL33 |
| C9orf123 |
| UHRF2 |
| FREM1 |
| KIAA2026 |
| C9orf150 |
| ZDHHC21 |
This is the comprehensive list of deleted genes in the wide peak for 13q14.2.
Table S68. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BRCA2 |
| CDX2 |
| FLT3 |
| LCP1 |
| RB1 |
| LHFP |
| hsa-mir-1297 |
| hsa-mir-15a |
| hsa-mir-621 |
| hsa-mir-320d-1 |
| PARP4 |
| ALOX5AP |
| ATP12A |
| ATP7B |
| CDK8 |
| RCBTB2 |
| CPB2 |
| DACH1 |
| ELF1 |
| ESD |
| FGF9 |
| FOXO1 |
| FLT1 |
| GJA3 |
| GJB2 |
| GPR12 |
| MLNR |
| GTF2F2 |
| GTF3A |
| GUCY1B2 |
| HMGB1 |
| HTR2A |
| PDX1 |
| KPNA3 |
| MAB21L1 |
| SMAD9 |
| MIPEP |
| NEK3 |
| PABPC3 |
| PCDH8 |
| PCDH9 |
| UBL3 |
| RFC3 |
| RFXAP |
| RNF6 |
| RPL21 |
| ATXN8OS |
| SGCG |
| SLC7A1 |
| TPT1 |
| TRPC4 |
| TUBA3C |
| ZMYM2 |
| IFT88 |
| TNFSF11 |
| SUCLA2 |
| DLEU2 |
| TSC22D1 |
| CCNA1 |
| MTMR6 |
| DCLK1 |
| ZMYM5 |
| KL |
| ITM2B |
| MTRF1 |
| UTP14C |
| NUPL1 |
| FRY |
| LPAR6 |
| SLC25A15 |
| TRIM13 |
| USPL1 |
| MRPS31 |
| SAP18 |
| DLEU1 |
| N4BP2L2 |
| OLFM4 |
| POSTN |
| GJB6 |
| HSPH1 |
| WASF3 |
| SUGT1 |
| LECT1 |
| WBP4 |
| AKAP11 |
| EXOSC8 |
| FNDC3A |
| PDS5B |
| KIAA0564 |
| ZC3H13 |
| SPG20 |
| LRCH1 |
| MTUS2 |
| SACS |
| INTS6 |
| LATS2 |
| CKAP2 |
| OR7E37P |
| NUFIP1 |
| SNORD102 |
| NBEA |
| PCDH17 |
| C13orf15 |
| MED4 |
| DNAJC15 |
| ALG5 |
| VPS36 |
| POLR1D |
| CRYL1 |
| PHF11 |
| POMP |
| UFM1 |
| ATP8A2 |
| IL17D |
| MPHOSPH8 |
| SOHLH2 |
| ENOX1 |
| RCBTB1 |
| PSPC1 |
| NUDT15 |
| KIAA1704 |
| TNFRSF19 |
| FAM48A |
| CENPJ |
| THSD1 |
| RNF17 |
| CYSLTR2 |
| C13orf1 |
| COG6 |
| KLHL1 |
| XPO4 |
| PCDH20 |
| MRP63 |
| NARG1L |
| RNASEH2B |
| DHRS12 |
| C13orf18 |
| C13orf23 |
| TDRD3 |
| CDADC1 |
| CAB39L |
| DIAPH3 |
| CCDC70 |
| COG3 |
| SETDB2 |
| KATNAL1 |
| KBTBD7 |
| EBPL |
| C13orf33 |
| KBTBD6 |
| STARD13 |
| N4BP2L1 |
| TPTE2 |
| EPSTI1 |
| ARL11 |
| WDFY2 |
| CG030 |
| LOC121838 |
| CSNK1A1L |
| RXFP2 |
| C13orf26 |
| PRR20A |
| C13orf30 |
| C13orf31 |
| HNRNPA1L2 |
| FAM10A4 |
| B3GALTL |
| DGKH |
| CCDC122 |
| STOML3 |
| EEF1DP3 |
| FAM123A |
| USP12 |
| MTIF3 |
| GSX1 |
| FAM194B |
| SPERT |
| DLEU7 |
| FAM124A |
| LOC220115 |
| LOC220429 |
| N6AMT2 |
| SKA3 |
| EFHA1 |
| SPATA13 |
| LNX2 |
| SLC25A30 |
| ZDHHC20 |
| PAN3 |
| DKFZp686A1627 |
| OR7E156P |
| SUGT1L1 |
| SIAH3 |
| KCNRG |
| FLJ37307 |
| SLC46A3 |
| LOC284232 |
| C1QTNF9 |
| FREM2 |
| NEK5 |
| LOC348021 |
| LOC374491 |
| THSD1P |
| KCTD4 |
| RASL11A |
| C1QTNF9B |
| SHISA2 |
| NHLRC3 |
| SERP2 |
| C13orf36 |
| MIR15A |
| MIR16-1 |
| ATP5EP2 |
| ALG11 |
| C13orf37 |
| PCOTH |
| SNORA27 |
| LOC646405 |
| PRHOXNB |
| ZAR1L |
| LOC646982 |
| SERPINE3 |
| SNORA31 |
| MIR621 |
| C13orf38 |
| PRR20B |
| PRR20C |
| PRR20D |
| PRR20E |
| LOC100101938 |
| RPL21P28 |
| LOC100188949 |
| LOC100190939 |
| LOC100288730 |
This is the comprehensive list of deleted genes in the wide peak for 15q21.1.
Table S69. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BUB1B |
| C15orf55 |
| C15orf21 |
| hsa-mir-1300 |
| hsa-mir-1266 |
| hsa-mir-147b |
| hsa-mir-1282 |
| hsa-mir-627 |
| hsa-mir-626 |
| hsa-mir-1233 |
| hsa-mir-1233 |
| hsa-mir-211 |
| hsa-mir-1268 |
| ACTC1 |
| APBA2 |
| B2M |
| BCL8 |
| CAPN3 |
| CHRM5 |
| CHRNA7 |
| CKMT1B |
| CYP19A1 |
| DUT |
| EPB42 |
| FBN1 |
| FGF7 |
| GABPB1 |
| GABRA5 |
| GABRB3 |
| GABRG3 |
| GALK2 |
| GANC |
| GATM |
| GCHFR |
| PDIA3 |
| HDC |
| ONECUT1 |
| IPW |
| ITPKA |
| IVD |
| LTK |
| MAP1A |
| MEIS2 |
| MFAP1 |
| TRPM1 |
| MYO5A |
| NDN |
| OCA2 |
| PLCB2 |
| MAPK6 |
| RAD51 |
| RYR3 |
| SCG5 |
| SLC12A1 |
| SNRPN |
| SORD |
| SPINT1 |
| SRP14 |
| THBS1 |
| TJP1 |
| TP53BP1 |
| TYRO3 |
| UBE3A |
| MKRN3 |
| SLC30A4 |
| PAR5 |
| EIF3J |
| JMJD7-PLA2G4B |
| SNAP23 |
| HERC2 |
| SNURF |
| USP8 |
| SLC28A2 |
| COPS2 |
| TGM5 |
| HISPPD2A |
| AQR |
| SECISBP2L |
| ARHGAP11A |
| LCMT2 |
| SLC12A6 |
| BCL2L10 |
| RASGRP1 |
| SERF2 |
| GNB5 |
| ARPP19 |
| SLC27A2 |
| GPR176 |
| CHP |
| OIP5 |
| BAHD1 |
| MTMR15 |
| CEP152 |
| MAPKBP1 |
| GOLGA8A |
| RTF1 |
| CYFIP1 |
| MGA |
| DMXL2 |
| VPS39 |
| FAM189A1 |
| AP4E1 |
| CCNDBP1 |
| EID1 |
| C15orf2 |
| C15orf63 |
| TMEM87A |
| RPAP1 |
| DKFZP434L187 |
| PLDN |
| GREM1 |
| RPUSD2 |
| TUBGCP4 |
| SCG3 |
| TMOD3 |
| TMOD2 |
| EHD4 |
| DUOX2 |
| MYEF2 |
| NDUFAF1 |
| NUSAP1 |
| TMEM85 |
| SPTBN5 |
| CTDSPL2 |
| KLF13 |
| DUOX1 |
| MAGEL2 |
| DLL4 |
| INO80 |
| TRPM7 |
| PPP1R14D |
| MTMR10 |
| ZNF770 |
| FLJ10038 |
| HAUS2 |
| FAM82A2 |
| DNAJC17 |
| NOP10 |
| MYO5C |
| NDNL2 |
| KIAA1370 |
| C15orf24 |
| PAK6 |
| DTWD1 |
| CASC5 |
| AVEN |
| ATP10A |
| GJD2 |
| VPS18 |
| SQRDL |
| ZFP106 |
| SPATA5L1 |
| CHAC1 |
| C15orf29 |
| ATP8B4 |
| WDR76 |
| TMEM62 |
| SEMA6D |
| SPG11 |
| ELL3 |
| NIPA2 |
| C15orf48 |
| C15orf41 |
| SPPL2A |
| ZFYVE19 |
| FRMD5 |
| DISP2 |
| CHRFAM7A |
| FAM7A3 |
| FAM7A1 |
| ARHGAP11B |
| ATPBD4 |
| C15orf57 |
| C15orf23 |
| BMF |
| SHF |
| DUOXA1 |
| SNORD107 |
| CHST14 |
| CASC4 |
| TUBGCP5 |
| TGM7 |
| CATSPER2 |
| LEO1 |
| NIPA1 |
| PLA2G4E |
| TRIM69 |
| PAR1 |
| C15orf43 |
| LOC145845 |
| TMCO5A |
| ZSCAN29 |
| TTBK2 |
| CDAN1 |
| STRC |
| CSNK1A1P |
| OTUD7A |
| SPRED1 |
| PGBD4 |
| ADAL |
| EXD1 |
| FSIP1 |
| RHOV |
| C15orf33 |
| UBR1 |
| PATL2 |
| LPCAT4 |
| PLA2G4F |
| LRRC57 |
| LYSMD2 |
| WDR72 |
| SLC24A5 |
| OR4N4 |
| FAM98B |
| PLA2G4D |
| GOLGA6L1 |
| GOLGA8G |
| GOLGA9P |
| SNORD108 |
| SNORD109A |
| SNORD109B |
| SNORD115-1 |
| WHAMML1 |
| POTEB |
| GLDN |
| FMN1 |
| SNORD64 |
| PAR4 |
| PAR-SN |
| MRPL42P5 |
| USP50 |
| C15orf52 |
| TNFAIP8L3 |
| GOLGA8E |
| OR4M2 |
| OR4N3P |
| SHC4 |
| CTXN2 |
| HERC2P2 |
| C15orf53 |
| C15orf54 |
| DUOXA2 |
| MIR211 |
| NF1P1 |
| WHAMML2 |
| GOLGA8B |
| EIF2AK4 |
| CATSPER2P1 |
| UNC13C |
| CKMT1A |
| SERINC4 |
| C15orf62 |
| C15orf56 |
| PHGR1 |
| LOC646214 |
| CXADRP2 |
| MIR548A3 |
| MIR626 |
| MIR627 |
| LOC723972 |
| SNORD116-19 |
| GOLGA6L6 |
| LOC727924 |
| LOC728758 |
| LOC729082 |
| GOLGA8C |
| PWRN1 |
| PWRN2 |
| SNORD116-1 |
| SNORD116-2 |
| SNORD116-3 |
| SNORD116-4 |
| SNORD116-5 |
| SNORD116-6 |
| SNORD116-7 |
| SNORD116-8 |
| SNORD116-9 |
| SNORD116-10 |
| SNORD116-11 |
| SNORD116-12 |
| SNORD116-13 |
| SNORD116-14 |
| SNORD116-15 |
| SNORD116-16 |
| SNORD116-17 |
| SNORD116-18 |
| SNORD116-20 |
| SNORD116-21 |
| SNORD116-22 |
| SNORD116-23 |
| SNORD116-24 |
| SNORD116-25 |
| SNORD115-2 |
| SNORD116-26 |
| SNORD116-27 |
| SNORD115-3 |
| SNORD115-4 |
| SNORD115-5 |
| SNORD115-6 |
| SNORD115-7 |
| SNORD115-8 |
| SNORD115-9 |
| SNORD115-10 |
| SNORD115-11 |
| SNORD115-12 |
| SNORD115-13 |
| SNORD115-14 |
| SNORD115-15 |
| SNORD115-16 |
| SNORD115-17 |
| SNORD115-18 |
| SNORD115-19 |
| SNORD115-20 |
| SNORD115-21 |
| SNORD115-22 |
| SNORD115-23 |
| SNORD115-25 |
| SNORD115-26 |
| SNORD115-29 |
| SNORD115-30 |
| SNORD115-31 |
| SNORD115-32 |
| SNORD115-33 |
| SNORD115-34 |
| SNORD115-35 |
| SNORD115-36 |
| SNORD115-37 |
| SNORD115-38 |
| SNORD115-39 |
| SNORD115-40 |
| SNORD115-41 |
| SNORD115-42 |
| SNORD115-43 |
| SNORD115-44 |
| SNORD116-28 |
| SNORD116-29 |
| SNORD115-48 |
| HBII-52-24 |
| HBII-52-27 |
| HBII-52-28 |
| HBII-52-45 |
| HBII-52-46 |
| MIR147B |
| LOC100129387 |
| GOLGA8F |
| LOC100132724 |
| GOLGA8D |
| JMJD7 |
| PLA2G4B |
| FAM7A2 |
This is the comprehensive list of deleted genes in the wide peak for 4q23.
Table S70. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| IL2 |
| RAP1GDS1 |
| TET2 |
| FBXW7 |
| hsa-mir-548g |
| hsa-mir-2054 |
| hsa-mir-1973 |
| hsa-mir-577 |
| hsa-mir-1243 |
| hsa-mir-302b |
| hsa-mir-297 |
| hsa-mir-576 |
| hsa-mir-1255a |
| ADH1A |
| ADH1B |
| ADH1C |
| ADH4 |
| ADH5 |
| ADH6 |
| ADH7 |
| ANK2 |
| ANXA2P1 |
| ANXA5 |
| BMPR1B |
| CAMK2D |
| CASP6 |
| CCNA2 |
| LRBA |
| CLGN |
| CENPE |
| CTSO |
| EDNRA |
| EGF |
| EIF4E |
| ELF2 |
| ENPEP |
| ETFDH |
| FABP2 |
| FGA |
| FGB |
| FGF2 |
| FGG |
| GAB1 |
| GLRB |
| GRIA2 |
| GUCY1A3 |
| GUCY1B3 |
| GYPA |
| GYPB |
| GYPE |
| H2AFZ |
| HADH |
| CFI |
| IL15 |
| MAD2L1 |
| SMAD1 |
| MANBA |
| MGST2 |
| NR3C2 |
| MTTP |
| NDUFC1 |
| NFKB1 |
| NPY2R |
| PDHA2 |
| PET112L |
| PITX2 |
| PLRG1 |
| EXOSC9 |
| POU4F2 |
| PPID |
| PPP3CA |
| ABCE1 |
| RPL34 |
| RPS3A |
| SFRP2 |
| TACR3 |
| TDO2 |
| TLR2 |
| TRPC3 |
| UBE2D3 |
| UCP1 |
| UGT8 |
| SMARCA5 |
| PRSS12 |
| UNC5C |
| MAPKSP1 |
| PDE5A |
| INPP4B |
| SNORD73A |
| PAPSS1 |
| LRAT |
| AIMP1 |
| NDST3 |
| RAPGEF2 |
| SEC24D |
| TSPAN5 |
| SPRY1 |
| ANAPC10 |
| PGRMC2 |
| SEC24B |
| MAB21L2 |
| RRH |
| PLK4 |
| PRDM5 |
| LSM6 |
| NUDT6 |
| HSPA4L |
| TBC1D9 |
| METAP1 |
| KIAA0922 |
| TRIM2 |
| SLC7A11 |
| CCRN4L |
| PPA2 |
| DAPP1 |
| DKK2 |
| INTU |
| ARFIP1 |
| ZNF330 |
| LEF1 |
| FAM198B |
| LARP7 |
| EMCN |
| MYOZ2 |
| ACCN5 |
| GAR1 |
| PCDH18 |
| USP53 |
| OTUD4 |
| DCHS2 |
| FLJ20184 |
| CCDC109B |
| BANK1 |
| LARP1B |
| BBS7 |
| TMEM144 |
| C4orf21 |
| AP1AR |
| MAML3 |
| TMEM184C |
| PDGFC |
| FSTL5 |
| BDH2 |
| INTS12 |
| ANKRD50 |
| RNF150 |
| PCDH10 |
| FNIP2 |
| METTL14 |
| OSTC |
| IL21 |
| RXFP1 |
| SCOC |
| NEUROG2 |
| SLC39A8 |
| HHIP |
| NDST4 |
| AGXT2L1 |
| ELOVL6 |
| C4orf31 |
| FAT4 |
| ARSJ |
| ARHGAP10 |
| GSTCD |
| MAP9 |
| TNIP3 |
| PHF17 |
| DNAJB14 |
| NARG1 |
| C4orf29 |
| ALPK1 |
| CXXC4 |
| SETD7 |
| PLA2G12A |
| SLC25A31 |
| RAB33B |
| TTC29 |
| MND1 |
| SLC10A7 |
| C4orf17 |
| QRFPR |
| KIAA1109 |
| COL25A1 |
| USP38 |
| C4orf49 |
| FHDC1 |
| PRMT10 |
| NAF1 |
| TIFA |
| RG9MTD2 |
| TBCK |
| CYP2U1 |
| DDIT4L |
| SCLT1 |
| C4orf33 |
| TMEM155 |
| PABPC4L |
| ADAD1 |
| C4orf32 |
| TRAM1L1 |
| NHEDC2 |
| NHEDC1 |
| ZNF827 |
| SH3D19 |
| C4orf45 |
| SPATA5 |
| BBS12 |
| DCLK2 |
| MMAA |
| RBM46 |
| SGMS2 |
| SYNPO2 |
| C4orf46 |
| TIGD4 |
| TMEM154 |
| ELMOD2 |
| NPNT |
| MFSD8 |
| LOC256880 |
| LOC285419 |
| LOC285456 |
| RNF175 |
| C4orf37 |
| LOC340017 |
| PRSS48 |
| LRIT3 |
| PCNAP1 |
| C4orf3 |
| MIR302A |
| EEF1AL7 |
| LOC441046 |
| MIR302B |
| MIR302C |
| MIR302D |
| MIR367 |
| CISD2 |
| LOC641518 |
| CEP170L |
| C4orf51 |
| SNORA24 |
| MIR577 |
| LOC729338 |
| FAM160A1 |
| SNHG8 |
| LOC100192379 |
This is the comprehensive list of deleted genes in the wide peak for 7q36.3.
Table S71. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BRAF |
| EZH2 |
| KIAA1549 |
| MLL3 |
| CREB3L2 |
| hsa-mir-595 |
| hsa-mir-153-2 |
| hsa-mir-671 |
| hsa-mir-1975 |
| hsa-mir-548f-4 |
| hsa-mir-490 |
| ABP1 |
| AKR1B1 |
| BPGM |
| CALD1 |
| CASP2 |
| CDK5 |
| CHRM2 |
| CLCN1 |
| DPP6 |
| EN2 |
| EPHA1 |
| EPHB6 |
| GBX1 |
| MNX1 |
| HTR5A |
| INSIG1 |
| KCNH2 |
| KEL |
| NDUFB2 |
| NOS3 |
| CNOT4 |
| PIP |
| PODXL |
| PRSS1 |
| PRSS2 |
| TAS2R38 |
| PTN |
| PTPRN2 |
| RARRES2 |
| RHEB |
| SHH |
| SLC4A2 |
| SMARCD3 |
| AKR1D1 |
| SSBP1 |
| TBXAS1 |
| VIPR2 |
| XRCC2 |
| ZYX |
| ARHGEF5 |
| ZNF212 |
| ZNF282 |
| CUL1 |
| TRIM24 |
| MGAM |
| DGKI |
| ACCN3 |
| PDIA4 |
| UBE3C |
| FAM131B |
| FAM115A |
| DNAJB6 |
| ABCF2 |
| FASTK |
| ABCB8 |
| PAXIP1 |
| SSPO |
| NUP205 |
| CLEC5A |
| MKRN1 |
| CNTNAP2 |
| GIMAP2 |
| OR2F1 |
| SLC13A4 |
| TPK1 |
| DENND2A |
| ZNF777 |
| TMEM176B |
| HIPK2 |
| WDR91 |
| REPIN1 |
| ATP6V0A4 |
| TAS2R3 |
| TAS2R4 |
| PRKAG2 |
| LUC7L2 |
| MRPS33 |
| NUB1 |
| TAS2R5 |
| CHPF2 |
| NCAPG2 |
| CHCHD3 |
| WDR60 |
| TMEM140 |
| GIMAP4 |
| GIMAP5 |
| TMEM176A |
| TRPV6 |
| AGK |
| TRPV5 |
| ZC3HAV1 |
| AKR1B10 |
| ACTR3B |
| KIAA1147 |
| ESYT2 |
| ZNF398 |
| EXOC4 |
| GALNT11 |
| LMBR1 |
| C7orf4 |
| NOM1 |
| PARP12 |
| LRRC61 |
| C7orf49 |
| ZNF767 |
| TTC26 |
| JHDM1D |
| TMUB1 |
| SLC37A3 |
| KRBA1 |
| SLC35B4 |
| OR6W1P |
| ADCK2 |
| PLXNA4 |
| ZC3HAV1L |
| LOC93432 |
| C7orf29 |
| AGAP3 |
| C7orf13 |
| OR9A4 |
| OR9A2 |
| C7orf34 |
| TMEM139 |
| NOBOX |
| OR2A14 |
| OR6B1 |
| OR2F2 |
| ZNF786 |
| PRSS37 |
| SVOPL |
| MTPN |
| LRGUK |
| ASB10 |
| TRYX3 |
| MOXD2 |
| RNF32 |
| TRY6 |
| LOC154761 |
| CLEC2L |
| C7orf55 |
| LOC154822 |
| TMEM213 |
| GIMAP8 |
| CRYGN |
| ZNF425 |
| ZNF783 |
| ZNF746 |
| ATP6V0E2 |
| RBM33 |
| GALNTL5 |
| GIMAP7 |
| ZNF467 |
| GIMAP1 |
| LOC202781 |
| C7orf33 |
| FABP5L3 |
| UBN2 |
| TAS2R39 |
| TAS2R40 |
| TAS2R41 |
| CNPY1 |
| FLJ40852 |
| FAM115C |
| ZNF775 |
| ATG9B |
| TAS2R60 |
| CTAGE6 |
| AGBL3 |
| OR6V1 |
| OR2A12 |
| OR2A1 |
| STRA8 |
| KLRG2 |
| WDR86 |
| GSTK1 |
| FAM180A |
| OR2A25 |
| OR2A5 |
| RAB19 |
| OR2A7 |
| OR2A20P |
| LOC401431 |
| OR2A42 |
| MIR153-2 |
| AKR1B15 |
| LOC441294 |
| OR2A9P |
| OR2A2 |
| ARHGEF5L |
| GIMAP6 |
| WEE2 |
| MIR490 |
| ZNF862 |
| PL-5283 |
| ACTR3C |
| MIR595 |
| LOC728743 |
| LUZP6 |
| MIR671 |
| LOC100124692 |
| LOC100128542 |
| CTAGE4 |
| LOC100128822 |
| LOC100132707 |
| LOC100134229 |
| LOC100134713 |
This is the comprehensive list of deleted genes in the wide peak for 19q13.33.
Table S72. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| AKT2 |
| BCL3 |
| CD79A |
| ERCC2 |
| KLK2 |
| PPP2R1A |
| CIC |
| CBLC |
| TFPT |
| ZNF331 |
| hsa-mir-1274b |
| hsa-mir-935 |
| hsa-mir-373 |
| hsa-mir-519a-2 |
| hsa-mir-643 |
| hsa-mir-125a |
| hsa-mir-150 |
| hsa-mir-220c |
| hsa-mir-769 |
| hsa-mir-642 |
| hsa-mir-330 |
| hsa-mir-641 |
| A1BG |
| ACTN4 |
| AP2A1 |
| APOC1 |
| APOC1P1 |
| APOC2 |
| APOC4 |
| APOE |
| KLK3 |
| ATP1A3 |
| AXL |
| BAX |
| BCAT2 |
| BCKDHA |
| CEACAM1 |
| BLVRB |
| C5AR1 |
| CA11 |
| CALM3 |
| CD33 |
| SIGLEC6 |
| CD37 |
| CEACAM5 |
| CGB |
| CEACAM3 |
| CEACAM7 |
| CEACAM8 |
| CEACAM4 |
| CKM |
| AP2S1 |
| CLC |
| CLPTM1 |
| CRX |
| CYP2A6 |
| CYP2A7 |
| CYP2A13 |
| CYP2B6 |
| CYP2B7P1 |
| CYP2F1 |
| DBP |
| DMPK |
| DMWD |
| ECH1 |
| MEGF8 |
| EMP3 |
| ERCC1 |
| ERF |
| FBL |
| ETFB |
| FCAR |
| FCGRT |
| FKBP1P1 |
| FLT3LG |
| FOSB |
| FPR1 |
| FPR2 |
| FPR3 |
| FTL |
| FUT1 |
| FUT2 |
| GIPR |
| GPR4 |
| GPR32 |
| GRIK5 |
| GRIN2D |
| GRLF1 |
| GSK3A |
| GYS1 |
| HAS1 |
| FOXA3 |
| HNRNPL |
| HRC |
| PRMT1 |
| IL11 |
| IRF3 |
| KCNA7 |
| KCNC3 |
| KCNJ14 |
| KCNN4 |
| KIR2DL1 |
| KIR2DL3 |
| KIR2DL4 |
| KIR2DS4 |
| KIR3DL1 |
| KIR3DL2 |
| KLK1 |
| LAIR1 |
| LAIR2 |
| LGALS4 |
| LGALS7 |
| LHB |
| LIG1 |
| LIM2 |
| LIPE |
| BCAM |
| MAP3K10 |
| MYBPC2 |
| CEACAM6 |
| NDUFA3 |
| NFKBIB |
| NKG7 |
| CNOT3 |
| NOVA2 |
| NPAS1 |
| NTF4 |
| NUCB1 |
| PAFAH1B3 |
| PEG3 |
| PLAUR |
| POLD1 |
| POU2F2 |
| PPP5C |
| PRKCG |
| PRRG2 |
| KLK7 |
| KLK6 |
| KLK10 |
| PSG1 |
| PSG2 |
| PSG3 |
| PSG4 |
| PSG5 |
| PSG6 |
| PSG7 |
| PSG9 |
| PSG11 |
| PSMC4 |
| PSMD8 |
| PTGIR |
| PTPRH |
| PVR |
| PVRL2 |
| RELB |
| RPL18 |
| RPL28 |
| MRPS12 |
| RPS5 |
| RPS9 |
| RPS11 |
| RPS16 |
| RPS19 |
| RRAS |
| RTN2 |
| RYR1 |
| CLEC11A |
| SEPW1 |
| SLC1A5 |
| SLC8A2 |
| SNRNP70 |
| SNRPA |
| SNRPD2 |
| SPIB |
| AURKC |
| SULT2B1 |
| SULT2A1 |
| SUPT5H |
| SYT5 |
| TGFB1 |
| TNNI3 |
| TNNT1 |
| TULP2 |
| NR1H2 |
| VASP |
| XRCC1 |
| ZFP36 |
| ZNF8 |
| ZNF17 |
| ZNF28 |
| MZF1 |
| ZNF45 |
| ZNF221 |
| ZNF222 |
| ZNF132 |
| ZNF134 |
| ZNF135 |
| ZNF137 |
| ZNF154 |
| ZNF155 |
| ZNF175 |
| ZNF180 |
| ZNF223 |
| ZNF224 |
| ZNF225 |
| ZNF226 |
| ZNF227 |
| ZFP112 |
| ZNF229 |
| ZNF230 |
| SYMPK |
| MIA |
| DPF1 |
| LTBP4 |
| TEAD2 |
| PPFIA3 |
| PLA2G4C |
| NAPA |
| SIGLEC5 |
| FCGBP |
| PGLYRP1 |
| UBE2M |
| ARHGEF1 |
| DYRK1B |
| NUMBL |
| CYTH2 |
| ZNF235 |
| ZNF264 |
| KCNK6 |
| NCR1 |
| NAPSA |
| GMFG |
| KLK4 |
| ZNF432 |
| DHX34 |
| SAE1 |
| TRIM28 |
| ZNF256 |
| LILRB2 |
| PAK4 |
| TOMM40 |
| ZNF211 |
| RABAC1 |
| TRAPPC2P1 |
| SPINT2 |
| DLL3 |
| ZNF234 |
| ZNF274 |
| ZNF460 |
| PPP1R13L |
| CD3EAP |
| RUVBL2 |
| LILRB1 |
| KDELR1 |
| LILRB5 |
| SLC27A5 |
| LILRB4 |
| KLK11 |
| LILRA1 |
| LILRB3 |
| LILRA3 |
| LILRA2 |
| HNRNPUL1 |
| SFRS16 |
| KPTN |
| MAP4K1 |
| KLK8 |
| PNKP |
| U2AF2 |
| ATF5 |
| ZFP30 |
| SAPS1 |
| CARD8 |
| SIRT2 |
| SIPA1L3 |
| ZC3H4 |
| FBXO46 |
| ETHE1 |
| RPL13A |
| SYNGR4 |
| LILRA4 |
| PRG1 |
| ZIM2 |
| NUP62 |
| HSPBP1 |
| PPP1R15A |
| PLD3 |
| EML2 |
| ZNF324 |
| KLK5 |
| PRKD2 |
| ZNF473 |
| KLK13 |
| CCDC9 |
| PRPF31 |
| IRF2BP1 |
| FGF21 |
| SNORD35A |
| SNORD34 |
| SNORD33 |
| SNORD32A |
| ZNF285A |
| SIGLEC7 |
| LYPD3 |
| BBC3 |
| DKKL1 |
| SIGLEC9 |
| SIGLEC8 |
| GPR77 |
| CHMP2A |
| DHDH |
| ZNF544 |
| EIF3K |
| UBE2S |
| SLC6A16 |
| LGALS13 |
| CYP2S1 |
| STRN4 |
| CCDC106 |
| EPN1 |
| SERTAD3 |
| SERTAD1 |
| GLTSCR2 |
| GLTSCR1 |
| EHD2 |
| KLK14 |
| KLK12 |
| SHANK1 |
| NOSIP |
| ZNF580 |
| HSD17B14 |
| GP6 |
| VRK3 |
| ZNF571 |
| ZNF581 |
| PTOV1 |
| RAB4B |
| PAF1 |
| PPP1R12C |
| TRPM4 |
| ZNF586 |
| QPCTL |
| FAM83E |
| EPS8L1 |
| RASIP1 |
| SARS2 |
| TMEM160 |
| PIH1D1 |
| SAMD4B |
| ATP5SL |
| C19orf73 |
| PNMAL1 |
| TMEM143 |
| ZNF444 |
| KLK15 |
| MED29 |
| NLRP2 |
| ZNF416 |
| ZNF446 |
| ZNF701 |
| ZNF83 |
| ZNF415 |
| C19orf61 |
| IRGC |
| CABP5 |
| SPHK2 |
| LGALS14 |
| EXOSC5 |
| MEIS3 |
| CEACAM19 |
| SLC17A7 |
| NAT14 |
| CD177 |
| VN1R1 |
| RCN3 |
| ZNF304 |
| TTYH1 |
| PNMAL2 |
| PRR12 |
| ZNF471 |
| LRFN1 |
| USP29 |
| PLEKHA4 |
| PRX |
| SPTBN4 |
| MARK4 |
| CATSPERG |
| ZNF71 |
| SCAF1 |
| CACNG8 |
| CACNG7 |
| CACNG6 |
| ZNF350 |
| TSKS |
| ZNF667 |
| DMRTC2 |
| C19orf33 |
| ELSPBP1 |
| LIN7B |
| HIF3A |
| ZNF574 |
| PLEKHG2 |
| ZNF649 |
| ZSCAN18 |
| MGC2752 |
| TSEN34 |
| TRAPPC6A |
| MBOAT7 |
| FKRP |
| ZSCAN5A |
| LENG1 |
| LILRP2 |
| LILRA6 |
| ZNF576 |
| ZNF329 |
| TBC1D17 |
| ZNF419 |
| GEMIN7 |
| ISOC2 |
| MYH14 |
| ZNF665 |
| ZNF552 |
| ZNF671 |
| ZNF613 |
| ADCK4 |
| CNTD2 |
| ZNF702P |
| ZNF606 |
| ZNF614 |
| FUZ |
| OPA3 |
| ITPKC |
| B9D2 |
| RSHL1 |
| ZNF611 |
| MED25 |
| BCL2L12 |
| TEX101 |
| GRWD1 |
| WDR87 |
| CCDC8 |
| ZNF541 |
| SYT3 |
| AKT1S1 |
| ZNF528 |
| BRSK1 |
| CNFN |
| SNORD35B |
| ZNF347 |
| ZNF577 |
| ZNF607 |
| SUV420H2 |
| C19orf48 |
| ZBTB45 |
| ZNF587 |
| FIZ1 |
| GALP |
| SIGLEC10 |
| SIGLEC12 |
| ZNF628 |
| KIR3DX1 |
| ZNF551 |
| CEACAM21 |
| ZNF616 |
| ZNF766 |
| CCDC97 |
| EXOC3L2 |
| ZNF468 |
| ZNF160 |
| CTU1 |
| ZNF835 |
| YIF1B |
| ZNF765 |
| NLRP12 |
| MYADM |
| ZNF845 |
| TIMM50 |
| SHKBP1 |
| CCDC114 |
| DKFZp434J0226 |
| ACPT |
| CGB5 |
| CGB7 |
| LRRC4B |
| LENG9 |
| CGB8 |
| GNG8 |
| PPP1R14A |
| EGLN2 |
| BIRC8 |
| FAM71E1 |
| RDH13 |
| PTH2 |
| ZIM3 |
| SIGLEC11 |
| CGB1 |
| CGB2 |
| LMTK3 |
| LENG8 |
| FBXO17 |
| KIR3DL3 |
| RASGRP4 |
| ZNF526 |
| ZNF837 |
| CLDND2 |
| ZNF816A |
| ZNF543 |
| CEACAM20 |
| COX6B2 |
| OSCAR |
| ZNF813 |
| JOSD2 |
| C19orf41 |
| CPT1C |
| ALDH16A1 |
| NTN5 |
| NLRP13 |
| NLRP8 |
| NLRP5 |
| ZNF787 |
| ZNF573 |
| EID2B |
| IRGQ |
| ZNF428 |
| RINL |
| FBXO27 |
| C19orf47 |
| ZFP28 |
| VSIG10L |
| NCRNA00085 |
| ZNF480 |
| ZNF534 |
| ZNF578 |
| C19orf18 |
| ZNF418 |
| ZNF417 |
| ZNF548 |
| FLJ40125 |
| KLC3 |
| LYPD4 |
| TMEM190 |
| HIPK4 |
| TMC4 |
| LOC147804 |
| ZNF524 |
| ZNF784 |
| CCDC155 |
| DACT3 |
| SIX5 |
| IGFL2 |
| NLRP4 |
| ZNF542 |
| ZNF582 |
| ZNF583 |
| FAM98C |
| CAPN12 |
| TTC9B |
| CDC42EP5 |
| ZNF569 |
| ZNF570 |
| ZNF836 |
| ZNF610 |
| ZNF600 |
| ZNF320 |
| ZNF497 |
| ZNF550 |
| ZNF296 |
| DEDD2 |
| ZNF579 |
| ZNF114 |
| ZNF781 |
| EID2 |
| ZNF780B |
| ZNF540 |
| ZNF525 |
| SPACA4 |
| NLRP7 |
| GGN |
| CADM4 |
| C19orf76 |
| ZNF584 |
| ZSCAN4 |
| NLRP11 |
| TMEM86B |
| PRR24 |
| ZNF549 |
| NAPSB |
| IL4I1 |
| IL28A |
| IL28B |
| IL29 |
| SSC5D |
| ZNF547 |
| ZIK1 |
| ZNF776 |
| ZSCAN1 |
| ZNF780A |
| C19orf54 |
| PRR19 |
| TMEM145 |
| CXCL17 |
| ZNF575 |
| LYPD5 |
| ZNF283 |
| NKPD1 |
| TPRX1 |
| MAMSTR |
| IZUMO1 |
| C19orf63 |
| KLK9 |
| SIGLECP3 |
| C19orf75 |
| ZNF615 |
| ZNF841 |
| LOC284379 |
| VSTM1 |
| TMEM150B |
| FAM71E2 |
| VN1R2 |
| VN1R4 |
| NLRP9 |
| ZNF546 |
| MYPOP |
| NANOS2 |
| NCCRP1 |
| SYCN |
| LEUTX |
| ZNF404 |
| ZNF284 |
| ZNF677 |
| RFPL4A |
| ZSCAN5B |
| ZSCAN22 |
| SELV |
| ZNF530 |
| C19orf51 |
| ZNF233 |
| LILRA5 |
| B3GNT8 |
| IGFL1 |
| ZNF773 |
| FLJ41856 |
| CEACAM16 |
| BLOC1S3 |
| IGFL3 |
| ZNF808 |
| ZNF761 |
| ZNF470 |
| ZNF749 |
| ZNF324B |
| ZNF793 |
| PAPL |
| ZNF805 |
| SPRED3 |
| ZNF321 |
| LOC400696 |
| SIGLEC16 |
| FLJ26850 |
| ZNF880 |
| ZNF772 |
| IGLON5 |
| MIRLET7E |
| MIR125A |
| MIR150 |
| MIR99B |
| PSG8 |
| OLT-2 |
| MIR330 |
| MIR371 |
| MIR372 |
| MIR373 |
| IGFL4 |
| NCRNA00181 |
| DPRX |
| DUXA |
| ASPDH |
| MIR512-1 |
| MIR512-2 |
| MIR498 |
| MIR520E |
| MIR515-1 |
| MIR519E |
| MIR520F |
| MIR515-2 |
| MIR519C |
| MIR520A |
| MIR526B |
| MIR519B |
| MIR525 |
| MIR523 |
| MIR518F |
| MIR520B |
| MIR518B |
| MIR526A1 |
| MIR520C |
| MIR518C |
| MIR524 |
| MIR517A |
| MIR519D |
| MIR521-2 |
| MIR520D |
| MIR517B |
| MIR520G |
| MIR516B2 |
| MIR526A2 |
| MIR518E |
| MIR518A1 |
| MIR518D |
| MIR516B1 |
| MIR518A2 |
| MIR517C |
| MIR520H |
| MIR521-1 |
| MIR522 |
| MIR519A1 |
| MIR527 |
| MIR516A1 |
| MIR516A2 |
| MIR519A2 |
| KLKP1 |
| TMEM91 |
| SBK2 |
| PSG10 |
| LGALS7B |
| PHLDB3 |
| SEC1 |
| SNORD23 |
| SNORD88A |
| SNORD88B |
| SNORD88C |
| MIR641 |
| MIR642 |
| MIR643 |
| RPL13AP5 |
| CCDC61 |
| CEACAM18 |
| SHISA7 |
| ZNF814 |
| MIR769 |
| KIR3DP1 |
| SIGLEC14 |
| MIMT1 |
| MIR935 |
| SNAR-G1 |
| SNAR-F |
| SNAR-A1 |
| SNAR-A2 |
| SNAR-A12 |
| LOC100129935 |
| BSPH1 |
| LOC100131691 |
| PEG3AS |
| SNAR-A3 |
| SNAR-A5 |
| SNAR-A7 |
| SNAR-A11 |
| SNAR-A9 |
| SNAR-A4 |
| SNAR-A6 |
| SNAR-A8 |
| SNAR-A13 |
| SNAR-A10 |
| SNAR-B2 |
| SNAR-C2 |
| SNAR-C4 |
| SNAR-E |
| SNAR-C5 |
| SNAR-B1 |
| SNAR-C1 |
| SNAR-C3 |
| SNAR-D |
| SNAR-G2 |
| SRRM5 |
| C19orf69 |
| SNAR-A14 |
This is the comprehensive list of deleted genes in the wide peak for 3q13.31.
Table S73. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BCL6 |
| FOXL2 |
| CBLB |
| CTNNB1 |
| EIF4A2 |
| ETV5 |
| FANCD2 |
| FHIT |
| GATA2 |
| LPP |
| MITF |
| MLF1 |
| MLH1 |
| MYD88 |
| PIK3CA |
| PPARG |
| RAF1 |
| RPN1 |
| SOX2 |
| TFRC |
| VHL |
| XPC |
| BAP1 |
| GMPS |
| SRGAP3 |
| TFG |
| FOXP1 |
| SETD2 |
| PBRM1 |
| hsa-mir-922 |
| hsa-mir-570 |
| hsa-mir-944 |
| hsa-mir-28 |
| hsa-mir-1248 |
| hsa-mir-1224 |
| hsa-mir-569 |
| hsa-mir-551b |
| hsa-mir-720 |
| hsa-mir-1263 |
| hsa-mir-16-2 |
| hsa-mir-1280 |
| hsa-mir-548i-1 |
| hsa-mir-198 |
| hsa-mir-568 |
| hsa-mir-567 |
| hsa-mir-1324 |
| hsa-mir-1284 |
| hsa-mir-135a-1 |
| hsa-let-7g |
| hsa-mir-566 |
| hsa-mir-191 |
| hsa-mir-1226 |
| hsa-mir-564 |
| hsa-mir-138-1 |
| hsa-mir-26a-1 |
| hsa-mir-128-2 |
| hsa-mir-563 |
| hsa-mir-885 |
| AADAC |
| ACAA1 |
| ACPP |
| ACTL6A |
| ACVR2B |
| ACY1 |
| ADCY5 |
| ADPRH |
| AGTR1 |
| AHSG |
| ALAS1 |
| ALCAM |
| AMT |
| APEH |
| APOD |
| ARF4 |
| RHOA |
| ATP1B3 |
| ATP2B2 |
| ATP6V1A |
| ATR |
| BCHE |
| BDH1 |
| BTD |
| C3orf51 |
| CACNA1D |
| SLC25A20 |
| CAMP |
| CASR |
| CAV3 |
| CCK |
| CD80 |
| CD86 |
| ENTPD3 |
| CD47 |
| CDC25A |
| CISH |
| AP2M1 |
| CLCN2 |
| CCR1 |
| CCR3 |
| CCR4 |
| CCR5 |
| CCR8 |
| CCBP2 |
| COL7A1 |
| COL8A1 |
| CP |
| CPA3 |
| CPB1 |
| CPN2 |
| CPOX |
| CRYGS |
| CSTA |
| CX3CR1 |
| CYP8B1 |
| DAG1 |
| DGKG |
| DAZL |
| DLG1 |
| DNASE1L3 |
| DOCK3 |
| DRD3 |
| DUSP7 |
| DVL3 |
| ECT2 |
| CELSR3 |
| EHHADH |
| EIF4G1 |
| EPHA3 |
| EPHB1 |
| EPHB3 |
| MECOM |
| FBLN2 |
| FGF12 |
| FLNB |
| GAP43 |
| GBE1 |
| GHSR |
| GLB1 |
| GNAI2 |
| GNAT1 |
| GOLGA4 |
| GOLGB1 |
| GP5 |
| GP9 |
| XCR1 |
| GPR15 |
| GPR27 |
| GPX1 |
| GRM2 |
| GRM7 |
| GSK3B |
| GTF2E1 |
| GYG1 |
| HCLS1 |
| HGD |
| HRH1 |
| HRG |
| HES1 |
| HTR1F |
| HYAL1 |
| IL1RAP |
| IL5RA |
| IL12A |
| IMPDH2 |
| IRAK2 |
| ITGA9 |
| ITGB5 |
| ITIH1 |
| ITIH3 |
| ITIH4 |
| ITPR1 |
| KNG1 |
| KPNA1 |
| KPNA4 |
| LAMB2 |
| RPSA |
| LSAMP |
| LTF |
| TM4SF1 |
| MAP4 |
| MBNL1 |
| MCM2 |
| MFI2 |
| MME |
| MOBP |
| CD200 |
| MST1 |
| MST1R |
| MUC4 |
| MYL3 |
| MYLK |
| NCK1 |
| NDUFB4 |
| NDUFB5 |
| NKTR |
| OGG1 |
| OPA1 |
| CLDN11 |
| OXTR |
| P2RY1 |
| PAK2 |
| CNTN3 |
| PCCB |
| PCYT1A |
| PDHB |
| PFKFB4 |
| PFN2 |
| SERPINI1 |
| SERPINI2 |
| PIK3CB |
| PLCD1 |
| PLD1 |
| PLOD2 |
| PLS1 |
| PLSCR1 |
| PLXNA1 |
| PLXNB1 |
| POLR2H |
| POU1F1 |
| PPP1R2 |
| PPP2R3A |
| PRKAR2A |
| PRKCD |
| PRKCI |
| PROS1 |
| MASP1 |
| PSMD2 |
| PTH1R |
| PTPRG |
| PTX3 |
| QARS |
| RAB5A |
| RAP2B |
| RARB |
| RARRES1 |
| RASA2 |
| RBP1 |
| RBP2 |
| RFC4 |
| RHO |
| SNORA63 |
| SNORA62 |
| ROBO1 |
| ROBO2 |
| RPL15 |
| RPL24 |
| RPL29 |
| RPL32 |
| RPL35A |
| RYK |
| SATB1 |
| ATXN7 |
| SCN5A |
| SCN10A |
| SEC13 |
| SEMA3F |
| SETMAR |
| TRA2B |
| SHOX2 |
| SI |
| SIAH2 |
| ST6GAL1 |
| SKIL |
| SLC2A2 |
| SLC6A1 |
| SLC6A6 |
| SLC6A11 |
| SLC15A2 |
| SLCO2A1 |
| HLTF |
| SMARCC1 |
| SSR3 |
| SST |
| STAC |
| NEK4 |
| SYN2 |
| TCTA |
| TDGF1 |
| TERC |
| TF |
| TFDP2 |
| TGM4 |
| TGFBR2 |
| THPO |
| THRB |
| TIMP4 |
| TKT |
| SEC62 |
| TM4SF4 |
| TMF1 |
| CLEC3B |
| TNNC1 |
| TOP2B |
| NR2C2 |
| TRH |
| TRPC1 |
| UBA7 |
| UBE2E1 |
| UBE2E2 |
| UBP1 |
| UPK1B |
| UMPS |
| USP4 |
| UQCRC1 |
| CLRN1 |
| VIPR1 |
| WNT5A |
| WNT7A |
| ZIC1 |
| CNBP |
| ZNF35 |
| ZNF80 |
| ZNF148 |
| BRPF1 |
| IFRD2 |
| MAPKAPK3 |
| SEMA3B |
| SLMAP |
| MANF |
| RAB7A |
| KCNAB1 |
| FXR1 |
| COLQ |
| ACOX2 |
| EOMES |
| HYAL3 |
| SOX14 |
| BFSP2 |
| CAMK1 |
| CGGBP1 |
| BHLHE40 |
| RUVBL1 |
| CADPS |
| TP63 |
| CHRD |
| HYAL2 |
| B4GALT4 |
| B3GALNT1 |
| SNX4 |
| TNFSF10 |
| SUCLG2 |
| HESX1 |
| KAT2B |
| NR1I2 |
| EIF2B5 |
| BSN |
| MBD4 |
| H1FX |
| USP13 |
| LIMD1 |
| KALRN |
| CCRL2 |
| UBA3 |
| RPL14 |
| CLDN1 |
| SEC22C |
| RRP9 |
| MAP3K13 |
| SLC33A1 |
| LRRFIP2 |
| MAGI1 |
| CACNA2D2 |
| COPB2 |
| ADIPOQ |
| SLC22A14 |
| SLC22A13 |
| CHST2 |
| SH3BP5 |
| SLC4A7 |
| STXBP5L |
| RNF7 |
| GUCA1C |
| IQCB1 |
| DZIP3 |
| VGLL4 |
| EDEM1 |
| KIAA0226 |
| ECE2 |
| VPRBP |
| TBC1D5 |
| TATDN2 |
| IP6K1 |
| TSC22D2 |
| EPM2AIP1 |
| PSMD6 |
| TOMM70A |
| TRANK1 |
| IQSEC1 |
| P2RY14 |
| DLEC1 |
| EXOG |
| XYLB |
| OXSR1 |
| NR1D2 |
| PDCD6IP |
| PARP3 |
| SMC4 |
| ABCC5 |
| COX17 |
| ARPC4 |
| ZNF197 |
| RBM6 |
| RBM5 |
| TNK2 |
| ALG3 |
| NME6 |
| CTDSPL |
| CD96 |
| KCNMB2 |
| STAG1 |
| EIF1B |
| TRAIP |
| ST3GAL6 |
| ARIH2 |
| TADA3 |
| CRTAP |
| ATG7 |
| ARL6IP5 |
| TUSC4 |
| IGF2BP2 |
| CXCR6 |
| CSPG5 |
| CLDN16 |
| POLQ |
| CHL1 |
| ARPP-21 |
| CCR9 |
| ALDH1L1 |
| USP19 |
| PDIA5 |
| SLC38A3 |
| CYB561D2 |
| TMEM115 |
| TOPBP1 |
| RPP14 |
| HHLA2 |
| FSTL1 |
| FAM107A |
| WDR6 |
| RASSF1 |
| NISCH |
| MRPL3 |
| PDCD10 |
| FILIP1L |
| TREX1 |
| SCN11A |
| TUSC2 |
| RNF13 |
| MGLL |
| TWF2 |
| MRAS |
| COPG |
| SLITRK3 |
| NLGN1 |
| TRAK1 |
| DHX30 |
| SACM1L |
| NCBP2 |
| SCAP |
| LAMB2L |
| MYH15 |
| PLCH1 |
| EXOSC7 |
| TMCC1 |
| PDZRN3 |
| TNIK |
| CAND2 |
| MCF2L2 |
| CLASP2 |
| PLXND1 |
| RAD54L2 |
| FRMD4B |
| STAB1 |
| GPD1L |
| RFTN1 |
| ATP11B |
| NBEAL2 |
| NUP210 |
| PLCL2 |
| ANKRD28 |
| C3orf63 |
| KLHL18 |
| DNAJC13 |
| SR140 |
| VPS8 |
| LARS2 |
| RYBP |
| C3orf27 |
| CAPN7 |
| ACAP2 |
| MKRN2 |
| NAT6 |
| FBXL2 |
| ANAPC13 |
| ARMC8 |
| ABHD14A |
| C3orf17 |
| WDR51A |
| ABI3BP |
| CNOT10 |
| TMEM158 |
| NDUFAF3 |
| THUMPD3 |
| MYRIP |
| PTPN23 |
| WWTR1 |
| PVRL3 |
| TIPARP |
| CHMP2B |
| DNAH1 |
| HIGD1A |
| LRIG1 |
| SUSD5 |
| UBXN7 |
| ERC2 |
| APPL1 |
| HACL1 |
| SGEF |
| ZBTB20 |
| TTLL3 |
| OR5K1 |
| OR5H1 |
| GNL3 |
| FAM162A |
| PCOLCE2 |
| SEC22A |
| GPR160 |
| FETUB |
| NPHP3 |
| ATP2C1 |
| LAMP3 |
| KCNMB3 |
| ZBTB11 |
| MORC1 |
| SERP1 |
| CNTN6 |
| LSM3 |
| RBMS3 |
| GOLIM4 |
| NKIRAS1 |
| SPCS1 |
| ACAD9 |
| ASTE1 |
| KLF15 |
| GTPBP8 |
| TAGLN3 |
| PRSS50 |
| RBM15B |
| GPR171 |
| GMPPB |
| SEC61A1 |
| LOH3CR2A |
| SCHIP1 |
| LMCD1 |
| PIK3R4 |
| PODXL2 |
| ARHGEF3 |
| TRAT1 |
| VILL |
| IMPG2 |
| C3orf32 |
| NMD3 |
| TRNT1 |
| ABHD5 |
| COMMD2 |
| DYNC1LI1 |
| A4GNT |
| C3orf18 |
| DBR1 |
| CRBN |
| SS18L2 |
| ZNF639 |
| CLDN18 |
| C3orf19 |
| SHISA5 |
| C3orf1 |
| ZDHHC3 |
| RSRC1 |
| ZMYND10 |
| PLA1A |
| TEX264 |
| CCDC72 |
| ZNF589 |
| HEMK1 |
| AMOTL2 |
| IP6K2 |
| SFMBT1 |
| NCKIPSD |
| PHF7 |
| CCRL1 |
| PEX5L |
| RAB6B |
| SELT |
| FBXO40 |
| DNAJB11 |
| GHRL |
| P2RY13 |
| IL20RB |
| GPR87 |
| TLR9 |
| DCUN1D1 |
| SEMA5B |
| XRN1 |
| WDR5B |
| LZTFL1 |
| PARP14 |
| P4HTM |
| SLC6A20 |
| IL17RD |
| GRAMD1C |
| ROPN1 |
| KLHL24 |
| SIDT1 |
| C3orf75 |
| SNRK |
| QRICH1 |
| PXK |
| CMTM6 |
| RG9MTD1 |
| SLC41A3 |
| PIGX |
| SLC25A38 |
| ULK4 |
| OXSM |
| SLC35A5 |
| TMEM45A |
| FEZF2 |
| IFT57 |
| FLJ10213 |
| ANO10 |
| DALRD3 |
| SHQ1 |
| MSL2 |
| TBCCD1 |
| FAIM |
| SLC25A36 |
| ARL8B |
| SETD5 |
| DPPA4 |
| LEPREL1 |
| TMEM39A |
| ZNF654 |
| TMEM40 |
| ABCF3 |
| LSG1 |
| ABHD10 |
| CHDH |
| PARL |
| IL17RB |
| CDV3 |
| MFN1 |
| YEATS2 |
| IFT122 |
| NGLY1 |
| TBC1D23 |
| WDR52 |
| CACNA2D3 |
| DCP1A |
| GLT8D1 |
| TMEM111 |
| EAF2 |
| C3orf10 |
| ZNF167 |
| MYNN |
| LMOD3 |
| EIF5A2 |
| CLDND1 |
| MUC13 |
| SUCNR1 |
| RAD18 |
| SEMA3G |
| MCCC1 |
| LXN |
| C3orf37 |
| MRPS22 |
| NIT2 |
| KTELC1 |
| BBX |
| KIF15 |
| ADAMTS9 |
| CCNL1 |
| PLSCR2 |
| PCBP4 |
| PLSCR4 |
| PCNP |
| HRASLS |
| MRPL47 |
| SENP7 |
| ABHD6 |
| LRTM1 |
| C3orf14 |
| KIAA1143 |
| ISY1 |
| HHATL |
| HEG1 |
| KIAA1257 |
| CDGAP |
| NCEH1 |
| IFT80 |
| KIAA1407 |
| WDR48 |
| LRRN1 |
| KIAA1524 |
| SLC7A14 |
| SRPRB |
| SELK |
| SENP2 |
| GNB4 |
| BPESC1 |
| PROK2 |
| EEFSEC |
| RNF123 |
| NSUN3 |
| CIDEC |
| CLSTN2 |
| POPDC2 |
| RTP4 |
| MAGEF1 |
| ZFYVE20 |
| KIF9 |
| NFKBIZ |
| AZI2 |
| ZMAT3 |
| MTMR14 |
| ATG3 |
| MRPS25 |
| CSRNP1 |
| GORASP1 |
| MFSD1 |
| CCDC14 |
| FNDC3B |
| P2RY12 |
| CDCP1 |
| CCDC71 |
| NT5DC2 |
| SLC26A6 |
| TMEM108 |
| CRELD1 |
| CAMKV |
| TMEM43 |
| OR5H6 |
| OR5H2 |
| ZXDC |
| ZBED2 |
| LRRC2 |
| FYCO1 |
| ATP13A3 |
| CEP97 |
| HSPBAP1 |
| C3orf52 |
| VEPH1 |
| QTRTD1 |
| CCDC51 |
| TBL1XR1 |
| ZBBX |
| ZNF385D |
| LRRC31 |
| CCDC48 |
| NEK11 |
| UBA5 |
| HDAC11 |
| MAP6D1 |
| PHC3 |
| C3orf36 |
| ARL14 |
| THOC7 |
| NAT13 |
| PIGZ |
| CEP63 |
| CEP70 |
| ABTB1 |
| WDR82 |
| TMEM22 |
| TSEN2 |
| GRIP2 |
| OR5AC2 |
| RTP3 |
| PARP9 |
| ESYT3 |
| SPATA16 |
| EIF2A |
| B3GNT5 |
| C3orf20 |
| ID2B |
| ARL6 |
| ZIC4 |
| ATRIP |
| ACAD11 |
| IQCG |
| ATP13A4 |
| FYTTD1 |
| NICN1 |
| CHCHD6 |
| MON1A |
| C3orf26 |
| HPS3 |
| ARPM1 |
| JAGN1 |
| KBTBD8 |
| SLC12A8 |
| C3orf42 |
| RETNLB |
| CCDC54 |
| MGC2889 |
| IL17RC |
| ABHD14B |
| LRCH3 |
| MINA |
| GPR128 |
| C3orf39 |
| DIRC2 |
| C3orf34 |
| EAF1 |
| DCLK3 |
| GFM1 |
| LMLN |
| KLHL6 |
| C3orf15 |
| PHLDB2 |
| VWA5B2 |
| UCN2 |
| LOC90246 |
| C3orf25 |
| TMEM41A |
| ZNF502 |
| BOC |
| FAM55C |
| RFT1 |
| OXNAD1 |
| SPSB4 |
| ACPL2 |
| ZBTB47 |
| TMEM44 |
| C3orf50 |
| ACTR8 |
| CAMK2N2 |
| CMTM7 |
| OSBPL10 |
| OSBPL11 |
| SLC25A26 |
| ZNF501 |
| C3orf24 |
| LRRC58 |
| LRRC3B |
| TM4SF19 |
| TM4SF18 |
| RPL39L |
| MED12L |
| CLRN1OS |
| GALNTL2 |
| GPR62 |
| CPNE4 |
| CCDC58 |
| KCNH8 |
| DNAJC19 |
| OTOL1 |
| FAM3D |
| ZPLD1 |
| LYZL4 |
| KBTBD5 |
| TRIM71 |
| FAM131A |
| CD200R1 |
| CHCHD4 |
| ZDHHC19 |
| CRYBG3 |
| DCBLD2 |
| LRRC15 |
| FAM43A |
| TPRA1 |
| TMEM42 |
| UROC1 |
| FAM194A |
| NUDT16 |
| COL6A6 |
| GRK7 |
| TMEM207 |
| METTL6 |
| C3orf31 |
| IL17RE |
| RTP1 |
| IQCF1 |
| GLYCTK |
| PPM1M |
| C3orf49 |
| SNTN |
| SYNPR |
| C3orf45 |
| RPL32P3 |
| H1FOO |
| PISRT1 |
| ASB14 |
| TTC14 |
| DTX3L |
| FAM19A4 |
| SGOL1 |
| C3orf48 |
| EFHB |
| LOC151658 |
| PPM1L |
| WDR49 |
| LRRC34 |
| CPNE9 |
| DPPA2 |
| CCDC80 |
| BTLA |
| CCDC12 |
| C3orf59 |
| PPP4R2 |
| C3orf21 |
| ROPN1B |
| LOC152024 |
| C3orf22 |
| C3orf55 |
| ZCWPW2 |
| CMC1 |
| NEK10 |
| C3orf79 |
| CCDC50 |
| PYDC2 |
| CCDC52 |
| CMTM8 |
| NUDT16P |
| CCDC13 |
| LOC152217 |
| LOC152225 |
| FGD5 |
| CIDECP |
| CNTN4 |
| IGSF11 |
| C3orf30 |
| PARP15 |
| C3orf57 |
| DNAJB8 |
| GPR156 |
| XIRP1 |
| RNF168 |
| CHST13 |
| PRICKLE2 |
| KBTBD12 |
| DHX36 |
| HTR3C |
| DZIP1L |
| TTC21A |
| ALG1L |
| C3orf67 |
| KCTD6 |
| LIPH |
| ARL13B |
| DHFRL1 |
| HTR3D |
| RPL22L1 |
| OSTalpha |
| FBXO45 |
| KLHDC8B |
| MUC20 |
| GABRR3 |
| PTPLB |
| STT3B |
| DNAH12 |
| PDE12 |
| FAM116A |
| TIGIT |
| LOC201651 |
| C3orf58 |
| SENP5 |
| KIAA2018 |
| LOC220729 |
| ZBTB38 |
| CADM2 |
| ZNF620 |
| NAALADL2 |
| ZDHHC23 |
| LOC255025 |
| TCTEX1D2 |
| C3orf43 |
| SDHALP1 |
| COL29A1 |
| GK5 |
| PLCXD2 |
| GCET2 |
| UTS2D |
| ALS2CL |
| TMIE |
| LOC285194 |
| SLC9A9 |
| C3orf64 |
| LOC285205 |
| EPHA6 |
| FBXW12 |
| C3orf38 |
| HTR3E |
| ZNF619 |
| ZNF621 |
| RABL3 |
| IGSF10 |
| C3orf33 |
| CCDC66 |
| SLC9A10 |
| C3orf23 |
| ZNF660 |
| LOC285359 |
| SUMF1 |
| RPUSD3 |
| PRRT3 |
| LOC285370 |
| LOC285375 |
| DPH3 |
| C3orf70 |
| TPRG1 |
| LOC285401 |
| ILDR1 |
| TRIM59 |
| TRIM42 |
| EIF4E3 |
| RAB43 |
| CCDC39 |
| CCDC36 |
| KY |
| C3orf35 |
| GADL1 |
| PRSS42 |
| C3orf47 |
| LOC344595 |
| LRRIQ4 |
| SAMD7 |
| AADACL2 |
| GPR149 |
| ZNF860 |
| TMPRSS7 |
| CD200R1L |
| PAQR9 |
| DVWA |
| RTP2 |
| OSTN |
| ATP13A5 |
| SOX2OT |
| TXNDC6 |
| WDR53 |
| LNP1 |
| CCDC37 |
| NCRNA00119 |
| TPRXL |
| LOC348840 |
| NMNAT3 |
| ZNF445 |
| SPATA12 |
| LHFPL4 |
| C3orf77 |
| C3orf62 |
| TMEM110 |
| LRRC33 |
| PRSS45 |
| AMIGO3 |
| ZNF662 |
| CDH29 |
| C3orf54 |
| IQCF2 |
| IQCF5 |
| MUSTN1 |
| VGLL3 |
| PRR23B |
| PRR23C |
| PLSCR5 |
| C3orf16 |
| LEKR1 |
| TMEM212 |
| VENTXP7 |
| LOC401052 |
| IQCF3 |
| FLJ25363 |
| C3orf72 |
| LOC401093 |
| FLJ42393 |
| OR5K2 |
| OR5H14 |
| OR5H15 |
| OR5K3 |
| OR5K4 |
| MIRLET7G |
| MIR128-2 |
| MIR135A1 |
| MIR138-1 |
| MIR15B |
| MIR16-2 |
| MIR191 |
| MIR198 |
| MIR26A1 |
| FAM19A1 |
| STX19 |
| LOC440944 |
| TMEM89 |
| IQCF6 |
| LOC440957 |
| MIR425 |
| ARGFX |
| SNORA6 |
| SNORA7A |
| SNORD2 |
| SNORA4 |
| TMPPE |
| TMEM30C |
| ALG1L2 |
| TXNRD3IT1 |
| TMEM14E |
| COL6A4P2 |
| SPINK8 |
| C3orf71 |
| LOC646498 |
| C3orf65 |
| PA2G4P4 |
| LOC647309 |
| IQCJ |
| SCARNA7 |
| C3orf66 |
| SNORA7B |
| SNORA58 |
| SNORA81 |
| SNORD19 |
| FAM86D |
| SNORD66 |
| SNORD69 |
| MIR551B |
| MIR563 |
| MIR564 |
| MIR567 |
| MIR568 |
| MIR569 |
| MIR570 |
| GLT8D4 |
| SDHALP2 |
| FAM157A |
| FAM198A |
| CCR2 |
| LOC729375 |
| PRR23A |
| HESRG |
| LOC100009676 |
| SNORD19B |
| LOC100125556 |
| MIR922 |
| MIR885 |
| MIR944 |
| EGOT |
| GHRLOS |
| LOC100128023 |
| LOC100128164 |
| C3orf74 |
| LOC100128640 |
| LOC100129354 |
| LOC100129550 |
| LOC100131551 |
| ZNF717 |
| SNAR-I |
| MIR1224 |
| LOC100287227 |
| FRG2C |
| LOC100302640 |
Table 3. Get Full Table Arm-level significance table - 25 significant results found.
| Arm | # Genes | Amp Frequency | Amp Z score | Amp Q value | Del Frequency | Del Z score | Del Q value |
|---|---|---|---|---|---|---|---|
| 1p | 1731 | 0.11 | -1.21 | 1 | 0.19 | 1.8 | 0.1 |
| 1q | 1572 | 0.29 | 5.5 | 1.81e-07 | 0.05 | -3.68 | 1 |
| 2p | 753 | 0.25 | 1.99 | 0.0752 | 0.03 | -5.36 | 1 |
| 2q | 1235 | 0.14 | -1.15 | 1 | 0.02 | -5.32 | 1 |
| 3p | 853 | 0.19 | -0.0103 | 1 | 0.54 | 12.7 | 0 |
| 3q | 917 | 0.64 | 15.5 | 0 | 0.35 | 4.34 | 2.32e-05 |
| 4p | 366 | 0.05 | -4.17 | 1 | 0.46 | 8.94 | 0 |
| 4q | 865 | 0.03 | -4.26 | 1 | 0.45 | 9.69 | 0 |
| 5p | 207 | 0.54 | 9.76 | 0 | 0.43 | 5.92 | 6.82e-09 |
| 5q | 1246 | 0.09 | -2.13 | 1 | 0.51 | 13 | 0 |
| 6p | 937 | 0.12 | -2.44 | 1 | 0.11 | -2.76 | 1 |
| 6q | 692 | 0.06 | -4.48 | 1 | 0.13 | -2.46 | 1 |
| 7p | 508 | 0.31 | 3.64 | 0.000694 | 0.11 | -2.96 | 1 |
| 7q | 1071 | 0.28 | 3.65 | 0.000694 | 0.08 | -3.29 | 1 |
| 8p | 495 | 0.19 | -0.367 | 1 | 0.48 | 9.37 | 0 |
| 8q | 697 | 0.39 | 6.82 | 5.96e-11 | 0.13 | -1.83 | 1 |
| 9p | 343 | 0.15 | -1.73 | 1 | 0.48 | 9.37 | 0 |
| 9q | 916 | 0.14 | -1.23 | 1 | 0.35 | 5.79 | 1.37e-08 |
| 10p | 312 | 0.07 | -4.56 | 1 | 0.23 | 0.63 | 0.607 |
| 10q | 1050 | 0.06 | -4.01 | 1 | 0.26 | 2.93 | 0.00501 |
| 11p | 731 | 0.10 | -3.1 | 1 | 0.17 | -0.769 | 1 |
| 11q | 1279 | 0.11 | -2.31 | 1 | 0.11 | -2.15 | 1 |
| 12p | 484 | 0.28 | 2.9 | 0.0067 | 0.00 | -6.22 | 1 |
| 12q | 1162 | 0.18 | 0.119 | 1 | 0.04 | -4.66 | 1 |
| 13q | 554 | 0.04 | -4.73 | 1 | 0.37 | 6.2 | 1.62e-09 |
| 14q | 1144 | 0.14 | -1.03 | 1 | 0.20 | 1.09 | 0.361 |
| 15q | 1132 | 0.13 | -1.64 | 1 | 0.13 | -1.48 | 1 |
| 16p | 719 | 0.09 | -3.52 | 1 | 0.16 | -1.03 | 1 |
| 16q | 562 | 0.08 | -4.01 | 1 | 0.18 | -0.8 | 1 |
| 17p | 575 | 0.10 | -2.86 | 1 | 0.38 | 6.17 | 1.69e-09 |
| 17q | 1321 | 0.21 | 1.71 | 0.131 | 0.16 | -0.117 | 1 |
| 18p | 117 | 0.18 | -1.42 | 1 | 0.17 | -1.71 | 1 |
| 18q | 340 | 0.13 | -2.66 | 1 | 0.24 | 0.889 | 0.456 |
| 19p | 870 | 0.21 | 0.769 | 0.616 | 0.20 | 0.3 | 0.828 |
| 19q | 1452 | 0.25 | 3.36 | 0.0017 | 0.14 | -0.696 | 1 |
| 20p | 295 | 0.31 | 3.32 | 0.00174 | 0.07 | -4.34 | 1 |
| 20q | 627 | 0.30 | 3.64 | 0.000694 | 0.07 | -3.85 | 1 |
| 21q | 422 | 0.08 | -3.6 | 1 | 0.35 | 4.87 | 1.95e-06 |
| 22q | 764 | 0.29 | 3.63 | 0.000694 | 0.11 | -2.55 | 1 |
Table 4. Get Full Table First 10 out of 211 Input Tumor Samples.
| Tumor Sample Names |
|---|
| TCGA-18-3406-01A-01D-0978-01 |
| TCGA-18-3408-01A-01D-0978-01 |
| TCGA-18-3409-01A-01D-0978-01 |
| TCGA-18-3412-01A-01D-0978-01 |
| TCGA-18-3414-01A-01D-0978-01 |
| TCGA-18-3415-01A-01D-0978-01 |
| TCGA-18-3416-01A-01D-0978-01 |
| TCGA-18-3417-01A-01D-1439-01 |
| TCGA-18-3419-01A-01D-0978-01 |
| TCGA-18-3421-01A-01D-0978-01 |
Figure 3. Segmented copy number profiles in the input data
GISTIC identifies genomic regions that are significantly gained or lost across a set of tumors. It takes segmented copy number ratios as input, separates arm-level events from focal events, and then performs two tests: (i) identifies significantly amplified/deleted chromosome arms; and (ii) identifies regions that are significantly focally amplified or deleted. For the focal analysis, the significance levels (Q values) are calculated by comparing the observed gains/losses at each locus to those obtained by randomly permuting the events along the genome to reflect the null hypothesis that they are all 'passengers' and could have occurred anywhere. The locus-specific significance levels are then corrected for multiple hypothesis testing. The arm-level significance is calculated by comparing the frequency of gains/losses of each arm to the expected rate given its size. The method outputs genomic views of significantly amplified and deleted regions, as well as a table of genes with gain or loss scores. A more in depth discussion of the GISTIC algorithm and its utility is given in [1], [3], and [5].
Regions of the genome that are prone to germ line variations in copy number are excluded from the GISTIC analysis using a list of germ line copy number variations (CNVs). A CNV is a DNA sequence that may be found at different copy numbers in the germ line of two different individuals. Such germ line variations can confound a GISTIC analysis, which finds significant somatic copy number variations in cancer. A more in depth discussion is provided in [6]. GISTIC currently uses two CNV exclusion lists. One is based on the literature describing copy number variation, and a second one comes from an analysis of significant variations among the blood normals in the TCGA data set.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.