GISTIC identifies genomic regions that are significantly gained or lost across a set of tumors. The pipeline first filters out normal samples from the segmented copy-number data by inspecting the TCGA barcodes and then executes GISTIC version 2.0.16 (cga svn revision 38839).
There were 144 tumor samples used in this analysis: 20 significant arm-level results, 6 significant focal amplifications, and 20 significant focal deletions were found.
Figure 1. Genomic positions of amplified regions: the X-axis represents the normalized amplification signals (top) and significance by Q value (bottom). The green line represents the significance cutoff at Q value=0.25.
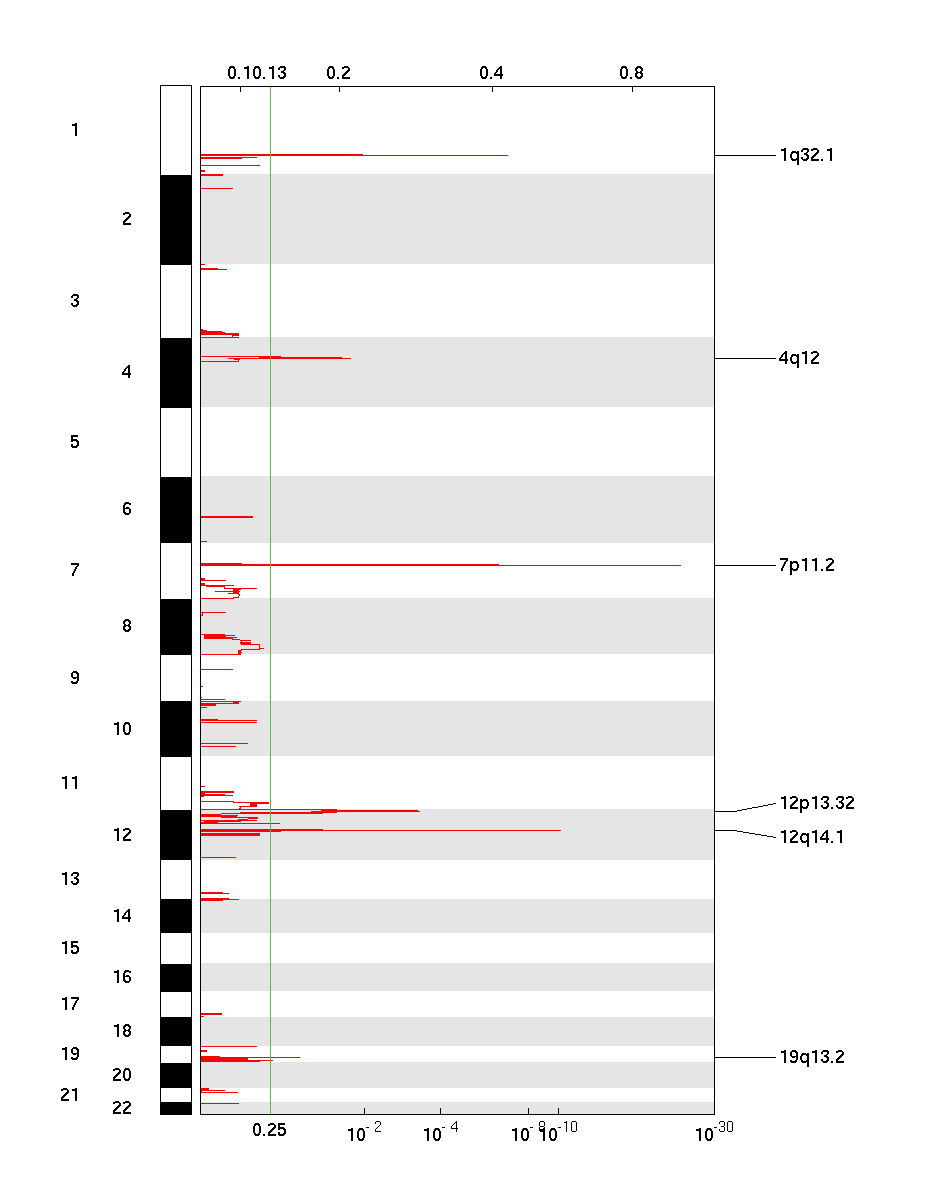
Table 1. Get Full Table Amplifications Table - 6 significant amplifications found. Click the link in the last column to view a comprehensive list of candidate genes. If no genes were identified within the peak, the nearest gene appears in brackets.
| Cytoband | Q value | Residual Q value | Wide Peak Boundaries | # Genes in Wide Peak |
|---|---|---|---|---|
| 7p11.2 | 1.6527e-24 | 1.6527e-24 | chr7:54942676-55107468 | 1 |
| 12q14.1 | 7.3432e-11 | 7.3432e-11 | chr12:58125396-58163019 | 6 |
| 1q32.1 | 1.4959e-07 | 1.4959e-07 | chr1:203710591-204630921 | 17 |
| 12p13.32 | 0.00045132 | 0.00045132 | chr12:3496687-4659158 | 9 |
| 4q12 | 0.018356 | 0.018356 | chr4:48068784-56491855 | 35 |
| 19q13.2 | 0.11431 | 0.11431 | chr19:40246077-40291097 | 1 |
This is the comprehensive list of amplified genes in the wide peak for 7p11.2.
Table S1. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| EGFR |
This is the comprehensive list of amplified genes in the wide peak for 12q14.1.
Table S2. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CDK4 |
| CYP27B1 |
| METTL1 |
| TSPAN31 |
| MARCH9 |
| AGAP2 |
This is the comprehensive list of amplified genes in the wide peak for 1q32.1.
Table S3. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| MDM4 |
| ATP2B4 |
| KISS1 |
| PIK3C2B |
| REN |
| SNRPE |
| SOX13 |
| ZC3H11A |
| LRRN2 |
| PLEKHA6 |
| LAX1 |
| ETNK2 |
| PPP1R15B |
| LOC127841 |
| GOLT1A |
| LINC00303 |
| ZBED6 |
This is the comprehensive list of amplified genes in the wide peak for 12p13.32.
Table S4. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CCND2 |
| FGF6 |
| FGF23 |
| RAD51AP1 |
| PRMT8 |
| PARP11 |
| C12orf4 |
| C12orf5 |
| EFCAB4B |
This is the comprehensive list of amplified genes in the wide peak for 4q12.
Table S5. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| KDR |
| KIT |
| PDGFRA |
| CHIC2 |
| FIP1L1 |
| SGCB |
| TEC |
| TXK |
| CLOCK |
| NMU |
| DCUN1D4 |
| OCIAD1 |
| TMEM165 |
| DANCR |
| SLAIN2 |
| USP46 |
| RASL11B |
| SRD5A3 |
| CWH43 |
| LNX1 |
| OCIAD2 |
| SPATA18 |
| PDCL2 |
| LOC152578 |
| SCFD2 |
| GSX2 |
| SLC10A4 |
| FRYL |
| ZAR1 |
| LRRC66 |
| RPL21P44 |
| SNORA26 |
| ERVMER34-1 |
| LOC100506462 |
| MIR4449 |
This is the comprehensive list of amplified genes in the wide peak for 19q13.2.
Table S6. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| LEUTX |
Figure 2. Genomic positions of deleted regions: the X-axis represents the normalized deletion signals (top) and significance by Q value (bottom). The green line represents the significance cutoff at Q value=0.25.

Table 2. Get Full Table Deletions Table - 20 significant deletions found. Click the link in the last column to view a comprehensive list of candidate genes. If no genes were identified within the peak, the nearest gene appears in brackets.
| Cytoband | Q value | Residual Q value | Wide Peak Boundaries | # Genes in Wide Peak |
|---|---|---|---|---|
| 9p21.3 | 6.9653e-37 | 6.9653e-37 | chr9:21991753-22448737 | 2 |
| 2q37.3 | 1.1914e-09 | 1.1914e-09 | chr2:233736245-243199373 | 106 |
| 19q13.43 | 3.3971e-10 | 1.5129e-08 | chr19:54037464-59128983 | 244 |
| 4q35.1 | 2.4912e-06 | 2.4912e-06 | chr4:169416144-191154276 | 99 |
| 10q25.3 | 0.0010176 | 0.0010176 | chr10:111624142-135534747 | 194 |
| 14q24.3 | 0.01465 | 0.014571 | chr14:58901446-80681882 | 185 |
| 11p15.5 | 0.020932 | 0.022809 | chr11:1-71303995 | 968 |
| 19q13.41 | 7.6233e-09 | 0.058439 | chr19:42881099-59128983 | 684 |
| 6p24.1 | 0.07614 | 0.07614 | chr6:1-15538684 | 91 |
| 22q13.31 | 0.076257 | 0.07614 | chr22:39424515-51304566 | 187 |
| 1p36.23 | 0.10174 | 0.10174 | chr1:1-28845930 | 471 |
| 1q44 | 0.10804 | 0.1081 | chr1:234036808-249250621 | 132 |
| 3q13.31 | 0.10804 | 0.1081 | chr3:89150943-138203274 | 308 |
| 6q12 | 0.13046 | 0.1275 | chr6:57206295-82464215 | 59 |
| 3q29 | 0.14672 | 0.14114 | chr3:194502405-198022430 | 50 |
| 11q14.1 | 0.18231 | 0.18746 | chr11:79148041-87846595 | 28 |
| 15q21.1 | 0.18592 | 0.18746 | chr15:1-65284370 | 471 |
| 17q25.3 | 0.19934 | 0.2067 | chr17:80379333-81195210 | 12 |
| 2p25.3 | 0.23562 | 0.24564 | chr2:1-243199373 | 1516 |
| 3p14.3 | 0.2449 | 0.24564 | chr3:1-198022430 | 1276 |
This is the comprehensive list of deleted genes in the wide peak for 9p21.3.
Table S7. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CDKN2B |
| CDKN2B-AS1 |
This is the comprehensive list of deleted genes in the wide peak for 2q37.3.
Table S8. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-3133 |
| hsa-mir-149 |
| hsa-mir-4269 |
| AGXT |
| KIF1A |
| BOK |
| COL6A3 |
| DTYMK |
| GBX2 |
| GPC1 |
| GPR35 |
| HDLBP |
| INPP5D |
| NDUFA10 |
| SEPT2 |
| NEU2 |
| PDCD1 |
| PPP1R7 |
| SAG |
| SPP2 |
| DGKD |
| PER2 |
| LRRFIP1 |
| HDAC4 |
| FARP2 |
| ARL4C |
| RAMP1 |
| STK25 |
| COPS8 |
| CAPN10 |
| PASK |
| ATG4B |
| SH3BP4 |
| NGEF |
| SNED1 |
| TRAF3IP1 |
| ANO7 |
| PRLH |
| THAP4 |
| ANKMY1 |
| SCLY |
| ASB1 |
| UGT1A10 |
| UGT1A8 |
| UGT1A7 |
| UGT1A6 |
| UGT1A5 |
| UGT1A9 |
| UGT1A4 |
| UGT1A1 |
| UGT1A3 |
| ATG16L1 |
| USP40 |
| HJURP |
| HES6 |
| CXCR7 |
| RNPEPL1 |
| GAL3ST2 |
| RAB17 |
| TRPM8 |
| MLPH |
| IQCA1 |
| C2orf54 |
| ILKAP |
| ING5 |
| MGC16025 |
| AGAP1 |
| TWIST2 |
| NEU4 |
| MTERFD2 |
| UBE2F |
| OTOS |
| MYEOV2 |
| OR6B3 |
| LOC150935 |
| LOC151171 |
| LOC151174 |
| MSL3P1 |
| LOC200772 |
| CXXC11 |
| DUSP28 |
| ESPNL |
| RBM44 |
| AQP12A |
| KLHL30 |
| OR6B2 |
| ASB18 |
| FLJ43879 |
| MIR149 |
| DNAJB3 |
| LOC643387 |
| PRR21 |
| AQP12B |
| SCARNA6 |
| SCARNA5 |
| D2HGDH |
| LOC728323 |
| PP14571 |
| LOC100286922 |
| BOK-AS1 |
| MIR4269 |
| UBE2F-SCLY |
| MIR2467 |
| MIR4440 |
| MIR4786 |
| MIR4441 |
This is the comprehensive list of deleted genes in the wide peak for 19q13.43.
Table S9. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| TFPT |
| ZNF331 |
| hsa-mir-1274b |
| hsa-mir-935 |
| hsa-mir-373 |
| hsa-mir-1283-2 |
| A1BG |
| FCAR |
| FKBP1AP1 |
| IL11 |
| KIR2DL1 |
| KIR2DL3 |
| KIR2DL4 |
| KIR2DS4 |
| KIR3DL1 |
| KIR3DL2 |
| LAIR1 |
| LAIR2 |
| NDUFA3 |
| CNOT3 |
| PEG3 |
| PRKCG |
| PTPRH |
| RPL28 |
| RPS5 |
| RPS9 |
| AURKC |
| SYT5 |
| TNNI3 |
| TNNT1 |
| ZNF8 |
| ZNF17 |
| MZF1 |
| ZNF132 |
| ZNF134 |
| ZNF135 |
| ZNF154 |
| UBE2M |
| ZNF264 |
| NCR1 |
| TRIM28 |
| ZNF256 |
| LILRB2 |
| ZNF211 |
| TRAPPC2P1 |
| ZNF274 |
| ZNF460 |
| LILRB1 |
| LILRB5 |
| SLC27A5 |
| LILRB4 |
| LILRA1 |
| LILRB3 |
| LILRA3 |
| LILRA2 |
| U2AF2 |
| PPP6R1 |
| LILRA4 |
| ZIM2 |
| HSPBP1 |
| ZNF324 |
| PRPF31 |
| CHMP2A |
| ZNF544 |
| UBE2S |
| CCDC106 |
| EPN1 |
| ZNF580 |
| GP6 |
| ZNF581 |
| PPP1R12C |
| ZNF586 |
| EPS8L1 |
| ZNF444 |
| NLRP2 |
| ZNF416 |
| ZNF446 |
| NAT14 |
| VN1R1 |
| ZNF304 |
| TTYH1 |
| ZNF471 |
| USP29 |
| ZNF71 |
| CACNG8 |
| CACNG7 |
| CACNG6 |
| ZNF667 |
| ZSCAN18 |
| MGC2752 |
| TSEN34 |
| MBOAT7 |
| ZSCAN5A |
| LENG1 |
| LILRP2 |
| LILRA6 |
| ZNF329 |
| ZNF419 |
| ISOC2 |
| ZNF552 |
| ZNF671 |
| ZNF606 |
| BRSK1 |
| SUV420H2 |
| ZBTB45 |
| ZNF587 |
| FIZ1 |
| GALP |
| ZNF628 |
| KIR3DX1 |
| ZNF551 |
| ZNF835 |
| NLRP12 |
| MYADM |
| LENG9 |
| RDH13 |
| ZIM3 |
| LENG8 |
| KIR3DL3 |
| ZNF837 |
| ZNF543 |
| COX6B2 |
| OSCAR |
| NLRP13 |
| NLRP8 |
| NLRP5 |
| ZNF787 |
| ZFP28 |
| LOC147670 |
| C19orf18 |
| ZNF418 |
| ZNF417 |
| ZNF548 |
| TMEM190 |
| TMC4 |
| ZNF524 |
| ZNF784 |
| NLRP4 |
| ZNF542 |
| ZNF582 |
| ZNF583 |
| CDC42EP5 |
| ZNF497 |
| ZNF550 |
| ZNF579 |
| NLRP7 |
| ZNF584 |
| ZSCAN4 |
| NLRP11 |
| TMEM86B |
| ZNF549 |
| SSC5D |
| ZNF547 |
| ZIK1 |
| ZNF776 |
| ZSCAN1 |
| LOC284379 |
| VSTM1 |
| TMEM150B |
| FAM71E2 |
| NLRP9 |
| RFPL4A |
| ZSCAN5B |
| ZSCAN22 |
| ZNF530 |
| C19orf51 |
| LILRA5 |
| ZNF773 |
| LOC386758 |
| TMEM238 |
| ZNF470 |
| ZNF749 |
| ZNF324B |
| ZNF805 |
| ZNF772 |
| TARM1 |
| MIR371A |
| MIR372 |
| MIR373 |
| A1BG-AS1 |
| DPRX |
| DUXA |
| MIR512-1 |
| MIR512-2 |
| MIR498 |
| MIR520E |
| MIR515-1 |
| MIR519E |
| MIR520F |
| MIR515-2 |
| MIR519C |
| MIR520A |
| MIR526B |
| MIR519B |
| MIR525 |
| MIR523 |
| MIR518F |
| MIR520B |
| MIR518B |
| MIR526A1 |
| MIR520C |
| MIR518C |
| MIR524 |
| MIR517A |
| MIR519D |
| MIR521-2 |
| MIR520D |
| MIR517B |
| MIR520G |
| MIR516B2 |
| MIR526A2 |
| MIR518E |
| MIR518A1 |
| MIR518D |
| MIR516B1 |
| MIR518A2 |
| MIR517C |
| MIR520H |
| MIR521-1 |
| MIR522 |
| MIR519A1 |
| MIR527 |
| MIR516A1 |
| MIR516A2 |
| MIR519A2 |
| SBK2 |
| LOC646862 |
| SHISA7 |
| ZNF814 |
| MIMT1 |
| MIR935 |
| LOC100128252 |
| LOC100128398 |
| SGK110 |
| LOC100131691 |
| PEG3-AS1 |
| ZNF587B |
| MIR1283-2 |
| MIR1323 |
| MIR1283-1 |
| ZNF865 |
| MIR4754 |
| MIR4752 |
| MIR371B |
This is the comprehensive list of deleted genes in the wide peak for 4q35.1.
Table S10. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| DUX4 |
| hsa-mir-1305 |
| hsa-mir-4276 |
| hsa-mir-548t |
| AGA |
| SLC25A4 |
| CASP3 |
| CLCN3 |
| DCTD |
| F11 |
| ACSL1 |
| FAT1 |
| FRG1 |
| GPM6A |
| HMGB2 |
| HPGD |
| HSP90AA4P |
| ING2 |
| IRF2 |
| KLKB1 |
| MTNR1A |
| NEK1 |
| TLR3 |
| VEGFC |
| GLRA3 |
| SORBS2 |
| SAP30 |
| HAND2 |
| MFAP3L |
| ADAM29 |
| SCRG1 |
| PALLD |
| FAM149A |
| FBXO8 |
| DUX2 |
| PDLIM3 |
| AADAT |
| GALNT7 |
| CLDN22 |
| C4orf27 |
| NEIL3 |
| UFSP2 |
| CDKN2AIP |
| ODZ3 |
| LRP2BP |
| STOX2 |
| KIAA1430 |
| SH3RF1 |
| SPCS3 |
| TRAPPC11 |
| MLF1IP |
| NBLA00301 |
| WWC2 |
| CEP44 |
| SNX25 |
| CBR4 |
| MGC45800 |
| WDR17 |
| ZFP42 |
| SPATA4 |
| ENPP6 |
| ASB5 |
| C4orf38 |
| RWDD4 |
| CCDC111 |
| TRIML2 |
| CCDC110 |
| CYP4V2 |
| LOC285441 |
| LOC285501 |
| LOC339975 |
| TRIML1 |
| ANKRD37 |
| LOC389247 |
| HELT |
| LOC401164 |
| FAM92A3 |
| HSP90AA6P |
| C4orf47 |
| DUX4L4 |
| GALNTL6 |
| FRG2 |
| SLED1 |
| FLJ38576 |
| DUX4L6 |
| DUX4L5 |
| DUX4L3 |
| LINC00290 |
| LOC728175 |
| DUX4L2 |
| LOC731424 |
| CLDN24 |
| LOC100288255 |
| MIR1305 |
| MIR4276 |
| MIR3945 |
| LOC100506085 |
| LOC100506122 |
| LOC100506229 |
This is the comprehensive list of deleted genes in the wide peak for 10q25.3.
Table S11. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| FGFR2 |
| DUX4 |
| hsa-mir-202 |
| hsa-mir-378c |
| hsa-mir-4297 |
| hsa-mir-4296 |
| hsa-mir-2110 |
| hsa-mir-4295 |
| hsa-mir-548e |
| ACADSB |
| ADAM8 |
| ADD3 |
| ADRA2A |
| ADRB1 |
| BNIP3 |
| CASP7 |
| CTBP2 |
| CYP2E1 |
| DMBT1 |
| DOCK1 |
| DUSP5 |
| ECHS1 |
| EMX2 |
| GFRA1 |
| PRLHR |
| GPR26 |
| GRK5 |
| HABP2 |
| HMX2 |
| INPP5A |
| ABLIM1 |
| MGMT |
| MKI67 |
| MXI1 |
| NRAP |
| OAT |
| PNLIP |
| PNLIPRP1 |
| PNLIPRP2 |
| HTRA1 |
| PTPRE |
| RGS10 |
| SLC18A2 |
| TCF7L2 |
| TECTB |
| TIAL1 |
| UROS |
| XPNPEP1 |
| SHOC2 |
| ADAM12 |
| UTF1 |
| EIF3A |
| SMC3 |
| BUB3 |
| BAG3 |
| FAM53B |
| DCLRE1A |
| SMNDC1 |
| GLRX3 |
| DPYSL4 |
| TACC2 |
| TUBGCP2 |
| PRDX3 |
| VAX1 |
| ATE1 |
| SEC23IP |
| RAB11FIP2 |
| INPP5F |
| FAM175B |
| ATRNL1 |
| C10orf137 |
| DUX2 |
| PDCD4 |
| VENTX |
| CUZD1 |
| CALY |
| CHST15 |
| ACSL5 |
| ZRANB1 |
| TTC40 |
| NSMCE4A |
| C10orf118 |
| WDR11 |
| DHX32 |
| PPP2R2D |
| FAM45B |
| TDRD1 |
| BCCIP |
| GPAM |
| KIAA1598 |
| FAM160B1 |
| PLEKHA1 |
| FAM204A |
| LHPP |
| IKZF5 |
| ZDHHC6 |
| MCMBP |
| C10orf81 |
| C10orf88 |
| LRRC27 |
| GPR123 |
| NKX6-2 |
| AFAP1L2 |
| KNDC1 |
| MTG1 |
| BBIP1 |
| FANK1 |
| SYCE1 |
| PRAP1 |
| ZNF511 |
| C10orf90 |
| BTBD16 |
| FAM24A |
| PSTK |
| MMP21 |
| PDZD8 |
| CTAGE7P |
| CLRN3 |
| PNLIPRP3 |
| SFXN4 |
| CPXM2 |
| TRUB1 |
| VTI1A |
| LOC143188 |
| C10orf82 |
| C10orf46 |
| C10orf91 |
| PWWP2B |
| EMX2OS |
| PPAPDC1A |
| PAOX |
| FAM24B |
| EBF3 |
| CASC2 |
| TCERG1L |
| HSPA12A |
| C10orf125 |
| JAKMIP3 |
| STK32C |
| RBM20 |
| LOC282997 |
| LOC283038 |
| LOC283089 |
| KCNK18 |
| VWA2 |
| NANOS1 |
| HMX3 |
| NHLRC2 |
| C10orf96 |
| FLJ46361 |
| ENO4 |
| ARMS2 |
| C10orf122 |
| LOC387723 |
| GUCY2GP |
| NKX1-2 |
| C10orf120 |
| LOC399815 |
| METTL10 |
| FLJ37035 |
| FOXI2 |
| FLJ46300 |
| LOC399829 |
| SPRNP1 |
| FAM45A |
| FRG2B |
| SPRN |
| MIR202 |
| NPS |
| LOC619207 |
| SNORA19 |
| FAM196A |
| RPL13AP6 |
| DUX4L7 |
| DUX4L6 |
| DUX4L5 |
| DUX4L3 |
| DUX4L2 |
| LOC100169752 |
| MIR2110 |
| MIR378C |
| MIR4297 |
| MIR4295 |
| MIR4296 |
| MIR3941 |
| MIR3663 |
| MIR3944 |
| LOC100505933 |
| FAM24B-CUZD1 |
| MIR4680 |
| MIR4483 |
| MIR4682 |
| MIR4484 |
| MIR4681 |
This is the comprehensive list of deleted genes in the wide peak for 14q24.3.
Table S12. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| GPHN |
| hsa-mir-1260 |
| hsa-mir-625 |
| hsa-mir-548h-1 |
| ACTN1 |
| ACYP1 |
| ARG2 |
| ZFP36L1 |
| ENTPD5 |
| DIO2 |
| DLST |
| EIF2S1 |
| ERH |
| ESR2 |
| ESRRB |
| FNTB |
| FOS |
| FUT8 |
| GPX2 |
| GSTZ1 |
| HIF1A |
| HSPA2 |
| LTBP2 |
| MAX |
| MAP3K9 |
| ALDH6A1 |
| MNAT1 |
| MTHFD1 |
| SIX6 |
| PGF |
| PIGH |
| PPM1A |
| PPP2R5E |
| PRKCH |
| PSEN1 |
| ABCD4 |
| RAD51B |
| RTN1 |
| SRSF5 |
| SIX1 |
| SLC8A3 |
| SLC10A1 |
| SNAPC1 |
| SPTB |
| TGFB3 |
| ZBTB25 |
| DPF3 |
| NUMB |
| ADAM21 |
| ADAM20 |
| DCAF5 |
| ALKBH1 |
| EIF2B2 |
| PNMA1 |
| NRXN3 |
| AKAP5 |
| SPTLC2 |
| RGS6 |
| KIAA0247 |
| KIAA0586 |
| KIAA0317 |
| MED6 |
| VTI1B |
| BATF |
| NPC2 |
| AHSA1 |
| ACOT2 |
| TMED10 |
| C14orf1 |
| VASH1 |
| ZBTB1 |
| SNW1 |
| PCNX |
| DAAM1 |
| TTLL5 |
| SYNE2 |
| ANGEL1 |
| ZFYVE26 |
| TTC9 |
| PLEKHG3 |
| SIPA1L1 |
| DCAF4 |
| PLEK2 |
| MLH3 |
| KCNH5 |
| POMT2 |
| COQ6 |
| FCF1 |
| RDH11 |
| COX16 |
| DACT1 |
| ATP6V1D |
| JKAMP |
| DHRS7 |
| SIX4 |
| ZFYVE1 |
| EXD2 |
| VRTN |
| SYNJ2BP |
| SLC39A9 |
| FLVCR2 |
| C14orf118 |
| YLPM1 |
| C14orf162 |
| ADCK1 |
| TMEM63C |
| RHOJ |
| GALNTL1 |
| PLEKHH1 |
| TRMT5 |
| ZNF410 |
| NGB |
| RBM25 |
| C14orf133 |
| SMOC1 |
| IRF2BPL |
| MPP5 |
| C14orf135 |
| GPR135 |
| ZC2HC1C |
| C14orf169 |
| C14orf45 |
| SGPP1 |
| SLIRP |
| DNAL1 |
| RPS6KL1 |
| SYT16 |
| KIAA1737 |
| PAPLN |
| CHURC1 |
| C14orf43 |
| LIN52 |
| NEK9 |
| IFT43 |
| WDR89 |
| C14orf149 |
| GPHB5 |
| NOXRED1 |
| JDP2 |
| ISCA2 |
| ACOT4 |
| RDH12 |
| ADAM21P1 |
| PPP1R36 |
| SLC38A6 |
| LOC145474 |
| PTGR2 |
| FAM161B |
| C14orf166B |
| ISM2 |
| FAM71D |
| TMEM229B |
| TMEM30B |
| SAMD15 |
| PROX2 |
| ZDHHC22 |
| TMED8 |
| C14orf178 |
| C14orf55 |
| C14orf39 |
| SNORD56B |
| VSX2 |
| RAB15 |
| HEATR4 |
| FLJ22447 |
| PLEKHD1 |
| LINC00238 |
| TEX21P |
| ACOT1 |
| ACOT6 |
| LOC645431 |
| FLJ43390 |
| SYNDIG1L |
| C14orf38 |
| LOC731223 |
| LOC100289511 |
| MIR1260A |
| LOC100506321 |
| SYNJ2BP-COX16 |
| CHURC1-FNTB |
| MIR4505 |
| MIR4708 |
| MIR4709 |
| MIR4706 |
| HIF1A-AS2 |
This is the comprehensive list of deleted genes in the wide peak for 11p15.5.
Table S13. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CCND1 |
| CARS |
| DDB2 |
| EXT2 |
| FANCF |
| HRAS |
| LMO1 |
| LMO2 |
| MEN1 |
| NUP98 |
| WT1 |
| CREB3L1 |
| hsa-mir-548k |
| hsa-mir-3164 |
| hsa-mir-3163 |
| hsa-mir-612 |
| hsa-mir-194-2 |
| hsa-mir-1237 |
| hsa-mir-1908 |
| hsa-mir-3162 |
| hsa-mir-130a |
| hsa-mir-3161 |
| hsa-mir-3160-2 |
| hsa-mir-129-2 |
| hsa-mir-610 |
| hsa-mir-3159 |
| hsa-mir-4299 |
| hsa-mir-302e |
| hsa-mir-483 |
| hsa-mir-675 |
| hsa-mir-4298 |
| hsa-mir-210 |
| ACP2 |
| ACTN3 |
| ADM |
| ADRBK1 |
| AP2A2 |
| APLNR |
| ALDH3B1 |
| ALDH3B2 |
| AMPD3 |
| APBB1 |
| RHOG |
| ARHGAP1 |
| ARL2 |
| ARNTL |
| ART1 |
| ASCL2 |
| BAD |
| BBS1 |
| BDNF |
| SERPING1 |
| C11orf2 |
| MRPL49 |
| ZNHIT2 |
| MPPED2 |
| C11orf9 |
| C11orf10 |
| DAGLA |
| CALCA |
| CALCB |
| CAPN1 |
| CAT |
| CCKBR |
| CD5 |
| CD6 |
| MS4A1 |
| MS4A3 |
| CD44 |
| CD59 |
| CD81 |
| CD151 |
| CDKN1C |
| CFL1 |
| CHKA |
| CHRM1 |
| CHRM4 |
| TPP1 |
| CNGA4 |
| CNTF |
| COPB1 |
| COX8A |
| CPT1A |
| CRY2 |
| CST6 |
| CSTF3 |
| CTNND1 |
| CTSD |
| CTSW |
| DDB1 |
| DHCR7 |
| DRD4 |
| DUSP8 |
| EEF1G |
| EIF4G2 |
| ELF5 |
| MARK2 |
| CTTN |
| ESRRA |
| F2 |
| FAU |
| MS4A2 |
| FEN1 |
| FGF3 |
| FGF4 |
| FKBP2 |
| FOLH1 |
| FSHB |
| FTH1 |
| GAS2 |
| GIF |
| GNG3 |
| GSTP1 |
| GTF2H1 |
| HBB |
| HBBP1 |
| HBD |
| HBE1 |
| HBG1 |
| HBG2 |
| SLC29A2 |
| HPX |
| DNAJC4 |
| IGF2 |
| IGHMBP2 |
| ILK |
| INCENP |
| INS |
| IRF7 |
| CD82 |
| KCNA4 |
| KCNC1 |
| KCNJ11 |
| KCNQ1 |
| KRTAP5-9 |
| LDHA |
| LDHC |
| FADS1 |
| FADS3 |
| LRP4 |
| LRP5 |
| LSP1 |
| LTBP3 |
| CAPRIN1 |
| MDK |
| SCGB2A1 |
| SCGB2A2 |
| MAP3K11 |
| MUC2 |
| MUC6 |
| MYBPC3 |
| MYOD1 |
| NAP1L4 |
| NDUFS3 |
| NDUFV1 |
| NDUFS8 |
| NELL1 |
| NUCB2 |
| SLC22A18 |
| SLC22A18AS |
| OSBP |
| OVOL1 |
| P2RX3 |
| PAX6 |
| PC |
| PDE3B |
| PGA5 |
| PIK3C2A |
| PLCB3 |
| POLR2G |
| POLR2L |
| PPP1CA |
| PPP2R5B |
| PRG2 |
| PSMA1 |
| PSMC3 |
| PSMD13 |
| PTH |
| PTPRCAP |
| PTPRJ |
| PYGM |
| RAB3IL1 |
| MAP4K2 |
| RAD9A |
| RAG1 |
| RAG2 |
| RAPSN |
| RARRES3 |
| RBM4 |
| RCN1 |
| RELA |
| DPF2 |
| RNH1 |
| ROM1 |
| MRPL23 |
| RPL27A |
| RPLP2 |
| RPS6KB2 |
| RPS13 |
| RRM1 |
| SAA1 |
| SAA2 |
| SAA3P |
| SAA4 |
| SCT |
| SIPA1 |
| SLC1A2 |
| SLC3A2 |
| SMPD1 |
| SPI1 |
| SPTBN2 |
| TRIM21 |
| SSRP1 |
| ST5 |
| STIM1 |
| STX3 |
| STX5 |
| ABCC8 |
| TAF10 |
| TALDO1 |
| TCN1 |
| TEAD1 |
| TH |
| TSPAN4 |
| TM7SF2 |
| TNNI2 |
| TNNT3 |
| TPH1 |
| TRAF6 |
| TRPC2 |
| TSG101 |
| PHLDA2 |
| TUB |
| SCGB1A1 |
| VEGFB |
| BEST1 |
| WEE1 |
| SF1 |
| ZFPL1 |
| ZNF143 |
| ZNF195 |
| ZNF214 |
| ZNF215 |
| RASSF7 |
| CSRP3 |
| PDHX |
| FOSL1 |
| BBOX1 |
| PPFIBP2 |
| PPFIA1 |
| SLC43A1 |
| IFITM1 |
| DGKZ |
| CHST1 |
| API5 |
| MADD |
| OR6A2 |
| DCHS1 |
| EIF3F |
| CTSF |
| FADD |
| BANF1 |
| RPS6KA4 |
| BRSK2 |
| AIP |
| SYT7 |
| SART1 |
| SLC6A5 |
| FIBP |
| MTA2 |
| UBE2L6 |
| SNORD29 |
| SNORD31 |
| SNORD30 |
| SNORD28 |
| SNORD27 |
| SNORD26 |
| SNORD25 |
| SNORD22 |
| SLC22A6 |
| SLC22A8 |
| NRXN2 |
| LPXN |
| PEX16 |
| FADS2 |
| MAPK8IP1 |
| TP53I11 |
| PITPNM1 |
| RIN1 |
| MTL5 |
| MICAL2 |
| CTR9 |
| ATG13 |
| CKAP5 |
| TRIM66 |
| FGF19 |
| CCS |
| RCE1 |
| NAALADL1 |
| NR1H3 |
| DPP3 |
| TSPAN32 |
| TSSC4 |
| USH1C |
| KCNK7 |
| HIPK3 |
| PRMT3 |
| RASGRP2 |
| GLYAT |
| CDK2AP2 |
| TCIRG1 |
| RTN3 |
| MRVI1 |
| TRIM22 |
| PRG3 |
| IFITM3 |
| SPON1 |
| RBM14 |
| CDC42EP2 |
| EIF3M |
| NXF1 |
| DEAF1 |
| KAT5 |
| IPO7 |
| SSSCA1 |
| HTATIP2 |
| IFITM2 |
| DRAP1 |
| TRIM3 |
| TAF6L |
| SCGB1D2 |
| SCGB1D1 |
| CELF1 |
| OR5I1 |
| OR7E14P |
| OR7E12P |
| LYVE1 |
| YIF1A |
| EHD1 |
| C11orf58 |
| STIP1 |
| CLP1 |
| KCNQ1OT1 |
| SF3B2 |
| CCDC85B |
| B3GNT1 |
| PLA2G16 |
| PKP3 |
| HPS5 |
| PTGDR2 |
| RRAS2 |
| ZP1 |
| SHANK2 |
| KDM2A |
| SWAP70 |
| ATG2A |
| GANAB |
| DTX4 |
| DENND5A |
| NUP160 |
| FNBP4 |
| RRP8 |
| SIRT3 |
| CLCF1 |
| OR52A1 |
| FAM89B |
| SNHG1 |
| ARFIP2 |
| POLA2 |
| FLRT1 |
| MTCH2 |
| FJX1 |
| C11orf41 |
| PRDX5 |
| ABTB2 |
| BRMS1 |
| C11orf20 |
| PAMR1 |
| ZDHHC5 |
| ATL3 |
| DAK |
| DKFZP434K028 |
| B3GAT3 |
| FBXO3 |
| SERGEF |
| EHF |
| OR5L2 |
| OR5E1P |
| PPP1R14B |
| OR10A3 |
| FXC1 |
| TIMM10 |
| MYEOV |
| BSCL2 |
| ELP4 |
| DKK3 |
| RBMXL2 |
| PGAP2 |
| PRPF19 |
| MACROD1 |
| SLC43A3 |
| COMMD9 |
| C11orf21 |
| PACSIN3 |
| TRPM5 |
| SAC3D1 |
| SNX15 |
| RHOD |
| EFEMP2 |
| UBQLN3 |
| KCNK4 |
| RNF141 |
| UBXN1 |
| APIP |
| TMX2 |
| GAL |
| SUV420H1 |
| HSD17B12 |
| IGF2-AS1 |
| TMEM216 |
| BET1L |
| CEND1 |
| SLC15A3 |
| PHF21A |
| MS4A4A |
| WT1-AS |
| CABP2 |
| TRMT112 |
| TMEM138 |
| CYB5R2 |
| C11orf24 |
| TRIM34 |
| CDHR5 |
| TOLLIP |
| ZDHHC13 |
| WDR74 |
| TRIM44 |
| MS4A12 |
| MRPL16 |
| SDHAF2 |
| SSH3 |
| TMEM132A |
| HRASLS2 |
| USP47 |
| VPS37C |
| ANO1 |
| TRIM68 |
| NADSYN1 |
| NAT10 |
| CCDC87 |
| PPP6R3 |
| UEVLD |
| LIN7C |
| SLC35C1 |
| TCP11L1 |
| LGR4 |
| PIDD |
| KCNQ1DN |
| SOX6 |
| CDC42BPG |
| OTUB1 |
| AMBRA1 |
| PACS1 |
| KBTBD4 |
| PARVA |
| TTC17 |
| SLC22A11 |
| MMP26 |
| AKIP1 |
| C11orf16 |
| TMEM9B |
| NRIP3 |
| ASCL3 |
| GPR137 |
| PRDM11 |
| CABP4 |
| CHRNA10 |
| SLC17A6 |
| PNPLA2 |
| CD248 |
| CORO1B |
| SCYL1 |
| CARNS1 |
| SYT13 |
| PHRF1 |
| LRRC4C |
| SCUBE2 |
| POLD4 |
| KRTAP5-8 |
| MS4A7 |
| ZBED5 |
| SIGIRR |
| ALX4 |
| RIC8A |
| MRPL17 |
| FAM111A |
| ANO3 |
| MS4A6A |
| MS4A5 |
| EPS8L2 |
| KLC2 |
| TUT1 |
| MRPL11 |
| STK33 |
| C11orf95 |
| CHID1 |
| LRFN4 |
| AHNAK |
| PRRG4 |
| TMEM223 |
| TMEM109 |
| CCDC86 |
| C11orf48 |
| C11orf49 |
| TRIM48 |
| OR51G1 |
| OR51B4 |
| OR51B2 |
| OR52N1 |
| RIC3 |
| C11orf80 |
| E2F8 |
| SLC25A22 |
| ZNF408 |
| NAA40 |
| QSER1 |
| AGBL2 |
| ZBTB3 |
| CPSF7 |
| PRR5L |
| ASRGL1 |
| ATHL1 |
| TMEM134 |
| MUS81 |
| INTS5 |
| ZFP91 |
| OR8J3 |
| OR51G2 |
| OR51E2 |
| OR4P4 |
| OR4C15 |
| OR4A5 |
| OR4A16 |
| OR4A15 |
| OR10W1 |
| OR10V2P |
| PTDSS2 |
| MOB2 |
| UNC93B1 |
| SBF2 |
| RAB1B |
| KIF18A |
| C11orf68 |
| MS4A8B |
| FERMT3 |
| TRPT1 |
| RBM4B |
| FRMD8 |
| FAM160A2 |
| RNASEH2C |
| FAR1 |
| BTBD10 |
| EIF1AD |
| NUDT22 |
| ARFGAP2 |
| SYVN1 |
| ACCS |
| MS4A14 |
| SPRYD5 |
| PTPN5 |
| MICALCL |
| LGALS12 |
| TRIM5 |
| TNKS1BP1 |
| GAL3ST3 |
| NAV2 |
| SYT8 |
| TSPAN18 |
| DKFZp761E198 |
| CCDC34 |
| SLC39A13 |
| DEPDC7 |
| SYT12 |
| ACY3 |
| GLYATL1 |
| PRKCDBP |
| CDCA5 |
| RPLP0P2 |
| SAAL1 |
| ODF3 |
| SLC22A9 |
| OSBPL5 |
| C1QTNF4 |
| PTPMT1 |
| LRRC56 |
| BATF2 |
| SLC22A12 |
| MRGPRD |
| MRGPRE |
| MRGPRF |
| ART5 |
| CATSPER1 |
| MRGPRX2 |
| MRGPRX3 |
| MRGPRX4 |
| HRASLS5 |
| TRIM78P |
| TRIM6 |
| OR52E2 |
| OR52J3 |
| OR51L1 |
| OR51A7 |
| OR51S1 |
| OR51F2 |
| OR52R1 |
| C11orf74 |
| OR4C46 |
| OR4X2 |
| OR4B1 |
| OR52M1 |
| OR52K2 |
| OR5P2 |
| OR5P3 |
| GYLTL1B |
| CYP2R1 |
| DBX1 |
| DNAJC24 |
| C11orf46 |
| OR8I2 |
| OR2D3 |
| OR2D2 |
| OR52W1 |
| OR56A4 |
| OR56A1 |
| LOC120824 |
| SYT9 |
| LDLRAD3 |
| OR52B4 |
| C11orf40 |
| OR52I2 |
| OR51E1 |
| UBQLNL |
| MUC15 |
| LOC143666 |
| C11orf94 |
| C11orf84 |
| PLEKHA7 |
| SPTY2D1 |
| TMEM86A |
| OR10A5 |
| OR2AG1 |
| DNHD1 |
| SCGB1C1 |
| SLC5A12 |
| LDHAL6A |
| C11orf42 |
| GPHA2 |
| NLRP6 |
| NS3BP |
| METTL15 |
| IMMP1L |
| OR56B4 |
| ANO5 |
| OR8U1 |
| OR4C16 |
| OR4C11 |
| OR4S2 |
| OR4C6 |
| OR5D14 |
| OR5L1 |
| OR5D18 |
| OR7E5P |
| OR5AS1 |
| OR8K5 |
| OR5T2 |
| OR8H1 |
| OR8K3 |
| OR8J1 |
| OR5R1 |
| OR5M3 |
| OR5M8 |
| OR5M11 |
| OR5AR1 |
| OR5AK4P |
| LRRC55 |
| SMTNL1 |
| YPEL4 |
| MED19 |
| MRPL21 |
| TPCN2 |
| OR6Q1 |
| OR9I1 |
| OR9Q1 |
| OR9Q2 |
| OR1S2 |
| OR1S1 |
| OR10Q1 |
| OR5B17 |
| OR5B21 |
| GLYATL2 |
| MPEG1 |
| OR5A2 |
| OR5A1 |
| OR4D6 |
| OR4D11 |
| PATL1 |
| PLAC1L |
| MS4A15 |
| VWCE |
| CYBASC3 |
| PPP1R32 |
| ORAOV1 |
| TIGD3 |
| LRRN4CL |
| HNRNPUL2 |
| ALKBH3 |
| LOC221122 |
| MS4A6E |
| PELI3 |
| LOC254100 |
| EHBP1L1 |
| SNX32 |
| TSGA10IP |
| CNIH2 |
| ZDHHC24 |
| C11orf86 |
| NUDT8 |
| LOC255512 |
| OR52B2 |
| OR4C3 |
| OR4S1 |
| C11orf35 |
| EML3 |
| TMEM151A |
| OR51F1 |
| SVIP |
| MRGPRX1 |
| NPAS4 |
| C11orf31 |
| OR51B5 |
| OR10AG1 |
| OR5J2 |
| OR4C13 |
| OR4C12 |
| KRT8P41 |
| LOC283104 |
| CSNK2A1P |
| OR51V1 |
| LOC283116 |
| H19 |
| C11orf85 |
| SLC25A45 |
| NEAT1 |
| OR9G4 |
| LOC283194 |
| LINC00301 |
| EFCAB4A |
| TMEM80 |
| CCDC88B |
| TTC9C |
| SLC22A24 |
| RCOR2 |
| HARBI1 |
| LINC00294 |
| IGSF22 |
| OR10A4 |
| OLFML1 |
| LOC283299 |
| C11orf36 |
| NLRP10 |
| NLRP14 |
| ANO9 |
| LUZP2 |
| LOC338651 |
| OR5F1 |
| OR5AP2 |
| ANKRD13D |
| B4GALNT4 |
| LOC338739 |
| OR52L1 |
| OR2AG2 |
| OR52B6 |
| DCDC1 |
| MS4A10 |
| OR10A2 |
| OVCH2 |
| TBX10 |
| PDDC1 |
| RTN4RL2 |
| GALNTL4 |
| B7H6 |
| DKFZp779M0652 |
| FAM111B |
| TMEM179B |
| TBC1D10C |
| MALAT1 |
| ZFP91-CNTF |
| MRGPRG |
| KRTAP5-1 |
| KRTAP5-3 |
| KRTAP5-4 |
| KRTAP5-10 |
| SLC22A25 |
| IFITM5 |
| FAM99A |
| OR56B1 |
| GVINP1 |
| INSC |
| FIBIN |
| C11orf96 |
| SLC22A10 |
| SPDYC |
| OR52K1 |
| OR52I1 |
| OR51D1 |
| OR52A5 |
| OR51B6 |
| OR51M1 |
| OR51Q1 |
| OR51I1 |
| OR51I2 |
| OR52D1 |
| OR52H1 |
| OR52N4 |
| OR52N5 |
| OR52N2 |
| OR52E6 |
| OR52E8 |
| OR52E4 |
| OR56A3 |
| OR56A5 |
| OR10A6 |
| ACCSL |
| OR4X1 |
| OR5D13 |
| OR5D16 |
| OR5W2 |
| OR8H2 |
| OR8H3 |
| OR5T3 |
| OR5T1 |
| OR8K1 |
| OR5M9 |
| OR5M10 |
| OR5M1 |
| OR9G1 |
| OR5AK2 |
| OR5B2 |
| OR5B12 |
| OR5AN1 |
| OR4D10 |
| OR4D9 |
| OR10V1 |
| LRRC10B |
| GPR152 |
| DOC2GP |
| NAV2-AS4 |
| HNRNPKP3 |
| FAM180B |
| PCNXL3 |
| FLJ42102 |
| OR51T1 |
| OR51A4 |
| OR51A2 |
| IFITM10 |
| OR4A47 |
| OR4C45 |
| SCGB1D4 |
| MIR129-2 |
| MIR130A |
| MIR192 |
| MIR194-2 |
| MIR210 |
| KRTAP5-5 |
| KRTAP5-2 |
| KRTAP5-6 |
| TMEM41B |
| LOC440028 |
| DKFZp686K1684 |
| LOC440040 |
| LOC440041 |
| SLC22A20 |
| KRTAP5-7 |
| KRTAP5-11 |
| LOC441601 |
| OR5B3 |
| TRIM6-TRIM34 |
| CCDC73 |
| LOC494141 |
| BDNF-AS1 |
| MS4A13 |
| OR8U8 |
| OR9G9 |
| MIR483 |
| SNORA3 |
| SNORA52 |
| BTBD18 |
| PGA3 |
| PGA4 |
| LOC644656 |
| RASSF10 |
| FAM86C2P |
| TRIM64C |
| LOC646813 |
| LOC650368 |
| LOC653486 |
| SNORA23 |
| SNORA45 |
| SNORA54 |
| SNORD67 |
| SNORA57 |
| SNORD97 |
| MIR610 |
| MIR611 |
| MIR612 |
| INS-IGF2 |
| MUC5B |
| LOC729013 |
| LOC729799 |
| METTL12 |
| C11orf83 |
| MIR675 |
| LOC100126784 |
| MRVI1-AS1 |
| LOC100130348 |
| LOC100130987 |
| C11orf91 |
| FAM99B |
| LOC100133161 |
| MRPL23-AS1 |
| MIR1908 |
| MIR1237 |
| MIR548K |
| MIR670 |
| MIR3160-2 |
| MIR3160-1 |
| MIR3162 |
| MIR4298 |
| MIR4299 |
| MTRNR2L8 |
| MIR3654 |
| MIR3664 |
| MIR3680-1 |
| LOC100500938 |
| MIR210HG |
| LOC100506305 |
| LOC100506540 |
| DCDC5 |
| LOC100507205 |
| LOC100507300 |
| LOC100507401 |
| RBM14-RBM4 |
| TMX2-CTNND1 |
| SAA2-SAA4 |
| ARL2-SNX15 |
| HNRNPUL2-BSCL2 |
| MIR4486 |
| MIR4686 |
| MIR4485 |
| MIR4489 |
| MIR4690 |
| MIR3973 |
| MIR4688 |
| MIR4691 |
| MIR4694 |
| MIR1343 |
| MIR4687 |
| MIR4488 |
This is the comprehensive list of deleted genes in the wide peak for 19q13.41.
Table S14. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BCL3 |
| ERCC2 |
| KLK2 |
| PPP2R1A |
| CBLC |
| TFPT |
| ZNF331 |
| hsa-mir-1274b |
| hsa-mir-935 |
| hsa-mir-373 |
| hsa-mir-1283-2 |
| hsa-mir-643 |
| hsa-mir-125a |
| hsa-mir-150 |
| hsa-mir-4324 |
| hsa-mir-220c |
| hsa-mir-3191 |
| hsa-mir-320e |
| hsa-mir-769 |
| hsa-mir-642 |
| A1BG |
| AP2A1 |
| APOC1 |
| APOC1P1 |
| APOC2 |
| APOC4 |
| APOE |
| KLK3 |
| BAX |
| BCAT2 |
| CEACAM1 |
| C5AR1 |
| CA11 |
| CALM3 |
| CD33 |
| SIGLEC6 |
| CD37 |
| CGB |
| CEACAM8 |
| CKM |
| AP2S1 |
| CLPTM1 |
| CRX |
| DBP |
| DMPK |
| DMWD |
| EMP3 |
| ERCC1 |
| ETFB |
| FCAR |
| FCGRT |
| FKBP1AP1 |
| FLT3LG |
| FOSB |
| FPR1 |
| FPR2 |
| FPR3 |
| FTL |
| FUT1 |
| FUT2 |
| GIPR |
| GPR4 |
| GPR32 |
| GRIN2D |
| ARHGAP35 |
| GYS1 |
| HAS1 |
| FOXA3 |
| HRC |
| PRMT1 |
| IL11 |
| IRF3 |
| KCNA7 |
| KCNC3 |
| KCNJ14 |
| KCNN4 |
| KIR2DL1 |
| KIR2DL3 |
| KIR2DL4 |
| KIR2DS4 |
| KIR3DL1 |
| KIR3DL2 |
| KLK1 |
| LAIR1 |
| LAIR2 |
| LHB |
| LIG1 |
| LIM2 |
| LIPE |
| BCAM |
| MYBPC2 |
| NDUFA3 |
| NKG7 |
| CNOT3 |
| NOVA2 |
| NPAS1 |
| NTF4 |
| NUCB1 |
| PEG3 |
| PLAUR |
| POLD1 |
| PPP5C |
| PRKCG |
| PRRG2 |
| KLK7 |
| KLK6 |
| KLK10 |
| PSG1 |
| PSG2 |
| PSG3 |
| PSG4 |
| PSG5 |
| PSG6 |
| PSG7 |
| PSG9 |
| PSG11 |
| PTGIR |
| PTPRH |
| PVR |
| PVRL2 |
| RELB |
| RPL18 |
| RPL28 |
| RPS5 |
| RPS9 |
| RPS11 |
| RRAS |
| RTN2 |
| CLEC11A |
| SEPW1 |
| SLC1A5 |
| SLC8A2 |
| SNRNP70 |
| SNRPD2 |
| SPIB |
| AURKC |
| SULT2B1 |
| SULT2A1 |
| SYT5 |
| TNNI3 |
| TNNT1 |
| TULP2 |
| NR1H2 |
| VASP |
| XRCC1 |
| ZNF8 |
| ZNF17 |
| ZNF28 |
| MZF1 |
| ZNF45 |
| ZNF221 |
| ZNF222 |
| ZNF132 |
| ZNF134 |
| ZNF135 |
| ZNF137P |
| ZNF154 |
| ZNF155 |
| ZNF175 |
| ZNF180 |
| ZNF223 |
| ZNF224 |
| ZNF225 |
| ZNF226 |
| ZNF227 |
| ZFP112 |
| ZNF229 |
| ZNF230 |
| SYMPK |
| TEAD2 |
| PPFIA3 |
| PLA2G4C |
| NAPA |
| SIGLEC5 |
| PGLYRP1 |
| UBE2M |
| CYTH2 |
| ZNF235 |
| ZNF264 |
| NCR1 |
| NAPSA |
| KLK4 |
| ZNF432 |
| DHX34 |
| SAE1 |
| TRIM28 |
| ZNF256 |
| LILRB2 |
| TOMM40 |
| ZNF211 |
| TRAPPC2P1 |
| ZNF234 |
| ZNF274 |
| ZNF460 |
| PPP1R13L |
| CD3EAP |
| RUVBL2 |
| LILRB1 |
| KDELR1 |
| LILRB5 |
| SLC27A5 |
| LILRB4 |
| KLK11 |
| LILRA1 |
| LILRB3 |
| LILRA3 |
| LILRA2 |
| CLASRP |
| KPTN |
| KLK8 |
| PNKP |
| U2AF2 |
| ATF5 |
| PPP6R1 |
| CARD8 |
| ZC3H4 |
| FBXO46 |
| ETHE1 |
| RPL13A |
| SYNGR4 |
| LILRA4 |
| PRG1 |
| ZIM2 |
| NUP62 |
| HSPBP1 |
| PPP1R15A |
| EML2 |
| ZNF324 |
| KLK5 |
| PRKD2 |
| ZNF473 |
| KLK13 |
| CCDC9 |
| PRPF31 |
| IRF2BP1 |
| FGF21 |
| SNORD35A |
| SNORD34 |
| SNORD33 |
| SNORD32A |
| ZNF285 |
| SIGLEC7 |
| LYPD3 |
| BBC3 |
| DKKL1 |
| SIGLEC9 |
| SIGLEC8 |
| GPR77 |
| CHMP2A |
| DHDH |
| ZNF544 |
| UBE2S |
| SLC6A16 |
| STRN4 |
| CCDC106 |
| EPN1 |
| GLTSCR2 |
| GLTSCR1 |
| EHD2 |
| KLK14 |
| KLK12 |
| SHANK1 |
| NOSIP |
| ZNF580 |
| HSD17B14 |
| GP6 |
| VRK3 |
| ZNF581 |
| PTOV1 |
| PPP1R12C |
| TRPM4 |
| ZNF586 |
| QPCTL |
| FAM83E |
| EPS8L1 |
| RASIP1 |
| TMEM160 |
| PIH1D1 |
| C19orf73 |
| PNMAL1 |
| TMEM143 |
| ZNF444 |
| KLK15 |
| NLRP2 |
| ZNF416 |
| ZNF446 |
| ZNF701 |
| ZNF83 |
| ZNF415 |
| SMG9 |
| IRGC |
| CABP5 |
| SPHK2 |
| MEIS3 |
| CEACAM19 |
| SLC17A7 |
| NAT14 |
| CD177 |
| VN1R1 |
| RCN3 |
| ZNF304 |
| TTYH1 |
| PNMAL2 |
| PRR12 |
| ZNF471 |
| USP29 |
| PLEKHA4 |
| MARK4 |
| ZNF71 |
| SCAF1 |
| CACNG8 |
| CACNG7 |
| CACNG6 |
| ZNF350 |
| TSKS |
| ZNF667 |
| ELSPBP1 |
| LIN7B |
| HIF3A |
| ZNF649 |
| ZSCAN18 |
| MGC2752 |
| TSEN34 |
| TRAPPC6A |
| MBOAT7 |
| FKRP |
| ZSCAN5A |
| LENG1 |
| LILRP2 |
| LILRA6 |
| ZNF576 |
| ZNF329 |
| TBC1D17 |
| ZNF419 |
| GEMIN7 |
| ISOC2 |
| MYH14 |
| ZNF665 |
| ZNF552 |
| ZNF671 |
| ZNF613 |
| ZNF702P |
| ZNF606 |
| ZNF614 |
| FUZ |
| OPA3 |
| RSPH6A |
| ZNF611 |
| MED25 |
| BCL2L12 |
| TEX101 |
| GRWD1 |
| CCDC8 |
| ZNF541 |
| SYT3 |
| AKT1S1 |
| ZNF528 |
| BRSK1 |
| CNFN |
| SNORD35B |
| ZNF347 |
| ZNF577 |
| SUV420H2 |
| C19orf48 |
| ZBTB45 |
| ZNF587 |
| FIZ1 |
| GALP |
| SIGLEC10 |
| SIGLEC12 |
| ZNF628 |
| KIR3DX1 |
| ZNF551 |
| ZNF616 |
| ZNF766 |
| EXOC3L2 |
| ZNF468 |
| ZNF160 |
| CTU1 |
| ZNF835 |
| ZNF765 |
| NLRP12 |
| MYADM |
| ZNF845 |
| CCDC114 |
| DKFZp434J0226 |
| ACPT |
| CGB5 |
| CGB7 |
| LRRC4B |
| LENG9 |
| CGB8 |
| GNG8 |
| BIRC8 |
| FAM71E1 |
| RDH13 |
| PTH2 |
| ZIM3 |
| SIGLEC11 |
| CGB1 |
| CGB2 |
| LMTK3 |
| LENG8 |
| KIR3DL3 |
| ZNF837 |
| CLDND2 |
| ZNF816 |
| ZNF543 |
| CEACAM20 |
| COX6B2 |
| OSCAR |
| ZNF813 |
| JOSD2 |
| IZUMO2 |
| CPT1C |
| ALDH16A1 |
| NTN5 |
| NLRP13 |
| NLRP8 |
| NLRP5 |
| ZNF787 |
| IRGQ |
| ZNF428 |
| ZFP28 |
| VSIG10L |
| LOC147646 |
| LINC00085 |
| ZNF480 |
| ZNF534 |
| ZNF578 |
| ERVV-1 |
| LOC147670 |
| C19orf18 |
| ZNF418 |
| ZNF417 |
| ZNF548 |
| PPM1N |
| KLC3 |
| IGSF23 |
| TMEM190 |
| TMC4 |
| LOC147804 |
| ZNF524 |
| ZNF784 |
| CCDC155 |
| DACT3 |
| SIX5 |
| IGFL2 |
| NLRP4 |
| ZNF542 |
| ZNF582 |
| ZNF583 |
| CDC42EP5 |
| ZNF836 |
| ZNF610 |
| ZNF600 |
| ZNF320 |
| ZNF497 |
| ZNF550 |
| ZNF296 |
| ZNF579 |
| ZNF114 |
| ZNF525 |
| SPACA4 |
| NLRP7 |
| CADM4 |
| C19orf76 |
| ZNF584 |
| ZSCAN4 |
| NLRP11 |
| TMEM86B |
| PRR24 |
| ZNF549 |
| NAPSB |
| IL4I1 |
| SSC5D |
| ZNF547 |
| ZIK1 |
| ZNF776 |
| ZSCAN1 |
| CXCL17 |
| LOC284344 |
| ZNF575 |
| LYPD5 |
| ZNF283 |
| PPP1R37 |
| NKPD1 |
| TPRX1 |
| MAMSTR |
| IZUMO1 |
| C19orf63 |
| MGC45922 |
| KLK9 |
| SIGLECP3 |
| C19orf75 |
| ZNF615 |
| ZNF841 |
| LOC284379 |
| VSTM1 |
| TMEM150B |
| FAM71E2 |
| VN1R2 |
| VN1R4 |
| NLRP9 |
| MYPOP |
| NANOS2 |
| ZNF404 |
| ZNF284 |
| C19orf81 |
| ZNF677 |
| RFPL4A |
| ZSCAN5B |
| ZSCAN22 |
| ZNF530 |
| C19orf51 |
| ZNF233 |
| LILRA5 |
| IGFL1 |
| ZNF773 |
| LOC386758 |
| CEACAM22P |
| CEACAM16 |
| BLOC1S3 |
| LOC388553 |
| IGFL3 |
| ZNF808 |
| ZNF761 |
| TMEM238 |
| ZNF470 |
| ZNF749 |
| ZNF324B |
| LOC390940 |
| ZNF805 |
| ZNF321P |
| SIGLEC16 |
| FLJ26850 |
| ZNF880 |
| ZNF772 |
| IGLON5 |
| MIRLET7E |
| MIR125A |
| MIR150 |
| MIR99B |
| PSG8 |
| TARM1 |
| MIR330 |
| MIR371A |
| MIR372 |
| MIR373 |
| IGFL4 |
| A1BG-AS1 |
| DPRX |
| DUXA |
| ASPDH |
| MIR512-1 |
| MIR512-2 |
| MIR498 |
| MIR520E |
| MIR515-1 |
| MIR519E |
| MIR520F |
| MIR515-2 |
| MIR519C |
| MIR520A |
| MIR526B |
| MIR519B |
| MIR525 |
| MIR523 |
| MIR518F |
| MIR520B |
| MIR518B |
| MIR526A1 |
| MIR520C |
| MIR518C |
| MIR524 |
| MIR517A |
| MIR519D |
| MIR521-2 |
| MIR520D |
| MIR517B |
| MIR520G |
| MIR516B2 |
| MIR526A2 |
| MIR518E |
| MIR518A1 |
| MIR518D |
| MIR516B1 |
| MIR518A2 |
| MIR517C |
| MIR520H |
| MIR521-1 |
| MIR522 |
| MIR519A1 |
| MIR527 |
| MIR516A1 |
| MIR516A2 |
| MIR519A2 |
| KLKP1 |
| LOC646508 |
| SBK2 |
| LOC646862 |
| PSG10P |
| PHLDB3 |
| SEC1 |
| SNORD23 |
| SNORD88A |
| SNORD88B |
| SNORD88C |
| MIR642A |
| MIR643 |
| RPL13AP5 |
| CCDC61 |
| CEACAM18 |
| SHISA7 |
| FLJ30403 |
| ZNF814 |
| MIR769 |
| SIGLEC14 |
| MIMT1 |
| MIR935 |
| SNAR-G1 |
| SNAR-F |
| SNAR-A1 |
| SNAR-A2 |
| SNAR-A12 |
| LOC100128252 |
| LOC100128398 |
| LOC100129083 |
| SGK110 |
| BSPH1 |
| LOC100131691 |
| PEG3-AS1 |
| SNAR-A3 |
| SNAR-A5 |
| SNAR-A7 |
| SNAR-A11 |
| SNAR-A9 |
| SNAR-A4 |
| SNAR-A6 |
| SNAR-A8 |
| SNAR-A13 |
| SNAR-A10 |
| SNAR-B2 |
| SNAR-C2 |
| SNAR-C4 |
| SNAR-E |
| SNAR-C5 |
| SNAR-B1 |
| SNAR-C1 |
| SNAR-C3 |
| SNAR-D |
| SNAR-G2 |
| SRRM5 |
| SNAR-A14 |
| ERVV-2 |
| LOC100287177 |
| LOC100289650 |
| ZNF587B |
| MIR1283-2 |
| MIR1323 |
| MIR1283-1 |
| LOC100379224 |
| MIR3191 |
| MIR3190 |
| MIR320E |
| MIR4324 |
| MIR642B |
| LOC100505681 |
| LOC100505715 |
| LOC100505812 |
| LOC100506012 |
| LOC100506033 |
| LOC100506068 |
| LOC100507003 |
| ZNF865 |
| ZNF816-ZNF321P |
| APOC4-APOC2 |
| MIR4754 |
| MIR4752 |
| MIR371B |
| MIR4749 |
| MIR4750 |
| MIR4531 |
| MIR4751 |
This is the comprehensive list of deleted genes in the wide peak for 6p24.1.
Table S15. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| IRF4 |
| BMP6 |
| BPHL |
| DSP |
| EDN1 |
| SERPINB1 |
| F13A1 |
| FOXF2 |
| FOXC1 |
| GCNT2 |
| GMDS |
| HIVEP1 |
| JARID2 |
| MAK |
| NEDD9 |
| NQO2 |
| SERPINB6 |
| SERPINB9 |
| RREB1 |
| SSR1 |
| TFAP2A |
| TUBB2A |
| RIPK1 |
| PRPF4B |
| GCM2 |
| CD83 |
| CDYL |
| LY86 |
| EEF1E1 |
| RANBP9 |
| ECI2 |
| FARS2 |
| RPP40 |
| SIRT5 |
| FAM50B |
| SLC35B3 |
| TBC1D7 |
| NRN1 |
| NOL7 |
| TMEM14C |
| GFOD1 |
| ELOVL2 |
| PAK1IP1 |
| EXOC2 |
| WRNIP1 |
| DUSP22 |
| LYRM4 |
| SLC22A23 |
| MUTED |
| CCDC90A |
| TXNDC5 |
| TMEM14B |
| RIOK1 |
| ADTRP |
| FOXQ1 |
| HUS1B |
| SNRNP48 |
| C6orf195 |
| PIP5K1P1 |
| RNF182 |
| PHACTR1 |
| C6orf228 |
| SYCP2L |
| LINC00518 |
| PXDC1 |
| MGC39372 |
| FAM217A |
| LOC285768 |
| LY86-AS1 |
| CAGE1 |
| MYLK4 |
| TUBB2B |
| C6orf52 |
| PSMG4 |
| DKFZP686I15217 |
| C6orf201 |
| ERVFRD-1 |
| PPP1R3G |
| HULC |
| TMEM170B |
| SCARNA27 |
| LOC100130275 |
| LOC100130357 |
| MIR3691 |
| LOC100506207 |
| LOC100506409 |
| LOC100507194 |
| LOC100508120 |
| MUTED-TXNDC5 |
| EEF1E1-MUTED |
| MIR4645 |
This is the comprehensive list of deleted genes in the wide peak for 22q13.31.
Table S16. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| EP300 |
| PDGFB |
| MKL1 |
| hsa-mir-3201 |
| hsa-let-7b |
| hsa-mir-1249 |
| hsa-mir-33a |
| hsa-mir-1281 |
| ACR |
| ACO2 |
| ADSL |
| ARSA |
| ATF4 |
| BIK |
| TSPO |
| MPPED1 |
| CHKB |
| CPT1B |
| CYP2D7P1 |
| CYP2D6 |
| CYB5R3 |
| TYMP |
| FBLN1 |
| XRCC6 |
| MCHR1 |
| MGAT3 |
| NAGA |
| NDUFA6 |
| NHP2L1 |
| PMM1 |
| PPARA |
| MAPK11 |
| RANGAP1 |
| RPL3 |
| MAPK12 |
| SBF1 |
| SREBF2 |
| ST13 |
| TCF20 |
| TEF |
| UPK3A |
| WNT7B |
| CACNA1I |
| SYNGR1 |
| GRAP2 |
| CELSR1 |
| PPP6R2 |
| ZBED4 |
| RBX1 |
| SCO2 |
| PKDREJ |
| TAB1 |
| SLC25A17 |
| NUP50 |
| TOB2 |
| RABL2B |
| PACSIN2 |
| TNRC6B |
| GRAMD4 |
| TTLL12 |
| MLC1 |
| ZC3H7B |
| KIAA0930 |
| CBX7 |
| MAPK8IP2 |
| PLXNB2 |
| BRD1 |
| ARHGAP8 |
| TBC1D22A |
| TTLL1 |
| SAMM50 |
| ATXN10 |
| FAM19A5 |
| SULT4A1 |
| RIBC2 |
| ARFGAP3 |
| SNORD43 |
| SMC1B |
| CSDC2 |
| RRP7A |
| MCAT |
| PPPDE2 |
| SGSM3 |
| PARVB |
| NCAPH2 |
| GTSE1 |
| A4GALT |
| MOV10L1 |
| SMCR7L |
| FAM118A |
| TTC38 |
| C22orf26 |
| MIOX |
| PRR5 |
| TRMU |
| SEPT3 |
| PANX2 |
| APOBEC3G |
| XPNPEP3 |
| PARVG |
| CERK |
| EFCAB6 |
| CENPM |
| ALG12 |
| CRELD2 |
| C22orf46 |
| CCDC134 |
| ADM2 |
| SCUBE1 |
| TRABD |
| PNPLA3 |
| SELO |
| L3MBTL2 |
| HDAC10 |
| LDOC1L |
| POLDIP3 |
| PHF5A |
| KIAA1644 |
| SHANK3 |
| TUBGCP6 |
| LOC90834 |
| LMF2 |
| RPS19BP1 |
| C22orf32 |
| RRP7B |
| SERHL |
| PHF21B |
| KLHDC7B |
| FAM83F |
| TNFRSF13C |
| RNU86 |
| SNORD83A |
| SNORD83B |
| ENTHD1 |
| DNAJB7 |
| CHADL |
| MEI1 |
| FAM109B |
| NFAM1 |
| PNPLA5 |
| LOC150381 |
| C22orf40 |
| CN5H6.4 |
| APOBEC3H |
| WBP2NL |
| POLR3H |
| APOBEC3F |
| SERHL2 |
| RNU12 |
| ATP5L2 |
| LOC284933 |
| RPL23AP82 |
| BK250D10.8 |
| LOC339685 |
| C22orf34 |
| CHKB-CPT1B |
| LOC388906 |
| LINC00207 |
| MIRLET7BHG |
| FLJ46257 |
| IL17REL |
| MIRLET7A3 |
| MIRLET7B |
| MIR33A |
| LINC00229 |
| FAM116B |
| PIM3 |
| SHISA8 |
| ODF3B |
| PRR5-ARHGAP8 |
| SYCE3 |
| LOC730668 |
| LOC100128946 |
| LOC100130899 |
| LOC100132273 |
| LOC100144603 |
| LOC100271722 |
| MIR1249 |
| MIR1281 |
| MIR3201 |
| MIR3619 |
| LOC100506472 |
| LOC100506714 |
| MIR4763 |
| MIR4762 |
| MIR4766 |
| MIR4535 |
This is the comprehensive list of deleted genes in the wide peak for 1p36.23.
Table S17. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| PAX7 |
| RPL22 |
| SDHB |
| ARID1A |
| TNFRSF14 |
| PRDM16 |
| MDS2 |
| hsa-mir-1976 |
| hsa-mir-3115 |
| hsa-mir-4253 |
| hsa-mir-1256 |
| hsa-mir-1290 |
| hsa-mir-1273d |
| hsa-mir-34a |
| hsa-mir-4252 |
| hsa-mir-551a |
| hsa-mir-4251 |
| hsa-mir-429 |
| hsa-mir-1302-2 |
| ALPL |
| RERE |
| C1QA |
| C1QB |
| C1QC |
| CA6 |
| CAPZB |
| CASP9 |
| RUNX3 |
| TNFRSF8 |
| CDA |
| CDK11B |
| CDC42 |
| CD52 |
| CLCN6 |
| CLCNKA |
| CLCNKB |
| CNR2 |
| CORT |
| DDOST |
| DFFA |
| DFFB |
| DVL1 |
| E2F2 |
| ECE1 |
| MEGF6 |
| EPHA2 |
| ENO1 |
| EPHA8 |
| EPHB2 |
| EXTL1 |
| EYA3 |
| FGR |
| MTOR |
| FUCA1 |
| IFI6 |
| GABRD |
| GALE |
| GNB1 |
| SFN |
| GPR3 |
| ZBTB48 |
| HMGN2 |
| HMGCL |
| HSPG2 |
| HTR1D |
| HTR6 |
| ID3 |
| TNFRSF9 |
| STMN1 |
| MFAP2 |
| MTHFR |
| NBL1 |
| NPPA |
| NPPB |
| PAFAH2 |
| PEX10 |
| PEX14 |
| PGD |
| PIK3CD |
| PLA2G2A |
| PLA2G5 |
| PLOD1 |
| EXOSC10 |
| PPP1R8 |
| PRKCZ |
| PTAFR |
| RAP1GAP |
| RHCE |
| RHD |
| RPA2 |
| RPL11 |
| RPS6KA1 |
| RSC1A1 |
| SCNN1D |
| SKI |
| SLC2A5 |
| SLC9A1 |
| SRM |
| TCEA3 |
| TCEB3 |
| TNFRSF1B |
| TP73 |
| TNFRSF4 |
| ZBTB17 |
| SLC30A2 |
| LUZP1 |
| PRDM2 |
| SNHG3 |
| NR0B2 |
| MMP23B |
| MMP23A |
| KCNAB2 |
| FCN3 |
| AKR7A2 |
| ALDH4A1 |
| EIF4G3 |
| TNFRSF25 |
| TNFRSF18 |
| PER3 |
| MAP3K6 |
| DHRS3 |
| VAMP3 |
| C1orf38 |
| H6PD |
| ISG15 |
| PLCH2 |
| CROCC |
| CEP104 |
| KLHL21 |
| SLC35E2 |
| ZBTB40 |
| MFN2 |
| CELA3A |
| WASF2 |
| ANGPTL7 |
| HNRNPR |
| SRRM1 |
| CNKSR1 |
| UBE4B |
| MAD2L2 |
| PDPN |
| NUDC |
| MASP2 |
| SRSF10 |
| UTS2 |
| RER1 |
| RCAN3 |
| MST1P2 |
| MST1P9 |
| PADI2 |
| LYPLA2 |
| PARK7 |
| CTRC |
| ACOT7 |
| DNAJC8 |
| CLSTN1 |
| AKR7A3 |
| SPEN |
| KDM1A |
| WDTC1 |
| KIAA0090 |
| KIF1B |
| PLEKHM2 |
| OTUD3 |
| KAZN |
| CAMTA1 |
| DNAJC16 |
| UBR4 |
| ATP13A2 |
| TARDBP |
| CELA3B |
| ICMT |
| PADI4 |
| TMEM50A |
| STX12 |
| CLIC4 |
| SYF2 |
| CHD5 |
| C1orf144 |
| LDLRAP1 |
| NOC2L |
| FBXO2 |
| FBXO6 |
| PLA2G2D |
| OR4F3 |
| HSPB7 |
| ARHGEF16 |
| AHDC1 |
| SMPDL3B |
| LINC00339 |
| SSU72 |
| UBIAD1 |
| PADI1 |
| PLA2G2E |
| WRAP73 |
| SLC45A1 |
| HP1BP3 |
| CELA2B |
| ZNF593 |
| SDF4 |
| MRTO4 |
| PADI3 |
| ERRFI1 |
| WNT4 |
| FBXO42 |
| RNF186 |
| MXRA8 |
| HES2 |
| GPN2 |
| FBLIM1 |
| MED18 |
| PQLC2 |
| CASZ1 |
| CPSF3L |
| C1orf159 |
| AURKAIP1 |
| MRPL20 |
| AIM1L |
| TMEM51 |
| XKR8 |
| ARHGEF10L |
| VPS13D |
| ATAD3A |
| TMEM57 |
| PANK4 |
| CAMK2N1 |
| ASAP3 |
| PNRC2 |
| PIGV |
| NBPF1 |
| NECAP2 |
| DNAJC11 |
| RCC2 |
| AJAP1 |
| FAM54B |
| CTNNBIP1 |
| C1orf63 |
| AGTRAP |
| PITHD1 |
| MAN1C1 |
| NIPAL3 |
| SEPN1 |
| TP73-AS1 |
| PLEKHG5 |
| LRRC47 |
| PTCHD2 |
| KIF17 |
| HES4 |
| GRHL3 |
| IL22RA1 |
| MIIP |
| CELA2A |
| GPATCH3 |
| PLA2G2F |
| CEP85 |
| NMNAT1 |
| VWA1 |
| PINK1 |
| PRAMEF1 |
| PRAMEF2 |
| NADK |
| PHACTR4 |
| C1orf135 |
| EFHD2 |
| MMEL1 |
| RSG1 |
| OR4F5 |
| MUL1 |
| NOL9 |
| LIN28A |
| AGMAT |
| LINC00115 |
| MORN1 |
| FAM110D |
| DHDDS |
| GPR157 |
| SPSB1 |
| GLTPD1 |
| ZNF436 |
| TAS1R2 |
| TAS1R1 |
| OR4F16 |
| ACTL8 |
| CCNL2 |
| SH3BGRL3 |
| SESN2 |
| ESPN |
| TAS1R3 |
| ATAD3B |
| TMEM222 |
| PLEKHN1 |
| USP48 |
| NBPF3 |
| ZDHHC18 |
| SLC25A33 |
| DDI2 |
| LZIC |
| TRIM63 |
| C1orf170 |
| CROCCP2 |
| SYTL1 |
| IGSF21 |
| KIAA1751 |
| KIAA2013 |
| THAP3 |
| C1orf201 |
| UBXN11 |
| C1orf158 |
| FBXO44 |
| ATPIF1 |
| CROCCP3 |
| FHAD1 |
| LOC115110 |
| FAM46B |
| RBP7 |
| ACAP3 |
| UBE2J2 |
| C1orf172 |
| LRRC38 |
| AADACL3 |
| PUSL1 |
| B3GALT6 |
| IFFO2 |
| TPRG1L |
| FAM213B |
| MYOM3 |
| KLHDC7A |
| VWA5B1 |
| UBXN10 |
| ARHGEF19 |
| ACTRT2 |
| MIB2 |
| C1orf127 |
| SAMD11 |
| LOC148413 |
| PHF13 |
| CCDC27 |
| C1orf213 |
| PDIK1L |
| C1orf64 |
| SLC2A7 |
| CALML6 |
| IL28RA |
| FAM43B |
| PAQR7 |
| FAM76A |
| TMEM201 |
| C1orf86 |
| C1orf126 |
| ATAD3C |
| AKR7L |
| LOC254099 |
| TTLL10 |
| TMCO4 |
| ZNF683 |
| NPHP4 |
| FAM41C |
| LOC284632 |
| LOC284661 |
| SLC25A34 |
| ESPNP |
| C1orf174 |
| KLHL17 |
| TMEM240 |
| TMEM52 |
| LOC339505 |
| AADACL4 |
| PRAMEF5 |
| HNRNPCL1 |
| PRAMEF9 |
| PRAMEF10 |
| FAM131C |
| PADI6 |
| C1orf187 |
| SPATA21 |
| AGRN |
| APITD1 |
| CATSPER4 |
| GPR153 |
| FAM132A |
| HES5 |
| LOC388588 |
| RNF207 |
| TMEM82 |
| TRNP1 |
| CD164L2 |
| HES3 |
| PRAMEF12 |
| PRAMEF21 |
| PRAMEF8 |
| PRAMEF18 |
| PRAMEF17 |
| PLA2G2C |
| PRAMEF4 |
| PRAMEF13 |
| SH2D5 |
| C1orf130 |
| RNF223 |
| PRAMEF3 |
| LDLRAD2 |
| MIR200A |
| MIR200B |
| MIR34A |
| FLJ42875 |
| PRAMEF11 |
| PRAMEF6 |
| LOC440563 |
| UQCRHL |
| MINOS1 |
| ANKRD65 |
| PRAMEF7 |
| MIR429 |
| FAM138F |
| LOC643837 |
| TMEM88B |
| C1orf233 |
| LOC644961 |
| C1orf200 |
| PRAMEF19 |
| PRAMEF20 |
| FAM138A |
| LOC646471 |
| LOC649330 |
| LOC653566 |
| PRAMEF22 |
| PRAMEF15 |
| WASH7P |
| PRAMEF16 |
| SCARNA1 |
| SNORA59B |
| SNORA59A |
| MIR551A |
| CDK11A |
| SLC35E2B |
| LOC728716 |
| LOC729059 |
| PRAMEF14 |
| FLJ37453 |
| LOC729737 |
| OR4F29 |
| LOC100129534 |
| LOC100130417 |
| LOC100132062 |
| LOC100132287 |
| LOC100133331 |
| LOC100133445 |
| LOC100133612 |
| DDX11L1 |
| TTC34 |
| LOC100288069 |
| MIR1976 |
| NPPA-AS1 |
| MIR3115 |
| MIR4253 |
| MIR4251 |
| MIR4252 |
| MIR3917 |
| MIR3675 |
| ENO1-AS1 |
| LOC100506730 |
| LOC100506801 |
| LOC100506963 |
| APITD1-CORT |
| C1orf151-NBL1 |
| MIR4695 |
| MIR4684 |
| MIR4689 |
| MIR4632 |
| MIR4417 |
| MIR378F |
| RCAN3AS |
This is the comprehensive list of deleted genes in the wide peak for 1q44.
Table S18. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| FH |
| hsa-mir-3124 |
| hsa-mir-3123 |
| hsa-mir-1537 |
| ACTN2 |
| ADSS |
| CHML |
| LYST |
| CHRM3 |
| GNG4 |
| HNRNPU |
| LGALS8 |
| MTR |
| NID1 |
| RGS7 |
| RYR2 |
| TARBP1 |
| TBCE |
| GPR137B |
| ZNF124 |
| KMO |
| EXO1 |
| GGPS1 |
| TOMM20 |
| CEP170 |
| AKT3 |
| ZNF238 |
| SDCCAG8 |
| RBM34 |
| OPN3 |
| TRIM58 |
| AHCTF1 |
| OR1C1 |
| OR2M4 |
| OR2L2 |
| OR2L1P |
| OR2T1 |
| PPPDE1 |
| SCCPDH |
| ARID4B |
| KIF26B |
| HEATR1 |
| ZNF692 |
| ERO1LB |
| FMN2 |
| ZNF695 |
| ZP4 |
| TFB2M |
| GREM2 |
| SMYD3 |
| ZNF669 |
| ZNF672 |
| SH3BP5L |
| OR2G3 |
| OR2G2 |
| OR2C3 |
| EFCAB2 |
| ZNF496 |
| ZNF670 |
| NLRP3 |
| FAM36A |
| OR2M5 |
| OR2M3 |
| OR2T12 |
| OR14C36 |
| OR2T34 |
| OR2T10 |
| OR2T4 |
| OR2T11 |
| OR2B11 |
| WDR64 |
| EDARADD |
| SLC35F3 |
| B3GALNT2 |
| C1orf150 |
| LOC148824 |
| LOC149134 |
| CNST |
| PLD5 |
| C1orf100 |
| OR2T6 |
| LOC255654 |
| C1orf101 |
| PGBD2 |
| OR2L13 |
| OR14A16 |
| HNRNPU-AS1 |
| VN1R5 |
| LOC339529 |
| LOC339535 |
| OR6F1 |
| OR2W3 |
| OR2T8 |
| OR2T3 |
| OR2T29 |
| IRF2BP2 |
| C1orf31 |
| C1orf229 |
| OR2M1P |
| OR11L1 |
| OR2L8 |
| OR2AK2 |
| OR2L3 |
| OR2M2 |
| OR2T33 |
| OR2M7 |
| OR2G6 |
| OR2T2 |
| OR2T5 |
| OR14I1 |
| OR2T27 |
| OR2T35 |
| MAP1LC3C |
| OR2W5 |
| OR13G1 |
| RPS7P5 |
| LOC646627 |
| SNORA14B |
| LOC731275 |
| LOC100130331 |
| LGALS8-AS1 |
| MIR1537 |
| LINC00184 |
| MIR3123 |
| MIR3124 |
| MIR3916 |
| LOC100506795 |
| LOC100506810 |
| ZNF670-ZNF695 |
| MIR4753 |
| MIR4677 |
| MIR4671 |
This is the comprehensive list of deleted genes in the wide peak for 3q13.31.
Table S19. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| CBLB |
| GATA2 |
| RPN1 |
| TFG |
| hsa-mir-1280 |
| hsa-mir-548i-1 |
| hsa-mir-544b |
| hsa-mir-198 |
| hsa-mir-568 |
| hsa-mir-567 |
| ACPP |
| ADCY5 |
| ADPRH |
| ALCAM |
| ATP6V1A |
| CASR |
| CD80 |
| CD86 |
| CD47 |
| COL8A1 |
| CPOX |
| CSTA |
| DRD3 |
| EPHA3 |
| EPHB1 |
| GAP43 |
| GOLGB1 |
| GP9 |
| GPR15 |
| GSK3B |
| GTF2E1 |
| HCLS1 |
| HGD |
| ITGB5 |
| KPNA1 |
| LSAMP |
| MCM2 |
| CD200 |
| MYLK |
| NCK1 |
| NDUFB4 |
| PCCB |
| PLXNA1 |
| PPP2R3A |
| PROS1 |
| RHO |
| RPL24 |
| RYK |
| SLC15A2 |
| SLCO2A1 |
| TF |
| TRH |
| UPK1B |
| UMPS |
| CNBP |
| ZNF80 |
| ZNF148 |
| RAB7A |
| SOX14 |
| BFSP2 |
| RUVBL1 |
| B4GALT4 |
| SNX4 |
| NR1I2 |
| MBD4 |
| H1FX |
| KALRN |
| STXBP5L |
| GUCA1C |
| IQCB1 |
| DZIP3 |
| TOMM70A |
| COX17 |
| CD96 |
| STAG1 |
| ST3GAL6 |
| POLQ |
| ALDH1L1 |
| PDIA5 |
| TOPBP1 |
| HHLA2 |
| FSTL1 |
| MRPL3 |
| FILIP1L |
| MGLL |
| MRAS |
| COPG |
| MYH15 |
| TMCC1 |
| PLXND1 |
| DNAJC13 |
| C3orf27 |
| ANAPC13 |
| ARMC8 |
| C3orf17 |
| ABI3BP |
| PVRL3 |
| ZBTB20 |
| OR5K1 |
| OR5H1 |
| FAM162A |
| SEC22A |
| NPHP3 |
| ATP2C1 |
| ZBTB11 |
| MORC1 |
| ACAD9 |
| ASTE1 |
| KLF15 |
| GTPBP8 |
| TAGLN3 |
| SEC61A1 |
| PIK3R4 |
| PODXL2 |
| TRAT1 |
| IMPG2 |
| A4GNT |
| DBR1 |
| CLDN18 |
| TIMMDC1 |
| PLA1A |
| AMOTL2 |
| CCRL1 |
| RAB6B |
| FBXO40 |
| IL20RB |
| SEMA5B |
| WDR5B |
| PARP14 |
| GRAMD1C |
| ROPN1 |
| SIDT1 |
| RG9MTD1 |
| SLC41A3 |
| SLC35A5 |
| TMEM45A |
| IFT57 |
| MSL2 |
| DPPA4 |
| TMEM39A |
| ABHD10 |
| CDV3 |
| IFT122 |
| TBC1D23 |
| WDR52 |
| EAF2 |
| CLDND1 |
| MUC13 |
| C3orf37 |
| NIT2 |
| POGLUT1 |
| BBX |
| PCNP |
| SENP7 |
| ISY1 |
| HEG1 |
| KIAA1257 |
| ARHGAP31 |
| KIAA1407 |
| KIAA1524 |
| SRPRB |
| EEFSEC |
| NSUN3 |
| POPDC2 |
| NFKBIZ |
| ATG3 |
| CCDC14 |
| TMEM108 |
| OR5H6 |
| OR5H2 |
| ZXDC |
| ZBED2 |
| CEP97 |
| HSPBAP1 |
| C3orf52 |
| QTRTD1 |
| CCDC48 |
| NEK11 |
| UBA5 |
| C3orf36 |
| NAA50 |
| CEP63 |
| ABTB1 |
| TMEM22 |
| OR5AC2 |
| PARP9 |
| ESYT3 |
| ARL6 |
| ACAD11 |
| CHCHD6 |
| C3orf26 |
| SLC12A8 |
| RETNLB |
| CCDC54 |
| MINA |
| GPR128 |
| DIRC2 |
| C3orf15 |
| PHLDB2 |
| LOC90246 |
| C3orf25 |
| BOC |
| FAM55C |
| TXNRD3 |
| OSBPL11 |
| LRRC58 |
| CPNE4 |
| CCDC58 |
| ZPLD1 |
| CD200R1 |
| DCBLD2 |
| TPRA1 |
| UROC1 |
| NUDT16 |
| COL6A6 |
| FAM172BP |
| RPL32P3 |
| H1FOO |
| DTX3L |
| LOC151658 |
| DPPA2 |
| CCDC80 |
| BTLA |
| ROPN1B |
| C3orf22 |
| SPICE1 |
| NUDT16P1 |
| LOC152225 |
| IGSF11 |
| C3orf30 |
| PARP15 |
| DNAJB8 |
| GPR156 |
| CHST13 |
| KBTBD12 |
| DZIP1L |
| ALG1L |
| ARL13B |
| DHFRL1 |
| GABRR3 |
| PTPLB |
| TIGIT |
| KIAA2018 |
| ZDHHC23 |
| LOC255025 |
| NUP210P1 |
| COL6A5 |
| PLCXD2 |
| GCET2 |
| LSAMP-AS3 |
| LOC285205 |
| EPHA6 |
| DNAJB8-AS1 |
| RABL3 |
| SLC9A10 |
| LOC285359 |
| ILDR1 |
| RAB43 |
| KY |
| LOC339874 |
| H1FX-AS1 |
| LOC344595 |
| TMPRSS7 |
| CD200R1L |
| NME9 |
| LNP1 |
| CCDC37 |
| NPHP3-AS1 |
| FLJ22763 |
| FLJ25363 |
| OR5K2 |
| OR5H14 |
| OR5H15 |
| OR5K3 |
| OR5K4 |
| MIR198 |
| STX19 |
| ARGFX |
| TMEM30C |
| ALG1L2 |
| TXNRD3NB |
| COL6A4P2 |
| LOC653712 |
| LINC00488 |
| SNORA7B |
| SNORA58 |
| MIR548A3 |
| MIR567 |
| MIR568 |
| FAM86HP |
| LOC100009676 |
| LOC100125556 |
| LOC100129550 |
| ZBTB20-AS1 |
| MIR1280 |
| MIR548I1 |
| LOC100302640 |
| MIR548G |
| MIR3921 |
| PVRL3-AS1 |
| IGSF11-AS1 |
| MYLK-AS1 |
| LOC100507032 |
| NPHP3-ACAD11 |
| ISY1-RAB43 |
| MIR4796 |
| MIR4788 |
| MIR4446 |
This is the comprehensive list of deleted genes in the wide peak for 6q12.
Table S20. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| hsa-mir-4282 |
| hsa-mir-30a |
| hsa-mir-30c-2 |
| hsa-mir-548u |
| BAI3 |
| BCKDHB |
| COL9A1 |
| COL12A1 |
| COL19A1 |
| COX7A2 |
| EEF1A1 |
| HTR1B |
| IMPG1 |
| MYO6 |
| RNY4 |
| ELOVL4 |
| TTK |
| PTP4A1 |
| HMGN3 |
| RIMS1 |
| PHF3 |
| MTO1 |
| SENP6 |
| SLC17A5 |
| FILIP1 |
| LGSN |
| PHIP |
| DDX43 |
| FAM46A |
| TMEM30A |
| LMBRD1 |
| KCNQ5 |
| FAM135A |
| SMAP1 |
| OGFRL1 |
| LINC00472 |
| KHDC1 |
| SH3BGRL2 |
| C6orf7 |
| MB21D1 |
| IRAK1BP1 |
| B3GAT2 |
| C6orf57 |
| CD109 |
| C6orf221 |
| LCA5 |
| KHDRBS2 |
| DPPA5 |
| EYS |
| GUSBP4 |
| C6orf147 |
| MIR30A |
| MIR30C2 |
| OOEP |
| MCART3P |
| KHDC1L |
| LOC100288198 |
| MIR4282 |
| LOC100506804 |
This is the comprehensive list of deleted genes in the wide peak for 3q29.
Table S21. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| TFRC |
| hsa-mir-922 |
| hsa-mir-570 |
| hsa-mir-3137 |
| APOD |
| BDH1 |
| DLG1 |
| MFI2 |
| MUC4 |
| PAK2 |
| PCYT1A |
| PPP1R2 |
| RPL35A |
| KIAA0226 |
| TNK2 |
| NCBP2 |
| ACAP2 |
| UBXN7 |
| PIGX |
| PIGZ |
| IQCG |
| FYTTD1 |
| LRCH3 |
| CEP19 |
| LMLN |
| TM4SF19 |
| ZDHHC19 |
| XXYLT1 |
| LOC152217 |
| RNF168 |
| OSTalpha |
| FBXO45 |
| MUC20 |
| SENP5 |
| LOC220729 |
| TCTEX1D2 |
| C3orf43 |
| SDHAP1 |
| WDR53 |
| ANKRD18DP |
| LRRC33 |
| LOC401109 |
| MIR570 |
| SDHAP2 |
| FAM157A |
| MIR922 |
| MFI2-AS1 |
| LOC100507086 |
| TM4SF19-TCTEX1D2 |
| MIR4797 |
This is the comprehensive list of deleted genes in the wide peak for 11q14.1.
Table S22. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| PICALM |
| hsa-mir-4300 |
| DLG2 |
| PRCP |
| FZD4 |
| OR7E2P |
| EED |
| ME3 |
| PRSS23 |
| RAB30 |
| C11orf73 |
| PCF11 |
| SYTL2 |
| TMEM126B |
| CREBZF |
| CCDC90B |
| CCDC81 |
| TMEM135 |
| TMEM126A |
| C11orf82 |
| CCDC83 |
| FAM181B |
| CCDC89 |
| ANKRD42 |
| SNORA70E |
| MIR4300 |
| LOC100506233 |
| LOC100506368 |
This is the comprehensive list of deleted genes in the wide peak for 15q21.1.
Table S23. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BUB1B |
| TCF12 |
| FLJ27352 |
| C15orf55 |
| hsa-mir-1272 |
| hsa-mir-422a |
| hsa-mir-190 |
| hsa-mir-2116 |
| hsa-mir-628 |
| hsa-mir-1266 |
| hsa-mir-147b |
| hsa-mir-1282 |
| hsa-mir-627 |
| hsa-mir-4310 |
| hsa-mir-626 |
| hsa-mir-1233-2 |
| hsa-mir-1233-1 |
| hsa-mir-211 |
| hsa-mir-1268 |
| hsa-mir-3118-6 |
| hsa-mir-3118-4 |
| ACTC1 |
| ADAM10 |
| ANXA2 |
| APBA2 |
| AQP9 |
| B2M |
| NBEAP1 |
| BNIP2 |
| CA12 |
| CAPN3 |
| CHRM5 |
| CHRNA7 |
| CKMT1B |
| CYP19A1 |
| DUT |
| EPB42 |
| FBN1 |
| FGF7 |
| GABPB1 |
| GABRA5 |
| GABRB3 |
| GABRG3 |
| GALK2 |
| GANC |
| GATM |
| GCHFR |
| PDIA3 |
| GTF2A2 |
| HDC |
| ONECUT1 |
| IPW |
| ITPKA |
| IVD |
| LIPC |
| LTK |
| MAP1A |
| MEIS2 |
| MFAP1 |
| TRPM1 |
| MYO1E |
| MYO5A |
| NDN |
| NEDD4 |
| OAZ2 |
| OCA2 |
| PLCB2 |
| PPIB |
| MAPK6 |
| RAB27A |
| RAD51 |
| RORA |
| RYR3 |
| SCG5 |
| SLC12A1 |
| SNRPN |
| SNX1 |
| SORD |
| SPINT1 |
| SRP14 |
| THBS1 |
| TJP1 |
| TP53BP1 |
| TPM1 |
| TYRO3 |
| UBE3A |
| MKRN3 |
| SLC30A4 |
| PAR5 |
| EIF3J |
| JMJD7-PLA2G4B |
| SNAP23 |
| ALDH1A2 |
| HERC2 |
| HERC1 |
| SNURF |
| USP8 |
| CCNB2 |
| SLC28A2 |
| CCPG1 |
| GCNT3 |
| COPS2 |
| TRIP4 |
| TGM5 |
| PIGB |
| PPIP5K1 |
| AQR |
| SECISBP2L |
| KIAA0101 |
| ARHGAP11A |
| LCMT2 |
| USP3 |
| SLC12A6 |
| BCL2L10 |
| RASGRP1 |
| SERF2 |
| GNB5 |
| ARPP19 |
| SLC27A2 |
| GPR176 |
| CHP |
| OIP5 |
| BAHD1 |
| FAN1 |
| CEP152 |
| MAPKBP1 |
| GOLGA8A |
| ZNF609 |
| RTF1 |
| CYFIP1 |
| MGA |
| DMXL2 |
| VPS39 |
| FAM189A1 |
| AP4E1 |
| CCNDBP1 |
| DAPK2 |
| EID1 |
| C15orf2 |
| C15orf63 |
| TMEM87A |
| RPAP1 |
| DKFZP434L187 |
| PYGO1 |
| PLDN |
| GREM1 |
| FOXB1 |
| RPUSD2 |
| TUBGCP4 |
| SCG3 |
| TMOD3 |
| TMOD2 |
| EHD4 |
| DUOX2 |
| MYEF2 |
| RPS27L |
| NDUFAF1 |
| RSL24D1 |
| NUSAP1 |
| TMEM85 |
| SPG21 |
| SPTBN5 |
| CTDSPL2 |
| KLF13 |
| RAB8B |
| DUOX1 |
| CSNK1G1 |
| MAGEL2 |
| DLL4 |
| INO80 |
| FAM63B |
| RNF111 |
| ZNF280D |
| TRPM7 |
| VPS13C |
| PPP1R14D |
| MTMR10 |
| ZNF770 |
| FLJ10038 |
| HAUS2 |
| FAM82A2 |
| DNAJC17 |
| MNS1 |
| NOP10 |
| MYO5C |
| NDNL2 |
| FAM214A |
| C15orf24 |
| PAK6 |
| DTWD1 |
| CASC5 |
| AVEN |
| ATP10A |
| GJD2 |
| STARD9 |
| VPS18 |
| SQRDL |
| ZFP106 |
| RFX7 |
| SPATA5L1 |
| CHAC1 |
| NARG2 |
| C15orf29 |
| SLTM |
| SNX22 |
| ATP8B4 |
| WDR76 |
| TMEM62 |
| SEMA6D |
| PIF1 |
| SPG11 |
| ELL3 |
| PLEKHO2 |
| POLR2M |
| NIPA2 |
| APH1B |
| TLN2 |
| FAM96A |
| C15orf48 |
| C15orf41 |
| SPPL2A |
| ZFYVE19 |
| CGNL1 |
| FRMD5 |
| DISP2 |
| CHRFAM7A |
| ULK4P3 |
| ULK4P1 |
| ARHGAP11B |
| ATPBD4 |
| C15orf57 |
| C15orf23 |
| BMF |
| SHF |
| DUOXA1 |
| SNORD107 |
| LDHAL6B |
| CHST14 |
| CASC4 |
| LACTB |
| TUBGCP5 |
| TGM7 |
| CATSPER2 |
| LEO1 |
| NIPA1 |
| PLA2G4E |
| TRIM69 |
| PAR1 |
| C15orf43 |
| LOC145663 |
| C2CD4A |
| FAM81A |
| GCOM1 |
| LOC145783 |
| LOC145845 |
| TMCO5A |
| ZSCAN29 |
| TTBK2 |
| CDAN1 |
| STRC |
| DYX1C1 |
| CSNK1A1P1 |
| OTUD7A |
| SPRED1 |
| PGBD4 |
| ADAL |
| EXD1 |
| FSIP1 |
| RHOV |
| C15orf33 |
| MGC15885 |
| UBR1 |
| PATL2 |
| LPCAT4 |
| PLA2G4F |
| LRRC57 |
| LYSMD2 |
| WDR72 |
| HMGN2P46 |
| SLC24A5 |
| PRTG |
| LOC283663 |
| LOC283683 |
| OR4N4 |
| LOC283710 |
| FAM98B |
| PLA2G4D |
| HERC2P3 |
| GOLGA6L1 |
| GOLGA8G |
| GOLGA8IP |
| FBXL22 |
| SNORD108 |
| SNORD109A |
| SNORD109B |
| SNORD115-1 |
| WHAMMP3 |
| POTEB |
| GLDN |
| FMN1 |
| SNORD64 |
| PAR4 |
| PAR-SN |
| RBPMS2 |
| ANKDD1A |
| LOC348120 |
| MRPL42P5 |
| USP50 |
| TEX9 |
| C15orf52 |
| TNFAIP8L3 |
| C2CD4B |
| GOLGA8E |
| OR4M2 |
| OR4N3P |
| SHC4 |
| CTXN2 |
| HERC2P2 |
| C15orf53 |
| C15orf54 |
| DUOXA2 |
| MIR190A |
| MIR211 |
| NF1P2 |
| HERC2P9 |
| WHAMMP2 |
| GOLGA8B |
| EIF2AK4 |
| CATSPER2P1 |
| UNC13C |
| LOC503519 |
| CKMT1A |
| SERINC4 |
| C15orf62 |
| C15orf56 |
| PHGR1 |
| LOC645212 |
| CHEK2P2 |
| LOC646214 |
| CXADRP2 |
| LOC646278 |
| REREP3 |
| LOC653061 |
| LOC653075 |
| HSP90AB4P |
| MIR626 |
| MIR627 |
| MIR628 |
| ANP32AP1 |
| SNORD116-19 |
| GOLGA6L6 |
| LOC727924 |
| LOC728758 |
| OIP5-AS1 |
| GOLGA8C |
| PWRN1 |
| PWRN2 |
| SNORD116-1 |
| SNORD116-2 |
| SNORD116-3 |
| SNORD116-4 |
| SNORD116-5 |
| SNORD116-6 |
| SNORD116-7 |
| SNORD116-8 |
| SNORD116-9 |
| SNORD116-10 |
| SNORD116-11 |
| SNORD116-12 |
| SNORD116-13 |
| SNORD116-14 |
| SNORD116-15 |
| SNORD116-16 |
| SNORD116-17 |
| SNORD116-18 |
| SNORD116-20 |
| SNORD116-21 |
| SNORD116-22 |
| SNORD116-23 |
| SNORD116-24 |
| SNORD116-25 |
| SNORD115-2 |
| SNORD116-26 |
| SNORD116-27 |
| SNORD115-3 |
| SNORD115-4 |
| SNORD115-5 |
| SNORD115-6 |
| SNORD115-7 |
| SNORD115-8 |
| SNORD115-9 |
| SNORD115-10 |
| SNORD115-11 |
| SNORD115-12 |
| SNORD115-13 |
| SNORD115-14 |
| SNORD115-15 |
| SNORD115-16 |
| SNORD115-17 |
| SNORD115-18 |
| SNORD115-19 |
| SNORD115-20 |
| SNORD115-21 |
| SNORD115-22 |
| SNORD115-23 |
| SNORD115-25 |
| SNORD115-26 |
| SNORD115-29 |
| SNORD115-30 |
| SNORD115-31 |
| SNORD115-32 |
| SNORD115-33 |
| SNORD115-34 |
| SNORD115-35 |
| SNORD115-36 |
| SNORD115-37 |
| SNORD115-38 |
| SNORD115-39 |
| SNORD115-40 |
| SNORD115-41 |
| SNORD115-42 |
| SNORD115-43 |
| SNORD115-44 |
| SNORD116-28 |
| SNORD116-29 |
| SNORD115-48 |
| SNORD115-24 |
| SNORD115-27 |
| SNORD115-28 |
| SNORD115-45 |
| SNORD115-47 |
| MIR147B |
| LOC100128714 |
| LOC100129387 |
| LOC100130855 |
| LOC100131089 |
| ANKRD63 |
| HERC2P7 |
| GOLGA8F |
| LOC100132724 |
| GOLGA8DP |
| JMJD7 |
| PLA2G4B |
| ULK4P2 |
| LOC100288615 |
| LOC100288637 |
| LOC100289656 |
| MIR1233-1 |
| MIR1272 |
| MIR1266 |
| MIR1282 |
| LOC100306975 |
| MIR2116 |
| MIR1233-2 |
| MIR4310 |
| MIR3942 |
| LOC100505648 |
| LOC100507466 |
| SERF2-C15ORF63 |
| DYX1C1-CCPG1 |
| MIR4509-1 |
| MIR4509-2 |
| MIR4508 |
| MIR4510 |
| MIR4716 |
| MIR4713 |
| MIR4509-3 |
| MIR4712 |
| MIR4715 |
| TMCO5B |
| MYZAP |
This is the comprehensive list of deleted genes in the wide peak for 17q25.3.
Table S24. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| FOXK2 |
| TBCD |
| RAB40B |
| NARF |
| WDR45L |
| FN3K |
| C17orf62 |
| FN3KRP |
| ZNF750 |
| B3GNTL1 |
| METRNL |
| FLJ43681 |
This is the comprehensive list of deleted genes in the wide peak for 2p25.3.
Table S25. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| ALK |
| ATIC |
| CHN1 |
| CREB1 |
| DNMT3A |
| ERCC3 |
| ACSL3 |
| MSH6 |
| HOXD11 |
| HOXD13 |
| IDH1 |
| MSH2 |
| MYCN |
| NFE2L2 |
| PAX3 |
| PMS1 |
| REL |
| PAX8 |
| NCOA1 |
| EML4 |
| BCL11A |
| FEV |
| TTL |
| hsa-mir-3133 |
| hsa-mir-149 |
| hsa-mir-4269 |
| hsa-mir-562 |
| hsa-mir-1471 |
| hsa-mir-1244-1 |
| hsa-mir-4268 |
| hsa-mir-3132 |
| hsa-mir-153-1 |
| hsa-mir-3131 |
| hsa-mir-26b |
| hsa-mir-548f-2 |
| hsa-mir-1302-4 |
| hsa-mir-2355 |
| hsa-mir-3130-4 |
| hsa-mir-3129 |
| hsa-mir-1245 |
| hsa-mir-561 |
| hsa-mir-1258 |
| hsa-mir-3128 |
| hsa-mir-1246 |
| hsa-mir-10b |
| hsa-mir-933 |
| hsa-mir-128-1 |
| hsa-mir-663b |
| hsa-mir-1302-3 |
| hsa-mir-4267 |
| hsa-mir-4266 |
| hsa-mir-4265 |
| hsa-mir-3127 |
| hsa-mir-4264 |
| hsa-mir-1285-2 |
| hsa-mir-3126 |
| hsa-mir-216b |
| hsa-mir-559 |
| hsa-mir-558 |
| hsa-mir-4263 |
| hsa-mir-1301 |
| hsa-mir-3125 |
| hsa-mir-4262 |
| hsa-mir-548s |
| hsa-mir-4261 |
| AAMP |
| ACADL |
| ACP1 |
| ACTG2 |
| ACVR1 |
| ACVR2A |
| ACYP2 |
| ADCY3 |
| ADD2 |
| ADRA2B |
| AGXT |
| ALPI |
| ALPP |
| ALPPL2 |
| BIN1 |
| ANXA4 |
| AOX1 |
| APOB |
| RHOB |
| RND3 |
| ATP5G3 |
| ATP6V1B1 |
| KIF1A |
| AUP1 |
| BARD1 |
| BCS1L |
| BMPR2 |
| BOK |
| ZFP36L2 |
| BUB1 |
| CACNB4 |
| CAD |
| CALM2 |
| CAPG |
| CASP8 |
| CASP10 |
| CCNT2 |
| CD8A |
| CD8B |
| CD28 |
| CENPA |
| CHRNA1 |
| CHRND |
| CHRNG |
| CLK1 |
| CNGA3 |
| COL3A1 |
| COL4A3 |
| COL4A4 |
| COL5A2 |
| COL6A3 |
| COX5B |
| CPS1 |
| ATF2 |
| CRYBA2 |
| CRYGA |
| CRYGB |
| CRYGC |
| CRYGD |
| CTLA4 |
| CTNNA2 |
| CYP1B1 |
| CYP27A1 |
| DARS |
| DBI |
| DCTN1 |
| DDX1 |
| DES |
| DGUOK |
| LOC1720 |
| DLX1 |
| DLX2 |
| DNAH6 |
| DYNC1I2 |
| DOK1 |
| DPP4 |
| DTNB |
| DTYMK |
| DUSP2 |
| E2F6 |
| EEF1B2 |
| EGR4 |
| EMX1 |
| EN1 |
| EPAS1 |
| EPHA4 |
| ERBB4 |
| FABP1 |
| FAP |
| EFEMP1 |
| FHL2 |
| FKBP1B |
| FN1 |
| FOSL2 |
| FRZB |
| FSHR |
| FTH1P3 |
| GAD1 |
| GALNT3 |
| GBX2 |
| GCG |
| GCKR |
| MSTN |
| GFPT1 |
| GGCX |
| GLI2 |
| GLS |
| GPC1 |
| GPD2 |
| GPR1 |
| GPR17 |
| GPR35 |
| GPR39 |
| GRB14 |
| GTF3C2 |
| GYPC |
| HADHA |
| HADHB |
| HDLBP |
| HK2 |
| HNMT |
| TLX2 |
| HOXD1 |
| HOXD3 |
| HOXD4 |
| HOXD8 |
| HOXD9 |
| HOXD10 |
| HOXD12 |
| HPCAL1 |
| AGFG1 |
| DNAJB2 |
| HSPD1 |
| HSPE1 |
| FOXN2 |
| HTR2B |
| ID2 |
| SP110 |
| IGFBP2 |
| IGFBP5 |
| IHH |
| IL1A |
| IL1B |
| IL1R1 |
| IL1RN |
| CXCR1 |
| CXCR2 |
| CXCR2P1 |
| INHA |
| INHBB |
| INPP1 |
| INPP4A |
| INPP5D |
| ITGA6 |
| IRS1 |
| ITGA4 |
| ITGAV |
| ITGB6 |
| KCNF1 |
| KCNJ3 |
| KCNJ13 |
| KCNK3 |
| KCNS3 |
| KHK |
| KIF3C |
| KIF5C |
| AFF3 |
| LCT |
| LHCGR |
| LIMS1 |
| LRP2 |
| LTBP1 |
| LY75 |
| EPCAM |
| MXD1 |
| MAL |
| MAP2 |
| MAT2A |
| MATN3 |
| MCM6 |
| MDH1 |
| MEIS1 |
| MGAT5 |
| MPV17 |
| MYO1B |
| MTIF2 |
| MYL1 |
| MYO7B |
| NAB1 |
| NCL |
| NEB |
| NDUFA10 |
| NDUFB3 |
| NDUFS1 |
| SEPT2 |
| NEU2 |
| NEUROD1 |
| NPAS2 |
| NPHP1 |
| NPPC |
| NR4A2 |
| ODC1 |
| ORC2 |
| ORC4 |
| OTX1 |
| REG3A |
| PCBP1 |
| PDCD1 |
| PDE1A |
| PDE6D |
| PDK1 |
| PEX13 |
| VIT |
| SERPINE2 |
| PIGF |
| PLCL1 |
| PLEK |
| PLGLB2 |
| PLGLB1 |
| POLR2D |
| POMC |
| POU3F3 |
| PPM1B |
| PPM1G |
| PPP1CB |
| PPP1R7 |
| PPP3R1 |
| PRKCE |
| EIF2AK2 |
| PROC |
| PSMD1 |
| PTH2R |
| PTMA |
| PTPN4 |
| PTPRN |
| RAB1A |
| RALB |
| RANBP2 |
| RBMS1 |
| REG1A |
| REG1B |
| REG1P |
| SNORD20 |
| RPE |
| RPL31 |
| RPL37A |
| RPS7 |
| RPS27A |
| RRM2 |
| RTKN |
| SAG |
| SCN1A |
| SCN2A |
| SCN3A |
| SCN7A |
| SCN9A |
| SCTR |
| CCL20 |
| SDC1 |
| SRSF7 |
| SFTPB |
| SIX3 |
| SLC4A3 |
| SLC1A4 |
| SLC3A1 |
| SLC8A1 |
| SLC9A2 |
| SLC11A1 |
| SLC20A1 |
| SNRPG |
| SOS1 |
| SOX11 |
| SP3 |
| SP100 |
| SPAST |
| SPP2 |
| SPR |
| SPTBN1 |
| SRD5A2 |
| SSB |
| SSFA2 |
| STAT1 |
| STAT4 |
| STRN |
| SULT1C2 |
| ADAM17 |
| TACR1 |
| GCFC2 |
| TFPI |
| TGFA |
| TIA1 |
| TNFAIP6 |
| TNP1 |
| TNS1 |
| TPO |
| TSN |
| TSSC1 |
| TTN |
| TUBA4A |
| SUMO1 |
| UCN |
| UGP2 |
| VIL1 |
| VRK2 |
| VSNL1 |
| WIPF1 |
| WNT6 |
| XDH |
| XPO1 |
| XRCC5 |
| ZAP70 |
| ZNF2 |
| ZNF142 |
| SLC30A3 |
| PXDN |
| ALMS1 |
| MOGS |
| RNF103 |
| IL1R2 |
| MALL |
| CXCR4 |
| FZD5 |
| SCG2 |
| DYSF |
| FZD7 |
| SDPR |
| NCK2 |
| DUSP11 |
| CUL3 |
| KLF11 |
| MAP4K3 |
| PKP4 |
| HAT1 |
| DGKD |
| AGPS |
| PRKRA |
| STK16 |
| SLC25A12 |
| KLF7 |
| ABCB11 |
| VAMP8 |
| MARCO |
| B3GALT1 |
| ADAM23 |
| SUCLG1 |
| IL18RAP |
| IL1RL2 |
| IL18R1 |
| NRP2 |
| CFLAR |
| ASAP2 |
| PER2 |
| ST3GAL5 |
| SLC5A6 |
| DDX18 |
| EIF2B4 |
| CDK5R2 |
| KYNU |
| TAF1B |
| NAT8 |
| NMI |
| RQCD1 |
| COX7A2L |
| TMSB10 |
| IL1RL1 |
| LRRFIP1 |
| STK17B |
| ITGB1BP1 |
| GPR55 |
| TRIP12 |
| GTF3C3 |
| PPIG |
| NRXN1 |
| OTOF |
| CIAO1 |
| TGFBRAP1 |
| HS6ST1 |
| CRIPT |
| ECEL1 |
| MAP4K4 |
| EIF2AK3 |
| EIF4E2 |
| ROCK2 |
| CHST10 |
| TP53I3 |
| CIR1 |
| MRPL33 |
| BRE |
| PREPL |
| CYTIP |
| FEZ2 |
| GCC2 |
| TTLL4 |
| SOCS5 |
| EIF5B |
| GREB1 |
| BZW1 |
| USP34 |
| LAPTM4A |
| HDAC4 |
| RNF144A |
| SNX17 |
| SERTAD2 |
| MRPL19 |
| ZEB2 |
| FARP2 |
| TLK1 |
| SUPT7L |
| CD302 |
| ARHGAP25 |
| TANK |
| BCL2L11 |
| FARSB |
| ABCB6 |
| ACTR3 |
| ACTR2 |
| ARPC2 |
| PREB |
| ACTR1B |
| ARL4C |
| LRPPRC |
| PDIA6 |
| ABI2 |
| CEBPZ |
| DHRS9 |
| TXNDC9 |
| MPHOSPH10 |
| CALCRL |
| PSMD14 |
| STAM2 |
| RAMP1 |
| SPEG |
| LANCL1 |
| NMUR1 |
| SMYD5 |
| KBTBD10 |
| MYCNOS |
| C1D |
| MERTK |
| UBE2E3 |
| STK25 |
| SEMA4F |
| CCT7 |
| CCT4 |
| GNLY |
| CDC42EP3 |
| STAMBP |
| TGOLN2 |
| MTX2 |
| CGREF1 |
| B3GNT2 |
| USP39 |
| TBR1 |
| SIX2 |
| MAP3K2 |
| NCKAP1 |
| VAMP5 |
| MTHFD2 |
| ARID5A |
| PROKR1 |
| RAB10 |
| EDAR |
| COPS8 |
| GPR75 |
| YWHAQ |
| IMMT |
| SNRNP27 |
| GTF2A1L |
| STON1 |
| RAPGEF4 |
| EMILIN1 |
| CAPN10 |
| TBC1D8 |
| RABL2A |
| GALNT5 |
| NXPH2 |
| GPR45 |
| SP140 |
| MGAT4A |
| GPN1 |
| IKZF2 |
| COBLL1 |
| AAK1 |
| FASTKD2 |
| MAPRE3 |
| PLA2R1 |
| RAB3GAP1 |
| RPIA |
| SLC4A1AP |
| EFR3B |
| SNRNP200 |
| MYT1L |
| WDR43 |
| LPIN1 |
| CEP68 |
| PASK |
| UBXN4 |
| ATG4B |
| PSME4 |
| EXOC6B |
| EHBP1 |
| SATB2 |
| CLASP1 |
| OBSL1 |
| PUM2 |
| NCAPH |
| RHOQ |
| SF3B1 |
| HAAO |
| TMEM131 |
| R3HDM1 |
| DNPEP |
| PSD4 |
| WBP1 |
| NTSR2 |
| TMEFF2 |
| SH3BP4 |
| PRKD3 |
| KCNE4 |
| RASGRP3 |
| NGEF |
| QPCT |
| GCA |
| VAX2 |
| SNORD82 |
| MOB4 |
| POLR1A |
| CNRIP1 |
| FAM98A |
| PNKD |
| UNC50 |
| SNED1 |
| GORASP2 |
| SPATS2L |
| RAB11FIP5 |
| GIGYF2 |
| EPC2 |
| TRAF3IP1 |
| ABCA12 |
| IFT172 |
| ARL5A |
| HIBCH |
| PTPN18 |
| CNNM4 |
| CNNM3 |
| IL36RN |
| SH3YL1 |
| SNORD53 |
| SNORD51 |
| CNPPD1 |
| STK36 |
| IL36B |
| IL37 |
| IL36A |
| SULT1C4 |
| NFU1 |
| ERLEC1 |
| MMADHC |
| BMP10 |
| ZNF638 |
| STK39 |
| HTRA2 |
| TRIB2 |
| CPS1-IT1 |
| METTL5 |
| LGALSL |
| OLA1 |
| C2orf27A |
| GRHL1 |
| TFCP2L1 |
| ICOS |
| GMPPA |
| NRBP1 |
| BAZ2B |
| SLC40A1 |
| KCNIP3 |
| EHD3 |
| SMARCAL1 |
| CD207 |
| ITSN2 |
| ANO7 |
| ARHGEF4 |
| PDE11A |
| TPRKB |
| FAHD2A |
| PRLH |
| WDPCP |
| MEMO1 |
| THAP4 |
| TRAPPC12 |
| ASB3 |
| INSIG2 |
| CRIM1 |
| ANKRD39 |
| FAM178B |
| RNF181 |
| MRPL30 |
| DNAJC27 |
| ANKMY1 |
| KRCC1 |
| MRPL35 |
| C2orf28 |
| PCYOX1 |
| GULP1 |
| REV1 |
| NAT8B |
| SCLY |
| VPS54 |
| NBAS |
| LIPT1 |
| NOP58 |
| DYNC2LI1 |
| SF3B14 |
| YPEL5 |
| CHMP3 |
| FKBP7 |
| ASB1 |
| CPSF3 |
| CAB39 |
| ZAK |
| LRP1B |
| PRKAG3 |
| PPIL3 |
| SNTG2 |
| DNAJC10 |
| ATAD2B |
| ETAA1 |
| HEATR5B |
| CCDC93 |
| ASNSD1 |
| UGT1A10 |
| UGT1A8 |
| UGT1A7 |
| UGT1A6 |
| UGT1A5 |
| UGT1A9 |
| UGT1A4 |
| UGT1A1 |
| UGT1A3 |
| AFTPH |
| MFSD6 |
| TMEM214 |
| RETSAT |
| INO80D |
| SEMA4C |
| C2orf18 |
| C2orf42 |
| TRMT61B |
| PID1 |
| PTCD3 |
| PLEKHB2 |
| ATG16L1 |
| FANCL |
| SRBD1 |
| FIGN |
| ANKZF1 |
| RIF1 |
| USP40 |
| MOB1A |
| STEAP3 |
| ASXL2 |
| ADI1 |
| THNSL2 |
| ACOXL |
| WDR33 |
| HJURP |
| STRADB |
| C2orf56 |
| HES6 |
| ACCN4 |
| C2orf29 |
| NAGK |
| DOCK10 |
| TTC27 |
| SMPD4 |
| TMEM127 |
| PRPF40A |
| SLC30A6 |
| IWS1 |
| LIMS2 |
| KANSL3 |
| MREG |
| CCDC88A |
| WDR12 |
| MBD5 |
| KDM3A |
| ALLC |
| PECR |
| ARHGAP15 |
| ZC3H15 |
| CFC1 |
| DNAH7 |
| GKN1 |
| IL36G |
| RPRM |
| CYP26B1 |
| POLE4 |
| KCNK12 |
| UGGT1 |
| KCMF1 |
| DPYSL5 |
| PNO1 |
| STARD7 |
| C2orf83 |
| MFF |
| RPL23AP32 |
| CXCR7 |
| RNPEPL1 |
| RTN4 |
| TRIM54 |
| PELI1 |
| SLC39A10 |
| TTC7A |
| SMEK2 |
| SLC4A10 |
| CYP20A1 |
| SPC25 |
| BIRC6 |
| ERMN |
| KIDINS220 |
| MTA3 |
| HECW2 |
| WDR35 |
| MARCH4 |
| WDFY1 |
| NYAP2 |
| DPP10 |
| RDH14 |
| EPB41L5 |
| ALS2 |
| ZDBF2 |
| USP37 |
| CWC22 |
| ANKRD36B |
| G6PC2 |
| SLC4A5 |
| CTDSP1 |
| NLRC4 |
| MPP4 |
| SLC5A7 |
| NIF3L1 |
| AGBL5 |
| C2orf43 |
| THADA |
| PCGEM1 |
| RBKS |
| GAL3ST2 |
| TMBIM1 |
| IFIH1 |
| OSGEPL1 |
| ATL2 |
| ABCG5 |
| ABCG8 |
| RAB17 |
| RNF25 |
| HS1BP3 |
| GMCL1 |
| TTC31 |
| ANAPC1 |
| COPS7B |
| RMND5A |
| FNDC4 |
| MARCH7 |
| OBFC2A |
| PAPOLG |
| MRPS9 |
| MRPS5 |
| REEP1 |
| RAPH1 |
| CDK15 |
| TMEM237 |
| NBEAL1 |
| CFLAR-AS1 |
| MRPL44 |
| SOWAHC |
| PLEKHA3 |
| TRAK2 |
| BOLL |
| COLEC11 |
| PDCL3 |
| TRPM8 |
| ATG9A |
| C2orf49 |
| MLPH |
| TMEM185B |
| FAM134A |
| CENPO |
| OR7E91P |
| GLB1L |
| C2orf47 |
| SPAG16 |
| CHPF |
| SAP130 |
| GALNT14 |
| SCRN3 |
| CCDC121 |
| FASTKD1 |
| SMC6 |
| GTDC1 |
| CLIP4 |
| IQCA1 |
| ALS2CR8 |
| TTC21B |
| CAMKMT |
| METTL8 |
| GEMIN6 |
| NHEJ1 |
| FAM124B |
| TM4SF20 |
| CYBRD1 |
| C2orf54 |
| NOL10 |
| ANKRD53 |
| CSRNP3 |
| PGAP1 |
| LRRTM4 |
| DCAF17 |
| TUBA4B |
| MZT2B |
| YSK4 |
| UXS1 |
| FBXO11 |
| ARMC9 |
| COQ10B |
| SLC35F5 |
| EFHD1 |
| C2orf44 |
| SPHKAP |
| WNT10A |
| SLC19A3 |
| TSGA10 |
| THSD7B |
| THUMPD2 |
| TMEM177 |
| KIAA1715 |
| ILKAP |
| FAM49A |
| LMAN2L |
| LBH |
| TMEM163 |
| ITM2C |
| TCF7L1 |
| INO80B |
| AMMECR1L |
| CDCA7 |
| WDR54 |
| ZRANB3 |
| RAB6C |
| WDR75 |
| FAM161A |
| FAM176A |
| ANTXR1 |
| POLR1B |
| ELMOD3 |
| RGPD5 |
| C2orf16 |
| RHBDD1 |
| CHCHD5 |
| YIPF4 |
| PRADC1 |
| C2orf88 |
| ING5 |
| CCDC115 |
| MKI67IP |
| C2orf40 |
| ZNF512 |
| ZC3H8 |
| KIAA1841 |
| ST6GAL2 |
| IL1F10 |
| DPY30 |
| MCEE |
| LOXL3 |
| ABHD1 |
| PCGF1 |
| DDX11L2 |
| MFSD9 |
| PLCD4 |
| SFT2D3 |
| CCDC142 |
| ZNF514 |
| FAM136A |
| TMEM87B |
| ATOH8 |
| LOC84931 |
| MGC16025 |
| TANC1 |
| EPT1 |
| LBX2 |
| PNPT1 |
| KCNH7 |
| FER1L5 |
| MCFD2 |
| ATP6V1E2 |
| LOC90499 |
| CCDC74A |
| LOC90784 |
| DHX57 |
| LOC91149 |
| SESTD1 |
| CCDC74B |
| PKDCC |
| ANKRD44 |
| RSAD2 |
| ZNF804A |
| TTC30A |
| DAPL1 |
| CAPN13 |
| TMEM169 |
| DNER |
| CCDC164 |
| IMP4 |
| HNRPLL |
| MARS2 |
| B3GNT7 |
| NT5C1B |
| NEURL3 |
| SP140L |
| SFXN5 |
| ORMDL1 |
| LIMS3 |
| LINC00152 |
| TUBA3E |
| CCDC104 |
| TEX261 |
| TUBA3D |
| STK11IP |
| FMNL2 |
| CCDC85A |
| GALNT13 |
| KLHL29 |
| OSBPL6 |
| NOSTRIN |
| DIRC1 |
| WDR92 |
| MOGAT1 |
| LYPD1 |
| MRPL53 |
| AGAP1 |
| TWIST2 |
| PARD3B |
| RPL23AP7 |
| PPP1R21 |
| C2orf89 |
| TMEM150A |
| NUP35 |
| XIRP2 |
| TYW5 |
| NMS |
| LYG1 |
| MITD1 |
| DIS3L2 |
| CMPK2 |
| MBOAT2 |
| CNTNAP5 |
| TMEM18 |
| FBLN7 |
| NEU4 |
| RBM45 |
| C2orf73 |
| TRIM43 |
| BBS5 |
| C2orf77 |
| ACMSD |
| ICA1L |
| FAM168B |
| CIB4 |
| REG3G |
| RFTN2 |
| C2orf63 |
| PLEKHH2 |
| AP1S3 |
| C2orf76 |
| SGPP2 |
| ACVR1C |
| OSR1 |
| TTC32 |
| UBR3 |
| KCTD18 |
| ALS2CR12 |
| ZNF513 |
| SPATA3 |
| LYPD6 |
| LYPD6B |
| GALM |
| TMEM198 |
| ZFAND2B |
| TMEM178 |
| CPO |
| MDH1B |
| C2orf50 |
| PQLC3 |
| TMEM182 |
| AHSA2 |
| FBXO36 |
| MTERFD2 |
| CCDC148 |
| C2orf65 |
| MYO3B |
| TMEM37 |
| UBE2F |
| DUSP19 |
| CKAP2L |
| CBWD2 |
| TEKT4 |
| LOC150527 |
| FLJ32063 |
| LOC150568 |
| SMYD1 |
| C2orf15 |
| LOC150622 |
| OTOS |
| MYEOV2 |
| OR6B3 |
| COMMD1 |
| PROM2 |
| ANKAR |
| FBXO41 |
| TTC30B |
| GPAT2 |
| ITPRIPL1 |
| LOC150776 |
| WTH3DI |
| FAM117B |
| TCF23 |
| LOC150935 |
| FAM59B |
| PUS10 |
| PKI55 |
| LINC00309 |
| LOC151009 |
| SEPT10 |
| C2orf67 |
| PLB1 |
| ZSWIM2 |
| ZNF385B |
| LOC151171 |
| LOC151174 |
| ARL6IP6 |
| METTL21A |
| CCNYL1 |
| KLHL23 |
| SULT1C2P1 |
| PPP1R1C |
| SGOL2 |
| ALS2CR11 |
| SLC38A11 |
| CCDC140 |
| SLC23A3 |
| LOC151300 |
| GPBAR1 |
| FAHD2B |
| MYADML |
| FAM84A |
| FAM82A1 |
| GDF7 |
| SLC16A14 |
| LOC151475 |
| LINC00471 |
| LOC151484 |
| MSL3P1 |
| ASPRV1 |
| WDSUB1 |
| UPP2 |
| LOC151534 |
| GPR155 |
| WDR69 |
| LONRF2 |
| CCDC138 |
| GPR113 |
| C2orf57 |
| OXER1 |
| FAM179A |
| FAM171B |
| C1QL2 |
| UBXN2A |
| CLEC4F |
| DQX1 |
| KCNG3 |
| CYS1 |
| FOXD4L1 |
| PCDP1 |
| VWA3B |
| CREG2 |
| ALMS1P |
| TET3 |
| GKN2 |
| C2orf51 |
| ANKRD23 |
| APLF |
| PIKFYVE |
| KRTCAP3 |
| LOC200726 |
| TMEM17 |
| SPRED2 |
| TIGD1 |
| LOC200772 |
| FAM123C |
| LINC00116 |
| C2orf69 |
| HNRNPA3 |
| SPDYA |
| ATP6V1C2 |
| RNASEH1 |
| LCLAT1 |
| CCDC75 |
| CERS6 |
| METAP1D |
| LOC254128 |
| LYG2 |
| CCDC108 |
| C2orf72 |
| SH2D6 |
| LOC284950 |
| CCDC150 |
| RNF149 |
| LOC284998 |
| LOC285000 |
| FAM150B |
| CCDC141 |
| LOC285033 |
| LINC00486 |
| C2orf61 |
| LOC285074 |
| LOC285084 |
| CXXC11 |
| LOC285103 |
| DNAJC5G |
| IAH1 |
| FLJ33534 |
| CYP1B1-AS1 |
| FAM126B |
| UNC80 |
| RUFY4 |
| PLGLA |
| RGPD4 |
| DUSP28 |
| STON1-GTF2A1L |
| SPOPL |
| MLK7-AS1 |
| CYP27C1 |
| ESPNL |
| C2orf70 |
| C2orf53 |
| LOC339788 |
| LINC00299 |
| LOC339803 |
| C2orf74 |
| LOC339807 |
| LOC339822 |
| MSGN1 |
| C2orf55 |
| FIGLA |
| NOTO |
| NCKAP5 |
| FOXI3 |
| EVX2 |
| CDKL4 |
| PRORSD1P |
| AOX2P |
| SH3RF3 |
| GPR148 |
| ECEL1P2 |
| LRRTM1 |
| GEN1 |
| C2orf48 |
| FONG |
| LOC348761 |
| PFN4 |
| LOC375190 |
| LOC375196 |
| ANKRD36 |
| WASH2P |
| RBM43 |
| LOC375295 |
| CERKL |
| C2orf62 |
| RBM44 |
| AQP12A |
| ZC3H6 |
| KLHL30 |
| LOC386597 |
| MFSD2B |
| C2orf71 |
| LOC388942 |
| LOC388946 |
| LOC388948 |
| TSPYL6 |
| C2orf78 |
| BOLA3 |
| C2orf81 |
| FUNDC2P2 |
| C2orf68 |
| SLC9A4 |
| LOC389023 |
| LOC389033 |
| LOC389043 |
| SP5 |
| PLEKHM3 |
| C2orf80 |
| RESP18 |
| C2orf82 |
| OR6B2 |
| PTRHD1 |
| SULT6B1 |
| DYTN |
| LOC400940 |
| LINC00487 |
| FLJ12334 |
| LOC400950 |
| UNQ6975 |
| EML6 |
| FLJ30838 |
| FLJ16341 |
| LOC400958 |
| PCBP1-AS1 |
| PAIP2B |
| RGPD1 |
| FLJ42351 |
| LOC401010 |
| DKFZp686O1327 |
| LOC401022 |
| FSIP2 |
| C2orf66 |
| ASB18 |
| FLJ43879 |
| VWC2L |
| MIR10B |
| MIR128-1 |
| MIR149 |
| MIR153-1 |
| MIR216A |
| MIR217 |
| MIR26B |
| C2orf27B |
| DNAJB3 |
| ASTL |
| CAPN14 |
| ACTR3BP2 |
| LOC440894 |
| LOC440895 |
| LOC440900 |
| LOC440905 |
| LOC440910 |
| POTEKP |
| LOC440925 |
| LOC442028 |
| SULT1C3 |
| POTEE |
| COA5 |
| PHOSPHO2 |
| CHAC2 |
| MIR375 |
| DFNB59 |
| LOC541471 |
| LOC554201 |
| ANKRD30BL |
| FBXO48 |
| SNORA41 |
| FAM110C |
| LOC643387 |
| PRR21 |
| LOC644838 |
| GGT8P |
| ANKRD36BP2 |
| LOC645949 |
| LOC646324 |
| LOC646736 |
| LOC646743 |
| PRSS56 |
| LOC647012 |
| C2orf84 |
| TRIM43B |
| CFC1B |
| AQP12B |
| RGPD3 |
| MZT2A |
| SNORA75 |
| LOC654342 |
| FAM138B |
| LOC654433 |
| SCARNA6 |
| SCARNA5 |
| SNORD11 |
| SNORD70 |
| SNORD89 |
| SNORD92 |
| SNORD94 |
| MIR558 |
| MIR559 |
| MIR561 |
| RGPD8 |
| LOC727982 |
| D2HGDH |
| LOC728323 |
| POTEF |
| LOC728537 |
| LOC728730 |
| PABPC1P2 |
| LOC728819 |
| LOC729121 |
| ANKRD20A8P |
| LOC729234 |
| RAD51AP2 |
| RGPD6 |
| DIRC3 |
| DNAJC27-AS1 |
| RGPD2 |
| MORN2 |
| LOC730811 |
| RFX8 |
| SNORD11B |
| SNORA36C |
| SNORA70B |
| MIR216B |
| MIR933 |
| LOC100128590 |
| OST4 |
| LOC100129175 |
| LOC100129726 |
| LOC100129961 |
| PP14571 |
| LOC100130451 |
| LOC100130452 |
| LOC100130691 |
| TMEM194B |
| LOC100131320 |
| SP9 |
| LOC100132215 |
| CYP4F30P |
| LOC100133985 |
| LOC100134259 |
| LOC100144595 |
| RNU4ATAC |
| DBIL5P2 |
| SNAR-H |
| LOC100189589 |
| LOC100216479 |
| ARHGEF33 |
| LOC100271832 |
| LIMS3-LOC440895 |
| LOC100286922 |
| LOC100286979 |
| LOC100287010 |
| LOC100287216 |
| LOC100288570 |
| LIMS3L |
| LOC100288911 |
| MIR1471 |
| MIR1246 |
| MIR548N |
| MIR1258 |
| MIR1245A |
| MIR1301 |
| MIR1244-1 |
| LOC100302650 |
| GPR75-ASB3 |
| SNORA80B |
| ZEB2-AS1 |
| MIR548F2 |
| MIR663B |
| LOC100329109 |
| SNORA70F |
| BOK-AS1 |
| MIR3128 |
| MIR4265 |
| MIR1244-3 |
| MIR1244-2 |
| MIR4264 |
| MIR3129 |
| MIR3127 |
| MIR4261 |
| MIR3131 |
| MIR4268 |
| MIR4263 |
| MIR3125 |
| MIR3130-1 |
| MIR4267 |
| MIR4262 |
| MIR3130-2 |
| MIR4266 |
| MIR3126 |
| MIR2355 |
| MIR3132 |
| MIR4269 |
| LOC100499194 |
| MIR3606 |
| MIR3682 |
| MIR3679 |
| LOC100505624 |
| LOC100505695 |
| LOC100505716 |
| LOC100505876 |
| LOC100505964 |
| LOC100506054 |
| LOC100506123 |
| LOC100506124 |
| LOC100506134 |
| LOC100506274 |
| LOC100506421 |
| LOC100506474 |
| LOC100506783 |
| LOC100506866 |
| LOC100507140 |
| BOLA3-AS1 |
| LOC100507334 |
| LOC100507443 |
| LOC100507600 |
| LY75-CD302 |
| RNF103-CHMP3 |
| NT5C1B-RDH14 |
| PHOSPHO2-KLHL23 |
| HSPE1-MOB4 |
| INO80B-WBP1 |
| UBE2F-SCLY |
| MIR4436B1 |
| MIR4772 |
| MIR4779 |
| MIR4783 |
| MIR4439 |
| MIR4782 |
| MIR4437 |
| MIR4765 |
| MIR4776-1 |
| MIR4757 |
| MIR4777 |
| MIR1245B |
| MIR4435-2 |
| MIR4426 |
| MIR4774 |
| MIR2467 |
| MIR4775 |
| MIR4785 |
| MIR4784 |
| MIR4773-1 |
| MIR4444-1 |
| MIR4440 |
| MIR4436A |
| MIR4786 |
| MIR4773-2 |
| MIR4434 |
| MIR4780 |
| MIR4778 |
| MIR4429 |
| MIR4776-2 |
| MIR4432 |
| MIR548AD |
| MIR4441 |
| MIR4435-1 |
| LOC100630918 |
| LOC100861402 |
This is the comprehensive list of deleted genes in the wide peak for 3p14.3.
Table S26. Genes in bold are cancer genes as defined by The Sanger Institute's Cancer Gene Census [7].
| Genes |
|---|
| BCL6 |
| FOXL2 |
| CBLB |
| CTNNB1 |
| EIF4A2 |
| ETV5 |
| FANCD2 |
| FHIT |
| GATA2 |
| LPP |
| MITF |
| MLF1 |
| MLH1 |
| MYD88 |
| PIK3CA |
| PPARG |
| RAF1 |
| RPN1 |
| SOX2 |
| TFRC |
| VHL |
| XPC |
| BAP1 |
| GMPS |
| SRGAP3 |
| TFG |
| FOXP1 |
| SETD2 |
| PBRM1 |
| hsa-mir-922 |
| hsa-mir-570 |
| hsa-mir-3137 |
| hsa-mir-944 |
| hsa-mir-28 |
| hsa-mir-1248 |
| hsa-mir-1224 |
| hsa-mir-569 |
| hsa-mir-551b |
| hsa-mir-720 |
| hsa-mir-1263 |
| hsa-mir-16-2 |
| hsa-mir-1280 |
| hsa-mir-548i-1 |
| hsa-mir-544b |
| hsa-mir-198 |
| hsa-mir-568 |
| hsa-mir-567 |
| hsa-mir-4273 |
| hsa-mir-1324 |
| hsa-mir-1284 |
| hsa-mir-3136 |
| hsa-mir-4272 |
| hsa-mir-135a-1 |
| hsa-mir-566 |
| hsa-mir-4271 |
| hsa-mir-191 |
| hsa-mir-711 |
| hsa-mir-2115 |
| hsa-mir-1226 |
| hsa-mir-564 |
| hsa-mir-138-1 |
| hsa-mir-26a-1 |
| hsa-mir-128-2 |
| hsa-mir-466 |
| hsa-mir-3135 |
| hsa-mir-563 |
| hsa-mir-3134 |
| hsa-mir-4270 |
| hsa-mir-885 |
| AADAC |
| ACAA1 |
| ACPP |
| ACTL6A |
| ACVR2B |
| ACY1 |
| ADCY5 |
| ADPRH |
| AGTR1 |
| AHSG |
| ALAS1 |
| ALCAM |
| AMT |
| APEH |
| APOD |
| ARF4 |
| RHOA |
| ATP1B3 |
| ATP2B2 |
| ATP6V1A |
| ATR |
| BCHE |
| BDH1 |
| BTD |
| C3orf51 |
| CACNA1D |
| SLC25A20 |
| CAMP |
| CASR |
| CAV3 |
| CCK |
| CD80 |
| CD86 |
| ENTPD3 |
| CD47 |
| CDC25A |
| CISH |
| AP2M1 |
| CLCN2 |
| CCR1 |
| CCR3 |
| CCR4 |
| CCR5 |
| CCR8 |
| CCBP2 |
| COL7A1 |
| COL8A1 |
| CP |
| CPA3 |
| CPB1 |
| CPN2 |
| CPOX |
| CRYGS |
| CSTA |
| CX3CR1 |
| CYP8B1 |
| DAG1 |
| DGKG |
| DAZL |
| DLG1 |
| DNASE1L3 |
| DOCK3 |
| DRD3 |
| DUSP7 |
| DVL3 |
| ECT2 |
| CELSR3 |
| EHHADH |
| EIF4G1 |
| EPHA3 |
| EPHB1 |
| EPHB3 |
| MECOM |
| FBLN2 |
| FGF12 |
| FLNB |
| GAP43 |
| GBE1 |
| GHSR |
| GLB1 |
| GNAI2 |
| GNAT1 |
| GOLGA4 |
| GOLGB1 |
| GP5 |
| GP9 |
| XCR1 |
| GPR15 |
| GPR27 |
| GPX1 |
| GRM2 |
| GRM7 |
| GSK3B |
| GTF2E1 |
| GYG1 |
| HCLS1 |
| HGD |
| HRH1 |
| HRG |
| HES1 |
| HTR1F |
| HYAL1 |
| IL1RAP |
| IL5RA |
| IL12A |
| IMPDH2 |
| IRAK2 |
| ITGA9 |
| ITGB5 |
| ITIH1 |
| ITIH3 |
| ITIH4 |
| ITPR1 |
| KNG1 |
| KPNA1 |
| KPNA4 |
| LAMB2 |
| RPSA |
| LSAMP |
| LTF |
| TM4SF1 |
| MAP4 |
| MBNL1 |
| MCM2 |
| MFI2 |
| MME |
| MOBP |
| CD200 |
| MST1 |
| MST1R |
| MUC4 |
| MYL3 |
| MYLK |
| NCK1 |
| NDUFB4 |
| NDUFB5 |
| NKTR |
| OGG1 |
| OPA1 |
| CLDN11 |
| OXTR |
| P2RY1 |
| PAK2 |
| CNTN3 |
| PCCB |
| PCYT1A |
| PDHB |
| PFKFB4 |
| PFN2 |
| SERPINI1 |
| SERPINI2 |
| PIK3CB |
| PLCD1 |
| PLD1 |
| PLOD2 |
| PLS1 |
| PLSCR1 |
| PLXNA1 |
| PLXNB1 |
| POLR2H |
| POU1F1 |
| PPP1R2 |
| PPP2R3A |
| PRKAR2A |
| PRKCD |
| PRKCI |
| PROS1 |
| MASP1 |
| PSMD2 |
| PTH1R |
| PTPRG |
| PTX3 |
| QARS |
| RAB5A |
| RAP2B |
| RARB |
| RARRES1 |
| RASA2 |
| RBP1 |
| RBP2 |
| RFC4 |
| RHO |
| SNORA63 |
| SNORA62 |
| ROBO1 |
| ROBO2 |
| RPL15 |
| RPL24 |
| RPL29 |
| RPL32 |
| RPL35A |
| RYK |
| SATB1 |
| ATXN7 |
| SCN5A |
| SCN10A |
| SEC13 |
| SEMA3F |
| SETMAR |
| TRA2B |
| SHOX2 |
| SI |
| SIAH2 |
| ST6GAL1 |
| SKIL |
| SLC2A2 |
| SLC6A1 |
| SLC6A6 |
| SLC6A11 |
| SLC15A2 |
| SLCO2A1 |
| HLTF |
| SMARCC1 |
| SSR3 |
| SST |
| STAC |
| NEK4 |
| SYN2 |
| TCTA |
| TDGF1 |
| TERC |
| TF |
| TFDP2 |
| TGM4 |
| TGFBR2 |
| THPO |
| THRB |
| TIMP4 |
| TKT |
| SEC62 |
| TM4SF4 |
| TMF1 |
| CLEC3B |
| TNNC1 |
| TOP2B |
| NR2C2 |
| TRH |
| TRPC1 |
| UBA7 |
| UBE2E1 |
| UBE2E2 |
| UBP1 |
| UPK1B |
| UMPS |
| USP4 |
| UQCRC1 |
| CLRN1 |
| VIPR1 |
| WNT5A |
| WNT7A |
| ZIC1 |
| CNBP |
| ZNF35 |
| ZNF80 |
| ZNF148 |
| BRPF1 |
| IFRD2 |
| MAPKAPK3 |
| SEMA3B |
| SLMAP |
| MANF |
| RAB7A |
| KCNAB1 |
| FXR1 |
| COLQ |
| ACOX2 |
| EOMES |
| HYAL3 |
| SOX14 |
| BFSP2 |
| CAMK1 |
| CGGBP1 |
| BHLHE40 |
| RUVBL1 |
| CADPS |
| TP63 |
| CHRD |
| HYAL2 |
| B4GALT4 |
| B3GALNT1 |
| SNX4 |
| TNFSF10 |
| SUCLG2 |
| HESX1 |
| KAT2B |
| NR1I2 |
| EIF2B5 |
| BSN |
| MBD4 |
| H1FX |
| USP13 |
| LIMD1 |
| KALRN |
| CCRL2 |
| UBA3 |
| RPL14 |
| CLDN1 |
| SEC22C |
| RRP9 |
| MAP3K13 |
| SLC33A1 |
| LRRFIP2 |
| MAGI1 |
| CACNA2D2 |
| COPB2 |
| ADIPOQ |
| SLC22A14 |
| SLC22A13 |
| CHST2 |
| SH3BP5 |
| SLC4A7 |
| STXBP5L |
| RNF7 |
| GUCA1C |
| IQCB1 |
| DZIP3 |
| VGLL4 |
| EDEM1 |
| KIAA0226 |
| ECE2 |
| VPRBP |
| TBC1D5 |
| TATDN2 |
| IP6K1 |
| TSC22D2 |
| EPM2AIP1 |
| PSMD6 |
| TOMM70A |
| TRANK1 |
| IQSEC1 |
| P2RY14 |
| DLEC1 |
| EXOG |
| XYLB |
| OXSR1 |
| NR1D2 |
| PDCD6IP |
| PARP3 |
| SMC4 |
| ABCC5 |
| COX17 |
| ARPC4 |
| ZNF197 |
| RBM6 |
| RBM5 |
| TNK2 |
| ALG3 |
| NME6 |
| CTDSPL |
| CD96 |
| KCNMB2 |
| STAG1 |
| EIF1B |
| TRAIP |
| ST3GAL6 |
| ARIH2 |
| TADA3 |
| CRTAP |
| ATG7 |
| ARL6IP5 |
| NPRL2 |
| IGF2BP2 |
| CXCR6 |
| CSPG5 |
| CLDN16 |
| POLQ |
| CHL1 |
| ARPP21 |
| CCR9 |
| ALDH1L1 |
| USP19 |
| PDIA5 |
| SLC38A3 |
| CYB561D2 |
| TMEM115 |
| TOPBP1 |
| RPP14 |
| HHLA2 |
| FSTL1 |
| FAM107A |
| WDR6 |
| RASSF1 |
| NISCH |
| MRPL3 |
| PDCD10 |
| FILIP1L |
| TREX1 |
| SCN11A |
| TUSC2 |
| RNF13 |
| MGLL |
| TWF2 |
| MRAS |
| COPG |
| SLITRK3 |
| NLGN1 |
| TRAK1 |
| DHX30 |
| SACM1L |
| NCBP2 |
| SCAP |
| LAMB2P1 |
| MYH15 |
| PLCH1 |
| EXOSC7 |
| TMCC1 |
| PDZRN3 |
| TNIK |
| CAND2 |
| MCF2L2 |
| CLASP2 |
| PLXND1 |
| RAD54L2 |
| FRMD4B |
| STAB1 |
| GPD1L |
| RFTN1 |
| ATP11B |
| NBEAL2 |
| NUP210 |
| PLCL2 |
| ANKRD28 |
| FAM208A |
| KLHL18 |
| DNAJC13 |
| U2SURP |
| VPS8 |
| LARS2 |
| RYBP |
| C3orf27 |
| CAPN7 |
| ACAP2 |
| MKRN2 |
| NAT6 |
| FBXL2 |
| ANAPC13 |
| ARMC8 |
| ABHD14A |
| C3orf17 |
| POC1A |
| ABI3BP |
| CNOT10 |
| TMEM158 |
| NDUFAF3 |
| THUMPD3 |
| MYRIP |
| PTPN23 |
| WWTR1 |
| PVRL3 |
| TIPARP |
| CHMP2B |
| DNAH1 |
| HIGD1A |
| LRIG1 |
| SUSD5 |
| UBXN7 |
| ERC2 |
| APPL1 |
| HACL1 |
| ARHGEF26 |
| ZBTB20 |
| TTLL3 |
| OR5K1 |
| OR5H1 |
| GNL3 |
| FAM162A |
| PCOLCE2 |
| SEC22A |
| GPR160 |
| FETUB |
| NPHP3 |
| ATP2C1 |
| LAMP3 |
| KCNMB3 |
| ZBTB11 |
| MORC1 |
| SERP1 |
| CNTN6 |
| LSM3 |
| RBMS3 |
| GOLIM4 |
| NKIRAS1 |
| SPCS1 |
| ACAD9 |
| ASTE1 |
| KLF15 |
| GTPBP8 |
| TAGLN3 |
| PRSS50 |
| RBM15B |
| GPR171 |
| GMPPB |
| SEC61A1 |
| LINC00312 |
| SCHIP1 |
| LMCD1 |
| PIK3R4 |
| PODXL2 |
| ARHGEF3 |
| TRAT1 |
| VILL |
| IMPG2 |
| C3orf32 |
| NMD3 |
| TRNT1 |
| ABHD5 |
| COMMD2 |
| DYNC1LI1 |
| A4GNT |
| C3orf18 |
| DBR1 |
| CRBN |
| SS18L2 |
| ZNF639 |
| CLDN18 |
| C3orf19 |
| SHISA5 |
| TIMMDC1 |
| ZDHHC3 |
| RSRC1 |
| ZMYND10 |
| PLA1A |
| TEX264 |
| CCDC72 |
| ZNF589 |
| HEMK1 |
| AMOTL2 |
| IP6K2 |
| SFMBT1 |
| NCKIPSD |
| PHF7 |
| CCRL1 |
| PEX5L |
| RAB6B |
| SELT |
| FBXO40 |
| DNAJB11 |
| GHRL |
| P2RY13 |
| IL20RB |
| GPR87 |
| TLR9 |
| DCUN1D1 |
| SEMA5B |
| XRN1 |
| WDR5B |
| LZTFL1 |
| PARP14 |
| P4HTM |
| SLC6A20 |
| IL17RD |
| GRAMD1C |
| ROPN1 |
| KLHL24 |
| SIDT1 |
| C3orf75 |
| SNRK |
| QRICH1 |
| PXK |
| CMTM6 |
| RG9MTD1 |
| SLC41A3 |
| PIGX |
| SLC25A38 |
| ULK4 |
| OXSM |
| SLC35A5 |
| TMEM45A |
| FEZF2 |
| IFT57 |
| EBLN2 |
| ANO10 |
| DALRD3 |
| SHQ1 |
| MSL2 |
| TBCCD1 |
| FAIM |
| SLC25A36 |
| ARL8B |
| SETD5 |
| DPPA4 |
| LEPREL1 |
| TMEM39A |
| ZNF654 |
| TMEM40 |
| ABCF3 |
| LSG1 |
| ABHD10 |
| CHDH |
| PARL |
| IL17RB |
| CDV3 |
| MFN1 |
| YEATS2 |
| IFT122 |
| NGLY1 |
| TBC1D23 |
| WDR52 |
| CACNA2D3 |
| DCP1A |
| GLT8D1 |
| TMEM111 |
| EAF2 |
| BRK1 |
| ZNF167 |
| MYNN |
| LMOD3 |
| EIF5A2 |
| CLDND1 |
| MUC13 |
| SUCNR1 |
| RAD18 |
| SEMA3G |
| MCCC1 |
| LXN |
| C3orf37 |
| MRPS22 |
| NIT2 |
| POGLUT1 |
| BBX |
| KIF15 |
| ADAMTS9 |
| CCNL1 |
| PLSCR2 |
| PCBP4 |
| PLSCR4 |
| PCNP |
| HRASLS |
| MRPL47 |
| SENP7 |
| ABHD6 |
| LRTM1 |
| C3orf14 |
| KIAA1143 |
| ISY1 |
| HHATL |
| HEG1 |
| KIAA1257 |
| ARHGAP31 |
| NCEH1 |
| IFT80 |
| KIAA1407 |
| WDR48 |
| LRRN1 |
| KIAA1524 |
| SLC7A14 |
| SRPRB |
| SELK |
| SENP2 |
| GNB4 |
| BPESC1 |
| PROK2 |
| EEFSEC |
| RNF123 |
| NSUN3 |
| CIDEC |
| CLSTN2 |
| POPDC2 |
| RTP4 |
| MAGEF1 |
| ZFYVE20 |
| KIF9 |
| NFKBIZ |
| AZI2 |
| ZMAT3 |
| MTMR14 |
| ATG3 |
| MRPS25 |
| CSRNP1 |
| GORASP1 |
| MFSD1 |
| CCDC14 |
| FNDC3B |
| P2RY12 |
| CDCP1 |
| CCDC71 |
| NT5DC2 |
| SLC26A6 |
| TMEM108 |
| CRELD1 |
| CAMKV |
| TMEM43 |
| OR5H6 |
| OR5H2 |
| ZXDC |
| ZBED2 |
| LRRC2 |
| FYCO1 |
| ATP13A3 |
| CEP97 |
| HSPBAP1 |
| C3orf52 |
| VEPH1 |
| QTRTD1 |
| CCDC51 |
| TBL1XR1 |
| ZBBX |
| ZNF385D |
| LRRC31 |
| CCDC48 |
| NEK11 |
| UBA5 |
| HDAC11 |
| MAP6D1 |
| PHC3 |
| C3orf36 |
| ARL14 |
| THOC7 |
| NAA50 |
| PIGZ |
| CEP63 |
| CEP70 |
| ABTB1 |
| WDR82 |
| TMEM22 |
| TSEN2 |
| GRIP2 |
| OR5AC2 |
| RTP3 |
| PARP9 |
| ESYT3 |
| SPATA16 |
| EIF2A |
| B3GNT5 |
| C3orf20 |
| ID2B |
| ARL6 |
| ZIC4 |
| ATRIP |
| ACAD11 |
| IQCG |
| ATP13A4 |
| FYTTD1 |
| NICN1 |
| CHCHD6 |
| MON1A |
| C3orf26 |
| HPS3 |
| ARPM1 |
| JAGN1 |
| KBTBD8 |
| SLC12A8 |
| GHRLOS2 |
| RETNLB |
| CCDC54 |
| MGC2889 |
| IL17RC |
| ABHD14B |
| LRCH3 |
| MINA |
| GPR128 |
| C3orf39 |
| DIRC2 |
| CEP19 |
| EAF1 |
| DCLK3 |
| GFM1 |
| LMLN |
| KLHL6 |
| C3orf15 |
| PHLDB2 |
| VWA5B2 |
| UCN2 |
| LOC90246 |
| C3orf25 |
| TMEM41A |
| ZNF502 |
| BOC |
| FAM55C |
| RFT1 |
| OXNAD1 |
| SPSB4 |
| ACPL2 |
| ZBTB47 |
| TMEM44 |
| EGFEM1P |
| ACTR8 |
| CAMK2N2 |
| CMTM7 |
| TXNRD3 |
| OSBPL10 |
| OSBPL11 |
| SLC25A26 |
| ZNF501 |
| C3orf24 |
| LRRC58 |
| LRRC3B |
| TM4SF19 |
| TM4SF18 |
| RPL39L |
| MED12L |
| CLRN1-AS1 |
| GALNTL2 |
| GPR62 |
| CPNE4 |
| CCDC58 |
| KCNH8 |
| DNAJC19 |
| OTOL1 |
| FAM3D |
| ZPLD1 |
| LYZL4 |
| KBTBD5 |
| TRIM71 |
| FAM131A |
| CD200R1 |
| CHCHD4 |
| ZDHHC19 |
| DCBLD2 |
| LRRC15 |
| FAM43A |
| TPRA1 |
| TMEM42 |
| UROC1 |
| FAM194A |
| NUDT16 |
| COL6A6 |
| GRK7 |
| FAM172BP |
| TMEM207 |
| METTL6 |
| TAMM41 |
| IL17RE |
| RTP1 |
| IQCF1 |
| GLYCTK |
| PPM1M |
| C3orf49 |
| SNTN |
| SYNPR |
| C3orf45 |
| RPL32P3 |
| H1FOO |
| PISRT1 |
| ASB14 |
| TTC14 |
| DTX3L |
| FAM19A4 |
| SGOL1 |
| PP2D1 |
| EFHB |
| LOC151658 |
| PPM1L |
| WDR49 |
| LRRC34 |
| CPNE9 |
| DPPA2 |
| CCDC80 |
| BTLA |
| CCDC12 |
| MB21D2 |
| PPP4R2 |
| XXYLT1 |
| ROPN1B |
| LOC152024 |
| C3orf22 |
| C3orf55 |
| ZCWPW2 |
| CMC1 |
| NEK10 |
| C3orf79 |
| CCDC50 |
| PYDC2 |
| SPICE1 |
| CMTM8 |
| NUDT16P1 |
| CCDC13 |
| LOC152217 |
| LOC152225 |
| FGD5 |
| CIDECP |
| CNTN4 |
| IGSF11 |
| C3orf30 |
| PARP15 |
| SPTSSB |
| DNAJB8 |
| GPR156 |
| XIRP1 |
| RNF168 |
| CHST13 |
| PRICKLE2 |
| KBTBD12 |
| DHX36 |
| HTR3C |
| DZIP1L |
| TTC21A |
| ALG1L |
| C3orf67 |
| KCTD6 |
| LIPH |
| ARL13B |
| DHFRL1 |
| HTR3D |
| RPL22L1 |
| OSTalpha |
| FBXO45 |
| KLHDC8B |
| MUC20 |
| GABRR3 |
| PTPLB |
| STT3B |
| LOC201617 |
| DNAH12 |
| PDE12 |
| FAM116A |
| TIGIT |
| LOC201651 |
| C3orf58 |
| SENP5 |
| KIAA2018 |
| LOC220729 |
| ZBTB38 |
| CADM2 |
| LOC253573 |
| ZNF620 |
| NAALADL2 |
| ZDHHC23 |
| LOC255025 |
| NUP210P1 |
| TCTEX1D2 |
| C3orf43 |
| SDHAP1 |
| COL6A5 |
| GK5 |
| PLCXD2 |
| GCET2 |
| UTS2D |
| ALS2CL |
| TMIE |
| LSAMP-AS3 |
| SLC9A9 |
| C3orf64 |
| LOC285205 |
| EPHA6 |
| DNAJB8-AS1 |
| FBXW12 |
| C3orf38 |
| HTR3E |
| ENTPD3-AS1 |
| ZNF619 |
| ZNF621 |
| RABL3 |
| IGSF10 |
| C3orf33 |
| LOC285326 |
| CCDC66 |
| SLC9A10 |
| C3orf23 |
| ZNF660 |
| FLJ39534 |
| LOC285359 |
| SUMF1 |
| RPUSD3 |
| PRRT3 |
| LOC285370 |
| LOC285375 |
| DPH3 |
| C3orf70 |
| TPRG1 |
| LOC285401 |
| ILDR1 |
| TRIM59 |
| TRIM42 |
| EIF4E3 |
| RAB43 |
| CCDC39 |
| CCDC36 |
| KY |
| LOC339862 |
| LOC339874 |
| C3orf35 |
| LOC339894 |
| GADL1 |
| PRSS42 |
| LOC339926 |
| LPP-AS2 |
| H1FX-AS1 |
| LOC344595 |
| LRRIQ4 |
| SAMD7 |
| AADACL2 |
| GPR149 |
| ZNF860 |
| TMPRSS7 |
| CD200R1L |
| PAQR9 |
| COL6A4P1 |
| LOC344887 |
| RTP2 |
| OSTN |
| ATP13A5 |
| SOX2-OT |
| NME9 |
| WDR53 |
| LNP1 |
| CCDC37 |
| NPHP3-AS1 |
| TPRXL |
| ANKRD18DP |
| NMNAT3 |
| ZNF445 |
| SPATA12 |
| LHFPL4 |
| C3orf77 |
| C3orf62 |
| TMEM110 |
| LRRC33 |
| PRSS45 |
| AMIGO3 |
| ZNF662 |
| CDHR4 |
| FAM212A |
| IQCF2 |
| IQCF5 |
| MUSTN1 |
| VGLL3 |
| PRR23B |
| PRR23C |
| PLSCR5 |
| ANKUB1 |
| LEKR1 |
| TMEM212 |
| VENTXP7 |
| LOC401052 |
| IQCF3 |
| LOC401074 |
| FLJ22763 |
| FLJ25363 |
| C3orf72 |
| LOC401093 |
| C3orf80 |
| FLJ46066 |
| FLJ42393 |
| FLJ34208 |
| LOC401109 |
| OR5K2 |
| OR5H14 |
| OR5H15 |
| OR5K3 |
| OR5K4 |
| MIRLET7G |
| MIR128-2 |
| MIR135A1 |
| MIR138-1 |
| MIR15B |
| MIR16-2 |
| MIR191 |
| MIR198 |
| MIR26A1 |
| FAM19A1 |
| STX19 |
| LOC440944 |
| FLJ33065 |
| TMEM89 |
| IQCF6 |
| C3orf78 |
| LOC440970 |
| MIR425 |
| ARGFX |
| SNORA6 |
| SNORA7A |
| SNORD2 |
| SNORA4 |
| TMPPE |
| TMEM30C |
| LOC644714 |
| ALG1L2 |
| LOC644990 |
| LOC645206 |
| TXNRD3NB |
| TMEM14E |
| LOC646168 |
| COL6A4P2 |
| SPINK8 |
| C3orf71 |
| LOC646498 |
| C3orf65 |
| LOC646903 |
| PA2G4P4 |
| LOC647107 |
| GMNC |
| LOC647323 |
| FLJ20518 |
| LOC653712 |
| IQCJ |
| SCARNA7 |
| LINC00488 |
| SNORA7B |
| SNORA58 |
| SNORA81 |
| SNORD19 |
| FAM86DP |
| SNORD66 |
| SNORD69 |
| MIR548A2 |
| MIR548A3 |
| MIR551B |
| MIR563 |
| MIR564 |
| MIR567 |
| MIR568 |
| MIR569 |
| MIR570 |
| GXYLT2 |
| SDHAP2 |
| FAM157A |
| FAM198A |
| CCR2 |
| FAM86HP |
| PRR23A |
| LOC730091 |
| ESRG |
| LOC100009676 |
| SNORD19B |
| LOC100125556 |
| MIR922 |
| MIR885 |
| MIR944 |
| EGOT |
| GHRLOS |
| LOC100128023 |
| WWTR1-AS1 |
| LOC100128164 |
| C3orf74 |
| LOC100128640 |
| NRADDP |
| LOC100129480 |
| LOC100129550 |
| ZBTB20-AS1 |
| LOC100131551 |
| LOC100131635 |
| ZNF717 |
| LOC100132146 |
| LOC100132526 |
| BSN-AS2 |
| SNAR-I |
| MIR1224 |
| TIPARP-AS1 |
| PRSS46 |
| LOC100287879 |
| LOC100288428 |
| FRG2C |
| LOC100289361 |
| MIR1284 |
| MIR1248 |
| MIR1280 |
| MIR548I1 |
| MIR1324 |
| MIR1226 |
| LOC100302640 |
| MIR548H2 |
| MIR711 |
| MIR548G |
| MIR3136 |
| MIR4270 |
| MIR4272 |
| MIR4271 |
| MIR4273 |
| LOC100498859 |
| MIR3919 |
| MIR3921 |
| MIR3938 |
| MIR3714 |
| IQCJ-SCHIP1 |
| FGD5-AS1 |
| LOC100505687 |
| LOC100505696 |
| KRBOX1 |
| PVRL3-AS1 |
| IGSF11-AS1 |
| MYLK-AS1 |
| IQCF4 |
| LOC100506994 |
| LOC100507032 |
| MFI2-AS1 |
| LOC100507062 |
| LOC100507086 |
| ADAMTS9-AS2 |
| LOC100507389 |
| LOC100507391 |
| ARHGEF26-AS1 |
| LOC100507537 |
| LOC100507582 |
| ARPC4-TTLL3 |
| ABHD14A-ACY1 |
| TMEM110-MUSTN1 |
| NPHP3-ACAD11 |
| ISY1-RAB43 |
| TM4SF19-TCTEX1D2 |
| MIR4793 |
| MIR4787 |
| MIR4795 |
| MIR4796 |
| MIR4797 |
| MIR4788 |
| MIR4791 |
| MIR4790 |
| MIR548AC |
| MIR4444-1 |
| MIR4789 |
| MIR4443 |
| MIR4792 |
| MIR4446 |
| MIR4442 |
| LOC100652759 |
| LUST |
Table 3. Get Full Table Arm-level significance table - 20 significant results found.
| Arm | # Genes | Amp Frequency | Amp Z score | Amp Q value | Del Frequency | Del Z score | Del Q value |
|---|---|---|---|---|---|---|---|
| 1p | 2121 | 0.05 | -0.431 | 0.996 | 0.29 | 11.9 | 0 |
| 1q | 1955 | 0.05 | -0.399 | 0.996 | 0.17 | 5.45 | 1.36e-07 |
| 2p | 924 | 0.01 | -2.52 | 0.996 | 0.01 | -2.16 | 0.998 |
| 2q | 1556 | 0.01 | -2.58 | 0.996 | 0.01 | -2.58 | 0.998 |
| 3p | 1062 | 0.01 | -2.51 | 0.996 | 0.03 | -1.45 | 0.998 |
| 3q | 1139 | 0.01 | -2.49 | 0.996 | 0.04 | -0.727 | 0.998 |
| 4p | 489 | 0.02 | -1.94 | 0.996 | 0.11 | 2.71 | 0.0102 |
| 4q | 1049 | 0.01 | -2.31 | 0.996 | 0.14 | 4.39 | 2.47e-05 |
| 5p | 270 | 0.01 | -2.35 | 0.996 | 0.08 | 1.61 | 0.132 |
| 5q | 1427 | 0.01 | -2.49 | 0.996 | 0.06 | -0.0382 | 0.998 |
| 6p | 1173 | 0.01 | -2.16 | 0.996 | 0.02 | -1.81 | 0.998 |
| 6q | 839 | 0.01 | -2.32 | 0.996 | 0.13 | 3.71 | 0.000374 |
| 7p | 641 | 0.20 | 7.8 | 6.06e-14 | 0.01 | -2.17 | 0.998 |
| 7q | 1277 | 0.30 | 12.7 | 0 | 0.01 | -2.02 | 0.998 |
| 8p | 580 | 0.09 | 1.97 | 0.188 | 0.02 | -1.59 | 0.998 |
| 8q | 859 | 0.15 | 5.17 | 1.55e-06 | 0.01 | -2.27 | 0.998 |
| 9p | 422 | 0.03 | -1.22 | 0.996 | 0.25 | 10.2 | 0 |
| 9q | 1113 | 0.01 | -2.12 | 0.996 | 0.04 | -0.706 | 0.998 |
| 10p | 409 | 0.14 | 4.21 | 0.000125 | 0.18 | 6.34 | 7.66e-10 |
| 10q | 1268 | 0.02 | -1.84 | 0.996 | 0.19 | 6.92 | 1.72e-11 |
| 11p | 862 | 0.04 | -0.818 | 0.996 | 0.11 | 2.72 | 0.0102 |
| 11q | 1515 | 0.07 | 0.695 | 0.996 | 0.01 | -2.1 | 0.998 |
| 12p | 575 | 0.06 | 0.14 | 0.996 | 0.03 | -1.29 | 0.998 |
| 12q | 1447 | 0.01 | -2.13 | 0.996 | 0.05 | -0.383 | 0.998 |
| 13q | 654 | 0.00 | -2.69 | 0.996 | 0.13 | 4.08 | 8.77e-05 |
| 14q | 1341 | 0.01 | -2.3 | 0.996 | 0.16 | 5.41 | 1.55e-07 |
| 15q | 1355 | 0.02 | -1.71 | 0.996 | 0.07 | 0.742 | 0.496 |
| 16p | 872 | 0.01 | -2.5 | 0.996 | 0.02 | -1.79 | 0.998 |
| 16q | 702 | 0.01 | -2.47 | 0.996 | 0.03 | -1.04 | 0.998 |
| 17p | 683 | 0.02 | -1.77 | 0.996 | 0.01 | -2.49 | 0.998 |
| 17q | 1592 | 0.03 | -1.16 | 0.996 | 0.00 | -2.9 | 0.998 |
| 18p | 143 | 0.03 | -1.18 | 0.996 | 0.09 | 1.71 | 0.114 |
| 18q | 446 | 0.02 | -1.95 | 0.996 | 0.10 | 2.34 | 0.0267 |
| 19p | 995 | 0.09 | 1.47 | 0.46 | 0.22 | 8.13 | 2.16e-15 |
| 19q | 1709 | 0.07 | 0.349 | 0.996 | 0.38 | 16.5 | 0 |
| 20p | 355 | 0.07 | 0.874 | 0.996 | 0.01 | -2 | 0.998 |
| 20q | 753 | 0.07 | 0.765 | 0.996 | 0.00 | -2.79 | 0.998 |
| 21q | 509 | 0.02 | -1.7 | 0.996 | 0.04 | -0.986 | 0.998 |
| 22q | 921 | 0.01 | -2.42 | 0.996 | 0.07 | 0.762 | 0.496 |
List of inputs used for this run of GISTIC2. All files listed should be included in the archived results.
-
Segmentation File = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_MergeDataFilesPipeline/LGG/1269597/2.GDAC_MergeDataFiles.Finished/LGG.snp__genome_wide_snp_6__broad_mit_edu__Level_3__segmented_scna_minus_germline_cnv_hg19__seg.seg.txt
-
Markers File = /xchip/tcga/CancerGenomeAnalysisData/trunk/copynumber/SNP6_annotations/genome.info.6.0_hg19.na31_minus_frequent_nan_probes_sorted_2.1.txt
-
Reference Genome = /xchip/gistic/variables/hg19/hg19_with_miR_20120227.mat
-
CNV Files = /xchip/gistic/CNV/blood_normals/CNV.hg19_111204/CNV.hg19.bypos.111213.txt
-
Amplification Threshold = 0.30
-
Deletion Threshold = 0.30
-
Cap Values = 2.0
-
Broad Length Cutoff = 0.5
-
Remove X-Chromosome = 1
-
Confidence Level = 0.99
-
Join Segment Size = 10
-
Arm Level Peel Off = 1
-
Maximum Sample Segments = 10000
Table 4. Get Full Table First 10 out of 144 Input Tumor Samples.
| Tumor Sample Names |
|---|
| TCGA-CS-4938-01B-11D-1892-01 |
| TCGA-CS-4941-01A-01D-1466-01 |
| TCGA-CS-4942-01A-01D-1466-01 |
| TCGA-CS-4943-01A-01D-1466-01 |
| TCGA-CS-4944-01A-01D-1466-01 |
| TCGA-CS-5390-01A-02D-1466-01 |
| TCGA-CS-5393-01A-01D-1466-01 |
| TCGA-CS-5394-01A-01D-1466-01 |
| TCGA-CS-5395-01A-01D-1466-01 |
| TCGA-CS-5396-01A-02D-1466-01 |
Figure 3. Segmented copy number profiles in the input data

GISTIC identifies genomic regions that are significantly gained or lost across a set of tumors. It takes segmented copy number ratios as input, separates arm-level events from focal events, and then performs two tests: (i) identifies significantly amplified/deleted chromosome arms; and (ii) identifies regions that are significantly focally amplified or deleted. For the focal analysis, the significance levels (Q values) are calculated by comparing the observed gains/losses at each locus to those obtained by randomly permuting the events along the genome to reflect the null hypothesis that they are all 'passengers' and could have occurred anywhere. The locus-specific significance levels are then corrected for multiple hypothesis testing. The arm-level significance is calculated by comparing the frequency of gains/losses of each arm to the expected rate given its size. The method outputs genomic views of significantly amplified and deleted regions, as well as a table of genes with gain or loss scores. A more in depth discussion of the GISTIC algorithm and its utility is given in [1], [3], and [5].
Regions of the genome that are prone to germ line variations in copy number are excluded from the GISTIC analysis using a list of germ line copy number variations (CNVs). A CNV is a DNA sequence that may be found at different copy numbers in the germ line of two different individuals. Such germ line variations can confound a GISTIC analysis, which finds significant somatic copy number variations in cancer. A more in depth discussion is provided in [6]. GISTIC currently uses two CNV exclusion lists. One is based on the literature describing copy number variation, and a second one comes from an analysis of significant variations among the blood normals in the TCGA data set.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.