This report serves to describe the mutational landscape and properties of a given individual set, as well as rank genes and genesets according to mutational significance. MutSig vS2N was used to generate the results found in this report.
-
Working with individual set: BLCA-TP
The input for this pipeline is a set of individuals with the following files associated for each:
-
An annotated .maf file describing the mutations called for the respective individual, and their properties.
-
A .wig file that contains information about the coverage of the sample.
-
MAF used for this analysis:BLCA-TP.final_analysis_set.maf
-
Significantly mutated genes (q ≤ 0.1): 29
Column Descriptions:
-
N = number of sequenced bases in this gene across the individual set
-
nnon = number of (nonsilent) mutations in this gene across the individual set
-
nnull = number of (nonsilent) null mutations in this gene across the individual set
-
nflank = number of noncoding mutations from this gene's flanking region, across the individual set
-
nsil = number of silent mutations in this gene across the individual set
-
p = p-value (overall)
-
q = q-value, False Discovery Rate (Benjamini-Hochberg procedure)
Table 1. Get Full Table A Ranked List of Significantly Mutated Genes. Number of significant genes found: 29. Number of genes displayed: 35. Click on a gene name to display its stick figure depicting the distribution of mutations and mutation types across the chosen gene (this feature may not be available for all significant genes).
| gene | N | nflank | nsil | nnon | nnull | p | q |
|---|---|---|---|---|---|---|---|
| TP53 | 3528 | 0 | 1 | 14 | 3 | 0 | 0 |
| KDM6A | 12824 | 0 | 2 | 6 | 6 | 1.6e-131 | 1.5e-127 |
| PRX | 9548 | 0 | 0 | 9 | 1 | 5.2e-61 | 3.3e-57 |
| ARID1A | 16296 | 0 | 1 | 8 | 5 | 1e-53 | 4.9e-50 |
| GOLGB1 | 34720 | 0 | 2 | 12 | 3 | 2e-28 | 7.5e-25 |
| ASXL2 | 12600 | 0 | 0 | 6 | 2 | 2.8e-18 | 8.8e-15 |
| FBXW7 | 8036 | 0 | 0 | 6 | 2 | 8.7e-17 | 2.3e-13 |
| MLL | 36344 | 0 | 1 | 9 | 2 | 1.4e-16 | 3.3e-13 |
| FLG | 30520 | 0 | 1 | 8 | 0 | 1.4e-15 | 2.9e-12 |
| HRNR | 15036 | 0 | 0 | 6 | 0 | 1.9e-15 | 3.7e-12 |
| HCN1 | 8148 | 0 | 1 | 5 | 3 | 1.1e-14 | 1.9e-11 |
| TTN | 356944 | 0 | 3 | 34 | 4 | 2.8e-14 | 4.4e-11 |
| BCLAF1 | 9072 | 0 | 0 | 5 | 1 | 2.9e-13 | 4.2e-10 |
| MACF1 | 85736 | 0 | 0 | 9 | 1 | 1.8e-11 | 2.4e-08 |
| RPAP1 | 10640 | 0 | 0 | 5 | 0 | 2e-11 | 2.5e-08 |
| HMCN1 | 55132 | 0 | 0 | 9 | 2 | 3.6e-11 | 4.2e-08 |
| ADAMTS12 | 15260 | 0 | 1 | 4 | 0 | 9.3e-10 | 1e-06 |
| DCC | 13580 | 0 | 1 | 5 | 0 | 4.1e-09 | 4.3e-06 |
| MLL2 | 35028 | 0 | 0 | 6 | 6 | 1.7e-08 | 0.000017 |
| ABCA10 | 17556 | 0 | 0 | 5 | 2 | 3.3e-07 | 0.00031 |
| XPR1 | 7476 | 0 | 1 | 5 | 2 | 9.3e-07 | 0.00084 |
| CREBBP | 20384 | 0 | 1 | 5 | 3 | 2.6e-06 | 0.0022 |
| SETD2 | 20664 | 0 | 0 | 5 | 3 | 3.1e-06 | 0.0025 |
| SACS | 47628 | 0 | 2 | 6 | 2 | 0.000023 | 0.017 |
| DNAH5 | 49336 | 0 | 1 | 7 | 1 | 0.000023 | 0.017 |
| SPTA1 | 24696 | 0 | 0 | 5 | 0 | 0.000024 | 0.018 |
| APOB | 49140 | 0 | 0 | 6 | 0 | 0.000031 | 0.022 |
| GPS2 | 3192 | 0 | 0 | 4 | 3 | 0.000033 | 0.022 |
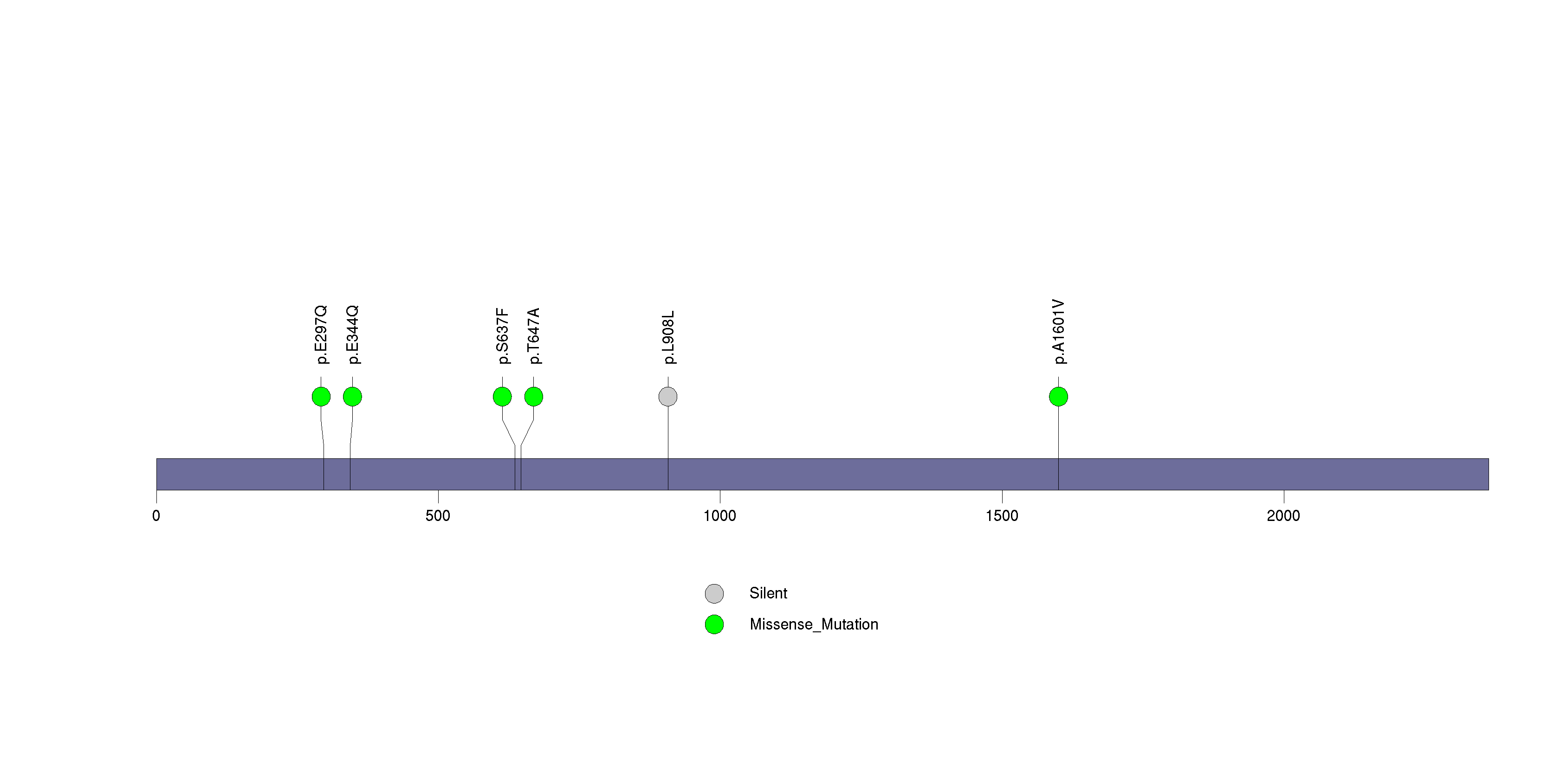
| TPR | 26152 | 0 | 1 | 5 | 0 | 0.000044 | 0.029 |
| CSMD3 | 37184 | 0 | 1 | 5 | 1 | 0.00019 | 0.12 |
| LAMA3 | 31220 | 0 | 0 | 5 | 2 | 0.00022 | 0.14 |
| SDPR | 4116 | 0 | 0 | 4 | 1 | 0.00031 | 0.18 |
| OTUD7A | 4956 | 0 | 0 | 4 | 0 | 0.0012 | 0.68 |
| SPATS2 | 5376 | 0 | 0 | 4 | 1 | 0.002 | 1 |
| KPNA3 | 5628 | 0 | 0 | 4 | 1 | 0.0026 | 1 |
Figure S1. This figure depicts the distribution of mutations and mutation types across the TP53 significant gene.
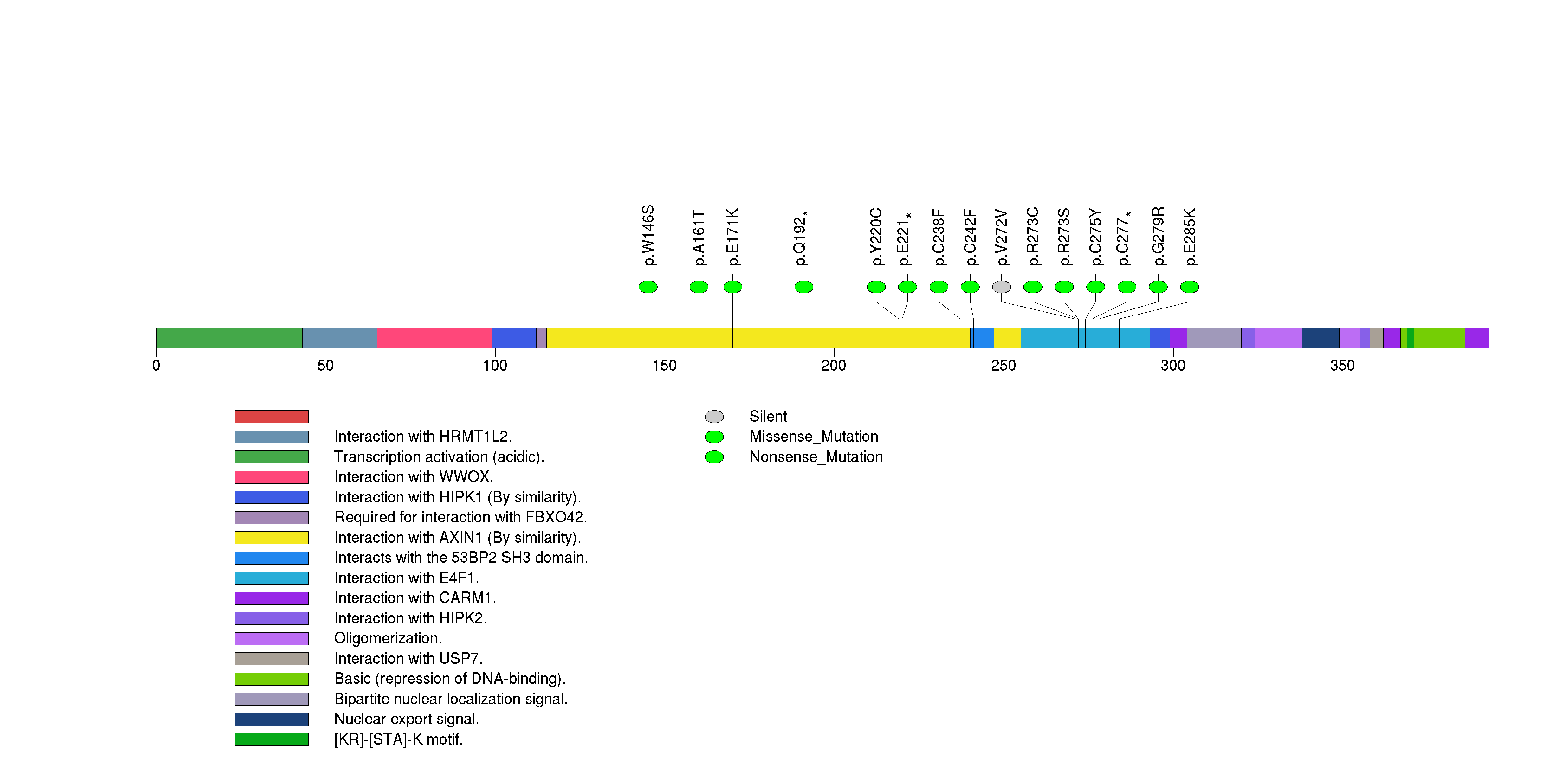
Figure S2. This figure depicts the distribution of mutations and mutation types across the KDM6A significant gene.
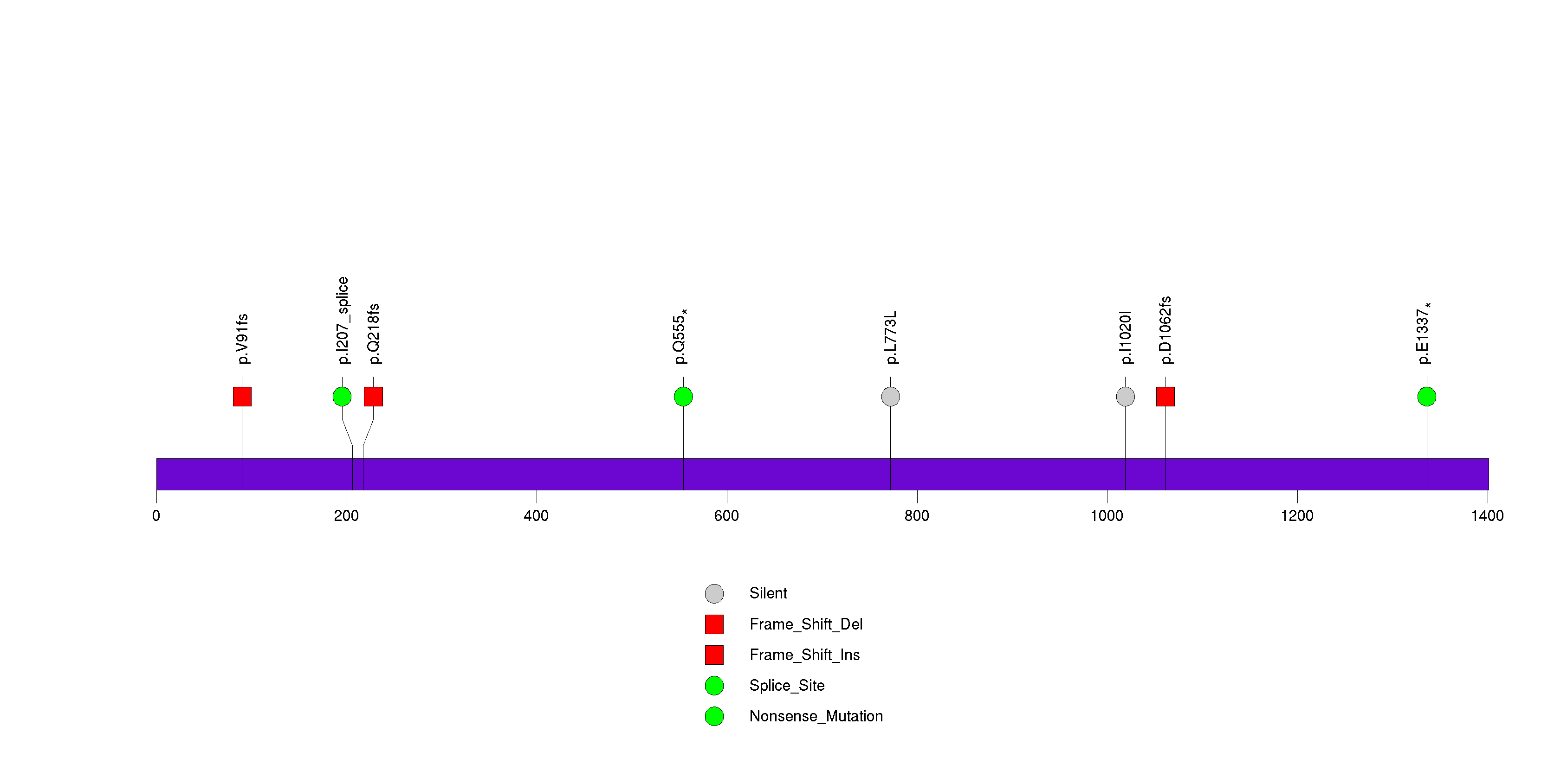
Figure S3. This figure depicts the distribution of mutations and mutation types across the PRX significant gene.
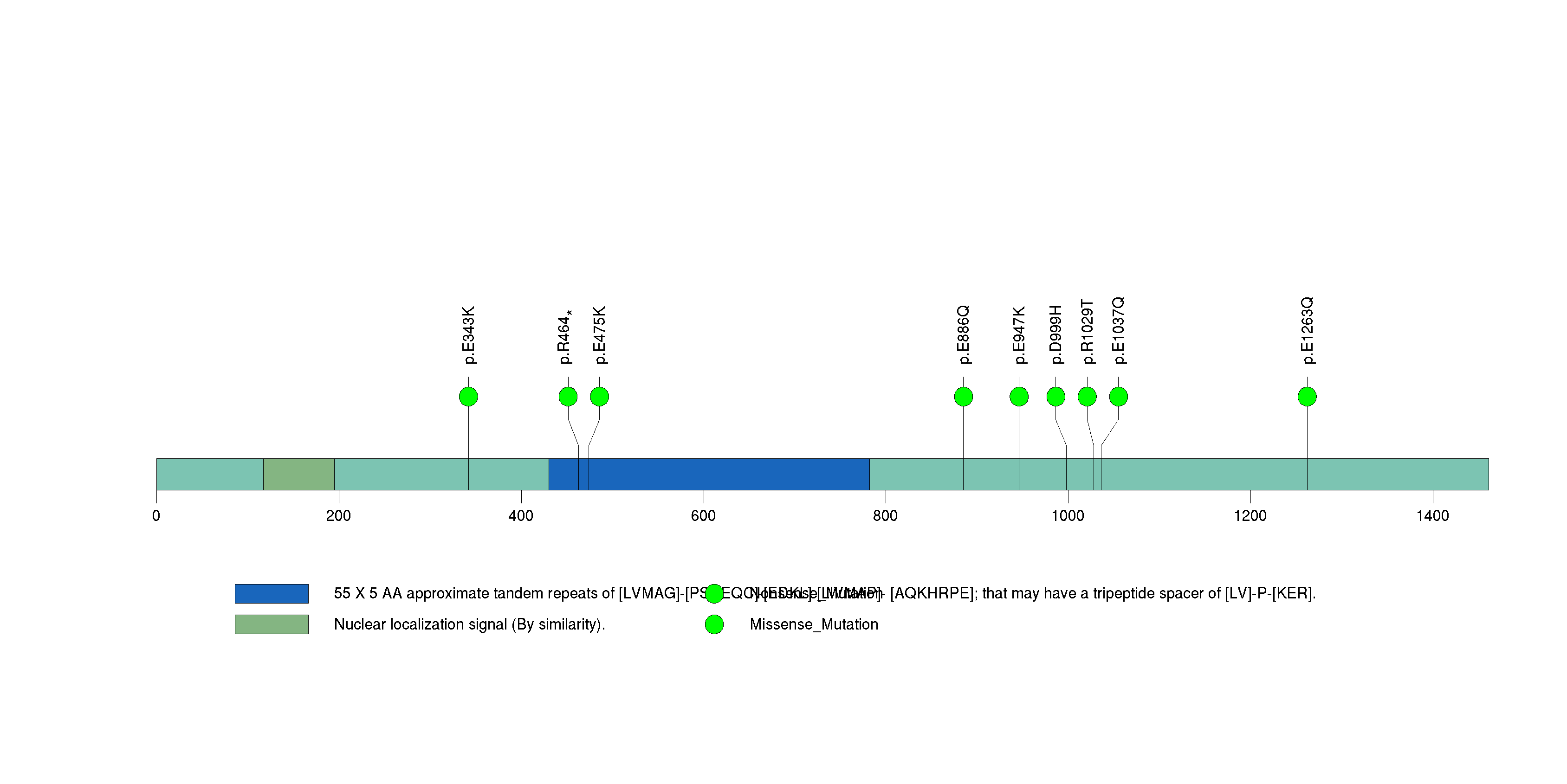
Figure S4. This figure depicts the distribution of mutations and mutation types across the ARID1A significant gene.
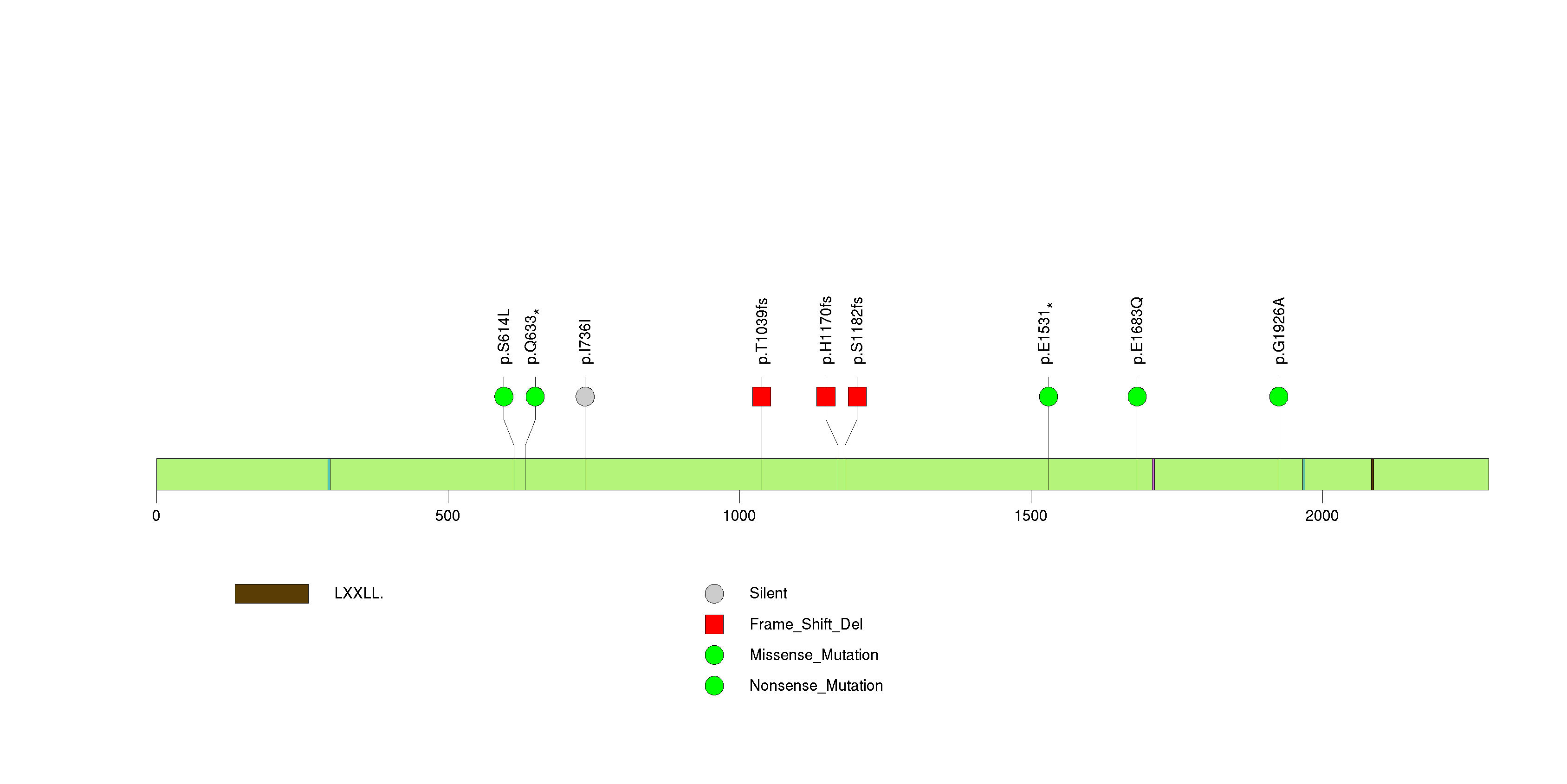
Figure S5. This figure depicts the distribution of mutations and mutation types across the GOLGB1 significant gene.
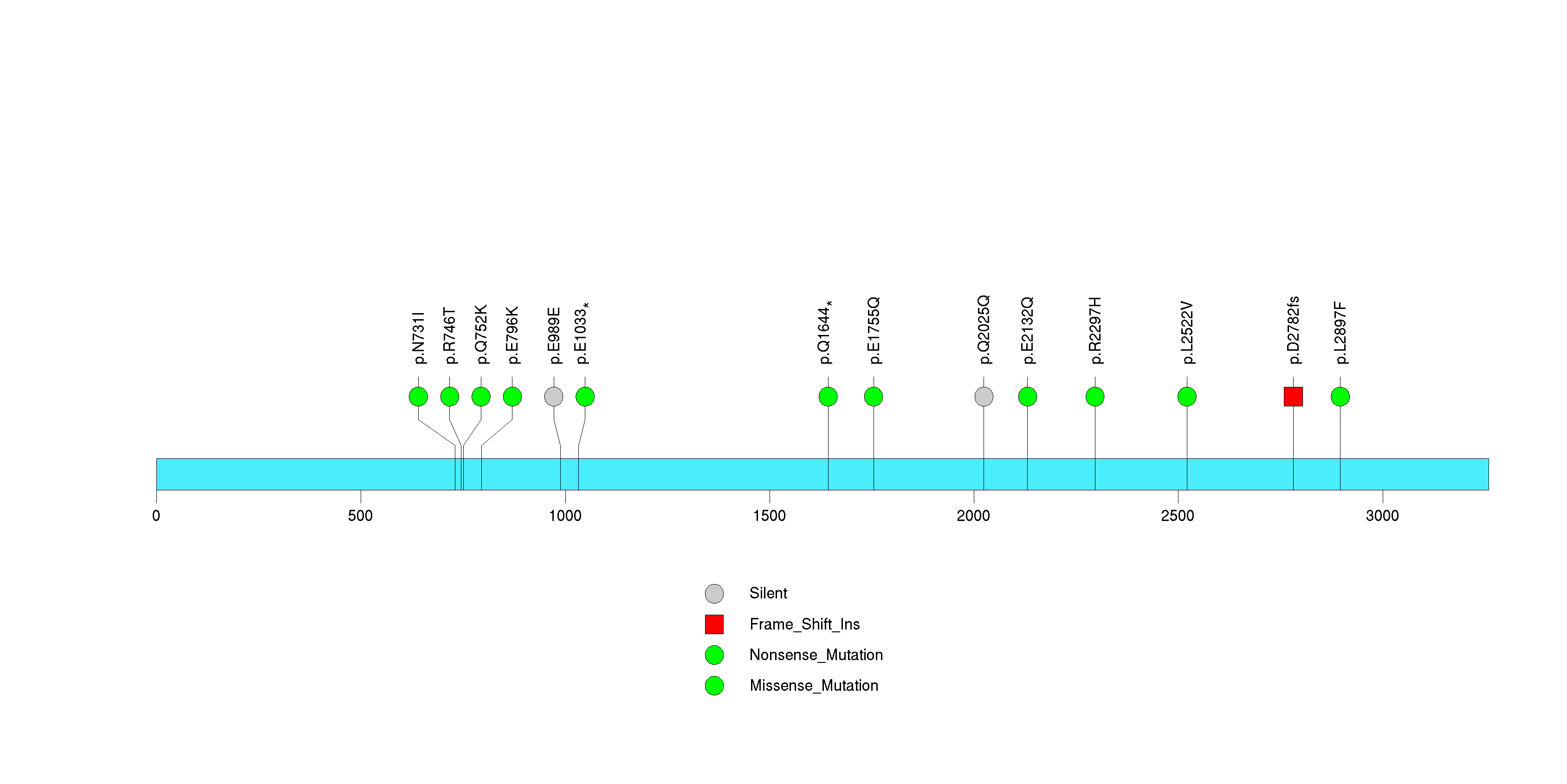
Figure S6. This figure depicts the distribution of mutations and mutation types across the ASXL2 significant gene.
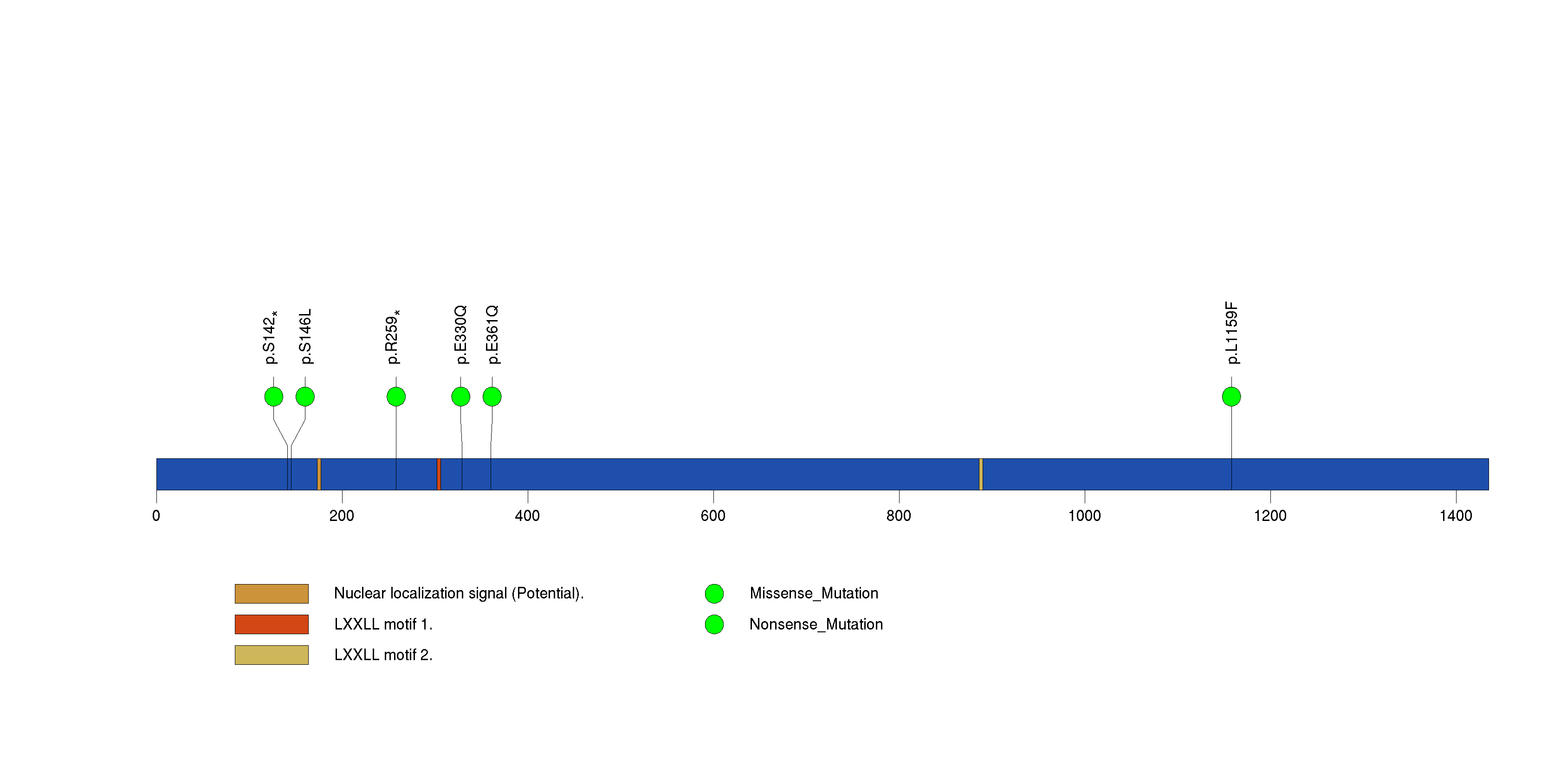
Figure S7. This figure depicts the distribution of mutations and mutation types across the FBXW7 significant gene.
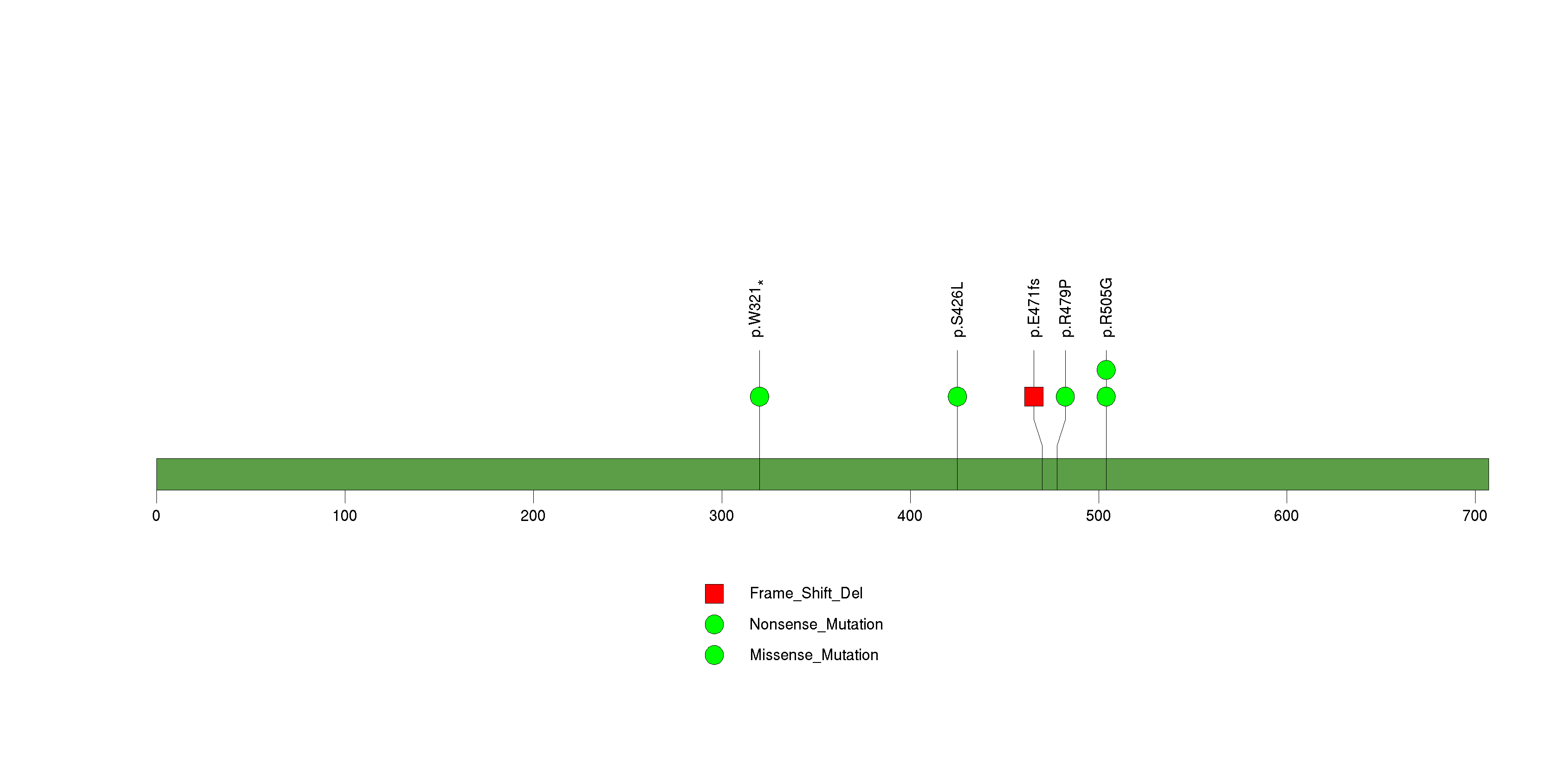
Figure S8. This figure depicts the distribution of mutations and mutation types across the MLL significant gene.
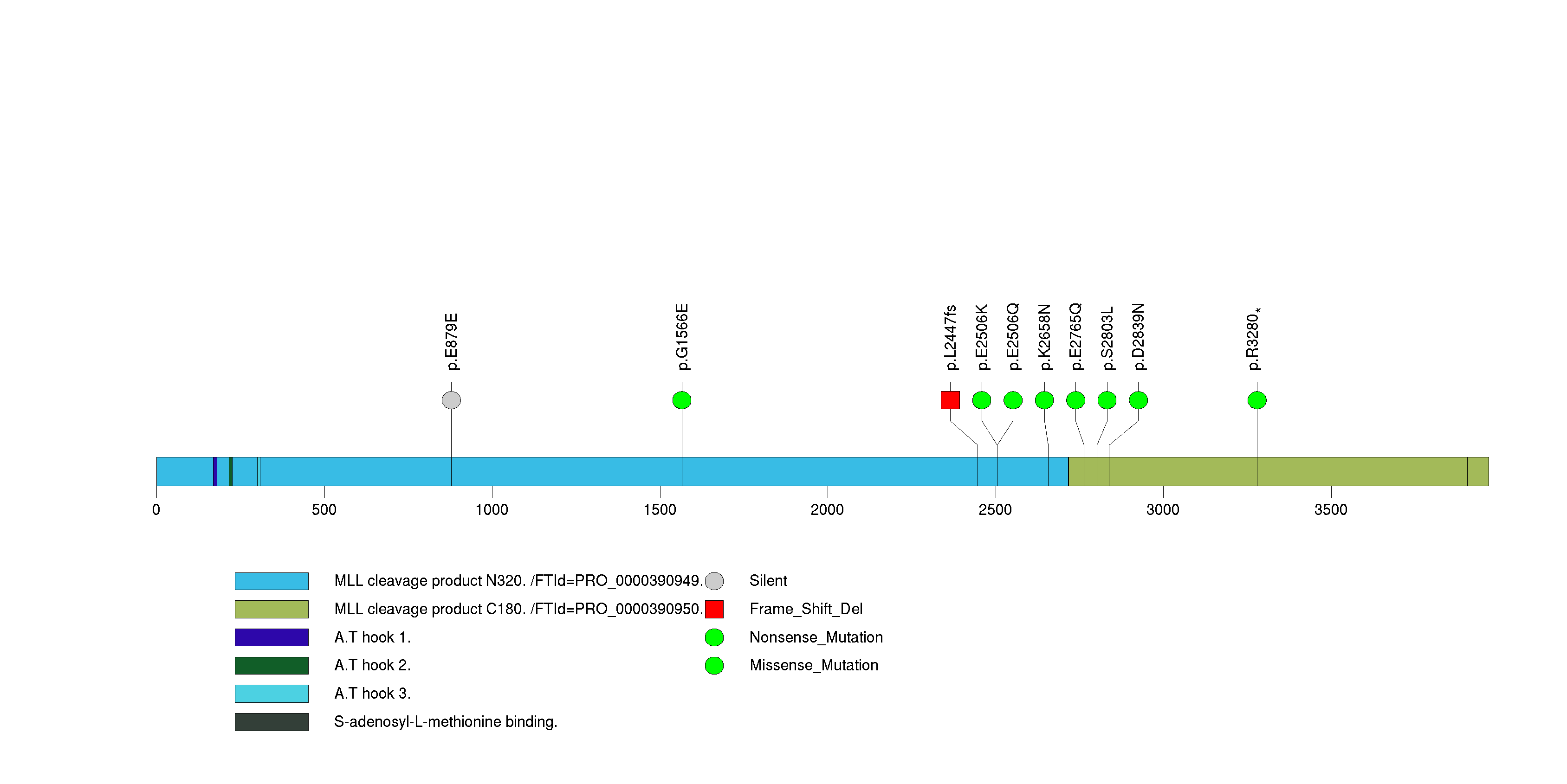
Figure S9. This figure depicts the distribution of mutations and mutation types across the FLG significant gene.
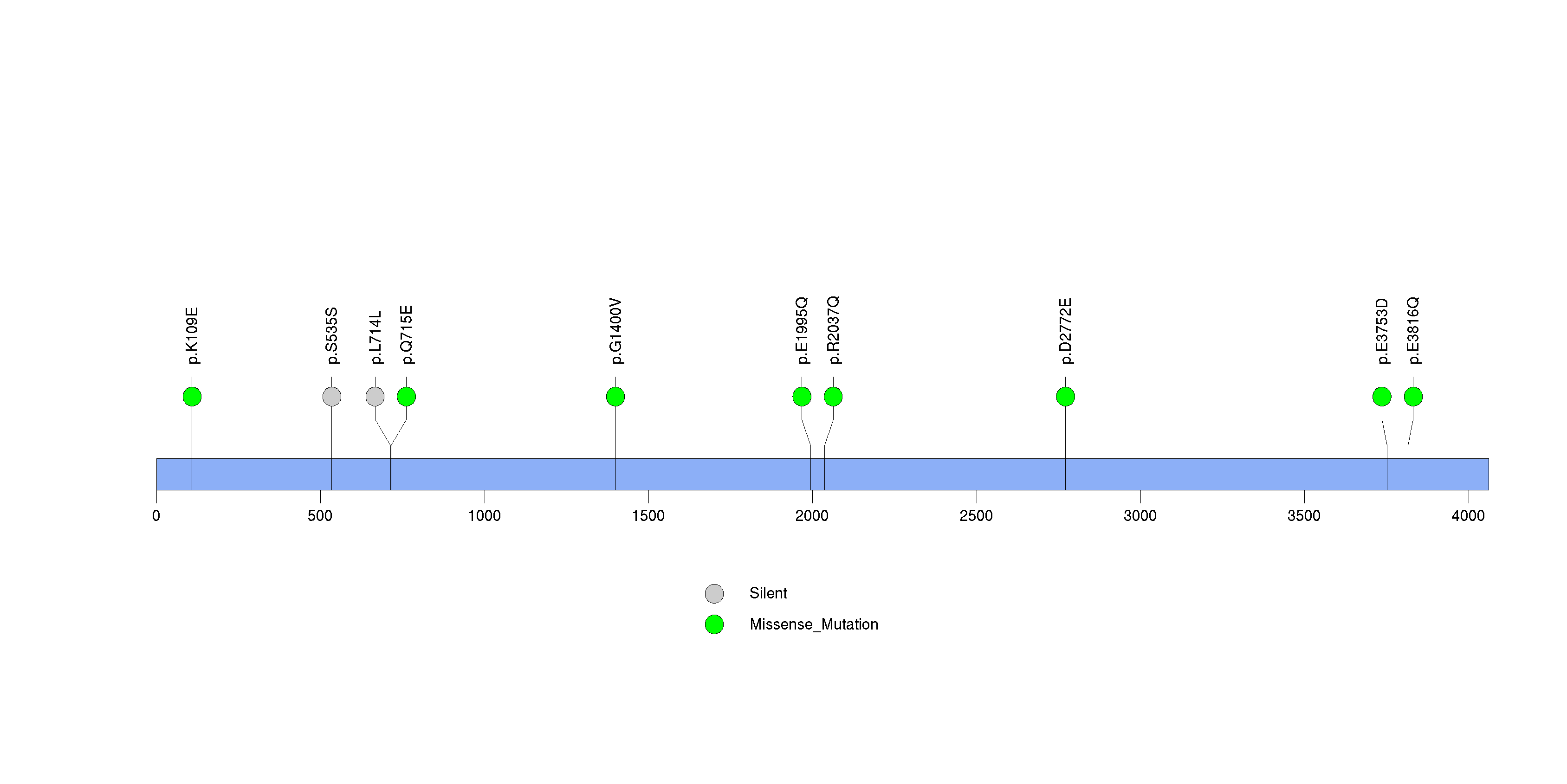
Figure S10. This figure depicts the distribution of mutations and mutation types across the HRNR significant gene.
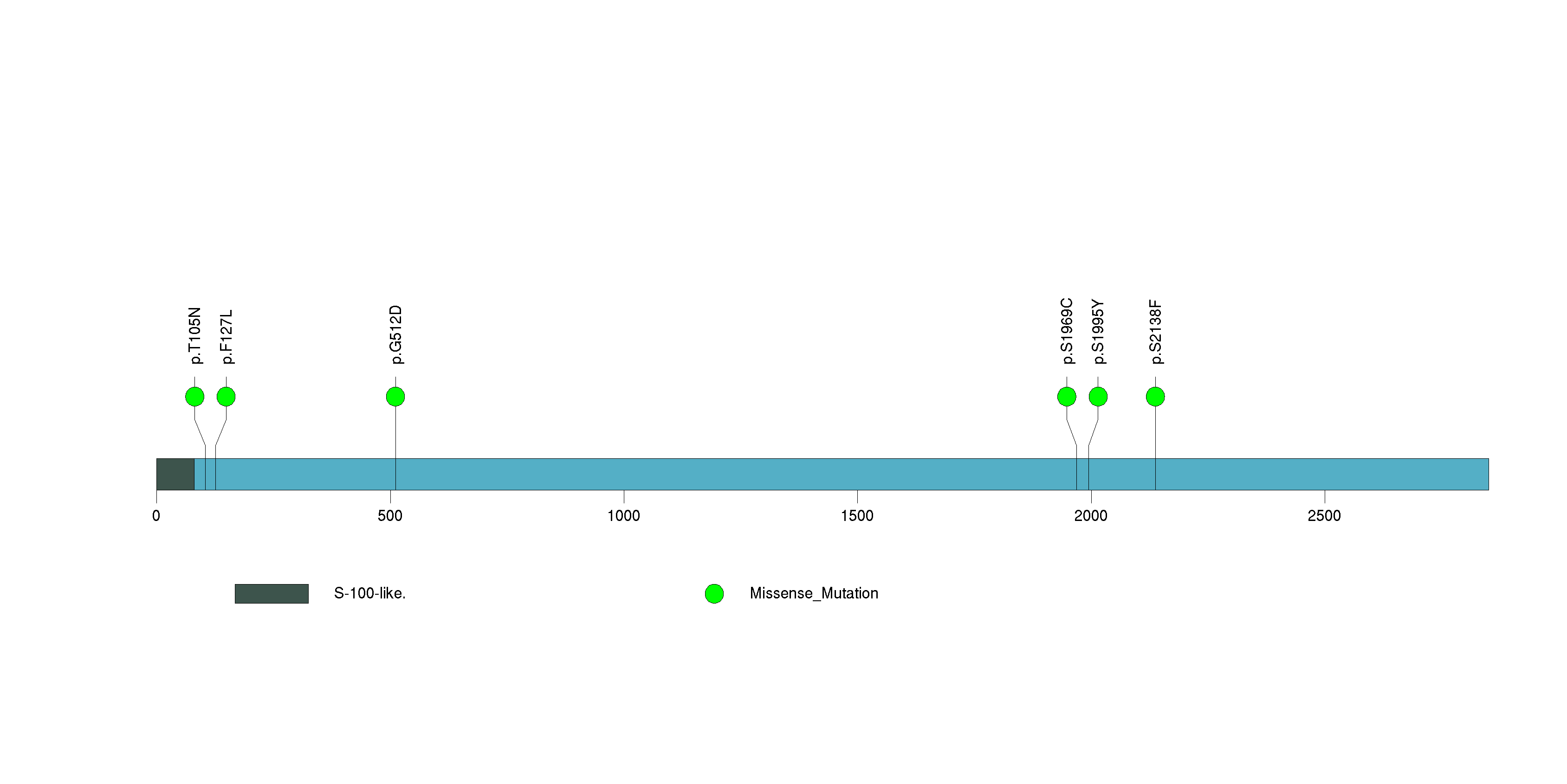
Figure S11. This figure depicts the distribution of mutations and mutation types across the HCN1 significant gene.
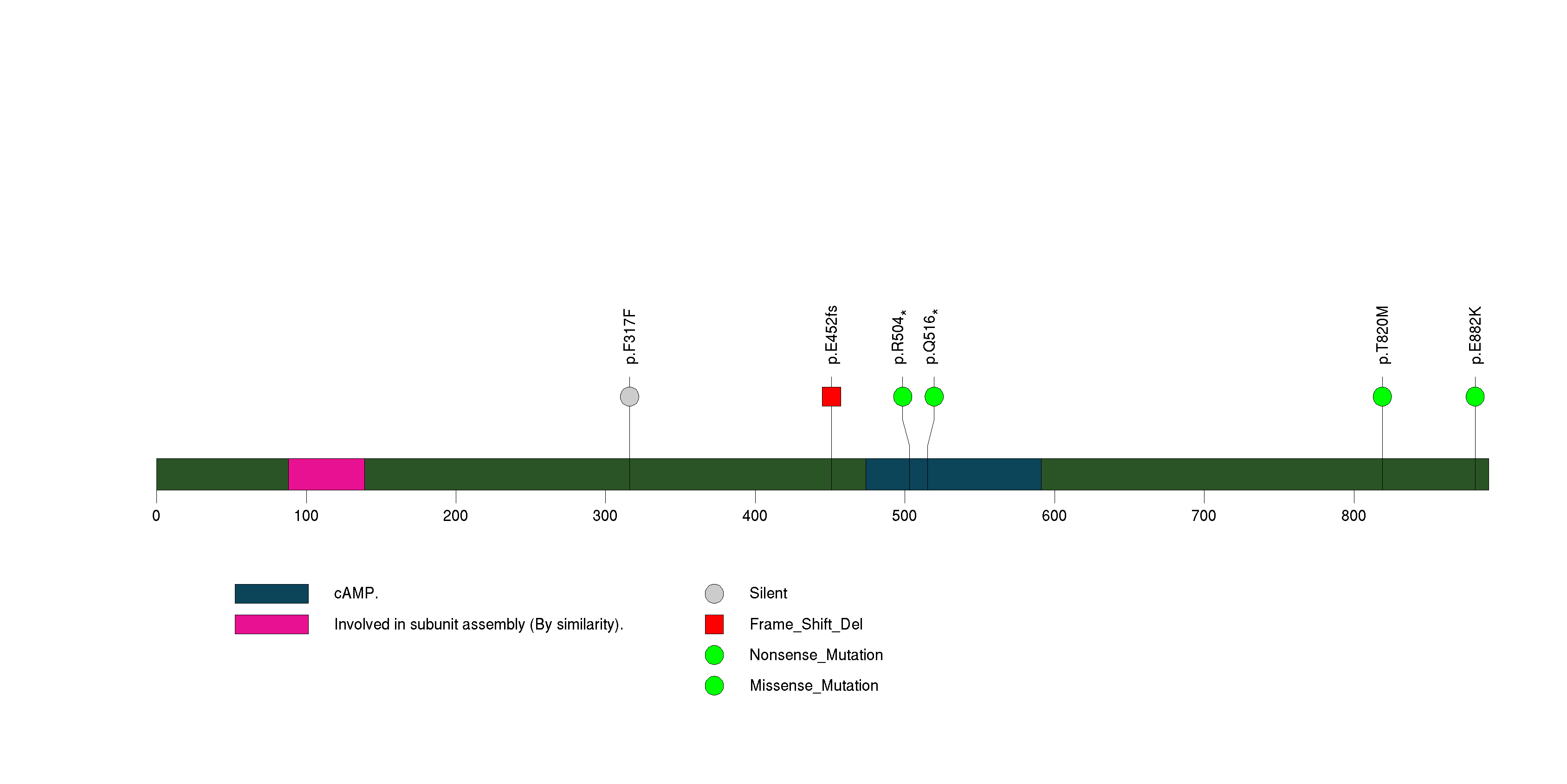
Figure S12. This figure depicts the distribution of mutations and mutation types across the TTN significant gene.
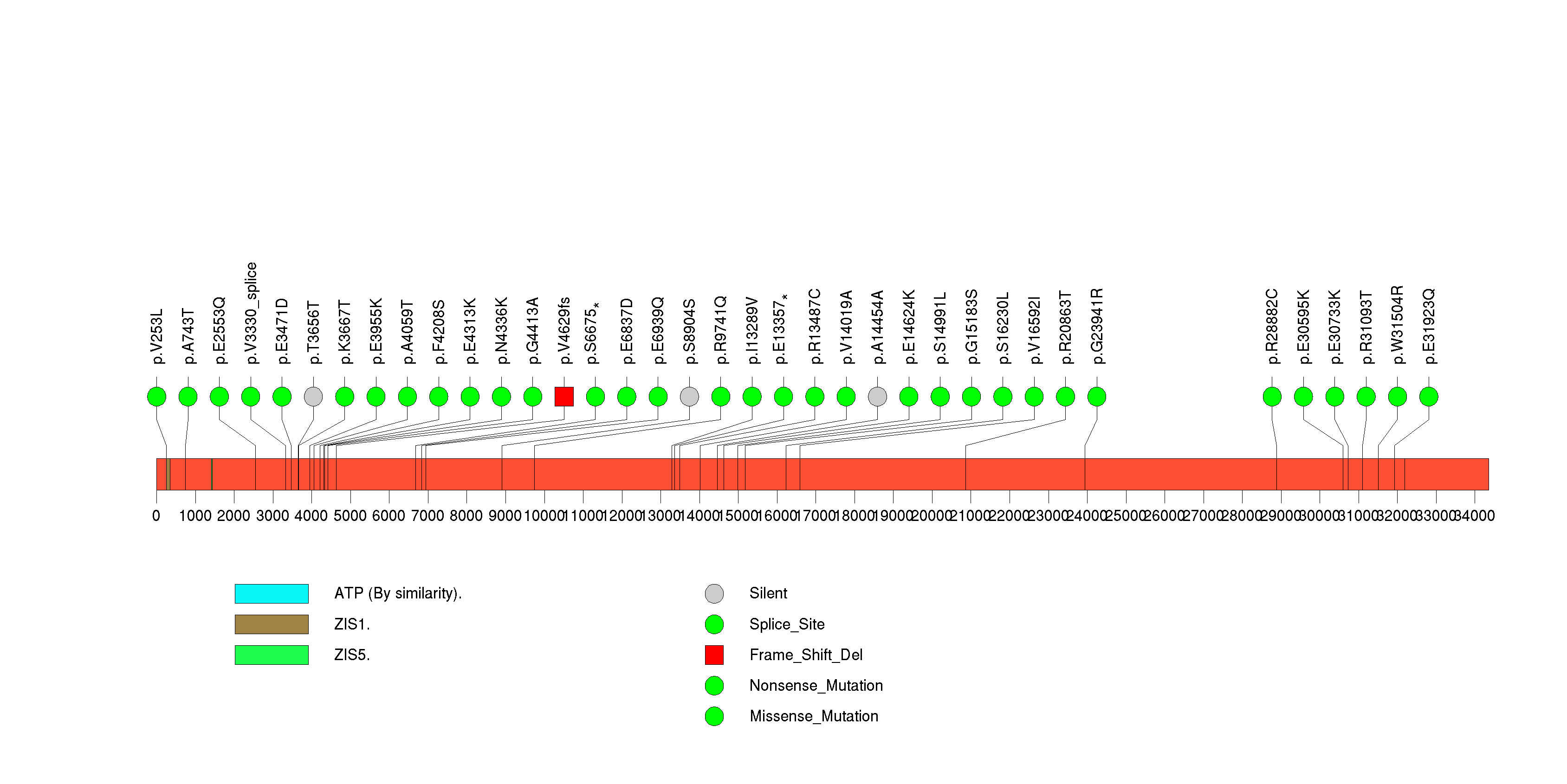
Figure S13. This figure depicts the distribution of mutations and mutation types across the BCLAF1 significant gene.
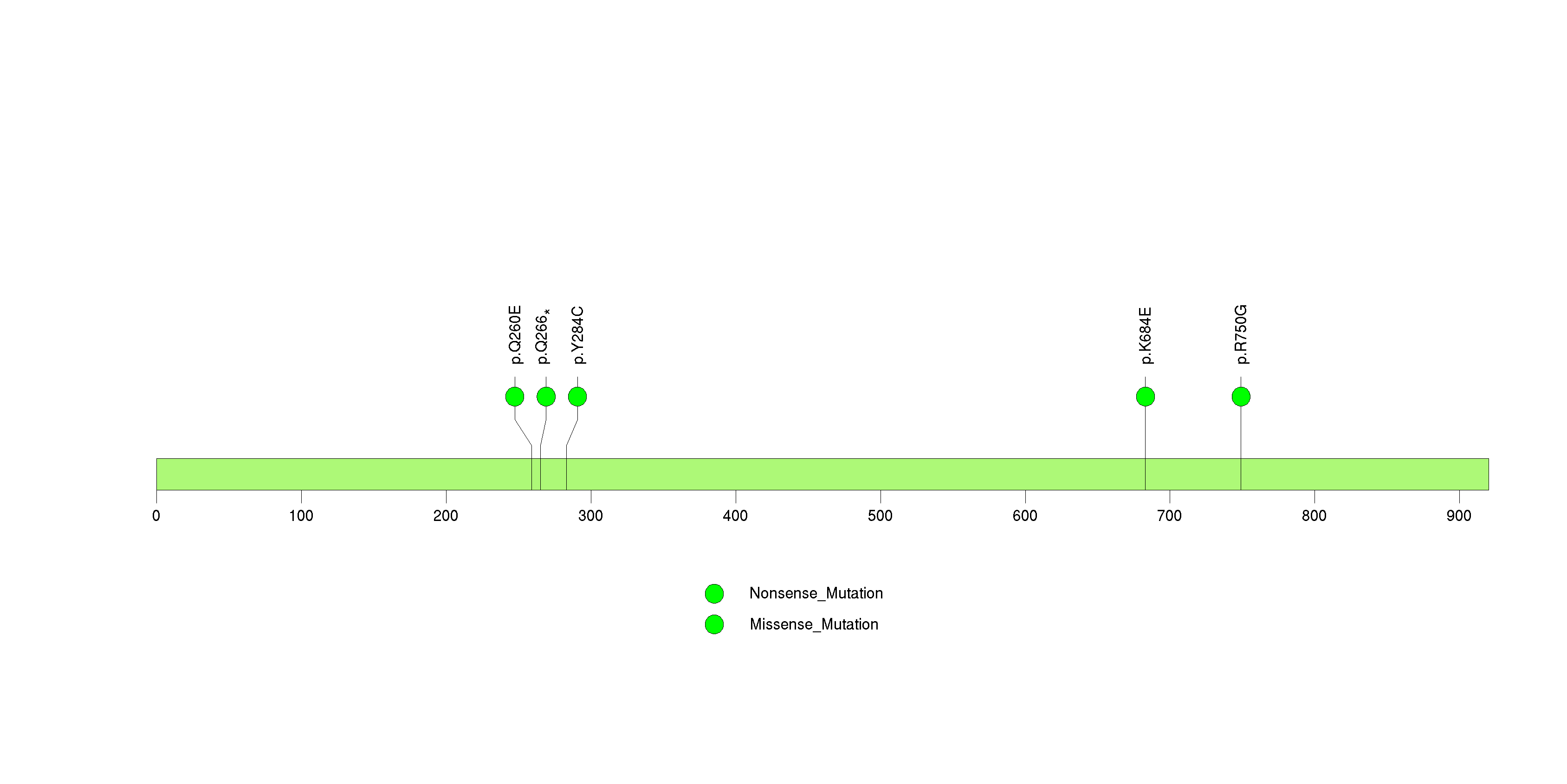
Figure S14. This figure depicts the distribution of mutations and mutation types across the MACF1 significant gene.
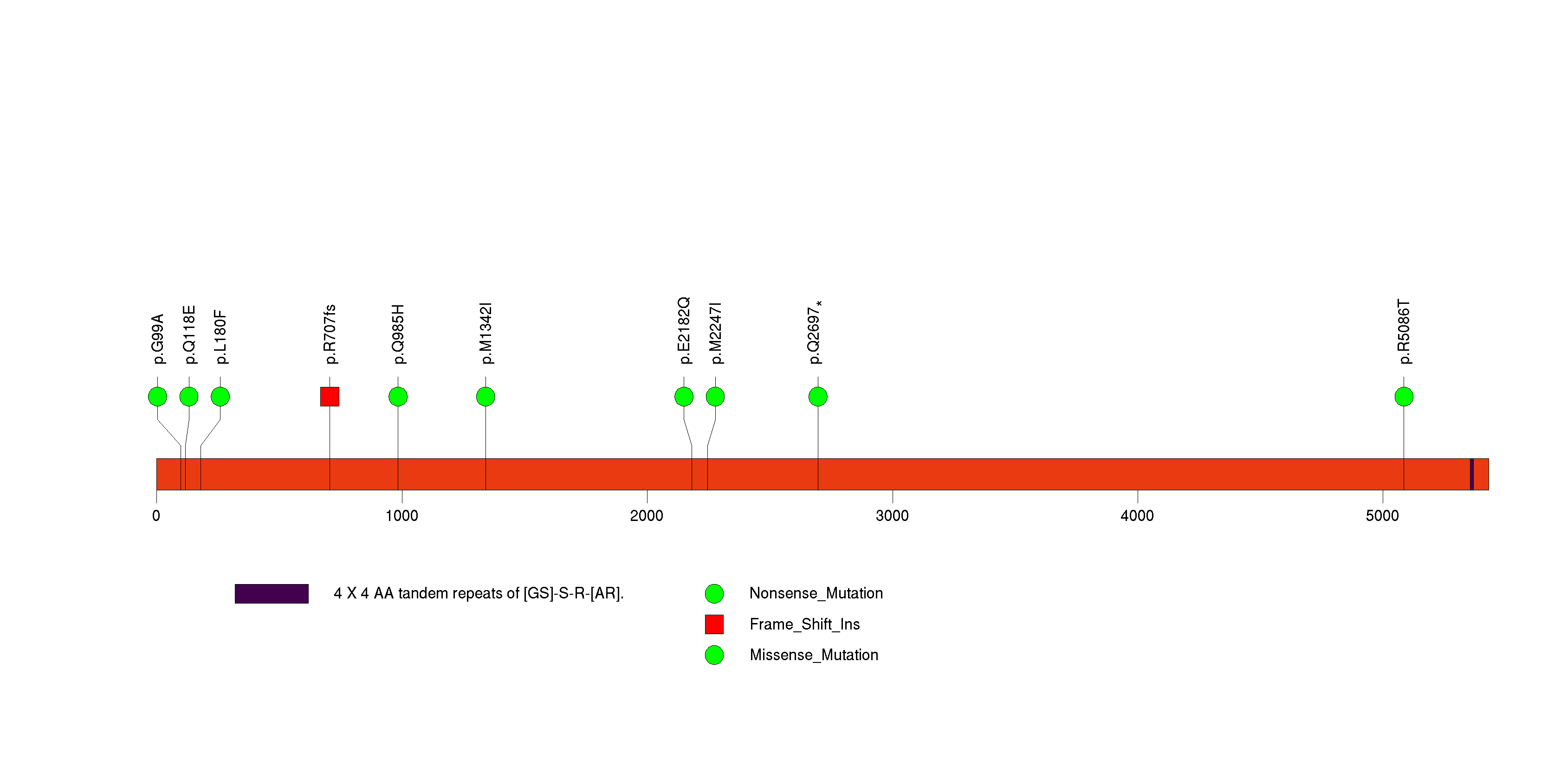
Figure S15. This figure depicts the distribution of mutations and mutation types across the RPAP1 significant gene.

Figure S16. This figure depicts the distribution of mutations and mutation types across the HMCN1 significant gene.
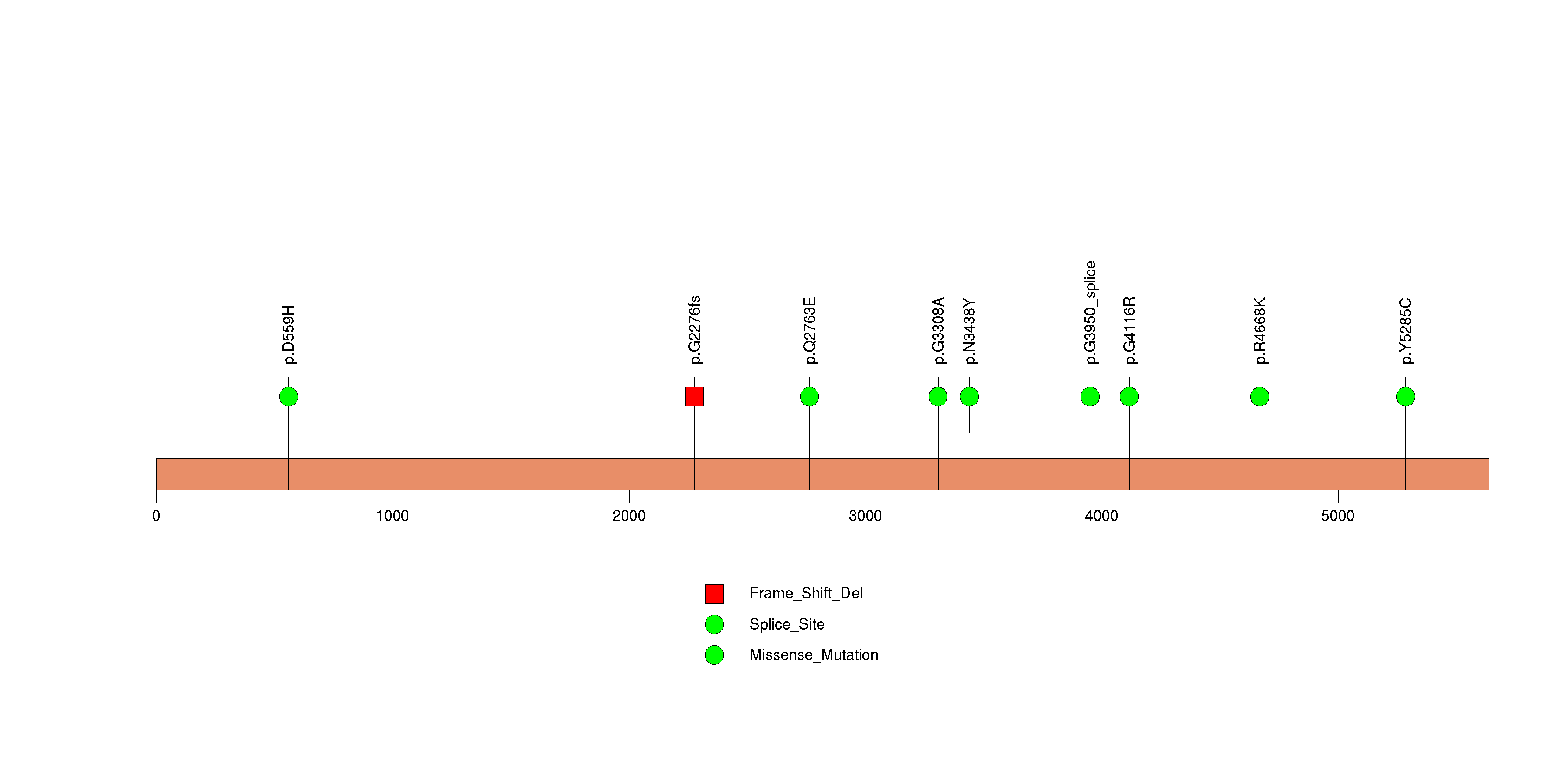
Figure S17. This figure depicts the distribution of mutations and mutation types across the ADAMTS12 significant gene.
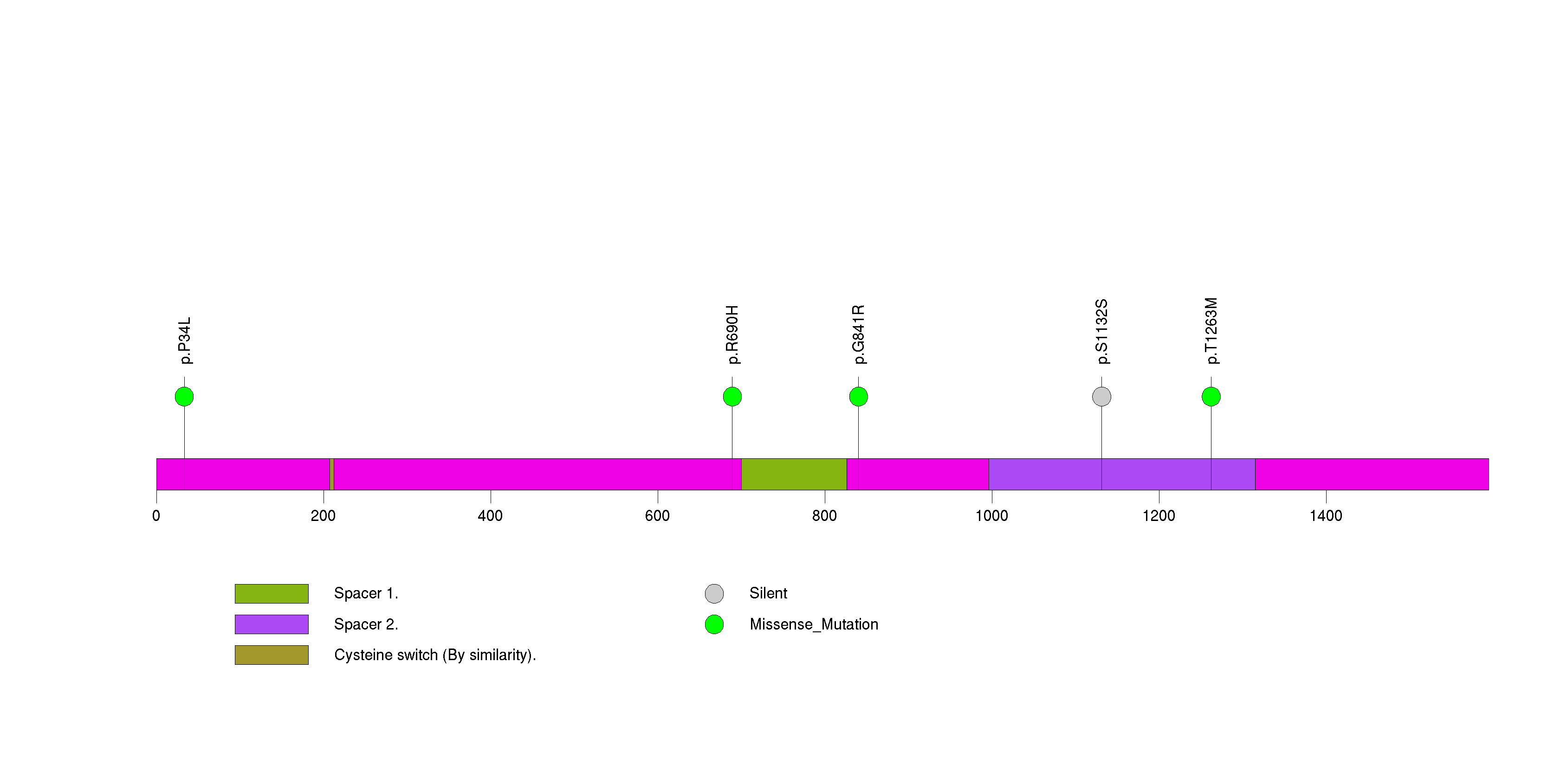
Figure S18. This figure depicts the distribution of mutations and mutation types across the DCC significant gene.
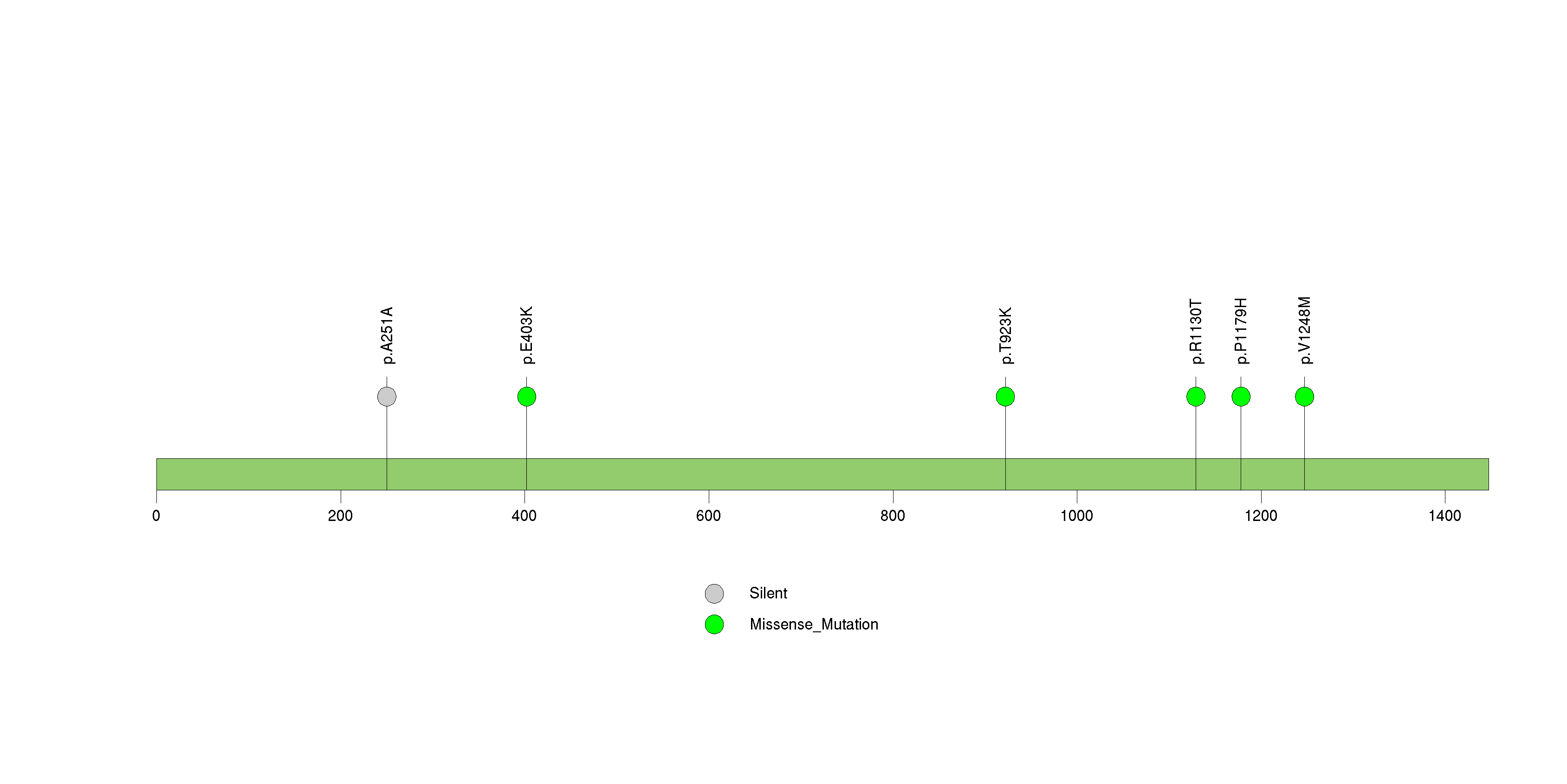
Figure S19. This figure depicts the distribution of mutations and mutation types across the MLL2 significant gene.
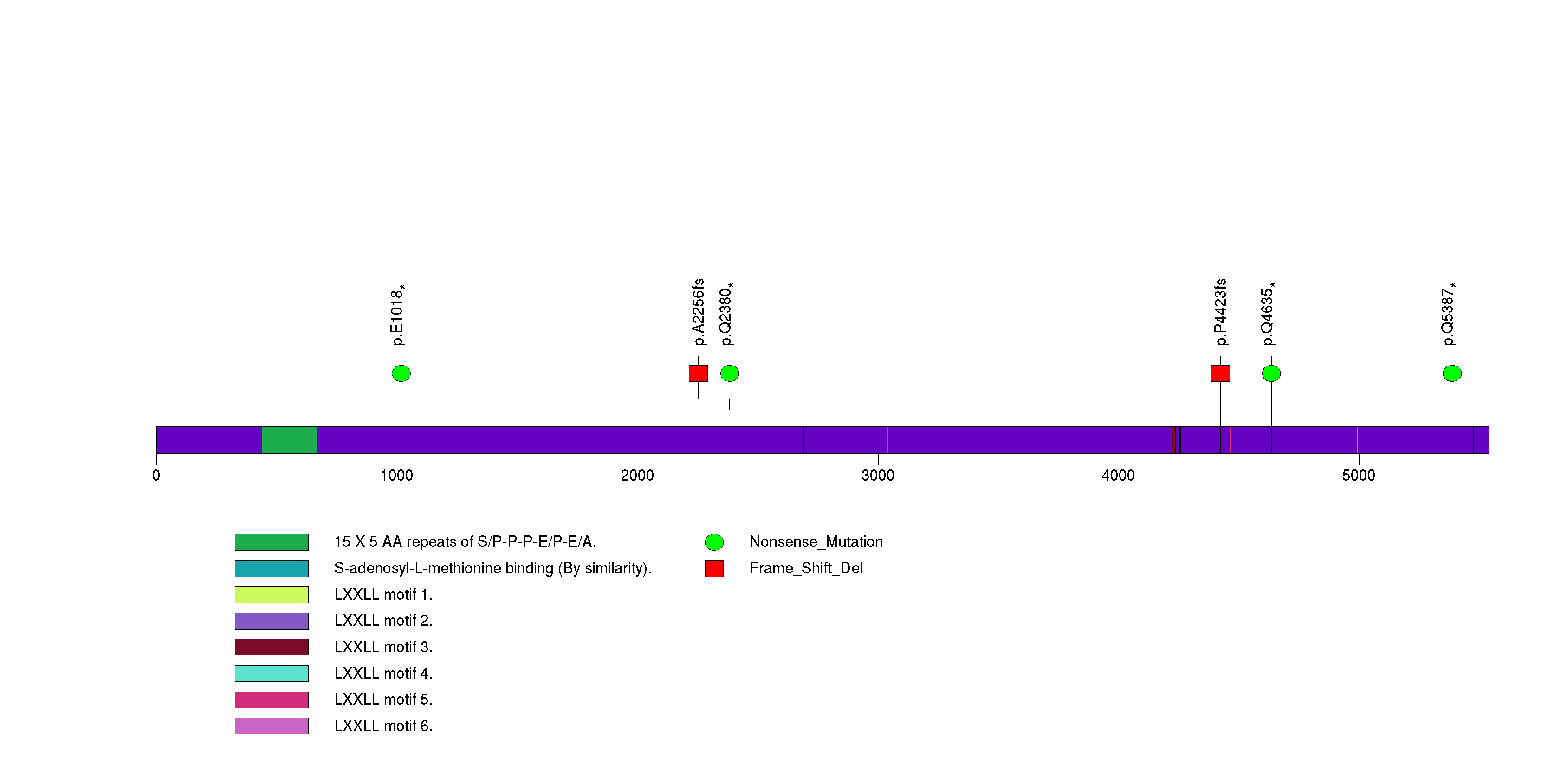
Figure S20. This figure depicts the distribution of mutations and mutation types across the ABCA10 significant gene.
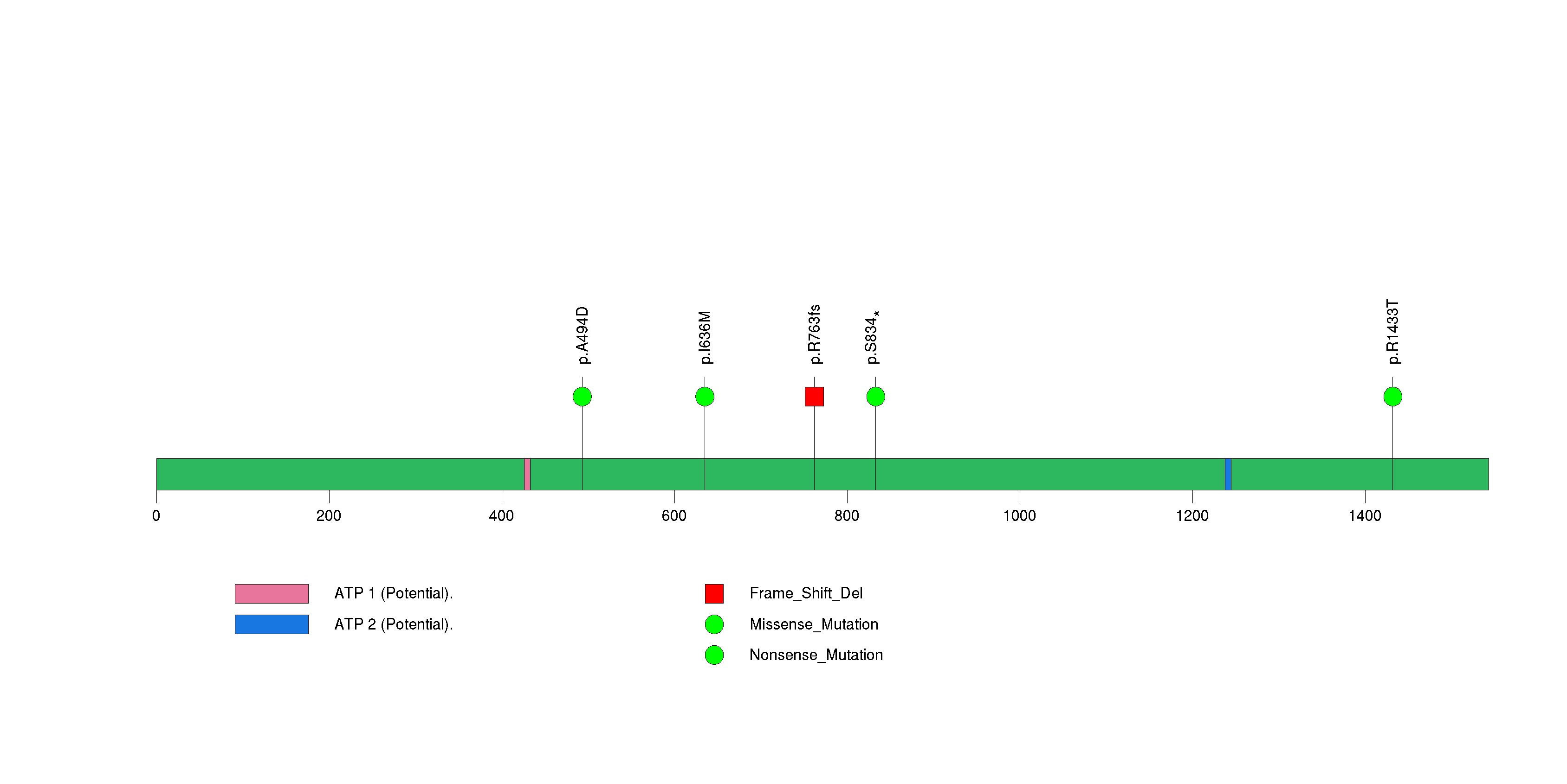
Figure S21. This figure depicts the distribution of mutations and mutation types across the XPR1 significant gene.
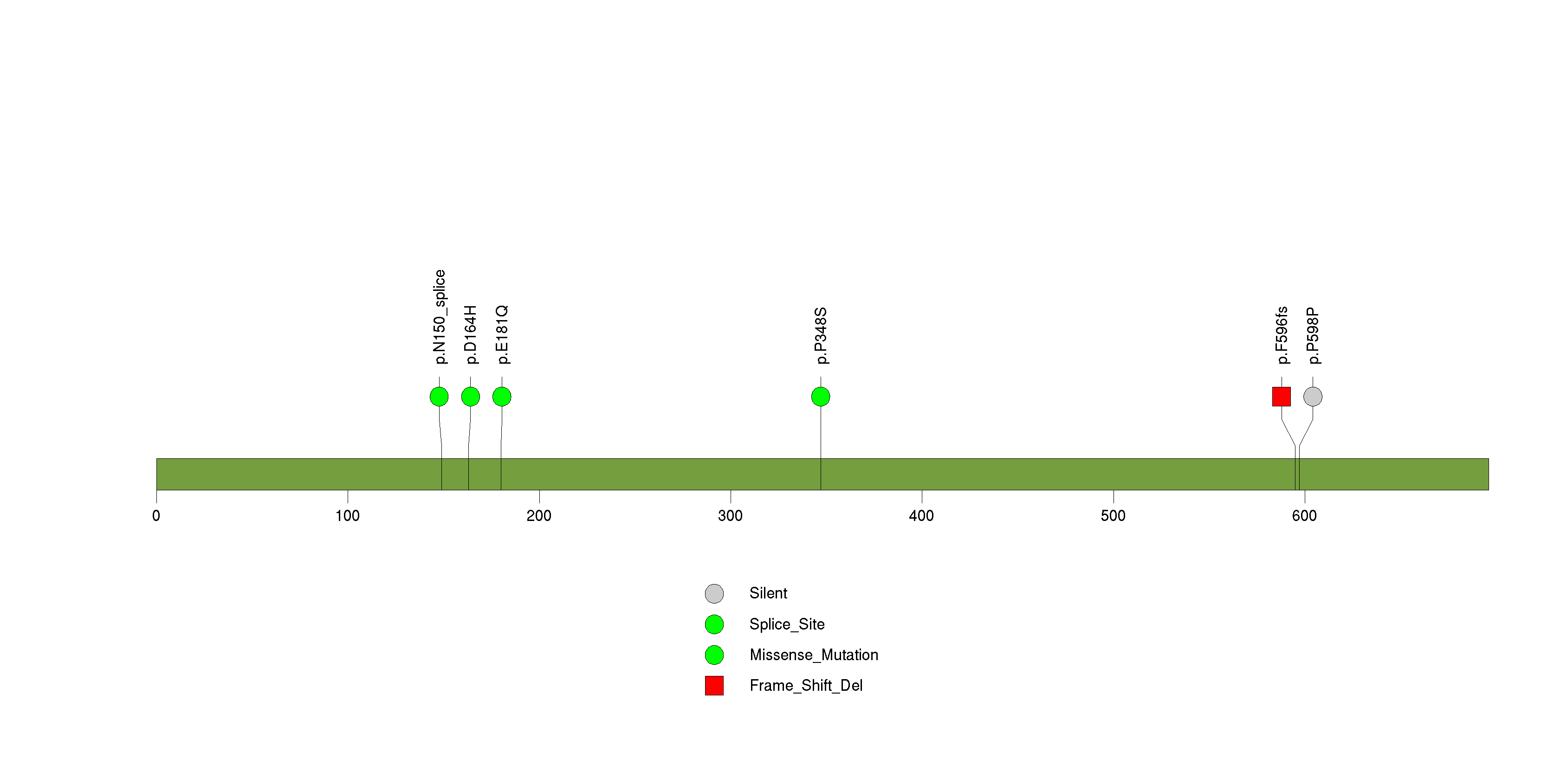
Figure S22. This figure depicts the distribution of mutations and mutation types across the CREBBP significant gene.
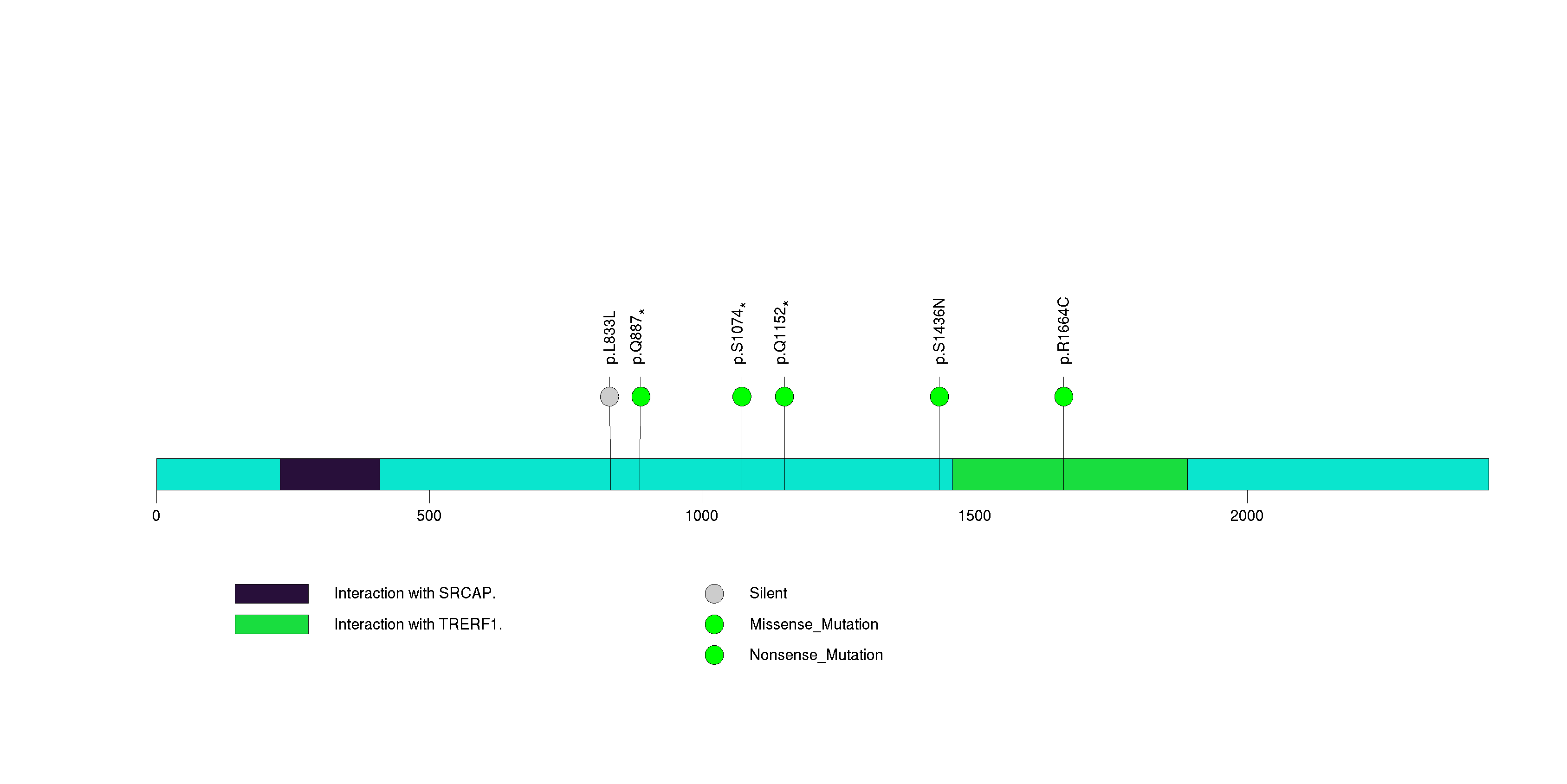
Figure S23. This figure depicts the distribution of mutations and mutation types across the SETD2 significant gene.
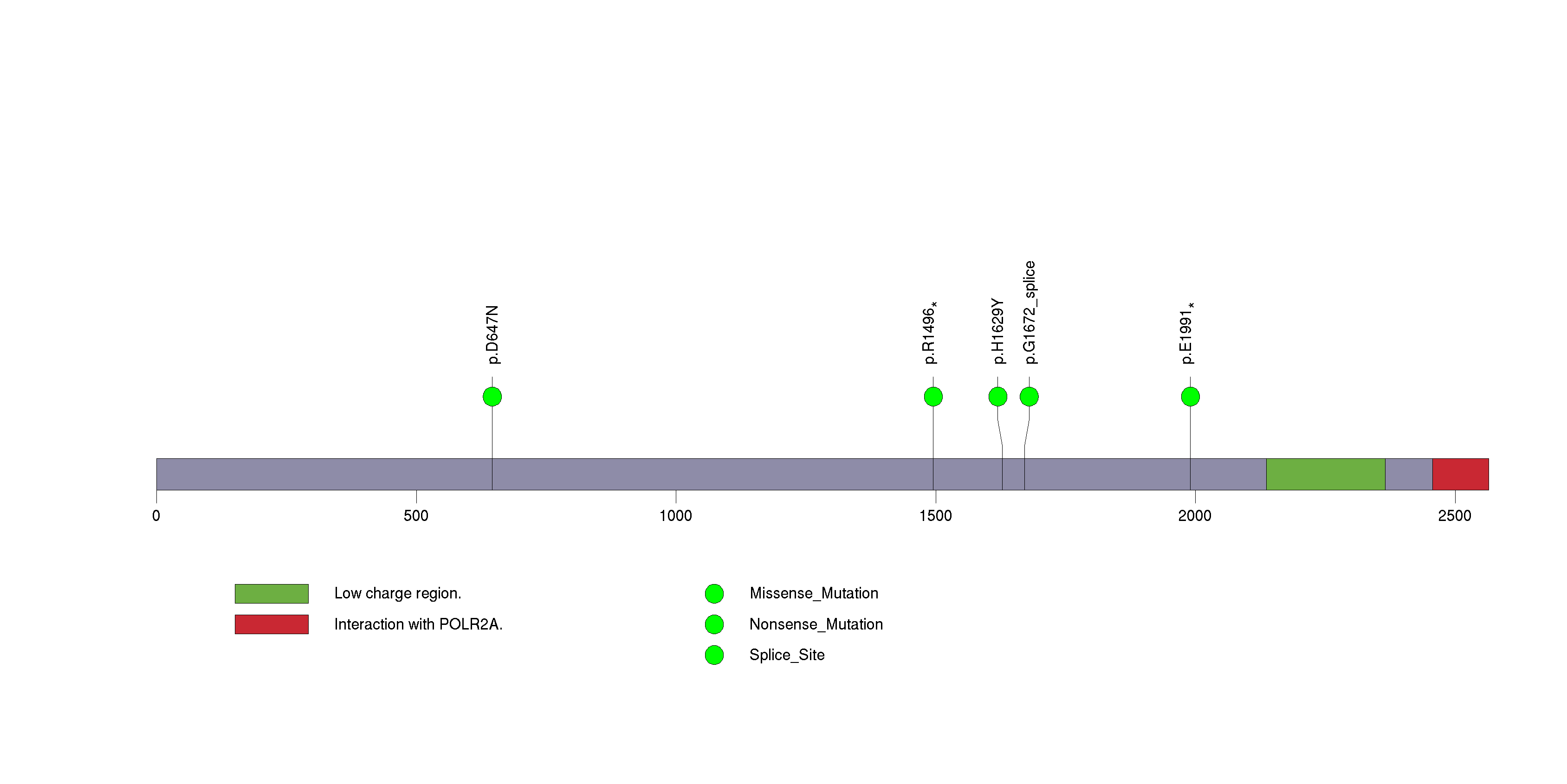
Figure S24. This figure depicts the distribution of mutations and mutation types across the SACS significant gene.

Figure S25. This figure depicts the distribution of mutations and mutation types across the DNAH5 significant gene.
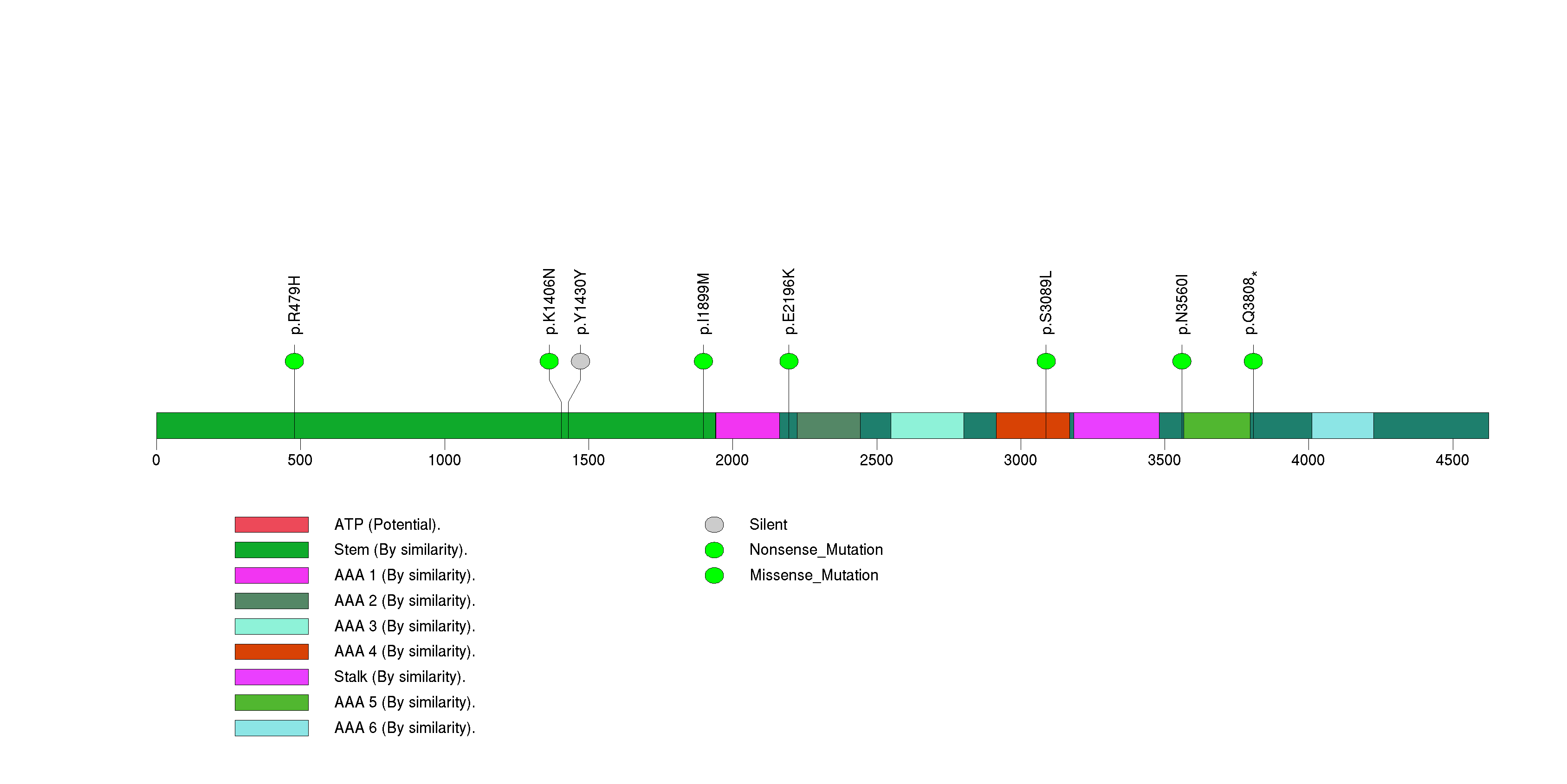
Figure S26. This figure depicts the distribution of mutations and mutation types across the SPTA1 significant gene.
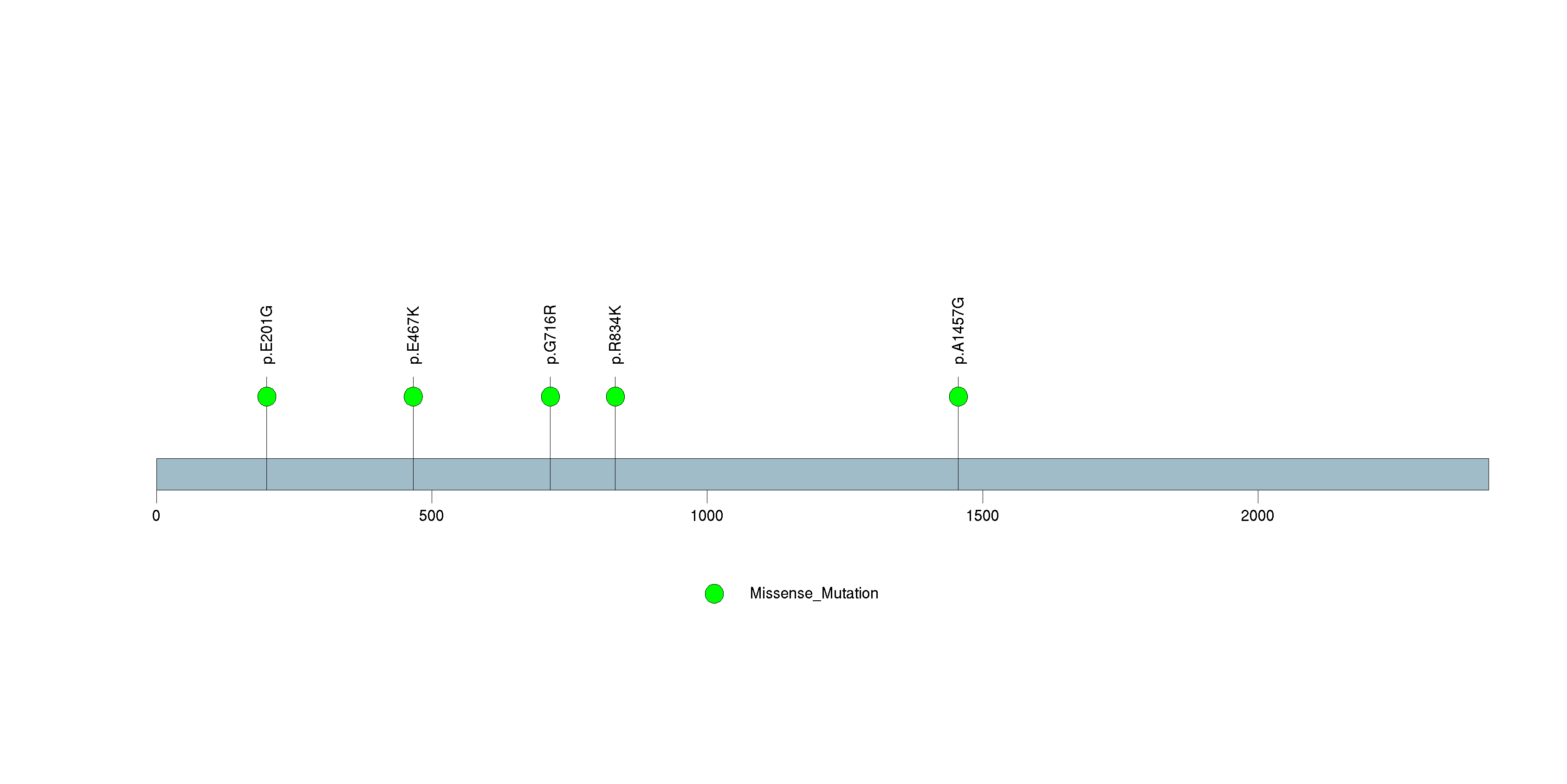
Figure S27. This figure depicts the distribution of mutations and mutation types across the APOB significant gene.
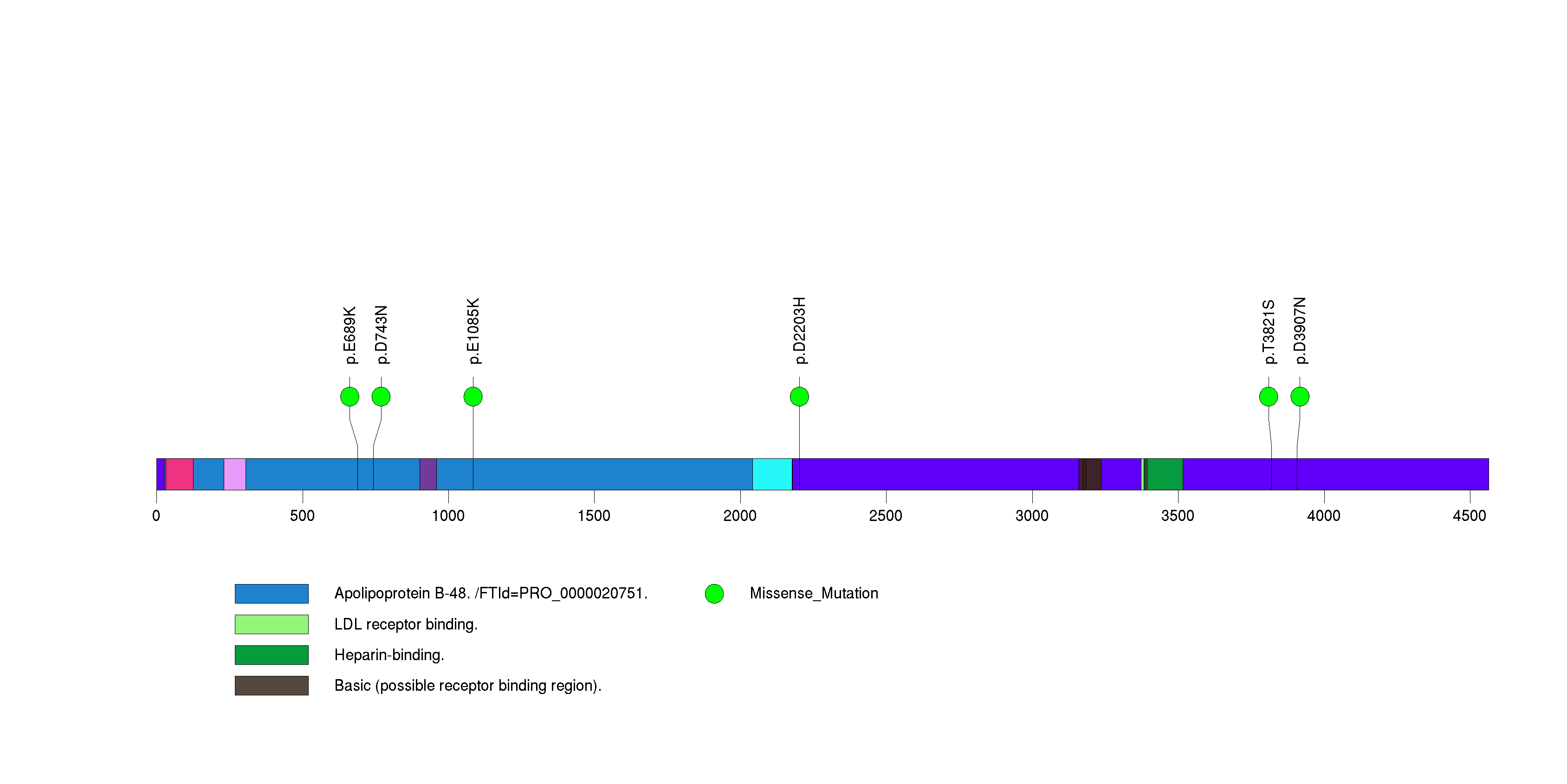
Figure S28. This figure depicts the distribution of mutations and mutation types across the GPS2 significant gene.
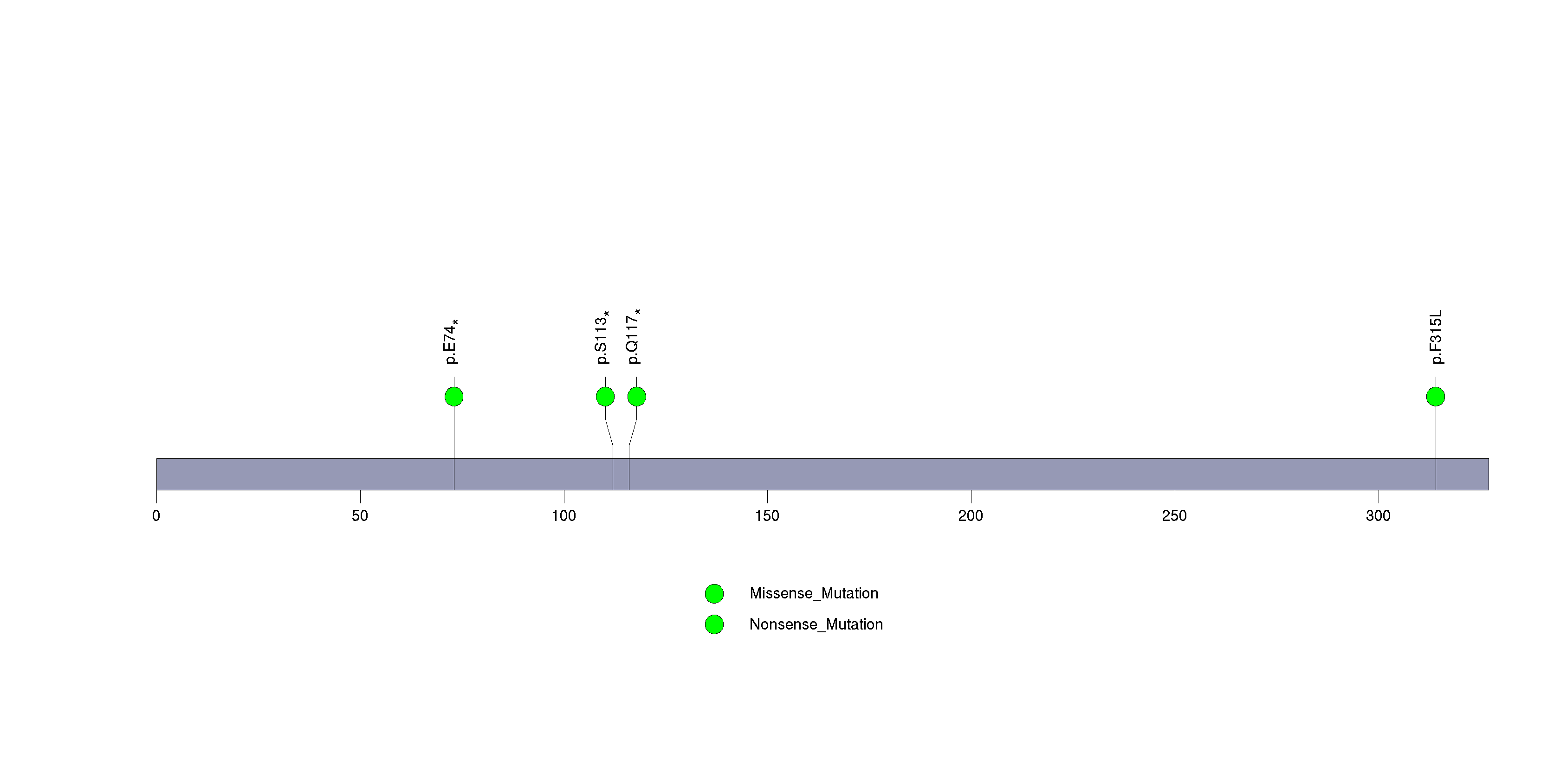
Figure S29. This figure depicts the distribution of mutations and mutation types across the TPR significant gene.
In brief, we tabulate the number of mutations and the number of covered bases for each gene. The counts are broken down by mutation context category: four context categories that are discovered by MutSig, and one for indel and 'null' mutations, which include indels, nonsense mutations, splice-site mutations, and non-stop (read-through) mutations. For each gene, we calculate the probability of seeing the observed constellation of mutations, i.e. the product P1 x P2 x ... x Pm, or a more extreme one, given the background mutation rates calculated across the dataset. [1]
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.