This pipeline computes the correlation between cancer subtypes identified by different molecular patterns and selected clinical features.
Testing the association between mutation status of 6 genes and 14 clinical features across 229 patients, 3 significant findings detected with P value < 0.05 and Q value < 0.25.
-
FUSION RET mutation correlated to 'AGE'.
-
MUTATION MUT HRAS mutation correlated to 'AGE' and 'DISTANT.METASTASIS'.
Table 1. Get Full Table Overview of the association between mutation status of 6 genes and 14 clinical features. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 3 significant findings detected.
|
Clinical Features |
AGE | GENDER |
HISTOLOGICAL TYPE |
RADIATIONS RADIATION REGIMENINDICATION |
RADIATIONEXPOSURE |
DISTANT METASTASIS |
EXTRATHYROIDAL EXTENSION |
LYMPH NODE METASTASIS |
COMPLETENESS OF RESECTION |
NUMBER OF LYMPH NODES |
TUMOR STAGECODE |
NEOPLASM DISEASESTAGE |
MULTIFOCALITY |
TUMOR SIZE |
||
| nMutated (%) | nWild-Type | t-test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Chi-square test | Fisher's exact test | t-test | t-test | Chi-square test | Fisher's exact test | t-test | |
| MUTATION MUT HRAS | 0 (NaN%) | 0 |
0.00121 (0.0918) |
1 (1.00) |
0.338 (1.00) |
0.599 (1.00) |
1 (1.00) |
0.00304 (0.225) |
0.384 (1.00) |
0.0227 (1.00) |
0.754 (1.00) |
0.5 (1.00) |
0.247 (1.00) |
0.745 (1.00) |
0.993 (1.00) |
|
| FUSION RET | 0 (NaN%) | 0 |
0.00143 (0.107) |
0.177 (1.00) |
0.714 (1.00) |
0.426 (1.00) |
1 (1.00) |
1 (1.00) |
0.721 (1.00) |
0.452 (1.00) |
0.309 (1.00) |
0.303 (1.00) |
0.144 (1.00) |
1 (1.00) |
0.816 (1.00) |
|
| FUSION PAX8 | 0 (NaN%) | 0 |
0.848 (1.00) |
0.255 (1.00) |
0.0274 (1.00) |
1 (1.00) |
1 (1.00) |
0.759 (1.00) |
0.712 (1.00) |
0.607 (1.00) |
0.726 (1.00) |
0.801 (1.00) |
0.757 (1.00) |
0.243 (1.00) |
0.0518 (1.00) |
|
| FUSION BRAF | 0 (NaN%) | 0 |
0.372 (1.00) |
0.553 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.595 (1.00) |
1 (1.00) |
0.725 (1.00) |
1 (1.00) |
0.618 (1.00) |
0.585 (1.00) |
|||
| MUTATION MUT BRAF | 0 (NaN%) | 0 |
0.507 (1.00) |
0.591 (1.00) |
0.0757 (1.00) |
0.212 (1.00) |
1 (1.00) |
0.445 (1.00) |
0.696 (1.00) |
0.315 (1.00) |
0.0292 (1.00) |
0.867 (1.00) |
0.268 (1.00) |
1 (1.00) |
0.0303 (1.00) |
|
| MUTATION MUT NRAS | 0 (NaN%) | 0 |
0.0416 (1.00) |
0.287 (1.00) |
0.219 (1.00) |
0.0582 (1.00) |
1 (1.00) |
0.399 (1.00) |
1 (1.00) |
0.0291 (1.00) |
0.766 (1.00) |
0.469 (1.00) |
0.16 (1.00) |
1 (1.00) |
0.638 (1.00) |
P value = 0.00143 (t-test), Q value = 0.11
Table S1. Gene #1: 'FUSION RET' versus Clinical Feature #1: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 171 | 47.5 (15.5) |
| subtype0 | 161 | 48.2 (15.6) |
| subtype1 | 10 | 36.1 (8.6) |
Figure S1. Get High-res Image Gene #1: 'FUSION RET' versus Clinical Feature #1: 'AGE'

P value = 0.177 (Fisher's exact test), Q value = 1
Table S2. Gene #1: 'FUSION RET' versus Clinical Feature #2: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 118 | 53 |
| subtype0 | 109 | 52 |
| subtype1 | 9 | 1 |
P value = 0.714 (Fisher's exact test), Q value = 1
Table S3. Gene #1: 'FUSION RET' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | OTHER | THYROID PAPILLARY CARCINOMA - CLASSICAL/USUAL | THYROID PAPILLARY CARCINOMA - FOLLICULAR (>= 99% FOLLICULAR PATTERNED) | THYROID PAPILLARY CARCINOMA - TALL CELL (>= 50% TALL CELL FEATURES) |
|---|---|---|---|---|
| ALL | 16 | 87 | 50 | 18 |
| subtype0 | 16 | 81 | 46 | 18 |
| subtype1 | 0 | 6 | 4 | 0 |
P value = 0.426 (Fisher's exact test), Q value = 1
Table S4. Gene #1: 'FUSION RET' versus Clinical Feature #4: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 9 | 162 |
| subtype0 | 8 | 153 |
| subtype1 | 1 | 9 |
P value = 1 (Fisher's exact test), Q value = 1
Table S5. Gene #1: 'FUSION RET' versus Clinical Feature #5: 'RADIATIONEXPOSURE'
| nPatients | NO | YES |
|---|---|---|
| ALL | 143 | 8 |
| subtype0 | 134 | 8 |
| subtype1 | 9 | 0 |
P value = 1 (Fisher's exact test), Q value = 1
Table S6. Gene #1: 'FUSION RET' versus Clinical Feature #6: 'DISTANT.METASTASIS'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 70 | 3 | 97 |
| subtype0 | 66 | 3 | 91 |
| subtype1 | 4 | 0 | 6 |
P value = 0.721 (Fisher's exact test), Q value = 1
Table S7. Gene #1: 'FUSION RET' versus Clinical Feature #7: 'EXTRATHYROIDAL.EXTENSION'
| nPatients | MINIMAL (T3) | MODERATE/ADVANCED (T4A) | NONE |
|---|---|---|---|
| ALL | 38 | 2 | 122 |
| subtype0 | 37 | 2 | 114 |
| subtype1 | 1 | 0 | 8 |
P value = 0.452 (Chi-square test), Q value = 1
Table S8. Gene #1: 'FUSION RET' versus Clinical Feature #8: 'LYMPH.NODE.METASTASIS'
| nPatients | N0 | N1 | N1A | N1B | NX |
|---|---|---|---|---|---|
| ALL | 84 | 7 | 35 | 24 | 21 |
| subtype0 | 81 | 6 | 33 | 21 | 20 |
| subtype1 | 3 | 1 | 2 | 3 | 1 |
P value = 0.309 (Fisher's exact test), Q value = 1
Table S9. Gene #1: 'FUSION RET' versus Clinical Feature #9: 'COMPLETENESS.OF.RESECTION'
| nPatients | R0 | R1 | RX |
|---|---|---|---|
| ALL | 131 | 14 | 12 |
| subtype0 | 124 | 12 | 12 |
| subtype1 | 7 | 2 | 0 |
P value = 0.303 (t-test), Q value = 1
Table S10. Gene #1: 'FUSION RET' versus Clinical Feature #10: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 135 | 2.7 (4.8) |
| subtype0 | 127 | 2.5 (4.3) |
| subtype1 | 8 | 6.1 (9.3) |
P value = 0.144 (Chi-square test), Q value = 1
Table S11. Gene #1: 'FUSION RET' versus Clinical Feature #12: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IVA | STAGE IVC |
|---|---|---|---|---|---|
| ALL | 91 | 25 | 38 | 13 | 3 |
| subtype0 | 82 | 25 | 38 | 12 | 3 |
| subtype1 | 9 | 0 | 0 | 1 | 0 |
P value = 1 (Fisher's exact test), Q value = 1
Table S12. Gene #1: 'FUSION RET' versus Clinical Feature #13: 'MULTIFOCALITY'
| nPatients | MULTIFOCAL | UNIFOCAL |
|---|---|---|
| ALL | 90 | 72 |
| subtype0 | 86 | 69 |
| subtype1 | 4 | 3 |
P value = 0.816 (t-test), Q value = 1
Table S13. Gene #1: 'FUSION RET' versus Clinical Feature #14: 'TUMOR.SIZE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 147 | 2.9 (1.5) |
| subtype0 | 141 | 2.9 (1.5) |
| subtype1 | 6 | 3.0 (1.6) |
P value = 0.848 (t-test), Q value = 1
Table S14. Gene #2: 'FUSION PAX8' versus Clinical Feature #1: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 171 | 47.5 (15.5) |
| subtype0 | 163 | 47.4 (15.6) |
| subtype1 | 8 | 48.5 (15.1) |
P value = 0.255 (Fisher's exact test), Q value = 1
Table S15. Gene #2: 'FUSION PAX8' versus Clinical Feature #2: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 118 | 53 |
| subtype0 | 114 | 49 |
| subtype1 | 4 | 4 |
P value = 0.0274 (Fisher's exact test), Q value = 1
Table S16. Gene #2: 'FUSION PAX8' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | OTHER | THYROID PAPILLARY CARCINOMA - CLASSICAL/USUAL | THYROID PAPILLARY CARCINOMA - FOLLICULAR (>= 99% FOLLICULAR PATTERNED) | THYROID PAPILLARY CARCINOMA - TALL CELL (>= 50% TALL CELL FEATURES) |
|---|---|---|---|---|
| ALL | 16 | 87 | 50 | 18 |
| subtype0 | 16 | 86 | 44 | 17 |
| subtype1 | 0 | 1 | 6 | 1 |
Figure S2. Get High-res Image Gene #2: 'FUSION PAX8' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
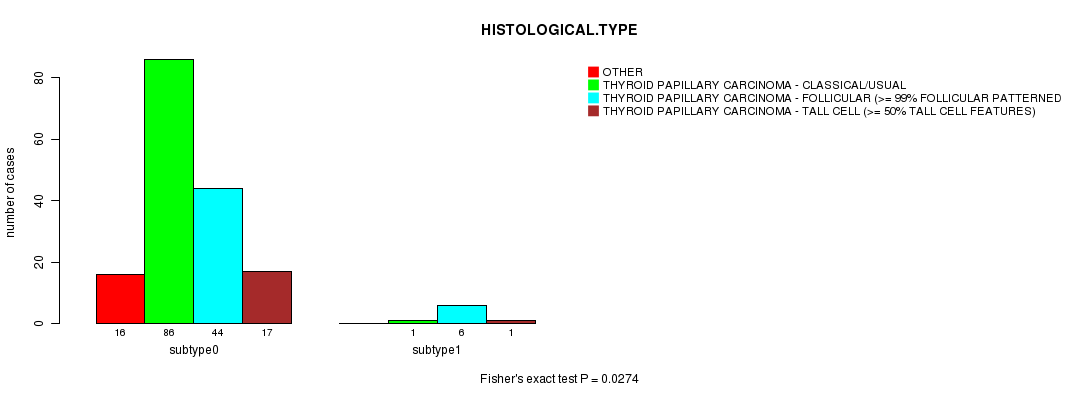
P value = 1 (Fisher's exact test), Q value = 1
Table S17. Gene #2: 'FUSION PAX8' versus Clinical Feature #4: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 9 | 162 |
| subtype0 | 9 | 154 |
| subtype1 | 0 | 8 |
P value = 1 (Fisher's exact test), Q value = 1
Table S18. Gene #2: 'FUSION PAX8' versus Clinical Feature #5: 'RADIATIONEXPOSURE'
| nPatients | NO | YES |
|---|---|---|
| ALL | 143 | 8 |
| subtype0 | 136 | 8 |
| subtype1 | 7 | 0 |
P value = 0.759 (Fisher's exact test), Q value = 1
Table S19. Gene #2: 'FUSION PAX8' versus Clinical Feature #6: 'DISTANT.METASTASIS'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 70 | 3 | 97 |
| subtype0 | 66 | 3 | 93 |
| subtype1 | 4 | 0 | 4 |
P value = 0.712 (Fisher's exact test), Q value = 1
Table S20. Gene #2: 'FUSION PAX8' versus Clinical Feature #7: 'EXTRATHYROIDAL.EXTENSION'
| nPatients | MINIMAL (T3) | MODERATE/ADVANCED (T4A) | NONE |
|---|---|---|---|
| ALL | 38 | 2 | 122 |
| subtype0 | 37 | 2 | 115 |
| subtype1 | 1 | 0 | 7 |
P value = 0.607 (Chi-square test), Q value = 1
Table S21. Gene #2: 'FUSION PAX8' versus Clinical Feature #8: 'LYMPH.NODE.METASTASIS'
| nPatients | N0 | N1 | N1A | N1B | NX |
|---|---|---|---|---|---|
| ALL | 84 | 7 | 35 | 24 | 21 |
| subtype0 | 80 | 7 | 33 | 24 | 19 |
| subtype1 | 4 | 0 | 2 | 0 | 2 |
P value = 0.726 (Fisher's exact test), Q value = 1
Table S22. Gene #2: 'FUSION PAX8' versus Clinical Feature #9: 'COMPLETENESS.OF.RESECTION'
| nPatients | R0 | R1 | RX |
|---|---|---|---|
| ALL | 131 | 14 | 12 |
| subtype0 | 125 | 13 | 12 |
| subtype1 | 6 | 1 | 0 |
P value = 0.801 (t-test), Q value = 1
Table S23. Gene #2: 'FUSION PAX8' versus Clinical Feature #10: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 135 | 2.7 (4.8) |
| subtype0 | 132 | 2.7 (4.8) |
| subtype1 | 3 | 2.3 (2.1) |
P value = 0.757 (Chi-square test), Q value = 1
Table S24. Gene #2: 'FUSION PAX8' versus Clinical Feature #12: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IVA | STAGE IVC |
|---|---|---|---|---|---|
| ALL | 91 | 25 | 38 | 13 | 3 |
| subtype0 | 86 | 23 | 37 | 13 | 3 |
| subtype1 | 5 | 2 | 1 | 0 | 0 |
P value = 0.243 (Fisher's exact test), Q value = 1
Table S25. Gene #2: 'FUSION PAX8' versus Clinical Feature #13: 'MULTIFOCALITY'
| nPatients | MULTIFOCAL | UNIFOCAL |
|---|---|---|
| ALL | 90 | 72 |
| subtype0 | 88 | 67 |
| subtype1 | 2 | 5 |
P value = 0.0518 (t-test), Q value = 1
Table S26. Gene #2: 'FUSION PAX8' versus Clinical Feature #14: 'TUMOR.SIZE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 147 | 2.9 (1.5) |
| subtype0 | 140 | 2.8 (1.6) |
| subtype1 | 7 | 3.7 (0.9) |
P value = 0.372 (t-test), Q value = 1
Table S27. Gene #3: 'FUSION BRAF' versus Clinical Feature #1: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 171 | 47.5 (15.5) |
| subtype0 | 168 | 47.6 (15.6) |
| subtype1 | 3 | 39.0 (13.1) |
P value = 0.553 (Fisher's exact test), Q value = 1
Table S28. Gene #3: 'FUSION BRAF' versus Clinical Feature #2: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 118 | 53 |
| subtype0 | 115 | 53 |
| subtype1 | 3 | 0 |
P value = 1 (Fisher's exact test), Q value = 1
Table S29. Gene #3: 'FUSION BRAF' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | OTHER | THYROID PAPILLARY CARCINOMA - CLASSICAL/USUAL | THYROID PAPILLARY CARCINOMA - FOLLICULAR (>= 99% FOLLICULAR PATTERNED) | THYROID PAPILLARY CARCINOMA - TALL CELL (>= 50% TALL CELL FEATURES) |
|---|---|---|---|---|
| ALL | 16 | 87 | 50 | 18 |
| subtype0 | 16 | 85 | 49 | 18 |
| subtype1 | 0 | 2 | 1 | 0 |
P value = 1 (Fisher's exact test), Q value = 1
Table S30. Gene #3: 'FUSION BRAF' versus Clinical Feature #4: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 9 | 162 |
| subtype0 | 9 | 159 |
| subtype1 | 0 | 3 |
P value = 1 (Fisher's exact test), Q value = 1
Table S31. Gene #3: 'FUSION BRAF' versus Clinical Feature #5: 'RADIATIONEXPOSURE'
| nPatients | NO | YES |
|---|---|---|
| ALL | 143 | 8 |
| subtype0 | 140 | 8 |
| subtype1 | 3 | 0 |
P value = 0.595 (Fisher's exact test), Q value = 1
Table S32. Gene #3: 'FUSION BRAF' versus Clinical Feature #6: 'DISTANT.METASTASIS'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 70 | 3 | 97 |
| subtype0 | 68 | 3 | 96 |
| subtype1 | 2 | 0 | 1 |
P value = 1 (Fisher's exact test), Q value = 1
Table S33. Gene #3: 'FUSION BRAF' versus Clinical Feature #7: 'EXTRATHYROIDAL.EXTENSION'
| nPatients | MINIMAL (T3) | MODERATE/ADVANCED (T4A) | NONE |
|---|---|---|---|
| ALL | 38 | 2 | 122 |
| subtype0 | 38 | 2 | 119 |
| subtype1 | 0 | 0 | 3 |
P value = 0.725 (Chi-square test), Q value = 1
Table S34. Gene #3: 'FUSION BRAF' versus Clinical Feature #8: 'LYMPH.NODE.METASTASIS'
| nPatients | N0 | N1 | N1A | N1B | NX |
|---|---|---|---|---|---|
| ALL | 84 | 7 | 35 | 24 | 21 |
| subtype0 | 83 | 7 | 34 | 24 | 20 |
| subtype1 | 1 | 0 | 1 | 0 | 1 |
P value = 1 (Fisher's exact test), Q value = 1
Table S35. Gene #3: 'FUSION BRAF' versus Clinical Feature #9: 'COMPLETENESS.OF.RESECTION'
| nPatients | R0 | R1 | RX |
|---|---|---|---|
| ALL | 131 | 14 | 12 |
| subtype0 | 128 | 14 | 12 |
| subtype1 | 3 | 0 | 0 |
P value = 0.618 (Chi-square test), Q value = 1
Table S36. Gene #3: 'FUSION BRAF' versus Clinical Feature #12: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IVA | STAGE IVC |
|---|---|---|---|---|---|
| ALL | 91 | 25 | 38 | 13 | 3 |
| subtype0 | 88 | 25 | 38 | 13 | 3 |
| subtype1 | 3 | 0 | 0 | 0 | 0 |
P value = 0.585 (Fisher's exact test), Q value = 1
Table S37. Gene #3: 'FUSION BRAF' versus Clinical Feature #13: 'MULTIFOCALITY'
| nPatients | MULTIFOCAL | UNIFOCAL |
|---|---|---|
| ALL | 90 | 72 |
| subtype0 | 89 | 70 |
| subtype1 | 1 | 2 |
P value = 0.507 (t-test), Q value = 1
Table S38. Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #1: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 136 | 46.0 (15.4) |
| subtype0 | 115 | 45.7 (15.7) |
| subtype1 | 21 | 47.9 (13.4) |
P value = 0.591 (Fisher's exact test), Q value = 1
Table S39. Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #2: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 100 | 36 |
| subtype0 | 83 | 32 |
| subtype1 | 17 | 4 |
P value = 0.0757 (Fisher's exact test), Q value = 1
Table S40. Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | OTHER | THYROID PAPILLARY CARCINOMA - CLASSICAL/USUAL | THYROID PAPILLARY CARCINOMA - FOLLICULAR (>= 99% FOLLICULAR PATTERNED) | THYROID PAPILLARY CARCINOMA - TALL CELL (>= 50% TALL CELL FEATURES) |
|---|---|---|---|---|
| ALL | 1 | 84 | 34 | 17 |
| subtype0 | 1 | 72 | 25 | 17 |
| subtype1 | 0 | 12 | 9 | 0 |
P value = 0.212 (Fisher's exact test), Q value = 1
Table S41. Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #4: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 12 | 124 |
| subtype0 | 12 | 103 |
| subtype1 | 0 | 21 |
P value = 1 (Fisher's exact test), Q value = 1
Table S42. Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #5: 'RADIATIONEXPOSURE'
| nPatients | NO | YES |
|---|---|---|
| ALL | 114 | 6 |
| subtype0 | 99 | 6 |
| subtype1 | 15 | 0 |
P value = 0.445 (Fisher's exact test), Q value = 1
Table S43. Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #6: 'DISTANT.METASTASIS'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 59 | 1 | 75 |
| subtype0 | 52 | 1 | 61 |
| subtype1 | 7 | 0 | 14 |
P value = 0.696 (Fisher's exact test), Q value = 1
Table S44. Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #7: 'EXTRATHYROIDAL.EXTENSION'
| nPatients | MINIMAL (T3) | MODERATE/ADVANCED (T4A) | NONE |
|---|---|---|---|
| ALL | 38 | 2 | 93 |
| subtype0 | 34 | 2 | 78 |
| subtype1 | 4 | 0 | 15 |
P value = 0.315 (Chi-square test), Q value = 1
Table S45. Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #8: 'LYMPH.NODE.METASTASIS'
| nPatients | N0 | N1 | N1A | N1B | NX |
|---|---|---|---|---|---|
| ALL | 66 | 8 | 31 | 17 | 14 |
| subtype0 | 53 | 8 | 29 | 14 | 11 |
| subtype1 | 13 | 0 | 2 | 3 | 3 |
P value = 0.0292 (Fisher's exact test), Q value = 1
Table S46. Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #9: 'COMPLETENESS.OF.RESECTION'
| nPatients | R0 | R1 | RX |
|---|---|---|---|
| ALL | 112 | 11 | 6 |
| subtype0 | 100 | 9 | 3 |
| subtype1 | 12 | 2 | 3 |
Figure S3. Get High-res Image Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #9: 'COMPLETENESS.OF.RESECTION'

P value = 0.867 (t-test), Q value = 1
Table S47. Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #10: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 106 | 2.8 (4.8) |
| subtype0 | 90 | 2.9 (4.8) |
| subtype1 | 16 | 2.6 (5.1) |
P value = 0.268 (Chi-square test), Q value = 1
Table S48. Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #12: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IVA | STAGE IVC |
|---|---|---|---|---|---|
| ALL | 75 | 18 | 31 | 10 | 1 |
| subtype0 | 65 | 12 | 27 | 9 | 1 |
| subtype1 | 10 | 6 | 4 | 1 | 0 |
P value = 1 (Fisher's exact test), Q value = 1
Table S49. Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #13: 'MULTIFOCALITY'
| nPatients | MULTIFOCAL | UNIFOCAL |
|---|---|---|
| ALL | 64 | 68 |
| subtype0 | 54 | 58 |
| subtype1 | 10 | 10 |
P value = 0.0303 (t-test), Q value = 1
Table S50. Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #14: 'TUMOR.SIZE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 98 | 2.8 (1.4) |
| subtype0 | 88 | 2.8 (1.4) |
| subtype1 | 10 | 2.2 (0.7) |
Figure S4. Get High-res Image Gene #4: 'MUTATION MUT BRAF' versus Clinical Feature #14: 'TUMOR.SIZE'
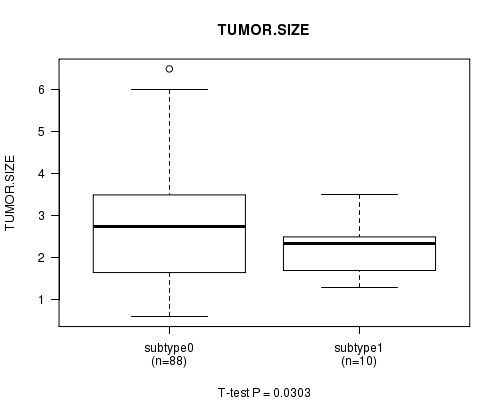
P value = 0.0416 (t-test), Q value = 1
Table S51. Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #1: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 136 | 46.0 (15.4) |
| subtype0 | 125 | 46.9 (15.2) |
| subtype1 | 11 | 36.2 (14.9) |
Figure S5. Get High-res Image Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #1: 'AGE'

P value = 0.287 (Fisher's exact test), Q value = 1
Table S52. Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #2: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 100 | 36 |
| subtype0 | 90 | 35 |
| subtype1 | 10 | 1 |
P value = 0.219 (Fisher's exact test), Q value = 1
Table S53. Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | OTHER | THYROID PAPILLARY CARCINOMA - CLASSICAL/USUAL | THYROID PAPILLARY CARCINOMA - FOLLICULAR (>= 99% FOLLICULAR PATTERNED) | THYROID PAPILLARY CARCINOMA - TALL CELL (>= 50% TALL CELL FEATURES) |
|---|---|---|---|---|
| ALL | 1 | 84 | 34 | 17 |
| subtype0 | 1 | 74 | 33 | 17 |
| subtype1 | 0 | 10 | 1 | 0 |
P value = 0.0582 (Fisher's exact test), Q value = 1
Table S54. Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #4: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 12 | 124 |
| subtype0 | 9 | 116 |
| subtype1 | 3 | 8 |
P value = 1 (Fisher's exact test), Q value = 1
Table S55. Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #5: 'RADIATIONEXPOSURE'
| nPatients | NO | YES |
|---|---|---|
| ALL | 114 | 6 |
| subtype0 | 103 | 6 |
| subtype1 | 11 | 0 |
P value = 0.399 (Fisher's exact test), Q value = 1
Table S56. Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #6: 'DISTANT.METASTASIS'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 59 | 1 | 75 |
| subtype0 | 56 | 1 | 67 |
| subtype1 | 3 | 0 | 8 |
P value = 1 (Fisher's exact test), Q value = 1
Table S57. Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #7: 'EXTRATHYROIDAL.EXTENSION'
| nPatients | MINIMAL (T3) | MODERATE/ADVANCED (T4A) | NONE |
|---|---|---|---|
| ALL | 38 | 2 | 93 |
| subtype0 | 35 | 2 | 85 |
| subtype1 | 3 | 0 | 8 |
P value = 0.0291 (Chi-square test), Q value = 1
Table S58. Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #8: 'LYMPH.NODE.METASTASIS'
| nPatients | N0 | N1 | N1A | N1B | NX |
|---|---|---|---|---|---|
| ALL | 66 | 8 | 31 | 17 | 14 |
| subtype0 | 61 | 5 | 29 | 16 | 14 |
| subtype1 | 5 | 3 | 2 | 1 | 0 |
Figure S6. Get High-res Image Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #8: 'LYMPH.NODE.METASTASIS'
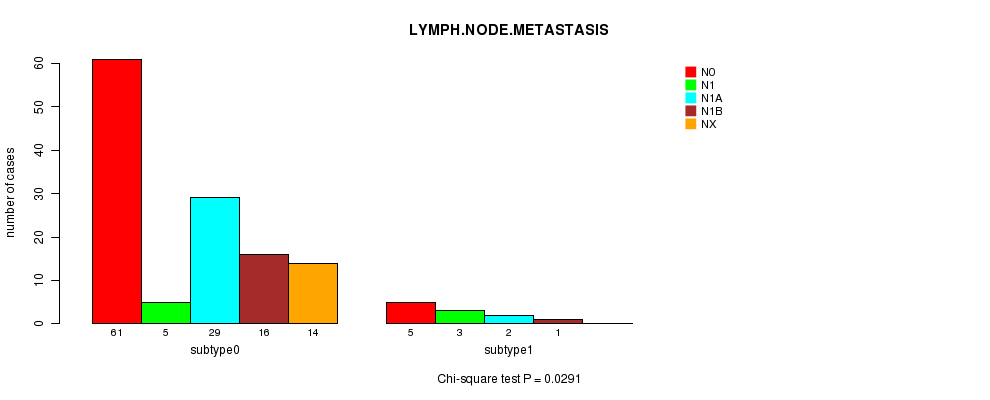
P value = 0.766 (Fisher's exact test), Q value = 1
Table S59. Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #9: 'COMPLETENESS.OF.RESECTION'
| nPatients | R0 | R1 | RX |
|---|---|---|---|
| ALL | 112 | 11 | 6 |
| subtype0 | 101 | 11 | 6 |
| subtype1 | 11 | 0 | 0 |
P value = 0.469 (t-test), Q value = 1
Table S60. Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #10: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 106 | 2.8 (4.8) |
| subtype0 | 97 | 2.7 (4.7) |
| subtype1 | 9 | 4.2 (5.9) |
P value = 0.16 (Chi-square test), Q value = 1
Table S61. Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #12: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IVA | STAGE IVC |
|---|---|---|---|---|---|
| ALL | 75 | 18 | 31 | 10 | 1 |
| subtype0 | 65 | 17 | 31 | 10 | 1 |
| subtype1 | 10 | 1 | 0 | 0 | 0 |
P value = 1 (Fisher's exact test), Q value = 1
Table S62. Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #13: 'MULTIFOCALITY'
| nPatients | MULTIFOCAL | UNIFOCAL |
|---|---|---|
| ALL | 64 | 68 |
| subtype0 | 59 | 62 |
| subtype1 | 5 | 6 |
P value = 0.638 (t-test), Q value = 1
Table S63. Gene #5: 'MUTATION MUT NRAS' versus Clinical Feature #14: 'TUMOR.SIZE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 98 | 2.8 (1.4) |
| subtype0 | 89 | 2.8 (1.4) |
| subtype1 | 9 | 2.6 (1.4) |
P value = 0.00121 (t-test), Q value = 0.092
Table S64. Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #1: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 136 | 46.0 (15.4) |
| subtype0 | 126 | 46.9 (15.4) |
| subtype1 | 10 | 34.6 (8.6) |
Figure S7. Get High-res Image Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #1: 'AGE'

P value = 1 (Fisher's exact test), Q value = 1
Table S65. Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #2: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 100 | 36 |
| subtype0 | 92 | 34 |
| subtype1 | 8 | 2 |
P value = 0.338 (Fisher's exact test), Q value = 1
Table S66. Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #3: 'HISTOLOGICAL.TYPE'
| nPatients | OTHER | THYROID PAPILLARY CARCINOMA - CLASSICAL/USUAL | THYROID PAPILLARY CARCINOMA - FOLLICULAR (>= 99% FOLLICULAR PATTERNED) | THYROID PAPILLARY CARCINOMA - TALL CELL (>= 50% TALL CELL FEATURES) |
|---|---|---|---|---|
| ALL | 1 | 84 | 34 | 17 |
| subtype0 | 1 | 75 | 33 | 17 |
| subtype1 | 0 | 9 | 1 | 0 |
P value = 0.599 (Fisher's exact test), Q value = 1
Table S67. Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #4: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 12 | 124 |
| subtype0 | 12 | 114 |
| subtype1 | 0 | 10 |
P value = 1 (Fisher's exact test), Q value = 1
Table S68. Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #5: 'RADIATIONEXPOSURE'
| nPatients | NO | YES |
|---|---|---|
| ALL | 114 | 6 |
| subtype0 | 104 | 6 |
| subtype1 | 10 | 0 |
P value = 0.00304 (Fisher's exact test), Q value = 0.22
Table S69. Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #6: 'DISTANT.METASTASIS'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 59 | 1 | 75 |
| subtype0 | 59 | 1 | 65 |
| subtype1 | 0 | 0 | 10 |
Figure S8. Get High-res Image Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #6: 'DISTANT.METASTASIS'

P value = 0.384 (Fisher's exact test), Q value = 1
Table S70. Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #7: 'EXTRATHYROIDAL.EXTENSION'
| nPatients | MINIMAL (T3) | MODERATE/ADVANCED (T4A) | NONE |
|---|---|---|---|
| ALL | 38 | 2 | 93 |
| subtype0 | 37 | 2 | 84 |
| subtype1 | 1 | 0 | 9 |
P value = 0.0227 (Chi-square test), Q value = 1
Table S71. Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #8: 'LYMPH.NODE.METASTASIS'
| nPatients | N0 | N1 | N1A | N1B | NX |
|---|---|---|---|---|---|
| ALL | 66 | 8 | 31 | 17 | 14 |
| subtype0 | 64 | 8 | 26 | 17 | 11 |
| subtype1 | 2 | 0 | 5 | 0 | 3 |
Figure S9. Get High-res Image Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #8: 'LYMPH.NODE.METASTASIS'

P value = 0.754 (Fisher's exact test), Q value = 1
Table S72. Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #9: 'COMPLETENESS.OF.RESECTION'
| nPatients | R0 | R1 | RX |
|---|---|---|---|
| ALL | 112 | 11 | 6 |
| subtype0 | 102 | 11 | 6 |
| subtype1 | 10 | 0 | 0 |
P value = 0.5 (t-test), Q value = 1
Table S73. Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #10: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 106 | 2.8 (4.8) |
| subtype0 | 99 | 2.9 (4.9) |
| subtype1 | 7 | 2.1 (2.4) |
P value = 0.247 (Chi-square test), Q value = 1
Table S74. Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #12: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IVA | STAGE IVC |
|---|---|---|---|---|---|
| ALL | 75 | 18 | 31 | 10 | 1 |
| subtype0 | 66 | 18 | 30 | 10 | 1 |
| subtype1 | 9 | 0 | 1 | 0 | 0 |
P value = 0.745 (Fisher's exact test), Q value = 1
Table S75. Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #13: 'MULTIFOCALITY'
| nPatients | MULTIFOCAL | UNIFOCAL |
|---|---|---|
| ALL | 64 | 68 |
| subtype0 | 60 | 62 |
| subtype1 | 4 | 6 |
P value = 0.993 (t-test), Q value = 1
Table S76. Gene #6: 'MUTATION MUT HRAS' versus Clinical Feature #14: 'TUMOR.SIZE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 98 | 2.8 (1.4) |
| subtype0 | 91 | 2.8 (1.4) |
| subtype1 | 7 | 2.8 (1.4) |
-
Mutation data file = THCA-TP.mergedCustomEvents_for_correlateion.txt
-
Clinical data file = THCA-TP.clin.merged.picked.txt
-
Number of patients = 229
-
Number of significantly mutated genes = 6
-
Number of selected clinical features = 14
-
Exclude genes that fewer than K tumors have mutations, K = 3
For continuous numerical clinical features, two-tailed Student's t test with unequal variance (Lehmann and Romano 2005) was applied to compare the clinical values between tumors with and without gene mutations using 't.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.