This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 9 genes and 8 molecular subtypes across 56 patients, 2 significant findings detected with P value < 0.05 and Q value < 0.25.
-
PTEN mutation correlated to 'METHLYATION_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between mutation status of 9 genes and 8 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 2 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Fisher's exact test | Chi-square test | Fisher's exact test | Chi-square test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| PTEN | 10 (18%) | 46 |
0.0264 (1.00) |
0.000529 (0.0381) |
0.00902 (0.632) |
0.000947 (0.0672) |
0.141 (1.00) |
0.0169 (1.00) |
0.038 (1.00) |
0.0442 (1.00) |
| FBXW7 | 21 (38%) | 35 |
0.0476 (1.00) |
0.697 (1.00) |
0.725 (1.00) |
0.715 (1.00) |
0.347 (1.00) |
1 (1.00) |
0.9 (1.00) |
0.671 (1.00) |
| KRAS | 7 (12%) | 49 |
0.322 (1.00) |
0.0205 (1.00) |
0.151 (1.00) |
0.0976 (1.00) |
0.464 (1.00) |
0.496 (1.00) |
0.54 (1.00) |
0.327 (1.00) |
| TP53 | 50 (89%) | 6 |
0.338 (1.00) |
0.54 (1.00) |
1 (1.00) |
0.752 (1.00) |
1 (1.00) |
0.694 (1.00) |
1 (1.00) |
1 (1.00) |
| PPP2R1A | 15 (27%) | 41 |
1 (1.00) |
0.807 (1.00) |
0.728 (1.00) |
0.973 (1.00) |
0.503 (1.00) |
0.72 (1.00) |
0.185 (1.00) |
1 (1.00) |
| PIK3CA | 19 (34%) | 37 |
0.33 (1.00) |
0.967 (1.00) |
0.328 (1.00) |
0.547 (1.00) |
0.142 (1.00) |
0.0374 (1.00) |
0.226 (1.00) |
0.0369 (1.00) |
| PCDHAC2 | 11 (20%) | 45 |
0.156 (1.00) |
0.126 (1.00) |
0.538 (1.00) |
0.896 (1.00) |
0.597 (1.00) |
0.586 (1.00) |
0.434 (1.00) |
0.435 (1.00) |
| PIK3R1 | 6 (11%) | 50 |
0.859 (1.00) |
0.952 (1.00) |
0.574 (1.00) |
0.689 (1.00) |
0.425 (1.00) |
0.706 (1.00) |
0.804 (1.00) |
0.464 (1.00) |
| ZBTB7B | 6 (11%) | 50 |
0.49 (1.00) |
0.952 (1.00) |
1 (1.00) |
0.844 (1.00) |
0.104 (1.00) |
0.194 (1.00) |
0.152 (1.00) |
0.287 (1.00) |
P value = 0.0476 (Fisher's exact test), Q value = 1
Table S1. Gene #1: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 23 | 10 |
| FBXW7 MUTATED | 12 | 8 | 1 |
| FBXW7 WILD-TYPE | 10 | 15 | 9 |
Figure S1. Get High-res Image Gene #1: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.697 (Chi-square test), Q value = 1
Table S2. Gene #1: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 9 | 13 | 13 |
| FBXW7 MUTATED | 6 | 4 | 3 | 3 | 5 |
| FBXW7 WILD-TYPE | 6 | 5 | 6 | 10 | 8 |
P value = 0.725 (Fisher's exact test), Q value = 1
Table S3. Gene #1: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 20 | 18 |
| FBXW7 MUTATED | 8 | 7 | 6 |
| FBXW7 WILD-TYPE | 9 | 13 | 12 |
P value = 0.715 (Chi-square test), Q value = 1
Table S4. Gene #1: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 4 | 3 | 15 | 12 | 21 |
| FBXW7 MUTATED | 1 | 1 | 8 | 4 | 7 |
| FBXW7 WILD-TYPE | 3 | 2 | 7 | 8 | 14 |
P value = 0.347 (Fisher's exact test), Q value = 1
Table S5. Gene #1: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 22 | 4 | 18 |
| FBXW7 MUTATED | 5 | 8 | 3 | 5 |
| FBXW7 WILD-TYPE | 6 | 14 | 1 | 13 |
P value = 1 (Fisher's exact test), Q value = 1
Table S6. Gene #1: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 26 | 28 |
| FBXW7 MUTATED | 0 | 10 | 11 |
| FBXW7 WILD-TYPE | 1 | 16 | 17 |
P value = 0.9 (Fisher's exact test), Q value = 1
Table S7. Gene #1: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 23 | 5 | 16 |
| FBXW7 MUTATED | 5 | 9 | 2 | 5 |
| FBXW7 WILD-TYPE | 6 | 14 | 3 | 11 |
P value = 0.671 (Fisher's exact test), Q value = 1
Table S8. Gene #1: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 25 | 29 |
| FBXW7 MUTATED | 0 | 11 | 10 |
| FBXW7 WILD-TYPE | 1 | 14 | 19 |
P value = 0.322 (Fisher's exact test), Q value = 1
Table S9. Gene #2: 'KRAS MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 23 | 10 |
| KRAS MUTATED | 2 | 5 | 0 |
| KRAS WILD-TYPE | 20 | 18 | 10 |
P value = 0.0205 (Chi-square test), Q value = 1
Table S10. Gene #2: 'KRAS MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 9 | 13 | 13 |
| KRAS MUTATED | 0 | 1 | 1 | 0 | 5 |
| KRAS WILD-TYPE | 12 | 8 | 8 | 13 | 8 |
Figure S2. Get High-res Image Gene #2: 'KRAS MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.151 (Fisher's exact test), Q value = 1
Table S11. Gene #2: 'KRAS MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 20 | 18 |
| KRAS MUTATED | 0 | 3 | 4 |
| KRAS WILD-TYPE | 17 | 17 | 14 |
P value = 0.0976 (Chi-square test), Q value = 1
Table S12. Gene #2: 'KRAS MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 4 | 3 | 15 | 12 | 21 |
| KRAS MUTATED | 0 | 0 | 0 | 4 | 3 |
| KRAS WILD-TYPE | 4 | 3 | 15 | 8 | 18 |
P value = 0.464 (Fisher's exact test), Q value = 1
Table S13. Gene #2: 'KRAS MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 22 | 4 | 18 |
| KRAS MUTATED | 1 | 4 | 1 | 1 |
| KRAS WILD-TYPE | 10 | 18 | 3 | 17 |
P value = 0.496 (Fisher's exact test), Q value = 1
Table S14. Gene #2: 'KRAS MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 26 | 28 |
| KRAS MUTATED | 0 | 2 | 5 |
| KRAS WILD-TYPE | 1 | 24 | 23 |
P value = 0.54 (Fisher's exact test), Q value = 1
Table S15. Gene #2: 'KRAS MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 23 | 5 | 16 |
| KRAS MUTATED | 1 | 5 | 0 | 1 |
| KRAS WILD-TYPE | 10 | 18 | 5 | 15 |
P value = 0.327 (Fisher's exact test), Q value = 1
Table S16. Gene #2: 'KRAS MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 25 | 29 |
| KRAS MUTATED | 0 | 5 | 2 |
| KRAS WILD-TYPE | 1 | 20 | 27 |
P value = 0.338 (Fisher's exact test), Q value = 1
Table S17. Gene #3: 'TP53 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 23 | 10 |
| TP53 MUTATED | 21 | 20 | 8 |
| TP53 WILD-TYPE | 1 | 3 | 2 |
P value = 0.54 (Chi-square test), Q value = 1
Table S18. Gene #3: 'TP53 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 9 | 13 | 13 |
| TP53 MUTATED | 12 | 8 | 7 | 12 | 11 |
| TP53 WILD-TYPE | 0 | 1 | 2 | 1 | 2 |
P value = 1 (Fisher's exact test), Q value = 1
Table S19. Gene #3: 'TP53 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 20 | 18 |
| TP53 MUTATED | 15 | 18 | 16 |
| TP53 WILD-TYPE | 2 | 2 | 2 |
P value = 0.752 (Chi-square test), Q value = 1
Table S20. Gene #3: 'TP53 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 4 | 3 | 15 | 12 | 21 |
| TP53 MUTATED | 3 | 3 | 14 | 10 | 19 |
| TP53 WILD-TYPE | 1 | 0 | 1 | 2 | 2 |
P value = 1 (Fisher's exact test), Q value = 1
Table S21. Gene #3: 'TP53 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 22 | 4 | 18 |
| TP53 MUTATED | 10 | 20 | 4 | 16 |
| TP53 WILD-TYPE | 1 | 2 | 0 | 2 |
P value = 0.694 (Fisher's exact test), Q value = 1
Table S22. Gene #3: 'TP53 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 26 | 28 |
| TP53 MUTATED | 1 | 23 | 26 |
| TP53 WILD-TYPE | 0 | 3 | 2 |
P value = 1 (Fisher's exact test), Q value = 1
Table S23. Gene #3: 'TP53 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 23 | 5 | 16 |
| TP53 MUTATED | 10 | 21 | 5 | 14 |
| TP53 WILD-TYPE | 1 | 2 | 0 | 2 |
P value = 1 (Fisher's exact test), Q value = 1
Table S24. Gene #3: 'TP53 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 25 | 29 |
| TP53 MUTATED | 1 | 23 | 26 |
| TP53 WILD-TYPE | 0 | 2 | 3 |
P value = 1 (Fisher's exact test), Q value = 1
Table S25. Gene #4: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 23 | 10 |
| PPP2R1A MUTATED | 6 | 6 | 3 |
| PPP2R1A WILD-TYPE | 16 | 17 | 7 |
P value = 0.807 (Chi-square test), Q value = 1
Table S26. Gene #4: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 9 | 13 | 13 |
| PPP2R1A MUTATED | 4 | 3 | 3 | 3 | 2 |
| PPP2R1A WILD-TYPE | 8 | 6 | 6 | 10 | 11 |
P value = 0.728 (Fisher's exact test), Q value = 1
Table S27. Gene #4: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 20 | 18 |
| PPP2R1A MUTATED | 6 | 5 | 4 |
| PPP2R1A WILD-TYPE | 11 | 15 | 14 |
P value = 0.973 (Chi-square test), Q value = 1
Table S28. Gene #4: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 4 | 3 | 15 | 12 | 21 |
| PPP2R1A MUTATED | 1 | 1 | 5 | 3 | 5 |
| PPP2R1A WILD-TYPE | 3 | 2 | 10 | 9 | 16 |
P value = 0.503 (Fisher's exact test), Q value = 1
Table S29. Gene #4: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 22 | 4 | 18 |
| PPP2R1A MUTATED | 5 | 5 | 1 | 4 |
| PPP2R1A WILD-TYPE | 6 | 17 | 3 | 14 |
P value = 0.72 (Fisher's exact test), Q value = 1
Table S30. Gene #4: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 26 | 28 |
| PPP2R1A MUTATED | 0 | 8 | 7 |
| PPP2R1A WILD-TYPE | 1 | 18 | 21 |
P value = 0.185 (Fisher's exact test), Q value = 1
Table S31. Gene #4: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 23 | 5 | 16 |
| PPP2R1A MUTATED | 6 | 5 | 1 | 3 |
| PPP2R1A WILD-TYPE | 5 | 18 | 4 | 13 |
P value = 1 (Fisher's exact test), Q value = 1
Table S32. Gene #4: 'PPP2R1A MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 25 | 29 |
| PPP2R1A MUTATED | 0 | 7 | 8 |
| PPP2R1A WILD-TYPE | 1 | 18 | 21 |
P value = 0.33 (Fisher's exact test), Q value = 1
Table S33. Gene #5: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 23 | 10 |
| PIK3CA MUTATED | 5 | 10 | 3 |
| PIK3CA WILD-TYPE | 17 | 13 | 7 |
P value = 0.967 (Chi-square test), Q value = 1
Table S34. Gene #5: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 9 | 13 | 13 |
| PIK3CA MUTATED | 4 | 3 | 4 | 4 | 4 |
| PIK3CA WILD-TYPE | 8 | 6 | 5 | 9 | 9 |
P value = 0.328 (Fisher's exact test), Q value = 1
Table S35. Gene #5: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 20 | 18 |
| PIK3CA MUTATED | 6 | 9 | 4 |
| PIK3CA WILD-TYPE | 11 | 11 | 14 |
P value = 0.547 (Chi-square test), Q value = 1
Table S36. Gene #5: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 4 | 3 | 15 | 12 | 21 |
| PIK3CA MUTATED | 1 | 0 | 6 | 3 | 9 |
| PIK3CA WILD-TYPE | 3 | 3 | 9 | 9 | 12 |
P value = 0.142 (Fisher's exact test), Q value = 1
Table S37. Gene #5: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 22 | 4 | 18 |
| PIK3CA MUTATED | 4 | 4 | 2 | 9 |
| PIK3CA WILD-TYPE | 7 | 18 | 2 | 9 |
P value = 0.0374 (Fisher's exact test), Q value = 1
Table S38. Gene #5: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 26 | 28 |
| PIK3CA MUTATED | 1 | 12 | 6 |
| PIK3CA WILD-TYPE | 0 | 14 | 22 |
Figure S3. Get High-res Image Gene #5: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
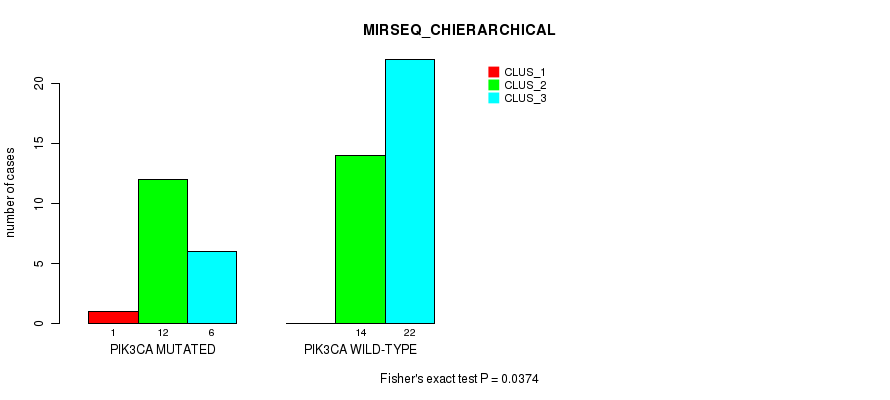
P value = 0.226 (Fisher's exact test), Q value = 1
Table S39. Gene #5: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 23 | 5 | 16 |
| PIK3CA MUTATED | 5 | 5 | 1 | 8 |
| PIK3CA WILD-TYPE | 6 | 18 | 4 | 8 |
P value = 0.0369 (Fisher's exact test), Q value = 1
Table S40. Gene #5: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 25 | 29 |
| PIK3CA MUTATED | 1 | 5 | 13 |
| PIK3CA WILD-TYPE | 0 | 20 | 16 |
Figure S4. Get High-res Image Gene #5: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 0.0264 (Fisher's exact test), Q value = 1
Table S41. Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 23 | 10 |
| PTEN MUTATED | 1 | 8 | 1 |
| PTEN WILD-TYPE | 21 | 15 | 9 |
Figure S5. Get High-res Image Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.000529 (Chi-square test), Q value = 0.038
Table S42. Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 9 | 13 | 13 |
| PTEN MUTATED | 0 | 0 | 5 | 5 | 0 |
| PTEN WILD-TYPE | 12 | 9 | 4 | 8 | 13 |
Figure S6. Get High-res Image Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.00902 (Fisher's exact test), Q value = 0.63
Table S43. Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 20 | 18 |
| PTEN MUTATED | 1 | 8 | 1 |
| PTEN WILD-TYPE | 16 | 12 | 17 |
Figure S7. Get High-res Image Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 0.000947 (Chi-square test), Q value = 0.067
Table S44. Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 4 | 3 | 15 | 12 | 21 |
| PTEN MUTATED | 3 | 0 | 0 | 0 | 7 |
| PTEN WILD-TYPE | 1 | 3 | 15 | 12 | 14 |
Figure S8. Get High-res Image Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
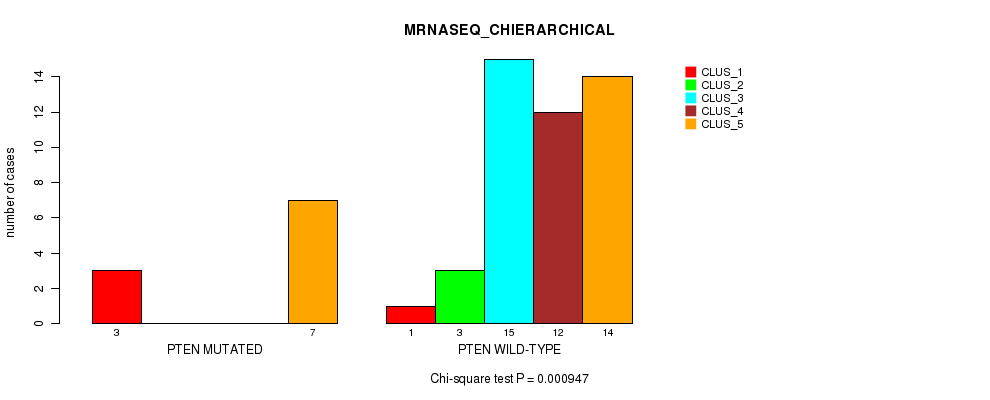
P value = 0.141 (Fisher's exact test), Q value = 1
Table S45. Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 22 | 4 | 18 |
| PTEN MUTATED | 2 | 1 | 1 | 5 |
| PTEN WILD-TYPE | 9 | 21 | 3 | 13 |
P value = 0.0169 (Fisher's exact test), Q value = 1
Table S46. Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 26 | 28 |
| PTEN MUTATED | 0 | 8 | 1 |
| PTEN WILD-TYPE | 1 | 18 | 27 |
Figure S9. Get High-res Image Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 0.038 (Fisher's exact test), Q value = 1
Table S47. Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 23 | 5 | 16 |
| PTEN MUTATED | 1 | 1 | 2 | 5 |
| PTEN WILD-TYPE | 10 | 22 | 3 | 11 |
Figure S10. Get High-res Image Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'

P value = 0.0442 (Fisher's exact test), Q value = 1
Table S48. Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 25 | 29 |
| PTEN MUTATED | 0 | 1 | 8 |
| PTEN WILD-TYPE | 1 | 24 | 21 |
Figure S11. Get High-res Image Gene #6: 'PTEN MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 0.156 (Fisher's exact test), Q value = 1
Table S49. Gene #7: 'PCDHAC2 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 23 | 10 |
| PCDHAC2 MUTATED | 2 | 7 | 1 |
| PCDHAC2 WILD-TYPE | 20 | 16 | 9 |
P value = 0.126 (Chi-square test), Q value = 1
Table S50. Gene #7: 'PCDHAC2 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 9 | 13 | 13 |
| PCDHAC2 MUTATED | 3 | 0 | 4 | 1 | 3 |
| PCDHAC2 WILD-TYPE | 9 | 9 | 5 | 12 | 10 |
P value = 0.538 (Fisher's exact test), Q value = 1
Table S51. Gene #7: 'PCDHAC2 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 20 | 18 |
| PCDHAC2 MUTATED | 4 | 5 | 2 |
| PCDHAC2 WILD-TYPE | 13 | 15 | 16 |
P value = 0.896 (Chi-square test), Q value = 1
Table S52. Gene #7: 'PCDHAC2 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 4 | 3 | 15 | 12 | 21 |
| PCDHAC2 MUTATED | 1 | 0 | 3 | 2 | 5 |
| PCDHAC2 WILD-TYPE | 3 | 3 | 12 | 10 | 16 |
P value = 0.597 (Fisher's exact test), Q value = 1
Table S53. Gene #7: 'PCDHAC2 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 22 | 4 | 18 |
| PCDHAC2 MUTATED | 2 | 3 | 0 | 5 |
| PCDHAC2 WILD-TYPE | 9 | 19 | 4 | 13 |
P value = 0.586 (Fisher's exact test), Q value = 1
Table S54. Gene #7: 'PCDHAC2 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 26 | 28 |
| PCDHAC2 MUTATED | 0 | 6 | 4 |
| PCDHAC2 WILD-TYPE | 1 | 20 | 24 |
P value = 0.434 (Fisher's exact test), Q value = 1
Table S55. Gene #7: 'PCDHAC2 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 23 | 5 | 16 |
| PCDHAC2 MUTATED | 2 | 3 | 0 | 5 |
| PCDHAC2 WILD-TYPE | 9 | 20 | 5 | 11 |
P value = 0.435 (Fisher's exact test), Q value = 1
Table S56. Gene #7: 'PCDHAC2 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 25 | 29 |
| PCDHAC2 MUTATED | 0 | 3 | 7 |
| PCDHAC2 WILD-TYPE | 1 | 22 | 22 |
P value = 0.859 (Fisher's exact test), Q value = 1
Table S57. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 23 | 10 |
| PIK3R1 MUTATED | 3 | 2 | 1 |
| PIK3R1 WILD-TYPE | 19 | 21 | 9 |
P value = 0.952 (Chi-square test), Q value = 1
Table S58. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 9 | 13 | 13 |
| PIK3R1 MUTATED | 2 | 1 | 1 | 1 | 1 |
| PIK3R1 WILD-TYPE | 10 | 8 | 8 | 12 | 12 |
P value = 0.574 (Fisher's exact test), Q value = 1
Table S59. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 20 | 18 |
| PIK3R1 MUTATED | 3 | 2 | 1 |
| PIK3R1 WILD-TYPE | 14 | 18 | 17 |
P value = 0.689 (Chi-square test), Q value = 1
Table S60. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 4 | 3 | 15 | 12 | 21 |
| PIK3R1 MUTATED | 0 | 0 | 3 | 1 | 2 |
| PIK3R1 WILD-TYPE | 4 | 3 | 12 | 11 | 19 |
P value = 0.425 (Fisher's exact test), Q value = 1
Table S61. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 22 | 4 | 18 |
| PIK3R1 MUTATED | 2 | 2 | 1 | 1 |
| PIK3R1 WILD-TYPE | 9 | 20 | 3 | 17 |
P value = 0.706 (Fisher's exact test), Q value = 1
Table S62. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 26 | 28 |
| PIK3R1 MUTATED | 0 | 2 | 4 |
| PIK3R1 WILD-TYPE | 1 | 24 | 24 |
P value = 0.804 (Fisher's exact test), Q value = 1
Table S63. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 23 | 5 | 16 |
| PIK3R1 MUTATED | 2 | 3 | 0 | 1 |
| PIK3R1 WILD-TYPE | 9 | 20 | 5 | 15 |
P value = 0.464 (Fisher's exact test), Q value = 1
Table S64. Gene #8: 'PIK3R1 MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 25 | 29 |
| PIK3R1 MUTATED | 0 | 4 | 2 |
| PIK3R1 WILD-TYPE | 1 | 21 | 27 |
P value = 0.49 (Fisher's exact test), Q value = 1
Table S65. Gene #9: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 22 | 23 | 10 |
| ZBTB7B MUTATED | 3 | 1 | 1 |
| ZBTB7B WILD-TYPE | 19 | 22 | 9 |
P value = 0.952 (Chi-square test), Q value = 1
Table S66. Gene #9: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 12 | 9 | 9 | 13 | 13 |
| ZBTB7B MUTATED | 2 | 1 | 1 | 1 | 1 |
| ZBTB7B WILD-TYPE | 10 | 8 | 8 | 12 | 12 |
P value = 1 (Fisher's exact test), Q value = 1
Table S67. Gene #9: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 17 | 20 | 18 |
| ZBTB7B MUTATED | 2 | 2 | 2 |
| ZBTB7B WILD-TYPE | 15 | 18 | 16 |
P value = 0.844 (Chi-square test), Q value = 1
Table S68. Gene #9: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 4 | 3 | 15 | 12 | 21 |
| ZBTB7B MUTATED | 1 | 0 | 2 | 1 | 2 |
| ZBTB7B WILD-TYPE | 3 | 3 | 13 | 11 | 19 |
P value = 0.104 (Fisher's exact test), Q value = 1
Table S69. Gene #9: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 22 | 4 | 18 |
| ZBTB7B MUTATED | 1 | 1 | 2 | 2 |
| ZBTB7B WILD-TYPE | 10 | 21 | 2 | 16 |
P value = 0.194 (Fisher's exact test), Q value = 1
Table S70. Gene #9: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 26 | 28 |
| ZBTB7B MUTATED | 0 | 5 | 1 |
| ZBTB7B WILD-TYPE | 1 | 21 | 27 |
P value = 0.152 (Fisher's exact test), Q value = 1
Table S71. Gene #9: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 11 | 23 | 5 | 16 |
| ZBTB7B MUTATED | 1 | 1 | 2 | 2 |
| ZBTB7B WILD-TYPE | 10 | 22 | 3 | 14 |
P value = 0.287 (Fisher's exact test), Q value = 1
Table S72. Gene #9: 'ZBTB7B MUTATION STATUS' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 25 | 29 |
| ZBTB7B MUTATED | 0 | 1 | 5 |
| ZBTB7B WILD-TYPE | 1 | 24 | 24 |
-
Mutation data file = transformed.cor.cli.txt
-
Molecular subtypes file = UCS-TP.transferedmergedcluster.txt
-
Number of patients = 56
-
Number of significantly mutated genes = 9
-
Number of Molecular subtypes = 8
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.