This pipeline computes the correlation between significant copy number variation (cnv focal) genes and selected clinical features.
Testing the association between copy number variation 62 focal events and 11 clinical features across 357 patients, 28 significant findings detected with Q value < 0.25.
-
amp_2p24.1 cnv correlated to 'GENDER'.
-
amp_2q33.1 cnv correlated to 'GENDER'.
-
amp_8q13.3 cnv correlated to 'GENDER'.
-
amp_8q24.21 cnv correlated to 'GENDER' and 'RACE'.
-
amp_17p11.2 cnv correlated to 'PATHOLOGY_T_STAGE' and 'COMPLETENESS_OF_RESECTION'.
-
amp_17q25.3 cnv correlated to 'PATHOLOGY_T_STAGE'.
-
amp_19p13.12 cnv correlated to 'GENDER'.
-
amp_xq28 cnv correlated to 'GENDER'.
-
del_3p13 cnv correlated to 'PATHOLOGY_T_STAGE' and 'HISTOLOGICAL_TYPE'.
-
del_4q21.3 cnv correlated to 'YEARS_TO_BIRTH' and 'RACE'.
-
del_4q22.3 cnv correlated to 'YEARS_TO_BIRTH' and 'RACE'.
-
del_4q35.1 cnv correlated to 'YEARS_TO_BIRTH' and 'RACE'.
-
del_6q27 cnv correlated to 'YEARS_TO_BIRTH'.
-
del_10q23.31 cnv correlated to 'RACE'.
-
del_12p12.1 cnv correlated to 'NEOPLASM_DISEASESTAGE'.
-
del_13q14.2 cnv correlated to 'PATHOLOGY_T_STAGE'.
-
del_16q23.1 cnv correlated to 'YEARS_TO_BIRTH' and 'RACE'.
-
del_17p11.2 cnv correlated to 'NEOPLASM_DISEASESTAGE' and 'PATHOLOGY_T_STAGE'.
-
del_22q13.32 cnv correlated to 'PATHOLOGY_T_STAGE' and 'RACE'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 62 focal events and 11 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 28 significant findings detected.
|
Clinical Features |
Time to Death |
YEARS TO BIRTH |
NEOPLASM DISEASESTAGE |
PATHOLOGY T STAGE |
PATHOLOGY N STAGE |
PATHOLOGY M STAGE |
GENDER |
HISTOLOGICAL TYPE |
COMPLETENESS OF RESECTION |
RACE | ETHNICITY | ||
| nCNV (%) | nWild-Type | logrank test | Wilcoxon-test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| amp 8q24 21 | 224 (63%) | 133 |
0.318 (0.782) |
0.0514 (0.428) |
0.104 (0.566) |
0.681 (0.927) |
0.237 (0.739) |
0.598 (0.886) |
0.00025 (0.0214) |
1 (1.00) |
0.609 (0.886) |
5e-05 (0.0112) |
0.541 (0.886) |
| amp 17p11 2 | 46 (13%) | 311 |
0.638 (0.889) |
0.311 (0.782) |
0.0125 (0.266) |
0.00361 (0.152) |
1 (1.00) |
0.455 (0.873) |
1 (1.00) |
1 (1.00) |
0.00988 (0.249) |
0.715 (0.94) |
0.143 (0.59) |
| del 3p13 | 69 (19%) | 288 |
0.864 (1.00) |
0.114 (0.566) |
0.0113 (0.256) |
0.00384 (0.152) |
0.471 (0.873) |
0.163 (0.613) |
0.0215 (0.326) |
0.00422 (0.152) |
0.578 (0.886) |
0.882 (1.00) |
1 (1.00) |
| del 4q21 3 | 165 (46%) | 192 |
0.942 (1.00) |
8.22e-05 (0.0112) |
0.468 (0.873) |
0.476 (0.873) |
0.606 (0.886) |
0.622 (0.886) |
0.495 (0.884) |
0.629 (0.886) |
0.729 (0.94) |
0.00162 (0.0921) |
0.121 (0.571) |
| del 4q22 3 | 166 (46%) | 191 |
0.855 (1.00) |
0.000248 (0.0214) |
0.543 (0.886) |
0.667 (0.913) |
0.609 (0.886) |
0.623 (0.886) |
0.427 (0.873) |
0.629 (0.886) |
0.732 (0.94) |
0.005 (0.155) |
0.122 (0.572) |
| del 4q35 1 | 168 (47%) | 189 |
0.0863 (0.531) |
0.000198 (0.0214) |
0.195 (0.675) |
0.102 (0.566) |
0.621 (0.886) |
0.623 (0.886) |
0.0687 (0.488) |
0.631 (0.886) |
0.467 (0.873) |
7e-05 (0.0112) |
0.764 (0.96) |
| del 16q23 1 | 159 (45%) | 198 |
0.739 (0.94) |
0.000578 (0.0394) |
0.171 (0.625) |
0.126 (0.579) |
0.609 (0.886) |
1 (1.00) |
0.209 (0.684) |
0.627 (0.886) |
0.597 (0.886) |
4e-05 (0.0112) |
0.554 (0.886) |
| del 17p11 2 | 186 (52%) | 171 |
0.0196 (0.309) |
0.712 (0.94) |
0.00849 (0.226) |
0.0086 (0.226) |
0.248 (0.756) |
0.623 (0.886) |
1 (1.00) |
0.727 (0.94) |
0.338 (0.79) |
0.791 (0.971) |
0.546 (0.886) |
| del 22q13 32 | 79 (22%) | 278 |
0.725 (0.94) |
0.404 (0.858) |
0.0379 (0.392) |
0.00496 (0.155) |
0.509 (0.886) |
1 (1.00) |
0.414 (0.867) |
0.0506 (0.428) |
0.0199 (0.309) |
0.00376 (0.152) |
0.0564 (0.437) |
| amp 2p24 1 | 64 (18%) | 293 |
0.102 (0.566) |
0.0975 (0.563) |
0.211 (0.684) |
0.0896 (0.541) |
0.0797 (0.531) |
0.131 (0.579) |
0.000963 (0.0597) |
0.739 (0.94) |
0.385 (0.841) |
0.125 (0.579) |
1 (1.00) |
| amp 2q33 1 | 62 (17%) | 295 |
0.184 (0.653) |
0.439 (0.873) |
0.0914 (0.546) |
0.0972 (0.563) |
0.471 (0.873) |
0.169 (0.624) |
3.65e-05 (0.0112) |
0.56 (0.886) |
0.546 (0.886) |
0.495 (0.884) |
0.418 (0.869) |
| amp 8q13 3 | 200 (56%) | 157 |
0.774 (0.966) |
0.0833 (0.531) |
0.158 (0.604) |
0.307 (0.782) |
0.569 (0.886) |
1 (1.00) |
0.000566 (0.0394) |
0.854 (1.00) |
0.786 (0.971) |
0.085 (0.531) |
1 (1.00) |
| amp 17q25 3 | 141 (39%) | 216 |
0.735 (0.94) |
0.872 (1.00) |
0.0174 (0.297) |
0.00447 (0.152) |
1 (1.00) |
1 (1.00) |
0.132 (0.579) |
0.622 (0.886) |
0.0337 (0.384) |
0.217 (0.697) |
0.537 (0.886) |
| amp 19p13 12 | 66 (18%) | 291 |
0.82 (0.987) |
0.578 (0.886) |
0.131 (0.579) |
0.716 (0.94) |
0.463 (0.873) |
0.0199 (0.309) |
0.0019 (0.0997) |
0.152 (0.597) |
0.3 (0.782) |
0.605 (0.886) |
0.133 (0.58) |
| amp xq28 | 83 (23%) | 274 |
0.306 (0.782) |
0.322 (0.782) |
0.928 (1.00) |
0.91 (1.00) |
1 (1.00) |
0.576 (0.886) |
0.00286 (0.139) |
0.305 (0.782) |
1 (1.00) |
0.288 (0.782) |
0.468 (0.873) |
| del 6q27 | 133 (37%) | 224 |
0.581 (0.886) |
0.00431 (0.152) |
0.71 (0.94) |
0.96 (1.00) |
0.559 (0.886) |
0.306 (0.782) |
0.29 (0.782) |
0.732 (0.94) |
0.441 (0.873) |
0.563 (0.886) |
0.219 (0.7) |
| del 10q23 31 | 95 (27%) | 262 |
0.38 (0.834) |
0.517 (0.886) |
0.105 (0.566) |
0.0237 (0.344) |
0.169 (0.624) |
0.577 (0.886) |
1 (1.00) |
0.151 (0.596) |
0.426 (0.873) |
0.00723 (0.213) |
1 (1.00) |
| del 12p12 1 | 73 (20%) | 284 |
0.615 (0.886) |
0.973 (1.00) |
0.00748 (0.213) |
0.669 (0.914) |
0.109 (0.566) |
0.205 (0.684) |
0.78 (0.966) |
0.466 (0.873) |
0.117 (0.57) |
0.577 (0.886) |
0.459 (0.873) |
| del 13q14 2 | 169 (47%) | 188 |
0.0537 (0.431) |
0.315 (0.782) |
0.332 (0.789) |
0.0102 (0.249) |
1 (1.00) |
1 (1.00) |
0.141 (0.59) |
0.855 (1.00) |
0.605 (0.886) |
0.0467 (0.428) |
1 (1.00) |
| amp 1p22 3 | 85 (24%) | 272 |
0.972 (1.00) |
0.0944 (0.555) |
0.0893 (0.541) |
0.0618 (0.458) |
0.573 (0.886) |
0.056 (0.437) |
0.425 (0.873) |
1 (1.00) |
0.0356 (0.386) |
0.157 (0.604) |
0.469 (0.873) |
| amp 1q22 | 272 (76%) | 85 |
0.203 (0.684) |
0.726 (0.94) |
0.915 (1.00) |
0.914 (1.00) |
0.538 (0.886) |
0.579 (0.886) |
0.505 (0.886) |
0.0293 (0.37) |
0.792 (0.971) |
0.244 (0.753) |
0.73 (0.94) |
| amp 1q42 3 | 261 (73%) | 96 |
0.803 (0.977) |
0.939 (1.00) |
0.397 (0.854) |
0.629 (0.886) |
0.575 (0.886) |
0.575 (0.886) |
0.608 (0.886) |
0.437 (0.873) |
0.9 (1.00) |
0.532 (0.886) |
0.501 (0.886) |
| amp 3q26 31 | 74 (21%) | 283 |
0.373 (0.826) |
0.428 (0.873) |
0.654 (0.901) |
0.632 (0.886) |
0.471 (0.873) |
1 (1.00) |
0.779 (0.966) |
0.776 (0.966) |
0.731 (0.94) |
0.192 (0.67) |
0.705 (0.94) |
| amp 4p14 | 52 (15%) | 305 |
0.0735 (0.509) |
0.0507 (0.428) |
0.479 (0.877) |
0.21 (0.684) |
1 (1.00) |
1 (1.00) |
0.262 (0.768) |
1 (1.00) |
0.432 (0.873) |
0.798 (0.973) |
1 (1.00) |
| amp 5p15 33 | 158 (44%) | 199 |
0.201 (0.684) |
0.499 (0.886) |
0.0747 (0.51) |
0.0254 (0.357) |
1 (1.00) |
0.632 (0.886) |
0.909 (1.00) |
0.626 (0.886) |
1 (1.00) |
0.367 (0.818) |
0.548 (0.886) |
| amp 5q35 3 | 131 (37%) | 226 |
0.377 (0.829) |
0.131 (0.579) |
0.743 (0.942) |
0.705 (0.94) |
0.555 (0.886) |
0.635 (0.888) |
0.345 (0.79) |
0.103 (0.566) |
0.113 (0.566) |
0.794 (0.971) |
0.538 (0.886) |
| amp 6p25 2 | 150 (42%) | 207 |
0.0699 (0.492) |
0.946 (1.00) |
0.111 (0.566) |
0.11 (0.566) |
1 (1.00) |
0.315 (0.782) |
0.302 (0.782) |
0.632 (0.886) |
0.734 (0.94) |
0.858 (1.00) |
1 (1.00) |
| amp 6p21 1 | 151 (42%) | 206 |
0.364 (0.817) |
0.0834 (0.531) |
0.324 (0.782) |
0.474 (0.873) |
0.27 (0.776) |
0.315 (0.782) |
0.107 (0.566) |
0.452 (0.873) |
0.694 (0.939) |
0.865 (1.00) |
0.766 (0.96) |
| amp 6q12 | 102 (29%) | 255 |
0.467 (0.873) |
0.551 (0.886) |
0.0273 (0.36) |
0.121 (0.571) |
0.183 (0.653) |
0.327 (0.782) |
0.258 (0.768) |
0.22 (0.7) |
0.317 (0.782) |
0.566 (0.886) |
0.508 (0.886) |
| amp 6q12 | 78 (22%) | 279 |
0.598 (0.886) |
0.476 (0.873) |
0.303 (0.782) |
0.208 (0.684) |
1 (1.00) |
0.193 (0.67) |
0.271 (0.776) |
0.28 (0.782) |
0.421 (0.872) |
0.486 (0.882) |
0.259 (0.768) |
| amp 7q31 2 | 115 (32%) | 242 |
0.82 (0.987) |
0.0789 (0.531) |
0.297 (0.782) |
0.698 (0.939) |
0.227 (0.711) |
1 (1.00) |
0.808 (0.979) |
0.494 (0.884) |
0.842 (1.00) |
0.263 (0.768) |
0.348 (0.793) |
| amp 9q34 2 | 44 (12%) | 313 |
0.616 (0.886) |
0.482 (0.879) |
0.028 (0.36) |
0.0167 (0.297) |
0.327 (0.782) |
0.416 (0.867) |
0.491 (0.884) |
1 (1.00) |
0.159 (0.605) |
0.584 (0.886) |
0.633 (0.886) |
| amp 11q13 3 | 56 (16%) | 301 |
0.434 (0.873) |
0.466 (0.873) |
0.051 (0.428) |
0.0524 (0.428) |
1 (1.00) |
0.52 (0.886) |
0.532 (0.886) |
0.717 (0.94) |
0.0468 (0.428) |
0.948 (1.00) |
1 (1.00) |
| amp 13q32 3 | 63 (18%) | 294 |
0.32 (0.782) |
0.287 (0.782) |
0.649 (0.901) |
0.495 (0.884) |
0.455 (0.873) |
0.571 (0.886) |
0.0379 (0.392) |
0.137 (0.586) |
0.538 (0.886) |
0.628 (0.886) |
0.698 (0.939) |
| amp 13q33 3 | 64 (18%) | 293 |
0.391 (0.844) |
0.41 (0.866) |
0.962 (1.00) |
0.974 (1.00) |
0.455 (0.873) |
0.571 (0.886) |
0.182 (0.653) |
0.141 (0.59) |
0.85 (1.00) |
0.151 (0.596) |
0.697 (0.939) |
| amp 15q26 3 | 53 (15%) | 304 |
0.652 (0.901) |
0.0589 (0.446) |
0.277 (0.782) |
0.261 (0.768) |
1 (1.00) |
0.502 (0.886) |
0.523 (0.886) |
0.325 (0.782) |
0.568 (0.886) |
0.816 (0.986) |
0.012 (0.264) |
| amp 19q12 | 75 (21%) | 282 |
0.799 (0.973) |
0.2 (0.684) |
0.289 (0.782) |
0.917 (1.00) |
0.494 (0.884) |
0.0279 (0.36) |
0.163 (0.613) |
0.265 (0.768) |
0.361 (0.815) |
0.592 (0.886) |
0.0628 (0.461) |
| amp 19q13 11 | 87 (24%) | 270 |
0.317 (0.782) |
0.372 (0.825) |
0.0528 (0.428) |
0.337 (0.79) |
0.524 (0.886) |
0.0378 (0.392) |
0.111 (0.566) |
0.404 (0.858) |
0.144 (0.593) |
0.209 (0.684) |
0.475 (0.873) |
| amp 20q13 13 | 118 (33%) | 239 |
0.0921 (0.546) |
0.0504 (0.428) |
0.0803 (0.531) |
0.0437 (0.428) |
0.253 (0.76) |
0.61 (0.886) |
0.117 (0.57) |
1 (1.00) |
0.278 (0.782) |
0.698 (0.939) |
1 (1.00) |
| amp 20q13 33 | 123 (34%) | 234 |
0.224 (0.705) |
0.142 (0.59) |
0.0333 (0.384) |
0.0865 (0.531) |
0.296 (0.782) |
0.146 (0.593) |
0.0319 (0.384) |
0.862 (1.00) |
0.608 (0.886) |
0.778 (0.966) |
0.755 (0.954) |
| del 1p36 31 | 159 (45%) | 198 |
0.738 (0.94) |
0.805 (0.977) |
0.971 (1.00) |
0.79 (0.971) |
0.591 (0.886) |
0.351 (0.797) |
0.138 (0.586) |
0.15 (0.596) |
0.186 (0.656) |
0.0824 (0.531) |
0.76 (0.958) |
| del 1p36 11 | 139 (39%) | 218 |
0.538 (0.886) |
0.723 (0.94) |
0.521 (0.886) |
0.343 (0.79) |
0.0671 (0.482) |
0.316 (0.782) |
0.486 (0.882) |
0.434 (0.873) |
0.184 (0.653) |
0.111 (0.566) |
0.536 (0.886) |
| del 2q22 1 | 51 (14%) | 306 |
0.254 (0.76) |
0.188 (0.66) |
0.976 (1.00) |
0.974 (1.00) |
1 (1.00) |
1 (1.00) |
0.748 (0.947) |
1 (1.00) |
0.299 (0.782) |
0.461 (0.873) |
0.654 (0.901) |
| del 2q37 3 | 60 (17%) | 297 |
0.559 (0.886) |
0.131 (0.579) |
0.822 (0.987) |
0.426 (0.873) |
0.389 (0.844) |
1 (1.00) |
0.763 (0.959) |
0.12 (0.571) |
0.364 (0.817) |
0.439 (0.873) |
0.391 (0.844) |
| del 4p16 3 | 80 (22%) | 277 |
0.302 (0.782) |
0.399 (0.856) |
0.0301 (0.373) |
0.367 (0.818) |
0.129 (0.579) |
0.242 (0.75) |
0.0136 (0.273) |
0.795 (0.971) |
0.39 (0.844) |
0.0352 (0.386) |
0.278 (0.782) |
| del 8p23 2 | 240 (67%) | 117 |
0.905 (1.00) |
0.0991 (0.566) |
0.222 (0.7) |
0.415 (0.867) |
0.553 (0.886) |
0.304 (0.782) |
0.336 (0.79) |
0.25 (0.757) |
0.919 (1.00) |
0.722 (0.94) |
1 (1.00) |
| del 8p12 | 234 (66%) | 123 |
0.659 (0.904) |
0.252 (0.76) |
0.469 (0.873) |
0.608 (0.886) |
0.553 (0.886) |
0.301 (0.782) |
0.475 (0.873) |
0.733 (0.94) |
0.876 (1.00) |
0.731 (0.94) |
0.342 (0.79) |
| del 9p21 3 | 146 (41%) | 211 |
0.43 (0.873) |
0.0235 (0.344) |
0.537 (0.886) |
0.473 (0.873) |
0.569 (0.886) |
1 (1.00) |
0.356 (0.806) |
0.732 (0.94) |
0.475 (0.873) |
0.104 (0.566) |
0.13 (0.579) |
| del 9q31 3 | 116 (32%) | 241 |
0.343 (0.79) |
0.909 (1.00) |
0.0578 (0.443) |
0.173 (0.627) |
0.0256 (0.357) |
0.0815 (0.531) |
0.717 (0.94) |
1 (1.00) |
0.162 (0.613) |
0.987 (1.00) |
0.345 (0.79) |
| del 10q26 13 | 102 (29%) | 255 |
0.206 (0.684) |
0.938 (1.00) |
0.279 (0.782) |
0.0545 (0.432) |
0.197 (0.679) |
0.58 (0.886) |
0.9 (1.00) |
0.137 (0.586) |
0.47 (0.873) |
0.0173 (0.297) |
1 (1.00) |
| del 11q14 1 | 72 (20%) | 285 |
0.965 (1.00) |
0.328 (0.783) |
0.052 (0.428) |
0.11 (0.566) |
0.121 (0.571) |
1 (1.00) |
1 (1.00) |
0.109 (0.566) |
0.55 (0.886) |
0.651 (0.901) |
0.0475 (0.428) |
| del 11q24 2 | 90 (25%) | 267 |
0.683 (0.928) |
0.222 (0.7) |
0.171 (0.625) |
0.375 (0.827) |
0.169 (0.624) |
1 (1.00) |
0.896 (1.00) |
0.245 (0.753) |
0.655 (0.901) |
0.323 (0.782) |
0.154 (0.597) |
| del 12q24 33 | 54 (15%) | 303 |
0.21 (0.684) |
0.338 (0.79) |
0.0161 (0.297) |
0.34 (0.79) |
1 (1.00) |
0.136 (0.586) |
0.266 (0.768) |
1 (1.00) |
0.879 (1.00) |
0.458 (0.873) |
0.678 (0.925) |
| del 14q23 3 | 126 (35%) | 231 |
0.819 (0.987) |
0.151 (0.596) |
0.288 (0.782) |
0.202 (0.684) |
1 (1.00) |
0.638 (0.889) |
0.405 (0.858) |
0.34 (0.79) |
0.874 (1.00) |
0.0168 (0.297) |
1 (1.00) |
| del 14q32 33 | 129 (36%) | 228 |
0.792 (0.971) |
0.0672 (0.482) |
0.153 (0.597) |
0.0355 (0.386) |
1 (1.00) |
0.632 (0.886) |
0.906 (1.00) |
0.147 (0.595) |
0.879 (1.00) |
0.14 (0.59) |
1 (1.00) |
| del 15q26 3 | 75 (21%) | 282 |
0.0618 (0.458) |
0.947 (1.00) |
0.0854 (0.531) |
0.027 (0.36) |
0.117 (0.57) |
0.575 (0.886) |
0.78 (0.966) |
0.264 (0.768) |
0.739 (0.94) |
0.453 (0.873) |
0.705 (0.94) |
| del 17p13 1 | 217 (61%) | 140 |
0.0327 (0.384) |
0.509 (0.886) |
0.0425 (0.428) |
0.0107 (0.251) |
0.284 (0.782) |
0.301 (0.782) |
0.561 (0.886) |
0.861 (1.00) |
0.588 (0.886) |
0.105 (0.566) |
0.538 (0.886) |
| del 18q21 2 | 88 (25%) | 269 |
0.872 (1.00) |
0.939 (1.00) |
0.741 (0.941) |
0.944 (1.00) |
1 (1.00) |
0.274 (0.781) |
0.292 (0.782) |
0.0519 (0.428) |
0.615 (0.886) |
0.921 (1.00) |
0.473 (0.873) |
| del 19p13 3 | 84 (24%) | 273 |
0.322 (0.782) |
0.114 (0.566) |
0.501 (0.886) |
0.215 (0.694) |
0.573 (0.886) |
1 (1.00) |
0.506 (0.886) |
1 (1.00) |
1 (1.00) |
0.0163 (0.297) |
0.733 (0.94) |
| del 20p12 1 | 27 (8%) | 330 |
0.32 (0.782) |
0.57 (0.886) |
0.317 (0.782) |
0.781 (0.966) |
1 (1.00) |
0.307 (0.782) |
0.525 (0.886) |
0.0739 (0.509) |
0.613 (0.886) |
0.557 (0.886) |
0.589 (0.886) |
| del 22q12 1 | 75 (21%) | 282 |
0.999 (1.00) |
0.247 (0.755) |
0.121 (0.571) |
0.0194 (0.309) |
1 (1.00) |
1 (1.00) |
0.402 (0.858) |
0.0427 (0.428) |
0.0502 (0.428) |
0.0135 (0.273) |
0.0504 (0.428) |
| del xq26 2 | 65 (18%) | 292 |
0.907 (1.00) |
0.0448 (0.428) |
0.603 (0.886) |
0.413 (0.867) |
0.471 (0.873) |
0.585 (0.886) |
0.556 (0.886) |
0.146 (0.593) |
0.326 (0.782) |
0.0337 (0.384) |
1 (1.00) |
P value = 0.000963 (Fisher's exact test), Q value = 0.06
Table S1. Gene #4: 'amp_2p24.1' versus Clinical Feature #7: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 113 | 244 |
| AMP PEAK 4(2P24.1) MUTATED | 32 | 32 |
| AMP PEAK 4(2P24.1) WILD-TYPE | 81 | 212 |
Figure S1. Get High-res Image Gene #4: 'amp_2p24.1' versus Clinical Feature #7: 'GENDER'

P value = 3.65e-05 (Fisher's exact test), Q value = 0.011
Table S2. Gene #5: 'amp_2q33.1' versus Clinical Feature #7: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 113 | 244 |
| AMP PEAK 5(2Q33.1) MUTATED | 34 | 28 |
| AMP PEAK 5(2Q33.1) WILD-TYPE | 79 | 216 |
Figure S2. Get High-res Image Gene #5: 'amp_2q33.1' versus Clinical Feature #7: 'GENDER'

P value = 0.000566 (Fisher's exact test), Q value = 0.039
Table S3. Gene #15: 'amp_8q13.3' versus Clinical Feature #7: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 113 | 244 |
| AMP PEAK 15(8Q13.3) MUTATED | 48 | 152 |
| AMP PEAK 15(8Q13.3) WILD-TYPE | 65 | 92 |
Figure S3. Get High-res Image Gene #15: 'amp_8q13.3' versus Clinical Feature #7: 'GENDER'

P value = 0.00025 (Fisher's exact test), Q value = 0.021
Table S4. Gene #16: 'amp_8q24.21' versus Clinical Feature #7: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 113 | 244 |
| AMP PEAK 16(8Q24.21) MUTATED | 55 | 169 |
| AMP PEAK 16(8Q24.21) WILD-TYPE | 58 | 75 |
Figure S4. Get High-res Image Gene #16: 'amp_8q24.21' versus Clinical Feature #7: 'GENDER'
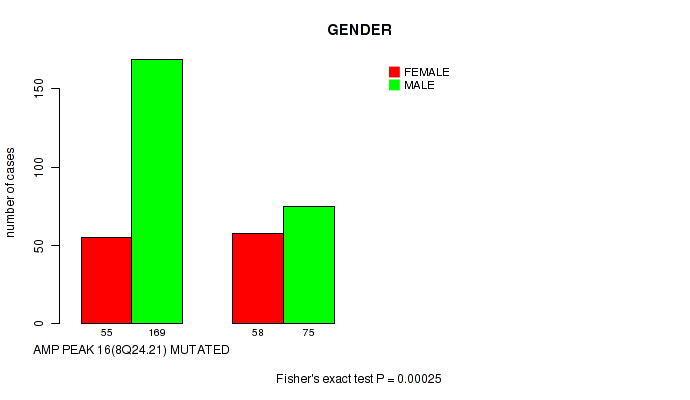
P value = 5e-05 (Fisher's exact test), Q value = 0.011
Table S5. Gene #16: 'amp_8q24.21' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | WHITE |
|---|---|---|---|---|
| ALL | 1 | 156 | 17 | 173 |
| AMP PEAK 16(8Q24.21) MUTATED | 1 | 117 | 11 | 88 |
| AMP PEAK 16(8Q24.21) WILD-TYPE | 0 | 39 | 6 | 85 |
Figure S5. Get High-res Image Gene #16: 'amp_8q24.21' versus Clinical Feature #10: 'RACE'

P value = 0.00361 (Fisher's exact test), Q value = 0.15
Table S6. Gene #22: 'amp_17p11.2' versus Clinical Feature #4: 'PATHOLOGY_T_STAGE'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 179 | 91 | 73 | 12 |
| AMP PEAK 22(17P11.2) MUTATED | 17 | 8 | 19 | 2 |
| AMP PEAK 22(17P11.2) WILD-TYPE | 162 | 83 | 54 | 10 |
Figure S6. Get High-res Image Gene #22: 'amp_17p11.2' versus Clinical Feature #4: 'PATHOLOGY_T_STAGE'

P value = 0.00988 (Fisher's exact test), Q value = 0.25
Table S7. Gene #22: 'amp_17p11.2' versus Clinical Feature #9: 'COMPLETENESS_OF_RESECTION'
| nPatients | R0 | R1 | R2 | RX |
|---|---|---|---|---|
| ALL | 315 | 15 | 1 | 19 |
| AMP PEAK 22(17P11.2) MUTATED | 37 | 5 | 1 | 3 |
| AMP PEAK 22(17P11.2) WILD-TYPE | 278 | 10 | 0 | 16 |
Figure S7. Get High-res Image Gene #22: 'amp_17p11.2' versus Clinical Feature #9: 'COMPLETENESS_OF_RESECTION'

P value = 0.00447 (Fisher's exact test), Q value = 0.15
Table S8. Gene #23: 'amp_17q25.3' versus Clinical Feature #4: 'PATHOLOGY_T_STAGE'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 179 | 91 | 73 | 12 |
| AMP PEAK 23(17Q25.3) MUTATED | 57 | 38 | 38 | 8 |
| AMP PEAK 23(17Q25.3) WILD-TYPE | 122 | 53 | 35 | 4 |
Figure S8. Get High-res Image Gene #23: 'amp_17q25.3' versus Clinical Feature #4: 'PATHOLOGY_T_STAGE'

P value = 0.0019 (Fisher's exact test), Q value = 0.1
Table S9. Gene #24: 'amp_19p13.12' versus Clinical Feature #7: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 113 | 244 |
| AMP PEAK 24(19P13.12) MUTATED | 32 | 34 |
| AMP PEAK 24(19P13.12) WILD-TYPE | 81 | 210 |
Figure S9. Get High-res Image Gene #24: 'amp_19p13.12' versus Clinical Feature #7: 'GENDER'
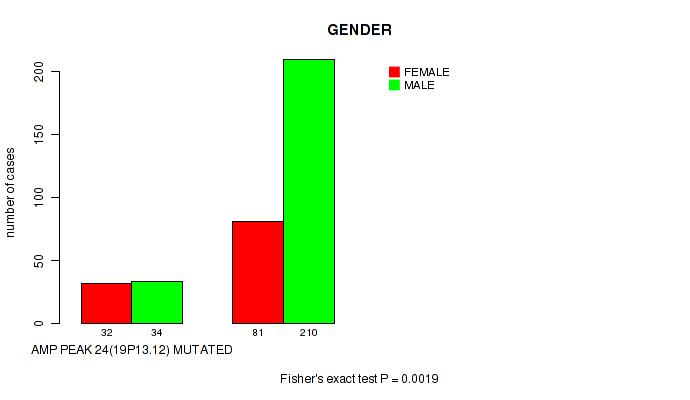
P value = 0.00286 (Fisher's exact test), Q value = 0.14
Table S10. Gene #29: 'amp_xq28' versus Clinical Feature #7: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 113 | 244 |
| AMP PEAK 29(XQ28) MUTATED | 15 | 68 |
| AMP PEAK 29(XQ28) WILD-TYPE | 98 | 176 |
Figure S10. Get High-res Image Gene #29: 'amp_xq28' versus Clinical Feature #7: 'GENDER'

P value = 0.00384 (Fisher's exact test), Q value = 0.15
Table S11. Gene #34: 'del_3p13' versus Clinical Feature #4: 'PATHOLOGY_T_STAGE'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 179 | 91 | 73 | 12 |
| DEL PEAK 5(3P13) MUTATED | 22 | 23 | 20 | 4 |
| DEL PEAK 5(3P13) WILD-TYPE | 157 | 68 | 53 | 8 |
Figure S11. Get High-res Image Gene #34: 'del_3p13' versus Clinical Feature #4: 'PATHOLOGY_T_STAGE'
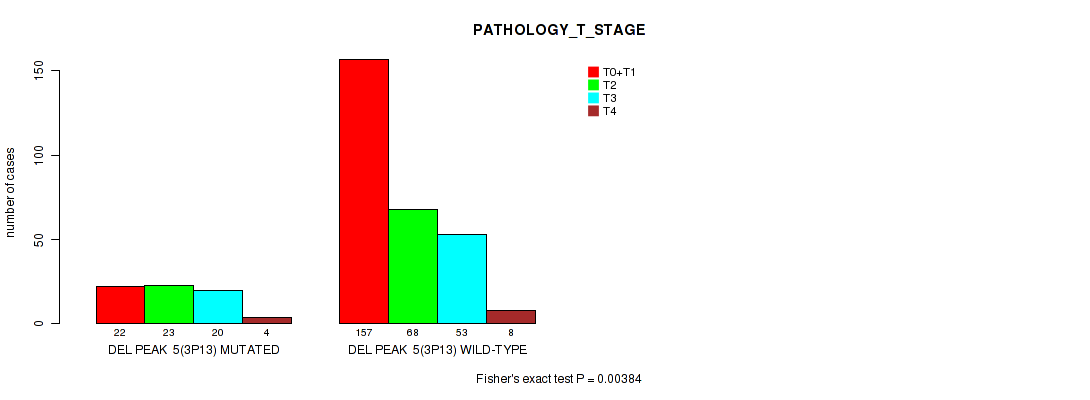
P value = 0.00422 (Fisher's exact test), Q value = 0.15
Table S12. Gene #34: 'del_3p13' versus Clinical Feature #8: 'HISTOLOGICAL_TYPE'
| nPatients | FIBROLAMELLAR CARCINOMA | HEPATOCELLULAR CARCINOMA | HEPATOCHOLANGIOCARCINOMA (MIXED) |
|---|---|---|---|
| ALL | 2 | 348 | 7 |
| DEL PEAK 5(3P13) MUTATED | 0 | 64 | 5 |
| DEL PEAK 5(3P13) WILD-TYPE | 2 | 284 | 2 |
Figure S12. Get High-res Image Gene #34: 'del_3p13' versus Clinical Feature #8: 'HISTOLOGICAL_TYPE'

P value = 8.22e-05 (Wilcoxon-test), Q value = 0.011
Table S13. Gene #36: 'del_4q21.3' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 354 | 59.8 (12.7) |
| DEL PEAK 7(4Q21.3) MUTATED | 163 | 57.2 (13.3) |
| DEL PEAK 7(4Q21.3) WILD-TYPE | 191 | 62.0 (11.8) |
Figure S13. Get High-res Image Gene #36: 'del_4q21.3' versus Clinical Feature #2: 'YEARS_TO_BIRTH'

P value = 0.00162 (Fisher's exact test), Q value = 0.092
Table S14. Gene #36: 'del_4q21.3' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | WHITE |
|---|---|---|---|---|
| ALL | 1 | 156 | 17 | 173 |
| DEL PEAK 7(4Q21.3) MUTATED | 0 | 88 | 9 | 64 |
| DEL PEAK 7(4Q21.3) WILD-TYPE | 1 | 68 | 8 | 109 |
Figure S14. Get High-res Image Gene #36: 'del_4q21.3' versus Clinical Feature #10: 'RACE'

P value = 0.000248 (Wilcoxon-test), Q value = 0.021
Table S15. Gene #37: 'del_4q22.3' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 354 | 59.8 (12.7) |
| DEL PEAK 8(4Q22.3) MUTATED | 164 | 57.3 (13.3) |
| DEL PEAK 8(4Q22.3) WILD-TYPE | 190 | 61.8 (11.8) |
Figure S15. Get High-res Image Gene #37: 'del_4q22.3' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
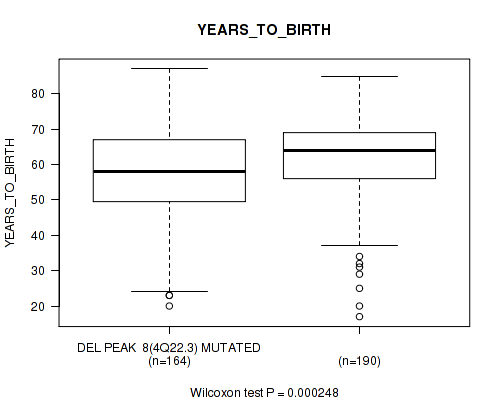
P value = 0.005 (Fisher's exact test), Q value = 0.15
Table S16. Gene #37: 'del_4q22.3' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | WHITE |
|---|---|---|---|---|
| ALL | 1 | 156 | 17 | 173 |
| DEL PEAK 8(4Q22.3) MUTATED | 0 | 87 | 9 | 66 |
| DEL PEAK 8(4Q22.3) WILD-TYPE | 1 | 69 | 8 | 107 |
Figure S16. Get High-res Image Gene #37: 'del_4q22.3' versus Clinical Feature #10: 'RACE'

P value = 0.000198 (Wilcoxon-test), Q value = 0.021
Table S17. Gene #38: 'del_4q35.1' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 354 | 59.8 (12.7) |
| DEL PEAK 9(4Q35.1) MUTATED | 165 | 57.3 (13.0) |
| DEL PEAK 9(4Q35.1) WILD-TYPE | 189 | 61.9 (12.1) |
Figure S17. Get High-res Image Gene #38: 'del_4q35.1' versus Clinical Feature #2: 'YEARS_TO_BIRTH'

P value = 7e-05 (Fisher's exact test), Q value = 0.011
Table S18. Gene #38: 'del_4q35.1' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | WHITE |
|---|---|---|---|---|
| ALL | 1 | 156 | 17 | 173 |
| DEL PEAK 9(4Q35.1) MUTATED | 0 | 95 | 10 | 63 |
| DEL PEAK 9(4Q35.1) WILD-TYPE | 1 | 61 | 7 | 110 |
Figure S18. Get High-res Image Gene #38: 'del_4q35.1' versus Clinical Feature #10: 'RACE'

P value = 0.00431 (Wilcoxon-test), Q value = 0.15
Table S19. Gene #39: 'del_6q27' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 354 | 59.8 (12.7) |
| DEL PEAK 10(6Q27) MUTATED | 132 | 62.4 (12.3) |
| DEL PEAK 10(6Q27) WILD-TYPE | 222 | 58.2 (12.7) |
Figure S19. Get High-res Image Gene #39: 'del_6q27' versus Clinical Feature #2: 'YEARS_TO_BIRTH'

P value = 0.00723 (Fisher's exact test), Q value = 0.21
Table S20. Gene #44: 'del_10q23.31' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | WHITE |
|---|---|---|---|---|
| ALL | 1 | 156 | 17 | 173 |
| DEL PEAK 15(10Q23.31) MUTATED | 0 | 46 | 10 | 38 |
| DEL PEAK 15(10Q23.31) WILD-TYPE | 1 | 110 | 7 | 135 |
Figure S20. Get High-res Image Gene #44: 'del_10q23.31' versus Clinical Feature #10: 'RACE'
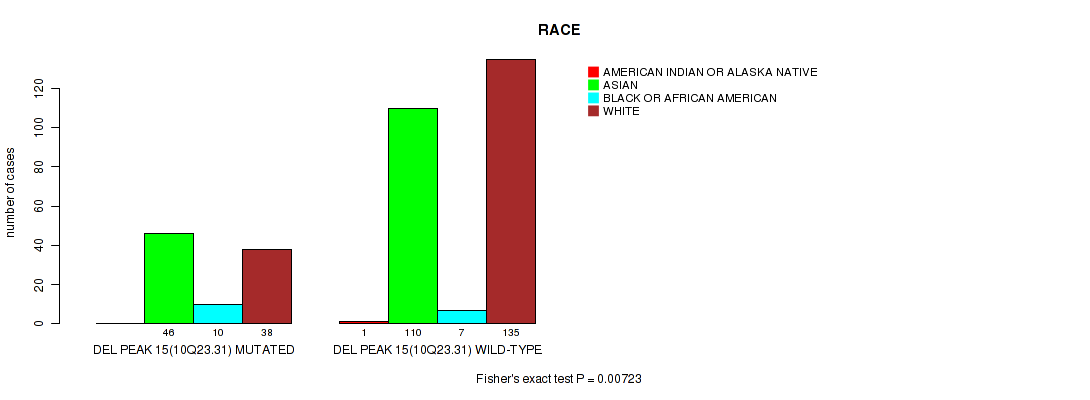
P value = 0.00748 (Fisher's exact test), Q value = 0.21
Table S21. Gene #48: 'del_12p12.1' versus Clinical Feature #3: 'NEOPLASM_DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IIIA | STAGE IIIB | STAGE IIIC | STAGE IV | STAGE IVA | STAGE IVB |
|---|---|---|---|---|---|---|---|---|---|
| ALL | 168 | 84 | 3 | 59 | 8 | 7 | 3 | 1 | 2 |
| DEL PEAK 19(12P12.1) MUTATED | 30 | 17 | 1 | 10 | 5 | 2 | 3 | 0 | 0 |
| DEL PEAK 19(12P12.1) WILD-TYPE | 138 | 67 | 2 | 49 | 3 | 5 | 0 | 1 | 2 |
Figure S21. Get High-res Image Gene #48: 'del_12p12.1' versus Clinical Feature #3: 'NEOPLASM_DISEASESTAGE'

P value = 0.0102 (Fisher's exact test), Q value = 0.25
Table S22. Gene #50: 'del_13q14.2' versus Clinical Feature #4: 'PATHOLOGY_T_STAGE'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 179 | 91 | 73 | 12 |
| DEL PEAK 21(13Q14.2) MUTATED | 75 | 51 | 33 | 10 |
| DEL PEAK 21(13Q14.2) WILD-TYPE | 104 | 40 | 40 | 2 |
Figure S22. Get High-res Image Gene #50: 'del_13q14.2' versus Clinical Feature #4: 'PATHOLOGY_T_STAGE'

P value = 0.000578 (Wilcoxon-test), Q value = 0.039
Table S23. Gene #54: 'del_16q23.1' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 354 | 59.8 (12.7) |
| DEL PEAK 25(16Q23.1) MUTATED | 158 | 57.4 (12.8) |
| DEL PEAK 25(16Q23.1) WILD-TYPE | 196 | 61.6 (12.3) |
Figure S23. Get High-res Image Gene #54: 'del_16q23.1' versus Clinical Feature #2: 'YEARS_TO_BIRTH'

P value = 4e-05 (Fisher's exact test), Q value = 0.011
Table S24. Gene #54: 'del_16q23.1' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | WHITE |
|---|---|---|---|---|
| ALL | 1 | 156 | 17 | 173 |
| DEL PEAK 25(16Q23.1) MUTATED | 0 | 90 | 7 | 58 |
| DEL PEAK 25(16Q23.1) WILD-TYPE | 1 | 66 | 10 | 115 |
Figure S24. Get High-res Image Gene #54: 'del_16q23.1' versus Clinical Feature #10: 'RACE'

P value = 0.00849 (Fisher's exact test), Q value = 0.23
Table S25. Gene #56: 'del_17p11.2' versus Clinical Feature #3: 'NEOPLASM_DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IIIA | STAGE IIIB | STAGE IIIC | STAGE IV | STAGE IVA | STAGE IVB |
|---|---|---|---|---|---|---|---|---|---|
| ALL | 168 | 84 | 3 | 59 | 8 | 7 | 3 | 1 | 2 |
| DEL PEAK 27(17P11.2) MUTATED | 71 | 49 | 1 | 35 | 6 | 6 | 3 | 1 | 1 |
| DEL PEAK 27(17P11.2) WILD-TYPE | 97 | 35 | 2 | 24 | 2 | 1 | 0 | 0 | 1 |
Figure S25. Get High-res Image Gene #56: 'del_17p11.2' versus Clinical Feature #3: 'NEOPLASM_DISEASESTAGE'

P value = 0.0086 (Fisher's exact test), Q value = 0.23
Table S26. Gene #56: 'del_17p11.2' versus Clinical Feature #4: 'PATHOLOGY_T_STAGE'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 179 | 91 | 73 | 12 |
| DEL PEAK 27(17P11.2) MUTATED | 78 | 55 | 43 | 9 |
| DEL PEAK 27(17P11.2) WILD-TYPE | 101 | 36 | 30 | 3 |
Figure S26. Get High-res Image Gene #56: 'del_17p11.2' versus Clinical Feature #4: 'PATHOLOGY_T_STAGE'

P value = 0.00496 (Fisher's exact test), Q value = 0.15
Table S27. Gene #61: 'del_22q13.32' versus Clinical Feature #4: 'PATHOLOGY_T_STAGE'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 179 | 91 | 73 | 12 |
| DEL PEAK 32(22Q13.32) MUTATED | 27 | 28 | 19 | 5 |
| DEL PEAK 32(22Q13.32) WILD-TYPE | 152 | 63 | 54 | 7 |
Figure S27. Get High-res Image Gene #61: 'del_22q13.32' versus Clinical Feature #4: 'PATHOLOGY_T_STAGE'

P value = 0.00376 (Fisher's exact test), Q value = 0.15
Table S28. Gene #61: 'del_22q13.32' versus Clinical Feature #10: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | WHITE |
|---|---|---|---|---|
| ALL | 1 | 156 | 17 | 173 |
| DEL PEAK 32(22Q13.32) MUTATED | 1 | 24 | 7 | 45 |
| DEL PEAK 32(22Q13.32) WILD-TYPE | 0 | 132 | 10 | 128 |
Figure S28. Get High-res Image Gene #61: 'del_22q13.32' versus Clinical Feature #10: 'RACE'

-
Copy number data file = all_lesions.txt from GISTIC pipeline
-
Processed Copy number data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_Correlate_Genomic_Events_Preprocess/LIHC-TP/15089885/transformed.cor.cli.txt
-
Clinical data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/Append_Data/LIHC-TP/15082975/LIHC-TP.merged_data.txt
-
Number of patients = 357
-
Number of significantly focal cnvs = 62
-
Number of selected clinical features = 11
-
Exclude genes that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.