This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and molecular subtypes.
Testing the association between copy number variation 32 arm-level events and 6 molecular subtypes across 191 patients, 79 significant findings detected with P value < 0.05 and Q value < 0.25.
-
1p gain cnv correlated to 'CN_CNMF'.
-
4p gain cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
4q gain cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
8p gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
8q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
10q gain cnv correlated to 'CN_CNMF'.
-
11p gain cnv correlated to 'CN_CNMF' and 'MIRSEQ_CHIERARCHICAL'.
-
11q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
13q gain cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
17q gain cnv correlated to 'CN_CNMF'.
-
19p gain cnv correlated to 'CN_CNMF'.
-
19q gain cnv correlated to 'CN_CNMF'.
-
21q gain cnv correlated to 'CN_CNMF'.
-
22q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
xp gain cnv correlated to 'CN_CNMF' and 'MIRSEQ_CHIERARCHICAL'.
-
xq gain cnv correlated to 'CN_CNMF' and 'MIRSEQ_CHIERARCHICAL'.
-
3p loss cnv correlated to 'CN_CNMF'.
-
3q loss cnv correlated to 'CN_CNMF'.
-
5q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
7p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
7q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
12p loss cnv correlated to 'CN_CNMF'.
-
15q loss cnv correlated to 'CN_CNMF' and 'MIRSEQ_CNMF'.
-
16q loss cnv correlated to 'CN_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
17p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
17q loss cnv correlated to 'CN_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
18p loss cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
18q loss cnv correlated to 'CN_CNMF'.
-
19p loss cnv correlated to 'CN_CNMF'.
-
19q loss cnv correlated to 'CN_CNMF'.
-
xp loss cnv correlated to 'CN_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
xq loss cnv correlated to 'CN_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 32 arm-level events and 6 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 79 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 8p gain | 22 (12%) | 169 |
0.00081 (0.00778) |
0.00025 (0.0032) |
0.0285 (0.0783) |
0.00088 (0.00805) |
0.00985 (0.035) |
0.0167 (0.05) |
| 8q gain | 23 (12%) | 168 |
0.00138 (0.0102) |
0.00039 (0.0044) |
0.014 (0.0453) |
0.00174 (0.0111) |
0.0069 (0.026) |
0.0138 (0.0453) |
| 7p loss | 17 (9%) | 174 |
4e-05 (0.000853) |
0.00019 (0.00261) |
0.00096 (0.00826) |
2e-05 (0.00048) |
0.00373 (0.0184) |
0.00346 (0.0184) |
| 7q loss | 20 (10%) | 171 |
1e-05 (0.00048) |
1e-05 (0.00048) |
0.00609 (0.0257) |
2e-05 (0.00048) |
0.00148 (0.0105) |
0.00078 (0.00778) |
| 22q gain | 9 (5%) | 182 |
0.00099 (0.00826) |
0.00384 (0.0184) |
0.137 (0.257) |
0.0161 (0.049) |
0.0555 (0.132) |
0.0131 (0.0448) |
| 5q loss | 6 (3%) | 185 |
5e-05 (0.00096) |
0.00641 (0.0257) |
0.226 (0.365) |
0.63 (0.768) |
0.0314 (0.0826) |
0.0322 (0.0835) |
| 17p loss | 13 (7%) | 178 |
1e-05 (0.00048) |
0.00169 (0.0111) |
0.154 (0.287) |
0.00164 (0.0111) |
0.161 (0.298) |
0.00359 (0.0184) |
| 11q gain | 7 (4%) | 184 |
2e-05 (0.00048) |
0.0308 (0.0822) |
0.0676 (0.149) |
0.0751 (0.162) |
0.291 (0.444) |
0.0155 (0.0482) |
| 16q loss | 4 (2%) | 187 |
0.0011 (0.0088) |
0.0565 (0.132) |
0.787 (0.922) |
1 (1.00) |
0.0152 (0.0477) |
0.0216 (0.0629) |
| 17q loss | 7 (4%) | 184 |
2e-05 (0.00048) |
0.113 (0.219) |
0.0669 (0.149) |
0.237 (0.38) |
0.0369 (0.0944) |
0.0472 (0.115) |
| xp loss | 5 (3%) | 186 |
0.0001 (0.00175) |
0.199 (0.345) |
0.224 (0.365) |
0.632 (0.768) |
0.00369 (0.0184) |
0.00483 (0.0216) |
| xq loss | 5 (3%) | 186 |
0.00011 (0.00176) |
0.202 (0.346) |
0.225 (0.365) |
0.631 (0.768) |
0.00313 (0.0172) |
0.00476 (0.0216) |
| 4p gain | 4 (2%) | 187 |
0.0276 (0.0769) |
0.0075 (0.0277) |
0.341 (0.5) |
0.834 (0.961) |
||
| 4q gain | 4 (2%) | 187 |
0.0292 (0.079) |
0.00795 (0.0288) |
0.341 (0.5) |
0.836 (0.961) |
||
| 11p gain | 4 (2%) | 187 |
0.00065 (0.00693) |
0.432 (0.596) |
0.467 (0.626) |
0.367 (0.53) |
0.523 (0.68) |
0.0222 (0.0636) |
| 13q gain | 6 (3%) | 185 |
0.042 (0.106) |
0.00667 (0.0257) |
0.123 (0.236) |
0.0832 (0.176) |
0.802 (0.933) |
0.0736 (0.161) |
| xp gain | 3 (2%) | 188 |
0.00196 (0.0121) |
0.704 (0.839) |
0.892 (0.978) |
0.047 (0.115) |
||
| xq gain | 3 (2%) | 188 |
0.00201 (0.0121) |
0.7 (0.839) |
0.89 (0.978) |
0.0455 (0.113) |
||
| 15q loss | 4 (2%) | 187 |
0.00027 (0.00324) |
0.549 (0.707) |
0.0666 (0.149) |
0.367 (0.53) |
0.0227 (0.064) |
0.183 (0.328) |
| 18p loss | 5 (3%) | 186 |
0.00012 (0.00177) |
0.0213 (0.0629) |
1 (1.00) |
0.865 (0.961) |
0.252 (0.387) |
0.185 (0.328) |
| 1p gain | 3 (2%) | 188 |
0.00394 (0.0185) |
0.209 (0.35) |
0.187 (0.328) |
0.589 (0.731) |
0.0927 (0.187) |
0.0937 (0.187) |
| 10q gain | 3 (2%) | 188 |
0.0142 (0.0453) |
0.209 (0.35) |
0.185 (0.328) |
0.59 (0.731) |
0.553 (0.708) |
0.0909 (0.187) |
| 17q gain | 3 (2%) | 188 |
0.00657 (0.0257) |
0.704 (0.839) |
1 (1.00) |
0.587 (0.731) |
0.483 (0.637) |
0.305 (0.461) |
| 19p gain | 5 (3%) | 186 |
0.0134 (0.0452) |
0.246 (0.385) |
1 (1.00) |
0.863 (0.961) |
0.58 (0.731) |
1 (1.00) |
| 19q gain | 5 (3%) | 186 |
0.0128 (0.0445) |
0.246 (0.385) |
1 (1.00) |
0.864 (0.961) |
0.579 (0.731) |
1 (1.00) |
| 21q gain | 8 (4%) | 183 |
1e-05 (0.00048) |
0.113 (0.219) |
0.125 (0.238) |
0.524 (0.68) |
0.0811 (0.173) |
0.412 (0.581) |
| 3p loss | 3 (2%) | 188 |
0.00668 (0.0257) |
0.21 (0.35) |
0.784 (0.922) |
1 (1.00) |
0.0633 (0.145) |
0.092 (0.187) |
| 3q loss | 3 (2%) | 188 |
0.00658 (0.0257) |
0.212 (0.351) |
0.785 (0.922) |
1 (1.00) |
0.0627 (0.145) |
0.0913 (0.187) |
| 12p loss | 4 (2%) | 187 |
0.00613 (0.0257) |
0.431 (0.596) |
0.374 (0.536) |
0.389 (0.553) |
0.484 (0.637) |
0.186 (0.328) |
| 18q loss | 4 (2%) | 187 |
0.00132 (0.0101) |
0.0557 (0.132) |
0.469 (0.626) |
0.862 (0.961) |
0.102 (0.203) |
0.188 (0.328) |
| 19p loss | 4 (2%) | 187 |
0.00295 (0.0172) |
0.435 (0.596) |
0.467 (0.626) |
0.863 (0.961) |
0.31 (0.461) |
0.248 (0.385) |
| 19q loss | 4 (2%) | 187 |
0.00309 (0.0172) |
0.431 (0.596) |
0.467 (0.626) |
0.866 (0.961) |
0.309 (0.461) |
0.249 (0.385) |
P value = 0.00394 (Fisher's exact test), Q value = 0.018
Table S1. Gene #1: '1p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 1P GAIN MUTATED | 0 | 2 | 1 | 0 |
| 1P GAIN WILD-TYPE | 148 | 12 | 26 | 2 |
Figure S1. Get High-res Image Gene #1: '1p gain' versus Molecular Subtype #1: 'CN_CNMF'
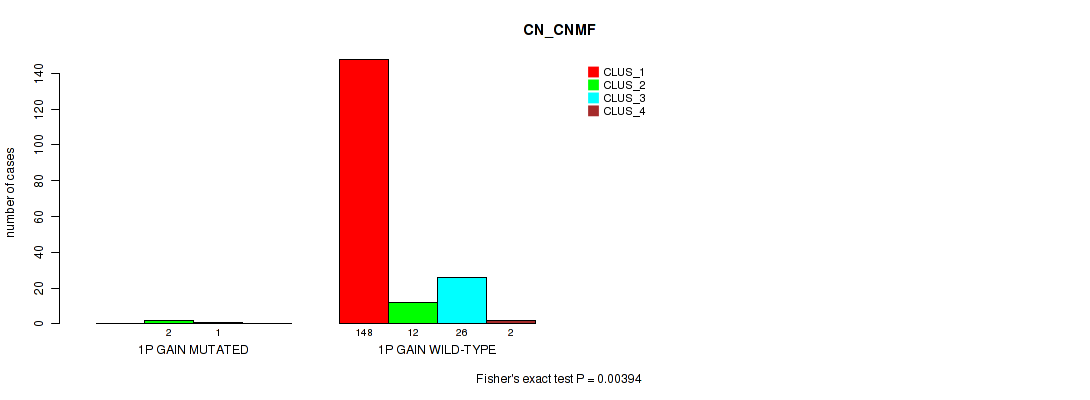
P value = 0.0276 (Fisher's exact test), Q value = 0.077
Table S2. Gene #2: '4p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 4P GAIN MUTATED | 1 | 0 | 3 | 0 |
| 4P GAIN WILD-TYPE | 147 | 14 | 24 | 2 |
Figure S2. Get High-res Image Gene #2: '4p gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.0075 (Fisher's exact test), Q value = 0.028
Table S3. Gene #2: '4p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| 4P GAIN MUTATED | 0 | 0 | 4 | 0 |
| 4P GAIN WILD-TYPE | 33 | 61 | 43 | 44 |
Figure S3. Get High-res Image Gene #2: '4p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
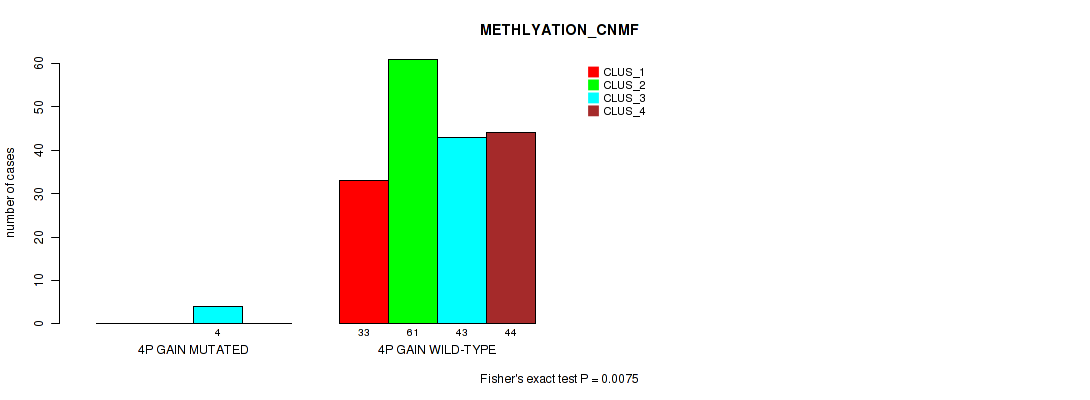
P value = 0.0292 (Fisher's exact test), Q value = 0.079
Table S4. Gene #3: '4q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 4Q GAIN MUTATED | 1 | 0 | 3 | 0 |
| 4Q GAIN WILD-TYPE | 147 | 14 | 24 | 2 |
Figure S4. Get High-res Image Gene #3: '4q gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00795 (Fisher's exact test), Q value = 0.029
Table S5. Gene #3: '4q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| 4Q GAIN MUTATED | 0 | 0 | 4 | 0 |
| 4Q GAIN WILD-TYPE | 33 | 61 | 43 | 44 |
Figure S5. Get High-res Image Gene #3: '4q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.00081 (Fisher's exact test), Q value = 0.0078
Table S6. Gene #4: '8p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 8P GAIN MUTATED | 11 | 1 | 10 | 0 |
| 8P GAIN WILD-TYPE | 137 | 13 | 17 | 2 |
Figure S6. Get High-res Image Gene #4: '8p gain' versus Molecular Subtype #1: 'CN_CNMF'
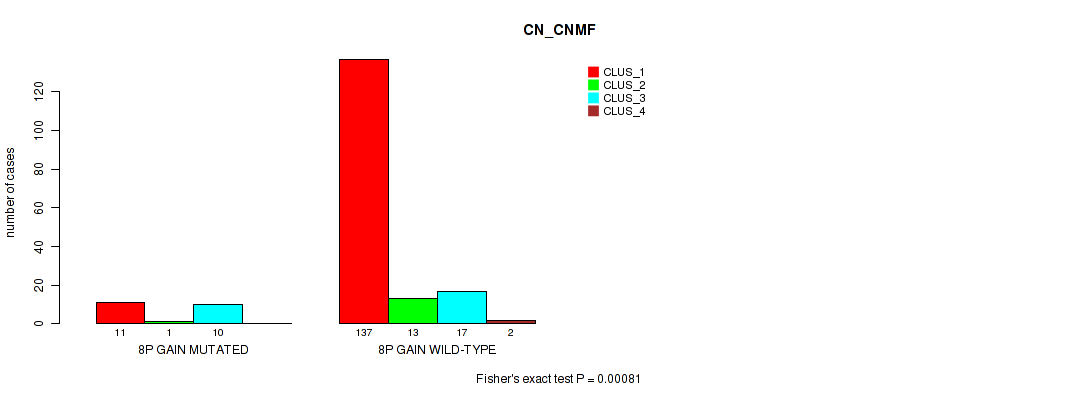
P value = 0.00025 (Fisher's exact test), Q value = 0.0032
Table S7. Gene #4: '8p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| 8P GAIN MUTATED | 1 | 16 | 3 | 1 |
| 8P GAIN WILD-TYPE | 32 | 45 | 44 | 43 |
Figure S7. Get High-res Image Gene #4: '8p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
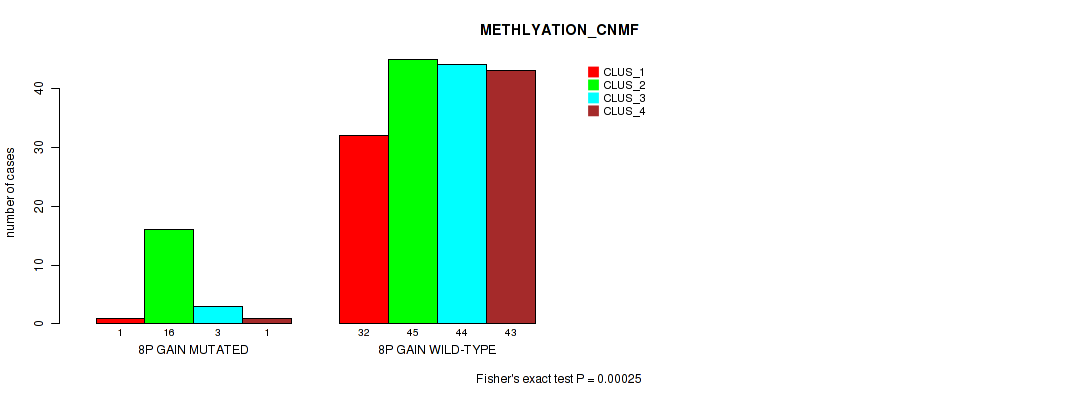
P value = 0.0285 (Fisher's exact test), Q value = 0.078
Table S8. Gene #4: '8p gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 70 | 52 | 44 |
| 8P GAIN MUTATED | 13 | 2 | 3 |
| 8P GAIN WILD-TYPE | 57 | 50 | 41 |
Figure S8. Get High-res Image Gene #4: '8p gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 0.00088 (Fisher's exact test), Q value = 0.008
Table S9. Gene #4: '8p gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| 8P GAIN MUTATED | 1 | 0 | 0 | 15 | 1 | 1 |
| 8P GAIN WILD-TYPE | 15 | 13 | 14 | 40 | 25 | 41 |
Figure S9. Get High-res Image Gene #4: '8p gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 0.00985 (Fisher's exact test), Q value = 0.035
Table S10. Gene #4: '8p gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| 8P GAIN MUTATED | 2 | 9 | 3 | 7 |
| 8P GAIN WILD-TYPE | 56 | 28 | 38 | 36 |
Figure S10. Get High-res Image Gene #4: '8p gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 0.0167 (Fisher's exact test), Q value = 0.05
Table S11. Gene #4: '8p gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 8P GAIN MUTATED | 3 | 8 | 10 |
| 8P GAIN WILD-TYPE | 64 | 26 | 68 |
Figure S11. Get High-res Image Gene #4: '8p gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 0.00138 (Fisher's exact test), Q value = 0.01
Table S12. Gene #5: '8q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 8Q GAIN MUTATED | 12 | 1 | 10 | 0 |
| 8Q GAIN WILD-TYPE | 136 | 13 | 17 | 2 |
Figure S12. Get High-res Image Gene #5: '8q gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00039 (Fisher's exact test), Q value = 0.0044
Table S13. Gene #5: '8q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| 8Q GAIN MUTATED | 1 | 16 | 4 | 1 |
| 8Q GAIN WILD-TYPE | 32 | 45 | 43 | 43 |
Figure S13. Get High-res Image Gene #5: '8q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
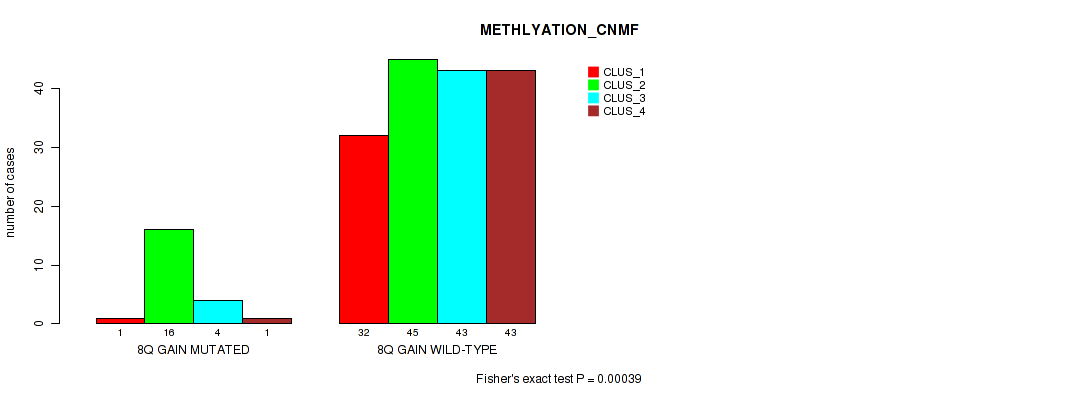
P value = 0.014 (Fisher's exact test), Q value = 0.045
Table S14. Gene #5: '8q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 70 | 52 | 44 |
| 8Q GAIN MUTATED | 14 | 2 | 3 |
| 8Q GAIN WILD-TYPE | 56 | 50 | 41 |
Figure S14. Get High-res Image Gene #5: '8q gain' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 0.00174 (Fisher's exact test), Q value = 0.011
Table S15. Gene #5: '8q gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| 8Q GAIN MUTATED | 1 | 0 | 1 | 15 | 1 | 1 |
| 8Q GAIN WILD-TYPE | 15 | 13 | 13 | 40 | 25 | 41 |
Figure S15. Get High-res Image Gene #5: '8q gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 0.0069 (Fisher's exact test), Q value = 0.026
Table S16. Gene #5: '8q gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| 8Q GAIN MUTATED | 2 | 9 | 3 | 8 |
| 8Q GAIN WILD-TYPE | 56 | 28 | 38 | 35 |
Figure S16. Get High-res Image Gene #5: '8q gain' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 0.0138 (Fisher's exact test), Q value = 0.045
Table S17. Gene #5: '8q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 8Q GAIN MUTATED | 3 | 8 | 11 |
| 8Q GAIN WILD-TYPE | 64 | 26 | 67 |
Figure S17. Get High-res Image Gene #5: '8q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 0.0142 (Fisher's exact test), Q value = 0.045
Table S18. Gene #6: '10q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 10Q GAIN MUTATED | 0 | 1 | 2 | 0 |
| 10Q GAIN WILD-TYPE | 148 | 13 | 25 | 2 |
Figure S18. Get High-res Image Gene #6: '10q gain' versus Molecular Subtype #1: 'CN_CNMF'
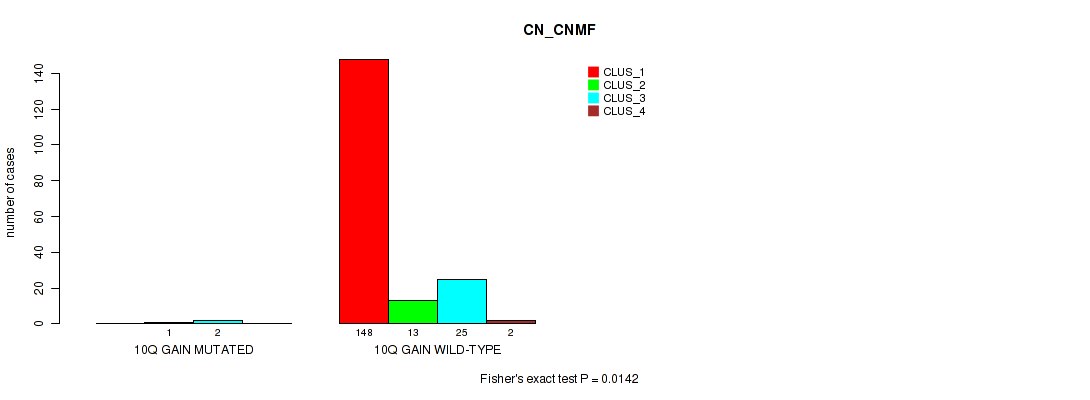
P value = 0.00065 (Fisher's exact test), Q value = 0.0069
Table S19. Gene #7: '11p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 11P GAIN MUTATED | 0 | 3 | 1 | 0 |
| 11P GAIN WILD-TYPE | 148 | 11 | 26 | 2 |
Figure S19. Get High-res Image Gene #7: '11p gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.0222 (Fisher's exact test), Q value = 0.064
Table S20. Gene #7: '11p gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 11P GAIN MUTATED | 0 | 3 | 1 |
| 11P GAIN WILD-TYPE | 67 | 31 | 77 |
Figure S20. Get High-res Image Gene #7: '11p gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
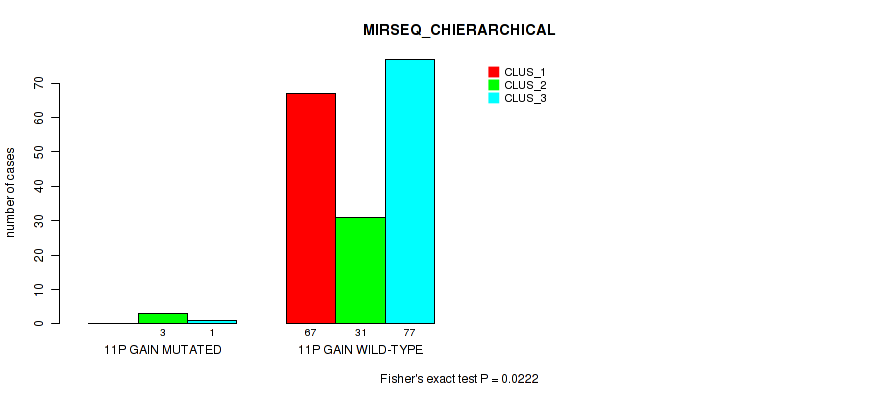
P value = 2e-05 (Fisher's exact test), Q value = 0.00048
Table S21. Gene #8: '11q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 11Q GAIN MUTATED | 0 | 4 | 3 | 0 |
| 11Q GAIN WILD-TYPE | 148 | 10 | 24 | 2 |
Figure S21. Get High-res Image Gene #8: '11q gain' versus Molecular Subtype #1: 'CN_CNMF'
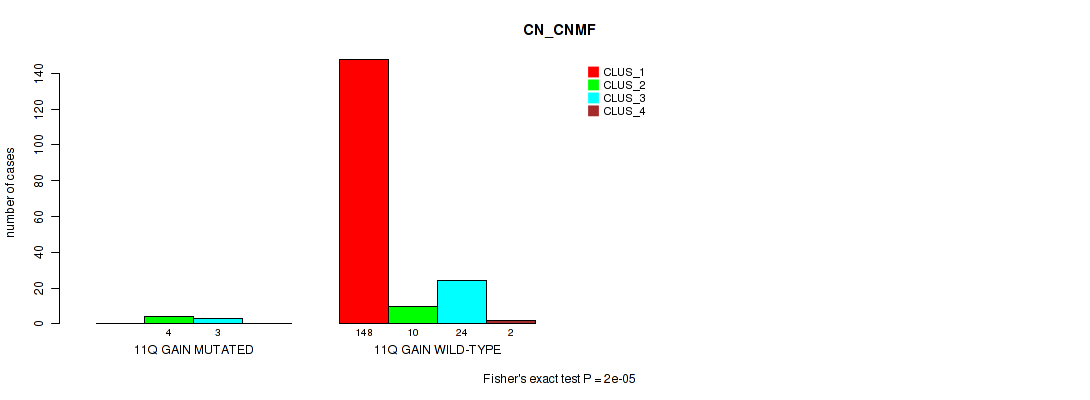
P value = 0.0308 (Fisher's exact test), Q value = 0.082
Table S22. Gene #8: '11q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| 11Q GAIN MUTATED | 0 | 6 | 1 | 0 |
| 11Q GAIN WILD-TYPE | 33 | 55 | 46 | 44 |
Figure S22. Get High-res Image Gene #8: '11q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.0155 (Fisher's exact test), Q value = 0.048
Table S23. Gene #8: '11q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 11Q GAIN MUTATED | 0 | 4 | 3 |
| 11Q GAIN WILD-TYPE | 67 | 30 | 75 |
Figure S23. Get High-res Image Gene #8: '11q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 0.042 (Fisher's exact test), Q value = 0.11
Table S24. Gene #9: '13q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 13Q GAIN MUTATED | 2 | 1 | 3 | 0 |
| 13Q GAIN WILD-TYPE | 146 | 13 | 24 | 2 |
Figure S24. Get High-res Image Gene #9: '13q gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00667 (Fisher's exact test), Q value = 0.026
Table S25. Gene #9: '13q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| 13Q GAIN MUTATED | 0 | 6 | 0 | 0 |
| 13Q GAIN WILD-TYPE | 33 | 55 | 47 | 44 |
Figure S25. Get High-res Image Gene #9: '13q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.00657 (Fisher's exact test), Q value = 0.026
Table S26. Gene #10: '17q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 17Q GAIN MUTATED | 0 | 0 | 3 | 0 |
| 17Q GAIN WILD-TYPE | 148 | 14 | 24 | 2 |
Figure S26. Get High-res Image Gene #10: '17q gain' versus Molecular Subtype #1: 'CN_CNMF'
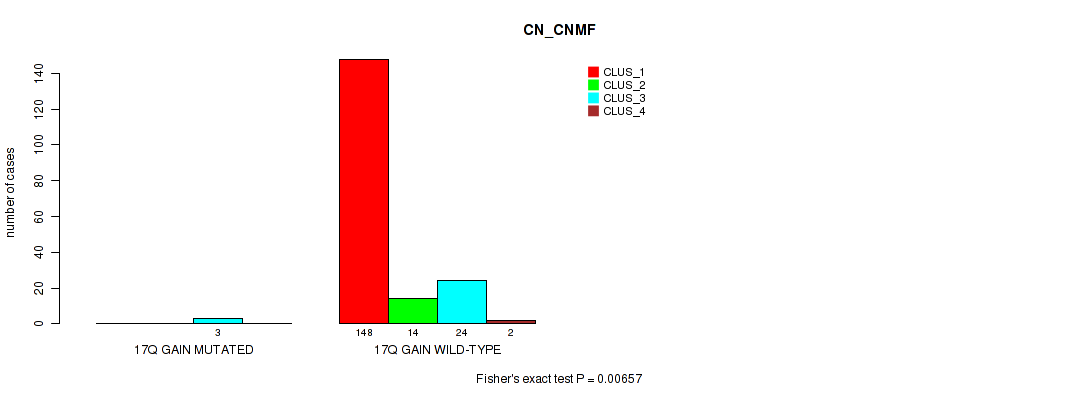
P value = 0.0134 (Fisher's exact test), Q value = 0.045
Table S27. Gene #11: '19p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 19P GAIN MUTATED | 1 | 1 | 3 | 0 |
| 19P GAIN WILD-TYPE | 147 | 13 | 24 | 2 |
Figure S27. Get High-res Image Gene #11: '19p gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.0128 (Fisher's exact test), Q value = 0.045
Table S28. Gene #12: '19q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 19Q GAIN MUTATED | 1 | 1 | 3 | 0 |
| 19Q GAIN WILD-TYPE | 147 | 13 | 24 | 2 |
Figure S28. Get High-res Image Gene #12: '19q gain' versus Molecular Subtype #1: 'CN_CNMF'
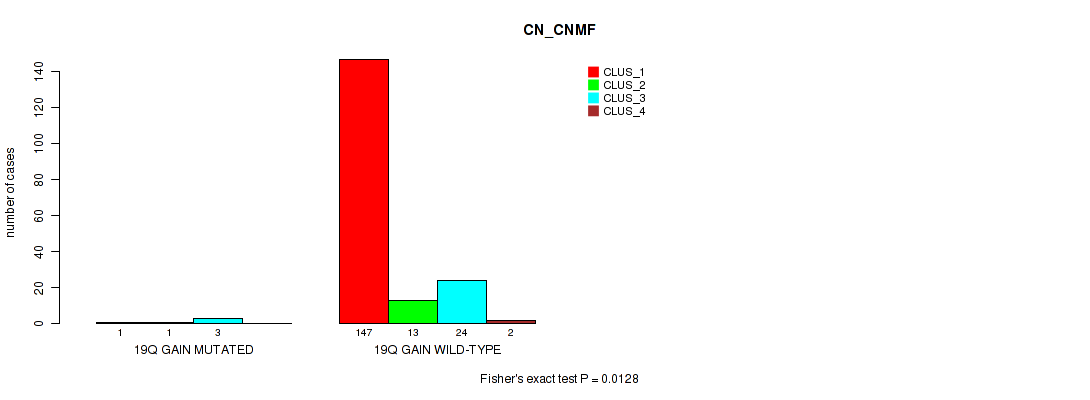
P value = 1e-05 (Fisher's exact test), Q value = 0.00048
Table S29. Gene #13: '21q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 21Q GAIN MUTATED | 0 | 0 | 8 | 0 |
| 21Q GAIN WILD-TYPE | 148 | 14 | 19 | 2 |
Figure S29. Get High-res Image Gene #13: '21q gain' versus Molecular Subtype #1: 'CN_CNMF'
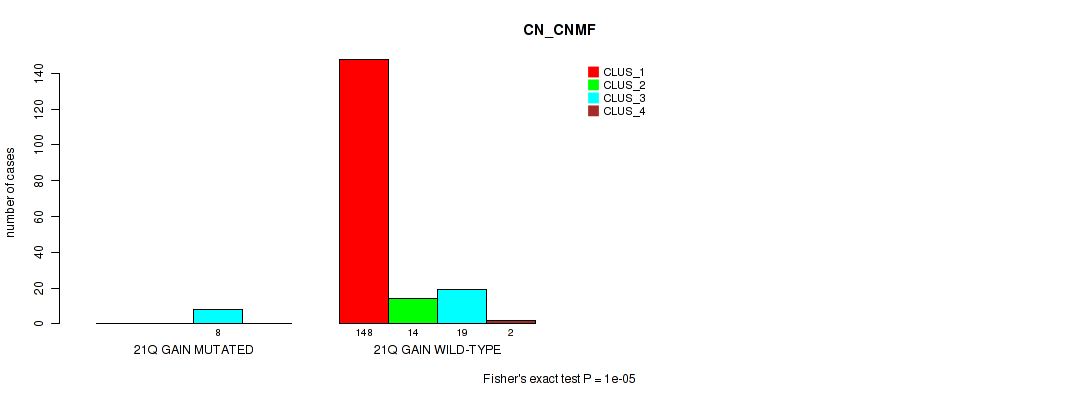
P value = 0.00099 (Fisher's exact test), Q value = 0.0083
Table S30. Gene #14: '22q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 22Q GAIN MUTATED | 2 | 2 | 5 | 0 |
| 22Q GAIN WILD-TYPE | 146 | 12 | 22 | 2 |
Figure S30. Get High-res Image Gene #14: '22q gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00384 (Fisher's exact test), Q value = 0.018
Table S31. Gene #14: '22q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| 22Q GAIN MUTATED | 0 | 8 | 1 | 0 |
| 22Q GAIN WILD-TYPE | 33 | 53 | 46 | 44 |
Figure S31. Get High-res Image Gene #14: '22q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.0161 (Fisher's exact test), Q value = 0.049
Table S32. Gene #14: '22q gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| 22Q GAIN MUTATED | 0 | 1 | 0 | 8 | 0 | 0 |
| 22Q GAIN WILD-TYPE | 16 | 12 | 14 | 47 | 26 | 42 |
Figure S32. Get High-res Image Gene #14: '22q gain' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
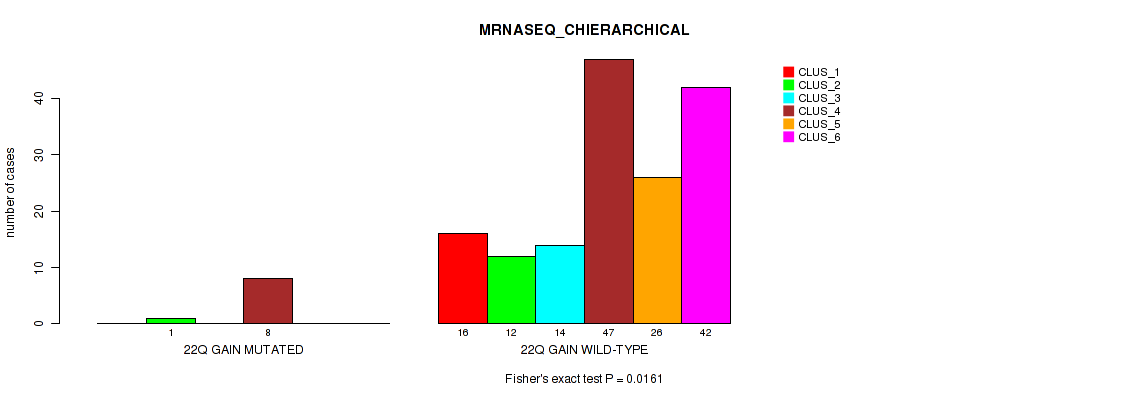
P value = 0.0131 (Fisher's exact test), Q value = 0.045
Table S33. Gene #14: '22q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 22Q GAIN MUTATED | 0 | 4 | 5 |
| 22Q GAIN WILD-TYPE | 67 | 30 | 73 |
Figure S33. Get High-res Image Gene #14: '22q gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 0.00196 (Fisher's exact test), Q value = 0.012
Table S34. Gene #15: 'xp gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| XP GAIN MUTATED | 0 | 1 | 1 | 1 |
| XP GAIN WILD-TYPE | 148 | 13 | 26 | 1 |
Figure S34. Get High-res Image Gene #15: 'xp gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.047 (Fisher's exact test), Q value = 0.11
Table S35. Gene #15: 'xp gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| XP GAIN MUTATED | 1 | 2 | 0 |
| XP GAIN WILD-TYPE | 66 | 32 | 78 |
Figure S35. Get High-res Image Gene #15: 'xp gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
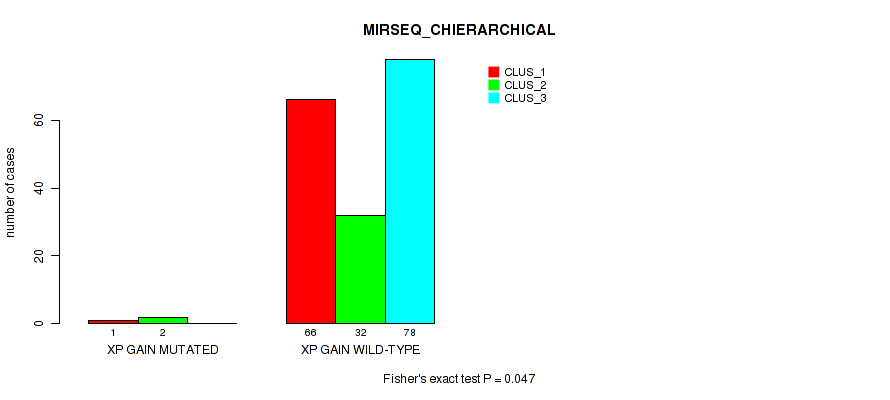
P value = 0.00201 (Fisher's exact test), Q value = 0.012
Table S36. Gene #16: 'xq gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| XQ GAIN MUTATED | 0 | 1 | 1 | 1 |
| XQ GAIN WILD-TYPE | 148 | 13 | 26 | 1 |
Figure S36. Get High-res Image Gene #16: 'xq gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.0455 (Fisher's exact test), Q value = 0.11
Table S37. Gene #16: 'xq gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| XQ GAIN MUTATED | 1 | 2 | 0 |
| XQ GAIN WILD-TYPE | 66 | 32 | 78 |
Figure S37. Get High-res Image Gene #16: 'xq gain' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
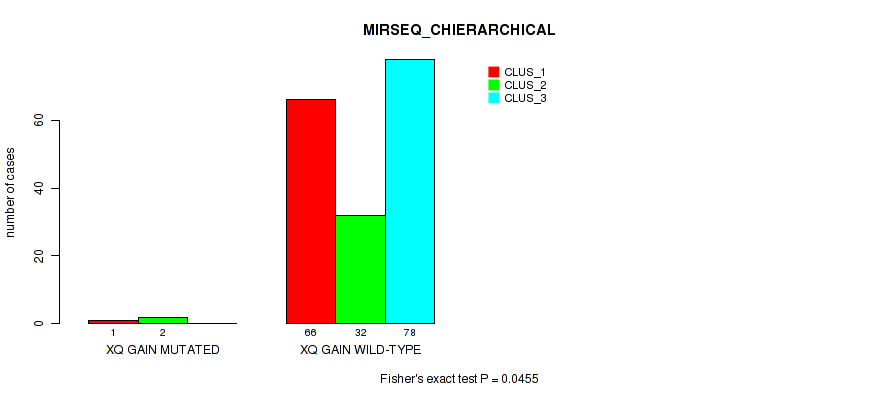
P value = 0.00668 (Fisher's exact test), Q value = 0.026
Table S38. Gene #17: '3p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 3P LOSS MUTATED | 0 | 0 | 3 | 0 |
| 3P LOSS WILD-TYPE | 148 | 14 | 24 | 2 |
Figure S38. Get High-res Image Gene #17: '3p loss' versus Molecular Subtype #1: 'CN_CNMF'
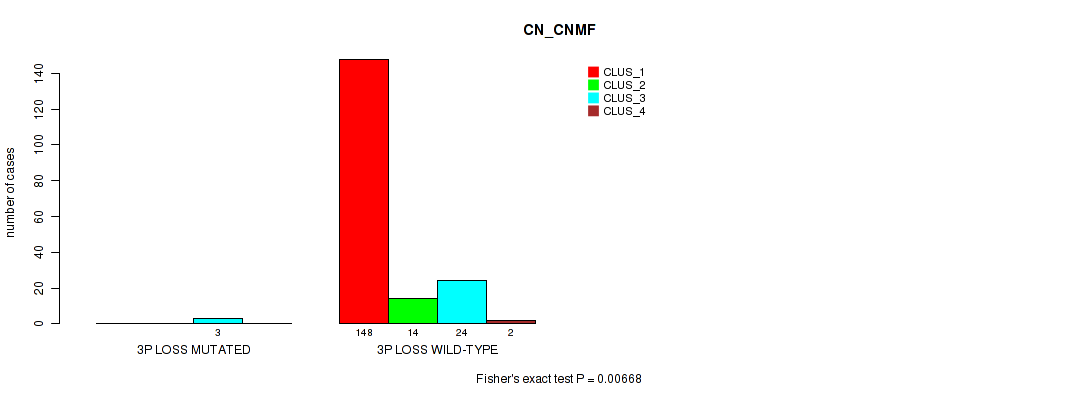
P value = 0.00658 (Fisher's exact test), Q value = 0.026
Table S39. Gene #18: '3q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 3Q LOSS MUTATED | 0 | 0 | 3 | 0 |
| 3Q LOSS WILD-TYPE | 148 | 14 | 24 | 2 |
Figure S39. Get High-res Image Gene #18: '3q loss' versus Molecular Subtype #1: 'CN_CNMF'
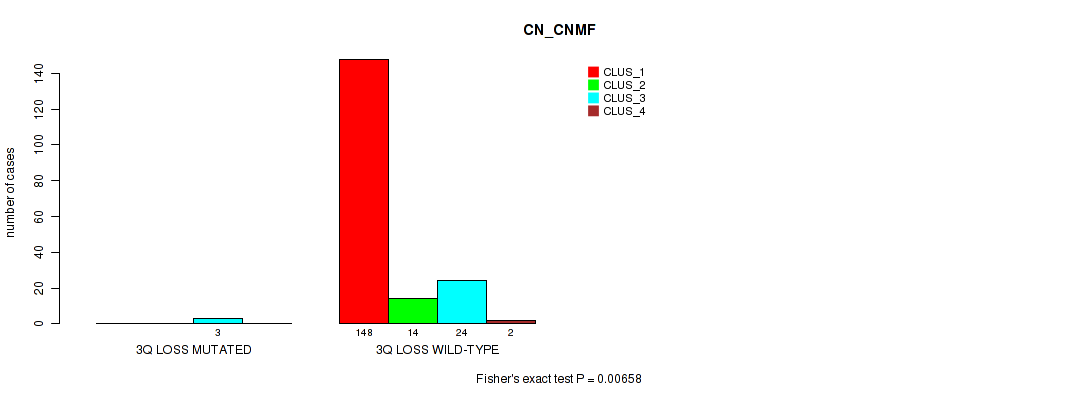
P value = 5e-05 (Fisher's exact test), Q value = 0.00096
Table S40. Gene #19: '5q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 5Q LOSS MUTATED | 0 | 0 | 6 | 0 |
| 5Q LOSS WILD-TYPE | 148 | 14 | 21 | 2 |
Figure S40. Get High-res Image Gene #19: '5q loss' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00641 (Fisher's exact test), Q value = 0.026
Table S41. Gene #19: '5q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| 5Q LOSS MUTATED | 0 | 6 | 0 | 0 |
| 5Q LOSS WILD-TYPE | 33 | 55 | 47 | 44 |
Figure S41. Get High-res Image Gene #19: '5q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.0314 (Fisher's exact test), Q value = 0.083
Table S42. Gene #19: '5q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| 5Q LOSS MUTATED | 0 | 3 | 2 | 0 |
| 5Q LOSS WILD-TYPE | 58 | 34 | 39 | 43 |
Figure S42. Get High-res Image Gene #19: '5q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

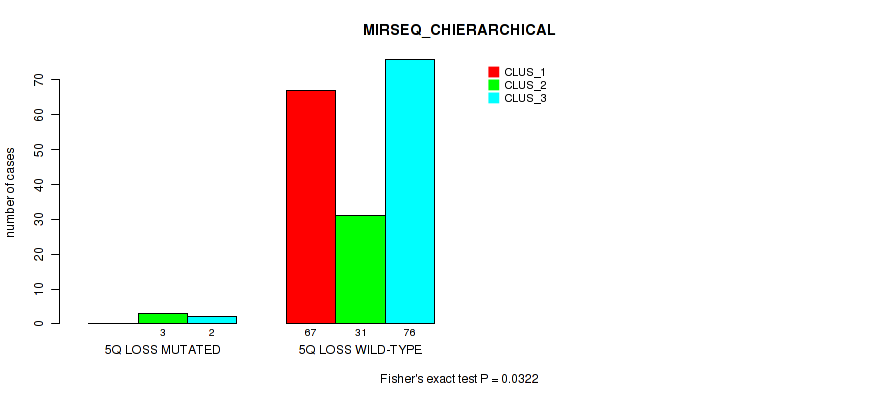
P value = 0.0322 (Fisher's exact test), Q value = 0.083
Table S43. Gene #19: '5q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 5Q LOSS MUTATED | 0 | 3 | 2 |
| 5Q LOSS WILD-TYPE | 67 | 31 | 76 |
Figure S43. Get High-res Image Gene #19: '5q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
P value = 4e-05 (Fisher's exact test), Q value = 0.00085
Table S44. Gene #20: '7p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 7P LOSS MUTATED | 7 | 0 | 9 | 1 |
| 7P LOSS WILD-TYPE | 141 | 14 | 18 | 1 |
Figure S44. Get High-res Image Gene #20: '7p loss' versus Molecular Subtype #1: 'CN_CNMF'
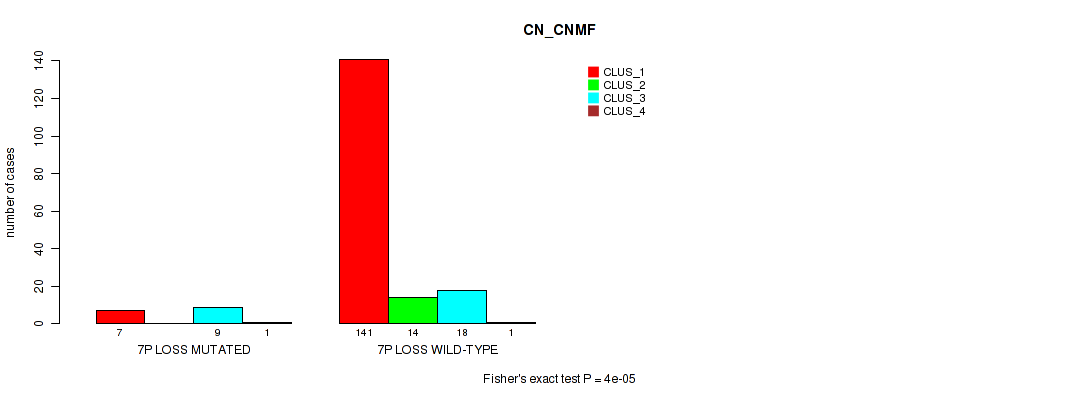
P value = 0.00019 (Fisher's exact test), Q value = 0.0026
Table S45. Gene #20: '7p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| 7P LOSS MUTATED | 0 | 14 | 1 | 2 |
| 7P LOSS WILD-TYPE | 33 | 47 | 46 | 42 |
Figure S45. Get High-res Image Gene #20: '7p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.00096 (Fisher's exact test), Q value = 0.0083
Table S46. Gene #20: '7p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 70 | 52 | 44 |
| 7P LOSS MUTATED | 13 | 0 | 3 |
| 7P LOSS WILD-TYPE | 57 | 52 | 41 |
Figure S46. Get High-res Image Gene #20: '7p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 2e-05 (Fisher's exact test), Q value = 0.00048
Table S47. Gene #20: '7p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| 7P LOSS MUTATED | 0 | 0 | 0 | 16 | 0 | 0 |
| 7P LOSS WILD-TYPE | 16 | 13 | 14 | 39 | 26 | 42 |
Figure S47. Get High-res Image Gene #20: '7p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 0.00373 (Fisher's exact test), Q value = 0.018
Table S48. Gene #20: '7p loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| 7P LOSS MUTATED | 1 | 9 | 3 | 4 |
| 7P LOSS WILD-TYPE | 57 | 28 | 38 | 39 |
Figure S48. Get High-res Image Gene #20: '7p loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
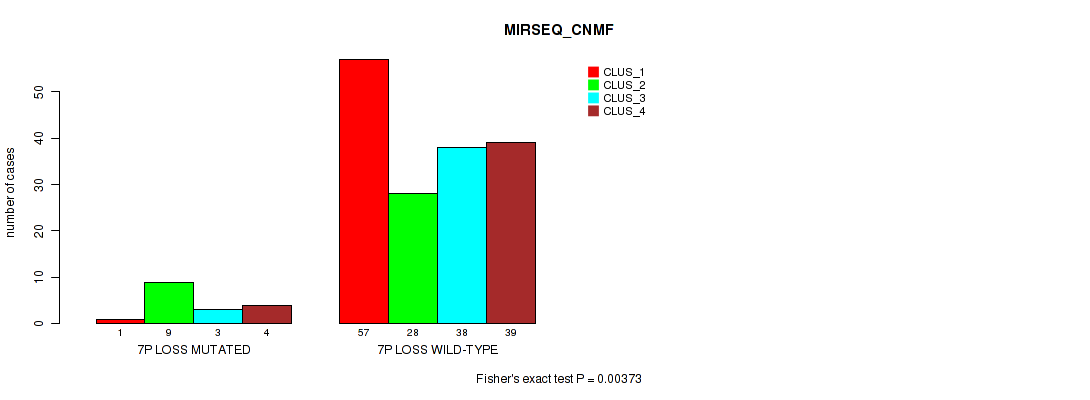
P value = 0.00346 (Fisher's exact test), Q value = 0.018
Table S49. Gene #20: '7p loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 7P LOSS MUTATED | 1 | 7 | 9 |
| 7P LOSS WILD-TYPE | 66 | 27 | 69 |
Figure S49. Get High-res Image Gene #20: '7p loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 1e-05 (Fisher's exact test), Q value = 0.00048
Table S50. Gene #21: '7q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 7Q LOSS MUTATED | 8 | 0 | 11 | 1 |
| 7Q LOSS WILD-TYPE | 140 | 14 | 16 | 1 |
Figure S50. Get High-res Image Gene #21: '7q loss' versus Molecular Subtype #1: 'CN_CNMF'
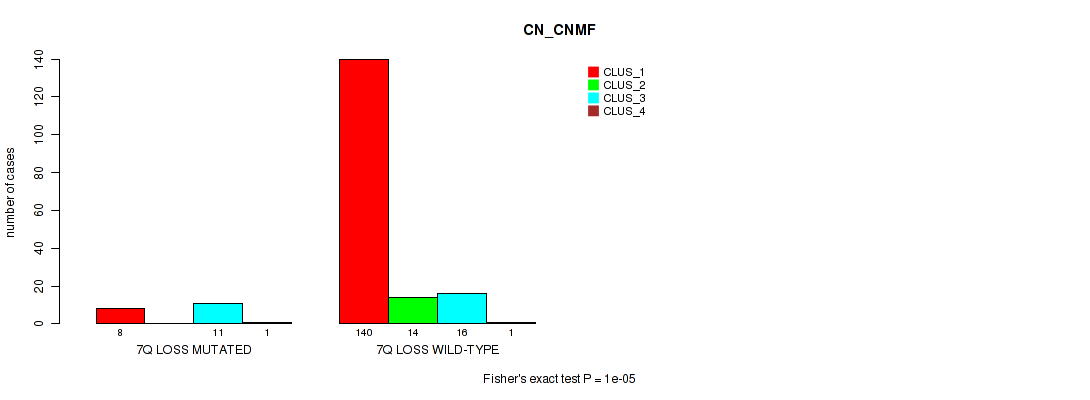
P value = 1e-05 (Fisher's exact test), Q value = 0.00048
Table S51. Gene #21: '7q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| 7Q LOSS MUTATED | 0 | 17 | 1 | 2 |
| 7Q LOSS WILD-TYPE | 33 | 44 | 46 | 42 |
Figure S51. Get High-res Image Gene #21: '7q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
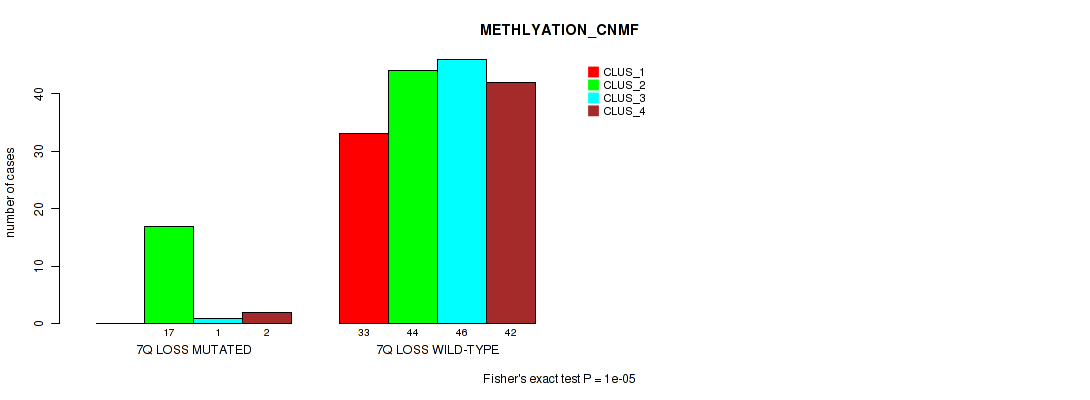
P value = 0.00609 (Fisher's exact test), Q value = 0.026
Table S52. Gene #21: '7q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 70 | 52 | 44 |
| 7Q LOSS MUTATED | 13 | 1 | 3 |
| 7Q LOSS WILD-TYPE | 57 | 51 | 41 |
Figure S52. Get High-res Image Gene #21: '7q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 2e-05 (Fisher's exact test), Q value = 0.00048
Table S53. Gene #21: '7q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| 7Q LOSS MUTATED | 0 | 0 | 0 | 17 | 0 | 0 |
| 7Q LOSS WILD-TYPE | 16 | 13 | 14 | 38 | 26 | 42 |
Figure S53. Get High-res Image Gene #21: '7q loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
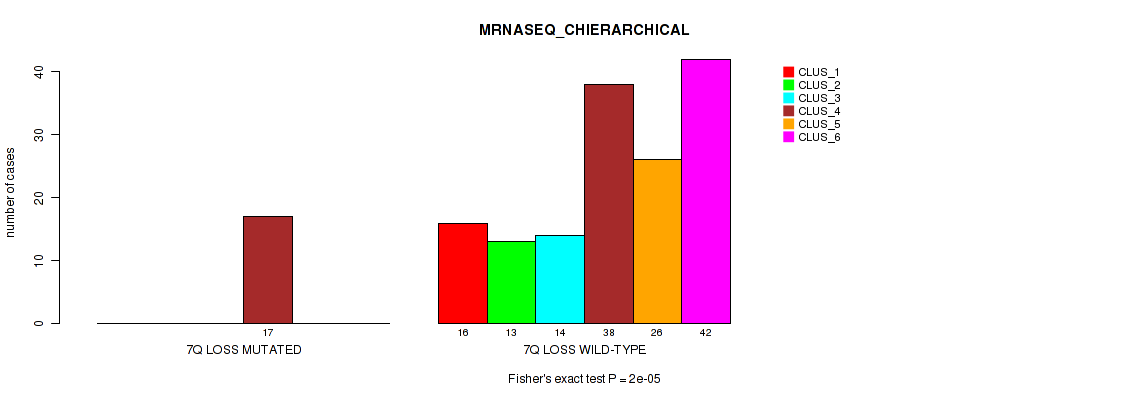
P value = 0.00148 (Fisher's exact test), Q value = 0.011
Table S54. Gene #21: '7q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| 7Q LOSS MUTATED | 1 | 10 | 4 | 4 |
| 7Q LOSS WILD-TYPE | 57 | 27 | 37 | 39 |
Figure S54. Get High-res Image Gene #21: '7q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
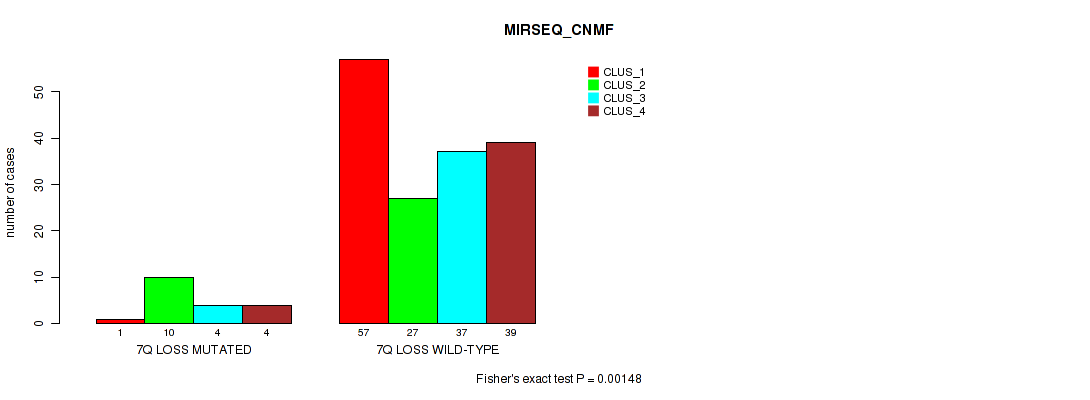
P value = 0.00078 (Fisher's exact test), Q value = 0.0078
Table S55. Gene #21: '7q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 7Q LOSS MUTATED | 1 | 8 | 10 |
| 7Q LOSS WILD-TYPE | 66 | 26 | 68 |
Figure S55. Get High-res Image Gene #21: '7q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
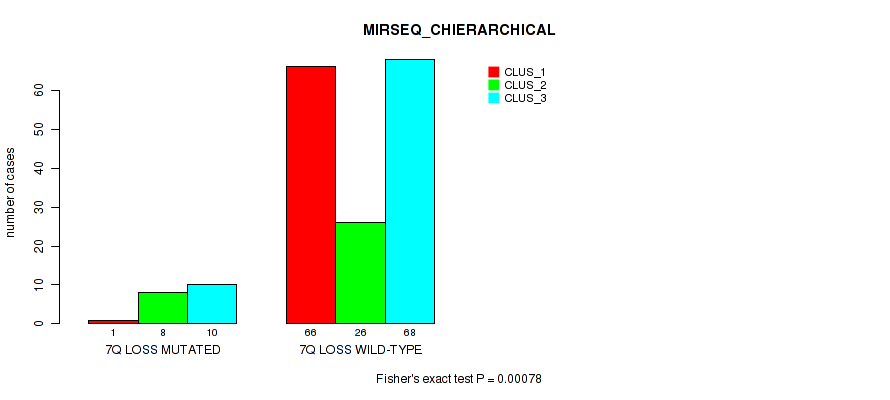
P value = 0.00613 (Fisher's exact test), Q value = 0.026
Table S56. Gene #22: '12p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 12P LOSS MUTATED | 1 | 0 | 2 | 1 |
| 12P LOSS WILD-TYPE | 147 | 14 | 25 | 1 |
Figure S56. Get High-res Image Gene #22: '12p loss' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00027 (Fisher's exact test), Q value = 0.0032
Table S57. Gene #23: '15q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 15Q LOSS MUTATED | 0 | 0 | 3 | 1 |
| 15Q LOSS WILD-TYPE | 148 | 14 | 24 | 1 |
Figure S57. Get High-res Image Gene #23: '15q loss' versus Molecular Subtype #1: 'CN_CNMF'
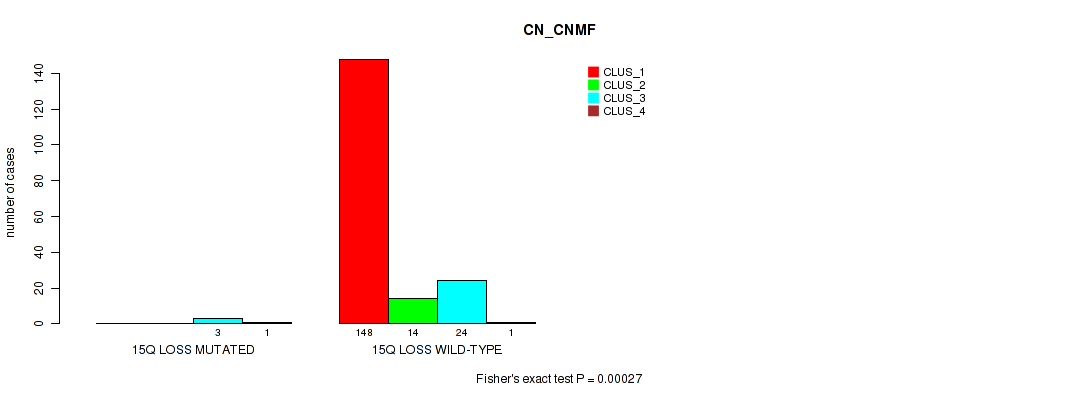
P value = 0.0227 (Fisher's exact test), Q value = 0.064
Table S58. Gene #23: '15q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| 15Q LOSS MUTATED | 0 | 3 | 0 | 1 |
| 15Q LOSS WILD-TYPE | 58 | 34 | 41 | 42 |
Figure S58. Get High-res Image Gene #23: '15q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
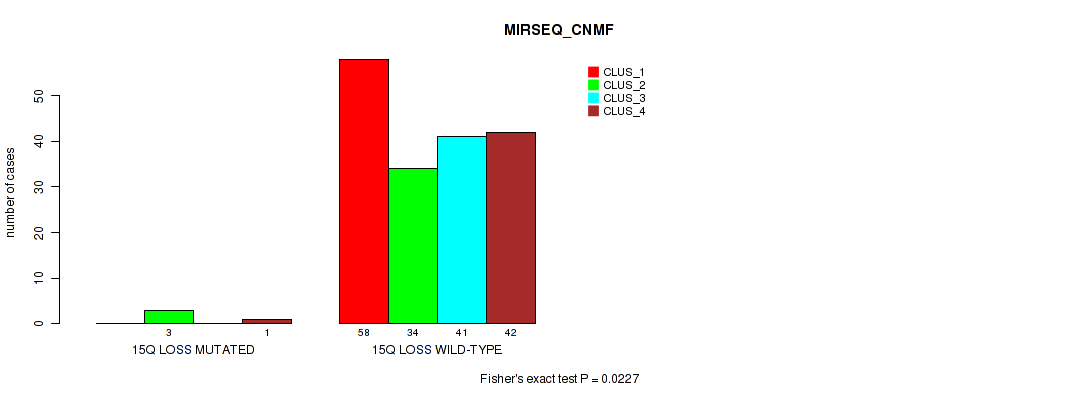
P value = 0.0011 (Fisher's exact test), Q value = 0.0088
Table S59. Gene #24: '16q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 16Q LOSS MUTATED | 0 | 0 | 4 | 0 |
| 16Q LOSS WILD-TYPE | 148 | 14 | 23 | 2 |
Figure S59. Get High-res Image Gene #24: '16q loss' versus Molecular Subtype #1: 'CN_CNMF'
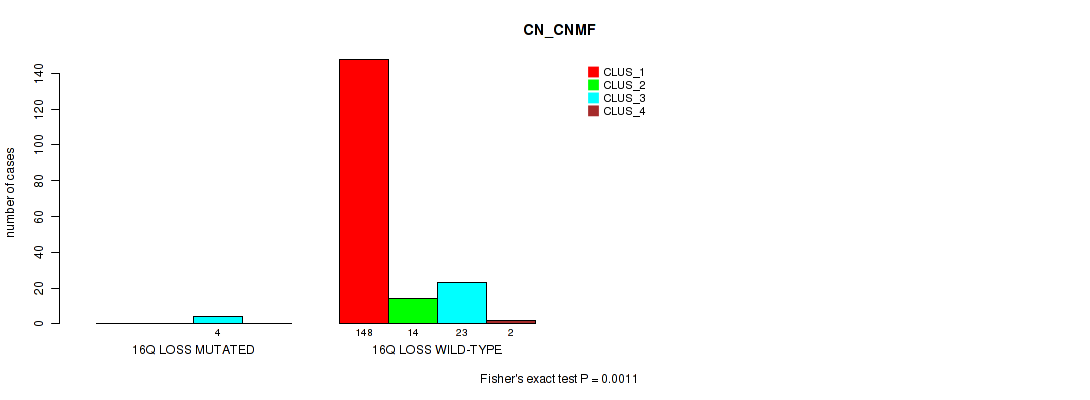
P value = 0.0152 (Fisher's exact test), Q value = 0.048
Table S60. Gene #24: '16q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| 16Q LOSS MUTATED | 0 | 3 | 1 | 0 |
| 16Q LOSS WILD-TYPE | 58 | 34 | 40 | 43 |
Figure S60. Get High-res Image Gene #24: '16q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
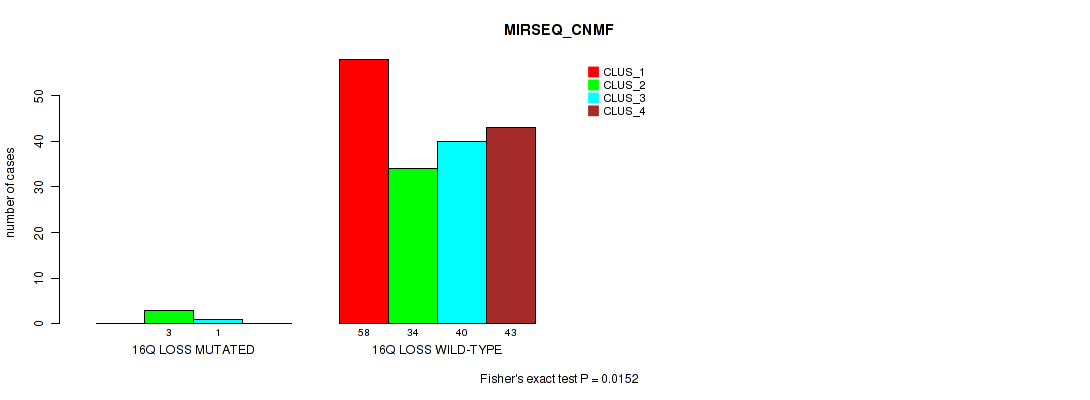
P value = 0.0216 (Fisher's exact test), Q value = 0.063
Table S61. Gene #24: '16q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 16Q LOSS MUTATED | 0 | 3 | 1 |
| 16Q LOSS WILD-TYPE | 67 | 31 | 77 |
Figure S61. Get High-res Image Gene #24: '16q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
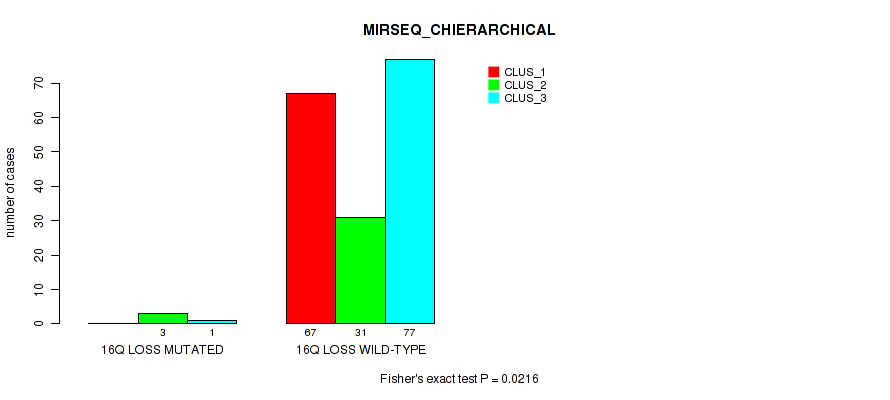
P value = 1e-05 (Fisher's exact test), Q value = 0.00048
Table S62. Gene #25: '17p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 17P LOSS MUTATED | 0 | 0 | 13 | 0 |
| 17P LOSS WILD-TYPE | 148 | 14 | 14 | 2 |
Figure S62. Get High-res Image Gene #25: '17p loss' versus Molecular Subtype #1: 'CN_CNMF'
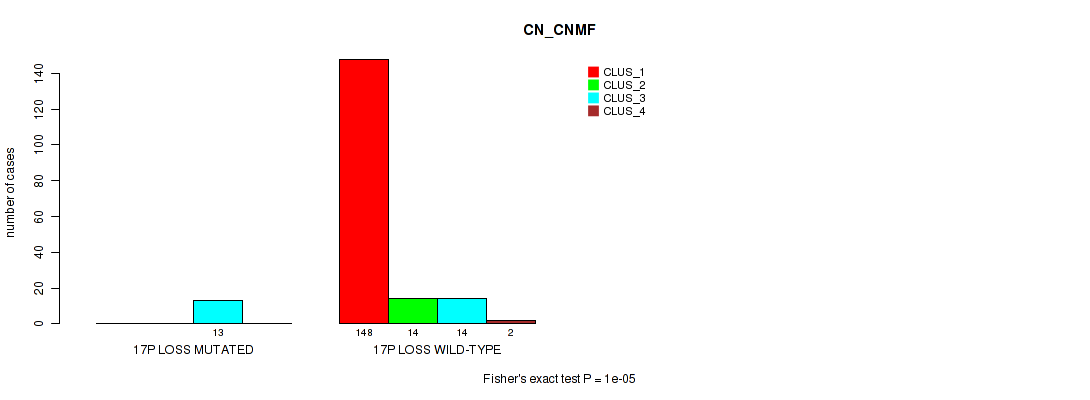
P value = 0.00169 (Fisher's exact test), Q value = 0.011
Table S63. Gene #25: '17p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| 17P LOSS MUTATED | 0 | 10 | 3 | 0 |
| 17P LOSS WILD-TYPE | 33 | 51 | 44 | 44 |
Figure S63. Get High-res Image Gene #25: '17p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.00164 (Fisher's exact test), Q value = 0.011
Table S64. Gene #25: '17p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| 17P LOSS MUTATED | 0 | 0 | 0 | 12 | 0 | 1 |
| 17P LOSS WILD-TYPE | 16 | 13 | 14 | 43 | 26 | 41 |
Figure S64. Get High-res Image Gene #25: '17p loss' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 0.00359 (Fisher's exact test), Q value = 0.018
Table S65. Gene #25: '17p loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 17P LOSS MUTATED | 0 | 4 | 9 |
| 17P LOSS WILD-TYPE | 67 | 30 | 69 |
Figure S65. Get High-res Image Gene #25: '17p loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
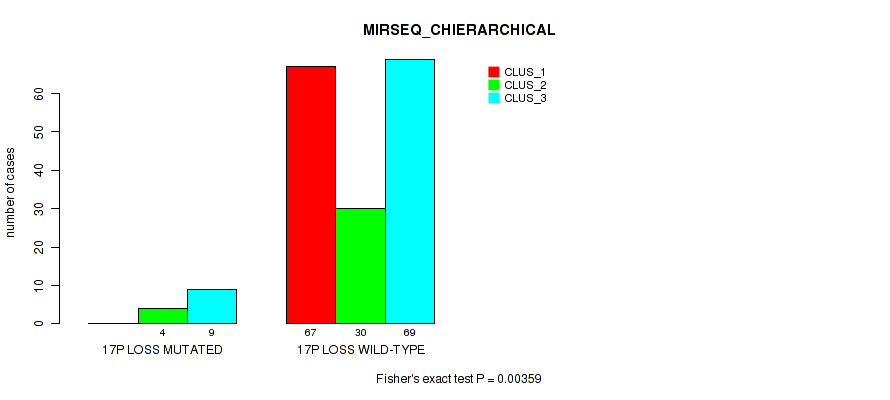
P value = 2e-05 (Fisher's exact test), Q value = 0.00048
Table S66. Gene #26: '17q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 17Q LOSS MUTATED | 0 | 0 | 7 | 0 |
| 17Q LOSS WILD-TYPE | 148 | 14 | 20 | 2 |
Figure S66. Get High-res Image Gene #26: '17q loss' versus Molecular Subtype #1: 'CN_CNMF'
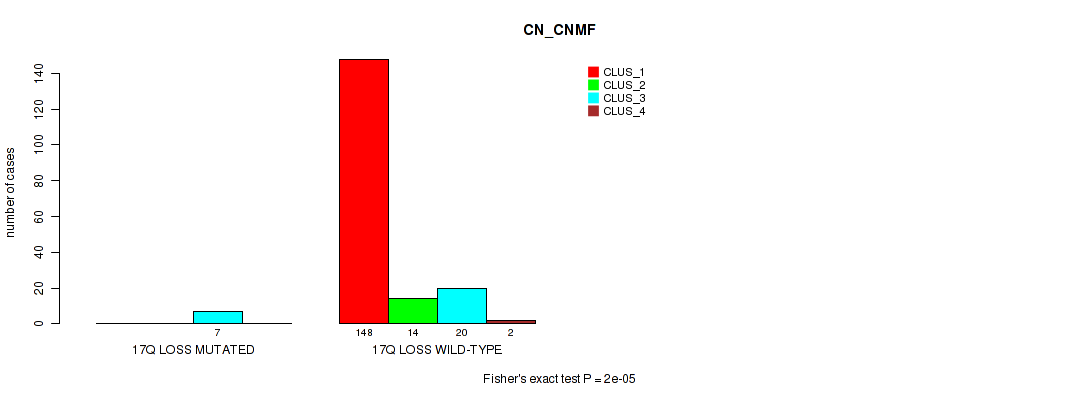
P value = 0.0369 (Fisher's exact test), Q value = 0.094
Table S67. Gene #26: '17q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| 17Q LOSS MUTATED | 0 | 4 | 1 | 2 |
| 17Q LOSS WILD-TYPE | 58 | 33 | 40 | 41 |
Figure S67. Get High-res Image Gene #26: '17q loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
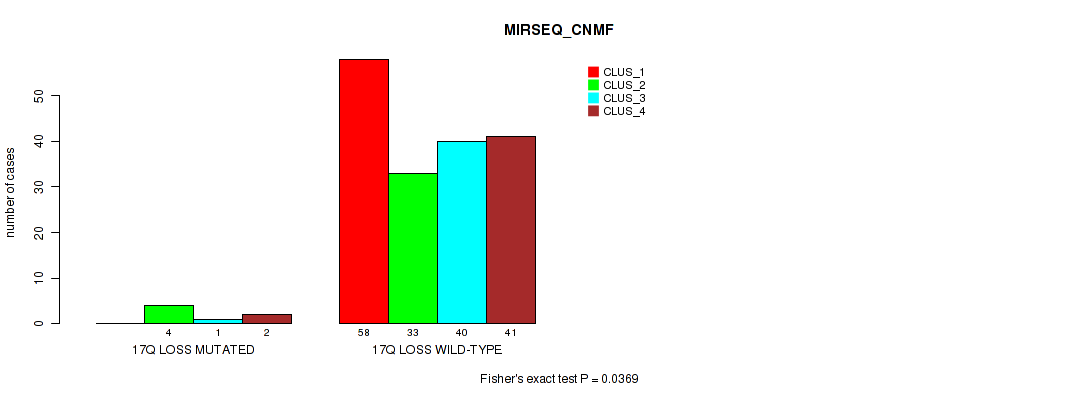
P value = 0.0472 (Fisher's exact test), Q value = 0.11
Table S68. Gene #26: '17q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| 17Q LOSS MUTATED | 0 | 3 | 4 |
| 17Q LOSS WILD-TYPE | 67 | 31 | 74 |
Figure S68. Get High-res Image Gene #26: '17q loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 0.00012 (Fisher's exact test), Q value = 0.0018
Table S69. Gene #27: '18p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 18P LOSS MUTATED | 0 | 0 | 5 | 0 |
| 18P LOSS WILD-TYPE | 148 | 14 | 22 | 2 |
Figure S69. Get High-res Image Gene #27: '18p loss' versus Molecular Subtype #1: 'CN_CNMF'
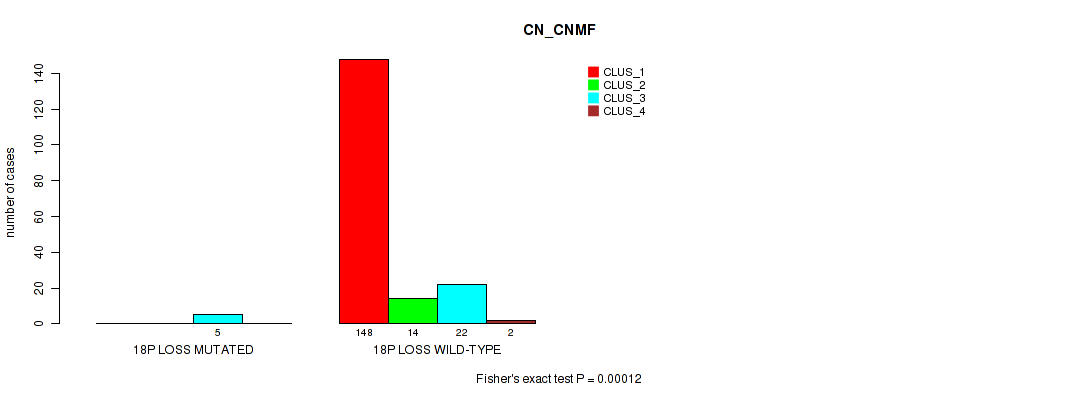
P value = 0.0213 (Fisher's exact test), Q value = 0.063
Table S70. Gene #27: '18p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| 18P LOSS MUTATED | 0 | 5 | 0 | 0 |
| 18P LOSS WILD-TYPE | 33 | 56 | 47 | 44 |
Figure S70. Get High-res Image Gene #27: '18p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
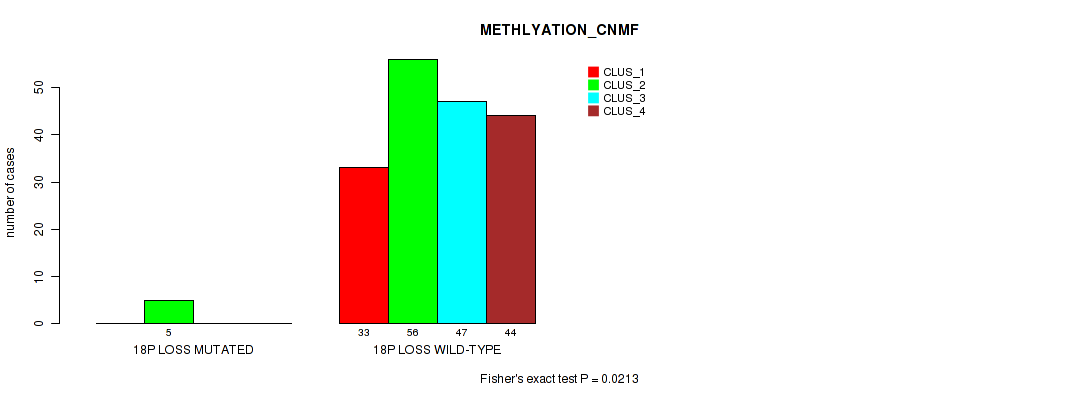
P value = 0.00132 (Fisher's exact test), Q value = 0.01
Table S71. Gene #28: '18q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 18Q LOSS MUTATED | 0 | 0 | 4 | 0 |
| 18Q LOSS WILD-TYPE | 148 | 14 | 23 | 2 |
Figure S71. Get High-res Image Gene #28: '18q loss' versus Molecular Subtype #1: 'CN_CNMF'
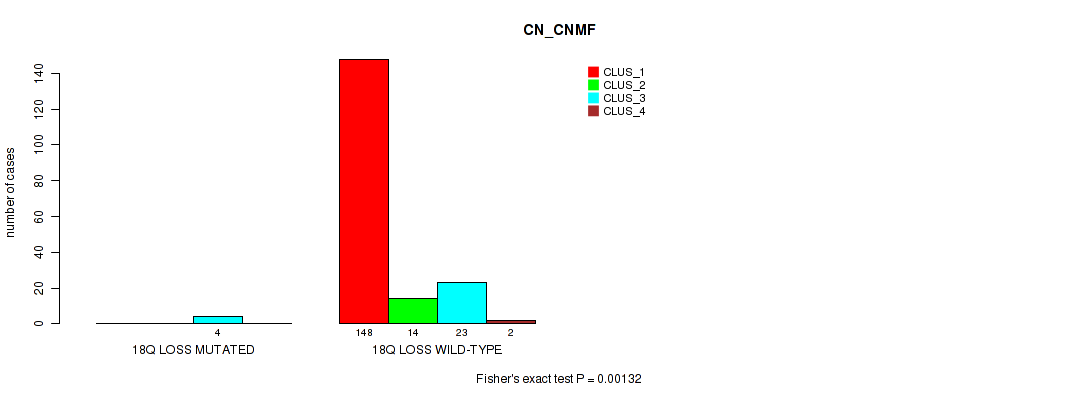
P value = 0.00295 (Fisher's exact test), Q value = 0.017
Table S72. Gene #29: '19p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 19P LOSS MUTATED | 0 | 1 | 3 | 0 |
| 19P LOSS WILD-TYPE | 148 | 13 | 24 | 2 |
Figure S72. Get High-res Image Gene #29: '19p loss' versus Molecular Subtype #1: 'CN_CNMF'
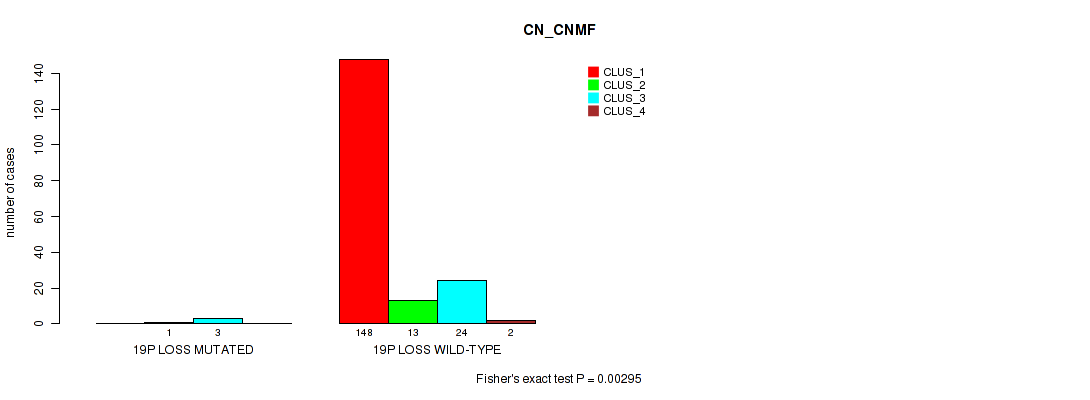
P value = 0.00309 (Fisher's exact test), Q value = 0.017
Table S73. Gene #30: '19q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| 19Q LOSS MUTATED | 0 | 1 | 3 | 0 |
| 19Q LOSS WILD-TYPE | 148 | 13 | 24 | 2 |
Figure S73. Get High-res Image Gene #30: '19q loss' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1e-04 (Fisher's exact test), Q value = 0.0017
Table S74. Gene #31: 'xp loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| XP LOSS MUTATED | 0 | 0 | 4 | 1 |
| XP LOSS WILD-TYPE | 148 | 14 | 23 | 1 |
Figure S74. Get High-res Image Gene #31: 'xp loss' versus Molecular Subtype #1: 'CN_CNMF'
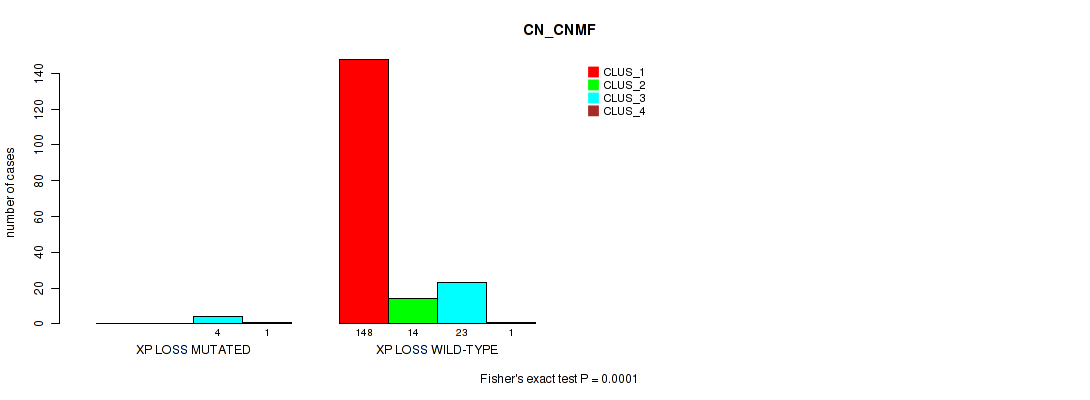
P value = 0.00369 (Fisher's exact test), Q value = 0.018
Table S75. Gene #31: 'xp loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| XP LOSS MUTATED | 0 | 4 | 1 | 0 |
| XP LOSS WILD-TYPE | 58 | 33 | 40 | 43 |
Figure S75. Get High-res Image Gene #31: 'xp loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 0.00483 (Fisher's exact test), Q value = 0.022
Table S76. Gene #31: 'xp loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| XP LOSS MUTATED | 0 | 4 | 1 |
| XP LOSS WILD-TYPE | 67 | 30 | 77 |
Figure S76. Get High-res Image Gene #31: 'xp loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 0.00011 (Fisher's exact test), Q value = 0.0018
Table S77. Gene #32: 'xq loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| XQ LOSS MUTATED | 0 | 0 | 4 | 1 |
| XQ LOSS WILD-TYPE | 148 | 14 | 23 | 1 |
Figure S77. Get High-res Image Gene #32: 'xq loss' versus Molecular Subtype #1: 'CN_CNMF'
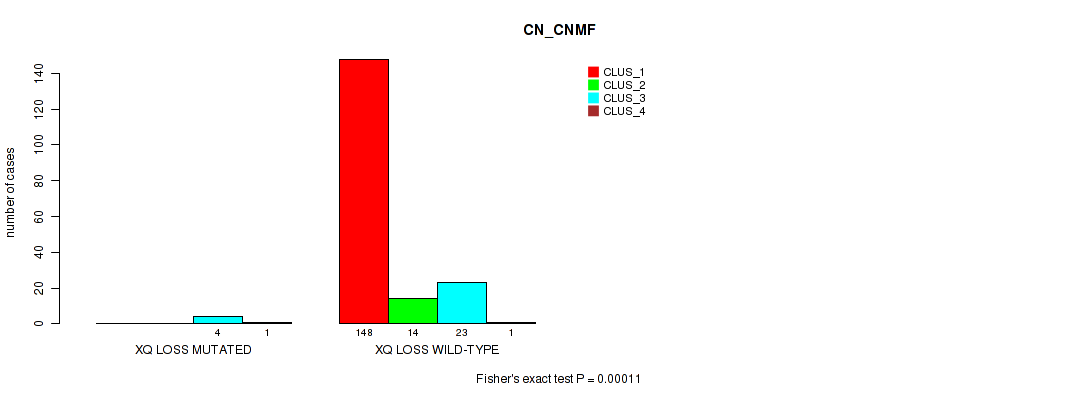
P value = 0.00313 (Fisher's exact test), Q value = 0.017
Table S78. Gene #32: 'xq loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| XQ LOSS MUTATED | 0 | 4 | 1 | 0 |
| XQ LOSS WILD-TYPE | 58 | 33 | 40 | 43 |
Figure S78. Get High-res Image Gene #32: 'xq loss' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

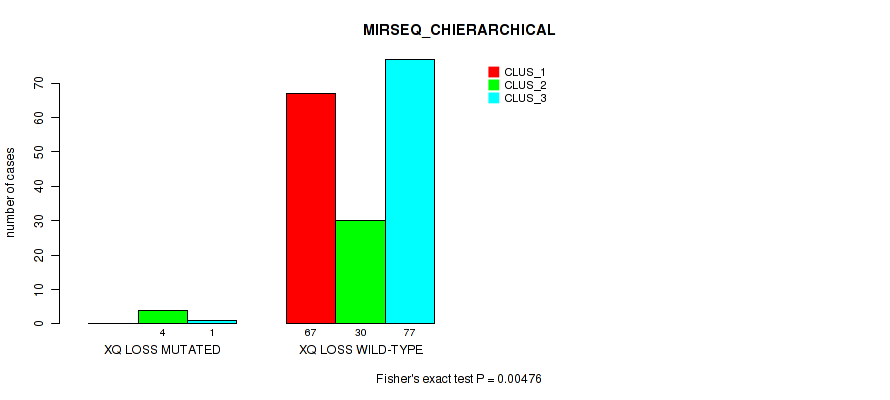
P value = 0.00476 (Fisher's exact test), Q value = 0.022
Table S79. Gene #32: 'xq loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| XQ LOSS MUTATED | 0 | 4 | 1 |
| XQ LOSS WILD-TYPE | 67 | 30 | 77 |
Figure S79. Get High-res Image Gene #32: 'xq loss' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
-
Copy number data file = broad_values_by_arm.txt from GISTIC pipeline
-
Processed Copy number data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_Correlate_Genomic_Events_Preprocess/LAML-TB/19782154/transformed.cor.cli.txt
-
Molecular subtypes file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_mergedClustering/LAML-TB/19785823/LAML-TB.transferedmergedcluster.txt
-
Number of patients = 191
-
Number of significantly arm-level cnvs = 32
-
Number of molecular subtypes = 6
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.