This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 19 genes and 2 molecular subtypes across 108 patients, 12 significant findings detected with P value < 0.05 and Q value < 0.25.
-
TP53 mutation correlated to 'LINCRNA_CHIERARCHICAL'.
-
KRAS mutation correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
STK11 mutation correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
EGFR mutation correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
LMO2 mutation correlated to 'MRNA_CHIERARCHICAL'.
-
C10ORF62 mutation correlated to 'MRNA_CHIERARCHICAL'.
-
BIRC6 mutation correlated to 'MRNA_CHIERARCHICAL'.
-
C16ORF82 mutation correlated to 'MRNA_CHIERARCHICAL'.
-
CLIP2 mutation correlated to 'MRNA_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between mutation status of 19 genes and 2 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 12 significant findings detected.
|
Clinical Features |
MRNA CHIERARCHICAL |
LINCRNA CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Fisher's exact test | Fisher's exact test | |
| KRAS | 34 (31%) | 74 |
2e-05 (0.000127) |
2e-05 (0.000127) |
| STK11 | 20 (19%) | 88 |
1e-05 (9.5e-05) |
1e-05 (9.5e-05) |
| EGFR | 37 (34%) | 71 |
1e-05 (9.5e-05) |
1e-05 (9.5e-05) |
| TP53 | 59 (55%) | 49 |
0.0665 (0.194) |
0.0362 (0.115) |
| LMO2 | 5 (5%) | 103 |
0.0353 (0.115) |
0.327 (0.547) |
| C10ORF62 | 4 (4%) | 104 |
0.0265 (0.112) |
0.688 (0.747) |
| BIRC6 | 6 (6%) | 102 |
0.0105 (0.0572) |
0.372 (0.547) |
| C16ORF82 | 5 (5%) | 103 |
0.0356 (0.115) |
0.0774 (0.21) |
| CLIP2 | 7 (6%) | 101 |
0.0244 (0.112) |
0.223 (0.474) |
| KEAP1 | 12 (11%) | 96 |
0.36 (0.547) |
0.684 (0.747) |
| RB1 | 8 (7%) | 100 |
0.643 (0.747) |
0.807 (0.852) |
| IL21R | 6 (6%) | 102 |
0.232 (0.474) |
0.372 (0.547) |
| EGFL6 | 5 (5%) | 103 |
0.511 (0.647) |
0.601 (0.737) |
| DKK3 | 4 (4%) | 104 |
0.459 (0.602) |
0.686 (0.747) |
| TDG | 3 (3%) | 105 |
0.237 (0.474) |
0.358 (0.547) |
| GRIK3 | 9 (8%) | 99 |
0.321 (0.547) |
0.416 (0.586) |
| AFAP1 | 5 (5%) | 103 |
0.374 (0.547) |
0.445 (0.602) |
| ELFN1 | 6 (6%) | 102 |
1 (1.00) |
1 (1.00) |
| TLR7 | 6 (6%) | 102 |
0.232 (0.474) |
0.0993 (0.252) |
P value = 0.0665 (Fisher's exact test), Q value = 0.19
Table S1. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| TP53 MUTATED | 18 | 19 | 22 |
| TP53 WILD-TYPE | 15 | 7 | 27 |
P value = 0.0362 (Fisher's exact test), Q value = 0.11
Table S2. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| TP53 MUTATED | 17 | 22 | 20 |
| TP53 WILD-TYPE | 15 | 8 | 26 |
Figure S1. Get High-res Image Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
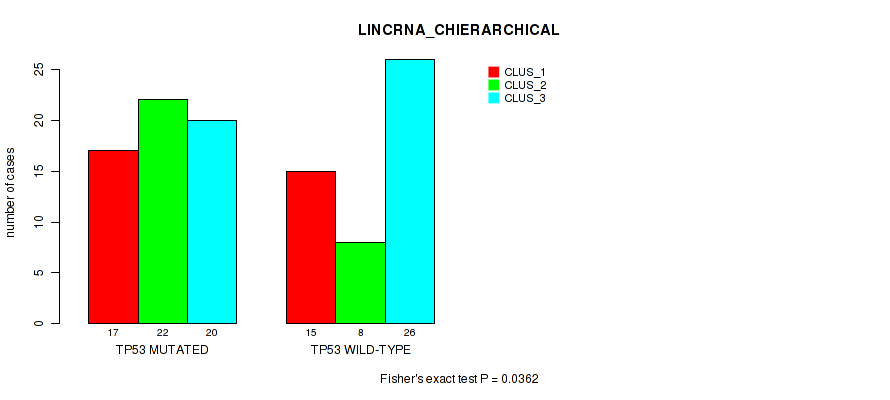
P value = 2e-05 (Fisher's exact test), Q value = 0.00013
Table S3. Gene #2: 'KRAS MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| KRAS MUTATED | 18 | 11 | 5 |
| KRAS WILD-TYPE | 15 | 15 | 44 |
Figure S2. Get High-res Image Gene #2: 'KRAS MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
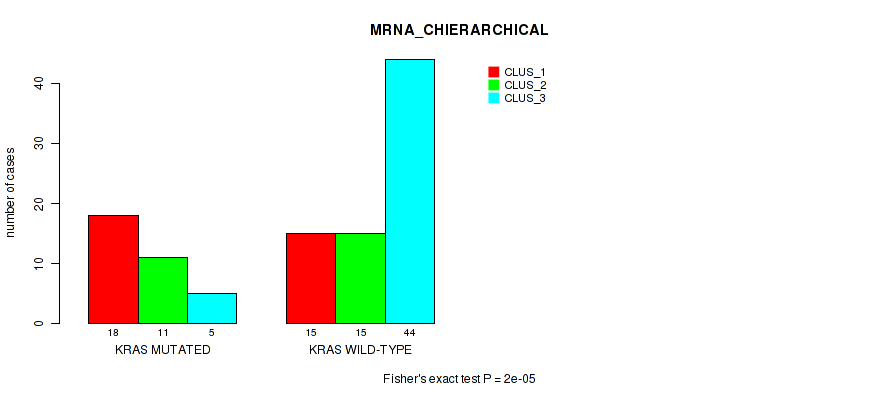
P value = 2e-05 (Fisher's exact test), Q value = 0.00013
Table S4. Gene #2: 'KRAS MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| KRAS MUTATED | 18 | 12 | 4 |
| KRAS WILD-TYPE | 14 | 18 | 42 |
Figure S3. Get High-res Image Gene #2: 'KRAS MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
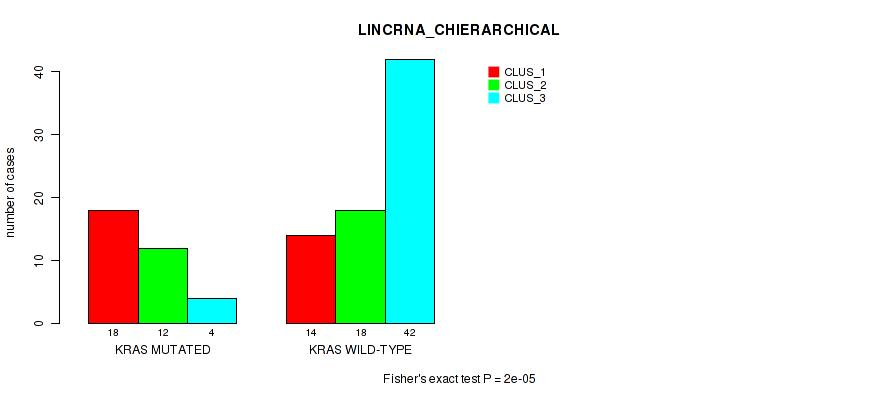
P value = 1e-05 (Fisher's exact test), Q value = 9.5e-05
Table S5. Gene #3: 'STK11 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| STK11 MUTATED | 18 | 1 | 1 |
| STK11 WILD-TYPE | 15 | 25 | 48 |
Figure S4. Get High-res Image Gene #3: 'STK11 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
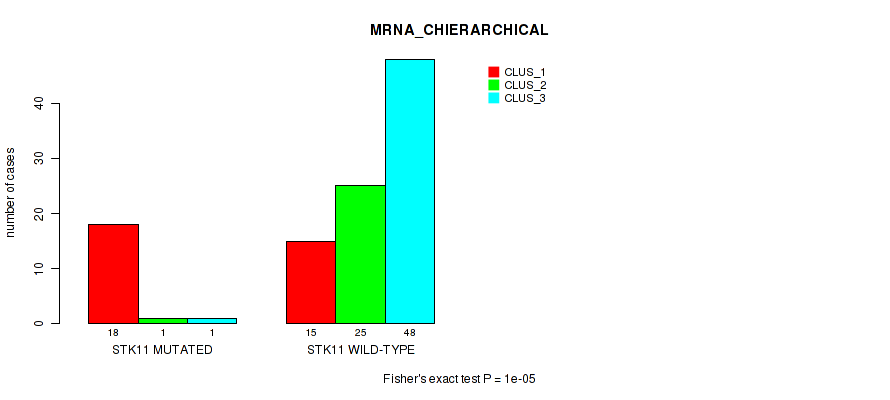
P value = 1e-05 (Fisher's exact test), Q value = 9.5e-05
Table S6. Gene #3: 'STK11 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| STK11 MUTATED | 19 | 0 | 1 |
| STK11 WILD-TYPE | 13 | 30 | 45 |
Figure S5. Get High-res Image Gene #3: 'STK11 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
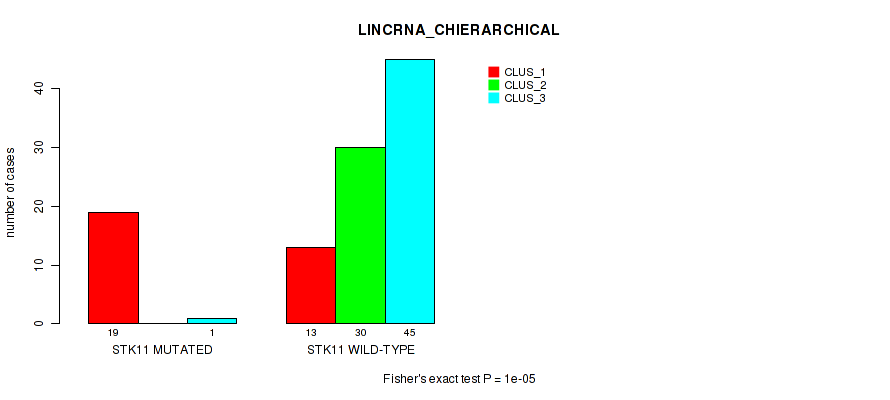
P value = 1e-05 (Fisher's exact test), Q value = 9.5e-05
Table S7. Gene #4: 'EGFR MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| EGFR MUTATED | 1 | 7 | 29 |
| EGFR WILD-TYPE | 32 | 19 | 20 |
Figure S6. Get High-res Image Gene #4: 'EGFR MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
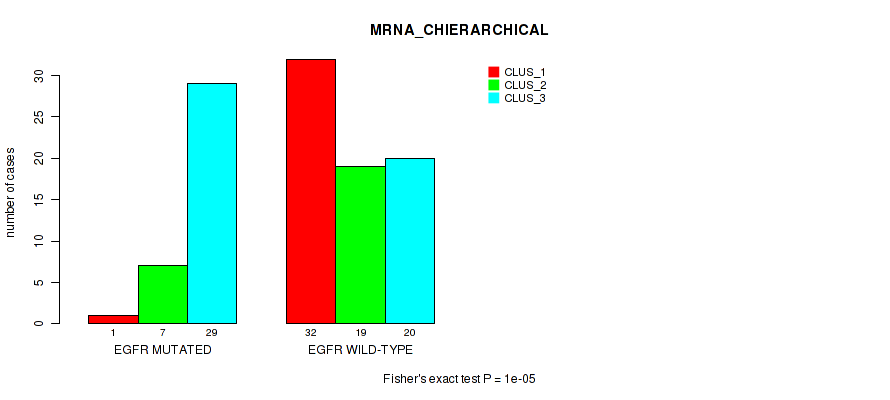
P value = 1e-05 (Fisher's exact test), Q value = 9.5e-05
Table S8. Gene #4: 'EGFR MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| EGFR MUTATED | 0 | 10 | 27 |
| EGFR WILD-TYPE | 32 | 20 | 19 |
Figure S7. Get High-res Image Gene #4: 'EGFR MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
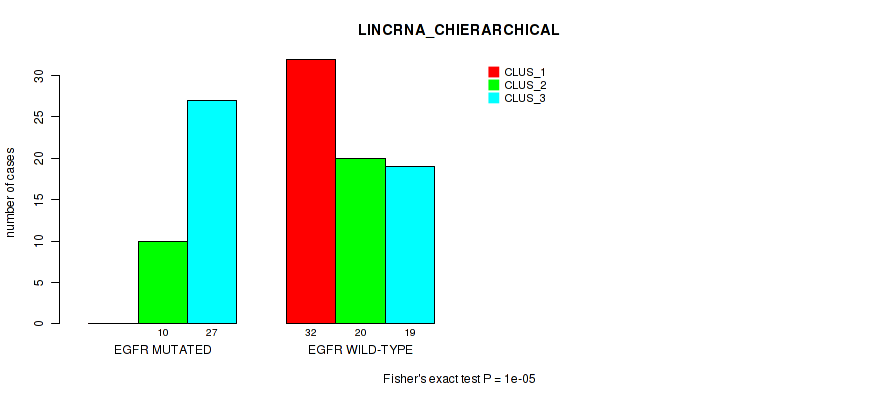
P value = 0.36 (Fisher's exact test), Q value = 0.55
Table S9. Gene #5: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| KEAP1 MUTATED | 6 | 2 | 4 |
| KEAP1 WILD-TYPE | 27 | 24 | 45 |
P value = 0.684 (Fisher's exact test), Q value = 0.75
Table S10. Gene #5: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| KEAP1 MUTATED | 5 | 3 | 4 |
| KEAP1 WILD-TYPE | 27 | 27 | 42 |
P value = 0.643 (Fisher's exact test), Q value = 0.75
Table S11. Gene #6: 'RB1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| RB1 MUTATED | 2 | 3 | 3 |
| RB1 WILD-TYPE | 31 | 23 | 46 |
P value = 0.807 (Fisher's exact test), Q value = 0.85
Table S12. Gene #6: 'RB1 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| RB1 MUTATED | 2 | 3 | 3 |
| RB1 WILD-TYPE | 30 | 27 | 43 |
P value = 0.232 (Fisher's exact test), Q value = 0.47
Table S13. Gene #7: 'IL21R MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| IL21R MUTATED | 2 | 3 | 1 |
| IL21R WILD-TYPE | 31 | 23 | 48 |
P value = 0.372 (Fisher's exact test), Q value = 0.55
Table S14. Gene #7: 'IL21R MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| IL21R MUTATED | 2 | 3 | 1 |
| IL21R WILD-TYPE | 30 | 27 | 45 |
P value = 0.511 (Fisher's exact test), Q value = 0.65
Table S15. Gene #8: 'EGFL6 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| EGFL6 MUTATED | 2 | 2 | 1 |
| EGFL6 WILD-TYPE | 31 | 24 | 48 |
P value = 0.601 (Fisher's exact test), Q value = 0.74
Table S16. Gene #8: 'EGFL6 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| EGFL6 MUTATED | 2 | 2 | 1 |
| EGFL6 WILD-TYPE | 30 | 28 | 45 |
P value = 0.0353 (Fisher's exact test), Q value = 0.11
Table S17. Gene #9: 'LMO2 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| LMO2 MUTATED | 2 | 3 | 0 |
| LMO2 WILD-TYPE | 31 | 23 | 49 |
Figure S8. Get High-res Image Gene #9: 'LMO2 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
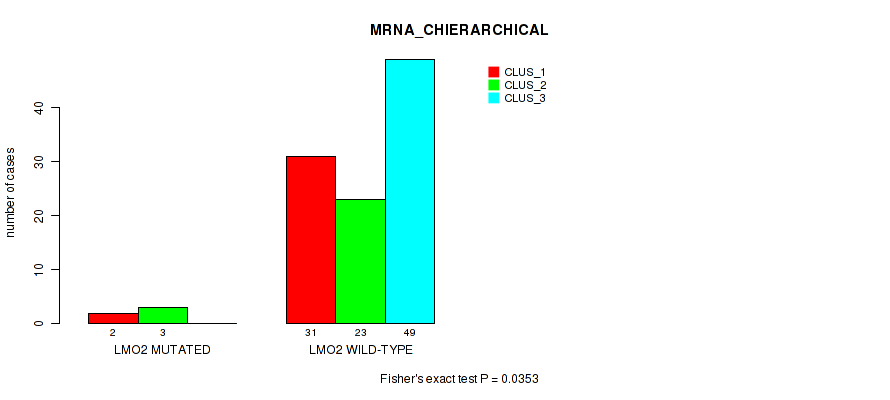
P value = 0.327 (Fisher's exact test), Q value = 0.55
Table S18. Gene #9: 'LMO2 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| LMO2 MUTATED | 1 | 3 | 1 |
| LMO2 WILD-TYPE | 31 | 27 | 45 |
P value = 0.0265 (Fisher's exact test), Q value = 0.11
Table S19. Gene #10: 'C10ORF62 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| C10ORF62 MUTATED | 1 | 3 | 0 |
| C10ORF62 WILD-TYPE | 32 | 23 | 49 |
Figure S9. Get High-res Image Gene #10: 'C10ORF62 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
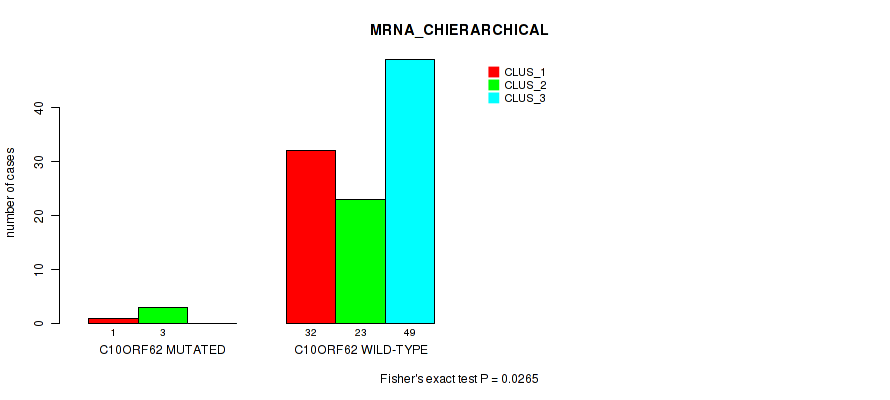
P value = 0.688 (Fisher's exact test), Q value = 0.75
Table S20. Gene #10: 'C10ORF62 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| C10ORF62 MUTATED | 1 | 2 | 1 |
| C10ORF62 WILD-TYPE | 31 | 28 | 45 |
P value = 0.459 (Fisher's exact test), Q value = 0.6
Table S21. Gene #11: 'DKK3 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| DKK3 MUTATED | 1 | 2 | 1 |
| DKK3 WILD-TYPE | 32 | 24 | 48 |
P value = 0.686 (Fisher's exact test), Q value = 0.75
Table S22. Gene #11: 'DKK3 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| DKK3 MUTATED | 1 | 2 | 1 |
| DKK3 WILD-TYPE | 31 | 28 | 45 |
P value = 0.0105 (Fisher's exact test), Q value = 0.057
Table S23. Gene #12: 'BIRC6 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| BIRC6 MUTATED | 2 | 4 | 0 |
| BIRC6 WILD-TYPE | 31 | 22 | 49 |
Figure S10. Get High-res Image Gene #12: 'BIRC6 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
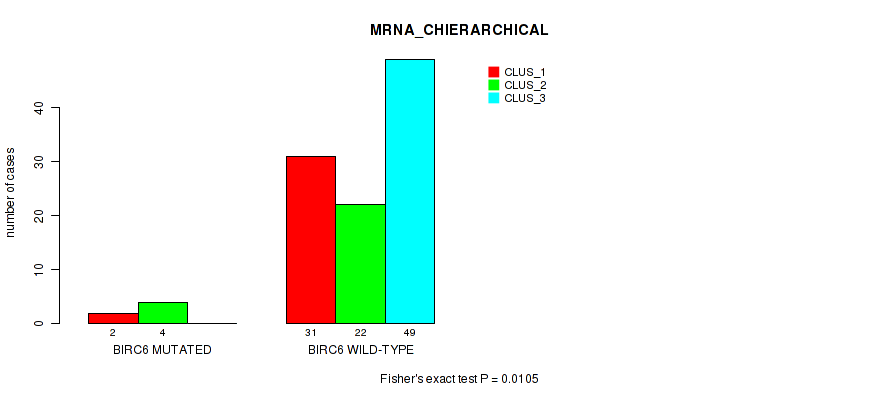
P value = 0.372 (Fisher's exact test), Q value = 0.55
Table S24. Gene #12: 'BIRC6 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| BIRC6 MUTATED | 2 | 3 | 1 |
| BIRC6 WILD-TYPE | 30 | 27 | 45 |
P value = 0.237 (Fisher's exact test), Q value = 0.47
Table S25. Gene #13: 'TDG MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| TDG MUTATED | 0 | 2 | 1 |
| TDG WILD-TYPE | 33 | 24 | 48 |
P value = 0.358 (Fisher's exact test), Q value = 0.55
Table S26. Gene #13: 'TDG MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| TDG MUTATED | 0 | 2 | 1 |
| TDG WILD-TYPE | 32 | 28 | 45 |
P value = 0.0356 (Fisher's exact test), Q value = 0.11
Table S27. Gene #14: 'C16ORF82 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| C16ORF82 MUTATED | 2 | 3 | 0 |
| C16ORF82 WILD-TYPE | 31 | 23 | 49 |
Figure S11. Get High-res Image Gene #14: 'C16ORF82 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
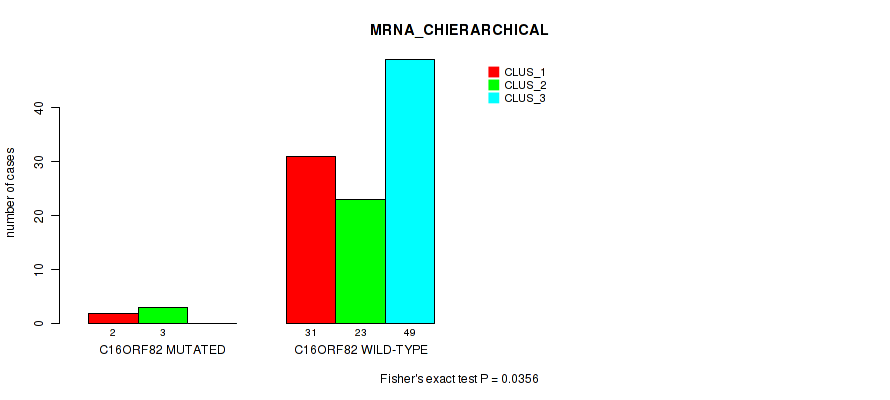
P value = 0.0774 (Fisher's exact test), Q value = 0.21
Table S28. Gene #14: 'C16ORF82 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| C16ORF82 MUTATED | 2 | 3 | 0 |
| C16ORF82 WILD-TYPE | 30 | 27 | 46 |
P value = 0.321 (Fisher's exact test), Q value = 0.55
Table S29. Gene #15: 'GRIK3 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| GRIK3 MUTATED | 4 | 3 | 2 |
| GRIK3 WILD-TYPE | 29 | 23 | 47 |
P value = 0.416 (Fisher's exact test), Q value = 0.59
Table S30. Gene #15: 'GRIK3 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| GRIK3 MUTATED | 4 | 3 | 2 |
| GRIK3 WILD-TYPE | 28 | 27 | 44 |
P value = 0.374 (Fisher's exact test), Q value = 0.55
Table S31. Gene #16: 'AFAP1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| AFAP1 MUTATED | 3 | 1 | 1 |
| AFAP1 WILD-TYPE | 30 | 25 | 48 |
P value = 0.445 (Fisher's exact test), Q value = 0.6
Table S32. Gene #16: 'AFAP1 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| AFAP1 MUTATED | 3 | 1 | 1 |
| AFAP1 WILD-TYPE | 29 | 29 | 45 |
P value = 0.0244 (Fisher's exact test), Q value = 0.11
Table S33. Gene #17: 'CLIP2 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| CLIP2 MUTATED | 4 | 3 | 0 |
| CLIP2 WILD-TYPE | 29 | 23 | 49 |
Figure S12. Get High-res Image Gene #17: 'CLIP2 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
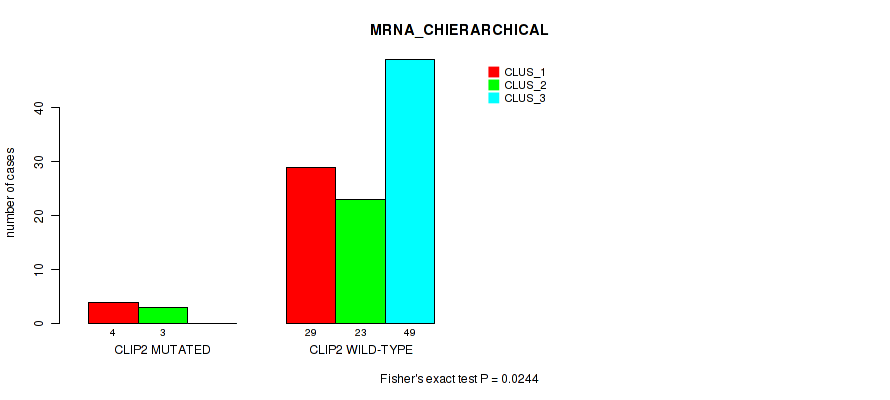
P value = 0.223 (Fisher's exact test), Q value = 0.47
Table S34. Gene #17: 'CLIP2 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| CLIP2 MUTATED | 4 | 2 | 1 |
| CLIP2 WILD-TYPE | 28 | 28 | 45 |
P value = 1 (Fisher's exact test), Q value = 1
Table S35. Gene #18: 'ELFN1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| ELFN1 MUTATED | 2 | 1 | 3 |
| ELFN1 WILD-TYPE | 31 | 25 | 46 |
P value = 1 (Fisher's exact test), Q value = 1
Table S36. Gene #18: 'ELFN1 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| ELFN1 MUTATED | 2 | 1 | 3 |
| ELFN1 WILD-TYPE | 30 | 29 | 43 |
P value = 0.232 (Fisher's exact test), Q value = 0.47
Table S37. Gene #19: 'TLR7 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 26 | 49 |
| TLR7 MUTATED | 2 | 3 | 1 |
| TLR7 WILD-TYPE | 31 | 23 | 48 |
P value = 0.0993 (Fisher's exact test), Q value = 0.25
Table S38. Gene #19: 'TLR7 MUTATION STATUS' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 30 | 46 |
| TLR7 MUTATED | 1 | 4 | 1 |
| TLR7 WILD-TYPE | 31 | 26 | 45 |
-
Mutation data file = sample_sig_gene_table.txt from Mutsig_2CV pipeline
-
Processed Mutation data file = /cromwell_root/fc-e7058367-eaa6-44b5-aab5-1ec08acf146a/03d5323d-b92b-4a22-90d3-f871afeb0c16/correlate_genomic_events_all/ee5cbd6a-2978-41ca-ada7-5146d30af8b4/call-preprocess_genomic_event/transformed.cor.cli.txt
-
Molecular subtypes file = /cromwell_root/fc-e7058367-eaa6-44b5-aab5-1ec08acf146a/69dd18b3-97fe-4143-b0f4-32e5d55e929d/aggregate_clusters_workflow/d44c315a-761b-4b72-be1f-cb969be36102/call-aggregate_clusters/CPTAC3-LUAD-TP.transposedmergedcluster.txt
-
Number of patients = 108
-
Number of significantly mutated genes = 19
-
Number of Molecular subtypes = 2
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.