(primary solid tumor cohort)
This pipeline computes the correlation between significantly recurrent gene mutations and selected clinical features.
Testing the association between mutation status of 181 genes and 5 clinical features across 276 patients, 2 significant findings detected with Q value < 0.25.
-
IDH1 mutation correlated to 'AGE'.
-
ANO2 mutation correlated to 'Time to Death'.
Table 1. Get Full Table Overview of the association between mutation status of 181 genes and 5 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 2 significant findings detected.
|
Clinical Features |
Time to Death |
AGE | GENDER |
KARNOFSKY PERFORMANCE SCORE |
RADIATIONS RADIATION REGIMENINDICATION |
||
| nMutated (%) | nWild-Type | logrank test | t-test | Fisher's exact test | t-test | Fisher's exact test | |
| IDH1 | 14 (5%) | 262 |
0.00261 (1.00) |
7.95e-05 (0.0686) |
0.269 (1.00) |
0.0319 (1.00) |
0.00287 (1.00) |
| ANO2 | 5 (2%) | 271 |
4.75e-09 (4.1e-06) |
0.0702 (1.00) |
0.0617 (1.00) |
0.363 (1.00) |
0.0512 (1.00) |
| EGFR | 59 (21%) | 217 |
0.59 (1.00) |
0.196 (1.00) |
0.0956 (1.00) |
0.34 (1.00) |
0.647 (1.00) |
| TP53 | 57 (21%) | 219 |
0.281 (1.00) |
0.666 (1.00) |
1 (1.00) |
0.102 (1.00) |
0.876 (1.00) |
| PTEN | 65 (24%) | 211 |
0.514 (1.00) |
0.24 (1.00) |
0.557 (1.00) |
0.244 (1.00) |
1 (1.00) |
| PIK3R1 | 30 (11%) | 246 |
0.775 (1.00) |
0.503 (1.00) |
0.548 (1.00) |
0.561 (1.00) |
0.686 (1.00) |
| RB1 | 18 (7%) | 258 |
0.407 (1.00) |
0.313 (1.00) |
1 (1.00) |
0.0189 (1.00) |
0.615 (1.00) |
| NF1 | 25 (9%) | 251 |
0.813 (1.00) |
0.263 (1.00) |
1 (1.00) |
0.449 (1.00) |
0.379 (1.00) |
| PIK3CA | 24 (9%) | 252 |
0.925 (1.00) |
0.676 (1.00) |
1 (1.00) |
0.922 (1.00) |
0.372 (1.00) |
| SPTA1 | 26 (9%) | 250 |
0.56 (1.00) |
0.228 (1.00) |
0.141 (1.00) |
0.572 (1.00) |
0.396 (1.00) |
| FLG | 23 (8%) | 253 |
0.262 (1.00) |
0.326 (1.00) |
1 (1.00) |
0.835 (1.00) |
0.494 (1.00) |
| GABRA6 | 10 (4%) | 266 |
0.848 (1.00) |
0.351 (1.00) |
1 (1.00) |
0.58 (1.00) |
1 (1.00) |
| RPL5 | 8 (3%) | 268 |
0.897 (1.00) |
0.846 (1.00) |
0.47 (1.00) |
0.881 (1.00) |
0.718 (1.00) |
| KEL | 11 (4%) | 265 |
0.796 (1.00) |
0.304 (1.00) |
1 (1.00) |
0.158 (1.00) |
1 (1.00) |
| RBM47 | 8 (3%) | 268 |
0.893 (1.00) |
0.525 (1.00) |
1 (1.00) |
0.414 (1.00) |
1 (1.00) |
| PCLO | 22 (8%) | 254 |
0.143 (1.00) |
0.967 (1.00) |
1 (1.00) |
0.104 (1.00) |
0.351 (1.00) |
| TCHH | 14 (5%) | 262 |
0.307 (1.00) |
0.277 (1.00) |
0.584 (1.00) |
0.322 (1.00) |
0.392 (1.00) |
| SEMA3C | 9 (3%) | 267 |
0.0616 (1.00) |
0.745 (1.00) |
0.494 (1.00) |
0.751 (1.00) |
0.284 (1.00) |
| OR8K3 | 6 (2%) | 270 |
0.704 (1.00) |
0.882 (1.00) |
0.42 (1.00) |
0.566 (1.00) |
0.668 (1.00) |
| PRB2 | 5 (2%) | 271 |
0.928 (1.00) |
0.17 (1.00) |
1 (1.00) |
0.000758 (0.653) |
1 (1.00) |
| RYR2 | 20 (7%) | 256 |
0.845 (1.00) |
0.645 (1.00) |
0.811 (1.00) |
0.0862 (1.00) |
0.466 (1.00) |
| TPTE2 | 7 (3%) | 269 |
0.191 (1.00) |
0.908 (1.00) |
0.709 (1.00) |
0.0356 (1.00) |
0.697 (1.00) |
| LZTR1 | 8 (3%) | 268 |
0.284 (1.00) |
0.496 (1.00) |
0.265 (1.00) |
0.579 (1.00) |
0.131 (1.00) |
| DCAF12L2 | 6 (2%) | 270 |
0.117 (1.00) |
0.972 (1.00) |
0.42 (1.00) |
0.629 (1.00) |
1 (1.00) |
| RIMS2 | 10 (4%) | 266 |
0.379 (1.00) |
0.00133 (1.00) |
0.0985 (1.00) |
0.0356 (1.00) |
1 (1.00) |
| PSPH | 5 (2%) | 271 |
0.219 (1.00) |
0.362 (1.00) |
0.655 (1.00) |
0.914 (1.00) |
0.661 (1.00) |
| PDGFRA | 9 (3%) | 267 |
0.378 (1.00) |
0.0637 (1.00) |
1 (1.00) |
0.507 (1.00) |
0.284 (1.00) |
| SPRYD5 | 5 (2%) | 271 |
0.555 (1.00) |
0.862 (1.00) |
1 (1.00) |
0.84 (1.00) |
1 (1.00) |
| LRFN5 | 7 (3%) | 269 |
0.0281 (1.00) |
0.399 (1.00) |
0.264 (1.00) |
0.323 (1.00) |
0.00797 (1.00) |
| MUC16 | 34 (12%) | 242 |
0.166 (1.00) |
0.925 (1.00) |
1 (1.00) |
0.939 (1.00) |
0.702 (1.00) |
| CALN1 | 5 (2%) | 271 |
0.152 (1.00) |
0.817 (1.00) |
1 (1.00) |
0.663 (1.00) |
1 (1.00) |
| HCN1 | 7 (3%) | 269 |
0.318 (1.00) |
0.897 (1.00) |
0.264 (1.00) |
0.661 (1.00) |
0.697 (1.00) |
| KIF2B | 7 (3%) | 269 |
0.271 (1.00) |
0.873 (1.00) |
0.709 (1.00) |
0.247 (1.00) |
0.242 (1.00) |
| AZGP1 | 4 (1%) | 272 |
0.644 (1.00) |
0.84 (1.00) |
1 (1.00) |
1 (1.00) |
|
| MUC17 | 16 (6%) | 260 |
0.997 (1.00) |
0.818 (1.00) |
0.597 (1.00) |
0.725 (1.00) |
1 (1.00) |
| AFM | 6 (2%) | 270 |
0.271 (1.00) |
0.398 (1.00) |
0.672 (1.00) |
0.363 (1.00) |
1 (1.00) |
| OR5D13 | 5 (2%) | 271 |
0.461 (1.00) |
0.434 (1.00) |
0.655 (1.00) |
0.661 (1.00) |
|
| DRD5 | 6 (2%) | 270 |
0.389 (1.00) |
0.425 (1.00) |
1 (1.00) |
0.668 (1.00) |
|
| SEMG1 | 6 (2%) | 270 |
0.487 (1.00) |
0.0963 (1.00) |
1 (1.00) |
0.0886 (1.00) |
0.187 (1.00) |
| SULT1B1 | 5 (2%) | 271 |
0.174 (1.00) |
0.635 (1.00) |
0.655 (1.00) |
0.981 (1.00) |
0.661 (1.00) |
| OR5D18 | 5 (2%) | 271 |
0.225 (1.00) |
0.768 (1.00) |
1 (1.00) |
0.323 (1.00) |
0.661 (1.00) |
| TRPV6 | 7 (3%) | 269 |
0.702 (1.00) |
0.217 (1.00) |
0.709 (1.00) |
0.479 (1.00) |
0.1 (1.00) |
| POM121L12 | 5 (2%) | 271 |
0.347 (1.00) |
0.216 (1.00) |
0.359 (1.00) |
0.661 (1.00) |
1 (1.00) |
| COL1A2 | 8 (3%) | 268 |
0.907 (1.00) |
0.528 (1.00) |
1 (1.00) |
0.0241 (1.00) |
0.131 (1.00) |
| LRRC55 | 5 (2%) | 271 |
0.0742 (1.00) |
0.963 (1.00) |
0.359 (1.00) |
0.113 (1.00) |
0.661 (1.00) |
| CNTNAP2 | 8 (3%) | 268 |
0.68 (1.00) |
0.516 (1.00) |
0.0545 (1.00) |
0.661 (1.00) |
0.718 (1.00) |
| HEATR7B2 | 9 (3%) | 267 |
0.655 (1.00) |
0.28 (1.00) |
1 (1.00) |
0.821 (1.00) |
0.284 (1.00) |
| OR4D5 | 5 (2%) | 271 |
0.601 (1.00) |
0.143 (1.00) |
0.162 (1.00) |
0.378 (1.00) |
0.346 (1.00) |
| WNT2 | 5 (2%) | 271 |
0.376 (1.00) |
0.237 (1.00) |
0.0617 (1.00) |
0.717 (1.00) |
1 (1.00) |
| GABRA1 | 4 (1%) | 272 |
0.947 (1.00) |
0.189 (1.00) |
1 (1.00) |
0.247 (1.00) |
1 (1.00) |
| OR5W2 | 4 (1%) | 272 |
0.324 (1.00) |
0.0738 (1.00) |
0.625 (1.00) |
0.302 (1.00) |
|
| SLC1A6 | 6 (2%) | 270 |
0.595 (1.00) |
0.806 (1.00) |
0.42 (1.00) |
0.0271 (1.00) |
0.668 (1.00) |
| DOCK5 | 10 (4%) | 266 |
0.175 (1.00) |
0.967 (1.00) |
0.335 (1.00) |
0.804 (1.00) |
0.743 (1.00) |
| HMCN1 | 15 (5%) | 261 |
0.62 (1.00) |
0.0247 (1.00) |
0.583 (1.00) |
0.635 (1.00) |
0.404 (1.00) |
| STAG2 | 8 (3%) | 268 |
0.000953 (0.814) |
0.866 (1.00) |
0.47 (1.00) |
0.0722 (1.00) |
0.455 (1.00) |
| UGT2A3 | 5 (2%) | 271 |
0.731 (1.00) |
0.0186 (1.00) |
0.655 (1.00) |
0.716 (1.00) |
0.167 (1.00) |
| CDKN2C | 3 (1%) | 273 |
0.731 (1.00) |
0.576 (1.00) |
0.301 (1.00) |
1 (1.00) |
|
| REN | 5 (2%) | 271 |
0.565 (1.00) |
0.969 (1.00) |
0.655 (1.00) |
0.146 (1.00) |
1 (1.00) |
| CDC27 | 5 (2%) | 271 |
0.201 (1.00) |
0.701 (1.00) |
0.359 (1.00) |
1 (1.00) |
|
| WBSCR17 | 5 (2%) | 271 |
0.00139 (1.00) |
0.117 (1.00) |
0.0617 (1.00) |
0.0512 (1.00) |
|
| GRM3 | 7 (3%) | 269 |
0.726 (1.00) |
0.638 (1.00) |
0.709 (1.00) |
0.447 (1.00) |
0.428 (1.00) |
| FOXG1 | 3 (1%) | 273 |
0.127 (1.00) |
0.697 (1.00) |
0.556 (1.00) |
1 (1.00) |
|
| KLF17 | 5 (2%) | 271 |
0.00218 (1.00) |
0.3 (1.00) |
0.162 (1.00) |
0.446 (1.00) |
1 (1.00) |
| SLCO6A1 | 6 (2%) | 270 |
0.513 (1.00) |
0.91 (1.00) |
0.42 (1.00) |
0.0486 (1.00) |
0.0952 (1.00) |
| UGT2B28 | 5 (2%) | 271 |
0.779 (1.00) |
0.889 (1.00) |
0.655 (1.00) |
0.414 (1.00) |
1 (1.00) |
| HIST1H2BE | 3 (1%) | 273 |
0.732 (1.00) |
0.192 (1.00) |
0.301 (1.00) |
0.5 (1.00) |
1 (1.00) |
| AP3S1 | 3 (1%) | 273 |
0.247 (1.00) |
0.351 (1.00) |
0.301 (1.00) |
0.247 (1.00) |
0.554 (1.00) |
| DSG3 | 7 (3%) | 269 |
0.233 (1.00) |
0.966 (1.00) |
0.0501 (1.00) |
0.342 (1.00) |
0.697 (1.00) |
| PROKR2 | 5 (2%) | 271 |
0.957 (1.00) |
0.0495 (1.00) |
1 (1.00) |
0.663 (1.00) |
0.167 (1.00) |
| QKI | 4 (1%) | 272 |
0.692 (1.00) |
0.836 (1.00) |
0.625 (1.00) |
0.014 (1.00) |
|
| TAS2R41 | 3 (1%) | 273 |
0.876 (1.00) |
0.13 (1.00) |
0.556 (1.00) |
0.278 (1.00) |
|
| PCDHA3 | 7 (3%) | 269 |
0.247 (1.00) |
0.275 (1.00) |
0.104 (1.00) |
0.0659 (1.00) |
0.697 (1.00) |
| PAN3 | 6 (2%) | 270 |
0.284 (1.00) |
0.47 (1.00) |
1 (1.00) |
0.763 (1.00) |
0.668 (1.00) |
| COL6A3 | 12 (4%) | 264 |
0.958 (1.00) |
0.264 (1.00) |
0.366 (1.00) |
0.192 (1.00) |
0.352 (1.00) |
| CYP3A5 | 5 (2%) | 271 |
0.313 (1.00) |
0.699 (1.00) |
0.359 (1.00) |
0.363 (1.00) |
1 (1.00) |
| OR5D16 | 4 (1%) | 272 |
0.0653 (1.00) |
0.205 (1.00) |
0.625 (1.00) |
0.837 (1.00) |
1 (1.00) |
| PLCXD3 | 4 (1%) | 272 |
0.0039 (1.00) |
0.721 (1.00) |
0.625 (1.00) |
0.612 (1.00) |
|
| NOX4 | 5 (2%) | 271 |
0.996 (1.00) |
0.426 (1.00) |
0.359 (1.00) |
0.566 (1.00) |
1 (1.00) |
| KLK6 | 3 (1%) | 273 |
0.248 (1.00) |
0.224 (1.00) |
0.556 (1.00) |
0.554 (1.00) |
|
| OR5AR1 | 3 (1%) | 273 |
0.817 (1.00) |
0.075 (1.00) |
0.556 (1.00) |
0.286 (1.00) |
1 (1.00) |
| PODNL1 | 4 (1%) | 272 |
0.102 (1.00) |
0.174 (1.00) |
0.625 (1.00) |
0.506 (1.00) |
0.123 (1.00) |
| TMPRSS6 | 6 (2%) | 270 |
0.828 (1.00) |
0.207 (1.00) |
0.672 (1.00) |
0.342 (1.00) |
0.668 (1.00) |
| CALCR | 5 (2%) | 271 |
0.0432 (1.00) |
0.0821 (1.00) |
0.655 (1.00) |
0.663 (1.00) |
0.661 (1.00) |
| PCDHA1 | 7 (3%) | 269 |
0.601 (1.00) |
0.311 (1.00) |
0.104 (1.00) |
0.981 (1.00) |
0.242 (1.00) |
| ADCY5 | 6 (2%) | 270 |
0.99 (1.00) |
0.346 (1.00) |
0.42 (1.00) |
0.422 (1.00) |
|
| TGFA | 3 (1%) | 273 |
0.501 (1.00) |
0.477 (1.00) |
0.556 (1.00) |
1 (1.00) |
|
| GABRB2 | 5 (2%) | 271 |
0.524 (1.00) |
0.0841 (1.00) |
0.359 (1.00) |
0.323 (1.00) |
0.346 (1.00) |
| ADAMTS16 | 6 (2%) | 270 |
0.482 (1.00) |
0.894 (1.00) |
0.42 (1.00) |
0.414 (1.00) |
0.668 (1.00) |
| POTEH | 3 (1%) | 273 |
0.0221 (1.00) |
0.334 (1.00) |
1 (1.00) |
0.000758 (0.653) |
0.554 (1.00) |
| OR52M1 | 4 (1%) | 272 |
0.966 (1.00) |
0.473 (1.00) |
1 (1.00) |
0.743 (1.00) |
1 (1.00) |
| BCOR | 6 (2%) | 270 |
0.183 (1.00) |
0.585 (1.00) |
0.672 (1.00) |
0.187 (1.00) |
|
| PSG1 | 3 (1%) | 273 |
0.0842 (1.00) |
0.283 (1.00) |
0.556 (1.00) |
1 (1.00) |
|
| TSHZ2 | 6 (2%) | 270 |
0.449 (1.00) |
0.656 (1.00) |
0.672 (1.00) |
0.629 (1.00) |
0.422 (1.00) |
| FGD5 | 7 (3%) | 269 |
0.0467 (1.00) |
0.962 (1.00) |
1 (1.00) |
0.0519 (1.00) |
|
| CDH18 | 6 (2%) | 270 |
0.0502 (1.00) |
0.592 (1.00) |
0.42 (1.00) |
0.479 (1.00) |
0.668 (1.00) |
| KDELC2 | 4 (1%) | 272 |
0.556 (1.00) |
0.904 (1.00) |
0.625 (1.00) |
0.548 (1.00) |
1 (1.00) |
| TREML2 | 3 (1%) | 273 |
0.412 (1.00) |
0.158 (1.00) |
1 (1.00) |
0.548 (1.00) |
0.278 (1.00) |
| FOXR2 | 4 (1%) | 272 |
0.124 (1.00) |
0.813 (1.00) |
0.625 (1.00) |
0.716 (1.00) |
1 (1.00) |
| ADAM29 | 6 (2%) | 270 |
0.734 (1.00) |
0.00818 (1.00) |
0.089 (1.00) |
0.000758 (0.653) |
0.668 (1.00) |
| DPP10 | 6 (2%) | 270 |
0.851 (1.00) |
0.737 (1.00) |
0.672 (1.00) |
0.167 (1.00) |
1 (1.00) |
| KPRP | 5 (2%) | 271 |
0.281 (1.00) |
0.956 (1.00) |
1 (1.00) |
0.84 (1.00) |
1 (1.00) |
| OR13C5 | 4 (1%) | 272 |
0.0612 (1.00) |
0.669 (1.00) |
0.141 (1.00) |
0.548 (1.00) |
0.302 (1.00) |
| OR5A1 | 4 (1%) | 272 |
0.837 (1.00) |
0.212 (1.00) |
0.141 (1.00) |
0.914 (1.00) |
0.302 (1.00) |
| SERPINB4 | 3 (1%) | 273 |
0.51 (1.00) |
0.869 (1.00) |
0.556 (1.00) |
0.278 (1.00) |
|
| LUM | 4 (1%) | 272 |
0.0826 (1.00) |
0.853 (1.00) |
1 (1.00) |
0.765 (1.00) |
0.612 (1.00) |
| GNAT3 | 4 (1%) | 272 |
0.685 (1.00) |
0.295 (1.00) |
0.3 (1.00) |
0.247 (1.00) |
0.302 (1.00) |
| PCDH19 | 6 (2%) | 270 |
0.618 (1.00) |
0.579 (1.00) |
0.196 (1.00) |
0.661 (1.00) |
0.422 (1.00) |
| OR5M3 | 4 (1%) | 272 |
0.659 (1.00) |
0.315 (1.00) |
1 (1.00) |
0.661 (1.00) |
0.612 (1.00) |
| ALPPL2 | 4 (1%) | 272 |
0.325 (1.00) |
0.141 (1.00) |
1 (1.00) |
1 (1.00) |
|
| RELN | 10 (4%) | 266 |
0.477 (1.00) |
0.851 (1.00) |
1 (1.00) |
0.84 (1.00) |
0.743 (1.00) |
| MSL3 | 3 (1%) | 273 |
0.899 (1.00) |
0.342 (1.00) |
1 (1.00) |
1 (1.00) |
|
| GLT1D1 | 3 (1%) | 273 |
0.419 (1.00) |
0.0397 (1.00) |
1 (1.00) |
1 (1.00) |
|
| ABCB1 | 7 (3%) | 269 |
0.493 (1.00) |
0.0432 (1.00) |
0.264 (1.00) |
0.0659 (1.00) |
0.242 (1.00) |
| ELTD1 | 5 (2%) | 271 |
0.908 (1.00) |
0.83 (1.00) |
0.655 (1.00) |
0.113 (1.00) |
1 (1.00) |
| TRAT1 | 3 (1%) | 273 |
0.0111 (1.00) |
0.084 (1.00) |
1 (1.00) |
0.278 (1.00) |
|
| MAGEB4 | 4 (1%) | 272 |
0.152 (1.00) |
0.114 (1.00) |
0.141 (1.00) |
0.166 (1.00) |
1 (1.00) |
| SLC5A7 | 4 (1%) | 272 |
0.0445 (1.00) |
0.896 (1.00) |
1 (1.00) |
0.0886 (1.00) |
1 (1.00) |
| BRAF | 3 (1%) | 273 |
0.879 (1.00) |
0.0474 (1.00) |
1 (1.00) |
0.000758 (0.653) |
0.278 (1.00) |
| DAO | 4 (1%) | 272 |
0.933 (1.00) |
0.975 (1.00) |
1 (1.00) |
0.765 (1.00) |
0.612 (1.00) |
| KRT37 | 4 (1%) | 272 |
0.947 (1.00) |
0.813 (1.00) |
1 (1.00) |
0.123 (1.00) |
|
| PRDM9 | 6 (2%) | 270 |
0.959 (1.00) |
0.0709 (1.00) |
0.672 (1.00) |
0.604 (1.00) |
1 (1.00) |
| ZNF181 | 4 (1%) | 272 |
0.378 (1.00) |
0.498 (1.00) |
0.141 (1.00) |
0.247 (1.00) |
1 (1.00) |
| IL18RAP | 4 (1%) | 272 |
0.945 (1.00) |
0.178 (1.00) |
0.141 (1.00) |
0.286 (1.00) |
1 (1.00) |
| POTEG | 4 (1%) | 272 |
0.874 (1.00) |
0.646 (1.00) |
0.141 (1.00) |
0.612 (1.00) |
|
| PCDH11Y | 6 (2%) | 270 |
0.282 (1.00) |
0.948 (1.00) |
0.089 (1.00) |
0.479 (1.00) |
0.422 (1.00) |
| RSPO3 | 3 (1%) | 273 |
0.397 (1.00) |
0.766 (1.00) |
0.556 (1.00) |
0.278 (1.00) |
|
| SEMA3E | 5 (2%) | 271 |
0.43 (1.00) |
0.416 (1.00) |
0.655 (1.00) |
0.166 (1.00) |
0.167 (1.00) |
| SLC27A2 | 5 (2%) | 271 |
0.374 (1.00) |
0.292 (1.00) |
0.359 (1.00) |
0.661 (1.00) |
0.661 (1.00) |
| CKMT1A | 3 (1%) | 273 |
0.491 (1.00) |
0.0428 (1.00) |
0.301 (1.00) |
0.861 (1.00) |
1 (1.00) |
| CYP2D6 | 4 (1%) | 272 |
0.0603 (1.00) |
0.201 (1.00) |
0.625 (1.00) |
0.123 (1.00) |
|
| GABRB3 | 4 (1%) | 272 |
0.939 (1.00) |
0.715 (1.00) |
0.141 (1.00) |
1 (1.00) |
|
| TAF1L | 8 (3%) | 268 |
0.12 (1.00) |
0.137 (1.00) |
0.0289 (1.00) |
0.137 (1.00) |
0.455 (1.00) |
| TNFSF9 | 3 (1%) | 273 |
0.816 (1.00) |
0.638 (1.00) |
0.301 (1.00) |
0.548 (1.00) |
0.554 (1.00) |
| ZDHHC4 | 3 (1%) | 273 |
0.802 (1.00) |
0.764 (1.00) |
1 (1.00) |
1 (1.00) |
|
| FBLN2 | 5 (2%) | 271 |
0.773 (1.00) |
0.0178 (1.00) |
1 (1.00) |
0.348 (1.00) |
0.661 (1.00) |
| SYT16 | 5 (2%) | 271 |
0.368 (1.00) |
0.367 (1.00) |
0.162 (1.00) |
0.116 (1.00) |
0.346 (1.00) |
| F9 | 4 (1%) | 272 |
0.0203 (1.00) |
0.559 (1.00) |
0.625 (1.00) |
1 (1.00) |
|
| CD33 | 4 (1%) | 272 |
0.52 (1.00) |
0.0328 (1.00) |
0.141 (1.00) |
1 (1.00) |
|
| CXORF22 | 6 (2%) | 270 |
0.856 (1.00) |
0.49 (1.00) |
0.026 (1.00) |
0.661 (1.00) |
0.668 (1.00) |
| CARD6 | 6 (2%) | 270 |
0.03 (1.00) |
0.00839 (1.00) |
0.42 (1.00) |
0.113 (1.00) |
0.422 (1.00) |
| PLEKHG4B | 6 (2%) | 270 |
0.36 (1.00) |
0.059 (1.00) |
0.196 (1.00) |
0.716 (1.00) |
0.422 (1.00) |
| CPNE8 | 4 (1%) | 272 |
0.901 (1.00) |
0.87 (1.00) |
1 (1.00) |
0.612 (1.00) |
|
| MRGPRE | 3 (1%) | 273 |
0.585 (1.00) |
0.447 (1.00) |
0.0481 (1.00) |
0.247 (1.00) |
0.554 (1.00) |
| RBMS3 | 4 (1%) | 272 |
0.556 (1.00) |
0.842 (1.00) |
0.625 (1.00) |
0.853 (1.00) |
0.302 (1.00) |
| NLRP12 | 6 (2%) | 270 |
0.985 (1.00) |
0.477 (1.00) |
1 (1.00) |
0.323 (1.00) |
0.187 (1.00) |
| OR4P4 | 3 (1%) | 273 |
0.749 (1.00) |
0.477 (1.00) |
1 (1.00) |
1 (1.00) |
|
| SDK1 | 9 (3%) | 267 |
0.502 (1.00) |
0.215 (1.00) |
0.494 (1.00) |
0.231 (1.00) |
0.0295 (1.00) |
| CFHR4 | 3 (1%) | 273 |
0.574 (1.00) |
0.838 (1.00) |
0.556 (1.00) |
0.716 (1.00) |
1 (1.00) |
| DPPA4 | 3 (1%) | 273 |
0.515 (1.00) |
0.552 (1.00) |
0.301 (1.00) |
0.716 (1.00) |
1 (1.00) |
| LPAR3 | 3 (1%) | 273 |
0.124 (1.00) |
0.847 (1.00) |
1 (1.00) |
0.0412 (1.00) |
|
| FBN3 | 10 (4%) | 266 |
0.246 (1.00) |
0.393 (1.00) |
0.505 (1.00) |
0.456 (1.00) |
1 (1.00) |
| CD1D | 3 (1%) | 273 |
0.0527 (1.00) |
0.235 (1.00) |
1 (1.00) |
0.000758 (0.653) |
0.554 (1.00) |
| POTEF | 5 (2%) | 271 |
0.971 (1.00) |
0.718 (1.00) |
1 (1.00) |
1 (1.00) |
|
| ACSM2B | 4 (1%) | 272 |
0.276 (1.00) |
0.377 (1.00) |
1 (1.00) |
0.716 (1.00) |
0.302 (1.00) |
| ZNF618 | 5 (2%) | 271 |
0.066 (1.00) |
0.356 (1.00) |
1 (1.00) |
0.505 (1.00) |
0.346 (1.00) |
| FBXL12 | 3 (1%) | 273 |
0.844 (1.00) |
0.71 (1.00) |
1 (1.00) |
1 (1.00) |
|
| CHD8 | 7 (3%) | 269 |
0.678 (1.00) |
0.424 (1.00) |
0.264 (1.00) |
0.539 (1.00) |
0.242 (1.00) |
| NLRP5 | 6 (2%) | 270 |
0.989 (1.00) |
0.318 (1.00) |
0.42 (1.00) |
0.283 (1.00) |
1 (1.00) |
| OR5P2 | 3 (1%) | 273 |
0.295 (1.00) |
0.32 (1.00) |
0.301 (1.00) |
0.000758 (0.653) |
1 (1.00) |
| CLYBL | 3 (1%) | 273 |
0.833 (1.00) |
0.356 (1.00) |
1 (1.00) |
1 (1.00) |
|
| MMP13 | 4 (1%) | 272 |
0.847 (1.00) |
0.307 (1.00) |
0.625 (1.00) |
0.116 (1.00) |
0.302 (1.00) |
| SLCO5A1 | 4 (1%) | 272 |
0.931 (1.00) |
0.724 (1.00) |
0.141 (1.00) |
0.612 (1.00) |
|
| OR4C11 | 3 (1%) | 273 |
0.481 (1.00) |
0.554 (1.00) |
1 (1.00) |
0.278 (1.00) |
|
| TRIML1 | 4 (1%) | 272 |
0.0651 (1.00) |
0.283 (1.00) |
0.141 (1.00) |
0.612 (1.00) |
|
| FRMD7 | 5 (2%) | 271 |
0.355 (1.00) |
0.527 (1.00) |
0.655 (1.00) |
0.921 (1.00) |
1 (1.00) |
| GRIN2A | 7 (3%) | 269 |
0.319 (1.00) |
0.614 (1.00) |
1 (1.00) |
0.566 (1.00) |
1 (1.00) |
| KCNB2 | 4 (1%) | 272 |
0.674 (1.00) |
0.8 (1.00) |
0.625 (1.00) |
0.123 (1.00) |
|
| SCN9A | 8 (3%) | 268 |
0.263 (1.00) |
0.538 (1.00) |
0.715 (1.00) |
0.84 (1.00) |
0.131 (1.00) |
| UGT2B10 | 4 (1%) | 272 |
0.685 (1.00) |
0.266 (1.00) |
1 (1.00) |
0.000758 (0.653) |
0.612 (1.00) |
| KDR | 6 (2%) | 270 |
0.14 (1.00) |
0.795 (1.00) |
0.42 (1.00) |
0.498 (1.00) |
0.668 (1.00) |
| NDST4 | 5 (2%) | 271 |
0.934 (1.00) |
0.327 (1.00) |
0.359 (1.00) |
0.0899 (1.00) |
0.167 (1.00) |
| PIK3C2G | 6 (2%) | 270 |
0.541 (1.00) |
0.869 (1.00) |
0.672 (1.00) |
0.661 (1.00) |
0.422 (1.00) |
| CDH9 | 5 (2%) | 271 |
0.763 (1.00) |
0.876 (1.00) |
0.359 (1.00) |
0.346 (1.00) |
|
| AHNAK2 | 14 (5%) | 262 |
0.859 (1.00) |
0.446 (1.00) |
0.0429 (1.00) |
0.735 (1.00) |
0.569 (1.00) |
| SH3RF2 | 5 (2%) | 271 |
0.392 (1.00) |
0.268 (1.00) |
1 (1.00) |
0.292 (1.00) |
0.167 (1.00) |
| USH2A | 14 (5%) | 262 |
0.598 (1.00) |
0.642 (1.00) |
0.269 (1.00) |
0.918 (1.00) |
0.0866 (1.00) |
| PCDHB7 | 4 (1%) | 272 |
0.686 (1.00) |
0.26 (1.00) |
0.3 (1.00) |
0.32 (1.00) |
1 (1.00) |
| TLR6 | 5 (2%) | 271 |
0.745 (1.00) |
0.667 (1.00) |
0.655 (1.00) |
1 (1.00) |
|
| PAK4 | 3 (1%) | 273 |
0.853 (1.00) |
0.337 (1.00) |
0.556 (1.00) |
0.765 (1.00) |
1 (1.00) |
| PTX4 | 4 (1%) | 272 |
0.643 (1.00) |
0.983 (1.00) |
0.625 (1.00) |
0.363 (1.00) |
0.612 (1.00) |
| SIGLEC8 | 3 (1%) | 273 |
0.861 (1.00) |
0.152 (1.00) |
1 (1.00) |
0.0469 (1.00) |
0.278 (1.00) |
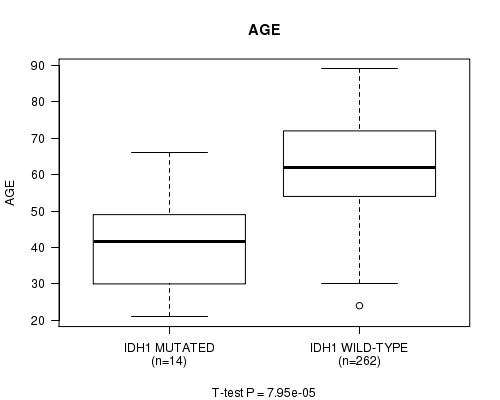
P value = 7.95e-05 (t-test), Q value = 0.069
Table S1. Gene #7: 'IDH1 MUTATION STATUS' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 276 | 61.1 (12.6) |
| IDH1 MUTATED | 14 | 40.7 (14.4) |
| IDH1 WILD-TYPE | 262 | 62.2 (11.5) |
Figure S1. Get High-res Image Gene #7: 'IDH1 MUTATION STATUS' versus Clinical Feature #2: 'AGE'
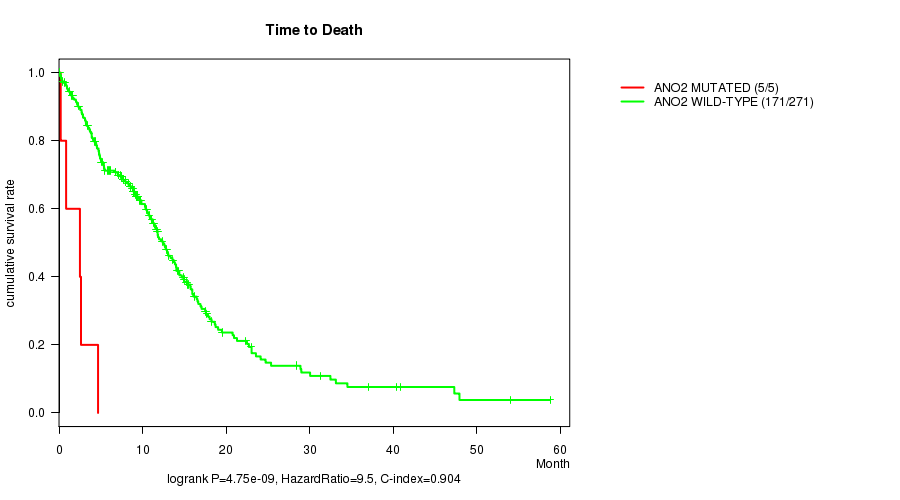
P value = 4.75e-09 (logrank test), Q value = 4.1e-06
Table S2. Gene #174: 'ANO2 MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 276 | 176 | 0.1 - 58.8 (8.4) |
| ANO2 MUTATED | 5 | 5 | 0.2 - 4.7 (2.5) |
| ANO2 WILD-TYPE | 271 | 171 | 0.1 - 58.8 (8.6) |
Figure S2. Get High-res Image Gene #174: 'ANO2 MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
-
Mutation data file = GBM-TP.mutsig.cluster.txt
-
Clinical data file = GBM-TP.clin.merged.picked.txt
-
Number of patients = 276
-
Number of significantly mutated genes = 181
-
Number of selected clinical features = 5
-
Exclude genes that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For continuous numerical clinical features, two-tailed Student's t test with unequal variance (Lehmann and Romano 2005) was applied to compare the clinical values between tumors with and without gene mutations using 't.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.