This is a summary of data mirrored from the Genomic Data Commons (GDC) and processed by the GDCtools package. Note that some sample data will be filtered as unsuitable for downstream pipelines, through one of three mechanisms: redactions, replicate filtering, and blacklisting. The report lists the counts and types of the sample data, in both hyperlinked tables and heatmap images; describes the three filtering mechanisms; lists the samples removed by filtering, why they were removed; and (eventually will) catalog how the data have been annotated by the respective projects that submitted them to the GDC.
There were 0 redactions, 0 replicate aliquots, 0 blacklisted aliquots, and 0 FFPE aliquots. The table below represents the sample counts for those samples that were ingested into firehose after filtering out redactions, replicates, and blacklisted data, and segregating FFPEs.
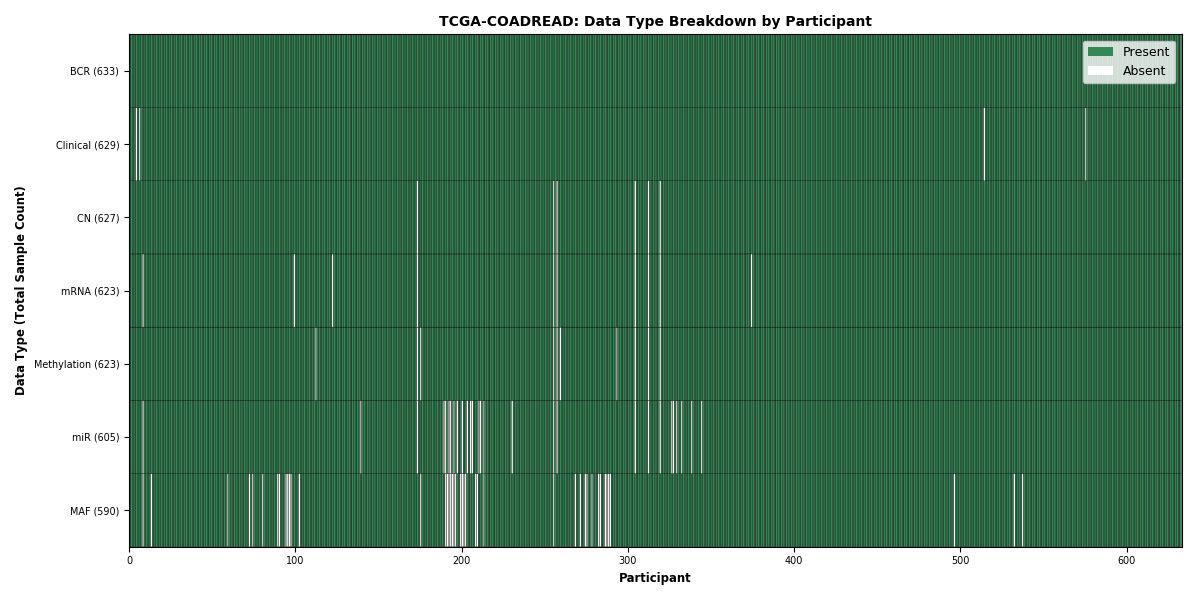
Table 1. This table provides a breakdown of sample counts on a per sample type and, if applicable, per subtype basis. Each count is a link to a table containing a list of the samples that comprise that count and details pertaining to each individual sample (e.g. platform, sequencing center, etc.). Please note, there are usually multiple protocols per data type, so there are typically many more rows than the count implies.
| Sample.Type | BCR | Clinical | CN | mRNA | miR | MAF | Methylation |
|---|---|---|---|---|---|---|---|
| FFPE | 13 | 13 | 12 | 13 | 7 | 12 | 12 |
| NB | 509 | 505 | 509 | 0 | 0 | 0 | 0 |
| TR | 2 | 2 | 2 | 2 | 2 | 2 | 1 |
| TP | 633 | 629 | 617 | 622 | 605 | 589 | 623 |
| TM | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| NT | 110 | 110 | 107 | 51 | 11 | 0 | 87 |
| Totals | 633 | 629 | 617 | 622 | 605 | 589 | 623 |
Table S1.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-A6-2672 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2674 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2677 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2684 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-3809 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-3810 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5656 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5659 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5661 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5665 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6650 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6780 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6781 | (TODO) -- GDC | clinical__biospecimen |
Table S2.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-A6-2672 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2674 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2677 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2684 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-3809 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-3810 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5656 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5659 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5661 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5665 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6650 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6780 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6781 | (TODO) -- GDC | clinical__primary |
Table S3.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-A6-2672-01B-03D-2297-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2672-01B-03D-2297-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2677-01B-02D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2677-01B-02D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2684-01C-08D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2684-01C-08D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-3809-01B-04D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-3809-01B-04D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-3810-01B-04D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-3810-01B-04D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5656-01B-02D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5656-01B-02D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5659-01B-04D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5659-01B-04D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5661-01B-05D-2297-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5661-01B-05D-2297-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5665-01B-03D-2297-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5665-01B-03D-2297-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6650-01B-02D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6650-01B-02D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6780-01B-04D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6780-01B-04D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6781-01B-06D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6781-01B-06D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
Table S4.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-A6-2672-01B-03R-2302-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2672-01B-03R-2302-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2672-01B-03R-2302-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2674-01B-04R-A277-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2674-01B-04R-A277-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2674-01B-04R-A277-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2677-01B-02R-A277-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2677-01B-02R-A277-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2677-01B-02R-A277-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2684-01C-08R-A277-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2684-01C-08R-A277-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2684-01C-08R-A277-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-3809-01B-04R-A277-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-3809-01B-04R-A277-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-3809-01B-04R-A277-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-3810-01B-04R-A277-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-3810-01B-04R-A277-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-3810-01B-04R-A277-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5656-01B-02R-A277-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5656-01B-02R-A277-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5656-01B-02R-A277-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5659-01B-04R-A277-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5659-01B-04R-A277-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5659-01B-04R-A277-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5661-01B-05R-2302-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5661-01B-05R-2302-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5661-01B-05R-2302-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5665-01B-03R-2302-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5665-01B-03R-2302-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5665-01B-03R-2302-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6650-01B-02R-A277-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6650-01B-02R-A277-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6650-01B-02R-A277-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6780-01B-04R-A277-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6780-01B-04R-A277-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6780-01B-04R-A277-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6781-01B-06R-A277-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6781-01B-06R-A277-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6781-01B-06R-A277-07 | (TODO) -- GDC | mRNA__counts__FPKM |
Table S5.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-A6-2672-01B-03R-2303-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2672-01B-03R-2303-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-5656-01B-02R-A27D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-5656-01B-02R-A27D-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-5661-01B-05R-2303-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-5661-01B-05R-2303-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-5665-01B-03R-2303-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-5665-01B-03R-2303-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6650-01B-02R-A27D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6650-01B-02R-A27D-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6780-01B-04R-A27D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6780-01B-04R-A27D-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6781-01B-06R-A27D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6781-01B-06R-A27D-13 | (TODO) -- GDC | miR__isoformExp |
Table S6.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-A6-2672-01B-03D-2298-08 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2672-01B-03D-2301-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2674-01B-04D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2677-01B-02D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2684-01C-08D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-3809-01B-04D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-3810-01B-04D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5656-01B-02D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5659-01B-04D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5665-01B-03D-2298-08 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6650-01B-02D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6780-01B-04D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6781-01B-06D-A270-10 | (TODO) -- GDC | SNV__mutect |
Table S7.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-A6-2672-01B-03D-2299-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2677-01B-02D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2684-01C-08D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-3809-01B-04D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-3810-01B-04D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5656-01B-02D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5659-01B-04D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5661-01B-05D-2299-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5665-01B-03D-2299-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6650-01B-02D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6780-01B-04D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6781-01B-06D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
Table S8.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-3L-AA1B | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-4N-A93T | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-4T-AA8H | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-5M-AAT4 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-5M-AAT5 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-5M-AAT6 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-5M-AATA | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-5M-AATE | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2670 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2671 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2672 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2674 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2675 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2676 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2677 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2678 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2679 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2680 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2681 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2682 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2683 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2684 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2685 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2686 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-3807 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-3808 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-3809 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-3810 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-4105 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-4107 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5656 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5657 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5659 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5660 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5661 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5662 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5664 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5665 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5666 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5667 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6137 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6138 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6140 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6141 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6142 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6648 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6649 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6650 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6651 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6652 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6653 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6654 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6780 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6781 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6782 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-A565 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-A566 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-A567 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-A56B | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-A5ZU | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3514 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3516 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3517 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3518 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3519 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3520 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3521 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3522 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3524 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3525 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3526 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3527 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3529 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3530 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3531 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3532 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3534 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3538 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3542 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3543 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3544 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3548 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3549 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3552 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3553 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3554 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3555 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3556 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3558 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3560 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3561 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3562 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3664 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3666 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3667 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3672 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3673 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3675 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3678 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3679 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3680 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3681 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3684 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3685 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3688 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3692 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3693 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3695 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3696 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3710 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3715 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3811 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3812 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3814 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3815 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3818 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3819 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3821 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3831 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3833 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3837 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3841 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3842 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3844 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3845 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3846 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3848 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3850 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3851 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3852 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3854 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3855 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3856 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3858 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3860 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3861 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3862 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3864 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3866 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3867 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3869 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3870 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3872 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3875 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3877 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3941 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3947 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3949 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3950 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3952 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3955 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3956 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3966 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3968 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3970 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3971 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3972 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3973 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3975 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3976 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3977 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3979 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3980 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3982 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3984 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3986 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3989 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3994 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A02K | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A02Y | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-5900 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6548 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6888 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6889 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6890 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6895 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6899 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6901 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6963 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6964 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6965 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-A5EJ | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-A5EK | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2687 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2689 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2690 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2691 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2692 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2693 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-3400 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-3911 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-3913 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-4110 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-5654 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-6136 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-6655 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-6672 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-A56K | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-A56L | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-A56N | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3574 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3575 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3578 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3580 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3581 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3582 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3583 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3584 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3586 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3587 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3591 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3592 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3593 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3594 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3598 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3599 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3600 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3602 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3608 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3609 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3612 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3726 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3727 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3728 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3878 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3881 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3882 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3883 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3885 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3887 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3890 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3892 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3893 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3894 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3896 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3898 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3901 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3902 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3999 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4001 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4005 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4007 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4008 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4015 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4021 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4022 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A026 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AH-6544 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AH-6549 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AH-6644 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AH-6897 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AH-6903 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AM-5820 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AM-5821 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AU-3779 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AU-6004 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-4070 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-4071 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-5543 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-6196 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-6197 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-6386 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-A69D | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-A71X | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-A8YK | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4308 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4313 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4315 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4323 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4614 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4615 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4616 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4681 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4682 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-5403 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-5407 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-BM-6198 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-5254 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-5255 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-5256 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-5796 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-5797 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-6715 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-6716 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-6717 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-6718 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-6719 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CI-6619 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CI-6620 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CI-6621 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CI-6622 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CI-6623 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CI-6624 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-4947 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-4950 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-4951 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-5912 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-5913 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-5915 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-5916 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-6746 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-6747 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-6748 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-6751 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CL-4957 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CL-5917 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CL-5918 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-4743 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-4746 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-4747 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-4748 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-4751 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-4752 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5341 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5344 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5348 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5349 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5860 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5861 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5862 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5863 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5864 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5868 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6161 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6162 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6163 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6164 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6166 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6167 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6168 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6169 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6170 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6171 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6172 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6674 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6675 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6676 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6677 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6678 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6679 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6680 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-5537 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-5538 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-5539 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-5540 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-5541 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6529 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6530 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6531 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6532 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6533 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6534 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6535 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6536 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6537 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6538 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6539 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6540 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6541 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6898 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6920 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6922 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6923 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6924 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6926 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6927 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6928 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6929 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6930 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6931 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6932 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-7000 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-4745 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-4749 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-5337 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-5869 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6154 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6155 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6156 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6157 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6158 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6681 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6682 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6683 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A0X9 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A0XD | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A0XF | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1D0 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1D4 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1D6 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1D7 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1D8 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1D9 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1DA | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1DB | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1HA | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1HB | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A280 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A285 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A288 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28A | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28C | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28E | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28F | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28G | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28H | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28K | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DT-5265 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DY-A0XA | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DY-A1DC | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DY-A1DD | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DY-A1DE | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DY-A1DF | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DY-A1DG | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DY-A1H8 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EF-5830 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EF-5831 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6506 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6507 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6508 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6509 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6510 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6511 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6512 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6513 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6514 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6881 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6882 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6883 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6884 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6885 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6917 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-7002 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-7004 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6459 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6460 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6461 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6463 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6569 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6570 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6703 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6805 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6806 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6807 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6808 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6809 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6854 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6855 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6856 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6464 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6465 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6571 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6702 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6810 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6812 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6813 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6814 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6861 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6863 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6864 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6293 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6294 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6295 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6297 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6298 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6299 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6302 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6303 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6304 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6306 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6307 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6309 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6310 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6311 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6314 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6315 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6317 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6320 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6321 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6322 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6323 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6586 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6588 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6626 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6627 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6628 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G5-6233 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G5-6235 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G5-6572 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G5-6641 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A50T | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A50U | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A50V | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A5IV | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A6GA | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A6GB | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A6GC | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A8F7 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A8F8 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-QG-A5YV | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-QG-A5YW | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-QG-A5YX | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-QG-A5Z1 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-QG-A5Z2 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-QL-A97D | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-RU-A8FL | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-SS-A7HO | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-T9-A92H | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-WS-AB45 | (TODO) -- GDC | clinical__biospecimen |
Table S9.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-3L-AA1B | (TODO) -- GDC | clinical__primary | |
| TCGA-4N-A93T | (TODO) -- GDC | clinical__primary | |
| TCGA-4T-AA8H | (TODO) -- GDC | clinical__primary | |
| TCGA-5M-AAT4 | (TODO) -- GDC | clinical__primary | |
| TCGA-5M-AAT6 | (TODO) -- GDC | clinical__primary | |
| TCGA-5M-AATE | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2670 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2671 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2672 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2674 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2675 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2676 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2677 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2678 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2679 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2680 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2681 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2682 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2683 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2684 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2685 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2686 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-3807 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-3808 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-3809 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-3810 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-4105 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-4107 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5656 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5657 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5659 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5660 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5661 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5662 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5664 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5665 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5666 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5667 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6137 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6138 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6140 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6141 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6142 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6648 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6649 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6650 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6651 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6652 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6653 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6654 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6780 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6781 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6782 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-A565 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-A566 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-A567 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-A56B | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-A5ZU | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3514 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3516 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3517 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3518 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3519 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3520 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3521 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3522 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3524 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3525 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3526 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3527 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3529 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3530 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3531 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3532 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3534 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3538 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3542 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3543 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3544 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3548 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3549 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3552 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3553 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3554 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3555 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3556 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3558 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3560 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3561 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3562 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3664 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3666 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3667 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3672 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3673 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3675 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3678 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3679 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3680 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3681 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3684 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3685 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3688 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3692 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3693 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3695 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3696 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3710 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3715 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3811 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3812 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3814 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3815 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3818 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3819 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3821 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3831 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3833 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3837 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3841 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3842 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3844 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3845 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3846 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3848 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3850 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3851 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3852 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3854 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3855 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3856 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3858 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3860 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3861 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3862 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3864 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3866 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3867 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3869 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3870 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3872 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3875 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3877 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3941 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3947 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3949 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3950 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3952 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3955 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3956 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3966 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3968 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3970 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3971 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3972 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3973 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3975 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3976 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3977 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3979 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3980 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3982 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3984 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3986 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3989 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3994 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A02K | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A02Y | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-5900 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6548 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6888 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6889 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6890 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6895 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6899 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6901 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6963 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6964 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6965 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-A5EJ | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-A5EK | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2687 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2689 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2690 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2691 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2692 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2693 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-3400 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-3911 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-3913 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-4110 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-5654 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-6136 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-6655 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-6672 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-A56K | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-A56L | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-A56N | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3574 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3575 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3578 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3580 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3581 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3582 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3583 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3584 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3586 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3587 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3591 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3592 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3593 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3594 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3598 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3599 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3600 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3602 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3608 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3609 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3612 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3726 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3727 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3728 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3878 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3881 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3882 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3883 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3885 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3887 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3890 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3892 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3893 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3894 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3896 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3898 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3901 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3902 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3999 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4001 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4005 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4007 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4008 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4015 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4021 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4022 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A026 | (TODO) -- GDC | clinical__primary | |
| TCGA-AH-6544 | (TODO) -- GDC | clinical__primary | |
| TCGA-AH-6549 | (TODO) -- GDC | clinical__primary | |
| TCGA-AH-6644 | (TODO) -- GDC | clinical__primary | |
| TCGA-AH-6897 | (TODO) -- GDC | clinical__primary | |
| TCGA-AH-6903 | (TODO) -- GDC | clinical__primary | |
| TCGA-AM-5820 | (TODO) -- GDC | clinical__primary | |
| TCGA-AM-5821 | (TODO) -- GDC | clinical__primary | |
| TCGA-AU-3779 | (TODO) -- GDC | clinical__primary | |
| TCGA-AU-6004 | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-4070 | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-4071 | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-5543 | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-6196 | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-6197 | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-6386 | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-A69D | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-A71X | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-A8YK | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4308 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4313 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4315 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4323 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4614 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4615 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4616 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4681 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4682 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-5403 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-5407 | (TODO) -- GDC | clinical__primary | |
| TCGA-BM-6198 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-5254 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-5255 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-5256 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-5796 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-5797 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-6715 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-6716 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-6717 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-6718 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-6719 | (TODO) -- GDC | clinical__primary | |
| TCGA-CI-6619 | (TODO) -- GDC | clinical__primary | |
| TCGA-CI-6620 | (TODO) -- GDC | clinical__primary | |
| TCGA-CI-6621 | (TODO) -- GDC | clinical__primary | |
| TCGA-CI-6622 | (TODO) -- GDC | clinical__primary | |
| TCGA-CI-6623 | (TODO) -- GDC | clinical__primary | |
| TCGA-CI-6624 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-4947 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-4950 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-4951 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-5912 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-5913 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-5915 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-5916 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-6746 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-6747 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-6748 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-6751 | (TODO) -- GDC | clinical__primary | |
| TCGA-CL-4957 | (TODO) -- GDC | clinical__primary | |
| TCGA-CL-5917 | (TODO) -- GDC | clinical__primary | |
| TCGA-CL-5918 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-4743 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-4746 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-4747 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-4748 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-4751 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-4752 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5341 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5344 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5348 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5349 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5860 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5861 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5862 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5863 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5864 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5868 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6161 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6162 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6163 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6164 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6166 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6167 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6168 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6169 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6170 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6171 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6172 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6674 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6675 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6676 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6677 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6678 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6679 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6680 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-5537 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-5538 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-5539 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-5540 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-5541 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6529 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6530 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6531 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6532 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6533 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6534 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6535 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6536 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6537 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6538 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6539 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6540 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6541 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6898 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6920 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6922 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6923 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6924 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6926 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6927 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6928 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6929 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6930 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6931 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6932 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-7000 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-4745 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-4749 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-5337 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-5869 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6154 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6155 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6156 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6157 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6158 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6681 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6682 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6683 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A0X9 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A0XD | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A0XF | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1D0 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1D4 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1D6 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1D7 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1D8 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1D9 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1DA | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1DB | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1HB | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A280 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A285 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A288 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28A | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28C | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28E | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28F | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28G | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28H | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28K | (TODO) -- GDC | clinical__primary | |
| TCGA-DT-5265 | (TODO) -- GDC | clinical__primary | |
| TCGA-DY-A0XA | (TODO) -- GDC | clinical__primary | |
| TCGA-DY-A1DC | (TODO) -- GDC | clinical__primary | |
| TCGA-DY-A1DD | (TODO) -- GDC | clinical__primary | |
| TCGA-DY-A1DE | (TODO) -- GDC | clinical__primary | |
| TCGA-DY-A1DF | (TODO) -- GDC | clinical__primary | |
| TCGA-DY-A1DG | (TODO) -- GDC | clinical__primary | |
| TCGA-DY-A1H8 | (TODO) -- GDC | clinical__primary | |
| TCGA-EF-5830 | (TODO) -- GDC | clinical__primary | |
| TCGA-EF-5831 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6506 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6507 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6508 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6509 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6510 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6511 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6512 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6513 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6514 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6881 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6882 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6883 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6884 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6885 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6917 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-7002 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-7004 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6459 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6460 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6461 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6463 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6569 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6570 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6703 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6805 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6806 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6807 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6808 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6809 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6854 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6855 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6856 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6464 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6465 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6571 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6702 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6812 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6813 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6814 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6861 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6863 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6864 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6293 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6294 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6295 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6297 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6298 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6299 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6302 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6303 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6304 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6306 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6307 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6309 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6310 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6311 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6314 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6315 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6317 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6320 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6321 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6322 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6323 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6586 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6588 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6626 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6627 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6628 | (TODO) -- GDC | clinical__primary | |
| TCGA-G5-6233 | (TODO) -- GDC | clinical__primary | |
| TCGA-G5-6235 | (TODO) -- GDC | clinical__primary | |
| TCGA-G5-6572 | (TODO) -- GDC | clinical__primary | |
| TCGA-G5-6641 | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A50T | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A50U | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A50V | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A5IV | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A6GA | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A6GB | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A6GC | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A8F7 | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A8F8 | (TODO) -- GDC | clinical__primary | |
| TCGA-QG-A5YV | (TODO) -- GDC | clinical__primary | |
| TCGA-QG-A5YW | (TODO) -- GDC | clinical__primary | |
| TCGA-QG-A5YX | (TODO) -- GDC | clinical__primary | |
| TCGA-QG-A5Z1 | (TODO) -- GDC | clinical__primary | |
| TCGA-QG-A5Z2 | (TODO) -- GDC | clinical__primary | |
| TCGA-QL-A97D | (TODO) -- GDC | clinical__primary | |
| TCGA-RU-A8FL | (TODO) -- GDC | clinical__primary | |
| TCGA-SS-A7HO | (TODO) -- GDC | clinical__primary | |
| TCGA-T9-A92H | (TODO) -- GDC | clinical__primary | |
| TCGA-WS-AB45 | (TODO) -- GDC | clinical__primary |
Table S10.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-3L-AA1B-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-3L-AA1B-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-4N-A93T-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-4N-A93T-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-4T-AA8H-10B-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-4T-AA8H-10B-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-5M-AAT4-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-5M-AAT4-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-5M-AAT5-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-5M-AAT5-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-5M-AAT6-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-5M-AAT6-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-5M-AATA-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-5M-AATA-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-5M-AATE-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-5M-AATE-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2670-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2670-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2671-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2671-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2672-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2672-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2674-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2674-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2675-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2675-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2676-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2676-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2677-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2677-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2678-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2678-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2679-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2679-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2680-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2680-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2681-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2681-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2682-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2682-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2683-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2683-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2684-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2684-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2684-10A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2684-10A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2685-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2685-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2686-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2686-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-3807-10A-01D-1450-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-3807-10A-01D-1450-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-3808-10A-01D-1450-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-3808-10A-01D-1450-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-3809-10A-01D-1450-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-3809-10A-01D-1450-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-3809-10A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-3809-10A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-3810-10A-01D-1450-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-3810-10A-01D-1450-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-4105-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-4105-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-4107-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-4107-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5656-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5656-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5657-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5657-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5659-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5659-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5659-10A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5659-10A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5660-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5660-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5661-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5661-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5662-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5662-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5664-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5664-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5665-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5665-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5666-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5666-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5667-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5667-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6137-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6137-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6138-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6138-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6140-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6140-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6141-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6141-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6142-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6142-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6648-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6648-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6649-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6649-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6650-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6650-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6651-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6651-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6652-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6652-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6653-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6653-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6654-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6654-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6780-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6780-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6781-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6781-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6781-10A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6781-10A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6782-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6782-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-A565-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-A565-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-A566-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-A566-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-A567-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-A567-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-A56B-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-A56B-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-A5ZU-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-A5ZU-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3514-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3514-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3516-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3516-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3517-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3517-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3518-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3518-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3519-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3519-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3520-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3520-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3521-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3521-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3522-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3522-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3524-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3524-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3525-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3525-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3526-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3526-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3527-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3527-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3529-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3529-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3530-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3530-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3531-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3531-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3532-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3532-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3534-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3534-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3538-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3538-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3542-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3542-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3543-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3543-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3544-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3544-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3548-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3548-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3549-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3549-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3552-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3552-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3553-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3553-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3554-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3554-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3555-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3555-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3556-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3556-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3558-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3558-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3560-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3560-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3561-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3561-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3562-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3562-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3664-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3664-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3666-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3666-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3667-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3667-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3672-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3672-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3673-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3673-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3675-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3675-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3678-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3678-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3679-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3679-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3680-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3680-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3681-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3681-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3684-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3684-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3685-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3685-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3688-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3688-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3692-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3692-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3693-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3693-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3695-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3695-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3696-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3696-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3710-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3710-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3715-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3715-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3811-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3811-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3812-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3812-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3814-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3814-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3815-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3815-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3818-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3818-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3819-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3819-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3821-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3821-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3831-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3831-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3833-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3833-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3837-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3837-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3841-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3841-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3842-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3842-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3844-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3844-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3845-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3845-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3846-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3846-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3848-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3848-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3850-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3850-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3851-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3851-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3852-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3852-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3854-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3854-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3855-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3855-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3856-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3856-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3858-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3858-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3860-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3860-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3861-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3861-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3862-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3862-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3864-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3864-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3866-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3866-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3867-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3867-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3869-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3869-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3870-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3870-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3872-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3872-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3875-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3875-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3877-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3877-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3941-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3941-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3947-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3947-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3949-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3949-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3950-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3950-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3952-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3952-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3955-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3955-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3956-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3956-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3966-10A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3966-10A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3968-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3968-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3970-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3970-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3971-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3971-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3972-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3972-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3973-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3973-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3975-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3975-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3976-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3976-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3977-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3977-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3979-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3979-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3980-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3980-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3982-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3982-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3984-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3984-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3986-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3986-10A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3989-10A-01D-1019-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3989-10A-01D-1019-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3994-10A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3994-10A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A02K-10A-01D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A02K-10A-01D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A02Y-10A-01D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A02Y-10A-01D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-5900-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-5900-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6548-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6548-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6888-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6888-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6889-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6889-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6890-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6890-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6895-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6895-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6899-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6899-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6901-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6901-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6963-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6963-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6964-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6964-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6965-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6965-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-A5EJ-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-A5EJ-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-A5EK-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-A5EK-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2687-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2687-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2689-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2689-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2690-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2690-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2691-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2691-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2692-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2692-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2693-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2693-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-3400-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-3400-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-3911-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-3911-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-3913-10A-01D-1450-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-3913-10A-01D-1450-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-4110-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-4110-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-5654-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-5654-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-6136-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-6136-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-6655-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-6655-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-6672-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-6672-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-A56K-10A-01D-A38W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-A56K-10A-01D-A38W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-A56L-10A-01D-A38W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-A56L-10A-01D-A38W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-A56N-10A-01D-A38W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-A56N-10A-01D-A38W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3574-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3574-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3575-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3575-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3578-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3578-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3580-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3580-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3581-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3581-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3582-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3582-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3583-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3583-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3584-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3584-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3586-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3586-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3587-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3587-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3591-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3591-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3592-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3592-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3593-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3593-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3594-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3594-10A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3598-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3598-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3599-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3599-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3600-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3600-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3602-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3602-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3608-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3608-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3609-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3609-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3612-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3612-10A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3726-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3726-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3727-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3727-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3728-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3728-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3878-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3878-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3881-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3881-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3882-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3882-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3883-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3883-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3885-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3885-10A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3887-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3887-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3890-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3890-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3892-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3892-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3893-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3893-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3894-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3894-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3896-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3896-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3898-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3898-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3901-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3901-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3902-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3902-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3999-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3999-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-4001-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-4001-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-4005-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-4005-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-4007-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-4007-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-4008-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-4008-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-4015-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-4015-10A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-4021-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-4021-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-4022-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-4022-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A026-10A-01D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A026-10A-01D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AH-6544-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AH-6544-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AH-6549-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AH-6549-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AH-6644-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AH-6644-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AH-6897-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AH-6897-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AH-6903-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AH-6903-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AM-5820-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AM-5820-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AM-5821-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AM-5821-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AU-3779-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AU-3779-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AU-6004-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AU-6004-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-4070-10A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-4070-10A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-4071-10A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-4071-10A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-5543-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-5543-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-6196-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-6196-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-6197-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-6197-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-6386-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-6386-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-A69D-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-A69D-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-A71X-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-A71X-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-A8YK-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-A8YK-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4308-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4308-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4313-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4313-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4315-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4315-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4323-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4323-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4614-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4614-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4615-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4615-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4616-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4616-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4681-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4681-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4682-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4682-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-5403-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-5403-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-5407-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-5407-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-BM-6198-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-BM-6198-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-5254-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-5254-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-5255-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-5255-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-5256-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-5256-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-5796-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-5796-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-5797-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-5797-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-6715-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-6715-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-6716-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-6716-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-6717-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-6717-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-6718-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-6718-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-6719-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-6719-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CI-6619-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CI-6619-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CI-6620-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CI-6620-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CI-6621-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CI-6621-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CI-6622-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CI-6622-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CI-6623-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CI-6623-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CI-6624-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CI-6624-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-4947-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-4947-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-4950-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-4950-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-4951-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-4951-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-5912-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-5912-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-5913-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-5913-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-5915-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-5915-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-5916-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-5916-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-6746-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-6746-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-6747-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-6747-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-6748-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-6748-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-6751-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-6751-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CL-4957-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CL-4957-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CL-5917-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CL-5917-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CL-5918-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CL-5918-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-4743-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-4743-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-4746-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-4746-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-4747-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-4747-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-4748-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-4748-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-4751-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-4751-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-4752-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-4752-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5341-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5341-10A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5344-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5344-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5348-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5348-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5349-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5349-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5860-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5860-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5861-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5861-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5862-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5862-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5863-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5863-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5864-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5864-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5868-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5868-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6161-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6161-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6162-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6162-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6163-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6163-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6164-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6164-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6166-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6166-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6167-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6167-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6168-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6168-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6169-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6169-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6170-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6170-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6171-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6171-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6172-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6172-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6674-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6674-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6675-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6675-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6676-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6676-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6677-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6677-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6678-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6678-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6679-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6679-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6680-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6680-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-5537-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-5537-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-5538-10A-02D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-5538-10A-02D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-5539-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-5539-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-5540-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-5540-10A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-5541-10A-02D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-5541-10A-02D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6529-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6529-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6530-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6530-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6531-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6531-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6532-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6532-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6533-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6533-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6534-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6534-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6535-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6535-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6536-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6536-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6537-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6537-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6538-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6538-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6539-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6539-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6540-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6540-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6541-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6541-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6898-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6898-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6920-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6920-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6922-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6922-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6923-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6923-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6924-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6924-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6926-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6926-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6927-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6927-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6928-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6928-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6929-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6929-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6930-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6930-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6931-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6931-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6932-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6932-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-7000-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-7000-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-4745-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-4745-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-4749-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-4749-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-5337-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-5337-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-5869-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-5869-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6154-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6154-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6155-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6155-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6156-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6156-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6157-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6157-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6158-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6158-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6681-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6681-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6682-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6682-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6683-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6683-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A0X9-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A0X9-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A0XD-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A0XD-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A0XF-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A0XF-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1D0-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1D0-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1D4-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1D4-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1D6-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1D6-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1D7-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1D7-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1D8-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1D8-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1D9-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1D9-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1DA-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1DA-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1DB-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1DB-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1HA-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1HA-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1HB-10A-01D-A182-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1HB-10A-01D-A182-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A280-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A280-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A285-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A285-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A288-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A288-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28A-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28A-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28C-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28C-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28E-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28E-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28F-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28F-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28G-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28G-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28H-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28H-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28K-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28K-10A-01D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DT-5265-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DT-5265-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DY-A0XA-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DY-A0XA-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DY-A1DC-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DY-A1DC-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DY-A1DD-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DY-A1DD-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DY-A1DE-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DY-A1DE-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DY-A1DF-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DY-A1DF-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DY-A1DG-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DY-A1DG-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DY-A1H8-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DY-A1H8-10A-01D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EF-5830-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EF-5830-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EF-5831-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EF-5831-10A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6506-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6506-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6507-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6507-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6508-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6508-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6509-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6509-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6510-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6510-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6511-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6511-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6512-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6512-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6513-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6513-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6514-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6514-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6881-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6881-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6882-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6882-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6883-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6883-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6884-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6884-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6885-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6885-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6917-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6917-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-7002-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-7002-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-7004-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-7004-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6459-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6459-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6460-10B-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6460-10B-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6461-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6461-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6463-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6463-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6569-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6569-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6570-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6570-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6703-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6703-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6805-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6805-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6806-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6806-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6807-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6807-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6808-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6808-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6809-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6809-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6854-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6854-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6855-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6855-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6856-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6856-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6464-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6464-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6465-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6465-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6571-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6571-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6702-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6702-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6810-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6810-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6812-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6812-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6813-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6813-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6814-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6814-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6861-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6861-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6863-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6863-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6864-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6864-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6293-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6293-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6294-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6294-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6295-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6295-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6297-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6297-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6298-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6298-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6299-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6299-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6302-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6302-10A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6303-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6303-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6304-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6304-10A-01D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6306-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6306-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6307-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6307-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6309-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6309-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6310-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6310-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6311-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6311-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6314-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6314-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6315-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6315-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6317-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6317-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6320-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6320-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6321-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6321-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6322-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6322-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6323-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6323-10A-01D-1718-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6586-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6586-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6588-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6588-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6626-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6626-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6627-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6627-10A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6628-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6628-10A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G5-6233-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G5-6233-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G5-6235-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G5-6235-10A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G5-6572-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G5-6572-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G5-6641-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G5-6641-10A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A50T-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A50T-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A50U-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A50U-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A50V-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A50V-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A5IV-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A5IV-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A6GA-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A6GA-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A6GB-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A6GB-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A6GC-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A6GC-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A8F7-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A8F7-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A8F8-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A8F8-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-QG-A5YV-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-QG-A5YV-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-QG-A5YW-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-QG-A5YW-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-QG-A5YX-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-QG-A5YX-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-QG-A5Z1-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-QG-A5Z1-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-QG-A5Z2-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-QG-A5Z2-10A-01D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-QL-A97D-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-QL-A97D-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-RU-A8FL-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-RU-A8FL-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-SS-A7HO-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-SS-A7HO-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-T9-A92H-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-T9-A92H-10A-01D-A36Z-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-WS-AB45-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-WS-AB45-10A-01D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
Table S11.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-G4-6317 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G5-6572 | (TODO) -- GDC | clinical__biospecimen |
Table S12.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-G4-6317 | (TODO) -- GDC | clinical__primary | |
| TCGA-G5-6572 | (TODO) -- GDC | clinical__primary |
Table S13.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-G4-6317-02A-11D-2062-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6317-02A-11D-2062-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G5-6572-02A-12D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G5-6572-02A-12D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
Table S14.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-G4-6317-02A-11R-2066-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6317-02A-11R-2066-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6317-02A-11R-2066-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G5-6572-02A-12R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G5-6572-02A-12R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G5-6572-02A-12R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM |
Table S15.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-G4-6317-02A-11H-2065-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6317-02A-11H-2065-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G5-6572-02A-12H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G5-6572-02A-12H-1829-13 | (TODO) -- GDC | miR__isoformExp |
Table S16.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-G4-6317-02A-11D-2067-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G5-6572-02A-12D-1826-10 | (TODO) -- GDC | SNV__mutect |
Table S17.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-G5-6572-02A-12D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
Table S18.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-3L-AA1B | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-4N-A93T | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-4T-AA8H | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-5M-AAT4 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-5M-AAT5 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-5M-AAT6 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-5M-AATA | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-5M-AATE | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2670 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2671 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2672 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2674 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2675 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2676 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2677 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2678 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2679 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2680 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2681 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2682 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2683 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2684 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2685 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2686 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-3807 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-3808 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-3809 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-3810 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-4105 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-4107 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5656 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5657 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5659 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5660 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5661 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5662 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5664 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5665 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5666 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5667 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6137 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6138 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6140 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6141 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6142 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6648 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6649 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6650 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6651 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6652 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6653 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6654 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6780 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6781 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-6782 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-A565 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-A566 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-A567 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-A56B | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-A5ZU | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3488 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3489 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3492 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3494 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3495 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3496 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3502 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3506 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3509 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3510 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3511 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3514 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3516 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3517 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3518 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3519 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3520 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3521 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3522 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3524 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3525 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3526 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3527 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3529 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3530 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3531 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3532 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3534 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3538 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3542 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3543 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3544 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3548 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3549 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3552 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3553 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3554 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3555 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3556 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3558 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3560 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3561 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3562 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3655 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3660 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3662 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3663 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3664 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3666 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3667 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3672 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3673 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3675 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3678 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3679 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3680 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3681 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3684 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3685 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3688 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3692 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3693 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3695 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3696 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3697 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3710 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3712 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3713 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3715 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3811 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3812 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3814 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3815 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3818 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3819 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3821 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3831 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3833 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3837 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3841 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3842 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3844 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3845 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3846 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3848 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3850 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3851 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3852 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3854 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3855 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3856 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3858 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3860 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3861 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3862 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3864 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3866 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3867 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3869 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3870 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3872 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3875 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3877 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3930 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3939 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3941 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3947 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3949 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3950 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3952 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3955 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3956 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3966 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3967 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3968 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3970 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3971 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3972 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3973 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3975 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3976 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3977 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3979 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3980 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3982 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3984 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3986 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3989 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3994 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A004 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A00A | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A00D | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A00E | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A00F | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A00J | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A00K | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A00L | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A00N | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A00O | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A00Q | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A00R | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A00U | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A00W | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A00Z | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A010 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A017 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01C | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01D | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01F | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01G | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01I | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01K | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01P | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01Q | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01R | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01S | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01T | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01V | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01X | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01Z | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A022 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A024 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A029 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A02E | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A02F | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A02H | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A02J | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A02K | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A02O | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A02R | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A02W | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A02Y | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A03F | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A03J | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-5900 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6548 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6888 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6889 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6890 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6895 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6899 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6901 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6963 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6964 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-6965 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-A5EJ | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AD-A5EK | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2687 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2689 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2690 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2691 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2692 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2693 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-3400 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-3911 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-3913 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-3914 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-4110 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-5654 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-6136 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-6655 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-6672 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-A56K | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-A56L | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-A56N | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3574 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3575 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3578 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3580 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3581 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3582 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3583 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3584 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3586 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3587 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3591 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3592 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3593 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3594 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3598 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3599 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3600 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3601 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3602 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3605 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3608 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3609 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3611 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3612 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3725 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3726 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3727 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3728 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3731 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3732 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3742 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3878 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3881 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3882 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3883 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3885 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3887 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3890 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3891 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3892 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3893 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3894 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3896 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3898 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3901 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3902 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3906 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3909 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3999 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4001 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4005 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4007 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4008 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4009 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4015 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4021 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-4022 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A002 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A008 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A00C | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A00H | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A00Y | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A011 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A014 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A015 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A016 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A01J | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A01L | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A01N | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A01W | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A01Y | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A020 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A023 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A025 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A026 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A02G | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A02N | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A02X | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A032 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A036 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AH-6544 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AH-6547 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AH-6549 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AH-6643 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AH-6644 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AH-6897 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AH-6903 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AM-5820 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AM-5821 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AU-3779 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AU-6004 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-4070 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-4071 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-5543 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-6196 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-6197 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-6386 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-A54L | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-A69D | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-A71X | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AY-A8YK | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4308 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4313 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4315 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4323 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4614 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4615 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4616 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4681 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4682 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-4684 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-5403 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-5407 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6598 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6599 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6600 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6601 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6603 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6605 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6606 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6607 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6608 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-BM-6198 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-5254 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-5255 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-5256 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-5796 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-5797 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-6715 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-6716 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-6717 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-6718 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CA-6719 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CI-6619 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CI-6620 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CI-6621 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CI-6622 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CI-6623 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CI-6624 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-4947 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-4948 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-4950 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-4951 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-4952 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-5912 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-5913 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-5914 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-5915 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-5916 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-6746 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-6747 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-6748 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CK-6751 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CL-4957 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CL-5917 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CL-5918 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-4743 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-4744 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-4746 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-4747 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-4748 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-4750 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-4751 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-4752 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5341 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5344 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5348 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5349 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5860 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5861 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5862 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5863 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5864 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-5868 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6161 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6162 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6163 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6164 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6165 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6166 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6167 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6168 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6169 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6170 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6171 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6172 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6674 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6675 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6676 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6677 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6678 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6679 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-CM-6680 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-5537 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-5538 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-5539 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-5540 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-5541 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6529 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6530 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6531 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6532 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6533 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6534 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6535 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6536 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6537 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6538 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6539 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6540 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6541 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6898 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6920 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6922 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6923 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6924 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6926 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6927 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6928 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6929 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6930 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6931 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-6932 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-D5-7000 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-4745 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-4749 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-5337 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-5869 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6154 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6155 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6156 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6157 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6158 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6160 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6681 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6682 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DC-6683 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A0X9 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A0XD | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A0XF | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1D0 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1D4 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1D6 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1D7 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1D8 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1D9 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1DA | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1DB | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1HA | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A1HB | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A280 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A282 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A285 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A288 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28A | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28C | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28E | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28F | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28G | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28H | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28K | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DM-A28M | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DT-5265 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DY-A0XA | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DY-A1DC | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DY-A1DD | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DY-A1DE | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DY-A1DF | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DY-A1DG | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-DY-A1H8 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EF-5830 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EF-5831 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6506 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6507 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6508 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6509 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6510 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6511 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6512 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6513 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6514 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6881 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6882 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6883 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6884 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6885 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-6917 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-7002 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-EI-7004 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6459 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6460 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6461 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6463 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6569 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6570 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6703 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6704 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6805 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6806 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6807 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6808 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6809 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6854 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6855 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6856 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6464 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6465 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6571 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6702 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6810 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6811 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6812 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6813 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6814 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6861 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6863 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F5-6864 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6293 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6294 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6295 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6297 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6298 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6299 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6302 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6303 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6304 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6306 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6307 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6309 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6310 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6311 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6314 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6315 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6317 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6320 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6321 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6322 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6323 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6586 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6588 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6625 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6626 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6627 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6628 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G5-6233 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G5-6235 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G5-6572 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G5-6641 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A50T | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A50U | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A50V | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A5IV | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A6GA | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A6GB | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A6GC | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A8F7 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-NH-A8F8 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-QG-A5YV | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-QG-A5YW | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-QG-A5YX | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-QG-A5Z1 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-QG-A5Z2 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-QL-A97D | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-RU-A8FL | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-SS-A7HO | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-T9-A92H | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-WS-AB45 | (TODO) -- GDC | clinical__biospecimen |
Table S19.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-3L-AA1B | (TODO) -- GDC | clinical__primary | |
| TCGA-4N-A93T | (TODO) -- GDC | clinical__primary | |
| TCGA-4T-AA8H | (TODO) -- GDC | clinical__primary | |
| TCGA-5M-AAT4 | (TODO) -- GDC | clinical__primary | |
| TCGA-5M-AAT6 | (TODO) -- GDC | clinical__primary | |
| TCGA-5M-AATE | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2670 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2671 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2672 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2674 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2675 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2676 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2677 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2678 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2679 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2680 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2681 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2682 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2683 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2684 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2685 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2686 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-3807 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-3808 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-3809 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-3810 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-4105 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-4107 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5656 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5657 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5659 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5660 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5661 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5662 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5664 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5665 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5666 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5667 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6137 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6138 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6140 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6141 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6142 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6648 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6649 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6650 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6651 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6652 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6653 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6654 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6780 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6781 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-6782 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-A565 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-A566 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-A567 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-A56B | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-A5ZU | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3488 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3489 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3492 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3494 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3495 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3496 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3502 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3506 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3509 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3510 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3511 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3514 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3516 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3517 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3518 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3519 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3520 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3521 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3522 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3524 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3525 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3526 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3527 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3529 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3530 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3531 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3532 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3534 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3538 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3542 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3543 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3544 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3548 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3549 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3552 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3553 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3554 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3555 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3556 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3558 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3560 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3561 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3562 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3655 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3660 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3662 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3663 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3664 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3666 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3667 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3672 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3673 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3675 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3678 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3679 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3680 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3681 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3684 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3685 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3688 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3692 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3693 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3695 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3696 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3697 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3710 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3712 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3713 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3715 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3811 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3812 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3814 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3815 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3818 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3819 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3821 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3831 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3833 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3837 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3841 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3842 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3844 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3845 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3846 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3848 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3850 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3851 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3852 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3854 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3855 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3856 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3858 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3860 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3861 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3862 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3864 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3866 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3867 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3869 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3870 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3872 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3875 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3877 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3930 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3939 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3941 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3947 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3949 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3950 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3952 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3955 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3956 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3966 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3967 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3968 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3970 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3971 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3972 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3973 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3975 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3976 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3977 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3979 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3980 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3982 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3984 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3986 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3989 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3994 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A004 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A00A | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A00D | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A00E | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A00F | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A00J | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A00K | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A00L | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A00N | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A00O | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A00Q | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A00R | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A00U | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A00W | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A00Z | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A010 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A017 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01C | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01D | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01F | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01G | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01I | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01K | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01P | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01Q | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01R | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01S | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01T | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01V | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01X | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01Z | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A022 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A024 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A029 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A02E | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A02F | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A02H | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A02J | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A02K | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A02O | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A02R | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A02W | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A02Y | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A03F | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A03J | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-5900 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6548 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6888 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6889 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6890 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6895 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6899 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6901 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6963 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6964 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-6965 | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-A5EJ | (TODO) -- GDC | clinical__primary | |
| TCGA-AD-A5EK | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2687 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2689 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2690 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2691 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2692 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2693 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-3400 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-3911 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-3913 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-3914 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-4110 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-5654 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-6136 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-6655 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-6672 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-A56K | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-A56L | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-A56N | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3574 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3575 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3578 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3580 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3581 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3582 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3583 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3584 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3586 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3587 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3591 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3592 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3593 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3594 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3598 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3599 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3600 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3601 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3602 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3605 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3608 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3609 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3611 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3612 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3725 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3726 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3727 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3728 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3731 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3732 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3742 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3878 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3881 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3882 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3883 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3885 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3887 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3890 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3891 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3892 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3893 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3894 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3896 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3898 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3901 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3902 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3906 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3909 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3999 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4001 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4005 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4007 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4008 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4009 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4015 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4021 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-4022 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A002 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A008 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A00C | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A00H | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A00Y | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A011 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A014 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A015 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A016 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A01J | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A01L | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A01N | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A01W | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A01Y | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A020 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A023 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A025 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A026 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A02G | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A02N | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A02X | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A032 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A036 | (TODO) -- GDC | clinical__primary | |
| TCGA-AH-6544 | (TODO) -- GDC | clinical__primary | |
| TCGA-AH-6547 | (TODO) -- GDC | clinical__primary | |
| TCGA-AH-6549 | (TODO) -- GDC | clinical__primary | |
| TCGA-AH-6643 | (TODO) -- GDC | clinical__primary | |
| TCGA-AH-6644 | (TODO) -- GDC | clinical__primary | |
| TCGA-AH-6897 | (TODO) -- GDC | clinical__primary | |
| TCGA-AH-6903 | (TODO) -- GDC | clinical__primary | |
| TCGA-AM-5820 | (TODO) -- GDC | clinical__primary | |
| TCGA-AM-5821 | (TODO) -- GDC | clinical__primary | |
| TCGA-AU-3779 | (TODO) -- GDC | clinical__primary | |
| TCGA-AU-6004 | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-4070 | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-4071 | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-5543 | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-6196 | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-6197 | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-6386 | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-A54L | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-A69D | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-A71X | (TODO) -- GDC | clinical__primary | |
| TCGA-AY-A8YK | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4308 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4313 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4315 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4323 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4614 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4615 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4616 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4681 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4682 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-4684 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-5403 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-5407 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6598 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6599 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6600 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6601 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6603 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6605 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6606 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6607 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6608 | (TODO) -- GDC | clinical__primary | |
| TCGA-BM-6198 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-5254 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-5255 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-5256 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-5796 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-5797 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-6715 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-6716 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-6717 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-6718 | (TODO) -- GDC | clinical__primary | |
| TCGA-CA-6719 | (TODO) -- GDC | clinical__primary | |
| TCGA-CI-6619 | (TODO) -- GDC | clinical__primary | |
| TCGA-CI-6620 | (TODO) -- GDC | clinical__primary | |
| TCGA-CI-6621 | (TODO) -- GDC | clinical__primary | |
| TCGA-CI-6622 | (TODO) -- GDC | clinical__primary | |
| TCGA-CI-6623 | (TODO) -- GDC | clinical__primary | |
| TCGA-CI-6624 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-4947 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-4948 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-4950 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-4951 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-4952 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-5912 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-5913 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-5914 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-5915 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-5916 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-6746 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-6747 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-6748 | (TODO) -- GDC | clinical__primary | |
| TCGA-CK-6751 | (TODO) -- GDC | clinical__primary | |
| TCGA-CL-4957 | (TODO) -- GDC | clinical__primary | |
| TCGA-CL-5917 | (TODO) -- GDC | clinical__primary | |
| TCGA-CL-5918 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-4743 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-4744 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-4746 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-4747 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-4748 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-4750 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-4751 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-4752 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5341 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5344 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5348 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5349 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5860 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5861 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5862 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5863 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5864 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-5868 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6161 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6162 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6163 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6164 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6165 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6166 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6167 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6168 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6169 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6170 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6171 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6172 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6674 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6675 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6676 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6677 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6678 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6679 | (TODO) -- GDC | clinical__primary | |
| TCGA-CM-6680 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-5537 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-5538 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-5539 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-5540 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-5541 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6529 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6530 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6531 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6532 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6533 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6534 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6535 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6536 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6537 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6538 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6539 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6540 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6541 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6898 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6920 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6922 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6923 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6924 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6926 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6927 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6928 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6929 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6930 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6931 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-6932 | (TODO) -- GDC | clinical__primary | |
| TCGA-D5-7000 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-4745 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-4749 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-5337 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-5869 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6154 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6155 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6156 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6157 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6158 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6160 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6681 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6682 | (TODO) -- GDC | clinical__primary | |
| TCGA-DC-6683 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A0X9 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A0XD | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A0XF | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1D0 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1D4 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1D6 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1D7 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1D8 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1D9 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1DA | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1DB | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A1HB | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A280 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A282 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A285 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A288 | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28A | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28C | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28E | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28F | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28G | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28H | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28K | (TODO) -- GDC | clinical__primary | |
| TCGA-DM-A28M | (TODO) -- GDC | clinical__primary | |
| TCGA-DT-5265 | (TODO) -- GDC | clinical__primary | |
| TCGA-DY-A0XA | (TODO) -- GDC | clinical__primary | |
| TCGA-DY-A1DC | (TODO) -- GDC | clinical__primary | |
| TCGA-DY-A1DD | (TODO) -- GDC | clinical__primary | |
| TCGA-DY-A1DE | (TODO) -- GDC | clinical__primary | |
| TCGA-DY-A1DF | (TODO) -- GDC | clinical__primary | |
| TCGA-DY-A1DG | (TODO) -- GDC | clinical__primary | |
| TCGA-DY-A1H8 | (TODO) -- GDC | clinical__primary | |
| TCGA-EF-5830 | (TODO) -- GDC | clinical__primary | |
| TCGA-EF-5831 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6506 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6507 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6508 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6509 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6510 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6511 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6512 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6513 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6514 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6881 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6882 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6883 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6884 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6885 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-6917 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-7002 | (TODO) -- GDC | clinical__primary | |
| TCGA-EI-7004 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6459 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6460 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6461 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6463 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6569 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6570 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6703 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6704 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6805 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6806 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6807 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6808 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6809 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6854 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6855 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6856 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6464 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6465 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6571 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6702 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6811 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6812 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6813 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6814 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6861 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6863 | (TODO) -- GDC | clinical__primary | |
| TCGA-F5-6864 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6293 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6294 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6295 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6297 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6298 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6299 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6302 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6303 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6304 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6306 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6307 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6309 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6310 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6311 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6314 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6315 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6317 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6320 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6321 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6322 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6323 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6586 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6588 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6625 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6626 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6627 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6628 | (TODO) -- GDC | clinical__primary | |
| TCGA-G5-6233 | (TODO) -- GDC | clinical__primary | |
| TCGA-G5-6235 | (TODO) -- GDC | clinical__primary | |
| TCGA-G5-6572 | (TODO) -- GDC | clinical__primary | |
| TCGA-G5-6641 | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A50T | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A50U | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A50V | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A5IV | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A6GA | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A6GB | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A6GC | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A8F7 | (TODO) -- GDC | clinical__primary | |
| TCGA-NH-A8F8 | (TODO) -- GDC | clinical__primary | |
| TCGA-QG-A5YV | (TODO) -- GDC | clinical__primary | |
| TCGA-QG-A5YW | (TODO) -- GDC | clinical__primary | |
| TCGA-QG-A5YX | (TODO) -- GDC | clinical__primary | |
| TCGA-QG-A5Z1 | (TODO) -- GDC | clinical__primary | |
| TCGA-QG-A5Z2 | (TODO) -- GDC | clinical__primary | |
| TCGA-QL-A97D | (TODO) -- GDC | clinical__primary | |
| TCGA-RU-A8FL | (TODO) -- GDC | clinical__primary | |
| TCGA-SS-A7HO | (TODO) -- GDC | clinical__primary | |
| TCGA-T9-A92H | (TODO) -- GDC | clinical__primary | |
| TCGA-WS-AB45 | (TODO) -- GDC | clinical__primary |
Table S20.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-3L-AA1B-01A-11D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-3L-AA1B-01A-11D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-4N-A93T-01A-11D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-4N-A93T-01A-11D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-4T-AA8H-01A-11D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-4T-AA8H-01A-11D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-5M-AAT4-01A-11D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-5M-AAT4-01A-11D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-5M-AAT5-01A-21D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-5M-AAT5-01A-21D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-5M-AAT6-01A-11D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-5M-AAT6-01A-11D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-5M-AATA-01A-31D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-5M-AATA-01A-31D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-5M-AATE-01A-11D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-5M-AATE-01A-11D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2670-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2670-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2671-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2671-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2671-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2671-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2672-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2672-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2672-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2672-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2674-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2674-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2674-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2674-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2675-01A-02D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2675-01A-02D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2676-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2676-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2676-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2676-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2677-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2677-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2677-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2677-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2677-01A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2677-01A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2678-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2678-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2678-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2678-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2679-01A-02D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2679-01A-02D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2679-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2679-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2680-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2680-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2680-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2680-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2681-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2681-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2681-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2681-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2682-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2682-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2682-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2682-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2683-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2683-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2683-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2683-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2684-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2684-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2684-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2684-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2684-01A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2684-01A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2685-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2685-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2685-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2685-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2686-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2686-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2686-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2686-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-3807-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-3807-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-3809-01A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-3809-01A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-3810-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-3810-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-3810-01A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-3810-01A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-4105-01A-02D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-4105-01A-02D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-4107-01A-02D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-4107-01A-02D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-4107-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-4107-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5656-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5656-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5656-01A-21D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5656-01A-21D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5657-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5657-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5659-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5659-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5659-01A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5659-01A-01D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5660-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5660-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5661-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5661-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5662-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5662-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5664-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5664-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5665-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5665-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5666-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5666-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5667-01A-21D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5667-01A-21D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6137-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6137-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6138-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6138-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6140-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6140-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6141-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6141-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6142-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6142-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6648-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6648-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6649-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6649-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6650-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6650-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6650-01A-11D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6650-01A-11D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6651-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6651-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6652-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6652-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6653-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6653-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6654-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6654-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6780-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6780-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6780-01A-11D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6780-01A-11D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6781-01A-22D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6781-01A-22D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6781-01A-22D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6781-01A-22D-A274-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-6782-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-6782-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-A565-01A-31D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-A565-01A-31D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-A566-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-A566-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-A567-01A-31D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-A567-01A-31D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-A56B-01A-31D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-A56B-01A-31D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-A5ZU-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-A5ZU-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3488-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3488-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3489-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3489-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3492-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3492-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3494-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3494-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3495-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3495-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3496-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3496-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3502-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3502-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3506-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3506-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3510-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3510-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3511-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3511-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3514-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3514-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3514-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3514-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3516-01A-02D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3516-01A-02D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3516-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3516-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3517-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3517-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3517-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3517-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3518-01A-02D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3518-01A-02D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3518-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3518-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3519-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3519-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3519-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3519-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3520-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3520-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3520-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3520-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3521-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3521-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3521-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3521-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3522-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3522-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3522-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3522-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3524-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3524-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3524-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3524-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3525-01A-02D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3525-01A-02D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3525-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3525-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3526-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3526-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3526-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3526-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3527-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3527-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3527-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3527-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3529-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3529-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3529-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3529-01A-02D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3530-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3530-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3530-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3530-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3531-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3531-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3531-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3531-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3532-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3532-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3532-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3532-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3534-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3534-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3534-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3534-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3538-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3538-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3542-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3542-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3543-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3543-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3544-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3544-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3548-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3548-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3549-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3549-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3552-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3552-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3553-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3553-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3554-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3554-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3555-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3555-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3556-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3556-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3558-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3558-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3560-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3560-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3561-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3561-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3562-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3562-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3655-01A-02D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3655-01A-02D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3660-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3660-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3662-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3662-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3663-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3663-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3664-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3664-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3666-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3666-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3667-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3667-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3672-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3672-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3673-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3673-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3675-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3675-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3678-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3678-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3679-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3679-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3680-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3680-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3681-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3681-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3684-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3684-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3685-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3685-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3688-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3688-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3692-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3692-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3693-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3693-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3695-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3695-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3696-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3696-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3697-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3697-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3710-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3710-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3712-01A-21D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3712-01A-21D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3713-01A-21D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3713-01A-21D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3715-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3715-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3811-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3811-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3812-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3812-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3814-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3814-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3818-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3818-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3819-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3819-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3821-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3821-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3831-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3831-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3833-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3833-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3837-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3837-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3841-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3841-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3842-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3842-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3844-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3844-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3845-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3845-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3846-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3846-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3848-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3848-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3850-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3850-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3851-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3851-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3852-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3852-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3854-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3854-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3855-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3855-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3856-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3856-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3858-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3858-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3860-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3860-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3864-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3864-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3866-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3866-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3867-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3867-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3869-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3869-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3870-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3870-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3872-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3872-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3875-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3875-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3877-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3877-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3930-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3930-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3939-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3939-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3941-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3941-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3947-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3947-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3949-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3949-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3950-01A-02D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3950-01A-02D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3952-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3952-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3955-01A-02D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3955-01A-02D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3956-01A-02D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3956-01A-02D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3966-01A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3966-01A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3968-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3968-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3971-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3971-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3972-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3972-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3973-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3973-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3975-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3975-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3976-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3976-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3977-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3977-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3979-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3979-01A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3982-01A-02D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3982-01A-02D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3984-01A-02D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3984-01A-02D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3986-01A-02D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3986-01A-02D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3989-01A-01D-1019-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3989-01A-01D-1019-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3994-01A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3994-01A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A004-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A004-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A00A-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A00A-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A00D-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A00D-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A00E-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A00E-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A00F-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A00F-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A00J-01A-02D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A00J-01A-02D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A00K-01A-02D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A00K-01A-02D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A00L-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A00L-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A00N-01A-02D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A00N-01A-02D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A00O-01A-02D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A00O-01A-02D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A00Q-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A00Q-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A00R-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A00R-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A00U-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A00U-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A00W-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A00W-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A00Z-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A00Z-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A010-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A010-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A017-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A017-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01C-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01C-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01D-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01D-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01F-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01F-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01G-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01G-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01I-01A-02D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01I-01A-02D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01K-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01K-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01P-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01P-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01Q-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01Q-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01R-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01R-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01S-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01S-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01T-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01T-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01V-01A-23D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01V-01A-23D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01X-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01X-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01Z-01A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01Z-01A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A022-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A022-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A024-01A-02D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A024-01A-02D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A029-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A029-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A02E-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A02E-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A02F-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A02F-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A02H-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A02H-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A02J-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A02J-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A02O-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A02O-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A02R-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A02R-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A02W-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A02W-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A02Y-01A-43D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A02Y-01A-43D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A03F-01A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A03F-01A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A03J-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A03J-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-5900-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-5900-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6548-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6548-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6888-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6888-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6889-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6889-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6890-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6890-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6895-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6895-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6899-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6899-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6901-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6901-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6963-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6963-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6964-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6964-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-6965-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-6965-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-A5EJ-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-A5EJ-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AD-A5EK-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AD-A5EK-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2687-01A-02D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2687-01A-02D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2689-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2689-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2689-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2689-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2690-01A-02D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2690-01A-02D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2691-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2691-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2691-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2691-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2692-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2692-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2693-01A-02D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2693-01A-02D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-3400-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-3400-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-3400-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-3400-01A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-3911-01A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-3911-01A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-3913-01A-02D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-3913-01A-02D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-4110-01A-02D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-4110-01A-02D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-5654-01A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-5654-01A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-6136-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-6136-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-6655-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-6655-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-6672-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-6672-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-A56K-01A-32D-A38W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-A56K-01A-32D-A38W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-A56L-01A-31D-A38W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-A56L-01A-31D-A38W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-A56N-01A-12D-A38W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-A56N-01A-12D-A38W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3574-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3574-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3575-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3575-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3578-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3578-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3580-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3580-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3581-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3581-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3582-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3582-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3583-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3583-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3584-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3584-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3586-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3586-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3587-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3587-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3591-01A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3591-01A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3592-01A-02D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3592-01A-02D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3593-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3593-01A-01D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3594-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3594-01A-02D-0819-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3598-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3598-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3599-01A-02D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3599-01A-02D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3600-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3600-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3601-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3601-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3602-01A-02D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3602-01A-02D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3605-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3605-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3608-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3608-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3609-01A-02D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3609-01A-02D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3611-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3611-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3612-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3612-01A-01D-0824-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3725-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3725-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3726-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3726-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3727-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3727-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3728-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3728-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3731-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3731-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3732-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3732-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3742-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3742-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3878-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3878-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3881-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3881-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3882-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3882-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3883-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3883-01A-02D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3885-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3885-01A-01D-0903-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3887-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3887-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3890-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3890-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3892-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3892-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3893-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3893-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3894-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3894-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3896-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3896-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3898-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3898-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3901-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3901-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3902-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3902-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3909-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3909-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3999-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3999-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-4001-01A-02D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-4001-01A-02D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-4005-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-4005-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-4007-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-4007-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-4008-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-4008-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-4015-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-4015-01A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-4021-01A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-4021-01A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-4022-01A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-4022-01A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A002-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A002-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A008-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A008-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A00C-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A00C-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A00H-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A00H-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A00Y-01A-02D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A00Y-01A-02D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A011-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A011-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A014-01A-02D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A014-01A-02D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A015-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A015-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A016-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A016-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A01J-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A01J-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A01L-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A01L-01A-01D-A003-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A01N-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A01N-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A01W-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A01W-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A01Y-01A-41D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A01Y-01A-41D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A020-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A020-01A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A023-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A023-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A025-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A025-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A026-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A026-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A026-01A-32D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A026-01A-32D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A02G-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A02G-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A02N-01A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A02N-01A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A02X-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A02X-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A032-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A032-01A-01D-A008-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AH-6544-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AH-6544-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AH-6547-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AH-6547-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AH-6643-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AH-6643-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AH-6644-01A-21D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AH-6644-01A-21D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AH-6897-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AH-6897-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AH-6903-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AH-6903-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AM-5820-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AM-5820-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AM-5821-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AM-5821-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AU-3779-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AU-3779-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AU-6004-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AU-6004-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-4070-01A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-4070-01A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-4071-01A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-4071-01A-01D-1427-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-5543-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-5543-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-6196-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-6196-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-6197-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-6197-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-6386-01A-21D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-6386-01A-21D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-A54L-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-A54L-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-A69D-01A-11D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-A69D-01A-11D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-A71X-01A-12D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-A71X-01A-12D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AY-A8YK-01A-11D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AY-A8YK-01A-11D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4308-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4308-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4313-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4313-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4315-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4315-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4323-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4323-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4614-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4614-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4615-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4615-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4616-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4616-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4681-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4681-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4682-01B-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4682-01B-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-4684-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-4684-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-5403-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-5403-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-5407-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-5407-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6598-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6598-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6599-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6599-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6600-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6600-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6601-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6601-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6603-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6603-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6605-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6605-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6606-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6606-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6607-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6607-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6608-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6608-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-BM-6198-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-BM-6198-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-5254-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-5254-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-5255-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-5255-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-5256-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-5256-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-5796-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-5796-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-5797-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-5797-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-6715-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-6715-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-6716-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-6716-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-6717-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-6717-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-6718-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-6718-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CA-6719-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CA-6719-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CI-6619-01B-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CI-6619-01B-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CI-6620-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CI-6620-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CI-6621-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CI-6621-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CI-6622-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CI-6623-01B-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CI-6623-01B-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CI-6624-01C-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CI-6624-01C-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-4947-01B-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-4947-01B-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-4948-01B-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-4948-01B-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-4950-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-4950-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-4951-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-4951-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-4952-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-4952-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-5912-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-5912-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-5913-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-5913-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-5914-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-5914-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-5915-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-5915-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-5916-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-5916-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-6746-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-6746-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-6747-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-6747-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-6748-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-6748-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CK-6751-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CK-6751-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CL-4957-01A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CL-4957-01A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CL-5917-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CL-5917-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CL-5918-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CL-5918-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-4743-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-4743-01A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-4744-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-4744-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-4746-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-4746-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-4747-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-4747-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-4748-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-4748-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-4750-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-4750-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-4751-01A-02D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-4751-01A-02D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-4752-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-4752-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5341-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5341-01A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5344-01A-21D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5344-01A-21D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5348-01A-21D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5348-01A-21D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5349-01A-21D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5349-01A-21D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5860-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5860-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5861-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5861-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5862-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5862-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5863-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5863-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5864-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5864-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-5868-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-5868-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6161-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6161-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6162-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6162-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6163-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6163-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6164-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6164-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6165-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6165-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6166-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6166-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6167-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6167-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6168-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6168-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6169-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6169-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6170-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6170-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6171-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6171-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6172-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6172-01A-11D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6674-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6674-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6675-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6675-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6676-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6676-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6677-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6677-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6678-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6678-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6679-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6679-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-CM-6680-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-CM-6680-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-5537-01A-21D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-5537-01A-21D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-5538-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-5538-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-5539-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-5539-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-5540-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-5540-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-5541-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-5541-01A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6529-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6529-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6530-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6530-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6531-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6531-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6532-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6532-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6533-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6533-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6534-01A-21D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6534-01A-21D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6535-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6535-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6536-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6536-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6537-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6537-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6538-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6538-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6539-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6539-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6540-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6540-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6541-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6541-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6898-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6898-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6920-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6920-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6922-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6922-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6923-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6923-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6924-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6924-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6926-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6926-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6927-01A-21D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6927-01A-21D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6928-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6928-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6929-01A-31D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6929-01A-31D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6930-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6930-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6931-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6931-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-6932-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-6932-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-D5-7000-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-D5-7000-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-4745-01A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-4745-01A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-4749-01A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-4749-01A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-5337-01A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-5337-01A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-5869-01A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-5869-01A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6154-01A-31D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6154-01A-31D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6155-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6155-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6156-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6156-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6157-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6157-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6158-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6158-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6160-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6160-01A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6681-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6681-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6682-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6682-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DC-6683-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DC-6683-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A0X9-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A0X9-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A0XD-01A-12D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A0XD-01A-12D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A0XF-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A0XF-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1D0-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1D0-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1D4-01A-21D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1D4-01A-21D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1D6-01A-21D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1D6-01A-21D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1D7-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1D7-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1D8-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1D8-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1D9-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1D9-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1DA-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1DA-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1DB-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1DB-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1HA-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1HA-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A1HB-01A-21D-A182-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A1HB-01A-21D-A182-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A280-01A-12D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A280-01A-12D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A282-01A-12D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A282-01A-12D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A285-01A-11D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A285-01A-11D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A288-01A-11D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A288-01A-11D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28A-01A-21D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28A-01A-21D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28C-01A-11D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28C-01A-11D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28E-01A-11D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28E-01A-11D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28F-01A-11D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28F-01A-11D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28G-01A-11D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28G-01A-11D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28H-01A-11D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28H-01A-11D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28K-01A-21D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28K-01A-21D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DM-A28M-01A-12D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DM-A28M-01A-12D-A16U-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DT-5265-01A-21D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DT-5265-01A-21D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DY-A0XA-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DY-A0XA-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DY-A1DC-01A-31D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DY-A1DC-01A-31D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DY-A1DD-01A-21D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DY-A1DD-01A-21D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DY-A1DE-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DY-A1DE-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DY-A1DF-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DY-A1DF-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DY-A1DG-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DY-A1DG-01A-11D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-DY-A1H8-01A-21D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-DY-A1H8-01A-21D-A151-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EF-5830-01A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EF-5830-01A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EF-5831-01A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EF-5831-01A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6506-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6506-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6507-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6507-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6508-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6508-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6509-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6509-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6510-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6510-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6511-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6511-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6512-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6512-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6513-01A-21D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6513-01A-21D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6514-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6514-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6881-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6881-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6882-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6882-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6883-01A-31D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6883-01A-31D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6884-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6884-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6885-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6885-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-6917-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-6917-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-7002-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-7002-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-EI-7004-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-EI-7004-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6459-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6459-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6460-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6460-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6461-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6461-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6463-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6463-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6569-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6569-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6570-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6570-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6703-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6703-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6704-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6704-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6805-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6805-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6806-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6806-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6807-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6807-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6808-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6808-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6809-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6809-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6854-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6854-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6855-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6855-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6856-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6856-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6464-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6464-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6465-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6465-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6571-01A-12D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6571-01A-12D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6702-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6702-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6810-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6810-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6811-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6811-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6812-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6812-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6813-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6813-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6814-01A-31D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6814-01A-31D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6861-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6861-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6863-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6863-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F5-6864-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F5-6864-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6293-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6293-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6294-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6294-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6295-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6295-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6297-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6297-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6298-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6298-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6299-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6299-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6302-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6302-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6303-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6303-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6304-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6304-01A-11D-1923-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6306-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6306-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6307-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6307-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6309-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6309-01A-21D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6310-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6310-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6311-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6311-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6314-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6314-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6315-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6315-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6317-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6317-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6320-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6320-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6321-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6321-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6322-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6322-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6323-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6323-01A-11D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6586-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6586-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6588-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6588-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6625-01A-21D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6625-01A-21D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6626-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6626-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6627-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6627-01A-11D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6628-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6628-01A-11D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G5-6233-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G5-6233-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G5-6235-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G5-6235-01A-11D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G5-6572-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G5-6572-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G5-6641-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G5-6641-01A-11D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A50T-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A50T-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A50U-01A-33D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A50U-01A-33D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A50V-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A50V-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A5IV-01A-42D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A5IV-01A-42D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A6GA-01A-11D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A6GA-01A-11D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A6GB-01A-11D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A6GB-01A-11D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A6GC-01A-12D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A6GC-01A-12D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A8F7-01A-11D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A8F7-01A-11D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-NH-A8F8-01A-72D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A8F8-01A-72D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-QG-A5YV-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-QG-A5YV-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-QG-A5YW-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-QG-A5YW-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-QG-A5YX-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-QG-A5YX-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-QG-A5Z1-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-QG-A5Z1-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-QG-A5Z2-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-QG-A5Z2-01A-11D-A28F-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-QL-A97D-01A-12D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-QL-A97D-01A-12D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-RU-A8FL-01A-11D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-RU-A8FL-01A-11D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-SS-A7HO-01A-21D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-SS-A7HO-01A-21D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-T9-A92H-01A-11D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-T9-A92H-01A-11D-A36W-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-WS-AB45-01A-11D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
Table S21.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-3L-AA1B-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-3L-AA1B-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-3L-AA1B-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-4N-A93T-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-4N-A93T-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-4N-A93T-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-4T-AA8H-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-4T-AA8H-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-4T-AA8H-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-5M-AAT4-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-5M-AAT4-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-5M-AAT4-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-5M-AAT5-01A-21R-A41B-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-5M-AAT5-01A-21R-A41B-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-5M-AAT5-01A-21R-A41B-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-5M-AAT6-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-5M-AAT6-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-5M-AAT6-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-5M-AATA-01A-31R-A41B-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-5M-AATA-01A-31R-A41B-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-5M-AATA-01A-31R-A41B-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-5M-AATE-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-5M-AATE-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-5M-AATE-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2671-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2671-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2671-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2672-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2672-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2672-01A-01R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2674-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2674-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2674-01A-02R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2674-01A-02R-A278-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2674-01A-02R-A278-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2674-01A-02R-A278-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2675-01A-02R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2675-01A-02R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2675-01A-02R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2676-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2676-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2676-01A-01R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2677-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2677-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2677-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2678-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2678-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2678-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2679-01A-02R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2679-01A-02R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2679-01A-02R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2680-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2680-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2680-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2681-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2681-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2681-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2682-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2682-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2682-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2683-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2683-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2683-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2684-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2684-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2684-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2684-01A-01R-A278-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2684-01A-01R-A278-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2684-01A-01R-A278-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2685-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2685-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2685-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2686-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2686-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2686-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-3807-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-3807-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-3807-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-3808-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-3808-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-3808-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-3809-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-3809-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-3809-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-3809-01A-01R-A278-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-3809-01A-01R-A278-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-3809-01A-01R-A278-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-3810-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-3810-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-3810-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-3810-01A-01R-A278-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-3810-01A-01R-A278-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-3810-01A-01R-A278-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-4105-01A-02R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-4105-01A-02R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-4105-01A-02R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-4107-01A-02R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-4107-01A-02R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-4107-01A-02R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5656-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5656-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5656-01A-21R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5656-01A-21R-A278-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5656-01A-21R-A278-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5656-01A-21R-A278-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5657-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5657-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5657-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5659-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5659-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5659-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5659-01A-01R-A278-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5659-01A-01R-A278-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5659-01A-01R-A278-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5660-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5660-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5660-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5661-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5661-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5661-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5662-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5662-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5662-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5664-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5664-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5664-01A-21R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5665-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5665-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5665-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5666-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5666-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5666-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5667-01A-21R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5667-01A-21R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5667-01A-21R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6137-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6137-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6137-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6138-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6138-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6138-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6140-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6140-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6140-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6141-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6141-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6141-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6142-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6142-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6142-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6648-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6648-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6648-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6649-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6649-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6649-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6650-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6650-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6650-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6650-01A-11R-A278-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6650-01A-11R-A278-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6650-01A-11R-A278-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6651-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6651-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6651-01A-21R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6652-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6652-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6652-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6653-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6653-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6653-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6654-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6654-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6654-01A-21R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6780-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6780-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6780-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6780-01A-11R-A278-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6780-01A-11R-A278-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6780-01A-11R-A278-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6781-01A-22R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6781-01A-22R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6781-01A-22R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6781-01A-22R-A278-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6781-01A-22R-A278-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6781-01A-22R-A278-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-6782-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-6782-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-6782-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-A565-01A-31R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-A565-01A-31R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-A565-01A-31R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-A566-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-A566-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-A566-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-A567-01A-31R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-A567-01A-31R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-A567-01A-31R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-A56B-01A-31R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-A56B-01A-31R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-A56B-01A-31R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-A5ZU-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-A5ZU-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-A5ZU-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3488-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3488-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3488-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3489-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3489-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3489-01A-21R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3492-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3492-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3492-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3494-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3494-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3494-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3495-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3495-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3495-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3496-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3496-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3496-01A-21R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3502-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3502-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3502-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3506-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3506-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3506-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3509-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3509-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3509-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3510-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3510-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3510-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3511-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3511-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3511-01A-21R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3514-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3514-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3514-01A-02R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3516-01A-02R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3516-01A-02R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3516-01A-02R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3517-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3517-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3517-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3518-01A-02R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3518-01A-02R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3518-01A-02R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3519-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3519-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3519-01A-02R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3520-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3520-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3520-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3521-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3521-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3521-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3522-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3522-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3522-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3524-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3524-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3524-01A-02R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3525-01A-02R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3525-01A-02R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3525-01A-02R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3526-01A-02R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3526-01A-02R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3526-01A-02R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3527-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3527-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3527-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3529-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3529-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3529-01A-02R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3530-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3530-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3530-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3531-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3531-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3531-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3532-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3532-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3532-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3534-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3534-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3534-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3538-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3538-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3538-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3542-01A-02R-1873-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3542-01A-02R-1873-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3542-01A-02R-1873-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3543-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3543-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3543-01A-01R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3544-01A-01R-1873-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3544-01A-01R-1873-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3544-01A-01R-1873-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3548-01A-01R-1873-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3548-01A-01R-1873-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3548-01A-01R-1873-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3549-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3549-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3549-01A-02R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3552-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3552-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3552-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3553-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3553-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3553-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3554-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3554-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3554-01A-01R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3555-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3555-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3555-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3556-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3556-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3556-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3560-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3560-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3560-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3561-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3561-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3561-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3562-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3562-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3562-01A-02R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3655-01A-02R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3655-01A-02R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3655-01A-02R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3660-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3660-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3660-01A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3662-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3662-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3662-01A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3663-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3663-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3663-01A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3664-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3664-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3664-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3666-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3666-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3666-01A-02R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3667-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3667-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3667-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3672-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3672-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3672-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3673-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3673-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3673-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3675-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3675-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3675-01A-02R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3678-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3678-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3678-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3679-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3679-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3679-01A-02R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3680-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3680-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3680-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3681-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3681-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3681-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3684-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3684-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3684-01A-02R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3685-01A-02R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3685-01A-02R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3685-01A-02R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3688-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3688-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3688-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3692-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3692-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3692-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3693-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3693-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3693-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3696-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3696-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3696-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3697-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3697-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3697-01A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3710-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3710-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3710-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3712-01A-21R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3712-01A-21R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3712-01A-21R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3713-01A-21R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3713-01A-21R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3713-01A-21R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3715-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3715-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3715-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3811-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3811-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3811-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3812-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3812-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3812-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3814-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3814-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3814-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3815-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3815-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3815-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3818-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3818-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3818-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3819-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3819-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3819-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3821-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3821-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3821-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3831-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3831-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3831-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3833-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3833-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3833-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3837-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3837-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3837-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3841-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3841-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3841-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3842-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3842-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3842-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3844-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3844-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3844-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3845-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3845-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3845-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3846-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3846-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3846-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3848-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3848-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3848-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3850-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3850-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3850-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3851-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3851-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3851-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3852-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3852-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3852-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3854-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3854-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3854-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3855-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3855-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3855-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3856-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3856-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3856-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3858-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3858-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3858-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3860-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3860-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3860-01A-02R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3861-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3861-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3861-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3862-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3862-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3862-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3864-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3864-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3864-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3866-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3866-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3866-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3867-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3867-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3867-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3869-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3869-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3869-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3870-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3870-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3870-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3872-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3872-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3872-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3875-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3875-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3875-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3877-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3877-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3877-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3930-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3930-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3930-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3939-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3939-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3939-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3941-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3941-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3941-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3947-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3947-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3947-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3949-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3949-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3949-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3950-01A-02R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3950-01A-02R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3950-01A-02R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3952-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3952-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3952-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3955-01A-02R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3955-01A-02R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3955-01A-02R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3956-01A-02R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3956-01A-02R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3956-01A-02R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3966-01A-01R-1113-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3966-01A-01R-1113-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3966-01A-01R-1113-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3968-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3968-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3968-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3970-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3970-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3970-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3971-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3971-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3971-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3972-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3972-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3972-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3973-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3973-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3973-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3975-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3975-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3975-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3976-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3976-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3976-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3977-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3977-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3977-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3979-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3979-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3979-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3980-01A-02R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3980-01A-02R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3980-01A-02R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3982-01A-02R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3982-01A-02R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3982-01A-02R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3984-01A-02R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3984-01A-02R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3984-01A-02R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3986-01A-02R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3986-01A-02R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3986-01A-02R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3989-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3989-01A-01R-1022-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3989-01A-01R-1022-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3994-01A-01R-1113-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3994-01A-01R-1113-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3994-01A-01R-1113-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A004-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A004-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A004-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A00A-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A00A-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A00A-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A00D-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A00D-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A00D-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A00E-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A00E-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A00E-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A00F-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A00F-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A00F-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A00J-01A-02R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A00J-01A-02R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A00J-01A-02R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A00K-01A-02R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A00K-01A-02R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A00K-01A-02R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A00L-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A00L-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A00L-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A00N-01A-02R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A00N-01A-02R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A00N-01A-02R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A00O-01A-02R-A089-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A00O-01A-02R-A089-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A00O-01A-02R-A089-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A00Q-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A00Q-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A00Q-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A00R-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A00R-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A00R-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A00U-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A00U-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A00U-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A00W-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A00W-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A00W-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A00Z-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A00Z-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A00Z-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A010-01A-01R-A089-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A010-01A-01R-A089-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A010-01A-01R-A089-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A017-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A017-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A017-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A01C-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A01C-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A01C-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A01D-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A01D-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A01D-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A01F-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A01F-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A01F-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A01G-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A01G-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A01G-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A01I-01A-02R-A089-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A01I-01A-02R-A089-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A01I-01A-02R-A089-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A01K-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A01K-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A01K-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A01P-01A-21R-A083-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A01P-01A-21R-A083-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A01P-01A-21R-A083-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A01Q-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A01Q-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A01Q-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A01R-01A-21R-A083-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A01R-01A-21R-A083-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A01R-01A-21R-A083-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A01S-01A-21R-A083-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A01S-01A-21R-A083-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A01S-01A-21R-A083-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A01T-01A-21R-A16W-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A01T-01A-21R-A16W-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A01T-01A-21R-A16W-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A01V-01A-23R-A083-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A01V-01A-23R-A083-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A01V-01A-23R-A083-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A01X-01A-21R-A083-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A01X-01A-21R-A083-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A01X-01A-21R-A083-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A01Z-01A-11R-A083-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A01Z-01A-11R-A083-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A01Z-01A-11R-A083-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A022-01A-21R-A16W-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A022-01A-21R-A16W-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A022-01A-21R-A16W-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A024-01A-02R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A024-01A-02R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A024-01A-02R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A029-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A029-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A029-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A02E-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A02E-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A02E-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A02F-01A-01R-A089-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A02F-01A-01R-A089-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A02F-01A-01R-A089-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A02H-01A-01R-A089-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A02H-01A-01R-A089-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A02H-01A-01R-A089-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A02J-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A02J-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A02J-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A02K-01A-03R-A32Y-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A02K-01A-03R-A32Y-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A02K-01A-03R-A32Y-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A02O-01A-21R-A16W-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A02O-01A-21R-A16W-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A02O-01A-21R-A16W-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A02R-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A02R-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A02R-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A02W-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A02W-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A02W-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A02Y-01A-43R-A32Y-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A02Y-01A-43R-A32Y-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A02Y-01A-43R-A32Y-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A03F-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A03F-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A03F-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-A03J-01A-21R-A16W-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-A03J-01A-21R-A16W-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-A03J-01A-21R-A16W-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AD-5900-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AD-5900-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AD-5900-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AD-6548-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AD-6548-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AD-6548-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AD-6888-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AD-6888-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AD-6888-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AD-6889-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AD-6889-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AD-6889-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AD-6890-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AD-6890-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AD-6890-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AD-6895-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AD-6895-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AD-6895-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AD-6899-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AD-6899-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AD-6899-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AD-6901-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AD-6901-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AD-6901-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AD-6963-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AD-6963-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AD-6963-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AD-6964-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AD-6964-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AD-6964-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AD-6965-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AD-6965-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AD-6965-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AD-A5EJ-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AD-A5EJ-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AD-A5EJ-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AD-A5EK-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AD-A5EK-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AD-A5EK-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-2687-01A-02R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-2687-01A-02R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-2687-01A-02R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-2690-01A-02R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-2690-01A-02R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-2690-01A-02R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-2691-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-2691-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-2691-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-2692-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-2692-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-2692-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-2693-01A-02R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-2693-01A-02R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-2693-01A-02R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-3400-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-3400-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-3400-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-3911-01A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-3911-01A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-3911-01A-01R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-3913-01A-02R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-3913-01A-02R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-3913-01A-02R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-4110-01A-02R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-4110-01A-02R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-4110-01A-02R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-5654-01A-01R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-5654-01A-01R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-5654-01A-01R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-6136-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-6136-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-6136-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-6655-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-6655-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-6655-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-6672-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-6672-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-6672-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-A56K-01A-32R-A39D-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-A56K-01A-32R-A39D-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-A56K-01A-32R-A39D-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-A56L-01A-31R-A39D-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-A56L-01A-31R-A39D-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-A56L-01A-31R-A39D-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-A56N-01A-12R-A39D-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-A56N-01A-12R-A39D-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-A56N-01A-12R-A39D-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3574-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3574-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3574-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3575-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3575-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3575-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3578-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3578-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3578-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3580-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3580-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3580-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3581-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3581-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3581-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3582-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3582-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3582-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3583-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3583-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3583-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3584-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3584-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3584-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3586-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3586-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3586-01A-02R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3587-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3587-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3587-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3591-01A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3591-01A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3591-01A-01R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3592-01A-02R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3592-01A-02R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3592-01A-02R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3593-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3593-01A-01R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3593-01A-01R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3594-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3594-01A-02R-0821-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3594-01A-02R-0821-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3598-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3598-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3598-01A-01R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3599-01A-02R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3599-01A-02R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3599-01A-02R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3600-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3600-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3600-01A-01R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3601-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3601-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3601-01A-01R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3602-01A-02R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3602-01A-02R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3602-01A-02R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3605-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3605-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3605-01A-01R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3608-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3608-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3608-01A-01R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3609-01A-02R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3609-01A-02R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3609-01A-02R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3611-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3611-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3611-01A-01R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3612-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3612-01A-01R-0826-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3612-01A-01R-0826-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3725-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3725-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3725-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3726-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3726-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3726-01A-02R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3727-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3727-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3727-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3728-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3728-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3728-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3731-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3731-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3731-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3732-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3732-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3732-01A-11R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3742-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3742-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3742-01A-11R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3878-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3878-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3878-01A-02R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3881-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3881-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3881-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3882-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3882-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3882-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3883-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3883-01A-02R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3883-01A-02R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3885-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3885-01A-01R-0905-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3885-01A-01R-0905-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3887-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3887-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3887-01A-01R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3890-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3890-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3890-01A-01R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3892-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3892-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3892-01A-01R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3893-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3893-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3893-01A-01R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3894-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3894-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3894-01A-01R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3896-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3896-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3896-01A-01R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3898-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3898-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3898-01A-01R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3901-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3901-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3901-01A-01R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3902-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3902-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3902-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3909-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3909-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3909-01A-01R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3999-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3999-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3999-01A-01R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-4001-01A-02R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-4001-01A-02R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-4001-01A-02R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-4005-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-4005-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-4005-01A-01R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-4007-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-4007-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-4007-01A-01R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-4008-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-4008-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-4008-01A-01R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-4015-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-4015-01A-01R-1119-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-4015-01A-01R-1119-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-4021-01A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-4021-01A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-4021-01A-01R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-4022-01A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-4022-01A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-4022-01A-01R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A002-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A002-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A002-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A008-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A008-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A008-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A00C-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A00C-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A00C-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A00H-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A00H-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A00H-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A00Y-01A-02R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A00Y-01A-02R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A00Y-01A-02R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A011-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A011-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A011-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A014-01A-02R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A014-01A-02R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A014-01A-02R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A015-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A015-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A015-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A016-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A016-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A016-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A01J-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A01J-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A01J-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A01L-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A01L-01A-01R-A002-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A01L-01A-01R-A002-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A01N-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A01N-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A01N-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A01W-01A-21R-A083-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A01W-01A-21R-A083-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A01W-01A-21R-A083-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A01Y-01A-41R-A083-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A01Y-01A-41R-A083-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A01Y-01A-41R-A083-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A020-01A-21R-A083-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A020-01A-21R-A083-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A020-01A-21R-A083-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A023-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A023-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A023-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A025-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A025-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A025-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A026-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A026-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A026-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A02G-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A02G-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A02G-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A02N-01A-11R-A083-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A02N-01A-11R-A083-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A02N-01A-11R-A083-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A02X-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A02X-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A02X-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A032-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A032-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A032-01A-01R-A00A-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-A036-01A-12R-A083-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-A036-01A-12R-A083-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-A036-01A-12R-A083-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AH-6544-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AH-6544-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AH-6544-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AH-6547-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AH-6547-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AH-6547-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AH-6549-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AH-6549-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AH-6549-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AH-6643-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AH-6643-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AH-6643-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AH-6644-01A-21R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AH-6644-01A-21R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AH-6644-01A-21R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AH-6897-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AH-6897-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AH-6897-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AH-6903-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AH-6903-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AH-6903-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AM-5820-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AM-5820-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AM-5820-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AM-5821-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AM-5821-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AM-5821-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AU-3779-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AU-3779-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AU-3779-01A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AU-6004-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AU-6004-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AU-6004-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AY-4070-01A-01R-1113-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AY-4070-01A-01R-1113-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AY-4070-01A-01R-1113-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AY-4071-01A-01R-1113-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AY-4071-01A-01R-1113-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AY-4071-01A-01R-1113-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AY-5543-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AY-5543-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AY-5543-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AY-6196-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AY-6196-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AY-6196-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AY-6197-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AY-6197-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AY-6197-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AY-6386-01A-21R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AY-6386-01A-21R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AY-6386-01A-21R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AY-A54L-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AY-A54L-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AY-A54L-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AY-A69D-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AY-A69D-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AY-A69D-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AY-A71X-01A-12R-A37K-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AY-A71X-01A-12R-A37K-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AY-A71X-01A-12R-A37K-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AY-A8YK-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AY-A8YK-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AY-A8YK-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-4308-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-4308-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-4308-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-4313-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-4313-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-4313-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-4315-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-4315-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-4315-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-4323-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-4323-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-4323-01A-21R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-4614-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-4614-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-4614-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-4615-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-4615-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-4615-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-4616-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-4616-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-4616-01A-21R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-4682-01B-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-4682-01B-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-4682-01B-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-4684-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-4684-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-4684-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-5403-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-5403-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-5403-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-5407-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-5407-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-5407-01A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6598-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6598-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6598-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6599-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6599-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6599-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6600-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6600-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6600-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6601-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6601-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6601-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6603-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6603-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6603-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6605-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6605-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6605-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6606-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6606-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6606-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6607-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6607-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6607-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6608-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6608-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6608-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-BM-6198-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-BM-6198-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-BM-6198-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CA-5254-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CA-5254-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CA-5254-01A-21R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CA-5255-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CA-5255-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CA-5255-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CA-5256-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CA-5256-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CA-5256-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CA-5796-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CA-5796-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CA-5796-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CA-5797-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CA-5797-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CA-5797-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CA-6715-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CA-6715-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CA-6715-01A-21R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CA-6716-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CA-6716-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CA-6716-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CA-6717-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CA-6717-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CA-6717-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CA-6718-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CA-6718-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CA-6718-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CA-6719-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CA-6719-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CA-6719-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CI-6619-01B-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CI-6619-01B-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CI-6619-01B-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CI-6620-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CI-6620-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CI-6620-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CI-6621-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CI-6621-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CI-6621-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CI-6622-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CI-6622-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CI-6622-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CI-6623-01B-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CI-6623-01B-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CI-6623-01B-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CI-6624-01C-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CI-6624-01C-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CI-6624-01C-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CK-4947-01B-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CK-4947-01B-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CK-4947-01B-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CK-4948-01B-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CK-4948-01B-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CK-4948-01B-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CK-4950-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CK-4950-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CK-4950-01A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CK-4951-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CK-4951-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CK-4951-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CK-4952-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CK-4952-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CK-4952-01A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CK-5912-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CK-5912-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CK-5912-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CK-5913-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CK-5913-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CK-5913-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CK-5914-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CK-5914-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CK-5914-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CK-5915-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CK-5915-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CK-5915-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CK-5916-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CK-5916-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CK-5916-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CK-6746-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CK-6746-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CK-6746-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CK-6747-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CK-6747-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CK-6747-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CK-6748-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CK-6748-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CK-6748-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CK-6751-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CK-6751-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CK-6751-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CL-4957-01A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CL-4957-01A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CL-4957-01A-01R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CL-5917-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CL-5917-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CL-5917-01A-11R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CL-5918-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CL-5918-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CL-5918-01A-11R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-4743-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-4743-01A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-4743-01A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-4744-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-4744-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-4744-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-4746-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-4746-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-4746-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-4747-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-4747-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-4747-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-4748-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-4748-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-4748-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-4750-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-4750-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-4750-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-4751-01A-02R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-4751-01A-02R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-4751-01A-02R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-4752-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-4752-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-4752-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-5341-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-5341-01A-01R-1410-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-5341-01A-01R-1410-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-5344-01A-21R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-5344-01A-21R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-5344-01A-21R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-5348-01A-21R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-5348-01A-21R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-5348-01A-21R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-5349-01A-21R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-5349-01A-21R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-5349-01A-21R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-5860-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-5860-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-5860-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-5861-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-5861-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-5861-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-5862-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-5862-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-5862-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-5863-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-5863-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-5863-01A-21R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-5864-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-5864-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-5864-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-5868-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-5868-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-5868-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6161-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6161-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6161-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6162-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6162-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6162-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6163-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6163-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6163-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6164-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6164-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6164-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6165-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6165-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6165-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6166-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6166-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6166-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6167-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6167-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6167-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6168-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6168-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6168-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6169-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6169-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6169-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6170-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6170-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6170-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6171-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6171-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6171-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6172-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6172-01A-11R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6172-01A-11R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6674-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6674-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6674-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6675-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6675-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6675-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6676-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6676-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6676-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6677-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6677-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6677-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6678-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6678-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6678-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6679-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6679-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6679-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-CM-6680-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-CM-6680-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-CM-6680-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-5537-01A-21R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-5537-01A-21R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-5537-01A-21R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-5538-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-5538-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-5538-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-5539-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-5539-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-5539-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-5540-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-5540-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-5540-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-5541-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-5541-01A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-5541-01A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6529-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6529-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6529-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6530-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6530-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6530-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6531-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6531-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6531-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6532-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6532-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6532-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6533-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6533-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6533-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6534-01A-21R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6534-01A-21R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6534-01A-21R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6535-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6535-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6535-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6536-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6536-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6536-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6537-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6537-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6537-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6538-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6538-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6538-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6539-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6539-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6539-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6540-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6540-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6540-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6541-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6541-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6541-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6898-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6898-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6898-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6920-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6920-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6920-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6922-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6922-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6922-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6923-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6923-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6923-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6924-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6924-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6924-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6926-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6926-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6926-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6927-01A-21R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6927-01A-21R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6927-01A-21R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6928-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6928-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6928-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6929-01A-31R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6929-01A-31R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6929-01A-31R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6930-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6930-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6930-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6931-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6931-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6931-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-6932-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-6932-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-6932-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-D5-7000-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-D5-7000-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-D5-7000-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DC-4745-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DC-4745-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DC-4745-01A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DC-4749-01A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DC-4749-01A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DC-4749-01A-01R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DC-5337-01A-01R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DC-5337-01A-01R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DC-5337-01A-01R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DC-5869-01A-01R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DC-5869-01A-01R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DC-5869-01A-01R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DC-6154-01A-31R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DC-6154-01A-31R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DC-6154-01A-31R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DC-6155-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DC-6155-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DC-6155-01A-11R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DC-6156-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DC-6156-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DC-6156-01A-11R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DC-6157-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DC-6157-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DC-6157-01A-11R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DC-6158-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DC-6158-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DC-6158-01A-11R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DC-6160-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DC-6160-01A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DC-6160-01A-11R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DC-6681-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DC-6681-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DC-6681-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DC-6682-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DC-6682-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DC-6682-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DC-6683-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DC-6683-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DC-6683-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A0X9-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A0X9-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A0X9-01A-11R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A0XD-01A-12R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A0XD-01A-12R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A0XD-01A-12R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A0XF-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A0XF-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A0XF-01A-11R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A1D0-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A1D0-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A1D0-01A-11R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A1D4-01A-21R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A1D4-01A-21R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A1D4-01A-21R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A1D6-01A-21R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A1D6-01A-21R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A1D6-01A-21R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A1D7-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A1D7-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A1D7-01A-11R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A1D8-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A1D8-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A1D8-01A-11R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A1D9-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A1D9-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A1D9-01A-11R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A1DA-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A1DA-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A1DA-01A-11R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A1DB-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A1DB-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A1DB-01A-11R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A1HA-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A1HA-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A1HA-01A-11R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A1HB-01A-21R-A180-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A1HB-01A-21R-A180-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A1HB-01A-21R-A180-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A280-01A-12R-A16W-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A280-01A-12R-A16W-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A280-01A-12R-A16W-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A282-01A-12R-A16W-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A282-01A-12R-A16W-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A282-01A-12R-A16W-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A285-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A285-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A285-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A288-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A288-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A288-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A28A-01A-21R-A32Y-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A28A-01A-21R-A32Y-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A28A-01A-21R-A32Y-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A28C-01A-11R-A32Y-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A28C-01A-11R-A32Y-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A28C-01A-11R-A32Y-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A28E-01A-11R-A32Y-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A28E-01A-11R-A32Y-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A28E-01A-11R-A32Y-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A28F-01A-11R-A32Y-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A28F-01A-11R-A32Y-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A28F-01A-11R-A32Y-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A28G-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A28G-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A28G-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A28H-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A28H-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A28H-01A-11R-A16W-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A28K-01A-21R-A32Y-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A28K-01A-21R-A32Y-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A28K-01A-21R-A32Y-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DM-A28M-01A-12R-A16W-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DM-A28M-01A-12R-A16W-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DM-A28M-01A-12R-A16W-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DT-5265-01A-21R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DT-5265-01A-21R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DT-5265-01A-21R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DY-A0XA-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DY-A0XA-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DY-A0XA-01A-11R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DY-A1DC-01A-31R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DY-A1DC-01A-31R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DY-A1DC-01A-31R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DY-A1DD-01A-21R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DY-A1DD-01A-21R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DY-A1DD-01A-21R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DY-A1DE-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DY-A1DE-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DY-A1DE-01A-11R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DY-A1DF-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DY-A1DF-01A-11R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DY-A1DF-01A-11R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DY-A1DG-01A-11R-A32Y-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DY-A1DG-01A-11R-A32Y-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DY-A1DG-01A-11R-A32Y-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-DY-A1H8-01A-21R-A155-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-DY-A1H8-01A-21R-A155-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-DY-A1H8-01A-21R-A155-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EF-5830-01A-01R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EF-5830-01A-01R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EF-5830-01A-01R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EF-5831-01A-01R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EF-5831-01A-01R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EF-5831-01A-01R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6506-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6506-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6506-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6507-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6507-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6507-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6508-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6508-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6508-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6509-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6509-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6509-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6510-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6510-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6510-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6511-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6511-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6511-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6512-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6512-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6512-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6513-01A-21R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6513-01A-21R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6513-01A-21R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6514-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6514-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6514-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6881-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6881-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6881-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6882-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6882-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6882-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6883-01A-31R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6883-01A-31R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6883-01A-31R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6884-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6884-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6884-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6885-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6885-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6885-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-6917-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-6917-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-6917-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-7002-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-7002-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-7002-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-EI-7004-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-EI-7004-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-EI-7004-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6459-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6459-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6459-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6460-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6460-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6460-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6461-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6461-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6461-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6463-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6463-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6463-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6569-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6569-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6569-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6570-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6570-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6570-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6703-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6703-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6703-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6704-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6704-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6704-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6805-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6805-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6805-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6806-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6806-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6806-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6807-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6807-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6807-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6808-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6808-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6808-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6809-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6809-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6809-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6854-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6854-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6854-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6855-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6855-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6855-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6856-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6856-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6856-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F5-6464-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F5-6464-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F5-6464-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F5-6465-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F5-6465-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F5-6465-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F5-6571-01A-12R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F5-6571-01A-12R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F5-6571-01A-12R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F5-6702-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F5-6702-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F5-6702-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F5-6810-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F5-6810-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F5-6810-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F5-6811-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F5-6811-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F5-6811-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F5-6812-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F5-6812-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F5-6812-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F5-6813-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F5-6813-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F5-6813-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F5-6814-01A-31R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F5-6814-01A-31R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F5-6814-01A-31R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F5-6861-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F5-6861-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F5-6861-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F5-6863-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F5-6863-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F5-6863-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F5-6864-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F5-6864-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F5-6864-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6293-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6293-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6293-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6294-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6294-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6294-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6295-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6295-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6295-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6297-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6297-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6297-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6298-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6298-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6298-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6299-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6299-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6299-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6302-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6302-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6302-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6303-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6303-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6303-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6304-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6304-01A-11R-1928-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6304-01A-11R-1928-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6306-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6306-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6306-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6307-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6307-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6307-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6309-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6309-01A-21R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6309-01A-21R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6310-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6310-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6310-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6311-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6311-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6311-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6314-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6314-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6314-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6315-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6315-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6315-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6317-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6317-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6317-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6320-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6320-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6320-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6321-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6321-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6321-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6322-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6322-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6322-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6323-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6323-01A-11R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6323-01A-11R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6586-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6586-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6586-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6588-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6588-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6588-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6625-01A-21R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6625-01A-21R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6625-01A-21R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6626-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6626-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6626-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6627-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6627-01A-11R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6627-01A-11R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G4-6628-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G4-6628-01A-11R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G4-6628-01A-11R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G5-6233-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G5-6233-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G5-6233-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G5-6235-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G5-6235-01A-11R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G5-6235-01A-11R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G5-6572-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G5-6572-01A-11R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G5-6572-01A-11R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-G5-6641-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-G5-6641-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-G5-6641-01A-11R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-NH-A50T-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-NH-A50T-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-NH-A50T-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-NH-A50U-01A-33R-A37K-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-NH-A50U-01A-33R-A37K-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-NH-A50U-01A-33R-A37K-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-NH-A50V-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-NH-A50V-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-NH-A50V-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-NH-A5IV-01A-42R-A37K-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-NH-A5IV-01A-42R-A37K-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-NH-A5IV-01A-42R-A37K-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-NH-A6GA-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-NH-A6GA-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-NH-A6GA-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-NH-A6GB-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-NH-A6GB-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-NH-A6GB-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-NH-A6GC-01A-12R-A41B-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-NH-A6GC-01A-12R-A41B-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-NH-A6GC-01A-12R-A41B-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-NH-A8F7-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-NH-A8F7-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-NH-A8F7-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-NH-A8F8-01A-72R-A41B-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-NH-A8F8-01A-72R-A41B-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-NH-A8F8-01A-72R-A41B-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-QG-A5YV-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-QG-A5YV-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-QG-A5YV-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-QG-A5YW-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-QG-A5YW-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-QG-A5YW-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-QG-A5YX-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-QG-A5YX-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-QG-A5YX-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-QG-A5Z1-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-QG-A5Z1-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-QG-A5Z1-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-QG-A5Z2-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-QG-A5Z2-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-QG-A5Z2-01A-11R-A28H-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-QL-A97D-01A-12R-A41B-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-QL-A97D-01A-12R-A41B-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-QL-A97D-01A-12R-A41B-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-RU-A8FL-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-RU-A8FL-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-RU-A8FL-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-SS-A7HO-01A-21R-A37K-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-SS-A7HO-01A-21R-A37K-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-SS-A7HO-01A-21R-A37K-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-T9-A92H-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-T9-A92H-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-T9-A92H-01A-11R-A37K-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-WS-AB45-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-WS-AB45-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-WS-AB45-01A-11R-A41B-07 | (TODO) -- GDC | mRNA__counts__FPKM |
Table S22.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-3L-AA1B-01A-11H-A37P-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-3L-AA1B-01A-11H-A37P-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-4N-A93T-01A-11H-A37P-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-4N-A93T-01A-11H-A37P-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-4T-AA8H-01A-11H-A41D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-4T-AA8H-01A-11H-A41D-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-5M-AAT4-01A-11H-A41D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-5M-AAT4-01A-11H-A41D-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-5M-AAT5-01A-21H-A41D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-5M-AAT5-01A-21H-A41D-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-5M-AAT6-01A-11H-A41D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-5M-AAT6-01A-11H-A41D-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-5M-AATA-01A-31H-A41D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-5M-AATA-01A-31H-A41D-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-5M-AATE-01A-11H-A41D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-5M-AATE-01A-11H-A41D-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2671-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2671-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2672-01A-01T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2672-01A-01T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2674-01A-02T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2674-01A-02T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2675-01A-02T-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2675-01A-02T-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2676-01A-01T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2676-01A-01T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2677-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2677-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2678-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2678-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2679-01A-02T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2679-01A-02T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2680-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2680-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2681-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2681-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2682-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2682-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2683-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2683-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2684-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2684-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2685-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2685-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2686-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2686-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-3807-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-3807-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-3808-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-3808-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-3809-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-3809-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-3810-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-3810-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-4105-01A-02T-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-4105-01A-02T-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-4107-01A-02T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-4107-01A-02T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-5656-01A-21H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-5656-01A-21H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-5656-01A-21H-A279-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-5656-01A-21H-A279-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-5657-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-5657-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-5659-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-5659-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-5660-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-5660-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-5661-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-5661-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-5662-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-5662-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-5664-01A-21H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-5664-01A-21H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-5665-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-5665-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-5666-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-5666-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-5667-01A-21H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-5667-01A-21H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6137-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6137-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6138-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6138-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6140-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6140-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6141-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6141-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6142-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6142-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6648-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6648-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6649-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6649-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6650-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6650-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6650-01A-11H-A279-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6650-01A-11H-A279-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6651-01A-21H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6651-01A-21H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6652-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6652-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6653-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6653-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6654-01A-21H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6654-01A-21H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6780-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6780-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6780-01A-11H-A279-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6780-01A-11H-A279-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6781-01A-22H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6781-01A-22H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6781-01A-22H-A279-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6781-01A-22H-A279-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-6782-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-6782-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-A565-01A-31H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-A565-01A-31H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-A566-01A-11H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-A566-01A-11H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-A567-01A-31H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-A567-01A-31H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-A56B-01A-31H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-A56B-01A-31H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-A5ZU-01A-11H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-A5ZU-01A-11H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3488-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3488-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3489-01A-21H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3489-01A-21H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3492-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3492-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3494-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3494-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3495-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3495-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3496-01A-21H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3496-01A-21H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3502-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3502-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3506-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3506-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3509-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3509-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3510-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3510-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3511-01A-21H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3511-01A-21H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3514-01A-02T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3514-01A-02T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3516-01A-02T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3516-01A-02T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3517-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3517-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3518-01A-02T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3518-01A-02T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3519-01A-02T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3519-01A-02T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3520-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3520-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3521-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3521-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3522-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3522-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3524-01A-02T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3524-01A-02T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3525-01A-02T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3525-01A-02T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3526-01A-02T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3526-01A-02T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3527-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3527-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3529-01A-02T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3529-01A-02T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3530-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3530-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3531-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3531-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3532-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3532-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3534-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3534-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3538-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3538-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3542-01A-02T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3542-01A-02T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3543-01A-01T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3543-01A-01T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3544-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3544-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3548-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3548-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3549-01A-02T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3549-01A-02T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3552-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3552-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3553-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3553-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3554-01A-01T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3554-01A-01T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3555-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3555-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3556-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3556-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3558-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3558-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3560-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3560-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3561-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3561-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3562-01A-02T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3562-01A-02T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3655-01A-02T-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3655-01A-02T-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3660-01A-01T-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3660-01A-01T-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3662-01A-01T-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3662-01A-01T-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3663-01A-01T-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3663-01A-01T-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3664-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3664-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3666-01A-02T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3666-01A-02T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3667-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3667-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3672-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3672-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3673-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3673-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3675-01A-02T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3675-01A-02T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3678-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3678-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3679-01A-02T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3679-01A-02T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3680-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3680-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3681-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3681-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3684-01A-02T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3684-01A-02T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3685-01A-02T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3685-01A-02T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3688-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3688-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3692-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3692-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3693-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3693-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3695-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3695-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3696-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3696-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3697-01A-01T-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3697-01A-01T-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3710-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3710-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3712-01A-21H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3712-01A-21H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3713-01A-21H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3713-01A-21H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3715-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3715-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3811-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3811-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3812-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3812-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3814-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3814-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3815-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3815-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3818-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3818-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3819-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3819-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3821-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3821-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3831-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3831-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3833-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3833-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3837-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3837-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3842-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3842-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3844-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3844-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3845-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3845-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3846-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3846-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3848-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3848-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3850-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3850-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3851-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3851-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3852-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3852-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3854-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3854-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3855-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3855-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3856-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3856-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3858-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3858-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3860-01A-02T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3860-01A-02T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3861-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3861-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3862-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3862-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3864-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3864-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3866-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3866-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3867-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3867-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3869-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3869-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3870-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3870-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3872-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3872-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3875-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3875-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3877-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3877-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3930-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3930-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3939-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3939-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3941-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3941-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3947-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3947-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3949-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3949-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3950-01A-02T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3950-01A-02T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3952-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3952-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3955-01A-02T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3955-01A-02T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3956-01A-02T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3956-01A-02T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3966-01A-01T-1114-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3966-01A-01T-1114-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3968-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3968-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3970-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3970-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3971-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3972-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3972-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3973-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3973-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3975-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3975-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3976-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3976-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3977-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3977-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3979-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3979-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3980-01A-02T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3980-01A-02T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3982-01A-02T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3982-01A-02T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3984-01A-02T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3984-01A-02T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3986-01A-02T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3986-01A-02T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3989-01A-01T-1021-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3989-01A-01T-1021-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3994-01A-01T-1114-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3994-01A-01T-1114-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A00D-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A00D-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A00J-01A-31R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A00J-01A-31R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A00L-01A-31R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A00L-01A-31R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A00O-01A-11R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A00O-01A-11R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A00Q-01A-31R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A00Q-01A-31R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A00U-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A00U-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A00W-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A00W-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A010-01A-54R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A010-01A-54R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A01D-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A01D-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A01F-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A01F-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A01G-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A01G-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A01P-01A-21R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A01P-01A-21R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A01R-01A-21R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A01R-01A-21R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A01S-01A-21R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A01S-01A-21R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A01T-01A-21R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A01T-01A-21R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A01V-01A-23R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A01V-01A-23R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A01X-01A-21R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A01X-01A-21R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A01Z-01A-11R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A01Z-01A-11R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A022-01A-21R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A022-01A-21R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A024-01A-11R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A024-01A-11R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A029-01A-22R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A029-01A-22R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A02E-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A02E-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A02F-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A02F-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A02H-01A-31R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A02H-01A-31R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A02J-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A02J-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A02K-01A-21R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A02K-01A-21R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A02O-01A-21R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A02O-01A-21R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A02R-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A02R-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A02Y-01A-43R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A02Y-01A-43R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A03F-01A-11R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A03F-01A-11R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-A03J-01A-21R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-A03J-01A-21R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AD-5900-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AD-5900-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AD-6548-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AD-6548-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AD-6888-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AD-6888-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AD-6889-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AD-6889-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AD-6890-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AD-6890-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AD-6895-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AD-6895-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AD-6899-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AD-6899-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AD-6901-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AD-6901-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AD-6963-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AD-6963-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AD-6964-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AD-6964-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AD-6965-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AD-6965-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AD-A5EJ-01A-11H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AD-A5EJ-01A-11H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AD-A5EK-01A-11H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AD-A5EK-01A-11H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-2687-01A-02T-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-2687-01A-02T-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-2689-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-2689-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-2690-01A-02T-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-2690-01A-02T-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-2691-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-2691-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-2692-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-2692-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-2693-01A-02T-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-2693-01A-02T-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-3400-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-3400-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-3911-01A-01T-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-3911-01A-01T-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-3913-01A-02T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-3913-01A-02T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-4110-01A-02T-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-4110-01A-02T-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-5654-01A-01T-1659-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-5654-01A-01T-1659-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-6136-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-6136-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-6655-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-6655-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-6672-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-6672-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-A56K-01A-32H-A39B-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-A56K-01A-32H-A39B-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-A56L-01A-31H-A39B-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-A56L-01A-31H-A39B-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-A56N-01A-12H-A39B-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-A56N-01A-12H-A39B-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3574-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3574-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3575-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3575-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3578-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3578-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3580-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3580-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3581-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3581-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3582-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3582-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3583-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3583-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3584-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3584-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3586-01A-02T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3586-01A-02T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3587-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3587-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3591-01A-01T-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3591-01A-01T-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3592-01A-02T-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3592-01A-02T-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3593-01A-01T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3593-01A-01T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3594-01A-02T-0822-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3594-01A-02T-0822-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3598-01A-01T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3598-01A-01T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3599-01A-02T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3599-01A-02T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3600-01A-01T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3600-01A-01T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3601-01A-01T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3601-01A-01T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3602-01A-02T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3602-01A-02T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3605-01A-01T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3605-01A-01T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3608-01A-01T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3608-01A-01T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3609-01A-02T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3609-01A-02T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3611-01A-01T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3611-01A-01T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3612-01A-01T-0827-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3612-01A-01T-0827-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3725-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3725-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3726-01A-02T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3726-01A-02T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3727-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3727-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3728-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3728-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3731-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3731-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3732-01A-11H-1659-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3732-01A-11H-1659-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3742-01A-11H-1659-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3742-01A-11H-1659-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3878-01A-02T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3878-01A-02T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3881-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3881-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3882-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3882-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3883-01A-02T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3883-01A-02T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3885-01A-01T-0906-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3885-01A-01T-0906-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3887-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3887-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3890-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3890-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3892-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3892-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3893-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3893-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3894-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3894-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3896-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3896-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3898-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3898-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3901-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3901-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3902-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3902-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3909-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3909-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-3999-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-3999-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-4001-01A-02T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-4001-01A-02T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-4005-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-4005-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-4007-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-4007-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-4008-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-4008-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-4015-01A-01T-1120-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-4015-01A-01T-1120-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-4021-01A-01T-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-4021-01A-01T-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-4022-01A-01T-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-4022-01A-01T-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A002-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A002-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A008-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A008-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A00C-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A00C-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A011-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A011-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A015-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A015-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A016-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A016-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A01L-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A01L-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A01N-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A01N-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A01W-01A-21R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A01W-01A-21R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A01Y-01A-41R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A01Y-01A-41R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A020-01A-21R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A020-01A-21R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A025-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A025-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A026-01A-32R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A026-01A-32R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A02G-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A02G-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A02N-01A-11R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A02N-01A-11R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A02X-01A-21R-A076-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A02X-01A-21R-A076-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AG-A036-01A-12R-A082-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AG-A036-01A-12R-A082-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AH-6544-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AH-6544-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AH-6547-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AH-6547-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AH-6549-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AH-6549-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AH-6643-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AH-6643-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AH-6644-01A-21H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AH-6644-01A-21H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AH-6897-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AH-6897-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AH-6903-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AH-6903-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AM-5820-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AM-5820-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AM-5821-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AM-5821-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AU-3779-01A-01T-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AU-3779-01A-01T-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AU-6004-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AU-6004-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AY-4070-01A-01T-1114-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AY-4070-01A-01T-1114-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AY-4071-01A-01T-1114-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AY-4071-01A-01T-1114-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AY-5543-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AY-5543-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AY-6196-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AY-6196-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AY-6197-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AY-6197-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AY-6386-01A-21H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AY-6386-01A-21H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AY-A54L-01A-11H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AY-A54L-01A-11H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AY-A69D-01A-11H-A37P-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AY-A69D-01A-11H-A37P-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AY-A71X-01A-12H-A37P-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AY-A71X-01A-12H-A37P-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AY-A8YK-01A-11H-A41D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AY-A8YK-01A-11H-A41D-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-4308-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-4308-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-4313-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-4313-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-4315-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-4315-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-4323-01A-21H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-4323-01A-21H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-4614-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-4614-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-4615-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-4615-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-4616-01A-21H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-4616-01A-21H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-4681-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-4681-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-4682-01B-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-4682-01B-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-4684-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-4684-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-5403-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-5403-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-5407-01A-01T-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-5407-01A-01T-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-6598-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-6598-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-6599-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-6599-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-6600-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-6600-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-6601-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-6601-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-6603-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-6603-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-6605-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-6605-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-6606-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-6606-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-6607-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-6607-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AZ-6608-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AZ-6608-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-BM-6198-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-BM-6198-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CA-5254-01A-21H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CA-5254-01A-21H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CA-5255-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CA-5255-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CA-5256-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CA-5256-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CA-5796-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CA-5796-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CA-5797-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CA-5797-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CA-6715-01A-21H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CA-6715-01A-21H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CA-6716-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CA-6716-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CA-6717-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CA-6717-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CA-6718-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CA-6718-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CA-6719-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CA-6719-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CI-6619-01B-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CI-6619-01B-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CI-6620-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CI-6620-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CI-6621-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CI-6621-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CI-6622-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CI-6622-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CI-6623-01B-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CI-6623-01B-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CI-6624-01C-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CI-6624-01C-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CK-4947-01B-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CK-4947-01B-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CK-4948-01B-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CK-4948-01B-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CK-4950-01A-01T-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CK-4950-01A-01T-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CK-4951-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CK-4951-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CK-4952-01A-01T-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CK-4952-01A-01T-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CK-5912-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CK-5912-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CK-5913-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CK-5913-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CK-5914-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CK-5914-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CK-5915-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CK-5915-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CK-5916-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CK-5916-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CK-6746-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CK-6746-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CK-6747-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CK-6747-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CK-6748-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CK-6748-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CK-6751-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CK-6751-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CL-4957-01A-01T-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CL-4957-01A-01T-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CL-5917-01A-11H-1659-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CL-5917-01A-11H-1659-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CL-5918-01A-11H-1659-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CL-5918-01A-11H-1659-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-4743-01A-01T-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-4743-01A-01T-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-4744-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-4744-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-4746-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-4746-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-4747-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-4747-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-4748-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-4748-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-4750-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-4750-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-4751-01A-02H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-4751-01A-02H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-4752-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-4752-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-5341-01A-01T-1409-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-5341-01A-01T-1409-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-5344-01A-21H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-5344-01A-21H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-5348-01A-21H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-5348-01A-21H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-5349-01A-21H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-5349-01A-21H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-5860-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-5860-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-5861-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-5861-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-5862-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-5862-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-5863-01A-21H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-5863-01A-21H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-5864-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-5864-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-5868-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-5868-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6161-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6161-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6162-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6162-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6163-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6163-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6164-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6164-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6165-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6165-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6166-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6166-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6167-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6167-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6168-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6168-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6169-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6169-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6170-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6170-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6171-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6171-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6172-01A-11H-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6172-01A-11H-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6674-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6674-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6675-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6675-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6676-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6676-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6677-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6677-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6678-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6678-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6679-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6679-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-CM-6680-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-CM-6680-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-5537-01A-21H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-5537-01A-21H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-5538-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-5538-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-5539-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-5539-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-5540-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-5540-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-5541-01A-01T-1652-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-5541-01A-01T-1652-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6529-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6529-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6530-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6530-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6531-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6531-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6532-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6532-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6533-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6533-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6534-01A-21H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6534-01A-21H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6535-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6535-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6536-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6536-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6537-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6537-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6538-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6538-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6539-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6539-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6540-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6540-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6541-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6541-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6898-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6898-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6920-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6920-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6922-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6922-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6923-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6923-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6924-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6924-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6926-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6926-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6927-01A-21H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6927-01A-21H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6928-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6928-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6929-01A-31H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6929-01A-31H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6930-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6930-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6931-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6931-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-6932-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-6932-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-D5-7000-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-D5-7000-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DC-4745-01A-01T-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DC-4745-01A-01T-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DC-4749-01A-01T-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DC-4749-01A-01T-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DC-5337-01A-01T-1659-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DC-5337-01A-01T-1659-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DC-5869-01A-01T-1659-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DC-5869-01A-01T-1659-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DC-6154-01A-31H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DC-6155-01A-11H-1659-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DC-6155-01A-11H-1659-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DC-6156-01A-11H-1659-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DC-6156-01A-11H-1659-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DC-6157-01A-11H-1659-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DC-6157-01A-11H-1659-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DC-6158-01A-11H-1659-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DC-6158-01A-11H-1659-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DC-6160-01A-11H-1659-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DC-6160-01A-11H-1659-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DC-6681-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DC-6681-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DC-6682-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DC-6682-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DC-6683-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DC-6683-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A0X9-01A-11H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A0X9-01A-11H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A0XD-01A-12H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A0XD-01A-12H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A0XF-01A-11H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A0XF-01A-11H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A1D0-01A-11H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A1D0-01A-11H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A1D4-01A-21H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A1D4-01A-21H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A1D6-01A-21H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A1D6-01A-21H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A1D7-01A-11H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A1D7-01A-11H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A1D8-01A-11H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A1D8-01A-11H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A1D9-01A-11H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A1D9-01A-11H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A1DA-01A-11H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A1DA-01A-11H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A1DB-01A-11H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A1DB-01A-11H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A1HA-01A-11H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A1HA-01A-11H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A1HB-01A-21H-A17X-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A1HB-01A-21H-A17X-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A280-01A-12H-A16S-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A280-01A-12H-A16S-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A282-01A-12H-A16S-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A282-01A-12H-A16S-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A285-01A-11H-A16S-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A285-01A-11H-A16S-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A288-01A-11H-A16S-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A288-01A-11H-A16S-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A28A-01A-21H-A16S-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A28A-01A-21H-A16S-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A28C-01A-11H-A16S-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A28C-01A-11H-A16S-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A28E-01A-11H-A16S-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A28E-01A-11H-A16S-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A28F-01A-11H-A16S-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A28F-01A-11H-A16S-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A28G-01A-11H-A16S-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A28G-01A-11H-A16S-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A28H-01A-11H-A16S-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A28H-01A-11H-A16S-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A28K-01A-21H-A16S-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A28K-01A-21H-A16S-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DM-A28M-01A-12H-A16S-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DM-A28M-01A-12H-A16S-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DT-5265-01A-21H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DT-5265-01A-21H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DY-A0XA-01A-11H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DY-A0XA-01A-11H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DY-A1DC-01A-31H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DY-A1DC-01A-31H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DY-A1DD-01A-21H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DY-A1DD-01A-21H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DY-A1DE-01A-11H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DY-A1DE-01A-11H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DY-A1DF-01A-11H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DY-A1DF-01A-11H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DY-A1DG-01A-11H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DY-A1DG-01A-11H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-DY-A1H8-01A-21H-A154-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-DY-A1H8-01A-21H-A154-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EF-5830-01A-01T-1659-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EF-5830-01A-01T-1659-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EF-5831-01A-01T-1659-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EF-5831-01A-01T-1659-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6506-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6506-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6507-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6507-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6508-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6508-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6509-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6509-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6510-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6510-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6511-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6511-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6512-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6512-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6513-01A-21H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6513-01A-21H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6514-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6514-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6881-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6881-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6882-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6882-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6883-01A-31H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6883-01A-31H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6884-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6884-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6885-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6885-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-6917-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-6917-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-7002-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-7002-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-EI-7004-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-EI-7004-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6459-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6459-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6460-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6460-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6461-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6461-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6463-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6463-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6569-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6569-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6570-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6570-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6703-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6703-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6704-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6704-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6805-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6805-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6806-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6806-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6807-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6807-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6808-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6808-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6809-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6809-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6854-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6854-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6855-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6855-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F4-6856-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F4-6856-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F5-6464-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F5-6464-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F5-6465-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F5-6465-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F5-6571-01A-12H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F5-6571-01A-12H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F5-6702-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F5-6702-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F5-6810-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F5-6810-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F5-6811-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F5-6811-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F5-6812-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F5-6812-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F5-6813-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F5-6813-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F5-6814-01A-31H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F5-6814-01A-31H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F5-6861-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F5-6861-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F5-6863-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F5-6863-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-F5-6864-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-F5-6864-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6293-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6293-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6294-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6294-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6295-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6295-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6297-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6297-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6298-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6298-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6299-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6299-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6302-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6302-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6303-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6303-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6304-01A-11H-1927-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6304-01A-11H-1927-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6306-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6306-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6307-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6307-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6309-01A-21H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6309-01A-21H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6310-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6310-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6311-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6311-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6314-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6314-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6315-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6315-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6317-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6317-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6320-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6320-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6321-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6321-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6322-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6322-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6323-01A-11H-1722-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6323-01A-11H-1722-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6586-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6586-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6588-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6588-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6625-01A-21H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6625-01A-21H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6626-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6626-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6627-01A-11H-1773-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6627-01A-11H-1773-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G4-6628-01A-11H-1838-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G4-6628-01A-11H-1838-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G5-6233-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G5-6233-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G5-6235-01A-11H-1735-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G5-6235-01A-11H-1735-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G5-6572-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G5-6572-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-G5-6641-01A-11H-1829-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-G5-6641-01A-11H-1829-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-NH-A50T-01A-11H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-NH-A50T-01A-11H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-NH-A50U-01A-33H-A37P-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-NH-A50U-01A-33H-A37P-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-NH-A50V-01A-11H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-NH-A50V-01A-11H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-NH-A5IV-01A-42H-A37P-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-NH-A5IV-01A-42H-A37P-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-NH-A6GA-01A-11H-A37P-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-NH-A6GA-01A-11H-A37P-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-NH-A6GB-01A-11H-A37P-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-NH-A6GB-01A-11H-A37P-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-NH-A6GC-01A-12H-A41D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-NH-A6GC-01A-12H-A41D-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-NH-A8F7-01A-11H-A41D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-NH-A8F7-01A-11H-A41D-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-NH-A8F8-01A-72H-A41D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-NH-A8F8-01A-72H-A41D-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-QG-A5YV-01A-11H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-QG-A5YV-01A-11H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-QG-A5YW-01A-11H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-QG-A5YW-01A-11H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-QG-A5YX-01A-11H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-QG-A5YX-01A-11H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-QG-A5Z1-01A-11H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-QG-A5Z1-01A-11H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-QG-A5Z2-01A-11H-A28I-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-QG-A5Z2-01A-11H-A28I-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-QL-A97D-01A-12H-A41D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-QL-A97D-01A-12H-A41D-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-RU-A8FL-01A-11H-A37P-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-RU-A8FL-01A-11H-A37P-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-SS-A7HO-01A-21H-A37P-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-SS-A7HO-01A-21H-A37P-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-T9-A92H-01A-11H-A37P-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-T9-A92H-01A-11H-A37P-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-WS-AB45-01A-11H-A41D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-WS-AB45-01A-11H-A41D-13 | (TODO) -- GDC | miR__isoformExp |
Table S23.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-3L-AA1B-01A-11D-A36X-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-4N-A93T-01A-11D-A36X-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-4T-AA8H-01A-11D-A40P-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-5M-AAT4-01A-11D-A40P-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-5M-AAT5-01A-21D-A40P-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-5M-AAT6-01A-11D-A40P-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-5M-AATA-01A-31D-A40P-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-5M-AATE-01A-11D-A40P-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2671-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2674-01A-02D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2674-01A-02D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2675-01A-02D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2677-01A-01D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2678-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2679-01A-02D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2680-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2681-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2682-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2683-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2684-01A-01D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2685-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-2686-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-3807-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-3808-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-3809-01A-01D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-3810-01A-01D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-4105-01A-02D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-4107-01A-02D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5656-01A-21D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5656-01A-21D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5657-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5659-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5659-01A-01D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5660-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5661-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5662-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5664-01A-21D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5665-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5666-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-5667-01A-21D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6137-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6138-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6140-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6141-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6142-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6648-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6649-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6650-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6650-01A-11D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6651-01A-21D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6652-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6653-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6654-01A-21D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6780-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6780-01A-11D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6781-01A-22D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6781-01A-22D-A270-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-6782-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-A565-01A-31D-A28G-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-A566-01A-11D-A28G-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-A567-01A-31D-A28G-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-A6-A56B-01A-31D-A28G-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3488-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3489-01A-21D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3492-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3494-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3495-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3496-01A-21D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3502-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3506-01A-01D-2188-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3509-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3510-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3511-01A-21D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3514-01A-02D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3517-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3519-01A-02D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3520-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3521-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3522-01A-01W-0831-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3524-01A-02D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3526-01A-02D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3527-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3529-01A-02D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3530-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3531-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3532-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3534-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3538-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3544-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3548-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3549-01A-02D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3556-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3558-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3560-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3561-01A-01D-1953-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3655-01A-02D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3660-01A-01D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3662-01A-01D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3663-01A-01D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3664-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3666-01A-02W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3667-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3672-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3673-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3675-01A-02W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3678-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3679-01A-02W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3680-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3681-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3684-01A-02W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3685-01A-02W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3688-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3692-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3693-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3695-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3696-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3697-01A-01D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3710-01A-01D-1981-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3712-01A-21D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3713-01A-21D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3715-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3811-01A-01D-1981-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3811-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3812-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3814-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3815-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3818-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3819-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3821-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3831-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3833-01A-01D-1981-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3833-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3837-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3841-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3842-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3844-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3845-01A-01D-1981-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3845-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3846-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3848-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3850-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3851-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3852-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3854-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3855-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3856-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3858-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3860-01A-02W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3861-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3862-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3864-01A-01D-1981-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3866-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3867-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3869-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3870-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3872-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3875-01A-01W-0900-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3877-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3930-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3939-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3941-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3947-01A-01D-1981-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3949-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3950-01A-02W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3952-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3955-01A-02W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3956-01A-02W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3966-01A-01D-1981-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3966-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3967-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3968-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3971-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3972-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3973-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3975-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3976-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3977-01A-01D-1981-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3977-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3979-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3980-01A-02W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3982-01A-02W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3984-01A-02D-1981-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3984-01A-02W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3986-01A-02W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3989-01A-01W-0995-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-3994-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A004-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A00N-01A-02D-A17O-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A00N-01A-02W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A00O-01A-02W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A00Z-01A-01D-A17O-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A010-01A-01D-A17O-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A010-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A017-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A01C-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A01D-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A01I-01A-02W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A01K-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A01P-01A-21D-A17O-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A01P-01A-21W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A01R-01A-21D-A17O-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A01R-01A-21W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A01S-01A-21W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A01T-01A-21W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A01V-01A-23W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A01X-01A-21W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A01Z-01A-11W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A022-01A-21D-A17O-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A022-01A-21W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A024-01A-02W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A029-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A02E-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A02F-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A02H-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A02J-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A02K-01A-21W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A02O-01A-21W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A02R-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A02W-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A02Y-01A-43W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A03F-01A-11W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AA-A03J-01A-21W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AD-5900-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AD-6548-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AD-6888-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AD-6889-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AD-6890-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AD-6895-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AD-6899-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AD-6901-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AD-6963-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AD-6964-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AD-6965-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AD-A5EJ-01A-11D-A28G-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AD-A5EK-01A-11D-A28G-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-2687-01A-02D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-2689-01A-01D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-2690-01A-02D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-2691-01A-01D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-2692-01A-01D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-2693-01A-02D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-3400-01A-01D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-3911-01A-01D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-3913-01A-02W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-3914-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-4110-01A-02D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-5654-01A-01D-1657-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-6136-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-6655-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-6672-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-A56K-01A-32D-A38X-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-A56L-01A-31D-A38X-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AF-A56N-01A-12D-A38X-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3574-01A-01D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3575-01A-01D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3580-01A-01D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3581-01A-01D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3583-01A-01D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3584-01A-01D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3591-01A-01D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3592-01A-02D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3594-01A-02D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3598-01A-01D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3599-01A-02D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3602-01A-02D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3605-01A-01D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3725-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3726-01A-02W-0899-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3727-01A-01W-0899-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3728-01A-01W-0899-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3731-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3732-01A-11D-1657-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3742-01A-11D-1657-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3878-01A-02W-0899-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3881-01A-01W-0899-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3882-01A-01W-0899-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3883-01A-02W-0899-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3885-01A-01W-0899-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3887-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3890-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3891-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3892-01A-01D-1989-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3892-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3893-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3894-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3896-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3898-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3901-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3902-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3906-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3909-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-3999-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-4001-01A-02W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-4005-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-4007-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-4008-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-4009-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-4015-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-4021-01A-01D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-4022-01A-01D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A002-01A-01W-A00K-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A008-01A-01D-A183-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A008-01A-01W-A00K-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A00C-01A-01D-A183-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A00C-01A-01W-A00K-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A00H-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A00Y-01A-02W-A00K-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A011-01A-01W-A00K-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A014-01A-02W-A00K-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A015-01A-01D-A183-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A015-01A-01W-A00K-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A016-01A-01W-A00K-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A01J-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A01L-01A-01W-A00K-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A01N-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A01W-01A-21W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A01Y-01A-41W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A020-01A-21W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A023-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A025-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A026-01A-32W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A02G-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A02N-01A-11D-A183-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A02N-01A-11W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A02X-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A032-01A-01W-A00E-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AG-A036-01A-12W-A096-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AH-6544-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AH-6547-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AH-6549-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AH-6643-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AH-6644-01A-21D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AH-6897-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AH-6903-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AM-5820-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AM-5821-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AU-3779-01A-01D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AU-6004-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AY-4070-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AY-4071-01A-01W-1073-09 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AY-5543-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AY-6196-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AY-6197-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AY-6386-01A-21D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AY-A54L-01A-11D-A28G-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AY-A69D-01A-11D-A36X-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AY-A71X-01A-12D-A36X-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AY-A8YK-01A-11D-A40P-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-4308-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-4313-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-4315-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-4323-01A-21D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-4614-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-4615-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-4616-01A-21D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-4681-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-4682-01B-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-4684-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-5403-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-5407-01A-01D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-6598-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-6599-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-6600-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-6601-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-6603-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-6605-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-6606-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-6607-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-AZ-6608-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-BM-6198-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CA-5254-01A-21D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CA-5255-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CA-5256-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CA-5796-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CA-5797-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CA-6715-01A-21D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CA-6716-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CA-6717-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CA-6718-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CA-6719-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CI-6619-01B-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CI-6620-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CI-6621-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CI-6622-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CI-6623-01B-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CI-6624-01C-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CK-4947-01B-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CK-4948-01B-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CK-4950-01A-01D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CK-4951-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CK-4952-01A-01D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CK-5912-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CK-5913-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CK-5914-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CK-5915-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CK-5916-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CK-6746-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CK-6747-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CK-6748-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CK-6751-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CL-4957-01A-01D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CL-5917-01A-11D-1657-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CL-5918-01A-11D-1657-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-4743-01A-01D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-4744-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-4746-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-4747-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-4748-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-4750-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-4751-01A-02D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-4752-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-5341-01A-01D-1408-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-5344-01A-21D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-5348-01A-21D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-5349-01A-21D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-5860-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-5861-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-5862-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-5863-01A-21D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-5864-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-5868-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6161-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6162-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6163-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6164-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6165-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6166-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6167-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6168-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6169-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6170-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6171-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6172-01A-11D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6674-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6675-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6676-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6677-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6678-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6679-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-CM-6680-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-5537-01A-21D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-5538-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-5539-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-5540-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-5541-01A-01D-1650-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6529-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6530-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6531-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6532-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6533-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6534-01A-21D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6535-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6536-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6537-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6538-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6539-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6540-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6541-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6898-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6920-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6922-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6923-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6924-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6926-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6927-01A-21D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6928-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6929-01A-31D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6930-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6931-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-6932-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-D5-7000-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DC-4745-01A-01D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DC-4749-01A-01D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DC-5337-01A-01D-1657-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DC-5869-01A-01D-1657-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DC-6154-01A-31D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DC-6155-01A-11D-1657-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DC-6157-01A-11D-1657-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DC-6158-01A-11D-1657-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DC-6160-01A-11D-1657-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DC-6681-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DC-6682-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DC-6683-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A0X9-01A-11D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A0XD-01A-12D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A0XF-01A-11D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A1D0-01A-11D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A1D4-01A-21D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A1D6-01A-21D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A1D7-01A-11D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A1D8-01A-11D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A1D9-01A-11D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A1DA-01A-11D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A1DB-01A-11D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A1HA-01A-11D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A1HB-01A-21D-A183-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A280-01A-12D-A16V-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A282-01A-12D-A16V-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A285-01A-11D-A16V-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A288-01A-11D-A16V-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A28A-01A-21D-A16V-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A28C-01A-11D-A16V-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A28E-01A-11D-A16V-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A28F-01A-11D-A16V-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A28G-01A-11D-A16V-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A28H-01A-11D-A16V-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A28K-01A-21D-A16V-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DM-A28M-01A-12D-A16V-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DT-5265-01A-21D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DY-A0XA-01A-11D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DY-A1DC-01A-31D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DY-A1DD-01A-21D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DY-A1DF-01A-11D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DY-A1DG-01A-11D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-DY-A1H8-01A-21D-A152-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EF-5830-01A-01D-1657-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6506-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6507-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6508-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6509-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6510-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6511-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6512-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6513-01A-21D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6514-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6881-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6882-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6883-01A-31D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6884-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6885-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-6917-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-7002-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-EI-7004-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6459-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6460-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6461-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6463-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6569-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6570-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6703-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6704-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6805-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6806-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6807-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6808-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6809-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6854-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6855-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F4-6856-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F5-6464-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F5-6465-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F5-6571-01A-12D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F5-6702-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F5-6810-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F5-6811-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F5-6812-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F5-6813-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F5-6814-01A-31D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F5-6861-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F5-6863-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-F5-6864-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6293-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6294-01A-11D-1806-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6295-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6297-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6298-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6299-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6302-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6303-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6304-01A-11D-1924-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6306-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6307-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6309-01A-21D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6310-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6311-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6314-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6315-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6317-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6320-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6321-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6322-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6323-01A-11D-1719-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6586-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6588-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6625-01A-21D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6626-01A-11D-1771-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6627-01A-11D-2188-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G4-6628-01A-11D-1835-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G5-6233-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G5-6235-01A-11D-1733-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G5-6572-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-G5-6641-01A-11D-1826-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-NH-A50T-01A-11D-A28G-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-NH-A50U-01A-33D-A36X-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-NH-A50V-01A-11D-A28G-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-NH-A5IV-01A-42D-A36X-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-NH-A6GA-01A-11D-A36X-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-NH-A6GB-01A-11D-A36X-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-NH-A6GC-01A-12D-A40P-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-NH-A8F7-01A-11D-A40P-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-NH-A8F8-01A-72D-A40P-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-QG-A5YV-01A-11D-A28G-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-QG-A5YW-01A-11D-A28G-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-QG-A5YX-01A-11D-A28G-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-QG-A5Z1-01A-11D-A28G-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-QG-A5Z2-01A-11D-A28G-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-QL-A97D-01A-12D-A40P-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-RU-A8FL-01A-11D-A36X-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-SS-A7HO-01A-21D-A36X-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-T9-A92H-01A-11D-A36X-10 | (TODO) -- GDC | SNV__mutect | |
| TCGA-WS-AB45-01A-11D-A40P-10 | (TODO) -- GDC | SNV__mutect |
Table S24.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-3L-AA1B-01A-11D-A36Y-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-4N-A93T-01A-11D-A36Y-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-4T-AA8H-01A-11D-A40X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-5M-AAT4-01A-11D-A40X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-5M-AAT5-01A-21D-A40X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-5M-AAT6-01A-11D-A40X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-5M-AATA-01A-31D-A40X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-5M-AATE-01A-11D-A40X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2670-01A-02D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-2671-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2672-01A-01D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-2674-01A-02D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-2675-01A-02D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2676-01A-01D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-2677-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-2677-01A-01D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2678-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-2679-01A-02D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2680-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2681-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2682-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2683-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-2684-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2684-01A-01D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2685-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2686-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-3807-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-3808-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-3809-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-3809-01A-01D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-3810-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-3810-01A-01D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-4105-01A-02D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-4107-01A-02D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5656-01A-21D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5656-01A-21D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5657-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5659-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5659-01A-01D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5660-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5661-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5662-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5664-01A-21D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5665-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5666-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5667-01A-21D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6137-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6138-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6140-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6141-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6142-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6648-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6649-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6650-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6650-01A-11D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6651-01A-21D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6652-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6653-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6654-01A-21D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6780-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6780-01A-11D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6781-01A-22D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6781-01A-22D-A27A-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-6782-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-A565-01A-31D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-A566-01A-11D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-A567-01A-31D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-A56B-01A-31D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-A5ZU-01A-11D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3488-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3489-01A-21D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3492-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3494-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3495-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3496-01A-21D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3502-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3506-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3509-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3510-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3511-01A-21D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3514-01A-02D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3516-01A-02D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3517-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3518-01A-02D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3519-01A-02D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3520-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3521-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3522-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3524-01A-02D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3525-01A-02D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3526-01A-02D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3527-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3529-01A-02D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3530-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3531-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3532-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3534-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3538-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3542-01A-02D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3543-01A-01D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3544-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3548-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3549-01A-02D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3552-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3553-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3554-01A-01D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3555-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3556-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3558-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3560-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3561-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3562-01A-02D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3655-01A-02D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3660-01A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3662-01A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3663-01A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3664-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3666-01A-02D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3667-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3672-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3673-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3678-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3679-01A-02D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3680-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3681-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3684-01A-02D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3685-01A-02D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3688-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3692-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3693-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3695-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3696-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3697-01A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3710-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3712-01A-21D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3713-01A-21D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3715-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3811-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3812-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3814-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3815-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3818-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3819-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3821-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3831-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3833-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3837-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3841-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3842-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3844-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3845-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3846-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3848-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3850-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3851-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3852-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3854-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3855-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3856-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3858-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3860-01A-02D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3861-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3862-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3864-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3866-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3867-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3869-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3870-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3872-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3875-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3877-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3930-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3939-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3941-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3947-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3949-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3950-01A-02D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3952-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3955-01A-02D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3956-01A-02D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3966-01A-01D-1110-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3968-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3971-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3972-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3973-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3975-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3976-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3977-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3979-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3980-01A-02D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3982-01A-02D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3984-01A-02D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3986-01A-02D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3989-01A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3994-01A-01D-1110-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A004-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A00A-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A00D-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A00E-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A00F-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A00J-01A-02D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A00K-01A-02D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A00L-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A00N-01A-02D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A00O-01A-02D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A00Q-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A00R-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A00U-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A00W-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A00Z-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A010-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A017-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01C-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01D-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01F-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01G-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01I-01A-02D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01K-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01P-01A-21D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01Q-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01R-01A-21D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01S-01A-21D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01T-01A-21D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01V-01A-23D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01X-01A-21D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01Z-01A-11D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A022-01A-21D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A024-01A-02D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A029-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A02E-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A02F-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A02H-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A02J-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A02K-01A-21D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A02O-01A-21D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A02R-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A02W-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A02Y-01A-43D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A03F-01A-11D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A03J-01A-21D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AD-5900-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AD-6548-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AD-6888-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AD-6889-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AD-6890-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AD-6895-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AD-6899-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AD-6901-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AD-6963-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AD-6964-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AD-6965-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AD-A5EJ-01A-11D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AD-A5EK-01A-11D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AF-2687-01A-02D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AF-2689-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AF-2690-01A-02D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AF-2691-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AF-2692-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AF-2693-01A-02D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AF-3400-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AF-3911-01A-01D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AF-3913-01A-02D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AF-4110-01A-02D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AF-6136-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AF-6655-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AF-6672-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AF-A56K-01A-32D-A39G-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AF-A56L-01A-31D-A39G-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AF-A56N-01A-12D-A39G-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-3574-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3575-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3578-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3580-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3581-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3582-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3583-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3584-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3586-01A-02D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3587-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3591-01A-01D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-3592-01A-02D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-3593-01A-01D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3594-01A-02D-0820-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3598-01A-01D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3599-01A-02D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3600-01A-01D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3601-01A-01D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3602-01A-02D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3605-01A-01D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3608-01A-01D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3609-01A-02D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3611-01A-01D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3612-01A-01D-0825-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3725-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-3726-01A-02D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3727-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3731-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-3732-01A-11D-1658-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-3742-01A-11D-1658-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-3878-01A-02D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3881-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3882-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3883-01A-02D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3885-01A-01D-0904-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3887-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3890-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3892-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3893-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3894-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3896-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3898-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3901-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3902-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3909-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3999-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-4001-01A-02D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-4005-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-4007-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-4008-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-4015-01A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-4021-01A-01D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-4022-01A-01D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-A002-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A008-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A00C-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A00H-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A00Y-01A-02D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A011-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A014-01A-02D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A015-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A016-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A01J-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A01L-01A-01D-A004-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A01N-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A01W-01A-21D-A081-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-A01Y-01A-41D-A081-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-A020-01A-21D-A081-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-A023-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A025-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A026-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A026-01A-32D-A081-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-A02G-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A02N-01A-11D-A081-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-A02X-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A032-01A-01D-A00B-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-A036-01A-12D-A081-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AH-6544-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AH-6547-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AH-6549-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AH-6643-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AH-6644-01A-21D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AH-6897-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AH-6903-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AM-5820-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AM-5821-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AU-3779-01A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AU-6004-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AY-4070-01A-01D-1110-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AY-4071-01A-01D-1110-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AY-5543-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AY-6196-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AY-6197-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AY-6386-01A-21D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AY-A54L-01A-11D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AY-A69D-01A-11D-A36Y-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AY-A71X-01A-12D-A36Y-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AY-A8YK-01A-11D-A40X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-4308-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-4313-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-4315-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-4323-01A-21D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-4614-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-4615-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-4616-01A-21D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-4681-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-4682-01B-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-4684-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-5403-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-5407-01A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-6598-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-6599-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-6600-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-6601-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-6603-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-6605-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-6606-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-6607-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-6608-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-BM-6198-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CA-5254-01A-21D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CA-5255-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CA-5256-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CA-5796-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CA-5797-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CA-6715-01A-21D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CA-6716-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CA-6717-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CA-6718-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CA-6719-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CI-6619-01B-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CI-6620-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CI-6621-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CI-6622-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CI-6623-01B-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CI-6624-01C-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CK-4947-01B-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CK-4948-01B-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CK-4950-01A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CK-4951-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CK-4952-01A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CK-5912-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CK-5913-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CK-5914-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CK-5915-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CK-5916-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CK-6746-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CK-6747-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CK-6748-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CK-6751-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CL-4957-01A-01D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CL-5917-01A-11D-1658-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CL-5918-01A-11D-1658-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-4743-01A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-4744-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-4746-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-4747-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-4748-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-4750-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-4751-01A-02D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-4752-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-5341-01A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-5344-01A-21D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-5348-01A-21D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-5349-01A-21D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-5860-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-5861-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-5862-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-5863-01A-21D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-5864-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-5868-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6161-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6162-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6163-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6164-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6165-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6166-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6167-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6168-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6169-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6170-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6171-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6172-01A-11D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6674-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6675-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6676-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6677-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6678-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6679-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-CM-6680-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-5537-01A-21D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-5538-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-5539-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-5540-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-5541-01A-01D-1651-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6529-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6530-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6531-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6532-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6533-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6534-01A-21D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6535-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6536-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6537-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6538-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6539-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6540-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6541-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6898-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6920-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6922-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6923-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6924-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6926-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6927-01A-21D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6928-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6929-01A-31D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6930-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6931-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-6932-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-D5-7000-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DC-4745-01A-01D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DC-4749-01A-01D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DC-5337-01A-01D-1658-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DC-5869-01A-01D-1658-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DC-6154-01A-31D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DC-6155-01A-11D-1658-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DC-6156-01A-11D-1658-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DC-6157-01A-11D-1658-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DC-6158-01A-11D-1658-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DC-6160-01A-11D-1658-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DC-6681-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DC-6682-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DC-6683-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A0X9-01A-11D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A0XD-01A-12D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A0XF-01A-11D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A1D0-01A-11D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A1D4-01A-21D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A1D6-01A-21D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A1D7-01A-11D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A1D8-01A-11D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A1D9-01A-11D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A1DA-01A-11D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A1DB-01A-11D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A1HA-01A-11D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A1HB-01A-22D-A17Z-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A280-01A-12D-A16X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A282-01A-12D-A16X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A285-01A-11D-A16X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A288-01A-11D-A16X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A28A-01A-21D-A16X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A28C-01A-11D-A16X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A28E-01A-11D-A16X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A28F-01A-11D-A16X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A28G-01A-11D-A16X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A28H-01A-11D-A16X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A28K-01A-21D-A16X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DM-A28M-01A-12D-A16X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DT-5265-01A-21D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DY-A0XA-01A-11D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DY-A1DC-01A-31D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DY-A1DD-01A-21D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DY-A1DE-01A-11D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DY-A1DF-01A-11D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DY-A1DG-01A-11D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-DY-A1H8-01A-21D-A153-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EF-5830-01A-01D-1658-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EF-5831-01A-01D-1658-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6506-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6507-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6508-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6509-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6510-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6511-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6512-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6513-01A-21D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6514-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6881-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6882-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6883-01A-31D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6884-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6885-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-6917-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-7002-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-EI-7004-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6459-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6460-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6461-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6463-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6569-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6570-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6703-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6704-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6805-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6806-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6807-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6808-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6809-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6854-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6855-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F4-6856-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F5-6464-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F5-6465-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F5-6571-01A-12D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F5-6702-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F5-6810-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F5-6811-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F5-6812-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F5-6813-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F5-6814-01A-31D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F5-6861-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F5-6863-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-F5-6864-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6293-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6294-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6295-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6297-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6298-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6299-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6302-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6303-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6304-01A-11D-1926-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6306-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6307-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6309-01A-21D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6310-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6311-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6314-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6315-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6317-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6320-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6321-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6322-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6323-01A-11D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6586-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6588-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6625-01A-21D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6626-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6627-01A-11D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6628-01A-11D-1837-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G5-6233-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G5-6235-01A-11D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G5-6572-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G5-6641-01A-11D-1828-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-NH-A50T-01A-11D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-NH-A50U-01A-33D-A36Y-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-NH-A50V-01A-11D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-NH-A5IV-01A-42D-A36Y-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-NH-A6GA-01A-11D-A36Y-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-NH-A6GB-01A-11D-A36Y-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-NH-A6GC-01A-12D-A40X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-NH-A8F7-01A-11D-A40X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-NH-A8F8-01A-72D-A40X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-QG-A5YV-01A-11D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-QG-A5YW-01A-11D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-QG-A5YX-01A-11D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-QG-A5Z1-01A-11D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-QG-A5Z2-01A-11D-A28O-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-QL-A97D-01A-12D-A40X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-RU-A8FL-01A-11D-A36Y-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-SS-A7HO-01A-21D-A36Y-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-T9-A92H-01A-11D-A36Y-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-WS-AB45-01A-11D-A40X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
Table S25.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-NH-A8F7 | (TODO) -- GDC | clinical__biospecimen |
Table S26.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-NH-A8F7 | (TODO) -- GDC | clinical__primary |
Table S27.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-NH-A8F7-06A-31D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-NH-A8F7-06A-31D-A40O-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
Table S28.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-NH-A8F7-06A-31R-A41B-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-NH-A8F7-06A-31R-A41B-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-NH-A8F7-06A-31R-A41B-07 | (TODO) -- GDC | mRNA__counts__FPKM |
Table S29.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-NH-A8F7-06A-31H-A41D-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-NH-A8F7-06A-31H-A41D-13 | (TODO) -- GDC | miR__isoformExp |
Table S30.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-NH-A8F7-06A-31D-A40P-10 | (TODO) -- GDC | SNV__mutect |
Table S31.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-NH-A8F7-06A-31D-A40X-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
Table S32.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-A6-2671 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2672 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2674 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2675 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2676 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2677 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2678 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2679 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2680 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2681 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2682 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2683 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2684 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2685 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-2686 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-3807 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-3808 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-3809 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-3810 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-4107 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5657 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5659 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5660 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5661 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5662 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5665 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5666 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-A6-5667 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3488 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3489 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3492 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3494 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3495 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3496 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3502 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3506 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3509 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3510 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3511 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3514 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3516 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3517 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3518 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3519 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3520 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3521 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3522 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3524 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3525 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3526 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3527 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3529 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3530 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3531 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3532 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3534 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3655 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3660 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3662 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3663 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3697 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3712 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-3713 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01P | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01R | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01S | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01T | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01V | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01X | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A01Z | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A022 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A02O | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A03F | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AA-A03J | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2689 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2691 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-2692 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-3400 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-3913 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AF-5654 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3725 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3731 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3732 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-3742 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A01W | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A01Y | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A020 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A02N | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AG-A036 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AH-6547 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AH-6643 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6598 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6599 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6600 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6601 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6603 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6605 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6606 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6607 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-AZ-6608 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-F4-6704 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6295 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6297 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6298 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6302 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6311 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6314 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6320 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6322 | (TODO) -- GDC | clinical__biospecimen | |
| TCGA-G4-6625 | (TODO) -- GDC | clinical__biospecimen |
Table S33.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-A6-2671 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2672 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2674 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2675 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2676 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2677 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2678 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2679 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2680 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2681 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2682 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2683 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2684 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2685 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-2686 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-3807 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-3808 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-3809 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-3810 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-4107 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5657 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5659 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5660 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5661 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5662 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5665 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5666 | (TODO) -- GDC | clinical__primary | |
| TCGA-A6-5667 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3488 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3489 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3492 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3494 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3495 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3496 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3502 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3506 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3509 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3510 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3511 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3514 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3516 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3517 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3518 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3519 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3520 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3521 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3522 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3524 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3525 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3526 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3527 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3529 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3530 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3531 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3532 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3534 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3655 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3660 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3662 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3663 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3697 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3712 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-3713 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01P | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01R | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01S | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01T | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01V | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01X | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A01Z | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A022 | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A02O | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A03F | (TODO) -- GDC | clinical__primary | |
| TCGA-AA-A03J | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2689 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2691 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-2692 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-3400 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-3913 | (TODO) -- GDC | clinical__primary | |
| TCGA-AF-5654 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3725 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3731 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3732 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-3742 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A01W | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A01Y | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A020 | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A02N | (TODO) -- GDC | clinical__primary | |
| TCGA-AG-A036 | (TODO) -- GDC | clinical__primary | |
| TCGA-AH-6547 | (TODO) -- GDC | clinical__primary | |
| TCGA-AH-6643 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6598 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6599 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6600 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6601 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6603 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6605 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6606 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6607 | (TODO) -- GDC | clinical__primary | |
| TCGA-AZ-6608 | (TODO) -- GDC | clinical__primary | |
| TCGA-F4-6704 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6295 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6297 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6298 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6302 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6311 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6314 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6320 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6322 | (TODO) -- GDC | clinical__primary | |
| TCGA-G4-6625 | (TODO) -- GDC | clinical__primary |
Table S34.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-A6-2671-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2671-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2672-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2672-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2674-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2674-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2675-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2675-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2676-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2676-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2677-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2677-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2678-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2678-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2679-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2679-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2680-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2680-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2681-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2681-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2682-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2682-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2683-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2683-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2684-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2684-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2685-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2685-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-2686-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-2686-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-3807-11A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-3807-11A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-3808-11A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-3808-11A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-3810-11A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-3810-11A-01D-1018-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-4107-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-4107-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5657-11A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5657-11A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5660-11A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5660-11A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5661-11A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5661-11A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5662-11A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5662-11A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5665-11A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5665-11A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5666-11A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5666-11A-01D-1649-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-A6-5667-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-A6-5667-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3488-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3488-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3489-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3489-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3492-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3492-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3494-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3494-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3495-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3495-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3496-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3496-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3502-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3502-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3506-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3506-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3509-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3509-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3510-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3510-11A-01D-1406-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3511-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3511-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3514-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3514-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3516-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3516-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3517-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3517-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3518-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3518-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3519-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3519-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3520-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3520-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3521-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3521-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3522-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3522-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3524-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3524-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3525-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3525-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3526-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3526-11A-01D-1549-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3527-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3527-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3529-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3529-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3530-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3530-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3532-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3532-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3534-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3534-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3655-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3655-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3660-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3660-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3662-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3662-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3663-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3663-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3697-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3697-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3712-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3712-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-3713-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-3713-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01P-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01P-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01R-11A-12D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01R-11A-12D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01S-11A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01S-11A-21D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01T-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01T-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01V-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01V-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01X-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01X-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A01Z-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A01Z-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A022-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A022-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A02O-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A02O-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A03F-11A-12D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A03F-11A-12D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AA-A03J-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AA-A03J-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2689-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2689-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2691-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2691-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-2692-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-2692-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-3400-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-3400-11A-01D-1550-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-3913-11A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-3913-11A-01D-1428-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AF-5654-11A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AF-5654-11A-11D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3725-11A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3725-11A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3731-11A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3731-11A-01D-1732-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3732-11A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3732-11A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-3742-11A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-3742-11A-01D-1656-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A01W-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A01W-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A01Y-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A01Y-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A020-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A020-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A02N-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A02N-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AG-A036-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AG-A036-11A-11D-A080-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AH-6547-11A-02D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AH-6547-11A-02D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AH-6643-11A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AH-6643-11A-01D-1825-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6598-11A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6598-11A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6599-11A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6599-11A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6600-11A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6600-11A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6601-11A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6601-11A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6603-11A-02D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6603-11A-02D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6605-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6605-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6606-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6606-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6607-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6607-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-AZ-6608-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-AZ-6608-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-F4-6704-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-F4-6704-11A-01D-1834-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6295-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6295-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6297-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6297-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6298-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6298-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6302-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6302-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6311-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6311-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6314-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6314-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6320-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6320-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6322-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6322-11A-01D-1717-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
| TCGA-G4-6625-11A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__unfiltered__snp6 |
| TCGA-G4-6625-11A-01D-1770-01 | Affymetrix SNP 6.0 | (TODO) -- GDC | CNV__snp6 |
Table S35.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-A6-2671-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2671-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2671-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2675-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2675-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2675-11A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2678-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2678-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2678-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2679-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2679-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2679-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2680-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2680-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2680-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2682-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2682-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2682-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2683-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2683-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2683-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2684-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2684-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2684-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2685-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2685-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2685-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-2686-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-2686-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-2686-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5659-11A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5659-11A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5659-11A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5662-11A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5662-11A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5662-11A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5665-11A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5665-11A-01R-1653-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5665-11A-01R-1653-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-A6-5667-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-A6-5667-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-A6-5667-11A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3489-11A-01R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3489-11A-01R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3489-11A-01R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3496-11A-01R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3496-11A-01R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3496-11A-01R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3511-11A-01R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3511-11A-01R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3511-11A-01R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3514-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3514-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3514-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3516-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3516-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3516-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3517-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3517-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3517-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3518-11A-01R-1672-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3518-11A-01R-1672-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3518-11A-01R-1672-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3520-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3520-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3520-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3522-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3522-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3522-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3525-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3525-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3525-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3527-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3527-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3527-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3531-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3531-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3531-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3534-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3534-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3534-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3655-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3655-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3655-11A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3660-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3660-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3660-11A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3662-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3662-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3662-11A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3663-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3663-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3663-11A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3697-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3697-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3697-11A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3712-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3712-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3712-11A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AA-3713-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AA-3713-11A-01R-1723-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AA-3713-11A-01R-1723-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-2689-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-2689-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-2689-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-2691-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-2691-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-2691-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-2692-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-2692-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-2692-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-3400-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-3400-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-3400-11A-01R-A32Z-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AF-5654-11A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AF-5654-11A-11R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AF-5654-11A-11R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3725-11A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3725-11A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3725-11A-01R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3731-11A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3731-11A-01R-1736-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3731-11A-01R-1736-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3732-11A-01R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3732-11A-01R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3732-11A-01R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AG-3742-11A-01R-1660-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AG-3742-11A-01R-1660-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AG-3742-11A-01R-1660-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AH-6643-11A-01R-1830-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AH-6643-11A-01R-1830-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AH-6643-11A-01R-1830-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6598-11A-01R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6598-11A-01R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6598-11A-01R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6599-11A-01R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6599-11A-01R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6599-11A-01R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6600-11A-01R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6600-11A-01R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6600-11A-01R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6601-11A-01R-1774-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6601-11A-01R-1774-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6601-11A-01R-1774-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6603-11A-02R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6603-11A-02R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6603-11A-02R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-AZ-6605-11A-01R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-AZ-6605-11A-01R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-AZ-6605-11A-01R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM | |
| TCGA-F4-6704-11A-01R-1839-07 | (TODO) -- GDC | mRNA__geneExpNormed__FPKM | |
| TCGA-F4-6704-11A-01R-1839-07 | (TODO) -- GDC | mRNA__geneExp__FPKM | |
| TCGA-F4-6704-11A-01R-1839-07 | (TODO) -- GDC | mRNA__counts__FPKM |
Table S36.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-A6-2671-11A-01R-1757-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2671-11A-01R-1757-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2680-11A-01R-1757-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2680-11A-01R-1757-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2683-11A-01R-1757-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2683-11A-01R-1757-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2684-11A-01R-1757-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2684-11A-01R-1757-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-A6-2685-11A-01R-1757-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-A6-2685-11A-01R-1757-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3520-11A-01R-1757-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3520-11A-01R-1757-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3525-11A-01R-1757-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3525-11A-01R-1757-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AA-3527-11A-01R-1757-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AA-3527-11A-01R-1757-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-2689-11A-01R-1757-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-2689-11A-01R-1757-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-2691-11A-01R-1757-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-2691-11A-01R-1757-13 | (TODO) -- GDC | miR__isoformExp | |
| TCGA-AF-2692-11A-01R-1757-13 | (TODO) -- GDC | miR__geneExp | |
| TCGA-AF-2692-11A-01R-1757-13 | (TODO) -- GDC | miR__isoformExp |
Table S37.
| TCGA Barcode | Platform | Center | Annotation |
|---|---|---|---|
| TCGA-A6-2671-11A-01D-1551-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2672-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-2674-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-2675-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2676-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-2677-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-2678-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-2679-11A-01D-1551-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2680-11A-01D-1551-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2681-11A-01D-1551-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2682-11A-01D-1551-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2683-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-2684-11A-01D-1551-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2685-11A-01D-1551-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-2686-11A-01D-1551-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-3807-11A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-3808-11A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-3809-11A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-3810-11A-01D-1020-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-A6-4107-11A-01D-1551-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-A6-5667-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3488-11A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3492-11A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3494-11A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3495-11A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3502-11A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3506-11A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3509-11A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3510-11A-01D-1407-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3514-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3516-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3517-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3518-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3519-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3520-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3521-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3522-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3524-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3525-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3526-11A-01D-1551-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3527-11A-01D-1552-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3529-11A-01D-1552-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3531-11A-01D-1552-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3532-11A-01D-1552-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3534-11A-01D-1552-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-3655-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3660-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3663-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3697-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3712-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-3713-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AA-A01P-11A-11D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01R-11A-12D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01S-11A-21D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01T-11A-11D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01V-11A-11D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01X-11A-11D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A01Z-11A-11D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A022-11A-11D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A02O-11A-11D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A03F-11A-12D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AA-A03J-11A-11D-A081-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AF-2689-11A-01D-1552-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AF-2691-11A-01D-1552-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AF-2692-11A-01D-1552-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AF-3400-11A-01D-1552-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AF-3913-11A-01D-1116-05 | Illumina Human Methylation 27 | (TODO) -- GDC | methylation__HM27 |
| TCGA-AG-3725-11A-01D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-3731-11A-01D-1734-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-A01W-11A-11D-A081-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-A01Y-11A-11D-A081-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-A020-11A-11D-A081-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-A02N-11A-11D-A081-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AG-A036-11A-11D-A081-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-6598-11A-01D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-6599-11A-01D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-6600-11A-01D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-AZ-6601-11A-01D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6295-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6297-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6298-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6302-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6311-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6314-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6320-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6322-11A-01D-1721-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
| TCGA-G4-6625-11A-01D-1772-05 | Illumina Human Methylation 450 | (TODO) -- GDC | methylation__HM450 |
The sample type short letter codes in the table above are defined in the following list.
-
TP: Primary Tumor
-
TR: Recurrent Tumor
-
TB: Primary Blood Derived Cancer - Peripheral Blood
-
TRBM: Recurrent Blood Derived Cancer - Bone Marrow
-
TAP: Additional - New Primary
-
TM: Metastatic
-
TAM: Additional Metastatic
-
THOC: Human Tumor Original Cells
-
TBM: Primary Blood Derived Cancer - Bone Marrow
-
NB: Blood Derived Normal
-
NT: Solid Tissue Normal
-
NBC: Buccal Cell Normal
-
NEBV: EBV Immortalized Normal
-
NBM: Bone Marrow Normal
-
CELLC: Control Analyte
-
TRB: Recurrent Blood Derived Cancer - Peripheral Blood
-
CELL: Cell Lines
-
XP: Primary Xenograft Tissue
-
XCL: Cell Line Derived Xenograft Tissue
Figure 1. Get High-res Image This figure depicts the distribution of available data on a per participant basis.