This pipeline attempts to aggregate clinical features and significant genomic events such as copy number alterations, significant mutation genes and molecular subtypes across ALL pipelines in the GDAC Firehose analysis workflow, into a single feature table. Figures of co-occurring and mutually-exclusive pairs for all copy number and mutation events are generated from this feature table using pairwise Fisher's exact test. For more details please visit the online documentation.
99 samples and 1161 features are included in this feature table. The figures below show which genomic pair events are co-occurring and which are mutually-exclusive.
Table 1. Get Full Table Part of feature table : 10 rows and 6 columns are showed below.
| participant_barcode | CLI_histological_type | CLI_clinical_stage | CLI_figo_grade | CLI_karnofsky_performance_score | CLI_msi |
|---|---|---|---|---|---|
| C3L-00006_TP | Endometrioid carcinoma | M0 | FIGO grade 1 | NA | MSI-L |
| C3L-00008_TP | Endometrioid carcinoma | M0 | FIGO grade 1 | NA | MSI-H |
| C3L-00032_TP | Endometrioid carcinoma | M0 | FIGO grade 2 | NA | NA |
| C3L-00090_TP | Endometrioid carcinoma | M0 | FIGO grade 2 | NA | NA |
| C3L-00098_TP | Endometrioid carcinoma | M0 | NA | NA | MSS |
| C3L-00136_TP | Endometrioid carcinoma | M0 | FIGO grade 1 | NA | MSS |
| C3L-00137_TP | Endometrioid carcinoma | M0 | FIGO grade 1 | NA | MSS |
| C3L-00139_TP | Serous carcinoma | M0 | NA | NA | MSS |
| C3L-00143_TP | Endometrioid carcinoma | M0 | FIGO grade 1 | NA | MSI-H |
| C3L-00145_TP | Endometrioid carcinoma | NA | FIGO grade 1 | NA | MSS |
Figure 1. Get High-res Image Co-occurring and mutually-exclusive pairs for copy number variations of arm level events. Blue means two events are co-occurring and red means two events are mutually-exclusive.
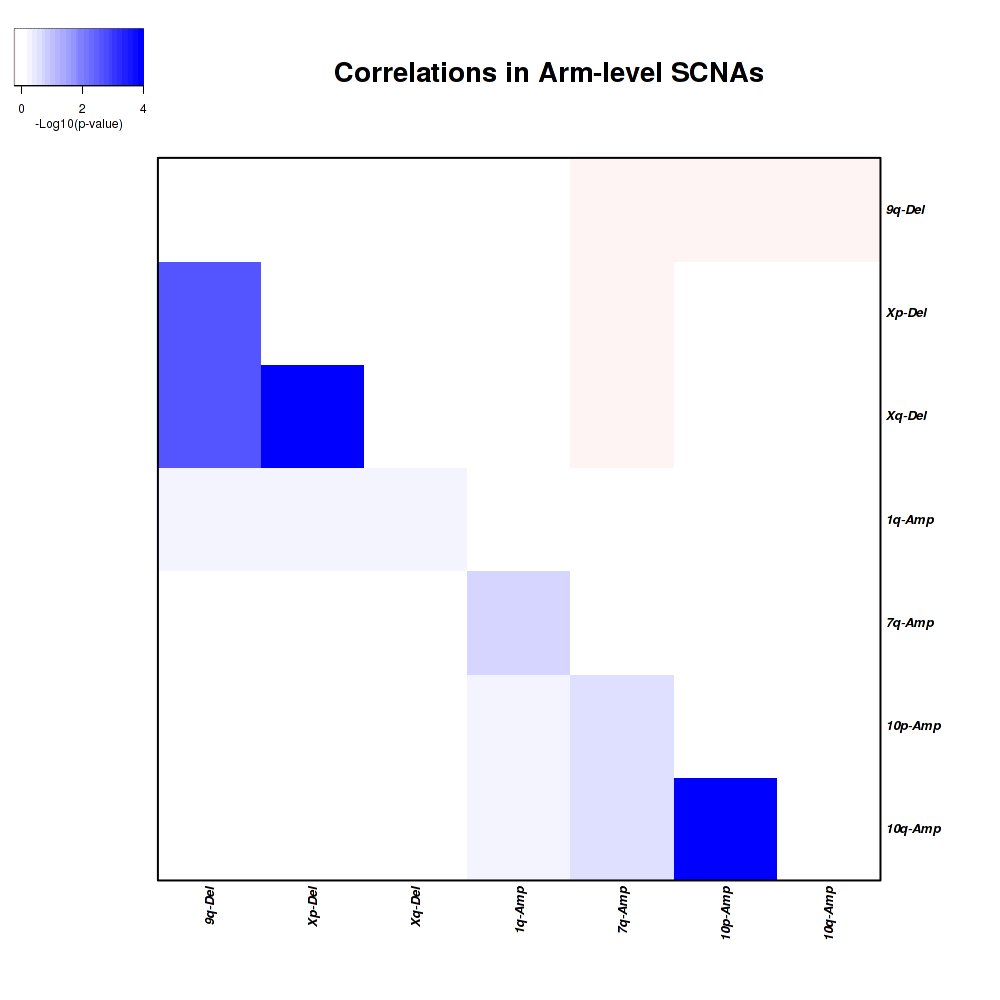
Figure 2. Get High-res Image Co-occurring and mutually-exclusive pairs for copy number variations of focal events. Blue means two events are co-occurring and red means two events are mutually-exclusive.
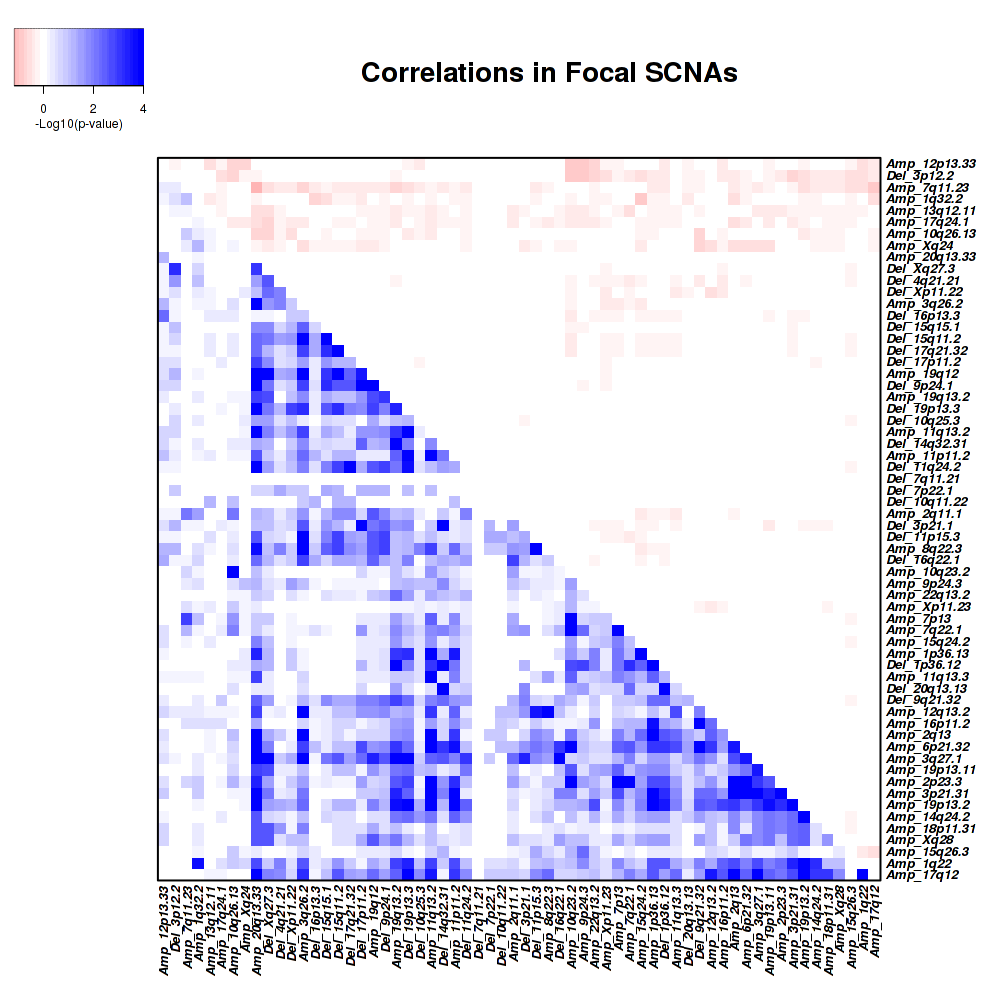
Figure 3. Get High-res Image Co-occurring and mutually-exclusive pairs for significantly mutated genes.
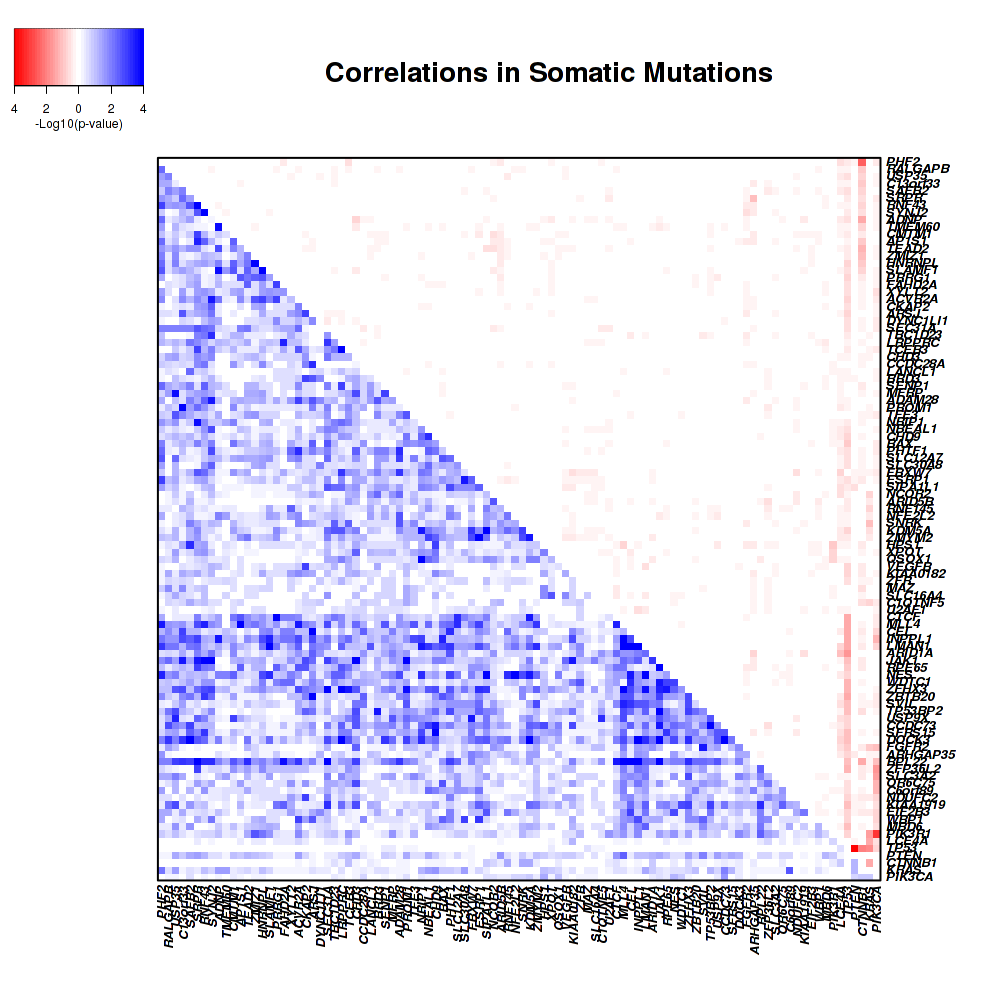
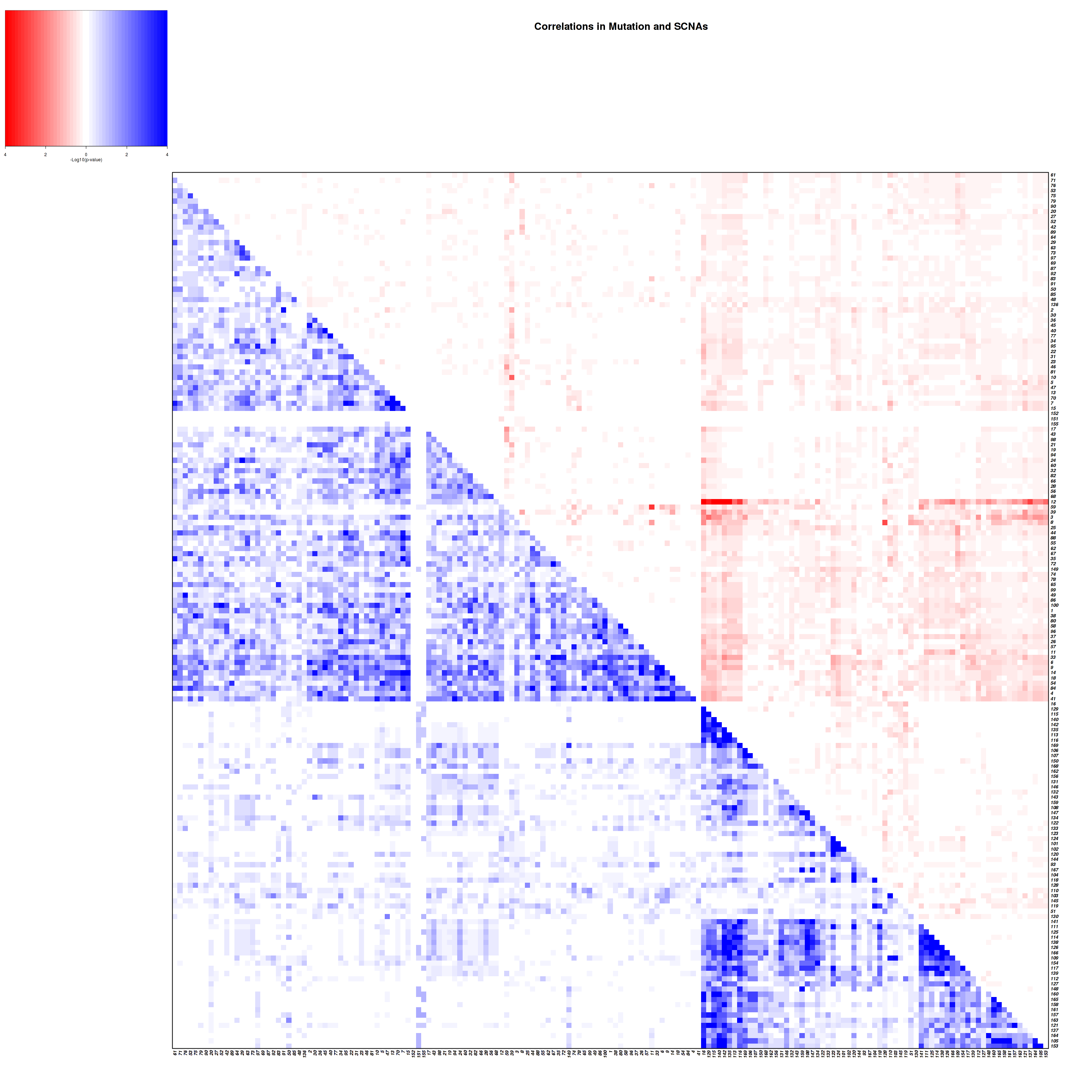
Figure 4. Get High-res Image Co-occurring and mutually-exclusive pairs for all copy number and mutation events. Blue means two events are co-occurring and red means two events are mutually-exclusive.
Download the latest R code for plotting these figures from here
-
Genelist : this file includes all copy number alteration genes from the copy number pancancer paper[1] and the census genes from Cosmic [2].
-
Clinical file :*.merged_data.txt from Append_CustomClinical.
-
Clustering file : *.mergedcluster.txt from Aggregate_Molecular_Subtype_Clusters.
-
Copy number file : broad_values_by_arm.txt, *.conf_99.txt and broad_significance_results.txt from CopyNumber_Gistic2.
-
Mutation file : *.maf, *.sig_genes.txt, *.cosmic_sig_genes.txt and patient_counts_and_rates.txt from MutSigRun2.0/CV/2CV.
-
mRNAseq expression file : *.subclassmarkers.txt and *.log2.txt from mRNAseq_Clustering_CNMF and mRNAseq_Preprocess.
A detailed description of the output files :
-
Clinical features : start with CLI_ followed by clinical feature name ex: CLI_age.
-
Clustering results : start with CLUS_ followed by platform_method ex: CLUS_mRNAseq_cHierarchical.
-
Somatic mutation genes : start with SMG_ followed by version number( mutsig2.0,cv,2cv)_gene name ex: SMG_mutsig.2CV_FAM47C.
-
Somatic mutation genes expression : start wit SMG_ followed by gene name_mRNA ex: SMG_KRT3_mRNA.
-
Marker genes in each mRNAseq clustering subtype : star with mRNA_ followed by CNMF_gene name_difference_cluster number ex: mRNA_CNMF_FAM66E_.0.6_2 (In each cluster, the top 5 up regulated and top 5 down regulated genes were selected).
-
Copy number alterations :
-
Copy number focal events : start with Amp_/Del_ followed by cytoband name ex: Amp_1q32.1/Del_1p36.32.
-
Copy number arm level events : start with CN_ followed by arm_Amp/Del ex: CN_10p_Amp/CN_10p_Del.
-
Copy number alterative gene with expression: start with Amp/Del_ followed by gene name, cytoband names which contains this alterative gene and _mRNA ex: Amp_SOX2_3q26.32_mRNA/Del_PARK2_6q24.3_mRNA.