This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and molecular subtypes.
Testing the association between copy number variation 72 arm-level events and 3 molecular subtypes across 100 patients, 67 significant findings detected with P value < 0.05 and Q value < 0.25.
-
1q gain cnv correlated to 'MRNA_CHIERARCHICAL', 'METHYLATION_CNMF', and 'METHYLATION_CHIERARCHICAL'.
-
3p gain cnv correlated to 'METHYLATION_CNMF'.
-
3q gain cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
6p gain cnv correlated to 'METHYLATION_CNMF'.
-
12p gain cnv correlated to 'MRNA_CHIERARCHICAL'.
-
12q gain cnv correlated to 'MRNA_CHIERARCHICAL'.
-
13q gain cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
14q gain cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
16p gain cnv correlated to 'METHYLATION_CNMF'.
-
18q gain cnv correlated to 'METHYLATION_CHIERARCHICAL'.
-
20p gain cnv correlated to 'MRNA_CHIERARCHICAL', 'METHYLATION_CNMF', and 'METHYLATION_CHIERARCHICAL'.
-
20q gain cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
21q gain cnv correlated to 'METHYLATION_CNMF'.
-
22q gain cnv correlated to 'METHYLATION_CNMF'.
-
1p loss cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
1q loss cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
3p loss cnv correlated to 'METHYLATION_CHIERARCHICAL'.
-
4p loss cnv correlated to 'MRNA_CHIERARCHICAL', 'METHYLATION_CNMF', and 'METHYLATION_CHIERARCHICAL'.
-
4q loss cnv correlated to 'MRNA_CHIERARCHICAL', 'METHYLATION_CNMF', and 'METHYLATION_CHIERARCHICAL'.
-
5p loss cnv correlated to 'METHYLATION_CNMF'.
-
5q loss cnv correlated to 'METHYLATION_CNMF'.
-
8p loss cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
8q loss cnv correlated to 'METHYLATION_CNMF'.
-
9p loss cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
9q loss cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
10q loss cnv correlated to 'METHYLATION_CHIERARCHICAL'.
-
11p loss cnv correlated to 'METHYLATION_CNMF'.
-
11q loss cnv correlated to 'METHYLATION_CNMF'.
-
12p loss cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
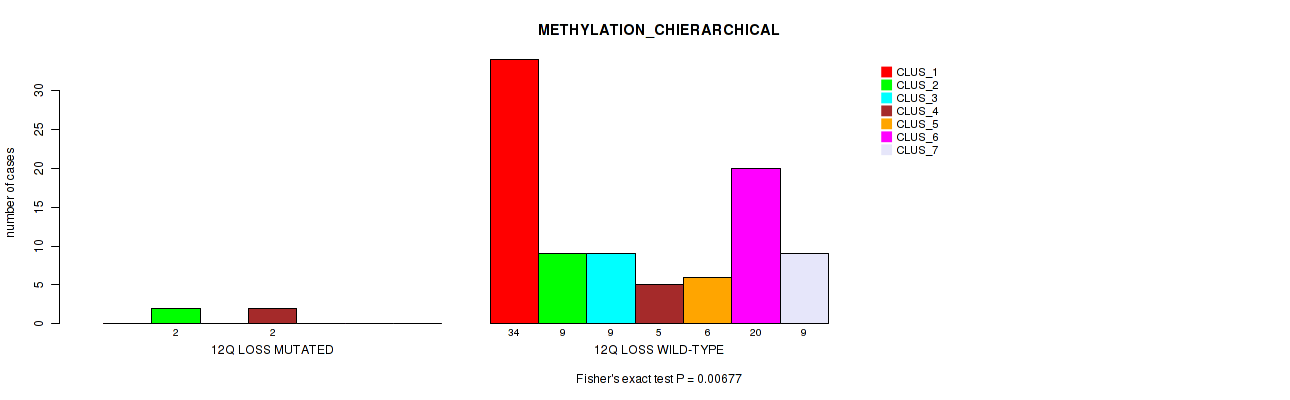
12q loss cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
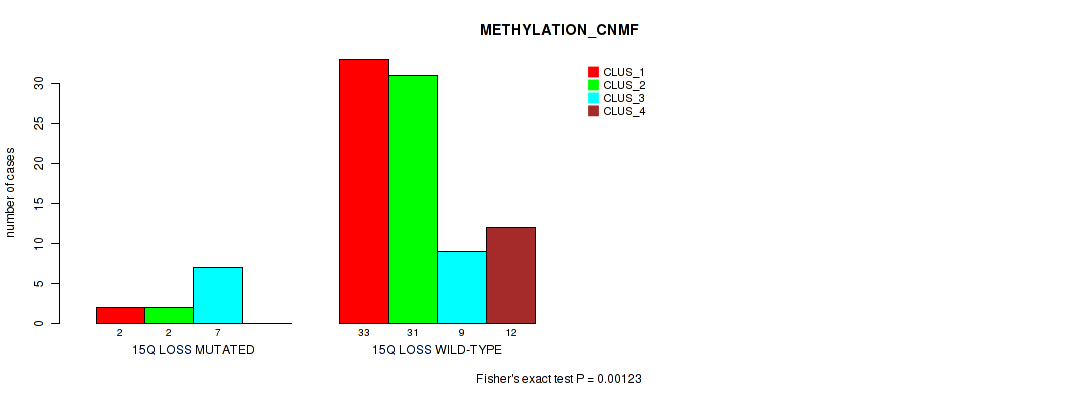
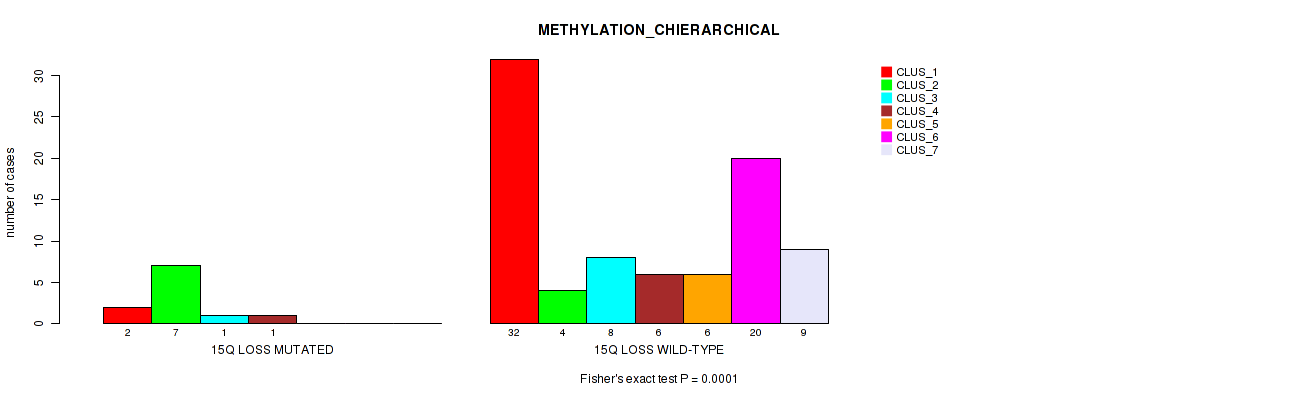
15q loss cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
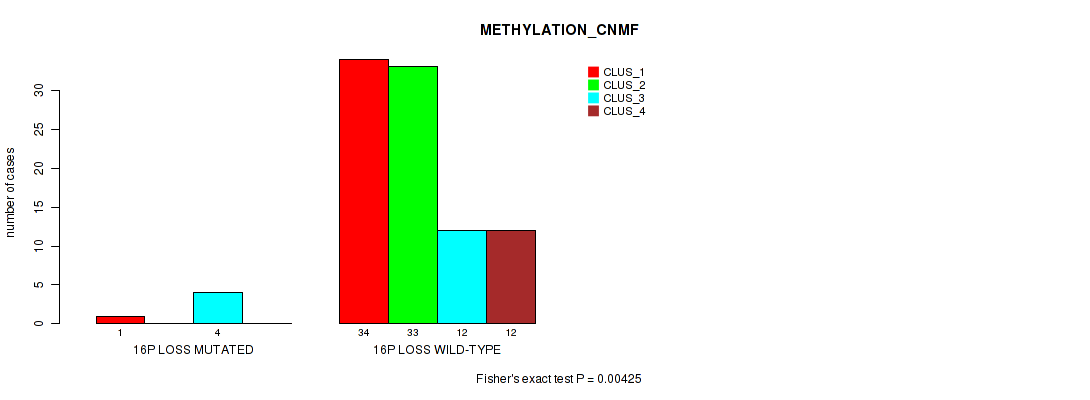
16p loss cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
16q loss cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
17p loss cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
17q loss cnv correlated to 'METHYLATION_CNMF'.
-
18p loss cnv correlated to 'METHYLATION_CNMF'.
-
19p loss cnv correlated to 'METHYLATION_CNMF'.
-
19q loss cnv correlated to 'METHYLATION_CNMF'.
-
22q loss cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
xp loss cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
-
xq loss cnv correlated to 'METHYLATION_CNMF' and 'METHYLATION_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 72 arm-level events and 3 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 67 significant findings detected.
|
Clinical Features |
MRNA CHIERARCHICAL |
METHYLATION CNMF |
METHYLATION CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 1q gain | 27 (27%) | 73 |
0.0237 (0.0968) |
1e-05 (0.00216) |
0.0007 (0.0126) |
| 20p gain | 22 (22%) | 78 |
0.0327 (0.124) |
0.00049 (0.0126) |
0.0482 (0.155) |
| 4p loss | 12 (12%) | 88 |
0.0154 (0.0773) |
0.00913 (0.0563) |
0.0176 (0.0793) |
| 4q loss | 12 (12%) | 88 |
0.0151 (0.0773) |
0.0085 (0.054) |
0.0165 (0.0774) |
| 3q gain | 16 (16%) | 84 |
0.248 (0.447) |
0.00526 (0.0437) |
0.00739 (0.0499) |
| 13q gain | 6 (6%) | 94 |
0.753 (0.86) |
0.00065 (0.0126) |
0.00777 (0.0509) |
| 14q gain | 7 (7%) | 93 |
0.46 (0.645) |
0.0039 (0.0366) |
0.034 (0.124) |
| 20q gain | 22 (22%) | 78 |
0.119 (0.283) |
5e-05 (0.0036) |
0.00592 (0.047) |
| 1p loss | 4 (4%) | 96 |
0.122 (0.285) |
0.0036 (0.0353) |
0.0334 (0.124) |
| 1q loss | 3 (3%) | 97 |
0.438 (0.634) |
0.00516 (0.0437) |
0.0352 (0.124) |
| 8p loss | 7 (7%) | 93 |
0.158 (0.341) |
4e-05 (0.0036) |
0.0301 (0.12) |
| 9p loss | 14 (14%) | 86 |
0.283 (0.494) |
0.0144 (0.0758) |
0.00249 (0.0299) |
| 9q loss | 16 (16%) | 84 |
0.127 (0.294) |
0.00733 (0.0499) |
0.011 (0.0658) |
| 12p loss | 4 (4%) | 96 |
0.939 (0.953) |
0.0159 (0.0774) |
0.00651 (0.0485) |
| 12q loss | 4 (4%) | 96 |
0.939 (0.953) |
0.0163 (0.0774) |
0.00677 (0.0487) |
| 15q loss | 13 (13%) | 87 |
0.547 (0.738) |
0.00123 (0.0204) |
0.0001 (0.0054) |
| 16p loss | 6 (6%) | 94 |
0.893 (0.937) |
0.00425 (0.0382) |
0.0455 (0.151) |
| 16q loss | 7 (7%) | 93 |
0.863 (0.927) |
0.00069 (0.0126) |
0.0144 (0.0758) |
| 17p loss | 13 (13%) | 87 |
0.101 (0.27) |
0.00232 (0.0299) |
0.00027 (0.0108) |
| 22q loss | 12 (12%) | 88 |
0.384 (0.604) |
0.0469 (0.153) |
0.0169 (0.0779) |
| xp loss | 19 (19%) | 81 |
0.651 (0.817) |
0.00046 (0.0126) |
0.00249 (0.0299) |
| xq loss | 19 (19%) | 81 |
0.648 (0.817) |
0.0003 (0.0108) |
0.00286 (0.0318) |
| 3p gain | 10 (10%) | 90 |
0.226 (0.428) |
0.0186 (0.0822) |
0.0943 (0.26) |
| 6p gain | 10 (10%) | 90 |
0.182 (0.367) |
0.0121 (0.0709) |
0.105 (0.273) |
| 12p gain | 10 (10%) | 90 |
0.0353 (0.124) |
0.492 (0.677) |
0.992 (0.992) |
| 12q gain | 10 (10%) | 90 |
0.0346 (0.124) |
0.492 (0.677) |
0.992 (0.992) |
| 16p gain | 6 (6%) | 94 |
0.163 (0.346) |
0.04 (0.135) |
0.326 (0.542) |
| 18q gain | 7 (7%) | 93 |
0.566 (0.755) |
0.148 (0.325) |
0.0386 (0.132) |
| 21q gain | 6 (6%) | 94 |
0.434 (0.634) |
0.00327 (0.0336) |
0.21 (0.408) |
| 22q gain | 8 (8%) | 92 |
0.766 (0.866) |
0.00232 (0.0299) |
0.165 (0.346) |
| 3p loss | 6 (6%) | 94 |
0.434 (0.634) |
0.149 (0.325) |
0.0327 (0.124) |
| 5p loss | 9 (9%) | 91 |
0.316 (0.529) |
0.0215 (0.0928) |
0.0951 (0.26) |
| 5q loss | 11 (11%) | 89 |
0.185 (0.37) |
0.00609 (0.047) |
0.101 (0.27) |
| 8q loss | 4 (4%) | 96 |
0.663 (0.819) |
0.00069 (0.0126) |
0.191 (0.379) |
| 10q loss | 4 (4%) | 96 |
0.662 (0.819) |
0.0843 (0.243) |
0.0357 (0.124) |
| 11p loss | 8 (8%) | 92 |
0.766 (0.866) |
0.0128 (0.0726) |
0.111 (0.276) |
| 11q loss | 7 (7%) | 93 |
0.426 (0.634) |
0.0131 (0.0726) |
0.113 (0.276) |
| 17q loss | 8 (8%) | 92 |
0.403 (0.61) |
0.00238 (0.0299) |
0.16 (0.343) |
| 18p loss | 4 (4%) | 96 |
0.94 (0.953) |
0.0223 (0.0944) |
0.369 (0.6) |
| 19p loss | 6 (6%) | 94 |
0.696 (0.835) |
0.00294 (0.0318) |
0.315 (0.529) |
| 19q loss | 6 (6%) | 94 |
0.278 (0.494) |
0.0231 (0.0961) |
0.56 (0.752) |
| 1p gain | 9 (9%) | 91 |
0.455 (0.643) |
0.0662 (0.199) |
0.239 (0.437) |
| 2p gain | 15 (15%) | 85 |
0.848 (0.921) |
0.0669 (0.199) |
0.282 (0.494) |
| 2q gain | 14 (14%) | 86 |
0.804 (0.885) |
0.0673 (0.199) |
0.282 (0.494) |
| 5p gain | 5 (5%) | 95 |
0.241 (0.437) |
0.811 (0.885) |
0.445 (0.634) |
| 6q gain | 8 (8%) | 92 |
0.383 (0.604) |
0.211 (0.408) |
0.721 (0.85) |
| 7p gain | 13 (13%) | 87 |
0.293 (0.502) |
0.586 (0.767) |
0.861 (0.927) |
| 7q gain | 14 (14%) | 86 |
0.143 (0.324) |
0.507 (0.693) |
0.79 (0.884) |
| 8p gain | 19 (19%) | 81 |
0.0613 (0.189) |
0.789 (0.884) |
0.67 (0.822) |
| 8q gain | 20 (20%) | 80 |
0.0547 (0.171) |
0.883 (0.937) |
0.6 (0.78) |
| 10p gain | 25 (25%) | 75 |
0.933 (0.953) |
0.748 (0.859) |
0.172 (0.357) |
| 10q gain | 24 (24%) | 76 |
0.739 (0.856) |
0.648 (0.817) |
0.105 (0.273) |
| 11p gain | 3 (3%) | 97 |
0.684 (0.829) |
0.109 (0.276) |
0.404 (0.61) |
| 11q gain | 4 (4%) | 96 |
0.381 (0.604) |
0.111 (0.276) |
0.404 (0.61) |
| 16q gain | 5 (5%) | 95 |
0.359 (0.587) |
0.148 (0.325) |
0.417 (0.625) |
| 17p gain | 7 (7%) | 93 |
0.0924 (0.26) |
0.136 (0.314) |
0.177 (0.365) |
| 17q gain | 6 (6%) | 94 |
0.234 (0.433) |
0.204 (0.401) |
0.181 (0.367) |
| 18p gain | 9 (9%) | 91 |
0.115 (0.277) |
0.0524 (0.166) |
0.0703 (0.205) |
| 19p gain | 7 (7%) | 93 |
0.918 (0.949) |
0.695 (0.835) |
0.301 (0.512) |
| 19q gain | 8 (8%) | 92 |
0.795 (0.885) |
0.335 (0.553) |
0.292 (0.502) |
| xp gain | 8 (8%) | 92 |
0.611 (0.79) |
0.872 (0.932) |
0.477 (0.665) |
| xq gain | 8 (8%) | 92 |
0.392 (0.61) |
0.439 (0.634) |
0.528 (0.718) |
| yp gain | 14 (14%) | 86 |
0.736 (0.856) |
0.386 (0.604) |
0.916 (0.949) |
| yq gain | 14 (14%) | 86 |
0.741 (0.856) |
0.384 (0.604) |
0.915 (0.949) |
| 7p loss | 5 (5%) | 95 |
0.724 (0.85) |
0.811 (0.885) |
0.444 (0.634) |
| 7q loss | 5 (5%) | 95 |
0.722 (0.85) |
0.811 (0.885) |
0.446 (0.634) |
| 10p loss | 5 (5%) | 95 |
0.892 (0.937) |
0.144 (0.324) |
0.0932 (0.26) |
| 13q loss | 10 (10%) | 90 |
0.214 (0.409) |
0.948 (0.957) |
0.664 (0.819) |
| 18q loss | 5 (5%) | 95 |
0.72 (0.85) |
0.114 (0.277) |
0.678 (0.827) |
| 21q loss | 5 (5%) | 95 |
0.891 (0.937) |
0.112 (0.276) |
0.4 (0.61) |
| yp loss | 47 (47%) | 53 |
0.645 (0.817) |
0.232 (0.433) |
0.575 (0.758) |
| yq loss | 47 (47%) | 53 |
0.646 (0.817) |
0.233 (0.433) |
0.575 (0.758) |
P value = 0.0237 (Fisher's exact test), Q value = 0.097
Table S1. Gene #2: '1q gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 28 | 20 | 11 | 20 | 21 |
| 1Q GAIN MUTATED | 6 | 2 | 1 | 8 | 10 |
| 1Q GAIN WILD-TYPE | 22 | 18 | 10 | 12 | 11 |
Figure S1. Get High-res Image Gene #2: '1q gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
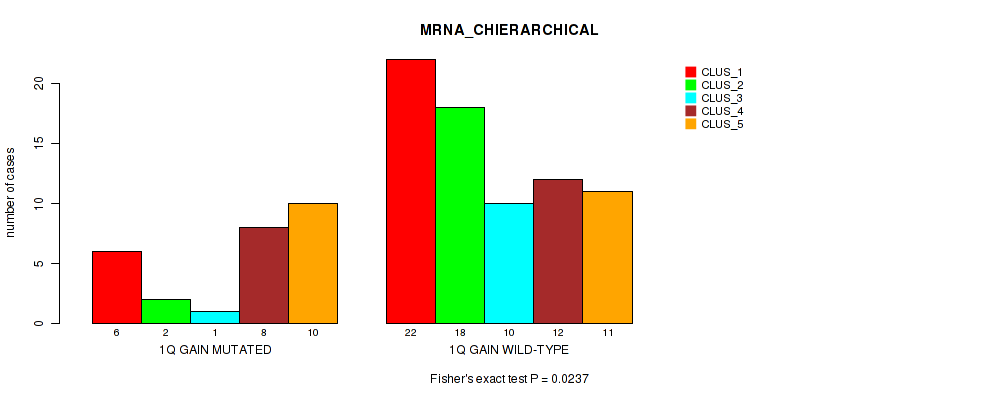
P value = 1e-05 (Fisher's exact test), Q value = 0.0022
Table S2. Gene #2: '1q gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 1Q GAIN MUTATED | 3 | 5 | 7 | 10 |
| 1Q GAIN WILD-TYPE | 32 | 28 | 9 | 2 |
Figure S2. Get High-res Image Gene #2: '1q gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
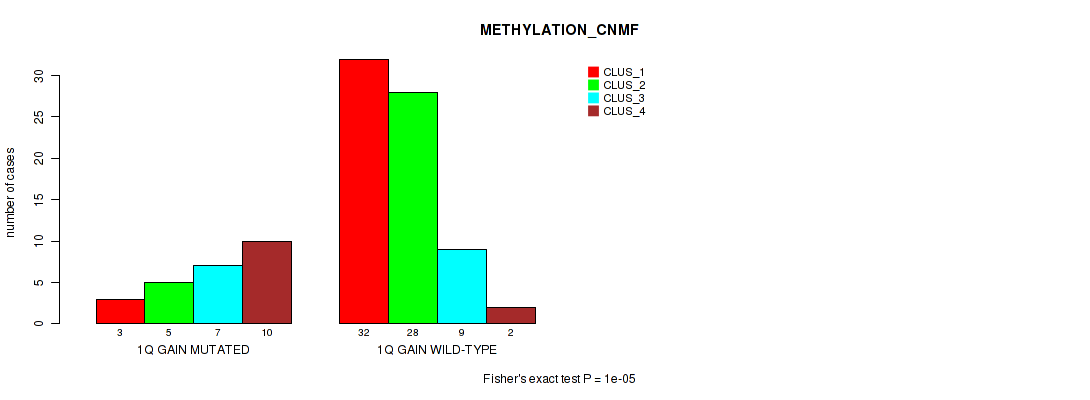
P value = 7e-04 (Fisher's exact test), Q value = 0.013
Table S3. Gene #2: '1q gain' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 1Q GAIN MUTATED | 7 | 4 | 2 | 3 | 6 | 3 | 0 |
| 1Q GAIN WILD-TYPE | 27 | 7 | 7 | 4 | 0 | 17 | 9 |
Figure S3. Get High-res Image Gene #2: '1q gain' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
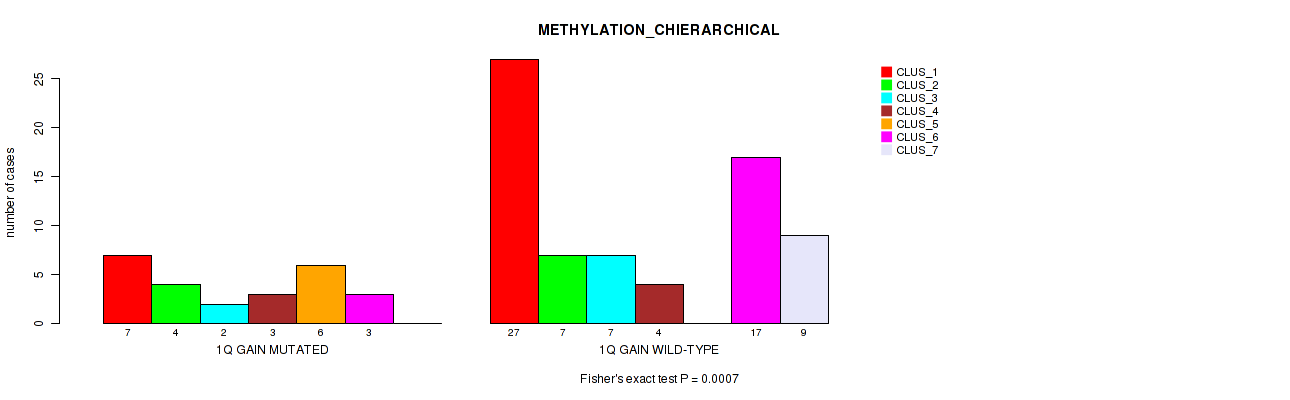
P value = 0.0186 (Fisher's exact test), Q value = 0.082
Table S4. Gene #5: '3p gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 3P GAIN MUTATED | 1 | 2 | 5 | 1 |
| 3P GAIN WILD-TYPE | 34 | 31 | 11 | 11 |
Figure S4. Get High-res Image Gene #5: '3p gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
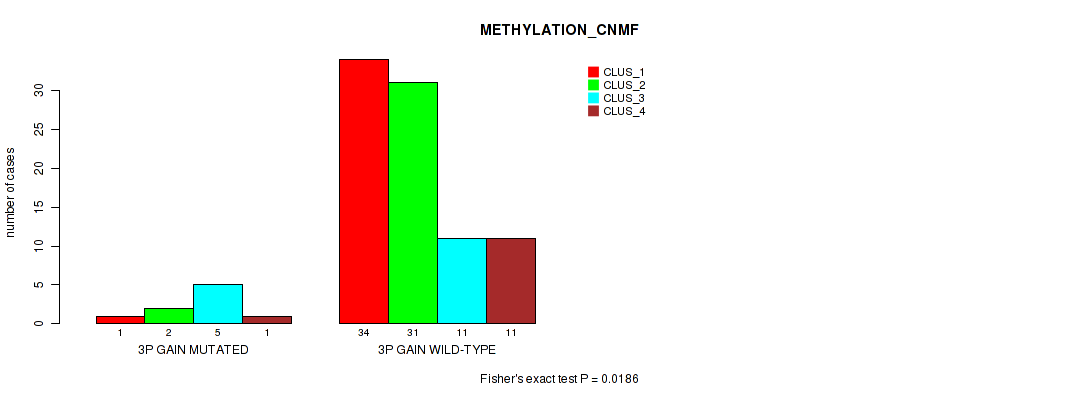
P value = 0.00526 (Fisher's exact test), Q value = 0.044
Table S5. Gene #6: '3q gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 3Q GAIN MUTATED | 2 | 3 | 7 | 1 |
| 3Q GAIN WILD-TYPE | 33 | 30 | 9 | 11 |
Figure S5. Get High-res Image Gene #6: '3q gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
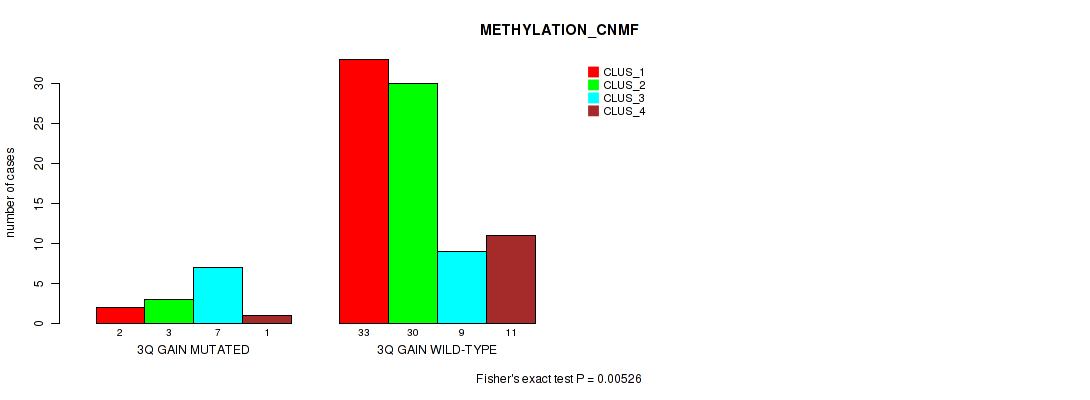
P value = 0.00739 (Fisher's exact test), Q value = 0.05
Table S6. Gene #6: '3q gain' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 3Q GAIN MUTATED | 4 | 5 | 0 | 3 | 0 | 1 | 0 |
| 3Q GAIN WILD-TYPE | 30 | 6 | 9 | 4 | 6 | 19 | 9 |
Figure S6. Get High-res Image Gene #6: '3q gain' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
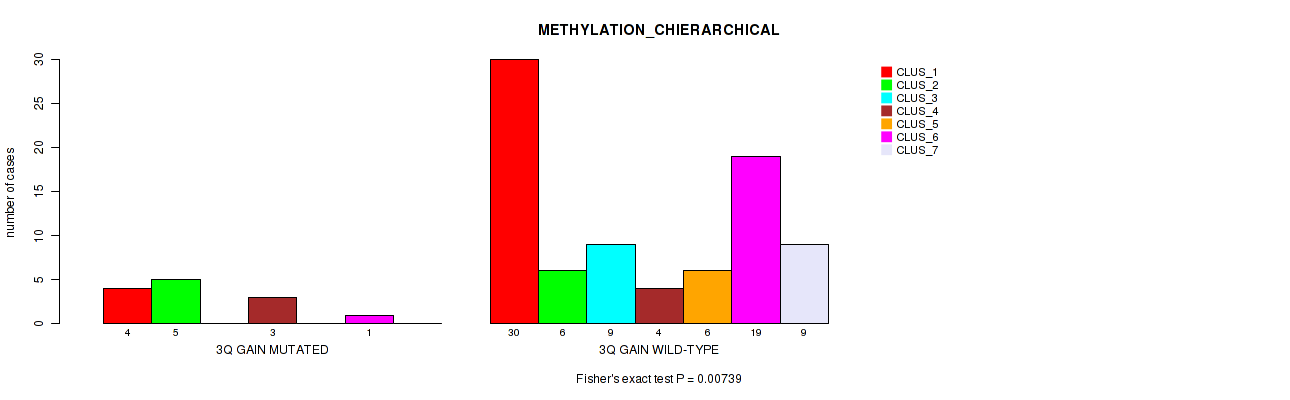
P value = 0.0121 (Fisher's exact test), Q value = 0.071
Table S7. Gene #8: '6p gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 6P GAIN MUTATED | 2 | 1 | 5 | 0 |
| 6P GAIN WILD-TYPE | 33 | 32 | 11 | 12 |
Figure S7. Get High-res Image Gene #8: '6p gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
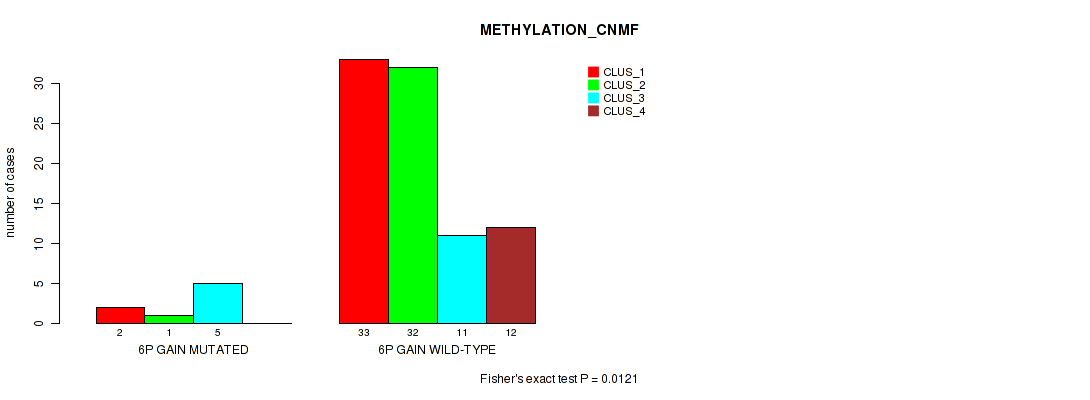
P value = 0.0353 (Fisher's exact test), Q value = 0.12
Table S8. Gene #18: '12p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 28 | 20 | 11 | 20 | 21 |
| 12P GAIN MUTATED | 1 | 1 | 0 | 6 | 2 |
| 12P GAIN WILD-TYPE | 27 | 19 | 11 | 14 | 19 |
Figure S8. Get High-res Image Gene #18: '12p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
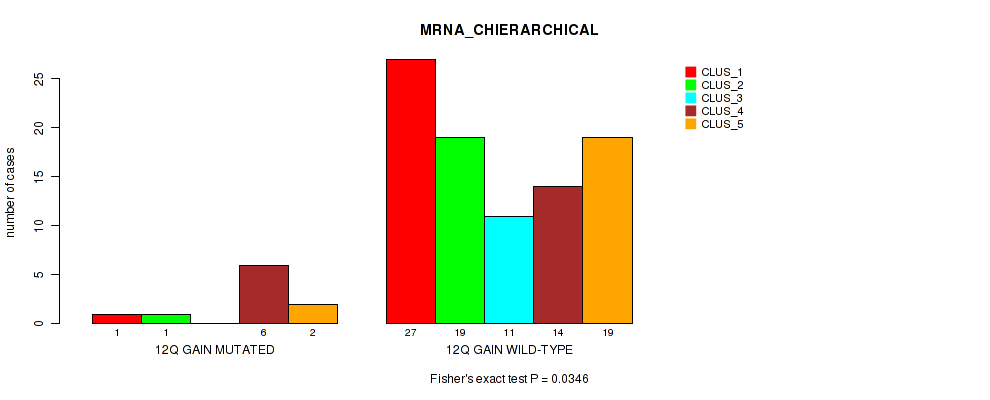
P value = 0.0346 (Fisher's exact test), Q value = 0.12
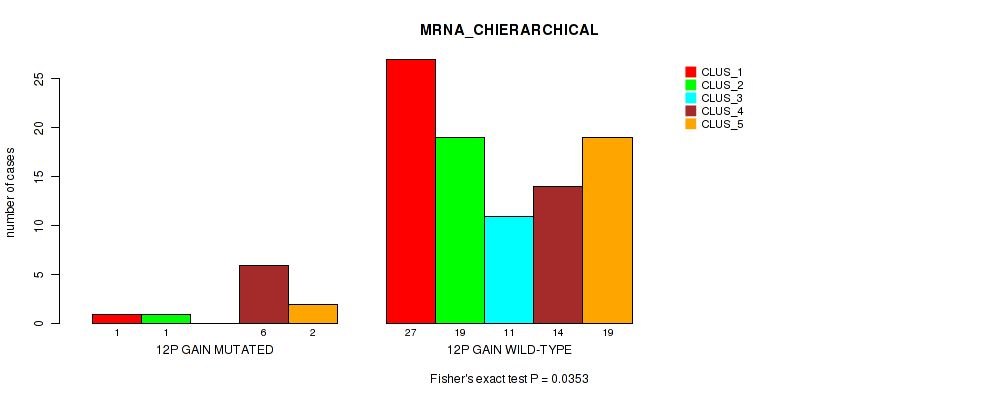
Table S9. Gene #19: '12q gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 28 | 20 | 11 | 20 | 21 |
| 12Q GAIN MUTATED | 1 | 1 | 0 | 6 | 2 |
| 12Q GAIN WILD-TYPE | 27 | 19 | 11 | 14 | 19 |
Figure S9. Get High-res Image Gene #19: '12q gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
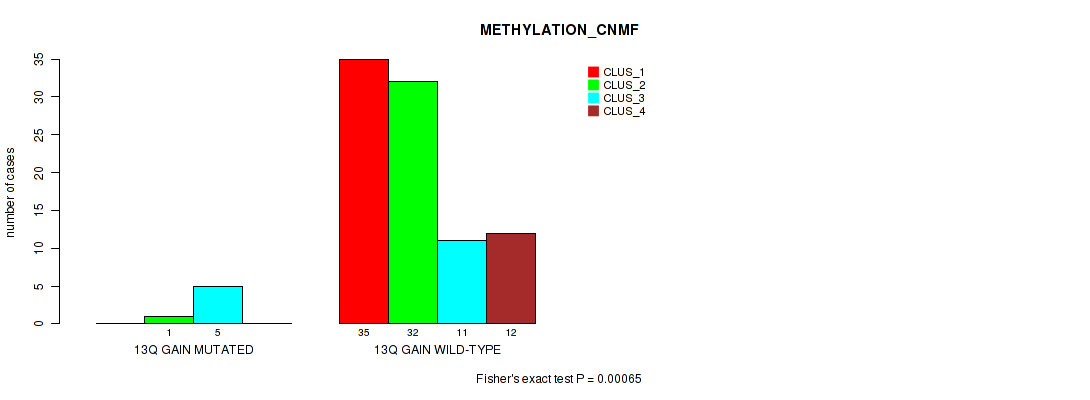
P value = 0.00065 (Fisher's exact test), Q value = 0.013
Table S10. Gene #20: '13q gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 13Q GAIN MUTATED | 0 | 1 | 5 | 0 |
| 13Q GAIN WILD-TYPE | 35 | 32 | 11 | 12 |
Figure S10. Get High-res Image Gene #20: '13q gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
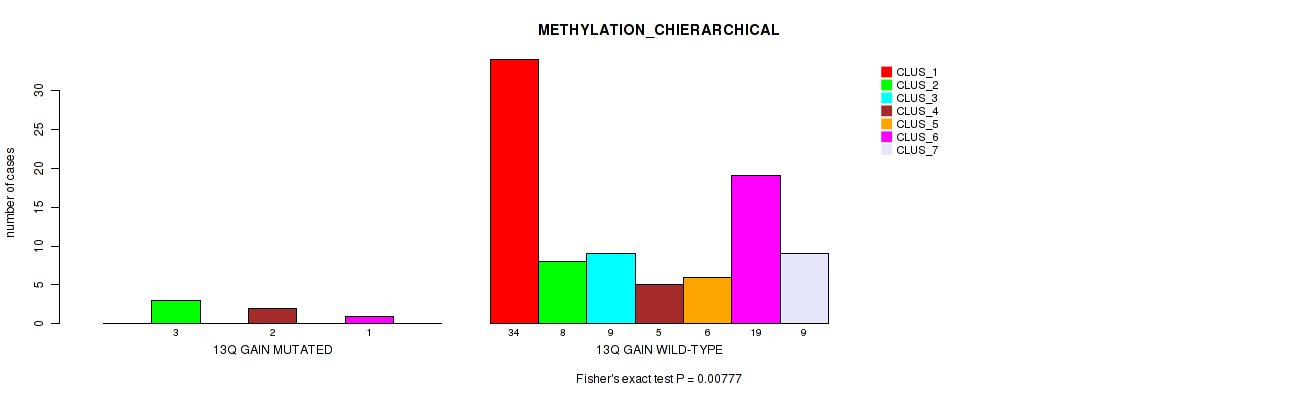
P value = 0.00777 (Fisher's exact test), Q value = 0.051
Table S11. Gene #20: '13q gain' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 13Q GAIN MUTATED | 0 | 3 | 0 | 2 | 0 | 1 | 0 |
| 13Q GAIN WILD-TYPE | 34 | 8 | 9 | 5 | 6 | 19 | 9 |
Figure S11. Get High-res Image Gene #20: '13q gain' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
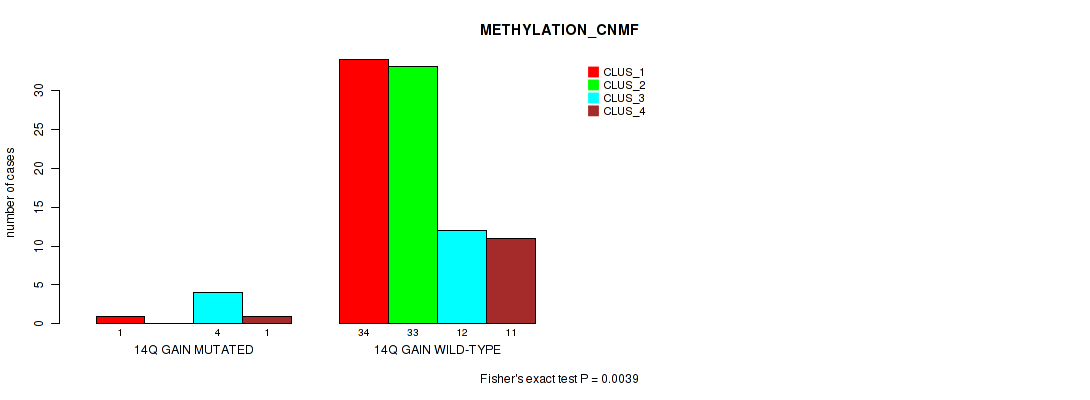
P value = 0.0039 (Fisher's exact test), Q value = 0.037
Table S12. Gene #21: '14q gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 14Q GAIN MUTATED | 1 | 0 | 4 | 1 |
| 14Q GAIN WILD-TYPE | 34 | 33 | 12 | 11 |
Figure S12. Get High-res Image Gene #21: '14q gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
P value = 0.034 (Fisher's exact test), Q value = 0.12
Table S13. Gene #21: '14q gain' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 14Q GAIN MUTATED | 1 | 2 | 0 | 2 | 1 | 0 | 0 |
| 14Q GAIN WILD-TYPE | 33 | 9 | 9 | 5 | 5 | 20 | 9 |
Figure S13. Get High-res Image Gene #21: '14q gain' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
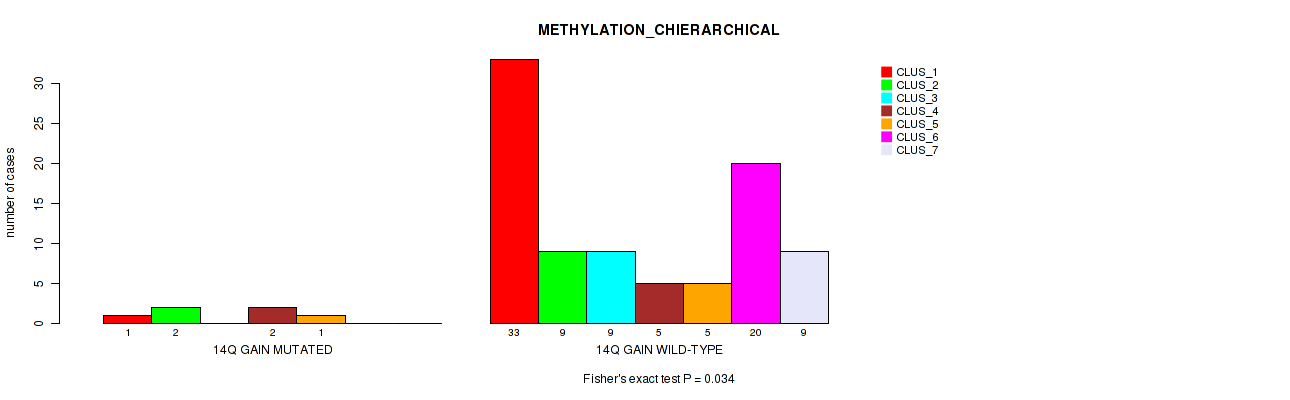
P value = 0.04 (Fisher's exact test), Q value = 0.13
Table S14. Gene #22: '16p gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 16P GAIN MUTATED | 0 | 2 | 3 | 1 |
| 16P GAIN WILD-TYPE | 35 | 31 | 13 | 11 |
Figure S14. Get High-res Image Gene #22: '16p gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
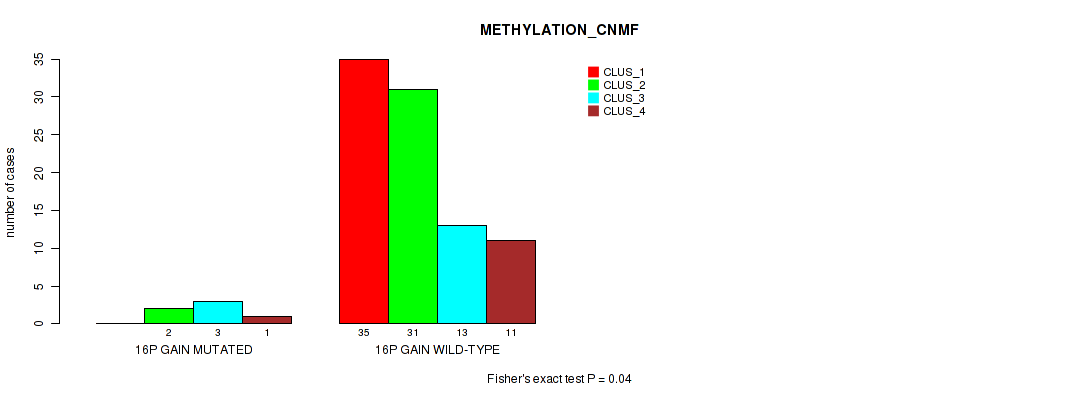
P value = 0.0386 (Fisher's exact test), Q value = 0.13
Table S15. Gene #27: '18q gain' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 18Q GAIN MUTATED | 0 | 0 | 1 | 2 | 0 | 1 | 1 |
| 18Q GAIN WILD-TYPE | 34 | 11 | 8 | 5 | 6 | 19 | 8 |
Figure S15. Get High-res Image Gene #27: '18q gain' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
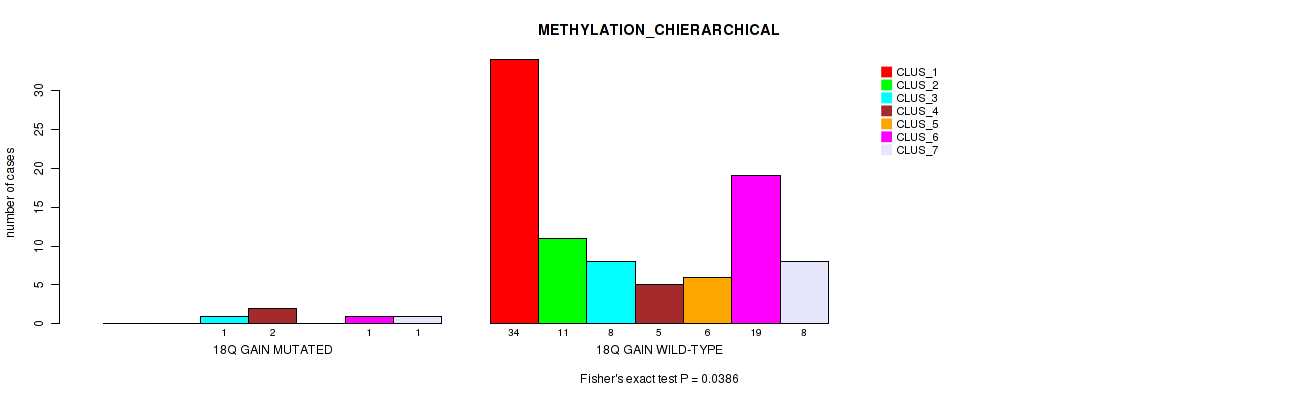
P value = 0.0327 (Fisher's exact test), Q value = 0.12
Table S16. Gene #30: '20p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 28 | 20 | 11 | 20 | 21 |
| 20P GAIN MUTATED | 6 | 2 | 0 | 9 | 5 |
| 20P GAIN WILD-TYPE | 22 | 18 | 11 | 11 | 16 |
Figure S16. Get High-res Image Gene #30: '20p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
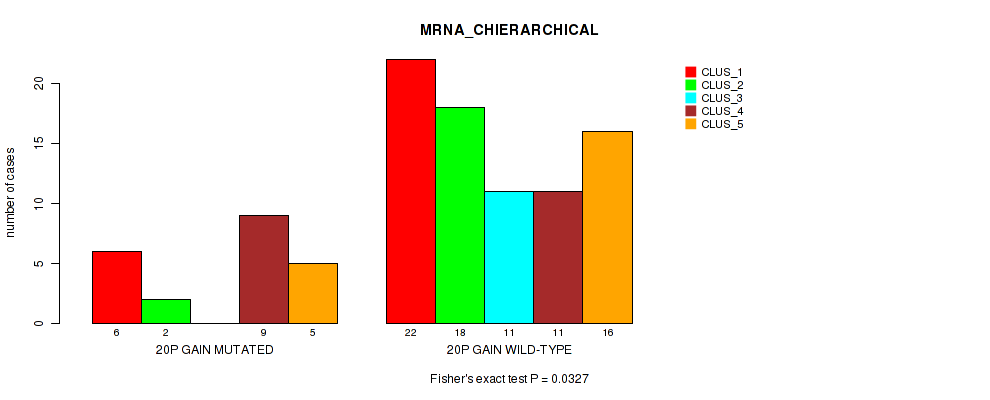
P value = 0.00049 (Fisher's exact test), Q value = 0.013
Table S17. Gene #30: '20p gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 20P GAIN MUTATED | 4 | 5 | 10 | 1 |
| 20P GAIN WILD-TYPE | 31 | 28 | 6 | 11 |
Figure S17. Get High-res Image Gene #30: '20p gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
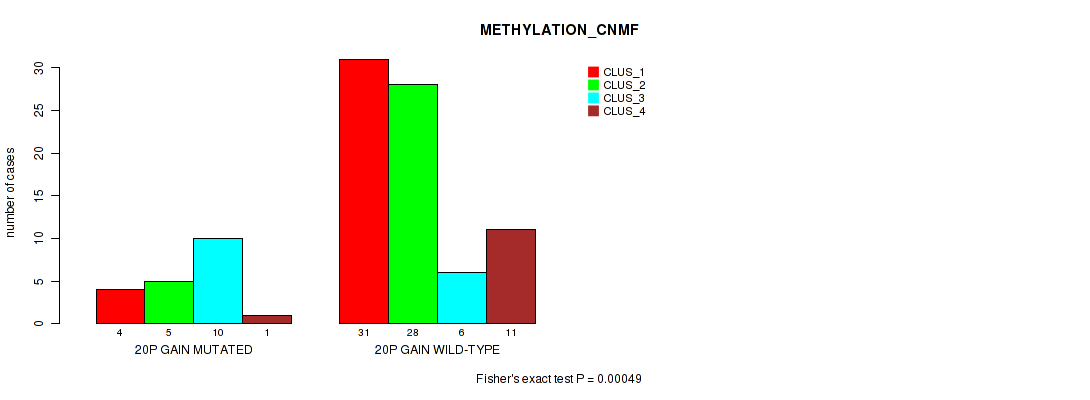
P value = 0.0482 (Fisher's exact test), Q value = 0.16
Table S18. Gene #30: '20p gain' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 20P GAIN MUTATED | 6 | 6 | 1 | 3 | 0 | 2 | 2 |
| 20P GAIN WILD-TYPE | 28 | 5 | 8 | 4 | 6 | 18 | 7 |
Figure S18. Get High-res Image Gene #30: '20p gain' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
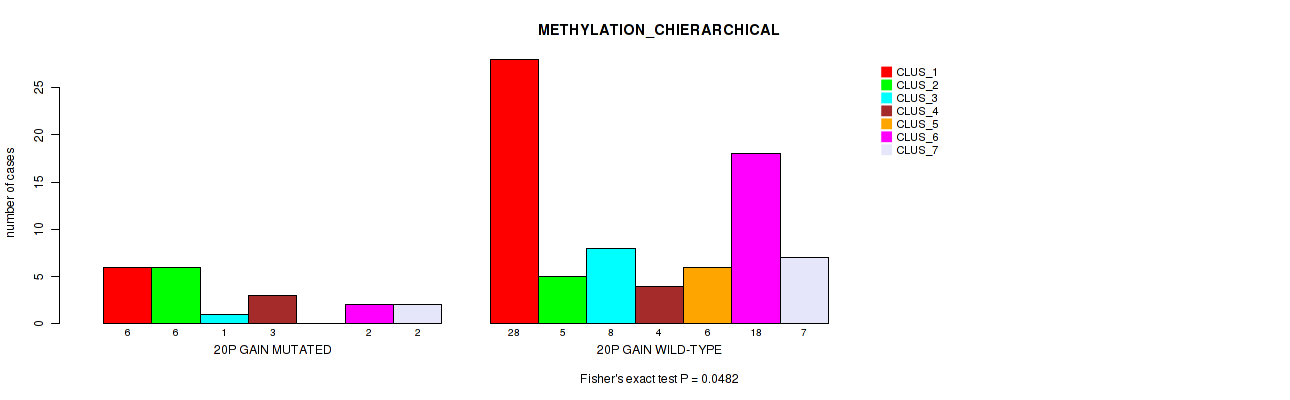
P value = 5e-05 (Fisher's exact test), Q value = 0.0036
Table S19. Gene #31: '20q gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 20Q GAIN MUTATED | 4 | 4 | 11 | 1 |
| 20Q GAIN WILD-TYPE | 31 | 29 | 5 | 11 |
Figure S19. Get High-res Image Gene #31: '20q gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
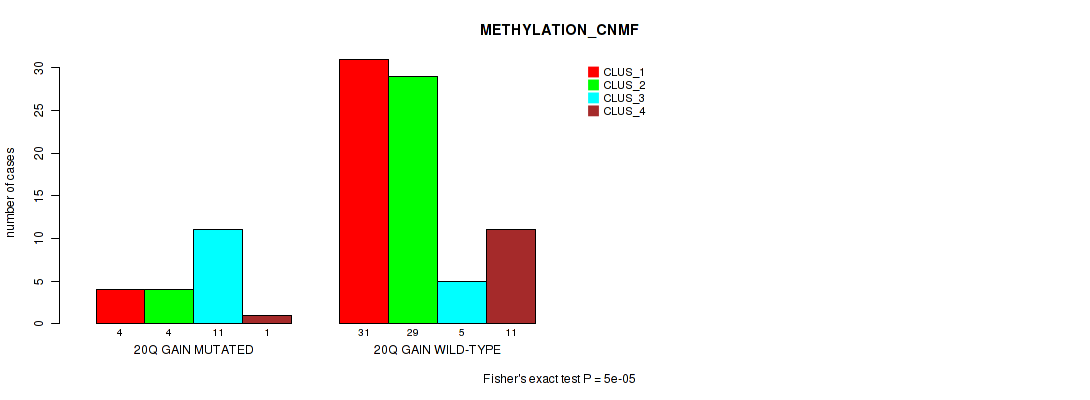
P value = 0.00592 (Fisher's exact test), Q value = 0.047
Table S20. Gene #31: '20q gain' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 20Q GAIN MUTATED | 6 | 6 | 1 | 4 | 0 | 1 | 2 |
| 20Q GAIN WILD-TYPE | 28 | 5 | 8 | 3 | 6 | 19 | 7 |
Figure S20. Get High-res Image Gene #31: '20q gain' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
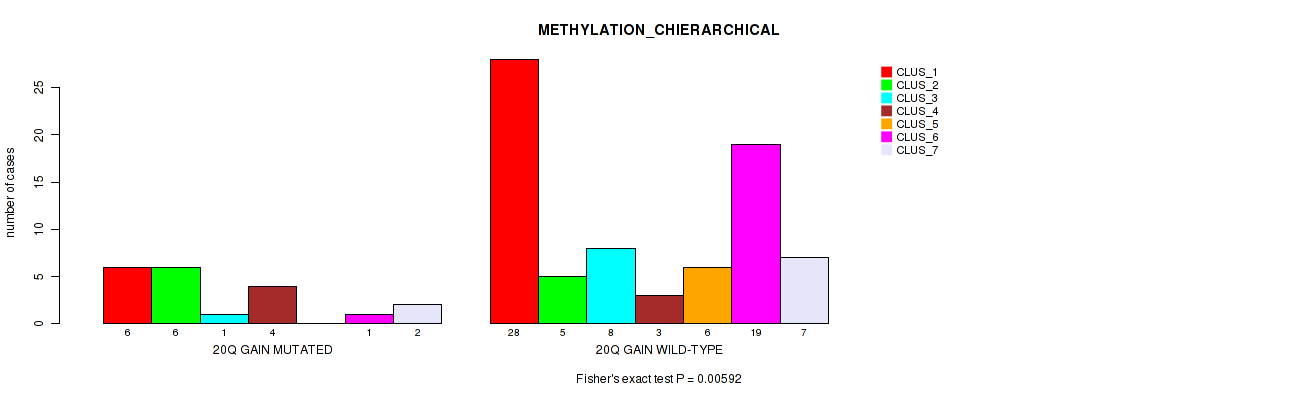
P value = 0.00327 (Fisher's exact test), Q value = 0.034
Table S21. Gene #32: '21q gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 21Q GAIN MUTATED | 0 | 1 | 4 | 1 |
| 21Q GAIN WILD-TYPE | 35 | 32 | 12 | 11 |
Figure S21. Get High-res Image Gene #32: '21q gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
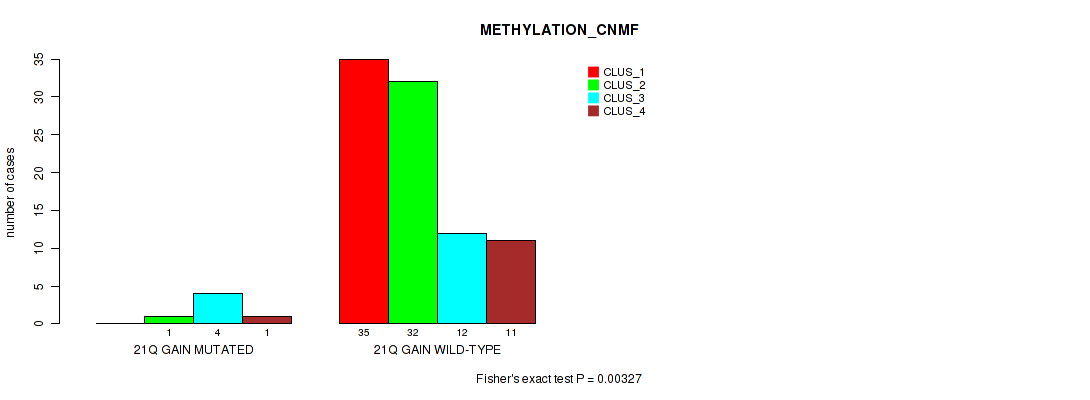
P value = 0.00232 (Fisher's exact test), Q value = 0.03
Table S22. Gene #33: '22q gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 22Q GAIN MUTATED | 0 | 2 | 5 | 1 |
| 22Q GAIN WILD-TYPE | 35 | 31 | 11 | 11 |
Figure S22. Get High-res Image Gene #33: '22q gain' versus Molecular Subtype #2: 'METHYLATION_CNMF'
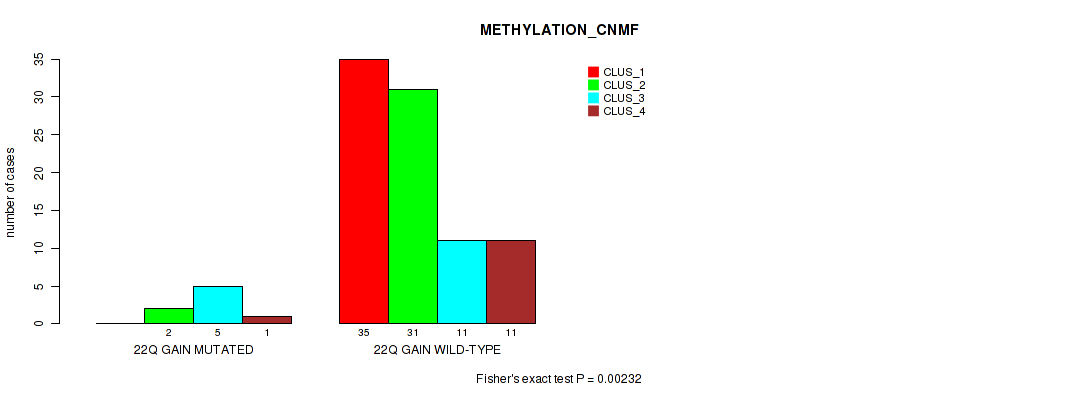
P value = 0.0036 (Fisher's exact test), Q value = 0.035
Table S23. Gene #38: '1p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 1P LOSS MUTATED | 0 | 0 | 3 | 1 |
| 1P LOSS WILD-TYPE | 35 | 33 | 13 | 11 |
Figure S23. Get High-res Image Gene #38: '1p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
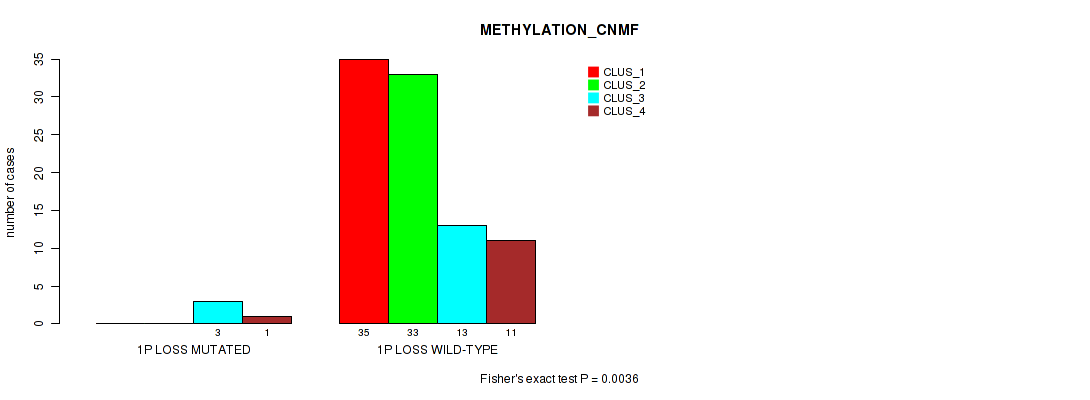
P value = 0.0334 (Fisher's exact test), Q value = 0.12
Table S24. Gene #38: '1p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 1P LOSS MUTATED | 0 | 2 | 1 | 1 | 0 | 0 | 0 |
| 1P LOSS WILD-TYPE | 34 | 9 | 8 | 6 | 6 | 20 | 9 |
Figure S24. Get High-res Image Gene #38: '1p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
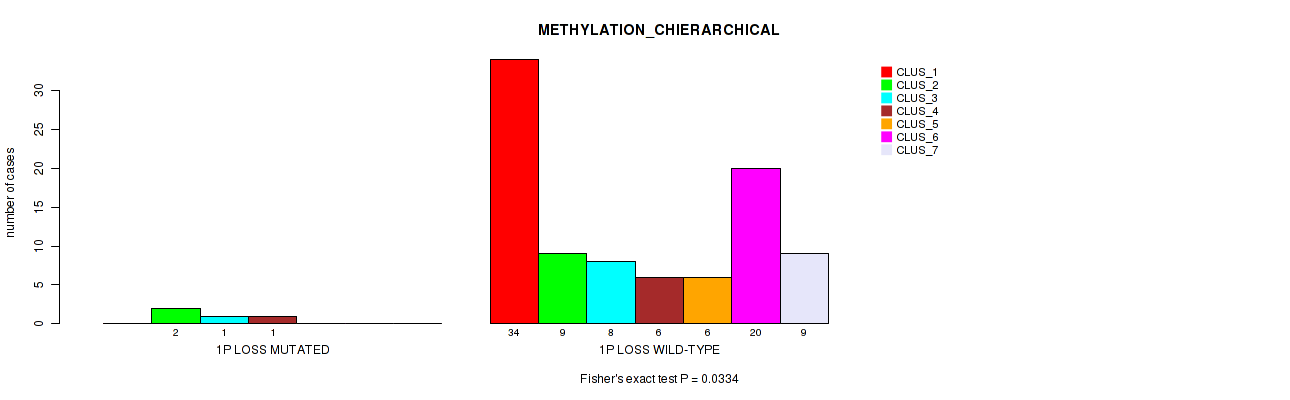
P value = 0.00516 (Fisher's exact test), Q value = 0.044
Table S25. Gene #39: '1q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 1Q LOSS MUTATED | 0 | 0 | 3 | 0 |
| 1Q LOSS WILD-TYPE | 35 | 33 | 13 | 12 |
Figure S25. Get High-res Image Gene #39: '1q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
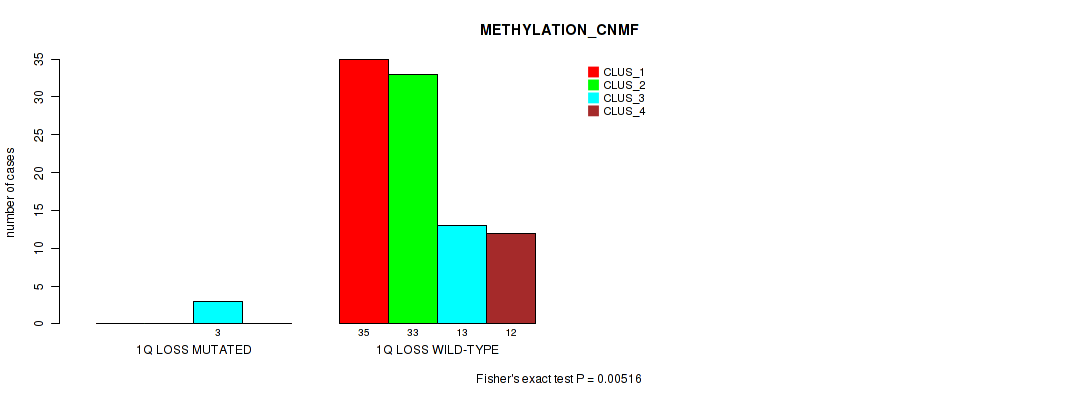
P value = 0.0352 (Fisher's exact test), Q value = 0.12
Table S26. Gene #39: '1q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 1Q LOSS MUTATED | 0 | 2 | 0 | 1 | 0 | 0 | 0 |
| 1Q LOSS WILD-TYPE | 34 | 9 | 9 | 6 | 6 | 20 | 9 |
Figure S26. Get High-res Image Gene #39: '1q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
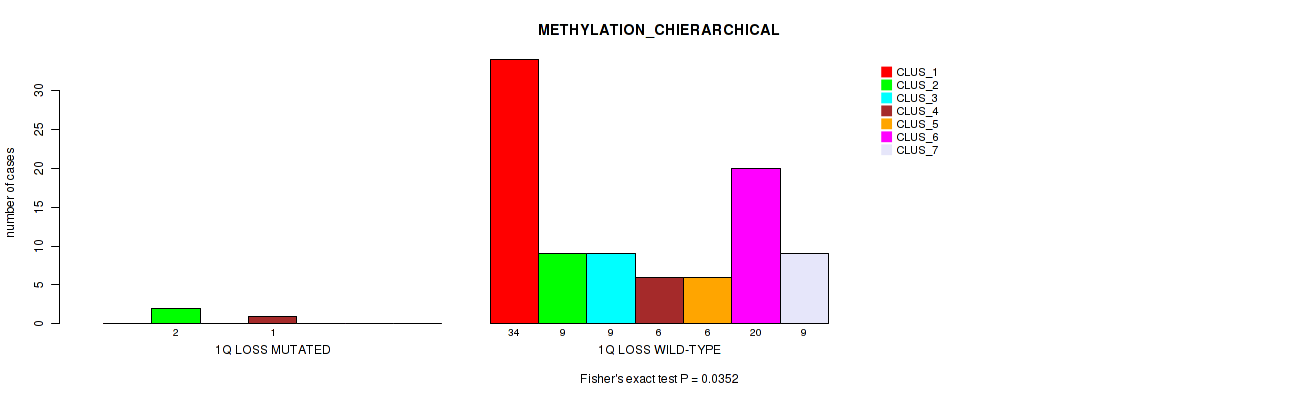
P value = 0.0327 (Fisher's exact test), Q value = 0.12
Table S27. Gene #40: '3p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 3P LOSS MUTATED | 0 | 3 | 1 | 0 | 0 | 1 | 0 |
| 3P LOSS WILD-TYPE | 34 | 8 | 8 | 7 | 6 | 19 | 9 |
Figure S27. Get High-res Image Gene #40: '3p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
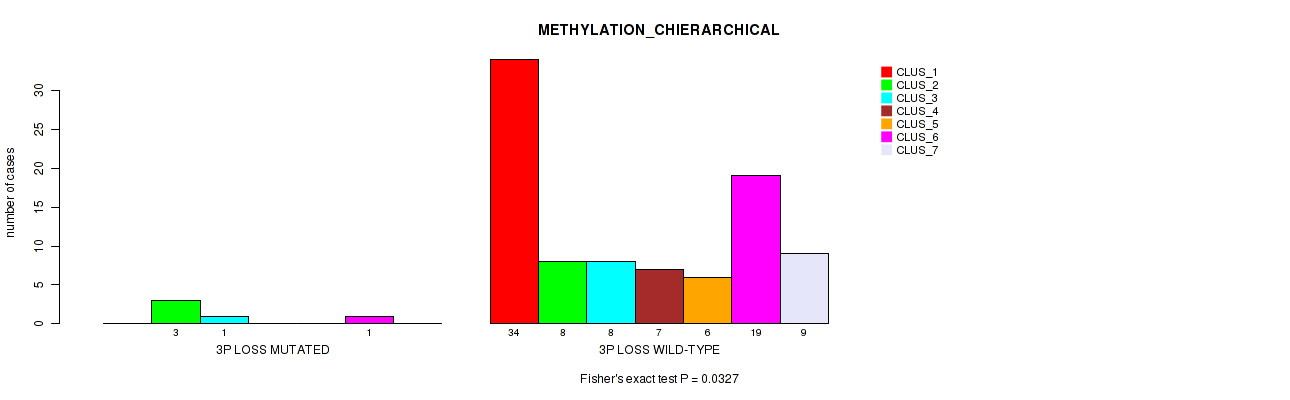
P value = 0.0154 (Fisher's exact test), Q value = 0.077
Table S28. Gene #41: '4p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 28 | 20 | 11 | 20 | 21 |
| 4P LOSS MUTATED | 5 | 0 | 0 | 6 | 1 |
| 4P LOSS WILD-TYPE | 23 | 20 | 11 | 14 | 20 |
Figure S28. Get High-res Image Gene #41: '4p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
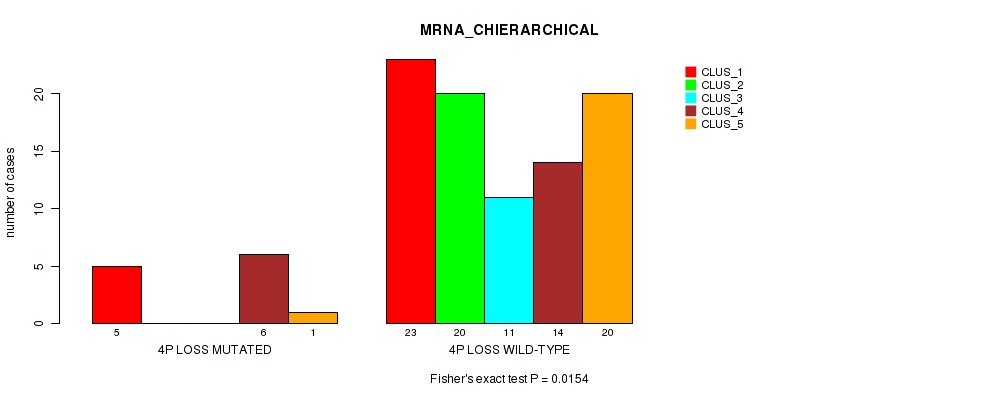
P value = 0.00913 (Fisher's exact test), Q value = 0.056
Table S29. Gene #41: '4p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 4P LOSS MUTATED | 3 | 1 | 6 | 1 |
| 4P LOSS WILD-TYPE | 32 | 32 | 10 | 11 |
Figure S29. Get High-res Image Gene #41: '4p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
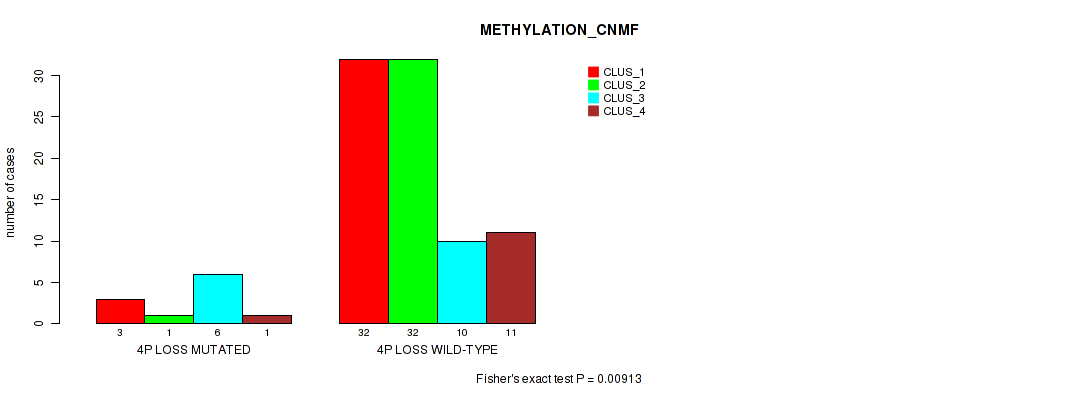
P value = 0.0176 (Fisher's exact test), Q value = 0.079
Table S30. Gene #41: '4p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 4P LOSS MUTATED | 2 | 5 | 2 | 1 | 0 | 1 | 0 |
| 4P LOSS WILD-TYPE | 32 | 6 | 7 | 6 | 6 | 19 | 9 |
Figure S30. Get High-res Image Gene #41: '4p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
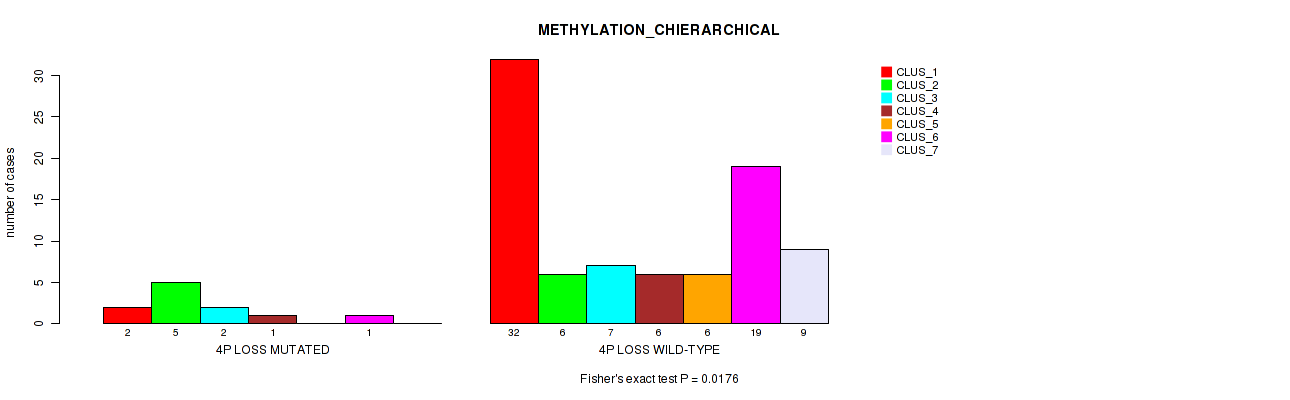
P value = 0.0151 (Fisher's exact test), Q value = 0.077
Table S31. Gene #42: '4q loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 28 | 20 | 11 | 20 | 21 |
| 4Q LOSS MUTATED | 5 | 0 | 0 | 6 | 1 |
| 4Q LOSS WILD-TYPE | 23 | 20 | 11 | 14 | 20 |
Figure S31. Get High-res Image Gene #42: '4q loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
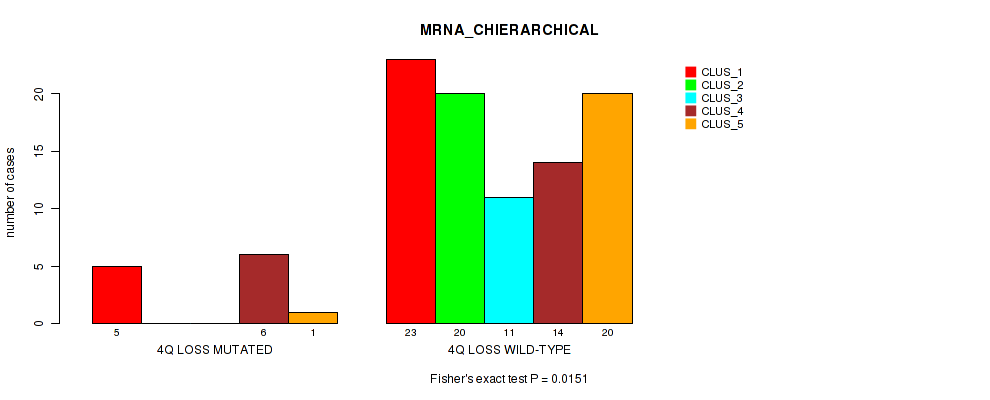
P value = 0.0085 (Fisher's exact test), Q value = 0.054
Table S32. Gene #42: '4q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 4Q LOSS MUTATED | 3 | 1 | 6 | 1 |
| 4Q LOSS WILD-TYPE | 32 | 32 | 10 | 11 |
Figure S32. Get High-res Image Gene #42: '4q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
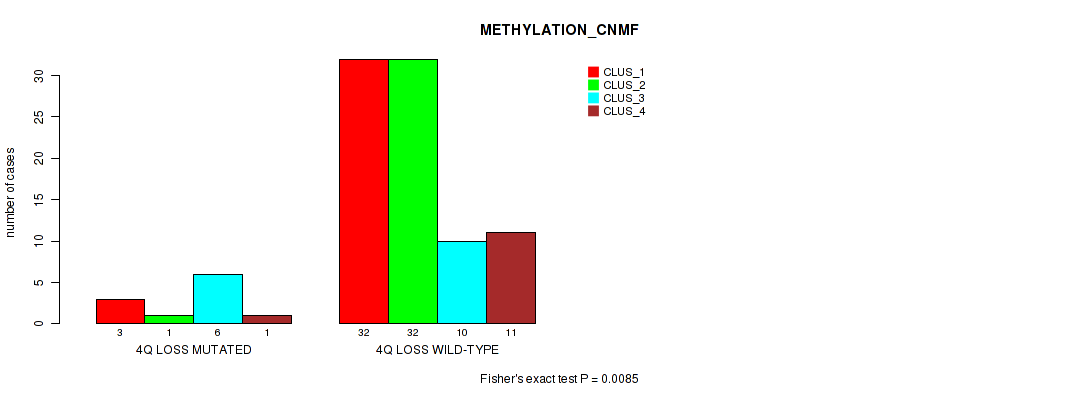
P value = 0.0165 (Fisher's exact test), Q value = 0.077
Table S33. Gene #42: '4q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 4Q LOSS MUTATED | 2 | 5 | 2 | 1 | 0 | 1 | 0 |
| 4Q LOSS WILD-TYPE | 32 | 6 | 7 | 6 | 6 | 19 | 9 |
Figure S33. Get High-res Image Gene #42: '4q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
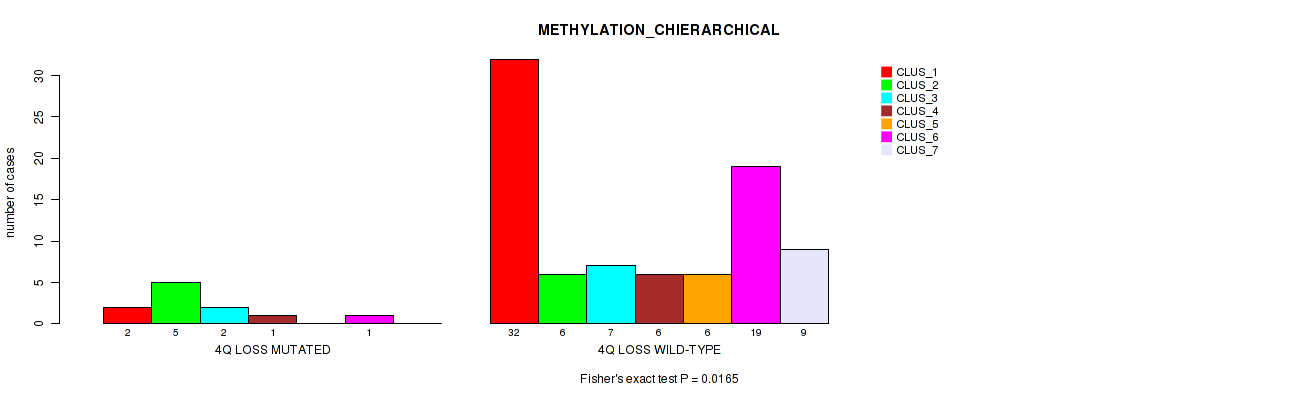
P value = 0.0215 (Fisher's exact test), Q value = 0.093
Table S34. Gene #43: '5p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 5P LOSS MUTATED | 3 | 0 | 4 | 1 |
| 5P LOSS WILD-TYPE | 32 | 33 | 12 | 11 |
Figure S34. Get High-res Image Gene #43: '5p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
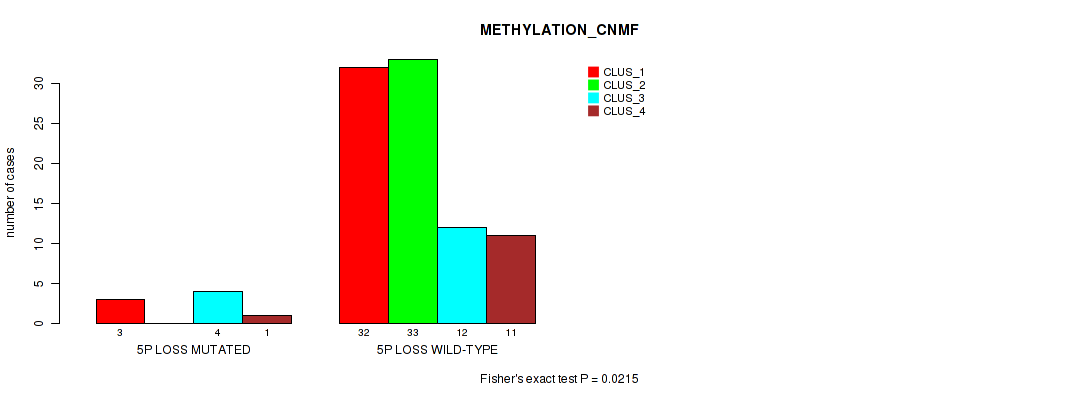
P value = 0.00609 (Fisher's exact test), Q value = 0.047
Table S35. Gene #44: '5q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 5Q LOSS MUTATED | 3 | 0 | 5 | 1 |
| 5Q LOSS WILD-TYPE | 32 | 33 | 11 | 11 |
Figure S35. Get High-res Image Gene #44: '5q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
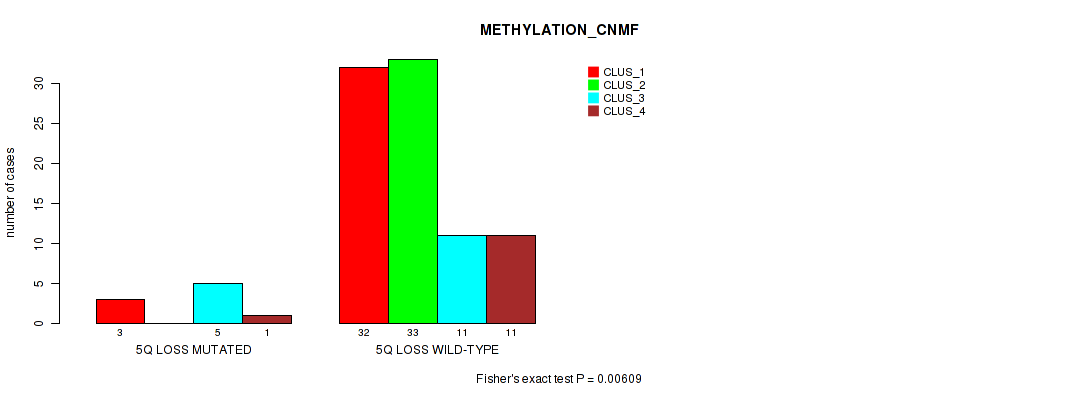
P value = 4e-05 (Fisher's exact test), Q value = 0.0036
Table S36. Gene #47: '8p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 8P LOSS MUTATED | 0 | 1 | 6 | 0 |
| 8P LOSS WILD-TYPE | 35 | 32 | 10 | 12 |
Figure S36. Get High-res Image Gene #47: '8p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
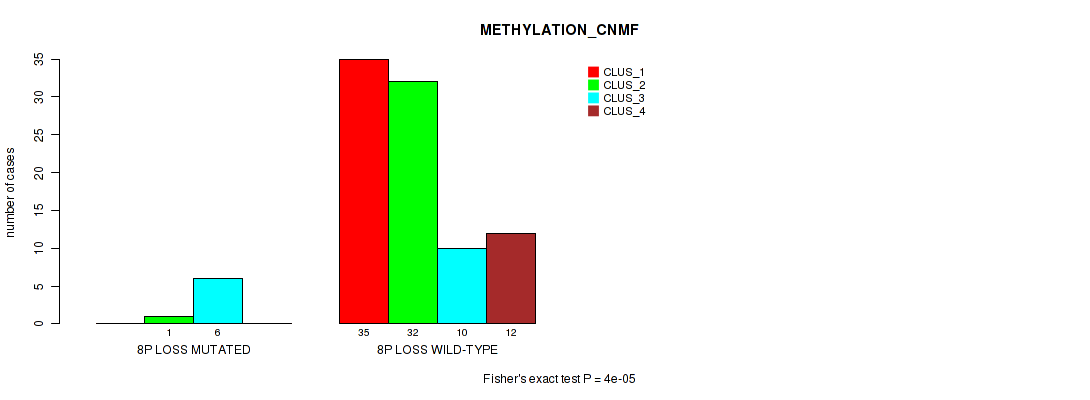
P value = 0.0301 (Fisher's exact test), Q value = 0.12
Table S37. Gene #47: '8p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 8P LOSS MUTATED | 1 | 4 | 0 | 1 | 0 | 1 | 0 |
| 8P LOSS WILD-TYPE | 33 | 7 | 9 | 6 | 6 | 19 | 9 |
Figure S37. Get High-res Image Gene #47: '8p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
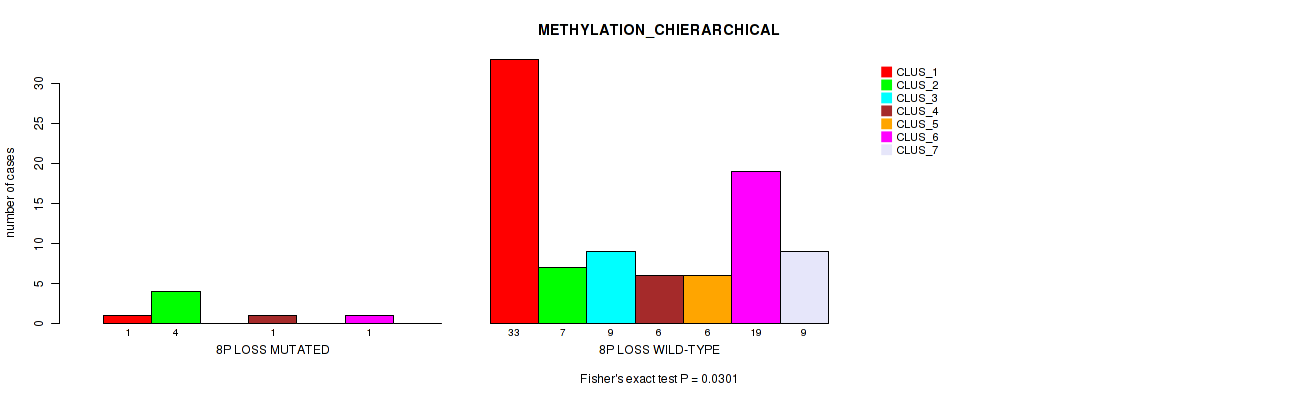
P value = 0.00069 (Fisher's exact test), Q value = 0.013
Table S38. Gene #48: '8q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 8Q LOSS MUTATED | 0 | 0 | 4 | 0 |
| 8Q LOSS WILD-TYPE | 35 | 33 | 12 | 12 |
Figure S38. Get High-res Image Gene #48: '8q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
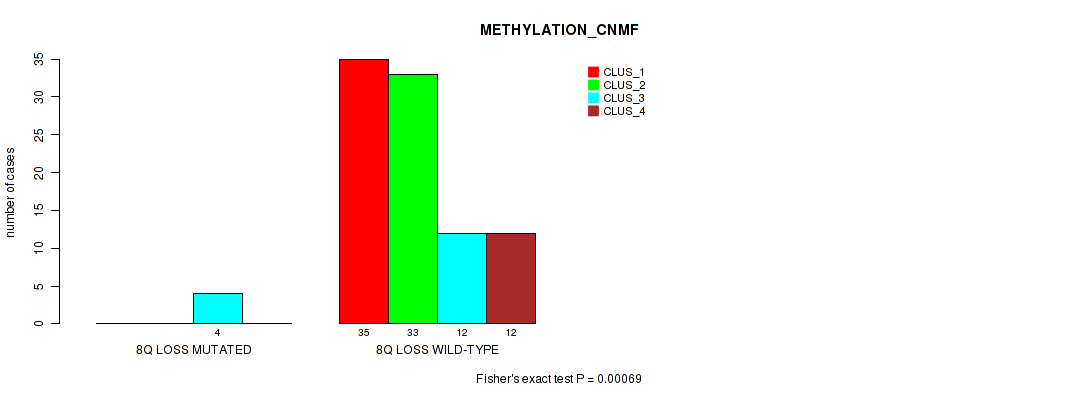
P value = 0.0144 (Fisher's exact test), Q value = 0.076
Table S39. Gene #49: '9p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 9P LOSS MUTATED | 2 | 4 | 6 | 0 |
| 9P LOSS WILD-TYPE | 33 | 29 | 10 | 12 |
Figure S39. Get High-res Image Gene #49: '9p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
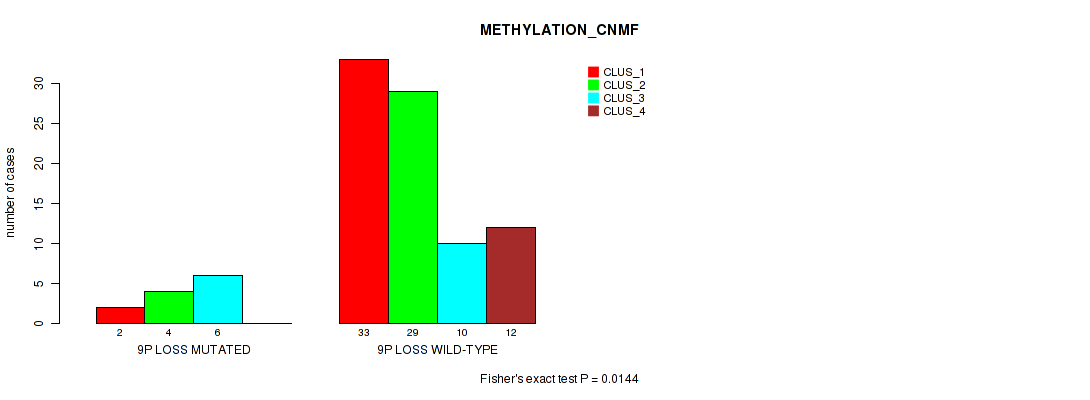
P value = 0.00249 (Fisher's exact test), Q value = 0.03
Table S40. Gene #49: '9p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 9P LOSS MUTATED | 2 | 6 | 0 | 2 | 0 | 2 | 0 |
| 9P LOSS WILD-TYPE | 32 | 5 | 9 | 5 | 6 | 18 | 9 |
Figure S40. Get High-res Image Gene #49: '9p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
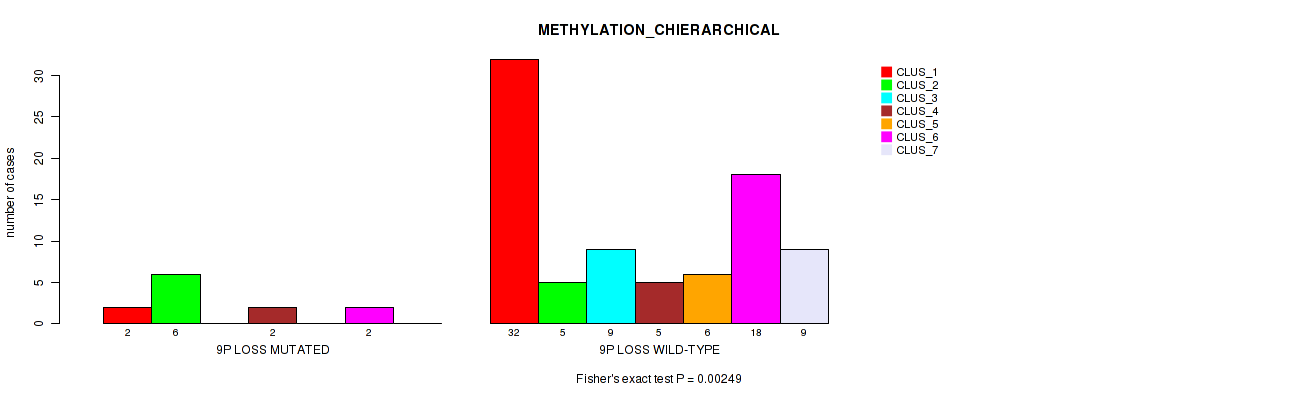
P value = 0.00733 (Fisher's exact test), Q value = 0.05
Table S41. Gene #50: '9q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 9Q LOSS MUTATED | 3 | 4 | 7 | 0 |
| 9Q LOSS WILD-TYPE | 32 | 29 | 9 | 12 |
Figure S41. Get High-res Image Gene #50: '9q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
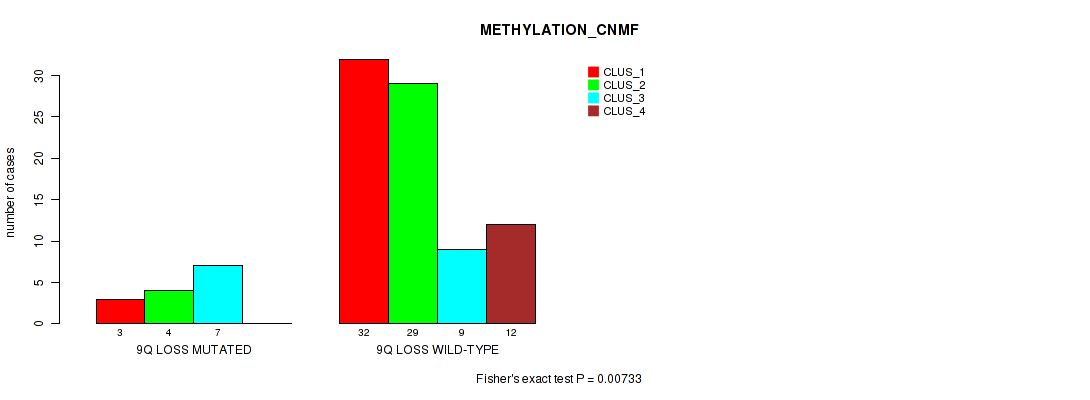
P value = 0.011 (Fisher's exact test), Q value = 0.066
Table S42. Gene #50: '9q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 9Q LOSS MUTATED | 3 | 6 | 1 | 2 | 0 | 2 | 0 |
| 9Q LOSS WILD-TYPE | 31 | 5 | 8 | 5 | 6 | 18 | 9 |
Figure S42. Get High-res Image Gene #50: '9q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
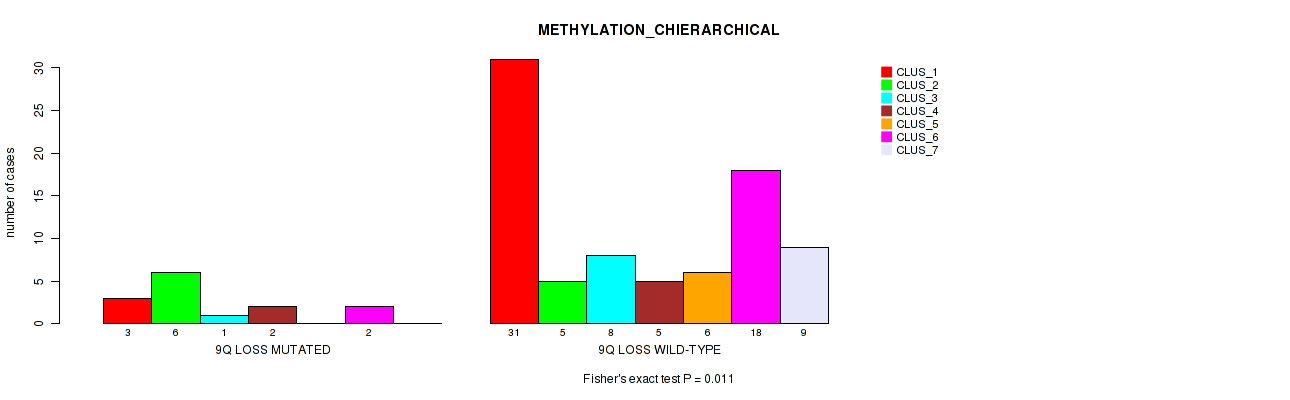
P value = 0.0357 (Fisher's exact test), Q value = 0.12
Table S43. Gene #52: '10q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 10Q LOSS MUTATED | 0 | 2 | 0 | 1 | 0 | 0 | 0 |
| 10Q LOSS WILD-TYPE | 34 | 9 | 9 | 6 | 6 | 20 | 9 |
Figure S43. Get High-res Image Gene #52: '10q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
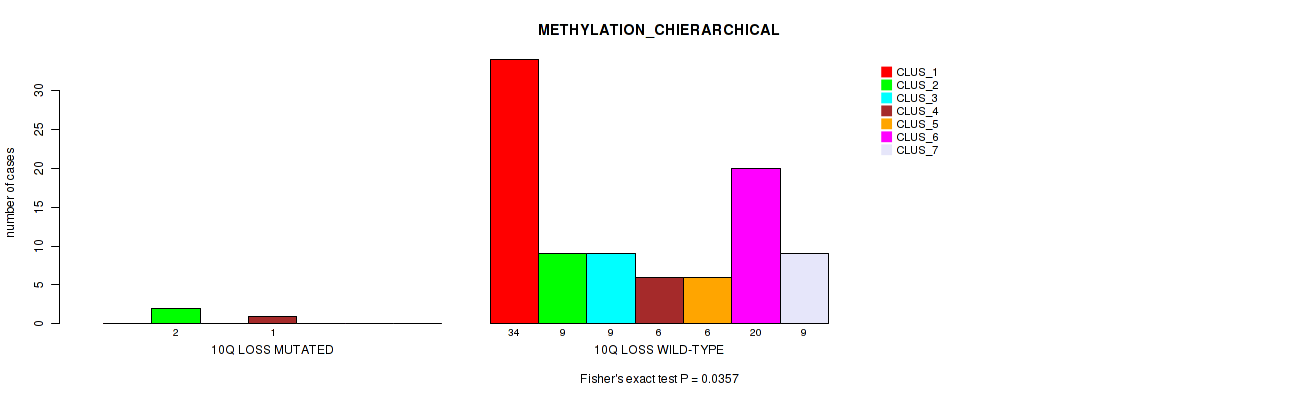
P value = 0.0128 (Fisher's exact test), Q value = 0.073
Table S44. Gene #53: '11p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 11P LOSS MUTATED | 2 | 0 | 4 | 1 |
| 11P LOSS WILD-TYPE | 33 | 33 | 12 | 11 |
Figure S44. Get High-res Image Gene #53: '11p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
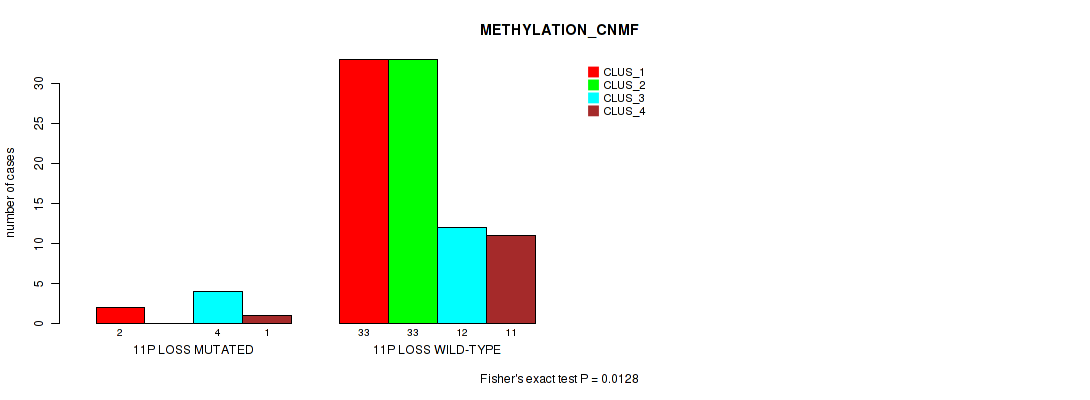
P value = 0.0131 (Fisher's exact test), Q value = 0.073
Table S45. Gene #54: '11q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 11Q LOSS MUTATED | 2 | 0 | 4 | 1 |
| 11Q LOSS WILD-TYPE | 33 | 33 | 12 | 11 |
Figure S45. Get High-res Image Gene #54: '11q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
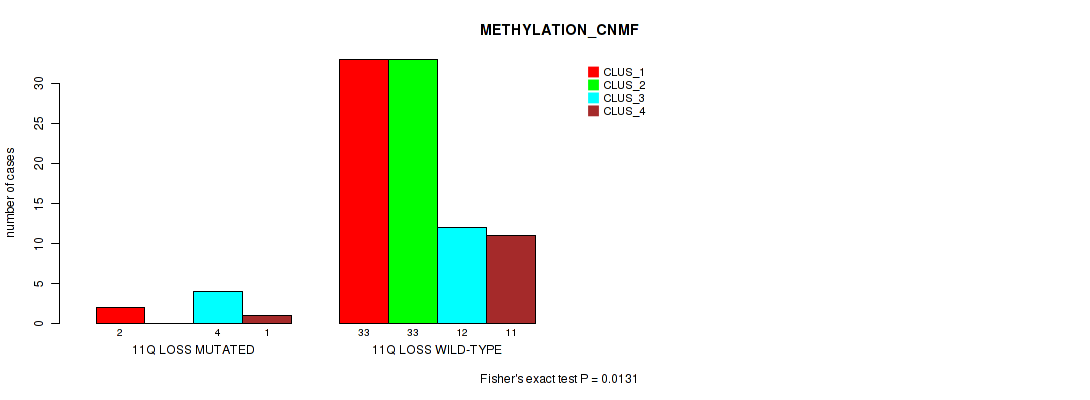
P value = 0.0159 (Fisher's exact test), Q value = 0.077
Table S46. Gene #55: '12p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 12P LOSS MUTATED | 0 | 1 | 3 | 0 |
| 12P LOSS WILD-TYPE | 35 | 32 | 13 | 12 |
Figure S46. Get High-res Image Gene #55: '12p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
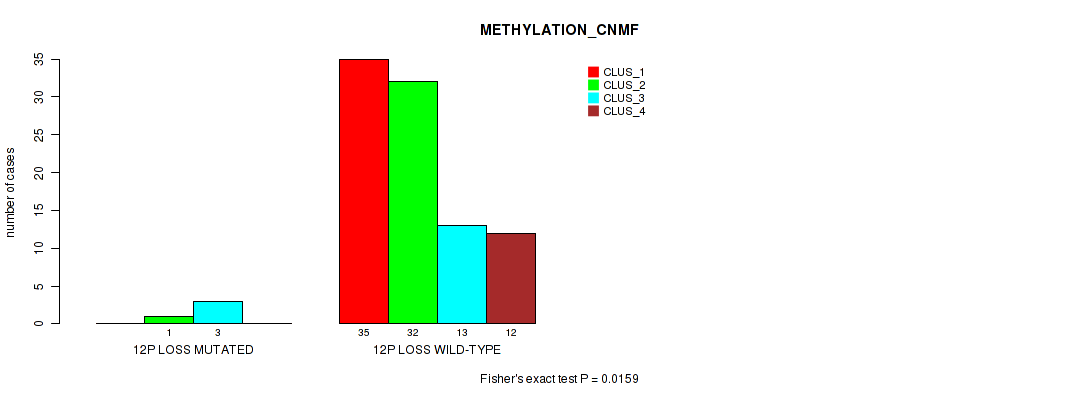
P value = 0.00651 (Fisher's exact test), Q value = 0.048
Table S47. Gene #55: '12p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 12P LOSS MUTATED | 0 | 2 | 0 | 2 | 0 | 0 | 0 |
| 12P LOSS WILD-TYPE | 34 | 9 | 9 | 5 | 6 | 20 | 9 |
Figure S47. Get High-res Image Gene #55: '12p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
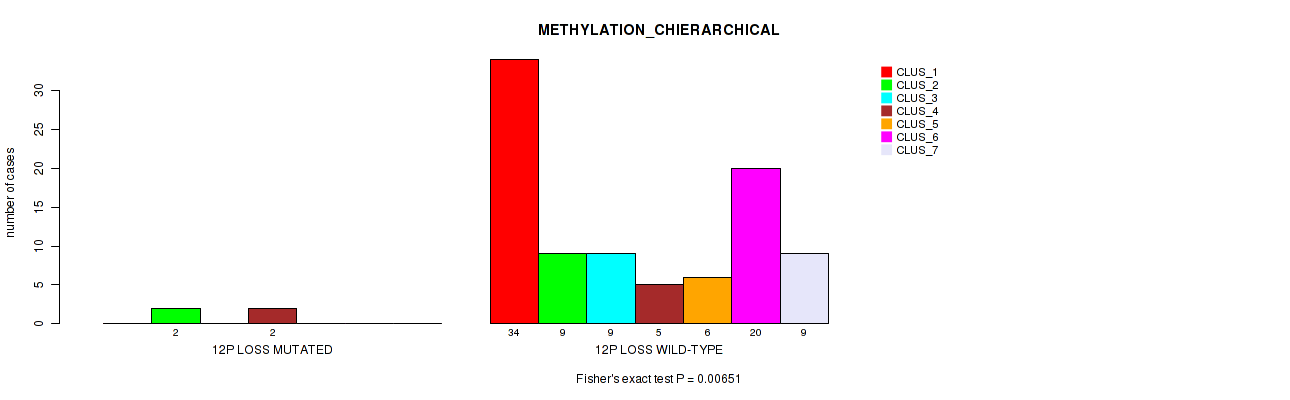
P value = 0.0163 (Fisher's exact test), Q value = 0.077
Table S48. Gene #56: '12q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 12Q LOSS MUTATED | 0 | 1 | 3 | 0 |
| 12Q LOSS WILD-TYPE | 35 | 32 | 13 | 12 |
Figure S48. Get High-res Image Gene #56: '12q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
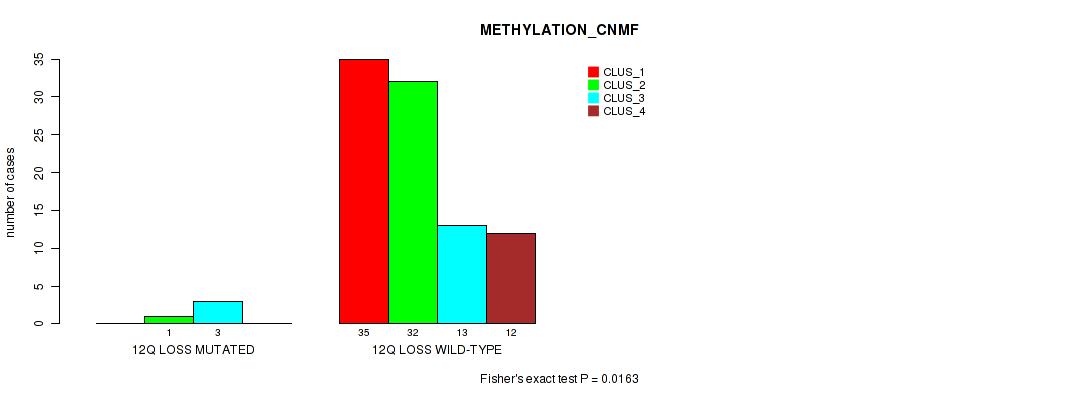
P value = 0.00677 (Fisher's exact test), Q value = 0.049
Table S49. Gene #56: '12q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 12Q LOSS MUTATED | 0 | 2 | 0 | 2 | 0 | 0 | 0 |
| 12Q LOSS WILD-TYPE | 34 | 9 | 9 | 5 | 6 | 20 | 9 |
Figure S49. Get High-res Image Gene #56: '12q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
P value = 0.00123 (Fisher's exact test), Q value = 0.02
Table S50. Gene #58: '15q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 15Q LOSS MUTATED | 2 | 2 | 7 | 0 |
| 15Q LOSS WILD-TYPE | 33 | 31 | 9 | 12 |
Figure S50. Get High-res Image Gene #58: '15q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
P value = 1e-04 (Fisher's exact test), Q value = 0.0054
Table S51. Gene #58: '15q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 15Q LOSS MUTATED | 2 | 7 | 1 | 1 | 0 | 0 | 0 |
| 15Q LOSS WILD-TYPE | 32 | 4 | 8 | 6 | 6 | 20 | 9 |
Figure S51. Get High-res Image Gene #58: '15q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
P value = 0.00425 (Fisher's exact test), Q value = 0.038
Table S52. Gene #59: '16p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 16P LOSS MUTATED | 1 | 0 | 4 | 0 |
| 16P LOSS WILD-TYPE | 34 | 33 | 12 | 12 |
Figure S52. Get High-res Image Gene #59: '16p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
P value = 0.0455 (Fisher's exact test), Q value = 0.15
Table S53. Gene #59: '16p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 16P LOSS MUTATED | 1 | 3 | 0 | 1 | 0 | 0 | 0 |
| 16P LOSS WILD-TYPE | 33 | 8 | 9 | 6 | 6 | 20 | 9 |
Figure S53. Get High-res Image Gene #59: '16p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
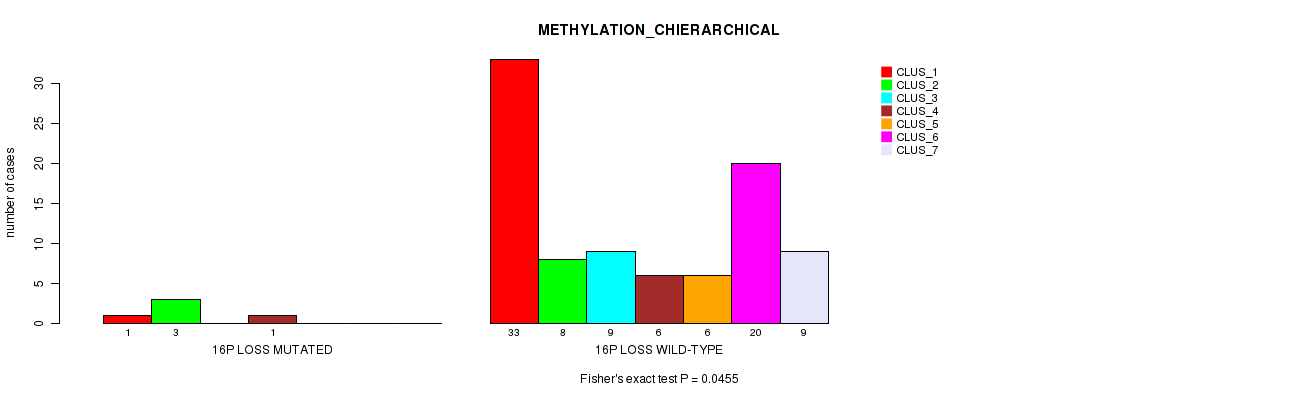
P value = 0.00069 (Fisher's exact test), Q value = 0.013
Table S54. Gene #60: '16q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 16Q LOSS MUTATED | 1 | 0 | 5 | 0 |
| 16Q LOSS WILD-TYPE | 34 | 33 | 11 | 12 |
Figure S54. Get High-res Image Gene #60: '16q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
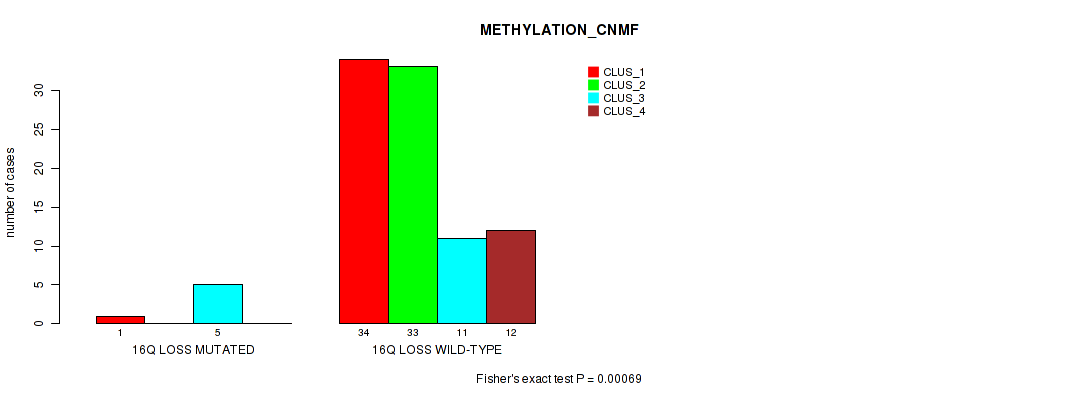
P value = 0.0144 (Fisher's exact test), Q value = 0.076
Table S55. Gene #60: '16q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 16Q LOSS MUTATED | 1 | 3 | 0 | 2 | 0 | 0 | 0 |
| 16Q LOSS WILD-TYPE | 33 | 8 | 9 | 5 | 6 | 20 | 9 |
Figure S55. Get High-res Image Gene #60: '16q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
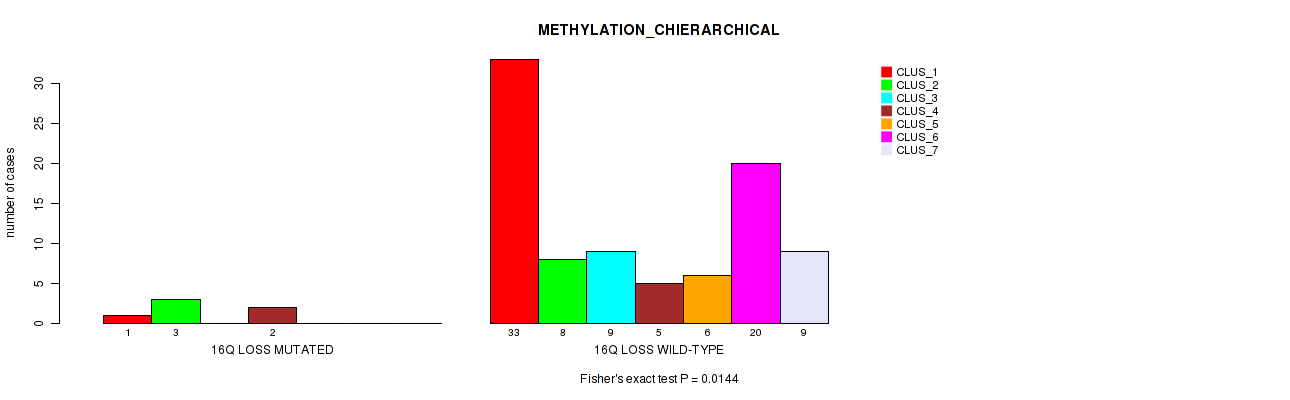
P value = 0.00232 (Fisher's exact test), Q value = 0.03
Table S56. Gene #61: '17p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 17P LOSS MUTATED | 3 | 2 | 7 | 0 |
| 17P LOSS WILD-TYPE | 32 | 31 | 9 | 12 |
Figure S56. Get High-res Image Gene #61: '17p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
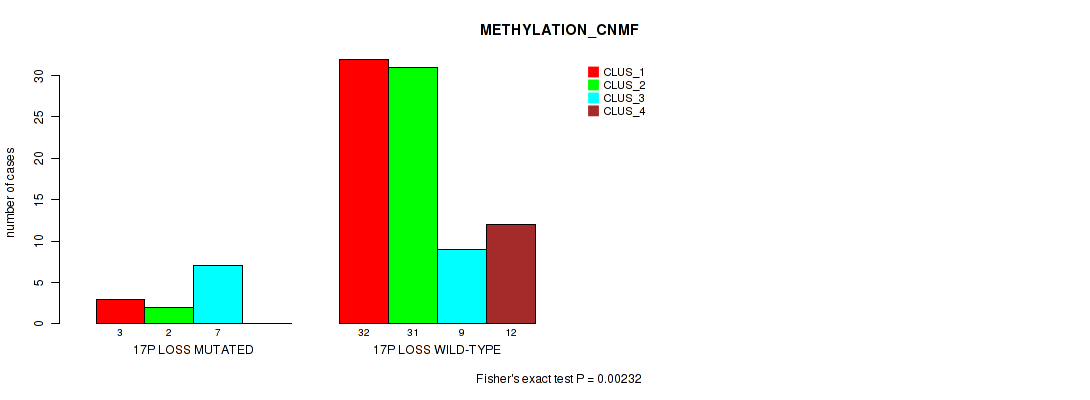
P value = 0.00027 (Fisher's exact test), Q value = 0.011
Table S57. Gene #61: '17p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 17P LOSS MUTATED | 3 | 7 | 1 | 1 | 0 | 0 | 0 |
| 17P LOSS WILD-TYPE | 31 | 4 | 8 | 6 | 6 | 20 | 9 |
Figure S57. Get High-res Image Gene #61: '17p loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
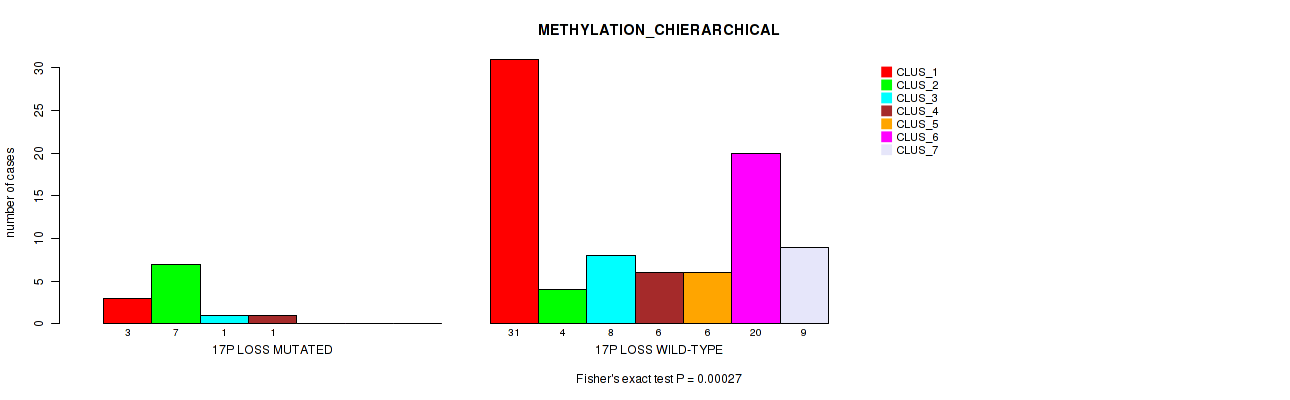
P value = 0.00238 (Fisher's exact test), Q value = 0.03
Table S58. Gene #62: '17q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 17Q LOSS MUTATED | 3 | 0 | 5 | 0 |
| 17Q LOSS WILD-TYPE | 32 | 33 | 11 | 12 |
Figure S58. Get High-res Image Gene #62: '17q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
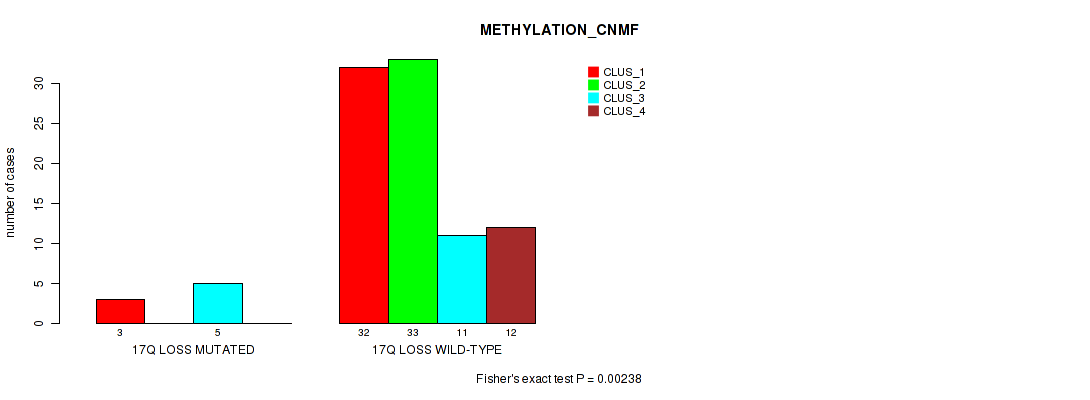
P value = 0.0223 (Fisher's exact test), Q value = 0.094
Table S59. Gene #63: '18p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 18P LOSS MUTATED | 1 | 0 | 3 | 0 |
| 18P LOSS WILD-TYPE | 34 | 33 | 13 | 12 |
Figure S59. Get High-res Image Gene #63: '18p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
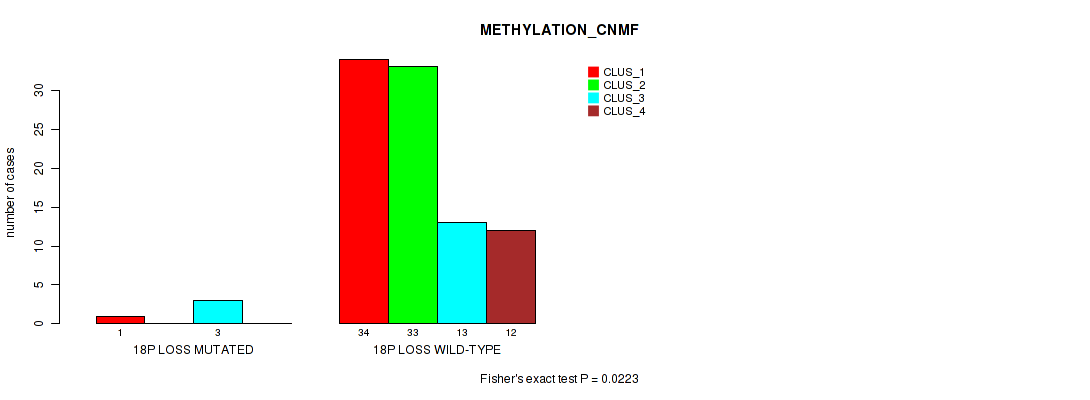
P value = 0.00294 (Fisher's exact test), Q value = 0.032
Table S60. Gene #65: '19p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 19P LOSS MUTATED | 0 | 1 | 4 | 0 |
| 19P LOSS WILD-TYPE | 35 | 32 | 12 | 12 |
Figure S60. Get High-res Image Gene #65: '19p loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
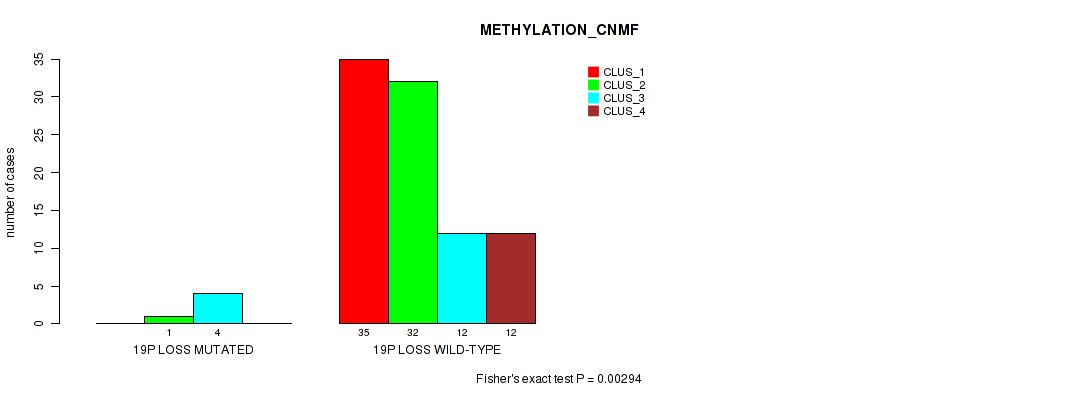
P value = 0.0231 (Fisher's exact test), Q value = 0.096
Table S61. Gene #66: '19q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 19Q LOSS MUTATED | 1 | 1 | 4 | 0 |
| 19Q LOSS WILD-TYPE | 34 | 32 | 12 | 12 |
Figure S61. Get High-res Image Gene #66: '19q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
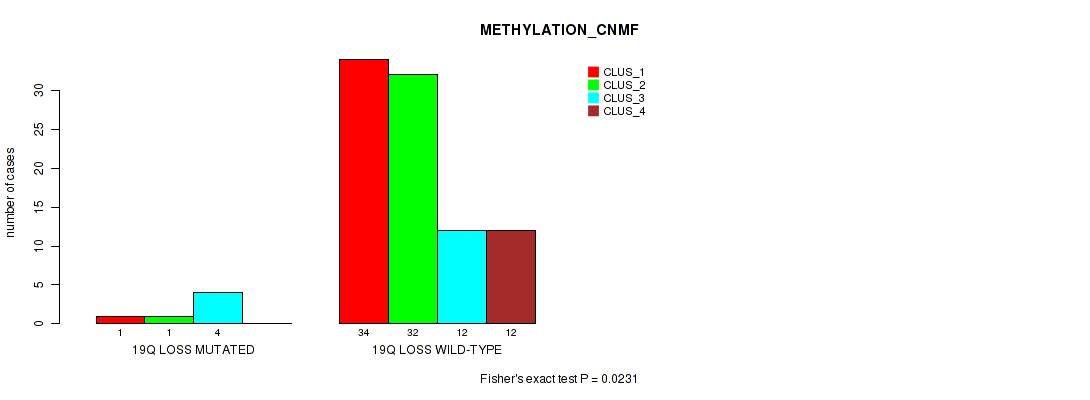
P value = 0.0469 (Fisher's exact test), Q value = 0.15
Table S62. Gene #68: '22q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| 22Q LOSS MUTATED | 4 | 2 | 5 | 0 |
| 22Q LOSS WILD-TYPE | 31 | 31 | 11 | 12 |
Figure S62. Get High-res Image Gene #68: '22q loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
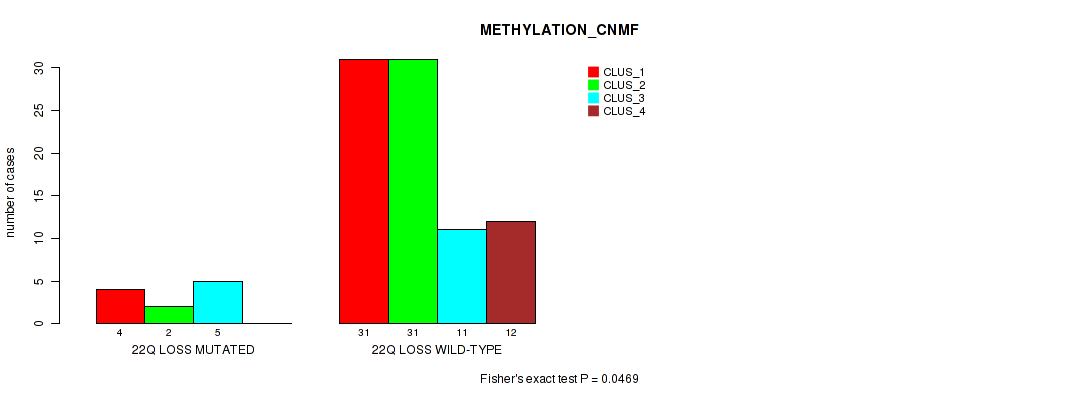
P value = 0.0169 (Fisher's exact test), Q value = 0.078
Table S63. Gene #68: '22q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| 22Q LOSS MUTATED | 4 | 5 | 1 | 1 | 0 | 0 | 0 |
| 22Q LOSS WILD-TYPE | 30 | 6 | 8 | 6 | 6 | 20 | 9 |
Figure S63. Get High-res Image Gene #68: '22q loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
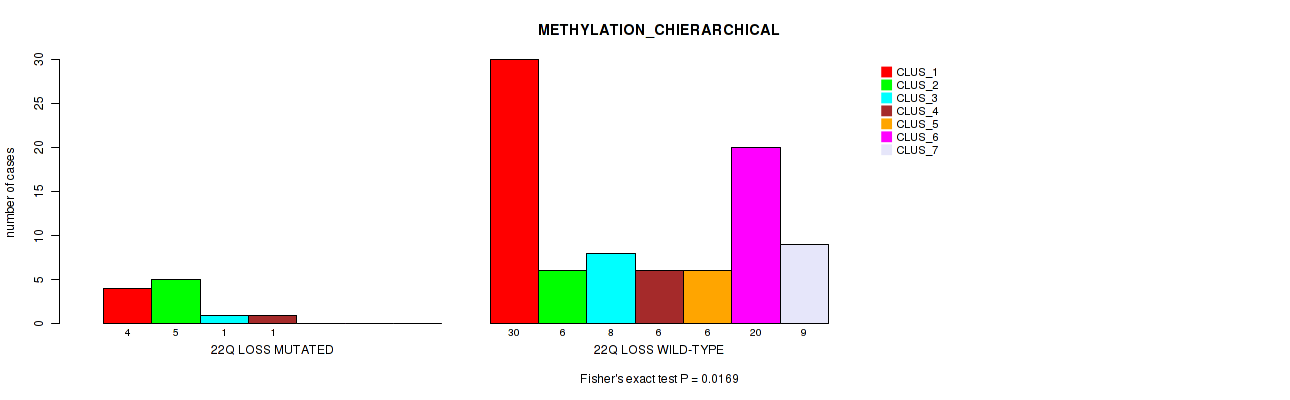
P value = 0.00046 (Fisher's exact test), Q value = 0.013
Table S64. Gene #69: 'xp loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| XP LOSS MUTATED | 3 | 5 | 9 | 0 |
| XP LOSS WILD-TYPE | 32 | 28 | 7 | 12 |
Figure S64. Get High-res Image Gene #69: 'xp loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
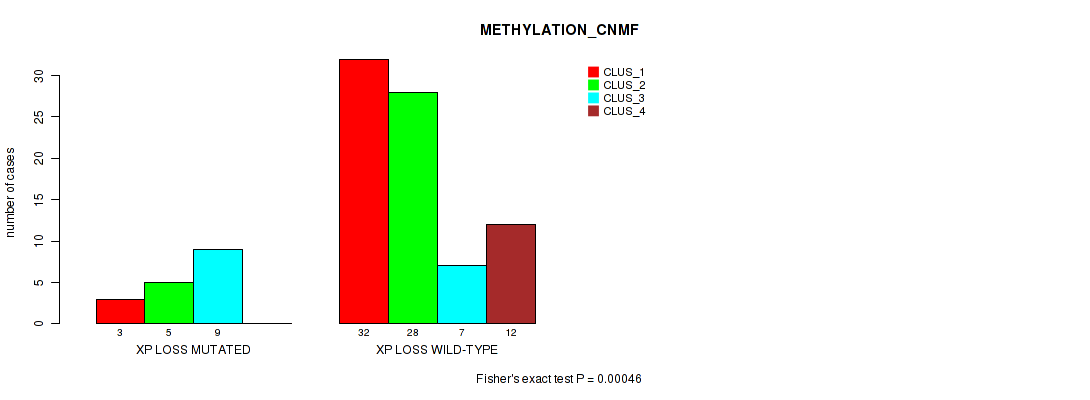
P value = 0.00249 (Fisher's exact test), Q value = 0.03
Table S65. Gene #69: 'xp loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| XP LOSS MUTATED | 3 | 6 | 1 | 3 | 0 | 1 | 3 |
| XP LOSS WILD-TYPE | 31 | 5 | 8 | 4 | 6 | 19 | 6 |
Figure S65. Get High-res Image Gene #69: 'xp loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
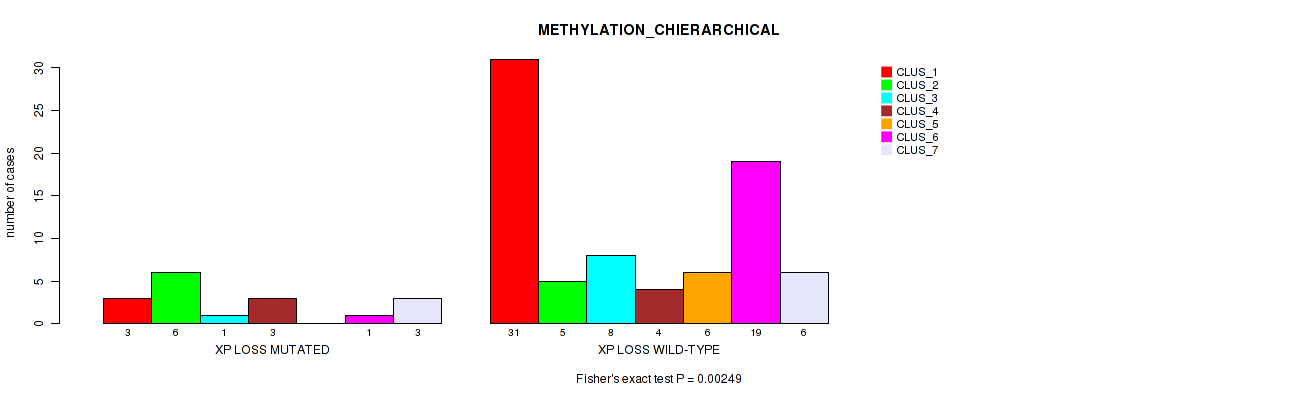
P value = 3e-04 (Fisher's exact test), Q value = 0.011
Table S66. Gene #70: 'xq loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 33 | 16 | 12 |
| XQ LOSS MUTATED | 3 | 5 | 9 | 0 |
| XQ LOSS WILD-TYPE | 32 | 28 | 7 | 12 |
Figure S66. Get High-res Image Gene #70: 'xq loss' versus Molecular Subtype #2: 'METHYLATION_CNMF'
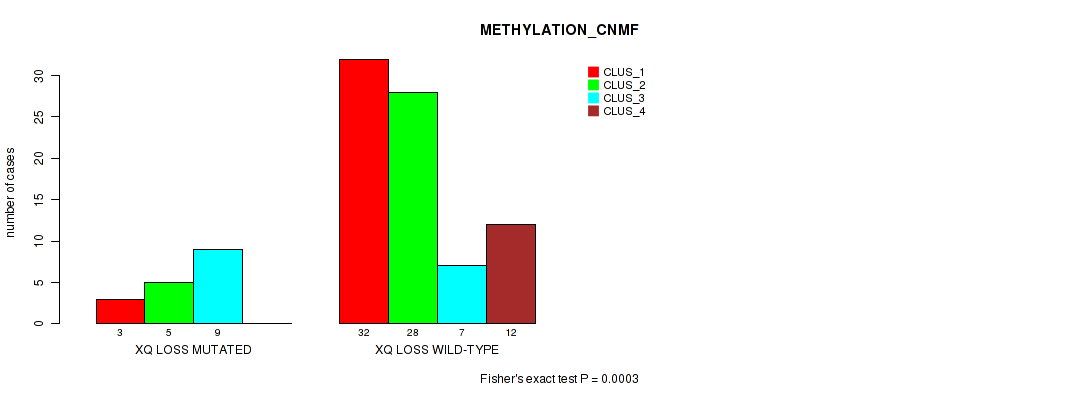
P value = 0.00286 (Fisher's exact test), Q value = 0.032
Table S67. Gene #70: 'xq loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 34 | 11 | 9 | 7 | 6 | 20 | 9 |
| XQ LOSS MUTATED | 3 | 6 | 1 | 3 | 0 | 1 | 3 |
| XQ LOSS WILD-TYPE | 31 | 5 | 8 | 4 | 6 | 19 | 6 |
Figure S67. Get High-res Image Gene #70: 'xq loss' versus Molecular Subtype #3: 'METHYLATION_CHIERARCHICAL'
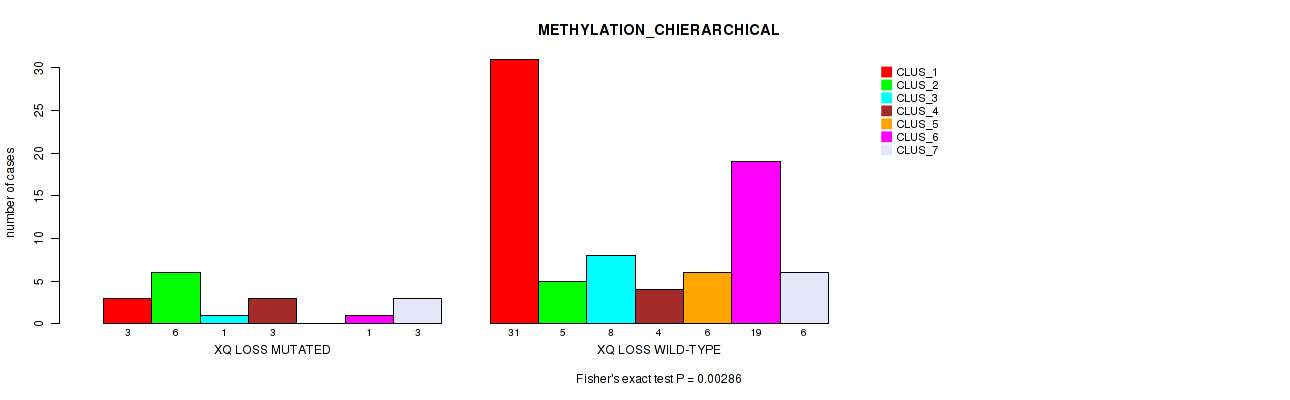
-
Copy number data file = broad_values_by_arm.txt from GISTIC pipeline
-
Processed Copy number data file = /cromwell_root/fc-8b2df640-93e1-40a2-b735-5b7a14ef6398/7d40bb96-d50a-43da-8092-a511a1314618/correlate_genomic_events_all/f2d23ad8-12fc-428e-bd64-52ab7a789763/call-preprocess_genomic_event/transformed.cor.cli.txt
-
Molecular subtypes file = /cromwell_root/fc-8b2df640-93e1-40a2-b735-5b7a14ef6398/10d02b45-07ea-40cc-ac82-013e5f89ebb2/aggregate_clusters_workflow/b0f8dcb2-0993-4424-ad42-552c0bd1a2d5/call-aggregate_clusters/CPTAC3-UCEC-TP.transposedmergedcluster.txt
-
Number of patients = 100
-
Number of significantly arm-level cnvs = 72
-
Number of molecular subtypes = 3
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.