This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and selected clinical features.
Testing the association between subtypes identified by 80 different clustering approaches and 21 clinical features across 4563 patients, 329 significant findings detected with Q value < 0.25.
-
2 subtypes identified in current cancer cohort by '1p gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', and 'PATHOLOGY.T'.
-
2 subtypes identified in current cancer cohort by '1q gain mutation analysis'. These subtypes correlate to 'Time to Death', 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', and 'PATHOLOGY.T'.
-
2 subtypes identified in current cancer cohort by '2p gain mutation analysis'. These subtypes correlate to 'AGE', 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'RADIATIONS.RADIATION.REGIMENINDICATION', and 'NEOPLASM.DISEASESTAGE'.
-
2 subtypes identified in current cancer cohort by '2q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '3p gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'TUMOR.STAGE', and 'NEOPLASM.DISEASESTAGE'.
-
2 subtypes identified in current cancer cohort by '3q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'KARNOFSKY.PERFORMANCE.SCORE', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'PATHOLOGY.N', 'TUMOR.STAGE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '4p gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '4q gain mutation analysis'. These subtypes do not correlate to any clinical features.
-
2 subtypes identified in current cancer cohort by '5p gain mutation analysis'. These subtypes correlate to 'AGE', 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '5q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '6p gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by '6q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by '7p gain mutation analysis'. These subtypes correlate to 'Time to Death', 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'PATHOLOGICSPREAD(M)', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '7q gain mutation analysis'. These subtypes correlate to 'Time to Death', 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'PATHOLOGICSPREAD(M)', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '8p gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'PATHOLOGY.T'.
-
2 subtypes identified in current cancer cohort by '8q gain mutation analysis'. These subtypes correlate to 'Time to Death', 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'PATHOLOGY.N', 'TUMOR.STAGE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '9p gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'PATHOLOGY.T'.
-
2 subtypes identified in current cancer cohort by '9q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by '10p gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'TUMOR.STAGE', and 'TOBACCOSMOKINGHISTORYINDICATOR'.
-
2 subtypes identified in current cancer cohort by '10q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by '11p gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '11q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'STOPPEDSMOKINGYEAR'.
-
2 subtypes identified in current cancer cohort by '12p gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'TUMOR.STAGE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '12q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '13q gain mutation analysis'. These subtypes correlate to 'AGE', 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'PATHOLOGY.N', 'PATHOLOGICSPREAD(M)', 'TUMOR.STAGE', 'RADIATIONS.RADIATION.REGIMENINDICATION', 'TOBACCOSMOKINGHISTORYINDICATOR', and 'COMPLETENESS.OF.RESECTION'.
-
2 subtypes identified in current cancer cohort by '14q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '15q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'PATHOLOGY.T'.
-
2 subtypes identified in current cancer cohort by '16p gain mutation analysis'. These subtypes correlate to 'Time to Death', 'PRIMARY.SITE.OF.DISEASE', 'GENDER', and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '16q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '17p gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '17q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '18p gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '18q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'PATHOLOGY.T'.
-
2 subtypes identified in current cancer cohort by '19p gain mutation analysis'. These subtypes correlate to 'Time to Death', 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '19q gain mutation analysis'. These subtypes correlate to 'Time to Death', 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '20p gain mutation analysis'. These subtypes correlate to 'Time to Death', 'AGE', 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'PATHOLOGICSPREAD(M)', 'TUMOR.STAGE', and 'COMPLETENESS.OF.RESECTION'.
-
2 subtypes identified in current cancer cohort by '20q gain mutation analysis'. These subtypes correlate to 'AGE', 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'TUMOR.STAGE', 'TOBACCOSMOKINGHISTORYINDICATOR', and 'COMPLETENESS.OF.RESECTION'.
-
2 subtypes identified in current cancer cohort by '21q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'TOBACCOSMOKINGHISTORYINDICATOR'.
-
2 subtypes identified in current cancer cohort by '22q gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', and 'PATHOLOGY.T'.
-
2 subtypes identified in current cancer cohort by 'Xq gain mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE'.
-
2 subtypes identified in current cancer cohort by '1p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'NEOPLASM.DISEASESTAGE'.
-
2 subtypes identified in current cancer cohort by '1q loss mutation analysis'. These subtypes do not correlate to any clinical features.
-
2 subtypes identified in current cancer cohort by '2p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'GENDER'.
-
2 subtypes identified in current cancer cohort by '2q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'TOBACCOSMOKINGHISTORYINDICATOR'.
-
2 subtypes identified in current cancer cohort by '3p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'PATHOLOGICSPREAD(M)', 'TUMOR.STAGE', 'RADIATIONS.RADIATION.REGIMENINDICATION', and 'TOBACCOSMOKINGHISTORYINDICATOR'.
-
2 subtypes identified in current cancer cohort by '3q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '4p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.N', 'TUMOR.STAGE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '4q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'TUMOR.STAGE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '5p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by '5q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by '6p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '6q loss mutation analysis'. These subtypes correlate to 'AGE', 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by '7p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by '7q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', and 'PATHOLOGY.T'.
-
2 subtypes identified in current cancer cohort by '8p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'TUMOR.STAGE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '8q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by '9p loss mutation analysis'. These subtypes correlate to 'Time to Death', 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'PATHOLOGY.T'.
-
2 subtypes identified in current cancer cohort by '9q loss mutation analysis'. These subtypes correlate to 'Time to Death', 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by '10p loss mutation analysis'. These subtypes correlate to 'Time to Death', 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '10q loss mutation analysis'. These subtypes correlate to 'Time to Death', 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '11p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.N', 'TUMOR.STAGE', and 'TOBACCOSMOKINGHISTORYINDICATOR'.
-
2 subtypes identified in current cancer cohort by '11q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by '12p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '12q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '13q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by '14q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'PATHOLOGY.N', and 'PATHOLOGICSPREAD(M)'.
-
2 subtypes identified in current cancer cohort by '15q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by '16p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'TUMOR.STAGE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '16q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'TUMOR.STAGE', 'COMPLETENESS.OF.RESECTION', and 'NEOPLASM.DISEASESTAGE'.
-
2 subtypes identified in current cancer cohort by '17p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'TUMOR.STAGE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '17q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', 'TUMOR.STAGE', 'RADIATIONS.RADIATION.REGIMENINDICATION', and 'COMPLETENESS.OF.RESECTION'.
-
2 subtypes identified in current cancer cohort by '18p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'PATHOLOGICSPREAD(M)', 'TUMOR.STAGE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '18q loss mutation analysis'. These subtypes correlate to 'AGE', 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'PATHOLOGY.N', 'PATHOLOGICSPREAD(M)', 'TUMOR.STAGE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '19p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'PATHOLOGY.T', 'TUMOR.STAGE', and 'RADIATIONS.RADIATION.REGIMENINDICATION'.
-
2 subtypes identified in current cancer cohort by '19q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by '20p loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '20q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE' and 'HISTOLOGICAL.TYPE'.
-
2 subtypes identified in current cancer cohort by '21q loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'HISTOLOGICAL.TYPE', 'TUMOR.STAGE', 'RADIATIONS.RADIATION.REGIMENINDICATION', and 'STOPPEDSMOKINGYEAR'.
-
2 subtypes identified in current cancer cohort by '22q loss mutation analysis'. These subtypes correlate to 'Time to Death', 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', and 'TUMOR.STAGE'.
-
2 subtypes identified in current cancer cohort by 'Xq loss mutation analysis'. These subtypes correlate to 'PRIMARY.SITE.OF.DISEASE', 'GENDER', 'HISTOLOGICAL.TYPE', and 'TUMOR.STAGE'.
Table 1. Get Full Table Overview of the association between subtypes identified by 80 different clustering approaches and 21 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 329 significant findings detected.
|
Clinical Features |
Time to Death |
AGE |
PRIMARY SITE OF DISEASE |
GENDER |
KARNOFSKY PERFORMANCE SCORE |
HISTOLOGICAL TYPE |
PATHOLOGY T |
PATHOLOGY N |
PATHOLOGICSPREAD(M) |
TUMOR STAGE |
RADIATIONS RADIATION REGIMENINDICATION |
NUMBERPACKYEARSSMOKED | STOPPEDSMOKINGYEAR | TOBACCOSMOKINGHISTORYINDICATOR | YEAROFTOBACCOSMOKINGONSET |
DISTANT METASTASIS |
LYMPH NODE METASTASIS |
COMPLETENESS OF RESECTION |
NUMBER OF LYMPH NODES |
TUMOR STAGECODE |
NEOPLASM DISEASESTAGE |
| Statistical Tests | logrank test | t-test | Chi-square test | Fisher's exact test | t-test | Chi-square test | Chi-square test | Chi-square test | Chi-square test | Chi-square test | Fisher's exact test | t-test | t-test | Chi-square test | t-test | Chi-square test | Chi-square test | Chi-square test | t-test | t-test | Chi-square test |
| 1p gain |
0.212 (1.00) |
0.0112 (1.00) |
1.5e-28 (2.24e-25) |
8.6e-07 (0.00116) |
0.457 (1.00) |
1.45e-29 (2.17e-26) |
0.000161 (0.205) |
0.0519 (1.00) |
0.253 (1.00) |
0.312 (1.00) |
0.568 (1.00) |
0.674 (1.00) |
0.726 (1.00) |
0.389 (1.00) |
0.669 (1.00) |
0.935 (1.00) |
0.915 (1.00) |
0.0161 (1.00) |
0.545 (1.00) |
0.315 (1.00) |
|
| 1q gain |
2.04e-10 (2.88e-07) |
0.841 (1.00) |
3.09e-135 (4.86e-132) |
1.89e-33 (2.85e-30) |
0.692 (1.00) |
2.19e-39 (3.33e-36) |
1.65e-05 (0.0217) |
0.329 (1.00) |
0.000871 (1.00) |
0.329 (1.00) |
0.965 (1.00) |
0.151 (1.00) |
0.304 (1.00) |
0.219 (1.00) |
0.71 (1.00) |
0.533 (1.00) |
0.37 (1.00) |
0.328 (1.00) |
0.229 (1.00) |
0.000266 (0.335) |
|
| 2p gain |
0.868 (1.00) |
0.000113 (0.145) |
1.25e-30 (1.88e-27) |
0.663 (1.00) |
0.011 (1.00) |
1.8e-13 (2.57e-10) |
0.0316 (1.00) |
0.0328 (1.00) |
0.845 (1.00) |
0.000208 (0.264) |
6.7e-06 (0.0089) |
0.596 (1.00) |
0.0145 (1.00) |
0.00544 (1.00) |
0.285 (1.00) |
0.576 (1.00) |
0.385 (1.00) |
0.868 (1.00) |
0.622 (1.00) |
2.47e-05 (0.0322) |
|
| 2q gain |
0.877 (1.00) |
0.000858 (1.00) |
1.17e-21 (1.73e-18) |
0.479 (1.00) |
0.785 (1.00) |
1e-08 (1.4e-05) |
0.186 (1.00) |
0.101 (1.00) |
0.627 (1.00) |
0.00618 (1.00) |
0.00163 (1.00) |
0.62 (1.00) |
0.119 (1.00) |
0.0754 (1.00) |
0.297 (1.00) |
0.501 (1.00) |
0.184 (1.00) |
0.82 (1.00) |
0.382 (1.00) |
0.0136 (1.00) |
|
| 3p gain |
0.489 (1.00) |
0.0841 (1.00) |
2.71e-30 (4.06e-27) |
0.0798 (1.00) |
0.0182 (1.00) |
6.17e-19 (9e-16) |
0.071 (1.00) |
0.336 (1.00) |
0.104 (1.00) |
7.88e-05 (0.101) |
0.0472 (1.00) |
0.817 (1.00) |
0.0811 (1.00) |
0.00405 (1.00) |
0.153 (1.00) |
0.504 (1.00) |
0.064 (1.00) |
0.0182 (1.00) |
0.587 (1.00) |
4.65e-05 (0.0604) |
|
| 3q gain |
0.152 (1.00) |
0.000438 (0.55) |
2.2e-87 (3.45e-84) |
0.837 (1.00) |
1.53e-05 (0.0201) |
1.25e-88 (1.97e-85) |
6.96e-07 (0.000944) |
9.15e-06 (0.0121) |
0.155 (1.00) |
1.34e-06 (0.00179) |
0.00013 (0.166) |
0.242 (1.00) |
0.0631 (1.00) |
0.818 (1.00) |
0.897 (1.00) |
0.407 (1.00) |
0.0233 (1.00) |
0.0631 (1.00) |
0.194 (1.00) |
0.000658 (0.82) |
|
| 4p gain |
0.244 (1.00) |
0.282 (1.00) |
1.01e-06 (0.00136) |
0.493 (1.00) |
0.0377 (1.00) |
1.31e-06 (0.00176) |
0.204 (1.00) |
0.924 (1.00) |
0.647 (1.00) |
0.277 (1.00) |
0.391 (1.00) |
0.971 (1.00) |
0.792 (1.00) |
0.508 (1.00) |
0.447 (1.00) |
0.258 (1.00) |
0.844 (1.00) |
0.388 (1.00) |
0.0778 (1.00) |
0.00579 (1.00) |
|
| 4q gain |
0.525 (1.00) |
0.364 (1.00) |
0.0341 (1.00) |
0.917 (1.00) |
0.476 (1.00) |
0.00527 (1.00) |
0.492 (1.00) |
0.962 (1.00) |
0.93 (1.00) |
0.275 (1.00) |
0.795 (1.00) |
0.748 (1.00) |
0.811 (1.00) |
0.266 (1.00) |
0.897 (1.00) |
0.378 (1.00) |
0.823 (1.00) |
0.296 (1.00) |
0.281 (1.00) |
0.0106 (1.00) |
|
| 5p gain |
0.317 (1.00) |
3.72e-05 (0.0484) |
7.92e-83 (1.24e-79) |
0.000467 (0.585) |
0.00649 (1.00) |
1.28e-52 (1.97e-49) |
8.46e-10 (1.19e-06) |
0.577 (1.00) |
0.191 (1.00) |
0.0171 (1.00) |
1.41e-08 (1.95e-05) |
0.511 (1.00) |
0.0706 (1.00) |
0.0157 (1.00) |
0.714 (1.00) |
0.732 (1.00) |
0.0942 (1.00) |
0.284 (1.00) |
0.726 (1.00) |
0.534 (1.00) |
|
| 5q gain |
0.00257 (1.00) |
0.00147 (1.00) |
9.05e-58 (1.4e-54) |
0.000301 (0.379) |
0.414 (1.00) |
4.57e-43 (6.96e-40) |
0.00575 (1.00) |
0.02 (1.00) |
0.0609 (1.00) |
0.00257 (1.00) |
0.00743 (1.00) |
0.863 (1.00) |
0.585 (1.00) |
0.764 (1.00) |
0.119 (1.00) |
0.214 (1.00) |
0.64 (1.00) |
0.368 (1.00) |
0.322 (1.00) |
0.938 (1.00) |
|
| 6p gain |
0.00365 (1.00) |
0.249 (1.00) |
1.08e-35 (1.63e-32) |
3.41e-07 (0.000465) |
0.444 (1.00) |
4.48e-22 (6.6e-19) |
0.114 (1.00) |
0.0261 (1.00) |
0.00247 (1.00) |
6.63e-07 (0.000899) |
0.00161 (1.00) |
0.783 (1.00) |
0.321 (1.00) |
0.76 (1.00) |
0.356 (1.00) |
0.706 (1.00) |
0.917 (1.00) |
0.0914 (1.00) |
0.418 (1.00) |
0.661 (1.00) |
|
| 6q gain |
0.0491 (1.00) |
0.0776 (1.00) |
1.31e-18 (1.9e-15) |
0.000741 (0.919) |
0.744 (1.00) |
2.41e-13 (3.44e-10) |
6.68e-05 (0.0861) |
0.129 (1.00) |
0.111 (1.00) |
1.85e-05 (0.0243) |
0.00972 (1.00) |
0.338 (1.00) |
0.793 (1.00) |
0.26 (1.00) |
0.249 (1.00) |
0.459 (1.00) |
0.601 (1.00) |
0.00978 (1.00) |
0.648 (1.00) |
0.697 (1.00) |
|
| 7p gain |
0 (0) |
0.0159 (1.00) |
1.14e-166 (1.8e-163) |
6.39e-27 (9.51e-24) |
0.148 (1.00) |
1.01e-51 (1.55e-48) |
1.49e-09 (2.09e-06) |
0.00457 (1.00) |
7.41e-05 (0.0954) |
0.204 (1.00) |
6.56e-14 (9.4e-11) |
0.75 (1.00) |
0.567 (1.00) |
0.92 (1.00) |
0.334 (1.00) |
0.539 (1.00) |
0.262 (1.00) |
0.00611 (1.00) |
0.0724 (1.00) |
0.44 (1.00) |
|
| 7q gain |
0 (0) |
0.106 (1.00) |
1.26e-182 (2e-179) |
8.98e-27 (1.33e-23) |
0.0433 (1.00) |
1.03e-38 (1.56e-35) |
1.08e-08 (1.5e-05) |
0.134 (1.00) |
9.32e-07 (0.00126) |
0.0824 (1.00) |
3.79e-17 (5.51e-14) |
0.372 (1.00) |
0.267 (1.00) |
0.618 (1.00) |
0.692 (1.00) |
0.0909 (1.00) |
0.164 (1.00) |
0.00826 (1.00) |
0.394 (1.00) |
0.265 (1.00) |
|
| 8p gain |
0.00356 (1.00) |
0.593 (1.00) |
4.18e-25 (6.19e-22) |
0.00149 (1.00) |
0.061 (1.00) |
7.81e-08 (0.000108) |
2.28e-06 (0.00304) |
0.0413 (1.00) |
0.737 (1.00) |
0.00782 (1.00) |
0.151 (1.00) |
0.741 (1.00) |
0.00391 (1.00) |
0.00578 (1.00) |
0.0413 (1.00) |
0.485 (1.00) |
0.781 (1.00) |
0.073 (1.00) |
0.591 (1.00) |
0.417 (1.00) |
|
| 8q gain |
0.000192 (0.244) |
0.576 (1.00) |
5.25e-86 (8.22e-83) |
9.07e-05 (0.116) |
0.00848 (1.00) |
8.16e-35 (1.23e-31) |
1.08e-14 (1.55e-11) |
1.34e-07 (0.000184) |
0.00811 (1.00) |
1.98e-09 (2.77e-06) |
7.22e-06 (0.00958) |
0.567 (1.00) |
0.0137 (1.00) |
0.0524 (1.00) |
0.167 (1.00) |
0.249 (1.00) |
0.124 (1.00) |
0.0946 (1.00) |
0.301 (1.00) |
0.138 (1.00) |
|
| 9p gain |
0.0084 (1.00) |
0.737 (1.00) |
8.2e-26 (1.22e-22) |
0.127 (1.00) |
0.382 (1.00) |
8.4e-17 (1.22e-13) |
3.08e-05 (0.0402) |
0.618 (1.00) |
0.199 (1.00) |
0.00172 (1.00) |
0.00153 (1.00) |
0.584 (1.00) |
0.668 (1.00) |
0.491 (1.00) |
0.743 (1.00) |
0.89 (1.00) |
0.19 (1.00) |
0.479 (1.00) |
0.283 (1.00) |
0.108 (1.00) |
|
| 9q gain |
0.69 (1.00) |
0.713 (1.00) |
1.21e-23 (1.79e-20) |
0.0157 (1.00) |
0.984 (1.00) |
1.32e-21 (1.94e-18) |
1.94e-07 (0.000267) |
0.603 (1.00) |
0.0581 (1.00) |
3.51e-07 (0.00048) |
0.811 (1.00) |
0.756 (1.00) |
0.304 (1.00) |
0.319 (1.00) |
0.742 (1.00) |
0.487 (1.00) |
0.212 (1.00) |
0.286 (1.00) |
0.576 (1.00) |
0.0674 (1.00) |
|
| 10p gain |
0.0283 (1.00) |
0.922 (1.00) |
1.17e-58 (1.81e-55) |
2.19e-20 (3.2e-17) |
0.915 (1.00) |
1.7e-37 (2.58e-34) |
0.0657 (1.00) |
0.213 (1.00) |
0.00992 (1.00) |
3.16e-13 (4.51e-10) |
0.123 (1.00) |
0.461 (1.00) |
0.577 (1.00) |
7.13e-05 (0.0918) |
0.409 (1.00) |
0.404 (1.00) |
0.483 (1.00) |
0.00132 (1.00) |
0.12 (1.00) |
0.00751 (1.00) |
|
| 10q gain |
0.0176 (1.00) |
0.847 (1.00) |
1.82e-46 (2.78e-43) |
3.74e-16 (5.41e-13) |
0.316 (1.00) |
3.63e-28 (5.42e-25) |
0.924 (1.00) |
0.0856 (1.00) |
0.394 (1.00) |
8.16e-07 (0.00111) |
0.8 (1.00) |
0.301 (1.00) |
0.838 (1.00) |
0.702 (1.00) |
0.838 (1.00) |
0.96 (1.00) |
0.957 (1.00) |
0.00735 (1.00) |
0.129 (1.00) |
0.0172 (1.00) |
|
| 11p gain |
0.086 (1.00) |
0.225 (1.00) |
4.94e-15 (7.12e-12) |
0.733 (1.00) |
0.346 (1.00) |
5.54e-06 (0.00737) |
0.0258 (1.00) |
0.222 (1.00) |
0.165 (1.00) |
0.261 (1.00) |
0.0164 (1.00) |
0.253 (1.00) |
0.0325 (1.00) |
0.733 (1.00) |
0.757 (1.00) |
0.865 (1.00) |
0.72 (1.00) |
0.673 (1.00) |
0.304 (1.00) |
0.0164 (1.00) |
|
| 11q gain |
0.732 (1.00) |
0.00297 (1.00) |
2.5e-16 (3.63e-13) |
0.793 (1.00) |
0.822 (1.00) |
5.8e-09 (8.1e-06) |
0.000622 (0.776) |
0.357 (1.00) |
0.228 (1.00) |
0.0331 (1.00) |
0.000705 (0.876) |
0.271 (1.00) |
8.26e-05 (0.106) |
0.144 (1.00) |
0.249 (1.00) |
0.731 (1.00) |
0.214 (1.00) |
0.422 (1.00) |
0.767 (1.00) |
0.0473 (1.00) |
|
| 12p gain |
0.812 (1.00) |
0.00149 (1.00) |
1.45e-43 (2.21e-40) |
0.367 (1.00) |
0.0505 (1.00) |
4.59e-25 (6.8e-22) |
0.00034 (0.428) |
0.697 (1.00) |
0.918 (1.00) |
9.68e-09 (1.35e-05) |
4.22e-07 (0.000575) |
0.378 (1.00) |
0.112 (1.00) |
0.213 (1.00) |
0.565 (1.00) |
0.876 (1.00) |
0.778 (1.00) |
0.498 (1.00) |
0.93 (1.00) |
0.921 (1.00) |
|
| 12q gain |
0.929 (1.00) |
0.00209 (1.00) |
7.1e-15 (1.02e-11) |
0.269 (1.00) |
0.258 (1.00) |
6.49e-05 (0.0837) |
0.0306 (1.00) |
0.688 (1.00) |
0.45 (1.00) |
0.000347 (0.436) |
0.000227 (0.288) |
0.38 (1.00) |
0.656 (1.00) |
0.929 (1.00) |
0.45 (1.00) |
0.0681 (1.00) |
0.948 (1.00) |
0.329 (1.00) |
0.0251 (1.00) |
0.983 (1.00) |
|
| 13q gain |
0.00394 (1.00) |
6.48e-11 (9.17e-08) |
3.03e-280 (4.83e-277) |
3.06e-05 (0.0399) |
0.116 (1.00) |
2.32e-211 (3.69e-208) |
1.2e-34 (1.81e-31) |
1.68e-06 (0.00225) |
1.36e-05 (0.0179) |
7.91e-08 (0.000109) |
9.57e-15 (1.38e-11) |
0.945 (1.00) |
0.77 (1.00) |
5.18e-07 (0.000705) |
0.129 (1.00) |
0.43 (1.00) |
0.239 (1.00) |
2.95e-05 (0.0385) |
0.869 (1.00) |
0.0352 (1.00) |
|
| 14q gain |
0.0299 (1.00) |
0.4 (1.00) |
7.71e-29 (1.15e-25) |
0.169 (1.00) |
0.0687 (1.00) |
4.79e-21 (7.03e-18) |
0.00301 (1.00) |
0.564 (1.00) |
0.00508 (1.00) |
0.552 (1.00) |
0.0185 (1.00) |
0.0554 (1.00) |
0.0673 (1.00) |
0.184 (1.00) |
0.283 (1.00) |
0.968 (1.00) |
0.614 (1.00) |
0.239 (1.00) |
0.0571 (1.00) |
0.14 (1.00) |
|
| 15q gain |
0.116 (1.00) |
0.98 (1.00) |
4.82e-08 (6.65e-05) |
1 (1.00) |
0.213 (1.00) |
5.1e-07 (0.000695) |
0.00018 (0.229) |
0.503 (1.00) |
0.478 (1.00) |
0.321 (1.00) |
0.056 (1.00) |
0.239 (1.00) |
0.686 (1.00) |
0.696 (1.00) |
0.302 (1.00) |
0.349 (1.00) |
0.6 (1.00) |
0.459 (1.00) |
0.306 (1.00) |
0.883 (1.00) |
|
| 16p gain |
2.01e-09 (2.82e-06) |
0.412 (1.00) |
1.31e-70 (2.04e-67) |
1.28e-06 (0.00172) |
0.0933 (1.00) |
4.74e-17 (6.88e-14) |
0.0296 (1.00) |
0.366 (1.00) |
0.669 (1.00) |
0.581 (1.00) |
0.0104 (1.00) |
0.497 (1.00) |
0.447 (1.00) |
0.36 (1.00) |
0.368 (1.00) |
0.0633 (1.00) |
0.182 (1.00) |
0.148 (1.00) |
0.104 (1.00) |
0.0039 (1.00) |
|
| 16q gain |
0.0677 (1.00) |
0.848 (1.00) |
2.54e-28 (3.79e-25) |
0.00263 (1.00) |
0.685 (1.00) |
1.06e-21 (1.56e-18) |
0.00998 (1.00) |
0.0626 (1.00) |
0.693 (1.00) |
0.428 (1.00) |
4.1e-06 (0.00546) |
0.477 (1.00) |
0.818 (1.00) |
0.221 (1.00) |
0.329 (1.00) |
0.0841 (1.00) |
0.551 (1.00) |
0.458 (1.00) |
0.965 (1.00) |
0.000739 (0.917) |
|
| 17p gain |
0.0936 (1.00) |
0.3 (1.00) |
1.5e-06 (0.00201) |
0.0403 (1.00) |
0.65 (1.00) |
9.33e-07 (0.00126) |
0.958 (1.00) |
0.36 (1.00) |
0.0267 (1.00) |
0.59 (1.00) |
0.837 (1.00) |
0.97 (1.00) |
0.262 (1.00) |
0.755 (1.00) |
0.0203 (1.00) |
0.851 (1.00) |
0.84 (1.00) |
0.24 (1.00) |
0.89 (1.00) |
0.628 (1.00) |
|
| 17q gain |
0.0147 (1.00) |
0.129 (1.00) |
3.22e-30 (4.82e-27) |
0.075 (1.00) |
0.604 (1.00) |
1.46e-19 (2.14e-16) |
0.357 (1.00) |
0.075 (1.00) |
0.0503 (1.00) |
0.0252 (1.00) |
0.176 (1.00) |
0.599 (1.00) |
0.0909 (1.00) |
0.168 (1.00) |
0.183 (1.00) |
0.0255 (1.00) |
0.24 (1.00) |
0.155 (1.00) |
0.873 (1.00) |
0.524 (1.00) |
|
| 18p gain |
0.19 (1.00) |
0.0722 (1.00) |
9.68e-18 (1.41e-14) |
0.559 (1.00) |
0.029 (1.00) |
2.74e-20 (4.01e-17) |
0.00168 (1.00) |
0.000984 (1.00) |
0.0368 (1.00) |
0.0237 (1.00) |
0.206 (1.00) |
0.00982 (1.00) |
0.258 (1.00) |
0.139 (1.00) |
0.795 (1.00) |
0.329 (1.00) |
0.682 (1.00) |
0.965 (1.00) |
0.166 (1.00) |
0.445 (1.00) |
|
| 18q gain |
0.142 (1.00) |
0.485 (1.00) |
6.25e-08 (8.62e-05) |
0.313 (1.00) |
0.0821 (1.00) |
4.22e-08 (5.84e-05) |
0.00019 (0.242) |
0.416 (1.00) |
0.365 (1.00) |
0.225 (1.00) |
0.43 (1.00) |
0.0888 (1.00) |
0.0925 (1.00) |
0.132 (1.00) |
0.135 (1.00) |
0.25 (1.00) |
0.966 (1.00) |
0.864 (1.00) |
0.954 (1.00) |
0.504 (1.00) |
|
| 19p gain |
1.12e-08 (1.56e-05) |
0.311 (1.00) |
5.15e-56 (7.95e-53) |
0.129 (1.00) |
0.464 (1.00) |
1.97e-20 (2.89e-17) |
0.136 (1.00) |
0.0519 (1.00) |
0.109 (1.00) |
0.00111 (1.00) |
3.56e-13 (5.08e-10) |
0.109 (1.00) |
0.135 (1.00) |
0.0228 (1.00) |
0.308 (1.00) |
0.437 (1.00) |
0.911 (1.00) |
0.641 (1.00) |
0.538 (1.00) |
0.814 (1.00) |
|
| 19q gain |
4.08e-08 (5.64e-05) |
0.0978 (1.00) |
3.05e-30 (4.58e-27) |
0.0566 (1.00) |
0.478 (1.00) |
1.2e-15 (1.73e-12) |
0.0924 (1.00) |
0.176 (1.00) |
0.572 (1.00) |
0.0328 (1.00) |
1.22e-05 (0.0161) |
0.279 (1.00) |
0.0424 (1.00) |
0.0037 (1.00) |
0.231 (1.00) |
0.664 (1.00) |
0.141 (1.00) |
0.513 (1.00) |
0.167 (1.00) |
0.149 (1.00) |
|
| 20p gain |
4.13e-05 (0.0537) |
7.61e-09 (1.06e-05) |
1.84e-64 (2.87e-61) |
0.0228 (1.00) |
0.965 (1.00) |
2.23e-68 (3.47e-65) |
6.36e-14 (9.12e-11) |
0.0267 (1.00) |
9.41e-06 (0.0124) |
2.76e-07 (0.000378) |
0.67 (1.00) |
0.304 (1.00) |
0.376 (1.00) |
0.00176 (1.00) |
0.0803 (1.00) |
0.0894 (1.00) |
0.222 (1.00) |
6.9e-06 (0.00916) |
0.055 (1.00) |
0.11 (1.00) |
|
| 20q gain |
0.0121 (1.00) |
3.39e-07 (0.000464) |
1.97e-130 (3.1e-127) |
0.203 (1.00) |
0.344 (1.00) |
1.03e-135 (1.63e-132) |
5.56e-29 (8.32e-26) |
0.000927 (1.00) |
0.000206 (0.261) |
2.83e-15 (4.09e-12) |
0.0142 (1.00) |
0.18 (1.00) |
0.0627 (1.00) |
8.01e-06 (0.0106) |
0.576 (1.00) |
0.435 (1.00) |
0.332 (1.00) |
9.3e-07 (0.00126) |
0.313 (1.00) |
0.0652 (1.00) |
|
| 21q gain |
0.454 (1.00) |
0.0303 (1.00) |
1.66e-15 (2.4e-12) |
0.00117 (1.00) |
0.307 (1.00) |
4.02e-10 (5.66e-07) |
0.0704 (1.00) |
0.229 (1.00) |
0.836 (1.00) |
0.0112 (1.00) |
0.772 (1.00) |
0.296 (1.00) |
0.471 (1.00) |
1.61e-05 (0.0212) |
0.742 (1.00) |
0.827 (1.00) |
0.0246 (1.00) |
0.135 (1.00) |
0.0824 (1.00) |
0.0112 (1.00) |
|
| 22q gain |
0.543 (1.00) |
0.0113 (1.00) |
4.2e-36 (6.35e-33) |
2.26e-09 (3.17e-06) |
0.0767 (1.00) |
1.24e-43 (1.9e-40) |
5.6e-07 (0.000761) |
0.0156 (1.00) |
0.807 (1.00) |
0.0143 (1.00) |
0.0313 (1.00) |
0.0683 (1.00) |
0.00222 (1.00) |
0.0541 (1.00) |
0.0265 (1.00) |
0.643 (1.00) |
0.345 (1.00) |
0.315 (1.00) |
0.431 (1.00) |
0.155 (1.00) |
|
| Xq gain |
0.0223 (1.00) |
0.0802 (1.00) |
2.22e-08 (3.08e-05) |
0.931 (1.00) |
0.512 (1.00) |
0.00714 (1.00) |
0.0974 (1.00) |
0.0219 (1.00) |
0.0181 (1.00) |
0.123 (1.00) |
0.0169 (1.00) |
0.982 (1.00) |
0.567 (1.00) |
0.627 (1.00) |
0.864 (1.00) |
0.118 (1.00) |
0.0134 (1.00) |
0.405 (1.00) |
0.0954 (1.00) |
0.128 (1.00) |
|
| 1p loss |
0.0632 (1.00) |
0.00323 (1.00) |
4.3e-20 (6.3e-17) |
0.527 (1.00) |
0.62 (1.00) |
4.74e-08 (6.56e-05) |
0.293 (1.00) |
0.0271 (1.00) |
0.205 (1.00) |
0.833 (1.00) |
0.0252 (1.00) |
0.00638 (1.00) |
0.331 (1.00) |
0.0311 (1.00) |
0.319 (1.00) |
0.14 (1.00) |
0.122 (1.00) |
0.165 (1.00) |
0.00378 (1.00) |
1.07e-05 (0.0142) |
|
| 1q loss |
0.734 (1.00) |
0.205 (1.00) |
0.000556 (0.696) |
0.137 (1.00) |
0.228 (1.00) |
0.188 (1.00) |
0.0921 (1.00) |
0.306 (1.00) |
0.799 (1.00) |
0.76 (1.00) |
0.458 (1.00) |
0.117 (1.00) |
0.199 (1.00) |
0.852 (1.00) |
0.993 (1.00) |
0.418 (1.00) |
0.973 (1.00) |
0.0296 (1.00) |
0.366 (1.00) |
0.936 (1.00) |
|
| 2p loss |
0.149 (1.00) |
0.319 (1.00) |
6.03e-17 (8.76e-14) |
6.09e-05 (0.0786) |
0.0276 (1.00) |
0.0848 (1.00) |
0.598 (1.00) |
0.889 (1.00) |
0.881 (1.00) |
0.439 (1.00) |
0.277 (1.00) |
0.539 (1.00) |
0.664 (1.00) |
0.0741 (1.00) |
0.626 (1.00) |
0.576 (1.00) |
0.665 (1.00) |
0.972 (1.00) |
0.463 (1.00) |
0.855 (1.00) |
|
| 2q loss |
0.162 (1.00) |
0.875 (1.00) |
2.8e-22 (4.12e-19) |
0.00258 (1.00) |
0.0276 (1.00) |
0.205 (1.00) |
0.927 (1.00) |
0.628 (1.00) |
0.153 (1.00) |
0.421 (1.00) |
1 (1.00) |
0.0444 (1.00) |
0.999 (1.00) |
1.65e-05 (0.0216) |
0.768 (1.00) |
0.339 (1.00) |
0.335 (1.00) |
0.358 (1.00) |
0.281 (1.00) |
0.236 (1.00) |
|
| 3p loss |
0.227 (1.00) |
0.637 (1.00) |
1.29e-218 (2.05e-215) |
7.17e-34 (1.08e-30) |
0.0732 (1.00) |
3.19e-144 (5.04e-141) |
5.56e-14 (7.98e-11) |
0.0222 (1.00) |
0.00016 (0.204) |
4.02e-13 (5.73e-10) |
2.88e-11 (4.08e-08) |
0.0372 (1.00) |
0.12 (1.00) |
5.35e-08 (7.38e-05) |
0.787 (1.00) |
0.771 (1.00) |
0.996 (1.00) |
0.399 (1.00) |
0.0729 (1.00) |
0.715 (1.00) |
|
| 3q loss |
0.446 (1.00) |
0.56 (1.00) |
6.51e-39 (9.9e-36) |
2.54e-09 (3.55e-06) |
0.963 (1.00) |
3.54e-28 (5.28e-25) |
0.0222 (1.00) |
0.304 (1.00) |
0.0873 (1.00) |
0.000443 (0.555) |
0.345 (1.00) |
0.108 (1.00) |
0.498 (1.00) |
0.0918 (1.00) |
0.0383 (1.00) |
0.914 (1.00) |
0.48 (1.00) |
0.716 (1.00) |
0.609 (1.00) |
0.629 (1.00) |
|
| 4p loss |
0.385 (1.00) |
0.664 (1.00) |
1.58e-69 (2.47e-66) |
0.0662 (1.00) |
0.00131 (1.00) |
5.22e-66 (8.13e-63) |
0.000198 (0.251) |
1.94e-05 (0.0254) |
0.78 (1.00) |
9.69e-14 (1.39e-10) |
3.67e-10 (5.17e-07) |
0.00449 (1.00) |
0.000486 (0.608) |
0.000563 (0.704) |
0.388 (1.00) |
0.743 (1.00) |
0.647 (1.00) |
0.552 (1.00) |
0.0064 (1.00) |
0.646 (1.00) |
|
| 4q loss |
0.281 (1.00) |
0.332 (1.00) |
2.93e-108 (4.6e-105) |
1.63e-06 (0.00218) |
0.00244 (1.00) |
6.11e-88 (9.59e-85) |
5.52e-05 (0.0714) |
0.166 (1.00) |
0.909 (1.00) |
6.53e-24 (9.66e-21) |
3.28e-13 (4.68e-10) |
0.0401 (1.00) |
0.0218 (1.00) |
0.0254 (1.00) |
0.116 (1.00) |
0.581 (1.00) |
0.139 (1.00) |
0.467 (1.00) |
0.0279 (1.00) |
0.628 (1.00) |
|
| 5p loss |
0.179 (1.00) |
0.944 (1.00) |
6.07e-16 (8.79e-13) |
0.000439 (0.551) |
0.827 (1.00) |
9.78e-19 (1.43e-15) |
0.164 (1.00) |
0.524 (1.00) |
0.663 (1.00) |
1.69e-06 (0.00226) |
0.464 (1.00) |
0.599 (1.00) |
0.969 (1.00) |
0.0939 (1.00) |
0.63 (1.00) |
0.835 (1.00) |
0.236 (1.00) |
0.668 (1.00) |
0.179 (1.00) |
0.641 (1.00) |
|
| 5q loss |
0.434 (1.00) |
0.212 (1.00) |
5.52e-60 (8.57e-57) |
0.86 (1.00) |
0.00434 (1.00) |
2.65e-59 (4.11e-56) |
5.38e-05 (0.0698) |
0.855 (1.00) |
0.152 (1.00) |
1.53e-05 (0.0201) |
0.000245 (0.31) |
0.0874 (1.00) |
0.505 (1.00) |
0.0051 (1.00) |
0.661 (1.00) |
0.46 (1.00) |
0.18 (1.00) |
0.563 (1.00) |
0.339 (1.00) |
0.0064 (1.00) |
|
| 6p loss |
0.0535 (1.00) |
0.001 (1.00) |
3.98e-28 (5.94e-25) |
1 (1.00) |
0.452 (1.00) |
4.31e-26 (6.4e-23) |
0.501 (1.00) |
0.79 (1.00) |
0.457 (1.00) |
0.000212 (0.269) |
0.797 (1.00) |
0.95 (1.00) |
0.683 (1.00) |
0.561 (1.00) |
0.246 (1.00) |
0.0332 (1.00) |
0.848 (1.00) |
0.000685 (0.852) |
0.351 (1.00) |
0.0844 (1.00) |
|
| 6q loss |
0.0275 (1.00) |
6.15e-07 (0.000835) |
1.12e-54 (1.73e-51) |
0.00173 (1.00) |
0.173 (1.00) |
7.11e-54 (1.1e-50) |
0.126 (1.00) |
0.853 (1.00) |
0.749 (1.00) |
2.29e-06 (0.00306) |
0.427 (1.00) |
0.00819 (1.00) |
0.94 (1.00) |
0.0348 (1.00) |
0.106 (1.00) |
0.0937 (1.00) |
0.951 (1.00) |
0.0249 (1.00) |
0.588 (1.00) |
0.055 (1.00) |
|
| 7p loss |
0.45 (1.00) |
0.253 (1.00) |
3.37e-48 (5.16e-45) |
1e-13 (1.43e-10) |
0.319 (1.00) |
1.84e-37 (2.79e-34) |
9e-05 (0.115) |
0.377 (1.00) |
0.735 (1.00) |
1.66e-14 (2.38e-11) |
0.0908 (1.00) |
0.135 (1.00) |
0.00903 (1.00) |
0.515 (1.00) |
0.0224 (1.00) |
0.649 (1.00) |
0.888 (1.00) |
0.42 (1.00) |
0.391 (1.00) |
0.0434 (1.00) |
|
| 7q loss |
0.745 (1.00) |
0.493 (1.00) |
4.47e-20 (6.54e-17) |
3.65e-11 (5.16e-08) |
0.029 (1.00) |
1.83e-23 (2.7e-20) |
3.89e-05 (0.0506) |
0.517 (1.00) |
0.55 (1.00) |
0.0189 (1.00) |
1 (1.00) |
0.769 (1.00) |
0.738 (1.00) |
0.69 (1.00) |
0.734 (1.00) |
0.0422 (1.00) |
0.99 (1.00) |
0.766 (1.00) |
0.221 (1.00) |
0.0454 (1.00) |
|
| 8p loss |
0.929 (1.00) |
0.041 (1.00) |
7.91e-53 (1.22e-49) |
0.00238 (1.00) |
0.0785 (1.00) |
8.98e-35 (1.35e-31) |
0.037 (1.00) |
0.141 (1.00) |
0.783 (1.00) |
1.33e-07 (0.000182) |
6.06e-10 (8.53e-07) |
0.068 (1.00) |
0.696 (1.00) |
0.134 (1.00) |
0.529 (1.00) |
0.586 (1.00) |
0.0967 (1.00) |
0.296 (1.00) |
0.00172 (1.00) |
0.865 (1.00) |
|
| 8q loss |
0.185 (1.00) |
0.203 (1.00) |
1.78e-12 (2.53e-09) |
1 (1.00) |
0.0058 (1.00) |
8.37e-10 (1.18e-06) |
0.0646 (1.00) |
0.404 (1.00) |
0.8 (1.00) |
0.000142 (0.182) |
0.719 (1.00) |
0.335 (1.00) |
0.902 (1.00) |
0.286 (1.00) |
0.818 (1.00) |
0.412 (1.00) |
0.525 (1.00) |
0.106 (1.00) |
0.656 (1.00) |
0.642 (1.00) |
|
| 9p loss |
2.53e-13 (3.61e-10) |
0.0669 (1.00) |
3.23e-49 (4.96e-46) |
0.00234 (1.00) |
0.22 (1.00) |
2.07e-49 (3.18e-46) |
9.7e-09 (1.35e-05) |
0.349 (1.00) |
0.742 (1.00) |
0.014 (1.00) |
0.127 (1.00) |
0.00247 (1.00) |
0.392 (1.00) |
0.187 (1.00) |
0.0454 (1.00) |
0.342 (1.00) |
0.0351 (1.00) |
0.0671 (1.00) |
0.274 (1.00) |
0.0254 (1.00) |
|
| 9q loss |
0.00016 (0.204) |
0.000739 (0.917) |
8.8e-58 (1.36e-54) |
0.00104 (1.00) |
0.787 (1.00) |
1.1e-56 (1.7e-53) |
0.00031 (0.39) |
0.00849 (1.00) |
0.434 (1.00) |
7.3e-09 (1.02e-05) |
0.00372 (1.00) |
0.151 (1.00) |
0.708 (1.00) |
0.0259 (1.00) |
0.0626 (1.00) |
0.305 (1.00) |
0.0552 (1.00) |
0.0286 (1.00) |
0.368 (1.00) |
0.0174 (1.00) |
|
| 10p loss |
0 (0) |
0.379 (1.00) |
0 (0) |
2.5e-22 (3.68e-19) |
0.00339 (1.00) |
9.96e-37 (1.51e-33) |
0.00377 (1.00) |
0.285 (1.00) |
0.85 (1.00) |
0.446 (1.00) |
9.03e-59 (1.4e-55) |
0.974 (1.00) |
0.0223 (1.00) |
0.593 (1.00) |
0.474 (1.00) |
0.884 (1.00) |
0.824 (1.00) |
0.66 (1.00) |
0.794 (1.00) |
0.514 (1.00) |
|
| 10q loss |
0 (0) |
0.0541 (1.00) |
0 (0) |
2.6e-17 (3.78e-14) |
0.000962 (1.00) |
5.71e-41 (8.7e-38) |
0.0807 (1.00) |
0.75 (1.00) |
0.633 (1.00) |
0.902 (1.00) |
2.56e-56 (3.96e-53) |
0.886 (1.00) |
0.104 (1.00) |
0.108 (1.00) |
0.503 (1.00) |
0.871 (1.00) |
0.277 (1.00) |
0.644 (1.00) |
0.972 (1.00) |
0.419 (1.00) |
|
| 11p loss |
0.00257 (1.00) |
0.469 (1.00) |
7.63e-41 (1.16e-37) |
1.95e-05 (0.0255) |
0.758 (1.00) |
1.17e-36 (1.76e-33) |
0.105 (1.00) |
4.77e-05 (0.0619) |
0.685 (1.00) |
2.7e-12 (3.84e-09) |
0.251 (1.00) |
0.344 (1.00) |
0.16 (1.00) |
2.25e-06 (0.00301) |
0.0494 (1.00) |
0.678 (1.00) |
0.0685 (1.00) |
0.815 (1.00) |
0.00279 (1.00) |
0.000208 (0.264) |
|
| 11q loss |
0.632 (1.00) |
0.974 (1.00) |
2.05e-44 (3.13e-41) |
5.76e-05 (0.0745) |
0.88 (1.00) |
4.7e-22 (6.92e-19) |
0.0392 (1.00) |
0.00121 (1.00) |
0.741 (1.00) |
1.04e-05 (0.0138) |
0.777 (1.00) |
0.899 (1.00) |
0.75 (1.00) |
0.069 (1.00) |
0.0573 (1.00) |
0.836 (1.00) |
0.129 (1.00) |
0.459 (1.00) |
0.0695 (1.00) |
0.0386 (1.00) |
|
| 12p loss |
0.0587 (1.00) |
0.206 (1.00) |
2.2e-07 (0.000301) |
0.00625 (1.00) |
0.961 (1.00) |
1.32e-10 (1.86e-07) |
0.00588 (1.00) |
0.0277 (1.00) |
0.152 (1.00) |
0.137 (1.00) |
0.155 (1.00) |
0.0079 (1.00) |
0.647 (1.00) |
0.764 (1.00) |
0.000571 (0.713) |
0.554 (1.00) |
0.555 (1.00) |
0.578 (1.00) |
0.883 (1.00) |
0.414 (1.00) |
|
| 12q loss |
0.163 (1.00) |
0.325 (1.00) |
3.31e-16 (4.79e-13) |
0.00183 (1.00) |
0.95 (1.00) |
7.04e-16 (1.02e-12) |
0.132 (1.00) |
0.075 (1.00) |
0.286 (1.00) |
0.00992 (1.00) |
0.862 (1.00) |
0.593 (1.00) |
0.626 (1.00) |
0.347 (1.00) |
0.0973 (1.00) |
0.703 (1.00) |
0.584 (1.00) |
0.615 (1.00) |
0.488 (1.00) |
0.129 (1.00) |
|
| 13q loss |
0.00165 (1.00) |
0.194 (1.00) |
7.7e-88 (1.21e-84) |
1.22e-06 (0.00165) |
0.173 (1.00) |
1.53e-83 (2.4e-80) |
9.98e-12 (1.42e-08) |
0.0228 (1.00) |
0.42 (1.00) |
1.38e-08 (1.92e-05) |
0.211 (1.00) |
0.27 (1.00) |
0.0886 (1.00) |
0.0203 (1.00) |
0.563 (1.00) |
0.913 (1.00) |
0.0971 (1.00) |
0.604 (1.00) |
0.139 (1.00) |
0.22 (1.00) |
|
| 14q loss |
0.00277 (1.00) |
0.205 (1.00) |
1.77e-38 (2.68e-35) |
0.0381 (1.00) |
0.823 (1.00) |
1.06e-38 (1.61e-35) |
2.02e-07 (0.000277) |
0.000151 (0.193) |
5.75e-05 (0.0744) |
0.000671 (0.836) |
0.0861 (1.00) |
0.339 (1.00) |
0.111 (1.00) |
0.283 (1.00) |
0.741 (1.00) |
0.942 (1.00) |
0.189 (1.00) |
0.109 (1.00) |
0.345 (1.00) |
0.248 (1.00) |
|
| 15q loss |
0.283 (1.00) |
0.00988 (1.00) |
3.19e-60 (4.96e-57) |
1.96e-12 (2.78e-09) |
0.936 (1.00) |
3.38e-63 (5.25e-60) |
1.85e-06 (0.00248) |
0.0822 (1.00) |
0.136 (1.00) |
3.04e-14 (4.37e-11) |
0.00398 (1.00) |
0.704 (1.00) |
0.362 (1.00) |
0.608 (1.00) |
0.454 (1.00) |
0.12 (1.00) |
0.539 (1.00) |
0.191 (1.00) |
0.0579 (1.00) |
0.704 (1.00) |
|
| 16p loss |
0.00995 (1.00) |
0.758 (1.00) |
3.63e-173 (5.75e-170) |
8.69e-20 (1.27e-16) |
0.304 (1.00) |
1.47e-134 (2.31e-131) |
0.00102 (1.00) |
0.813 (1.00) |
0.774 (1.00) |
9.54e-28 (1.42e-24) |
3.42e-06 (0.00456) |
0.283 (1.00) |
0.941 (1.00) |
0.0613 (1.00) |
0.44 (1.00) |
0.939 (1.00) |
0.83 (1.00) |
0.0073 (1.00) |
0.966 (1.00) |
0.601 (1.00) |
|
| 16q loss |
0.00145 (1.00) |
0.668 (1.00) |
1.19e-229 (1.89e-226) |
7.3e-72 (1.14e-68) |
0.0846 (1.00) |
1.95e-195 (3.1e-192) |
1.01e-06 (0.00136) |
0.392 (1.00) |
0.643 (1.00) |
3.38e-42 (5.15e-39) |
0.000689 (0.857) |
0.229 (1.00) |
0.387 (1.00) |
0.032 (1.00) |
0.797 (1.00) |
0.146 (1.00) |
0.0779 (1.00) |
1.07e-05 (0.0141) |
0.221 (1.00) |
9.99e-06 (0.0132) |
|
| 17p loss |
0.0253 (1.00) |
0.362 (1.00) |
2.2e-195 (3.49e-192) |
1.96e-32 (2.94e-29) |
0.00321 (1.00) |
5.37e-162 (8.48e-159) |
2.22e-07 (0.000304) |
0.00366 (1.00) |
0.0556 (1.00) |
2.74e-31 (4.12e-28) |
3.23e-23 (4.76e-20) |
0.292 (1.00) |
0.483 (1.00) |
0.259 (1.00) |
0.714 (1.00) |
0.61 (1.00) |
0.77 (1.00) |
0.00132 (1.00) |
0.0512 (1.00) |
0.0524 (1.00) |
|
| 17q loss |
0.869 (1.00) |
0.00759 (1.00) |
9.79e-204 (1.56e-200) |
8.72e-49 (1.34e-45) |
0.945 (1.00) |
1.38e-170 (2.19e-167) |
0.544 (1.00) |
0.0672 (1.00) |
0.277 (1.00) |
2.14e-45 (3.27e-42) |
2.1e-08 (2.91e-05) |
0.5 (1.00) |
0.831 (1.00) |
0.164 (1.00) |
0.39 (1.00) |
0.502 (1.00) |
0.367 (1.00) |
8.95e-05 (0.115) |
0.467 (1.00) |
0.13 (1.00) |
|
| 18p loss |
0.805 (1.00) |
0.000808 (0.999) |
1.86e-147 (2.94e-144) |
0.057 (1.00) |
0.916 (1.00) |
1.74e-130 (2.74e-127) |
1.75e-26 (2.6e-23) |
0.000217 (0.275) |
4.8e-09 (6.72e-06) |
5e-12 (7.09e-09) |
4.81e-12 (6.83e-09) |
0.00564 (1.00) |
0.449 (1.00) |
0.701 (1.00) |
0.00913 (1.00) |
0.328 (1.00) |
0.36 (1.00) |
0.000346 (0.435) |
0.0572 (1.00) |
0.41 (1.00) |
|
| 18q loss |
0.842 (1.00) |
3.22e-05 (0.042) |
6.45e-148 (1.02e-144) |
0.887 (1.00) |
0.373 (1.00) |
1.9e-112 (2.98e-109) |
3.09e-24 (4.58e-21) |
8.37e-07 (0.00113) |
2.29e-05 (0.0299) |
5.26e-14 (7.55e-11) |
2.46e-19 (3.59e-16) |
0.299 (1.00) |
0.733 (1.00) |
0.0675 (1.00) |
0.0135 (1.00) |
0.395 (1.00) |
0.387 (1.00) |
0.00296 (1.00) |
0.0708 (1.00) |
0.096 (1.00) |
|
| 19p loss |
0.00671 (1.00) |
0.832 (1.00) |
6.43e-67 (1e-63) |
0.00229 (1.00) |
0.669 (1.00) |
3.63e-51 (5.59e-48) |
2.22e-07 (0.000304) |
0.307 (1.00) |
0.0399 (1.00) |
3.85e-06 (0.00513) |
2.75e-06 (0.00367) |
0.72 (1.00) |
0.648 (1.00) |
0.00738 (1.00) |
0.0133 (1.00) |
0.128 (1.00) |
0.0726 (1.00) |
0.3 (1.00) |
0.318 (1.00) |
0.153 (1.00) |
|
| 19q loss |
0.0664 (1.00) |
0.273 (1.00) |
5.1e-64 (7.94e-61) |
5.16e-07 (0.000703) |
0.873 (1.00) |
1.28e-50 (1.97e-47) |
0.00706 (1.00) |
0.971 (1.00) |
0.167 (1.00) |
8.01e-10 (1.13e-06) |
0.00171 (1.00) |
0.188 (1.00) |
0.572 (1.00) |
0.145 (1.00) |
0.0111 (1.00) |
0.695 (1.00) |
0.109 (1.00) |
0.00413 (1.00) |
0.864 (1.00) |
0.116 (1.00) |
|
| 20p loss |
0.724 (1.00) |
0.779 (1.00) |
1.02e-10 (1.45e-07) |
0.422 (1.00) |
0.963 (1.00) |
5.75e-09 (8.03e-06) |
0.00253 (1.00) |
0.044 (1.00) |
0.401 (1.00) |
0.179 (1.00) |
0.0187 (1.00) |
0.994 (1.00) |
0.457 (1.00) |
0.629 (1.00) |
0.0616 (1.00) |
0.527 (1.00) |
0.561 (1.00) |
0.906 (1.00) |
0.261 (1.00) |
0.215 (1.00) |
|
| 20q loss |
0.631 (1.00) |
0.534 (1.00) |
9.63e-09 (1.34e-05) |
0.92 (1.00) |
0.335 (1.00) |
8.42e-07 (0.00114) |
0.0205 (1.00) |
0.472 (1.00) |
0.617 (1.00) |
0.165 (1.00) |
0.0577 (1.00) |
0.644 (1.00) |
0.722 (1.00) |
0.623 (1.00) |
0.159 (1.00) |
0.546 (1.00) |
0.928 (1.00) |
0.336 (1.00) |
0.459 (1.00) |
0.666 (1.00) |
|
| 21q loss |
0.817 (1.00) |
0.342 (1.00) |
2.1e-46 (3.21e-43) |
0.147 (1.00) |
0.0337 (1.00) |
1.61e-27 (2.4e-24) |
0.0199 (1.00) |
0.00555 (1.00) |
0.726 (1.00) |
5.65e-06 (0.00751) |
1.08e-07 (0.000149) |
0.11 (1.00) |
5.24e-05 (0.068) |
0.0646 (1.00) |
0.996 (1.00) |
0.0811 (1.00) |
0.677 (1.00) |
0.83 (1.00) |
0.0647 (1.00) |
0.0334 (1.00) |
|
| 22q loss |
5.47e-05 (0.0708) |
0.266 (1.00) |
2.28e-167 (3.61e-164) |
5.59e-34 (8.42e-31) |
0.017 (1.00) |
8.78e-170 (1.39e-166) |
0.000676 (0.842) |
0.000747 (0.925) |
0.53 (1.00) |
3.92e-53 (6.04e-50) |
0.659 (1.00) |
0.401 (1.00) |
0.863 (1.00) |
0.102 (1.00) |
0.0119 (1.00) |
0.658 (1.00) |
0.116 (1.00) |
0.000853 (1.00) |
0.182 (1.00) |
0.188 (1.00) |
|
| Xq loss |
0.394 (1.00) |
0.0846 (1.00) |
5.04e-48 (7.72e-45) |
4.06e-07 (0.000553) |
0.838 (1.00) |
5.42e-35 (8.18e-32) |
0.82 (1.00) |
0.556 (1.00) |
0.975 (1.00) |
2.78e-10 (3.92e-07) |
0.00345 (1.00) |
0.136 (1.00) |
0.246 (1.00) |
0.585 (1.00) |
0.925 (1.00) |
0.684 (1.00) |
1 (1.00) |
0.102 (1.00) |
0.444 (1.00) |
0.000281 (0.354) |
Table S1. Get Full Table Description of clustering approach #1: '1p gain mutation analysis'
| Cluster Labels | 1P GAIN MUTATED | 1P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 359 | 4204 |
P value = 1.5e-28 (Chi-square test), Q value = 2.2e-25
Table S2. Clustering Approach #1: '1p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 1P GAIN MUTATED | 15 | 38 | 97 | 9 | 17 | 11 | 5 | 76 | 1 | 85 | 1 | 4 |
| 1P GAIN WILD-TYPE | 110 | 506 | 749 | 403 | 430 | 308 | 488 | 586 | 1 | 463 | 1 | 155 |
Figure S1. Get High-res Image Clustering Approach #1: '1p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
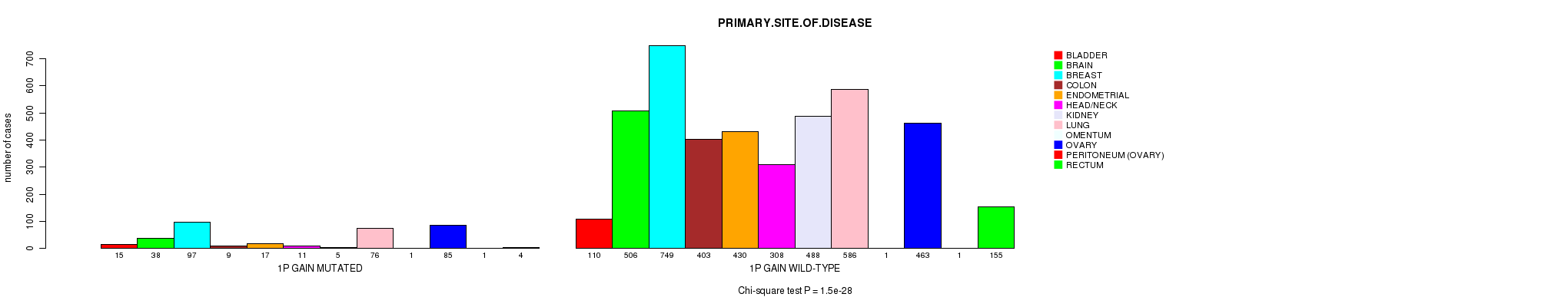
P value = 8.6e-07 (Fisher's exact test), Q value = 0.0012
Table S3. Clustering Approach #1: '1p gain mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 1P GAIN MUTATED | 269 | 90 |
| 1P GAIN WILD-TYPE | 2612 | 1592 |
Figure S2. Get High-res Image Clustering Approach #1: '1p gain mutation analysis' versus Clinical Feature #4: 'GENDER'
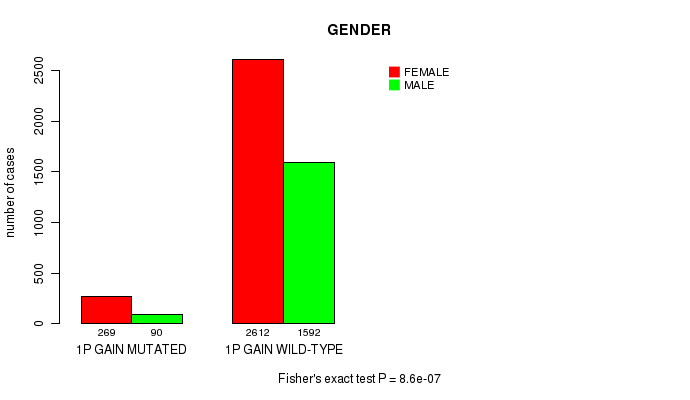
P value = 1.45e-29 (Chi-square test), Q value = 2.2e-26
Table S4. Clustering Approach #1: '1p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 1P GAIN MUTATED | 8 | 1 | 10 | 11 | 5 | 1 | 10 | 30 | 4 | 0 | 3 | 0 | 1 | 0 | 2 | 0 | 0 | 1 | 0 | 23 | 0 | 1 | 3 | 1 | 87 | 7 | 0 | 1 |
| 1P GAIN WILD-TYPE | 348 | 53 | 340 | 308 | 488 | 9 | 65 | 175 | 3 | 3 | 12 | 2 | 2 | 2 | 11 | 1 | 1 | 0 | 2 | 292 | 18 | 6 | 140 | 12 | 465 | 72 | 1 | 19 |
Figure S3. Get High-res Image Clustering Approach #1: '1p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 0.000161 (Chi-square test), Q value = 0.21
Table S5. Clustering Approach #1: '1p gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 1P GAIN MUTATED | 19 | 53 | 24 | 7 |
| 1P GAIN WILD-TYPE | 447 | 579 | 670 | 202 |
Figure S4. Get High-res Image Clustering Approach #1: '1p gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'

Table S6. Get Full Table Description of clustering approach #2: '1q gain mutation analysis'
| Cluster Labels | 1Q GAIN MUTATED | 1Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 1174 | 3389 |
P value = 2.04e-10 (logrank test), Q value = 2.9e-07
Table S7. Clustering Approach #2: '1q gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 4296 | 1401 | 0.0 - 224.0 (14.9) |
| 1Q GAIN MUTATED | 1116 | 275 | 0.0 - 223.4 (15.2) |
| 1Q GAIN WILD-TYPE | 3180 | 1126 | 0.0 - 224.0 (14.5) |
Figure S5. Get High-res Image Clustering Approach #2: '1q gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
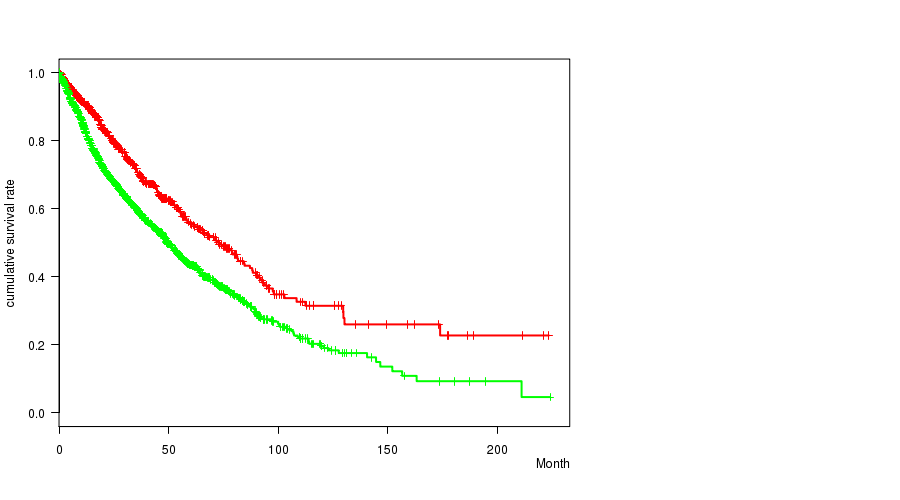
P value = 3.09e-135 (Chi-square test), Q value = 4.9e-132
Table S8. Clustering Approach #2: '1q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 1Q GAIN MUTATED | 29 | 42 | 464 | 51 | 133 | 39 | 31 | 206 | 1 | 150 | 2 | 25 |
| 1Q GAIN WILD-TYPE | 96 | 502 | 382 | 361 | 314 | 280 | 462 | 456 | 1 | 398 | 0 | 134 |
Figure S6. Get High-res Image Clustering Approach #2: '1q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
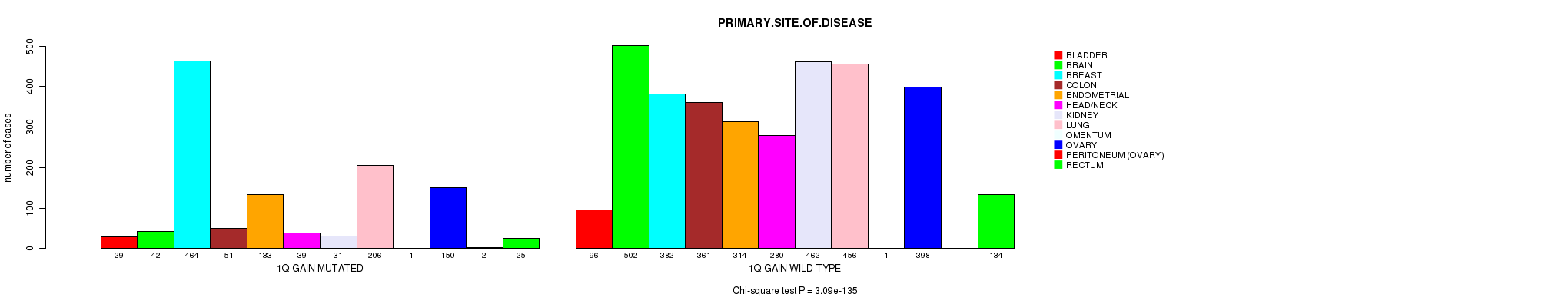
P value = 1.89e-33 (Fisher's exact test), Q value = 2.8e-30
Table S9. Clustering Approach #2: '1q gain mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 1Q GAIN MUTATED | 909 | 265 |
| 1Q GAIN WILD-TYPE | 1972 | 1417 |
Figure S7. Get High-res Image Clustering Approach #2: '1q gain mutation analysis' versus Clinical Feature #4: 'GENDER'

P value = 2.19e-39 (Chi-square test), Q value = 3.3e-36
Table S10. Clustering Approach #2: '1q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 1Q GAIN MUTATED | 48 | 3 | 116 | 39 | 31 | 4 | 34 | 75 | 6 | 0 | 7 | 1 | 2 | 0 | 7 | 0 | 0 | 1 | 1 | 66 | 3 | 2 | 21 | 4 | 153 | 14 | 0 | 1 |
| 1Q GAIN WILD-TYPE | 308 | 51 | 234 | 280 | 462 | 6 | 41 | 130 | 1 | 3 | 8 | 1 | 1 | 2 | 6 | 1 | 1 | 0 | 1 | 249 | 15 | 5 | 122 | 9 | 399 | 65 | 1 | 19 |
Figure S8. Get High-res Image Clustering Approach #2: '1q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.65e-05 (Chi-square test), Q value = 0.022
Table S11. Clustering Approach #2: '1q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 1Q GAIN MUTATED | 71 | 140 | 88 | 45 |
| 1Q GAIN WILD-TYPE | 395 | 492 | 606 | 164 |
Figure S9. Get High-res Image Clustering Approach #2: '1q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
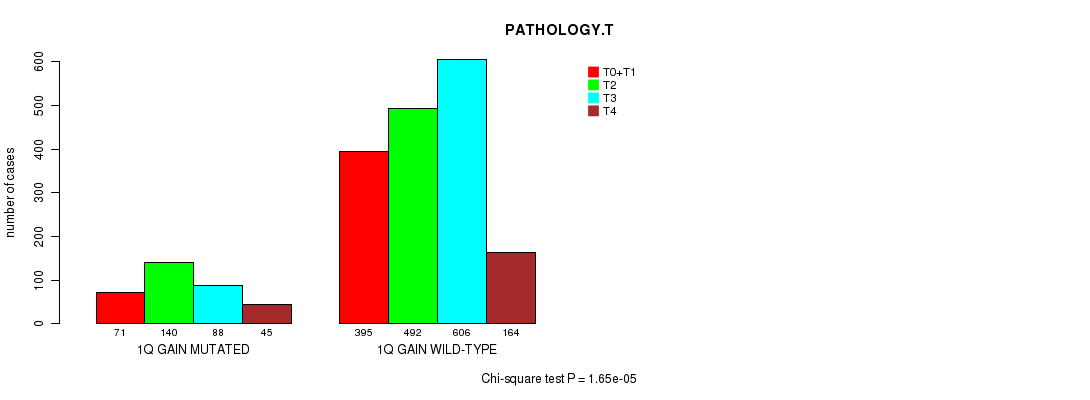
Table S12. Get Full Table Description of clustering approach #3: '2p gain mutation analysis'
| Cluster Labels | 2P GAIN MUTATED | 2P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 518 | 4045 |
P value = 0.000113 (t-test), Q value = 0.15
Table S13. Clustering Approach #3: '2p gain mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 4507 | 61.9 (12.6) |
| 2P GAIN MUTATED | 507 | 63.8 (12.1) |
| 2P GAIN WILD-TYPE | 4000 | 61.6 (12.7) |
Figure S10. Get High-res Image Clustering Approach #3: '2p gain mutation analysis' versus Clinical Feature #2: 'AGE'

P value = 1.25e-30 (Chi-square test), Q value = 1.9e-27
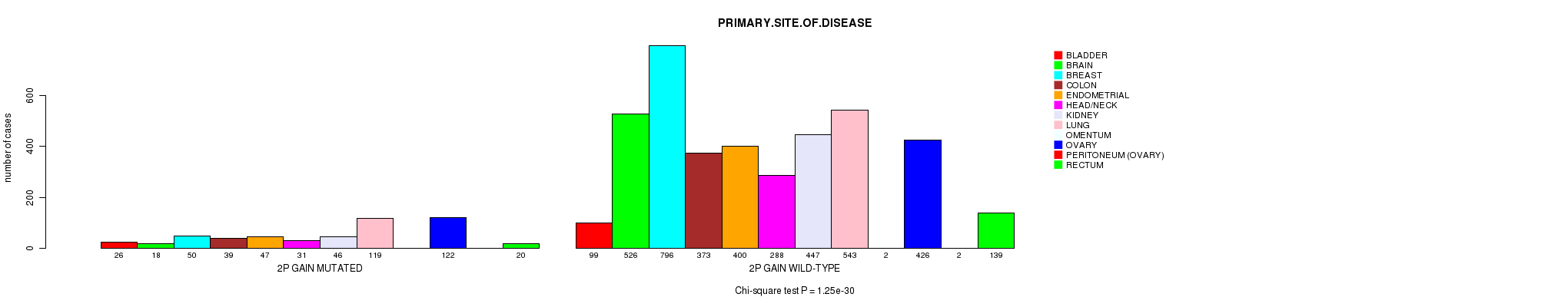
Table S14. Clustering Approach #3: '2p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 2P GAIN MUTATED | 26 | 18 | 50 | 39 | 47 | 31 | 46 | 119 | 0 | 122 | 0 | 20 |
| 2P GAIN WILD-TYPE | 99 | 526 | 796 | 373 | 400 | 288 | 447 | 543 | 2 | 426 | 2 | 139 |
Figure S11. Get High-res Image Clustering Approach #3: '2p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
P value = 1.8e-13 (Chi-square test), Q value = 2.6e-10
Table S15. Clustering Approach #3: '2p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 2P GAIN MUTATED | 38 | 1 | 31 | 31 | 46 | 1 | 11 | 26 | 5 | 0 | 1 | 1 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 70 | 3 | 0 | 20 | 0 | 122 | 13 | 0 | 0 |
| 2P GAIN WILD-TYPE | 318 | 53 | 319 | 288 | 447 | 9 | 64 | 179 | 2 | 3 | 14 | 1 | 3 | 2 | 10 | 0 | 1 | 1 | 2 | 245 | 15 | 7 | 123 | 13 | 430 | 66 | 1 | 20 |
Figure S12. Get High-res Image Clustering Approach #3: '2p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 6.7e-06 (Fisher's exact test), Q value = 0.0089
Table S16. Clustering Approach #3: '2p gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 2P GAIN MUTATED | 58 | 460 |
| 2P GAIN WILD-TYPE | 768 | 3277 |
Figure S13. Get High-res Image Clustering Approach #3: '2p gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'

P value = 2.47e-05 (Chi-square test), Q value = 0.032
Table S17. Clustering Approach #3: '2p gain mutation analysis' versus Clinical Feature #21: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE IA | STAGE IB | STAGE II | STAGE IIA | STAGE IIB | STAGE III | STAGE IIIA | STAGE IIIB | STAGE IIIC | STAGE IV | STAGE TIS | STAGE X |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 72 | 62 | 7 | 43 | 287 | 186 | 42 | 124 | 25 | 41 | 59 | 1 | 16 |
| 2P GAIN MUTATED | 1 | 3 | 0 | 8 | 21 | 10 | 7 | 6 | 0 | 2 | 13 | 0 | 3 |
| 2P GAIN WILD-TYPE | 71 | 59 | 7 | 35 | 266 | 176 | 35 | 118 | 25 | 39 | 46 | 1 | 13 |
Figure S14. Get High-res Image Clustering Approach #3: '2p gain mutation analysis' versus Clinical Feature #21: 'NEOPLASM.DISEASESTAGE'
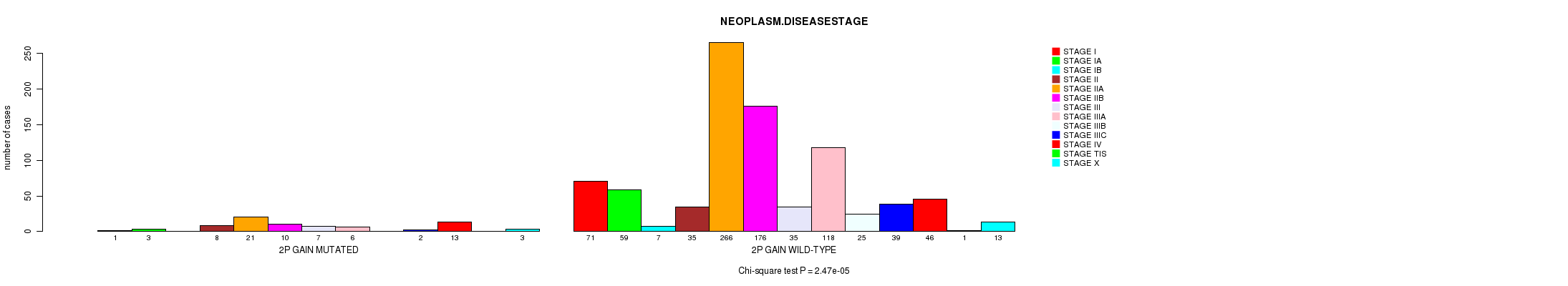
Table S18. Get Full Table Description of clustering approach #4: '2q gain mutation analysis'
| Cluster Labels | 2Q GAIN MUTATED | 2Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 396 | 4167 |
P value = 1.17e-21 (Chi-square test), Q value = 1.7e-18
Table S19. Clustering Approach #4: '2q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 2Q GAIN MUTATED | 14 | 15 | 27 | 38 | 37 | 22 | 48 | 78 | 0 | 96 | 0 | 21 |
| 2Q GAIN WILD-TYPE | 111 | 529 | 819 | 374 | 410 | 297 | 445 | 584 | 2 | 452 | 2 | 138 |
Figure S15. Get High-res Image Clustering Approach #4: '2q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
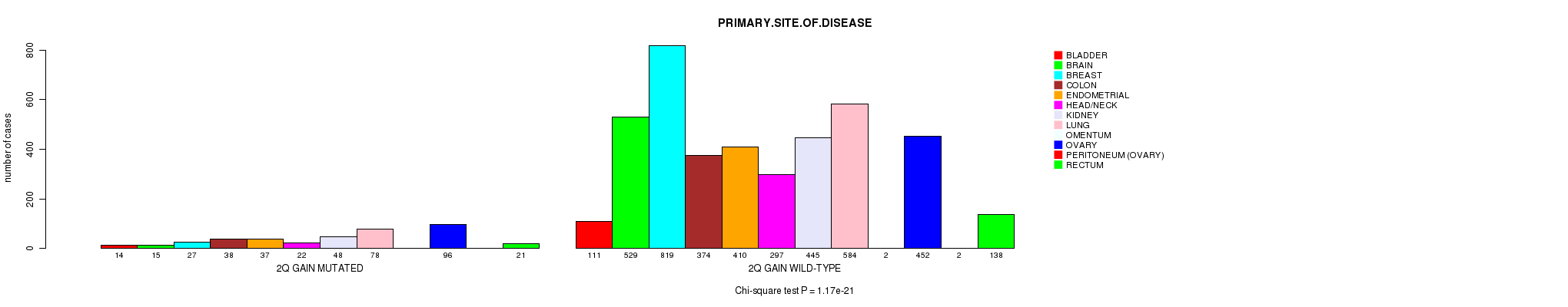
P value = 1e-08 (Chi-square test), Q value = 1.4e-05
Table S20. Clustering Approach #4: '2q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 2Q GAIN MUTATED | 37 | 1 | 25 | 22 | 48 | 1 | 8 | 25 | 5 | 0 | 0 | 0 | 0 | 0 | 4 | 1 | 0 | 0 | 0 | 34 | 1 | 0 | 21 | 0 | 96 | 11 | 0 | 0 |
| 2Q GAIN WILD-TYPE | 319 | 53 | 325 | 297 | 445 | 9 | 67 | 180 | 2 | 3 | 15 | 2 | 3 | 2 | 9 | 0 | 1 | 1 | 2 | 281 | 17 | 7 | 122 | 13 | 456 | 68 | 1 | 20 |
Figure S16. Get High-res Image Clustering Approach #4: '2q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S21. Get Full Table Description of clustering approach #5: '3p gain mutation analysis'
| Cluster Labels | 3P GAIN MUTATED | 3P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 365 | 4198 |
P value = 2.71e-30 (Chi-square test), Q value = 4.1e-27
Table S22. Clustering Approach #5: '3p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 3P GAIN MUTATED | 28 | 28 | 58 | 19 | 24 | 29 | 5 | 58 | 0 | 99 | 1 | 15 |
| 3P GAIN WILD-TYPE | 97 | 516 | 788 | 393 | 423 | 290 | 488 | 604 | 2 | 449 | 1 | 144 |
Figure S17. Get High-res Image Clustering Approach #5: '3p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
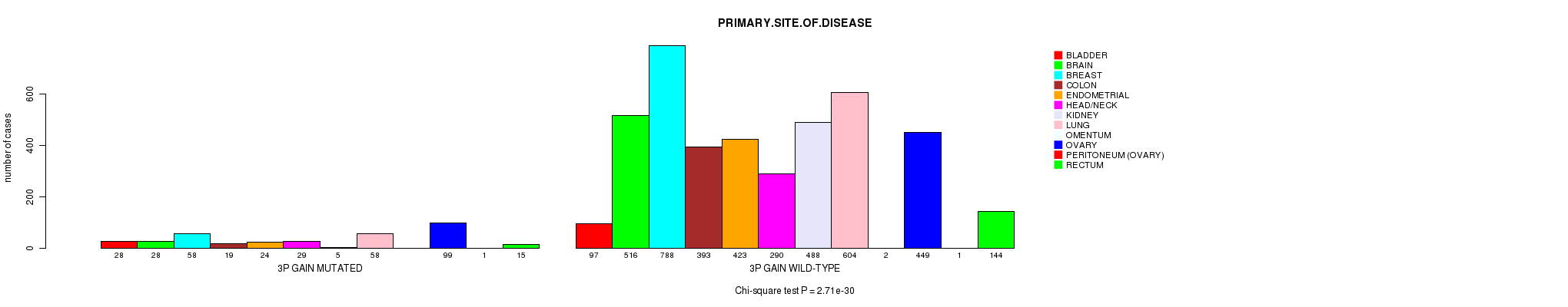
P value = 6.17e-19 (Chi-square test), Q value = 9e-16
Table S23. Clustering Approach #5: '3p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 3P GAIN MUTATED | 18 | 1 | 18 | 29 | 5 | 1 | 2 | 8 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 45 | 0 | 0 | 15 | 0 | 100 | 6 | 0 | 1 |
| 3P GAIN WILD-TYPE | 338 | 53 | 332 | 290 | 488 | 9 | 73 | 197 | 6 | 3 | 15 | 2 | 3 | 2 | 12 | 1 | 1 | 1 | 2 | 270 | 18 | 7 | 128 | 13 | 452 | 73 | 1 | 19 |
Figure S18. Get High-res Image Clustering Approach #5: '3p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 7.88e-05 (Chi-square test), Q value = 0.1
Table S24. Clustering Approach #5: '3p gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 3P GAIN MUTATED | 37 | 39 | 103 | 37 |
| 3P GAIN WILD-TYPE | 675 | 459 | 778 | 386 |
Figure S19. Get High-res Image Clustering Approach #5: '3p gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
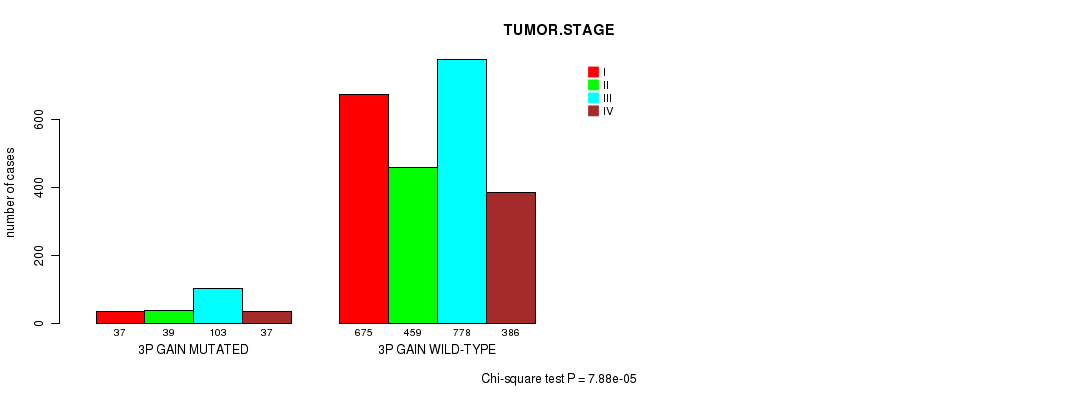
P value = 4.65e-05 (Chi-square test), Q value = 0.06
Table S25. Clustering Approach #5: '3p gain mutation analysis' versus Clinical Feature #21: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE IA | STAGE IB | STAGE II | STAGE IIA | STAGE IIB | STAGE III | STAGE IIIA | STAGE IIIB | STAGE IIIC | STAGE IV | STAGE TIS | STAGE X |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 72 | 62 | 7 | 43 | 287 | 186 | 42 | 124 | 25 | 41 | 59 | 1 | 16 |
| 3P GAIN MUTATED | 4 | 1 | 0 | 9 | 25 | 7 | 5 | 13 | 4 | 2 | 14 | 0 | 0 |
| 3P GAIN WILD-TYPE | 68 | 61 | 7 | 34 | 262 | 179 | 37 | 111 | 21 | 39 | 45 | 1 | 16 |
Figure S20. Get High-res Image Clustering Approach #5: '3p gain mutation analysis' versus Clinical Feature #21: 'NEOPLASM.DISEASESTAGE'
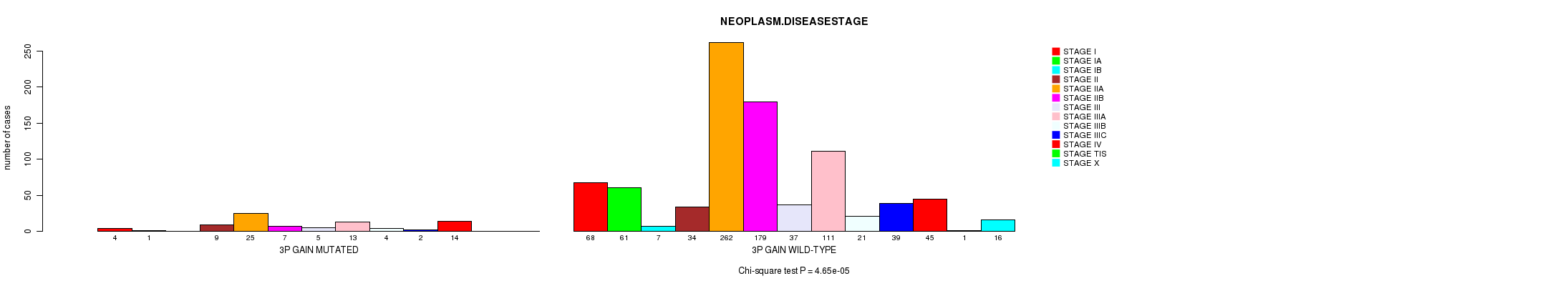
Table S26. Get Full Table Description of clustering approach #6: '3q gain mutation analysis'
| Cluster Labels | 3Q GAIN MUTATED | 3Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 758 | 3805 |
P value = 2.2e-87 (Chi-square test), Q value = 3.5e-84
Table S27. Clustering Approach #6: '3q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 3Q GAIN MUTATED | 36 | 28 | 99 | 28 | 44 | 104 | 20 | 180 | 0 | 193 | 2 | 23 |
| 3Q GAIN WILD-TYPE | 89 | 516 | 747 | 384 | 403 | 215 | 473 | 482 | 2 | 355 | 0 | 136 |
Figure S21. Get High-res Image Clustering Approach #6: '3q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'

P value = 1.53e-05 (t-test), Q value = 0.02
Table S28. Clustering Approach #6: '3q gain mutation analysis' versus Clinical Feature #5: 'KARNOFSKY.PERFORMANCE.SCORE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 640 | 73.6 (23.9) |
| 3Q GAIN MUTATED | 90 | 58.8 (34.9) |
| 3Q GAIN WILD-TYPE | 550 | 76.0 (20.7) |
Figure S22. Get High-res Image Clustering Approach #6: '3q gain mutation analysis' versus Clinical Feature #5: 'KARNOFSKY.PERFORMANCE.SCORE'
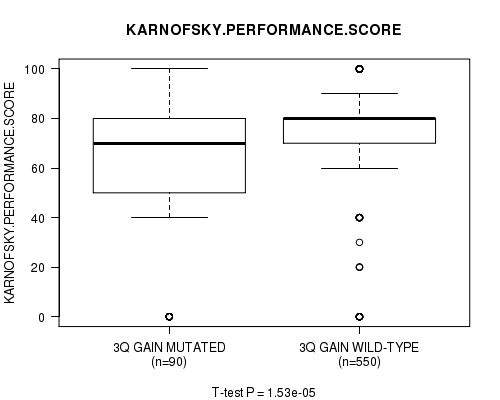
P value = 1.25e-88 (Chi-square test), Q value = 2e-85
Table S29. Clustering Approach #6: '3q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 3Q GAIN MUTATED | 26 | 2 | 24 | 104 | 20 | 1 | 4 | 16 | 5 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 152 | 2 | 0 | 23 | 0 | 195 | 18 | 0 | 1 |
| 3Q GAIN WILD-TYPE | 330 | 52 | 326 | 215 | 473 | 9 | 71 | 189 | 2 | 3 | 15 | 2 | 3 | 2 | 11 | 1 | 1 | 1 | 2 | 163 | 16 | 7 | 120 | 13 | 357 | 61 | 1 | 19 |
Figure S23. Get High-res Image Clustering Approach #6: '3q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 6.96e-07 (Chi-square test), Q value = 0.00094
Table S30. Clustering Approach #6: '3q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 3Q GAIN MUTATED | 72 | 138 | 80 | 48 |
| 3Q GAIN WILD-TYPE | 394 | 494 | 614 | 161 |
Figure S24. Get High-res Image Clustering Approach #6: '3q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
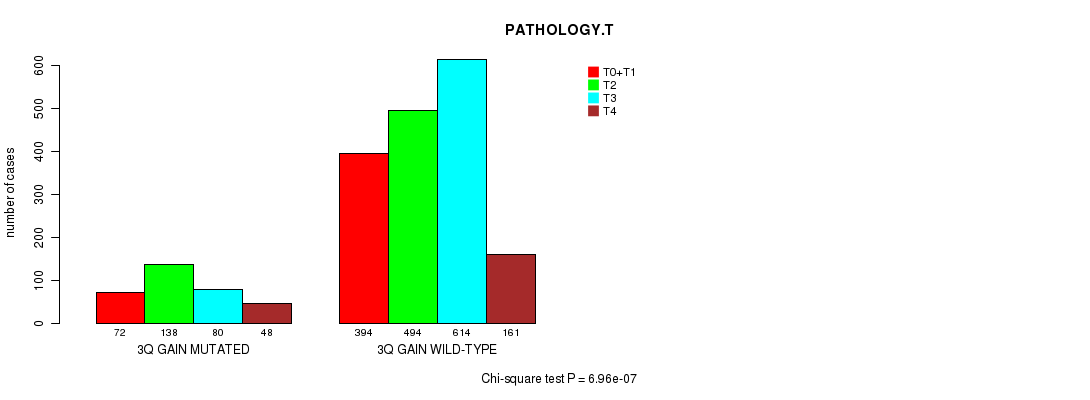
P value = 9.15e-06 (Chi-square test), Q value = 0.012
Table S31. Clustering Approach #6: '3q gain mutation analysis' versus Clinical Feature #8: 'PATHOLOGY.N'
| nPatients | N0 | N1 | N2 | N3 |
|---|---|---|---|---|
| ALL | 1067 | 353 | 282 | 11 |
| 3Q GAIN MUTATED | 166 | 86 | 63 | 6 |
| 3Q GAIN WILD-TYPE | 901 | 267 | 219 | 5 |
Figure S25. Get High-res Image Clustering Approach #6: '3q gain mutation analysis' versus Clinical Feature #8: 'PATHOLOGY.N'
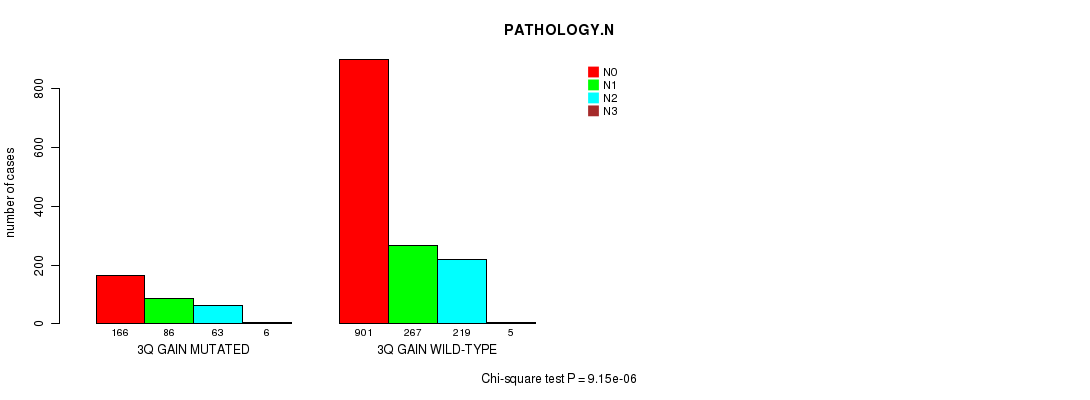
P value = 1.34e-06 (Chi-square test), Q value = 0.0018
Table S32. Clustering Approach #6: '3q gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 3Q GAIN MUTATED | 112 | 86 | 228 | 98 |
| 3Q GAIN WILD-TYPE | 600 | 412 | 653 | 325 |
Figure S26. Get High-res Image Clustering Approach #6: '3q gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
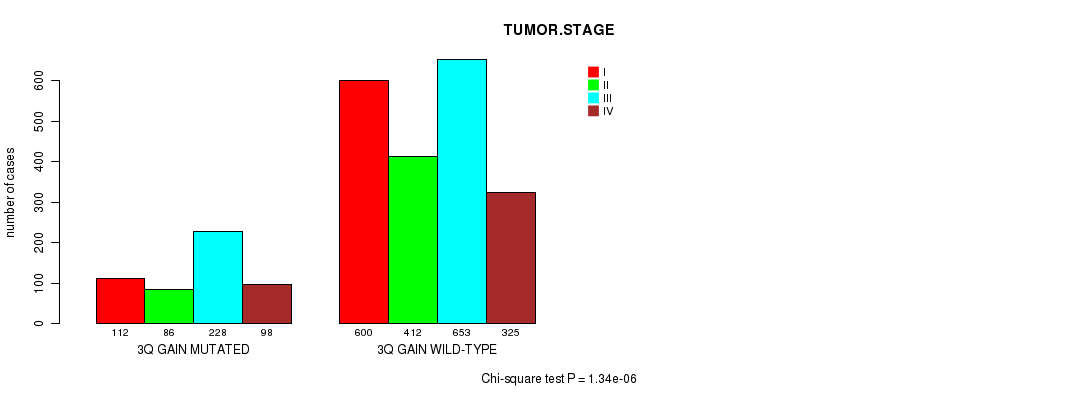
P value = 0.00013 (Fisher's exact test), Q value = 0.17
Table S33. Clustering Approach #6: '3q gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 3Q GAIN MUTATED | 101 | 657 |
| 3Q GAIN WILD-TYPE | 725 | 3080 |
Figure S27. Get High-res Image Clustering Approach #6: '3q gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
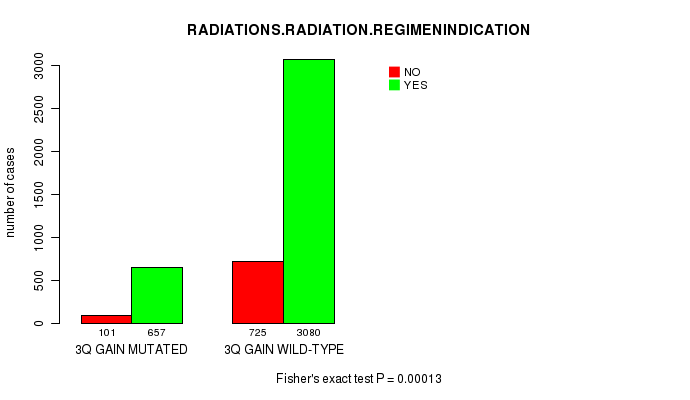
Table S34. Get Full Table Description of clustering approach #7: '4p gain mutation analysis'
| Cluster Labels | 4P GAIN MUTATED | 4P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 151 | 4412 |
P value = 1.01e-06 (Chi-square test), Q value = 0.0014
Table S35. Clustering Approach #7: '4p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 4P GAIN MUTATED | 8 | 11 | 32 | 2 | 5 | 14 | 8 | 37 | 0 | 30 | 0 | 4 |
| 4P GAIN WILD-TYPE | 117 | 533 | 814 | 410 | 442 | 305 | 485 | 625 | 2 | 518 | 2 | 155 |
Figure S28. Get High-res Image Clustering Approach #7: '4p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
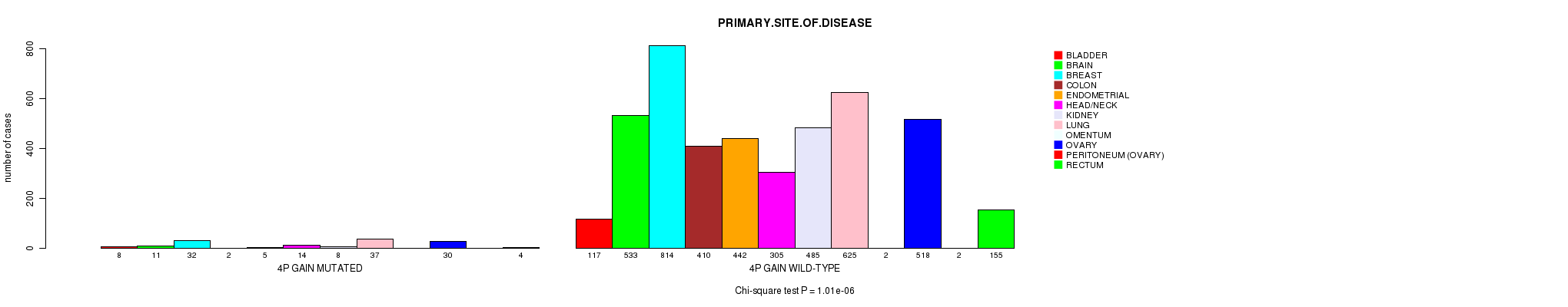
P value = 1.31e-06 (Chi-square test), Q value = 0.0018
Table S36. Clustering Approach #7: '4p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 4P GAIN MUTATED | 2 | 0 | 1 | 14 | 8 | 1 | 3 | 18 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 13 | 0 | 0 | 4 | 0 | 30 | 4 | 0 | 0 |
| 4P GAIN WILD-TYPE | 354 | 54 | 349 | 305 | 485 | 9 | 72 | 187 | 6 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 1 | 302 | 18 | 7 | 139 | 13 | 522 | 75 | 1 | 20 |
Figure S29. Get High-res Image Clustering Approach #7: '4p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S37. Get Full Table Description of clustering approach #8: '4q gain mutation analysis'
| Cluster Labels | 4Q GAIN MUTATED | 4Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 101 | 4462 |
Table S38. Get Full Table Description of clustering approach #9: '5p gain mutation analysis'
| Cluster Labels | 5P GAIN MUTATED | 5P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 963 | 3600 |
P value = 3.72e-05 (t-test), Q value = 0.048
Table S39. Clustering Approach #9: '5p gain mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 4507 | 61.9 (12.6) |
| 5P GAIN MUTATED | 949 | 63.3 (11.7) |
| 5P GAIN WILD-TYPE | 3558 | 61.5 (12.8) |
Figure S30. Get High-res Image Clustering Approach #9: '5p gain mutation analysis' versus Clinical Feature #2: 'AGE'
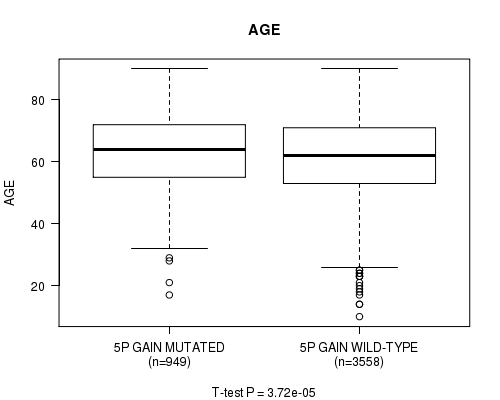
P value = 7.92e-83 (Chi-square test), Q value = 1.2e-79
Table S40. Clustering Approach #9: '5p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 5P GAIN MUTATED | 38 | 27 | 166 | 37 | 29 | 81 | 133 | 287 | 0 | 142 | 2 | 21 |
| 5P GAIN WILD-TYPE | 87 | 517 | 680 | 375 | 418 | 238 | 360 | 375 | 2 | 406 | 0 | 138 |
Figure S31. Get High-res Image Clustering Approach #9: '5p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
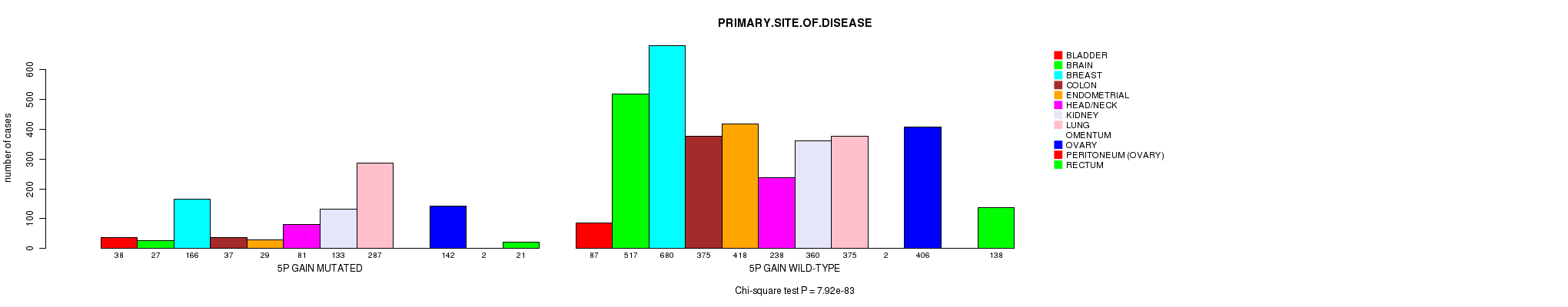
P value = 1.28e-52 (Chi-square test), Q value = 2e-49
Table S41. Clustering Approach #9: '5p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 5P GAIN MUTATED | 34 | 3 | 11 | 81 | 133 | 2 | 31 | 82 | 4 | 1 | 6 | 2 | 2 | 0 | 4 | 0 | 0 | 1 | 1 | 151 | 1 | 0 | 21 | 0 | 144 | 17 | 1 | 1 |
| 5P GAIN WILD-TYPE | 322 | 51 | 339 | 238 | 360 | 8 | 44 | 123 | 3 | 2 | 9 | 0 | 1 | 2 | 9 | 1 | 1 | 0 | 1 | 164 | 17 | 7 | 122 | 13 | 408 | 62 | 0 | 19 |
Figure S32. Get High-res Image Clustering Approach #9: '5p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 8.46e-10 (Chi-square test), Q value = 1.2e-06
Table S42. Clustering Approach #9: '5p gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 5P GAIN MUTATED | 133 | 225 | 135 | 50 |
| 5P GAIN WILD-TYPE | 333 | 407 | 559 | 159 |
Figure S33. Get High-res Image Clustering Approach #9: '5p gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
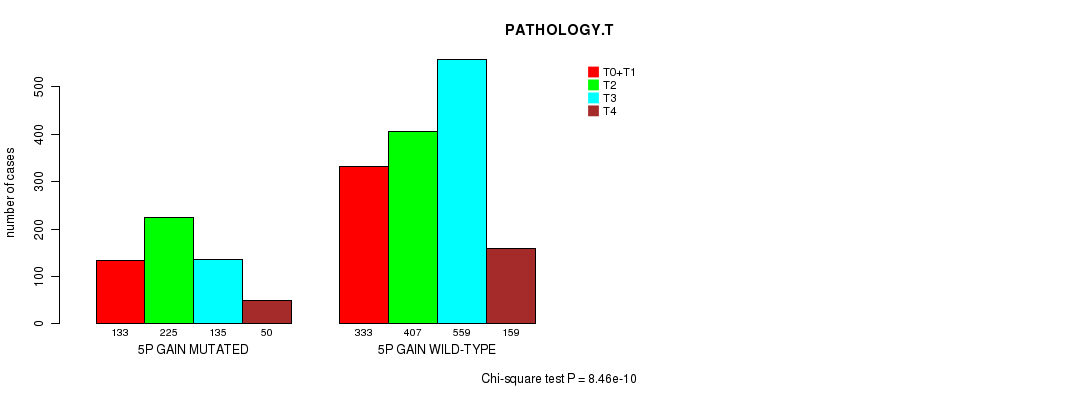
P value = 1.41e-08 (Fisher's exact test), Q value = 2e-05
Table S43. Clustering Approach #9: '5p gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 5P GAIN MUTATED | 116 | 847 |
| 5P GAIN WILD-TYPE | 710 | 2890 |
Figure S34. Get High-res Image Clustering Approach #9: '5p gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
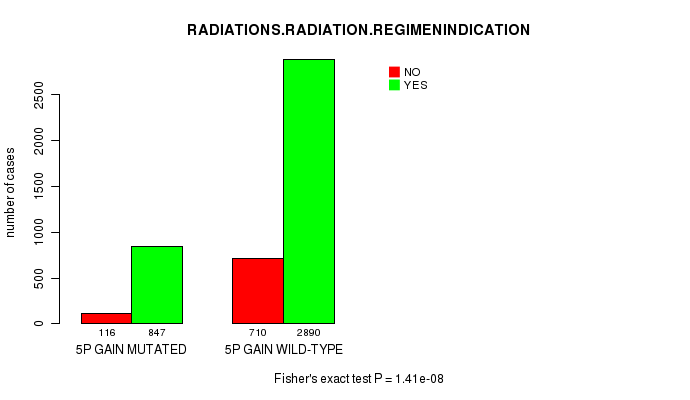
Table S44. Get Full Table Description of clustering approach #10: '5q gain mutation analysis'
| Cluster Labels | 5Q GAIN MUTATED | 5Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 463 | 4100 |
P value = 9.05e-58 (Chi-square test), Q value = 1.4e-54
Table S45. Clustering Approach #10: '5q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 5Q GAIN MUTATED | 16 | 20 | 100 | 20 | 8 | 30 | 145 | 83 | 0 | 29 | 1 | 11 |
| 5Q GAIN WILD-TYPE | 109 | 524 | 746 | 392 | 439 | 289 | 348 | 579 | 2 | 519 | 1 | 148 |
Figure S35. Get High-res Image Clustering Approach #10: '5q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
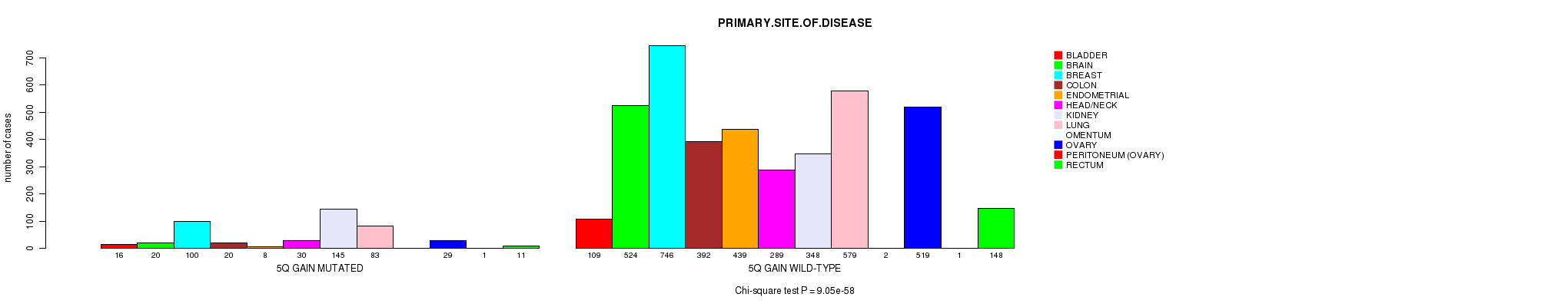
P value = 4.57e-43 (Chi-square test), Q value = 7e-40
Table S46. Clustering Approach #10: '5q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 5Q GAIN MUTATED | 19 | 1 | 3 | 30 | 145 | 1 | 11 | 28 | 1 | 1 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 37 | 0 | 0 | 11 | 0 | 30 | 5 | 1 | 0 |
| 5Q GAIN WILD-TYPE | 337 | 53 | 347 | 289 | 348 | 9 | 64 | 177 | 6 | 2 | 14 | 1 | 3 | 2 | 11 | 1 | 1 | 1 | 2 | 278 | 18 | 7 | 132 | 13 | 522 | 74 | 0 | 20 |
Figure S36. Get High-res Image Clustering Approach #10: '5q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S47. Get Full Table Description of clustering approach #11: '6p gain mutation analysis'
| Cluster Labels | 6P GAIN MUTATED | 6P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 453 | 4110 |
P value = 1.08e-35 (Chi-square test), Q value = 1.6e-32
Table S48. Clustering Approach #11: '6p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 6P GAIN MUTATED | 10 | 5 | 100 | 47 | 34 | 26 | 5 | 83 | 0 | 115 | 1 | 27 |
| 6P GAIN WILD-TYPE | 115 | 539 | 746 | 365 | 413 | 293 | 488 | 579 | 2 | 433 | 1 | 132 |
Figure S37. Get High-res Image Clustering Approach #11: '6p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
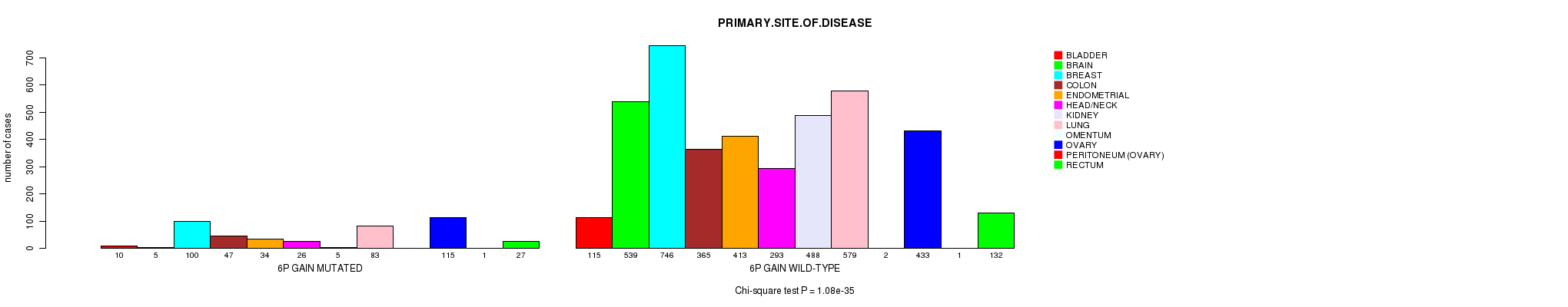
P value = 3.41e-07 (Fisher's exact test), Q value = 0.00047
Table S49. Clustering Approach #11: '6p gain mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 6P GAIN MUTATED | 335 | 118 |
| 6P GAIN WILD-TYPE | 2546 | 1564 |
Figure S38. Get High-res Image Clustering Approach #11: '6p gain mutation analysis' versus Clinical Feature #4: 'GENDER'

P value = 4.48e-22 (Chi-square test), Q value = 6.6e-19
Table S50. Clustering Approach #11: '6p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 6P GAIN MUTATED | 40 | 7 | 20 | 26 | 5 | 3 | 13 | 28 | 0 | 0 | 4 | 1 | 1 | 0 | 2 | 0 | 0 | 1 | 0 | 30 | 0 | 0 | 27 | 0 | 116 | 14 | 0 | 0 |
| 6P GAIN WILD-TYPE | 316 | 47 | 330 | 293 | 488 | 7 | 62 | 177 | 7 | 3 | 11 | 1 | 2 | 2 | 11 | 1 | 1 | 0 | 2 | 285 | 18 | 7 | 116 | 13 | 436 | 65 | 1 | 20 |
Figure S39. Get High-res Image Clustering Approach #11: '6p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 6.63e-07 (Chi-square test), Q value = 9e-04
Table S51. Clustering Approach #11: '6p gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 6P GAIN MUTATED | 53 | 54 | 144 | 45 |
| 6P GAIN WILD-TYPE | 659 | 444 | 737 | 378 |
Figure S40. Get High-res Image Clustering Approach #11: '6p gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
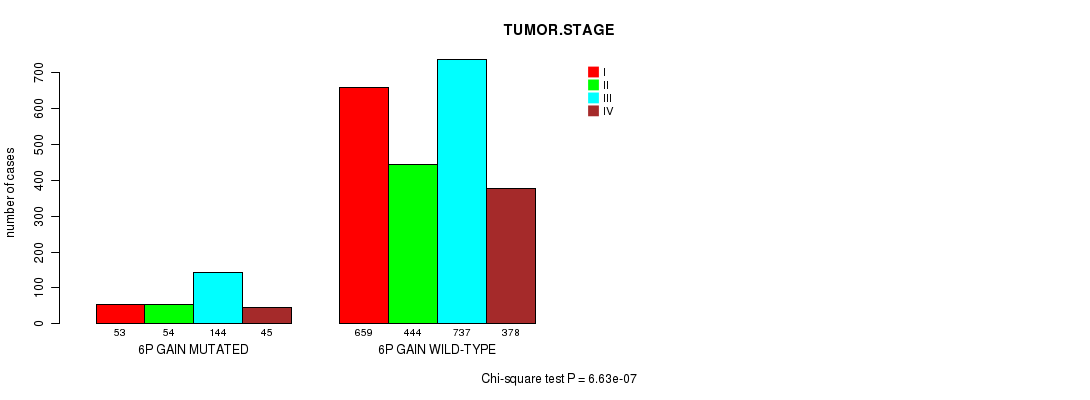
Table S52. Get Full Table Description of clustering approach #12: '6q gain mutation analysis'
| Cluster Labels | 6Q GAIN MUTATED | 6Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 295 | 4268 |
P value = 1.31e-18 (Chi-square test), Q value = 1.9e-15
Table S53. Clustering Approach #12: '6q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 6Q GAIN MUTATED | 6 | 7 | 65 | 43 | 31 | 19 | 4 | 33 | 0 | 60 | 0 | 26 |
| 6Q GAIN WILD-TYPE | 119 | 537 | 781 | 369 | 416 | 300 | 489 | 629 | 2 | 488 | 2 | 133 |
Figure S41. Get High-res Image Clustering Approach #12: '6q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
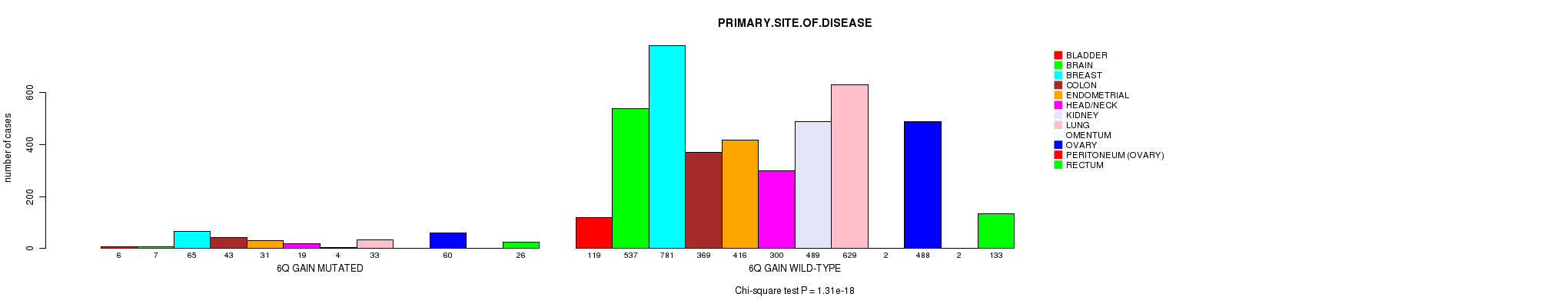
P value = 2.41e-13 (Chi-square test), Q value = 3.4e-10
Table S54. Clustering Approach #12: '6q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 6Q GAIN MUTATED | 38 | 5 | 17 | 19 | 4 | 0 | 2 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 18 | 0 | 0 | 26 | 0 | 60 | 14 | 0 | 0 |
| 6Q GAIN WILD-TYPE | 318 | 49 | 333 | 300 | 489 | 10 | 73 | 194 | 7 | 3 | 15 | 2 | 3 | 2 | 12 | 1 | 1 | 0 | 2 | 297 | 18 | 7 | 117 | 13 | 492 | 65 | 1 | 20 |
Figure S42. Get High-res Image Clustering Approach #12: '6q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 6.68e-05 (Chi-square test), Q value = 0.086
Table S55. Clustering Approach #12: '6q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 6Q GAIN MUTATED | 13 | 31 | 63 | 16 |
| 6Q GAIN WILD-TYPE | 453 | 601 | 631 | 193 |
Figure S43. Get High-res Image Clustering Approach #12: '6q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
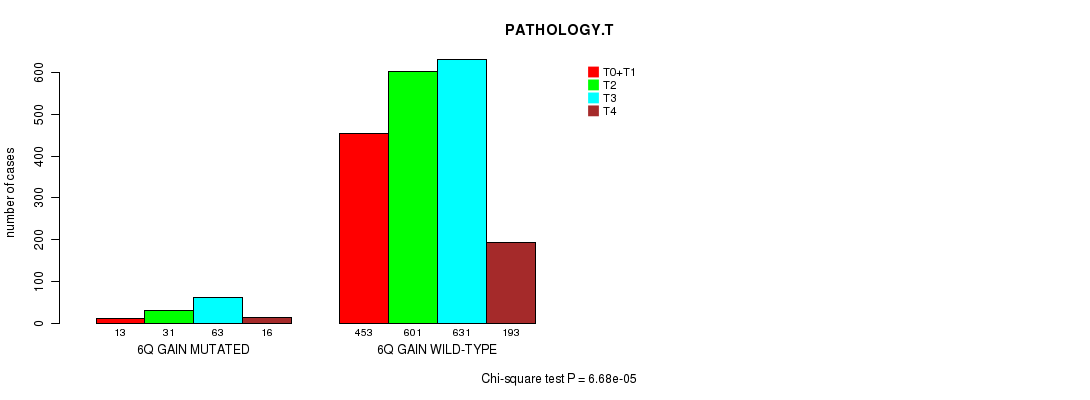
P value = 1.85e-05 (Chi-square test), Q value = 0.024
Table S56. Clustering Approach #12: '6q gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 6Q GAIN MUTATED | 24 | 38 | 86 | 32 |
| 6Q GAIN WILD-TYPE | 688 | 460 | 795 | 391 |
Figure S44. Get High-res Image Clustering Approach #12: '6q gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
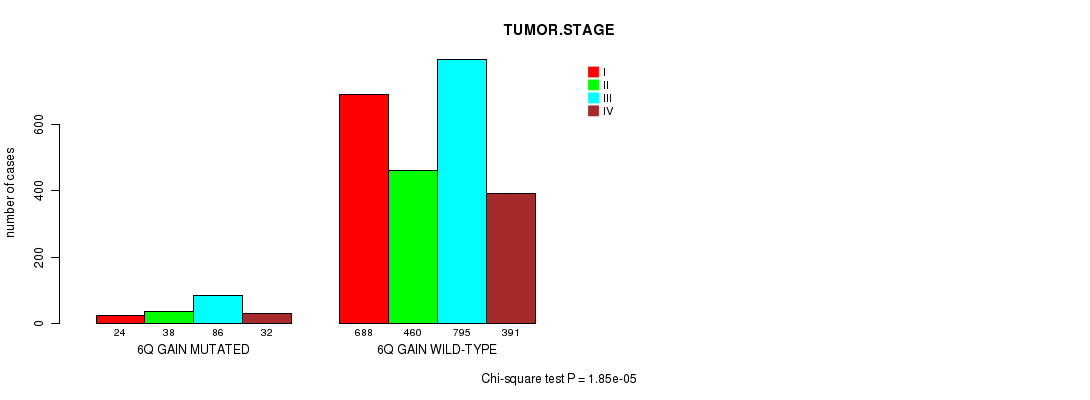
Table S57. Get Full Table Description of clustering approach #13: '7p gain mutation analysis'
| Cluster Labels | 7P GAIN MUTATED | 7P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 1455 | 3108 |
P value = 0 (logrank test), Q value = 0
Table S58. Clustering Approach #13: '7p gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 4296 | 1401 | 0.0 - 224.0 (14.9) |
| 7P GAIN MUTATED | 1360 | 572 | 0.0 - 224.0 (11.0) |
| 7P GAIN WILD-TYPE | 2936 | 829 | 0.0 - 223.4 (17.5) |
Figure S45. Get High-res Image Clustering Approach #13: '7p gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
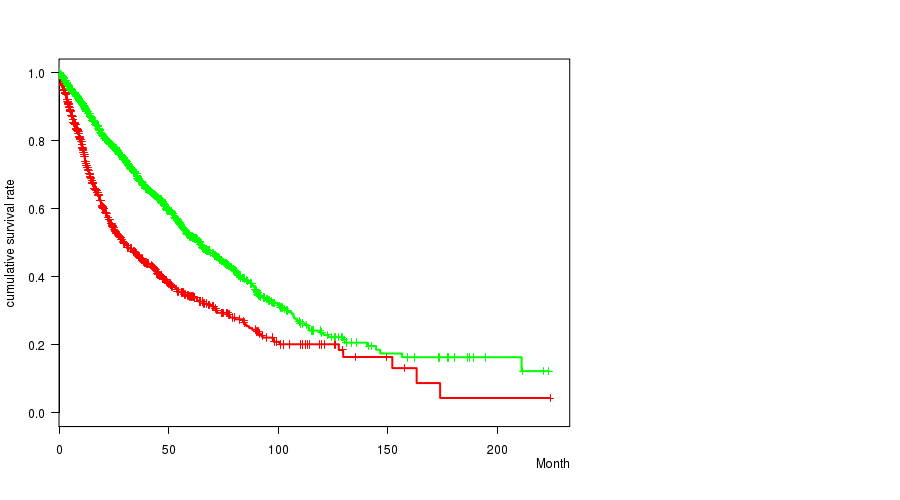
P value = 1.14e-166 (Chi-square test), Q value = 1.8e-163
Table S59. Clustering Approach #13: '7p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 7P GAIN MUTATED | 35 | 412 | 156 | 199 | 40 | 73 | 127 | 210 | 0 | 112 | 0 | 89 |
| 7P GAIN WILD-TYPE | 90 | 132 | 690 | 213 | 407 | 246 | 366 | 452 | 2 | 436 | 2 | 70 |
Figure S46. Get High-res Image Clustering Approach #13: '7p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
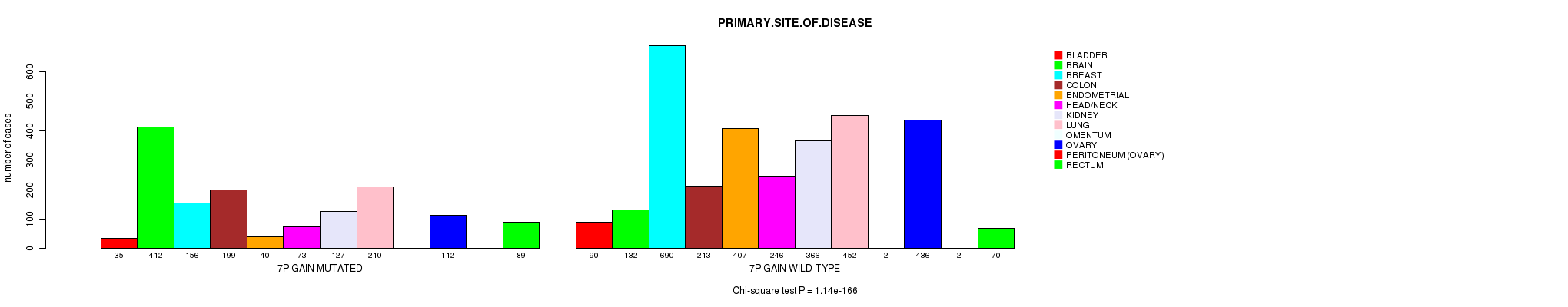
P value = 6.39e-27 (Fisher's exact test), Q value = 9.5e-24
Table S60. Clustering Approach #13: '7p gain mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 7P GAIN MUTATED | 754 | 701 |
| 7P GAIN WILD-TYPE | 2127 | 981 |
Figure S47. Get High-res Image Clustering Approach #13: '7p gain mutation analysis' versus Clinical Feature #4: 'GENDER'
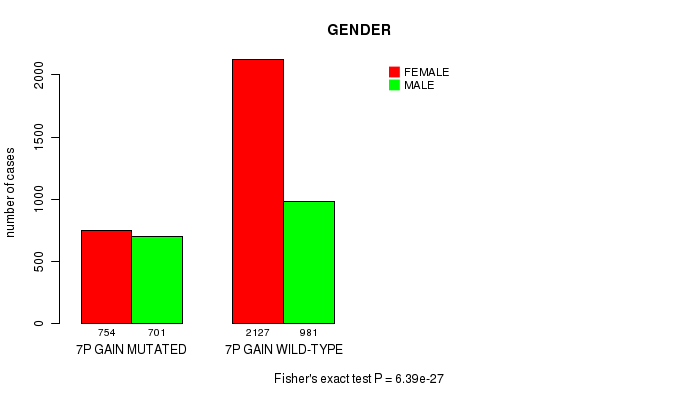
P value = 1.01e-51 (Chi-square test), Q value = 1.6e-48
Table S61. Clustering Approach #13: '7p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 7P GAIN MUTATED | 184 | 15 | 27 | 73 | 127 | 5 | 20 | 78 | 3 | 2 | 4 | 0 | 1 | 0 | 5 | 1 | 0 | 0 | 1 | 89 | 1 | 1 | 87 | 2 | 112 | 12 | 0 | 14 |
| 7P GAIN WILD-TYPE | 172 | 39 | 323 | 246 | 366 | 5 | 55 | 127 | 4 | 1 | 11 | 2 | 2 | 2 | 8 | 0 | 1 | 1 | 1 | 226 | 17 | 6 | 56 | 11 | 440 | 67 | 1 | 6 |
Figure S48. Get High-res Image Clustering Approach #13: '7p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.49e-09 (Chi-square test), Q value = 2.1e-06
Table S62. Clustering Approach #13: '7p gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 7P GAIN MUTATED | 110 | 221 | 294 | 67 |
| 7P GAIN WILD-TYPE | 356 | 411 | 400 | 142 |
Figure S49. Get High-res Image Clustering Approach #13: '7p gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'

P value = 7.41e-05 (Chi-square test), Q value = 0.095
Table S63. Clustering Approach #13: '7p gain mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
| nPatients | M0 | M1 | M1A | M1B | MX |
|---|---|---|---|---|---|
| ALL | 1424 | 168 | 9 | 3 | 165 |
| 7P GAIN MUTATED | 479 | 80 | 8 | 1 | 59 |
| 7P GAIN WILD-TYPE | 945 | 88 | 1 | 2 | 106 |
Figure S50. Get High-res Image Clustering Approach #13: '7p gain mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
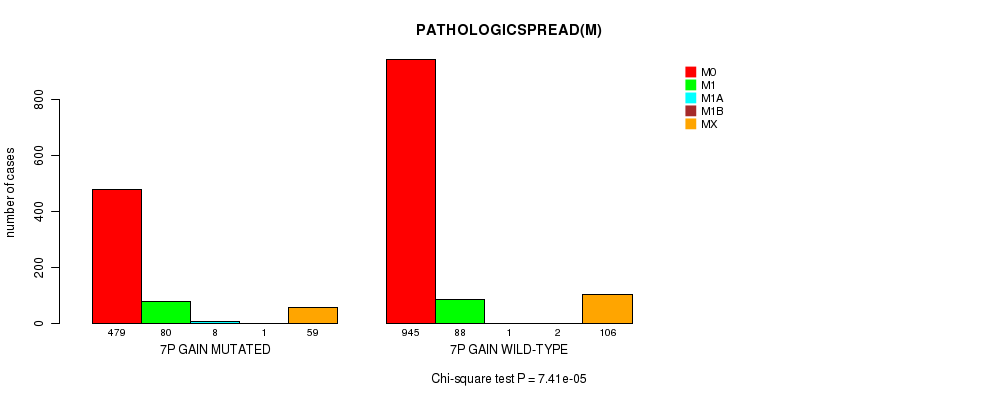
P value = 6.56e-14 (Fisher's exact test), Q value = 9.4e-11
Table S64. Clustering Approach #13: '7p gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 7P GAIN MUTATED | 356 | 1099 |
| 7P GAIN WILD-TYPE | 470 | 2638 |
Figure S51. Get High-res Image Clustering Approach #13: '7p gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
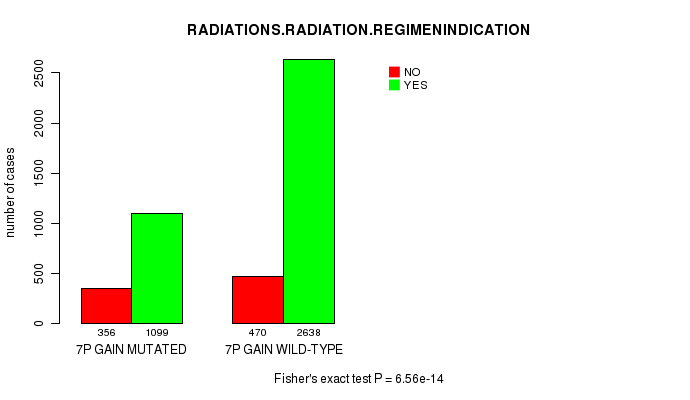
Table S65. Get Full Table Description of clustering approach #14: '7q gain mutation analysis'
| Cluster Labels | 7Q GAIN MUTATED | 7Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 1359 | 3204 |
P value = 0 (logrank test), Q value = 0
Table S66. Clustering Approach #14: '7q gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 4296 | 1401 | 0.0 - 224.0 (14.9) |
| 7Q GAIN MUTATED | 1274 | 559 | 0.0 - 224.0 (11.1) |
| 7Q GAIN WILD-TYPE | 3022 | 842 | 0.0 - 223.4 (17.2) |
Figure S52. Get High-res Image Clustering Approach #14: '7q gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
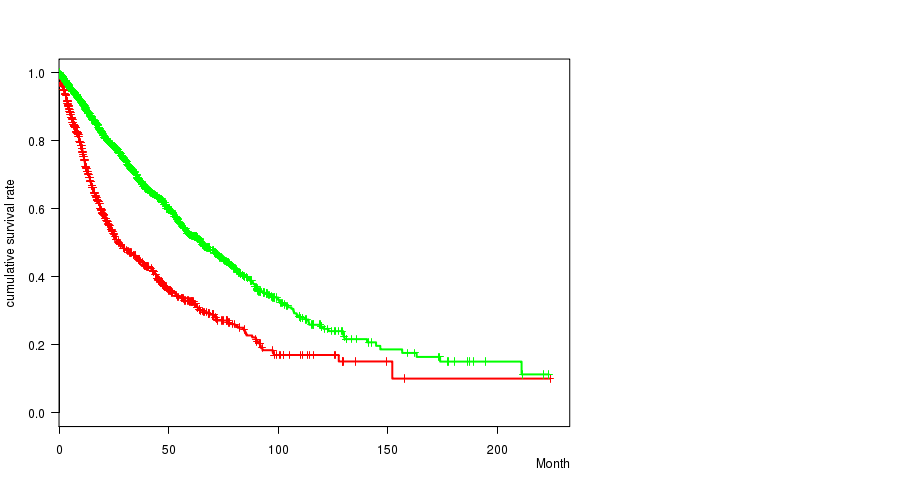
P value = 1.26e-182 (Chi-square test), Q value = 2e-179
Table S67. Clustering Approach #14: '7q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 7Q GAIN MUTATED | 35 | 419 | 115 | 175 | 39 | 50 | 130 | 177 | 0 | 138 | 1 | 78 |
| 7Q GAIN WILD-TYPE | 90 | 125 | 731 | 237 | 408 | 269 | 363 | 485 | 2 | 410 | 1 | 81 |
Figure S53. Get High-res Image Clustering Approach #14: '7q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
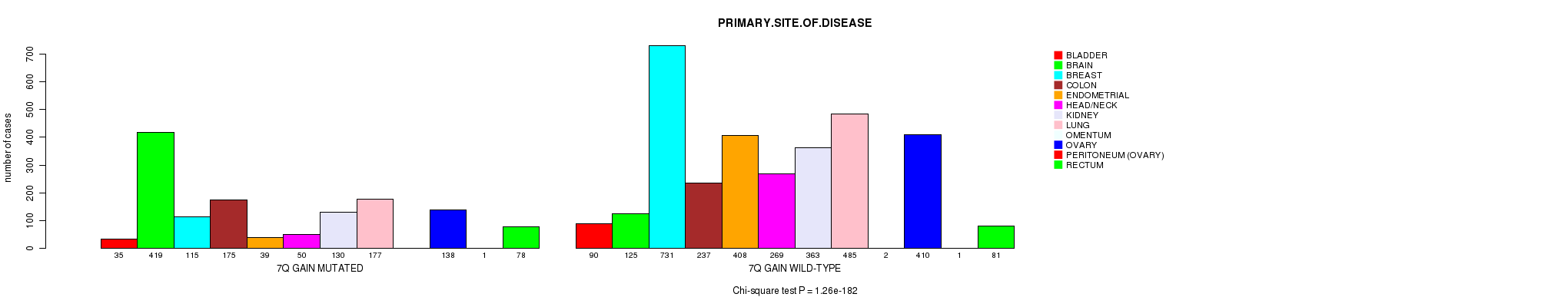
P value = 8.98e-27 (Fisher's exact test), Q value = 1.3e-23
Table S68. Clustering Approach #14: '7q gain mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 7Q GAIN MUTATED | 697 | 662 |
| 7Q GAIN WILD-TYPE | 2184 | 1020 |
Figure S54. Get High-res Image Clustering Approach #14: '7q gain mutation analysis' versus Clinical Feature #4: 'GENDER'
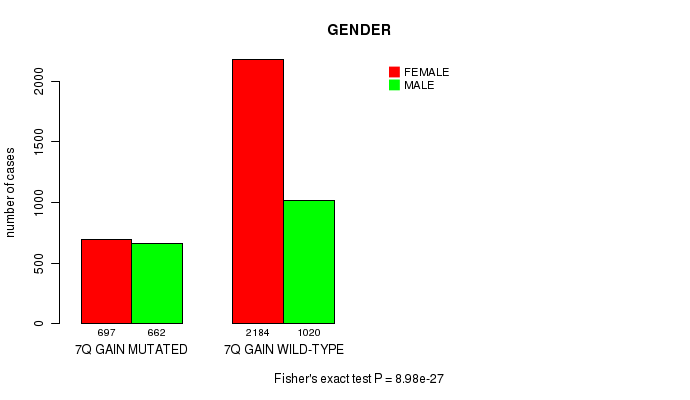
P value = 1.03e-38 (Chi-square test), Q value = 1.6e-35
Table S69. Clustering Approach #14: '7q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 7Q GAIN MUTATED | 161 | 14 | 29 | 50 | 130 | 3 | 19 | 56 | 2 | 2 | 3 | 0 | 1 | 0 | 4 | 1 | 0 | 0 | 0 | 86 | 1 | 0 | 77 | 1 | 139 | 9 | 1 | 15 |
| 7Q GAIN WILD-TYPE | 195 | 40 | 321 | 269 | 363 | 7 | 56 | 149 | 5 | 1 | 12 | 2 | 2 | 2 | 9 | 0 | 1 | 1 | 2 | 229 | 17 | 7 | 66 | 12 | 413 | 70 | 0 | 5 |
Figure S55. Get High-res Image Clustering Approach #14: '7q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.08e-08 (Chi-square test), Q value = 1.5e-05
Table S70. Clustering Approach #14: '7q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 7Q GAIN MUTATED | 99 | 186 | 265 | 56 |
| 7Q GAIN WILD-TYPE | 367 | 446 | 429 | 153 |
Figure S56. Get High-res Image Clustering Approach #14: '7q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
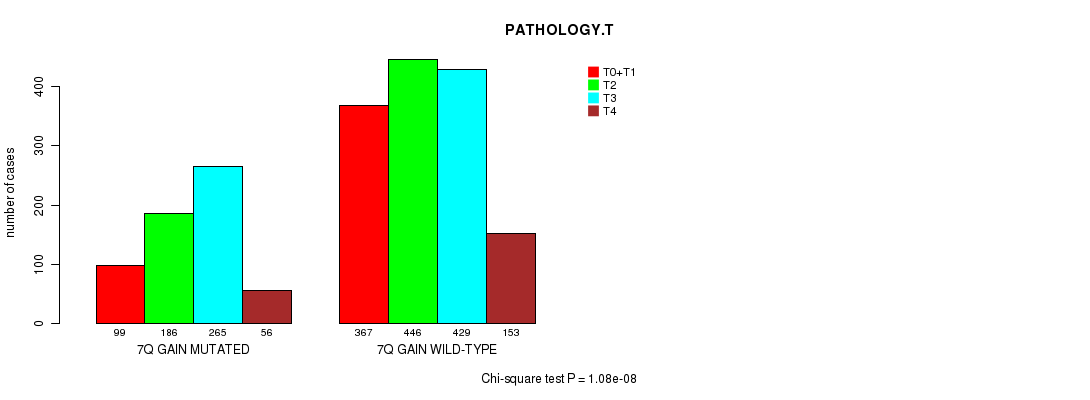
P value = 9.32e-07 (Chi-square test), Q value = 0.0013
Table S71. Clustering Approach #14: '7q gain mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
| nPatients | M0 | M1 | M1A | M1B | MX |
|---|---|---|---|---|---|
| ALL | 1424 | 168 | 9 | 3 | 165 |
| 7Q GAIN MUTATED | 421 | 78 | 8 | 1 | 51 |
| 7Q GAIN WILD-TYPE | 1003 | 90 | 1 | 2 | 114 |
Figure S57. Get High-res Image Clustering Approach #14: '7q gain mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
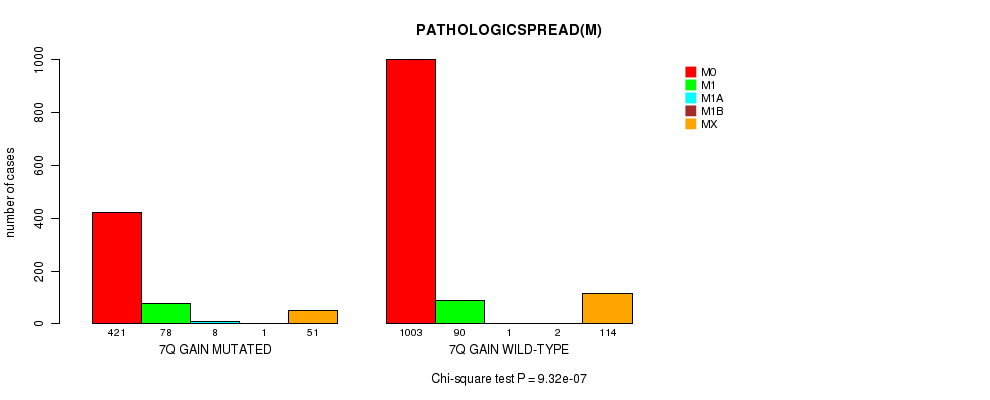
P value = 3.79e-17 (Fisher's exact test), Q value = 5.5e-14
Table S72. Clustering Approach #14: '7q gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 7Q GAIN MUTATED | 349 | 1010 |
| 7Q GAIN WILD-TYPE | 477 | 2727 |
Figure S58. Get High-res Image Clustering Approach #14: '7q gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
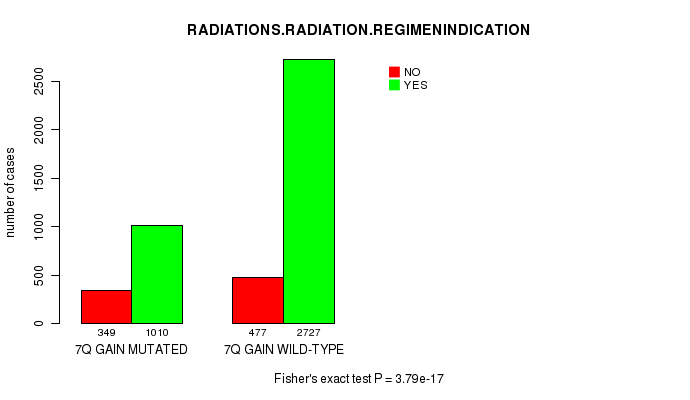
Table S73. Get Full Table Description of clustering approach #15: '8p gain mutation analysis'
| Cluster Labels | 8P GAIN MUTATED | 8P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 643 | 3920 |
P value = 4.18e-25 (Chi-square test), Q value = 6.2e-22
Table S74. Clustering Approach #15: '8p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 8P GAIN MUTATED | 17 | 24 | 178 | 74 | 80 | 54 | 14 | 96 | 0 | 80 | 1 | 25 |
| 8P GAIN WILD-TYPE | 108 | 520 | 668 | 338 | 367 | 265 | 479 | 566 | 2 | 468 | 1 | 134 |
Figure S59. Get High-res Image Clustering Approach #15: '8p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
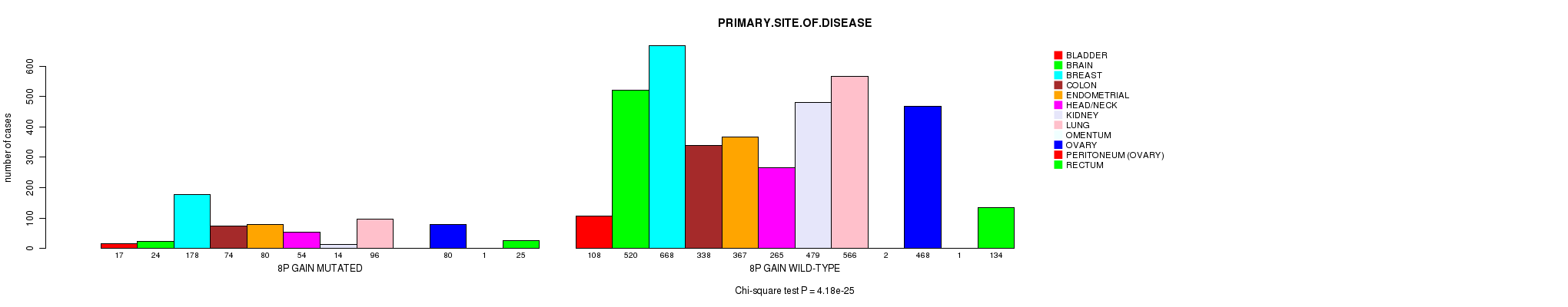
P value = 7.81e-08 (Chi-square test), Q value = 0.00011
Table S75. Clustering Approach #15: '8p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 8P GAIN MUTATED | 64 | 10 | 61 | 54 | 14 | 3 | 11 | 28 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 49 | 4 | 0 | 23 | 2 | 81 | 15 | 0 | 0 |
| 8P GAIN WILD-TYPE | 292 | 44 | 289 | 265 | 479 | 7 | 64 | 177 | 5 | 3 | 14 | 2 | 3 | 2 | 12 | 1 | 1 | 0 | 2 | 266 | 14 | 7 | 120 | 11 | 471 | 64 | 1 | 20 |
Figure S60. Get High-res Image Clustering Approach #15: '8p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 2.28e-06 (Chi-square test), Q value = 0.003
Table S76. Clustering Approach #15: '8p gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 8P GAIN MUTATED | 26 | 96 | 97 | 35 |
| 8P GAIN WILD-TYPE | 440 | 536 | 597 | 174 |
Figure S61. Get High-res Image Clustering Approach #15: '8p gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
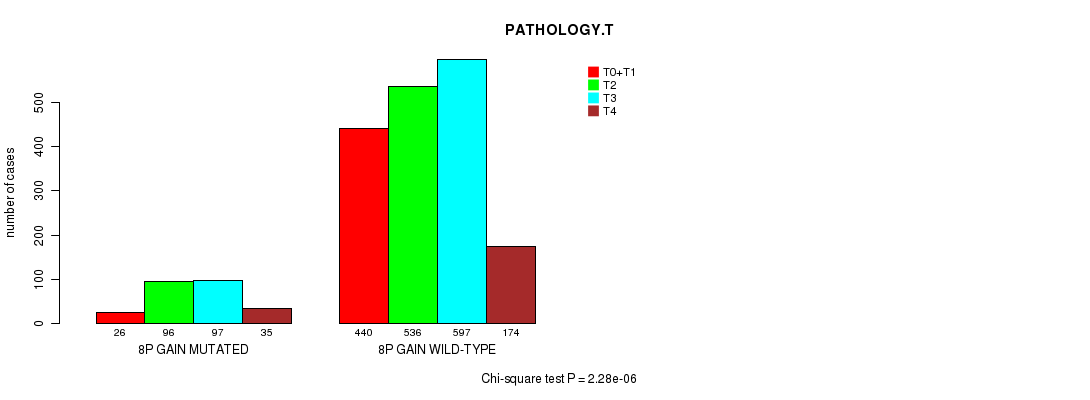
Table S77. Get Full Table Description of clustering approach #16: '8q gain mutation analysis'
| Cluster Labels | 8Q GAIN MUTATED | 8Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 1335 | 3228 |
P value = 0.000192 (logrank test), Q value = 0.24
Table S78. Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 4296 | 1401 | 0.0 - 224.0 (14.9) |
| 8Q GAIN MUTATED | 1223 | 345 | 0.0 - 220.9 (15.1) |
| 8Q GAIN WILD-TYPE | 3073 | 1056 | 0.0 - 224.0 (14.7) |
Figure S62. Get High-res Image Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
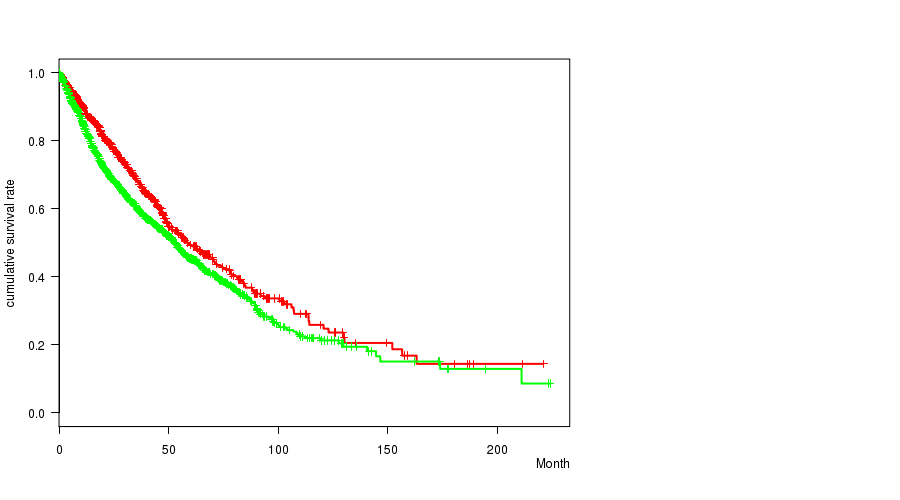
P value = 5.25e-86 (Chi-square test), Q value = 8.2e-83
Table S79. Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 8Q GAIN MUTATED | 35 | 28 | 368 | 163 | 103 | 132 | 36 | 208 | 1 | 189 | 2 | 70 |
| 8Q GAIN WILD-TYPE | 90 | 516 | 478 | 249 | 344 | 187 | 457 | 454 | 1 | 359 | 0 | 89 |
Figure S63. Get High-res Image Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
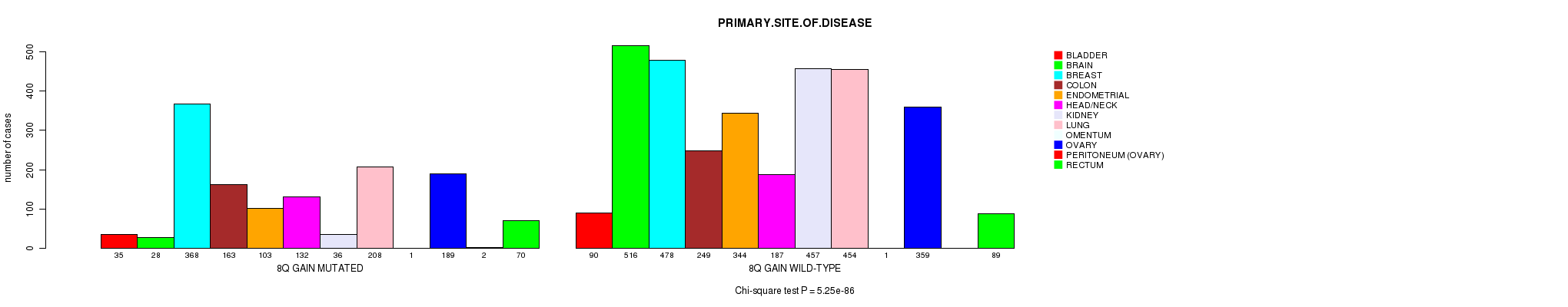
P value = 9.07e-05 (Fisher's exact test), Q value = 0.12
Table S80. Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 8Q GAIN MUTATED | 901 | 434 |
| 8Q GAIN WILD-TYPE | 1980 | 1248 |
Figure S64. Get High-res Image Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #4: 'GENDER'
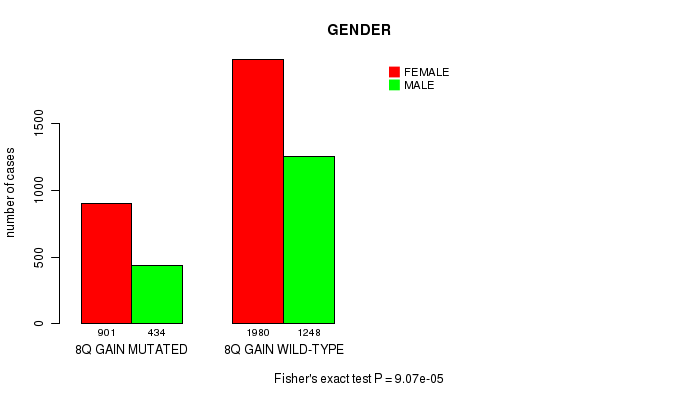
P value = 8.16e-35 (Chi-square test), Q value = 1.2e-31
Table S81. Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 8Q GAIN MUTATED | 149 | 13 | 70 | 132 | 36 | 5 | 25 | 65 | 3 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 100 | 5 | 0 | 65 | 3 | 192 | 28 | 0 | 0 |
| 8Q GAIN WILD-TYPE | 207 | 41 | 280 | 187 | 457 | 5 | 50 | 140 | 4 | 3 | 11 | 2 | 3 | 2 | 9 | 1 | 1 | 1 | 0 | 215 | 13 | 7 | 78 | 10 | 360 | 51 | 1 | 20 |
Figure S65. Get High-res Image Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.08e-14 (Chi-square test), Q value = 1.6e-11
Table S82. Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 8Q GAIN MUTATED | 70 | 207 | 226 | 87 |
| 8Q GAIN WILD-TYPE | 396 | 425 | 468 | 122 |
Figure S66. Get High-res Image Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
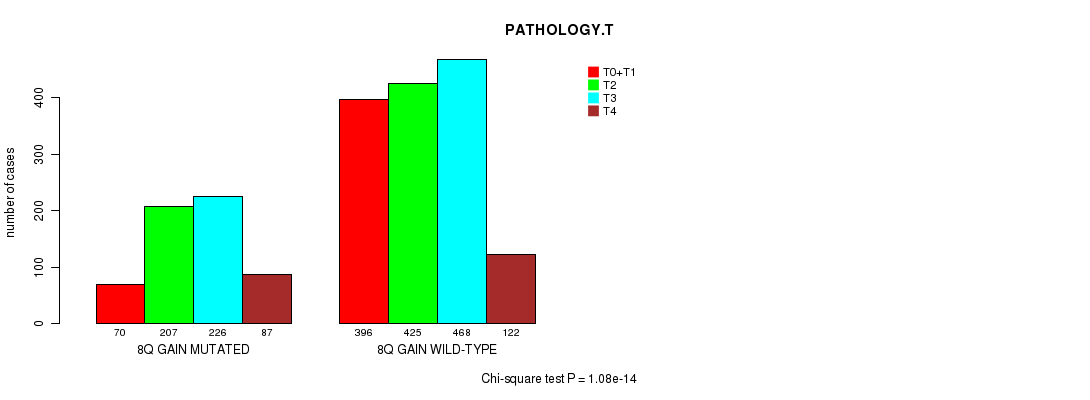
P value = 1.34e-07 (Chi-square test), Q value = 0.00018
Table S83. Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #8: 'PATHOLOGY.N'
| nPatients | N0 | N1 | N2 | N3 |
|---|---|---|---|---|
| ALL | 1067 | 353 | 282 | 11 |
| 8Q GAIN MUTATED | 300 | 123 | 130 | 5 |
| 8Q GAIN WILD-TYPE | 767 | 230 | 152 | 6 |
Figure S67. Get High-res Image Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #8: 'PATHOLOGY.N'
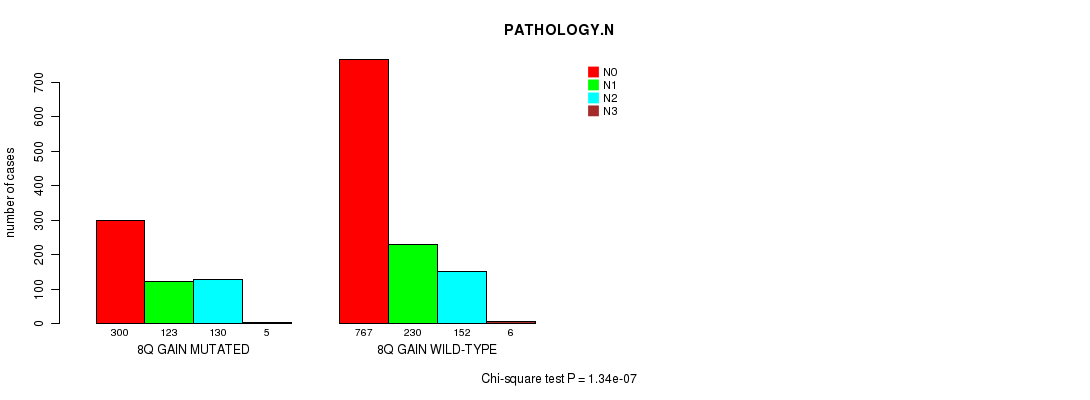
P value = 1.98e-09 (Chi-square test), Q value = 2.8e-06
Table S84. Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 8Q GAIN MUTATED | 152 | 163 | 297 | 159 |
| 8Q GAIN WILD-TYPE | 560 | 335 | 584 | 264 |
Figure S68. Get High-res Image Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
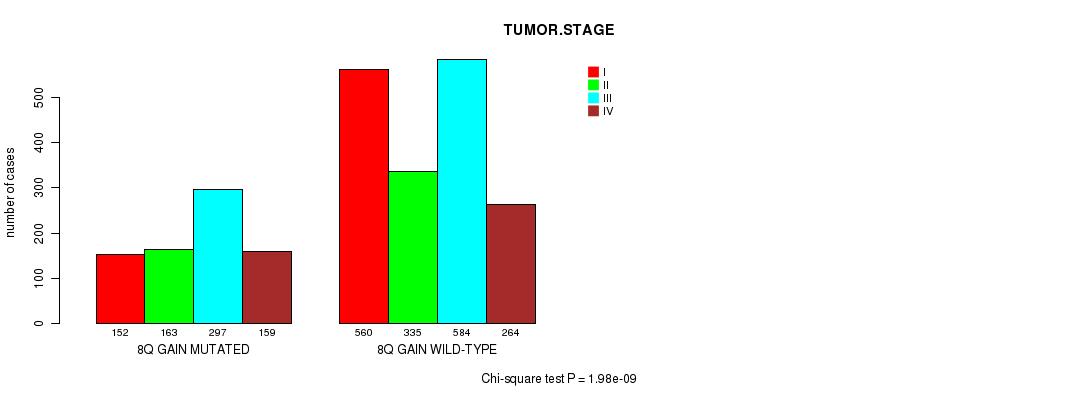
P value = 7.22e-06 (Fisher's exact test), Q value = 0.0096
Table S85. Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 8Q GAIN MUTATED | 189 | 1146 |
| 8Q GAIN WILD-TYPE | 637 | 2591 |
Figure S69. Get High-res Image Clustering Approach #16: '8q gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
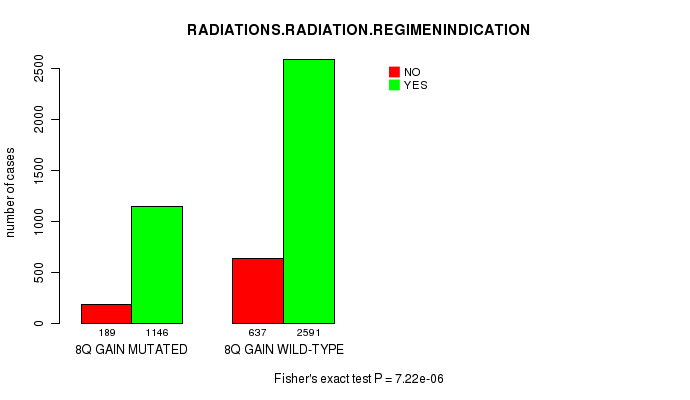
Table S86. Get Full Table Description of clustering approach #17: '9p gain mutation analysis'
| Cluster Labels | 9P GAIN MUTATED | 9P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 337 | 4226 |
P value = 8.2e-26 (Chi-square test), Q value = 1.2e-22
Table S87. Clustering Approach #17: '9p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 9P GAIN MUTATED | 14 | 15 | 71 | 48 | 13 | 38 | 8 | 33 | 0 | 62 | 0 | 35 |
| 9P GAIN WILD-TYPE | 111 | 529 | 775 | 364 | 434 | 281 | 485 | 629 | 2 | 486 | 2 | 124 |
Figure S70. Get High-res Image Clustering Approach #17: '9p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
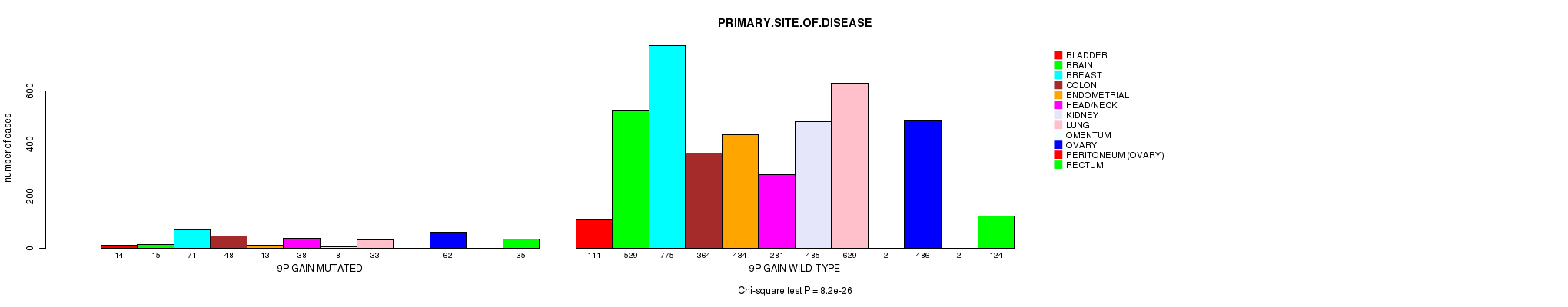
P value = 8.4e-17 (Chi-square test), Q value = 1.2e-13
Table S88. Clustering Approach #17: '9p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 9P GAIN MUTATED | 40 | 8 | 7 | 38 | 8 | 1 | 4 | 6 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19 | 1 | 0 | 31 | 3 | 62 | 5 | 0 | 1 |
| 9P GAIN WILD-TYPE | 316 | 46 | 343 | 281 | 485 | 9 | 71 | 199 | 4 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 296 | 17 | 7 | 112 | 10 | 490 | 74 | 1 | 19 |
Figure S71. Get High-res Image Clustering Approach #17: '9p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 3.08e-05 (Chi-square test), Q value = 0.04
Table S89. Clustering Approach #17: '9p gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 9P GAIN MUTATED | 16 | 46 | 72 | 25 |
| 9P GAIN WILD-TYPE | 450 | 586 | 622 | 184 |
Figure S72. Get High-res Image Clustering Approach #17: '9p gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
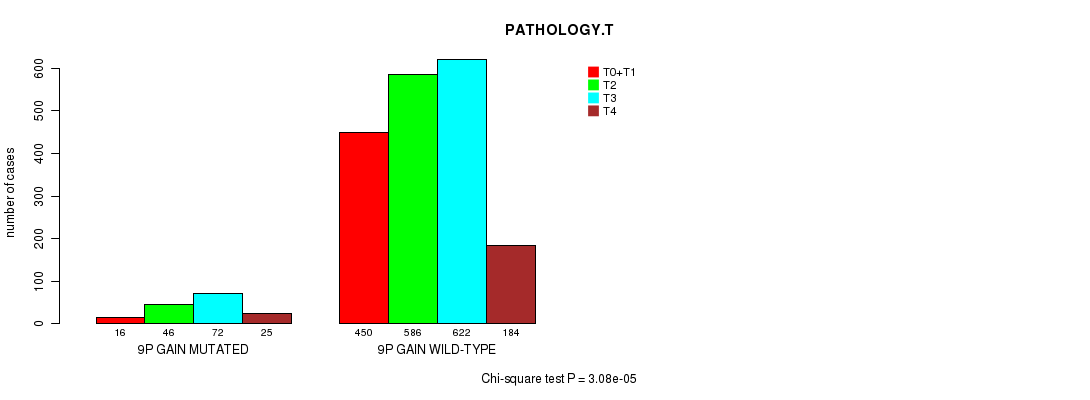
Table S90. Get Full Table Description of clustering approach #18: '9q gain mutation analysis'
| Cluster Labels | 9Q GAIN MUTATED | 9Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 284 | 4279 |
P value = 1.21e-23 (Chi-square test), Q value = 1.8e-20
Table S91. Clustering Approach #18: '9q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 9Q GAIN MUTATED | 13 | 26 | 57 | 40 | 4 | 50 | 7 | 34 | 0 | 27 | 0 | 26 |
| 9Q GAIN WILD-TYPE | 112 | 518 | 789 | 372 | 443 | 269 | 486 | 628 | 2 | 521 | 2 | 133 |
Figure S73. Get High-res Image Clustering Approach #18: '9q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
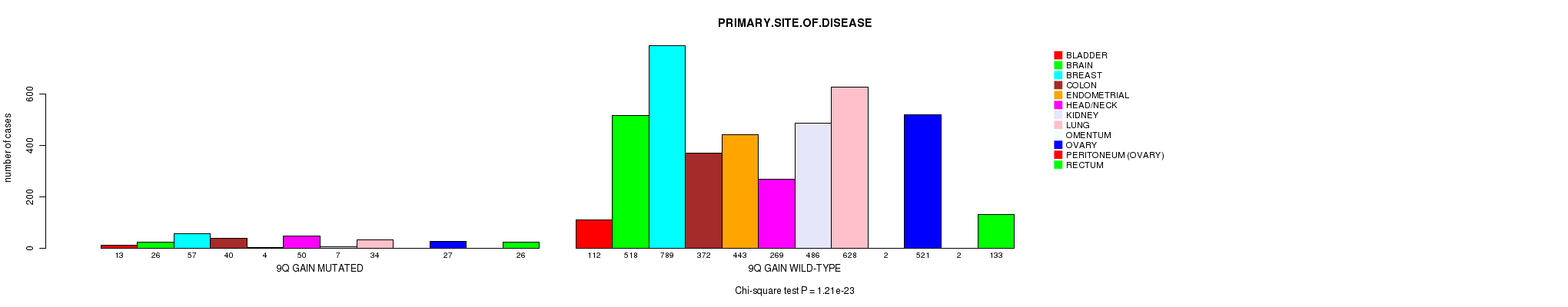
P value = 1.32e-21 (Chi-square test), Q value = 1.9e-18
Table S92. Clustering Approach #18: '9q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 9Q GAIN MUTATED | 34 | 6 | 1 | 50 | 7 | 0 | 2 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 26 | 1 | 0 | 23 | 2 | 27 | 2 | 1 | 1 |
| 9Q GAIN WILD-TYPE | 322 | 48 | 349 | 269 | 486 | 10 | 73 | 201 | 5 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 289 | 17 | 7 | 120 | 11 | 525 | 77 | 0 | 19 |
Figure S74. Get High-res Image Clustering Approach #18: '9q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.94e-07 (Chi-square test), Q value = 0.00027
Table S93. Clustering Approach #18: '9q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 9Q GAIN MUTATED | 14 | 44 | 63 | 32 |
| 9Q GAIN WILD-TYPE | 452 | 588 | 631 | 177 |
Figure S75. Get High-res Image Clustering Approach #18: '9q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
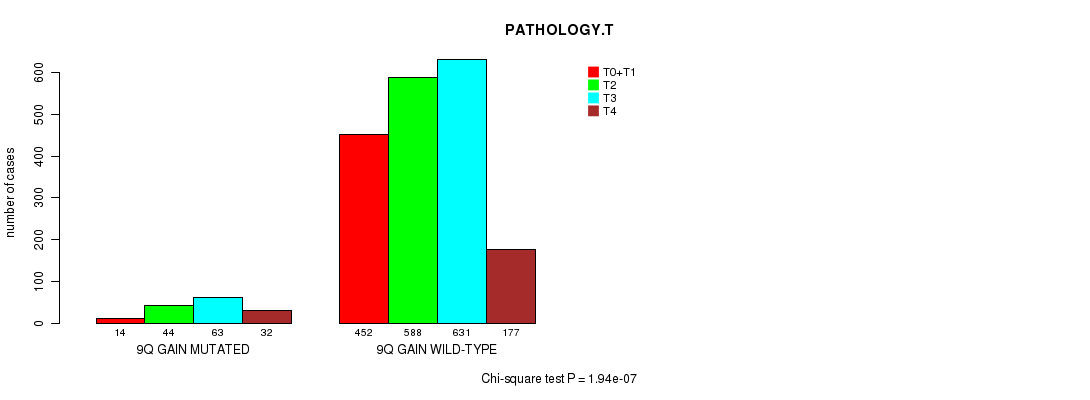
P value = 3.51e-07 (Chi-square test), Q value = 0.00048
Table S94. Clustering Approach #18: '9q gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 9Q GAIN MUTATED | 30 | 55 | 45 | 43 |
| 9Q GAIN WILD-TYPE | 682 | 443 | 836 | 380 |
Figure S76. Get High-res Image Clustering Approach #18: '9q gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
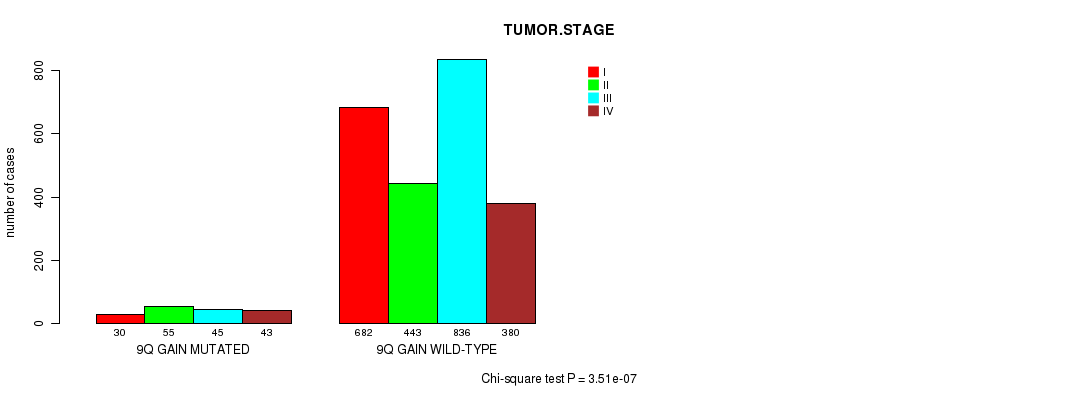
Table S95. Get Full Table Description of clustering approach #19: '10p gain mutation analysis'
| Cluster Labels | 10P GAIN MUTATED | 10P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 454 | 4109 |
P value = 1.17e-58 (Chi-square test), Q value = 1.8e-55
Table S96. Clustering Approach #19: '10p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 10P GAIN MUTATED | 25 | 10 | 108 | 19 | 82 | 13 | 5 | 53 | 0 | 129 | 2 | 8 |
| 10P GAIN WILD-TYPE | 100 | 534 | 738 | 393 | 365 | 306 | 488 | 609 | 2 | 419 | 0 | 151 |
Figure S77. Get High-res Image Clustering Approach #19: '10p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
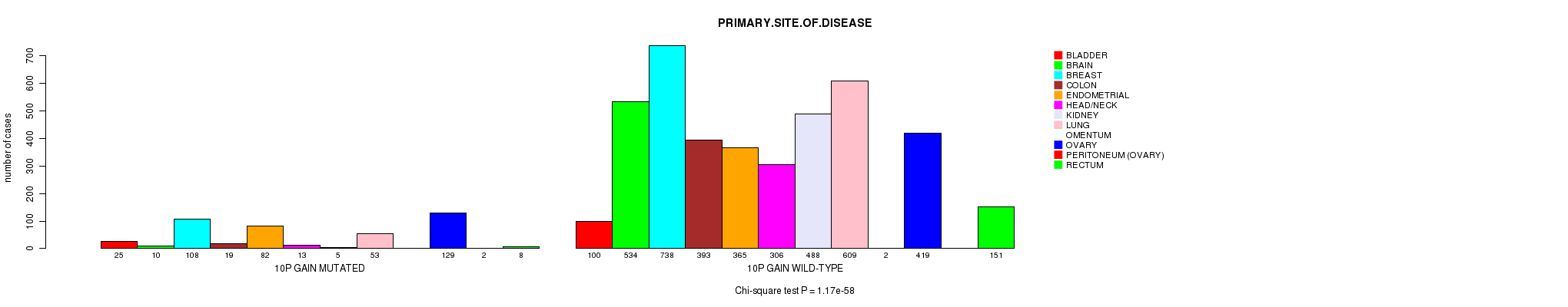
P value = 2.19e-20 (Fisher's exact test), Q value = 3.2e-17
Table S97. Clustering Approach #19: '10p gain mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 10P GAIN MUTATED | 373 | 81 |
| 10P GAIN WILD-TYPE | 2508 | 1601 |
Figure S78. Get High-res Image Clustering Approach #19: '10p gain mutation analysis' versus Clinical Feature #4: 'GENDER'
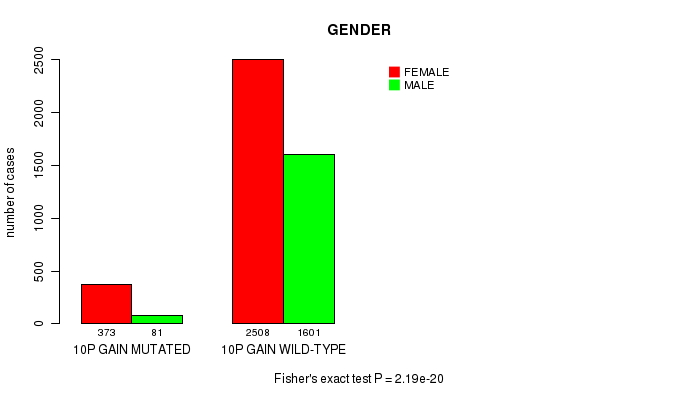
P value = 1.7e-37 (Chi-square test), Q value = 2.6e-34
Table S98. Clustering Approach #19: '10p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 10P GAIN MUTATED | 17 | 1 | 69 | 13 | 5 | 1 | 10 | 22 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 0 | 16 | 2 | 0 | 8 | 0 | 131 | 11 | 0 | 0 |
| 10P GAIN WILD-TYPE | 339 | 53 | 281 | 306 | 488 | 9 | 65 | 183 | 7 | 3 | 15 | 2 | 2 | 2 | 10 | 1 | 1 | 1 | 2 | 299 | 16 | 7 | 135 | 13 | 421 | 68 | 1 | 20 |
Figure S79. Get High-res Image Clustering Approach #19: '10p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 3.16e-13 (Chi-square test), Q value = 4.5e-10
Table S99. Clustering Approach #19: '10p gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 10P GAIN MUTATED | 35 | 25 | 130 | 33 |
| 10P GAIN WILD-TYPE | 677 | 473 | 751 | 390 |
Figure S80. Get High-res Image Clustering Approach #19: '10p gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
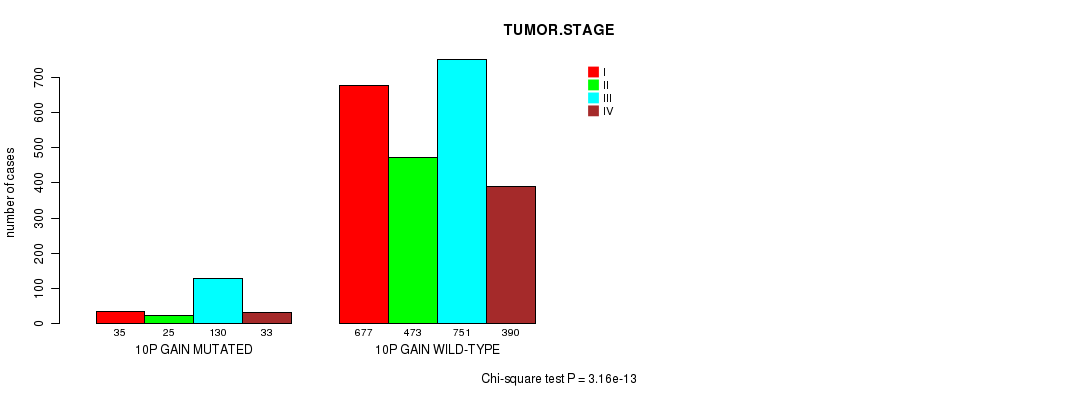
P value = 7.13e-05 (Chi-square test), Q value = 0.092
Table S100. Clustering Approach #19: '10p gain mutation analysis' versus Clinical Feature #14: 'TOBACCOSMOKINGHISTORYINDICATOR'
| nPatients | 1 | 2 | 3 | 4 | 5 | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|---|---|---|---|
| ALL | 28 | 23 | 37 | 29 | 2 | 355 | 217 | 250 | 128 |
| 10P GAIN MUTATED | 5 | 4 | 10 | 4 | 0 | 28 | 17 | 18 | 2 |
| 10P GAIN WILD-TYPE | 23 | 19 | 27 | 25 | 2 | 327 | 200 | 232 | 126 |
Figure S81. Get High-res Image Clustering Approach #19: '10p gain mutation analysis' versus Clinical Feature #14: 'TOBACCOSMOKINGHISTORYINDICATOR'
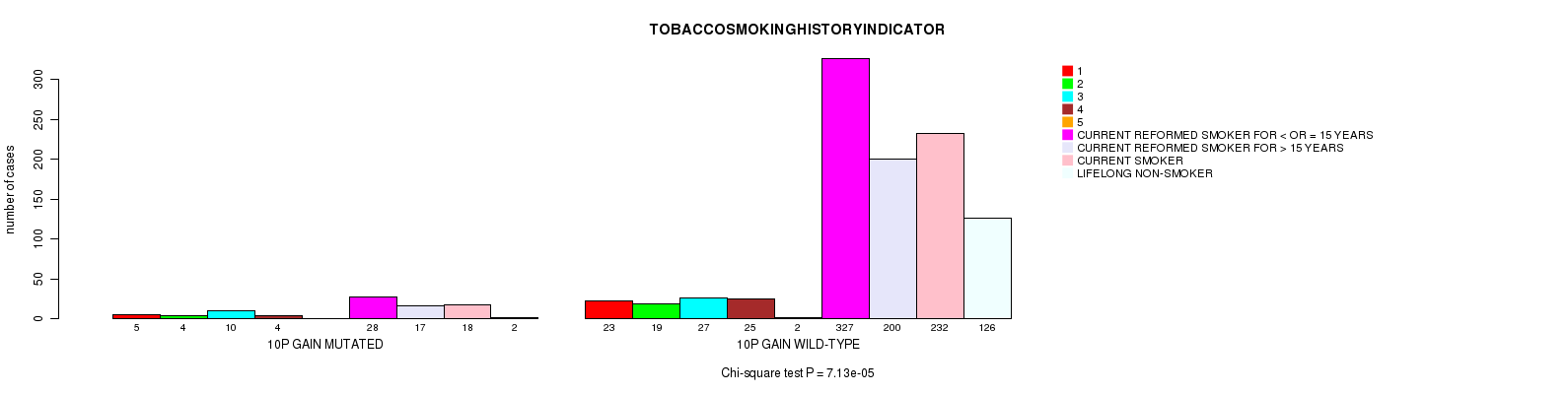
Table S101. Get Full Table Description of clustering approach #20: '10q gain mutation analysis'
| Cluster Labels | 10Q GAIN MUTATED | 10Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 249 | 4314 |
P value = 1.82e-46 (Chi-square test), Q value = 2.8e-43
Table S102. Clustering Approach #20: '10q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 10Q GAIN MUTATED | 7 | 0 | 48 | 8 | 78 | 6 | 3 | 31 | 0 | 63 | 1 | 4 |
| 10Q GAIN WILD-TYPE | 118 | 544 | 798 | 404 | 369 | 313 | 490 | 631 | 2 | 485 | 1 | 155 |
Figure S82. Get High-res Image Clustering Approach #20: '10q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
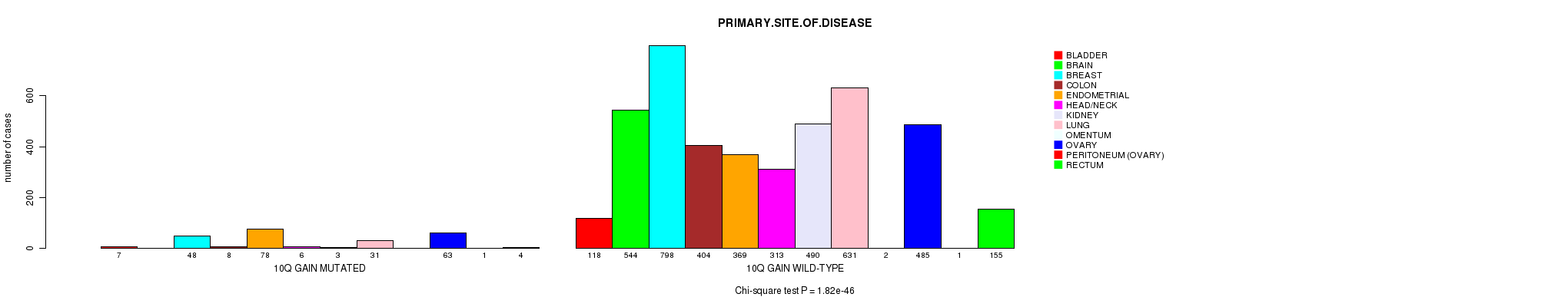
P value = 3.74e-16 (Fisher's exact test), Q value = 5.4e-13
Table S103. Clustering Approach #20: '10q gain mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 10Q GAIN MUTATED | 214 | 35 |
| 10Q GAIN WILD-TYPE | 2667 | 1647 |
Figure S83. Get High-res Image Clustering Approach #20: '10q gain mutation analysis' versus Clinical Feature #4: 'GENDER'
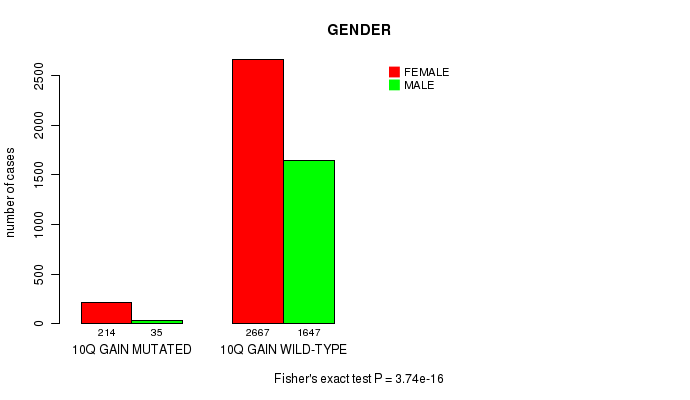
P value = 3.63e-28 (Chi-square test), Q value = 5.4e-25
Table S104. Clustering Approach #20: '10q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 10Q GAIN MUTATED | 6 | 2 | 66 | 6 | 3 | 1 | 7 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 8 | 2 | 0 | 4 | 0 | 64 | 10 | 0 | 0 |
| 10Q GAIN WILD-TYPE | 350 | 52 | 284 | 313 | 490 | 9 | 68 | 193 | 7 | 3 | 15 | 2 | 3 | 2 | 10 | 1 | 1 | 1 | 2 | 307 | 16 | 7 | 139 | 13 | 488 | 69 | 1 | 20 |
Figure S84. Get High-res Image Clustering Approach #20: '10q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 8.16e-07 (Chi-square test), Q value = 0.0011
Table S105. Clustering Approach #20: '10q gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 10Q GAIN MUTATED | 23 | 12 | 67 | 11 |
| 10Q GAIN WILD-TYPE | 689 | 486 | 814 | 412 |
Figure S85. Get High-res Image Clustering Approach #20: '10q gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'

Table S106. Get Full Table Description of clustering approach #21: '11p gain mutation analysis'
| Cluster Labels | 11P GAIN MUTATED | 11P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 244 | 4319 |
P value = 4.94e-15 (Chi-square test), Q value = 7.1e-12
Table S107. Clustering Approach #21: '11p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 11P GAIN MUTATED | 6 | 3 | 66 | 19 | 6 | 16 | 17 | 50 | 0 | 38 | 1 | 22 |
| 11P GAIN WILD-TYPE | 119 | 541 | 780 | 393 | 441 | 303 | 476 | 612 | 2 | 510 | 1 | 137 |
Figure S86. Get High-res Image Clustering Approach #21: '11p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
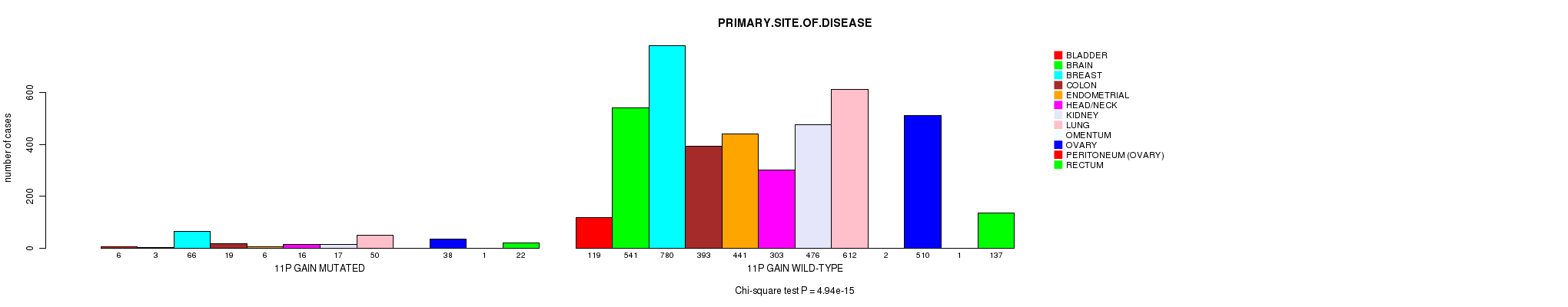
P value = 5.54e-06 (Chi-square test), Q value = 0.0074
Table S108. Clustering Approach #21: '11p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 11P GAIN MUTATED | 19 | 0 | 2 | 16 | 17 | 1 | 7 | 17 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 22 | 0 | 0 | 22 | 0 | 39 | 4 | 0 | 0 |
| 11P GAIN WILD-TYPE | 337 | 54 | 348 | 303 | 476 | 9 | 68 | 188 | 7 | 2 | 15 | 2 | 3 | 2 | 11 | 1 | 1 | 1 | 2 | 293 | 18 | 7 | 121 | 13 | 513 | 75 | 1 | 20 |
Figure S87. Get High-res Image Clustering Approach #21: '11p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S109. Get Full Table Description of clustering approach #22: '11q gain mutation analysis'
| Cluster Labels | 11Q GAIN MUTATED | 11Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 264 | 4299 |
P value = 2.5e-16 (Chi-square test), Q value = 3.6e-13
Table S110. Clustering Approach #22: '11q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 11Q GAIN MUTATED | 6 | 6 | 49 | 22 | 7 | 16 | 15 | 66 | 0 | 60 | 0 | 17 |
| 11Q GAIN WILD-TYPE | 119 | 538 | 797 | 390 | 440 | 303 | 478 | 596 | 2 | 488 | 2 | 142 |
Figure S88. Get High-res Image Clustering Approach #22: '11q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
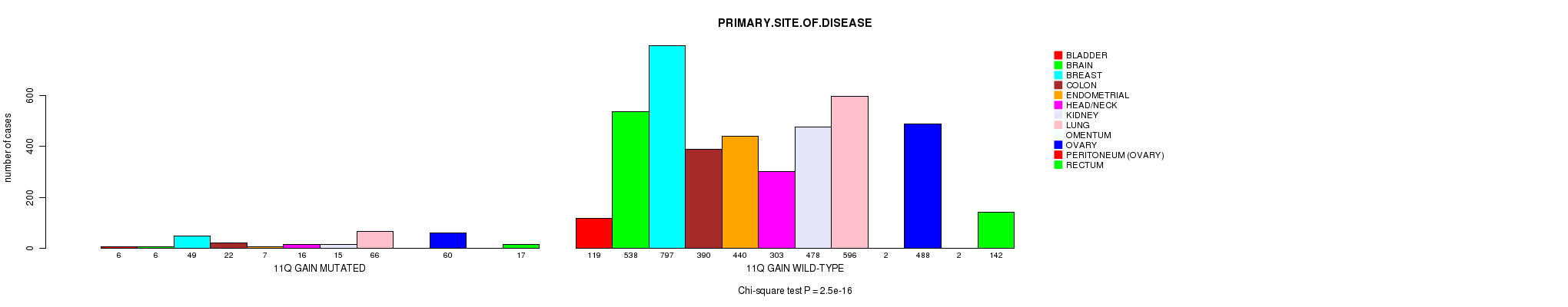
P value = 5.8e-09 (Chi-square test), Q value = 8.1e-06
Table S111. Clustering Approach #22: '11q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 11Q GAIN MUTATED | 22 | 0 | 4 | 16 | 15 | 1 | 7 | 31 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 24 | 0 | 0 | 17 | 0 | 60 | 3 | 0 | 0 |
| 11Q GAIN WILD-TYPE | 334 | 54 | 346 | 303 | 478 | 9 | 68 | 174 | 6 | 2 | 15 | 2 | 3 | 2 | 12 | 1 | 1 | 1 | 2 | 291 | 18 | 7 | 126 | 13 | 492 | 76 | 1 | 20 |
Figure S89. Get High-res Image Clustering Approach #22: '11q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 8.26e-05 (t-test), Q value = 0.11
Table S112. Clustering Approach #22: '11q gain mutation analysis' versus Clinical Feature #13: 'STOPPEDSMOKINGYEAR'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 568 | 1995.2 (13.5) |
| 11Q GAIN MUTATED | 51 | 2001.3 (10.6) |
| 11Q GAIN WILD-TYPE | 517 | 1994.6 (13.6) |
Figure S90. Get High-res Image Clustering Approach #22: '11q gain mutation analysis' versus Clinical Feature #13: 'STOPPEDSMOKINGYEAR'
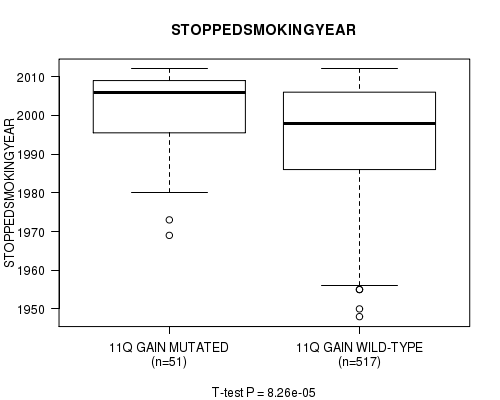
Table S113. Get Full Table Description of clustering approach #23: '12p gain mutation analysis'
| Cluster Labels | 12P GAIN MUTATED | 12P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 765 | 3798 |
P value = 1.45e-43 (Chi-square test), Q value = 2.2e-40
Table S114. Clustering Approach #23: '12p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 12P GAIN MUTATED | 22 | 35 | 113 | 67 | 33 | 61 | 78 | 131 | 0 | 195 | 2 | 28 |
| 12P GAIN WILD-TYPE | 103 | 509 | 733 | 345 | 414 | 258 | 415 | 531 | 2 | 353 | 0 | 131 |
Figure S91. Get High-res Image Clustering Approach #23: '12p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
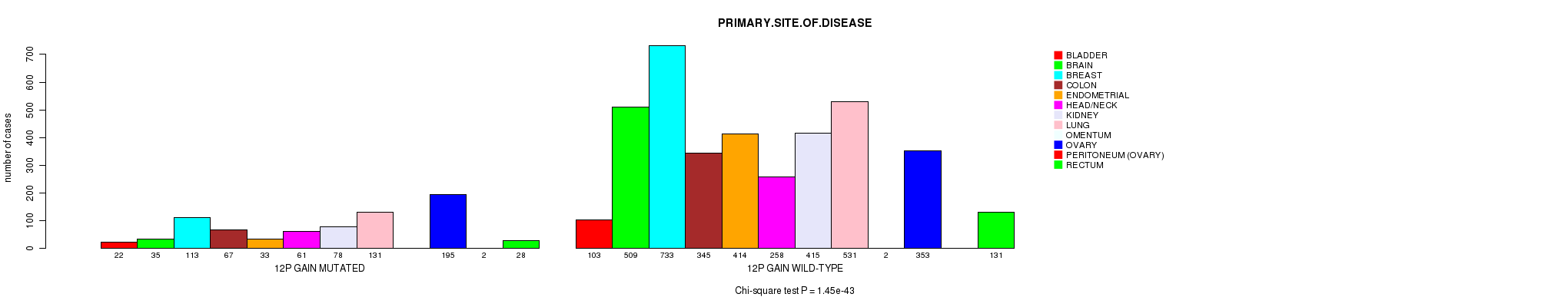
P value = 4.59e-25 (Chi-square test), Q value = 6.8e-22
Table S115. Clustering Approach #23: '12p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 12P GAIN MUTATED | 58 | 8 | 19 | 61 | 78 | 0 | 9 | 29 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 88 | 1 | 1 | 26 | 2 | 197 | 13 | 0 | 1 |
| 12P GAIN WILD-TYPE | 298 | 46 | 331 | 258 | 415 | 10 | 66 | 176 | 5 | 3 | 15 | 1 | 3 | 2 | 12 | 1 | 1 | 1 | 2 | 227 | 17 | 6 | 117 | 11 | 355 | 66 | 1 | 19 |
Figure S92. Get High-res Image Clustering Approach #23: '12p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 9.68e-09 (Chi-square test), Q value = 1.3e-05
Table S116. Clustering Approach #23: '12p gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 12P GAIN MUTATED | 109 | 111 | 248 | 81 |
| 12P GAIN WILD-TYPE | 603 | 387 | 633 | 342 |
Figure S93. Get High-res Image Clustering Approach #23: '12p gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
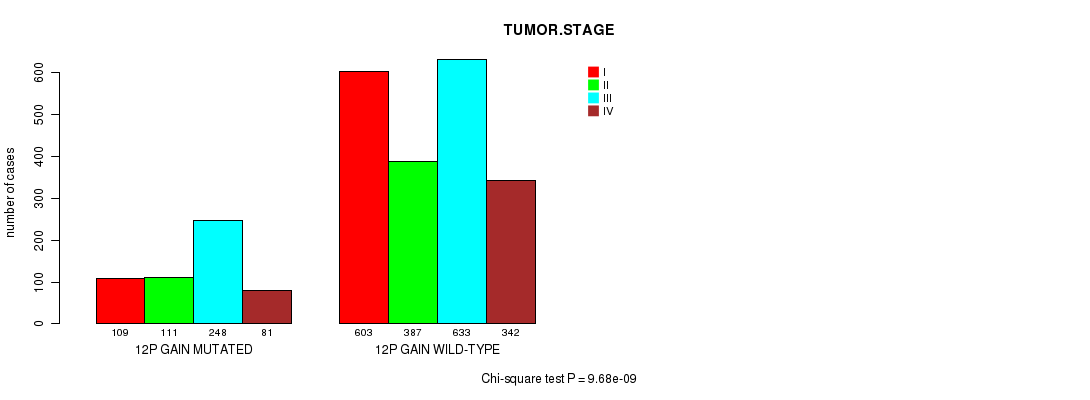
P value = 4.22e-07 (Fisher's exact test), Q value = 0.00057
Table S117. Clustering Approach #23: '12p gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 12P GAIN MUTATED | 91 | 674 |
| 12P GAIN WILD-TYPE | 735 | 3063 |
Figure S94. Get High-res Image Clustering Approach #23: '12p gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
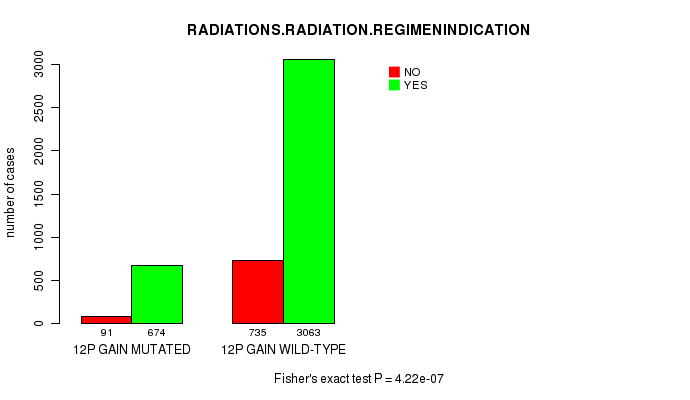
Table S118. Get Full Table Description of clustering approach #24: '12q gain mutation analysis'
| Cluster Labels | 12Q GAIN MUTATED | 12Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 525 | 4038 |
P value = 7.1e-15 (Chi-square test), Q value = 1e-11
Table S119. Clustering Approach #24: '12q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 12Q GAIN MUTATED | 17 | 26 | 88 | 60 | 26 | 26 | 79 | 79 | 0 | 105 | 1 | 18 |
| 12Q GAIN WILD-TYPE | 108 | 518 | 758 | 352 | 421 | 293 | 414 | 583 | 2 | 443 | 1 | 141 |
Figure S95. Get High-res Image Clustering Approach #24: '12q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
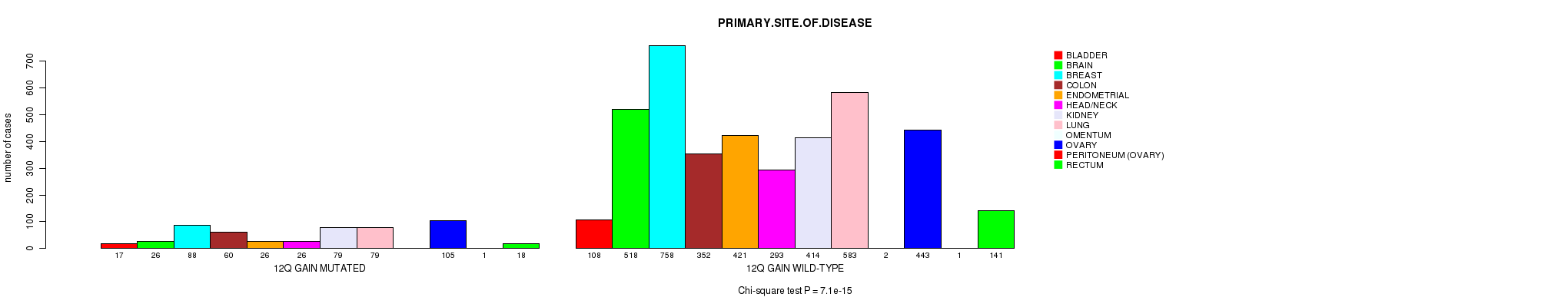
P value = 6.49e-05 (Chi-square test), Q value = 0.084
Table S120. Clustering Approach #24: '12q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 12Q GAIN MUTATED | 50 | 9 | 18 | 26 | 79 | 1 | 7 | 23 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 43 | 1 | 1 | 17 | 1 | 106 | 7 | 0 | 1 |
| 12Q GAIN WILD-TYPE | 306 | 45 | 332 | 293 | 414 | 9 | 68 | 182 | 6 | 3 | 15 | 1 | 3 | 2 | 12 | 1 | 1 | 1 | 1 | 272 | 17 | 6 | 126 | 12 | 446 | 72 | 1 | 19 |
Figure S96. Get High-res Image Clustering Approach #24: '12q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S121. Get Full Table Description of clustering approach #25: '13q gain mutation analysis'
| Cluster Labels | 13Q GAIN MUTATED | 13Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 491 | 4072 |
P value = 6.48e-11 (t-test), Q value = 9.2e-08
Table S122. Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 4507 | 61.9 (12.6) |
| 13Q GAIN MUTATED | 489 | 65.2 (11.9) |
| 13Q GAIN WILD-TYPE | 4018 | 61.4 (12.7) |
Figure S97. Get High-res Image Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #2: 'AGE'
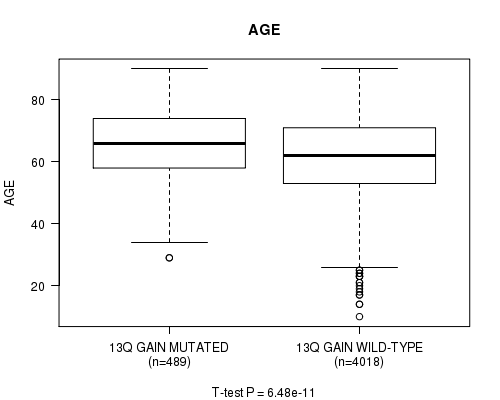
P value = 3.03e-280 (Chi-square test), Q value = 4.8e-277
Table S123. Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 13Q GAIN MUTATED | 21 | 1 | 46 | 211 | 19 | 18 | 12 | 24 | 0 | 38 | 0 | 98 |
| 13Q GAIN WILD-TYPE | 104 | 543 | 800 | 201 | 428 | 301 | 481 | 638 | 2 | 510 | 2 | 61 |
Figure S98. Get High-res Image Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'

P value = 3.06e-05 (Fisher's exact test), Q value = 0.04
Table S124. Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 13Q GAIN MUTATED | 267 | 224 |
| 13Q GAIN WILD-TYPE | 2614 | 1458 |
Figure S99. Get High-res Image Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #4: 'GENDER'
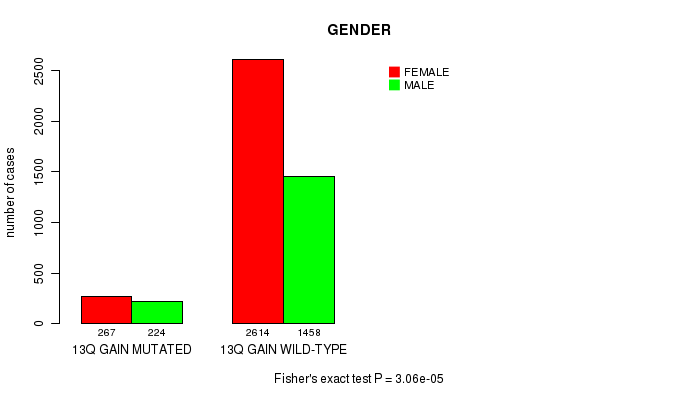
P value = 2.32e-211 (Chi-square test), Q value = 3.7e-208
Table S125. Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 13Q GAIN MUTATED | 199 | 12 | 6 | 18 | 12 | 0 | 4 | 8 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 9 | 1 | 0 | 95 | 2 | 38 | 12 | 0 | 0 |
| 13Q GAIN WILD-TYPE | 157 | 42 | 344 | 301 | 481 | 10 | 71 | 197 | 7 | 3 | 15 | 1 | 3 | 2 | 12 | 1 | 1 | 0 | 2 | 306 | 17 | 7 | 48 | 11 | 514 | 67 | 1 | 20 |
Figure S100. Get High-res Image Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.2e-34 (Chi-square test), Q value = 1.8e-31
Table S126. Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 13Q GAIN MUTATED | 20 | 79 | 220 | 44 |
| 13Q GAIN WILD-TYPE | 446 | 553 | 474 | 165 |
Figure S101. Get High-res Image Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'

P value = 1.68e-06 (Chi-square test), Q value = 0.0023
Table S127. Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #8: 'PATHOLOGY.N'
| nPatients | N0 | N1 | N2 | N3 |
|---|---|---|---|---|
| ALL | 1067 | 353 | 282 | 11 |
| 13Q GAIN MUTATED | 177 | 101 | 68 | 0 |
| 13Q GAIN WILD-TYPE | 890 | 252 | 214 | 11 |
Figure S102. Get High-res Image Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #8: 'PATHOLOGY.N'

P value = 1.36e-05 (Chi-square test), Q value = 0.018
Table S128. Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
| nPatients | M0 | M1 | M1A | M1B | MX |
|---|---|---|---|---|---|
| ALL | 1424 | 168 | 9 | 3 | 165 |
| 13Q GAIN MUTATED | 258 | 51 | 6 | 0 | 31 |
| 13Q GAIN WILD-TYPE | 1166 | 117 | 3 | 3 | 134 |
Figure S103. Get High-res Image Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
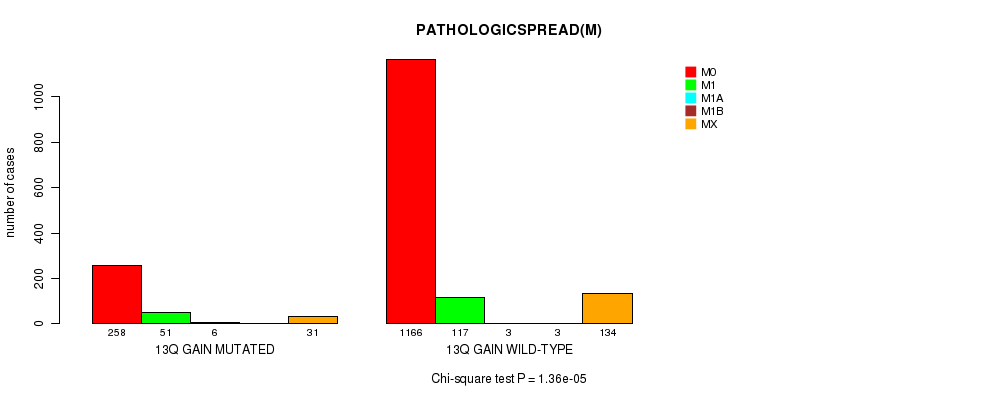
P value = 7.91e-08 (Chi-square test), Q value = 0.00011
Table S129. Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 13Q GAIN MUTATED | 66 | 107 | 147 | 67 |
| 13Q GAIN WILD-TYPE | 646 | 391 | 734 | 356 |
Figure S104. Get High-res Image Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
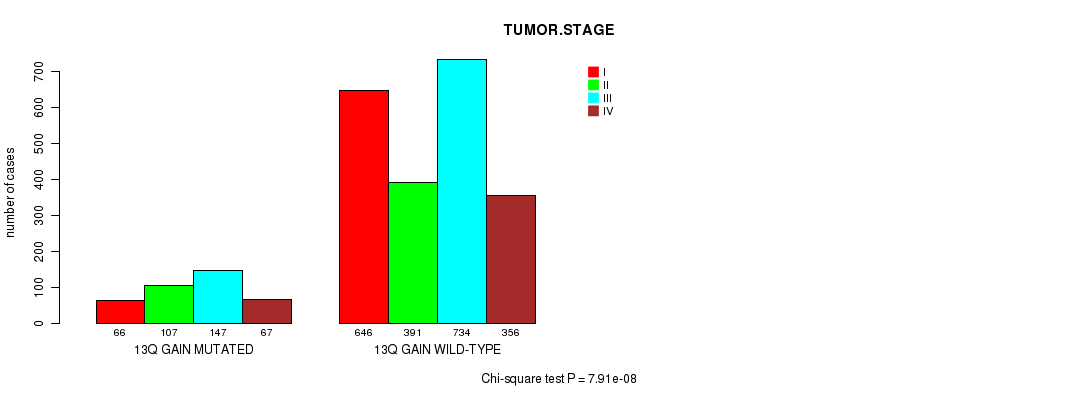
P value = 9.57e-15 (Fisher's exact test), Q value = 1.4e-11
Table S130. Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 13Q GAIN MUTATED | 32 | 459 |
| 13Q GAIN WILD-TYPE | 794 | 3278 |
Figure S105. Get High-res Image Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
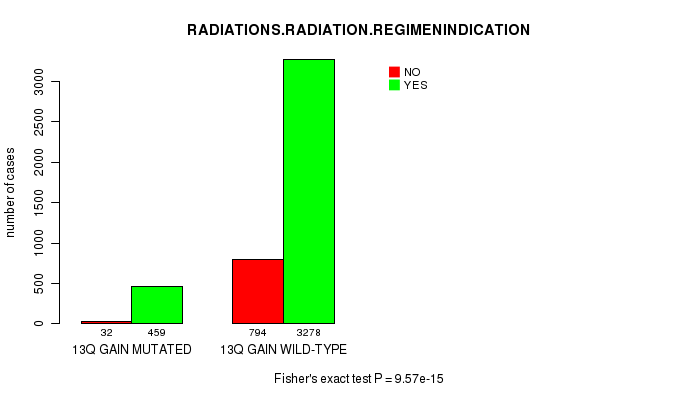
P value = 5.18e-07 (Chi-square test), Q value = 7e-04
Table S131. Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #14: 'TOBACCOSMOKINGHISTORYINDICATOR'
| nPatients | 1 | 2 | 3 | 4 | 5 | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|---|---|---|---|
| ALL | 28 | 23 | 37 | 29 | 2 | 355 | 217 | 250 | 128 |
| 13Q GAIN MUTATED | 4 | 6 | 6 | 4 | 1 | 13 | 13 | 10 | 5 |
| 13Q GAIN WILD-TYPE | 24 | 17 | 31 | 25 | 1 | 342 | 204 | 240 | 123 |
Figure S106. Get High-res Image Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #14: 'TOBACCOSMOKINGHISTORYINDICATOR'
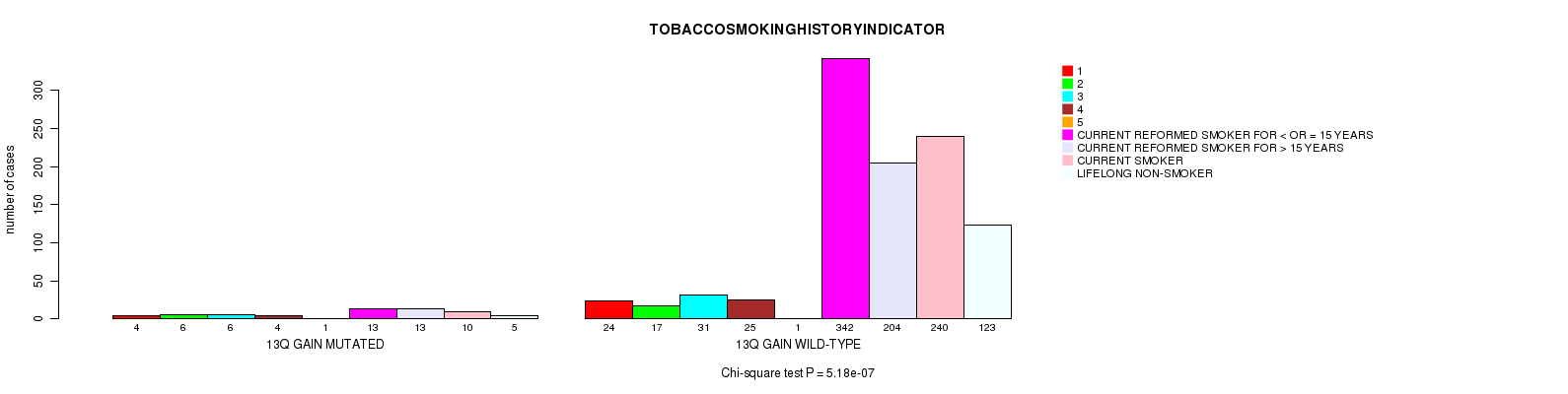
P value = 2.95e-05 (Chi-square test), Q value = 0.038
Table S132. Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #18: 'COMPLETENESS.OF.RESECTION'
| nPatients | R0 | R1 | R2 | RX |
|---|---|---|---|---|
| ALL | 1171 | 69 | 59 | 77 |
| 13Q GAIN MUTATED | 254 | 5 | 24 | 11 |
| 13Q GAIN WILD-TYPE | 917 | 64 | 35 | 66 |
Figure S107. Get High-res Image Clustering Approach #25: '13q gain mutation analysis' versus Clinical Feature #18: 'COMPLETENESS.OF.RESECTION'
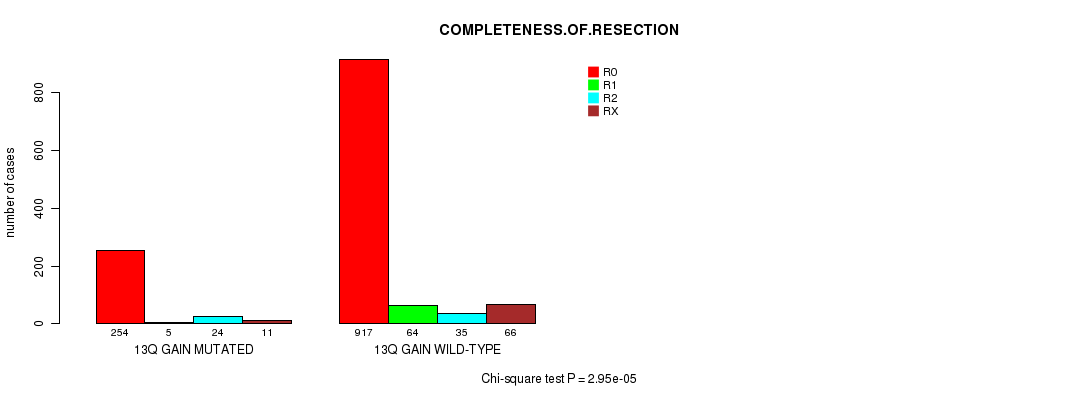
Table S133. Get Full Table Description of clustering approach #26: '14q gain mutation analysis'
| Cluster Labels | 14Q GAIN MUTATED | 14Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 294 | 4269 |
P value = 7.71e-29 (Chi-square test), Q value = 1.2e-25
Table S134. Clustering Approach #26: '14q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 14Q GAIN MUTATED | 10 | 4 | 77 | 15 | 16 | 53 | 5 | 76 | 0 | 33 | 0 | 5 |
| 14Q GAIN WILD-TYPE | 115 | 540 | 769 | 397 | 431 | 266 | 488 | 586 | 2 | 515 | 2 | 154 |
Figure S108. Get High-res Image Clustering Approach #26: '14q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'

P value = 4.79e-21 (Chi-square test), Q value = 7e-18
Table S135. Clustering Approach #26: '14q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 14Q GAIN MUTATED | 12 | 3 | 9 | 53 | 5 | 2 | 5 | 30 | 1 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 33 | 0 | 0 | 5 | 0 | 33 | 7 | 1 | 0 |
| 14Q GAIN WILD-TYPE | 344 | 51 | 341 | 266 | 488 | 8 | 70 | 175 | 6 | 2 | 15 | 2 | 2 | 2 | 11 | 1 | 1 | 1 | 1 | 282 | 18 | 7 | 138 | 13 | 519 | 72 | 0 | 20 |
Figure S109. Get High-res Image Clustering Approach #26: '14q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S136. Get Full Table Description of clustering approach #27: '15q gain mutation analysis'
| Cluster Labels | 15Q GAIN MUTATED | 15Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 155 | 4408 |
P value = 4.82e-08 (Chi-square test), Q value = 6.7e-05
Table S137. Clustering Approach #27: '15q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 15Q GAIN MUTATED | 4 | 3 | 43 | 5 | 5 | 14 | 14 | 43 | 0 | 21 | 0 | 3 |
| 15Q GAIN WILD-TYPE | 121 | 541 | 803 | 407 | 442 | 305 | 479 | 619 | 2 | 527 | 2 | 156 |
Figure S110. Get High-res Image Clustering Approach #27: '15q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
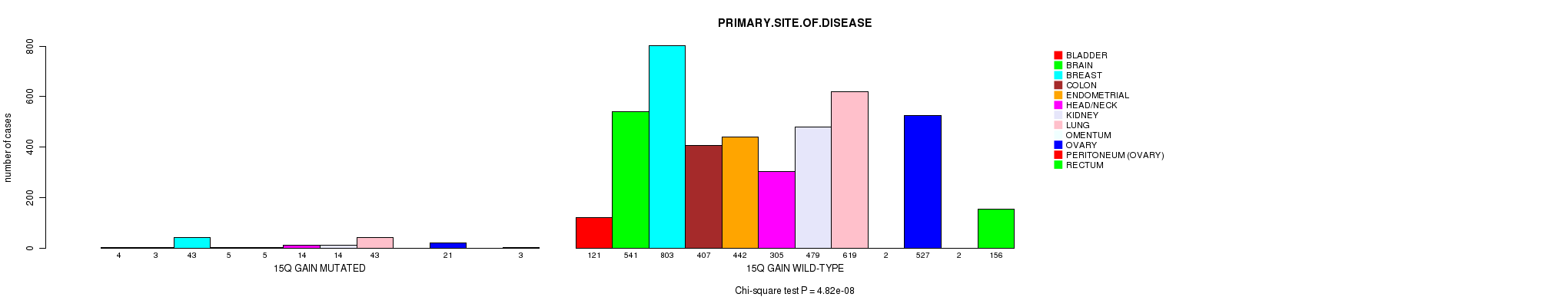
P value = 5.1e-07 (Chi-square test), Q value = 7e-04
Table S138. Clustering Approach #27: '15q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 15Q GAIN MUTATED | 5 | 0 | 4 | 14 | 14 | 1 | 3 | 5 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 30 | 0 | 0 | 2 | 1 | 21 | 1 | 0 | 0 |
| 15Q GAIN WILD-TYPE | 351 | 54 | 346 | 305 | 479 | 9 | 72 | 200 | 5 | 2 | 15 | 2 | 3 | 2 | 12 | 1 | 1 | 1 | 2 | 285 | 18 | 7 | 141 | 12 | 531 | 78 | 1 | 20 |
Figure S111. Get High-res Image Clustering Approach #27: '15q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 0.00018 (Chi-square test), Q value = 0.23
Table S139. Clustering Approach #27: '15q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 15Q GAIN MUTATED | 15 | 40 | 12 | 8 |
| 15Q GAIN WILD-TYPE | 451 | 592 | 682 | 201 |
Figure S112. Get High-res Image Clustering Approach #27: '15q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
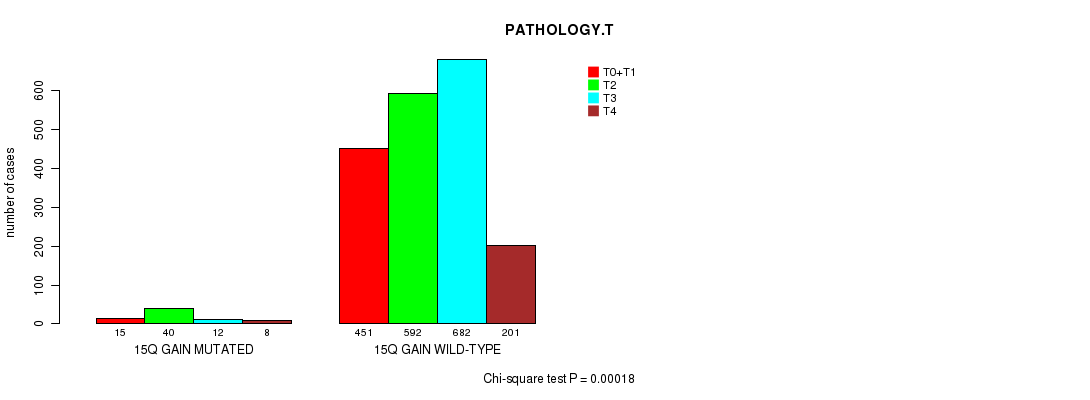
Table S140. Get Full Table Description of clustering approach #28: '16p gain mutation analysis'
| Cluster Labels | 16P GAIN MUTATED | 16P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 569 | 3994 |
P value = 2.01e-09 (logrank test), Q value = 2.8e-06
Table S141. Clustering Approach #28: '16p gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 4296 | 1401 | 0.0 - 224.0 (14.9) |
| 16P GAIN MUTATED | 529 | 109 | 0.0 - 224.0 (15.0) |
| 16P GAIN WILD-TYPE | 3767 | 1292 | 0.0 - 220.9 (14.9) |
Figure S113. Get High-res Image Clustering Approach #28: '16p gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
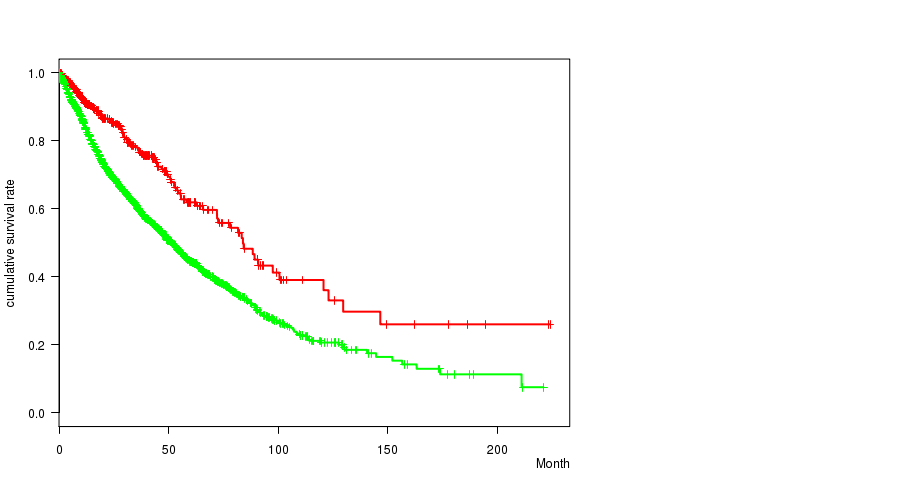
P value = 1.31e-70 (Chi-square test), Q value = 2e-67
Table S142. Clustering Approach #28: '16p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 16P GAIN MUTATED | 11 | 14 | 249 | 68 | 11 | 24 | 65 | 66 | 0 | 31 | 0 | 29 |
| 16P GAIN WILD-TYPE | 114 | 530 | 597 | 344 | 436 | 295 | 428 | 596 | 2 | 517 | 2 | 130 |
Figure S114. Get High-res Image Clustering Approach #28: '16p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
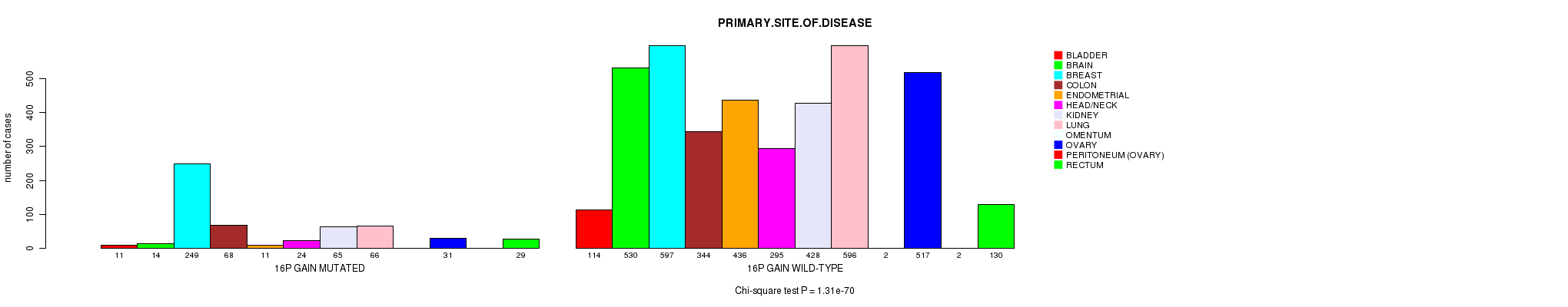
P value = 1.28e-06 (Fisher's exact test), Q value = 0.0017
Table S143. Clustering Approach #28: '16p gain mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 16P GAIN MUTATED | 411 | 158 |
| 16P GAIN WILD-TYPE | 2470 | 1524 |
Figure S115. Get High-res Image Clustering Approach #28: '16p gain mutation analysis' versus Clinical Feature #4: 'GENDER'
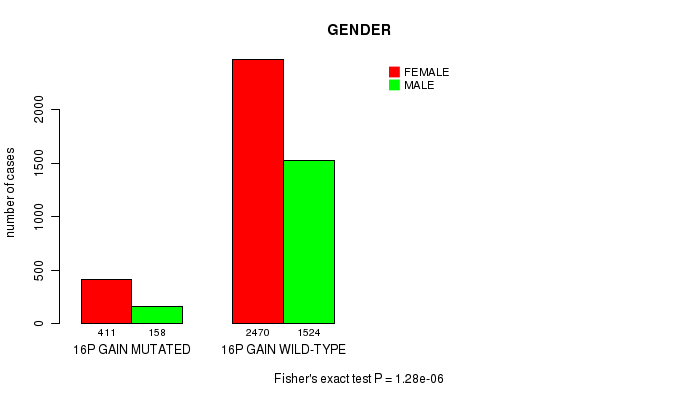
P value = 4.74e-17 (Chi-square test), Q value = 6.9e-14
Table S144. Clustering Approach #28: '16p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 16P GAIN MUTATED | 65 | 3 | 5 | 24 | 65 | 3 | 7 | 34 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 16 | 0 | 1 | 29 | 0 | 31 | 6 | 0 | 0 |
| 16P GAIN WILD-TYPE | 291 | 51 | 345 | 295 | 428 | 7 | 68 | 171 | 6 | 3 | 14 | 2 | 2 | 2 | 12 | 1 | 1 | 0 | 2 | 299 | 18 | 6 | 114 | 13 | 521 | 73 | 1 | 20 |
Figure S116. Get High-res Image Clustering Approach #28: '16p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S145. Get Full Table Description of clustering approach #29: '16q gain mutation analysis'
| Cluster Labels | 16Q GAIN MUTATED | 16Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 334 | 4229 |
P value = 2.54e-28 (Chi-square test), Q value = 3.8e-25
Table S146. Clustering Approach #29: '16q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 16Q GAIN MUTATED | 13 | 14 | 53 | 66 | 4 | 27 | 58 | 53 | 0 | 15 | 0 | 30 |
| 16Q GAIN WILD-TYPE | 112 | 530 | 793 | 346 | 443 | 292 | 435 | 609 | 2 | 533 | 2 | 129 |
Figure S117. Get High-res Image Clustering Approach #29: '16q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
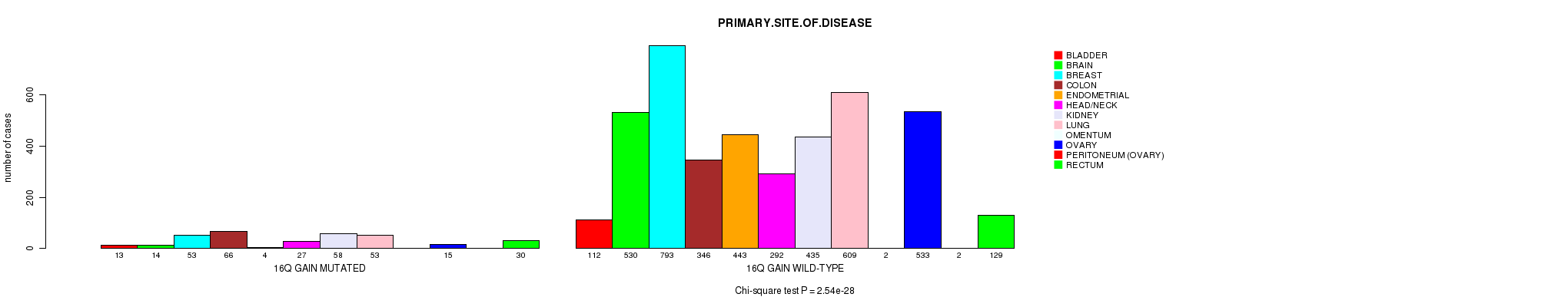
P value = 1.06e-21 (Chi-square test), Q value = 1.6e-18
Table S147. Clustering Approach #29: '16q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 16Q GAIN MUTATED | 63 | 3 | 3 | 27 | 58 | 2 | 1 | 24 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 21 | 0 | 1 | 30 | 0 | 15 | 1 | 0 | 0 |
| 16Q GAIN WILD-TYPE | 293 | 51 | 347 | 292 | 435 | 8 | 74 | 181 | 7 | 3 | 14 | 2 | 2 | 2 | 12 | 1 | 1 | 0 | 2 | 294 | 18 | 6 | 113 | 13 | 537 | 78 | 1 | 20 |
Figure S118. Get High-res Image Clustering Approach #29: '16q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 4.1e-06 (Fisher's exact test), Q value = 0.0055
Table S148. Clustering Approach #29: '16q gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 16Q GAIN MUTATED | 31 | 303 |
| 16Q GAIN WILD-TYPE | 795 | 3434 |
Figure S119. Get High-res Image Clustering Approach #29: '16q gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'

Table S149. Get Full Table Description of clustering approach #30: '17p gain mutation analysis'
| Cluster Labels | 17P GAIN MUTATED | 17P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 166 | 4397 |
P value = 1.5e-06 (Chi-square test), Q value = 0.002
Table S150. Clustering Approach #30: '17p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 17P GAIN MUTATED | 10 | 9 | 50 | 11 | 6 | 19 | 16 | 32 | 0 | 9 | 0 | 4 |
| 17P GAIN WILD-TYPE | 115 | 535 | 796 | 401 | 441 | 300 | 477 | 630 | 2 | 539 | 2 | 155 |
Figure S120. Get High-res Image Clustering Approach #30: '17p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
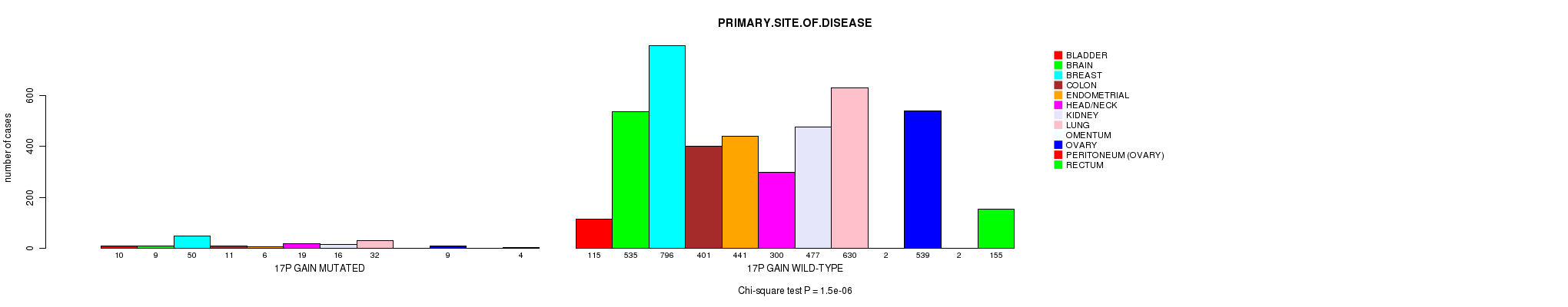
P value = 9.33e-07 (Chi-square test), Q value = 0.0013
Table S151. Clustering Approach #30: '17p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 17P GAIN MUTATED | 11 | 0 | 4 | 19 | 16 | 1 | 5 | 13 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 11 | 0 | 0 | 4 | 0 | 9 | 2 | 0 | 1 |
| 17P GAIN WILD-TYPE | 345 | 54 | 346 | 300 | 477 | 9 | 70 | 192 | 7 | 3 | 15 | 2 | 3 | 1 | 13 | 0 | 1 | 1 | 2 | 304 | 18 | 7 | 139 | 13 | 543 | 77 | 1 | 19 |
Figure S121. Get High-res Image Clustering Approach #30: '17p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S152. Get Full Table Description of clustering approach #31: '17q gain mutation analysis'
| Cluster Labels | 17Q GAIN MUTATED | 17Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 405 | 4158 |
P value = 3.22e-30 (Chi-square test), Q value = 4.8e-27
Table S153. Clustering Approach #31: '17q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 17Q GAIN MUTATED | 23 | 17 | 126 | 48 | 10 | 23 | 22 | 100 | 0 | 19 | 0 | 17 |
| 17Q GAIN WILD-TYPE | 102 | 527 | 720 | 364 | 437 | 296 | 471 | 562 | 2 | 529 | 2 | 142 |
Figure S122. Get High-res Image Clustering Approach #31: '17q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'

P value = 1.46e-19 (Chi-square test), Q value = 2.1e-16
Table S154. Clustering Approach #31: '17q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 17Q GAIN MUTATED | 45 | 2 | 7 | 23 | 22 | 2 | 14 | 40 | 2 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 38 | 0 | 0 | 17 | 0 | 19 | 3 | 1 | 2 |
| 17Q GAIN WILD-TYPE | 311 | 52 | 343 | 296 | 471 | 8 | 61 | 165 | 5 | 3 | 15 | 1 | 3 | 1 | 12 | 1 | 1 | 1 | 1 | 277 | 18 | 7 | 126 | 13 | 533 | 76 | 0 | 18 |
Figure S123. Get High-res Image Clustering Approach #31: '17q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S155. Get Full Table Description of clustering approach #32: '18p gain mutation analysis'
| Cluster Labels | 18P GAIN MUTATED | 18P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 419 | 4144 |
P value = 9.68e-18 (Chi-square test), Q value = 1.4e-14
Table S156. Clustering Approach #32: '18p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 18P GAIN MUTATED | 19 | 26 | 85 | 15 | 33 | 53 | 18 | 88 | 0 | 75 | 0 | 7 |
| 18P GAIN WILD-TYPE | 106 | 518 | 761 | 397 | 414 | 266 | 475 | 574 | 2 | 473 | 2 | 152 |
Figure S124. Get High-res Image Clustering Approach #32: '18p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
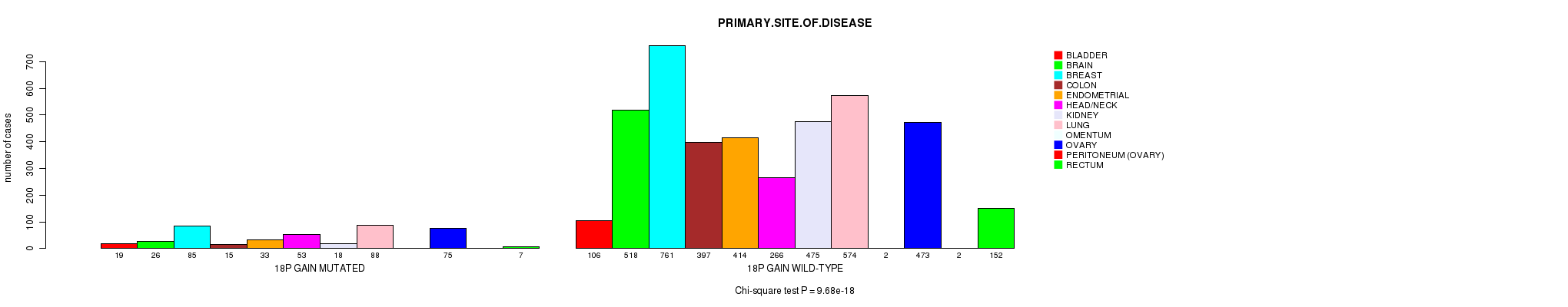
P value = 2.74e-20 (Chi-square test), Q value = 4e-17
Table S157. Clustering Approach #32: '18p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 18P GAIN MUTATED | 10 | 5 | 11 | 53 | 18 | 4 | 8 | 18 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 53 | 2 | 0 | 6 | 1 | 75 | 20 | 0 | 1 |
| 18P GAIN WILD-TYPE | 346 | 49 | 339 | 266 | 475 | 6 | 67 | 187 | 4 | 3 | 15 | 2 | 3 | 2 | 11 | 1 | 1 | 1 | 2 | 262 | 16 | 7 | 137 | 12 | 477 | 59 | 1 | 19 |
Figure S125. Get High-res Image Clustering Approach #32: '18p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S158. Get Full Table Description of clustering approach #33: '18q gain mutation analysis'
| Cluster Labels | 18Q GAIN MUTATED | 18Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 288 | 4275 |
P value = 6.25e-08 (Chi-square test), Q value = 8.6e-05
Table S159. Clustering Approach #33: '18q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 18Q GAIN MUTATED | 7 | 27 | 78 | 9 | 16 | 23 | 18 | 63 | 0 | 43 | 0 | 4 |
| 18Q GAIN WILD-TYPE | 118 | 517 | 768 | 403 | 431 | 296 | 475 | 599 | 2 | 505 | 2 | 155 |
Figure S126. Get High-res Image Clustering Approach #33: '18q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
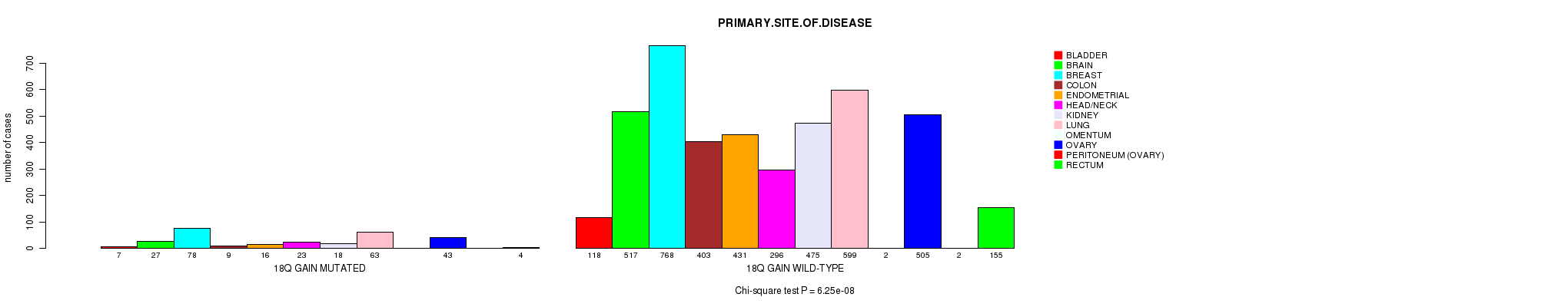
P value = 4.22e-08 (Chi-square test), Q value = 5.8e-05
Table S160. Clustering Approach #33: '18q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 18Q GAIN MUTATED | 6 | 3 | 7 | 23 | 18 | 2 | 5 | 12 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 39 | 2 | 0 | 4 | 0 | 43 | 7 | 0 | 1 |
| 18Q GAIN WILD-TYPE | 350 | 51 | 343 | 296 | 475 | 8 | 70 | 193 | 4 | 3 | 15 | 2 | 3 | 2 | 11 | 1 | 1 | 1 | 2 | 276 | 16 | 7 | 139 | 13 | 509 | 72 | 1 | 19 |
Figure S127. Get High-res Image Clustering Approach #33: '18q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 0.00019 (Chi-square test), Q value = 0.24
Table S161. Clustering Approach #33: '18q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 18Q GAIN MUTATED | 23 | 54 | 21 | 12 |
| 18Q GAIN WILD-TYPE | 443 | 578 | 673 | 197 |
Figure S128. Get High-res Image Clustering Approach #33: '18q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
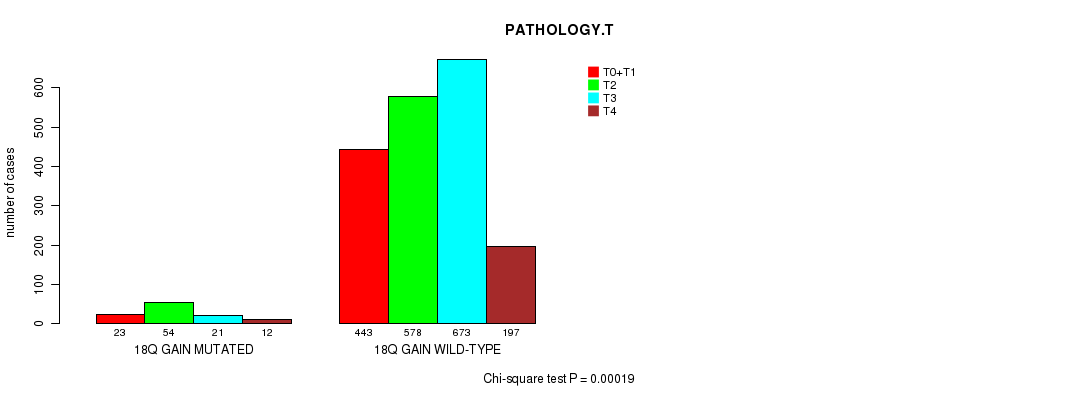
Table S162. Get Full Table Description of clustering approach #34: '19p gain mutation analysis'
| Cluster Labels | 19P GAIN MUTATED | 19P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 503 | 4060 |
P value = 1.12e-08 (logrank test), Q value = 1.6e-05
Table S163. Clustering Approach #34: '19p gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 4296 | 1401 | 0.0 - 224.0 (14.9) |
| 19P GAIN MUTATED | 485 | 210 | 0.0 - 180.2 (13.3) |
| 19P GAIN WILD-TYPE | 3811 | 1191 | 0.0 - 224.0 (15.0) |
Figure S129. Get High-res Image Clustering Approach #34: '19p gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
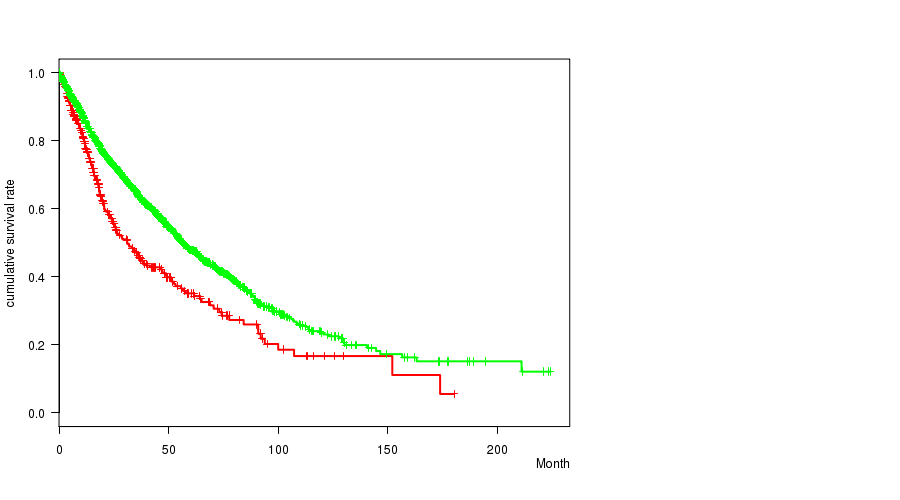
P value = 5.15e-56 (Chi-square test), Q value = 8e-53
Table S164. Clustering Approach #34: '19p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 19P GAIN MUTATED | 13 | 167 | 69 | 34 | 27 | 15 | 24 | 48 | 0 | 87 | 0 | 19 |
| 19P GAIN WILD-TYPE | 112 | 377 | 777 | 378 | 420 | 304 | 469 | 614 | 2 | 461 | 2 | 140 |
Figure S130. Get High-res Image Clustering Approach #34: '19p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
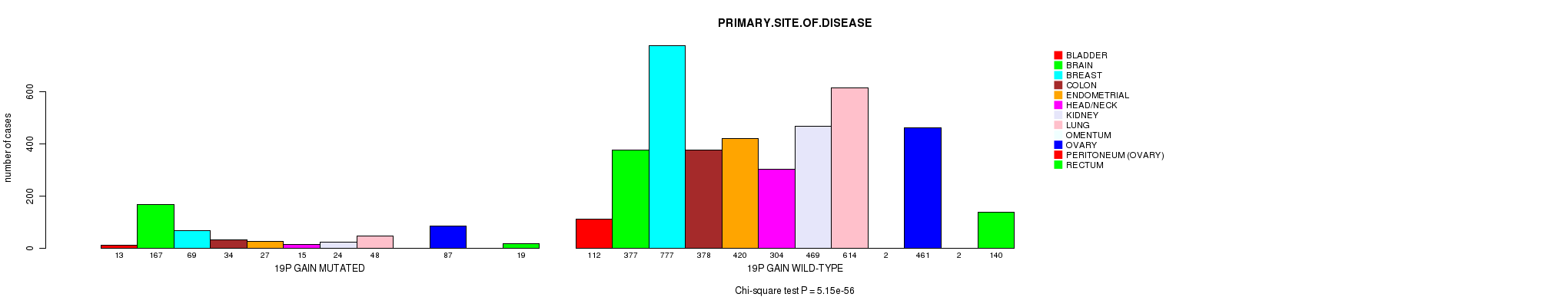
P value = 1.97e-20 (Chi-square test), Q value = 2.9e-17
Table S165. Clustering Approach #34: '19p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 19P GAIN MUTATED | 33 | 1 | 12 | 15 | 24 | 0 | 1 | 10 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 32 | 0 | 0 | 19 | 0 | 87 | 15 | 1 | 6 |
| 19P GAIN WILD-TYPE | 323 | 53 | 338 | 304 | 469 | 10 | 74 | 195 | 2 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 283 | 18 | 7 | 124 | 13 | 465 | 64 | 0 | 14 |
Figure S131. Get High-res Image Clustering Approach #34: '19p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 3.56e-13 (Fisher's exact test), Q value = 5.1e-10
Table S166. Clustering Approach #34: '19p gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 19P GAIN MUTATED | 154 | 349 |
| 19P GAIN WILD-TYPE | 672 | 3388 |
Figure S132. Get High-res Image Clustering Approach #34: '19p gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
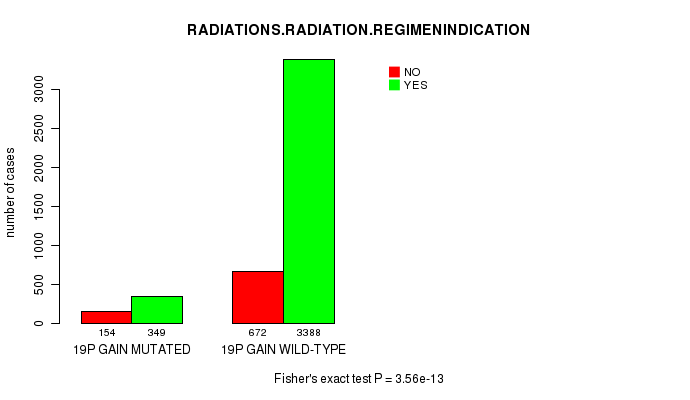
Table S167. Get Full Table Description of clustering approach #35: '19q gain mutation analysis'
| Cluster Labels | 19Q GAIN MUTATED | 19Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 536 | 4027 |
P value = 4.08e-08 (logrank test), Q value = 5.6e-05
Table S168. Clustering Approach #35: '19q gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 4296 | 1401 | 0.0 - 224.0 (14.9) |
| 19Q GAIN MUTATED | 513 | 210 | 0.0 - 180.2 (12.6) |
| 19Q GAIN WILD-TYPE | 3783 | 1191 | 0.0 - 224.0 (15.1) |
Figure S133. Get High-res Image Clustering Approach #35: '19q gain mutation analysis' versus Clinical Feature #1: 'Time to Death'

P value = 3.05e-30 (Chi-square test), Q value = 4.6e-27
Table S169. Clustering Approach #35: '19q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 19Q GAIN MUTATED | 25 | 142 | 84 | 41 | 24 | 21 | 28 | 72 | 0 | 76 | 0 | 23 |
| 19Q GAIN WILD-TYPE | 100 | 402 | 762 | 371 | 423 | 298 | 465 | 590 | 2 | 472 | 2 | 136 |
Figure S134. Get High-res Image Clustering Approach #35: '19q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
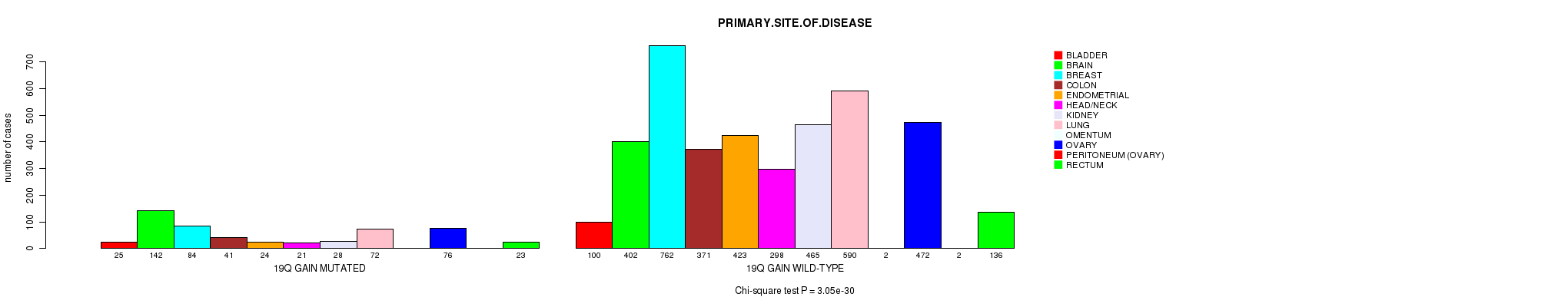
P value = 1.2e-15 (Chi-square test), Q value = 1.7e-12
Table S170. Clustering Approach #35: '19q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 19Q GAIN MUTATED | 37 | 4 | 7 | 21 | 28 | 0 | 3 | 18 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 46 | 0 | 0 | 23 | 0 | 76 | 17 | 1 | 3 |
| 19Q GAIN WILD-TYPE | 319 | 50 | 343 | 298 | 465 | 10 | 72 | 187 | 2 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 269 | 18 | 7 | 120 | 13 | 476 | 62 | 0 | 17 |
Figure S135. Get High-res Image Clustering Approach #35: '19q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.22e-05 (Fisher's exact test), Q value = 0.016
Table S171. Clustering Approach #35: '19q gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 19Q GAIN MUTATED | 135 | 401 |
| 19Q GAIN WILD-TYPE | 691 | 3336 |
Figure S136. Get High-res Image Clustering Approach #35: '19q gain mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
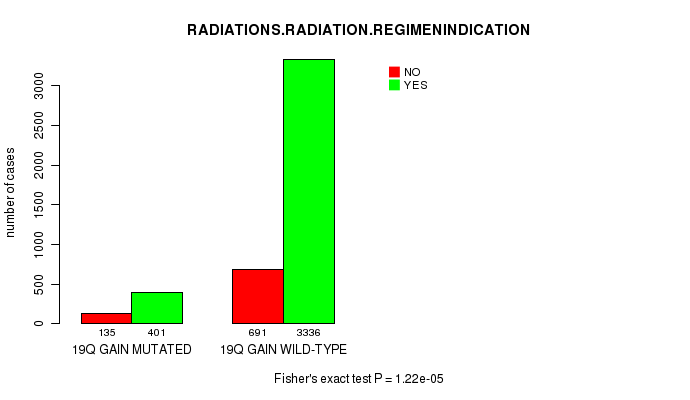
Table S172. Get Full Table Description of clustering approach #36: '20p gain mutation analysis'
| Cluster Labels | 20P GAIN MUTATED | 20P GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 1300 | 3263 |
P value = 4.13e-05 (logrank test), Q value = 0.054
Table S173. Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 4296 | 1401 | 0.0 - 224.0 (14.9) |
| 20P GAIN MUTATED | 1200 | 433 | 0.0 - 224.0 (13.1) |
| 20P GAIN WILD-TYPE | 3096 | 968 | 0.0 - 223.4 (15.1) |
Figure S137. Get High-res Image Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #1: 'Time to Death'
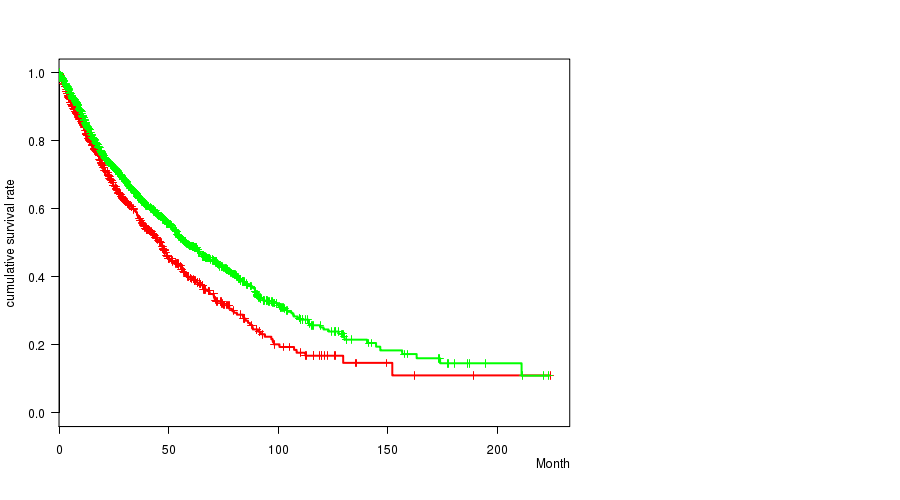
P value = 7.61e-09 (t-test), Q value = 1.1e-05
Table S174. Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 4507 | 61.9 (12.6) |
| 20P GAIN MUTATED | 1289 | 63.5 (12.3) |
| 20P GAIN WILD-TYPE | 3218 | 61.2 (12.7) |
Figure S138. Get High-res Image Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #2: 'AGE'
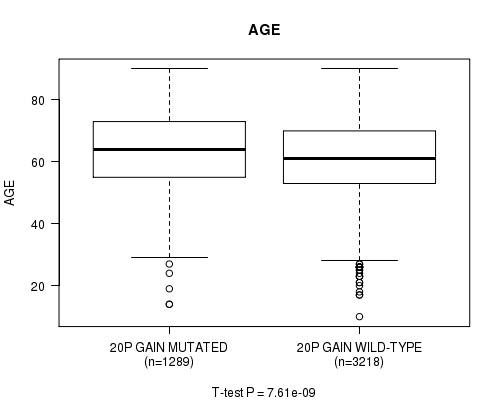
P value = 1.84e-64 (Chi-square test), Q value = 2.9e-61
Table S175. Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 20P GAIN MUTATED | 48 | 174 | 233 | 197 | 52 | 64 | 66 | 148 | 1 | 223 | 1 | 91 |
| 20P GAIN WILD-TYPE | 77 | 370 | 613 | 215 | 395 | 255 | 427 | 514 | 1 | 325 | 1 | 68 |
Figure S139. Get High-res Image Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
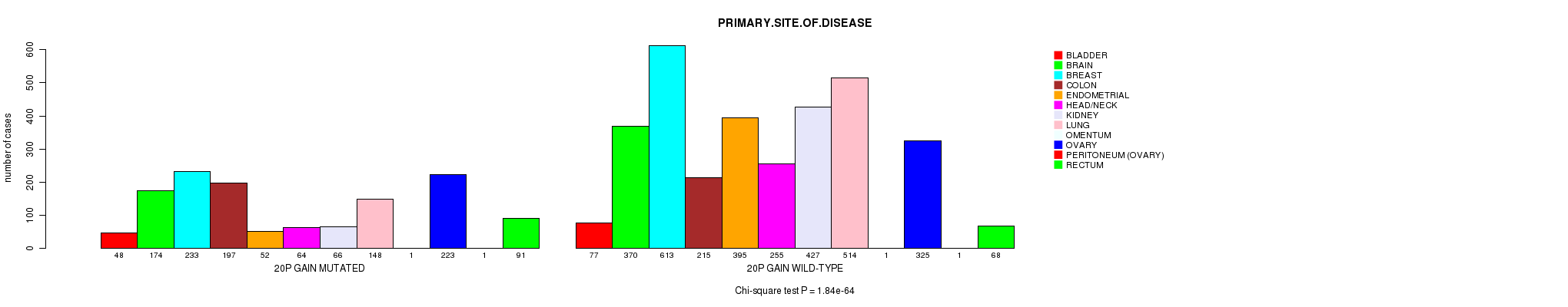
P value = 2.23e-68 (Chi-square test), Q value = 3.5e-65
Table S176. Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 20P GAIN MUTATED | 182 | 13 | 26 | 64 | 66 | 2 | 11 | 34 | 5 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 1 | 0 | 91 | 3 | 0 | 87 | 2 | 225 | 23 | 1 | 7 |
| 20P GAIN WILD-TYPE | 174 | 41 | 324 | 255 | 427 | 8 | 64 | 171 | 2 | 2 | 15 | 2 | 2 | 2 | 11 | 1 | 1 | 0 | 2 | 224 | 15 | 7 | 56 | 11 | 327 | 56 | 0 | 13 |
Figure S140. Get High-res Image Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 6.36e-14 (Chi-square test), Q value = 9.1e-11
Table S177. Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 20P GAIN MUTATED | 71 | 171 | 255 | 59 |
| 20P GAIN WILD-TYPE | 395 | 461 | 439 | 150 |
Figure S141. Get High-res Image Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
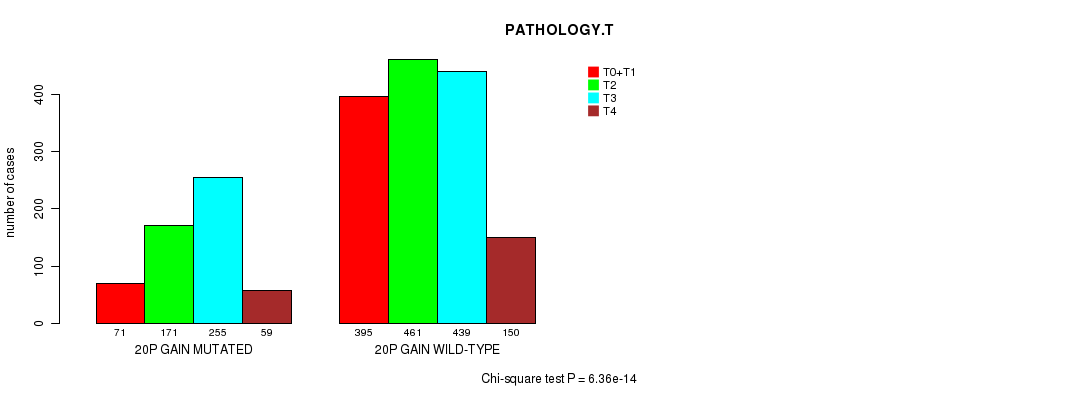
P value = 9.41e-06 (Chi-square test), Q value = 0.012
Table S178. Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
| nPatients | M0 | M1 | M1A | M1B | MX |
|---|---|---|---|---|---|
| ALL | 1424 | 168 | 9 | 3 | 165 |
| 20P GAIN MUTATED | 387 | 66 | 8 | 0 | 43 |
| 20P GAIN WILD-TYPE | 1037 | 102 | 1 | 3 | 122 |
Figure S142. Get High-res Image Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
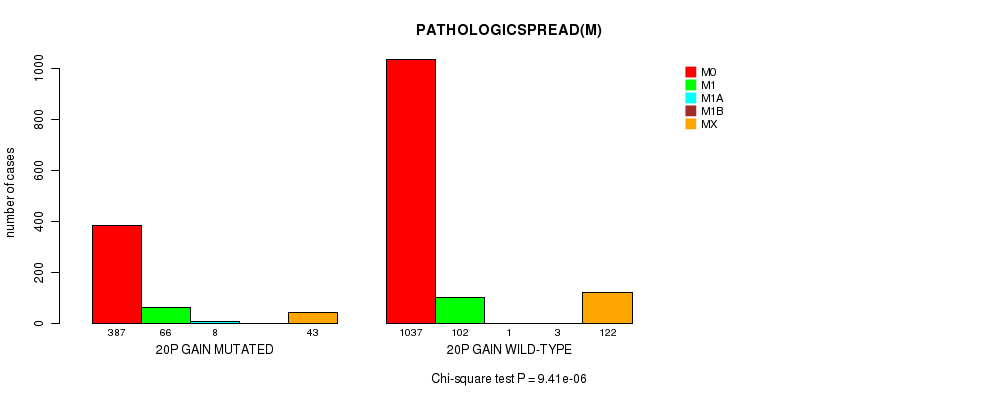
P value = 2.76e-07 (Chi-square test), Q value = 0.00038
Table S179. Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 20P GAIN MUTATED | 160 | 153 | 315 | 132 |
| 20P GAIN WILD-TYPE | 552 | 345 | 566 | 291 |
Figure S143. Get High-res Image Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
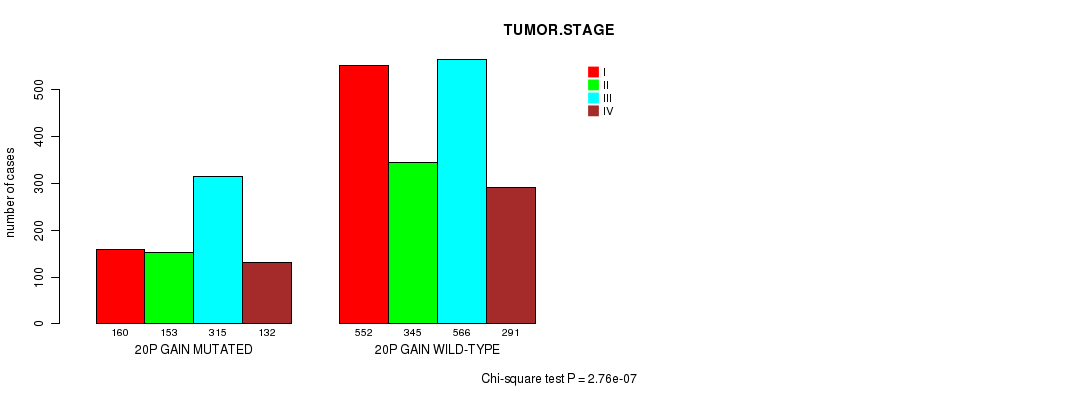
P value = 6.9e-06 (Chi-square test), Q value = 0.0092
Table S180. Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #18: 'COMPLETENESS.OF.RESECTION'
| nPatients | R0 | R1 | R2 | RX |
|---|---|---|---|---|
| ALL | 1171 | 69 | 59 | 77 |
| 20P GAIN MUTATED | 342 | 15 | 33 | 14 |
| 20P GAIN WILD-TYPE | 829 | 54 | 26 | 63 |
Figure S144. Get High-res Image Clustering Approach #36: '20p gain mutation analysis' versus Clinical Feature #18: 'COMPLETENESS.OF.RESECTION'
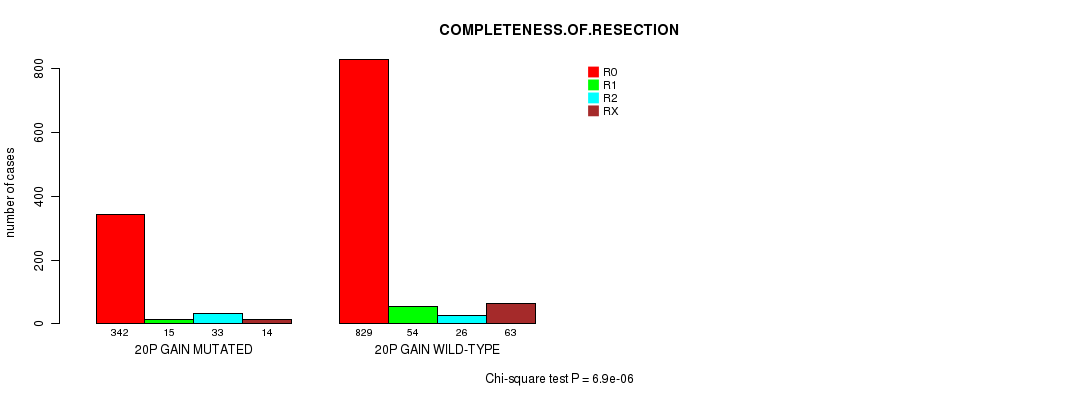
Table S181. Get Full Table Description of clustering approach #37: '20q gain mutation analysis'
| Cluster Labels | 20Q GAIN MUTATED | 20Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 1509 | 3054 |
P value = 3.39e-07 (t-test), Q value = 0.00046
Table S182. Clustering Approach #37: '20q gain mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 4507 | 61.9 (12.6) |
| 20Q GAIN MUTATED | 1498 | 63.2 (12.4) |
| 20Q GAIN WILD-TYPE | 3009 | 61.2 (12.7) |
Figure S145. Get High-res Image Clustering Approach #37: '20q gain mutation analysis' versus Clinical Feature #2: 'AGE'

P value = 1.97e-130 (Chi-square test), Q value = 3.1e-127
Table S183. Clustering Approach #37: '20q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 20Q GAIN MUTATED | 52 | 171 | 271 | 267 | 60 | 68 | 68 | 151 | 1 | 264 | 1 | 132 |
| 20Q GAIN WILD-TYPE | 73 | 373 | 575 | 145 | 387 | 251 | 425 | 511 | 1 | 284 | 1 | 27 |
Figure S146. Get High-res Image Clustering Approach #37: '20q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
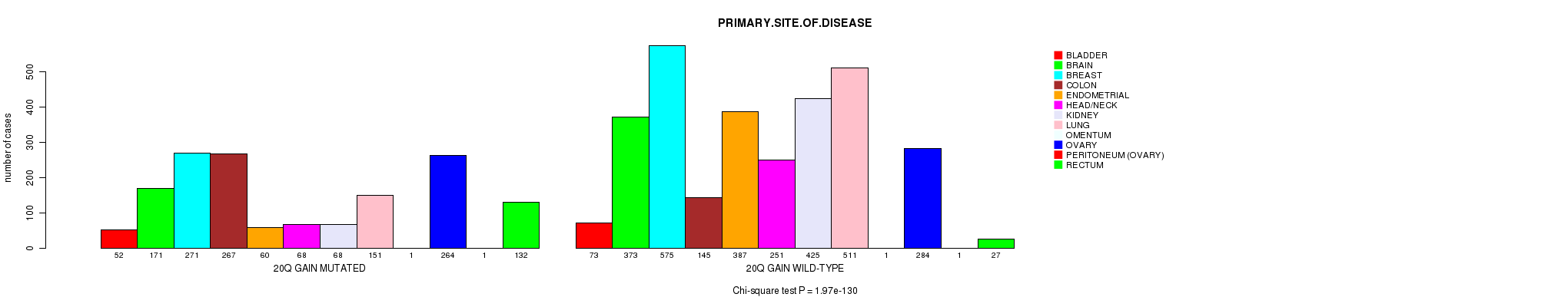
P value = 1.03e-135 (Chi-square test), Q value = 1.6e-132
Table S184. Clustering Approach #37: '20q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 20Q GAIN MUTATED | 249 | 16 | 27 | 68 | 68 | 3 | 14 | 44 | 5 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 1 | 1 | 79 | 3 | 0 | 125 | 5 | 266 | 30 | 1 | 6 |
| 20Q GAIN WILD-TYPE | 107 | 38 | 323 | 251 | 425 | 7 | 61 | 161 | 2 | 2 | 15 | 2 | 2 | 2 | 11 | 1 | 1 | 0 | 1 | 236 | 15 | 7 | 18 | 8 | 286 | 49 | 0 | 14 |
Figure S147. Get High-res Image Clustering Approach #37: '20q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 5.56e-29 (Chi-square test), Q value = 8.3e-26
Table S185. Clustering Approach #37: '20q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 20Q GAIN MUTATED | 74 | 198 | 336 | 71 |
| 20Q GAIN WILD-TYPE | 392 | 434 | 358 | 138 |
Figure S148. Get High-res Image Clustering Approach #37: '20q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
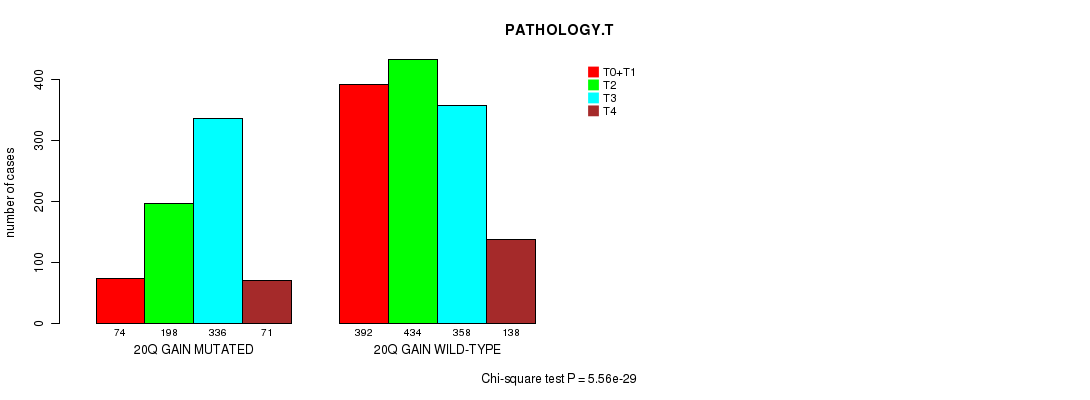
P value = 2.83e-15 (Chi-square test), Q value = 4.1e-12
Table S186. Clustering Approach #37: '20q gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 20Q GAIN MUTATED | 176 | 199 | 394 | 152 |
| 20Q GAIN WILD-TYPE | 536 | 299 | 487 | 271 |
Figure S149. Get High-res Image Clustering Approach #37: '20q gain mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
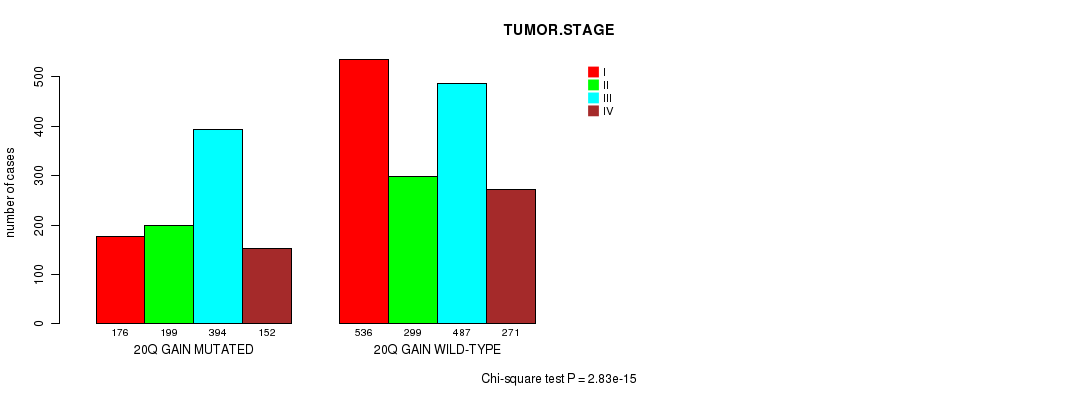
P value = 8.01e-06 (Chi-square test), Q value = 0.011
Table S187. Clustering Approach #37: '20q gain mutation analysis' versus Clinical Feature #14: 'TOBACCOSMOKINGHISTORYINDICATOR'
| nPatients | 1 | 2 | 3 | 4 | 5 | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|---|---|---|---|
| ALL | 28 | 23 | 37 | 29 | 2 | 355 | 217 | 250 | 128 |
| 20Q GAIN MUTATED | 7 | 12 | 15 | 17 | 0 | 91 | 43 | 54 | 27 |
| 20Q GAIN WILD-TYPE | 21 | 11 | 22 | 12 | 2 | 264 | 174 | 196 | 101 |
Figure S150. Get High-res Image Clustering Approach #37: '20q gain mutation analysis' versus Clinical Feature #14: 'TOBACCOSMOKINGHISTORYINDICATOR'
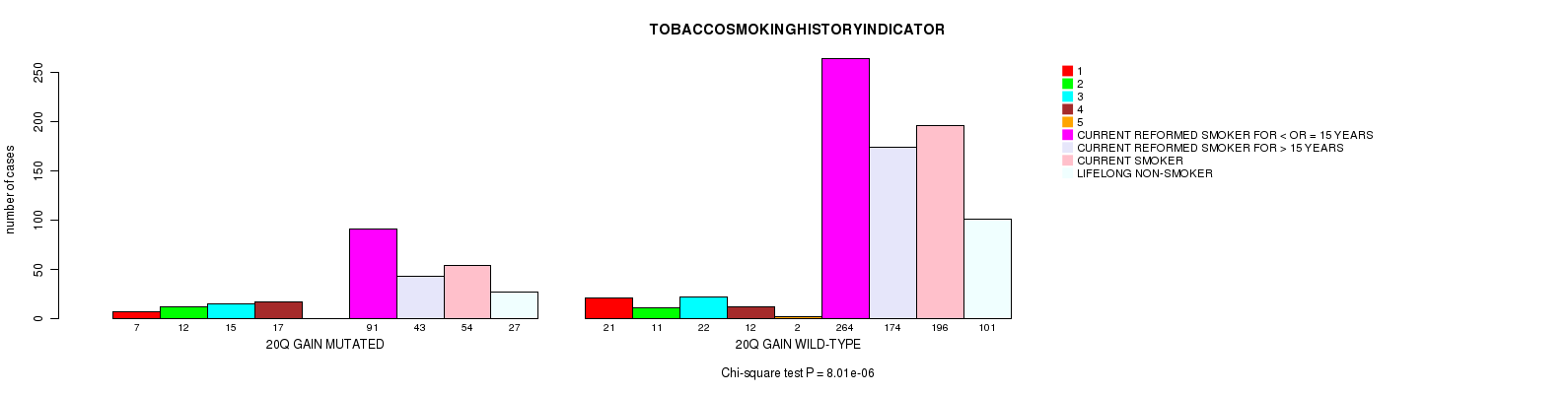
P value = 9.3e-07 (Chi-square test), Q value = 0.0013
Table S188. Clustering Approach #37: '20q gain mutation analysis' versus Clinical Feature #18: 'COMPLETENESS.OF.RESECTION'
| nPatients | R0 | R1 | R2 | RX |
|---|---|---|---|---|
| ALL | 1171 | 69 | 59 | 77 |
| 20Q GAIN MUTATED | 428 | 19 | 40 | 20 |
| 20Q GAIN WILD-TYPE | 743 | 50 | 19 | 57 |
Figure S151. Get High-res Image Clustering Approach #37: '20q gain mutation analysis' versus Clinical Feature #18: 'COMPLETENESS.OF.RESECTION'
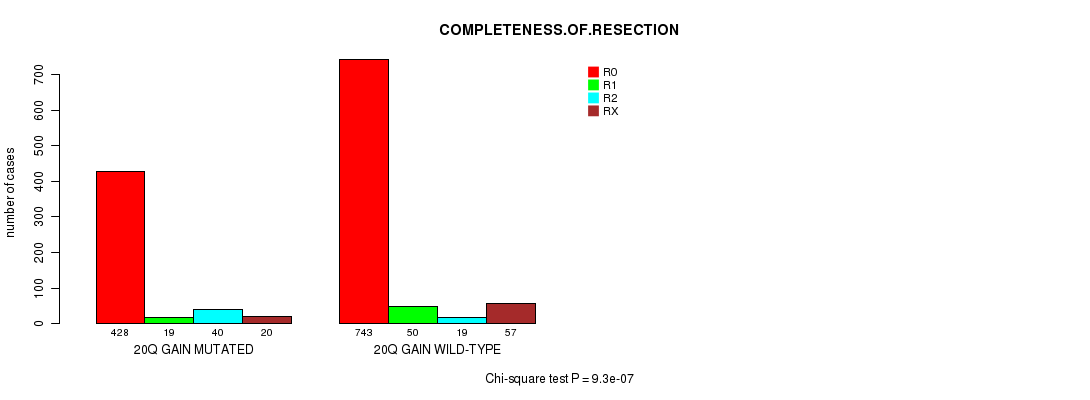
Table S189. Get Full Table Description of clustering approach #38: '21q gain mutation analysis'
| Cluster Labels | 21Q GAIN MUTATED | 21Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 347 | 4216 |
P value = 1.66e-15 (Chi-square test), Q value = 2.4e-12
Table S190. Clustering Approach #38: '21q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 21Q GAIN MUTATED | 23 | 31 | 98 | 11 | 20 | 8 | 33 | 51 | 0 | 63 | 1 | 7 |
| 21Q GAIN WILD-TYPE | 102 | 513 | 748 | 401 | 427 | 311 | 460 | 611 | 2 | 485 | 1 | 152 |
Figure S152. Get High-res Image Clustering Approach #38: '21q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
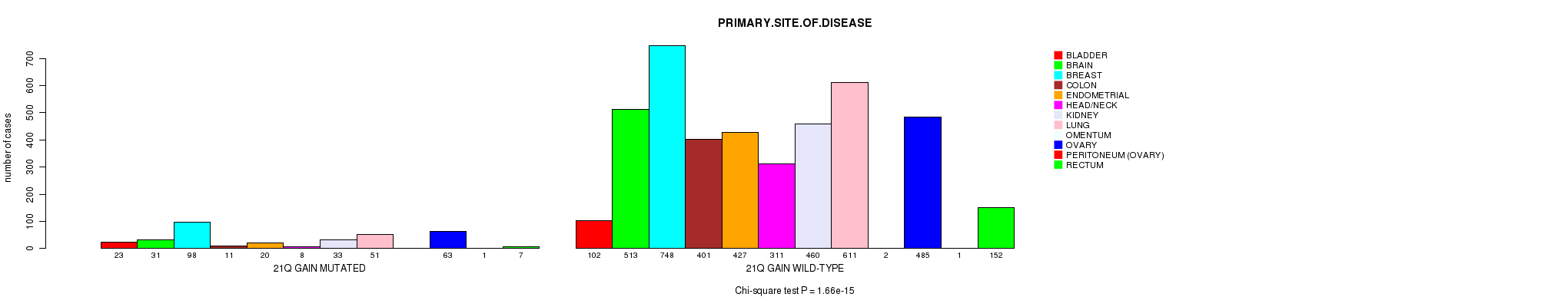
P value = 4.02e-10 (Chi-square test), Q value = 5.7e-07
Table S191. Clustering Approach #38: '21q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 21Q GAIN MUTATED | 10 | 1 | 9 | 8 | 33 | 1 | 5 | 24 | 2 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 16 | 0 | 0 | 7 | 0 | 64 | 11 | 0 | 1 |
| 21Q GAIN WILD-TYPE | 346 | 53 | 341 | 311 | 460 | 9 | 70 | 181 | 5 | 3 | 15 | 2 | 2 | 2 | 12 | 1 | 1 | 0 | 2 | 299 | 18 | 7 | 136 | 13 | 488 | 68 | 1 | 19 |
Figure S153. Get High-res Image Clustering Approach #38: '21q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.61e-05 (Chi-square test), Q value = 0.021
Table S192. Clustering Approach #38: '21q gain mutation analysis' versus Clinical Feature #14: 'TOBACCOSMOKINGHISTORYINDICATOR'
| nPatients | 1 | 2 | 3 | 4 | 5 | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|---|---|---|---|
| ALL | 28 | 23 | 37 | 29 | 2 | 355 | 217 | 250 | 128 |
| 21Q GAIN MUTATED | 6 | 2 | 8 | 7 | 0 | 23 | 13 | 10 | 12 |
| 21Q GAIN WILD-TYPE | 22 | 21 | 29 | 22 | 2 | 332 | 204 | 240 | 116 |
Figure S154. Get High-res Image Clustering Approach #38: '21q gain mutation analysis' versus Clinical Feature #14: 'TOBACCOSMOKINGHISTORYINDICATOR'
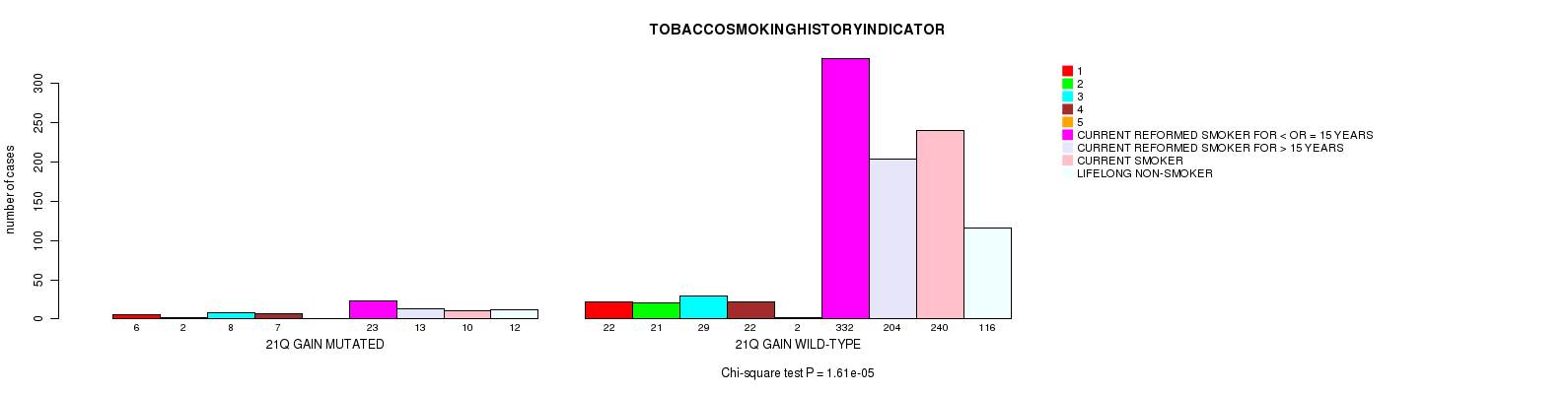
Table S193. Get Full Table Description of clustering approach #39: '22q gain mutation analysis'
| Cluster Labels | 22Q GAIN MUTATED | 22Q GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 240 | 4323 |
P value = 4.2e-36 (Chi-square test), Q value = 6.3e-33
Table S194. Clustering Approach #39: '22q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 22Q GAIN MUTATED | 11 | 11 | 38 | 2 | 6 | 39 | 25 | 93 | 0 | 10 | 0 | 5 |
| 22Q GAIN WILD-TYPE | 114 | 533 | 808 | 410 | 441 | 280 | 468 | 569 | 2 | 538 | 2 | 154 |
Figure S155. Get High-res Image Clustering Approach #39: '22q gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
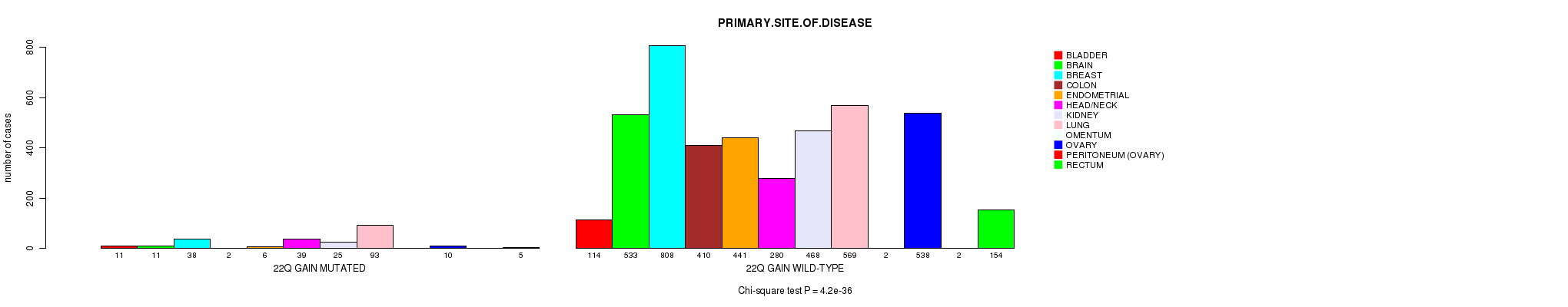
P value = 2.26e-09 (Fisher's exact test), Q value = 3.2e-06
Table S195. Clustering Approach #39: '22q gain mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 22Q GAIN MUTATED | 107 | 133 |
| 22Q GAIN WILD-TYPE | 2774 | 1549 |
Figure S156. Get High-res Image Clustering Approach #39: '22q gain mutation analysis' versus Clinical Feature #4: 'GENDER'

P value = 1.24e-43 (Chi-square test), Q value = 1.9e-40
Table S196. Clustering Approach #39: '22q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 22Q GAIN MUTATED | 2 | 0 | 2 | 39 | 25 | 0 | 1 | 15 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 74 | 0 | 0 | 5 | 0 | 10 | 4 | 0 | 0 |
| 22Q GAIN WILD-TYPE | 354 | 54 | 348 | 280 | 468 | 10 | 74 | 190 | 5 | 3 | 15 | 2 | 2 | 2 | 13 | 1 | 1 | 1 | 2 | 241 | 18 | 7 | 138 | 13 | 542 | 75 | 1 | 20 |
Figure S157. Get High-res Image Clustering Approach #39: '22q gain mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 5.6e-07 (Chi-square test), Q value = 0.00076
Table S197. Clustering Approach #39: '22q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 22Q GAIN MUTATED | 26 | 79 | 33 | 19 |
| 22Q GAIN WILD-TYPE | 440 | 553 | 661 | 190 |
Figure S158. Get High-res Image Clustering Approach #39: '22q gain mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
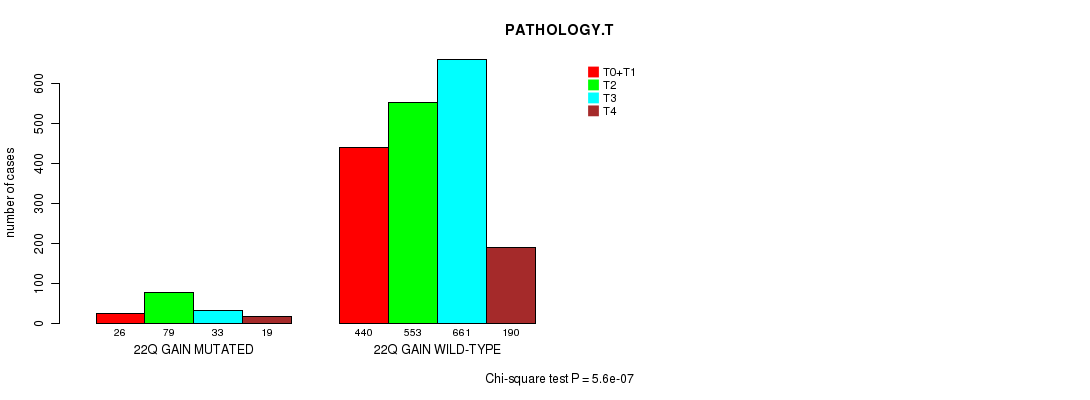
Table S198. Get Full Table Description of clustering approach #40: 'Xq gain mutation analysis'
| Cluster Labels | XQ GAIN MUTATED | XQ GAIN WILD-TYPE |
|---|---|---|
| Number of samples | 149 | 4414 |
P value = 2.22e-08 (Chi-square test), Q value = 3.1e-05
Table S199. Clustering Approach #40: 'Xq gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| XQ GAIN MUTATED | 8 | 3 | 21 | 11 | 8 | 20 | 7 | 29 | 0 | 36 | 0 | 6 |
| XQ GAIN WILD-TYPE | 117 | 541 | 825 | 401 | 439 | 299 | 486 | 633 | 2 | 512 | 2 | 153 |
Figure S159. Get High-res Image Clustering Approach #40: 'Xq gain mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
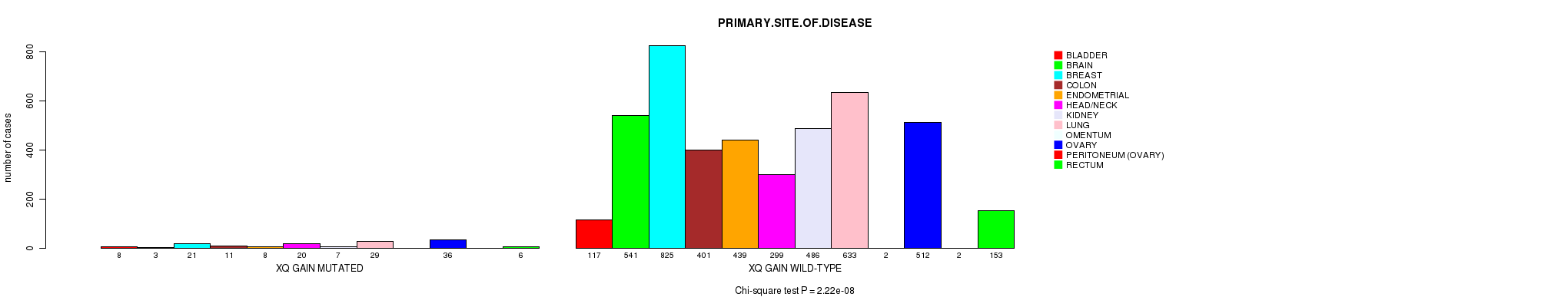
Table S200. Get Full Table Description of clustering approach #41: '1p loss mutation analysis'
| Cluster Labels | 1P LOSS MUTATED | 1P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 351 | 4212 |
P value = 4.3e-20 (Chi-square test), Q value = 6.3e-17
Table S201. Clustering Approach #41: '1p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 1P LOSS MUTATED | 2 | 9 | 113 | 49 | 11 | 15 | 31 | 62 | 0 | 36 | 0 | 22 |
| 1P LOSS WILD-TYPE | 123 | 535 | 733 | 363 | 436 | 304 | 462 | 600 | 2 | 512 | 2 | 137 |
Figure S160. Get High-res Image Clustering Approach #41: '1p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'

P value = 4.74e-08 (Chi-square test), Q value = 6.6e-05
Table S202. Clustering Approach #41: '1p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 1P LOSS MUTATED | 46 | 3 | 5 | 15 | 31 | 1 | 6 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 40 | 2 | 1 | 21 | 0 | 36 | 4 | 0 | 0 |
| 1P LOSS WILD-TYPE | 310 | 51 | 345 | 304 | 462 | 9 | 69 | 194 | 7 | 3 | 15 | 2 | 3 | 2 | 11 | 0 | 1 | 1 | 2 | 275 | 16 | 6 | 122 | 13 | 516 | 75 | 1 | 20 |
Figure S161. Get High-res Image Clustering Approach #41: '1p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.07e-05 (Chi-square test), Q value = 0.014
Table S203. Clustering Approach #41: '1p loss mutation analysis' versus Clinical Feature #21: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE IA | STAGE IB | STAGE II | STAGE IIA | STAGE IIB | STAGE III | STAGE IIIA | STAGE IIIB | STAGE IIIC | STAGE IV | STAGE TIS | STAGE X |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 72 | 62 | 7 | 43 | 287 | 186 | 42 | 124 | 25 | 41 | 59 | 1 | 16 |
| 1P LOSS MUTATED | 4 | 5 | 0 | 1 | 36 | 19 | 1 | 21 | 11 | 7 | 5 | 0 | 4 |
| 1P LOSS WILD-TYPE | 68 | 57 | 7 | 42 | 251 | 167 | 41 | 103 | 14 | 34 | 54 | 1 | 12 |
Figure S162. Get High-res Image Clustering Approach #41: '1p loss mutation analysis' versus Clinical Feature #21: 'NEOPLASM.DISEASESTAGE'
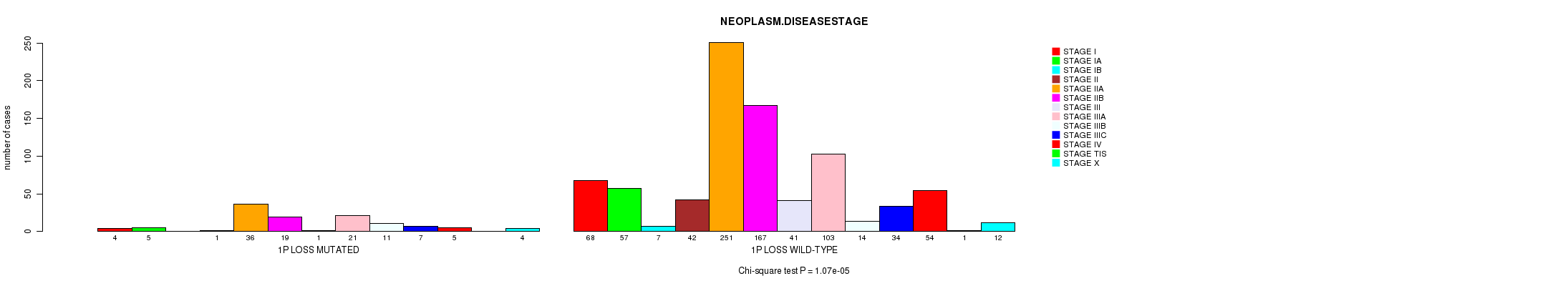
Table S204. Get Full Table Description of clustering approach #42: '1q loss mutation analysis'
| Cluster Labels | 1Q LOSS MUTATED | 1Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 112 | 4451 |
Table S205. Get Full Table Description of clustering approach #43: '2p loss mutation analysis'
| Cluster Labels | 2P LOSS MUTATED | 2P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 148 | 4415 |
P value = 6.03e-17 (Chi-square test), Q value = 8.8e-14
Table S206. Clustering Approach #43: '2p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 2P LOSS MUTATED | 6 | 13 | 69 | 4 | 5 | 6 | 10 | 4 | 0 | 24 | 0 | 7 |
| 2P LOSS WILD-TYPE | 119 | 531 | 777 | 408 | 442 | 313 | 483 | 658 | 2 | 524 | 2 | 152 |
Figure S163. Get High-res Image Clustering Approach #43: '2p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'

P value = 6.09e-05 (Fisher's exact test), Q value = 0.079
Table S207. Clustering Approach #43: '2p loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 2P LOSS MUTATED | 116 | 32 |
| 2P LOSS WILD-TYPE | 2765 | 1650 |
Figure S164. Get High-res Image Clustering Approach #43: '2p loss mutation analysis' versus Clinical Feature #4: 'GENDER'
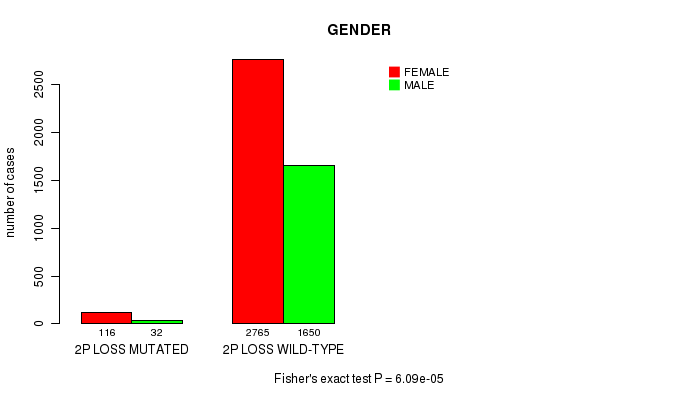
Table S208. Get Full Table Description of clustering approach #44: '2q loss mutation analysis'
| Cluster Labels | 2Q LOSS MUTATED | 2Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 188 | 4375 |
P value = 2.8e-22 (Chi-square test), Q value = 4.1e-19
Table S209. Clustering Approach #44: '2q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 2Q LOSS MUTATED | 13 | 12 | 83 | 6 | 7 | 11 | 11 | 11 | 0 | 28 | 1 | 5 |
| 2Q LOSS WILD-TYPE | 112 | 532 | 763 | 406 | 440 | 308 | 482 | 651 | 2 | 520 | 1 | 154 |
Figure S165. Get High-res Image Clustering Approach #44: '2q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
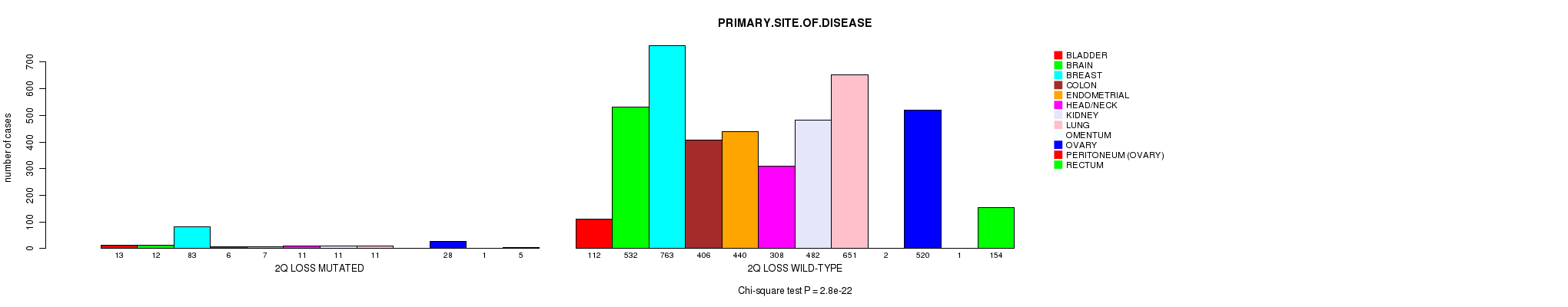
P value = 1.65e-05 (Chi-square test), Q value = 0.022
Table S210. Clustering Approach #44: '2q loss mutation analysis' versus Clinical Feature #14: 'TOBACCOSMOKINGHISTORYINDICATOR'
| nPatients | 1 | 2 | 3 | 4 | 5 | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|---|---|---|---|
| ALL | 28 | 23 | 37 | 29 | 2 | 355 | 217 | 250 | 128 |
| 2Q LOSS MUTATED | 3 | 1 | 4 | 4 | 0 | 4 | 1 | 11 | 4 |
| 2Q LOSS WILD-TYPE | 25 | 22 | 33 | 25 | 2 | 351 | 216 | 239 | 124 |
Figure S166. Get High-res Image Clustering Approach #44: '2q loss mutation analysis' versus Clinical Feature #14: 'TOBACCOSMOKINGHISTORYINDICATOR'
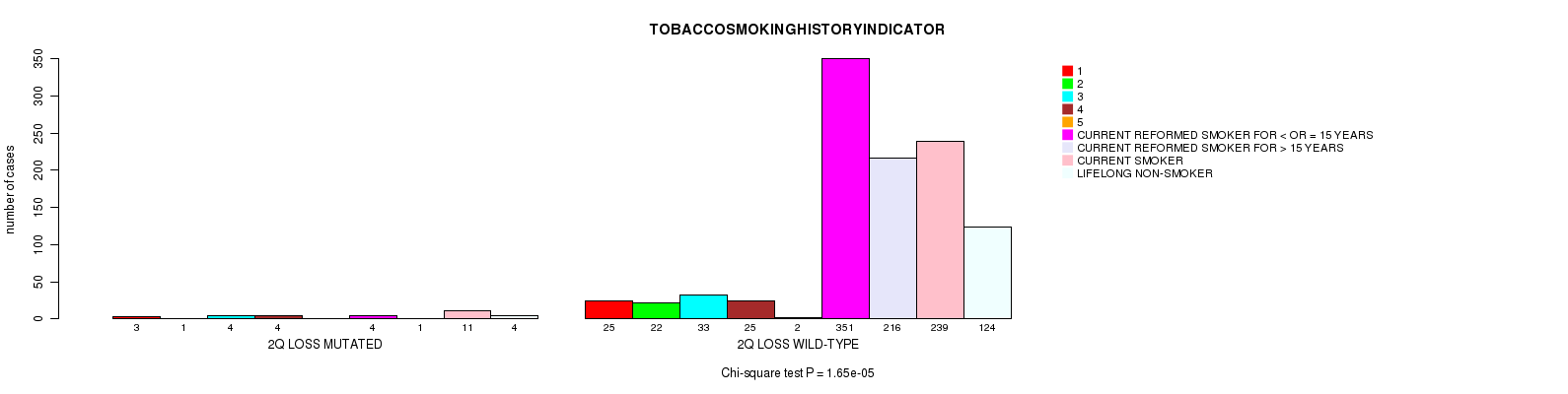
Table S211. Get Full Table Description of clustering approach #45: '3p loss mutation analysis'
| Cluster Labels | 3P LOSS MUTATED | 3P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 868 | 3695 |
P value = 1.29e-218 (Chi-square test), Q value = 2.1e-215
Table S212. Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 3P LOSS MUTATED | 7 | 25 | 85 | 28 | 26 | 121 | 318 | 193 | 1 | 56 | 0 | 8 |
| 3P LOSS WILD-TYPE | 118 | 519 | 761 | 384 | 421 | 198 | 175 | 469 | 1 | 492 | 2 | 151 |
Figure S167. Get High-res Image Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
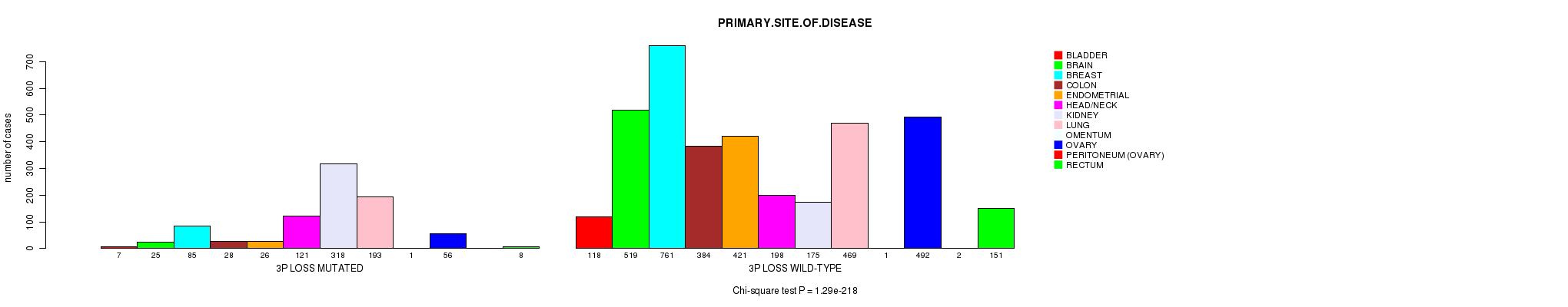
P value = 7.17e-34 (Fisher's exact test), Q value = 1.1e-30
Table S213. Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 3P LOSS MUTATED | 390 | 478 |
| 3P LOSS WILD-TYPE | 2491 | 1204 |
Figure S168. Get High-res Image Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #4: 'GENDER'
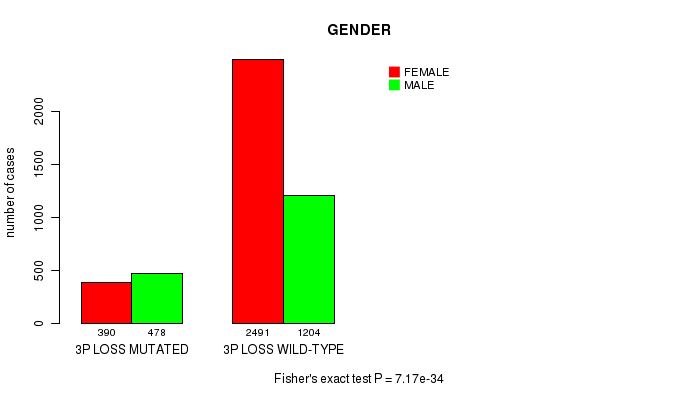
P value = 3.19e-144 (Chi-square test), Q value = 5e-141
Table S214. Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 3P LOSS MUTATED | 27 | 1 | 11 | 121 | 318 | 3 | 17 | 44 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 122 | 4 | 1 | 5 | 2 | 57 | 11 | 0 | 1 |
| 3P LOSS WILD-TYPE | 329 | 53 | 339 | 198 | 175 | 7 | 58 | 161 | 6 | 3 | 14 | 2 | 3 | 2 | 11 | 0 | 1 | 0 | 2 | 193 | 14 | 6 | 138 | 11 | 495 | 68 | 1 | 19 |
Figure S169. Get High-res Image Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 5.56e-14 (Chi-square test), Q value = 8e-11
Table S215. Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 3P LOSS MUTATED | 222 | 183 | 189 | 55 |
| 3P LOSS WILD-TYPE | 244 | 449 | 505 | 154 |
Figure S170. Get High-res Image Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
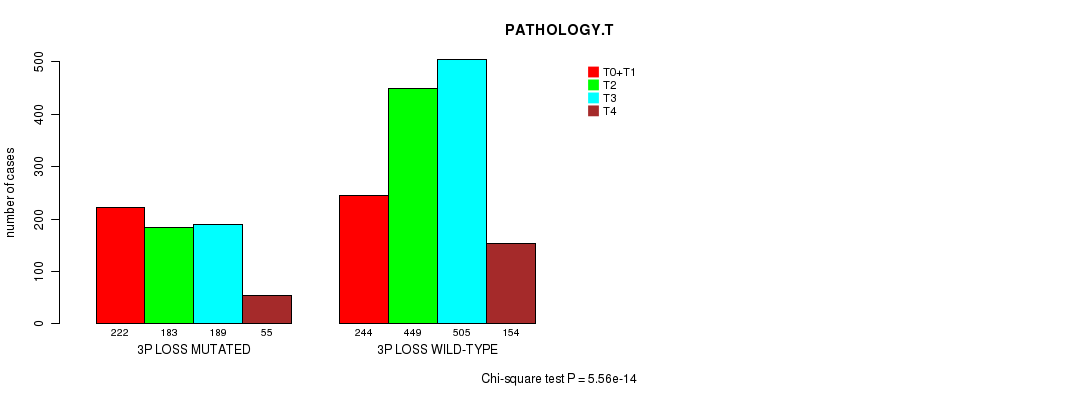
P value = 0.00016 (Chi-square test), Q value = 0.2
Table S216. Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
| nPatients | M0 | M1 | M1A | M1B | MX |
|---|---|---|---|---|---|
| ALL | 1424 | 168 | 9 | 3 | 165 |
| 3P LOSS MUTATED | 469 | 57 | 1 | 0 | 27 |
| 3P LOSS WILD-TYPE | 955 | 111 | 8 | 3 | 138 |
Figure S171. Get High-res Image Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
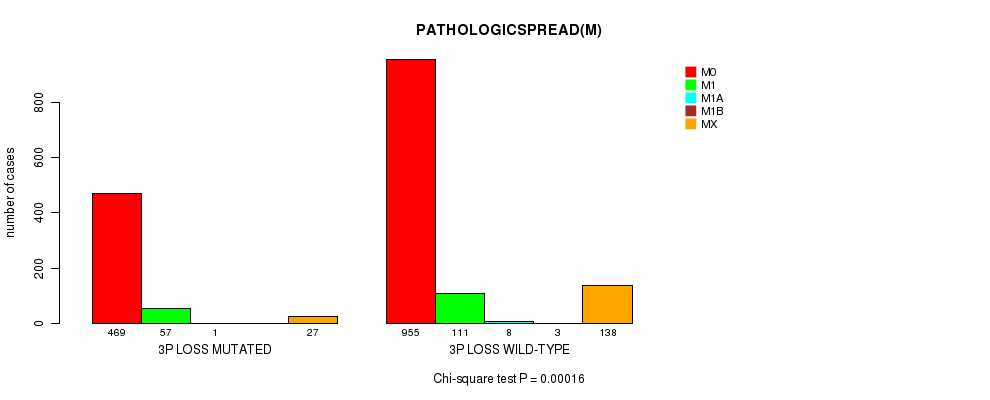
P value = 4.02e-13 (Chi-square test), Q value = 5.7e-10
Table S217. Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 3P LOSS MUTATED | 268 | 116 | 186 | 130 |
| 3P LOSS WILD-TYPE | 444 | 382 | 695 | 293 |
Figure S172. Get High-res Image Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'

P value = 2.88e-11 (Fisher's exact test), Q value = 4.1e-08
Table S218. Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 3P LOSS MUTATED | 92 | 776 |
| 3P LOSS WILD-TYPE | 734 | 2961 |
Figure S173. Get High-res Image Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
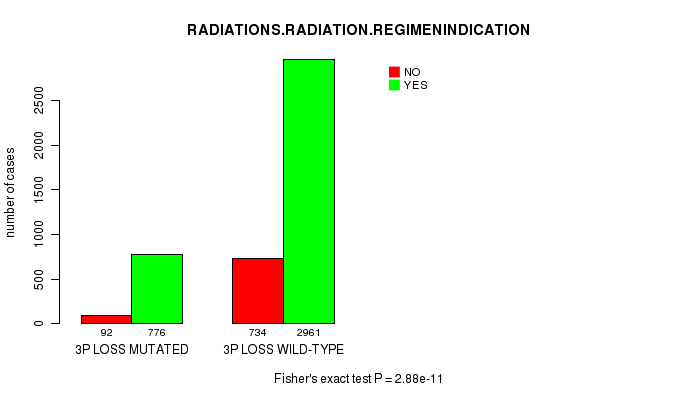
P value = 5.35e-08 (Chi-square test), Q value = 7.4e-05
Table S219. Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #14: 'TOBACCOSMOKINGHISTORYINDICATOR'
| nPatients | 1 | 2 | 3 | 4 | 5 | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|---|---|---|---|
| ALL | 28 | 23 | 37 | 29 | 2 | 355 | 217 | 250 | 128 |
| 3P LOSS MUTATED | 0 | 1 | 4 | 1 | 0 | 121 | 61 | 93 | 29 |
| 3P LOSS WILD-TYPE | 28 | 22 | 33 | 28 | 2 | 234 | 156 | 157 | 99 |
Figure S174. Get High-res Image Clustering Approach #45: '3p loss mutation analysis' versus Clinical Feature #14: 'TOBACCOSMOKINGHISTORYINDICATOR'
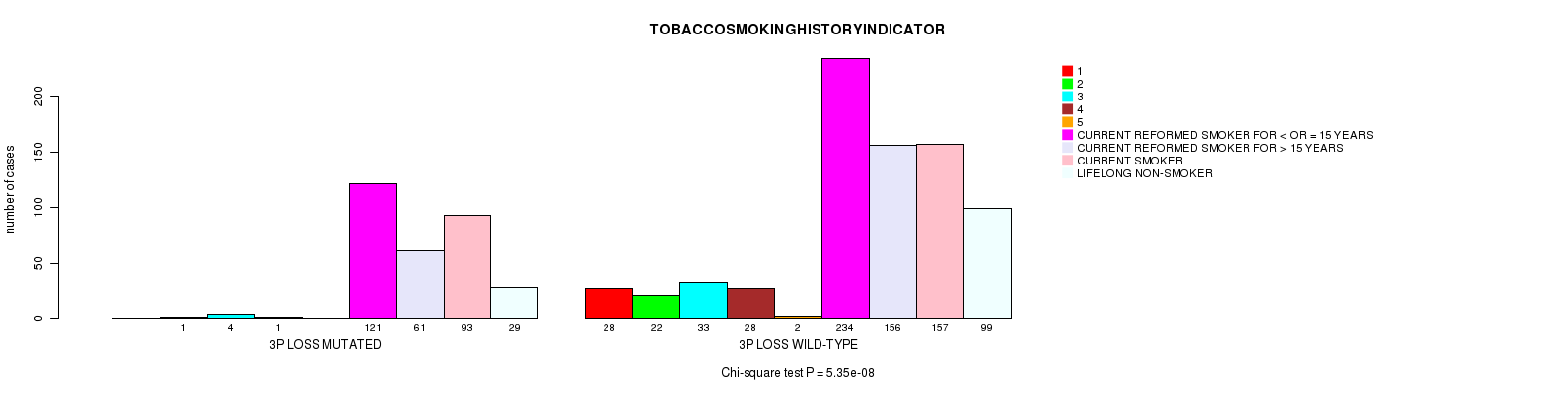
Table S220. Get Full Table Description of clustering approach #46: '3q loss mutation analysis'
| Cluster Labels | 3Q LOSS MUTATED | 3Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 291 | 4272 |
P value = 6.51e-39 (Chi-square test), Q value = 9.9e-36
Table S221. Clustering Approach #46: '3q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 3Q LOSS MUTATED | 1 | 20 | 41 | 13 | 12 | 21 | 97 | 63 | 1 | 18 | 0 | 4 |
| 3Q LOSS WILD-TYPE | 124 | 524 | 805 | 399 | 435 | 298 | 396 | 599 | 1 | 530 | 2 | 155 |
Figure S175. Get High-res Image Clustering Approach #46: '3q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
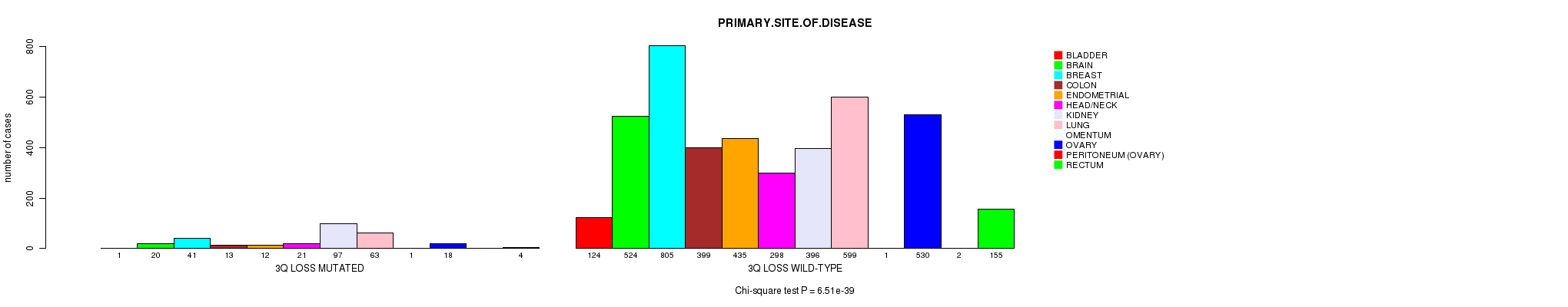
P value = 2.54e-09 (Fisher's exact test), Q value = 3.6e-06
Table S222. Clustering Approach #46: '3q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 3Q LOSS MUTATED | 135 | 156 |
| 3Q LOSS WILD-TYPE | 2746 | 1526 |
Figure S176. Get High-res Image Clustering Approach #46: '3q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
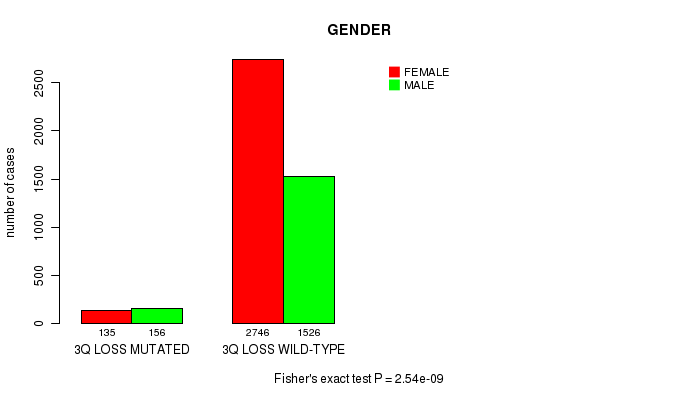
P value = 3.54e-28 (Chi-square test), Q value = 5.3e-25
Table S223. Clustering Approach #46: '3q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 3Q LOSS MUTATED | 13 | 0 | 6 | 21 | 97 | 3 | 10 | 26 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 21 | 0 | 0 | 1 | 3 | 19 | 6 | 0 | 0 |
| 3Q LOSS WILD-TYPE | 343 | 54 | 344 | 298 | 396 | 7 | 65 | 179 | 7 | 3 | 14 | 2 | 3 | 2 | 12 | 0 | 1 | 1 | 2 | 294 | 18 | 7 | 142 | 10 | 533 | 73 | 1 | 20 |
Figure S177. Get High-res Image Clustering Approach #46: '3q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S224. Get Full Table Description of clustering approach #47: '4p loss mutation analysis'
| Cluster Labels | 4P LOSS MUTATED | 4P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 917 | 3646 |
P value = 1.58e-69 (Chi-square test), Q value = 2.5e-66
Table S225. Clustering Approach #47: '4p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 4P LOSS MUTATED | 22 | 29 | 174 | 85 | 47 | 77 | 35 | 169 | 2 | 235 | 0 | 39 |
| 4P LOSS WILD-TYPE | 103 | 515 | 672 | 327 | 400 | 242 | 458 | 493 | 0 | 313 | 2 | 120 |
Figure S178. Get High-res Image Clustering Approach #47: '4p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
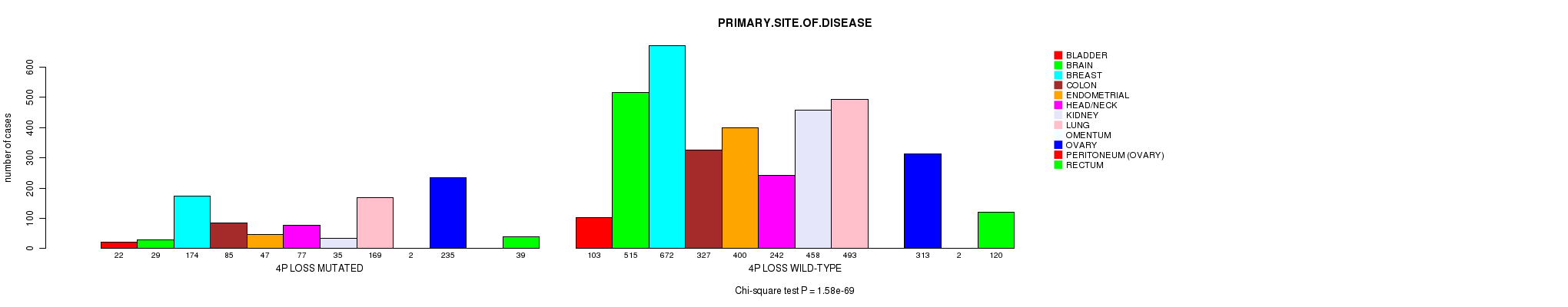
P value = 5.22e-66 (Chi-square test), Q value = 8.1e-63
Table S226. Clustering Approach #47: '4p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 4P LOSS MUTATED | 81 | 4 | 18 | 77 | 35 | 2 | 7 | 19 | 2 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 133 | 3 | 1 | 37 | 1 | 237 | 26 | 0 | 2 |
| 4P LOSS WILD-TYPE | 275 | 50 | 332 | 242 | 458 | 8 | 68 | 186 | 5 | 3 | 14 | 2 | 3 | 1 | 12 | 0 | 1 | 0 | 2 | 182 | 15 | 6 | 106 | 12 | 315 | 53 | 1 | 18 |
Figure S179. Get High-res Image Clustering Approach #47: '4p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.94e-05 (Chi-square test), Q value = 0.025
Table S227. Clustering Approach #47: '4p loss mutation analysis' versus Clinical Feature #8: 'PATHOLOGY.N'
| nPatients | N0 | N1 | N2 | N3 |
|---|---|---|---|---|
| ALL | 1067 | 353 | 282 | 11 |
| 4P LOSS MUTATED | 199 | 84 | 84 | 6 |
| 4P LOSS WILD-TYPE | 868 | 269 | 198 | 5 |
Figure S180. Get High-res Image Clustering Approach #47: '4p loss mutation analysis' versus Clinical Feature #8: 'PATHOLOGY.N'

P value = 9.69e-14 (Chi-square test), Q value = 1.4e-10
Table S228. Clustering Approach #47: '4p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 4P LOSS MUTATED | 125 | 92 | 290 | 117 |
| 4P LOSS WILD-TYPE | 587 | 406 | 591 | 306 |
Figure S181. Get High-res Image Clustering Approach #47: '4p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'

P value = 3.67e-10 (Fisher's exact test), Q value = 5.2e-07
Table S229. Clustering Approach #47: '4p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 4P LOSS MUTATED | 103 | 814 |
| 4P LOSS WILD-TYPE | 723 | 2923 |
Figure S182. Get High-res Image Clustering Approach #47: '4p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'

Table S230. Get Full Table Description of clustering approach #48: '4q loss mutation analysis'
| Cluster Labels | 4Q LOSS MUTATED | 4Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 859 | 3704 |
P value = 2.93e-108 (Chi-square test), Q value = 4.6e-105
Table S231. Clustering Approach #48: '4q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 4Q LOSS MUTATED | 18 | 28 | 139 | 70 | 45 | 50 | 29 | 154 | 2 | 277 | 1 | 42 |
| 4Q LOSS WILD-TYPE | 107 | 516 | 707 | 342 | 402 | 269 | 464 | 508 | 0 | 271 | 1 | 117 |
Figure S183. Get High-res Image Clustering Approach #48: '4q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
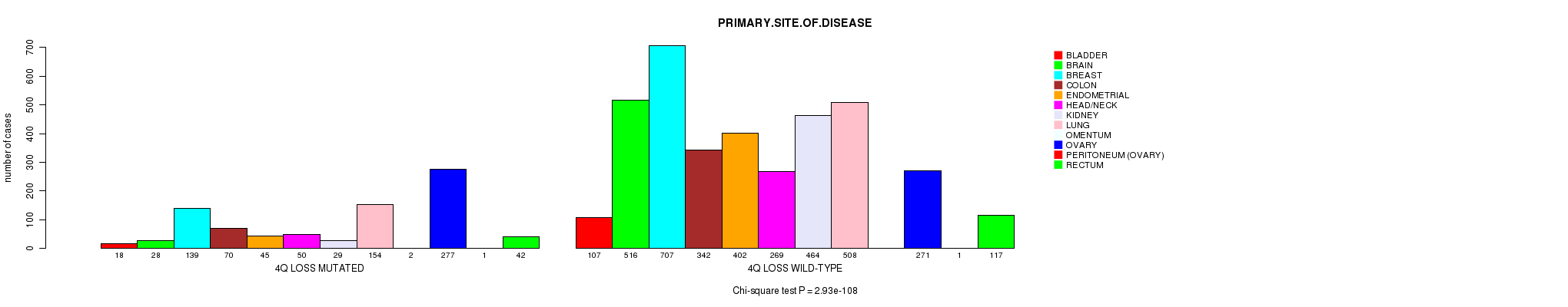
P value = 1.63e-06 (Fisher's exact test), Q value = 0.0022
Table S232. Clustering Approach #48: '4q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 4Q LOSS MUTATED | 603 | 256 |
| 4Q LOSS WILD-TYPE | 2278 | 1426 |
Figure S184. Get High-res Image Clustering Approach #48: '4q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
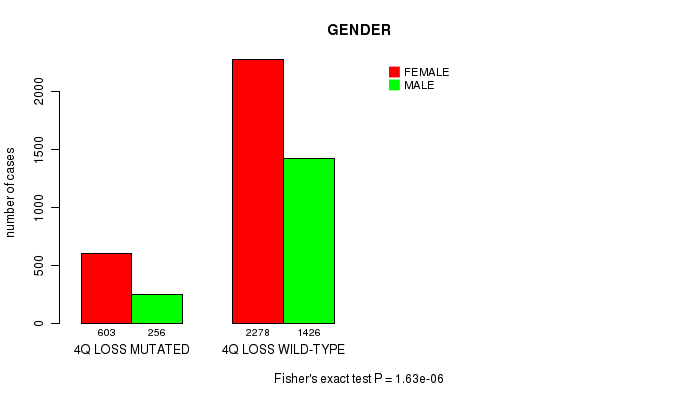
P value = 6.11e-88 (Chi-square test), Q value = 9.6e-85
Table S233. Clustering Approach #48: '4q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 4Q LOSS MUTATED | 67 | 3 | 19 | 50 | 29 | 2 | 8 | 22 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 114 | 4 | 1 | 40 | 1 | 280 | 22 | 0 | 1 |
| 4Q LOSS WILD-TYPE | 289 | 51 | 331 | 269 | 464 | 8 | 67 | 183 | 6 | 3 | 13 | 2 | 3 | 2 | 12 | 0 | 1 | 0 | 1 | 201 | 14 | 6 | 103 | 12 | 272 | 57 | 1 | 19 |
Figure S185. Get High-res Image Clustering Approach #48: '4q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 5.52e-05 (Chi-square test), Q value = 0.071
Table S234. Clustering Approach #48: '4q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 4Q LOSS MUTATED | 47 | 126 | 123 | 44 |
| 4Q LOSS WILD-TYPE | 419 | 506 | 571 | 165 |
Figure S186. Get High-res Image Clustering Approach #48: '4q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
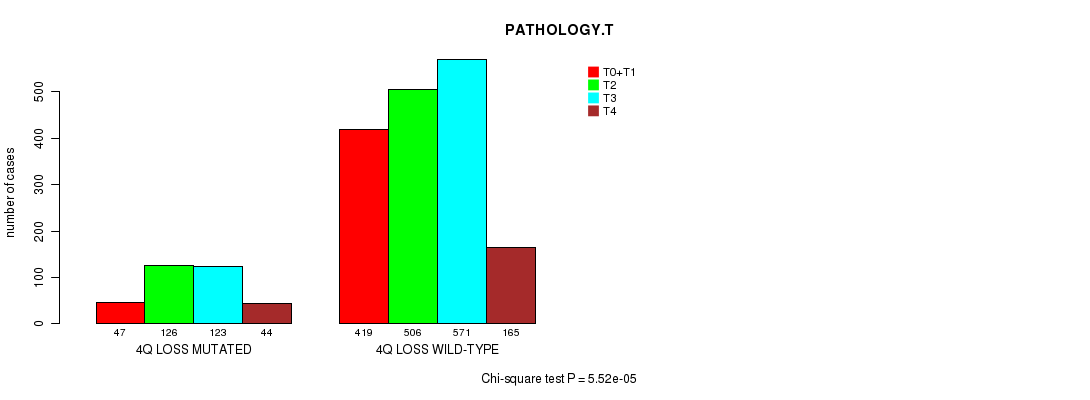
P value = 6.53e-24 (Chi-square test), Q value = 9.7e-21
Table S235. Clustering Approach #48: '4q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 4Q LOSS MUTATED | 120 | 79 | 320 | 93 |
| 4Q LOSS WILD-TYPE | 592 | 419 | 561 | 330 |
Figure S187. Get High-res Image Clustering Approach #48: '4q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'

P value = 3.28e-13 (Fisher's exact test), Q value = 4.7e-10
Table S236. Clustering Approach #48: '4q loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 4Q LOSS MUTATED | 85 | 774 |
| 4Q LOSS WILD-TYPE | 741 | 2963 |
Figure S188. Get High-res Image Clustering Approach #48: '4q loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
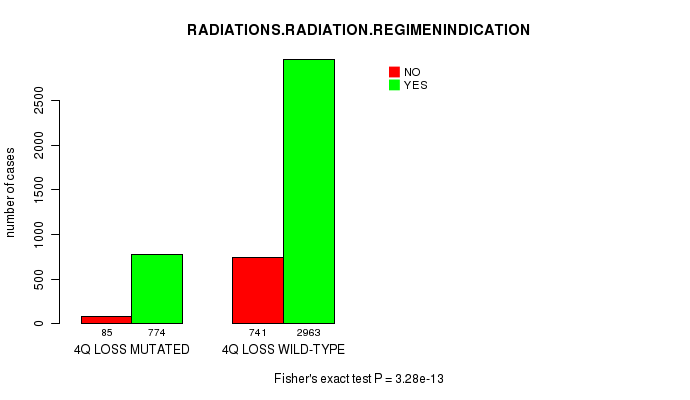
Table S237. Get Full Table Description of clustering approach #49: '5p loss mutation analysis'
| Cluster Labels | 5P LOSS MUTATED | 5P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 271 | 4292 |
P value = 6.07e-16 (Chi-square test), Q value = 8.8e-13
Table S238. Clustering Approach #49: '5p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 5P LOSS MUTATED | 12 | 24 | 59 | 27 | 21 | 9 | 2 | 34 | 1 | 71 | 0 | 11 |
| 5P LOSS WILD-TYPE | 113 | 520 | 787 | 385 | 426 | 310 | 491 | 628 | 1 | 477 | 2 | 148 |
Figure S189. Get High-res Image Clustering Approach #49: '5p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
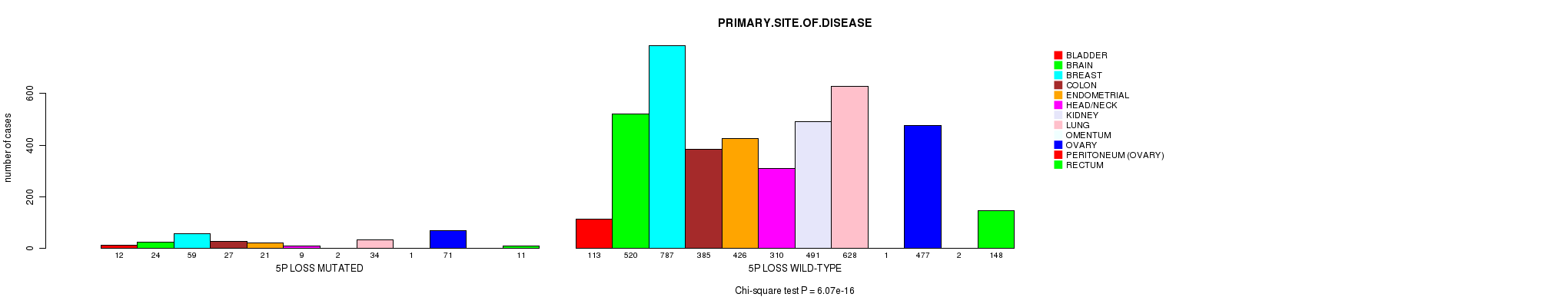
P value = 9.78e-19 (Chi-square test), Q value = 1.4e-15
Table S239. Clustering Approach #49: '5p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 5P LOSS MUTATED | 26 | 1 | 8 | 9 | 2 | 2 | 4 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 21 | 0 | 0 | 11 | 0 | 72 | 13 | 0 | 0 |
| 5P LOSS WILD-TYPE | 330 | 53 | 342 | 310 | 491 | 8 | 71 | 202 | 7 | 3 | 14 | 2 | 3 | 2 | 11 | 0 | 1 | 1 | 2 | 294 | 18 | 7 | 132 | 13 | 480 | 66 | 1 | 20 |
Figure S190. Get High-res Image Clustering Approach #49: '5p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.69e-06 (Chi-square test), Q value = 0.0023
Table S240. Clustering Approach #49: '5p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 5P LOSS MUTATED | 22 | 25 | 82 | 21 |
| 5P LOSS WILD-TYPE | 690 | 473 | 799 | 402 |
Figure S191. Get High-res Image Clustering Approach #49: '5p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
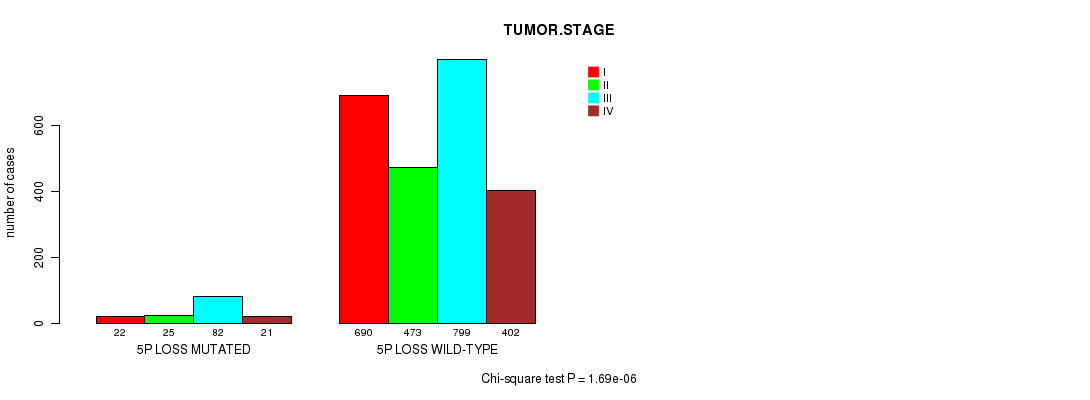
Table S241. Get Full Table Description of clustering approach #50: '5q loss mutation analysis'
| Cluster Labels | 5Q LOSS MUTATED | 5Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 638 | 3925 |
P value = 5.52e-60 (Chi-square test), Q value = 8.6e-57
Table S242. Clustering Approach #50: '5q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 5Q LOSS MUTATED | 30 | 24 | 110 | 39 | 35 | 59 | 2 | 158 | 1 | 157 | 1 | 22 |
| 5Q LOSS WILD-TYPE | 95 | 520 | 736 | 373 | 412 | 260 | 491 | 504 | 1 | 391 | 1 | 137 |
Figure S192. Get High-res Image Clustering Approach #50: '5q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
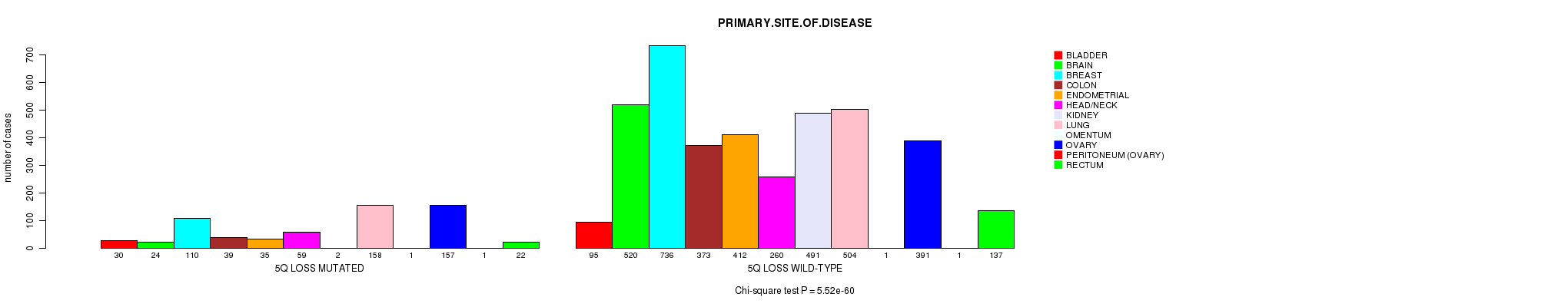
P value = 2.65e-59 (Chi-square test), Q value = 4.1e-56
Table S243. Clustering Approach #50: '5q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 5Q LOSS MUTATED | 38 | 1 | 12 | 59 | 2 | 2 | 13 | 19 | 1 | 0 | 3 | 0 | 2 | 1 | 3 | 1 | 0 | 1 | 0 | 112 | 1 | 0 | 20 | 1 | 159 | 22 | 0 | 0 |
| 5Q LOSS WILD-TYPE | 318 | 53 | 338 | 260 | 491 | 8 | 62 | 186 | 6 | 3 | 12 | 2 | 1 | 1 | 10 | 0 | 1 | 0 | 2 | 203 | 17 | 7 | 123 | 12 | 393 | 57 | 1 | 20 |
Figure S193. Get High-res Image Clustering Approach #50: '5q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 5.38e-05 (Chi-square test), Q value = 0.07
Table S244. Clustering Approach #50: '5q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 5Q LOSS MUTATED | 46 | 110 | 77 | 40 |
| 5Q LOSS WILD-TYPE | 420 | 522 | 617 | 169 |
Figure S194. Get High-res Image Clustering Approach #50: '5q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'

P value = 1.53e-05 (Chi-square test), Q value = 0.02
Table S245. Clustering Approach #50: '5q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 5Q LOSS MUTATED | 97 | 70 | 193 | 64 |
| 5Q LOSS WILD-TYPE | 615 | 428 | 688 | 359 |
Figure S195. Get High-res Image Clustering Approach #50: '5q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
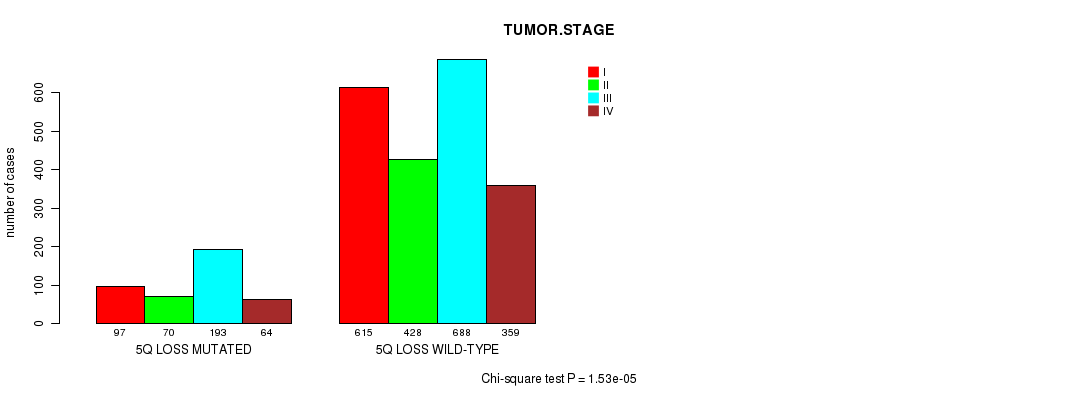
Table S246. Get Full Table Description of clustering approach #51: '6p loss mutation analysis'
| Cluster Labels | 6P LOSS MUTATED | 6P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 450 | 4113 |
P value = 3.98e-28 (Chi-square test), Q value = 5.9e-25
Table S247. Clustering Approach #51: '6p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 6P LOSS MUTATED | 12 | 60 | 90 | 13 | 8 | 21 | 61 | 59 | 1 | 118 | 0 | 6 |
| 6P LOSS WILD-TYPE | 113 | 484 | 756 | 399 | 439 | 298 | 432 | 603 | 1 | 430 | 2 | 153 |
Figure S196. Get High-res Image Clustering Approach #51: '6p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
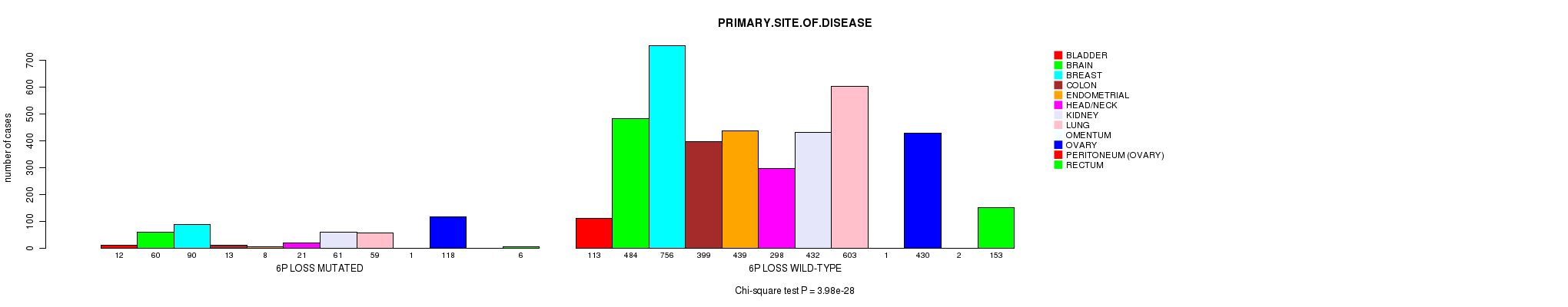
P value = 4.31e-26 (Chi-square test), Q value = 6.4e-23
Table S248. Clustering Approach #51: '6p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 6P LOSS MUTATED | 12 | 1 | 3 | 21 | 61 | 2 | 9 | 10 | 2 | 0 | 1 | 0 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 30 | 0 | 1 | 4 | 2 | 119 | 5 | 0 | 4 |
| 6P LOSS WILD-TYPE | 344 | 53 | 347 | 298 | 432 | 8 | 66 | 195 | 5 | 3 | 14 | 2 | 3 | 2 | 10 | 0 | 1 | 1 | 2 | 285 | 18 | 6 | 139 | 11 | 433 | 74 | 1 | 16 |
Figure S197. Get High-res Image Clustering Approach #51: '6p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S249. Get Full Table Description of clustering approach #52: '6q loss mutation analysis'
| Cluster Labels | 6Q LOSS MUTATED | 6Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 711 | 3852 |
P value = 6.15e-07 (t-test), Q value = 0.00083
Table S250. Clustering Approach #52: '6q loss mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 4507 | 61.9 (12.6) |
| 6Q LOSS MUTATED | 698 | 59.7 (12.4) |
| 6Q LOSS WILD-TYPE | 3809 | 62.2 (12.6) |
Figure S198. Get High-res Image Clustering Approach #52: '6q loss mutation analysis' versus Clinical Feature #2: 'AGE'
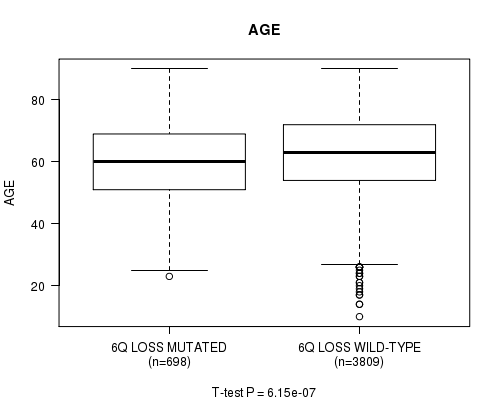
P value = 1.12e-54 (Chi-square test), Q value = 1.7e-51
Table S251. Clustering Approach #52: '6q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 6Q LOSS MUTATED | 24 | 90 | 151 | 21 | 9 | 22 | 84 | 108 | 2 | 187 | 0 | 11 |
| 6Q LOSS WILD-TYPE | 101 | 454 | 695 | 391 | 438 | 297 | 409 | 554 | 0 | 361 | 2 | 148 |
Figure S199. Get High-res Image Clustering Approach #52: '6q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
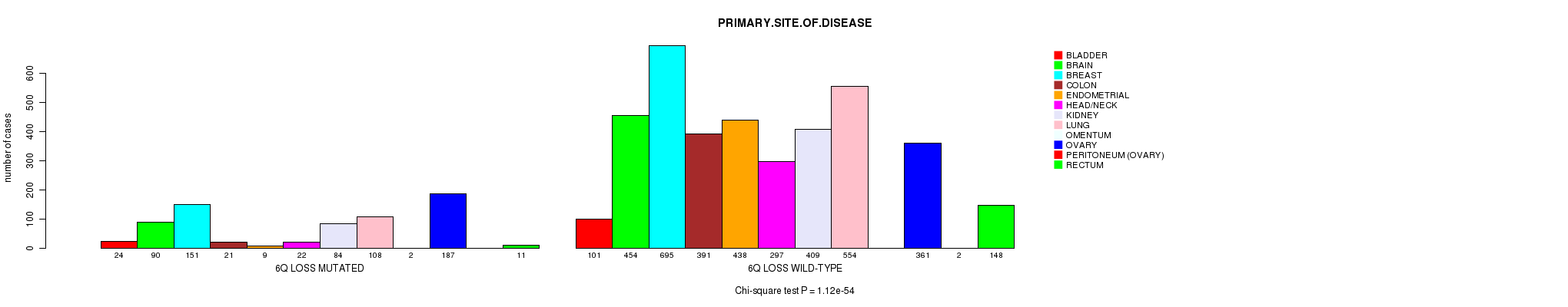
P value = 7.11e-54 (Chi-square test), Q value = 1.1e-50
Table S252. Clustering Approach #52: '6q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 6Q LOSS MUTATED | 18 | 3 | 4 | 22 | 84 | 4 | 19 | 39 | 2 | 2 | 3 | 0 | 0 | 0 | 5 | 1 | 0 | 0 | 0 | 32 | 0 | 1 | 8 | 3 | 189 | 5 | 0 | 6 |
| 6Q LOSS WILD-TYPE | 338 | 51 | 346 | 297 | 409 | 6 | 56 | 166 | 5 | 1 | 12 | 2 | 3 | 2 | 8 | 0 | 1 | 1 | 2 | 283 | 18 | 6 | 135 | 10 | 363 | 74 | 1 | 14 |
Figure S200. Get High-res Image Clustering Approach #52: '6q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 2.29e-06 (Chi-square test), Q value = 0.0031
Table S253. Clustering Approach #52: '6q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 6Q LOSS MUTATED | 104 | 60 | 195 | 65 |
| 6Q LOSS WILD-TYPE | 608 | 438 | 686 | 358 |
Figure S201. Get High-res Image Clustering Approach #52: '6q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'

Table S254. Get Full Table Description of clustering approach #53: '7p loss mutation analysis'
| Cluster Labels | 7P LOSS MUTATED | 7P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 200 | 4363 |
P value = 3.37e-48 (Chi-square test), Q value = 5.2e-45
Table S255. Clustering Approach #53: '7p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 7P LOSS MUTATED | 0 | 4 | 44 | 0 | 28 | 5 | 1 | 29 | 0 | 86 | 1 | 2 |
| 7P LOSS WILD-TYPE | 125 | 540 | 802 | 412 | 419 | 314 | 492 | 633 | 2 | 462 | 1 | 157 |
Figure S202. Get High-res Image Clustering Approach #53: '7p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
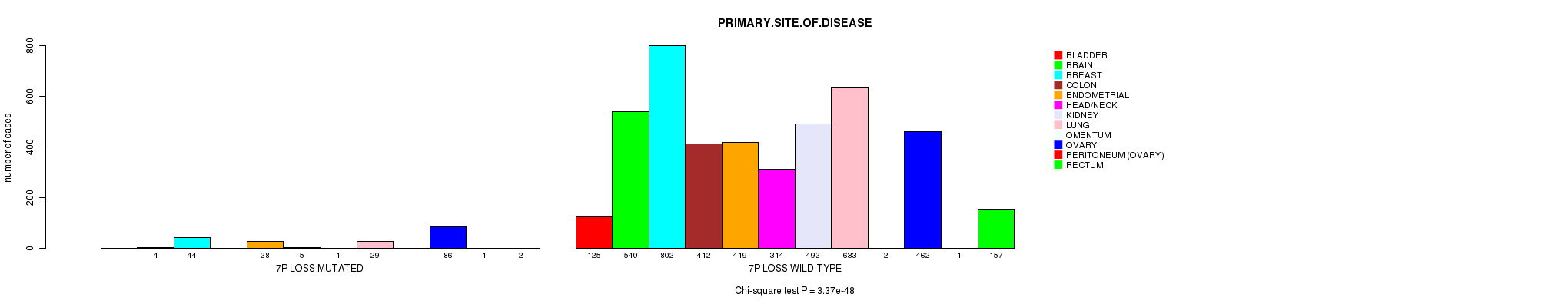
P value = 1e-13 (Fisher's exact test), Q value = 1.4e-10
Table S256. Clustering Approach #53: '7p loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 7P LOSS MUTATED | 173 | 27 |
| 7P LOSS WILD-TYPE | 2708 | 1655 |
Figure S203. Get High-res Image Clustering Approach #53: '7p loss mutation analysis' versus Clinical Feature #4: 'GENDER'

P value = 1.84e-37 (Chi-square test), Q value = 2.8e-34
Table S257. Clustering Approach #53: '7p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 7P LOSS MUTATED | 0 | 0 | 11 | 5 | 1 | 0 | 1 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 19 | 1 | 0 | 1 | 1 | 87 | 16 | 0 | 0 |
| 7P LOSS WILD-TYPE | 356 | 54 | 339 | 314 | 492 | 10 | 74 | 198 | 7 | 3 | 15 | 2 | 3 | 2 | 11 | 1 | 1 | 1 | 2 | 296 | 17 | 7 | 142 | 12 | 465 | 63 | 1 | 20 |
Figure S204. Get High-res Image Clustering Approach #53: '7p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 9e-05 (Chi-square test), Q value = 0.12
Table S258. Clustering Approach #53: '7p loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 7P LOSS MUTATED | 7 | 23 | 2 | 4 |
| 7P LOSS WILD-TYPE | 459 | 609 | 692 | 205 |
Figure S205. Get High-res Image Clustering Approach #53: '7p loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
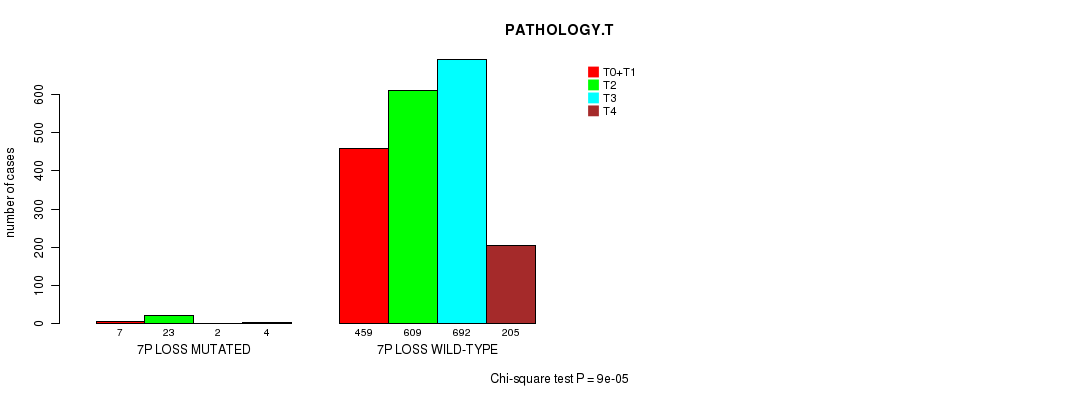
P value = 1.66e-14 (Chi-square test), Q value = 2.4e-11
Table S259. Clustering Approach #53: '7p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 7P LOSS MUTATED | 20 | 7 | 85 | 11 |
| 7P LOSS WILD-TYPE | 692 | 491 | 796 | 412 |
Figure S206. Get High-res Image Clustering Approach #53: '7p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'

Table S260. Get Full Table Description of clustering approach #54: '7q loss mutation analysis'
| Cluster Labels | 7Q LOSS MUTATED | 7Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 162 | 4401 |
P value = 4.47e-20 (Chi-square test), Q value = 6.5e-17
Table S261. Clustering Approach #54: '7q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 7Q LOSS MUTATED | 2 | 3 | 61 | 1 | 23 | 8 | 1 | 21 | 0 | 39 | 1 | 2 |
| 7Q LOSS WILD-TYPE | 123 | 541 | 785 | 411 | 424 | 311 | 492 | 641 | 2 | 509 | 1 | 157 |
Figure S207. Get High-res Image Clustering Approach #54: '7q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
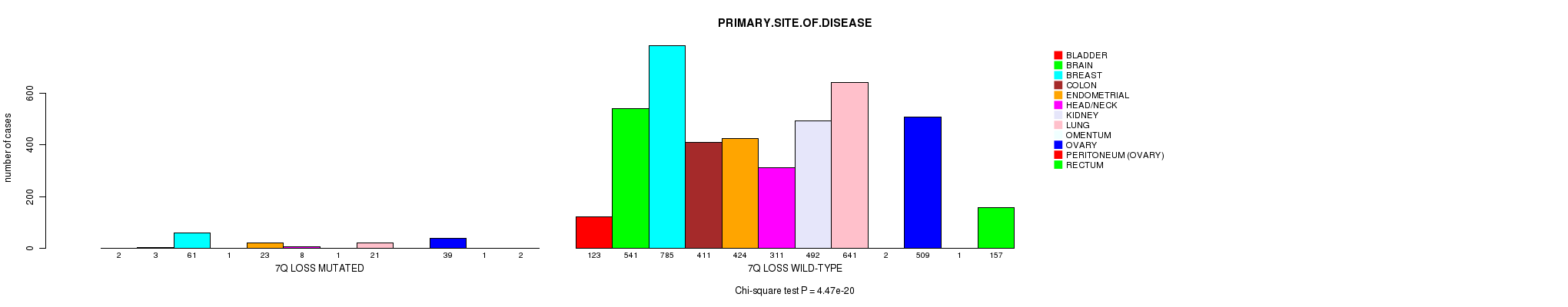
P value = 3.65e-11 (Fisher's exact test), Q value = 5.2e-08
Table S262. Clustering Approach #54: '7q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 7Q LOSS MUTATED | 140 | 22 |
| 7Q LOSS WILD-TYPE | 2741 | 1660 |
Figure S208. Get High-res Image Clustering Approach #54: '7q loss mutation analysis' versus Clinical Feature #4: 'GENDER'

P value = 1.83e-23 (Chi-square test), Q value = 2.7e-20
Table S263. Clustering Approach #54: '7q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 7Q LOSS MUTATED | 1 | 0 | 6 | 8 | 1 | 0 | 0 | 8 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 9 | 1 | 2 | 1 | 1 | 40 | 16 | 0 | 0 |
| 7Q LOSS WILD-TYPE | 355 | 54 | 344 | 311 | 492 | 10 | 75 | 197 | 7 | 3 | 15 | 1 | 3 | 2 | 12 | 1 | 1 | 1 | 2 | 306 | 17 | 5 | 142 | 12 | 512 | 63 | 1 | 20 |
Figure S209. Get High-res Image Clustering Approach #54: '7q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 3.89e-05 (Chi-square test), Q value = 0.051
Table S264. Clustering Approach #54: '7q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 7Q LOSS MUTATED | 4 | 22 | 3 | 2 |
| 7Q LOSS WILD-TYPE | 462 | 610 | 691 | 207 |
Figure S210. Get High-res Image Clustering Approach #54: '7q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
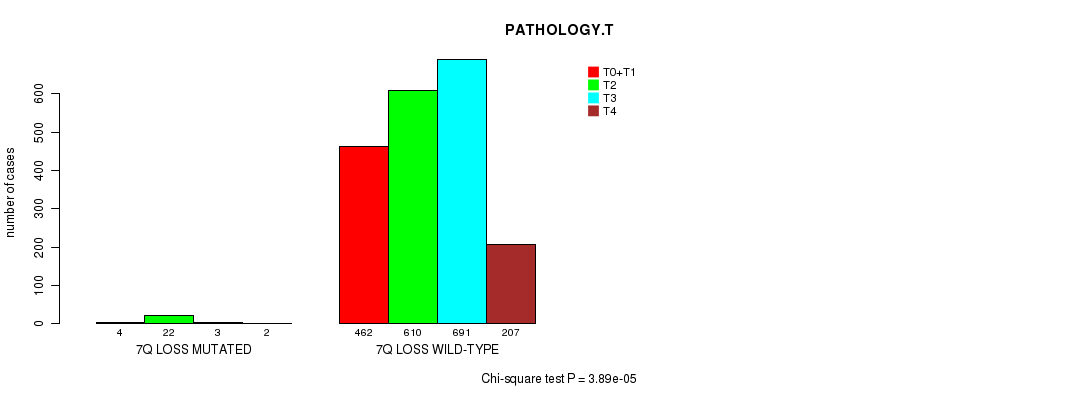
Table S265. Get Full Table Description of clustering approach #55: '8p loss mutation analysis'
| Cluster Labels | 8P LOSS MUTATED | 8P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 1074 | 3489 |
P value = 7.91e-53 (Chi-square test), Q value = 1.2e-49
Table S266. Clustering Approach #55: '8p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 8P LOSS MUTATED | 40 | 39 | 249 | 93 | 43 | 57 | 94 | 175 | 2 | 225 | 0 | 55 |
| 8P LOSS WILD-TYPE | 85 | 505 | 597 | 319 | 404 | 262 | 399 | 487 | 0 | 323 | 2 | 104 |
Figure S211. Get High-res Image Clustering Approach #55: '8p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
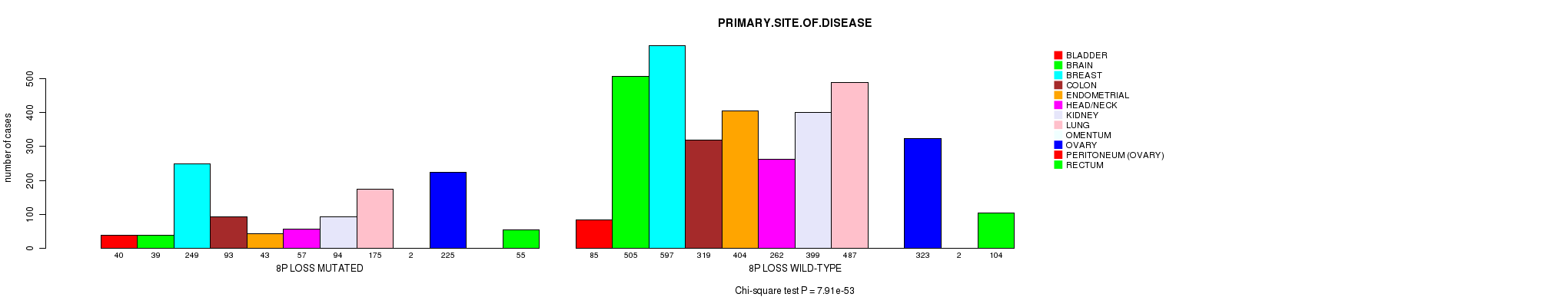
P value = 8.98e-35 (Chi-square test), Q value = 1.4e-31
Table S267. Clustering Approach #55: '8p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 8P LOSS MUTATED | 86 | 6 | 11 | 57 | 94 | 2 | 16 | 56 | 2 | 0 | 5 | 1 | 3 | 0 | 2 | 1 | 0 | 0 | 0 | 87 | 2 | 0 | 51 | 4 | 227 | 30 | 0 | 3 |
| 8P LOSS WILD-TYPE | 270 | 48 | 339 | 262 | 399 | 8 | 59 | 149 | 5 | 3 | 10 | 1 | 0 | 2 | 11 | 0 | 1 | 1 | 2 | 228 | 16 | 7 | 92 | 9 | 325 | 49 | 1 | 17 |
Figure S212. Get High-res Image Clustering Approach #55: '8p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.33e-07 (Chi-square test), Q value = 0.00018
Table S268. Clustering Approach #55: '8p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 8P LOSS MUTATED | 162 | 116 | 303 | 106 |
| 8P LOSS WILD-TYPE | 550 | 382 | 578 | 317 |
Figure S213. Get High-res Image Clustering Approach #55: '8p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
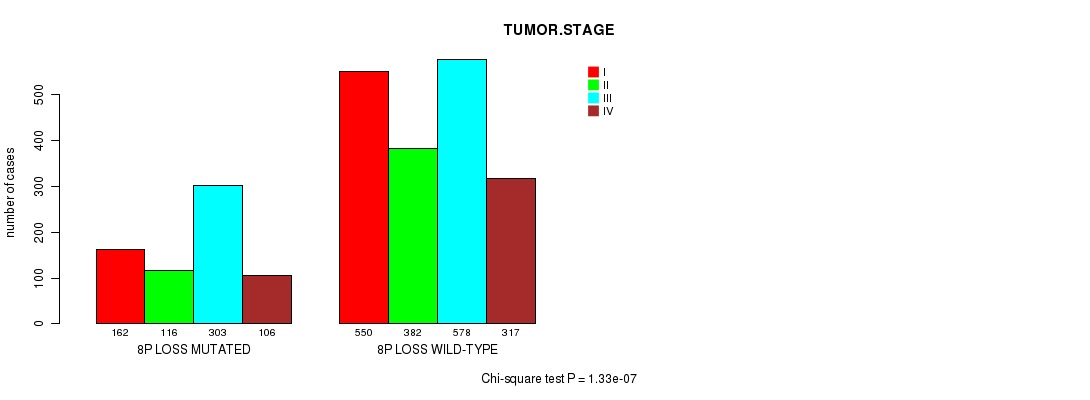
P value = 6.06e-10 (Fisher's exact test), Q value = 8.5e-07
Table S269. Clustering Approach #55: '8p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 8P LOSS MUTATED | 128 | 946 |
| 8P LOSS WILD-TYPE | 698 | 2791 |
Figure S214. Get High-res Image Clustering Approach #55: '8p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
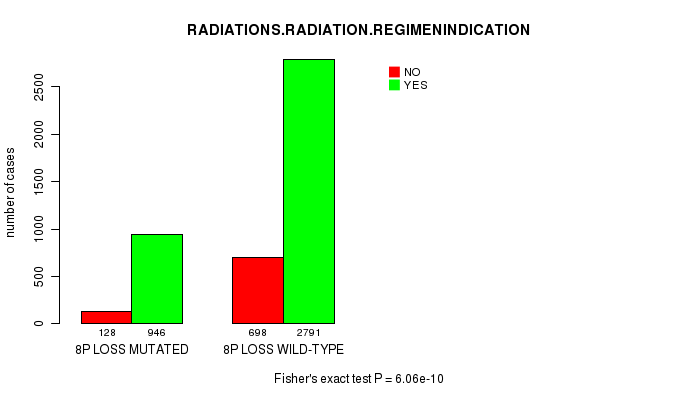
Table S270. Get Full Table Description of clustering approach #56: '8q loss mutation analysis'
| Cluster Labels | 8Q LOSS MUTATED | 8Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 219 | 4344 |
P value = 1.78e-12 (Chi-square test), Q value = 2.5e-09
Table S271. Clustering Approach #56: '8q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 8Q LOSS MUTATED | 5 | 28 | 39 | 9 | 8 | 6 | 40 | 19 | 0 | 58 | 0 | 6 |
| 8Q LOSS WILD-TYPE | 120 | 516 | 807 | 403 | 439 | 313 | 453 | 643 | 2 | 490 | 2 | 153 |
Figure S215. Get High-res Image Clustering Approach #56: '8q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'

P value = 8.37e-10 (Chi-square test), Q value = 1.2e-06
Table S272. Clustering Approach #56: '8q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 8Q LOSS MUTATED | 8 | 1 | 1 | 6 | 40 | 0 | 1 | 8 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 6 | 0 | 58 | 7 | 0 | 1 |
| 8Q LOSS WILD-TYPE | 348 | 53 | 349 | 313 | 453 | 10 | 74 | 197 | 7 | 3 | 15 | 2 | 2 | 2 | 13 | 1 | 1 | 1 | 2 | 306 | 18 | 7 | 137 | 13 | 494 | 72 | 1 | 19 |
Figure S216. Get High-res Image Clustering Approach #56: '8q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 0.000142 (Chi-square test), Q value = 0.18
Table S273. Clustering Approach #56: '8q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 8Q LOSS MUTATED | 32 | 19 | 71 | 13 |
| 8Q LOSS WILD-TYPE | 680 | 479 | 810 | 410 |
Figure S217. Get High-res Image Clustering Approach #56: '8q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'

Table S274. Get Full Table Description of clustering approach #57: '9p loss mutation analysis'
| Cluster Labels | 9P LOSS MUTATED | 9P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 1105 | 3458 |
P value = 2.53e-13 (logrank test), Q value = 3.6e-10
Table S275. Clustering Approach #57: '9p loss mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 4296 | 1401 | 0.0 - 224.0 (14.9) |
| 9P LOSS MUTATED | 1051 | 438 | 0.0 - 162.0 (14.3) |
| 9P LOSS WILD-TYPE | 3245 | 963 | 0.0 - 224.0 (14.9) |
Figure S218. Get High-res Image Clustering Approach #57: '9p loss mutation analysis' versus Clinical Feature #1: 'Time to Death'
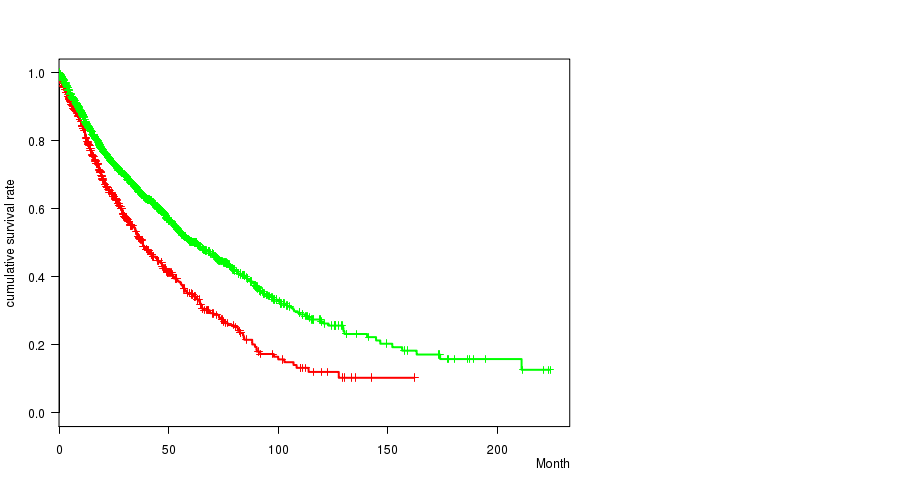
P value = 3.23e-49 (Chi-square test), Q value = 5e-46
Table S276. Clustering Approach #57: '9p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 9P LOSS MUTATED | 38 | 180 | 178 | 27 | 61 | 86 | 88 | 234 | 0 | 199 | 2 | 12 |
| 9P LOSS WILD-TYPE | 87 | 364 | 668 | 385 | 386 | 233 | 405 | 428 | 2 | 349 | 0 | 147 |
Figure S219. Get High-res Image Clustering Approach #57: '9p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
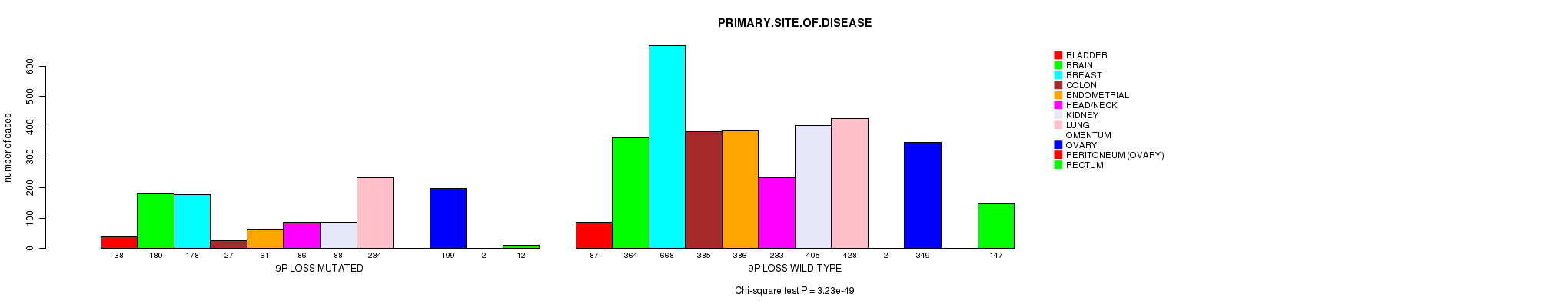
P value = 2.07e-49 (Chi-square test), Q value = 3.2e-46
Table S277. Clustering Approach #57: '9p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 9P LOSS MUTATED | 25 | 2 | 23 | 86 | 88 | 5 | 24 | 57 | 2 | 1 | 3 | 0 | 1 | 1 | 4 | 1 | 0 | 1 | 2 | 130 | 4 | 2 | 11 | 1 | 201 | 34 | 0 | 9 |
| 9P LOSS WILD-TYPE | 331 | 52 | 327 | 233 | 405 | 5 | 51 | 148 | 5 | 2 | 12 | 2 | 2 | 1 | 9 | 0 | 1 | 0 | 0 | 185 | 14 | 5 | 132 | 12 | 351 | 45 | 1 | 11 |
Figure S220. Get High-res Image Clustering Approach #57: '9p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 9.7e-09 (Chi-square test), Q value = 1.4e-05
Table S278. Clustering Approach #57: '9p loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 9P LOSS MUTATED | 101 | 186 | 105 | 42 |
| 9P LOSS WILD-TYPE | 365 | 446 | 589 | 167 |
Figure S221. Get High-res Image Clustering Approach #57: '9p loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
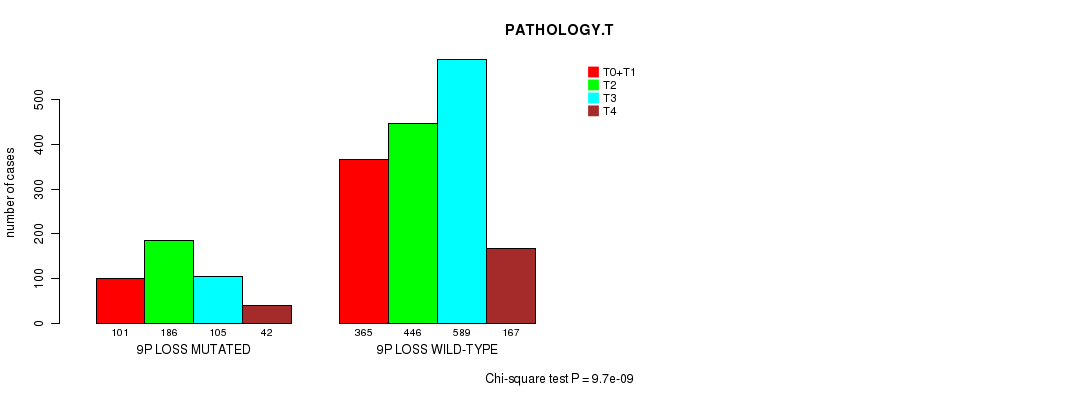
Table S279. Get Full Table Description of clustering approach #58: '9q loss mutation analysis'
| Cluster Labels | 9Q LOSS MUTATED | 9Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 900 | 3663 |
P value = 0.00016 (logrank test), Q value = 0.2
Table S280. Clustering Approach #58: '9q loss mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 4296 | 1401 | 0.0 - 224.0 (14.9) |
| 9Q LOSS MUTATED | 853 | 343 | 0.1 - 162.0 (17.9) |
| 9Q LOSS WILD-TYPE | 3443 | 1058 | 0.0 - 224.0 (14.2) |
Figure S222. Get High-res Image Clustering Approach #58: '9q loss mutation analysis' versus Clinical Feature #1: 'Time to Death'
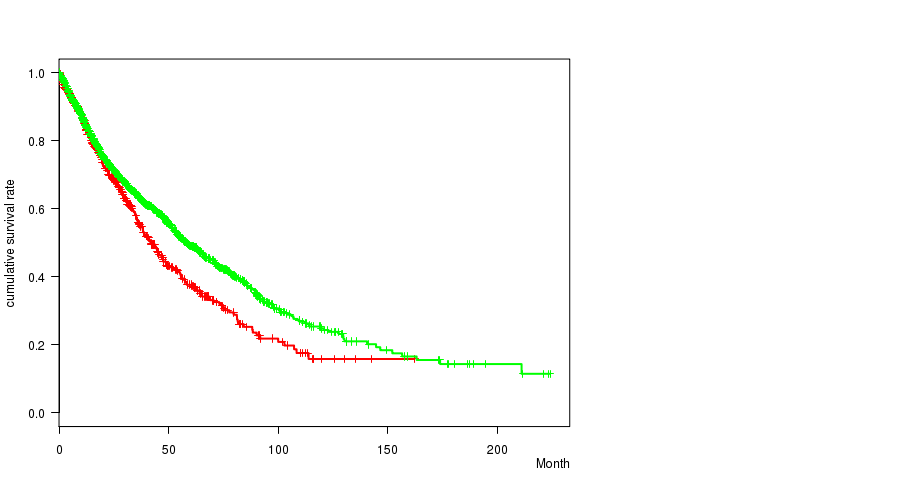
P value = 8.8e-58 (Chi-square test), Q value = 1.4e-54
Table S281. Clustering Approach #58: '9q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 9Q LOSS MUTATED | 36 | 92 | 134 | 24 | 80 | 29 | 92 | 171 | 0 | 228 | 2 | 10 |
| 9Q LOSS WILD-TYPE | 89 | 452 | 712 | 388 | 367 | 290 | 401 | 491 | 2 | 320 | 0 | 149 |
Figure S223. Get High-res Image Clustering Approach #58: '9q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'

P value = 1.1e-56 (Chi-square test), Q value = 1.7e-53
Table S282. Clustering Approach #58: '9q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 9Q LOSS MUTATED | 23 | 1 | 36 | 29 | 92 | 5 | 17 | 52 | 1 | 0 | 3 | 0 | 1 | 0 | 4 | 1 | 0 | 1 | 2 | 83 | 5 | 1 | 9 | 1 | 230 | 39 | 0 | 5 |
| 9Q LOSS WILD-TYPE | 333 | 53 | 314 | 290 | 401 | 5 | 58 | 153 | 6 | 3 | 12 | 2 | 2 | 2 | 9 | 0 | 1 | 0 | 0 | 232 | 13 | 6 | 134 | 12 | 322 | 40 | 1 | 15 |
Figure S224. Get High-res Image Clustering Approach #58: '9q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 7.3e-09 (Chi-square test), Q value = 1e-05
Table S283. Clustering Approach #58: '9q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 9Q LOSS MUTATED | 132 | 82 | 254 | 79 |
| 9Q LOSS WILD-TYPE | 580 | 416 | 627 | 344 |
Figure S225. Get High-res Image Clustering Approach #58: '9q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
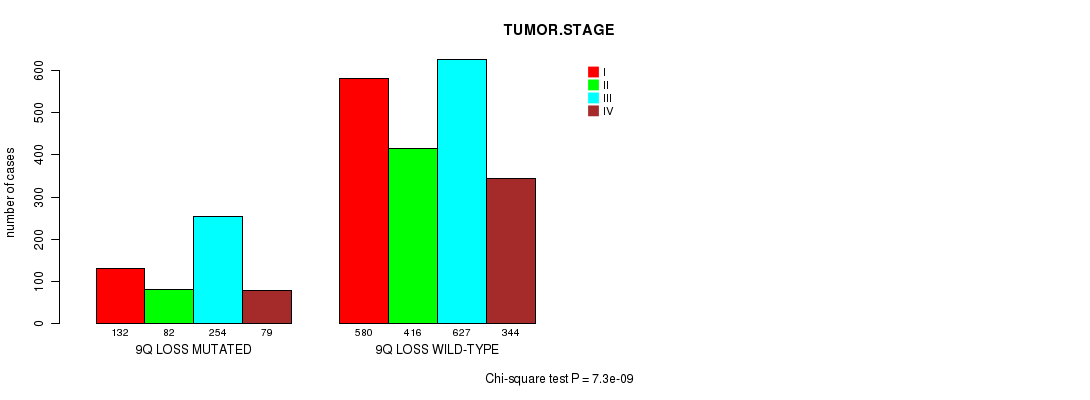
Table S284. Get Full Table Description of clustering approach #59: '10p loss mutation analysis'
| Cluster Labels | 10P LOSS MUTATED | 10P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 808 | 3755 |
P value = 0 (logrank test), Q value = 0
Table S285. Clustering Approach #59: '10p loss mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 4296 | 1401 | 0.0 - 224.0 (14.9) |
| 10P LOSS MUTATED | 787 | 439 | 0.0 - 131.2 (10.6) |
| 10P LOSS WILD-TYPE | 3509 | 962 | 0.0 - 224.0 (16.6) |
Figure S226. Get High-res Image Clustering Approach #59: '10p loss mutation analysis' versus Clinical Feature #1: 'Time to Death'
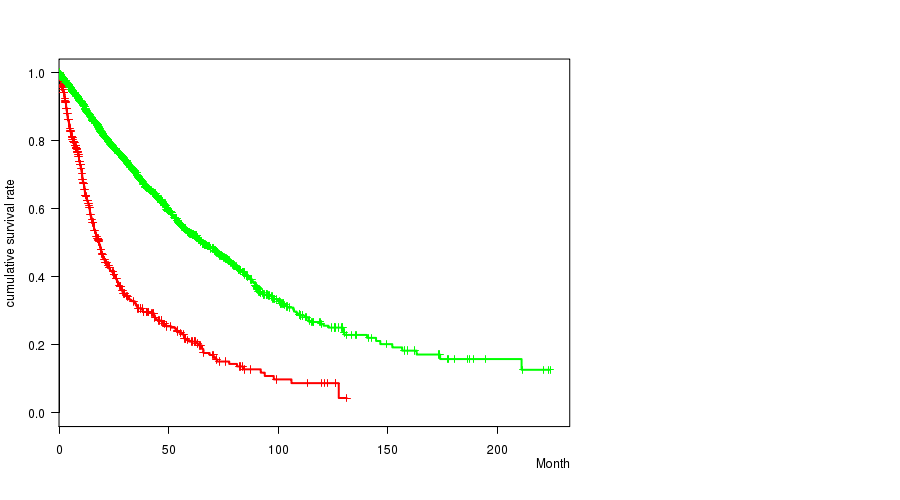
P value = 0 (Chi-square test), Q value = 0
Table S286. Clustering Approach #59: '10p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 10P LOSS MUTATED | 16 | 432 | 65 | 23 | 18 | 48 | 32 | 97 | 0 | 59 | 0 | 17 |
| 10P LOSS WILD-TYPE | 109 | 112 | 781 | 389 | 429 | 271 | 461 | 565 | 2 | 489 | 2 | 142 |
Figure S227. Get High-res Image Clustering Approach #59: '10p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
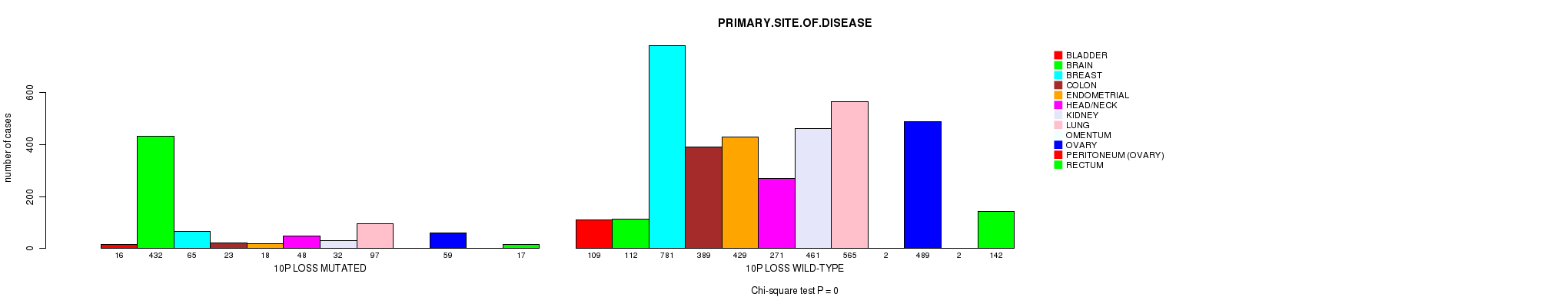
P value = 2.5e-22 (Fisher's exact test), Q value = 3.7e-19
Table S287. Clustering Approach #59: '10p loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 10P LOSS MUTATED | 387 | 421 |
| 10P LOSS WILD-TYPE | 2494 | 1261 |
Figure S228. Get High-res Image Clustering Approach #59: '10p loss mutation analysis' versus Clinical Feature #4: 'GENDER'
P value = 9.96e-37 (Chi-square test), Q value = 1.5e-33
Table S288. Clustering Approach #59: '10p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 10P LOSS MUTATED | 20 | 3 | 7 | 48 | 32 | 2 | 2 | 19 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 68 | 1 | 1 | 13 | 4 | 59 | 10 | 1 | 16 |
| 10P LOSS WILD-TYPE | 336 | 51 | 343 | 271 | 461 | 8 | 73 | 186 | 5 | 2 | 15 | 2 | 2 | 2 | 13 | 0 | 1 | 1 | 2 | 247 | 17 | 6 | 130 | 9 | 493 | 69 | 0 | 4 |
Figure S229. Get High-res Image Clustering Approach #59: '10p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 9.03e-59 (Fisher's exact test), Q value = 1.4e-55
Table S289. Clustering Approach #59: '10p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 10P LOSS MUTATED | 320 | 488 |
| 10P LOSS WILD-TYPE | 506 | 3249 |
Figure S230. Get High-res Image Clustering Approach #59: '10p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'

Table S290. Get Full Table Description of clustering approach #60: '10q loss mutation analysis'
| Cluster Labels | 10Q LOSS MUTATED | 10Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 895 | 3668 |
P value = 0 (logrank test), Q value = 0
Table S291. Clustering Approach #60: '10q loss mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 4296 | 1401 | 0.0 - 224.0 (14.9) |
| 10Q LOSS MUTATED | 865 | 463 | 0.0 - 131.2 (11.2) |
| 10Q LOSS WILD-TYPE | 3431 | 938 | 0.0 - 224.0 (16.7) |
Figure S231. Get High-res Image Clustering Approach #60: '10q loss mutation analysis' versus Clinical Feature #1: 'Time to Death'
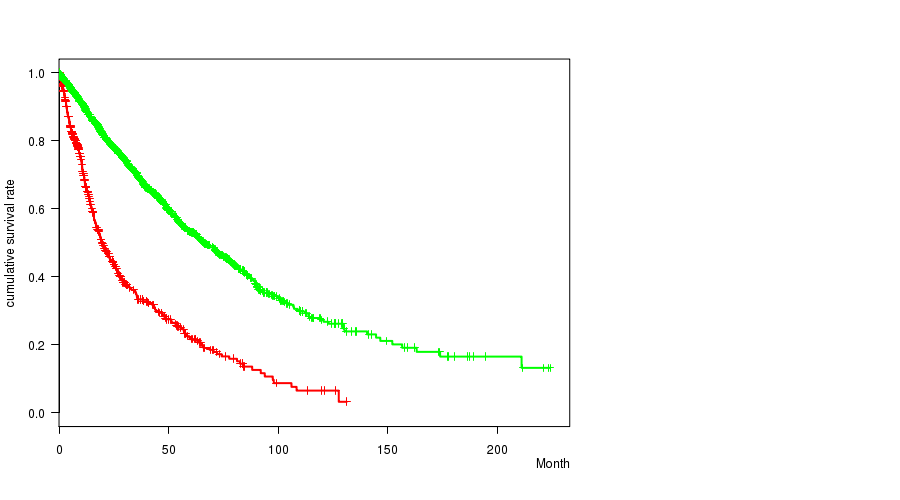
P value = 0 (Chi-square test), Q value = 0
Table S292. Clustering Approach #60: '10q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 10Q LOSS MUTATED | 24 | 448 | 102 | 24 | 17 | 29 | 48 | 99 | 0 | 81 | 0 | 22 |
| 10Q LOSS WILD-TYPE | 101 | 96 | 744 | 388 | 430 | 290 | 445 | 563 | 2 | 467 | 2 | 137 |
Figure S232. Get High-res Image Clustering Approach #60: '10q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'

P value = 2.6e-17 (Fisher's exact test), Q value = 3.8e-14
Table S293. Clustering Approach #60: '10q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 10Q LOSS MUTATED | 454 | 441 |
| 10Q LOSS WILD-TYPE | 2427 | 1241 |
Figure S233. Get High-res Image Clustering Approach #60: '10q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
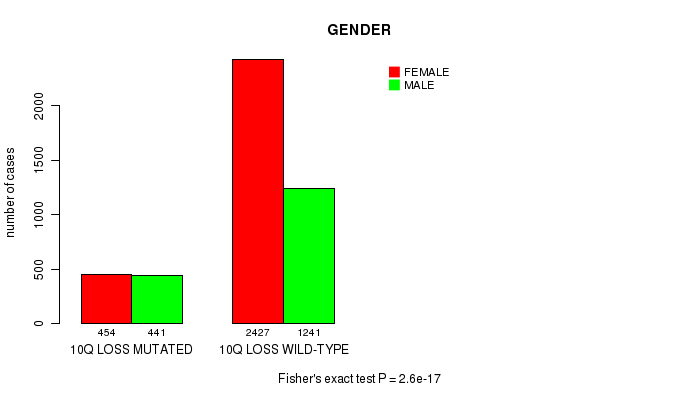
P value = 5.71e-41 (Chi-square test), Q value = 8.7e-38
Table S294. Clustering Approach #60: '10q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 10Q LOSS MUTATED | 21 | 3 | 9 | 29 | 48 | 1 | 5 | 17 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 2 | 68 | 1 | 1 | 18 | 4 | 81 | 7 | 1 | 18 |
| 10Q LOSS WILD-TYPE | 335 | 51 | 341 | 290 | 445 | 9 | 70 | 188 | 6 | 2 | 15 | 2 | 1 | 2 | 13 | 0 | 1 | 1 | 0 | 247 | 17 | 6 | 125 | 9 | 471 | 72 | 0 | 2 |
Figure S234. Get High-res Image Clustering Approach #60: '10q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 2.56e-56 (Fisher's exact test), Q value = 4e-53
Table S295. Clustering Approach #60: '10q loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 10Q LOSS MUTATED | 337 | 558 |
| 10Q LOSS WILD-TYPE | 489 | 3179 |
Figure S235. Get High-res Image Clustering Approach #60: '10q loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
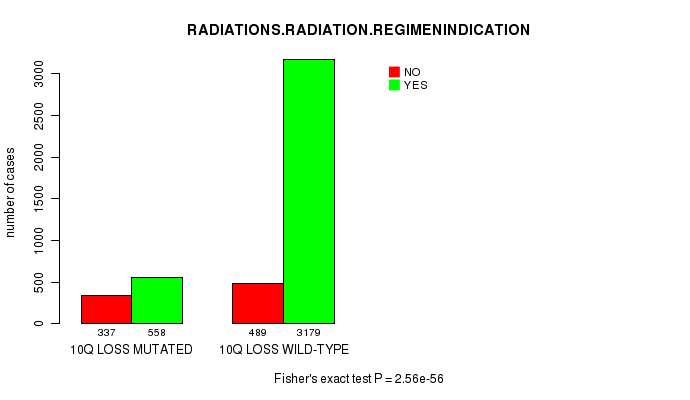
Table S296. Get Full Table Description of clustering approach #61: '11p loss mutation analysis'
| Cluster Labels | 11P LOSS MUTATED | 11P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 587 | 3976 |
P value = 7.63e-41 (Chi-square test), Q value = 1.2e-37
Table S297. Clustering Approach #61: '11p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 11P LOSS MUTATED | 39 | 66 | 133 | 21 | 45 | 43 | 5 | 73 | 0 | 143 | 0 | 19 |
| 11P LOSS WILD-TYPE | 86 | 478 | 713 | 391 | 402 | 276 | 488 | 589 | 2 | 405 | 2 | 140 |
Figure S236. Get High-res Image Clustering Approach #61: '11p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
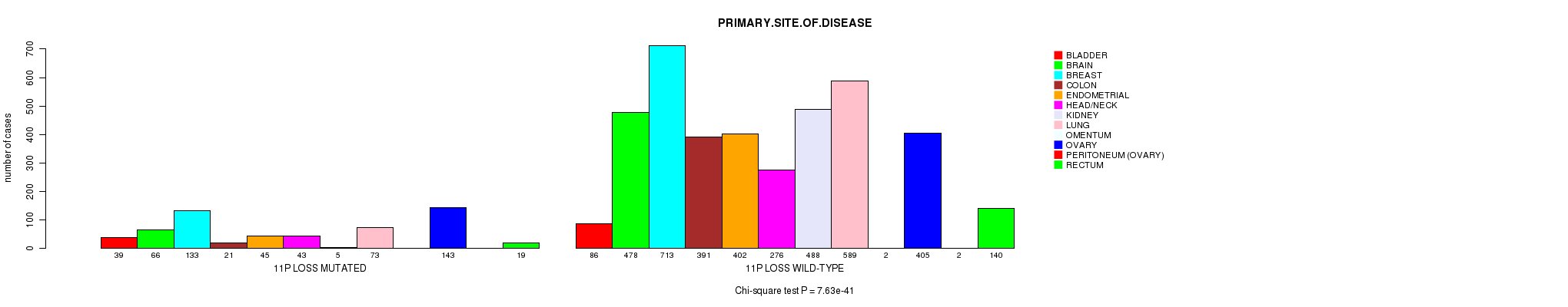
P value = 1.95e-05 (Fisher's exact test), Q value = 0.026
Table S298. Clustering Approach #61: '11p loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 11P LOSS MUTATED | 417 | 170 |
| 11P LOSS WILD-TYPE | 2464 | 1512 |
Figure S237. Get High-res Image Clustering Approach #61: '11p loss mutation analysis' versus Clinical Feature #4: 'GENDER'
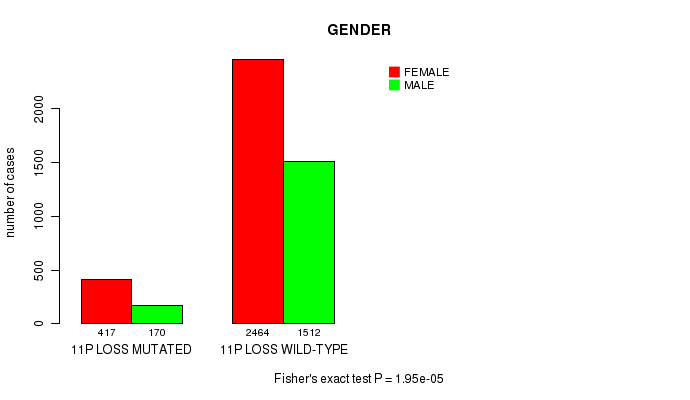
P value = 1.17e-36 (Chi-square test), Q value = 1.8e-33
Table S299. Clustering Approach #61: '11p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 11P LOSS MUTATED | 21 | 0 | 21 | 43 | 5 | 0 | 4 | 16 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 48 | 4 | 0 | 14 | 4 | 143 | 20 | 0 | 3 |
| 11P LOSS WILD-TYPE | 335 | 54 | 329 | 276 | 488 | 10 | 71 | 189 | 6 | 3 | 14 | 1 | 3 | 2 | 12 | 1 | 1 | 0 | 2 | 267 | 14 | 7 | 129 | 9 | 409 | 59 | 1 | 17 |
Figure S238. Get High-res Image Clustering Approach #61: '11p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 4.77e-05 (Chi-square test), Q value = 0.062
Table S300. Clustering Approach #61: '11p loss mutation analysis' versus Clinical Feature #8: 'PATHOLOGY.N'
| nPatients | N0 | N1 | N2 | N3 |
|---|---|---|---|---|
| ALL | 1067 | 353 | 282 | 11 |
| 11P LOSS MUTATED | 68 | 47 | 29 | 3 |
| 11P LOSS WILD-TYPE | 999 | 306 | 253 | 8 |
Figure S239. Get High-res Image Clustering Approach #61: '11p loss mutation analysis' versus Clinical Feature #8: 'PATHOLOGY.N'

P value = 2.7e-12 (Chi-square test), Q value = 3.8e-09
Table S301. Clustering Approach #61: '11p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 11P LOSS MUTATED | 48 | 40 | 158 | 47 |
| 11P LOSS WILD-TYPE | 664 | 458 | 723 | 376 |
Figure S240. Get High-res Image Clustering Approach #61: '11p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'

P value = 2.25e-06 (Chi-square test), Q value = 0.003
Table S302. Clustering Approach #61: '11p loss mutation analysis' versus Clinical Feature #14: 'TOBACCOSMOKINGHISTORYINDICATOR'
| nPatients | 1 | 2 | 3 | 4 | 5 | CURRENT REFORMED SMOKER FOR < OR = 15 YEARS | CURRENT REFORMED SMOKER FOR > 15 YEARS | CURRENT SMOKER | LIFELONG NON-SMOKER |
|---|---|---|---|---|---|---|---|---|---|
| ALL | 28 | 23 | 37 | 29 | 2 | 355 | 217 | 250 | 128 |
| 11P LOSS MUTATED | 10 | 6 | 10 | 9 | 1 | 49 | 18 | 38 | 8 |
| 11P LOSS WILD-TYPE | 18 | 17 | 27 | 20 | 1 | 306 | 199 | 212 | 120 |
Figure S241. Get High-res Image Clustering Approach #61: '11p loss mutation analysis' versus Clinical Feature #14: 'TOBACCOSMOKINGHISTORYINDICATOR'
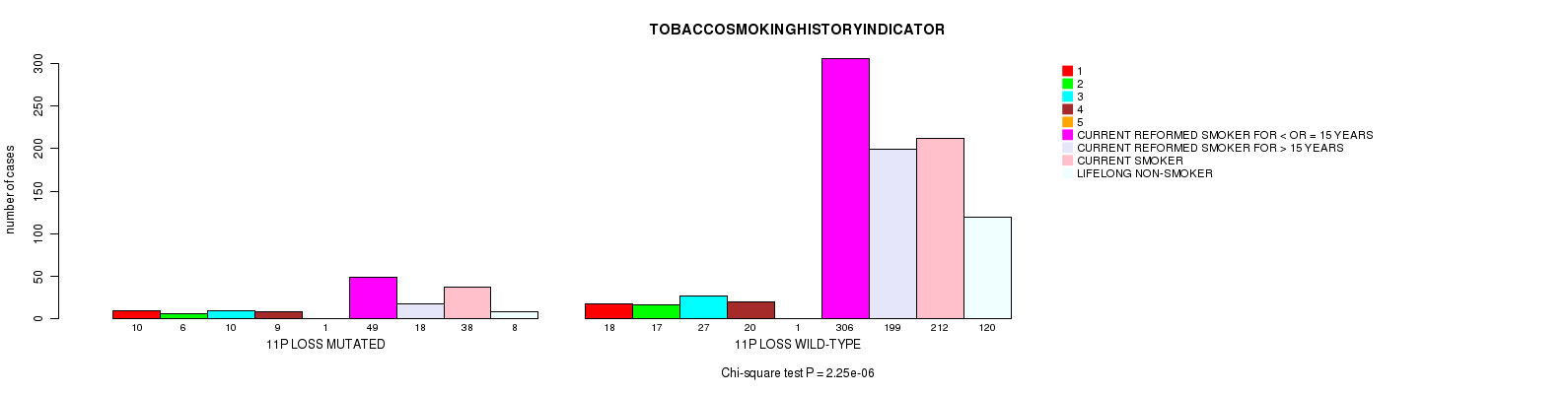
Table S303. Get Full Table Description of clustering approach #62: '11q loss mutation analysis'
| Cluster Labels | 11Q LOSS MUTATED | 11Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 609 | 3954 |
P value = 2.05e-44 (Chi-square test), Q value = 3.1e-41
Table S304. Clustering Approach #62: '11q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 11Q LOSS MUTATED | 29 | 56 | 211 | 27 | 37 | 61 | 8 | 54 | 0 | 101 | 0 | 23 |
| 11Q LOSS WILD-TYPE | 96 | 488 | 635 | 385 | 410 | 258 | 485 | 608 | 2 | 447 | 2 | 136 |
Figure S242. Get High-res Image Clustering Approach #62: '11q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
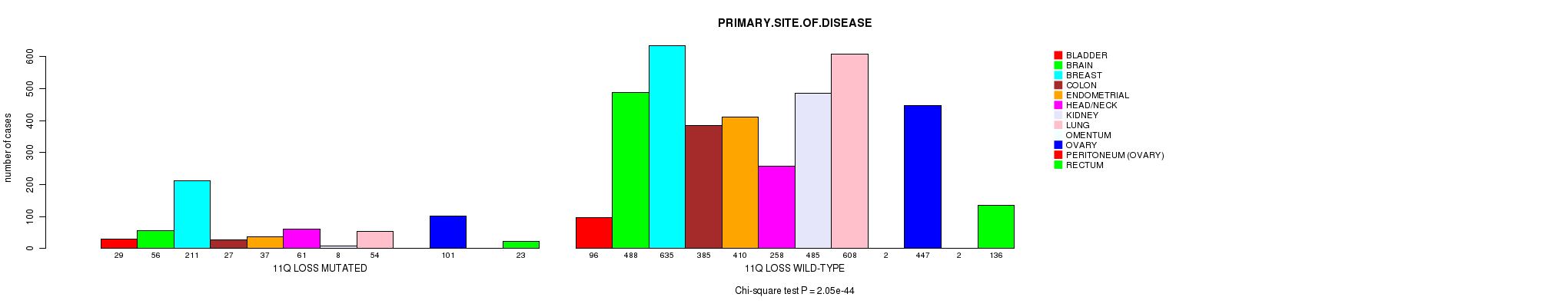
P value = 5.76e-05 (Fisher's exact test), Q value = 0.074
Table S305. Clustering Approach #62: '11q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 11Q LOSS MUTATED | 429 | 180 |
| 11Q LOSS WILD-TYPE | 2452 | 1502 |
Figure S243. Get High-res Image Clustering Approach #62: '11q loss mutation analysis' versus Clinical Feature #4: 'GENDER'

P value = 4.7e-22 (Chi-square test), Q value = 6.9e-19
Table S306. Clustering Approach #62: '11q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 11Q LOSS MUTATED | 25 | 2 | 16 | 61 | 8 | 1 | 5 | 10 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 33 | 5 | 0 | 17 | 5 | 101 | 16 | 0 | 3 |
| 11Q LOSS WILD-TYPE | 331 | 52 | 334 | 258 | 485 | 9 | 70 | 195 | 7 | 3 | 13 | 1 | 3 | 2 | 11 | 1 | 1 | 1 | 2 | 282 | 13 | 7 | 126 | 8 | 451 | 63 | 1 | 17 |
Figure S244. Get High-res Image Clustering Approach #62: '11q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.04e-05 (Chi-square test), Q value = 0.014
Table S307. Clustering Approach #62: '11q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 11Q LOSS MUTATED | 43 | 46 | 119 | 51 |
| 11Q LOSS WILD-TYPE | 669 | 452 | 762 | 372 |
Figure S245. Get High-res Image Clustering Approach #62: '11q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
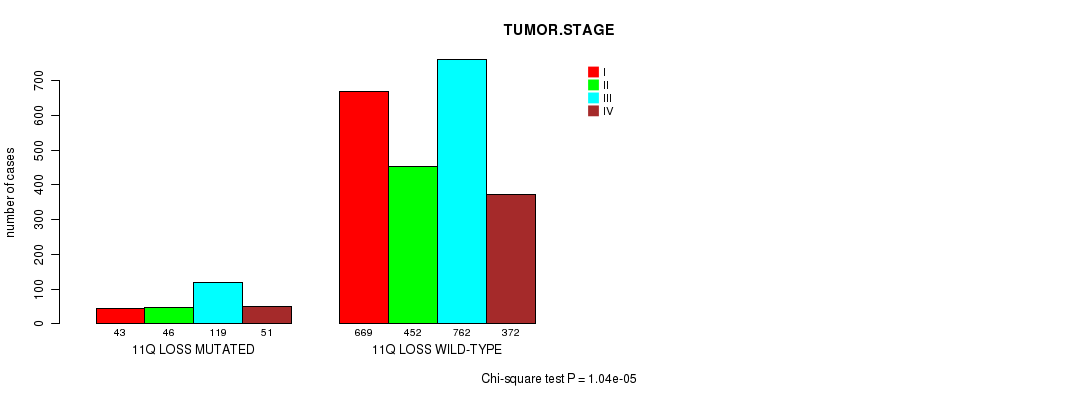
Table S308. Get Full Table Description of clustering approach #63: '12p loss mutation analysis'
| Cluster Labels | 12P LOSS MUTATED | 12P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 256 | 4307 |
P value = 2.2e-07 (Chi-square test), Q value = 3e-04
Table S309. Clustering Approach #63: '12p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 12P LOSS MUTATED | 6 | 47 | 62 | 21 | 18 | 11 | 2 | 33 | 0 | 44 | 0 | 12 |
| 12P LOSS WILD-TYPE | 119 | 497 | 784 | 391 | 429 | 308 | 491 | 629 | 2 | 504 | 2 | 147 |
Figure S246. Get High-res Image Clustering Approach #63: '12p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
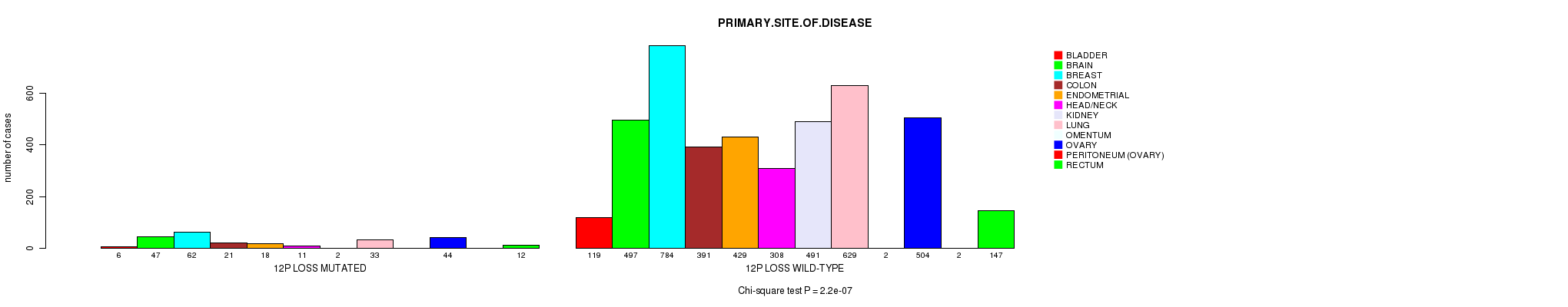
P value = 1.32e-10 (Chi-square test), Q value = 1.9e-07
Table S310. Clustering Approach #63: '12p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 12P LOSS MUTATED | 21 | 0 | 6 | 11 | 2 | 0 | 6 | 18 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 4 | 1 | 0 | 12 | 0 | 44 | 11 | 0 | 2 |
| 12P LOSS WILD-TYPE | 335 | 54 | 344 | 308 | 491 | 10 | 69 | 187 | 7 | 3 | 13 | 2 | 3 | 2 | 11 | 1 | 1 | 1 | 1 | 311 | 17 | 7 | 131 | 13 | 508 | 68 | 1 | 18 |
Figure S247. Get High-res Image Clustering Approach #63: '12p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S311. Get Full Table Description of clustering approach #64: '12q loss mutation analysis'
| Cluster Labels | 12Q LOSS MUTATED | 12Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 237 | 4326 |
P value = 3.31e-16 (Chi-square test), Q value = 4.8e-13
Table S312. Clustering Approach #64: '12q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 12Q LOSS MUTATED | 8 | 45 | 48 | 20 | 12 | 7 | 2 | 21 | 0 | 64 | 0 | 10 |
| 12Q LOSS WILD-TYPE | 117 | 499 | 798 | 392 | 435 | 312 | 491 | 641 | 2 | 484 | 2 | 149 |
Figure S248. Get High-res Image Clustering Approach #64: '12q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'

P value = 7.04e-16 (Chi-square test), Q value = 1e-12
Table S313. Clustering Approach #64: '12q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 12Q LOSS MUTATED | 20 | 0 | 4 | 7 | 2 | 0 | 6 | 10 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 10 | 0 | 64 | 7 | 0 | 1 |
| 12Q LOSS WILD-TYPE | 336 | 54 | 346 | 312 | 491 | 10 | 69 | 195 | 7 | 3 | 14 | 2 | 3 | 2 | 12 | 1 | 1 | 1 | 1 | 313 | 17 | 7 | 133 | 13 | 488 | 72 | 1 | 19 |
Figure S249. Get High-res Image Clustering Approach #64: '12q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S314. Get Full Table Description of clustering approach #65: '13q loss mutation analysis'
| Cluster Labels | 13Q LOSS MUTATED | 13Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 1007 | 3556 |
P value = 7.7e-88 (Chi-square test), Q value = 1.2e-84
Table S315. Clustering Approach #65: '13q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 13Q LOSS MUTATED | 18 | 148 | 248 | 7 | 52 | 61 | 33 | 194 | 1 | 238 | 2 | 5 |
| 13Q LOSS WILD-TYPE | 107 | 396 | 598 | 405 | 395 | 258 | 460 | 468 | 1 | 310 | 0 | 154 |
Figure S250. Get High-res Image Clustering Approach #65: '13q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'

P value = 1.22e-06 (Fisher's exact test), Q value = 0.0016
Table S316. Clustering Approach #65: '13q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 13Q LOSS MUTATED | 701 | 306 |
| 13Q LOSS WILD-TYPE | 2180 | 1376 |
Figure S251. Get High-res Image Clustering Approach #65: '13q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
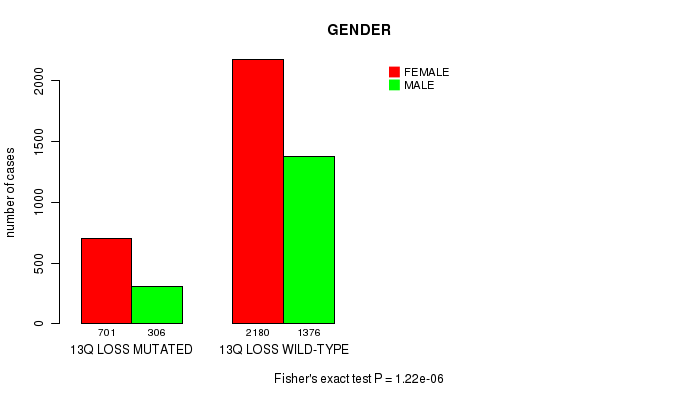
P value = 1.53e-83 (Chi-square test), Q value = 2.4e-80
Table S317. Clustering Approach #65: '13q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 13Q LOSS MUTATED | 6 | 1 | 30 | 61 | 33 | 6 | 15 | 47 | 0 | 0 | 5 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 116 | 3 | 1 | 4 | 1 | 241 | 19 | 0 | 6 |
| 13Q LOSS WILD-TYPE | 350 | 53 | 320 | 258 | 460 | 4 | 60 | 158 | 7 | 3 | 10 | 1 | 2 | 2 | 12 | 1 | 1 | 1 | 1 | 199 | 15 | 6 | 139 | 12 | 311 | 60 | 1 | 14 |
Figure S252. Get High-res Image Clustering Approach #65: '13q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 9.98e-12 (Chi-square test), Q value = 1.4e-08
Table S318. Clustering Approach #65: '13q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 13Q LOSS MUTATED | 66 | 136 | 52 | 37 |
| 13Q LOSS WILD-TYPE | 400 | 496 | 642 | 172 |
Figure S253. Get High-res Image Clustering Approach #65: '13q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
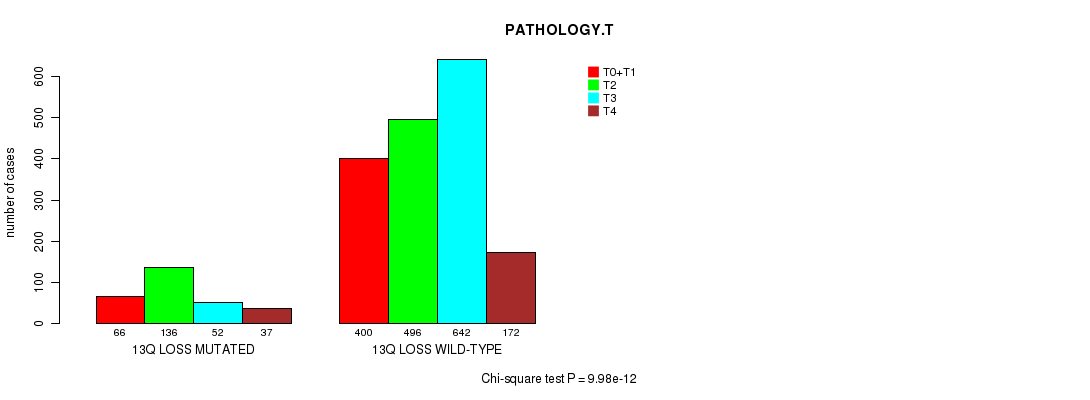
P value = 1.38e-08 (Chi-square test), Q value = 1.9e-05
Table S319. Clustering Approach #65: '13q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 13Q LOSS MUTATED | 112 | 85 | 243 | 90 |
| 13Q LOSS WILD-TYPE | 600 | 413 | 638 | 333 |
Figure S254. Get High-res Image Clustering Approach #65: '13q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
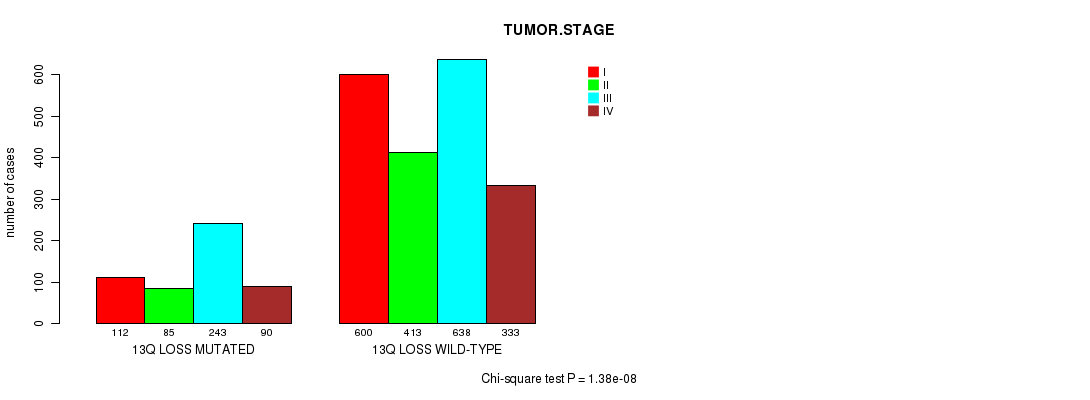
Table S320. Get Full Table Description of clustering approach #66: '14q loss mutation analysis'
| Cluster Labels | 14Q LOSS MUTATED | 14Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 866 | 3697 |
P value = 1.77e-38 (Chi-square test), Q value = 2.7e-35
Table S321. Clustering Approach #66: '14q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 14Q LOSS MUTATED | 19 | 123 | 120 | 94 | 39 | 17 | 159 | 89 | 0 | 155 | 0 | 49 |
| 14Q LOSS WILD-TYPE | 106 | 421 | 726 | 318 | 408 | 302 | 334 | 573 | 2 | 393 | 2 | 110 |
Figure S255. Get High-res Image Clustering Approach #66: '14q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
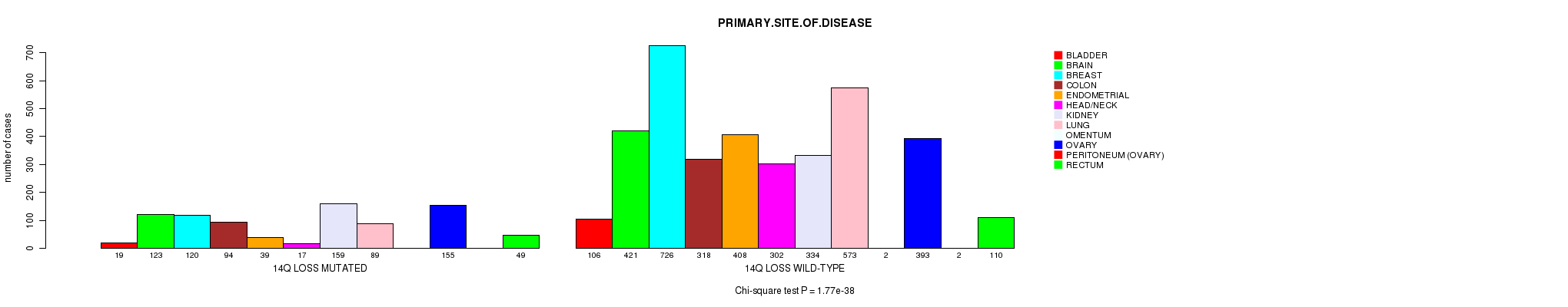
P value = 1.06e-38 (Chi-square test), Q value = 1.6e-35
Table S322. Clustering Approach #66: '14q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 14Q LOSS MUTATED | 91 | 3 | 13 | 17 | 159 | 2 | 7 | 14 | 0 | 0 | 3 | 1 | 0 | 1 | 2 | 0 | 0 | 1 | 0 | 58 | 3 | 0 | 49 | 0 | 155 | 23 | 0 | 2 |
| 14Q LOSS WILD-TYPE | 265 | 51 | 337 | 302 | 334 | 8 | 68 | 191 | 7 | 3 | 12 | 1 | 3 | 1 | 11 | 1 | 1 | 0 | 2 | 257 | 15 | 7 | 94 | 13 | 397 | 56 | 1 | 18 |
Figure S256. Get High-res Image Clustering Approach #66: '14q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 2.02e-07 (Chi-square test), Q value = 0.00028
Table S323. Clustering Approach #66: '14q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 14Q LOSS MUTATED | 101 | 108 | 177 | 18 |
| 14Q LOSS WILD-TYPE | 365 | 524 | 517 | 191 |
Figure S257. Get High-res Image Clustering Approach #66: '14q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
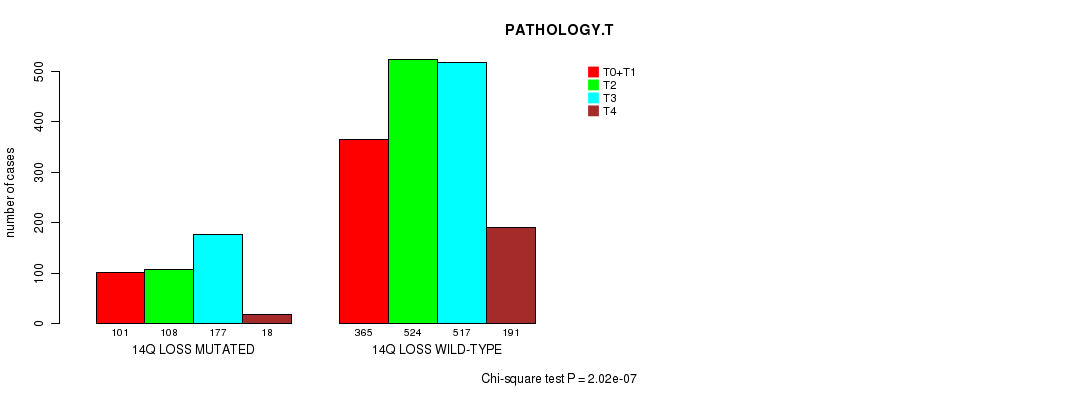
P value = 0.000151 (Chi-square test), Q value = 0.19
Table S324. Clustering Approach #66: '14q loss mutation analysis' versus Clinical Feature #8: 'PATHOLOGY.N'
| nPatients | N0 | N1 | N2 | N3 |
|---|---|---|---|---|
| ALL | 1067 | 353 | 282 | 11 |
| 14Q LOSS MUTATED | 214 | 82 | 30 | 0 |
| 14Q LOSS WILD-TYPE | 853 | 271 | 252 | 11 |
Figure S258. Get High-res Image Clustering Approach #66: '14q loss mutation analysis' versus Clinical Feature #8: 'PATHOLOGY.N'
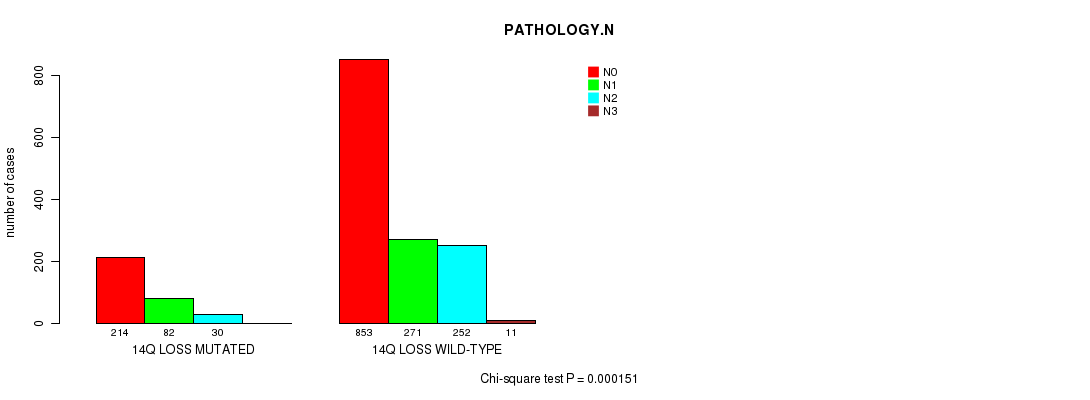
P value = 5.75e-05 (Chi-square test), Q value = 0.074
Table S325. Clustering Approach #66: '14q loss mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
| nPatients | M0 | M1 | M1A | M1B | MX |
|---|---|---|---|---|---|
| ALL | 1424 | 168 | 9 | 3 | 165 |
| 14Q LOSS MUTATED | 317 | 54 | 3 | 0 | 17 |
| 14Q LOSS WILD-TYPE | 1107 | 114 | 6 | 3 | 148 |
Figure S259. Get High-res Image Clustering Approach #66: '14q loss mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
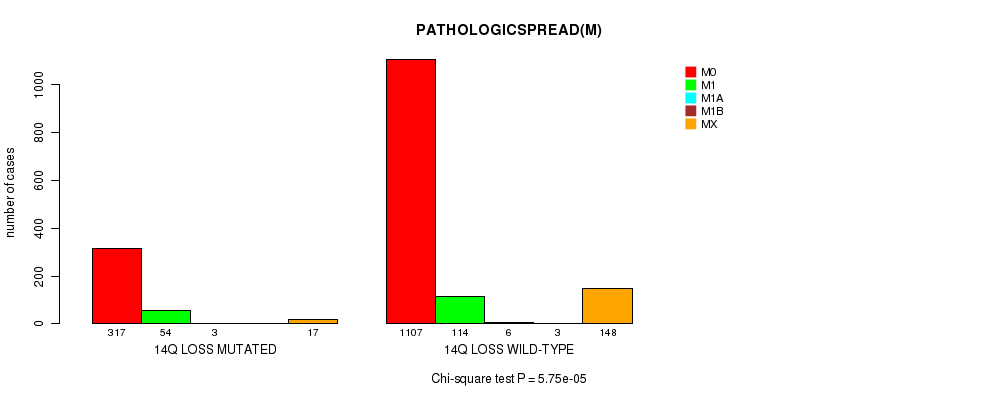
Table S326. Get Full Table Description of clustering approach #67: '15q loss mutation analysis'
| Cluster Labels | 15Q LOSS MUTATED | 15Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 803 | 3760 |
P value = 3.19e-60 (Chi-square test), Q value = 5e-57
Table S327. Clustering Approach #67: '15q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 15Q LOSS MUTATED | 15 | 73 | 151 | 99 | 64 | 23 | 12 | 103 | 1 | 202 | 2 | 55 |
| 15Q LOSS WILD-TYPE | 110 | 471 | 695 | 313 | 383 | 296 | 481 | 559 | 1 | 346 | 0 | 104 |
Figure S260. Get High-res Image Clustering Approach #67: '15q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'

P value = 1.96e-12 (Fisher's exact test), Q value = 2.8e-09
Table S328. Clustering Approach #67: '15q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 15Q LOSS MUTATED | 593 | 210 |
| 15Q LOSS WILD-TYPE | 2288 | 1472 |
Figure S261. Get High-res Image Clustering Approach #67: '15q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
P value = 3.38e-63 (Chi-square test), Q value = 5.3e-60
Table S329. Clustering Approach #67: '15q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 15Q LOSS MUTATED | 95 | 4 | 27 | 23 | 12 | 1 | 10 | 47 | 0 | 0 | 3 | 0 | 2 | 0 | 5 | 0 | 0 | 1 | 1 | 33 | 6 | 0 | 50 | 4 | 205 | 31 | 0 | 5 |
| 15Q LOSS WILD-TYPE | 261 | 50 | 323 | 296 | 481 | 9 | 65 | 158 | 7 | 3 | 12 | 2 | 1 | 2 | 8 | 1 | 1 | 0 | 1 | 282 | 12 | 7 | 93 | 9 | 347 | 48 | 1 | 15 |
Figure S262. Get High-res Image Clustering Approach #67: '15q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.85e-06 (Chi-square test), Q value = 0.0025
Table S330. Clustering Approach #67: '15q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 15Q LOSS MUTATED | 41 | 83 | 138 | 29 |
| 15Q LOSS WILD-TYPE | 425 | 549 | 556 | 180 |
Figure S263. Get High-res Image Clustering Approach #67: '15q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
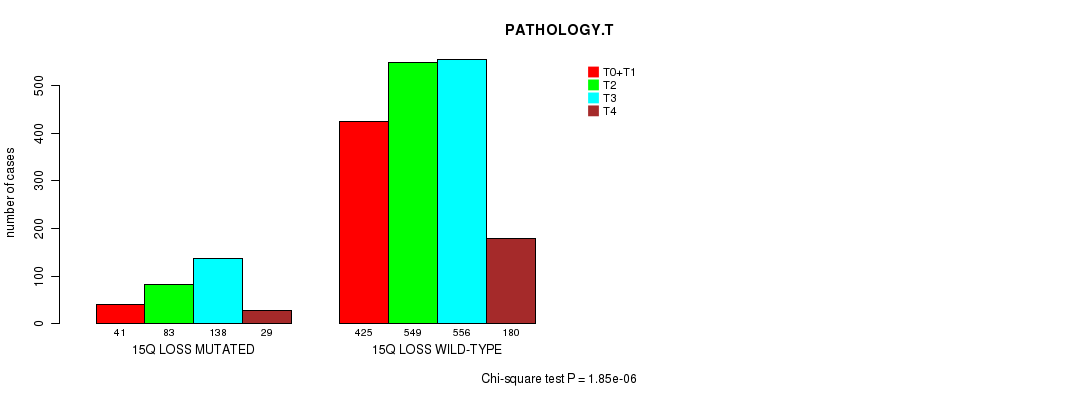
P value = 3.04e-14 (Chi-square test), Q value = 4.4e-11
Table S331. Clustering Approach #67: '15q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 15Q LOSS MUTATED | 74 | 97 | 234 | 81 |
| 15Q LOSS WILD-TYPE | 638 | 401 | 647 | 342 |
Figure S264. Get High-res Image Clustering Approach #67: '15q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
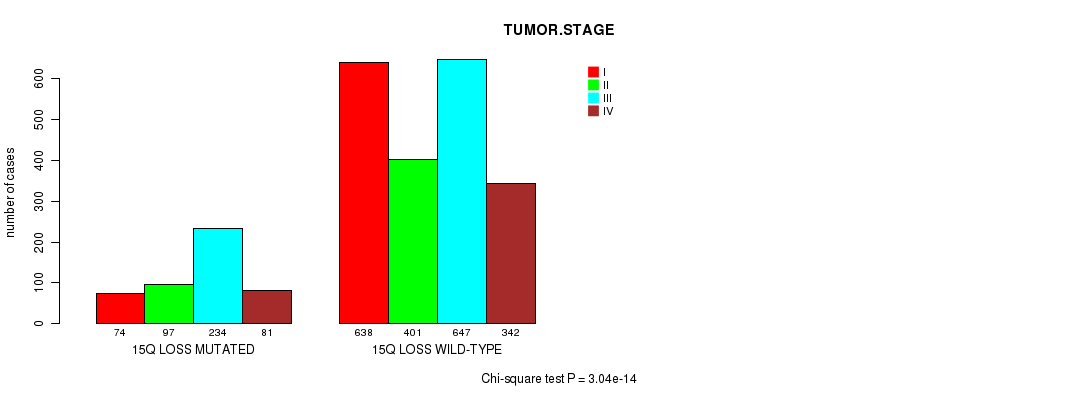
Table S332. Get Full Table Description of clustering approach #68: '16p loss mutation analysis'
| Cluster Labels | 16P LOSS MUTATED | 16P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 499 | 4064 |
P value = 3.63e-173 (Chi-square test), Q value = 5.8e-170
Table S333. Clustering Approach #68: '16p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 16P LOSS MUTATED | 14 | 30 | 47 | 7 | 57 | 12 | 2 | 72 | 2 | 248 | 1 | 7 |
| 16P LOSS WILD-TYPE | 111 | 514 | 799 | 405 | 390 | 307 | 491 | 590 | 0 | 300 | 1 | 152 |
Figure S265. Get High-res Image Clustering Approach #68: '16p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
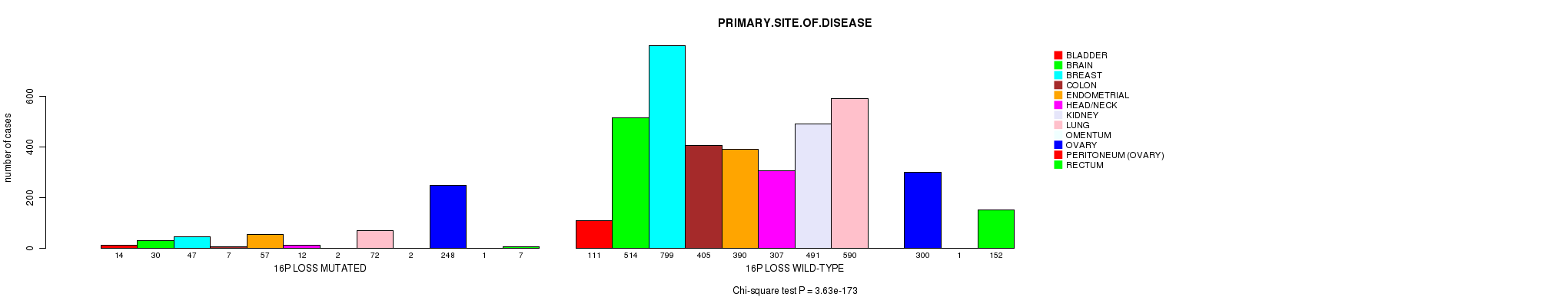
P value = 8.69e-20 (Fisher's exact test), Q value = 1.3e-16
Table S334. Clustering Approach #68: '16p loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 16P LOSS MUTATED | 404 | 95 |
| 16P LOSS WILD-TYPE | 2477 | 1587 |
Figure S266. Get High-res Image Clustering Approach #68: '16p loss mutation analysis' versus Clinical Feature #4: 'GENDER'
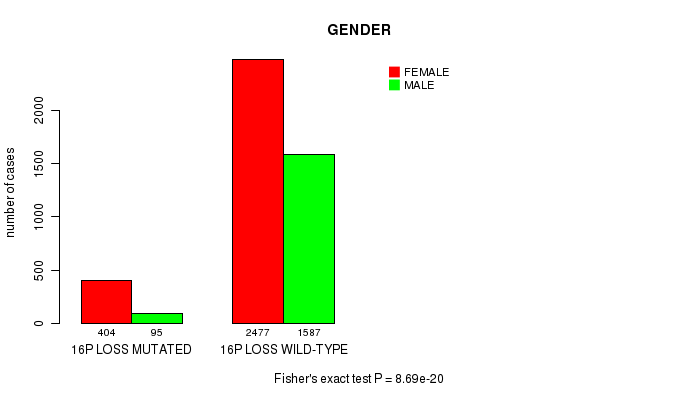
P value = 1.47e-134 (Chi-square test), Q value = 2.3e-131
Table S335. Clustering Approach #68: '16p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 16P LOSS MUTATED | 5 | 2 | 24 | 12 | 2 | 1 | 5 | 14 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 46 | 5 | 1 | 5 | 1 | 251 | 28 | 0 | 1 |
| 16P LOSS WILD-TYPE | 351 | 52 | 326 | 307 | 491 | 9 | 70 | 191 | 7 | 3 | 14 | 1 | 2 | 2 | 12 | 1 | 1 | 1 | 1 | 269 | 13 | 6 | 138 | 12 | 301 | 51 | 1 | 19 |
Figure S267. Get High-res Image Clustering Approach #68: '16p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 9.54e-28 (Chi-square test), Q value = 1.4e-24
Table S336. Clustering Approach #68: '16p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 16P LOSS MUTATED | 46 | 32 | 209 | 57 |
| 16P LOSS WILD-TYPE | 666 | 466 | 672 | 366 |
Figure S268. Get High-res Image Clustering Approach #68: '16p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
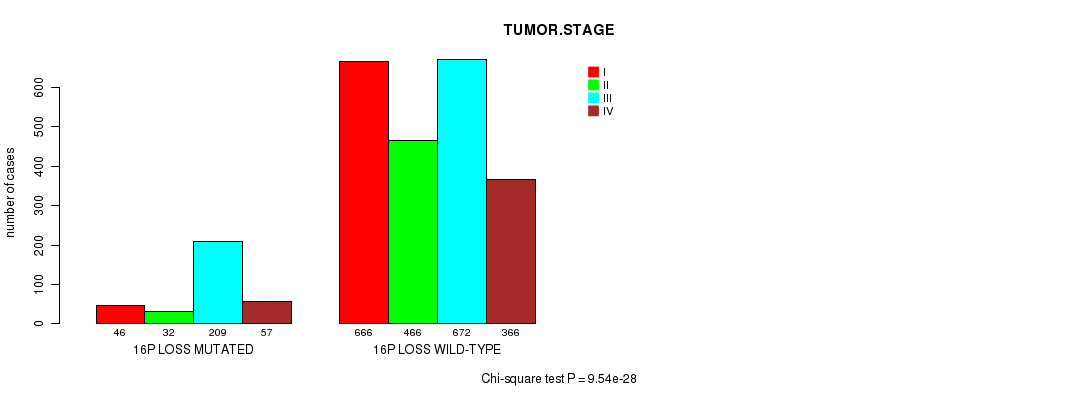
P value = 3.42e-06 (Fisher's exact test), Q value = 0.0046
Table S337. Clustering Approach #68: '16p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 16P LOSS MUTATED | 54 | 445 |
| 16P LOSS WILD-TYPE | 772 | 3292 |
Figure S269. Get High-res Image Clustering Approach #68: '16p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
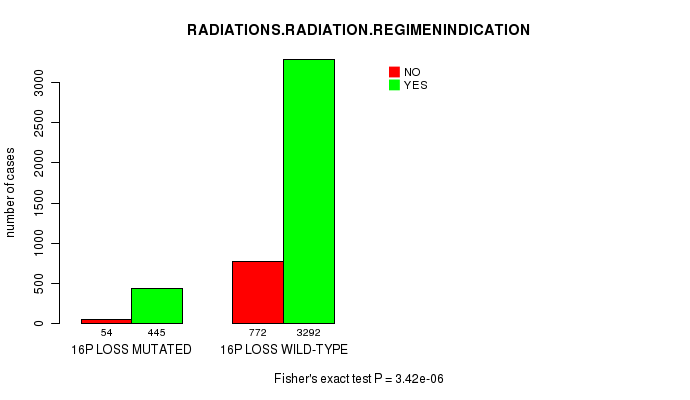
Table S338. Get Full Table Description of clustering approach #69: '16q loss mutation analysis'
| Cluster Labels | 16Q LOSS MUTATED | 16Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 1000 | 3563 |
P value = 1.19e-229 (Chi-square test), Q value = 1.9e-226
Table S339. Clustering Approach #69: '16q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 16Q LOSS MUTATED | 15 | 45 | 351 | 5 | 94 | 24 | 5 | 104 | 1 | 344 | 2 | 10 |
| 16Q LOSS WILD-TYPE | 110 | 499 | 495 | 407 | 353 | 295 | 488 | 558 | 1 | 204 | 0 | 149 |
Figure S270. Get High-res Image Clustering Approach #69: '16q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
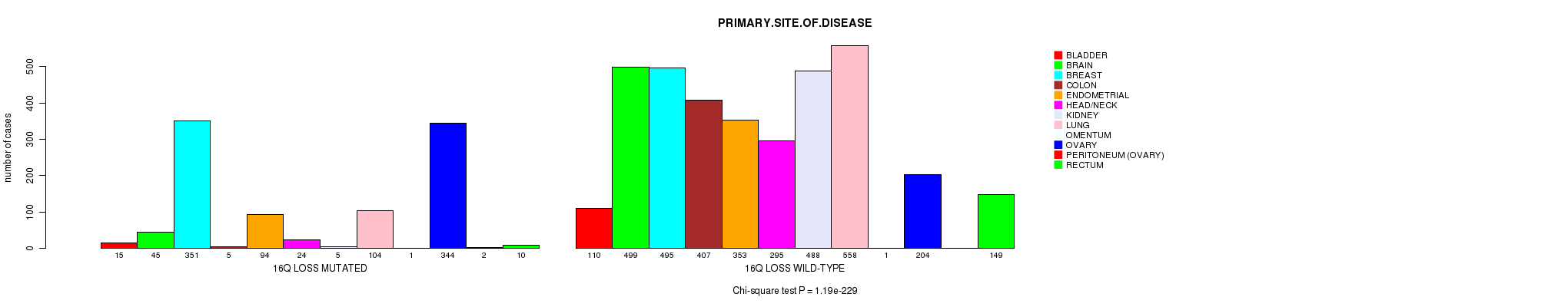
P value = 7.3e-72 (Fisher's exact test), Q value = 1.1e-68
Table S340. Clustering Approach #69: '16q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 16Q LOSS MUTATED | 860 | 140 |
| 16Q LOSS WILD-TYPE | 2021 | 1542 |
Figure S271. Get High-res Image Clustering Approach #69: '16q loss mutation analysis' versus Clinical Feature #4: 'GENDER'

P value = 1.95e-195 (Chi-square test), Q value = 3.1e-192
Table S341. Clustering Approach #69: '16q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 16Q LOSS MUTATED | 3 | 2 | 45 | 24 | 5 | 3 | 9 | 24 | 0 | 0 | 3 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 59 | 6 | 1 | 8 | 1 | 347 | 43 | 0 | 4 |
| 16Q LOSS WILD-TYPE | 353 | 52 | 305 | 295 | 488 | 7 | 66 | 181 | 7 | 3 | 12 | 0 | 2 | 2 | 12 | 1 | 1 | 1 | 1 | 256 | 12 | 6 | 135 | 12 | 205 | 36 | 1 | 16 |
Figure S272. Get High-res Image Clustering Approach #69: '16q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.01e-06 (Chi-square test), Q value = 0.0014
Table S342. Clustering Approach #69: '16q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 16Q LOSS MUTATED | 29 | 71 | 26 | 11 |
| 16Q LOSS WILD-TYPE | 437 | 561 | 668 | 198 |
Figure S273. Get High-res Image Clustering Approach #69: '16q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
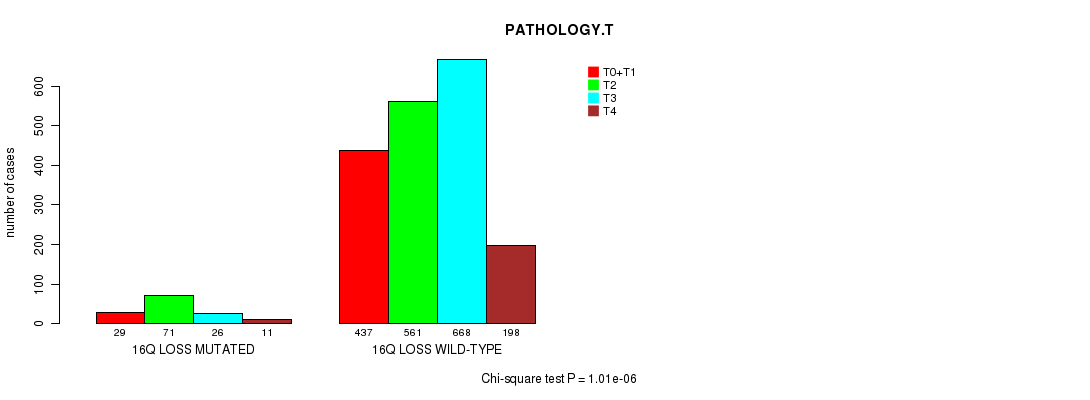
P value = 3.38e-42 (Chi-square test), Q value = 5.1e-39
Table S343. Clustering Approach #69: '16q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 16Q LOSS MUTATED | 69 | 44 | 295 | 69 |
| 16Q LOSS WILD-TYPE | 643 | 454 | 586 | 354 |
Figure S274. Get High-res Image Clustering Approach #69: '16q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
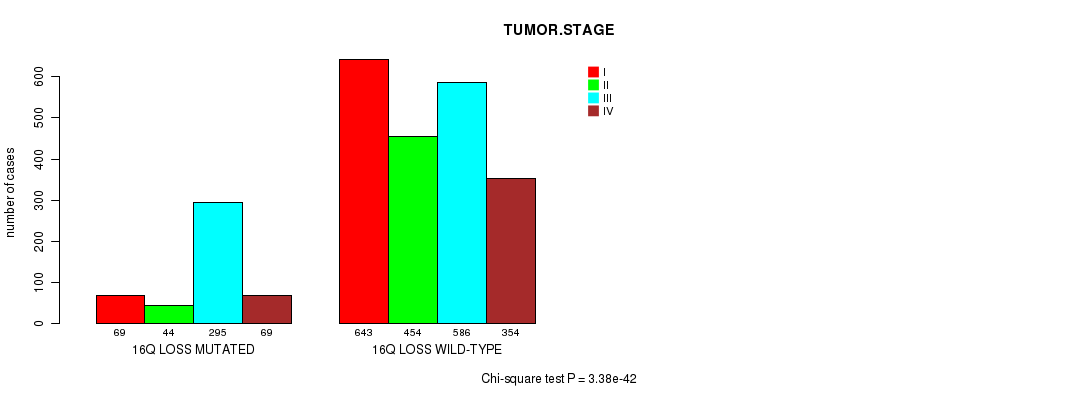
P value = 1.07e-05 (Chi-square test), Q value = 0.014
Table S344. Clustering Approach #69: '16q loss mutation analysis' versus Clinical Feature #18: 'COMPLETENESS.OF.RESECTION'
| nPatients | R0 | R1 | R2 | RX |
|---|---|---|---|---|
| ALL | 1171 | 69 | 59 | 77 |
| 16Q LOSS MUTATED | 141 | 23 | 9 | 11 |
| 16Q LOSS WILD-TYPE | 1030 | 46 | 50 | 66 |
Figure S275. Get High-res Image Clustering Approach #69: '16q loss mutation analysis' versus Clinical Feature #18: 'COMPLETENESS.OF.RESECTION'
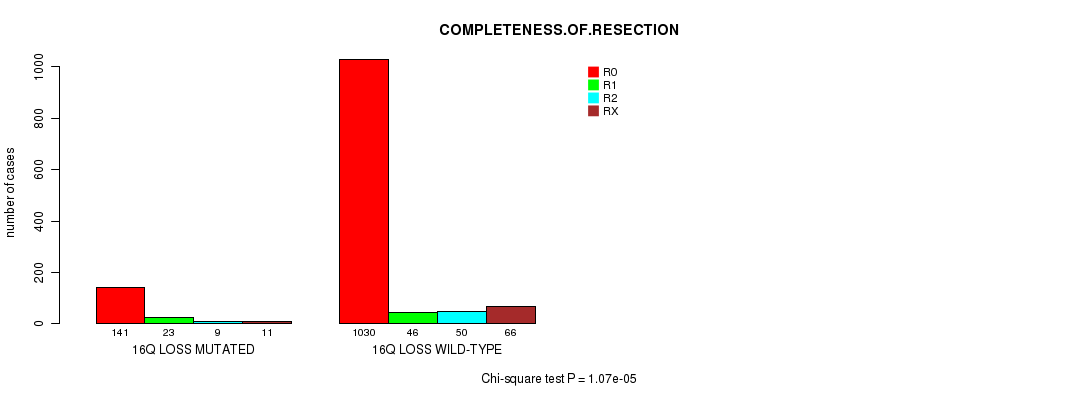
P value = 9.99e-06 (Chi-square test), Q value = 0.013
Table S345. Clustering Approach #69: '16q loss mutation analysis' versus Clinical Feature #21: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE IA | STAGE IB | STAGE II | STAGE IIA | STAGE IIB | STAGE III | STAGE IIIA | STAGE IIIB | STAGE IIIC | STAGE IV | STAGE TIS | STAGE X |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 72 | 62 | 7 | 43 | 287 | 186 | 42 | 124 | 25 | 41 | 59 | 1 | 16 |
| 16Q LOSS MUTATED | 32 | 30 | 1 | 7 | 131 | 67 | 4 | 48 | 10 | 16 | 12 | 0 | 7 |
| 16Q LOSS WILD-TYPE | 40 | 32 | 6 | 36 | 156 | 119 | 38 | 76 | 15 | 25 | 47 | 1 | 9 |
Figure S276. Get High-res Image Clustering Approach #69: '16q loss mutation analysis' versus Clinical Feature #21: 'NEOPLASM.DISEASESTAGE'
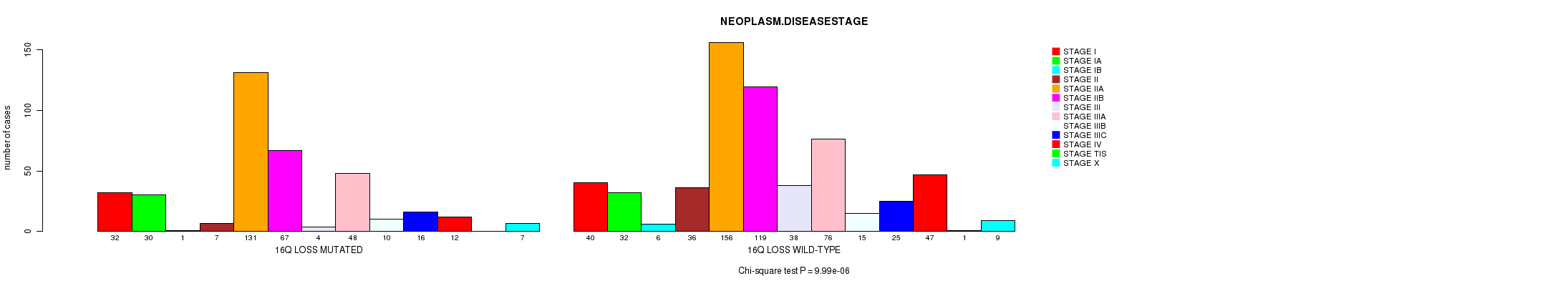
Table S346. Get Full Table Description of clustering approach #70: '17p loss mutation analysis'
| Cluster Labels | 17P LOSS MUTATED | 17P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 1418 | 3145 |
P value = 2.2e-195 (Chi-square test), Q value = 3.5e-192
Table S347. Clustering Approach #70: '17p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 17P LOSS MUTATED | 35 | 38 | 352 | 162 | 86 | 41 | 24 | 187 | 1 | 403 | 1 | 85 |
| 17P LOSS WILD-TYPE | 90 | 506 | 494 | 250 | 361 | 278 | 469 | 475 | 1 | 145 | 1 | 74 |
Figure S277. Get High-res Image Clustering Approach #70: '17p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
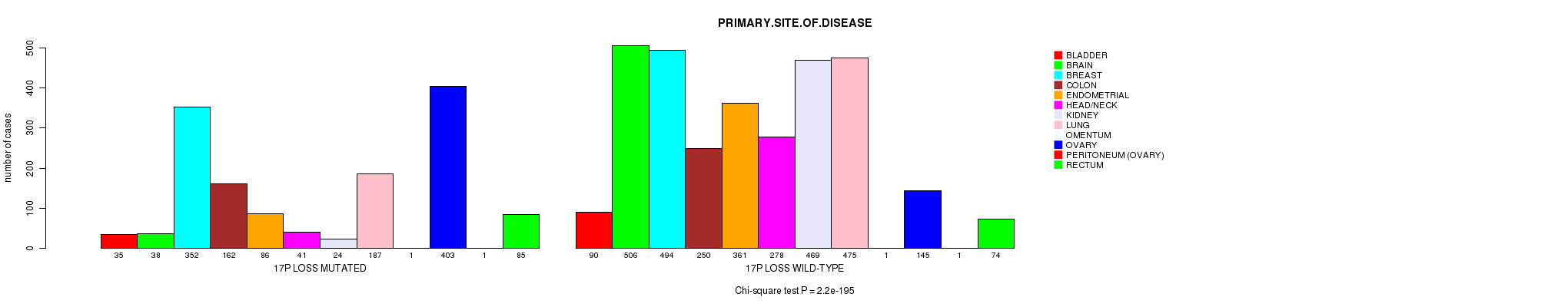
P value = 1.96e-32 (Fisher's exact test), Q value = 2.9e-29
Table S348. Clustering Approach #70: '17p loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 17P LOSS MUTATED | 1071 | 347 |
| 17P LOSS WILD-TYPE | 1810 | 1335 |
Figure S278. Get High-res Image Clustering Approach #70: '17p loss mutation analysis' versus Clinical Feature #4: 'GENDER'
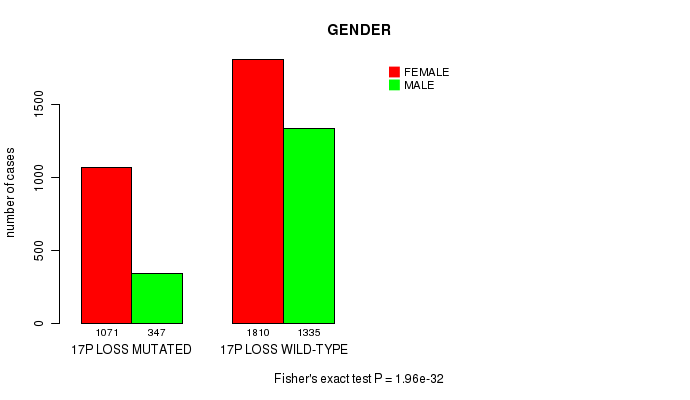
P value = 5.37e-162 (Chi-square test), Q value = 8.5e-159
Table S349. Clustering Approach #70: '17p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 17P LOSS MUTATED | 155 | 7 | 33 | 41 | 24 | 2 | 15 | 54 | 3 | 1 | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 104 | 8 | 2 | 78 | 6 | 405 | 45 | 0 | 0 |
| 17P LOSS WILD-TYPE | 201 | 47 | 317 | 278 | 469 | 8 | 60 | 151 | 4 | 2 | 13 | 1 | 2 | 2 | 12 | 1 | 1 | 0 | 2 | 211 | 10 | 5 | 65 | 7 | 147 | 34 | 1 | 20 |
Figure S279. Get High-res Image Clustering Approach #70: '17p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 2.22e-07 (Chi-square test), Q value = 3e-04
Table S350. Clustering Approach #70: '17p loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 17P LOSS MUTATED | 75 | 164 | 212 | 43 |
| 17P LOSS WILD-TYPE | 391 | 468 | 482 | 166 |
Figure S280. Get High-res Image Clustering Approach #70: '17p loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'

P value = 2.74e-31 (Chi-square test), Q value = 4.1e-28
Table S351. Clustering Approach #70: '17p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 17P LOSS MUTATED | 154 | 162 | 440 | 133 |
| 17P LOSS WILD-TYPE | 558 | 336 | 441 | 290 |
Figure S281. Get High-res Image Clustering Approach #70: '17p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
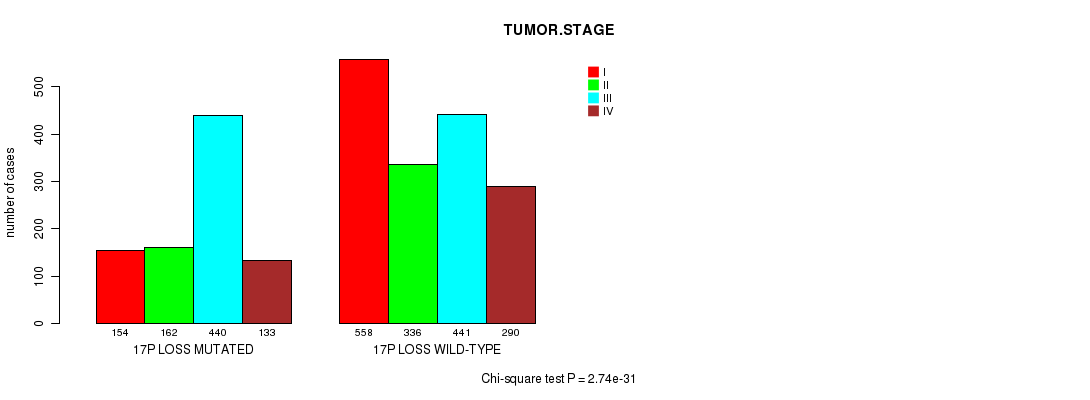
P value = 3.23e-23 (Fisher's exact test), Q value = 4.8e-20
Table S352. Clustering Approach #70: '17p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 17P LOSS MUTATED | 142 | 1276 |
| 17P LOSS WILD-TYPE | 684 | 2461 |
Figure S282. Get High-res Image Clustering Approach #70: '17p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
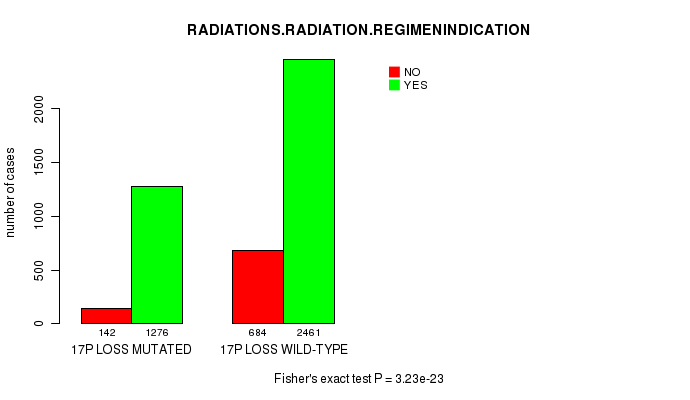
Table S353. Get Full Table Description of clustering approach #71: '17q loss mutation analysis'
| Cluster Labels | 17Q LOSS MUTATED | 17Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 629 | 3934 |
P value = 9.79e-204 (Chi-square test), Q value = 1.6e-200
Table S354. Clustering Approach #71: '17q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 17Q LOSS MUTATED | 6 | 25 | 134 | 35 | 57 | 5 | 12 | 32 | 1 | 301 | 1 | 20 |
| 17Q LOSS WILD-TYPE | 119 | 519 | 712 | 377 | 390 | 314 | 481 | 630 | 1 | 247 | 1 | 139 |
Figure S283. Get High-res Image Clustering Approach #71: '17q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
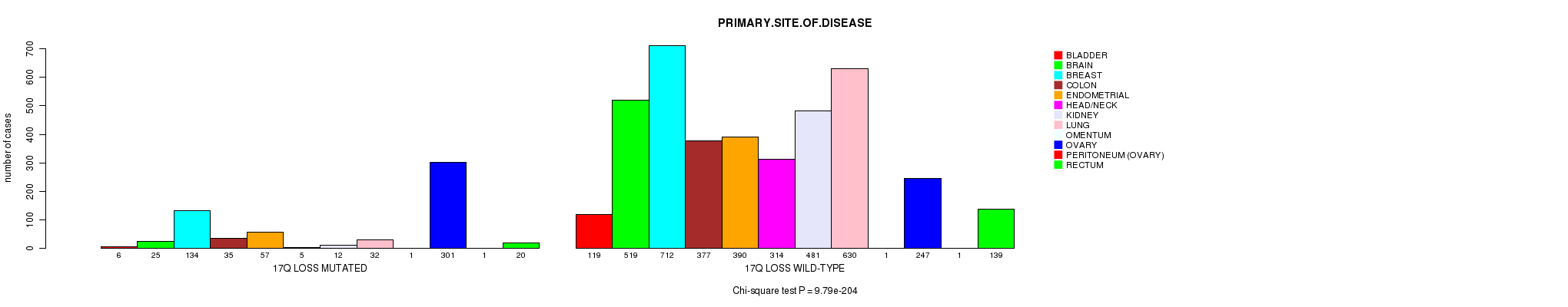
P value = 8.72e-49 (Fisher's exact test), Q value = 1.3e-45
Table S355. Clustering Approach #71: '17q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 17Q LOSS MUTATED | 551 | 78 |
| 17Q LOSS WILD-TYPE | 2330 | 1604 |
Figure S284. Get High-res Image Clustering Approach #71: '17q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
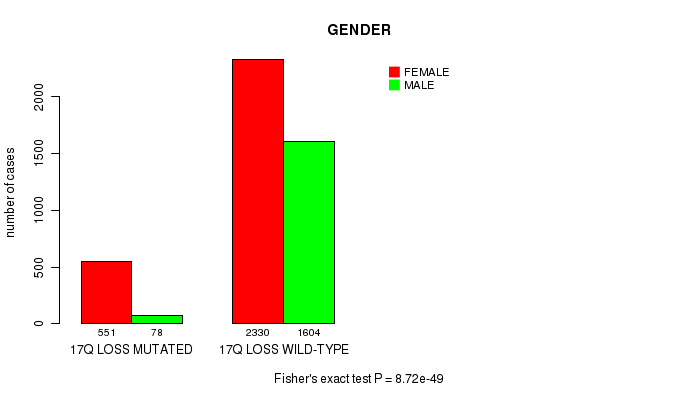
P value = 1.38e-170 (Chi-square test), Q value = 2.2e-167
Table S356. Clustering Approach #71: '17q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 17Q LOSS MUTATED | 33 | 2 | 29 | 5 | 12 | 0 | 3 | 9 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 18 | 6 | 0 | 17 | 2 | 303 | 22 | 0 | 0 |
| 17Q LOSS WILD-TYPE | 323 | 52 | 321 | 314 | 481 | 10 | 72 | 196 | 7 | 3 | 15 | 2 | 2 | 2 | 12 | 1 | 1 | 1 | 2 | 297 | 12 | 7 | 126 | 11 | 249 | 57 | 1 | 20 |
Figure S285. Get High-res Image Clustering Approach #71: '17q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 2.14e-45 (Chi-square test), Q value = 3.3e-42
Table S357. Clustering Approach #71: '17q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 17Q LOSS MUTATED | 36 | 41 | 262 | 62 |
| 17Q LOSS WILD-TYPE | 676 | 457 | 619 | 361 |
Figure S286. Get High-res Image Clustering Approach #71: '17q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'

P value = 2.1e-08 (Fisher's exact test), Q value = 2.9e-05
Table S358. Clustering Approach #71: '17q loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 17Q LOSS MUTATED | 66 | 563 |
| 17Q LOSS WILD-TYPE | 760 | 3174 |
Figure S287. Get High-res Image Clustering Approach #71: '17q loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
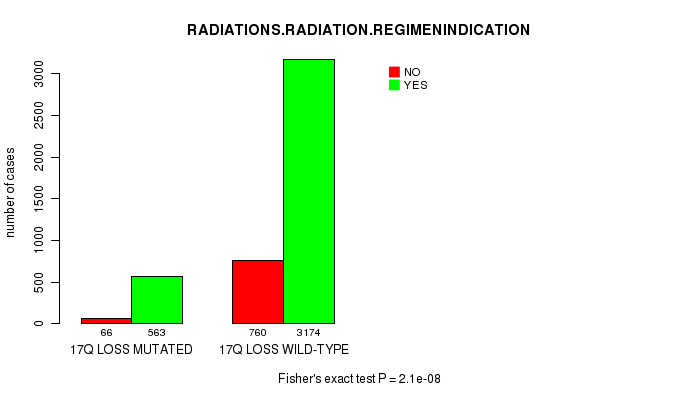
P value = 8.95e-05 (Chi-square test), Q value = 0.11
Table S359. Clustering Approach #71: '17q loss mutation analysis' versus Clinical Feature #18: 'COMPLETENESS.OF.RESECTION'
| nPatients | R0 | R1 | R2 | RX |
|---|---|---|---|---|
| ALL | 1171 | 69 | 59 | 77 |
| 17Q LOSS MUTATED | 102 | 17 | 6 | 12 |
| 17Q LOSS WILD-TYPE | 1069 | 52 | 53 | 65 |
Figure S288. Get High-res Image Clustering Approach #71: '17q loss mutation analysis' versus Clinical Feature #18: 'COMPLETENESS.OF.RESECTION'
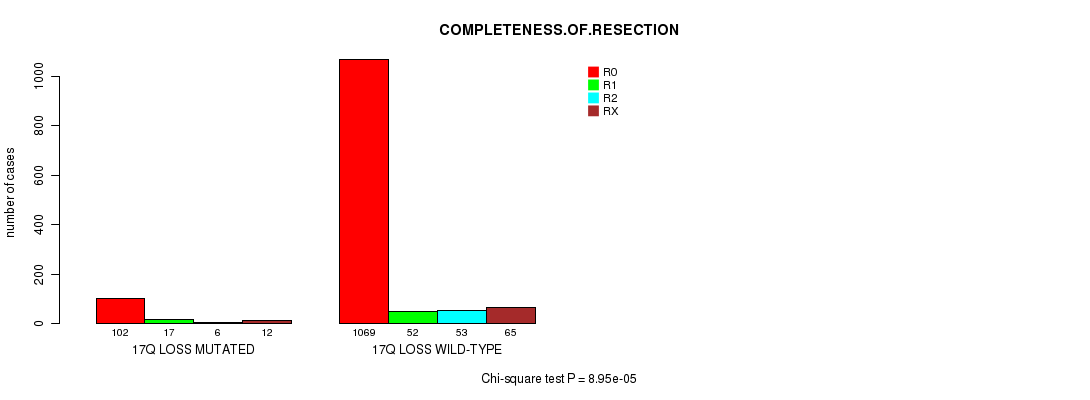
Table S360. Get Full Table Description of clustering approach #72: '18p loss mutation analysis'
| Cluster Labels | 18P LOSS MUTATED | 18P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 931 | 3632 |
P value = 1.86e-147 (Chi-square test), Q value = 2.9e-144
Table S361. Clustering Approach #72: '18p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 18P LOSS MUTATED | 18 | 50 | 168 | 212 | 28 | 27 | 46 | 91 | 1 | 175 | 0 | 112 |
| 18P LOSS WILD-TYPE | 107 | 494 | 678 | 200 | 419 | 292 | 447 | 571 | 1 | 373 | 2 | 47 |
Figure S289. Get High-res Image Clustering Approach #72: '18p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'

P value = 1.74e-130 (Chi-square test), Q value = 2.7e-127
Table S362. Clustering Approach #72: '18p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 18P LOSS MUTATED | 204 | 7 | 16 | 27 | 46 | 2 | 15 | 30 | 0 | 0 | 3 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 34 | 5 | 3 | 107 | 5 | 176 | 7 | 0 | 4 |
| 18P LOSS WILD-TYPE | 152 | 47 | 334 | 292 | 447 | 8 | 60 | 175 | 7 | 3 | 12 | 2 | 1 | 2 | 11 | 1 | 1 | 1 | 2 | 281 | 13 | 4 | 36 | 8 | 376 | 72 | 1 | 16 |
Figure S290. Get High-res Image Clustering Approach #72: '18p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 1.75e-26 (Chi-square test), Q value = 2.6e-23
Table S363. Clustering Approach #72: '18p loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 18P LOSS MUTATED | 44 | 131 | 258 | 54 |
| 18P LOSS WILD-TYPE | 422 | 501 | 436 | 155 |
Figure S291. Get High-res Image Clustering Approach #72: '18p loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
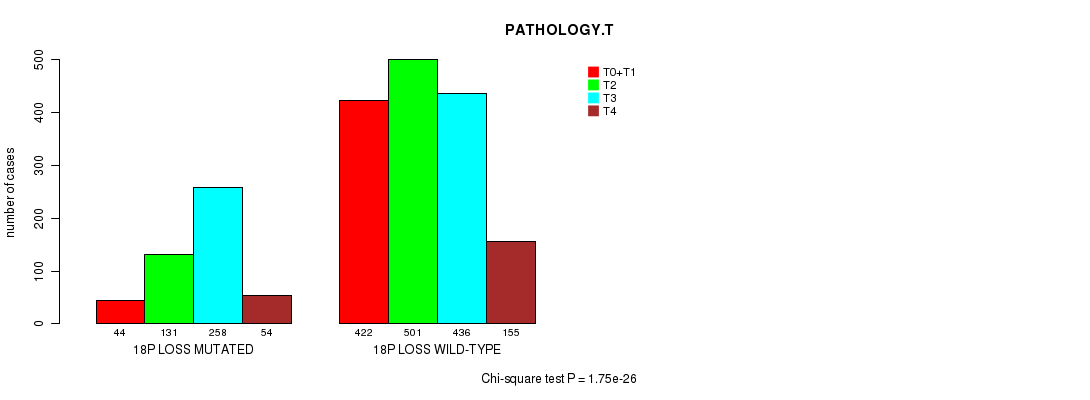
P value = 4.8e-09 (Chi-square test), Q value = 6.7e-06
Table S364. Clustering Approach #72: '18p loss mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
| nPatients | M0 | M1 | M1A | M1B | MX |
|---|---|---|---|---|---|
| ALL | 1424 | 168 | 9 | 3 | 165 |
| 18P LOSS MUTATED | 336 | 73 | 7 | 0 | 42 |
| 18P LOSS WILD-TYPE | 1088 | 95 | 2 | 3 | 123 |
Figure S292. Get High-res Image Clustering Approach #72: '18p loss mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
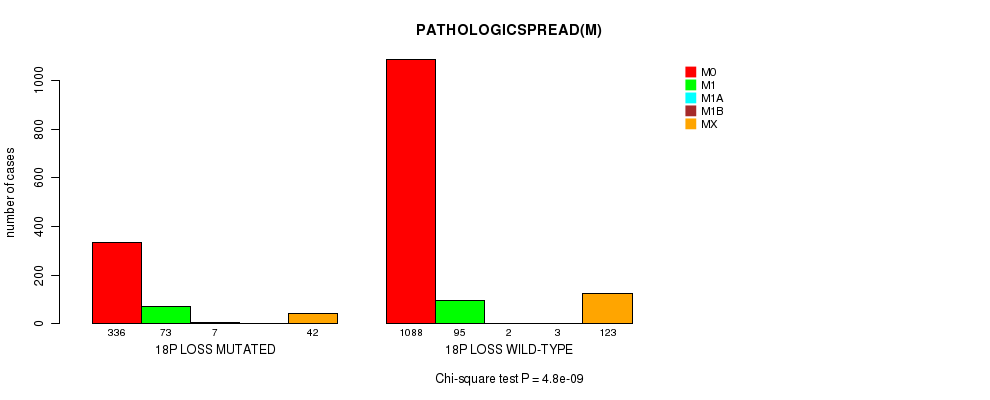
P value = 5e-12 (Chi-square test), Q value = 7.1e-09
Table S365. Clustering Approach #72: '18p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 18P LOSS MUTATED | 110 | 139 | 274 | 120 |
| 18P LOSS WILD-TYPE | 602 | 359 | 607 | 303 |
Figure S293. Get High-res Image Clustering Approach #72: '18p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
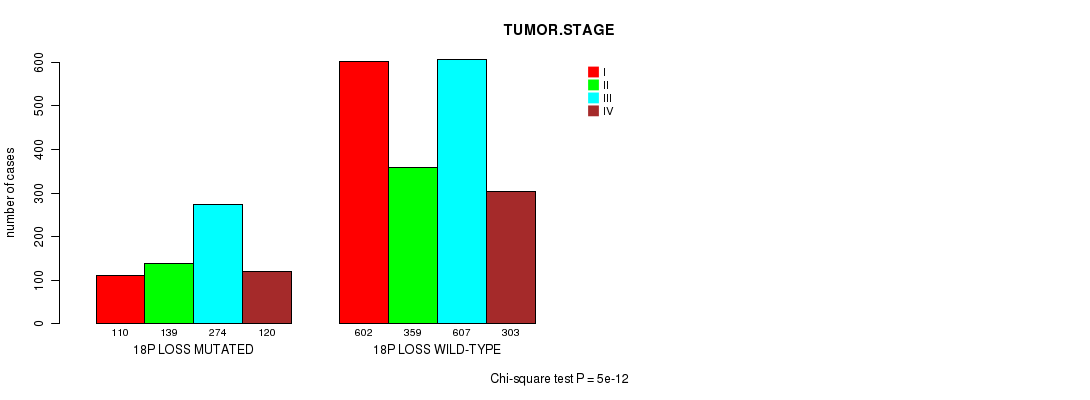
P value = 4.81e-12 (Fisher's exact test), Q value = 6.8e-09
Table S366. Clustering Approach #72: '18p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 18P LOSS MUTATED | 99 | 832 |
| 18P LOSS WILD-TYPE | 727 | 2905 |
Figure S294. Get High-res Image Clustering Approach #72: '18p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
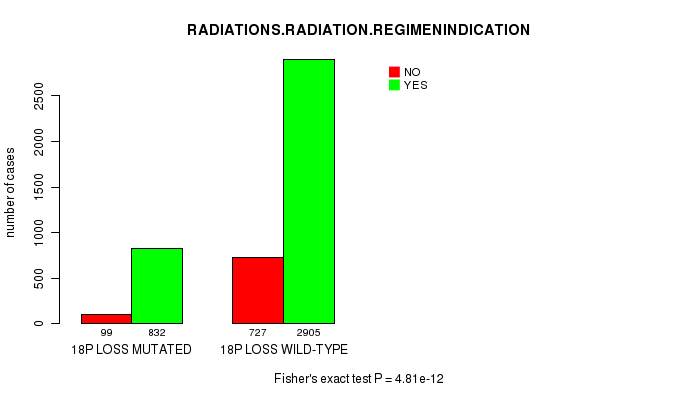
Table S367. Get Full Table Description of clustering approach #73: '18q loss mutation analysis'
| Cluster Labels | 18Q LOSS MUTATED | 18Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 1126 | 3437 |
P value = 3.22e-05 (t-test), Q value = 0.042
Table S368. Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 4507 | 61.9 (12.6) |
| 18Q LOSS MUTATED | 1109 | 63.2 (12.2) |
| 18Q LOSS WILD-TYPE | 3398 | 61.4 (12.7) |
Figure S295. Get High-res Image Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #2: 'AGE'
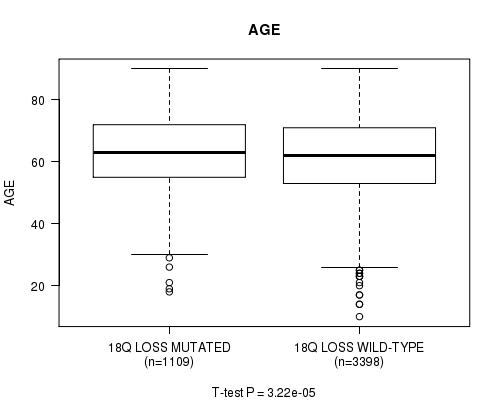
P value = 6.45e-148 (Chi-square test), Q value = 1e-144
Table S369. Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 18Q LOSS MUTATED | 31 | 43 | 168 | 226 | 41 | 85 | 48 | 139 | 1 | 217 | 0 | 123 |
| 18Q LOSS WILD-TYPE | 94 | 501 | 678 | 186 | 406 | 234 | 445 | 523 | 1 | 331 | 2 | 36 |
Figure S296. Get High-res Image Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
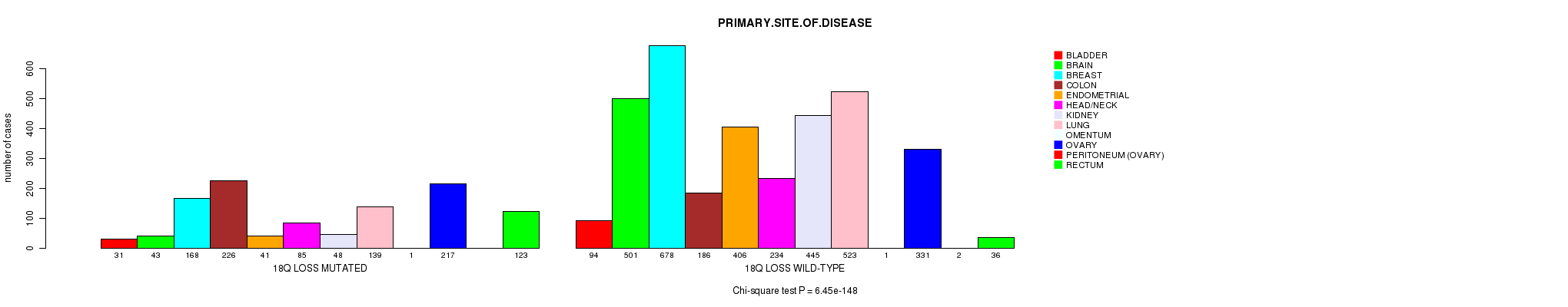
P value = 1.9e-112 (Chi-square test), Q value = 3e-109
Table S370. Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 18Q LOSS MUTATED | 215 | 10 | 20 | 85 | 48 | 3 | 23 | 42 | 0 | 0 | 5 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 57 | 6 | 3 | 113 | 9 | 218 | 15 | 0 | 5 |
| 18Q LOSS WILD-TYPE | 141 | 44 | 330 | 234 | 445 | 7 | 52 | 163 | 7 | 3 | 10 | 1 | 1 | 1 | 12 | 0 | 1 | 1 | 2 | 258 | 12 | 4 | 30 | 4 | 334 | 64 | 1 | 15 |
Figure S297. Get High-res Image Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 3.09e-24 (Chi-square test), Q value = 4.6e-21
Table S371. Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 18Q LOSS MUTATED | 67 | 177 | 298 | 77 |
| 18Q LOSS WILD-TYPE | 399 | 455 | 396 | 132 |
Figure S298. Get High-res Image Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'

P value = 8.37e-07 (Chi-square test), Q value = 0.0011
Table S372. Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #8: 'PATHOLOGY.N'
| nPatients | N0 | N1 | N2 | N3 |
|---|---|---|---|---|
| ALL | 1067 | 353 | 282 | 11 |
| 18Q LOSS MUTATED | 311 | 148 | 121 | 4 |
| 18Q LOSS WILD-TYPE | 756 | 205 | 161 | 7 |
Figure S299. Get High-res Image Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #8: 'PATHOLOGY.N'
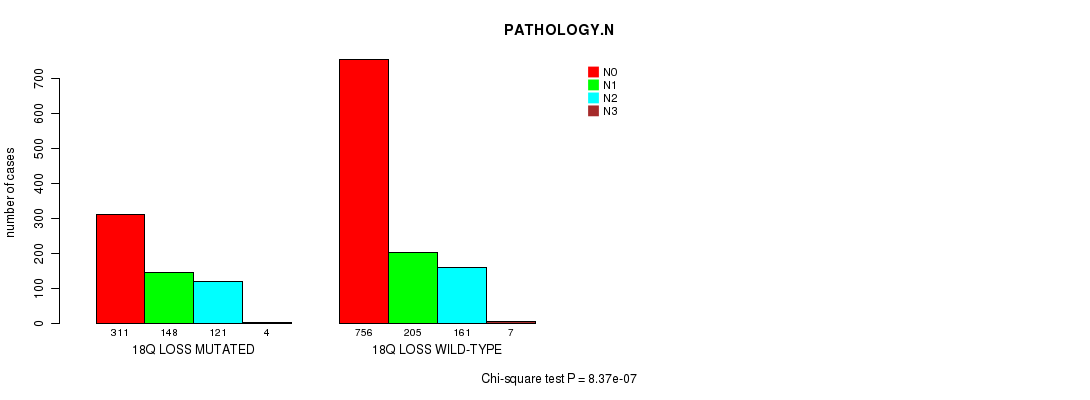
P value = 2.29e-05 (Chi-square test), Q value = 0.03
Table S373. Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
| nPatients | M0 | M1 | M1A | M1B | MX |
|---|---|---|---|---|---|
| ALL | 1424 | 168 | 9 | 3 | 165 |
| 18Q LOSS MUTATED | 408 | 73 | 7 | 0 | 54 |
| 18Q LOSS WILD-TYPE | 1016 | 95 | 2 | 3 | 111 |
Figure S300. Get High-res Image Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #9: 'PATHOLOGICSPREAD(M)'
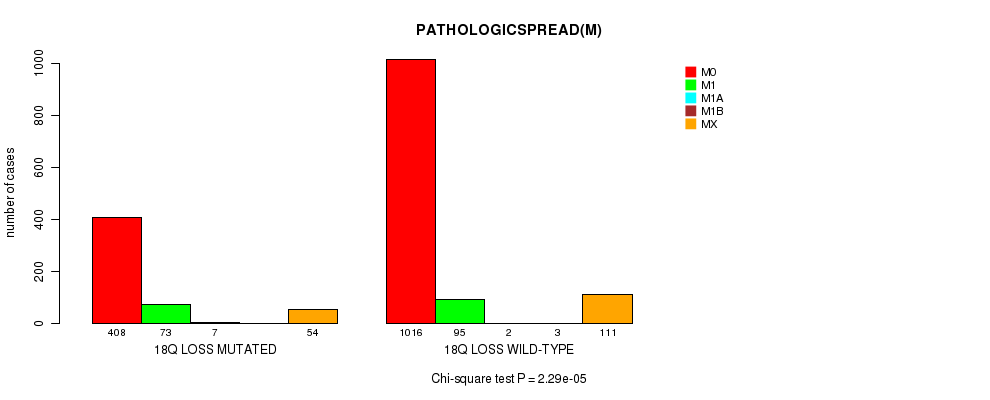
P value = 5.26e-14 (Chi-square test), Q value = 7.5e-11
Table S374. Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 18Q LOSS MUTATED | 146 | 182 | 328 | 160 |
| 18Q LOSS WILD-TYPE | 566 | 316 | 553 | 263 |
Figure S301. Get High-res Image Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
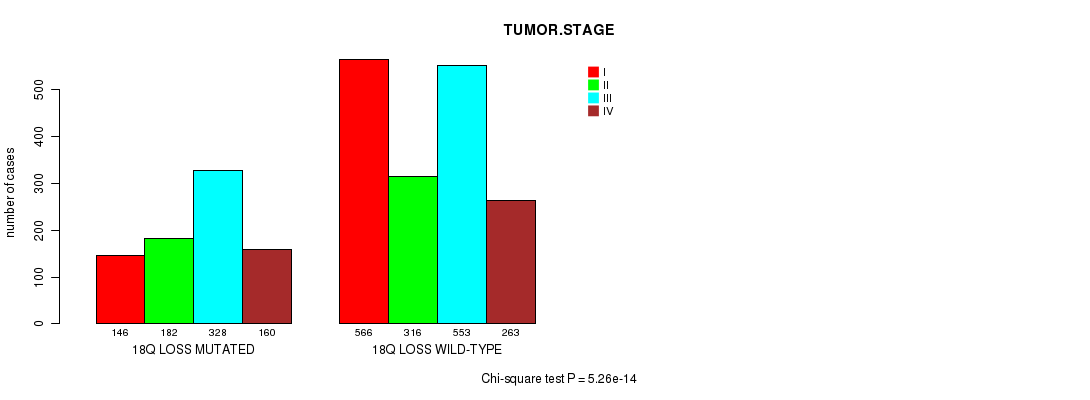
P value = 2.46e-19 (Fisher's exact test), Q value = 3.6e-16
Table S375. Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 18Q LOSS MUTATED | 108 | 1018 |
| 18Q LOSS WILD-TYPE | 718 | 2719 |
Figure S302. Get High-res Image Clustering Approach #73: '18q loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
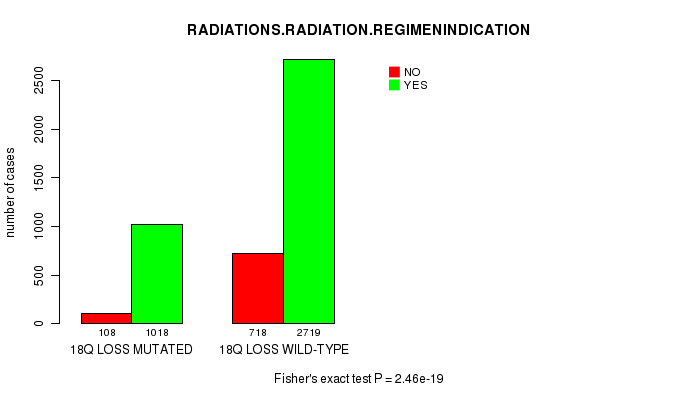
Table S376. Get Full Table Description of clustering approach #74: '19p loss mutation analysis'
| Cluster Labels | 19P LOSS MUTATED | 19P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 446 | 4117 |
P value = 6.43e-67 (Chi-square test), Q value = 1e-63
Table S377. Clustering Approach #74: '19p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 19P LOSS MUTATED | 8 | 17 | 66 | 16 | 30 | 23 | 4 | 137 | 1 | 137 | 0 | 7 |
| 19P LOSS WILD-TYPE | 117 | 527 | 780 | 396 | 417 | 296 | 489 | 525 | 1 | 411 | 2 | 152 |
Figure S303. Get High-res Image Clustering Approach #74: '19p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
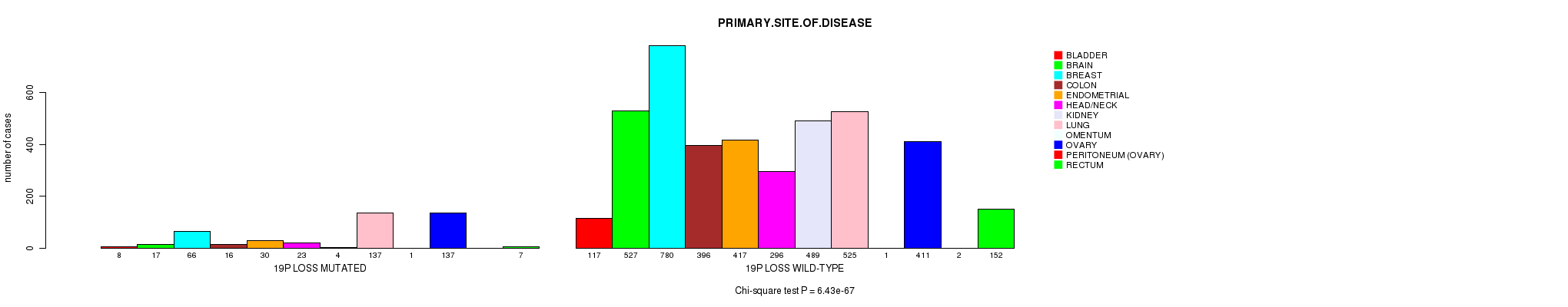
P value = 3.63e-51 (Chi-square test), Q value = 5.6e-48
Table S378. Clustering Approach #74: '19p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 19P LOSS MUTATED | 15 | 1 | 10 | 23 | 4 | 2 | 24 | 47 | 0 | 0 | 5 | 0 | 1 | 1 | 3 | 0 | 0 | 1 | 1 | 50 | 2 | 2 | 6 | 0 | 138 | 18 | 0 | 2 |
| 19P LOSS WILD-TYPE | 341 | 53 | 340 | 296 | 489 | 8 | 51 | 158 | 7 | 3 | 10 | 2 | 2 | 1 | 10 | 1 | 1 | 0 | 1 | 265 | 16 | 5 | 137 | 13 | 414 | 61 | 1 | 18 |
Figure S304. Get High-res Image Clustering Approach #74: '19p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 2.22e-07 (Chi-square test), Q value = 3e-04
Table S379. Clustering Approach #74: '19p loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
| nPatients | T0+T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| ALL | 466 | 632 | 694 | 209 |
| 19P LOSS MUTATED | 37 | 90 | 36 | 22 |
| 19P LOSS WILD-TYPE | 429 | 542 | 658 | 187 |
Figure S305. Get High-res Image Clustering Approach #74: '19p loss mutation analysis' versus Clinical Feature #7: 'PATHOLOGY.T'
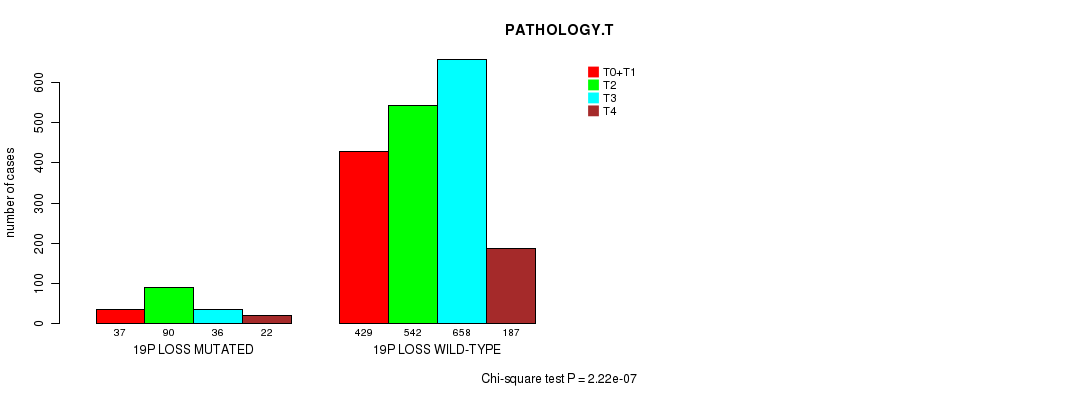
P value = 3.85e-06 (Chi-square test), Q value = 0.0051
Table S380. Clustering Approach #74: '19p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 19P LOSS MUTATED | 71 | 54 | 154 | 41 |
| 19P LOSS WILD-TYPE | 641 | 444 | 727 | 382 |
Figure S306. Get High-res Image Clustering Approach #74: '19p loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
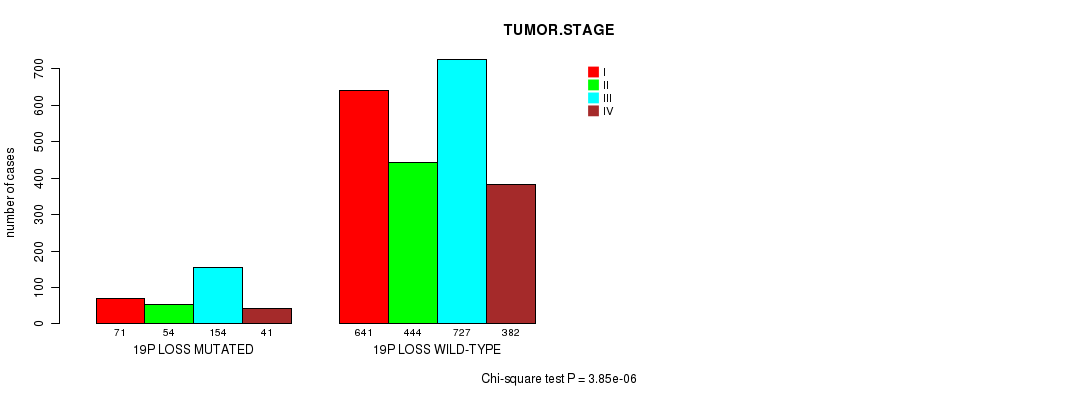
P value = 2.75e-06 (Fisher's exact test), Q value = 0.0037
Table S381. Clustering Approach #74: '19p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 19P LOSS MUTATED | 46 | 400 |
| 19P LOSS WILD-TYPE | 780 | 3337 |
Figure S307. Get High-res Image Clustering Approach #74: '19p loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
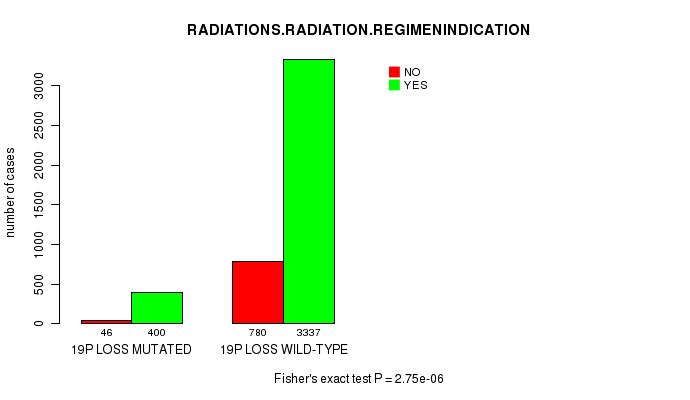
Table S382. Get Full Table Description of clustering approach #75: '19q loss mutation analysis'
| Cluster Labels | 19Q LOSS MUTATED | 19Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 362 | 4201 |
P value = 5.1e-64 (Chi-square test), Q value = 7.9e-61
Table S383. Clustering Approach #75: '19q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 19Q LOSS MUTATED | 3 | 22 | 51 | 14 | 23 | 17 | 1 | 86 | 1 | 137 | 0 | 7 |
| 19Q LOSS WILD-TYPE | 122 | 522 | 795 | 398 | 424 | 302 | 492 | 576 | 1 | 411 | 2 | 152 |
Figure S308. Get High-res Image Clustering Approach #75: '19q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
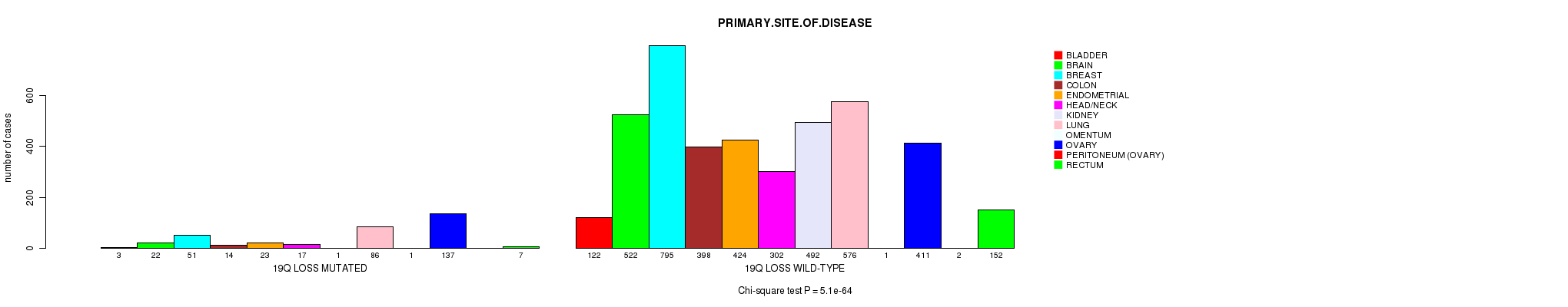
P value = 5.16e-07 (Fisher's exact test), Q value = 7e-04
Table S384. Clustering Approach #75: '19q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 19Q LOSS MUTATED | 272 | 90 |
| 19Q LOSS WILD-TYPE | 2609 | 1592 |
Figure S309. Get High-res Image Clustering Approach #75: '19q loss mutation analysis' versus Clinical Feature #4: 'GENDER'

P value = 1.28e-50 (Chi-square test), Q value = 2e-47
Table S385. Clustering Approach #75: '19q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 19Q LOSS MUTATED | 13 | 1 | 9 | 17 | 1 | 2 | 13 | 27 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 1 | 0 | 1 | 2 | 34 | 2 | 1 | 6 | 0 | 138 | 12 | 0 | 3 |
| 19Q LOSS WILD-TYPE | 343 | 53 | 341 | 302 | 492 | 8 | 62 | 178 | 7 | 3 | 13 | 2 | 3 | 2 | 10 | 0 | 1 | 0 | 0 | 281 | 16 | 6 | 137 | 13 | 414 | 67 | 1 | 17 |
Figure S310. Get High-res Image Clustering Approach #75: '19q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 8.01e-10 (Chi-square test), Q value = 1.1e-06
Table S386. Clustering Approach #75: '19q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 19Q LOSS MUTATED | 48 | 37 | 139 | 34 |
| 19Q LOSS WILD-TYPE | 664 | 461 | 742 | 389 |
Figure S311. Get High-res Image Clustering Approach #75: '19q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
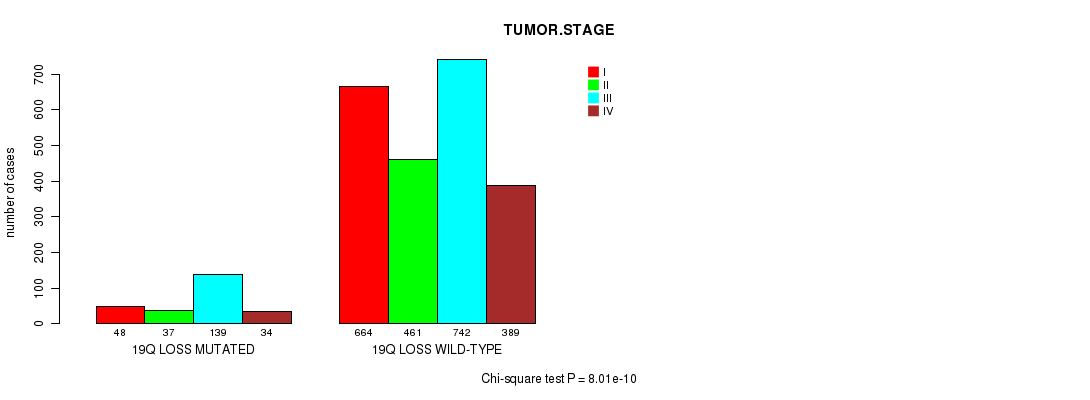
Table S387. Get Full Table Description of clustering approach #76: '20p loss mutation analysis'
| Cluster Labels | 20P LOSS MUTATED | 20P LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 253 | 4310 |
P value = 1.02e-10 (Chi-square test), Q value = 1.4e-07
Table S388. Clustering Approach #76: '20p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 20P LOSS MUTATED | 3 | 12 | 48 | 33 | 15 | 21 | 6 | 60 | 0 | 35 | 0 | 19 |
| 20P LOSS WILD-TYPE | 122 | 532 | 798 | 379 | 432 | 298 | 487 | 602 | 2 | 513 | 2 | 140 |
Figure S312. Get High-res Image Clustering Approach #76: '20p loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
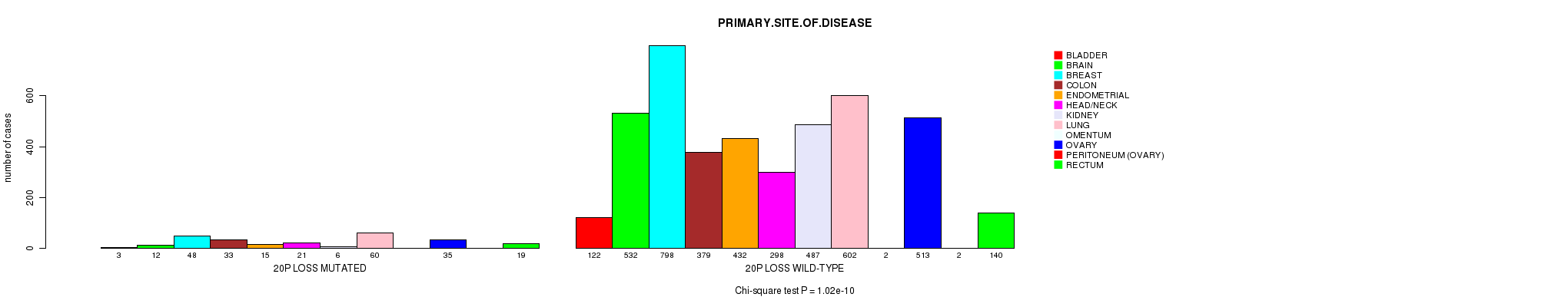
P value = 5.75e-09 (Chi-square test), Q value = 8e-06
Table S389. Clustering Approach #76: '20p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 20P LOSS MUTATED | 31 | 2 | 7 | 21 | 6 | 1 | 11 | 28 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 16 | 1 | 1 | 17 | 2 | 35 | 7 | 0 | 0 |
| 20P LOSS WILD-TYPE | 325 | 52 | 343 | 298 | 487 | 9 | 64 | 177 | 7 | 3 | 14 | 2 | 2 | 2 | 13 | 1 | 1 | 1 | 1 | 299 | 17 | 6 | 126 | 11 | 517 | 72 | 1 | 20 |
Figure S313. Get High-res Image Clustering Approach #76: '20p loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S390. Get Full Table Description of clustering approach #77: '20q loss mutation analysis'
| Cluster Labels | 20Q LOSS MUTATED | 20Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 108 | 4455 |
P value = 9.63e-09 (Chi-square test), Q value = 1.3e-05
Table S391. Clustering Approach #77: '20q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 20Q LOSS MUTATED | 1 | 11 | 22 | 2 | 5 | 10 | 0 | 36 | 0 | 21 | 0 | 0 |
| 20Q LOSS WILD-TYPE | 124 | 533 | 824 | 410 | 442 | 309 | 493 | 626 | 2 | 527 | 2 | 159 |
Figure S314. Get High-res Image Clustering Approach #77: '20q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
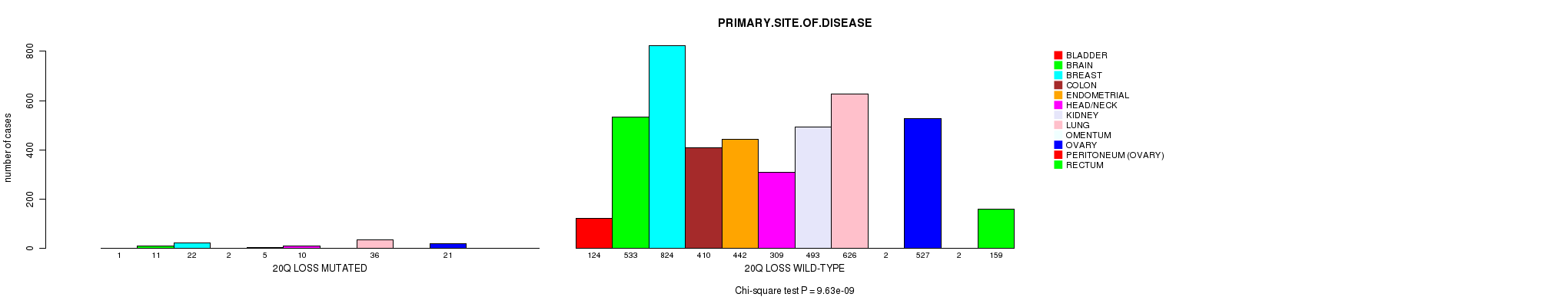
P value = 8.42e-07 (Chi-square test), Q value = 0.0011
Table S392. Clustering Approach #77: '20q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 20Q LOSS MUTATED | 2 | 0 | 3 | 10 | 0 | 0 | 6 | 10 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 17 | 1 | 1 | 0 | 0 | 21 | 1 | 0 | 1 |
| 20Q LOSS WILD-TYPE | 354 | 54 | 347 | 309 | 493 | 10 | 69 | 195 | 7 | 3 | 14 | 2 | 2 | 2 | 13 | 1 | 1 | 1 | 2 | 298 | 17 | 6 | 143 | 13 | 531 | 78 | 1 | 19 |
Figure S315. Get High-res Image Clustering Approach #77: '20q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

Table S393. Get Full Table Description of clustering approach #78: '21q loss mutation analysis'
| Cluster Labels | 21Q LOSS MUTATED | 21Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 647 | 3916 |
P value = 2.1e-46 (Chi-square test), Q value = 3.2e-43
Table S394. Clustering Approach #78: '21q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 21Q LOSS MUTATED | 13 | 27 | 80 | 71 | 26 | 58 | 33 | 143 | 0 | 151 | 0 | 44 |
| 21Q LOSS WILD-TYPE | 112 | 517 | 766 | 341 | 421 | 261 | 460 | 519 | 2 | 397 | 2 | 115 |
Figure S316. Get High-res Image Clustering Approach #78: '21q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
P value = 1.61e-27 (Chi-square test), Q value = 2.4e-24
Table S395. Clustering Approach #78: '21q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 21Q LOSS MUTATED | 67 | 4 | 10 | 58 | 33 | 3 | 11 | 27 | 2 | 0 | 1 | 0 | 1 | 0 | 3 | 1 | 0 | 0 | 1 | 93 | 3 | 0 | 40 | 2 | 151 | 13 | 0 | 2 |
| 21Q LOSS WILD-TYPE | 289 | 50 | 340 | 261 | 460 | 7 | 64 | 178 | 5 | 3 | 14 | 2 | 2 | 2 | 10 | 0 | 1 | 1 | 1 | 222 | 15 | 7 | 103 | 11 | 401 | 66 | 1 | 18 |
Figure S317. Get High-res Image Clustering Approach #78: '21q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 5.65e-06 (Chi-square test), Q value = 0.0075
Table S396. Clustering Approach #78: '21q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 21Q LOSS MUTATED | 115 | 78 | 219 | 74 |
| 21Q LOSS WILD-TYPE | 597 | 420 | 662 | 349 |
Figure S318. Get High-res Image Clustering Approach #78: '21q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
P value = 1.08e-07 (Fisher's exact test), Q value = 0.00015
Table S397. Clustering Approach #78: '21q loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 826 | 3737 |
| 21Q LOSS MUTATED | 71 | 576 |
| 21Q LOSS WILD-TYPE | 755 | 3161 |
Figure S319. Get High-res Image Clustering Approach #78: '21q loss mutation analysis' versus Clinical Feature #11: 'RADIATIONS.RADIATION.REGIMENINDICATION'
P value = 5.24e-05 (t-test), Q value = 0.068
Table S398. Clustering Approach #78: '21q loss mutation analysis' versus Clinical Feature #13: 'STOPPEDSMOKINGYEAR'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 568 | 1995.2 (13.5) |
| 21Q LOSS MUTATED | 112 | 1999.2 (10.8) |
| 21Q LOSS WILD-TYPE | 456 | 1994.2 (13.9) |
Figure S320. Get High-res Image Clustering Approach #78: '21q loss mutation analysis' versus Clinical Feature #13: 'STOPPEDSMOKINGYEAR'

Table S399. Get Full Table Description of clustering approach #79: '22q loss mutation analysis'
| Cluster Labels | 22Q LOSS MUTATED | 22Q LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 1165 | 3398 |
P value = 5.47e-05 (logrank test), Q value = 0.071
Table S400. Clustering Approach #79: '22q loss mutation analysis' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 4296 | 1401 | 0.0 - 224.0 (14.9) |
| 22Q LOSS MUTATED | 1090 | 427 | 0.0 - 224.0 (16.1) |
| 22Q LOSS WILD-TYPE | 3206 | 974 | 0.0 - 223.4 (14.2) |
Figure S321. Get High-res Image Clustering Approach #79: '22q loss mutation analysis' versus Clinical Feature #1: 'Time to Death'
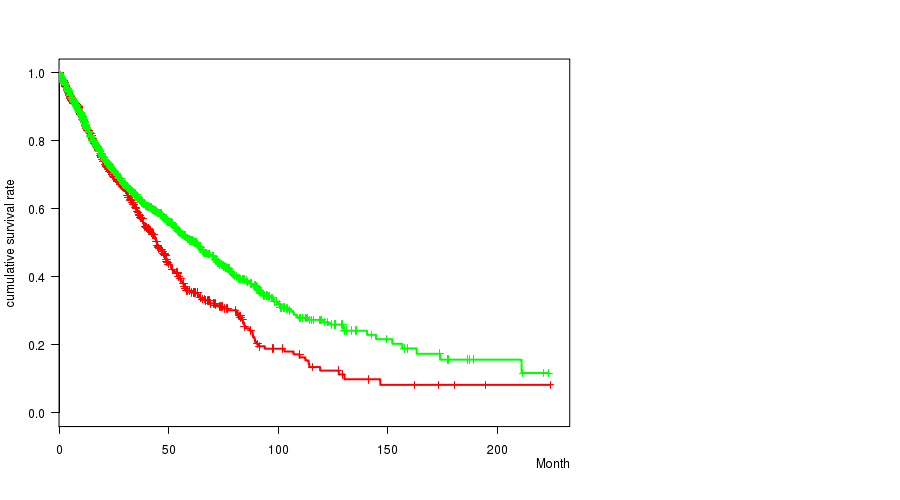
P value = 2.28e-167 (Chi-square test), Q value = 3.6e-164
Table S401. Clustering Approach #79: '22q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| 22Q LOSS MUTATED | 23 | 146 | 287 | 80 | 72 | 24 | 11 | 106 | 1 | 373 | 1 | 40 |
| 22Q LOSS WILD-TYPE | 102 | 398 | 559 | 332 | 375 | 295 | 482 | 556 | 1 | 175 | 1 | 119 |
Figure S322. Get High-res Image Clustering Approach #79: '22q loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
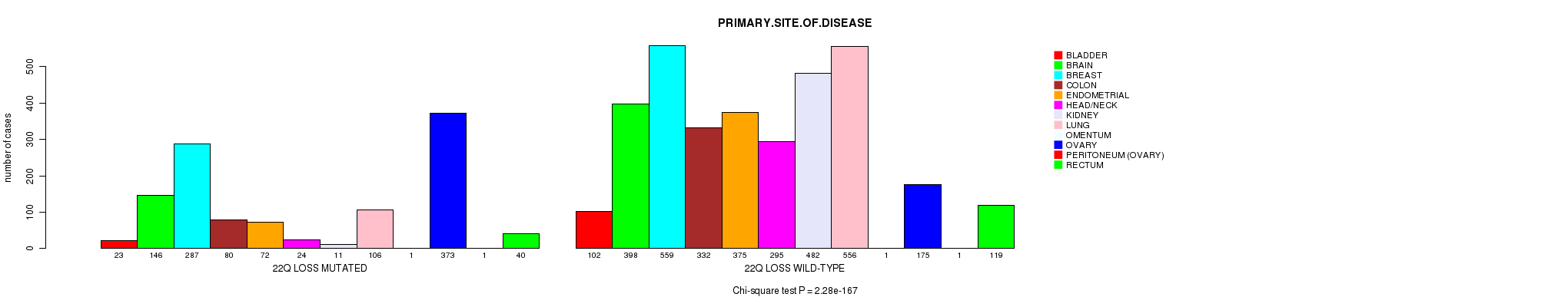
P value = 5.59e-34 (Fisher's exact test), Q value = 8.4e-31
Table S402. Clustering Approach #79: '22q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| 22Q LOSS MUTATED | 904 | 261 |
| 22Q LOSS WILD-TYPE | 1977 | 1421 |
Figure S323. Get High-res Image Clustering Approach #79: '22q loss mutation analysis' versus Clinical Feature #4: 'GENDER'
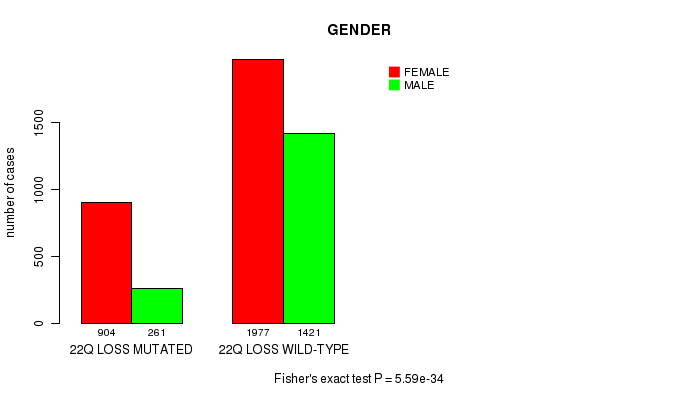
P value = 8.78e-170 (Chi-square test), Q value = 1.4e-166
Table S403. Clustering Approach #79: '22q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| 22Q LOSS MUTATED | 79 | 1 | 36 | 24 | 11 | 4 | 15 | 44 | 1 | 0 | 5 | 0 | 1 | 0 | 2 | 1 | 0 | 1 | 1 | 30 | 6 | 1 | 36 | 4 | 375 | 30 | 0 | 6 |
| 22Q LOSS WILD-TYPE | 277 | 53 | 314 | 295 | 482 | 6 | 60 | 161 | 6 | 3 | 10 | 2 | 2 | 2 | 11 | 0 | 1 | 0 | 1 | 285 | 12 | 6 | 107 | 9 | 177 | 49 | 1 | 14 |
Figure S324. Get High-res Image Clustering Approach #79: '22q loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 3.92e-53 (Chi-square test), Q value = 6e-50
Table S404. Clustering Approach #79: '22q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| 22Q LOSS MUTATED | 75 | 83 | 374 | 89 |
| 22Q LOSS WILD-TYPE | 637 | 415 | 507 | 334 |
Figure S325. Get High-res Image Clustering Approach #79: '22q loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'

Table S405. Get Full Table Description of clustering approach #80: 'Xq loss mutation analysis'
| Cluster Labels | XQ LOSS MUTATED | XQ LOSS WILD-TYPE |
|---|---|---|
| Number of samples | 200 | 4363 |
P value = 5.04e-48 (Chi-square test), Q value = 7.7e-45
Table S406. Clustering Approach #80: 'Xq loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
| nPatients | BLADDER | BRAIN | BREAST | COLON | ENDOMETRIAL | HEAD/NECK | KIDNEY | LUNG | OMENTUM | OVARY | PERITONEUM (OVARY) | RECTUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 125 | 544 | 846 | 412 | 447 | 319 | 493 | 662 | 2 | 548 | 2 | 159 |
| XQ LOSS MUTATED | 2 | 10 | 30 | 5 | 13 | 8 | 11 | 21 | 1 | 93 | 0 | 5 |
| XQ LOSS WILD-TYPE | 123 | 534 | 816 | 407 | 434 | 311 | 482 | 641 | 1 | 455 | 2 | 154 |
Figure S326. Get High-res Image Clustering Approach #80: 'Xq loss mutation analysis' versus Clinical Feature #3: 'PRIMARY.SITE.OF.DISEASE'
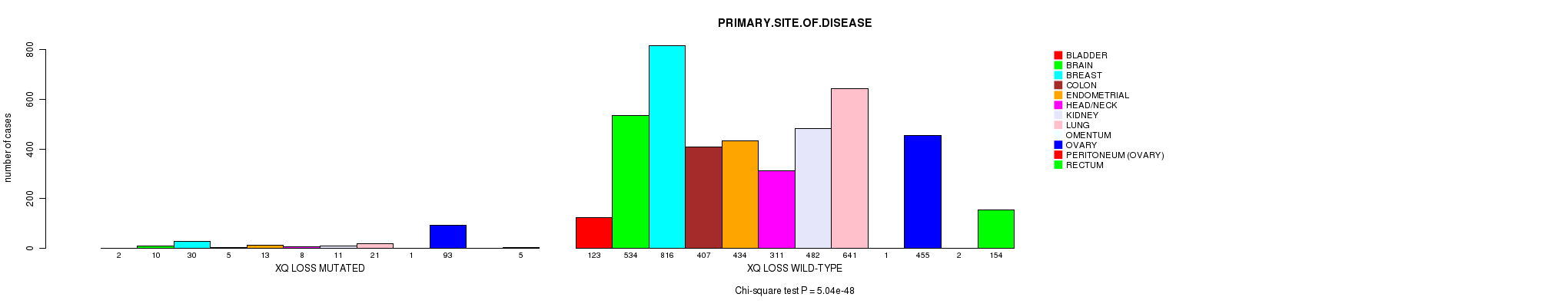
P value = 4.06e-07 (Fisher's exact test), Q value = 0.00055
Table S407. Clustering Approach #80: 'Xq loss mutation analysis' versus Clinical Feature #4: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 2881 | 1682 |
| XQ LOSS MUTATED | 159 | 41 |
| XQ LOSS WILD-TYPE | 2722 | 1641 |
Figure S327. Get High-res Image Clustering Approach #80: 'Xq loss mutation analysis' versus Clinical Feature #4: 'GENDER'
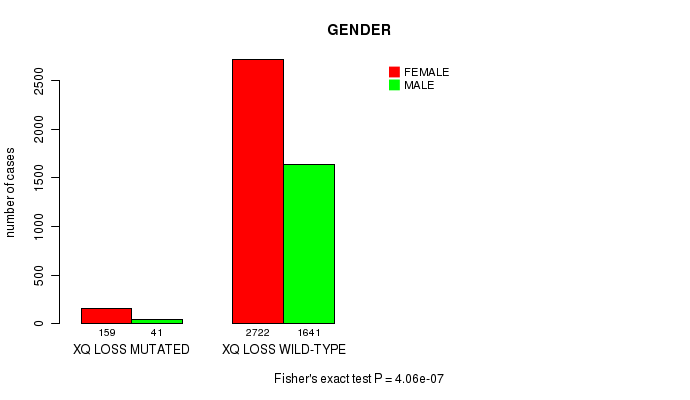
P value = 5.42e-35 (Chi-square test), Q value = 8.2e-32
Table S408. Clustering Approach #80: 'Xq loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'
| nPatients | COLON ADENOCARCINOMA | COLON MUCINOUS ADENOCARCINOMA | ENDOMETRIOID ENDOMETRIAL ADENOCARCINOMA | HEAD & NECK SQUAMOUS CELL CARCINOMA | KIDNEY CLEAR CELL RENAL CARCINOMA | LUNG ACINAR ADENOCARCINOMA | LUNG ADENOCARCINOMA MIXED SUBTYPE | LUNG ADENOCARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | LUNG BASALOID SQUAMOUS CELL CARCINOMA | LUNG BRONCHIOLOALVEOLAR CARCINOMA MUCINOUS | LUNG BRONCHIOLOALVEOLAR CARCINOMA NONMUCINOUS | LUNG CLEAR CELL ADENOCARCINOMA | LUNG MICROPAPILLARY ADENOCARCINOMA | LUNG MUCINOUS ADENOCARCINOMA | LUNG PAPILLARY ADENOCARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARCINOMA | LUNG PAPILLARY SQUAMOUS CELL CARICNOMA | LUNG SMALL CELL SQUAMOUS CELL CARCINOMA | LUNG SOLID PATTERN PREDOMINANT ADENOCARCINOMA | LUNG SQUAMOUS CELL CARCINOMA- NOT OTHERWISE SPECIFIED (NOS) | MIXED SEROUS AND ENDOMETRIOID | MUCINOUS (COLLOID) ADENOCARCINOMA | RECTAL ADENOCARCINOMA | RECTAL MUCINOUS ADENOCARCINOMA | SEROUS CYSTADENOCARCINOMA | SEROUS ENDOMETRIAL ADENOCARCINOMA | TREATED PRIMARY GBM | UNTREATED PRIMARY (DE NOVO) GBM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 356 | 54 | 350 | 319 | 493 | 10 | 75 | 205 | 7 | 3 | 15 | 2 | 3 | 2 | 13 | 1 | 1 | 1 | 2 | 315 | 18 | 7 | 143 | 13 | 552 | 79 | 1 | 20 |
| XQ LOSS MUTATED | 4 | 1 | 6 | 8 | 11 | 1 | 1 | 7 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 10 | 1 | 0 | 5 | 0 | 94 | 6 | 0 | 0 |
| XQ LOSS WILD-TYPE | 352 | 53 | 344 | 311 | 482 | 9 | 74 | 198 | 7 | 3 | 15 | 1 | 3 | 2 | 13 | 1 | 1 | 0 | 2 | 305 | 17 | 7 | 138 | 13 | 458 | 73 | 1 | 20 |
Figure S328. Get High-res Image Clustering Approach #80: 'Xq loss mutation analysis' versus Clinical Feature #6: 'HISTOLOGICAL.TYPE'

P value = 2.78e-10 (Chi-square test), Q value = 3.9e-07
Table S409. Clustering Approach #80: 'Xq loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
| nPatients | I | II | III | IV |
|---|---|---|---|---|
| ALL | 712 | 498 | 881 | 423 |
| XQ LOSS MUTATED | 21 | 14 | 87 | 20 |
| XQ LOSS WILD-TYPE | 691 | 484 | 794 | 403 |
Figure S329. Get High-res Image Clustering Approach #80: 'Xq loss mutation analysis' versus Clinical Feature #10: 'TUMOR.STAGE'
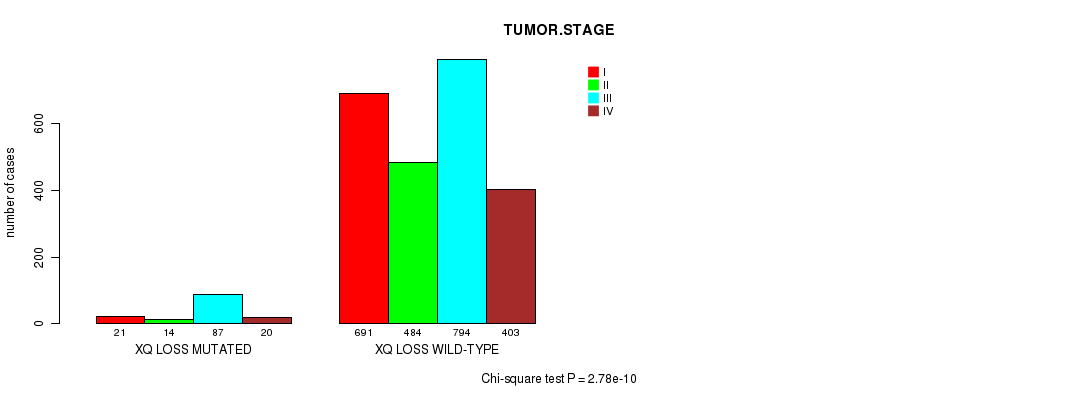
-
Cluster data file = broad_values_by_arm.mutsig.cluster.txt
-
Clinical data file = PANCAN12-TP.clin.merged.picked.txt
-
Number of patients = 4563
-
Number of clustering approaches = 80
-
Number of selected clinical features = 21
-
Exclude small clusters that include fewer than K patients, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For continuous numerical clinical features, two-tailed Student's t test with unequal variance (Lehmann and Romano 2005) was applied to compare the clinical values between two tumor subtypes using 't.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For binary clinical features, two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.