(primary solid tumor cohort)
This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 41 genes and 8 molecular subtypes across 323 patients, 20 significant findings detected with P value < 0.05 and Q value < 0.25.
-
BRAF mutation correlated to 'METHLYATION_CNMF', 'RPPA_CNMF', 'RPPA_CHIERARCHICAL', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
HRAS mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
NRAS mutation correlated to 'METHLYATION_CNMF', 'RPPA_CHIERARCHICAL', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between mutation status of 41 genes and 8 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 20 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| BRAF | 183 (57%) | 140 |
0.0194 (1.00) |
3.62e-38 (9.99e-36) |
9.4e-07 (0.000252) |
2.51e-11 (6.84e-09) |
4.03e-46 (1.13e-43) |
1.56e-39 (4.31e-37) |
9.88e-42 (2.75e-39) |
1.03e-43 (2.86e-41) |
| HRAS | 12 (4%) | 311 |
0.000222 (0.0582) |
1.48e-06 (0.000393) |
9.23e-05 (0.0243) |
0.00529 (1.00) |
1.37e-05 (0.00362) |
6.79e-06 (0.0018) |
1.31e-06 (0.00035) |
3.85e-07 (0.000104) |
| NRAS | 26 (8%) | 297 |
0.00692 (1.00) |
6.06e-14 (1.67e-11) |
1 (1.00) |
0.000583 (0.152) |
4.7e-11 (1.27e-08) |
4.03e-11 (1.09e-08) |
9.19e-13 (2.51e-10) |
2.47e-13 (6.78e-11) |
| EMG1 | 6 (2%) | 317 |
0.548 (1.00) |
1 (1.00) |
0.327 (1.00) |
0.0846 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.778 (1.00) |
| PTTG1IP | 4 (1%) | 319 |
1 (1.00) |
1 (1.00) |
0.242 (1.00) |
0.252 (1.00) |
0.469 (1.00) |
0.393 (1.00) |
||
| RPTN | 8 (2%) | 315 |
0.805 (1.00) |
0.442 (1.00) |
0.265 (1.00) |
0.801 (1.00) |
0.0249 (1.00) |
0.415 (1.00) |
0.82 (1.00) |
0.666 (1.00) |
| EIF1AX | 5 (2%) | 318 |
0.731 (1.00) |
0.0278 (1.00) |
0.444 (1.00) |
0.132 (1.00) |
0.0621 (1.00) |
0.0109 (1.00) |
0.0581 (1.00) |
0.236 (1.00) |
| CCDC15 | 5 (2%) | 318 |
0.731 (1.00) |
0.115 (1.00) |
1 (1.00) |
0.381 (1.00) |
0.132 (1.00) |
0.136 (1.00) |
0.63 (1.00) |
0.53 (1.00) |
| ZNF845 | 6 (2%) | 317 |
0.42 (1.00) |
0.637 (1.00) |
0.639 (1.00) |
0.125 (1.00) |
0.943 (1.00) |
0.166 (1.00) |
0.777 (1.00) |
0.328 (1.00) |
| ZNF878 | 4 (1%) | 319 |
1 (1.00) |
0.487 (1.00) |
0.0454 (1.00) |
0.833 (1.00) |
0.691 (1.00) |
1 (1.00) |
||
| TG | 16 (5%) | 307 |
1 (1.00) |
0.0205 (1.00) |
0.34 (1.00) |
0.492 (1.00) |
0.406 (1.00) |
0.293 (1.00) |
0.0452 (1.00) |
0.0253 (1.00) |
| PRB2 | 6 (2%) | 317 |
1 (1.00) |
0.637 (1.00) |
0.0801 (1.00) |
0.851 (1.00) |
0.474 (1.00) |
0.723 (1.00) |
0.522 (1.00) |
0.598 (1.00) |
| R3HDM2 | 4 (1%) | 319 |
0.0124 (1.00) |
0.063 (1.00) |
0.0699 (1.00) |
0.0388 (1.00) |
0.0191 (1.00) |
0.00822 (1.00) |
||
| IL32 | 3 (1%) | 320 |
1 (1.00) |
0.742 (1.00) |
1 (1.00) |
0.78 (1.00) |
0.202 (1.00) |
0.645 (1.00) |
||
| TMCO2 | 3 (1%) | 320 |
1 (1.00) |
0.416 (1.00) |
0.778 (1.00) |
0.2 (1.00) |
0.492 (1.00) |
0.303 (1.00) |
0.401 (1.00) |
0.778 (1.00) |
| PPTC7 | 3 (1%) | 320 |
0.0791 (1.00) |
0.0627 (1.00) |
0.172 (1.00) |
0.0916 (1.00) |
0.299 (1.00) |
0.403 (1.00) |
||
| MUC7 | 5 (2%) | 318 |
0.731 (1.00) |
0.372 (1.00) |
0.683 (1.00) |
0.26 (1.00) |
0.805 (1.00) |
0.848 (1.00) |
0.866 (1.00) |
0.629 (1.00) |
| LYPD3 | 3 (1%) | 320 |
0.547 (1.00) |
0.416 (1.00) |
0.492 (1.00) |
0.303 (1.00) |
0.644 (1.00) |
0.778 (1.00) |
||
| TMEM90B | 3 (1%) | 320 |
0.135 (1.00) |
1 (1.00) |
1 (1.00) |
0.78 (1.00) |
1 (1.00) |
1 (1.00) |
||
| ZNF799 | 5 (2%) | 318 |
0.465 (1.00) |
1 (1.00) |
0.505 (1.00) |
1 (1.00) |
0.592 (1.00) |
0.848 (1.00) |
0.451 (1.00) |
1 (1.00) |
| PPM1D | 5 (2%) | 318 |
1 (1.00) |
1 (1.00) |
0.778 (1.00) |
0.125 (1.00) |
0.693 (1.00) |
0.848 (1.00) |
0.866 (1.00) |
0.329 (1.00) |
| MAP3K3 | 4 (1%) | 319 |
0.061 (1.00) |
0.224 (1.00) |
0.459 (1.00) |
0.0846 (1.00) |
0.274 (1.00) |
0.184 (1.00) |
0.469 (1.00) |
0.555 (1.00) |
| TROAP | 3 (1%) | 320 |
1 (1.00) |
0.742 (1.00) |
0.778 (1.00) |
0.535 (1.00) |
0.746 (1.00) |
1 (1.00) |
0.519 (1.00) |
0.403 (1.00) |
| SYNPO2L | 3 (1%) | 320 |
0.547 (1.00) |
1 (1.00) |
1 (1.00) |
0.78 (1.00) |
1 (1.00) |
1 (1.00) |
||
| ATAD2 | 4 (1%) | 319 |
1 (1.00) |
0.823 (1.00) |
0.808 (1.00) |
1 (1.00) |
0.691 (1.00) |
1 (1.00) |
||
| KRAS | 3 (1%) | 320 |
1 (1.00) |
0.0627 (1.00) |
0.172 (1.00) |
0.0916 (1.00) |
0.0616 (1.00) |
0.0276 (1.00) |
||
| SLC5A11 | 3 (1%) | 320 |
0.547 (1.00) |
0.742 (1.00) |
0.299 (1.00) |
0.403 (1.00) |
||||
| PRG4 | 4 (1%) | 319 |
0.23 (1.00) |
0.104 (1.00) |
0.0128 (1.00) |
0.142 (1.00) |
0.469 (1.00) |
0.555 (1.00) |
||
| SCUBE2 | 3 (1%) | 320 |
1 (1.00) |
0.281 (1.00) |
0.314 (1.00) |
0.303 (1.00) |
0.644 (1.00) |
0.522 (1.00) |
||
| ZNF479 | 3 (1%) | 320 |
1 (1.00) |
0.742 (1.00) |
1 (1.00) |
0.78 (1.00) |
0.519 (1.00) |
1 (1.00) |
||
| SREBF2 | 3 (1%) | 320 |
1 (1.00) |
0.742 (1.00) |
0.568 (1.00) |
1 (1.00) |
0.401 (1.00) |
0.778 (1.00) |
||
| SLC26A11 | 3 (1%) | 320 |
0.0791 (1.00) |
1 (1.00) |
0.368 (1.00) |
0.78 (1.00) |
0.778 (1.00) |
0.301 (1.00) |
||
| TSC22D1 | 3 (1%) | 320 |
1 (1.00) |
1 (1.00) |
0.746 (1.00) |
1 (1.00) |
0.778 (1.00) |
0.645 (1.00) |
||
| ANKRD30A | 5 (2%) | 318 |
0.731 (1.00) |
0.462 (1.00) |
0.749 (1.00) |
0.507 (1.00) |
0.74 (1.00) |
0.865 (1.00) |
||
| FAM155A | 4 (1%) | 319 |
0.144 (1.00) |
0.156 (1.00) |
0.778 (1.00) |
0.535 (1.00) |
0.0372 (1.00) |
0.555 (1.00) |
||
| SLA | 3 (1%) | 320 |
0.547 (1.00) |
0.0627 (1.00) |
0.286 (1.00) |
0.285 (1.00) |
0.172 (1.00) |
0.0916 (1.00) |
0.0616 (1.00) |
0.0276 (1.00) |
| ZFHX3 | 10 (3%) | 313 |
0.171 (1.00) |
0.616 (1.00) |
0.48 (1.00) |
0.632 (1.00) |
0.794 (1.00) |
0.437 (1.00) |
0.613 (1.00) |
0.723 (1.00) |
| ARMCX3 | 3 (1%) | 320 |
0.547 (1.00) |
0.742 (1.00) |
0.646 (1.00) |
0.213 (1.00) |
0.519 (1.00) |
0.202 (1.00) |
||
| SLC25A45 | 3 (1%) | 320 |
0.135 (1.00) |
1 (1.00) |
0.202 (1.00) |
0.403 (1.00) |
||||
| COL5A3 | 6 (2%) | 317 |
0.752 (1.00) |
0.317 (1.00) |
0.778 (1.00) |
0.758 (1.00) |
0.044 (1.00) |
0.753 (1.00) |
0.877 (1.00) |
0.523 (1.00) |
| CDC27 | 3 (1%) | 320 |
1 (1.00) |
1 (1.00) |
0.368 (1.00) |
0.0446 (1.00) |
0.778 (1.00) |
0.301 (1.00) |
P value = 3.62e-38 (Fisher's exact test), Q value = 1e-35
Table S1. Gene #1: 'BRAF MUTATION STATUS' versus Clinical Feature #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 105 | 52 | 166 |
| BRAF MUTATED | 10 | 30 | 143 |
| BRAF WILD-TYPE | 95 | 22 | 23 |
Figure S1. Get High-res Image Gene #1: 'BRAF MUTATION STATUS' versus Clinical Feature #2: 'METHLYATION_CNMF'
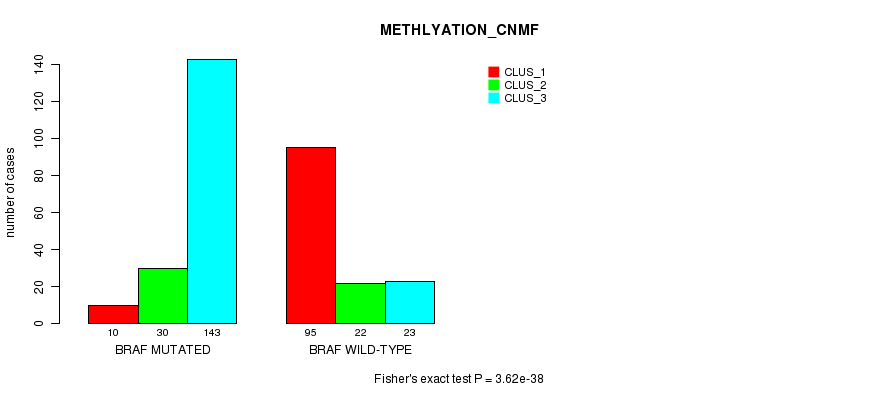
P value = 9.4e-07 (Fisher's exact test), Q value = 0.00025
Table S2. Gene #1: 'BRAF MUTATION STATUS' versus Clinical Feature #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 62 | 76 | 79 |
| BRAF MUTATED | 19 | 57 | 42 |
| BRAF WILD-TYPE | 43 | 19 | 37 |
Figure S2. Get High-res Image Gene #1: 'BRAF MUTATION STATUS' versus Clinical Feature #3: 'RPPA_CNMF'
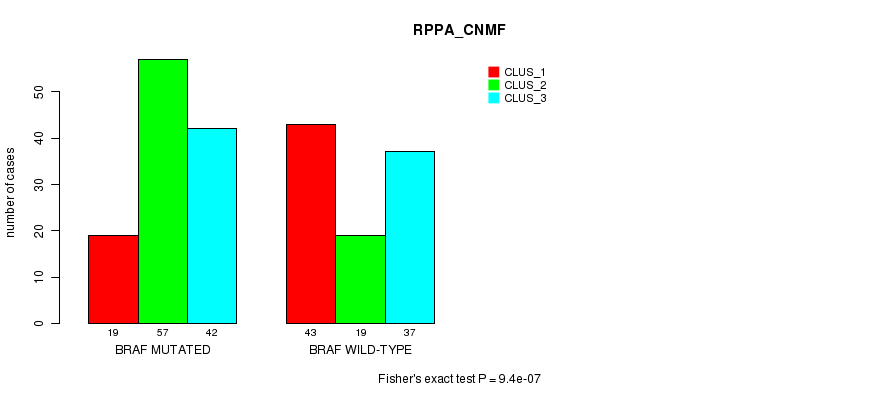
P value = 2.51e-11 (Fisher's exact test), Q value = 6.8e-09
Table S3. Gene #1: 'BRAF MUTATION STATUS' versus Clinical Feature #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 96 | 32 | 89 |
| BRAF MUTATED | 30 | 16 | 72 |
| BRAF WILD-TYPE | 66 | 16 | 17 |
Figure S3. Get High-res Image Gene #1: 'BRAF MUTATION STATUS' versus Clinical Feature #4: 'RPPA_CHIERARCHICAL'
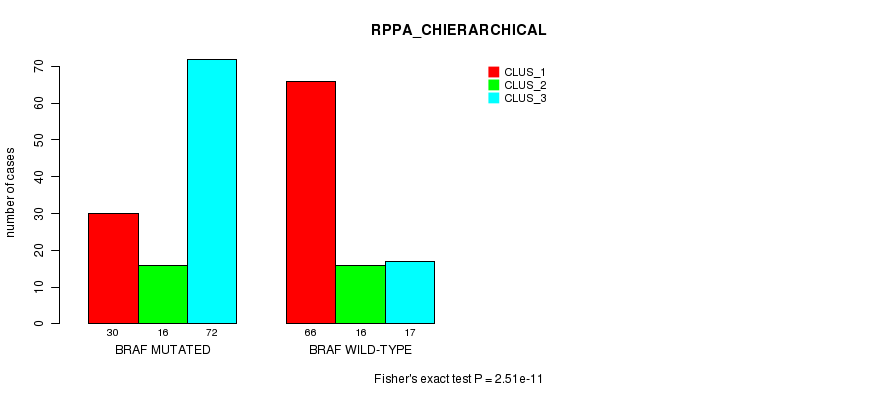
P value = 4.03e-46 (Fisher's exact test), Q value = 1.1e-43
Table S4. Gene #1: 'BRAF MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 102 | 36 | 70 | 95 |
| BRAF MUTATED | 6 | 20 | 57 | 90 |
| BRAF WILD-TYPE | 96 | 16 | 13 | 5 |
Figure S4. Get High-res Image Gene #1: 'BRAF MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
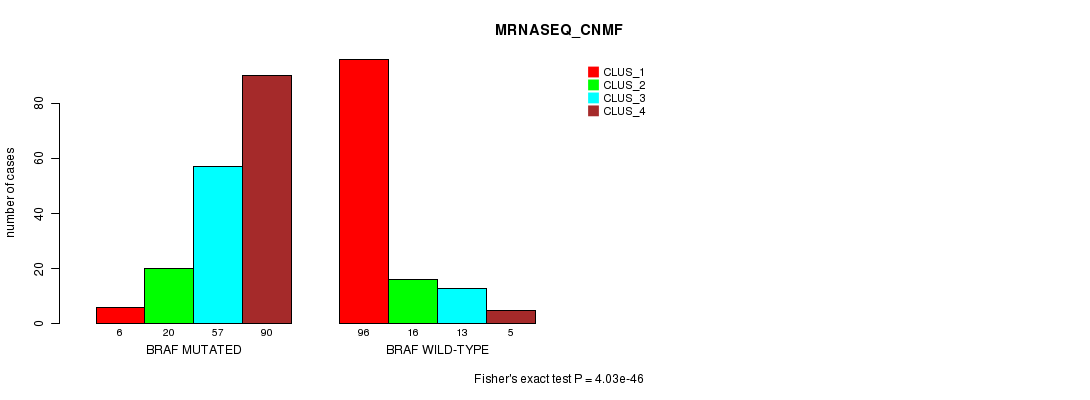
P value = 1.56e-39 (Fisher's exact test), Q value = 4.3e-37
Table S5. Gene #1: 'BRAF MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 110 | 136 | 57 |
| BRAF MUTATED | 11 | 120 | 42 |
| BRAF WILD-TYPE | 99 | 16 | 15 |
Figure S5. Get High-res Image Gene #1: 'BRAF MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
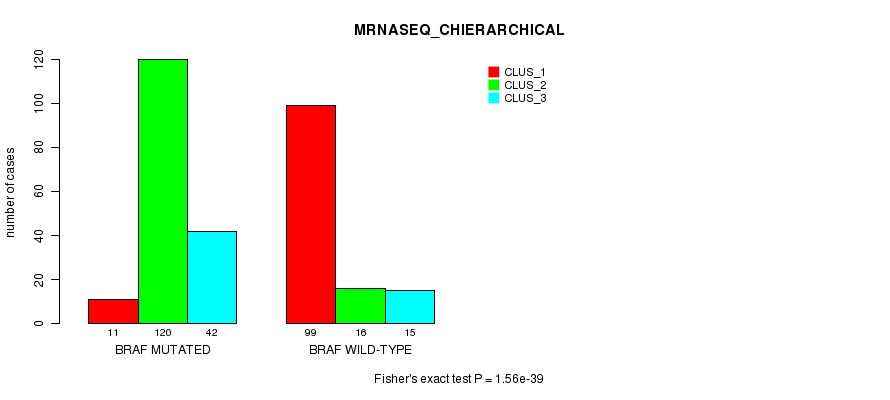
P value = 9.88e-42 (Fisher's exact test), Q value = 2.7e-39
Table S6. Gene #1: 'BRAF MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 105 | 119 | 98 |
| BRAF MUTATED | 6 | 103 | 73 |
| BRAF WILD-TYPE | 99 | 16 | 25 |
Figure S6. Get High-res Image Gene #1: 'BRAF MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
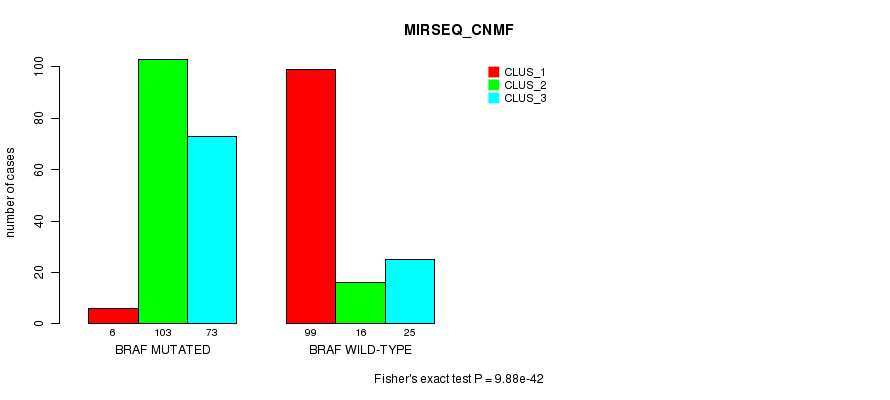
P value = 1.03e-43 (Fisher's exact test), Q value = 2.9e-41
Table S7. Gene #1: 'BRAF MUTATION STATUS' versus Clinical Feature #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 106 | 98 |
| BRAF MUTATED | 94 | 86 | 2 |
| BRAF WILD-TYPE | 24 | 20 | 96 |
Figure S7. Get High-res Image Gene #1: 'BRAF MUTATION STATUS' versus Clinical Feature #8: 'MIRSEQ_CHIERARCHICAL'
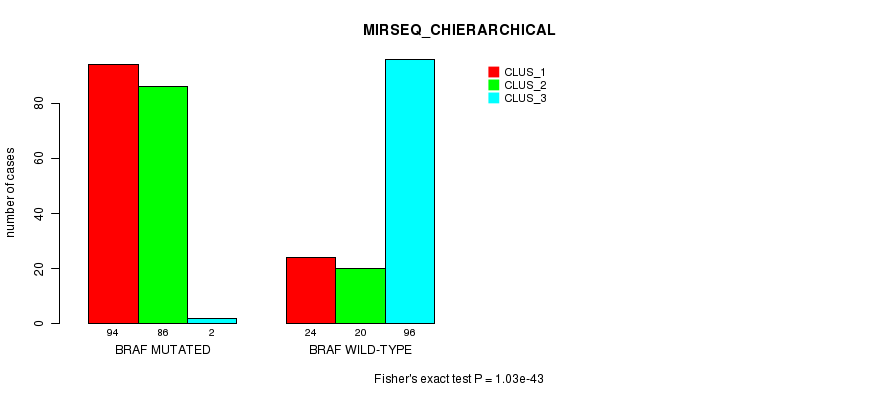
P value = 0.000222 (Fisher's exact test), Q value = 0.058
Table S8. Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 25 | 246 | 49 |
| HRAS MUTATED | 0 | 4 | 8 |
| HRAS WILD-TYPE | 25 | 242 | 41 |
Figure S8. Get High-res Image Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #1: 'CN_CNMF'
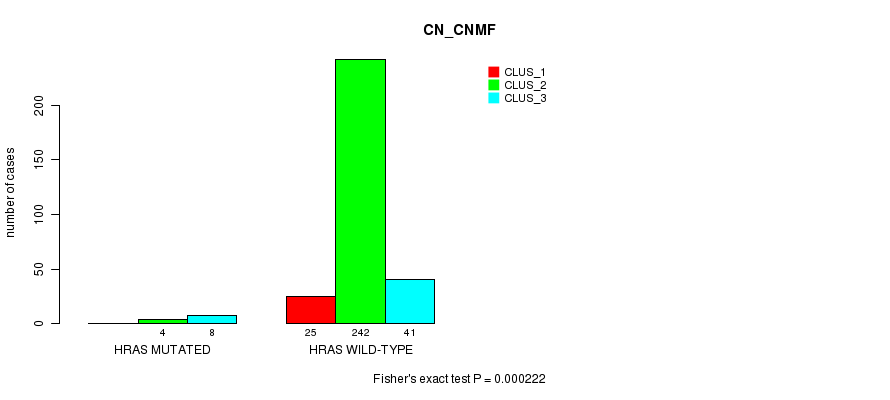
P value = 1.48e-06 (Fisher's exact test), Q value = 0.00039
Table S9. Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 105 | 52 | 166 |
| HRAS MUTATED | 12 | 0 | 0 |
| HRAS WILD-TYPE | 93 | 52 | 166 |
Figure S9. Get High-res Image Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #2: 'METHLYATION_CNMF'
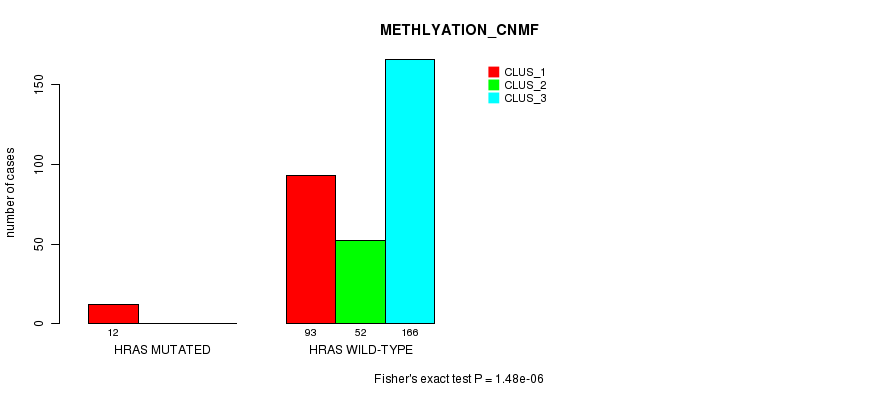
P value = 9.23e-05 (Fisher's exact test), Q value = 0.024
Table S10. Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 62 | 76 | 79 |
| HRAS MUTATED | 9 | 2 | 0 |
| HRAS WILD-TYPE | 53 | 74 | 79 |
Figure S10. Get High-res Image Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #3: 'RPPA_CNMF'
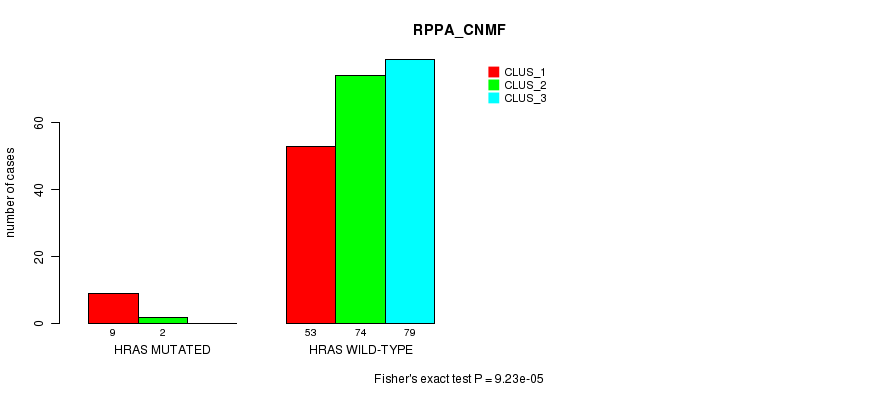
P value = 1.37e-05 (Fisher's exact test), Q value = 0.0036
Table S11. Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 102 | 36 | 70 | 95 |
| HRAS MUTATED | 12 | 0 | 0 | 0 |
| HRAS WILD-TYPE | 90 | 36 | 70 | 95 |
Figure S11. Get High-res Image Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
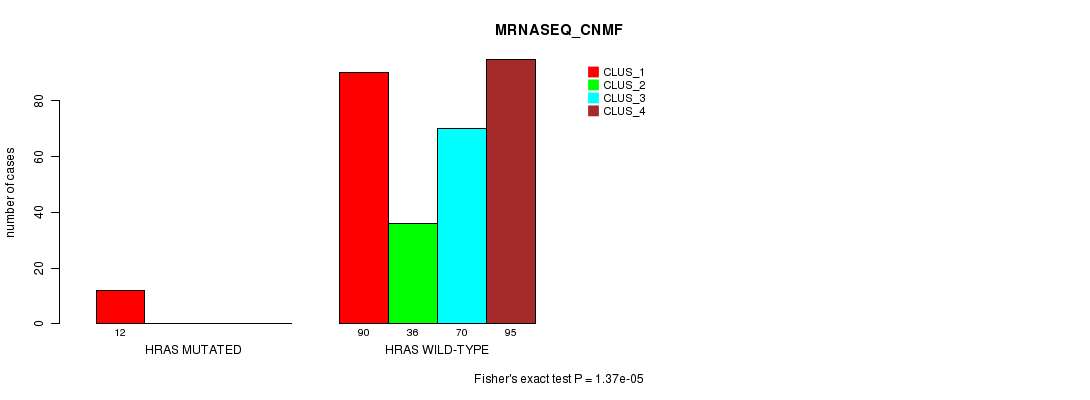
P value = 6.79e-06 (Fisher's exact test), Q value = 0.0018
Table S12. Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 110 | 136 | 57 |
| HRAS MUTATED | 12 | 0 | 0 |
| HRAS WILD-TYPE | 98 | 136 | 57 |
Figure S12. Get High-res Image Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
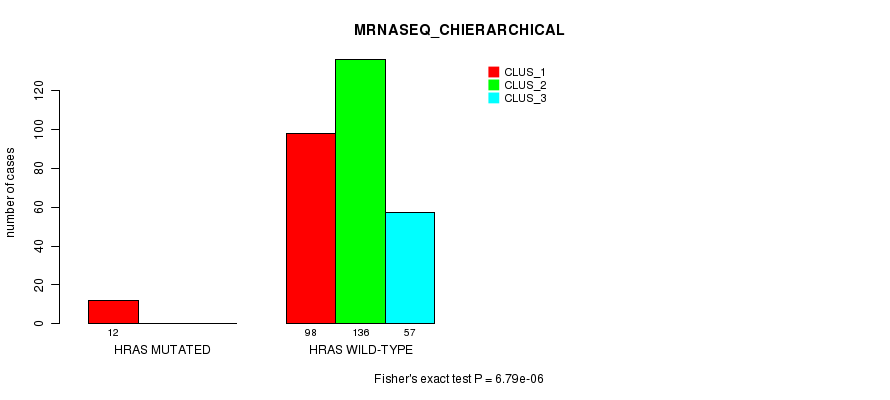
P value = 1.31e-06 (Fisher's exact test), Q value = 0.00035
Table S13. Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 105 | 119 | 98 |
| HRAS MUTATED | 12 | 0 | 0 |
| HRAS WILD-TYPE | 93 | 119 | 98 |
Figure S13. Get High-res Image Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
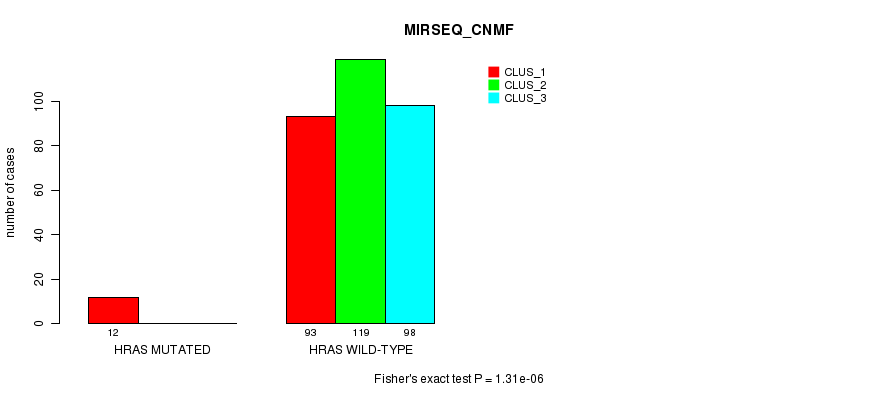
P value = 3.85e-07 (Fisher's exact test), Q value = 1e-04
Table S14. Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 106 | 98 |
| HRAS MUTATED | 0 | 0 | 12 |
| HRAS WILD-TYPE | 118 | 106 | 86 |
Figure S14. Get High-res Image Gene #3: 'HRAS MUTATION STATUS' versus Clinical Feature #8: 'MIRSEQ_CHIERARCHICAL'
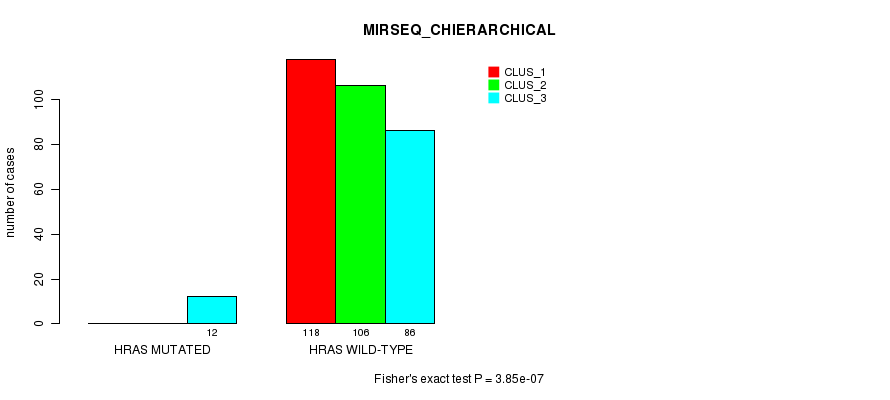
P value = 6.06e-14 (Fisher's exact test), Q value = 1.7e-11
Table S15. Gene #4: 'NRAS MUTATION STATUS' versus Clinical Feature #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 105 | 52 | 166 |
| NRAS MUTATED | 26 | 0 | 0 |
| NRAS WILD-TYPE | 79 | 52 | 166 |
Figure S15. Get High-res Image Gene #4: 'NRAS MUTATION STATUS' versus Clinical Feature #2: 'METHLYATION_CNMF'
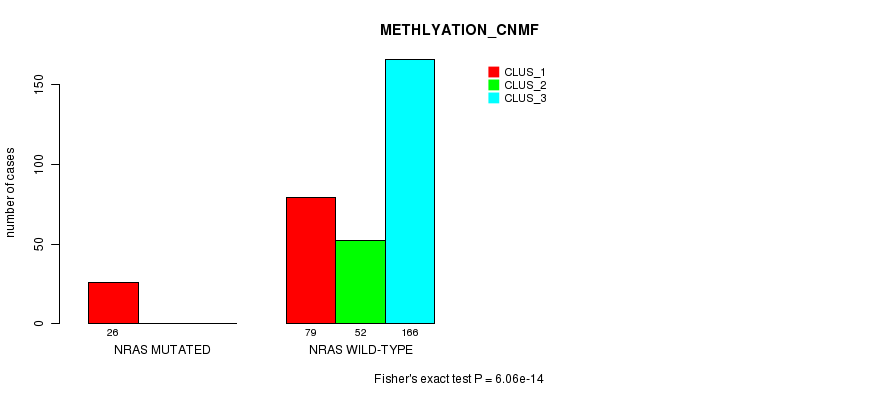
P value = 0.000583 (Fisher's exact test), Q value = 0.15
Table S16. Gene #4: 'NRAS MUTATION STATUS' versus Clinical Feature #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 96 | 32 | 89 |
| NRAS MUTATED | 9 | 5 | 0 |
| NRAS WILD-TYPE | 87 | 27 | 89 |
Figure S16. Get High-res Image Gene #4: 'NRAS MUTATION STATUS' versus Clinical Feature #4: 'RPPA_CHIERARCHICAL'
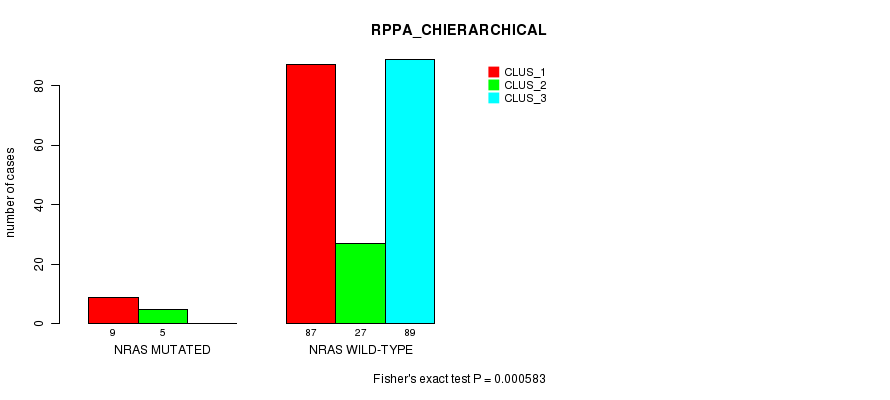
P value = 4.7e-11 (Fisher's exact test), Q value = 1.3e-08
Table S17. Gene #4: 'NRAS MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 102 | 36 | 70 | 95 |
| NRAS MUTATED | 23 | 0 | 0 | 0 |
| NRAS WILD-TYPE | 79 | 36 | 70 | 95 |
Figure S17. Get High-res Image Gene #4: 'NRAS MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
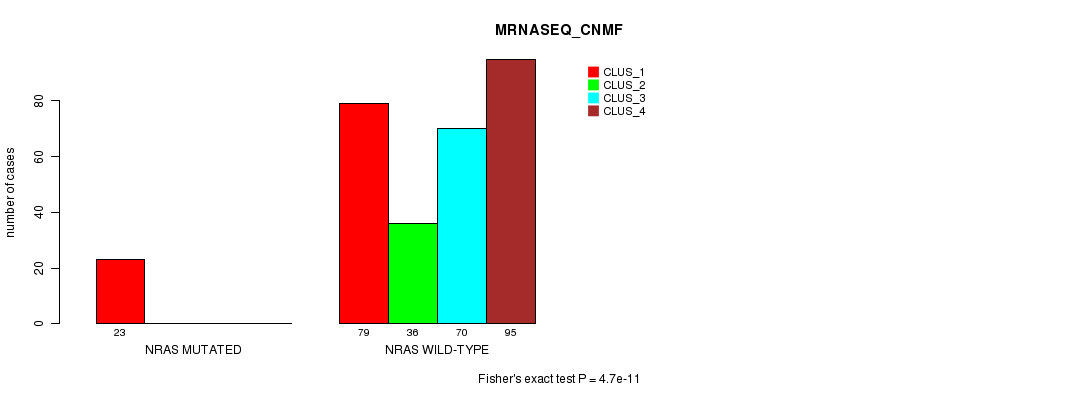
P value = 4.03e-11 (Fisher's exact test), Q value = 1.1e-08
Table S18. Gene #4: 'NRAS MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 110 | 136 | 57 |
| NRAS MUTATED | 23 | 0 | 0 |
| NRAS WILD-TYPE | 87 | 136 | 57 |
Figure S18. Get High-res Image Gene #4: 'NRAS MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
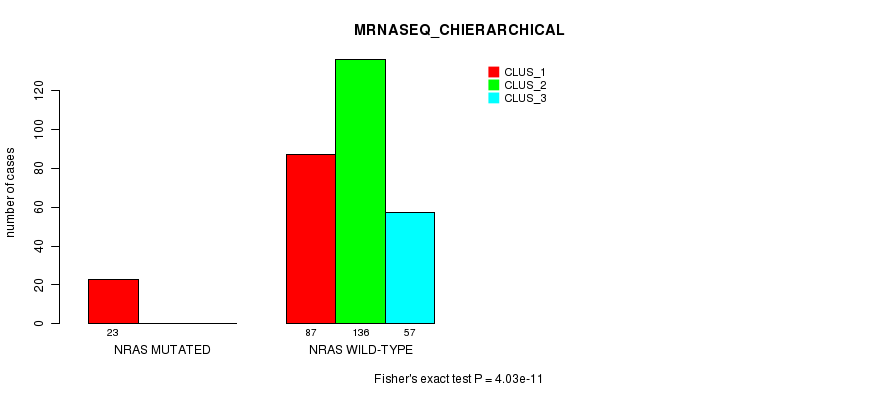
P value = 9.19e-13 (Fisher's exact test), Q value = 2.5e-10
Table S19. Gene #4: 'NRAS MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 105 | 119 | 98 |
| NRAS MUTATED | 25 | 0 | 1 |
| NRAS WILD-TYPE | 80 | 119 | 97 |
Figure S19. Get High-res Image Gene #4: 'NRAS MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
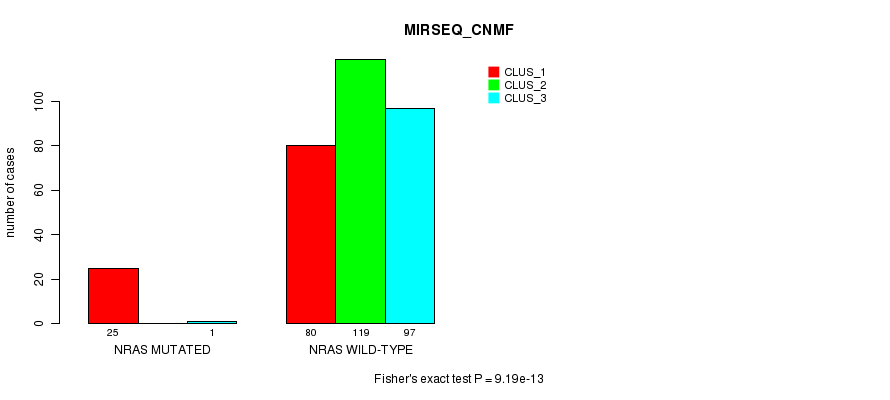
P value = 2.47e-13 (Fisher's exact test), Q value = 6.8e-11
Table S20. Gene #4: 'NRAS MUTATION STATUS' versus Clinical Feature #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 106 | 98 |
| NRAS MUTATED | 1 | 0 | 25 |
| NRAS WILD-TYPE | 117 | 106 | 73 |
Figure S20. Get High-res Image Gene #4: 'NRAS MUTATION STATUS' versus Clinical Feature #8: 'MIRSEQ_CHIERARCHICAL'
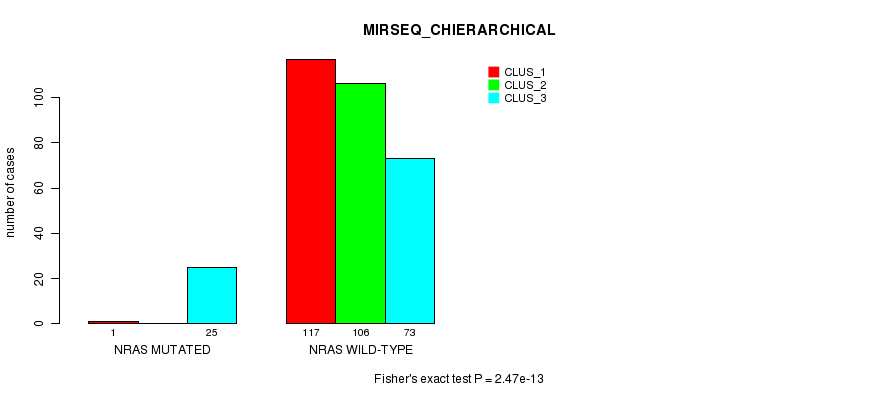
-
Mutation data file = THCA-TP.mutsig.cluster.txt
-
Molecular subtypes file = THCA-TP.transferedmergedcluster.txt
-
Number of patients = 323
-
Number of significantly mutated genes = 41
-
Number of Molecular subtypes = 8
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.